Abstract
Nucleoporins (nups) in the nuclear pore complex (NPC) form a selective barrier that suppresses the diffusion of most macromolecules while enabling rapid transport of nuclear transport receptor (NTR)–bound cargos. Recent studies have shown that the NPC may dilate and constrict, but how altering the NPC diameter affects its selective barrier properties remains unclear. Here, we build DNA nanopores with programmable diameters and nup arrangements to model the constricted and dilated NPCs. We find that Nup62 proteins form a dynamic cross-channel barrier impermeable to hepatitis B virus (HBV) capsids when grafted inside 60-nm-wide nanopores but not in 79-nm pores, where Nup62 cluster locally. Furthermore, importin-β1 substantially changes the dynamics of Nup62 assemblies and facilitates the passage of HBV capsids through the 60-nm NPC mimics containing Nup62 and Nup153. Our study shows that transport channel width is critical to the permeability of nup barriers and underscores NTRs’ role in dynamically remodeling nup assemblies and mediating the nuclear entry of viruses.
DNA-based nanopores illustrate how changes in nuclear pore width may affect the transport of ultralarge cargos like virus cores.
INTRODUCTION
Nuclear pore complexes (NPCs)—massive (~50 to 120 MDa) protein channels spanning the nuclear membranes—control molecular transport between the nucleus and cytoplasm (1, 2). In an NPC, hundreds of nucleoporins (nups) containing unstructured Phe-Gly (FG) domains densely decorate a central channel (also known as the central transporter) as narrow as ~40 nm. These spatially confined FG-nups generate a diffusion barrier that prevents inert molecules larger than a few nanometers (~40 kDa) from freely crossing but allows nuclear transport receptors (NTRs) to rapidly ferry cargos up to tens of nanometers in size (several megadaltons). The exact physicochemical state of the FG-nups in the NPC is still under debate and has been depicted as a hydrogel (3, 4), polymer brush (5, 6), and phase-separated liquid droplet (7). Nevertheless, the multivalent and transient nup-nup and nup-NTR interactions have long been recognized as the key contributors underlying the NPC’s function as a selective barrier (8–11). In recent years, the structural flexibility of the NPC has been increasingly appreciated via in cellulo cryo–electron tomography, demonstrating central channels with diameters ranging from ~40 to 70 nm depending on the cell state (12–14). These structural studies raise the notion that the width of a nuclear pore may modulate its barrier function.
In addition to the central role the nucleocytoplasmic transport machinery plays in maintaining the life cycle of healthy cells, it is also exploited by viruses to deliver viral genomes to the nucleus of infected cells (15). A common strategy viruses use is to disguise their capsids—the major structural component of the virus core—as cargos for nuclear transport. To achieve this, virus capsids incorporate nup-binding features to directly engage with NPCs (e.g., HIV-1) (16–19) and/or display sequences that recruit NTRs to mediate their nuclear import [e.g., hepatitis B virus (HBV)] (19). Notably, nearly intact capsids of certain viruses (such as HIV-1 and HBV) can cross the NPC (20, 21). With the widths (30 to 65 nm) of these virus cores approaching that of the NPC’s central channel, we consider the possibility that dilated nuclear pores are preferred or even required for the transport of such ultralarge foreign objects.
The challenges to deterministically manipulate the NPC structure and function have made it difficult to establish a clear-cut correlation between the central channel width and NPC permeability. Because of the structural complexity of the NPC and the functional redundancy of FG domains, there remains no obvious way to manipulate the NPC diameter genetically. Structures of NPCs derived from purified nuclear envelopes are in a constricted state (22). By contrast, biomimetic nanopores can be built with predefined geometry and nup composition while recapitulating the NPC’s basic barrier and transport functionalities (23–27). Promisingly, a recent study showed that the transport selectivity of FG-nup–functionalized solid-state nanopores diminishes with increasing diameter beyond 55 nm (28). However, the permeability of the NPC mimics in this study was established using relatively small proteins (<100 kDa). Furthermore, the distribution of FG domains as a function of pore diameter was not experimentally probed. In a separate study, we showed a minimal width requirement for insertion of HIV-1 capsids deep into DNA origami–based NPC mimics (termed NuPODs for nups organized on DNA) (29). However, such a requirement was attributed to size compatibility (i.e., a NuPOD must be at least as wide as the capsid to permit insertion) rather than altered nup behaviors in channels of different widths. Therefore, how the nanopore width regulates FG-nup morphology, dynamics, and selectivity for large (>10 nm) cargos remains an open question.
To address this question, here, we build NuPODs with two defined widths to mimic the central channels of constricted and dilated NPCs. As a reductionist model system, we grafted Nup62, a major constituent (along with Nup58 and Nup54) of the central transport channel, and Nup153, an FG-nup at the nuclear basket with known affinity for HBV (30), to the interior of DNA origami nanopores to mimic their stoichiometry and localization in the human NPC (2, 31). Transmission electron microscopy (TEM) and high-speed atomic force microscopy (HS-AFM) reveal drastically different FG-nup morphology and dynamics in NuPODs that differ by ~20 nm in width. The cross-channel nup meshwork and the central plug–like Nup62/importin-β1 complexes are unique to the narrower NuPOD; the absence of these features in the wider NuPOD suggests a less stringent barrier. Corroborating these findings, the passage of HBV capsids (~30 nm in diameter) through the narrower Nup62-Nup153 NuPOD necessitates importin-β1, an NTR known to bind both nups (32) and the HBV core (19, 33), whereas the capsids can readily cross the wider pore housing the same nups without importin-β1. The reductionist NuPOD system thus provides compelling evidence supporting a potential regulatory role for central channel width in modulating the NPC’s permeability to large-size objects, suggesting that geometrical (e.g., pore diameter) and biochemical (e.g., NTR concentration) properties codetermine the barrier-transport activities of the NPC.
RESULTS
Two NuPODs mimic NPC central channels of different widths
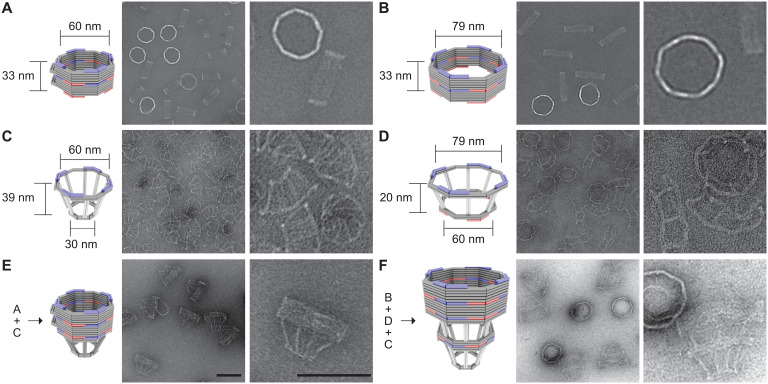
To model variations in NPC diameter, we designed two DNA origami nanopores that differ chiefly in width (60 nm versus 79 nm; Fig. 1, A and B). Both nanopores are 33-nm-deep dimers, have an octagonal cross section, and display single-stranded DNA (ssDNA) extensions (handles) on the interior for attaching nups. Accounting for the lengths of DNA handles (typically 21 bases or ~7 nm), the nup anchoring points on the opposite faces of the channel are theoretically 46 and 65 nm apart in the two design variants, approximating the diameters of a constricted and dilated NPC central channel, respectively. A typical DNA nanopore carries 32 handles distributed symmetrically in the middle of the channel for anchoring Nup62 and another 32 handles near one end for Nup153, closely mimicking these FG-nups’ stoichiometry and respective residence in the central channel and nuclear basket of a native NPC (2, 31). The two sets of handles have orthogonal sequences, and their length, copy number, and location can be easily redesigned. We name NuPODs based on width and nup composition, for example, “60-nm Nup62 NuPOD” (designed nup copy number = 32 unless otherwise specified). We further designed frustum-like DNA baskets (Fig. 1, C and D) that can heterodimerize with one another and dock onto one end of the nanopores via shape-complementary interfaces (Fig. 1, E and F, and fig. S1). Besides providing a visual marker for the “nucleoplasm-facing” end of the NuPODs where Nup153 is attached, the narrow opening of the basket precludes the entry of objects exceeding 30 nm in diameter, paving the way for a TEM-based HBV penetration assay.
Fig. 1. DNA origami nanostructures used to build NuPODs.
