Abstract
Background
The gut microbiota (GM) has proven to be essential for both physical health and mental wellbeing, yet the forces that ultimately shape its composition remain opaque. One critical force known to affect the GM is the social environment. Prior work in humans and free-ranging non-human primates has shown that cohabitation and frequent social interaction can lead to changes in GM composition. However, it is difficult to assess the direction of causation in these studies, and interpretations are complicated by the influence of uncontrolled but correlated factors, such as shared diet.
Results
We performed a 15-month longitudinal investigation wherein we disentangled the impacts of diet and social living conditions on GM composition in a captive cohort of 13 male cynomolgus macaques. The animals were in single housing for the first 3 months of the study initially with a variable diet. After baseline data collection they were placed on a controlled diet for the remainder of the study. Following this diet shift the animals were moved to paired housing for 6 months, enabling enhanced social interaction, and then subsequently returned to single housing at the end of our study. This structured sequencing of diet and housing changes allowed us to assess their distinct impacts on GM composition. We found that the early dietary adjustments led to GM changes in both alpha and beta diversity, whereas changes in social living conditions only altered beta diversity. With respect to the latter, we found that two particular bacterial families — Lactobacillaceae and Clostridiaceae — demonstrated significant shifts in abundance during the transition from single housing to paired housing, which was distinct from the shifts we observed based on a change in diet. Conversely, we found that other bacteria previously associated with sociality were not altered based on changes in social living conditions but rather only by changes in diet.
Conclusions
Together, these findings decouple the influences that diet and social living have on GM composition and reconcile previous observations in the human and animal literatures. Moreover, the results indicate biological alterations of the gut that may, in part, mediate the relationship between sociality and wellbeing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-024-00355-y.
Keywords: Social environment, Gut microbiota, Primates, Social living, Monkeys, Metagenomics
Introduction
The mammalian gastrointestinal tract is home to trillions of microbes that make up the gut microbiota (GM). These bacteria play many crucial roles in the health and wellbeing of their host, including training the immune system [1, 2], defending the body against disease [3–5], and contributing to the harvest, storage, and use of energy from diet [5–7]. A key signature of the GM is its variable composition across individuals. Recent work has begun to establish relationships between this variation and an individual’s susceptibility to disease, including health disorders such as heart disease [8, 9], cancer [10, 11], diabetes [5, 9], autoimmune disorders [12], obesity [7, 9], anxiety [13, 14], and depression [14–16]. However, apart from these associations, the factors that actually shape the composition of the GM remain opaque.
One of the critical factors that has been shown to shape the GM is one’s social environment. Social interactions in bumblebees, termites, and ants have been found to bestow significant health benefits to the individual (e.g., protecting the host against virulent gut parasites [17, 18]), and have been shown to aid conspecific recognition and immune synchronization [19–22]. In contrast, other work has indicated that social relationships can confer disease risks associated with the microbiota [23]. Evidence from rodent models, for instance, has shown that cohabitation encourages the spread of bacterial communities contributing to inflammatory bowel disease [24]. Similarly, stressful social situations (e.g., social defeat) can disrupt the GM and significantly reduce microbial diversity [25–30]. In humans, stressful social experiences reportedly increase the inflammatory immune response, as well as symptoms of anxiety and depression [31]. Collectively, these findings suggest that sociality confers both fitness benefits and costs to the individual, consistent with the view that the GM has a heretofore under-recognized relationship with the social environment [32–35]. This relationship may underlie, in part, the particularly strong correlation between longevity and the prevalence of social interactions in animal species with higher-level cognition and complex social systems, like primates [36–40].
Studies in humans and non-human primates (NHPs) have identified three key relationships between social environment and GM composition [23, 41–43]. First, the GM of cohabiting individuals often resembles one another. For instance, cohabiting humans (e.g., spouses) and group members in free-ranging NHPs (e.g., chimpanzees, baboons, and lemurs) exhibit greater similarity in GM composition than those not sharing a home [23, 43–45] or social group [19, 21, 41, 46], respectively. Second, when individuals migrate from living together to apart (or vice versa), concomitant shifts in the microbiota follow. Microbiota similarities in wild NHPs decrease rapidly amongst group members who relocate to different locations [47] or to a new social group [19, 48, 49]. Finally, sociality has been found to be closely linked to beneficial gut bacteria, which changes social behaviour corresponding to shifts in the abundance of these microbes, and vice versa. For example, research indicates that greater social engagement leads to an increase in beneficial gut bacteria (e.g., Faecalibacterium [50]), as well as a higher overall GM diversity [44]. These changes can provide significant advantages to the host, including enhanced protection against pathogens and increased longevity. In addition, attenuation of impaired social behaviours has been frequently linked to the presence and increased abundance of beneficial bacteria (e.g., Lactobacillus [33, 51–53]). Together, these findings identify important changes in GM diversity with respect to one’s social environment. However, the studies highlighting these relationships have not controlled for changes in dietary habits, which itself is known to have a direct impact on GM composition [23, 41, 42, 54, 55].
Diet is a strong external determinant of GM composition [56–58]. It is perhaps the fastest and most effective way to trigger immediate changes to the GM. Evidence suggests that dietary influences on the composition and function of one’s GM can be incredibly fast acting (on the scale of days) [59–61]. In humans, diet has even been shown to change factors of GM composition in as little as 24-hours [62]. Diet has also been known to have an effect on various biological systems such as the metabolic and immune systems [63, 64]. Similar patterns are also observed in NHPs, with diet being a critical driver of GM composition [65, 66]. Not only can diet rapidly change the GM composition, but in many ways, diet is linked to the social environment. For instance, humans that live together often share dietary habits [67–69]; the same is also true for NHPs in shared social groups [70]. Given the speed with which diet alters the GM, and the close connection between dietary and social factors, it is difficult then to disentangle dietary influences from social influences on the GM, particularly during periods of social change (e.g., moving social groups). Thus, studies to date in wild or free-ranging NHPs have struggled to show sociality-specific changes in GM composition when unable to control diet.
Disentangling the microbial effects of one’s social environment from diet is immensely challenging in humans, as it requires the ability to causally manipulate the social living conditions of individuals while also controlling their food. However, for laboratory-housed NHPs, which serve as a critical translational model for understanding human social cognition and neurobiology [71–74], such manipulations are more feasible. Various NHPs, including cynomolgus macaques—the model species selected for this study—frequently engage in social behaviours, such as grooming [75–78]. These animals’ tendency for frequent social activity is critical for enabling us to assess social influences on GM composition, as physical social contact provides a key vector for microbial transmission [21, 79–81].
Here, we performed a longitudinal study in a colony of 13 male cynomolgus macaques wherein we manipulated the animals’ diet and social living conditions over the course of 15 months. The macaques were first housed individually with a variable diet. Following this, they were maintained on a regulated diet for 3 months without changing their solitary housing arrangement. Importantly, this first feature of the study allowed us to directly measure the impact of a change in diet on GM composition. After the first 3 months, while maintaining the regulated diet, the NHPs were moved to protected pairwise housing for 6 months to facilitate increased social interaction. Finally, they were returned to single housing at the conclusion of our study. This second feature of the study, focusing on social housing conditions, allowed us to uniquely measure the impact of a change in social living conditions on GM composition, which could then be separately compared to the magnitude of the effect related to a change in diet. For each dietary and housing condition, we collected fecal samples from the animals. This longitudinal and systematized approach allowed us to control for various extraneous factors, and directly assess how changes in both diet and social living conditions differentially affect GM composition.
Methods
Subjects
Thirteen unrelated male Mauritian cynomolgus macaques (Macaca fascicularis) were housed continually at Queen’s University under the care of lab animal technicians and the university veterinarian. The animals ranged in age from 6 to 11 years old (mean age: 7.5yrs) and weighed between 6.3 and 12.8 kg. All animals were naive prior to our study. All procedures and animal experiments were approved by the Queen’s University Animal Care Committee and were conducted in full compliance with the guidelines and policies of the Canadian Council on Animal Care and the Animals for Research Act.
Study design
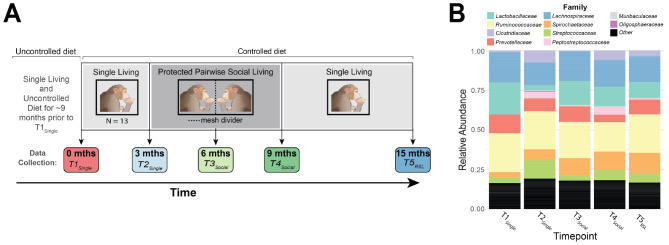
The study spanned 15 months, and was segmented into three separate living condition phases (Fig. 1A): Single Living (3 months duration), Protected Pairwise Social Living (6 months duration), and Return to Single Living (6 months duration). Phase lengths were selected such that we could examine more long-term, chronic changes in the gut microbiota as a result of our diet and sociality manipulations. Importantly, these phase lengths also allowed us to sync-up data collection with several other neurobiological measures that were collected as part of our larger study. In each of the first two phases, two data collection sessions occurred approximately 3 months apart, whereas in the last phase, we collected data only at the 6-month timepoint. We hereby refer to these as T1Single, T2Single, T3Social, T4Social and T5RSL data collection timepoints. Note that within the phases, the animals’ social environment was held constant between data collection sessions. During data collection sessions, stool samples were gathered from each animal. Importantly, prior to T1Single data collection, animals were maintained in single housing with a variable diet.
