Abstract
PROTACs are an emerging therapeutic approach towards targeted protein degradation. This article examines the leading examples of this modality that are in clinical development through the prism of their physicochemical properties. In particular, the optimisation of the various components of PROTACs together with the difficulties faced by medicinal chemists seeking to achieve oral bioavailability in this challenging space are outlined. Guidance, opinion and advice based on the authors' own experiences in this area are offered in the hope this may be useful to others working in this fascinating frontier of drug discovery.
PROTACs are an emerging therapeutic approach towards targeted protein degradation. This article examines the leading examples of this modality that are in clinical development through the prism of their physicochemical properties.
Introduction
PROteolysis TArgeting Chimeras (PROTACs), are an emerging therapeutic modality.1,2 They are molecules which bring a protein of interest (POI) into close proximity with an E3 ligase and engage the ubiquitin-proteasome system (UPS) to facilitate targeted protein degradation. The ability to selectively remove proteins from a cell on an event driven basis, rather than a more traditional occupancy-based inhibition of protein function, offers several advantages. In terms of pharmacology, this has the potential to offer a more complete blockade of protein function (e.g. removal of scaffolding activity) and is therefore more likely to recapitulate the effects of a genetic knock-down experiment. In simple terms, this not only blocks signalling but removes the ability of a cell to broadcast that signal by dismantling the machinery required to do so. The cell can only regain function by generating a new protein, therefore the pharmacology is influenced by the resynthesis rate of the protein as well as the pharmacodynamics of the drug, offering the potential for extended duration of action. Additionally, the requisite interplay between the POI and E3 components in the PROTAC mechanism offer additional opportunities for selectivity between isoforms (e.g. depending on the ternary complex formation and/or available surface lysine residues)3,4 and within tissues (e.g. depending on E3 expression levels).5 Consequently the PROTAC paradigm has attracted immense interest within the medicinal chemistry community and has been the subject of several excellent recent reviews.6–8 The first examples of this modality have been demonstrated to be well tolerated and efficacious in patients and have advanced into late stage clinical trials. This opinion article focusses on the physicochemical and drug-like properties of PROTACs and discusses factors relevant to their optimisation. It is hoped that this will be useful for those working in this rapidly evolving field.
Clinical PROTACs
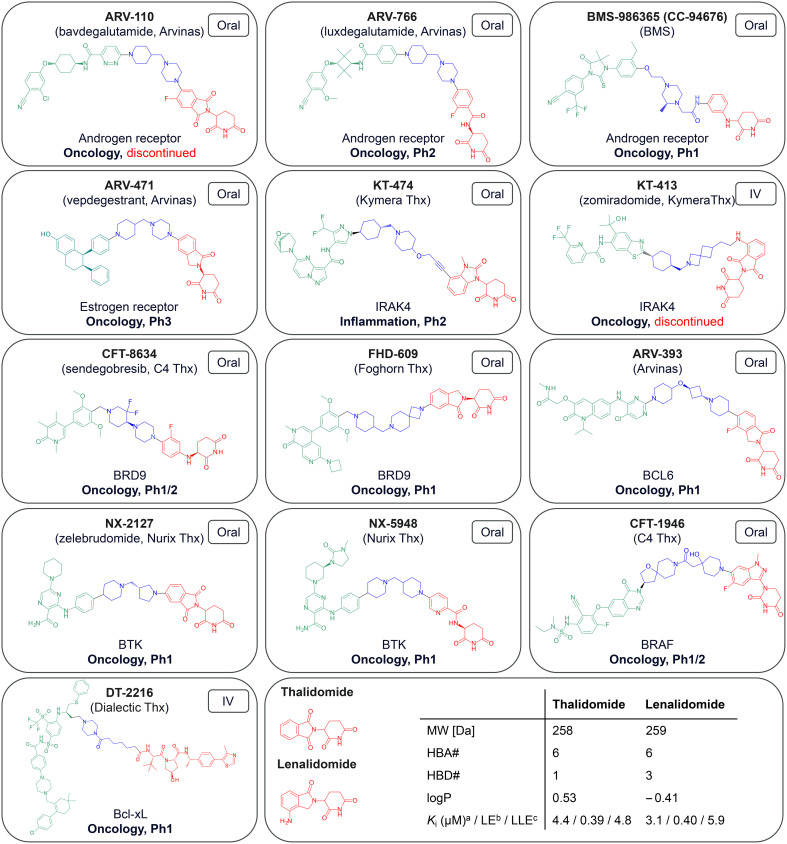
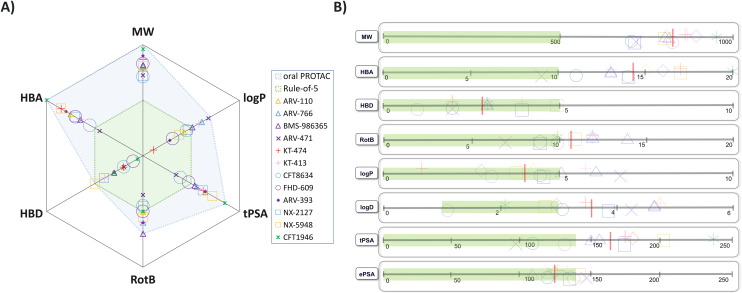
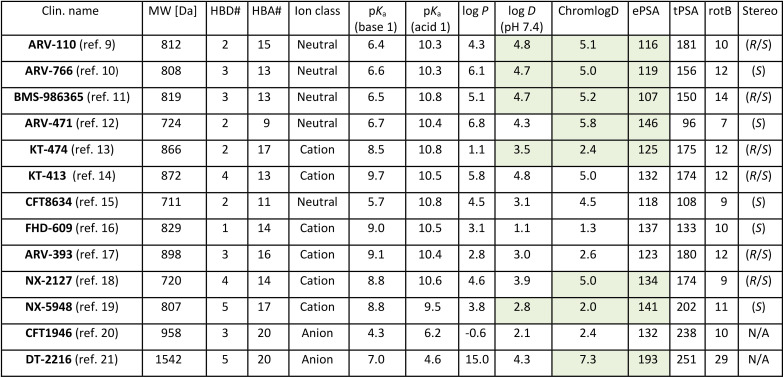
As of July 2024 there were 13 PROTACs whose structures have been disclosed that have progressed to clinical trials, as described in Fig. 1.9–21 The majority are for oncology related indications, with one treatment for inflammation. Two of these (ARV-110, KT-413) have subsequently been discontinued for strategic reasons. Twelve of them engage the E3 ligase cereblon (CRBN) and only the IV-administered DT-2216 carries a von-Hippel–Lindau (VHL) E3 ligase-binding motif. The physicochemical properties of these compounds are detailed in Table 1. The CRBN-binding PROTACs range in molecular weight (MW) from 711 to 958 Da, in lipophilicity (log D at pH 7.4) from 1.1 to 4.8, and in chromatographic polar surface area (ePSA) from 107 to 146 Å2 (Table 1). Calculated predictions of molecular lipophilicity (log P) and polar surface area (tPSA) scatter much wider with log P ranging from −0.6 to 6.8 and tPSA from 96 to 238 Å2. Formal hydrogen bond acceptors (HBA) range from 9 to 20, while formal hydrogen bond donors (HBD) are all ≤5. Fig. 2A highlights that the property space of clinical oral PROTAC (blue area) expands beyond Lipinski's ‘Rule-of-5’ (Ro5)22 and Veber's guidelines regarding rotatable bonds (RotB) and polar surface area (PSA)23 (green area) in all properties, with the notable exception of HBD. Fig. 2B shows bar charts of properties with accepted property ranges for small molecules shown as green bars. In terms of lipophilicity, several of the PROTACs push beyond the limits of log P (≤5) and log D (1–3) established for small molecules. Notably the values for ePSA (107 to 146 Å2) occupy a much narrower range than tPSA (96 to 238 Å2) suggesting this may be a more valuable guideline for design of oral PROTACs.
