Abstract
Three species of cecidomyiid midges (Diptera: Cecidomyiidae) cause significant yield losses on wheat in Europe: Sitodiplosis mosellana (Géhin), Contarinia tritici (Kirby) and Haplodiplosis marginata (von Roser). Eggs and young larvae may be parasitised by a complex of hymenopteran parasitoids belonging to the Pteromalidae and Platygastridae families which contributes to natural pest control. We have developed molecular tools for detecting and identifying seven parasitoid species previously encountered in Belgium inside individual wheat midge larvae. Barcode DNA sequences from COI, 18S and 28S genes were obtained from the midges and parasitoid species. Each of the three genes allowed all the species to be distinguished although 18S was the only one displaying a barcoding gap, both between parasitoids and midges, and at the species level. Based on the 18S gene, we developed a TaqMan assay to assess parasitism in midge larvae, regardless of the midge and parasitoid species. Next, two group-specific PCR primer pairs were generated, allowing the separate amplification of midge DNA or parasitoid DNA in parasitised individuals and subsequent identification by Sanger sequencing. Finally, species-specific primers were designed to identify six parasitoid species by simple PCR amplification. These tools were successfully applied to assess the parasitism rate of S. mosellana larvae in seven Belgian fields.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79398-9.
Keywords: DNA barcoding, TaqMan, Wheat midge parasitoids, Cecidomyiidae, Pterolamidae, Platygastridae
Subject terms: Entomology, Ecosystem ecology, Molecular ecology
Introduction
The orange wheat blossom midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae), is an important pest that damages wheat kernels in the northern hemisphere with consequences on the yield and quality of the harvest1–3. Several outbreaks have occurred in Europe since the early 2000s4–6 but also in North America where it was introduced in the 1800s7. Alongside this species, other wheat pest midges also attack florets of emerging wheat heads such as the yellow wheat blossom midge, Contarinia tritici (Kirby) or the stem such as the saddle gall midge, Haplodiplosis marginata (von Roser). Notably, a resurgence of this latter species has been observed since 2010 across various European countries8,9 with significant yield losses of up to 13% in Belgium10. All these species overwinter in the soil as larvae and adults emerge in the spring. Mated females lay their eggs within wheat ears for S. mosellana and C. tritici while eggs are deposited on the leaves for H. marginata. The eggs hatch a few days after being laid. The larvae of S. mosellana and C. tritici feed on developing kernels and flower parts, respectively. The young larvae of H. marginata move under the leaf sheath to feed on the stem, where they induce the formation of saddle-shaped galls. After rainfall between mid-June and mid-July, the larvae drop to the ground, bury themselves and initiate diapause.
Eggs and young larvae may be parasitised by a complex of around 27 hymenopteran parasitoid species11, contributing to natural biological control. Inside parasitised midge larvae, the parasitoid wasps accomplish their development the following spring after diapause by killing their host at the L3 stage. In Belgium, a total of ten hymenopteran species belonging to two families and six genera have been reported in midge species. Among them, eight hymenopteran species were observed as parasitoids of S. mosellana12: Macroglenes penetrans (Kirby, 1800) belonging to the Pteromalidae family accounted for 23 to 100% of the occurrences of the parasitoid complex depending on the sampled field. The Platygastridae family (0 to 77% of the occurrences of the parasitoid complex) was mainly represented by three species: Euxestonotus error (Fitch, 1865), Platygaster tuberosula Kieffer, 1926 and an undescribed species Euxestonotus sp. The four other Platygastridae species were rare: Platygaster gracilipes Huggert, 1975, Platygaster nisus Walker, 1836, Amblyaspis tritici (Walker, 1836), and another undescribed species called Leptacis sp. In contrast, only one species of parasitoid was reported for H. marginata: Platygaster equestris Spittler, 1969, and another for C. tritici: Synopeas myles (Walker, 1836)12.
Traditionally, the parasitism assessment was made by rearing host larvae until parasitoids emerged. This method is time-consuming and carries a risk of failure due to host and/or parasitoid mortality during larvae rearing. Moreover, accurate morphological identification of parasitoids requires expert skills, sometimes needing specimens to be sent to researchers with appropriate taxonomic expertise12,13. To overcome the limitations of conventional techniques (host rearing and dissection), molecular tools can facilitate studies on host-parasitoid associations14. This could help us to detect parasitism and accurately identify the parasitoid species attacking the wheat midges. In addition, these tools could prove a precious aid for investigating the population dynamics of parasitoids and the parasitism rate in different landscape contexts. In light of these elements, biological control of wheat midges could be enhanced to reduce chemical spraying and limit yield losses at harvest.
The aim of this study was to devise molecular tools for assessing the parasitism of three wheat midges: S. mosellana, C. tritici and H. marginata. As DNA sequence data were scarce or absent for most of the parasitoid species, we firstly generated DNA barcode sequences from mitochondrial COI, and the 18S and 28S nuclear ribosomal RNA genes. Next, we evaluated the suitability of the different barcodes for the development of molecular markers to detect and identify parasitoids in their respective hosts. Finally, we developed different molecular tools enabling: (1) the detection of parasitism in the hosts whatever the parasitoid species: TaqMan assay as well as group-specific PCR assay, (2) the identification of the parasitoid species: species-specific PCR assay and a multiplex PCR assay for the three most common parasitoids of S. mosellana.
Material and methods
Biological material
Samples for DNA barcoding
Specimens of the three wheat midge species and of the most frequent parasitoid species in Belgium12 were used for DNA barcoding. All metadata are available in the BOLD (Barcode of Life Data System) project “Midge parasitoids (PARCE)” (Dataset: DS-MMDIWMP). The geographical locations and dates of collection can be found in Supplementary Table S1.
Wheat midges
Sitodiplosis mosellana, H. marginata and C. tritici larvae were collected in 2015 from fields in Wallonia (Belgium) by water spraying the wheat ears to mimic a rainfall. They were identified with morphological keys for Cecidomyiidae15,16.
