Abstract
Variably Protease Sensitive Prionopathy (VPSPr) is a rare human prion disease that, like Creutzfeldt-Jakob disease (CJD), results in the deposition of abnormally folded prion protein aggregates in the brain and is ultimately fatal. Neuropathology and clinical features of VPSPr are heterogeneous. However, the key discriminating feature is the relative sensitivity of the pathological prion protein to proteinase digestion compared to that typically seen in other human prion cases. Three major fragments of 23, 17 and 7 kDa are characteristic of the disease following digestion with proteinase K. We recently reported the utility of the highly adaptive and reproducible ProteinSimple™ capillary electrophoresis (CE) system to perform protein separation of PK digested prion protein in CJD. Consequently, we explored capillary-based electrophoresis (CE) technology as a sensitive method to detect and characterize VPSPr in a cohort of 29 cases. The unique 7 kDa fragment has high intensity, particularly in cases with the codon 129 VV genotype, but can be missed by regular Western blotting due to the small size. However, this fragment is readily detected by CE in all cases. In addition, the flexibility of CE produced highly reproducible, semi-quantitative data for determining relative proteinase K sensitivity and epitope mapping of representative cases from each codon 129 genotype (VV, MV and MM).
Subject terms: Diseases of the nervous system, Clinical microbiology, Prions
Introduction
Human prion diseases are fatal neurodegenerative disorders caused by misfolded cellular prion proteins (PrPC) that aggregate to form brain deposits, resulting in spongiform vacuolation and neuronal loss. The most prevalent human prion disease is Creutzfeldt-Jakob disease (CJD), with a worldwide annual incidence of 1–2 cases per million individuals1. Most commonly, CJD occurs sporadically, although genetic and acquired forms also occur. In disease, PrPC, a primarily α-helical prion protein, spontaneously misfolds, taking on a pathological conformation highly enriched in β-sheet secondary structure, PrPSc, that can self-propagate2,3. Uniquely, the C-terminus of PrPSc is highly resistant to proteases, and this characteristic is exploited for diagnosing CJD, as digestion with proteinase K (PK) leads to distinctive fragment sizes based on tertiary structure and glycosylation site occupancy. A molecular classification scheme for CJD that considers the apparent molecular weight of the PK-resistant unglycosylated fragment of PrPSc (21 or 19 kDa) and the methionine (M)/valine (V) polymorphism at codon-129 (MM, VV or MV) on the prion protein gene (PRNP) has been adopted worldwide4–6. The resulting six subtypes largely correspond with disease phenotypes in sporadic CJD. However, clinicopathological features may not always correlate, and variations in PK-resistant fragment sizes and glycosylation are occasionally identified. Evidently, atypical cases representing alternative prion strains or unique subtypes exist7–11.
One such novel sporadic human prion disease is variably protease-sensitive prionopathy (VPSPr) that was first described in 2008, and cases have since been identified in USA10, UK12–16, the Netherlands17, Spain18 and Canada19. This disease is typified by an extended disease course and the presence of micro plaques in the brain as well as distinctive biochemistry, where PrPSc is more susceptible to proteolytic treatment10,17,20. A hallmark low molecular weight (7-8 kDa) band is detected by Western blot in VPSPr cases that is similar to that seen in the genetic prionopathy Gerstmann-Straussler-Scheinker syndrome (GSS); however, no mutations have been associated to VPSPr10. VPSPr is also unique from CJD in that di-glycosylated VPSPr PrPC does not convert to PrPSc, and therefore, di-glycosylated fragments are notably absent on Western blots in these cases20–22. Subsequent studies confirmed the transmissibility of VPSPr in humanized mice, albeit at reduced efficiency compared to CJD, supporting the direct role of VPSPr-derived PrPSc in disease pathology23.
We previously described a capillary-based electrophoresis system capable of high-throughput PrPSc characterization in CJD cases, which demonstrated improved sensitivity compared to traditional Western blot methods and an enhanced capacity to detect cases of VPSPr19. Here, we expand on this work to illustrate the utility of capillary electrophoresis methods in characterizing VPSPr. Using a large VPSPr cohort (29 cases), we have identified and characterized the biochemical profile of VPSPr by capillary electrophoresis across all codon 129 genotypes and indirectly investigated the structure of VPSPr-derived PrPSc by epitope mapping of select cases.
Results
Characterization of a cohort of 29 cases of VPSPr human sporadic prionopathies
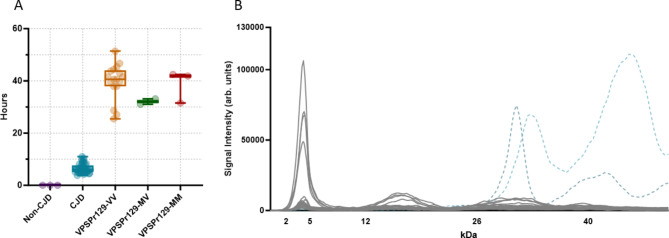
A cohort of 29 VPSPr cases from the United States of America (n = 20), the United Kingdom (n = 7), and Canada (n = 2) were used in this study. All VPSPr cases were diagnosed and defined by genotype in their respective countries before being sent to our laboratory for this study, Table 1. We confirmed that all cases contained prion seeding activity by real-time quaking-induced conversion (RT-QuIC) assays using diluted brain homogenate (10− 4). VPSPr is associated with an extended disease duration in patients and following transmission to animal models10,23. We, therefore, compared the lag phase (the time from the start of the assay to the point at which seed-dependent amyloid formation reached a threshold for detection) with equivalent data from sporadic CJD patients. Seeding was uniformly slow in all 29 VPSPr patients tested (25–50 h) versus 5–11 h for sCJD, Fig. 1A.
Table 1.
Cohort codon 129 genotype and disease duration.
| Case ID Number | Originating region | Codon129 genotype | Disease duration (months) |
|---|---|---|---|
| 1 | Canada | VV | 45 |
| 2 | Canada | VV | 14 |
| 3 | UK | VV | 41 |
| 4 | UK | VV | ~ 90 |
| 5 | UK | VV | ~ 5 |
| 6 | UK | MV | 12 |
| 7 | UK | VV | 41 |
| 8 | UK | VV | 26 |
| 9 | UK | VV | 38 |
| 10 | USA | MV | 24 |
| 11 | USA | VV | 48 |
| 12 | USA | MV | 8 |
| 13 | USA | VV | 11 |
| 14 | USA | VV | 15 |
| 15 | USA | VV | 21 |
| 16 | USA | VV | 6 |
| 17 | USA | MM | 45 |
| 18 | USA | VV | 8 |
| 19 | USA | MM | 30 |
| 20 | USA | VV | 15 |
| 21 | USA | VV | 9 |
| 22 | USA | MM | Unknown |
| 23 | USA | VV | 17 |
| 24 | USA | VV | 14 |
| 25 | USA | VV | 8 |
| 26 | USA | VV | 10 |
| 27 | USA | VV | 9 |
| 28 | USA | VV | Unknown |
| 29 | USA | VV | Unknown |
Fig. 1.
