Summary
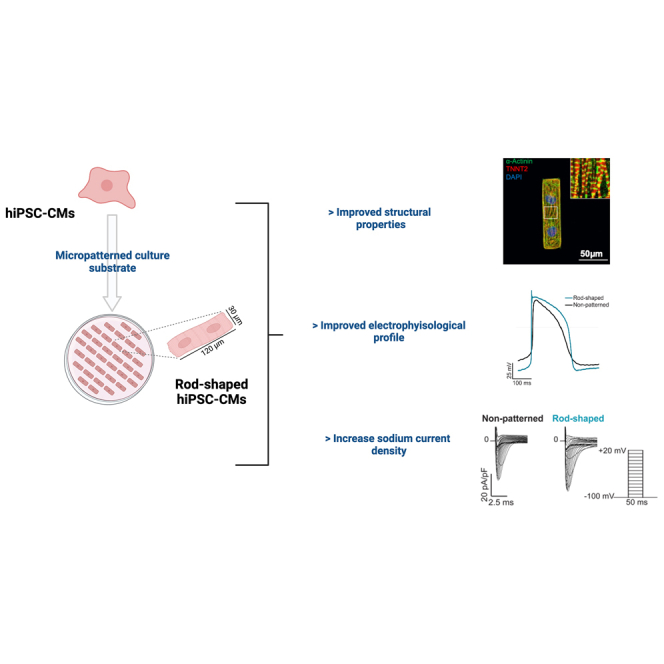
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) offer great potential for drug screening and disease modeling. However, hiPSC-CMs remain immature compared to the adult cardiac cells. Cardiomyocytes isolated from adult human hearts have a typical rod-shaped morphology. Here, we sought to develop a simple method to improve the architectural maturity of hiPSC-CMs by using a rod-shaped cell micropatterned substrate consisting of repeated rectangles (120 μm long × 30 μm wide) surrounded by a chemical cell repellent. The generated hiPSC-CMs exhibit numerous characteristics similar to adult human cardiomyocytes, including elongated cell shape, well-organized sarcomeres, and increased myofibril density. The improvement in structural properties correlates with the enrichment of late ventricular action potentials characterized by a more hyperpolarized resting membrane potential and an enhanced depolarization consistent with an increased sodium current density. The more mature hiPSC-CMs generated by this method may serve as a useful in vitro platform for characterizing cardiovascular disease.
Graphical abstract
Highlights
-
•
Culture on micropatterned substrates improves architectural maturity of hiPSC-CMs
-
•
Forcing a rod-shaped pattern favors morphological and electrophysiological properties
-
•
Rod-shaped hiPSC-CMs could serve as a platform for studying cardiovascular disease
Culture of human induced pluripotent stem cells-derived cardiomyocytes on rod-shaped micropatterned substrates improves their morphological and electrophysiological properties associated with increased sodium current density. The more mature hiPSC-CMs could serve as a useful in vitro platform for characterizing cardiovascular disease.
Introduction
Over the past decade, the use of human pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) for cardiac disease modeling and drug screening has grown tremendously. Unlike the other in vitro models, hiPSC-CMs provide an unlimited supply of patient-specific cardiomyocytes. These cells develop a typical action potential (AP) and contractile properties after a few days in culture (Thomas et al., 2022; Zhao et al., 2018). However, hiPSC-CMs, typically derived from chemically induced differentiation protocols, exhibit a fetal-like phenotype with immature architectural and electrophysiological properties compared to their adult cardiomyocyte counterparts (Koivumaki et al., 2018; Thomas et al., 2022),which limits the modeling of complex adult cardiac diseases.
Within the myocardium, cardiomyocytes are embedded in a highly structured microenvironment that imposes specific constraints (Andres-Delgado and Mercader, 2016). These geometric cues play an essential role in configuring the orientation of myofibrils, the alignment of sarcomeres, the intracellular architecture, and the polarization of cardiomyocytes (Bray et al., 2008). Experimental studies of cardiac development have suggested that the acquisition of the rod-shaped morphology of cardiomyocytes results from extrinsic mechanical and hemodynamic forces (Auman et al., 2007; Lin et al., 2012). Progressive sarcomere orientation may also contribute to cardiomyocyte shape. However, the exact mechanisms underlying the geometric changes of cardiomyocytes and their typical rod-shaped morphology are unclear. The conventional culture techniques of hiPSC-CMs do not impose physical constraints. During differentiation, pluripotent stem cells are typically seeded on a uniform adhesion substrate, allowing them to expand in all directions, resulting in hiPSC-CMs with circular shapes, disorganized structures, and incomplete expression of cytoskeletal proteins and ion channels (Koivumaki et al., 2018). Various maturation techniques have been proposed in order to improve the maturation of generated hiPSC-CMs, including thyroid hormone stimulation (Parikh et al., 2017), metabolic maturation media (Feyen et al., 2020), and Matrigel mattress (Feaster et al., 2015). In response to these chemical-stimulation-based maturation techniques, some hiPSC-CMs can adopt an elongated shape (Feaster et al., 2015; Parikh et al., 2017). It has also been reported that the structural maturation of hiPSC-CMs depends on interactions with the extracellular matrix and activation of integrin signaling (Herron et al., 2016). However, fewer maturation techniques impose geometric constraints as seen in the tissue (Jimenez-Vazquez et al., 2022a, 2022b).
Micropatterning methods allow the reconstitution of tissue-like conditions for in vitro cell culture. They involve the design of a culture substrate featuring microscopic characteristics that dictate a precise cell adhesion pattern (Thery, 2010). The patterned monolayers are easy to prepare and reproducible, and the cells can be maintained in culture over extended periods. This technique has been previously used with various cell types, including stem cell-derived cells, in order to dictate their spatial organization (Herron et al., 2016; Pioner et al., 2022; Ribeiro et al., 2017). In particular, it was shown that sarcomere alignment in neonatal rat ventricular myocytes is shape dependent and influenced by external physical constraints (Bray et al., 2008; Parker et al., 2008). Micropatterning offers advantages but is not the most commonly used method for maturation. A culture technique based on Matrigel-coated plates and a reusable micropatterned silicone stamp was recently reported to improve the anisotropy of hiPSC-CMs (Jimenez-Vazquez et al., 2022a, 2022b).
Here, we aimed to develop and evaluate a method to phenotype cardiac cells, optimally at the single-cell level. This method involves culturing cells on pre-made and ready-to-use micropatterned substrates, specifically tailored to exhibit a rod-shaped morphology, aiming to promote hiPSC-CMs elongation and anisotropy.
Results
Structural features of hiPSC-CMs upon rod-shaped micropatterning
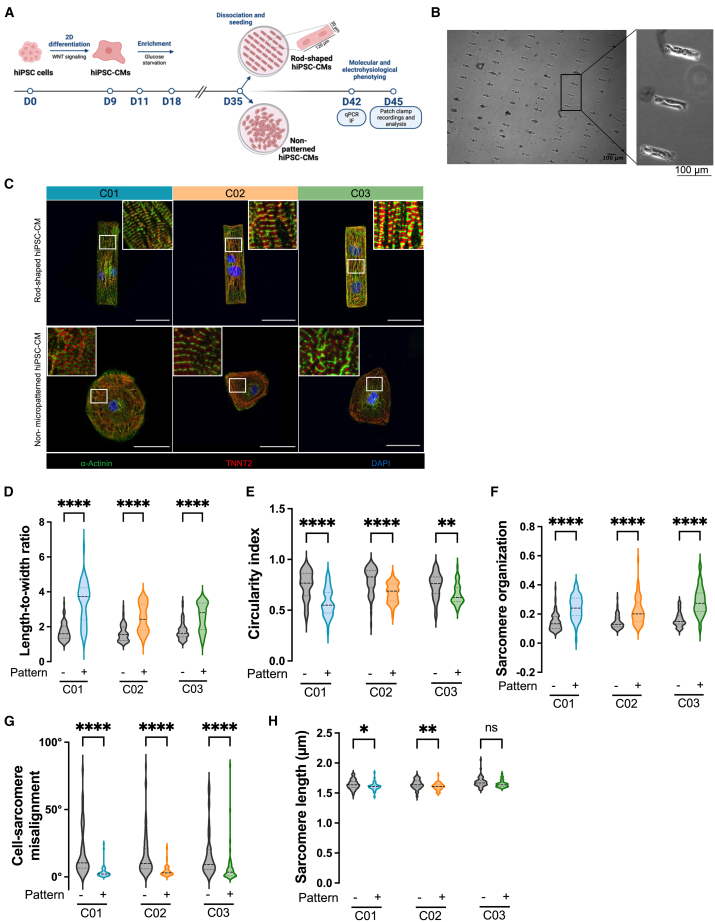
To investigate whether rod-shaped micropatterning could promote hiPSC-CMs maturation, we generated ventricular CMs from three independent hiPSC lines, using the Wnt modulation protocol (Figure 1A). Expression of pluripotency markers was validated by immunostaining (Figure S1A). After around 10 days of cardiac induction, cultures exhibited a contractile phenotype, and flow cytometry analysis on day 21 showed a cardiomyocyte purity of 95.3% ± 4.5% as measured by TNNT2-positive cells (Figures S1B and S1C). After 35 days of differentiation, the cells were enzymatically dispersed and replated for up to 7 to 10 days on specially designed individual micropatterned slides with a rod-shaped morphology or on control culture slides (referred to as non-patterned). Briefly, the micropatterned slides were stamped with Matrigel-coated rectangles (120 μm long × 30 μm wide) surrounded by an anti-adhesive reagent (Figure 1A). In preliminary experiments, we found this size to provide the optimal yield among various length-to-width ratios tested (i.e., 3:1, 4:1, and 5:1, corresponding to 90, 120, and 150 μm long for a width of 30 μm). In the following days, hiPSC-CMs from the 3 different hiPSC lines grew in the predesigned areas and developed into rod-shaped cardiomyocytes (Figures 1B and S2A). We found that roughly half of the rectangular patterns contained one cardiomyocyte while the other half contained 2 or more (Figures S2B and S2C). Seven days after seeding, we assessed the cell morphology and sarcomere organization by troponin T and α-actinin immunostaining (Figure 1C). As compared to hiPSC-CMs derived in the standard non-patterned condition, hiPSC-CMs plated on rod-shaped micropattern exhibited enhanced organization of the contractile machinery, with increased myofibrils density and alignment throughout the cytoplasm of the cell (Figure 1C). hiPSC-CMs seeded in the standard non-patterned condition showed round to polygonal shapes with lower degree of alignment (Figure 1C), consistent with previous reports (Lundy et al., 2013).
