Abstract
Background:
Staphylococcus aureus has emerged as a major public health concern. It is a common pathogen in animal and human medicine.
Aim:
Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) was used to fingerprint 10 strains of S. aureus obtained from nasal swabs in domesticated animals and humans to ascertain how comparable the different strains’ genetic makeup was.
Methods:
These isolates were previously identified using standard molecular and microbiological methods. ERIC primers were amplified for all isolates. The dendrogram was generated using PGMA and the dice similarity coefficient. The strains were genotyped according to the diversity of sample source (human or animal), and the geographic source.
Results:
Ten S. aureus strains were classified into eight “ERIC” kinds (genotypes) using “ERIC-PCR genotyping”, in which the two most common clones were genotypes 8 and 2, which were represented by one strain from humans, one from cows, and two strains from sheep. Two strains derived from separate geographic areas and from different sample sources (human and cow) were determined to share the same genotype. Another two strains from different geographic areas but from the same sample source (sheep) were categorized under the same genotype. All the remaining strains were classified as a singular genotype.
Conclusion:
This study supports the possible bacterial transmission from animal to human and from animals themselves that usually happens during live animal marketing. Recognizing the interconnected nature of transmission systems and implementing the required approaches to disease prevention and control is essential for mitigating the risks posed by bacterial pathogens.
Keywords: Livestock-associated Staphylococcus aureus, Nasal strains, Clonal analysis, ERIC-PCR
Introduction
The “Livestock-associated Staphylococcus aureus (LA-SA)” is gaining attention as a significant zoonotic pathogen (Crespo-Piazuelo and Lawlor, 2012). LA-SA strains, primarily harbored in the nasal passages of livestock, have been increasingly identified as a source of infections in both animal and human populations (Cuny et al., 2015). People in activities that involve frequent animal interaction are especially susceptible to contracting LA-SA infections or becoming colonized by the pathogen, and further human-to-human transmission cannot be ruled out (Graveland et al., 2011). There is no host specificity for LA-SA in animals, and it can readily overcome species boundaries to colonize or infect a variety of other animals, including humans and animals including cattle, sheep, goats, poultry, dogs, cats, horses, rabbits, rats, and mink (Silva et al., 2023).
Understanding the genetic diversity of LA-SA strains circulating among animals and humans is crucial for elucidating transmission dynamics, assessing the risk of zoonotic transmission, and developing effective control strategies. This diversity analysis aims to investigate the genetic relatedness and distribution of LA-SA strains isolated from various animal species and humans (Bal et al., 2016). This can be employed using molecular typing techniques such as “whole-genome sequencing (WGS) or multi-locus sequence typing (MLST)” and “enterobacterial repetitive intergenic consensus PCR” (Sabat et al., 2013). This study aimed to enhance our understanding of the epidemiology and transmission dynamics of LA-SA in animals and humans.
Material and Methods
Bacterial isolates and “genomic DNA purification”
Using enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) fingerprinting, the genetic diversity and clonal relatedness of 10 isolates of S. aureus were examined in this study. These strains have previously been identified from the nasal cavities of human and domestic animals, including sheep, goats, cattle, and buffalo (Rahma, 2024; Rahma and Jwher, 2024). These strains were had been isolated from different geographic sources (Gogjalii, Wana, Jeleokhan, Mosul Falls, Hammam al-Alil, Badush, Baashiqah) in Mosul city, Iraq. Generally, in Mosul city, there is only one animal selling and buying live animal marketing with high-density animal gathering.
By using phenotypic techniques such as Grams’ stain, mannitol fermenter (yellow) colonies in “mannitol salt agar”, and double zone of hemolysis on “blood agar”, the isolates were identified as S. aureus (Rasol and Abdulrahman, 2023). While the typical greenish-colored colonies on “HiCrome Staph Selective Agar (HI Media, India)” was supported the identification process. The “thermonuclease (nuc)” gene, a particular target gene for S. aureus, was amplified by PCR as the final step in verifying these isolates (Abdulrahman, 2020).
According to Taha and Yassin (2019) thermal extraction process was used to extract the “genomic DNA”. The blood agar plates were inoculated with 100 µl of stock culture, and the incubation process was carried out as previously mentioned. Two to three pure colonies were chosen and put in a 1.5 ml tube along with 200 µl of sterile double-distilled water. Following an initial period of 15 seconds of vortexing, the mixture was heated to 95ºC for an interval of 10–12 minutes. After that, the mixture was quickly cooled with ice for 5 minutes, then centrifugation was applied to the cooled solution. For the “polymerase chain reaction (PCR)”, 150 µl of supernatant was utilized as a “template of DNA”. Through a nanodrop device, the quantity and clarity of DNA were evaluated.
