Abstract
PARP inhibitors (PARPi) represent a game-changing treatment for patients with ovarian cancer with tumors deficient for the homologous recombination (HR) pathway treated with platinum (Pt)–based therapy. PARPi exert their cytotoxic effect by both trapping PARP1 on the damaged DNA and by restraining its enzymatic activity (PARylation). How PARP1 is recruited and trapped at the DNA damage sites and how resistance to PARPi could be overcome are still matters of investigation. Here, we described PARP1 as a substrate of the deubiquitinase USP1. At molecular level, USP1 binds PARP1 to remove its K63-linked polyubiquitination and controls PARP1 chromatin trapping and PARylation activity, regulating sensitivity to PARPi. In both Pt/PARPi-sensitive and -resistant cells, USP1/PARP1 combined blockade enhances replicative stress, DNA damage, and cell death. Our work dissected the biological interaction between USP1 and PARP1 and recommended this axis as a promising and powerful therapeutic choice for not only sensitive but also chemoresistant patients with ovarian cancer irrespective of their HR status.
USP1 targeting improves PARP inhibitors effectiveness in ovarian cancer by regulating PARP1 trapping and catalytic activity.
INTRODUCTION
Since the first approval of olaparib by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) in 2014 for the treatment of advanced ovarian cancers associated with defective BReast CAncer gene 1/2 (BRCA1/2), four poly-[adenosine 5′-diphosphate (ADP)–ribose] polymerase inhibitors (PARPi) (olaparib, niraparib, rucaparib, and talazoparib) are now currently used in clinical practice for different types of cancers, showing impressive and hopeful survival-changing results, in particular for patients with ovarian cancer (1, 2). Epithelial ovarian cancer (EOC) is considered a devastating disease with an overall survival rate of ~40%, making it the most lethal gynecologic malignancy (3, 4). High-grade serous ovarian cancers (HGSOCs), which represent approximately 70 to 80% of all EOCs, are characterized by marked genomic instability and molecular alterations associated with increased replicative stress (RS) (4). Almost 50% of HGSOCs are defective in the homologous recombination (HR) DNA repair pathway making these HR-deficient (HRD) patients initially highly sensitive to Pt-based chemotherapy and to PARPi (5). Yet, the development of progressive Pt/PARPi-resistant relapses, also in HRD patients, has been consistently reported and profoundly affects on subsequent treatments and patients’ survival (6, 7). Pt resistance is a strong predictor for PARPi resistance, indicating that they probably share common mechanisms (7). Elucidating these biologic mechanisms is crucial to find new and effective therapeutic options for advanced patients with EOC.
In this context, we have recently demonstrated that ubiquitin-specific protease 1 (USP1) mediates EOC cell response to Pt, pointing USP1 as a promising target for anticancer therapies for patients with EOC (8). USP1 belongs to the family of deubiquitinases (DUBs) balancing several key ubiquitination processes, in particular in response to DNA damage. It deubiquitinates the Fanconi Anemia (FA) proteins, FANCD2 and FANCI to regulate the FA pathway (9, 10). USP1 also plays an important role in regulating translesion synthesis and replication fork protection events via the deubiquitination of proliferating cell nuclear antigen (PCNA) (11–13). Moreover, it has been reported that USP1 inhibition is efficacious in killing BRCA1-deficient cells and in a subset of BRCA1/2 wild-type (WT) tumors (13, 14). Yet, it is still poorly clarified whether targeting USP1 in combination with PARPi could represent a powerful strategy to treat cancer cells regardless of their BRCA1/2 mutational status and a novel hopeful therapy option for Pt/PARPi-resistant patients. Here, we filled this knowledge gap and demonstrated why and how USP1-PARP1 interaction is a previously undisclosed vulnerability to effectively target treatment-resistant EOC.
RESULTS
USP1 inhibition potentiates the efficacy of PARPi irrespective of HR status
On the basis of the notion that USP1 has a relevant role in replication fork protection (12, 13, 15), we explored the role of USP1 in RS and the subsequent activation of DNA repair machinery in the context of the response to chemotherapy in EOC. We used a panel of seven EOC cell lines as a model of high RS and genomic instability (16), all harboring mutant p53 but with different BRCA1/2 mutational and HR status (Fig. 1A). USP1 inhibition, using the selective inhibitor SJB3-019A (hereafter SJB), induced a marked activation of DNA damage response and of genomic instability as displayed by Comet assay, phosphorylation of H2AX and micronuclei (MN) formation in both BRCA-deficient (HRD) and BRCA-proficient (HRP) cell lines (Fig. 1, B to D, and fig. S1, A and B). Increased DNA damage and genomic instability were associated with the induction of stalled replication fork (Fig. 1E) and activation of DNA replication checkpoint, biochemically identified by the phosphorylation of the Replication Protein A2, RPA32 and Checkpoint Kinase 1 (CHK1) proteins and by the autophosphorylation of the Ataxia Telangiectasia And Rad3-Related protein (ATR) (Fig. 1F and fig. S1C). These biochemical modifications could be prevented by the concomitant use of the ATR inhibitor, AZD-6738 (Fig. 1F and fig. S1C) and were accompanied by increased formation of nuclear foci for single-stranded DNA as revealed by analyses of 5-bromo-2′-deoxyuridine (BrdU), RPA32, and TP53-Binding Protein 1 (53BP1) proteins (Fig. 1G and fig. S1, D and E), overall indicating that USP1 inhibition activated an ATR-dependent DNA replication checkpoint independently of the HR status of EOC cells.
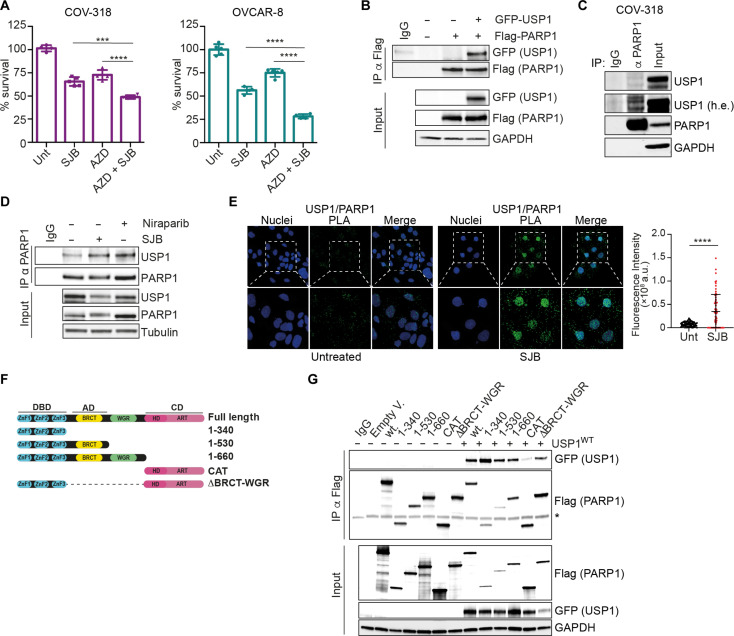
Fig. 1. USP1 inhibition increases RS.
(A) Table reporting EOC cell lines used. p53 and BRCA1/2 mutational status are indicated according to literature data and HR status according to experimental analysis using AmoyDx HRD focus panel. (B and C) USP1 inhibitor SJB (500 nM) increases DNA strand breaks as demonstrated by alkaline comet assay (B) and Western blot (WB) of H2AX phosphorylation (γH2AX) (C). In (B) data are presented as the mean ± SD of the percentage of DNA content in the tails (>50 cells and >30 cells counted/condition for COV-318 and OVCAR-8, respectively; n = 3). Representative images of the comets were reported. (D) Graphs reporting the percentage of micronuclei (MN) in EOC cells treated or not with SJB (500 nM) for 3 hours. Data are presented as mean ± SD (>500 cells and >300 cells counted/condition for COV-318 and OVCAR-8 respectively; n = 3). (E) Graphs reporting fork symmetry in COV-318 and OVCAR-8 cells treated with SJB 500 nM for 3 hours according to the label scheme reported over the graphs. Data are presented as the mean ± SD of CldU/IdU ratio of DNA fibers. Representative images of the fibers were reported under the graphs (>50 fibers counted/condition, n = 3). (F) WB analysis of replicative stress (RS) markers in COV-318 cells treated with SJB (300 nM) and ATRi (5 μM) (AZD-6378) for the indicated time points. (G) OVCAR-8 cells were treated or not with SJB 500 nM for 3 hours and 53BP1 nuclear bodies (NBs) were detected by immunofluorescence (IF). The graph reports the percentage of cells positive for 53BP1 (NBs) (two biological replicates performed in duplicate). (H) Graph reporting fork symmetry in the indicated OVCAR-8 cells according to the label scheme reported above. Data are presented as the mean ± SD of CldU/IdU ratio of DNA fibers. Representative images of the fibers were reported (>40 fibers counted/condition, n = 3). Two-tailed, unpaired Student’s t test (*P < 0.05, **P < 0.01, and ****P < 0.0001).
Then, by generating different USP1 knockout (USP1KO) EOC cell lines using the CRISPR-Cas9 technology, we confirmed that also genetic inhibition of USP1 resulted in the enhancement of RPA32 and CHK1 phosphorylation, accompanied by a stalling of the replication fork (Fig. 1H and fig. S1, F and G). Stalled replication fork observed in USP1KO cells could be prevented by the re-expression of USP1WT protein but not by the expression of the catalytic inactive USP1C90S mutant (Fig. 1H), in line with the observation made using the SJB inhibitor. Yet, phenotypically, the abrogation of USP1 expression resulted in reduced cell proliferation rather than in increased DNA strand break formation (fig. S1, H and I), as expected for a chronic rather than acute USP1 inhibition. Overall, these results demonstrated that, in both HRD and HRP EOC cell lines, USP1 inhibition induced a high RS accompanied by stalled replication fork and activation of DNA replication checkpoint.