Fig. 1.
Study design and the relative abundance of GM bacterial families across timepoints. (A) Illustration of our 15-month longitudinal study design. The social living manipulation phases are denoted by gray boxes and labels. The data collection sessions during these living conditions are identified with downward pointing arrows and different coloured bubbles (red: T1Single, light blue: T2Single, light green: T3Social, dark green: T4Social, dark blue: T5RSL). Diet manipulation is indicated at the top with bars and labels: “Uncontrolled diet” and “Controlled diet”. (B) Average relative abundance of the 10 most abundant GM bacterial families across all timepoints and animals
Note that this manuscript leverages a subset of data (fecal samples) originally gathered for an extensive biomarker investigation of the multifaceted neurobiological impacts of alterations in social environment (including data collection related to changes in brain structure/function, systemic cortisol concentrations (via hair samples), various blood and cerebrospinal fluid biomarkers, and dominance ratings). Here, we specifically focus on the variations in gut microbiota composition observed within this dataset.
Living conditions
Single Living
Animals began the study in Single Living, where they were singly-housed in individual, non-contiguous cages (2.6 m height x 1.3 m width x 1.3 m length) within one larger room (i.e., all 13 animals were in the same room, cages set ~ 1 m apart). This allowed for each macaque to see, hear, and smell the others (i.e., auditory, visual, and olfactory social cues/interactions were present), but this did not allow for any physical social contact. During this first single living phase, T1Single and T2Single data were collected. Note that the social environment condition was unchanged between data collection sessions.
Protected Pairwise Social Living
At the 3-month mark, the Protected Pairwise Social Living phase was implemented. In this phase, groupings of 2 animals, with the exception of one group of 3 animals, were created. Animals in a grouping were housed in single occupancy, adjoining pens (dimensions: 2 m height x 1.5 m width x 2.5 m length). These adjoined pens were separated by a protective metal-mesh divider (dimensions: 53 cm width x 182.5 cm height) such that some, but not all, physical social interactions were permitted between animals in a grouping. The mesh squares (3 cm x 3 cm) were large enough for the animals to reach their fingers through to touch one another, but not so large that full grooming was possible. Animals in different groupings were separated by solid panels that prohibited any physical social contact. As with Single Living, the close proximity of the animals allowed them to see, hear, and smell one another, regardless of grouping, and thus close-up auditory, olfactory, and visual social cues/interactions were also present. In addition, the animals were rotated in their pairs between cages every 14 days when the pens were cleaned by animal care staff; as such, the groupings shared living spaces over the course of this phase, but not at the same time. Rotation through the pens ensured that pen location was not a confound for observed gut changes. The Protected Pairwise Social Living condition lasted for 6 months, during which T3Social and T4Social data were collected.
Return to Single Living
At the 9-month mark, the Return to Single Living phase was implemented, wherein the macaques were returned to their previous single housing conditions (as in Single Living). The final data collection session, referred to as T5RSL, took place 6 months into this phase.
Diet
All NHPs were fed 2–4% of their body weight in standard chow (code: 5050, LabDiet, St Louis, MO) [82] for the entirety of the study. This diet was fiber and protein rich to promote healthy weight management in captive primates. A full list of ingredients can be found at the LabDiet website [82]. Ad libitum municipal water was available to the animals at all times throughout the study. No antibiotics or supplements were given to the animals at any point.
Uncontrolled diet
In order to quantify the effect of a change in diet on the GM, we allowed some enrichment elements of our NHPs’ diet to vary for approximately 9 months prior to beginning the study (while living in the single living conditions) and the data collection at T1Single (moving forward we refer to this period as “uncontrolled diet”; Fig. 1A). Aside from the animals receiving 2–4% of their body weight in standard chow, their food intake also included LabDiet Monkey Jumble foraging mix (code: 5L0N, LabDiet, St Louis, MO) [83], seasonal fruit, and occasional food rewards. It was these three enrichment elements that varied during the uncontrolled diet period of our study. Specifically, the foraging mix, which was a blend of fruit, seeds, nuts, and vegetables, was given to animals in variable quantities (¼ - ½ cup) each day. A full list of the ingredients in the Monkey Jumble foraging mix can be found at the LabDiet website [83]. Seasonal fruit (melon, banana, strawberries) was given randomly to animals each day such that not all animals got the same fruit each day, though typically it was a ¼ piece for each animal. Food rewards (additional amounts of Monkey Jumble) were also given variably across animals, such that not all animals received a reward and when they did, the exact quantity of additional Monkey Jumble was not controlled between animals.
Controlled diet
Following T1Single data collection, at the beginning of the Single Living phase, we strictly controlled the enrichment elements of our NHP’s diet for the remainder of the study (referred to as “controlled” diet from here on out; Fig. 1A). Specifically, this involved all animals receiving the exact same proportions and type of food each day. Fruit, foraging mix, and any additional food rewards were given equally to all animals each day. This controlled diet approach not only enabled us to (1) directly investigate how diet alterations impact GM composition during a period of stable single living, i.e., comparing the transition from uncontrolled (T1Single) to controlled diet (T2Single) during the Single Living phase, but also (2) established a foundation for contrasting these dietary GM changes with those stemming from our social manipulation, i.e., comparing T2Single data to the data collected during the Protected Pairwise Social Living phase (T3Social, T4Social) and the Return to Single Living phase (T5RSL).
Fecal sample collection and processing
Gut microbiota composition was characterized using fecal samples collected from the animals at the data collection timepoints highlighted in Fig. 1. Fecal matter is regarded as the gold-standard for assessing the richness and diversity of the gastrointestinal microbiome [84], and is preferred due its stable biological characteristics that allow for long-term sample storage [85]. In addition to stool samples, we also collected several biomarker samples (i.e., CSF, blood, and hair) at each timepoint in accordance with the larger overarching project objectives. Animals were briefly anesthetized under ketamine (5-15 mg/kg, intramuscular) after which fresh stool samples were immediately collected from each animal’s home cage pan. Labeled stool samples were preserved in 95% ethanol in a 15 mL falcon tube and promptly stored at -80 °C until sample analysis [86]. On the same day, while anesthetized, the animals were weighed, set into lateral recumbency and their lumbar area (L4/L5) was shaved (~ 2 in x 2 in). Shaved hair was collected into pre-folded tinfoil pouches for storage and later analysis. The shaved region was aseptically cleaned, a lumbar puncture was performed, and 1 mL of CSF was collected. From the femoral vein, 5 mL of blood were collected into EDTA vacutainers and 6 mL of blood were collected in serum separator tubes (note that the additional biomarker data: CSF, blood, and hair, are not reported in the current paper). Sample collection was conducted between 9 a.m. − 12 p.m. for all animals.
16S rRNA library construction and sequencing
DNA was extracted from 50 mg of fecal matter using the E.Z.N.A. DNA extraction kit for stool (OMEGA Bio-Tek Inc., Norcross, GA, USA), following the manufacturer’s instructions. 1 µL of extracted DNA (including a nuclease-free water negative control and an Escherichia coli positive control) were amplified using custom DNA primers with overhang adapters from Invitrogen (Carlsbad, California, USA) that were designed based off of the Illumina recommendations [87, 88] for 16S next-generation sequencing (16S Amplicon PCR Forward Primer: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3’, 16S Amplicon PCR Reverse Primer: 5’- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’) for the bacterial 16S ribosomal RNA (16 S rRNA) gene, specifically variable region 3 (V3) and 4 (V4). Illumina flow cell adapter sequences and a 12 bp barcode were then incorporated into the PCR primers using the Nextera XT Index Kit (Illumina, FC-131‐1001I). DNA amplicon purity and concentration were quantified on a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, California, USA) and Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Next-generation sequencing was performed using the Illumina MiSeq system.
16S rRNA sequence data processing
Raw sequencing reads were assessed and processed to remove low-quality nucleotides and adapter sequences using Cutadapt v3.4 [89]. The remaining high-quality sequences were processed in R (v4.3.0) [90] using the DADA2 (v1.28.0) [91] package, including the following parameters: left-trim (trimLeft = 15), truncation of forward and reverse reads (285 bp and 250 bp respectively), and removal of chimeric reads. DADA2’s Naïve Bayesian classifier and Ribosomal Database Project taxonomic database, version 18 (RDP18) [92, 93] were used for taxonomic annotation, where the classifier allowed reverse complement matching. Using this process, operational taxonomic units (OTU’s), genetic clusters that represent a group of microorganisms, were assigned at 97% sequence identity using the Greengenes database [94] and then translated to known taxonomic classifications. These classifications were used to determine the relative abundance of each OTU in each sample. For generation of weighted UniFrac distances, a phylogenetic tree was constructed using the phangorn (v2.11.1) [95] package, using a maximum-likelihood method and nucleotide-based similarity model. Distance values were then calculated using the vegan (v2.6.4) [96] package. All figures were generated using the ggplot2 (v3.4.4) [97] package.
