Abstract
Simian virus 40 (SV40) is a monkey virus with tumorigenic potential in rodents and is associated with several types of human cancers, including lymphomas. A related Merkel cell polyomavirus causes carcinoma in humans by expressing truncated large tumor antigen (LT), with truncations caused by APOBEC family of cytidine deaminase-induced mutations. AID (activation-induced cytidine deaminase), a member of the APOBEC family, is the initiator of the antibody diversification process known as somatic hypermutation and its aberrant expression and targeting is a frequent source of lymphomagenesis. In this study, we investigated whether AID could cause mutations in SV40 LT. We demonstrate that the SV40 enhancer has strong somatic hypermutation targeting activity in several cell types and that AID-induced mutations accumulate in SV40 LT in B cells and kidney cells and cause truncated LT expression in B cells. Our results argue that the ability of the SV40 enhancer to target somatic hypermutation to LT is a potential source of LT truncation events that could contribute to tumorigenesis in various cell types, thereby linking SV40 infection with malignant development through a novel mutagenic pathway.
Keywords: Somatic hypermutation, SV40, Enhancer, AID, Large tumor antigen, Tumorigenesis
1. Introduction
Simian Virus 40 (SV40) is a member of polyomavirus family out of which Merkel cell polyomavirus (MCPyV), BK polyomavirus (BKPyV), JC polyomavirus (JCPyV) and trichodysplasia spinulosa associated polyomavirus (TSPyV) are known to cause disease in humans [1]. SV40 naturally infects monkeys and became widely known as a contaminant of Polio vaccine in 1950s–60s (reviewed in Ref. [2]). It was then discovered that SV40 transforms rodent cells efficiently and can transform cultured human cells, which led to a fear of a cancer outburst due to contaminated vaccine. This outburst never happened. Evidence of SV40 association is strongest with mesotheliomas, brain cancers, bone tumors, and in non-Hodgkin lymphomas [[3], [4], [5], [6], [7], [8]]. However, the role of SV40 in human cancers has remained controversial as numerous studies have failed to demonstrate the presence of SV40 or its incidence was low [[9], [10], [11], [12], [13], [14]].
SV40 Large Tumor Antigen (LT) can transform cells by binding to retinoblastoma (RB) and p53 proteins, which interferes with cell cycle regulation and apoptosis [[15], [16], [17]]. In MCPyV-induced Merkel cell carcinoma (MCC) mutations of LT causing LT truncation have a major role in oncogenesis [[18], [19], [20]]. Truncated forms of LT with a disrupted C-terminal helicase domain are unable to bind to the viral origin of replication and initiate viral replication. Efficient replication of both MCPyV and SV40 viruses have cytotoxic effects in human cells and therefore disruption of viral replication is crucial for malignant transformation [18,[21], [22], [23]]. Mutations also accumulate in the SV40 LT C-terminus [[24], [25], [26]] and truncated SV40 LT retains its capability to transform cells, even when its p53 interaction domain is lost [27]. While the significance of LT truncation events has not been demonstrated for SV40-induced tumorigenesis, it is probable that, as with truncated MCPyV LT, truncated SV40 LT triggers less DNA damage response and is less immunogenic than full-length LT, which would be beneficial for virus persistence in host cells and provide the cells a transformative advantage [18,19].
Somatic hypermutation (SHM) is a process that introduces point mutations into the variable region of Ig loci and is necessary for fine modification of antibody specificity and production of high-affinity antibodies [[28], [29], [30]]. DNA subjected to SHM is deaminated at cytidines by activation-induced cytidine deaminase (AID; encoded by AICDA). The resulting deoxyuridine is recognized and processed by error-prone base excision repair and mismatch repair resulting in mutation not only at the original site of deamination but also at flanking residues [29]. Little is known about the mechanism of SHM targeting to specific genes. Ig loci contain “mutation enhancer elements” that overlap with their classical transcriptional enhancer elements. Such mutation enhancers have the ability to increase SHM of a nearby transcribed gene by two orders of magnitude or more, highlighting the importance of cis-acting regulatory elements for SHM targeting [31,32]. The SHM-targeting activity of these elements relies on a number of transcription factor binding sites (TFBSs), including NF-kB, octamer, MEF, and IRF-Ets composite sites [[32], [33], [34]]. All known Ig mutation enhancer elements contain E-boxes (binding sites for E protein transcription factors that have important roles in B and T cell development) that were found to be important but not sufficient elements of mutation enhancers [35]. In most cases, no single TFBS was critical for the activity indicating both cooperative and redundant roles [32]. The mutation enhancer activity of Ig enhancers is conserved through avian and mammalian species [32].
SHM is also detected at a subset of non-Ig genes, both in human B cell tumors and normal germinal center B cells [[36], [37], [38], [39]]. Recently, we identified a number of non-Ig enhancers in the human genome with varying SHM-targeting activity [40]. This finding is of particular relevance given that SHM targeting especially to proto-oncogenes or tumor suppressors, is a known source of lymphomagenesis [37,41]. In addition, we showed weak SHM targeting activity in a MCPyV non-coding regulatory region, but the majority of LT mutations were likely caused by APOBEC3 enzymes rather than AID-induced SHM [42]. Together these findings indicate that mutation enhancer activity is not an exclusive property of Ig enhancers but is more widely spread in biology than previously appreciated.
SV40 can infect several human cell types including B cells, where AID is naturally expressed during B cell development and activation [4,43,44]. SV40 is frequently associated with B cell-derived non-Hodgkin’s lymphoma [6,8,[45], [46], [47], [48]], with one study finding 62 out of 91 lymphomas to be SV40-positive [7]. Association of SV40 with B cells and lymphomas together with observed mutations and truncations of SV40 LT indicates that SHM in antigen-activated B cells might potentiate SV40 tumorigenicity.
Inflammatory signals can induce AID expression in non-B cells and AID has been detected in some non-B cell malignancies [49,50]. In addition, infection with hepatitis C virus was shown to increase the rate of SHM in B cells and hepatocellular carcinoma cells implying a role of viral infection in mutation [51]. Together with the ability of SV40 to infect various cell types, these data suggest that SHM could promote SV40-mediated tumorigenesis beyond B cells. Given the requirement of a targeting element for efficient mutation of a transcribed region and the association of SHM-targeting activity with enhancer elements, we investigated the ability of SV40 enhancer element to target SHM to the adjacent LT region and its potential to generate the truncated form of LT.
Here, we show that the SV40 enhancer has strong SHM targeting activity and that this activity can result in mutation of SV40 LT in B and non-B cancer cell lines. A subset of these mutations forms truncated LTs that could contribute to transformation of human cells. We propose that this mutation targeting activity provides a rationale for how SV40 might contribute to tumorigenesis of B cell-derived as well as non-B cell-derived malignancies.
2. Materials and methods
2.1. Cell culture
DT40 cells were cultured at +40 °C, 5 % CO2, 90 % humidity. Growth media included RPMI 1640 HEPES modification (Sigma) with 10 % FBS (HyClone), 1 % NCS (Biowest), 1x penicillin-streptomycin antibiotic (Gibco), 1x Glutamax (Gibco), and 50 μM β-mercaptoethanol. Ramos cells were cultured at +37 °C, 5 % CO2, 90 % humidity. Growth media included RPMI 1640 HEPES modification (Sigma) with 10 % FBS (Gibco), 1x penicillin-streptomycin antibiotic (Gibco) and 1x Glutamax (Gibco). UO-31, 293T, U2OS and MDA-MB-231 cells were cultured at +37 °C, 5 % CO2, 90 % humidity. Growth media included Dulbecco’s Modified Eagle’s Medium – high glucose (Sigma) supplemented with 10 % FBS (Gibco) and 1 % penicillin-Streptomycin (Sigma).
2.2. Cloning to GFP vectors
SV40 enhancer was amplified with PCR using Q5 High-Fidelity DNA Polymerase (New England Biolabs, NEB) from plasmid template. The following regions of human and mouse genomes were amplified using Phusion High-Fidelity DNA polymerase (NEB): GRCh38 chr8:11,536,697–11,539,424 (hBLKe); GRCh38 chr6:14,093,401–14,096,308 (hCD83e); GRCm38 chr7:24,903,876–24,904,597 (mArfgef1); GRCm38 chr9:51,225,574–51,226,089 (mPou2af1e). Enhancers were cloned into GFP2, GFP4, and GFP7 expressing vector using In-Fusion cloning kit (Takara) according to manufacturer’s protocol. NheI/SpeI site was used for cloning to GFP2 and GFP4 vectors and HpaI site was used for cloning to GFP7 vector. The mutated SV40 2x72bp enhancer sequences were created with QuickChange site-directed mutagenesis kit (Stratagene) or using In-Fusion site-directed mutagenesis and verified by sequencing. The cloned plasmids were isolated with GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific) or QIAprep Spin Miniprep Kit (QIAGEN). For GFP2 and GFP4 vectors successfully cloned enhancer constructs were further isolated with ZymoPure II Maxiprep kit (Zymo Research).
2.3. GFP loss assay (DT40)
Plasmids containing SV40 enhancer and modifications were transfected to chicken B cell line DT40 (RRID:CVCL_0249) cells with modifications (UNG-/-AICDAR/puro). Assay performance is described in detail in Refs. [32,42]. At the end of the assay cells were analyzed using Accuri C6 cytometer (BD Bioscience) and Novocyte (Agilent) cytometers. Results were further analyzed with FlowJo software (RRID:SCR_008520) and GraphPad Prism 9 software (RRID:SCR_002798).
2.4. GFP loss assay (Ramos and other cell lines)
The GFP loss assay with the GFP7 reporter was done as previously described [40]. Plasmids containing SV40 enhancer and modifications were transduced to Ramos B cells, UO-31 cells, U2OS cells, MDA-MB-231 cells, NIH3T3 cells and HEK293T cells. GFP expression was assessed by analysis with an LSRII cytometer (Becton Dickinson). Results were further analyzed with FlowJo software and GraphPad Prism 9 software.
2.5. Ectopic AID expression
MSCV-based retroviral vector expressing AID-mCherry fusion protein were used for introduction of AID expression cassette into the studied cell lines. AID-mCherry expression together with hygromycin resistance gene under IRES control in the same transcription unit was controlled by the MSCV LTR promoter. We prepared vectors containing AID-mCherry fusion proteins with human, mouse and chicken AID coding sequences. Cell line clones containing GFP reporter vectors were transduced with AID-mCherry expressing vector and selected with hygromycin two days after the transduction. Three weeks after the transduction, GFP fluorescence was analyzed using Symphony or LSRII cytometers (Becton Dickinson). GFP fluorescence of mCherry-positive cells was analyzed using FlowJo software.
