Abstract
Heterozygosity for inverted formin-2 (INF2) mutations causes focal segmental glomerulosclerosis (FSGS) with or without Charcot-Marie-Tooth disease. A key question is whether the disease is caused by gain-of-function effects on INF2 or loss of function (haploinsufficiency). Despite established roles in multiple cellular processes, neither INF2 knockout mice nor mice with a disease-associated point mutation display an evident kidney or neurologic phenotype. Here, we compared responses to puromycin aminonucleoside (PAN)–induced kidney injury between INF2 R218Q and INF2 knockout mice. R218Q INF2 mice are susceptible to glomerular disease, in contrast to INF2 knockout mice. Colocalization, coimmunoprecipitation analyses, and cellular actin measurements showed that INF2 R218Q confers a gain-of-function effect on the actin cytoskeleton. RNA expression analysis showed that adhesion and mitochondria-related pathways were enriched in the PAN-treated R218Q mice. Both podocytes from INF2 R218Q mice and human kidney organoids with an INF2 mutation (S186P) recapitulate adhesion and mitochondrial phenotypes. Thus, gain-of-function mechanisms drive INF2-related FSGS and explain this disease’s autosomal dominant inheritance.
Mutations in the actin regulatory molecule INF2 cause kidney disease through a dominant, gain-of-function mechanism.
INTRODUCTION
Chronic kidney disease is a serious global health burden, affecting about 10% of the world’s population. A large fraction of cases originate in the kidney’s glomerulus due to the dysfunction of a specific cell type: the podocyte (1–4). Glomerular epithelial cells, or podocytes, are highly specialized cells that wrap around the outer surface of glomerular capillaries (1, 2). They exhibit a unique morphology, wherein the cell body gives rise to long extensions that branch into primary processes and secondary foot processes. These foot processes interdigitate with the foot processes of neighboring podocytes to form filtration slit diaphragms. This complex morphology depends on the underlying actin cytoskeletal arrangement, which, if impaired, can lead to podocyte injury and kidney disease (3, 4).
Focal segmental glomerulosclerosis (FSGS) is a histological pattern of injury defined by the presence of scarring (sclerosis) in some parts (segmental) of certain glomeruli (focal) within the kidney (5). This pathology occurs when podocytes are dedifferentiat ed or reduced in number due to injury (5, 6). To date, mutations in more than 50 genes have been described as the causative events affecting the podocytes’ structure directly or indirectly (7, 8). Of these, mutations in an actin regulatory gene, inverted formin 2 (INF2), are relatively common, accounting for 17% of familial FSGS and 1% of sporadic cases (9–12). Over 60 different disease-associated INF2 mutations have been identified (12). These mutations lead to a kidney phenotype characterized by proteinuria, progressive kidney dysfunction, and FSGS with or without Charcot-Marie-Tooth disease (11, 13).
INF2 is one of the 15-member formin family of proteins that are primarily involved in polymerizing monomeric “globular” actin (G-actin) into actin filaments (F-actin) (14, 15). The defining feature of these formins, including INF2, is a formin homology domain (FH2) responsible for actin nucleation and elongation. In addition, they contain other domains, such as formin homology 1 (FH1) and diaphanous autoregulatory domain (DAD), working in tandem with FH2 to promote actin assembly. The N-terminal region of INF2 contains a diaphanous inhibitory domain (DID) that can interact with the DAD in the C-terminal region of INF2 and enable the protein to fold to an autoinhibited state (12, 14, 16), which tightly regulates actin assembly. All identified FSGS-causing mutations in INF2 localize to within INF2-DID, leading to the release of the DAD and other domains from this inhibition (12).
Over the past several years, multiple studies have shown that INF2 regulates various processes in cells, such as mitochondrial calcium uptake, mitochondrial fission, cell spreading, vesicle trafficking, T cell polarization, and placental implantation, among other processes (12, 17–21). In addition, we have observed defects in lamellipodium in primary podocytes derived from a mouse model with a point mutant of INF2, known as R218Q knock-in (22). Despite these findings, there are still gaps in understanding how INF2 mutations lead to FSGS. Despite the large list of different disease-causing INF2 mutations, it is notable that none are clear loss-of-function alleles.
Our goal in this study was to identify specific underlying events related to the in vivo disease rather than broad INF2 function in cells. Therefore, to better define the mechanism(s) of INF2-related kidney disease, we interrogated R218Q INF2 point mutant (knock-in) and INF2 knockout mouse models in response to puromycin aminonucleoside (PAN) stress and compared their phenotypic responses. We report that, in contrast to the INF2 knockout mouse model, the R218Q knock-in mutant mouse model is susceptible to developing proteinuria and FSGS in response to PAN injury. Furthermore, the ability of R218Q INF2 to alter the localization of wild-type INF2 and induce defects in actin arrangements, podocyte adhesion, and mitochondria indicates that a gain-of-function mechanism drives the development of disease. These data help explain the autosomal dominant inheritance of this disease and have potential implications for treating INF2-associated disease and for understanding the potential role of INF2 as a drug target for both INF2-mediated and non–INF2-mediated disease.
RESULTS
INF2-mediated FSGS is a gain-of-function disease
Prior human genetic studies have described pathogenic mutations at multiple positions within the INF2 DID (12). All of these FSGS-causing INF2 mutations are found in the heterozygous state, consistent with the dominant inheritance of the disease, and all encode missense amino acid changes. None are predicted to cause total protein loss. Mice with complete knockout of INF2 are grossly normal and lack any apparent kidney disease phenotype (23). On the basis of these observations, we reasoned that the INF2-mediated FSGS disease likely occurs via pure gain-of-function effect of mutant INF2.
To examine this experimentally, we compared heterozygous INF2 R218Q knock-in (point mutant) mice and heterozygous INF2 knockout mice. We stressed these mice with PAN and compared the development of kidney disease as a function of genotype. We chose PAN because, among the various stressors we tested, it was the only one that selectively induced disease in knock-in models (fig. S1). When we examined the development of albuminuria (measured as urinary albumin to creatinine ratio), we observed albuminuria at day 3 in the heterozygous knock-in mouse model. The proteinuria peaked at day 11 post-PAN injury, after which levels of urine albumin declined but did not return to the pre-injury baseline level. In contrast, the heterozygous knockout mouse model did not develop any albuminuria within the indicated time frames post-PAN injury (Fig. 1, A and B).
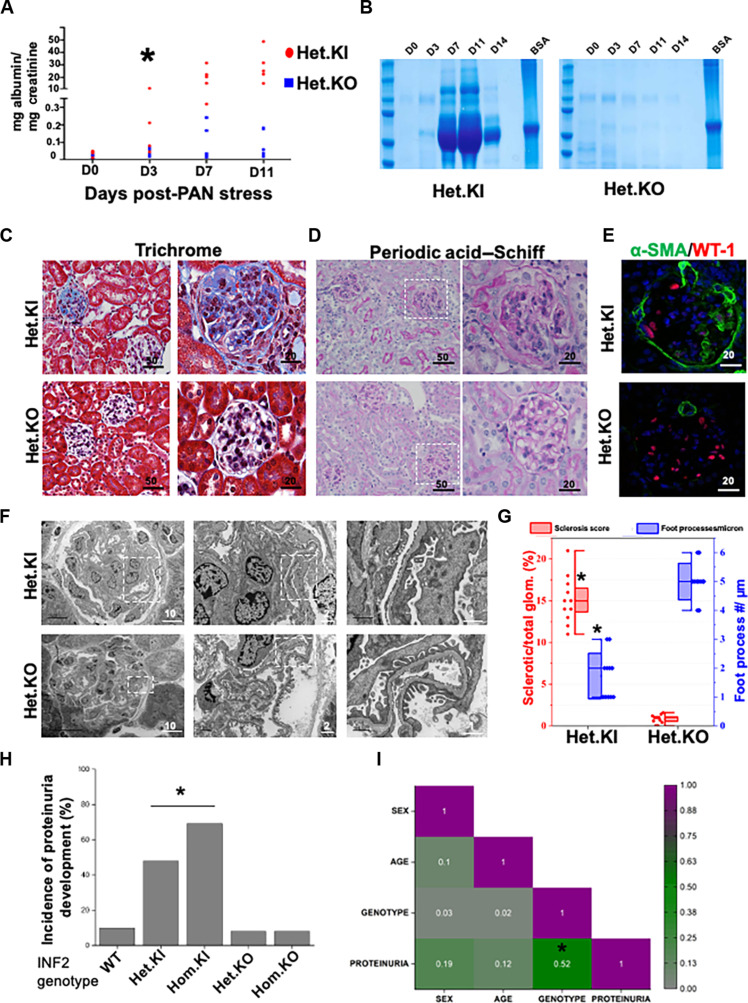
Fig. 1. INF2 R218Q knock-in mutant and INF2 knockout mouse models differ in disease development.
[(A) to (G)] Heterozygous INF2 R218Q knock-in and heterozygous INF2 knockout mouse models were stressed with puromycin aminonucleoside (PAN) and assessed for development of kidney disease. (A) Quantification of urinary albumin to creatinine ratio. Statistical significant differences were seen between knock-in versus knockout mice from day 3 (*P < 0.01; Student’s t test). (B) Urine-gel electrophoresis. Urine samples from different time points of a representative PAN-stressed heterozygous knock-in and heterozygous knockout mouse underwent electrophoresis in an SDS-PAGE gel. [(C) and (D)] Histology analysis. PAN-stressed mice were examined after eight weeks for histological changes. (C) Trichrome staining and (D) periodic acid–Schiff staining. Fibrotic lesions were noted in heterozygous knock-in but not in heterozygous knockout mice. Higher magnification images of the highlighted regions are shown in series. (E) Immunofluorescence analysis of WT-1 (podocyte marker) and α–smooth muscle actin (α-SMA; fibrosis marker). Increased smooth muscle actin expression was noted in PAN-stressed heterozygous knock-in mice but not in heterozygous knockout mice. (F) Ultrastructural analysis of podocytes. Higher magnification images of highlighted regions were shown in series. (G) Quantification of sclerosis and foot processes effacement (*P < 0.01; Student’s t test). (H) Disease development among various INF2 genotypes. Mice from all INF2 genotypes were PAN stressed and evaluated for proteinuria development. Incidence of proteinuria incidences was significantly higher in heterozygous and homozygous knock-in genotype mice (*P < 0.01; one-way ANOVA). (I) Correlogram representing the Pearson correlation coefficient matrix between various mice features. Colors indicate the values of the correlation coefficient, as indicated in the scale bar. A significant correlation was present between genotype and proteinuria but not with other mice features (*P < 0.05; r = 0.52). All scale bars are in micrometers.
