Abstract
Background
Tryptophan is an essential amino acid involved in critical cellular processes in vertebrates, serving as a precursor for serotonin and kynurenine, which are key neuromodulators to influence neural and immune functions. Systematic and quantitative measurement of tryptophan is vital to understanding these processes.
Results
Here, we utilized a robust and highly responsive green ratiometric indicator for tryptophan (GRIT) to quantitatively measure tryptophan dynamics in bacteria, mitochondria of mammalian cell cultures, human serum, and intact zebrafish. At the cellular scale, these quantitative analyses uncovered differences in tryptophan dynamics across cell types and organelles. At the whole-organism scale, we revealed that inflammation-induced tryptophan concentration increases in zebrafish brain led to elevated serotonin and kynurenine levels, prolonged sleep duration, suggesting a novel metabolic connection between immune response and behavior. Moreover, GRIT’s application in detecting reduced serum tryptophan levels in patients with inflammation symptoms suggests its potential as a high-throughput diagnostic tool.
Conclusions
In summary, this study introduces GRIT as a powerful method for studying tryptophan metabolism and its broader physiological implications, paving the way for new insights into the metabolic regulation of health and disease across multiple biological scales.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-024-02058-x.
Keywords: Tryptophan quantitative imaging, Bacteria, Mitochondria, Zebrafish, Human serum
Background
Tryptophan, an essential amino acid exclusively obtained from food, plays a crucial role in cellular protein synthesis and survival. Its absorption, transportation, and metabolic pathways within all vertebrates are highly conserved, leading to the production of various active molecules through pathways involving kynurenine, serotonin, and indole [1–3]. Thus, tryptophan metabolism serves as a pivotal metabolic regulator for processes such as immune responses, neural activities, and intestinal functions across vertebrates. Given the wide-ranging influence of tryptophan metabolism, abnormalities in its regulation can trigger systemic effects, potentially exacerbating diseases like neurodegeneration, infections, and cancer [4]. Notably, plasma tryptophan levels are closely linked to disease progression and prognosis [5], underscoring the need for precise, quantitative measurement of tryptophan dynamics across biological systems.
Traditional methods for measuring tryptophan, such as high-performance liquid chromatography (HPLC), fall short in revealing the minor metabolic differences between cell types and subcellular compartments [6, 7], and they lack the temporal resolution necessary to track rapid metabolic changes during cellular stress [8]. This limitation has hindered a comprehensive understanding of tryptophan’s role in various physiological and pathological contexts.
In contrast, our recently developed green ratiometric indicator for tryptophan, GRIT [9], represents a significant advancement in the field of metabolic measurement. GRIT surpasses existing FRET-based tryptophan indicators (FLIPW-CTYT) [10], offering a substantially larger dynamic range (9.55 versus 0.3), a smaller molecular size (39.7 kDa versus 80.1 kDa), and an excitation ratio that is independent of photobleaching [10, 11]. These improvements make GRIT a highly effective tool for the quantitative measurement of tryptophan.
In this study, we demonstrate GRIT allows a new paradigm offering high spatiotemporal resolution, systematically quantifying dynamic changes in quantitatively mapping tryptophan dynamics in bacteria, mitochondria, zebrafish, and human serum. Such advancements uncovered previously unrecognized differences in tryptophan dynamics across cell types and organelles, and revealed that inflammation-induced tryptophan redistribution in the zebrafish brain is associated with increased serotonin and kynurenine levels, linking immune response to behavioral changes. Additionally, we found that reduced serum tryptophan levels in patients with inflammation could serve as a potential biochemical marker for inflammatory conditions.
The introduction of GRIT as a robust, versatile, and quantitative tool for studying tryptophan metabolism marks a significant step forward in our ability to systematically measure and understand this critical amino acid’s role in health and disease. This work opens new avenues for research into the metabolic regulation of immune responses, neural function, and disease progression, with broad implications for clinical diagnostics and therapeutic strategies.
Results
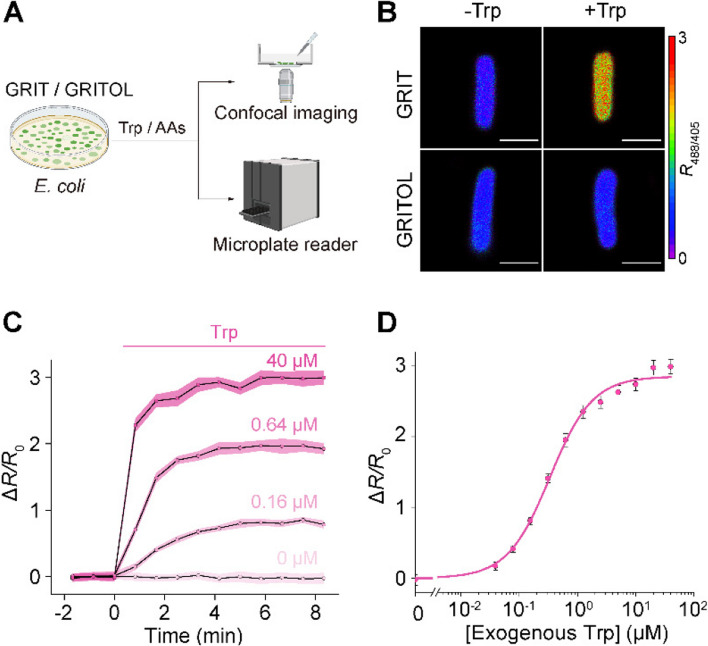
Monitoring tryptophan dynamics in living bacteria
Bacteria possess a tight regulatory mechanism on tryptophan metabolism [12]. We investigated tryptophan uptake into Escherichia coli cells by expressing the GRIT sensor in the cytoplasm (Fig. 1A, B). Exogenous tryptophan addition induced a strong and rapid fluorescence response in GRIT-positive bacteria, with a maximum ~ threefold increase (Fig. 1B–D), but not in GRITOL (GRIT control sensor)-expressing bacteria (Fig. 1B). GRIT could respond to 0.1–100 μM exogenous tryptophan in a dose-dependent manner (Fig. 1D, Additional file 1: Fig. S1A) with an absorption rate of ~ 16.5 μM/s (Additional file 1: Table S1). The Michaelis constant (Km) of tryptophan uptake (~ 0.58 μM) is similar to the reported biophysical properties of aroP (~ 0.4 μM) [13], a tryptophan transporter that is highly expressed in bacteria according to the transcriptome [14] (Additional file 1: Fig. S1B). The free concentrations of intracellular tryptophan in tryptophan deprivation and tryptophan exposure conditions were measured to be 10.6 ± 3.7 μM (similar to the reported level 12 μM [15]) and 192 ± 23.9 μM, respectively (Additional file 1: Table S1). In addition, no significant fluorescence change was observed when 19 other tested amino acids were added in the absence or presence of tryptophan (Additional file 1: Fig. S1C). These results suggest that GRIT sensor can monitor the uptake and concentration changes of tryptophan in living bacteria in real-time and with high sensitivity (Fig. 1C–D, Additional file 1: Fig. S1A) and specificity (Additional file 1: Fig. S1C), thereby aiding in the study of the regulatory mechanisms of bacterial tryptophan metabolism.