Figure 1.
Improved structural organization through rod-shaped micropatterning
(A) Schematic overview of the experimental study. On day 35 of differentiation, hiPSC-CMs were seeded onto rod-shaped micropattern or non-patterned slides for 7 to 10 days before molecular (real-time qPCR and immunofluorescence [IF]) and electrophysiological (patch-clamp and transient calcium recordings) phenotyping.
(B) Representative phase-contrast microscopy images of rod-shaped hiPSC-CMs. Images were taken with ×4 (left) and ×10 (right) objectives, scale bar, 100 μm.
(C) Representative IF images of hiPSC-CMs from three different hiPSCs lines (C01, C02, and C03), for alpha-sarcomeric actinin (green) and cardiac troponin T2 (red) under rod-shaped micropatterning (top) vs. non-patterned conditions (down), 7 days after seeding. DAPI (blue) was used to stain nuclei. Scale bar, 50 μm.
(D–H) Violin plots showing quantification data for cell length/width ratio (D), cell circularity index (E), sarcomere organization (F), cell-sarcomere misalignment (G), and sarcomere length (H). Non-patterned conditions, N = 88 for C01, N = 70 for C02, N = 58 for C03; rod-shaped micropattern conditions, N = 47 for C01, N = 58 for C02, N = 47 for C03.
Data are expressed as mean of 3 independent experiments from three different hiPSCs lines. Mann-Whitney: ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗∗p < 0.0001.
Additional analyses revealed that the cell elongation (as measured by the ratio of the major axis length to the minor axis length) increased in rod-shaped hiPSC-CMs (p < 0.0001 for the 3 lines, Figure 1D). To gain deeper insight into the cardiomyocyte shape, we assessed the cell circularity index (Circularity = 4π × Area × Perimeter−2), where “0.5” signifies a rectangular cell and “1” represents a perfectly circular cell. Rod-shaped micropatterning led to a decrease in the cell circularity index across all three lines (Figure 1E), with a higher proportion of hiPSC-CMs showing a circularity index of 0.5 (Figure S2D), underscoring the changes in shape and reproducibility of the rod-shaped micropatterning approach. Importantly, rod-shaped cardiomyocytes showed a notably higher alignment of myofibrils, as evidenced by an increase in sarcomere organization score (p < 0.0001 for the 3 lines, Figure 1F) and a decrease in the cell-sarcomere misalignment (p < 0.0001 for the 3 lines, Figure 1G). We also found significant difference in resting sarcomere lengths between the two conditions for 2 lines, mainly attributed to reduced intercellular variability within the micropatterned conditions and possible differences in the contraction/relaxation state between cells in the absence of drugs used to stop the cells beating before fixation (Figure 1H). There was no significant difference in the expression of sarcomeric genes (Figure S2E), nor in spontaneous beating rates (Figure S2F; Videos S1 and S2). Taken together, these data suggest that hiPSC-CMs obtained with the rod-shaped micropatterning technique display significant structural changes that make them more similar to mature cardiomyocytes.
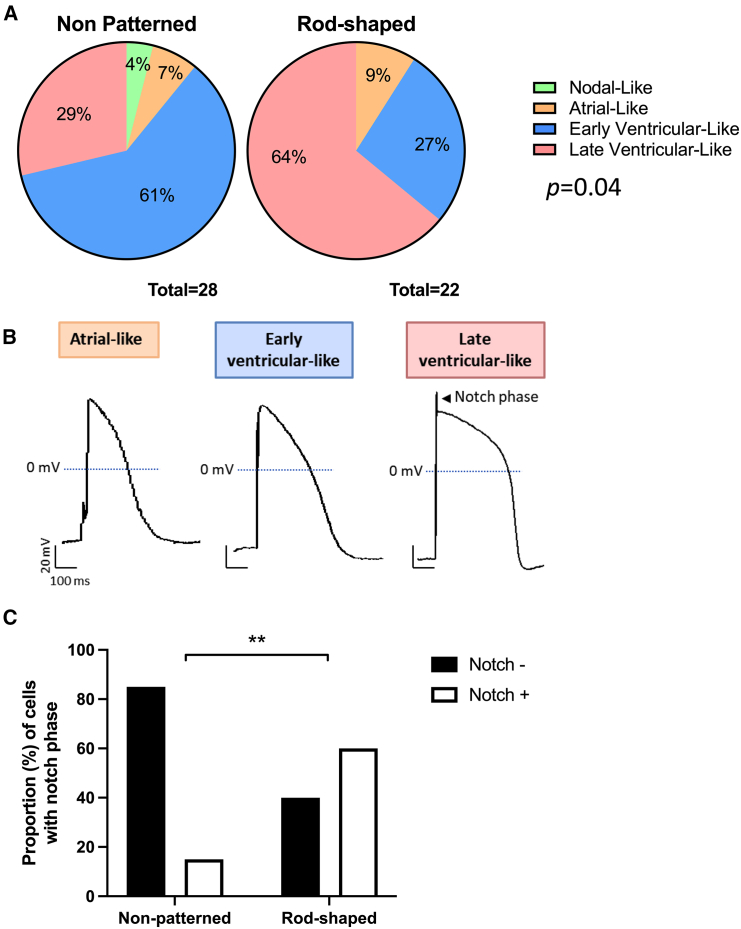
Enrichment in the late ventricular-like type cardiomyocytes in rod-shaped hiPSC-CMs
We then evaluated the electrophysiological properties recorded by patch-clamp technique by comparing the morphology of APs from hiPSC-CMs cultured under both conditions (Figure S3A). First, we used the ratio (Action potential direction) APD30-40/APD70-80, indicative of the plateau phase, to discriminate between ventricular and non-ventricular-like hiPSC-CMs. Consistent with the use of a ventricular differentiation protocol, APs from both non-patterned and rod-shaped hiPSC-CMs were predominantly categorized as ventricular-like (average 90% ± 1%; Figures 2A and 2B). Then, ventricular-like hiPSC-CMs were classified into early ventricular-like (dV/dtmax < 30 V/s) or late ventricular-like (faster upstroke defined by dV/dtmax > 30 V/s) CMs based on their depolarization velocity. The percentage of late ventricular-like APs was increased in hiPSC-CMs cultured in rod-shaped micropatterns (29% vs. 64% in non-patterned vs. rod-shaped micropatterns, respectively), and the percentage of early ventricular-like APs was decreased (61% vs. 27% in non-patterned and rod-shaped micropatterns, respectively) (p = 0.04, Figure 2A). Furthermore, the number of APs with a typical mature notch, indicating of greater involvement of Ito, was increased to up to 60% in rod-shaped hiPSC-CMs (Figure 2C). These results suggest that the acquisition of adult-like rod-shaped morphology stimulates electrophysiological configuration to a more mature ventricular hiPSC-CMs.
Figure 2.
Generation of hiPSC-CMs with late ventricular-like action potentials favored by rod-shaped micropatterning
(A) Pie charts showing the distribution of early ventricular, late ventricular, atrial, and nodal-like APs in non-patterned (n = 28) and rod-shaped (n = 22) hiPSC-CMs. p = 0.04 by Fisher’s exact test.