We then assessed whether podocyte injury and related histological changes follow the pattern of albuminuria in the heterozygous knock-in and knockout mouse models. We observed a fragmented pattern for marker proteins nephrin (a podocyte slit diaphragm protein) and endomucin (an endothelial cell marker), reduced WT-1 positive cells (podocytes), and increased α–smooth muscle actin (fibrosis) expression in PAN-injured heterozygous knock-in mouse glomeruli. These alterations were not present in the glomeruli of the heterozygous knockout mouse model (Fig. 1C and fig. S1). To examine the disruption in the foot process/slit diaphragm and transition toward fibrosis, we assessed the kidney sections using periodic acid–Schiff and Masson’s trichrome staining and electron microscopy. We observed fibrotic lesions and foot process effacement in PAN-stressed heterozygous R218Q knock-in mice, characteristic of an FSGS pattern of injury (Fig. 1, D to G). In contrast, the PAN-stressed heterozygous INF2 knockout mouse did not develop these aberrations in marker protein distribution and glomerular histology (fig. S2 and Fig. 1, C to G).
Next, we tested whether disease development in heterozygous knock-in mice was driven solely by the mutant allele’s gain-of-function effects. For this purpose, we applied PAN stress to mice spanning a range of INF2 genotypes—wild-type, heterozygous, and homozygous forms of knock-in and knockout alleles, genders, and adult age groups. We observed a high incidence of proteinuria in both heterozygous and homozygous knock-in mice, whereas both the heterozygous and homozygous knockout mice had infrequent proteinuria, similar to wild-type mice (Fig. 1H). Pearson correlation coefficients revealed a positive correlation between disease development and mutant allele genotype but not with other gender- and age-related features of the mice (Fig. 1I). These observations indicate that INF2-mediated kidney disease occurs as a consequence of the gain-of-function effects of mutant INF2.
Mutation in INF2-DID confers gain-of-function effect by altering the localization of wild-type INF2 and INF2 activity
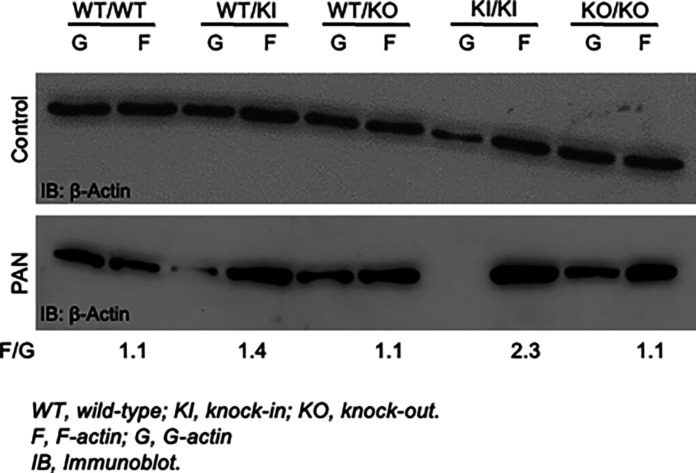
Since INF2 is a member of the formin family of proteins that are primarily involved in regulating actin and the actin cytoskeleton, we reasoned that a DID mutation–induced gain-of-function would likely manifest in alterations to the actin arrangement in cells. To assess this, we compared the F-actin/G-actin ratio in cells derived from mice of various genotypes in both untreated and PAN-treated podocytes. We observed that the knock-in mouse podocytes, particularly in homozygous conditions, exhibited a higher F-actin/G-actin ratio compared to wild-type and knockout podocytes (Fig. 2). This trend is further amplified with PAN treatment, causing a substantial change in both heterozygous and homozygous knock-in podocytes.
Fig. 2. R218Q pathogenic mutation alters the content of actin arrangement in podocytes.
Image shows an F-actin/G-actin assay. Control and PAN-stressed podocytes were examined for F-actin and G-actin content. Homozygous R218Q INF2 knock-in podocytes showed mild alteration in the basal condition in F-actin/G-actin distribution. In PAN-stressed conditions, both heterozygous and homozygous R218Q INF2 knock-in podocytes showed a prominent F-actin/G-actin distribution alteration. A representative immunoblot of three independent experiments was shown. The numerical value represents the mean F-actin/G-actin ratio for the PAN-stressed condition from three independent experiments. F/G, F-actin/G-actin.
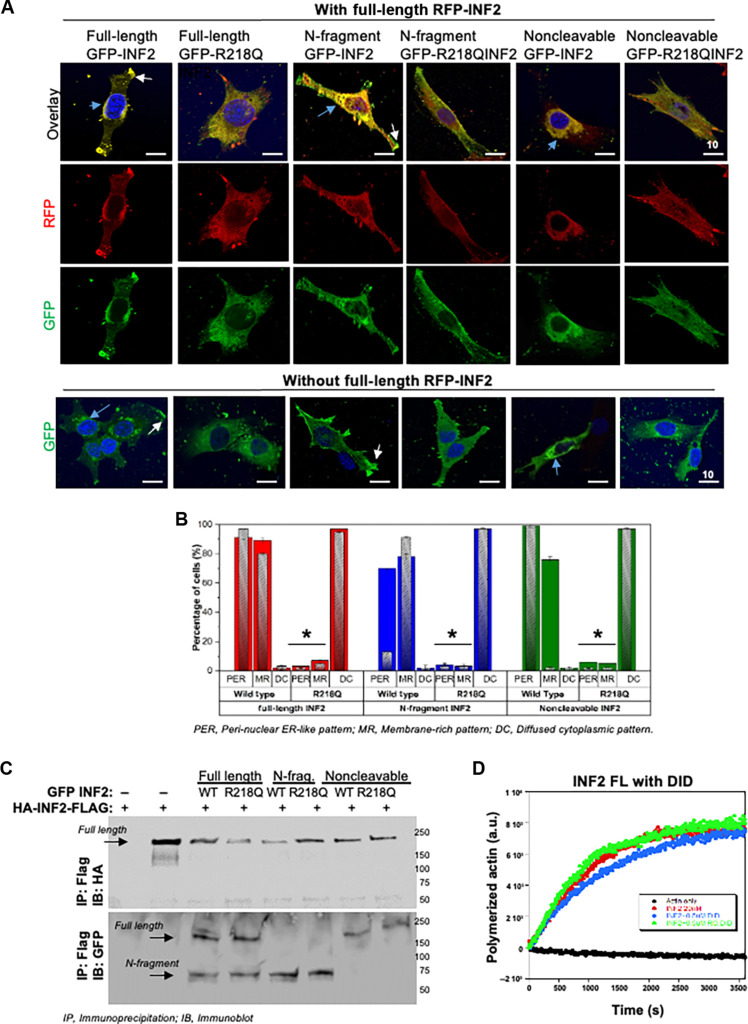
To determine how INF2-DID mutations lead to this aberrant actin distribution, we examined the localization (Fig. 3, A and B) and activity of mutant INF2 in podocytes (Fig. 3, C and D). In podocytes, INF2-DID protein is present in a full-length INF2-CAAX isoform and in an INF2 N-terminal cleavage product fragment (23). Therefore, we examined the ability of these forms of mutant INF2-DID to localize and interact with wild-type full-length INF2-CAAX in podocytes. We transiently overexpressed red fluorescent protein (RFP)–tagged (wild type) full-length INF2-CAAX, green fluorescent protein (GFP)–tagged (wild-type and R218Q mutant) N-terminal fragment, full-length INF2-CAAX, and noncleavable full-length INF2-CAAX in mouse INF2 KO podocytes. When GFP-tagged wild-type INF2 is present, the RFP-tagged wild-type full-length INF2-CAAX isoform was localized mainly at the endoplasmic reticulum (ER)–rich cell body and some cell boundary membrane regions (MR). In contrast, when GFP-tagged R218Q mutant INF2 isoforms are present, the RFP-tagged wild-type INF2 shifts to a diffuse cytoplasmic localization pattern (Fig. 3, A and B). We further probed for their interaction to confirm that these localization patterns are indeed due to the interaction between the wild-type and mutant forms of INF2 proteins. We coexpressed wild-type hemagglutinin (HA)–INF2–FLAG with GFP-tagged wild-type or mutant INF2 forms in 293T cells and examined their interactions through coimmunoprecipitation assays. We found that the N-fragment, full-length, and noncleavable forms of R218Q INF2 all interact with full-length wild-type INF2-CAAX, as is the case with the equivalent wild-type forms (Fig. 3C).
Fig. 3. R218Q pathogenic mutation leads to altered localization and activity of wild-type INF2.
