Abstract
The cryo-EM structure of the human rhinovirus B14 determined in this study reveals 13-bp RNA duplexes symmetrically bound to regions around each of the 30 two-fold axes in the icosahedral viral capsid. The RNA duplexes (~12% of the ssRNA genome) define a quasi-dodecahedral cage that line a substantial part of the capsid interior surface. The RNA duplexes establish a complex network of non-covalent interactions with pockets in the capsid inner wall, including coulombic interactions with a cluster of basic amino acid residues that surround each RNA duplex. A direct comparison was made between the cryo-EM structure of RNA-filled virions and that of RNA-free (empty) capsids that resulted from genome release from a small fraction of viruses. The comparison reveals that some specific residues involved in capsid-duplex RNA interactions in the virion undergo remarkable conformational rearrangements upon RNA release from the capsid. RNA release is also associated with the asynchronous opening of channels at the 30 two-fold axes. The results provide further insights into the molecular mechanisms leading to assembly of rhinovirus particles and their genome uncoating during infection. They may also contribute to development of novel antiviral strategies aimed at interfering with viral capsid-genome interactions during the infectious cycle.
Subject terms: Cryoelectron microscopy, Virus structures
Cryo-EM structure of human rhinovirus B-14 reveals genomic RNA duplexes that delineate a dodecahedral cage associated with the capsid inner surface, and an asynchronous opening of capsid channels that facilitate the release of the genome.
Introduction
Respiratory viruses are the leading cause of human disease1. Human rhinoviruses (RVs) are highly transmissible respiratory viruses that cause most common colds, leading to huge economic losses due to expenditure on palliative medication and lost work/school days2,3. Moreover, RVs cause or exacerbate severe respiratory diseases including asthma or chronic obstructive pulmonary disease (the third leading cause of death worldwide according to the WHO Global Health Estimates for 2019)4–6. There are currently no approved vaccines or drugs for the prevention or treatment of RV infection7.
RVs belong to the Enterovirus genus within the Picornaviridae family. The RV icosahedral pseudo-T = 3 capsid (∼300 Å in diameter) is built from 60 copies of four capsid proteins (CP, termed VP1 to VP4) that encapsidate a ~7.5 kb, positive-sense, single-stranded (ss) RNA genome. VP1, VP2, and VP3 share a similar protein topology, with two four-stranded β-sheets facing each other almost in parallel to the capsid surface. The 60 copies of the small VP4 polypeptide are extended across the capsid inner surface8,9.
RVs are classified into three species, RV-A, RV-B, and RV-C, that include 169 serotypes10. All RV-B types and most RV-A types use intercellular adhesion molecule 1 (ICAM-1) as their receptor. Other RV-A types attach to and enter cells via the low-density lipoprotein receptor11, whilst RV-C types bind cadherin related family member 3 (CDHR3) as receptor12. Genome uncoating in the host cell is mediated by receptor binding (in the case of ICAM-binding RVs) and/or by acidification in the endosomes, that promote irreversible and extensive conformational changes in the virion13–16. These rearrangements lead first to the externalization of the amphipathic VP1 N-termini, and the release of VP4 that permeabilizes membranes through pore formation, resulting in an intermediate state of the viral particle (120S particle)17–19. In a second step, important changes occur in the interactions between RNA and the inner capsid surface leading to the organization of an RNA layer that facilitates exit of the genome, leaving an empty protein capsid (80S particle)20. The viral RNA probably exits the 120S intermediate via one of the transient channels formed at the two-fold symmetry axes of the capsid and is translocated through the endosomal membrane to the cytosol21,22.
Picornavirus morphogenesis involves the assembly of 5S protomers each containing one copy of VP0 (a covalently-linked VP4 + VP2), VP1, and VP3, followed by the association of protomers to form 14S pentameric intermediates. Pentamers bind the viral RNA in a co-assembly process that yields an immature virion23. Picornavirus maturation usually involves the autocatalytic cleavage of VP0 to VP4 and VP2 to yield an infectious virion24.
Co-assembly of capsid and nucleic acid occurs not only in picornaviruses, but also in many other ssRNA viruses25,26. The co-assembly process is mediated by sequence/structure elements in the genomic RNA (called packaging signals, PS). PSs may be distributed along the nucleotide sequence and bind specific sites on the inner surface of CP subunits or capsid assembly intermediates. For example, a study on Ljungan virus (a picornavirus of the Parechovirus genus), showed that stem-loop structural elements in the genomic RNA may be critical for RNA-guided virus assembly27. Interactions between CP and RNA have also been observed in human parechovirus 125,28, where multiple RNA stem-loops were identified across the genome, all presenting a GxU motif in their loop that was essential for interaction with the CP. In enterovirus-E, stem-loops presenting stacked purines in contact with CP were identified that may be conserved in other enteroviruses such as polioviruses29. The above results suggest that a common PS-mediated co-assembly mechanism based on stem-loop elements in the viral genome is shared by picornaviruses belonging to different genera30.
RNA-capsid interactions may also play a role during enterovirus uncoating. In human RV B14 (RV-B14), RNA octanucleotide segments of the genome are bound to the capsid inner wall18. Those interactions could be modulated during receptor or pH-induced conformational changes, which would facilitate genome release18.
Additional structural studies on enteroviral capsid-genome interactions could facilitate the understanding of the role of those interactions in the regulation of virion morphogenesis and genome uncoating30,31. Many crystallographic and cryogenic electron microscopy (cryo-EM) structures are available for different enterovirus or other picornavirus capsids under different conditions [see the VIPER database, https://viperdb.org/32]. However, the detailed structure and organization of the encapsidated genomic RNA in most virions, including capsid-bound RNA elements, remains to be established.
In the present study we have determined by high-resolution cryo-EM the detailed structure of capsid-bound genomic RNA elements in the RV-B14 virion. The results reveal a complex network of interactions between the viral genome and the capsid inner wall. About 12% of the genomic ssRNA is organized into 30 RNA duplexes each composed of 13 bp with two pendant bases at each end. These duplexes are bound to the capsid protein-layer at particle two-fold axes and delineate a quasi-dodecahedral cage in each virion. This network of RNA-capsid subunit interactions inter-connects different assembly intermediates and may promote capsid protein-RNA co-assembly as well as stabilizing the resulting virion. Moreover, comparison of the cryo-EM structures of RNA-filled virions with the genome-free capsids also determined here suggests that the disruption of the interactions between the capsid and the different RNA duplexes is not a simultaneous process. Channels at the capsid two-fold axes are opened asynchronously, which during the uncoating process may facilitate the selective release of the genome through one of the early opened channels.
