Abstract
Abstract
Cattle manure or its digestate, which often contains antibiotic residues, can be used as an organic fertilizer and copper (Cu) as a fungicide in agriculture. Consequently, both antibiotics and Cu are considered soil contaminants. In this work, microcosms were performed with soil amended with either manure or digestate with Cu and an antibiotic (sulfamethoxazole, SMX) co-presence and the planting of Lactuca sativa. After the addition of the organic amendments, a prompt increase in the microbial activity and at the same time of the sul1 and intI1 genes was observed, although ARGs generally decreased over time. In the amended and spiked microcosms, the microbial community was able to remove more than 99% of SMX in 36 days and the antibiotic did not bioaccumulate in the lettuce. Interestingly, where Cu and SMX were co-present, ARGs (particularly sul2) increased, showing how copper had a strong effect on resistance persistence in the soil. Copper also had a detrimental effect on the plant-microbiome system, affecting plant biomass and microbial activity in all conditions except in a digestate presence. When adding digestate microbial activity, biodiversity and lettuce biomass increased, with or without copper present. Not only did the microbial community favour plant growth, but lettuce also positively influenced its composition by increasing bacterial diversity and classes (e.g., Alphaproteobacteria) and genera (e.g., Bacillus), thus indicating a good-quality soil.
Key points
• Cattle digestate promoted the highest microbial activity, diversity, and plant growth
• Cattle digestate counteracted detrimental contaminant effects
• Cu presence promoted antibiotic cross-resistance in soil
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-024-13324-x.
Keywords: Lettuce, Plant-microbiome system, Antibiotics, Cattle manure, Cattle manure digestate, ARGs
Introduction
Antibiotics (ABs) and heavy metals (HMs) are widely used in agriculture for different purposes, such as treating animal disease and crop pests, respectively. It has been generally recognized that ABs, owing to their incomplete metabolism in treated organisms, are excreted through feces and urine and can be found as active residues in wastewater, surface water, sediment, and soils (Zhang et al. 2018a; Barra Caracciolo et al. 2020b). Consequently, the application of manure or biosolids as organic fertilizers to agricultural soils can unintentionally introduce antibiotics, often in mixtures with other contaminants such as heavy metals (Kuppusamy et al. 2018; Bünemann et al. 2024).
AB occurrence in environments represents a significant concern for ecosystems and human health, as highlighted by the “One Health Concept” (White and Hughes 2019; Prata et al. 2022) which states that animal and human health is linked to that of their environment. In fact, AB residues in ecosystems can promote the spread of antibiotic resistance between resistant bacteria and natural microorganisms (Ben et al. 2019; Samreen et al. 2021; Narciso et al. 2023a) and from them to animal and human microbiomes through contaminated water or edible vegetal species (Gudda et al. 2020; Soto-Giron et al. 2021; Barra Caracciolo et al. 2022).
ABs are emerging and ubiquitous micropollutants and, owing to their biocide properties, they can kill or inhibit natural microbial communities involved in key ecosystem functioning (Grenni et al. 2018; Cycoń et al. 2019; Sodhi et al. 2021; Narciso et al. 2023a). In addition, particularly at below minimum inhibitory concentrations, they act as selective agents which can increase antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in natural ecosystems (Berendsen et al. 2018; Barra Caracciolo et al. 2020b, 2022; Singh et al. 2021).
Sulfamethoxazole (SMX) is a sulfonamide antibiotic among the most widely used in both human and veterinary medicine, and its residues are frequently detected in organic fertilizers and agricultural soils (Conde-Cid et al. 2020; Vieublé Gonod et al. 2022; Visca et al. 2022; Patyra et al. 2024). SMX is not an intrinsically persistent compound (e.g., its halving in soil is ca. 7 days, Rauseo et al. 2019; Garbini et al. 2022) but its continuous use and release determine its pseudo-persistence in water (Patrolecco et al. 2018) and soil (Wu et al. 2024). Indeed, sulfonamide resistance genes are very common in the environment and for this reason are considered resistance determinants in water, livestock waste, and manure-amended agricultural soils (Ben et al. 2019; He et al. 2020; Radu et al. 2021).
Most antibiotic resistance genes (ARGs), including sul genes, are located on mobile genetic elements, such as plasmids, which favour their horizontal transfer between different species (Pinilla-Redondo et al. 2018; Dimitriu 2022). Plasmids can contain gene cassettes, which confer resistance not only to ABs but also to other contaminants (e.g., HMs) or environmental stresses (Chaturvedi et al. 2021; Huang et al. 2021). For this reason, not only ABs but also other factors can favour ARG persistence among natural bacterial populations (Feng et al. 2021). Indeed, antibiotic resistance has been frequently found in metal-tolerant bacteria, and this can contribute to a reservoir of transferable antibiotic resistance genes in the environment (Nath et al. 2019; Glibota et al. 2019).
Several HMs are essential micronutrients for biota, but their occurrence at concentrations higher than the natural ones (due to anthropogenic pollution) can exert toxic effects (Masmoudi et al. 2013; Ali et al. 2019; Kumar et al. 2021; Barra Caracciolo and Terenzi 2021).
As a trace element, Cu is a co-factor in several animal, plant, and bacterial enzymes (Hänsch and Mendel 2009; Kaur et al. 2023). Cu is also a biocide in the high amounts widely applied on vineyards and horticultural crops (up to 4 kg/ha/y) for preventing fungal attacks and other plant disease (Fagnano et al. 2020). Cu concentrations in agroecosystems can range from 20 up to 800 mg/kg, as, for example, in South European agricultural soils impacted by long-term fungicide applications (Panagos et al. 2018; Fagnano et al. 2020; Zamulina et al. 2022; Tamm et al. 2022). The EU Reg. 1981/2018 limits the use of copper compounds in agriculture to an average of 4 kg/ha per year, and an Italian decree (DM 46/2019) indicates thresholds of 200 mg/kg in soils (Tóth et al. 2016).
