Abstract
The brain helps us survive by forming internal representations of the external world1,2. Excitatory cortical neurons are often precisely tuned to specific external stimuli3,4. However, inhibitory neurons, such as parvalbumin-positive (PV) interneurons, are generally less selective5. PV interneurons differ from excitatory neurons in their neurotransmitter receptor subtypes, including AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptors (AMPARs)6,7. Excitatory neurons express calcium-impermeable AMPARs that contain the GluA2 subunit (encoded by GRIA2), whereas PV interneurons express receptors that lack the GluA2 subunit and are calcium-permeable (CP-AMPARs). Here we demonstrate a causal relationship between CP-AMPAR expression and the low feature selectivity of PV interneurons. We find low expression stoichiometry of GRIA2 mRNA relative to other subunits in PV interneurons that is conserved across ferrets, rodents, marmosets and humans, and causes abundant CP-AMPAR expression. Replacing CP-AMPARs in PV interneurons with calcium-impermeable AMPARs increased their orientation selectivity in the visual cortex. Manipulations to induce sparse CP-AMPAR expression demonstrated that this increase was cell-autonomous and could occur with changes beyond development. Notably, excitatory–PV interneuron connectivity rates and unitary synaptic strength were unaltered by CP-AMPAR removal, which suggested that the selectivity of PV interneurons can be altered without markedly changing connectivity. In Gria2-knockout mice, in which all AMPARs are calcium-permeable, excitatory neurons showed significantly degraded orientation selectivity, which suggested that CP-AMPARs are sufficient to drive lower selectivity regardless of cell type. Moreover, hippocampal PV interneurons, which usually exhibit low spatial tuning, became more spatially selective after removing CP-AMPARs, which indicated that CP-AMPARs suppress the feature selectivity of PV interneurons independent of modality. These results reveal a new role of CP-AMPARs in maintaining low-selectivity sensory representation in PV interneurons and implicate a conserved molecular mechanism that distinguishes this cell type in the neocortex.
Subject terms: Neural circuits, Molecular neuroscience, Neural encoding, Neurophysiology, Computational biophysics
Calcium-permeable AMPA receptors are identified to have a role in maintaining low feature selectivity in a specific population of inhibitory interneurons, and this function is conserved across ferrets, rodents, marmosets and humans.
Main
Genes dictate the response of a neuron to synaptic input and thereby program its biophysical computations6,8. For instance, neurotransmitter receptor profiles can determine the influx of Ca2+ to dendrites, which triggers neuronal changes that enable information storage2,9,10. Notably, gene expression varies widely across neuron types, which results in specialized roles within a given network and distinct responses to the same sensory input11,12. A key gene expression difference among the cardinal neuron types lies in their synaptic receptor composition13,14. Despite significant advances in understanding the role of synaptic protein genes, how these genes affect computations in the native brain is underexplored.
Neurons in the neocortex compute and represent features of the outside world through sparse, decorrelated activity1. This capability is particularly well characterized in the hippocampus, where place cells fire strongly to specific locations3, and in the primary visual cortex (V1), where neurons are highly tuned to oriented edges or movement directions in the visual receptive field4. In the visual cortex, the response selectivity of a neuron for orientation, spatial frequency, colour or speed is conferred through organized synaptic inputs that arise from thalamocortical neurons and excitatory and inhibitory neurons from within the cortex4,15. However, the genes, receptors and plasticity programs that give rise to this finely tuned circuit organization are largely unknown. A crucial clue to this question arises from the natural diversity of feature tuning expressed by different cell types16. Excitatory glutamatergic neurons display high orientation and direction selectivity, whereas PV GABAergic interneurons show low orientation selectivity17–21 (but see ref. 22). This distinction seems consistent across various modalities, with PV neurons typically displaying less selectivity. For instance, in the hippocampus, PV cells show much lower spatial selectivity than adjacent excitatory place cells5.
PV basket cells are highly adapted to fast spiking and provide strong and rapid feedback inhibition to nearby neurons5. They possess a distinct glutamate receptor profile, with small NMDA receptor (NMDAR) currents but large CP-AMPAR currents7,23,24. AMPARs are tetrameric glutamate receptors that mediate the majority of fast excitatory synaptic transmission in the brain. CP-AMPARs lack the voltage-dependent block of NMDARs by Mg2+, which allows them to induce distinct Ca2+ dynamics and forms of plasticity at synapses25,26. This characteristic suggests that CP-AMPARs in PV neurons have the potential to continually shape and adjust the relative strengths of inputs that carry diverse types of information in a manner distinct from nearby excitatory neurons. Here we explore the consequences of high CP-AMPAR levels and reveal that they play an important part in maintaining the low orientation selectivity of PV interneurons.
Conserved AMPAR subunit mRNA stoichiometry
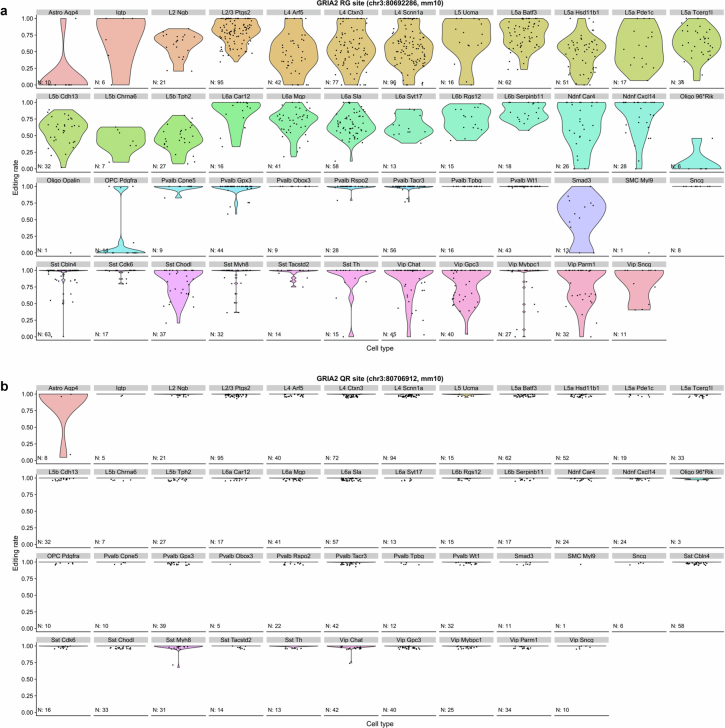
The calcium permeability of an AMPAR arises from two distinct routes, both involving GluA2: the lack of a GluA2 subunit (GluA2-lacking AMPAR) or the lack of RNA editing in the Gria2 site that encodes a crucial pore amino acid of GluA2 (unedited GluA2)23. The lack of GluA2 in turn could be due to transcriptional or translational regulation. We analysed SmartSeq-based high-coverage single-cell RNA sequencing (scRNA-seq) data from the mouse cortex14. The results revealed that RNA editing at the GluA2 Q/R site was uniformly complete (>95%) in all neuronal cell types (Extended Data Fig. 1), which suggested that unedited GluA2 does not significantly contribute to CP-AMPAR expression in PV neurons. Validated antibodies against the major AMPAR subunits GluA1 and GluA2 (Extended Data Fig. 2) showed that PV neurons express GluA2 at roughly 60% of the levels of neighbouring CaMKIIα+ excitatory neurons in both mice (Extended Data Fig. 3a,b) and marmosets (Extended Data Fig. 3c,d). Conversely, GluA1 was expressed about 1.7-fold higher in PV neurons, which contributed to a reduced GluA2 to GluA1 ratio and abundant GluA2-deficient CP-AMPARs (Extended Data Fig. 3e–g).
Extended Data Fig. 1. RNA editing at the Gria2 Q/R site is largely complete across many cortical cell types.
a, A-to-I RNA editing rates at the Gria2 R/G site. Editing rates [G/(A + G)] were stratified according to the cell types defined in Tasic et al.14 Each dot represents the editing rate in a single cell. The number of samples (cells) is noted in each panel. b, A-to-I RNA editing rates at the Gria2 Q/R editing site. Due to high concentration of data points near 1 (complete RNA editing) in this panel, many violin symbols were not visible, and single data points were jittered by 0.05 along the y-axis to aid visualization.
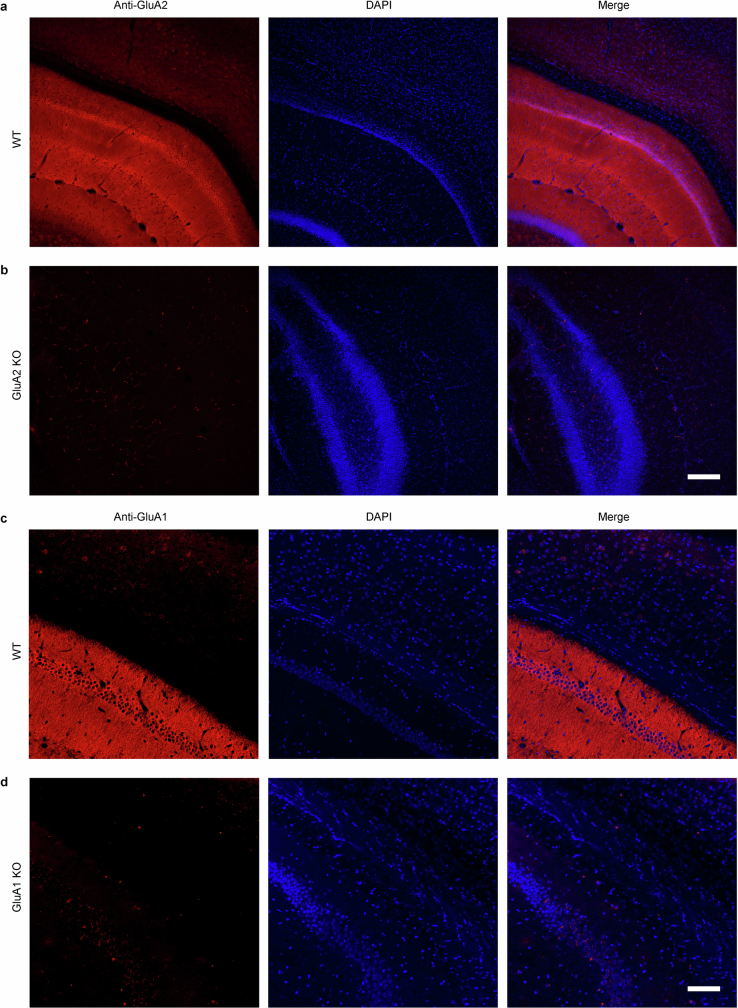
Extended Data Fig. 2. Immunohistochemical validation of anti-GluA2 and anti-GluA1 antibodies.
a-d, Immunohistochemical staining of GluA2 in GluA2−/− knockout mice (KO, b), wild-type littermates (WT, a), staining of GluA1 in GluA1−/− knockout mice (KO, d), and wild-type littermates (WT, c). Images of hippocampus and visual cortex were taken at 20x magnification. Scale bars, 200 μm.
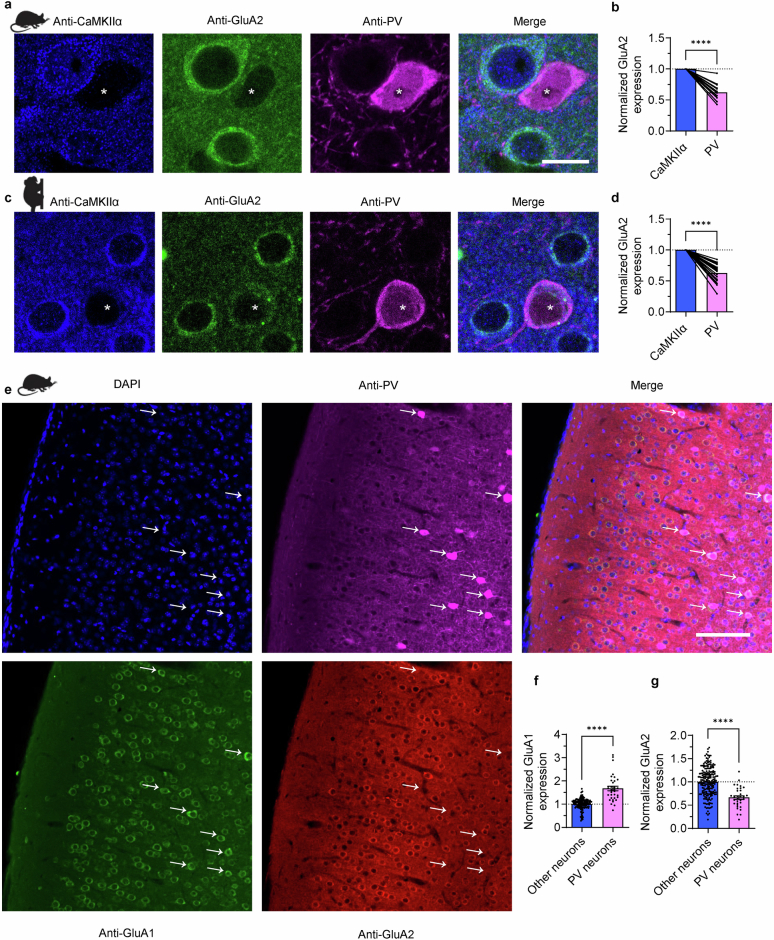
Extended Data Fig. 3. Selective low expression of GluA2 and Gria2 in PV and SST interneurons in mice, marmosets, and humans.
a, Immunohistochemical staining of PV and GluA2 in visual cortex layer 2/3. PV interneurons (asterisks) show markedly lower GluA2 expression compared to CaMKIIα excitatory counterparts. Layer 2/3 of visual cortex, scale bars, 10 μm. b, Quantification of relative GluA2 expression as a ratio of PV/CaMKIIα neurons (mean ± SEM) shows that PV interneurons express significantly less GluA2 (PV: 0.62 ± 0.03-fold vs CaMKIIα; n = 15 neuron pairs from 3 slices, 3 mice; P = 1.605x10−8, 1-sample t-test). c, GluA2 expression in PV interneurons and CaMKIIα excitatory neurons in the marmoset cortex. Scale bars, 10 μm. d, Marmoset PV interneurons express significantly less GluA2 compared to nearby CaMKIIα neurons (PV: 0.63 ± 0.03-fold vs CaMKIIα; n = 22 pairs from 7 slices, 3 marmosets, P = 1.062x10−10, one sample t-test). Bars and error bars denote mean ± SEM. e-g. High expression of GluA1 and low expression of GluA2 protein in PV interneurons. e, Immunohistochemical staining of PV, GluA1, and GluA2 in layer 2/3 of mouse visual cortex. PV interneurons (arrows) show markedly lower GluA2 expression, and higher GluA1 expression compared to all other neurons (all GluA1+ or GluA2+ and DAPI+ cells). Scale bar, 100 μm. f, Quantification of GluA1 expression (mean ± SEM) shows that PV interneurons express significantly more GluA1 (other neurons: 1.00 ± 0.02, n = 203 neurons/3 slices/3 mice; PV interneurons: 1.68 ± 0.10, n = 33 neurons; P = 2.319−25, unpaired t-test). g, Quantification of GluA2 expression (mean ± SEM) shows that PV interneurons express significantly less GluA2 (other neurons: 1.00 ± 0.02; PV interneurons: 0.66 ± 0.04; P = 5.861x10−9, unpaired t-test).
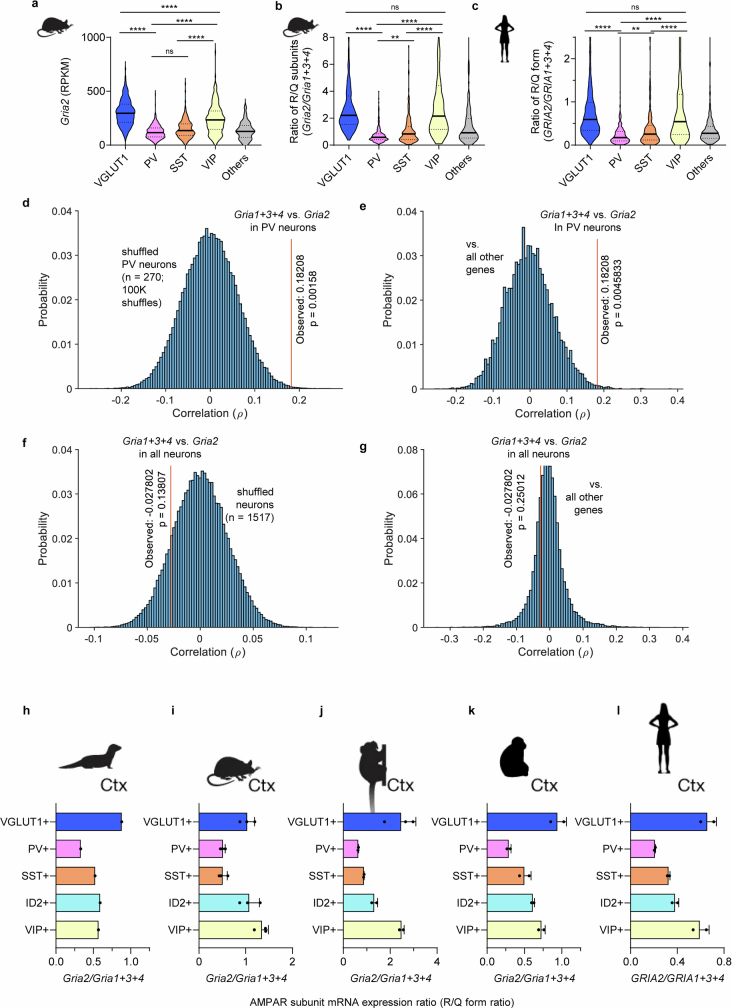
Single-cell transcriptomics data14 of AMPAR subunits matched the protein-level data, with PV neurons expressing GRIA2 mRNA at about 40% of its level in excitatory neurons (VGLUT1+; Extended Data Fig. 4a), which indicated the occurence of transcriptional regulation of CP-AMPARs. SST interneurons, which share the same developmental origin (medial ganglionic eminence), also expressed reduced GluA2 levels7. However, VIP+ interneurons, which originate separately from the caudal ganglionic eminence, had GRIA2 mRNA levels similar to excitatory neurons (Extended Data Fig. 4a). Motivated by the tightly correlated mRNA expression of AMPAR subunits (Extended Data Fig. 4d–g), we calculated the ratio of GluA2 to GluA1–GluA3–GluA4 subunits, which reflects the relative levels of calcium impermeable to permeable (R:Q) subunits, and found a similar ratio profile (Extended Data Fig. 4b). Notably, this ratio was always lowest in PV neurons across ferret, mouse, marmoset, macaque and human cortex datasets27,28 (Extended Data Fig. 4h–l), which suggests that there is evolutionary pressure towards lower R:Q ratios specifically in these neurons. These results show that PV neurons across these species express GluA2 at a tightly regulated low expression stoichiometry, probably through a transcriptional mechanism that is strongest in PV neurons.
Extended Data Fig. 4. Conserved low expression of Gria2 mRNA in PV/SST interneurons across mammalian species, and potential co-regulation of Gria1-4 mRNA expression in PV interneurons.
a, Analysis of Smart-seq single-cell RNA-seq data14 from the visual cortex of p56 mice shows distinctly lower expression of Gria2 mRNA in PV and SST interneurons (n = 756/270/178/185/118 neurons from VGLUT1/PV/SST/VIP/Other cell types, respectively, = 610.9, P < 1.000x10−15, KW 1-way ANOVA; P < 0.0001 for all VGLUT1 post-hoc comparisons, Dunn’s multiple comparison correction). A fraction of outlier cells was omitted for visualization. Conventional marker protein names are adapted to denote cardinal neuronal cell classes (VGLUT1 neurons and CaMKIIα neurons both refer to forebrain excitatory neurons). Post-hoc comparisons with the ‘others’ group are omitted for brevity. b, c, This low expression of Gria2 contributes to the lower ratio of calcium impermeable/calcium permeable AMPAR subunits (R/Q subunit ratio) both in mice (b) and in humans27 (c). In both (b) and (c), a KW 1-way ANOVA test reveals a significant difference (mice: = 593.6, P < 1.000x10−15; humans: = 491.9, P < 1.000x10−15), and post-hoc comparisons demonstrate significant differences between all non-‘others’ pairs except VGLUT vs. VIP (panel c shows human data from n = 2151/235/193/282/181 neurons from VGLUT1/PV/SST/VIP/Other cell types, respectively). Post-hoc comparisons with the ‘others’ group are omitted for brevity. Thick center lines and dotted lines in violin plots represent median and 25–75% interquartile range, respectively. d-g, Potential co-regulation of Gria1-4 mRNA expression in PV interneurons. d, Single-cell mRNA expression14 of Gria2 in PV neurons showed a strong correlation (ρ = 0.18208, n = 270 cells) with the sum of Gria1, Gria3, Gria4 mRNA expression, which was highly significant compared to a bootstrap randomized distribution (100,000 shuffles across PV neurons, P = 0.0019). The Monte Carlo P-value was determined by comparing the observed correlation statistic to the simulated distribution. e, The correlation between Gria2 expression and the sum of Gria1, Gria3, Gria4 expression in PV neurons was also highly significant compared to the distribution of correlations of Gria1 + 2 + 3 with all other genes (top 0.45 percentile, P = 0.0046). f, g, This correlation was not present in the entire neuron population (n = 1517 cells, ρ = −0.027802; P = 0.1381 in comparison to shuffled neuron data; P = 0.25012 in comparison to the correlation of Gria1 + 2 + 3 to all other genes beyond Gria2). These results suggest a tight co-regulation of Gria2 vs. Gria1 + 3 + 4 mRNA expression ratio unique to PV interneurons. h-l, Conserved low expression of Gria2 mRNA in PV/SST interneurons across mammalian species. Analysis of Drop-seq single-cell RNA-seq data28 from the cortex of (h) ferrets (n = 1 replicates), (i) mice (n = 3 replicates), (j) marmosets (n = 3/2/2/2/2 replicates), (k) macaques (n = 2 replicates), and (l) humans (n = 2 replicates) shows a conserved lower ratio of calcium impermeable/calcium permeable AMPAR subunits (R/Q form ratio) in PV and SST interneurons. VGLUT1 cells correspond to cortical excitatory neurons (CaMKIIα), and ID2 correspond to neurogliaform cells. Bars and error bars denote mean ± SD of Drop-seq samples, which were averaged within replicates.