Results
Cryo-EM structure of the RV-B14 virion
A wealth of structural data is available for RV capsids. However, information relative to the structural organization of the nucleic acid inside these virions and its interactions with the viral capsid is limited18–20. To determine the genome organization and RNA-capsid interactions in a RV virion, purified RV-B14 virus particles were visualized in a 200-kV Talos Arctica electron microscope. Most isometric particles in the preparation used were homogeneously electrodense (Fig. 1A) and corresponded to RNA-filled virions. Only a few of them were empty or partially empty capsids (Fig. 1A, white arrows). We selected ~474,000 particles that were initially 2D classified and subjected to a 3D classification with icosahedral symmetry to render ~313,000 full particles and ~8400 empty particles, and their 3D reconstructions were calculated separately. The final resolution of the reconstructions for RV-B14 full virions and empty capsids was 2.9 Å and 3.8 Å, respectively, as estimated by the standard of a 0.143 Fourier shell correlation (FSC) coefficient criterion (Supplementary Fig. 1A). The quality of different regions of the full and empty capsid cryo-EM map was determined using MonoRes. Whereas the full map was highly homogeneous in resolution (Supplementary Fig. 1B), the empty map exhibited lowest resolution regions around the 5-fold axes (Supplementary Fig. 1C).
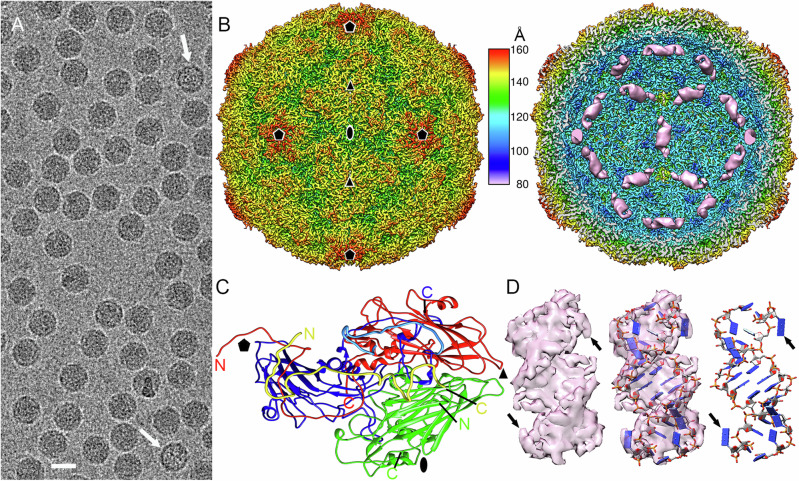
Fig. 1. Cryo-EM structure of RV-B14 virions at 2.9 Å resolution.
A Cryo-EM image of RV-B14. Arrows indicate empty capsids. Bar = 250 Å. B Radially color-coded surface representation of RV-B14 outer (left) and inner (right) surfaces viewed along a two-fold axis. Ordered, genomic RNA at the inner surface of the RV-B14 capsid beneath the two-fold axes (pink). The color key (center) indicates the radial distance (in Å) from the particle center. In these images the capsid density is sharpened, whereas dsRNA density is unsharpened. Symbols indicate icosahedral symmetry axes. C A 5S protomer (or asymmetric unit) viewed from the inside of the capsid, showing the ordered regions of VP1 in blue, VP2 in green, VP3 in red, and VP4 in yellow. Ordered N and C termini are indicated. The N-terminal segment of VP1 Glu7-Lys30 is highlighted in light blue. D Sharpened cryo-EM density of a dsRNA helical segment (left), fitted with an atomic model of a 13bp-dsRNA segment and two pendant bases (blue slabs for bases) (center & right images). Arrows indicate the two terminal unpaired nucleotides at each end of the dsRNA.
The cryo-EM map of full RV-B14 particles is 318 Å in diameter and the protein shell is ~46 Å thick, as determined from the spherically-averaged radial density plot of the 3D reconstruction (3DR). The capsid shares the typical topological features of EVs; i.e., the star-shaped “mesas” at the five-fold axes, and the “canyons” surrounding these mesas that contain the ICAM-1 binding sites, with three-bladed “propeller” folds at the three-fold axes (Fig. 1B, left). In addition, a helical density compatible with a double-stranded (ds) RNA region was clearly detected on the capsid interior at each two-fold axis (Fig. 1B, right, pink densities).
Using the crystal structure of the RV-B14 capsid [PDB ID: 1NCQ33] as an initial template, we built with Coot (as described in Methods) the four polypeptide chains for the structural proteins VP1, VP2, VP3, and VP4 of the asymmetric unit (Supplementary Table 1). The atomic model obtained for VP1 includes the sequence Glu7-Tyr289, for VP2 the sequence Cys7-Gln262, for VP3 the complete sequence (Gly1-Glu236), and for VP4 the sequence Ser23-Lys68; i.e., only small N-terminal segments are invisible (or disordered) in VP1, VP2, and VP4. The model showed the expected features of the RV capsid9. VP1-VP3 are folded as single β-barrels. The VP2 and VP3 β-barrels are parallel to the capsid surface, whilst that of VP1 is tilted. VP4 lacks secondary structure elements and can be seen as a cleaved N-terminal extension of VP2 (Fig. 1C and Supplementary Fig. 2). Notably, the polypeptide chain of VP3 has limited access to the capsid contents (the viral RNA), because the extended VP1 N-terminal region (Glu7-Asp66) sits on the inner side of the VP3 β-barrel (Fig. 1C, indicated as a cyan peptide). Importantly, the atomic model obtained reveals unprecedented detail of the capsid-RNA genome interactions in RV.
Structure of duplex RNA elements inside the RV-B14 virion
Sharpening the map of the RNA-filled virion, including the internal helical densities ascribed to intrachain double-stranded (ds) RNA, allowed us to build an atomic model for the RNA elements bound at each capsid two-fold axis, using initially a dsRNA type-A segment generated de novo with Coot as described in Methods. The observed density corresponds to a 13 bp dsRNA duplex, in which the terminal nucleotides at both ends are non-matched (Fig. 1D, arrows). These dsRNA elements account for ~12% of the full-length genomic ssRNA (Supplementary Movie 1).
Although the resolution of these duplex densities is limited to ~3.8 Å (Supplementary Fig. 1B), the data imply that the flanking nucleotide bases of the RNA duplexes are purines (see below). This observation is consistent with previous structures of RV-B14 that allowed the modeling of capsid-bound octanucleotide segments, and pointed to the presence of a ss guanine in the equivalent positions18. In the present study, the unambiguous location of sufficient RNA density after imposing icosahedral symmetry and sharpening indicates a high occupancy at the 30 two-fold symmetry axes, which delineate an almost complete, dodecahedral cage of dsRNA filaments immediately under the capsid inner surface (Supplementary Movie 1). The occupancy of the dsRNA at the two-fold axes is ~100% as a focused classification on the dsRNA regions resulted in five classes containing a density that corresponded to a dsRNA segment (Supplementary Movie 2). In addition, different 3D classifications without imposing icosahedral symmetry resolved these genome duplexes in the resulting asymmetric maps (Supplementary Fig. 3, red boxes; see Materials and Methods for details).