ERIC PCR
Using primer sequences “(ERIC1: 5’-ATGTAAGCTCCTGGGGATTCAC-3’ and ERIC2: 5’-AAGTAAGTGACTGGGGTGAGCG-3’)”, which Ahmed (2023) recently adopted, this was used to genetically separate within strains. In a total volume of 25 µl, the PCR experiments were performed. A total of 12 µl of “hot start premix (Genedirex, Taiwan)”, 1 µl of primers (10 pmol), 2 µl of sample DNA (30–100 ng/µl), and up to 25 µl of “nuclease-free water (Qiagen, Germany)” were used in each reaction. Following the PCR program used by Taha (2022), the “9700 GeneAmp PCR machine (Applied Biosystem, USA)” was used to perform the PCR reactions. The initial denaturation lasted 5 minutes at 94°C. Following that, a total of 35 cycles of similar steps: 94°C for 1 minute, 52°C for 1 minute of altered annealing, and 72°C for 5 minutes. Finally, a 10-minute “post-PCR elongation” was conducted at 72°C. The amplified PCR products were loaded onto a 1.5% agarose gel and stained with “red safe DNA staining solution (GeNetBio, Korea)”. A 100-bp molecular size “standard DNA ladder (Genedirex, Taiwan)” was employed. In order to analyze the data, a photograph was taken.
“ERIC PCR” data analysis
In this investigation, 10 wells were analyzed, each representing one strain of S. aureus. This was done to determine whether or not there were any DNA bands on the “ERIC-PCR gel”. Following that, a software analysis was performed through “GelJ version 2.0” (available at https://sourceforge.net/projects/gelj/) to build a dendrogram. This approach is compatible with earlier research carried out by Heras et al. (2015). Using the “Dice similarity coefficient and the Unweighted Pair Group Method with Arithmetic Mean (UPGMA)”, the strains were separated accordingly. Strains were assigned to a single genotype if they showed a “correlation coefficient” of 90% or above, which is equivalent to a 90% similarity requirement (Taha, 2021). The geographic source (particularly, where the isolates derived from the same or distinct geographic places) and the variety of specimen origins (human or animal) were used to categorize the strains.
Ethical approval
Not needed for this study.
Results
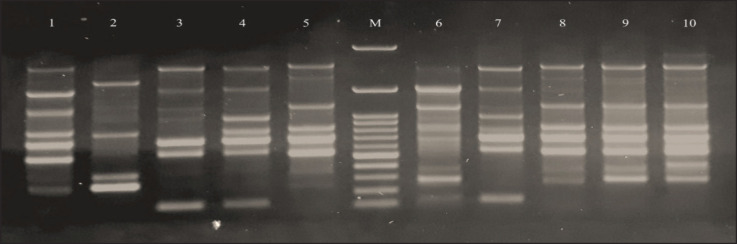
The findings of the “ERIC-PCR fingerprinting” analysis (Fig. 1) demonstrated that there was a genetic similarity range of 60%–94% between the S. aureus strains. The degree of the difference depended on the differences that were noticed in the number and size of “ERIC sequences” in each strain. Eight distinct genotypes were found as a consequence of this “ERIC fingerprinting”.
Fig. 1. ERIC-PCR DNA fingerprint patterns of 10 S. aureus strains isolated from nasal swabs from different animal species and human. Lane M: 100 bp DNA ladder, Lane 1–10 represents studied samples.
However, it was shown that two strains (strain numbers 9 and 8) that originated from distinct specimen sources (cow and human) as well as distinct geographical regions shared an identical genotype (genotype 2). Additionally, another 2 strains (genotype 8; strain number 1 and 2) from different geographic areas but from the same sample source (sheep) were categorized under the same genotype. The other strains (n = 6; 60%) were categorized as a singular genotype because of the significant genetic variation found in them, even though they came from the same geographic area and were collected from an identical source (Fig. 2).
Fig. 2. The ERIC-PCR dendrogram displays the banding patterns of 10 S. aureus strains isolated from nasal swabs from different animal species and human sources.