Parasitoids
Adult Macroglenes penetrans specimens were collected with an insect net during swarm flights in wheat fields in 2015 in Wallonia. Adult specimens of Euxestonotus error, Platygaster tuberosula, P. gracilipes, P. nisus, P. equestris and Synopeas myles were harvested during a study on soil samples collected in Belgium12 and determined by Dr Peter Neerup Buhl (IT University of Copenhagen, Copenhagen, Denmark). DNA could unfortunately not be retrieved from the rare specimens belonging to the three other species reported in Belgium (Euxestonotus sp., Amblyaspis tritici and Leptacis sp.) because a few specimens only were captured and submitted for morphological identification. After loans and manipulations by taxonomists, DNA content was too low or too degraded. In total, they represented only 3.1% of the specimens harvested. M. penetrans was identified using the key for Pteromalidae by Graham17 and the description given by Johansson18. Platygaster spp. and Synopeas myles were identified with the key for Platygaster by Buhl19 and the specific descriptions of each species: P. tuberosula and S. myles with Kieffer20 and Johansson18, P. gracilipes with Huggert21, P. nisus with Vlug22 and P. equestris with Spittler23. Euxestonotus error was identified using the key for Platygastridae by Kozlov24 and the description given by Gahan25.
The individuals were stored in 70% denatured ethyl alcohol (with 3% diethyl ether) (ethanol, CAS 64-17-5) at room temperature until DNA extraction and subsequent deposition at the Royal Belgian Institute of Natural Sciences (RBINS).
M. penetrans and Platygaster ssp bulk DNA extraction
To obtain enough DNA to ensure reliable quantification and repeated adjustment tests without wasting DNA from reference specimens intended for conservation, bulk DNA was extracted from pools of 10 Macroglenes penetrans adults and 10 Platygaster spp. adults. The Platygaster spp. specimens came from the S. mosellana rearing at CRA-W and were not identified to the species level.
Field parasitism monitoring
In 2015, the larvae of S. mosellana were gathered in seven wheat fields in the Walloon region, Belgium. In each field sampled, one hundred wheat heads were collected before the larvae dropped to the soil. Wheat heads were put on a grid above a water tray. Next, they were sprayed with water to mimic rainfall and collect larvae. The larvae were morphologically identified and stored in pure isopropyl alcohol (100%) (Propan-2-ol, CAS 67-63-0, Fisher Chemical, P/7500/21) at − 20 °C.
DNA extraction
Total genomic DNA was extracted from each individual sample (adults, larvae) using the NucleoSpin Tissue kit (MACHEREY-NAGEL). The specimens were crushed in the lysis buffer, except for the reference specimens designated for preservation, which were soaked intact in the solution. The lysis step was performed for 3 h at 56 °C with shaking at 450 rpm. DNA was eluted in 35μL elution buffer and stored at − 20 °C.
PCR amplifications and sequencing of COI, 18S and 28S barcodes
Preliminary tests on M. penetrans and Platygaster spp. bulk individuals failed to amplify the mitochondrial cytochrome c oxidase subunit I gene (COI) fragments with the classically used primer pairs LCO1490 + HCO219826 or C_LepFolF + C_LepFolR27. Therefore, a shorter COI fragment was sequenced (463 to 472 bp) using C1-J-1718F and C1-N-2191 primers28 which succeeded in producing amplification on all seven species analysed in this study. PCR were carried out in a total volume of 25 µl consisting of 1 µl genomic DNA, 1× Taq buffer (Platinum Taq DNA Polymerase, Invitrogen), 4 mM MgCl2, 0.2 mM of each dNTP, 0.2 µM of each primer, 0.5 mg/ml BSA (Bovine Serum Albumine) and 0.5 U Taq polymerase. The initial denaturation step at 94 °C for 2 min was followed by 35 cycles of 30 s at 94 °C, 30 s at 47 °C and 45 s at 72 °C and a final extension at 72 °C for 10 min. Part of the 18S ribosomal RNA region (18S rDNA) and 28S ribosomal RNA region (28S rDNA) were amplified with primers 18S-441 F and 18S-1299 R29 as well as with primers 28S-F and 28S-R30 respectively. PCR mixtures contained 1 µl genomic DNA, 1 × Taq buffer (GoTaq G2 DNA Polymerase, Promega), 1 mM MgCl2, 0.2 mM of each dNTP, 0.2 µM of each primer and 0.15 U Taq polymerase. Cycling conditions were as follows: initial denaturation at 94 °C for 3 min followed by 35 cycles of 1 min at 94 °C, 1 min at 50 °C (18S) or 55 °C (28S), 1 min at 72 °C and a final extension at 72 °C for 10 min. Amplicons were Sanger sequenced in both directions at Eurofins Genomics (Germany). The sequences were assembled using Vector NTI suite 6.0, Informax Inc., Bethesda, MD, USA and deposited in BOLD project (PARCE project) and GeneBank (accession no. PP213884 to PP213977 (COI); PP213978 to PP214056 (28S) and PP214062 to PP214130 (18S)).
Distance analyses
All publicly available COI, 18S and 28S sequences for the parasitoid and midge species were downloaded from GenBank and BOLD databases (accessed on 8 August 2023) and aligned with the sequences obtained in the present study using ClustalW31 in Vector NTI (suite 6.0).
Within species distances (WSD) and between species distances (BSD) were calculated using the Kimura 2-parameter model32 in MEGA X33 with the pairwise deletion option for gaps and missing data treatment.
For each barcode and for each species pair, maximum WSD was plotted against minimum BSD to assess the suitability of the different barcodes to distinguish the studied species. To assess the suitability for group-specific detection, two groups were formed: (i) the three midge species and (ii) the seven parasitoid species and maximum within group distance (WGD) was plotted against minimum between group distance (BGD). Neighbour-Joining trees34 were constructed based on the Kimura 2-parameter distances using MEGA X with 1000 bootstraps. All ambiguous positions, gaps and missing data were removed for the calculation of the distances between each sequence pair (pairwise deletion option).
Primers and probe design
Primers and a TaqMan probe were designed based on the alignment of the 18S consensus sequences from the 10 species studied (Table 1). Primer-BLAST was used for the primer design35. The primers and the TaqMan probe were synthesised at Eurogentec (Belgium).
Table 1.