Secondary structure prediction for the RV-B14 genome with the RNAfold server (University of Vienna, http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) identifies 31 duplex segments at least 12–16 bp long, i.e., matching the lengths of the RNA duplexes bound at the 30 two-fold capsid symmetry axes. Of these 31 duplexes, 17 contain two pendant purines, as determined in the RV-B14 map (Supplementary Fig. 4). The RV-B14 genomic sequence may have evolved to have such base paired regions as a result of a biological advantage conferred by their interaction with the capsid proteins.
Interactions between the RNA duplexes and the capsid
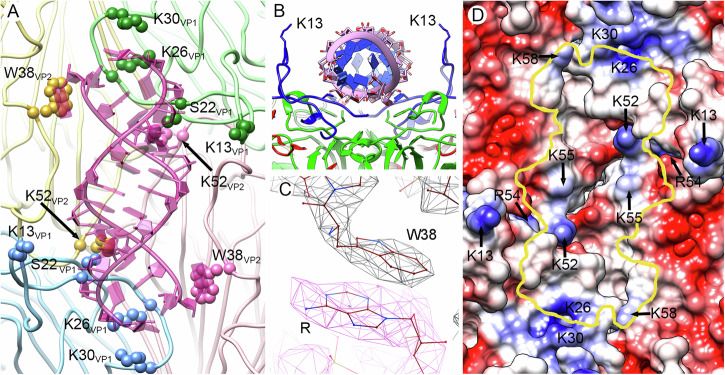
Analysis of viral RNA-protein attractive interactions reveals the involvement of 11 VP1 residues, three VP2 residues and one VP4 residue (Supplementary Table 2, Fig. 2A and B). Most of these residues are highly conserved in RV B sequences, consistent with them playing important roles in virion structure/maturation (Supplementary Table 2). The two terminal unpaired bases at the ends of the base-paired helices form stacking interactions with the side-chain of Trp38 of VP2 (Fig. 2C). Fitting of a purine (R) in these flanking densities has a higher occupancy than that of a pyrimidine (Y) (Fig. 2C, Supplementary Fig. 5). Our sequence alignment of 123 RV-A, RV-B or RV-C serotypes shows that Trp38 of VP2 is fully conserved.
Fig. 2. Capsid protein-dsRNA interactions in RV-B14 virions.
A Interactions of Trp38 and Lys52 of VP2, and Lys13, Ser22, Lys26, and Lys 30 of VP1 with the genomic RNA duplex (pink) viewed from the inside of the capsid. These interacting residues are highlighted (spheres). The four 5S protomers that interact with a dsRNA segment are colored in pale yellow, green, blue, and pink. Protomers in yellow and blue belong to the same pentamer, and protomers in green and pink to an adjacent pentamer. B Transverse view of the structure (rotated 90° relative to A). The RNA duplex backbone is shown as pink coils with blue slabs for the bases. The side-chains of two Lys13 residues, one from each of the neighboring VP1 act as “guide posts” for duplex placement. C The stacking contact between the aromatic rings of Trp38 in VP2 and the purine at the end of the dsRNA segment. D The RV-B14 capsid inner surface is represented as an electrostatic potential surface, showing the distribution of negative (red) and positive (blue) charges, around a two-fold position. The yellow line indicates the outline for a bound dsRNA duplex, indicating the basic residues that interact with it.
Many basic residues from the capsid proteins (Lys13, Lys26, His27, Lys30, and Arg54 of VP1, Lys52, and Lys 55 of VP2, and Lys58 of VP4) make electrostatic contacts with phosphates in the RNA duplexes. Lys26 of VP1; Lys52 and Lys55 of VP2; and Lys58 of VP4 are located at the interface between the dsRNA and the capsid internal surface (Fig. 2D, Supplementary Movie 3). Sequence alignment of strains of the 29 RV-B serotypes identified revealed that 5 of these 8 basic residues are absolutely conserved, whilst another one is 90% conserved. In addition, eight acidic residues (Glu7, Glu12, and Glu52 of VP1, Glu40, Asp44, Asp57, and Glu250 of VP2 and Asp59 of VP4) are located close to each RNA duplex and may modulate the local electrostatic potential. Again, 5 out of 8 of those acidic residues are absolutely conserved in the 29 RV-B serotypes, whilst another one is 83% conserved (Supplementary Table 2). The 13 bp duplexes fit in small depressions at the two-fold axes, and the side-chains of Lys13 from two VP1 act as guide posts stabilizing the locations of the duplexes (Fig. 2B, D). Four protomers (VP1-VP4) from two adjacent pentamers (14S assembly subunits) participate in capsid-RNA interactions. Two of them contribute with their VP1 molecules and two others contribute with their VP2 (Fig. 2A). Thus, each genomic dsRNA duplex mediates the positioning of four protomers and two pentamers (Supplementary Fig. 6).
Cryo-EM structure of the empty RV-B14 capsid
In the purified RV preparation used, a fraction of particles (~1.5%) corresponded to empty capsids. Picornavirus capsid assembly is mediated by the viral RNA, which can be readily released from the RV virion by mild heating19. The empty capsids observed in our purified RV preparations are therefore likely the result of the spontaneous release of the RNA from a small fraction of the purified virions (see also below).
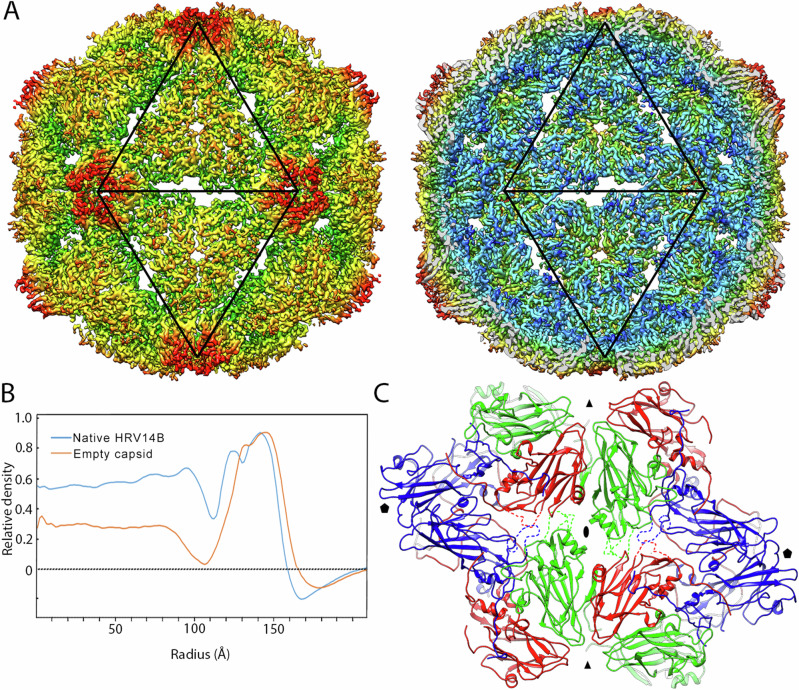
In order to define the conformational changes in the RV-B14 capsid after genome release, these empty capsids (that were imaged together with full virions) were analyzed separately (Fig. 3A). The cryo-EM map of the empty capsid reveals opening of pores at the three-fold axes (~12 Å in diameter), and of elongated Z-shaped apertures at the two-fold axes (~12 Å x 29 Å in size), relative to the RNA-containing virion. The diameters of full and empty particles, determined from spherically-averaged, radial density plots of the 3DR, were 318 Å and 332 Å, respectively (Fig. 3B). The ~4% size increase of the RNA-free capsid relative to the virion concurs with similar results with full and empty shells from RV-A219 and other EVs. VP4 is not detected in the empty RV-B14 capsid structure, as expected, since previous observations suggest it is released during genome uncoating10,14. The lack of VP4 confirms that the empty capsids are the result of RNA release from virions, and not of their RNA-independent assembly.
