Abstract
Background:
Porcine epidemic diarrhea virus (PEDV) is a recurring coronavirus that causes severe diarrhea in pigs with high mortality and morbidity rates, especially in neonatal pigs. Despite the availability of vaccines, their efficacy is limited owing to antigenic differences between the vaccine and field strains, which poses a challenge to infection control. Antiviral drugs targeting conserved PEDV proteins show promise for complementing vaccination strategies. PEDV Nsp3 (PL2Pro) and Nsp5 (3CLPro) are essential proteases vital for viral replication, making them attractive targets for drug development against PEDV.
Aim:
To address the lack of therapeutics against recurring PEDV outbreaks and bridge the gap in the application of bioinformatics in veterinary drug discovery, this study aimed to discover compounds that inhibit PEDV proteases from Philippine medicinal plants by applying a modified virtual screening methodology that considers the physiology of swine hosts.
Methods:
This study employed a library of 690 metabolites from Philippine medicinal plants to screen for potential protease inhibitors targeting PEDV PL2Pro and 3CLPro. This includes evaluating the binding affinity, pharmacokinetics, dynamic stability, and critical binding site residues. Compounds demonstrating high affinity underwent a modified ADMET analysis, considering the enteric localization of the virus and potential toxicity to swine hosts. Furthermore, molecular dynamics simulations assessed compound stability under physiological swine conditions.
Results:
The study identified Bisandrographolide from Andrographis paniculata, CID 162866964 from Euphorbia neriifolia, and betulinic acid from Vitex negundo and Ocimum basilicum as metabolites that bind favorably and selectively to PEDV 3CLPro and have excellent pharmacokinetic properties and dynamic stability. In contrast, no selective inhibitor for PL2pro passed the same criteria.
Conclusion:
Employing the modified virtual screening protocol tailored for swine host considerations, the compounds identified in this study are anticipated to exert inhibitory effects against PEDV without off-target binding to analogous swine proteases and receptors. CID 162866964, bisandrographolide, and betulinic acid show promise for developing potent antivirals against PEDV.
Keywords: Antiviral, Drug discovery, PEDV, Veterinary drug, Virtual screening
Introduction
Porcine epidemic diarrhea is a viral enteric disease that affects pigs at various growth stages but is especially lethal to young and neonatal pigs (Lee, 2015; Paraguison-AliLee, 2015 Domingo, 2016; Salamat et al., 2021; Orosco, 2023). The disease is caused by the porcine epidemic diarrhea virus (PEDV), an alpha coronavirus belonging to the family Coronaviridae, which localizes in the swine intestinal mucosa and causes severe diarrhea, vomiting, dehydration, wasting, and decreased reproductive performance (Lee, 2015). Mortality and morbidity rates associated with the disease have caused significant economic losses in the global livestock industry, particularly in swine-producing countries in Southeast Asia, such as the Philippines (Lee, 2015; Paraguison-AliLee, 2015 Domingo, 2016). Viral disease recurrence and the emergence of new recombinant strains make treatment and control initiatives imperative.
Due to the historically recurring nature and the ever-present threat this viral infection poses to the swine industry, various control efforts have been implemented, including biosecurity measures, intentional exposure of sows for feedback immunization, and vaccination. Despite the availability of vaccines, their effectiveness is often limited by various factors, including variations between the viral strains used in the vaccine and field epidemic strains (Lee, 2015; Zhang et al., 2022). Hence, managing the viral infection must not rely entirely on vaccines. Although vaccines protect healthy hogs from exposure, no treatment options are currently available for hogs with active infections, which are, in most cases, deemed futile. This necessitates discovering PEDV-specific antiviral drugs that can be used in active cases and serve as a complementary strategy to vaccination.
Traditional drug discovery pipelines are notorious for being lengthy and prone to failure in clinical trials, which not only costs billions of years for the industry but also sets back drug development to years (Anderson, 2003; DiMasi et al., 2016; Wang and Zhu, 2016; Batool et al., 2019; Orosco, 2023). Consequently, modern pharmaceutical research and development prioritizes the early and cost-effective identification of promising candidates (DiMasi et al., 2016). Computational methods have emerged as a solution, enabling researchers to screen existing drug libraries for repurposing, evaluate natural products for bioactivity, and predict pharmacokinetic behavior (Anderson, 2003; Cheng et al., 2012; DiMasi et al., 2016; Wang and Zhu, 2016; Batool et al., 2019). Structure-based drug discovery is a common approach employed in the computational screening of potential drugs. This method involves the identification of compounds with high affinity for known viral targets that are essential in the viral replication cycle, thereby inhibiting viral proliferation (Anderson, 2003; Cheng et al., 2012; Batool et al., 2019).
Among the various proteins associated with PEDV, proteases play vital roles in viral replication and pathogenesis. Hence, targeting proteases is a promising strategy for developing antiviral drugs. Notably, two essential proteases, 3CLpro and PL2pro, are desirable targets because of their crucial function in the viral life cycle.
Philippine medicinal plants possess a wide range of biological activities, including antiviral effects (Dayrit et al., 2021) and good pharmacokinetic properties, making them a valuable source of potential lead compounds in drug discovery. Veterinary informatics (or vetinformatics) is an emerging interdisciplinary research area that utilizes bioinformatics to solve challenging veterinary science issues (Sujatha et al., 2018; Pathak and Kim, 2022; Kim and Pathak, 2023). One issue that vetinformatics aims to address is the gap in the use of computational drug discovery in developing veterinary drugs (Pathak and Kim, 2022). However, despite developments in computational drug design and biochemical simulations, there is still a scarcity of tools adapted to animal models, as the tools focus more on drugs against human diseases and pathogens (Sujatha et al., 2018; Pathak and Kim, 2022; Kim and Pathak, 2023). Even so, several in silico drug screening studies have been applied to swine pathogens, such as PEDV (Pathak et al., 2023), porcine reproductive and respiratory syndrome virus (Pathak et al., 2022a,b, 2023), and African Swine Fever Virus (Macalalad and Orosco, 2024), albeit lacking modifications in the protocol for animal host considerations.
To address the lack of PEDV-specific antiviral drugs to complement vaccination strategies and take advantage of the bioactivity of the Philippines’ medicinal plant resources, this study applied a modified computational drug discovery protocol to screen 690 secondary metabolites from Philippine medicinal plants for potential inhibitory activity against PEDV 3CLPro and PL2Pro. Conversely, a modified virtual screening protocol adapted to swine host conditions bridges the gap in vet-informatics and veterinary drug discovery to study the compounds’ pharmacokinetics and stability in swine host conditions.
Materials and Methods
Modified virtual screening protocol for veterinary application
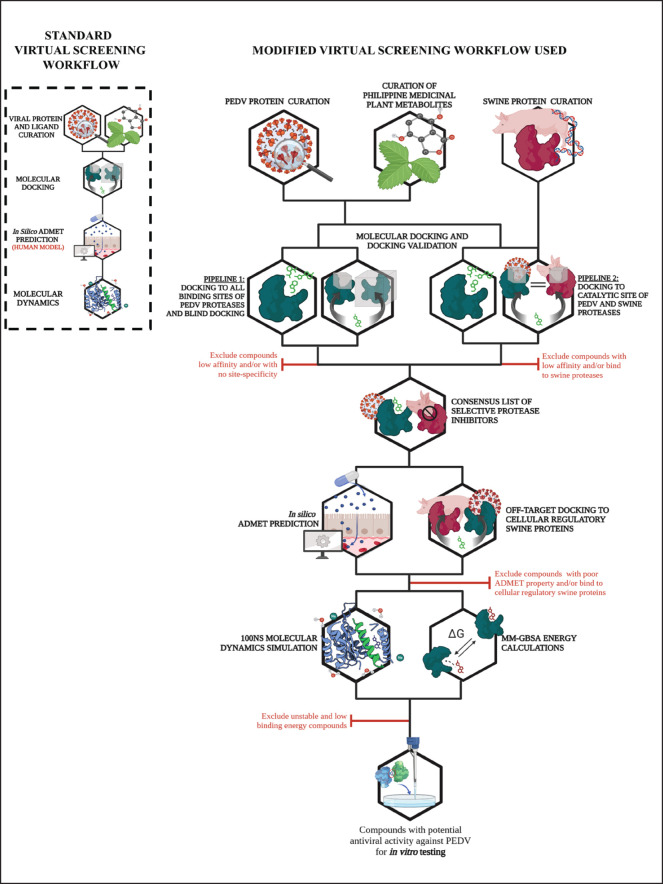
A modified multi-level virtual screening protocol was used to screen the metabolites of Philippine medicinal plants for potential antiviral activity against PEDV 3CLPro and PL2Pro. Each level of the virtual screening protocol has strict criteria and validations for the exclusion of poor-performing candidates from further analyses. Modifications were made to the standard virtual screening setup to consider the different physiological conditions of pigs from those of standard human-centered protocols. Figure 1 presents the methodological framework for virtual screening used in this study, contrasting with the standard virtual screening protocol.
Fig. 1. Modified virtual screening protocol adjusted for veterinary (i.e., swine) drug discovery compared to the standard protocol (enclosed in box). Created with BioRender.com.
The standard virtual screening protocol enclosed in a box shown in Figure 1 is primarily designed for human drug discovery studies, focusing on human physiological and metabolic parameters. The tools needed for the protocol all use human models; hence, the protocol is much more straightforward. However, when applying these methods to veterinary applications, such as developing antiviral drugs for swine, it is crucial to adapt the screening process to account for species-specific differences in physiology, biochemistry, and drug metabolism.
In this study, modifications were made to the standard virtual screening protocol to accommodate the physiological conditions of pigs. These modifications included the addition of cross-docking experiments to determine the preference of the ligands for viral protease as opposed to the analogous swine proteases, off-target docking to cellular regulatory proteins of swine hosts to determine promiscuity and unwanted interactions, adjusted physicochemical property thresholds to account for enteric localization in swine host, and finally, simulation of the ligand-protease complex using swine physiological conditions. All the modifications were made to ensure the identified compounds were safe and effective for veterinary use.
Ligand curation and standardization
Dayrit et al. (2021) identified 10 Philippine medicinal plants (Allium sativum, Andrographis paniculata, Cocos nucifera, Euphorbia hirta, Euphorbia neriifolia, Moringa oreifera, Ocimum basilicum, Piper nigrum, Zingiber officinale, Vitex negundo)) as sources of antiviral compounds for drug discovery. The initiative aims to inform research efforts seeking potential solutions against viral pathogens such as SARS-CoV-2 and the Nipah virus; hence, it was employed in this study.
Six hundred ninety secondary metabolites (Table S1) from the ten Philippine medicinal plants mentioned earlier were curated by sourcing information from chemical repositories. Data on these compounds were obtained from PubChem (www.pubchem.com) (Kim and Pathak, 2023), FooDB version 1.0 (www.foodb.ca), and Dr. Duke’s Phytochemical and Ethnobotanical Databases of the United States Department of Agriculture (www.phytochem.nal.usda.gov).
