Abstract
Viral infection leads to heterogeneous cellular outcomes ranging from refractory to abortive and fully productive states. Single cell transcriptomics enables a high resolution view of these distinct post-infection states. Here, we have interrogated the host-pathogen dynamics following reactivation of Epstein-Barr virus (EBV). While benign in most people, EBV is responsible for infectious mononucleosis, up to 2% of human cancers, and is a trigger for the development of multiple sclerosis. Following latency establishment in B cells, EBV reactivates and is shed in saliva to enable infection of new hosts. Beyond its importance for transmission, the lytic cycle is also implicated in EBV-associated oncogenesis. Conversely, induction of lytic reactivation in latent EBV-positive tumors presents a novel therapeutic opportunity. Therefore, defining the dynamics and heterogeneity of EBV lytic reactivation is a high priority to better understand pathogenesis and therapeutic potential. In this study, we applied single-cell techniques to analyze diverse fate trajectories during lytic reactivation in three B cell models. Consistent with prior work, we find that cell cycle and MYC expression correlate with cells refractory to lytic reactivation. We further found that lytic induction yields a continuum from abortive to complete reactivation. Abortive lytic cells upregulate NFκB and IRF3 pathway target genes, while cells that proceed through the full lytic cycle exhibit unexpected expression of genes associated with cellular reprogramming. Distinct subpopulations of lytic cells further displayed variable profiles for transcripts known to escape virus-mediated host shutoff. These data reveal previously unknown and promiscuous outcomes of lytic reactivation with broad implications for viral replication and EBV-associated oncogenesis.
Author summary
Viral infections profoundly alter host cell biological programming in ways that potentiate disease. Epstein-Barr virus (EBV) is a particularly prevalent human pathogen associated with diverse cancers and several autoimmune disorders. EBV predominantly establishes latent infection in B cells and can promote B cell malignancies through functions of well-characterized latent oncoproteins. Aspects of the viral lytic cycle also clearly contribute to EBV-associated diseases, although pathologic roles of lytic reactivation are incompletely understood. Here, we use single-cell techniques to examine cellular responses to EBV lytic reactivation in multiple B cell models. Consistent with prior studies, reactivation from latency is incomplete (abortive) in some cells and successful in others. Abortive and full lytic trajectories exhibit distinct biological responses that each may promote pathogenesis and reinforce bimodal latent-lytic control. Intriguingly, a portion of cells that proceed through the lytic cycle exhibits unexpected and striking expression of genes associated with cellular reprogramming, pluripotency, and self-renewal. Collectively, this study provides a valuable resource to understand diverse host-virus dynamics and fates during viral reactivation and identifies multiple modes of EBV lytic pathogenesis to investigate in future research.
Introduction
Viral infections lead to heterogeneous cell fate outcomes including resistance, abortive infection, latency, or full virion amplification often leading to cell death. Cells that resist viral infection often display elevated pre-existing anti-viral responses [1–4]. Likewise, cell responses that enable survival following virus replication can prime for further anti-viral responses [5,6]. Herpesviruses are large double-stranded DNA viruses that provide a unique and complex infection paradigm to model the heterogeneity of viral infection as they reactivate from a latent state in response to diverse stimuli.
Epstein-Barr virus (EBV) was the first oncogenic human virus to be discovered [7]. Since its isolation from endemic Burkitt Lymphoma (BL) cells in 1964, EBV infection has been linked to an expansive set of human cancers and, more recently, autoimmune diseases [8–11]. EBV infection in immunosuppressed individuals can lead to post-transplant lymphoproliferative disease (PTLD) [12] and HIV-related diffuse large B cell lymphomas (DLBCL) [13] as well as up to 40% of Hodgkin Lymphoma (HL) [14]. and rare individuals with chronic active EBV (CAEBV) can develop T and NK cell lymphomas [15,16]. Beyond these hematologic malignancies, EBV infection is associated with epithelial cancers such as nasopharyngeal carcinoma (NPC) [17] and gastric carcinomas [18]. Collectively, EBV causes is or otherwise associated with nearly 2% of all cancers diagnosed annually [8].
This prevalence in malignant disease vastly underrepresents the success of EBV as a human pathogen. Globally, it is estimated that over 95% of adults are infected with EBV [19]. EBV is transmitted via saliva, which enables the virus to traverse oral epithelial tissues and infect B lymphocytes within the tonsils [20]. EBV infects B cells via the surface receptor CD21 (CR2) [21,22] and rapidly induces B cell adaptive immune programs to mimic germinal center (GC)-like dynamics [23–26]. Successful evasion of antiviral defenses, immune tolerance checkpoints, and growth-induced damage [27–29] allows memory B cells latently infected with EBV to exit from this virus-manipulated GC reaction. Viral latency establishment within the memory B cell compartment yields lifelong persistence [30, 31]. Lytic reactivation from this latent state triggers the production of new virions and is essential to the replicative cycle and transmission between hosts. The lytic gene program is transcriptionally orchestrated by two immediate early (IE) lytic genes: BZLF1 (encodes for the transcription factor Zta / Z / ZEBRA) and BRLF1 (encodes for the transcription factor Rta / R) [32–34]. While Zta and Rta both play essential roles in lytic reactivation, Zta is the master lytic transactivator in B cells. BZLF1 expression is induced upon cell differentiation and stress [35,36], a prototypical example being post-GC B cell differentiation into plasmablasts [37]. Host cell transcriptional regulators of plasma cell generation including XBP1 and BLIMP1 (PRDM1) induce EBV lytic reactivation via direct transactivation of the BZLF1 promoter [38–40]. Zta then transactivates subsequent expression of early and late lytic genes by binding at Z-responsive elements (ZREs) throughout the viral genome [41]. As an AP-1 family homolog [33], Zta also binds loci throughout the host genome [42] and has characteristics of a ‘pioneer’ transcription factor. Consistent with this, BZLF1 expression and the early stages of EBV reactivation cause considerable alterations to the host cell epigenome and resulting gene expression [43,44].
Prior work suggests that lytic gene expression is functionally important for tumorigenesis. Notably, viral strains that carry the NFATc1-responsive Z promoter variant Zp-V3 exhibit increased lytic replication and are enriched in EBV-associated cancers relative to strains with prototypical Zp [45]. In SCID and NSG mouse models with reconstituted human immune systems, significantly fewer animals developed EBV+ lymphomas after infection with BZLF1 knockout virus versus a wild-type (WT) control strain [46]. Further, infection with a Zta-overexpressing strain that failed to complete reactivation (i.e., abortive lytic) promoted tumor growth in mice similar to WT EBV [47]. Recent experiments in immunocompromised mice confirmed the tumorigenic role of abortive lytic infection by using EBV lacking the BALF5 gene, which encodes a viral DNA polymerase subunit essential for lytic replication [48]. These studies demonstrated that expression of BZLF1 (and possibly other early lytic genes) contributes to tumorigenesis in vivo regardless of the potential for horizontal infection of bystander cells by new virions. While detailed insights regarding the oncogenic effects of successful or abortive lytic replication are limited, tumor microenvironment inflammatory conditioning by cytokines secreted from reactivating cells has been proposed [49–53].
Another complication in the relation between viral reactivation and oncogenicity stems from observations that a significant proportion of EBV-infected tumor cells are resistant or otherwise refractory to lytic reactivation. In BL-derived P3HR1 and Akata cells, high expression of the oncoprotein c-Myc promotes viral latency maintenance and suppresses lytic reactivation via direct interaction with the origin of lytic replication (oriLyt) and inhibition of chromatin looping to activate BZLF1 expression [54]. Accordingly, MYC suppression facilitates BZLF1 expression and the subsequent induction of viral lytic genes. It is noteworthy that constitutive oncogene expression favors viral genome propagation through proliferation of latently infected host cells whereas lytic replication becomes a more advantageous strategy in its absence. Similarly, BL-derived cells refractory to lytic reactivation have also been found to express high levels of STAT3 [55–57], which functions as an oncogene in B cells and inhibits apoptosis via induction of BCL2 expression. Beyond simply being expressed by refractory cells, STAT3 antagonizes lytic reactivation of EBV+ cells through the functions of its transcriptional targets [56]. In fact, LCLs derived from patients with autosomal dominant hyper-IgE syndrome (AD-HIES), a disease that leads to non-functional STAT3 activity, went lytic at a higher rate than LCLs derived from healthy donors [58]. Given the therapeutic potential of drug-induced lytic reactivation followed by viral DNA synthesis inhibition to treat EBV-latent cancers, investigators are actively exploring means to make refractory cells more sensitive to lytic induction [59–61]. However, such efforts should be weighed against the known associations between the EBV lytic cycle and oncogenesis, which remain to be fully elucidated.
Many EBV gene products contribute to virus-driven malignancies by mediating functions associated with cancer hallmarks including uncontrolled proliferation, tumor suppressor inhibition, epigenetic reprogramming, genome instability, apoptotic resistance, and immune evasion [62]. EBV+ cells with cancer stem cell (CSC) features have also been reported in NPC and gastric carcinoma [63, 64], suggesting the potential for cellular self-renewal associated with infection. In the CSC model, a small subset of tumor cells retain the capacity for self-renewal and proliferation through activation of signaling pathways (e.g., Wnt, Notch), transactivators of the epithelial-to-mesenchymal (EMT) transition, and critical regulators of pluripotency (e.g., SOX2, OCT4). CSCs may serve as progenitors for other tumor cells, especially in lymphoid malignancies that are derived from cells of origin that intrinsically retain self-renewal properties to support immunologic memory [65–67]. Aberrant expression of self-renewal genes and other CSC biomarkers [68] may originate from significant (epi)genomic reprogramming and result in cellular phenotypic plasticity. Lytic replication of EBV (and DNA viruses from several other families [69]) clearly constitutes a major reprogramming event for the host cell. Nuclear chromatin is globally disrupted by IE gene expression, the formation of viral replication compartments, and the accumulation of viral DNA [43,70]. Moreover, preferential binding of BZLF1 to methylated promoters can reverse epigenetic silencing of both EBV and cellular genes through nucleosome eviction, resulting in heterochromatin-to-euchromatin conversion [44,71–73]. While evidence for stem-like reprogramming and CSC gene expression during the EBV lytic cycle has not been reported to our knowledge, it is noteworthy that reactivation of HSV-1 (another herpesvirus) induces embryonic development programs including Wnt/β-catenin activity that licenses late viral gene expression [74].
These previous studies demonstrate that EBV reactivation from latency is a complex process that culminates in heterogeneous host cell responses germane to the progression of virus-associated cancers. Single-cell sequencing techniques are particularly well suited to dissect the inherent complexity of host-virus interactions and their effects on cell fate [74–77]. In recent studies of early EBV infection [25,26] and established latency [78,79], we have used single-cell sequencing to successfully resolve and study diverse phenotypes arising from complex host-pathogen dynamics. We reasoned that a similar high-resolution experimental and informatic approach would clarify distinct courses of lytic reactivation, provide essential data for future studies of viral pathogenesis, and inform potential therapeutic strategies to address EBV-driven oncogenesis. To this end, we performed time-resolved single-cell RNA sequencing (scRNA-seq), flow cytometry, and RNA Flow-FISH (fluorescence in situ hybridization) in P3HR1-ZHT cells to define initial cell state diversity, differential fate trajectories, and previously unknown lytic response phenotypes. We further extended and validated these studies with scRNA-seq in the B958-ZHT LCL and the Akata BL cell line thereby defining common mechanisms across diverse viral strains and reactivation stimuli.
Results
Heterogeneous responses to EBV lytic reactivation in individual cells
P3HR1-ZHT cells are an inducible model of EBV lytic reactivation (Fig 1A). This model system constitutively expresses the EBV immediate early lytic transactivator Zta (encoded by the BZLF1 gene) fused with a modified murine estrogen receptor hormone binding domain. While the encoded fusion protein is normally rapidly degraded, addition of 4-hydroxytamoxifen (4HT) stabilizes it and promotes its nuclear translocation, whereupon the Zta domain binds and transactivates Zta-responsive elements (ZREs) in both host and viral genomes. Because Zta has positive regulatory control of its own promoter via ZRE binding [80], 4HT treatment also leads to expression of endogenous BZLF1, thus initiating viral lytic reactivation. Although all cells in the P3HR1-ZHT line express the inducible construct, it has been observed that complete EBV lytic reactivation occurs only in a subset of 4HT treated cells [81,82].
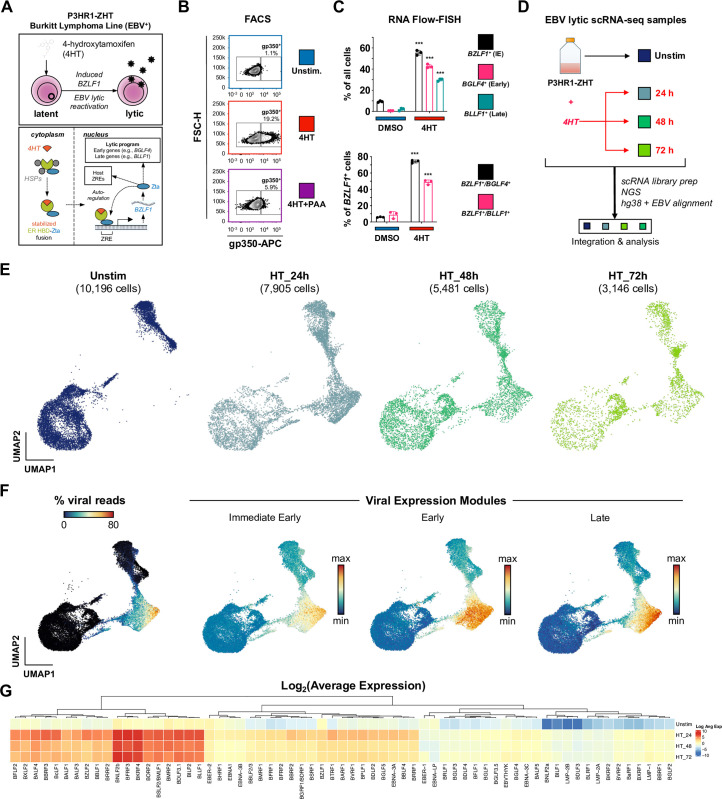
Fig 1. EBV lytic reactivation in the P3HR1-ZHT Burkitt Lymphoma line at single-cell resolution.