Fig. 3. Cryo-EM structure of empty RV-B14 capsids at 3.8 Å resolution.
A Radially color-coded surface representation of empty RV-B14 outer (left) and inner (right) surfaces viewed along a two-fold axis. Black triangles outline two adjacent icosahedral facets and indicate the icosahedral symmetry axes that expand to create pores. B Comparison of radial density profiles of the cryo-EM maps of virions (blue) and empty capsids (orange). Ordered RNA is located at radii < 113 Å in the virion. C Atomic models around the two-fold, Z-shaped pores viewed from the outside of the capsid. VP1 is shown in blue, VP2 in green, and VP3 in red. Highly flexible (or invisible) loops are indicated by dashed lines in the conformation of closed virions. Symbols indicate icosahedral symmetry axes.
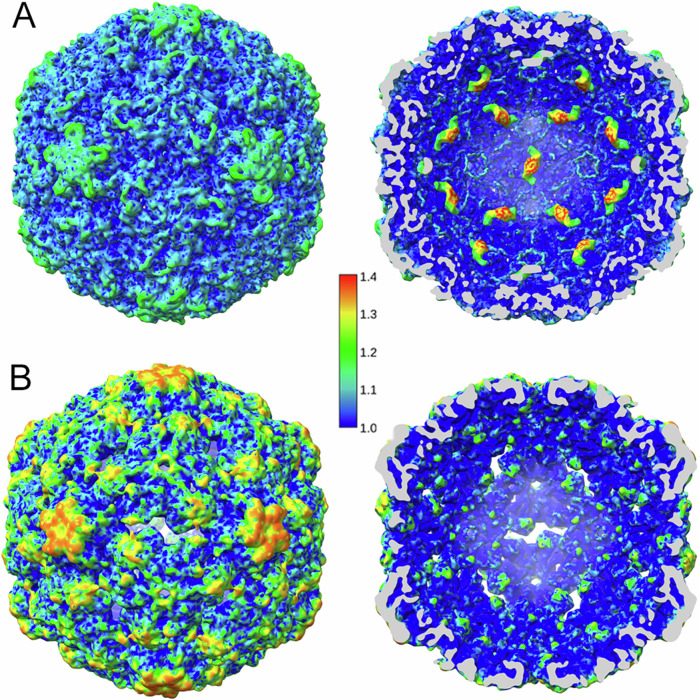
This capsid expansion is the result of changes in the relative positions of the VP monomers. Their superposition shows that only small changes in conformation occur, with an rmsd of 2.4 Å for VP1, 2.1 Å for VP2 and 1.8 Å for VP3 (Supplementary Fig. 7). In addition, the VP1-VP3 regions that form the edges of the aperture at each two-fold axis in the empty capsid (segments Tyr204-Val217 of VP1, Val50-Pro56 and Ser252-Gln262 of VP2, and Gln172-Asp177 of VP3), could not be traced in empty particles because of a lack of density. This would be consistent with much higher flexibility of the segments of polypeptide chain around the two-fold axes in the empty capsid compared to the virion (Fig. 3C, dashed lines). The relative local resolution in the cryoEM maps provides a signature of local equilibrium dynamics34. Relative local resolution analysis reveals that the outer surface and the regions around the two-fold, three-fold and (especially) five-fold axes are more dynamic in the RV-B14 empty capsid than in the virion (Fig. 4). Thus, the capsid protein-genome interactions stiffen the capsid that becomes more dynamic after the genome is released.
Fig. 4. Relative local resolution of cryo-EM maps of full and empty RV-B14 maps.
Relative local resolution assessment for full virion (A) and empty capsid (B) RV-B14 maps was calculated by dividing the absolute local resolution values by the global (averaged), FSC-derived resolution of the map. In the virion the density corresponding to RNA duplexes at each 2-fold axis has been included. Surface-shaded representations of the RV-B14 particle outer (left) and inner (right) surfaces viewed along an icosahedral two-fold axis (scale color at center). Cryo-EM density regions with relative local resolution values > 1.1 (i.e., lower resolution) indicate high conformational dynamics. Regions with the highest resolution (relative local resolution values < 1.1; deep blue shades) correspond to low dynamic conformational states.
Rearrangements of some specific residues involved in the binding of the RNA duplexes were detected by comparing the virion and the empty capsid structures. Lys52 of VP2 interacts with the genome in the full particle, but reorientates towards the capsid exterior in the empty particle. In addition, Trp38 of VP2 in the empty capsid moves away from its position in the virion, where it interacts with the genome. The interactions between these residues and the genomic RNA duplexes may promote virion assembly and stabilize the capsid. In addition, their rearrangements during the uncoating process could guide the release of the genome. Many amino acid side chains located towards the N-terminus of VP1 (between Glu7 and Lys30) would also be expected to help bind the nucleic acid to the capsid proteins. During RV uncoating, the VP1 N-terminus is externalized, necessarily disrupting the interactions between its side-chains and the RNA (Supplementary Movie 4). The disruption of contacts between the RNA molecule and the capsid inner wall may facilitate the release of the viral genome. Translocation of the RNA would be expected to occur via one of the capsid channels that are opened at the two-fold axes upon particle expansion.