[(A) and (B)] Localization analysis of wild-type INF2 in the presence of R218Q INF2. Wild-type and R218Q GFP-tagged INF2 forms were coexpressed with or without RFP-tagged wild-type full-length INF2. Wild-type full-length RFP-INF2 showed a peri-nuclear ER-like pattern (blue arrow) with some membrane regions (white arrow, membrane region localization). The coexistence of R218Q GFP INF2 forms (full-length, N-fragment, and noncleavable) alters this localization to a diffused cytoplasmic pattern. (A) Quantification of wild-type full-length RFP INF2 localization pattern in podocytes. Color bars correspond to the coexpression of GFP- and RFP-tagged INF2 conditions. Red bars, coexpression with full-length GFP-INF2; blue bars, coexpression with N-fragment GFP-INF2; green bars, coexpression with a noncleavable GFP-INF2. Inlet gray bars represent their respective controls (without RFP-tagged INF2 expression). The peri-nuclear ER-like and membrane-rich localization pattern of wild-type RFP-INF2 is altered to a diffused cytosolic pattern of localization by the R218Q GFP-INF2 presence (*P < 0.01; Student’s t test). (B) Representative cell images for wild-type full-length RFP-INF2 localization with various GFP-INF2 forms. Scale bar, 10 μm. (C) Interaction analysis of wild-type INF2 with R218Q INF2. Coimmunoprecipitation of wild-type HA-INF-FLAG with different GFP-tagged INF2 expression forms. GFP-tagged and HA-tagged INF2 forms were cotransfected in 293T cells. Cell lysates were pulled down using the FLAG tag and blotted for HA. HA immunoblot confirmed the immunoprecipitation of wild-type full-length INF2. GFP immunoblot showed an interaction of wild-type full-length INF2 with both wild-type and R218Q mutant forms of N-fragment, full-length, and noncleavable INF2. Each immunoblot is representative of three independent experiments with similar results. (D) Pyrene actin polymerization assays. Actin polymerizing activity of 20 nM full-length wild-type INF2 was assessed with 0.5 μM wild-type and R218Q DID regions, respectively. Neither the wild-type nor the R218Q-DID region had any effect in the actin polymerizing activity of full-length INF2.
We also tested whether this interaction was due to a DID-DAD interaction or dimerization between N-fragment and full-length INF2. To determine that, we examined the biochemical activity of full-length wild-type INF2 with the wild-type and R218Q mutant N-terminal region. Our results showed that neither the wild-type nor the mutant form of DID regions affected the biochemical activities of wild-type full-length INF2 (Fig. 3D), suggesting that the interaction is due to dimerization and the DAD activity of the wild-type full-length INF2 can remain functional postinteraction. Together, these results show that both the N-fragment and full-length forms of INF2 are able to interact with each other and that this interaction is not noticeably altered by disease-causing mutations. However, the presence of a pathogenic mutation markedly alters the localization of the wild-type INF2-CAAX, leading to INF2 actin polymerizing activity at abnormal sites.
RNA sequencing analysis identifies cellular processes that are altered in INF2-mediated FSGS
The gain-of-function effect of mutant INF2 underscores the sensitivity of podocytes to the development of functional alterations, leading to their structural and functional collapse. Therefore, we performed glomerular RNA sequencing (RNA-seq) transcriptome-based gene set enrichment analysis (GSEA) of PAN-stressed heterozygous knock-in and heterozygous knockout mouse models to identify the altered downstream processes. We conducted our study at an early postinjury time (day 3 post-PAN stress) to better focus on identifying direct changes due to mutant INF2 activity rather than more generalized effects reflecting advanced stages of injury. RNA-seq analysis of unstressed heterozygous knock-in and heterozygous knockout mouse glomeruli was also performed to elucidate the changes happening under the basal conditions in which podocyte morphology and glomerular function are grossly normal.
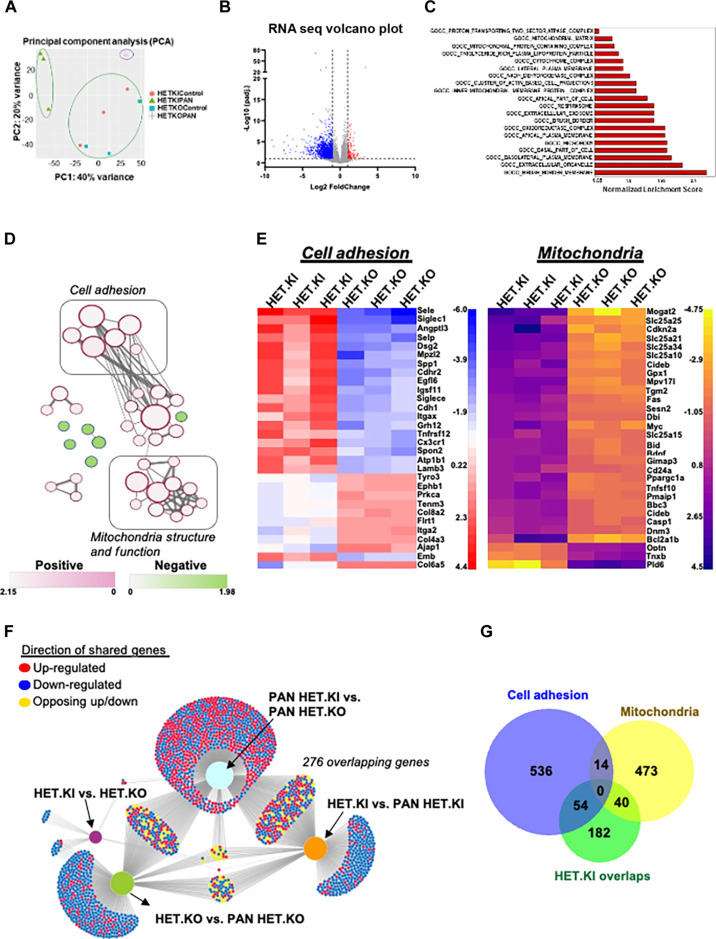
Principal components analysis of the total RNA-seq data of all groups displayed distinct clustering of transcriptomic signature by genotype and phenotype (Fig. 4A). The total number of differentially expressed genes reaching genome-wide statistical significance was ~30 between heterozygous knock-in and heterozygous knockout at the basal condition. In contrast, in the PAN-stressed condition, this number increased to ~1500. This notable difference in the differentially expressed gene count suggests that heterozygous knock-in and heterozygous knockout mouse models exhibit similar functional activity under basal conditions. However, with PAN stress, they differ substantially in their response, leading to an overt disease phenotype in heterozygous knock-in mice but not in heterozygous knockout mice. Therefore, to understand the specific signatures associated with disease phenotype development, we conducted a GSEA analysis of the PAN-stressed heterozygous knock-in and heterozygous knockout comparison (Fig. 4, B and C). The enrichment results included multiple actin arrangement–related cellular function pathways, such as the “cluster of actin-based cell projections” within the 20 most enriched pathways (Fig. 4C and table S1). Integration of these pathways as a network revealed the enrichment of related cellular function pathways as clusters of shared genes. We noted at least two distinct clusters of pathways that are broadly related to (i) actin and cell membrane arrangement (adhesion) and (ii) mitochondria structure and function (Fig. 4, D and E).
Fig. 4. RNA-sequencing analysis identifies pathogenic processes present in INF2-related FSGS.
Total RNA-seq of mouse glomeruli compared by INF2 genotype. (A) Principal components analysis plot of total RNA-seq results. [(B) to (E)] GSEA of differentially expressed genes of PAN-stressed heterozygous R218Q INF2 knock-in and heterozygous INF2 knockout mouse comparison. (B) Volcano plot of differentially expressed genes. (C) Top enrichment pathways (false discovery rate, q value < 0.05). The pathways were considered down-regulated or up-regulated based on the gene expression patterns of the pathway genes. (D) Enrichment network. Pathways were plotted as a network in the Cytoscape application (48). Color nodes indicate the up-regulated and down-regulated pathways (table S1). Connecting lines indicate the gene overlap between pathways. Nodes were manually laid out to form a clearer picture of gene overlaps between pathways. Node clusters were identified using an AutoAnnotate Cytoscape application. Individual node labels were removed for clarity and to emphasize clustering. Network with individual node labeling was provided in fig. S2. (E) The 30 most up-regulated and down-regulated differentially expressed genes of cell adhesion and mitochondria clusters identified through GSEA. (F) Network visualization of differentially expressed genes (DEGs) using the DiVenn application (49). DEGs from different comparison sets were mapped as a network to identify overlapping genes between them. A total of 276 genes were common between comparison sets that involved disease development (PAN-HET KI). (G) Venn diagram showing consensus between two methods of analysis: (i) gene enrichment analysis between PAN-stressed heterozygous knock-in versus heterozygous knockout and (ii) DEGs shared between comparison sets involving disease phenotype: HET-KI versus PAN-HET KI and PAN-HET KO versus PAN-HET KI. The 14 genes that are shared between cell adhesion and mitochondria gene ontology pathways are Atp6ap2, Atp6v1e1, Atp6v0d1, Tcirg1, Atp6v1a, Atp6v1b2, Nox4, Atp6v0d2, Atp6v1b1, Cyba, Atp6v0a4, Hspa1b, Hax1, and Lrrk2.
To further confirm that these changes are robust and meaningfully associated with disease, we wished to determine whether a similar conclusion is obtained with an alternate approach to RNA-seq analysis. To perform this analysis, we also included the differentially expressed genes from two other comparisons: (i) heterozygous knock-in versus PAN-stressed heterozygous knock-in and (ii) heterozygous knockout versus PAN-stressed heterozygous knockout, and collectively analyzed all the four comparison groups to identify the overlapping gene expression changes common to these different comparisons. We reasoned that the overlapping genes between the comparison groups involving the disease phenotype are likely important in the disease process, while others may not be relevant to disease pathology. We noted 276 overlapping genes between the comparison groups involving the PAN-stressed heterozygous knock-in mouse model (Fig. 4F). Of these, 94 genes (one-third) overlap with the cell adhesion and mitochondria-related pathways identified from the GSEA (Fig. 4G). Together, this observation suggests that these processes could be compromised due to INF2 mutation and likely play an important role in disease progression.
Mutant mouse podocytes have altered cell adhesion and mitochondrial morphology
We next evaluated whether the alterations in cell adhesion and mitochondria-related processes identified through RNA-seq analysis are observed in the podocytes of these mice. Our previous studies have shown that both the small interfering RNA–silenced INF2 human podocytes and mutant podocytes from R218Q knock-in mice exhibit impaired cell spreading (23, 24). More recently, we have shown that N-fragment (DID region) mediates the cell-spreading function of INF2, and that this spreading may be altered in the presence of a mutation (23). Similarly, INF2-mediated actin arrangements have been implicated in mitochondrial fission (25). While these studies suggest a role for INF2 in cell spreading and mitochondria, how they compare in basal versus stressed (or disease) conditions and how disease mutations alter these processes has not been defined.