Information about the primers and probe used in this study for DNA barcoding and to specifically detect parasitoids in wheat midges.
| Application | Gene | Specificity | Name | Sequence | Thybr | Size (pb) | References |
|---|---|---|---|---|---|---|---|
| DNA barcoding | COI | – | C1-J-1718F | GGAGGATTTGGAAATTGATTAGTTCC | 47 °C | 463–472 | Simon et al.28 |
| – | C1-N-2191 | CCCGGTAAAATTAAAATATAAACTTC | Simon et al.28 | ||||
| 18S | – | 18S-441 F | AAATTACCCACTCCCGGCA | 50 °C | 774–828 | Heraty et al.29 | |
| – | 18S-1299 R | TGGTGAGGTTTCCCGTGT T | Heraty et al.29 | ||||
| 28S | – | 28S-F | AGAGAGAGTTCAAGAGTACGTG | 55 °C | 367–489 | Dowton and Austin30 | |
| – | 28S-R | TTGGTCCGTGTTTCAAGACGGG | Dowton and Austin30 | ||||
| Taqman detection of parasitism | 18S | Parasitoid-group | Taqman probe/ParTaq536 | FAM- CTTGGATCGTCGCAAG-MGB | 60 °C | 145 | This study |
| Parasitoid-group | Par500F | CCGAG(G/A)TAATGATTAATAGGGACAGA | |||||
| - | Par643R | CGAACCTCTAACTTTCGTTCTTGA | |||||
| Group specific detection | 18S | Parasitoid-group | Par164F | AAGCTCGTAGTTGAATCTGTG | 60 °C | 302–358 | This study |
| Parasitoid-group | Par473R | CCCCCATCTGTCCCTATTA | |||||
| Midge-group | Cec164F | ACGTTCGTAGTTGAACTTGTG | 60 °C | 326 | This study | ||
| Midge-group | Cec473R | CCCCCAATTGCCTCCATTA | |||||
| Species specific detection | 18S | Parasitoid-group | Par164F | AAGCTCGTAGTTGAATCTGTG | This study | ||
| M. penetrans | macro382R | CAGTATTCAGGCGAACATAG | 60 °C | 214 | |||
| E. error | euxe382R | GATTTTTCAGGCTTTTGTTAGG | 60 °C | 246 | |||
| P. tuberosula | tuber295R | TAAAGCTCCCAACGAGACGA | 60 °C | 123 | |||
| P. equestris | Equation 382R | GAATTTTCAGGCTTTTCAATTG | 55 °C | 239 | |||
| P. gracilipes | graci382R | GAATTTTCAGGCGTATATTTTG | 55 °C | 233 | |||
| S. myles | sinop382R | GATAATTCAGGCTTGTTGTAGG | 55 °C | 270 |
Thybr: optimal hybridisation temperature.
A group-specific TaqMan assay to detect parasitoids in their hosts was developed by designing a pair of primers (Par500F + Par643R) amplifying a 145 bp fragment and a TaqMan probe (ParTaq536). The probe and the forward primer were chosen so that they hybridise in regions differentiating the parasitoid group from the wheat midge group but remain conserved within the groups.
Two pairs of primers enabling a group-specific PCR amplification of either the parasitoids or the midges were designed (Par164F + Par473R and Cec164F + Cec473R respectively). These primers were chosen in regions that were conserved within each group, and allowed, depending on the species, the amplification of a 302 to 358 bp sequence covering a hypervariable region. This aims to achieve species verification by Sanger sequencing.
Finally, species-specific reverse primers were designed to allow the specific detection of M. penetrans (macro382R), E. error (euxe382R), P. equestris (equ382R), P. gracilipes (graci382R), S. myles (sinop382R) and P. tuberosula (tuber295R) in parasitised hosts. Although the 18S sequence for P.nisus looks suitable for designing a species specific reverse primer, this has not been done due to the lack of enough DNA for testing.
TaqMan assays and PCR amplifications
Amplifications with TaqMan probes were carried out in a total volume of 20 µl using 10 μl of Takyon No ROX Probe 2X Mastermix Blue dTTP (Eurogentec, Belgium), 300 nM of each primer, 200 nM of the TaqMan probe labelled with the FAM fluorophore and a 3’ Eclipse quencher (EQ) attached to a minor groove binder (MGB) molecule (Eurofins Genomics, Germany) and 1 µl of genomic DNA. Serial dilutions of M. penetrans genomic DNA were used as positive controls and water was used as a negative control. PCR amplifications were performed with the C1000 Touch Thermal Cycler coupled to the CFX96 Touch Real-Time Detection System (Bio-Rad). The following thermal cycling protocol was used: initial denaturation at 95 °C for 10 min followed by 40 cycles at 95 °C for 10 s and 60 °C for 45 s.
Group-specific and species-specific PCR amplifications were performed in a total reaction volume of 25 μL containing 1 µl of genomic DNA, 0.2 mM of each dNTP, 1 mM MgCl2, 0.2 μM of each primer and 0.03 U Taq DNA polymerase (GoTaq G2 Hot Start Taq Polymerase, Promega). The initial denaturation step at 94 °C for 3 min was followed by 35 cycles of 1 min at 94 °C, 1 min at optimal hybridisation T° (as reported in Table 1) and 1 min at 72 °C with a final elongation of 10 min at 72 °C.
A parasitoid species-specific multiplex PCR assay was developed to target the three most common species in S. mosellana using 0.2 µM of primer Par164F and 0.07 µM of each of the following primers macro382R, euxe382R and tuber295R and 1 µl of genomic DNA. The Phusion U Multiplex PCR Master Mix (Thermo Scientific) was used with the following cycling conditions: initial denaturation at 98 °C for 30 s followed by 35 cycles at 98 °C for 10 s, 60 °C for 30 s and 72 °C for 15 s and a final elongation at 72 °C for 5 min. Amplification products were visualised by electrophoresis on a 1.5% agarose gel.
Assessment of the sensitivity and specificity
The sensitivity of the TaqMan method was evaluated on bulk DNA extracted from pools of 10 Macroglenes penetrans adults or 10 Platygaster spp. adults. DNA was quantified using a Qubit 4 fluorometer (Thermo Fisher Scientific). TaqMan amplifications were carried out on five serial dilutions (from 1 ng to 100 fg DNA). The dilutions and TaqMan amplifications were repeated three times. The specificity of the TaqMan method (parasitoids vs. midges) was assessed by amplification on DNA extracted from the three midge species (C. tritici, S. mosellana, H. marginata). Two larvae were tested for each species with three replicate amplifications for each DNA extract.