Fig. 1. Chemical structures of clinical stage PROTACs coloured by POI-ligand (green), linker (blue) and E3-ligand (red) and labelled with company, molecular target, disease area and current clinical status. aKi values taken from Heim et al.24bLE = ligand efficiency, expressed in kcal mol−1 per heavy atom count; cLLE = lipophilic ligand efficiency = pKi – log P; LE, LLE not disclosed in original publication.
Physicochemical properties of clinical-stage PROTACs.
|
Fig. 2. Physicochemical properties of clinical CRBN PROTACs coloured by compound name. A) Spider plot of properties with Ro5 property space shaded green and PROTAC shaded blue; B) bar charts of properties with accepted property ranges for small molecules shown as green bars, mean values of PROTACs shown as red lines and compounds jittered on y-axis for ease of visualisation.
Despite the extension beyond previously established physicochemical space constraints, oral administration remains the preferred route of administration, with the exception of IV-administered DT-2216 and KT-413 specifically developed for haematological cancers.
In the sum of the parts, there is only the parts
Each of the PROTACs described above have been the subject of an extensive optimisation campaign to achieve the optimal balance of pharmacological and physicochemical properties. With their modular, tri-component nature, the skill in PROTAC design lies in counterbalancing the features of the individual pieces so that the overall properties of the resultant PROTAC are in an acceptable space.26
POI-ligand: potent, with few exposed HBD
Many of the PROTACs utilise modified inhibitors/antagonists against the target POI. These are often highly potent and ligand efficient motifs from which an exit vector to solvent has been identified allowing linkage to the E3 binding motif. Most are neutral, rather than acids or bases, and have ≤1 HBD. ARV-393 has 3 formal donors and although NX-2127, KT-413 and NX-5948 have 4, 4 and 5 formal donors respectively, intramolecular hydrogen bonding likely shields several of them.
E3: CRBN leading the way27
Taking into consideration recent reports of novel ligands and corresponding PROTACs for a growing number of E3 ligases,28,29 the preponderance of CRBN-harnessing PROTACs in clinical trials is notable. Ligands of CRBN (Fig. 1, red 2D structures), including those derived from the immunomodulatory imide drugs (IMiDs) thalidomide and lenalidomide,30 are highly polar, efficient, have low MW and only one HBD within the essential binding pharmacophore (glutarimide or dihydrouracil ring). This, together with their lack of susceptibility towards efflux transporters, makes them ideal for incorporating into PROTACs, especially if oral bioavailability is to be achieved. The safety concerns associated with the IMiD class of ligands (e.g. teratogenicity, haematotoxicity)31 may explain why the initial therapeutic indications for PROTACs have predominantly been in oncology.31 Perhaps in response to this, PROTACs composed of novel CRBN ligands, distinct from the IMiD class are now starting to emerge. These ligands include both epimeric mixtures (e.g.KT-474, BMS-986365) and chirally pure glutarimides (e.g.ARV-766, CFT-8634, NX-5948). Of note is the dihydrouracil (DHU) functional group present in CFT-1946. This achiral motif represents an intriguing replacement for the ubiquitous glutarimide, not least because of recent reports of its improved chemical and metabolic stability.32–34
Linker: bases and ring-rich motifs predominate
A common feature of the linkers is the presence of a single tertiary amine base, either piperidine, piperazine or azetidine, with CFT-1946 being a notable exception to this observation. In some cases, the basicity of a piperidine appears to have been tempered with the addition of fluoro (CFT-8634) or oxygen (ARV-393, KT-474) substituents with basicities not exceeding a predicted pKa of 9.1 for all orally administered PROTACs (Table 1). The intravenously dosed PROTAC DT-2216 has a very high predicted lipophilicity (log P) of 15.0, however, this is reduced to a much lower log D of 4.3, probably due to its zwitterionic charge state. Its high experimental polarity of 193 Å2, which exceeds the ePSA values of the oral PROTACs by 47 Å2 and the ePSA of the related bRo5 orally administered drug venetoclax by 31 Å2,35 appears to reflect this zwitterionic state.