Fig. 1.
Detection of tryptophan uptake in bacteria with GRIT sensor. A Schematic representation illustrating the fluorescence detection of tryptophan (Trp) dynamics with the GRIT or GRITOL sensor in Escherichia coli cells. B Fluorescence images of bacteria expressing GRIT and GRITOL upon the addition of 0.1 mM Trp. C Changes in the fluorescence of GRIT in response to various concentrations of Trp in Escherichia coli. D The dose–response curve of GRIT-positive bacteria treated with exogenous Trp. Data are from Fig. S1A. The curve was fitted using Eq. 1 provided in the “Methods” section, with the binding constant being ~ 0.33 μM. Scale bars, 1 μm. Data shown as mean ± s.e.m., n = 3 independent experiments in C and D. See also Additional file 1: Fig. S1 and Table S1
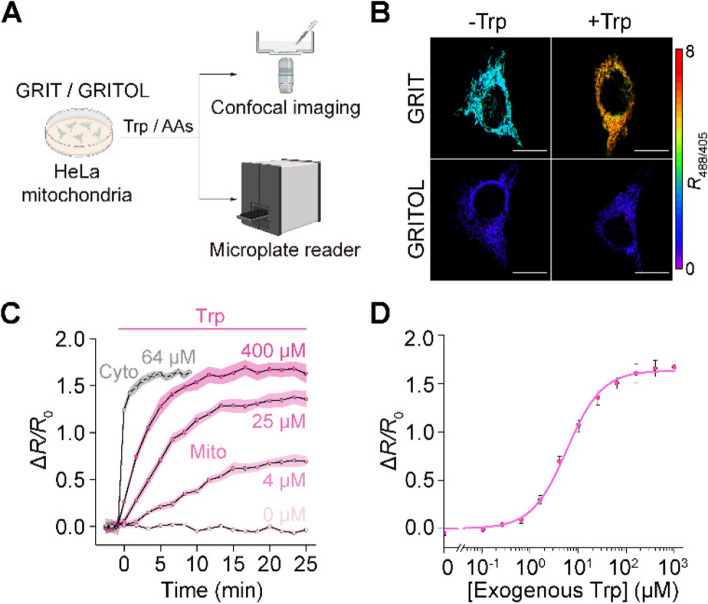
Detection of mitochondrial tryptophan dynamics in mammalian cells
Mitochondria are semiautonomous subcellular organelles with their own genome and require tryptophan transport from the cytosol to synthesize mitochondrial proteins. However, due to the lack of specific detection methods, reports on mitochondrial tryptophan metabolism have been sparse [16]. We selectively targeted the GRIT sensor in the mitochondrial matrix (Fig. 2A, B and Additional file 1: Note S1), resulting in a marked increase in fluorescence signals compared to the control cells (Additional file 1: Fig. S2A, B). This enhancement enables effective fluorescence measurement using both microplate readers and microscopy (Fig. 2A). Mito-GRIT showed an ~ 1.7-fold fluorescence increases upon the addition of 0.5 mM tryptophan (Fig. 2B). The mitochondrial tryptophan concentration was determined to be 112.4 ± 10.0 μM (Additional file 1: Table S1), which was close to the HPLC measurements (108.0 ± 10.3 μM, Additional file 1: Table S1). However, the tryptophan uptake rate of mitochondria (~ 9.6 μM/min) was only ~ 18% of that in the cytoplasm (~ 52.6 μM/min) (Fig. 2C, Additional file 1: Table S1). Upon tryptophan deprivation, the mitochondrial tryptophan level was relatively stable in the first 20 min and decreased linearly at a rate of ~ 0.8 μM/min (Additional file 1: Fig. S2D, E and Table S2). This suggests that the mitochondrial tryptophan pool is predominantly regulated by cytosolic supply and the mitochondrial consumption, such as protein synthesis.
Fig. 2.
GRIT sensor reports mitochondrial tryptophan dynamics in cultured HeLa cells. A Schematic showing the fluorescence detection with mito-GRIT or mito-GRITOL sensor in HeLa cells. B Fluorescence responses of HeLa cells expressing GRIT (upper) and GRITOL (bottom) in mitochondria upon the addition of exogenous 0.5 mM Trp in HBSS buffer. The traces (C) and dose-dependence curve (D) of mitochondrial GRIT sensor in response to varying concentrations of added tryptophan. The gray trace in C represents the fluorescence response kinetics of cytosolic GRIT to indicated Trp concentration (64 μM) in HBSS buffer, suggesting a significantly faster tryptophan uptake into the cytosol compared to the mitochondria. The curve in D was fitted using Eq. 1 provided in the “Methods” section, with the binding constant being ~ 5.84 μM. Scale bars, 10 μm. Data shown as mean ± s.e.m. n = 3 independent experiments. See also Additional file 1: Fig. S2 and Tables S1 and S2
We have demonstrated that cytosolic tryptophan pool is finely regulated by the supply of external amino acids, which can be classified into three groups, class I (large and neutral amino acids, classic LAT1 substrate), class II (five uncharged amino acids), and class III (six charged or nonpolar amino acids) [9]. To further understand the characteristics of mitochondrial tryptophan transportation, we detected mitochondrial tryptophan dynamics in response to all other 19 amino acids. The six amino acids from class III, which have no effect on cytosolic tryptophan flux, also did not affect mitochondrial tryptophan dynamics (Additional file 1: Fig. S2C–D, F and Table S2). In contrast, the 13 amino acids from class I and class II, despite their varied effects on cytosolic tryptophan efflux, displayed similar impacts on mitochondrial tryptophan dynamics, both in the absence or presence of tryptophan (Additional file 1: Fig. S2C–D, F and Table S2). Collectively, the biophysical properties of mitochondrial tryptophan metabolism suggest specific and unidirectional mitochondrial tryptophan transporters, which are different from the bidirectional transportation of cell surface SLC7A5 (LAT1) [9].
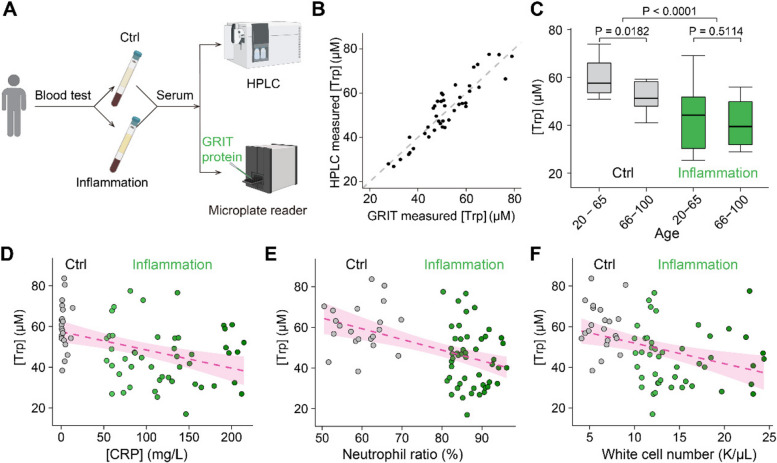
Detection of serum tryptophan in patients with inflammation syndrome
The clinical relevance of tryptophan metabolism lies in its association with various diseases and inflammatory responses, particularly evident in the variations in serum levels [5]. Therefore, a rapid and efficient in vitro detection method could significantly advance diagnostic technologies. We demonstrated that purified GRIT sensor could show large fluorescence ratio changes to tryptophan ranging from 20 μM to 1 mM (Additional file 1: Fig. S3A). This broad detection range is line with the theoretical prediction of an established R package “SensorOverlord” [17] (Additional file 1: Fig. S3B), which overlaps with the concentration fluctuations of tryptophan in most physiological and pathological conditions.
