Abstract
CD8+ T cell exhaustion hampers control of cancer and chronic infections and limits chimeric antigen receptor (CAR) T cell efficacy. Targeting TET2 in CAR T cells provides therapeutic benefit; however, TET2’s role in exhausted T cell (TEX) development is unclear. In chronic lymphocytic choriomeningitis virus (LCMV) infection, TET2 drove conversion from stem cell–like TEX progenitors toward terminally differentiated and effector (TEFF)–like TEX. TET2 also enforced a terminally differentiated state in the early bifurcation between TEFF and TEX, indicating broad roles for TET2 in acquisition of effector biology. To exploit the therapeutic potential of TET2, we developed clinically actionable TET2-targeted CAR T cells by disrupting TET2 via knock-in of a safety switch alongside CAR knock-in at the TRAC locus. TET2-targeted CAR T cells exhibited restrained terminal exhaustion in vitro and enhanced antitumor responses in vivo. Thus, TET2 regulates fate transitions in TEX differentiation and can be targeted with a safety mechanism in CAR T cells for improved tumor control.
Modulation of exhausted CD8+ T cell differentiation by targeting TET2 improves therapeutic potential of CAR T cells in cancer.
INTRODUCTION
T cell exhaustion limits disease control in cancer and chronic viral infections. In the context of persistent antigen exposure, exhausted CD8+ T cells (TEX) coexpress multiple inhibitory receptors (IRs) and exhibit altered cytokine secretion, impaired proliferation, and metabolic deficiencies compared to memory T cells (TMEM) and effector T cells (TEFF) (1). While TEX hold substantial clinical relevance, our understanding of TEX development and the fundamental cellular and molecular mechanisms governing TEX formation, maintenance, and activity, particularly in the setting of immunotherapies, is incomplete. Addressing these knowledge gaps could offer new strategies for enhancing patient outcomes.
Cellular immunotherapies such as chimeric antigen receptor (CAR) T cells have transformed the treatment of cancer. However, T cell exhaustion compromises the persistence and antitumor effector function of CAR T cells in vivo, resulting in relapses in hematological malignancies and limited efficacy against solid tumors (2). Strategies including genetic modification of CAR T cells to avert exhaustion (3–6) or use of immunotherapies such as programmed cell death protein 1 (PD1) blockade to reinvigorate TEX have been proposed to enhance treatment efficacy (7). However, current reinvigoration strategies are insufficient to permanently reverse exhaustion (8), limiting therapeutic potential.
TEX are a distinct epigenetic lineage, and the discovery of regulators that govern the epigenetic remodeling events underpinning TEX development holds promise for improving cell-based immunotherapies (9, 10). We previously reported massive clonal expansion of a single CAR T cell in a patient undergoing therapy for chronic lymphocytic leukemia (CLL) that resulted in enhanced antitumor activity and subsequent complete and sustained disease remission (10). This unique clone had the CAR transgene integrated into Tet methylcytosine dioxygenase 2 (TET2), accompanied by a pre-existing hypomorphic mutation in the patient’s second TET2 allele. As a methylcytosine dioxygenase, TET2 plays a pivotal role in active DNA demethylation by initiating the conversion of 5-methylcytosine into 5-hydroxymethylcytosine, a first step in removal of the methyl group (11, 12). This finding suggested that modulation of TET2 could be used to alter the epigenetic landscape of TEX and CAR T cells for therapeutic benefit. Accordingly, biallelic disruption of TET2, with concomitantly sustained expression of basic leucine zipper ATF-like transcription factor 3 (BATF3), resulted in clonal proliferation of CAR T cells with altered effector function (13). These data are consistent with skewed TMEM versus TEFF differentiation of TET2-deficient T cells observed following acute viral infection (14). However, the role of TET2 in the precise T cell fate transitions that govern TEX differentiation remains unknown. Given that TET2 loss augments CAR T cell efficacy (10, 13), unraveling the diverse roles of TET2 in the differentiation of CD8+ T cells, particularly the epigenetic programming of TEX across key developmental checkpoints, is critical for deciphering the underlying mechanisms of T cell exhaustion and further enhancing the effectiveness of immunotherapies.
RESULTS
TET2 is a frequent locus of transgene integration in CAR T cell–treated patients
Our previous report on TET2 disruption driving enhanced proliferation and sustained tumor clearance mediated by a single CAR T cell in one patient (10) led us to investigate other potential occurrences of lentiviral integration into TET2 in additional patients who underwent CAR T cell therapy for CLL and acute lymphocytic leukemia (ALL) (3) (tables S1 and S2). Within these cohorts, 36% of patients with CLL and 51% of patients with ALL had at least one instance of lentiviral integration into TET2 (fig. S1A). In total, 33 and 75 unique sites of integration were identified within CLL and ALL cohorts, respectively (fig. S1B). Most of these integration sites were of low abundance (fig. S1C), reaching less than 1% abundance at later time points (tables S1 and S2). In comparison, in our previous case study (10), a TET2-deficient clone reached upward of 90% relative clonal abundance at 4 months after infusion and near 25% relative clonal abundance around 1 year after infusion. Furthermore, the majority of these integrations occurred only once (tables S1 and S2); however, one patient with CLL (p04409-09) exhibited a CAR transgene insertion at chr4+105190185, which was observed at multiple time points, at 2 weeks following adoptive transfer in purified CAR T cells and at 1 month in whole blood (table S1). Lentiviruses favor integration into actively transcribed sites (15), and tend to integrate within the gene body rather than near promoters, suggesting low risk for oncogenic transformation (16). Of the integration events we identified within 50 kB of TET2, ~94% of CLL sites and ~ 93% of ALL sites occurred within the transcriptional boundary of the TET2 transcriptional unit (fig. S1D). The repeated lentiviral integration of a CAR transgene into TET2 motivates deeper analysis of the role of TET2 in T cell biology and especially in CAR T cell differentiation.
TET2-deleted CAR T cells adopt a central memory–like state after manufacturing
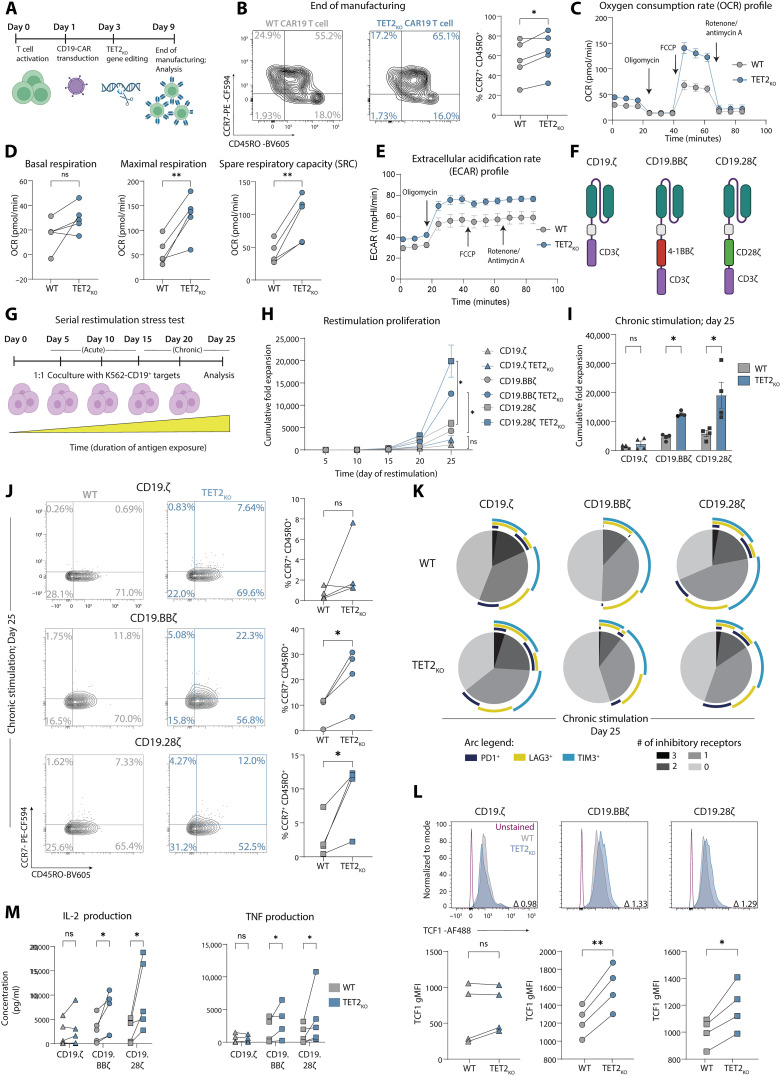
TET2 loss correlates with clinical response to CAR T cell therapy (10), and its deletion promotes acquisition of a memory CD8+ T cell fate in the setting of acute infection (14). To investigate the impact of TET2 loss in human CAR T cell differentiation, we generated TET2-deficient CAR T cells (TET2KO) through CRISPR-Cas9 gene editing (Fig. 1A and fig. S2, A to C). Following lentiviral CAR transduction and primary expansion, we observed a slightly elevated proportion of central memory CAR T cells in the TET2-deficient condition (Fig. 1B and fig. S2D). Mitochondrial respiration profiling indicated that, after production, TET2KO CAR T cells were programmed for enhanced oxidative phosphorylation (Fig. 1C), with increased basal respiration, maximal respiration, and spare respiratory capacity (SRC) (Fig. 1D). TET2KO CAR T cells also had increased aerobic glycolysis (Fig. 1E). Thus, TET2 deficiency augments cellular metabolism, potentially providing a greater adenosine triphosphate (ATP) reserve during heightened cellular activity or metabolic stress, aligning with the bioenergetic advantage and rapid recall capability of memory CD8+ T cells (17).
Fig. 1. TET2-deleted CAR T cells adopt a central memory–like state following manufacturing and exhibit increased expansion and reduced IR expression under chronic stimulation across multiple CAR constructs.
(A) CAR T cell manufacturing and TET2 gene editing schematic. (B) Example plots and data after CAR T cell expansion (day 9), highlighting CCR7+ CD45RO+ central memory subset, n = 5. (C and D) Longitudinal oxygen consumption rate (OCR) (C), basal respiration, maximal respiration, and SRC (D) of TET2-disrupted cells at end of expansion (day 9) after oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) and antimycin A/rotenone administration as indicated in (C), n = 5, run in triplicate. (E) Extracellular acidification rate (ECAR) of the same TET2-disrupted cells from (C) and (D). (F and G) Schematic of CD19.CD3ζ, CD19.BBζ, and CD19.CD28ζ CAR constructs ± TET2KO (F) placed in a serial restimulation stress test (G). (H and I) Cumulative fold expansion of serially restimulated CAR T cells ± TET2KO throughout (H) and at day 25 (I), n = 4. (J) Example plots and data of serially restimulated CAR T cells ± TET2KO showing distribution of CCR7+ CD45RO+ central memory–associated markers in CD8+ CAR T cell populations after five stimulations, n = 4. (K) SPICE plots showing distribution of IR coexpression in serially restimulated CD8+ CAR T cells ± TET2KO after five stimulations (chronic, day 25) with K562-CD19+ cells as depicted in (G), n = 4. (L) Example histograms (top) and data (bottom) of TCF1 gMFI in serially restimulated CD8+ CAR T cells ± TET2KO after five stimulations, with indicated fold change, n = 4. (M) IL-2 and TNF production from supernatant collected 24 hours after fifth stimulation, n = 5. Data shown as means ± SEM [(C), (E), and (H)] or individual values [(B), (D), (I), (J), (L), and (M)] from independent donors. ns P > 0.05; *P < 0.05; **P < 0.01 by paired t test. Schematics [(A), (F), and (G)] created with BioRender.com.
TET2 loss increases expansion and reduces IR expression after chronic stimulation for multiple CAR constructs
CD8+ T cell exhaustion is characterized by bioenergetic insufficiencies and altered glycolysis (18, 19). The metabolic profile of TET2KO CAR T cells suggested that targeting TET2 might improve CD8+ T cell survival and function in the setting of chronic antigen stimulation. To examine the role of TET2 in the long-term persistence of CAR T cells and to investigate the role of specific costimulatory domains (Fig. 1F), we used an in vitro “stress test” incorporating chronic antigen stimulation that recapitulates several features of progressive T cell exhaustion (Fig. 1G) (4, 6, 20). Both TET2KO 41BB-costimulated CAR T cells (CD19.BBζ) and TET2KO CD28-costimulated CAR T cells (CD19.28ζ) demonstrated greater proliferative capacity following repeated antigen stimulation when compared to AAVS1KO control CAR T cells and CAR T cells lacking a costimulatory domain (CD19.ζ) (Fig. 1, H and I). Given this increased proliferative capacity, we next investigated the differentiation and phenotype of chronically stimulated TET2KO CAR T cells. After chronic stimulation, CD8+ TET2KO CAR T cells skewed toward a CCR7+ CD45RO+ central memory–like population (Fig. 1J and fig. S2E). High IR expression and decreased expression of the transcription factor (TF) T cell factor 1 (TCF1) is associated with terminal differentiation of CD8+ T cells (21). However, CD8+ TET2KO CAR T cells had decreased coexpression of IRs including PD1 and T cell immunoglobulin and mucin-domain containing-3 (TIM3) (Fig. 1K) and increased expression of TCF1 (Fig. 1L) compared to AAVS1KO control CAR T cells, suggesting that, in the absence of TET2, CAR T cells were less terminally differentiated. Moreover, TET2 loss resulted in increased production of cytokines including interleukin-2 (IL-2) and tumor necrosis factor (TNF) from the total CAR T cell product following overnight restimulation with tumor cells after chronic antigen stimulation (Fig. 1M). However, other cytokines such as interferon-γ (IFN-γ) were not affected by TET2 deficiency (fig. S2F). Last, TET2 knockout similarly affected the phenotype of both 41BB- and CD28-costimulated CAR T cells, suggesting that the role of TET2 is independent of the costimulatory domain used. Together, these data implied that loss of TET2 restrained CAR T cell terminal differentiation during chronic antigen stimulation.
TET2 loss enhances CAR T cell efficacy in a tonic CAR signaling model
To further investigate the potential role of TET2 in CAR T cell terminal differentiation, we next knocked out TET2 in HA.28ζ-CAR T cells (fig. S2G), which exhibit robust tonic signaling and attain functional, transcriptomic, and epigenetic features of exhaustion by day 11 of culture (3). Expression of progenitor/stem cell–associated markers CCR7, CD27, and CD62L (fig. S2H) were increased on TET2KO compared to AAVS1KO HA.28ζ-CAR T cells, supporting the idea that TET2KO CD8+ T cells are less terminally differentiated. Next, we tested whether the phenotypic reprogramming of exhausted CAR T cells induced by TET2 loss would confer enhanced efficacy. TET2KO HA.28ζ-CAR T cells exhibited superior expansion (fig. S2I), increased cytotoxicity by the bulk CAR T cell product (fig. S2, J and K), and enhanced cytokine secretion (fig. S2L) compared to AAVS1KO HA.28ζ-CAR T cells when cocultured with 143B-GL osteosarcoma or with NALM-6-GD2 leukemia (fig. S2, M and N) cells. Together, these data indicate that TET2 deletion may improve CAR T cell efficacy in the setting of chronic antigen exposure and tonic CAR signaling, potentially by limiting terminal differentiation and increasing expression of proteins involved in T cell survival/persistence.
TET2 mediates the transition out of the TEX progenitor pool and toward terminal exhaustion in chronic viral infection
In CAR T cell exhaustion models using chronic antigen stimulation or tonic antigen receptor signaling, TET2 disruption limited acquisition of some features of exhaustion, such as IR expression, and enriched for expression of proteins associated with cell renewal, including TCF1. Despite the utility of CAR T cell exhaustion models, the distinct and complex developmental trajectory of CD8+ T cell exhaustion is likely incompletely recapitulated in reductionist in vitro systems (22). Furthermore, CAR T cell systems require T cell activation for CRISPR-Cas9–mediated TET2KO and CAR transduction, preventing the study of TET2-deficient TEX generated from naïve T cells. To further investigate the role of TET2 in the developmental trajectory of TEX, we used the well-characterized lymphocytic choriomeningitis virus (LCMV) clone 13 chronic infection model (23–26). T cells from Tet2fl/fl Cd4Cre+ mice delete TET2 during development in the thymus, resulting in mice that lack TET2 expression in both CD4+ and CD8+ T cells. These Tet2fl/fl Cd4Cre+ mice were crossed with T cell receptor (TCR) transgenic P14 mice that express a major histocompatibility complex class I–restricted TCR specific for LCMV DbGP33–41 to generate TET2KO P14 mice (Tet2fl/fl Cd4Cre+ P14) where TET2 is knocked out in LCMV antigen-specific CD8+ T cells. CD8+ TET2KO P14 cells were adoptively cotransferred with CD8+ wild-type (WT) P14 cells at a 1:1 ratio into recipient mice. Recipient mice were then infected with LCMV clone 13, and cotransferred P14 cells were analyzed throughout chronic infection (Fig. 2A and fig. S3A).