Asymmetric opening of apertures in the empty RV-B14 capsid
If the apertures at the 30 two-fold axes were opened synchronously, the genome could be pulled towards 30 different openings, effectively impairing its egress. We therefore sought to determine whether these channels open simultaneously or are formed in an asynchronous process. The capsid regions around individual two-fold axes were analyzed independently, by extracting them from the original cryo-EM images as independent subparticles and then subjecting them to a three-dimensional classification without alignment. In addition to a fully closed state and a fully open state, as detected after icosahedral averaging (Fig. 5, bottom), this procedure identifies at least five intermediate states with progressively opened channels (Fig. 5, states I−V). “State I” accounted for 14% of the total two-fold regions analyzed that were at least partially opened, “state II” for another 14%; “state III” for 19%; “state IV” for 17%; and “state V” for 36%.
Fig. 5. Analysis of pores in empty RV-B14 capsids.
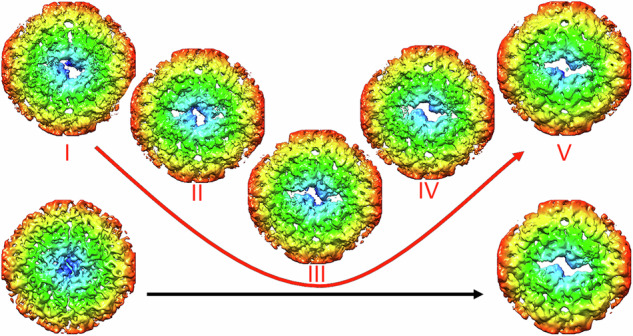
Asymmetric classification of pores at the two-fold axes in the five classes of the expanded virus particles, termed I−V (red), viewed from the inside. Localized reconstructions were calculated around the two-fold axes within a radius of 81 Å. Aperture sizes increase from left to right, starting with a closed structure (left, bottom) and leading to a capsid state in which the two-fold axis is fully opened (right, bottom). These conformational states were derived from virions and empty capsid particles, respectively, after applying icosahedral symmetry; arrows indicate aperture sizes in each case.
The distribution of aperture conformations in each empty particle was also determined, and showed that all five aperture intermediate states are simultaneously present in each particle. “State V” was the most abundant, with an average number of ~12 apertures/capsid, whilst the other “states (I-IV)” were present with a copy number of 4−5 per particle (Supplementary Fig. 8). This result suggests that opening of the capsid channels is not synchronized, and that the viral RNA may exit through one of the channels formed earlier in the capsid during the uncoating process.
Discussion
This study provides novel insights into the structural determinants of virus assembly and stability, and viral genome uncoating for RV-B14, an important human pathogen. In the cryo-EM structure of RV-B14 determined here, the general conformation of capsid-bound, 13 bp-long, type A RNA duplexes, and several details of their structure (such as the helix grooves) were clearly discernible. In addition, unpaired flanking bases, most probably purines, were identified at both ends of each dsRNA segment. We found a remarkable coincidence between the number of 12–16 bp dsRNA regions predicted to occur in the secondary structure of the free ssRNA genome (31 such regions) and the number of 13 bp duplexes visualized inside the RV-B14 virion structure (30 such duplexes). The dsRNA segments in the predicted secondary structure of the RV-B14 genome could correspond to those seen to interact with the RV capsid, consistent with a coassembly process in which the RNA secondary structure would not be substantially altered. In at least another ssRNA virus, cowpea chlorotic mottle virus (CCMV), the structure of a genomic 2774 nucleotide-long segment may be largely conserved upon encapsidation, as the ssRNA molecule in its “free state” is almost as compact as when encapsidated35. However, the possibility that genomic ssRNA-capsid protein interactions may influence the acquisition of secondary and tertiary RNA structures within the particle cannot be ruled out. Furthermore, a recent experimental study on the complete secondary structure of the enterovirus D68 RNA genome revealed a dynamic situation with alternative conformations36. An in-depth analysis is needed to determine whether the predicted intrachain duplexes in the RV-B14 genome are stable and can be predicted also for other RV serotypes and species and for other enteroviruses, and whether they may actually be present in the RNA packaged inside the virion (under study).
The secondary structure prediction does not identify a consensus nucleotide sequence. Although the conformations of the sugar-phosphate backbone in the RNA duplexes are clearly defined, symmetry-averaging completely blurs the identity of every base except for the terminal purines. Note, the electron density for these purines was high enough to suggest that they are present at both ends of each RNA duplex, and therefore bound to the two Trp38 residues at both sides of each icosahedral two-fold axis. There is no hint of a hairpin loop in the electron density connecting the two strands in the capsid-bound RNA duplexes in our structure, but this could be due to differences in loop size and conformation at different edges.
These observations do not exclude the possibility that at least some of the observed RNA duplexes in RV-B14 may correspond to the stem regions of stem-loops present in the encapsidated ssRNA genome. However, they suggest that, in RV-B14, capsid-RNA genome interactions directly involve complementary nucleotide sequences in dsRNA duplexes, and not ssRNA loops defining a consensus sequence. These duplexes provide an extensive, symmetrical network of interactions between a dodecahedral cage of dsRNA and equivalent pockets in the inner surfaces of the capsid proteins.
The dodecahedral cage defined by RNA duplexes in RV-B14 and its interactions with the inner capsid wall are particularly reminiscent of those previously unveiled for viruses of the nodavirus family37–39. Encapsidation of heterologous, non-viral RNA in the nodaviral capsid still led to the formation of a dodecahedral dsRNA cage, suggesting that the RNA-capsid interactions driving coassembly of nodaviruses are basically independent of the sequences of the capsid-bound RNA segments38. However, the increased susceptibility to proteolysis of such nodavirus particles suggests that RNA sequence-specificity mediates their stability, at least in part. This possibility remains to be investigated for RV. How are the dsRNA filaments in the dodecahedral cage connected in the linear ssRNA molecule inside the RV virion? Weak, unassigned trefoil-shaped electron density around each capsid three-fold axis in the RV cryo-EM maps (Supplementary Movie 1) may correspond to the base of RNA “stalactites” similar to those that may contribute to the organization of the full-length RNA genome in nodaviruses39,40.
The basic residues that define positively charged “boxes”, where the RNA duplexes are accommodated, partly neutralize the negatively-charged RNA and most are fully conserved in the RV B species. These observations suggest that the positively-charged box around each two-fold axis may be essential not only for RNA binding, but also for the correct positioning of genomic RNA duplexes to drive virus assembly and genome packaging41.
Each RNA duplex in the RV-B14 virion links four protomer assembly intermediates that belong to two adjacent pentamers. Thus, a likely possibility is that the RNA duplexes in the RV genome may promote two successive capsid assembly stages: the binding of protomers to form pentamers, and the binding of pentamers to form the capsid. In addition, the many bridging interactions between each RNA duplex and its neighboring protein pentamers will be directly responsible for the observed substantial stabilization of the RV virion relative to the empty capsid42. Such stabilization may make the viral particle more resistant to dissociation into pentamers and, perhaps, also less prone to untimely capsid conformational rearrangements that compromise the infection process.