Discussion
Studying bacterial genetic diversity is crucial for tracking the spread of infectious diseases. Genetic diversity studies help identify the sources of outbreaks, track transmission routes, and implement effective control measures (Bentley and Parkhill, 2015). The current study has proven the existence of diversity in the genotypes of methicillin-resistant S. aureus isolated from different regions and sources, due to differences in hosts, type of culture, geographical location, diversity of genotypes of S. aureus and its unlimited spread, as well as the methods used in using medications to treat the infection, and this is what the researchers (Hidron et al., 2005; Shekarabi et al., 2017) indicated.
For these reasons, this study was undertaken to assess the extent of the genetic diversity and similarity between S. aureus strains from different animal species in the same and different geographical source areas and to find out the possibility of bacterial transmission between animals and humans.
In this study, a high genotypic pattern (8 genotypes out of 10 strains) was observed. This shows that the bacteria have a high degree of genetic diversity and that many S. aureus clones are present in these flocks of animals, which may originate from different geographical areas as Iraq imports animals from neighboring countries. Point mutations or the insertion or deletion of particular DNA sequences are generally responsible for producing microbial genetic variation. Because of this, microbial communities will exhibit strain differences, which may happen after they have passed through different hosts or habitats (Jerome et al. 2011). This is a good explanation of why there is such genetic diversity seen among S. aureus strains in this study.
However, two strains from separate geographic areas and from different sample sources (Gokjali and Glikhan), which were sourced from humans and cows, and another two isolates from sheep, which were from the areas of Wana and Gokjali, were categorized under the same genotype. This suggests that these strains share very similar genetic characteristics despite their geographical and environmental differences. This could be attributed to a cross-transmission between these flocks, and this could be linked to the fact that the mixing of animals and their passage through common grazing areas between the study areas, in addition to the mixing of animals and humans in markets selling live animals (Shekarabi et al., 2017; Rahmani et al., 2023). However, there are many factors that could support the transmission of bacterial species from animals to humans; one of these is the close proximity of humans and various species of live animals in these markets, which facilitates the direct transmission of pathogens through contact with animal feces, saliva, and urine (Fong, 2017). The market environment, including surfaces and equipment, can become contaminated with bacteria shed by animals, increasing the risk of indirect transmission to humans who come into contact with these contaminated surfaces (Bloomfield and Ackerley, 2023).
Conclusion
Bacterial transmission between animals and humans, as well as among animals themselves, is a complex and multifaceted phenomenon with far-reaching implications for public health, agriculture, and conservation. Recognizing the interconnected nature of these systems and implementing integrated approaches to disease prevention and control is essential for mitigating the risks posed by bacterial pathogens and promoting health and well-being across species boundaries.
Acknowledgments
The authors are grateful for the support provided by the Duhok Research Centre, College of Veterinary Medicine, Duhok, Iraq, and the College of Veterinary Medicine, Mosul, Iraq.
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this research.
Funding
Self-funding.
Data availability
All data are provided in the manuscript.
Authors’ contributions
The experiment was designed by the first and third researchers, and the other researchers participated in the work, analyzing the results, and writing the manuscript, including the financial costs.
References
- Abdulrahman R.F. Detection of Staphylococcus aureus from local and imported chicken in Duhok Province/Kurdistan region of Iraq using conventional and molecular methods. Bas. J. Vet. Res. 2020;19(1):134–146. [Google Scholar]