We collected serum samples from patients with inflammatory symptoms characterized by following specific clinical criteria [17]: hypersensitive C-reactive protein (hs-CRP) concentration > 50 mg/L, neutrophil ratio > 80%, and white cell count > 10,000/μL (Fig. 3A). We first analyzed serum tryptophan levels using HPLC assay, followed by GRIT sensor detection of the remaining portion of the same sample (Fig. 3A). Despite the autofluorescence of blood may interfere with the fluorescence assays, our results showed that GRIT sensor possesses much higher fluorescence signal than the serum background (Additional file 1: Fig. S3C–F). In addition, the quantified tryptophan concentration was highly consistent between HPLC and GRIT sensor measurements (Fig. 3B), demonstrating the robustness of tryptophan measurement with purified GRIT sensor in human serum samples. The serum tryptophan level in inflamed patients (44.7 ± 4.7 μM) decreased ~ 25% compared with that of control (59.6 ± 3.6 μM) (Additional file 1: Table S1). This reduction in serum tryptophan was consistently observed under various conditions, including different sexes, ages, and extents of inflammation (Fig. 3C–F, Additional file 1: Fig. S3G–I). These results validate the GRIT sensor as an alternative high-throughput in vitro diagnostic method and a decrease in serum tryptophan level could be a biochemical marker of inflammation in patients.
Fig. 3.
Decreased serum tryptophan concentration is a biochemical mark of inflamed patients. A Schematic of the experimental procedures for the quantification of human serum tryptophan. B A comparative analysis of quantitative serum tryptophan level in identical samples measured by GRIT sensor assay or HPLC measurement (n = 42). C Tryptophan concentrations of human serum samples in the control group (gray, n = 12 in 20–65 subgroup, n = 13 in the 66–100 subgroup) and inflammation group (green, n = 17 in 20–65 subgroup, n = 35 in the 66–100 subgroup) based on age. Note that the total n number (42) in B is smaller than the n number (77) in C, due to the volume of some serum samples were not enough for GRIT sensor assay after HPLC (< 0.3 mL). Relationships between tryptophan concentrations in human serum samples and concentrations of hypersensitive C-reactive protein D, neutrophil ratio E, and white cell number F. The dashed magenta line represents a linear fit of the data, with the shaded area indicating the fitting confidence interval. Two-tailed Student’s unpaired t-test for C. See also Additional file 1: Fig. S3 and Table S1
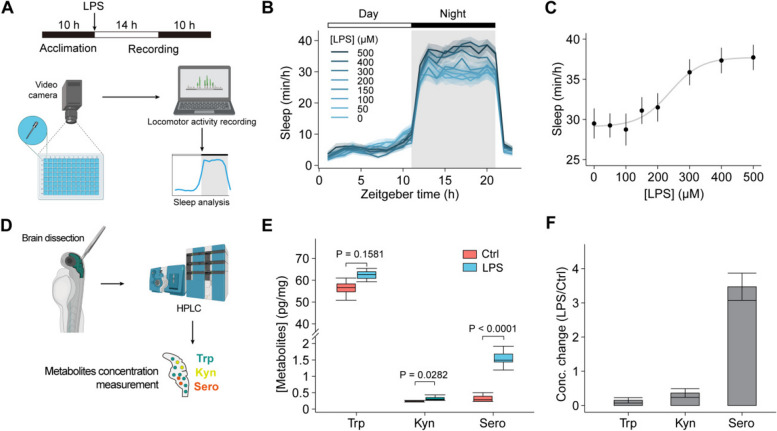
Detection of tryptophan metabolites and behaviors during inflammation in animal model
In the central nervous system, tryptophan is catabolized into serotonin and kynurenine, both of which modulate neural activities and are related to sleep, depressive behaviors, and fatigue symptoms [18, 19]. To further explore the pathological implications of the documented inflammation-triggered tryptophan redistribution phenomenon [9], we subsequently exposed zebrafish to lipopolysaccharide (LPS). This resulted in a dose-dependent increase in sleep duration (Fig. 4A–C, Additional file 1: Fig. S4A), consistent with the sustained elevation of tryptophan levels in the brain following LPS treatment (Additional file 1: Fig. S4B, C). Next, we measured the concentrations of tryptophan, serotonin, and kynurenine in isolated zebrafish brains. Inflamed zebrafish showed a slight but non-significant increase in total tryptophan, a significant over threefold increase in serotonin, and a 36% increase in the kynurenine, compared to control individuals (Fig. 4E, F). The rises of tryptophan metabolites in the brain may contribute to the increased sleep behaviors during inflammation. Furthermore, inhibiting these metabolic pathways resulted in developmental deformities, leading to their demise (Additional file 1: Fig. S4D, E), suggesting that disrupting tryptophan metabolism leads to severe pathological consequences.
Fig. 4.
Detection of metabolite levels in zebrafish larvae and zebrafish sleep changes during inflammation. A Schematic of the zebrafish sleep behavior measurement during inflammation. B Changes in the sleep time of zebrafish in response to various concentrations of LPS (n = 60 fishes from 5 to 7 independent experiments). LPS was added at 0 h with indicated concentrations. C The dose–response curve of mean sleep time at night of zebrafish treated with different concentrations of LPS. The curve was fitted to a sigmoidal function. D Schematic of the experimental procedures for the quantification of zebrafish brain tryptophan. The absolute levels (pg/mg) (E) and relative changes (F) of tryptophan, kynurenine, and serotonin in isolated zebrafish brain with (n = 7) or without LPS treatment (n = 6). Data shown as mean ± s.e.m. Two-tailed unpaired Student’s t-test for D. See also Additional file 1: Fig. S4
In summary, our results demonstrate that inflammatory responses modulate tryptophan levels across tissues, with altered metabolic levels in the brain influencing behavior. This indicates a metabolic link between the inflammatory response and animal behavior.
Discussion
In this study, we have demonstrated the versatility of using GRIT biosensor for quantitatively measuring tryptophan levels across divers biological models, including bacteria, subcellular organelles, and human serum. The GRIT, a superior genetically encoded tool, supports multiple applications and long-term recordings of tryptophan.
