Abstract
For patients with advanced non-small-cell lung cancer (NSCLC), dual immune checkpoint blockade (ICB) with CTLA4 inhibitors and PD-1 or PD-L1 inhibitors (hereafter, PD-(L)1 inhibitors) is associated with higher rates of anti-tumour activity and immune-related toxicities, when compared with treatment with PD-(L)1 inhibitors alone. However, there are currently no validated biomarkers to identify which patients will benefit from dual ICB1,2. Here we show that patients with NSCLC who have mutations in the STK11 and/or KEAP1 tumour suppressor genes derived clinical benefit from dual ICB with the PD-L1 inhibitor durvalumab and the CTLA4 inhibitor tremelimumab, but not from durvalumab alone, when added to chemotherapy in the randomized phase III POSEIDON trial3. Unbiased genetic screens identified loss of both of these tumour suppressor genes as independent drivers of resistance to PD-(L)1 inhibition, and showed that loss of Keap1 was the strongest genomic predictor of dual ICB efficacy—a finding that was confirmed in several mouse models of Kras-driven NSCLC. In both mouse models and patients, KEAP1 and STK11 alterations were associated with an adverse tumour microenvironment, which was characterized by a preponderance of suppressive myeloid cells and the depletion of CD8+ cytotoxic T cells, but relative sparing of CD4+ effector subsets. Dual ICB potently engaged CD4+ effector cells and reprogrammed the tumour myeloid cell compartment towards inducible nitric oxide synthase (iNOS)-expressing tumoricidal phenotypes that—together with CD4+ and CD8+ T cells—contributed to anti-tumour efficacy. These data support the use of chemo-immunotherapy with dual ICB to mitigate resistance to PD-(L)1 inhibition in patients with NSCLC who have STK11 and/or KEAP1 alterations.
Subject terms: Non-small-cell lung cancer, Immunotherapy, Predictive markers, Immunogenetics
Alterations in the tumour suppressor genes STK11 and/or KEAP1 can identify patients with advanced non-small-cell lung cancer who are likely to benefit from combinations of PD-(L)1 and CTLA4 immune checkpoint inhibitors added to chemotherapy.
Main
Immunotherapy using antibodies that inhibit the PD-1 and PD-L1 immune checkpoint (PD-(L)1i) has been shown to prolong survival in patients with advanced NSCLC, and is frequently given in combination with platinum-based chemotherapy (CT), particularly for patients who have tumours that express PD-L1 at low (less than 1% of tumour cells) or intermediate (1–49%) levels4. Several lines of evidence suggest that response to PD-(L)1i is determined not only by tumour PD-L1 levels, but also by the presence of genomic alterations in tumour oncogenes, such as activating EGFR mutations, as well as tumour suppressor genes (TSGs). In particular, alterations in two common NSCLC TSGs have been associated with a lack of responsiveness to PD-(L)1i: STK11, which encodes the LKB1 protein that functions as a master regulator of tumour cell metabolism, growth and polarity; and KEAP1, which encodes an adaptor protein that is crucial for the ubiquitination and proteasomal degradation of NRF2 and is thus a key regulator of antioxidant and cytoprotective responses5–11. We and others have previously observed that alterations in STK11 and KEAP1 can promote an immunosuppressive tumour microenvironment and together might be responsible for half or more of the primary resistance to PD-(L)1i among patients with nonsquamous NSCLC (nsNSCLC) when given as monotherapy5,12–17. Inactivating somatic mutations in these TSGs commonly co-occur and are enriched in tumours that contain KRAS oncogenic mutations. Loss of LKB1 and KEAP1 function occurs mainly through somatic mutations, but can also happen through genomic copy number loss and non-genetic mechanisms. Loss of LKB1 and KEAP1 function cooperatively promotes malignant tumour phenotypes; for example, loss of LKB1 upregulates the KEAP1–NRF2 pathway, which drives glutamine dependence and resistance to radiotherapy and ferroptosis18,19. So far, no therapies in the clinic have been shown to be effective in overcoming the immunosuppressive phenotype associated with inactivation of these TSGs.
Inhibitors of a second immune checkpoint, the CTLA4 pathway (CTLA4i), have also improved clinical outcomes for some patients with NSCLC when combined with PD-(L)1i (dual immune checkpoint blockade or ICB), with or without chemotherapy1,3,20. Although the use of PD-(L)1i is guided, at least in part, by tumour PD-L1 levels, there are at present no biomarkers in routine use for selecting patients who are more likely to benefit from dual ICB compared with PD-(L)1i alone. Identifying which patients might benefit from dual ICB is particularly important given that dual ICB causes more immune-related adverse events than do regimens containing only PD-(L)1i.
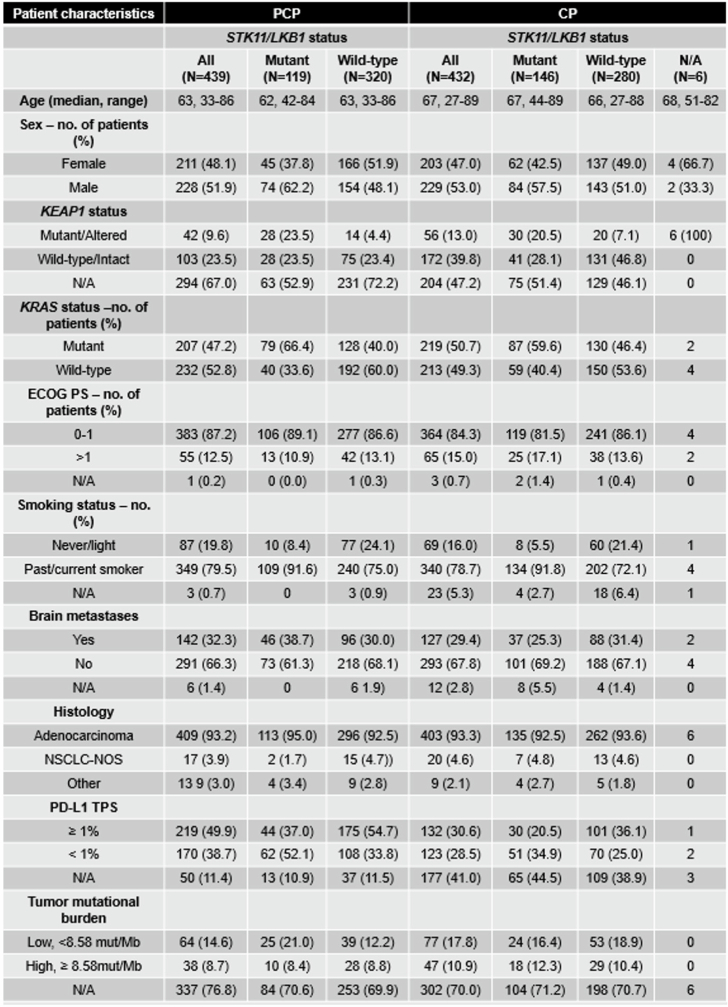
Although STK11 and KEAP1 mutations are associated with limited benefit from PD-(L)1i, the association between these mutations and benefit from combinations of PD-(L)1i with chemotherapy is not yet as well established. To address this question, we first conducted a retrospective analysis of a multicentre cohort of 871 patients with molecularly annotated advanced nsNSCLC who were treated with standard chemotherapy (carboplatin or cisplatin plus pemetrexed, or CP; n = 432) or CP in combination with the PD-1 inhibitor pembrolizumab (PCP; n = 439) as part of standard-of-care treatment (Extended Data Table 1). Among patients treated with PCP, those who had tumours with STK11 mutations (STK11MUT) had significantly shorter progression-free survival (PFS) and overall survival (OS) compared with those who had STK11 wild-type (STK11WT) tumours (PFS hazard ratio (HR) = 1.60, 95% confidence interval (CI): 1.24–2.07, P = 0.002 by log-rank test, median PFS (mPFS) 4.8 and 7.0 months, respectively; OS HR 1.55, 95% CI: 1.18–2.05, P = 0.014; median OS (mOS) 11.1 and 16.7 months, respectively (Fig. 1a and Extended Data Fig. 1). This association with worse outcomes occurred regardless of KRAS mutation status or tumour mutational burden (TMB), but was dependent on tumour PD-L1 expression (Extended Data Fig. 1). KEAP1 mutations (KEAP1MUT) had an even stronger negative association with outcome, with significantly shorter PFS (HR 2.07, 95% CI: 1.35–3.17, mPFS 2.7 versus 5.7 months for KEAP1MUT and KEAP1WT, respectively, P < 0.0001 by log-rank test) and OS (HR 2.24, 95% CI: 1.42–3.54, mOS 7.6 versus 16.6 months, P < 0.001) and a lower objective response rate (ORR; 14.3% versus 43.0%, P < 0.0001) (Fig. 1a). This outcome was independent of KRAS mutational status, TMB and tumour cell PD-L1 expression (Extended Data Fig. 1a). The combination of the two alterations resulted in a lower ORR to PCP than did either alteration individually, with an ORR of 48.6% for tumours lacking either alteration, 29.6% or 28.6% for those with either STK11 or KEAP1 mutations alone, respectively, and 7.1% for patients who had both mutations (Extended Data Fig. 1c). Notably, although both STK11 and KEAP1 alterations were associated with inferior clinical outcomes with both PCP chemo-immunotherapy (Fig. 1a and Extended Data Fig. 1b,c) and CP chemotherapy (Extended Data Fig. 1d–f), an analysis of their individual effect revealed that KEAP1 alterations had a dominant role in mediating poor outcomes with platinum-based chemotherapy, whereas STK11 mutations—in the absence of concurrent KEAP1 alteration—were associated with only a modest effect on PFS and ORR with CP (Extended Data Fig. 1e,f). Inferior clinical outcomes with chemo-immunotherapy for patients who have STK11- and/or KEAP1-mutant NSCLC were also reported in a second retrospective study as well as in the KEYNOTE-189 randomized phase III clinical trial, further supporting the validity of these findings11,21.
Extended Data Table 1.
Clinicogenomic characteristics of patients with advanced nsNSCLC treated with carboplatin or cisplatin and pemetrexed (CP) or CP plus pembrolizumab (PCP)
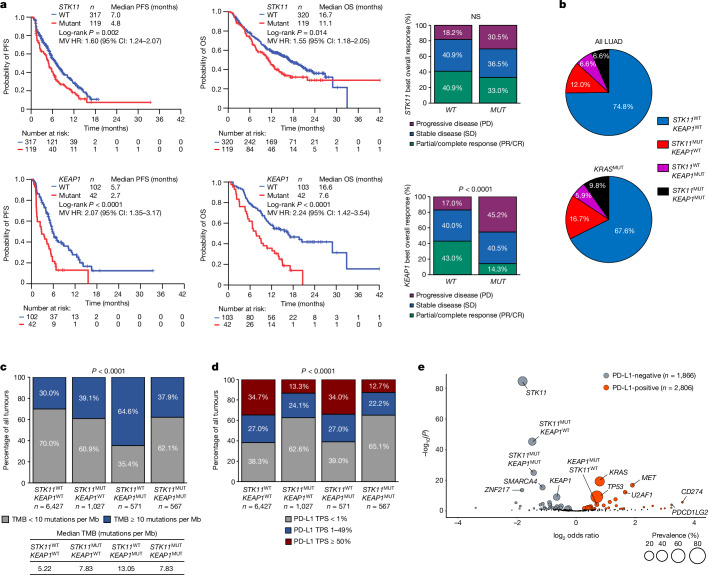
Fig. 1. Immunogenomic correlates and clinical outcomes with PCP chemo-immunotherapy in patients with STK11- and/or KEAP1-mutant advanced nsNSCLC.
a, PFS, OS and ORR with pemetrexed, carboplatin or cisplatin and pembrolizumab (PCP) in patients with STK11MUT (n = 119) versus STK11WT (n = 320) (top) or with KEAP1MUT (n = 42) versus KEAP1WT (n = 103) (bottom) advanced nsNSCLC. The comparison of ORR (partial response (PR)/complete response (CR)) in patients with STK11MUT versus STK11WT and KEAP1MUT versus KEAP1WT tumours was based on the chi-squared test. Log-rank test was used for comparisons of PFS and OS. Multivariate (MV) HRs (adjusted for age, ECOG performance status and presence of brain metastases) and 95% CIs were estimated using a Cox proportional hazards model. P ≤ 0.05 was considered statistically significant. NS, not significant. b, Prevalence of individual and combined STK11 and KEAP1 alterations in advanced LUAD (left; n = 8,592) or KRAS-mutated LUAD (right; n = 3,224) from the Foundation Medicine (FMI) database. c, TMB in single and double STK11MUT and/or KEAP1MUT and STK11WT/KEAP1WT LUAD in the FMI dataset. Median TMB (table insert) and fraction of tumours with a TMB of 10 or more mutations per Mb or fewer than 10 mutations per Mb (bar chart) in each subgroup are indicated. d, PD-L1 tumour proportion score (TPS) in single and double STK11MUT and/or KEAP1MUT and STK11WT/KEAP1WT LUAD in the FMI dataset (n = 8,836). The chi-squared test from a 2 × 4 contingency table was used to compare the distribution of PD-L1-positive (TPS ≥ 1%) and -negative (TPS < 1%) tumours across the four oncogenotypes. e, Volcano plot of enriched somatic genomic alterations in PD-L1-negative (TPS < 1%, grey circles) versus PD-L1-positive (TPS ≥ 1%, red circles) LUAD with an intermediate or high TMB (TMBI/H; six or more mutations per Mb; n = 4,672). The size of individual circles is proportional to the prevalence of the corresponding alteration. Two-sided Fisher’s exact test was used for statistical comparisons and statistical significance was established at false discovery rate (FDR)-adjusted P ≤ 0.05.
Extended Data Fig. 1. Clinical outcomes in patients with STK11- and/or KEAP1-mutated nsNSCLC treated with PCP or CP.
a. PFS and OS with PCP chemo-immunotherapy in patient subgroups with advanced (i) STK11MUT vs STK11WT (top panel) and (ii) KEAP1MUT vs KEAP1WT (bottom panel) nsNSCLC. HRs and 95% CIs were estimated using a Cox proportional hazards model. b. Best overall response with PCP in the indicated patient subgroups. The analysis was limited to the subset of response-evaluable patients with available comprehensive NGS profiling that included both STK11 and KEAP1 (N = 141). c. Kaplan–Meier estimates of PFS (left) and OS (right) with PCP in patients with advanced (i) STK11WT;KEAP1WT (WT,WT; indicated in blue); (ii) STK11MUT;KEAP1WT (MUT,WT; indicated in red); (iii) STK11WT;KEAP1MUT (WT,MUT; indicated in purple) and (iv) STK11MUT;KEAP1MUT (MUT, MUT; indicated in black) nsNSCLC. The analysis was limited to the subset of patients with available comprehensive NGS profiling that included both STK11 and KEAP1 (N = 145). d. Kaplan–Meier estimates of PFS (left) and OS (right) with CP in patients with advanced (i) STK11MUT vs STK11WT and (ii) KEAP1MUT vs KEAP1WT nsNSCLC. e. Kaplan–Meier estimates of PFS (left) and OS (right) with CP in patients with advanced (i) STK11WT;KEAP1WT (WT,WT; indicated in blue); (ii) STK11MUT;KEAP1WT (MUT,WT; indicated in red); (iii) STK11WT;KEAP1MUT (WT,MUT; indicated in purple) and (iv) STK11MUT;KEAP1MUT (MUT, MUT; indicated in black) nsNSCLC. The analysis was limited to the subset of patients with available comprehensive NGS profiling that included both STK11 and KEAP1 (N = 222). f. Best overall response with CP in the indicated patient subgroups. The analysis was limited to the subset of response-evaluable patients with available ORR data and comprehensive NGS profiling that included both STK11 and KEAP1 (N = 180).
Immune correlates of STK11MUT and KEAP1MUT NSCLC
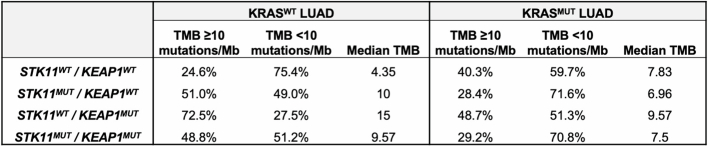
To further investigate the mechanisms that might underlie the resistance of STK11MUT and KEAP1MUT tumours to PD-(L)1i, we investigated the immune and genomic profiles of 8,592 patients with lung adenocarcinoma (LUAD) with these and other alterations (Foundation Medicine (FMI) cohort). STK11 and/or KEAP1 mutations were observed in 25.2% and 32.4% of the overall LUAD and KRAS-mutant LUAD populations, respectively (Fig. 1b). For other subsets of NSCLC that are poorly responsive to PD-(L)1i, such as tumours containing EGFR mutations or ALK fusions, a lower TMB and/or lower levels of PD-L1 are thought to contribute to their immunologically cold phenotype22–24. A high TMB, by contrast, has been associated with improved responsiveness to both PD-(L)1i and dual ICB25,26. We therefore investigated whether the lack of PD-(L)1i response could be due to a low TMB and/or low levels of PD-L1. Notably, we observed that, compared with tumours that were wild type for both genes (median TMB 5.22 mutations per Mb), tumours with mutations in STK11 or KEAP1 had a higher TMB. This was particularly notable for KEAP1MUT tumours (median TMB 13.05 mutations per Mb) (Fig. 1c), especially in the absence of concurrent KRAS mutations (Extended Data Table 2). Therefore, lower TMB does not seem to account for the reduced responsiveness of STK11MUT and KEAP1MUT NSCLC tumours to immunotherapy.
Extended Data Table 2.
Tumour mutational burden in subgroups of KRAS-mutated and KRAS wild-type advanced LUAD in the FMI dataset
We next investigated the association between these alterations and PD-L1 levels. STK11MUT tumours, in the absence or presence of KEAP1 mutations, had significantly lower levels of PD-L1, compared with tumours that were wild type for both genes, consistent with previous observations5,9,11–13,27,28 whereas KEAP1MUT tumours (in the absence of STK11 mutations) did not have lower levels of PD-L1 (Fig. 1d). This suggests that lower levels of PD-L1, indicative of a lack of immune engagement, contribute to the lack of PD-(L)1i responsiveness in STK11MUT—but not in KEAP1MUT—NSCLC, and that STK11 and KEAP1 have overlapping as well as distinct mechanisms by which they promote an immunosuppressive tumour microenvironment.
Next, we investigated whether other genomic alterations were also associated with lower PD-L1 levels and how these alterations compared with STK11. Given the possible association between a low TMB and low PD-L1 levels29, we limited the analysis to tumours with an intermediate or high TMB (TMBI/H, six or more mutations per Mb; n = 4,672). We observed that STK11 was the most significantly enriched gene in PD-L1-negative tumours (Fig. 1e). Of note, KEAP1MUT LUAD tumours without concurrent STK11 alterations (KEAP1MUTSTK11WT) were not enriched in PD-L1-negative tumours, and exhibited similar levels of PD-L1 expression to those of LUAD tumours that were wild type for both genes (STK11WTKEAP1WT).
Dual ICB in STK11MUT and KEAP1MUT nsNSCLC
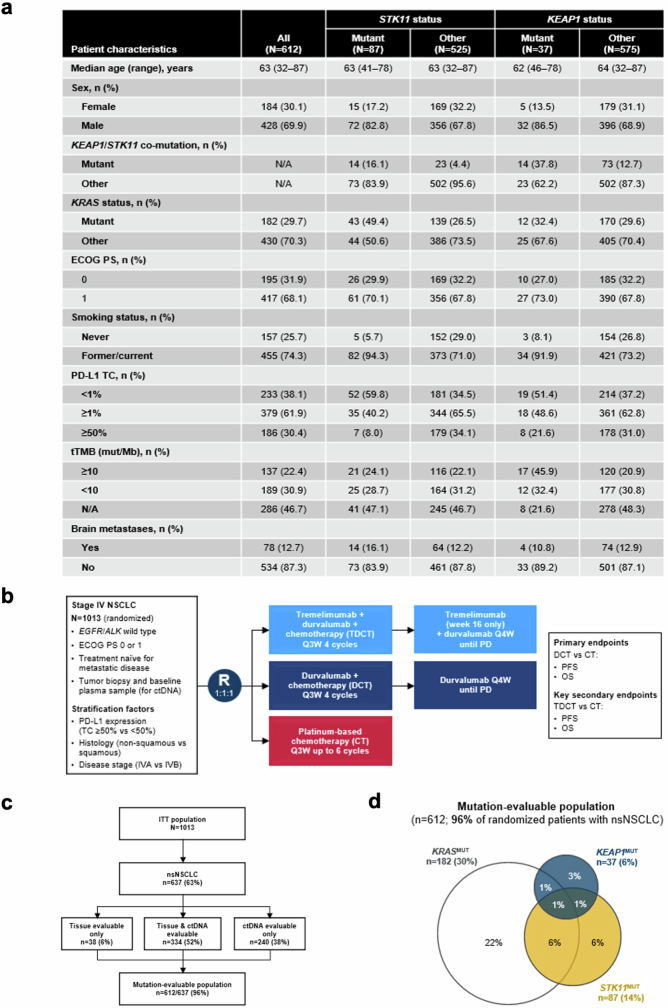
Given the correlation between STK11 and KEAP1 mutations and poor outcomes in patients with metastatic nsNSCLC, and previous evidence suggesting that these alterations promote PD-(L)1i resistance and an immunologically ‘cold’ TME, we hypothesized that dual ICB provides greater benefit than PD-(L)1i monotherapy in this setting, as has been suggested in both NSCLC and other tumour types30. To investigate this further, we evaluated patients treated in the POSEIDON study, a randomized phase III study of 1,013 patients with metastatic NSCLC comparing standard-of-care chemotherapy alone (CT) and CT combined with the PD-L1i durvalumab (DCT) or with the combination of durvalumab and the CTLA4 inhibitor tremelimumab (TDCT) (Extended Data Fig. 11). The primary clinical results from this study were previously reported3. A total of 637 patients had nonsquamous histology, among whom 612 (96%) were evaluable for mutations; KRAS, STK11 and KEAP1 mutations were present in 30%, 14% and 6% of this population, respectively (Extended Data Fig. 11). Differences in the prevalence of STK11 and KEAP1 mutations between patients from different racial backgrounds, distinct assay platforms and variable inclusion criteria for variants of unknown significance might have accounted for the observed minor variations in the reported prevalence of individual alterations compared with the FMI and retrospective real-world cohorts. The baseline clinical and molecular characteristics of these subgroups are shown in Extended Data Fig. 11.