Simplified Molecular Input Line-Entry System identifiers were used to construct 3D structures using OpenBabel (O’Boyle et al., 2011), which was then standardized in alvaMolecule (Mauri and Bertola, 2022). Standardization procedures include eliminating duplicates, correcting non-standard valences, and neutralizing charged atoms or molecules. For any corrections necessitating the removal or isolation of segments within multi-molecular structures, manual adjustments were carried out using BIOVIA Discovery Studio Visualizer version 21.1.0.20298.
Subsequently, the corrected 3D structures were minimized using Avogadro (Hanwell et al., 2012) utilizing a 5,000-step conjugate gradient minimization protocol with Merck Molecular Forcefield 94 (Halgren, 1996). These minimized structures were exported as PDBQT files for molecular docking studies.
Protein curation and preparation
The experimental structures of PEDV 3-chymotrypsin-like (3CLpro) (6W81) and papain-like (PL2pro) (7F0U) proteases were obtained from the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics (RCSB-PDB) (Berman et al., 2000).
For proteins with incomplete structures, corrective measures such as amputation of available structures, rotamer addition (Shapovalov and Dunbrack, 2011), and hydrogen addition was undertaken, followed by minimization using ANTECHAMBER (Wang et al., 2004). The resultant prepared protein structures were then converted to the PDBQT format using AutoDock Tools (Forli et al., 2016).
The co-crystalized ligand of PEDV 3CLpro, X77 or N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide, was isolated from the structure and used for re-docking experiment to validate the docking protocol. On the other hand, only the protein ubiquitin and no small molecule ligands were bound to the PL2pro structure and hence were not used in re-docking.
Crystal structures were visualized using UCSF Chimera to identify the binding sites of these proteins, as reported in the relevant literature, and their close homologs (Pettersen et al., 2004). Molecular docking grid box parameters were generated by covering the identified binding sites with a cubic search space of suitable dimensions. Additionally, a separate grid box covering the entire protein was employed for blind molecular docking to confirm preferred binding at the identified sites. The coordinates and dimensions of each grid box were determined using the AutoDock Vina (Trott and Olson, 2010) plugin of UCSF Chimera. Six grid boxes were used to examine ligand binding to PEDV proteases. The coordinates and dimensions of the grid boxes, including the key interacting residues from the literature, are presented in Table 1.
Table 1. Grid boxes used for site-specific and blind docking protocols.
| Protein | Coverage | Coordinates | Dimensions | Key residues | ||||
| X | Y | Z | X | Y | Z | |||
| 3CLpro | Whole (Blind) | −10.0521 | −5.12531 | 12.9697 | 50 | 75 | 45 | -- |
| 3CLpro | Catalytic Site | 3.125 | 9.46471 | 13.3693 | 25 | 25 | 25 |
His-Cys Catalytic Dyad HIS41, CYS144 |
| 3CLpro | Dimerization Site | −4.80755 | −8.71609 | −0.06256 | 30 | 50 | 25 |
Dimer interface ALA1, GLY2, ARG4, ALA7, SER10, GLY11, GLU14, GLY26, GLU286, SER138, PHE139. GLU165, GLY278, THR281, GLN295 |
| PL2pro | Whole(Blind) | −3.48628 | −19.8137 | 12.7294 | 45 | 70 | 45 | -- |
| PL2pro | Ubiquitin binding site | −3.817 | −22.1078 | 21.9595 | 25 | 25 | 25 |
Core ASN103, ASN138, ASN100 Tail ASP97, GLN94, GLY202, SER96, ASN43, SER44, GLY202, HIS203 |
| PL2pro | Catalytic Site | 10.7159 | −7.39618 | 18.321 | 20 | 20 | 15 |
Asp-His-Ser Catalytic Triad ASP216, HIS203, SER44 |
In addition to viral proteases, binding to off-targets, including analogous swine proteases (Table S2) and cellular and regulatory proteins (Table S3), was also considered as a descriptor of toxicity and to accommodate potential veterinary applications. Homology models of these off-target swine receptors and proteases were generated from homologous proteins with available crystal structures from the RCSB-PDB using SWISS-MODEL (Schwede et al., 2003). The co-crystallized ligands of the reference protein homologs used for receptor modeling served as positive controls for assessing ligand affinity. The resulting homology models were converted to PDBQT files using AutoDock tools for subsequent analyses.
Molecular docking
This study aimed to identify potential inhibitors targeting PEDV proteases while ensuring they do not bind to swine proteases. Given the inherent unpredictability of the ligand dataset and the possibility of yielding zero specific inhibitors, a conservative approach is necessary. Therefore, parallel analyses were conducted using these two pipelines.
The first pipeline involved standard targeted docking to catalytic, oligomerization, and substrate-binding sites, followed by blind docking validation.
The second pipeline utilized the results of targeted docking to the catalytic sites from pipeline one and compared them to the affinities of ligands for analogous swine proteases in cross-docking analysis.
Notably, only a single instance of site-specific docking was performed to target all binding sites. A critical distinction between the pipelines lies in analyzing the results; the first pipeline examined all grid box results, whereas the second pipeline only utilized the results from the catalytic site docking.
The top 10 compounds from both pipelines were compared to obtain a consensus list of high-affinity (≤ −7.0 Kcal mol−1), site-specific, and selective protease inhibitors. This parallel approach allows for a comprehensive assessment while mitigating the risk of overlooking potential inhibitors or encountering false positives.
General docking protocol
An in-house Python script was used for automated molecular docking using AutoDock Vina 1.1 (Trott and Olson, 2010) and incorporating clustering across multiple runs. Triplicate static or rigid-receptor docking was conducted using predefined grid box coordinates (refer to Table 1 for PEDV proteases, Table S2 for swine proteases, Table S3 for cellular regulatory swine proteins) covering either a specific binding site (targeted) or the entire protein (blind), with a search exhaustiveness set to 32. Poses from these three runs were clustered based on the AutoDock 4.0 protocol (Morris et al., 2009), employing a root mean square deviation (RMSD) cut-off of 2.0 Å. Binding affinity was computed as the average per cluster, with the top pose from the top clusters designated as the reference pose. A threshold of −7.0 Kcal mol−1 (Wong et al., 2022) was adapted to identify ligands with high affinity (≤ −7.0 Kcal mol−1), which are more likely to exhibit bioactivity.
Validation of docking protocol
Before data collection using the two docking pipelines, the general Vina docking protocol was validated by re-docking the co-crystalized ligands of the target proteases, 3CLpro and PL2pro. As mentioned in the previous sections, only 3CLpro has an available small molecule ligand, X77, since the ligand of PL2pro is ubiquitin. Hence, only molecular docking to 3CLpro was validated by re-docking.
The structure of X77 obtained from the previous section was reparametrized using the same ligand preparation protocol previously discussed and was docked to 3CLpro using the same Vina protocol. The resulting docked pose was then compared to its native pose by calculating the heavy-atom (hydrogens were not considered) RMSD. Re-docking is deemed successful when the heavy atom RMSD between the crystal pose and the docked pose is less than or equal to 1.5 Å (Hevener et al., 2009).
Standard docking pipeline and blind docking validation for site-specificity
Two standard docking setups were used in the first docking pipeline of the data collection proper. The first setup involves site-specific docking to the catalytic, oligomerization, and protein interaction interfaces of PEDV proteases. This approach allows for broad coverage evaluation of ligand interactions and the discovery of additional binding modes. Subsequently, a blind docking setup was employed to validate the preferential binding of the metabolites to specific active sites. An RMSD cut-off of 1.5 Å, as adapted from Hevener et al. (2009), was utilized as a reference value for the similarity between poses from site-specific and blind molecular docking. To be deemed valid, poses generated from blind docking must closely resemble those generated from targeted docking despite the larger search space.
Only metabolites meeting the RMSD cut-off and exhibiting binding affinities ≤ −7.0 Kcal mol−1 (Wong et al., 2022) were considered valid and subjected to further analysis. The conformations of the enzyme-ligand complexes were visualized and analyzed using UCSF Chimera and Biovia Discovery Studios v. 4.1.
Cross-docking pipeline for protease selectivity
In the second pipeline, cross-docking experiments were conducted to ascertain ligand selectivity and binding preferences toward the catalytic site of PEDV proteases compared to structurally analogous swine proteases. Notably, PEDV 3CLpro and PL2pro have structural counterparts in swine hosts. Specifically, PEDV 3CLpro demonstrates a structural resemblance to other proteases in similar family such as elastase (3HGN), trypsin (1Z7K), and kallikrein (1HIA), whereas PL2pro shares similarities with Cathepsin B (3PBH), Cathepsin H (8CPH), and Cathepsin L (1CS8). The cross-docking experiment rigorously evaluated the affinity of the ligands towards all six analogous swine proteases, shedding light on their potential promiscuity.
Selective antivirals against PEDV 3CLPro and PL2Pro catalytic sites were determined by first clustering the phytochemicals through either Butina clustering with ECFP4-like Morgan fingerprinting (r = 2, cutoff = 0.5) in the rdkit Python library (O’Boyle and Sayle, 2016) or Scaffold Hunter (Wetzel et al., 2009). In both methods, the selected compound clusters must have an average binding score against the catalytic sites stronger than the −7.0 Kcal mol−1 threshold. The binding score to swine proteases was strongly correlated (R2 = 0.88 to 0.92) with binding to viral protease, which led to inflation of the difference in protease binding at stronger binding regimes. Selectivity was measured by calculating a particular cluster’s residual (>0.3) in the regression model of the binding score against the viral protease to the average binding score against the respective swine proteases. The residual threshold was validated to correspond to a standardized residual greater than one, significantly greater than the bootstrapped mean standardized residual (0) at α = 0.05.
Consensus of parallel docking pipelines
The integration of parallel docking pipelines has facilitated the development of a robust consensus list of protease inhibitors, addressing potential challenges, such as obtaining no compound that meets all the criteria, while ensuring comprehensive exploration. Employing stringent criteria, including a binding affinity threshold of −7 Kcal mol−1 (Wong et al., 2022), guided the selection process, ensuring the inclusion of compounds with robust ligand-protein interactions. Additionally, the compounds identified through the initial pipeline demonstrated validated site-specificity. In contrast, those from the secondary pipeline exhibited confirmed selectivity for PEDV proteases, thus minimizing the risk of off-target effects on swine host proteases. A carefully curated consensus list was established by consolidating the top candidates from both pipelines, providing a solid foundation for subsequent analyses and further characterization.
In silico ADMET analysis
The standard in silico ADMET workflow was adapted to integrate considerations specific to swine hosts. Modifications encompass cross-docking experiments involving analogous swine proteases, off-target docking to cellular regulatory swine proteins for potential toxicity, and adjustment of ADMET thresholds to account for enteric localization of PEDV in swine hosts.
