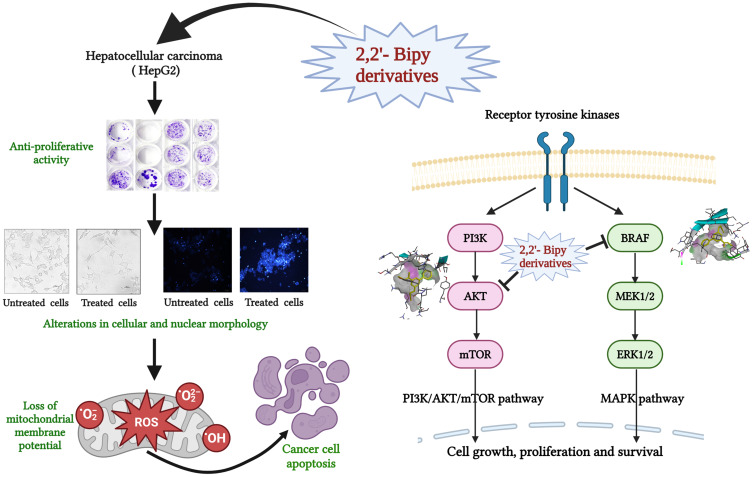
Abstract
Purpose
To elucidate the therapeutic potential of 2,2’-bipyridine derivatives [NPS (1–6)] on hepatocellular carcinoma HepG2 cells.
Methods
The effects on cell survival, colony formation, cellular and nuclear morphology, generation of reactive oxygen species (ROS), change in the integrity of mitochondrial membrane potential (MMP), and apoptosis were investigated. Additionally, docking studies were conducted to analyze and elucidate the interactions between the derivatives and AKT and BRAF proteins.
Results
NPS derivatives (1, 2, 5 and 6) significantly impaired cell viability of HepG2 cell lines at nanogram range concentrations - 72.11 ng/mL, 154.42 ng/mL, 71.78 ng/mL, and 71.43 ng/mL, while other derivatives were also effective at concentrations below 1 µg/mL. These compounds reduced the colony formation capacity of HepG2 cells in a dose-dependent manner following treatment. Mechanistic studies revealed that these derivatives induce reactive oxygen species (ROS) accumulation and cause mitochondrial membrane depolarization, ultimately triggering apoptosis in HepG2 cells. In the presence of these derivatives, cells demonstrated that 75% of cells underwent apoptosis, compared to 25% in the control group. Additionally, there was a marked increase in mitochondrial depolarization (95% cells) and a threefold rise in ROS levels compared to the controls. Docking studies revealed interactions between the derivatives and the signaling proteins AKT (PDB ID: 6HHF) and BRAF (PDB ID: 8C7Y) with binding affinities ranging from −7.10 to −9.91, highlighting their pivotal role in targeting key players in hepatocellular carcinoma progression.
Conclusion
The findings of this study underscore the therapeutic potential of these derivatives against HepG2 cells and offer valuable insights for further experimental validation of their efficacy as inhibitors targeting AKT or BRAF signaling pathways.
Keywords: hepatocellular carcinoma, ROS, MMP, apoptosis, AKT, BRAF
Graphical Abstract
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common form of liver cancer, accounting for 75–85% of cases worldwide, and the second leading cause of death, affecting millions of people every year, leading to a high mortality rate owing to late diagnosis and limited treatment options.1,2 Cancer patients who reach advanced stage are typically unsuitable for surgery and are treated with chemotherapy or radiotherapy.3 Conventional treatments can prolong lives, but their adverse effects often present significant challenges. Sorafenib, an FDA-approved drug for advanced liver cancer treatment, has a low patient survival rate, highlighting the need for new and more effective treatment options.4 Genetic and epigenetic shifts in HCC progression promote malignant transformation and abnormal activation, making them crucial targets for therapeutic intervention.5,6 AKT and BRAF inhibitors demonstrate promising potential in preclinical and clinical research, offering new therapeutic options for HCC through precision medicine. These inhibitors specifically target the PI3K/AKT/mTOR and RAF-MAPK signaling pathways, providing a strategic approach to managing the disease.7–10 AKT, a key effector in the PI3K/AKT/mTOR signaling pathway, induced apoptosis, halted HCC cell proliferation and suppressed tumor growth in animal models.11,12 However, early studies with ipatasertib, MK-2206, and perifosine have shown modest clinical activity, primarily due to drug resistance and off-target effects.13–15 The FDA has recently approved AstraZeneca’s Capivasertib, as the first AKT inhibitor, following promising results from the Phase III CAPItello-291 trial involving 708 patients with HR-positive, HER2-negative breast cancer.16,17 BRAF, a kinase protein, promotes an oncogenic cascade in HCC by cell proliferation, invasion, and angiogenesis.18,19 BRAF mutations are relatively rare in HCC, but dysregulation of the RAF/MAPK signaling pathway is frequently observed. BRAF inhibitors, such as sorafenib and vemurafenib, show efficacy in suppressing HCC cell proliferation and tumor growth.20,21 However, clinical trials have demonstrated only partial effectiveness of BRAF inhibitors due to adaptive resistance mechanisms and the complexity of the RAF/MAPK signaling pathway. As a result, combination strategies targeting multiple nodes of this pathway are currently being investigated.21,22
2,2’-Bipyridine (2,2’-Bipy) derivatives are used as chelating ligands in bioactive natural products such as caerulomycins, and collismycins.23,24 They form complexes with metal ions and exhibit antiviral, antifungal, antimicrobial, and potent antitumor activities.25–27 Recent studies have explored the potential of metal complexes in targeted anticancer therapies; for example, Ru(II) polypyridine complexes have shown promising encapsulation,28 while half-sandwich Rhodium(III) complexes show cytotoxic activity against HT-29 and MCF-7 cancer cells, though with high IC50 values.29 Additionally, Gold (III) complexes induced apoptosis in cancer cells, underscoring their potential for use in chemotherapy.30 Cyanine-functionalized 2,2′-bipyridine compounds are also being investigated for photocatalytic cancer therapy, showing anticancer activity through intracellular ROS accumulation.31 These derivatives also exhibit immunosuppressive properties against iron-related autoimmune diseases and reduce cellular iron levels in iron overload diseases.32–35 In our lab, we previously synthesized and characterized novel 2,2’-Bipy derivatives assessed for antituberculosis potential in in vitro.36,37 Some of these novel derivatives, NPS 1 to NPS 6, have been evaluated for their drug-likeness, ADMET studies, and safety against the normal murine fibroblast cell line 3T3L-1 and C57BL/6 mice, as well as for their antibacterial activity in cell lines and mice (data under review from our lab). While 2,2’-Bipy complexes have been extensively studied in various tumor types, the use of 2,2’ -Bipy derivatives for combating hepatocellular carcinoma remains unexplored in the literature. Therefore, the current study investigated the effects of these six derivatives (NPS1-6) on cell viability, cell proliferation, induction of apoptosis, and ROS generation in hepatocellular carcinoma. Using molecular docking to assess the virtual binding mechanism to AKT and BRAF as molecular targets of inhibition, we postulated that these compounds may act as inhibitors of PI3K/AKT/mTOR and RAS-BRAF-ERK, prototypical survival signaling in HCC carcinogenesis.
Materials and Method
Compounds
Six 2,2’-Bipy derivatives (2,2’-Bipy), namely NPS1, NPS2, NPS3, NPS4, NPS5, and NPS6, were used in this study. All these derivatives were synthesized and characterized previously in our lab.36 Dimethyl sulfoxide (DMSO) (Himedia Laboratories, Catalogue number-TC185-250ML), cell culture tested, was used to dissolve these derivatives at 10 mM concentration and kept at −20 °C until use. DMSO was used to dissolve compounds, serving as the negative control in each experiment. Doxorubicin (Sigma-Aldrich, Catalogue number-D1515-10MG) (1 µM) was used as reference drug. The final concentration of DMSO was maintained below 0.1% of the total medium volume.
Culture of HepG2 Cells
The hepatocellular carcinoma cell line of human origin (HepG2) was initially procured from the National Centre for Cell Sciences (NCCS) in Pune, a reputable and accredited source of cell lines. The cell line has been maintained in the institute’s tissue culture labs and was a kind gift from Dr. Manoj Raje from CSIR-IMTECH, Chandigarh. The authenticity of the cell line has been confirmed by numerous publications that have utilized it.38–40 HepG2 cell line was incubated at 37°C in an incubator maintained at 95% humidity and 5% CO2 in Roswell Park Memorial Institute, RPMI-1640 medium (Gibco, Thermo Scientific, Catalogue number-118775135) complemented with 10% fetal bovine serum (FBS, US origin, Gibco, Thermo Scientific, Catalogue number-26140079) and 1% penicillin-streptomycin (Gibco, Thermo Scientific, Catalogue number-15140122).