Manipulation of CP-AMPARs in PV neurons
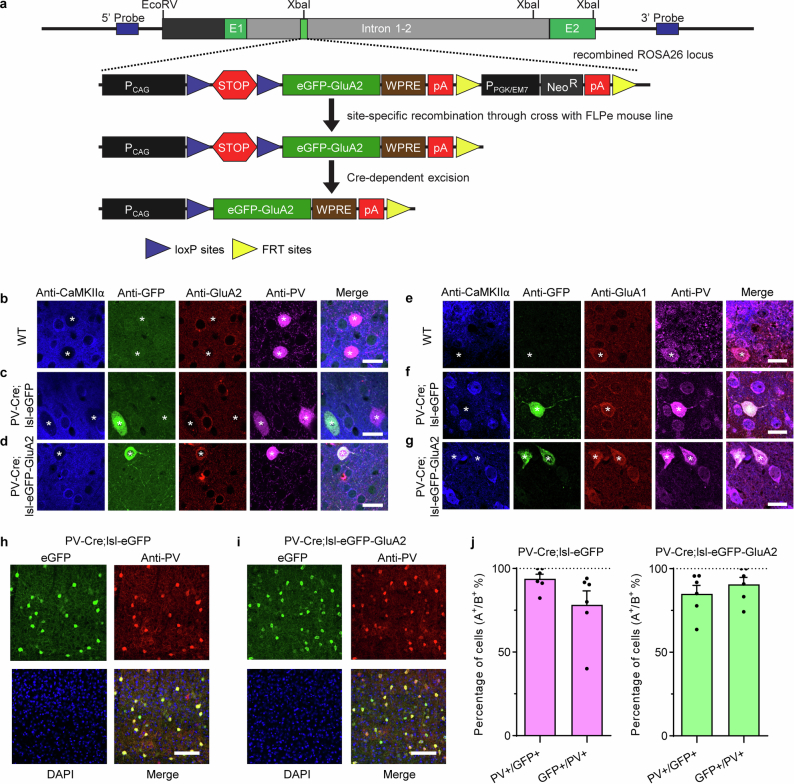
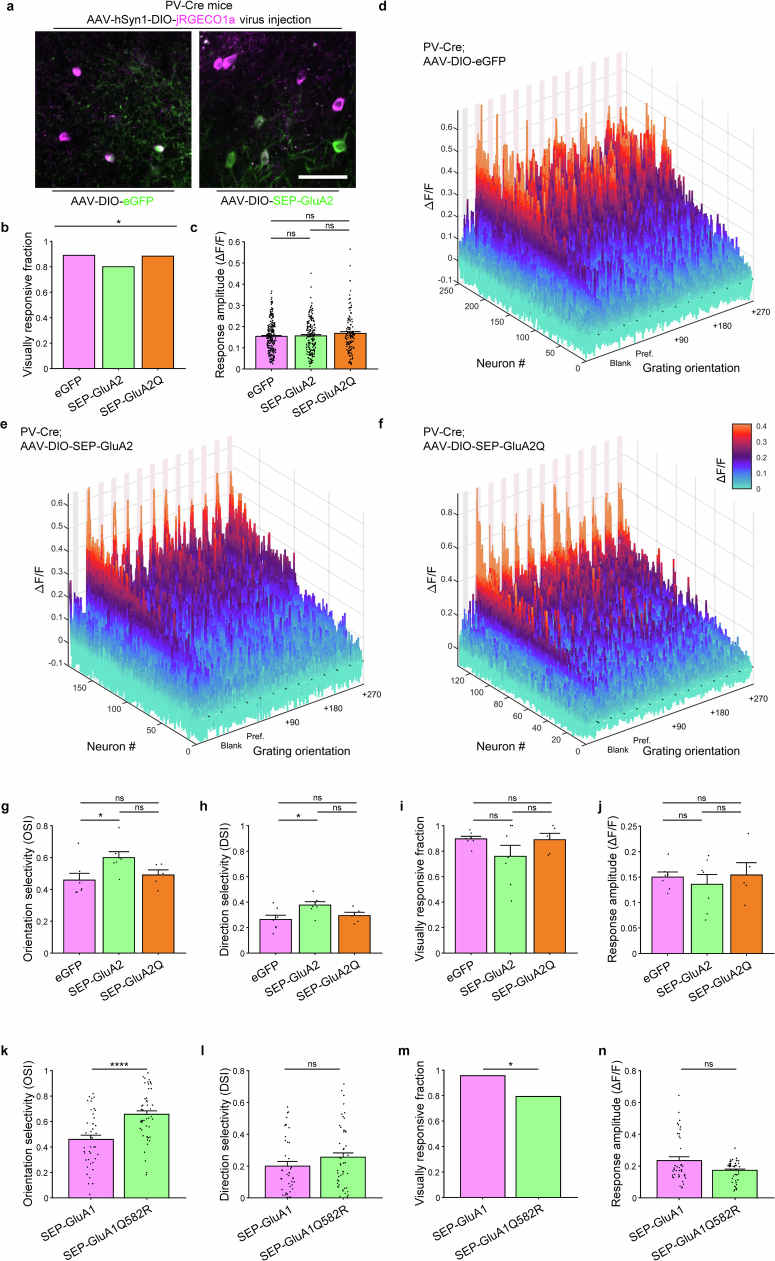
To test the functional significance of low GluA2 and high CP-AMPAR expression, we used a recently generated transgenic mouse line29 to express additional GluA2 with an eGFP tag in a Cre-dependent fashion (Extended Data Fig. 5). We crossed this transgenic mouse line with PV-Cre knock-in mice30 to generate PV-Cre;lsl-eGFP-GluA2 mice (Fig. 1a). These mice strongly expressed GluA2 at the cell soma and along the dendrites of PV neurons (Extended Data Fig. 5b–d and Fig. 1b) at high concordance with PV immunofluorescence (Extended Data Fig. 5h–j). The transgenic expression of GluA2 led to PV interneurons with levels of GluA2 comparable to excitatory neurons, roughly twice the level of PV-Cre;lsl-eGFP control mice. This increased expression occurred both at the mRNA level (221.6 ± 26.9%) and protein level (217.2 ± 11.2%; Fig. 1b and Extended Data Fig. 5b–d), as revealed by FACS-assisted PV neuron RNA-seq (Methods) and immunohistochemistry, respectively. Notably, GluA1 protein (but not mRNA) expression was reduced in PV neurons expressing exogenous GluA2 (Fig. 1c and Extended Data Fig. 5e–g), similar to excitatory neuron levels, which suggested a post-transcriptional homeostatic or displacing effect. Transgenic expression of GluA2 in PV cells did not produce significant changes in PV or SST neuron density in the visual cortex (Extended Data Fig. 6).
Extended Data Fig. 5. Development and characterization of a Rosa26 knock-in mouse to conditionally express eGFP-tagged GluA2 in a Cre-dependent manner.
a, To enable robust expression of eGFP-GluA2, we used a strong ubiquitous CMV-βactin hybrid (CAG) promoter (consisting of three gene regulatory elements: 5′ cytomegalovirus early enhancer element, chicken β-actin promoter and rabbit β-globin intron) and added a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) at the 3′ end of the eGFP-GluA2 coding sequence. The WPRE sequence allows rapid exit of mRNA from the nucleus and increases the mRNA stability in the cytosol. For inducible expression of eGFP-GluA2, a “stopper” cassette consisting of loxP-flanked 3X SV40 polyA (loxP-STOP-loxP, “lsl”) was placed upstream of the coding sequence, preventing expression until cyclic recombinase (Cre)-dependent excision. A Neomycin resistance cassette (NeoR) flanked by Flippase Recognition Target sequences (FRT) was present in the targeting vector to allow for selection. To prevent gene-silencing effects and ensure consistent and long-term expression of these transgenes in all cell types, the CAG-driven inducible eGFP-GluA2 transgenic constructs were targeted to the ubiquitously expressed Rosa26 locus. For homologous recombination in mouse embryonic stem (ES) cells, the gene-targeting vector was assembled into a ROSA26 targeting plasmid containing a 1.2 kb 5′ homology arm, 4.3 kb 3′ homology arm, and PGK-DTA (Diphtheria toxin fragment A, downstream of 3′ homology arm) for negative selection. ES cells, derived from a SV129 mouse strain, were electroporated with the AsiSI-linearized targeting vectors. A nested PCR screening strategy along the 5′ homology arm was used to identify ES cell clones harboring the correct genomic targeting event. After verification of homologous recombination by Southern blot analysis and confirmation of the karyotypes, correctly targeted ES cell clones were used to generate chimeric mice by injection into blastocysts derived from SV129 females at the Johns Hopkins University Transgenic Core. Germline transmission was achieved by breeding male chimeric founders to C57BL/6 N wild-type female mice. The FRT-NeoR cassette was removed by breeding to a transgenic FLPe mouse line101. b-j, Transgenic expression of GluA2 in PV interneurons mimics excitatory neuron GluA2 expression levels (related to Fig. 2b,c). b-d, Representative data for Fig. 1b. Immunohistochemical staining of GluA2 expression in PV interneurons and excitatory neurons. PV interneurons are marked by white asterisks and display negative CaMKIIα staining. Images were acquired in layer 2/3 of visual cortex. Scale bars, 15 μm. e-g, Representative data for Fig. 1c. Immunohistochemical staining of GluA1 expression in PV interneurons and excitatory neurons. Scale bars, 15 μm. h-j, Concordance of conditionally expressed eGFP and eGFP-GluA2 with PV immunostaining. h, i, Immunohistochemical staining of PV interneurons in mouse visual cortex. Scale bars, 100 μm. j, Quantification of conditional expression concordance in PV interneurons. In both PV-Cre;lsl-eGFP and PV-Cre; lsl-eGFP-GluA2 mouse lines, the ratio of PV+ cells among GFP+ cells and GFP+ cells among PV+ cells was high (n = 6/6 slices for each genotype; PV-Cre;lsl-eGFP mice: PV + /GFP + = 93.8 ± 2.7%, GFP + /PV + = 78.3 ± 8.3%; PV-Cre;lsl-eGFP-GluA2 mice: PV + /GFP + = 85.0 ± 5.1%, GFP + /PV + = 90.7 ± 4.0%). Bars and error bars denote mean ± SEM.
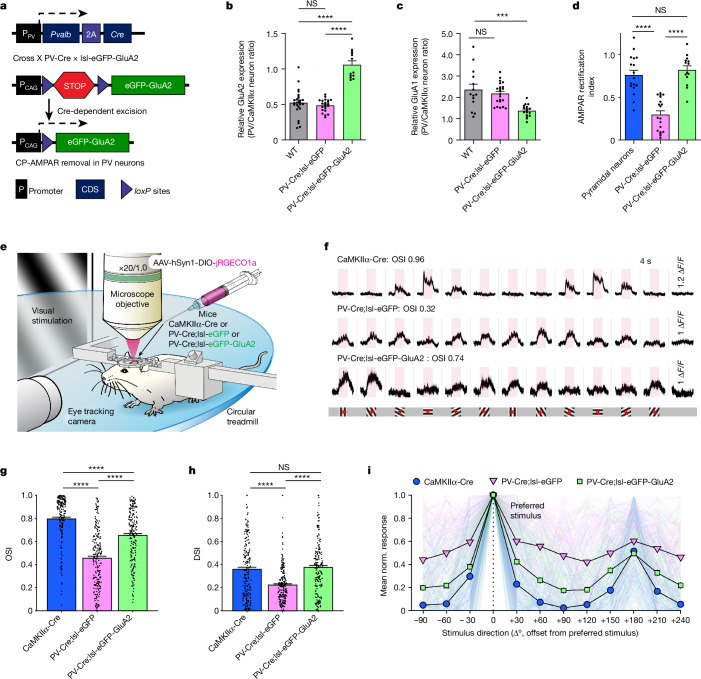
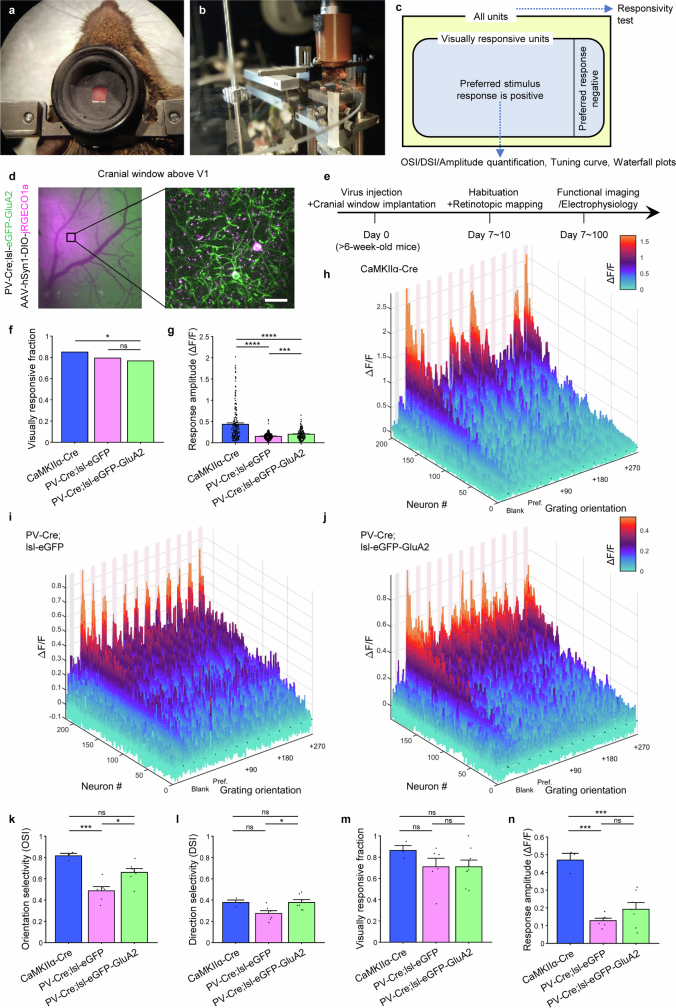
Fig. 1. GluA2 expression in PV interneurons alters orientation selectivity in the L2/3 of the mouse visual cortex.
a, Strategy to selectively remove CP-AMPARs in PV neurons. CDS, coding sequence. b, Relative GluA2 protein expression in PV and CaMKIIα neurons (left to right: n = 25, 22 and 13 pairs from 4, 4 and 4 slices, 3, 3 and 3 mice, respectively; P = 3.961 × 10−7, Kruskal–Wallis one-way ANOVA; P < 0.0001 for all PV-Cre;lsl-eGFP-GluA2 post hoc comparisons, Dunn’s multiple comparison correction). NS, not significant; WT, wild type. c, Relative GluA1 expression (left to right: n = 14, 22 and 17 pairs from 3, 3 and 3 slices, 3, 3 and 3 mice, respectively; P = 4.845 × 10−5, one-way ANOVA; P < 0.001 for all eGFP-GluA2 group post hoc comparisons, Tukey’s multiple comparison correction). Data are mean ± s.e.m. d, The low AMPAR rectification index in PV control neurons (lsl-eGFP, 0.298 ± 0.044) is increased in PV-Cre;lsl-eGFP-GluA2 mice (0.823 ± 0.047) to levels comparable with pyramidal neurons (0.763 ± 0.056) recorded for comparison (left to right: n = 17, 19 and 14 cells from 4, 3 and 2 mice, respectively; P = 4.056 × 10−10, one-way ANOVA test; P < 0.0001 for all post hoc comparisons with PV-Cre;lsl-eGFP mice, Tukey’s multiple comparison correction), thereby indicating the removal of CP-AMPARs. e, Mice were head-fixed and visually stimulated during 2P imaging of the V1. f, Neuronal soma activity traces. Pink rectangles denote the 4-s visual stimulation period and 1.2, 1.0 and 1.0 ΔF/F for each group. Grey shading corresponds to s.e.m. Whole screen drifting grating stimulation in 12 different directions was used to assess orientation selectivity. Red arrows mark the drifting direction. g, CaMKIIα neurons in CaMKIIα-Cre mice and PV neurons in PV-Cre;lsl-eGFP-GluA2 group displayed higher OSI values than in PV neurons in PV-Cre;lsl-eGFP controls (left to right: n = 202, 215 and 197 visually positive responsive neurons of 291, 316 and 395 total neurons from 3, 6 and 7 mice, respectively; = 175.67, P = 7.154 × 10−39, Kruskal–Wallis one-way ANOVA; P < 0.0001, Dunn’s multiple comparison correction). h, The CaMKIIα-Cre group and the PV-Cre;lsl-eGFP-GluA2 group displayed higher direction selectivity index (DSI) values than the PV-Cre;lsl-eGFP controls ( = 50.76, P = 9.517 × 10−12, Kruskal–Wallis one-way ANOVA; P < 0.0001). i, Normalized (norm.) average response profile of all positively responding neurons from each group aligned to their preferred stimulus direction (0°). A prominent peak is also present at +180° owing to the orientation selective nature of V1 neurons. Responses are plotted as the mean ± s.e.m.
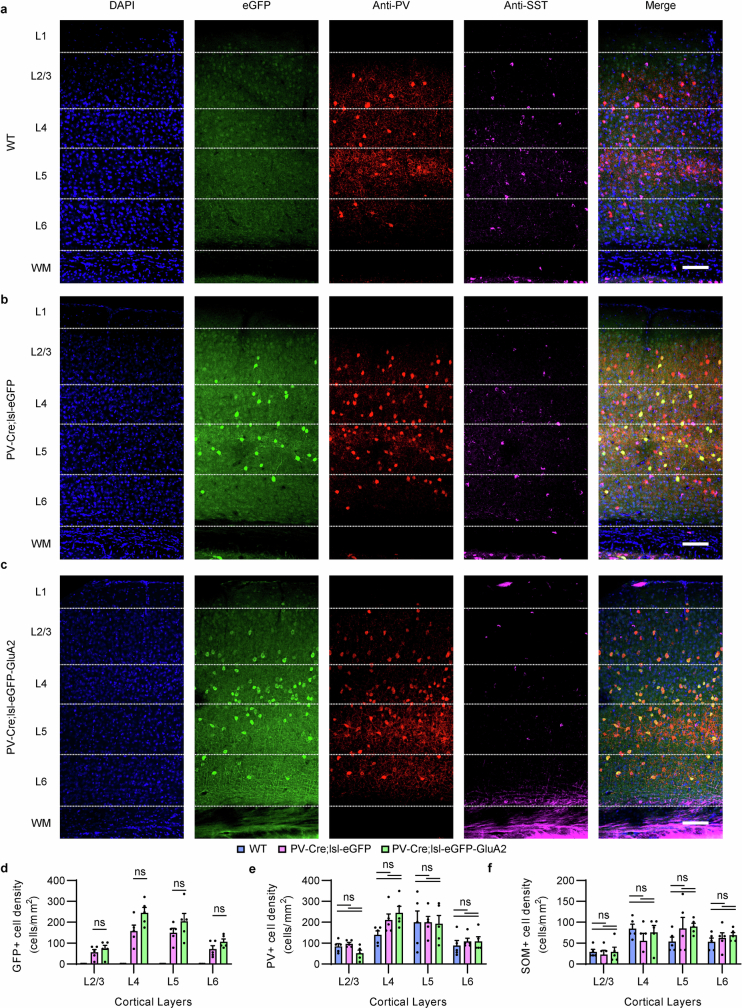
Extended Data Fig. 6. Transgenic expression of GluA2 in PV interneurons does not alter PV/SST interneuron density.
a-c, Visual cortex immunohistochemical staining of PV/SST interneurons. Layer segmentation was based on marker gene expression staining in V1 of internal and Allen Brain Atlas mouse brain sections. Scale bars, 100 μm. d-f, Quantification of GFP +, PV +, and SST+ cell density in cortical layers. PV and SST neuron density did not significantly change in PV-Cre;lsl-eGFP or PV-Cre;lsl-eGFP-GluA2 mice (n = 5/5/5 slices, 3/3/3 mice, 2-way ANOVA, P > 0.05 for all post-hoc comparisons, Šídák’s multiple comparison correction). Bars and error bars denote mean ± SEM.
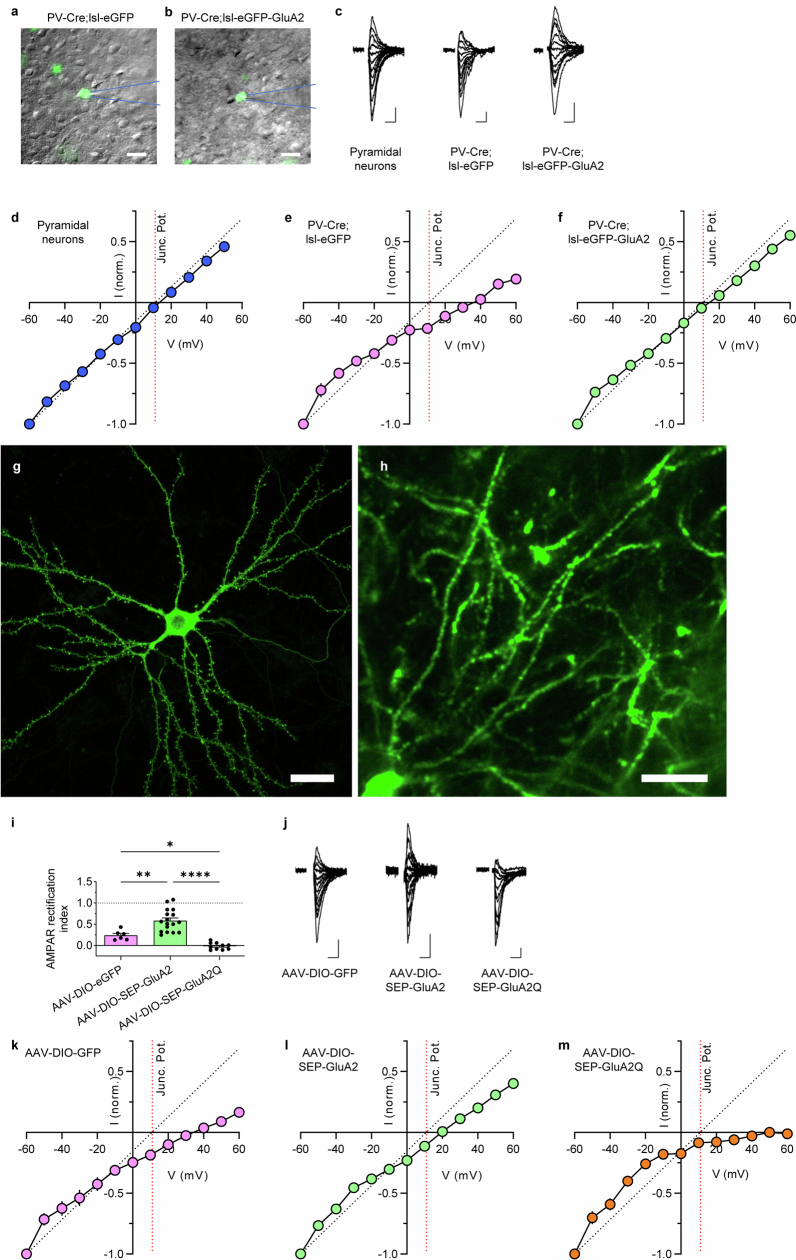
Electrophysiological recordings revealed that the signature inward rectification of CP-AMPARs typical of PV neurons was absent in PV-Cre;lsl-eGFP-GluA2 mice (Fig. 1d and Extended Data Fig. 7a–f), which showed synaptic incorporation of calcium-impermeable AMPARs. These results confirm that the PV-Cre;lsl-eGFP-GluA2 mouse model is suitable for testing the functional role of CP-AMPARs in PV interneurons.