Compounds from the consensus list of high affinity(≤ −7.0 Kcal mol−1), site-specific, and selective protease inhibitors resulting from parallel pipelines underwent thorough evaluation of their predicted ADMET properties using a range of metrics. These metrics include the total count of violations based on Lipinski’s Rule of Five, as well as ADMET parameters obtained from ADMETLab 2.0 (https://admetmesh.scbdd.com/) (Xiong et al., 2021) and SWISS-ADME (http://www.swissadme.ch) (Daina et al., 2017) web servers. The calculated parameters and expected values are presented in Table 2.
Table 2. ADMET endpoints examined and their respective thresholds.
| Property | Server | Output | Ideal result |
| Lipinski’s rule of five | |||
| MW HBA HBD Log P |
ADMETLab 2.0 | g/mol count count Log P value |
≤500 ≤10 HBA ≤ 5 HBD 2–5 Log P* |
| Absorption | |||
| Caco-2 permeability | ADMETLab 2.0 admetSAR |
log cm/second +/- |
>−5.15 log cm/second + |
| Gastrointestinal absorption | ADMETLab 2.0 SwissADME admetSAR |
p (0 to1) High/Low +/- |
p < 0.3 High + |
| P-glycoprotein (P-gp) inhibitor/substrate | ADMETLab 2.0 SwissADME admetSAR |
p (0 to1) Yes/No +/- |
p < 0.7* No - |
| Distribution | |||
| Blood-brain barrier permeation | ADMETLab 2.0 SwissADME admetSAR |
p (0 to1) Yes/No +/- |
p < 0.7 No - |
| Plasma protein binding | ADMETLab 2.0 admetSAR |
% bound | <90% |
| Metabolism and excretion | |||
| Cytochrome P450 (3A4, 1A2, 2C19, 2C9, 2D6) substrate/inhibitor | ADMETLab 2.0 SwissADME admetSAR |
p (0 to1) Yes/No +/- |
p < 0.7* No - |
| Toxicity | |||
| AMES mutagenicity | ADMETLab 2.0 admetSAR |
p (0 to1) +/- |
p < 0.7 - |
| hERG cardiotoxicity | |||
| Hepatotoxicity | |||
| Carcinogenicity |
The threshold of the parameters marked with a star (*) was adjusted for maximum penetration and lower first-pass metabolism, leading to increased bioavailability in the gut. Compounds with no more than one violation were selected as lead compounds and analyzed for dynamic stability in molecular dynamics (MD) simulations.
Off-target docking
In silico ADMET analyses rely on computational models to simulate the pharmacokinetic and pharmacodynamic properties of human drugs (Daina et al., 2017; Guan et al., 2018; Xiong et al., 2021). Although human and swine ADME properties share similarities and are often extrapolated in preclinical studies (Henze et al., 2019), it is essential to highlight that the toxicities reported by these computational tools and servers are specific to humans and human receptors.
To mitigate this disparity, we investigated the potential off-target effects of ligands by docking them into receptors associated with vital cellular functions in swine. Ligand docking to X-linked inhibitor of apoptosis protein, mouse double minute 2 homolog, and Bcl-2-associated X protein was performed to assess their influence on apoptotic pathways to identify perturbations in programmed cell death regulation. DNA Polymerase epsilon, Topoisomerase I, and dihydrofolate reductase were used to evaluate their impact on DNA replication fidelity and nucleotide metabolism. Docking analysis of eukaryotic initiation factor 4E and the complex mammalian target of rapamycin and FK506-binding protein-rapamycin-associated protein explored alterations in protein synthesis and cellular signaling pathways, respectively. Tubulin and F0F1 ATP synthase (F0F1 ATPs) were examined to understand the potential disruptions in energy metabolism and cytoskeletal dynamics. Additionally, interactions with methionyl-tRNA synthetase, pyruvate dehydrogenase complex component E1, and acyl-CoA dehydrogenase were explored to assess their effects on metabolic processes. Through additional off-target analysis, we elucidated the binding affinity and mode of interaction between ligands and these diverse protein targets, providing insights into their potential toxicity or off-target effects on swine physiology and health.
Subsequently, compounds exhibiting favorable results in the protease selectivity test, standard ADME metrics, and swine off-target interactions were chosen for stability analysis through MD simulations.
MD simulations
MD simulations were performed using GPU-enabled GROMACS 2023.3 (Abraham et al., 2015). Protein parameterization utilized the July 2022 release of the CHARMM36 force field (Best et al., 2012), whereas ligands were parameterized using the CHARMM General Force Field (CGenFF) (Vanommeslaeghe et al., 2010).
The systems were enclosed within a cubic box with 15 Å padding and solvated using the TIP-3P solvent model. Sodium (Na+) and chloride ions (Cl-) were introduced into the system at a concentration of 0.150 M. Simulation parameters included a leapfrog integrator, LINCS for controlling covalent bonds, a maximum distance of 12 Å for nonbonded interactions, a modified Berendsen thermostat for temperature coupling, and active periodic boundary conditions.
The system’s energy minimization was conducted until the maximum force converged below 10 KJ mol−1 or reached a maximum of 50,000 steps of steepest descent. Isochoric equilibration ensembles were performed with 100 ps of relaxed pressure coupling at 312 K. Isobaric equilibration ensembles were executed through 100 ps of isotropic Berendsen pressure coupling in NPT (1 ps timestep, 4.5 × 10−5 bar−1 compressibility) at 312 K.
Subsequently, production runs were conducted in triplicate runs of isotropic Parrinello-Rahman pressure coupling for 100 ns, with frames generated every 0.1 ns (1,000 frames) at 312 K. The same preparation, minimization, equilibration, and production run protocols were applied uniformly across all analyses, including runs for drug complexes as positive controls and apoproteins as negative controls.
Post-MD analysis
The stability of the complexes was assessed using several parameters, including the RMSD, which was computed using Equation 1, the root-mean-square fluctuation (RMSF) described by Equation 2, the radius of gyration (Rg), and the hydrogen bond count from analysis tools available in GROMACS.
| (1) |
| (2) |
The gmxMMPBSA tool (Valdés-Tresanco et al. 2021) was used to determine the binding energies of the complexes. The gmxMMPBSA tool can use two methods in calculating binding free energies: Molecular Mechanics-Poisson-Boltzmann Surface Area and Molecular Mechanics-Generalized Born Surface Area (MM-GBSA). The calculation method used for the set of complexes is the MM-GBSA method, which uses the GB-Neck2 model (Su et al., 2015) and the recommended mbondi3 radii set alongside default parameters, a relatively simpler model and results in shorter calculation times.
The binding and total free energies were calculated using Equations 3, 4, and 5. However, it is important to note that the calculation of the entropic term in Equation 5 is computationally intensive and prone to high uncertainty. As a result, most virtual screening studies rely solely on binding free energy to measure relative stability (Duan et al., 2016).
| (3) |
| (4) |
| (5) |
Energy decomposition of stable complexes was also conducted on residues within 6 Å of the ligand to discern the residues contributing to binding affinity and type of interaction.
Statistical analysis
Statistical analysis were conducted using the R software. Most statistical calculations, such as the Wilcoxon rank sum test, were carried out using basic packages, and the pheatmap package was used to generate the heat maps.
Results
Re-docking experiment validated the docking protocol
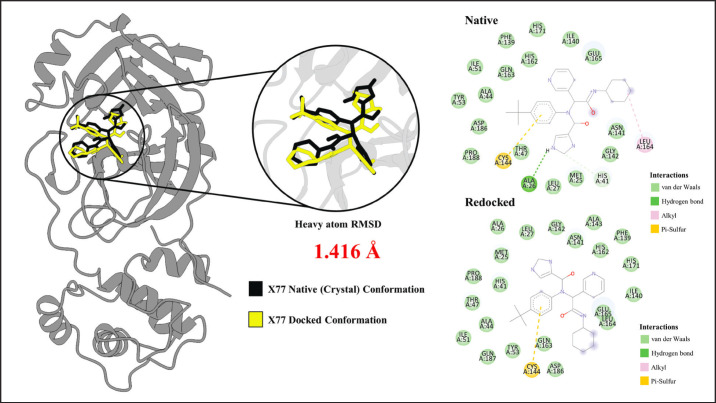
Using the docking parameters used in the study, re-docking the X77 control ligand back to 3CLpro yielded a similar pose as its native conformation. The result of the re-docking experiment is visualized in Figure 2.
Fig. 2. Redocked Pose of X77 compared to the native conformation. The docked pose shown in yellow has high similarity (RMSD = 1.416 Å) with the native crystal confirmation. Analysis of binding interactions show similar interactions with the HIS41-CYS144 catalytic dyad. Similar pi-sulfur interaction between the aromatic ring of X77 and CYS144 in both the native and redocked conformations were observed.
Figure 2 shows the high similarity between the native pose (black) and the re-docked pose (yellow) of X77. The heavy atom RMSD between the two poses was found to be 1.416 Å, which is less than the 1.5 Å threshold for a valid docking protocol (Hevener et al., 2009). Furthermore, examination of the interacting residues for the native and docked complexes shows similar interacting residues. Both the native and redocked complexes show identical interaction with the catalytic dyad HIS41 and CYS144. Specifically, the ligands in both complexes form van der Waals interaction with HIS41 and a pi-sulfur electrostatic interaction between the aromatic ring and the CYS144. All the points discussed imply that the protocol reproduces the crystal pose successfully and that the docking parameters are valid.
First docking pipeline: targeted docking and blind docking validation
Targeted Docking to Catalytic, Substrate-binding, and Oligomerization sites
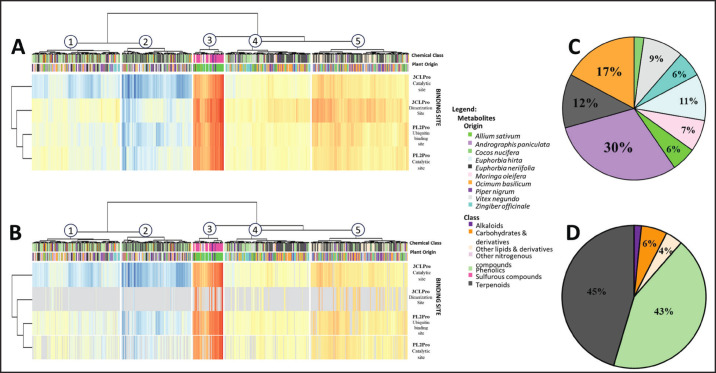
Docking 690 metabolites to the known binding sites of PEDV 3CLPro and PL2pro yielded 2760 docked poses in a diverse range of activities, with docking scores spanning from −2.10 to −9.604 Kcal mol−1. The results of site-specific docking are depicted in Figure 3A.
Fig. 3. Results of site-specific and blind docking analyses from the first pipeline. (A) Heatmap of site-specific molecular docking (B) Filtered heatmap of blind docking with greyed-out areas indicating docked poses from site-specific docking that were not validated in blind docking. (C, D) Pie charts showing the source plant (C) and chemical class (D) of screened ligands.
