Abstract
There is growing interest in research on segmentation for the vestibular schwannoma (VS) and cochlea using high-resolution T2 (hrT2) imaging over contrast-enhanced T1 (ceT1) imaging due to the contrast agent side effects. However, the hrT2 imaging remains a problem of insufficient annotated data, which is fatal for building more robust segmentation models. To address the issue, recent studies have adopted unsupervised domain adaptation approaches that translate ceT1 images to hrT2 images. However, previous studies did not consider the size and visual characteristics of the target objects, such as VS and cochlea, during image translation. Specifically, those works simply performed normalization on the entire image without considering its significant impact on the quality of the translated images. These approaches tend to erase the small target objects, making it difficult to preserve the structure of these objects when generating pseudo-target images. Furthermore, they may also struggle to accurately reflect the unique style of the target objects within the images. Therefore, we propose a target-aware unsupervised domain adaptation framework, designed for translating target objects, each tailored to their unique visual characteristics and size using target-aware normalization. We demonstrate the superiority of the proposed framework on a publicly available challenge dataset. Codes are available at https://github.com/Bokyeong-Kang/TANQ.
Subject terms: Brain imaging, Computer science
Introduction
A vestibular schwannoma (VS) is a benign tumor that develops in the nerve sheath cells of the vestibular nerve1. As the VS grows, it compresses the adjacent cranial nerves and blood vessels, leading to the deterioration of hearing and vestibular functions2. Moreover, it can cause hydrocephalus by obstructing the circulation of the cerebrospinal fluid3 and develop severe adhesions with adjacent organs, such as the cochlea. Hence, it is essential to diagnose and treat a VS promptly to prevent adhesions and potential damage to associated organs4. In the diagnosis of a VS, techniques that accurately segment VS and surrounding organs5, especially the cochleas, play a key role in determining the severity of VS6.
Magnetic resonance imaging has been widely used for VS segmentation because it can clearly determine the degree of adhesion with surrounding organs, such as the cochleas4. In particular, contrast-enhanced () magnetic resonance imaging has traditionally been used for its capability to precisely visualize neural structures using a gadolinium-containing contrast agent7. Recently, owing to the potential risks associated with gadolinium-based contrast agents8, high-resolution () imaging has gained attention as an alternative to imaging with far lower associated costs and risks8.
However, because the imaging modality has been introduced for VS segmentation recently, it suffers from a relative lack of annotation labels for VS and cochleas compared with the imaging modality1. Notably, acquiring the annotation labels for images incurs additional costs because of the time-consuming and labor-intensive nature of manual annotation conducted by radiologists and physicians9. This issue significantly limits the use of deep learning (DL)-based models for automatic VS segmentation with images because the small amount of annotated data used for model training significantly degrades model performance and reliability10.
To address the label scarcity in imaging, numerous studies11–14 have been proposed; these employ unsupervised domain adaptation (UDA) by translating imaging modalities from a label-rich source domain (i.e., ) to a label-poor target domain (i.e., ). This approach allows the annotation labels from the source domain to be directly utilized for the target domain to train a segmentation model for VS and cochleas based on target images. Specifically, in this approach, pseudo- images from real images are first generated by translating the visual characteristics (or style) of images (i.e., source domain) to those of images (i.e., target domain) while preserving the anatomical structures of the source images. Subsequently, pairs of the generated pseudo- images and annotation labels on images are used to train a DL-based segmentation model for VS and cochleas based on images. Finally, the VS and cochleas in the real images are segmented with the trained segmentation model.
Earlier studies11,12 utilized basic image translation models e.g., NiceGAN15 and CUT16. However, these methods focused on generating pseudo-images that simply resembled the style of the target domain, rather than performing translation with a focus on the target object e.g., the VS and cochlea. Shin et al.13 and Dong et al.14 have enhanced these methods by integrating a segmentation decoder into the generator. This refined the generator training through the segmentation output, thereby effectively preserving the structural integrity of the VS and cochlea. In our previous study17, we further developed the UDA approach to preserve the VS and cochlea. Our method employed the query-selected attention (QS-Attn) model18, which was designed to focus on the informative regions of the source images during translation. Our method successfully preserved the structural fidelity of the VS and cochlea during image translation with contrastive learning. Our approach is effective in focusing on small and critical target objects during translation. Additionally, we adopted a multi-view approach to generate diverse images by leveraging two constraint models with distinct strengths to enhance the performance of the segmentation model. As a result, our method achieved second place in the CrossMoDA challenge 2022 with the best and second-best segmentation results for VS and cochleas, respectively.
Despite their success, existing methods11–14,17 do not consider the size and visual characteristics of the target object during image translation, both of which are important for successful image translation from to . Most existing methods11–13, including our primary work17, have adopted instance normalization (IN)19, which performs normalization within the entire image. While IN can be effective for image synthesis tasks, it has limitations for our specific task due to the small size and distinct visual characteristics of the VS and cochlea in and images. In contrast to the target objects in other medical imaging datasets such as Multi-Modality Whole Heart Segmentation (MMWHS)20, the VS and cochlea in and are extremely small, accounting for only 0.028% and 0.002% of the total volume, respectively10. Furthermore, there are significant differences in the image characteristics of and for these objects. As show in Fig. 1 (a), in images, the VS voxels appear with higher intensity compared to other tissues due to the contrast agent. Conversely, as show in Fig. 1 (b), in images, the VS is less distinguishable from surrounding tissues since the voxels of the VS have low intensity similar to surrounding tissues1,17,21. Additionally, the cochlea, which is even smaller, appears at a very low intensity in images (Fig. 1 (a)) compared with the images (Fig. 1 (b))1,17,21. For these reasons, IN contains two major limitations for our task of translating to images. Firstly, it tends to “wash away”22 the structural information of extremely small-sized target objects such as the VS and cochlea, with the cochlea being particularly affected due to its low intensity. This “wash away” effect is a significant limitation due to the failure in generating pseudo- images that preserve the detailed structures of the VS and cochlea. Secondly, it is challenging to reflect the low-intensity style of VS that appears in real images. Considering the significant style differences, such as intensity, between the VS in source and target images, tailored adjustments are necessary to reflect the low-intensity style of the VS in pseudo- images. Therefore, only applying IN without these specific adjustments can lead to a continuous presence of the high-intensity style of VS, originally observed in the source images, which is not desirable for the target images.
Fig. 1.