Cytotoxicity Assay
MTT assay was performed to ascertain the cytotoxicity potential of the derivatives.33 HepG2 cells were cultured, and 8000 cells/well were added to 96-well tissue culture flat-bottom microtiter plates overnight and then incubated with synthesized derivatives, NPS (1–6) at a concentration ranging from 0 µM–10 µM for 12–48 h at 37°C in a CO2 incubator.41 The derivative-containing media was replaced with 100 µL of diluted Thiazolyl Blue Tetrazolium Bromide (MTT, Sigma-Aldrich, Catalogue number-M2128-5G) solution in fresh media (0.5 mg/mL in media). Following 4 h of treatment, 100 µL of stop solution [50% Dimethylformamide (DMF) and 20% Sodium Do-decyl Sulphate (SDS)] was added to the wells to dissolve formazan crystals. The plate was further incubated for an additional hour. A BioTEK Synergy multi-plate reader recorded an absorbance at 570 nm.42 The Following formula was used to calculate the percent cell viability:
 |
The experiments were repeated 3 times in triplicates. Graph Pad Prism 9.2 software was used to calculate IC50 value of each derivative.
Colony Formation Assay
Cultured HepG2 cells (200 cells) were seeded into 24 well plates. These cells were treated for 48 h at three different doses (0.5 x IC50, IC50, and 2 × IC50) of each derivative and incubated at 37°C in 5% CO2. The growth medium was discarded and replaced with a fresh medium without any compound. HepG2 cells were cultured for an additional 14 days, changing the medium every two days. Following a PBS (Phosphate buffer saline) rinse, cells were fixed for 15 minutes with methanol and stained with 0.2% crystal violet for 30 minutes.43,44 ImageJ software was used to count the colonies, and plating efficiency (PE) was calculated using Franken’s method.44
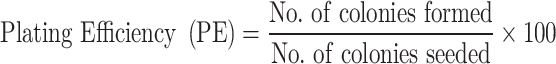 |
Microscopy
Cell Morphology
The effects of 2,2’-Bipy derivatives on the cytomorphology of HepG2 cells were investigated and documented using light (phase-contrast) microscopic techniques. In 6-well plates, 1×105 cells were seeded, kept overnight, and treated with derivatives for 48 h. After washing cells twice with 1X PBS, the cells were fixed with 2% paraformaldehyde solution. The fixed cells were visualized under the bright field microscope at 40X resolution.45
Nuclear Morphology
To further evaluate the effect of 2,2’-Bipy derivatives on the nuclear morphology of the cancer cells. Cells were treated as mentioned above for cellular morphology. After fixation, cells were washed with PBS twice, stained with 1 mg/mL of Hoechst 33258 (Sigma, Catalogue number-B1155), and incubated for 10 minutes at room temperature in the dark. Cells were washed with PBS thrice to remove excess stains. Fluorescent microscopy was used to examine any changes in nuclear morphology of cells.46,47
Annexin V/ Propidium Iodide Assay
Annexin V-FITC Apoptosis Kit (Catalogue No. BMS500FI-300, eBioscience, Invitrogen) was used to determine apoptosis based on phosphatidylserine (PS) exposure according to instructions of the manufacturer.48 Briefly, 2×105 HepG2 cells were seeded in six-well plates kept overnight and the cells were then treated with IC50 values of the derivatives. After 48 h incubation, cells were harvested, washed with ice-cold 1X PBS, and resuspended in ice-cold 1X Annexin binding buffer, and finally, Annexin V and PI dye were used for staining of the cells. Cells were processed using BD FACSVerse Flow Cytometer, and data was analysed using FlowJo software version 10.8.1.49 In all cases, apoptosis was identified by PS exposure labelled with Annexin V FITC, and cells that accumulated only PI dye were necrotic cells.
Mitochondrial Membrane Depolarization Assay
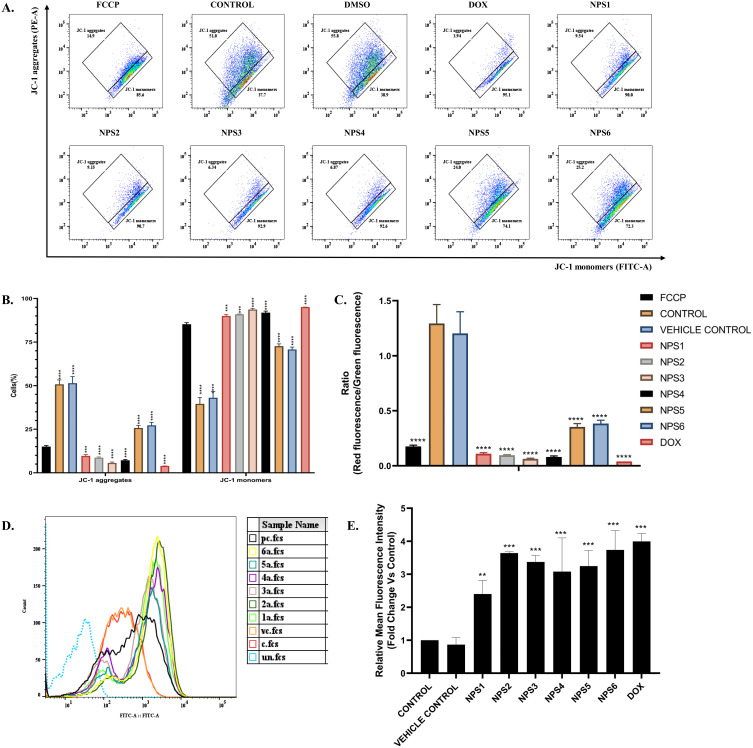
To determine the mitochondrial membrane potential (ΔψM), JC-1 (5,5,6,6’-tetrachloro-1,1’, 3,3’ tetraethylbenzimi-dazoylcarbocyanine iodide) dye (PromokKine, Catalogue number- PK-CA707-70011) was used.49 Briefly, 2×105 HepG2 cells were seeded in 6 well plates kept overnight followed by incubation with IC50 concentrations of the derivatives. FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, Sigma-Aldrich, Catalogue number- C2920-10MG), a protonophore that uncouples the mitochondrial oxidative phosphorylation, was used as a reference control to treat HepG2 cells at a concentration of 50 µM for 30 minutes. The cells were then collected, rinsed twice with 1X PBS, and stained with 2 µM JC-1 dye for 30 minutes. Excess stain was removed with 1X PBS. The BD FACSVerse Flow Cytometer was used to process the prepared samples, and FlowJo software version 10.8.1 was used for analysis. This experiment used FCCP to gate the cell population; healthy mitochondria was represented by aggregation of red JC-1, whereas green JC-1 monomers indicated unhealthy mitochondria with disturbed membrane potential.50
Reactive Oxygen Species (ROS) Quantification
The fluorophore 2’-7’-Dichlorodihydrofluorescein diacetate (DCFDA, Sigma-Aldrich, Catalogue number- D6883-50MG) was utilized to quantify reactive oxygen species.51,52 Upon entering the cell, DCFDA combines with reactive oxygen species to produce dichlorofluorescein (DCF), a green fluorescent molecule. Both fluorometry and microscopy were used to quantify the intracellular ROS. A 6-well plate was seeded with roughly 2×105 cells per well, which were grown overnight, and subsequently, cells were treated with IC50 doses of derivatives for 24 h. Cells were harvested and washed with 1X PBS twice, and then incubated at 37°C in the dark with 10 μM DCFDA for 30 minutes; washing was done using PBS and data was analyzed in a flow cytometer.
DNA Ladder Assay
A DNA ladder assay was executed to check the impact of 2,2’-Bipy derivatives on DNA integrity.53 After treating HepG2 cells with the derivatives for 48 h, cells were harvested, and DNA was extracted from both untreated and treated cells. Quantification was done using Nanodrop, and 5 µg of DNA was then loaded for electrophoresis (60V) on agarose gel (1.5%) with 0.5 mg/mL EtBr. A UV transilluminator was used to observe the electrophoresis gel, and an image was taken.54
Ligand Library Preparation
A library of 2,2’-Bipy derivatives was designed as potential anticancer agents using the bioisosteric substitution method based on research from a wide range of literature.55–57 The bipyridine nucleus has a long history of diverse therapeutic potential, including anticancer effects. In order to find effective anticancer drugs and understand the physiological mechanisms behind their therapeutic benefits, this tailored ligand library was screened against different cancer targets.57–60
Macromolecular Target Selection and Preparation
The inhibition of AKT and BRAF is related to suppressing tumor growth in various cancers, including activation of the apoptotic mechanism. Thus, the ligand library was designed for molecular docking analysis against these concerned anticancer targets to develop multi-targeting therapeutics and novel cancer treatments with a broad spectrum of activity against diverse types of cancer.61–63 RCSB PDB database was used to retrieve a 3D model of above-mentioned targets.64,65 The downloaded protein models were modified by removing the complex ligands, adding polar hydrogen and ADT atom types, and preparing them for docking analysis.
Docking Studies
Using the Autodock-4.2 program, the AKT and BRAF targets in the ongoing docking investigation were re-docked with the divided reference ligands.66,67 The docking parameters for each target were verified by equating the configuration and chemical comparison of corresponding ligand with active site of macromolecule. After verification, the created ligand library was computationally screened against the cancer targets using the restrictions.68–70
Statistical Analysis
The results are expressed as the mean ± SD. The Shapiro–Wilk and Kolmogorov–Smirnov tests validated the normality of the data, and the homogeneity of variance was confirmed using Brown-Forsythe and Welch tests. All statistical analyses were done using one-way and two-way ANOVA followed by the student unpaired t-test. The p-values of 0.001 (***) and 0.0001 (****) considered highly significant.