Recent studies suggest that Cu can increase the toxic effects of ABs and/or favour ARB, but the conditions and amounts causing this phenomenon need to be investigated (Perković et al. 2022). For example, a recent work showed that a mixture of copper (3 mg/L) and SMX (0.7 mg/L) displayed an additive ecotoxicological effect on the A. fischeri bacterium, compared with that obtained from single solutions (with only copper or sulfamethoxazole) (Narciso et al. 2023b). In another work, Kang et al. (2018) demonstrated that ARGs (comprising those involved in sulfonamide resistance) and mobile genetic elements (MGEs, as intI1) increased significantly in a soil microbial community treated with different copper concentrations (from 50 to 1000 mg/kg).
Microbial community structure (biodiversity) and activity, including the capability to respond to contamination with different strategies (e.g., developing resistance and/or capability to degrade and remove toxic compounds), depend on a plethora of site-specific environmental conditions. Among them, temperature is a critical variable modulating microbial development and activity (Burman and Bengtsson-Palme 2021) and affecting contaminant fate and dissipation. For example, sulfonamide degradation in the composting of chicken manure was found to increase when passing from mesophilic (30–40 °C) to thermophilic (50–60 °C) conditions (Lin et al. 2017). Indeed increasing temperatures not only influenced microbial community activity and composition but also favoured AB dissipation by promoting abiotic hydrolysis processes (Mitchell et al. 2014; Yang et al. 2021).
Light is another abiotic factor, which can influence AB transformation, especially for photosensitive molecules such as SMX. Although the primary role of microbial degradation in SMX removal has been demonstrated, sulfamethoxazole dissipation increases significantly in the presence of light (Lai et al. 2011; Poirier-Larabie et al. 2016; Patrolecco et al. 2018).
For evaluating AB and ARG environmental fate and persistence, it is therefore necessary to take into consideration different abiotic and biotic factors, including possible sudden temperature changes, due to ever more frequent extreme weather events (Li et al. 2023; Jampani et al. 2024). For example, recent studies have reported that, if high temperatures unexpectedly persist, they can both promote the survival of gut bacterial populations and increase the possibility of ARG spreading from pathogens to natural bacterial populations (MacFadden et al. 2018). On the other hand, temperatures higher than the gut bacteria optimal ones (ca. 37 °C) can select the mesophilic and thermophilic bacteria involved in composting and anaerobic digestion processes and possibly in AB degradation (Mitchell et al. 2015; Lin et al. 2017; Visca et al. 2022).
Another complex aspect to be considered is the relationships between AB concentrations and their ARGs. For example, Selvam et al. (2012b) reported that sulfadiazine and chlortetracycline degraded completely, in 3 and 14 days, respectively, of composting and this process decreased tet genes during the mesophilic stage. On the contrary, sulfamethoxazole gene (sul1, sul2, dfrA1, dfrA7) abundances were not influenced by a sulfadiazine presence (initially spiked at 2–20 mg/kg and then completely removed), and these genes were still present after its degradation (up to 21–28 days). The authors hypothesized that the high sul persistence might be due to the possible presence of antibiotic metabolites or high mobility of these genes among bacteria. The authors concluded that temperature is essential for reducing ARGs during the thermophilic phase because it selects for different microbial populations and resistance mechanisms (Selvam et al. 2012a). In a biogas plant, operating in mesophilic conditions (33–35 °C) and fed with zootechnical waste, it was found that both antibiotics (sulfamethoxazole, ciprofloxacin, and enrofloxacin) and their ARGs decreased during the 45-day anaerobic digestion process. In particular, SMX was removed by up to 100%, ENR up to 84%, and CIP up to 92%. At the same time, ARGs (including sul1, sul2, and intI1) declined significantly (up to 80%) in the digestate samples (Visca et al. 2021).
Overall, sulfamethoxazole ARGs (such as sul1 and sul2) can be influenced by temperature variations. Some authors simulated a biological treatment of dairy lagoon water by incubating some samples at 4° and 20 °C in both aerobic and anaerobic conditions. In aerobic conditions, sul1 had the highest abundance at 20 °C and at day 10 and then decreased significantly; on the other hand, sul1 at 4 °C (although with lower values) persisted for 100 days. In anaerobic conditions, a similar temperature effect was found, with a general lower ARG presence than in aerobic conditions. Interestingly, sul2 increased over time at 20 °C, with the highest level at 60 days, in both aerobic and anaerobic conditions. This phenomenon was not found at 4 °C in anaerobic conditions (Pei et al. 2007; Lin et al. 2017).
Another key aspect to be considered in environmental studies is contaminant bioavailability. In fact, not only ABs but also heavy metals (including copper) can have different effects on microorganisms depending on their bioavailability in a contaminated matrix. Heavy metals can form stable complexes with soil organic matter (Kim et al. 2015; Barra Caracciolo and Terenzi 2021), and soluble or non-soluble fractions of organic carbon (OC) can influence antibiotic-soil and copper-soil interactions. For example, copper can bound strongly to clay minerals and form both insoluble and soluble organic complexes with OC (Shao et al. 2020). Moreover, in the presence of an external carbon source, increases in adsorption and complexation of Cu can occur (Bolan et al. 2003). Interestingly, some authors found that fulvic acids resulted particularly in decreasing plant uptake and toxicity of heavy metals at high concentrations (25 mg/kg), because of their strong binding affinity (Shahid et al. 2012; Canellas et al. 2015).
AB and ARG persistence in the environment is a very complex issue, which depends on the combination of various biotic and abiotic factors, including multicontamination, which can act as opposite selective forces on natural microbial communities. In this context, this work aimed at evaluating the possible effects of SMX alone or in a copper co-presence on a natural soil microbial community in the presence/absence of organic amendments (manure or digestate) and lettuce plants. The soil microbial community was assessed in terms of abundance (DAPI counts), activity (dehydrogenase), structure (NGS), and ARGs (qPCR). Moreover, SMX dissipation in the soil and lettuce biomass was also evaluated at the end of the experiment (36 days).
Materials and methods
Sampling site and organic amendments
An agricultural sandy-clay-loam (60% sandy, 18% clay, 22% loam, USDA 1987) soil was collected from an organic farm close to Rome. The samples were collected from the surface layer (0–20 cm) using a shovel, and an initial characterization for organic carbon, total nitrogen, heavy metals, and microbial abundance was performed.