(A) Schematic of 4HT-inducible BZLF1 (Zta) expression initiating lytic reactivation in the Burkitt Lymphoma-derived P3HR1-ZHT cell line. (B) Flow cytometry validation 24 h after 4HT-induced lytic reactivation and inhibition of complete reactivation by phosphonoacetic acid (PAA) in P3HR1-ZHT. Cellular expression of the viral glycoprotein gp350 (encoded by the late lytic gene BLLF1) serves as a proxy for successful reactivation. Co-treatment with the viral DNA polymerase inhibitor PAA prevents complete reactivation by blocking viral DNA replication, which is required for expression of late viral genes / gene products including gp350. (C) RNA Flow-FISH validation of select immediate early (IE), early, and late lytic gene expression in P3HR1-ZHT. The majority of cells express detectable BZLF1 24 h after 4HT treatment. Substantial fractions express early genes including the EBV DNA polymerase (BGLF4) and late genes including BLLF1. However, not all BZLF1+ cells exhibit early and late gene expression, indicating variable progression of reactivation in individual cells. Asterisks denote significantly higher expression in 4HT-treated samples versus DMSO controls (n = 3 per condition; two-tailed Welch’s t-test; ***p<0.001). (D) Experimental design schematic for time-resolved scRNA-seq study of EBV reactivation in P3HR1-ZHT. Single-cell libraries were prepared from unstimulated cells and from cells at three timepoints (24 h, 48 h, and 72 h) after 4HT treatment. Libraries were sequenced, mapped to a multispecies reference genome, integrated into a single data object, and analyzed. (E) UMAP representation of single cells captured across the experimental timecourse. Plots display the number of cells in each library after QC filtering. (F) EBV gene expression overview in merged timecourse scRNA-seq data. (From left to right) Viral fraction of captured transcripts per cell; scores for an immediate early (IE) expression module (BZLF1, BRLF1); scores for an early gene expression module (BRRF1, BBLF4, BALF1, LF3, BARF1, BaRF1, BVLF1, and BALF3); scores for a late gene expression module (BZLF2, BLLF1, BILF2, BBRF3, BcLF1, BRRF2, BSRF1, BCRF1, and BBRF1). Modules were curated based on viral expression kinetics determined by CAGE-seq [162]. (G) Hierarchically clustered average expression of all detected viral genes by timepoint.
We confirmed inducible yet non-uniform viral reactivation of P3HR1-ZHT cells in response to 4HT treatment using FACS staining for the viral glycoprotein gp350, which was expressed in cells that reached the late stage of lytic reactivation. Unstimulated P3HR1-ZHT cells expressed minimal gp350 (1.1%), but treatment with 100 nM 4HT for 24 hours resulted in gp350 expression in 19.2% of cells. When we simultaneously treated cells with 4HT and PAA, an inhibitor of viral DNA replication, we observed a significant reduction in gp350 expression by 24 hours (Figs 1B and S1). These results indicated that cells exhibited heterogenous responses to viral lytic reactivation and that completion of the full lytic cycle was dependent upon successful viral DNA replication, which has been previously described in herpesviruses [83–87]. We expanded upon these gp350 FACS results using RNA Flow-FISH assays to detect viral RNAs from genes expressed at different stages of the lytic cycle: the immediate early lytic gene BZLF1, the early lytic gene BGLF4, and the late lytic gene BLLF1. After 24 hours of 4HT treatment, we observed a significant increase in expression of all three lytic transcripts compared to mock treated cells. However, there was a stepwise decrease in expression level between early and late lytic genes (Figs 1C and S2). These results confirmed that a significant proportion of Z-HT induced P3HR1 cells were refractory to full lytic reactivation.
Since we observed heterogeneous responses upon lytic reactivation, we applied time-resolved single-cell RNA sequencing (scRNA-seq) to study the concurrent cellular responses in the P3HR1-ZHT system after 24, 48, and 72 hours of 4HT treatment compared to untreated cells (Fig 1D). UMAP projection of samples by timepoint demonstrated that substantial transcriptomic changes occurred after 4HT stimulation (Fig 1E). Cells expressing high levels of viral reads clustered together, however there was a distinction between cells expressing immediate early, early, and late viral transcripts (Fig 1F). Analysis of all EBV transcripts identified genes with high, moderate, and low expression; however, all 4HT-treated samples expressed more viral transcripts compared to untreated cells (Fig 1G). These results confirmed heterogeneous responses to lytic reactivation observed by flow cytometry and enabled subsequent genome-wide analyses.
Identification of distinct EBV reactivation response clusters
Cells from integrated timecourse scRNA-seq libraries were hierarchically clustered by host and viral transcriptome similarity, which led to the identification of five main clusters (Fig 2A). Unstimulated cells were mostly present in clusters A and B, while clusters C, D, and E primarily comprised 4HT-treated cells across the experimental time course (Fig 2B) and displayed elevated viral gene expression compared to clusters A and B (S3 Fig). Further examination of these clusters revealed differences in the number of total and unique RNAs, the percentage of viral RNAs, and the percentage of mitochondrial RNAs (Fig 2C). These differences in unique and total RNA features suggested major phenotypic differences both in unstimulated and reactivated cells. Therefore, we scored the clusters based on cell cycle state and found that there was a decrease in G2/M specific gene expression and an increase in G1 gene expression after 24 hours of 4HT treatment, consistent with EBV lytic reactivation occurring in a pseudo-S phase [88, 89] (S4A Fig). We confirmed this finding using BrdU/7-AAD staining of untreated versus 4HT-treated cells (S4B Fig). Consistent with induced cell cycle arrest, lytic reactivation upon 4HT treatment led to a reduction of S phase cells (43.2% vs. 54.3%) and modest increase in G0/G1 cells. Because pulsed BrdU staining does not discriminate cellular and viral DNA synthesis, a portion of S phase 4HT-treated cells were likely undergoing viral but not cellular DNA synthesis. This was further evidenced by a significant fraction of gp350+ cells within the gated S phase population (S4B Fig). We also assayed MitoTracker signal stratified by gp350 expression and found that gp350+ cells had lower mitochondrial content (S4C Fig).
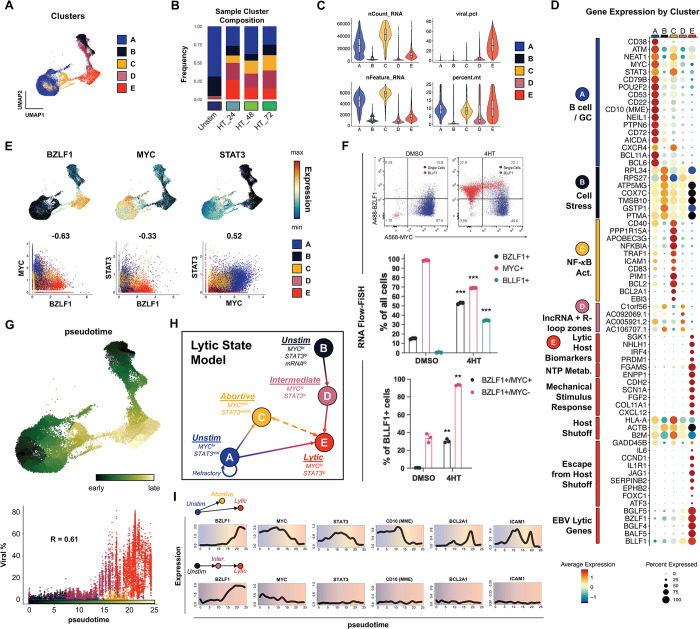
Fig 2. P3HR1-ZHT phenotypic heterogeneity and response trajectories during lytic induction.
(A) P3HR1-ZHT cell clusters identified in merged timecourse data via unsupervised methods. (B) Cluster composition of cells from individual timepoints. Cluster colors are coded as in 2A. (C) QC feature distributions by cluster. The total number of mapped reads per cell is given by nCount_RNA. The number of unique RNA features (i.e., genes, lncRNAs) per cell is given by nFeature_RNA. The viral fraction of mapped reads per cell (viral.pct) and mitochondrial transcript fractions (percent.mt) were calculated using the PercentageFeatureSet() function in Seurat [154]. (D) Differential gene expression by cluster. Genes are annotated by their known biological roles and functions derived from gene ontology (GO) analysis and primary literature. Dot size represents the percentage of cells in each cluster that express a given gene and color encodes average expression across the cluster. (E) UMAP expression profiles (top row) and pairwise correlation plots (bottom row, Pearson R) for BZLF1, MYC, and STAT3. Correlation plots depict individual cells colored by cluster. (F) RNA Flow-FISH validation of reduced MYC expression in BZLF1+BLLF1+ cells (top panel). Asterisks in the middle panel bar plot denote significantly reduced frequency of MYC+ P3HR1-ZHT cells and increased frequencies of BZLF1+ and BLLF1+ cells after 4HT treatment (n = 3 per condition; two-tailed Welch’s t-test; ***p<0.001). Asterisks in the bottom panel bar plot denote significantly increased frequencies of BZLF1+MYC+ and BZLF1+MYC- cells after 4HT treatment (n = 3 per condition; two-tailed Welch’s t-test; **p<0.01). (G) UMAP of graph-based pseudotime trajectory calculation for timecourse-merged scRNA-seq data. Trajectory root cells were selected from both clusters A and B, which were present in the unstimulated (day 0) P3HR1-ZHT library (top panel). Viral read content in individual cells ordered by pseudotime and coded by cluster (bottom panel). (H) Cluster- and pseudotime-informed annotated cell state model of EBV lytic reactivation in P3HR1-ZHT. Solid line arrows denote cell response trajectories supported by time-resolved scRNA-seq data. The dashed line denotes a putative state interconversion. (I) Gene expression dynamics along distinct pseudotime trajectories in the lytic reactivation timecourse. Highlighted genes were selected from those differentially expressed across unstimulated, abortive, and fully lytic cells.
Cells traverse heterogeneous biological response trajectories during lytic reactivation
Next, we analyzed differentially expressed genes by cluster and grouped them by ontology using a combined approach with software-based annotation tools [90] and primary literature searches (Fig 2D). Unstimulated cells were almost exclusively present in clusters A and B, which were distinguished from each other by total transcripts and unique features per cell (Fig 2B and 2C). Unstimulated cells with high RNA and feature counts (cluster A) exhibited a germinal center (GC) B cell profile including MME (CD10) [91], BCL6 [92,93], BCL11A [94], POU2F2 (OCT2) [95], and AICDA (AID) [96,97]. Along with high MYC expression, this phenotype is consistent with the profile of endemic BL from which P3HR1-ZHT is derived. In contrast, unstimulated cells with low RNA and features counts (cluster B) exhibited a cell stress expression signature that included slight enrichment of genes for ribosomal subunits (RPL34, RPS27), nuclear-encoded components of mitochondrial respiratory complexes (COX7C), and the apoptotic resistance genes PTMA [98] and GSTP1, the latter of which also mediates oxidative stress [99]. Cluster C, which was comprised of 4HT-treated samples, displayed antiviral restriction (APOBEC3G, PPP1R15A, TRIM14, FURIN), inflammatory (CCL4L2, CCL3L1, NKG7), and NF-κB signaling (NFKBIA, ICAM1, CD83, BCL2, BCL2A1) signatures. Cluster D had a similar gene expression pattern to cluster B with the addition of lytic transcripts and several long noncoding RNAs from R-loop “hot spots” (C1orf56, AC092069.1, AC005921.2, AC106707.1) associated with genomic instability related to unscheduled gene expression or DNA synthesis (in contexts including herpesviral reactivation) [100–104]. Finally, cluster E primarily contained cells that had entered the lytic cycle after 4HT treatment. Lytic cells expressed known host biomarkers of reactivation (SGK1, NHLH1, PRDM1) [105], downregulation of genes targeted by virus-induced host shutoff (HLA-A, ACTB, B2M) [106] mediated by EBV BGLF5 [107], expression of genes that escape host shutoff (e.g., GADD45B, IL6, CCND1, JAG1, SERPINB2, FOXC1, ATF3) [108–110], and numerous IE, early, and late lytic genes. Furthermore, an extended list of EBV genes expressed across all clusters highlights that most lytic transcripts are highly expressed in cluster E, while there is some limited expression of select genes in clusters C and D (S6 Fig).
We next focused on individual genes that are differentially expressed between the clusters. We specifically chose STAT3 and MYC because they have been established as key regulators of EBV lytic reactivation [54,56,57,59] (Fig 2E). In line with these published results, MYC expression was strongly anti-correlated with BZLF1 induction (Fig 2E, bottom left panel). STAT3 expression, which has been previously shown to be upregulated in cells refractory to lytic reactivation [59], was likewise anti-correlated with expression of BZLF1 (Fig 2E, bottom middle panel). STAT3 and MYC expression were positively correlated and highest in unstimulated (cluster A) and abortive (cluster C) cells (Fig 2E, bottom right panel). Prediction of transcription factor activities based on gene regulatory network (GRN) enrichment likewise identified enhanced STAT3 (and NF-κB) target expression in cluster C (S5 Fig). RNA Flow-FISH detection of BZLF1 and MYC validated scRNA-seq data and provided additional insight with respect to partial versus complete reactivation indicated by expression of the late lytic gene BLLF1 (Fig 2F). Specifically, 4HT treatment induced significant increases in BZLF1+and BLLF1+ cells and a concomitant decrease in MYC+ cells relative to DMSO-treated controls (Fig 2F, top and middle panels). Moreover, the majority of BLLF1+ cells were BZLF1+/MYC- (Fig 2F, bottom panel).