(B) Representative APs classified as atrial, early ventricular, and late ventricular-like APs from hiPSC-CMs cultured in rod-shaped micropatterns. hiPSC-CMs were paced at 1 Hz frequency. The arrow indicates the notch phase.
(C) Percentage of ventricular-like cardiomyocytes showing a notch phase from at least 3 independent experiments. Fisher’s exact ∗∗p < 0.01.
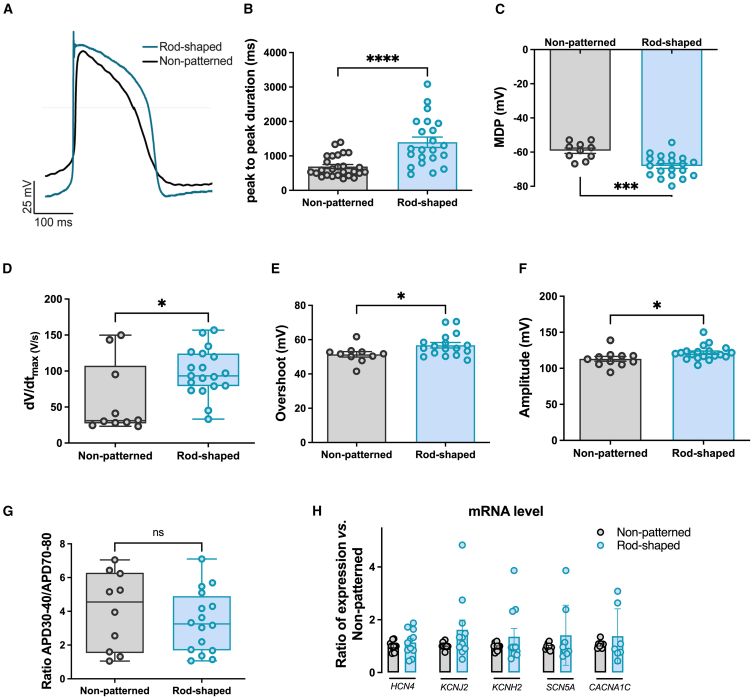
Improved electrophysiological profile of rod-shaped hiPSC-CMs
We next examined the effect of the rod-shaped micropatterning on spontaneous electrical activity and AP characteristics. Representative AP traces are shown in Figure 3A. First, we found that rod-shaped hiPSC-CMs exhibited larger peak-to-peak intervals, reflecting a slower beating rate and less spontaneous activity typical to the adult phenotype (Figure 3B). Another marker of maturation is the maximal diastolic potential (MDP), which was more hyperpolarized in rod-shaped hiPSC-CMs as compared to the non-patterned ones (−68.12 ± 6.2mV vs. −59.18 ± 4.9 mV, respectively, p = 0.0046) (Figure 3C). Depolarization parameters were also improved in the rod-shaped micropattern condition. Indeed, at 1 Hz pacing frequency, rod-shaped hiPSC-CMs exhibited 1.65 times faster upstroke velocity (Figure 3D) and increased depolarization parameters (Figures 3E and 3F) compared to non-patterned hiPSC-CMs. There were no significant changes in repolarization duration (Figure S3B), or in APD30-40/APD70-80 ratio in rod-shaped hiPSC-CMs (Figure 3G). To ascertain whether this shift toward a more mature electrical phenotype stemmed from a differential expression of key ion channels involved in the cardiomyocyte AP, we employed quantitative real-time PCR (real-time qPCR). Analysis of key ion channels transcripts involved in the generation of If, IK1, IKr, INa, and ICa,L (HCN4, KCNJ2, KCNH2, SCN5A, and CACNA1C, respectively) showed no statistically significant difference between rod-shaped and non-patterned CMs (Figure 3H). In summary, the AP in rod-shaped hiPSC-CMs demonstrated improvements that brought it closer to the characteristic profile seen in adult cardiomyocytes without significant change in the main ion channels transcripts level.
Figure 3.
Improved electrophysiological properties of rod-shaped micropatterned hiPSC-CMs
(A) Representative overlayed ventricular-like AP traces recorded from non-patterned and rod-shaped micropatterned hiPSC-CMs 10 days after seeding.
(B and C) Bar graphs quantifying (B) peak-to-peak interval duration averaged from spontaneously beating hiPSC-CMs and (C) membrane diastolic potential measured in non-patterned and rod-shaped CMs, paced at 1 Hz.
(D) Box whisker blot showing maximum upstroke velocity dV/dtmax calculated in non-patterned and rod-shaped hiPSC-CMs, paced at 1 Hz.
(E and F) Bar graphs quantifying (E) upstroke overshoot analyzed from APs recorded in non-patterned and rod-shaped hiPSC-CMs which were paced at 1 Hz and (F) depolarization amplitude averaged from APs paced at 1 Hz and recorded in non-patterned and rod-shaped hiPSC-CMs.
(G) APD30-40/APD70-80 ratio, which reflects the plateau phase ratio in non-patterned and rod-shaped hiPSC-CMs paced at 1 Hz.
(H) RNA expression of HCN4, KCNJ2, KCNH2, SCN5A, and CACNA1C measured by SYBR green in non-patterned and rod-shaped hiPSC-CMs. CTs were normalized to RPL32, and the ratio of rod-shaped to non-patterned was calculated for each cardiac differentiation. CTs, threshold cycle.
Data are expressed as mean ± SEM. For (B–G), n = 10–26 from 4 independent experiments. For (H), n = 3 independent experiments from 7 differentiations per condition. Mann-Whitney: ns, p > 0.05, ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 vs. non-patterned hiPSC-CMs.
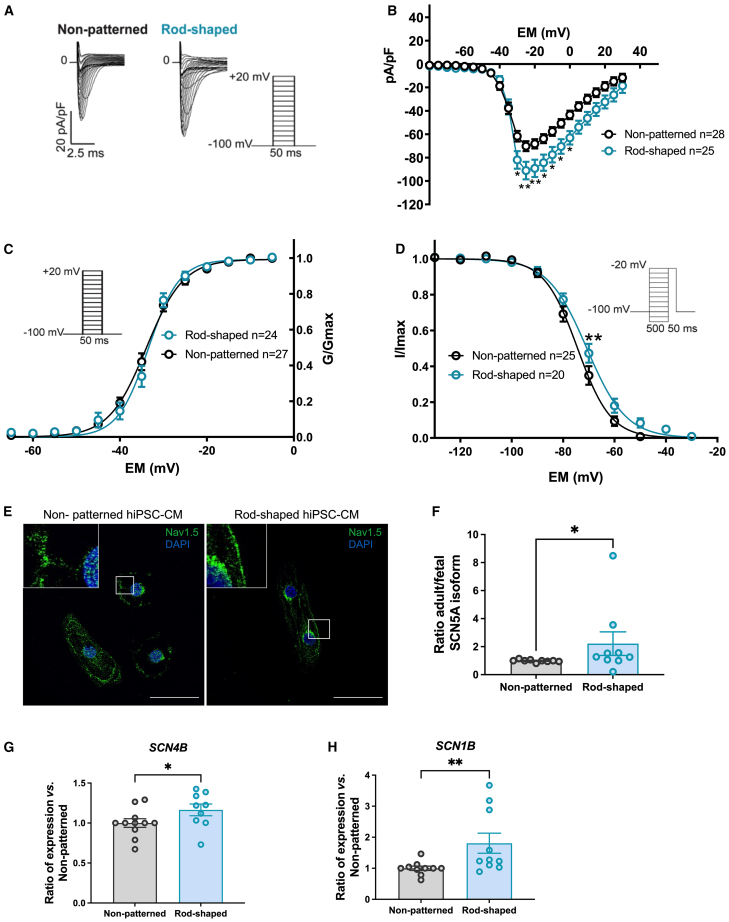
Increase in INa density in rod-shaped hiPSC-CMs
The faster AP depolarization in rod-shaped hiPSC-CMs may be the result of either a greater contribution of INa at more negative MDP or an alteration in ion channel expression and gating properties. INa drives the depolarization in adult ventricular cardiomyocytes and is often deficient in hiPSC-CM with fetal-like phenotype (Al Sayed et al., 2021a, 2021b; Zhu et al., 2021). Representative INa traces (with the corresponding voltage protocol) in rod-shaped micropatterned and non-patterned hiPSC-CMs are shown in Figure 4A. In rod-shaped hiPSC-CMs, the mean peak INa density was significantly higher than that in non-patterned hiPSC-CMs (Figures 4B and S3C). However, cell size, as measured by cell capacitance, did not show a statistically significant difference between the two culture conditions (Figure S3D), indicating that the differences were due to changes in cell shape rather than cell size. The voltage dependence of activation was not altered between rod-shaped and non-patterned hiPSC-CMs (Figure 4C; Table 1). However, the voltage dependence of inactivation of the rod-shaped hiPSC-CMs was shifted toward depolarized potential by 3.6 mV when compared with that of the non-patterned hiPSC-CMs (Figure 4D). The slope values in the inactivation curves were not changed, but the V1/2 was significantly (p = 0.0495) more positive in the rod-shaped hiPSC-CMs (Table 1).
