This is a correction to: Diptendu Chatterjee, Meena Fatah, Deniz Akdis, Danna A Spears, Tamara T Koopmann, Kirti Mittal, Muhammad A Rafiq, Bruce M Cattanach, Qili Zhao, Jeff S Healey, Michael J Ackerman, Johan Martijn Bos, Yu Sun, Jason T Maynes, Corinna Brunckhorst, Argelia Medeiros-Domingo, Firat Duru, Ardan M Saguner, Robert M Hamilton, An autoantibody identifies arrhythmogenic right ventricular cardiomyopathy and participates in its pathogenesis, European Heart Journal, Volume 39, Issue 44, 21 November 2018, Pages 3932–3944, https://doi.org/10.1093/eurheartj/ehy567
In the original published version of this manuscript, authors regret that errors were made reusing a single molecular weight marker in the article figures and they did not adequately describe the re-probing of some blot images for the evaluation of more than one serum.
To correct these errors, authors have provided new figures for figure 1A, 1B, 1C, 2A, 2B, and 3, including an individual protein ladder to each blot. Authors have also re-scanned autoradiographs of the original blots on a clean scanner bed using a new high-resolution scanner to remove scanner artefacts, replacing blot images published within the article. The “Western blot analysis” section of the Methods text on page 3933, column 2 has been updated to include details of re-probing membranes and provide further clarity for readers. Figure legend text for figure 1, 2, and 3 has also been updated to identify applicable blot images. These changes do not affect the overall scientific message of the paper.
These details have been corrected only in this correction notice to preserve the published version of record.
Western blot analysis text addition:
“For individual exposures to sera, western blots from 10-lanes (one molecular weight ladder and 9 proteins) were first cut into groups of 3 proteins (N-cadherin, Desmocollin-2and Desmoglein-2). Stripping and re-probing of some membranes were performed up to three times for exposures to sequential patient or control sera. These are identified in each figure legend.”
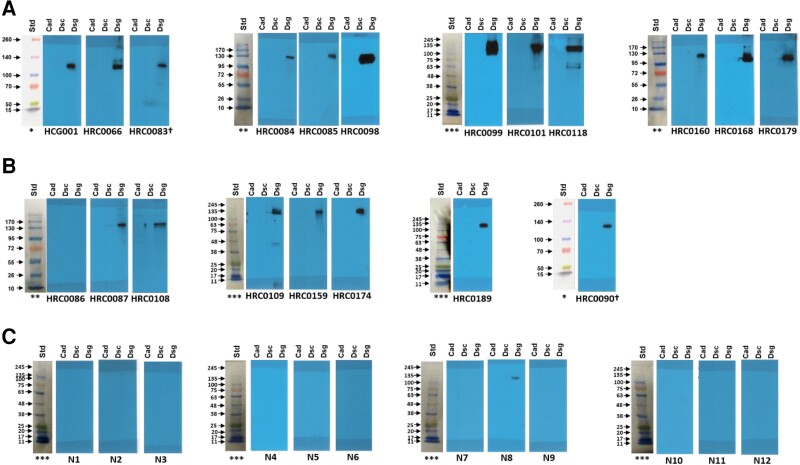
Corrected Figure 1:
Updated figure legend:
Figure 1: Discovery Cohort (with protein ladders for each set of blots):
Row A. Definite ARVC blots; Row B. Borderline ARVC blots; Row C. First set of control blots.
In a 10-lane mini protein gel, protein ladders were loaded in lane #1 and Cad, Dsc and Dsg2 proteins were loaded sequentially in 9 lanes. After running the gel, lane 1 (the protein ladder) was cut and the remaining 9 lanes were cut into 3 pieces, each with Cad, Dsc and Dsg2 proteins), each set incubated with different sera.
† Membrane HRC0083 was stripped of antibodies and re-probed with HRC0090 antibody (sera).
Different protein ladders were used for different gel runs:
* Thermo Scientific™ Spectra™ Multicolor Broad Range Protein Ladder. Cat # 26623
**Fisher BioReagents™ EZ-Run™ Prestained Rec Protein Ladder, Fisher BioReagents. Cat # 10638393
*** FroggaBio BLUeye Prestained Protein Ladder. Cat # PM007-0500
All blots are the original blots performed but have been rescanned to include all original ladders and remove scanner-induced repetitive artifacts.