Given the observed heterogeneity of phenotypic states before and after lytic induction, we aimed to better understand the distinct response trajectories of EBV-infected cells using pseudotemporal ordering (Fig 2G). Pseudotime analyses [111] are preferable over purely chronologic sampling for studying biological state transitions due to initial state variability and asynchronous responses to infection among individual cells [26]. Root cells (pseudotime = 0) for the reactivation trajectory graph were chosen within clusters A and B since both of these phenotypes were represented by unstimulated cells (Fig 2G, top panel). As shown by per cell viral fractions of captured mRNA transcripts, reactivation generally progresses in pseudotime, with limited viral expression in abortive cells at intermediate coordinates and high viral expression in fully lytic cells in late pseudotime (Fig 2G, bottom panel). Notably, trajectories from both clusters A and B pass through incomplete reactivation states (C and D, respectively) before convening within the lytic phenotype (cluster E) at late pseudotime (Fig 2G).
Collectively, cluster-resolved expression, MYC and STAT3 profiles, and pseudotime trajectory analysis enabled us to construct a state model for lytic reactivation in the P3HR1-ZHT system (Fig 2H). Unstimulated cells express elevated MYC and STAT3 and may undergo abortive reactivation in response to 4HT in which BZLF1 expression is minimal while MYC and STAT3 levels are largely maintained. Alternatively, cells may proceed to lytic reactivation, during which both MYC and STAT3 expression are severely diminished. Although clusters C and E were connected by a bridge of cells in the UMAP embedding, we cannot make definitive conclusions from these data alone regarding possible interconversion between abortive and lytic states. While global mRNA levels decrease along the transition from A to E consistent with host shutoff, the trajectory from cluster B (unstimulated) through D (intermediate) toward E (lytic) was characterized by relative increases in total and unique host and viral mRNA content. However, reduced MYC expression was also observed along the B to E trajectory. Overall, these results indicated that heterogeneity in unstimulated cells and differential responses to BZLF1 induction each contributed to the generation of distinct cell states during lytic reactivation. Analysis of gene expression along state-specific pseudotime trajectories captured these distinct biological response coordinates (Fig 2I). For example, trajectories starting from clusters A and B both exhibited upregulated BZLF1 and net MYC reduction. However, STAT3 expression was consistently low across B, D, and E while STAT3 increased from A to C and decreased from A to E. Likewise, dynamic expression of GC B cell and NF-κB signature genes along A➔(C)➔E were not observed along B➔D➔E.
Abortive lytic cells are characterized by high NF-κB pathway gene expression
Abortive lytic replication, or the initiation of the lytic cycle without expression of late lytic genes / proteins, has been identified in various systems [47,112]. We sought to characterize this replication sub-state further through analysis of the abortive lytic cells in the cluster C phenotype. Using markers identified in Fig 2D we were able to clearly distinguish unstimulated, abortive lytic, and lytic cells using CD38, BCL2A1, and BLLF1 expression, respectively (Fig 3A). STAT3+ cells in the BZLF1+ abortive lytic state (cluster C) notably co-expressed BCL2A1 and other NF-κB pathway target genes (Fig 3B). RNA Flow-FISH for CD38, BCL2A1, and BLLF1 in cells treated with DMSO (control), 4HT (lytic), and 4HT + PAA (an abortive lytic model due to inhibited viral DNA synthesis) confirmed these distinct response states (Figs 3C and S7). This experiment confirmed that CD38 RNA was primarily expressed in unstimulated cells and decreased upon 4HT treatment. BLLF1 (gp350) RNA was almost exclusively expressed in 4HT treated cells, and its expression was blocked upon PAA treatment as expected. BCL2A1 RNA was significantly elevated in 4HT + PAA-treated cells, especially by 48 hours post-treatment (Fig 3D). Thus, these markers reliably delineated latent, abortive, and lytic phenotypes identified from scRNA-seq as clusters A, C, and E.
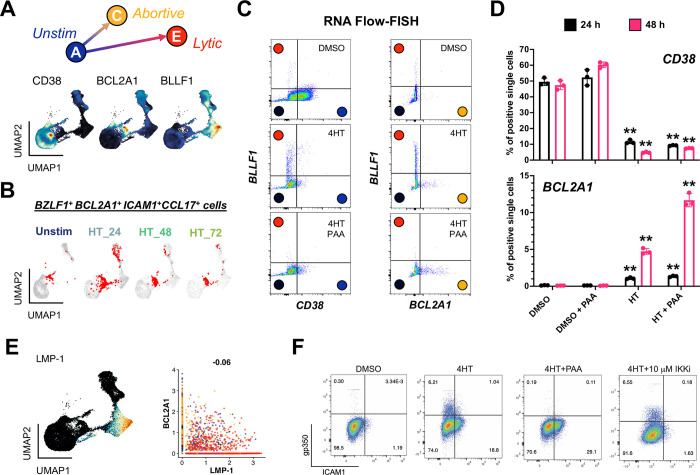
Fig 3. Validation of an abortive response with elevated NF-κB activity distinct from full lytic reactivation.
(A) Identification of CD38, BCL2A1, and BLLF1 as respective biomarkers for unstimulated, abortive, and lytic P3HR1-ZHT cells. (B) Co-detection of BZLF1 and NF-κB pathway transcriptional targets in abortive cells (co-positive cells in red) by timepoint. (C) RNA Flow-FISH validation of full (BLLF1+) and abortive (BCL2A1+) reactivation as orthogonal responses at 48 h post 4HT treatment. DMSO control-treated cells are predominantly CD38+ and exhibit minimal spontaneously lytic (full or abortive) cells (top panel). 4HT treatment induces distinct full lytic and abortive subsets (middle panels). Inhibition of viral DNA synthesis with PAA blocks full lytic reactivation and increases the frequency of BCL2A1+ abortive cells (bottom panels). Colored circles denote predicted corresponding model states defined from scRNA-seq. (D) Frequencies of CD38+ and BCL2A1+ cells presented in 3C by treatment condition at 24 h and 48 h. Asterisks denote significantly decreased frequencies of CD38+ cells and increased frequencies of BCL2A1+ cells upon 4HT and 4HT+PAA treatment versus respective control treatments (n = 3 per condition; two-tailed Welch’s t-test; **p<0.01). (E) EBV LMP-1, which encodes a potent activator of NF-κB signaling, is expressed in late lytic cells (left panel) but not associated with abortive cells that exhibit upregulated NF-κB transcriptomic signature including BCL2A1 (right panel, Pearson R = -0.06). (F) Flow cytometry analysis of protein biomarkers of full lytic reactivation (gp350) and NF-κB activity (ICAM1) at 48 h post 4HT treatment. Consistent with mRNA measurements, separate gp350+ and ICAM1+ populations are induced following 4HT treatment. Co-treatment with PAA reduces gp350+ cell frequency and increases ICAM1+ fractions. IKK inhibitor co-treatment reduces ICAM1+ cell frequency but does not substantially affect gp350+ cell frequency.
Because EBV LMP-1 partially mimics the activated CD40 receptor that induces NF-κB signaling, we reasoned that LMP-1 might be associated with the abortive lytic phenotype. However, LMP-1 expression was largely restricted to cluster E (Fig 3E), consistent with its transcription during the lytic cycle [113,114]. This observation suggested that the abortive lytic phenotype and associated NF-κB signaling was not dependent upon LMP-1 expression. We confirmed this finding through FACS detection of gp350 (lytic cells) and ICAM1, a surface-expressed proxy for NF-κB pathway transcriptional activation (and in this context, abortive reactivation). Untreated P3HR1-ZHT cells did not express gp350 or ICAM1 (Figs 3F and S8). Treatment with 4HT induced expression of both gp350 and ICAM1; notably, expression of these proteins was observed in distinct cell subpopulations, supporting our finding that NF-κB signaling was primarily active in cells that had not entered the full lytic cycle. Accordingly, co-treatment with 4HT + PAA to induce an abortive lytic state by blocking viral DNA synthesis led to increased ICAM1+ cell frequency consistent with the BCL2A1 upregulation observed in Fig 3D. Conversely, co-treatment with 4HT and an inhibitor of IKKβ (a key component of NF-κB signaling) eliminated ICAM1 expression, but did not increase gp350 expression. These results demonstrated that NF-κB signaling is a feature of abortive lytic cells that is independent of LMP-1 activity, but does not restrict late viral gene expression.
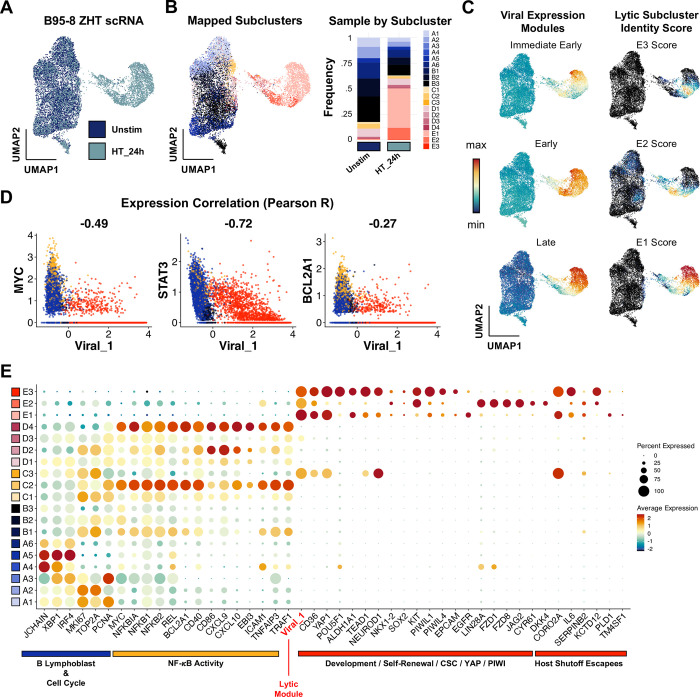
Lytic subpopulations are reprogrammed to stem-like plasticity during EBV reactivation
We next focused on the lytic fate by analyzing cells in cluster E. Paradoxically, lytic cells in cluster E collectively expressed the most unique genes (i.e., transcript diversity) of any cluster despite having low mRNA density per cell consistent with host shutoff (Fig 4A). In addition to differences in early and late lytic gene expression across cluster E (Fig 1F), this observation was consistent with enhanced cell-to-cell variability in gene expression. We therefore subclustered cells at higher resolution to examine heterogeneity among lytic subpopulations (Fig 4B). This yielded three subclusters of BZLF1+ cells–one with high late gene expression corresponding to complete reactivation (cluster E1) and two with comparatively lower late gene expression (clusters E2 and E3). Differential expression analysis by subcluster revealed remarkably broad cellular plasticity and developmental pluripotency signatures in E2 and E3 (Fig 4C). Although MYC was downregulated, the master pluripotency regulators POU5F1 (OCT4), SOX2, KLF4, NANOG, and LIN28A were expressed in E2 and E3 [115–120]. Intriguingly, many essential transcriptional regulators of pluripotency exit and germ layer specification were also co-expressed with BZLF1+ in E2 and E3 lytic subpopulations. Expression of ALDH1A1, ALPL, ITGA6, CD44, PROM1 (CD133), LGR5, and YAP1 upregulated in the E2 and E3 phenotypes was consistent with cancer hallmarks including cell plasticity, self-renewal, and drug-tolerant persistence [68,121–126]. Related to YAP1 expression, we identified distinct Hedgehog [127], Notch [128], and Wnt [129, 130] signaling pathway signatures in E2 and E3 lytic phenotypes as well as Hippo-independent YAP pathway [131] components reported in cancer. E3 cells also expressed genes encoding several PIWI-like family proteins, which protect germline cell genomes from transposable element insertion, maintain stemness, and are upregulated in some cancers [132–135].
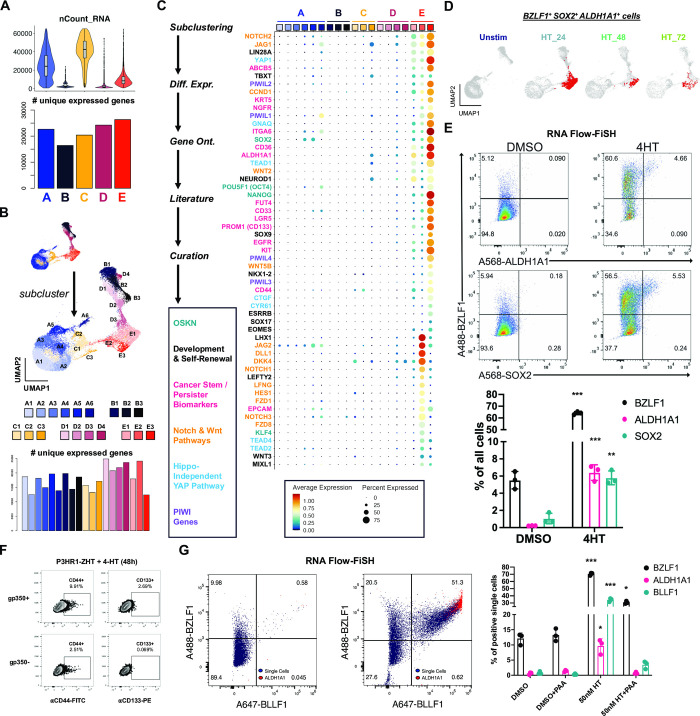
Fig 4. Cancer-associated cellular plasticity and self-renewal signature identification in EBV lytic cell subsets.