Results
Cytotoxic Effects of 2,2’-Bipy Derivatives Against HepG2 Cell Line
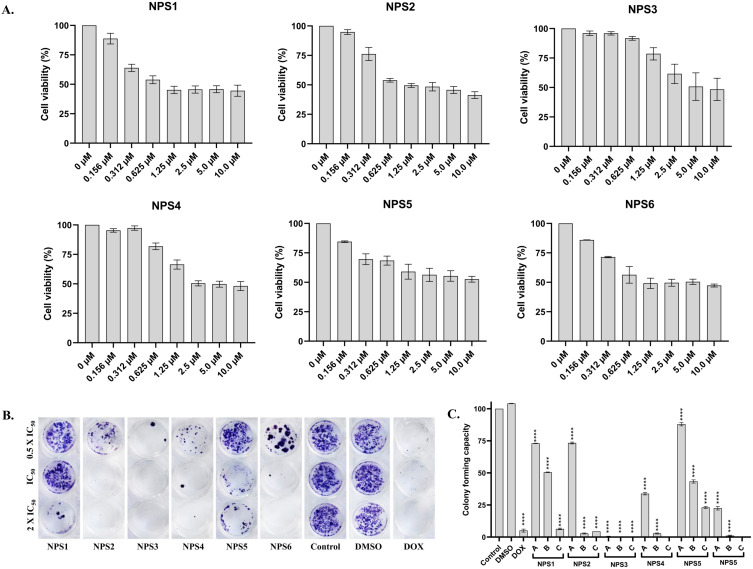
The cytotoxic potential of derivatives on HepG2 cells was examined in vitro using the MTT assay. A dose-dependent and time-dependent decrease in cell viability was observed in HepG2 cells following treatment with the derivatives (Figure S1; Cell viability Assay in Supplementary materials). Specifically, after 48 h of treatment, the viability of HepG2 was significantly reduced by 80%, 60%, and 40% as the concentration increased from 0 to 10 µM (Figure 1A). After 48 h, the inhibitory concentration (IC50) of derivatives (NPS (1–6)) has been represented in Table 1. The IC50 value is lower and almost similar for NPS1, NPS5, and NPS6, with values of 0.27, 0.25, and 0.28 µM, respectively. For NPS2, it is 0.43 µM, whereas NPS3 and NPS4 have a higher inhibitory concentration of 2.66 µM and 1.37 µM respectively. Derivatives NPS1, NPS2, NPS5, and NPS6 were effective at nanogram concentrations, ie, 72.11 ng/mL, 154.42ng/mL, 71.78 ng/mL, and 71.43 ng/mL (Table 1) while the rest of the derivatives (NPS3 and 4) were also effective at less than 1 µg/mL concentration.
Figure 1.
2,2’-Bipy derivatives NPS (1–6) inhibited the viability and proliferation of HepG2 cell lines. (A) Effects of various doses of the derivatives NPS (1–6) on the cell viability of HepG2 cells. The total percentage of cells was measured using an MTT assay. A dose-dependent decrease was observed in cell viability after treatment at 48 h; (B) Effects of the derivatives NPS (1–6) at three doses 0.5 × IC50, IC50, and 2 × IC50 on the cell proliferation of HepG2 cells by colony formation assay using crystal violet dye. A dose-dependent decrease in colony numbers appears with an increase in concentration. (C) Colony formation capacity of HepG2 cells after treatment with the derivatives. The asterisks shown are significant when calculated for their colony formation capacity when compared to the control. The data from three independent experiments were presented as mean ± SD, with statistical differences indicated compared to the control group. The p-value ≤ 0.0001 (****) indicates significance.
Table 1.
IC50 Values of 2,2’-Bipy derivatives Against HepG2 Cell Lines
| Derivatives | IC50 (Mean ± SD) | |
|---|---|---|
| (µM) | (ng/mL) | |
| NPS1 | 0.27 ± 0.01 | 72.11 ± 2.67 |
| NPS2 | 0.43 ± 0.03 | 154.42 ± 10.77 |
| NPS3 | 2.66 ± 0.17 | 0.97 ± 0.6* |
| NPS4 | 1.37 ± 0.26 | 0.45 ± 0.08* |
| NPS5 | 0.25 ± 0.01 | 71.78 ± 2.87 |
| NPS6 | 0.28 ± 0.01 | 71.43 ± 2.55 |
Notes: * Signifies value expressed in (µg/mL).
Effect on Colony Formation Activity
A dose-dependent decrease in the ability of HepG2 cells to form colonies was the outcome of exposure to NPS (1–6) derivatives, suggesting that the derivatives could affect cell growth and viability. A concentration of 2 × IC50 of the derivatives, greater than the corresponding IC50 value, caused an almost zero-fold change in colony numbers in both cell types compared to controls (Figure 1B and C).
Derivatives Alter the Cellular Morphology of Cancer Cells
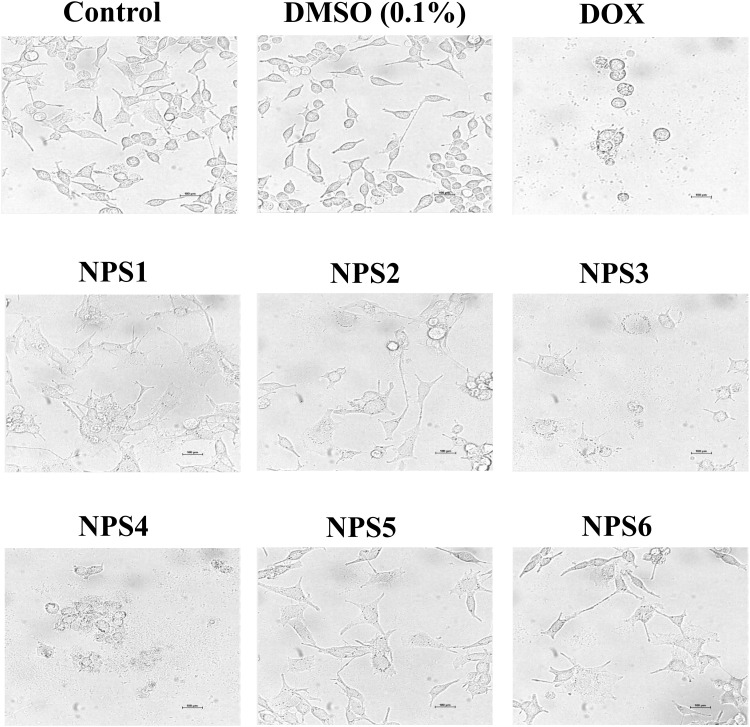
Cell apoptosis is a special form of cell death to remove damaged cells from normal tissue. Although both necrosis and apoptosis are types of cell death, they have distinct physical traits. Light microscopy revealed cytomorphologic alterations in HepG2 cells associated with triggering death (Figure 2). Fragmentation of cells into apoptotic entities, extensive membrane blebbing, and shrinkage of the cells occurred. In untreated HepG2 cells, confluent aggregates of polygons were observed, while upon exposure to the derivatives for 48 h, they turned round out, contracted with membrane blebbing, and disengaged from the culture dish surface.
Figure 2.
2,2’-Bipy derivatives NPS (1–6) induce apoptosis-related cytomorphological changes in HepG2 cells.
Derivatives Alter the Nuclear Morphology of HepG2 Cell
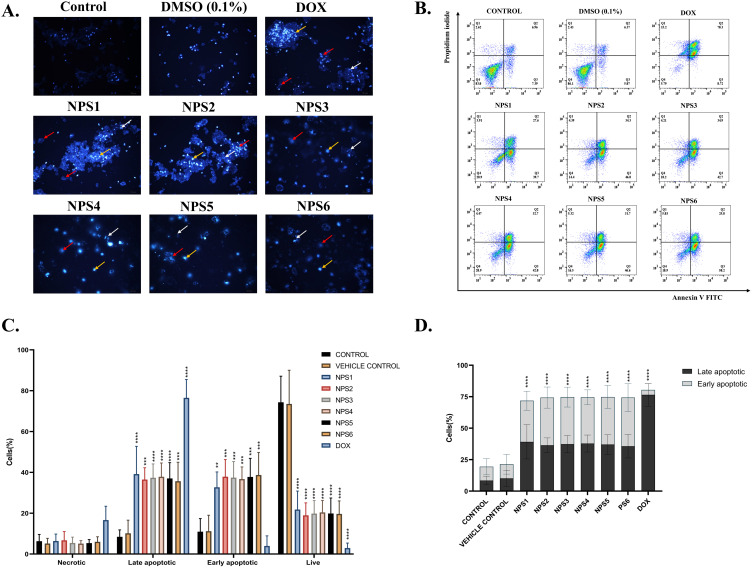
Using fluorescence microscopy, a concentration-dependent increase in cell blebbing and several apoptotic cells were observed, indicating that the derivatives induce toxicity. Fluorescence microscopy revealed a fraction of cells with bright blue, abridged, and shattered and fragmented nuclei in contrast to homogeneously blue-stained nuclei in vehicle-treated and untreated controls (DMSO) of derivative-treated cells stained with Hoechst 33258, a DNA binding dye. Following a 48h exposure to IC50 values, most cells displayed significant Hoechst signaling, chromatin condensation, and nuclear fragmentation (Figure 3A). These findings show apoptosis is primarily responsible for cell death.
Figure 3.