Extended Data Fig. 11. The randomized phase III POSEIDON clinical trial.
a, Clinicogenomic characteristics of mutation-evaluable patients with advanced nsNSCLC in the POSEIDON clinical trial. b, POSEIDON clinical trial schema. c, Mutation-evaluable population among patients with nsNSCLC. d, Venn diagram indicating overlap of somatic mutations in KRAS, STK11 and KEAP1 in the POSEIDON dataset.
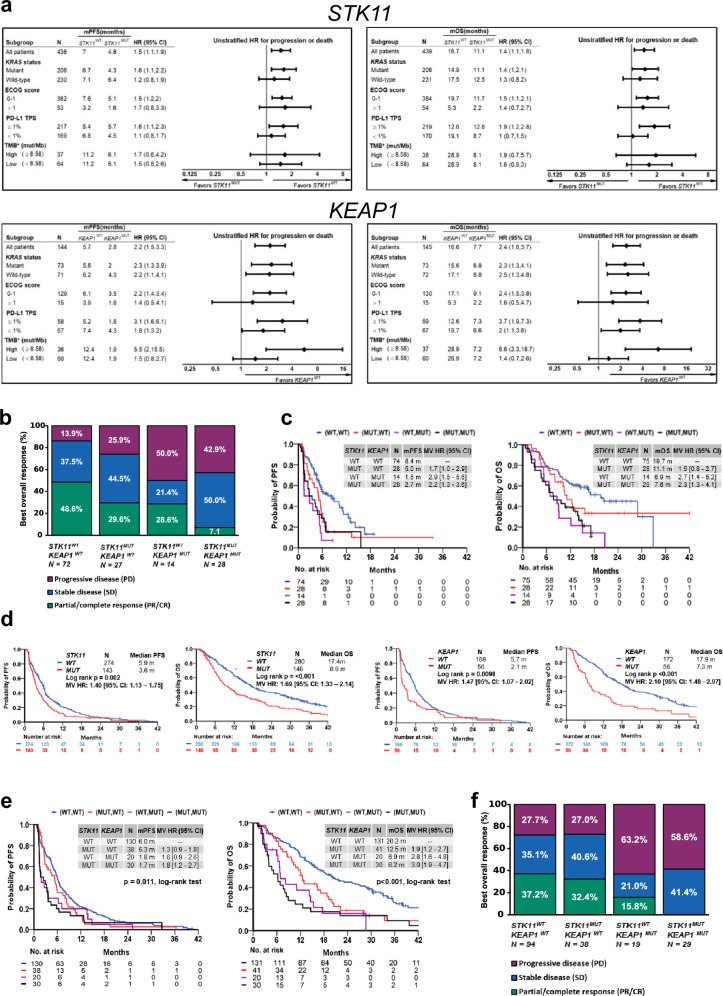
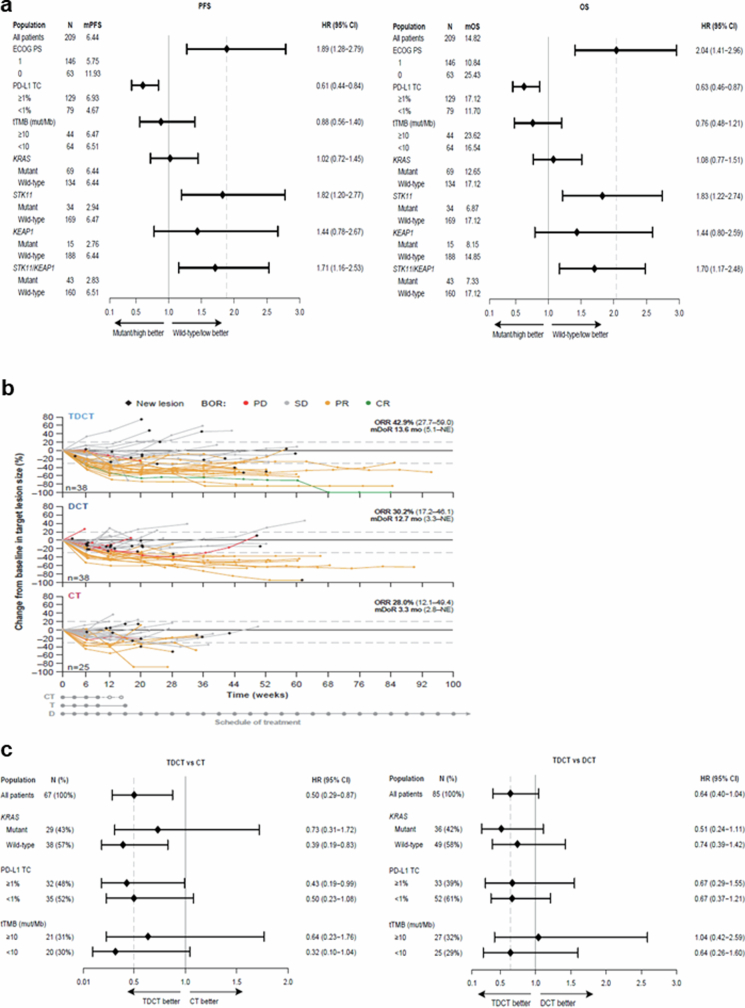
Consistent with the previous observations with pembrolizumab plus chemotherapy, STK11 and KEAP1 mutations were associated with worse outcomes in the DCT arm, compared with patients with wild-type tumours. The HR for OS was 1.83 (95% CI: 1.22–2.74) in patients with STK11MUT versus STK11WT tumours; 1.44 (95% CI: 0.80–2.59) for those with KEAP1MUT versus KEAP1WT tumours; and 1.70 (95% CI: 1.17–2.48) for those with either gene mutated versus those that were wild type for both genes. Similar effects were noted for PFS (Extended Data Fig. 2a).
Extended Data Fig. 2. Clinical outcomes in patient subgroups in the POSEIDON clinical trial.
a. PFS (left) and OS (right) with DCT in patient subgroups defined by clinical and molecular characteristics. HRs and 95% CIs were estimated using unstratified Cox proportional hazards models. The analysis of PFS was based on a data cut-off date of July 24, 2019 and the analysis of OS was based on a data cut-off date of March 12, 2021. b. Spider plots, depicting patient-level % change compared to baseline in the size of target lesion(s) (per RECIST v1.1) in patients with STK11 and/or KEAP1-mutated nsNSCLC treated with TDCT (top), DCT (middle) and CT (bottom). Individual trajectories are colour-coded based on best overall response. Only patients with both a baseline and at least one available post-baseline target lesion measurement are included. ORR and mDoR are based on confirmed objective responses by BICR. The analysis was based on a data cut-off date of July 24, 2019. c. OS in molecularly defined subgroups of patients with STK11MUT and/or KEAP1MUT metastatic nsNSCLC treated with TDCT vs CT (left) and TDCT vs DCT (right). HRs and 95% CIs were estimated using unstratified Cox proportional hazards models. The analysis was based on a data cut-off date of March 12, 2021.
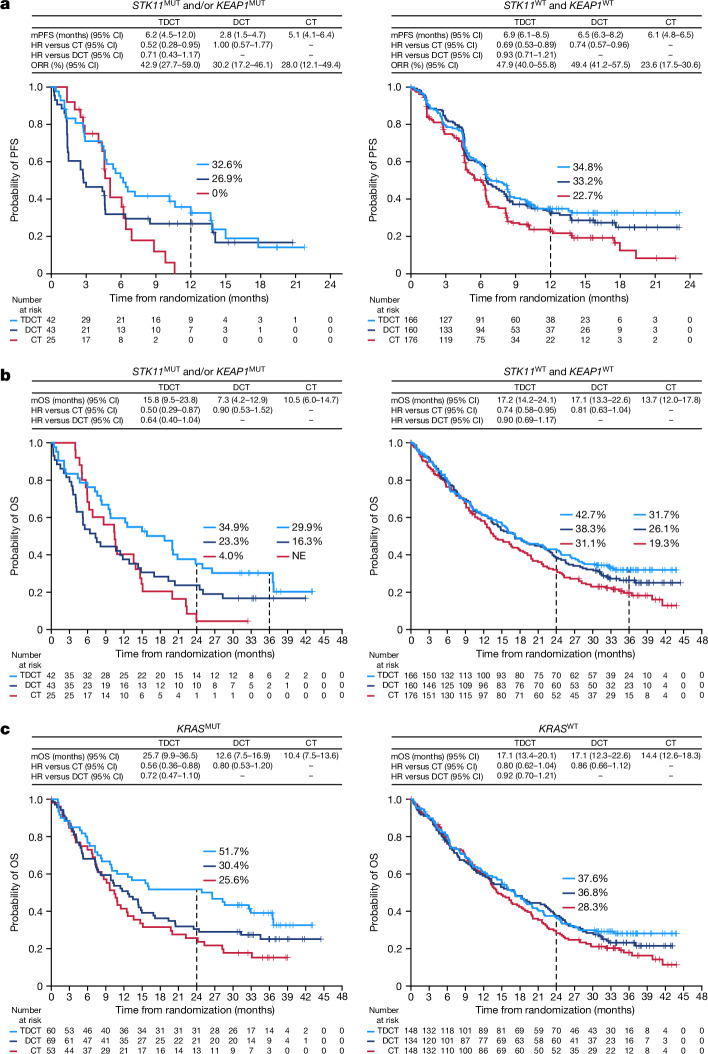
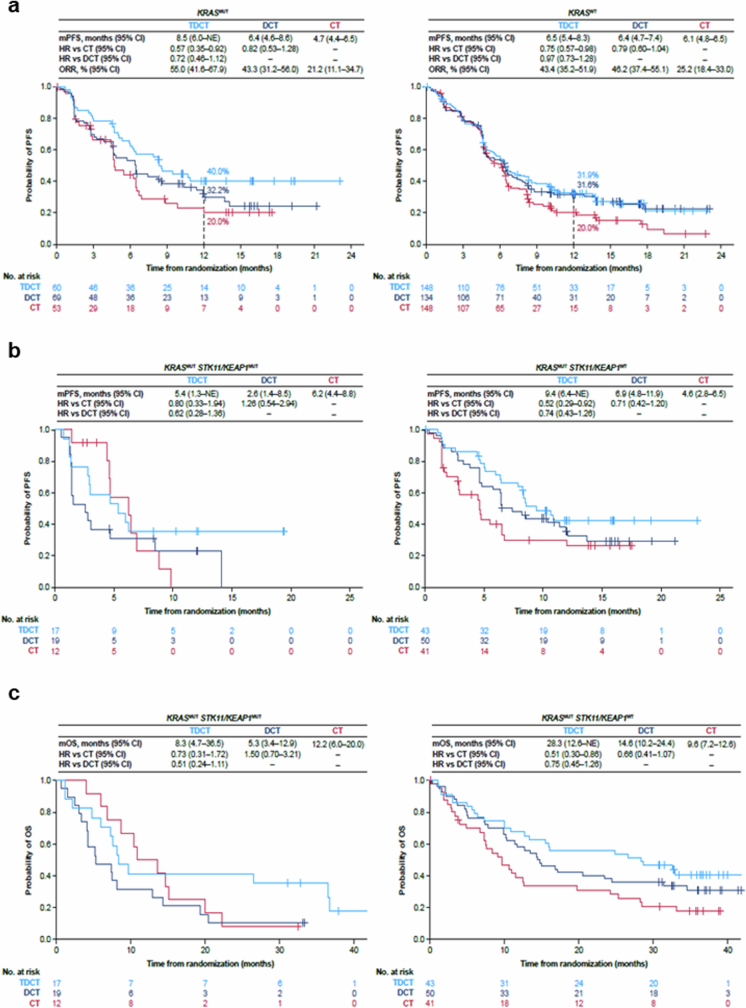
Next, we investigated the relative benefit of DCT compared with CT alone. In patients with STK11 and/or KEAP1 mutations (referred to as the STK11/KEAP1 subgroup), the addition of durvalumab to chemotherapy provided no benefit in prolonging PFS (HR 1.00, 95% CI: 0.57–1.77) for DCT versus CT; notably, adding durvalumab to chemotherapy prolonged PFS for patients without either of these alterations (STK11/KEAP1 WT, HR 0.74, 95% CI: 0.57–0.96) (Fig. 2a). A similar trend was observed for OS (Fig. 2b). This indicates that patients with STK11 and/or KEAP1 mutations derive little, if any, benefit from the addition of durvalumab to chemotherapy and that the benefit of DCT is largely confined to patients who lack mutations in either gene.
Fig. 2. Clinical outcomes in molecularly defined patient subgroups in the phase III POSEIDON clinical trial.
a,b, Kaplan–Meier estimates of PFS according to blinded independent central review (BICR) per RECIST v.1.1 (a) and OS (b) with tremelimumab, durvalumab and platinum chemotherapy (TDCT, light blue curve), durvalumab plus platinum chemotherapy (DCT, dark blue curve) or platinum doublet chemotherapy alone (CT, red curve) in patients with (i) STK11MUT and/or KEAP1MUT (left) and (ii) STK11WT and KEAP1WT (right) metastatic nsNSCLC. Landmark 12-month PFS rates and 24-month and 36-month OS rates in each of the treatment arms are also shown (dotted lines). NE, not evaluable. c, Kaplan–Meier estimates of OS with TDCT, DCT or CT in patients with KRASMUT (left) and KRASWT (right) metastatic nsNSCLC. PFS and ORR analyses were based on a data cut-off date of 24 July 2019. OS analyses were based on a data cut-off date of 12 March 2021. HRs and 95% CIs were estimated using unstratified Cox proportional hazards models.
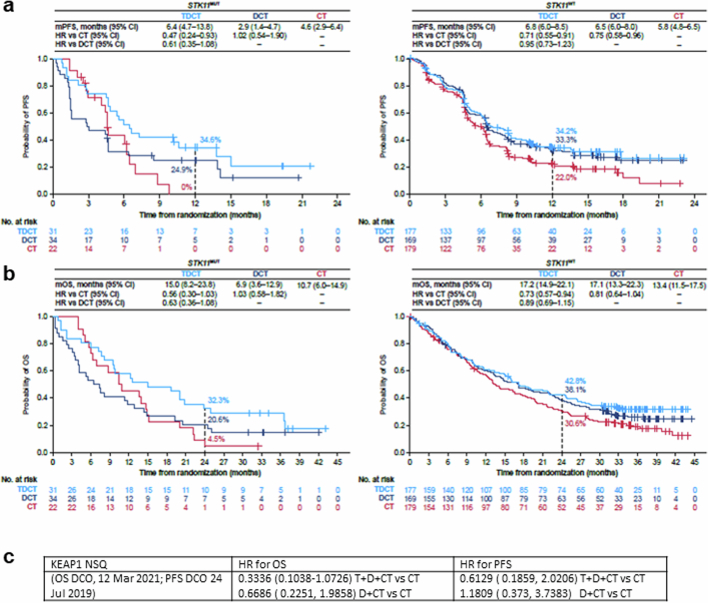
We then investigated whether the addition of tremelimumab to DCT could mitigate the PD-(L)1i resistance associated with the STK11/KEAP1 subgroup by comparing the TDCT arm with the DCT and CT arms. For the STK11/KEAP1 subgroup, the ORR was higher in the TDCT arm (42.9%) than in the DCT (30.2%) or CT (28%) arms (Extended Data Fig. 2b). An analysis of the duration of response (DoR), illustrated using spider plots (Extended Data Fig. 2b), supports the finding that the addition of dual ICB not only increased the number of patients with STK11/KEAP1-mutant tumours that responded to treatment, but enhanced their median DoR, which was only 3.3 months in the CT arm, 12.7 months in the DCT arm and 13.6 months in the TDCT arm. In the STK11/KEAP1 group, TDCT improved PFS and OS compared with the CT arm (HR 0.52 (95% CI: 0.28–0.95) and 0.50 (95% CI: 0.29–0.87)), independently of KRAS mutation status, tumour cell PD-L1 expression or TMB (Fig. 2a,b and Extended Data Fig. 2c). To determine the clinical benefit from the addition of CTLA4 blockade, we performed a comparative analysis of survival in the TDCT versus the DCT arms. In the STK11/KEAP1 subgroup, median OS was more than doubled for the TDCT arm (15.8 versus 7.3 months, HR 0.64, 95% CI: 0.40–1.04; Fig. 2b), whereas in the group lacking mutations in either gene, little benefit was observed from the addition of tremelimumab (17.2 versus 17.1 months, HR 0.90, 95% CI: 0.69–1.17). OS was favourable in the TDCT arm regardless of KRAS mutation status or tumour cell PD-L1 expression (Extended Data Fig. 2c). Similar trends were observed for PFS (Fig. 2a). Concordant results were also observed when STK11 and KEAP1 alterations were assessed individually, although this analysis was limited by the modest number of patients with KEAP1MUT tumours (Extended Data Fig. 3). KRAS mutations were also associated with tremelimumab benefit, in part because of the higher frequency of STK11 and KEAP1 mutations in this subgroup (Fig. 2c and Extended Data Fig. 4). Notably, a trend towards improved outcomes with the combination of the PD-1 inhibitor nivolumab and the CTLA4 inhibitor ipilimumab, with platinum-based chemotherapy versus chemotherapy alone, in patients with STK11- and/or KEAP1-mutant NSCLC was also observed in the CheckMate 9LA phase III randomized clinical trial31. These data support the hypothesis that CTLA4 inhibition can mitigate the resistance to chemotherapy plus PD-(L)1i observed in patients who have STK11 and/or KEAP1 mutations, and suggest that this group of patients derives greater benefit from CTLA4 inhibition than do patients who lack either alteration.
Extended Data Fig. 3. Clinical outcomes in patients with STK11-mutated or KEAP1-mutated nsNSCLC in the phase III POSEIDON clinical trial.
a,b. Kaplan–Meier estimates of PFS according to BICR per RECIST v1.1 (a) and OS (b) with TDCT (light blue curve) vs DCT (dark blue curve) vs CT (red curve) in patients bearing STK11MUT (left panel) and STK11WT (right panel) metastatic nsNSCLC. Landmark 12-month PFS rates and 24-month OS rates in each of the treatment arms are also shown (dotted lines). PFS analyses were based on a data cut-off date of July 24, 2019. OS analyses were based on a data cut-off date of March 12, 2021. HRs and 95% CIs were estimated using unstratified Cox proportional hazards models. c. HR for OS and PFS with D + T + CT or D + CT versus CT in nsNSCLC subgroup with KEAP1 alterations.
Extended Data Fig. 4. Clinical outcomes in patients with KRAS-mutated nsNSCLC in the phase III POSEIDON clinical trial.
a. Kaplan–Meier estimates of PFS according to BICR per RECIST v1.1 with TDCT (light blue curve) vs DCT (dark blue curve) vs CT (red curve) in patients bearing KRASMUT (left panel) and KRASWT (right panel) metastatic nsNSCLC. Landmark 12-month PFS rates in each of the treatment arms are also shown (dotted lines). b,c. Kaplan–Meier estimates of PFS according to BICR per RECIST v1.1 (b) and OS (c) with TDCT vs DCT vs CT in patients bearing KRASMUT; STK11MUT and/or KEAP1MUT (left panel) and KRASMUT; STK11WT and KEAP1WT (right panel) metastatic nsNSCLC. PFS and ORR analyses were based on a data cut-off date of July 24, 2019. OS analyses were based on a data cut-off date of March 12, 2021. HRs and 95% CIs were estimated using unstratified Cox proportional hazards models.
Mechanisms of dual ICB efficacy
In light of the observed clinical efficacy of chemo-immunotherapy with dual ICB, but not PD-(L)1i, in patients with STK11- and/or KEAP1-mutant NSCLC, we next tested whether inactivation of Stk11, Keap1 or other TSGs that are frequently mutated in NSCLC directly affect sensitivity to PD-(L)1i or dual ICB using immune-competent mouse models of NSCLC.
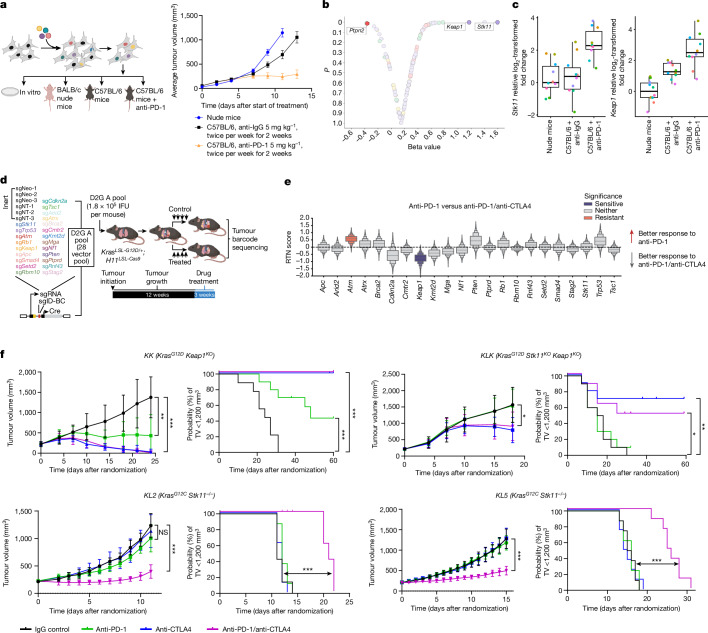
We first performed a pooled TSG-focused in vivo CRISPR–Cas9 genetic screen designed to identify candidate cancer-cell-intrinsic mediators of immune evasion and PD-1i resistance. C57BL/6 mice were implanted with library-transduced Lewis lung carcinoma 3LL cells and treated with anti-PD-1 or with isotype control IgG (Fig. 3a). In this unbiased screen, Stk11 and Keap1 emerged as two of the three most significantly enriched TSGs (Fig. 3b). Progressive enrichment of multiple unique single-guide RNAs (sgRNAs) targeting Stk11 or Keap1 in subcutaneous tumours grown in (i) nude mice, (ii) syngeneic, immune-competent C57BL/6 mice treated with IgG isotype control antibody and (iii) C57BL/6 mice treated with anti-PD-1 reflects the enhanced fitness of Stk11- or Keap1- deficient cells under increasing immune pressure (Fig. 3c). Thus, inactivations of Stk11 and Keap1 constitute direct, independent mediators of immune evasion and de novo resistance to PD-(L)1 axis blockade in NSCLC models—in agreement with previous reports12,13, our retrospective NSCLC cohort (Fig. 1) and the clinical data from the POSEIDON study (Fig. 2).