The manure and digestate used as organic amendments came from a cattle manure biogas plant located in a livestock farm in central Italy, and their characteristics are described in detail in previous works (Visca et al. 2021, 2022; Mazzurco Miritana et al. 2022). All the parameters analyzed showed values which met the European Regulation limits for fertilizing products (EU 1009/2019).
Microcosm set-up
Soil microcosms (plant pots containing 1-kg soil each) were set up with different experimental conditions. The pots contained soil spiked with the antibiotic sulfamethoxazole (7 mg/kg) and copper sulfate (30 mg/kg) and were amended with manure (1% w/w) or digestate (1% w/w). Half of the experimental conditions were performed in the presence of lettuce plants (Lactuca sativa). Moreover, 36 microcosms were performed without the SMX antibiotic (in the presence/absence of the organic amendments) in order to assess the possible influence of copper alone on the soil microbial community.
The 18 experimental conditions performed are summarised as follows in Table 1.
Table 1.
Summary of the 18 experimental conditions. Cu copper, SMX sulfamethoxazole, P Lactuca sativa, M manure, D digestate
| Experimental condition | Acronym |
|---|---|
| Soil + manure + Cu + SMX | SMCA |
| Soil + digestate + Cu + SMX | SDCA |
| Soil + Cu + SMX | SCA |
| Soil + manure + Cu | SCM |
| Soil + digestate + Cu | SCD |
| Soil + Cu | SC |
| Soil + manure | SM |
| Soil + digestate | SD |
| Control soil | S |
| Soil + manure + Cu + SMX + L. sativa | SMCA_P |
| Soil + digestate + Cu + SMX + L. sativa | SDCA_P |
| Soil + Cu + SMX + L. sativa | SCA_P |
| Soil + manure + Cu + L. sativa | SCM_P |
| Soil + digestate + Cu + L. sativa | SCD_P |
| Soil + Cu + L. sativa | SC_P |
| Soil + manure + L. sativa | SM_P |
| Soil + digestate + L. sativa | SD_P |
| Control soil + L. sativa | S_P |
Each experimental condition was performed in three replicates. The experiment was maintained for 36 days in a greenhouse (from 1st May 2022), and each microcosm was watered daily to keep the same soil moisture. The initial temperature was 23.7 ± 2.8 °C, in line with average spring values at this latitude. From day 14, the temperature rapidly increased to 30.4 ± 15.5 °C in the following days, reaching an average temperature of 32.8 °C with variations between a minimum temperature of 22.2 °C and a maximum one of 43.3 °C, at day 18. The average temperature remained at 30 ± 2 °C until the end of the experiment (day 36).
Two samplings were performed, at the start (3 h) and at the end (36 days) of the experiment.
At the end of the experiment, plant roots developed throughout the soil inside each pot, and, consequently, the soil results refer to “rhizosphere soil.”
For each condition, destructive samplings were performed in three replicates. Different aliquots of soil were collected and used for microbiological analyses or chemical determinations.
Soil physicochemical characterization and amendment organic carbon analyses
The soil pH and electrical conductivity (EC) were measured in a water solution (1:2.5 and 1:5 soil to distilled water (w/v) ratio, respectively) (Allison and Moodie 1965). The soil water content was determined with a gravimetric analysis, following official methods (DM 13/09/1999 and s.m.i.). Aliquots of air-dried samples (15–20 mg each) were used for the total organic carbon (OC) and nitrogen (N) analyses, using an elemental carbon analyser (Carlo Erba NA 1500 series 2 C/H/N/O/S). Further details are described in previous works (Barra Caracciolo et al. 2020a).
The water-soluble organic carbon was determined in water extracts of 1:10 (w/v) and 1:50 (w/v) of soil samples and amendments respectively, after filtration through a synthetic filter with a pore diameter of 0.45 μm, by using an automatic analyser for liquid samples (TOC-V CSN Analyzer, Shimadzu) (Bustamante et al. 2012). The extractable organic carbon (CEX) was determined in organic amendments (manure and digestate) by dissolving sample aliquots in 0.1 M NaOH. The fulvic acid–like carbon (CFA) amount after the precipitation of the humic acid–like carbon (CHA) one was found to be pH 2.0 (Sánchez-Monedero et al. 1996). The CHA was calculated by subtracting the CFA from the CEX.
Chemicals and reagents
Pure solvents (HPLC grade), such as methanol (MeOH), acetone (ACT), acetonitrile (ACN), and hydrochloric acid (37%, HCl), were purchased from VWR (Radnor, PA, USA). Formic acid (98–100%) for LC–MS LiChropur™, used to acidify solvents and composing the mobile phase for the analytic determinations, was purchased from Sigma-Aldrich (Steinheim, Germany). The pH of the mobile phase was adjusted with a portable pH meter (HANNA Instruments, Woonsocket, RI USA). A Milli-Q Millipore system (Bedford, MA, USA) produced the ultrapure water (18 MΩ/cm quality).
SMX vetranal analytical standards were from Merck KGaA (Darmstadt, Germany). Deuterated SMX (SMX-d4, Clearsynth) was used as the internal standard.
CuSO4·5H2O was purchased from Sigma-Aldrich, Germany, and used to prepare a copper solution (30 mg/L) in ultrapure water.
An individual stock solution (100 mg/L) was prepared by dissolving the AB standard powder in methanol and storing it in the dark at − 20 °C. A working standard solution was prepared at each sampling time by diluting the stock solution with water and then stored at 4 °C.
The mixture of SMX and CuSO4 was prepared by dissolving 2.5 mg of SMX in MeOH (50 ml) to obtain a final concentration of 50 mg/L and stored at − 20 °C. Daily working standard solutions of the antibiotic were obtained by dilution of the stock solution with a mixture of ultrapure water: MeOH (1:1 v/v) and stored at 4 °C. Waters Oasis Hydrophilic–Lipophilic Balance (HLB) cartridges (6 ml, 1 g) were from Waters (Milford, MA, USA). The inert material used to fill the extraction cells was diatomaceous earth (Dionex™ ASE™ Prep DE) purchased from Thermo Scientific (Waltham, MA, USA).