The results of this study indicate that the capsid-bound RNA duplexes may also have a major function during genome uncoating. In poliovirus there is clear evidence that the RNA genome exits the viral particle through one of the two-fold axes that are opened during the uncoating process43, and the same may occur in RV19,44. The RNA duplexes in the RV virion are bound to the inner face of the capsid at the two-fold symmetry axes, where Z-shaped pores (~12 Å x 29 Å) are observed in the empty capsids after the RNA was released from the virion. The average diameter of A-form dsRNA (~20 nm) is too large to allow its exit through a 12 Å x 29 Å opening. However, local resolution analyses indicate that the capsid regions around the two-fold axes have a relatively high conformational flexibility. Moreover, many of the protein loops around the pore were found structurally disorganized in the empty capsid. Thus, the capsid-bound RNA duplexes and other dsRNA regions in the ssRNA genome may not need to be fully unfolded during uncoating.
Remarkably, many of the numerous basic capsid residues that establish charge-charge interactions with the RNA duplexes in the RV virion are located within the extended N-terminal segment of VP1. During the uncoating process, the capsid conformational rearrangements induced by receptor (ICAM-1) binding and/or acidification at the endosomes lead to the externalization of this region. Its exit must involve the gradual disruption of its interactions with the negatively charged phosphate backbone of the RNA. The RNA duplex elements would, thus, be gradually freed from steric restrictions, and could even be destabilized by the establishment of electrostatic repulsions between phosphates. Together with the conformational rearrangement of Lys52 of VP2 (Supplementary Movie 4), the N-terminal end of VP1 might play an active role in “dragging” the resulting ssRNA segments towards the multiple channels observed in the empty capsid. Similarly, the rotation of Trp38 of VP2 (Supplementary Movie 4) would help push the RNA outwards of the capsid before its interaction with the unpaired base at each duplex end is fully disrupted.
A previous cryo-EM study of RV-B1418 detected, instead of 13 bp-long RNA duplexes, pairs of octanucleotide ssRNA strands bound by their ends through two bp interactions only. Like the RNA duplexes observed in the present study, those octanucleotides were bound to W2038 residues through flipped purine bases (Fig. 6). The structures of these linked ssRNA oligonucleotides18 and those of the RNA duplexes are shown superimposed in Fig. 6. The comparison suggests the possibility that the RV-B14 virion fluctuates between two conformational states that differ in the structural organization of the genomic RNA. The basal state would contain a dodecahedral cage of RNA duplexes. Upon supply of a very modest amount of energy, the RNA duplexes could partially unfold leading to the observed linked octanucleotides18 as a preliminary stage for genome uncoating. Together, these observations are consistent with the view that the RV genome may exit the capsid in a totally or partially unfolded state after the melting of capsid-bound RNA duplexes.
Fig. 6. Comparisons of different organization states for the ordered dsRNA in RV-B14.
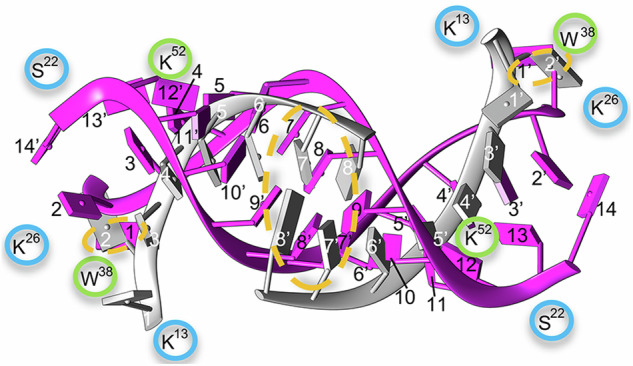
Ordered RNA regions in two initial states of the RV-B14 virion. Our model of dsRNA (pink,13 bp indicated with black numbers) is superimposed onto PDB 7BG618 (gray, 8 bp indicated using white numbers). Similar areas are highlighted by orange dashed ovals: two paired bps at the center and the non-matched base interacting with Trp38 of VP2 at the end of the RNA segments. Some interacting residues of VP1 and VP2 are shown (encircled in blue and green, respectively).
Structural classification of single channels in the ensemble of empty capsids analyzed showed that only one-third of them were (almost) fully open, while other channels were partially opened to different degrees. In our study, we considered only several discrete conformational states of the channels to obtain localized reconstructions at medium resolution, ~6–8 Å. However, a continuum of channels opened to different degrees likely exist in each individual capsid. The opening of channels and, probably, the conformational changes around each capsid two-fold axis during genome uncoating may not be a synchronous, symmetric process. Even the opening of each pore is not symmetric, as intermediate states are observed where only one side of the Z-shaped pore was open, while the other side remained closed. These results suggest an asynchronous process in which one of the pores would form more rapidly to serve as the preferential exit for the genome. Based on theoretical calculations, it had been previously proposed that, during RV genome uncoating a preferential, early transition may occur around the capsid two-fold axis closer to the 3’ end of the viral RNA, which leaves the capsid first45. Disassembly intermediates of nodaviruses (FHV) and picornaviruses may share important features: disassembly-related conformational changes are minimally global and largely local. They mainly occur at a unique two-fold axis, resulting in asymmetric particles and the release of the viral genome through a particular capsid pore without disintegration of the capsid46,47. An alternative possibility is that those asynchronous conformational changes facilitate the dissociation of one or a few capsid pentamers, opening a hole through which the RNA may exit, as found for another enterovirus (echovirus)48. Absence of one pentamer in the empty capsid (after RNA release) would lead to a reduction in electron density of 8%, which may remain undetected after icosahedral averaging.
To recapitulate, the dodecahedral cage formed by capsid-bound RNA duplexes in RV may electrostatically glue together capsid subunits, drive their association during virion assembly, facilitate the structural organization and efficient packaging of the viral genome, and stabilize the virion. In addition, a complex network of interactions between the capsid inner surface and the RNA dodecahedral cage may prevent the untimely release of the viral RNA molecule. The asynchronous opening of capsid pores may lead to the early formation of a preferential pore through which the full-length RNA molecule will be extruded.
Methods
Purification of RV-B14 virions
Virions were purified from H1 HeLa cells (American Type Culture Collection [ATCC CRL-1958]) infected with RV-B14 at a multiplicity of infection of 10 PFU/cell. Cells and supernatant were harvested at 24 h post infection, and the cell component was washed twice in PBS buffer. The cellular pellet was resuspended in 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1.5 mM MgCl2 containing 0.5% Nonidet P-40. The lysate was clarified by centrifugation (16,000 x g, 10 min, 4 °C) and the resulting supernatant was adjusted to 1% sarkosyl (sodium lauroyl sarcosinate) and 0.1% β-mercaptoethanol. This extract was mixed with the infection supernatant and processed on a 30% (wt/vol) sucrose cushion containing 0.01% BSA (285,000 x g, 130 min, 4 °C), and the resulting pellet was resuspended in PBS with 0.01% BSA and centrifuged (164,000 x g, 5 min, 4 °C). The supernatant was centrifuged in a linear (7.5–45%) sucrose gradient (285,000 x g, 105 min, 4 °C). Gradient fractions containing viral particles were dialyzed against the PBS.