(A) Total mapped RNA reads per cell (top panel) versus total unique genes expressed across each cluster (bottom panel). (B) Unsupervised identification of high-resolution subclusters across P3HR1-ZHT time course scRNA-seq data. (C) Differentially expressed genes upregulated in lytic subclusters (E1, E2, and E3). Genes were identified by comparing each subclusters versus all others, summarized by gene ontology methods, cross-referenced against primary literature, and curated by biological annotation. (D) Co-expression of BZLF1 and genes associated with cellular pluripotency and cancer stemness (SOX2, ALDH1A1) in single cells (co-positive cells in red) by timepoint. (E) RNA Flow-FISH validation of ALDH1A1 and SOX2 expression in BZLF1+ cells (top panel) at 24 h post 4HT treatment. Frequencies of ALDH1A1+ and SOX2+ cells significantly increase in response to 4HT induction of the lytic cycle versus DMSO control treatment (bottom panel; n = 3 per condition; two-tailed Welch’s t-test; ***p<0.001; **p<0.01). (F) Flow cytometry protein level validation of elevated CD44 and CD133 expression in gp350+ versus gp350- P3HR1-ZHT cells. (G) RNA Flow-FISH analysis of ALDH1A1 expression by lytic cycle progression at 24 h post 4HT treatment. Rare spontaneously reactivated BZLF1+BLLF1+ cells express ALDH1A1 without lytic induction treatment (left panel). The frequency of BZLF1+BLLF1+ALDH1A1+ cells increases upon 4HT treatment (middle panel). ALDH1A1+ P3HR1-ZHT cells are significantly enriched after 4HT treatment but not in the context of co-treatment with PAA to block viral DNA synthesis (right panel; n = 3 per condition; two-tailed Welch’s t-test; ***p<0.001; **p<0.01; *p<0.05).
In total, 6,900 of 26,728 cells (25.8%) across all sampled timepoints expressed BZLF1 transcripts. Co-expression of genes including ALDH1A1 and SOX2 in a subset of BZLF1+ cells demonstrated an association between cellular plasticity and EBV lytic reactivation (Fig 4D). GRN analysis further supported a role for SOX2 transcriptional activity in a fraction of lytic cells (S10 Fig). RNA Flow-FISH validated ALDH1A1 and SOX2 expression in BZLF1+ cells in 4HT treated P3HR1-ZHT cultures (Figs 4E and S9). To validate these findings at the protein level, we performed intracellular flow cytometry for the SOX2 protein and BMRF1 and confirmed that a small percentage of lytic cells express SOX2 (S10 Fig). We also used flow cytometry to validate increased expression of the CSC biomarkers CD44, CD133 (PROM1), and CD166 (ALCAM) at the protein level in gp350+ cells (late lytic) relative to gp350- subsets across treatment conditions (Figs 4F and S11).
We next examined whether lytic cycle initiation was sufficient to induce CSC-associated pluripotency expression or if successful viral DNA synthesis was required. To do so, we used RNA Flow-FISH to detect BZLF1, ALDH1A1, and BLLF1. ALDH1A1 was expressed in BZLF1+BLLF1+ cells following 4HT treatment, consistent with its expression in late stages of lytic reactivation (Fig 4G, left and middle panels). Consistent with a role for viral DNA replication in CSC gene induction, co-treatment with PAA and 4HT diminished BZLF1+BLLF1+ cell frequency and ablated ALDH1A1 expression (Fig 4G, right panel). Collectively, these data support a unique program of cellular plasticity induced in the late phase of EBV lytic reactivation.
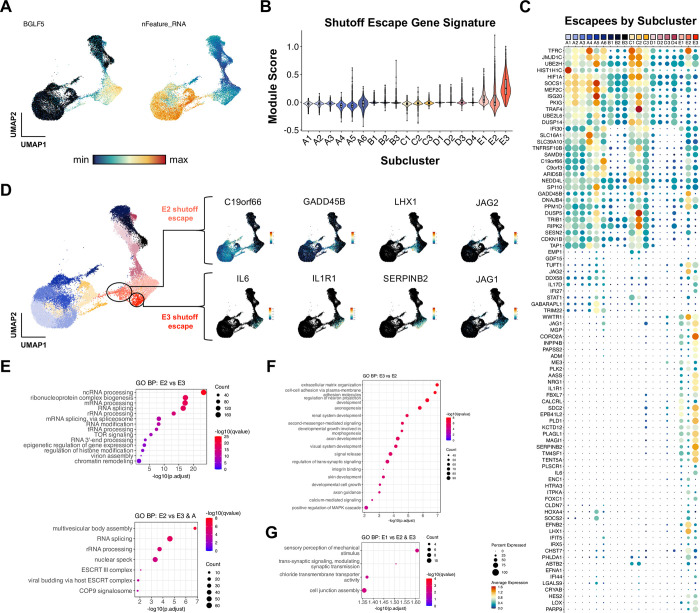
Host shutoff escapees in lytic subclusters exhibit distinct ontologies
Because lytic subclusters identified at high resolution displayed distinct cellular transcriptomes, we asked whether host shutoff responses differed among lytic cells. RNA for BGLF5, an early EBV lytic gene that mediates host shutoff [107], was detected at variable levels across lytic cells and inversely correlated with per cell mRNA feature density as expected (Fig 5A). We also observed an increase in expression of BSLF2/BMLF1 (EBV SM) across lytic cells (S6 Fig). EBV SM mediates a change from the preferential export of spliced mRNAs to non-spliced viral mRNAs, which leads to an overall reduction in functional cellular gene product expression [136]. Moreover, transcripts for genes previously found to escape host shutoff [109, 110] were identified in each lytic subcluster (E1, E2, and E3) (Fig 5B and 5C). Host shutoff escapee expression could be broadly categorized by two patterns–some escapees (e.g., C19orf66, CDKN1B) were expressed in unstimulated P3HR1-ZHT cells and retained across abortive and lytic cells, whereas other escapees (e.g., IL6, SERPINB2, LHX1, JAG1) were exclusively expressed in lytic cells (Fig 5C). Intriguingly, lytic subclusters exhibited different host shutoff escapee profiles. Anecdotally, we also noted that several escapees in clusters E2 and E3 were related to inflammatory responses and overlapped with CSC and developmental pluripotency signatures (Fig 5D). We applied gene ontology (GO) analyses to differentially expressed genes among lytic subclusters to further investigate potential biological differences. Cells in E2 displayed significant enrichment of GO terms related to mRNA splicing and post-transcriptional regulation and epigenetic regulation versus cells in E3 (Fig 5E, top panel). RNA processing GO terms were also upregulated in E2 when compared jointly against clusters E3 and A to filter out differences related to transcripts basally expressed in unstimulated cells (Fig 5E, bottom panel). Conversely, the top enriched GO terms in cluster E3 versus E2 were related to cell-cell adhesion, morphogenesis, and diverse tissue-specific developmental programs (Fig 5F). Relatively few cellular GO terms were enriched in fully lytic cells (E1), consistent with extensive host shutoff and predominantly viral gene expression (Fig 5G).
Fig 5. Distinct virus-mediated host shutoff responses and escapees in lytic subclusters.
(A) UMAP representation of host shutoff mediator BGLF5 expression (left panel) and per cell feature RNA (right panel) in P3HR1-ZHT timecourse scRNA-seq data. (B) Module scores for a curated set of genes that escape host shutoff (GADD45B, IL6, CCND1, IL1R1, JAG1, SERPINB2, EPHB2, FOXC1, ATF3, ZNF526, P2RY11, and HES4) by high resolution cluster. (C) Subcluster-level expression of host shutoff escapees curated from primary literature. (D) Detail of distinct host shutoff escapee signatures in two lytic subclusters (E2 and E3). (E) Biological process gene ontology (GO) analysis for genes upregulated in lytic subcluster E2 versus E3 (top panel) and E2 versus E3 + A (unstimulated cells). (F) Biological process GO analysis for genes upregulated in lytic subcluster E3 versus E2. (G) Biological process GO analysis for genes upregulated in lytic subcluster E1 versus E2 and E3.
Phenotype validation across viral strain and host background
Finally, we confirmed key findings through additional independent scRNA-seq experiments capturing responses of B958-ZHT cell lines to 4HT treatment (Fig 6), the BL line Akata to anti-Ig stimulation (Fig 7), and technical replication in P3HR1-ZHT (S14 Fig). Unstimulated and 24 h post-4HT B958-ZHT cell libraries were generated and analyzed as in previous experiments (Fig 6A). High-resolution cluster annotations from P3HR1-ZHT scRNA-libraries were mapped to B958-ZHT cells by anchor feature identification and transfer to evaluate the preservation of biological phenotypes across cell systems (Fig 6B). Cells corresponding to each high-resolution cluster were identified in the B958-ZHT dataset. Viral IE, early, and late gene expression modules were also scored across B958-ZHT cells and compared against scores for the three lytic subclusters (Fig 6C). As in the P3HR1-ZHT system, E1 cells exhibited high late gene expression consistent with complete reactivation while E2 and E3 cells displayed reduced late gene scores. In B958-ZHT, the E3 cluster most closely associated with plasticity and self-renewal signatures had the lowest IE, early, and late expression relative to other cells in lytic clusters. Prior findings of viral gene anticorrelation with MYC, STAT3, and BCL2A1 (Fig 6D) and lytic cell upregulation of cancer-associated stem-like pluripotency and host shutoff escapees were conserved in B958-ZHT (Figs 6E, S12, and S13).
Fig 6. Lytic subset reprogramming and host shutoff escape signatures are conserved in B958-ZHT lymphoblastoid cells.
(A) UMAP representation of scRNA-seq data from the inducible lytic marmoset lymphoblastoid cell line B958-ZHT before (Unstim) and 24 h after 4HT treatment. (B) Mapping of cell subclusters defined from P3HR1-ZHT analyses to B958-ZHT scRNA-seq data via transfer anchor integration (left panel). Subcluster composition is presented for unstimulated and 4HT-treated cell libraries (right panel). (C) Viral expression module (IE, early, late) and mapped lytic subcluster scores in timecourse merged B958-ZHT data. Of note, the assigned subclusters in 6B represent qualitative classifications based on maximum annotation signature scores for each cell. Accordingly, a given cell may score highly for more than one related signature while being assigned to a single classification. The underlying quantitative signature scores for E1, E2, and E3 presented here thus reflect a lytic phenotypic continuum rather than purely discrete states. (D) Conserved anticorrelation between EBV gene expression (Viral_1 module score) and genes characteristic of unstimulated and abortive phenotypes (MYC, STAT3, BCL2A1). Values denote pairwise Pearson R coefficients. (E) Conservation of key gene expression signatures identified from P3HR1-ZHT (a BL cell line) within B958-ZHT (a lymphoblastoid cell line) during EBV lytic reactivation.
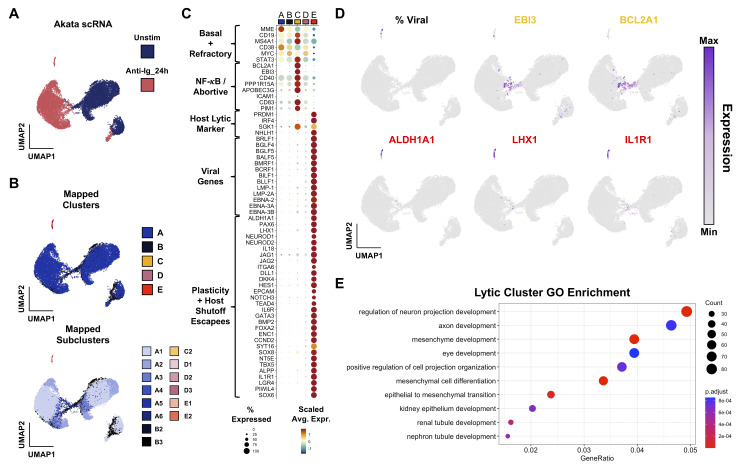
Fig 7. Validation of abortive and reprogramming gene expression signatures in Akata BL cells stimulated by anti-Ig.
(A) Integrated scRNA-seq data from Akata cells without stimulation (blue) and 24 h after anti-Ig treatment to induce lytic reactivation (rose). (B) Cluster and subcluster mapping from P3HR1-ZHT data to Akata data via transfer anchor integration. Clusters are colored as for P3HR1-ZHT and B958-ZHT datasets. Colors represent the reference phenotype with the maximal integration score for each cell. (C) Cluster-resolved expression in integrated Akata scRNA-seq dataset. (D) UMAP visualization of viral read fraction and representative markers of abortive (yellow) and reprogramming (red) signatures. LHX1 and IL1R1 are known escapees of host shutoff. (E) GO enrichment of DE genes upregulated in anti-Ig-induced Akata lytic cells.
In Akata cells, unstimulated and 24 h post anti-Ig cell libraries were generated as in previous experiments (Fig 7A). We again mapped annotations from the P3HR1-ZHT scRNA-libraries to Akata cells by anchor feature identification, both at the cluster and subcluster level (Fig 7B). As in the P3HR1-ZHT and B958-ZHT systems, STAT3, MYC, and BCL2A1 expression were anticorrelated with successful lytic reactivation (Fig 7C and 7D). Viral gene expression and expression of plasticity markers like ALDH1A1 were co-expressed (Fig 7C and 7D). Gene ontology enrichment for differentially expressed genes in the lytic cluster also highlights the presence of plasticity and developmental factors (Fig 7E). Thus, our findings in the P3HR1-ZHT system are applicable across EBV strains, host cell genetic backgrounds, and reactivation stimuli.