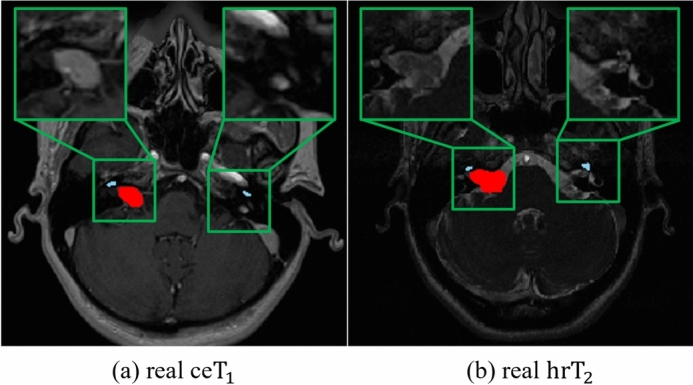
Difference in characteristics of target objects (VS and cochlea) between and images. (a) represents a real image, and (b) is a real image, with the VS highlighted in red and the cochlea in sky-blue by overlapping the segmentation map. In (a) real , the segmentation map was overlaid using the real annotation labels, while in (b) real image, since real annotation labels were not available, we used pseudo-labels for the overlap.
To alleviate these issues, we propose a novel target-aware, UDA framework that integrates a Target-Aware Normalization-based QS-Attn(TANQ). The TANQ is designed to facilitate image translation of target objects, each tailored to their distinct visual characteristics in and images. Specifically, it is designed to prevent the “wash away” effect that erases details in the small and low-intensity cochlea. We separate the cochlea region from others and perform individual normalization of its features within the region. This approach ensures the preservation of the cochlea’s fine structural details. Additionally, to address the issue of the high intensity of the VS within pseudo- images, we inject the style information from surrounding tissues into the VS during its normalization, reflecting its low-intensity in pseudo- images. This adjustment aims to match the VS’s style to the lower intensity observed in real target images. Based on target-aware normalization techniques, TANQ can enhance its ability to: 1) preserve the structural information of each target object, even for very small and low-intensity cochleas; and 2) convert the target objects to possess specific visual characteristics such as textures and intensities typically observed in real images. The TANQ model enables the generation of realistic pseudo- images with accurately preserved structures of the target objects while ensuring that the VS exhibits low-intensity characteristics, resulting in a direct enhancement of the segmentation model performance. TANQ is then integrated with our primary framework comprising multi-view image translation and self-training-based segmentation17 to further improve the performance of VS/cochlea segmentation based on unannotated images.
The key contributions of our study are outlined as follows:
We propose a target-aware unsupervised domain adaptation framework, aimed at translating target objects, with each tailored to their specific visual characteristics and size through target-aware normalization.
Based on our in-depth analysis of visual characteristics between source and target modalities, we prevented structural loss of target objects and generated higher quality pseudo- images, thereby enhancing the performance of the VS and cochlea segmentation models.
We demonstrate the superiority of the proposed method compared with other existing methods and the effectiveness of our framework through quantitative and qualitative analysis using a publicly available challenge dataset.
Related work
Unsupervised domain adaptation addresses the domain shift between source and target domains, enabling it to perform a task better in the target domain without the need for annotation labels. Several studies have adopted feature alignment23,24, which mitigates domain discrepancies by mapping the source and target images to a shared latent space and learning domain-invariant representations through adversarial learning. Chen et al.25 utilized a low-level feature refinement module and prediction map alignment to mitigate the domain gap at the feature level. In contrast, image-level alignment has been also adopted to translate the source domain into the target domain using a generative model and reducing the differences between the source and target domains in pixel-level space26,27. Kang et al.28 applied mutual information and texture co-occurrence loss to convert the source domain into the target domain, successfully performing domain adaptation for segmentation tasks. Moreover, several research studies have attempted to combine image- and feature-level alignments by generating pseudo-target images first and then applying feature-level techniques to minimize the domain gap between pseudo- and real target images29,30. Hu et al.31 proposed a domain-specific convolution module to extract domain-invariant features and employed a high-frequency reconstruction module to generate target images, thereby achieving domain adaptation.
In the medical domain, domain adaptation has been conducted to enhance the performance of downstream tasks such as segmentation, with a particular emphasis on preserving the structure of small target objects to achieve better results. Chen et al.29 leveraged synergistic learning to apply both image and feature alignment, enabling bidirectional cross-modality domain adaptation between MRI and CT images. Jiang et al.32 introduced a structure discriminator to model the co-dependency between images and their corresponding segmentations using joint probability, which helped focus on generating accurate representations of target objects. However, the target objects in these studies, such as cardiac structures and abdominal organs, are significantly larger than our target objects such as VS and cochlea. Given these distinct characteristics, there is growing recognition of the necessity for more intricate and refined processing through image-level alignment, which is now being preferred over feature-level23,24 or combination approaches29,30 in ongoing research.
In the CrossMoDA challenge, various image-level alignment approaches were utilized to translate images to images11–13. Specifically, Dong et al.11 and Choi et al.12 generated target pseudo- images from source images by utilizing NiceGAN15 and CycleGAN33, respectively. Shin et al.13 and PAST14 appended a segmentation decoder to a decoder within a CycleGAN-based generator and utilized the segmentation results in the image translation process to train the generator, thus improving the structural preservation of the VS and cochlea during image translation. Furthermore, several studies17,21,34 have enhanced the richness of pseudo-target images using a range of image translation methods for augmentation. Fgh36521 utilized cross-site and cross-modality image translation approaches using CycleGAN33 to generate diverse pseudo- images with a rule-based offline augmentation technique for domain gap mitigation. Our prior multi-view approach17 also attempted to mitigate the domain gap between and images by generating pseudo- images using two parallel constraint models. TBA34 employed CycleGAN and tumor-blending augmentation with SinGAN35 to enhance the appearance diversity of the regions of interest (e.g., the VS and cochlea) and achieve effective domain adaptation. However, despite these efforts, the structural information of the VS and cochlea was not perfectly preserved, and there still remains room for improvement in reflecting the style of VS and cochlea in real images.
Proposed method
As illustrated in Fig. 2, our proposed framework consists of three main parts: 1) TANQ-based image translation from to images, 2) multi-view pseudo- representation obtained using additional CycleGAN-based image translation in parallel, and 3) a VS/cochlea segmentation model construction based on multi-view pseudo- images and self-training with real- images.
Fig. 2.