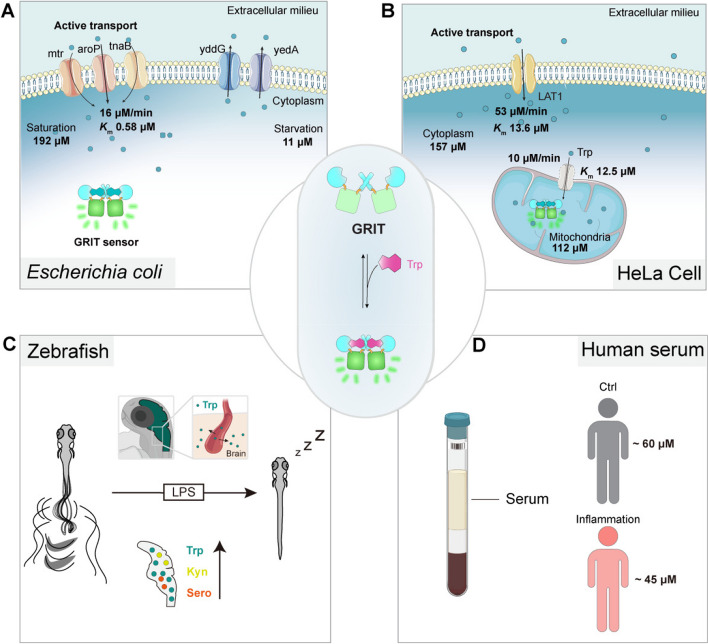
We have diagrammed tryptophan metabolism and functions in bacteria, HeLa cells, intact zebrafish, and human serum for reference (Fig. 5). Our findings show that intracellular tryptophan levels of different cell types and animal models typically lie in the sub-millimolar range with distinct transportation properties (Additional file 1: Table S1), which are in relevance to their specific functions in respective cells. In Escherichia coli, tryptophan anabolism stands out as one of the most energy-intensive metabolic pathways [20], which is activated exclusively under tryptophan deprivation conditions (lower than sub-micromolar), along with a significant increase in its half-proliferation time [21]. The high affinity and rapid response to tryptophan facilitate rapid growth to adapt variable environments [21] and allow gut microbes to continually metabolize tryptophan into indole derivatives, influencing host the enteric nervous systems [22, 23] (Fig. 5A).
Fig. 5.
The proposed biophysical models of tryptophan dynamics in bacteria, mammalian cells, zebrafish, and human serum. A Pathways contributing to tryptophan metabolism in bacteria. Bacteria absorb external tryptophan mainly by aroP and export excess tryptophan with yddG and yedA. Gut microbe could utilize tryptophan to synthesize indole compounds to regulate neuron activities. B Proposed model for the tryptophan metabolism in the mitochondria of mammalian cells. Tryptophan enters mitochondria as the speed of 10 μM/min. Mitochondrial tryptophan pool depends on the cytosolic influx and endogenous consumption. C Proposed model of tryptophan dynamics in zebrafish larvae under LPS-induced inflammation. During inflammation, plasma tryptophan enters the brain and increases the levels of kynurenine and serotonin, leading to prolonged sleep time of zebrafish larvae. D The serum tryptophan levels in inflamed patients and control individuals. All the uptake rates or efflux rates are calculated at the Trp concentration around the Michaelis constant (Km) of relative transporters
In mammal system, reprogramming tryptophan metabolism has become attractive for tumor therapy, such as targeting LAT1 or indoleamine 2,3-dioxygenase (IDO) [5, 24]. In contrast to the bidirectional and rapid turnover of the cytosolic tryptophan pool, mitochondrial tryptophan transportation exhibits a slower rate and higher specificity. The stringent regulation of mitochondrial tryptophan pool may benefit protein synthesis and quality control within mitochondria. Considering that the intense, ratiometric GRIT sensor is suitable for high-throughput screening, it offers an opportunity to identify the mitochondrial transporter [25], and an optical method and paradigm for studying the metabolism of subcellular organelles (Fig. 5B). Unlike the invasive method like HPLC, genetically encoded GRIT sensor can monitor the mitochondrial tryptophan dynamics in living cells, thus eliminating the need for sample preparation and inaccurate mitochondrial volume estimation for quantification [26]. This could potentially lead to accurate tryptophan concentration in situ.
Quantitative measurements by GRIT sensor in intact animal and human serum could reveal systemic tryptophan dynamics and its functions (Fig. 5C–D). Tryptophan metabolism regulates immune systems and neural behaviors through kynurenine and serotonin pathways [5, 19, 27], making the redistribution of tryptophan crucial for balancing the physiological functions under pathological conditions [5, 28]. Our measurements suggest that a flow of plasma tryptophan to the brain and the consequent increase of tryptophan metabolites (serotonin and kynurenine) during inflammation resulted in prolonged sleep time in zebrafish (Fig. 5C). These results align with the elevated serotonin concentration in the brain and fatigue and depression behaviors in LPS-treated mice [29] and patients with inflammation symptoms, suggesting a potential metabolic link between immune response and animal behavior [1].
In vitro diagnostics plays a pivotal role in disease detection, monitoring, and personalized medicine, ultimately contributing to better patient outcomes and overall public health [30, 31]. The simple, fast, and accurate GRIT probe allows high throughput quantification of tryptophan concentrations in a patient’s serum, which marks a significant advancement in the clinical applications of fluorescent probes (Fig. 5D). The reductions in serum tryptophan levels during inflammatory responses exhibit remarkable consistency between zebrafish and human patients. Interestingly, a similar redistribution of tryptophan has been observed in glioblastoma [32]. Moreover, our results suggest a correlation between patients’ ages and serum tryptophan levels (Fig. 3C, Additional file 1: Fig. S3H, I), but not with gender, implying a potential role of tryptophan in aging, which are consistent with a previous report that tryptophan supplement could extend worm lifespan via mitochondria NAD de novo synthesis [33]. These results underscore the highly conserved nature of tryptophan metabolism and circulation among vertebrates, emphasizing the relevance of our observations across different species. Considering inflammation is a hallmark of various diseases, such as cancer, aging, obesity, and cardiovascular disease, the systemic and quantitative measurement of tryptophan dynamics on zebrafish offers new insights into the metabolic regulation mechanisms of these diseases [4, 5, 34].
Limitations of study
The GRIT sensor can detect intracellular tryptophan dynamics in a wide range of scenarios, including tryptophan deprivation conditions (~ 5 μM) [35]. However, when the direct detection of low extracellular tryptophan is desirable to evaluate microenvironments, such as in tumor tissues, new versions of GRIT sensors with tight affinities should be designed.
Sensors with different emission spectra could further broaden the applications. A red fluorescent control sensor can be developed as a reference channel to enable cell-by-cell corrections in situ [36]. In addition, far-red versions of the tryptophan sensors could be developed to overcome the low penetration depth of visible light in deep tissue imaging of non-transparent organisms.
Conclusions
Overall, the systemic quantification of tryptophan metabolism using the GRIT sensor represents a major advancement in our understanding of this essential amino acid. By enabling precise measurements across different biological contexts, GRIT has the potential to significantly impact both basic research and clinical practice, offering new insights into the metabolic regulation of health and disease. Quantitative imaging paradigms in model animals also establish a foundation for future studies aimed at uncovering the broader implications of amino acid metabolism in human diseases.