Fig. 3. Efficacy of anti-PD-1 monotherapy and dual anti-PD-1/anti-CTLA4 therapy in immune-competent models of STK11- and/or KEAP1-deficient NSCLC.
a, Left, in vivo TSG-focused CRISPR–Cas9 screening platform for identifying drivers of PD-1 inhibitor resistance. Right, the gradual decrease in tumour growth from untreated nude mice (n = 18) to C57BL/6 mice treated with anti-IgG (n = 18) to C57BL/6 mice treated with anti-PD-1 (n = 20) reflects increased anti-tumour immunity. Data are mean ± s.e.m. b, Volcano plot of relative sgRNA enrichment or depletion in tumours from C57BL/6 mice that were treated with anti-PD-1 (5 mg per kg; n = 20; 10 mg per kg; n = 25; total n = 45) versus anti-IgG isotype control (n = 18). c, Box plots of relative log2-transformed fold change for sgRNAs targeting Stk11 (left) or Keap1 (right) across treatment groups (nude, n = 18; IgG, n = 20; anti-PD-1 5 mg per kg, n = 20). Individual sgRNAs are represented by coloured circles (10 sgRNAs per gene). Median (central line), interquartile range (box plot edges) and data range (whiskers) are indicated. d, PGx-Tuba-seq experimental strategy for in vivo multiplexed quantitative evaluation of the effect of TSG depletion on immunotherapy responses in an autochthonous, genetically engineered KrasG12D-driven LUAD model (see Methods). IFU, infectious units. e, Relative tumour number (RTN) score reflecting differential sensitivity to dual anti-PD-1/anti-CTLA4 blockade compared with anti-PD-1 monotherapy. Significant effects are highlighted in colour (n = 16–19 mice per group). f, Sensitivity to dual ICB across several STK11 and/or KEAP1-deficient syngeneic models of KrasG12D (KK, KLK) and KrasG12C (KL2, KL5) mutant NSCLC (n = 8–10 mice per group). Comparison of tumour volume (TV) was performed at the time point at which the first mouse in any treatment arm reached end-point (tumour volume ≥ 1,500 mm3) and was based on the Mann–Whitney U test. Data are mean ± s.d. Time to tumour volume (TTV) ≥ 1,200 mm3 was used as a surrogate for survival. Comparison of TTV between treatment groups was based on the log-rank test. Statistical significance is indicated (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
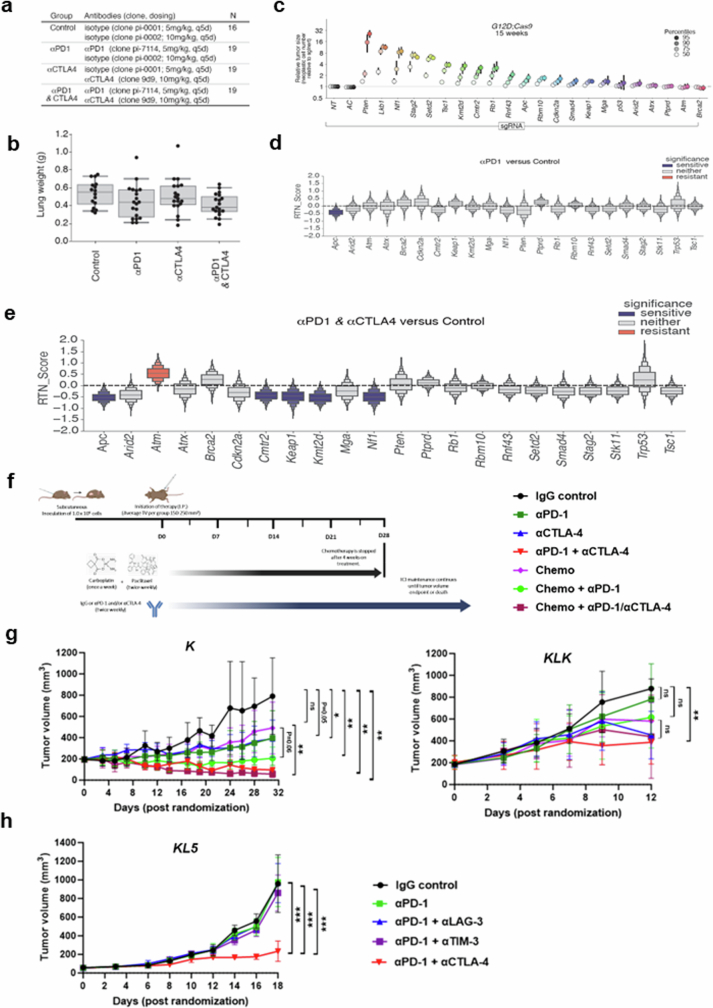
Next, we investigated whether specific genomic alterations could promote differential sensitivity to dual ICB compared with PD-(L)1i with a genetically engineered autochthonous lung cancer model. Specifically, we used tumour barcoding coupled with high-throughput barcode sequencing (Tuba-seq32) and multiplexed somatic CRISPR–Cas9 genome editing to quantitatively assess the effect of inactivating 22 TSGs on the efficacy of dual ICB relative to anti-PD-1 monotherapy32 (Fig. 3d,e and Extended Data Fig. 5a–e). Twelve weeks after the initiation of tumours with a pool of barcoded Lenti-sgRNA/Cre vectors, KrasLSL-G12D/+H11LSL-Cas9 mice (n = 16–19 per group) were randomly assigned to treatment groups (i) anti-PD-1 and anti-CTLA4 (anti-PD-1/anti-CTLA4); (ii) anti-PD-1 and isotype control (anti-PD-1); (iii) anti-CTLA4 and isotype control (anti-CTLA4); or (iv) isotype controls (Extended Data Fig. 5a). After three weeks of treatment, genomic DNA was extracted from bulk tumour-bearing lungs for Tuba-seq library generation and analysis (Fig. 3d). Inactivation of several TSGs—including Stk11 and, to a lesser extent, Keap1—promoted in vivo tumour growth in the absence of therapy, as reported previously33–35 (Extended Data Fig. 5c). Notably, inactivation of Keap1 significantly and markedly enhanced sensitivity to the dual ICB compared with anti-PD-1 monotherapy (Fig. 3e and Extended Data Fig. 5d,e).
Extended Data Fig. 5. Effect of distinct co-mutations on tumour growth and immune checkpoint inhibitor response in immune-competent models of KRAS-mutant NSCLC.
a. Therapeutic schedule and cohort size of ICB treatment study experimental groups. b. Mouse lung weight in the four treatment arms. Dots represent individual mice. c. Tumour growth effects of individual co-alterations. Tumours at the indicated percentiles of the tumour size distribution for each barcoded Lenti-sgRNA/Cre vector are shown, with 95% CIs. d,e. RTN score reflecting sensitivity to anti-PD-1 monotherapy (d) and dual anti-PD-1/anti-CTLA4 therapy (e) compared with isotype control IgG-treated mice. Significant effects are highlighted in colour. f. Experimental strategy to evaluate the anti-tumour activity of single or dual ICB with or without platinum doublet chemotherapy in the K and KLK (clone 17) isogenic allograft models. g. Efficacy of (chemo)-immunotherapy encompassing single (anti-PD-1) or dual (anti-PD-1/anti-CTLA4) ICB in the K and KLK isogenic models (N = 6-7 mice/group). Comparison of tumour volume (TV) between treatment arms in the KLK model was performed when the first mouse in any treatment group reached a TV of ≥ 1200 mm3. In the K model, comparison of tumour volume was performed when the second mouse across the entire cohort reached a TV of ≥ 1200 mm3, to account for the presence of a single allograft tumour with an atypical growth pattern in the IgG control group (rapid tumour growth over a 2-day interval – this mouse was included in the analysis and censored at the time of death). h. Evaluation of anti-tumour activity of distinct combination immunotherapies in the KL5 allograft model (N = 7 mice/group). Comparison of tumour volume was performed at the time point where the first mouse in any treatment group reached a TV ≥ 1200 mm3. The Mann–Whitney U test was used for pairwise statistical comparisons. Error bars represent standard deviation from the mean. Statistical significance is indicated at the P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) levels.
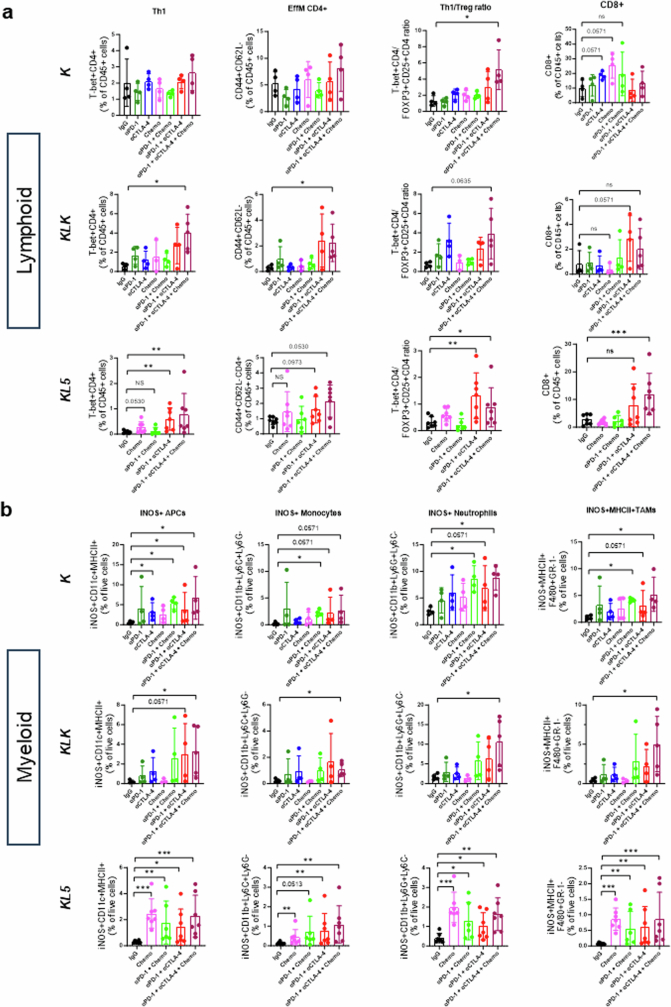
To further validate these findings, we used several syngeneic models driven by either KrasG12C (the most prevalent KRAS mutant allele in NSCLC) or KrasG12D and concurrent inactivation of Stk11 and/or Keap1. We surmised that owing to the partially shared biology between Keap1 and Stk11 loss, including the upregulation of a NRF2-driven transcriptional program18,36, dual ICB might represent an effective therapeutic strategy that applies to both oncogenotypes, as supported by subgroup analyses from POSEIDON. In several immune-competent mouse models of KrasG12C or KrasG12D-driven NSCLC with Keap1 and/or Stk11 inactivation with varying degrees of resistance to anti-PD-1 therapy, combined anti-PD-1/anti-CTLA4 significantly inhibited the growth of subcutaneous allograft tumours and prolonged survival (defined as time to a tumour volume of at least 1,200 mm3, or death or loss of condition), compared with isotype IgG control (Fig. 3f). Stk11-deficient models included the genetically engineered mouse KG12CL2 (KL2) and KG12CL5 (KL5) tumour-derived LUAD models as well as an isogenic derivative of the LKR13 KrasG12D-driven LUAD cell line with CRISPR–Cas9-mediated inactivation of Stk11 and Keap1 (KLK, clone 18). Although Keap1-deficient models also showed relative resistance to PD-1i monotherapy, this effect was more variable and ranged from complete to partial insensitivity; by contrast, the KK (KrasG12D-mutant and Keap1-deficient) and KLK LKR13 models exhibited marked sensitivity to anti-CTLA4—both in combination with anti-PD-1 and also as monotherapy—and this resulted in long-term tumour regression in a subset of treated mice (Fig. 3f). Similar results were obtained when the preclinical trial was expanded to incorporate LKR13 allograft cohorts (originating from the distinct single-cell-derived clone 17) treated with platinum doublet chemotherapy either alone or in combination with anti-PD-1 or anti-PD-1/anti-CTLA4, to better emulate the arms of the POSEIDON clinical trial (Extended Data Fig. 5f,g). Of note, although both anti-PD-1 therapy (with or without concurrent platinum-based doublet chemotherapy) and dual ICB (with or without chemotherapy) curtailed in vivo tumour growth in the K allograft model, only dual ICB (with or without chemotherapy) exhibited robust anti-tumour activity in the isogenic KLK model (Extended Data Fig. 5g). Furthermore, reinvigoration of anti-tumour immunity in the KLK allograft model was achieved selectively with PD-1/CTLA4 co-inhibition and was not observed with concurrent inhibition of PD-1 and either TIM-3 or LAG-3 (Extended Data Fig. 5h). Together, these findings provide evidence that dual ICB encompassing anti-PD-(L)1 in combination with anti-CTLA4 can mitigate primary resistance to PD-1i therapy in Stk11-deficient and, even more markedly, in Keap1-deficient NSCLC models.
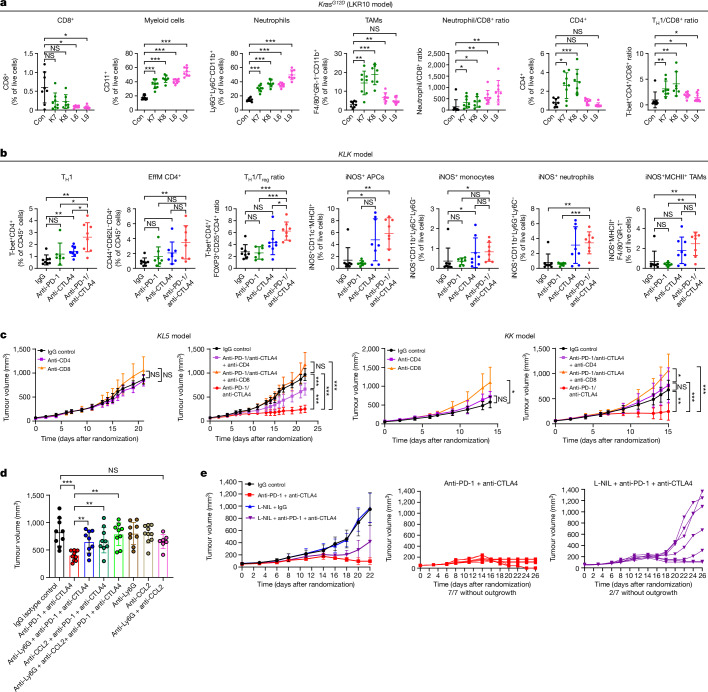
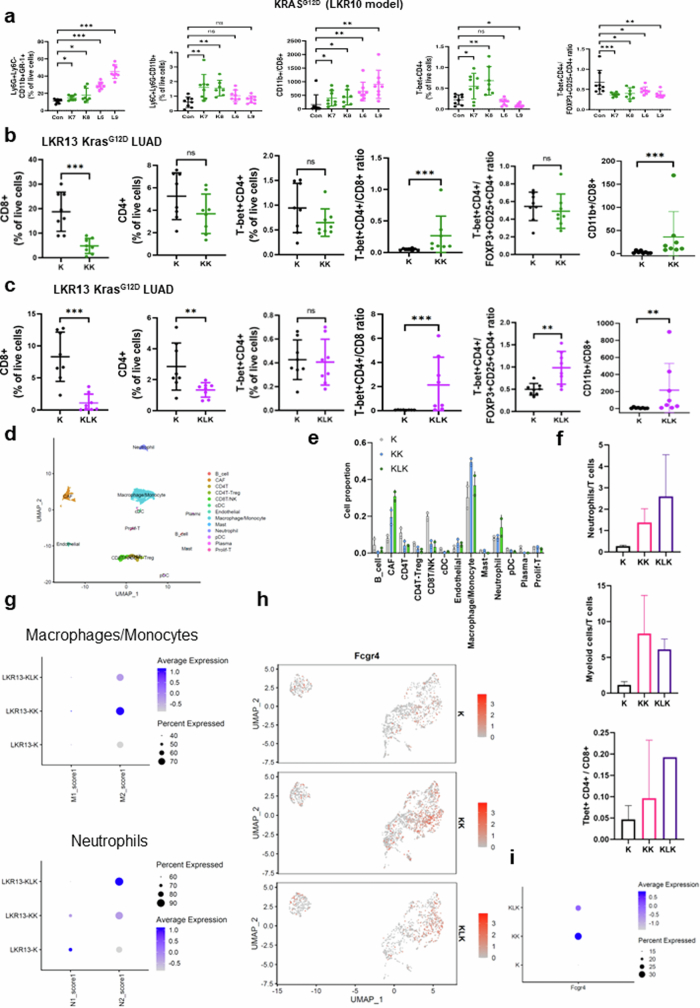
To interrogate the mechanistic underpinnings of increased sensitivity to dual ICB in STK11 and/or KEAP1-mutant NSCLC, we next investigated the immune phenotype of Stk11 and/or Keap1-deficient mouse tumours and the effect of blockade of the PD-(L)1 and/or CTLA4 pathways using fluorescence-activated cell sorting (FACS)-based immune profiling. The tumour immune microenvironment (TIME) of both Stk11- and Keap1-deficient tumours before treatment was characterized by an accumulation of myeloid cell subsets and a paucity of CD8+ cytotoxic T cells, resulting in a markedly increased CD11b+/CD8+ ratio (Fig. 4a and Extended Data Fig. 6a–c), consistent with their previously reported immunosuppressive phenotype5,12,13,37. Notably, Stk11 inactivation predominantly boosted the polymorphonuclear neutrophil compartment and triggered a prominent increase in the neutrophil/CD8+ ratio, whereas loss of Keap1 fostered a TIME rich in tumour-associated macrophages and monocytes as well as neutrophils (Fig. 4a and Extended Data Fig. 6a). These results were further supported by single-cell RNA sequencing (scRNA-seq) analysis, which revealed an enrichment of immune-suppressive transcriptional states in myeloid cells retrieved from Stk11- and/or Keap1-deficient tumours (Extended Data Fig. 6d–i). This imbalance in the relative abundance of myeloid and lymphoid subsets in STK11MUT and/or KEAP1MUT tumours was recapitulated in three independent cohorts of surgically resected nsNSCLC and was further validated using functional signatures for LKB1 loss or NRF2 transcriptional activation38 (Extended Data Fig. 7a–c). Despite a substantial reduction in T cells as a fraction of the total immune infiltrate, as compared with wild-type controls, a more detailed analysis revealed non-uniform changes in distinct populations of effector T cells. Keap1-deficient tumours (K7 and K8) had a trend towards fewer CD8+ cytotoxic T cells but a retention of or increase in CD4+ T cell subsets, including T helper 1 (TH1) cells (T-bet+CD4+), resulting in a significantly increased TH1/CD8+ ratio in all tested models (Fig. 4a and Extended Data Fig. 6a–c). Stk11-deficient tumours had a profound depletion of CD8+ cells (Fig. 4a and Extended Data Fig. 6c) and relative retention of CD4+ T cells, also resulting in an increased TH1/CD8+ ratio (Fig. 4a and Extended Data Fig. 6a,c). Our analysis of the Immunogenomic Profiling of Non-small cell lung cancer (ICON) dataset of surgically resected nsNSCLC revealed broadly consistent patterns: both STK11MUTKEAP1MUT or WT and STK11WTKEAP1MUT nsNSCLC exhibited a relative enrichment in TH1 CD4+ T cells (Extended Data Fig. 7d). Similar findings were observed in The Cancer Genome Atlas (TCGA) dataset (Extended Data Fig. 7e).
Fig. 4. Innate immune cells and CD4+ effectors are crucial mediators of dual anti-PD-1/anti-CTLA4 efficacy in Stk11- and/or Keap1-deficient models of KRAS-mutant NSCLC.
a, FACS-based enumeration of immune cell subsets in Keap1-deficient (K7, K8), Stk11-deficient (L6, L9) or isogenic Keap1 and Stk11-proficient LKR10 (control, Con) allograft tumours reveals a myeloid-cell-enriched and CD8+ T-cell-depleted suppressive TIME with relative sparing of TH1 CD4+ cells. Data are mean ± s.d. (n = 7–8 mice per group). TAMs, tumour-associated macrophages. b, FACS-based assessment of single and dual ICB-induced changes in the abundance of distinct T cell (left panels) and myeloid cell (right panels) subsets in the microenvironment of the Stk11- and Keap1-deficient KLK model. Data are mean ± s.d. (n = 7–8 mice per group). EffM, effector memory cells. c, Effect of CD4+ or CD8+ depletion on the in vivo growth kinetics of KL5 and KK models in the absence of treatment or with dual anti-PD-1/anti-CTLA4 therapy (n = 7–8 mice per group). Comparison of tumour volume was performed at the time point at which the first mouse in any treatment group reached a tumour volume ≥ 1,500 mm3. d, The anti-tumour activity of dual PD-1/CTLA4 ICB in the KL5 model is dependent on innate immune cells (n = 7–10 mice per group). Tumour volume is shown for the indicated treatment arms. e, iNOS inhibition curtails the anti-tumour efficacy of dual ICB in the KLK model (n = 7–8 mice per group). Comparison of tumour volume was performed at the time point at which the first mouse in any treatment group reached a tumour volume ≥ 1,200 mm3. Individual tumour growth trajectories in the anti-PD-1 + anti-CTLA4 (red) and L-NIL + anti-PD-1 + anti-CTLA4 (purple) treatment arms are also shown. Mann–Whitney U test was used for all pairwise statistical comparisons. Data are mean ± s.d. Statistical significance is indicated (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Extended Data Fig. 6. Characterization of the STK11MUT and/or KEAP1MUT NSCLC TIME.
a. FACS-based enumeration of immune cell subsets in Keap1- deficient (K7,K8), Stk11-deficient (L6,L9) or isogenic Keap1 and Stk11-proficient LKR10 allograft tumours. Error bars indicate standard deviation from the mean (N = 7-8 mice/group). b,c. FACS-based assessment of T cell subsets in the immune microenvironment of syngeneic Keap1-deficient (KK) (b) or Keap1- and Stk11-deficient (KLK) (c) KrasG12D-mutant allograft tumours compared with isogenic Keap1 and Stk11-proficient (K) tumours (all models were derived from the LKR13 mouse LUAD cell line, with CRISPR/Cas9-mediated editing of the corresponding genomic loci). Error bars indicate standard deviation from the mean (N = 8 mice/group). The Mann–Whitney U test was used for comparison of the abundance of immune cell subsets. Statistical significance in all panels is indicated at the P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) levels. d. Uniform manifold approximation and projection (UMAP) plot of immune cells derived from untreated K, KK and KLK allograft tumours and processed for scRNA-seq (N = 2 mice/group). e. Bar plot depicting the relative abundance of distinct immune cell subsets (as a % of all non-tumour cells) in each isogenic model. f. Representation of scRNA-seq-derived neutrophil/T cell (top panel), myeloid/T cell (middle panel) and TH1 CD4+/CD8+ T cell (bottom panel) ratios across the K, KK and KLK isogenic models. g. Proportions and average M1 or M2 scores (top panel) and N1 or N2 scores (bottom panel) in the monocyte/macrophage or neutrophil compartments in the K, KK and KLK models. h,i. UMAP visualization (h) and average expression levels (i) of Fcgr4 mRNA expression in the myeloid compartment demonstrating enrichment in Stk11 and/or Keap1-deficient models.