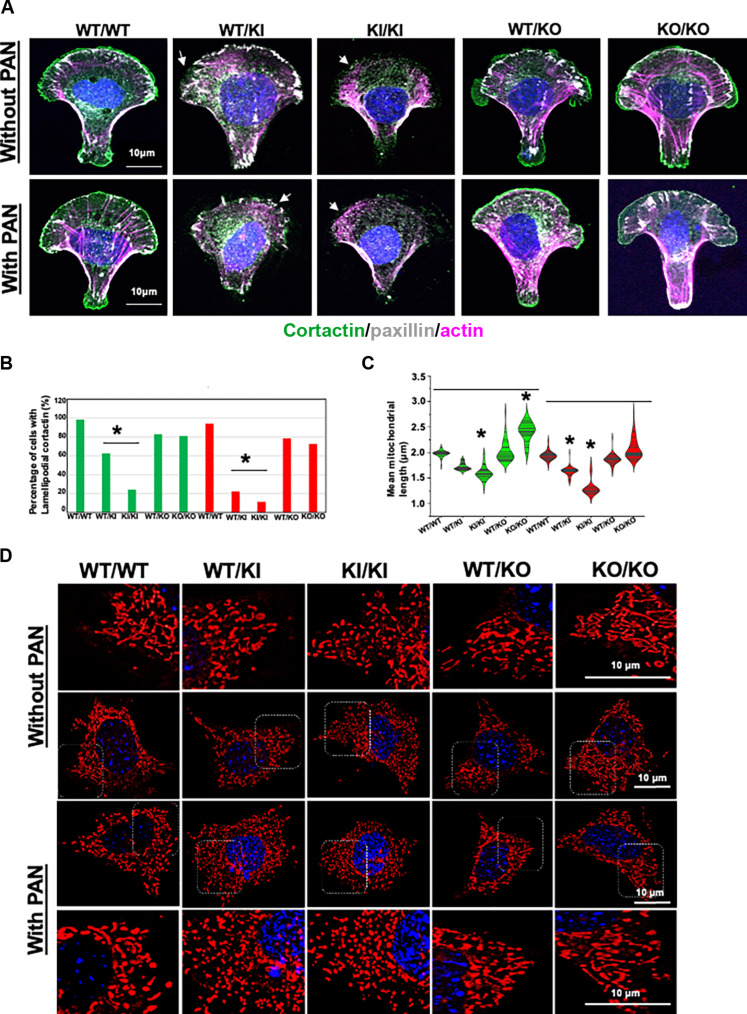
To examine this, we first tested the cell adhesion properties of mouse podocytes of various INF2 genotypes in both basal and PAN-stressed conditions using cortactin, paxillin, and F-actin staining. We conducted this analysis on a micropatterned substrate to have uniformity in cell dimensions and shape (Fig. 5A). Under basal conditions, wild-type podocyte morphology on crossbow-shaped micropatterns showed cortactin present in lamellipodium and paxillin present in focal adhesions, with F-actin emanating from them. This distribution pattern is retained in heterozygous and homozygous INF2 knockout podocytes but was altered in both heterozygous knock-in and homozygous knock-in podocytes. We noted a loss of both lamellipodium-cortactin and focal adhesion-paxillin in homozygous knock-in podocytes, whereas in heterozygous knock-in podocytes, this is limited to the loss of cortactin in lamellipodium. We also examined how these trends manifest in response to PAN stress. We noted that the wild-type, heterozygous knockout, and homozygous knockout podocytes retained their characteristic lamellipodium-cortactin, focal-adhesion paxillin, and F-actin distribution pattern. By contrast to the basal condition, heterozygous knock-in podocytes in the PAN-stressed condition were similar to homozygous knock-in podocytes but with more pronounced effect with loss of both lamellipodium-cortactin and focal adhesion-paxillin (Fig. 5B).
Fig. 5. Cell adhesion and mitochondria characterization in normal and mutant mouse podocytes.
[(A) and (B)] Cell adhesion assays in crossbow micropatterns. (A) Mouse podocytes from different INF2 genotypes were compared for their cell adhesion qualities in both basal and PAN-stressed conditions using cortactin, paxillin, and F-actin staining (n, 100 cells for each group). Lack of lamellipodium cortactin, focal adhesion paxillin, and diffused cytoplasmic F-actin were associated with defective cell spreading and adhesion. R218Q knock-in podocytes showed defective cell adhesion qualities in basal and PAN-stressed conditions (white arrow). Scale bars, 10 μm. (B) Quantification for defective cell adhesion (as determined by lamellipodium cortactin). Green bars, untreated; and red bars, PAN-treated. Heterozygous and homozygous R218Q knock-in podocytes showed a significant defect in cell adhesion (*P > 0.001 R218Q INF2 knock-in podocytes versus other groups; one-way ANOVA and Tukey’s multiple comparison test; n = 100 for each group). [(C) and (D)] Mitochondria assessment using mitotracker staining. Mouse podocytes from all INF2 genotypes were compared for mitochondrial length (n, 100 cells for each group). Live cells were imaged as z stacks and maximum-intensity projection images were generated to visualize the entire mitochondrial network in cells. (C) Quantification of mean mitochondrial length. Green, untreated; and red, PAN-treated. Under basal conditions, R218Q INF2 homozygous knock-in podocytes had shorter mitochondria, whereas homozygous knockout podocytes had longer mitochondria (*P > 0.001, R218Q INF2 homozygous knock-in podocytes versus other groups and homozygous knockout podocytes versus other groups; one-way ANOVA and Tukey’s multiple comparison test; n = 100 for each group). In PAN condition, heterozygous and homozygous R218Q INF2 knock-in podocytes showed shorter mitochondria (*P > 0.001, R218Q INF2 heterozygous and homozygous knock-in podocytes versus other groups; one-way ANOVA and Tukey’s multiple comparison test; n, 100 for each group). (D) Representative mitochondria images from all genotypes in basal and PAN-treated conditions. Higher-magnification images of highlighted regions were shown in a separate panel. Scale bars, 10 μm.
To examine mitochondria in podocytes, we compared the mitochondria filament length in basal and PAN-stressed conditions. We observed differences as a function of INF2 genotype (Fig. 4C). Under basal conditions, homozygous knock-in podocytes showed shorter mitochondrial filaments than wild-type podocytes. Homozygous knockout podocytes had longer mitochondrial filaments. The PAN stress further enhanced the trends in knock-in podocytes, making the mitochondria filaments in both heterozygous knock-in and homozygous knock-in podocytes significantly shorter than the PAN-stressed wild-type podocytes. In contrast, despite the PAN stress, heterozygous and homozygous knockout podocytes exhibited mitochondrial filament length similar to wild-type podocytes (Fig. 5, C and D).
Last, to confirm that the mitochondrial defects are not adaptive secondary changes due to PAN stress and can contribute to phenotype development, we conducted Seahorse bioenergetics analyses under basal conditions (fig. S4). We found that podocytes with the R218Q mutation, especially in the homozygous state, showed a significant increase in both basal respiration and maximum respiration [oxygen consumption rate (OCR) values], as well as glycolysis and glycolytic capacity [extracellular acidification rate (ECAR) values] compared to wild-type podocytes. In contrast, homozygous knockout podocytes exhibited a reduction in OCR and ECAR values compared to wild-type podocytes. These findings suggest that the mutation can lead to changes in some, if not all, of the mitochondrial bioenergetics. The opposing trends between knock-in and knockout mitochondrial bioenergetics align with the opposing trends in fission, indicating that the changes in INF2-mediated fission drive the changes in mitochondrial bioenergetics. Together, these observations confirm that the presence of mutant alleles in podocytes leads to cell adhesion and mitochondria defects, making them vulnerable to developing disease in response to PAN stress.
INF2-patient kidney organoid–derived podocytes recapitulate mouse disease phenotypes
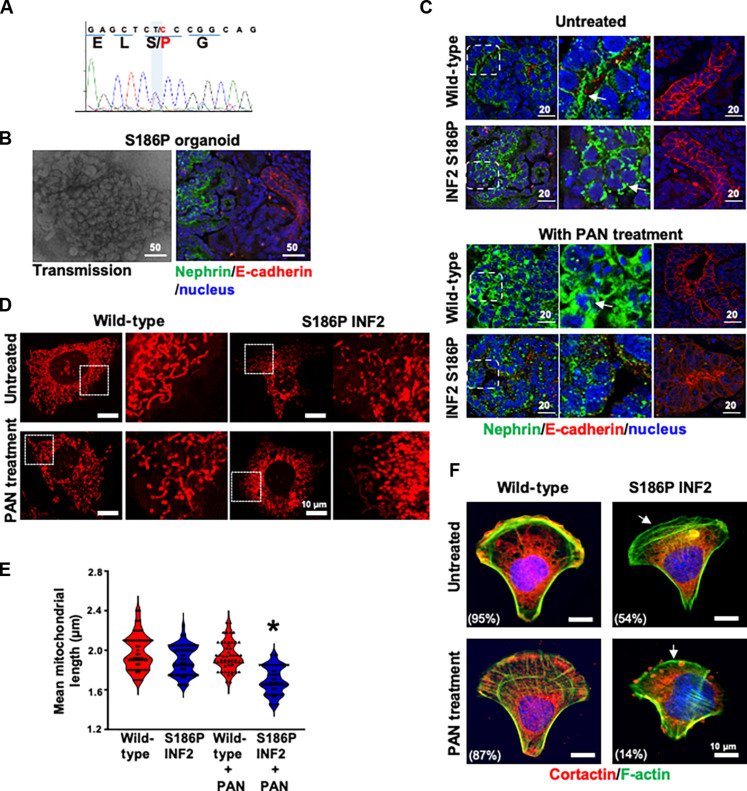
To examine whether our results correlate with human conditions, we prepared induced pluripotent stem cells (IPSCs) from an individual with INF2-related FSGS. Sequence analysis confirmed that the IPSCs retained the heterozygous S186P INF2 mutation (Fig. 6A). We then used these cells to prepare kidney organoids and assessed their morphology using marker proteins. The results showed that the S186P INF2 IPSCs formed kidney organoids with a typical distribution of glomeruli (20%) and tubular structures (70%) determined by their respective marker proteins, nephrin, and E-cadherin (Fig. 6B). However, when we examined the subcellular localization of these proteins, we noted that the podocytes of wild-type organoids displayed a basolateral localization for nephrin, whereas in S186P INF2 organoids, nephrin had an altered punctate pattern mislocalizing to peri-nuclear regions. The PAN injury further altered these patterns, exhibiting a punctate cytoplasmic distribution in normal organoids and a loss of nephrin staining intensity in SI86P organoids. In contrast, the localization of E-cadherin in tubules remained similar between wild-type and S186P organoids, both in basal and PAN-treated conditions (Fig. 6C).