Figure 4.
Sodium channel function and expression were improved in rod-shaped hiPSC-CMs
(A) Representative INa densities in both conditions. Inset shows voltage-clamp protocol.
(B) Average peak INa densities (pA/pF) versus membrane potential (Em) in non-patterned and rod-shaped hiPSC-CMs. pA/pF, peak current amplitude to cell membrane capacitance.
(C) INa voltage dependence of activation in both conditions. GNa (as INa/(Vm−ENa), where ENa is the equilibrium potential for Na+ ions) was normalized to its maximum value (Gmax) and plotted as a function of the potential of the test pulse (inset: voltage protocol).
(D) INa voltage dependence of inactivation in both conditions. INa was normalized to its maximum value and plotted as a function of the potential of the conditioning pulse preceding the −20mV test pulse (inset: voltage protocol).
(E) Representative images showing Nav1.5 immunostaining in non-patterned and rod-shaped hiPSC-CMs at day 7 of seeding. Nuclei are shown in blue (DAPI). Scale bar, 50 μm.
(F) Ratio of adult vs. fetal SCN5A mRNA expression in non-patterned and rod-shaped hiPSC-CMs measured by SYBR green. Relative mRNA levels were normalized to non-patterned samples.
(G and H) RNA expression of SCN1B and SCN4B measured by SYBR green in non-patterned and rod-shaped hiPSC-CMs. Cts were normalized to RPL32, and the ratio of rod-shaped to non-patterned was calculated for each cardiac differentiation.
Data are expressed as mean ± SEM. For (B–D), n = 20–28 cells from 4 independent experiments. Two-way ANOVA with Bonferroni: ∗p < 0.05, ∗∗p < 0.01. For (F–H), n = 3 independent experiments from 5 to 7 differentiations per condition. Mann-Whitney, ∗p < 0.05, ∗∗p < 0.01.
Table 1.
Biophysical parameters of INa in hiPSC-CMs
| Non-patterned | n | Rod-shaped | n | p value | |
|---|---|---|---|---|---|
| Cell capacitance (pF) | 35.70 ± 1.97 | 30 | 35.89 ± 2.35 | 27 | 0.7659 |
| INa density (pA/pF) at −20 mV | −64.92 ± 4.24 | 26 | −89.30 ± 5.02 | 25 | 0.0005 |
| Steady-state activation | 25 | 20 | |||
| V1/2 (mV) | −33.94 ± 0.69 | −33.72 ± 0.98 | 0.5867 | ||
| Kact | 3.39 ± 0.26 | 2.70 ± 0.28 | 0.0858 | ||
| Steady-state inactivation | 25 | 19 | |||
| V1/2 (mV) | −74.15 ± 1.21 | −70.34 ± 1.47 | 0.0384 | ||
| Kinact | −5.76 ± 0.45 | −7.15 ± 0.82 | 0.3592 |
INa, sodium current density measured at −20 mV from −100 mV holding potential; V1/2, voltage of half-maximal (in)activation; K, slope factor of voltage dependence of (in)activation. Values are mean ± SEM. Mann-Whitney test.
We performed Nav1.5 immunostaining and detected the sodium channel at the plasma membrane in hiPSC-CMs under both conditions (Figure 4E). Considering the improvements in INa density, we compared the expression levels of the fetal versus adult isoforms of α-subunit 5 of the sodium voltage-gated ion channel (SCN5A) using real-time qPCR. The analysis revealed a significant increase of the adult/fetal isoform ratio in the rod-shaped hiPSC-CMs (Figures 4F, S4A, and S4B), indicating a shift toward more mature channel forms. In addition, the expression of the Na+ channel β subunits β1 (SCN1B) and β4 (SCN4B) was significantly increased in the rod-shaped hiPSC-CMs (Figures 4G and 4H), whereas SCN2B and SCN3B were similarly expressed (Figures S4C and S4D). Overall, these findings suggest that micropatterning led to a change in the expression of the different sodium channel subunits, resulting in increased sodium current density in the rod-shaped hiPSC-CMs.
Rod-shaped micropatterning does not impact calcium handling properties
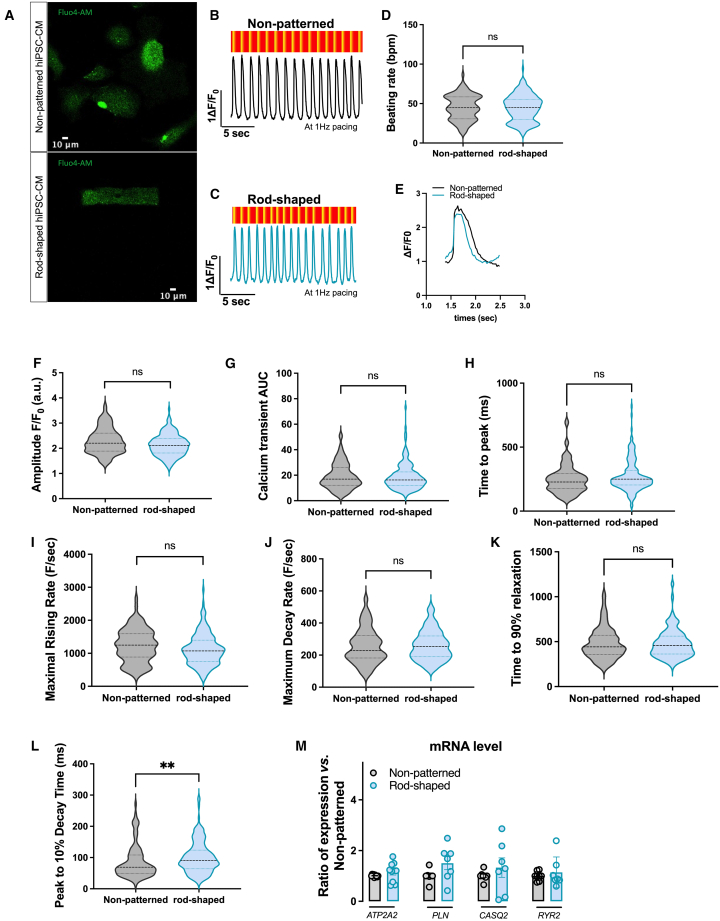
Finally, we compared Ca2+ handling between rod-shaped and non-patterned hiPSC-CMs using the intracellular calcium dye Fluo-4-AM. Despite the difference in cell shape, staining with the calcium dye showed that both non-patterned and rod-shaped hiPSC-CMs have robust intracellular Ca2+ stores (Figure 5A). Representative traces for both conditions are shown in Figures 5B and 5C. Non-patterned and rod-shaped CMs showed similar beating rates and calcium amplitude under electrical stimulation (Figures 5D–5F), but with an important intercellular variability, suggesting that not all cells correctly followed the imposed stimulation. The calcium transient area under the curve (AUC) and time to peak were not significantly changed between rod-shaped and non-patterned hiPSC-CMs (Figures 5G and 5H; Table 2). We found that the maximum rising rate and the maximum decay rate of the rod-shaped CMs were not significantly different from those of the non-patterned CMs (Figures 5I–5K), while the time to 10% transient decay was significantly increased in the rod-shaped hiPSC-CMs (Figure 5L). Similar results were found in the non-paced cells, however with a significantly smaller amplitude and maximal rising rate in the rod-shaped hiPSC-CMs (Figures S5A–S5G). These data indicate that hiPSC-CMs from rod-shaped conditions did not exhibit enhanced calcium handling properties as compared to non-patterned standard hiPSC-CMs. We also found a lack of significant differences in the transcript levels of key calcium handling genes (CACN1C, RYR2, ATP2A2, PLN, and CASQ2; Figures 3H and 5M).
Figure 5.
Calcium handling profile
(A) Representative confocal images of non-patterned and rod-shaped CMs loaded with Fluo-4-AM Ca indicator.
(B and C) Representative recordings of field stimulation-induced calcium transient (1 Hz) in non-patterned (B) and rod-shaped hiPSC-CMs (C) and their corresponding fluorescence map.
(D) Violin plot showing beating rates from non-patterned and rod-shaped hiPSC-CMs, stimulated at 1 Hz.
(E) Representative superimposed calcium recording traces from non-patterned and rod-shaped hiPSC-CMs.
(F–L) Violin plots of (F) calcium transient amplitude, (G) calcium transient area under the curve, (H) time to peak, (I) maximum rising rate, (J) maximum decay rate, (K) time to 90% relaxation, and (L) peak to 10% decay time.