Cryo-EM data collection
Purified virions (~4 μL) were applied onto R2/2 300 mesh copper grids with continuous carbon layer that was glow discharged for 15 s at 25 mA (K100X, Emitech), and vitrified using a Vitrobot Mark IV cryofixation unit (Thermo Fisher Scientific). Data were collected on a Talos Arctica electron microscope (Thermo Fisher Scientific) operated at 200 kV, and images recorded with a Falcon III detector operating in linear mode, using the EPU Automated Data Acquisition Software for Single Particle Analysis (Thermo Fisher Scientific). The total number of recorded movies, nominal magnification, calibrated pixel size at the specimen level, total exposure, exposure per frame, and defocus range for each specimen are described in Supplementary Table 1.
Image processing
All image-processing steps were performed within the Scipion software framework49. Movies were motion-corrected and dose-weighted with Motioncor250. Aligned, non-dose-weighted micrographs were then used to estimate the contrast transfer function with Ctffind451. All subsequent image processing steps were performed using Relion 2.152,53. Using an automated particle picking with Xmipp, a total of 473,724 particles were extracted and normalized. 2D classification was performed with Relion and 313,305 and 8381 RV-B14 full and empty particles were selected to perform a 3D classification imposing icosahedral symmetry, and using a model obtained from preliminary datasets low-pass filtered to 40 Å resolution as initial model. The best particles were included in 3D auto-refinement imposing icosahedral symmetry; yielding maps with an overall resolution at 2.89 Å (full), and 3.77 Å (empty) based on the gold-standard (FSC = 0.143) criterion. Local resolution was estimated using MonoRes54.
The 3D map of full particles, obtained after imposing icosahedral symmetry, showed well-defined densities at the two-fold axes compatible with ds RNA segments. To check the authenticity of these dsRNA segments the following classifications without imposing icosahedral symmetry were performed: (1) C1 classification of original images using as initial model the icosahedral capsid, in which the RNA densities were removed computationally with a mask based on the capsid contour; (2) C1 classification of difference images obtained after capsid subtraction, and using as initial model the 3D map of the genome without capsid, which was obtained in the C1 classification described in (1) that corresponds to 61% of the particles; and (3) C1 classification of difference images obtained after capsid subtraction, using as initial model only the map corresponding to the RNA density obtained in the icosahedral reconstruction. In the three different classifications, similar densities assigned to dsRNA are observed in the asymmetric maps (Supplementary Fig. 3).
To evaluate the pores in the 3D map of the empty particles, local areas of these regions in the original images were extracted and treated as asymmetric single particles (or subparticles) following the localized reconstruction method55 using the LocalRec plugin available in the Scipion software framework56. The extracted subparticles were classified into 5 classes without angular assignment.
Model building and refinement
The crystal structure of RV-B14 (PDB ID: 1NCQ) that includes the four capsid proteins VP1, VP2, VP3, and VP4 was used as template to build a homology model. The model was first manually docked as a rigid body into the density and followed by real space fitting with the Fit in Map routine in UCSF Chimera57. The model was then manually adjusted in Coot58 to optimize the fit to the density. Then, real space refinement was performed in Phenix59 with global minimization, local grid search and atomic displacement parameter options selected. These refinements were made first with the icosahedral asymmetric unit and then with the whole capsid. The quality of the obtained model was assessed with Molprobity60 as implemented in Phenix61 and with the Worldwide PDB (wwPDB) OneDep System (https://deposit-pdbe.wwpdb.org/deposition). Refinement statistics are listed in Supplementary Table 1. Graphics were produced using UCSF Chimera (52).
Model analysis
The electrostatic potential for the RV-B14 capsid was calculated using the Coulombic surface coloring tool available within UCSF Chimera. RNA-capsid interactions were analyzed with the Find clashes/contacts tool within UCSF Chimera. Secondary structure prediction for the ssRNA genome of RV-B14 was done with the server RNAfold of University of Vienna (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
We thank Rocío Arranz and Javier Chichón of the Cryo-EM CNB/CIB-CSIC facility (Madrid), in the context of the CRIOMECORR project (ESFRI-2019-01-CSIC-16), for help with cryo-EM data acquisition. We thank José Luis Vilas and Juan Manuel Martínez-Romero for their help in the analysis of the pores in the empty RV-B14. This work was supported by grants from the Spanish Ministry of Science and Innovation (PID2020-113287RB-I00 and PID2023-146143NB-I00) and the Comunidad Autónoma de Madrid (P2018/NMT-4389) to J.R.C., and by grants of the Spanish Ministry of Science and Innovation (RTI2018-096635-B-100 and PID2021-126973OB-I00) to M.G.M. M.G.M. acknowledges also an institutional grant from Fundación Ramón Areces. M.G.M. is an associate member of the Center for Biocomputation and Physics of Complex Systems. R.T. acknowledges funding from an EPSRC Fellowship (EP/R023204/1) and a Royal Society Wolfson Fellowship (RSWF/R1/180009). P.G.S. and R.T. were also funded by two joint Wellcome Trust Investigator Awards (224509/Z/21/Z & 224509/A/21/Z and 110145 & 110146, respectively) that also provided funding for C.P.M. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions
A.R.-H. and L.V. purified viruses; D.G.-C. acquired the cryo-EM images; D.G.-C. and C.P.M. processed images and built the models; D.G.-C., C.P.M., L.V., A.V., R.T., P.G.S., M.G.M. and J.R.C. analyzed data; M.G.M. and J.R.C. conceived, coordinated the project, obtained funding for the study, and wrote the paper with inputs from all authors.
Peer review
Peer review information
Communications Biology thanks Vidya Mangala Prasad, Antonio Real-Hohn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Manidipa Banerjee and Mengtan Xing. A peer review file is available.
Data availability
The atomic coordinates and cryo-EM density maps were deposited in the Protein Data Bank and EM Data Bank with codes 8PNF and EMD-17781 for the full RV-B14 virion, and 8PNB and EMD-17780 for the empty RV-B14 capsid.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mauricio G. Mateu, Email: mgarcia@cbm.csic.es
José R. Castón, Email: jrcaston@cnb.csic.es
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07213-2.
References
- 1.Weston, M. & Frieman, M. B. In Encyclopedia of microbiology (ed. Schmidt, T.) 4th edn 85-101 (Elsevier, 2019).