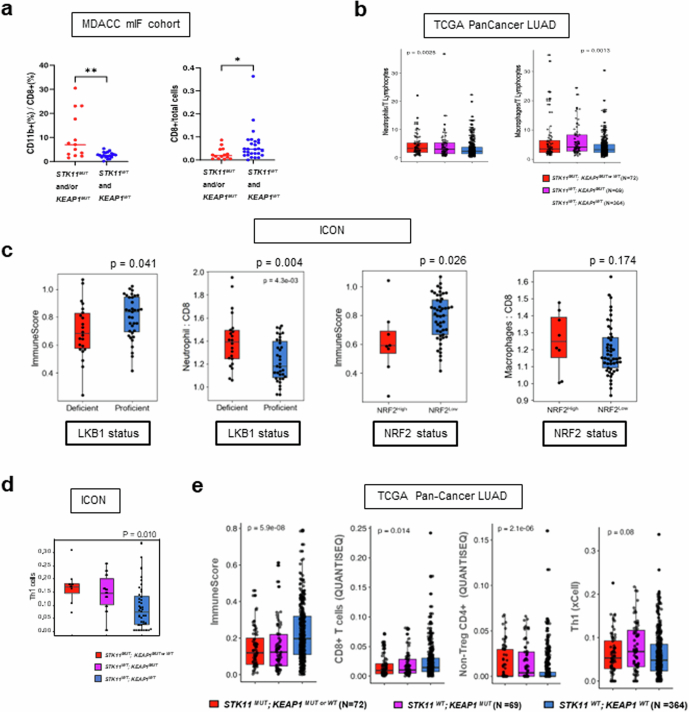
Extended Data Fig. 7. Characterization of the STK11MUT and/or KEAP1MUT NSCLC TIME in clinical cohorts.
a. Multicolour immunofluorescence (mIF) analysis of surgically resected early-stage human nsNSCLC confirms a higher ratio of CD11b+/CD8+ cells (N = 13, STK11MUT and/or KEAP1MUT and N = 19, STK11WT and KEAP1WT) and lower abundance of CD8+ T cells (N = 14; STK11MUT and/or KEAP1MUT and N = 27; STK11WT and KEAP1WT) in STK11MUT and/or KEAP1MUT NSCLC. The Mann–Whitney U test was used for statistical comparisons. b. Inferred neutrophil: T lymphocyte and macrophage: T lymphocyte ratios in STK11 and/or KEAP1-mutant LUAD in the TCGA dataset. The Kruskal–Wallis test was used for the three-group statistical comparisons. c. Immune contexture of LKB1-deficient (N = 22) versus LKB1-proficient (N = 35) (top row) and NRF2High (N = 8) versus NRF2Low (N = 49) (bottom row) nsNSCLC in the ICON cohort, based on previously validated gene expression signatures. The Mann–Whitney U test was used for statistical comparisons and P ≤ 0.05 was considered statistically significant. d. xCell-based digital deconvolution of the tumour immune microenvironment in the ICON cohort of surgically resected nsNSCLC (N = 8, STK11MUT;KEAP1MUTorWT; N = 10, STK11WT;KEAP1MUT;N = 39, STK11WT;KEAP1WT). Each box indicates the interquartile range (IQR) with the median and whiskers indicate the upper and lower values within 1.5 times the IQR. The Kruskal–Wallis test was used for the three-group statistical comparison and P ≤ 0.05 was considered statistically significant. e. RNA-seq-based deconvolution of the STK11MUT and or KEAP1MUT TIME in the TCGA PanCancer Atlas lung adenocarcinoma cohort. Left to right: ImmuneScore (assessed by xCell); CD8+ T cells (evaluated by QuanTIseq); non-Treg CD4+ T cells (QuanTIseq); and TH1 signature (xCell). Each box indicates the interquartile range (IQR) with the median and whiskers indicate the upper and lower values within 1.5 times the IQR. The Kruskal–Wallis H test was used for statistical comparisons.
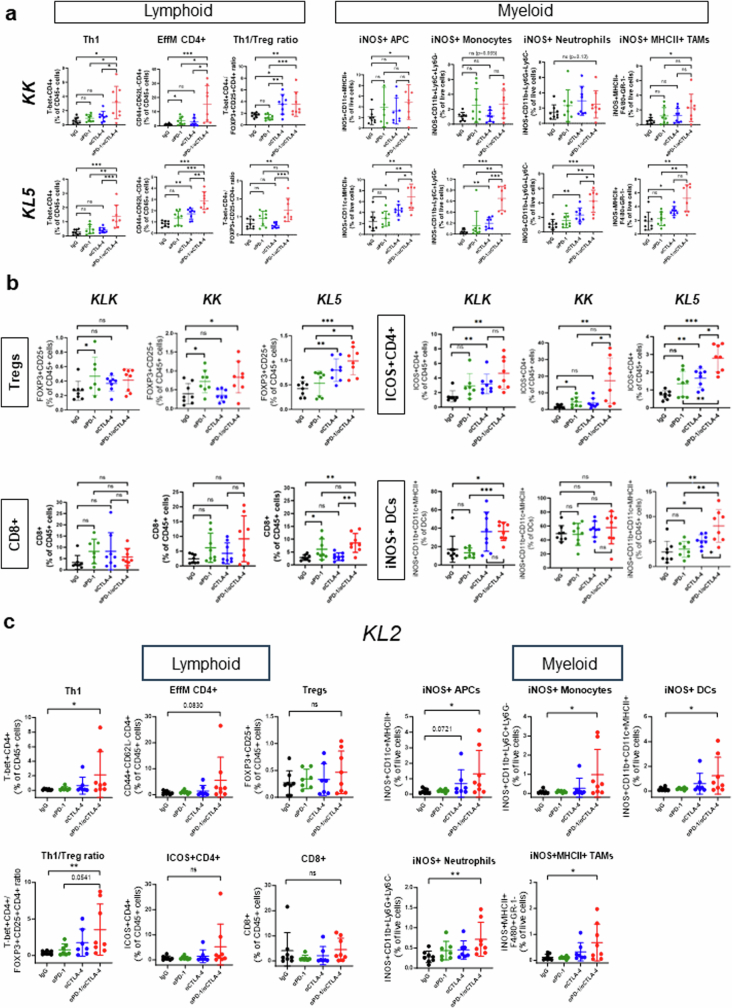
To further investigate the sensitivity of Keap1- and Stk11-mutant tumours to CTLA4 blockade and dual ICB, we treated immune-competent mice, bearing syngeneic Kras-mutant flank tumours with inactivation of Keap1 (KK; LKR13 KrasG12DK), Stk11 (KL5; KG12CL5) or both (KLK; LKR13 KrasG12D, clone 18), with anti-PD-1, anti-CTLA4, dual ICB or IgG isotype controls. Tumours were collected two weeks after the start of treatment, and the modulation of distinct myeloid and T cell populations was subsequently evaluated by FACS-based profiling (Fig. 4b and Extended Data Fig. 8a,b). Dual ICB resulted in a robust increase in CD4+ subsets, including TH1 T cells (T-bet+CD4+), and effector memory CD4+ T cells (CD4+CD44+CD62L−), as a percentage of the CD45+ population across all tested models (Fig. 4b and Extended Data Fig. 8a). This finding is consistent with previous reports of CD4+ T cell phenotypic expansion in response to CTLA4 pathway blockade39,40. Notably, an increased frequency of ICOS+CD4+ cells in tumour tissue in response to combined PD-1/CTLA4 blockade (which was also evident in our models; Extended Data Fig. 8b) was previously associated with improved efficacy in clinical studies, and with increased cytokine production, which might promote the recruitment and activation of CD8+ T cells40,41. Regulatory T (Treg) cells were not depleted in response to dual ICB in our study, and in all cases the TH1/Treg ratio was substantially enhanced (Fig. 4b and Extended Data Fig. 8a,b). Similar findings were observed in another Stk11-deficient model (KL2; KG12CL2) (Extended Data Fig. 8c). The effects of dual therapy on CD8+ cell subsets were less prominent and varied across individual models, although a modest increase was generally observed with dual ICB (Extended Data Fig. 8b,c). Of note, dual ICB considerably remodelled the myeloid cell compartment and triggered a marked induction of iNOS+ antigen-presenting cells, monocytes, neutrophils and MHCII+ tumour-associated macrophages across several tested models (Fig. 4b and Extended Data Fig. 8a–c). Comparable results were obtained in a distinct single-cell-derived LKR13 model (KLK clone 17) in response to dual ICB, with or without chemotherapy (Extended Data Fig. 9a,b). Consistent with their recalcitrance to PD-1 inhibition (as monotherapy or in combination with platinum doublet chemotherapy), KLK allograft tumours exhibited increased CD4+ TH1 cells and an accumulation of iNOS-expressing tumoricidal myeloid cell subsets only in the treatment arms that included concurrent CTLA4 inhibition (Extended Data Fig. 9a,b).
Extended Data Fig. 8. Dual anti-PD-1/anti-CTLA4 blockade reshapes the immune contexture of Stk11- and/or Keap1-deficient models of KRAS-mutant NSCLC.
a. FACS-based assessment of single and dual ICB-induced changes in the abundance of distinct T cell (left panels) and myeloid cell (right panels) subsets in the microenvironment of Stk11-deficient (KL5) and Keap1-deficient (KK) models. Error bars indicate standard deviation from the mean (N = 7-8 mice/group). The Mann–Whitney U test was used for pairwise comparisons of the abundance of immune cell subsets. Statistical significance is indicated at the P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) levels. b. ICB-induced changes in the abundance of distinct T cell and dendritic cell subsets in the microenvironment of KLK, KK and KL5 models assessed by FACS. Error bars indicate standard deviation from the mean (N = 7-8 mice/group). c. FACS-based quantification of lymphoid and myeloid cell subsets in KL2 allograft tumours in response to single or dual ICB. Error bars indicate standard deviation from the mean (N = 7-8 mice/group). The Mann–Whitney U test was used for comparison of the abundance of immune cell subsets. Statistical significance in all panels is indicated at the P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) levels.
Extended Data Fig. 9. Treatment-induced immune modulation in the TIME of Stk11- and/or Keap1-deficient models of KRAS-mutant NSCLC.
a,b, FACS-based enumeration of lymphoid (a) and myeloid cell (b) subsets in isogenic K (LKR13 KrasG12D-mutant; Stk11/Keap1 WT) and KLK (KrasG12D-mutant, Stk11/Keap1 knockout, clone 17) allograft models as well as in the KL5 (KrasG12C-mutant Stk11-deficient) allograft model (N = 3-5 mice/group for the K and KLK models and N = 6-7 mice/group for the KL5 model). The Mann–Whitney U test was used for pairwise statistical comparisons. Error bars represent standard deviation from the mean. Statistical significance is indicated at the P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) levels.
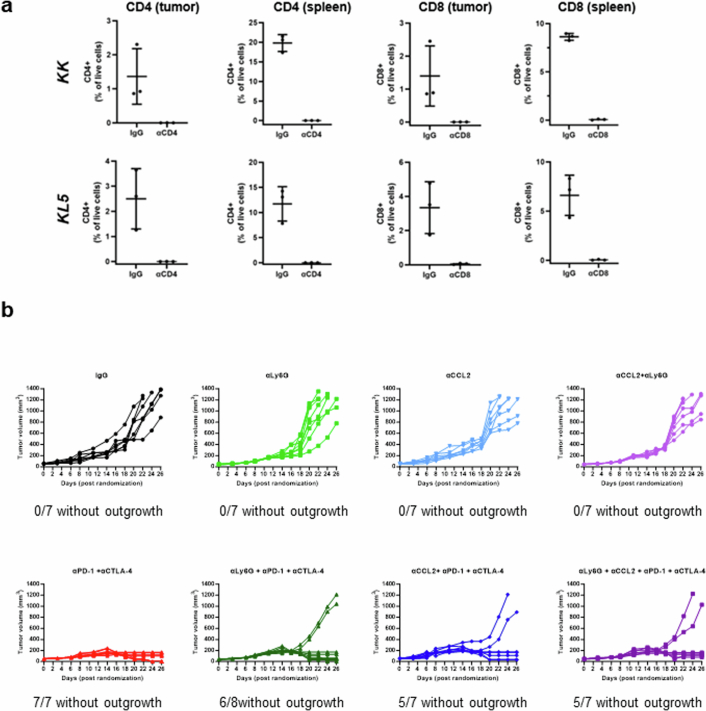
These data suggest that innate immune cells, including one or more subpopulations of myeloid cells, as well as CD4+—and potentially CD8+—T cell effector subsets, could be important contributors to the response of these tumours to dual ICB. To directly test this hypothesis, we initially specifically depleted CD4+ or CD8+ T cells using anti-CD4 or anti-CD8 antibodies and tested the effect of dual ICB on the growth of the aforementioned KK and KL5 syngeneic models in immune-competent mice. Selective depletion of CD4+ or CD8+ T cells was confirmed by FACS analysis of splenocytes and tumours (Extended Data Fig. 10a). Growth of KL5 tumours in the absence of ICB was not affected by CD4+ or CD8+ T cell depletion, consistent with their inert immune phenotype at baseline (Fig. 4c). Depletion of CD8+ (but not CD4+) T cells fostered accelerated tumour growth in the KK model, indicative of more active immune surveillance in this oncogenotype, whereas depletion of CD4+ T cells had a minor effect. Dual ICB significantly curtailed in vivo tumour growth compared with isotype-control-treated mice in both models (Fig. 4c). Depletion of CD8+ T cells nullified the anti-tumour activity of combination therapy in both models. Crucially, the efficacy of dual ICB was also strictly dependent on CD4+ T cells in the Keap1-deficient KK model, whereas Stk11-deficient KL5 tumours exhibited a partial dependence on CD4+ T cells. This effect was observed despite co-depletion of both effector and suppressive (that is, Treg) CD4+ T cell subsets (Fig. 4c). We next examined the potential contribution of myeloid cell subsets to the anti-tumour activity of dual ICB. Treatment with anti-Ly6G, anti-CCL2 or their combination (anti-Ly6G + anti-CCL2), before and during dual ICB, completely abolished the efficacy of combined anti-PD-1/anti-CTLA4 inhibition in the KL5 model (Fig. 4d), whereas a more modest effect was observed in the KLK model (Extended Data Fig. 10b). Notably, chemical inhibition of iNOS with the highly specific inhibitor L-NIL (N6-(1-iminoethyl)-l-lysine) in the KLK model partially abrogated the eradication of tumours in response to dual ICB (Fig. 4e). The dependence on CD4+ T cells, myeloid cells and iNOS for the immune-mediated killing of these tumours is noteworthy in light of a previous report, which found that CD4+ T-cell-induced inflammatory cell death can have a key role in the eradication of immunologically inert, MHC class I-deficient melanoma tumours that escape direct CD8+ targeting42. In that study, it was observed that relatively sparse CD4+ TH1 cells present at tumour margins reprogram myeloid cells towards a more phagocytic and tumoridical phenotype and trigger distal inflammatory cell death. Neutrophil-mediated, iNOS-dependent eradication of tumour cells in response to dual ICB was also reported to be crucial for the clearance of antigenically heterogeneous tumours43,44; this dependence might thus be particularly relevant for STK11MUT tumours that are characterized by defects in antigen processing and/or presentation16.
Extended Data Fig. 10. Immune depletion studies.
a. Effective immune depletion of CD4+ or CD8+ T cells with anti-CD4+ and anti-CD8+ antibodies, respectively. Error bars indicate standard deviation from the mean (N = 3 mice/group). b. Spider plots indicating individual LKR13 KLK allograft tumour volume trajectories in response to the indicated therapies (N = 7-8 mice/group).
Together, this study provides evidence that STK11 and KEAP1 mutations are associated with a relative lack of benefit from PD-(L)1 inhibitors in combination with chemotherapy in patients with metastatic NSCLC, and that this resistance can be mitigated by the addition of CTLA4 blockade, as observed in the TDCT arm of the POSEIDON study. Unbiased genetic screens in several immune-competent mouse models confirmed that the inactivation of Stk11 and Keap1 promotes resistance to PD-(L)1 inhibitors and confers selective sensitivity to dual ICB. Mechanistically, combined inhibition of PD-(L)1 and CTLA4 exploits at least two cardinal features of the suppressive STK11MUT and/or KEAP1MUT NSCLC TIME: (a) the relative retention of certain anti-CTLA4-responsive CD4+ T cell subsets, including TH1 T cells (T-bet+CD4+); and (b) a myeloid-cell-rich tumour ecosystem that can be reprogrammed in response to dual ICB towards iNOS-expressing tumoricidal phenotypes. This distinct imbalance in the immune contexture of STK11MUT and/or KEAP1MUT NSCLC could, at least partially, underpin the clinical utility of dual ICB in combination with chemotherapy for this difficult-to-treat population of patients. Ostensibly, the recruitment of circulating neutrophils might also contribute to the anti-tumour activity of dual ICB, and it is notable that patients with STK11MUT NSCLC have previously been reported to have a higher circulating neutrophil-to-lymphocyte ratio45. Furthermore, although our experiments were not designed to address the possible contribution of direct FcγR-mediated innate immune remodelling, this might, at least partially, affect anti-tumour efficacy in clinical settings46–48. In support of this possibility, an analysis of single myeloid cell transcriptomes from KK or KLK allograft tumours revealed increased expression of Fcgr4 (Extended Data Fig. 6h,i). Several possible further effects of dual ICB, including de novo CD8+ T cell priming in lymphoid organs and expansion of the T cell receptor repertoire, might not be oncogenotype specific but might rather apply broadly to immunologically ‘cold’ tumours across different cancer types. Additional downstream mediators of this immune phenotype and CTLA4 inhibitor responsiveness are an area of active investigation. Previous studies have highlighted STING pathway suppression15,49, impaired antigen presentation16 and enhanced lactate37 or IL-6 secretion13,50 as potential mediators of PD-(L)1 inhibitor resistance in STK11MUT tumours. Finally, the observation that dual ICB mitigates STK11MUT and KEAP1MUT-associated PD-(L)1 inhibitor resistance suggests that these genes could serve as biomarkers for identifying patients with NSCLC who will benefit from the addition of CTLA4 blockade. Given the inherent limitations of interpreting post-hoc analyses in patient subgroups, prospective clinical studies that directly compare dual ICB and PD-(L)1 inhibition in combination with chemotherapy in these subgroups—such as the actively recruiting phase IIIb TRITON clinical trial (NCT06008093)—are warranted, and these findings could ultimately change how patients with NSCLC who have these most recalcitrant tumour genotypes are cared for.
Methods
Retrospective multi-institutional nsNSCLC patient cohort
Study population
Reviews of electronic medical records were performed for patients from 22 academic institutions in North America and Europe, including MD Anderson Cancer Center, Memorial Sloan Kettering Cancer Center, Ohio State University, Dana Farber Cancer Institute, Massachusetts General Hospital, Cleveland Clinic, University of Chicago, Yale University, University of Pennsylvania, University of Colorado, University of Cologne, UHN Research, Columbia University Medical Center, Gustave Roussy, John’s Hopkins, Stanford University, University of Torino–Orbassano, University of California Davis, University of California Los Angeles, University of California San Francisco, Moffitt Cancer Center and University of North Carolina at Chapel Hill. Patients with stage IV nsNSCLC who received treatment with either (a) carboplatin or cisplatin, pemetrexed and pembrolizumab (PCP cohort) or (b) carboplatin or cisplatin and pemetrexed before the regulatory approval of PCP at each medical jurisdiction (CP cohort), were alive for 14 or more days after the start of treatment and had genomic profiling results that included STK11 (for patients in the PCP cohort) and STK11 and/or KEAP1 (for patients in the CP cohort) available from tumour or blood before starting first-line systemic therapy were eligible. Patients who had tumours with sensitizing EGFR mutations or ALK rearrangements were excluded. Patients treated with bevacizumab as part of first-line systemic therapy were excluded. Prior immunotherapy was not allowed; prior neoadjuvant or adjuvant chemotherapy for early-stage surgically resectable disease or as part of concurrent chemoradiation for locally advanced disease was allowed if completed at least five months before the initiation of systemic therapy for stage IV disease. The dataset was locked on 31 December 2018 for the PFS analysis and on 31 August 2019 for the OS analysis. Patient information was collected through chart review. Tumour cell PD-L1 expression was available for 88.6% of patients in the PCP cohort and was determined with the Dako 22C3 pharmDx or 28-8 pharmDx, Ventana SP263 or Ventana SP142 and E1L3N assays (data regarding the used assay were not available for 15 patients). The study was approved by an Institutional Review Board (IRB) at participating centres and included a waiver of patient informed consent. This study was conducted in accordance with ethical guidelines including the Declaration of Helsinki and US Common Rule.
Genomic profiling
Patients must have had genomic profiling results including STK11 (for patients in the PCP cohort) and STK11 and/or KEAP1 (for patients in the CP cohort) from tumour and/or plasma prior to starting first-line systemic therapy to be included in the analysis. Only tests performed through commercially approved assays or in a CLIA-certified laboratory were allowed. When available, we integrated results from tumour and plasma profiling for the analysis. All nonsynonymous STK11 and KEAP1 missense mutations and bi-allelic deletions were considered pathogenic. Cross-platform TMB harmonization was performed by applying a normal transformation followed by standardization to z-scores as previously described51. A threshold of 8.58 mutations per Mb for harmonized TMB (representing the whole-exome sequencing equivalent of 10 mutations per Mb in the FoundationOne CDx assay) was used as a cut-off to separate patients with low (<8.58 mutations per Mb) versus high (≥8.58 mutations per Mb) TMB.