The specificity of group-specific and species-specific PCR primers was tested on DNA from the 10 studied species (7 parasitoids and 3 midges).
Field parasitism monitoring
Genomic DNA was extracted as described above from 132 larvae of S. mosellana. The presence of DNA and the absence of potential inhibitors that could prevent PCR amplification were checked by amplification of a COI fragment using C1-J-1718F and C1-N-2191 primers. The presence of parasitism was tested using the TaqMan assay on each of these larvae; water was used as negative control. Individuals were considered as positive according to the “call” of the Bio-Rad CFX Manager Software (version 3.1). The parasitoid group-specific PCR (Par164F + Par473R) amplification was tested on all the positive individuals. The parasitoid species were identified by species-specific multiplex PCR and/or Sanger sequencing and confirmed by the corresponding species-specific primer pairs.
Results
Species delimitation using COI, 18S and 28S
GenBank and BOLD databases were searched for sequences of the seven parasitoids and three midge species considered in this study. On August 8 2023, 521 COI sequences were available to download, mainly from S. mosellana and S. myles (Table 2). COI sequences were absent for three of the parasitoid species: P. gracilipes, P. nisus and P. equestris. Only one 28S sequence was available (S. mosellana) and no 18S sequence was available for the ten species considered here. The DNA fragments targeted here will therefore complement the databases. In total, 94, 69 and 79 sequences were generated in this study for COI, 18S and 28S respectively (Table 2).
Table 2.
COI, 28S and 18S barcoding of wheat midges and their parasitoid species: studied species, number of sequences obtained from specimens collected in this study, length of the sequences, number of sequences available to download from GenBank and BOLD, average within species distances (WSD) calculated using the Kimura 2-parameter model.
| Order | Family | Genus | Species | COI sequences | 28S sequences | 18S sequences | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| This study | Size (bp) | BOLD/NCBI | WSD (mean) | This study | size (bp) | BOLD/NCBI | WSD (mean) | This study | Size (bp) | BOLD/NCBI | WSD (mean) | ||||
| Hymenoptera | Pteromalidae | Macroglenes | penetrans | 25 | 466 | 4 | 0.000 | 23 | 483 | 0 | 0.000 | 18 | 774 | 0 | 0.000 |
| Hymenoptera | Platygastridae | Euxestonotus | error | 20 | 463 | 6 | 0.013 | 9 | 483 | 0 | 0.000 | 11 | 804 | 0 | 0.000 |
| Hymenoptera | Platygastridae | Platygaster | tuberosula | 12 | 463 | 6 | 0.001 | 8 | 462–470 | 0 | 0.001 | 8 | 827 | 0 | 0.000 |
| Hymenoptera | Platygastridae | Platygaster | gracilipes | 4 | 463 | 0 | 0.004 | 4 | 443 | 0 | 0.000 | 5 | 794 | 0 | 0.000 |
| Hymenoptera | Platygastridae | Platygaster | nisus | 3 | 463 | 0 | 0.007 | 4 | 471–475 | 0 | 0.004 | 4 | 817 | 0 | 0.001 |
| Hymenoptera | Platygastridae | Platygaster | equestris | 13 | 463 | 0 | 0.001 | 17 | 451 | 0 | 0.001 | 10 | 798 | 0 | 0.002 |
| Hymenoptera | Platygastridae | Synopeas | myles | 8 | 463 | 106 | 0.003 | 8 | 489 | 0 | 0.000 | 5 | 828 | 0 | 0.000 |
| Diptera | Cecidomyiinae | Sitodiplosis | mosellana | 3 | 472 | 396 | 0.005 | 3 | 367 | 1 | 0.004 | 3 | 801 | 0 | 0.000 |
| Diptera | Cecidomyiidae | Haplodiplosis | marginata | 3 | 472 | 1 | 0.006 | 2 | 370 | 0 | 0.000 | 2 | 801 | 0 | 0.001 |
| Diptera | Cecidomyiidae | Contarinia | tritici | 3 | 472 | 2 | 0.006 | 1 | 368 | 0 | n/c | 3 | 803 | 0 | 0.002 |
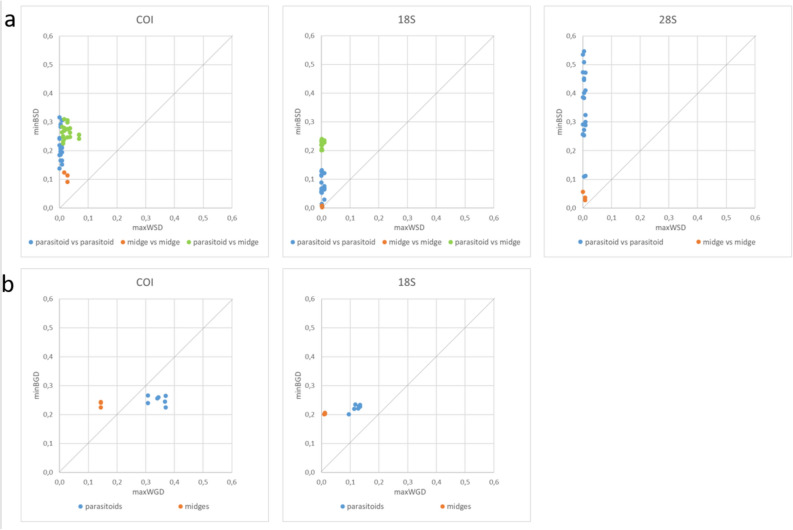
Each of the three barcodes enabled the 10 species to be distinguished. A barcoding gap was found between all species for COI and 18S, meaning that the minimum distance between species was greater than the maximum distance within species whatever the pair of species considered (Fig. 1a). The COI and 18S sequences of the different species formed distinct clusters in the Neighbour-Joining trees (Supplementary Fig. S1).
Fig. 1.
‘Barcode gap’ for COI, 18S and 28S. (a) between species pairwise distances: minimum between species distances (BSD) were plotted against maximum within species distances (WSD), (b) between groups (parasitoids vs. midges): for each species, the minimum between group distance (BGD) was plotted against the maximum within group distance (WGD). Due to high variability in 28S, BGD could not be calculated for this marker.