RS is a key vulnerability of cancer cells that provides a potential target of antitumor treatment sensitization as for instance demonstrated by the successful clinical use of PARPi (16). We thus evaluated whether USP1 inhibition could improve the therapeutic benefit of PARPi in a broad panel of EOC cell lines. To this aim, we first treated OVCAR-8 USP1KO cells with increasing doses of SJB alone to confirm the specificity of the inhibitor and to choose the dose able to reduce by 25% cell survival (IC25) avoiding off-target toxicities (fig. S2A). This IC25 dose was then used in combination with PARPi. Treatment with SJB enhanced the sensitivity to PARPi of both HRP and HRD cells and significantly reduced niraparib IC50 in all tested EOC cell lines (Fig. 2A and fig. S2B). Similar results were observed using the USP1-specific inhibitor, KSQ-4279 (KSQ), which is now being tested in an interventional clinical trial in patients with advanced solid tumors (NCT05240898). Both SJB and KSQ increased mono-ubiquitination of FANCD2 and PCNA, two of the most validated targets of USP1 and also KSQ significantly improved the activity of niraparib in both HRD and HRP EOC models (fig. S2, C to E). Moreover, fluorescence-activated cell sorting (FACS) analysis on cells treated with SJB for 16/24/48/72 hours, showed no significant alterations in cell cycle distribution upon USP1 pharmacological inhibition (fig. S2F), suggesting that the effect on cell viability after USP1 inhibition was not due to an altered distribution of cell cycle phases, and likely not dependent on its role on PCNA or FANCD2 proteins activity. We then tested whether USP1i/PARPi combination treatment was synergic in killing OVCAR-8 and COV-318 cells. We used equipotent doses of SJB and niraparib (IC10:10, IC25:25, and IC50:50 cytotoxic ratio) or an excess of either agent (IC10:25, IC10:50, and IC25:50 cytotoxic ratio) for 72 hours and calculated the corresponding highest single agent (HSA) score. As reported by the synergy maps shown in Fig. 2B, we observed that USP1 and PARP1 pharmacological inhibition was strongly synergic in both HRP and HRD cells (Fig. 2B). More importantly, USP1KO cells treated with PARPi in combination or not with SJB demonstrated that all tested USP1KO models are more sensitive to the PARPi niraparib, confirming that USP1 inhibition was synthetic lethal with PARPi, independently on the HR status of the used models and that SJB had not significant off-target effects in our models (Fig. 2C and fig. S3, A and B).
Fig. 2. USP1 inhibition potentiates the efficacy of PARPi in both HRD and HRP EOC cells.
(A) Nonlinear regression analysis of cell viability assays in COV-318 and OVCAR-8 cells treated for 16 hours with increasing doses of PARPi, niraparib (Nir.), in combination, or not with SJB (COV-318 200 nM; OVCAR-8 100 nM). (B) Two-dimensional synergy maps highlighting synergistic and antagonistic dose regions in red and green colors, respectively, as indicated on the right. The darker red boxes represent the most synergic area. Highest single agent (HSA) scores are reported (HSA score > 10 = synergism; HSA score < −10 = antagonism). (C) Nonlinear regression analysis of cell viability assays in COV-318 and OVCAR-8 USP1WT and USP1KO cells treated for 72 hours with increasing doses of niraparib. (D) Nonlinear regression analysis of cell viability assays in OVCAR-8 Pt-sensitive and Pt-res cells treated for 72 hours with increasing doses of niraparib. (E) Nonlinear regression analysis of cell viability assays in OVCAR-8 Pt-res cells treated for 72 hours with increasing doses of niraparib, in combination, or not with SJB (200 nM). In (A) and (C) to (E), data are expressed as percentage of viable cells with respect to the untreated cells and represent the mean (±SD) of three biological replicates. Fisher’s exact test was used to calculate the global P value reported in the graphs. The tables below the graphs show the IC50 and the confidence interval (CI) of each condition.
Resistance to Pt therapy is strongly predictive of resistance to PARPi therapy consistent with overlapping biologic mechanisms (6). We generated several isogenic Pt-resistant (Pt-res) models of EOC (17–19) that all exhibited cross-resistance to PARPi (Fig. 2D and fig. S3C). Using these models, we found that the combined regimen of both USP1 and PARP inhibitors in Pt-res clones significantly reduced cell viability respect to the single treatment with niraparib (Fig. 2E and fig. S3D), emphasizing that USP1/PARPi axis could be a powerful option of therapy for both Pt-sensitive and, more importantly, patients with Pt-res EOC regardless of their HR status.
USP1 regulates K63-linked PARP1 deubiquitination
The main target of PARPi is PARP1, the founding member of PARP family (1, 2). Therefore, we tested, by cell viability assay, whether USP1 inhibition potentiated the efficacy of AZD9574, a PARP1 selective inhibitor (with an 8000-fold selectivity for PARP1 compared to the other members of the PARP family) (20), indicating that the observed synergistic effect of USP1i and PARPi is principally mediated by the inhibition of PARP1 (Fig. 3A). Accordingly, in PARP1-silenced cells, SJB and AZD9574 used alone or in combination had no effects on cell viability, indicating that USP1i/PARPi-induced cell death is mediated by PARP1 expression (fig. S4A). Together, these observations implied that USP1 could increase PARPi activity by directly and specifically acting on PARP1. Accordingly, we found that USP1 interacted with PARP1 both when exogenously overexpressed and at endogenous level in all tested models (Fig. 3, B and C, and fig. S4, B and C). We also observed that this interaction occurred preferentially into the nucleus and increased upon USP1 or PARP1 inhibition (Fig. 3, D and E, and fig.S4D). Since PARP1 and USP1 are DNA binding proteins, we performed coimmunoprecipitation (co-IP) analysis in OVCAR-8 whole lysates treated with benzonase to remove nucleic acid contaminants and we observed that USP1 and PARP1 interact also in absence of DNA, supporting their direct interaction rather than a DNA bridging event (fig. S4E). USP1/PARP1 interaction was observed also in G1-blocked cells, although with lesser efficiency (fig. S4, F and G), again suggesting that the synergistic effect observed with their combined inhibition was not cell cycle specific and likely not linked to the activity of USP1 on PCNA or FANCD2 proteins.
Fig. 3. PARP1 is a target of USP1.
(A) Graphs representing cell viability of COV-318 and OVCAR-8 cells after 16 hours of treatment with the specific PARP1 inhibitor AZD-9574, SJB, or the combination (COV-318: SJB 200 nM and AZD 500 μM; OVCAR-8: SJB 100 nM and AZD 250 μM). Data are expressed as percentage of viable cells with respect to the untreated conditions and represented the mean (±SD). (B and C) Coimmunoprecipitation (co-IP) analysis of exogenous in 293T17 (B) or endogenous in COV-318 cells (C) interaction between USP1 and PARP1 proteins. (D) Co-IP analysis of endogenous USP1 and PARP1 interaction in OVCAR-8 cells treated or not with SJB (500 nM) or niraparib (1μM) for 3 hours. (E) Representative confocal microscopy images of endogenous PARP1/USP1 proximity ligation assay (PLA) in OVCAR-8 cells treated or not with SJB (500 nM) for 3 hours. White dashed boxes highlight the areas magnified in the panels below. The graph on the right report the fluorescence intensity/cells for each condition (>60 cells analyzed per condition, n = 3). (F) Schematic representation of PARP1-truncated mutants used in this work. (G) Co-IP analysis performed in whole lysates of 293T17 cells transfected with FLAG-tagged plasmids for PARP1 described in (F) together with plasmid encoding GFP-tagged USP1 (* = aspecific band). In (B), (C), (D), and (G), input indicates the expression of PARP1 and USP1 in cell lysates. Immunoglobulin G (IgG) represents the control IP using an unrelated antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or tubulin was used as loading controls. In (A) and (E), two-tailed, unpaired Student’s t test was used (***P < 0.001 and ****P < 0.0001).
To map PARP1 domain(s) necessary for its interaction with USP1, we used different truncated mutants of PARP1 and demonstrated that the DNA binding domain (DBD) of PARP1 was necessary for the interaction with USP1 (Fig. 3, F and G). Accordingly, in silico docking analysis supported this observation (fig. S4H).
On the basis of the notion that PARP1 could undergo to ubiquitin-mediated degradation (21), we tested whether USP1 could act on PARP1 protein stability. Consistently with its DUB role, USP1 depleted cells had an increased amount of polyubiquitinated form of PARP1, which was completely abolished after treatment with the ubiquitin-activating enzyme inhibitor MLN-7243 (fig. S5, A and B). Moreover, the overexpression of USP1WT but not of the inactive form USP1C90S (9) reduced PARP1 polyubiquitination (Fig. 4A). Nevertheless, by treating USP1WT and USP1KO cells with cycloheximide (CHX), we observed that USP1 inhibition did not decrease PARP1 protein half-life (fig. S5C). Together, these data suggested that USP1 deubiquitinates PARP1, but it is not involved in the regulation of its protein stability that, conversely, seems to be linked to the activity of USP15 in triple-negative breast cancer model (22). Polyubiquitination could have different outcome on the modified substrate depending on the type of specific linkage. In particular, K48 polyubiquitination targets protein substrates to proteasome degradation, while K63 linkage-specific ubiquitination regulates a variety of nonproteolytic cellular functions, including DNA damage repair (fig. S5D) (23). Using specific antibodies that distinguish between these two different polyubiquitin chains, we demonstrated that K63 but not the K48-linked polyubiquitination of PARP1 was regulated by USP1 expression (Fig. 4B and fig. S5E), supporting the possibility that USP1 regulates PARP1 activity rather than its stability.
Fig. 4. USP1 regulates PARP1 K63-linked ubiquitination.
(A) His-Ubiquitin pull down assay of Flag-PARP1 in 293T17 cells co-transfected with His-tagged ubiquitin, PARP1, USP1WT, or USP1C90S. (B) Endogenous PARP1 ubiquitination analysis in OVCAR-8 USP1WT or KO cells transfected with His-tagged ubiquitin. Whole-cell lysates were subjected to IP with antibody against PARP1 and immunoblotted for specific K63-linked ubiquitin chains. (C) Co-IP analysis on 293T17 cells lysates transfected with His-tagged ubiquitin and treated with the inhibitors as indicated for 3 hours (SJB 800 nM and Nir./ML-792/MLN-7243 1 μM). α-PARP1 antibody was used for the immunoprecipitation. (D) Nonlinear regression analysis of cell viability assay in OVCAR-8 cells transfected with Flag-tagged PARP1 full-length (FL) or ΔBRCT/WGR deletion mutant and treated with increasing doses of niraparib together or not with SJB (50 nM). Overexpression was checked by WB analysis. Data and statistical analysis were calculated and expressed as described in Fig. 2. The table below the graph shows the IC50 and the CI of each condition. In (A) to (C), input indicates the expression of PARP1, USP1, and ubiquitin in cell lysates. IgG represents the control IP using an unrelated antibody. GAPDH or tubulin was used as loading controls.