Fig. 6. FSGS patient’s IPSC-kidney organoid-derived podocytes exhibit defective cell adhesion and mitochondria.
(A) FSGS patient’s IPSC characterization. DNA sequencing analysis of the INF2-DID region showed a heterozygous missense mutation causing serine (S) to proline (P) at amino acid position 186 in INF2-CAAX. (B) S186P INF2 IPSCs formed kidney organoids with glomeruli and tubular structures. There is no apparent difference in the distribution of glomeruli (nephrin stained) and tubules (E-cadherin stained). (C) Marker protein analysis of S186P organoids in basal and PAN-treated condition. The basolateral localization pattern of nephrin is altered to a punctate pattern by the S186P mutation. PAN injury affected nephrin staining pattern in normal and S186P organoids. The E-cadherin staining pattern remained unaffected in tubular structures. (D and E) Mitochondria assessments. Outgrown podocytes were examined for mitochondrial filament length. PAN-treated S186P podocytes showed a shorter filament length (*P > 0.001, PAN-treated heterozygous knock-in S186P INF2 podocytes versus other groups; one-way ANOVA and Tukey’s multiple comparison test; n, 100 for each group). (F) Cell adhesion assessments. Heterozygous S186P knock-in podocytes lack lamellipodial cortactin (white arrow) and are defective in cell adhesion on a crossbow micropattern in basal and PAN-treated conditions. The percentage of cells with lamellipodial cortactin was indicated (*P > 0.001, percentage of heterozygous knock-in S186P INF2 podocytes versus other groups; one-way ANOVA and Tukey’s multiple comparison test; n, 100 for each group). Scale bars, 10 μm.
We also examined whether the alterations in cell adhesion and mitochondrial processes noted in mutant mouse podocytes are present in human S186P INF2 podocytes. To evaluate this, we used podocytes outgrown from glomeruli of normal and S186P organoids and assessed their cell adhesion and mitochondrial qualities. We found that the average mitochondrial filament length in S186P podocytes was shorter than the average mitochondria filament length in wild-type podocytes in both basal and PAN-stressed conditions (Fig. 6D and E). Similarly, we found that wild-type podocytes were able to establish cortactin in the lamellipodium and spread normally in the crossbow micropattern in both basal and PAN-treated conditions, whereas S186P INF2 podocytes were defective in both conditions (Fig. 6F). These results confirm that defects in cell adhesion and mitochondria are consistent cellular features of INF2-related FSGS.
DISCUSSION
In this study, by comparing the injury responses of INF2 knock-in and knockout mouse models, we have demonstrated that kidney disease occurs as a result of mutant INF2’s gain-of-function effects. Given that all of the clearly pathological INF2 mutations that have been identified to date are missense variants, this observation explains the disease’s autosomal dominant inheritance pattern. In addition, we observed that the mutant INF2 acquires gain of function by altering the localization of wild-type INF2, thereby leading to INF2 activity at abnormal sites. These results are consistent with earlier findings on wild-type INF2 and mutant INF2 localization patterns (23, 26, 27). In this model, INF2 dimers composed of wild-type and mutant INF2 will differ in their localization from wild-type INF2 dimers and induce abnormal actin cytoskeletal arrangements. By doing so, mutant INF2 can cause defects in various processes integral to glomerular structure and function.
However, the described model, in particular in the context of gain of function, leaves some questions that still need to be clarified. For instance, the point mutant mouse model needs a stress injury to develop a disease phenotype. At endogenous levels, we have noted that mutant protein appears unstable, reducing levels in the knock-in podocytes. These observations are intriguing in that it would be reasonable to hypothesize that INF2 is a stress response protein, requiring a stress-induced injury for its function to become essential. Under such a model, we would expect reduced or absent INF2 to cause disease due to loss of some necessary INF2 function. However, to the contrary, the INF2 knockout mouse model does not develop kidney disease, arguing against a loss-of-function mechanism. However, if the human (and mouse) INF2-associated disease is driven solely by gain of function, the evolutionary pressure (physiological need) to maintain high INF2 expression in podocytes remains unclear.
The observation that disease-causing mutations are all located in the N-terminal region requires explanation. Previously, we showed that INF2 is cleaved, and postcleavage, the INF2 N-terminal region can regulate the DAD activity of mDIA formins (23). While this provides one possible mechanism in which the INF2 N-terminal region may function independently of the C terminus, other regulatory mechanisms still need to be described. In this study, our analysis of INF2 localization indicates that both the N-fragment and full-length INF2, when mutated, can alter the localization of full-length wild-type INF2. This suggests that the INF2 N-terminal region has an essential function in regulating INF2 localization. Together, the results indicate at least two unique functions to the N-terminal region: (i) determining the localization of INF2-forms in cells and (ii) regulating the DAD activity. The pathogenic mutations appear to alter both of these activities.
A major goal of our RNA-seq studies was to identify the downstream events in which the mutation-induced alterations have an effect. Our findings from the GSEA indicated actin and cell-membrane arrangements (grouped as cell adhesion) and mitochondria-related genes are heavily altered during the disease. These data align with the clinical observation that podocytes in many forms of progressive glomerular diseases are often defective in their adhesion with capillary tuft and manifest mitochondrial dysfunction (28, 29). However, we note that adhesion- and mitochondria-related processes are not the only enrichment identified and, thus, may not be the only driving factors for INF2-related disease onset and progression. We noted many other processes, related or unrelated to INF2, in our gene enrichment analysis. For example, Madrid et al. (30) reported that INF2 interacts with MAL2, a vesicular protein, and regulates basolateral-to-apical transcytosis in kidney epithelial cells. In our gene enrichment analysis, we have also identified processes relating to vesicle transport being down-regulated. It is thus interesting to note that other processes beyond cell adhesion and mitochondria cluster may also contribute to the disease progression. Future studies of such processes will establish their relation with INF2 and their significance in disease progression.
Our previous studies have led us to hypothesize that INF2 functions downstream of Rho A and inhibits mDIA-related DAD activity through the N-terminal region (22–24, 31). By doing so, INF2 mediates cell adhesion by promoting lamellipodial structure formation. The results shown in this study agree with this hypothesis in that the presence of mutation affects cell spreading in micropatterns. However, INF2 knockout podocytes did not manifest similar defects in the micropatterns, suggesting that mutant INF2-driven activities are essential for the defective phenotype. Together, we propose that mutant podocytes may exhibit multiple alterations; in one aspect, the absence of INF2 at proper localization leads to alterations, as noted with mDIA regulation, and with the other, the presence of mutant INF2 at aberrant sites causes unwarranted alterations, which is evident in abnormal F-actin/G-actin distribution in cells (Fig. 2A). Future comparative assessment of actin distribution among various subcellular compartments of INF2 mutant and INF2 knockout cells will further confirm this point and help us to identify the aberrant sites where mutant INF2 may cause alterations within the cells.
Several studies have shown that INF2 and mitochondrial fission are related and how DID mutations may affect this relationship (25, 32–34). However, this work presents the clear evidence for gain-of-function effects in mitochondria fission by pathogenic R218Q INF2 in podocytes. On the one hand, it is conceivable that the alteration in fission may lead to mitochondrial dysfunction and related FSGS; on the other hand, it raises the question as to why the R218Q INF2 mouse model does not develop FSGS without additional toxic injury. As discussed above, one explanation is that mutant protein clearance may limit the extent to which mitochondrial fission occurs, preventing FSGS development. It is worth noting that the knockout podocyte manifests decreased mitochondrial fission and has altered mitochondria structure. However, our mouse data align with a model in which podocyte dysfunction is enhanced in the presence of the excessive fission due to point mutant R218Q rather than the decreased fission conditions due to the loss of INF2. Future studies on the podocyte mitochondrial function with excessive or reduced fission can clarify these questions.
We have found that S186P INF2 kidney organoids, derived from the INF2 patient’s IPSC, exhibit an altered nephrin distribution from wild-type kidney organoids. Yoshimura et al. and others (35, 36) have shown that nephrin exhibits a basolateral distribution in normal organoids, and this pattern may be altered as a consequence of mutations in various genes. Meanwhile, in mouse models, nephrin distribution remains unaltered in basal conditions in both R218Q INF2 knock-in and INF2 knockout models (22, 23). However, R218Q INF2 becomes defective when recycling nephrin to the plasma membrane when injured (37). Reconciling these disparate results will require assessments from a matured vascularized organoid system with in vivo physiology. Our findings suggest that the effect of an S186P INF2 mutation is similar to the effect of R218Q in causing impaired cell adhesion and mitochondria structure.
In summary, we have shown that mutant INF2’s gain-of-function activity drives INF2-related FSGS (Fig. 7). The muta nt INF2 acquires this quality by altering the localization of wild-type INF2, thereby making INF2 exert activity at aberrant sites. In addition, we have observed that cell adhesion and mitochondrial structures are significantly affected by pathogenic mutations in INF2. These data provide a plausible explanation for the autosomal dominant inheritance pattern of the disease and highlight the molecular events driving the INF2-related FSGS. Future studies tailored to define the full range of INF2-related pathways and their significance in maintaining the podocyte’s unique structure will increase our understanding of INF2 in normal physiology and disease pathology.