2,2’-Bipy derivatives (NPS (1–6) induced apoptosis in HepG2 cells. (A) Nuclear morphological alterations in HepG2 cells are associated with apoptosis using Hoechst 33258 staining, and cells treated with derivatives show condensed nuclei with respect to control. Yellow arrow show Hoechst-stained cells, and red and white arrows represent chromatin condensation and nuclear fragmentation, respectively. (B) Schematic representation of flow cytometric analysis of cells stained with Annexin V FITC/PI staining. (C) The percentages of live, cells undergoing apoptosis and necrotic cells the total number of cells were used to calculate data. (D) In the overall apoptotic population, these derivatives induce significant apoptosis, and the asterisks shown are significant when calculated for their colony formation capacity compared to the control. The data from three independent experiments were presented as mean ± SD, with statistical differences compared to the control group. The p-values of 0.01 (**), 0.001 (***) and 0.0001 (****) considered significant.
Derivatives Induce Apoptosis of HepG2 Cell
Nuclear morphology and cytomorphology suggest that apoptosis may be the cause of cell death. The annexin V FITC/ Propidium iodide assay was performed with the cells treated with derivatives. One of the hallmarks of apoptosis is the presence of phosphatidylserine (PS) on the cell surface, which is usually restricted to the inner membrane layer. FITC-tagged annexin V binds to externalized PS in a calcium-dependent environment, allowing its quantification and detection with a flow cytometer. Since PI can only interact with DNA whose membrane integrity is damaged, co-staining of necrotic/late apoptotic cells with annexin V facilitates the differentiation of these cells from early apoptotic cells.71 When PS externalization was measured in the cells treated with derivatives, the percentage of apoptotic cells increased threefold (Figure 3B-D).
Derivatives Depolarize the MMP of HepG2 Cell
The previous experiment confirmed that 2,2’-Bipy derivatives induce cell death by apoptosis. Apoptosis occurs via two primary signaling mechanisms: intrinsic mitochondrial signaling and external death receptor signaling. Mitochondrial function, a critical indicator of cell health, and changes in MMPs are linked to the intrinsic pathway of apoptosis. To explore the mechanisms of apoptosis induction by the derivatives, an MMP assay was performed using the cationic carbocyanine dye JC-1. Carbonyl cyanide p- (trifluoromethoxy)phenylhydrazone, or FCCP, a potent inhibitor of mitochondrial oxidative phosphorylation and a positive regulator of mitochondrial membrane depolarization or apoptosis, served as a reference control (Figure 4A). In cells treated with derivatives, the intensity of green fluorescence was higher than that of red fluorescence (Figure 4B). In healthy mitochondria, the ratio of red to green should be 1 or > 1; if it is lower, then the mitochondria are damaged. As shown in Figure 4C, the ratio was considerably lower in the group treated with the derivatives than the control group (P<0.001). These outcomes suggest that NPS (1–6) depolarizes the MMPs and is one of the pathways through which cell death is initiated.
Figure 4.
2,2’-Bipy depolarizes mitochondrial membrane depolarization and generates intracellular ROS in HepG2 Cells. (A) Schematic representation of flow cytometric analysis of HepG2 cells stained with JC-1 dye, X-axis, and Y-axis denotes JC-1 aggregates and JC-1 monomers. (B) The percentage of JC-1 aggregates and monomers and the increase in the monomeric population indicate mitochondrial damage. (C) The ratio is less than one in the ratiometric presentation of the JC-1 aggregates and monomers, indicating a significant difference from the controls. (D) Histogram representing Change in DCFDA fluorescence after treatment using flow cytometry, there is a shift in the fluorescence towards the right side with an increase in the intensity (c-untreated control, vc- solvent control, pc- doxorubicin treated, 1a-6a represents the cells treated by derivatives NPS1-6) and (E) The ROS activity shows mean representative data obtained from flow cytometry in comparison to control indicating a significant increase in the ROS activity. The data from three independent experiments were presented as mean ± SD, with statistical differences compared to the control group. The p-values of 0.01 (**), 0.001 (***) and 0.0001 (****) considered significant.
Derivatives Generate Oxidative Stress in HepG2 Cell
Next, we investigated whether 2,2’-Bipy derivatives stimulate ROS production in HepG2 cells, as there is evidence that intracellular ROS can affect cell signaling and trigger cell death. ROS levels are elevated in almost all malignancies and promote various aspects of tumor formation and progression. Novel therapeutic approaches will depend on how well intracellular ROS signaling is controlled to prevent ROS-induced tumor promoting actions in cells leading to a shift in balance in favour of ROS-induced apoptotic signaling. Apoptosis is interrelated to augmented mitochondrial oxidative stress and release of cytochrome C, an irreversible process that promotes caspase activation and cell death. Cell cycle arrest, senescence and ultimately cell death in cancer can result from disproportionately increase in intracellular ROS. Thus, DCFH-DA, a cell permeable, non-fluorescent probe that was first oxidized to membrane-impermeable H2DCF before being converted back to highly fluorescent DCF, was used by cellular esterases to measure intracellular ROS. Increased ROS activity (%) was observed in Figure 4D and E, indicating the potential of the derivatives in inducing oxidative pressure by uncoupling oxidative phosphorylation.
Derivatives Damage the DNA of HepG2 Cell
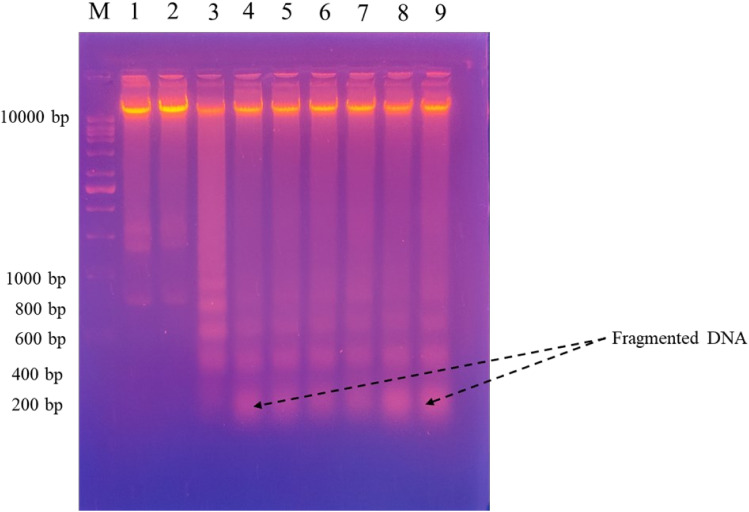
DNA fragmentation is a hallmark feature of apoptosis. The disintegration of nuclear DNA into nucleosomal fragments is a clear indication of apoptotic cell death.72 Many apoptotic triggers in a variety of cell types lead to this result. In Figure 5, panels 4–9 show HeG2 cells treated with derivatives in which the DNA ladder has formed compared to the untreated cells.
Figure 5.
DNA ladder assay presenting damaged DNA in the HepG2 cells after treatment by 2,2’-Bipy NPS (1–6); M- marker, 1- Control (Untreated), 2- Solvent control (DMSO), 3-Positive control (Dox), (4–9) represents the derivatives treated sections.
Design of Ligand Library
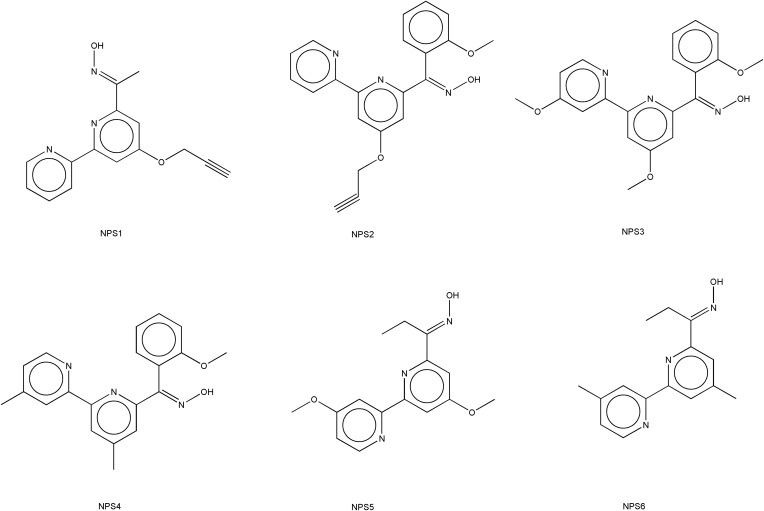
Six ligands belonging to the class of 2,2’-bipyridines were prepared to evaluate for anticancer potential.73,74 Structures of specific ligands considered in the current study are shown in Figure 6. The two-dimensional structures of each ligand were constructed using ChemDraw 9.0 software.75,76 Subsequently, energy minimization was performed utilizing the MM2 force field within the Chem3D tool to generate the corresponding three-dimensional structures. These 3-D structures were then utilized for computer-assisted screening against the anticancer target of interest.
Figure 6.
Chemical structures of studied 2,2’-bipyridine derivatives NPS (1–6).