Another common aspect of oral PROTACs is the use of cyclic rings as a conformational constraint with all examples containing between one and three rings in the linker. In terms of flexibility, the majority of examples contain a methylene spacer between rings with only BMS-986365 having an ethoxy 3-atom spacer. CFT-1946 represents the most conformationally rigid linker with an amide linking a piperidine with a spirocycle. KT-474 has the largest acyclic component with a 4-atom propargylic ether spacer but even this is relatively rigid suggesting that conformational constraint is an important design consideration for PROTACs. This notion is supported by systematic linker optimisation of preclinical AR degraders revealing that ring-rich, rigid linkers can yield higher cellular target degradation potencies and higher metabolic stability.36,37 Consequently, rotatable bonds are only marginally above the upper limits recommended for Ro5 compliant oral drugs.23 As a stark outlier, DT-2216 with 29 rotatable bonds shows that highly flexible linkers can still lead to potent degraders and are suitable for IV-administered PROTACs.
The challenge of oral bioavailability
As outlined above, the tripartite composition makes it challenging to build a PROTAC with a MW below ∼700 Da. This violation of Lipinski's rule on MW is often accompanied by challenges of maintaining other Ro5 criteria (HBA, HBD, log P) and physicochemical descriptor limits (PSA, rotatable bonds).38 Despite this, PROTACs have been shown to be bioavailable in preclinical species and, most importantly, to achieve efficacious exposure levels in humans.39 As medicinal chemists have experimented with this modality, datasets are now emerging that are providing insights into the key physicochemical parameters to control. Recent publications from Arvinas (fa × fg)40 and AstraZeneca (F%)25 have highlighted the importance of restricting the number of ‘unsatisfied’ or solvent-exposed HBDs (eHBDs) for oral absorption. Both of these publications independently conclude that, whilst physicochemical properties, such as MW, lipophilicity, molecular polarity and HBAs can exceed the Ro5 limits for oral drugs, a criteria of eHBD ≤ 2 is a useful design criterion. Related investigations on the nature of HBDs in macrocyclic bRo5 drugs found an upper limit of 2 for amide HBDs, but a much more permissive limit of 7 formal HBDs in total.41 This may explain why the orally administered PROTACs that have progressed to the clinic have harnessed CRBN-engaging motifs which are small, efficient and with only a single eHBD.
Oral bioavailability in preclinical species and humans appears challenging yet achievable for these bRo5 molecules. The concept of a molecular properties ‘budget’ that must be judiciously spent feels a key aspect of PROTAC design, especially when it comes to the most restrictive aspect, namely HBDs.
The three-body problem
A PROTAC can only perform its function of targeted protein degradation if it is able to efficiently form a ternary complex that facilitates ubiquitination of surface lysines. Modelling this trimeric complex is challenging and, although progress has been made, the fact that multiple ternary complex conformations may exist and that not all of them may lead to efficient degradation has frustrated structure-based design of PROTACs.42,43
The molecular size of PROTACs together with the conformational and physicochemical property interplay between POI-linker-E3 components means there is more ‘chemical equity’ to optimise than for traditional small molecule programmes. The complexity of generating an ‘active’ PROTAC, given the uncertainty surrounding what constitutes a productive ternary complex, and which lies in the correct physicochemical space, leads to a significant design and synthesis hurdle to overcome.
In order to tackle this challenge, medicinal chemists have developed ways of generating ‘libraries’ for exploring a broad chemical space and thus attempt to practically address the colossally large combinatorial options that exist in an empirical fashion.44–48 For a given target's ligand, ‘PROTAC toolboxes’ of versatile intermediates can be employed in a hit generation campaign and afford numerous heterobifunctional molecules bearing diverse combinations of E3 ligase ligands and linkers for screening. Active PROTACs are subsequently optimised for degradation efficiency and physicochemical properties. As accounts of the discovery of clinical-quality PROTACs appear in the chemical literature, common themes are emerging. For example, linear linkers present in PROTACs are often evolved towards ring-rich, conformationally constrained versions often containing a basic motif with a modulated pKa. This may reflect the efforts of medicinal chemists to overcome the dual challenges of achieving acceptable solubility whilst maintaining permeability. Although the synthesis of bespoke single use libraries per target is a pragmatic approach to this challenge, it can be arduous, costly, wasteful and lead to bottlenecks necessitating a compromise with respect to the number of compounds made. In order to address these drawbacks in PROTAC hit generation, researchers have developed ‘direct to biology’ or ‘nano-SAR’ paradigms49,50 to generate nanomole-scale, non-purified libraries that can be screened directly in cells. These methodologies offer a more viable, rapid, cost-effective and sustainable approach to PROTAC hit discovery.