Seeding lag phase and electrophoretogram of VPSPr PrPSc immunoprobed with antibody 3F4. (A) Seeding lag phases of PrPSc from non-CJD, sCJD, and VPSPr (all genotypes) 10% (w/v) brain homogenates at a 10− 4 dilution were seeded with recombinant full-length hamster PrPC using the real-time quaking-induced conversion (RT-QuIC) assay. Time from the assay start time to when a pre-defined amyloid formation threshold was reached (lag phase time) was calculated and plotted along with bars showing the min–max values. sCJD (n=33) displayed with a lag phase time ranging from 5 to 11 hours. VPSPr (n=20) displayed a longer lag phase, ranging from 25 to over 50 hours to reach the threshold. (B) Overlay of electrophoretograms from 29 VPSPr Cases (grey) and individual representative type 1 and 2 CJD cases (light and dark blue dotted lines, respectively) PK-digested (25 µg/ml) and immunoprobed with antibody 3F4 using the CE immunoassay.
We previously described the capillary electrophoresis assay as a convenient and accurate way to detect the low molecular weight 7 kDa PK-resistant fragment that is stereotypical of VPSPr cases19. As previously noted, the relative molecular weights of fragments determined by the CE immunoassay are not directly comparable to those determined by western immunoblotting19,24–26. However, bands corresponding to fragments confirmed by western blot can be deduced by observing trends in fragmentation patterns and establishing the glycosylation state of observed PK-resistant species. Phosphotungstate anion (PTA)-precipitated and PK-digested brain homogenates from 29 VPSPr cases were analyzed using antibody 3F4, which recognizes amino acids 106–112 of human PrP. The small fragment with an apparent molecular weight of 4 kDa on capillary electrophoresis (corresponding to the 7 kDa fragment by western blot) was consistently detected in all 29 cases, Fig. 1B. This was independent of the codon 129 genotype, of which 23 cases were VV, 3 were MV, and 3 were MM. Treatment of PK-digested samples with PNGase confirmed the identity of higher molecular weight mono- and unglycosylated species, Figs. 2A, 3A, 4A.
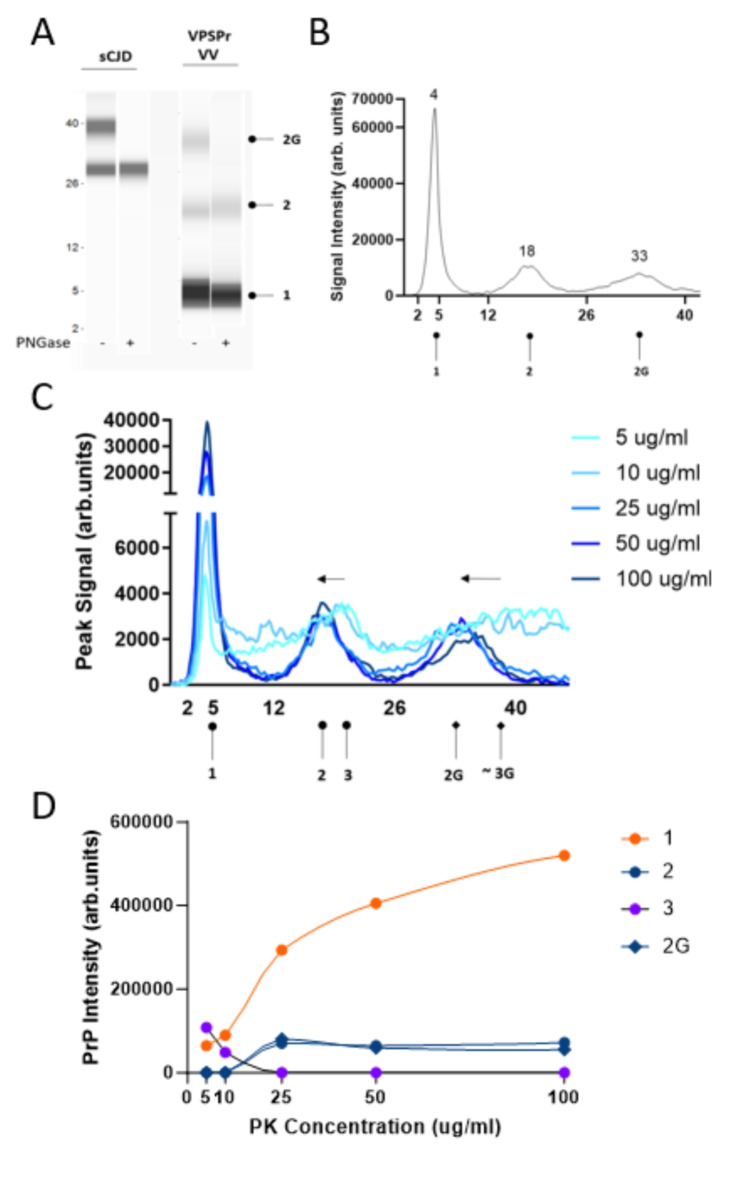
Fig. 2.
Electrophoretic profiles of VPSPr129-VV fragments and their sensitivity to proteinase-K digestion. (A) Mock “western blot view” of sCJD and VPSPr129-VV digested with 25 µg/ml of PK treated with or without PNGase using the CE assay. Circle heads and labels are used to annotate VPSPr fragments: “1” is the C-terminal truncated unglycosylated fragment, “2” is the larger C-terminal truncated unglycosylated fragment, “2G” is the monoglycosylated form of “2”, “3” is the full-length unglycosylated fragment (not detected at 25 µg/ml, shown in Panel C), and “3G” is the monoglycosylated form of “3” (not detected at 25 µg/ml, shown in Panel C) (B) Electrophoretogram of VPSPr129-VV digested with 25 µg/ml of PK (C) Electrophoretogram of PK-titration assay of a VPSPr129-VV case. The arrows demonstrate the shift from a higher molecular weight to a lower molecular weight due to the truncation of fragments as PK concentrations increase. While we did observe a rough signal where the band “3G” was anticipated to be located, the signal was too faint to indicate an exact molecular weight. (D) Graph showing the PrP signal intensity change of each VPSPr fragment throughout the PK titration assay.
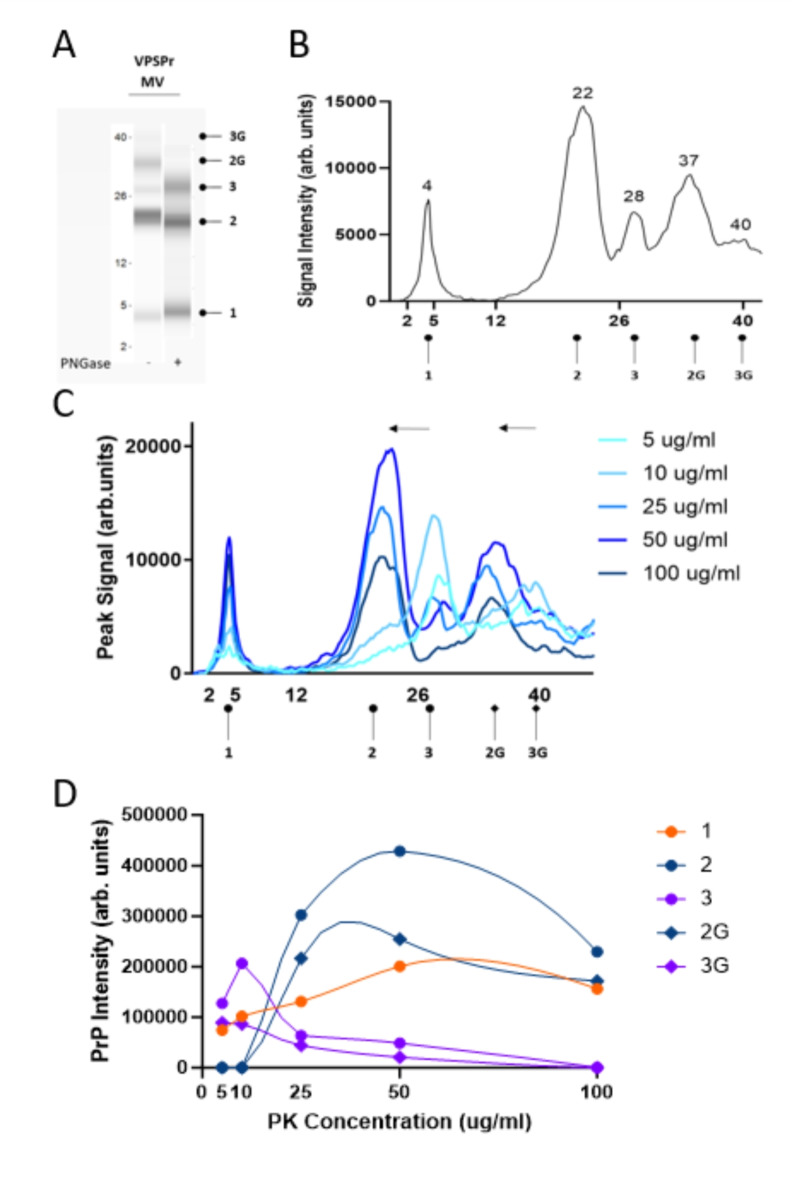
Fig. 3.