The apparent low affinity observed towards the proteins was anticipated because docking was conducted without prior knowledge of the specific metabolite types exhibiting activity against the proteases. Moreover, the compounds were selected based on their plant sources’ potential antiviral activities, thus including inactive compounds within the docking results. To discern the relevant protein-ligand interactions from the docking results, an energy threshold of −7 Kcal mol−1 (Wong et al., 2022) was utilized to filter out compounds with low affinity (> −7 Kcal mol−1) for PEDV proteases. This filter yielded 438 (15.87%) docked poses with high affinity (<−7 Kcal mol−1) to PEDV proteases, which are more likely to translate to in vitro and in vivo activity.
Blind docking validation
Blind molecular docking was used to confirm the specificity of the docked compounds within their respective binding sites. The blind docking analysis successfully replicated 361 of the 438 (82.42%) site-specific poses of high affinity (≤ −7 Kcal mol−1) compounds with binding scores ranging from −7.001 to −9.60 Kcal mol−1. The results of the blind docking are shown in Figure 3B.
In Figure 3B, areas shaded in grey represent poses from site-specific docking that were not reproduced by blind molecular docking, indicating their invalidity. From the 96 validated docking poses, the top 10 highest-scoring compounds per grid box meeting the adjusted −7 Kcal mol−1 binding energy threshold were selected. This rigorous selection procedure identified 43 compounds with high binding affinity to PEDV proteases, displaying docking scores ranging from −7.34 to −9.604 Kcal mol−1 and confirmed site-specificity.
Subsequently, 35 compounds passed the site-specific and blind docking validation of the first pipeline. The 35 compounds were subsequently compared with the results of the second pipeline to obtain a consensus list of selective protease inhibitors for further in-depth analysis based on their pharmacokinetic properties using in silico ADMET analysis.
Docking validation studies identified the most promising binding sites, plant sources, and metabolite classes
Heatmap analysis revealed a distinct trend in protease preferences for specific binding sites. The docking results show that the catalytic site of PEDV 3CLpro displays a notably higher affinity for ligands than its dimer interface. Although the dimer interface showed numerous hits in targeted docking (Fig. 3A), blind docking indicated lower specificity, with ligands favoring the catalytic site (Fig. 3B).
In contrast, PL2pro demonstrates greater receptivity at the ubiquitin-binding site than at its catalytic site. This observation suggests a need for tailored targeting strategies. Although competitive inhibition may prove effective for 3CLpro, disrupting protein-protein interactions might be more suitable for inhibiting PL2pro. This nuanced understanding of binding site preferences informs the development of effective antiviral agents, providing insights into the divergent targeting strategies needed for the optimal inhibition of PEDV proteases.
To facilitate the discussion of the most promising plant sources and metabolite classes, the heatmap was divided into five columns. Each column within the heatmap was assigned numbers 1 to 5. The clustered blind-validated heatmap (Fig. 3B) identified columns 1 and 2 as the most promising interaction clusters. From the data in columns 1 and 2, the most promising plant sources (Fig. 3C), chemical classes (Fig. 3D), and viral targets were identified. Examination of top columns 1 and 2 identified Andrographis paniculata, Euphorbia neriifolia, and Ocimum basilicum as the best-performing plant sources (Fig. 3C), which were all plants identified in prior literature to contain compounds with antiviral potential (Dayrit et al., 2021; Khursheed et al., 2022; Orosco and Wong, 2023). At the same time, terpenoids and phenolics are the best-performing chemical classes (Fig. 3D), which are chemical classes known for their antiviral activities (Masyita et al., 2022; Saidu et al., 2022; Orosco and Quimque, 2024).
Second docking pipeline: targeted docking to catalytic sites and cross-docking experiments
Cross-docking to analogous swine proteases
The binding affinities of the ligands to the catalytic sites of PEDV 3CLpro and PL2pro were obtained from the site-specific docking results in the first pipeline. The same set of ligands was docked onto analogous swine proteases. Selective antivirals against PEDV 3CLPro and PL2Pro catalytic sites were determined by first clustering of the phytochemicals through either Butina clustering with ECFP4-like Morgan fingerprinting (r = 2, cutoff = 0.5) in the rdkit Python library (O’Boyle and Sayle, 2016) or through Scaffold Hunter (Wetzel et al., 2009). In both methods, the selected compound clusters must have an average binding score against the catalytic sites higher than the −7.0 Kcal mol−1 threshold. The binding score to swine proteases was strongly correlated (R2 = 0.88 to 0.92) to binding to viral protease, which leads to inflation of the difference in protease binding. As such, selectivity was measured by calculating the residual (>0.3) of the particular cluster in the regression model of the binding score against the viral protease to the average binding score against the respective swine proteases. The residual threshold was validated to correspond to a standardized residual greater than one, significantly greater than the bootstrapped mean standardized residual (0) at α = 0.05. Compounds with lower promiscuity from cross-target docking and subsequent clustering were compared with the results of the first pipeline.
Consensus list of selective protease inhibitors
The results from both pipelines were compared to form a consensus list of inhibitors targeting PEDV protease. Analysis of the results from both pipelines yielded 35 ligands that performed well in both the cross-docking and blind-docking analyses and were selected for further evaluation. The resulting consensus list of the ligands is presented in Table 3.
Table 3. Consensus list of high-affinity selective proteases from the two docking pipelines employed.
| Code | Compound | Target | Site | Binding energy | Protease specificity |
| O7443 | beta-Cryptoxanthin | PL2pro | Catalytic Site | −8.8685 | + |
| O7407 | Zeaxanthin | PL2pro | Catalytic Site | −8.4565 | + |
| P5490 | Dipiperamide A, B | PL2pro | Catalytic Site | −8.365 | + |
| O7444 | Cryptoxanthin | PL2pro | Catalytic Site | −8.35575 | + |
| P5489 | Dipiperamide C | PL2pro | Catalytic Site | −8.1512 | + |
| P5493 | Dipiperamide D | PL2pro | Catalytic Site | −7.997 | + |
| P5491 | Dipiperamide E | PL2pro | Catalytic Site | −7.887 | + |
| M5288 | Pterygospermin | PL2pro | Catalytic Site | −7.29667 | + |
| A7152 | Ergosterol peroxide | PL2pro | Catalytic Site | −7.07067 | + |
| O7407 | Zeaxanthin | PL2pro | Ubiquitin binding site | −8.8995 | - |
| O7408 | Antheraxanthin | PL2pro | Ubiquitin binding site | −8.40544 | - |
| N7236 | Brein | PL2pro | Ubiquitin binding site | −8.32733 | - |
| V5568 | Neobignonoside | PL2pro | Ubiquitin binding site | −8.30033 | - |
| P5491 | Dipiperamide E | PL2pro | Ubiquitin binding site | −8.269 | - |
| PO7157 | Beta-Sitosterol | PL2pro | Ubiquitin binding site | −8.24344 | - |
| O5332 | Secoisolariciresinol | PL2pro | Ubiquitin binding site | −8.1703 | - |
| P7538 | Phytosterols | PL2pro | Ubiquitin binding site | −8.106 | - |
| A7147 | Bisandrographolide | PL2pro | Ubiquitin binding site | −8.047 | - |
| P5494 | Pipercyclobutanamide B | PL2pro | Ubiquitin binding site | −8.018 | - |
| A7147 | Bisandrographolide | 3CLpro | Catalytic Site | −9.604 | + |
| N7247 | Eurifoloid E | 3CLpro | Catalytic Site | −8.52617 | + |
| N5233 | Eurifoloid F | 3CLpro | Catalytic Site | −7.93144 | + |
| N7240 | CID 101570868 | 3CLpro | Catalytic Site | −8.14257 | + |
| N7248 | CID 118716354 | 3CLpro | Catalytic Site | −7.698 | + |
| Z3629 | Galanolactone | 3CLpro | Catalytic Site | −7.49188 | + |
| Z7680 | Aframodial | 3CLpro | Catalytic Site | −7.06792 | + |
| N3229 | CID 162866962 | 3CLpro | Catalytic Site | −7.35633 | + |
| N3230 | CID 162866964 | 3CLpro | Catalytic Site | −7.18 | + |
| E7206 | Ingenol triacetate | 3CLpro | Catalytic Site | −7.24442 | + |
| B788 | Saponins | 3CLpro | Catalytic Site | −7.72657 | + |
| O7407 | Zeaxanthin | 3CLpro | Dimerization Site | −8.0922 | - |
| P5491 | Dipiperamide E | 3CLpro | Dimerization Site | −7.7095 | - |
| P5489 | Dipiperamide C | 3CLpro | Dimerization Site | −7.70038 | - |
| N7252 | CID 118716358 | 3CLpro | Dimerization Site | −7.66 | - |
| VO7603 | Betulinic acid | 3CLpro | Dimerization Site | −7.34525 | - |
The 35 ligands from the parallel docking pipelines were expected to have high affinity, site-specific, and selective protease inhibitors. They were subsequently analyzed for in silico ADMET properties and swine off-target binding to balance veterinary safety per their expected potency.
In silico ADMET prediction and off-target docking
The compounds identified in the consensus list of specific protease inhibitors were analyzed for in silico ADMET properties using various web servers. A consensus pass/fail was obtained based on the agreement of the web server results. The total count of violations was computed based on the number of ADMET parameters violated, including Lipinski’s rule of five (Ro5). The rule of five only counts as one violation in the total score if more than two Ro5 parameters are violated. The results of the ADMET property predictions are summarized in Table 4 (for a complete list of ADMET results, refer to Table S4).
Table 4. ADMET violations of the compounds from the consensus list.
| Code | Lipinski violations* | Absorption violations | Distribution violations | CYP450 violations | Toxicity violations | Total | Pass/Fail | |
| O7443 | 2 | 1 | 1 | 0 | 1 | 4 | FAIL | |
| O7407 | 2 | 1 | 1 | 0 | 1 | 4 | FAIL | |
| P5490 | 1 | 1 | 0 | 1 | 1 | 3 | FAIL | |
| O7444 | 2 | 1 | 1 | 0 | 1 | 4 | FAIL | |
| P5489 | 1 | 1 | 0 | 1 | 1 | 3 | FAIL | |
| P5493 | 1 | 1 | 0 | 1 | 2 | 4 | FAIL | |
| P5491 | 1 | 1 | 0 | 1 | 1 | 3 | FAIL | |
| M5288 | 0 | 1 | 1 | 2 | 1 | 5 | FAIL | |
| A7152 | 1 | 0 | 1 | 0 | 0 | 1 | PASS | |
| O7408 | 2 | 1 | 0 | 0 | 1 | 3 | FAIL | |
| N7236 | 1 | 0 | 0 | 0 | 0 | 0 | PASS | |
| V5568 | 3 | 1 | 1 | 0 | 0 | 3 | FAIL | |
| PO7157 | 1 | 0 | 1 | 0 | 0 | 1 | PASS | |
| O5332 | 0 | 0 | 0 | 0 | 0 | 0 | PASS | |
| P7538 | 1 | 0 | 1 | 0 | 0 | 1 | PASS | |
| A7147 | 1 | 0 | 0 | 0 | 0 | 0 | PASS | |
| P5494 | 1 | 0 | 1 | 1 | 0 | 2 | FAIL | |
| N7247 | 0 | 0 | 1 | 0 | 0 | 1 | PASS | |
| N5233 | 0 | 0 | 0 | 0 | 0 | 0 | PASS | |
| N7240 | 0 | 0 | 1 | 0 | 0 | 1 | PASS | |
| N7248 | 0 | 0 | 1 | 0 | 0 | 1 | PASS | |
| Z3629 | 0 | 0 | 0 | 0 | 0 | 0 | PASS | |
| Z7680 | 0 | 0 | 1 | 0 | 0 | 1 | PASS | |
| N3229 | 0 | 0 | 0 | 0 | 0 | 0 | PASS | |
| N3230 | 0 | 0 | 0 | 0 | 0 | 0 | PASS | |
| E7206 | 0 | 1 | 0 | 0 | 1 | 2 | FAIL | |
| B788 | 3 | 1 | 0 | 0 | 0 | 2 | FAIL | |
| N7252 | 0 | 0 | 0 | 0 | 0 | 0 | PASS | |
| VO7603 | 1 | 0 | 1 | 0 | 0 | 1 | PASS | |
* Counted as one in “TOTAL” if more than two drug-likeness rules are violated.