Analyses of microbiota diversity and community composition
We examined the alpha and beta diversity of the gut microbiota to investigate the impact of our diet control and our social manipulation. Alpha diversity measures the distribution of species within a single sample (i.e., species richness and species evenness), whereas beta diversity measures the differences in community composition between samples. For these analyses, the samples were rarefied without replacement through random sub-sampling in phyloseq (v1.4.4) [98]. Without rarefaction, uneven sequence counts can artificially inflate alpha diversity metrics with higher read counts [99]; rarefaction was chosen as opposed to other methods, such as a negative binomial distribution, because the variance in read counts between samples was low (range: 6,125 − 85,410, mean: 38,639, IQR: 13,046.75). We set our rarefaction value to 25,000, which resulted in two NHP samples (Quinn at T1Single and Eli at T5RSL) being removed due to their low read depths (< 10,000). These samples had less than half of the reads for all other samples. Dropping samples with low read depths is a standard procedure as smaller biomass samples are often lower quality and contain a higher presence of contaminating sequences [100, 101]. One of our animals (Popeye) was also removed from T1Single analyses due to an inability to collect a sample at that time. Note that all statistical tests that are significant with our rarefied data were also significant with non-rarefied data. See supplemental, Fig. S1, for description of each animal’s available timepoints.
Microbial diversity
To assess alpha diversity, we used the Chao1 index based on family-level OTUs (though see supplemental material, Figs. S2 and S3, for diversity calculated using the Fisher, Shannon, and Faith’s Phylogenetic Diversity (PD) indices). For this metric, timepoints were compared using a Wilcoxon signed-rank test (for the effect of diet) and a Friedman test (for the social living effect). The Wilcoxon signed-rank tests included a “paired = TRUE” argument to ensure the repeated measures nature of this study was accounted for. To assess beta diversity across timepoints, we performed Permutational Multivariate Analysis of Variance (PERMANOVA) tests with the family-level OTUs calculated in vegan (v2.6.4) [96] using weighted UniFrac distance matrices [102, 103] (see supplemental material, Figs. S5 and S6, for the results of PERMANOVAs calculated using the Bray-Curtis distance method [104], which returned qualitatively identical results; see also supplemental materials for the results using genus-level OTUs). In these PERMANOVA tests we specified that the permutations be conducted within animal such that the repeated measures design of this study was accounted for. An advantage of the PERMANOVA approach is that it did not require us to make any assumptions about the underlying data distribution (e.g., assumption of normality). All analyses conducted were repeated measures to account for the repeated measures nature of this study. We used weighted UniFac distances because they consider both the phylogenetic-relatedness of the taxa and their relative abundances within the samples, and we provide the Bray-Curtis results in our supplemental material because it is another common dissimilarity metric used in the literature. PERMANOVAs were conducted on the distance matrices using 1,000 permutations. All significant PERMANOVA results were followed up with a dispersion analysis via the vegan (v.2.6.4) [96] package to ensure that results were not skewed by unequal group dispersions. Like our PERMANOVA tests, our dispersion analyses also permute within animal. To meaningfully depict beta diversity differences, non-metric multidimensional scaling (NMDS) was used for each of the distance matrices via the vegan (v.2.6.4) [96] package. NMDS is the most appropriate method here as it allows for the use of various distance metrics and maps onto a low-dimensional space, with high explanatory power for relative differences between samples [105].
Linear discriminant analysis
To follow up a significant PERMANOVA effect, we assessed bacterial differences at the family level across timepoints using the linear discriminant analysis (LDA) effect size (LEfSe) method [106] (see supplemental material, Fig. S7, for LEfSe results at the genus level). LEfSe combines statistical significance tests with biological consistency tests to identify genomic features most likely to explain the differences between communities. Importantly, the analysis produces estimates of the magnitude of variation among OTUs, thus allowing us to differentiate the phenotypes of interest. Differences in OTUs are first assessed with two-tailed nonparametric Kruskal-Wallis rank-sum tests. This is followed by pairwise tests across groups (Wilcoxon rank-sum), and lastly, an LDA is performed to estimate the effect size of each differentially abundant feature [106]. LEfSe tests for differentially abundant taxa were conducted with the microbiomeMarker (v. 1.6.0) [107] package in R. The significance criteria for the LEfSe tests were set to an LDA score of less than 10− 3, with a Wilcoxon and Kruskal-Wallis p-value less than 0.05. For completeness, we have also conducted an ANCOMBC2 [108] analysis on our data as it is a common analysis method in the literature. We have included the results of these analyses in the supplemental material (see Tables S6 and S7).
Results
Changes in diet cause changes in gut microbiota composition
In the current study, we sought to disentangle the effects of a change in diet from the effects of a change in social environment (i.e., from single living to cohabitation). To do this, we implemented a diet change while maintaining animals’ living conditions (and prior to beginning our social living manipulations; Fig. 1A), thereby allowing us to directly compare GM composition during a period of controlled diet (T2Single) to uncontrolled diet (T1Single). This established a baseline ‘diet effect’, to which we could subsequently compare the effects of changes in social living conditions.
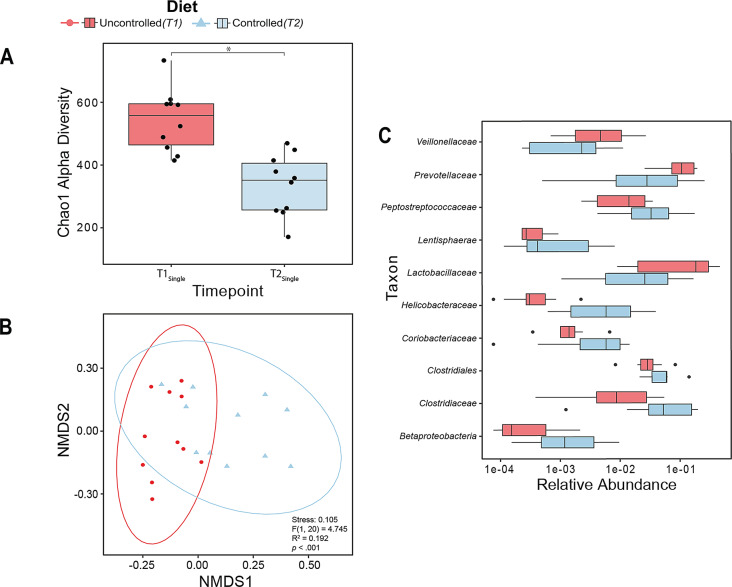
Visual inspection of our data at the group level suggested a shift in relative abundance from T1Single to T2Single, potentially mediated by a change in diet (Fig. 1B, see also supplemental material, Fig. S1, for individual animal profiles of relative abundance over time). To more directly quantify this observation, we examined our alpha and beta measures of GM diversity. Note that all analyses conducted take into account the repeated measures nature of our study design. A non-parametric Wilcoxon signed-rank test on our Chao1 alpha diversity measure revealed a significant difference between T1Single and T2Single, (W = 55, p = .002; Fig. 2A) such that alpha diversity decreased from T1Single to T2Single. Next, we performed a pairwise PERMANOVA test on the beta diversity data using weighted UniFrac distances. This analysis also revealed a significant effect from T1Single to T2Single (F(1,20) = 4.745, p < .001, R2 = 0.192, stress = 0.105; Fig. 2B), indicating that changes in diet not only resulted in changes to GM species richness, but also changed diversity between samples. The group dispersions for T1Single and T2Single were not significantly different from one another (F(1,20) = 2.501, p = .118; see supplemental material, Table S5, for a full list of values).
Fig. 2.
Changes in diet result in changes in GM composition. (A) Chao1 alpha diversity differs significantly between the diet-uncontrolled condition (red, T1Single) and the diet-controlled condition (light blue, T2Single), * p < .05. In the boxplots, the ends of the boxes denote the first (25%) and third (75%) quartiles, the center line represents the median, and the whiskers represent the range of values falling within the interquartile range (IQR) x 1.5. Single black data points represent individuals. (B) NMDS plot with weighted UniFrac distances depicting the effect of diet on GM diversity. Diet-uncontrolled condition (red, T1Single) is significantly different from diet-controlled condition (light blue, T2Single), p < .001. (C) Differential relative abundance of bacterial families exhibiting the diet effect. LEfSe analysis identified 10 bacterial families that significantly changed between diet-uncontrolled (red, T1Single) and diet-controlled (light blue, T2Single) conditions. In the horizontal boxplots, the ends of the boxes denote the first (25%) and third (75%) quartiles, the center line represents the median, and the whiskers represent the range of values falling within the interquartile range (IQR) x 1.5. Single black data points represent individuals outside this range
To examine which families of bacteria might be contributing to these above patterns of effects, we employed the LEfSe [106] method (at the family level), which identified significant differences in bacterial abundances for several families (e.g., Lactobacillaceae and Clostridiaceae) between the diet-controlled and diet-uncontrolled conditions (Fig. 2C). Notably, some of the bacterial families identified through this LEfSe analysis have been previously implicated in primate and rodent social behaviour (e.g., Helicobacteraceae [109, 110], Lactobacillaceae [51], Prevotellacaeae [50]), underscoring the importance of being able to delineate effects related to changes in diet from changes in sociality when studying GM composition.