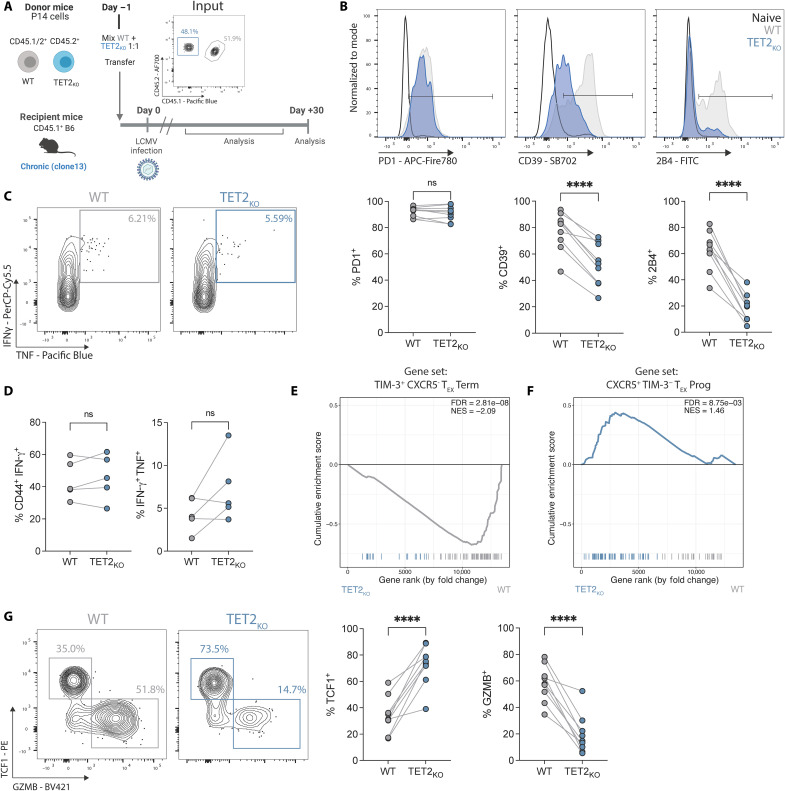
Fig. 2. TET2 mediates the transition out of the progenitor TEX subset toward terminal exhaustion.
(A) Cotransfer experimental schematic. Inset plot shows initial P14 cotransfer. (B) Example plots and data for IR expression on TET2KO P14 cells compared to WT P14 cells. (C and D) Example plots (C) and data (D) comparing IFN-γ and TNF expression following peptide restimulation for WT and TET2KO P14 cells. (E and F) GSEA of terminal TEX (E) and TEX progenitor (F) signatures between WT and TET2KO P14 cells at day 15 p.i. with LCMV clone 13 (Gene sets from GSE84105). (G) Example plots and data comparing TCF1+ TEX progenitor and GZMB+ terminally differentiated TEX frequencies within WT and TET2KO P14 cells. (B and G) n = 9, spleen at day 30 p.i. with LCMV clone 13. Data for individual mice shown; representative of >4 independent experiments. (D) n = 5, spleen at day 37 p.i. with LCMV clone 13. Data for individual mice shown; representative of three independent experiments. [(B), (D), and (G)] ns P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by paired t test. Schematic (A) created with BioRender.com.
The impact of TET2 loss on frequencies of antigen-specific CD8+ T cells over the course of chronic infection was variable and influenced by factors in the LCMV model. For example, when recipient WT CD4+ T cells were present (i.e., no CD4+ T cell depletion before infection), WT P14 outnumbered TET2KO P14 cells by ~7:1 in blood during the early stage of chronic infection [~day 8 postinfection (p.i.); fig. S3B], despite initial transfer at a 1:1 ratio. However, once exhaustion was established (27) TET2KO P14 cells expanded, outcompeting WT P14 cells by ~1.8:1 at ~day 60 p.i. (fig. S3B) and reflecting the expansion seen in human CAR T cell models. In contrast, when CD4+ T cells were depleted by in vivo administration of anti-CD4 antibody GK1.5, TET2KO P14 cells often did not rebound and remained underrepresented compared to WT P14 cells in blood and spleen (fig. S3, C and D). Together, these data suggest that the role of TET2 in TEX proliferation/survival may be affected by CD8+ T cell–extrinsic pressures that regulate exhaustion, such as chronic antigen burden/vial load and CD4+ T cell help.
CD8+ T cell exhaustion is characterized by high IR expression and decreased production of effector cytokines [reviewed in (1)]. We first asked whether TET2KO P14 cells retained core features of exhaustion in the LCMV chronic infection model. Expression of some IRs such as PD1 (Fig. 2B) and lymphocyte activation gene 3 (LAG3) (fig. S3E) remained high on TET2KO P14 cells and was comparable to cotransferred WT P14 cells. In contrast, TET2 loss strongly decreased expression of other IRs including CD39 and 2B4 (Fig. 2B). For example, at day 30 p.i. in spleen, only ~20% of TET2KO P14 cells expressed 2B4 compared to ~63% of WT P14 (Fig. 2B). However, TET2KO P14 cells did not acquire surface characteristics of classical TEFF and TMEM that arise during acute resolving infections (28, 29). Rather, TET2KO killer cell lectin-like receptor G1–positive (KLRG1+) TEFF-like P14 cells were effectively absent from the spleen at day 30 p.i., whereas WT KLRG1+ P14 cells were detectable at low frequencies as expected (fig. S3, F and G). Although TET2KO P14 cells had moderately increased expression of the IL-7 receptor CD127, which is associated with memory-like differentiation, protein levels remained low and consistent with expression in chronic rather than acute infection (fig. S3, F and G). Furthermore, despite decreased expression of some key IRs, TET2KO P14 cells remained functionally exhausted, as a similar frequency of TET2KO P14 cells produced IFN-γ or coproduced IFN-γ and TNF following in vitro restimulation with LCMV peptide as WT P14 cells (Fig. 2, C and D).
To investigate transcriptional changes associated with loss of TET2 during chronic infection, we next performed bulk RNA sequencing (RNA-seq) on WT and TET2KO P14 cells at day 15 p.i. with LCMV clone 13. TET2KO P14 cells had a distinct transcriptional profile, with ~1750 genes differentially expressed between WT and TET2KO P14 cells [false discovery rate (FDR) < 0.05; fig. S3H and table S3], including decreased expression of the IRs Entpd1 (CD39) and Cd244a (2B4), supporting protein expression data. The TEX lineage is functionally diverse. TEX progenitors retain proliferative potential, express TCF1, and have decreased expression of specific IRs such as CD39 and TIM3 (30–36) despite high expression of the exhaustion-associated TF TOX (37–42). TEX progenitors differentiate into a terminal TEFF-like subset that have reacquired some effector functions and contribute to viral control (36, 43–47) or into terminally exhausted TEX with increased IR expression and decreased proliferative capacity (44). Loss of TET2 decreased expression of genes associated with terminal differentiation and effector biology, including Zeb2 (48, 49), Id2 (50), Klrg1 (51, 52), Runx3 (53, 54), Prdm1 (BLIMP-1) (55–57), Nkg7 (58, 59), and Gzma and Gzmk (60) (fig. S3H). Furthermore, Runx3 and Prdm1 were hypermethylated when TET2 was depleted in CD8+ T cells during acute LCMV infection, resulting in shift away from TEFF and toward TMEM (14). The loss of effector-associated TFs and molecules, along with decreased expression of certain IRs, as well as our initial CAR T cell data, provoked the hypothesis that TET2 may have a role in terminal TEX differentiation. Gene Set Enrichment Analysis (GSEA) revealed depletion of a terminally differentiated TEX gene set in TET2KO P14 cells (Fig. 2E), suggesting a decrease in terminal exhaustion compared to WT controls. Conversely, a TEX progenitor gene set was enriched in TET2KO P14 cells (Fig. 2F); indeed, genes associated with TEX progenitor biology including Slamf6 (LY108) and Tnfsf4 (OX40L) were up-regulated when TET2 was knocked out (fig. S3H). These gene expression differences were associated with robust changes in TEX subset distribution. TET2KO P14 cells had a relative increase in the proportion of TEX progenitor cells (TCF1+ GZMB−) with a marked reduction in terminally differentiated TEX (TCF1− GZMB+) (Fig. 2G). These observations suggest that TET2 regulates differentiation into terminally differentiated TEX subsets, including TEFF-like TEX, during chronic infection.
Loss of TET2 limits terminal differentiation of exhausted CD8+ T cells
Bulk RNA-seq comparing WT and TET2KO P14 cells suggested that TET2 acts at a checkpoint in the epigenetic transition between TEX progenitor and terminally differentiated TEX fates, driving terminal exhaustion at the expense of a stem cell–like state. To determine whether TET2 functions at the transition between TEX subsets or regulates TEX differentiation within these subsets, we isolated LY108+ TEX progenitor and LY108neg terminally differentiated TEX from WT and TET2KO P14 cells at day 15 p.i. with LCMV clone 13 and performed RNA-seq and assay for transposase-accessible chromatin sequencing (ATAC-seq) on the isolated subsets (Fig. 3, A and B). TEX progenitor and terminally differentiated TEX are transcriptionally and epigenetically distinct (35). Principal component analysis (PCA) revealed that cell subset (LY108+ versus LY108neg TEX) was the major contributor to sample-to-sample variation and separated samples along PC1 regardless of genotype (Fig. 3, C and D). In contrast, PC2 was driven by genotype, with all isolated TET2KO P14 populations localized in distinct regions compared to WT P14 subsets (Fig. 3, C and D). Therefore, although TET2KO TEX retain key features of WT TEX (Fig 2, B to D, and fig. S3H), loss of TET2 may affect differentiation within TEX subsets.
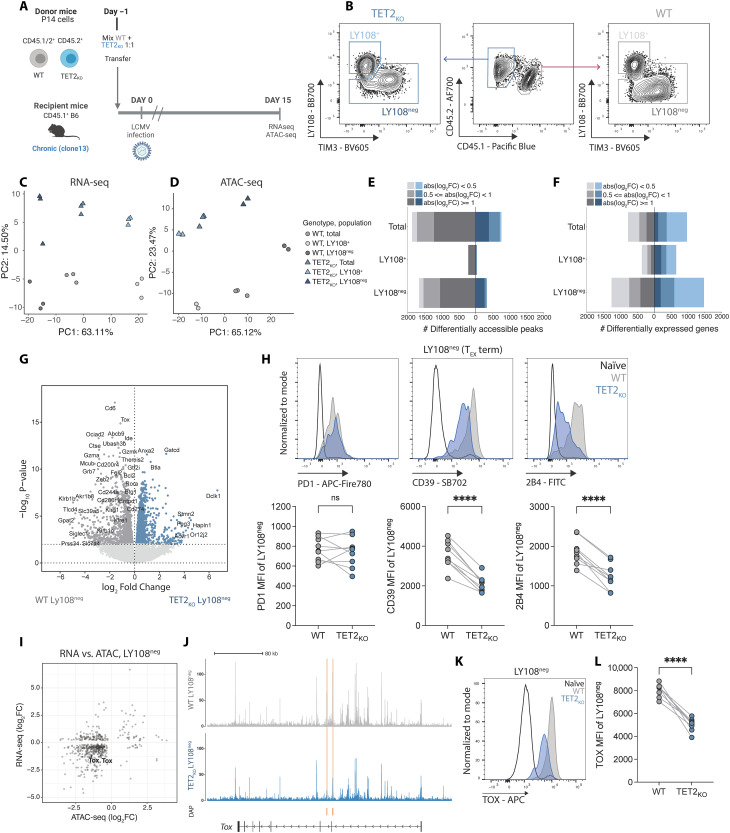
Fig. 3. Loss of TET2 limits terminal differentiation of exhausted CD8 T cells.
(A) Experiment schematic for RNA-seq and ATAC-seq. WT and TET2KO P14 cells were analyzed at day 15 p.i. with LCMV clone 13. (B) Sorting strategy for TEX subsets for RNA and ATAC-seq. (C and D) PCA of RNA-seq (C) and ATAC-seq (D) data for WT and TET2KO TEX subsets. (E and F) Number of DACRs (E) or DEGs (F) for each pairwise comparison between WT and TET2KO TEX subsets {FDR < 0.05, with variable absolute log2 fold changes [abs(log2FC)] indicated}. (G) Volcano plot highlighting DEG in WT compared to TET2KO LY108neg TEX. (H) Example plots and data comparing expression of PD1, CD39, and 2B4 on TET2KO LY108neg TEX to WT LY108neg TEX. (I) Correlation plot of differential gene expression and peak accessibility in TET2KO LY108neg TEX compared to WT LY108neg TEX with TOX labelled. (J) Example tracks showing accessibility at the Tox locus in LY108neg TEX. Differentially Accessible Peaks (DAPs) are indicated in orange. (K and L) Example plots (K) and data (L) comparing TOX expression in TET2KO LY108neg TEX to WT LY108neg TEX. [(H) and (L)] n = 9, spleen at day 30 p.i. with LCMV clone 13. Data for individual mice shown; representative of three independent experiments. ns P > 0.05; ****P < 0.0001 by paired t test. Schematic (A) created with BioRender.com.
To examine the potential role of TET2 within TEX subsets, we directly compared WT and TET2KO TEX within isolated TEX subsets and identified differentially expressed genes (DEG) and differentially accessible chromatin regions (DACRs). These analyses revealed two major features of TET2 function in TEX differentiation. First, 1855 DACRs and 2744 DEGs distinguished WT from TET2KO LY108neg terminally differentiated TEX compared to only 255 DACRs and 1018 DEGs between WT and TET2KO LY108+ TEX progenitors (Fig. 3, E and F; fig. S4, A and B; and tables S3 and S4). Reflecting these relative differences, WT and TET2KO LY108+ TEX progenitors were closer to each other in PCA space than WT and TET2KO terminally differentiated TEX (fig. S4, C and D), indicating that terminally differentiated TEX are more transcriptionally and epigenetically distinct following loss of TET2 than the TEX progenitor subset. Second, within the terminally differentiated LY108neg TEX subset, the majority of DACR (82.7%; FDR < 0.05, log2 FC > 0.5) were less accessible following TET2 knockout. Together, these data suggest that TET2 is required to sustain and/or increase chromatin accessibility at, and expression of, genes associated with terminal TEX differentiation.
We next asked which genes were unable to be up-regulated in LY108neg terminally differentiated TEX in the absence of TET2. Multiple genes associated with effector functions, including KLR family members (Klrg1, Klrb1b, Klrb1c, and Klre1), cytotoxic markers Gzma and Gzmk and the TF-encoding genes Zeb2, Btg1, and Rora were decreased in expression in TET2KO compared to WT terminally differentiated TEX (Fig. 3G) (61–64). Furthermore, TF motif analysis revealed that binding sites for effector-associated TFs including RUNX and TBET were less accessible in terminally differentiated TEX following removal of TET2 (fig. S4E). Thus, in terminally differentiated TEX lacking TET2, binding sites for key effector-driving TFs are less accessible and expression of effector genes is diminished. These data support the notion that TET2 promotes the acquisition of effector-associated genes in the terminally differentiated TEX subset.
RNA and protein expression of key IRs including CD39 (Entpd1) was lower on total TET2KO P14 cells than WT P14 cells (Fig. 2B and fig. S3H). To determine whether this change in IR expression reflected the population shift toward TEX populations with decreased IR expression (TEX progenitor cells) or differential regulation of IRs within terminally differentiated subsets, we next assessed IR expression within isolated TEX subsets. IR expression, including Cd200r1 (CD200R), Entpd1 (CD39), Cd274 (PDL1), and Cd244a (2B4), was decreased in TET2KO terminally differentiated TEX (Fig. 3, G and H), whereas levels of these IRs were low and more comparable to WT controls for TET2KO TEX progenitors (fig. S4, F and G). LY108 (Slamf6) decreases in expression as TEX terminally differentiate (35); however, LY108 expression remained high in TET2KO terminally exhausted TEX (fig. S4H). Together, these data support the hypothesis that TET2 regulates loss of TEX progenitor biology and is required for complete differentiation within the terminally exhausted TEX population.
The TF TOX has been proposed to regulate terminal exhaustion (35). Therefore, we interrogated whether the decreased differentiation with the terminal TEX subset following TET2KO was associated with changes in TOX. Tox expression was decreased within TET2KO LY108neg terminal TEX, and this decreased expression was associated with reduced chromatin accessibility at the Tox locus (Fig. 3, I and J). These changes in RNA expression and chromatin accessibility translated to markedly reduced TOX protein expression in TET2KO terminal TEX compared to WT terminal TEX (Fig. 3, K and L). Therefore, TET2 may coordinate with TOX to regulate the terminal differentiation of TEX.
TET2 regulates early bifurcation of TEX from TEFF-like cells
At least three major epigenetic remodeling events underpin the developmental trajectory of TEX. The first occurs immediately following naïve CD8+ T cell activation. The second occurs early, within days of initial activation, when terminally differentiated TEFF-like cells bifurcate from TCF1+ TEX precursors. Analysis of TET2KO P14 cells late in chronic infection suggested that TET2 regulates the third major rewiring event occurring in established TEX, when TEX progenitors transition into TEFF-like and terminally exhausted TEX subsets (35). RNA-seq and ATAC-seq analysis of terminally differentiated TEX suggested a role in coordinating the reacquisition of effector-like biology. To interrogate the role of TET2 in the second bifurcation event before fate commitment to exhaustion, we set up an adoptive cotransfer of WT and TET2KO P14 cells as described above and then analyzed CD8+ T cell responses to chronic LCMV infection at early time points (Fig. 4A). At days 6 and 8 p.i., the proportion and absolute frequency of PD1low KLRG1+ TEFF were markedly reduced in the absence of TET2 (Fig. 4, B and C, and fig. S4I). Furthermore, TET2KO P14 cells skewed toward TCF1+ TEX precursors at the expense of granzyme B–expressing TEFF-like cells (Fig. 4D). Together, these findings imply that TET2 regulates the acquisition of effector-like biology at multiple steps in TEX differentiation, both in the early bifurcation between TEX precursors and TEFF and in established exhaustion.