Recent evidences demonstrated that a stepwise SUMOylation-ubiquitination modification of PARP1 trapped on the DNA was necessary to remove cytotoxic bound PARP1 from chromatin through the recruitment of the p97 ATPase (24). We thus tested whether USP1 is a DUB enzyme possibly involved in this pathway. To this aim, we treated OVCAR-8 cells with SJB and niraparib in combination or not with the SUMOylation inhibitor ML-792 or with the ubiquitin-activating enzyme inhibitor MLN-7243. We confirmed that USP1 inhibition, alone and better in combination with niraparib, enhanced PARP1 ubiquitination (Fig. 4C and fig. S5F). These modifications induced by USP1 inhibition were almost completely abolished when cells were treated with MLN-7243, confirming that USP1 regulates PARP1 polyubiquitination. Yet, the use of ML-792 (SUMOylation inhibitor) had no/little effects on SJB-induced PARP1 ubiquitination (Fig. 4C), suggesting that USP1-mediated deubiquitination of PARP1 is not primarily involved in the described Protein Inhibitor Of Activated STAT Protein 4 (PIAS4)/Ring Finger Protein 4 (RNF4)/p97 axis and does not require the PARP1 SUMOylation step.
Next, using both His-ubiquitin pull down assays and in silico protein structure analysis, we mapped the USP1-induced deubiquitination of PARP1 into the region comprising the BRCT and the tryptophane (W)–, glycine (G)–, arginine (R)–rich domain (WGR domain), which, together with Zn1/3, binds to DNA, forming interdomain contacts to link the DNA damage interface to the catalytic domain (CAT) (fig. S5, G and H) (25). We found that USP1-mediated deubiquitination of PARP1 had biological consequences as demonstrated by overexpression of PARP1 full length but not of its ΔBRCT/WGR mutant (which lacks WGR domain but maintains both DNA binding and CAT domains) in cells then treated with niraparib, in combination or not with SJB. Cell viability assay and Western blot analysis showed that, as expected, ΔBRCT/WGR-deleted mutant was more resistant to PARPi and “insensitive” to niraparib-induced DNA trapping, since the lack of WGR domain presumably disrupted the allosteric communication required for PARP1 trapping, contributing to PARPi resistance (Fig. 4D and fig. S5I). Cells expressing the ΔBRCT/WGR mutant showed unresponsiveness also to the PARPi + USP1 inhibitor combination (Fig. 4D), reinforcing the concept that USP1-mediated deubiquitination of PARP1 has a relevant role in regulating the sensitivity of EOC cells to PARPi.
USP1 inhibition induces PARP1 trapping
As mentioned above, the Zn1, Zn3, and WGR domains of PARP1 are essential to support DNA-dependent PARP1 enzymatic activity, since they lead, after DNA binding, to a marked conformational change that facilitates the access of the substrate nicotinamide adenine dinucleotide (NAD+) and activates the CAT (25). Mutations/alterations in these domains, by disrupting interdomain allosteric communication, reduced the poly-ADP-ribosylation (PARylation) activity (25–27). The data collected so far indicated that USP1 is necessary to regulate PARP1 deubiquitination on its BRCT-WGR.Consistently, USP1 inhibition, inducing a persistent K63-linked chain [which predominantly forms an extended conformation (28)] could prevent the “activation” of the CAT domain due to steric hindrance of the polyubiquitin chain.
Therefore, we tested whether USP1 inhibition could affect global PARylation activity of PARP1 and observed that USP1 impairment resulted in a massive abrogation of PARylation in all tested EOC cells, in both dose- and time-dependent manner independently on their BRCA1/2 status (Fig. 5, A and B, and fig. S6, A and B). Of note, using an enzyme-linked immunosorbent assay (ELISA) kit measuring PARP1 activity on histone proteins, we observed that the amount of PAR chains deposited by PARP1 was significantly reduced by both SJB and niraparib treatments with no significant differences between the two inhibitors (Fig. 5C and fig. S6C). Accordingly, in USP1KO cells, niraparib had a ~10 fold higher activity in inhibiting PARylation respect to USP1WT cells (Fig. 5D and fig. S6D). In line with the above experiments, we also observed that USP1 inhibition prevented PARP1 auto-PARylation and that it was regulated by the overexpression of USP1WT but not of the catalytic inactive form USP1C90S (Fig. 5E and fig. S6E). Together, these data confirmed that USP1-mediated deubiquitination of PARP1 is fundamental to regulate its catalytic activity.
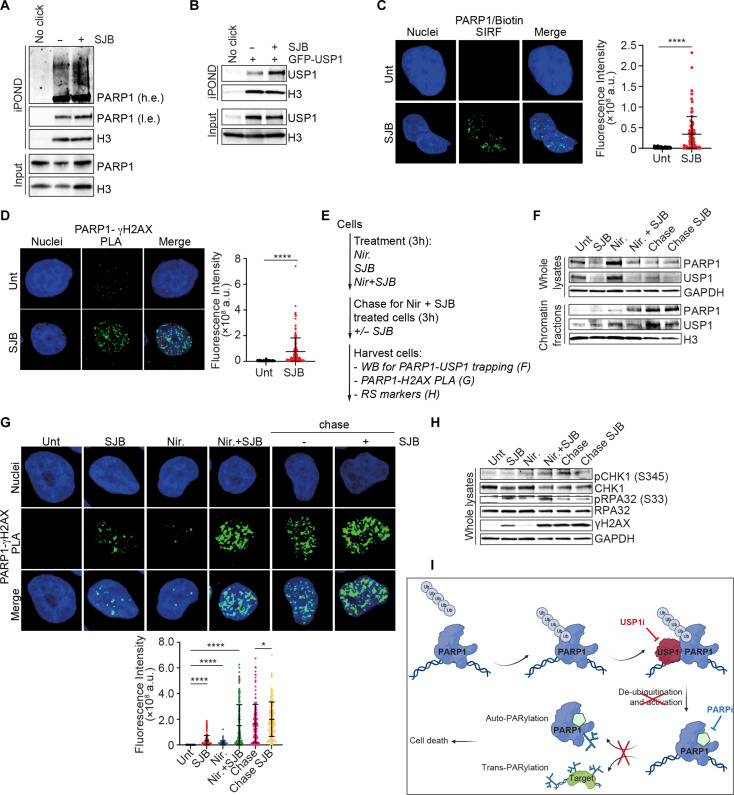
Fig. 5. USP1 inhibition impairs PARylation and induces PARP1 trapping.
(A and B) WB analysis of PAR content in EOC cells treated with SJB (500 nM for OVCAR-8 and 300 nM for COV-318) for 3 hours (A) or in COV-318 as indicated (B). (C) PARP1 activity analyzed by ELISA in COV-318 cells treated with SJB (300 nM) or niraparib (Nir) (1 μM) for 16 hours. Results are expressed as mUnits PARP/ng protein (mean ± SD of three biological determinations). (D) WB analysis of PAR content in whole lysates of OVCAR-8 USP1WT or USP1KO cells treated or not with niraparib as indicated for 24 hours. (E) Co-IP analysis of endogenous PARP1 auto-PARylation in OVCAR-8 cells treated with SJB (500 nM) or niraparib (1 μM) for 3 hours. (F) WB analysis of USP1 and PARP1 trapping in whole lysates and chromatin fractions of COV-318 cells treated with SJB (300 nM) as indicated. (G) Representative images showing GFP-PARP1 accumulation at laser-induced DNA damage (dashed boxes) at the indicated time points in OVCAR-8 cells treated or not with SJB (500 nM, 3 hours). (H and I) Graphs representing the percentage of recovery of the damaged area over time (H) and at last recorded frame (4 min) (I) (>40 cells were considered/condition). Two-tailed unpaired t test with Welch’s correction was used (***P < 0.001). (J and L) Co-IP analysis of endogenous USP1 and PARP1 interaction (J) and PARP1 polyubiquitination (L) on chromatin of OVCAR-8 cells treated with SJB (500 nM) for 3 hours. (* = aspecific band). (K) Representative confocal microscopy images of endogenous PARP1 and ubiquitin (Ubi) PLA in OVCAR-8 cells overexpressing His-ubiquitin and treated or not with SJB (500 nM, 3 hours). White dashed boxes highlight the magnified areas. The graphs on the right report the fluorescence intensity/cells for each condition (>100 cells analyzed, n = 3). (M) WB analysis of PARP1 expression on whole lysates and chromatin fractions of OVCAR-8 overexpressing Flag-PARP1 FL or ΔBRCT/WGR mutant and treated as in (J). In (C) and (K), two-tailed, unpaired Student’s t test was used (*P < 0.05 and ****P < 0.0001)
PARP1 catalyses auto-PARylation to lead its release from DNA and allow DNA repair and replication to proceed (2). In line with this notion, we found that SJB treatment induced not only USP1 but also PARP1 trapping in both dose- and time-dependent manner (Fig. 5F and fig. S6, F to H). To support the involvement of USP1 in PARP1 trapping and check whether USP1 inhibition impairs also the recruitment of PARP1 to DNA damage sites, we performed laser microirradiation on OVCAR-8 cells transfected with green fluorescent protein (GFP)–tagged PARP1. As expected, we found that GFP-PARP1 quickly localized to DNA lesions, but its recruitment was not affected by SJB treatment (Fig. 5, G and H, and movies S1 and S2). Conversely, we observed that, 4 min after laser-induced DNA damage, USP1 inhibition kept a persistent PARP1 localization on the lesions respect to the untreated cells (Fig. 5I and movies S1 and S2), indicating that USP1 did not affect PARP1 recruitment to the site of damage but regulated PARP1 trapping on damaged DNA.
All the data described above supported the possibility that USP1/PARP1 interaction occurred preferentially on DNA and that USP1 inhibition prolonged PARP1 presence on the damaged DNA. Using co-IP on chromatin fraction, chromatin immunoprecipitation (ChIP) assay and in silico docking analysis, we confirmed this hypothesis (Fig. 5J and fig. S6, I and J). Moreover, in USP1KO cells, we found that PARP1 increased in the chromatin fraction with a smeared banding pattern respect to the USP1WT cells (fig. S6K). Accordingly, proximity ligation assay (PLA) and co-IP analysis confirmed that the increment of PARP1 polyubiquitination after USP1 inhibition occurred preferentially in the nucleus and specifically on chromatin-bound PARP1 (Fig. 5, K and L, and fig. S6L). Moreover, the ΔBRCT/WGR-deleted mutant, insensitive to USP1 regulation (Fig. 4D and fig. S5I), was more bound on chromatin respect to PARP1 full-length protein and its trapping was not affected by SJB treatment (Fig. 5M), suggesting that USP1-mediated deubiquitination of PARP1 may regulate its attachment/detachment balance on DNA.