(M) RNA expression of ATP2A2, PLN, CASQ2, and RYR2 measured by SYBR green in non-patterned and rod-shaped hiPSC-CMs. Cts were normalized to RPL32, and the ratio of rod-shaped vs. non-patterned was calculated for each cardiac differentiation. For (D–K), n = 100–175 cells from 3 independent experiments. Adjusted linear mixed models: ns p > 0.05, ∗∗p < 0.01 vs. non-patterned hiPSC-CMs. For (M), n = 3 independent experiments of 7 differentiations per condition. Mann-Whitney: ns, p > 0.05 vs. non-patterned hiPSC-CMs.
Table 2.
Calcium handling, related to Figure 5
| Parameter | Non-patterned | Rod-shaped | p value |
|---|---|---|---|
| Ca transient amplitude (F/F0) | 2.296 ± 0.506 | 2.125 ± 0.425 | 0.078 |
| Time to peak (ms) | 250.7 ± 114.8 | 274.1 ± 122.7 | 0.089 |
| Area under the curve (AU) | 19.23 ± 10.11 | 18.88 ± 10.32 | 0.71 |
| Beating rate (bpm) | 45.05 ± 15.66 | 44.52 ± 16.55 | 0.69 |
| Maximal rising rate (F/s) | 1230 ± 497.6 | 1115 ± 465.5 | 0.21 |
| Maximal decay rate (F/s) | 256.1 ± 107.8 | 259.9 ± 92.55 | 0.56 |
| Peak to 10% decay time (ms) | 86.80 ± 53.30 | 100.6 ± 50.62 | 0.0016 |
Ca2+ kinetic measurement of rod-shaped vs. non-patterned hiPSC paced at 1 Hz. Data presented are mean ± SD (n = 100 for non-patterned and n = 175 for rod-shaped). Adjusted mixed linear models.
Ca2+ kinetic measurement of rod-shaped vs. non-patterned hiPSC paced at 1 Hz. Data presented are mean ± SD (n = 100 for non-patterned and n = 175 for rod-shaped). Linear mixed models with adjustment on clustered data (according to differentiation and experiment).
Discussion
Human hiPSC-CMs provide a powerful cell source for disease modeling, drug screening, and toxicity testing. A major limitation of the hiPSC-CMs is that they can exhibit immature properties, which limits their utility in disease modeling and designing therapeutic applications (Koivumaki et al., 2018; Yang et al., 2014a, 2014b). Therefore, promoting the maturation of hiPSC-CMs has become an important topic in the field. To date, several approaches have been proposed to improve the maturation status of hiPSC-CMs, such as prolonged culture (Lundy et al., 2013), the use of fatty acid medium (Feyen et al., 2020; Yang et al., 2019), engineered heart tissue (Cho et al., 2022), electrical stimulation (Chan et al., 2013), or the addition of various soluble factors during the differentiation process (Yang et al., 2014a, 2014b; Yoshida et al., 2018). However, these techniques do not primarily address a key feature of mature cardiomyocytes, which is their genuine rectangular shape. Here, we directly addressed this issue by forcing hiPSC-CMs to grow under geometric constraints imposed by a micropatterned substrate made of repeated rectangles 120 μm long and 30 μm wide surrounded by a chemical cell repellent. Of note, this approach does not involve the addition of biochemical stimuli to induce changes in cell shape but rather relies on a physically induced modeling of cell architecture. We first found that the different hiPSC-CMs lines used in this study grew well on the substrate, rapidly and reproducibly, and mostly exhibited a rod-shaped morphology with improved anisotropy and sarcomere organization and alignment. In contrast, hiPSC-CMs cultured in non-patterned conditions mostly showed a circular shape. The significant change in cell shape between the two conditions is likely the predominant reason for the electrophysiological findings, underscoring the impact of cell shape on cardiomyocyte properties, including cytoskeletal structure, as previously suggested (Chen et al., 1997, 1998).
A pivotal finding of our study centers on the remarkable enhancement of the electrophysiological profile in response to the geometric constraints. The morphological changes were accompanied by several key electrophysiological changes that are characteristic of an enhanced maturation of hiPSC-CMs, such as a more hyperpolarized resting membrane potential, closer to the adult human ventricular value of −80mV (Horvath et al., 2018), and the increased proportion of cells with a late ventricular-like AP. We also found an increase in sodium current and faster depolarization velocity, in line with a shift in the expression profile of ion channels from fetal to adult isoforms. This latter finding suggests cytoskeletal changes that signal to the nucleus and influence gene expression. Prior studies have suggested that the development of contractility is dependent on the biomechanical environment (Ribeiro et al., 2015; Tsan et al., 2021). Notably, a previous study underscored how micropatterning controls cell shape and facilitates electrophysiological measurements (Jimenez-Vazquez et al., 2022a, 2022b). Our direct comparison between non-patterned standard hiPSC-CMs and rod-shaped micropatterned hiPSC-CMs provides a critical description of the achieved electrophysiological maturation that emerges from imposing geometrical constraints.
Previous reports have highlighted that the gene expression pattern of hiPSC-CMs is immature and resembles the gene expression of first-trimester fetal heart (van den Berg et al., 2015). Typically, hiPSC-CMs exhibit lower levels of sodium currents when compared to fully developed adult cardiomyocytes. The study of many cardiac arrhythmias, notably those linked to an impaired sodium current, has been hampered by the immature traits of hiPSC-CMs (Al Sayed et al., 2021a, 2021b; Campostrini et al., 2023; Zhu et al., 2021). Consistent with a 2-fold increase previously observed in hiPSC-CMs cultured on a Matrigel mattress (Feaster et al., 2015), our investigation revealed an enhancement in sodium current. This observation was substantiated by notable shifts in the expression of the adult SCN5A isoform and elevated expression of the auxiliary β subunits (i.e., SCN1B and 4B). These auxiliary subunits interact with the alpha subunit of the sodium channel, thereby influencing its kinetics and ultimately generating a more ample current, consistent with our results. As the understanding of Navβ subunit contributions to cardiac arrhythmias evolves, the micropatterning technique is poised to contribute to modeling-related hereditary channelopathies (Martinez-Moreno et al., 2020; Medeiros-Domingo et al., 2007; Ransdell et al., 2022).
Conversely, the calcium handling properties were not improved in the rod-shaped micropatterned hiPSC-CMs. The expressions of sarcomeric and calcium genes were not different, suggesting that micropatterning does not affect the gene expression of these important cardiac systems. The direct comparison of the generated hiPSC-CMs between the two culture conditions has some limitations that may have influenced our results. While patch clamping was performed at the single-cell level, calcium imaging was performed directly on the wells capturing the signal of multiple cells at a time. While our micropatterning technique includes a cell repellent that prevents cells from spreading and forming contacts with other cells, hiPSC-CMs grown under standard non-patterned conditions can connect with each other, which may promote maturation of the excitation-contraction coupling machinery. Another limitation is that our micropatterning design limits cell-to-cell connections and does not reproduce the 3D structure found in the native myocardium. This approach allows the production of hiPSC-CMs with morphology closer to that observed in mature isolated adult cardiomyocytes, but this does not perfectly reflect the in situ environment by recreating a cardiac myofibril for instance. Lastly, the large intercellular variability also suggests that not all cells correctly followed the imposed stimulation, which could have affected our results.
Other micropatterning designs may be useful to better mimic the myofibril organization. However, we specifically chose to use a 120 × 30 μm rectangular design because rod-shaped iPSC-CMs are more suitable for a variety of techniques performed on isolated cardiomyocytes, including stretch assays, ion imaging, or patch clamping. Of note, other format might be more appropriate to study the contractile function, such as ring-shaped cardiac organoids as recently developed (Seguret et al., 2024; Vermersch et al., 2024).
Our study has some limitations. First, the vast majority of cells in the non-patterned conditions spontaneously adopt a circular shape, but this was not imposed by, for example, a circular micropatterned substrate. Thus, we cannot exclude that factors other than cell shape change contribute to the electrophysiological maturation observed in the rod-shaped micropatterned hiPSC-CMs. Second, the analysis of contractility parameters from raw image sequences of video recordings obtained with this assay will require further development to detect and record signals at the single-cell level. It should also be noted that approximately half of the rectangular patterns contained 2 or more cells, a feature that could not be controlled for in the assay. These issues appear to be a limitation of the assay. Third, the responses to different pharmacological compounds (including ionotropic drugs) have not yet been tested.