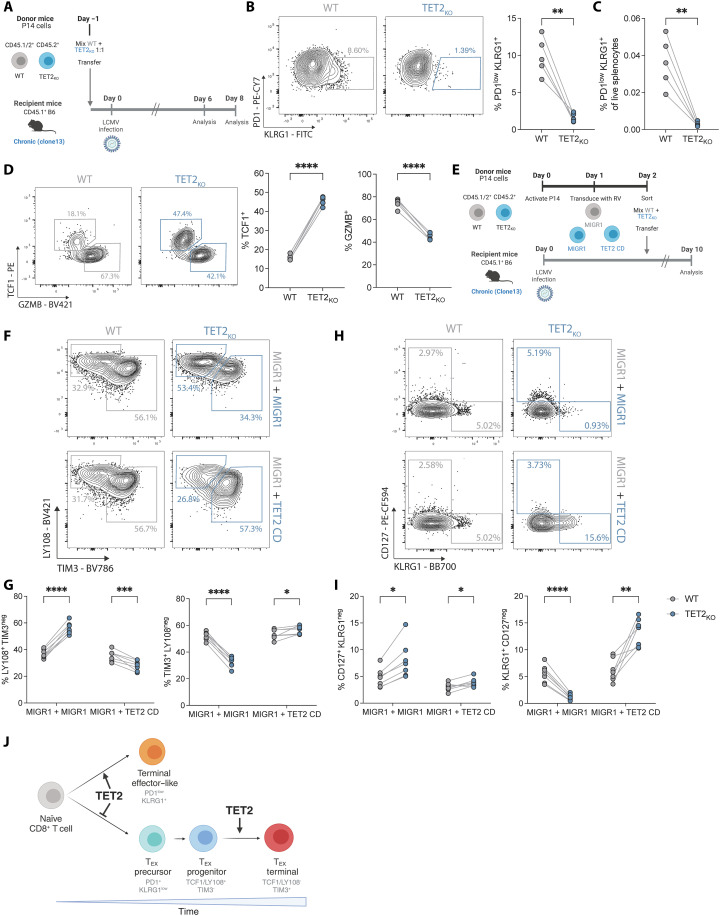
Fig. 4. TET2 regulates early bifurcation of TEX from TEFF-like cells.
(A) Experiment schematic for analysis of TET2KO P14 cells early (days 6 to 8) of LCMV clone 13 infection. (B) Example plots and data comparing frequency of early TEFF-like cells (KLRG1+ PD1low) within WT and TET2KO P14 cells at day 8 p.i. (C) Frequencies of WT and TET2KO KLRG1+ PD1low TEFF-like cells from total live splenocytes at day 8 p.i. (D) Example plots and data comparing expression of TCF1 and GZMB for WT and TET2KO P14 cells. (E) Experiment schematic for rescue of TET2 function in TET2KO P14 cells. (F and G) Example plots (F) and data (G) comparing frequencies of LY108+ TEX and TIM3+ TEX for WT and TET2KO P14 cells with or without overexpression of the TET2 catalytic domain (TET2 CD versus MIGR1). (H and I) Example plots (H) and data (I) comparing frequencies of CD127+ TMEM-like and KLRG1+ TEFF-like cells for WT and TET2KO P14 cells with or without overexpression of the TET2 catalytic domain (TET2 CD). (J) Model for TET2 role at major bifurcation events in chronic infection. [(B) to (D)] n = 5, spleen at day 8 p.i. with LCMV clone 13. Data for individual mice shown, representative of two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by paired t test. [(G) and (I)] n = 7, spleen at day 10 p.i. with LCMV clone 13. Data for individual mice shown, representative of two independent experiments. ns P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by multiple paired t test with Holm-Šídák posttest correction. Schematics [(A), (E), and (J)] created with BioRender.com.
These data suggested that TET2 deficiency limits acquisition of effector–like biology. To test whether forced TET2 expression rescues the phenotypes observed TET2KO P14 cells or promotes T cell effector biology, we overexpressed the TET2 catalytic domain (TET2 CD) in TET2KO P14 and compared the impact of TET2 “rescue” to TET2KO P14 cells, as well as WT P14 cells transduced with an empty vector (MIGR1) (Fig. 4E). As previously observed, TEX subset distribution in TET2KO P14 was skewed toward TEX progenitors at the expense of terminal TEX differentiation (Fig. 4, F and G). In contrast, expression of the TET2 catalytic domain normalized the proportions of TEX subsets and pushed TEX slightly toward terminal differentiation (Fig. 4, F and G). Furthermore, whereas KLRG1+ TET2KO P14 cells were effectively absent, expression of the TET2 catalytic domain increased KLRG1 expression by ~2-fold compared to WT P14 cells and ~11.6-fold compared to TET2KO P14 cells (Fig. 4, H and I). Therefore, TET2 promotes terminal TEX differentiation and the TET2 catalytic domain is sufficient for this activity.
Together, these data imply that TET2 acts a rheostat to regulate terminal differentiation and the acquisition of effector-like biology at multiple checkpoints in CD8+ T cell differentiation (Fig. 4J). In acute infection, TET2 modulates the bifurcation between classical short-lived TEFF and memory precursors (14), enforcing terminal differentiation and the acquisition of effector functions. TET2 plays parallel roles in chronic infection. In the initial stages of chronic infection, TET2 drives CD8+ T cells toward TEFF-like cells and away from the formation of the TEX precursor pool (65). Once exhaustion is established, TET2 pushes TEX progenitors toward a terminally differentiated and TEFF-like TEX state, again mirroring the role of TET2 as an enforcer of differentiation. These data further provoke the hypothesis that similar epigenetic programs are used and reused throughout CD8+ T cell differentiation to regulate function in distinct contexts (66).
TET2-edited dual knock-in allogeneic CAR T cells resist terminal differentiation, allowing enhanced tumor control
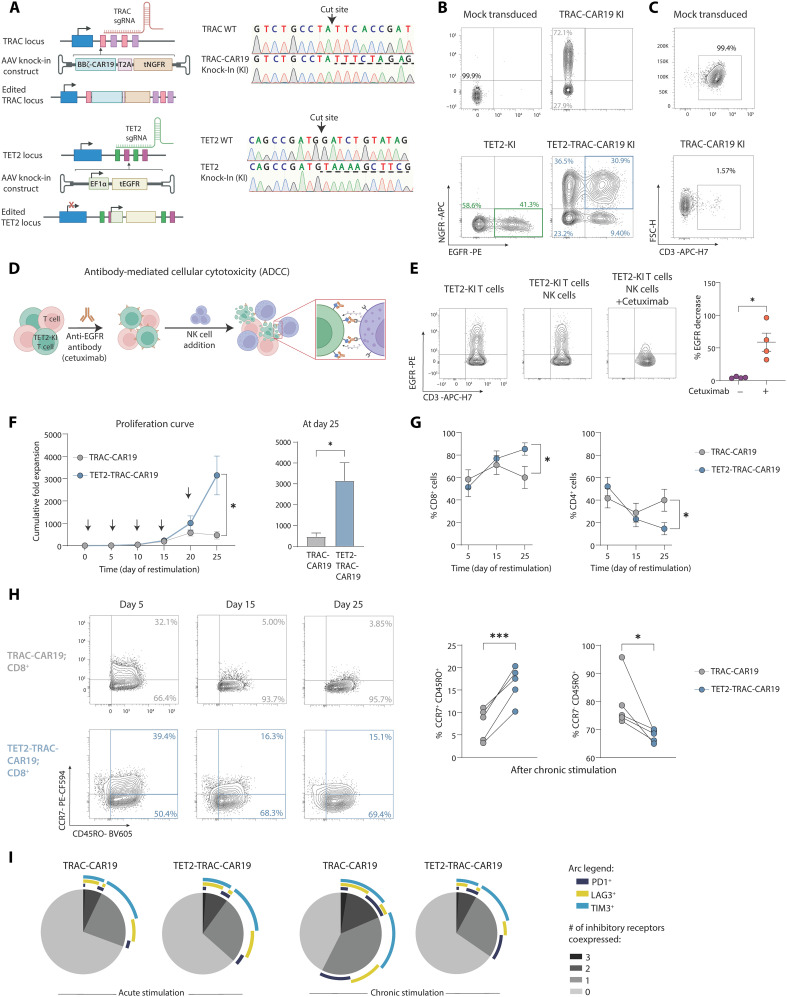
Data from the chronic infection model suggested that TET2 regulates CD8+ T cell differentiation and that loss of TET2 limits terminal exhaustion. Our initial TET2KO CAR T cell data implied that targeting TET2 to restrain terminal differentiation could improve CAR T cell expansion and efficacy. However, biallelic loss of TET2 with BATF3 expression led to clonal proliferation of CAR T cells (13) and highlighted the additional considerations required to safely manipulate epigenetic regulators in the clinic. Therefore, we next applied synthetic biology principles to design a clinically actionable CAR T cell with disrupted TET2 for improved efficacy. A key element of our strategy involved a dual knock-in (KI), simultaneously editing the TRAC and TET2 loci using CRISPR-Cas9 and introducing new genetic templates at these sites. First, we designed a single-guide RNA (sgRNA) to target the 5′ end of the first exon of TRAC. This enabled integration of an anti-CD19 CAR from an adeno-associated virus (AAV) donor DNA cassette into the TRAC locus and simultaneously resulted in TCR knockout (Fig. 5A). The KI construct (TRAC-CAR19) featured a 41BB costimulatory endodomain and coexpressed a truncated nerve growth factor receptor (tNGFR) for selection (Fig. 5B). This approach was designed to delay effector T cell differentiation and exhaustion through CAR insertion at the TRAC locus as previously described (67), while leveraging the potential benefits of 41BB costimulation (4). Furthermore, this TCR knockout strategy could enhance therapeutic safety by reducing risks of TCR-induced autoimmunity and alloreactivity. In addition, expression of the TRAC-CAR19 construct is controlled by the endogenous TRAC promoter, thus driving physiological CAR expression on the cell surface. The TRAC-CAR19 KI efficiency was proportional to the AAV dosage, achieving over 70% efficiency at a multiplicity of infection (MOI) of 50,000 (Fig. 5B and fig. S5A) and between 72 and 98% of CAR+ T cells were CD3 negative (Fig. 5C and fig. S5A), validating this dual knockout and KI strategy.
Fig. 5. Dual KI TET2-TRAC-CAR19 T cells have enhanced proliferation and maintain memory-associated marker expression.
(A) Left: Schematic of TRAC (top) and TET2 (bottom) loci alongside rAAV6 KI vectors. Right: Sanger sequencing electropherogram confirming integration of TRAC and TET2 KI constructs, underlined with dashed line. (B) Example plots of TET2 and TRAC-CAR19 single KI or dual TET2-TRAC-CAR19 KI T cells. (C) Example plots of CD3 loss detected by flow in TRAC-CAR19-KI T cells. (D and E) Schematic of in vitro ADCC assay (D) to deplete CRISPR-edited TET2-KI T cells. Example plots and data (E) of EGFR expression on TET2-KI T cells alone or in an NK cell coculture ± cetuximab incubation, gated on CD56− populations, n = 4. (F) Cumulative fold expansion of TRAC-CAR19 and TET2-TRAC-CAR19 T cells during restimulation and at day 25, arrows represent addition of irradiated K562-CD19+ target cells, n = 5. (G) Proportions of CD4+ versus CD8+ T cells in TRAC-CAR19 and TET2-TRAC-CAR19 T cells after stimulation, n = 7. (H) Example plots showing distribution of central (CCR7+ CD45RO+) and effector (CCR7− CD45RO+) memory-associated markers in CD8+ CAR T cell populations after restimulation, with summary after five stimulations, n = 5. (I) SPICE plot showing distribution of IR coexpression in CD8+ TRAC-CAR19 and TET2-TRAC-CAR19 T cells after 1 (acute) and 5 (chronic) stimulations, n = 6. (J) Data shown as means ± SEM [(F) and (G)] or individual values [(E) and (H)] from independent donors. ns P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001 by paired t test. Schematics [(A and (D)] created with BioRender.com.
In parallel with targeted modifications at the TRAC locus, we used CRISPR-Cas9 and a second AAV vector repair matrix to both disrupt TET2 (TET2-TRAC-CAR19) and integrate a truncated human epidermal growth factor receptor (tEGFR) cDNA at the TET2 locus (Fig. 5A and fig. S5A), under the regulation of an exogenous human EF1α promoter. The successful incorporation and functionality of the tEGFR enabled in vitro selection of TET2-edited cells (fig. S5B). We next tested whether tEGFR expression could function as a “safety switch” and allow targeted elimination of TET2-disrupted CAR T cells. TET2-TRAC-CAR19 T cells were cultured in vitro with natural killer (NK) cells and the Food and Drug Administration–approved antibody cetuximab. Cetuximab targets EGFR and induces antibody-dependent cellular cytotoxicity (ADCC) (Fig. 5D). Following coculture, EGFR-expressing CAR T cells were selectively depleted (Fig. 5E). Thus, tEGFR provides a critical safety switch that allows for controlled depletion of CRISPR-edited cells.
We next confirmed that our dual CRISPR editing and AAV KI CAR T cell engineering approach did not negatively affect manufacturing. TRAC-CAR19 T cells expanded as expected (fig. S5C), and TET2-TRAC-CAR19 T cells exhibited similar metabolic potency enhancements as TET2KO CAR T cells (fig. S5D) during manufacturing.
To test whether targeting TET2 in TRAC-CAR19 T cells could provide an advantage in settings of chronic antigen, we isolated edited (tEGFR+ and tNGFR+; fig. S5B) TRAC-CAR19 and TET2-TRAC-CAR19 T cells and subjected them to the in vitro restimulation assay described above to recapitulate features of progressive T cell exhaustion (Fig. 1G). During restimulation, TET2-TRAC-CAR19 T cells demonstrated substantial proliferative potential, with a sixfold increase in cumulative expansion by day 25 compared to TRAC-CAR19 T cells (Fig. 5F) that was antigen dependent (fig. S5E). This increased expansion only became apparent by the fourth round of stimulation (day 20), suggesting that this proliferative advantage was associated with chronic antigen exposure. The proliferative advantage of TET2 disruption was most apparent for CD8+ T cells, as CD8+ TET2-TRAC-CAR19 T cells expanded more than the CD4+ T cell equivalent (Fig. 5G). Concurrently, TET2-TRAC-CAR19 CD8+ T cells maintained a higher expression of the progenitor-associated receptor CCR7 (Fig. 5H) and exhibited lower frequencies of IR coexpression (PD1, LAG3, and TIM3) (Fig. 5I) than control TRAC-CAR19 CD8+ T cells following chronic antigen stimulation. Together, these data demonstrate that TET2 deficiency improves maintenance of a less terminally differentiated TRAC-CAR19 CD8+ T cell pool under conditions of chronic antigen stimulation, supporting our findings with TET2KO CAR T cells and TET2KO in chronic infection.