Discussion
The single-cell data presented herein substantially expand and refine transcriptome-wide contours of host-virus dynamics during the EBV lytic cycle. Prior studies discovered that EBV-infected BL cells are prone versus resistant to reactivation dependent on STAT3 expression, activity, and functions of its downstream transcriptional targets [56,57,59]. A population of STAT3-/lo cells in unstimulated P3HR1-ZHT revealed by scRNA-seq (cluster B), which exhibits globally reduced mRNA levels consistent with cellular quiescence, may be more permissive to successful reactivation than cells with basally elevated STAT3 (cluster A). Additionally, cells that undergo abortive lytic reactivation retain STAT3 expression (and predicted transcriptional activity) after stimulation, while STAT3 and host transcript loads are drastically reduced in fully lytic cells, consistent with host shutoff functions exhibited by diverse viruses [137–141]. Single-cell data are also consistent with the functional importance of c-MYC in regulating EBV latency versus lytic reactivation [54]. MYC expression exhibits cluster-level patterns similar to STAT3, with the notable exception that MYC is more strongly expressed in cluster B cells–likely due to constitutive expression resulting from the chr8:chr14 (Ig-MYC) translocation in BL. Single-cell sequencing and RNA-FiSH results further identify unique upregulation of NF-κB and IRF3 pathway transcriptional targets in abortive lytic cells. Inhibition of viral DNA synthesis by PAA treatment accentuates this abortive phenotype, though we recognize that this model may not truly recapitulate an abortive lytic state in vivo. Regardless, paired with STAT3 and MYC activity, we speculate that this concerted response might sustain viability and reinforce latency in cells that fail to meet the lytic switch threshold.
Acquisition of cellular plasticity within lytic cell subsets in multiple EBV+ B cell models is particularly striking. Several aspects of the lytic cycle could conceivably contribute to host cell plasticity through reversing epigenetic repression of lineage-ectopic genes. As observed across several DNA virus families, EBV genome replication within intranuclear compartments induces dramatic reorganization of host chromatin [69,70,73,142]. Along with this alteration to nuclear architecture, Zta binding at accessible AP-1 recognition sequences [33] (particularly methylated sites [71,72]) may reverse epigenetic silencing through supporting nucleosome eviction, enhancement of chromatin accessibility, and recruitment of transactivators to facilitate aberrant gene expression [43, 44]. ChIP-seq for Zta has revealed many such potential sites throughout the host genome, including POU5F1 (Oct-4) [42,43]. From the viral perspective, Zta binding across the cellular genome may function as a “sink” that supports bimodal control of the switch between latency (Zta absence or noise-level expression) and lytic reactivation (high Zta) [43]. From the host perspective, our findings suggest that these BZLF1 interactions with cellular DNA and nuclear chromatin remodeling during later stages the lytic cycle have substantial–and potentially pathogenic–collateral effects on biological reprogramming. Along these lines, developmental reprogramming associated with Wnt/β-catenin signaling has been observed in a single-cell study of HSV-1 lytic infection [74].
Additionally, DNA damage, antiviral nucleic acid sensing, cytoskeletal rearrangements, and other major mechanobiological changes that manifest during reactivation may activate intrinsic responses to cellular injury leading to NF-κB and IRF3 signaling in the abortive population [143–145]. Paired with lytic-mediated growth arrest [146,147], we speculate that this process may engage cellular senescence and injury responses that promote autocrine and paracrine cellular reprogramming. An essential feature of damage-associated induction of cellular pluripotency is upregulation of pro-inflammatory cytokines such as IL-6 [148]. In both P3HR1-ZHT and B958-ZHT scRNA-seq datasets, IL6 expression was exclusive to fully lytic cell subsets. However, IL6R was expressed in abortive cells in P3HR1-ZHT and most latently infected cells in B958-ZHT. In the Akata scRNA-seq dataset, IL6R was exclusively expressed in the lytic cluster. Expression of JAK1/2 and STAT3 in latently infected cells from both lines was suggestive of an IL-6 response axis (IL6(R)/JAK/STAT3) known to be activated in hematologic malignancies [149]. This raises the intriguing possibility that cells from one reactivation trajectory and viral replication mode (fully lytic cells) might reinforce the survival and proliferation of tumor cells resulting from an alternative response (abortive, latently infected) through paracrine mechanisms. In addition to its escape from host shutoff [110], IL-6 autocrine support for latent EBV+ B cell proliferation and its depletion in BZLF1- and BRLF1-deificient tumors in murine models of EBV-driven lymphoproliferative disease are especially noteworthy [53,150,151]. A similar effect has been observed during infection with KSHV, which encodes a viral IL-6 homolog. Thus, the developmental pluripotency profiles and responses of lytic cell subsets may be associated with cellular DNA damage responses that have inadvertent pathogenic effects in EBV+ tumors. Notably, cytokine production by EBV-infected tumor cells (including abortive lytic cells) has also been proposed to support oncogenesis through microenvironment conditioning, polarization of tumor infiltrating lymphocytes, and evasion of T-cell surveillance [49,50].
In summary, our findings support a model of differential response trajectories to EBV lytic induction. The first determinant in this model is initial cell state, where ground-state STAT3 and MYC expression and activity predict a ‘high-resistance’, low-probability path to full reactivation. Conversely, cells with globally reduced transcription and reduced expression of STAT3 (and MYC) at the time of lytic reactivation traverse a ‘low-resistance’ path with high probability of complete reactivation. These data have potentially important clinical implications, as they suggest that quiescent EBV+ tumor cells may be more sensitive to lytic induction therapies. However, a critical second fate determinant that manifests in lytic cells may complicate this pursuit. To this point, our scRNA-seq and RNA Flow-FISH results are consistent with the previously identified role of lytic cycle induction in tumorigenesis [46,47,53]. Most cells that undergo full reactivation and new virion release are likely to die. However, some lytic cells undergo profound reprogramming to plastic CSC-like states that may promote malignancy through multiple mechanisms, even independent of their own survival. For example, we found transcript-level evidence that lytic cells could reinforce viral latency and survival of abortive or refractory cells via IL6/JAK2/STAT3 signaling. Additional studies are necessary to explore, dissect, and therapeutically perturb the IL-6/JAK/STAT3 pathway in EBV+ lymphomas.
Furthermore, it is important to note that while there were many overlapping findings between our three datasets, there are key differences to be acknowledged for studies moving forward. The P3HR1-ZHT model is a BL-derived line with constitutive MYC expression that lacks functional EBNA2. The B958-ZHT line was transformed in vitro with a strain of EBV that contains functional EBNA2. These differences will lead to natural variation in phenotypes present in each model. Additionally, both lines are engineered to express a mutant estrogen receptor binding domain fused to the Zta protein. While this model is useful for studying lytic reactivation, we recognize that it is not a true physiologic trigger for lytic reactivation. To add breadth to our observations, we compared Z-HT to stimulation of the Akata BL cell line with anti-Ig, which leads to reactivation through BCR signaling. While reactivation efficiency was relatively low in the Akata system, we confirmed our overarching findings in all three models.
Given these findings, subsequent examinations of the epigenetic consequences of early EBV reactivation at high resolution should be prioritized, and the possibility of double-edged consequences of oncolytic therapies should be specifically examined in detail. Future single-cell approaches should interrogate the frequency of viable abortive lytic cells [152] and the particular changes in chromatin accessibility as well as other epigenetic features of this phenotype. Similar experimental approaches should be applied to study clinical EBV+ tumor specimens to understand oncogenic correlates of lytic reactivation in situ.
Materials and methods
Ethics statement
Human cell lines used in this study were not accompanied with HIPAA identifiers or PHI. All experiments were thus categorized as non-human subjects research and approved by a Duke University IRB “Mechanisms of Epstein-Barr virus transformation using adult peripheral blood from the Red Cross” (eIRB #Pro00006262).
Cell lines, culture, and treatments
P3HR1-ZHT cells (derived from the Type 2 EBV+ [P3 strain] Jijoye eBL line) and B958-ZHT (a marmoset lymphoblastoid cell line transformed with Type 1 EBV [B95-8 strain]) were used in this study. Each cell line was cultured at 37°C with 5% CO2 in RPMI + 10% FBS (R10) media (Gibco RPMI 1640, ThermoFisher). To induce lytic gene expression, 4x105 cells/mL for a given cell line in log-phase growth were treated with 25 nM, 50 nM, or 100 nM 4-hydroxytamoxifen (4HT) in methanol (4HT, Millipore Sigma). Phosphonoacetic acid (PAA, 1 μM) was included in parallel with lytic induction treatments to inhibit viral DNA synthesis and prevent complete reactivation in separate experimental groups (i.e., abortive lytic replication). Control groups were prepared via treatment with 0.1% DMSO (and DMSO + PAA). All treatments for flow cytometry and RNA Flow-FISH experiments described below were performed in triplicate (technical replicates) in 6-, 12-, or 24-well culture plates.
Flow cytometry
Flow cytometric cell cycle analysis of unstimulated and 4HT-treated P3HR1-ZHT cells was performed using pulsed BrdU incorporation (20 min) and nuclear staining with 7-AAD in fixed cells (Invitrogen eBioscience BrdU staining kit, cat #8811-6600-42; 7-AAD, cat #00-6993-50) in addition to surface staining for gp350 (mouse anti-gp350 antibody clone 72A1 prepared in house then conjugated to Alexa 647 by Columbia Biosciences). Mitochondrial content versus gp350 expression in 4HT-induced cells was assayed using MitoTracker Green (ThermoFisher, cat #M46750). Flow cytometry was also used to assay surface expression of gp350, CD44, CD133 (PROM1), and CD166 (ALCAM). With the exception of the gp350 antibody, antibodies were purchased from BioLegend (anti-CD44_FITC, cat #397517; anti-CD133_PE, cat #397903; anti-CD166_PE-Cy7, cat #343911). In these experiments, removal of lytic inducing and control treatments at 6, 12, or 24 h via media replacement all yielded similar results. Cell were also stained and gated by viability (ZombieAqua, ThermoFisher, cat #L34965).
RNA Flow-FISH
RNA Flow FISH analysis of unstimulated and 4HT-induced P3HR1-ZHT cells (24 and 48 h post-treatment) was performed using RNA PrimeFlow reagents (ThermoFisher RNA PrimeFlow Kit Catalog #: 88-18005-210) and validated RNA probes (ThermoFisher. Type 1 probes: BLLF1_A647. Type 4 probes: BZLF1_A488, BCL2A1_A488. Type 10 probes: BGLF4_A568, CD38_A568, ALDH1A1_A568, SOX2_A568). PrimeFlow sample preparation was completed per ThermoFisher protocol with no adjustments. Briefly, cells were washed, fixed, and permeabilized. Cells were then incubated with target probes for 2 h in a 40°C water bath. Cells were washed and stored overnight at 4°C and then incubated with a Pre-amplification buffer for 1.5 h in a 40°C water bath followed by a 1.5 h incubation in amplification buffer. Cells were then incubated in label probes for 1 h in a 40°C water bath, washed with FACS buffer and subsequently analyzed on a Cytek Aurora. Spectral flow unmixing was performed with SpectroFlo software and uniformly applied to all samples. Further analysis and gating was completed in FlowJo.
Single-cell sample and library preparation
P3HR1-ZHT cells were plated at 4 x 105 cells/ mL in 5 mL R10 then treated with methanol (mock- 0 h) or with 25 nM 4HT (4-hydroxytamoxifen). The cells incubated in 4HT for 72, 48, and 24 hours then all cells were harvested for library preparation at the same time. The viabilities of the 0, 24, 48, and 72 h samples at time of collection were approximately 90%, 80%, 75%, and 75%, respectively. Harvested cells were resuspended at the recommended concentration to collect approximately 10,000 cells per sample during GEM generation. Single-cell transcriptomes from all four samples were captured and reverse transcribed into cDNA libraries using the 10x Genomics Chromium Next GEM Single Cell 3’ gene expression kit with v3.1 chemistry and Chromium microfluidic controller according to recommended protocols (10x Genomics, Pleasanton, CA). All cDNA gene expression libraries were pooled for sequencing.
Sequencing, read alignment, and QC
Pooled single-cell libraries were sequenced across two lanes of an S2 flow cell on a NovaSeq6000 (Illumina, San Diego, CA) with 50 bp paired-end reads at a target sequencing depth of 50,000 reads per cell. Output base calls (.bcl) were assembled into sample-demultiplexed reads (.fastq) using cellranger mkfastq with default settings (10x Genomics, Pleasanton, CA). Reads were mapped to a concatenated reference genome package (hg38 + NC_009334 [type 2 EBV]; prepared via cellranger mkref) to generate single-cell expression matrices by running cellranger count (10x Genomics, Pleasanton, CA). Cellranger output files (genes.tsv, barcodes.tsv, matrix.mtx) were used to create Seurat data objects in R [153–155], which were subsequently pre-processed using QC filters. Cells and features were included if they met the following criteria: feature (gene) expression in a minimum of three cells; mitochondrial genes accounting for < 25% of all transcripts; a minimum of 200 unique expressed genes; < 65,000 total transcripts to exclude non-singlets. The elevated mitochondrial transcript and total transcript cutoffs relative to those used for resting PBMC samples [156] were chosen because of the highly proliferative nature of the P3HR1 cell line, the expectation of apoptosis as one outcome to lytic reactivation, and the implementation of viability enrichment prior to library preparation described above. A total of 26,728 cells across the timecourse passed all QC filters (nuntreated = 10,196; n24h = 7,905; n48h = 5,841; n72h = 3146).