(A) A 60-nm-wide channel built by homodimerizing DNA nanopores with shape-complementary docking interfaces (blue and red). (B) A 79-nm-wide channel built by homodimerizing DNA nanopores with shape-complementary interfaces. (C) Small DNA basket with a docking interface (blue) compatible with the 60-nm channel. (D) Large DNA basket with a top docking interface (blue) compatible with the 79-nm channel and a bottom interface (red) compatible with the small basket. (E) A 60-nm channel capped by the small basket on one end. (F) A 79-nm channel capped by the two baskets on one end. For each panel, a cartoon model (DNA double helices are represented by rods) is shown next to representative TEM images. The experiment was repeated three times (technical replicates) with similar folding results. Scale bars, 100 nm.
We adapted our established workflow for assembling NuPODs (29). All DNA origami components (monomeric nanopores, and baskets) folded with good yield and expected geometry, as confirmed by agarose gel electrophoresis and negative-stain TEM (Fig. 1, C and D, and fig. S2). Oligomerizing selected monomeric structures yielded open-ended (Fig. 1, A and B) or capped nanopores (Fig. 1, E and F, and fig. S2). The purified nanopores display inward facing handles that guide the subsequent nup attachment. We recombinantly expressed human Nup62FL (full length, amino acid 1 to 522) and Nup153CTD (C-terminal domain, amino acid 896 to 1485) as maltose-binding protein (MBP; for solubility and stability) and SNAP-tag (for DNA conjugation) fusion proteins (fig. S3). Both nups conjugated efficiently with O6-benzylguanine (BG)–labeled DNA oligonucleotides complementary to the handles (termed anti-handles; Fig. 2A and fig. S3). Adding the purified nup–anti-handle conjugates to DNA nanopores bearing cognate handles generated NuPODs with designated channel widths and nup configurations. As the expected result of nup attachment, all NuPODs migrated slower than the corresponding empty nanopores during electrophoresis in agarose gels (fig. S3). Using rate-zone ultracentrifugation to remove unbound nups and Western blots to determine the amount of nanopore-attached nups, we estimated an average of ~31 copies of Nup62 in a NuPOD, closely matching the designed number (32 copies) of Nup62 anchor points (fig. S4).
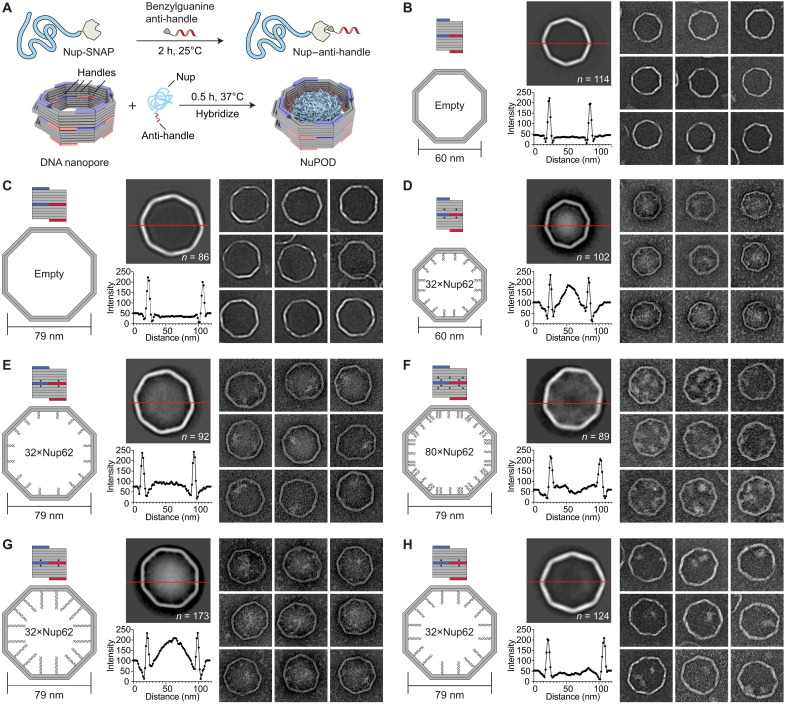
Fig. 2. Morphology of Nup62 inside DNA origami nanopores of different widths.
(A) Schematic diagrams showing the process of attaching nups inside DNA nanopores to form NuPODs. h, hours. (B) Empty 60-nm DNA channel. (C) Empty 79-nm DNA channel. (D) A 60-nm NuPOD with 32 copies of Nup62 grafted on 21-nt handles. (E) A 79-nm NuPOD with 32 copies of Nup62 grafted on 21-nt handles. (F) A 79-nm NuPOD with 80 copies of Nup62 grafted on 21-nt handles. (G) A 79-nm NuPOD with 32 copies of Nup62 grafted on 51-nt handles. (H) A 79-nm NuPOD with 32 copies of Nup62 grafted on 38-nt handles. For (B) to (H), (left) schematics showing an interior face (top; nup-grafting handle positions denoted by black dots) and the top view (bottom; handle/anti-handle pairs shown as double helices) of a DNA channel; (middle) class average negative-stain TEM image (top) and intensity profile across the center of the DNA channel (red line); (right) representative TEM images of the DNA channel. All images are 120 by 120 nm2.
NuPOD width modulates FG-nup morphology
To probe the collective morphology of nanopore-confined FG-nups, we imaged NuPODs housing Nup62 under negative-stain TEM (Fig. 2 and fig. S5). Nup62 is one of the most abundant FG-nups in the central channel of the human NPC where it plays an essential role in nuclear transport. Both 60- and 79-nm NuPODs were assembled without baskets to allow unobstructed view of the nups. Empty nanopores showed a minimal signal in their lumens (Fig. 2, B and C). Anchoring up to 32 copies of Nup62 in the 60-nm nanopore gave rise to strong protein signals (amorphous light-colored bodies against a dark background) near the center of the nanopore (Fig. 2D), suggesting that the Nup62 bridged the channel to occlude the nanopore. Such a cross-channel meshwork is missing from the 79-nm NuPOD bearing 32 copies of Nup62, where one to two off-center clusters formed along the channel rim (Fig. 2E). To test if the different morphology is due to a lower nup density, which could change the conformations of the intrinsically disordered domain of Nup62 from brush-like to mushroom-like, or because the polypeptide chains of Nup62 are not long enough to reach across the wider channel, we performed two sets of control experiments. We first built a 79-nm NuPOD with a maximum of 80 Nup62 proteins with higher grafting density (1/41 nm−2) than the 60-nm 32×Nup62 NuPOD (1/62 nm−2). The elevated nup density only increased the number of clusters near the rim (three to four) but did not reconstitute the pore-filling meshwork (Fig. 2F). Notably, averaged micrographs revealed a ~20-nm-wide void in the center, suggesting that very few Nup62 molecules could reach this area. Second, we used longer handles (51 bases, compared to 21 bases in typical NuPODs) to project Nup62 further inside the lumen of the 79-nm NuPODs. The additional ~10-nm linker length appeared to help Nup62 congregate in the center (Fig. 2G), whereas Nup62 anchored via a medium-length linker (38 bp) still predominantly formed local clusters (Fig. 2H). The FG domains of Nsp1 (amino acid 2 to 603), the yeast ortholog of Nup62, showed similar width-dependent morphologies in NuPODs as Nup62, albeit with a generally more diffuse appearance, perhaps because Nsp1 has weaker cohesiveness compared to Nup62 as suggested by our previous findings (29) (fig. S6). Therefore, we propose that the movement and interactions of nups anchored inside an NPC-like nanopore, hence their abilities to form effective pore-occluding protein bodies, are limited by nup length and the channel width as defined by the nup anchoring points.