In conclusion, we propose a simple method to boost the architectural maturity of hiPSC-CMs. This involves culturing these cells on a rod-shaped cell micropatterned substrate formed by repeated rectangles with a size ratio of 1:4. This induces a shape similar to adult cardiomyocytes in a short time frame, all without the use of supplements that may impact cardiomyocyte behavior. The resulting hiPSC-CMs exhibit numerous structural and electrical characteristics that align them more closely with adult human cardiomyocytes. However, other parameters, like sarcomere length and calcium amplitude, were not improved. This model holds potential to replicate the features and function of adult cardiomyocytes in vitro more faithfully and reproducibly, advancing thereby our capability to comprehend diverse cardiovascular diseases and drug discovery at the single-cell level.
Experimental procedures
hiPSC reprograming and maintenance
All hiPSC lines were reprogrammed from skin fibroblasts. The three hiPSC lines used in this study were SKiPSC-31.3 (C01), which was derived from a healthy adult male volunteer (Karakikes et al., 2014), and two lines (P11007 (C02) and P11014 (C03)) which were derived from healthy male and female donors, respectively (Stillitano et al., 2017). HiPSCs were seeded onto stem cell-qualified Matrigel-coated (354277, Corning) plates and maintained in mTeSR Plus medium (100-0276, STEMCELL Technologies) in a humidified incubator (5% CO2, 37°C). At 70%–80% confluence, cells were passaged in clumps by scraping with a pipette tip. Pluripotency was assessed by immunofluorescence and routinely tested for mycoplasma every 1 month according to the manufacturer’s recommendations (Lonza, LT07-318). Genome stability was assessed by detection of recurrent genetic abnormalities using the iCS-digital PSC test, provided as a service by Stem Genomics (https://www.stemgenomics.com/), as described previously (Assou et al., 2020).
hiPSC-CMs differentiation and culture
Once confluent, hiPSC were differentiated into cardiomyocytes using a chemically defined cardiac differentiation protocol (adapted from Garg et al., 2018). Briefly, mTeSR Plus was replaced with RPMI-1640 supplemented with B27 supplement minus insulin (A1895601, Thermo Fisher Scientific) and 6 μM CHIR-99021 (120890, Abcam) and maintained in a 5% CO2/air environment for 48 h. The medium was then changed to RPMI-1640/B27 minus insulin for 24 h and switched to RPMI-B27 minus insulin supplemented with 5 μM IWR-1 (I0161, Sigma-Aldrich) for 48 h. On day 5, the medium was changed back to RPMI-1640/B27 minus insulin for 48 h. Starting on day 7, the cells were placed in RPMI-1640 with B27 supplement plus insulin (17505044, Thermo Fisher Scientific). On day 11, beating hiPSC-CMs were subjected to glucose starvation in RPMI-1640/B27 plus insulin without glucose (11879020, Thermo Fischer Scientific) for 3 days. The cells were then dissociated using 0.05% trypsin (25300104, Thermo Fisher Scientific) for 10 min and seeded at 0.3 × 106 cells/cm2 in RPMI-B1640/B27 plus insulin for 24 h. The cells were then subjected to a second round of glucose starvation for 3 days. These two rounds of glucose have been shown to dramatically increase the percentage of cardiomyocytes obtained (Sharma et al., 2015). Until day 18, the cells were cultured in RPMI-1640/B27 plus insulin and the medium was changed every two days.
Growing cells on rod-shaped micropatterned surface
Single-cell micropatterned glass slides are coated with a cell repellent, based on a modified Polydymethylsiloxane polymer, which is then burned by ultraviolet light with a mask designed to contain repeating rectangles 120 μm long and 30 μm wide, with each rectangle being separated by 300 μm. The mask was designed by our team, and the micropatterns were fabricated by the small bioengineering company 4Dcell (Montreuil, France). Prior to cell seeding, micropatterned glass slides were coated with Matrigel. At day 35 after differentiation, hiPSC-CMs were seeded onto the Matrigel-coated micropatterned glass slides at a density of approximately 18 × 103 cells/cm2 and allowed to grow for 7 to 10 days in RPMI-1640/B27 plus insulin at 37°C in 5% CO2/air environment. hiPSC-CMs cultured under standard unconstrained conditions were used as a control (non-patterned condition).
Flow cytometry analysis
On day 21 of differentiation, hiPSC-CMs were dissociated with 0.05% trypsin-EDTA (130-110-204, Miltenyi Biotec) and stained with the fixable Zombie NIR viability kit (423105, BioLegend). The collected cell pellets were then fixed and permeabilized with the Inside Stain kit (130-090-477, Miltenyi Biotec) for 10 min at room temperature. Cells were incubated with either Allophycocyanin (APC) anti-cardiac troponin T (cTnT) antibody (130-120-403, Miltenyi Biotec; 1:100) or APC isotype control (130-120-709, Miltenyi Biotec; 1:100) for 10 min at room temperature. Cells were analyzed on a BD Biosciences fluorescence-activated cell sorting LSRFortessa X-20 instrument with a minimum of 30,000 cells. Results were processed using FlowJo v10 (FlowJo, LLC).
Immunostaining
HiPSC-CMs were cultured on Matrigel-coated rod-shaped or non-patterned glass slides. After one week, the cells were sequentially fixed with 4% paraformaldehyde (1573590, Electron Microscopy Sciences) for 10 min at room temperature and then permeabilized and blocked with a blocking solution containing 0.5% Triton X-100 (T-8787, Sigma) and 2% bovine serum albumin (001-000-162, Jackson ImmunoResearch) in PBS for 1 h. Subsequently, primary antibody incubation was performed overnight at 4°C in 1:10 diluted blocking solution: anti-cTnT (ab45932, Abcam; 1:500), anti-alpha-actinin (A7811, Sigma-Aldrich; 1:1000), or anti-Nav1.5 (ASC-005, Alomone; 1:100). After washing, cells were incubated with secondary antibodies goat anti-mouse or anti-rabbit immunoglobulin G conjugated to Alexa Fluor 488 (A10680, Thermo Fisher Scientific; 1:500) or 546 (A11010, Thermo Fisher Scientific; 1:500)/DAPI and mounted with Dako Faramount aqueous mounting (S3025, Agilent).
Beating rate analysis
Analysis of spontaneous beating rates was performed on hiPSC-CMs after 7 days of culture on rod-shaped or non-patterned slides using Fiji. Briefly, 30-s videos were recorded and converted to raw .avi files for analysis and then imported into Fiji. Region of interest and multi measure were used to obtain peak contraction over time. Peak-to-peak duration was used to calculate beating rates.
Imaging and morphological analysis
Fluorescent images were acquired using a camera mounted on a Leica TSC SP8 microscope, and confocal images were processed using Fiji. Morphological features and internal organization of sarcomeres were analyzed with an adapted MATLAB interface of SarcOmere Texture Analysis (SOTA) coded by Sutcliffe et al., 2018. A total of 50–100 cells were analyzed per condition from three independent experiments.
RNA extraction and qPCR
Total RNA preparations were extracted from hiPSC-CMs after 7 days of culture on rod-shaped or non-patterned slides using the NucleoSpin RNA XS kit (740902.50, MACHEREY-NAGEL) and retrotranscribed using the SuperScript IV VILO kit (11756050, Thermo Fisher Scientific) as per the manufacturer’s instructions. Real-time qPCR was performed using amplified cDNA, gene-specific primers (Table S1), and an SYBR select master mix (Thermo Fisher Scientific, 4472908) on the QuantStudio 3 real-time PCR system (Applied Biosystems). RPL32 was used as the housekeeping gene. Relative mRNA levels were normalized to non-patterned samples.
Patch clamp
Cardiomyocytes were enzymatically dissociated into single cells on day 35 of differentiation using 0.05% trypsin-EDTA for 10 min at 37°C. Single-cell cardiomyocytes were then plated on Matrigel-coated slides with vs. without the rectangular rod-shaped micropattern. Ten days after seeding, patch-clamp recordings were performed using an Axopatch 200B amplifier (Molecular Devices) controlled by Axon pCLAMP 11 software through an A/D converter (Digidata 1550B; Molecular Devices). All recordings were made at 37°C. Data were collected from at least three independent differentiations and analyzed using the pCLAMP software (Molecular Devices).
AP recording
Using an amphotericin-B perforated-patch configuration, APs were recorded from hiPSC-CMs bathed in a Tyrode’s solution containing (in mM): 140 NaCl, 4 KCl, 1 CaCl2, 0.5 MgCl2, 10 glucose, and 10 HEPES (pH 7.4 adjusted with NaOH). Borosilicate glass pipettes (4–5 MΩ of tip resistance) were filled with a solution containing (in mM): 125 K-Gluconate, 20 KCl, 5 NaCl, 5 HEPES, and 0.22 amphotericin-B (pH 7.2 adjusted with KOH). The beating rate was calculated from spontaneous APs. Cells were paced with 1 ms 30–50 pA/pF stimulation pulse at 1,000 ms (1 Hz frequency) of cycle length using custom software running on RT-Linux and an A/D converter (National Instruments PCI-6221) connected to the current command of the amplifier (Al Sayed et al., 2021a; 2021b). AP characterization was calculated from an average of 7 consecutive APs. APs with APD30-40/APD70-80 <1.5 were classified as either nodal (dV/dtmax < 10 V/s) or atrial (dV/dtmax > 10 V/s); those with a ratio APD30-40/APD70-80 >1.5 were classified as early ventricular-like (dV/dtmax < 30 V/s) or late ventricular-like type (dV/dtmax > 30 V/s) (Fukushima et al., 2020).