PARPi exerts their cytotoxic effects mainly by trapping PARP1 on DNA (29). Yet, using three different PARPi, specifically olaparib, veliparib, and talazoparib alone or in combination with USP1 inhibitor, we observed that SJB treatment not only strengthened the killing efficacy of all PARPi used but also boosted their trapping capacity and DNA damage activation, irrespective of their trapping potency (fig. S7, A and B).
Data collected so far indicated that USP1 inhibition, like PARPi treatment, could induce USP1 and PARP1 trapping on DNA specifically at the replication fork, thus explaining why it induced stalled replication fork and activation of replication stress (Fig. 1). To verify this possibility, we used iPOND (isolation of proteins on native DNA) and SIRF (single-cell assay for in situ protein interaction with nascent DNA replication forks) analyses and demonstrated the presence of both USP1 and PARP1 on nascent DNA in basal condition and their enrichment upon SJB treatment, in line with the observed stalled replication fork (Fig. 6, A to C and fig. S7C). Moreover, detecting the proximity of PARP1 and USP1 to phosphorylated H2AX (γH2AX) by PLA after SJB treatment, we confirmed that they both localize at the sites of DNA damage, likely as a consequence of fork collapse (Fig. 6D and fig. S7, D to F).
Fig. 6. USP1-PARP1 interaction is localized on replication fork.
(A and B) iPOND analyses of endogenous PARP1 (A) and exogenous USP1 (B) at replication forks in OVCAR-8 cells transfected (B) or not (A) with GFP-tagged USP1 and treated with SJB (500 nM) for 3 hours. Inputs represent 2% of the total cellular protein content. (C) Representative confocal microscopy images of SIRF analysis in OVCAR-8 cells treated with SJB (500 nM) for 3 hours. Green dots represent PARP1/biotin PLA foci. (D) Representative confocal microscopy images of PLA analysis between USP1-γH2AX in COV-318 cells treated or not with SJB (300 nM) for 3 hours. (E to H) WB analysis of USP1 and PARP1 (F) or RS markers (H) expression or images from PARP1/γH2AX PLA (G) (>150 cells counted in each condition, n = 3) in OVCAR-8 cells used for a trapping–chase experiment according to the experiments scheme reported in (E) (500 nM SJB, 1 μM Nir). In (C), (D), and (G), the graphs report the fluorescence intensity/cells for each condition expressed with mean ± SD of quantifications of green dots for each cell. Two-tailed, unpaired Student’s t test was used (*P < 0.05 and ****P < 0.0001). (I) Schematic representation of the proposed USP1-mediated processing of PARP1 deubiquitination. Created with BioRender.com. In (A) and (B), Input indicates the expression of PARP1 and USP1 in cell lysates. IgG represents the control IP using an unrelated antibody. GAPDH was used as loading control for whole lysates, Histone H3 for chromatin fractions.
It has been observed that PARPi binding to the CAT of PARP1 causes its reversible inhibition (30). This observation has been confirmed in our EOC models treated with niraparib for 3 hours and then released in PARPi-free medium. After niraparib withdrawal, chromatin-bound PARP1 was almost completely removed and its PARylation activity restored (fig. S7G). Conversely, when niraparib was used in combination with the USP1 inhibitor, we observed not only that the combined regimen boosted PARP1 and USP1 trapping on DNA, but, importantly, that SJB prevented the reversible effect of niraparib keeping PARP1 trapped on DNA damage sites even after PARPi replacement, resulting in the activation of RS and DNA damage (Fig. 6, E to H, and fig. S7H). Overall, the data collected here indicated that USP1 activity is necessary to deubiquitinate PARP1 at the site of DNA damage thereby modulating PARP1 dynamic activity necessary for the repair of damaged DNA (Fig. 6I).
USP1 inhibitor strengthens the efficacy of PARPi in Pt/PARPi-resistant in vivo models
To further investigate whether our in vitro findings translated to a clinically relevant context, we first tested the efficacy of USP1/PARP1 inhibitors combination in six primary cultures isolated from the ascites of four EOC with HGSOC (fig. S8A and table S1). The treatment with USP1 inhibitor effectively increased the efficacy of PARPi in all tested models among which we also included primary cultures from PARPi-relapsed patients (Fig. 7A).
Fig. 7. USP1/PARP1 inhibitors combination is effective also in in vivo models.
(A) Cell viability of HGSOC patient–derived primary cells treated for 72 hours with increasing doses of niraparib in combination or not with SJB (OV-213 = 300 nM; OV-194, OV-196, OV-202, OV-206, and OV-225 = 200 nM). (B) Nonlinear regression analysis of cell viability assays in ID8 p53−/−PTEN−/− cells treated as in (A) (SJB = 300 nM). In (A) and (B), data are expressed as percentage of viable cells with respect to the untreated cells and represent the mean (±SD) of three biological replicates. (C to E) Graph (D) and radar plot (E) reporting the number of tumor cells present in ascites and the distribution of abdominal metastasis in C57Bl/6 mice treated according to the scheme depicted in (C) (n = 4 mice for conditions). (F to H) Graph (G) and radar plot (H) showing the number and the distribution in abdominal organs of tumor masses in NSG mice injected with PDX#OV218.3 and treated as described in (F) (n = 5 mice for conditions). In (H), metastasis with >100 tumor cells were considered. In (E) and (H), colored lines indicate the number [0 to 4 in (E); 0 to 5 in (H)] of mice affected for each district. (I) Typical images of hematoxylin and eosin (H&E) staining of liver metastasis in mice described in (F) to (H). 5× and 10× (dashed boxes indicate magnified areas) images of the same field are shown. (J and K) WB analysis of RS markers on tumor cells collected from ascites (J) and IF of γH2AX on tumor masses in abdominal cavity (K) collected from mice in (F) to (H). On the right, γH2AX foci positive cells (>5 foci/cell) were counted: Data are presented as the mean ± SD of the percentage of positive cells for each field (>280 cells/field counted in each condition). Representative images were reported in (K). Two-tailed, unpaired Student’s t test was used (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
To the best of our knowledge, SJB inhibitor has never been used in mouse models. We thus first tested its tolerability in vivo in C57Bl/6 mice. Mice treated with SJB used alone or in combination with PARPi showed no sign of suffering and blood tests revealed that it was not toxic and neither increased niraparib hematological toxicity (fig. S8B), supporting the feasibility of using this therapeutic regimen in vivo.
Pt- and PARPi-resistant tumors represent the most clinically reasonable EOC diseases to test the usefulness of USP1/PARPi combination. We thus evaluated the effect of SJB treatment in combination with niraparib in CRISPR-Cas9–modified murine model of HGSOC, ID8 double knockout (KO) for the Tumor Protein P53 (TP53) and the Phosphatase and tensin homolog (PTEN) genes (ID8 p53−/−/PTEN−/−) (31, 32), which displayed increased resistance to both Pt and PARPi treatments respect to ID8 TP53 KO cells (fig. S8, C and D). Also, in this model, USP1 inhibition significantly resensitized PTEN KO cells to niraparib treatment, recapitulating what observed in human-derived cell lines (Fig. 7B).
We next assessed the intraperitoneal (i.p.) growth and spreading of ID8 p53−/−/PTEN−/− in female C57Bl/6 mice treated with niraparib in combination with USP1 inhibitors, SJB or KSQ following the schemes reported in Fig. 7C. These cells displayed a very aggressive behavior when injected i.p. in mice, inducing the formation of huge volume of hemorrhagic ascites and high tumor deposits on abdominal organs, in particular on peritoneal wall and diaphragm surface (fig. S8E) (32). In this scenario, we demonstrated that mice treated with the combination of USP1 and PARP inhibitors presented lower amount of ascites with a reduced number of tumor cells (Fig. 7D and fig. S8F). Moreover, histopathological analyses strongly pointed out that SJB/KSQ-niraparib combined regimen limited the metastatic dissemination of ID8 p53−/−/PTEN−/− cells (Fig. 7E and fig. S8G).
Similar results were obtained in nonobese diabetic severe combined immunodeficient gamma (NSG) mice injecting i.p. a patient-derived xenograft model (PDX#OV218.3) established in our laboratory from a patient with BRCA1-mutated HGSOC with acquired Pt/PARPi resistance according to her clinical history (Fig. 7F, fig. S9A, and table S1). Macroscopic measure of tumor spreading clearly revealed that the cotreatment with both SJB and niraparib led to a significant reduction, relative to single-agent therapies, in the amount of tumor masses and in the number of tumor cells in ascitic fluids (Fig. 7G and fig. S9B). Tumor infiltration of the organs in the peritoneal cavity and the number of metastases were also significantly reduced in combo-treated mice (Fig. 7, H and I, and fig. S9C). Last, in accordance with the in vitro data, USP1 inhibition alone, and better in combination with PARPi, activated the replication stress pathway and increased the presence of DNA damage in vivo (Fig. 7, J and K, and fig. S9D).
Together, these data supported the importance to restrain USP1/PARP1 axis to not only improve response to chemotherapy and overcome resistance but also lessen the associated tumor aggressiveness and invasion capabilities.
DISCUSSION
PARPi have drastically transformed the management of patients with EOC, especially for those harboring defects in genes involved in HR pathway. Unfortunately, despite the initial exciting responsiveness to PARPi, increasing numbers of patients experience disease progression due to the development of resistance. In this context, combination strategies involving PARPi are currently tested in multiple clinical trials to enhance efficacy and overcome resistance through synergistic activity. In this scenario, different groups have suggested also USP1 as a potential therapeutic target to increase PARPi response but only in a subset of Pt-sensitive and BRCA1/2-deficient tumors (13, 14, 33).
In our work, we clearly demonstrated that USP1 inhibition/abrogation potentiates the response against PARPi irrespective of HR status and also in EOC recurrent resistant disease, which is currently uncurable. Our in vitro and in vivo data pointed the extreme efficacy of the combination of USP1/PARP1 inhibitors that might merit to be tested in future clinical trials in particular for patients with Pt/PARPi-resistant EOC who, until now, do not have valid therapeutic opportunities. Resistance to PARPi could depend on multiple causes, first of all, on restoration of HR DNA repair pathway, for instance, by BRCA1/2 mutation reversion (34). The fact that USP1i improves PARPi activity in both HRP and HRD models and in ovarian cancer models that become resistant to Pt and PARPi suggests that in these settings, the combination treatment we propose here could have promising clinical applications.