2.6. Luciferase assay
Tested SV40 enhancer and modifications were cloned to SalI/BamHI site at pGL4.23 vector. 20 μg of the plasmid was co-transfected with 2.5 μg of pGL4.75 Renilla luciferase control vector (Promega) into DT40 UNG-/-AICDAR/puro cells. Transfection was performed using the Amaxa Nucleofector kit V program B-023 (Lonza) or Xcell PBS protocol. The Dual-Glo Luciferase Assay System (Promega) was used to determine the relative activity of firefly luciferase to Renilla luciferase. Assay was performed according to manufacturer’s protocol.
2.7. Statistical analysis
Mann–Whitney U test was performed to evaluate statistical significance of differences in the medians of GFP loss and mean fluorescence intensity. Unpaired t-test was used to evaluate statistical significance of relative luciferase activity. Fisher’s exact test was used to evaluate statistical significance of WRC mutations compared to overall C targeting mutations. Statistical analyses were performed using GraphPad Prism 9 software.
2.8. LT area insertion to DT40 genome
SV40 LT area was amplified with PCR using Q5 High-Fidelity DNA Polymerase (NEB). Template was SV40 genome WT4 plasmid which was received as a gift from James DeCaprio. Primers were designed according to In-Fusion primer design protocol. Cloning to vectors was performed with In-Fusion cloning (Takara) kit according to the manufacturer’s protocol. Cloning sites were NheI/SpeI (GFP2).
GFP2 vector containing SV40 LT area was transfected to DT40 IgL(−) cells. Transfection was conducted as in GFP loss assay protocol [32]. Day after transfection blasticidin selection was added to kill the cells which had not taken the plasmid in. Final blasticidin concentration was 20 μg/ml. After 7–9 days, single clones were picked and expression cassette integration the right locus was confirmed with puromycin selection (final concentration 1 μg/ml). After this, targeted clones were cultured for 12 weeks. Genomic DNA and proteins were extracted at certain time points and the end of 12-week culture period. gDNA miniprep kit (Zymo Research) was used for DNA extraction. Protein extraction is described separately.
LT area was amplified from 12-week gDNA with PCR using Q5 High-Fidelity DNA Polymerase (NEB) and cloned to pUC19 vector to BamHI/HindIII site. Primers were designed according to In- Fusion primer design protocol. Cloning was performed with In-Fusion cloning (Takara) kit according to the manufacturer’s protocol. The cloned plasmids were isolated with GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific). Sequencing was performed at Eurofins Genomics, Germany. Sequences were analyzed using SnapGene software (RRID: SCR_015052).
2.9. LT area insertion to Ramos and UO-31
SV40 LT area was amplified with PCR using Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Scientific). Template was SV40 genome WT4 plasmid which was received as a gift from James DeCaprio. The PCR product was cloned via In-Fusion cloning system (Takara) into HpaI restriction site in GFP7 vector. Virus production was performed as described previously [40]. Ramos WT and UO-31 cell lines were transduced with the vector in multiplicity resulting in 1–2% of GFP-positive cells in culture to achieve one copy of the vector per cell. Three days after the infection, the GFP-positive cells were sorted, expanded and cultured for 12 weeks. Genomic DNA and proteins were extracted at the end of culture.
2.10. Mutation analysis of LT in Ramos and UO-31 cells
LT area was divided to three subregions for sequencing. From Ramos we obtained 35 sequences from first region (16–810bp), 191 sequences from second region (694–1521bp) and 190 sequences from third region (1467–2456bp). From UO-31 we obtained 94 sequences from first region, 190 sequences from second region and 192 sequences from third region. Sequences were aligned to SV40 reference genome using SnapGene software. Single-base substitutions, insertions, and deletions relative to the reference genome were included in the analysis. To avoid overrepresentation of SV40 strain variants or PCR-induced mutations, each mutation was calculated once despite its occurrence in multiple sequences. Mutation distribution, type, frequency of AID hotspot WRC mutations, and mutations leading to STOP-codons were calculated. As LT was sequenced in three regions to cover entire area, but different amounts of sequences were obtained, mutation frequency was calculated using 200 bp bins except for the last bin which was 273bp. The number of mutations was divided by the number of sequences analyzed. Graphs were made with GraphPad Prism software.
2.11. Protein extraction and western blot analysis in DT40 cells
10M cells in a suspension were centrifuged at 500×g, 5 min at +4 °C. Supernatant was discarded and cells were resuspended to 500 μl of PBS and centrifuged at 2000×g, 3 min at +4 °C. Supernatant was discarded and cells were resuspended to 100 μl ice-cold RIPA buffer with 1x protease inhibitor cocktail and incubated for 1 h on ice. After this, lysate was centrifuged at 10 000×g, 10 min at +4 °C. Supernatant was transferred to clean Eppendorf tube and 33.3 μl of 4x NuPage LDS Sample buffer with 0.2M DTT was added. Lysate was incubated at +70 °C for 10 min and stored at −80 °C.
10 μl of protein lysate and 5 μl of Spectra BR marker (Thermo Fisher Scientific) was loaded to NuPAge Bis-Tris 4–12 % gel with MOPS buffer in Xcell system (Thermo Fisher Scientific). Samples were run at 180V for 1 h. After this, proteins were transferred to nitrocellulose membrane using Xcell blotting system using manufacturers protocol in RT, 30V for 1 h. Membrane was blocked with 5%-BSA-TBS for 2–3 h at RT. Primary antibody incubations were performed overnight at +4 °C in rocker (LT and AID) or at RT, 15 min in rocker (GAPDH). Secondary antibody incubations were performed at dark, RT, 2–3 h (LT and AID) or 15 min (GAPDH), in a rocker. Membrane was imaged using Odyssey Fc machine (LICOR). Membrane was washed between blocking, changing the antibodies and before imaging for 3x5min with TBS.
Primary antibodies used in western blot were αAID 30F12 Rabbit (Cell Signaling) 1:1000 dilution in 5%-BSA-TBS, αSV40-LT pAb419 Mouse (Genetex) 1:100 dilution in 5%-BSA-TBS and αGAPDH 5G4 Mouse (Hytest) 1:10000 dilution in 5%-BSA-TBS. Secondary antibodies used were IRdye αRabbit 680RD 1:2500 in 5%-BSA-TBS for detecting AID expression, IRdye αMouse 800CW 1:5000 in 5%-BSA-TBS for detecting LT expression and IRdye αMouse 680RD 1:10000 in 5%-BSA-TBS for detecting GAPDH expression.
2.12. Protein extraction and western blot in Ramos B cells and UO-31 cells
Cells were washed with ice-cold PBS and the supernatant was removed by aspiration then 1× Laemmli sample buffer (0.5 mL per 5×106 cells/60 mm dish) was added. The cell lysate was boiled for 5min and sheared through a needle 10×.
5μl of protein lysate (Ramos WT and AID -/- cells), 30 μl of (UO-31), and 5 μl of color protein standard marker (NEB) was loaded to 13 % (for AID) or 12 % (for LT) SDS-PAGE Gel with Elfo buffer in Xcell system. Samples were run at 100V 10min and then 150V 1h. After this, proteins were transferred to PVDF membrane (pre-washed in 100 % methanol and incubate in buffer blotting 10min) using trans-blot SD semi dry transfer system (Biorad) in RT, 15V for 30min. The membrane was blocked with 5%-BSA-TBS (for AID detection) or 5 % milk (for LT detection) for 1h at RT. Primary antibody incubations were performed overnight at +4 °C in rocker. Secondary antibody incubations were performed at dark, RT, 1h. The chemiluminescent Detection Substrate-West Pico Plus (Thermo Fisher Scientific) was used for the detection. The images were obtained using the UVITEC Cambridge imaging system. The membrane was washed between blocking, changing the antibodies, and before imaging for 3×5min with TBS.
Used primary antibodies were αAID Monoclonal Antibody ZA001 Mouse (Thermo Fisher Scientific) 1:1000 dilution in 5%-BSA-TBS, αSV40-Tag pAb419 Mouse (Genetex) 1:100 dilution in 5%-BSA-TBS and αGAPDH Mouse (Thermo Fisher Scientific) 1:2000 dilution. αMouse IgG HRP linked antibody (Cell Signaling) 1:3000 dilution in 5%-BSA-TBS and goat αMouse IgG HRP linked antibody (NEB) 1:3000 dilution were used as secondary antibodies.
2.13. Immunohistochemistry
Ramos WT, Ramos AID-/- and UO-31 were seeded on microscope cover glasses (3 × 105/well in the 6-well plates with the cover glass with poly-L-Lysine). After overnight incubation, the cells were fixed with 4 % paraformaldehyde for 15min; washed 3× with PBS, permeabilized with 0,1 % Triton-1% BSA in PBS for 10 min; and washed 3× with PBS. The blocking was performed using 2 % BSA-0.15 % Glycine-10 % FCS in PBS at RT for 45 min; washed 3x with PBS. The slides were incubated for 1h at RT with primary antibody (1:20) Mouse Monoclonal Ab anti-AID (ZA001, Invitrogen), in PBS-0.1 % BSA-0.4 % Tween20, washed, and incubated with secondary antibodies anti-mouse-Cy3 (1:500) for 1 h at room temperature. Hoechst 33258 for 15min at RT was added to visualize the cell nucleus, and washed 3x with PBS. Images were taken using a Leica SP8 confocal microscope.
2.14. Leica SP8 confocal microscopy acquisition
Samples were acquired using a Leica SP8 FLIM confocal microscope, equipped with 405 nm, 445 nm, 552 nm, and white light laser. The 63x oil immersion objective was used (HC PL APO CS2 63x Oil, NA 1.4, WD 0.14 mm, DIC), zoom 4 for Ramos cells and zoom 2 for UO-31 cells. Hoechst 550 was detected using the laser 405 and detector PMT1 (420–480), and Cy3 was detected using laser 552 and detector PTM4 (554–620). The acquisition of Hoechst and Cy3 were separate in two different channels to avoid background. The laser intensities were adjusted to avoid bleed-through between channels. Data were collected with a resolution of 1024 × 1024 pixels and step every 0.45μm. Images were deconvoluted by Huygens software and the contrast was enhanced using ImageJ software. Deconvoluted images were subsequently exported for analysis using Fiji J.
2.15. Reverse transcription and quantitative PCR
Different cell lines were analyzed: Ramos WT, Ramos AID-/-, UO-31, U2OS, and MDA-MB-231 for AID and GAPDH. The reagent RNAzol® RT was used to isolate total RNA. H2Odd was added to the sample and incubated at RT for 15min, samples were centrifuged at RT at 16 000G for 15min. The supernatant was transferred and 75 % ethanol was added to precipitate RNA 10min at RT. The pellets were kept after centrifugation for 10 min at 12 000G at 15 °C and washed 2x with ethanol. H2Odd was added to the dry pellet and heated at 55 °C for 10min.
DNAase treatment using the kit RQ1 Rnase-free Dnase (Promega, ref. M6101) was performed. The prepared RNA was reversely transcribed with Protoscript II Reverse Transcriptase (NEB, M0368) using random primers from the kit.