Methods
Cell cultures
HeLa and HEK293T cell lines, obtained from the Cell Bank of the Chinese Academy of Science, were confirmed to be free of mycoplasma contamination. These cell lines were cultured in high glucose DMEM (BBI), supplemented with 10% FBS (ExCell Bio) and 1% penicillin–streptomycin (10,000 U/mL). The cells were maintained at 37 °C in a humidified atmosphere composed of 95% air and 5% CO2.
Zebrafish
Colonies of zebrafish (Danio rerio), specifically the Nacre strain (mitfaw2/w2), were maintained following standard procedures. The experimental protocols involving zebrafish were approved by the Chinese Academy of Sciences (Approval no. NA-046–2019). The zebrafish were kept under a light–dark cycle of 14 h light to 10 h dark, and the water temperature was consistently maintained at 28 °C.
Human plasma samples
All the population studied were from patients of Taizhou Hospital of Zhejiang Province affiliated to Wenzhou Medical University. The experiments were performed according to the medical ethics rules (no. K20211202).
Technical details for GRIT and GRITOL indicators
GRIT sensor was engineered via inserting circularly permuted superfolder YFP (cpSFYFP) into TrpR followed by several cycles of iterative sensor design and optimization [8]. As a single fluorophore dual channel sensor, GRIT sensor exhibits two excitation peaks near 420 nm and 500 nm and one emission peak near 515 nm. In practical experiments, we measure the fluorescence ratio (R485/420 or R488/405) of the sensor at similar wavelengths (microplate reader: Ex1 420/10, Ex2 485/20, Em 528/20; confocal microscope: Ex1 405 nm, Ex2 488 nm, Em 500–550 nm) to measure tryptophan concentration. In tryptophan quantification experiments, it is necessary to conduct parallel control experiments with tryptophan-insensitive control sensor GRITOL to correct for the effects of other environmental factors, such as pH. The correction is achieved by dividing the fluorescence ratios of GRIT and GRITOL from parallel experiments, resulting in the normalized R488/405 or R485/420.
Plasmid construction
For mitochondrial expression, the mitochondrial signal peptide from CoxVIII (cytochrome c oxidase subunit VIII) was duplicated and fused at the N-terminus of tryptophan biosensors [8]. To make lentiviral vectors, genes of mito-GRIT and mito-GRITOL were subcloned into pLVX-IRES-Puro backbone by NheI and PmeI.
Cell culture, transfections, and establishment of stable cell lines
To generate lentiviral particles, HEK293T cells were transfected with lentiviral plasmids, including two packaging vectors (pMD2.G and psPAX2), using Hieff Trans (YEASEN). Lentivirus-containing supernatants were collected at 48 and 72 h post transfection and subsequently applied to HeLa cells cultured in 6-well tissue culture plates supplemented with 8 μg/mL polybrene. Stable HeLa cell lines were established by cultivating the cells with 1 μg/mL puromycin for 1 week, followed by sorting using MoFlo Astrios EQ (Beckman Coulter) with a 488-nm laser line. The resulting stable HeLa cells were used for subsequent fluorescence measurement experiments.
Fluorescence measurement of living cells with microplate reader
For bacterial experiment, fluorescent bacteria cells were centrifugated at 4000 rpm for 2 min, followed by washing three times with modified PBS buffer containing 10 mM HEPES, pH 7.3. The optical density (OD600) of bacterial suspensions was diluted to ~ 0.01, followed by a 30-min pre-starvation before measurements. Three points were detected as the steady state, followed by addition of different concentrations of tryptophan and a 10-min continuous reading on the BioTek SynergyNeo Multi-Mode microplate reader. The fluorescence ratios (R485/420) of GRIT and GRITOL were divided to fit with titration curve for calibration.
For mammalian cell experiments, the HeLa cell lines stably expressing GRIT sensors were trypsinized, resuspended, and plated in a black 96-well flat-bottom plate at a density of 26,000 cells per well, allowing them to cultivate for 10–12 h. Before the fluorescence measurements, the DMEM media was removed, and cells were washed twice with freshly prepared HEPES buffer. Various amino acids were then introduced into the detection buffer, each at specified absolute concentrations. To correct the fluorescence values of GRIT sensors, the signals from HeLa cell samples were background-subtracted.
Fluorescence microscopy
For bacterial imaging, fluorescent bacteria cells were collected and diluted to the OD600 ~ 0.2 with modified PBS buffer with or without 0.1 mM tryptophan. Bacteria solution (10 μL) was added on the 0.17 mm coverslips (Nunc), then the coverslip was put upside down on a glass slide for imaging.
For cell culture imaging, cultured HeLa cells expressing Mito-GRIT and Mito-GRITOL were transferred to an 8-well glass-bottomed dish (Cellvis) where the culture medium was replaced with HEPES buffer with or without 0.5 mM tryptophan prior to experimentation. Utilizing an inverted confocal Olympus FV3000 automatic microscope with a UPlanSApo 40 × Sil objective (N.A. 1.25), dual-excitation ratio imaging was performed.
For in vivo imaging, zebrafish larvae were paralyzed and immobilized dorsal side up in 1.5% low-melting agarose on a glass-bottom dish under a stereoscopic microscope. The larvae were immersed in egg water for 20 min, followed by imaging on an Olympus FV3000 upright confocal microscope with a XLUMPlanFL N 20X water-immersion objective (N.A. 1.0).
The GRIT sensor was excited using 405-nm and 488-nm lasers, capturing emission figures between 500 and 550 nm with a photomultiplier tube (PMT) in a 1024 × 1024 format and 12-bit depth. For subsequent data analysis, a semiautomatic analysis was carried out using the FIQA code implemented in ImageJ Fiji. This analysis involved several crucial steps, including background subtraction to eliminate unwanted noise, skin removal for enhanced clarity, generation of ratiometric pseudocolored images to visualize variations, and batch measurement of fluorescence intensities.
In vitro transcription and mRNA purification
The mRNA preparation involved the utilization of a T7 mMESSAGE mMACHINE kit (Invitrogen). We amplified the cDNAs corresponding to GRIT, GRITOL, or Tol2 and performed in vitro transcription following the manufacturer’s guidelines. To obtain purified mRNA, a 2-h incubation in lithium chloride at − 20 °C was performed, followed by dilution with DEPC-treated nuclease-free water. This approach facilitated the generation of high-quality mRNA samples for further experimental applications.
Zebrafish sleep and locomotion analysis
Sleep analysis was performed based on tracking results of individual larval zebrafish. Zebrafish larvae were raised with a light (14 h) and dark (10 h) cycle. The daytime starts at 9 am and the night-time starts at 11 pm. When experiments started, individual larvae were transferred into wells of a 96-well plate, which was placed inside Zebrabox (Viewpoint Life Sciences, Lyon, France). Their locomotor activity was recorded using the quantization mode in a video-tracking system (Viewpoint Life Sciences). Parameters for detecting different movement patterns were determined empirically: detection thresholds, 15; burst, 29; freeze, 3; and bin size, 60 s. PBS or different doses of LPS were added into the bath at 9 am on the second morning and maintained throughout the experiment.