Chemical determinations
The antibiotic extraction procedure was performed using a SPE (solid-phase extraction) equipped with Oasis Hydrophilic-Lipophilic Balance (HLB, Waters, Milford, USA) cartridges (6 ml, 200 mg), in line with previous work (Spataro et al. 2019). Briefly, the cartridges were activated with 5 ml of MeOH and 5 ml of ultrapure water (18 MΩ/cm quality, Millipore, Bedford, USA) passed at a flow rate of 5 ml/min. Soil water extracts were passed into the cartridges, and their elution was performed with a mixture of MeOH, acetonitrile (ACN, HPLC grade, VWR-Radnor, USA), and acetic acid (Sigma Aldrich, Germany) (40:40:20 v/v/v). The samples were then evaporated with a gentle nitrogen stream and reconstituted with 0.5 ml of a mixture of ACN and ultrapure water (50:50 v/v) acidified with formic acid at pH 3. SMX analytical determination was performed with high-performance liquid chromatography (HPLC, column oven mod. LC-100, micropump series 200, Perkin Elmer, USA) interfaced with a triple quadrupole mass spectrometer (MS/MS, mod. API 3000, AB Sciex, Germany). The operative conditions were as follows: injection loop of 20 μl and the chromatographic column was Gemini column (150 × 4.6 mm, 5 μm RP C 18, Phenomenex, France) (Spataro et al. 2019). SMX elution was carried out in gradient mode at a flow rate of 0.3 ml/min. The mobile phase was composed of MeOH (phase A) and ultrapure water (phase B), both acidified with 0.1% of formic acid. The gradient profile started with phase A at 10%. It then increased from 10 to 90% in 10 min and finally returned to 10% in 15 min. The ESI (electrospray ionization) operated with 12 units of curtain gas, 14 units of nebulizer gas, and source temperature of 400 °C. Nitrogen was both collision and drying gas. The AB quantification was based on the m/z ion ratios and a comparison of the retention times (RTs) of product ions/fragments in the samples with those of each standard, considering criteria differences of 0.2 min. Two calibration curves (five points) at lower (0.5, 1.0, 2.0, 2.5, and 5.0 ng/L) and higher (5, 20, 50, 100, and 500 ng/L) concentration ranges were considered, indicating the good linearity of the method (R2 > 0.98). The SMX recoveries were also evaluated using ultrapure water artificially spiked at different concentrations (50, 200, and 500 ng/L) and ranging between 82 and 99%. The detection limits (LODs) for SMX were calculated following the IUPAC method (Thompson et al. 2002) and were 2.1, 5, and 4 ng/L, respectively. Quantification limits were set at three times the LODs.
Copper concentrations were measured using an inductively coupled plasma optical emission spectrometer (ICP-OES, 5800 Agilent technologies, USA). The quantification was performed by interpolation using a calibration curve obtained by diluting a 1 g/L stock solution into six standard solutions. The wavelengths (nm) used for Cu were 327 and 395 (Barreto et al. 2021; Iannelli et al. 2022).
Total microbial number, cell viability, and dehydrogenase activity
The total microbial number (N. cells/g soil) of each sample was analyzed in formaldehyde-fixed soil aliquots (1 g each), using the epifluorescence direct count method, which relies on DAPI (4′,6-diamidino-2-phenylindole), as the DNA fluorescent intercalant. Samples were processed as detailed in previous works (Barra Caracciolo et al. 2005, 2013, Barra Caracciolo et al. 2015b).
Cell viability (% live cells/live + dead) was assessed in non-fixed soil samples using two fluorescent dyes, SYBR Green II and propidium iodide (Sigma-Aldrich Germany), to discriminate between viable (green) and dead (red) cells, as previously detailed (Grenni et al. 2014).
Both the total microbial cell number and the percentage of viable cells were counted under a Leica DM 4000B fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Finally, the N. of live cell abundance (N. live cells/g soil) was obtained by multiplying each total microbial abundance datum (N. total cell/g) by the corresponding cell viability one (% live cells/live + dead), as reported in other works (Amalfitano et al. 2008; Garbini et al. 2022).
Dehydrogenase activity (DHA) was evaluated in 6-g soil samples, using a colorimetric method based on the quantification of 2,3,5-triphenyl formazan (TPF) compound produced from the reduction of 2,3,5-triphenylte-trazoliumchloride (Grenni et al. 2009, 2012). The microbial activity (expressed as TPF/g soil) was measured using a Multiskan Sky Microplate Spectrophotometer (Thermo Scientific, Waltham, MA, USA).
All the microbiological analyses were performed in at least three replicates for each experimental condition and sampling.
Microbial DNA extraction and quantification of ARGs
Total DNA was extracted from each soil sample using a DNeasy PowerSoil kit (Qiagen, USA) following the procedure described by Barra Caracciolo et al. (2022). Further details are provided in the Supplementary Information. The DNA extracted was used for both the qPCR and NGS analyses.
Quantitative PCR was used to quantify two SMX-resistance genes (sul1 and sul2) and the class 1 integron-integrase gene (intl1). The 16S rRNA gene copy numbers were determined for calculating the relative abundance of the resistance genes targeted in the samples. The qPCR procedure used is described in detail by Visca et al. (2022). Further description of the method is reported in the Supplementary Information. The quantitative PCR data were expressed as the ratio of ARG or intI1 gene copy number per 16S copy number to evaluate the relative abundance of each target gene in the bacterial community.
Phylogenetic characterization of the microbial community
Aliquots of the DNA extracted (15 ng/μl) from each replicate were used for NGS. The V3–V4 region of 16S rRNA genes was amplified with the 341F and 805R primers (Table S1, Supplementary Information) for the identification of the prokaryotic community (Bacteria and Archaea). The method used followed the procedure previously described (Callahan et al. 2016; Bolyen et al. 2019; Mazzurco Miritana et al. 2022; Garbini et al. 2023). Further details are provided in Supplementary Information. Alpha-diversity indices (Shannon, Evenness, and Chao1) were also estimated.