An amount of 2 μL of cDNA was then used for quantitative PCR with the MESA GREEN qPCR MasterMix Plus (Eurogentec) and primers for AID (Forward 5′ AATTCAAAAATGTCCGCTGGGC∗T3′, Reverse 5′AGCGGAGGAAGAGCAATTC∗C3′) or GAPDH (Forward 5′AGGGCTGCTTTTAACTCTGG∗T3′, Reverse 5‘CCCCACTTGATTTTGGAGGG∗A3‘).
To generate the standard curve for absolute quantification of gene expression, we used serial dilution of Ramos WT. The samples (in triplicates) were run in a Bio-Rad CFX96TM Real-Time instrument with a 3-step protocol: one cycle of 3 min at 95 °C, then 40 cycles of 15 s at 95 °C, 20 s at 60 °C, and 20 s at 72 °C and final polymerization at 72 °C for 10 min. Cycles of quantification (Cq) values were generated by the CFX Manager software.
3. Results
3.1. SV40 enhancer has strong SHM targeting activity in DT40 and Ramos B cell lines
SHM is targeted to Ig genes by their enhancers and enhancer-like sequences during antibody affinity maturation [32]. The mechanism of targeting is yet to be elucidated but is thought to involve modulation of RNA Pol II progression [52]. Similar to Ig enhancers from various species, the SV40 enhancer contains an octamer, E-box sequences, an IRF-Ets composite site, and NF-kB binding sites, all demonstrated to be important for SHM targeting activity in B cells (Fig. 1A–D) [[32], [33], [34], [35]]. We measured the SHM targeting activity of the SV40 enhancer using established GFP loss assays by measuring the percentage of GFP-negative cells (GFP loss) based on reporter vectors expressing GFP (GFP2 assay) [31] or a hypermutation target sequence (HTS)-GFP fusion gene (GFP4 and GFP7 assays) (Fig. S1) [32,40]. HTS contains numerous SHM hotspot motifs designed to yield stop codons upon mutation of cytidine, allowing the vector to report SHM activity with high sensitivity by virtue of the loss of GFP fluorescence. These assays differ in sensitivity in detecting SHM and were performed in different host cells, thus the percentages of GFP loss with different assays (GFP2, GFP4 and GFP7) are not directly comparable. GFP loss assays have been extensively used to identify and characterize both Ig and non-Ig SHM-targeting enhancers as well as for mapping SHM-susceptible regions in the human B cell genome [[31], [32], [33],35,40,52]. An important and unifying feature of these reporter vectors is that GFP expression is driven by a strong, constitutively active promoter and the addition of a strong enhancer increases GFP transcription and fluorescence levels modestly or, in some cases, not at all—thereby allowing effects on SHM to be distinguished from those on transcription levels [32,40].
Fig. 1.
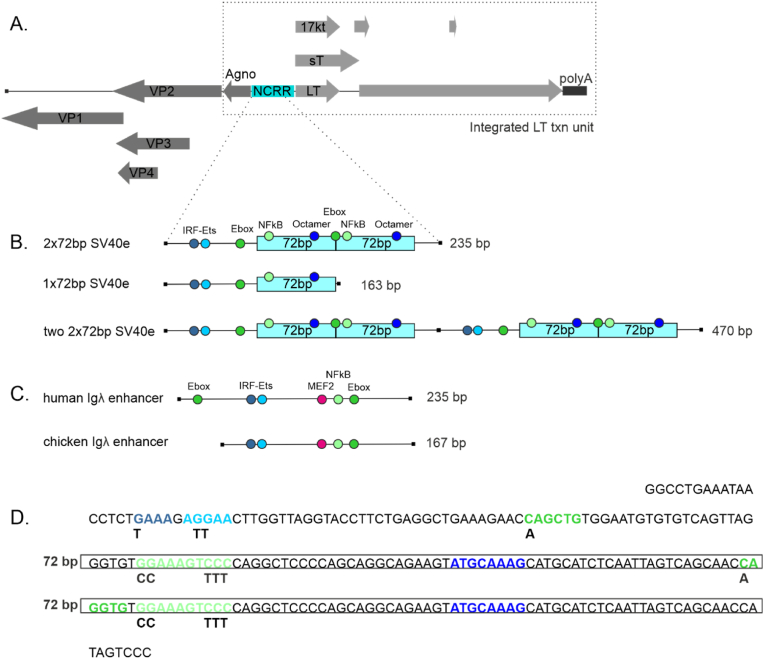
Diagram of SV40 genome, SV40 enhancer and Ig enhancers used in this study.
A. Genome organization of SV40. SV40 LT transcription unit integrated to Ramos, UO-31 and DT40 genome is indicated with rectangle with dashed lines. NCRR: non-coding regulatory region, LT: large tumor antigen, sT: small tumor antigen, 17 kT: 17 kDa protein, polyA: SV40 polyA signal, agno: agno protein: VP: viral structural proteins. Dashed lines to panel B indicate the SV40 enhancer region measured with GFP loss assay.
B. SV40 enhancer region and its modifications measured in GFP loss assays. Transcription factor binding sites which are important for SHM targeting in Ig enhancers are indicated in different colors.72bp: 72 base pair repeat sequence.
C. TFBS composition of chicken and human Igλ enhancers.
D. Sequence of SV40 enhancer. Locations of TFBSs are indicated in different colors (see panel B for coloring). 72bp repeats are highlighted with boxes. Changed nucleotides in mutated TFBSs are shown under the original sequence. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We found that the SV40 enhancer is a strong mutation enhancer in the chicken bursal lymphoma cell line DT40. The SV40 enhancer naturally contains one or two direct 72 bp sequence repeats depending on the strain of the virus [2]. The archetype SV40 enhancer contains one direct repeat of a 72bp sequence and, e.g., the reference laboratory strain 776 contains two repeats [2,53]. In DT40, the two-repeat enhancer (2×72bp) increases SHM-mediated GFP loss 25-fold (GFP4 loss 1.89 % in empty reporter vs 47.5 %, p < 0.0001) while the enhancer with two tandem copies (two 2x72bp) increased GFP loss 39-fold (74.3 %, p < 0.0001, respectively, Fig. 2A). This SHM targeting activity of the SV40 enhancer is higher than of the human Igλ sequence (GFP4 loss 20.2%, p=0,0033 compared to two 2x72bp enhancer and p=0.0002 compared to 2x72bp enhancer) and comparable to that of the chicken Igλ enhancer 3'core sequence (GFP4 loss 66.5%, p=0.0082 compared to two 2x72bp enhancer and p=0.5748 compared to 2x72bp enhancer) (Fig. 2A) [32]. The SV40 enhancer also increases SHM in the human Burkitt’s lymphoma cell line Ramos, driving a 40-fold and 85-fold increase in GFP7 loss when present in one or two copies (2x72bp or two 2x72bp), respectively (0.021 % in empty vector vs. 0.855 %, p < 0.0001 and 1.755 %, p < 0.0001 respectively, Fig. 2B). In Ramos cells, the SHM targeting activity of the two 2x72bp enhancer and one 2x72bp enhancer is comparable to human Ig heavy chain intronic enhancer core sequence (GFP7 loss 1,18%, p=0.2073 compared to two 2x72bp enhancer and p=0.1580 compared to 2x72bp enhancer (Fig. 2B). The consistency of the somatic hypermutation activity of the SV40 enhancer in Ramos and DT40 cells indicates that its mutation enhancer activity is evolutionarily conserved in vertebrates. This is in agreement with data reporting human and mouse Ig enhancer mutation activity in chicken DT40 B cell line or chicken enhancer element mutation activity in the human Ramos B cell line [32,40].
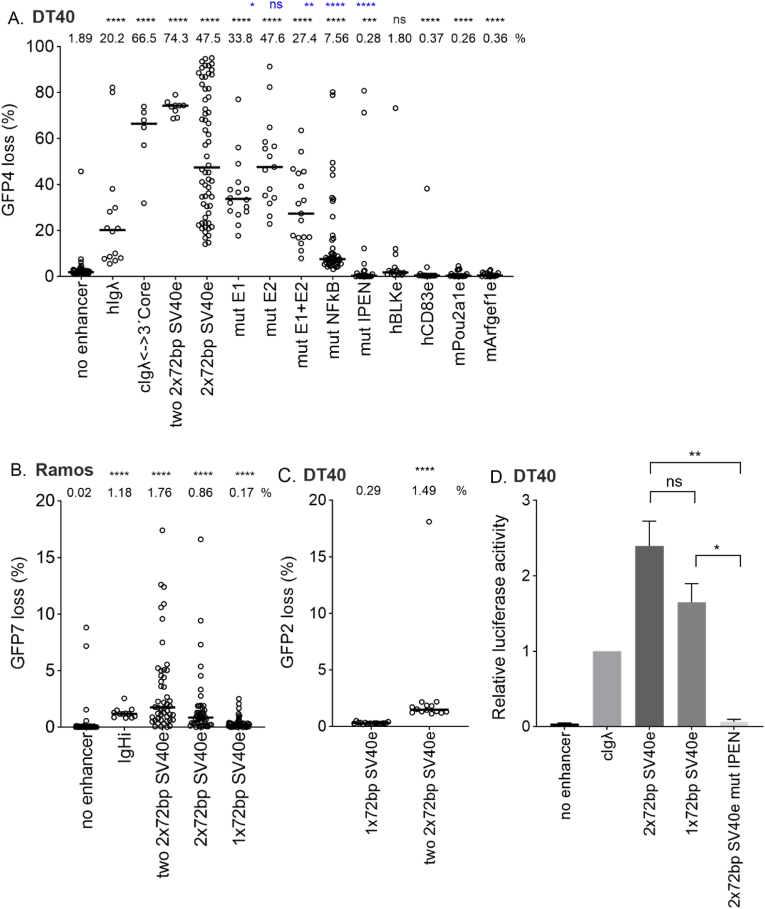
Fig. 2.
SV40 enhancer SHM targeting activity in chicken DT40 cells and human Ramos B cells.
A. SHM targeting activity of two 2x72bp SV40 enhancer, 2x72bp SV40 enhancer and its TFBS mutants as well as four genomic enhancers (human BLKe, human CD83, mouse Obf1 and mouse Cd79) measured in GFP loss assay in GFP4 reporter in chicken DT40 cell line. Empty GFP4 reporter vector was used as negative control and human Igλ (hIgλ) enhancer core and chicken Igλ enhancer 3′ core (cIgλ<−>3′core) were used as positive controls. Median GFP loss (%) values are shown above the graph. Statistical significance compared to negative control is indicated in black and statistical significance compared to 2x72bp enhancer is indicated in blue. Mann-Whitney U test was used to calculate statistical significances. Mutated TFBS binding site(s) are indicated in x-axes naming. IPEN: IRF-Ets, E-box and NF-KB binding site mutated.
B. SHM targeting activity of two 2x72bp SV40 enhancer, 2x72bp SV40 enhancer and 1x72bp SV40 enhancer in Ramos human B cell line. Empty GFP7 reporter vector was used as negative control and human Ig heavy chain intronic enhancer core (IgHi) was used as positive control. Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significances.