Electrophoretic profiles of VPSPr129-MV fragments and their sensitivity to proteinase-K digestion. (A) Mock “western blot view” of VPSPr129-MV digested with 25 µg/ml of PK treated with or without PNGase using the CE assay. Fragments are labelled according to convention described in Figure 2. (B) Electrophoretogram of VPSPr129-MV digested with 25 µg/ml of PK (C) Electrophoretogram of PK-titration assay of a VPSPr129-MV case. The arrows demonstrate the shift from a higher molecular weight to a lower molecular weight due to the truncation of fragments as PK concentrations increase (3 → 2 and 3G → 2G). (D) Graph showing the PrP signal intensity change of each VPSPr fragment throughout the PK titration assay.
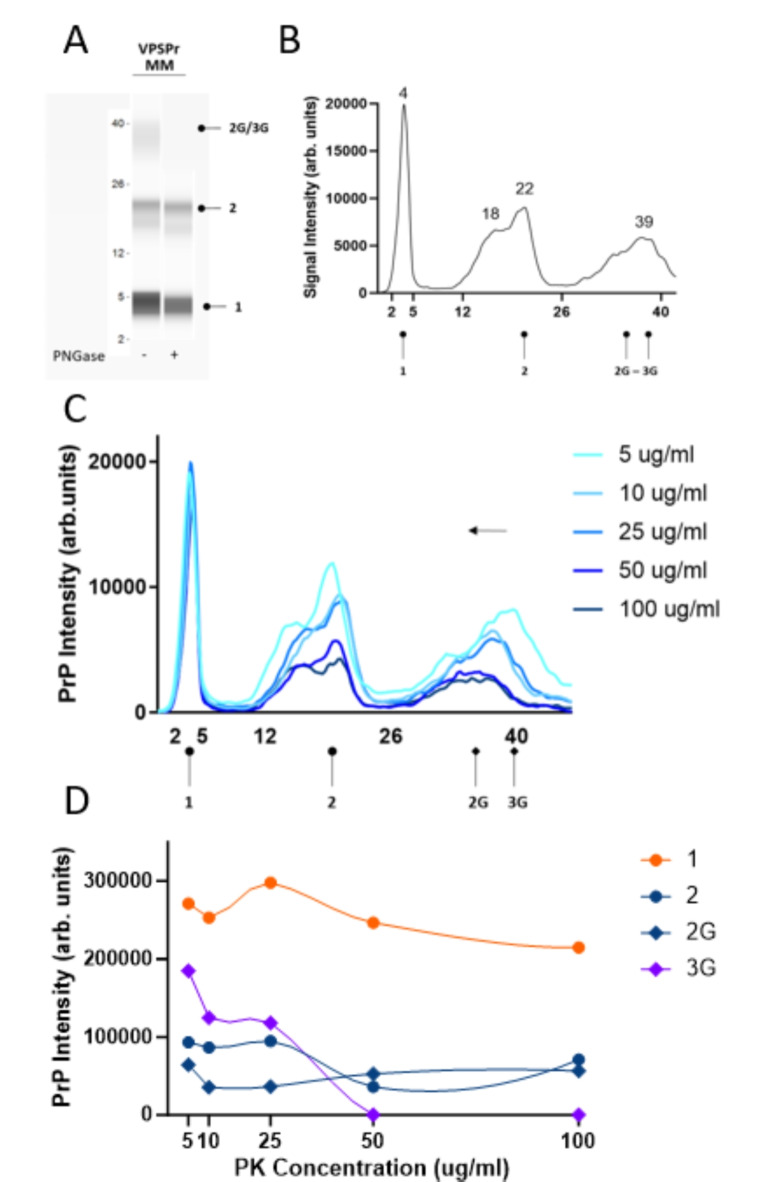
Fig. 4.
Electrophoretic profiles of VPSPr129-MM fragments and their sensitivity to proteinase-K digestion. (A) Mock “western blot view” of VPSPr129-MM digested with 25 µg/ml of PK treated with or without PNGase using the CE assay. Fragments are labelled according to convention described in Figure 2. (B) Electrophoretogram of VPSPr129-MM digested with 25 µg/ml of PK (C) Electrophoretogram of PK-titration assay of a VPSPr129-MM case. The arrows demonstrate the shift from a higher molecular weight to a lower molecular weight due to the truncation of fragments as PK concentrations increase (3G → 2G) (D) Graph showing the PrP signal intensity change of each VPSPr fragment throughout the PK titration assay.
Characterization of VPSPr fragments according to genotype VPSPr129-VV
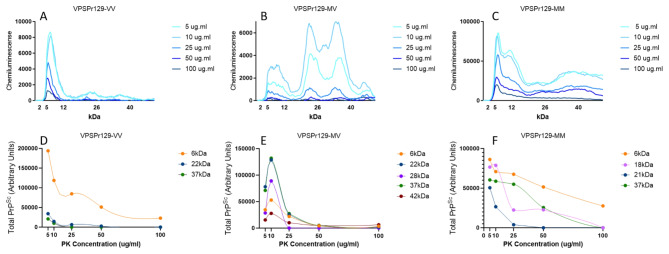
VPSPr129-VV cases presented with three bands at apparent molecular weights by CE of 30-33 kDa, 18 kDa, and 4 kDa when probed with antibody 1E4, corresponding to N-terminal truncated monoglycosylated and unglycosylated fragments, and the smallest truncated unglycosylated fragment at 4 kDa, Fig. 2A,B. The truncated 1E4 reactive mono- and un-glycosylated fragments are faint or absent in the VPSPr129-VV cases when treated with a final PK concentration of 25 µg/ml10,21. To more carefully assess the protease sensitivity of VPSPr129-VV fragments, titration assays were performed by treating PTA-precipitated brain homogenate with PK at final concentrations ranging from 5 µg/ml to 100 µg/ml and, immunoprobing with 1E4, Fig. 2C. The 4 kDa band increased in intensity as the PK concentration increased. The higher molecular weight peaks at 18 and 33 kDa, representing un- and mono-glycosylated truncated species, respectively, were undetectable until PK concentrations reached 25 µg/ml, where the signal intensity remained relatively constant as PK concentration increased. An additional faint peak at 20 kDa was detectable at very low PK concentrations before becoming undetectable at 25 µg/ml. It is likely this PK-sensitive 20 kDa fragment represents the full-length form of the unglycosylated VPSPr fragment before shifting to the lower molecular weight seen at 17 kDa, with a complete shift to the truncated version at 25 µg/ml of PK, Fig. 2D.