Docking Studies
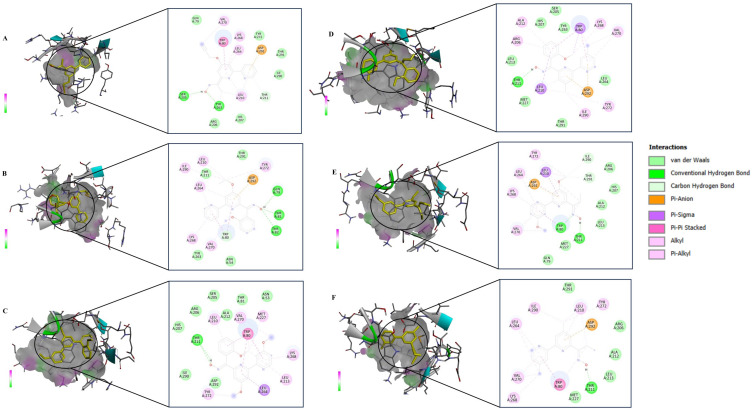
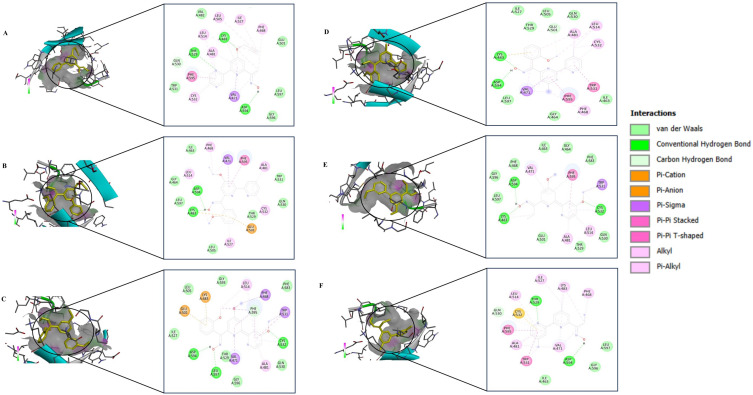
The structural models of the derivative targets were attained from the PDB and converted into monomeric nascent receptors by removing other chains and complex ligands. The generated monomeric receptors were modified by addition of polar hydrogen atoms, achieving an even dispersal of Gasteiger charge among the residues prepared for docking analysis. Crystal structures of AKT (PDB ID:6HHF) and BRAF (PDB ID: 8C7Y) were taken from Protein Data Bank. 2,2’-Bipy derivatives were prepared as a ligand using standard molecular modelling tools. Docking was performed using AutoDock Vina software, employing a grid-based approach to investigate potential binding sites to AKT and BRAF. Favourable binding interactions were found between 2,2’-Bipy derivatives and the active site of AKT and BRAF. The binding energies of these derivatives were calculated against AKT and BRAF, and a negative binding energy indicates a strong interaction between the ligand and the receptor as shown in Table 2. 2,2’-Bipy derivatives were found to modulate AKT activity and BRAF activity in HepG2 cells. Docking studies showed hydrophobic and hydrogen bonding interactions, suggesting that they can form complexes with amino acid residues within the binding pocket (Figures 7 and 8). The findings suggest the role of these derivatives in inhibiting AKT and BRAF activity, which is a key driver in the progression of HCC and requires further experimental validation.
Table 2.
Binding Energies and Interacting Residues of Derivatives Docked with AKT and BRAF
| Protein (PDB ID) |
Derivatives | Binding Energies | Interacting Residues |
|---|---|---|---|
| AKT (6HHF) |
NPS1 | −7.63 | GLN79, VAL270, TRP80, LYS268, LEU264, ASP292, THR291, ILE290, THR211, LEU210, HIS207, ARG206, TYR263, SER205 |
| NPS2 | −8.36 | ILE290, LEU210, THR211, LEU264, THR291, ASP292, TYR272, GLN79, THR81, ASN54, VAL270, LYS268, TYR263, THR82, TRP80 | |
| NPS3 | −8.53 | HIS207, ARG206, THR211, LEU210, SER205, ALA212, VAL270, TRP80, MET227, ASN53, TRP80, LYS268, LEU213, LEU 264, TYR272, ASP292, ILE290 | |
| NPS4 | −9.64 | LEU210, MET227, THR211, LEU213, ARG206, ALA212, HIS207, SER205, TYR263, TRP80, LYS268, VAL270, LEU264, ASP292, TYR272, ILE290, THR291 | |
| NPS5 | −7.48 | VAL270, LYS268, LEU264, TYR272, LEU210, ASP292, ILE290, THR291, ARG206, HIS207, ALA212, LEU213, TRP80, THR211, MET227, GLN79 | |
| NPS6 | −8.10 | LEU264, ILE290, THR291, LEU210, TYR272, ASP292, ARG206, ALA212, LEU213, THR211, MET227, TRP80, VAL270, LYS268 | |
| BRAF (8C7Y) |
NPS1 | −8.28 | VAL482, LEU505, ILE527, PHE468, GLU501, LEU514, LYS483, THR529, ALA481, GLN530, PHE595, TRP531, CYS532, VAL471, ASP594, GLY596, LEU597 |
| NPS2 | −9.54 | ILE463, PHE468, VAL471, PHE595, ALA481, TRP531, GLN530, CYS532, THR529, GLU501, ILE527, LEU505, LYS483, ASP594, LEU597, GLY464, LEU514 | |
| NPS3 | −9.62 | ILE527, GLU501, LEU505, LYS483, GLY593, LEU514, PHE468, PHE595, PHE583, TRP531, CYS532, GLN530, ALA481, GLY596, VAL471, THR529, LEU597, ASP594 | |
| NPS4 | −9.91 | ILE527, THR529, LEU505, GLU501, GLN530, ALA481, LEU514, CYS532, TRP531, ILE463, PHE595, PHE468, GLY464, VAL471, LEU597, ASP594, LYS483 | |
| NPS5 | −7.13 | LYS483, LEU597, GLY596, ASP594, PHE468, VAL471, ILE463, GLY464, PHE595, PHE583, TRP531, CYS532, GLN530, LEU514, THR529, ALA481, GLU501 | |
| NPS6 | −9.07 | LEU597, GLY596, ASP594, VAL471, ILE463, TRP531, ALA481, PHE595, CYS532, GLN530, THR529, LEU514, ILE527, LYS483, PHE468 |
Figure 7.
Inhibition of AKT (PDB: 6HHF) by 2,2- bipyridine derivatives. In silico molecular docking 2D and 3D interactions of (A) NPS1, (B) NPS2, (C) NPS3, (D) NPS4, (E) NPS5 and, (F) NPS6.
Figure 8.
BRAF (PDB:8C7Y) inhibition by 2,2- bipyridine derivatives. In silico molecular docking 2D and 3D interactions of (A) NPS1, (B) NPS2, (C) NPS3, (D) NPS4, (E) NPS5 and, (F) NPS6.
Discussion
Hepatocellular carcinoma (HCC) is the most prevalent form of primary liver cancer. It typically develops in the setting of chronic liver diseases such as hepatitis B or C virus infections, non-alcoholic steatohepatitis (NASH), or other conditions that lead to liver cirrhosis. These underlying liver conditions create an environment conducive to the development of HCC, making it a significant concern in patients with chronic liver disease.77 2,2’-Bipyridines are crucial bidentate ligands in organometallic and inorganic chemistry due to their high redox stability and simplicity of functionalization, which makes them ideal metal-chelating ligands.23 2,2’- Bipy ligands form complexes with metals that exhibit cytotoxic effects against various cancer cell lines and have been explored for therapeutic and diagnostic applications.78–81 They frequently exhibit significant toxicity and inadequate pharmacological characteristics, and factors such as the choice of metal, coordination geometry, and ligand type influence their biophysical properties.82 Here, we studied 2,2’-Bipy derivatives (with different functional groups) for their cytotoxic activity against HepG2 cells. The MTT assay was conducted to evaluate the toxicity of the derivatives on HepG2 cells. Our findings indicate that these derivatives effectively inhibit the viability of HepG2 cells. The anticancer drug demonstrates cytostatic and cytotoxic effects on cancer cells, inhibiting their growth and inducing cell death. This was confirmed through a clonogenic assay.44 We found that NPS2, NPS3, NPS4, and NPS6 showed antiproliferative activity at a concentration of half the IC50, and NPS1 and NPS5 were also antiproliferative at increasing concentrations (Figure 1).
Cell death, particularly through programmed cell suicide, plays a critical role in maintaining physiological balance by removing damaged cells. Additionally, it can serve as an abnormal pathological response to harmful stimuli.83 Apoptosis represents a form of programmed cell death characterized by consistent morphological changes across various cell types and species, albeit with timing variations contingent upon the specific cell type, stimulus, and apoptotic pathway involved.84,85 Nucleus hallmarks include chromatin condensation, nuclear fragmentation, cell rounding, cellular volume reduction, and retraction of pseudopods.86 Cells were examined for cellular and nuclear morphology under bright field and fluorescence microscopy and exhibited apoptosis, membrane blebbing, shrinkage, and apoptosis, with some fraction showing bright blue, abridged and fragmented nuclei, compared to vehicle-treated and untreated controls and Annexin-V FITC staining was employed to confirm apoptosis. Apoptotic cells induced by these derivatives are detected by double staining with Annexin-V/PI.87 As shown in Figure 3D, the study indicates that treatment of HepG2 cells with the derivatives led to a notable increase in both early and late apoptotic cell populations, suggesting that these compounds primarily impede HepG2 cell proliferation via apoptosis.