The Illumina Miseq sequencing raw data are deposited in the NCBI (National Center for Biotechnology Information) database (accession number: PRJNA1153537).
Lettuce biomass
L. sativa plants were sampled at the end of the experiment, washed with sterile MilliQ water to remove any soil particles, weighed (aerial parts + roots), and put in an air-forced oven at 60 °C for 72 h to evaluate the dry biomass.
Statistical analyses
All the statistical analyses were performed using R (4.3.1version https://www.r-project.org).
The microbiological and chemical results are reported as average values ± standard errors of at least three replicates and were compared with each other with ANOVA using the aov function. More specifically, if the ANOVA test indicated the significative difference among results obtained from different conditions, the Tukey honestly significant difference test (TukeyHSD function) was applied to evaluate the paired comparison of experimental condition values. A significance threshold of 0.05 was considered (p < 0.05). To detect any significant differences between the MGE and ARGs in the experimental conditions, a non-parametric one-way ANOVA (Kruskal–Wallis) was performed using the kruskal.test function combined with the pairwise.wilcox.test function as a post hoc test (Benavoli et al. 2016). The microbial community alpha diversity was analyzed using the Evenness and Shannon diversity indices, while the Chao1 index (Chao et al. 2005) was used as an estimator of potential richness.
The effect of the amendments, SMX and Cu, and Lactuca sativa on the prokaryotic communities in the different conditions was evaluated using the principal coordinate analysis (PCoA) based on the Bray–Curtis distance estimated using the function vegdist (Vegan package of R, 10.32614/CRAN.package.vegan).
A multivariate ANOVA with permutations (PERMANOVA) was applied to assess significance. Pairwise PERMANOVA was performed using the function pairwise.perm.manova from the RVAideMemoire package to evaluate the significance of ASV changes in the prokaryotic composition in the different experimental conditions.
Most abundant classes and genera from each experimental condition were selected, and their relative abundance was estimated using the Phyloseq package (https://code.bioconductor.org/browse/phyloseq/). ASV abundances of the selected genera were normalized by z-score and displayed in a heatmap generated by the ComplexHeatmap package. In the heatmap, genera and experimental conditions were grouped in accordance with hierarchical clustering dendrograms.
Network analyses were performed to visualize correlations between ARGs, MGE, SMX, and microbial genera relative abundances. Spearman’s rank correlations (ρ > 0.75 and p < 0.01) were used to build a correlation matrix on which to base the network analysis (Hmisc r package). The network visualization was carried out on the interactive “Gephi” platform (Gephi Version 0.10.1, https://gephi.org/).
Results
Manure, digestate, and soil physicochemical characterization
Table 2 reports the main characteristics of the manure and digestate (e.g., pH, electrical conductivity, total nitrogen (N), total organic carbon (OC), water-soluble C, extractable C (fulvic and humic acids)) used in this experiment. Both organic amendments contained residual concentrations of SMX (ca. 0.045 mg/kg).
Table 2.
Physicochemical and chemical characterization of the cattle manure and digestate
| Manure | Digestate | |
|---|---|---|
| pH | 6.8 ± 0.3 | 7.6 ± 0.0 |
| Electrical conductivity (µS/cm) | 4.5 ± 0.0 | 1.4 ± 0.0 |
| Total N (%) | 1.8 ± 0.1 | 1.5 ± 0.1 |
| Total organic C (%) | 41.3 ± 0.7 | 32.8 ± 0.3 |
| Water-soluble C (%) | 6.7 ± 0.0 | 1.2 ± 0.4 |
| Humic acid–like C (%) | 12.7 ± 0.1 | 6.7 ± 0.0 |
| Fulvic acid–like C (%) | 1.1 ± 0.0 | 3.5 ± 0.1 |
The agricultural soil used for the experiment had an organic carbon content of 1.05% ± 0.05 and nitrogen of 0.33% ± 0.00 and a pH of 6.8 (Table S2). The highest increase in soil organic carbon and pH was observed in the case of digestate-amended soils (SD, SCD, and SDCA) (Table S2).
Chemical determinations
The SMX antibiotic almost disappeared over the 36 experimental days in all conditions, from 99.4% in SCA to 99.5% in SDCA. Adding plant did not promote any significant further antibiotic removal (Table 3).
Table 3.
SMX residues (%) in soil (3 h and 36 days) and in leaves (36 days) and the corresponding bioaccumulation factor (BAF)
| 3 h (mg/kg) | Soil 36 days (mg/kg) | Leaves 36 days (mg/kg) | BAF | |
|---|---|---|---|---|
| SMCA | 7.01 ± 0.01 | 0.0430 ± 0.001 | - | - |
| SDCA | 7.01 ± 0.03 | 0.0329 ± 0.00 | - | - |
| SCA | 7.01 ± 0.01 | 0.0450 ± 0.001 | - | - |
| SMCA_P | 7.01 ± 0.01 | 0.0419 ± 0.0004 | 0.263 ± 0.000 | 0.038 |
| SDCA_P | 7.01 ± 0.03 | 0.0377 ± 0.0004 | 0.096 ± 0.000 | 0.014 |
| SCA_P | 7.01 ± 0.01 | 0.0425 ± 0.001 | 0.399 ± 0.000 | 0.057 |
Considering the bioaccumulation factors (BAF = SMXleaves/SMXsoil3h), negligible residual concentrations of sulfamethoxazole were found in the lettuce leaves. The BAF values were always lower than 1, excluding SMX bioaccumulation in the edible part of the lettuce (Table 3).
The initial amount of copper in the soil was 21.97 ± 1.7 mg/kg; with addition of the copper solution, its concentration increased to 30.66 ± 1.8 mg/kg. Interestingly, in the planted microcosms, higher amounts (average values 43.31 ± 0.7 mg/kg) of copper than in the corresponding un-planted conditions (SCA, SMCA, SDCA) were observed at 36 days, showing how lettuce, through its roots, concentrated it inside its rhizosphere.