The analysis identified 16 compounds with no more than 1 violation. Subsequently, these 16 compounds were scrutinized for potential off-target effects by molecular docking to cellular regulatory proteins, and their promiscuity was ranked accordingly (Table S5).
Molecular docking to cellular regulatory swine proteins yielded 11 compounds (Table 5) with minimal off-target interactions, which were further analyzed for dynamic stability through MD.
Table 5. Finalized list of selective protease inhibitors to be analyzed for dynamic stability.
| Compound | Target protein | Target site | Affinity |
| A7147 | 3CLpro | Catalytic site | −9.604 |
| N7247 | 3CLpro | Catalytic site | −8.52617 |
| N7240 | 3CLpro | Catalytic site | −8.14257 |
| A7147 | PL2pro | Ubiquitin binding site | −8.047 |
| N5233 | 3CLpro | Catalytic site | −7.93144 |
| N7248 | 3CLpro | Catalytic site | −7.698 |
| Z3629 | 3CLpro | Catalytic site | −7.49188 |
| N3229 | 3CLpro | Catalytic site | −7.35633 |
| VO7603 | 3CLpro | Dimerization site | −7.34525 |
| N3230 | 3CLpro | Catalytic site | −7.18 |
| Z7680 | 3CLpro | Catalytic site | −7.06792 |
Stability of complexes from MD
During the 100-nanosecond runs of MD, the stability of the ligands within the binding sites was elucidated. Various stability metrics, including RMSD, RMSF, RoG, and H-bond count, were used to assess the relative stability of the complexes. As part of the analysis, apoproteins served as negative controls for protein-centered metrics (RMSF, Rg), whereas the complex of X77 or N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide, a known inhibitor of PEDV 3CLpro co-crystallized at the catalytic site, was used as a positive control.
Root mean square deviation
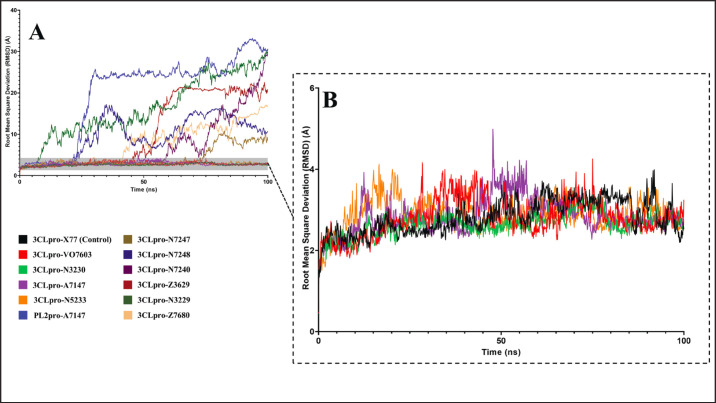
The study examined the complexes’ RMSD to evaluate any structural deviations observed among the system components during the 100 ns simulation. The RMSD analysis results for the 11 complexes are depicted in Figure 4.
Fig. 4. RMSD profiles of 11 complexes in the 100 ns MD simulations. (A) All complexes (B) Contain relatively stable complexes with a maximum RMSD of less than 5 Å and fluctuations of < 2 Å.
The higher temperature setting used in the MD simulations, which is representative of the swine internal temperature, posed challenges for ligand stabilization. Among the initial 11 compounds studied, only 4 demonstrated relatively stable behavior within their binding sites, as observed by mean RMSD values of less than 5 Å and fluctuations within 2 Å. In contrast, the remaining 7 exhibited unbinding events and mean RMSD values of 15.20, 4.17, 9.55, 10.86, 7.60, and 20.07 Å for 3CLpro-N3229, 3CLpro-N7247, 3CLpro-N7248, 3CLpro-Z3620, 3CLpro-Z7680, and PL2pro-A7147, respectively.
The complexes 3CLpro-VO7603, 3CLpro-N5233, 3CLpro-A7147, and 3CLpro-N3230 exhibited relative stability with average RMSD values of 2.81, 2.89, 2.86, and 2.64 Å, which are much less than those of the unstable complexes. Although the RMSD values for these complexes fluctuated, the fluctuations remained within a 2 Å range, indicating stability. Visualization of the trajectories revealed that these ligands remained within their binding pockets despite exploring a larger conformational space owing to the higher temperature.
A known inhibitor of PEDV 3CLpro, X77, was also examined under the same conditions to validate the findings. Figure 4B shows that the X77 control exhibited similar fluctuations within a 2 Å range and an average RMSD value of 2.81 Å. The four experimental ligands that demonstrated relative stability exhibited RMSD profiles comparable to those of the X77 control, confirming their stability.
Principal component analysis (PCA) of trajectories
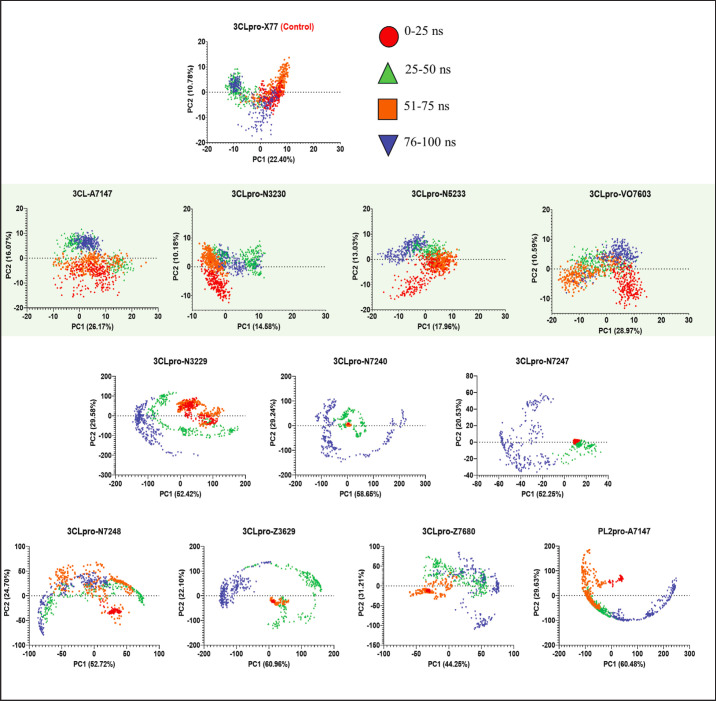
To further assess the stability demonstrated through the RMSD analysis, PCA of the simulated trajectories was performed. The first two principal components with the highest contributions to the total variance were used to plot the 2D projection of the trajectory. The PCA plots for all simulated trajectories are shown in Figure 5.
Fig. 5. 2D Projection of the MD trajectories using the two principal components with highest eigenvalues from PCA. Stable complexes are enclosed in a green shaded box.
The PCA plots are used to examine the fluctuations in the trajectory and confirm whether a convergence occurs in different time intervals. A scattered projection indicates structural fluctuations, while clustering of projections indicates structural convergence and stability. The projections plotted in Figure 5 are divided into 25 ns intervals. Ideally, structures are expected to converge at the last 25 ns of the simulation.
Based on the PCA plot, scattered projections are observed within the first 25 ns of the simulation for all complexes. This is an expected behavior as the system is still equilibrating and relaxing into the solvated system. The trajectory should, however, show visible convergence in the following intervals, and clustering should be observed within the last 25 ns of the simulation, shown as blue markers.
The stable complexes from the RMSD analyses (enclosed in green box), 3CLpro-VO7603, 3CLpro-N5233, 3CLpro-A7147, and 3CLpro-N3230, showed the characteristic scattering, albeit minimal, within the first 25 ns as the systems equilibrate but has observed reduced scattering in the 26–50 ns, 51–75 ns, and more prominently, 76–100 ns region or the last 25 ns. This indicates convergence in the structural dynamics and stabilization of the complex.
On the other hand, the less stable complexes, 3CLpro-N3229, 3CLpro-N7247, 3CLpro-N7248, 3CLpro-Z3620, 3CLpro-Z7680, and PL2pro-A7147, show highly scattered projections that become more apparent as the simulation progresses which reflects the unbinding events observed for said ligands. Periodic boundary conditions confined the unbound ligands, forming a circular pattern in the trajectory projection.
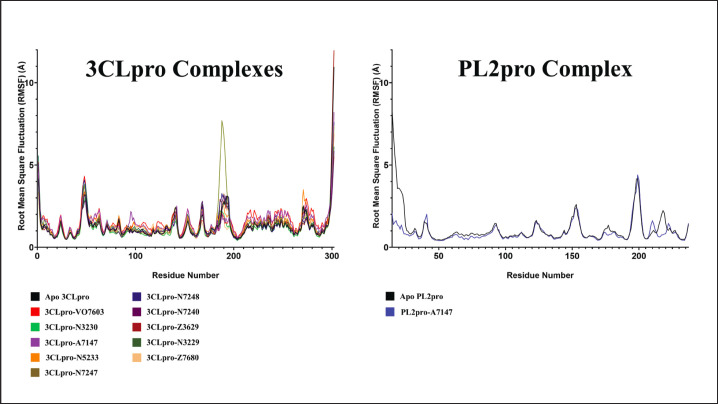
Root mean square fluctuation
This study also examined the RMSF of the complexes to assess the flexibility of the amino acid residues. Figure 6 illustrates the RMSF of the protein or protomer interacting with ligands.
Fig. 6. RMSF profiles of the 11 complexes.