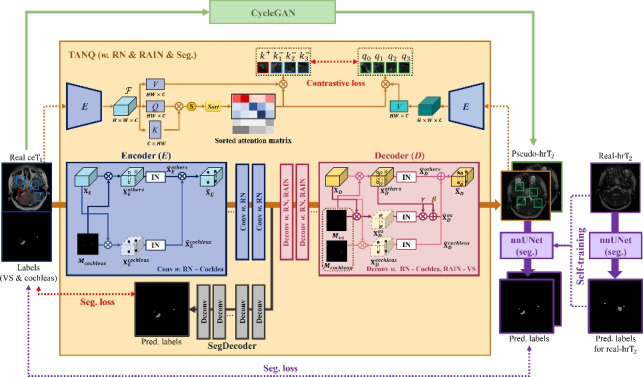
Our proposed framework consists of three main parts: 1) TANQ-based image translation from to images, 2) Multi-view pseudo- representation via CycleGAN, 3) Construction of a VS/cochlea segmentation model using multi-view pseudo- images and self-training with real- images. Specifically, TANQ divides the features based on the labels in both the encoder and decoder, applying target-aware normalization. Furthermore, it includes an additional decoder called SegDecoder. The Encoder E extracts features from both the real images and pseudo- images and then calculates the contrastive loss between selected features using a sorted attention matrix.
TANQ-based image translation
Target-aware encoder–decoder construction
In the proposed TANQ, we constructed a ResNet-based encoder–decoder architecture, followed by the original QS-Attn18. Here, instead of IN19 used in the original QS-Attn, which performs normalization using the mean and variance of the feature maps of each convolution layer in the architecture, we design a target-aware normalization approach in TANQ, which normalizes the feature maps of each convolution layer by focusing on the target objects (i.e., regions of VS and cochleas) in different processes according to their inherent visual characteristics in and images, as mentioned in Section 1. Specifically, as shown in Fig. 2, we first perform region normalization (RN)36 for cochleas to prevent the loss of their anatomical structures, that is, the cochlear regions are normalized independently of other tissues as they exhibit relatively low-intensity textures and blurry boundaries in images. By contrast, for the VS, we use region-aware adaptive instance normalization (RAIN)37 in each decoder layer to convert a relatively high-intensity style of the VS in images to the VS in the images, which exhibits an intensity similar to that of other tissues.
Each feature map passing through the convolution in each encoder layer of TANQ can be described as: , where H, W, and C denote the height, width, and number of channels of each feature map, respectively. Based on the annotation mask of cochleas in image, can be divided into two categorical regions as follows:
| 1 |
where represents the masked feature map of each categorical region of r. By adopting RN36, each masked feature map is separately normalized by region-wise IN, as shown in equation (2).
| 2 |
where and denote the mean and standard deviation of each masked feature map , respectively. The normalized feature maps from all the regions are merged into as equation (3) and then passed as input to the next encoder layer.
| 3 |
According to RN, which can retain the distinct characteristics of the cochleas in images, the anatomical structures of the cochleas can be precisely preserved during the encoding process, and the information is passed to the subsequent decoding process as well as the contrastive learning process of QS-Attn18 to generate more realistic pseudo- images.
In the decoding process, each feature map , which is passed through deconvolution in each decoder layer of TANQ, is divided into three different categorical regions based on the annotation masks of , that is, and for VS and cochleas, respectively:
| 4 |
where represents the masked feature map of each categorical region of s. Each region-wise feature map is also separately normalized based on region-wise IN, as shown in equation (5).
| 5 |
where and denote the mean and standard deviation of each masked feature map , respectively. For VS, we further adopt RAIN37 to render VS with an intensity similar to that of other tissues in pseudo- images as follows:
| 6 |
where and denote the mean and standard deviation of , respectively.
The normalized feature maps from all the regions are merged into as equation (7) and then passed as input to the next decoder layer, as shown in Fig. 2.
| 7 |
Furthermore, in TANQ, we introduce an additional decoding process to enhance its ability to preserve the structural information of the target objects in the encoding process, that is, the SegDecoder in Fig. 2. SegDecoder is responsible for generating segmentation outputs for the target objects, (VS and cochlea) using encoded feature maps. By adding SegDecoder, the attention of the encoder is directed toward the target objects, facilitating a more efficient extraction of feature maps while simultaneously preserving their structures. SegDecoder minimizes cross-entropy loss, which is calculated by comparing the output of the SegDecoder with the ground-truth annotation labels of image, as presented in the following equation38:
| 8 |
where N is the total number of pixels, C is the total number of classes, is the ground-truth label for the pixel and class, which is one-hot encoded, and represents the predicted probability of the pixel belonging to the class.
Contrastive and adversarial learning in TANQ
In QS-Attn18, contrastive learning forces a specific patch from the translated image to be close to the patch located at the same position in the source image and farther away from patches at different positions in the source image. This process enables the generator to preserve its structure while being insensitive to style differences. Unlike CUT16, which selects patches randomly, QS-Attn identifies the informative patches used for contrastive learning according to the entropy of the features from the source images. To calculate the entropy of the features, the feature is extracted from the images using encoder E in QS-Attn. The feature is reshaped into a 2D matrix Q , multiplied by its transpose matrix K , and converted into the final attention matrix by applying the softmax function. The entropy of each row in is then computed using equation (9), which allows us to measure the degree of similarity to other features18.
| 9 |
A low entropy value indicates that only a few features are similar to the query. This assumes a significant role in the calculation of contrastive loss as it distinctly sets them apart from other features. These features are prioritized by sorting the rows of based on entropy, and the N smallest rows are used for the contrastive loss18.
This contrastive learning of QS-Attn is further enhanced by combining it with the target-aware encoder–decoder architecture introduced in Section3.1 within a unified framework. Specifically, in TANQ, based on the target-aware encoder–decoder architecture designed to preserve the structures of target objects more precisely, patches around the target objects are more frequently selected as informative patches compared to the original QS-Attn (refer to Fig. 5 for more details). Consequently, contrastive learning applied to the selected patches around the target objects can attempt to restore their information in the source to pseudo- images during image translation in the proposed TANQ method, as shown in Fig. 2.
The full objective loss of TANQ is as follows:
| 10 |
where is the adversarial loss39 used to generate pseudo-images similar to real images16. refers to the contrastive loss between the source image and the translated image, and denotes the identity loss, which is calculated as the contrastive loss between the real target image and the translated image16.