For the 18S barcode, the minimum Kimura P2 distances separating the parasitoid from the midge species (from 0.201 to 0.241 according to the species pair; see green dots in Fig. 1a) are clearly higher than the minimum distances between parasitoid species (0.014 to 0.132) or between midge species (0.003 to 0.008). For COI, this was not the case (minimum distances from 0.225 to 0.310 between parasitoids and midges versus 0.138 to 0.317 between parasitoids and 0.091 to 0.124 between midges).
The 28S barcode showed a low within species variability (mean WSD of 0.000 to 0.004, Table 2) but very high variations between species sequences and sequence lengths (from 367 to 489 bp). Consequently, aligning the 28S sequences and calculating distances between parasitoid and midge species was not possible, even when using longer reference sequences to map it.
Our first objective was to develop a DNA-based parasitoid group-specific tool to detect the presence of parasitoids in their hosts, regardless of the species. If we consider two groups, the first one including the seven parasitoid species and the second one containing the three midge species, Fig. 1b indicates that a barcoding gap was found between groups for all species for the 18S barcode. In contrast, no barcoding gap was revealed for the parasitoid species using the COI barcode: the maximum distance between M. penetrans sequences and all other parasitoid species (from 0.308 to 0.370) exceeded the minimum distances between H. marginata and those same species (0.240 to 0.266). The Neighbour-Joining trees showed all the parasitoid sequences clustering apart from the midge species for 18S while Macroglenes sequences clustered midway between the two groups for the COI barcode (Supplementary Fig. S1).
Consequently, the 18S barcode seems better suited for developing DNA tools to distinguish between parasitoid and midge groups while presenting sufficient variability for species level identification.
Group specific TaqMan assay
A group-specific TaqMan assay was designed to detect the presence of parasitoids in their hosts, regardless of the species of parasitoids and wheat midges. The TaqMan probe hybridises in a conserved region for the seven parasitoid species targeted (Fig. 2).
Fig. 2.
Location of the primers and probe developed in this study on the alignment of part of the 18S barcode sequences. In green: primers and probe of the parasitoid-specific TaqMan assay. In blue: location of the group-specific primers allowing amplification of only the parasitoid DNA (Par) in the host or only the host DNA (Cec). In orange: location of the species-specific primers; tuber295R: P. tuberosula, spec382R: M. penetrans (macro382R), E. error (euxe382R), P. equestris (Eq. 382R), P. gracilipes (graci382R) and S. myles (sinop382R). Figure created with Geneious version 2022.1 (https://www.geneious.com).
In order to set the fluorescence threshold line, we performed specificity tests on midges of the three species: the average fluorescence emission at 40 cycles was 498 RFU (2 individuals per species, 3 replicates) with a maximum of 1300 RFU for one H. marginata individual. However, in an assay based on serial dilutions (Supplementary Fig. S2), the RFU was higher (ca. 2000 RFU) for some midges. Therefore, the fluorescence threshold line was set at 2500 RFU.
The TaqMan assay was evaluated on serial dilutions of genomic DNA from Macroglenes penetrans (10 pooled adults) and Platygaster spp. (10 pooled adults). Standard curves were established based on the corresponding qPCRs (see Table 3) and the R2 scores were > 0.999 for both genera. Limits of detection (LOD) were evaluated at 1 pg genomic DNA for M. penetrans and at 100 fg for Platygaster spp.
Table 3.
TaqMan assay for the detection of wheat midges parasitoids: metrics inferred from standard curves (linear regression) and limit of detection (LOD) for Macroglenes penetrans and Platygaster spp. genomic DNA.
| Species | Equation | R2 | CqLOD | LOD |
|---|---|---|---|---|
| Macroglenes penetrans | − 3.72x + 47.99 | 0.999 | 36.81 ± 0.44 | 1 pg |
| Platygaster spp. | − 3.70x + 45.29 | 0.999 | 37.82 ± 0.31 | 100 fg |
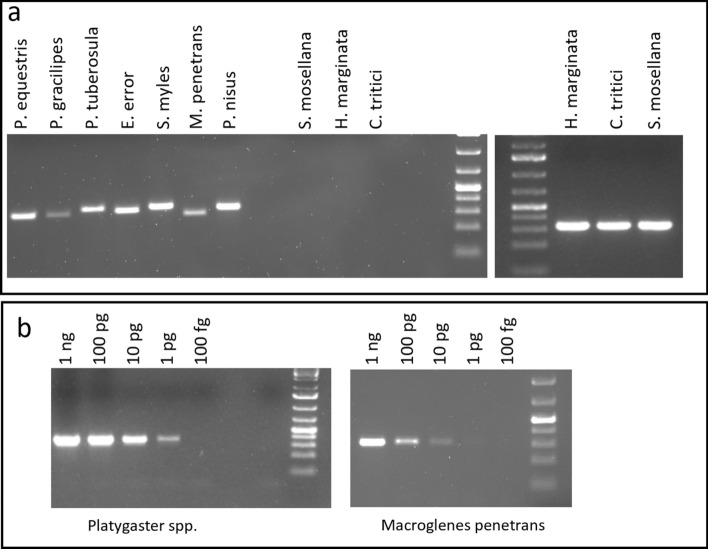
Group-specific and species-specific PCR
A group-specific PCR method was developed to amplify a hypervariable part of the 18S sequence in M. penetrans, P. nisus, P. gracilipes, P. tuberosula, S. myles, E. error and P. equestris but not in their hosts (S. mosellana, C. tritici and H. marginata). Primers Par164F + Par473R allowed the amplification of a single fragment of 302 to 358 bp according to the parasitoid species while no PCR product was obtained for the three wheat midge species (Fig. 3a). Evidence of multiple parasitism could be revealed with this pair of primers if the size difference between the amplicons is detected. Conversely, primers Cec164F + Cec473R amplified a single fragment of 326 bp for the 3 midge species (Fig. 3a) while no amplification was observed for the parasitoids. This PCR method allowed the detection of an amount of 1 pg genomic DNA of Platygaster spp. (Fig. 3b). For M. penetrans, 10 pg genomic DNA was clearly detectable while 1 pg is weakly noticeable. These limits of detection are higher than for the TaqMan assay. Nevertheless, group-specific PCR detection enables subsequent species determination by amplicon sequencing.
Fig. 3.