Data pre-processing, dropout imputation, analysis, and visualization
A complete list of loaded packages and versions (RStudio sessionInfo() output) is provided as a supplementary file (S1 File). Single-cell expression data were analyzed and visualized with R (v4.0.5) / RStudio (v2022.07.1+554) using Seurat v4.1.0. Data from each timepoint were analyzed, separated, and merged into a single object to support time-resolved analysis. Raw count data were normalized and scaled prior to feature identification (NormalizeData and ScaleData functions). Cell cycle scores and phases were assigned based on annotated gene sets provided in the Seurat package (CellCycleScoring function). Expression data were dimensionally reduced using principal component analysis of identified variable features (RunPCA), and the first 30 principal components were used for subsequent UMAP dimensional reduction (FindNeighbors, RunUMAP). Cell clusters were identified at multiple resolutions for phenotype identification and comparative analysis (FindClusters).
Biological zero-preserving imputation was applied to correct technical read dropout using adaptive low-rank approximation (ALRA) of the RNA count matrix [157]. Data presented throughout this study was generated from imputed read data. Differential gene expression analysis of the merged timecourse RNA and imputed (ALRA) assays was performed at multiple clustering resolutions. Outputs from this analysis are provided as supplementary tables (S1–S4 Tables). Single-cell gene expression, co-expression, and cluster-averaged expression were visualized with Seurat functions (e.g., DimPlot, FeaturePlot, FeatureScatter, VlnPlot, DotPlot, DoHeatmap). Additional visualization of multi-gene co-expression was generated with the UpSetR package [158].
Pseudotime analysis
Pseudotime trajectories were calculated for day 0 and merged timecourse datasets using Monocle3 [111,159]. Briefly, Seurat objects were adapted as cell dataset objects and used to learn and order cells along pseudotime graphs anchored at manually determined root cells. Calculated pseudotime values were added as a feature to original Seurat objects and used for subsequent gene expression analyses. Pseudotime-gene correlation was plotted and fit via smoothing splines to visualize expression dynamics across clusters (cell phenotypes).
Gene ontology and gene regulatory network analyses
Low and high-resolution cluster gene ontology (GO) enrichment analysis for biological processes was performed using the enrichGO function in clusterProfiler [90]. Statistically significant enrichment results were visualized using the barplot, pairwise_termism, and emapplot functions. Cluster-level gene regulatory network (GRN) inference of transcription factor activities was conducted using CollecTRI in the R package decoupleR [160,161].
Statistical analyses
Raw and adjusted p values (Bonferroni correction) were calculated and provided for all identified differentially expressed genes from scRNA-seq data (S1–S4 Tables). For conventional flow cytometry and RNA flow-FISH experiments, statistically significant differences between treatment groups were determined via two-tailed Welch’s t test (n = 3 replicates per condition).
Supporting information
(A) Lymphocyte, singlet, live-cell, and gp350+ gating for unstimulated cells. (B) The same gating strategy as above applied for 4HT-treated cells. (C) The same gating strategy as above applied for cells co-treated with 4HT and PAA.
(TIF)
(A) Co-expression of BZLF1 with BGLF4 or BLLF1 in DMSO control treatment and 4HT-induced reactivation. (B) Co-expression of BZLF1, BGLF4, and BLLF1 (red cells) in DMSO control treatment and 4HT-induced reactivation.
(TIF)
(TIF)
(A) Cell cycle phase annotations in P3HR1-ZHT scRNA-seq data. (B) Flow cytometry cell cycle analysis in unstimulated and 4HT-treated P3HR1-ZHT cells with gp350+ cells highlighted. (C) MitoTracker staining by gp350 status in 4HT-treated P3HR1-ZHT cells.
(TIF)
(TIF)
(TIF)
(A) Technical controls, 24 h, and 48 h responses to DMSO, 4HT, and 4HT+PAA for BCL2A1 versus BLLF1 expression. (B) Technical controls, 24 h, and 48 h responses to DMSO, 4HT, and 4HT+PAA for CD38 versus BLLF1 expression. (C) Technical controls, 24 h, and 48 h responses to DMSO, 4HT, and 4HT+PAA for BCL2A1 versus CD38 expression.
(TIF)
Statistical comparisons between groups (n = 3 replicates per treatment condition) were evaluated via Welch’s two-tailed t tests (***p<0.001)
(TIF)
(TIF)
(A) SOX2 scRNA-seq expression and gene regulatory network activity. (B) Hierarchical clustering of predicted TF activities by P3HR1-ZHT subcluster. (C) Flow plots of untreated and treated P3HR1-ZHT cells 24 h post stimulation. Flow plots show that a small percentage of induced cells express SOX2 and that most of these cells are lytic (BMRF1+). (D) Bar graph depicting the percentage of cells that expressed SOX2 between treatment groups, in biological triplicate.
(TIF)
(A) Controls, gating, and stemness biomarker expression by gp350 status in unstimulated cells. (B) Controls, gating, and stemness biomarker expression by gp350 status in 4HT-treated cells. (C) Controls, gating, and stemness biomarker expression by gp350 status in cells co-treated with 4HT and PAA.
(TIF)
(A) Controls, gating, and gp350 expression in unstimulated cells. (B) Controls, gating, and gp350 expression in 4HT-treated cells. (C) Controls, gating, and gp350 expression in cells co-treated with 4HT and PAA.
(TIF)
(A) Controls, gating, and stemness biomarker expression by gp350 status in unstimulated cells. (B) Controls, gating, and stemness biomarker expression by gp350 status in 4HT-treated cells. (C) Controls, gating, and stemness biomarker expression by gp350 status in cells co-treated with 4HT and PAA.
(TIF)
(A) Overview of P3HR1-ZHT replicate experiment treatments (methanol control and 4HT) and identified clusters. (B) UMAP visualization of global QC metrics (top row), differential abortive and lytic responses correlated with STAT3 and MYC levels (2nd and 3rd rows), and upregulated pluripotency signature in lytic cell subsets (4th and 5th rows).
(TIF)
List of differentially expressed genes separated by each unique cluster. Each sheet is a separate cluster and includes gene name, average log2 fold change, percentages of cells expressing the gene in the cluster versus all other clusters, and adjusted p value.
(XLSX)
List of differentially expressed genes separated by each unique cluster and data was imputed with ALRA in Seurat. Each sheet is a separate cluster and includes gene name, average log2 fold change, percentages of cells expressing the gene in the cluster versus all other clusters, and adjusted p value.
(XLSX)
List of GO terms separated by each unique cluster. Each sheet is a separate cluster and includes GO ID, description, adjusted p value, counts, and gene IDs.
(XLSX)
List of GO terms separated by each unique cluster and data was imputed with ALRA in Seurat. Each sheet is a separate cluster and includes GO ID, description, adjusted p value, and gene IDs.
(XLSX)
(R)
Acknowledgments
We would like to acknowledge the Heaton Lab at Duke for their assistance with preparing the 10X single cell libraries. We also thank the staff of the Duke Center for Genomic and Computational Biology (GCB) for sequencing support.
Data Availability
We have deposited our single-cell RNA sequencing data with NIH GEO under GSE272763. All other relevant data are in the manuscript and its supporting information files.
Funding Statement
o E.D.S. wishes to acknowledge support from the Department of Molecular Genetics and Microbiology Viral Oncology Training Grant (NIH T32 #T32CA009111) and a postdoctoral fellowship from the American Cancer Society – Charlotte County, Virginia TPAC (PF-21-084-01-DMC). L.E.H. wishes to acknowledge NIH F31 support from the National Institute of Dental and Craniofacial Research (NIDCR; award #1F31DE033216). K.A.W. wishes to acknowledge funding from the National Science Foundation Graduate Research Fellowship Program (NSF GRFP #1644868). J.C. wishes to acknowledge support from St. Baldrick’s Foundation. This work was supported by NIH R01 funding from the National Institute of Dental and Craniofacial Research (NIDCR; award #R01DE025994; M.A.L.) and the National Cancer Institute (NCI; award #R01CA215185; H.C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wu X, Thi VLD, Huang Y, Billerbeck E, Saha D, Hoffmann H-H, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172(3):423–38. e25. doi: 10.1016/j.cell.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell AB, Elshina E, Kowalsky JR, Te Velthuis AJ, Bloom JD. Single-cell virus sequencing of influenza infections that trigger innate immunity. Journal of virology. 2019;93(14):e00500–19. doi: 10.1128/JVI.00500-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510(7505):363–9. doi: 10.1038/nature13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–40. doi: 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heaton NS, Langlois RA, Sachs D, Lim JK, Palese P, tenOever BR. Long-term survival of influenza virus infected club cells drives immunopathology. Journal of Experimental Medicine. 2014;211(9):1707–14. doi: 10.1084/jem.20140488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers BS, Heaton BE, Rausch K, Dumm RE, Hamilton JR, Cherry S, et al. DNA mismatch repair is required for the host innate response and controls cellular fate after influenza virus infection. Nature microbiology. 2019;4(11):1964–77. doi: 10.1038/s41564-019-0509-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein M, Achong B, Barr Y. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964:702–3. doi: 10.1016/s0140-6736(64)91524-7 [DOI] [PubMed] [Google Scholar]
- 8.Shannon-Lowe C, Rickinson A. The global landscape of EBV-associated tumors. Frontiers in oncology. 2019;9:713. doi: 10.3389/fonc.2019.00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–68. Epub 2004/10/29. doi: 10.1038/nrc1452 . [DOI] [PubMed] [Google Scholar]
- 10.Bjornevik K, Münz C, Cohen JI, Ascherio A. Epstein–Barr virus as a leading cause of multiple sclerosis: mechanisms and implications. Nature Reviews Neurology. 2023;19(3):160–71. doi: 10.1038/s41582-023-00775-5 [DOI] [PubMed] [Google Scholar]
- 11.Ascherio A, Munger KL. EBV and autoimmunity. Epstein Barr Virus Volume 1. 2015:365–85. doi: 10.1007/978-3-319-22822-8_15 [DOI] [PubMed] [Google Scholar]
- 12.Gottschalk S, Rooney CM, Heslop HE. Post-transplant lymphoproliferative disorders. Annu Rev Med. 2005;56:29–44. doi: 10.1146/annurev.med.56.082103.104727 [DOI] [PubMed] [Google Scholar]
- 13.Healy JA, Dave SS. The role of EBV in the pathogenesis of diffuse large B cell lymphoma. Epstein Barr Virus Volume 1: One Herpes Virus: Many Diseases. 2015:315–37. doi: 10.1007/978-3-319-22822-8_13 [DOI] [PubMed] [Google Scholar]
- 14.Gandhi MK, Tellam JT, Khanna R. Epstein–Barr virus-associated Hodgkin’s lymphoma. British journal of haematology. 2004;125(3):267–81. doi: 10.1111/j.1365-2141.2004.04902.x [DOI] [PubMed] [Google Scholar]
- 15.Chan JK, Sin V, Wong K, Ng C, Tsang WY, Chan C, et al. Nonnasal lymphoma expressing the natural killer cell marker CD56: a clinicopathologic study of 49 cases of an uncommon aggressive neoplasm. Blood, The Journal of the American Society of Hematology. 1997;89(12):4501–13. [PubMed] [Google Scholar]
- 16.Ko Y, Cho EY, Kim JE, Lee SS, Huh JR, Chang HK, et al. NK and NK-like T-cell lymphoma in extranasal sites: a comparative clinicopathological study according to site and EBV status. Histopathology. 2004;44(5):480–9. doi: 10.1111/j.1365-2559.2004.01867.x [DOI] [PubMed] [Google Scholar]
- 17.Raab-Traub N, editor Epstein–Barr virus in the pathogenesis of NPC. Seminars in cancer biology; 2002: Elsevier. [DOI] [PubMed] [Google Scholar]
- 18.Takada K. Epstein-Barr virus and gastric carcinoma. Molecular Pathology. 2000;53(5):255. doi: 10.1136/mp.53.5.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickinson A, Kieff E. Epstein-Barr virus, p 2655–2700. Fields virology. 2007;2. [Google Scholar]
- 20.Thorley-Lawson DA, Hawkins JB, Tracy SI, Shapiro M. The pathogenesis of Epstein–Barr virus persistent infection. Current opinion in virology. 2013;3(3):227–32. doi: 10.1016/j.coviro.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proceedings of the National Academy of Sciences. 1984;81(14):4510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemerow G, Mold C, Schwend VK, Tollefson V, Cooper N. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. Journal of virology. 1987;61(5):1416–20. doi: 10.1128/JVI.61.5.1416-1420.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roughan JE, Thorley-Lawson DA. The intersection of Epstein-Barr virus with the germinal center. Journal of virology. 2009;83(8):3968–76. doi: 10.1128/JVI.02609-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babcock GJ, Hochberg D, Thorley-Lawson DA. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity. 2000;13(4):497–506. doi: 10.1016/s1074-7613(00)00049-2 [DOI] [PubMed] [Google Scholar]
- 25.SoRelle ED, Reinoso-Vizcaino NM, Dai J, Barry AP, Chan C, Luftig MA. Epstein-Barr virus evades restrictive host chromatin closure by subverting B cell activation and germinal center regulatory loci. Cell Reports. 2023:112958. doi: 10.1016/j.celrep.2023.112958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SoRelle ED, Dai J, Reinoso-Vizcaino NM, Barry AP, Chan C, Luftig MA. Time-resolved transcriptomes reveal diverse B cell fate trajectories in the early response to Epstein-Barr virus infection. Cell Reports. 2022;40(9):111286. doi: 10.1016/j.celrep.2022.111286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AM, Dai J, Bazot Q, Patel L, Nikitin PA, Djavadian R, et al. Epstein-Barr virus ensures B cell survival by uniquely modulating apoptosis at early and late times after infection. Elife. 2017;6. Epub 2017/04/21. doi: 10.7554/eLife.22509 ; PubMed Central PMCID: PMC5425254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikitin PA, Price AM, McFadden K, Yan CM, Luftig MA. Mitogen-induced B-cell proliferation activates Chk2-dependent G1/S cell cycle arrest. PLoS One. 2014;9(1):e87299. doi: 10.1371/journal.pone.0087299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell host & microbe. 2010;8(6):510–22. doi: 10.1016/j.chom.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babcock GJD L.L.; Volk M.; Thorley-Lawson D.A. EBV Persistence in Memory B Cells In Vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6 [DOI] [PubMed] [Google Scholar]