Extended Data Fig. 7. Conditional transgenic/AAV expression of GluA2 in PV interneurons reduces calcium-permeable AMPARs.
Related to Figs. 1d and 2. a-b, Epi-fluorescence microscopy overlaid upon IR-DIC images of PV-Cre;lsl-eGFP and PV-Cre;lsl-eGFP-GluA2 mice. Scale bars, 20 μm. Layer 2/3 eGFP+ neurons in the visual cortex of PV-Cre;lsl-eGFP and PV-Cre;lsl-eGFP-GluA2 mice were targeted for whole cell patch clamp and rectification measurement. c, Example AMPAR-EPSC traces (Vh = −60 mV to 60 mV in 10 mV increments and in the presence of 100 μM AP5) from eGFP or eGFP-GluA2-expressing PV interneurons. Scale bars, 20 pA, 10 ms. d-f, Plot of the average I-V relationship for the AMPAR EPSC peak amplitudes of all recorded neurons (mean ± SEM). Red dotted lines denote the uncorrected junction potential (~11 mV). Black dotted lines indicate the expected linear I-V relationship from non-rectifying AMPARs to reveal deviations from linearity. Pyramidal neurons were recorded identically for comparison except for a subset which were measured from Vh = −60 mV to 50 mV. Note that PV control neurons (PV-Cre;lsl-eGFP) display inwardly rectifying AMPAR currents partially reminiscent of the doubly-rectifying CP-AMPAR currents reported previously in heterologous cells102,103, and additionally display a higher reversal potential, suggesting an alteration of AMPAR channel pore selectivity in cortical PV interneurons. g-m, AAV-mediated Cre-dependent expression of SEP-GluA2 and SEP-GluA2Q in PV interneurons bidirectionally regulates calcium-permeable AMPAR expression (related to Fig. 2). g, Confocal micrograph of a primary cultured excitatory cortical neuron transfected with FUW-Cre and pAAV-hSyn1-DIO-SEP-GluA2 plasmids. h, In vivo two-photon micrograph of a cortical PV interneuron infected with AAV2/9-hSyn1-DIO-SEP-GluA2. Scale bars: 20 µm. i, In mice injected with 3 different AAVs, layer 2/3 GFP+ visual cortex neurons were targeted for whole cell patch clamp and AMPAR rectification measurement. Data are presented as mean values ± SEM. Expression of the calcium-impermeable (wild-type) GluA2 subunit (AAV-DIO-SEP-GluA2) relieved AMPAR rectification and increased the rectification index compared to the control group (AAV-DIO-GFP). In comparison, the expression of the calcium-permeable mutant GluA2Q subunit (AAV-DIO-SEP-GluA2Q) resulted in further rectifying AMPAR currents and a lower rectification index (n = 6/17/10 cells from 2/2/3 mice, P < 0.0001, 1-way ANOVA test; P = 0.003 for AAV-DIO-eGFP vs. AAV-DIO-SEP-GluA2, P = 0.048 for AAV-DIO-eGFP vs. AAV-DIO-SEP-GluA2Q, P < 0.0001 for AAV-DIO-SEP-GluA2 vs. AAV-DIO-SEP-GluA2Q, Tukey’s multiple comparison correction). j, Example AMPAR-EPSC traces (Vh = −60 mV to 60 mV in 10 mV increments and in the presence of 100 μM AP5) from AAV-expressing PV interneurons. Scale bars, 20 pA, 10 ms. k-m, Plot of the average I-V relationship for the AMPAR EPSC peak amplitudes of all recorded neurons. Red dotted lines denote the uncorrected junction potential (~11 mV). Black dotted lines indicate the expected linear I-V relationship from non-rectifying AMPARs to reveal deviations from linearity.
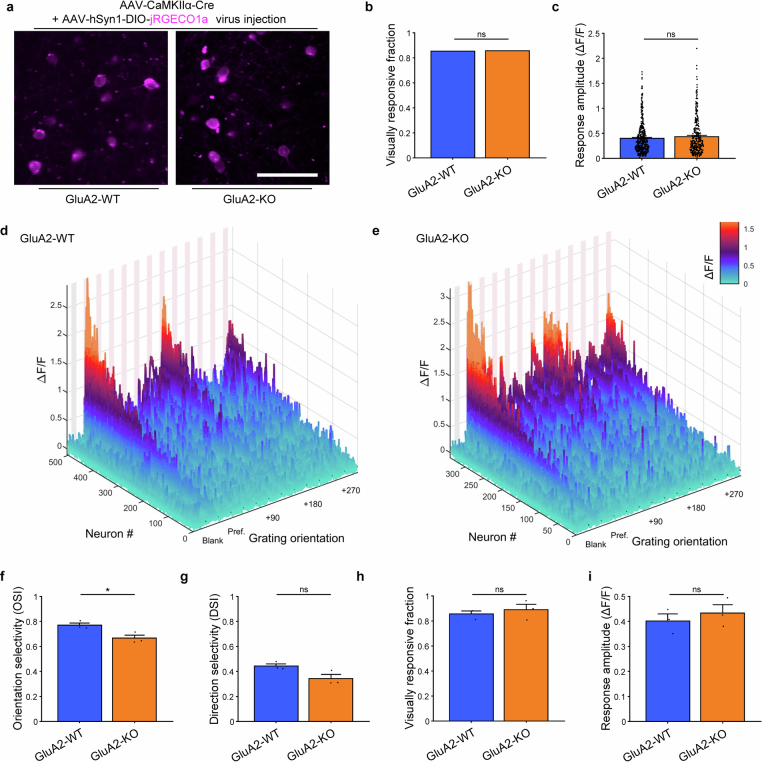
CP-AMPARs suppress PV selectivity
To test the role of CP-AMPARs in PV interneurons on sensory representation in awake mice, we assessed the orientation preference of layer 2/3 (L2/3) neurons in the V1 with in vivo two-photon (2P) calcium imaging (Fig. 1e). We injected PV-Cre;lsl-eGFP-GluA2 mice, PV-Cre;lsl-eGFP mice (controls) and CaMKIIα-Cre mice (as comparators) with a Cre-dependent jRGECO1a adeno-associated virus (AAV) targeting L2/3 of the visual cortex. Cranial windows were implanted on these mice and retinotopic mapping was performed to map the monocular V1 for 2P imaging (Extended Data Fig. 8a–e). We imaged somatic Ca2+ activity as a proxy for action potential activity during drifting grating presentation, focusing on the effect of neuronal firing rather than dendritic calcium dynamics. A proportion of PV neurons was visually responsive (Extended Data Fig. 8f) and displayed strong activity towards the 4-s presentations of full-field drifting gratings but not to blank isoluminant grey screen control trials (Fig. 1f). Consistent with previous observations17–21, the orientation selectivity of L2/3 PV interneurons was lower than excitatory neurons (with considerable variability in both populations4,22). Selectivity was significantly enhanced when GluA2 expression was increased to match the level in excitatory neurons (Fig. 1g; = 175.67, P < 0.0001, Kruskal–Wallis one-way analysis of variance (ANOVA); P < 0.0001 for CaMKIIα-Cre versus PV-Cre;lsl-eGFP and for PV-Cre;lsl-eGFP versus PV-Cre;lsl-eGFP-GluA2, Dunn’s multiple comparison correction). Despite distinct circuit underpinnings4,15,16,19,31–33, direction selectivity was similarly enhanced, which indicated a general increase in selectivity as a result of CP-AMPAR removal (Fig. 1h; = 50.76, P < 0.0001, Kruskal–Wallis one-way ANOVA; P = 0.0001 for CaMKIIα-Cre versus PV-Cre;lsl-eGFP and for PV-Cre;lsl-eGFP versus PV-Cre;lsl-eGFP-GluA2). These observations remained significant in more stringent animal-level statistics (Extended Data Fig. 8k,l). The average response amplitude was not significantly changed in neuron-level comparisons (Extended Data Fig. 8g,n). The average tuning curve demonstrated how relative non-preferred stimuli responses were reduced in PV neurons without CP-AMPARs, which produced increased orientation and direction selectivity (Fig. 1i and Extended Data Fig. 8h–j).
Extended Data Fig. 8. Awake head-fixed two photon imaging of the visual cortex reveals visual representation changes induced by Cre-dependent eGFP-GluA2 expression in PV interneurons.
a, Reinforced headposts with light-proofing rings to allow visual stimulation during two photon imaging. b, Circular treadmill for head-fixed awake imaging. The frame is mounted on a pair of goniometers to allow arbitrary tilt correction. c, Venn diagram displaying the hierarchical grouping of visual cortex units based on the responsivity to drifting grating stimuli and valence of largest response. Blue arrows indicate the level at which each analysis or visualization is carried out. d, Epifluorescence image of mouse visual cortex through an implanted cranial window (left) and two-photon imaging within the monocular V1 area (right). Scale bar, 20 μm. e, Experimental schedule. f, Fraction of neurons with statistically significant visual response in CaMKIIα or PV interneurons in each group (0.85, 0.79, 0.76; n = 395/316/355 neurons from 3/6/7 mice, = 8.4242, dF = 2, P = 1.487x10−2, Chi-square test). g, Average response amplitudes (∆F/F, mean ± SEM) of each group. CaMKIIα+ neurons in the CaMKIIα-Cre mice displayed higher amplitude preferred responses compared to PV interneurons in the PV-Cre;lsl-eGFP and PV-Cre;lsl-eGFP-GluA2 group (n = 202/215/197 neurons, = 96.78, P = 9.663×10−22, KW 1-way ANOVA; P < 0.0001 for all post-hoc comparisons with CaMKIIα-Cre, Dunn’s multiple comparison correction). h-j, Waterfall plots displaying the overall visual response profile of the population of visually responsive neurons with positive preferred stimulus responses in the (h) CaMKIIα-Cre, (i) PV-Cre;lsl-eGFP, (j) PV-Cre;lsl-eGFP-GluA2 groups. k-n, Mouse-level statistics confirm that removal of CP-AMPARs in PV interneurons increases feature selectivity in layer 2/3 of mouse visual cortex. k, CaMKIIα neurons in the CaMKIIα-Cre mice displayed higher orientation selectivity compared to PV interneurons in PV-Cre;lsl-eGFP mice, and the PV-Cre;lsl-eGFP-GluA2 group showed higher OSI than PV-Cre;lsl-eGFP controls (n = 3/6/7 mice, P = 0.0005, one-way ANOVA; P = 0.0111 for PV-Cre;lsl-eGFP vs. PV-Cre;lsl-eGFP-GluA2 and P = 0.0005 for CaMKIIα-Cre vs. PV-Cre;lsl-eGFP, Tukey’s multiple comparison correction). l, The PV-Cre;lsl-eGFP-GluA2 group showed higher DSI than PV-Cre;lsl-eGFP controls (P = 0.0278, one-way ANOVA; P = 0.0315 for PV-Cre;lsl-eGFP vs. PV-Cre;lsl-eGFP-GluA2). m, Fraction of neurons with statistically significant visual response in CaMKIIα or PV interneurons in each group were not different (0.86, 0.71, 0.71; n = 3/6/7 mice, P = 0.4016, one-way ANOVA). n, Average response amplitudes (∆F/F) of each group. CaMKIIα+ neurons in the CaMKIIα-Cre mice displayed higher amplitude preferred responses compared to PV interneurons in the PV-Cre;lsl-eGFP and PV-Cre;lsl-eGFP-GluA2 group (n = 3/6/7 mice, P < 0.0001, one-way ANOVA; P = 0.0001 for CaMKIIα-Cre vs. PV-Cre;lsl-eGFP and P = 0.0004 for CaMKIIα-Cre vs. PV-Cre;lsl-eGFP-GluA2, Tukey’s multiple comparison correction).
Cell-autonomous effect of CP-AMPAR removal
These results suggested that CP-AMPARs help reduce the visual feature selectivity of PV interneurons. To ask whether this effect arose from the systemic expression of GluA2 in PV neurons or from a cell-autonomous effect, we developed an AAV vector that sparsely expresses super-ecliptic pHluorin (SEP)-tagged GluA2 in a Cre-dependent fashion (Methods). Using this AAV-DIO-SEP-GluA2 virus, we observed strong expression of SEP-GluA2 in cultured neurons and in vivo (Extended Data Fig. 7g,h). As expected, rectification measurements demonstrated strong removal of CP-AMPARs in PV neurons compared with the control AAV-DIO-eGFP (Extended Data Fig. 7i–l). To further control for increased GluA2 expression, we used the calcium-permeable form of GluA2 (AAV-DIO-SEP-GluA2Q). This AAV similarly supplements GluA2 but should increase the proportion of CP-AMPARs. Indeed, this virus increased the rectification in PV neurons (Extended Data Fig. 7m).
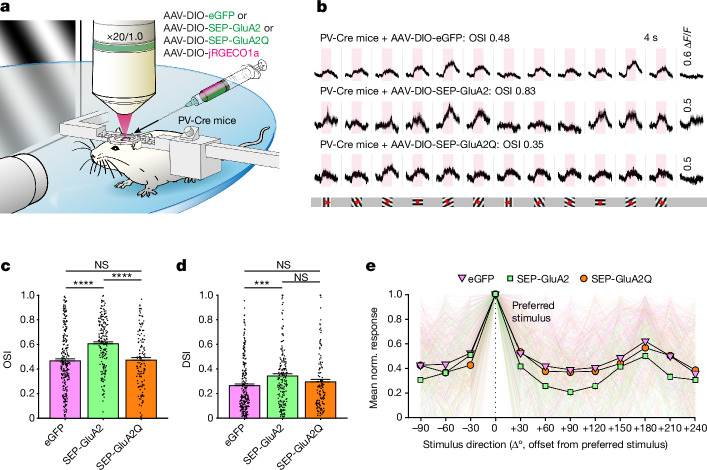
Sparsely expressing GluA2 in PV neurons increased orientation selectivity, which suggested that the effect of CP-AMPAR removal is cell-autonomous (Fig. 2a–c and Extended Data Fig. 9; = 41.68, P < 0.0001, Kruskal–Wallis one-way ANOVA; P < 0.0001 for eGFP versus SEP-GluA2 orientation selectivity index (OSI), Dunn’s multiple comparison correction). Notably, this effect was observed even in a >8-month-old mouse (P < 0.0001, Mann–Whitney U-test), a finding consistent with an ongoing role of CP-AMPARs in suppressing orientation selectivity. The calcium-permeable form of GluA2 (SEP-GluA2Q) did not increase PV neuron orientation selectivity (P = 0.9863 for eGFP versus SEP-GluA2Q), which suggested that channel pore calcium permeability has a specific role. Moreover, SEP-GluA2Q expression did not lead to a decrease in orientation selectivity, which indicated a saturation or a floor effect.
Fig. 2. Sparse GluA2 expression in L2/3 PV interneurons increases their orientation and direction selectivity.
a, Pre-injected mice were head-fixed and visually stimulated during 2P imaging of the V1 to reveal differences in tuning. b, Representative traces of neurons infected with AAV-DIO-eGFP or AAV-DIO-SEP-GluA2. Pink regions denote the 4-s visual stimulation period and 0.6, 0.5 and 0.5 ΔF/F for each group. Drifting grating stimulation was used to assess orientation selectivity. Red arrows mark the drifting direction. c, The SEP-GluA2 group displayed higher orientation selectivity than eGFP controls, whereas the SEP-GluA2Q group did not show increased OSI (left to right: n = 252, 184 and 132 visually responsive neurons of 325, 267 and 161 total neurons from 7, 7 and 5 mice, respectively; = 41.68, P = 8.892 × 10−10, one-way ANOVA; P < 0.0001 for eGFP versus SEP-GluA2; P = 0.9863 for eGFP versus SEP-GluA2Q, Dunn’s multiple comparison correction). d, Similarly, the SEP-GluA2 group displayed higher direction selectivity than the eGFP control group, whereas the SEP-GluA2Q group did not show higher DSI values (P = 9.843 × 10−4, = 13.85, Kruskal–Wallis one-way ANOVA; P = 0.0006 for eGFP versus SEP-GluA2; P = 0.5195 for eGFP versus SEP-GluA2Q). e, Normalized average response profile of all positively responding neurons aligned to their preferred stimulation (0°). Responses are plotted as the mean ± s.e.m.
Extended Data Fig. 9. Visual representation changes induced by sparse viral SEP-GluA2 expression in PV interneurons.
a, AAV-infected layer 2/3 PV interneurons within the monocular V1 area. Scale bar, 50 μm. b, The fraction of neurons with significant visual responses in each group (n = 325/267/161 total neurons from 7/7/5 mice, χ2 = 11.09, dF = 2, P = 0.0039, Chi-square test). c, The average response amplitude (∆F/F, mean ± SEM) of each group was not statistically different (n = 252/184/132 neurons, = 0.5550, P = 0.7578, KW 1-way ANOVA). d-f, Waterfall plots displaying the overall visual response profile of the population of visually responsive neurons with positive preferred stimulus responses in the (d) PV-Cre;AAV-DIO-eGFP, (e) PV-Cre;AAV-DIO-SEP-GluA2, (f) PV-Cre;AAV-DIO-SEP-GluA2Q groups. g-j, Mouse-level statistics confirm that removal of CP-AMPARs in PV interneurons increases feature selectivity in layer 2/3 of mouse visual cortex. g, The SEP-GluA2 group displayed higher orientation selectivity compared to eGFP controls, whereas the SEP-GluA2Q group did not show increased OSI (n = 7/7/5 mice, P = 0.0401, one-way ANOVA; P = 0.0385 for eGFP vs. SEP-GluA2, P = 0.8307 for eGFP vs. SEP-GluA2Q, Tukey’s multiple comparison correction). h, Similarly, the SEP-GluA2 group displayed higher direction selectivity compared to the eGFP control group, whereas the SEP-GluA2Q group failed to show higher DSI (n = 7/7/5 mice, P = 0.0348, one-way ANOVA; P = 0.0318 for eGFP vs. SEP-GluA2, P = 0.7699 for eGFP vs. SEP-GluA2Q). i, Fraction of neurons with statistically significant visual response in each group were not different (0.86, 0.76, 0.89; n = 7/7/5 mice, P = 0.2247, one-way ANOVA). j, Average preferred response amplitudes (∆F/F) of each group were not statistically different (n = 7/7/5 mice, P = 0.7319, one-way ANOVA). k, The SEP-GluA1Q582R group showed a significantly higher OSI (mean ± SEM) compared to the SEP-GluA1 group (n = 42/49 neurons from 44/68 total neurons from 3/3 mice, P < 0.0001, unpaired t-test). l, The DSI (mean ± SEM) was not significantly different (P < 0.1648, Mann-Whitney U-test). m, The fraction of neurons with significant visual responses in each group was different (0.95/0.79; n = 44/68 total neurons from n = 3/3 mice, χ2 = 5.615, dF = 1, P = 0.0178, Chi-square test). n, The average response amplitude (∆F/F) of each group (mean ± SEM) was not significantly different (n = 42/49 neurons from 3/3 mice, P = 0.6076, Mann-Whitney U-test).
Sparse SEP-GluA2 expression increased direction selectivity, whereas SEP-GluA2Q did not (Fig. 2d; P = 0.0009, = 13.85, Kruskal–Wallis one-way ANOVA; P = 0.0006 for eGFP versus SEP-GluA2; P = 0.5195 for eGFP versus SEP-GluA2Q), which suggested that CP-AMPARs are necessary for the low selectivity of PV neurons to various visual features. These changes in orientation and direction selectivity were replicated in stringent animal-level statistics (Extended Data Fig. 9g,h), but the proportion of visually responsive neurons and average response amplitude were not consistently different (Extended Data Fig. 9b,c,i,j). The average tuning curve confirmed reduced responses to non-preferred stimuli in SEP-GluA2-expressing PV neurons (Fig. 2e). Expression of the calcium-impermeable mutant GluA1 (GluA1(Q582R)) led to significantly higher orientation selectivity (but not direction selectivity) compared with wild-type GluA1 (Extended Data Fig. 9k). This result shows that R-form AMPAR subunits commonly increase orientation selectivity in PV neurons.