Group-specific PCR amplification of a part of the 18S sequence. (a) parasitoids were detected using the specific primer pair Par164F + Par473R (left part); midges were detected using Cec164F + Cec164R (right part), (b) the detection threshold of Par164 + Par473 was estimated on serial dilutions of 10 pooled adults M. penetrans and Platygaster spp. DNA. 1 kb ladder was used. Original gels are presented in Supplementary Fig. S4.
The species-specific reverse primers (Table 1) used with Par164F allowed amplifying fragments from 123 to 270 bp for the corresponding species while no cross-amplification was noted with the other species. The multiplex PCR assay enabled the amplification of fragments from P. tuberosula, M. penetrans and E. error (both separately in separate DNA extracts or simultaneously in a mixture of DNA extracts from the three parasitoids). The sizes of the amplicons were easily distinguishable on agarose gel (Fig. 4).
Fig. 4.
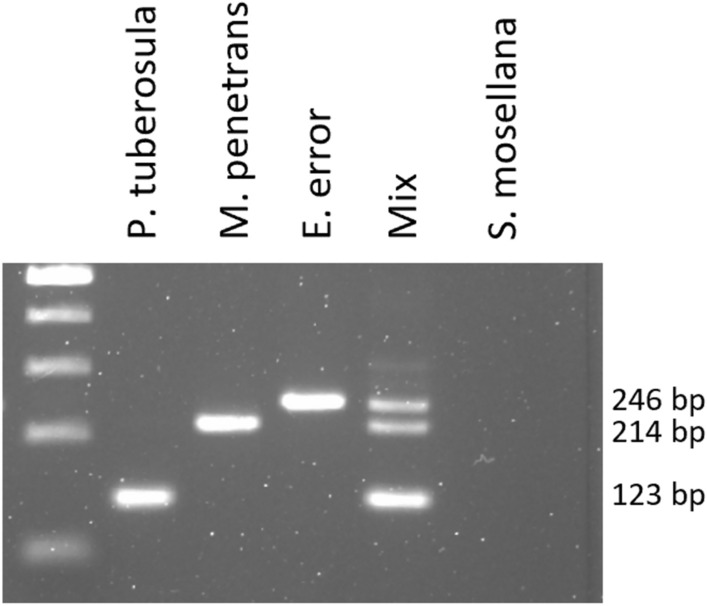
Multiplex PCR assay for the targeted detection of P. tuberosula, M. penetrans and E. error. A part of the 18S sequence is amplified using the primers Par164F, macro382R, euxe382R and tuber295R. Mix: a mixture of 1 µl of each genomic DNA from each species. 1 kb ladder was used. Original gel is presented in Supplementary Fig. S4.
Field parasitism
The molecular tools developed in this study were tested on 132 S. mosellana larvae collected in seven fields located in the Walloon region, Belgium (Table 4 and Supplementary Fig. S3). All 132 larvae allowed the PCR amplification of a COI fragment (C1-J-1718F + C1-N-2191 primer pair) indicating that the DNA extraction was successful and that potential inhibitors in the DNA extract could not prevent amplification. Out of the 132 larvae analysed, the TaqMan analysis revealed that 20 larvae were parasitised (15.2%). We observed a high variability in the amount of parasitoid DNA estimated according to the equations in Table 3. Estimated amounts ranged from 0.8 to 476 pg per larva for M. penetrans (n = 13) and from 0.3 to 50 pg per larva for P. tuberosula (n = 3) (Supplementary Table S2). The level of parasitism varied from site to site, ranging from 0% (n = 15 larvae analysed) to 33% (n = 15). This result was confirmed using the group-specific primer pair (Par164 + Par473): an amplification was obtained for the 20 individuals, although repetitions were sometimes necessary, mostly for those with the lowest estimated DNA amounts with the TaqMan assay (Supplementary Table S2). This is consistent with the difference in detection thresholds between both tools.
Table 4.
Detection of midge parasitism at seven locations in Belgium.
| Location | Latitude N | Longitude E | N | Parasitised | % | M. Penetrans | P. tuberosula | E. error | M. Penetrans + E. error |
|---|---|---|---|---|---|---|---|---|---|
| Ciney | 50,30725252 | 5,06835821 | 27 | 4 | 14.8 | 3 | 0 | 0 | 1 |
| Ernage | 50,60288404 | 4,67571176 | 27 | 4 | 14.8 | 0 | 1 | 3 | 0 |
| Gembloux | 50,58368337 | 4,69034771 | 16 | 4 | 25.0 | 3 | 1 | 0 | 0 |
| Juprelle | 50,70804044 | 5,58654127 | 16 | 2 | 12.5 | 1 | 1 | 0 | 0 |
| Lillois | 50,62194227 | 4,37769566 | 15 | 0 | 0.0 | 0 | 0 | 0 | 0 |
| Limont | 50,66634385 | 5,32071588 | 15 | 5 | 33.3 | 5 | 0 | 0 | 0 |
| Sorée | 50,40837572 | 5,12478659 | 16 | 1 | 6.3 | 1 | 0 | 0 | 0 |
| Total | 132 | 20 | 15.2 | 13 | 3 | 3 | 1 |
Parasitism of S. mosellana larvae was assessed by using the TaqMan method and the group specific PCR assay developed in this study. Species were identified by Sanger sequencing and species-specific PCR amplification. N: sample size.
The species were identified by multiplex PCR and/or Sanger sequencing and confirmed by species-specific PCR. The most commonly identified species was M. penetrans (13 detections, observed at 5 sites), followed by P. tuberosula (3 larvae in 3 sites) and E. error (3 larvae found at 1 site). At the Ciney site, a single larva exhibited a faint double amplification with primers Par164 + Par473 resulting in failed Sanger sequencing. Species-specific amplifications identified two distinct parasitoid species (M. penetrans and E. error), suggesting double parasitism (See supplementary Fig. S3).