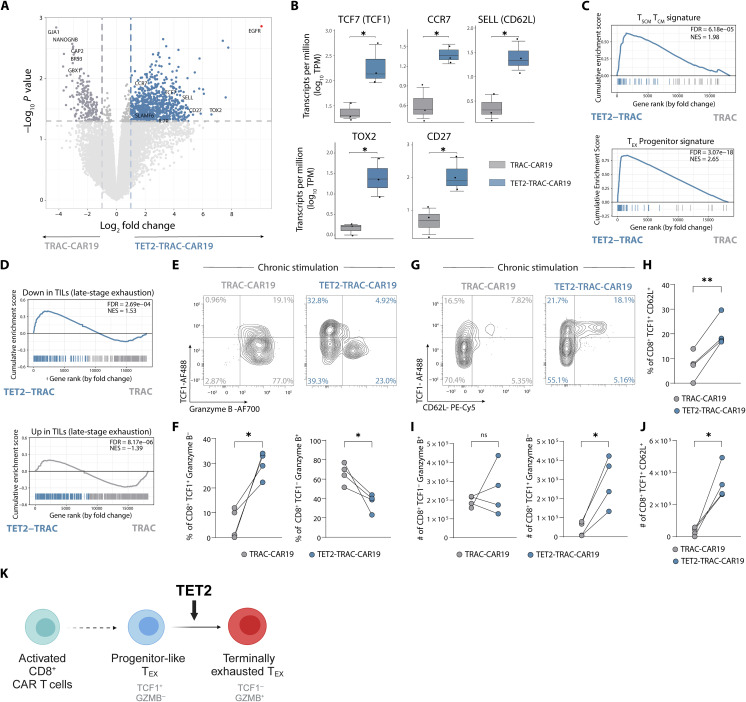
To gain a more detailed understanding of the impact of TET2 loss on CAR T cell responses to chronic antigen stimulation, we performed bulk RNA-seq on isolated TET2-TRAC-CAR19 CD8+ T cells following four rounds of in vitro restimulation. TET2-TRAC-CAR19 cells up-regulated progenitor- and memory-associated genes typically lowly expressed in terminally exhausted CD8+ T cells including TCF7, CCR7, SELL, and TOX2 (68) (Fig. 6, A and B, and table S5) compared to control TRAC-CAR19 cells. This increase in expression of progenitor-associated genes was accompanied by a down-regulation of genes related to calcium signaling and TF activity including BRS3, GJA1, CAP2, NANOGNB, and GBX1 (Fig. 6A). GSEA further supported the notion that TET2-TRAC-CAR19 cells retained features of more stem-like cells, with an enrichment for both a TEX progenitor signature and a stem cell/central memory CD8+ T cell signature (TSCM/TCM) (Fig. 6C). In contrast, a terminally exhausted tumor-infiltrating lymphocyte signature (69) was negatively enriched in TET2-TRAC-CAR19 (Fig. 6D). In line with GSEA results, chronic stimulation led to an increase in the proportion of CD8+ TET2-TRAC-CAR19 cells expressing the progenitor-associated TF TCF1 (70) and a decrease in the proportion of cells expressing the terminal differentiation-associated effector molecule granzyme B (71) compared to TRAC-CAR19 control cells (Fig. 6, E and F, and fig. S5F). Furthermore, TET2-TRAC-CAR19 cells retained the CD8+ TCF1+ CD62L+ population suggested to be essential for TEX progenitor proliferative responses (72) (Fig. 6, G and H, and fig. S5G). Although the frequency of TCF1− Granzyme B+ CD8+ T cells within TET2-TRAC-CAR19 cells was reduced after chronic stimulation, the number of TCF1− Granzyme B+ TET2KO cells remained comparable to TRAC-CAR19 control cells (Fig. 6I), because of enhanced expansion of TET2-TRAC-CAR19 cells compared to TRAC-CAR19 controls during chronic restimulation (Fig. 5F). Furthermore, the number of TCF1-expressing TET2-TRAC-CAR19 cells that were either granzyme B− (Fig. 6I) or CD62L+ (Fig. 6J) was higher than TRAC-CAR19 control cells at the same time point, suggesting that TET2 loss promotes expansion of progenitor-like TCF1+ CD8+ CAR T cells. Together, these data further suggest that the role of TET2 in chronically stimulated TRAC-CAR19 CD8+ T cells mirrors that identified in chronic LCMV infection, whereby TET2 drives terminal differentiation, and loss of TET2 supports maintenance of a progenitor-like phenotype (Fig. 6K).
Fig. 6. TET2 disruption limits terminal differentiation of TRAC-CAR19 T cells.
(A) Volcano plot showing differentially regulated genes in CD8+ TET2-TRAC-CAR19 and TRAC-CAR19 T cells after four stimulations; select markers highlighted. EGFR highlighted in red as TET2-KI construct positive control. Graph axes represent log2 fold change and -log10 P value, n = 3. (B) Box plots of individual gene log10 Transcripts per Million (TPM) for TET2-TRAC-CAR19 and TRAC-CAR19 T cells from (A), n = 3. (C and D) GSEA for signatures of stem-cell central memory and central memory T cells (TSCM and TCM) and TEX progenitors (gene sets from GSE147398) (C) and from human tumor infiltrating lymphocytes (TILs) [from (69)]. (D) Normalized enrichment score (NES) and FDR indicated in panel. (E and F) Example plots (E) and data (F) of CD8+ CAR T cell TCF1/Granzyme B subpopulation frequencies after four stimulations, n = 4. (G and H) Example plots (G) and data (H) of CD8+ CAR T cell TCF1/CD62L subpopulation frequencies after four stimulations, n = 4. (I and J) Number of CD8+ TCF1− Granzyme B+, CD8+ TCF1+ Granzyme B− (I) and CD8+ TCF1+ CD62L+ (J) CAR T cells from (F) and (H), n = 4. (K) Model for TET2 role during CAR T cell chronic stimulation. Data shown as means ± SEM (B) or individual values [(F) and (H) to (J)] from independent donors. ns P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001 by paired t test. Schematic (K) created with BioRender.com.
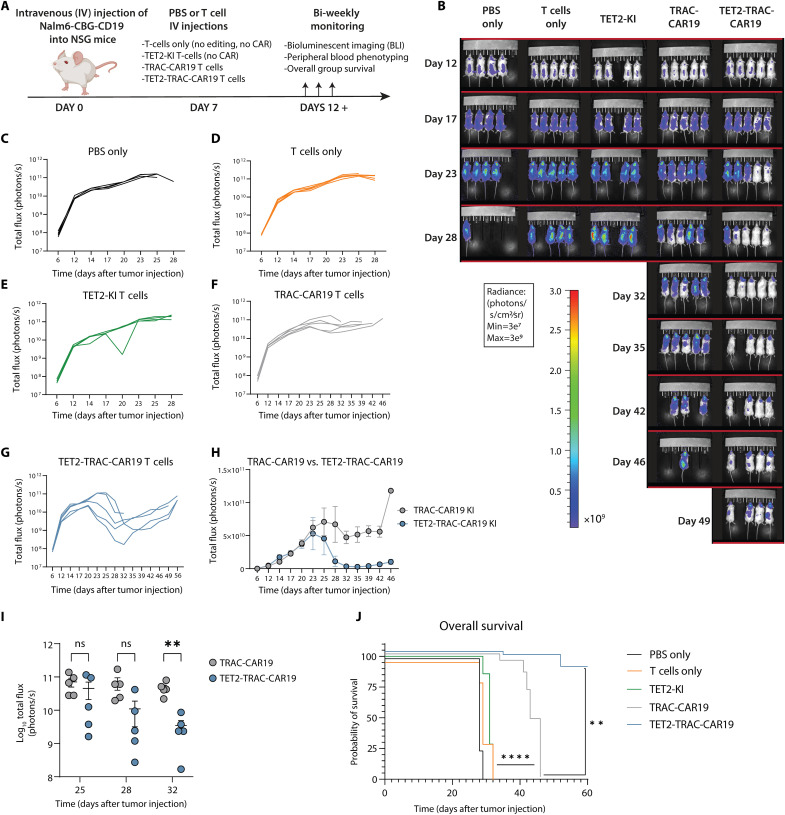
Dual KI CAR T cells display enhanced tumor control in aggressive B-ALL
Last, we tested the in vivo antitumor function of CAR T cells lacking TET2 in an NSG xenograft mouse model for aggressive B cell ALL (NALM-6) (Fig. 7A). NALM-6 cells expressing CD19 and click beetle green luciferase (CBG) were engrafted into recipient mice, and tumor growth was tracked through luminescent imaging. First, we found that TET2KO CAR T cells (fig. S6A) mediated superior antitumor control (fig. S6B) and increased survival compared to AAVS1KO CAR T cells (fig. S6C). We next evaluated whether TET2-TRAC-CAR KI also provided improved tumor control. TRAC-CAR19 T cells or TET2-TRAC-CAR19 T cells were administered 7 days after tumor injection. Phosphate-buffered saline (PBS) only (i.e., no cells), unedited T cells (without CRISPR editing or CAR transduction), and TET2-KI–only T cells (without CAR) were administered as controls. All control groups had rapid disease progression and succumbed to B-ALL by day 31 (Fig. 7, B to E), whereas both CAR T cell experimental groups demonstrated considerable tumor control (Fig. 7, B, F, and G). TET2-TRAC-CAR19 T cells showed enhanced efficacy compared to TRAC-CAR19 T cells (Fig. 7H) that was reflected in a substantial reduction in tumor burden by day 32 after tumor injection (Fig. 7I). Sustained tumor control following TET2-TRAC-CAR19 T cell administration translated into improved survival of mice compared to both the control and TRAC-CAR19 T cell groups (Fig. 7J). Thus, precise TET2 disruption under the control of a safety switch in allogenic CAR T cells enhances tumor control and animal survival.
Fig. 7. TET2 KI enhances the antitumor activity of TRAC-CAR19 T cells in vivo.
(A) Overview of in vivo experimental design, n = 4 to 5 mice per experimental group; one experiment. (B) Longitudinal tumor burden of all experimental groups by bioluminescent imaging (BLI). (C to G) Tumor outgrowth for PBS only (C), T cell (D), TET2-KI T cell (E), TRAC-CAR19 T cell (F), and TET2-TRAC-CAR19 T cell (G) groups, with individual mice shown. (H) Longitudinal comparison of TRAC-CAR19 and TET2-TRAC-CAR19 T cell group tumor burden. (I) BLI comparison at days during and immediately after peak CAR T cell response in TRAC-CAR19 and TET2-TRAC-CAR19 groups, line at mean with SEM; ns P > 0.05; *P < 0.05; **P < 0.01 by multiple paired t tests with Holm-Šídák correction. (J) Overall group survival, **P < 0.01; ****P < 0.0001 by Mantel-Cox log-rank test. Data shown as means ± SEM (H) or individual values [(C) to (G) and I)] from each mouse. Schematic (A) created with BioRender.com.
DISCUSSION
We previously reported a case of CAR transgene integration at the TET2 locus, which shifted T cell differentiation toward a central memory–like phenotype (10). Here, we identified similar insertions among multiple patients with leukemia receiving CAR T cell therapy. The prevalence of these integrations and disruption of TET2 prompted further investigation into the role of TET2 in regulating T cell fate and suggested a mechanism through which TET2 could be manipulated to improve CAR T cell efficacy. Using both in vitro and in vivo approaches, we identified a role for TET2, a key enzyme in active DNA demethylation, in driving the epigenetic transitions of CD8+ T cells toward terminal differentiation, at the expense of retaining the stem cell–like characteristics of progenitor cells. We took advantage of this function to engineer TET2-disrupted CD19 CAR T cells with improved tumor control. Crucially, the addition of a functional safety switch into this CAR T cell design provides a therapeutically viable approach to alter T cell fate for potential patient benefit.
TET2 deletion restrained terminal differentiation in both chronic LCMV infection in vivo and in two in vitro CAR T cell models of exhaustion. In CAR T cell exhaustion models, this decrease in terminal differentiation resulted in improved proliferative capacity and cytokine production and decreased expression of IRs. A previous study showed that disruption of TET2 enhanced the in vivo expansion of 41BB-costimulated but not CD28-costimulated CAR T cells (13). However, here, loss of TET2 affected CAR-T cell phenotype comparably for both 41BBζ- and CD28ζ- CD19 CAR T cell constructs in vitro. The difference in these findings likely reflects differences in the timing of the measured responses and/or context of antigen stimulation. Thus, the role of costimulatory domains in TET2-deficient CAR T cells warrants further investigation, as our data suggest that modulation of TET2 could be used to improve expansion for CAR T cell products that typically have shorter persistence (73).
The decreased terminal differentiation observed in chronically stimulated TET2-deficient CAR T cells was accompanied by elevated expression of several stem cell– and memory-associated genes, including TCF1. Expression of stem cell–like features is a hallmark of the TEX progenitor cells that sustain exhausted CD8+ T cell responses during cancer and chronic viral infection (30–36). To investigate where TET2 might function in the developmental trajectory of CD8+ T cell exhaustion, we turned to the well-characterized LCMV chronic infection model. TET2 deficiency in this setting also protected the TEX progenitor population and restrained development of terminally exhausted TEX. Thus, disruption of TET2 enabled maintenance of a progenitor-like population in CAR T cells and CD8+ T cells exposed to chronic viral infection. Furthermore, in chronic viral infection, the terminally exhausted TEX that escaped this differentiation block retained key features of TEX progenitors, including decreased expression of several IRs and effector-associated genes. TET2 ablation limited chromatin accessibility for TEFF-associated TFs in terminally differentiated TEX and reduced expression of TOX. TOX has been proposed to regulate terminal exhaustion (35); thus, together, these data suggest that TET2 may coordinate with TOX and TEFF-associated TFs to initiate and/or maintain the terminal differentiation of TEX.
In transitioning to a progenitor-like memory state and away from terminal differentiation, T cells undergo metabolic reprogramming, including a shift to oxidative metabolism (74), that augments proliferation and function. Accordingly, TCM and TSCM demonstrate superior antitumor potency compared to effector-like cells in CAR T cell therapies (10, 75–77). Metabolic programming also shifts during CD8+ T cell exhaustion. Metabolic fitness, including glucose uptake capacity, may decrease as TEX progenitors increase PD1 expression and terminally differentiate (18, 78). Together, these findings highlight commonalities in how metabolic programming is associated with both progenitor-associated biology and terminal differentiation across multiple contexts. TET2KO CAR T cells displayed enhanced metabolic fitness after manufacturing, potentially providing a growth and/or survival advantage. Our finding that TET2 loss enhanced glycolysis of CAR T cells is supported by previous reports suggesting that TET2 suppresses glycolytic metabolism in a broad range of cell types, including myeloid cells, cancer cells, and embryotic stem cells, through multiple pathways (79–81). Furthermore, in myeloid cells, TET2 loss is also associated with enhanced oxidative metabolism (80), consistent with the increase in oxidative metabolism in CAR T cells we observed following TET2 disruption. Loss of TET2 could drive increased oxidative metabolism by either a shift toward fatty acid oxidation as altered fuel utilization (82) or changes in mitochondrial volume due to enhanced mitochondrial biogenesis or morphological crista reorganization (74, 83). The DNMT family of DNA methyltransferases has previously been reported to shape the metabolic phenotype of CAR T cells, at least in part through regulation of mitochondrial biogenesis (9, 84) adding weight to the hypothesis that epigenetic regulators may modulate mitochondrial function with consequences for cellular metabolism. As the metabolic changes seen following TET2 disruption were accompanied by an increase in frequency of TCM, it will be important to disentangle how TET2 may regulate metabolism independently of memory differentiation. However, these data imply first that TET2 could play a critical role in regulating metabolic reprogramming during early T cell differentiation, and second, that loss of TET2 promotes more progenitor-like metabolism. Therefore, TET2 loss may limit terminal exhaustion, at least in part, through regulation of metabolic function either early in CD8+ T cell differentiation before establishment of exhaustion or as exhaustion progresses.
In settings of both acute and chronic antigen exposure in vitro and in vivo, TET2 regulated the acquisition (or reacquisition) of effector-like biology and drove terminal differentiation. Moreover, within the TEX program, TET2 promoted effector biology and TEFF differentiation before fate commitment to exhaustion, driving development of short-lived TEFF-like cells and limiting formation of the TEX precursor pool (65). Together, these findings suggest that TET2 orchestrates the development of a core effector and terminal differentiation program that coordinates with context-dependent TFs and the epigenetic landscape to result in distinct CD8+ T cell fates. Supporting this hypothesis, we identified TFs that were regulated by TET2 in both the acute and chronic infection settings, including Prdm1 and Runx3 (14), suggesting that shared TET2-regulated gene expression programs likely drive TEFF formation at the expense of TMEM in acute infection and enforce terminal exhaustion at the expense of TEX progenitors during chronic infection. Thus, TET2 coordinates terminal differentiation and the acquisition of effector-like biology at multiple checkpoints in CD8+ T cell differentiation, implying that analogous epigenetic and transcriptional programs, including those regulated by TET2, are reused throughout CD8+ T cell differentiation (61). However, as an epigenetic enzyme and mediator of active DNA demethylation, it is likely that TET2 coordinates the expression of not one but multiple genes and pathways to support terminal TEX differentiation. TET2 regulated the RNA expression and predicted motif accessibility of many key TFs in exhausted CD8+ T cells (figs. S3H and S4E). Thus, our data are consistent with the hypothesis that multiple TET2-regulated epigenetic and transcriptional changes are required to orchestrate TEX differentiation (85, 86). Further work is needed to understand how TET2 may cooperate with lineage-specific TFs and other epigenetic regulators to coordinate these epigenetic transitions.
TET2 disruption decreased expression of effector-associated molecules, including granzymes, both in chronic-stimulation CAR-T cell models and in chronic infection. However, loss of TET2 did not negatively affect CAR T cell antitumor efficacy. Rather, TET2-TRAC-CAR19 T cells exhibited improved in vivo tumor control compared to TRAC-CAR19 T cells. One possible explanation for our findings is that although the frequency of granzyme B+ TET2KO CD8+ CAR T cells was reduced following chronic stimulation, TET2KO cells underwent greater expansion and had an outgrowth of CD8+ cells, resulting in similar numbers of granzyme B+ TET2KO CAR T cells as control TRAC-CAR19 T cells. Concurrently, TET2KO CAR T cell cultures had both higher frequencies and numbers of TCF1+ granzyme B− cells after chronic stimulation compared to TRAC-CAR19 cultures. This TCF1+ granzyme B− progenitor population sustains effective CD8+ T cell responses during chronic infection and cancer (34, 72, 87, 88), provides the proliferative burst in response to checkpoint immunotherapy (30, 88), and maintains the pool of terminally differentiated TEX. Therefore, increased antitumor efficacy of TET2KO CAR T cells likely stemmed from enhanced CD8+ T cell proliferation and/or survival, driven by improved maintenance of the self-renewing stem cell–like progenitor population. Moreover, TEX progenitors retain the ability to produce cytokines, including IL-2, despite low expression of cytotoxic markers (33). Thus, strategies that increase the maintenance of progenitor populations, in addition to enhancing effector function, may have potential as a means to improve CAR T cell efficacy.