Apoptosis occurs via different pathways, and to test whether apoptosis is mediated by mitochondrial involvement, a depolarization assay using the dye JC-1 dye was performed. Red or green fluorescence is generated by JC-1 aggregations in the mitochondrial matrix caused by high ΔΨm. This transition is a useful marker for apoptosis and for measuring a decrease in membrane potential.88 Our research shows that these derivatives strongly reduced mitochondrial membrane potential, which in turn encouraged the death of HepG2 cells. These derivatives can be inferred to induce apoptosis in HepG2 cells by mitigating mitochondrial dysfunction. Ninety percent of cellular reactive oxygen species are generated within the mitochondrial membrane,89,90 and ROS are potentially carcinogenic due to their involvement in the regulation of tumor formation; excessive accumulation of ROS in the cell has been shown to have detrimental effects.91,92 Dichlorodihydrofluorescein, a fluorescent substance, can penetrate cell membranes and be hydrolyzed to dichlorodihydrofluorescein (DCFH), while DCFH cannot breach the membrane of the cell and remains in cell.51,52 Since these derivatives cannot penetrate the cell membrane, they increase the DCFH content in HepG2 cells, promote ROS accumulation, and induce apoptotic processes. A Hoechst staining procedure was employed to validate the induction of apoptosis further. DNA fragmentation, a critical biochemical feature of apoptotic cell death, was observed as a confirming hallmark. A DNA ladder experiment was performed, and the results showed that these derivatives damage the DNA in the cells as DNA fragments are formed.
The PI3K-AKT-mTOR signaling, recognized as a pivotal biochemical reaction in cancer, is commonly dysregulated and influences various cellular processes, including cell cycle progression, differentiation metabolism, survival, and migration.93–95 Due to its robust connectivity with the RAS-RAF-MEK-ERK pathway, its ability to activate the MYC oncogene, and its potential as a druggable target, it is regarded as an appealing candidate for cancer therapy.96,97 BRAF, a kinase protein, phosphorylates and activates downstream effectors that promote an oncogenic cascade that results in the progression of HCC through the proliferation of cells, invasion, and angiogenesis.18,19 Genetic alterations in tumors activate the oncoprotein BRAF and promote dysregulation of the RAF-MEK-ERK signaling, making BRAF inhibitors a valid therapeutic strategy for many cancer patients.20,98 The MAPK and AKT/mTOR signaling pathways are stimulated in over 50% of cases of HCC and are linked to aggressive proliferative tumors of the class.99 Therefore, docking studies were performed with BRAF and AKT, and all derivatives showed interactions. Our findings suggest the role of these derivatives in inhibiting AKT and BRAF activity, which is a critical reason for the progression of HCC and requires further experimental validation.
Conclusion
In conclusion, 2,2’-Bipy derivatives have a potential therapeutic role in hepatocellular carcinoma. All six derivatives exhibit cytotoxicity, antiproliferative, and apoptotic activity against HCC through ROS generation and mitochondrial membrane depolarization. In addition, molecular docking studies suggest that it exerts its effect through interaction with AKT and BRAF, two essential signaling proteins implicated in HCC progression. These findings underline the promising therapeutic potential of the investigated derivatives for the therapy of HCC. However, further studies are warranted to validate these findings and to explore the therapeutic potential of 2,2’-Bipy as a targeted therapy for HCC. Future research will concentrate on bridging the gap between in vitro findings and clinical applications by validating the therapeutic potential of these compounds through in vivo investigations, including the use of sophisticated animal models. We will also investigate the impact of these derivatives on important enzymes that regulate mitochondrial membrane potential and ROS production, aiming to gain a deeper understanding of the underlying molecular mechanisms. These efforts are essential for evaluating the long-term safety, efficacy, and translational potential of these compounds for clinical development.
Acknowledgments
Priyanka and Ram Bharti acknowledge CSIR for fellowships. The authors acknowledge consistent support from the faculty and staff of IMTECH.
Funding Statement
This work was supported and funded by CSIR-IMTECH, Grant No.OLP-0168 to NK.
Data Sharing Statement
All data supporting the findings of this study are available within the article.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/CAAC.21660 [DOI] [PubMed] [Google Scholar]
- 3.Balogh J, Iii DV, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daher S, Massarwa M, Benson AA, Khoury T. Current and future treatment of hepatocellular carcinoma: an updated comprehensive review. J Clin Transl Hepatol. 2018;6(1):69. doi: 10.14218/JCTH.2017.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doghish AS, Ali MA, Elyan SS, et al. miRNAs role in cervical cancer pathogenesis and targeted therapy: signaling pathways interplay. Pathol Res Pract. 2023;244:154386. doi: 10.1016/J.PRP.2023.154386 [DOI] [PubMed] [Google Scholar]
- 6.Glaviano A, Foo ASC, Lam HY, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22(1):1–37. doi: 10.1186/S12943-023-01827-6/TABLES/1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahar ME, Kim HJ, Kim DR. Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies. Signal Transduction and Targeted Therapy. 2023;8(1):1–38. doi: 10.1038/s41392-023-01705-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paskeh MDA, Ghadyani F, Hashemi M, et al. Biological impact and therapeutic perspective of targeting PI3K/Akt signaling in hepatocellular carcinoma: promises and Challenges. Pharmacol Res. 2023;187:106553. doi: 10.1016/J.PHRS.2022.106553 [DOI] [PubMed] [Google Scholar]
- 9.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12(7):408–424. doi: 10.1038/nrclinonc.2015.103 [DOI] [PubMed] [Google Scholar]
- 10.Villanueva A. Hepatocellular Carcinoma. New Eng J Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMRA1713263 [DOI] [PubMed] [Google Scholar]
- 11.Bang J, Jun M, Lee S, Moon H, Ro SW. Targeting EGFR/PI3K/AKT/mTOR Signaling in Hepatocellular Carcinoma. Pharmaceutics. 2023;15(8):2130. doi: 10.3390/PHARMACEUTICS15082130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nitulescu GM, Margina D, Juzenas P, et al. Akt inhibitors in cancer treatment: the long journey from drug discovery to clinical use (Review). Int J Oncol. 2016;48(3):869. doi: 10.3892/IJO.2015.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckingham L, Hao T, O’Donnell J, et al. Ipatasertib, an oral AKT inhibitor, inhibits cell proliferation and migration, and induces apoptosis in serous endometrial cancer. Am J Cancer Res. 2022;12(6):2850. [PMC free article] [PubMed] [Google Scholar]
- 14.Savill KMZ, Lee BB, Oeh J, et al. Distinct resistance mechanisms arise to allosteric vs. ATP-competitive AKT inhibitors. Nat Commun. 2022;13(1):1–17. doi: 10.1038/s41467-022-29655-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing Y, Lin NU, Maurer MA, et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019;21(1):1–12. doi: 10.1186/S13058-019-1154-8/FIGURES/3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullard A. FDA approves first-in-class AKT inhibitor. Nat Rev Drug Discov. 2023;22(11):862. doi: 10.1038/D41573-023-00202-W. [DOI] [PubMed] [Google Scholar]
- 17.Turner NC, Oliveira M, Howell SJ, et al. Capivasertib in hormone receptor–positive advanced breast cancer. N Engl J Med. 2023;388(22):2058–2070. doi: 10.1056/NEJMOA2214131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- 19.Singh AK, Sonawane P, Kumar A, et al. Challenges and opportunities in the crusade of BRAF inhibitors: from 2002 to 2022. ACS Omega. 2023;8(31):27819–27844. doi: 10.1021/ACSOMEGA.3C00332/ASSET/IMAGES/LARGE/AO3C00332_0015.JPEG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- 21.Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label Phase 2 trial. Lancet Oncol. 2018;19(7):940–952. doi: 10.1016/S1470-2045(18)30351-6 [DOI] [PubMed] [Google Scholar]
- 22.Rustgi N, Maria A, Toumbacaris N, et al. Combined RAF and MEK inhibition to treat activated non-V600 BRAF-altered advanced cancers. Oncologist. 2024;29(1):15–24. doi: 10.1093/ONCOLO/OYAD247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaes C, Katz A, Hosseini MW. Bipyridine: the most widely used ligand. A review of molecules comprising at least two 2,2′-bipyridine units. Chem Rev. 2000;100(10):3553–3590. doi: 10.1021/cr990376z [DOI] [PubMed] [Google Scholar]