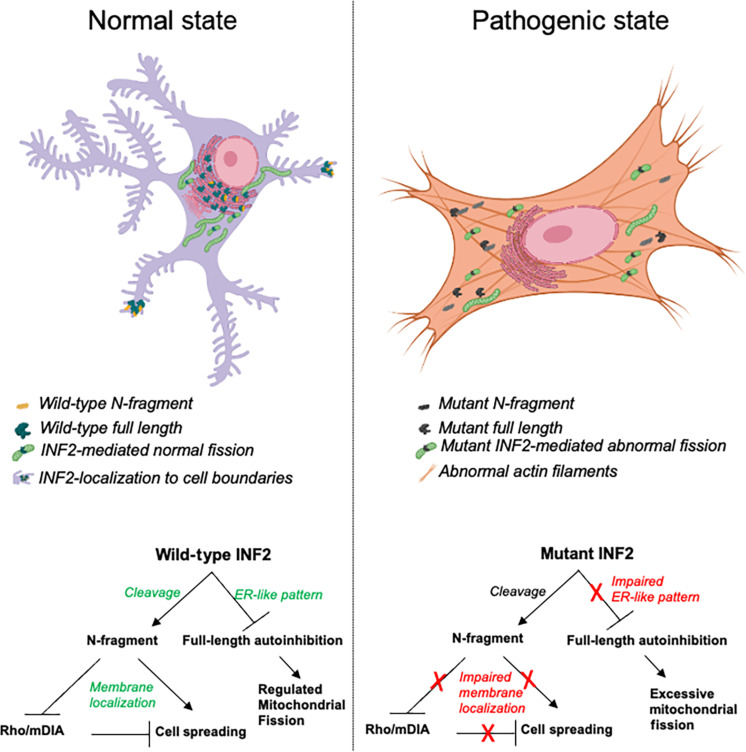
Fig. 7. Summary model for INF2-related FSGS.
INF2-DID region exists in two INF2 forms: full-length INF2 and N-fragment INF2. Full-length INF2 predominantly localizes to peri-nuclear ER-rich regions, while N-fragment preferentially localizes to membrane regions. However, with the pathogenic mutations, their localization is altered to a diffused cytoplasmic pattern. In one possible model for disease development, the pathogenic mutation may cause (i) abnormal actin distribution due to INF2’s activity at altered localization sites and (ii) excessive mitochondria fission. With an extraneous insult that causes podocytes to lose their structural integrity, the N-fragment may not localize properly, perhaps unable to counteract mDIA signaling, causing defective adhesion and impaired recovery. This combination of mutant-driven predisposition and postinjury defect may lead to chronic pathology and FSGS.
MATERIALS AND METHODS
Ethical statement
All studies involving mice were performed in compliance with an animal care protocol approved by the Institute Animal Care and Use Committee (IACUC) at Beth Israel Deaconess Medical Center. Human IPSC lines were derived under the aegis of a protocol approved by the Beth Israel Deaconess Medical Center Human Research Committee.
IACUC approval
All animal experiments were approved by the Beth Israel Deaconess Center’s IACUC (protocol no. 004-2022). The studies were performed in accordance with institutional and national guidelines for Animal Care and Use. The study is compliant with all relevant ethical regulations regarding animal research.
IRB approval
The study was approved by the Institutional Review Board of Beth Israel Deaconess Medical Center (protocol no. 2009P000430). Blood samples were collected and handled according to the guidelines described in the Code of Conduct for Proper Use of Human Tissue, with written informed consent from the patients before sampling.
Mouse experiments
The development of INF2 mutant mice is described in (22). We conducted podocyte injury analysis in INF2 mouse models using various stressors as described previously (38–41). For the PAN injury response, mice were injected with PAN intraperitoneally at a dosage of 490 mg/kg, and urine samples were collected on days 0 (before the PAN injection), 3, 7, 11, and 14 for analysis. For other stressors, albumin (10 mg/g daily intraperitoneal injection for 5 days), adriamycin (15 mg/kg intravenous injection), lipopolysaccharide (10 mg/g intraperitoneal injection), and angiotensin II (36 mg/kg intraperitoneal injection), urine samples were collected on days 0 (baseline level before injection), 1 (except for albumin stress, where 24 hours after all the injections), 3, 5, and 7 for analysis. The SDS–polyacrylamide gel electrophoresis (PAGE) method was used to separate the urine samples, which were then stained with Coomassie dye to detect albuminuria. Albumin and creatinine levels in the urine were quantified using an albumin enzyme-linked immunosorbent assay kit (Bethyl Labs, TX) and a colorimetric kit, respectively, following the manufacturers’ protocols.
Histology
Mice kidneys were harvested at indicated time frames and processed to examine glomerulosclerosis and podocyte foot process arrangement. For analyzing sclerosis, kidneys were fixed in 10% buffered formalin and paraffin-embedded to prepare formalin-fixed paraffin-embedded blocks. These blocks were then cut into 5-μm sections and processed for periodic acid–Schiff and Masson’s trichrome staining. The ratio of sclerotic glomeruli to total glomeruli was calculated and plotted. For podocyte ultrastructure analysis, kidneys were fixed in a modified Karnovsky’s fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4), processed using 1% osmium tetroxide and Epon embedded for sectioning. Sections collected on grids were imaged by the JEOL 1400 Transmission electron microscope. The number of foot processes per micron was quantified and plotted.
Mouse podocyte preparations
Podocytes from mice were obtained from eight-week-old INF2 mouse models (wild-type, heterozygous R218Q knock-in, heterozygous knockout, homozygous R218Q knock-in, and homozygous knockout) as previously described (42). In brief, podocytes were outgrown from glomeruli on cell culture dishes and immortalized with a temperature-sensitive T antigen. All podocyte preparations were then checked for marker proteins (e.g., podocin), and only cell preparations expressing marker proteins in more than 90% of cells were used for subsequent experiments. Before conducting experiments with them, podocytes were differentiated by culturing them at 37°C.
Cell culture
Podocytes were maintained in RPMI 1640 (Corning, MA), supplemented with 1% insulin-transferrin-selenium (ITS), 1% penicillin-streptomycin, and 10% fetal bovine serum (FBS) (23). Human embryonic kidney epithelial cells (HEK293T cells) were maintained in Dulbecco’s minimum essential medium (Corning, MA) supplemented with 10% FBS and 1% penicillin-streptomycin.
INF2 plasmid constructs and expression
For the RFP-tagged construct, the full-length human INF2-CAAX isoform was cloned into a turbo RFP plasmid (N-terminal RFP) (OriGene, MD). For GFP-tagged constructs, human INF2-CAAX isoform sequences corresponding to amino acids 1 to 547 (N-fragment) and 1 to 1249 (full-length) were cloned into an EGFPC1 plasmid (N-terminal GFP tag) (Takara Bio, CA). For noncleavable INF2, amino acids 543 to 548 of INF2 were replaced with hexa-alanine in full-length CAAX constructs. For HA- and Flag-tagged construct, full-length INF2-CAAX isoform was cloned into pCMV6 vector. HA tag was included at the N terminus, and Flag tag was added at the C terminus of INF2. For transient expression, plasmids were transfected with Lipofectamine 3000 (for podocytes) or 2000 (for 293T cells) as per the manufacturer’s guidelines (Thermo Fisher Scientific, MA). All interaction and imaging analyses were performed at 48 hours post-transfection. For INF2 constructs for the pyrene-actin assays, human INF2-FL-nonCAAX cDNA was cloned into an EGFPC1 vector containing Strep-tag II (IBA Life Sciences) and an HRV3C cleavage site that was located N-terminal to the INF2 start codon. For INF2 DID proteins (wild-type and R218Q), human INF2 N-terminal region (amino acids 1 to 267) was cloned into pGEX-KT vector.
Coimmunoprecipitation
HEK293T cells were transiently transfected with the indicated INF2 constructs using Lipofectamine 2000 (Invitrogen, CA). After 48 hours, the cells were lysed in 1% NP-40 lysis buffer (1% NP-40, 50 mM tris-HCl, 150 mM NaCl, and 5 mM EDTA, pH 7.4) supplemented with protease inhibitors. Cell lysates were then incubated with mouse anti-FLAG beads (MA5-15256; Thermo Fisher Scientific, MA) for 4 hours, followed by their magnetic sorting and suspension in premixed Laemmli buffer (Bio-Rad, CA). The coimmunoprecipitation of full-length HA-INF2-FLAG with various GFP-tagged INF2 forms was then analyzed. Samples were probed for HA or GFP using the antibodies for GFP (1:500) (ab13970; Abcam, MA) or HA (1:500) (2367; Cell Signaling Technology, MA). The membranes were then incubated with IRDye secondary antibodies and imaged using an Odyssey Clx Infrared system (LI-COR, NE).
Immunoblotting
Cell lysates were prepared in an RIPA buffer (Boston BioProducts, MA) supplemented with a cocktail of protease and phosphatase inhibitors (Roche, CA). Equal protein loads were electrophoresed on a 4 to 20% gradient gel, quick-transferred to a polyvinylidene difluoride membrane (Bio-Rad, CA), and probed with respective primary and secondary antibodies as follows: INF2 (1:500) (A303-427A; Bethyl Laboratories, TX), INF2 (C) (1:500) (20466-1-AP; Proteintech, IL), podocin (1:1000) (P0372; Sigma-Aldrich, MO), and anti-rabbit (1:4000) (Santa Cruz Biotechnology, CA). The membranes were then developed using a chemiluminescent-based substrate (Super Signal West Dura). Total β-actin (1:4000) (sc47778; Santa Cruz Biotechnology, CA) level was used as a loading control.