- 31.Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. Journal of virology. 1997;71(7):4882–91. doi: 10.1128/JVI.71.7.4882-4891.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rooney C, Rowe D, Ragot T, Farrell P. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. Journal of virology. 1989;63(7):3109–16. doi: 10.1128/JVI.63.7.3109-3116.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrell PJ, Rowe DT, Rooney C, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. The EMBO journal. 1989;8(1):127–32. doi: 10.1002/j.1460-2075.1989.tb03356.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, et al. The Epstein–Barr virus lytic program is controlled by the co-operative functions of two transactivators. The EMBO journal. 2000;19(12):3080–9. doi: 10.1093/emboj/19.12.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lassoued S, Gargouri B, El Feki AeF, Attia H, Van Pelt J. Transcription of the Epstein–Barr virus lytic cycle activator BZLF-1 during oxidative stress induction. Biological trace element research. 2010;137(1):13–22. doi: 10.1007/s12011-009-8555-y [DOI] [PubMed] [Google Scholar]
- 36.Taylor GM, Raghuwanshi SK, Rowe DT, Wadowsky RM, Rosendorff A. Endoplasmic reticulum stress causes EBV lytic replication. Blood, The Journal of the American Society of Hematology. 2011;118(20):5528–39. doi: 10.1182/blood-2011-04-347112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J Virol. 2005;79(2):1296–307. Epub 2004/12/23. doi: 10.1128/JVI.79.2.1296-1307.2005 ; PubMed Central PMCID: PMC538585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhende PM, Dickerson SJ, Sun X, Feng WH, Kenney SC. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J Virol. 2007;81(14):7363–70. Epub 2007/05/12. doi: 10.1128/JVI.00154-07 ; PubMed Central PMCID: PMC1933364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun CC, Thorley-Lawson DA. Plasma cell-specific transcription factor XBP-1s binds to and transactivates the Epstein-Barr virus BZLF1 promoter. J Virol. 2007;81(24):13566–77. Epub 2007/09/28. doi: 10.1128/JVI.01055-07 ; PubMed Central PMCID: PMC2168822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reusch JA, Nawandar DM, Wright KL, Kenney SC, Mertz JE. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. Journal of virology. 2015;89(3):1731–43. doi: 10.1128/JVI.02781-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramasubramanyan S, Kanhere A, Osborn K, Flower K, Jenner RG, Sinclair AJ. Genome-wide analyses of Zta binding to the Epstein-Barr virus genome reveals interactions in both early and late lytic cycles and an epigenetic switch leading to an altered binding profile. Journal of virology. 2012;86(23):12494–502. doi: 10.1128/JVI.01705-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godfrey A, Osborn K, Sinclair AJ. Interaction sites of the Epstein–Barr virus Zta transcription factor with the host genome in epithelial cells. Access Microbiology. 2021;3(11). doi: 10.1099/acmi.0.000282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buschle A, Mrozek-Gorska P, Cernilogar FM, Ettinger A, Pich D, Krebs S, et al. Epstein-Barr virus inactivates the transcriptome and disrupts the chromatin architecture of its host cell in the first phase of lytic reactivation. Nucleic acids research. 2021;49(6):3217–41. doi: 10.1093/nar/gkab099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaeffner M, Mrozek-Gorska P, Buschle A, Woellmer A, Tagawa T, Cernilogar FM, et al. BZLF1 interacts with chromatin remodelers promoting escape from latent infections with EBV. Life science alliance. 2019;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bristol JA, Djavadian R, Albright ER, Coleman CB, Ohashi M, Hayes M, et al. A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS pathogens. 2018;14(7):e1007179. doi: 10.1371/journal.ppat.1007179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma S-D, Hegde S, Young KH, Sullivan R, Rajesh D, Zhou Y, et al. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. Journal of virology. 2011;85(1):165–77. doi: 10.1128/JVI.01512-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma S-D, Yu X, Mertz JE, Gumperz JE, Reinheim E, Zhou Y, et al. An Epstein-Barr Virus (EBV) mutant with enhanced BZLF1 expression causes lymphomas with abortive lytic EBV infection in a humanized mouse model. Journal of virology. 2012;86(15):7976–87. doi: 10.1128/JVI.00770-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okuno Y, Murata T, Sato Y, Muramatsu H, Ito Y, Watanabe T, et al. Defective Epstein–Barr virus in chronic active infection and haematological malignancy. Nature microbiology. 2019;4(3):404–13. doi: 10.1038/s41564-018-0334-0 [DOI] [PubMed] [Google Scholar]
- 49.Münz C. Latency and lytic replication in Epstein–Barr virus-associated oncogenesis. Nature Reviews Microbiology. 2019;17(11):691–700. doi: 10.1038/s41579-019-0249-7 [DOI] [PubMed] [Google Scholar]
- 50.Münz C. Tumor microenvironment conditioning by abortive lytic replication of oncogenic γ-Herpesviruses. Tumor Microenvironment: Recent Advances. 2020:127–35. [DOI] [PubMed] [Google Scholar]
- 51.Dolcetti R, Dal Col J, Martorelli D, Carbone A, Klein E, editors. Interplay among viral antigens, cellular pathways and tumor microenvironment in the pathogenesis of EBV-driven lymphomas. Seminars in cancer biology; 2013: Elsevier. [DOI] [PubMed] [Google Scholar]
- 52.Hong GK, Kumar P, Wang L, Damania B, Gulley ML, Delecluse H-J, et al. Epstein-Barr virus lytic infection is required for efficient production of the angiogenesis factor vascular endothelial growth factor in lymphoblastoid cell lines. Journal of virology. 2005;79(22):13984–92. doi: 10.1128/JVI.79.22.13984-13992.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong GK, Gulley ML, Feng W-H, Delecluse H-J, Holley-Guthrie E, Kenney SC. Epstein-Barr Virus Lytic Infection Contributes to Lymphoproliferative Disease in a SCID Mouse Model. Journal of Virology. 2005;79(22):13993–4003. doi: 10.1128/JVI.79.22.13993-14003.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo R, Jiang C, Zhang Y, Govande A, Trudeau SJ, Chen F, et al. MYC controls the Epstein-Barr virus lytic switch. Molecular cell. 2020;78(4):653–69. e8. doi: 10.1016/j.molcel.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frey TR, Brathwaite J, Li X, Burgula S, Akinyemi IA, Agarwal S, et al. Nascent Transcriptomics Reveal Cellular Prolytic Factors Upregulated Upstream of the Latent-to-Lytic Switch Protein of Epstein-Barr Virus. Journal of Virology. 2020;94(7). doi: 10.1128/JVI.01966-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koganti S, Clark C, Zhi J, Li X, Chen EI, Chakrabortty S, et al. Cellular STAT3 Functions via PCBP2 To Restrain Epstein-Barr Virus Lytic Activation in B Lymphocytes. Journal of Virology. 2015;89(9):5002–11. doi: 10.1128/JVI.00121-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill ER, Koganti S, Zhi J, Megyola C, Freeman AF, Palendira U, et al. Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes. Journal of virology. 2013;87(21):11438–46. doi: 10.1128/JVI.01762-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koganti S, de la Paz A, Freeman AF, Bhaduri-McIntosh S. B lymphocytes from patients with a hypomorphic mutation in STAT3 resist Epstein-Barr virus-driven cell proliferation. Journal of virology. 2014;88(1):516–24. doi: 10.1128/JVI.02601-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, et al. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. Journal of virology. 2010;84(2):993–1004. doi: 10.1128/JVI.01745-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh SK, Perrine SP, Williams RM, Faller DV. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood, The Journal of the American Society of Hematology. 2012;119(4):1008–17. doi: 10.1182/blood-2011-06-362434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kerr JR. Epstein-Barr virus (EBV) reactivation and therapeutic inhibitors. Journal of clinical pathology. 2019;72(10):651–8. doi: 10.1136/jclinpath-2019-205822 [DOI] [PubMed] [Google Scholar]
- 62.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discovery. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 63.Lun SW-M, Cheung ST, Cheung PFY, To K-F, Woo JK-S, Choy K-W, et al. CD44+ cancer stem-like cells in EBV-associated nasopharyngeal carcinoma. PloS one. 2012;7(12):e52426. doi: 10.1371/journal.pone.0052426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yasui M, Kunita A, Numakura S, Uozaki H, Ushiku T, Fukayama M. Cancer stem cells in Epstein-Barr virus-associated gastric carcinoma. Cancer Science. 2020;111(7):2598–607. doi: 10.1111/cas.14435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. doi: 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- 66.Batlle E, Clevers H. Cancer stem cells revisited. Nature Medicine. 2017;23(10):1124–34. doi: 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 67.Park C, Bergsagel D, McCulloch E. Mouse myeloma tumor stem cells: a primary cell culture assay. Journal of the National Cancer Institute. 1971;46(2):411–22. [PubMed] [Google Scholar]
- 68.Walcher L, Kistenmacher A-K, Suo H, Kitte R, Dluczek S, Strauß A, et al. Cancer stem cells—origins and biomarkers: perspectives for targeted personalized therapies. Frontiers in immunology. 2020;11:1280. doi: 10.3389/fimmu.2020.01280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosemarie Q, Sugden B. Five families of diverse DNA viruses comprehensively restructure the nucleus. PLoS Biology. 2023;21(11):e3002347. doi: 10.1371/journal.pbio.3002347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosemarie Q, Kirschstein E, Sugden B. How Epstein-Barr Virus Induces the Reorganization of Cellular Chromatin. mBio. 2023;0(0):e02686–22. doi: 10.1128/mbio.02686-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woellmer A, Arteaga-Salas JM, Hammerschmidt W. BZLF1 governs CpG-methylated chromatin of Epstein-Barr Virus reversing epigenetic repression. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet. 2004;36(10):1099–104. Epub 2004/09/14. doi: 10.1038/ng1424 . [DOI] [PubMed] [Google Scholar]
- 73.Chiu Y-F, Sugden AU, Sugden B. Epstein-Barr viral productive amplification reprograms nuclear architecture, DNA replication, and histone deposition. Cell host & microbe. 2013;14(6):607–18. doi: 10.1016/j.chom.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drayman N, Patel P, Vistain L, Tay S. HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations. Elife. 2019;8:e46339. doi: 10.7554/eLife.46339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Russell AB, Trapnell C, Bloom JD. Extreme heterogeneity of influenza virus infection in single cells. Elife. 2018;7. Epub 2018/02/17. doi: 10.7554/eLife.32303 ; PubMed Central PMCID: PMC5826275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwartz M, Shnayder M, Nachshon A, Arazi T, Kitsberg Y, Levi Samia R, et al. Molecular characterization of human cytomegalovirus infection with single-cell transcriptomics. Nature microbiology. 2023;8(3):455–68. doi: 10.1038/s41564-023-01325-x [DOI] [PubMed] [Google Scholar]
- 77.Shnayder M, Nachshon A, Krishna B, Poole E, Boshkov A, Binyamin A, et al. Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio. 2018;9(2):e00013–18. doi: 10.1128/mBio.00013-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.SoRelle ED, Reinoso-Vizcaino NM, Horn GQ, Luftig MA. Epstein-Barr virus perpetuates B cell germinal center dynamics and generation of autoimmune-associated phenotypes in vitro. Frontiers in Immunology. 2022;13. doi: 10.3389/fimmu.2022.1001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.SoRelle ED, Dai J, Bonglack EN, Heckenberg EM, Zhou JY, Giamberardino SN, et al. Single-cell RNA-seq reveals transcriptomic heterogeneity mediated by host–pathogen dynamics in lymphoblastoid cell lines. Elife. 2021;10:e62586. doi: 10.7554/eLife.62586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flemington E, Speck SH. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. Journal of virology. 1990;64(3):1227–32. doi: 10.1128/JVI.64.3.1227-1232.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ersing I, Nobre L, Wang LW, Soday L, Ma Y, Paulo JA, et al. A Temporal Proteomic Map of Epstein-Barr Virus Lytic Replication in B Cells. Cell Reports. 2017;19(7):1479–93. Epub 2017/05/18. doi: 10.1016/j.celrep.2017.04.062 ; PubMed Central PMCID: PMC5446956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klein G, Sugden B, Leibold W, Menezes J. Infection of EBV-Genome-Negative and–Positive Human Lymphoblastoid Cell Lines with Biologically Different Preparations of EBV. Intervirology. 1974;3(4):232–44. doi: 10.1159/000149760 [DOI] [PubMed] [Google Scholar]
- 83.Chiu Y-F, Sugden B. Epstein-Barr Virus: The Path from Latent to Productive Infection. Annual Review of Virology. 2016;3(1):359–72. doi: 10.1146/annurev-virology-110615-042358 . [DOI] [PubMed] [Google Scholar]
- 84.Boehmer PE, Lehman I. Herpes simplex virus DNA replication. Annual review of biochemistry. 1997;66(1):347–84. doi: 10.1146/annurev.biochem.66.1.347 [DOI] [PubMed] [Google Scholar]
- 85.Purushothaman P, Uppal T, Verma SC. Molecular biology of KSHV lytic reactivation. Viruses. 2015;7(1):116–53. doi: 10.3390/v7010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reichelt M, Brady J, Arvin AM. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. Journal of virology. 2009;83(8):3904–18. doi: 10.1128/JVI.02137-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emery VC, Cope AV, Bowen EF, Gor D, Griffiths PD. The dynamics of human cytomegalovirus replication in vivo. The Journal of experimental medicine. 1999;190(2):177–82. doi: 10.1084/jem.190.2.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kudoh A, Fujita M, Kiyono T, Kuzushima K, Sugaya Y, Izuta S, et al. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. Journal of virology. 2003;77(2):851–61. doi: 10.1128/jvi.77.2.851-861.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, et al. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. Journal of biological chemistry. 2005;280(9):8156–63. doi: 10.1074/jbc.M411405200 [DOI] [PubMed] [Google Scholar]