Discussion
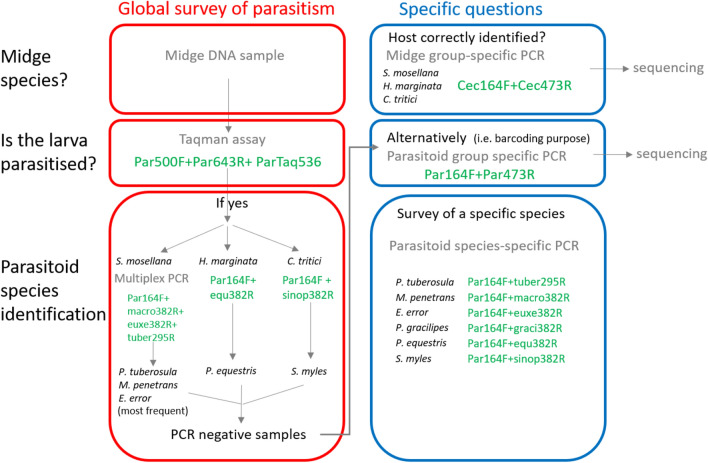
In this work, we developed molecular tools for identifying parasitoids known to occur in Belgium within individual wheat midge larvae. In order to achieve this, we first delivered COI, 18S and 28S reference sequences for seven parasitoid species occurring in Belgian wheat midges and representing 96.9% of the specimens harvested by Chavalle et al.12. These sequences were obtained from specimens carefully identified morphologically. These three genes were evaluated for their suitability to detect and identify the parasitoids within the hosts. Although each of the three genes allowed all the species to be distinguished, we found that the 18S gene was the only one displaying a barcoding gap at the species level and between the two groups (parasitoids vs. midges). The suitability of 18S for group-specific identification of parasitoids had already been pointed out for the detection of cereal aphids parasitoids36. The same authors also remarked on the insufficient resolution of 18S for identification at species level. For the midge parasitoids studied here, we found a highly variable zone in the 18S sequence allowing identification to species level, either by Sanger sequencing of a group-specific PCR amplification or by species-specific amplification with primers localised in the variable zone. The group-specific primers used here can also be used more broadly for detecting parasitism by Hymenopterans in other agricultural pests, but the validity of species detection and identification will have to be tested.
For a global survey of parasitism, we propose the following sorting method (Fig. 5). Midge larvae are generally identified on a morphological basis; this identification can be confirmed by midge group-specific PCR followed by Sanger sequencing. The TaqMan assay that we developed can then be used to answer the question “are the larvae parasitised?”, regardless of the midge and parasitoid species among those studied in this work. Next, a multiplex PCR assay was used to identify S. mosellana parasitoids from the 3 most represented species (90% according to12) according to the sizes of the amplicons. Regarding H. marginata and C. tritici, species-specific PCR can be used to specifically detect the only parasitoid known for each of these species (P. equestris and S. myles respectively). Lastly, rare parasitoid species that do not amplify using these techniques can be identified by parasitoid group-specific PCR followed by Sanger sequencing. Note that the undescribed Euxestonotus sp. could potentially be detected by the E. error species-specific primers pair. In future field studies where E. error is identified by species-specific primers, Sanger sequencing of the corresponding group-specific PCR amplicon could be performed to check for potential sequence differences.
Fig. 5.
Molecular approaches for detecting parasitism in wheat midges and for the species identification of the parasitoids.
The usefulness of our molecular tools was assessed using S. mosellana larvae collected in the field. A parasitism rate of 0 to 33% was found depending on the location. Three species were identified: M. penetrans (65%), E. error (15%) and P. tuberosula (15%). This dominance of M. penetrans over E. error and P. tuberosula is in agreement with a Belgian survey conducted through rearing and morphological identification12.
Given the low number of available sequences for some species, we cannot rule out the possibility of missing some parasitoids whose sequences may display polymorphism in the regions targeted by the primers. In our field survey, however, the results obtained were in agreement using two different techniques (TaqMan assay and group-specific PCR) whose primers target different sequences.
Molecular tools for detecting parasitism and identificating parasitoid species offer several advantages over traditional morphological-based methods37. Firstly, they eliminate the need for time-consuming and costly laboratory larvae rearing. The tools described here will aid rapid responses for determining parasitism levels in the context of agricultural warnings. Secondly, larvae samples can be stored in alcohol or a freezer until molecular analysis, providing flexibility and allowing a large number of samples to be analysed in a batch. Thirdly, no taxonomic expertise based on morphology is required. Fourthly and finally, unlike the rearing approach, molecular tools can detect multiparasitism and cases of unsuccessful parasitoid emergence.
The environmental resources requirement for midge parasitoids is poorly understood. These molecular tools could be invaluable for studying the parasitoid population dynamics, food webs and parasitism rates in various landscape contexts. With these insights, biological control of wheat midges could be enhanced, reducing the need for chemical spraying and minimising yield losses at harvest.
Supplementary Information
Acknowledgements
The Authors thank Roberte Baleux and Tiffany Herbiet for their technical assistance.
Author contributions
LH conceived the work. SC collected the specimens. PB morphologically identified the reference specimens. DM designed the molecular biology developments and performed the experiments. DM and LH analysed and interpreted the results. BD provided assistance in sequence analysis. GS participated in the design of the molecular developments and the deposition of the specimens. LH and DM wrote the manuscript. BD and GS significantly improved the manuscript. All Authors reviewed the manuscript and approved the final submitted version.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chavalle, S., Censier, F., San Martin y Gomez, G. & De Proft, M. Protection of winter wheat against orange wheat blossom midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae): efficacy of insecticides and cultivar resistance: Control of Sitodiplosis mosellana using insecticides and cultivar resistance. Pest. Manag. Sci.71, 783–790 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Dexter, J. E. et al. The influence of orange wheat blossom midge (Sitodiplosis mosellana Géhin) damage on hard red spring wheat quality and the effectiveness of insecticide treatments. Can. J. Plant Sci.67, 697–712 (1987). [Google Scholar]
- 3.Olfert, O. O., Mukerji, M. K. & Doane, J. F. Relationship between infestation levels and yield loss caused by wheat midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae), in spring wheat in Saskatchewan. Can. Entomol.117, 593–598 (1985). [Google Scholar]
- 4.Gaafar, N., El-Wakeil, N. & Volkmar, C. Assessment of wheat ear insects in winter wheat varieties in central Germany. J. Pest. Sci.84, 49–59 (2011). [Google Scholar]
- 5.Jacquemin, G., Chavalle, S. & De Proft, M. Forecasting the emergence of the adult orange wheat blossom midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae) in Belgium. Crop Protect.58, 6–13 (2014). [Google Scholar]
- 6.Oakley, J. N. et al. Integrated control of wheat blossom midge: Variety choice, use of pheromone traps and treatment thresholds. Project Report No. 363, HGCA, Kenilworth, Warks, UK (2005).