Multi-view representation
We employed a multi-view image translation approach based on our prior study17. Specifically, we improved the segmentation model performance by augmenting the pseudo- images using TANQ and CycleGAN33 in parallel, as shown in Fig. 2. CycleGAN is well-suited for generating pseudo- images that reliably reflect the characteristics observed in real images owing to its use of a pixel-level reconstruction constraint17. Specifically, CycleGAN preserves the structural information of the image by enforcing cycle-consistency loss33 as follows:
| 11 |
where G and F indicate the generators from the target to the source images, and vice versa33. When a source image , for example, image, is fed into the generator G and subsequently reconstructed by the generator F, the structure of the image is preserved by minimizing the discrepancy between and . Similarly, the structural information of the target image , for example, the image, is preserved by minimizing the discrepancy between the reconstructed image and original . Consequently, CycleGAN also facilitates image translation from to the pseudo- images with the overall loss function of CycleGAN as follows:
| 12 |
where represents the adversarial loss39 used during the generation of images from images, while represents the adversarial loss employed during the generation of images from images in the reverse process33.
By employing our multi-view image translation approach, which combines the pseudo- images generated using TANQ and CycleGAN, we enhance the variability and richness of the pseudo- images. This approach enables a subsequent segmentation model to effectively learn the diverse characteristics present within images, thereby facilitating effective learning.
Segmentation model construction and self-training
We first construct a nnUNet40-based model for VS/cochlea segmentation with pseudo- images. The choice of nnUNet is based on its established performance in numerous medical image segmentation challenges as it automatically configures a UNet-based segmentation pipeline based on the analysis of the provided training cases40. We directly employed all multi-view pseudo- images, along with their associated annotation labels from the source images, to train the VS/cochlea segmentation model.
The segmentation model trained on the pseudo- images enhances its generalization ability for the unseen real images based on self-training41. The self-training process consists of the following four steps. 1) We train the segmentation model using multi-view pseudo- scans along with the ground-truth labels from the scans. 2) The segmentation model trained in step 1 is then utilized to generate pseudo-labels for the unlabeled real scans. 3) The segmentation model is retrained using both the pseudo- scans with ground-truth labels and the real scans with pseudo-labels. 4) To further enhance the performance of the segmentation model, we repeat steps 2 and 3 four times. This self-training process aims to refine the performance of the segmentation model by leveraging a combination of pseudo- and real images, that is, as the segmentation model is retrained, the pseudo-annotation labels for the real images are improved, and the results can be used again to further improve the model performance41. Although we employed a basic self-training approach, it is widely recognized that even simple self-training methods can enhance the performance and generalization of segmentation models42.
Experimental settings
Dataset and preprocessing
We used the CrossMoDA dataset1,4 to evaluate the effectiveness of our framework for VS/cochlea segmentation in unseen real scans. The dataset comprised 210 annotated and 210 unannotated scans to be used for training. We evaluated our framework using the 64 scans provided as the CrossMoDA validation dataset, and the evaluation was conducted using the official CrossMoDA 2022 website. The segmentation results of the dataset comprised the VS, cochlea, and background classes. The CrossMoDA dataset consisted of two distinct datasets: London SC-GK data and Tilburg SC-GK data1, both of which provide an equal number of and scans. We resampled all scans to 0.410.411.5 voxel sizes to ensure consistency. The 3D scans were converted into a series of 2D images along the axial plane. The images were then center-cropped and resized to 256256 pixels for image translation. Following image translation, the translated images were reconstructed into 3D volumes and fed into the proposed segmentation model.
Competing methods and ablation study
The competing methods13,14,17,21,34,43 were derived from the top-ranked models of the CrossMoDA 2021, 2022 challenge, including our previous work, which secured the second position in the challenge. Furthermore, we conducted an ablation study to examine the effectiveness of the individual components of image translation within our framework. We trained segmentation models for the VS and cochlea using pseudo- images generated using each image translation method and evaluated their performance without conducting self-training. We also demonstrate the effect of target-aware normalization through the image translation results of target objects and visualization of informative patch selection in Section 5.3. The results were evaluated on the validation set using two evaluation metrics: the Dice score44 and average symmetric surface distance (ASSD)45. Furthermore, we conducted a paired t-test to assess the statistical significance of our results, particularly focusing on the Dice score and ASSD metrics.
Implementation details
For the cross-modality image translation from to images, we employed CycleGAN33 and TANQ, where both architectures consisted of a ResNet-based generator46 and a PatchGAN-based discriminator47. We trained CycleGAN and TANQ with a batch size of 4 for 200 epochs and 400 epochs, respectively, using the Adam48 optimizer with an initial learning rate of 0.0002. Following the completion of half of the total epochs, the learning rate linearly decayed to 0 over the remaining epochs. In TANQ, a global attention matrix was computed, and 256 features based on it were selected18. The dimensions of the anchor, query, and key features used in the contrastive loss were set to 25618. We adopted multilayer feature extraction with five layers, and the QS-Attn module was applied in the last two layers18. During the segmentation and self-training phases, we used a semantic segmentation method based on nnUNet40. The nnUNet model was trained using both Dice and cross-entropy losses, and the batch size was 8 for 1000 epochs using the SGD optimizer40 with an initial learning rate of 0.01. The learning rate was decayed according to the ’poly’ learning rate policy49 throughout the training phase40. Finally, we performed an ensemble of predictions from the 2D and 3D segmentation models through the ensemble selection process within nnUNet40.
For the image translation network, we strictly followed the default parameters and training protocols of CycleGAN33 and QS-Attn18, as provided in their respective GitHub repositories. The training procedures were adhered to exactly as outlined in their repositories18,33. We selected the final model from the last epoch, consistent with the methodology used in these baseline models18,33. For the segmentation network, we strictly followed the default parameters of nnUNet40 and selected the final model using five-fold cross-validation.
Our TANQ consists of three main components, namely the Generator, Discriminator, and Feature Projector, which have 8.219M, 2.763M, and 0.560M parameters, respectively. CycleGAN33 requires two Generators and two Discriminators for reverse mapping. Each Generator in CycleGAN33 consists of 11.366M parameters, while each Discriminator contains 2.763M parameters. For the segmentation process, we utilize nnUNet40, which comprises a total of 30.76 million parameters. Regarding training duration, both TANQ and CycleGAN33 required a total of 72 GPU hours using a single RTX 3090 GPU. On the other hand, the Segmentation network required 24 GPU hours using 5 RTX 3090 GPUs. For inference time, TANQ takes approximately 0.0669 seconds per 2D image, while CycleGAN33 performs inference in 0.0043 seconds per 2D image. For the nnUNet40 model, inference on a 3D scan takes about 12.104 seconds.