From a molecular point of view, similarly to what observed in other cancer models (22, 35), we identified PARP1 as a key substrate of USP1. We demonstrated that USP1 binds to the DBD of PARP1 to promote the removal of K63-linked polyubiquitin chains from its BRCT-WGR domain, and regulate its enzymatic activity rather than its protein stability. Accordingly, loss of PARP1 deubiquitination due to USP1 inhibition could induce in the BRCT-WGR domain of PARP1, a steric hindrance of the persistent polyubiquitin chains that could prevent the “activation” of the CAT domain retaining, in turn, PARP1 trapped on chromatin, impairing its PARylation activity and inducing cell death (Fig. 6I). While this manuscript was in preparation, others demonstrated that USP1 regulates PARP1 stability in cholangiocarcinoma cells (35). Therefore, it would be interesting to understand whether this activity of USP1 on PARP1, not observed in ovarian cancer (this manuscript) or in triple-negative breast cancer (which is known to be genetically more similar to high-grade ovarian cancer) (22), is dependent on the cancer model tested or on some USP1 posttranslational modification (e.g., acetylation) associated with the progression of different tumor types.
Acquired PARPi resistance has been linked to point mutations in PARP1 that decrease the trapping ability of PARPi (27). Our observations that USP1 contributed to the trapping of PARP1 on the damage lesions and that USP1i prolonged the presence of PARP1 at the damage sites suggested that USP1 inhibitors could also be active in the presence of decreased trapping abilities of PARPi. This possibility is in line with our provided evidences on the fact that USP1i increased the sensitivity to different PARPi, irrespective of their trapping potency and in multiple PARPi-sensitive and PARPi-resistant models. Yet, whether USP1i could restore the sensitivity to PARPi in cells with acquired PARP1 mutations (e.g., PARP1R591C) is something that should be proved in future works.
Recently, Lord’s group described a new molecular cascade mediated by PIAS4/RNF4/p97 axis essential for the removal of trapped PARP1 from DNA (24). Our data suggest that USP1 is not involved in this stepwise mechanism, even if these differentially mediated posttranslational modifications could cooperate to regulate PARP1 activity and, in turn, influence the cellular response to PARPi. Further analysis will be useful to elucidate this point.
Both USP1 and PARP1 are involved in replication fork protection (12, 13, 15, 36, 37). We demonstrated that USP1/PARP1 interaction occurs preferentially and specifically on the replication fork, where USP1-mediated PARP1 deubiquitination seems to be necessary for its full activity in DNA damages repair. Laser microirradiation results suggested that USP1-mediated K63-linked deubiquitination on PARP1 was not important to drive PARP1 to the DNA damage site but only to regulate its dynamic attachment/detachment on the lesions (Fig. 6I). The identification of the lysine residue in the BRCT-WGR domains involved in USP1-mediated deubiquitination, together with structural and crystallography analyses, could help, in the future, to better clarify this point. Moreover, we have not defined here which are the ubiquitin ligases involved in PARP1 K63-linked ubiquitination. Yet, it has been shown that the E3 ubiquitin ligase, Rad18, is involved in the regulation of stalled replication forks by monoubiquitinating PCNA (38) and several reports disclosed the importance of USP1/Rad18/PCNA axis in fork protection during RS and in cell sensitivity after USP1 inhibitors treatment (13, 14). Accordingly, RAD18 was identified as a novel gene important for cell survival after PARPi (29). Thus, it would be worth in the future to verify whether Rad18 could be the E3 ligase involved in PARP1 K63-linked ubiquitination that balance USP1-mediated regulation of PARP1 trapping and activity.
In conclusion, our work elucidates a previously unknown mechanism of USP1 in the regulation of DNA damage repair and RS by targeting PARP1 to coordinate its DNA trapping and PARylation activity. This USP1/PARP1 axis could explain the synergistic activity of USP1 and PARP1 inhibitors when used in combination and could be an encouraging new therapeutic choice for patients with Pt/PARPi-resistant EOC regardless their HRD status.
MATERIALS AND METHODS
Human samples
Biospecimens were obtained from patients who gave their informed consent, under protocols approved on 7 October 2019 by the Ethics Committee (OutCoME protocol, CRO-2019-53 approval CEUR 2019-Sper-084). Patients’ data were pseudonymized and collected in a prospective database. Patient-derived primary cells were obtained from ascites of patients with EOC collected at Centro di Riferimento Oncologico of Aviano (CRO; table S1). First, primary tumor cells were separated from ascitic fluid by centrifugation at 1200 rpm for 10 min. Then, if necessary, red blood cells were removed from the pellet by applying the lysis buffer (lysing buffer, BD Bioscience) for 5 min at 37°C and recentrifuged at 1200 rpm for 5 min. Primary cells were plated in 150-cm2 flasks and maintained in OCMI medium [M199 medium + F12-HAM’s medium supplemented with insulin (20 μg/ml), hydrocortisone (500 ng/ml), epidermal growth factor (EGF) (10 ng/μl), 2% fetal bovine serum (FBS), 1% penicillin/streptomycin, cholera toxin (25 ng/ml), and 2% ascitic liquid].
Mice
Animal experimentation was reviewed and approved by CRO Institutional Organism for Animal Wellbeing and by the Italian Ministry of Health (aut. no.1261/2015-PR, released to G.B. and aut. no. 655/2023 released to M.So.). Female C57BL/6 or NSG (3/4 weeks old) were acquired from Charles River Laboratories and maintained in the animal facility of CRO in standard conditions.
C57Bl/6 and NSG mice were injected intraperitoneally with 2 × 106 ID8 P53−/− PTEN−/− or PDX (PDX#OV218.3), respectively. In each experiment, mice were randomly divided into four groups (four mice per group for C57Bl/6 and five mice per group for NSG) and treated by i.p. injection with SJB (20 mg/kg) or by oral gavage of KSQ (30 mg/kg) in combination or not with niraparib (30 mg/kg) by oral gavage according to the scheme reported in figures. Ascitic fluids, tumor masses, and abdominal tissues were collected for analysis of tumor dissemination. Formalin-fixed tissues were then dehydrated in ethanol and xylene before paraffin embedding and then stained with hematoxylin and eosin.
Cell cultures
OVCAR-8 cells were purchased from National Cancer Institute Developmental Therapeutics Program Tumor Repository; KURAMOCHI (JCRB0098) and OVSAHO (JCRB1046) cells were obtained from the Japanese Collection of Research Bioresources Cell Bank; COV-362 (07071910) and COV-318 (07081903) were from European Collection of Authenticated Cell Cultures; and TOV-112D (CRL-11731) and NIH:OVCAR-3 (HTB-161) were from American Type Culture Collection. All OC cell lines were maintained in RPMI-1640 medium (Gibco, no. 61870) containing 10% heat-inactivated FBS (Gibco, no. A5256701) and 1% penicillin/streptomycin. COV-318 are maintained in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, no. 61965) complete medium. Cisplatin-resistant (PT-res) EOC cells and USP1 KO cells establishment was described previously (8, 17). Human embryonic kidney (HEK) 293/T17 cells (ATC, CRL-11268) that have been used for overexpression strategies were grown in DMEM high-glucose complete medium. ID8 P53−/− PTEN−/− murine cells were provided by I. McNeish (Hammersmith Hospital, London) and grown in DMEM high-glucose complete medium added with ITS [insulin (5 μg/ml), transferrin (5 μg/ml), and sodium selenite (5 ng/ml)]. All cell lines were grown in standard conditions at 37°C and 5% CO2. All cell lines were routinely authenticated in our laboratory using the Cell ID TM System (Promega) protocol and using the PowerPlex 16 HS System to identify DNA short tandem repeat profiles. HR status of EOC cells used was verified using the AmoyDx HRD Focus panel (AmoyDx Diagnostics) according to the manufacturer’s protocol.
Growth curves
OVCAR-8 and COV-318 USP1WT and USP1KO cells were seeded in 12-well plates (50,000 and 30,000 cells per well, respectively). Viable cells were counted daily in triplicate for 7 days by the trypan-blue dye exclusion method.
Compounds and drug treatments
Dose-response curves were performed essentially as previously described (8). In brief, EOC cells were seeded in 96-well culture plates and, when necessary, transfected with overexpressing vectors as indicated. The day after seeding (or 72 hours after transfection), cells were treated with USP1 inhibitor SJB (MedChemExpress, no. HY-80012) or KSQ (Selleckchem, no. E1214) and/or with different PARPi: niraparib (Selleckchem, no. S2741), olaparib (Selleckchem, no. S1060), veliparib (Selleckchem, no. S1004), talazoparib (Selleckchem, no. S7048), and AZD-9574 (Selleckchem, no. E2147) as indicated. Cell viability was analyzed with the CellTiter 96 AQueous cell proliferation assay (MTS) (Promega, no. G358C) following the manufacturer’s instructions. To evaluate PARP1 protein stability, OVCAR-8 USP1WT and USP1KO cells were treated with CHX (Sigma-Aldrich, no. C4859, 30 μg/ml) for 4, 8, and 16 hours as indicated. SUMOylation and ubiquitination were analyzed using specific inhibitors, 1 μM ML-792 (MedChemExpress, no. HY-108702) and 1 μM MLN7243 (Selleckchem, no. S8341), respectively. ATR-mediated DNA replication checkpoint was analyzed using the specific ATR inhibitor AZD-6738 (ceralasertib) 5 μM (Selleckchem, no. S7693).
Synergy measurements
The synergism of the USP1i/PARPi combined treatment was calculated using the HSA metric by the online tool SynergyFinder (39, 40). For HSA, a combination of drug X and drug Y is classified as synergistic if the effect of the combination is larger than the effect of either drug X alone or drug Y alone. HSA score larger than 10 indicates that the interaction between two drugs is likely to be synergistic. To calculate HSA score in our model, OVCAR8 and COV-318 cells were seeded in 96-well culture plates and treated with equipotent doses of SJB and niraparib (IC10:10, IC25:25, and IC50:50 cytotoxic ratio) or an excess of either agent (IC10:25, IC10:50, and IC25:50 cytotoxic ratio) for 72 hours. Cell viability was measured through MTS assay as indicated above.