Immunofluorescence
Cells were phosphate-buffered saline (PBS) rinsed and fixed with 4% paraformaldehyde for 15 minutes. The fixed cells were then quenched, permeabilized with 0.5% Triton X-100, and blocked using 5% bovine serum albumin solution. The primary and secondary antibody preparations were sequentially incubated for 2 hours and counterstained cell nuclei with Hoechst (dsDNA) (Invitrogen). For organoid and mouse glomeruli, samples were processed into paraffin blocks, and 5-μm sections were then cut and processed for staining analysis. The following primary antibodies with indicated dilutions were prepared in blocking solution and used for staining analysis: nephrin (BP5030, 1:200; OriGene, MD), INF2 (DT-157, 1:100), WT-1 (ab89901, 1:100; Abcam, MA), α–smooth muscle actin (ab56941, 100; Abcam, MA), paxillin (ab32084, 1:100; Abcam, MA), synaptopodin (AP33487SU-N, 1:200; OriGene, MD), cortactin (MA5-15831, 1:100; Thermo Fisher Scientific, MA), and F-actin (rhodamine-phalloidin, 1:100; Invitrogen, CA). For mitochondrial imaging, cells were incubated with deep red Mito tracker dye (8778; Cell Signaling Technology, MA) per the manufacturer’s instructions and imaged as live cells. All samples were imaged for fluorescence using laser-scanning confocal microscopy (LSM 510; Zeiss) using ZEN lite Black edition software.
Micropatterning cells
Crossbow micropatterns, custom prepared in Chromemask (Advance Repro, MA), were used to program cells to reach uniform shape and size. Micropatterning of cells using the mask was performed as described previously (43). In brief, poly-l-lysine–grafted polyethylene glycol was coated on a cover glass, placed under a chrome mask containing the micropatterns, and exposed to an ultraviolet light through the chrome mask. The exposed parts of the PLL-PEG were washed in distilled water, dried, and coated with fibronectin (250 ng/ml). Cells were plated as single suspensions. After 1 hour, cover glasses were washed away to remove unattached cells. For PAN treatments, cells were pretreated with PAN overnight, trypsinized, and replated with PAN on micropatterns and maintained through cell attachment. All analyses were performed after 6 hours of cell spreading in micropatterns.
Actin assays
Mouse podocytes were lysed and examined for their total G-actin and F-actin levels using a G-actin/F-actin in vivo biochemical assay kit (Cytoskeleton, CO). In brief, 100 μl of precleared lysates were ultracentrifuged for 100,000g at 37°C for 1 hour. The supernatant containing the G-actin was collected and processed for immunoblotting. The residual pellet containing the F-actin was incubated with 100 μl of depolymerizing buffer in ice for 1 hour and then processed further for immunoblotting. The ratio of F-actin to G-actin was calculated from their band intensity.
Biochemical actin polymerization assays
Actin polymerizing activity of full-length INF2 with wild-type and R218Q mutant DID regions was assessed using pyrene actin polymerization assay as described previously (44). In brief, rabbit skeletal muscle actin in G-buffer (6 μM actin and 10% pyrene) were converted to Mg2 salt by the addition of EGTA and MgCl2 (to 1 and 0.1 mM, respectively) for 2 min at 23°C immediately before polymerization. Polymerization was induced by two volumes of 1.5× polymerization buffer [75 mM KCl, 1.5 mM MgCl2, 1.5 mM EGTA, 15 mM Hepes (pH 7.4), 2 mM dithiothreitol, 2 mM tris-HCl, 0.2 mM adenosine 5′-triphosphate, 0.1 mM CaCl2, and 0.01% (w/v) NaN3] containing other proteins (20 nM INF2-FL, 6 μM profilin, 50 nM capping protein, and/or 0.5 μM DID region). Pyrene fluorescence (365/410 nm) was monitored in a 96-well fluorescence plate reader within 1 min of inducing polymerization and plotted versus time.
RNA-seq and GSEA
Total RNA from mouse glomeruli was sequenced on a HiSeq 4000 following standard workflow using an NEB Next Ultra II RNA Library Preparation kit (New Englab Biolabs, MA) (Illumina, CA) and paired-end sequencing (Genewiz, MA). The samples had 78 million pass filter reads, with 89% of bases above the quality score of Q30 and a mean quality score of 37.32. Adapters and low-quality bases were trimmed and aligned with the mouse reference genome (GRCm38) and GENCODE annotation using STAR 2.7. The average mapping rate of all samples was 90%.
Genes showing differential expression were analyzed for (i) HET.KI versus HET.KO, (ii) PAN-treated HET.KI versus PAN-treated HET.KO, (iii) HET.KI versus PAN-treated HET.KI, and (iv) HET.KO versus PAN-treated HET.KO groups. The common and unique genes between these analyses were identified using DiVenn 1.2. GSEA was conducted between PAN-treated HET.KI versus PAN-treated HET.KO (GSEA version 4.3.3 https://gsea-msigdb.org/gsea/license_terms_list.jsp). The enriched gene ontology cellular component gene sets (P < 0.05) were identified and visualized as a network in Cytoscape using EnrichmentMap, AutoAnnotate, and clusterMaker2 application. The overlap of the number of genes between GSEA and DiVenn analysis was plotted using Venny 2.1.
Human kidney organoids and outgrown podocyte analysis
Human IPSC lines were generated from a normal and patient with INF2-related FSGS using CytoTune-iPS 2.0 Sendai reprogramming kit (Thermo Fisher Scientific, MA) at Harvard Stem Cell Institute IPS Core Facility. Kidney organoids were then prepared as described previously (45), with minor modifications. In each step where animal product free medium was called for, APELII (StemCell Technologies) supplemented with 5% protein-free hybridomia medium II (PFHMII) (Thermo Fisher Scientific, MA) was used. In addition, the initial 9 days of the protocol were carried out in three-dimensional (3D) gel suspension to enhance homogeneity in differentiation. In brief, 6 μl of the IPSC suspension was plated in ~175 μl of collagen matrix in 3D [222 μl of rat tail collagen (~10 mg/ml) or 667 μl of rat tail collagen (3 mg/ml), 227 μl of geltrex, 100 μl of 10× PBS, 4 μl of 1 mol/liter NaOH] in 24-well plates with 1 ml of mTeSR (StemCell Technologies) for 48 hours. The media was then changed to 0.5 ml of APELII medium supplemented with 5% PFHMII and 8 μM CHIR99021 for 4 days with media changes every 2 days. On day 4, media was switched to 1 ml of APELII supplemented with 5% PFHMII and FGF9 (200 ng ml−1) and heparin (1 μg ml−1) until it has been 7 days since the cells were first introduced to CHIR99021. The collagen mix was then digested with ~200 μl of collagenase A (10 mg/ml) for 30 min. The released spheroids were spun down into aggregates with 1 ml of APELII with 5% PFHMII, and all further steps were performed in accordance with the Takasato et al. (45) protocol. For PAN treatments, mature organoids (5+ days post-aggregate formation) were given APELII media with 5% PFHMII and PAN (1 mg/ml) dissolved in PBS. For harvesting podocytes, 10 organoid cultures were pooled, and glomeruli extraction was performed using the sieve method, as described previously (46). The isolated glomerular structures were then maintained in a media composition of RPMI 1640 supplemented with 10% FBS and 1% ITS and podocytes were outgrown on a collagen I–coated surface. Cell preparations were then stained for podocyte marker proteins including podocin and synaptopodin (fig. S5). Cells that tested positive for at least two podocyte marker proteins were classified as podocytes and cell batches with over 95% positivity were used in the cell adhesion and mitochondria assessments.
Seahorse analysis
Seahorse extracellular flux (XF) analysis was performed as described previously (47). In brief, podocytes derived from INF2 mouse models were plated and cultured for 24 hours in Seahorse culture plates before measurements. ECAR and OCR were measured in basal conditions using the Seahorse XF Glycolysis Stress Test and Seahorse XF Mito stress test following the manufacturer’s instruction (Agilent Technologies, CA).
Plotting and statistical analysis
All plotting and statistical analyses were performed using OriginPro software. All values are reported as mean ± SD. Analyses were performed using one-way analysis of variance (ANOVA) between test groups. Tukey’s multiple comparison test was used to find group differences. Statistical significance was set at a minimal value of P < 0.05. For evaluating the correlation between mice features and disease development, PAN-stressed mice were categorized on the basis of sex (male and female), age [mature adult (>6 months) and middle aged (>12 month)], and INF2 genotypes (wild-type, heterozygous knock-in, heterozygous knockout, homozygous knock-in, and homozygous knockout) and analyzed using the Pearson correlation coefficient method for their correlation with phenotype development (proteinuria). Features with r values >0.5 are selected as having a strong positive correlation with disease development.
Acknowledgments
We thank BIDMC’s confocal imaging and histology facility for technical assistance with experiments. We also thank Harvard stem cell core facility for IPSC preparation and characterization.
Funding: This work was supported by NIH grants R01DK088826 (M.R.P. and H.N.H.) and RC2DK122397 (M.R.P.) and the Peer Reviewed Medical Research Program (PRMRP)-Department of Defense (DoD) grant W81XWH2010320 (B.S.).