Live cell abundance and dehydrogenase activity
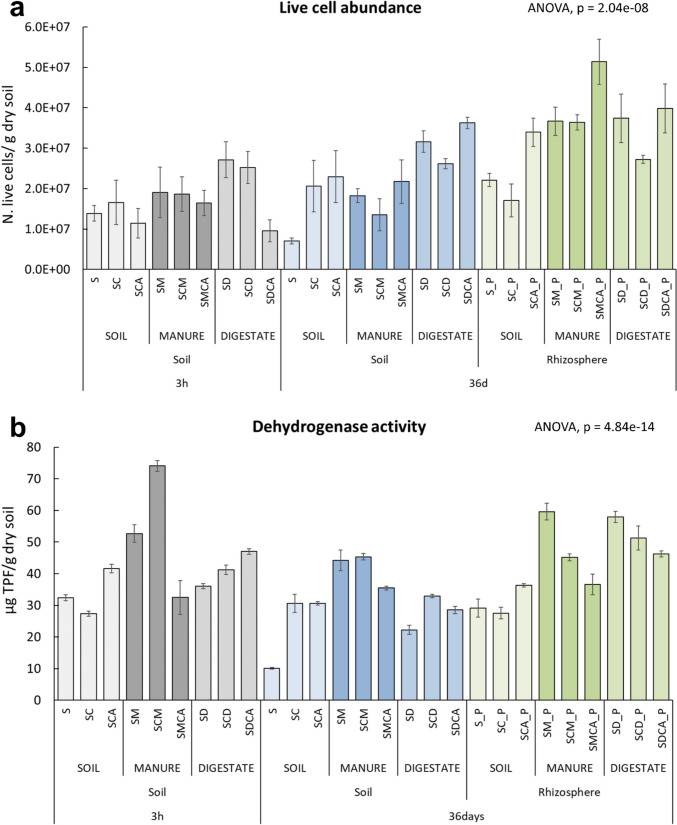
Figure 1a shows how amendments promoted live cell abundance (N. of live cells/g soil). However, copper and SMX had an initial detrimental effect on cell numbers (p < 0.05, Table S3) in all conditions where they were co-present (SCA, SMCA, and SDCA). At day 36, microbial abundances were significantly higher than in the control (S) in all conditions (p < 0.05), with the highest values in the amended rhizosphere. The amendments also promoted overall dehydrogenase activity, and this effect was amplified by the plant presence (36 days) (Fig. 1b). However, only in the digestate planted microcosms (SD_P, SCD_P, and SDCA_P) did DHA show values that were always significantly higher (p < 0.05, Table S4) than in the corresponding un-planted conditions.
Fig. 1.
a Live cell abundance (N. live cells/g dry soil). b Dehydrogenase activity (µg TPF/ g dry soil). Data are means of three independent replicates. The vertical bars represent the standard errors. The post hoc tests are reported in detail in Supplementary Material (Table S3 and Table S4)
Quantification of ARGs
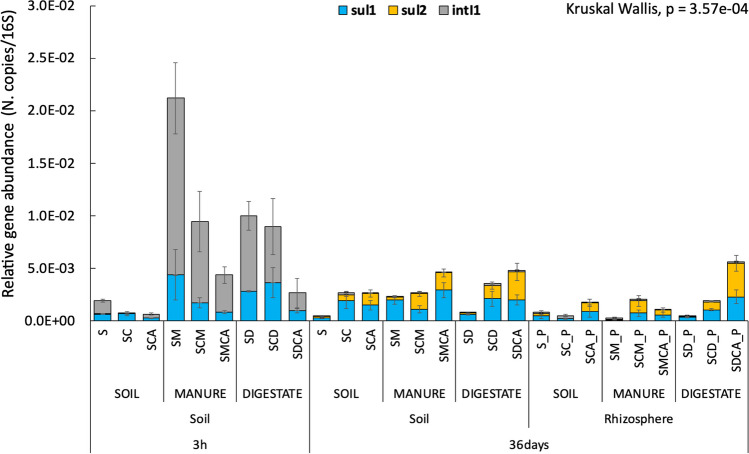
Manure and digestate introduced both intI1 and sul1 genes to the amended soils. Overall, when adding manure, a significantly higher (Kruskal Wallis p < 0.05, Table S5) ARG abundance was found than in digestate-amended conditions. The intI1 gene was initially the most abundant, but at 36 days, it had almost disappeared in all conditions (Fig. 2). At the same time, a general decrease in sul1 was also observed (except in the conditions where copper was present), and a positive correlation (p < 0.05) between sul1 and intI1 was found.
Fig. 2.
Relative gene abundances (ARGs or MGE/16S) in the various experimental conditions. The post hoc test is reported in detail in Supplementary Material (Table S5)
Indeed, the relative highest values (p < 0.05) of ARGs (sul1 + sul2) were with a copper and sulfamethoxazole co-presence in both the soil (SMCA and SDCA) and rhizosphere (SMCA_P, SDCA_P at 36 days). Moreover, comparing to the control soil (S), a higher relative abundance of sul1 + sul2 was found where copper was present (SC and SCA at 36 days).
Biodiversity and phylogenetic characterization of the microbial community
The prokaryotic community was characterized by sequencing the 16S gene in the various experimental conditions. Table 4 reports the diversity (Shannon, Evenness, and Chao1) indices, based on ASV abundance.
Table 4.