Results and discussion
Performance evaluation
Table 1 presents the comparative results, including statistical analysis, of the proposed and competing methods for the validation set provided by the CrossMoDA 2021, 2022 . Our proposed method demonstrated the best performance, with a mean Dice score of 0.8650 (), outperforming all the other methods. In particular, it demonstrated outstanding performance in cochlear segmentation, achieving a remarkable Dice score of 0.8750 and an ASSD of 0.1553 , with statistically highly significant () improvements in both metrics compared to all other methods. In the case of VS segmentation, the proposed method demonstrated the second-best performance, with a Dice score of 0.8550 and ASSD of 0.4643 after the TBA34 method on the validation set. Additionally, our framework showed statistically highly significant improvements in ASSD when compared to MSF-Net43 and PAST14, while also exhibiting significant differences () in both Dice score and ASSD against the COSMOS13 andFgh36521 methods. On the other hand, when compared to TBA34, the Dice score was found to be statistically significantly lower , but there was no statistically significant difference in the ASSD metric. Furthermore, our previous model (i.e., Multi-view17) which was adopted as the backbone framework in this study, demonstrated the best VS segmentation performance on the test dataset of the CrossMoDA 2022 As listed in Table 1, our proposed method achieved better performance in VS segmentation than our previous method in terms of both the Dice score and ASSD. Based on these results, we expected that our proposed method would also exhibit excellent performance in VS segmentation on the test dataset. The comparative results, including statistical analysis, highlight the effectiveness of considering the image characteristics of the target objects (the VS and cochlea) during image translation from to images. By normalizing these target regions separately, the proposed method enables the generation of more stable and realistic pseudo- images, ultimately leading to improved segmentation performance.
Table 1.
Comparison of segmentation results of the unsupervised domain adaptation methods from the top-ranked methods of the CrossMoDA 2021, 2022 challenge. The paired t-test was conducted to assess the statistical significance of our results. .
| Method | Dice score () | ASSD () | |||
|---|---|---|---|---|---|
| VS | Cochlea | Mean | VS | Cochlea | |
| COSMOS13 | 0.6104 (±0.3065)** | 0.8184 (±0.0257)** | 0.7144 (±0.1480) | 3.8170 (±5.0257)** | 0.2293 (±0.1633)** |
| Fgh36521 | 0.8178 (±0.0803)** | 0.8433 (±0.0293)** | 0.8306 (±0.0420) | 0.6673 (±0.2713)** | 0.2053 (±0.1489)** |
| MSF-Net43 | 0.8493 (±0.0683) | 0.8294 (±0.0268)** | 0.8394 (±0.0368) | 0.5202 (±0.2288)** | 0.2454 (±0.2102)** |
| PAST14 | 0.8473 (±0.0633) | 0.8547 (±0.0283)** | 0.8511 (±0.0322) | 0.5513 (±0.3026)** | 0.1874 (±0.1478)** |
| Multi-view17 | 0.8520 (±0.0889) | 0.8488 (±0.0235)** | 0.8504 (±0.0466) | 0.4748 (±0.2072) | 0.1992 (±0.1524)** |
| TBA34 | 0.8682 (±0.0601)* | 0.8506 (±0.0294)** | 0.8594 (±0.0347) | 0.4302 (±0.1780) | 0.1892 (±0.1457)** |
| Ours | 0.8550 (±0.0731) | 0.8750 (±0.0217) | 0.8650 (±0.0370) | 0.4643 (±0.2000) | 0.1553 (±0.1448) |
Ablation study results
Table 2 presents the segmentation results of the ablation study according to the individual components of the image translation method within our framework. Here, in order to directly compare the segmentation results according to the image translation methods, self-training was not applied. First, in Table2, we can observe that QS-Attn18, which adopts entropy-based informative patch selection in contrastive learning, achieved better VS/cochlea segmentation performance than CUT16, which conducts contrastive learning with randomly selected patches. This result implies that selecting informative patches from images is beneficial for preserving the structures of the VS and cochleas with contrastive learning. Second, upon comparing the results of Multi-view17 with those of CycleGAN33 and QS-Attn, we can assume that the multi-view image translation approach, which uses both CycleGAN and QS-Attn in parallel, leads to enhanced segmentation performance with its enriched pseudo- image representations. Finally, based on the proposed target-aware normalization techniques for the VS and cochleas, the proposed method further achieved enhanced segmentation performance in the ablation study. Specifically, when only RN was applied to our multi-view approach (i.e., Ours (w/o. RAIN)), we can observe from Table 2 that the segmentation performance of the cochleas had improved in terms of both the Dice score and ASSD. This observation suggests that RN is effective in preserving the structural information of the cochlea. Ours (w/o. SegDecoder) represents the method that incorporates the proposed target-aware normalization technique while excluding solely the SegDecoder. Compared to the Ours (w/o. RAIN), Ours (w/o. SegDecoder) exhibited an improvement in the VS and cochlea Dice scores. This indicates that both RN and RAIN in our target-aware normalization effectively preserve the structural information of the target objects, VS and cochlea, and accurately reflect these visual characteristics in real hrT2 images. Moreover, as shown in Table 2, Ours, which adopts both target-aware normalization and the SegDecoder, further improved the segmentation performance of VS and cochlea across all metrics compared to other methods. This result highlights Ours as a tailored approach for cross-modality image translation, specifically designed for the VS and cochlea. We also performed a statistical evaluation for our ablation study. In cochlear segmentation, Ours showed statistically highly significant differences compared to CycleGAN33, CUT16, QS-Attn18, and Multi-view17. For Ours (w/o. RAIN), statistically significant differences were observed in cochlear segmentation. In VS segmentation, Ours demonstrated statistically significant differences in both Dice score and ASSD compared to CycleGAN33, CUT16, and QS-Attn18. Notably, compared to the baseline Multi-view17 and Ours (w/o. SegDecoder), a statistically significant improvement in ASSD for VS segmentation was achieved. These statistical anaylses highlight the importance of target-aware normalization during image translation when considering the different image characteristics of the target objects.
Table 2.