Survival and objective response analyses
For the PFS analysis, data for patients who were alive and had no evidence of progression at the time of dataset lock or who were lost to follow-up were censored at the time of the last radiologic tumour assessment. For the OS analysis, data for patients who were alive or lost to follow-up at the time of dataset lock were censored at the time of the last documented patient contact. The Kaplan–Meier method was used to estimate PFS and OS, and differences were assessed by log-rank test. HRs and corresponding CIs were estimated with the use of a stratified Cox proportional hazards model adjusted for clinical variables (age, history of brain metastasis, performance status (0−1 versus >1)). Best response was determined through investigator-assessed RECIST v.1.1. ORR was defined as the percentage of response-evaluable patients who achieved a complete or partial response. Patients who died 14 days or more after the start of first-line systemic therapy, but before the first restaging scan, were considered to have progressive disease. Attribution of stable disease as best overall response to therapy required a minimum interval of 30 days between the first day of the first cycle of treatment (C1D1) and radiologic evaluation. Differences in categorical variables were assessed by two-sided Fisher’s exact test. Significance was established at P ≤ 0.05. Statistical analysis was performed in IBM SPSS Statistics v.24.0, R v.4.1.2 (2021-11-01) and SAS v.9.4.
POSEIDON phase III randomized clinical trial dataset
Study population
Study design and patient eligibility criteria for the POSEIDON global phase III randomized clinical trial (ClinicalTrials.gov identifier: NCT03164616) have been previously described3. In brief, patients were eligible if they were at least 18 years old, were diagnosed with stage IV NSCLC and had not previously received systemic therapy for metastatic NSCLC; had ECOG PS 0 or 1; and had measurable disease according to RECIST v.1.1. Tumour PD-L1 expression assessment at a central laboratory using the VENTANA PD-L1 (SP263) immunohistochemistry assay (Ventana Medical Systems) was required before random assignment. Patients who had tumours with sensitizing EGFR mutations or ALK rearrangements were not eligible to participate in the study. Patients with treated and stable brain metastases were eligible.
Treatment
Patients were randomly assigned (1:1:1) with stratification by PD-L1 expression (≥50% versus <50% of tumour cells), disease stage (IVA versus IVB, per the International Association for the Study of Lung Cancer Staging Manual in Thoracic Oncology v.8) and histology (squamous versus nonsquamous) to one of three treatment arms: (1) tremelimumab 75 mg plus durvalumab 1,500 mg and chemotherapy for up to four 21-day cycles, followed by durvalumab 1,500 mg once every 4 weeks until disease progression (PD), with one additional tremelimumab dose after chemotherapy at week 16/cycle 6 (fifth dose) (TDCT arm); (2) durvalumab 1,500 mg plus chemotherapy for up to four 21-day cycles, followed by durvalumab 1,500 mg once every 4 weeks until PD (DCT arm); or (3) platinum doublet chemotherapy for up to six 21-day cycles (CT arm). Only patients with nsNSCLC enrolled in POSEIDON were included in the analysis. Chemotherapy options for patients with nonsquamous histology included cisplatin or carboplatin and pemetrexed or, alternatively, carboplatin and nab-paclitaxel. Patients with nonsquamous histology who received pemetrexed–platinum doublet could receive pemetrexed maintenance therapy if eligible. Patients continued treatment until PD, unacceptable toxicity or withdrawal of consent. In-study crossover was not allowed.
The study was performed in accordance with the Declaration of Helsinki and the International Conference on Harmonisation Good Clinical Practice guidelines. The protocol and all modifications were approved by relevant ethics committees and regulatory authorities. All patients provided written informed consent.
End-points and statistical analysis
The primary end-points were progression-free survival (PFS), evaluated by blinded independent central review (BICR) per RECIST v.1.1, and overall survival (OS) for DCT versus CT. PFS was defined as the time from random assignment to objective PD or death from any cause in the absence of progression and OS as the time from random assignment to death from any cause. Key alpha-controlled secondary end-points were PFS and OS for TDCT versus CT.
A hierarchical multiple testing procedure with a gatekeeping strategy was used across the primary end-points and alpha-controlled secondary end-points. First, 1% alpha and 4% alpha were allocated to PFS and OS, respectively, for the DCT versus CT comparison. Positivity for either primary end-point enabled alpha recycling to the key secondary PFS and OS end-points (TDCT versus CT). If either of the key secondary PFS or OS end-points was met, the alpha could be recycled to the other key secondary end-point. The primary and key secondary PFS and OS analyses were performed using a stratified log-rank test adjusted for the stratification variables of tumour PD-L1 expression, disease stage and histology, with HRs and 95% CIs estimated using a stratified Cox proportional hazards model. ORR was analysed using a logistic regression model, adjusted for the same factors as the primary end-points, and odds ratios and 95% CIs calculated. The Kaplan–Meier method was used to calculate median OS, PFS and DoR. Efficacy data were analysed in the biomarker-evaluable nsNSCLC population.
POSEIDON biomarker analyses
Pre-treatment tumour tissue or plasma samples underwent next-generation sequencing (NGS) of DNA through Foundation Medicine’s FoundationOne CDx assay for tissue and Guardant Health’s GuardantOMNI assay for ctDNA. Pre-treatment tumour tissue samples were scored for tumour cell expression of PD-L1 using the VENTANA PD-L1 (SP263) immunohistochemistry assay, with a 1% or 50% cut-off for positivity. TMB was assessed as described previously52. Samples were considered altered in STK11 or KEAP1 if there was detection of a known nonsynonymous somatic mutation, as described in OncoKB53, any truncating alteration, including frameshift insertions and deletions, splice site mutations within 2 bp of exon, or homozygous deletion of one or more exons; and altered for KRAS if there was detection of a known missense somatic hotspot mutation.
Exploratory analyses of PFS (by (BICR) per RECIST v.1.1) and OS were performed using Kaplan–Meier estimates, with HRs and 95% CIs estimated using unstratified Cox proportional hazards models. ORRs were calculated using confirmed responses (at least one visit response of complete response or partial response and a confirmatory scan no sooner than four weeks after the initial response) by BICR per RECIST v.1.1. Kaplan–Meier estimates were used to describe the duration of response, defined as the time from the first documentation of complete response/partial response until the date of progression, death in absence of progression or the last evaluable RECIST assessment for patients who progressed or died after two or more missed visits. Analyses were performed using SAS v.9.4 and R v.4.2.0.
Foundation Medicine cohort
A large cohort of 8,592 unselected patients with LUAD who submitted samples to FMI for hybrid capture-based comprehensive genomic profiling were included in an integrated analysis of TMB, PD-L1 expression and genomic alterations of individual cancer-related genes. Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (protocol no. 20152817). Samples submitted to Foundation Medicine were processed at a CLIA-certified laboratory as described previously54. TMB was measured by Foundation Medicine as described previously52. Raw TMB values were measured in units of mutations per Mb sequenced and characterized as low (TMB < 6), intermediate (6 ≤ TMB < 20) or high (TMB ≥ 20). Assessment of tumour cell PD-L1 expression in the FMI cohort was based on the Dako 22C3 pharmDx assay. Tumours were characterized as PD-L1 negative (PD-L1 TPS < 1%), low positive (PD-L1 TPS 1-49%) or high positive (PD-L1 TPS ≥ 50%). In the FMI cohort, a LUAD sample was considered altered in STK11 or KEAP1 if there was detection of a known or likely pathogenic nonsynonymous somatic mutation, any truncating alteration or bi-allelic loss. Statistical analyses were assessed by two-sided Fisher’s exact test and significance was established at FDR-adjusted P ≤ 0.05.
MD Anderson Cancer Center mIF cohort
Surgically resected early-stage human NSCLC specimens obtained at the MD Anderson Cancer Center were evaluated by multiplex immunofluorescence (mIF), following a similar methodology to that previously described55. In brief, 4-µm-thick formalin-fixed, paraffin-embedded tumour sections were stained using an automated staining system (BOND-RX; Leica Microsystems), applying previously validated mIF antibody panels55. Staining was performed sequentially according to each corresponding fluorophore in the Opal 7 colour IHC kit (NEL797001KT; Akoya Biosciences), including DAPI and Opal Polaris 520, 540, 570, 620, 650, 690 and coumarin. Stained tissue microarray (TMA) slides were scanned using the multispectral microscope, PhenoImager HT (formerly Vectra Polaris) 1.0.13 imaging system (Akoya Biosciences), under fluorescence at low magnification at 10× and high magnification at 20×. Each core from the TMAs was analysed using InForm 2.8.2 image analysis software (Akoya Biosciences). Marker colocalization was used to identify different cellular phenotypes and quantify the number of cells per mm2. Data were consolidated using RStudio 3.5.3 (Phenopter 0.2.2; https://rdrr.io/github/akoyabio/phenoptrReports/f/, Akoya Biosciences). NGS-based genomic profiling was obtained by either whole-exome sequencing (WES) or CLIA-certified clinical NGS assays as part of routine standard of care and was accessed as part of an IRB-approved research protocol.
Analysis of the ICON NSCLC cohort
Tumour samples from patients enrolled in the ICON cohort56 with nsNSCLC histology, no prior neoadjuvant chemotherapy and available transcriptional profiling data were considered eligible (n = 57). Raw RNA-seq data underwent normalization and transformation using the vsd function from DESeq2. Z-scores and average gene expression data were used for signature-based evaluation of immune cell subsets. TH1 levels and overall immune infiltrate (ImmuneScore) were estimated as gene signatures using XCell57 and plotted according to the presence of STK11 and/or KEAP1 somatic mutations or functional status using previously validated signatures for LKB1 deficiency58 or NRF2 transcriptional activation8. Neutrophil, macrophage and CD8+ T cell signature data were assessed with MCPCounter to obtain ratios per sample. All analyses were performed using Python v3.9 and R v.4.3.1.
Analysis of the TCGA PanCancer Atlas lung adenocarcinoma dataset
Paired WES, RNA-seq and clinical data from patients with LUAD from the TCGA PanCancer Atlas cohort were downloaded from the cBioPortal for Cancer Genomics repository59. Immune cell subsets were estimated using aggregated data from TIMER2.060, which provides standardized output from six different immune estimation methods. Neutrophil, macrophage and CD8+ T cell signature data were assessed with MCPCounter to obtain ratios per sample. QuanTIseq was used to compare T cell and nonregulatory CD4 levels across mutation groups. Overall immune infiltrate (ImmuneScore) and TH1 levels were estimated as gene signatures using XCell57. Comparisons across groups were performed using the Kruskal–Wallis H test. Analyses were performed using R v.4.0.3.
In vivo CRISPR–Cas9 screen for drivers of immune evasion and PD-1 inhibitor resistance
Library design and construction
We established a library of 1,813 sgRNAs targeting 162 genes, with a focus on genes with loss-of-function mutations or deletions in cancer. In addition, we included known essential genes, immune resistance genes and immune sensitizing genes, to assess and validate the in vivo screening platform. Eleven per cent of the sgRNAs included in the library represented non-targeting controls. sgRNA oligos were cloned into a lentiCRISPR v2 plasmid61, using Gibson assembly.
Cell line engineering
The 3LL Lewis lung carcinoma cell line was purchased from the JCRB cell bank. 3LL cells were grown in RPMI-1640 with 10% heat-inactivated fetal bovine serum (FBS) at 37 °C with 5% CO2. 293FT cells were purchased from Invitrogen and were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated FBS. A lentiviral vector library was produced in 293FT cells with transfection of library plasmids plus packaging helper plasmids psPAX2 and pVSVG using lipofectamine 3000 (Invitrogen). Virus-containing supernatant was collected 66 h after transfection and concentrated by ultracentrifugation. 3LL cells were transduced with the sgRNA library at an infection rate of 30% and subjected to puromycin selection four days after infection to generate stable cells. Cells were passaged several times in culture, to allow for effective gene editing.
In vivo CRISPR screening
The in vivo screen was conducted at WuXi AppTec, China. The study design was approved by the WuXi AppTec Institutional Animal Care and Use Committee (IACUC). Six-to-eight-week-old BALBc nude mice and C57BL/6J mice were purchased from Shanghai SLAC Laboratory Animal Co. For screening, 2 × 106 library-transduced 3LL cells, resuspended in 0.1 ml phosphate-buffered saline (PBS), were injected subcutaneously into the right flank. Mice were grouped for treatment when average tumour volume reached approximately 40–60 mm3 on day 0. C57BL/6J mice were treated by intraperitoneal injection with anti-mPD-1 (BioXCell, BE0146) or anti-IgG2a (BioXCell, BE0089) at 5 mg per kg or 10 mg per kg, twice per week for two weeks. Tumours were measured every other day and tumour volume was estimated with the formula (L × W2)/2. Mice were euthanized and tumours collected for genomic DNA extraction when the average tumour volume for the group reached around 1,000 mm3. Tumours from the nude mice group and the C57BL/6 groups were collected on day 11 and day 13, respectively. For genomic DNA extraction, tumour tissues were ground by FreezerMill in liquid nitrogen. Tissue powders were used for DNA extraction using the Blood & Cell Culture DNA Midi kit or Mini kit (Qiagen), according to the manufacturer’s protocol.
Data analysis
Genomic DNA isolation and sequencing were performed as previously described62. To normalize the raw counts of sgRNA guides in each sample, a sample-specific size factor was determined by the ratio of the median count of the non-targeting control guides to the one of the plasmid pool. The raw counts were scaled by multiplying the sample-specific size factors for each sample and adding one to all the values. The log2-transformed fold change for each sgRNA in each perturbed arm was calculated using the control arm as a reference and averaged over biological replicates. The gene-level log2-transformed fold change was computed by averaging the sgRNAs targeting the same gene. Raw sgRNA counts were also used as input into MAGeCK to call significantly enriched and depleted genes using the MAGeCK Python package v.0.5.763,64. The median ratio of non-targeting sgRNAs was chosen as the normalization method when running MAGeCK. Gene-level score matrices included log2-transformed fold change, FDR and P values.
Multiplexed in vivo assessment of TSG loss-selective vulnerability to single and dual ICB in autochthonous KrasG12D-driven LUAD using Tuba-seq
Design and generation of Lenti-sgRNA/Cre vectors
We generated lentiviral vectors encoding Cre (expressed from a PGK promoter65) and an sgRNA (expressed from a human U6 promoter) targeting each of the following 22 genes, which are known or putative tumour suppressors that are recurrently mutated in LUAD (or pan-carcinoma) and represent diverse cancer pathways66,67: Apc, Arid2, Atm, Atrx, Brca2, Cdkn2a, Cmtr2, Keap1, Kmt2d, Mga, Nf1, Pten, Ptprd, Rb1, Rbm10, Rnf43, Setd2, Smad4, Stag2, Stk11, Trp53 and Tsc1. Vectors encoding ‘inert’ sgRNAs were also generated: sgRosa26-1, sgRosa26-2, sgRosa26-3, sgNT-1, sgNT-2 and sgNT-3.
sgRNAs were designed and selected as follows. First, all possible 20-bp sgRNAs (using an NGG PAM) targeting each gene of interest were identified and scored for predicted on-target cutting efficiency using an available sgRNA design/scoring algorithm68. For each TSG, we then selected the sgRNA predicted to be the most likely to produce null alleles: preference was given to sgRNAs that were previously validated in vivo32,69,70, had the highest predicted on-target cutting efficiencies, targeted exons conserved in all known splice isoforms (ENSEMBL), targeted splice acceptor/splice donor sites, were positioned earliest in the gene coding region, occurred upstream of or within annotated functional domains (InterPro; UniProt), and occurred upstream of or at known recurrent mutation sites in human LUAD. The sgRNA sequences for each target are listed in Supplementary Table 1.
To generate Lenti-sgRNA/Cre vectors containing each sgRNA, a double-stranded DNA fragment (IDT gBlock) containing a U6-sgRNA-tracrRNA cassette flanked by restriction sites (AscI and SbfI) was synthesized and digested by AscI and SbfI. This digested DNA fragment was then cloned into an AscI/SbfI-digested parental lentivector encoding Cre to produce each circularized Lenti-sgRNA/Cre vector.
Barcode diversification of Lenti-sgRNA/Cre
To enable quantification of the number of cancer cells in individual tumours in parallel using high-throughput sequencing, we diversified the Lenti-sgRNA/Cre vectors with a 46-bp multi-component barcode cassette that would be unique to each tumour by virtue of stable integration of the lentiviral vector into the initial transduced cell. This 46-bp DNA barcode cassette consisted of a known 6-nucleotide ID specific to the vector backbone (vectorID), a 10-nucleotide ID specific to each individual sgRNA (sgID) and a 30-nucleotide random barcode containing 20 degenerate bases (random BC).
The 46-bp barcode cassette for each sgRNA was flanked by universal Illumina TruSeq adapter sequences and synthesized as single-stranded DNA oligos. Forward and reverse primers complimentary to the universal TruSeq sequences and containing 5’ tails with restriction enzyme sites (AscI and NotI) were used in a PCR reaction to generate and amplify double-stranded barcode cassettes for cloning. Each Lenti-sgRNA/Cre vector and its matching insert barcode PCR product was digested with AscI and NotI.
To generate a large number of uniquely barcoded vectors, we ligated 1 µg of linearize vector and 50 ng of insert with T4 DNA ligase in a 100 µl ligation reaction. Four to five hours after incubation at room temperature, ligated DNA was precipitated by centrifugation at 14,000 rpm for 12 min after adding 5 µl glycogen (5 mg ml−1) and 280 µl 100% ethanol into the ligation reaction. The DNA pellet was washed with 80% ethanol and air-dried before being resuspended with 10 µl water. This 10 µl well-dissolved DNA was transformed into 100 µl of SURE electrical competent cells using a Bio-Rad electroporation system following the manufacturer’s instructions. Electroporation-transformed cells were immediately recovered by adding into 5 ml pre-warmed SOC medium. From these 5 ml of bacteria, 10 µl were further diluted with LB ampicillin broth, and a final dilution of 1:200,000 was plated on an LB ampicillin plate for incubation at 37 °C. The remaining bacteria were mixed gently and thoroughly before being inoculated into 100 ml LB ampicillin broth, shaking at 220 rpm at 37 °C overnight. The next day, colony numbers on the LB ampicillin plate were counted to estimate the complexity of each library and the 100 ml bacterial culture was pelleted for plasmid purification.
Eight colonies from each library were picked and PCR screened for verification of the specific sgRNA sequence and corresponding barcode sequence among these eight colonies. The final purified library plasmid for each library was again sequence verified.
Production, purification and titration of lentivirus
Twenty-four hours before transfection, 2.4 × 107 293T cells were plated on a 15-cm tissue culture plate. Thirty micrograms of pPack (packaging plasmid mix) and 15 µg of library plasmid DNA were mixed well in 1.5 ml serum-free DMEM before an equal volume of serum-free DMEM containing 90 µl LipoD293 was added. The resulting mixture was incubated at room temperature for 10–20 min before adding into 293T cells. At 24 h after transfection, the medium containing complexes was replaced with 30 ml of fresh DMEM supplemented with 10% FBS, DNase I (1 unit per ml), MgCl2 (5 mM) and 20 mM HEPES, pH 7.4. The entire virus-containing medium from each plate was collected and filtered through a 0.2-µm PES filter (Nalgene) at 48 h after transfection. The viruses were further concentrated by centrifugation at 18,500 rpm, 4 °C for 2 h, and the pellet was resuspended in 500 µl PBS buffer. Virus aliquots of 50 µl were stored at −80 °C.
To quantify the titre of packaged library constructs, 105 LSL-YFP MEF cells71 were transduced with 1 µl viruses in 1 ml culture medium containing 5 µg ml−1 polybrene. Transduced cells were incubated for 72 h before being collected for FACS analysis to measure the percentage of YFP-positive cells. Control viruses were used in parallel to normalize the virus titres.
Pooling of Lenti-sgRNA/Cre vectors
To generate a pool of barcoded Lenti-sgRNA/Cre vectors for the initiation of multiple tumour genotypes within individual mice, barcoded Lenti-sgRNA/Cre vectors targeting the 22 genes described above (Apc, Arid2, Atm, Atrx, Brca2, Cdkn2a, Cmtr2, Keap1, Kmt2d, Mga, Nf1, Pten, Ptprd, Rb1, Rbm10, Rnf43, Setd2, Smad4, Stag2, Stk11, Trp53 and Tsc1), and those containing the inert, negative control sgRNAs, were combined such that the viruses would be at equal ratios in relation to their estimated in vitro or in vivo titres. The virus pool was diluted with 1× Dulbecco’s phosphate buffered saline (DPBS) to reach a titre of 180,000 transduction units (TU).
Mice, tumour initiation and tissue collection for in vivo study
KrasLSL-G12D and H11LSL-Cas9 alleles have been described previously71,72. A total of 125 male and female KrasLSL-G12D/+H11LSL-Cas9/LSL-Cas9 (KC) mice in a BL6 background (B6J), ranging from 14 to 22 weeks of age, were received from Jackson Laboratories on 19 February 2020. Mice were housed five per cage in Explora Biolabs and acclimated before initiation of the study. Housing was in a barrier, pathogen-free ventilated rack with autoclaved tap-water. Food and water were given ad libitum. Mice were identified individually by ear tag. The study performed by Pionyr Immunotherapeutics was under the guidance of and approved by the IACUC of Explora BioLabs.
Lung tumours were initiated in mice at 15–23 weeks of age via intratracheal delivery under isoflurane anaesthesia of 2 × 104–5 × 105 TU of a lentivirus pool containing barcoded Lenti-sgRNA/Cre vectors targeting the 22 different genes described above (Apc, Arid2, Atm, Atrx, Brca2, Cdkn2a, Cmtr2, Keap1, Kmt2d, Mga, Nf1, Pten, Ptprd, Rb1, Rbm10, Rnf43, Setd2, Smad4, Stag2, Stk11, Trp53 and Tsc1), as well as three sgRNAs targeting the NeoR gene (sgNeo-1, sgNeo-2 and sgNeo-3) and three non-targeting sgRNAs (sgNT-1, sgNT-2 and sgNT-3).
After lung tumours developed for 12 weeks after transduction, mice were randomized into 4 groups of 20 mice each (equally distributed across sex) before initiation of treatment. Mice were dosed every five days intraperitoneally with the indicated antibodies on the basis of the average body weight of each group. Throughout the in-life portion of the study, individual body weights and health monitoring were checked twice per week.