- 24.Pang B, Liao R, Tang Z, Guo S, Wu Z, Liu W. Caerulomycin and collismycin antibiotics share a trans-acting flavoprotein-dependent assembly line for 2,2’-bipyridine formation. Nature Communications. 2021;12(1):1–12. doi: 10.1038/s41467-021-23475-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgart Y, Shchegolkov E, Shchur I, et al. Promising antifungal and antibacterial agents based on 5-Aryl-2,2′-bipyridines and their heteroligand salicylate metal complexes: synthesis, bioevaluation, molecular docking. ChemMedChem. 2022;17(3). doi: 10.1002/cmdc.202100577 [DOI] [PubMed] [Google Scholar]
- 26.Nasiri Sovari S, Kolly I, Schindler K, et al. Synthesis, characterization, and in vivo evaluation of the anticancer activity of a series of 5- and 6-(halomethyl)-2,2′-bipyridine rhenium tricarbonyl complexes. Dalton Trans. 2023;52(20):6934–6944. doi: 10.1039/D2DT04041G [DOI] [PubMed] [Google Scholar]
- 27.Omoregie HO, Eseola AO, Akong RA. Mixed ligand complexes of copper(II) with benzoyltrifluoroacetone, 1,10-phenanthroline and 2,2′-bipyridine: structure, spectroscopic and antimicrobial properties. J Mol Struct. 2022;1250:131826. doi: 10.1016/J.MOLSTRUC.2021.131826 [DOI] [Google Scholar]
- 28.Karges J. Encapsulation of Ru(II) polypyridine complexes for tumor-targeted anticancer therapy. BME Front. 2023;4. doi: 10.34133/BMEF.0024/ASSET/14E346C3-8543-4D30-9CBC-EA9EBCA483CB/ASSETS/GRAPHIC/BMEF.0024.FIG.0010.JPG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graf M, Ochs J, Mayer P, Metzler-Nolte N, Böttcher HC. Cytotoxic activity of some half-sandwich rhodium(III) complexes containing 4,4’-disubstituted-2,2’-bipyridine ligands. Z Anorg Allg Chem. 2024;650(1):e202300195. doi: 10.1002/ZAAC.202300195 [DOI] [Google Scholar]
- 30.Alhoshani A, Sulaiman AAA, Sobeai HMA, et al. Anticancer activity and apoptosis induction of gold(III) complexes containING 2,2′-bipyridine-3,3′-Dicarboxylic acid and dithiocarbamates. Molecules. 2021;26(13):3973. doi: 10.3390/MOLECULES26133973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Wei L, Yadav AK, et al. Cyanine-functionalized 2,2′-bipyridine compounds for photocatalytic cancer therapy. J Org Chem. 2023;88(1):626–631. doi: 10.1021/ACS.JOC.2C00956/ASSET/IMAGES/LARGE/JO2C00956_0003.JPEG [DOI] [PubMed] [Google Scholar]
- 32.Kaur S, Srivastava G, Sharma AN, Jolly RS. Novel immunosuppressive agent caerulomycin A exerts its effect by depleting cellular iron content. Br J Pharmacol. 2015;172(9):2286–2299. doi: 10.1111/bph.13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kujur W, Gurram RK, Haleem N, Maurya SK, Agrewala JN. Caerulomycin A inhibits Th2 cell activity: a possible role in the management of asthma. Sci Rep. 2015;5. doi: 10.1038/srep15396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kujur W, Gurram RK, Maurya SK, et al. Caerulomycin A suppresses the differentiation of naïve T cells and alleviates the symptoms of experimental autoimmune encephalomyelitis. Autoimmunity. 2017;50(5):317–328. doi: 10.1080/08916934.2017.1332185 [DOI] [PubMed] [Google Scholar]
- 35.Singla AK, Gurram RK, Chauhan A, et al. Caerulomycin a suppresses immunity by inhibiting T cell activity. PLoS One. 2014;9(10):e107051. doi: 10.1371/journal.pone.0107051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhavna V. Synthesis and Evaluation of 2,2’-Bipyridine Derivatives for Therapeutic Applications. CSIR- Institute of Microbial Technology; 2018. [Google Scholar]
- 37.Jolly RS, Sharma AN, Mishra P, Vad B, Khatri N. (12) INTERNATIONAL APPLICATION PUBLISHED UNDER the PATENT COOPERATION TREATY (Pct) (19). World Intellectual Property Organization International Bureau.; 2018. [Google Scholar]
- 38.Bhagyaraj E, Ahuja N, Kumar S, et al. TGF-β induced chemoresistance in liver cancer is modulated by xenobiotic nuclear receptor PXR. Cell Cycle. 2019;18(24):3589–3602. doi: 10.1080/15384101.2019.1693120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Arora R, Gupta S, et al. Nuclear receptor Rev-erbα role in fine-tuning erythropoietin gene expression. Blood Adv. 2024;8(14):3705–3717. doi: 10.1182/bloodadvances.2023012228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tambat R, Jangra M, Mahey N, et al. Microbe-derived indole metabolite demonstrates potent multidrug efflux pump inhibition in staphylococcus aureus. Front Microbiol. 2019;10:10. doi: 10.3389/fmicb.2019.02153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- 42.Međedović M, Rilak Simović A, Ćoćić D, et al. New ruthenium(II) complexes with quinone diimine and substituted bipyridine as inert ligands: synthesis, characterization, mechanism of action, DNA/HSA binding affinity and cytotoxic activity. Dalton Trans. 2023;52(5):1323–1344. doi: 10.1039/D2DT02993F [DOI] [PubMed] [Google Scholar]
- 43.Côrte-Real L, Karas B, Brás AR, et al. Ruthenium-cyclopentadienyl bipyridine-biotin based compounds: synthesis and biological effect. Inorg Chem. 2019;58(14):9135–9149. doi: 10.1021/ACS.INORGCHEM.9B00735/ASSET/IMAGES/LARGE/IC-2019-00735C_0008.JPEG [DOI] [PubMed] [Google Scholar]
- 44.Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nature Protocols. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339 [DOI] [PubMed] [Google Scholar]
- 45.Soumya T, Lakshmipriya T, Klika KD, Jayasree PR, Manish Kumar PR. Anticancer potential of rhizome extract and a labdane diterpenoid from curcuma mutabilis plant endemic to western ghats of India. Sci Rep. 2021;11(1). doi: 10.1038/S41598-020-79414-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y, Zhang M, Zhang Z, Jia Y, Zhang C, Peng L. Hydrazinocurcumin and 5-fluorouracil enhance apoptosis and restrain tumorigenicity of HepG2 cells via disrupting the PTEN-mediated PI3K/Akt signaling pathway. Biomed Pharmacother. 2020;129:109851. doi: 10.1016/J.BIOPHA.2020.109851 [DOI] [PubMed] [Google Scholar]
- 47.Xie H, Mao L, Fan G, et al. Design and synthesis of cabotegravir derivatives bearing 1,2,3-triazole and evaluation of anti-liver cancer activity. Front Pharmacol. 2023:14. doi: 10.3389/FPHAR.2023.1265289/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Liu Y, Cui J, et al. Effects of Scutellaria strigillosa Hemsl. extract on HepG2 cell proliferation and apoptosis through binding to aspartate β-hydroxylase. Biochem Biophys Res Commun. 2023;668:62–69. doi: 10.1016/J.BBRC.2023.05.077 [DOI] [PubMed] [Google Scholar]
- 49.Yan J, Xie B, Tian Y, et al. MicroRNA-5195-3p mediated malignant biological behaviour of insulin-resistant liver cancer cells via SOX9 and TPM4. BMC Cancer. 2023;23(1):1–13. doi: 10.1186/S12885-023-11068-X/FIGURES/7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sivandzade F, Bhalerao A, Cucullo L. Analysis of the mitochondrial membrane potential using the cationic JC-1 Dye a sensitive fluorescent probe. Biol Protoc. 2019;9(1). doi: 10.21769/BIOPROTOC.3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiao Y, Xiang Q, Yuan L, Xu L, Liu Z, Liu X. Herbacetin induces apoptosis in HepG2 cells: involvements of ROS and PI3K/Akt pathway. Food and Chemical Toxicology. 2013;51(1):426–433. doi: 10.1016/j.fct.2012.09.036 [DOI] [PubMed] [Google Scholar]
- 52.Yuan L, Wang J, Xiao H, Xiao C, Wang Y, Liu X. Isoorientin induces apoptosis through mitochondrial dysfunction and inhibition of PI3K/Akt signaling pathway in HepG2 cancer cells. Toxicol Appl Pharmacol. 2012;265(1):83–92. doi: 10.1016/j.taap.2012.09.022 [DOI] [PubMed] [Google Scholar]
- 53.Pavlina M, Jan C, Filip P, Jiri H, Tomas R. Quantitative spectrofluorometric assay detecting nuclear condensation and fragmentation in intact cells. Sci Rep. 2021;11(1):1–13. doi: 10.1038/s41598-021-91380-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdraboh ME, Essa ZS, Abdelrazzak AB, et al. Radio-sensitizing effect of a cocktail of phytochemicals on HepG2 cell proliferation, motility and survival. Biomed Pharmacother. 2020;131:110620. doi: 10.1016/J.BIOPHA.2020.110620 [DOI] [PubMed] [Google Scholar]
- 55.Mujwar S. Computational bioprospecting of andrographolide derivatives as potent cyclooxygenase-2 inhibitors. Biomed Biotechnol Res J. 2021;5(4):446–450. doi: 10.4103/BBRJ.BBRJ_56_21 [DOI] [Google Scholar]
- 56.Mujwar S, Harwansh RK. In silico bioprospecting of taraxerol as a main protease inhibitor of SARS-CoV-2 to develop therapy against COVID-19. Struct Chem. 2022;33(5):1517–1528. doi: 10.1007/S11224-022-01943-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mujwar S, Sun L, Fidan O. In silico evaluation of food-derived carotenoids against SARS-CoV-2 drug targets: crocin is a promising dietary supplement candidate for COVID-19. J Food Biochem. 2022;46(9). doi: 10.1111/JFBC.14219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh NK, Mujwar S, Garabadu D. in silico anti-cholinestarase activity of flavonoids: a computational approach. Asian J Chem. 2019;31(12):2859–2864. doi: 10.14233/AJCHEM.2019.22153 [DOI] [Google Scholar]