VPSPr129-MV
VPSPr129-MV cases presented with 5 peaks on CE at 40 kDa, 37 kDa, 28 kDa, 22 kDa and 4 kDa corresponding to full-length and truncated monoglycosylated, full-length and truncated unglycosylated and the smallest truncated fragment, Fig. 3A,B. The shift to N-terminal truncated fragments derived from the GPI-anchored (full-length) form is evident in the case of VPSPr129-MV, Fig. 3C. At PK concentrations between 25 and 50 µg/ml, the bands previously seen at 40 kDa (monoglycosylated) and 28 kDa (unglycosylated) are replaced with bands at 37 and 22 kDa, respectively, and like the trend seen in the VPSPr129-VV sample, the 4 kDa signal intensifies slightly as the PK concentration increases, reaching its maximum intensity at 50 µg/ml of PK, Fig. 3D.
VPSPr129-MM
PK digestion of VPSPr129-MM cases produced 4 bands on CE at 37 kDa, 21 kDa, 18 kDa and 4 kDa, Fig. 4A,B. As previously described, the CE assay cannot discriminate between bands within a 1–2 kDa range of one another and instead will present as a wider peak19. Notably, the band at 37 kDa, corresponding to the monoglycosylated fragment that resolves at 23–26 kDa on traditional western blotting, presents with a broad signal and jagged peak, which may reflect multiple fragments that are close in size or heterogeneous glycan chain lengths. Similar to VPSPr129-MV, these fragments migrated slightly slower than those detected in VPSPr129-VV cases20.
PK titration experiments with VPRSr129-MM revealed that, unlike the other genotypes, the intensity of the 4 kDa peak remains relatively constant throughout the PK titration, Fig. 4C. A shift from the mono-glycosylated fragment at 40 kDa to the truncated 37 kDa form is apparent, with the 40 kDa band becoming nearly undetectable at the low PK concentration of 10 µg/ml. Interestingly, throughout the range of PK concentrations, two partially overlapping peaks are detected at 18 and 22 kDa, which both decrease in intensity as PK concentration is increased. Overall, in agreement with other findings, VPSPr129-MM appears to be the most resistant to PK degradation 11.
Epitope mapping of VPSPr using capillary electrophoresis
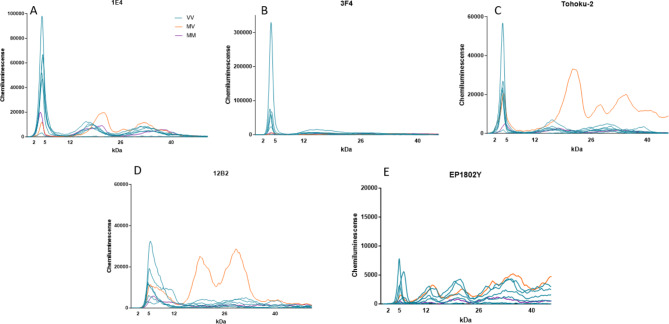
To characterize the PrPSc species present in VPSPr cases, we measured the electrophoretic profiles of a smaller group of 8 VPSPr cases for which sufficient tissue was available (6 VV129; 1 MV129, and 1 MM129) using a panel of antibodies, Fig. 5. Following PK digestion (50 µg/ml), antibodies 1E4, 3F4, and Tohoku-2 showed similar fragment migration patterns. However, the relative proportions of each fragment detected differ, which may be due to differences in antibody binding affinities or relative abundances of PK-digested fragments Fig. 6. Antibody 3F4 bound preferentially to VPSPr129-VV cases with barely any detection of the other fragments from the VPSPr129-MV and VPSPr129-MM cases. Antibodies 1E4 and Tohoku-2 have epitopes beginning at amino acids 96 and 97, respectively; therefore, it is unsurprising that these antibodies behave similarly. Notably, all three antibodies universally detected the stereotypical 4 kDa fragment. Importantly, Tohoku-2 recognizes a PrP epitope beginning precisely at residue Ser9727, which is exposed following PK digestion of typical type 2 sCJD cases, suggesting that PrPSc of VPSPr adopts conformations that at least partially resemble those present in type-2 sCJD.
Fig. 5.
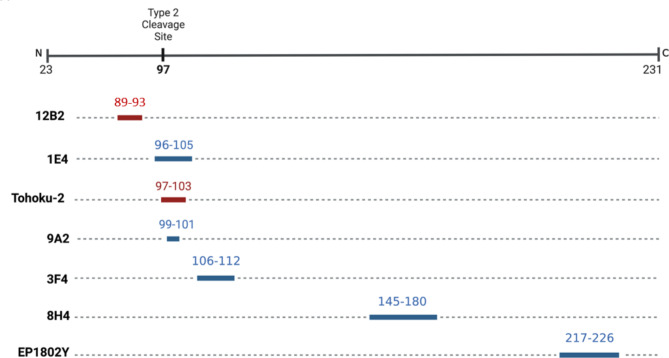
PrP-Specific Antibody Epitope Binding Regions to the Human Prion Protein. Antibodies recognizing PrP epitopes depicted in red are sCJD type-specific, 12B2 is type-1 CJD specific47 and Tohoku-2 is type-2 CJD specific48. Antibodies recognizing epitopes depicted in blue have a binding affinity to both Type 1 and 2 CJD.
Fig. 6.
Epitope Mapping of VPSPr Cases with PrP-specific Antibodies. VPSPr129-VV, -MV and -MM cases were PTA precipitated and PK digested at a final concentration of 50 µg/ml before being probed with PrP-specific antibodies using the CE assay. Electrophoretogram data was exported and graphed on Graphpad Prism to provide signal overlays (orange MV, blue VV, and purple MM) (A) 1E4 (residues 96-105), (B) 3F4 (residues 106-112), (C) Tohoku-2 (residues 97-103), (D) 12B2 (residues 89-93), and (E) EP1802Y (residues 217-226).
When probed with antibody 12B2, which recognizes residues 89–93, the 4 kDa bands observed with 1E4, 3F4 and Tohoku-2 are replaced with a slightly larger species at 6–7 kDa for all VPSPr genotypes, Fig. 6D. Since this species does not cross-react with 1E4, 3F4 and Tohoku-2 antibodies, it is likely that the N-terminal region of the 6 kDa fragment retains secondary structure during electrophoresis and occludes the 1E4/3F4/Tohoku-2 epitopes while retaining reactivity towards 12B2.
To probe the C-terminal region of VPSPr-derived PrPSc, samples were also analyzed using antibody EP1802Y, Fig. 6E. VPSPr129-VV cases probed with EP1802Y showed bands at 5 kDa, 11 kDa, 20 kDa, 27 kDa, and 37 kDa. This antibody binds to an additional fragment at 11 kDa (corresponding to the 12/13 kDa fragment observed with western blotting21,22). The VPSPr129-MV and -MM cases exhibited similar binding profiles; however, in the MV case, the 20 kDa fragment seen in the VV case was replaced by a slightly larger fragment at 22/23 kDa. EP1802Y binds all C-terminal VPSPr fragments and is therefore available to bind to the C-terminal fragment (cleavage site of a.a 158), that lacks the N-terminal epitopes the other antibodies probe for22. We also probed VPSPr cases with antibody 8H4, which recognizes an epitope central in the prion protein, binding at 145–18028. No signal was detected with this antibody in any of the VPSPr cases (data not shown). 8H4 is not commonly used to probe human prion diseases, and it is therefore possible that this antibody has a poor affinity for human prion diseases altogether. Another possible reason as to why we did not observe a signal with 8H4, is that VPSPr PrPSc may retain conformational properties that prevent the epitope region of this antibody from binding.