Quantitative results from the ablation study. The results were obtained without conducting self-training. The paired t-test was conducted to assess the statistical significance of our results. .
| Method | Dice score () | ASSD () | |||
|---|---|---|---|---|---|
| VS | Cochlea | Mean | VS | Cochlea | |
| CycleGAN33 | 0.7798 (±0.1901)* | 0.8066 (±0.0323)** | 0.7932 (±0.0972) | 0.8750 (±0.9222)* | 0.2422 (±0.1608)** |
| CUT16 | 0.7693 (±0.2097)* | 0.7950 (±0.0337)** | 0.7822 (±0.1095) | 0.7126 (±0.6179)* | 0.2586 (±0.1668)** |
| QS-Attn18 | 0.7779 (±0.1825)* | 0.8158 (±0.0287)** | 0.7968 (±0.0929) | 0.6667 (±0.3891)* | 0.2365 (±0.1573)** |
| Multi-view17 | 0.8043 (±0.1656) | 0.8158 (±0.0289)** | 0.8101 (±0.0863) | 0.5742 (±0.2461)* | 0.2387 (±0.1581)** |
| Ours (w/o. RAIN) | 0.8017 (±0.1696) | 0.8372 (±0.0304)* | 0.8195 (±0.0896) | 1.3809 (±6.5339) | 0.2070 (±0.1608)* |
| Ours (w/o. SegDecoder) | 0.8057 (±0.1356) | 0.8393 (±0.0299) | 0.8225 (±0.0716) | 1.7198 (±6.5030)* | 0.2064 (±0.1597) |
| Ours | 0.8097 (±0.1635) | 0.8397 (±0.0293) | 0.8247 (±0.0863) | 0.5708 (±0.2919) | 0.2042 (±0.1610) |
Fig. 3 exhibits the segmentation results of the real images obtained from segmentation models trained on psuedo- images from different image translation methods adopted for the ablation study. Notably, our approach excels in segmenting the VS and cochleas compared to the other models, as shown in Fig. 3, demonstrating exceptional performance by achieving significantly finer and more precise segmentation than the other methods.
Fig. 3.
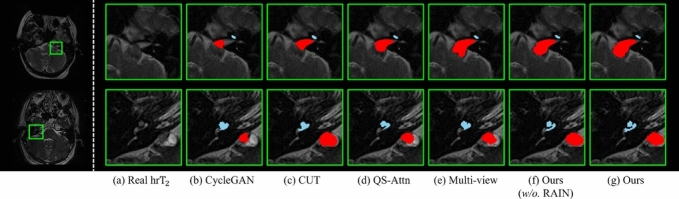
Qualitative comparison of segmentation results for VS (red) and cochleas (sky-blue) on the ablation study. (a) is the real image, while (b) to (g) show the segmentation results overlaid on the real image (a). These results are obtained from segmentation models trained using pseudo- images generated by the corresponding translation models for (b) to (g). Note that the ground truths of the real scans are not accessible in the CrossMoDA challenge.
Effects of target-aware normalization
Visual characteristics of target objects
Fig. 4 shows the image translation results under conditions where RN36 and RAIN37 are included or excluded for each of the target objects. Fig. 4(a) and (e) show samples of real and images, respectively, while (b)-(d) illustrate the pseudo- images generated from the real images based on the proposed TANQ with different RN/RAIN configurations.
Fig. 4.
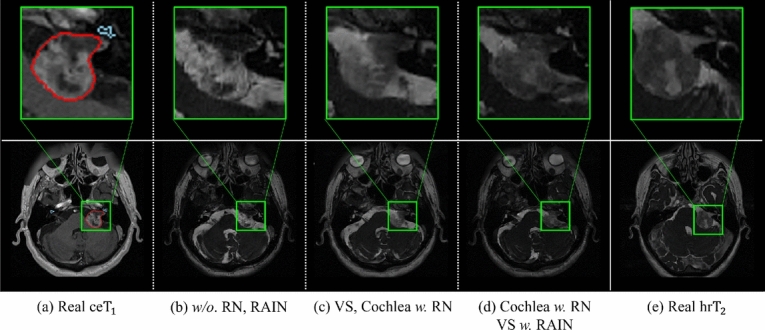
Visualization of the effectiveness of RN and RAIN for VS (red) and cochleas (sky-blue) according to different normalization configurations of TANQ; (a) real images, (b)-(d) pseudo- images from images generated by TANQ with different normalization configurations, and (e) real images. In the real images, the intensity of the VS is higher than that of other tissues due to the contrast agent, while the VS in the real images shows similar intensity to other tissues. Conversely, the cochlea exhibits very low intensity in real images (a), whereas it appears relatively distinct in real images (e).
From the results, it can first be observed that the pseudo- images from (b) TANQ w/o. RN and RAIN represent the VS with significantly high intensity, indicating a failure to reflect the low-intensity characteristic of VS in (e) the real images, and the structure of the cochlea is poorly preserved and appears blurred. By contrast, the pseudo- images from (c) TANQ with RN applied to both the VS and cochleas better preserve the information of these structures. This implies that preserving the information of each region using RN is essential for maintaining the integrity of the VS and cochlear structures. Finally, the pseudo- images from (d) TANQ with RN and RAIN for cochleas and VS show a lower intensity for the VS, similar to (e) the real images, as well as better preserved cochlear structures. These observations suggest that the use of the RN and RAIN is essential for accurately reflecting the distinctive characteristics of the target objects and for maintaining the integrity of the VS and cochlear structures. Based on the pseudo- images generated with the advantages of the proposed TANQ, the segmentation model can successfully learn the realistic characteristics of the VS and cochleas from the given images for VS and cochlea segmentation.
Informative patch selection
The locations of the selected patches for contrastive learning in (a) CUT16, (b) QS-Attn18, and (c) our TANQ are visualized with green indicators in Fig.5. It can be observed from Fig. 5(a) that CUT selects patches arbitrarily. This visualization demonstrates that such random selection tends to select patches from the background area as well, rather than focusing on specific anatomical structures of the brain, resulting in the lack of crucial information regarding brain structure, including the VS and cochlea. Numerous studies have also noted that using these “easy” negative samples for contrastive learning can result in poor performance in image translation tasks18,50,51.
Fig. 5.
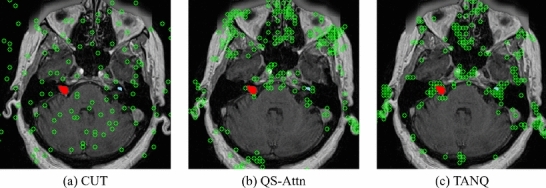
Visualization of the locations(green circle) of selected features when calculating the contrastive loss. The red indicates VS, and sky-blue represents cochlea in the image.