NuPOD width modulates the dynamics of the Nup62/importin-β1 complex
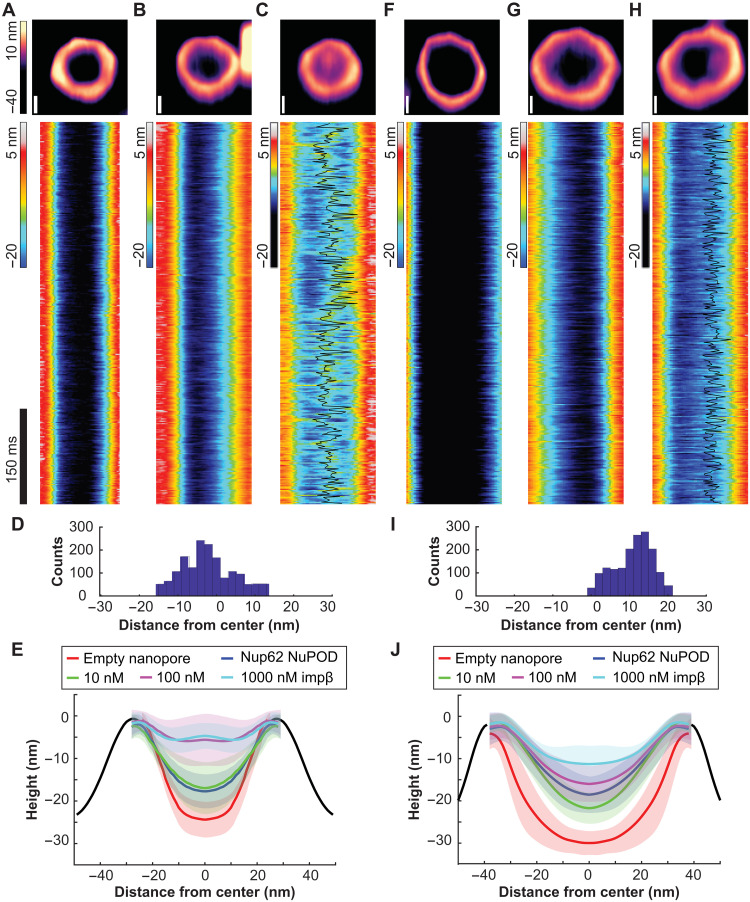
HS-AFM studies on NPCs and NPC mimics have revealed protein dynamics in the central channel on the timescale of nuclear transport events (i.e., milliseconds), which were interpreted as the hallmark of a functional barrier transporter (34–36). We therefore imaged NuPODs by HS-AFM to understand how the pore diameter modulates nup dynamics (Fig. 3). In keeping with our previous studies, we removed MBP tags from NuPODs by tobacco etch virus (TEV) protease to better resolve the FG domains (25, 34, 36). HS-AFM images and cross-sectional height profiles that were quantified from kymographs derived from rapid HS-AFM line scans (~1.9 ms per line) showed height increases in the lumen of 60-nm Nup62 NuPODs compared with empty nanopores and further topographic elevation upon the addition of increasing concentrations (1 to 1000 nM) of importin-β1, confirming that the nanopore-confined Nup62 retained their native affinity to the NTR (Fig. 3, A to C and E, and fig. S7). Adding 100 nM importin-β1 to the 60-nm Nup62 NuPODs formed a dense yet mobile structure resembling the “central plug” in the NPC patrolling the entire nanopore (Fig. 3, C and D, and movie S1) (36). While still dynamic and capable of importin-β1 binding, Nup62 in the 79-nm NuPODs behaved very differently (Fig. 3, F to J). The movement of Nup62/importin-β1 complexes was largely peripheral inside the 79-nm channels (Fig. 3, H and I, and movie S2), leaving a large portion of the nanopore seemingly empty most of the time. These findings are in qualitative agreement with our TEM studies and reinforce the role of channel width in regulating the collective nup behaviors.
Fig. 3. Protein dynamics inside Nup62 NuPODs of different widths.
(A) Empty 60-nm DNA channel. (B) A 60-nm Nup62 NuPOD. (C) A 60-nm Nup62 NuPOD with 100 nM importin-β1 (impβ). In (A) to (C), (top row) representative AFM images, scale bars, 20 nm; (middle row) representative kymographs derived from HS-AFM line scans (1.875 ms per line), with movement tracking of the central plug–like cluster overlaid (black line) for the 100 nM impβ condition. (D) Histogram summarizing the positions of the central plug–like cluster in the 60-nm Nup62 NuPOD with 100 nM impβ. (E) Height profiles of empty nanopores and NuPODs with different concentrations of importin-β1. Solid curves and shadows represent the means and SDs of the line scan height profiles, respectively. Number of nanopores measured: 6 (empty pore), 10 (NuPOD), 7 (NuPOD+10 nM impβ), 8 (NuPOD+100 nM impβ), and 8 (NuPOD+1 μM impβ). (F to J) Same as (A) to (E), except for the 79-nm pores. Number of nanopores measured: 12 (empty pore), 7 (NuPOD), 6 (NuPOD+10 nM impβ), 12 (NuPOD+100 nM impβ), and 11 (NuPOD+1 μM impβ). DNA handles are 21-nt long (~7 nm upon hybridization with anti-handles).
NuPOD width modulates Nup62 barrier permeability against HBV capsids
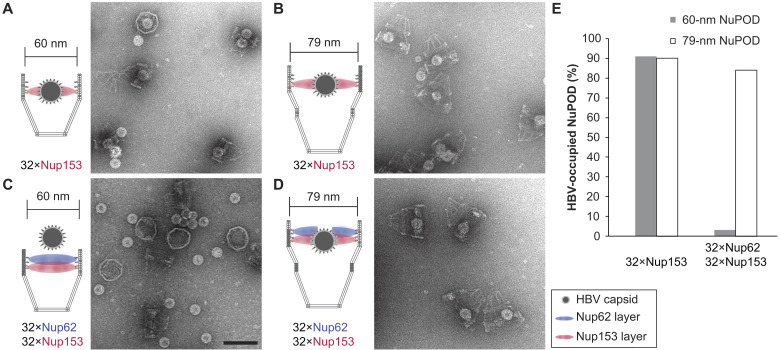
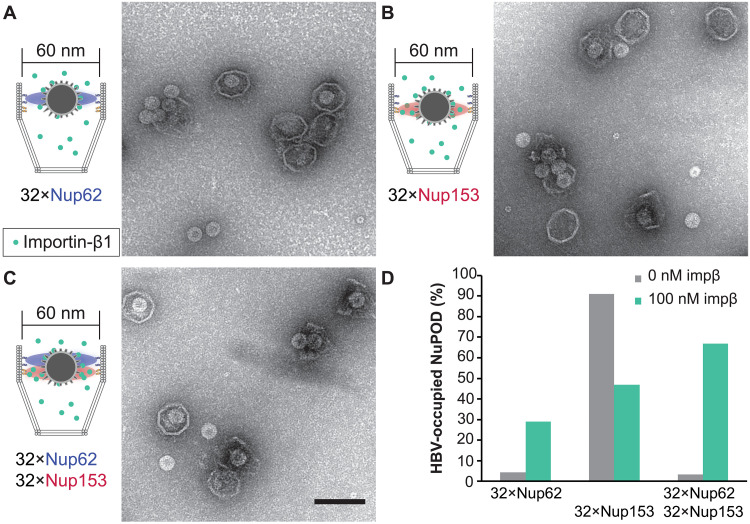
The absence of a central plug–like feature that dynamically occludes the 79-nm NuPODs suggests that the FG-nup barriers in the wider NuPODs may be less stringent (i.e., leakier) than their narrower counterparts. We therefore wanted to compare the permeability of 60- and 79-nm-wide NuPODs against HBV capsids. In cells, the binding of HBV capsids with importins and Nup153, a nucleoplasm-facing FG-nup, have been proposed to mediate the translocation of the virus core through the NPC (37, 38). We assembled HBV capsids in vitro from HBV core protein (HBc) expressed in cultured mammalian cells, generating near-spherical protein shells devoid of viral genome with an average diameter of ~31.0 ± 1.7 nm (fig. S8). Using an ultracentrifugation-based co-migration assay (see Materials and Methods), we confirmed that Nup153CTD, but not Nup62FL, strongly binds HBV capsids (fig. S9). Consistent with these results, HBV capsids entered basket-bearing Nup153 NuPODs efficiently regardless of the pore diameter, with ~90% NuPODs occupied by least one capsid (Fig. 4, A, B, and E), while the Nup62 NuPODs showed negligible capsid binding (fig. S8). Moreover, HBV capsids were largely excluded from 60-nm Nup62-Nup153 NuPODs (~3% occupancy), suggesting that the Nup62 layer effectively blocked the Nup153-capsid interaction (Fig. 4C). This is in stark contrast to the 79-nm Nup62-Nup153 NuPOD, whose capsid binding (~84% occupancy) was on par with the Nup153 only NuPODs (Fig. 4, D and E). From the side views of these HBV-occupied NuPODs, we found a distribution of capsid penetration depths centering around the expected location of the Nup153 anchors (fig. S10), reflecting Nup153-HBV binding and the extended conformation of Nup153. Because capsids must first encounter with Nup62 to engage Nup153 in the dual-nup NuPODs (the Nup153 end is capped), the increased capsid binding indicates higher permeability of the Nup62 barrier, supporting our hypothesis that wider channels lead to leakier FG-nup barriers that permit the passage of otherwise inadmissible large objects.
Fig. 4. HBV capsid interactions with NuPODs of different widths.
(A) HBV capsids mixed with capped 60-nm Nup153 NuPODs. (B) HBV capsids mixed with capped 79-nm Nup153 NuPODs. (C) HBV capsids mixed with capped 60-nm Nup62-Nup153 NuPODs. (D) HBV capsids mixed with capped 79-nm Nup62-Nup153 NuPODs. For (A) to (D), schematic diagrams of the binding experiments are shown next to representative TEM images. Scale bar, 100 nm. (E) Percentages of NuPODs occupied by HBV capsids in experiments (A) to (D). The experiments were repeated two to three times (technical replicates) with similar results. NuPODs counted in each experiment are (from left to right) 168, 247, 279, and 215.