Understanding the epigenetic regulation of TEX development holds promise for advancing immunotherapies like CAR T cells. The hyperproliferative phenotype of TET2-deficient CAR T cells underscores the efficacy of epigenetic reprogramming yet raises substantial long-term safety concerns (10, 13). Individual mutations implicated in T cell lymphoma alone typically do not lead to lymphomagenesis directly; instead, they are often detected in aberrant cells contributing to autoinflammatory or autoimmune disorders (89). In addition, prior studies leveraging gene knockout strategies targeting T cell lymphoma tumor suppressors have shown no signs of malignant transformation (90). However, deliberate disruption of TET2 for CAR T cell therapy warrants caution, especially in elderly patients susceptible to acquiring DNMT3A mutations (91), which can cooperate with TET2 loss, potentially leading to T cell oncogenesis (92). To address these concerns, we used synthetic biology to develop a next-generation cell therapy product. This approach improves the antitumor efficacy of CAR T cells, modulating TET2 to limit terminal exhaustion, while also reducing the risk of lymphomagenesis, autoimmunity, or graft-versus-host disease. First, disrupting TRAC mitigates the risk of pathological signaling from the endogenous TCR. Second, our dual KI strategy ensures precise insertion of transgenes at specific loci, averting random genomic integration. Third, incorporating a safety switch allows for depletion of TET2-disrupted cells if necessary. Such depletion strategies provide assurance against the theoretical risk of T cell lymphoma following CD19 CAR administration (93). Furthermore, additional mitigation strategies could be applied, such as screening for pre-existing mutations predisposing engineered cell products to hyperproliferation or transformation (93, 94), administering corticosteroids, which TET2KO T cells are highly sensitive to (10, 13), and transient or partial suppression of TET2 during CAR T cell production and/or after infusion. Our findings thus underscore the practical significance and feasibility of targeted epigenetic reprogramming to shape CAR T cell differentiation, highlighting the potential of TET2 modulation to redirect TEX fate.
MATERIALS AND METHODS
Study design
The study investigated the role of TET2 in regulating exhausted CD8+ T cell differentiation (TEX) in cancer and chronic viral infection, using human CAR T cells and a murine LCMV model. We evaluated the impact of TET2 disruption on TEX phenotypes, differentiation fate, and functional outcomes both in vitro and in vivo. Flow cytometry assessed protein expression related to T cell exhaustion and memory differentiation, while ATAC-seq explored chromatin accessibility landscapes across TEX subsets. Transcriptomic analysis elucidated underlying pathways and transcriptional regulation by TET2. In addition, we developed a CRISPR-Cas9–based genome editing approach to engineer allogeneic CAR T cells for enhanced antitumor responses through TET2 modulation of TEX. Sample sizes were estimated on the basis of preliminary experiments, with in vitro functional assays performed at least three times. Investigators were not blinded during experiments or outcome assessment. All in vivo mouse work was performed under approved Institutional Animal Care and Use Committee protocols no. 803978, no. 805082, and no. 806619.
Primary human cells
CAR T cells and control samples were generated from healthy donor peripheral blood mononuclear cells (PBMCs) through leukapheresis, following University of Pennsylvania Institutional Review Board–approved protocols. Written informed consent was obtained from all participants, consistent with the principles outlined in the Declaration of Helsinki, International Conference on Harmonization Guidelines for Good Clinical Practice, and the US Common Rule.
Cell lines
For viral vector production, human embryonic kidney (HEK) 293 T cells and GP2-293, a HEK 293–derived retroviral packaging cell line, were cultured in hR10 medium (RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2% Hepes buffer, 1% GlutaMAX, and 1% penicillin-streptomycin). SUP-T1 cells were used to determine lentiviral titers. HEK 293 T cells were sourced from the American Type Culture Collection and GP2-293 from Takara Bio. NALM-6 cells expressing CBG and green fluorescent protein (GFP), provided by M. Ruella at the University of Pennsylvania, were used. K562 human leukemia cell lines including a variant expressing the extracellular domain of the CD19 protein were obtained from C. H. June at the University of Pennsylvania and maintained in hR10 medium. 143B osteosarcoma cells modified to express GFP and firefly luciferase, along with NALM-6 cells engineered for GD2 expression, were also cultured in hR10 medium. Cell line authenticity was confirmed via short-tandem-repeat profiling meeting the International Cell Line Authentication Committee’s guidelines with more than 80% match, conducted by the University of Arizona Genetics Core. Regular mycoplasma screenings were performed to ensure cell line health and purity before and after genetic modifications.
Analysis of TET2 integration sites in CAR T cell–treated patients with leukemia
Genomic DNA from patient cell sources (whole blood, bone marrow, PBMCs, or T cells; before and after infusion) was isolated for library preparation followed by paired-end Illumina sequencing, as previously described (10, 95). CLL and ALL human integration site data were aligned to the human genome hg38 and analyzed using a previously published integration analysis pipeline (96). Sample time points were grouped into four categories (day 0, days 1 to 15, days 16 to 31, and day 31+). Percent relative abundance represents the estimated proportion of cells with integration in a sample. Visualization code for TET2 patient integration site data analysis is available at https://github.com/helixscript/TET2_ALL_CLL.
Lentiviral packaging
Briefly, HEK 293 T cells were transfected with 7 μg of pVSV-G glycoprotein envelope plasmid, 18 μg of pMDLg/p.RRE Gag/Pol plasmid and 18 μg of pRSV.Rev. plasmid alongside 15 μg of transfer vector plasmid encoding for CAR of interest using Lipofectamine 2000 (Thermo Fisher Scientific) and Opti-MEM (Gibco). Cell culture supernatant was harvested 24 and 48 hours after transfection, centrifuged at 900 RCF for 10 min at 4°C, and filtered through a 0.45 μM vacuum filter. Following filtration, 24-hour supernatant was concentrated by ultracentrifugation at 8877 RCF overnight at 4°C, while 48-hour supernatant was concentrated overtop of the overnight viral pellet at 76,790 RCF for 2 hours at 4°C. Concentrated virus was stored at −80°C.
T cell culture and lentiviral transduction
T cells were isolated from healthy donor PBMCs using the Pan T Cell Isolation Kit following manufacturer’s instructions (Miltenyi Biotec). Isolated T cells were activated using anti-CD3/CD28 antibody-coated Dynabeads (Thermo Fisher Scientific) at a 3:1 bead-to-cell ratio in T cell media consisting of OpTmizer CTS SFM media (Thermo Fisher Scientific) supplemented with 5% human AB serum and human IL-2 (100 U/ml; PeproTech). After a 24-hour incubation, lentivirus containing the appropriate CAR construct was introduced to the culture at an MOI of 2.5. CAR T cell expansion proceeded following established protocols (19).
AAV construct design
DNA sequences containing either a truncated EGFR (tEGFR) sequence driven by an EF1α promoter (for TET2-KI), or a tNGFR sequence, T2A sequence and an anti-CD19 single-chain variable fragment (scFv) fused to 4-1BB and CD3ζ stimulatory endodomains (for TRAC-CAR19-KI) were subcloned into recombinant AAV6 plasmids (GenScript). DNA sequences were flanked with 400-bp homology arms immediately upstream and downstream of the TET2 gRNA or TRAC gRNA cut sites, respectfully. Large-scale packaging of AAV6 virus was done by cotransfection of a packaging cell line with the rAAV6 transgene plasmid of interest, a rep- and cap-encoding plasmid and an adenovirus-derived replication helper plasmid (Charles River Laboratories).
CRISPR-Cas9–mediated editing and AAV transduction
TET2 and TRAC editing via CRISPR-Cas9 was conducted 72 hours after T cell activation. sgRNA reagents from Integrated DNA Technologies targeted the TET2 and TRAC loci. The sgRNA sequences with protospacer-adjacent motif sequences are indicated as follows: TET2 5′-CGGGGATACCTATACAGATCCAT-3′ and TRAC 5′-AGGGAGAATCAAAATCGGTGAAT-3′. The control AAVS1 targeted sequence is: 5′-CCATCGTAAGCAAACCTTAGAGG-3′.
Activated T cells were debeaded magnetically, washed with 1× PBS at 300g for 5 min, and resuspended in P3 4D-nucleofection buffer (Lonza). TrueCut Cas9 Protein v2 (Thermo Fisher Scientific). sgRNAs targeting TET2 and/or TRAC were individually complexed at 6 μg:3.2 μg for 10 min at room temperature (RT) to form ribonucleoprotein complexes before nucleofection. Nucleofection into T cells was performed using a Lonza 4D Nucleofector X Unit with high-fidelity program EO-115, followed by a 10-min resting period. For AAV-mediated KI, cells were transduced with AAV viral vectors carrying TRAC-CAR19-tNGFR and/or TET2-tEGFR constructs (Charles River Laboratories) at an MOI of 50,000.
TET2 knockout efficiency was confirmed by isolating genomic DNA from CAR T cells at day 7 using the dNeasy Blood & Tissue Kit (Qiagen). Polymerase chain reaction (PCR) of genomic DNA was performed with TET2 forward primer 5′-TCCCTGAGTCCCAGTCCATC-3′ and reverse primer 5′-TCAGGAATGGCCAGGTTCTG-3′ using MyTaq Red 2X Mix (Meridian Bioscience). Purified control and edited PCR products underwent Sanger sequencing (Azenta), and editing efficiency was determined by Tracking of Indels by DEcomposition (TIDE) through comparison of control and edited Sanger sequence electropherogram files.
To confirm tEGFR and tNGFR-CAR19 construct KIs, genomic DNA was isolated from end-of-expansion transduced CAR T cells. PCR of genomic DNA was carried out with the following primer sets: TET2 (unedited) forward primer 5′-TCCCTGAGTCCCAGTCCATC-3′, TET2 (unedited) reverse primer 5′-TCAGGAATGGCCAGGTTCTG-3′, TET2 (edited) forward primer 5′-CATCACGAGCAGCTGGTTTC-3′, TET2 (edited) reverse primer 5′-GGCAATTGAACCGGTGCCTA-3′, TRAC (unedited) forward primer 5′-TCCCTGAGTCCCAGTCCATC-3′, TRAC (unedited) everse primer 5′-CTTCATGCCCTGCATCTCCA-3′, TRAC (edited) forward primer 5′-CATCACGAGCAGCTGGTTTC-3′, TRAC (edited) reverse primer 5′-CATCAGTTGCAGGGCAAGTC-3′. Edited and unedited PCR products underwent purification and Sanger sequencing.
Western blot analysis of TET2 knockout
CAR T cells were lysed in 1× lysis buffer (Cell Signaling Technology) and supernatants were collected after centrifugation. Cell lysate samples (30 μg) were separated on a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen) and transferred onto a membrane using the iBlot 2 Dry Blotting System (Invitrogen). The membrane was blocked with 5% skim milk and probed with primary antibodies overnight at 4°C: either rabbit monoclonal anti-TET2 (Cell Signaling Technology) or monoclonal mouse anti–glyceraldehyde phosphate dehydrogenase (Thermo Fisher Scientific). Primary antibodies were diluted in 1× PBS with 0.2% Tween and 5% bovine serum albumin. After washing, the membrane was incubated with goat anti-mouse or anti-rabbit horseradish peroxidase–linked secondary antibody (Thermo Fisher Scientific) for 1 hour at RT. Last, the membrane was treated with equal parts of Pierce ECL Western blotting substrate (Thermo Fisher Scientific) and visualized.
Flow cytometry of human immune cells
Cells were collected and stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen) for 20 min at RT. After washing with hFACS buffer (PBS + 2% FBS + 0.05% sodium azide), surface antibodies were incubated with cells for 30 min at 4°C in hFACS buffer and Brilliant Stain Buffer (BD Biosciences). For intracellular staining, samples were fixed and permeabilized using the FoxP3 Transcription Factor Staining Buffer Kit (Thermo Fisher Scientific) for 30 min, followed by staining with intracellular antibodies for an additional 30 min. Data acquisition was performed using a BD LSRFortessa and analyzed with FlowJo software (BD Life Sciences). Compensation setup used Anti-Mouse Ig, κ and Anti-Rat/Hamster Ig, κ CompBeads (BD Biosciences) along with Fluorescence Minus One controls to establish gating boundaries. SPICE plots were generated from single gated IRs, grouped using Boolean “AND” gates, and plotted using SPICE 6.1 software (https://niaid.github.io/spice/). Refer to table S6 for antibody details.
Seahorse metabolic flux assay
Using a Seahorse xFe96 Analyzer (Agilent), we conducted the Seahorse Mitochondrial Stress Test. The xFe96 Pro sensor cartridge (Agilent) was hydrated overnight with sterile water at 37°C in a non-CO2 incubator, followed by XF Calibrant (Agilent) hydration. Thawed T cells were rested overnight, washed with 1× PBS, and treated with Seahorse Assay Media. Cells (1 × 105 to 2 × 105) were plated in poly-d lysine–coated 96-well microplates (Agilent) with four to five technical replicates. The microplate was centrifuged and incubated to facilitate cell attachment. Drug solutions (oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone/antimycin A) were prepared in the sensor cartridge. The assay measured basal oxygen consumption rate (OCR) and ECAR in triplicate at baseline and after each drug addition using WAVE software (Agilent). Basal respiration is defined as the OCR value before any drug additions. Maximal respiration is defined as the OCR value after addition of FCCP. SRC is defined as the difference in OCR between maximal and basal respiration levels.
CAR T cell serial restimulation assay
CAR+ T cells were purified using a biotin-conjugated AffiniPure Goat Anti-Mouse IgG F(ab’)₂ fragment specific antibody and antibiotin microbeads (Miltenyi Biotec). In the case of dual KI TET2-TRAC CAR T cells, EGFR+, NGFR+, and EGFR/NGFR-dual positive cells were purified using phycoerythrin (PE) and allophycocyanin (APC)–conjugated antibodies alongside anti-PE, anti-APC, or anti-PE MultiSort microbeads as per the manufacturer’s instructions (Miltenyi Biotec). Purity was assessed by flow staining against KI markers tEGFR and tNGFR. K562-CD19+ cells were exposed to 100-Gy ionizing radiation using the xRad320 (Precision X-Ray). CAR T cells were cocultured with irradiated K562-CD19+ cells at a 1:1 ratio with 1 million CAR T cells per 1 million K562 cells in hR10. Coculture supernatants were harvested 24 hours after each stimulation and frozen at −20°C. At 5 days after stimulation (defined as acute stimulation), absolute CAR T cell counts were assessed with a LUNA-FL Dual Fluorescence Cell Counter (Logos Biosystems) and recultured at a 1:1 ratio with fresh hR10 media and newly irradiated K562 cells for four to five additional stimulations (defined as chronic stimulation). CAR T cells were then cryopreserved for phenotyping and transcriptomic profiling.
Cytokine analysis
Supernatant cytokines were quantified flow cytometrically using the LEGENDplex Human CD8/NK Panel as per the manufacturer’s instructions (BioLegend). Data were acquired on LSRFortessa, and data analysis was performed with BioLegend LEGENDplex Data Analysis Software Suites (BioLegend Qognit Cloud Platform).
Assay for exhaustion in CAR T cells with high tonic signaling
CAR T cells were transduced with retrovirus on days 2 and 3 after activation. Briefly, 12- or 24-well plates, non-tissue-culture-treated, were coated with 1 ml or 500 μl, respectively, of RetroNectin (25 μg/ml; Takara) in PBS and incubated at 4°C overnight. The following day, plates were washed with PBS and then blocked with 2% BSA in PBS for 10 min. Retroviral supernatants were added, and plates were centrifuged at 32°C for 2 hours at 2500 RCF. After centrifugation, viral supernatants were removed, and T cells were seeded into each virus-coated well at a density of 1 × 106 T cells per well for 12-well plates and 0.5 × 106 T cells per well for 24-well plates. CRISPR knockout of TET2 (or AAVS1 as a control) was performed 2 to 4 days after T cell activation to achieve maximal editing efficiency, using the EH115 program on a Lonza 4D Nucleofector. Cells were immediately recovered in 260 μl of warm complete AIM-V media supplemented with IL-2 (500 U/ml) in round-bottom 96-well plates and expanded into 1 ml of fresh medium after 24 hours. Cells were maintained at densities of 0.5 × 106 to 2 × 106 cells/ml in well plates until days 14 to 16 for functional and phenotypic characterization. Editing efficiency was assessed using TIDE as described previously. Immunophenotyping of CAR T cells via flow cytometry was performed on days 11 and 15 of expansion. Cytotoxicity of HA.28ζ CAR T cells was evaluated using an Incucyte Live-Cell Analysis System at day 15 at the end of expansion. In brief, 25 × 105 GFP+ 143b-GL osteosarcoma tumor cells were seeded in triplicate in 96-well plates and cocultured with T cells at effector:target ratios of 1:1, 1:2, 1:4, 1:8, and/or 1:16 in 300 μl of T cell medium without IL-2 in 96-well flat-bottom plates. Plates were imaged at 10× zoom with four to nine images per well every 2 to 4 hours for 96 hours using the IncuCyte ZOOM Live-Cell analysis system. Total integrated GFP intensity per well or total GFP area (square micrometer per well) were used to analyze expansion or contraction of 143B cells, with four images captured per well at each time point. Total tumor GFP fluorescence (normalized to the initial t = 0 time point) was recorded, and the normalized tumor GFP signal was used as the cytolysis threshold.