Cohorts of mice were treated as follows:
Group 1: Mouse IgG1 isotype control (clone MOPC-21) and mouse IgG2a (clone C1.18.4 in an afucosylated format) delivered intraperitoneally (IP), at 5 mg per kg with once daily dosing, every 5 days for 2 weeks until takedown at 14 weeks after tumour initiation.
Group 2: Anti-PD-1 (clone RMP1-14 recombinantly produced as mouse IgG1 D265A format) delivered IP, at 5 mg per kg with once daily dosing, every 5 days for 2 weeks until takedown at 14 weeks after tumour initiation.
Group 3: Anti-CTLA4 (clone 9D9 in mouse IgG2b format produced at BioXcell, BP0164) delivered IP, at 10 mg per kg once daily dosing, every 5 days for 2 weeks until takedown at 14 weeks after tumour initiation.
Group 4: Anti-PD-1 delivered IP, at 5 mg per kg and anti-CTLA4 delivered IP, at 10 mg per kg with once daily dosing, every 5 days for 2 weeks until takedown at 14 weeks after tumour initiation.
After treatment, whole lung tissue was extracted from euthanized mice as previously described32. Lung mass measurements were recorded as a proxy for overall lung tumour burden and were then stored at −80 °C before subsequent processing for NGS (see sections below).
Generation of double-stranded DNA spike-in controls
DNA barcode cassettes comprising 46-bp barcode cassettes and flanked by universal Illumina TruSeq adapter sequences as well as additional buffer sequences to extend their total length to more than 400 bp were generated either by direct synthesis of the double-stranded DNA fragments (GeneWiz, IDT) or by synthesis of single-stranded DNA oligos (GeneWiz, IDT) with overlapping complementary regions that were extended and amplified by PCR to create double-stranded DNA products that were then purified. Aliquots of these stock double-stranded DNA fragments were diluted to the desired copy numbers using DNase-free ultra-pure H2O and stored at −20 °C.
Generation of cell spike-in controls
DNA barcode cassettes comprising known 46-bp sequences were flanked by universal Illumina TruSeq adapter sequences and synthesized as single-stranded DNA oligos. Forward and reverse primers complimentary to the universal TruSeq sequences and containing 5′ tails with restriction enzyme sites (Xba1 and BstB1) were used in a PCR reaction to generate and amplify double-stranded barcode cassettes for cloning. A lentivector pRCMERP-CMV-MCS-EF1-TagR-Puro and each of the barcode insert PCR products were digested by Xba1 and BstB1 restriction enzymes.
Each digested barcode insert was cloned into a linearized vector by T4 DNA ligase and transformed into OmniMax chemical competent cells (Invitrogen). Colonies from each transformation plate were screened by PCR and sequencing. One positive clone from each barcode-containing construct was cultured for plasmid DNA extraction. Virus was packaged from each of the barcoded pRCMERP constructs in six-well plates using pPack packaging mix and LipoD293 reagent. Virus-containing medium was collected at 48 h after transfection and filtered with a Nalgene 0.2-μm PES filter before being frozen down in aliquots at −80 °C. Small aliquots of frozen viruses were thawed and added into HEK293 cells in 12-well plates for measuring titre by FACS analysis 72 h after transduction.
To generate individual cell lines containing each barcode construct, virus-containing medium was added to HEK293 cells at a multiplicity of infection of 0.1 in 10-cm plates. After overnight incubation, cells were recovered in fresh EMEM complete medium for 48 h before splitting into a new plate containing 1 μg ml−1 puro in complete EMEM medium for puro selection. After three days of puro selection, barcode-containing HEK293 cells were recovered in fresh EMEM complete medium without puro for another three days before being further expanded in 10-cm plates. Each established cell line was quality controlled by PCR amplification of the barcode region from genomic DNA to confirm the integration of the correct barcode sequences.
After cell expansion, cells from each barcoded HEK293 cell line were collected and diluted in PBS buffer containing 0.1% BSA to the desired concentrations. These cell suspensions were aliquoted and frozen down at −80 °C.
Isolation of genomic DNA from mouse lungs
Whole lungs were removed from the freezer and allowed to thaw at room temperature. Spike-ins were added to each whole lung sample. Qiagen cell lysis buffer and proteinase K from the Qiagen Gentra PureGene Tissue kit (158689) was added as described in the manufacturer protocol. Whole lungs plus spike-ins from each mouse were homogenized in the cell lysis buffer and proteinase K solution using a tissue homogenizer (FastPrep-24 5G, MP Biomedicals 116005500). Homogenized tissue was incubated at 55 °C overnight. To remove RNA from each tissue samples, RNase A was added with additional spike-ins to whole homogenized tissue. To maintain an accurate representation of all tumours, DNA was extracted, and alcohol precipitated from the entire lung lysate using the Qiagen Gentra PureGene kit as described in the manufacturer’s protocol. More spike-ins were added to the resuspended DNA.
Preparation of barcode libraries for sequencing
Libraries were prepared by amplifying the barcode region from 32 µg of genomic DNA per mouse. The barcode region of the integrated Lenti-sgRNA/Cre vectors was PCR amplified using primer pairs that bound to the universal Illumina TruSeq adapters and contained dual unique multiplexing tags. We used a single-step PCR amplification of barcode regions, which we found to be a highly reproducible and quantitative method to determine the number of cancer cells in each tumour. We performed eight 100-µl PCR reactions per mouse (4 µg DNA per reaction) using Q5 HF HS 2X Master Mix (NEB M0515) with the PCR program described in Supplementary Table 2.
The concentration of amplified barcode product in each PCR was determined by TapeStation (Agilent Technologies). Sets of 20–60 PCRs were pooled at equal molar ratios of barcode product, normalized to the estimated burden of tumours in each mouse lung sample (measured lung mass minus an estimated normal lung mass of between 0.15 and 0.18 g) associated with the PCRs. Pooled PCRs were cleaned up using a two-sided SPRI bead purification. Samples were sequenced on an Illumina NextSeq.
Analysis of sequencing data
Paired-end sequencing reads were demultiplexed via unique dual indexes using BCLConvert (v.3.8.2) and adapter sequences were trimmed using CutAdapt (v.4.1). CutAdapt was used in paired-end mode with the following parameters: minimum-length=0, error-rate=0.1, overlap=3. Paired-end alignments were constructed between mate-paired reads and library-specific databases of the expected oligonucleotide spike-in and tumour barcode insert sequences using Bowtie2 (v.2.4.4). These alignments were stringently filtered from downstream analysis if they failed to meet any of several quality criteria, including:
No mismatches between the two mate pairs, which fully overlap one another, at any location.
No mismatches between the mate-paired reads and expected constant regions of the barcode or spike-in to which they best align.
No indels in alignments between mate-paired reads and the barcode or spike-in to which they best align.
After alignment, errors in paired-end reads were corrected using a simple greedy clustering algorithm:
Reads were dereplicated into read sequence/count tuples, (si, ri).
These tuples were reordered from highest to lowest on the basis of their read abundances, (ri) (refs. 6,13,49,63).
- This list of tuples was traversed from i = 1…N, taking one of the following actions for each tuple (si, ri):
- If si is not within a Hamming distance of 1 from any sj with j < i, then (si, ri) initiates a new cluster.
- If sj is within a Hamming distance of 1 from some sj with j < i, then it joins the cluster of sj.
The resulting clusters are each considered to represent an error-corrected sequence equal to that of the sequence that founded the cluster with read count equal to the sum of the read counts of the dereplicated reads that are members of the cluster.
A second stage of error correction was performed to remove additional errors. Hamming distance D(si, sj) was computed on all pairs of error-corrected sequences. Then, each sequence si (with ri reads) was absorbed into the most abundant sequence sj (with rj > ri reads) if either of the following criteria were met:
D(si, sj) ≤ 3
D(si, sj) ≤ 5 and rj/ri ≤ 5 or ri ≤ 3.
These heuristics were established on the basis of internal control data. After applying both rounds of error correction, there were 0 false positives out of the 18,889,362 reads assigned to spike-in oligonucleotide sequences (which have no degenerate bases) that were not added to the samples. After error correction, a filter was applied to remove sequences that could have originated from cross-contamination: barcodes were compared across samples in the same study, and any exact sequences that were found in more than one library were removed.
After error correction and cross-contamination removal, the read counts of each unique barcode were converted to neoplastic cell number by dividing the number of reads of the spike-in oligonucleotide added to the sample before tissue homogenization and lysis at a fixed, known concentration.
Removal of mice that did not get sufficient viral titre during transduction
After the sequence processing, mice were removed if they did not reach a lower bound of total neoplastic cells. Mice were removed if they had fewer than 106 total neoplastic cells. This threshold was chosen using by examining the distribution of total neoplastic cells per mouse across each study. Most mice fell within around two orders of magnitude of each other, and any outliers fell at least an order of magnitude below the rest of the distribution.
Calculation of pharmacogenomic interactions
From these collections of tumour sizes across paired groups of drug-treated and vehicle-treated mice, the relative tumour number (RTN) metric was computed as previously described73. Namely, shrinkage of inert tumours was estimated by finding the S that matches the median number of tumours larger than a cut-off L in such paired groups after the vehicle-treated tumour sizes are multiplied by S (S < 1 when the drug works to shrink tumours). Subsequently, for each non-inert tumour genotype, the ratio of the number of tumours with this genotype larger than L in the control mice to the number of tumours larger than L × S in the treated mice was computed. The resulting ratio was divided by the same ratio computed for the inert tumours, and the log2(•) of this ratio of ratios was determined. This metric, RTNscore is expected to be greater than 0 for resistant genotypes and less than 0 for sensitive genotypes. For example, RTNscore = 1 corresponds to a 2× change in tumour number (larger than each cut-off) relative to the change in untreated versus treated for oncogene-only tumours, whereas RTNscore = −1 corresponds to a 0.5× change and drug sensitivity.
To generate CIs for RTNscore, bootstrap resamplings were made by (1) sampling mice with replacement from the control and therapy arms to match the original group sizes, and (2) sampling tumours (of all sizes) with replacement from each mouse. For each mouse/tumour bootstrap, the RTNscore was recomputed. A genotype was then considered sensitive if the 95th percentile of these bootstrap RTNscore values fell below 0, or resistant if the 5th percentile exceeded 0. We performed this bootstrapping procedure at tumour size cut-offs ranging from L = 300 cells to L = 30,000 cells.
RTNscores were also generated comparing anti-PD-1 and anti-CTLA4 combination treatment to anti-PD-1 monotherapy, using the same method as for the drug-treated versus vehicle comparison.
Calculation of tumour size percentiles
First, tumours were pooled across all mice in the vehicle group, and separated into tumours that map to each Lenti-sgRNA/Cre guide. Tumours from Lenti-sgInert/Cre were pooled (sgNT-1, sgNT-2, sgNT-3, sgRosa26-1, sgRosa26-2, sgRosa26-3) to create one pool of sgInert tumours. For this analysis, we used all tumours that had at least 500 neoplastic cells, which was the minimum tumour size we could reliably quantify in this study.
For each set of Lenti-sgRNA/Cre tumours in each oncogene–background pair, size percentiles of tumours above the 500 cell cut-off were computed and divided by the same size percentiles for the sgInert tumours in the same context with the same cut-off. Mice and tumours were bootstrapped 200 times and the calculation was repeated each time. A 95% CI from these bootstraps was reported.
Animal cohorts and generation of immune-competent Stk11- and/or Keap1-deficient syngeneic NSCLC models
Mice were housed in the Research Animal Support Facility at the University of Texas MD Anderson Cancer Center. All animal studies described here were performed according to protocols approved by the University of Texas MD Anderson Cancer Center IACUC, and according to pre-specified criteria for euthanasia, tumour size limit and animal cohort sample size. Investigators were not blinded to treatment arm allocation.
The KL5 and KL2 polyclonal KrasG12C-mutant Stk11-deficient LUAD cell lines were established from autochthonous lung tumours in compound conditional KrasLSL-G12C+/WTStk11/Lkb1Fl/Fl male mice (on a C57Bl/6 genetic background), after lung tumour induction with intranasal instillation of of adenoviral Cre recombinase (University of Iowa Viral Vector Core) as previously described74 and were used to establish subcutaneous allograft tumours in sex-matched (male) syngeneic recipient C57Bl/6 mice.
LKR10 and LKR13 KrasG12D-mutant LUAD cells (on a 129/Sv genetic background, previously generated in the laboratory of T. Jacks) were transiently transfected with Keap1 CRISPR–Cas9 KO plasmid (sc-424513-KO-2) or Stk11/Lkb1 CRISPR–Cas9 KO plasmid (sc-423192) from Santa Cruz Biotechnology (LKR10 derivative models) or pSpCas9(BB)-2A-GFP (PX458) plasmid with specific sgRNAs (LKR13 derivative models). The plasmid vectors for CRISPR–Cas9-mediated Keap1 and Stk11/Lkb1 KO were verified by DNA sequencing. The transfected cells were sorted by BD FACSAria (BD Biosciences) for GFP+ into 96-well plates and single-cell-derived colonies were screened for KEAP1 and LKB1 loss by western blotting.
In vivo preclinical studies with anti-PD-1 and anti-CTLA4 in syngeneic tumour models
Six-to-twelve week-old male C57BL/6 mice (for experiments with KL2 and KL5 cell lines) and male 129/Sv mice (for experiments with LKR10 and LKR13-derived isogenic cell lines) were injected subcutaneously with 1.5 × 106 cells of the indicated genotypes in a 100 μl suspension, consisting of 50% Matrigel basement membrane matrix (Corning) and 50% HBSS. Mice bearing subcutaneous tumours with an average tumour volume (TV) ranging from 150 to 250 mm3 (for long-term efficacy studies) or 50 to 100 mm3 (for immune profiling, immune depletion and iNOS chemical inhibition studies) were randomly assigned to receive treatment with (i) anti-PD-1 200 µg (clone 29F.1A12, BioXCell); (ii) anti-CTLA4 200 µg (clone 9H10, BioXCell); (iii) anti-PD-1 + anti-CTLA4; or (iv) corresponding isotype IgG control antibodies (BioXCell; BE0089 and BE0087) by IP injection twice weekly. Mice were monitored daily and TV was calculated three times weekly on the basis of bidirectional caliper tumour measurements, using the formula V = W2L/2. Comparison of average TV across treatment groups was performed at the time that the first mouse in any treatment arm reached end-point (defined as TV ≥ 1,200 mm3 or 1,500 mm3 depending on the individual experiment), but treatment continued in remaining mice to enable comparisons of time to a tumour volume of 1,200 mm3 or greater.
For dual anti-PD-1/anti-TIM-3 and anti-PD-1/anti-LAG-3 ICB experiments, mice bearing KL5 allograft tumours (TV 50–100 mm3, day 5 after tumour implantation) were randomized to treatment with (i) anti-PD-1 200 µg (clone 29F.1A12, BioXCell) plus anti-CTLA4 200 µg (clone 9H10, BioXCell); (ii) anti-PD-1 plus anti-TIM-3 200 µg (clone B8.2C12, BioXCell); or (iii) anti-PD-1 plus anti-LAG-3 200 µg (clone C9B7W, BioXCell) twice weekly by IP injection and were monitored as described above.
Immune cell depletion and INOS chemical inhibition studies
CD4+ and CD8+ T cell depletion was performed by intraperitoneal administration of 400 μg of anti-CD4 (clone GK1.5, BioXCell) or anti-CD8 (clone 2.43, BioXCell) depleting monoclonal antibodies twice weekly, starting on day 4 after subcutaneous implantation of 1.5 × 106 KK or KL5 tumour cells. Neutrophil depletion and/or CCL2 neutralization was achieved by intraperitoneal administration of 400 µg of anti-Ly6G antibody (clone 1A8, BioXCell) and/or 200 µg of anti-CCL2 antibody (clone 2H5, BioXCell) starting at day −3 before tumour cell injection and followed by administration of 100 µg of anti-Ly6G or 200 µg of anti-CCL2 twice weekly. Inhibition of iNOS was performed by daily IP injection of 200 µg of L-NIL (Cayman Chemicals) diluted in 100 µl of PBS as previously described42. Treatment with anti-PD-1/ anti-CTLA4 commenced on day 5 when mouse cohorts exhibited average tumour volumes of 50–100 mm3 and followed the same schedule as described above.
Preclinical experiments with chemo-immunotherapy
Cohorts of male 129/Sv mice bearing subcutaneous allograft K (LKR13 KrasG12D mutant; Keap1/Stk11 WT) or isogenic KLK (Keap1- and Stk11-deficient, clone 17) allograft tumours were randomly assigned to (chemo) ICI therapy arms at an average tumour volume of around 210 mm3. Chemotherapy consisted of carboplatin 12.5 mg per kg (institutional pharmacy, NDC 61703-339-56) administered by IP injection every 7 days (for a maximum of 4 doses) and paclitaxel 5 mg per kg (NDC 70860-200-50) administered by IP injection twice weekly (for a maximum of 8 doses). Anti-PD-1 (clone RMP1-14, BioXCell; 8 mg per kg) and anti-CTLA4 (clone 9H10, BioXCell; 8 mg per kg) were administered twice weekly by IP injection until the study end-point was reached.
Flow cytometry
For studies of immune modulation, treatment with single or dual ICB commenced on day 5 after subcutaneous tumour implantation and followed the same dosing regimen as previously described for the efficacy experiments. Tumour samples were collectd after five doses of single or dual ICB, on day 18. For chemo-immunotherapy experiments with the K and KLK (clone 17) models, mice received a total of three doses of chemotherapy and/or immunotherapy (D1: carboplatin + paclitaxel + ICB); D4 (paclitaxel + ICB); D7 (carboplatin + paclitaxel + ICB), and tumours were collected for FACS-based immune profiling 48 h later. Tumours were immediately processed into 2–4-mm3 pieces and transferred to a gentleMACS Octo Dissociator tube containing a solution with liberase (25 μg ml−1), DNase I type 1 (30 U ml−1) and hyaluronidase (0.01%) in serum-free RPMI-1640 medium and incubated for 45 min at 37 °C. Enzyme reactions were stopped by the addition of cold RPMI-1640 (10% FBS) and suspensions were dispersed through a 70-μm cell strainer twice. After red blood cell lysis (using RBC lysis buffer, Biolegend), single-cell suspensions (around 1 × 106 cells in 50 μl total volume) were incubated with FcR-blocking reagent rat anti-mouse CD16/32 (2.4G2, BD Biosciences) for 15 min on ice. For extracellular staining, cells were stained with a mixture of conjugated antibodies in FACS buffer, including Ghost Dye Violet 510 (Thermo Fisher Scientific) for 1 h at room temperature in the dark. For intracellular cytokine and transcription factor staining, single-cell suspensions were fixed and permeabilized using the eBioescience FoxP3/Transcription Factor Staining kit (Life Technologies) according to the manufacturer’s instructions. Fluorochrome-conjugated monoclonal antibodies were purchased from Biolegend: Pacific Blue-anti-CD45 (30-F11), PE/Dazzle 594 anti-CD3 (17A2), APC/Cy7–anti-CD4 (RM4-5), PE/Cy7–anti-CD8 (53-6.7), APC–anti-T-bet (4B10), PE–anti-ICOS (7E.17G9), BV711–anti-CD44 (IM7), BV605–anti-PD-1 (29F.1A12), BV711–anti-Gr-1 (RB6-8C5), BV785–anti-CD11c (N418), BV650–anti-CD11b (M1/70), PE/Cy7–anti-I-A/I-E (MHC class II, M5/114.15.2), BV605–anti-Ly6C (HK1.4), PerCP/Cy5.5–anti-Ly6G (1A8); from BD Biosciences: BUV395–anti-CD25 (PC61), BV605–anti-PD-1 (RMP1-30); from Life Technologies: PerCP–Cy5.5–anti-FOXP3 (FJK-16s); from Invitrogen PE–anti-iNOS (CXNFT); and from Tonbo Bioscience FITC–anti-CD62L (MEL-14), APC–anti-F4/80 (BM8.1). Flow cytometry data were acquired on an LSR Fortessa flow cytometer (BD Biosciences) and were analysed using FlowJo v.10.8.1 software (BD Biosciences).
scRNA-seq
Single-cell isolation, library preparation and sequencing
scRNA-seq of all samples was conducted on the 10x Chromium system (10x Genomics), in collaboration with the MD Anderson Advanced Technology Genomics Core facility. Tumours derived from syngeneic K, KK and KLK LKR13 mouse models were collected when the tumour volume reached 500–700 mm3. Whole tumours were processed and dissociated in a solution containing collagenase A, hyaluronidase, dispase and DNase I. Cell suspensions were strained through a 70-μm cell strainer, submitted to RBC lysis and resuspended in PBS with 0.04% BSA. After viability assessment, isolated cells were submitted to single-cell capture, barcoding and library preparation, following the 10x Genomics Single Cell Chromium 5’ V2 protocols. Pooled samples underwent sequencing with the NovaSeq6000 sequencer, performed at the ATGC core at the MD Anderson Cancer Center.
scRNA-seq data analysis
Raw scRNA-seq data were preprocessed (demultiplex cellular barcodes, read alignment and generation of gene count matrix) using the Cell Ranger Single Cell pipeline provided by 10x Genomics, using the mouse reference transcriptome GRCm38 (mm10). Data merging, filtering, doublet removal, batch-effect evaluation and data normalization were performed following standard protocols as previously described75. The Seurat R package (v.4.3) was used to analyse the normalized gene–cell matrix and Harmony (v.0.1.1) was applied for batch-effect correction. Each cluster was determined according to highly variable genes identified applying the FindClusters Seurat function and validated with SingleR (v.2.2.0). Dimensionality reduction and two-dimensional visualization of cell clusters was performed using uniform manifold approximation and projection (UMAP) with the Seurat function RunUMAP. Differentially expressed genes were identified with the FindMarkers function. Gene signature scores were obtained with AddModuleScore function.
Graphical illustrations
Graphical elements used to illustrate experimental design schemes were created with BioRender.com, licensed by the University of Texas MD Anderson Cancer Center.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-07943-7.