- 7.Dufton, S. V. et al. A global review of orange wheat blossom midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae), and integrated pest management strategies for its management. Can. Entomol.154, e30 (2022). [Google Scholar]
- 8.Censier, F., De Proft, M. & Bodson, B. The saddle gall midge, Haplodiplosis marginata (von Roser) (Diptera: Cecidomyiidae): Population dynamics and integrated management. Crop Prot.78, 137–145 (2015). [Google Scholar]
- 9.Dewar, A. M. Ecology and control of saddle gall midge, Haplodiplosis marginata von Roser (Diptera; Cecidomyiidae). HGCA Res. Rev. (76) (2012).
- 10.Censier, F., Chavalle, S., San Martin y Gomez, G., De Proft, M. & Bodson, B. Targeted control of the saddle gall midge, Haplodiplosis marginata (von Roser) (Diptera: Cecidomyiidae), and the benefits of good control of this pest to winter wheat yield: Harmful effects of Haplodiplosis marginata and targeted chemical control. Pest. Manag. Sci.72, 731–737 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Affolter, F. Structure and dynamics of the parasitoid complex of the wheat midges Sitodiplosis mosellana (Géhin) and Contarinia tritici (Kirby) (Dipt. Cecidomyiidae). In 108 pp. (C.A.B. International Institute of Biological Control, Delemont, Switzerland, 1990).
- 12.Chavalle, S., Buhl, P. N., San Martin y Gomez, G. & De Proft, M. Parasitism rates and parasitoid complexes of the wheat midges, Sitodiplosis mosellana, Contarinia tritici and Haplodiplosis marginata. BioControl63, 641–653 (2018). [Google Scholar]
- 13.Dufton, S. V., Laird, R. A., Floate, K. D. & Otani, J. K. Diversity, rate, and distribution of wheat midge parasitism in the Peace River region of Alberta, Canada. Can. Entomol.153, 461–469 (2021). [Google Scholar]
- 14.Gariepy, T., Kuhlmann, U., Gillott, C. & Erlandson, M. A large-scale comparison of conventional and molecular methods for the evaluation of host–parasitoid associations in non-target risk-assessment studies. J. Appl. Ecol.45, 708–715 (2008). [Google Scholar]
- 15.Harris, K. M. Gall midge genera of economic importance (Diptera: Cecidomyiidae) part 1: Introduction and subfamily Cecidomyiinae; supertribe Cecidomyiidi. Trans. R. Entomol. Soc. Lond.118, 313–358 (1966). [Google Scholar]
- 16.Skuhrava, M. Family Cecidomyiidae. In Contributions to a Manual of Palaearctic Diptera Vol. 1 71–204 (Science Herald, 1997). [Google Scholar]
- 17.Graham, M. W. R. D. V. The Pteromalidae of North-Western Europe (Hymenoptera: Chalcidea) (British museum, Natural history, 1969). [Google Scholar]
- 18.Johansson, E. Studier och forsök rörande vetemyggorna, Contarinia tritici Kirby och Clinodiplosis mosellana Géh. samt deras bekämpande. IV. Undersökning av vetemyggornas parasiter: 1. I Svalöf och Weibullsholm åren 1932–1935 anträffade arter. Statens Växtskyddsanstalt Meddelanden15, 1–19 (1936). [Google Scholar]
- 19.Buhl, P. N. Key to Platygaster (Hymenoptera, Platygastridae) from Denmark, with descriptions of new species. Steenstrupia29, 127–168 (2006). [Google Scholar]
- 20.Kieffer, J. J. Scelionidae Vol. 48 (W de Gruyter & Co, 1926). [Google Scholar]
- 21.Huggert, L. Descriptions of Platygastrinae from Sweden (Hymenoptera: Platygastridae). Insect Syst. Evol.6, 61–66 (1975). [Google Scholar]
- 22.Vlug, H. J. Catalogue of the Platygastridae (Platygastroidea) (Insecta: Hymenoptera). Vol. 19 (London, SPB Academic Publishing, 1995). [Google Scholar]
- 23.Spittler, H. Beiträge zur morphologie, biologie und ökologie des sattelmückenparasiten Platygaster equestris nov. spec. (Hymenoptera, Proctotrupoidea, Scelionidae) unter besonderer berücksichtigung seines abundanzdynamischen einflusses auf Haplodiplosis equestris Wagner (Diptera, Cecidomyiidae. Z. Für Angew. Entomol.64, 1–34 (1969). [Google Scholar]
- 24.Kozlov, M. A. Superfamily Proctotrupoidea. Determination of insects of the European portion of the USSR3, 538–664 (1978).
- 25.Gahan, A. B. The Serphoid and Chalcidoid Parasites of the Hessian Fly. (US Dept. of Agriculture, 1933).
- 26.Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol.3, 294–299 (1994). [PubMed] [Google Scholar]
- 27.Hernández-Triana, L. M. et al. Recovery of DNA barcodes from blackfly museum specimens (Diptera: Simuliidae) using primer sets that target a variety of sequence lengths. Mol. Ecol. Resour.14, 508–518 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Simon, C. et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am.87, 651–701 (1994). [Google Scholar]
- 29.Heraty, J. et al. Evolution of the hymenopteran megaradiation. Mol. Phylogenet. Evolut.60, 73–88 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Dowton, M. & Austin, A. D. Phylogenetic relationships among the microgastroid wasps (Hymenoptera: Braconidae): Combined analysis of 16S and 28S rDNA genes and morphological data. Mol. Phylogenet. Evolut.10, 354–366 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res.22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol.16, 111–120 (1980). [DOI] [PubMed] [Google Scholar]
- 33.Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evolut.35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou, N. & Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evolut.4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Ye, J. et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform.13, 134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye, Z., Vollhardt, I. M. G., Tomanovic, Z. & Traugott, M. Evaluation of three molecular markers for identification of European primary parasitoids of cereal aphids and their hyperparasitoids. PLoS ONE12, e0177376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nisole, A. et al. Identification of spruce budworm natural enemies using a qPCR-based molecular sorting approach. Forests11, 621 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).