Importin-β1 mediates deep penetration of HBV capsids into NuPODs
In native NPCs, NTRs are an integral part of the selective barrier and are responsible for the transport of cellular and foreign cargos. Having established that 60-nm Nup62 NuPODs interact with importin-β1 to form central plug–like structures, we asked if such morphology translates to NPC-like selectivity. We continued to use the HBV capsid as a model cargo. Although nuclear import of mature HBV cores requires both importin-α and importin-β (39), empty HBV capsids bind directly to importin-β1 (33), thereby simplifying our setup (fig. S9). Importin-β1 substantially increased the binding between 60-nm Nup62 NuPODs and HBV capsids (Fig. 5, A and D, left). Preincubating Nup153 NuPODs with 100 nM importin-β1 resulted in a partial loss of the NuPOD’s affinity to capsids (Fig. 5, B and D, middle). This is expected, considering that both importin-β1 and HBV capsids compete for the Nup153 FG domain binding (30). In the presence of importin-β1, about two-thirds of 60-nm Nup62-Nup153 NuPODs were occupied by capsids, a nearly 20-fold increase compared to the importin-free condition (Fig. 5, C and D, right). There appeared to be three populations of capsids in these dual-nup NuPODs, with the first two groups around the Nup62 and Nup153 layer, respectively, and the third group near the bottom of the DNA basket (fig. S10). Because importin-β1 is known to collapse Nup153 to its anchoring point (6), we interpret the deep penetration events (i.e., the third group) as the result of capsids being released from Nup153, consistent with the reduced HBV binding of Nup153 NuPODs caused by importin-β1. In contrast to the capsids assembled from the wild-type (WT) HBc, similarly sized capsids formed by HBc with a truncated C terminus (fig. S8), which bind to Nup153 but not importins (fig. S9) (30), largely failed to enter the 60-nm Nup62-Nup153 NuPODs with or without importin-β1 (fig. S11). Thus, HBV capsids penetrate the Nup62 layer in the 60-nm NuPODs in an importin-β1–dependent manner, demonstrating that these NuPODs not only form passive diffusion barriers but also support selective, NTR-mediated transport.
Fig. 5. Importin-mediated HBV capsid interaction with NuPODs.
(A) HBV capsids mixed with capped 60-nm Nup62 NuPODs in the presence of 100 nM importin-β1 (impβ). (B) HBV capsids mixed with capped 60-nm Nup153 NuPODs in the presence of 100 nM impβ. (C) HBV capsids mixed with capped 60-nm Nup62-Nup153 NuPODs in the presence of 100 nM impβ. For (A) to (C), schematic diagrams of the binding experiments are shown next to representative TEM images. The experiments were repeated twice (technical replicates) with similar results. Scale bar, 100 nm. (D) Percentages of NuPODs occupied by HBV capsids with (green) or without (gray) 100 nM impβ. NuPODs counted in each experiment (from left to right): 288, 206, 168, 380, 279, and 203. Occupancies of importin-free Nup153 and Nup62-Nup153 NuPODs (Fig. 4E) are shown here for comparison.
DISCUSSION
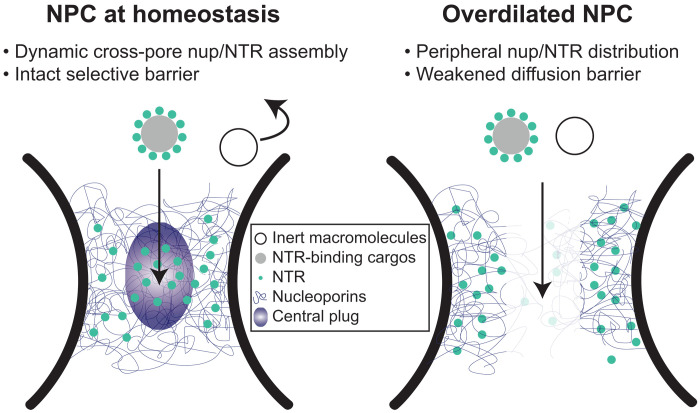
Regulation of nucleocytoplasmic transport has been attributed to biochemical determinants, such as the charge and hydrophobicity of FG-nups (40–42), NTR-nup association (10, 11, 32), and the Ran-GTP (guanosine triphosphate) cycle (43). Intrigued by the heterogeneous nuclear pore diameters observed in cells (12–14) and the increased nuclear import of a transcription factor caused by stretching nuclei (44), we asked how the width of the central transport channel, a geometrical attribute of the NPC, may regulate the behavior of resident FG-nups. In this study, we showed distinct Nup62 morphologies and dynamics in DNA nanopores of different widths, which we correlated to their contrasting permeability to HBV capsids. We found that increasing the channel width can limit the cross-channel FG-nup interactions, hinder the formation of a dynamic central cluster in the presence of importin-β1, and leave the channel less guarded (Fig. 6). The pore-spanning FG-nup meshwork could not be rescued by increasing the Nup62 grafting density in the wider nanopore but was restored by extending the Nup62-anchoring DNA handles, confirming that the pore diameter is a fundamental determinant of cross-channel FG-nup interactions. The width-dependent morphology is shared by Nsp1 and Nup62 despite their different tendency to self-interact, suggesting that nuclear pore dilation–induced barrier permeabilization may be universal, although the exact behavior of the other FG-nups and their subcomplexes warrant further study.
Fig. 6. Proposed model of diameter-modulated NPC selectivity.
(Left) Cells maintain a range of nuclear pore sizes, under which a dynamic central plug consisting of nups and NTRs define the selectivity of nucleocytoplasmic transport. (Right) When nuclear pores dilate beyond the homeostatic range, the central channel of the NPC contains less gatekeeping molecules (nups and NTRs), leading to a weaker barrier that allows nonselective transport events.
The NuPOD penetrating behaviors of HBV capsids are consistent with the previously established roles of Nup153 and importin-β in mediating the HBV nuclear import (37, 38). However, organizing FG-nups in the DNA nanopores revealed emerging properties not readily observable by canonical in vitro assays. For example, although free Nup62 showed moderate affinity toward HBV capsids, the Nup62 NuPODs almost always failed to capture capsids without the help of importin-β1. Combined with the Nup62 clustering in NuPODs, this indicates that the cohesive interactions among nanopore-confined Nup62 molecules altered their binding capabilities with virus capsids, consistent with their HIV-binding behaviors in our prior work (29). HBV capsids, which require importin-β1 to breach the Nup62 barrier in the 60-nm nanopore, went through the Nup62-decorated 79-nm nanopores independently of importin, suggesting that nuclear pore dilation, in addition to NTRs, may aid the nuclear import of the virus and possibly other large objects (Fig. 6).
Although our current penetration assay only captures the equilibrium state (typically after 15 min; see fig. S12 for a time-course study) of the capsid-NuPOD interaction, the TEM study still provides a glimpse of the dynamicity of the nuclear transport process. In the absence of importin-β1, interactions between NuPODs and HBV capsids were binary: Nearly the entire NuPOD population was either occupied by or devoid of HBV capsids (Fig. 4), reflecting the FG-nups’ barrier formation and HBV binding activities. Introducing importin-β1 gave rise to a range of NuPOD occupancy by capsids (Fig. 5), suggesting that the importin-associated cargos are translocated across the NuPODs with comparable on and off rates. These observations further support the role of NTRs in creating a fluid yet selective barrier, in which the transient nup-NTR interactions mediate the cargo transport (10, 32, 36, 45).
Powered by DNA nanotechnology (46, 47), the NuPOD platform has enabled the reconstitution of precisely engineered NPC mimics that form selective barriers against macromolecules (25, 27, 29, 48), affording the opportunity to dissect the sophisticated nuclear transport machinery in a reduced-complexity system. This work not only expands our toolbox by introducing more structural components (e.g., wider nanopore and baskets) but also demonstrates the value of NuPODs for the mechanistic study of nuclear import. The NuPODs complement other synthetic nuclear pore mimics, most notably the nup-functionalized solid-state nanopores (23, 24, 28, 40), by allowing the precise placement of multiple FG-nup types and high-resolution study of the FG-nup–filled channel by TEM and AFM. In the future, coupling NuPODs to solid-support nanopores (49, 50) or incorporating them into lipid bilayers (48, 51, 52) would enable the reconstitution of energy-dependent active transport and the measurement of molecular transport rates. Such setups would help determine the impact of transport channel geometry on the kinetics of passive and active transport.