Shape shifting chimeras
In addition to the complexities relating to the generation of productive ternary complexes to effect degradation, the tripartite composition and molecular size of the PROTAC makes them more conformationally challenging to understand than traditional small molecules. The tactic of pre-organising a molecule to adopt a bio-active conformation in order to reduce the entropic penalty upon binding is a well-established strategy in medicinal chemistry.51 With PROTACs however, the greater degree of conformational freedom accessible to the ligand coupled with the conformational plasticity and uncertainty surrounding the ternary complex makes it challenging to establish exactly what ‘bio-active’ conformation to attempt to mimic.
In relation to physicochemical properties, the application of NMR techniques coupled with molecular dynamics (MD) simulations, to understand the conformational behaviour and influence on permeability, have been reported for PROTACs.52,53 Observed differences in both polar (e.g. water, or DMSO-d6 if aqueous solubility is low) and non-polar (e.g. CDCl3) solvents provided insights into conformational ensembles as well as end-to-end backfolding and potential intramolecular hydrogen bonds (IMHB). This identified solvent-dependent (designated ‘chameleonic’) intramolecular folding for both CRBN- and VHL-engaging PROTACs that increased their passive cell permeability compared with unfolded matched pairs.52,53
In contrast, MD and NMR investigation of ARV-110, ARV-766, ARV-471 and KT-474 with substantially more rigid linker motifs revealed that extended conformers prevail in both solvent environments for these clinical PROTACs.25
The world must be measured by eye
An additional challenge faced by those working in the PROTAC field has been the ability to generate meaningful data using in vitro assays configured for traditional small molecule optimisation programmes.54 These large, often lipophilic bases, have a tendency to adhere to labware/proteins/membranes leading to poor recoveries in permeability and protein binding assays. Traditional shake-flask lipophilicity assays often struggle with log D values > 4 and solubilities are often below the limit of detection making triaging candidates in the absence of data extremely challenging. However, in the authors' experience, metabolic stability assays (hepatocyte and microsomal incubations) generate robust values that are invaluable in weeding out unstable PROTACs.
Considerable overlap with efforts in the bRo5 arena have spurred progress in this area.55 Chromatographic methods (chromlogD and ePSA) have expanded the measurable range of lipophilicity and exposed polarity and allow for ranking of compounds within a series.56 Moreover, the combination of chromatographic methods with in silico methods to generate more relevant 3D-descriptors has been proposed as a way to increase the success of designing orally available PROTACs.57 Measuring solubilities in more bio-relevant fluids (e.g. FaSSIF, FeSSIF) and ‘pre-saturation’ methods have allowed for more accurate estimations of free levels.58 Nevertheless, experimental log D values ≥ 3 for 10 of 13 clinical PROTACs (Table 1) reveals that this equity sits outside more traditional ranges of lipophilicity (log D 1–3) for small molecule, oral drugs.59 An analogous drift into higher lipophilicity space has been noted for other synthetic bRo5 drugs and an upper limit of neutral form molecular polarity has been proposed for achieving high permeability with bRo5 drugs.60
Notwithstanding the aforementioned progress with computational and in vitro techniques,61 the definitive assessment of pharmacokinetic profile for advancing a PROTAC into higher-species animal models, toxicity studies and ultimately progression towards human currently remains an in vivo assessment.
Pitfalls to avoid
Integration of CRBN-binding entities, in particular IMiDs, within a PROTAC carries associated safety risks (vide supra) in need of monitoring. The risks are linked to the possibility that, much like IMiDs, the corresponding PROTACs can act as molecular glue degraders (MGD) and result in the degradation of a number of proteins (neosubstrates) which in turn induces a toxicological effect.31 Of note is the neosubstrate GSPT1, a translation termination factor. Inadvertent degradation of GSPT1, reduces the rate of protein synthesis and results in cytotoxicity accompanied by an apparent and misleading degradation of the targeted POI.62 This added complication likely extends beyond CRBN-harnessing PROTACs, as ligands to other E3 ligases, including VHL, have reported MGD-activity.63 On a similar theme, recently there have been reports of the MG capability of PROTACs stemming from the POI ligand motif. A JQ1-derived PROTAC, designed to degrade BRD4 by harnessing the DCAF15 E3 ligase, was shown to degrade BRD4 in a DCAF15 independent manner.64 The degradation is in fact driven by the POI ligand gluing BRD4 to a related ligase, DCAF16. Collectively, these findings highlight the need for meticulous control experiments to interrogate the mechanism of action (MoA) of PROTACs and thus facilitate their optimisation into efficacious and safe drugs.
Moving from the discovery phase into the evaluation of PROTACs in vivo, a key learning from the authors' own experience has been the importance of understanding their metabolic fate. Even with relatively metabolically stable PROTACs, fragments can be generated that are often more polar with lower lipophilicities and unbound clearance. These can then compete in binding to the POI or E3 and reduce the effectiveness of the ternary complex formation and hence the in vivo efficacy.65 Moreover, a metabolite of even a meticulously designed CRBN-harnessing PROTAC with no MGD activity, could in theory act as a MGD and thus introduce MGD-associated safety liabilities.31 Assessment of such risks during the execution of the current clinical trials will thus undoubtedly inform the scientific community as to the suitability of CRBN-harnessing PROTACs for non-oncology indications.
Conclusions
The PROTAC modality has delivered upon the initial promise of targeted protein degradation with a number of high-quality degraders currently under clinical evaluation. This initial cohort has been primarily focussed on oncology indications and has utilised CRBN as a favoured E3 ligase. Preliminary data is emerging that biological efficacy and oral bioavailability in humans is achievable, further validating the promise of PROTACs. Common structural features (e.g. bases and ring-rich linkers) and physicochemical parameters (e.g. eHBD ≤ 2) are highlighted for those PROTACs with disclosed structures in the hope this will provide utilisable design inputs for those working in this area.