Diversity indices (Shannon, Evenness, and Chao1)
| Experimental condition | Average values | Standard errors | ||||||
|---|---|---|---|---|---|---|---|---|
| Shannon | Evenness | Chao1 | Shannon | Evenness | Chao1 | |||
| 3 h | Soil | S | 9.57 | 0.927 | 1410.35 | 0.23 | 0.00 | 296.66 |
| SC | 9.06 | 0.895 | 1166.26 | 0.41 | 0.03 | 98.28 | ||
| SCA | 9.61 | 0.929 | 1365.05 | 0.09 | 0.00 | 105.70 | ||
| SM | 9.69 | 0.929 | 1409.55 | 0.03 | 0.00 | 45.59 | ||
| SCM | 9.01 | 0.921 | 902.75 | 0.12 | 0.00 | 82.25 | ||
| SMCA | 9.51 | 0.924 | 1314.21 | 0.13 | 0.00 | 160.84 | ||
| SD | 9.43 | 0.922 | 1236.46 | 0.15 | 0.00 | 132.34 | ||
| SCD | 9.61 | 0.928 | 1354.93 | 0.10 | 0.00 | 114.90 | ||
| SDCA | 9.55 | 0.927 | 1315.32 | 0.06 | 0.00 | 54.66 | ||
| 36 days | Soil | S | 8.54 | 0.915 | 675.77 | 0.19 | 0.00 | 93.22 |
| SC | 8.07 | 0.921 | 443.99 | 0.07 | 0.00 | 24.37 | ||
| SCA | 8.50 | 0.920 | 618.61 | 0.08 | 0.00 | 48.30 | ||
| SM | 8.10 | 0.879 | 625.98 | 0.20 | 0.00 | 103.74 | ||
| SCM | 8.53 | 0.909 | 742.21 | 0.38 | 0.00 | 178.20 | ||
| SMCA | 8.89 | 0.916 | 855.74 | 0.06 | 0.00 | 47.48 | ||
| SD | 8.18 | 0.921 | 494.19 | 0.24 | 0.00 | 78.30 | ||
| SCD | 8.66 | 0.906 | 785.57 | 0.19 | 0.00 | 106.86 | ||
| SDCA | 8.88 | 0.923 | 813.74 | 0.13 | 0.00 | 73.76 | ||
| Rhizosphere | S_P | 9.42 | 0.908 | 1396.23 | 0.09 | 0.00 | 130.58 | |
| SC_P | 9.29 | 0.917 | 1212.35 | 0.25 | 0.00 | 277.56 | ||
| SCA_P | 9.30 | 0.912 | 1302.72 | 0.32 | 0.00 | 302.46 | ||
| SM_P | 9.37 | 0.918 | 1219.18 | 0.06 | 0.00 | 48.78 | ||
| SCM_P | 9.14 | 0.897 | 1240.20 | 0.19 | 0.00 | 196.34 | ||
| SMCA_P | 9.29 | 0.918 | 1151.64 | 0.05 | 0.00 | 44.66 | ||
| SD_P | 9.47 | 0.917 | 1322.11 | 0.11 | 0.00 | 103.92 | ||
| SCD_P | 9.28 | 0.911 | 1203.83 | 0.12 | 0.00 | 154.68 | ||
| SDCA_P | 9.55 | 0.922 | 1378.43 | 0.14 | 0.00 | 181.18 | ||
The Shannon and Evenness values were significantly different (p < 0.05) between the SMX and Cu co-presence conditions (SCA, SMCA, and SDCA) and the other conditions (S, SC, SM, SMC, SD, SDC) and between the initial and the final sampling (p < 0.05). The Evenness values were also significantly different (p < 0.05), between the amended (manure or digestate) conditions in co-presence of SMX and Cu (SMCA and SDCA) and the non-amended ones (SCA).
The Chao1 index values showed a significant difference between the initial and final sampling (p < 0.05) for all conditions. Interestingly, Chao1 was found to be lower (p < 0.05) in the SCM condition at 3 h in comparison with S and SM at 3 h. At 36 days, the SC value was significantly lower than those found in S, SCA, SMCA, SCD, and SDCA.
Finally, the diversity values were generally higher in a plant presence (rhizosphere) in all conditions, confirming the positive effect of the rhizosphere on microbial community structure and activity.
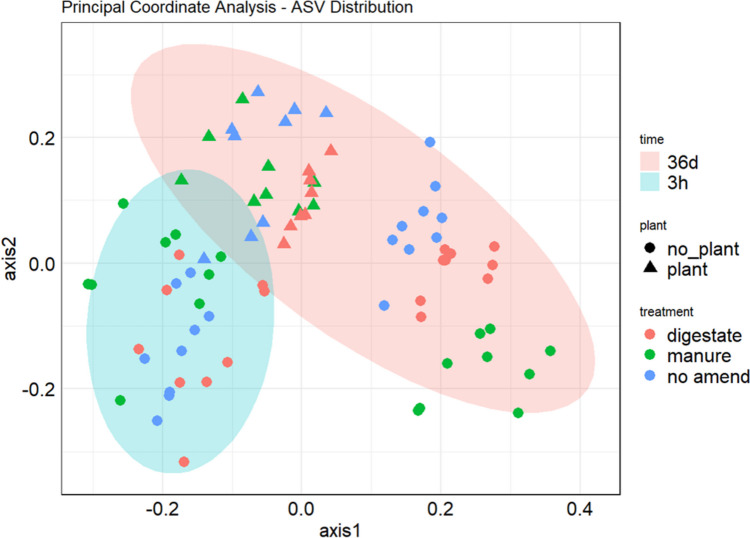
The principal coordinate analysis (PCoA) of the ASV distribution did not show significant differences between the presence and absence of SMX and/or Cu. At 36 days, there were significant differences (PERMANOVA, p-value < 0.001, R = 0.89, Table S6) between amended and un-amended conditions and between plant and un-planted ones (PERMANOVA, p-value < 0.001, R = 0.94). In the absence of plants, the soil microbial communities were significantly different (PERMANOVA, p-value < 0.001, R = 0.84, Table S7) between manure and digestate at 36 days. In a plant presence, the effect of the amendments was not observed (Fig. 3).
Fig. 3.