CP-AMPARs shape excitability but not connectivity
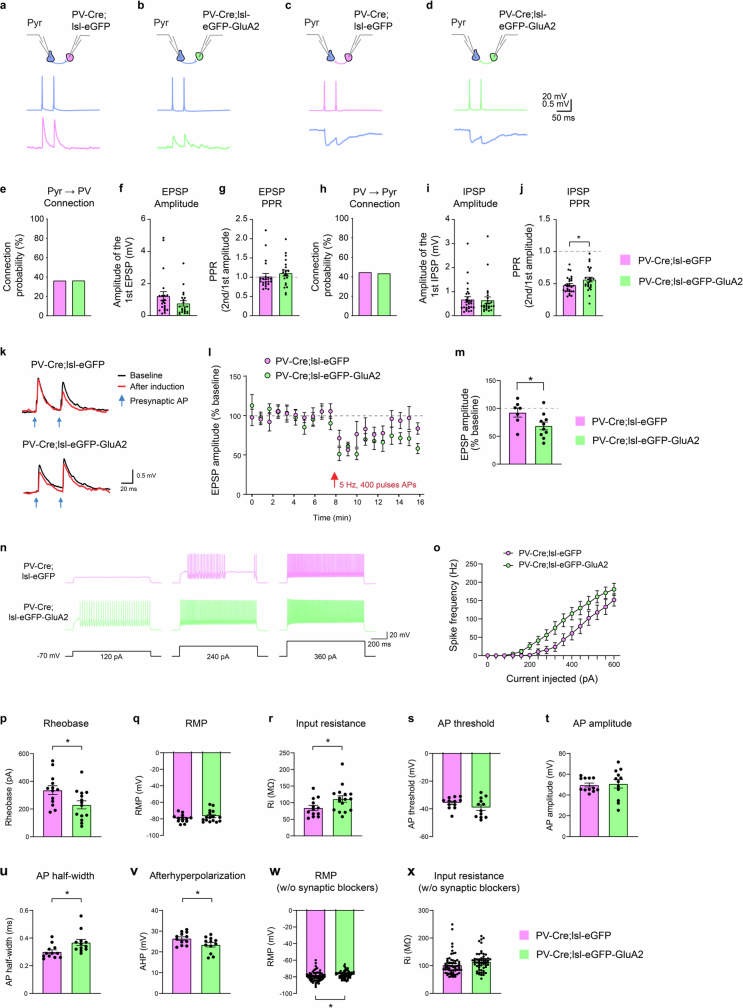
Excitatory neurons have a sparser input connectivity with local neurons than with PV neurons, which may underlie their higher orientation selectivity33–36. Thus, it is possible that dense local excitatory input connections of PV neurons become sparser following removal of CP-AMPARs to resemble excitatory neurons. Alternatively, the nominal connectivity rate may be unchanged, thereby implying that more specific mechanisms account for selectivity change. We used paired whole-cell patch-clamp recordings in acute brain slices to test the connectivity of L2/3 PV neurons and nearby excitatory neurons. We found no significant changes in the excitatory input or in reciprocal inhibitory output connection probability (Extended Data Fig. 10a–h). In connected pairs, the excitatory-to-PV neuron unitary excitatory postsynaptic potential (EPSP) amplitudes were not significantly reduced (Extended Data Fig. 10f), despite the loss of high-conductance CP-AMPARs, which suggests that there is a feedback or homeostatic mechanism that preserves synaptic strength. These results reject the idea that a large change in excitatory input connectivity is a mechanism for increasing orientation selectivity. However, they do not exclude the possibility that presynaptic input reorganization could lead to higher selectivity.
Extended Data Fig. 10. Synaptic connectivity, plasticity, and intrinsic excitability of PV interneurons in PV-Cre;lsl-eGFP and PV-Cre;lsl-eGFP-GluA2 mice.
Recording configurations (top), representative action potential traces in presynaptic cells (middle) and voltage response traces in postsynaptic cells (bottom) for Pyr→control PV (a), Pyr→GluA2 PV (b), control PV→Pyr (c), and GluA2 PV→Pyr (d) pairs. e, The probability of connection for tested Pyr→control PV and Pyr→ GluA2 PV connections (Pyr→control PV: 36.7%, n = 22 of 60 tested connections; Pyr→GluA2 PV: 36.8%, n = 21 of 57 tested connections, P = 1, Fisher’s exact test). f, The amplitudes of the unitary excitatory postsynaptic potentials (uEPSPs) of connected pairs (Pyr→control PV: 1.22 ± 0.30 mV, n = 22 pairs; Pyr→GluA2 PV: 0.76 ± 0.17 mV, n = 21 pairs; P = 0.17702, Mann-Whitney U test). g, The paired-pulse ratio (PPR) for connected pairs (Pyr→control PV: 1.02 ± 0.08, n = 22 pairs; Pyr→GluA2 PV: 1.11 ± 0.08, n = 21 pairs; P = 0.13104, Mann-Whitney U test). h, The probability of connection for tested control PV→Pyr and GluA2 PV→Pyr connections (PV→Pyr: 45%, n = 27 of 60 tested connections; GluA2 PV→Pyr: 43.9%, n = 25 of 57 tested connections, P = 1, Fisher exact test). i, The amplitudes of the unitary inhibitory postsynaptic potentials (uIPSPs) of connected pairs (PV→Pyr: 0.68 ± 0.12 mV, n = 27 pairs; GluA2 PV→Pyr: 0.64 ± 0.14 mV, n = 25 pairs; P = 0.75656, Mann-Whitney U test). j, The paired-pulse ratio (PPR) for connected pairs (PV→Pyr: 0.48 ± 0.03, n = 27 pairs; GluA2 PV→Pyr: 0.56 ± 0.04, n = 25 pairs; P = 0.0394, Mann-Whitney U test). k, Representative EPSP traces measured before (black, average of 50 traces) and after (red, average of last 20 traces) anti-Hebbian (AH) plasticity induction (400 presynaptic action potentials at 5 Hz paired with hyperpolarization of postsynaptic PV interneurons to −90 mV). The time points of the presynaptic action potentials for measuring EPSPs are marked with blue arrows. l, Normalized EPSP amplitude before and after AH plasticity induction (eGFP, n = 7 pairs from 4 mice; eGFP-GluA2, n = 10 pairs from 9 mice). Red arrow indicates the time point of AH plasticity induction. m, A summary graph showing normalized EPSP amplitude of the average of last 20 traces after AH plasticity induction (n = 7/10; eGFP, 92.6 ± 8.1%; eGFP-GluA2, 68.6 ± 6.9%, P = 0.03968, t-test). Representative voltage traces (n), and the current–spike (mean ± SEM) frequency relationship (o) recorded from control and eGFP-GluA2-expressing PV interneurons (for panels n-v, control eGFP: n = 13 cells from 3 mice; eGFP-GluA2: n = 14 cells from 3 mice; P = 0.0025, 2-way ANOVA interaction effect). p, The rheobase measured in GluA2-overexpressing PV interneurons was substantially lower than in control PV interneurons (control, 336.31 ± 31.69 pA; GluA2, 230.00 ± 29.35 pA; P = 0.0209, Mann-Whitney U-test). q, Resting membrane potential (RMP, control, −79.5 ± 1.37 mV; GluA2, −76.9 ± 1.73 mV; P = 0.4413, Mann-Whitney U-test), in the presence of glutamate and GABA receptor blockers (5 µM NBQX, 5 µM (RS)-CPP, and 10 µM SR95531; applies to panels n-v). r, Input resistance (Ri, control, 84.3 ± 7.5 MΩ; GluA2, 111.1 ± 9.7 MΩ; P = 0.0453, Mann-Whitney U-test). s, Action potential threshold (control, −35.7 ± 1.2 mV; GluA2, −39.2 ± 2.0 mV; P = 0.1501, Mann-Whitney U-test). t, Action potential amplitude (control, 49.7 ± 1.9 mV; GluA2, 50.8 ± 4.1 mV; P = 0.509, Mann-Whitney U-test). u, Action potential half-width (control, 0.30 ± 0.01 ms; GluA2, 0.37 ± 0.02 ms; P = 0.01, Mann-Whitney U-test). v, Afterhyperpolarization (AHP, control, 26.4 ± 0.8 mV; GluA2, 23.4 ± 1.1 mV; P = 0.0387, Mann-Whitney U-test). w, Resting membrane potential measured without synaptic glutamate and GABA receptor blockers (RMP; for panels w and x, control eGFP: n = 61 cells; eGFP-GluA2: n = 57 cells; control, −79.5 ± 0.75 mV; GluA2, −77.2 ± 0.58 mV; P = 0.0185, Mann-Whitney U-test). x, Input resistance measured without synaptic glutamate and GABA receptor blockers (Ri, control, 102.5 ± 4.7 MΩ; GluA2, 114.3 ± 4.5 MΩ; P = 0.0762, Mann-Whitney U-test). Data are presented as mean values ± SEM.
Input reorganization could arise from altered synaptic plasticity in PV neurons due to the removal of CP-AMPARs, which mediate an anti-Hebbian form of long-term potentiation (LTP) in hippocampal PV neurons through their ability to allow calcium influx at polarized potentials25. Synaptic plasticity has been underexplored in cortical PV neurons and generally presents as long-term depression (LTD) or smaller LTP compared with hippocampal neurons and excitatory neurons37,38. Consistently, several anti-Hebbian LTP induction protocols failed to result in potentiation in visual cortex PV neurons, instead leading to depression (Extended Data Fig. 10k–m). This LTD was exaggerated in PV-Cre;lsl-eGFP-GluA2 mice compared with control mice (P = 0.03968, unpaired t-test), which suggested that CP-AMPARs regulate the expression of non-Hebbian LTD in PV neurons.
Notably, PV neurons displayed markedly higher intrinsic excitability after CP-AMPAR removal, with a substantial increase in current-injected spike frequency, input resistance and action potential half-width, along with a decrease in rheobase and afterhyperpolarization (Extended Data Fig. 10n–v). Short action potential half-width, low input resistance and large afterhyperpolarization are all canonical features of PV neurons5. This finding suggests that removing CP-AMPARs led to a shift towards excitatory neuron-like intrinsic excitability characteristics. The resting membrane potential was also increased in PV-Cre;lsl-eGFP-GluA2 mice when measured without synaptic glutamate and GABA receptor blockers (Extended Data Fig. 10w), which suggested that there was a change in the balance of tonically active excitatory and inhibitory inputs (extrinsic synaptic excitability). Together, these results show that there is intact connectivity but altered synaptic plasticity and intrinsic excitability in PV neurons after removing CP-AMPARs.
Transcriptional response to CP-AMPAR removal
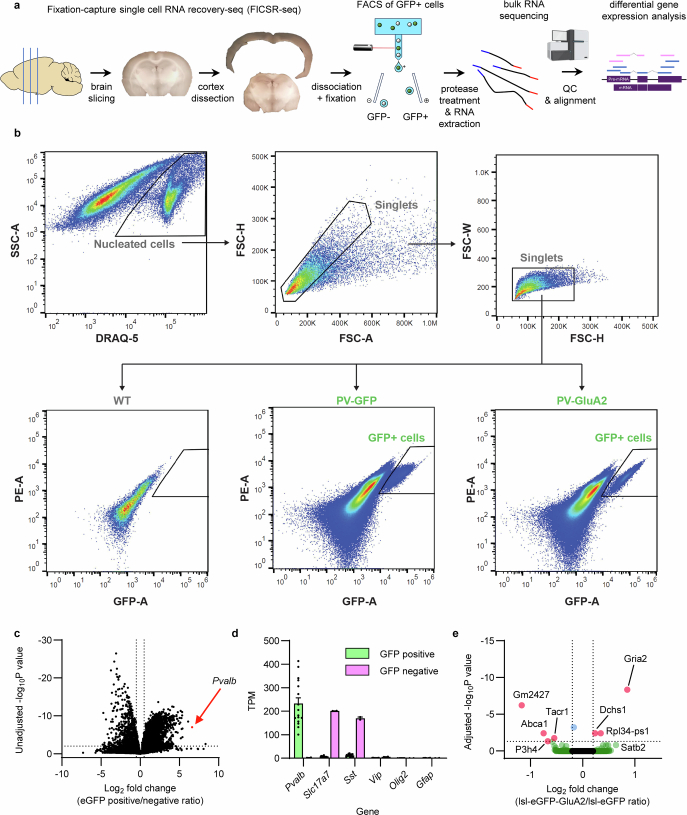
To investigate the new link between CP-AMPARs and excitability, we assessed global PV neuron transcriptome changes with fixation-capture single-cell RNA recovery sequencing (FICSR-seq; Methods) on forebrain PV neurons (Extended Data Fig. 11). FACS-assisted PV neuron bulk RNA-seq of PV-Cre;lsl-eGFP-GluA2 mice and PV-Cre;lsl-eGFP control mice showed no changes in expression in 278 out of 279 genes that constitute the major classes of ion channels and excitatory and inhibitory synapse proteins (Extended Data Figs. 12 and 13). This result points towards a post-transcriptional regulation of intrinsic and extrinsic (synaptic) excitability. The exception was Gria2 mRNA, which was expressed about twofold higher than in control PV neurons (Padjusted = 4.63 × 10−9, Benjamini–Hochberg correction; Extended Data Fig. 12), a result in agreement with protein measurements (Fig. 1b). GluA1, although downregulated at the protein level (Fig. 1c), was unchanged at the mRNA level. These transcriptomics results suggest that the substantial changes in PV neuron excitability after CP-AMPAR removal are not caused by changes in gene expression but probably reflect post-transcriptional regulation.
Extended Data Fig. 11. FACS-assisted bulk RNA-seq of cortical PV interneurons.
a, Overview of the workflow isolating and analyzing mouse cortex PV interneuron mRNA expression. Fixation-capture single cell RNA recovery-seq (FICSR-seq) was used to recover PV interneurons without substantial loss of PV cells during dissociation. After brain slicing, enzymatic dissociation was followed with fixation in 4% PFA and mechanical dissociation. GFP+/DRAQ5+ cells were isolated using FACS and were treated with proteinase K before RNA extraction which removes RNA-binding proteins and increases the yield of intact RNA. The resulting mRNA was sequenced with paired-end Illumina sequencing and analyzed for differential gene expression. b, Representative gating diagrams and FACS flowchart. DRAQ-5 was used to sort nuclei-containing cells from debris, and singlets are further sorted into GFP+ and GFP- cells. c, d, Bulk RNA-seq reveals ~100-fold enrichment of Pvalb mRNA in GFP+ vs GFP- cell samples (from n = 16/2 mice, mean ± SEM), validating FACS-based isolation of PV interneuron population. TPM stands for transcripts per million. e, Differential gene expression in PV-Cre;lsl-eGFP-GluA2 mice vs. PV-Cre;lsl-eGFP mice (n = 7/9 mice). Gria2 transgenic overexpression is observed (Padj = 4.63×10−9, Benjamini-Hochberg correction), together with other regulated genes (red).
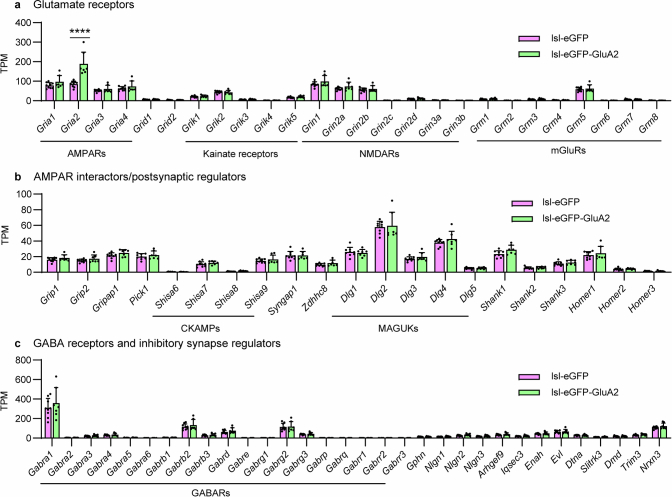
Extended Data Fig. 12. FACS-assisted bulk RNA-seq of cortical PV interneurons: synaptic gene expression.
a, Gene expression in PV interneurons of each genotype was statistically analyzed with DESeq2 and plotted using TPM (transcripts per million) to visualize expression levels. PV interneurons in PV-Cre;lsl-GFP-GluA2 mice display largely unchanged expression (mean values ± SEM) of major glutamate receptor genes compared to PV-Cre;lsl-eGFP control mice, with the exception of Gria2, which is overexpressed by roughly 2-fold in these mice (n = 9/7 mice; Wald test, Benjamini-Hochberg adjusted P < 0.0001). This is in line with the protein level increases (Fig. 1b) and matches the Gria2 expression level of typical excitatory neurons. b-c, AMPAR/GABAR interactor/postsynaptic regulator genes also display unaltered expression.
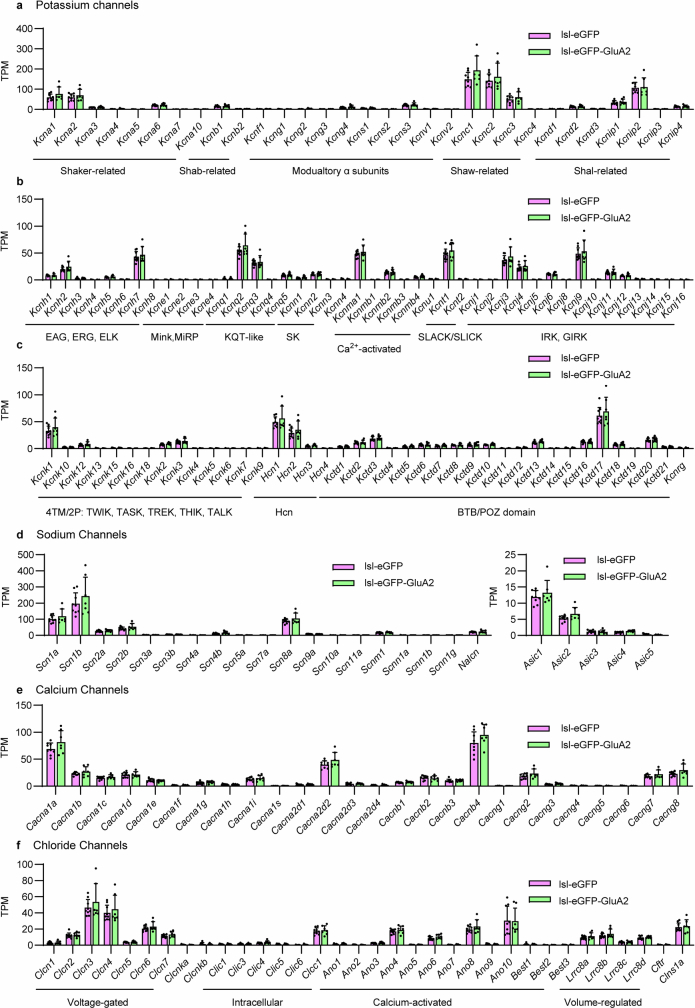
Extended Data Fig. 13. FACS-assisted bulk RNA-seq of cortical PV interneurons: intrinsic excitability genes including K+/Na+/Ca2+/Cl- channel gene expression.
Gene expression in PV interneurons of each genotype was statistically analyzed with DESeq2 and plotted using TPM (transcripts per million) to visualize expression levels. PV interneurons in PV-Cre;lsl-GFP-GluA2 mice display largely unchanged expression of (a-c) major K+ channel genes, (d), Ca2+ channel genes (e), and Cl- channel genes (f), compared to PV-Cre;lsl-eGFP control mice compared to PV-Cre;lsl-eGFP control mice (n = 9/7 mice). Data are presented as mean values ± SEM.
CP-AMPARs blunt excitatory selectivity
Removing CP-AMPARs from PV neurons renders them more selective. Therefore, we wondered whether introducing CP-AMPARs to excitatory neurons would reduce their orientation selectivity. To test this hypothesis, we assessed visual representation in GluA2 homozygous knockout mice, in which even excitatory neurons express abundant amounts of CP-AMPARs. Earlier studies39,40 had established that hippocampal neurons of these mice have reduced AMPAR currents with complete inward rectification and altered synaptic plasticity, but the impact on sensory representation has not been reported.
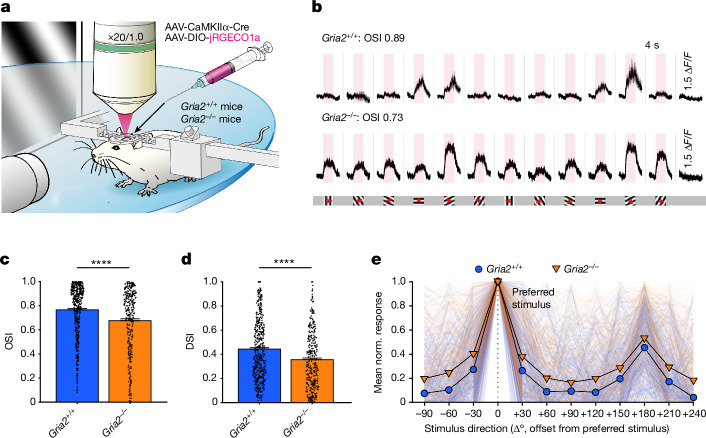
Using a dual virus approach (Fig. 3a), we measured the visual responses of excitatory neurons (which typically have low CP-AMPAR levels) in GluA2-knockout mice (Gria2–/–) and littermate controls (Gria2+/+). Excitatory neurons in the Gria2–/– mice displayed substantially reduced orientation selectivity (Fig. 3b,c,e; P < 0.0001, Mann–Whitney U-test) and direction selectivity (Fig. 3d,e; P < 0.0001, Mann–Whitney U-test). The proportion of visually responsive neurons and average response amplitude were not significantly different (Extended Data Fig. 14b,c). The change in direction selectivity did not reach significance in animal-level statistics, which suggests that the causal impact of CP-AMPARs on direction selectivity may be weaker than on orientation selectivity (Extended Data Fig. 14f,g). Non-preferred stimuli responses were relatively increased in excitatory neurons in the Gria2–/– mice, which in turn led to decreased orientation and direction selectivity (Fig. 3e and Extended Data Fig. 14d,e). These results suggest that CP-AMPAR expression is sufficient to reduce selectivity regardless of neuron type.