- 59.Kciuk M, Mujwar S, Rani I, et al. Computational bioprospecting guggulsterone against ADP ribose phosphatase of SARS-CoV-2. Molecules. 2022;27(23):8287. doi: 10.3390/MOLECULES27238287/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mountzios G, Kostopoulos I, Kotoula V, et al. INSILICO ANALYSIS OF DIETARY AGENTS AS ANTICANCER INHIBITORS OF INSULIN LIKE GROWTH FACTOR 1 RECEPTOR (IGF1R). Int J Pharm Sci. 2015;8(1):191–196. doi: 10.1371/JOURNAL.PONE.0054048 [DOI] [Google Scholar]
- 61.Yang H, Wei L, Xun Y, Yang A, You H. BRD4: an emerging prospective therapeutic target in glioma. Mol Ther Oncolytics. 2021;21:1. doi: 10.1016/J.OMTO.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu J, Pan D, Li G, Chen K, Hu X. Regulation of programmed cell death by Brd4. Cell Death Dis. 2022;13(12):1–15. doi: 10.1038/s41419-022-05505-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang N, Wu R, Tang D, Kang R. The BET family in immunity and disease. Signal Transduction and Targeted Therapy. 2021;6(1):1–22. doi: 10.1038/s41392-020-00384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/NAR/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deshpande N, Addess KJ, Bluhm WF, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33(Database Issue):D233. doi: 10.1093/NAR/GKI057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta N, Qayum A, Singh S, Mujwar S, Sangwan PL. Isolation, anticancer evaluation, molecular docking, drug likeness and ADMET studies of secondary metabolites from psoralea corylifolia seeds. Chemistry Select. 2022;7(41):e202202115. doi: 10.1002/SLCT.202202115 [DOI] [Google Scholar]
- 67.Morris GM, Ruth H, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785. doi: 10.1002/JCC.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jain R, Mujwar S. Repurposing metocurine as main protease inhibitor to develop novel antiviral therapy for COVID-19. Struct Chem. 2020;31(6):2487. doi: 10.1007/S11224-020-01605-W [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mujwar S, Kumar V. Computational drug repurposing approach to identify potential fatty acid-binding protein-4 inhibitors to develop novel antiobesity therapy. Assay Drug Dev Technol. 2020;18(7):318–327. doi: 10.1089/ADT.2020.976 [DOI] [PubMed] [Google Scholar]
- 70.Rani I, Goyal A, Sharma M. Computational design of phosphatidylinositol 3-Kinase Inhibitors. Assay Drug Dev Technol. 2022;20(7):317–337. doi: 10.1089/ADT.2022.057 [DOI] [PubMed] [Google Scholar]
- 71.Koç E, Çelik-uzuner S, Uzuner U, Çakmak R. The detailed comparison of cell death detected by annexin v-pi counterstain using fluorescence microscope, flow cytometry and automated cell counter in mammalian and microalgae cells. J Fluoresc. 2018;28(6):1393–1404. doi: 10.1007/S10895-018-2306-4/METRICS [DOI] [PubMed] [Google Scholar]
- 72.Wong RSY. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30(1):1–14. doi: 10.1186/1756-9966-30-87/TABLES/3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yue YJ, Xu S, ning SY, Xu Y, long GQ, bin WL. Novel CDK9 inhibitor oroxylin A promotes wild-type P53 stability and prevents hepatocellular carcinoma progression by disrupting both MDM2 and SIRT1 signaling. Acta Pharmacol Sin. 2022;43(4):1033. doi: 10.1038/S41401-021-00708-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan H. Medicinal plants in light of history: recognized therapeutic modality. J Evid Based Complementary Altern Med. 2014;19(3):216–219. doi: 10.1177/2156587214533346 [DOI] [PubMed] [Google Scholar]
- 75.Cousins KR. Computer review of ChemDraw Ultra 12.0. J Am Chem Soc. 2011;133(21):8388. doi: 10.1021/JA204075S [DOI] [PubMed] [Google Scholar]
- 76.Cousins KR. ChemDraw Ultra 9.0. CambridgeSoft, 100 CambridgePark Drive, Cambridge, MA 02140. www. cambridgesoft.com. See Web site for pricing options. J Am Chem Soc. 2005;127(11):4115–4116. doi: 10.1021/JA0410237 [DOI] [Google Scholar]
- 77.Coffman-D’annibale K, Xie C, Hrones DM, Ghabra S, Greten TF, Monge C. The current landscape of therapies for hepatocellular carcinoma. Carcinogenesis. 2023;44(7):537–548. doi: 10.1093/carcin/bgad052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.El-bendary MM, Akhdhar A, Ali EMM, et al. Synthesis, structure characterization and antitumor activities of copper and cobalt thiocyanate complexes with 3-acetylpyridine ligand. Polyhedron. 2023:242. doi: 10.1016/j.poly.2023.116511 [DOI] [Google Scholar]
- 79.El-Shafey ES, Elsherbiny ES. Therapeutic potential of a 2,2’-bipyridine-based vanadium(IV) complex on HepG2 cells: cytotoxic effects and molecular targeting. Egypt J Basic Appl Sci. 2023;10(1):204–217. doi: 10.1080/2314808X.2023.2176969 [DOI] [Google Scholar]
- 80.Rodrigues JHV, de Carvalho AB, Silva VR, et al. Copper(II)/diiminic complexes based on 2-hydroxybenzophenones: DNA- and BSA-binding studies and antitumor activity against HCT116 and HepG2 tumor cells. Polyhedron. 2023:239. doi: 10.1016/j.poly.2023.116431 [DOI] [Google Scholar]
- 81.Zhang HQ, Lu X, Wu JL, et al. Discovery of mitochondrion-targeting copper(ii)-plumbagin and -bipyridine complexes as chemodynamic therapy agents with enhanced antitumor activity. Dalton Trans. 2024;53(7):3244–3253. doi: 10.1039/d3dt03806h [DOI] [PubMed] [Google Scholar]
- 82.Karges J, Stokes RW, Cohen SM. Metal complexes for therapeutic applications. Trends Chem. 2021;3(7):523–534. doi: 10.1016/j.trechm.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang H, Zhou X, Li C, et al. The emerging role of pyroptosis in pediatric cancers: from mechanism to therapy. J Hematol Oncol. 2022;15(1). doi: 10.1186/s13045-022-01365-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Häcker G. The morphology of apoptosis. Cell Tissue Res. 2000;301(1):5–17. doi: 10.1007/S004410000193/METRICS [DOI] [PubMed] [Google Scholar]
- 85.Saraste A, Pulkki K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res. 2000;45(3):528–537. doi: 10.1016/S0008-6363(99)00384-3/2/45-3-528-FIG2.GIF [DOI] [PubMed] [Google Scholar]
- 86.Kroemer G, El-Deiry WS, Golstein P, et al. Classification of cell death: recommendations of the nomenclature committee on cell death. Cell Death Differ. 2005;12(S2):1463–1467. doi: 10.1038/sj.cdd.4401724 [DOI] [PubMed] [Google Scholar]
- 87.Lin YC, Chipot C, Scheuring S. Annexin-V stabilizes membrane defects by inducing lipid phase transition. Nat Commun. 2020;11(1):1–13. doi: 10.1038/s41467-019-14045-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elefantova K, Lakatos B, Kubickova J, Sulova Z, Breier A. Detection of the mitochondrial membrane potential by the cationic dye JC-1 in L1210 cells with massive overexpression of the plasma membrane ABCB1 drug transporter. Int J Mol Sci. 2018;19(7):1985. doi: 10.3390/IJMS19071985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tirichen H, Yaigoub H, Xu W, Wu C, Li R, Li Y. Mitochondrial reactive oxygen species and their contribution in chronic kidney disease progression through oxidative stress. Front Physiol. 2021;12:627837. doi: 10.3389/FPHYS.2021.627837/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Giorgio M, Migliaccio E, Orsini F, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122(2):221–233. doi: 10.1016/j.cell.2005.05.011 [DOI] [PubMed] [Google Scholar]
- 91.Niu D, Zhang J, Ren Y, Feng H, Chen WN. HBx genotype D represses GSTP1 expression and increases the oxidative level and apoptosis in HepG2 cells. Mol Oncol. 2009;3(1):67–76. doi: 10.1016/J.MOLONC.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dröge W. Oxidative Stress and Aging. Adv Exp Med Biol. 2003;543:191–200. doi: 10.1007/978-1-4419-8997-0_14 [DOI] [PubMed] [Google Scholar]
- 93.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–156. doi: 10.1038/nrd4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol. 2018;15(5):273–291. doi: 10.1038/nrclinonc.2018.28 [DOI] [PubMed] [Google Scholar]
- 95.Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18(1):1–28. doi: 10.1186/S12943-019-0954-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee T, Yao G, Nevins J, You L. Sensing and integration of Erk and PI3K signals by Myc. PLoS Comput Biol. 2008;4(2):e1000013. doi: 10.1371/journal.pcbi.1000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dey N, Leyland-Jones B, De P. MYC-Xing It up with PIK3CA Mutation and Resistance to PI3K Inhibitors: summit of two giants in breast cancers. Am J Cancer Res 2015. 5:1. [PMC free article] [PubMed] [Google Scholar]
- 98.Karoulia Z, Gavathiotis E, Poulikakos PI. New perspectives for targeting RAF kinase in human cancer. Nat Rev Cancer. 2017;17(11):676–691. doi: 10.1038/nrc.2017.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Calderaro J, Ziol M, Paradis V, Zucman-Rossi J Molecular and Histological Correlations in Liver Cancer.; 2019. Available from: https://gco.iarc.fr/. Accessed November 06, 2024. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are available within the article.