Protease resistance profiles of 12B2-binding PrPSc fragments
Our epitope mapping studies using antibody 12B2 identified a small 6 kDa molecular weight fragment in all VPSPr genotypes, which, to our knowledge, has not been identified in previous studies. To further investigate these findings, we performed PK-titration assays on representative cases of VPSPr from each genotype using antibody 12B2.
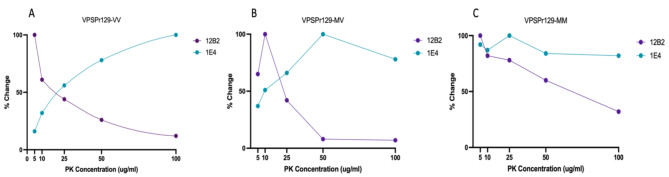
Overall, we found the fragments with in-tact epitopes binding to 12B2 were highly sensitive to PK digestion, with minimal detection of VPSPr fragments at 100 µg/ml of PK for all genotypes, Fig. 7. This differs drastically from the PK digestion pattern observed in the VPSPr fragments using antibody 1E4. This difference is most pronounced in VPSPr129-VV cases when analyzing the lowest molecular weight fragments (4 kDa for 1E4 and 6 kDa for 12B2). The rates at which the 4 kDa intensity increases as PK intensifies and the rate at which the 6 kDa intensity decreases as PK concentrations increase are inversely associated with one another, Fig. 8A.
Fig. 7.
Analysis from PK-Titration of PrPSc Fragments from VPSPr129-VV, -MV, and -MM Genotypes Immunoprobed with PrP-Antibody 12B2. Equal concentrations of PTA-precipitated 10% brain homogenate were treated with PK concentrations ranging from 5 µg/ml to 100 µg/ml followed by capillary-electrophoresis to analyze conformational stability of the different VPSPr fragments from (A) VPSPr129-VV, (B) VPSPr129-MV and (C) VPSPr-MM. (D), (E), & (C) Signal intensity changes for each VPSPr fragment was evaluated for each genotype.
Fig. 8.
Rate of Change During PK-Titration of Low Molecular Weight VPSPr Fragments Bound to PrP-Specific 1E4 and 12B2. Rates of change were calculated for the smallest VPSPr fragment for (A) VPSPr129-VV (B) VPSPr129-MV and (C) VPSPr129-MM. Each graph shows the rates of change when the smallest molecular weight VPSPr fragment was immunoprobed with 1E4 (residues 96–106) and 12B2 (89–93) for a single representative sample.
Interestingly, this association appears to be genotype-specific. The VPSPr129-MV case appeared to behave similarly to the rates of change observed with VPSPr129-VV. However, in VPSPr129-MV, both low molecular weight fragments increased at low PK concentrations before displaying an inverse relationship, Fig. 8B. Conversely, the VPSPr129-MM case showed nearly identical rates of change of the lowest molecular weight fragments (4 kDa and 6 kDa), Fig. 8C. The inverse association of these low molecular weight fragments in VPSPr129-VV and MV cases suggests that in these genotypes the 6 kDa fragment with epitope availability to 12B2 is digested by PK (N-terminal digestion), resulting in a secondary structure change that exposes the 1E4 epitope in a truncated version at 4 kDa. The 12B2-reactive epitope of VPSPr, therefore, appears to exist transiently and is rapidly digested in the presence of PK.
Discussion
Classification of proteinase-K-resistant prion proteins through western blotting is the gold standard assay for providing diagnostic information in post-mortem CJD cases. We previously evaluated the utility of a capillary-electrophoresis system to classify CJD post-mortem and found the assay accurately described CJD cases with remarkable sensitivity19. The benefits of this assay include a short preparation and running time (~ 4 h total), a small volume of sample required for analysis (maximum of 3 µl sample per capillary), and a semi-quantitative read-out allowing for a reproducible, comparative analysis across samples29. While western blotting and capillary electrophoresis both involve the immunodetection of denatured proteins separated by size, key differences may affect the appearance of banding patterns. In western blotting, an abundance of protein is loaded into each well of a gel and probed with a small concentration of antibodies to avoid over-saturation of signal on the membrane. The CE assay is the exact opposite, where a minute amount of sample is loaded into a capillary, along with an excess of antibody to probe the samples. These differences will alter the kinetics of immunodetection, with signal detection more representative of the amount of protein loaded rather than the relative affinity of each antibody as with Western blotting. Therefore, while it is expected that both techniques should identify the same fragments, the relative intensities of each fragment may appear different with each methodology. Given the high sensitivity of the CE assay, proteins and peptides at low concentrations may be detected more readily than by Western blot. Each assay has advantages and disadvantages. Whilst CE can provide semi-quantitative data with greater consistency than the western blot, the lack of resolution between proteins that are very close in molecular weight can be a caveat to the CE method29–32.
VPSPr was first reported in 2008 and is the latest human prion disease to be identified. However, given its rarity, clinical and histopathologic heterogeneity, and the prion protein’s unusual sensitivity to protease digestion, the detection and characterization of this disease is challenging. VPSPr can mimic many other neurodegenerative diseases, and a diagnosis requires post-mortem immunohistochemistry and biochemical analysis of the pathogenic prion protein (PrPSc) through western blotting16,33. However, traditional western blotting techniques that can be used to characterize other human prion diseases are inadequate to study VPSPr PrPSc, and stringent conditions are typically applied15,20,21. This is primarily due to its PrPSc sensitivity to protease-K and the small size of its fragments (< 25 kDa on western blot). It is, therefore, expected that VPSPr cases have been underdiagnosed in the past20,34. Given the sensitivity of the CE assay, we explored the utility of this assay to characterize a large cohort of VPSPr cases from multiple centers with this new methodology.
To supplement the characterization of VPSPr cases in this study using CE, we first evaluated the seeding lag phases of all cases. Consistent with previous studies15,21,35, the lag phases of PrPSc from VPSPr patients is slower than that observed with sCJD PrPSc. The slow PrPSc seeding observed in VPSPr cases support the theory that VPSPr prions represent an “incompetent” prion strain in humans and that the relatively slow aggregation results in extended disease durations seen in patients36. Furthermore, previous research has shown inefficient VPSPr transmission to transgenic mice and bank voles, which may be due to the inability or reduced capacity of VPSPr prions to seed aggregation in a new host23,37.
When analyzing VPSPr using the CE assay, VPSPr fragments were consistently detected, demonstrating its robustness in this context. Notably, the smallest VPSPr fragment at 4 kDa (7 kDa on western blot) was prominently detected in all cases, regardless of PRNP genotype or PK concentration. Also, the 4 kDa fragment was easily detectable with the antibody 3F4 in all samples, which contrasts with previous reports using western blotting techniques, where fragments were barely detectable or undetectable with the same antibody21. This underscores one of the key advantages of CE detection, as the excess of the 3F4 antibody applied to the capillary enhances the detection of the small fragment, despite its lower affinity compared to other antibodies.