Fig. 3. GluA2 homozygous knockout leads to decrease of selectivity in excitatory neurons.
a, Pre-injected GluA2-knockout (Gria2–/–) and littermate wild-type (Gria2+/+) mice were head-fixed and visually stimulated during 2P imaging of the V1 to reveal differences in tuning. b, Representative traces of Gria2+/+ and Gria2–/– excitatory neurons. Pink regions denote the 4-s visual stimulation period and 1.5 ΔF/F for both groups. Whole screen drifting grating stimulation with 12 different orientations was used to assess orientation selectivity. Red arrows mark the drifting direction. c, Quantification of orientation selectivity shows a significantly lower OSI in Gria2–/– neurons than in wild-type controls (left to right: n = 504 and 340 from 3 and 3 mice, respectively; P = 8.733 × 10−7, Mann–Whitney U-test). d, The Gria2–/– group also displayed lower direction selectivity (P = 3.552 × 10−7, Mann–Whitney U-test). e, Normalized average response profile of all positively responding neurons aligned to their preferred stimulation (0°). Responses are plotted as the mean ± s.e.m.
Extended Data Fig. 14. Visual representation changes in excitatory neurons induced by global GluA2 homozygous knockout.
a, Two-photon micrograph of AAV-infected layer 2/3 excitatory neurons within monocular V1. Scale bar, 50 μm. b, The fraction of neurons with statistically significant visual responses in each group was not different (0.85/0.86; n = 739/500 neurons from n = 3/3 mice, χ2 = 0.0292, dF = 1, P = 0.8644, Chi-square test). c, The average response amplitude (∆F/F) of each group was not significantly different (n = 504/340 neurons; P = 0.4060, Mann-Whitney U-test). Data are presented as mean values ± SEM. d, e, Waterfall plots displaying the overall visual response profile of the population of visually responsive neurons with positive preferred stimulus responses in the (d) GluA2-WT (+/+), (e) GluA2-KO (−/−) groups. f-i, Mouse-level statistics confirm that homozygous knockout of GluA2 lowers excitatory neuron orientation selectivity in layer 2/3 of mouse visual cortex. f, Quantification of orientation selectivity shows a significantly lower OSI in GluA2 knockouts compared to littermate wildtype (WT) controls (n = 3/3 mice, P = 0.0288, unpaired t-test). g, The GluA2-KO group displays a trend towards lower direction selectivity (n = 3/3 mice, P = 0.0540, unpaired t-test). h, The fraction of neurons with statistically significant visual responses in each group was not different. (n = 3/3 mice, P = 0.5605, unpaired t-test). i, The average response amplitude (∆F/F) of each group was not significantly different (n = 3/3 mice, P = 0.5144, unpaired t-test). Responses are plotted as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., not significant, P ≥ 0.05.
Spatial selectivity of CA1 PV interneurons
We asked whether CP-AMPARs regulate PV interneuron selectivity beyond the visual cortex. PV neurons in the hippocampus have lower spatial selectivity than their neighbouring pyramidal cells3,5, but the molecular mechanisms underlying this reduced selectivity are unknown.
Using a virtual navigation task in head-fixed animals41 (Fig. 4a and Methods), in which mice are running on a 4-m-long virtual linear track, we imaged hundreds of PV neurons in the CA1 of PV-Cre mice transfected with Cre-dependent AAV expressing SEP-GluA2 or eGFP as a control (Fig. 4b). We previously measured reliable place fields of excitatory neurons with this experimental set up41, which indicated that the hippocampus forms a strong internal representation of the virtual environment.
Fig. 4. Increased spatial tuning of hippocampal PV interneurons after expression of GluA2.
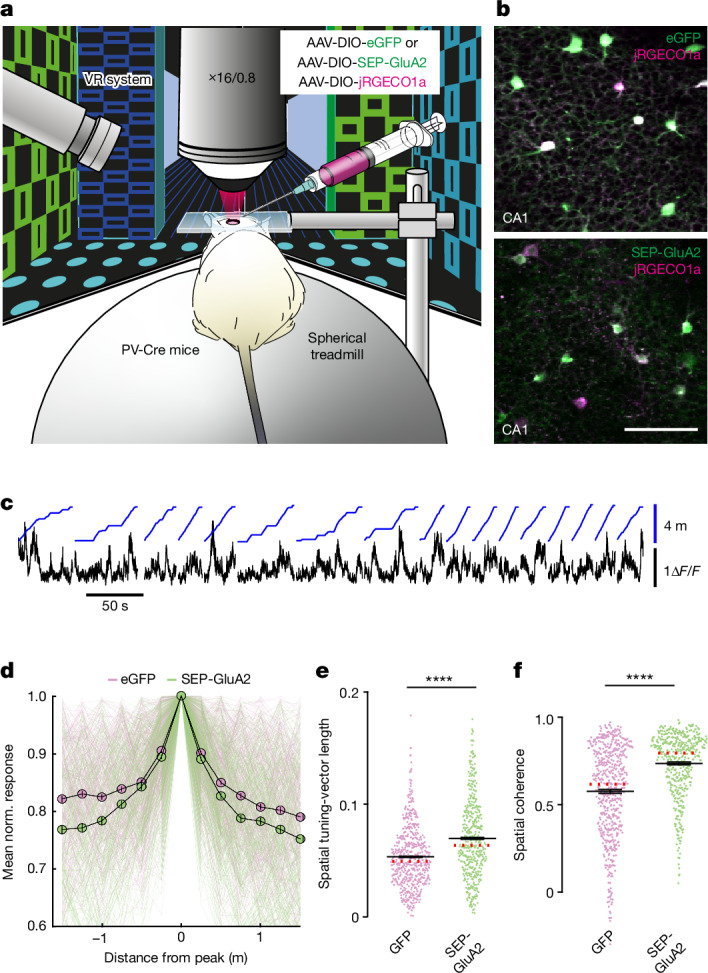
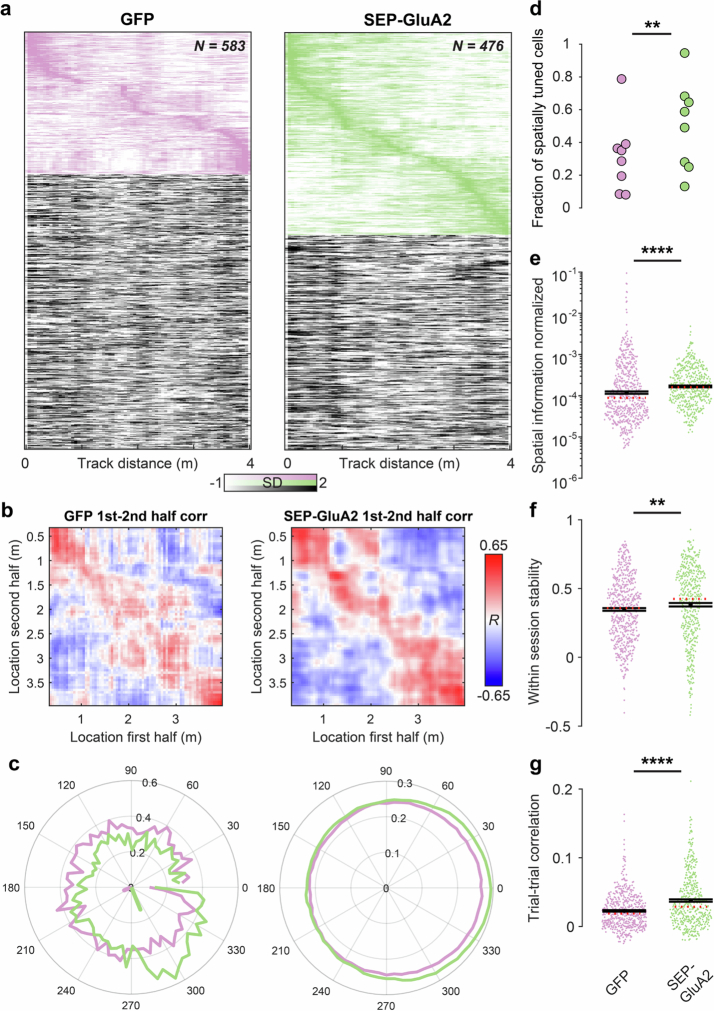
a, Experimental schematic of the virtual reality (VR) system. b, Time average of fluorescence acquired in vivo for jRGECO1a and SEP-GluA2 or eGFP. Scale bar, 100 μm. c, Ca2+ activity traces (black) and mouse position in virtual reality linear track (blue) over time. d, Normalized average spatial response profile of hippocampal CA1 PV neurons expressing SEP-GluA2 (green) or eGFP (magenta) aligned to the location of their peak activation. Responses are plotted as the mean ± s.e.m. Thin lines denote individual cells. e, Spatial tuning-vector lengths (Extended Data Fig. 15c) of PV neurons transfected with SEP-GluA2 were significantly higher than GFP controls (left to right: n = 583 and 476 cells from 4 and 4 mice, respectively; P = 1.472 × 10−14, Wilcoxon rank-sum test). f, Spatial coherence was also higher in the SEP-GluA2 group (P = 1.532 × 10−26, Wilcoxon rank-sum test). Black lines in e and f denote the mean ± s.e.m., and the red dotted line denotes the median. Dots denote values for individual cells.
Notably, we observed that the activity of SEP-GluA2-expressing PV neurons (Fig. 4c) was more sharply tuned to the preferred location than GFP-expressing control PV neurons (Fig. 4d). This was reflected in increases in spatial tuning-vector length (Fig. 4e and Extended Data Fig. 15c), spatial coherence (local smoothness of the spatial tuning curve; Fig. 4f) spatial information and within-session stability of spatial tuning curves, and decreases in variability of spatial responses between trials (Extended Data Fig. 15b,f,e). In summary, these data suggest that GluA2-deficient CP-AMPARs reduce the selectivity of PV neurons regardless of modality and have a broad role in sensory representation beyond the neocortex.
Extended Data Fig. 15. Increased spatial tuning of hippocampal PV interneurons after removal of CP-AMPARs.
a, Raster plots of Ca2+ activity over distance (normalized) for PV interneurons expressing GFP (left) or SEP-GluA2 (right). Activity for cells with significant spatial tuning is shown on top (magenta, green, respectively) and sorted by the peak location of activation. Activity of non-spatially tuned cells is shown below (greyscale). b, Average cross correlation of spatial activity profiles of cells between the first and second half of recording sessions (10 runs each). Note the higher and more consistent correlations of spatial tuning in cells expressing GluA2. c, Left, two representative examples of spatial vector tuning for a GFP (magenta) and SEP-GluA2 expressing (green) PV interneuron. The cell’s activity along the track is projected into a circular coordinate system. The orientation and length of the calculated tuning vector in the center correspond to the tuning direction and specificity, respectively. Right, Average activity profile for all GFP (magenta) or SEP-GluA2 expressing (green) PV interneurons aligned to their preferred orientation (0°). Note the higher asymmetry and stronger vectorial tuning for GluA2 expressing cells. d, Fraction of cells with significant spatial tuning in GFP (magenta) and SEP-GluA2 (green) expressing animals. Dots denote fractions in individual experiments. (n = 8/8 experiments from n = 4/4 mice, P = 0.001, t-test). e, Spatial information normalized to activity (see Methods) for GFP (magenta) and SEP-GluA2 (green) expressing cells. Dots denote individual cells. (n = 583/476 cells from n = 4/4 mice, P = 2.23×10−8, Wilcoxon rank-sum test). f, Same as in (e) but for spatial map stability (correlation) between the first and second half of the recording session (P = 0.0026, Wilcoxon rank-sum test). g, Same as in (e) but for spatial activity map correlations between individual trials of a session (P = 2.28×10−9, Wilcoxon rank-sum test). Note the consistently higher stability of spatial representation in PV interneurons expressing GluA2. Black lines in (e-g) denote mean ± SEM, and the red dotted line denotes the median. Dots denote values for individual cells.
Computational modelling of CP-AMPAR removal
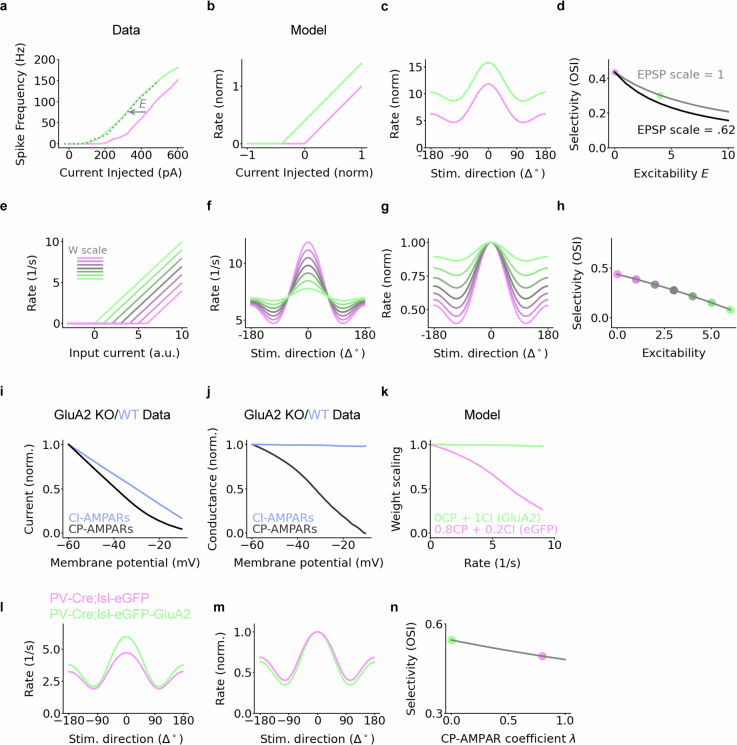
Given the electrophysiological changes we observed with CP-AMPAR removal, we used computational models to identify which mechanisms are consistent with the increase in feature selectivity. We explored the following three electrophysiological circuit changes: (1) increased intrinsic excitability, (2) the loss of inward-rectifying AMPAR current and (3) enhanced LTD. We used a model comprising a single PV neuron receiving a set of inputs from presynaptic excitatory neurons with predefined stimulus tuning (Fig. 5a). To endow the PV neuron with stimulus tuning, the strength of excitatory–PV synapses was modelled as bell-shaped around the preferred orientation of PV neurons (Fig. 5b), which enabled PV neurons to inherit their tuning from pyramidal cells (Fig. 5c).
Fig. 5. Mathematical models of the impact of CP-AMPAR removal on selectivity.
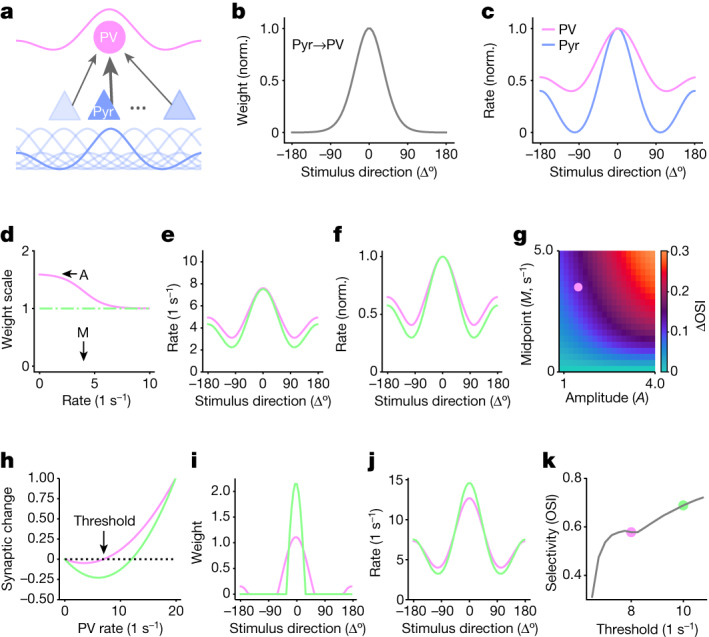
a, Feed-forward network architecture of a PV neuron (circle) receiving inputs from pyramidal cells (Pyr; triangles, n = 64). Insets depict tuning curves. b, Pyramidal–PV connectivity depends on the difference between the preferred orientation of the PV neuron and the pyramidal cell in question. c, Tuning of pyramidal and PV responses. OSI: 0.73 (Pyr) and 0.44 (PV). Rates are normalized by their maximum for visual comparison. d, Rate-dependent weight reduction as a model of CP-AMPAR-dependent inward rectification in control PV-Cre;lsl-eGFP neurons (magenta), parametrized by a maximum amplitude A and a midpoint M. The removal of CP-AMPARs and inward rectification in PV-Cre;Isl-eGFP-GluA2 mice is modelled by removing the rate dependence of the synaptic weights (green). e, Removal of CP-AMPARs decreases responses to non-preferred stimuli but not to preferred stimuli, thereby increasing stimulus selectivity. OSI with and without CP-AMPARs: 0.48 and 0.59, respectively. f, As for e, but normalized. g, Orientation selectivity increases for most combinations of amplification and midpoint. Dot shows the example shown in e and f (A = 1.6, M = 4). h, Bienenstock–Cooper–Munro plasticity rule. PV rates below the threshold cause LTD, whereas PV rates above the threshold cause LTP. Exaggerated LTD is modelled by increasing the threshold (magenta, 8 Hz; green, 12 Hz). i,j Increased LTD sharpens the pyramidal–PV connectivity (i) and increases PV selectivity (j). OSI with baseline and increased LTD: 0.58 and 0.69, respectively. k, Orientation selectivity increases with the threshold as long as this threshold is within the range of PV responses (between about 6 and 11 Hz).
First, we found that implementing increased PV neuron intrinsic excitability alone could not account for the observed increase in stimulus selectivity. In fact, it reduced selectivity by increasing the response of the neuron to all stimuli (Extended Data Fig. 16a–h). Notably, the inward-rectifying nature of the CP-AMPAR ion channel effectively dampened excitatory transmission during strong responses compared with weak responses across a wide range of conditions (Fig. 5d–g and Extended Data Fig. 16i–n). Last, when we incorporated exaggerated LTD, this preferentially weakened synapses from excitatory neurons that elicited weak responses in the PV neuron (Fig. 5i). Both the removal of inward rectification and the exaggerated LTD reduced responses to non-preferred stimuli relative to the preferred stimuli (Figs. 2e and 5e,j), which accounted for the increases in orientation selectivity. These modelling studies suggest that both acute rectification and cumulative plasticity triggered by resident CP-AMPARs in PV neuron dendrites may sufficiently account for their role in maintaining low selectivity (Extended Data Fig. 17).
Extended Data Fig. 16. Computational modeling: increased intrinsic excitability and CP-AMPAR inward rectification decreases selectivity.
a, PV-Cre;lsl-eGFP-GluA2 mice show intrinsic excitability increase in the form of a shift of the FI curve to lower input currents (Extended Data Fig. 10o). Dashed line: FI curve of PV-Cre;lsl-eGFP mice, shifted by E = 106.03 pA (fit to data). b, Model of increased intrinsic excitability: a shift of the PV cell’s FI-curve. c, Increased excitability causes a uniform increase in stimulus responses, decreasing stimulus selectivity (OSI: 0.44 vs OSI: 0.3). d, Orientation selectivity decreases for any increase in excitability. The decrease is even more pronounced in the presence of downscaled weights, mimicking a potentially decreased weight scale (black line; cf. Extended Data Fig. 10f). Magenta and green dots indicate examples shown in (b), (c). e, A range of increases in excitability, each compensated by a homeostatic synaptic scaling to preserve the mean rate (inset). f, Tuning curves for different levels of increased excitability. g, As (f), but normalized to the maximum response. h, Orientation selectivity monotonically decreases with increasing excitability, even for commensurately decreasing synaptic scaling. i-n, Alternative models of inward rectification. i, Average I-V relationship in calcium permeable (CP) and calcium-impermeable (CI) receptors, measured in excitatory neurons of wild-type and GluA2 knockout mice, respectively, derived from Lu et al.96 j, Conductance measured from k, estimated as current / voltage. k, Model of voltage or rate-dependent desensitization of inputs, parametrized by the relative portion α of CP versus CI conductances. The conductance in PV-Cre;lsl-eGFP-GluA2 neurons was modeled as CI-AMPARs only (green, CP-AMPAR coefficient λ = 0). The conductance in PV-Cre;lsl-eGFP neurons was modeled as a combination of CI and CP receptors (magenta, CP-AMPAR coefficient λ = 0.8). l, Increased responses to preferred stimuli in simulated PV-Cre;lsl-eGFP-GluA2 neurons. Orientation selectivity index (OSI): 0.49 (eGFP) and 0.55 (GluA2). m, As (d), but normalized by maximum response. n, Orientation selectivity monotonically decreases as the portion of CP-AMPARs λ increases. Dots indicate values used in panels k, l, m.
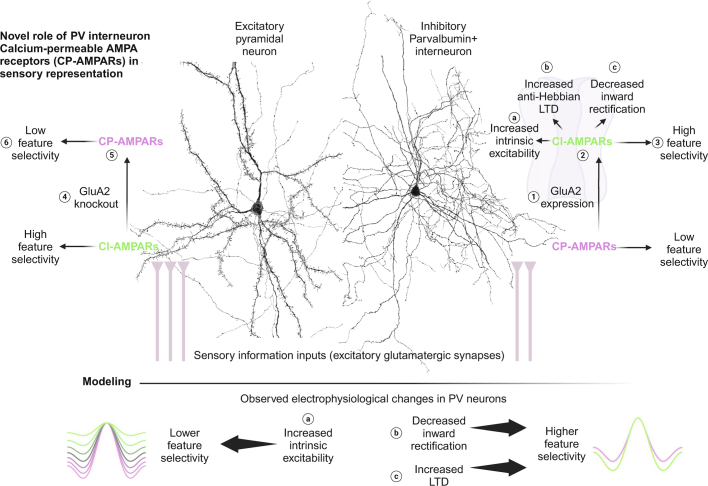
Extended Data Fig. 17. Summary of findings.
The abundant CP-AMPARs in PV interneurons were removed by ① targeted expression of GluA2, which ② replaces them with CI-AMPARs. This causes several electrophysiological changes (ⓐ-ⓒ) and ③ increases orientation selectivity in the visual cortex or spatial selectivity in the hippocampus. In excitatory forebrain neurons, which primarily have CI-AMPARs, ④ knocking out GluA2 makes all AMPARs ⑤ calcium permeable. This leads to ⑥ lower orientation selectivity. These results collectively demonstrate a strong role of CP-AMPARs in deciding the feature selectivity of a neuron. Computational modeling reveals that of the three cardinal electrophysiological changes we detect with CP-AMPAR removal (green traces), ⓐ increased intrinsic excitability is likely to decrease selectivity (opposite to our findings), whereas ⓑ decreased inward rectification and ⓒ increased anti-Hebbian LTD are likely to increase feature selectivity (consistent with our findings), providing a potential mechanism of the increased orientation selectivity and spatial selectivity after CP-AMPAR removal observed.