- 90.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: a journal of integrative biology. 2012;16(5):284–7. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dogan A, Bagdi E, Munson P, Isaacson PG. CD10 and BCL-6 expression in paraffin sections of normal lymphoid tissue and B-cell lymphomas. The American journal of surgical pathology. 2000;24(6):846–52. doi: 10.1097/00000478-200006000-00010 [DOI] [PubMed] [Google Scholar]
- 92.Cattoretti G, Chang C-C, Cechova K, Zhang J, Ye B, Falini B, et al. BCL-6 protein is expressed in germinal-center B cells. 1995. [PubMed] [Google Scholar]
- 93.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Advances in immunology. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8 [DOI] [PubMed] [Google Scholar]
- 94.Liu H, Ippolito GC, Wall JK, Niu T, Probst L, Lee B-S, et al. Functional studies of BCL11A: characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells. Molecular cancer. 2006;5:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hodson DJ, Shaffer AL, Xiao W, Wright GW, Schmitz R, Phelan JD, et al. Regulation of normal B-cell differentiation and malignant B-cell survival by OCT2. Proceedings of the National Academy of Sciences. 2016;113(14):E2039–E46. doi: 10.1073/pnas.1600557113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, et al. AID is required for germinal center–derived lymphomagenesis. Nature genetics. 2008;40(1):108–12. doi: 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- 97.Muramatsu M, Sankaranand V, Anant S, Sugai M, Kinoshita K, Davidson NO, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. Journal of Biological Chemistry. 1999;274(26):18470–6. doi: 10.1074/jbc.274.26.18470 [DOI] [PubMed] [Google Scholar]
- 98.Malicet C, Giroux V, Vasseur S, Dagorn JC, Neira JL, Iovanna JL. Regulation of apoptosis by the p8/prothymosin α complex. Proceedings of the National Academy of Sciences. 2006;103(8):2671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cumming RC, Lightfoot J, Beard K, Youssoufian H, O’Brien PJ, Buchwald M. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nature medicine. 2001;7(7):814–20. doi: 10.1038/89937 [DOI] [PubMed] [Google Scholar]
- 100.Lin R, Zhong X, Zhou Y, Geng H, Hu Q, Huang Z, et al. R-loopBase: a knowledgebase for genome-wide R-loop formation and regulation. Nucleic acids research. 2022;50(D1):D303–D15. doi: 10.1093/nar/gkab1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.García-Muse T, Aguilera A. R loops: from physiological to pathological roles. Cell. 2019;179(3):604–18. doi: 10.1016/j.cell.2019.08.055 [DOI] [PubMed] [Google Scholar]
- 102.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell. 2017;170(4):774–86. e19. doi: 10.1016/j.cell.2017.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jackson BR, Noerenberg M, Whitehouse A. A novel mechanism inducing genome instability in Kaposi’s sarcoma-associated herpesvirus infected cells. PLoS pathogens. 2014;10(5):e1004098. doi: 10.1371/journal.ppat.1004098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rennekamp AJ, Lieberman PM. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. Journal of virology. 2011;85(6):2837–50. doi: 10.1128/JVI.02175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arvey A, Tempera I, Tsai K, Chen H-S, Tikhmyanova N, Klichinsky M, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell host & microbe. 2012;12(2):233–45. doi: 10.1016/j.chom.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Richner JM, Clyde K, Pezda AC, Cheng BYH, Wang T, Kumar GR, et al. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS pathogens. 2011;7(7):e1002150. doi: 10.1371/journal.ppat.1002150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, et al. Host shutoff during productive Epstein–Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proceedings of the National Academy of Sciences. 2007;104(9):3366–71. doi: 10.1073/pnas.0611128104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Muller M, Glaunsinger BA. Nuclease escape elements protect messenger RNA against cleavage by multiple viral endonucleases. PLoS pathogens. 2017;13(8):e1006593. doi: 10.1371/journal.ppat.1006593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chandriani S, Ganem D. Host transcript accumulation during lytic KSHV infection reveals several classes of host responses. PLoS One. 2007;2(8):e811. doi: 10.1371/journal.pone.0000811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Glaunsinger B, Ganem D. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. The Journal of experimental medicine. 2004;200(3):391–8. doi: 10.1084/jem.20031881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32(4):381. doi: 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scholz BA, Harth-Hertle ML, Malterer G, Haas J, Ellwart J, Schulz TF, et al. Abortive lytic reactivation of KSHV in CBF1/CSL deficient human B cell lines. PLoS pathogens. 2013;9(5):e1003336. doi: 10.1371/journal.ppat.1003336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ahsan N, Kanda T, Nagashima K, Takada K. Epstein-Barr virus transforming protein LMP1 plays a critical role in virus production. Journal of virology. 2005;79(7):4415–24. doi: 10.1128/JVI.79.7.4415-4424.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nawandar DM, Ohashi M, Djavadian R, Barlow E, Makielski K, Ali A, et al. Differentiation-dependent LMP1 expression is required for efficient lytic Epstein-Barr virus reactivation in epithelial cells. Journal of virology. 2017;91(8): doi: 10.1128/JVI.02438-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 116.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–55. doi: 10.1016/s0092-8674(03)00392-1 [DOI] [PubMed] [Google Scholar]
- 117.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–42. doi: 10.1016/s0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- 118.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–37. doi: 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell stem cell. 2016;19(1):66–80. doi: 10.1016/j.stem.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 120.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature genetics. 2009;41(7):843–8. doi: 10.1038/ng.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7(10):11018. doi: 10.18632/oncotarget.6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cariati M, Naderi A, Brown JP, Smalley MJ, Pinder SE, Caldas C, et al. Alpha-6 integrin is necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line. International journal of cancer. 2008;122(2):298–304. doi: 10.1002/ijc.23103 [DOI] [PubMed] [Google Scholar]
- 123.Leushacke M, Barker N. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene. 2012;31(25):3009–22. doi: 10.1038/onc.2011.479 [DOI] [PubMed] [Google Scholar]
- 124.Barker N, Van Es JH, Kuipers J, Kujala P, Van Den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. doi: 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- 125.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer cell. 2016;29(6):783–803. doi: 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes & development. 2010;24(11):1106–18. doi: 10.1101/gad.1903310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fernandez-L A, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes & development. 2009;23(23):2729–41. doi: 10.1101/gad.1824509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Totaro A, Castellan M, Di Biagio D, Piccolo S. Crosstalk between YAP/TAZ and Notch signaling. Trends in cell biology. 2018;28(7):560–73. doi: 10.1016/j.tcb.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Park HW, Kim YC, Yu B, Moroishi T, Mo J-S, Plouffe SW, et al. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162(4):780–94. doi: 10.1016/j.cell.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158(1):157–70. [DOI] [PubMed] [Google Scholar]
- 131.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer cell. 2014;25(6):831–45. doi: 10.1016/j.ccr.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee JH, Schütte D, Wulf G, Füzesi L, Radzun H-J, Schweyer S, et al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Human molecular genetics. 2006;15(2):201–11. doi: 10.1093/hmg/ddi430 [DOI] [PubMed] [Google Scholar]
- 133.Huang H, Yu X, Han X, Hao J, Zhao J, Bebek G, et al. Piwil1 regulates glioma stem cell maintenance and glioblastoma progression. Cell reports. 2021;34(1). doi: 10.1016/j.celrep.2020.108522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bamezai S, Pulikkottil AJ, Yadav T, Vegi NM, Mueller J, Mark J, et al. A noncanonical enzymatic function of PIWIL4 maintains genomic integrity and leukemic growth in AML. Blood, The Journal of the American Society of Hematology. 2023;142(1):90–105. [DOI] [PubMed] [Google Scholar]
- 135.Gomes Fernandes M, He N, Wang F, Van Iperen L, Eguizabal C, Matorras R, et al. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Human reproduction. 2018;33(2):258–69. doi: 10.1093/humrep/dex365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Casco A, Johannsen E. EBV Reactivation from Latency Is a Degrading Experience for the Host. Viruses. 2023;15(3):726. doi: 10.3390/v15030726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Abernathy E, Glaunsinger B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology. 2015;479:600–8. doi: 10.1016/j.virol.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. A common strategy for host RNA degradation by divergent viruses. Journal of virology. 2012;86(17):9527–30. doi: 10.1128/JVI.01230-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. Journal of virology. 2009;83(18):9554–66. doi: 10.1128/JVI.01051-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Molecular cell. 2004;13(5):713–23. doi: 10.1016/s1097-2765(04)00091-7 [DOI] [PubMed] [Google Scholar]
- 141.Kwong A, Frenkel N. The herpes simplex virus virion host shutoff function. Journal of virology. 1989;63(11):4834–9. doi: 10.1128/JVI.63.11.4834-4839.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Procario MC, Sexton JZ, Halligan BS, Imperiale MJ. Single-Cell, High-Content Microscopy Analysis of BK Polyomavirus Infection. Microbiology Spectrum. 2023;11(3):e00873–23. doi: 10.1128/spectrum.00873-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kawanishi M. Epstein-Barr virus induces fragmentation of chromosomal DNA during lytic infection. Journal of virology. 1993;67(12):7654–8. doi: 10.1128/JVI.67.12.7654-7658.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang’Ondu R, Teal S, Park R, Heston L, Delecluse H, Miller G. DNA damage signaling is induced in the absence of Epstein—Barr virus (EBV) lytic DNA replication and in response to expression of ZEBRA. PLoS One. 2015;10(5):e0126088. doi: 10.1371/journal.pone.0126088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bailey SG, Verrall E, Schelcher C, Rhie A, Doherty AJ, Sinclair AJ. Functional interaction between Epstein-Barr virus replication protein Zta and host DNA damage response protein 53BP1. Journal of virology. 2009;83(21):11116–22. doi: 10.1128/JVI.00512-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Flemington EK. Herpesvirus lytic replication and the cell cycle: arresting new developments. Journal of virology. 2001;75(10):4475–81. doi: 10.1128/JVI.75.10.4475-4481.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rodriguez A, Armstrong M, Dwyer D, Flemington E. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. Journal of virology. 1999;73(11):9029. doi: 10.1128/JVI.73.11.9029-9038.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mosteiro L, Pantoja C, de Martino A, Serrano M. Senescence promotes in vivo reprogramming through p16 INK 4a and IL-6. Aging cell. 2018;17(2):e12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nature reviews Clinical oncology. 2018;15(4):234–48. doi: 10.1038/nrclinonc.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tosato G, Jones K, Breinig M, McWilliams H, McKnight J. Interleukin-6 production in posttransplant lymphoproliferative disease. The Journal of clinical investigation. 1993;91(6):2806–14. doi: 10.1172/JCI116523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tosato G, Tanner J, Jones KD, Revel M, Pike SE. Identification of interleukin-6 as an autocrine growth factor for Epstein-Barr virus-immortalized B cells. Journal of Virology. 1990;64(6):3033–41. doi: 10.1128/JVI.64.6.3033-3041.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Willard KA, Barry AP, Oduor CI, Ong’echa JM, Bailey JA, Moormann AM, et al. Viral and host factors drive a type 1 Epstein–Barr virus spontaneous lytic phenotype. Mbio. 2023;14(6):e02204–23. doi: 10.1128/mbio.02204-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902. e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome biology. 2019;20(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015;33(5):495–502. doi: 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Osorio D, Cai JJ. Systematic determination of the mitochondrial proportion in human and mice tissues for single-cell RNA-sequencing data quality control. Bioinformatics. 2021;37(7):963–7. doi: 10.1093/bioinformatics/btaa751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Linderman GC, Zhao J, Roulis M, Bielecki P, Flavell RA, Nadler B, et al. Zero-preserving imputation of single-cell RNA-seq data. Nature Communications. 2022;13(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–40. doi: 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nature methods. 2017;14(10):979. doi: 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Müller-Dott S, Tsirvouli E, Vazquez M, Ramirez Flores RO, Badia-i-Mompel P, Fallegger R, et al. Expanding the coverage of regulons from high-confidence prior knowledge for accurate estimation of transcription factor activities. Nucleic Acids Research. 2023;51(20):10934–49. doi: 10.1093/nar/gkad841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Badia-i-Mompel P, Vélez Santiago J, Braunger J, Geiss C, Dimitrov D, Müller-Dott S, et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinformatics Advances. 2022;2(1):vbac016. doi: 10.1093/bioadv/vbac016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Djavadian R, Hayes M, Johannsen E. CAGE-seq analysis of Epstein-Barr virus lytic gene transcription: 3 kinetic classes from 2 mechanisms. PLoS Pathog. 2018;14(6):e1007114. Epub 2018/06/05. doi: 10.1371/journal.ppat.1007114 ; PubMed Central PMCID: PMC6005644. [DOI] [PMC free article] [PubMed] [Google Scholar]