Plasmids, vectors and transductions
pEGFP-C1 USP1WT plasmid was provided by R. Bernards; pEGFP-C1 USP1 mutant C90S was generated ad described (8). pCMV-PARP1-3xFlag-WT was from T. Muir (provided from Addgene no. 111575). PARP1 deletion mutants 1-340, 1-530, 1-660, CAT, and ΔBRCT-WGR were generated using a QuickChange XL site-directed mutagenesis kit (Agilent, no. 200516). Human GFP-tagged PARP1 was provided from OriGene (no. RG207085). pCI-His-hUbi was from Astar Winoto (provided from Addgene no. 31815). Plasmids were transfected in HEK293/T17 or OVCAR-8 cells using FuGENE HD Transfection Reagent (Promega) according to the manufacturer’s indications. COV-318 PARP1-silenced cells were generated through lentiviral transduction particles purchased from Sigma-Aldrich (clone ID: TRCN0000007928). Seventy-two hours after transduction, cells were selected with puromycin (1.5 μg/ml) for stable silencing.
Preparation of cell lysates, immunoblotting, and immunoprecipitation
Cells were lysed in cold RIPA buffer [150 mM NaCl, 50 mM tris-HCl (pH 8), 0.1% SDS, 1% Igepal, and 0.5% NP-40] added with protease inhibitors cocktail (Roche), phosphatase inhibitors (1 mM Na3VO4 and 10 nM NaF) and 1 mM dithiothreitol. To evaluate PARylation levels, by Western blot analysis Poly(ADP-ribose) glycohydrolase (PARG) inhibitor ADP-HPD dihydrate ammonium salt (no. 118415, Sigma-Aldrich) was added to RIPA lysis buffer complemented with protease inhibitors listed before. To obtain chromatin fractions, residual pellets after lysis were resuspended in SDS 1% and sonicated. Proteins’ quantification was performed by Bradford colorimetric assay (Bio-Rad, no. 5000006). Proteins were separated by 4 to 20% SDS–polyacrylamide gel electrophoresis (Criterion Precast gels, Bio-Rad) and blotted on nitrocellulose membrane (GE Healthcare). Immunoprecipitations were performed using cell lysates or chromatin fractions diluted in HNTG buffer, with the addition of the indicated primary antibody (anti-Flag M2, Sigma-Aldrich, no. F3165; anti PARP1, Cell Signaling Technology, no. 9532). After overnight rotation at 4°C, the immunocomplexes were precipitated adding protein G or protein A (GE Healthcare, no. 17061801 and no. 17528001) agarose conjugates for an additional 1 hour and 30 min at 4°C, then washed in HNTG buffer and resuspended in 3× Laemmli sample buffer. Membranes were blotted overnight at 4°C with primary antibodies. The following primary antibodies were used: USP1 (no. HPA028440, 1:1000) and Flag M2 (no. F3165, 1:500) from Sigma-Aldrich; PARP1 (sc-8007, 1:1000), tubulin (sc-9104, 1:400), fibrillarin (sc-25397, 1:500), and CHK1 (sc-8408, 1:500) from Santa Cruz Biotechnology; ubiquitin (no. 3936, 1:1000), pCHK1 S345 (no. 2341, 1:800), RPA32 (no. 2208, 1:1000), H3 (no. 4499, 1:500), and ATR (no. 2790, 1:500) from Cell Signaling Technology; γH2AX (no. 05-636, 1:1000), Ub-K63 (no. 05-1308, 1:1000), Ub-K48 (no. 05-1307, 1:1000), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; no. CB1001, 1:1000) from Millipore; pRPA32 S33 (no. A300-246A, 1:1000) from Bethyl Lab; RAD51 (no. PA5-27195, 1:1000) from Invitrogen; PAR (no. 4335-MC, 1:1000) from R&D; and pATR T1989 (no. GTX128145, 1:500) from GeneTex. Horseradish peroxidase–conjugated (Bethyl Lab) or Alexa-conjugated (Invitrogen) secondary antibodies were used for detection.
Benzonase treatment
Benzonase treatment was performed according to the protocol published by Cattoglio et al. (41) to evaluate DNA contribution in USP1-PARP1 interaction. In brief, cells were seeded in 150-mm culture dishes and treated or not with 500 nM SJB for 3 hours. Cells were washed twice and harvested with ice-cold phosphate-buffered saline (PBS) 1× added of protease inhibitors, centrifuged, and the pellet were fast-frozen and stored at −80°C. The pellet was resuspended in freshly prepared cell lysis buffer with protease inhibitors and rotated for 8 min, then centrifuged obtaining the cytoplasmic fraction. The insoluble part was further resuspended and sonicated; the samples were then equally split in two parts, one to be left untreated and one to digest with 500 U of benzonase (no. E1014, Sigma-Aldrich), rotating 4 hours at 4°C. Samples were spun and the supernatant quantified by Bradford assay, then used to immunoprecipitate the protein of interest.
Ubiquitin pulldown
Ubiquitin pulldown was performed on cell lysates using Ub-affinity beads (Cytoskeleton, UBA01-beads) according to the manufacturer’s instructions. In brief, 20 μl of beads suspension was added to 600 μg of cell lysates carried out in 50% RIPA buffer and incubated on a rotating platform at 4°C for 2 hours. Beads were washed three times with 50% RIPA buffer, then resuspended in 2× nonreducing SDS sample buffer and 1 μl of β-mercaptoethanol.
Isolation of proteins from nascent chromatin (iPOND) assay
The iPOND assays was performed essentially as described (42). In brief, about 9 to 10 × 107 proliferating OVCAR-8 cells were pulse labeled with 10 μM EdU (Thermo Fisher Scientific, no. A10044) for 10 min, then lysed with ice-cold nuclei extraction buffer [20 mM Hepes (pH 7.2), 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% IGEPAL CA630] and nuclei were harvested. The suspension was centrifuged and the pellet resuspended with ice-cold click reaction mix to conjugate biotin-azide (no. B10184, Thermo Fisher Scientific) to EdU-labeled DNA. Click reaction mix was composed by biotin-azide (or dimethyl sulfoxide as “no-click” control), sodium ascorbate (no. A4034, Sigma-Aldrich), and copper sulfate (no. 209198, Sigma-Aldrich) added in this order to cold PBS. Samples were rotated at 4°C for 1 hour, then centrifuged and washed with ice-cold PBS: If the click reactions have been successful, the pellet should be slightly blue. After washing, samples were resuspended in lysis buffer and sonicated twice for 3 s at 10% amplitude, then 12 times for 10 s at 10% amplitude. Pulldown samples underwent to streptavidine capture adding 100 μl of streptavidine-agarose beads (Sigma-Aldrich, no. 69203-3) in B2 buffer [150 mM NaCl, 2 mM EDTA, 50 mM tris-HCl (pH 8.0), and 0.5% IGEPAL CA630] to each sample and rotating them overnight at 4°C. Proteins were resolved on SDS-PAGE and detected by Western blotting.
Chromatin immunoprecipitation assay
OVCAR-8 cells (~1 × 107 for each treatment condition) were treated for 3 hours with SJB, then cross-linked with 1% formaldehyde for 10 min and chromatin was prepared via MNase enzymatic digestion according to the protocol. ChIP was performed using SimpleChIP Enzymatic Chromatin IP kit (Magnetic Beads) no. 9003 from Cell Signaling Technology, using PARP1 primary antibody (anti PARP1, Cell Signaling Technology). Protein G magnetic beads (30 μl) were added to each sample and rotated for 2 hours at 4°C, then washed four times. Binding was analyzed by Western blot.
Immunofluorescence
Cells were seeded on coverslips and treated with SJB as indicated in the figure legends. Cells were fixed in 4% paraformaldehyde (PFA) for 20 min, permeabilized with 0.5% Triton X-100 for 10 min and blocked in 1% bovine serum albumin (BSA) for 1 hour. Slides were incubated overnight at 4°C in humid chamber with primary antibodies in 1% BSA: γH2AX (Millipore, 1:500) and 53BP1 (Novus Biologicals, 1:50). Alexa Fluor secondary antibodies (1:200) and TO-PRO-3 iodide (Invitrogen) (1:500) were incubated 1 hour at room temperature, then coverslips were placed on slides with MOWIOL with DABCO 2.5% and images were acquired through the TCS-SP8 Confocal Systems (Leica Microsystems) interfaced with the Leica Confocal Software (version 3.5.5.19976) or the Leica Application Suite software (version 6.1.1).
BrdU foci immunostaining
Cells were seeded on coverslips at low confluence and treated with 10 μM BrdU (Roche, no. 1170376) combined with SJB for 3 hours. Following fixation in methanol for 10 min, slides were rehydrated in PBS and then DNA denatured in 2 mol/liter HCl for 1 hour at 37°C. Acid was neutralized with 0.1 M borate buffer (pH 8.5), changing buffer twice. Slides were incubated 1 hour at room temperature with anti-BrdU primary antibody (BrdU labeling and detection kit, Roche) diluted 1:10 in PBS with 0.1% BSA. Alexa Fluor488 anti-mouse secondary antibody was incubated at room temperature for 1 hour. Final images were acquired as described in the Immunofluorescence section.
RPA32 foci immunostaining
Cells were seeded on coverslips and the day after they were treated with SJB for 1 hour. The slides were processed according to the protocol described (43). In brief, slides were incubated first with extraction buffer 1 [10 mM PIPES (pH 7.0), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100] and then with extraction buffer 2 [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 1% Tween 40, and 0.5% sodium deoxycholate], both for 10 min. Cells were fixed in PFA 4% for 20 min, permeabilized with Triton X-100 0.5% for 10 min and blocked in BSA 5% for 20 min. RPA32 primary antibody (Cell Signaling no. 2208, 1:500) was incubated in BSA 1% overnight at 4°C and detected with Alexa Fluor secondary antibody. Final images were acquired as described in the Immunofluorescence section.
Proximity ligation assay (PLA)
A Duolink in situ PLA kit (Sigma-Aldrich) was used to perform the assay following the indicated protocol. In brief, cells, seeded on coverslips, were treated as indicated in the figure legends. Cells were then fixed in PFA 4%, permeabilized in Triton X-100 0.5% for 10 min and blocked in blocking buffer. Primary antibodies (anti-PARP1, no. 436400 from Invitrogen, 1:50, or from Cell Signaling, 1:50; anti-USP1, no. 8033 from Cell Signaling, 1:100; anti-ubiquitin, Santa Cruz Biotechnology, 1:50; and anti-γH2AX, Millipore, 1:500) were incubated overnight at 4°C. For PLA-negative controls only one primary antibody was incubated. PLUS (Merck, no. DUO92002) and MINUS (Merck, no. DUO92004) PLA probes diluted in antibody diluent were incubated for 1 hour at 37°C; ligation and amplification steps (Merck, no. DUO92014) were conducted at 37°C for 30 and 100 min, respectively. In all wash steps, PLA wash buffers A and B (Merck, no. DUO82049) were used. Before mounting with Mowiol, slides were incubated for 30 min with TO-PRO-3 iodide (Invitrogen) (1:500) in 0.01× wash buffer B. Final images were acquired as described in the Immunofluorescence section. Total cell fluorescence intensity (CTCF) for each cell in different treatment conditions were measured using ImageJ program and calculated using the formula: CTCF = integrated density − (area of selected cell × mean fluorescence of background readings).