The final concentration of PK applied to samples significantly influenced the appearance of VPSPr fragments across PRNP genotypes. Consistent with previous reports, we found VPSPr129-MM PrPSc to be the most resistant to protease K digestion, VPSPr129-VV the most sensitive, and VPSPr129-MV an intermediate between the two10,20. The 4 kDa fragment in the VPSPr129-VV case increased in intensity as PK concentrations increased, while the other fragments were barely detectable, even at low PK concentrations. The 4 kDa fragment of the VPSPr129-MV case behaved similarly to the VV genotype; however, unlike VPSPr129-VV, the higher molecular weight fragments were generally more prominent than the 4 kDa fragment, and these fragments migrated to slightly lower molecular weights as PK increased, presumably from fragment truncation. The VPSPr129-MM case presented unique characteristics, with the signal intensity of the 4 kDa fragment seemingly unaffected by increasing PK concentrations and the higher unglycosylated fragment decreasing slightly in intensity as PK intensified, but with no apparent truncation in response to increasing PK concentrations. Lastly, the monoglycosylated fragment in VPSPr129-MM shifted to a lower molecular weight, indicating truncation. Interestingly, each band seemed to exhibit independent PK sensitivities within this genotype.
Using an epitope mapping protocol adapted to the CE system, we characterized the population of PrPSc fragments present following PK digestion of brain tissue from confirmed cases of VPSPr. We found that for all genotypes of VPSPr, PK-resistant fragments that were reactive to 1E4 were also reactive with the type-2 CJD specific Tohoku-2 antibody. While both of these antibodies recognize overlapping PrP epitopes (Fig. 5), only Tohoku-2 recognizes the specific N-terminal PK-cleavage site exposed in type-2 CJD. Therefore, this study demonstrates that PK-resistant PrPSc fragments isolated from all genotypes of VPSPr patients at least in-part resemble those fragments found in type-2 CJD.
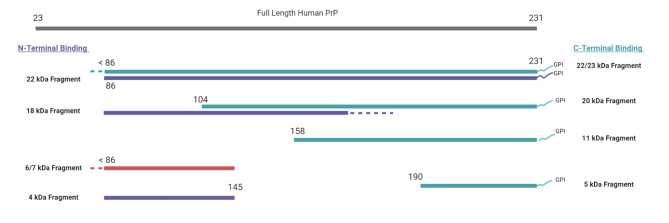
Perhaps the most surprising finding from this research is the detection of a novel VPSPr fragment which exclusively bound to antibody 12B221. Interestingly, the 4 kDa band observed with all other antibodies (except for EP1802Y) was replaced with a slightly higher fragment at 6 kDa when probing with 12B2. When we exposed this fragment to increasing PK concentrations, the fragment showed sensitivity to PK and was barely or un-detectable at final PK concentrations of over 25 µg/ml. Furthermore, in VPSPr129-VV and –MV, the rate at which the 4 kDa fragment (bound to 1E4) increased in intensity as PK concentrations increased was roughly inverse to the rate at which the 6 kDa fragment (bound to 12B2) disappeared. However, this trend was not observed with VPSPr129-MM, which further emphasizes the unique features of PrPSc from VPSPr129-MM cases. Based on this data, we propose this 6 kDa fragment may be a precursor to the 4 kDa fragment prior to truncation. The generation of VPSPr fragments has previously been linked to differences in C-terminal truncation from PK21, yet our findings suggest additional truncation N-terminal to amino acid 86 also occurs, as the N-terminal 12B2 epitope is lost as PK concentrations intensify. To validate this theory, SAF70 (residues 156–161) was used to probe the C-terminal region adjacent to the 4 kDa cleavage site (residue 145) of VPSPr PrPSc (data not shown). There was no detection of the 6 kDa fragment regardless of PK concentration, thus supporting the N-terminal extension in the 6 kDa VPSPr fragment. To our knowledge, this is the first report of a precursor to the lowest VPSPr fragment, and it is only detectable with antibody 12B2. While 12B2 has previously been applied to VPSPr21,22, it is possible that slight differences at the extreme end of the molecular weight range may not be detectable using standard western blotting protocols, or due to the extreme PK-sensitivity of these fragments, western blotting may lack adequate sensitivity for detection38. Based on these results, we generated an updated diagram of the proposed PK-resistant fragments of VPSPr, building on the work conducted by Zhang et al.21, Fig. 9.
Fig. 9.
Schematic Diagram of VPSPr-Associated PK-resistant PrPSc Fragments, Adapted from Zhang et al21. The gray bar at the top represents full length human PrP (23–231). The purple bars represent fragments bound to N-terminal antibody 1E4 (96–105), the green bars represent fragments bound to C-terminal antibody EP1802Y (217–226), and the red fragment represents the fragment exclusively bound to N-terminal antibody 12B2 (89–93). VPSPr fragments with known cleavage sites are labelled with an amino acid cleavage site number21, whereas dotted lines represent the regions of currently unknown cleavage sites.
Taken together, this study provides insights into the utility of using the CE assay for analyzing VPSPr PrPSc. This system provides an automated method for the reliable detection of VPSPr fragments, particularly those with very low abundances. While additional experiments are necessary to validate this assay as a robust diagnostic tool, the results presented here can serve as preliminary data to support further experiments using the CE system to analyze VPSPr heterogeneity and molecular mechanisms.
Methods and materials
Samples and ethics approval
Confirmed VPSPr cases were sampled from the United States, United Kingdom, and Canada. The specimens were collected from frozen brain tissue with the appropriate consent for use in research (Health Canada-Public Health Agency of Canada Research Ethics Board reference numbers REB 2009-036P and REB 2017-009P). All UK human tissue samples examined in this study were provided by the National CJD Research & Surveillance Unit (NCJDRSU), which is part of the MRC Edinburgh Brain & Tissue Bank (Edinburgh Brain Bank 16-ES-0084).
Tissue preparation
100 mg of brain tissue was placed in an OMNI International™ Biomasher filter tube and homogenized using a disposable homogenizer column and pestle. The column was centrifuged at 14,500 rpm for 30 s to filter out solid particulate. Following the removal of the filter of the column, 900µls of homogenization buffer (100 mM NaCl, 10 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate, 100 mM Tris-HCl, pH 8.0) was added to the tissue to yield a 10% brain homogenate. The homogenate was vortexed for one minute and centrifuged at 15,000xg for 10 min. The supernatant was removed and stored at −80 °C.
Antibodies
The following monoclonal antibodies were used: 3F4 which recognizes residues 106–112 (dilution 1:20)39, 1E4 which recognizes residues 96–105 (dilution 1:20)40, 12B2 which recognizes residues 89–93 (dilution 1:20)41, 8H4 which recognizes residues 145–180 (dilution 1:10)42, EP1802Y which recognizes residues 217–226 (dilution 1:20)43 and the antibody Tohoku-2, which was graciously gifted to our lab by Dr. Tetsuyuki Kitamoto from the University of Tohoku, this antibody binds to human PrP residues 97–103 (dilution 1:10 dilution)27 .