- 2.Andrup, L., Krogfelt, K. A., Hansen, K. S. & Madsen, A. M. Transmission route of rhinovirus - the causative agent for common cold. A systematic review. Am. J. Infect. Control51, 938–957 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Jacobs, S. E. et al. Clinical and molecular epidemiology of human rhinovirus infections in patients with hematologic malignancy. J. Clin. Virol.71, 51–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershenson, M. B. In The Picornaviruses (eds Ehrenfeld, E., Domingo, E., & Roos, R.) 369-381 (ASM Press, 2010).
- 5.Jacobs, S. E., Lamson, D. M., St George, K. & Walsh, T. J. Human rhinoviruses. Clin. Microbiol. Rev.26, 135–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Royston, L. & Tapparel, C. Rhinoviruses and respiratory enteroviruses: not as simple as ABC. Viruses8. 10.3390/v8010016 (2016) [DOI] [PMC free article] [PubMed]
- 7.Ljubin-Sternak, S. & Mestrovic, T. Rhinovirus–a true respiratory threat or a common inconvenience of childhood? Viruses15. 10.3390/v15040825 (2023) [DOI] [PMC free article] [PubMed]
- 8.Rossmann, M. G. et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature317, 145–153 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Arnold, E. & Rossmann, M. G. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 A. J. Mol. Biol.211, 763–801 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Waris, M. & Ruuskanen, O. In Encyclopedia of virology Vol. 2 (eds Bamford, D. H. & Zuckerman, M.) 757−764 (Elsevier., 2021).
- 11.Fuchs, R. & Blaas, D. Productive entry pathways of human rhinoviruses. Adv. Virol.2012, 826301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bochkov, Y. A. et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc. Natl. Acad. Sci. USA112, 5485–5490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossmann, M. G. Viral cell recognition and entry. Protein Sci.3, 1712–1725 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casasnovas, J. M. In Structure and physics of viruses, subcellular biochemistry Vol. 68 (ed Mateu, M. G.) 441-466 (Springer, 2013).
- 15.Blaas, D. Viral entry pathways: the example of common cold viruses. Wien. Med. Wochenschr.166, 211–226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neubauer, C., Frasel, L., Kuechler, E. & Blaas, D. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology158, 255–258 (1987). [DOI] [PubMed] [Google Scholar]
- 17.Casasnovas, J. M. & Springer, T. A. Pathway of rhinovirus disruption by soluble intercellular adhesion molecule 1 (ICAM-1): an intermediate in which ICAM-1 is bound and RNA is released. J. Virol.68, 5882–5889 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrebik, D. et al. ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating. Proc. Natl. Acad. Sci. USA118, 10.1073/pnas.2024251118 (2021) [DOI] [PMC free article] [PubMed]
- 19.Garriga, D. et al. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog.8, e1002473 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickl-Herk, A. et al. Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle. Proc. Natl. Acad. Sci. USA110, 20063–20068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilek, G. et al. Liposomal nanocontainers as models for viral infection: monitoring viral genomic RNA transfer through lipid membranes. J. Virol.85, 8368–8375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panjwani, A. et al. Capsid protein VP4 of human rhinovirus induces membrane permeability by the formation of a size-selective multimeric pore. PLoS Pathog.10, e1004294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nugent, C. I. & Kirkegaard, K. RNA binding properties of poliovirus subviral particles. J. Virol.69, 13–22 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, P., Liu, Y., Ma, H. C., Paul, A. V. & Wimmer, E. Picornavirus morphogenesis. Microbiol. Mol. Biol. Rev.78, 418–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shakeel, S. et al. Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis. Nat. Commun.7, 11387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchida, T., Sasaki, M., Tanaka, Y. & Ishimori, K. A dye-decolorizing peroxidase from vibrio cholerae. Biochemistry54, 6610–6621 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Zhu, L. et al. Structure of Ljungan virus provides insight into genome packaging of this picornavirus. Nat. Commun.6, 8316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shakeel, S. et al. Genomic RNA folding mediates assembly of human parechovirus. Nat. Commun.8, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandler-Bostock, R. et al. Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts. PLoS Pathog.16, e1009146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Twarock, R. & Stockley, P. G. RNA-mediated virus assembly: mechanisms and consequences for viral evolution and therapy. Annu. Rev. Biophys.48, 495–514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luque, D. & Caston, J. R. Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol.16, 231–239 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Montiel-Garcia, D. et al. VIPERdb v3.0: a structure-based data analytics platform for viral capsids. Nucleic Acids Res.49, D809–D816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Y. et al. Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds. J. Virol.78, 11061–11069 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luque, D. et al. Equilibrium dynamics of a biomolecular complex analyzed at single-amino acid resolution by cryo-electron microscopy. J. Mol. Biol.435, 168024 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Knobler, C. M. & Gelbart, W. M. How and why RNA genomes are (partially) ordered in viral capsids. Curr. Opin. Virol.52, 203–210 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Zhang, M. et al. Optimized photochemistry enables efficient analysis of dynamic RNA structuromes and interactomes in genetic and infectious diseases. Nat. Commun.12, 2344 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher, A. J. & Johnson, J. E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature361, 176–179 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Tihova, M. et al. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J. Virol.78, 2897–2905 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, L. et al. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol.8, 77–83 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Schneemann, A. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol.60, 51–67 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Perlmutter, J. D., Qiao, C. & Hagan, M. F. Viral genome structures are optimal for capsid assembly. Elife2, e00632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valbuena, A. et al. Single-molecule analysis of genome uncoating from individual human rhinovirus particles, and modulation by antiviral drugs. Small.20, e2304722 (2024). [DOI] [PubMed] [Google Scholar]
- 43.Levy, H. C., Bostina, M., Filman, D. J. & Hogle, J. M. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol.84, 4426–4441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, X. et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol.19, 424–429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harutyunyan, S. et al. Viral uncoating is directional: exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3’-end. PLoS Pathog.9, e1003270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bostina, M., Levy, H., Filman, D. J. & Hogle, J. M. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J. Virol.85, 776–783 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azad, K. & Banerjee, M. Structural dynamics of nonenveloped virus disassembly intermediates. J. Virol.93, 10.1128/JVI.01115-19 (2019) [DOI] [PMC free article] [PubMed]
- 48.Buchta, D. et al. Enterovirus particles expel capsid pentamers to enable genome release. Nat. Commun.10, 1138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Rosa-Trevin, J. M. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol.195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D. Struct. Biol.73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilas, J. L. et al. MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure26, 337–344.e334 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Ilca, S. L. et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun.6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abrishami, V. et al. Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Prog. Biophys. Mol. Biol.160, 43–52 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Pettersen, E. F. et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr.60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Adams, P. D. et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr.66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr.66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci.27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
The atomic coordinates and cryo-EM density maps were deposited in the Protein Data Bank and EM Data Bank with codes 8PNF and EMD-17781 for the full RV-B14 virion, and 8PNB and EMD-17780 for the empty RV-B14 capsid.