Sodium current measurement
INa measurements were recorded in a ruptured patch configuration and low-pass filtered at 5 kHz. Cells were bathed with a Tyrode’s solution containing (in mM): 130 NaCl, 10 CsCl, 1.8 CaCl2, 1.2 MgCl2, 11 glucose, and 5 HEPES (pH 7.4 adjusted with NaOH). Pipettes with 2–4 MΩ tip resistance were filled with a solution containing (in mM): 90 CsGluconate, 5 NaCl, 45 CsCl, 1 EGTA, 10 HEPES, and 2 MgATP; (pH 7.2 adjusted with CsOH). The holding potential was set to −100 mV. Series resistance was compensated. Leakage current was subtracted. Peak current was normalized to cell capacitance (Cm) and plotted against voltage to generate the peak current density-voltage curve. Steady-state activation and inactivation curves were fitted using a Boltzmann equation: I/Imax = A/{1.0+exp [(V1/2 – V)/k} where k > 0 for the activation curve and k < 0 for the inactivation curve.
Calcium transient imaging
Following a 10-day recovery period after seeding, cells were loaded with 2 μM Fluo-4-AM Direct calcium assay kit (F10471, Thermo Fisher Scientific) for 15 min at 37°C. To allow complete de-esterification of the Ca2+ probe, the medium was then replaced with a solution containing 140 mM NaCl, 5 mM KCl, 10 mM glucose, 10 mM HEPES, 1 mM MgCl2, and 1.8 mM CaCl2 (pH 7.4 adjusted with NaOH) for 30 min. Fluorescence videos of calcium transients were captured at 44 frames per second using a Leica TSC SP8 imaging system at 37°C incubation. Each recording was made using electric field stimulation at 1 Hz with custom-made platinum electrodes connected to a stimulus generator (STG1002, Multi Channel Systems). Raw imaging data were analyzed using CalTrack MATLAB script (MathWorks) (Psaras et al., 2021).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 10.0.2 and the R environment. Data are presented as mean ± standard error of the mean (SEM). The Mann-Whitney U test was used for comparing continuous variables between two groups. Two-way analysis ANOVA with Bonferroni post hoc test was used for analysis of sodium current densities and activation and inactivation curves. For calcium transients’ data, the comparison of variables between groups was tested using linear mixed models adjusted for clustered data (i.e., differentiation and seeding preparation). Fisher’s exact test or χ2 test was used for discrete variables as appropriate. p values <0.05 were considered statistically significant.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jean-Sébastien Hulot (jean-sebastien.hulot@inserm.fr).
Materials availability
The rod-shaped cell micropatterned substrate generated in this study is marketed by 4DCell (Montreuil, France). Other materials generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and code availability
This study did not generate new datasets.
Acknowledgments
We thank Camille Knops and Yunling Xu from the Flow Cytometry and the Microscopy platforms, respectively, from Université de Paris, Paris Cardiovascular Research Center, Paris, France. We thank 4Dcell staff for discussion about the cell repellant and design of the rod-shaped micropatterns. This work was supported by grants from the Leducq Foundation (CVD18-05), from the Fondation pour la Recherche Médicale (EQU201903007852), and from ANR (CORRECT_LMNA ANR-19-CE17-0013-02).
Author contributions
Conceptualization, J.-S.H., C.J., and Z.R.A.S.; funding acquisition, J.-S.H.; investigations, Z.R.A.S., C.J., M.S., C.P., and A.R.-V.; methodology, J.-S.H. and D.-A.T.; project administration, J.-S.H.; resources, C.J.; supervision, J.-S.H.; writing – original draft, J.-S.H., C.J., and Z.R.A.S.; writing – review and editing, M.S., A.R.-V., C.P., and D.-A.T.
Declaration of interests
J.-S.H. reports research grants from Sanofi and Pliant Therapeutics unrelated to the present work and speaker, advisory board, or consultancy fees from Alnylam, Amgen, AstraZeneca, Bayer Pharma, BioSerenity, Boehringer Ingelheim, Bristol-Myers Squibb, Novartis, Novo Nordisk, and Vifor Pharma all unrelated to the present work.
The rod-shaped cell micropatterned substrate is marketed by 4DCell (Montreuil, France).
Published: September 19, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.08.005.
Supplemental information
References
- Andres-Delgado L., Mercader N. Interplay between cardiac function and heart development. Biochim. Biophys. Acta. 2016;1863:1707–1716. doi: 10.1016/j.bbamcr.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Sayed Z.R., Canac R., Cimarosti B., Bonnard C., Gourraud J.B., Hamamy H., Kayserili H., Girardeau A., Jouni M., Jacob N., et al. Human model of IRX5 mutations reveals key role for this transcription factor in ventricular conduction. Cardiovasc. Res. 2021;117:2092–2107. doi: 10.1093/cvr/cvaa259. [DOI] [PubMed] [Google Scholar]
- Al Sayed Z.R., Jouni M., Gourraud J.B., Belbachir N., Barc J., Girardeau A., Forest V., Derevier A., Gaignerie A., Chariau C., et al. A consistent arrhythmogenic trait in Brugada syndrome cellular phenotype. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S., Girault N., Plinet M., Bouckenheimer J., Sansac C., Combe M., Mianné J., Bourguignon C., Fieldes M., Ahmed E., et al. Recurrent Genetic Abnormalities in Human Pluripotent Stem Cells: Definition and Routine Detection in Culture Supernatant by Targeted Droplet Digital PCR. Stem Cell Rep. 2020;14:1–8. doi: 10.1016/j.stemcr.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman H.J., Coleman H., Riley H.E., Olale F., Tsai H.J., Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M.A., Sheehy S.P., Parker K.K. Sarcomere alignment is regulated by myocyte shape. Cell Motil Cytoskeleton. 2008;65:641–651. doi: 10.1002/cm.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campostrini G., Kosmidis G., Ward-van Oostwaard D., Davis R.P., Yiangou L., Ottaviani D., Veerman C.C., Mei H., Orlova V.V., Wilde A.A.M., et al. Maturation of hiPSC-derived cardiomyocytes promotes adult alternative splicing of SCN5A and reveals changes in sodium current associated with cardiac arrhythmia. Cardiovasc. Res. 2023;119:167–182. doi: 10.1093/cvr/cvac059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.C., Ting S., Lee Y.K., Ng K.M., Zhang J., Chen Z., Siu C.W., Oh S.K.W., Tse H.F. Electrical stimulation promotes maturation of cardiomyocytes derived from human embryonic stem cells. J. Cardiovasc. Transl. Res. 2013;6:989–999. doi: 10.1007/s12265-013-9510-z. [DOI] [PubMed] [Google Scholar]
- Chen C.S., Mrksich M., Huang S., Whitesides G.M., Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Chen C.S., Mrksich M., Huang S., Whitesides G.M., Ingber D.E. Micropatterned surfaces for control of cell shape, position, and function. Biotechnol. Prog. 1998;14:356–363. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- Cho S., Discher D.E., Leong K.W., Vunjak-Novakovic G., Wu J.C. Challenges and opportunities for the next generation of cardiovascular tissue engineering. Nat. Methods. 2022;19:1064–1071. doi: 10.1038/s41592-022-01591-3. [DOI] [PubMed] [Google Scholar]
- Feaster T.K., Cadar A.G., Wang L., Williams C.H., Chun Y.W., Hempel J.E., Bloodworth N., Merryman W.D., Lim C.C., Wu J.C., et al. Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyen D.A.M., McKeithan W.L., Bruyneel A.A.N., Spiering S., Hörmann L., Ulmer B., Zhang H., Briganti F., Schweizer M., Hegyi B., et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H., Yoshioka M., Kawatou M., López-Dávila V., Takeda M., Kanda Y., Sekino Y., Yoshida Y., Yamashita J.K. Specific induction and long-term maintenance of high purity ventricular cardiomyocytes from human induced pluripotent stem cells. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P., Oikonomopoulos A., Chen H., Li Y., Lam C.K., Sallam K., Perez M., Lux R.L., Sanguinetti M.C., Wu J.C. Genome Editing of Induced Pluripotent Stem Cells to Decipher Cardiac Channelopathy Variant. J. Am. Coll. Cardiol. 2018;72:62–75. doi: 10.1016/j.jacc.2018.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron T.J., Rocha A.M.D., Campbell K.F., Ponce-Balbuena D., Willis B.C., Guerrero-Serna G., Liu Q., Klos M., Musa H., Zarzoso M., et al. Extracellular Matrix-Mediated Maturation of Human Pluripotent Stem Cell-Derived Cardiac Monolayer Structure and Electrophysiological Function. Circ. Arrhythm. Electrophysiol. 