Human plasma sample preparation
Venous blood samples of patients were collected in tubes early in the morning, after an overnight fast for a standard blood test, which are divided into control group and inflammation group based on the blood test with clinical criteria [37]. For control group: (1) the number of white cells is between 4000 and 9000/μL; (2) the concentration of hypersensitive C-reactive protein (hs-CRP) is less than 20 mg/L; and (3) the neutrophil ratio is between 50 and 70%. For inflammation group: (1) the number of white cells is over 10,000/μL; (2) the concentration of hs-CRP is more than 50 mg/L; and (3) the neutrophil ratio is over 80%. The studied population consisted of 77 adult subjects, which were randomly selected independent of gender and age. Selected plasma samples were centrifuged at 3500 rpm for 2 min and kept at 4 °C until analysis.
Quantifying measurement accuracy using sensor overlord
To evaluate the quantitative capability of the GRIT sensor for measuring tryptophan concentration, we utilized the “SensorOverlord” R package to model the GRIT sensor and predict its accuracy. Based on our previous research [9], we calculated the midpoint potential (pKd) to be 3.37, derived from a dissociation constant (Kd) of 430 μM, a critical parameter indicative of the sensor’s affinity. Using our fluorescence spectral data and two excitation peaks, we determined the values of Rmin, Rmax, and delta2 to be 0.52, 7.73, and 0.25, respectively. For microscopy error, we determined it to be 0.02, based on our experimental findings demonstrating that 95% of observed ratios had a relative error of no more than 2% [16]. Additionally, we set an error threshold 0.05–0.2 to represent the detection range of tryptophan concentrations by GRIT as previously described.
Principle and equations for quantitative measurement of tryptophan in vitro and in vivo
The fundamental assumption of quantitative measurement with the ratiometric biosensor GRIT is that the properties of the purified sensor could be used as a reference to measure tryptophan levels in cells and in vivo [38].
(1) The binding affinity of the GRIT sensor follows the Hill equation [39]:
| 1 |
where Kd is the apparent dissociation constant, which is equal to the tryptophan concentration when half of the sensor is bound, and n is the Hill coefficient. R represents the excitation fluorescence ratio of the biosensor (F485/F420). Rmin and Rmax correspond to the 0% and 100% fraction bound, respectively.
(2) The GRIT sensor is purified from the bacterial expression system, so we can calibrate tryptophan levels in bacteria by fitting the excitation fluorescence ratios of cells with the in vitro titration curve. Intracellular pH could change the excitation ratio of the GRIT sensor and mislead the calibration of tryptophan concentration; thus, the GRTIOL sensor should be used as a control in the parallel experiment for correction.
(3) It has been proven that the Kd values of biosensors in vitro and in cells are often comparable [40]. Thus, we could measure the Rmin and Rmax of the GRIT sensor in situ to calibrate the tryptophan concentration in HeLa cell cultures. LAT1 is highly expressed in HeLa cells and can import and export tryptophan upon the addition of tryptophan and histidine, respectively. Therefore, we could approximate the maximal and minimal fluorescence ratios of the GRIT sensor in living cells by those treatments, and the dynamic range of the GRIT sensor in cells (9.6-fold) is comparable with that of purified protein (12.9-fold). The control experiment with the GRITOL sensor should also be performed in parallel experiments. With the two points Rmin and Rmax, we can calculate the sensor occupancy using the equation below:
| 2 |
where θ is defined as the sensor occupancy, obtained by dividing the fluorescence ratio of bound GRIT to the fluorescence ratio of total GRIT. The sensor occupancy θ could be converted to tryptophan concentration by fitting with the in vitro titration curve.
| 3 |
(4) To calculate the biophysical properties of tryptophan metabolism, we measured the parameters of Km and tryptophan concentration change rate. Km is defined as the concentration of substrate that is transported at half the maximal velocity (Vmax) of transport. We calculated Km with the Michaelis–Menten equation.
| 4 |
where V is defined as the change in the excitation ratio of the intracellular GRIT sensor per min and can be calculated with the equation below.
| 5 |
The tryptophan concentration change rate is defined as the changes in intracellular tryptophan concentrations per min, including the tryptophan uptake rate, tryptophan export rate, and endogenous tryptophan consumption rate. These rates can be calculated with the equation below:
| 6 |
where the [Trp] concentrations are calculated with Eq. 3 using the sensor occupancy at a specific timepoint (t0 and t1), and Δt is the interval between t0 and t1.
(5) For the quantitative measurement of tryptophan in zebrafish, the ratiometric GRIT sensor provides built-in self-calibration for signal correction, which is not affected by sensor expression levels. We isolated primary zebrafish cells expressing the GRIT sensor from zebrafish larvae. We measured the Rmin and Rmax of the GRIT sensor by adding tryptophan and histidine because they also express the LAT1 transporter [41]. The dynamic range of the GRIT sensor in primary zebrafish cells (7.5-fold) is comparable to that of HeLa cells (9.6-fold). Because we used the same imaging conditions and confocal microscopy, the Rmin and Rmax of primary cells in vitro can be used to calculate the mean GRIT sensor occupancy in tissue with Eq. 2.
The tryptophan concentrations in a given tissue were calibrated by normalizing the GRIT signals by the mean GRITOL signal from massive cells. Thus, the corrected mean GRIT sensor occupancy is independent of the minor heterogeneity among tissue cells (such as pH) and represents the mean tryptophan concentration in this tissue with Eq. 3. This GRITOL-based calibration is applicable at the tissue level but not at the single-cell level.
Quantitative measurement of tryptophan with GRIT sensor in vitro and in vivo
Accurate and quantitative assessments of tryptophan levels in cells and serum samples are achievable through the calculation of normalized sensor fluorescence ratios and comparing these ratios with those obtained from purified GRIT protein in vitro, as described previously [9].
For the quantification of serum tryptophan concentrations with GRIT sensor, serum sample was equally mixed with 1 μM GRIT or GRITOL sensor in 100 mM HEPES buffer, 100 mM KCl (pH 7.3), followed by routine fluorescence measurement similar to protein titration experiments.
For quantification bacterial tryptophan, we measured the excitation fluorescence ratios of cells expressing the GRIT sensor with or without 40 μM tryptophan as the intracellular tryptophan levels in tryptophan-fed cells and tryptophan-starved cells, respectively. To account for the pH effect, the excitation ratios of cells expressing GRIT were corrected by those of cells expressing GRITOL measured in parallel and fitted with an in vitro protein titration curve at pH 7.3.
For in situ live-cell calibration, the unbound and saturated states of the mito-GRIT sensor in HeLa cells could be approximated by treatment with 5 mM histidine and 0.5 mM tryptophan in modified HEPES buffer (pH 7.4). The detection conditions were similar to those in live-cell fluorescence measurements. The mitochondrial tryptophan levels were estimated by fitting with purified protein titration curves at pH 8.0.