Changes in social living conditions impact the gut microbiota composition
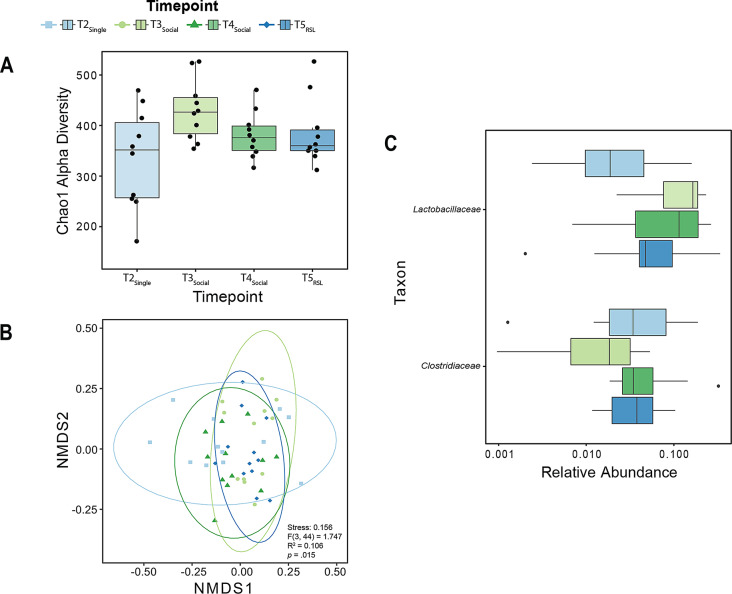
We next sought to determine whether changes in social living conditions impacted the GM above and beyond the aforementioned effects of diet (i.e., compare timepoints T2Single, T3Social, T4Social and T5RSL). To this end, we again assessed alpha and beta diversity of our socially manipulated (but diet-controlled) samples (again, all analyses take into account the repeated measures nature of our study design). Notably, as a departure from that observed for the change in diet, when we analyzed alpha diversity, we found no significant changes across the four timepoints (Friedman test: 𝛸2(3) = 5.88, p = .12, Fig. 3A). However, when we examined our beta diversity measure (using a repeated measures PERMANOVA with weighted UniFrac distances) we did find a significant effect of our social living manipulation (F(3,44) = 1.747, p = .01, R2 = 0.106, stress = 0.156; Fig. 3B). The group dispersions for T2Single, T3Social, T4Social and T5RSL were not significantly different from one another (F(4,53) = 1.733, p = .139; see supplemental material, Table S5, for a full list of values). Follow-up pairwise comparisons using a False Discovery Rate (FDR) correction revealed that this significant effect was primarily driven by a difference between the T2Single and T3Social timepoints, and not among the other timepoints (see Table S1 and S2 in supplemental materials for a full list of values and comparisons). This suggests that changes in the composition of the GM only occurred after the transition from single to pairwise living.
Fig. 3.
Changes in social living conditions result in changes in GM composition. (A) Chao1 alpha diversity did not differ across our diet-controlled, social living conditions. In the boxplots, the ends of the boxes denote the first (25%) and third (75%) quartiles, the center line represents the median, and the whiskers represent the range of values falling within the interquartile range (IQR) x 1.5. Single black data points represent individuals. (B) NMDS plot with weighted UniFrac distances depicting the effect of social living on beta diversity. A significant effect due to changes in social living conditions was detected via PERMANOVA analysis, p < .05. (C) Differential relative abundance of bacterial families for the effect of social living. Our LEfSe analysis identified 2 bacterial families, Lactobacillaceae and Clostridiaceae, that significantly changed across our social living conditions. In the horizontal boxplots, the ends of the boxes denote the first (25%) and third (75%) quartiles, the center line represents the median, and the whiskers represent the range of values falling within the interquartile range (IQR) x 1.5. Single black data points represent individuals outside that range
To examine which families of bacteria may contribute to these changes in community composition across timepoints, we performed another LEfSe [106] analysis (at the family level). This analysis identified significant differences in abundance within the Lactobacillaceae and Clostridiaceae families across our social living conditions (Fig. 3C). Notably, Clostridiaceae and Lactobacillaceae taxa were also both identified as being impacted by changes in diet (Fig. 2C). This suggests that both diet and social environment have distinct modulatory effects on these bacteria.
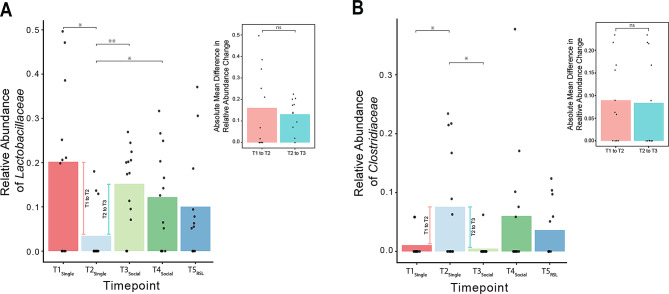
In order to quantify these differential effects, for each bacteria, we conducted a series of Wilcoxon signed-rank tests between all timepoints (Fig. 4), again using the argument “paired = TRUE” to ensure the repeated measures design of the study was accounted for, and found significant differences in relative abundance between various time points (e.g., T1Single vs. T2Single and T2Single vs. T3Social; Fig. 4A and B, and supplemental materials, Table S3 and S4, for a full list of p-values). Note that, like our analysis of beta diversity, we did not find evidence of a change in GM composition from T4Social to T5RSL. Next, we assessed whether the magnitude of change in these two bacteria differed between the diet transition versus the single to pairwise living transition by running another Wilcoxon signed-rank test on the absolute (unsigned) change in relative abundance from T1Single to T2Single (diet effect) compared to the absolute change from T2Single to T3Social (social living effect), again using the argument “paired = TRUE”. Note that our use of “absolute” here is in reference to the mathematical sense of the word. We found no significant difference between the diet effect and social living effect for either Lactobacillaceae (p = .969; Fig. 4A inset) or Clostridiaceae (p = .423; Fig. 4B inset). This suggests that both the change in diet and the change in social living conditions had a similar magnitude of effect on the relative abundance of Clostridiaceae and Lactobacillaceae in the GM. Notably, however, these effects were observed in opposite directions: Whereas Lactobacillaceae decreased in relative abundance when diet was controlled for, and then increased and remained constant when the animals moved into pairwise housing (Fig. 4A), Clostridiaceae was low in abundance during the uncontrolled-diet period, and then increased when diet was controlled for, and then decreased again throughout the remaining timepoints (Fig. 4B). Notably, our ANCOMBC2 analysis also identified Lactobacillaceae as a significant bacterial family with changes in abundance across our social living timepoints (i.e., T2Single, see supplemental materials Table S6 and S7).
Fig. 4.
Lactobacillaceae and Clostridaceae appear to be equally modulated by a change in diet and a change in social living conditions. (A) Changes in the relative abundance of Lactobacillaceae across all timepoints. Wilcoxon signed-rank tests were conducted across all timepoints to identify differences between timepoints, as indicated by significance bars. Data points indicate the values for individual animals. Bars denote the average relative abundance for each timepoint. The pink and blue lines to the left and right of T2Single, respectively, highlight the change in relative abundance from T1Single to T2Single (effect of diet) and T2Single to T3Social (effect of moving from single to pairwise living). The inset graph at the top right denotes absolute differences in the relative abundance change from T1Single to T2Single (pink) and T2Single to T3Social (blue). Bars indicate the mean values for each group. Data points indicate the absolute values of change across the respective timepoints for each animal. * p < .05, ** p < .001, ns = non-significant. (B) Changes in the relative abundance of Clostridiaceae across all timepoints. This figure is set up in the same manner as (A)
Discussion
Prior work has indicated that one’s social environment can shape GM composition [19, 21, 23, 50]. However, these effects are often confounded by the fact that close social groups tend to share similar dietary habits, which itself is known to directly influence the GM [23, 41, 42, 54, 65]. To disentangle these different factors, here we performed a 15-month longitudinal study in a colony of 13 male NHPs where we manipulated the animals’ social living conditions while controlling their diet. A key feature of our social manipulation paradigm was that we maintained animals’ living conditions for a duration of 3 months (Single Living) after controlling their diet. This allowed us to (1) directly assess the effects of a change in diet on GM composition (between T1Single and T2Single) while also (2) providing a critical foundation for examining, and interpreting, any subsequent changes in the GM as a result of our social manipulations, i.e., moving from Single Living (T2Single) to Protected Pairwise Social Living (T3Social, T4Social) then back to single living (T5RSL).