The Ro5 has been an invaluable guiding principle for a generation of medicinal chemists grappling with the challenge of oral bioavailability. However, the emergence of orally administered PROTACs, >800 Da molecular glues (e.g.RMC-6291),66 >1500 Da cyclic peptidomimetic antagonists (MK-0616)67 and other clinical candidates that violate these principles, have increasingly challenged this dogma and established that some ‘rules’ are able to be at least ‘challenged’ and occasionally ‘broken’.68 PROTACs, together with these trailblazing examples, have redefined the concept of what constitutes ‘orally bioavailable chemical space’. That said, in our efforts to understand the boundaries of this new frontier, we need to remain open minded about what is possible and cautious about simply establishing new constraints on our chemical creativity.
Data availability
No primary research results, software or code have been included and no new data were generated as part of this review.
Author contributions
JS, IM & MS conceptualised, wrote and reviewed the manuscript jointly.
Conflicts of interest
JS, IM & MS are employees and shareholders in AstraZeneca.
Acknowledgments
Ian Storer, Andy Pike, Maximillian Lee and David Wilson are thanked for their comments on the manuscript.
Notes and references
- Li K. Crews C. M. Chem. Soc. Rev. 2022;51:5214–5236. doi: 10.1039/D2CS00193D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M. Langley D. R. Crews C. M. Nat. Rev. Drug Discovery. 2022;21:181–200. doi: 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofink C. Trainor N. Mair B. Wöhrle S. Wurm M. Mischerikow N. Roy M. J. Bader G. Greb P. Garavel G. Diers E. McLennan R. Whitworth C. Vetma V. Rumpel K. Scharnweber M. Fuchs J. E. Gerstberger T. Cui Y. Gremel G. Chetta P. Hopf S. Budano N. Rinnenthal J. Gmaschitz G. Mayer M. Koegl M. Ciulli A. Weinstabl H. Farnaby W. Nat. Commun. 2022;13:5969. doi: 10.1038/s41467-022-33430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D. Pal P. Liu X. Jia Y. Thummuri D. Zhang P. Hu W. Pei J. Zhang Q. Zhou S. Khan S. Zhang X. Hua N. Yang Q. Arango S. Zhang W. Nayak D. Olsen S. K. Weintraub S. T. Hromas R. Konopleva M. Yuan Y. Zheng G. Zhou D. Nat. Commun. 2021;12:6896. doi: 10.1038/s41467-021-27210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. Zhang X. Lv D. Zhang Q. He Y. Zhang P. Liu X. Thummuri D. Yuan Y. Wiegand J. S. Pei J. Zhang W. Sharma A. McCurdy C. R. Kuruvilla V. M. Baran N. Ferrando A. A. Kim Y. M. Rogojina A. Houghton P. J. Huang G. Hromas R. Konopleva M. Zheng G. Zhou D. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien Laramy M. N. Luthra S. Brown M. F. Bartlett D. W. Nat. Rev. Drug Discovery. 2023;22:410–427. doi: 10.1038/s41573-023-00652-2. [DOI] [PubMed] [Google Scholar]
- Chirnomas D. Hornberger K. R. Crews C. M. Nat. Rev. Clin. Oncol. 2023;20:265–278. doi: 10.1038/s41571-023-00736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Liu M. Yang Z. Li J. Gao Y. Tan R. Molecules. 2022;27:8828. doi: 10.3390/molecules27248828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L. B. Neklesa T. K. Chen X. Dong H. Ferraro C. Gordon D. A. Macaluso J. Pizzano J. Wang J. Willard R. R. Vitale N. Peck R. Moore M. D. Crews C. M. Houston J. Crew A. P. Taylor I. Cancer Res. 2021;81:43. doi: 10.1158/1538-7445.AM2021-43. [DOI] [Google Scholar]
- Snyder L. Lee S. H. Neklesa T. K. Chen X. Dong H. Ferraro C. Gordon D. A. Macaluso J. Pizzano J. Wang J. Willard R. R. Vitale N. Peck R. Moore M. D. Crews C. M. Houston J. Crew A. P. Taylor I. Cancer Res. 2023;83:ND03. doi: 10.1158/1538-7445.AM2023-ND03. [DOI] [Google Scholar]
- Xu S. Nayak S. Norris J. D. Ammirante M. Rychak E. Wardell S. E. Tsuji T. Liu K. Meiring J. Piccotti J. R. Dalvie D. Liao D. Kandimalla R. Guerrero N. Sapinoso L. Baker J. G. Bae Y. Baughman J. Toyama B. Fontanillo C. F. Norris S. Horn E. J. Plantevin-Krenitsky V. Mortensen D. Cathers B. Nguyen M. H. Hensen J. D. Hamann L. G. McDonnell D. P. Narla R. K. Rolfe M. Cancer Res. 2024;84:ND02. doi: 10.1158/1538-7445.AM2024-ND02. [DOI] [Google Scholar]