Cell culture supernatants from 1:1 E:T cocultures were used to determine IL-2 and IFN-γ concentrations via ELISA. Specifically, 5 × 104 CAR T cells were cocultured with 5 × 104 tumor cells in 200 μl of complete T cell medium (AIM-V or RPMI) without IL-2 in a 96-well plate, all in triplicate. After 24 hours of coculture, culture supernatants were collected, diluted 20 to 100-fold, and analyzed for IL-2 and IFN-γ using ELISA MAX kits and Nunc Maxisorp 96-well ELISA plates. Absorbance readings were obtained using a Spark plate reader (Tecan Life Sciences).
LCMV mouse studies
Mice were maintained in a specific pathogen–free facility at the University of Pennsylvania, in accordance with the Institutional Animal Care and Use Committee. B6;129S-Tet2tm1.1Iaai/J (Tet2fl/fl) mice and Cd4Cre+ mice were obtained from the Jackson Laboratory (JAX). Cd4Cre+ mice induced deletion of Tet2 during thymocyte development resulting in CD4+ and CD8+ T cells that lack TET2 expression. TCR transgenic P14 C57BL/6 mice expressing a TCR specific for LCMV peptide DbGP33–41 (97, 98) were bred in house and served as a source of LCMV-specific CD8+ T cells (P14 cells). All mice were backcrossed to and maintained on a C57BL/6 J background. P14 mice were bred to Tet2+/+ Cd4Cre+ or Tet2fl/fl Cd4Cre+ mice to generate WT (Tet2+/+ Cd4Cre+ P14+) and TET2KO (Tet2fl/fl Cd4Cre+ P14+) P14 donor mice. For all experiments, WT and TET2KO donor mice were age and sex matched. For P14 cotransfer experiments, sex-matched recipient C57BL/6 mice were purchased from JAX at 5 to 8 weeks of age.
Chronic LCMV infection
Recipient mice were infected intravenously with 4 × 106 PFU of LCMV clone 13. LCMV titers were determined via plaque assay as described (99).
Naïve P14 cell cotransfer
Adoptive transfer of P14 cells was performed as described (8). P14 cells were isolated from the peripheral blood of naïve congenically distinct WT and TET2KO donor mice using a histopaque 1083 gradient (Sigma-Aldrich). WT and TET2KO P14 cells were mixed at a 1:1 ratio and a total of 500 P14 cells (250 WT and 250 TET2KO) were adoptively transferred intravenously into recipient mice of a third congenic background. The 1:1 ratio was confirmed by flow cytometry (BD LSRII). One day after adoptive transfer, recipient mice were infected with LCMV clone 13 (day 0). Unless otherwise indicated, recipient mice were treated with CD4-depleting antibody (GK1.5, 200 mg per injection) on day −1 and day +1 relative to infection with LCMV clone 13.
Retroviral transduction of the TET2 catalytic domain
The FLAG-tagged murine TET2 catalytic domain in pMXs was provided by R. Kohli (University of Pennsylvania) and subsequently inserted into MIGR1 courtesy of W. Pear (University of Pennsylvania), with an expanded multiple cloning site introduced. Empty MIGR plasmid was used as a control. Retroviruses (RV) were generated in HEK 293 T cells. P14 cells from either WT or TET2KO donor mice were activated, and retroviral transduction performed as previously described (100, 101). CD8+ T cells were isolated from spleens of P14 donor mice by negative selection using the EasySep Mouse CD8+ T cell isolation kit (STEMCELL Technologies). P14 cells were activated in vitro for 24 to 28 hours with recombinant IL-2 (100 U/ml), LEAF anti-mouse CD3e (1 mg/ml), and LEAF anti-mouse CD28 in mouse R10 media (0.5 mg/ml) (mR10: RPMI-1640 supplemented with 10% FCS, penicillin and streptomycin (50 U/ml), l-glutamine, 20 mM Hepes, nonessential amino acids (1:100), 1 mM sodium pyruvate, and 50 mM b-mercaptoethanol). Activated P14 cells were transduced by spinfection at 2000g for 90 min at 32°C in mR10 + IL-2 (100 U/ml) and polybrene (0.5 mg/ml). WT P14 cells were transduced with MIGR1, while TET2KO P14 cells from donor mice of a distinct congenic were transduced with either MIGR1 or TET2 CD. After 24 hours of rest, P14 cells expressing the retroviral reporter GFP were sorted (BD FACS Aria, 37°C) and WT and TET2KO P14 cells mixed in a 1:1 ratio before adoptive transfer intravenously into recipient mice of a third congenic background. The 1:1 ratio was confirmed by flow cytometry (BD LSRII). A total of 4 × 104 to 5 × 104 total P14 cells were transferred per LCMV clone 13–infected recipient mouse. Recipient mice were infected with LCMV clone 13 on the same day as P14 cell activation.
Peptide stimulation, flow cytometry, and sorting of murine immune cells
PBMCs were isolated from peripheral blood by repeated lysis with Ammonium-Chloride-Potassium (ACK) lysis buffer and immediately stained in mouse FACS Buffer (mFACS Buffer; PBS + 3% FCS + 2 mM EDTA). Splenocytes were processed to a single-cell suspension by mechanical disruption over a 70-μm filter, followed by ACK lysis and then counted.
Splentocyte samples were either aliquoted in mFACS Buffer for staining or resuspended in mR10 and stimulated ex vivo with LCMV peptide DbGP33 (0.2 μg/ml) in the presence of Golgi Plug and Golgi Stop (BD Bioscience) for 5 hours at 37°C. Following stimulation, samples were washed in PBS, incubated with a viability dye (15 min at RT), and stained with an antibody cocktail targeting surface markers in mFACS Buffer + Brilliant Stain buffer for 30 min at 4°C or 1 hour at RT. In some panels, samples were stained with gp33 tetramer for 1 hour at 37°C in mR10 before surface staining. Biotinylated primary antibodies were detected with streptavidin-conjugated secondary antibody for 30 min at 4°C. For intracellular staining, samples were permeabilized using the eBioscience FoxP3 Transcription Factor Staining Buffer Kit or BD Cytofix/Cytoperm Fixation/Permeabilization kit and incubated with intracellular antibodies for 30 min at 4°C or 1 hour at RT then washed and stored in BD Stabilizing Fixative until acquisition. Samples were acquired on a BD LSRII or a BD FACSymphony A5 and analyzed in FlowJo v10.8 software. Voltages on flow cytometry machines were standardized using fluorescent targets and Spherotech rainbow beads.
For cell sorting for sequencing, splenocytes were stained with a surface antibody cocktail in mR10 + Brilliant Stain Buffer for 30 min at 4°C. Samples were sorted on a BD FACSAria at 4°C into mR10 media with 50% FCS. Sorting accuracy was confirmed through postsort purity checks. See table S7 for antibody information.
Bulk RNA-seq
For in vivo LCMV sample preparation, WT and TET2KO P14 cells were cotransferred into recipient mice as described in “Naïve P14 cell cotransfer”. Total WT and TET2KO P14 cells and TEX subsets were isolated at day 15 p.i. with LCMV clone 13 with >95% purity, as detailed above. A total of 2 × 104 cells were sorted in triplicate per sample and stored at −80°C in RLT buffer (Qiagen). RNA was isolated using the Qiagen RNeasy Micro Kit, and cDNA libraries were generated following the manufacturer’s instructions with the SMART-Seq v4 Ultra Low Input RNA Kit and Nextera XT DNA library kit. After quantification with the KAPA Library Quant Kit, cDNA libraries were pooled and diluted to 1.8 pg/ml and paired-end sequencing was conducted on a NextSeq 550 (Illumina) using a NextSeq 500/550 Mid Output Kit v2.5 (150 cycles).
For in vitro CAR T cell sample preparation, CD8+ T cells were enriched via positive selection (Miltenyi Biotec), resuspended in TRIzol (Thermo Fisher Scientific), and stored at −80°C. Upon thawing, total RNA was extracted, treated with Dnase, and further processed using an RNA Clean and Concentrator Kit (Zymo Research). Bulk RNA-seq was performed by Novogene using the NovaSeq 6000 system with a paired-end 150-bp approach, generating 6 GB of sequencing read data per sample.
Mouse reads were aligned to transcriptome mm39 using STAR with quantification via cufflinks, while human reads were pseudoaligned to the human transcriptome GRCh38 using kallisto. Data was imported into R, transformed into log2 counts per million, and normalized using Trimmed Mean of M-values (TMM) with EdgeR. Differential Gene Expression was determined with linear modeling and adjusted P values using the Benjamini-Hochberg correction method with limma and EdgeR.
GSEA was conducted using GSEA software from UC San Diego and Broad Institute developers. Filtered, normalized expression data were used as input, with parameters including a weighted enrichment statistic and Signal2Noise metric for gene rankings. PCA plots were generated using the function Prcomp in R (www.rdocumentation.org/packages/stats/versions/3.6.2/topics/prcomp) (102).
Assay for transposase-accessible chromatin sequencing (ATAC-seq)
For each LCMV TEX sample, 1 × 104 to 2 × 104 P14 cells were sorted in duplicate or triplicate and processed as previously described (103) with minor modifications (101). Briefly, P14 cells were washed with cold PBS, resuspended in 50 μl of cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630), and centrifuged (750g, 10 min, 4°C) to remove lysates. Nuclei were immediately resuspended in 25 μl of the transposition reaction mix (12.5 μl of 2x TD Buffer (Illumina), 1.25 μl of Tn5 Transposases, 11.25 μl of nuclease-free H2O) and incubated at 37°C for 45 min. Transposed DNA fragments were purified using the QIAGEN Reaction MiniElute Kit, barcoded with NEXTERA dual indexes (Illumina), and PCR amplified with NEBNext High Fidelity 2x PCR Master Mix (New England Biolabs). Following purification with the PCR Purification Kit (QIAGEN), fragment sizes were confirmed using the 2200 TapeStation and High Sensitivity D1000 ScreenTapes (Agilent). ATAC-seq libraries were quantified, pooled, and sequenced as described above for RNA-seq. Alignment to the mm39 genome was performed using bwa-mem, and peak calling was performed using MACS2. Differential peak analysis was performed using limma-voom, and motif enrichment was performed using HOMER.
ADCC coculture
TET2-KI T cells were rested overnight in hR10 media, while donor-matched NK cells were isolated using an NK cell isolation kit (Miltenyi Biotec) and cultured overnight in hR10 supplemented with IL-15 at 10 ng/ml (PeproTech). The following day, TET2 KI T cells were incubated with a cetuximab biosimilar (R&D Systems, no. MAB9577) at a concentration of 2000 ng/ml for 20 min. T cells were then cocultured at a 1:10 ratio with NK cells, with T cell numbers normalized to EGFR+ expression. After 16 hours, cocultures were harvested and stained with LIVE-DEAD Aqua, CD3, CD56, and a human IgG PE-conjugated secondary antibody. EGFR expression on TET2-KI T cells alone compared to NK coculture with or without cetuximab (gated on live, CD56− CD3+ EGFR+) was used to calculate percent EGFR decrease as a readout for ADCC.
Mouse xenograft studies
Male NOD/SCID/IL-2Rγ-null (NSG) mice, aged 7 weeks, were used for xenograft studies. Mice received an intravenous injection of 3 × 105 NALM-6 tumor cells expressing a CBG luciferase reporter suspended in 200 μl of PBS. Tumor engraftment was confirmed on day 6 via intraperitoneal injection of IVISbrite D-Luciferin Potassium Salt Bioluminescent Substrate (XenoLight, PerkinElmer), followed by bioluminescent imaging (BLI) using the IVIS Lumina III In Vivo Imaging System (PerkinElmer). On day 7, mice were administered with 5 × 105 control T cells, experimental CAR T cells, or PBS alone. Biweekly tumor imaging was conducted using the IVIS Lumina system after IP luciferin injection to monitor tumor growth or reduction. Regions of interest (ROIs) were delineated around mice for the calculation of bioluminescent tumor burden. Peripheral blood samples were collected via cheek bleeding at the peak of CAR T cell expansion and lysed with ACK Lysing Buffer (Gibco) to obtain T cells for immunophenotyping via flow cytometry. Absolute cell counts were determined using 123count eBeads Counting Beads (Invitrogen). Kaplan-Meier survival curves were generated on the basis of endpoint survival data.
Statistical analyses
Summary data are presented as means ± SEM, as indicated in the figure legends, alongside corresponding P values. Pairwise sample comparisons were evaluated using a paired t test. For multiple pairwise comparisons, multiple paired t tests with Holm-Šídák correction were used. One-way analysis of variance (ANOVA) analysis was used for comparisons involving multiple groups, initially assessing differences in mean values with a global omnibus F-test, followed by post hoc analysis for multiple comparisons if the initial test yielded significance (P < 0.05). Mouse survival was analyzed using the Mantel-Cox log-rank test. Number of donors, animals and experiments are indicated in the figure legends. All statistical analyses were conducted using Prism 9 or 10 (GraphPad Software), and significance was defined as P < 0.05.
Acknowledgments
We are grateful to the Human Immunology Core at the University of Pennsylvania (RRID SCR_022380) and the Hospital of the University of Pennsylvania Apheresis Unit for provision of peripheral blood mononuclear cells. We acknowledge the Stem Cell and Xenograft Core at the University of Pennsylvania (RRID SCR_010035) for husbandry services and support with in vivo mouse studies, the Cell and Animal Radiation Core at the University of Pennsylvania (RRID SCR_022377) for access to the xRad irradiator, and the Penn Cytomics and Cell Sorting Resource Laboratory (RRID SCR_022376). We thank F. Zhang from the Translational and Correlative Sciences Laboratory (TCSL) for helpful advice regarding gene set enrichment analysis and acknowledge the contributions of the TCSL and the Product Development Laboratory (PDL) from the University of Pennsylvania Center for Cellular Immunotherapies. Special thanks to M. Davis (Director of PDL) and K. Alexander from the University of Pennsylvania Epigenetics Institute for invaluable discussions. We also extend our gratitude to C. June (University of Pennsylvania) for insightful advice and input on translational aspects of TET2 modulation in T cell therapy. Schematic illustrations were created with BioRender.
Funding: This study was supported by grants T32 AI007632 (awarded to A.J.D.), F31 CA274961 (to C.R.H.), and EEC1648035 from the National Science Foundation Engineering Research Center for Cell Manufacturing Technologies (to B.L.L. and J.A.F.). Additional support came from the Bob Levis Funding Group (to B.L.L. and J.A.F.), R21 AI144732 and an Emerson Collective grant (to M.S.J.), an Alliance for Cancer Gene Therapy Investigator Award in Cell and Gene Therapy for Cancer (to J.A.F.), an American Society of Hematology Scholar award (to S.A.C.) and the Parker Institute for Cancer Immunotherapy (to E.J.W. and E.W.W.). Additional support from National Institutes of Health (NIH) grants came from U54 CA244711 (with a bench-to-bedside supplement) and P01 CA214278 (to J.A.F.); AI155577, AI115712, AI117950, AI108545, AI082630, and CA210944 (to E.J.W.); and 1RO1CA241762-01 (to J.J.M. and F.D.B.). Additional funding and support came from the Parker Institute for Cancer Immunotherapy (to E.J.W. and E.W.W.). The study also received funding from U01 AG066100 via the Samuel Waxman Cancer Research Foundation (to J.A.F.), along with support from an ACC P30 Core Grant P30 CA016520 (to J.A.F.).
Author contributions: Conceptualization: M.S.J., J.R.G., J.J.M., Z.C., F.H., S.A.C., J.A.F., A.J.D., B.L.L., M.H.P., G.T.R., V.W., S.A.G., E.J.W., R.M.Y., A.E.B., and F.D.B. Data curation: A.J.D. and G.M.C. Formal analysis: S.S.-M., J.A.F., A.J.D., G.T.R., G.M.C., K.A., J.K.E., H.H., S.D., A.E.B., and S.C. Funding acquisition: M.S.J., J.J.M., S.A.C., J.A.F., A.J.D., S.A.G., and E.J.W. Investigation: C.R.H., A.C., Z.C., I.-Y.J., A.J.D., N.G., A.T.D., N.V.F., C.H.H., R.B., G.T.R., K.D., W.K., O.M.K., S.D., A.E.B., and F.D.B. Methodology: M.S.J., J.R.G., J.J.M., Z.C., R.O., F.H., J.A.F., A.J.D., E.W.W., B.L.L., M.H.P., G.T.R., W.K., V.W., E.J.W., and A.E.B. Project administration: M.S.J., S.A.C., J.A.F., A.J.D., E.W.W., S.L.B., S.A.G., E.J.W., and A.E.B. Resources: M.S.J., J.R.G., Z.C., R.O., S.A.C., D.L.P., A.J.D., N.V.F., C.H.H., B.L.L., G.T.R., V.W., S.A.G., E.J.W., and S.L.M. Software: S.S.-M., G.M.C., K.A., J.K.E., and H.H. Supervision: M.S.J., J.A.F., E.W.W., S.L.B., S.A.G., E.J.W., A.E.B., and G.V. Validation: M.S.J., J.J.M., A.C., S.A.C., S.S.-M., J.A.F., A.J.D., E.W.W., R.B., G.T.R., J.K.E., E.J.W., and S.D. Visualization: S.S.-M., J.A.F., A.J.D., E.W.W., G.T.R., G.M.C., K.A., and A.E.B. Writing—original draft: J.A.F., A.J.D., and A.E.B. Writing—review and editing: C.R.H., M.S.J., J.R.G., J.J.M., F.H., I.-Y.J., D.L.P., J.A.F., A.J.D., N.V.F., B.L.L., G.M.C., J.K.J., E.J.W., and A.E.B.