Author contributions: B.S. and M.R.P. conceived the idea and wrote the paper. B.S., S.W., S.K., M.L., S.J., M.-F.H., and C.V.R. performed the experiments and analyzed the data. B.S., H.N.H., S.L.A., and M.R.P. analyzed the data and edited the drafts.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Original RNA-seq data files have been uploaded to the Sequence Read Archive (SRA; PRJNA1119066).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.Scott R. P., Quaggin S. E., Review series: The cell biology of renal filtration. J. Cell Biol. 209, 199–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollak M. R., Quaggin S. E., Hoenig M. P., Dworkin L. D., The glomerulus: The sphere of influence. Clin. J. Am. Soc. Nephrol. 9, 1461–1469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perico L., Conti S., Benigni A., Remuzzi G., Podocyte-actin dynamics in health and disease. Nat. Rev. Nephrol. 12, 692–710 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Reiser J., Sever S., Podocyte biology and pathogenesis of kidney disease. Annu. Rev. Med. 64, 357–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg A. Z., Kopp J. B., Focal segmental glomerulosclerosis. Clin. J. Am. Soc. Nephrol. 12, 502–517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp J. B., Anders H. J., Susztak K., Podesta M. A., Remuzzi G., Hildebrandt F., Romagnani P., Podocytopathies. Nat. Rev. Dis. Primers. 6, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollak M. R., Familial FSGS. Adv. Chronic Kidney Dis. 21, 422–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akchurin O., Reidy K. J., Genetic causes of proteinuria and nephrotic syndrome: Impact on podocyte pathobiology. Pediatr. Nephrol. 30, 221–233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barua M., Brown E. J., Charoonratana V. T., Genovese G., Sun H., Pollak M. R., Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int. 83, 316–322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gbadegesin R. A., Lavin P. J., Hall G., Bartkowiak B., Homstad A., Jiang R., Wu G., Byrd A., Lynn K., Wolfish N., Ottati C., Stevens P., Howell D., Conlon P., Winn M. P., Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int. 81, 94–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown E. J., Schlondorff J. S., Becker D. J., Tsukaguchi H., Tonna S. J., Uscinski A. L., Higgs H. N., Henderson J. M., Pollak M. R., Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat. Genet. 42, 72–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labat-de-Hoz L., Alonso M. A., The formin INF2 in disease: Progress from 10 years of research. Cell. Mol. Life Sci. 77, 4581–4600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer O., Nevo F., Plaisier E., Funalot B., Gribouval O., Benoit G., Huynh Cong E., Arrondel C., Tete M. J., Montjean R., Richard L., Karras A., Pouteil-Noble C., Balafrej L., Bonnardeaux A., Canaud G., Charasse C., Dantal J., Deschenes G., Deteix P., Dubourg O., Petiot P., Pouthier D., Leguern E., Guiochon-Mantel A., Broutin I., Gubler M. C., Saunier S., Ronco P., Vallat J. M., Alonso M. A., Antignac C., Mollet G., INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N. Engl. J. Med. 365, 2377–2388 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Hegsted A., Yingling C. V., Pruyne D., Inverted formins: A subfamily of atypical formins. Cytoskeleton (Hoboken) 74, 405–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labat-de-Hoz L., Alonso M. A., Formins in human disease. Cells 10, 2554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramabhadran V., Hatch A. L., Higgs H. N., Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. J. Biol. Chem. 288, 26847–26855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panzer L., Trube L., Klose M., Joosten B., Slotman J., Cambi A., Linder S., The formins FHOD1 and INF2 regulate inter- and intra-structural contractility of podosomes. J. Cell Sci. 129, 298–313 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Andres-Delgado L., Anton O. M., Bartolini F., Ruiz-Saenz A., Correas I., Gundersen G. G., Alonso M. A., INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. J. Cell Biol. 198, 1025–1037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andres-Delgado L., Anton O. M., Madrid R., Byrne J. A., Alonso M. A., Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood 116, 5919–5929 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Lamm K. Y. B., Johnson M. L., Baker Phillips J., Muntifering M. B., James J. M., Jones H. N., Redline R. W., Rokas A., Muglia L. J., Inverted formin 2 regulates intracellular trafficking, placentation, and pregnancy outcome. eLife 7, e31150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti R., Ji W. K., Stan R. V., de Juan Sanz J., Ryan T. A., Higgs H. N., INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 217, 251–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian B., Sun H., Yan P., Charoonratana V. T., Higgs H. N., Wang F., Lai K. V., Valenzuela D. M., Brown E. J., Schlondorff J. S., Pollak M. R., Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int. 90, 363–372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian B., Chun J., Perez-Gill C., Yan P., Stillman I. E., Higgs H. N., Alper S. L., Schlondorff J. S., Pollak M. R., FSGS-causing INF2 mutation impairs cleaved INF2 N-fragment functions in podocytes. J. Am. Soc. Nephrol. 31, 374–391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H., Schlondorff J., Higgs H. N., Pollak M. R., Inverted formin 2 regulates actin dynamics by antagonizing Rho/diaphanous-related formin signaling. J. Am. Soc. Nephrol. 24, 917–929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korobova F., Ramabhadran V., Higgs H. N., An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramabhadran V., Korobova F., Rahme G. J., Higgs H. N., Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell 22, 4822–4833 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayraktar S., Nehrig J., Menis E., Karli K., Janning A., Struk T., Halbritter J., Michgehl U., Krahn M. P., Schuberth C. E., Pavenstadt H., Wedlich-Soldner R., A deregulated stress response underlies distinct INF2-associated disease profiles. J. Am. Soc. Nephrol. 31, 1296–1313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagata M., Podocyte injury and its consequences. Kidney Int. 89, 1221–1230 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Fan J., Zhu W., Niu Y., Wu M., Zhang A., Therapeutic potential targeting podocyte mitochondrial dysfunction in focal segmental glomerulosclerosis. Kidney Dis. (Basel) 9, 254–264 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madrid R., Aranda J. F., Rodriguez-Fraticelli A. E., Ventimiglia L., Andres-Delgado L., Shehata M., Fanayan S., Shahheydari H., Gomez S., Jimenez A., Martin-Belmonte F., Byrne J. A., Alonso M. A., The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev. Cell 18, 814–827 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Sun H., Schlondorff J. S., Brown E. J., Higgs H. N., Pollak M. R., Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2). Proc. Natl. Acad. Sci. U.S.A. 108, 2933–2938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung T. S., Chakrabarti R., Higgs H. N., The multiple links between actin and mitochondria. Nat. Rev. Mol. Cell Biol. 24, 651–667 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin X., Wang J., Gao K., Zhang P., Yao L., Tang Y., Tang L., Ma J., Xiao J., Zhang E., Zhu J., Zhang B., Zhao S. M., Li Y., Ren S., Huang H., Yu L., Wang C., Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. PLOS Genet. 13, e1006748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Y., Lv Z., Cao W., Shi W., He Q., Gao K., Phosphorylation of INF2 by AMPK promotes mitochondrial fission and oncogenic function in endometrial cancer. Cell Death Dis. 15, 65 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimura Y., Taguchi A., Tanigawa S., Yatsuda J., Kamba T., Takahashi S., Kurihara H., Mukoyama M., Nishinakamura R., Manipulation of nephron-patterning signals enables selective induction of podocytes from human pluripotent stem cells. J. Am. Soc. Nephrol. 30, 304–321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorison A., Ghobrial I., Graham A., Peiris T., Forbes T. A., See M., Das M., Saleem M. A., Quinlan C., Lawlor K. T., Ramialison M., Howden S. E., Little M. H., Kidney organoids generated using an allelic series of NPHS2 point variants reveal distinct intracellular podocin mistrafficking. J. Am. Soc. Nephrol. 34, 88–109 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H., Perez-Gill C., Schlondorff J. S., Subramanian B., Pollak M. R., Dysregulated dynein-mediated trafficking of nephrin causes INF2-related podocytopathy. J. Am. Soc. Nephrol. 32, 307–322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown B. J., Boekell K. L., Stotter B. R., Talbot B. E., Schlondorff J. S., Gain-of-function, focal segmental glomerulosclerosis Trpc6 mutation minimally affects susceptibility to renal injury in several mouse models. PLOS ONE 17, e0272313 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zubcevic J., Santisteban M. M., Perez P. D., Arocha R., Hiller H., Malphurs W. L., Colon-Perez L. M., Sharma R. K., de Kloet A., Krause E. G., Febo M., Raizada M. K., A single angiotensin ii hypertensive stimulus is associated with prolonged neuronal and immune system activation in Wistar-Kyoto rats. Front. Physiol. 8, 592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryant C., Cianciolo R., Govindarajan R., Agrawal S., Adriamycin-induced nephropathy is robust in N and modest in J substrain of C57BL/6. Front. Cell Dev. Biol. 10, 924751 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altintas M. M., Moriwaki K., Wei C., Moller C. C., Flesche J., Li J., Yaddanapudi S., Faridi M. H., Godel M., Huber T. B., Preston R. A., Jiang J. X., Kerjaschki D., Sever S., Reiser J., Reduction of proteinuria through podocyte alkalinization. J. Biol. Chem. 289, 17454–17467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy G. M., Blasio A., Donovan O. G., Schaller L. B., Bock-Hughes A., Magraner J. M., Suh J. H., Tattersfield C. F., Stillman I. E., Shah S. S., Zsengeller Z. K., Subramanian B., Friedman D. J., Pollak M. R., Recessive, gain-of-function toxicity in an APOL1 BAC transgenic mouse model mirrors human APOL1 kidney disease. Dis. Model. Mech. 14, dmm048952 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao L., McBeath R., Chen C. S., Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells 28, 564–572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu A., Fung T. S., Francomacaro L. M., Huynh T., Kotila T., Svindrych Z., Higgs H. N., Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself. Proc. Natl. Acad. Sci. U.S.A. 117, 439–447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takasato M., Er P. X., Chiu H. S., Little M. H., Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 11, 1681–1692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hale L. J., Howden S. E., Phipson B., Lonsdale A., Er P. X., Ghobrial I., Hosawi S., Wilson S., Lawlor K. T., Khan S., Oshlack A., Quinlan C., Lennon R., Little M. H., 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 9, 5167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abe Y., Sakairi T., Kajiyama H., Shrivastav S., Beeson C., Kopp J. B., Bioenergetic characterization of mouse podocytes. Am. J. Physiol. Cell Physiol. 299, C464–C476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C., Merico D., Bader G. D., Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, cytoscape and enrichmentMap. Nat. Protoc. 14, 482–517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L., Dong S., Ge Y., Fonseca J. P., Robinson Z. T., Mysore K. S., Mehta P., DiVenn: An interactive and integrated web-based visualization tool for comparing gene lists. Front. Genet. 10, 421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1