Discussion
Our results showed that CP-AMPARs are both necessary for low orientation selectivity in PV interneurons and sufficient to induce reduced selectivity in excitatory neurons, which typically have few CP-AMPARs. The function of postsynaptic AMPARs in synaptic transmission and defining synaptic strength is well understood9,10,24. Our new results suggest that their biophysical properties can control neuronal response tuning, thereby expanding their active role in computation. Whereas CP-AMPARs have been extensively studied in excitatory neuron synapses, our results attributed a new function to forebrain CP-AMPARs, which overwhelmingly reside in inhibitory neurons.
Our findings have broad implications for understanding inhibitory architecture. Inhibitory PV interneurons provide rapid feedback inhibition to local excitatory neurons. This lateral inhibition constrains the timing and extent of their firing while reducing informational redundancy5,12. The selectivity of PV neuron activity compared with excitatory neurons17–22 and the tuning bias of their outputs on local excitatory neurons31,34,35,42–44 have been under debate. Here we showed that the reduced selectivity of these PV neurons is biophysically implemented with a well-conserved molecular mechanism, including transcriptional Gria2 downregulation.
Whether other mammalian and non-mammalian organisms share such molecular or computational architecture is a question that needs investigation, and answers may reveal how molecular motifs that enable computation have evolved. It is notable that even PV-like GABAergic neurons in evolutionarily distant lizards, which lack Pvalb expression, also display low Gria2 expression45. Conversely, the importance of low CP-AMPAR expression in excitatory neurons has been highlighted by the discovery of human heterozygous de novo GRIA2 mutations through whole-genome sequencing efforts46. Mutations that lead to GRIA2 loss of function or remove the calcium-blocking pore residue of GRIA2 are often associated with intellectual disability and autistic behaviours, which suggests that the tight control of AMPAR calcium permeability is essential for human cognition.
In a given brain area, neurons display varying levels of selectivity to their preferred stimulus set. This selectivity can be stratified along the line of neuronal cell types12,16,47. This diversity indicates that gene expression can significantly affect the selectivity and sensory representation of a given neuron. Because synaptic inputs are summed in space and time to trigger neuronal activity, the selectivity of a neuron is dictated by the functional bias of the synaptic input pool31, the organization of the input synapses along the dendritic structure and the intrinsic excitability of each neuron.
Previous studies have suggested that PV neurons have low selectivity because they receive high-density excitatory input from cells with diverse tuning features and low overall functional bias20,33–36. Our paired recordings showed that gross input connectivity rates are intact in mice when PV neuron CP-AMPARs are removed, thereby demonstrating that PV neuron orientation selectivity can increase without significantly changing connection rates. However, these recordings did not address whether the functional bias of input connections or the clustering of such synapses throughout the dendritic tree is altered by the lack of CP-AMPARs. Thus, anti-Hebbian plasticity may have a role in the dendritic organization of functionally tuned synapses. The inward rectification of CP-AMPARs may limit strong functional summation from a particular dendrite or presynaptic partner, thereby further contributing to broader tuning. Meanwhile, our results showed that the intrinsic excitability of PV neurons is tightly coupled to the AMPAR profile, which implicates interleaved and coordinated mechanisms that define the computation of a given neuron. It is possible that blocking a key Ca2+ input source in dendrites by removing CP-AMPARs leads to a homeostatic response in PV neurons to upregulate excitability48. Intrinsic excitability is specifically adapted in PV neurons5, and how this tightly regulated feature of PV neurons affects its computations remains an essential question for future studies.
It is interesting to consider what these results tell us about biological and in silico intelligent circuits. Hebbian plasticity in neuronal networks is predicted to increase the correlation between neuronal activity and degrade total information content2. Anti-Hebbian plasticity is a possible mechanism to counteract this by reducing redundancy and keeping the representation more independent. However, researchers have traditionally thought that anti-Hebbian plasticity is implemented at the output GABAergic synapses of the inhibitory network2,49. By contrast, our work showed that CP-AMPARs at the input of the inhibitory network reduce GABAergic selectivity, thereby allowing PV neurons to broadly inhibit correlated activity through lateral inhibition. This characteristic adds to the flexibility of the network and may contribute to the canonical normalization computation that PV neurons are thought to carry out50. How the increased feature selectivity of PV neurons after CP-AMPAR removal affects local neuronal population coding and broader cognition are important outstanding questions.
We described a mechanism that commonly governs PV neuron selectivity across multiple modalities, from orientation and direction selectivity in the visual cortex to spatial selectivity in the hippocampus. By no means did we exhaustively assess the selectivity of these neurons in other domains, such as colour, ocular dominance and speed4,15,19,21,31. Selectivity to some visual features emerges before visual experience at eye-opening and can be driven by genetically determined circuit formation15. Future studies will show how experience-dependent synaptic regulation through CP-AMPARs and other synaptic molecules interacts with genetically determined mechanisms to fine-tune sensory representation.
Methods
Mice and marmosets
All procedures were approved by the Johns Hopkins Animal Care and Use Committee and conducted per the guidelines of the National Institutes of Health and the Society for Neuroscience. Hippocampal imaging experiments were carried out according to German national and institutional guidelines and approved by the ‘Tierversuchskommission’ of the Regierungspräsidium Freiburg (license number G16/037). Marmoset post-mortem tissue was obtained from terminal experiments approved by NIH Institutional Animal Care and Use Committees. The following mouse lines were used: PV-Cre30 (Jackson Laboratory (JAX), 008069), lsl-eGFP51 (JAX, 010701), lsl-eGFP-GluA2 (Extended Data Fig. 5), GluA2 KO39 (JAX, 002913), and GluA1 KO52 (JAX, 024422). We generated the ROSA26-lsl-eGFP-GluA2 mouse line by electroporating mouse embryonic stem (ES) cells with an engineered construct containing ROSA26-CAG-loxP-STOP-loxP-eGFP-Gria2-WPRE (adapted from targeting vector used to generate Ai14 mice53) and homologous recombination (Extended Data Fig. 5). We generated PV-Cre;lsl-eGFP-GluA2 (and PV-Cre;lsl-eGFP) mice from crosses with PV-Cre mice, born at Mendelian ratios. GluA2–/– pups displayed lower body weight compared with wild-type littermates. They displayed occasional mortality, mitigated by separating the littermates from the parents to reduce litter sizes39. All lines were maintained on a mixed background composed primarily of C57BL/6J, and mice of both sexes were used for experiments. We maintained all animals on a 12-h light–dark cycle at 20–26 °C and 30–70% relative humidity.
Constructs
We used Q/R and R/G RNA-edited flip-isoform short c-tail rat Gria2 cDNA sequences for mutant animal generation and viruses unless otherwise stated. SEP-GluA2 and GFP-GluA2 fusion constructs were generated by amino-terminal insertion of SEP or GFP at four amino acids after the signal peptide padded with linker sequences, as in previously published constructs54. We generated the FUW-Cre construct by replacing the eGFP in FUGW with the Cre recombinase gene.
pAAV.Syn.Flex.NES-jRGECO1a.WPRE.SV40 (ref. 55) was a gift from D. Kim and the GENIE Project (Addgene, plasmid 100853). The loxP/lox2272 sequences in the Flex cassette were inverted or exchanged with lox511/loxFAS to mitigate compatibility with other DIO AAVs. pAAV-CW3SL-eGFP56 was a gift from B.-K. Kaang (Addgene, plasmid 61463).
To deliver large genes, such as the SEP-GluA2 fusion gene, with the high tropism and low cytotoxicity provided by AAV vectors, we heavily optimized vector components to allow larger transgene size. Using the short hSyn1 promoter (469 bp), abbreviated linker sequences and DIO sequences and an optimized WPRE+polyA signal (CW3SL, 425 bp)56, we generated a pan-neuronal Cre-dependent AAV expression vector with a minimal backbone (1,350 bp from inverted terminal repeat (ITR) to ITR without cargo) and large cargo capacity size (about 3.65 kb; based on an earlier estimation of 5 kb AAV genome size limit57; 3.85 kb when Cre dependency is not required). The loxP/lox2272 sites were spaced by a minimal 64 bp (5′ end-to-5′ end) to set the second recombination event distance (128 bp) above 118 bp, at which inefficient recombination has been reported, but at an exact multiple of the helical repeat length (10.6 bp). This repeat length allowed better-aligned loxP sites after DNA looping, thereby maximizing the efficiency of Cre-mediated excision58.
As proof of principle, this study showed that SEP-GluA2 (3,378 bp), a large fusion protein previously only expressed through electroporation or lentiviral transfection, can be strongly expressed with this vector both in vitro and in vivo (Extended Data Fig. 7). The DIO-SEP-GluA2Q vector harboured Gria2 cDNA unedited at the Q/R editing site (R607Q)59. GluA2 Q/R RNA editing occurs at the pre-mRNA stage and requires a hairpin structure in the adjacent intron, which is absent in this vector. This structure bypasses RNA editing and expression of a calcium-permeable GluA2Q subunit. The DIO-eGFP control virus was similarly generated, replacing SEP-GluA2 with eGFP, for use as a control. These plasmids have been deposited to Addgene for distribution to the scientific community.
AAV was produced by HHMI-Janelia Viral Tools using a PEI triple transfection protocol into AAV293T cells (an ITR-containing plasmid, 2/9 capsid helper from UPenn Vector Core and the E1-deleted pHelper plasmid from Agilent). The cells were grown under serum-free conditions (three 150 mm culture dishes at about 3 × 107 cells per dish for each 100 µl batch), purified by two rounds of CsCl density gradient centrifugation and exchanged into storage buffer (1× PBS, 5% sorbitol and 350 mM NaCl). Virus titres (GC per ml) were determined by qPCR targeting the AAV ITRs.
Stereotaxic cranial surgeries
We used stereotaxic surgery to inject viruses and to implant 4 mm square cranial windows over the left V1. Mice of mixed sex (>6 weeks old) were given carprofen (5 mg kg–1) or buprenorphine (sustained release; 0.5–1.0 mg kg–1) and dexamethasone (4 mg kg–1) for analgesia and were anaesthetized using avertin or isoflurane (1.5–2.5%). We made a craniotomy with a number 11 scalpel blade centred at 2.5 mm lateral and 3.4 mm posterior to bregma.
For AAV injections, viruses were diluted with sterile PBS to 1–5 × 1013 GC per ml. We injected the solution at 5–10 sites spanning the posterior central area of the craniotomy (corresponding to the V1) with about 100 nl injections at each site at 250 μm below the dura surface. Injections were made using a bevelled glass pipette and a custom mineral oil-based injection system over 2–4 min. We left the pipette in place for another 2–3 min to allow diffusion and to prevent backflow.
We placed a 4 mm square glass coverslip over the craniotomy and attached a stainless-steel head bar to the skull during surgery to allow rigid head-fixation during imaging. We allowed mice to recover for 1–2 weeks before imaging and handled them extensively to alleviate experiment-related stress.
For hippocampal experiments, virus injections and cortical excavation or window implantation were done in separate surgeries. We made a small craniotomy over the hippocampus and injected 500 nl of AAV into the CA1 (anterior–posterior (AP): −2.0 mm; medial–lateral (ML) 2.0 mm; dorsal–ventral (DV): −1.4 mm). In the same surgical session, we implanted mice with a stainless-steel head plate (25 × 10 × 0.8 mm with an 8 mm central aperture) horizontally. We allowed mice to recover from surgery for at least 5 days before training sessions. We continued postoperative analgesic treatment with carprofen (5 mg kg–1 body weight) for 3 days after surgery.
Cortical excavation and hippocampal imaging window implantation were performed >10 days after the initial virus injection per published protocols41. We made a craniotomy (diameter 3 mm) centred at AP −1.5 mm and ML −1.5 mm. Parts of the somatosensory cortex and posterior parietal association cortex were gently aspirated while irrigating with chilled saline. We continued aspiration until the external capsule was exposed. We then gently peeled away the outer part of the external capsule using fine forceps, leaving the inner capsule and the hippocampus undamaged. The imaging window implant consisted of a 3 mm diameter coverslip (CS-3R, Warner Instruments) glued to the bottom of a stainless-steel cannula (3 mm diameter 1.2–1.5 mm height). The window was gradually lowered into the craniotomy using forceps until the glass was in contact with the external capsule. The implant was then affixed to the skull using cyanoacrylate. We allowed mice to recover from window implantation for 2–3 days.
Awake in vivo 2P fluorescence imaging
We performed retinotopic mapping60,61 to verify the location of the V1 using optimized protocols and software (https://github.com/ingiehong/retinotopy). We conducted awake in vivo 2P imaging with a custom-built, resonant/galvo 2P laser-scanning microscope (Sutter Instrument) controlled by ScanImage (Vidrio Technologies) and light-proofed to allow imaging in ambient light during visual stimulation. The designs for the head-fixed imaging platform and lightproofing apparatus are available online (https://github.com/ingiehong/StackGPS). We imaged neurons in the L2/3 of monocular V1 expressing eGFP or SEP and jRGECO1a using a ×20/1.0 NA water-immersion objective (Zeiss) and a Ti:Sapphire laser (Coherent Chameleon Ultra; Spectra-Physics Insight X3) tuned at 930 nm or 1,040 nm, respectively, with 20–100 mW of power delivered to the back-aperture of the objective.
We corrected the lateral motion of acquired image sequences using a rigid motion correction algorithm (NoRMCorre62). Neuronal somata with calcium transients were segmented using a constrained non-negative matrix factorization algorithm63. The source-separated GCaMP or jRGECO1a signal from each neuron was used to estimate various visual response properties of L2/3 neurons.
Visual stimulation
Visual stimuli were presented on a gamma-corrected 27″ LED monitor placed 22 cm in front of the centre of the eye contralateral to the hemisphere in which imaging was performed. The visual stimuli consisted of full-screen drifting gratings (4 s of duration, sinusoidal, 0.05 cycles per degree, 1 Hz, 100% contrast) following a 4-s iso-luminant grey screen. Six orientation gratings spaced at 30° were presented drifting in both directions orthogonal to the gratings (total of 12 directions) in a pseudo-randomized order to characterize sensory tuning using Psychtoolbox-3 (ref. 64) and FocusStack/Stimserver65. We used the average response during the 4 s of stimuli across 9–11 presentations to calculate visual responsiveness and orientation and direction selectivity. Visually responsive neurons were defined as cells with significant stimulus-related fluorescence changes (ANOVA across blank and 12 direction periods, P < 0.05)66.
The orientation and direction tuning curve was constructed by measuring the mean ΔF/F, averaged over the stimulus period for each grating drifting direction θ, denoted as R(θ). The OSI was calculated for visually responsive units21,66,67 with slight modifications on previous definitions67 to avoid values outside the intended interval ([0 1]) and to accommodate occasional bona fide negative responses68–70. The preferred drifting direction (θpref) of the cell was determined as the stimuli that induced the greatest responses, and , as a sum where , . The OSI was defined as follows:
where θorth+ = θpref+90°, θorth– = θpref–90°. All response values were subtracted by the most negative R(θ) when negative responses were present (Rcorrected), which effectively ensured that the relative dynamic range of responses were reflected in the index for which they would otherwise distort the index (leading to values outside [0 1]), or be clipped (when negative values were discarded). Formally,
Empirically, this modified index correlates tightly with the OSI calculated using the previous definition67 of orientation index and OSI, is bounded by [0 1] and accommodates tuning curves that are partially or entirely negative. Notably, the trends and results of statistical comparisons in this work did not change with the choice of index definition. The DSI, global OSI (gOSI) and global DSI (gDSI) were defined as follows:
gOSI and gDSI gave the same conclusions as OSI and DSI (data not shown). Note that can also be used in gOSI and gDSI, with the same benefits.
Head-fixed navigation and hippocampal imaging
Mice implanted with hippocampal imaging windows were subjected to a custom head-fixed virtual reality environment as previously described41. It consisted of a spherical treadmill monitored by an optical sensor that translated motion on the treadmill into forward motion through the virtual environment. We adjusted the forward gain so that 4 m of distance travelled along the circumference of the treadmill equalled one full traversal along a simulated linear track displayed on monitors surrounding the mouse. The track consisted of textured walls, floors and other 3D-rendered objects at the sides of the track as visual cues. To motivate consistent behaviour, we administered soy-milk rewards (4 µl) when the animal traversed certain locations that were spread at fixed distances along the track, and animals were trained for 5–10 days until they displayed consistent running behaviour before commencing imaging experiments.
Imaging was performed using a resonant/galvo high-speed laser scanning 2P microscope (Neurolabware) with a frame rate of 30 Hz for bidirectional scanning and a power of 5–20 mW measured at the objective front aperture. The microscope had an electrically tunable, fast z-focusing lens (Optotune, Edmund optics) to switch between z planes within less than a millisecond. Images were acquired through a ×16 objective (Nikon, 0.8 N.A., 3 mm WD). eGFP and jRGECO1a were excited at 930 nm or 1,040 nm, respectively, with a femtosecond-pulsed 2P laser (Mai Tai DeepSee, Spectra-Physics). We scanned 3 imaging planes (about 25 µm z spacing between planes) in rapid alternation so that each plane was sampled at 10 Hz. The planes spanned 300–500 µm in the x/y direction and were placed so that as many labelled neurons as possible were captured. We attached the animal’s head plate to the bottom of an opaque imaging chamber before each experiment to block ambient light from the photodetectors. We fixed the chamber in the behavioural apparatus with the animal. A ring of black foam rubber between the imaging chamber and the microscope objective blocked any remaining stray light.
Spatial tuning analysis
We motion-corrected all imaging data line-by-line71 with a 2D hidden Markov model using the software package SIMA71 or with block-wise non-rigid registration through the software package Suite2P72. If no suitable motion correction could be achieved, we discarded the data. To segment interneuron somata, regions of interest (ROIs) were manually drawn using ImageJ (NIH) or automatically drawn by applying Suite2P72. For automated ROI settings, the experimenter subsequently inspected individual ROIs. The average jRGECO1a signal over time was then obtained from each ROI for all runs. We restricted our analysis to mouse running periods with a minimum speed of 5 cm s−1. To obtain baseline-normalized ΔF/F calcium traces, we examined the fluorescence value distribution of the jRGECO1a signal and subtracted and divided the entire trace by the eighth percentile value of this distribution73. In rare instances, individual datapoints were below zero after baseline subtraction, and we set these negative values to zero for further calculations.
To compute spatial vector tuning, we plotted the mean activity (ΔF/F) of each spatial bin at its respective angle from the start position on the circular track into a polar coordinate system (Fig. 4e and Extended Data Fig. 15c). We then computed the circular mean of this distribution to obtain the mean tuning vector length and angle of the cell. Spatial coherence (Fig. 4f) was determined as the correlation (Pearson’s R) between the mean fluorescence value in each 5-cm bin on the track and its two nearest neighbours, measuring the local smoothness of the spatial tuning curve74. To calculate spatial information (SI; Extended Data Fig. 15e), we computed the average calcium activity (mean ΔF/F) for each 5-cm-wide bin along the linear track to approximate the average firing rate of neurons in that location. SI was then calculated for each cell as / λ, where λi and pi are the average calcium activity and fraction of time spent in the ith bin, respectively, λ is the overall calcium activity averaged over the entire linear track, and N is the number of bins on the track. Given the distribution of the underlying values, we plotted the log10 of SI values and compared them statistically (Extended Data Fig. 15e).
To assess the stability of the spatial representation of a cell within a session, we divided the track into 5-cm bins and calculated the mean ΔF/F value for each bin while the animal was moving on the track with a speed >5 cm s–1 to obtain activity maps for each individual cell. This mapping was done separately for the first and second half of the recording session. We then computed the within-session stability as the cross-correlation between the mean activity maps of the first and second half of the session (Extended Data Fig. 15b,f). We also computed population vector correlations as a function of position in the first and second half of the recording (Extended Data Fig. 15g) to visualize the local similarity of population activity across time. Before computing these correlations, we re-normalized the map of each neuron by subtracting the mean over space and dividing by the standard deviation (z scoring) to mitigate the potential effects of mean rate differences between cells on assessing local population vector similarity.
Quantification of Gria2 mRNA A-to-I editing rates
We mapped the raw sequencing reads from a mouse brain scRNA-seq dataset (n = 1,679)14 to the mouse reference genome (GRCm38) with a gene annotation, GENCODE (v.M16)75, using STAR76. The uniquely mapped reads whose sequencing qualities (Phred score) were greater than 20 were counted for the QR and RG RNA-editing sites in Gria2. We filtered out samples if the proportions of the sequencing read with A or G alleles together accounted for less than 95% to avoid potential sequencing errors. We defined the RNA-editing rate for a given site as a ratio of the number of sequencing reads showing G relative to the number of reads with either A or G.
FACS-assisted RNA-seq of PV interneurons
To assess transcriptional changes specifically in PV interneurons after removing CP-AMPARs with RNA-seq, we sorted dissociated cortical PV interneurons by their GFP fluorescence using FACS. Dissociation of adult mouse brain neurons leads to a rapid decimation of viable PV interneurons77–79, which potentially biases downstream analyses to a select subpopulation of PV interneurons. Various proposed methods to mitigate PV interneuron loss failed to recover them at native cell frequencies in adult mice80. Several fixation-based FACS approaches have been proposed to target immune cells and neurons, but crosslinking leads to lower RNA yield for RNA-seq.
We developed and used a brain-slice optimized workflow, FICSR-seq (Extended Data Fig. 11a), which recovers PV interneurons vulnerable to dissociation at native cell frequencies. We cut brain slices from adult mice (113.1 ± 11.6 days old) in NMDG cutting solution + trehalose77 and diced them into small pieces <1 mm3. Extracellular proteins were digested with pronase (2 mg ml–1; 8 U µl–1) at 34–37 °C, after which the slice pieces were fixed in 4% paraformaldehyde (PFA) in PBS (with 0.1 U ml–1 RNase inhibitor, Promega) for 15 min and dissociated into single cells through careful trituration. We filtered the single cells through a 40-μm filter, labelled them with the cell-permeable nuclear dye DRAQ5 (1:1,000 dilution) to identify nuclei-containing cells and then subjected them to FACS. DRAQ5+GFP+ or DRAQ5+GFP– cells were sorted, and more than 20,000 cells were collected per mouse cortex to provide extensive coverage of low-expressing PV interneuron transcripts.