Supplementary information
Source data
Source Data Figs. 3, 4 and Source Data Extended Data Fig. 5–10
Acknowledgements
We acknowledge support from funding sources including the Lung SPORE NCI P50 CA070907 (University of Texas), NCI 1R01 CA262469-01 (F.S.), NCI 1R01 CA279452-01A1 (F.S.), NCI R01 CA205150 (J.V.H. and K.K.W.), NCI CA016672/NIH NIH 1S10OD024977-01 (ATGC core of MD Anderson Cancer Center), NCI P30 CA016672 (University of Texas MD Anderson Cancer Center), NCI P30 CA016058 and the Biospecimen Services Shared Resource (the Ohio State University Comprehensive Cancer Center), and NCI P30 CA008748 (Memorial Sloan Kettering Cancer Center), as well as CPRIT RP160652 (J.V.H.); Stand Up To Cancer, the Mark Foundation, the Cain Foundation, the Gunnigar Fund, the Andrea Mugnaini Lung Cancer Reseach Fund, the Ford Petrin Fund, Rexanna’s Foundation for Fighting Lung Cancer, and David Bruton, Jr. Chair Endowments. This research was also supported by philanthropic contributions to the University of Texas MD Anderson Lung Moon Shot Program. We thank L. Blair, G. Wall and W. Nie for experimental and technical assistance, L. Morris and M. Nilsson for editorial assistance and D. Petrov for his contribution to the development of analytical methods for autochthonous lung cancer studies.
Extended data figures and tables
Author contributions
F.S. and J.V.H. conceived the study, developed the study methodology, directed and supervised all analyses, analysed data, provided resources and study administration, acquired funding for the study and wrote the original draft of the manuscript. T.C. contributed to study methodology and data analysis, supervised experiments outlined in Extended Data Fig. 5g,h that were performed by X.S. and J.T.L., provided resources and contributed to study funding. H.A.A. performed preclinical studies, curated and analysed data from the multi-institutional retrospective cohort and preclinical studies and contributed to developing the study methodology and writing the original draft. M.T.D., Y.Q., X.S., A.G.C. and J.T.L. performed preclinical experiments, analysed data and contributed to the study methodology, with assistance from D.M., R.L., T.Z., A.P., Y.K., T.T. and A.G.P. M.M. and L.A. provided access to and analysed the FMI dataset. R.P., N.J., J.M.J., M.M.W., M.J.R. and I.P.W. developed the methodology, performed all experiments and analysed data related to the in vivo Tuba-seq based screen in Fig. 3d–e and Extended Data Fig. 9. C.M., Y.Y., X.P. and A.H. developed methodology, performed all experiments and analysed data related to the in vivo CRISPR–Cas9 screen in Fig. 3a–c. K.K.W. contributed to the development of the mouse models of Kras mutant NSCLC. N.V. and S.T.S. performed the immune deconvolution analysis in the TCGA and ICON datasets. K.C.A., D.H.O., R.M., P.D.P., M.E.M., M.M.A., J.C.M., J.A.H., J.F.G., A.D., C. M. Bestvina, C.A.S., J.W.R., C. M. Blakely, C.V.P., L.M., F.T., M.S., S.D. M.J.M., A.G.S., S.C.M.L., A.N.S., J.R., R.P.J., C.L., T.S., S.B.G., J.K., Z.W., K.S., K.D.D., M.G.W., V.A., K.A.M., P.M.F., B.R., D.V., E.M.V.A., A.L.C., J.W.G., H.S., M.K., S.K., C.A., Y.N., J.T.A., J.S., L.R., J.W.N., L.L., Y.Y.E., M.V.N., X.L., V.K.L., W.E.L., H.N.K., B.C., J.A.R., S.S., E.R.P.C., C.B., I.I.W., J.Z., G.R.B., C.G., L.A.B., D.L.G., A.T., T.G.B., D.R.C., J.E.G., N.L., B.L., J.R.B., M.C.G., D.R.G., E.B.G., N.A.R., G.V.S., J.W., D.P., B.B., R.S.H., H.A.W., N.A.P., A.T.S., P.A.J., D.P.C., M.D.H., C.M.R. and V.A.P. provided resources and contributed to clinical data collection and curation as part of the multi-institutional retrospective cohort. J.J.L. oversaw statistical analyses in the multi-institutional retrospective clinical cohort. M.D.H., H.M., Z.Z., Z.L., R.S. and P.J. contributed data and performed all analyses in the POSEIDON clinical trial. S.P. and M.L.J. were the lead clinical investigators in POSEIDON. All co-authors contributed to writing, review and editing of the final manuscript.
Peer review
Peer review information
Nature thanks Fred Hirsch, Daniel Tan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Data generated in this study are available in the article and its supplementary files. scRNA-seq raw and processed data reported in this article are available upon request and have been deposited to the NCBI Gene Expression Omnibus (GEO) under the accession number GSE267321. Data underlying the clinical trial findings may be requested in accordance with AstraZeneca’s data sharing policy, described in further detail at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. All other individual de-identified participant data supporting the retrospective clinical data results reported in this article will be available on request according to General Data Protection Regulation (GDPR) standards. Only summary clinical data may be shared; patient-level image or genetic data are not available for access in our repository in the interest of protecting patient privacy. Materials, reagents or other experimental data are available upon reasonable request from the corresponding authors. Source data are provided with this paper.
Competing interests
F.S. reports consulting for AstraZeneca, Amgen, Revolution Medicines, Novartis, BridgeBio, Beigene, BergenBio, Guardant Health, Calithera Biosciences, Tango Therapeutics, Hookipa Pharma, Novocure, Merck Sharp & Dohme, Roche; grant or research support from Amgen, Mirati Therapeutics, Revolution Medicines, Pfizer, Novartis, Merck & Co; stockholder in BioNTech, Moderna; and honoraria from ESMO, Japanese Lung Cancer Society, Medscape, Intellisphere, VSPO McGill Universite de Montreal, RV Mais Promocao Eventos, MJH Life Sciences, IDEOlogy Health, MI&T, PER, CURIO, DAVA Oncology, the American Association for Cancer Research and the International Association for the Study of Lung Cancer. M.M. reports stockholder in Roche Holdings. N.J. reports shareholder and former employee of Pionyr Immunotherapeutics. J.M.J. reports stock of D2G Oncology. C.M., Y.Y., X.P. and A.H. report employee of Tango Therapeutics. K.C.A. reports personal fees from Sanofi Genzyme and other support from Revolution Medicines, Genentech, and Mirati outside the submitted work. N.V. receives consulting fees from Sanofi Genzyme, Oncocyte, Eli Lilly, Regeneron, and research funding to the institution from Mirati, Oncocyte, and Circulogene, outside the submitted work. P.D.P. reports advisory fees from AstraZeneca and Jazz Pharmaceuticals. M.E.M. reports research funding from Eli Lilly (Inst), AstraZeneca (Inst), Merck (Inst), Genentech (Inst); consulting role with AstraZeneca, Novocure, Boehringer Ingelheim, Janssen, Takeda, Blueprint Pharmaceuticals, Bayer, Bristol Myers Squibb, Ikena; honorarium from Thermo Fisher Scientific; and stock in Merck, Johnson & Johnson. M.M.A. reports grants and personal fees from Genentech, Bristol Myers Squibb and AstraZeneca; grants from Lilly; and personal fees from Maverick, Blueprint Medicine, Syndax, Nektar, Gritstone, ArcherDX, Mirati, NextCure, Novartis, EMD Serono and Panvaxal/NovaRX, outside of the submitted work. J.C.M. reports consulting or honoraria: MJH Life Sciences, Johnson & Johnson and Doximity; and research funding (to institution): Merck via the Conquer Cancer Foundation. J.F.G. reports served as a compensated consultant or received honoraria from Bristol Myers Squibb, Genentech (Roche), Takeda, Loxo (Lilly), Blueprint Medicine, Gilead, Moderna, AstraZeneca, Mariana Therapeutics, Mirati, Jounce, Merus Pharmacueticals, Nuvalent, Pfizer, Novocure, AI Proteins, Novartis, Merck, iTeos, Karyopharm and Silverback Therapeutics; research support from Novartis, Genentech (Roche) and Takeda; institutional research support from Bristol Myers Squibb, Palleon, Tesaro, Moderna, Blueprint, Jounce, Array Biopharma, Merck, Adaptimmune, Novartis and Alexo; has equity in AI Proteins; and has an immediate family member who is an employee with equity at Ironwood Pharmaceuticals. A.D. reports honoraria: Intellisphere, Roche (Genentech); ad board: TP Therapeutics, Guardant Health, AnHeart Therapeutics, ChromaCode; clinical trial support: Syntrix Pharmaceuticals, Novartis, Merck, AnHeart Therapeutics, Sorrento Therapeutics, Guardant Health and Philogen. C. M. Bestvina reports consulting or advisory role: AstraZeneca, Genentech, AbbVie, Curio Science, OncLive Clinical Congress Consultants, Seagen, Creative Educational Concepts, Takeda, Janssen, CVS, Bristol Myers Squibb (Celgene), Jazz Pharmaceuticals, Novartis, Sanofi–Regeneron and Novocure; speakers’ bureau: Merck; research funding: Bristol Myers Squibb and AstraZeneca. C.A.S. reports advisory boards to AstraZeneca, Gilead, Janssen, Genentech, Mirati, Takeda and Arcus Biosciences. J.W.R. reports honoraria or consulting (self): Boehringer Ingelheim, BMS, Turning Point Roche (Genentech), Biodesix, EMD Serono, Daiichi Sankyo, Blueprint, Novartis, Regeneron, Sanofi, Janssen, Jazz Pharmaceuticals, Bayer, Beigene, Merus and Seagen; research support (to institution): ArriVent, Merck, Novartis, AstraZeneca, Spectrum, Revolution Medicines, IO Biotech and Kinnate. C. M. Blakely reports research funding: AstraZeneca, Novartis, Mirati, Spectrum, Takeda, Puma and Pfizer; and consulting: Janssen and Bayer. C.V.P. reports a founder and equity and intellectual property in EnFuego Therapeutics. F.T. reports speaker bureau or honoraria: Roche, AstraZeneca, Novartis and Takeda. M.S. reports institutional support: Dracen Pharmaceuticals; advisory board: Amgen, AstraZeneca, Boehringer Ingelheim, Janssen Pharmaceuticals, Novartis, Pfizer, Roche, Sanofi-Aventis, Siemens Healthineers, Takeda Pharmaceuticals and Bristol Myers Squibb; leadership or fiduciary role: ESMO and EORTC. S.D. reports independent image analysis for hospital-contracted clinical research trials programs for Merck, Pfizer, Bristol Myers Squibb, Novartis, Roche, Polaris, Cascadian, Abbvie, Gradalis, Bayer, Zai laboratories, Biengen, Resonance and Analise; research grants from Lunit, GE, Qure AI; and an honorarium from Siemens. M.J.M. reports AstraZeneca, Bristol Myers Squibb and Istari Oncology & Regeneron. A.G.S. reports AstraZeneca, Genentech (Roche) and Bristol Myers Squibb. A.N.S. reports serving as a consultant and/or advisory board member for Eli Lilly, Daiichi Sankyo and Zymeworks; and research funding from Eli Lilly, Novartis, Daiichi Sankyo, Genentech, AstraZeneca, Memgen, Mersana, Turning Point Therapeutics, BioAtla and Genmab to his institution. J.R. reports consulting fees or honoraria from AstraZeneca, Takeda, Sanofi Genzyme, Genentech, Guardant Health, Summit, G1 Therapeutics, BioAtla, Jazz, Amgen, and Janssen; contracted for research (institutional) with AstraZeneca, BioAtla, Blueprint Medicines, EpimAb Biotherapeutics, LOXO Oncology, Enliven, Redcloud and ORIC. T.S. reports advisory board or consulting fees for Seagen, Astellas and AstraZeneca; and research grant GRAIL. S.B.G. reports research funding from AstraZeneca, Boehringer Ingelheim and Mirati; and consulting or advisory board member for AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Amgen, Blueprint Medicine, Sanofi Genzyme, Daiichi Sankyo, Takeda, Janssen, Summit Therapeutics, Merck and Regeneron. J.K. reports for Arterys. K.S. reports consultant, advisor or speaker for Clinica Alemana Santiago, Shattuck Labs, AstraZeneca, EMD Serono, Takeda, Torque/Repertoire Therapeutics, CSRlife, Agenus, Genmab, OnCusp, Parthenon Therapeutics, Bristol Myers Squibb, Roche, Molecular Templates, Abbvie, Sanofi, Merck, PeerView, PER and Forefront Collaborative; research funding from Navigate BP, Tesaro/GSK, Moderna, Takeda, Surface Oncology, Pierre-Fabre, Merck, Bristol Myers Squibb, AstraZeneca, Ribon Therapeutics, Eli Lilly, Boehringer Ingelheim, Genentech (Roche) and Akoya Biosciences. K.D.D. reports consulting fees or other remuneration from Rain Therapeutics. V.A. reports research funding to Johns Hopkins University from AstraZeneca and Personal Genome Diagnostics, has received research funding to Johns Hopkins University from Bristol Myers Squibb and Delfi Diagnostics in the past five years and is an advisory board member for Neogenomics and AstraZeneca. V.A. is an inventor on patent applications (63/276,525, 17/779,936, 16/312,152, 16/341,862, 17/047,006 and 17/598,690) submitted by Johns Hopkins University related to cancer genomic analyses, ctDNA therapeutic response monitoring and immunogenomic features of response to immunotherapy that have been licensed to one or more entities. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. K.A.M. reports consulting or advisory fees from AstraZeneca, Amgen, Janssen, Mirati Therapeutics, Daiichi Sankyo (Lilly) and Puma Biotechnology, as well as honoraria from AstraZeneca. K.A.M. receives research funding to Johns Hopkins University from Bristol Myers Squibb and Mirati Therapeutics. P.M.F. reports consulting fee (for example, advisory board): Ascendis, AstraZeneca, BMS, Curevac, Novartis, Regeneron, G1, Genelux, Genentech, Gritstone, Merck, Janssen, F-Star, Sanofi, Amgen, Fosun, Teva, Synthekine, Flame, Iteos, Tavotek and Teva; contracted research: AstraZeneca, BMS, Novartis, Regeneron and BioNTech. B.R. reports advisory board or consultant: Regeneron, AstraZeneca, Amgen; honoraria: Targeted Oncology. E.M.V.A reports advisory or consulting: Tango Therapeutics, Genome Medical, Genomic Life, Enara Bio, Manifold Bio, Monte Rosa, Novartis Institute for Biomedical Research, Riva Therapeutics and Serinus Bio; research support: Novartis, BMS and Sanofi; equity: Tango Therapeutics, Genome Medical, Genomic Life, Syapse, Enara Bio, Manifold Bio, Microsoft, Monte Rosa, Riva Therapeutics and Serinus Bio; patents: institutional patents filed on chromatin mutations and immunotherapy response, and methods for clinical interpretation; intermittent legal consulting on patents for Foaley & Hoag; editorial boards: JCO Precision Oncology and Science Advances. A.L.C. reports consulting for AstraZeneca, Merus and Tempus; research funds from AstraZeneca, Genentech (Roche), Merck and Novartis. J.W.G. reports research grants: AstraZeneca, BMS, Eli Lilly, Genentech, Merck and Pfizer; consulting fees: AstraZeneca, BMS, Genentech, Gilead, Gritstone, Eli Lilly, Pfizer and Regeneron. C.A. reports consulting or advisory role: Genentech, Lilly, Celgene, Merck, AstraZeneca, Blueprint Genetics, Shionogi, Daiichi Sankyo–AstraZeneca, Sanofi/Regeneron, Eisai, BeiGene, Turning Point Therapeutics, Pfizer, Janssen, Boehringer Ingelheim; research funding: Genentech/Roche (Inst), Incyte (Inst), Macrogenics (Inst), Merck Sharp & Dohme (Inst) and AstraZeneca–MedImmune (Inst). L.R. reports honoraria: Merck, AstraZeneca, Genzyme, EMD Serono and Astellas Pharmas; consulting or advisory role: Amgen. J.W.N. reports honoraria: CME Matters, Clinical Care Options CME, Research to Practice CME, Medscape CME, Biomedical Learning Institute CME, MLI Peerview CME, Prime Oncology CME, Projects in Knowledge CME, Rockpointe CME, MJH Life Sciences CME, Medical Educator Consortium, HMP education consulting or advisory role: AstraZeneca, Genentech (Roche), Exelixis, Takeda Pharmaceuticals, Eli Lilly, Amgen, Iovance Biotherapeutics, Blueprint Pharmaceuticals, Regeneron Pharmaceuticals, Natera, Sanofi, D2G Oncology, Surface Oncology, Turning Point Therapeutics, Mirati Therapeutics, Gilead Sciences, Abbive, Summit Therapeutics, Novartis, Novocure, Janssen Oncology, Anheart Therapeutics Research Funding: Genentech (Roche), Merck, Novartis, Boehringer Ingelheim, Exelixis, Nektart Therapeutics, Takeda Pharmaceuticals, Adaptimmune, GlaxoSmithKline, Janssen Pharmaceuticals, Abbvie Pharmaceuticals and Novocure. Y.Y.E. reports honoraria: CME Matters, Clinical Care Options CME, Research to Practice CME, Medscape CME, Biomedical Learning Institute CME, MLI Peerview CME, Prime Oncology CME, Projects in Knowledge CME, Rockpointe CME, MJH Life Sciences CME, Medical Educator Consortium, HMP education consulting or advisory role: AstraZeneca, Genentech (Roche), Exelixis, Takeda Pharmaceuticals, Eli Lilly, Amgen, Iovance Biotherapeutics, Blueprint Pharmaceuticals, Regeneron Pharmaceuticals, Natera, Sanofi, D2G Oncology, Surface Oncology, Turning Point Therapeutics, Mirati Therapeutics, Gilead Sciences, Abbive, Summit Therapeutics, Novartis, Novocure, Janssen Oncology, Anheart Therapeutics Reserach Funding: Genentech/Roche, Merck, Novartis, Boehringer Ingelheim, Exelixis, Nektart Therapeutics, Takeda Pharmaceuticals, Adaptimmune, GlaxoSmithKline, Janssen Pharmaceuticals, Abbvie Pharmaceuticals and Novocure. M.V.N. reports research funding to institution: Mirati, Novartis, Checkmate (ended), Alaunos, AstraZeneca, Pfizer, Genentech, Navire; consultant or advisory board: Mirati, Merck/MSD, Novartis and Genentech. X.L. reports receiving consulting or advisory fees from EMD Serono (Merck KGaA), AstraZeneca, Spectrum Pharmaceutics, Novartis, Eli Lilly, Boehringer Ingelheim, Hengrui Therapeutics, Janssen, Blueprint Medicines, Regeneron, Sensei Biotherapeutics, Abion and Abbvie, and research funding from Eli Lilly, EMD Serono, Janssen, ArriVent, Teligene, BlackDiamond, Regeneron and Boehringer Ingelheim. V.K.L. reports consultant or advisory role for Takeda, Seattle Genetics, BMS, AstraZeneca, Guardant Health and AnHeart Therapeutics and has received research funding from GSK, BMS, AstraZeneca, Merck and Seattle Genetics. W.E.L. reports advisory boards: Eisai and Takeda, Grand Rounds and BMS for repotrectinib. J.A.R. reports consultancy, stock, Genprex; patents issued and pending. T.C. reports speaker fees or honoraria from the Society for Immunotherapy of Cancer (SITC), Mark Foundation for Cancer Research, Bristol Myers Squibb, Roche, Medscape, IDEOlogy Health, Physicians’ Education Resource (PER), OncLive and PeerView; travel and/or food/beverage expenses from SITC, International Association for the Study of Lung Cancer, Parker Institute for Cancer Immunotherapy, PER, Dava Oncology, IDEOlogy Health, AstraZeneca and Bristol Myers Squibb; advisory role or consulting fees from MedImmune–AstraZeneca, Bristol Myers Squibb, Merck, Genentech, Arrowhead Pharmaceuticals, Pfizer and Regeneron; and institutional research funding from MedImmune–AstraZeneca, Bristol Myers Squibb and EMD Serono. J.Z. reports grants from Merck, grants and personal fees from Johnson & Johnson and Novartis, personal fees from Bristol Myers Squibb, AstraZeneca, GenePlus, Innovent and Hengrui outside the submitted work. G.R.B. reports grant or contract for clinical trial from MacroGenics; grants from Amgen, Bayer, Adaptimmune, Exelixis, Daiichi Sankyo, GlaxoSmithKline, Immatics, Immunocore, Incyte, Kite Pharma, MacroGenics, Torque, AstraZeneca, Bristol Myers Squibb, Celgene, Genentech, MedImmune, Merck, Novartis, Roche, Sanofi, Xcovery, Tmunity Therapeutics, Regeneron, BeiGene, Repertoire Immune Medicines, Verastem, CytomX Therapeutics and Duality Biologics; consulting fees from AbbVie, Adicet, Amgen, Ariad, Bayer, Clovis Oncology, AstraZeneca, Bristol Myers Squibb, Celgene, Daiichi Sankyo, Instil Bio, Genentech, Genzyme, Gilead, Eli Lilly, Janssen, MedImmune, Merck, Novartis, Roche, Sanofi, Tyme Oncology, Xcovery, Virogin Biotech, Maverick Therapeutics, BeiGene, Regeneron, Cytomx Therapeutics, Intervenn Biosciences and Onconova Therapeutics; participation in advisory boards at Virogin Biotech and Maverick Therapeutics; stock in Virogin Biotech; and employment of family member at Johnson & Johnson/Janssen. C.G. reports member of the advisory board at Jazz Pharmaceuticals, AstraZeneca and Bristol Myers Squibb and served as speaker for AstraZeneca and BeiGene. L.A.B. reports consulting fees and research funding from AstraZeneca, GenMab and Sierra Oncology and research funding from Tolero Pharmaceuticals and served as advisor or consultant for PharmaMar, AbbVie, Bristol Myers Squibb, Alethia, Merck, Pfizer, Jazz Pharmaceuticals, Genentech and Debiopharm Group. D.L.G. reports AstraZeneca, Eli Lilly, Menarini Richerche, 4D Pharma, Onconova Therapeutics and Sanofi. A.T. reports Ariad, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Eli Lilly, EMD Serono, Genentech, GlaxoSmithKline, Merck, Novartis, Pfizer, Roche, Seattle Genetics, Gilead Sciences and Summit Therapeutics. T.G.B. reports Chan-Zuckerberg Biohub. D.R.C. reports CoI Company sponsored trials at institution (PI roles): AZ, Roche (Genentech) advisory role: ad hoc advisory boards or consultations 2023: Abbvie (steering committee), Anheart, Beigene (IDMC), Eli Lilly (IDMC), Imagene, Immunocore, Janssen, Mirati (IDMC), Prelude, Seattle Genetics, Valencee; 2022: Abbvie, Anheart, Appolomics (SRC), AstraZeneca–Daiichi (ILD adjudication committee), Beigene (DSMB) Dizal, EMD Serono, Elevation, Hengrui, (DSMB), Hummingbird, Janssen, Medtronic, Mersana (ILD adjudication committee), Mirati, Nalo Therapeutics, Onkure, Regeneron, Roche, Sanofi, Takeda, Theseus, Xcovery; 2021: Abbvie, Amgen, Anheart, Apollomics (SRC), AstraZeneca (SRC/SC), Beigene (DSMC), Bio-Thera (DSMB), Blueprint, Daiichi Sankyo (ILD adjudication committee), Elevation (SRC), Eli Lilly (DSMB and NCCN), EMD Serono, Helsinn (DSMB), Hengrui (DSMC), Janssen, Kestrel (SAB, Shares), Mersana, Nuvalent (SAB), Puma (NCCN), Ribon, Roche (Genentech), Sanofi, Seattle Genetics, Takeda and Turning Point. J.E.G. reports honoraria: Jazz Pharmaceuticals, Merck and OncoCyte; consulting or advisory role: AstraZeneca, Blueprint Medicines, Bristol Myers Squibb, EMD Serono, Lilly, Sanofi, Merck Sharp & Dohme, Loxo, Jazz Pharmaceuticals, Novartis, AstraZeneca–MedImmune, Janssen Scientific Affairs, National Comprehensive Cancer Network, AbbVie and Regeneron; research funding: Merck (Inst), AstraZeneca (Inst), Bristol Myers Squibb (Inst), Boehringer Ingelheim (Inst), Genentech (Roche) (Inst), G1 Therapeutics (Inst), Novartis (Inst), Pfizer (Inst), Ludwig Institute for Cancer Research (Inst), SWOG (Inst), Array BioPharma (Inst) and ECOG-ACRIN (Inst). N.L. reports CME and research funding: AstraZeneca. B.L. reports Genentech (Roche), Eli Lilly, Merck, Pfizer, Janssen, Daiichi Sankyo, Takeda, Mirati, Novartis and Guardant 360. J.R.B. reports grant funding: AstraZeneca; advisory board: AstraZeneca, Amgen, Novartis, Incyte, Bristol Myers Squibb, Genentech, Janssen, Sanofi, Regeneron, Merck, GlaxoSmithKline; DSMB: Sanofi, Janssen and GlaxoSmithKline. M.C.G. reports AstraZeneca, Abion, MSD International, Bayer, BMS, Boehringer Ingelheim Italia S.p.A, Celgene, Eli Lilly, Incyte, Novartis, Pfizer, Roche, Takeda, Seattle