- Gough S. M. Flanagan J. J. Teh J. Andreoli M. Rousseau E. Pannone M. Bookbinder M. Willard R. Davenport K. Bortolon E. Cadelina G. Gordon D. Pizzano J. Macaluso J. Soto L. Corradi J. Digianantonio K. Drulyte I. Morgan A. Quinn C. Békés M. Ferraro C. Chen X. Wang G. Dong H. Wang J. Langley D. R. Houston J. Gedrich R. Taylor I. C. Clin. Cancer Res. 2024;30:3549–3563. doi: 10.1158/1078-0432.CCR-23-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. Ji N. Campbell V. Slavin A. Zhu X. Chen D. Rong H. Enerson B. Mayo M. Sharma K. Browne C. M. Klaus C. R. Li H. Massa G. McDonald A. A. Shi Y. Sintchak M. Skouras S. Walther D. M. Yuan K. Zhang Y. Kelleher J. Liu G. Luo X. Mainolfi N. Weiss M. M. J. Med. Chem. 2024;67:18022–18037. doi: 10.1021/acs.jmedchem.4c01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. M. Zheng X. Ji N. Browne C. M. Campbell V. Chen D. Enerson B. Fei X. Huang X. Klaus C. R. Li H. Mayo M. McDonald A. A. Paul A. Rong H. Sharma K. Shi Y. Slavin A. Walther D. M. Yuan K. Zhang Y. Zhu X. Kelleher J. Walker D. Mainolfi N. J. Med. Chem. 2024;67:10548–10566. doi: 10.1021/acs.jmedchem.3c01823. [DOI] [PubMed] [Google Scholar]
- Jackson K. L. Agafonov R. V. Carlson M. W. Chaturvedi P. Cocozziello D. Cole K. Deibler R. Eron S. J. Good A. Hart A. A. He M. Henderson C. S. Huang H. Isasa M. Kirby R. J. Lee L. Mahler M. Moustakim M. Nasveschuk C. G. Palmer M. Poling L. L. Pollock R. M. Schnaderbeck M. Spence S. Veits G. K. Yap J. L. Yin N. Zeid R. Crystal A. S. Phillips A. J. Fisher S. L. Cancer Res. 2022;82:ND09. doi: 10.1158/1538-7445.AM2022-ND09. [DOI] [Google Scholar]
- Netherton M., Drug Discovery Chemistry, San Diego, USA, 13th of April 2023. Presentation title: Discovery of FHD-609: A Potent and Selective Heterobifunctional Degrader of BRD9
- Sherman D. Cancer Res. 2024;84:ND05. doi: 10.1158/1538-7445.AM2024-ND05. [DOI] [Google Scholar]
- Robbins D. W. Noviski M. A. Tan Y. S. Konst Z. A. Kelly A. Auger P. Brathaban N. Cass R. Chan M. L. Cherala G. Clifton M. C. Gajewski S. Ingallinera T. G. Karr D. Kato D. Ma J. McKinnell J. McIntosh J. Mihalic J. Murphy B. Panga J. R. Peng G. Powers J. Perez L. Rountree R. Tenn-McClellan A. Sands A. T. Weiss D. R. Wu J. Ye J. Guiducci C. Hansen G. Cohen F. J. Med. Chem. 2024;67:2321–2336. doi: 10.1021/acs.jmedchem.3c01007. [DOI] [PubMed] [Google Scholar]
- Mihalic J. T., Auger P., Brathaban N., Bousquet H., Gajewski S., Ingallinera T., Iuliano J., Kato D., Konst Z., Kumar A., Lu H., Ma J., McIntosh J., Mukerji R., Noviski M., Panga J., Peng G., Perez L., Robbins D., Rountree R., Tan M., Tenn-McClellan A., Wu J., Ye J., Yung S., Guiducci C., Hansen G. and Cohen F., Abstracts of Papers, ACS Fall 2023, San Francisco, CA, United States, 2023 [Google Scholar]
- Liang Y. Sowa M. E. Jackson K. L. Simard J. R. Kreger B. Li P. Poling L. Baddour J. Good A. Huang H. Eron S. Nasveschuk C. G. Yu R. Fitzgerald M. Garza V. O'Shea M. W. Veits G. Yap J. Y. Moustakim M. Hart A. Agafonov R. V. Sarkissian G. Patel J. S. Deibler R. Cole K. S. Cocozziello D. Rahman F. Phillips A. J. Norton E. Crystal A. S. Pollock R. M. Fisher S. L. Cancer Res. 2023;83:3425. doi: 10.1158/1538-7445.AM2023-3425. [DOI] [Google Scholar]
- Khan S. Zhang X. Lv D. Zhang Q. He Y. Zhang P. Liu X. Thummuri D. Yuan Y. Wiegand J. S. Pei J. Zhang W. Sharma A. McCurdy C. R. Kuruvilla V. M. Baran N. Ferrando A. A. Kim Y.-m. Rogojina A. Houghton P. J. Huang G. Hromas R. Konopleva M. Zheng G. Zhou D. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A. Lombardo F. Dominy B. W. Feeney P. J. Adv. Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Veber D. F. Johnson S. R. Cheng H. Y. Smith B. R. Ward K. W. Kopple K. D. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Heim C. Pliatsika D. Mousavizadeh F. Bär K. Hernandez Alvarez B. Giannis A. Hartmann M. D. J. Med. Chem. 2019;62:6615–6629. doi: 10.1021/acs.jmedchem.9b00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade M. Scott J. S. Hayhow T. G. Pike A. Terstiege I. Ahlqvist M. Johansson J. R. Diene C. R. Fallan C. Balazs A. Y. S. Chiarparin E. Wilson D. J. Med. Chem. 2024;67:13106–13116. doi: 10.1021/acs.jmedchem.4c01017. [DOI] [PubMed] [Google Scholar]