Our study revealed two primary findings, first is the effect of diet changes on the GM composition. Dietary changes significantly altered both the alpha and beta diversity of the GM (Fig. 2A and B). These findings were to be expected, as diet is a well-established modulator of GM composition. However, shifts in relative abundance identified for this diet effect were observed in several bacterial families, including some that have been previously linked to sociality (e.g., Helicobacteraceae [109, 110], Prevotellacaeae [50]) (Fig. 2C). This finding emphasizes the need to disentangle dietary and social effects on the GM. Where prior studies have claimed certain bacteria to be influenced by sociality, our findings suggest that in fact such changes may be diet related. Our social living manipulation allows us to tease these effects apart.
Our second finding is the effect of social living conditions on the GM composition. Contrasting the diet effect, we found that changes in social living conditions (single vs. pairwise living) only influenced beta diversity (Fig. 3B). Notably, other bacteria previously linked to sociality (e.g., Helicobacteraceae [109, 110], Prevotellacaeae [50]), which were altered by diet (Fig. 2C), did not exhibit significant changes with our social manipulation. However, we did find that two bacterial families involved in driving this social living effect — Lactobacillaceae and Clostridiaceae (Fig. 3C) — also responded significantly to dietary changes (Fig. 2C). This indicates an interplay between sociality and GM composition. This result demonstrates the distinct effects of diet and social living on GM composition, that is, we see sociality is impacting relative abundance of bacterial families, above and beyond the effects of a diet change. Collectively, our findings highlight a nuanced relationship between social environment and the GM which we discuss in more depth next.
While diet profoundly shapes gut microbial community composition, our findings demonstrate that an individual’s social environment also plays a unique role. Specifically, comparing the GM composition between T2Single and T3Social (when animal diet was held constant) reveals a distinct shift driven by the transition from single to social living. We note two key observations here. First, we found that bacteria previously implicated in sociality (e.g., Prevotella [50]) did not show significant changes in accordance with our social manipulations once we controlled for diet. This suggests that some of the previously identified sociality-related bacterial changes in the literature may actually relate to the effects of a shared diet (in cohabiting individuals). Second, we found that a small subset of bacteria — namely Lactobacillaceae and Clostridiaceae — exhibited both a diet effect and a separate social living effect (Fig. 4). Notably, for these bacteria, we found that the magnitude of the social effect (i.e., absolute difference in relative abundance between T2Single and T3Social) was not significantly different from the magnitude of the diet effect (i.e., absolute difference in relative abundance between T1Single and T2Single). This is an important observation, as it suggests separate but equal effects of diet and sociality on certain bacteria. We also found that the changes in Lactobacillaceae and Clostridiaceae across timepoints were inversely related. For reasons we will discuss next, this may not be surprising given that the gut is a hostile environment where opportunistic bacteria can thrive when homeostasis is disrupted (i.e., dysbiosis [111, 112]), like from a change in diet. Thus, these findings may be highlighting a “balancing out” interaction between these bacteria as the microbiota strives to return to a stable condition [113].
In the prior literature, the Lactobacillaceae family is recognized for its beneficial properties in both human and animal gut microbiomes. Notably, Lactobacillaceae has been linked to immune system enhancement and helping to prevent or mitigate infectious diseases by controlling opportunistic pathogens [114–117]. In contrast, the Clostridiaceae family is often associated with negative health outcomes; its overgrowth is connected to dysbiosis, including inflammatory bowel diseases [118], and has been implicated in certain cancers, such as breast cancer [119]. Given these distinct associations, it may not be surprising to observe an inverse relationship between these two bacterial families in our study (Fig. 4). After dietary control (from T1Single to T2Single), we noted a decrease in Lactobacillaceae and an increase in Clostridiaceae. This suggests that diet change disrupted the microbiota. However, this trend reversed upon introduction to social living (from T2Single to T3Social), with an increase in Lactobacillaceae and a decrease in Clostridiaceae. This shift may underscore the potential impact of social interactions on these different bacterial families: The advantageous effects of social living may foster the growth of beneficial gut bacteria like Lactobacillaceae, which in turn helps to regulate and diminish the prevalence of harmful bacteria such as Clostridiaceae. While clearly such speculation represents an oversimplification of the complex mechanistic interactions between competing bacteria, it nevertheless suggests intriguing avenues to be explored in future research.
Notably, when the animals returned to single living conditions at the end of the study (T5RSL), we found that their GM composition did not fully revert back to the initial state observed at T2Single. This contrasts with studies on free-ranging NHPs, where relocation typically leads to swift GM changes [19, 47–49]. This result seems particularly noteworthy considering that the animals spent a substantial six-month period in the T5RSL condition before data collection. As such, it suggests that microbial alterations due to social living (in T3Social and T4Social) can have a relatively long-lasting impact on GM composition (i.e., at least 6 months duration). Furthermore, it suggests that the swift changes in GM composition observed in prior field observations could reflect dietary changes associated with relocation, rather than purely social factors. In future research, it will be useful to examine whether an even longer period in the T5RSL condition, perhaps up to 12 months, results in a full return to the T2Single GM composition.
Although the current study has demonstrated that alterations in the social environment can change the GM, it remains unclear the specific mechanisms that mediate these changes. Recent advances in GM research have implicated the microbiota-gut-brain axis (MGBA) as a key bi-directional communication pathway between the gut and brain [110] that can significantly influence host social behaviour [28, 33, 52, 120, 121]. For instance, prior work suggests that this axis plays an important role in mediating sociality-related changes [122] in both the brain [123] and gut [21, 50]. When considering the features of the animals’ social environment that could potentially drive GM changes in our study, it is crucial to note that physical touch (e.g., grooming), a primary form of sociality in humans and NHPs [75, 124, 125], was relatively limited due to the use of cage dividers. Our findings, therefore, highlight two possible (albeit not mutually exclusive) explanations for the GM changes we observe: (1) the GM may have been influenced via other social modalities through the MGBA, such as chemical and auditory cues or, (2) though limited, the physical touch permitted during our social living condition had a strong enough effect on the MGBA to alter the GM.
Chemical signals, such as odorant bacterial metabolites from bodily microbiota (e.g., skin), function as important social cues (e.g., indicating relatedness) and can affect social behaviour [121, 126]. For instance, in fruit flies, microorganisms of the GM have been found to mediate the production and release of chemical scents associated with social signaling, such as attraction [127], kin recognition [128, 129], and mating preferences [130]. This implies that social communication via the GM not only involves the transmission of signals but, through the MGBA, may also influence the receiver’s behaviour and subsequently affect their GM composition, even in the absence of physical contact. Similarly, auditory social cues in primates, conveying information about sex, identity, kinship, and group membership [131, 132], may also impact the GM. Previous investigations have found that chronic stress on the auditory system and fear learning can cause anatomical alterations in the auditory cortex [133]. Considering this and the known link between chronic social stress, host social behaviour, and resulting changes to GM composition [19, 21, 33, 35, 45], it is plausible then that changes in the auditory social environment may lead to brain changes with downstream effects on the GM through the MGBA. These findings highlight the complexity of social interactions and their multi-sensory nature, offering avenues for exploring how non-physical social cues can influence the gut microbiota.
In addition to the alternative sensory modalities noted above, physical touch stands out as a compelling mechanism for driving the observed changes in GM composition. Touch is more than a microbial transmission route; it is fundamental to social communication and bonding, offering intrinsic rewards for both the giver and receiver [75, 124, 125, 134]. Since neurotransmitters can influence the gut via the MGBA [110], the neurobiological rewards of social touch — for example, the release of endogenous opioids [134] — raise the possibility that touch could modify the GM through this axis. In any case, when taken together, the degree of GM change from single to pairwise social living suggests a surprising responsiveness to even limited physical contact and indirect sensory (odor, audition) social cues.