In situ analysis of protein interactions at DNA replication forks (SIRF)
SIRF assay was performed in accordance to the protocol described (44). OVCAR-8 cells, seeded on coverslips, were pulsed with 125 μM EdU, and then treated or not with SJB for 3 hours. Cells were then fixed in PFA 2% for 15 min and permeabilized with Triton X-100 0.25% for 15 min. Click reaction was performed directly on slides with 30 μl of fresh click reaction cocktail: 20 μM biotin-azide, 200 mM sodium ascorbate, and 4 mM copper sulfate added in this order to PBS. Slides were incubated for 1 hour at room temperature, followed by blocking in PLA blocking buffer. For the following steps, PLA protocol and reagents were used: the same primary antibodies and the same reagents concentrations and incubation timing were used. Final images were acquired as described in the Immunofluorescence section.
DNA fiber assay
The protocol described in Schwab and Niedzwiedz was used for DNA fiber assay (45). OVCAR-8 and COV-318 cells, EOC USP1WT or USP1KO cells, and OVCAR8 USP1KO overexpressing USP1WT or USP1C90S were seeded at low confluence and labeled according to the schemes reported in the figures: Cells were incubated first with 25 μM IdU (no. I7125, Sigma-Aldrich) for 20 min, then with 250 μM CldU (no. C6891, Sigma-Aldrich) for 20 min, or together with SJB treatment for 3 hours. Then, cells were collected in cold PBS and counted to have a final concentration of 300,000 cells/ml. Cells were spotted on glass slides and lysed with fiber lysis solution (50 mM EDTA and 0.5% SDS in 200 mM tris-HCl, pH 7.5). Slides were then tilted to allow the fibers to spread. Fibers were fixed for 10 min in methanol/acetic acid, denatured in HCl 2.5 M for 1 hour, and then blocked in BSA 5% for 1 hour. Anti-BrdU (mouse) (no. 347580, BD Bioscience) and anti-BrdU (rat) (no. ab6326, Abcam) primary antibodies were incubated overnight at 4°C, while Alexa Fluor 568 and Alexa Fluor 488 secondary antibodies were incubated 1 hour at room temperature. Final images were acquired as described in the Immunofluorescence section. We measured the length of about 100 fibers for each condition.
Comet assay
Comet Assay Kit (R&D, no. 4250-050-K) was used following alkaline protocol according to the manufacturer’s indications. OVCAR-8 and COV-318 cells or EOC USP1WT or USP1KO cells were seeded 60-mm dishes and, the day after, treated or not with SJB for 3 hours. After treatment (or after 24 hours from seeding for USP1WT and USP1KO cells), cells were collected and counted to have a suspension with a final concentration of 4 × 105 cells/ml in PBS. Cell suspension (50 μl) was added to 500 μl of LM agarose and then 50 μl of this mix were spread onto a prewarmed slide. Cells were lysed and DNA denatured according to the protocol. DNA was labeled for the detection with Midori Green DNA stain (Nippon Genetics, no. MG04) 1:1500 in TE buffer for 30 min at room temperature. Images were acquired through Leica TCS confocal, and Comet fluorescence intensity was measured with ImageJ software.
HT chemiluminescent PARP/apoptosis assay
The PARP/Apoptosis Chemiluminescent Assay is ideal for measuring PARP activity. This ELISA assay (R&D, no. 4685-096-K) detects semiquantitatively the amount of PAR chains deposited onto immobilized histone proteins in a 96-well format and was performed following provider’s indications. Cell lysates of COV-318 cells treated with SJB or niraparib as indicated or of EOC USP1WT or USP1KO were used as samples. After rehydration of histone-coated plate, samples, standards, and controls were distributed. Ribosylation reaction was activated by the addition of PARP substrate cocktail. After primary and secondary antibody incubation, chemiluminescent signal detection was performed adding PeroxyGlow A + B solution and reading it through Infinite M1000 Tecan microplate reader.
Laser microirradiation
Laser-induced DNA damage was exploited to evaluate PARP1 recruitment on the lesion site. OVCAR-8 cells were seeded in glass-bottom dishes and transfected with GFP-tagged PARP1 vector. Forty-eight hours after transfection, cells were presensitized with 10 μM BrdU for 24 hours and then treated or not with 500 nM SJB for 3 hours. DNA damage was induced using a 405-nm laser at maximum power (100% laser power). Cells were recorded every 2 s for 2 min; after 8 s, a single region of interest (0.47 μm width) was bleached; and then the cells were monitored for 4 min more acquiring images every 5 s. Temperature, CO2 concentration, and humidity are controlled using OKOlab incubator. Time-lapse movies were recorded using a Confocal Spinning Disk microscope (Olympus) equipped with IX83 inverted microscope provided with an IXON 897 Ultra camera (Andor), using a 60× UPlanSApo 1.35–numerical aperture objective. The system is driven by the Olympus CellSens Dimension 1.18 software (Build 16686). The data collected were analyzed through Fiji software.
Flow cytometry (FACS analysis)
To evaluate cell cycle distribution, OVCAR-8 cells were seeded in 60-mm tissue culture plates and, when indicated, starved for 72 hours in serum-free medium. Cells were harvested at the indicated time points, washed with PBS, fixed in ice-cold 70% ethanol, and stored at −20°C overnight. The fixed cells were washed twice with ice-cold PBS and the resuspended in propidium iodide (50 μg/ml) (Invitrogen, no. P3566) supplemented with RNAse (200 μg/ml) in PBS. Stained cells were subjected to flow cytometry analysis using FACs LSFortessa (BD Bioscience). Distribution of cells in G1, S, and G2/M phases of the cell cycle was calculated using the ModFit software.
In silico structural analysis
Crystal structures of PAPR1 (4AV1) (46) and USP1 (7ZH3) (47) were retrieved from Protein Data Bank. Full-length PARP1 was retrieved from Alphafold (48) and polyubiquitin chain was built on AlphaFold-Multimer (49). Protein-protein docking were performed in HADDOCK (50) and visualized in Chimera (51).
Statistical analysis
Statistical analysis was performed using GraphPad PRISM (version 9, GraphPad, Inc.) software using the most appropriate test, as specified in each figure. When two datasets were compared, significance was determined by a two-tailed Students t test. Difference was considered significant at P < 0.05 (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001).
Acknowledgments
We thank the patients who donated their samples and all members of the M.O. Unit for helpful discussions and feedback. We thank L. Cesaratto and M. Coan for technical support with CRISPR-Cas9 technology. We thank D. Parazzoli and S. Magni from IFOM (Milan) for technical and analysis support with Laser microirradiation experiment.
Funding: This work was supported by grants from: CRO Aviano Ricerca Corrente core grant (linea 1) of Ministero della Salute (G.B.); Associazione Italiana Ricerca sul Cancro (AIRC) (IG 26253) (G.B.); CRO-Aviano 5‰ Grant (G. Baldassarre); Alleanza Contro il Cancro (ACC) (RCR-2022-23682287) (G.B.); Ministero della Salute (GR-2016-02361041) (M.So.); and Associazione Italiana Ricerca sul Cancro (AIRC) (MFAG-2020: 24321) (M.So.).
Author contributions: Conceptualization: M.So., G.B., and A.N. Formal analysis: A.N., L.S., I.P., A.G., J.K., E.L., M.B., M.So., and G.B. Investigation: M.So., A.N., L.S., A.G., G.L.R.V., I.P., I.S., S.D., S.B., and F.O. Methodology: A.N., L.S., A.G., R.S., and B.B. Visualization: A.N., L.S., I.P., and J.K. Resources: M.So., J.K., R.S., M.S.N., M.B., E.L., G.G., V.C., and F.P. Supervision: B.B., F.P., M.So., and G.B. Writing–original draft: M.So., G.B., and A.N. Writing–review and editing: A.N., I.P., B.B., M.Sc., F.P., V.C., M.S.N., M.So., and G.B. Project administration: M.So. and GB. Validation: A.N., L.S., M.So., and M.Sc. Funding acquisition: M.So. and G.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in this paper are present in the paper and/or the Supplementary Materials. Any materials described in the manuscript can be provided by G.B. pending scientific review and a completed material transfer agreement. Request for the materials should be submitted to the corresponding authors M.So. (msonego@cro.it) and/or G.B. (gbaldassarre@cro.it).