C. SHM targeting activity of 1x72bp SV40 enhancer and two 2x72bp SV40 enhancer measured in GFP loss assay in GFP2 reporter in chicken DT40 cell line. Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significance. The percentages of GFP loss in panels A, B and C are not directly comparable with each other due to the differences in sensitivity and host cell between GFP2, GFP4 and GFP7 reporters.
D. Enhancer activity of 2x72bp SV40 enhancer, 1x72bp SV40 enhancer and 2x72bp SV40 IPEN mutant enhancer measured in luciferase assay. Chicken Igλ enhancer (cIgλ) was used as positive control and empty vector as negative control. Two replicate measurements were performed. Fold changes over cIgλE are shown. Unpaired t-test was used to calculate statistical significances. ∗∗∗∗ p<0.0001, ∗∗∗ p<0.001, ∗∗ p<0.01, ∗ p<0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To determine whether SV40 mutation enhancer activity depends on similar TFBSs compared to Ig enhancer, we created TFBS mutants from the single enhancer (2x72bp), where binding sites were mutated individually (E-box1, E-box2, NF-kB) or in various combinations (both E-boxes, or E-boxes together with IRF-Ets and NF-kB binding sites, termed IPEN) and tested them in the GFP4 loss assay in DT40 cells (Fig. 1, Fig. 2A). Mutation of E-box1 decreases GFP loss by 28.8 % compared to WT 2x72bp enhancer (33.8 % vs. 47.5 %, p = 0.0423) (Fig. 2A). Mutation of E-box2 has no or little effect on 2x72bp SV40 enhancer SHM targeting activity alone (GFP loss 47.6 % vs. 47.5 %, p = 0.7605), but has a small additive effect with the mutation of the other E-box (GFP loss 27.4 % vs. 47.5 %, p = 0.0012). Mutating the NF-kB site decreases SHM targeting activity by 84.1 % (GFP loss 7.56 % vs. 47.5 %, p < 0.0001). Mutating IRF-Ets both E-boxes, and NF-kB binding sites abolishes SHM targeting activity completely (GFP loss 0.28 % vs. 47.5 %, p < 0.0001). This indicates that the SHM-targeting activity of the SV40 enhancer in B cells is dependent on the same transcription factor binding sites as Ig enhancers [32]. Together these data demonstrate that the SV40 enhancer is a bona fide mutation enhancer.
We also determined SHM targeting activity of four genomic enhancers: human BLK and CD83 enhancers (hBLKe and hCD83e) and mouse Obf1 and Arfgef1 enhancers (mObf1e and mArfgef1e). None of these elements exhibited mutation enhancer activity as their GFP loss percentage was equal to or significantly lower than GFP loss of the negative control (GFP loss 1.89 % vs. 1.80 %, p = 0.8869; 0.37 %, p < 0.0001; 0.26 %, p < 0.0001 and 0.36 %, p < 0.0001, respectively) demonstrating that not all enhancers are mutation enhancers.
The number of 72 bp repeats affects virus properties such as virus growth and replication efficiency [2,3,54]. To test if the number of repeats affect mutation enhancer activity, we tested the one (1x72bp) and two-repeat (2x72bp) SV40 enhancers for their SHM targeting activity using GFP loss assays. SHM targeting activity was 5-fold decreased in 1x72bp enhancer when compared to 2x72bp enhancer in Ramos (GFP7 loss 0.17 % vs. 0.86 %, p < 0.0001, Fig. 2B). Comparison of 1x72bp enhancer with two 2x72bp SV40 enhancer (containing total of four 72bp repeats) revealed 5-fold decrease of SHM targeting activity in DT40 (GFP2 loss 0.29 % vs. 1.49 %, p < 0.0001, Fig. 2C) and 10-fold decrease in Ramos (GFP7 loss 0.17 % vs. 1.76 %, p < 0.0001, Fig. 2B). Comparison of single 2x72bp enhancer with two 2x72bp SV40 enhancer showed 1.6-fold decrease in DT40 (GFP4 loss 47.5 % vs. 74.3 %, p = 0.0666, Fig. 2A) and .1-fold decrease in Ramos (GFP7 loss 0.86 % vs. 1.76 %, p = 0.0034, Fig. 2B).
As in previous studies [34,40], we used GFP mean fluorescence intensity (MFI) of the GFP positive population to assess the effects of various enhancers on levels of transcription. The mean GFP MFI of cells containing a vector with two 2x72bp SV40 enhancers was 1.5-fold higher than 1x72bp SV40 enhancer in DT40 (p < 0.0001) (Fig. S2A). We also measured the gene expression induced by 2x72bp SV40 enhancer, 1x72bp SV40 enhancer, and 2x72bp SV40 enhancer IPEN mutant with a luciferase assay (Fig. 2D). There was no significant difference in fold changes between 2x72bp enhancer and 1x72bp enhancer (p = 0.1256). The fold change of activity of 2x72bp and 1x72bp enhancers compared to 2x72bp enhancer IPEN mutant was significantly greater (p = 0.0100 and p = 0.0121, respectively). In Ramos cells, the two 2x72bp SV40 enhancer increased GFP mean fluorescence intensity while the single 2x72bp SV40 enhancer and single 1x72bp SV40 enhancer had significantly smaller effect (Fig. S2B). The fluorescence intensity of GFP driven by a single 2x72bp SV40 enhancer was not affected by the loss of one 72bp repeat in Ramos, further highlighting the role of two 72bp repeats for mutation activity (Fig. S2B).
The expression of AID in Ramos and DT40 was confirmed using qPCR and Western blot methods (Fig. 3, Fig. 5B). To confirm the dependency of the observed GFP loss on AID activity, we performed GFP loss experiments in AID-deficient Ramos cells (Ramos AID-/-). SHM targeting activity of SV40 enhancer in Ramos AID-/- cells was restored to Ramos WT cell level by transducing cells with a retroviral vector bearing an AID-mCherry expression cassette (Fig. 3B). SHM targeting activity of the SV40 enhancer was entirely dependent on AID (GFP loss 0 %; Fig. 3C). Reconstitution of AID expression in the AID-deficient cells by transduction with a retroviral vector bearing an AID-mCherry expression cassette fully restored SHM and GFP loss with a 60-fold GFP loss increase upon SV40 enhancer insertion (Fig. 3D). Similarly, by reconstituting species-specific AID to AID-deficient DT40 cells by transducing with human, mouse or chicken AID-mCherry vector and measuring GFP loss of two 2x72bp SV40 enhancer we showed that human and chicken AID are equally active (GFP loss 2.25 % vs. 2.01, p = 0.8485) and the activity of mouse AID is around 2-fold higher than human or chicken AID (GFP loss 5.1 % vs. 2.25 % or 2.01, p = 0.0087 and 00152, respectively (Fig. S3A).
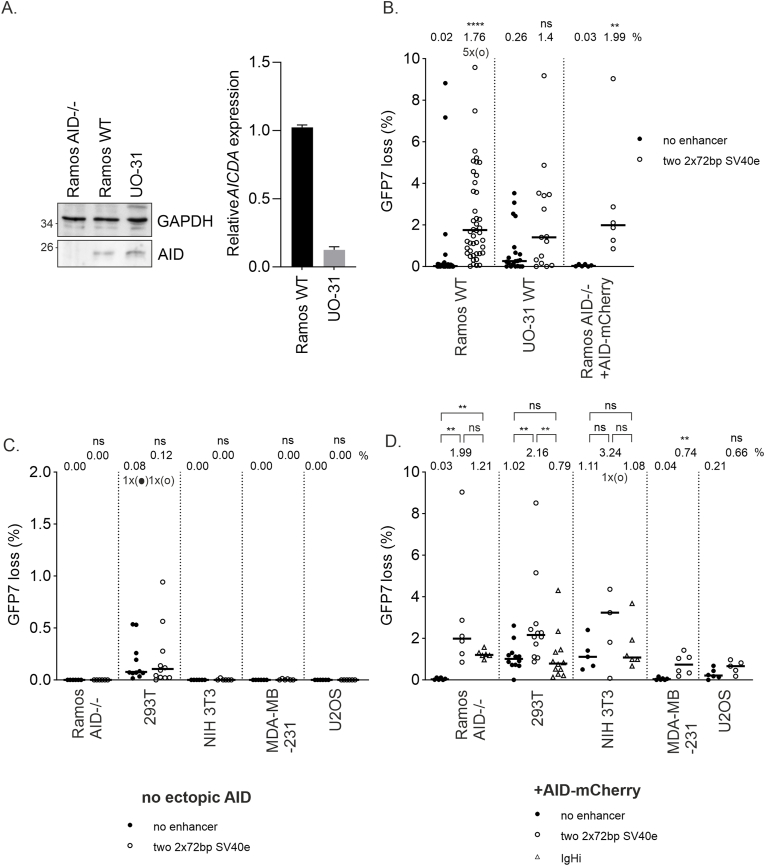
Fig. 3.
SV40 enhancer SHM targeting activity in non-B cells.
A. Endogenous AID and GAPDH expression of Ramos AID-/-, Ramos WT, and UO-31 WT cells measured with western blot (left) and endogenous AICDA expression of Ramos WT and UO-31 WT cells measured with RT-qPCR (right).
B. GFP loss of two 2x72bp SV40 enhancer (open circles) measured in GFP loss assay in GFP7 reporter in Ramos WT and UO-31 WT cell lines with endogenous AID. AID expression is restored to Ramos AID-/- cells by transducing with AID-mCherry vector which also restores SHM targeting activity. Ramos AID-/- + AID-mCherry data were replicated from Fig. 3D to facilitate comparison. GFP7 vector without enhancer is used as negative control (black circles). Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significance compared to negative control.
C. GFP loss of two 2x72bp SV40 enhancer (open circles) measured in GFP loss assay in GFP7 reporter in Ramos AID-/-, 293T, NIH 3T3, MDA-MB-231 and U2OS cell lines without exogenous AID expression. GFP7 vector without enhancer is used as negative control (black circles). Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significance compared to negative control. D. GFP loss of two 2x72bp SV40 enhancer (open circles) measured in GFP loss assay in GFP7 reporter in Ramos AID-/-, 293T, NIH 3T3, MDA-MB-231 and U2OS cell lines with exogenous AID expression from AID-mCherry expression vector. GFP loss of IgHi (open triangles) was also measured in Ramos AID-/-, 293T and NIH 3T3 cells with exogenous AID expression. GFP7 vector without enhancer is used as negative control (black circles). Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significance compared to negative control (black) and two 2x72bp SV40 enhancer compared to IgHi enhancer (blue).
Data points outside y-axis limits are marked with circles in brackets (o).