Competing interests: W.K. is currently affiliated with Teva Pharmaceutical Industries. R.O. holds patents licensed to Novartis in Biomedical Research and has equity interests in Nucleus Biologics and Stoic Bio, while also serving as a scientific advisor to Nucleus Biologics. J.J.M. has received fees from IASO Biotherapeutics, Poseida Therapeutics, and Kite Pharma, unrelated to this work. J.J.M. also holds patents related to enhancing immune cell efficacy and predicting chimeric antigen responsiveness, issued to Novartis. S.L.M. has received clinical trial support and advisory roles from Novartis and Wugen and holds a pending patent with Novartis. S.A.G. and D.L.P. are involved with various pharmaceutical companies, receiving research funding, holding equity, and serving in advisory roles. D.L.P. also benefits from patents and royalties with Tmunity Therapeutics and Wiley and Sons Publishing. B.L.L. reports personal fees from Avectas, Akron Bio, Immusoft, In8bio, Immunell, Ori Biotech, Oxford Biomedica, and Vycellix. B.L.L. also maintains consultancy and advisory positions with Terumo and GSK and holds equity in Tmunity Therapeutics and Capstan Therapeutics. S.S.-M. and N.V.F. are engaged with Sana Biotechnology. N.V.F. additionally consults for Novartis and Syndax Pharmaceuticals and receives funding from Kite Pharma. S.A.G. has disclosed receiving support and serving in advisory capacities for multiple entities, including Novartis and Servier. M.H.P. is on the Board of Directors at Graphite Bio, serves on the Scientific Advisory Board of Allogene Therapeutics (where they receive cash compensation and hold equity), and advises Versant Ventures. M.H.P. also holds equity in CRISPR Therapeutics and is a founder and holds equity in Kamau Therapeutics. E.W.W. is a consultant to and holds equity in Lyell Immunopharma, consults for Umoja Immunopharma, and serves on the Scientific Advisory Board and holds equity in MedGene Therapeutics. E.J.W. is an advisor for Danger Bio, Marengo, Janssen, New Limit, Pluto Immunotherapeutics Related Sciences, Rubius Therapeutics, Santa Ana Bio, Synthekine, and Surface Oncology. E.J.W. is a founder of and holds stock in Surface Oncology, Danger Bio, and Arsenal Biosciences. J.A.F. receives research funding from Tceleron (formerly Tmunity Therapeutics) and Danaher Corporation, consults for Retro Biosciences, and is a member of the scientific advisory boards of Cartography Biosciences, Shennon Biotechnologies, and OverT Bio with cash compensation and equity. J.R.G. is a consultant for Arsenal Biosciences and Cellanome. J.K.J. has received payment for consulting services from UTC Therapeutics Inc. and BlueWhale Bio. R.M.Y. and J.A.F. hold patents and intellectual property in T cell–based cancer immunotherapy, from which they have received royalties. E.J.W. and M.S.J. have a patent pending related to regulators of T cell exhaustion. B.L.L. holds multiple patents related to cancer treatment and CAR T cell therapy, issued, licensed, and with royalties paid from the University of Pennsylvania. The remaining authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data are deposited in Gene Expression Omnibus (GEO) under accession number GSE261093 and integration site sequencing data in the Sequence Read Archive (SRA BioProject PRJNA510570), both hosted by the National Center for Biotechnology Information (NCBI).
Supplementary Materials
The PDF file includes:
Figs. S1 to S6
Tables S1 and S2, S6 and S7
Legends for tables S3 to S5
Other Supplementary Material for this manuscript includes the following:
Tables S3 to S5
REFERENCES AND NOTES
- 1.Blank C. U., Haining W. N., Held W., Hogan P. G., Kallies A., Lugli E., Lynn R. C., Philip M., Rao A., Restifo N. P., Schietinger A., Schumacher T. N., Schwartzberg P. L., Sharpe A. H., Speiser D. E., Wherry E. J., Youngblood B. A., Zehn D., Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albelda S. M., CAR T cell therapy for patients with solid tumours: Key lessons to learn and unlearn. Nat. Rev. Clin. Oncol. 21, 47–66 (2024). [DOI] [PubMed] [Google Scholar]
- 3.Lynn R. C., Weber E. W., Sotillo E., Gennert D., Xu P., Good Z., Anbunathan H., Lattin J., Jones R., Tieu V., Nagaraja S., Granja J., de Bourcy C. F. A., Majzner R., Satpathy A. T., Quake S. R., Monje M., Chang H. Y., Mackall C. L., c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long A. H., Haso W. M., Shern J. F., Wanhainen K. M., Murgai M., Ingaramo M., Smith J. P., Walker A. J., Kohler M. E., Venkateshwara V. R., Kaplan R. N., Patterson G. H., Fry T. J., Orentas R. J., Mackall C. L., 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., López-Moyado I. F., Seo H., Lio C.-W. J., Hempleman L. J., Sekiya T., Yoshimura A., Scott-Browne J. P., Rao A., NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung I. Y., Narayan V., McDonald S., Rech A. J., Bartoszek R., Hong G., Davis M. M., Xu J., Boesteanu A. C., Barber-Rotenberg J. S., Plesa G., Lacey S. F., Jadlowsky J. K., Siegel D. L., Hammill D. M., Cho-Park P. F., Berger S. L., Haas N. B., Fraietta J. A., BLIMP1 and NR4A3 transcription factors reciprocally regulate antitumor CAR T cell stemness and exhaustion. Sci. Transl. Med. 14, eabn7336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adusumilli P. S., Zauderer M. G., Riviere I., Solomon S. B., Rusch V. W., O’Cearbhaill R. E., Zhu A., Cheema W., Chintala N. K., Halton E., Pineda J., Perez-Johnston R., Tan K. S., Daly B., Araujo Filho J. A., Ngai D., McGee E., Vincent A., Diamonte C., Sauter J. L., Modi S., Sikder D., Senechal B., Wang X., Travis W. D., Gonen M., Rudin C. M., Brentjens R. J., Jones D. R., Sadelain M., A phase I trial of regional mesothelin-targeted CAR T-cell therapy in patients with malignant pleural disease, in combination with the anti–PD-1 agent pembrolizumab. Cancer Discov. 11, 2748–2763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauken K. E., Sammons M. A., Odorizzi P. M., Manne S., Godec J., Khan O., Drake A. M., Chen Z., Sen D. R., Kurachi M., Barnitz R. A., Bartman C., Bengsch B., Huang A. C., Schenkel J. M., Vahedi G., Haining W. N., Berger S. L., Wherry E. J., Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prinzing B., Zebley C. C., Petersen C. T., Fan Y., Anido A. A., Yi Z., Nguyen P., Houke H., Bell M., Haydar D., Brown C., Boi S. K., Alli S., Crawford J. C., Riberdy J. M., Park J. J., Zhou S., Velasquez M. P., DeRenzo C., Lazzarotto C. R., Tsai S. Q., Vogel P., Pruett-Miller S. M., Langfitt D. M., Gottschalk S., Youngblood B., Krenciute G., Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci. Transl. Med. 13, eabh0272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraietta J. A., Nobles C. L., Sammons M. A., Lundh S., Carty S. A., Reich T. J., Cogdill A. P., Morrissette J. J. D., Denizio J. E., Reddy S., Hwang Y., Gohil M., Kulikovskaya I., Nazimuddin F., Gupta M., Chen F., Everett J. K., Alexander K. A., Lin-Shiao E., Liu T. G., Young L. M., Ambrose R. A., Wang L., Levine B. W., Siegel D., Levine A. S., Lacey M. L., Carreno K. J., Lamontagne M. L., Fesnak J. H., Siegel L. C., Gill R. M., Clarke J. D. A., Melenhorst D. T., Porter D. L., June C. H., Melenhorst J. J., Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature, 558, 307–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen K. D., Helin K., Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan L., Shi Y. G., Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139, 1895–1902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain N., Zhao Z., Feucht J., Koche R., Iyer A., Dobrin A., Mansilla-Soto J., Yang J., Zhan Y., Lopez M., Gunset G., Sadelain M., TET2 guards against unchecked BATF3-induced CAR T cell expansion. Nature 615, 315–322 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carty S. A., Gohil M., Banks L. B., Cotton R. M., Johnson M. E., Stelekati E., Wells A. D., Wherry E. J., Koretzky G. A., Jordan M. S., The loss of TET2 promotes CD8+ T cell memory differentiation. J. Immunol. 200, 82–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irving M., Lanitis E., Migliorini D., Ivics Z., Guedan S., Choosing the right tool for genetic engineering: Clinical lessons from chimeric antigen receptor-T Cells. Hum. Gene Ther. 32, 1044–1058 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. K. MacLachlan, in Nonclinical Development of Novel Biologics, Biosimilars, Vaccines and Specialty Biologics, L. M. Plitnick, D. J. Herzyk, Eds. (Academic Press, 2013), pp. 259–285. [Google Scholar]
- 17.van der Windt G. J., O’Sullivan D., Everts B., Huang S. C., Buck M. D., Curtis J. D., Chang C. H., Smith A. M., Ai T., Faubert B., Jones R. G., Pearce E. J., Pearce E. L., CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. U.S.A. 110, 14336–14341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bengsch B., Johnson A. L., Kurachi M., Odorizzi P. M., Pauken K. E., Attanasio J., Stelekati E., McLane L. M., Paley M. A., Delgoffe G. M., Wherry E. J., Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 45, 358–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong W., Dimitri A., Wang W., Jung I. Y., Ott C. J., Fasolino M., Wang Y., Kulikovskaya I., Gupta M., Yoder T., DeNizio J. E., Everett J. K., Williams E. F., Xu J., Scholler J., Reich T. J., Bhoj V. G., Haines K. M., Maus M. V., Melenhorst J. J., Young R. M., Jadlowsky J. K., Marcucci K. T., Bradner J. E., Levine B. L., Porter D. L., Bushman F. D., Kohli R. M., June C. H., Davis M. M., Lacey S. F., Vahedi G., Fraietta J. A., BET bromodomain protein inhibition reverses chimeric antigen receptor extinction and reinvigorates exhausted T cells in chronic lymphocytic leukemia. J. Clin. Invest. 131, e145459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung I. Y., Bartoszek R. L., Rech A. J., Collins S. M., Ooi S. K., Williams E. F., Hopkins C. R., Narayan V., Haas N. B., Frey N. V., Hexner E. O., Siegel D. L., Plesa G., Porter D. L., Cantu A., Everett J. K., Guedan S., Berger S. L., Bushman F. D., Herbst F., Fraietta J. A., Type I interferon signaling via the EGR2 transcriptional regulator potentiates CAR T cell-intrinsic dysfunction. Cancer Discov. 13, 1636–1655 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escobar G., Mangani D., Anderson A. C., T cell factor 1: A master regulator of the T cell response in disease. Sci. Immunol., 5, eabb9726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J. E., Manne S., Ngiow S. F., Baxter A. E., Huang H., Freilich E., Clark M. L., Lee J. H., Chen Z., Khan O., Staupe R. P., Huang Y. J., Shi J., Giles J. R., Wherry E. J., In vitro modeling of CD8+ T cell exhaustion enables CRISPR screening to reveal a role for BHLHE40. Sci. Immunol. 8, eade3369 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Wherry E. J., Ahmed R., Memory CD8 T-cell differentiation during viral infection. J. Virol. 78, 5535–5545 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallimore A., Glithero A., Godkin A., Tissot A. C., Pluckthun A., Elliott T., Hengartner H., Zinkernagel R., Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187, 1383–1393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zajac A. J., Blattman J. N., Murali-Krishna K., Sourdive D. J., Suresh M., Altman J. D., Ahmed R., Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188, 2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moskophidis D., Lechner F., Pircher H., Zinkernagel R. M., Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature 362, 758–761 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Angelosanto J. M., Blackburn S. D., Crawford A., Wherry E. J., Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 86, 8161–8170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wherry E. J., Teichgraber V., Becker T. C., Masopust D., Kaech S. M., Antia R., von Andrian U. H., Ahmed R., Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4, 225–234 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Kaech S. M., Tan J. T., Wherry E. J., Konieczny B. T., Surh C. D., Ahmed R., Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Blackburn S. D., Shin H., Freeman G. J., Wherry E. J., Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc. Natl. Acad. Sci. U.S.A. 105, 15016–15021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller B. C., Sen D. R., Al Abosy R., Bi K., Virkud Y. V., LaFleur M. W., Yates K. B., Lako A., Felt K., Naik G. S., Manos M., Gjini E., Kuchroo J. R., Ishizuka J. J., Collier J. L., Griffin G. K., Maleri S., Comstock D. E., Weiss S. A., Brown F. D., Panda A., Zimmer M. D., Manguso R. T., Hodi F. S., Rodig S. J., Sharpe A. H., Haining W. N., Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paley M. A., Kroy D. C., Odorizzi P. M., Johnnidis J. B., Dolfi D. V., Barnett B. E., Bikoff E. K., Robertson E. J., Lauer G. M., Reiner S. L., Wherry E. J., Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science, 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Im S. J., Hashimoto M., Gerner M. Y., Lee J., Kissick H. T., Burger M. C., Shan Q., Hale J. S., Lee J., Nasti T. H., Sharpe A. H., Freeman G. J., Germain R. N., Nakaya H. I., Xue H. H., Ahmed R., Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utzschneider D. T., Charmoy M., Chennupati V., Pousse L., Ferreira D. P., Calderon-Copete S., Danilo M., Alfei F., Hofmann M., Wieland D., Pradervand S., Thimme R., Zehn D., Held W., T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity, 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Beltra J. C., Manne S., Abdel-Hakeem M. S., Kurachi M., Giles J. R., Chen Z., Casella V., Ngiow S. F., Khan O., Huang Y. J., Yan P., Nzingha K., Xu W., Amaravadi R. K., Xu X., Karakousis G. C., Mitchell T. C., Schuchter L. M., Huang A. C., Wherry E. J., Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqui I., Schaeuble K., Chennupati V., Fuertes Marraco S. A., Calderon-Copete S., Pais Ferreira D., Carmona S. J., Scarpellino L., Gfeller D., Pradervand S., Luther S. A., Speiser D. E., Held W., Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Khan O., Giles J. R., McDonald S., Manne S., Ngiow S. F., Patel K. P., Werner M. T., Huang A. C., Alexander K. A., Wu J. E., Attanasio J., Yan P., George S. M., Bengsch B., Staupe R. P., Donahue G., Xu W., Amaravadi R. K., Xu X., Karakousis G. C., Mitchell T. C., Schuchter L. M., Kaye J., Berger S. L., Wherry E. J., TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo H., Chen J., Gonzalez-Avalos E., Samaniego-Castruita D., Das A., Wang Y. H., Lopez-Moyado I. F., Georges R. O., Zhang W., Onodera A., Wu C. J., Lu L. F., Hogan P. G., Bhandoola A., Rao A., TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U.S.A. 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfei F., Kanev K., Hofmann M., Wu M., Ghoneim H. E., Roelli P., Utzschneider D. T., von Hoesslin M., Cullen J. G., Fan Y., Eisenberg V., Wohlleber D., Steiger K., Merkler D., Delorenzi M., Knolle P. A., Cohen C. J., Thimme R., Youngblood B., Zehn D., TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Scott A. C., Dundar F., Zumbo P., Chandran S. S., Klebanoff C. A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., Zamarin D., Walther T., Snyder A., Femia M. R., Comen E. A., Wen H. Y., Hellmann M. D., Anandasabapathy N., Liu Y., Altorki N. K., Lauer P., Levy O., Glickman M. S., Kaye J., Betel D., Philip M., Schietinger A., TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao C., Sun H.-W., Lacey N. E., Ji Y., Moseman E. A., Shih H. Y., Heuston E. F., Kirby M., Anderson S., Cheng J., Khan O., Handon R., Reilley J., Fioravanti J., Hu J., Gossa S., Wherry E. J., Gattinoni L., McGavern D. B., O’Shea J. J., Schwartzberg P. L., Wu T., Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., He Q., Shen H., Xia A., Tian W., Yu W., Sun B., TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J. Hepatol. 71, 731–741 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Hudson W. H., Gensheimer J., Hashimoto M., Wieland A., Valanparambil R. M., Li P., Lin J. X., Konieczny B. T., Im S. J., Freeman G. J., Leonard W. J., Kissick H. T., Ahmed R., Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zander R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., Cui W., CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51, 1028–1042.