The 60-nm NuPODs formed a dynamic central plug–like Nup62/importin-β1 assembly and permitted importin-β1–dependent HBV capsid insertion, thus faithfully reproducing key structural and functional aspects of the natural NPC (36, 53, 54). However, as with any biomimetic system, one needs to be cognizant of differences between DNA origami–based NPC mimics and proteinaceous NPCs. For example, NuPODs used in this study consist of only 1 or 2 FG-nup species of more than 10 types of FG-nups in human NPCs. Consequently, the overall FG repeat concentration in the DNA channel is lower than in the nuclear pore despite the realistic stoichiometry and grafting density of each nup species. The DNA handles are likely different in size and flexibility compared to the protein linkers that attach FG-nups to the NPC scaffold. Therefore, while we expect the channel width–permeability relationship to qualitatively hold true in cells, some quantitative measurements of the NuPODs, such as the exact pore diameter and capsid occupancy, may not perfectly match biological parameters. Building more realistic NPC mimics containing additional FG-nup species would unveil their individual and synergistic behaviors in modulating transport and remains an important objective. Another difference between a NuPOD and the NPC arises from the negatively charged DNA channel. However, varying the sodium ion concentration (4 to 400 mM) did not noticeably change NuPOD occupancy by HBV capsids (fig. S13), suggesting that the electrostatic property of the DNA nanopore had a minor effect on the capsid-NuPOD interaction. In future studies, cationic block copolymers can be used to further mitigate electrostatic interactions with DNA when necessary (55, 56). Last, mechanical forces are thought to affect nucleocytoplasmic transport by inducing nuclear pore diameter changes and, in turn, leading to spatially reorganized nups (12–14, 57). It would therefore be useful to build flexible, mechanosensitive DNA nanostructures that dynamically expand and contract to mimic the structural heterogeneity and alterations of the NPC. We expect these dynamic NuPODs to unravel mechanistic details that may have gone unnoticed in conventional cell-free experiments, such as the deformation/disintegration of virus capsids and the central channel dilation during the nuclear transport.
MATERIALS AND METHODS
Nup expression and DNA conjugation
Genes encoding Homo sapiens Nup62FL (amino acid 1 to 522) and Nup153CTD (amino acid 896 to 1475), both with MBP and SNAP-tag, were cloned into a pET-28a–derived vector (Novagen). To express the nups, BL21DE3 cells (New England Biolabs) were cultured at 37°C in a shaking incubator until the optical density at 600 nm (OD600) reaches 0.5 to 1. Isopropyl-β-d-thiogalactopyranoside (1 mM) was added to induce expression of target genes; the culture was kept on a shaker at 20°C overnight. The bacteria cells were then pelleted at 4500 rpm using a Beckman Coulter JS-5.0 rotor for 15 min. A cell homogenizer (EmulsiFlex-C3, Avestin) was used to break the bacteria in a lysis buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and 1× Roche cOmplete protease inhibitor cocktail]. The cell lysate was then centrifuged at 38,000 rpm using a Beckman Coulter SW 45 Ti for 30 min to remove cell debris. The lysate containing Nup153 was then loaded to a 5-ml HisTrap column (Cytiva) on an ÄKTA system (Cytiva) and eluted using a 0 to 400 mM imidazole gradient. The lysate containing Nup62 was loaded to 5-ml MBPTrap HP columns (Cytiva) and eluted by 10 mM maltose. The eluted proteins were loaded to a Superdex 200 pg 16/600 column (Cytiva) to further purify target proteins by size exclusion chromatography. The protein purity is confirmed by SDS–polyacrylamide gel electrophoresis (PAGE) and Coomassie stain (Thermo Fisher Scientific).
To prepare BG-modified oligonucleotides (anti-handles), BG-GLA-NHS (New England Biolabs) was dissolved in dimethyl sulfoxide for a final concentration of 20 mM. Dried 5′-amine labeled anti-handles (Integrated DNA Technologies) were resuspended in Milli-Q ultrapure water at the stock concentration of 2 mM. The amine-DNA (0.33 mM) and the N-hydroxysuccinimide (NHS) ester cross-linker (10 mM) were allowed to react in a Hepes (67 mM, pH 8.5) buffer for 30 min in room temperature (RT). The BG-DNA was purified by ethanol precipitation to remove unreacted cross-linkers and resuspended in Milli-Q ultrapure water. Purified BG-DNA (10 μM) was mixed with nups with a SNAP-tag protein (5 μM) in a 50 mM Tris (pH 8.0) buffer and incubated at RT for 2 hours. The reaction mixture was then loaded to a Superdex 200 pg 16/600 column (Cytiva) to purify the anti-handle–conjugated nups from unreacted BG-DNA.
Importin-β1 preparation
Importin-β was expressed and purified according to established protocols (58), with the addition that the importin-β1–containing fraction was dialyzed against the TEV protease cleavage buffer [10 mM tris-HCl (pH 8), 100 mM NaCl, 1 mM dithiothreitol (DTT), and 1 mM EDTA]. After the His6-tag cleavage by TEV protease (expressed in our lab), importin-β1 was purified again using the Ni-NTA column (Roche). This was followed by additional purification using a Superdex 200 size exclusion column. The collected fraction was validated using a 12% SDS gel and flash frozen at −80°C.
DNA origami design and preparation
DNA structures were designed using caDNAno (59). DNA nanopores with handles extended from the 3′ end of staple strands were shown in fig. S1. The handle sequences for attaching Nup62 and Nup153 are 5′-CTACCATCTCTCCTAAACTCA-3′ and 5′-AAATTATCTACCACAACTCAC-3′, respectively. DNA origami three-dimensional (3D) models were rendered using Maya (Autodesk). To fold a monomeric DNA origami structure, a mixture of the M13mp18-derived bacteriophage circular ssDNA (8064 nt, 20 nM) and staple oligonucleotides (120 nM each) in a 1× TE buffer [5 mM Tris and 1 mM EDTA (pH 8.0)] with 14 mM MgCl2 was annealed using a 36-hour, 85° to 25°C protocol. The folding products were characterized by agarose gel electrophoresis (fig. S2). The desired combinations of unpurified monomers were then mixed at equimolar ratio (10 nM each) and incubated at RT with adjusted magnesium concentrations for overnight to obtain dimers (14 mM MgCl2), trimers (14 mM MgCl2), or tetramers (24 mM MgCl2). The oligomers (open-ended and capped nanopores) were purified by rate-zonal centrifugation purification as previously reported (60). Briefly, to make the glycerol gradient, two layers of glycerol solutions (15 and 45%) containing the 1× TE buffer and 14 mM Mg2+ were load to a 13- by 51-mm centrifuge tube (Beckman Coulter). The quasi-linear glycerol gradient was built using a gradient station (BioComp Instruments). The unpurified DNA nanopores were loaded to the top of the gradient media and centrifuged at 45,000 rpm for 1.5 hours followed by fraction collection. Fractions containing the correctly assembled nanopores (determined by agarose gel electrophoresis) were combined. Glycerol was subsequently removed by buffer exchange to 1× TE and 15 mM MgCl2 (pH 8.0) buffer using Amicon Ultra centrifugal filters with a nominal molecular weight limit (NMWL) of 10 kDa (MilliporeSigma).
Assembly of NuPODs
The NuPODs were assembled from purified DNA nanopores and nup-DNA conjugates as previously described (29). Anti-handle–conjugated nups [50 mM Tris (pH 8.0), 300 mM NaCl, and 0.2 mM tris(2-carboxyethyl)phosphine] were added to nanopores (1× TE and 15 mM MgCl2) at a 2:1 (anti-handle:handle) ratio. For example, the mixture for assembling 60-nm Nup62 NuPODs contained a 5 nM 60-nm DNA nanopore carrying 32× handles and 320 nM anti-handle–conjugated Nup62. The mixtures were incubated for 0.5 to 2 hours at 37°C. The formation of NuPODs was validated by the retardation of NuPOD bands compared with the corresponding empty nanopores in SDS-agarose gels (fig. S3, H to J). The same protocol was used for assembling NuPODs for the HS-AFM study with two exceptions: (i) MgCl2 was adjusted to 20 mM, and (ii) immediately before HS-AFM analyses, MBP was cleaved from NuPODs by incubating with TEV protease (1:50 ratio to the substrate) and DTT (1 mM) for 1.5 hours at 30°C.
Determining the nup copy number in NuPODs
The copy number of Nup62 in NuPODs was determined using a protocol described in (25) with the results summarized in fig. S4.
First, the Nup62 NuPODs were purified by rate-zonal centrifugation. Glycerol gradients were built in 5- by 41-mm centrifuge tubes by sequentially layering 45, 40, 35, 30, 25, 20, and 15% glycerol solutions in the 1× TE buffer containing 14 mM MgCl2 (90 μl each). The Nup62 NuPODs were loaded to the top of the glycerol gradient and centrifuged at 45,000 rpm for 1.5 hours. After centrifugation, nine consecutive 75-μl solution fractions were collected from top to bottom; the last fraction (<75 μl) contained aggregates. All nine fractions were analyzed by Western blot (see below). The first eight fractions were loaded into separate lanes of an SDS-agarose gel alongside the empty nanopore as a control. The amount of NuPOD was estimated by comparing band intensities (stained by ethidium bromide) with the control sample (empty nanopore), the concentration of which was measured by OD260. Fractions 6, 7, and 8, which contained NuPODs, were combined.