Since the RMSF primarily characterizes the protein’s flexibility, the RMSF calculations for the experimental complexes were compared to the apo form of their respective proteins. Upon close examination of the RMSF plot in Figure 6, it was evident that for ligands bound to 3CLpro, only the complexes with VO7603, N5233, A7147, and N3230 exhibited lower RMSF profiles than those of apo 3CLpro. This reduction in the RMSF profile suggested decreased protein flexibility during complex formation, indicating stability.
On the other hand, for PL2pro, the lone complex with A7147 displayed RMSF profiles similar to the Apo PL2pro, indicating that the ligand had no discernible effect on the stability of the protein complex. In summary, the results of the RMSF analysis confirmed the relative stability of the VO7603, N5233, A7147, and N3230 complexes bound to 3CLpro.
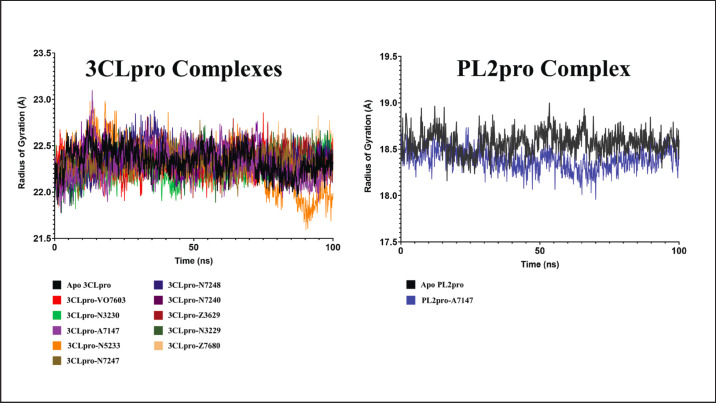
Radius of gyration
The Rg was computed as a measure of protein compactness throughout the simulation. Fluctuations in the Rg values signify the typical expansion characteristics observed in free proteins. However, this expansion is typically restricted when a ligand binds, decreasing Rg. Figure 7 shows the Rg profiles of the 11 complexes.
Fig. 7. Rg profiles of the 11 complexes in the 100 ns MD simulations.
Consistent with the RMSF calculations, the stability of the complexes formed between 3CLpro and the ligands VO7603, N5233, A7147, and N3230 relative to Apo 3CLpro is discernible from the Rg profiles. Among all ligands bound to 3CLpro, only these four compounds exhibited Rg profiles lower than the apoprotein, indicating constrained protein movement, restricted expansion, and a stabilized complex.
In contrast, although the PL2pro-A7147 complex initially displayed a lower Rg profile compared to apo PL2pro, it is noteworthy that this decrease occurred between 30 ns and gradually increased back to 90 ns. Beyond this period, the Rg profile of the complex converged with that of the apoprotein, suggesting an instability in ligand binding. In summary, the Rg profiles further support the stability of VO7603, N5233, A7147, and N3230 in complexes with 3CLpro.
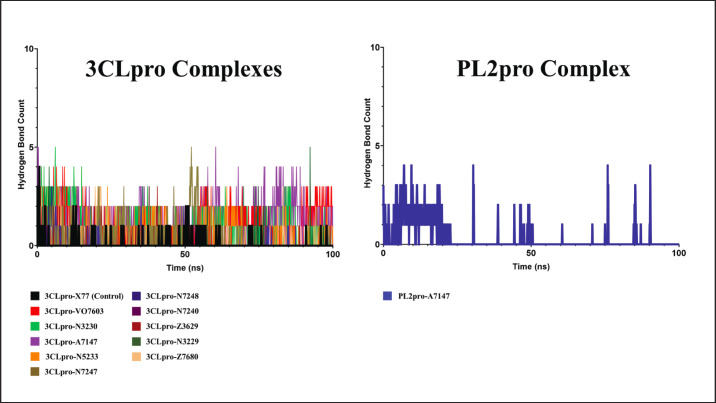
Hydrogen bond count
Finally, the number of hydrogen bond contacts was measured throughout the 100 ns MD simulation to estimate the relative stability of the attractive force holding the ligands in place. The hydrogen bond count analysis summary throughout the 100 ns runs is shown in Figure 8.
Fig. 8. Hydrogen bond count between the eleven ligands and PEDV proteases in the 100 ns MD simulations.
In the control complex of X77 with PEDV 3CLpro, while it remained stable in the binding pocket, fluctuating hydrogen bond counts were observed, with an average of only 0.10, suggesting a nuanced interplay between the ligand and protein over time. Although hydrogen bonds are pivotal in stabilizing complexes, their transient nature hints at a dynamic binding landscape, where interactions are formed and broken throughout the simulation.
Interestingly, this behavior extends beyond the control complex to the experimental ligands, exhibiting fluctuating hydrogen-bond patterns. This suggests that the observed dynamics might not be unique to a specific ligand but a characteristic feature of the binding event itself.
Considering this information, Figure 8 shows that ligands VO7603, N5233, A7147, and N3230 were the only ligands with sustained, albeit fluctuating, hydrogen bonds. Their average hydrogen bond counts were 0.91, 0.81, 0.98, and 1.35, respectively, which are much higher than those of the control ligand. This indicates continued interaction with the complex, further affirming the trends already presented by other stability metrics such as RMSD, RMSF, and Rg.
While stable hydrogen bonds are desirable for robust ligand binding, transient interactions may also play significant roles in ligand recognition and binding kinetics. Thus, a comprehensive analysis requires more concrete measures of ligand stability, such as the binding energy and decomposition analysis.
Binding analysis, energy calculation, and energy decomposition
MM-GBSA energies of the stable 3CLpro complexes
Binding free energy calculations were conducted using MMGBSA to validate the relative stability of the complexes. The binding energy of the native PEDV 3CLpro ligand, X77, served as a positive control and underwent a similar analysis. Given the scarcity of experimentally validated compounds against PEDV proteins, only X77, a control for the catalytic site, was prepared. A summary of the free-energy calculations is shown in Figure 9.
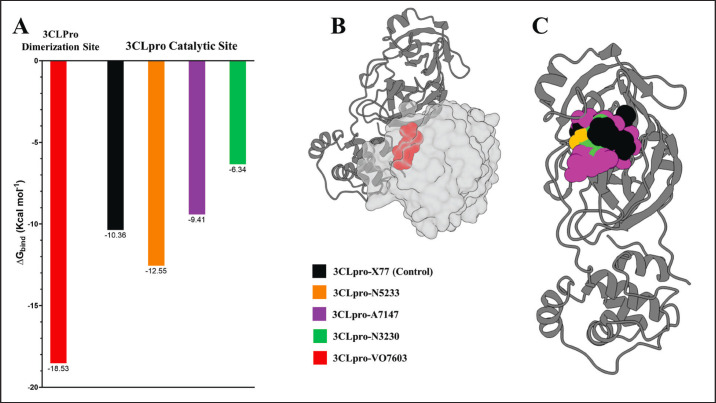
Fig. 9. Binding energy calculations of the four stable 3Lpro complexes (A) Binding energy graph of the four experimental complexes and X77 control (B) VO7603 bound to 3CLpro dimerization site (C) X77 control, N5233, A7147, and N3230 inside the 3CLpro catalytic site.
Examination of the binding free energy calculations revealed that two of the three compounds bound to the 3CLpro catalytic site with binding energies similar to or greater than that of the X77 control (−10.36 Kcal mol−1). N5233 and A7147, which have binding energies of −12.55 and −9.41 Kcal mol−1 are within 1 Kcal mol−1 difference with the control and were thus considered suitable candidates for further drug development of inhibitors targeting the catalytic site of PEDV 3CLpro.
On the other hand, while no small molecule control is available for the dimerization site, VO7603 exhibited a high affinity for the dimerization interface with a binding energy of −18.53 Kcal mol−1. In cases of protein-protein interaction inhibitors such as VO7603, especially when no control is available to compare the energy, it is more important to determine whether the ligand interacts and occupies the hotspot residues to destabilize the dimer formation. To achieve this, energy decomposition and binding interaction analysis were performed. The results of these analyses are detailed in the succeeding sections.
Inhibition of catalytically active 3CLpro dimer formation
VO7603 exhibits a calculated binding energy of −18.53 Kcal mol−1 according to MM-GBSA analysis. Despite surpassing the binding energy of the control complex, it is noteworthy that VO7603 binds to the dimerization site, in contrast to the catalytic site where X77 binds. Therefore, the difference in binding energy may not robustly indicate whether the compound serves as a potent inhibitor.
For compounds binding to the protein-protein interaction interface, such as oligomerization inhibitors, determining the binding site residues utilized by the ligand pose is crucial in assessing their inhibitory potential. A ligand must interact with identical residues within the hotspots of protein interactions.
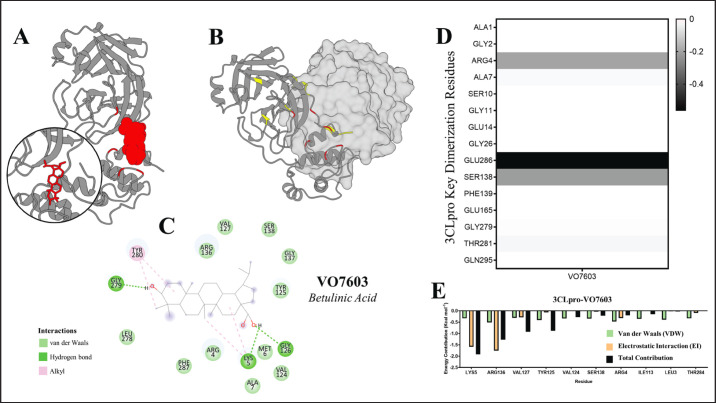
The dimerization site of PEDV 3CLPro utilizes the residues ALA1, GLY2, ARG4, ALA7, SER10, GLY11, GLU14, GLY26, GLU286, SER138, PHE139, GLU165, GLY279, THR281, and GLN295 (Ye et al., 2016) to form a catalytically active dimer. To ascertain whether VO7603 engages with identical residues for binding interactions, energy decomposition analysis was conducted to identify the critical binding site residues utilized by the ligand. The detailed results of energy decomposition and binding analysis of VO7603 at the PEDV 3CLpro dimerization site are presented in Figure 10.
Fig. 10. Energy decomposition and binding analysis of VO7603 at the PEDV 3CLpro dimerization interface. (A) VO703 inside the dimer interface (B) The dimer interface with key residues highlighted yellow and the VO7603 binding residues highlighted red (C) Structure and interaction map of VO7603 (betulinic acid) (D) Heatmap showing dimerization hotspot residues used by VO603 (E) Energy decomposition analysis of the 10 residues with the highest contribution to VO7603 binding.
Energy decomposition analysis revealed that VO7603 interacts with six residues from the dimer interface hotspot, potentially inhibiting the formation of a catalytically active dimer. The heatmap in Figure 10D shows that VO7603 employs ARG4, ALA7, SER10, GLU286, GLY279, and THR281. However, the energy decomposition of the 10 residues with the highest contribution (Fig. 10D) indicated that only ARG4 significantly influenced the binding energy. Additionally, most residues involved in VO7603 binding participated in electrostatic interactions with residues adjacent to the dimerization hotspot. Nevertheless, using key residues in binding and the strong electrostatic forces with residues near the dimer hotspot suggest promising potential as 3CLpro dimerization inhibitors.