- Li M. Zhi Y. Liu B. Yao Q. J. Med. Chem. 2023;66:2308–2329. doi: 10.1021/acs.jmedchem.2c01555. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S. Hoff O. Muellner M. K. Molecules. 2022;27:8119. doi: 10.3390/molecules27238119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T. Ciulli A. SLAS Discovery. 2021;26:484–502. doi: 10.1177/2472555220965528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelides I. N. Collie G. W. J. Med. Chem. 2023;66:3173–3194. doi: 10.1021/acs.jmedchem.2c01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B. Lentzsch S. Pharmacol. Ther. 2012;136:56–68. doi: 10.1016/j.pharmthera.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Moreau K. Coen M. Zhang A. X. Pachl F. Castaldi M. P. Dahl G. Boyd H. Scott C. Newham P. Br. J. Pharmacol. 2020;177:1709–1718. doi: 10.1111/bph.15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H. Li C. Tang H. Tandon I. Liao J. Roberts B. L. Zhao Y. Tang W. J. Med. Chem. 2023;66:2904–2917. doi: 10.1021/acs.jmedchem.2c01941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarusiewicz J. A. Yoshimura S. Mayasundari A. Actis M. Aggarwal A. McGowan K. Yang L. Li Y. Fu X. Mishra V. Heath R. Narina S. Pruett-Miller S. M. Nishiguchi G. Yang J. J. Rankovic Z. ACS Med. Chem. Lett. 2023;14:141–145. doi: 10.1021/acsmedchemlett.2c00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borjesson U., Perry M. W. D., Grebner C., Michaelides I. N., Hayhow T. G. C., Kettle J. G., Collie G. W., Storer R. I., Bagal S. K. and Fallan C., Compounds and their use in treating cancer, WO2022069520A1, AstraZeneca AB, 2022 [Google Scholar]
- David L. Wenlock M. Barton P. Ritzén A. ChemMedChem. 2021;16:2669–2685. doi: 10.1002/cmdc.202100306. [DOI] [PubMed] [Google Scholar]
- Han X. Wang C. Qin C. Xiang W. Fernandez-Salas E. Yang C.-Y. Wang M. Zhao L. Xu T. Chinnaswamy K. Delproposto J. Stuckey J. Wang S. J. Med. Chem. 2019;62:941–964. doi: 10.1021/acs.jmedchem.8b01631. [DOI] [PubMed] [Google Scholar]
- Xiang W. Zhao L. Han X. Qin C. Miao B. McEachern D. Wang Y. Metwally H. Kirchhoff P. D. Wang L. Matvekas A. He M. Wen B. Sun D. Wang S. J. Med. Chem. 2021;64:13487–13509. doi: 10.1021/acs.jmedchem.1c00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike A. Williamson B. Harlfinger S. Martin S. McGinnity D. F. Drug Discovery Today. 2020;25:1793–1800. doi: 10.1016/j.drudis.2020.07.013. [DOI] [PubMed] [Google Scholar]
- Petrylak D. P. Gao X. Vogelzang N. J. Garfield M. H. Taylor I. Moore M. D. Peck R. A. Burris III H. A. J. Clin. Oncol. 2020;38:3500. [Google Scholar]
- Hornberger K. R. Araujo E. M. V. J. Med. Chem. 2023;66:8281–8287. doi: 10.1021/acs.jmedchem.3c00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Jimenez D. Poongavanam V. Kihlberg J. J. Med. Chem. 2023;66:5377–5396. doi: 10.1021/acs.jmedchem.3c00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. ChemMedChem. 2024;19:e202400171. doi: 10.1002/cmdc.202400171. [DOI] [PubMed] [Google Scholar]
- Rovers E. Schapira M. J. Chem. Inf. Model. 2024;64:6162–6173. doi: 10.1021/acs.jcim.4c00426. [DOI] [PubMed] [Google Scholar]
- Wurz R. P. Dellamaggiore K. Dou H. Javier N. Lo M. C. McCarter J. D. Mohl D. Sastri C. Lipford J. R. Cee V. J. J. Med. Chem. 2018;61:453–461. doi: 10.1021/acs.jmedchem.6b01781. [DOI] [PubMed] [Google Scholar]
- Brownsey D. K. Rowley B. C. Gorobets E. Gelfand B. S. Derksen D. J. Chem. Sci. 2021;12:4519–4525. doi: 10.1039/D0SC05442A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhela I. P. Ranza A. Balestrero F. C. Serafini M. Aprile S. Di Martino R. M. C. Condorelli F. Pirali T. J. Med. Chem. 2022;65:15282–15299. doi: 10.1021/acs.jmedchem.2c01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis T. A. La Clair J. J. Burkart M. D. Chem. Commun. 2021;57:1026–1029. doi: 10.1039/D0CC05395C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovicova S. Jorda R. Hendrychova D. Krystof V. Soural M. Chem. Commun. 2019;55:929–932. doi: 10.1039/C8CC08716D. [DOI] [PubMed] [Google Scholar]
- Plesniak M. P. Taylor E. K. Eisele F. Kourra C. M. B. K. Michaelides I. N. Oram A. Wernevik J. Valencia Z. S. Rowbottom H. Mann N. Fredlund L. Pivnytska V. Novén A. Pirmoradian M. Lundbäck T. Storer R. I. Pettersson M. De Donatis G. M. Rehnström M. ACS Med. Chem. Lett. 2023;14:1882–1890. doi: 10.1021/acsmedchemlett.3c00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R. Bendito-Moll E. Battersby D. J. Miah A. H. Wellaway N. Law R. P. Stacey P. Klimaszewska D. Macina J. M. Burley G. A. Harling J. D. J. Med. Chem. 2023;66:15437–15452. doi: 10.1021/acs.jmedchem.3c01604. [DOI] [PubMed] [Google Scholar]