Next, the purified 60-nm Nup62 NuPOD and a series of Nup62 solutions with known concentrations (16.6 to 498 nM) were loaded into separate lanes of a 4 to 12% acrylamide gel (Thermo Fisher Scientific). Proteins were separated by SDS-PAGE and then transferred to a nitrocellulose (Bio-Rad). Immunoblotting was performed using Nup62 polyclonal antibody (Bethyl Laboratories Inc., A304-942A-T). Goat anti-rabbit horseradish peroxidase–conjugated secondary antibody (Jackson ImmunoResearch, catalog no. 111-035-003) was used for detection. Band intensities on the Western blot were measured using ImageJ, from which a calibration curve of band intensity versus Nup62 concentration was generated. The band intensity of the purified NuPOD in the Western blot was compared to the calibration curve to determine the Nup62 concentration. The average Nup62 copy number was calculated as [Nup62]/[NuPOD].
Negative-stain TEM and data analyses
TEM specimens were prepared by depositing samples onto glow discharged Formvar/carbon-coated copper grids (Electron Microscopy Sciences) followed by staining with 2% (w/v) uranyl formate solutions. After wicking away the staining solution, the air-dried grids were imaged using a JEOL JEM-1400Plus microscope with a bottom-mount 4k × 3k charge-coupled device camera (Advanced Microscopy Technologies). Particles (nanopores and NuPODs) were manually picked from TEM images (fig. S5). Class averages were generated by RELION (61). The intensity profiles were measured using Fiji (62).
Sample preparation for HS-AFM measurements
Supported lipid bilayers (SLBs) were prepared as previously described (25). Dipalmitoylphosphatidylcholine (Avanti Polar Lipids) and didodecyldimethylammonium bromide (Avanti Polar Lipids) lipids were dissolved in chloroform and mixed in a 3:1 molar ratio. The solvent was initially allowed to evaporate under a gentle nitrogen stream within a fume hood for 1 hour and then placed in a desiccator for at least 4 hours. Milli-Q water was added to the dry lipids to reach a final concentration of ~1 mg/ml. The lipid suspension was then treated in an ultrasonic bath (Elmasonic p30H, Elma Schmidbauer GmbH) at ~65°C, using a pulse setting at 80 kHz for 15 min. The lipid solution was then loaded into syringes of an Avanti mini-extruder kit (Avanti Polar Lipids), maintaining a temperature of ~65°C, and passed through a Nuclepore Track-Etched Membrane (100 nm in pore size) that is supported by a polyester drain disc on both sides (Global Lifesciences Solutions UK Ltd., Buckinghamshire, United Kingdom). The process was repeated at least 20 times, resulting in the formation of small unilamellar vesicles (SUVs).
A 2.7-μl droplet of a vesicle buffer (1 μl of 1 M MgCl2, 1 μl of 1 M CaCl2, and 16 μl of Milli-Q water) was deposited onto an HS-AFM sample stage, which consisted of a freshly cleaved mica sheet glued to a glass cylinder. SUVs (0.3 μl, described above) were then added to the droplet. The sample stage was then exposed to a temperature of ~65°C for 20 min inside a humid petri dish, followed by a gradual reduction to RT over 20 min. This thermal treatment induced vesicle rupture, leading to the formation of a positively charged SLB. Any excess vesicles in solution were removed by rinsing with water, followed by exchange with a phosphate buffer with Mg2+ [10 mM sodium phosphate and 40 mM MgCl2 (pH 7.0)]. This washing step was repeated four times before introducing 3 μl of TEV-treated NuPODs (2 nM). Free Nup62 and MBP in solution were removed by gently washing the stage with the 1× TE buffer and 40 mM MgCl2 and buffer exchange with the PB-Mg buffer before HS-AFM imaging.
After depositing the NuPODs on the lipid bilayer, importin-β1 was added to the sample in the imaging buffer at specified concentrations (10 nM, 100 nM, or 1 μM) and incubated for 30 min prior to the measurements.
HS-AFM measurements on NuPODs
HS-AFM measurements and data processing were performed as described elsewhere (36). The acquisition of all HS-AFM data was conducted using the HS-AFM 1.0 system (RIBM, Japan). The system used a standard scanner operating in tapping mode. Throughout the experiments, pristine QUANTUM-AC10-SuperSharp and QUANTUM-AC10-SuperSharp Enhanced cantilevers (nanotools GmbH) were used, featuring a tip radius of ≤2 nm, a nominal spring constant of 0.1 N/m, a resonant frequency of ~0.5 MHz, and a quality factor of around 2 in water. The typical set point amplitude (Aset) was maintained at 80 to 90% of the free cantilever oscillation amplitude (Afree), set within the range of 2 to 3 nm (63, 64).
HS-AFM line scanning measurements on NuPODs
In HS-AFM line scanning (HS-AFM-LS), the slow-scan axis was deactivated, and solely the fast-scan axis was engaged (65). A NuPOD was first imaged and centered within a region of interest prior to commencing line scanning. This ensures that the NuPOD’s center is scanned repetitively, generating a kymograph as a function of the distance from the pore center. Line scans were captured over 80 pixels in the fast-scan axis at a rate of 1.875 ms per line.
HS-AFM data processing
All 2D images acquired through HS-AFM were drift corrected using the in-house Python-based software, which concurrently transformed the files into TIFF format (35, 36). Subsequent analysis of HS-AFM images was done using ImageJ along with custom analysis routines written in Python. Various image processing tasks, such as filtering, contrast adjustment, and height/diameter measurements, were also performed using ImageJ. Initially, all HS-AFM images underwent correction for xy tilting through the application of a flattening filter using a first-order polynomial plane. Following this, the heights of the HS-AFM images were normalized in relation to the scaffold by subtracting the average scaffold height. Zoomed-in HS-AFM images were treated using a 2D Gaussian filter with a 1-pixel SD.
HS-AFM-LS data were processed using custom analysis routines written in Python as follows. Full kymographs were spliced into 600-ms segments. Corrections for drift in the z axis and tilt in the fast-scan axis were applied, and the heights were adjusted relative to the NuPOD scaffold, provided that the top surface of the scaffold was discernible within the same kymograph. Furthermore, a fast Fourier transform filter was applied to eliminate periodic noise at frequencies of 50 and 150 Hz, with root mean square amplitudes of ~0.09 and 0.08 nm, respectively. Given their negligible size, these amplitudes had no discernible impact on the dynamic analysis of HS-AFM-LS. The height values obtained from kymographs were averaged along the time axis at corresponding radii and then overlaid over the average profile of the scaffold (Fig. 3, E and J).
Particle tracking and positional distribution
A custom Python routine tracked the displacement of the central plug–like feature in HS-AFM kymographs on a line-by-line basis along the fast-scan (x) axis (36). Initially, a low-pass filter was applied to the kymographs along this axis to enhance the signal-to-noise ratio. Detection was specifically limited to the central region, excluding the NuPOD scaffold. If displacement of the central plug–like cluster was detected within a line, then the algorithm selected the position of the highest local maximum; otherwise, the line was skipped. Trajectories from all lines containing the cluster were then compiled into a histogram to display the distribution of the cluster relative to the center of a NuPOD (referenced in Fig. 3, D and I).
In vitro assembly of HBV capsids of mammalian cell origin
Expi293F cells (part of the Expi293 expression system kit, Thermo Fisher Scientific, A14635) were transfected with WT HBc cloned in the pcDNA3.4 mammalian vector (Thermo Fisher Scientific). After culturing at 37°C for 72 hours, the cells were harvested. The cell pellet was resuspended in a lysis buffer [50 mM tris-HCl (pH 8.5), 150 mM NaCl, 1% NP-40, and protease inhibitors] and chilled at 4°C for ~1 hour. In cases of incomplete lysis, cells were manually disrupted using a Dounce homogenizer until complete lysis was observed under a light microscope.
The cell lysate was then centrifuged at 17,000g for 45 min at 4°C. The supernatant was collected and loaded onto a 20 to 60% glycerol gradient [20 mM Tris (pH 8.0) and 150 mM NaCl]. This gradient was centrifuged at 4°C for ~5 hours at 153,700g in an SW 32 Ti rotor. After spinning for 5 hours, the entire gradient was fractionated into 1.5-ml aliquots, which were analyzed using 4 to 12% SDS-PAGE. After buffer exchange to remove glycerol, the fractions with the most abundant full-length HBc were imaged by negative-stain TEM. On the basis of TEM analysis, the fractions containing the most intact HBV capsids were concentrated and stored at −80°C before being used for binding and penetration assays. The absorption (220 to 350 nm, with 1-nm increment) spectrum of the capsids was measured using a NanoDrop spectrometer (Thermo Fisher Scientific).
In vitro assembly of HBV capsids of bacterial origin
Plasmids encoding HBc (WT or ΔC) were transfected to BL21(DE3) cells and expressed as described in Nup purification and DNA conjugation. After lysing the bacteria in a buffer [50 mM Tris (pH 8) and 300 mM NaCl], bacterial debris were removed by centrifugation at 12,000g for 1 hour. HBc proteins were pelleted from the supernatant by adding 30% of (NH4)2SO4 and centrifuging for 1 hour at 12,000g. The pellets were resuspended in 50 mM tris-HCl (pH 8.0) before being dialyzed overnight against a buffer [50 mM Tris (pH 8)]. After centrifugation at 12,000g for 1 hour to remove aggregates, the dialyzed sample was loaded to a HiPrep Q FF 16/10 anion-exchange chromatography column (Cytiva) and the flow through from the column, which contained HBc, was collected. The flow through was subjected to rate-zonal centrifugation at 30,000 rpm (SW 32 Ti rotor) and 4°C for 3 hours in a 20 to 60% glycerol gradient [20 mM Tris (pH 8.0) and 150 mM NaCl]. Fractions collected from the gradient were analyzed by SDS-PAGE. Fractions containing HBc of the expected molecular weight were collected and concentrated by Amicon Ultra centrifugal filters with an NMWL of 100 kDa. The fractions with the most abundant HBc were imaged by negative-stain TEM to screen for intact HBV capsids. The capsids were stored at −80°C before being used for binding and penetration assays. The absorption (220 to 350 nm, with 1-nm increment) spectra of the capsids were measured using a NanoDrop spectrometer (Thermo Fisher Scientific).
HBV penetration assay
Purified HBVs (50 μg/ml HBc) and NuPODs (5 nM) were mixed to achieve an ~1:1 capsid:NuPOD molar ratio and incubated at RT for 15 min. To test the effect of NTRs on HBV-NuPOD interactions, NuPODs were preincubated with 100 nM importin-β1 for 15 min before the addition of HBV capsids. The mixtures were then imaged by negative-stain TEM. Electron micrographs were analyzed by ImageJ to determine the HBV occupancy (number of HBV-bound NuPODs/total number of NuPODs) and penetration depth (defined as the distance from the NuPOD entrance to the center of the capsid). Practically, the penetration depth was calculated by subtracting the average radius of the HBV capsids (15 nm) from the distance measured from the NuPOD entrance to the bottom of the HBVs. All NuPOD-bound capsids were measured as long as the capsid center passed the entrance of the NuPOD.The penetration depth data were plotted as normalized histograms and fitted by multiple normal distributions using MATLAB (MathWorks).
Co-migration–based binding assays
WT HBV (3 μM HBc) and ΔC HBV (3 μM HBc) were incubated separately with ~1 μM Nup62, Nup153, or importin-β1 at RT for 15 min. The mixtures were each loaded onto a linear glycerol gradient [15 to 45%, in 1× TE (pH 8) and 15 mM MgCl2] and spun for 90 min at 48,000 rpm using an SW 55 Ti rotor. The collected fractions (12 to 13 fractions in total, 50 μl each) were characterized by 4 to 12% SDS-PAGE to determine the co-migrating proteins (fig. S9). HBV capsids, nups, and importin-β1 were individually subjected to the same procedures as controls.
Data analysis and statistics
Scatterplots are presented as means ± SD using the software Prism 8.0.0 (GraphPad). The two-tailed t test was used to compare the diameters of the WT HBV and ΔC HBV, assuming normal distribution with equal variances.
Acknowledgments
Funding: This work is supported by an NIH grant R01AI162260 to C.P.L., Y.X., and C.L., an NIH grant R01GM117386 to M.M., and a Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation) grant no. 310030_201062 to R.Y.H.L. T.K. acknowledges the support of a Swiss Nanoscience Institute PhD Fellowship.
Author contributions: Q.F. and C.L. conceived and designed the project. Q.F., M.S., W.Z., E.C., A.Z., P.B., C.W., X.L., Q.S., and L.E.K. performed experiments. Q.F. and M.S. analyzed data. Q.F., M.S., M.M., C.P.L., Y.X., R.Y.H.L., and C.L. interpreted data. T.K. developed software tools. Q.F. and C.L. prepared the manuscript. All authors participated in the discussions and reviewed and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Figs. S1 to S13
Legends for movies S1 and S2
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
Data S1
REFERENCES AND NOTES
- 1.Hampoelz B., Andres-Pons A., Kastritis P., Beck M., Structure and assembly of the nuclear pore complex. Annu. Rev. Biophys. 48, 515–536 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Lin D. H., Hoelz A., The structure of the nuclear pore complex (an update). Annu. Rev. Biochem. 88, 725–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey S., Gorlich D., A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Hulsmann B. B., Labokha A. A., Gorlich D., The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150, 738–751 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T., Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 13, 622–628 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Lim R. Y., Fahrenkrog B., Koser J., Schwarz-Herion K., Deng J., Aebi U., Nanomechanical basis of selective gating by the nuclear pore complex. Science 318, 640–643 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Celetti G., Paci G., Caria J., VanDelinder V., Bachand G., Lemke E. A., The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J. Cell Biol. 219, e201907157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel S. S., Belmont B. J., Sante J. M., Rexach M. F., Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Eisele N. B., Labokha A. A., Frey S., Gorlich D., Richter R. P., Cohesiveness tunes assembly and morphology of FG nucleoporin domain meshworks—Implications for nuclear pore permeability. Biophys. J. 105, 1860–1870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapinos L. E., Schoch R. L., Wagner R. S., Schleicher K. D., Lim R. Y. H., Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J. 106, 1751–1762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn R., Osmanovic D., Ehret S., Araya Callis C., Frey S., Stewart M., You C., Gorlich D., Hoogenboom B. W., Richter R. P., A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies. eLife 5, e14119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmerli C. E., Allegretti M., Rantos V., Goetz S. K., Obarska-Kosinska A., Zagoriy I., Halavatyi A., Hummer G., Mahamid J., Kosinski J., Beck M., Nuclear pores dilate and constrict in cellulo. Science 374, eabd9776 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Schuller A. P., Wojtynek M., Mankus D., Tatli M., Kronenberg-Tenga R., Regmi S. G., Dip P. V., Lytton-Jean A. K. R., Brignole E. J., Dasso M., Weis K., Medalia O., Schwartz T. U., The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akey C. W., Singh D., Ouch C., Echeverria I., Nudelman I., Varberg J. M., Yu Z., Fang F., Shi Y., Wang J., Salzberg D., Song K., Xu C., Gumbart J. C., Suslov S., Unruh J., Jaspersen S. L., Chait B. T., Sali A., Fernandez-Martinez J., Ludtke S. J., Villa E., Rout M. P., Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 185, 361–378.e25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fay N., Pante N., Nuclear entry of DNA viruses. Front. Microbiol. 6, 467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu L., Weiskopf E. N., Akkermans O., Swanson N. A., Cheng S., Schwartz T. U., Gorlich D., HIV-1 capsids enter the FG phase of nuclear pores like a transport receptor. Nature 626, 843–851 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickson C. F., Hertel S., Tuckwell A. J., Li N., Ruan J., Al-Izzi S. C., Ariotti N., Sierecki E., Gambin Y., Morris R. G., Towers G. J., Bocking T., Jacques D. A., The HIV capsid mimics karyopherin engagement of FG-nucleoporins. Nature 626, 836–842 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue G., Yu H. J., Buffone C., Huang S. W., Lee K., Goh S. L., Gres A. T., Guney M. H., Sarafianos S. G., Luban J., Diaz-Griffero F., KewalRamani V. N., The HIV-1 capsid core is an opportunistic nuclear import receptor. Nat. Commun. 14, 3782 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R., Ko Y.-H., Li F., Lokareddy R. K., Hou C.-F. D., Kim C., Klein S., Antolinez S., Marin J. F., Perez-Segura C., Jarrold M. F., Zlotnick A., Hadden-Perilla J. A., Cingolani G., Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core. Sci. Adv. 10, eadi7606 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zila V., Margiotta E., Turonova B., Muller T. G., Zimmerli C. E., Mattei S., Allegretti M., Borner K., Rada J., Muller B., Lusic M., Krausslich H. G., Beck M., Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032–1046.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabe B., Vlachou A., Pante N., Helenius A., Kann M., Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc. Natl. Acad. Sci. U.S.A. 100, 9849–9854 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Q., Rout M. P., Akey C. W., Three-dimensional architecture of the isolated yeast nuclear pore complex: Functional and evolutionary implications. Mol. Cell 1, 223–234 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Jovanovic-Talisman T., Tetenbaum-Novatt J., McKenney A. S., Zilman A., Peters R., Rout M. P., Chait B. T., Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457, 1023–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalczyk S. W., Kapinos L., Blosser T. R., Magalhaes T., van Nies P., Lim R. Y., Dekker C., Single-molecule transport across an individual biomimetic nuclear pore complex. Nat. Nanotechnol. 