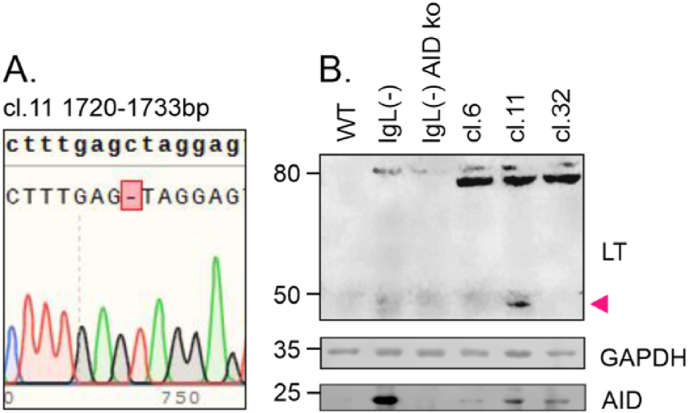
Fig. 5.
SV40 LT STOP codon mutation and truncated LT expression in DT40 cells.
A. Sequencing chromatogram from clone 11 lined with reference SV40 LT sequence. Deletion of C base at position 1727 results in-frame TAG STOP codon immediately after deletion.
B. SV40 LT, GAPDH and AID protein expression in DT40 cell lines measured with western blot. SV40 LT and GAPDH expression was measured from DT40 wild-type, IgL(−) and IgL(−) AID knock-out cell lines. Clones (cl.) 6, 11 and 32 are IgL(−) cells where SV40 LT expression cassette is integrated in to the deleted Ig light chain locus that lacks endogenous Ig enhancers (described previously in Refs. [31,33]). Protein size is indicated in left. Truncated LT is marked with arrow.
3.2. SV40 enhancer has SHM targeting activity in non-B cells
SV40 can infect various cell types and is linked to non-B cell malignancies such as mesotheliomas and bone and brain tumors [55]. Therefore, we also investigated SHM targeting activity of SV40 enhancer (2x72bp) in non-B cell lines. Human renal cell carcinoma UO-31 cells had detectable AICDA mRNA expression corresponding to 13 % of AICDA expression in Ramos (Fig. 3A) as measured with RT-qPCR. AID expression in UO-31 cells was further confirmed using western blot and immunohistochemistry (Fig. 3A and S3B). We measured the SHM targeting activity in UO-31 cells using the retroviral GFP7 reporter. The GFP loss was increased by SV40 enhancer more than 5-fold (Fig. 3B). Sequencing of the GFP7 coding sequence revealed 16 mutations out of which 10 (62.6 %) were targeted to Cs confirming the SHM-induced mutations in the GFP7 reporter (Fig. S5).
Non-B cell lines expressing AID are rare. We selected four cell lines of different tissue and species origin and transduced them with the GFP7 reporter. Analysis of clones from transduced mouse embryonic fibroblast NIH3T3, human embryonic kidney epithelial cell 293T, human osteosarcoma U2OS, and human breast adenocarcinoma MDA-MB-231 found no significant GFP loss, confirming that these lines do not have active SHM without induced AID expression (Fig. 3C). Ramos AID-/- cells were used as a control cell line for inactive SHM. Only 293T cells exhibited a significant percentage of GFP-negative cells. This was due to low sensitivity of 293T cell line to blasticidin selection, which did not fully remove cells with transcriptionally silenced GFP7 vector (Fig. 3C, data not shown).
We then introduced ectopic AID expression to the four parental cell lines 293T, NIH3T3, U2OS, and MDA-MB-231 using the AID-mCherry expression vector. Ramos AID-/- was used as a control to verify that this vector is able to restore the pattern of GFP loss we observed in wild-type Ramos cells. RT-qPCR analysis of AICDA expression upon AID-mCherry transduction into human non-B cell lines shows that the ectopic AICDA expression was achieved in all cell types (Fig. S4). GFP loss did not correlate with AID-mCherry expression, consistent with previous reports inficating that AID expression alone does not determine mutation frequency. Instead, factors such as DNA repair, especially in cancer cells, significantly influence AID-dependent mutagenicity [[29], [77]]. The panel of cells was then transduced with the GFP7 reporter vector lacking or bearing the SV40 enhancer. The SV40 enhancer increased the GFP loss 2–20-fold over the no enhancer control (Fig. 3D). Most of these non-B cell lines cells exhibited significant GFP loss even in the control vector. GFP loss did not correlate strongly with increased GFP fluorescence intensity indicating that the induction of SHM is not solely caused by an increase in transcription (Fig. S2C). Thus, the SV40 enhancer is a versatile SHM-targeting element able to target mutations in various cell types provided they express AID. Notably, this property of the SV40 enhancer is distinct from the well-characterized IgH intronic enhancer, which is a potent SHM-targeting element in B cell-derived cell lines (Ramos, DT40) but has neither SHM-targeting nor enhancer activity in non-B cells (Fig. 3D–S2C).
3.3. SHM targeting activity of SV40 enhancer leads to mutation accumulation in the SV40 LT region in human B cells and can lead to truncated LT expression
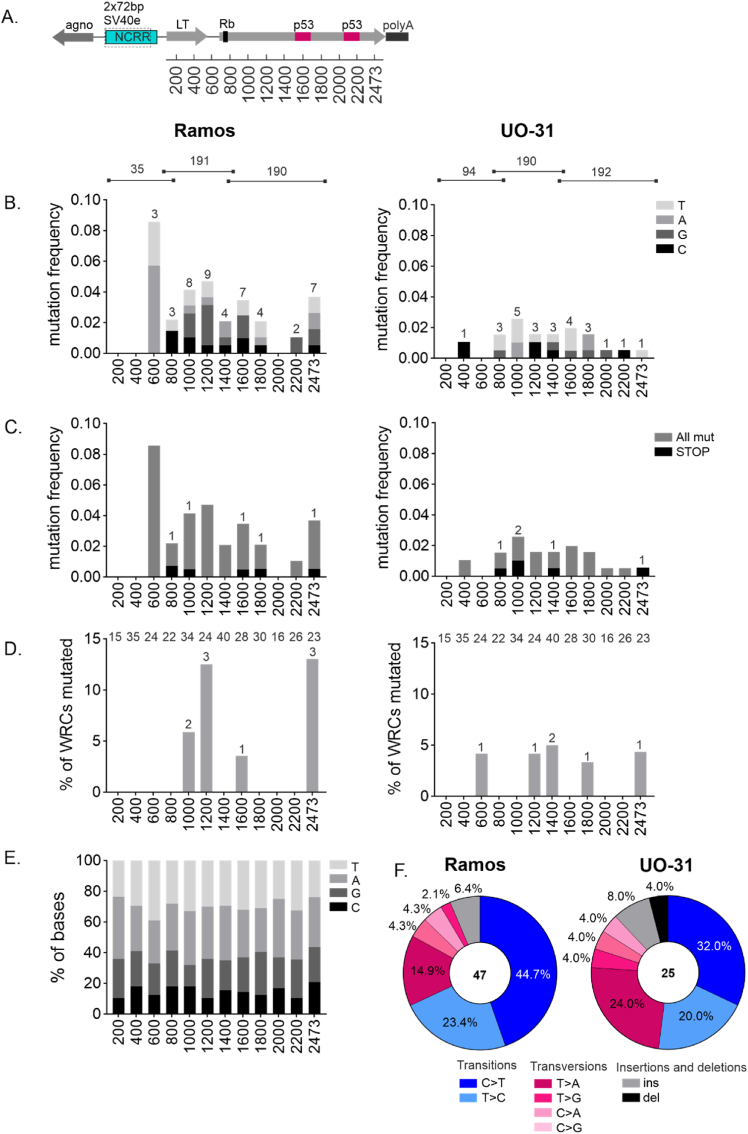
Our experiments demonstrate that the SV40 enhancer is able to target SHM to the GFP transcription units of the GFP2, GFP4, and GFP7 reporters. To determine whether the SV40 enhancer is able to target SHM to the SV40 LT region, we introduced the SV40 LT transcription unit including the enhancer region into Ramos B cells (Fig. 1, Fig. 4A). We also included the upstream SV40 agnoprotein coding sequence, which is transcribed in the reverse orientation from the same promoter-enhancer as LT (Fig. 1A) to mimic the natural genomic context of the SV40 enhancer. Following a 12-week culture period, we sequenced the LT region for mutations in three amplicons with 35, 191 and 190 sequences and grouped the mutations in bins (Fig. 4B–D). The LT region is uniformly A/T rich in all bins (Fig. 4E). We found 47 mutations out of which 26 (55.3 %) were targeted to Cs (Fig. 4F). 19.1 % of all mutations and 34.6 % of C-targeting mutations were at AID hotspots (WRC, Fig. 4D), with C-targeting mutations being significantly enriched at hotspots (p = 0.0025). 27.3 % and 72.2 % of substitutions were transversions and transitions, respectively (Fig. 4F). Three of the mutations introduced insertions or deletions causing frameshifts and two of the substitutions generated STOP codons (Fig. 4C). In a parallel experiment in DT40 cells, we also detected generation of a STOP codon in the LT coding sequence in one clone (cl. 11, Fig. 5A). In this clone, we also detected an approximately 50 kDa truncated LT protein (Fig. 5B) corresponding to the deletion of C at position +1727 downstream of the transcription start site, resulting in an in-frame STOP codon at position +1728 (Fig. 5A). These results further confirm that SV40 enhancer-induced SHM can cause truncation events in LT transcription unit in B cells.
Fig. 4.
Mutation type and distribution of LT region in Ramos and UO-31 cells.
A. A diagram of genome-inserted SV40 LT transcription unit with locations of Rb1-binding site and two-part p53 binding site. Location of bins is shown below the figure
B. Overall mutation distribution along SV40 LT region. Mutations targeting different bases are indicated in different colors. Mutation frequency (number of mutations/number of sequences per bin) is shown in the y-axis. Number of mutations observed in each bin is shown. Number of sequences obtained from Ramos and UO-31 LT region is shown above the graphs and concerns panels B–D.
C. Overall and STOP-codon forming mutation distribution along SV40 LT region. Overall and STOP-mutation are indicated in different colors. Number of STOP codon forming mutations is shown. Mutation frequency (number of mutations/number of sequences per bin) is shown in the y-axis.
D. % of mutations targeting AID hotspot WRC per bin along SV40 LT. Number of mutated WRC per bin is shown. Number of WRC motifs per bin in LT region is indicated on the top of the graphs.
E. Each bin of SV40 LT presented by its base content.
F. Mutation types in LT area. Substitutions are presented in six substitution classes: C > A, C > T, C > G, T > A, T > C, T > G. Substitutions are also divided to transitions (blue) and transversions (red) and are arranged accordingly. In addition, deletions and insertions (grey and black) are shown.