We treated the fixed cells with proteinase K before RNA extraction (RecoverAll Total Nucleic Acid Isolation kit for FFPE, Thermo Fisher Scientific) to liberate RNA from protein–protein and protein–nucleic acid crosslinks generated by fixation. We prepared cDNA libraries from GFP+ and GFP– samples (NEBNext Ultra RNA Library Prep kit for Illumina, NEB) from RNA enriched with mRNA through bead-based polyA selection. cDNA libraries were barcoded and sequenced together on an Illumina Hiseq 2500 sequencer, generating 150-bp paired-end reads. We processed RNA-seq reads with bcbio-nextgen (v.1.2.3; 10.5281/zenodo.3564938)81, aligning to GRCm38 with the STAR aligner76 and quantifying counts per gene with Sailfish82 using the Ensembl annotation. We used DESeq2 (ref. 83) to analyse differential expression.
Brain slice preparation and whole-cell patch-clamp recordings
To test post-critical period electrophysiological properties and to maintain consistency within experiments, we used mice of either sex, aged postnatal day 32 (P32)–P62 for studies of synaptic properties and aged P69–P77 for studies of intrinsic properties. We first anaesthetized mice of either sex using isoflurane. We rapidly removed their brains in an ice-cold sucrose solution containing the following (in mM): 76 NaCl, 25 NaHCO3, 25 glucose, 75 sucrose, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2 and 7 MgSO4, pH 7.3, 315 mOsm. We hemisected the brain along the midline and mounted one or both hemispheres on a 30° ramp. We then sectioned acute parasagittal slices of the visual cortex, 300-μm thick, in the same ice-cold sucrose-cutting solution using a vibratome (VT-1200s, Leica). Slices were incubated in warm (32–35 °C) sucrose solution for 30 min and then transferred to warm (32–35 °C) artificial cerebrospinal fluid (aCSF) composed of the following (in mM): 125 NaCl, 26 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 1 MgSO4, 20 d-(+)-glucose, 2 CaCl2, 0.4 ascorbic acid, 2 pyruvic acid and 4 l-lactic acid, pH 7.3, 315 mOsm. Slices were then allowed to cool to room temperature. For rectification measurements, we cut coronal slices with a NMDG-based cutting solution and incubated them for >15 min. Then we transferred them to aCSF (see the section ‘Analysis of AMPAR rectification’). All solutions were continuously equilibrated with 95% O2 and 5% CO2.
We transferred slices to a submersion chamber on an upright microscope (Zeiss AxioExaminer; ×40 objective, 1.0 NA) and continuously superfused (2–4 ml min–1) them with warm (about 32–34 °C) oxygenated aCSF. We visualized neurons with a CCD camera (Sensicam QE, Cooke) using either infrared differential interference contrast (IR-DIC) microscopy or epifluorescence. The visual cortex was identified based on the relative position of the cortex and hippocampus and the anatomical borderline between the visual cortex and retrosplenial dysgranular cortex. We selected slices in which the apical dendrites of infragranular pyramidal neurons ran parallel to the plane of the slice up through L2/3 in the area targeted for recording. PV interneurons were targeted for recording based on eGFP or SEP-GluA2 expression along with unlabelled L2/3 pyramidal neurons. We filled patch pipettes (2–4 MΩ) pulled (P-97, Sutter Instrument) from borosilicate capillary glass (Sutter Instrument) with an internal solution containing (in mM): 2.7 KCl, 120 KMeSO3, 9 HEPES, 0.18 EGTA, 4 ATP magnesium salt, 0.3 GTP sodium salt and 20 phosphocreatine disodium salt, adjusted to pH 7.3, 295 mOsm. For recordings of PV interneurons, the internal solution included 0.25% w/v biocytin. Whole-cell patch-clamp recordings were obtained using Multiclamp 700B amplifiers (Molecular Devices) and digitized using an Instrutech ITC-18 (HEKA) and software written in Igor Pro (Wavemetrics). All signals were low-pass filtered at 10 kHz and sampled at 20–100 kHz. Neurons with an access resistance >30 MΩ or a resting membrane potential greater than −60 mV were not used for further recordings or analyses. The access resistance was not compensated in current clamp, and recordings were not corrected for the liquid junction potential.
Analysis of intrinsic excitability, synaptic connectivity and synaptic plasticity
We measured the resting membrane potential (RMP) shortly after establishing the whole-cell current-clamp recording configuration. A 1-s hyperpolarizing current (−100 pA) pulse was used to calculate the input resistance of recorded neurons. To assess the spiking behaviour of the cell, we injected 1-s depolarizing current steps into the recorded neurons. We measured the current–spike frequency relationship with a range of depolarizing current steps presented in pseudorandom order (1-s long, 40-pA increments, 5-s inter-stimulus intervals). Each current intensity was tested three times. For each current intensity, we counted the total number of action potentials exceeding an amplitude of 0 mV generated during each current step, then averaged the values across the three trials. We determined the rheobase by first probing the response of the neuron with 1-s-long depolarizing steps (5-s inter-stimulus intervals) to define a small range of current steps that bounded the rheobase. We then tested the neuron response within this range using 1-s-long depolarizing steps with 1-pA increments. We measured action potential properties from single spikes evoked by rheobase current injections. To compare the current–spike frequency relationship and rheobase between cells from the same baseline, we held cell membrane potentials at −70 mV when injecting depolarizing current steps. We performed all electrophysiological recordings that were assessing the intrinsic properties of PV interneurons in the presence of the following blockers of glutamate and GABA receptors (all from Tocris Bioscience): 5 µM NBQX (AMPA receptor antagonist); 5 µM (RS)-3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (NMDA receptor antagonist); and 10 µM 6-imino-3-(4-methoxyphenyl)-1(6H)-pyridazinebutanoic acid hydrobromide (SR95531; GABAA receptor antagonist).
To determine the properties of unitary synaptic connections among neurons, we generated two action potentials in the presynaptic neuron by injecting short, depolarizing current steps (3-ms pulse duration, 20 Hz, 10-s inter-trial interval). We held pyramidal neurons and PV interneurons at approximately −55 mV and −70 mV during synaptic connectivity tests to detect inhibitory postsynaptic potentials (IPSPs) and EPSPs, respectively. We assessed synaptic connectivity (EPSP or IPSP) with an average of 10–50 trials. A synaptic connection was detected if the first response amplitude of the average trace was >3 times the root mean squared of the average trace during baseline conditions and visually verified. We calculated the paired-pulse ratio by dividing the amplitude of the second postsynaptic potential by the first.
We subjected a subset of connected pyramidal→PV pairs, all of which exhibited an average EPSP amplitude of >0.3 mV at baseline, to an anti-Hebbian protocol. After recording 50 traces (6 Hz) as a baseline, we induced synaptic plasticity by pairing 400 presynaptic action potentials delivered at 5 Hz with continuous hyperpolarization of the postsynaptic PV interneuron to –90 mV25,84. After induction, EPSPs were recorded under the same conditions as the baseline measurement (50 traces in response to presynaptic action potentials, 6 Hz).
Analysis of AMPAR rectification
To measure AMPAR rectification85–88, we cut coronal brain slices in ice-cold cutting solution containing (in mM) 96 NMDG, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 d-(+)-glucose, 10 MgSO4, 0.5 CaCl2, 96 HCl, 20 HEPES, 12 N-acetylcysteine and 5 sodium l-ascorbate, and oxygenated with carbogen gas (95% O2 and 5% CO2). The 300-µm-thick slices were kept in aCSF (125 NaCl, 2.5 KCl, 2 MgCl2, 2 CaCl2, 1.0 NaH2PO4, 26.2 NaHCO3 and 11 glucose) and oxygenated with carbogen gas at 23–25 °C until they were transferred for recording to a submerged chamber superfused with aCSF (1–3 ml min–1) supplemented with about 50 µM picrotoxin and 100 μM APV (2-amino-5-phosphonovaleric acid) to isolate AMPAR-mediated excitatory synaptic transmission.
We made targeted whole-cell recordings of eGFP/SEP-GluA2-positive L2/3 PV interneurons using pipettes of 3–5 MΩ resistance. The intracellular solution contained (in mM): 115 CsMeSO4, 0.4 EGTA, 5.0 TEA-Cl, 1 QX314, 2.8 NaCl, 20 HEPES, 3.0 ATP magnesium salt, 0.5 GTP sodium salt, 10 phosphocreatine disodium salt and 0.1 spermine and was adjusted to pH 7.2, 285–290 mOsm. When we achieved whole-cell mode, we allowed >5 min for dialysis of the intracellular solution before collecting data. We held cells at −70 mV holding potential and recorded them at room temperature. We left the junction potential (about 11 mV) uncorrected. Signals were measured with a MultiClamp 700B amplifier, digitized using a Digidata 1440A digitizer (Molecular Devices) at 20 kHz and acquired with pClamp 10 software (Molecular Devices). We recorded AMPAR currents at 11 membrane potentials to construct a current–voltage (I–V) plot (Vh = −60 to +60 mV, except for a subset of pyramidal neurons recorded for comparison up to +50 mV). We calculated the rectification index as a weighted ratio of negative (−60 mV) and positive (+60 mV) currents. We compensated for the junction potential (11 mV): rectification index (RI) = (I–60 mV/–71)/(I+60 mV/49). An AMPAR rectification index of 1 represented perfect linearity, whereas values <1 indicate inward rectification. We estimated the reversal potential (Erev) by cubic polynomial regression that fitted the linear, rectifying and double-rectifying AMPAR I–V curves well.
Immunohistochemistry
We deeply anaesthetized mice with isoflurane then transcardially perfused them with PBS and 4% PFA. We removed and post-fixed the brain in 4% PFA–PBS for >2 h. We sectioned the brain coronally into 25 μm slices using a vibratome (VT-1000, Leica). We acquired marmoset brains post-mortem from terminal experiments and sliced them into 40 µm sections. Free-floating sections underwent antigen retrieval using LAB solution (Polysciences) when necessary and were blocked and permeabilized in 3% BSA with 0.3% Triton X-100 in PBS for 1 h at room temperature. We incubated sections with primary antibodies overnight at 4 °C, washed them with PBS 3 times for 5 min, and then incubated them with secondary antibodies for 2 h at room temperature. After another round of washes, we mounted the slices on glass slides in PermaFluor mounting medium (Thermo Fisher Scientific) and imaged them using a laser scanning confocal microscope (Zeiss LSM880). Controls were carefully carried out, including antibody staining of homozygous knockout mice (Extended Data Fig. 2) to ensure antibody specificity. For GluA1 and GluA2 quantification, ROIs were made around cell somas, and the background signal was subtracted to estimate protein levels.
The following primary antibodies were used: rabbit anti-parvalbumin (1:2,000, PV25, Swant); goat anti-parvalbumin (1:1,000, PVG-213, Swant); rat anti-somatostatin (1:200, MAB354, Chemicon); mouse anti-CaMKIIα (1:1,000, sc-32288, Santa Cruz); rabbit anti-GluA1 (1:1,000, JH4294, generated in-house); mouse anti-GluA2 (1:5,000; clone 15F1, gift from E. Gouaux); chicken anti-GFP (1:1,000, GFP-1020, Aves); and rabbit anti-dsRed2 (1:1,000, 632496, Clontech). The following secondary antibodies were used: Alexa Fluor 405 donkey anti-goat (1:1,000, ab175665, Abcam); Dylight 405 goat anti-mouse IgG2a (1:1,000, 115-477-186 Jackson ImmunoResearch); Alexa Fluor 488 goat anti-mouse IgG2a (1:1,000, A-21131, Thermo Fisher Scientific); Alexa Fluor 488 goat anti-chicken (1:1,000, A-11039, Thermo Fisher Scientific); Alexa Fluor 546 goat anti-rabbit (1:1,000, A-11035, Thermo Fisher Scientific); Alexa Fluor 568 goat anti-mouse IgG1 (1:1,000, A-21124, Thermo Fisher Scientific); Alexa Fluor 568 goat anti-rabbit (1:500, Thermo Fisher Scientific); Texas Red donkey anti-goat (1:1,000, SAB3700332, Millipore Sigma); Alexa Fluor 647 goat anti-rabbit (1:1,000, A-21245, Thermo Fisher Scientific); Alexa Fluor 647 goat anti-mouse IgG2a (1:1,000, A-21241, Thermo Fisher Scientific); Alexa Fluor 647 donkey anti-goat (1:1,000, A-21447, Thermo Fisher Scientific); and Alexa Fluor 647 goat anti-rat (1:500, A-21247, Thermo Fisher Scientific).
Computational modelling
The low feature selectivity of PV neurons17–21,89 (but see refs. 22,90–92) and the enhancement in PV-Cre;lsl-eGFP-GluA2 mice could result from several mechanisms. We used computational models to identify which mechanisms are consistent with the observed link between CP-AMPARs and feature selectivity. We examined the impact of three observed electrophysiological circuit changes: (1) increased intrinsic excitability (Extended Data Fig. 10o); (2) the loss of inward-rectifying AMPARs (Extended Data Fig. 7e,f); and (3) enhanced LTD (Extended Data Fig. 10l). Each mechanism was incorporated into a variation of a common base model. This model comprises a single PV neuron receiving excitatory inputs from a set of presynaptic pyramidal neurons with predefined stimulus tuning (Fig. 5a). The output of the PV neuron is a firing rate that is computed as a weighted sum of the inputs. Negative inputs are rectified to ensure a positive firing rate. To endow the PV neuron with stimulus tuning, pyramidal–PV connectivity was modelled as bell-shaped around the preferred orientation of the PV neuron (Fig. 5b), which enabled these neurons to inherit their tuning from pyramidal cells (Fig. 5c). We adjusted the parameters of pyramidal selectivity and connectivity to match the observed PV (and pyramidal) selectivity in the data.
Modelling increased intrinsic excitability
PV interneurons without CP-AMPARs showed increased intrinsic excitability (Extended Data Fig. 10o). PV neuron activation typically requires the coincident activation of multiple excitatory synaptic inputs93,94. However, the reduced rheobase, and increased RMP and input resistance in PV-Cre;lsl-eGFP-GluA2 mice suggested some strong synapses may reach the activation threshold unilaterally, which may increase selectivity95. To test whether this alone could account for the increased stimulus selectivity of PV interneurons, we increased the excitability of the PV model neuron by introducing a positive baseline current to the PV cell, mirroring the empirical shift of the frequency–current (F–I) curve (Extended Data Fig. 16a,b). We discovered that increased excitability reduced stimulus selectivity, contradicting the experimental observation. The response of the PV neuron was increased for all stimuli, thereby reducing the relative magnitude of the preferred response when compared with non-preferred responses (Extended Data Fig. 16c). This held for any rise in intrinsic excitability, regardless of a potential reduction in unitary EPSP amplitude (Extended Data Fig. 10f) when implemented as synaptic scaling (Extended Data Fig. 16d). We also simulated a scenario whereby enhanced intrinsic excitability was adjusted such that it homeostatically maintained the mean rate of the neuron by compensating for a multiplicative decrease in EPSPs (Extended Data Fig. 16e). In this scenario, stimulus selectivity was also reduced (Extended Data Fig. 16f–h). In conclusion, nonselective mechanisms such as increased intrinsic excitability and synaptic downscaling are insufficient to increase stimulus selectivity in the model.
Modelling removal of inward-rectifying AMPAR current
CP-AMPARs are inward-rectifying, which means that their conductance decreases with increasing postsynaptic potential (Extended Data Fig. 7e,f). This implies that they could become less effective for coincident stimuli that induce a strong postsynaptic response. To model this effect, we introduced a dependence of synaptic weights on the postsynaptic potential of the PV interneuron. In this model, we used conductance instead of current-based synapses to allow for a better comparison with experimentally measured current–voltage relationships. We modelled each synaptic weight as the sum of two components (Fig. 5d). The first represents CP-AMPARs and weakens with increasing postsynaptic potential. The second symbolizes other calcium-impermeable AMPARs unaffected by postsynaptic potential (Fig. 5d, dashed line), except due to changes in synaptic drive. We systematically varied the amount of CP-AMPARs relative to calcium-impermeable AMPARs and the membrane potential at which they inactivate (inactivation threshold). The intuition behind CP-AMPARs influencing stimulus selectivity is that they should remain open for weak (that is, non-preferred) stimuli but deactivate for strong (that is, preferred) stimuli. PV neurons fire at high frequencies, which makes this more relevant, and compartmentalized dendritic depolarizations could further exacerbate this effect. This would selectively enhance the response to non-preferred stimuli, thus reducing stimulus selectivity. Conversely, eliminating CP-AMPARs would enhance stimulus selectivity. Indeed, we observed that removing the CP-AMPAR component reduced the response to non-preferred stimuli without affecting preferred stimuli, thereby increasing stimulus selectivity (Fig. 5e,f and compare with Extended Data Fig. 9c and Fig. 2e). This effect was robust to variations in the relative abundance of the CP-AMPARs and their inactivation threshold (Fig. 5g).
A qualitatively similar outcome emerged from applying a previously measured empirical I–V curve from Gria2–/– mice96 to estimate inward rectification (Extended Data Figs. 16 and 7e,f). Systematically varying the proportion of CP-AMPARs in the PV neuron model revealed that orientation selectivity monotonically decreases as the proportion of CP-AMPARs increases (Extended Data Fig. 16f). Two previous papers have examined the potential impact of CP-AMPARs on postsynaptic activation from slightly different perspectives of EPSC kinetics and dendritic summation sublinearity93,97, and both arrived at conclusions similar to ours. In conclusion, increased stimulus selectivity may be due to the removal of CP-AMPAR-mediated inward rectification.
Modelling increased LTD
Pyramidal–PV connections exhibited exaggerated LTD in PV-Cre;lsl-eGFP-GluA2 mice compared with control mice (Extended Data Fig. 10l). This could enhance selectivity by weakening synaptic inputs from pyramidal cells tuned to non-preferred stimuli. We modelled this scenario by introducing synaptic plasticity in the pyramidal–PV synapses. Synaptic weights changed according to a Bienenstock–Cooper–Munro (BCM) rule, which has been broadly studied as a model for the development of stimulus selectivity98. The BCM learning rule is an associative rule that changes synapses when the presynaptic (pyramidal) neuron and the postsynaptic (PV) neuron are simultaneously active. However, the direction of the change is determined by the postsynaptic firing rate. When PV activity is below a threshold, synaptic efficacy decreases. If PV activity surpasses the threshold, synaptic efficacy increases (Fig. 5h). Here we used a fixed instead of the typical activity-dependent threshold in the classical BCM model. This allowed us to test the effect of increased LTD by varying the threshold. Specifically, we increased the LTP–LTD threshold to model the exaggerated LTD in PV-Cre;lsl-eGFP-GluA2 mice (Fig. 5h and Extended Data Fig. 10l). This weakened synapses from pyramidal cells activated for stimuli that elicit only a weak response in the PV cell (Fig. 5i). The exaggerated LTD consequently reduced the PV response to non-preferred stimuli (Fig. 5j) while enhancing its response to preferred stimuli. The resulting boost in selectivity was observable across a wide range of LTD–LTP thresholds as long as the threshold was within the range of PV responses (Fig. 5k). We conclude that increased selectivity could arise from changes in synaptic plasticity if this plasticity, in a BCM-like manner, can generate both potentiation and depression, and if depression is exaggerated after the removal of CP-AMPARs.
Conclusions of modelling studies
These modelling studies demonstrate that the inward-rectifying nature of the CP-AMPAR ion channel and the exaggerated LTD observed in PV-Cre;lsl-eGFP-GluA2 mice can both effectively reduce responses to non-preferred stimuli, thereby accounting for the increases in orientation selectivity. However, neither the rise in intrinsic excitability nor a potential general reduction in excitatory input in PV interneurons due to GluA2 expression can explain the increase in orientation selectivity. These modelling findings imply that acute rectification and cumulative plasticity triggered by resident CP-AMPARs may sufficiently account for their role in maintaining low selectivity. Determining the extent of contribution of these two mechanisms to sensory selectivity in vivo poses a challenging question, which will necessitate rigorous empirical investigation in the future.
Network modelling architecture
The model was a feed-forward rate network of n presynaptic pyramidal neurons and a single postsynaptic PV neuron. We first describe the base model and then its elaborations. The presynaptic pyramidal neurons were tuned to stimulus direction and orientation according to a mixture of von Mises distributions. Specifically, the response of the ith pyramidal cell to a moving grating with direction θ was given by the following:
The proportionality sign indicates a normalization between a minimum of 0 and a maximum of 1 across stimuli. Here θi is the preferred direction of the cell, κ determines its tuning width and α controls the strength of direction tuning (κ = 2 and α = 0.5). The preferred directions of the pyramidal cells were equally spaced in the interval [0,2π). The tuning of the PV cell was determined by the pyramidal tuning and the pyramidal-to-PV connectivity. Without loss of generality, we defined the preferred orientation of the PV cell to be 0°. The connectivity from the ith pyramidal cell onto the PV cell was given by a single von Mises distribution:
Weights were normalized across presynaptic cells, such that the minimum and maximum weights were equal to 0 and 1, respectively. The connectivity and pyramidal response together defined the PV voltage and rate using the following equations:
Here, τ = 10 ms denotes the membrane time constant. To simulate the PV activity from these equations, we used forward Euler discretization with a time step Δt = 1 ms. We simulated a time T = 100 ms unless specified otherwise and confirmed that the system had reached its steady state. This steady-state activity was used to compute tuning curves.