PTA precipitation, quantification, and PK digestion
Prion proteins were precipitated with sodium phosphotungstate anion (NaPTA) as described44,45, with modifications. Briefly, 10% brain homogenate is combined with benzonase (Millipore) at a final concentration of 0.5% and sarkosyl at a final concentration of 2%. The solution was incubated at 37 °C, shaking at 850 rpm for 2 h. 10% PTA was prepared in 18Ω water and adjusted to a pH of 7.0 with NaOH. NaPTA was added to the brain homogenate at a final concentration of 2% (v/v) and returned to incubate under the same conditions previously stated overnight. Next, the sample was centrifuged at 13,200 x g for 30 min. The supernatant was removed and the pellet was resuspended in 2% sarksoyl and 2% NaPTA. The sample was then incubated at 37 °C shaking at 850 rpm for 1 h, followed by centrifugation for 30 min at 13,200 × g. Finally, the supernatant was removed, and the pellet was resuspended in 1X DPBS. PTA-precipitated prion proteins were quantified using Invitrogen’s Qubit Fluorimeter assay. Prion proteins were diluted to a final concentration of 1.25 µg/µL in DPBS. Samples were then digested with varying Proteinase K (PK) concentrations for 1 h of shaking at 850 rpm. Protease digestion activity was stopped using EDTA-free protease inhibitor cocktail tablets (Roche). For samples that were subject to PNGase treatment, N-linked glycans were removed using the peptide-N-glycosidase F (PNGase F) kit (New England Biolabs) according to the manufacturer’s instructions.
Capillary Western assay
Samples were analysed using the JESS capillary electrophoresis assay (ProteinSimple) as previously described19. Briefly, digested PTA precipitated homogenates were denatured with the ProteinSimple 5X Fluorescent Master Mix (final concentration of 1% SDS) at 95 °C for 5 min with agitation at 1000 rpm. To prepare the JESS electrophoresis plate, 5µls of prepared ladder solution and 3µls of denatured sample were loaded in row A. Rows B-E were filled with Antibody Diluent, Primary antibody, Streptavidin-HRP for detection of the ladder, Secondary antibody, and Luminol-Peroxide solution for chemiluminescent detection. Each plate can load 12 or 24 samples, depending on the Separation Module ordered. The loaded plates were centrifuged at 1000 x g for 5 min to eliminate bubbles from the wells. Plates and capillaries (2–40 kDa) were loaded into the JESS equipment, and the appropriate protocol was selected using Compass for SW software. The running time of the JESS takes approximately 3 h. Data collected by Compass for SW is presented as chemiluminescent arbitrary units vs. apparent molecular weight (MW).
Real-time quaking-induced conversion assay (RT-QuIC)
10% (w/v) brain homogenate was serially diluted 9-fold in PBS enhanced with 0.04% (w/v) SDS and 0.4% (w/v) N2 supplement (Gibco). Reactions were prepared in a 96-well plate by combining 2 µl of diluted brain homogenate with 98 µl of prepared master mix for reactions with final concentrations of 1mM EDTA, 10nM ThT, 0.002% (w/v) SDS and 10 µg of hamster rPrP substrate (residues 23–231). The substrate was prepared in-house using recombinant expression in Escherichia coli, followed by purification with histidine affinity chromatography46. Brain homogenate dilutions, 10− 4 to 10− 9, were used to seed RT-QuIC reactions with 4 replicates per dilution for each sample. Brain homogenate from hamster 263 K and the PBS (mock)--infected hamsters were included for each RT-QuIC reaction plate as positive and negative technical controls, respectively. Reaction plates were sealed with optical adhesive film and then ran on FLUOstar Omega microplate readers (BMG). ThT fluorescence (450 nm excitation and 480 nm emission) was measured in 15-minute cycles of double-orbital shaking for 50 h. The threshold for ThT amplification was calculated as follows:
Average [Absorbance reading at 30 minutes, 60 minutes, and 1 hour 30 minutes] + 5 x Standard Deviation [Reading at 30 minutes, 60 minutes, and 1 hour 30 minutes]
Data was analyzed using Microsoft Excel (2016) and visualized on GraphPad (Version 9.5.1).
Acknowledgements
The authors would like to sincerely thank the Canadian Creutzfeldt-Jakob Disease Surveillance System (CJDSS) and all patients, their families, neurologists, and referring physicians for providing tissues and clinical information for research. The authors would also like to thank the Department of Pathology and Laboratory Medicine at the University of Ottawa. The National Prion Disease Pathology Surveillance Center (U.S.A) is funded by the CDC (NU38CK000486).
Author contributions
J.M. and S.B. conceptualized the study. J.M. and K.A. performed all experiments. B.B-E provided input, expertise, and assistance with experimental design and data analysis. J.M. analysed the data and prepared manuscript figures. J.M. and B.B-E wrote the manuscript draft. M.B, D.R, M.C and B.A reviewed and selected VPSPr cases for this study and provided clinical data for each case. All authors read, edited and approved the final version of this manuscript.
Data availability
All data analysis for this study is included in the manuscript. Raw RT-QuIC data or electrophoretograms of VPSPr cases are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sitammagari, K. K. & Masood, W. Creutzfeldt Jakob Disease, In: StatPearls, StatPearls Publishing, Treasure Island (FL), http://www.ncbi.nlm.nih.gov/books/NBK507860/ (Accessed 25 July 2023). [PubMed]
- 2.Kupfer, L., Hinrichs, W., Groschup, M. H. & Misfolding, P. P. Curr. Mol. Med.9 826–835. 10.2174/156652409789105543. (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Requena, J. R. & Wille, H. The structure of the infectious prion protein. Prion8, 60–66. 10.4161/pri.28368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill, A. F. et al. Molecular classification of sporadic Creutzfeldt-Jakob disease. Brain J. Neurol.126, 1333–1346. 10.1093/brain/awg125 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Gambetti, P., Kong, Q., Zou, W., Parchi, P. & Chen, S. G. Sporadic and familial CJD: classification and characterisation. Br. Med. Bull.66, 213–239. 10.1093/bmb/66.1.213 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Parchi, P. et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol.46, 224–233 (1999). [PubMed] [Google Scholar]
- 7.Cali, I. et al. Classification of sporadic Creutzfeldt-Jakob disease revisited. Brain J. Neurol.129, 2266–2277. 10.1093/brain/awl224 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Parchi, P. et al. Incidence and spectrum of sporadic Creutzfeldt–Jakob disease variants with mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol. (Berlin). 118, 659–671. 10.1007/s00401-009-0585-1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parchi, P. et al. Consensus classification of human prion disease histotypes allows reliable identification of molecular subtypes: an inter-rater study among surveillance centres in Europe and USA. Acta Neuropathol. (Berlin). 124, 517–529. 10.1007/s00401-012-1002-8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou, W. Q. et al. Variably protease-sensitive prionopathy: a new sporadic disease of the prion protein. Ann. Neurol.68, 162–172. 10.1002/ana.22094 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baiardi, S. et al. Defining the phenotypic spectrum of sporadic Creutzfeldt–Jakob disease MV2K: the Kuru plaque type. Brain awad074. 10.1093/brain/awad074 (2023). [DOI] [PMC free article] [PubMed]
- 12.Head, M. W. et al. A case of protease sensitive prionopathy in a patient in the UK. Neuropathol. Appl. Neurobiol.35, 628–632. 10.1111/j.1365-2990.2009.01040.x (2009). [DOI] [PubMed] [Google Scholar]
- 13.Head, M. W. et al. Variably protease-sensitive prionopathy in a PRNP codon 129 heterozygous UK patient with co-existing tau, α synuclein and Aβ pathology. Acta Neuropathol. (Berlin)120, 821–823. 10.1007/s00401-010-0766-y (2010). [DOI] [PubMed] [Google Scholar]