For panels BE LT area is divided to 200 bp bins except final bin being 273 bp. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. SHM targeting activity of SV40 enhancer induces mutations in LT in a non-B cell line
Since SV40 enhancer targets SHM both in B cells and in non-B cells, we also introduced the SV40 enhancer in the context of LT transcription unit (Fig. 1, Fig. 4A) into UO-31 cells which exhibit endogenous AID expression and SHM targeting. After 12 weeks of culture, we again sequenced the three amplicons (94, 190 and 192 sequences) covering the LT coding area (Fig. 4B–D). We detected 25 mutations out of which 10 (40.0 %) were targeted to Cs. Out of substitution mutations 40.5 % were transversions and 59.1 % were transitions (Fig. 4F). The mutation load was lower than in Ramos cells, which likely reflects the lower AID expression in UO-31 cells. Six mutations were in AID hotspots (Fig. 4D) and 60.0 % of all C targeting mutations were at WRC, indicating significant AID hotspot enrichment in C targeting mutations (p = 0.0012). Three of the mutations introduced insertions or deletions causing frameshifts and two of the substitutions generated STOP codons (Fig. 4C).
4. Discussion
In this study, we characterized the SHM-targeting properties of the SV40 enhancer in various cell types, which has implications for both tumorigenesis and virus fitness. We found that the SV40 enhancer has SHM targeting activity comparable to Ig enhancers in B cells and that unlike Ig enhancers, the SV40 enhancer is also active in non-B cells where it can stimulate SHM up to 20-fold. Furthermore, in cells expressing AID, the SV40 enhancer can substantially increase the probability of LT truncation mutations. LT truncation has been implicated as the event necessary for human cell malignant transformation of a related MCPyV polyomavirus [18] and truncation events have been observed with SV40 in human cell lines [56,57]. Comparison of SV40 enhancer SHM-targeting activity in various cell types revealed that it is most potent in B cells, where it shows a one order of magnitude higher stimulation of SHM than in non-B cells. We found that the SV40 enhancer has SHM targeting activity in human, chicken, and mouse cells, arguing for an evolutionary conserved mechanism of its SHM targeting activity.
Somatic hypermutation is targeted in the genome by a combination of mechanisms and the targeting mechanism has been an active topic of research for more than two decades. Cis-acting DNA elements are the major players in the specificity-determining mechanism, but the mechanism by which these elements operate is not well understood and the specific protein factors that target Ig SHM have not been definitively identified. The analysis of the mechanism has proven to be difficult, partly hampered by redundancy of factors and binding sites and in some cases, the essential nature of the protein factors for cell viability. Our findings argue that the mechanism by which the SV40 enhancer and Ig enhancers target SHM is similar. The same B cell TFBSs are important for the SHM-targeting activity of the SV40 enhancer and Ig enhancers in B cells (Fig. 2A) [32,35]. Distinct from B cell specific Ig enhancers, the SV40 enhancer has the ability to target SHM in non-B cells. This argues that certain non-B cell factors contribute to SHM targeting in non-B cells. Such factors might include transcription factors AP-2 and AP-1 which bind the SV40 enhancer [58]. AP-2 and AP-1 are expressed in kidney cells and have been implicated in early carcinogenesis of renal cell carcinomas [[59], [60], [61], [62]] and are thus potential contributors to the targeting of SHM to SV40 LT in kidney cells. AID targets different genes depending on cell type [63] implicating lineage appropriate factor contribution in the targeting activity in non-B cells. Elucidating the mechanism of action of Ig and SV40 mutation enhancers is a future task.
SV40 can integrate into the host genome and depending on host cell permissiveness (permissive or non-permissive), it can replicate. In semi-permissive human cells, where SV40 replicates with variable efficiencies depending on the cell type, virus replication results in host cell death. Stable infection of human cells, which is necessary for cell transformation, requires restriction of virus replication efficiency. One mechanism to decrease replication efficiency is LT truncation, which is found in human SV40-transformed cell lines [56,57]. This is a major difference compared to rodent cells which are non-permissive and thus do not require LT truncation events to limit virus replication and propagation. Thus it is not surprising that truncation of LT has no clear benefits but rather is a disadvantage in the transformation of rodent cells [24,[64], [65], [66]]. In human cells, SV40 small T antigen (sT) and LT are widely used for cell transformation and in such experiments LT truncation is not required in the absence of the entire SV40 genome. SHM can introduce stop codons via substitution or insertion/deletion-induced frame shift. Of the SV40 enhancer-induced mutations identified in this study, only a small fraction led to premature stop codon formation. While these mutations are uncommon, our data demonstrate that stop codons can be generated in the LT gene, which can lead to the production of truncated LT protein. Thus, truncation of LT in AID-expressing cells is a possible route for preserving the virus genome in host cells and for immune evasion and might predispose to cellular transformation in vivo.
Kidney cells are SV40’s natural host in monkeys, and lymphocytes and kidney cells are proposed reservoirs of SV40 in humans [55]. Among lymphomas, SV40 is most frequently found in diffuse large B cell lymphoma and follicular lymphoma, both of which have been linked to aberrant SHM [41,67]. Furthermore, traces of SV40 have been detected in human renal cell carcinomas [68,69]. It should be noted that evidence of SV40 found in these and other cancer types might reflect the ability of SV40 to infect and persist in various cell types, rather than reflecting a role in human oncogenesis. Nevertheless, our results show that the SV40 enhancer can facilitate mutation of LT via SHM in B cells, and at a lower rate in kidney cells, suggesting that these reservoir cells could serve as the site of truncation of LT and provide a model of how polyomaviruses could acquire mutations beneficial for their life cycle in these reservoir cells.
Although many studies have found associations between SV40 and human cancers, several studies have reported none or infrequent presence of SV40 at least in mesotheliomas and non-Hodgkin lymphomas [9,[11], [12], [13],[70], [71], [72]]. The detected presence of SV40 genetic material in human cancers might have originated from contaminated polio vaccine. Since SV40 is widely used in different molecular biology applications, the tested tumor samples might have been contaminated in the laboratory [70]. In addition, early anti-SV40 antibodies were cross-reactive and recognized parts of JCPyV and BKPyV, which frequently infect humans, possibly creating false-positive results [14]. Importantly, those individuals who received contaminated polio vaccines did not show increased incidence of cancer [10,14]. Thus, there is currently little direct evidence for the involvement of SV40 in human oncogenesis and the issue remains unresolved.
AID expression is a requirement for SHM and is usually limited to B cells. However, AID expression can be induced in non-B cells by inflammatory signals [[73], [74], [75]], which we simulated by expressing AID-mCherry in the MDA-MB-231, U2OS, NIH 3T3 and HEK293T cell lines. Since the SV40 enhancer can recruit SHM in various cell types upon AID expression, it is attractive to think that SHM contributes to SV40-mediated oncogenesis. Given that SV40 LT is sufficient to transform cells and maintain the transformed phenotype but might not be enough to lead to full cancer progression [2], additional factors such as inflammation might be needed. SV40-associated cancers are more frequent in immunocompromised individuals [58] further arguing that overcoming SV40 virus immunogenicity, either by immune system malfunction or restriction of virus replication, can contribute to the development of cancer. However, it should be noted that cancers arise more frequently in immunocompromised individuals regardless of viral status due to reduced detection and/or clearance of malignant cells. The link between truncation and/or other mutations of SV40 LT and malignant transformation remains to be addressed.
Ramos and DT40 are model cell lines for studying SHM. Their ability to perform AID-dependent SHM, detected by the reporter assays used here, is well established [[31], [32], [33],35,40,76] and we show here that SHM does not occur without AID expression (Fig. 3C and D and S3A). Mutations target mostly cytosines and present high amount of C > T (and T > A) transitions, which is characteristic to AID-induced mutations [77,78]. We also saw enrichment of C targeting mutations to AID hotspot sequences (WRC) in Ramos and UO-31 cells. Therefore, our analysis of mutations in LT and earlier observations of these model cell lines expressing AID implicates AID as the main cause of these mutations despite the absence of AID-/- controls in our sequencing experiments.
Our results show that 2x72bp enhancer has stronger SHM targeting activity than 1x72bp enhancer (Fig. 2). However, duplication of the 72bp repeat can occur in some cell types [79] and two 72bp repeats can significantly increase transcription of the target gene [80]. In addition, 2x72bp repeat virus quickly replaces one-repeat virus in culture when co-transfected, which indicates a growth advantage of the two-repeat variant [81]. Nevertheless, 1x72bp variant has been detected in tumors more often than 2x72bp [2], which may be attributable to the higher prevalence of the 1x72pb variant, unstable nature of polyomavirus regulatory region [58], or loss of one repeat by AID-mediated recombination during cellular passage [32]. From the perspective of preserving the virus in the genome, 1x72bp might be favored as it limits transcription/replication and thus immunogenicity of the virus.
In rodents, the SV40 early region (enhancer and LT) is usually found intact and integrated into the genome without apparent integration site preference [2]. Research into sites of SV40 integration in the human genome is limited, but integration has been shown at least in osteosarcomas [82] and in immortalized human fibroblasts [83]. It has been proposed that polyomavirus LT expression is needed in early cell transformation but becomes dispensable later in tumor development (hit-and-run mechanism) [84,85] in which case integration is not necessary. However, if integrated into the genome, the SV40 enhancer could target SHM to nearby tumor-suppressor genes or proto-oncogenes, thus creating an additional mechanism by which SV40 could contribute to tumorigenesis. Our study reveals the first viral enhancer exhibiting strong SHM-targeting properties and demonstrates that this activity can introduce LT truncating mutations which could play a role in SV40-induced tumorigenesis in various cell types. The significance of the model in which SV40 enhancer contributes to human malignancies via SHM in vivo remains to be demonstrated.
CRediT authorship contribution statement
Filip Šenigl: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization. Anni I. Soikkeli: Writing – review & editing, Writing – original draft, Visualization, Investigation, Funding acquisition, Formal analysis. Salomé Prost: Investigation. David G. Schatz: Writing – review & editing, Supervision, Resources, Funding acquisition. Martina Slavková: Investigation. Jiří Hejnar: Supervision, Resources, Funding acquisition. Jukka Alinikula: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization.
Funding
This work was supported by the Czech Science Foundation (project 22-30384S to F.Š.); Czech Academy of Sciences (Premium Academiae Award 2018 to J.H.); the Finnish Cultural Foundation (to J.A.); the Sigrid Juselius Foundation (to J.A.), the Turku University Foundation (to J.A.); the Finnish Cultural Foundation Kymenlaakso Regional Fund (to A.S.); the Alfred Kordelin Foundation (to A.S.); and Cancer Society of Southwest Finland (to A.S. and J.A.); and National Institutes of Health (R01 AI 127642 to D.G.S.). FŠ and J.H. were supported by the project National Institute of Virology and Bacteriology (program EXCELES, no. LX22NPO5103) funded by the European Union–Next Generation EU; we also acknowledge institutional support from the project RVO (68378050).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tvr.2024.200293.
Contributor Information
Filip Šenigl, Email: filip.senigl@img.cas.cz.