Inhibition of 3CLpro catalytic activity
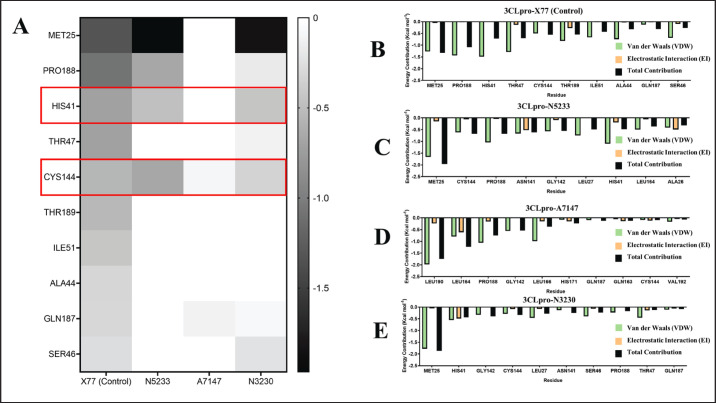
From Figure 9A, the metabolites bound to the 3CLpro catalytic site have calculated binding energies of −12.55, −9.41, and −6.34 Kcal mol−1 for N5233, A7147, and N3230, respectively. Of the three ligands, only N5233 and A7147 have binding energies within 1 Kcal mol−1 difference from the control ligand X77, which has −10.36 Kcal mol−1. To compare the metabolites’ performance to that of the control inhibitor, an energy decomposition analysis was performed to determine the highest contributing residues to the overall binding energy and see if the metabolites use the same residues as that of the known inhibitor. The results of energy decomposition analysis for the 3CLpro catalytic site inhibitors are detailed in Figure 11.
Fig. 11. Energy decomposition per residue of 3CLpro catalytic site inhibitor complexes. (A) Heatmap comparing the energy contributions of residues used by the control ligand and those used by the experimental ligands. Enclosed in red boxes are catalytic residues of PEDV 3CLpro. (B–E) Energy decomposition analysis of 10 residues with the highest energy contributions to 3CLpro complexes with X77 control (B), N3230 (C), A7147 (D), and N5233 (E).
A comparison of the binding site residues in the heatmap (Fig. 11A) revealed that N5233 uses both residues in the 3CLpro catalytic dyad, HIS41 and CYS144, and shares four similar binding site residues with the X77 control: MET25, PRO188, HIS41, and CYS144, exhibiting relatively strong energy contributions (Fig. 11B). The energy decomposition of the ten highest contributing residues (Fig. 11C) also includes these four residues, characterized by a mix of strong van der Waals and electrostatic interactions.
In contrast, A7147 uses only one residue from the 3CLpro catalytic dyad, CYS144, and shares two residues with the X77 control (Fig. 11A), displaying relatively weak energy contributions. The energy decomposition of the highest contribution (Fig. 11D) includes these two residues. However, most energy contributions come from strong van der Waals and electrostatic interactions involving LEU190, LEU164, PRO188, GLY142, and LEU166. Despite this, the high-energy residue interactions and the use of a catalytic residue make it a potential candidate for a PEDV 3CLpro inhibitor.
Finally, N3230 uses catalytic dyad residues and shares seven residues with the X77 control (Fig. 11A), the highest of the three experimental ligands, with relatively high contributions. However, the energy decomposition of the ten highest contributing residues (Fig. 11E) shows that the interactions are primarily van der Waals interactions, leading to binding energy (−6.34 Kcal mol−1) significantly smaller than that of the X77 control (−10.36 Kcal mol−1), rendering it an unsuitable candidate. The findings regarding the residues used to interact with the 3CLpro catalytic site indicate the potential for derivatization that could improve its binding interaction with the binding site and lead to a more potent antiviral form.
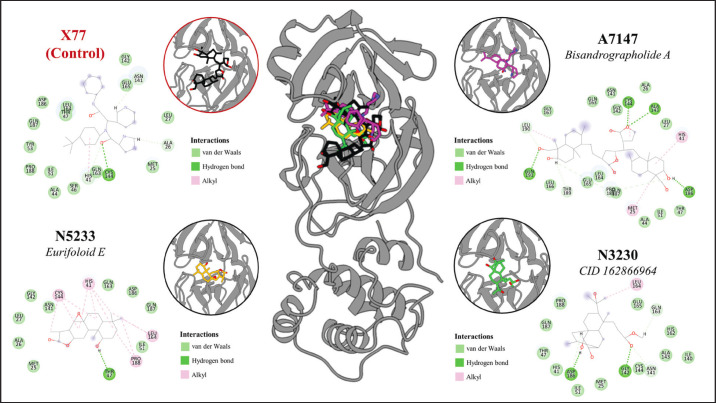
The energy calculations and binding analysis results identified N5233 and A7147 as promising inhibitors of the PEDV 3CLpro catalytic site. Aside from performing better or similar to the control, the ligands also interact with the catalytic dyad, HIS41 and CYS144, further solidifying their potential as inhibitors of catalytic activity. However, while N3230 failed to meet the energy threshold set by the control and was not considered a promising candidate, the ligand also interacted with the catalytic dyad with even higher contributions than the two leads. This signifies that although it performs poorly in this study, derivatization may improve its binding affinity to 3CLpro. The binding interaction plot of the 3CLpro-binding compounds is detailed in Figure 12, which shows a mix of van der Waals, hydrogen bond, and alkyl interactions reflected on their energy decomposition analysis.
Fig. 12. Binding interaction analysis of 3CLpro catalytic site inhibitors compared to the control ligand, the X77 inhibitor.
Discussion
This study delved into various facets of antiviral drug discovery against PEDV, shedding light on critical insights from molecular docking, in silico ADMET screening, and MD simulations.
Molecular docking analyses revealed intriguing nuances in targeting strategies for PEDV proteases, highlighting distinct preferences for ligand binding between the catalytic site and protein interaction interfaces. Notably, the study observed increased receptivity of the catalytic site within PEDV 3CLpro, in contrast to its dimer interface. In comparison, ligands docked to PL2pro displayed a stronger affinity for the ubiquitin-binding site than its catalytic site. This observation is consistent with findings from research on related coronavirus, SARS-CoV-2, and their structurally similar proteases, where challenges in identifying effective compounds for the catalytic site of PL2pro have been encountered ( Štekláč et al., 2021; Verma et al., 2021), whereas ligands have shown greater success in binding to the ubiquitin-binding site (Chu et al., 2018), indicating susceptibility to disruption of protein-protein interactions. Interestingly, although considerable attention has been devoted to targeting the catalytic site of 3CLpro in previous studies (Vergoten and Bailly, 2021; Zhang et al., 2021; Lam et al., 2022; Pathak et al., 2023; Li et al., 2024), its dimerization site has been relatively overlooked (Ye et al., 2016). Such distinctions suggest the need for tailored targeting strategies, promoting competitive inhibition of 3CLpro and disrupting protein-protein interactions for PL2pro. These insights offer an invaluable direction for optimizing drug design strategies to combat PEDV.
Furthermore, the docking results yielded valuable insights into the most promising plant sources and metabolite classes for PEDV proteases. Among these, Andrographis paniculata emerged as a promising plant source, while terpenoids emerged as the most promising chemical class. Similar research targeting the 3CLpro and PL2pro of structurally related SARS-CoV-2 has also highlighted the strong binding affinity of a terpenoid derived from Andrographis paniculata (Verma et al., 2021). These findings validate the potential of natural compounds, particularly those from Andrographis paniculata and compounds belonging to the terpenoid class, as valuable candidates for further exploration in the quest for effective antiviral agents against PEDV and related coronaviruses.
Beyond molecular docking simulations and determination of binding affinity, the integration of swine-adapted in silico ADMET screening provides a robust framework for balancing drug potency with veterinary safety. Many computational drug discovery studies have been applied to veterinary pathogens, such as PEDV (Vergoten and Bailly, 2021; Verma et al., 2021; Pathak et al., 2022a). However, no drug discovery protocol has been adjusted for veterinary use. Hence, this study is the first to report a veterinary informatics approach to antiviral drug discovery against an animal pathogen. By incorporating off-target docking into swine-specific receptors and proteases, this approach identified promising candidates with favorable ADMET profiles, laying the groundwork for developing safe and effective veterinary antiviral drugs against PEDV. This interdisciplinary effort bridges computational pharmacology with veterinary medicine, thereby facilitating the translation of in silico discoveries into tangible veterinary applications.
Finally, MD simulations offered invaluable insights into compound stability under swine physiological conditions, essential for optimizing drug efficacy and safety. The observed fluctuating values of the stability metrics, including RMSD, RMSF, Rg, and H-bonds, from the MD simulations, were consistent with findings from previous studies that encountered similar dynamic behaviors (Štekláč et al., 2021). Notably, ligand binding to proteases tends to exhibit fluctuating behaviors characteristic of their binding modes. This phenomenon becomes particularly pronounced when simulating higher temperatures to better mimic host physiology. Despite the higher temperature settings reflective of swine internal environments, certain compounds demonstrated remarkable stability within their binding sites, indicating rational drug design efforts and enhancing the prospects of veterinary drug development against PEDV. These findings highlight the significance of interdisciplinary approaches in advancing veterinary informatics-driven drug discovery, offering a pathway toward effective antiviral interventions in veterinary medicine.
In conclusion, this study represents a pioneering effort in antiviral drug discovery against PEDV, leveraging cutting-edge computational methodologies and interdisciplinary approaches. By elucidating the preferences of PEDV proteases for ligand binding and identifying promising plant sources and chemical classes, this study provides valuable insights for tailored veterinary drug design strategies. The integration of swine-adapted in silico ADMET screening bridges the gap between computational pharmacology and veterinary medicine, thereby facilitating the development of safe and effective veterinary antiviral drugs. Additionally, MD simulations offer critical insights into compound stability under physiological conditions in swine, further enhancing drug design efforts. Overall, this interdisciplinary study lays the groundwork for the advancement of veterinary informatics-driven drug discovery, offering promising prospects for combating PEDV in veterinary medicine.
Acknowledgments
The authors extend their appreciation to the Advanced Science and Technology Institute (DOST-ASTI) of the Philippines’ Department of Science and Technology for providing access to their High-Performance Computing (HPC) units through the Computing and Archiving Research Environment (COARE), which facilitated the execution of the analyses and data collection for this study.
Conflict of interest
The authors declare no conflicts of interest.
Funding
The authors gratefully acknowledge the support of the Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (DOST-PCAARRD) of the Philippines’ Department of Science and Technology, which funded the project under the Virology and Vaccine Research and Development Program.