Methodological considerations and future directions
While the current study has disentangled GM effects related to diet and sociality, there remain several questions to be addressed through future research. Firstly, while our use of laboratory-housed NHPs enabled a controlled manipulation of diet and social conditions, this approach may not capture the full spectrum of gut bacteria present in free-ranging macaque groups. For instance, prior work has shown that wild macaques exhibit greater GM diversity (both alpha and beta) in comparison to laboratory-housed NHPs [54, 135]. Despite this limitation, our controlled environment was necessary to isolate sociality-related effects, as field studies pose considerable practical challenges in this regard and make assessments of causality more difficult [21]. Secondly, due to our sample size consisting of 13 male NHPs, it is unclear whether (1) we would have observed more diverse changes (e.g., other bacteria previously implicated in sociality) had the sample size been larger, and (2) whether our effects also generalize to females. These are important questions, and we have plans to examine these relationships in our future work. Thirdly, the 3-month intervals between most data collection points in our 15-month study may have limited our ability to detect much more subtle, short-term changes in the GM. Indeed, while our approach may have been effective for observing broad dietary and social effects, it is possible that this time frame might have obscured finer-scale microbial fluctuations. Given that previous GM studies have often been conducted with shorter data collection intervals (over days or weeks [136, 137]), future research could benefit from a more compressed timeline to help capture these nuances. Finally, it is possible that the changes in GM composition observed from T2Single to T3Social could reflect residual effects of the dietary manipulation at T1Single, rather than the direct influence of social living conditions (as we have interpreted). While dietary changes are often rapid, with microbial composition adjusting within days to weeks [61], there is evidence, including from our own findings, that dietary effects can also manifest over longer periods. For instance, we observe significant changes in GM composition from T1Single to T2Single over a 3-month period, following a controlled dietary change, indicating that diet can have sustained effects on microbial composition [138]. However, the patterns of microbial change we observe from T2Single to T3Social suggest that other factors, particularly social living conditions, are more likely driving the shifts during the Protected Pairwise Social Living phase. If prolonged dietary effects were the primary driver, we would expect to see a more continuous progression of changes across all timepoints. Instead, we see non-linear, time-specific shifts, such as the saw-tooth (opposing) fluctuations in Lactobacillaceae and Clostridiaceae abundances (from T1Single to T2Single and from T2Single to T3Social, see Fig. 4). These non-linear patterns are difficult to explain solely by the passage of time after dietary changes and are more consistent with the introduction of a new environmental factor—social living conditions—affecting the GM at T2Single and beyond. Moreover, the documented impact of social interactions on the GM [35] lends further support to this interpretation. While we acknowledge that disentangling the longer-term effects of diet from the effects of sociality can be challenging, the distinct pattern of shifts we observe across time points strongly suggests that social factors, rather than lingering dietary effects, are the primary drivers of the microbial changes between T2Single to T3Social.
Conclusions
We found that changes in diet resulted in shifts in both the alpha and beta diversity of the GM, whereas changes in social living conditions only affected beta diversity of the GM. Notably, two bacterial families — Lactobacillaceae and Clostridiaceae — demonstrated significant abundance changes that were attributable to the transition from single to pairwise social living, and that were distinct from those caused by a change in diet. Together, these findings uncover the separate impacts of diet and social factors on GM composition, highlighting a complex interaction between social environments and gut health.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the Animal Care Staff at Queen’s University, specifically Brittney Armitage-Brown, Kim Moore, Jessica Freeman, Angie Thompson, Darlene Potterton, and Kurt Scrutton, for their assistance with this work.
Abbreviations
- ANCOMBC2
Analysis of compositions of microbiomes with biase correction 2
- FDR
False discovery rate
- GM
Gut microbiota
- LEfSe
Linear discriminant analysis effect size
- MGBA
Microbiota-gut-brain axis
- NHPs
Non-human primates
- NMDS
Non-metric multidimensional scaling
- OTU
Operational taxonomic unit
- T5RSL
Return to single living
- T1Single
Timepoint 1, single living
- T2Single
Timepoint 2, single living
- T3Single
Timepoint 3, social living
- T4Single
Timepoint 4, social living
Author contributions
CSP collected samples, analyzed the data, and prepared the manuscript. JPG, SB, AW, conceived of this study, collected samples, and contributed resources. DB, KD, JG performed laboratory assays. CSP, AK and CPS analyzed the data. DG, JYN, VK, MAS, GB, FGDF, MP, DJC, SHS, DPM, and AT contributed resources. All authors edited and approved the final manuscript draft.
Funding
Funding was provided by the New Frontiers in Research Fund (NFRFE-2021-00936).
Data availability
All sequence data supporting the findings of this study were deposited in NCBI’s repository (SRA) under accession number PRJNA1073535 and are available at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/1073535.
Declarations
Ethics approval and consent to participate
All procedures and animal experiments were approved by the Queen’s University Animal Care Committee and were conducted in full compliance with the guidelines and policies of the Canadian Council on Animal Care and the Animals for Research Act (reference number: 2020–2024).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Belkaid Y, Hand T. Role of the Microbiota in Immunity and inflammation. Cell. 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the Microbiota and the Immune System. Science. 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maciel-Fiuza MF, Muller GC, Campos DMS, do, Socorro Silva Costa P, Peruzzo J, Bonamigo RR et al. Role of gut microbiota in infectious and inflammatory diseases. Front Microbiol. 2023;14:1098386. [DOI] [PMC free article] [PubMed]
- 4.Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013;29:51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanz Y, Olivares M, Moya-Pérez Á, Agostoni C. Understanding the role of gut microbiome in metabolic disease risk. Pediatr Res. 2015;77:236–44. [DOI] [PubMed] [Google Scholar]
- 6.Krajmalnik-Brown R, Ilhan Z-E, Kang D-W, DiBaise JK. Effects of Gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27:201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 8.Trøseid M, Andersen GØ, Broch K, Hov JR. The gut microbiome in coronary artery disease and heart failure: current knowledge and future directions. eBioMedicine. 2020;52. [DOI] [PMC free article] [PubMed]
- 9.Chen Y, Zhou J, Wang L. Role and mechanism of Gut Microbiota in Human Disease. Front Cell Infect Microbiol. 2021;11. [DOI] [PMC free article] [PubMed]
- 10.Pouncey AL, Scott AJ, Alexander JL, Marchesi J, Kinross J. Gut microbiota, chemotherapy and the host: the influence of the gut microbiota on cancer treatment. Ecancermedicalscience. 2018;12:868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the Anticancer Immune effects of Cyclophosphamide. Science. 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaheen WA, Quraishi MN, Iqbal TH. Gut microbiome and autoimmune disorders. Clin Exp Immunol. 2022;209:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohajeri MH, La Fata G, Steinert RE, Weber P. Relationship between the gut microbiome and brain function. Nutr Rev. 2018;76:481–96. [DOI] [PubMed] [Google Scholar]
- 14.Kelly JR, Borre Y, O’ Brien C, Patterson E, El Aidy S, Deane J, et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–18. [DOI] [PubMed] [Google Scholar]
- 15.Heym N, Heasman BC, Hunter K, Blanco SR, Wang GY, Siegert R, et al. The role of microbiota and inflammation in self-judgement and empathy: implications for understanding the brain-gut-microbiome axis in depression. Psychopharmacology. 2019;236:1459–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21:786–96. [DOI] [PubMed] [Google Scholar]
- 17.Engel P, Moran NA. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. [DOI] [PubMed] [Google Scholar]
- 18.Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proceedings of the National Academy of Sciences. 2011;108:19288–92. [DOI] [PMC free article] [PubMed]
- 19.Perofsky AC, Lewis RJ, Abondano LA, Di Fiore A, Meyers LA. Hierarchical social networks shape gut microbial composition in wild Verreaux’s sifaka. Proceedings of the Royal Society B: Biological Sciences. 2017;284:20172274. [DOI] [PMC free article] [PubMed]
- 20.Archie EA. Bat microbiomes are socially synchronized. Nat Ecol Evol. 2019;3:18–9. [DOI] [PubMed] [Google Scholar]
- 21.Tung J, Barreiro LB, Burns MB, Grenier J-C, Lynch J, Grieneisen LE, et al. Social networks predict gut microbiome composition in wild baboons. eLife. 2015;4:e05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archie EA, Theis KR. Animal behaviour meets microbial ecology. Anim Behav. 2011;82:425–36. [Google Scholar]
- 23.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae Act in Concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey KL, Bloomsmith MA, Michopoulos V, Remillard CM, Young LA. Influence of female coalitionary aggressive behavior on the success of male introductions to female groups of rhesus macaques (Macaca Mulatta). Appl Anim Behav Sci. 2021;237:105293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharwani A, Mian MF, Foster JA, Surette MG, Bienenstock J, Forsythe P. Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Psychoneuroendocrinology. 2016;63:217–27. [DOI] [PubMed] [Google Scholar]