6, 433–438 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Fisher P. D. E., Shen Q., Akpinar B., Davis L. K., Chung K. K. H., Baddeley D., Saric A., Melia T. J., Hoogenboom B. W., Lin C., Lusk C. P., A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano 12, 1508–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ketterer P., Ananth A. N., Laman Trip D. S., Mishra A., Bertosin E., Ganji M., van der Torre J., Onck P., Dietz H., Dekker C., DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat. Commun. 9, 902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Q., Tian T., Xiong Q., Ellis Fisher P. D., Xiong Y., Melia T. J., Lusk C. P., Lin C., DNA-origami nanotrap for studying the selective barriers formed by phenylalanine-glycine-rich nucleoporins. J. Am. Chem. Soc. 143, 12294–12303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klughammer N., Barth A., Dekker M., Fragasso A., Onck P. R., Dekker C., Diameter dependence of transport through nuclear pore complex mimics studied using optical nanopores. eLife 12, RP87174 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Q., Feng Q., Wu C., Xiong Q., Tian T., Yuan S., Shi J., Bedwell G. J., Yang R., Aiken C., Engelman A. N., Lusk C. P., Lin C., Xiong Y., Modeling HIV-1 nuclear entry with nucleoporin-gated DNA-origami channels. Nat. Struct. Mol. Biol. 30, 425–435 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz A., Schwarz A., Foss M., Zhou L., Rabe B., Hoellenriegel J., Stoeber M., Pante N., Kann M., Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLOS Pathog. 6, e1000741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosalaganti S., Obarska-Kosinska A., Siggel M., Taniguchi R., Turonova B., Zimmerli C. E., Buczak K., Schmidt F. H., Margiotta E., Mackmull M. T., Hagen W. J. H., Hummer G., Kosinski J., Beck M., AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376, eabm9506 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Kapinos L. E., Huang B., Rencurel C., Lim R. Y. H., Karyopherins regulate nuclear pore complex barrier and transport function. J. Cell Biol. 216, 3609–3624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C., Wang J. C., Pierson E. E., Keifer D. Z., Delaleau M., Gallucci L., Cazenave C., Kann M., Jarrold M. F., Zlotnick A., Importin β can bind hepatitis B virus core protein and empty core-like particles and induce structural changes. PLOS Pathog. 12, e1005802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley G. J., Akpinar B., Shen Q., Fisher P. D. E., Lusk C. P., Lin C., Hoogenboom B. W., Quantification of biomolecular dynamics inside real and synthetic nuclear pore complexes using time-resolved atomic force microscopy. ACS Nano 13, 7949–7956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakiyama Y., Mazur A., Kapinos L. E., Lim R. Y. H., Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat. Nanotechnol. 11, 719–723 (2016). [DOI] [PubMed] [Google Scholar]
- 36.T. Kozai, J. Fernandez-Martinez, T. van Eeuwen, P. Gallardo, L. E. Kapinos, A. Mazur, W. Zhang, J. Tempkin, R. Panatala, M. Delgado-Izquierdo, B. Raveh, A. Sali, B. T. Chait, L. M. Veenhoff, M. P. Rout, R. Y. H. Lim, Dynamic molecular mechanism of the nuclear pore complex permeability barrier. bioRxiv 535055 [Preprint] (2023). 10.1101/2023.03.31.535055. [DOI]
- 37.Diogo Dias J., Sarica N., Neuveut C., Early steps of hepatitis B life cycle: From capsid nuclear import to cccDNA formation. Viruses 13, 757 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallucci L., Kann M., Nuclear import of hepatitis B virus capsids and genome. Viruses 9, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kann M., Sodeik B., Vlachou A., Gerlich W. H., Helenius A., Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145, 45–55 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fragasso A., de Vries H. W., Andersson J., van der Sluis E. O., van der Giessen E., Dahlin A., Onck P. R., Dekker C., A designer FG-Nup that reconstitutes the selective transport barrier of the nuclear pore complex. Nat. Commun. 12, 2010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng S. C., Biswas A., Huyton T., Schunemann J., Reber S., Gorlich D., Barrier properties of Nup98 FG phases ruled by FG motif identity and inter-FG spacer length. Nat. Commun. 14, 747 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labokha A. A., Gradmann S., Frey S., Hulsmann B. B., Urlaub H., Baldus M., Gorlich D., Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32, 204–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A. E., Slepchenko B. M., Schaff J. C., Loew L. M., Macara I. G., Systems analysis of Ran transport. Science 295, 488–491 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Elosegui-Artola A., Andreu I., Beedle A. E. M., Lezamiz A., Uroz M., Kosmalska A. J., Oria R., Kechagia J. Z., Rico-Lastres P., Le Roux A. L., Shanahan C. M., Trepat X., Navajas D., Garcia-Manyes S., Roca-Cusachs P., Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Schoch R. L., Kapinos L. E., Lim R. Y. H., Nuclear transport receptor binding avidity triggers a self-healing collapse transition in FG-nucleoporin molecular brushes. Proc. Natl. Acad. Sci. U.S.A. 109, 16911–16916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeman N. C., Sleiman H. F., DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2018). [Google Scholar]
- 47.Dey S., Fan C., Gothelf K. V., Li J., Lin C., Liu L., Liu N., Nijenhuis M. A. D., Saccà B., Simmel F. C., Yan H., Zhan P., DNA origami. Nat. Rev. Methods Primers 1, 13 (2021). [Google Scholar]
- 48.Shen Q., Xiong Q., Zhou K., Feng Q., Liu L., Tian T., Wu C., Xiong Y., Melia T. J., Lusk C. P., Lin C., Functionalized DNA-origami-protein nanopores generate large transmembrane channels with programmable size-selectivity. J. Am. Chem. Soc. 145, 1292–1300 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bell N. A., Engst C. R., Ablay M., Divitini G., Ducati C., Liedl T., Keyser U. F., DNA origami nanopores. Nano Lett. 12, 512–517 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Wei R., Martin T. G., Rant U., Dietz H., DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. Engl. 51, 4864–4867 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Fragasso A., De Franceschi N., Stommer P., van der Sluis E. O., Dietz H., Dekker C., Reconstitution of ultrawide DNA origami pores in liposomes for transmembrane transport of macromolecules. ACS Nano 15, 12768–12779 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing Y., Dorey A., Jayasinghe L., Howorka S., Highly shape- and size-tunable membrane nanopores made with DNA. Nat. Nanotechnol. 17, 708–713 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Pante N., Kann M., Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13, 425–434 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paci G., Zheng T., Caria J., Zilman A., Lemke E. A., Molecular determinants of large cargo transport into the nucleus. eLife 9, e55963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agarwal N. P., Matthies M., Gur F. N., Osada K., Schmidt T. L., Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem. Int. Ed. Engl. 56, 5460–5464 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Ponnuswamy N., Bastings M. M. C., Nathwani B., Ryu J. H., Chou L. Y. T., Vinther M., Li W. A., Anastassacos F. M., Mooney D. J., Shih W. M., Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8, 15654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koh J., Blobel G., Allosteric regulation in gating the central channel of the nuclear pore complex. Cell 161, 1361–1373 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Kalita J., Kapinos L. E., Zheng T., Rencurel C., Zilman A., Lim R. Y. H., Karyopherin enrichment and compensation fortifies the nuclear pore complex against nucleocytoplasmic leakage. J. Cell Biol. 221, e202108107 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Douglas S. M., Marblestone A. H., Teerapittayanon S., Vazquez A., Church G. M., Shih W. M., Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin C., Perrault S. D., Kwak M., Graf F., Shih W. M., Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 41, e40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ando T., High-speed atomic force microscopy and its future prospects. Biophys. Rev. 10, 285–292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uchihashi T., Kodera N., Ando T., Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Matin T. R., Heath G. R., Huysmans G. H. M., Boudker O., Scheuring S., Millisecond dynamics of an unlabeled amino acid transporter. Nat. Commun. 11, 5016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Legends for movies S1 and S2
Legend for data S1
Movies S1 and S2
Data S1