Data availability
The primary data supporting this study’s findings are included in the manuscript. All supplementary Tables (S1-S5) are available and can be requested from the Corresponding Author.
Authors’ contributions
FLO conceptualized and acquired funding for the study. JCCDG and ANGD conceptualized and optimized the workflow in the paper. JCCDG and ANGD performed the dataset curation. JCCDG conducted most of the experiments, performed the post-experiment analyses, and drafted the paper. ANGD conducted the cross-docking experiments and the related statistical tests. JCCDG, ANGD, and FLO revised the paper. All authors approved the submission of the final version of the manuscript.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- Anderson A.C. The process of structure-based drug design. Chem. Biol. 2003;10:787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Batool M., Ahmad B., Choi S. A structure-based drug discovery paradigm. Int. J. Mol. Sci. 2019;20:2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R.B., Zhu X., Shim J., Lopes P.E.M., Mittal J., Feig M., Jr MacKerell A.D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 Dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Li Q., Zhou Z., Wang Y., Bryant S.H. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 2012;14:133–141. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.F., Chen C.C., Moses D.C., Chen Y.H., Lin C.H., Tsai Y.C., Chou C.Y. Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine. Antiviral Res. 2018;158:199–205. doi: 10.1016/j.antiviral.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayrit F., Guidote A., Gloriani N., de Paz-Silava S.L., Villaseñor I., Macahig R.A., Tan M., Chua J.R., Sia I. Philippine medicinal plants with potential immunomodulatory and anti-SARS-CoV-2 activities. Philipp. J. Sci. 2021;150:999–1015. [Google Scholar]
- DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of RandD costs. J. Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Duan L., Liu X., Zhang J.Z.H. Interaction entropy: a new paradigm for highly efficient and reliable computation of protein–ligand binding free energy. J. Am. Chem. Soc. 2016;138:5722–5728. doi: 10.1021/jacs.6b02682. [DOI] [PubMed] [Google Scholar]
- Forli S., Huey R., Pique M.E., Sanner M., Goodsell D.S., Olson A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Yang H., Cai Y., Sun L., Di P., Li W., Liu G., Tang Y. ADMET-score—a comprehensive scoring function for evaluation of chemical drug-likeness. Med. Chem. Comm. 2018;10:148–157. doi: 10.1039/c8md00472b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996;17:616–641. [Google Scholar]
- Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze L.J., Koehl N.J., O’Shea J.P., Kostewicz E.S., Holm R., Griffin B.T. The pig as a preclinical model for predicting oral bioavailability and in vivo performance of pharmaceutical oral dosage forms: a PEARRL review. J. Pharm. Pharmacol. 2019;71:581–602. doi: 10.1111/jphp.12912. [DOI] [PubMed] [Google Scholar]
- Hevener K.E., Zhao W., Ball D.M., Babaoglu K., Qi J., White S.W., Lee R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009;49:444–460. doi: 10.1021/ci800293n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursheed A., Jain V., Wani A.R. Euphorbia hirta as a gold mine of high-value phytochemicals: a comprehensive review of its pharmacological activities and possible role against SARS-CoV-2. Biomed. Res. Ther. 2022;9:4930–4949. [Google Scholar]
- Kim J.M., Pathak R.K. Editorial: vetinformatics: an insight for decoding livestock systems through in silico biology. Front. Vet. Sci. 2023;10:1292733. doi: 10.3389/fvets.2023.1292733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.P., Nguyen D.N., Mai T.T., Tran T.D., Le M.T., Huynh P.N.H., Nguyen D.T., Tran V.H., Trinh D.T.T., Truong P., Vo C.V.T., Thai K.M. Exploration of chalcones as 3-chymotrypsin-like protease (3CLpro) inhibitors of SARS-CoV-2 using computational approaches. Struct. Chem. 2022;33:1707–1725. doi: 10.1007/s11224-022-02000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhu L., Wang L., Huang Y., Zhang Y., Zhao D., Wang L., Yi D., Hou Y., Wu T. Identification of two flavonoids antiviral inhibitors targeting 3C-like protease of porcine epidemic diarrhea virus. Front. Microbiol. 2024;15:1357470. doi: 10.3389/fmicb.2024.1357470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalalad M.A.B., Orosco F.L. In silico identification of multi-target inhibitors from medicinal fungal metabolites against the base excision repair pathway proteins of African swine fever virus. RSC Adv. 2024;14:10039–10055. doi: 10.1039/d4ra00819g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyita A., Mustika Sari R., Dwi Astuti A., Yasir B., Rahma Rumata N., Emran T.B., Nainu F., Simal-Gandara J. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives. Food Chem. X. 2022;13:100217. doi: 10.1016/j.fochx.2022.100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri A., Bertola M. Alvascience: a new software suite for the QSAR workflow applied to the blood–brain barrier permeability. Int. J. Mol. Sci. 2022;23:12882. doi: 10.3390/ijms232112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminformatics. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N.M., Sayle R.A. Comparing structural fingerprints using a literature-based similarity benchmark. J. Cheminf. 2016;8:36. doi: 10.1186/s13321-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosco F.L. Current advances in antiviral potential of artemisia against major viral infections. J. Bacteriol. Virol. 2023;53:61–73. [Google Scholar]
- Orosco F., Quimque M. Antiviral potential of terpenoids against major viral infections: recent advances, challenges, and opportunities. J. Adv. Biotechnol. Exp. Ther. 2024;7:221. [Google Scholar]
- Orosco F.L., Wong J.E. Andrographolides as antiviral agents: insights into mechanisms, modifications, and delivery innovations. Trop. J. Nat. Prod. Res. 2023;7:5341–5354. [Google Scholar]
- Paraguison-Alili R., Domingo C.Y.J. Phylogenetic tracking of current porcine epidemic diarrhea virus (PEDV) strains in the Philippines. Arch. Virol. 2016;161:2601–2604. doi: 10.1007/s00705-016-2938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R.K., Kim J.M. Vetinformatics from functional genomics to drug discovery: insights into decoding complex molecular mechanisms of livestock systems in veterinary science. Front. Vet. Sci. 2022;9:1008728. doi: 10.3389/fvets.2022.1008728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R.K., Kim W.I., Kim J.M. Targeting the PEDV 3CL protease for identification of small molecule inhibitors: an insight from virtual screening ADMET prediction, molecular dynamics, free energy landscape, and binding energy calculations. J. Biol. Eng. 2023;17:29. doi: 10.1186/s13036-023-00342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R.K., Kim D.Y., Lim B., Kim J.M. Investigating multi-target antiviral compounds by screening of phytochemicals from neem (Azadirachta indica) against PRRSV: a vetinformatics approach. Front. Vet. Sci. 2022a;9:854528. doi: 10.3389/fvets.2022.854528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R.K., Seo Y.J., Kim J.M. Structural insights into inhibition of PRRSV Nsp4 revealed by structure-based virtual screening, molecular dynamics, and MM-PBSA studies. J. Biol. Eng. 2022b;16:4. doi: 10.1186/s13036-022-00284-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Saidu M.B., Kúsz N., Tsai Y.C., Vágvölgyi M., Berkecz R., Kókai D., Burián K., Hohmann J., Rédei D. Triterpenes and phenolic compounds from Euphorbia deightonii with antiviral activity against herpes simplex virus type-2. Plants. 2022;11:764. doi: 10.3390/plants11060764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamat S.E.A., Collantes T.M.A., Lumbera W.M.L., Tablizo F.A., Mutia C.T.M., Ong J.D.P., Bandoy D.J.D.R. Sequence analysis of new variants of porcine epidemic diarrhea virus in Luzon, Philippines, in 2017. Arch. Virol. 2021;166:1859–1867. doi: 10.1007/s00705-021-05068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapovalov M.V., Dunbrack R.L. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Struct. Lond. Engl. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štekláč M., Zajaček D., Bučinský L. 3CLpro and PLpro affinity, a docking study to fight COVID19 based on 900 compounds from PubChem and literature. Are there new drugs to be found? J. Mol. Struct. 2021;1245:130968. doi: 10.1016/j.molstruc.2021.130968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P.C., Tsai C.C., Mehboob S., Hevener K.E., Johnson M.E. Comparison of radii sets, entropy QM methods, and sampling on MM-PBSA, MM-GBSA, and QM/MM-GBSA ligand binding energies of F. tularensis enoyl-ACP reductase (FabI) J. Comput. Chem. 2015;36:1859–1873. doi: 10.1002/jcc.24011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujatha P.L, Kumarasamy P, Preetha S.P., Balachandran P. Vetinformatics: a new paradigm for quality veterinary services. Res. Rev. J. Vet. Sci. Technol. 2018;5:16–19. [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-Tresanco M.S., Valdés-Tresanco M.E., Valiente P.A., Moreno E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 2021;17:6281–6291. doi: 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., MacKerell A.D. CHARMM general force field (CGenFF): a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergoten G., Bailly C. In silico analysis of echinocandins binding to the main proteases of coronaviruses PEDV (3CLpro) and SARS-CoV-2 (Mpro) Silico Pharmacol. 2021;9:41. doi: 10.1007/s40203-021-00101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D., Mitra D., Paul M., Chaudhary P., Kamboj A., Thatoi H., Janmeda P., Jain D., Panneerselvam P., Shrivastav R., Pant K., Das Mohapatra P.K. Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/ 3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components. Curr. Res. Pharmacol. Drug Discov. 2021;2:100038. doi: 10.1016/j.crphar.2021.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wang G., Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med. Chem. 2016;8:1707–1710. doi: 10.4155/fmc-2016-0143. [DOI] [PubMed] [Google Scholar]
- Wetzel S., Klein K., Renner S., Rauh D., Oprea T.I., Mutzel P., Waldmann H. Interactive exploration of chemical space with Scaffold Hunter. Nat. Chem. Biol. 2009;5(8):581–583. doi: 10.1038/nchembio.187. [DOI] [PubMed] [Google Scholar]
- Wong F., Krishnan A., Zheng E.J., Stärk H., Manson A.L., Earl A.M., Jaakkola T., Collins J.J. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 2022;18:e11081. doi: 10.15252/msb.202211081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A., Chen X., Hou T., Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G., Deng F., Shen Z., Luo R., Zhao L., Xiao S., Fu Z.F., Peng G. Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease. Virology. 2016;494:225–235. doi: 10.1016/j.virol.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Zhou J., Wang X., Ma L., Li J., Yang L., Yuan H., Pang D., Ouyang H. Porcine epidemic diarrhea virus: an updated overview of virus epidemiology, virulence variation patterns and virus–host interactions. Viruses. 2022;14:2434. doi: 10.3390/v14112434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen H., Zou M., Oerlemans R., Shao C., Ren Y., Zhang R., Huang X., Li G., Cong Y. Hypericin inhibit alpha-coronavirus replication by targeting 3CL protease. Viruses. 2021;13:1825. doi: 10.3390/v13091825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The primary data supporting this study’s findings are included in the manuscript. All supplementary Tables (S1-S5) are available and can be requested from the Corresponding Author.