In contrast, Fig. 5(b) illustrates that QS-Attn effectively calculates the contrastive loss by selecting features from regions that represent the structural information of the brain. Despite these advantages, Fig. 5(b) indicates that QS-Attn tends to struggle with feature selection in smaller VS (red) and cochlear (sky-blue) regions. In contrast, Fig. 5(c) demostrates a significant increase in the patch selection related to the regions of VS and cochlea, indicating a stronger focus on the VS and cochlea than in QS-Attn. This proves that our TANQ is remarkably effective with its target-aware normalization in preventing information loss in the VS and cochleas, guaranteeing the selection of patches specifically associated with these regions. Our framework generates more realistic pseudo- images by focusing on the target objects during image translation for more efficient and effective contrastive learning techniques. Consequently, this approach enhances the performance of the segmentation model by obtaining suitable pseudo- images.
Limitations and future works
Our current framework employs two independent stages: the image translation stage and the segmentation stage. This independent approach does not allow for interaction between the image translation models, such as CycleGAN33 and TANQ, and the segmentation model, nnUNet40. As a result, the performance of the segmentation model is highly dependent on the quality of the image translation model. Additionally, despite the significant impact of the initial quality of the pseudo-labels on the final segmentation performance52, our current work did not involve any refinement of the pseudo-labels.
In our future work, we will aim to develop a framework that enables bidirectional interaction between the image translation and segmentation stages. This will allow for more effective interaction between the two stages, potentially enhancing the overall performance of both tasks. Furthermore, we plan to incorporate pseudo-label refinement techniques to enhance the effectiveness of the self-training process, ultimately contributing to improved performance of the segmentation model.
Conclusion
In this study, we propose a target-aware, unsupervised domain adaptation framework for VS and cochlea segmentation. Specifically, we acquire pseudo- images that reliably reflect the characteristics of real images while preserving the structural information of small-sized VS and cochleas by using target-aware normalization according to the specific characteristics of the VS and cochlea. By applying this target-aware normalization to QS-Attn and integrating multi-view representation, we can construct a robust VS and cochlea segmentation model in real images using these pseudo- images. Our approach demonstrates its effectiveness by significantly surpassing the performance of state-of-the-art cross-modality VS and cochlea segmentation methods.
Acknowledgements
This work was supported by the Institute of Information & Communications Technology Planning & Evaluation (IITP) grant (No. 2019-0-00079, the Artificial Intelligence Graduate School Program (Korea University)) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS202300212498). This work was supported by Institute of Information & communications Technology Planning & Evaluation (IITP) grant funded by the Korea government(MSIT) (No.RS-2021-II212068, Artificial Intelligence Innovation Hub).
Author contributions
B.K. and T.-E.K. conceived the study and designed the methodology. B.K. implemented the source code and performed the related experiments. B.K. and T.-E.K. conducted the analysis. H.N. supported B.K. in drafting the manuscript under the supervision of T.-E.K. M.K supported B.K. in designing the methodology. H.N., M.K., K.-S.H., M.L., and J.-H.O. reviewed and edited the manuscript under the supervision of T.-E.K.
Data availability
The CrossMoDA dataset is publicly available, and more information can be found at the following link: https://crossmoda-challenge.ml/challenge2022/
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dorent, R. et al. Crossmoda 2021 challenge: Benchmark of cross-modality domain adaptation techniques for vestibular schwnannoma and cochlea segmentation. arXiv preprint arXiv:2201.02831 (2022). [DOI] [PMC free article] [PubMed]
- 2.Kentala, E. & Pyykkö, I. Clinical picture of vestibular schwannoma. Auris Nasus Larynx28, 15–22 (2001). [DOI] [PubMed] [Google Scholar]
- 3.di Russo, P. et al. Characteristics and management of hydrocephalus associated with vestibular schwannomas: a systematic review. Neurosurgical Review44, 687–698 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Shapey, J. et al. Segmentation of vestibular schwannoma from mri, an open annotated dataset and baseline algorithm. Scientific Data8, 286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGrath, H. et al. Manual segmentation versus semi-automated segmentation for quantifying vestibular schwannoma volume on mri. International Journal of Computer Assisted Radiology and Surgery15, 1445–1455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahara, D. et al. Variations in cochlear size of cochlear implant candidates. International archives of otorhinolaryngology23, 184–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marasini, R., Thanh Nguyen, T. D. & Aryal, S. Integration of gadolinium in nanostructure for contrast enhanced-magnetic resonance imaging. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology12, e1580 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Khawaja, A. Z. et al. Revisiting the risks of mri with gadolinium based contrast agents—review of literature and guidelines. Insights into imaging6, 553–558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao, K. et al. A novel 3d unsupervised domain adaptation framework for cross-modality medical image segmentation. IEEE Journal of Biomedical and Health Informatics26, 4976–4986 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Shin, H. et al. Sdc-uda: Volumetric unsupervised domain adaptation framework for slice-direction continuous cross-modality medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 7412–7421 (2023).
- 11.Dong, H., Yu, F., Zhao, J., Dong, B. & Zhang, L. Unsupervised domain adaptation in semantic segmentation based on pixel alignment and self-training. arXiv preprint arXiv:2109.14219 (2021).
- 12.Choi, J. W. Using out-of-the-box frameworks for unpaired image translation and image segmentation for the crossmoda challenge. arXiv e-prints arXiv–2110 (2021).
- 13.Shin, H. et al. Cosmos: Cross-modality unsupervised domain adaptation for 3d medical image segmentation based on target-aware domain translation and iterative self-training. arXiv preprint arXiv:2203.16557 (2022).
- 14.Hexin, D. et al. Unsupervised domain adaptation in semantic segmentation based on pixel alignment and self-training (past). CrossMoDA 2022 Challenge.
- 15.Chen, R., Huang, W., Huang, B., Sun, F. & Fang, B. Reusing discriminators for encoding: Towards unsupervised image-to-image translation. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 8168–8177 (2020).
- 16.Park, T., Efros, A. A., Zhang, R. & Zhu, J.-Y. Contrastive learning for unpaired image-to-image translation. In European conference on computer vision, 319–345 (Springer, 2020).
- 17.Kang, B., Nam, H., Han, J.-W., Heo, K.-S. & Kam, T.-E. Multi-view cross-modality mr image translation for vestibular schwannoma and cochlea segmentation. arXiv preprint arXiv:2303.14998 (2023).
- 18.Hu, X. et al. Qs-attn: Query-selected attention for contrastive learning in i2i translation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 18291–18300 (2022).