Supplementary Materials
The PDF file includes:
Figs. S1 to S9
Table S1
Legends for movies S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Evans T., Matulonis U., PARP inhibitors in ovarian cancer: Evidence, experience and clinical potential. Ther. Adv. Med. Oncol. 9, 253–267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matulonis U. A., Sood A. K., Fallowfield L., Howitt B. E., Sehouli J., Karlan B. Y., Ovarian cancer. Nat. Rev. Dis. Primers. 2, 16061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowtell D. D., Böhm S., Ahmed A. A., Aspuria P.-J., Bast R. C. Jr., Beral V., Berek J. S., Birrer M. J., Blagden S., Bookman M. A., Brenton J. D., Chiappinelli K. B., Martins F. C., Coukos G., Drapkin R., Edmondson R., Fotopoulou C., Gabra H., Galon J., Gourley C., Heong V., Huntsman D. G., Iwanicki M., Karlan B. Y., Kaye A., Lengyel E., Levine D. A., Lu K. H., McNeish I. A., Menon U., Narod S. A., Nelson B. H., Nephew K. P., Pharoah P., Powell D. J., Ramos P., Romero I. L., Scott C. L., Sood A. K., Stronach E. A., Balkwill F. R., Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 15, 668–679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konstantinopoulos P. A., Ceccaldi R., Shapiro G. I., D’Andrea A. D., Homologous recombination deficiency: Exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 5, 1137–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Summey R., Uyar D., Ovarian cancer resistance to PARPi and platinum-containing chemotherapy. Cancer Drug Resist. 5, 637–646 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMullen M., Karakasis K., Madariaga A., Oza A. M., Overcoming platinum and PARP-inhibitor resistance in ovarian cancer. Cancer 12, 1607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonego M., Pellarin I., Costa A., Vinciguerra G. L. R., Coan M., Kraut A., D’Andrea S., Dall’Acqua A., Castillo-Tong D. C., Califano D., Losito S., Spizzo R., Couté Y., Vecchione A., Belletti B., Schiappacassi M., Baldassarre G., USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci. Adv. 5, eaav3235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijman S. M. B., Huang T. T., Dirac A. M. G., Brummelkamp T. R., Kerkhoven R. M., D’Andrea A. D., Bernards R., The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Sims A. E., Spiteri E., Sims R. J., Arita A. G., Lach F. P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., Auerbach A. D., Huang T. T., FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 14, 564–567 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Huang T. T., Nijman S. M. B., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D’Andrea A. D., Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 341–347 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Jones M. J. K., Colnaghi L., Huang T. T., Dysregulation of DNA polymerase κ recruitment to replication forks results in genomic instability. EMBO J. 31, 908–918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim K. S., Li H., Roberts E. A., Gaudiano E. F., Clairmont C., Sambel L. A., Ponnienselvan K., Liu J. C., Yang C., Kozono D., Parmar K., Yusufzai T., Zheng N., D’Andrea A. D., USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol. Cell 72, 925–941.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simoneau A., Engel J. L., Bandi M., Lazarides K., Liu S., Meier S. R., Choi A. H., Zhang H., Shen B., Martires L., Gotur D., Pham T. V., Li F., Gu L., Gong S., Zhang M., Wilker E., Pan X., Whittington D. A., Throner S., Maxwell J. P., Chen Y., Yu Y., Huang A., Andersen J. N., Feng T., Ubiquitinated PCNA drives USP1 synthetic lethality in cancer. Mol. Cancer Ther. 22, 215–226 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman K. E., Yin Y., Lui S. K. L., Keegan S., Fenyo D., Smith D. J., Rothenberg E., Huang T. T., USP1-trapping lesions as a source of DNA replication stress and genomic instability. Nat. Commun. 13, 1740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Da Costa A. A. B. A., Chowdhury D., Shapiro G. I., D’Andrea A. D., Konstantinopoulos P. A., Targeting replication stress in cancer therapy. Nat. Rev. Drug Discov. 22, 38–58 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonego M., Pellizzari I., Dall’Acqua A., Pivetta E., Lorenzon I., Benevol S., Bomben R., Spessotto P., Sorio R., Gattei V., Belletti B., Schiappacassi M., Baldassarre G., Common biological phenotypes characterize the acquisition of platinum-resistance in epithelial ovarian cancer cells. Sci. Rep. 7, 7104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonego M., Poletto E., Pivetta E., Nicoloso M. S., Pellicani R., Rampioni Vinciguerra G. L., Citron F., Sorio R., Mongiat M., Baldassarre G., TIMP-1 is overexpressed and secreted by platinum resistant epithelial ovarian cancer cells. Cells 9, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenzon I., Pellarin I., Pellizzari I., D’Andrea S., Belletti B., Sonego M., Baldassarre G., Schiappacassi M., Identification and characterization of a new platinum-induced TP53 mutation in MDAH ovarian cancer cells. Cells 9, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamal K., Staniszewska A., Gordon J., Wen S., McGrath F., Dowdell G., Kabbabe D., Illuzzi G., Griffin M., Davies B. R., Hamerlik P., Abstract 2609: AZD9574 is a novel, brain penetrant PARP-1 selective inhibitor with activity in an orthotopic, intracranial xenograft model with aberrant DNA repair. Cancer Res. 82, 2609–2609 (2022). [Google Scholar]
- 21.Zhu T., Zheng J.-Y., Huang L.-L., Wang Y.-H., Yao D.-F., Dai H.-B., Human PARP1 substrates and regulators of its catalytic activity: An updated overview. Front. Pharmacol. 14, 1137151 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X., Tang H., Chen Y., Chen Z., Hu Z., Cui Z., Tao Y., Yuan J., Fu Y., Zhuang Z., He Q., Li Q., Xu X., Wan X., Jiang Y., Mao Z., Loss of the receptors ER, PR and HER2 promotes USP15-dependent stabilization of PARP1 in triple-negative breast cancer. Nat. Cancer 4, 716–733 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Dammer E. B., Na C. H., Xu P., Seyfried N. T., Duong D. M., Cheng D., Gearing M., Rees H., Lah J. J., Levey A. I., Rush J., Peng J., Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 286, 10457–10465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krastev D. B., Li S., Sun Y., Wicks A. J., Hoslett G., Weekes D., Badder L. M., Knight E. G., Marlow R., Pardo M. C., Yu L., Talele T. T., Bartek J., Choudhary J. S., Pommier Y., Pettitt S. J., Tutt A. N. J., Ramadan K., Lord C. J., The ubiquitin-dependent ATPase p97 removes cytotoxic trapped PARP1 from chromatin. Nat. Cell Biol. 24, 62–73 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langelier M.-F., Planck J. L., Roy S., Pascal J. M., Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffen J. D., Tholey R. M., Langelier M.-F., Planck J. L., Schiewer M. J., Lal S., Bildzukewicz N. A., Yeo C. J., Knudsen K. E., Brody J. R., Pascal J. M., Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res. 74, 31–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettitt S. J., Krastev D. B., Brandsma I., Dréan A., Song F., Aleksandrov R., Harrell M. I., Menon M., Brough R., Campbell J., Frankum J., Ranes M., Pemberton H. N., Rafiq R., Fenwick K., Swain A., Guettler S., Lee J.-M., Swisher E. M., Stoynov S., Yusa K., Ashworth A., Lord C. J., Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes-Turcu F. E., Wilkinson K. D., Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 109, 1495–1508 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai J., Huang S. N., Das B. B., Renaud A., Zhang Y., Doroshow J. H., Ji J., Takeda S., Pommier Y., Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toss A., Cortesi L., Molecular mechanisms of PARP inhibitors in BRCA-related ovarian cancer. J. Cancer Sci. Ther. 05, 409–416 (2013). [Google Scholar]
- 31.Walton J. B., Farquharson M., Mason S., Port J., Kruspig B., Dowson S., Stevenson D., Murphy D., Matzuk M., Kim J., Coffelt S., Blyth K., McNeish I. A., CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Sci. Rep. 7, 16827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walton J., Blagih J., Ennis D., Leung E., Dowson S., Farquharson M., Tookman L. A., Orange C., Athineos D., Mason S., Stevenson D., Blyth K., Strathdee D., Balkwill F. R., Vousden K., Lockley M., McNeish I. A., CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res. 76, 6118–6129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Da Costa A. A. B. A., Bose A., Martignetti D., Ayala-Zambrano C., Ravindranathan R., Sadatrezaei G., Jiao Y., Kochupurakkal B., Nguyen H., Lazaro J.-B., Parmar K., Shapiro G. I., D’Andrea A. D., Abstract 5725: The USP1 inhibitor I-138 kills BRCA1-deficient tumor cells and overcomes PARP inhibitor resistance. Cancer Res. 83, 5725–5725 (2023). [Google Scholar]
- 34.Dias M. P., Moser S. C., Ganesan S., Jonkers J., Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 18, 773–791 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Zhang D. Y., Zhu Y., Wu Q., Ma S., Ma Y., Shen Z. C., Wang Z., Sun W., Zhou Y. C., Wang D., Zhou S., Liu Z., Kwong L. N., Lu Z., USP1 promotes cholangiocarcinoma progression by deubiquitinating PARP1 to prevent its proteasomal degradation. Cell Death Dis. 14, 669 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ying S., Hamdy F. C., Helleday T., Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72, 2814–2821 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Ronson G. E., Piberger A. L., Higgs M. R., Olsen A. L., Stewart G. S., McHugh P. J., Petermann E., Lakin N. D., PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 9, 746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M., Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23, 3886–3896 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ianevski A., Giri A. K., Aittokallio T., SynergyFinder 3.0: An interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 50, W739–W743 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehár J., Zimmermann G. R., Krueger A. S., Molnar R. A., Ledell J. T., Heilbut A. M., Short G. F., Giusti L. C., Nolan G. P., Magid O. A., Lee M. S., Borisy A. A., Stockwell B. R., Keith C. T., Chemical combination effects predict connectivity in biological systems. Mol. Syst. Biol. 3, 80 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cattoglio C., Pustova I., Darzacq X., Tjian R., Hansen A., Assessing self-interaction of mammalian nuclear proteins by co-immunoprecipitation. Bio Protoc. 10, e3526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung K. H. T., El Hassan M. A., Bremner R., A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques 55, 204–206 (2013). [DOI] [PubMed] [Google Scholar]
- 43.B. Mukherjee, N. Tomimatsu, S. Burma, “Immunofluorescence-based methods to monitor DNA end resection” in Stress Responses, C. M. Oslowski, Ed. (Springer New York, New York, NY, 2015) vol. 1292 of Methods in Molecular Biology, pp. 67–75. [DOI] [PMC free article] [PubMed]
- 44.Roy S., Luzwick J. W., Schlacher K., SIRF: Quantitative in situ analysis of protein interactions at DNA replication forks. J. Cell Biol. 217, 1521–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwab R. A. V., Niedzwiedz W., Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J. Vis. Exp., e3255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali A. A. E., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P. O., Hassler M., Ladurner A. G., Pearl L. H., Oliver A. W., The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 19, 685–692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rennie M. L., Arkinson C., Chaugule V. K., Walden H., Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site. Sci. Adv. 8, eabq6353 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. Evans, M. O’Neill, A. Pritzel, N. Antropova, A. Senior, T. Green, A. Žídek, R. Bates, S. Blackwell, J. Yim, O. Ronneberger, S. Bodenstein, M. Zielinski, A. Bridgland, A. Potapenko, A. Cowie, K. Tunyasuvunakool, R. Jain, E. Clancy, P. Kohli, J. Jumper, D. Hassabis, “Protein complex prediction with AlphaFold-Multimer” (preprint, Bioinformatics, 2021). bioRxiv 10.1101/2021.10.04.463034.
- 50.Dominguez C., Boelens R., Bonvin A. M. J. J., HADDOCK: A protein−protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Meng E. C., Pettersen E. F., Couch G. S., Huang C. C., Ferrin T. E., Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7, 339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1
Legends for movies S1 and S2
Movies S1 and S2