- 27.Partrick KA, Chassaing B, Beach LQ, McCann KE, Gewirtz AT, Huhman KL. Acute and repeated exposure to social stress reduces gut microbiota diversity in Syrian hamsters. Behav Brain Res. 2018;345:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkar A, Harty S, Johnson KV-A, Moeller AH, Carmody RN, Lehto SM, et al. The role of the microbiome in the neurobiology of social behaviour. Biol Rev. 2020;95:1131–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szyszkowicz JK, Wong A, Anisman H, Merali Z, Audet M-C. Implications of the gut microbiota in vulnerability to the social avoidance effects of chronic social defeat in male mice. Brain Behav Immun. 2017;66:45–55. [DOI] [PubMed] [Google Scholar]
- 30.Galley JD, Nelson MC, Yu Z, Dowd SE, Walter J, Kumar PS, et al. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 2014;14:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim C-S, Shin G-E, Cheong Y, Shin J-H, Shin D-M, Chun WY. Experiencing social exclusion changes gut microbiota composition. Transl Psychiatry. 2022;12:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montiel-Castro A, González-Cervantes R, Bravo-Ruiseco G, Pacheco-Lopez G. The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality. Front Integr Nuerosci. 2013;7. [DOI] [PMC free article] [PubMed]
- 33.Münger E, Montiel-Castro AJ, Langhans W, Pacheco-López G. Reciprocal interactions between gut microbiota and host Social Behavior. Front Integr Nuerosci. 2018;12. [DOI] [PMC free article] [PubMed]
- 34.Lombardo MP. Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav Ecol Sociobiol. 2008;62:479–97. [Google Scholar]
- 35.Archie EA, Tung J. Social behavior and the microbiome. Curr Opin Behav Sci. 2015;6:28–34. [Google Scholar]
- 36.Sapolsky RM. Social Status and Health in humans and other animals. Annu Rev Anthropol. 2004;33:393–418. [Google Scholar]
- 37.Berkman LF, Syme SL. Social networks, host resistance, and mortality: a nine-year follow-up study of Alameda County residents. Am J Epidemiol. 1979;109:186–204. [DOI] [PubMed] [Google Scholar]
- 38.House JS, Landis KR, Umberson D. Social relationships and health. Science. 1988;241:540–5. [DOI] [PubMed] [Google Scholar]
- 39.Holt-Lunstad J, Smith TB, Layton JB. Social relationships and Mortality Risk: a Meta-analytic review. PLoS Med. 2010;7:e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bzdok D, Dunbar RIM. Social isolation and the brain in the pandemic era. Nat Hum Behav. 2022;6:1333–43. [DOI] [PubMed] [Google Scholar]
- 41.Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML et al. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proceedings of the National Academy of Sciences. 2012;109:13034–9. [DOI] [PMC free article] [PubMed]
- 42.Kinross J, Nicholson JK. Dietary and social modulation of gut microbiota in the elderly. Nat Rev Gastroenterol Hepatol. 2012;9:563–4. [DOI] [PubMed] [Google Scholar]
- 43.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2:e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dill-McFarland KA, Tang Z-Z, Kemis JH, Kerby RL, Chen G, Palloni A, et al. Close social relationships correlate with human gut microbiota composition. Sci Rep. 2019;9:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raulo A, Ruokolainen L, Lane A, Amato K, Knight R, Leigh S, et al. Social behaviour and gut microbiota in red-bellied Lemurs (Eulemur rubriventer): in search of the role of immunity in the evolution of sociality. J Anim Ecol. 2018;87:388–99. [DOI] [PubMed] [Google Scholar]
- 46.Rudolph K, Schneider D, Fichtel C, Daniel R, Heistermann M, Kappeler PM. Drivers of gut microbiome variation within and between groups of a wild Malagasy primate. Microbiome. 2022;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grieneisen LE, Livermore J, Alberts S, Tung J, Archie EA. Group Living and male dispersal predict the core gut microbiome in wild baboons. Integr Comp Biol. 2017;57:770–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodfellow CK, Whitney T, Christie DM, Sicotte P, Wikberg EC, Ting N. Divergence in gut microbial communities mirrors a social group fission event in a black-and-white colobus monkey (Colobus vellerosus). Am J Primatol. 2019;81:e22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perofsky AC, Ancel Meyers L, Abondano LA, Di Fiore A, Lewis RJ. Social groups constrain the spatiotemporal dynamics of wild sifaka gut microbiomes. Mol Ecol. 2021;30:6759–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson KV-A, Watson KK, Dunbar RIM, Burnet PWJ. Sociability in a non-captive macaque population is associated with beneficial gut bacteria. Front Microbiol. 2022;13. [DOI] [PMC free article] [PubMed]
- 51.Bharwani A, Mian MF, Surette MG, Bienenstock J, Forsythe P. Oral treatment with Lactobacillus rhamnosus attenuates behavioural deficits and immune changes in chronic social stress. BMC Med. 2017;15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherwin E, Bordenstein SR, Quinn JL, Dinan TG, Cryan JF. Microbiota and the social brain. Science. 2019;366:eaar2016. [DOI] [PubMed] [Google Scholar]
- 53.Mazzone L, Dooling SW, Volpe E, Uljarević M, Waters JL, Sabatini A, et al. Precision microbial intervention improves social behavior but not autism severity: a pilot double-blind randomized placebo-controlled trial. Cell Host Microbe. 2024;32:106–e1166. [DOI] [PubMed] [Google Scholar]
- 54.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA et al. Captivity humanizes the primate microbiome. Proceedings of the National Academy of Sciences. 2016;113:10376–81. [DOI] [PMC free article] [PubMed]
- 55.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, et al. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol. 2015;69:434–43. [DOI] [PubMed] [Google Scholar]
- 56.Ross FC, Patangia D, Grimaud G, Lavelle A, Dempsey EM, Ross RP et al. The interplay between diet and the gut microbiome: implications for health and disease. Nat Rev Microbiol. 2024;:1–16. [DOI] [PubMed]
- 57.Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776–80. [DOI] [PubMed] [Google Scholar]
- 58.Rinninella E, Cintoni M, Raoul P, Lopetuso LR, Scaldaferri F, Pulcini G, et al. Food Components and Dietary habits: Keys for a healthy gut microbiota composition. Nutrients. 2019;11:2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leeming ER, Johnson AJ, Spector TD, Le Roy CI. Effect of Diet on the gut microbiota: rethinking intervention duration. Nutrients. 2019;11:2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bourdeau-Julien I, Castonguay-Paradis S, Rochefort G, Perron J, Lamarche B, Flamand N, et al. The diet rapidly and differentially affects the gut microbiota and host lipid mediators in a healthy population. Microbiome. 2023;11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, et al. Linking long-term dietary patterns with gut Microbial Enterotypes. Science. 2011;334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnenburg JL, Bäckhed F. Diet-Microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng D, Liwinski T, Elinav E. Interaction between Microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clayton JB, Gomez A, Amato K, Knights D, Travis DA, Blekhman R, et al. The gut microbiome of nonhuman primates: lessons in ecology and evolution. Am J Primatol. 2018;80:e22867. [DOI] [PubMed] [Google Scholar]
- 66.Clayton JB, Al-Ghalith GA, Long HT, Tuan BV, Cabana F, Huang H, et al. Associations between Nutrition, Gut Microbiome, and Health in A Novel Nonhuman Primate Model. Sci Rep. 2018;8:11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartmann C, Dohle S, Siegrist M. Time for change? Food choices in the transition to cohabitation and parenthood. Public Health Nutr. 2014;17:2730–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Craig PL, Truswell AS. Dynamics of food habits of newly married couples: who makes changes in the foods consumed? J Hum Nutr Dietetics. 1994;7:347–61. [Google Scholar]
- 69.Bove CF, Sobal J, Rauschenbach BS. Food choices among newly married couples: convergence, conflict, individualism, and projects. Appetite. 2003;40:25–41. [DOI] [PubMed] [Google Scholar]
- 70.Yue L, Wang C, Meng B, Xie B, Cao H, Su H, et al. The Food Niche Overlap and interspecific relationship between the Sympatric Tibetan Macaque and Grey Snub-Nosed Monkey. Anim (Basel). 2023;13:2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rushworth MFS, Mars RB, Sallet J. Are there specialized circuits for social cognition and are they unique to humans? Curr Opin Neurobiol. 2013;23:436–42. [DOI] [PubMed] [Google Scholar]
- 72.Camus S, Ko WKD, Pioli E, Bezard E. Why bother using non-human primate models of cognitive disorders in translational research? Neurobiol Learn Mem. 2015;124:123–9. [DOI] [PubMed] [Google Scholar]
- 73.Phillips KA, Bales KL, Capitanio JP, Conley A, Czoty PW, ‘t Hart BA, et al. Why primate models matter. Am J Primatol. 2014;76:801–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuthyar S, Manus MB, Amato KR. Leveraging non-human primates for exploring the social transmission of microbes. Curr Opin Microbiol. 2019;50:8–14. [DOI] [PubMed] [Google Scholar]
- 75.Jablonski NG. Social and affective touch in primates and its role in the evolution of social cohesion. Neuroscience. 2021;464:117–25. [DOI] [PubMed] [Google Scholar]
- 76.Cooper MA, Bernstein IS. Social grooming in assamese macaques (Macaca assamensis). Am J Primatol. 2000;50:77–85. [DOI] [PubMed] [Google Scholar]
- 77.Gumert MD, Ho M-HR. The trade balance of grooming and its coordination of reciprocation and tolerance in Indonesian long-tailed macaques (Macaca fascicularis). Primates. 2008;49:176–85. [DOI] [PubMed] [Google Scholar]
- 78.Disarbois E, Duhamel J-R. Virtual social grooming in macaques and its psychophysiological effects. Sci Rep. 2024;14:11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valles-Colomer M, Blanco-Míguez A, Manghi P, Asnicar F, Dubois L, Golzato D, et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature. 2023;614:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brito IL, Gurry T, Zhao S, Huang K, Young SK, Shea TP, et al. Transmission of human-associated microbiota along family and social networks. Nat Microbiol. 2019;4:964–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2:e1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.5050 - Laboratory Fiber-Plus® Monkey Diet Jumbo. 2020. https://www.labdiet.com/product/detail/5050-laboratory-fiber-plus-monkey-diet-jumbo. Accessed 28 Feb 2024.