Tryptophan quantification with high-performance liquid chromatography
HPLC method was used to accurately quantify intracellular tryptophan levels in HeLa cells. We extracted the intracellular metabolites using 80% pre-cold methanol, evaporated the supernatant, and dissolved the samples in 100 µL of 90% acetonitrile to remove proteins before LC/MS quantification. The quantification process was carried out using a UHPLC system (1290 series; Agilent Technologies, USA) coupled with a quadruple time-of-flight mass spectrometer (TripleTOF 6600, AB SCIEX, USA). The results were analyzed using the Agilent MassHunter Qualitative Analysis Navigator (B.08) software.
Mitochondria were extracted by a mitochondria isolation kit (Beyotime) and protein concentration was quantified with a BCA kit (Mxbioscience LLC), followed by LC/MS measurement. The protein amount was converted to mitochondria volume as reported [8].
The plasma tryptophan level was quantified with pre-column derivatization and reverse phase ultra-performance liquid chromatography (RP-HPLC) [42]. In brief, protein was precipitated by acetonitrile treatment in serum samples and centrifuged at 13,000 rpm for 20 min. Derivatization of the supernatant was carried out at 55 °C for 10 min, followed by separation and quantitative analysis by external standard method with RP-HPLC.
Statistical analysis
Data are presented as the mean ± s.e.m. from at least three experiments. All statistical analyses and graphical representations were performed using R and Python. The figures were compiled and finalized using Adobe Illustrator 2020.
The normality of the data was tested using the Kolmogorov–Smirnov test. For comparisons between two groups, the two-tailed unpaired Student’s t-test was employed for data with a normal distribution in the majority of results, while the Mann–Whitney test was used for data with a non-normal distribution. The specific sample size (n numbers) and P values of each figure in the zebrafish and human serum sample experiments are listed in the corresponding figures or legends.
Supplementary Information
Additional file 1: Figures S1-S4, Tables S1-S2 and Note S1. Figure S1 [14]. Detection of tryptophan uptake in living bacteria. Figure S2. Detection of tryptophan dynamics in mitochondria of HeLa cells. Figure S3. Detection of tryptophan levels in the serum samples from inflamed individuals. Figure S4. Zebrafish sleep changes during inflammation, detection of metabolites levels in zebrafish larvae, and the death rate post tryptophan metabolism related inhibitors treatment. Table S1 [9, 10, 15, 26, 43]. Quantifications of tryptophan level in bacteria, mammalian cells, zebrafish, and human serum. Table S2. The effect of 19 amino acids on the tryptophan dynamics in mitochondria of HeLa cells. Note S1. The amino acid sequence information of Mito-GRIT and Mito-GRITOL sensor.
Acknowledgements
We thank Dongjian Wang, Hongbo Chi, Haifei Xiang, Zhehang Zhou, Xiandong Shao, Guo Yu, Rui Lu, and Yuqian Cai for their kind supports and suggestions of patient inflammation test. We appreciate the technical assistance from Dr. Hanyang Hu, Dr. Xufei Du, Hao Deng, and Dr. Weixi Feng. We thank Jiwen Bu for secretarial assistance. We thank Chemical Biology Core Facility and Molecular Biology Core Facility in CEMCS, CAS, for technical support. We thank Dr. Qian Hu and the Optical Imaging Facility at CEBSIT (Institute of Neuroscience), CAS, for support with the image analysis.
Abbreviations
- GRIT
Green ratiometric indicator for tryptophan
- GRITOL
GRIT control sensor
- Mito-GRIT
Mitochondrial expressed GRIT
- HPLC
High-performance liquid chromatography
- FLIPW
Fluorescent indicator protein for tryptophan
- LAT1
L-type amino acid transporter 1
- LPS
Lipopolysaccharide
- hs-CRP
High-sensitivity C-reactive protein
- IDO
Indoleamine 2,3-dioxygenase
Authors’ contributions
R. T. and Y. M. conceived the project. R. T., K. W., T. C., J. C., X. Z., and W. P. performed experiments. R. T., Y. M., K. W., T. C., and X. Z. analyzed data and made figures. C. J. and W. M. provided the human serum samples. R. T., Y. M., and J. D. wrote the draft and revised the manuscript. All authors read and approved of the final manuscript.
Authors’ Twitter handles
Twitter handles: @vivianstats (Wei Vivian Li).
Funding
This work was supported by grants from National Science and Technology Innovation 2030 Major Program (2021ZD0202203, SQ2021AAA010882, 2021ZD0204500, 2021ZD0203704), National Natural Science Foundation of China (32171090, 32171026, 32321003), Shanghai Science and Technology Commission (21ZR1482600), Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), 2023 Youth Innovation Promotion Association CAS, Strategic Priority Research Program of the Chinese Academy of Sciences (XDA27010403), Scientific Instrument Developing Project of the Chinese Academy of Sciences (YJKYYQ20210029), and Basic Public Welfare Research Project of Zhejiang Province (LGD20H090005).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The cytoplasmic version of the GRIT or GRITOL sensor sequences used for bacterial expression can be found in our previously published article [9]. The Mito-GRIT and Mito-GRITOL sensor amino acid sequences are listed in Additional file 1: Note S1. All constructs are available upon requests.
Declarations
Ethics approval and consent to participate
The experimental protocols involving zebrafish were approved by the Chinese Academy of Sciences (Approval no. NA-046–2019). All the population studied were from patients of Taizhou Hospital of Zhejiang Province affiliated to Wenzhou Medical University. The experiments were performed according to the medical ethics rules (no. K20211202).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kui Wang, Tian-lun Chen, Xin-xin Zhang and Jian-bin Cao contributed equally to this paper.
Contributor Information
Jiu-lin Du, Email: forestdu@ion.ac.cn.
Yu Mu, Email: my@ion.ac.cn.
Rongkun Tao, Email: rktao@ion.ac.cn.