Genetics, Mirati, Daiichi Sankyo, Regeneron, Merck, Blueprint, Janssen, Sanofi, AbbVie, BeiGenius, Oncohost and Medscape. D.R.G. reports institutional research grants from Amgen, AstraZeneca, Genentech and Merck; consultant or advisory board for Adagene, AstraZeneca, Roche (Genentech), Guardant, IO Biotech, Oncocyte, OncoHost, Lilly, Merck and Novartis. E.B.G. reports consulting or advisory role: Novartis, GlaxoSmithKline, Merck, Boehringer Ingelheim, Shionogi, Eisai, Bristol Myers Squibb, ABL Bio, Xilio Therapeutics, Natera, Sanofi–Regeneron, Lilly, Personalis, Gilead Sciences, AstraZeneca, AbbVie–Abbott, Arcus Biosciences, Seagan and Summit Therapeutics; research funding: Merck (Inst), Genentech (Inst), AstraZeneca (Inst), Novartis (Inst), Lilly (Inst), Bristol Myers Squibb (Inst), Mirati Therapeutics (Inst), Dynavax Technologies (Inst), Iovance Biotherapeutics (Inst), Neon Therapeutics (Inst), EMD Serono (Inst), ABL Bio (Inst) and Daiichi Sankyo–UCB Japan (Inst); patents, royalties, other intellectual property: diagnosistic and therapeutic use of ‘Motif Neoepitopes’ as defined by Cummings et al.76. N.A.R. reports an employee at Synthekine and holds equity at Synthekine and Gritstone. G.V.S. reports honoraria, research funding and personal fees from AstraZeneca, Bayer, BeiGene Switzerland, F. Hoffman–La Roche, Merck Sharpe & Dohme, Eli Lilly, Johnson & Johnson, Pfizer, Takeda Oncology, Tesaro and Verastem outside the submitted work. J.W. reports advisory boards and lecture fees: Amgen, AstraZeneca, Bayer, Blueprint, BMS, Boehringer Ingelheim, Chugai, Daiichi Sankyo, Ignyta, Janssen, Lilly, Loxo, Merck, Mirati, MSD (Merck Sharp & Dohme), Novartis, Nuvalent, Pfizer, Roche, Seattle Genetics, Takeda and Turning Point; research support (to institution): BMS, Janssen, Novartis, Pfizer and AstraZeneca. B.B. reports receiving grants from AbbVie, Amgen, AstraZeneca, Chugai, Daiichi Sankyo, Ellipse, EISAI, Genmab, Genzyme Corporation, Hedera Dx, Inivata, IPSEN, Janssen, MSD, PharmaMar, Roche (Genentech), Sanofi, Socar Research, Tahio Oncology and Turning Point Therapeutics. R.S.H. reports consulting roles with AbbVie Pharmaceuticals, ARMO Biosciences, AstraZeneca, Biodesix, Bolt Biotherapeutics, Bristol Myers Squibb, Cybrexa Therapeutics, eFFECTOR Therapeutics, Eli Lilly, EMD Serono, Genentech (Roche), Genmab, Halozyme Therapeutics, Heat Biologics, I-Mab Biopharma, Immunocore, Infinity Pharmaceuticals, Loxo Oncology, Merck, Mirati Therapeutics, Nektar, Neon Therapeutics, NextCure, Novartis, Oncternal Therapeutics, Pfizer, Sanofi, Seattle Genetics, Shire, Spectrum Pharmaceuticals, STCube Pharmaceuticals, Symphogen, Takeda, Tesaro, Tocagen and WindMIL Therapeutics; advisory board roles with AstraZeneca, Bolt Biotherapeutics, Cybrexa Therapeutics, EMD Serono, I-Mab Biopharma, Immunocore, Infinity Pharmaceuticals, Neon Therapeutics, Novartis and STCube Pharmaceuticals; research support from AstraZeneca, Eli Lilly, Genentech (Roche) and Merck; and non-executive board membership for Junshi Pharmaceuticals and Immunocore. H.A.W. reports grants or contracts from any entity: Bayer, Arrys Therapeutics, AstraZeneca–Medimmune, BMS, Clovis Oncology, Genentech (Roche), Merck, Novartis, Seagen, Xcovery and Helsinn; advisory board: Mirati, Merck and Genentech (Roche); leadership: International Association for the Study of Lung Cancer (IASLC), President; ECOG-ACRIN, executive committee. N.A.P. reports an employee at Synthekine and holds equity at Synthekine and Gritstone. A.T.S. is currently an employee of Novartis. P.A.J. reports stock and other ownership interests: Gatekeeper Pharmaceuticals and Loxo; consulting or advisory role: Pfizer, Boehringer Ingelheim, AstraZeneca, Merrimack, Chugai, Roche (Genentech), Loxo, Mirati Therapeutics, Araxes Pharma, Ignyta, Lilly, Takeda, Novartis, Biocartis, Voronoi Health Analytics, SFJ Pharmaceuticals Group, Sanofi, Daiichi Sankyo, Silicon Therapeutics, Nuvalent, Eisai, Bayer, Syndax, AbbVie, Allorion Therapeutics, Accutar Biotech, Transcenta, Monte Rosa Therapeutics, Scorpion Therapeutics, Merus, Frontier Medicines, Hongyun Biotech and Duality Biologics; research funding: AstraZeneca (Inst), Astellas Pharma (Inst), Daiichi Sankyo (Inst), Lilly (Inst), Boehringer Ingelheim (Inst), Puma Biotechnology (Inst), Takeda (Inst) and Revolution Medicines (Inst); patents, royalties, other intellectual property: I am a co-inventor on a DFCI owned patent on EGFR mutations licensed to Lab Corp. I receive post-marketing royalties from this invention. D.P.C. reports advisory boards or consulting: Abbvie, Arcus Biosciences, AstraZeneca, BMS Israel, G1 Therapeutics, Genentech, GlaxoSmithKline, InThought, Iovance Biotherapeutics, Janssen, Jazz, JNJ, Merck–EMD Serono, Merck KGaA, Mirati, MSD, Novartis, Novocure, OncoHost, Pfizer Egypt, Regeneron, Roche and Sanofi; KOL/presentation/education/forum: AstraZeneca, Curio Science, Intellisphere, Merck US, OncLive, Pfizer, PPD Development and Roche Taiwan. M.D.H. is an employee of and stockholder in AstraZeneca. C.M.R. reports consulted regarding oncology drug development with AbbVie, Amgen, AstraZeneca, D2G, Daiichi Sankyo, Epizyme, Genentech (Roche), Ipsen, Jazz, Kowa, Lilly, Merck and Syros. He serves on the scientific advisory boards of Auron, Bridge Medicines, DISCO, Earli and Harpoon Therapeutics. L.A. is an employee of Foundation Medicine and a stockholder of Roche Holding AG. H.M. reports full-time employee of AstraZeneca and stock/shares. Z.Z. reports employee of AstraZeneca as well as stockholder of AstraZeneca and Pfizer. Z.L. reports full-time employee and stock owner of AstraZeneca. R.S. reports other support from AstraZeneca during the conduct of the study, as well as other support from Pfizer outside the submitted work. S.P. reports consultation or advisory role: AbbVie, Amgen, Arcus, AstraZeneca, Bayer, Beigene, BerGenBio, Biocartis, BioInvent, Blueprint Medicines, Boehringer Ingelheim, Bristol Myers Squibb, Clovis, Daiichi Sankyo, Debiopharm, Eli Lilly, F-Star, Fishawack, Foundation Medicine, Genzyme, Gilead, GSK, Hutchmed, Illumina, Incyte, Ipsen, iTeos, Janssen, Merck Sharp and Dohme, Merck Serono, Merrimack, Mirati, Nykode Therapeutics, Novartis, Novocure, PharmaMar, Promontory Therapeutics, Pfizer, Regeneron, Roche (Genentech), Sanofi, Seattle Genetics and Takeda; Board of Directors position: Galenica; talk in a company’s organized public event: AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Eli Lilly, Foundation Medicine, GSK, Illumina, Ipsen, Merck Sharp and Dohme, Mirati, Novartis, Pfizer, Roche (Genentech), Sanofi and Takeda; receipt of grants or research support: principal investigator in trials (institutional financial support for clinical trials) sponsored by Amgen, Arcus, AstraZeneca, Beigene, Bristol Myers Squibb, GSK, iTeos, Merck Sharp and Dohme, Mirati, PharmaMar, Promontory Therapeutics, Roche (Genentech) and Seattle Genetics. J.M.J. and M.J.R. are employees and shareholders of D2G Oncology. I.P.W. is a co-founder, employee and shareholder of D2G Oncology. M.M.W. is a co-founder, shareholder, member of the board of directors and compensated scientific advisor of D2G Oncology. I.P.W. and M.M.W. are co-inventors of patents relating to technologies for an autochthonous mouse model of human cancer, which D2G Oncology has exclusively licensed from Stanford University. V.A.P. reports Pfizer employee and stockholder. K.K.W. is a founder and equity holder of G1 Therapeutics and has sponsored research agreements with Takeda, TargImmune, Bristol Myers Squibb, Mirati, Merus and Alkermes, and consulting and sponsored research agreements with AstraZeneca, Janssen, Pfizer, Novartis, Merck, Zentalis, BridgeBio and Blueprint. P.J. reports employee and stock or stock options of AstraZeneca. I.I.W. reports honoraria from Genentech (Roche), Bayer, Bristol Myers Squibb, AstraZeneca, Pfizer, Merck, Guardant Health, Flame, Novartis, Sanofi, Daiichi Sankyo, Amgen, Janssen, Merus, G1 Therapeutics, Abbvie, Catalyst Therapeutics, Regeneron, Oncocyte, Medscape, Platform Health and Physicians’ Education Resources; research support from Genentech, Merck, Bristol Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis and Akoya. A.G.P. reports Pfizer. J.V.H. reports Advisory Committees: Genentech, Mirati Therapeutics, Eli Lilly, Janssen, Boehringer Ingelheim, Regeneron, Takeda, BerGenBio, Jazz, Curio Science, Novartis, AstraZeneca, BioAlta, Sanofi, Spectrum, GlaxoSmithKline, EMD Serono, BluePrint Medicine and Chugai; research support: AstraZeneca, Boehringer Ingelheim, Spectrum, Mirati, Bristol Myers Squibb and Takeda; licensing or royalties: Spectrum. H.A.A., M.T.D., A.G.C., Y.Q., R.P., D.M., D.H.O., R.M., J.A.H., L.M., S.C.M.L., R.P.J., C.L., Z.W., M.G.W., D.V., H.S., M.K., S.K., Y.N., J.T.A., J.S., L.L., H.N.K., B.C., S.S., R.L., T.Z., J.J.L., D.P., M.L.J. T.T., A.P., Y.K. and J.L.: no disclosures were reported by these authors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Haniel A. Araujo, Minh Truong Do
These authors jointly supervised this work: Ferdinandos Skoulidis, John V. Heymach
Contributor Information
Ferdinandos Skoulidis, Email: fskoulidis@mdanderson.org.
John V. Heymach, Email: jheymach@mdanderson.org
Extended data
is available for this paper at 10.1038/s41586-024-07943-7.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-07943-7.
References
- 1.Brahmer, J. R. et al. Five-year survival outcomes with nivolumab plus ipilimumab versus chemotherapy as first-line treatment for metastatic non–small-cell lung cancer in CheckMate 227. J. Clin. Oncol.41, 1200–1212 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Castro, G. et al. Five-year outcomes with pembrolizumab versus chemotherapy asfirst-line therapy in patients with non–small-cell lung cancer and programmed death ligand-1 tumor proportion score ≥ 1% in the KEYNOTE-042 study. J. Clin. Oncol.41, 1986–1991 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson, M. L. et al. Durvalumab with or without tremelimumab in combination with chemotherapy as first-line therapy for metastatic non–small-cell lung cancer: the phase III POSEIDON study. J. Clin. Oncol.41, 1213–1227 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med.378, 2078–2092 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Skoulidis, F. et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov.8, 822–835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbour, K. C. et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin. Cancer Res.24, 334–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristescu, R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh, A. et al. NRF2 activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin. Cancer Res.27, 877–888 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricciuti, B. et al. Diminished efficacy of Programmed Death-(Ligand) 1 inhibition in STK11- and KEAP1-mutant lung adenocarcinoma is affected by KRAS mutation status. J. Thorac. Oncol.17, 399–410 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West, H. J. et al. Clinical efficacy of atezolizumab plus bevacizumab and chemotherapy in KRAS-mutated non-small cell lung cancer with STK11, KEAP1, or TP53 comutations: subgroup results from the phase III IMpower150 trial. J. Immunother. Cancer10, e003027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alessi, J. V. et al. Clinicopathologic and genomic factors impacting efficacy of first-line chemoimmunotherapy in advanced NSCLC. J. Thorac. Oncol.18, 731–743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoulidis, F. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov.5, 860–877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyama, S. et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res.76, 999–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Best, S. A. et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metab.27, 935–943 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Kitajima, S. et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov.9, 34–45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, J. et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1 mutant lung cancer. Nat. Cancer2, 503–514 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzio, A. et al. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion. Cell185, 169–183 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sitthideatphaiboon, P. et al. STK11/LKB1 mutations in NSCLC are associated with KEAP1/NRF2-dependent radiotherapy resistance targetable by glutaminase inhibition. Clin. Cancer Res.27, 1720–1733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlhieter, C. A. et al. Concurrent mutations in STK11 and KEAP1 promote ferroptosis protection and SCD1 dependence in lung cancer. Cell Rep.33, 108444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paz-Ares, L. et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol.22, 198–211 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Garassino, M. C. et al. Associations of tissue tumor mutational burden and mutational status with clinical outcomes with pembrolizumab plus chemotherapy versus chemotherapy for metastatic NSCLC. JTO Clin. Res. Rep.4, 100431 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gainor, J. F. et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin. Cancer Res.22, 4585–4593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazieres, J. et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann. Oncol.30, 1321–1328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negrao, M. V. et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J. Immunother. Cancer9, e002891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizvi, N. A. et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellmann, M. D. et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med.378, 2093–2104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biton, J. et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin. Cancer Res.24, 5710–5723 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Papillon-Cavanagh, S., Doshi, P., Dobrin, R., Szustakowski, J. & Walsh, A. M. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open5, e000706 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarchoan, M. et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight4, e126908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson, D. J. et al. Pembrolizumab plus Ipilimumab following anti-PD-1/L1 failure in melanoma. J. Clin. Oncol.39, 2647–2655 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paz-Ares, L. G. et al. First-Line nivolumab plus ipilimumab with chemotherapy versus chemotherapy alone for metastatic NSCLC in CheckMate 9LA: 3-year clinical update and outcomes in patients with brain metastases or select somatic mutations. J. Thorac. Oncol.18, 204–222 (2023). [Google Scholar]
- 32.Rogers, Z. N. et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nat. Methods14, 737–742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji, H. et al. LKB1 modulates lung cancer differentiation and metastasis. Nature448, 807–810 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Hollstein, P. E. et al. The AMPK-related kinases SIK1 and SIK3 mediate key tumor-suppressive effects of LKB1 in NSCLC. Cancer Discov.9, 1606–1627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, C. W. et al. An LKB1–SIK axis suppresses lung tumor growth and controls differentiation. Cancer Discov.9, 1590–1605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galan-Cobo, A. et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res.79, 3251–3267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian, Y. et al. MCT4-dependent lactate secretion suppresses antitumor immunity in LKB1-deficient lung adenocarcinoma. Cancer Cell41, 1363–1380 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parra, E. R. et al. Immune cellular patterns of distribution affect outcomes of patients with non-small cell lung cancer. Nat. Commun.14, 2364 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, S. C. et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell170, 1120–1133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei, S. C. et al. Negative co-stimulation constrains t cell differentiation by imposing boundaries on possible cell states. Immunity50, 1084–1098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carthon, B. C. et al. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin. Cancer Res.16, 2861–2871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruse, B. et al. CD4+ T cell-induced inflammatory cell death controls immune-evasive tumours. Nature618, 1033–1040 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirschhorn, D. et al. T cell immunotherapies engage neutrophils to eliminate tumor antigen escape variants. Cell186, 1432–1447 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gungabeesoon, J. et al. A neutrophil response linked to tumor control in immunotherapy. Cell186, 1448–1464 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proulx-Rocray, F. et al. The prognostic impact of KRAS, TP53, STK11 and KEAP1 mutations and their influence on the NLR in NSCLC patients treated with immunotherapy. Cancer Treat. Res. Commun.37, 100767 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Yofe, I. et al. Anti-CTLA-4 antibodies drive myeloid activation and reprogram the tumor microenvironment through FcγR engagement and type I interferon signaling. Nat. Cancer3, 1336–1350 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Waight, J. D. et al. Selective FcγR Co-engagement on APCs Modulates the activity of therapeutic antibodies targeting T cell antigens. Cancer Cell33, 1033–1047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arce Vargas, F. et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell33, 649–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitajima, S. et al. MPS1 inhibition primes immunogenicity of KRAS-LKB1 mutant lung cancer. Cancer Cell40, 1128–1144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, R. et al. Inhibition of granulocytic myeloid-derived suppressor cells overcomes resistance to immune checkpoint inhibition in LKB1-deficient non-small cell lung cancer. Cancer Res.81, 3295–3308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vokes, N. I. et al. Harmonization of tumor mutational burden quantification and association with response to immune checkpoint blockade in non-small-cell lung cancer. JCO Precis. Oncol.3, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chalmers, Z. R. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med.9, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol.1, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol.31, 1023–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parra, E. R. et al. Immuno-profiling and cellular spatial analysis using five immune oncology multiplex immunofluorescence panels for paraffin tumor tissue. Sci. Rep.11, 8511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt, S. T. Shared nearest neighbors approach and interactive browser for network analysis of a comprehensive non–small-cell lung cancer data set. JCO Clin. Cancer Inform.6, e2200040 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaufman, J. M. et al. LKB1 loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT. J. Thorac. Oncol.9, 794–804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal.6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res.48, W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manguso, R. T. et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li, W. et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol. 16, 281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiou, S. H. et al. A conditional system to specifically link disruption of protein-coding function with reporter expression in mice. Cell Rep. 7, 2078–2086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.AACR Project Genie Consortium. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov.7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westcott, P. M. K. & To, M. D. The genetics and biology of KRAS in lung cancer. Chin. J. Cancer32, 63–70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol.34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogers, Z. N. et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat. Genet.50, 483–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winters, I. P. et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat. Commun.8, 2053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiou, S.-H. et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev.29, 1576–1585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev.15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li, C. et al. Quantitative in vivo analyses reveal a complex pharmacogenomic landscape in lung adenocarcinoma. Cancer Res.81, 4570–4580 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc.4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, R. et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat. Med.27, 141–151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cummings, A. L. et al. Mutational landscape influences immunotherapy outcomes among patients with non-small-cell lung cancer with human leukocyte antigen supertype B44. Nat. Cancer 1, 1167–1175 (2020). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data Figs. 3, 4 and Source Data Extended Data Fig. 5–10
Data Availability Statement
Data generated in this study are available in the article and its supplementary files. scRNA-seq raw and processed data reported in this article are available upon request and have been deposited to the NCBI Gene Expression Omnibus (GEO) under the accession number GSE267321. Data underlying the clinical trial findings may be requested in accordance with AstraZeneca’s data sharing policy, described in further detail at https://astrazenecagrouptrials.pharmacm.com/ST/Submission/Disclosure. All other individual de-identified participant data supporting the retrospective clinical data results reported in this article will be available on request according to General Data Protection Regulation (GDPR) standards. Only summary clinical data may be shared; patient-level image or genetic data are not available for access in our repository in the interest of protecting patient privacy. Materials, reagents or other experimental data are available upon reasonable request from the corresponding authors. Source data are provided with this paper.