Jukka Alinikula, Email: jukka.alinikula@utu.fi.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Decaprio J.A., Garcea R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013;11(4):264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butel J.S., Lednicky J.A. Response to more about: cell and molecular biology of simian virus 40: implications for human infections and disease. J. Natl. Cancer Inst. 2000;92(6):496–497. doi: 10.1093/jnci/92.6.496. [DOI] [PubMed] [Google Scholar]
- 3.Vilchez R.A., Kozinetz C.A., Butel J.S. Conventional epidemiology and the link between SV40 and human cancers. Lancet Oncol. 2003;4(3):188–191. doi: 10.1016/s1470-2045(03)01024-6. [DOI] [PubMed] [Google Scholar]
- 4.Dolcetti R., Martini F., Quaia M., Gloghini A., Vignocchi B., Cariati R., et al. Simian virus 40 sequences in human lymphoblastoid B-cell lines. J. Virol. 2003;77(2):1595–1597. doi: 10.1128/JVI.77.2.1595-1597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbone M., Gazdar A., Butel J.S. SV40 and human mesothelioma. Transl. Lung Cancer Res. 2020;9(Suppl 1):S47–S59. doi: 10.21037/tlcr.2020.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butel J.S., Vilchez R.A., Jorgensen J.L., Kozinetz C.A. Association between SV40 and non-Hodgkin’s lymphoma. Leuk. Lymphoma. 2003;44(Suppl 3):S33–S39. doi: 10.1080/10428190310001623784. [DOI] [PubMed] [Google Scholar]
- 7.Heinsohn S., Scholz R., Kabisch H. SV40 and p53 as team players in childhood lymphoproliferative disorders. Int. J. Oncol. 2011;38(5):1307–1317. doi: 10.3892/ijo.2011.967. [DOI] [PubMed] [Google Scholar]
- 8.Vilchez R., Butel J. SV40 in human brain cancers and non-Hodgkin’s lymphoma. Oncogene. 2003;22(33):5164–5172. doi: 10.1038/sj.onc.1206547. [DOI] [PubMed] [Google Scholar]
- 9.Samaka R.M., Aiad H.A., Kandil M.A., Asaad N.Y., Holah N.S. The prognostic role and relationship between E2F1 and SV40 in diffuse large B-cell lymphoma of Egyptian patients. Anal. Cell Pathol. 2015;2015 doi: 10.1155/2015/919834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah K.V. SV40 and human cancer: a review of recent data. Int. J. Cancer. 2007;120(2):215–223. doi: 10.1002/ijc.22425. [DOI] [PubMed] [Google Scholar]
- 11.De Rienzo A., Tor M., Sterman D.H., Aksoy F., Albelda S.M., Testa J.R. Detection of SV40 DNA sequences in malignant mesothelioma specimens from the United States, but not from Turkey. J. Cell. Biochem. 2002;84(3):455–459. [PubMed] [Google Scholar]
- 12.Hirvonen A., Mattson K., Karjalainen A., Ollikainen T., Tammilehto L., Hovi T., et al. Simian virus 40 (SV40)-like DNA sequences not detectable in Finnish mesothelioma patients not exposed to SV40-contaminated polio vaccines. Mol. Carcinog. 1999;26(2):93–99. [PubMed] [Google Scholar]
- 13.Manfredi J.J., Dong J., Liu W.J., Resnick-Silverman L., Qiao R., Chahinian P., et al. Evidence against a role for SV40 in human mesothelioma. Cancer Res. 2005;65(7):2602–2609. doi: 10.1158/0008-5472.CAN-04-2461. [DOI] [PubMed] [Google Scholar]
- 14.Poulin D.L., DeCaprio J.A. Is there a role for SV40 in human cancer? J. Clin. Oncol. 2006;24(26):4356–4365. doi: 10.1200/JCO.2005.03.7101. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N., Bernards R., Friend S.H., Gooding L.R., Hassell J.A., Major E.O., et al. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J. Virol. 1990;64(3):1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane D.P.C.L. T antigen is bound to a host protein in SV 40-transformed cells. Nature. 1979;278(March):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 17.McCormick F., Harlow E. Association of a murine 53,000-dalton phosphoprotein with simian virus 40 large-T antigen in transformed cells. J. Virol. 1980;34(1):213–224. doi: 10.1128/jvi.34.1.213-224.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuda M., Feng H., Kwun H.J., Rosen S.T., Gjoerup O., Moore P.S., et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA. 2008;105(42):16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., Wang X., Diaz J., Tsang S.H., Buck C.B., You J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 2013;87(16):9173–9188. doi: 10.1128/JVI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hesbacher S., Pfitzer L., Wiedorfer K., Angermeyer S., Borst A., Haferkamp S., et al. RB1 is the crucial target of the Merkel cell polyomavirus Large T antigen in Merkel cell carcinoma cells. Oncotarget. 2016;7(22):32956–32968. doi: 10.18632/oncotarget.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt M., Wieland U., Kreuter A., Pawlita M. C-terminal deletions of Merkel cell polyomavirus large T-antigen, a highly specific surrogate marker for virally induced malignancy. Int. J. Cancer. 2012;131(12):2863–2868. doi: 10.1002/ijc.27607. [DOI] [PubMed] [Google Scholar]
- 22.Duncavage E.J., Magrini V., Becker N., Armstrong J.R., Demeter R.T., Wylie T., et al. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J. Mol. Diagnost. 2011;13(3):325–333. doi: 10.1016/j.jmoldx.2011.01.006. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carbone M., Rdzanek M.A., Rudzinski J.J., De Marco M.A., Bocchetta M., Niño M.R., et al. SV40 detection in human tumor specimens. Cancer Res. 2005;65(21):10120–10121. doi: 10.1158/0008-5472.CAN-05-1911. [DOI] [PubMed] [Google Scholar]
- 24.Peden K.W.C., Srinivasan A., Vartikar J.V., Pipas J.M. Effects of mutations within the SV40 large T antigen ATPase/p53 binding domain on viral replication and transformation. Virus Gene. 1998;16(2):153–165. doi: 10.1023/a:1007941622680. [DOI] [PubMed] [Google Scholar]
- 25.Lednicky J.A., Stewart A.R., Jenkins J.J., Finegold M.J., Butel J.S. SV40 DNA in human osteosarcomas shows sequence variation among T- antigen genes. Int. J. Cancer. 1997;72(5):791–800. doi: 10.1002/(sici)1097-0215(19970904)72:5<791::aid-ijc15>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 26.Lednicky J.A., Garcea R.L., Bergsagel D.J., Butel J.S. Natural Simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology. 1995;212:710–717. doi: 10.1006/viro.1995.1529. [DOI] [PubMed] [Google Scholar]
- 27.Truckenmiller M.E., Tornatore C., Wright R.D., Dillon-Carter O., Meiners S., Geller H.M., et al. A truncated SV40 large T antigen lacking the p53 binding domain overcomes p53-induced growth arrest and immortalizes primary mesencephalic cells. Cell Tissue Res. 1998;291(2):175–189. doi: 10.1007/s004410050989. [DOI] [PubMed] [Google Scholar]
- 28.Casellas R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., Di Noia J.M. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 2016;16(3):164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76(1):1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 30.Methot S.P., Di Noia J.M. Chapter two – molecular mechanisms of somatic hypermutation and class switch recombination. Adv. Immunol. Elsevier Inc. 2017:37–87. doi: 10.1016/bs.ai.2016.11.002. Vol. 133. [DOI] [PubMed] [Google Scholar]
- 31.Blagodatski A., Batrak V., Schmidl S., Schoetz U., Caldwell R.B., Arakawa H., et al. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS Genet. 2009;5(1):1–11. doi: 10.1371/journal.pgen.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buerstedde J.M., Alinikula J., Arakawa H., McDonald J.J., Schatz D.G. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 2014;12(4) doi: 10.1371/journal.pbio.1001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohler K.M., McDonald J.J., Duke J.L., Arakawa H., Tan S., Kleinstein S.H., et al. Identification of core DNA elements that target somatic hypermutation. J. Immunol. 2012;189(11):5314–5326. doi: 10.4049/jimmunol.1202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinesh R.K., Barnhill B., Ilanges A., Wu L., Michelson D.A., Senigl F., et al. Transcription factor binding at Ig enhancers is linked to somatic hypermutation targeting. Eur. J. Immunol. 2020;50(3):380–395. doi: 10.1002/eji.201948357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald J.J., Alinikula J., Buerstedde J.-M., Schatz D.G. A critical context-dependent role for E boxes in the targeting of somatic hypermutation. J. Immunol. 2013;191(4):1556–1566. doi: 10.4049/jimmunol.1300969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müschen M., Re D., Jungnickel B., Diehl V., Rajewsky K., Küppers R. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J. Exp. Med. 2000;192(12):1833–1839. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasqualucci L., Migliazza A., Fracchiolla N., William C., Neri A., Baldini L., et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. U.S.A. 1998;95(20):11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen H.M., Peters A., Baron B., Zhu X., Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science (80-) 1998;280(5370):1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 39.Álvarez-Prado Á.F., Pérez-Durán P., Pérez-García A., Benguria A., Torroja C., de Yébenes V.G., et al. A broad atlas of somatic hypermutation allows prediction of activation-induced deaminase targets. J. Exp. Med. 2018;215(3):761–771. doi: 10.1084/jem.20171738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senigl F., Maman Y., Dinesh R.K., Alinikula J., Seth R.B., Pecnova L., et al. Topologically associated domains delineate susceptibility to somatic hypermutation. Cell Rep. [Internet] 2019;29(12):3902–3915. doi: 10.1016/j.celrep.2019.11.039. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasqualucci L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S.K., Küppers R., et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. 2001;412(July):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 42.Soikkeli A.I., Kyläniemi M.K., Sihto H., Alinikula J. Oncogenic Merkel cell polyomavirus T antigen truncating mutations are mediated by APOBEC3 activity in Merkel cell carcinoma. Cancer Res. Commun. 2022;2(November):1344–1354. doi: 10.1158/2767-9764.CRC-22-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alaribe F.N., Mazzoni E., Rigolin G.M., Rizzotto L., Maniero S., Pancaldi C., et al. Extended lifespan of normal human B lymphocytes experimentally infected by SV40 or transfected by SV40 large T antigen expression vector. Leuk Res. [Internet] 2013;37(6):681–689. doi: 10.1016/j.leukres.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 44.McNees A.L., Harrigal L.J., Kelly A., Minard C.G., Wong C., Butel J.S. Viral microRNA effects on persistent infection of human lymphoid cells by polyomavirus SV40. PLoS One. 2019;13:1–31. doi: 10.1371/journal.pone.0192799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tognon M., Luppi M., Corallini A., Taronna A., Barozzi P., Rotondo J.C., et al. Immunologic evidence of a strong association between non-Hodgkin lymphoma and simian virus 40. Cancer. 2015;121(15):2618–2626. doi: 10.1002/cncr.29404. [DOI] [PubMed] [Google Scholar]