References
- 1.Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science. 2017;357:eaaf9794. [DOI] [PubMed] [Google Scholar]
- 2.Fernstrom JD, Wurtman RJ. Brain serotonin content: physiological regulation by plasma neutral amino acids. Science. 1972;178:414–6. [DOI] [PubMed] [Google Scholar]
- 3.Gheorghe CE, Martin JA, Manriquez FV, Dinan TG, Cryan JF, Clarke G. Focus on the essentials: tryptophan metabolism and the microbiome-gut-brain axis. Curr Opin Pharmacol. 2019;48:137–45. [DOI] [PubMed] [Google Scholar]
- 4.Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019;18:379–401. [DOI] [PubMed] [Google Scholar]
- 5.Xue C, Li G, Zheng Q, Gu X, Shi Q, Su Y, et al. Tryptophan metabolism in health and disease. Cell Metab. 2023;35:1304–26. [DOI] [PubMed] [Google Scholar]
- 6.Lan-gan P, Ai-guo T, Xi-ming M, Xi-bo L, Xiang A. More rapid and sensitive method for simultaneous determination of tryptophan and kynurenic acid by HPLC. Clin Biochem. 2009;42:420–5. [DOI] [PubMed] [Google Scholar]
- 7.Knott PJ, Curzon G. Free tryptophan in plasma and brain tryptophan metabolism. Nature. 1972;239:452–3. [DOI] [PubMed] [Google Scholar]
- 8.Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat Methods. 2017;14:720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao R, Wang K, Chen T, Zhang X, Cao J, Zhao W, et al. A genetically encoded ratiometric indicator for tryptophan. Cell Discov. 2023;9:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaper T, Looger LL, Takanaga H, Platten M, Steinman L, Frommer WB. Nanosensor detection of an immunoregulatory tryptophan influx/kynurenine efflux cycle. PLoS Biol. 2007;5:e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piston DW, Kremers G-J. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–14. [DOI] [PubMed] [Google Scholar]
- 12.Crawford IP, Stauffer GV. Regulation of tryptophan biosynthesis. Annu Rev Biochem. 1980;49:163–95. [DOI] [PubMed] [Google Scholar]
- 13.Pittard J, Yang J. Biosynthesis of the aromatic amino acids. EcoSal Plus. 2008;3:10.1128/ecosalplus.3.6.1.8. [DOI] [PubMed]
- 14.Sastry AV, Gao Y, Szubin R, Hefner Y, Xu S, Kim D, et al. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat Commun. 2019;10:5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hewton KG, Johal AS, Parker SJ. Transporters at the interface between cytosolic and mitochondrial amino acid metabolism. Metabolites. 2021;11:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanley JA, Johnsen SB, Apfeld J. The SensorOverlord predicts the accuracy of measurements with ratiometric biosensors. Sci Rep. 2020;10:16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikonomou G, Altermatt M, Zhang R, Coughlin GM, Montz C, Gradinaru V, et al. The serotonergic raphe promote sleep in zebrafish and mice. Neuron. 2019;103:686-701.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarcz R, Bruno JP, Muchowski PJ, Wu H-Q. Kynurenines in the mammalian brain: when physiology meets pathology. Nat Rev Neurosci. 2012;13:465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci. 2002;99:3695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shehata TE, Marr AG. Effect of nutrient concentration on the growth of Escherichia coli. J Bacteriol. 1971;107:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye L, Bae M, Cassilly CD, Jabba SV, Thorpe DW, Martin AM, et al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe. 2021;29:179-196.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao K, Mu C, Farzi A, Zhu W. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr. 2020;11:709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Löb S, Königsrainer A, Rammensee H-G, Opelz G, Terness P. Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: can we see the wood for the trees? Nat Rev Cancer. 2009;9:445–52. [DOI] [PubMed] [Google Scholar]
- 25.Qiu Z, Dubin AE, Mathur J, Tu B, Reddy K, Miraglia LJ, et al. SWELL1, a plasma membrane protein, is an essential component of volume-regulated anion channel. Cell. 2014;157:447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166:1324-1337.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemos H, Huang L, Prendergast GC, Mellor AL. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat Rev Cancer. 2019;19:162–75. [DOI] [PubMed] [Google Scholar]
- 28.Mondanelli G, Volpi C. The double life of serotonin metabolites: in the mood for joining neuronal and immune systems. Curr Opin Immunol. 2021;70:1–6. [DOI] [PubMed] [Google Scholar]
- 29.O’Connor JC, Lawson MA, André C, Moreau M, Lestage J, Castanon N, et al. Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Mol Psychiatry. 2009;14:511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou Y, Wang A, Huang L, Zhu X, Hu Q, Zhang Y, et al. Illuminating NAD+ metabolism in live cells and in vivo using a genetically encoded fluorescent sensor. Dev Cell. 2020;53:240-252.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McRae MP, Rajsri KS, Alcorn TM, McDevitt JT. Smart diagnostics: combining artificial intelligence and in vitro diagnostics. Sensors. 2022;22:6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panitz V, Končarević S, Sadik A, Friedel D, Bausbacher T, Trump S, et al. Tryptophan metabolism is inversely regulated in the tumor and blood of patients with glioblastoma. Theranostics. 2021;11:9217–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castro-Portuguez R, Sutphin GL. Kynurenine pathway, NAD+ synthesis, and mitochondrial function: targeting tryptophan metabolism to promote longevity and healthspan. Exp Gerontol. 2020;132:110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsyuba E, Mottis A, Zietak M, De Franco F, Van Der Velpen V, Gariani K, et al. De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature. 2018;563:354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timosenko E, Ghadbane H, Silk JD, Shepherd D, Gileadi U, Howson LJ, et al. Nutritional stress induced by tryptophan-degrading enzymes results in ATF4-dependent reprogramming of the amino acid transporter profile in tumor cells. Cancer Res. 2016;76:6193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao R, Shi M, Zou Y, Cheng D, Wang Q, Liu R, et al. Multicoloured fluorescent indicators for live-cell and in vivo imaging of inorganic mercury dynamics. Free Radic Biol Med. 2018;121:26–37. [DOI] [PubMed] [Google Scholar]
- 37.Brenner DR, Scherer D, Muir K, Schildkraut J, Boffetta P, Spitz MR, et al. A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol Biomarkers Prev. 2014;23:1729–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JG, Palmer AE. Quantitative measurement of Ca2+ and Zn2+ in mammalian cells using genetically encoded fluorescent biosensors. In: Zhang J, Ni Q, Newman RH, editors. Fluorescent protein-based biosensors: methods and protocols. Totowa, NJ: Humana Press; 2014. p. 29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, et al. Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–7. [DOI] [PubMed] [Google Scholar]
- 40.Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods. 2009;6:737–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellingsen S, Narawane S, Fjose A, Verri T, Rønnestad I. The zebrafish cationic amino acid transporter/glycoprotein-associated family: sequence and spatiotemporal distribution during development of the transport system b0,+ (slc3a1/slc7a9). Fish Physiol Biochem. 2021;47:1507–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Çevikkalp SA, Löker GB, Yaman M, Amoutzopoulos B. A simplified HPLC method for determination of tryptophan in some cereals and legumes. Food Chem. 2016;193:26–9. [DOI] [PubMed] [Google Scholar]
- 43.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB the Human Metabolome Database. Nucleic Acids Res. 2007;35 Database:D521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figures S1-S4, Tables S1-S2 and Note S1. Figure S1 [14]. Detection of tryptophan uptake in living bacteria. Figure S2. Detection of tryptophan dynamics in mitochondria of HeLa cells. Figure S3. Detection of tryptophan levels in the serum samples from inflamed individuals. Figure S4. Zebrafish sleep changes during inflammation, detection of metabolites levels in zebrafish larvae, and the death rate post tryptophan metabolism related inhibitors treatment. Table S1 [9, 10, 15, 26, 43]. Quantifications of tryptophan level in bacteria, mammalian cells, zebrafish, and human serum. Table S2. The effect of 19 amino acids on the tryptophan dynamics in mitochondria of HeLa cells. Note S1. The amino acid sequence information of Mito-GRIT and Mito-GRITOL sensor.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The cytoplasmic version of the GRIT or GRITOL sensor sequences used for bacterial expression can be found in our previously published article [9]. The Mito-GRIT and Mito-GRITOL sensor amino acid sequences are listed in Additional file 1: Note S1. All constructs are available upon requests.