2016;9 doi: 10.1161/CIRCEP.113.003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A., Lemoine M.D., Loser A., Mannhardt I., Flenner F., Uzun A.U., Neuber C., Breckwoldt K., Hansen A., Girdauskas E., et al. Low Resting Membrane Potential and Low Inward Rectifier Potassium Currents Are Not Inherent Features of hiPSC-Derived Cardiomyocytes. Stem Cell Rep. 2018;10:822–833. doi: 10.1016/j.stemcr.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vazquez E.N., Arad M., Macias A., Vera-Pedrosa M.L., Cruz F.M., Gutierrez L.K., Cuttitta A.J., Monteiro da Rocha A., Herron T.J., Ponce-Balbuena D., et al. SNTA1 gene rescues ion channel function and is antiarrhythmic in cardiomyocytes derived from induced pluripotent stem cells from muscular dystrophy patients. Elife. 2022;11 doi: 10.7554/eLife.76576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vazquez E.N., Jain A., Jones D.K. Enhancing iPSC-CM Maturation Using a Matrigel-Coated Micropatterned PDMS Substrate. Curr. Protoc. 2022;2 doi: 10.1002/cpz1.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I., Senyei G.D., Hansen J., Kong C.W., Azeloglu E.U., Stillitano F., Lieu D.K., Wang J., Ren L., Hulot J.S., et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl. Med. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivumaki J.T., Naumenko N., Tuomainen T., Takalo J., Oksanen M., Puttonen K.A., Lehtonen Š., Kuusisto J., Laakso M., Koistinaho J., Tavi P. Structural Immaturity of Human iPSC-Derived Cardiomyocytes: In Silico Investigation of Effects on Function and Disease Modeling. Front. Physiol. 2018;9:80. doi: 10.3389/fphys.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.F., Swinburne I., Yelon D. Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev. Biol. 2012;362:242–253. doi: 10.1016/j.ydbio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy S.D., Zhu W.Z., Regnier M., Laflamme M.A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moreno R., Selga E., Riuró H., Carreras D., Parnes M., Srinivasan C., Wangler M.F., Pérez G.J., Scornik F.S., Brugada R. An SCN1B Variant Affects Both Cardiac-Type (Na(V)1.5) and Brain-Type (Na(V)1.1) Sodium Currents and Contributes to Complex Concomitant Brain and Cardiac Disorders. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.528742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros-Domingo A., Kaku T., Tester D.J., Iturralde-Torres P., Itty A., Ye B., Valdivia C., Ueda K., Canizales-Quinteros S., Tusié-Luna M.T., et al. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S.S., Blackwell D.J., Gomez-Hurtado N., Frisk M., Wang L., Kim K., Dahl C.P., Fiane A., Tønnessen T., Kryshtal D.O., et al. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 2017;121:1323–1330. doi: 10.1161/CIRCRESAHA.117.311920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K.K., Tan J., Chen C.S., Tung L. Myofibrillar architecture in engineered cardiac myocytes. Circ. Res. 2008;103:340–342. doi: 10.1161/CIRCRESAHA.108.182469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioner J.M., Santini L., Palandri C., Langione M., Grandinetti B., Querceto S., Martella D., Mazzantini C., Scellini B., Giammarino L., et al. Calcium handling maturation and adaptation to increased substrate stiffness in human iPSC-derived cardiomyocytes: The impact of full-length dystrophin deficiency. Front. Physiol. 2022;13 doi: 10.3389/fphys.2022.1030920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaras Y., Margara F., Cicconet M., Sparrow A.J., Repetti G.G., Schmid M., Steeples V., Wilcox J.A.L., Bueno-Orovio A., Redwood C.S., et al. CalTrack: High-Throughput Automated Calcium Transient Analysis in Cardiomyocytes. Circ. Res. 2021;129:326–341. doi: 10.1161/CIRCRESAHA.121.318868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransdell J.L., Moreno J.D., Bhagavan D., Silva J.R., Nerbonne J.M. Intrinsic mechanisms in the gating of resurgent Na(+) currents. Elife. 2022;11 doi: 10.7554/eLife.70173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A.J.S., Ang Y.S., Fu J.D., Rivas R.N., Mohamed T.M.A., Higgs G.C., Srivastava D., Pruitt B.L. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. USA. 2015;112:12705–12710. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A.J.S., Schwab O., Mandegar M.A., Ang Y.S., Conklin B.R., Srivastava D., Pruitt B.L. Multi-Imaging Method to Assay the Contractile Mechanical Output of Micropatterned Human iPSC-Derived Cardiac Myocytes. Circ. Res. 2017;120:1572–1583. doi: 10.1161/CIRCRESAHA.116.310363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguret M., Davidson P., Robben S., Jouve C., Pereira C., Lelong Q., Deshayes L., Cerveau C., Le Berre M., Rodrigues Ribeiro R.S., Hulot J.S. A versatile high-throughput assay based on 3D ring-shaped cardiac tissues generated from human induced pluripotent stem cell derived cardiomyocytes. Elife. 2024;12 doi: 10.7554/eLife.87739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Li G., Rajarajan K., Hamaguchi R., Burridge P.W., Wu S.M. Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J. Vis. Exp. 2015;18 doi: 10.3791/52628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillitano F., Hansen J., Kong C.W., Karakikes I., Funck-Brentano C., Geng L., Scott S., Reynier S., Wu M., Valogne Y., et al. Modeling susceptibility to drug-induced long QT with a panel of subject-specific induced pluripotent stem cells. Elife. 2017;6 doi: 10.7554/eLife.19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe M.D., Tan P.M., Fernandez-Perez A., Nam Y.J., Munshi N.V., Saucerman J.J. High content analysis identifies unique morphological features of reprogrammed cardiomyocytes. Sci. Rep. 2018;8:1258. doi: 10.1038/s41598-018-19539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 2010;123:4201–4213. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- Thomas D., Cunningham N.J., Shenoy S., Wu J.C. Human-induced pluripotent stem cells in cardiovascular research: current approaches in cardiac differentiation, maturation strategies, and scalable production. Cardiovasc. Res. 2022;118:20–36. doi: 10.1093/cvr/cvab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsan Y.C., DePalma S.J., Zhao Y.T., Capilnasiu A., Wu Y.W., Elder B., Panse I., Ufford K., Matera D.L., Friedline S., et al. Physiologic biomechanics enhance reproducible contractile development in a stem cell derived cardiac muscle platform. Nat. Commun. 2021;12:6167. doi: 10.1038/s41467-021-26496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C.W., Okawa S., Chuva de Sousa Lopes S.M., van Iperen L., Passier R., Braam S.R., Tertoolen L.G., del Sol A., Davis R.P., Mummery C.L. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development. 2015;142:3231–3238. doi: 10.1242/dev.123810. [DOI] [PubMed] [Google Scholar]
- Vermersch E., Neuvendel S., Jouve C., Ruiz-Velasco A., Pereira C., Seguret M., Cattin-Messaoudi M.E., Lotfi S., Dorval T., Berson P., Hulot J.S. hsa-miR-548v controls the viscoelastic properties of human cardiomyocytes and improves their relaxation rates. JCI Insight. 2024;9 doi: 10.1172/jci.insight.161356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Pabon L., Murry C.E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Rodriguez M., Pabon L., Fischer K.A., Reinecke H., Regnier M., Sniadecki N.J., Ruohola-Baker H., Murry C.E. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Rodriguez M.L., Leonard A., Sun L., Fischer K.A., Wang Y., Ritterhoff J., Zhao L., Kolwicz S.C., Jr., Pabon L., et al. Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2019;13:657–668. doi: 10.1016/j.stemcr.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Miyagawa S., Fukushima S., Kawamura T., Kashiyama N., Ohashi F., Toyofuku T., Toda K., Sawa Y. Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Mol. Ther. 2018;26:2681–2695. doi: 10.1016/j.ymthe.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Lan H., El-Battrawy I., Li X., Buljubasic F., Sattler K., Yücel G., Lang S., Tiburcy M., Zimmermann W.H., et al. Ion Channel Expression and Characterization in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cells Int. 2018;2018 doi: 10.1155/2018/6067096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang L., Cui C., Qin H., Chen H., Chen S., Lin Y., Cheng H., Jiang X., Chen M. Pathogenesis and drug response of iPSC-derived cardiomyocytes from two Brugada syndrome patients with different Na (v)1.5-subunit mutations. J. Biomed. Res. 2021;35:395–407. doi: 10.7555/JBR.35.20210045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets.