Abstract
Purpose
Small cell lung cancer (SCLC) has an extremely poor prognosis. Despite high initial response rates to chemotherapy and modest survival improvements with the addition of immune checkpoint inhibitors (ICI), almost all patients experience relapse and fatal outcomes. Recent genomic insights uncovered extensive molecular heterogeneity in addition to the almost uniform loss of RB1 and TRP53. Additionally, defective DNA mismatch repair (MMR) has recently been described in some SCLC cases. Here, we generated a novel SCLC mouse model capturing MMR deficiency and assessed immunotherapy responses.
Methods
We developed an MMR-deficient genetically engineered mouse model (GEMM) of SCLC by introducing a conditional Msh2 gene, crucial for maintaining MMR integrity, into the standard Rb1fl/fl;Trp53fl/fl (RP) model. Genomic characteristics and preclinical therapy responses were evaluated by focusing on overall survival and whole exome sequencing (WES) analyses.
Results
MMR-defective SCLC tumors (Rb1fl/fl;Trp53fl/fl;Msh2fl/fl (RPM)) developed later than tumors in MMR-proficient mice. However, the time from tumor manifestation to death of the affected animals was substantially shortened (median survival 55 days in RP vs. 46.5 days in RPM), indicating increased aggressiveness of MMR-defective tumors. RPM tumors exhibited MMR deficiency, high tumor mutational burden (TMB), and an elevated load of candidate neoantigens, compared to RP lesions (p = 0.0106), suggesting increased immunogenicity. Importantly, the overall survival of RPM animals was significantly improved when exposed to ICI.
Conclusion
We propose a novel RPM mouse model as a suitable system to mimic MMR-defective SCLC and tumors with high TMB. We provide in vivo evidence that Msh2 deficiency enhances ICI sensitivity. These findings could contribute to stratifying SCLC patients to immunotherapy, thereby improving treatment outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00432-024-05942-9.
Keywords: Small cell lung cancer, Immune checkpoint inhibitor, Tumor mutational burden, Mismatch repair deficiency, Genetically engineered mouse model
Introduction
Small cell lung cancer (SCLC) comprises 15% of lung tumors and is characterized by rapid proliferation and early dissemination of metastases. While patients with SCLC are almost uniformly responsive to initial chemotherapy, the 5 year survival rate remains < 10% since most patients will inevitably relapse (Nicholson et al. 2016). Despite recent advancements in improving therapeutic strategies, such as the addition of immune checkpoint inhibitors (ICI), atezolizumab or durvalumab, to chemotherapy as the firstline treatment, only a modest increase in overall survival by approximately two months was achieved (Horn et al. 2018; Dingemans et al. 2021; Paz-Ares Lancet 2019). Additionally, the clinical response varies substantially among patients, and it is still unclear which patients may benefit from the addition of ICI.
Comprehensive studies on deciphering the genomic complexity of SCLC have identified highly recurrent alterations in the RB1 and TRP53 tumor suppressor genes, which occur in almost all SCLC cases (George et al. 2015, 2024; Sivakumar et al. 2023). Although SCLC is typically associated with massive tobacco smoke exposure (Wang et al. 2023), emerging evidence strongly suggests that approx. 15% of clinical cases harbor a predominant DNA mismatch repair (MMR) mutational signature (Liu et al. 2024). MMR defects result in genomic instability and a high tumor mutational burden (TMB) (Dietlein et al. 2014a). MMR has been shown to influence not only tumorigenesis, but also the response to immunotherapy by increasing the sensitivity of solid tumors to immune checkpoint inhibition (Le et al. 2017). In fact, the FDA has approved ICI therapy for cancers with MMR deficiency, underscoring the clinical significance of this biomarker (fda & cder, n.d.; Marcus et al. 2019). Similarly, recent efficacy data observed in patients with SCLC revealed that a high TMB provided greater clinical benefit when treated with a combination of two ICI agents (Hellmann et al. 2018). At this point, further validation of the impact of the mutational burden is needed to understand mechanisms mediating ICI efficacy.
Identifying mutationally-defined SCLC subtypes that respond distinctly to immunotherapy is, at this point, challenging since most patients present with an extensive stage disease that complicates in-depth studies on tumors. Furthermore, all patients are currently treated with a combination of chemotherapy and ICI, which complicates dissecting the effect of ICI (Rudin et al. 2021). Although genetically modified mice have provided important insights into the molecular features of the malignancy, current SCLC preclinical models do not fully recapitulate the human disease (Oser et al. 2024). The most commonly used mouse models mirror the conditional loss of Rb1 and Trp53 (referred to as the RP model, for Rb1fl/fl;Trp53fl/fl) and key histological and genome alterations found in patients (Meuwissen et al. 2003; McFadden et al. 2014), but fail to capture the high TMB that is typically observed in human SCLC.
Here, we introduce a novel SCLC mouse model harboring an MMR deficiency mutational signature and explore the impact of this genetic makeup on tumor aggressiveness and response to checkpoint blockade. We adapted the RP mouse model by breeding in a conditional Msh2 knockout gene (Rb1fl/fl;Trp53fl/fl;Msh2fl/fl (RPM)) using a previously published Msh2 allele (Kucherlapati et al. 2010). Msh2 is crucial to the MMR pathway by recognizing and binding to mismatched nucleotides during DNA replication, recruiting other MMR proteins to excise the error, and thus maintaining genomic stability. Upon deletion of Msh2, microsatellite instability (MSI) is induced (De Wind et al. 1995; Dietlein et al. 2014a). MSI induction leads to increased genomic instability and has significant implications for cancer treatment. Specifically, an MSI-high status in colorectal cancer was strongly associated with favorable survival outcomes in patients treated with pembrolizumab (a PD-1 blocking antibody) (Le et al. 2015). This finding prompted the FDA to approve MSI/MMR as a predictive biomarker for the pembrolizumab treatment of adult and pediatric patients with metastatic or unresectable solid tumors (Marcus et al. 2019; Wang et al. 2021). Through genomic and survival analyses, we investigated the impact of Msh2 deletion on SCLC tumor development, mutational patterns, and ICI response. We detected higher tumor aggressiveness, increased TMB, a predominant MMR mutational signature, and more candidate neoantigens in RPM tumors compared to tumors derived from the RP model. Importantly, in vivo treatment uncovered an increased ICI sensitivity in RPM tumor-bearing mice compared to the RP parental strain. Our findings offer insights into potential patient stratification strategies and might ultimately help clinicians design informed patient-tailored clinical studies.
Materials and methods
Experimental mice
Animal experiments in this study were approved by the local Ethics Committee of Animal Experiments authorities (LANUV, North Rhine-Westphalia, Germany) under license number 81-02.04.2019-A491. All mice were maintained according to FELASA recommendations and in compliance with the European Union and German guidelines. Mice were bred and housed up to five per cage in individually ventilated cages (IVC), with a 12-hour light/dark cycle and a temperature of 20–22 °C. This study utilized the standard SCLC mouse line harboring Rb1fl/fl, where exons 18 and 19 are flanked by loxP sites, and Trp53fl/fl, where loxP sites enclose exons 2–10. This model is well-established for recapitulating SCLC and is commonly referred to as the RP model (Meuwissen et al. 2003). To induce MMR deficiency in the RP background, we introduced a conditional Msh2 gene, generating the RPM model. This was achieved by mating the RP mice with Msh2LoxP/LoxP mice, an Msh2-deficient intestinal cancer mouse carrying an Msh2 genomic fragment with a flanked exon 12 (Kucherlapati et al. 2010), and then intercrossing to obtain the Rb1fl/fl;Trp53fl/fl;Msh2fl/fl genetic background.
Processing of whole exome sequencing (WES) data
Sequencing reads were aligned to the ensembl mouse reference GRCm39 using BWA v.0.7.17 (Li and Durbin 2009). PICARD v2.26 and samtools v 1.13 were used to mark and exclude PCR duplicates. Mutect2 from the Genome Analysis ToolKit (GATK) v4.2.1.0 was used to call somatic mutations using a panel of normals generated with 14 healthy control samples (Depristo et al. 2011). The resulting variants were filtered using GATK FilterMutectCalls, but without the filter for slippage. Variants identified in more than one sample or in dbsnp were excluded. Loci covered at a depth of less than 30 were also excluded. The TMB was calculated as the number of mutations/million bases covered at a depth of 30 or more.
Estimation of the Msh2 recombination efficiency
The read counts for each exon were calculated using GATK CollectReadCounts over the exons included in the GENCDE vM27 reference. The counts of the floxed Msh2 exon 12 were first normalized to the counts of the other Msh2 exons and this ratio was then normalized to that of control mice of the RP genotype to derive the estimated recombination efficiency as the copy number of exon 12. For visualization (Extended Data Fig. 1a–f), the coverage of individual bases within the Msh2 gene was extracted using GATK pileup v4.2.10. The predicted coverage profile of exon 12 in the absence of recombination (copy number = 2) was calculated using control mice of the RP genotype by dividing the coverage of individual exon 12 bases to the median coverage of bases within other exons. For each sample, this ratio was then multiplied by the median coverage of bases within non-floxed exons in the same sample.
Mutational signature analysis
WES data was used to extract the genomic context of the single nucleotide variants (SNV) (i.e., the triplets) utilizing a customized Python script and the Mus_musculus.GRCm39 assembly as the reference genome. Mutational signature analysis was performed using SigProfilerAssignment (v0.0.24) and the mouse signature COSMIC_v3 as a reference set panel (Alexandrov et al. 2020). All resulting signatures with an activity > 0 across all samples were selected. To assess the activity of MMR deficiency signatures, SBS15 and SBS21 mutational signatures were considered. Statistical analyses were conducted using the Kruskal-Wallis statistical test.
HLA-I binding prediction
The WES output was used to create a library of mutant coding sequences using an in-house pipeline, including missense, in-frame, and frameshift mutations. For missense and in-frame mutations, mutant sequences were trimmed to contain the mutation in a centered position flanked by 3’ and 5’ short sequences of 13 basepairs. Frameshift mutant sequences were trimmed only in the 3’ direction while preserving the whole frameshift in the 5’ end. This trimming strategy allows for the prediction of HLA-I binders of all possible sizes fitting the HLA-I peptide pocket (8–14 mers). Trimmed peptide sequences were used as input into the immune epitope database (IEDB) resource NetMHCpan (ver. 4.1) tool for HLA-I binding prediction (Reynisson et al. 2021). For simplicity, HLA-I binders were predicted only for the murine H2kb allele and 9 bp peptide length (9 mers), the most common binder size. Binders were selected based on a percentage rank below 2 and categorized as strong binders (rank < 0.5) and weak binders (0.5 < rank < 2). Statistical analyses were performed using the Kruskal-Wallis statistical test.
Treatment cohorts
After MRI confirmation of tumor growth and sufficient tumor burden within the 5–20 mm3 range, mice were randomly assigned to different therapy regimens. Compound solutions were administered as follows: The anti-PD-1 antibody RMP1-14 (BioXCell) was administered intraperitoneally (i.p.) at 200 mg/mouse twice a week for three weeks or three times per week until termination criteria were met. Etoposide (Hexal) was injected i.p. at 8 mg/kg on days 1, 2, and 3 of a 14-day cycle, while Cisplatin (Accord) was given i.p. at 4 mg/kg on day 1 of a 14-day cycle. Phosphate-buffered saline (PBS) was used as the vehicle control for comparison. Survival analysis was recorded by classifying as events only the animals that succumbed to the disease or were euthanized due to predefined termination criteria. Animals terminated for unrelated reasons (e.g., non-malignancy weight loss or injuries caused by cage mates) were excluded from the analysis. The p-values were calculated using the Mann-Whitney statistical test.
Results
MMR deficiency is associated with enhanced SCLC aggressiveness in vivo
To date, translational studies in SCLC are mainly restricted to the RP mouse model, driven by conditional biallelic inactivation of the Rb1 and Trp53 tumor suppressor genes (Meuwissen et al. 2003), which are almost universally mutated across SCLC patients (George et al. 2015, 2024). However, existing preclinical tools do not fully reflect the genomic complexity of human disease, which includes high TMB, among other characteristics. Moreover, the current models only partially cover the emerging transcriptomic and genetic subtypes of SCLC. For instance, recent research efforts have suggested that SCLC comprises distinct molecular subtypes based on the expression of ASCL1, NEUROD1, POUF2F3, or YAP1 transcription factors translating into potentially distinct therapeutic vulnerabilities (Rudin et al. 2019; Ireland et al. 2020). Additionally, a largescale gene expression analysis of human tumors classified patients into smoking-dominant, MMR-dominant, and APOBEC-dominant subtypes (Liu et al. 2024).
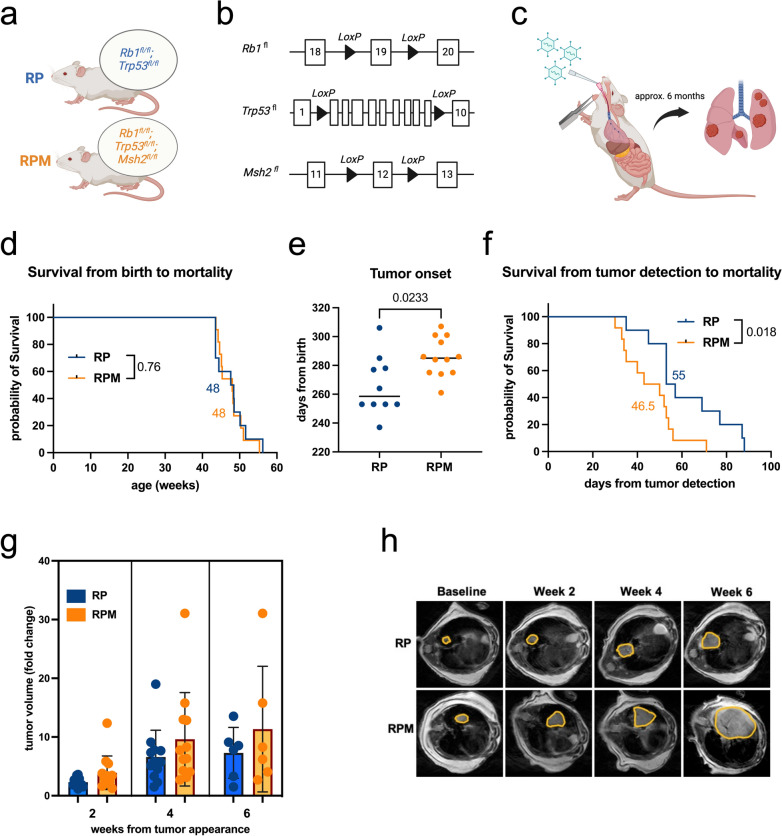
We, therefore, aimed to model a tumor with a high mutational burden. Since Msh2 is a critical component of the MMR pathway, we introduced a conditional Msh2 knockout allele into the RP background (Kucherlapati et al. 2010) (Fig. 1a, b). Lung tumors were induced by intratracheal instillation of Adeno-CMV-Cre, which led to the combined loss of all Rb1, Trp53, and Msh2 alleles in the infected cells (Fig. 1c). Tumor manifestation was detected approximately six months following Adeno-CMV-Cre exposure and bi-weekly MRI scanning was utilized to track tumor development. We confirmed the loss of Msh2 in tumors through coverage plots of WES data, which demonstrated successful Cre-mediated recombination of Msh2 exon 12 in RPM mice, consistent with the expected coverage profiles for heterozygously and homozygously recombined alleles (Extended Data Fig. 1a–f). Histopathological examination of harvested lung lesions confirmed typical histological features of human SCLC in RPM tumors, such as dense tissue with small round to fusiform cells with scant cytoplasm and granular nuclear chromatin, which is also similar to tumors from RP mice (Raso et al. 2021; Meuwissen et al. 2003) (Extended Data Fig. 1g). To evaluate the impact of MMR deficiency on tumor development, we monitored the overall survival and tumor volume progression of RPM and RP animals. We observed no survival differences (Fig. 1d); however, tumor onset in RPM animals was significantly delayed compared to the RP parental strain with a median onset of 285 days in RPM animals vs. 258.5 days in RP animals (p = 0.0233) (Fig. 1e). Notably, RPM mice exhibited significantly decreased survival following initial tumor onset with a median survival of 46.5 days compared to 55 days in RP mice (p = 0.018) (Fig. 1f). MRI scans revealed no significant tumor volume differences, although RPM tumors tended to be larger than RP lesions at any given time (Fig. 1g, h and Extended Data Fig. 1h). Both models rarely developed metastases, which, when present, were confined to the liver (Extended Data Fig. 1i). Altogether, our findings indicate that incorporating an Msh2 deletion into the RP background enhances the aggressiveness of SCLC tumors.
Fig. 1.
RPM tumors reveal enhanced aggressiveness compared to RP lesions. a Schematic of the RP and RPM mice with different genetic backgrounds. b Schematic of Rb1, Trp53, and Msh2 alleles in the mouse models. c Induction of lung tumors via intratracheal inhalation of Adeno-Cre virus. d Survival curves from birth of RP (n = 10, median 48 weeks) and RPM mice (n = 11, median 48 weeks). e Tumor onset in RP (n = 10, median 258.5 days) and RPM mice (n = 12, median 285 days). f Overall survival determined from the timepoint of tumor onset of RP (n = 10, median 55 weeks) and RPM mice (n = 12, median 46.5 weeks). g Fold change in tumor volumes determined by quantifying segmented tumors in MRI scans at week 2 (n = 12 RP, n = 13 RPM), week 4 (n = 12 RP, n = 12 RPM), and week 6 (n = 6 RP, n = 6 RPM) after tumor detection. h Exemplary MRI scans of RP and RPM lung tumors as in (g). The figure was produced using licensed Biorender.com. Logrank (MantelCox) statistical test (d, f) and Mann-Whitney statistical t-test (e, g)
Msh2 deletion in the RP background results in high TMB and genomic instability
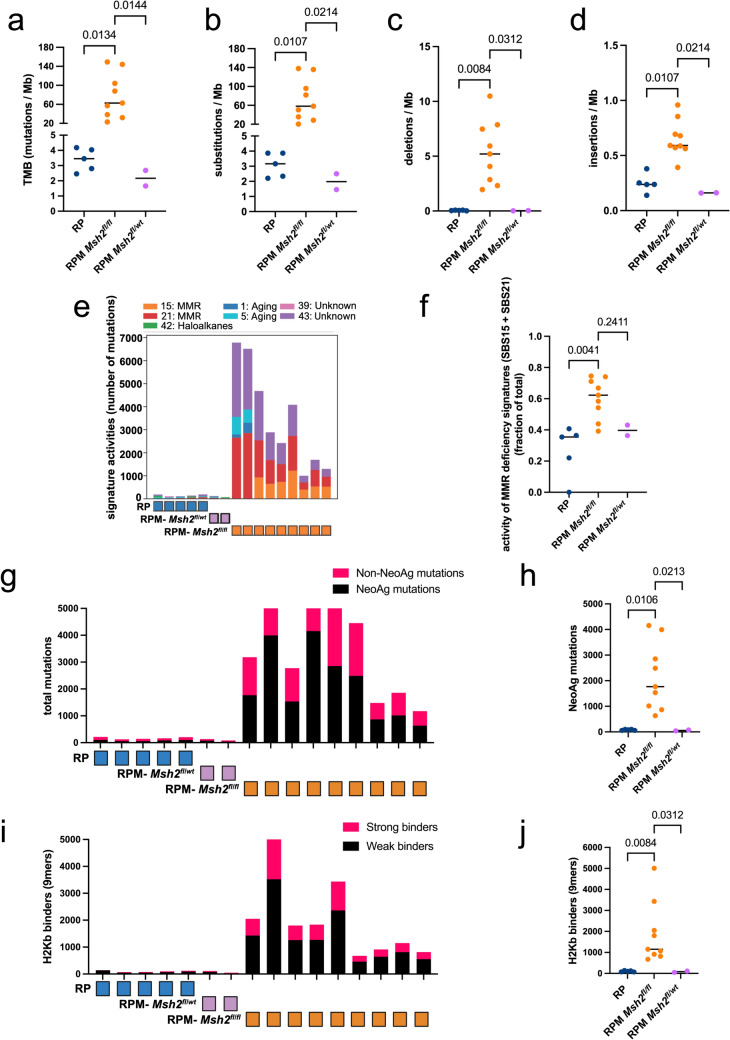
To assess the impact of Msh2 loss on the genomic landscape of RPM tumors and to verify the presence of the desired engineered deletions, we conducted WES on harvested lung lesions. Among 11 RPM subjects derived from mating RP and Msh2LoxP/LoxP mice (Meuwissen et al. 2003; Kucherlapati et al. 2010), 9 exhibited a homozygous loss of Msh2 (Msh2fl/fl), while 2 had a mono-allelic loss (Msh2fl/wt) (Extended Data Fig. 2a). We determined the TMB by measuring the total number of mutations per million bases for each tumor. RPM tumors with Msh2fl/fl showed an 18-fold greater TMB than the RP (p = 0.0134) and RPM tumors with heterozygous loss of Msh2 (p = 0.0144), indicating that a complete loss of Msh2 is necessary to increase the mutational load (Fig. 2a). Additionally, frameshift substitutions, deletions, and insertions were more common in RPM vs. RP tumors (Fig. 2b–d). To identify mutational MMR deficiency signatures, we performed mutational signature analysis based on the Catalogue of Somatic Mutations in Cancer (COSMIC). MMR deficiency signatures indicate the presence of deficiencies in DNA mismatch repair, typically represented by SBS15 and SBS21 mutational signatures, while MMR signature activities refers to the relative contribution of mutations associated with mismatch repair deficiency signatures among all mutations detected in a given tumor. We confirmed a predominant MMR deficiency signature in RPM mice in which Msh2 was homozygously deleted. Of note, Msh2 loss was not evident in tumors from RP mice (Extended Fig. 1a–b) and the MMR deficiency signature was substantially less dominant (p = 0.0041) (Fig. 2e, f). Yet, the detection of the MMR deficiency signature activities even in RP tumors, further indicates that MMR deficiency might be a selected event in SCLC. RPM tumors with heterozygous loss of Msh2 displayed an MMR deficiency signature activity level comparable to RP tumors (Fig. 2e, f). Subsequently, to assess the immunogenic potential of mutations, we performed neoantigen prediction using the HLA-I binding tool from the immune epitope database (Reynisson et al. 2021). Analysis of mutant peptide sequences derived from WES revealed that RPM subjects had significantly more HLA-I binders than RP samples, with heterozygous RPM samples falling within the RP range. This was evidenced by a significantly higher number of neoantigenic mutations (p = 0.0106) and both strong and weak H2Kb binders (p = 0.0084) (Fig. 2g–j and Extended Data Fig. 2b). These results strongly suggest higher immunogenicity of RPM tumors, compared to RP lesions or subjects with a heterozygous loss of Msh2. Overall, WES analysis demonstrated that the homozygous loss of Msh2 in the RP background significantly increased the TMB, MMR mutational signature, and candidate neoantigenic load, resembling the MMR-defective subset of human SCLC (Liu et al. 2024).
Fig. 2.
Msh2 deletion on the RP background increases the TMB, exhibits an MMR mutation signature, and harbors significantly more neoantigens. a Analysis of the TMB extent by WES. Identification of frameshift b substitutions, c deletions, and d insertions in the RP, RPM Msh2fl/fl , and RPM Msh2fl/wt background. e Assessment of mutational signatures by WES in RP and RPM animals based on COSMIC. f Statistical analysis of the activity of MMR deficiency signatures (SBS15 and SBS21) demonstrated as a fraction of the total in RP, RPM Msh2fl/fl , and RPM Msh2fl/wt lesions. g Assessment of neoantigen and non-neoantigen mutations, and h the corresponding statistical analysis. i Assessment of strong and weak H2Kb binders, and j the corresponding statistical analysis. All analyses are performed in n = 5 RP, n = 9 RPM with Msh2fl/fl, and n = 2 RPM Msh2fl/wt. The figure was produced using licensed Biorender.com. Kruskal-Wallis statistical test
RPM tumors display increased sensitivity to immune checkpoint inhibitors
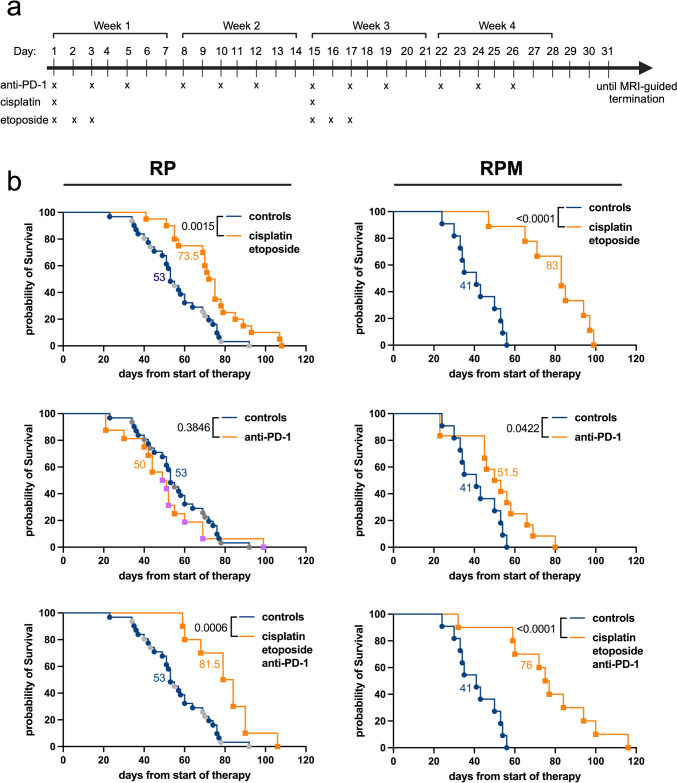
To determine the effects of the Msh2 deletion on ICI sensitivity, tumor-bearing animals were treated with an anti-PD-1 inhibitor upon MRI scanning-confirmed tumor manifestation. A pre-established preclinical dose was administered, with treatments continuing for three weeks or until predefined termination criteria were met. Additionally, treatments with cisplatin/etoposide alone and combined with ICI were included, representing the first-line treatment for patients with SCLC (Fig. 3a). As expected, since SCLC is initially highly responsive to chemotherapy, chemotherapy alone was effective in both RP and RPM models. Although tumors in both models responded well when exposed to cisplatin/etoposide, RPM tumors demonstrated a longer tumor response with a median survival of 83 days compared to 41 days in controls (p < 0.0001), while tumor-bearing RP mice had a median survival of 73.5 days, as compared to 53 days in controls (p = 0.0015). Strikingly, while ICI alone did not affect survival in RP mice, it significantly improved survival in RPM mice, with a median survival of 51.5 days compared to 41 days in controls (p = 0.0422). Similar to chemotherapy alone, the combination of cisplatin/etoposide with an anti-PD-1 antibody was effective in both mouse models, with RPM tumors showing higher sensitivity (Fig. 3b). Our findings suggest that MMR deficiency sensitizes SCLC to ICI therapy.
Fig. 3.
RPM tumors display increased sensitivity toward ICI treatment. a Preclinical treatment schedule for RP and RPM tumor-bearing mice. b Kaplan-Meier survival curves for RP mice (left column) and RPM mice (right column) treated with: cisplatin/etoposide (n = 20 RP and n = 9 RPM), anti-PD-1 antibody (n = 16 RP and n = 12 RPM), a combination of cisplatin/etoposide and anti-PD-1 antibody (n = 10 RP and n = 10 RPM), and vehicle control (n = 31 RP and n = 11 RPM). Cisplatin was dosed at 4 mg/kg, i.p., and etoposide was dosed at 8 mg/kg, i.p. Two dosing schedules were applied for the anti-PD-1 antibody: three times/week until termination criteria (orange) and twice/week for a total of three weeks (purple). The figure was produced using licensed Biorender.com. Log-rank (Mantel-Cox) statistical test
Discussion
Our study assesses the role of Msh2 deletion in driving tumorigenesis, genomic instability, and response to ICI in a novel mouse model of SCLC. By incorporating a conditional Msh2 knockout allele into the RP model, we generated a preclinical model (RPM) that mimics the MMR mutational signature and high TMB as observed in 15% of SCLC clinical cases in a recent study (Liu et al. 2024). Unlike previous preclinical tools, our system enables the study of therapeutic response dynamics in the presence of substantial mutational rates driven by an MMR defect, which is also detected in a subset of human SCLC (Liu et al. 2024).
Our findings uncovered that RPM tumors exhibit accelerated aggressiveness compared to RP lesions. Despite a delayed onset of the tumor formation, RPM-depleted tumors led to earlier death. We speculate that Msh2 deletion accelerates tumor progression, a finding that aligns with the role of MMR deficiency in promoting genomic instability (Dietlein et al. 2014a). This insight, combined with existing literature on the contribution of MMR deficiency to poor prognosis and aggressive tumor behavior in various cancers, such as colorectal malignancies (Oliveira et al. 2019), underscores the potential clinical relevance of our research.
WES analysis on micro-dissected lung tumors revealed a marked increase in TMB and mutational signatures associated with MMR in RPM tumors with homozygous Msh2 deletion (Msh2fl/fl). The relationship between MMR deficiency and high TMB is well documented, with studies showing that MSI, a form of genetic hypermutability resulting from impaired Msh2, causes the accumulation of very high numbers of somatic mutations (Popat et al. 2005b). Moreover, the Msh2 deletion in the RP background significantly amplified the load of predicted neoantigens, suggesting a stronger potential to elicit antitumor immune responses and heightened susceptibility to ICI.
Furthermore, as TMB and MSI-high have been proposed as potential predictive biomarkers for ICI response in SCLC (Taniguchi et al. 2020), we designed in vivo treatment regimens with a PD-1 blocking antibody alone or in combination with platinum-based chemotherapy, mirroring the clinical standard of care. We observed that while chemotherapy alone or in combination with ICI was beneficial in both mouse systems, only the RPM mice displayed increased overall survival when exposed to ICI alone. This notable response rate, which could be attributed to the elevated mutational load, aligns with recent clinical data indicating that high TMB bolsters the efficacy of immune checkpoint inhibition in SCLC patients (Hellmann et al. 2018). Previous studies have also reported a correlation between MMR-deficient/MSI-high mutational landscapes and enhanced ICI benefits in patients with colon cancer (Le et al. 2015; Olivares-Hernández et al. 2022). Notably, the role of MMR genes has been recently explored in non-small cell lung cancer (NSCLC), and alterations were associated with improved efficacy of nivolumab (a PD-1 checkpoint inhibitor) (Rizvi et al. 2015). However, evidence supporting the inclusion of TMB and MMR defects as ICI predictors in clinical practice is insufficient and contradictory. To illustrate, Westcott et al. (2023) reported that MMR deficiency developed in similar in vivo lung and colon cancer models failed to display increased ICI response due to pronounced intratumor heterogeneity of mutations. In addition, in our study, combining an anti-PD-1 antibody with chemotherapy in RPM tumors did not outperform chemotherapy alone (Fig. 3b). There are a number of possible explanations for this. First, the chemotherapy backbone may simply induce toxicities in the T cell compartment, which prevent efficient T cell-mediated tumor eradication. However, given that numerous chemotherapy/immune checkpoint inhibitor combinations were shown to increase efficacy, compared to chemotherapy alone (for instance cisplatin/etoposide/durvalumab in SCLC (Paz-Ares et al. 2019) or carboplatin/pemetrexed/pembrolizumab in NSCLC (Garassino et al. 2023), we believe that this explanation is unlikely. We rather expect that addition of immune therapy to the cisplatin/etoposide backbone increases overall toxicity, which may cover a potential survival benefit brought about by the addition of an immune checkpoint inhibitor. While we did not investigate any potential immune-mediated toxicities in our experimental animals, the occurrence of immune-related adverse events (irAEs) could restrict the effectiveness of the combined treatment (Postow et al. 2018).
While this study provides insights into the role of MMR deficiency on SCLC tumorigenesis and response to immunotherapy, there are several limitations to acknowledge. Despite effectively mirroring MMR deficiency observed in some human SCLC tumors, the RPM mouse model may not fully capture the genetic and phenotypic diversity of human SCLC. The artificial genetic manipulations in mouse models might not entirely reflect the spontaneous development of mutations in human cancers. In fact, our experimental mice are not exposed to cigarette smoke, which is the dominant carcinogen that drives human SCLC development. In addition, while Msh2 deficiency clearly promotes and MMR deficiency phenotype, there are numerous additional genetic aberrations that drive an MMR deficiency phenotype (Reinhardt and Yaffe 2013; Dietlein et al. 2014b; Dietlein and Reinhardt 2014). Whether all of these genomic aberrations produce an identical genome maintenance defect is, however, unclear. Thus, our RPM approach likely only mimics a subset of MMR-defective SCLC cases. Furthermore, while our findings suggest enhanced sensitivity to immune checkpoint inhibitors in MMR-deficient SCLC, further studies are needed to validate our findings and assess potential toxicities of such a therapeutic approach and evaluate the long-term efficacy and potential resistance mechanisms in a clinical setting. It is worth noting that the experimental schemes used in this study, including the genetic engineering of mice and specific immunotherapy regimens, do not comply with standard clinical practice. This presents a challenge in translating our results to human patients. These limitations highlight the need to deepen our understanding of the role of MMR deficiency in SCLC by using diverse models and complementary methodologies.
All together, our research suggests that our novel high TMB SCLC preclinical model, RPM, is a valuable tool for investigating therapeutic response dynamics in vivo. Particularly, this model holds the potential to deepen our understanding of the complex interplay among the Msh2 deletion, elevated mutational load, and response to ICI. These insights could pave the way for potential stratification strategies for patients who would benefit from such treatment while minimizing unnecessary toxic effects on the remaining patients. Additionally, the RPM model may be valuable in developing and testing new treatment strategies targeting tumors with MSI/TMB-high tumors. Future efforts to validate our observations and identify the underlying mechanisms could significantly contribute towards personalized treatment approaches in SCLC, potentially improving treatment outcomes and optimizing therapeutic strategies.
Conclusion
Our study introduces a novel genetically engineered mouse model that recapitulates MMR deficiency in SCLC (RPM). This model uncovers that MMR deficiency leads to increased tumor aggressiveness, elevated TMB, and enhanced response to ICI therapy. These results suggest that MMR deficiency could be considered a stratifying criterion for ICI treatment of SCLC patients. Further research and clinical validation are necessary to fully understand the implications of MMR deficiency in SCLC and improve patient outcomes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Bianca Göbel, Yagmur Sahbaz, and Melanie Nelles from the University Hospital Cologne for their outstanding technical support. We also thank the CMMC Microscopy Facility for the use of light microscopes and the Cologne Center for Genomics for their sequencing services. We furthermore thank the Regional Computing Center of the University of Cologne (RRZK) for providing computing time on the DFG-funded (Funding number: INST 216/512/1FUGG) High Performance Computing (HPC) system CHEOPS as well as support.
Abbreviations
- COSMIC
Catalogue Of Somatic Mutations In Cancer
- FELASA
Federation of European Laboratory Animal Science Associations
- GATK
Genome Analysis ToolKit
- GEMM
Genetically Engineered Mouse Model
- HLA-I
Human Leukocyte Antigen Class I
- ICI
Immune Checkpoint Inhibitor
- IEDB
Immune Epitope Database
- irAEs
Immune-related adverse events
- IVC
Individually Ventilated Cages
- LANUV
North Rhine-Westphalia State Agency for Nature, Environment, and Consumer Protection
- MMR
Mismatch Repair
- MRI
Magnetic Resonance Imaging
- MSI
Microsatellite instability
- PBS
Phosphate-Buffered Saline
- RP
Rb1fl/fl;Trp53fl/fl
- RPM
Rb1fl/fl;Trp53fl/fl;Msh2fl/fl
- SCLC
Small Cell Lung Cancer
- SNV
Single Nucleotide Variant
- TMB
Tumor Mutational Burden
- WES
Whole Exome Sequencing
Author contributions
G.S.H-S., H.C.R designed the study. O.I., F.R., F.B., A.S., J.M.H., H.W., H.G., R.B., G.B., J.G., R.K.T., K.B., G.S.H-S., H.C.R designed the experiments. O.I., A.S., G.L., J.S., D.B., M.K., G.G., L.L., J.C., L.N., M.I. performed the experiments. O.I., F.R., F.B., M.C., L.T.F., G.L. analyzed the data. O.I., F.B., L.T.F. generated the figures. O.I., H.C.R., G.S.H-S. wrote the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the German Research Foundation (DFG) (SFB1399- grant no. 413326622, to H.C.R., G.S.H.-S., J.G., R.K.T., M.C., F.B., H.W., G.L., H.G.; SFB1430 – grant no. 424228829, to H.C.R. and SFB1530 – grant no. 455784452, to H.C.R., HE 6897/2 − 1 to G.S.H.-S., HE 6810/3 − 1 to J.M.H.), the Else Kröner-Fresenius Stiftung (EKFS-2014-A06 to H.C.R., 2016_Kolleg.19 to H.C.R.), the Deutsche Krebshilfe (1117240 to H.C.R., 70113041 to H.C.R. and F.B. and the Mildred Scheel Nachwuchszentrum Grant 70113307 to F.B.), the German Ministry of Education and Research (BMBF e: Med Consortium InCa, grant 01ZX1901 and 01ZX2201A to R.K.T., J.G. and H.C.R.), the Center for Molecular Medicine Cologne (CAP-13 to J.M.H., CAP-16 to G.S.H.S., B02 to G.S.H.-S. and J.M.H.), and the Faculty of Medicine, University of Cologne: Koeln Fortune Program (272/2019) and Gusyk family support grant (to G.S.H.-S.). G.L. is funded by the Center for Biochemistry, Univeristy of Cologne – 956400, Köln Fortune, CANcer TARgeting (CANTAR) project NW21-062 A, two collaborative research center grants: SFB1399-413326622 Project C06, SFB1530-455784452 Project A03 both funded by the Deutsche Forschungsgemeinschaft (DFG) and associated to the collaborative SFB1403 also funded by the DFG. HW is funded by the Alexander von Humboldt Foundation, a Wellcome Trust Investigator Award (214342/Z/18/Z), a Medical Research Council Grant (MR/S00811X/1), a Cancer Research UK Programme Grant (A27323) and three collaborative research center grants (SFB1399, Project C06, SFB1530-455784452, Project A03 and SFB1403–414786233) funded by the Deutsche Forschungsgemeinschaft (DFG) and CANcer TARgeting (CANTAR) funded by Netzwerke 2021 Additional funding was received from the program “Netzwerke 2021”, an initiative of the Ministry of Culture and Science of the State of North Rhine-Westphalia for the CANTAR project to H.C.R., J.G. and R.K.T.
Data availability
The datasets generated and/or analyzed in the current study are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
H.C.R. received consulting and lecture fees from Abbvie, AstraZeneca, Vertex, and Merck. H.C.R. received research funding from AstraZeneca and Gilead Pharmaceuticals. H.C.R. is a co-founder of CDL Therapeutics GmbH. R.K.T. is a founder of Disco Pharmaceuticals GmbH, PearlRiver Bio (now part of Centessa), a shareholder of Centessa, founder and shareholder of Epiphanes Inc. and a consultant to PearlRiver Bio and Epiphanes Inc. R.K.T. has received research support from Roche. The remaining authors declare no competing financial interest.
Animal ethics declaration
Animal experiments in this study were approved by the local Ethics Committee of Animal Experiments authorities (LANUV, North Rhine-Westphalia, Germany) under license number 81-02.04.2019-A491. All mice were maintained according to FELASA recommendations and in compliance with the European Union and German guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Olta Ibruli and France Rose have contributed equally to this work.
Kasia Bozek, Hans Christian Reinhardt and Grit S. Herter-Sprie have contributed equally to this work.
References
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng T, Wu AW, Boot Y, Covington A, Gordenin KR, Bergstrom DA, Islam ENA, Lopez-Bigas SM, Klimczak N, McPherson LJ, Morganella JR, Sabarinathan S, Wheeler R, Mustonen DA, Getz V, Yu G, W (2020) The repertoire of mutational signatures in human cancer. Nature 578(7793):94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wind N, Dekker M, Berns A, Radman M, Riele TH (1995) Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell 82 [DOI] [PubMed]
- Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43(5):491–501. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietlein F, Reinhardt HC (2014) Molecular pathways: exploiting tumor-specific molecular defects in DNA repair pathways for precision cancer therapy. Clin Cancer Res 20(23):5882–5887. 10.1158/1078-0432.CCR-14-1165 [DOI] [PubMed] [Google Scholar]
- Dietlein F, Thelen L, Reinhardt HC (2014a) Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genetics 30:326–339. 10.1016/j.tig.2014.06.003. (Elsevier Ltd.) [DOI] [PubMed] [Google Scholar]
- Dietlein F, Thelen L, Reinhardt HC (2014b) Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genetics 30:326–339. 10.1016/j.tig.2014.06.003. (Elsevier Ltd.) [DOI] [PubMed] [Google Scholar]
- Dingemans AMC, Früh M, Ardizzoni A, Besse B, Faivre-Finn C, Hendriks LE, Lantuejoul S, Peters S, Reguart N, Rudin CM, De Ruysscher D, Van Schil PE, Vansteenkiste J, Reck M (2021) Small-cell lung cancer: ESMO Clinical Practice guidelines for diagnosis, treatment and follow-up☆. Ann Oncol 32(7):839–853. 10.1016/j.annonc.2021.03.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- fda, & cder. (n.d.). Highlights of prescribing information. www.fda.gov/medwatch
- Garassino MC, Gadgeel S, Speranza G, Felip E, Esteban E, Manuel OD, Hochmair MJ, Steven PF, Bischoff HG, Peled N, Grossi F, Ross JR, Reck M, Hui R, Garon EB, Kurata T et al (2023) Pembrolizumab Plus Pemetrexed and Platinum in Nonsquamous Non-small-cell Lung Cancer: 5-Year outcomes from the phase 3 KEYNOTE-189 study. J Clin Oncol 41:1992–1998. 10.1200/JCO.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretia L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, Müller C, Dahmen I, Jahchan NS, Park KS, Yang D, Karnezis AN, Vaka D, Torres A, Wang MS, Thomas RK (2015) Comprehensive genomic profiles of small cell lung cancer. Nature 524(7563):47–53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Maas L, Abedpour N, Cartolano M, Kaiser L, Fischer RN, Scheel AH, Weber JP, Hellmich M, Bosco G, Volz C, Mueller C, Dahmen I, John F, Alves CP, Werr L, Panse JP, Kirschner M, Engel-Riedel W et al (2024) Evolutionary trajectories of small cell lung cancer under therapy. Nature 627(8005):880–889. 10.1038/s41586-024-07177-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, Rizvi NA, Hirsch FR, Selvaggi G, Szustakowski JD, Sasson A, Golhar R, Vitazka P, Chang H, Geese WJ, Antonia SJ (2018) Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 33(5):853-861e4. 10.1016/j.ccell.2018.04.001. (e4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn L, Mansfield AS, Szczęsna A, Havel L, Krzakowski M, Hochmair MJ, Huemer F, Losonczy G, Johnson ML, Nishio M, Reck M, Mok T, Lam S, Shames DS, Liu J, Ding B, Lopez-Chavez A, Kabbinavar F, Lin W, Liu SV (2018) First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 379(23):2220–2229. 10.1056/nejmoa1809064 [DOI] [PubMed] [Google Scholar]
- Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, Conley CC, Chen OS, Guthrie MR, Soltero D, Qiao Y, Huang X, Tarapcsák S, Devarakonda S, Chalishazar MD, Gertz J, Moser JC, Marth G, Puri S, Oliver TG (2020) MYC drives temporal evolution of small cell Lung Cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell 38(1):60–78e12. 10.1016/j.ccell.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati MH, Lee K, Nguyen AA, Clark AB, Hou H, Rosulek A, Li H, Yang K, Fan K, Lipkin M, Bronson RT, Jelicks L, Kunkel TA, Kucherlapati R, Edelmann W (2010) An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology. 10.1053/j.gastro.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Diaz LA (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372(26):2509–2520. 10.1056/nejmoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Diaz LA (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics (Oxford, England) 25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang J, Guo C, Wang M, Wang C, Yan Y, Sun L, Wang D, Zhang L, Yu H, Hou L, Wu C, Zhu Y, Jiang G, Zhu H, Zhou Y, Fang S, Zhang T, Hu L, Zhang P (2024) Proteogenomic characterization of small cell lung cancer identifies biological insights and subtype-specific therapeutic strategies. Cell 187(1):184–203e28. 10.1016/j.cell.2023.12.004 [DOI] [PubMed] [Google Scholar]