- Ahmed M.S., Abdulrahman Z.F.A., Taha Z.M.A. Risk factors of clonally related, multi, and extensively drug-resistant Acinetobacter baumannii in severely Ill COVID-19 patients. Can. J. Infect. Dis. Med. Microbiol. 2023;2023:3139270. doi: 10.1155/2023/3139270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal A.M., Coombs G.W., Holden M.T., Lindsay. J.A., Nimmo G.R., Tattevin P., Skov R.L. Genomic insights into the emergence and spread of international clones of healthcare-, community-and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J. Glob. Antimicrob. Resist. 2016;6:95–101. doi: 10.1016/j.jgar.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Bentley S.D., Parkhill J. Genomic perspectives on the evolution and spread of bacterial pathogens. Proceedings of the Royal Society B: Biol. Sci. 2015;282:20150488. doi: 10.1098/rspb.2015.0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield S.F., Ackerley L.M. Developing resilience against the threat of infectious diseases and anti-microbial resistance: putting targeted hygiene into practice in home and everyday lives. Public Health Pract. (Oxf). 2023;5:100362. doi: 10.1016/j.puhip.2023.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Piazuelo D., Lawlor P.G. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Irish Vet. J. 2012;74:1–2. doi: 10.1186/s13620-021-00200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny C., Wieler L.H., Witte W. Livestock-associated MRSA: the impact on humans. Antibio. 2015;4(4):521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong I.W. Animals and mechanisms of disease transmission. Emerg Zoonoses. 2017;8:15–38. [Google Scholar]
- Graveland H., Duim B., van Duijkeren E., Heederik D., Wagenaar J.A. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med. Microbiol. 2011;301:630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Heras J., Domínguez C., Mata E., Pascual V. GelJ—a tool for analyzing DNA fingerprint gel images. BMC Bio. 2015;16(1):1–8. doi: 10.1186/s12859-015-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron A.I., Kourbatova E.V., Halvosa J.S., Terrell B.J., McDougal L.K., Tenover F.C., Blumberg H.M., King M.D. Risk factors for colonization with methicillin-resistant Staphylococcus aureus (MRSA) in patients admitted to an Urban Hospital: emergence of community-associated MRSA Nasal Carriage. Clin. Inf. Dis. 2005;41(2):159–166. doi: 10.1086/430910. [DOI] [PubMed] [Google Scholar]
- Jerome J.P., Bell J.A., Plovanich-Jones A.E., Barrick J.E. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One. 2011;6(1):1–11. doi: 10.1371/journal.pone.0016399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahma H., Jwher Dh. Detection of clfA, clfB and coa genes in methicillin-resistant Staphylococcus aureus (MRSA) isolated from Nasal Cavity of Cows, Buffalo and their Breeders in Nineveh Governorate, Iraq. J. App. Vet. Sci. 2024;9(3):1–10. [Google Scholar]
- Rahma H.Y. Isolation and identification of methicillin-resistant Staphylococcus aureus (MRSA) in Nasal Carriage from domestic animals and those handling them in Nineveh governorate. Unpublished Master Thesis, College of Veterinary Medicine, University of Mosul, Mosul, Iraq. 2024 [Google Scholar]
- Rahmani Z., Hosseini S.S., Bagheri P., Goudarzi M. Genetic diversity of Staphylococcus aureus isolated from ear infections in Iran: emergence of CC8/ST239-SCCmec III as major genotype. Acta Microbiol Immunol Hung. 2023;70(3):231–238. doi: 10.1556/030.2023.02081. [DOI] [PubMed] [Google Scholar]
- Rasol V.A., Abdulrahman R.F. Detection and molecular characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) nasal carriage isolates from healthy domestic animal in Duhok Province. Egy. J. Vet. Sci. 2023;54(2):263–273. [Google Scholar]
- Sabat A.J., Budimir A., Nashev D., Sá-Leão R., Van Dijl J.M., Laurent F., Grundmann H., Friedrich A.W. ESCMID Study Group of Epidemiological Markers (ESGEM. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eurosurveillance. 2013;18(4):20380. doi: 10.2807/ese.18.04.20380-en. [DOI] [PubMed] [Google Scholar]
- Shekarabi M., Hajikhani B., Salimi Chirani A., Fazeli M., Goudarzi M. Molecular characterization of vancomycin-resistant Staphylococcus aureus strains isolated from clinical samples: A three-year study in Tehran, Iran. PLoS One. 2017;12(8):e0183607. doi: 10.1371/journal.pone.0183607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva V., Araújo S., Monteiro A., Eira J., Pereira J.E., Maltez L., Igrejas G., Lemsaddek T.S., Poeta P. Staphylococcus aureus and MRSA in livestock: antimicrobial resistance and genetic lineages. Micro. 2023;11:124. doi: 10.3390/microorganisms11010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha Z.M. Genetic diversity and clonal relatedness of Aeromonas hydrophila strains isolated from hemorrhagic septicemia’s cases in common Carp (Cyprinus carpio) farms. Iraqi J. Vet. Sci. 2021;35(4):643–648. [Google Scholar]
- Taha Z.M. Genotyping and genetic diversity of corynebacterium pseudotuberculosis strains isolated from Caseous Lymphadenitis in sheep and goat. Exp. Ani. Med. Res. 2022;12(1):85–90. [Google Scholar]
- Taha Z.M., Yassin N.A. Prevalence of diarrheagenic Escherichia coli in animal products in Duhok province, Iraq. Iran J. Vet .Res. 2019;20(4):255–262. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in the manuscript.