- Hargreaves D. Carbajo R. J. Bodnarchuk M. S. Embrey K. Rawlins P. B. Packer M. Degorce S. L. Hird A. W. Johannes J. W. Chiarparin E. Schade M. Proc. Natl. Acad. Sci. U. S. A. 2023;120(21):e2221967120. doi: 10.1073/pnas.2221967120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilaw Y. Poongavanam V. Svensson Nilsson C. Nguyen D. Giese A. Meibom D. Erdelyi M. Kihlberg J. ACS Med. Chem. Lett. 2021;12:107–114. doi: 10.1021/acsmedchemlett.0c00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poongavanam V. Atilaw Y. Siegel S. Giese A. Lehmann L. Meibom D. Erdelyi M. Kihlberg J. J. Med. Chem. 2022;65:13029–13040. doi: 10.1021/acs.jmedchem.2c00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volak L. P. Duevel H. M. Humphreys S. Nettleton D. Phipps C. Pike A. Rynn C. Scott-Stevens P. Zhang D. Zientek M. Drug Metab. Dispos. 2023;51:792–803. doi: 10.1124/dmd.122.001154. [DOI] [PubMed] [Google Scholar]
- Poongavanam V. Doak B. C. Kihlberg J. Curr. Opin. Chem. Biol. 2018;44:23–29. doi: 10.1016/j.cbpa.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Price E. Weinheimer M. Rivkin A. Jenkins G. Nijsen M. Cox P. B. DeGoey D. J. Med. Chem. 2024;67:5683–5698. doi: 10.1021/acs.jmedchem.3c02332. [DOI] [PubMed] [Google Scholar]
- Apprato G. Poongavanam V. Garcia Jimenez D. Atilaw Y. Erdelyi M. Ermondi G. Caron G. Kihlberg J. Drug Discov. Today. 2024;29(4):103917. doi: 10.1016/j.drudis.2024.103917. [DOI] [PubMed] [Google Scholar]
- Riccardi K. Cawley S. Yates P. D. Chang C. Funk C. Niosi M. Lin J. Di L. J. Pharm. Sci. 2015;104:2627–2636. doi: 10.1002/jps.24506. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Expert Opin. Drug Discovery. 2010;5:235–248. doi: 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- Möbitz H. ChemMedChem. 2024;19:e202300395. doi: 10.1002/cmdc.202300395. [DOI] [PubMed] [Google Scholar]
- Rossi Sebastiano M. Doak B. C. Backlund M. Poongavanam V. Over B. Ermondi G. Caron G. Matsson P. Kihlberg J. J. Med. Chem. 2018;61:4189–4202. doi: 10.1021/acs.jmedchem.8b00347. [DOI] [PubMed] [Google Scholar]
- Vetma V. Perez L. C. Eliaš J. Stingu A. Kombara A. Gmaschitz T. Braun N. Ciftci T. Dahmann G. Diers E. Gerstberger T. Greb P. Kidd G. Kofink C. Puoti I. Spiteri V. Trainor N. Weinstabl H. Westermaier Y. Whitworth C. Ciulli A. Farnaby W. McAulay K. Frost A. B. Chessum N. Koegl M. ACS Chem. Biol. 2024;19:1484–1494. doi: 10.1021/acschembio.4c00152. [DOI] [PubMed] [Google Scholar]
- Tutter A., Buckley D., Golosov A. A., Ma X., Shu W., McKay D. J. J., Darsigny V., Dovala D., Beckwith R., Solomon J., Rao P., Xu L., Fazal A., Lingel A., Wartchow C., Cobb J. S., Hachey A., Tullai J. and Michaud G. A., bioRxiv, 2024, preprint, 10.1101/2024.01.25.576086 [DOI]
- Hsia O. Hinterndorfer M. Cowan A. D. Iso K. Ishida T. Sundaramoorthy R. Nakasone M. A. Imrichova H. Schätz C. Rukavina A. Husnjak K. Wegner M. Correa-Sáez A. Craigon C. Casement R. Maniaci C. Testa A. Kaulich M. Dikic I. Winter G. E. Ciulli A. Nature. 2024;627:204–211. doi: 10.1038/s41586-024-07089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhow T. G. Williamson B. Lawson M. Cureton N. Braybrooke E. L. Campbell A. Carbajo R. J. Cheraghchi-Bashi A. Chiarparin E. Diène C. R. Fallan C. Fisher D. I. Goldberg F. W. Hopcroft L. Hopcroft P. Jackson A. Kettle J. G. Klinowska T. Künzel U. Lamont G. Lewis H. J. Maglennon G. Martin S. Gutierrez P. M. Morrow C. J. Nikolaou M. Nissink J. W. M. O'Shea P. Polanski R. Schade M. Scott J. S. Smith A. Weber J. Wilson J. Yang B. Crafter C. Commun. Biol. 2024;7:563. doi: 10.1038/s42003-024-06238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. Nichols R. J. Yang Y. C. Schulze C. J. Wang Z. Dua R. Jiang J. Nasholm N. Knox J. E. Seamon K. Longhi M. Tomlinson A. Chou K. Li S. Wildes D. P. Singh M. Koltun E. S. Gill A. L. Smith J. A. Cancer Res. 2023;83:ND07. doi: 10.1158/1538-7445.AM2023-ND07. [DOI] [Google Scholar]
- Iskandar S. E. Bowers A. A. ACS Med. Chem. Lett. 2022;13:1379–1383. doi: 10.1021/acsmedchemlett.2c00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung I. V. Huck B. R. Crespo A. Nat. Rev. Chem. 2023;7:3–4. doi: 10.1038/s41570-022-00451-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No primary research results, software or code have been included and no new data were generated as part of this review.