- Marcus L, Lemery SJ, Keegan P, Pazdur R (2019) FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res 25(13):3753–3758. 10.1158/1078-0432.CCR-18-4070 [DOI] [PubMed] [Google Scholar]
- McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, Bhutkar A, McKenna A, Dooley A, Vernon A, Sougnez C, Malstrom S, Heimann M, Park J, Chen F, Farago AF, Dayton T, Shefler E, Gabriel S, Jacks T (2014) Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156(6):1298–1311. 10.1016/j.cell.2014.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A (2003) Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model [DOI] [PubMed]
- Nicholson AG, Chansky K, Crowley J, Beyruti R, Kubota K, Turrisi A, Eberhardt WEE, Van Meerbeeck J, Rami-Porta R (2016) The international association for the study of lung cancer lung cancer staging project: proposals for the revision of the clinical and pathologic staging of small cell lung cancer in the forthcoming eighth edition of the tnm classification for lung cancer. J Thorac Oncol 11(3):300–311. 10.1016/j.jtho.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Olivares-Hernández A, Del Barco Morillo E, Parra Pérez C, Miramontes-González JP, Figuero-Pérez L, Martín-Gómez T, Escala-Cornejo R, Bellido Hernández L, González Sarmiento R, Cruz-Hernández JJ, de la Ludeña MD (2022) Influence of DNA Mismatch Repair (MMR) System in Survival and Response to Immune Checkpoint Inhibitors (ICIs) in Non-Small Cell Lung Cancer (NSCLC): retrospective analysis. Biomedicines. 10.3390/biomedicines10020360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AF, Bretes L, Furtado I (2019) Review of PD-1/PD-L1 inhibitors in metastatic DMMR/MSI-H colorectal cancer. Front Oncol. 10.3389/fonc.2019.00396. (Frontiers Media S.A.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser MG, MacPherson D, Oliver TG, Sage J, Park KS (2024) Genetically-engineered mouse models of small cell lung cancer: the next generation. Oncogene 43:457–469. 10.1038/s41388-023-02929-7. (Springer Nature) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, Hochmair MJ, Özgüroğlu M, Ji JH, Voitko O, Poltoratskiy A, Ponce S, Verderame F, Havel L, Bondarenko I, Kazarnowicz A, Losonczy G, Conev NV et al (2019) Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 394(10212):1929–1939. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- Popat S, Hubner R, Houlston RS (2005a) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol (Vol 23:609–618. 10.1200/JCO.2005.01.086 [DOI] [PubMed] [Google Scholar]
- Popat S, Hubner R, Houlston RS (2005b) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23:609–618. 10.1200/JCO.2005.01.086 [DOI] [PubMed] [Google Scholar]
- Postow MA, Sidlow R, Hellmann MD (2018) Immune-related adverse events Associated with Immune Checkpoint Blockade. N Engl J Med 378(2):158–168. 10.1056/nejmra1703481 [DOI] [PubMed] [Google Scholar]
- Raso MG, Bota-Rabassedas N, Wistuba II (2021) Pathology and classification of SCLC. Cancers 13(4):1–11 (MDPI AG.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt C, Yaffe MB (2013) Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Reviews Mol Cell Biol 14(9):563–580. 10.1038/nrm3640 [DOI] [PubMed] [Google Scholar]
- Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M (2021) NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48(W1):W449–W454. 10.1093/NAR/GKAA379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C et al (2015) Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348(6230):124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers A, Dive L, Dowlati C, George A, Heymach J, Johnson JV, Lehman JE, Macpherson JM, Massion D, Minna PP, Oliver JD, Quaranta TG, Sage V, Thomas J, Vakoc RK, C. R., Gazdar AF (2019) Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data HHS Public Access. Nat Rev Cancer 19(5):289–297. 10.1038/s41574-XXX-XXXX-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Brambilla E, Faivre-Finn C, Sage J (2021) Small-cell lung cancer. Nat Rev Dis Primers. 10.1038/s41572-020-00235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Moore JA, Montesion M, Sharaf R, Lin DI, Colón CI, Fleishmann Z, Ebot EM, Newberg JY, Mills JM, Hegde PS, Pan Q, Dowlati A, Frampton GM, Sage J, Lovly CM (2023) Integrative Analysis of a large real-world cohort of small cell Lung Cancer identifies distinct genetic subtypes and insights into histologic Transformation. Cancer Discov 13(7):1572–1591. 10.1158/2159-8290.CD-22-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Sen T, Rudin CM (2020) Targeted therapies and biomarkers in small cell lung cancer. Front Oncol. 10.3389/fonc.2020.00741. (Frontiers Media S.A.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tong Z, Zhang W, Zhang W, Buzdin A, Mu X, Yan Q, Zhao X, Chang HH, Duhon M, Zhou X, Zhao G, Chen H, Li X (2021) FDA-approved and emerging next generation predictive biomarkers for immune checkpoint inhibitors in cancer patients. Front Oncol. 10.3389/fonc.2021.683419. (Frontiers Media S.A.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Gümüş ZH, Colarossi C, Memeo L, Wang X, Kong CY, Boffetta P (2023) SCLC: epidemiology, risk factors, genetic susceptibility, Molecular Pathology, Screening, and early detection. J Thoracic Oncol 18(31–46):1. 10.1016/j.jtho.2022.10.002. (Elsevier Inc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PMK, Muyas F, Hauck H, Smith OC, Sacks NJ, Ely ZA, Jaeger AM, Rideout WM, Zhang D, Bhutkar A, Beytagh MC, Canner DA, Jaramillo GC, Bronson RT, Naranjo S, Jin A, Patten JJ, Cruz AM, Shanahan SL, Jacks T (2023) Mismatch repair deficiency is not sufficient to elicit tumor immunogenicity. Nat Genet 55(10):1686–1695. 10.1038/s41588-023-01499-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in the current study are available from the corresponding author upon reasonable request.