- 46.Malkin D. Simian virus 40 and non-Hodgkin lymphoma. Lancet. 2002;359(9309):812–813. doi: 10.1016/S0140-6736(02)07957-6. [DOI] [PubMed] [Google Scholar]
- 47.Vilchez R., Butel J. Polyomavirus SV40 infection and lymphomas in Spain. Int. J. Cancer. 2003;107(3):505–508. doi: 10.1002/ijc.11394. [DOI] [PubMed] [Google Scholar]
- 48.Martini F., Dolcetti R., Gloghini A., Iaccheri L., Carbone A., Boiocchi M., et al. Simian-virus-40 footprints in human lymphoproliferative disorders of HIV- and HIV+ patients. Int. J. Cancer. 1998;78(6):669–674. doi: 10.1002/(sici)1097-0215(19981209)78:6<669::aid-ijc1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 49.Nonaka T., Toda Y., Hiai H., Uemura M., Nakamura M., Yamamoto N., et al. Involvement of activation-induced cytidine deaminase in skin cancer development. J. Clin. Invest. 2016;126(4):1367–1382. doi: 10.1172/JCI81522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okura R., Yoshioka H., Yoshioka M., Hiromasa K., Nishio D., Nakamura M. 2014. Letter to the Editor Expression of AID in Malignant Melanoma with BRAF V600E Mutation; pp. 2013–2014. [DOI] [PubMed] [Google Scholar]
- 51.Machida K., Cheng K.T.N., Sung V.M.H., Shimodaira S., Lindsay K.L., Levinet A.M., et al. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. U.S.A. 2004;101(12):4262–4267. doi: 10.1073/pnas.0303971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarsalainen A., Maman Y., Meng F.-L., Kyläniemi M.K., Soikkeli A., Budzyńska P., et al. Ig enhancers increase RNA polymerase II stalling at somatic hypermutation target sequences. J. Immunol. 2022;208(1):143–154. doi: 10.4049/jimmunol.2100923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmiter R.D., Chent H.Y., Messing A., Brinstert R.L.X. SV40 enhancer and large-T antigen are instrumental in development of choroid plexus tumours in transgenic mice. Nature. 1985;316(1):7–10. doi: 10.1038/316457a0. [DOI] [PubMed] [Google Scholar]
- 54.Lednicky J.A., Arrington A.S., Stewart A.R., Dai X.M., Wong C., Jafar S., et al. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 1998;72(5):3980–3990. doi: 10.1128/jvi.72.5.3980-3990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotondo J.C., Mazzoni E., Bononi I., Tognon M., Martini F. Association between simian virus 40 and human tumors. Front. Oncol. 2019;9(July):1–19. doi: 10.3389/fonc.2019.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gish W.R., Botchan M.R. Simian virus 40-transformed human cells that express large T antigens defective for viral DNA replication. J. Virol. 1987;61(9):2864–2876. doi: 10.1128/jvi.61.9.2864-2876.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kao C., Hauser P., Reznikoff W.S., Reznikoff C.A. Simian virus 40 (SV40) T-antigen mutations in tumorigenic transformation of SV40-immortalized human uroepithelial cells. J. Virol. 1993;67(4):1987–1995. doi: 10.1128/jvi.67.4.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J.F., You J. Regulation of polyomavirus transcription by viral and cellular factors. Viruses. 2020;12(10):1–18. doi: 10.3390/v12101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dalgin G.S., Drever M., Williams T., King T., DeLisi C., Liou L.S. Identification of novel epigenetic markers for clear cell renal cell carcinoma. J. Urol. 2008;180(3):1126–1130. doi: 10.1016/j.juro.2008.04.137. [DOI] [PubMed] [Google Scholar]
- 60.Urakami S., Tsuchiya H., Orimoto K., Kobayashi T., Igawa M.H.O. Overexpression of AP-1 transcriptional factor family and inhibition of cell growth by AP-1 genes antisense oligonucleotides in the Tsc2 gene mutant (Eker) rat renal carcinomas. Br. J. Urol. 1997;80(SUPPL. 2):133. doi: 10.1006/bbrc.1997.7731. [DOI] [PubMed] [Google Scholar]
- 61.Zhu H., Wang S., Shen H., Zheng X., Xu X. SP1/AKT/FOXO3 signaling is involved in miR-362-3p-mediated inhibition of cell-cycle pathway and EMT progression in renal cell carcinoma. Front. Cell Dev. Biol. 2020;8(May):1–9. doi: 10.3389/fcell.2020.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oya M., Mikami S., Mizuno R., Miyajima A., Horiguchi Y., Nakashima J., et al. Differential expression of activator protein-2 isoforms in renal cell carcinoma. Urology. 2004;64(1):162–167. doi: 10.1016/j.urology.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 63.Qian J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.R., Resch W., et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159(7):1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seneca N.T.M., Sáenz Robles M.T., Pipas J.M. Removal of a small C-terminal region of JCV and SV40 large T antigens has differential effects on transformation. Virology. 2014;468–470(412):47–56. doi: 10.1016/j.virol.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Comerford S.A., Schultz N., Hinnant E.A., Klapproth S., Hammer R.E. Comparative analysis of SV40 17kT and LT function in vivo demonstrates that LTs C-terminus re-programs hepatic gene expression and is necessary for tumorigenesis in the liver. Oncogenesis. 2012;1(9):1–9. doi: 10.1038/oncsis.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hermannstädter A., Ziegler C., Kühl M., Deppert W., Tolstonog G.V. Wild-type p53 enhances efficiency of simian virus 40 large-T-antigen-induced cellular transformation. J. Virol. 2009;83(19):10106–10118. doi: 10.1128/JVI.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vilchez R.A., Madden C.R., Kozinetz C.A., Halvorson S.J., White Z.S., Jorgensen J.L., et al. Association between simian virus 40 and non-Hodgkin lymphoma. Lancet. 2002;359(9309):817–823. doi: 10.1016/S0140-6736(02)07950-3. [DOI] [PubMed] [Google Scholar]
- 68.Malkin D., Chilton-Macneill S., Meister L.A., Sexsmith E., Diller L., Garcea R.L. Tissue-specific expression of SV40 in tumors associated with the Li-Fraumeni syndrome. Oncogene. 2001;20(33):4441–4449. doi: 10.1038/sj.onc.1204583. [DOI] [PubMed] [Google Scholar]
- 69.Shein H.M., Enders J.F. JDL. Transformation induced by simian virus 40 in human renal cell cultures. II. Cell-virus relationships. Proc. Natl. Acad. Sci. U.S.A. 1962;48:1350–1357. doi: 10.1073/pnas.48.8.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.López-Ríos F., Illei P.B., Rusch V., Ladanyi M. Evidence against a role for SV40 infection in human mesotheliomas and high risk of false-positive PCR results owing to presence of SV40 sequences in common laboratory plasmids. Lancet. 2004;364(9440):1157–1166. doi: 10.1016/S0140-6736(04)17102-X. [DOI] [PubMed] [Google Scholar]
- 71.Rizzo P., Carbone M., Fisher S.G., Matker C., Swinnen L.J., Powers A., et al. Simian virus 40 is present in most United States human mesotheliomas, but it is rarely present in non-Hodgkin’s lymphoma. Chest. 1999;116(6 SUPPL):470S–473S. doi: 10.1378/chest.116.suppl_3.470S. [Internet] [DOI] [PubMed] [Google Scholar]
- 72.Krainer M., Schenk T., Zielinski Cm C.C. Failure to confirm presence of SV40 sequences in human tumours. Eur. J. Cancer. 1995;31(11):1893–1895. doi: 10.1016/0959-8049(95)00284-p. [DOI] [PubMed] [Google Scholar]
- 73.Nonaka T., Minato N., Kinoshita K., Nonaka T., Toda Y., Hiai H., et al. Involvement of activation-induced cytidine deaminase in skin cancer development Find the latest version : involvement of activation-induced cytidine deaminase in skin cancer development. 2016;126(4):1367–1382. doi: 10.1172/JCI81522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Endo Y., Marusawa H., Kou T., Nakase H., Fujii S., Fujimori T., et al. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 2008;135(3):889–898. doi: 10.1053/j.gastro.2008.06.091. [DOI] [PubMed] [Google Scholar]
- 75.Endo Y., Marusawa H., Kinoshita K., Morisawa T., Sakurai T., Okazaki I.M., et al. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-κB signaling. Oncogene. 2007;26(38):5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- 76.Kothapalli N., Norton D.D., Fugmann S.D. Cutting edge: a cis -acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J. Immunol. 2008;180(4):2019–2023. doi: 10.4049/jimmunol.180.4.2019. [DOI] [PubMed] [Google Scholar]
- 77.Gu X., Booth C.J., Liu Z., Strout M.P. AID-associated DNA repair pathways regulate malignant transformation in a murine model of BCL6-driven diffuse large. B-cell lymphoma. 2019;127(1):102–113. doi: 10.1182/blood-2015-02-628164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen H.M., Tanaka A., Bozek G., Nicolae D., Storb U. Somatic hypermutation and class switch recombination in Msh6−/−Ung−/− double-knockout mice. J. Immunol. 2006;177(8):5386–5392. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- 79.O'Neill F.J., Greenlee J.E., Carney H. The archetype enhancer of simian virus 40 DNA is duplicated during virus growth in human cells and rhesus monkey kidney cells but not in green monkey kidney cells. Virology. 2003;310(1):173–182. doi: 10.1016/s0042-6822(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 80.Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmidt K., Keiser S., Günther V., Georgiev O., Hirsch H.H., Schaffner W., et al. Transcription enhancers as major determinants of SV40 polyomavirus growth efficiency and host cell tropism. J. Gen. Virol. 2016;97(7):1597–1603. doi: 10.1099/jgv.0.000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mendoza S.M., Konishi T., Miller C.W. Integration of SV40 in human osteosarcoma DNA. Oncogene. 1998;17(19):2457–2462. doi: 10.1038/sj.onc.1202179. [DOI] [PubMed] [Google Scholar]
- 83.Hara H., Kaji H. Random integration of SV40 in SV40-transformed, immortalized human fibroblasts. Exp. Cell Res. 1987;168(2):531–538. doi: 10.1016/0014-4827(87)90025-5. [DOI] [PubMed] [Google Scholar]
- 84.Liu J., Li H., Nomura K., Dofuku R., Kitagawa T. Cytogenetic analysis of hepatic cell lines derived from SV40-T antigen gene-harboring transgenic mice. Cancer Genet. Cytogenet. 1991;55(2):207–216. doi: 10.1016/0165-4608(91)90079-a. [DOI] [PubMed] [Google Scholar]
- 85.Baker S.C., Mason A.S., Slip R.G., Skinner K.T., Macdonald A., Masood O., et al. Induction of APOBEC3-mediated genomic damage in urothelium implicates BK polyomavirus (BKPyV) as a hit-and-run driver for bladder cancer. Oncogene. 2022;41(15):2139–2151. doi: 10.1038/s41388-022-02235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.