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Zander R. A., Wu X., Schauder D. M., Kasmani M. Y., Shen J., Zheng S., Burns R., Taparowsky E. J., Cui W., BATF regulates progenitor to cytolytic effector CD8+ T cell transition during chronic viral infection. Nat. Immunol., 22, 996–1007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rome K. S., Stein S. J., Kurachi M., Petrovic J., Schwartz G. W., Mack E. A., Uljon S., Wu W. W., DeHart A. G., McClory S. E., Xu L., Gimotty P. A., Blacklow S. C., Faryabi R. B., Wherry E. J., Jordan M. S., Pear W. S., Trib1 regulates T cell differentiation during chronic infection by restraining the effector program. J. Exp. Med. 217, e20190888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClory S. E., Bardhan O., Rome K. S., Giles J. R., Baxter A. E., Xu L., Gimotty P. A., Faryabi R. B., Wherry E. J., Pear W. S., Jordan M. S., The pseudokinase Trib1 regulates the transition of exhausted T cells to a KLR+ CD8+ effector state, and its deletion improves checkpoint blockade. Cell Rep. 42, 112905 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez C. X., Amezquita R. A., Guan T., Marshall H. D., Joshi N. S., Kleinstein S. H., Kaech S. M., The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan T., Dominguez C. X., Amezquita R. A., Laidlaw B. J., Cheng J., Henao-Mejia J., Williams A., Flavell R. A., Lu J., Kaech S. M., ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J. Exp. Med. 215, 1153–1168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C. Y., Best J. A., Knell J., Yang E., Sheridan A. D., Jesionek A. K., Li H. S., Rivera R. R., Lind K. C., D’Cruz L. M., Watowich S. S., Murre C., Goldrath A. W., The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herndler-Brandstetter D., Ishigame H., Shinnakasu R., Plajer V., Stecher C., Zhao J., Lietzenmayer M., Kroehling L., Takumi A., Kometani K., Inoue T., Kluger Y., Kaech S. M., Kurosaki T., Okada T., Flavell R. A., KLRG1+ Effector CD8+ T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity 48, 716–729.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renkema K. R., Huggins M. A., Borges da Silva H., Knutson T. P., Henzler C. M., Hamilton S. E., KLRG1+ memory CD8 T cells combine properties of short-lived effectors and long-lived memory. J. Immunol. 205, 1059–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruz-Guilloty F., Pipkin M. E., Djuretic I. M., Levanon D., Lotem J., Lichtenheld M. G., Groner Y., Rao A., Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206, 51–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Begona Ruiz M., Prado A., Goni F. M., Alonso A., An assessment of the biochemical applications of the non-ionic surfactant Hecameg. Biochim. Biophys. Acta 1193, 301–306 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Shin H., Blackburn S. D., Intlekofer A. M., Kao C., Angelosanto J. M., Reiner S. L., Wherry E. J., A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Q., Cai D., Liu D., Zhao X., Li R., Xu W., Xie B., Gou M., Wei K., Li Y., Huang J., Chi X., Wei P., Hao J., Guo X., Pan B., Fu Y., Ni L., Dong C., BCL6 promotes a stem-like CD8+ T cell program in cancer via antagonizing BLIMP1. Sci. Immunol. 8, eadh1306 (2023). [DOI] [PubMed] [Google Scholar]
- 57.Crotty S., Johnston R. J., Schoenberger S. P., Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11, 114–120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turman M. A., Yabe T., McSherry C., Bach F. H., Houchins J. P., Characterization of a novel gene (NKG7) on human chromosome 19 that is expressed in natural killer cells and T cells. Hum. Immunol. 36, 34–40 (1993). [DOI] [PubMed] [Google Scholar]
- 59.Li X. Y., Corvino D., Nowlan B., Aguilera A. R., Ng S. S., Braun M., Cillo A. R., Bald T., Smyth M. J., Engwerda C. R., NKG7 is required for optimal antitumor T-cell immunity. Cancer Immunol. Res. 10, 154–161 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Sarkar S., Kalia V., Haining W. N., Konieczny B. T., Subramaniam S., Ahmed R., Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205, 625–640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giles J. R., Ngiow S. F., Manne S., Baxter A. E., Khan O., Wang P., Staupe R., Abdel-Hakeem M. S., Huang H., Mathew D., Painter M. M., Wu J. E., Huang Y. J., Goel R. R., Yan P. K., Karakousis G. C., Xu X., Mitchell T. C., Huang A. C., Wherry E. J., Shared and distinct biological circuits in effector, memory and exhausted CD8+ T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat. Immunol. 23, 1600–1613 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daniel B., Yost K. E., Hsiung S., Sandor K., Xia Y., Qi Y., Hiam-Galvez K. J., Black M., Raposo C. J., Shi Q., Meier S. L., Belk J. A., Giles J. R., Wherry E. J., Chang H. Y., Egawa T., Satpathy A. T., Divergent clonal differentiation trajectories of T cell exhaustion. Nat. Immunol. 23, 1614–1627 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Good C. R., Aznar M. A., Kuramitsu S., Samareh P., Agarwal S., Donahue G., Ishiyama K., Wellhausen N., Rennels A. K., Ma Y., Tian L., Guedan S., Alexander K. A., Zhang Z., Rommel P. C., Singh N., Glastad K. M., Richardson M. W., Watanabe K., Tanyi J. L., O’Hara M. H., Ruella M., Lacey S. F., Moon E. K., Schuster S. J., Albelda S. M., Lanier L. L., Young R. M., Berger S. L., June C. H., An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 184, 6081–6100.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasmani M. Y., Zander R., Chung H. K., Chen Y., Khatun A., Damo M., Topchyan P., Johnson K. E., Levashova D., Burns R., Lorenz U. M., Tarakanova V. L., Joshi N. S., Kaech S. M., Cui W., Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection. J. Exp. Med. 220, e20220679 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Z., Ji Z., Ngiow S. F., Manne S., Cai Z., Huang A. C., Johnson J., Staupe R. P., Bengsch B., Xu C., Yu S., Kurachi M., Herati R. S., Vella L. A., Baxter A. E., Wu J. E., Khan O., Beltra J. C., Giles J. R., Stelekati E., McLane L. M., Lau C. W., Yang X., Berger S. L., Vahedi G., Ji H., Wherry E. J., TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51, 840–855.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giles J. R., Manne S., Freilich E., Oldridge D. A., Baxter A. E., George S., Chen Z., Huang H., Chilukuri L., Carberry M., Giles L., Weng N. P., Young R. M., June C. H., Schuchter L. M., Amaravadi R. K., Xu X., Karakousis G. C., Mitchell T. C., Huang A. C., Shi J., Wherry E. J., Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 55, 557–574.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S. J. C., Hamieh M., Cunanan K. M., Odak A., Gönen M., Sadelain M., Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collins S. M., Alexander K. A., Lundh S., Dimitri A. J., Zhang Z., Good C. R., Fraietta J. A., Berger S. L., TOX2 coordinates with TET2 to positively regulate central memory differentiation in human CAR T cells. Sci. Adv. 9, eadh2605 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saleh R., Sasidharan Nair V., Toor S. M., Taha R. Z., Murshed K., Al-Dhaheri M., Khawar M., Petkar M. A., Abu Nada M., Al-Ejeh F., Elkord E., Differential gene expression of tumor-infiltrating CD8+ T cells in advanced versus early-stage colorectal cancer and identification of a gene signature of poor prognosis. J. Immunother. Cancer 8, e001294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng W., Wei J., Zebley C. C., Jones L. L., Dhungana Y., Wang Y. D., Mavuluri J., Long L., Fan Y., Youngblood B., Chi H., Geiger T. L., Regnase-1 suppresses TCF-1+ precursor exhausted T-cell formation to limit CAR-T-cell responses against ALL. Blood 138, 122–135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zangari B., Tsuji T., Matsuzaki J., Mohammadpour H., Eppolito C., Battaglia S., Ito F., Chodon T., Koya R., Robert McGray A. J., Odunsi K., Tcf-1 protects anti-tumor TCR-engineered CD8+ T-cells from GzmB mediated self-destruction. Cancer Immunol. Immunother. 71, 2881–2898 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsui C., Kretschmer L., Rapelius S., Gabriel S. S., Chisanga D., Knopper K., Utzschneider D. T., Nussing S., Liao Y., Mason T., Torres S. V., Wilcox S. A., Kanev K., Jarosch S., Leube J., Nutt S. L., Zehn D., Parish I. A., Kastenmuller W., Shi W., Buchholz V. R., Kallies A., MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature 609, 354–360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopez-Cantillo G., Uruena C., Camacho B. A., Ramirez-Segura C., CAR-T cell performance: How to improve their persistence? Front. Immunol. 13, 878209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buck M. D., O’Sullivan D., Klein Geltink R. I., Curtis J. D., Chang C.-H. C., Sanin D. E., Qiu J., Kretz O., Braas D., van der Windt G. J., Chen Q., Huang S. C., O’Neill C. M., Edelson B. T., Pearce E. J., Sesaki H., Huber T. B., Rambold A. S., Pearce E. L., Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gattinoni L., Lugli E., Ji Y., Pos Z., Paulos C. M., Quigley M. F., Almeida J. R., Gostick E., Yu Z., Carpenito C., Wang E., Douek D. C., Price D. A., June C. H., Marincola F. M., Roederer M., Restifo N. P., A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Popplewell L. L., Wagner J. R., Naranjo A., Blanchard M. S., Mott M. R., Norris A. P., Wong C. W., Urak R. Z., Chang W. C., Khaled S. K., Siddiqi T., Budde L. E., Xu J., Chang B., Gidwaney N., Thomas S. H., Cooper L. J., Riddell S. R., Brown C. E., Jensen M. C., Forman S. J., Phase 1 studies of central memory-derived CD19 CAR T-cell therapy following autologous HSCT in patients with B-cell NHL. Blood 127, 2980–2990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biasco L., Izotova N., Rivat C., Ghorashian S., Richardson R., Guvenel A., Hough R., Wynn R., Popova B., Lopes A., Pule M., Thrasher A. J., Amrolia P. J., Clonal expansion of T memory stem cells determines early anti-leukemic responses and long-term CAR T cell persistence in patients. Nat. Cancer 2, 629–642 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scharping N. E., Menk A. V., Moreci R. S., Whetstone R. D., Dadey R. E., Watkins S. C., Ferris R. L., Delgoffe G. M., The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X., Yang J., Shi D., Cao Z., TET2 suppresses nasopharyngeal carcinoma progression by inhibiting glycolysis metabolism. Cancer Cell Int. 20, 363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tengesdal I. W., Mills T. S., Elder A., Patel S. B., May M., Schleicher W., Marchetti C., Lyons T., Dinarello C., Pietras E. M., Metabolic alterations associated with loss of TET2 potentiates inflammation in myeloid cells driving breast cancer progression. Blood, 142, 2536 (2023). [Google Scholar]
- 81.Yang Y., Cavalier M., Suris A., Chen K., An C., Fan J., Rivera L., Fang S., Guo L., Zhou Y., Huang Y., Enhanced glucose metabolism in Tet-deficient mouse embryonic stem cells. Front. Epigenet. Epigenom. 2, 1245823 (2024). [Google Scholar]
- 82.O’Sullivan D., van der Windt G. J., Huang S. C.-C., Curtis J. D., Chang C.-H., Buck M. D., Qiu J., Smith A. M., Lam W. Y., DiPlato L. M., Hsu F.-F., Birnbaum M. J., Pearce E. J., Pearce E. L., Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawalekar O. U., O’Connor R. S., Fraietta J. A., Guo L., McGettigan S. E., Posey A. D. Jr., Patel P. R., Guedan S., Scholler J., Keith B., Snyder N. W., Blair I. A., Milone M. C., June C. H., Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Marin T. L., Gongol B., Zhang F., Martin M., Johnson D. A., Xiao H., Wang Y., Subramaniam S., Chien S., Shyy J. Y., AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci. Signal. 10, eaaf7478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ichiyama K., Chen T., Wang X., Yan X., Kim B. S., Tanaka S., Ndiaye-Lobry D., Deng Y., Zou Y., Zheng P., Tian Q., Aifantis I., Wei L., Dong C., The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 42, 613–626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lio C. W., Zhang J., Gonzalez-Avalos E., Hogan P. G., Chang X., Rao A., Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 5, e18290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu T., Ji Y., Moseman E. A., Xu H. C., Manglani M., Kirby M., Anderson S. M., Handon R., Kenyon E., Elkahloun A., Wu W., Lang P. A., Gattinoni L., McGavern D. B., Schwartzberg P. L., The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1, eaai8593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y., Hu J., Li Y., Xiao M., Wang H., Tian Q., Li Z., Tang J., Hu L., Tan Y., Zhou X., He R., Wu Y., Ye L., Yin Z., Huang Q., Xu L., The transcription factor TCF1 preserves the effector function of exhausted CD8 T cells during chronic viral infection. Front. Immunol. 10, 169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mustjoki S., Young N. S., Somatic mutations in “Benign” disease. N. Engl. J. Med. 384, 2039–2052 (2021). [DOI] [PubMed] [Google Scholar]
- 90.Stadtmauer E. A., Fraietta J. A., Davis M. M., Cohen A. D., Weber K. L., Lancaster E., Mangan P. A., Kulikovskaya I., Gupta M., Chen F., Tian L., Gonzalez V. E., Xu J., Jung I. Y., Melenhorst J. J., Plesa G., Shea J., Matlawski T., Cervini A., Gaymon A. L., Desjardins S., Lamontagne A., Salas-Mckee J., Fesnak A., Siegel D. L., Levine B. L., Jadlowsky J. K., Young R. M., Chew A., Hwang W. T., Hexner E. O., Carreno B. M., Nobles C. L., Bushman F. D., Parker K. R., Qi Y., Satpathy A. T., Chang H. Y., Zhao Y., Lacey S. F., June C. H., CRISPR-engineered T cells in patients with refractory cancer. Science 367, eaba7365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Couronne L., Bastard C., Bernard O. A., TET2 and DNMT3A mutations in human T-cell lymphoma. N. Engl. J. Med. 366, 95–96 (2012). [DOI] [PubMed] [Google Scholar]
- 92.Zhang X., Su J., Jeong M., Ko M., Huang Y., Park H. J., Guzman A., Lei Y., Huang Y. H., Rao A., Li W., Goodell M. A., DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 48, 1014–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghilardi G., Fraietta J. A., Gerson J. N., Van Deerlin V. M., Morrissette J. J. D., Caponetti G. C., Paruzzo L., Harris J. C., Chong E. A., Susanibar Adaniya S. P., Svoboda J., Nasta S. D., Ugwuanyi O. H., Landsburg D. J., Fardella E., Waxman A. J., Chong E. R., Patel V., Pajarillo R., Kulikovskaya I., Lieberman D. B., Cohen A. D., Levine B. L., Stadtmauer E. A., Frey N. V., Vogl D. T., Hexner E. O., Barta S. K., Porter D. L., Garfall A. L., Schuster S. J., June C. H., Ruella M., T cell lymphoma and secondary primary malignancy risk after commercial CAR T cell therapy. Nat. Med. 30, 984–989 (2024). [DOI] [PubMed] [Google Scholar]
- 94.Garcia J., Daniels J., Lee Y., Zhu I., Cheng K., Liu Q., Goodman D., Burnett C., Law C., Thienpont C., Alavi J., Azimi C., Montgomery G., Roybal K. T., Choi J., Naturally occurring T cell mutations enhance engineered T cell therapies. Nature, 626, 626–634 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nobles C. L., Sherrill-Mix S., Everett J. K., Reddy S., Fraietta J. A., Porter D. L., Frey N., Gill S. I., Grupp S. A., Maude S. L., Siegel D. L., Levine B. L., June C. H., Lacey S. F., Melenhorst J. J., Bushman F. D., CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J. Clin. Invest. 130, 673–685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sherman E., Nobles C., Berry C. C., Six E., Wu Y., Dryga A., Malani N., Male F., Reddy S., Bailey A., Bittinger K., Everett J. K., Caccavelli L., Drake M. J., Bates P., Hacein-Bey-Abina S., Cavazzana M., Bushman F. D., INSPIIRED: A pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol. Ther. Methods Clin. Dev. 4, 39–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pircher H., Baenziger J., Schilham M., Sado T., Kamisaku H., Hengartner H., Zinkernagel R. M., Characterization of virus-specific cytotoxic T cell clones from allogeneic bone marrow chimeras. Eur. J. Immunol. 17, 159–166 (1987). [DOI] [PubMed] [Google Scholar]
- 98.Pircher H., Michalopoulos E. E., Iwamoto A., Ohashi P. S., Baenziger J., Hengartner H., Zinkernagel R. M., Mak T. W., Molecular analysis of the antigen receptor of virus-specific cytotoxic T cells and identification of a new V alpha family. Eur. J. Immunol. 17, 1843–1846 (1987). [DOI] [PubMed] [Google Scholar]
- 99.Odorizzi P. M., Pauken K. E., Paley M. A., Sharpe A., Wherry E. J., Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212, 1125–1137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kurachi M., Kurachi J., Chen Z., Johnson J., Khan O., Bengsch B., Stelekati E., Attanasio J., McLane L. M., Tomura M., Ueha S., Wherry E. J., Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function. Nat. Protoc. 12, 1980–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baxter A. E., Huang H., Giles J. R., Chen Z., Wu J. E., Drury S., Dalton K., Park S. L., Torres L., Simone B. W., Klapholz M., Ngiow S. F., Freilich E., Manne S., Alcalde V., Ekshyyan V., Berger S. L., Shi J., Jordan M. S., Wherry E. J., The SWI/SNF chromatin remodeling complexes BAF and PBAF differentially regulate epigenetic transitions in exhausted CD8+ T cells. Immunity 56, 1320–1340.e10 (2023). [DOI] [PubMed] [Google Scholar]
- 102.Lever J., Krzywinski M., Altman N., Principal component analysis. Nat. Methods 14, 641–642 (2017). [Google Scholar]
- 103.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2, S6 and S7
Legends for tables S3 to S5
Tables S3 to S5