- 14.Head, M. W. et al. Variably protease-sensitive prionopathy in the UK: a retrospective review 1991–2008. Brain136, 1102–1115. 10.1093/brain/aws366 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Peden, A. H. et al. The prion protein protease sensitivity, stability and seeding activity in variably protease sensitive prionopathy brain tissue suggests molecular overlaps with sporadic Creutzfeldt-Jakob disease. Acta Neuropathol. Commun.2, 152. 10.1186/s40478-014-0152-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aizpurua, M. et al. Variably protease-sensitive prionopathy mimicking frontotemporal dementia. Neuropathology39, 135–140. 10.1111/neup.12538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen, C. et al. The first case of protease-sensitive prionopathy (PSPr) in the Netherlands: a patient with an unusual GSS-like clinical phenotype. J. Neurol. Neurosurg. Psychiatry81, 1052–1055. 10.1136/jnnp.2009.175646 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Martínez, A. B. et al. A novel form of human disease with a protease-sensitive prion protein and heterozygosity methionine/valine at codon 129: case report. BMC Neurol.10, 99. 10.1186/1471-2377-10-99 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myskiw, J. et al. Development of an automated capillary immunoassay to detect prion glycotypes in Creutzfeldt-Jakob disease. Lab. Invest. 100029. 10.1016/j.labinv.2022.100029 (2023). [DOI] [PubMed]
- 20.Baiardi, S. et al. Phenotypic heterogeneity of variably protease-sensitive prionopathy: a report of three cases carrying different genotypes at PRNP Codon 129. Viruses14, 367. 10.3390/v14020367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, W. et al. Further characterization of glycoform-selective prions of variably protease-sensitive prionopathy. Pathogens10, 513. 10.3390/pathogens10050513 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou, W. Q. et al. Prions in variably protease-sensitive prionopathy: an update. Pathogens2, 457–471. 10.3390/pathogens2030457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notari, S. et al. Transmission characteristics of variably protease-sensitive prionopathy. Emerg. Infect. Dis.20, 2006–2014. 10.3201/eid2012.140548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castle, A. R., Daude, N., Gilch, S. & Westaway, D. Application of high-throughput, capillary-based western analysis to modulated cleavage of the cellular prion protein. J. Biol. Chem.10.1074/jbc.RA118.006367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fourier, A., Dorey, A., Perret-Liaudet, A. & Quadrio, I. Detection of CSF 14-3-3 protein in sporadic Creutzfeldt-Jakob disease patients using a new Automated capillary Western assay. Mol. Neurobiol.55, 3537–3545. 10.1007/s12035-017-0607-2 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Fourier, A. et al. Development of an automated capillary nano-immunoassay-simple western assay-to quantify total TDP43 protein in human platelet samples. Anal. Bioanal. Chem.411, 267–275. 10.1007/s00216-018-1437-4 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, A. et al. Experimental verification of a traceback phenomenon in prion infection. J. Virol.84, 3230–3238. 10.1128/JVI.02387-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanusso, G. et al. Prion protein expression in different species: Analysis with a panel of new mAbs. Proc. Natl. Acad. Sci.95 8812–8816. 10.1073/pnas.95.15.8812 (1998). [DOI] [PMC free article] [PubMed]
- 29.Sormunen, A., Koivulehto, E., Alitalo, K., Saksela, K. & Laham-Karam, N. Ylä-Herttuala. Comparison of automated and traditional Western blotting methods. Methods Protoc.6, 43. 10.3390/mps6020043 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, S. C. & Posch, A. The design of a quantitative Western blot experiment. BioMed. Res. Int.2014, 361590. 10.1155/2014/361590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra, M., Tiwari, S. & Gomes, A. V. Protein purification and analysis: next generation Western blotting techniques. Expert Rev. Proteom.14, 1037–1053. 10.1080/14789450.2017.1388167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai-Kastoori, L., Schutz-Geschwender, A. R. & Harford, J. A. A systematic approach to quantitative Western blot analysis. Anal. Biochem.593, 113608. 10.1016/j.ab.2020.113608 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Gambetti, P. et al. A Novel Human Disease with abnormal prion protein sensitive to protease. Ann. Neurol.63, 697–708. 10.1002/ana.21420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou, W. Q. et al. Gambetti, variably protease-sensitive prionopathy: a new sporadic disease of the prion protein. Ann. Neurol.68, 162–172. 10.1002/ana.22094 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cannon, A. et al. Concurrent variably protease-sensitive prionopathy and amyotrophic lateral sclerosis. Acta Neuropathol. (Berlin)128, 313–315. 10.1007/s00401-014-1309-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notari, S., Appleby, B. S. & Gambetti, P. Chap. 10 - Variably protease-sensitive prionopathy, In: M. Pocchiari, J. Manson (Eds.), Handb. Clin. Neurol. 175–190. 10.1016/B978-0-444-63945-5.00010-6 (Elsevier, 2018). [DOI] [PubMed]
- 37.Diack, A. B. et al. Variably protease-sensitive prionopathy, a unique prion variant with inefficient transmission properties. Emerg. Infect. Dis.20, 1969–1979. 10.3201/eid2012.140214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bass, J. et al. An overview of technical considerations for Western blotting applications to physiological research. Scand. J. Med. Sci. Sports27, 4–25. 10.1111/sms.12702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou, W. Q. et al. PrP conformational transitions alter species preference of a PrP-specific antibody. J. Biol. Chem.285, 13874–13884. 10.1074/jbc.M109.088831 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, J. et al. Accessibility of a critical prion protein region involved in strain recognition and its implications for the early detection of prions. Cell. Mol. Life Sci. CMLS65, 631–643. 10.1007/s00018-007-7478-z (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notari, S. et al. A refined method for molecular typing reveals that co-occurrence of PrPSc types in Creutzfeldt–Jakob disease is not the rule. Lab. Invest.87, 1103–1112. 10.1038/labinvest.3700676 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Sigurdsson, E. M. et al. Anti-prion antibodies for prophylaxis following prion exposure in mice. Neurosci. Lett.336, 185–187. 10.1016/s0304-3940(02)01192-8 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Doolan, K. M. & Colby, D. W. Conformation-dependent epitopes recognized by prion protein antibodies probed using mutational scanning and deep sequencing. J. Mol. Biol.427, 328–340. 10.1016/j.jmb.2014.10.024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine, D. J. et al. Mechanism of scrapie prion precipitation with phosphotungstate anions. ACS Chem. Biol.10, 1269–1277. 10.1021/cb5006239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woerman, A. L. et al. Propagation of prions causing synucleinopathies in cultured cells. Proc. Natl. Acad. Sci. U. S. A.112 E4949-4958. 10.1073/pnas.1513426112 (2015). [DOI] [PMC free article] [PubMed]
- 46.Orrù, C. D. et al. RT-QuIC assays for prion disease detection and diagnostics, methods. Mol. Biol. Clifton NJ1658, 185–203. 10.1007/978-1-4939-7244-9_14 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Cali, I. et al. Co-existence of PrP D types 1 and 2 in sporadic Creutzfeldt-Jakob disease of the VV subgroup: phenotypic and prion protein characteristics. Sci. Rep.10, 1503. 10.1038/s41598-020-58446-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi, A. et al. Co-occurrence of types 1 and 2 PrPres in sporadic Creutzfeldt-Jakob Disease MM1, am. J. Pathol.178, 1309–1315. 10.1016/j.ajpath.2010.11.069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analysis for this study is included in the manuscript. Raw RT-QuIC data or electrophoretograms of VPSPr cases are available from the corresponding author upon reasonable request.