- 83.5L0N - Monkey Jumble Lab Diet Details. 2020. https://www.labdiet.com/product/detail/5l0n-monkey-jumble. Accessed 23 Oct 2023.
- 84.Thomas V, Clark J, Doré J. Fecal microbiota analysis: an overview of sample collection methods and sequencing strategies. Future Microbiol. 2015;10:1485–504. [DOI] [PubMed] [Google Scholar]
- 85.Tang Q, Jin G, Wang G, Liu T, Liu X, Wang B et al. Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol. 2020;10. [DOI] [PMC free article] [PubMed]
- 86.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, et al. Preservation methods Differ in Fecal Microbiome Stability, affecting suitability for Field studies. mSystems. 2016;1:e00021–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.16S Metagenomic Sequencing Library Preparation. 2013. https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html. Accessed 9 Sep 2024.
- 89.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. [Google Scholar]
- 90.R Core Team. R: a Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2023. [Google Scholar]
- 91.Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, Gulati AS, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Callahan B. RDP taxonomic training data formatted for DADA2 (RDP trainset 18/release 11.5). 2020.
- 93.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:633–42. Database issue:D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3. [DOI] [PMC free article] [PubMed]
- 96.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara B et al. Vegan: Community Ecology Package. R Package Version 22 – 1. 2022;2:1–2.
- 97.Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016.
- 98.McMurdie PJ, Holmes S. Phyloseq: an R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schloss PD. Rarefaction is currently the best approach to control for uneven sequencing effort in amplicon sequence analyses. mSphere. 2024;0:e00354–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kennedy K, Hall MW, Lynch MDJ, Moreno-Hagelsieb G, Neufeld JD. Evaluating Bias of Illumina-based bacterial 16S rRNA gene profiles. Appl Environ Microbiol. 2014;80:5717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing Microbial communities. Appl Environ Microbiol. 2005;71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bray JR, Curtis JT. An ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr. 1957;27:325–49. [Google Scholar]
- 105.Zhu C, Yu J. Nonmetric Multidimensional Scaling corrects for Population structure in Association Mapping with different sample types. Genetics. 2009;182:875–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cao Y, Dong Q, Wang D, Zhang P, Liu Y, Niu C. microbiomeMarker: an R/Bioconductor package for microbiome marker identification and visualization. Bioinformatics. 2022;38:4027–9. [DOI] [PubMed] [Google Scholar]
- 108.Lin H, Peddada SD. Multigroup analysis of compositions of microbiomes with covariate adjustments and repeated measures. Nat Methods. 2024;21:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo G, Jia K-R, Shi Y, Liu X-F, Liu K-Y, Qi W, et al. Psychological stress enhances the colonization of the stomach by Helicobacter pylori in the BALB/c mouse. Stress. 2009;12:478–85. [DOI] [PubMed] [Google Scholar]
- 110.Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The Microbiota-Gut-Brain Axis. Physiol Rev. 2019;99:1877–2013. [DOI] [PubMed] [Google Scholar]
- 111.DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis. 2016;22:1137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wallen ZD, Appah M, Dean MN, Sesler CL, Factor SA, Molho E, et al. Characterizing dysbiosis of gut microbiome in PD: evidence for overabundance of opportunistic pathogens. NPJ Parkinsons Dis. 2020;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dogra SK, Doré J, Damak S. Gut microbiota resilience: definition, link to health and strategies for intervention. Front Microbiol. 2020;11. [DOI] [PMC free article] [PubMed]
- 114.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–85. [DOI] [PubMed] [Google Scholar]
- 115.Kamdar K, Khakpour S, Chen J, Leone V, Brulc J, Mangatu T, et al. Genetic and metabolic signals during Acute Enteric bacterial infection alter the Microbiota and Drive Progression to Chronic Inflammatory Disease. Cell Host Microbe. 2016;19:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, et al. Gut-resident Lactobacillus Abundance Associates with IDO1 inhibition and Th17 dynamics in SIV-Infected macaques. Cell Rep. 2015;13:1589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–40. [DOI] [PubMed] [Google Scholar]
- 118.Muñiz Pedrogo DA, Chen J, Hillmann B, Jeraldo P, Al-Ghalith G, Taneja V, et al. An increased abundance of Clostridiaceae characterizes arthritis in inflammatory bowel disease and rheumatoid arthritis: a cross-sectional study. Inflamm Bowel Dis. 2019;25:902–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu Q, Zhang Y, Zhang Y, Xia C, Lai Q, Dong Z, et al. Potential effects of antibiotic-induced gut microbiome alteration on blood–brain barrier permeability compromise in rhesus monkeys. Ann N Y Acad Sci. 2020;1470:14–24. [DOI] [PubMed] [Google Scholar]
- 120.Agranyoni O, Meninger-Mordechay S, Uzan A, Ziv O, Salmon-Divon M, Rodin D, et al. Gut microbiota determines the social behavior of mice and induces metabolic and inflammatory changes in their adipose tissue. Npj Biofilms Microbiomes. 2021;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.MacRae M, Kenkel WM, Kentner AC. Social rejection following neonatal inflammation is mediated by olfactory scent cues. Brain Behav Immun. 2015;49:43–8. [DOI] [PubMed] [Google Scholar]
- 122.Pinacho-Guendulain B, Montiel-Castro AJ, Ramos-Fernández G, Pacheco-López G. Social complexity as a driving force of gut microbiota exchange among conspecific hosts in non-human primates. Front Integr Nuerosci. 2022. 10.3389/fnint.2022.876849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sallet J, Mars RB, Noonan MP, Andersson JL, O’Reilly JX, Jbabdi S, et al. Social Network size affects neural circuits in macaques. Science. 2011;334:697–700. [DOI] [PubMed] [Google Scholar]
- 124.Simpson EA, Maylott SE, Lazo RJ, Leonard KA, Kaburu SSK, Suomi SJ, et al. Social touch alters newborn monkey behavior. Infant Behav Dev. 2019;57:101368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cascio CJ, Moore D, McGlone F. Social touch and human development. Dev Cogn Neurosci. 2018;35:5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bienenstock J, Kunze WA, Forsythe P. Disruptive physiology: olfaction and the microbiome-gut-brain axis. Biol Rev Camb Philos Soc. 2018;93:390–403. [DOI] [PubMed] [Google Scholar]
- 127.Venu I, Durisko Z, Xu J, Dukas R. Social attraction mediated by fruit flies’ microbiome. J Exp Biol. 2014;217:1346–52. [DOI] [PubMed] [Google Scholar]
- 128.Lizé A, McKay R, Lewis Z. Kin recognition in Drosophila: the importance of ecology and gut microbiota. ISME J. 2014;8:469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lewis Z, Heys C, Prescott M, Lizé A. You are what you eat. Gut Microbes. 2014;5:541–3. [DOI] [PubMed] [Google Scholar]
- 130.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proceedings of the National Academy of Sciences. 2010;107:20051–6. [DOI] [PMC free article] [PubMed]
- 131.Ghazanfar AA, Hauser MD. The auditory behaviour of primates: a neuroethological perspective. Curr Opin Neurobiol. 2001;11:712–20. [DOI] [PubMed] [Google Scholar]
- 132.Lewis RJ. Subordination signals improve the quality of social relationships in Verreaux’s Sifaka: implications for the evolution of power structures and social complexity. Am J Phys Anthropol. 2019;169:599–607. [DOI] [PubMed] [Google Scholar]
- 133.Dagnino-Subiabre A. Effects of chronic stress on the auditory system and fear learning: an evolutionary approach. Rev Neurosci. 2013;24:227–37. [DOI] [PubMed] [Google Scholar]
- 134.McGlone F, Wessberg J, Olausson H. Discriminative and affective touch: sensing and feeling. Neuron. 2014;82:737–55. [DOI] [PubMed] [Google Scholar]
- 135.Wang Y, Yang X, Zhang M, Pan H. Comparative analysis of Gut Microbiota between Wild and Captive Golden Snub-Nosed monkeys. Animals. 2023;13:1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schlomann BH, Parthasarathy R. Timescales of gut microbiome dynamics. Curr Opin Microbiol. 2019;50:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gerber GK. The dynamic microbiome. FEBS Lett. 2014;588:4131–9. [DOI] [PubMed] [Google Scholar]
- 138.Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146:1564–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data supporting the findings of this study were deposited in NCBI’s repository (SRA) under accession number PRJNA1073535 and are available at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/1073535.