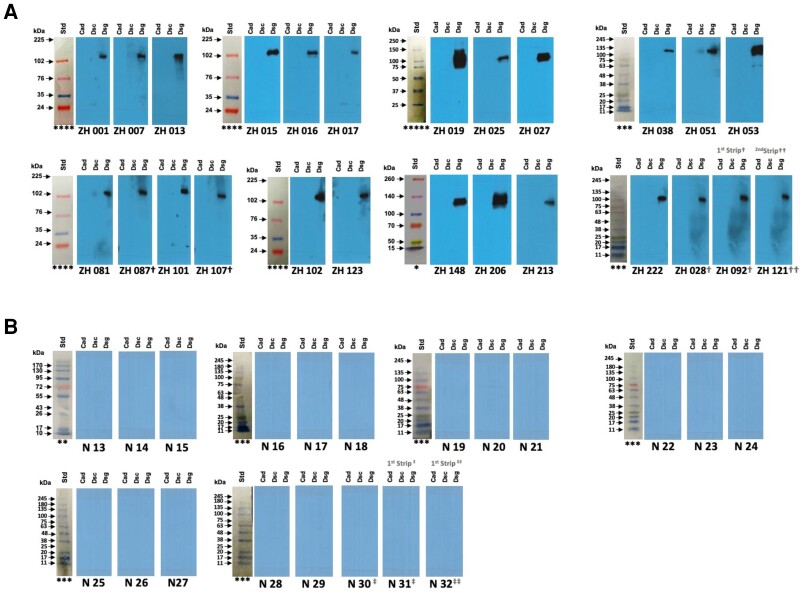
Corrected Figure 2:
Updated figure legend:
Figure 2. Validation Cohort (with protein ladders for each set of blots):
In a 10-lane mini protein gel, protein ladders were loaded in lane 1 and N-cadherin (Cad), Desmocollin-2 (Dsc) and Desmoglein-2 (Dsg2) proteins were loaded sequentially in 9 lanes. After running the gel, lane 1 (the protein ladder) was cut and the remaining 9 lanes were cut into 3 pieces, each with Cad, Dsc and Dsg2 proteins), each set incubated with different sera. In some cases, membranes were stripped and re-probed with another serum as indicated in the figure. The membrane was stripped from ZH 087† and re-probed with ZH 107†. For ZH 092† and ZH 121††, the membrane was stripped from ZH 028† and re-probed first with ZH 092†; Membrane from ZH 092† was stripped again and re-probed with ZH121††. N30‡, N31‡ and N32‡‡: Membrane was stripped from N30‡ and re-probed with N31‡. Membrane was stripped from N31‡ again and re-probed with N32‡‡.
Protein Ladders: Different protein ladders were used for different gel runs:
* Thermo Scientific™ Spectra™ Multicolor Broad Range Protein Ladder. Cat # 26623;
** Fisher BioReagents™ EZ-Run™ Prestained Rec Protein Ladder, Fisher BioReagents. Cat # 10638393
*** FroggaBio BLUeye Prestained Protein Ladder. Cat # PM007-0500
**** Cytiva Rainbow™ Molecular Weight Markers, Cytiva RPN800E, Fisher Scientific. Cat # 11580684;
***** Precision Plus Protein™ All Blue Prestained Protein Standards, BioRad. Cat #1610393.
All blots are the original blots performed but have been rescanned to include all original ladders and remove scanner-induced repetitive artifacts.
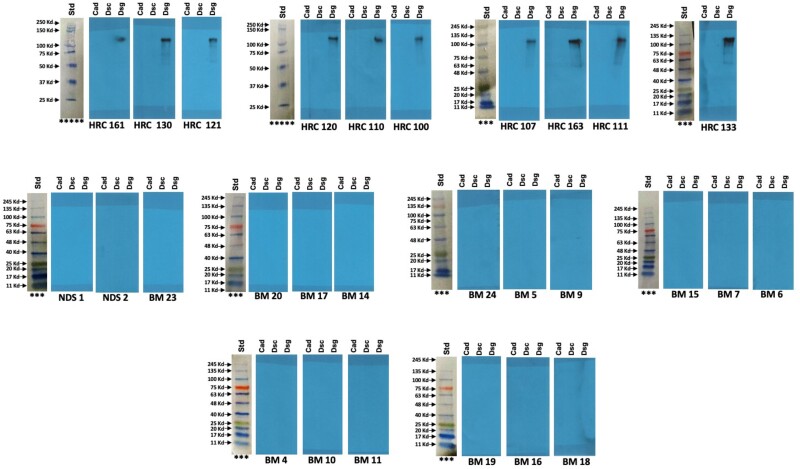
Corrected Figure 3:
Updated figure legend:
Figure 3: Boxer Dog Cohort (with protein ladders for each set of blots):
In a 10-lane mini protein gel, protein ladders were loaded in lane #1 and Cad, Dsc and Dsg2 proteins were loaded sequentially in 9 lanes. After running the gel, lane 1 (the protein ladder) was cut and the remaining 9 lanes were cut into 3 pieces, each with Cad, Dsc and Dsg2 proteins), each set incubated with different sera.
Different protein ladders were used for different gel runs:
*** FroggaBio BLUeye Prestained Protein Ladder. Cat # PM007-0500
*****Precision Plus Protein™ All Blue Prestained Protein Standards, BioRad. Cat #1610393
All blots are the original blots performed but have been rescanned to include all original ladders and remove scanner-induced repetitive artifacts.