- 19.Ulyanov, D., Vedaldi, A. & Lempitsky, V. Instance normalization: The missing ingredient for fast stylization. arXiv preprint arXiv:1607.08022 (2016).
- 20.Zhuang, X. & Shen, J. Multi-scale patch and multi-modality atlases for whole heart segmentation of mri. Medical image analysis31, 77–87 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Liu, H., Fan, Y. & Dawant, B. M. Enhancing data diversity for self-training based unsupervised cross-modality vestibular schwannoma and cochlea segmentation. arXiv preprint arXiv:2209.11879 (2022).
- 22.Park, T., Liu, M.-Y., Wang, T.-C. & Zhu, J.-Y. Semantic image synthesis with spatially-adaptive normalization. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2337–2346 (2019).
- 23.Dou, Q. et al. Pnp-adanet: Plug-and-play adversarial domain adaptation network at unpaired cross-modality cardiac segmentation. IEEE Access7, 99065–99076 (2019). [Google Scholar]
- 24.Yan, W., Wang, Y., Xia, M. & Tao, Q. Edge-guided output adaptor: Highly efficient adaptation module for cross-vendor medical image segmentation. IEEE Signal Processing Letters26, 1593–1597 (2019). [Google Scholar]
- 25.Chen, Z., Pan, Y. & Xia, Y. Reconstruction-driven dynamic refinement based unsupervised domain adaptation for joint optic disc and cup segmentation. IEEE Journal of Biomedical and Health Informatics27, 3537–3548 (2023). [DOI] [PubMed] [Google Scholar]
- 26.Cai, J., Zhang, Z., Cui, L., Zheng, Y. & Yang, L. Towards cross-modal organ translation and segmentation: A cycle-and shape-consistent generative adversarial network. Medical image analysis52, 174–184 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Jue, J. et al. Integrating cross-modality hallucinated mri with ct to aid mediastinal lung tumor segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part VI 22, 221–229 (Springer, 2019). [DOI] [PMC free article] [PubMed]
- 28.Kang, M., Chikontwe, P., Won, D., Luna, M. & Park, S. H. Structure-preserving image translation for multi-source medical image domain adaptation. Pattern Recognition144, 109840 (2023). [Google Scholar]
- 29.Chen, C., Dou, Q., Chen, H., Qin, J. & Heng, P. A. Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation. IEEE transactions on medical imaging39, 2494–2505 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Chen, C., Dou, Q., Chen, H., Qin, J. & Heng, P.-A. Synergistic image and feature adaptation: Towards cross-modality domain adaptation for medical image segmentation. In Proceedings of the AAAI conference on artificial intelligence33, 865–872 (2019). [Google Scholar]
- 31.Hu, S., Liao, Z. & Xia, Y. Domain specific convolution and high frequency reconstruction based unsupervised domain adaptation for medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 650–659 (Springer, 2022).
- 32.Jiang, J. et al. Psigan: Joint probabilistic segmentation and image distribution matching for unpaired cross-modality adaptation-based mri segmentation. IEEE transactions on medical imaging39, 4071–4084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE international conference on computer vision, 2223–2232 (2017).
- 34.Sallé, G. et al. Cross-modal tumor segmentation using generative blending augmentation and self training. arXiv preprint arXiv:2304.01705 (2023). [DOI] [PubMed]
- 35.Shaham, T. R., Dekel, T. & Michaeli, T. Singan: Learning a generative model from a single natural image. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 4570–4580 (2019).
- 36.Yu, T. et al. Region normalization for image inpainting. In Proceedings of the AAAI conference on artificial intelligence34, 12733–12740 (2020). [Google Scholar]
- 37.Ling, J., Xue, H., Song, L., Xie, R. & Gu, X. Region-aware adaptive instance normalization for image harmonization. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 9361–9370 (2021).
- 38.Yi-de, M., Qing, L. & Zhi-Bai, Q. Automated image segmentation using improved pcnn model based on cross-entropy. In Proceedings of 2004 International Symposium on Intelligent Multimedia, Video and Speech Processing, 2004., 743–746 (IEEE, 2004).
- 39.Goodfellow, I. et al. Generative adversarial nets. Advances in Neural Information Processing Systems27 (2014).
- 40.Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J. & Maier-Hein, K. H. nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods18, 203–211 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Xie, Q., Luong, M.-T., Hovy, E. & Le, Q. V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 10687–10698 (2020).
- 42.Yang, N., Rongione, C., Jacquemart, A.-L., Draye, X. & Vleeschouwer, C. D. On the importance of diversity when training deep learning segmentation models with error-prone pseudo-labels. Applied Sciences14, 5156 (2024). [Google Scholar]
- 43.Han, L., Huang, Y., Tan, T. & Mann, R. Unsupervised cross-modality domain adaptation for vestibular schwannoma segmentation and koos grade prediction based on semi-supervised contrastive learning. arXiv preprint arXiv:2210.04255 (2022).
- 44.Dice, L. R. Measures of the amount of ecologic association between species. Ecology26, 297–302 (1945). [Google Scholar]
- 45.Huttenlocher, D. P., Klanderman, G. A. & Rucklidge, W. J. Comparing images using the hausdorff distance. IEEE Transactions on pattern analysis and machine intelligence15, 850–863 (1993). [Google Scholar]
- 46.Johnson, J., Alahi, A. & Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11-14, 2016, Proceedings, Part II 14, 694–711 (Springer, 2016).
- 47.Isola, P., Zhu, J.-Y., Zhou, T. & Efros, A. A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 1125–1134 (2017).
- 48.Kinga, D., Adam, J. B. et al. A method for stochastic optimization. In International conference on learning representations (ICLR), vol. 5, 6 (San Diego, California;, 2015).
- 49.Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K. & Yuille, A. L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence40, 834–848 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Jung, C., Kwon, G. & Ye, J. C. Exploring patch-wise semantic relation for contrastive learning in image-to-image translation tasks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 18260–18269 (2022).
- 51.Wang, W., Zhou, W., Bao, J., Chen, D. & Li, H. Instance-wise hard negative example generation for contrastive learning in unpaired image-to-image translation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 14020–14029 (2021).
- 52.Higashimoto, R., Yoshida, S., Horihata, T. & Muneyasu, M. Unbiased pseudo-labeling for learning with noisy labels. IEICE TRANSACTIONS on Information and Systems107, 44–48 (2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CrossMoDA dataset is publicly available, and more information can be found at the following link: https://crossmoda-challenge.ml/challenge2022/