Intrinsic excitability
We fitted the change in the empirical I–F curve by numerically finding the shift that minimized the squared difference between the PV-Cre;lsl-eGFP-GluA2 and the PV-Cre;lsl-eGFP mean values. This was done using the minimize_scalar method of SciPy99 with the shift as the optimization parameter. In the model, we increased the intrinsic excitability by adding an untuned positive baseline input I0:
We varied I0 between 0 and 10. Note that firing rates, membrane potential, weights and currents are unitless in our model. This does not alter the results, because orientation tuning is assessed based on relative rates. Decreases in unitary EPSPs were modelled by downscaling the synaptic weights with a factor p:
We downscaled the weights in two different ways. In Extended Data Fig. 16d, we used p = 0.62, reflecting the mean empirical reduction in EPSPs (Extended Data Fig. 10f). To investigate the effect of homeostatic increases in excitability, we used the minimize_scalar function to find the scaling that would keep the average PV rate constant given a specific increase in its excitability I0.
Inward rectification
We modelled the inward-rectifying calcium currents by adding a voltage-dependent weight scaling p(u) to the PV dynamics. We also introduced conductance-based synapses to allow for a better comparison with experimental data:
Here, u0 = 30 is the reversal potential. In our simulations, the precise value of u0 and the choice for conductance versus current-based synapses scale the postsynaptic responses without strongly affecting relative stimulus tuning in different conditions. The scale p smoothly increases for decreasing voltages:
This is a decreasing sigmoid function between 1 and A, with a slope β and a midpoint M. The midpoint M describes the threshold potential at which the CP-AMPARs deactivate, and β how sensitive the inactivation is to the membrane potential. A quantifies the abundance of rectifying AMPARs relative to the number of non-rectifying AMPARs. We varied A between 0 and 3 and M between 0 and 5; we fixed β to 0.5. The removal of CP-AMPARs was modelled by fixing p to 1. We increased the width of the presynaptic tuning to κ = 3.6 to achieve approximately equal selectivity in the presence of rectification.
In addition to this idealized model of inward rectification, we also simulated a data-driven model. Our starting point were previously measured current–voltage relationships96 (Extended Data Fig. 16i). These data were collected in excitatory cells of wild-type and Gria2–/– mice, which allowed for a direct comparison of calcium permeable (CP) and calcium-impermeable (CI) receptors. Specifically, we used these published measurements96 to estimate the normalized conductance at each voltage as the ratio I/V (Extended Data Fig. 16j). We did this for both wild-type and GluA2 traces, and normalized each between 0 and 1. This resulted in scaling factors and that represent the strength of CP and CI receptors, respectively, in our model (Extended Data Fig. 16k). Their convex sum determined the total synaptic rectification:
We found that orientation selectivity slowly but monotonically decreased with increasing λ (Extended data Fig. 16l–n). In the data-driven model, neurons with a larger relative abundance of CP receptors therefore have a weaker orientation selectivity, consistent with the idealized model and with our experimental findings.
Plasticity
We modelled synaptic plasticity using a plasticity rule inspired by BCM theory98. According to BCM, the change in synaptic efficacy is given by:
Here η = 0.02 is a small learning rate that controls the speed of learning but does not affect the outcome. rpre and rpost are the presynaptic and postsynaptic rates, respectively, and θBCM is the threshold between LTD and LTP. In most applications of the BCM rule, this threshold is adaptive and depends on the recent PV activity. Here we fixed it to a single value per experiment to allow full control over the amount of LTD. Specifically, LTD was implemented by increasing the threshold from 8 to 10 Hz. We further varied the threshold between 6.5 and 11 Hz. As the empirical response distribution seems to be largely unaffected by CP-AMPAR removal, we added synaptic scaling100 to keep the mean postsynaptic rate constant:
Here r* is the target mean rate, which we fixed to the mean rate across stimuli before the onset of plasticity. The mean rate was computed after every weight update by averaging across all stimuli. In the plasticity experiments, we first simulated T = 100 time steps without plasticity to allow the system to reach a steady state. At subsequent time steps, we computed Δw for each individual stimulus, and used the average Δw across stimuli to update the weights. This continued until the weights and rates converged to a new steady state.
Statistical analysis and reproducibility
We performed statistical tests in Matlab (Mathworks), Prism (GraphPad) or R. Data distributions were tested for normality using Shapiro–Wilk test. We used parametric tests if the data were normally distributed and nonparametric otherwise, as detailed in the text describing each comparison. For parametric tests, we used unpaired or paired t-tests and one-way or two-way ANOVA tests with Tukey’s post hoc multiple comparison correction (all two-sided). For data that did not follow normal or log-normal distributions, we used the following statistical tests where appropriate: Mann–Whitney U-test (Wilcoxon rank-sum test), Kruskal–Wallis one-way ANOVA with Dunn’s post hoc multiple comparison correction (all two-sided). For categorical data, we used Fisher’s test or χ2 with or without Yates correction according to degrees of freedom and sample size. We report centre and spread values as the mean ± s.e.m. or median ± interquartile range unless otherwise stated. We did not use statistical methods to plan sample sizes, but used sample sizes similar to those frequently used in the field. The text or figure legends include the number of animals and cells. We did not use randomization, and data collection and analyses were not performed blind to the conditions of the experiments unless otherwise stated. P values < 0.05 were considered to be significant. When we used a statistical test, the P value is noted either in the manuscript text or depicted in figures and legends as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS, not significant, P ≥ 0.05. Representative examples such as traces and micrographs were chosen from at least three or more independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08027-2.
Supplementary information
Acknowledgements
We would like to thank A. Bygrave, R. Roth, E. Lopez-Ortega, H. Tan, A. Graves, A. Irving, S. Richardson, M. L. Pucak, M. Ayad, S. Das, J. Kufera and J. Moran for technical assistance and scientific discussions; L. Hamm for administrative support; S. SheikhBahaei for assistance in processing marmoset tissue; T.-w. Chen, H. Dana and S. Van Hooser for assistance in acquiring and analysing visual responses; D. B. Arnold, Y. Li and S. Hestrin for sharing tools; C. Bone for scientific illustrations; A. N. Connor for editing the manuscript; and Janelia AAV core for packaging services. The MICrONS Consortium, Neuroglancer and BioRender were used for parts of the summary figure. This work was supported by R37NS036715 (R.L.H.), R01NS085121 (S.B.), KBRI Basic Research Program funded by the Ministry of Science and ICT (24-BR-02-02 to J. Kim), CRC-TRR 384/1 2024-514483642 (M.B. and H.S.), and U01DA056556 (I.H. and R.L.H.).
Extended data figures and tables
Author contributions
Generation of 2P imaging data: I.H., T. Hainmueller and T.C. Generation of electrophysiology data: I.H. and J. Kim. Generation of RNA-seq data: I.H., D.W.K., R.C.J., N.L., T. Hwang and F.M.K. Generation and validation of mice: I.H., Z.Y., D.C. and A.A. Generation of marmoset data: I.H., S.H.P. and D.A.L. Computational modelling: J. Keijser, H.S. and I.H. Data analyses: I.H., J. Kim, T. Hainmueller, D.W.K., R.C.J., S.H.P., N.L., Z.Y., D.C., T. Hwang, A.A., T.C., F.M.K., S.A.M., X.D., D.A.L., S.B., D.E.B., M.B., S.P.B. and R.L.H. Data interpretation: I.H., J. Kim, T. Hainmueller, D.W.K., S.H.P., S.B., D.E.B., M.B., S.P.B. and R.L.H. Writing the manuscript: I.H., J. Kim, T. Hainmueller, J. Keijser, H.S., D.A.L., D.E.B., M.B., S.P.B. and R.L.H.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
The data supporting the findings in this work are presented in the main figures and source data. Sequencing data included in this manuscript can be accessed at NCBI’s Gene Expression Omnibus (GEO) under accession GSE262872. Other data of this study are available from the corresponding authors upon reasonable request. We used the following previously published datasets: adult mouse cortical cell taxonomy by single-cell transcriptomics14 (GEO accession GSE71585); neuronal subtypes and diversity revealed by scRNA-seq the human brain27 (dbGaP study accession phs000833.v3.p1); and innovations in primate interneuron repertoire28 (GEO accession GSE151761).
Code availability
Computer codes used to acquire data and analyse results of the study are available at GitHub (https://github.com/ingiehong/retinotopy, https://github.com/ingiehong/StackGPS and https://github.com/JoramKeijser/pv_selectivity) and from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ingie Hong, Juhyun Kim, Thomas Hainmueller
Contributor Information
Ingie Hong, Email: ingiehong@jhmi.edu.
Richard L. Huganir, Email: rhuganir@jhmi.edu
Extended data
is available for this paper at 10.1038/s41586-024-08027-2.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08027-2.
References
- 1.Olshausen, B. A. & Field, D. J. Sparse coding of sensory inputs. Curr. Opin. Neurobiol.14, 481–487 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Destexhe, A. & Marder, E. Plasticity in single neuron and circuit computations. Nature431, 789–795 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Moser, M. B., Rowland, D. C. & Moser, E. I. Place cells, grid cells, and memory. Cold Spring Harb. Perspect. Biol.7, a021808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niell, C. M. Cell types, circuits, and receptive fields in the mouse visual cortex. Annu. Rev. Neurosci.38, 413–431 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Hu, H., Gan, J. & Jonas, P. Interneurons. Fast-spiking, parvalbumin+ GABAergic interneurons: from cellular design to microcircuit function. Science345, 1255263 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Huang, Z. J. & Paul, A. The diversity of GABAergic neurons and neural communication elements. Nat. Rev. Neurosci.20, 563–572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matta, J. A. et al. Developmental origin dictates interneuron AMPA and NMDA receptor subunit composition and plasticity. Nat. Neurosci.16, 1032–1041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen, C. C. & Crochet, S. Synaptic computation and sensory processing in neocortical layer 2/3. Neuron78, 28–48 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Huganir, R. L. & Nicoll, R. A. AMPARs and synaptic plasticity: the last 25 years. Neuron80, 704–717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malinow, R. & Malenka, R. C. AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci.25, 103–126 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Zeng, H. & Sanes, J. R. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci.18, 530–546 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Fishell, G. & Kepecs, A. Interneuron types as attractors and controllers. Annu. Rev. Neurosci.43, 1–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul, A. et al. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell171, 522–539.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasic, B. et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci.19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seabrook, T. A., Burbridge, T. J., Crair, M. C. & Huberman, A. D. Architecture, function, and assembly of the mouse visual system. Annu. Rev. Neurosci.40, 499–538 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Isaacson, J. S. & Scanziani, M. How inhibition shapes cortical activity. Neuron72, 231–243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohya, K., Kameyama, K., Yanagawa, Y., Obata, K. & Tsumoto, T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J. Neurosci.27, 2145–2149 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowak, L. G., Sanchez-Vives, M. V. & McCormick, D. A. Lack of orientation and direction selectivity in a subgroup of fast-spiking inhibitory interneurons: cellular and synaptic mechanisms and comparison with other electrophysiological cell types. Cereb. Cortex18, 1058–1078 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, B. H. et al. Visual receptive field structure of cortical inhibitory neurons revealed by two-photon imaging guided recording. J. Neurosci.29, 10520–10532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerlin, A. M., Andermann, M. L., Berezovskii, V. K. & Reid, R. C. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron67, 858–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niell, C. M. & Stryker, M. P. Highly selective receptive fields in mouse visual cortex. J. Neurosci.28, 7520–7536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Runyan, C. A. et al. Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron67, 847–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger, J. R. et al. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron15, 193–204 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Traynelis, S. F. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev.62, 405–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamsa, K. P., Heeroma, J. H., Somogyi, P., Rusakov, D. A. & Kullmann, D. M. Anti-Hebbian long-term potentiation in the hippocampal feedback inhibitory circuit. Science315, 1262–1266 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, J. G., Albuquerque, C., Lee, C. J. & MacDermott, A. B. Synaptic strengthening through activation of Ca2+-permeable AMPA receptors. Nature381, 793–796 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Lake, B. B. et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science352, 1586–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krienen, F. M. et al. Innovations present in the primate interneuron repertoire. Nature586, 262–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khawaja, R. R. et al. GluA2 overexpression in oligodendrocyte progenitors promotes postinjury oligodendrocyte regeneration. Cell Rep.35, 109147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hippenmeyer, S. et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol.3, e159 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholl, B., Thomas, C. I., Ryan, M. A., Kamasawa, N. & Fitzpatrick, D. Cortical response selectivity derives from strength in numbers of synapses. Nature590, 111–114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi, L. F., Harris, K. D. & Carandini, M. Spatial connectivity matches direction selectivity in visual cortex. Nature588, 648–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholl, B., Pattadkal, J. J., Dilly, G. A., Priebe, N. J. & Zemelman, B. V. Local integration accounts for weak selectivity of mouse neocortical parvalbumin interneurons. Neuron87, 424–436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofer, S. B. et al. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat. Neurosci.14, 1045–1052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bock, D. D. et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature471, 177–182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Packer, A. M. & Yuste, R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J. Neurosci.31, 13260–13271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, J. T., Li, C. Y., Zhao, J. P., Poo, M. M. & Zhang, X. H. Spike-timing-dependent plasticity of neocortical excitatory synapses on inhibitory interneurons depends on target cell type. J. Neurosci.27, 9711–9720 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Duigou, C., Savary, E., Kullmann, D. M. & Miles, R. Induction of anti-Hebbian LTP in CA1 stratum oriens interneurons: interactions between group I metabotropic glutamate receptors and M1 muscarinic receptors. J. Neurosci.35, 13542–13554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia, Z. et al. Enhanced LTP in mice deficient in the AMPA receptor GluR2. Neuron17, 945–956 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Meng, Y., Zhang, Y. & Jia, Z. Synaptic transmission and plasticity in the absence of AMPA glutamate receptor GluR2 and GluR3. Neuron39, 163–176 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Hainmueller, T. & Bartos, M. Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature558, 292–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Znamenskiy, P. et al. Functional specificity of recurrent inhibition in visual cortex. Neuron112, 991–1000.e8 (2024). [DOI] [PubMed] [Google Scholar]
- 43.Karnani, M. M., Agetsuma, M. & Yuste, R. A blanket of inhibition: functional inferences from dense inhibitory connectivity. Curr. Opin. Neurobiol.26, 96–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura, Y. & Callaway, E. M. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat. Neurosci.8, 1552–1559 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Tosches, M. A. et al. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science360, 881–888 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Salpietro, V. et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat. Commun.10, 3094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bugeon, S. et al. A transcriptomic axis predicts state modulation of cortical interneurons. Nature607, 330–338 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Debanne, D., Inglebert, Y. & Russier, M. Plasticity of intrinsic neuronal excitability. Curr. Opin. Neurobiol.54, 73–82 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Pehlevan, C., Hu, T. & Chklovskii, D. B. A Hebbian/anti-Hebbian neural network for linear subspace learning: a derivation from multidimensional scaling of streaming data. Neural Comput.27, 1461–1495 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Carandini, M. & Heeger, D. J. Normalization as a canonical neural computation. Nat. Rev. Neurosci.13, 51–62 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sousa, V. H., Miyoshi, G., Hjerling-Leffler, J., Karayannis, T. & Fishell, G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb. Cortex19, i1–i10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zamanillo, D. et al. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science284, 1805–1811 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci.13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekine-Aizawa, Y. & Huganir, R. L. Imaging of receptor trafficking by using α-bungarotoxin-binding-site-tagged receptors. Proc. Natl Acad. Sci. USA101, 17114–17119 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dana, H. et al. Sensitive red protein calcium indicators for imaging neural activity. eLife5, e12727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi, J. H. et al. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain7, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong, J. Y., Fan, P. D. & Frizzell, R. A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther.7, 2101–2112 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Hoess, R., Wierzbicki, A. & Abremski, K. Formation of small circular DNA molecules via an in vitro site-specific recombination system. Gene40, 325–329 (1985). [DOI] [PubMed] [Google Scholar]
- 59.Araki, Y., Lin, D. T. & Huganir, R. L. Plasma membrane insertion of the AMPA receptor GluA2 subunit is regulated by NSF binding and Q/R editing of the ion pore. Proc. Natl Acad. Sci. USA107, 11080–11085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wekselblatt, J. B., Flister, E. D., Piscopo, D. M. & Niell, C. M. Large-scale imaging of cortical dynamics during sensory perception and behavior. J. Neurophysiol.115, 2852–2866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Juavinett, A. L., Nauhaus, I., Garrett, M. E., Zhuang, J. & Callaway, E. M. Automated identification of mouse visual areas with intrinsic signal imaging. Nat. Protoc.12, 32–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods291, 83–94 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Giovannucci, A. et al. CaImAn an open source tool for scalable calcium imaging data analysis. eLife8, e38173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleiner, M. et al. What’s new in psychtoolbox-3. Perception36, 1–16 (2007). [Google Scholar]
- 65.Muir, D. R. & Kampa, B. M. FocusStack and StimServer: a new open source MATLAB toolchain for visual stimulation and analysis of two-photon calcium neuronal imaging data. Front. Neuroinform.8, 85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazurek, M., Kager, M. & Van Hooser, S. D. Robust quantification of orientation selectivity and direction selectivity. Front. Neural Circuits8, 92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Valois, R. L., Yund, E. W. & Hepler, N. The orientation and direction selectivity of cells in macaque visual cortex. Vision Res.22, 531–544 (1982). [DOI] [PubMed] [Google Scholar]
- 69.Li, Y., Van Hooser, S. D., Mazurek, M., White, L. E. & Fitzpatrick, D. Experience with moving visual stimuli drives the early development of cortical direction selectivity. Nature456, 952–956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sclar, G. & Freeman, R. D. Orientation selectivity in the cat’s striate cortex is invariant with stimulus contrast. Exp. Brain Res.46, 457–461 (1982). [DOI] [PubMed] [Google Scholar]
- 71.Kaifosh, P., Zaremba, J. D., Danielson, N. B. & Losonczy, A. SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform.8, 80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pachitariu, M. et al. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. Preprint at BioRxiv10.1101/061507 (2016).
- 73.Sheffield, M. E. J., Adoff, M. D. & Dombeck, D. A. Increased prevalence of calcium transients across the dendritic arbor during place field formation. Neuron96, 490–504.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kubie, J. L., Muller, R. U. & Bostock, E. Spatial firing properties of hippocampal theta cells. J. Neurosci.10, 1110–1123 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res.47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joseph, D. J. et al. Protocol for isolating young adult parvalbumin interneurons from the mouse brain for extraction of high-quality RNA. STAR Protoc.2, 100714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Saxena, A. et al. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques52, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chapman, B. et al. bcbio/bcbio-nextgen v.1.2.9. Zenodo10.5281/zenodo.3564938 (2021).
- 82.Patro, R., Mount, S. M. & Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol.32, 462–464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le Roux, N., Cabezas, C., Bohm, U. L. & Poncer, J. C. Input-specific learning rules at excitatory synapses onto hippocampal parvalbumin-expressing interneurons. J. Physiol.591, 1809–1822 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hong, I. et al. AMPA receptor exchange underlies transient memory destabilization on retrieval. Proc. Natl Acad. Sci. USA110, 8218–8223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clem, R. L. & Huganir, R. L. Calcium-permeable AMPA receptor dynamics mediate fear memory erasure. Science330, 1108–1112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sommer, B., Kohler, M., Sprengel, R. & Seeburg, P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell67, 11–19 (1991). [DOI] [PubMed] [Google Scholar]
- 88.Isaac, J. T., Ashby, M. C. & McBain, C. J. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron54, 859–871 (2007). [DOI] [PubMed] [Google Scholar]
- 89.Li, L. Y. et al. Differential receptive field properties of parvalbumin and somatostatin inhibitory neurons in mouse auditory cortex. Cereb. Cortex25, 1782–1791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Runyan, C. A. & Sur, M. Response selectivity is correlated to dendritic structure in parvalbumin-expressing inhibitory neurons in visual cortex. J. Neurosci.33, 11724–11733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Najafi, F. et al. Excitatory and inhibitory subnetworks are equally selective during decision-making and emerge simultaneously during learning. Neuron105, 165–179.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moore, A. K. & Wehr, M. Parvalbumin-expressing inhibitory interneurons in auditory cortex are well-tuned for frequency. J. Neurosci.33, 13713–13723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geiger, J. R., Lubke, J., Roth, A., Frotscher, M. & Jonas, P. Submillisecond AMPA receptor-mediated signaling at a principal neuron–interneuron synapse. Neuron18, 1009–1023 (1997). [DOI] [PubMed] [Google Scholar]
- 94.Galarreta, M. & Hestrin, S. Spike transmission and synchrony detection in networks of GABAergic interneurons. Science292, 2295–2299 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Goetz, L., Roth, A. & Hausser, M. Active dendrites enable strong but sparse inputs to determine orientation selectivity. Proc. Natl Acad. Sci. USA118, e2017339118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu, W. et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron62, 254–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cornford, J. Computational Aspects of Parvalbumin-Positive Interneuron Function. PhD thesis, Univ. College London (2017).
- 98.Bienenstock, E. L., Cooper, L. N. & Munro, P. W. Theory for the development of neuron selectivity: orientation specificity and binocular interaction in visual cortex. J. Neurosci.2, 32–48 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turrigiano, G. G. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell135, 422–435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rodriguez, C. I. et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet.25, 139–140 (2000). [DOI] [PubMed] [Google Scholar]
- 102.Hollmann, M., Hartley, M. & Heinemann, S. Ca2+ permeability of KA-AMPA-gated glutamate receptor channels depends on subunit composition. Science252, 851–853 (1991). [DOI] [PubMed] [Google Scholar]
- 103.Verdoorn, T. A., Burnashev, N., Monyer, H., Seeburg, P. H. & Sakmann, B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science252, 1715–1718 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings in this work are presented in the main figures and source data. Sequencing data included in this manuscript can be accessed at NCBI’s Gene Expression Omnibus (GEO) under accession GSE262872. Other data of this study are available from the corresponding authors upon reasonable request. We used the following previously published datasets: adult mouse cortical cell taxonomy by single-cell transcriptomics14 (GEO accession GSE71585); neuronal subtypes and diversity revealed by scRNA-seq the human brain27 (dbGaP study accession phs000833.v3.p1); and innovations in primate interneuron repertoire28 (GEO accession GSE151761).
Computer codes used to acquire data and analyse results of the study are available at GitHub (https://github.com/ingiehong/retinotopy, https://github.com/ingiehong/StackGPS and https://github.com/JoramKeijser/pv_selectivity) and from the corresponding author upon request.