Principal coordinate analysis (PCoA) based on the Bray–Curtis distance matrix and calculated on ASV distribution. Red points indicate the digestate-amended conditions, green points the manure-amended conditions, and blue ones the no amended conditions. The circle shape indicates no plant presence. The pink ellipse indicates the initial sampling time (3 h) and the light blue one the finale sampling time (36 days)
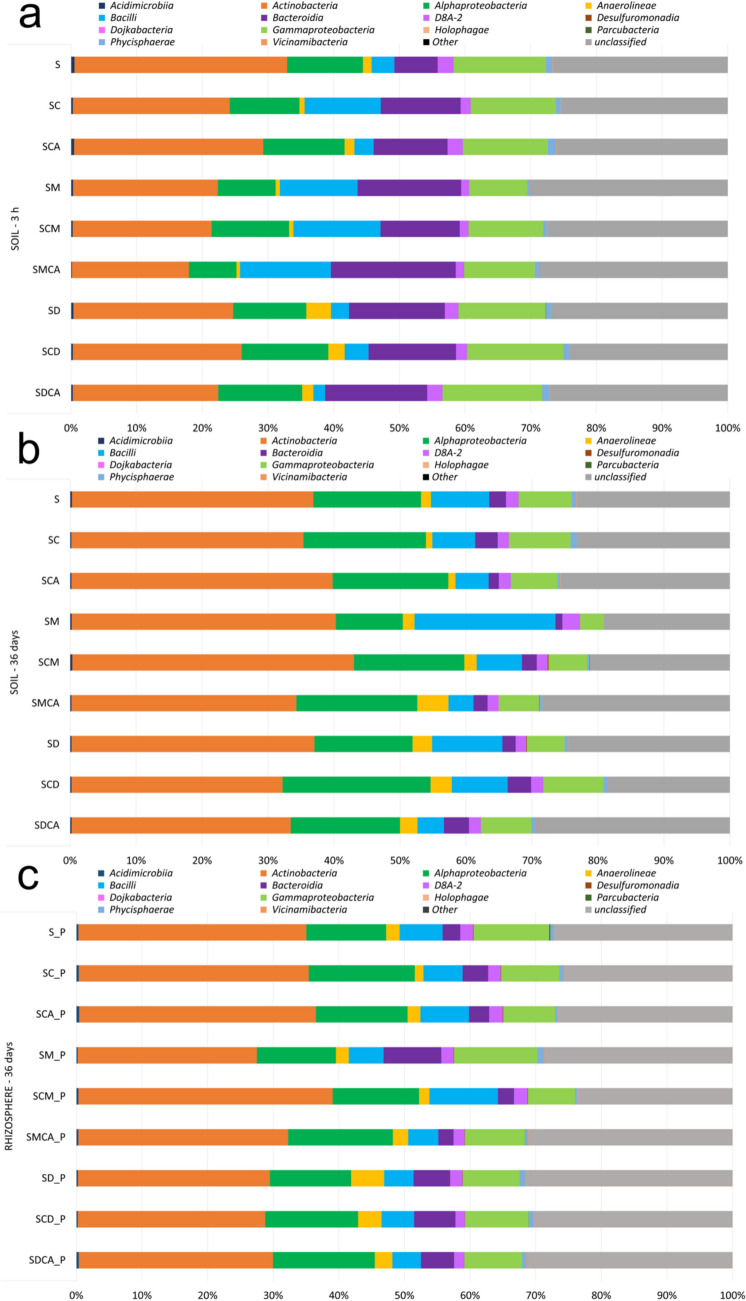
As regards the microbial community at class level, Actinobacteria (32.4%), Gammaproteobacteria (14.01%), and Alphaproteobacteria (11.5%) were predominant in the control soil (S at 3 h). Bacteroidia were found to be relatively more abundant in SC (12.2%) and SCA (11.3) than in the S microcosms, whereas Bacilli only in SC (11.6%). In manure-amended conditions, Bacteroidia were also in a relatively high number and in the co-presence of SMX and Cu (SMCA) were the dominant group (19%). An overall description of the classes (%) in the various conditions can be found in Fig. 4 and Table S8.
Fig. 4.
Class relative abundances (% ASV) with an average presence > 1% in each experimental condition at 3 h (a), soil at 36 days (b), and rhizosphere at 36 days (c). Data are means of three independent replicates
At 36 days, a general increase in Actinobacteria was observed in all experimental conditions (Fig. 4b, c), with relative percentages ranging from 27.3% (SM_P) and 28.5% (SCD_P) in a plant presence to 40.04% (SM) and 42.7% (SCM). Moreover, a sharp decrease in Bacteroidia, with the lowest values in SM and SCA (1.05% and 1.54%, respectively), was observed.
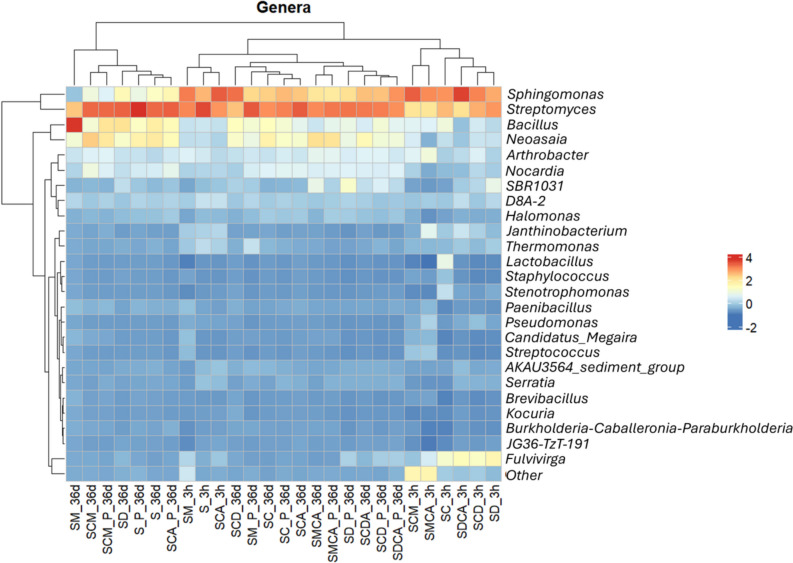
At the genus level (Fig. 5), S was initially (3 h) characterized by a high abundance of the Streptomyces (10.53%) and Sphingomonas (8.05%) genera; at 36 days, Streptomyces increased and Sphingomonas decreased in both the soil and rhizosphere (S and S_P). Copper spiking (SC) caused a prompt increase in Bacillus (Bacilli), Fulvivirga (Bacteroidia), and Lactobacillus (Bacilli) and a decrease in Streptomyces and Sphingomonas. In a similar way, an SMX and Cu co-presence (SCA) negatively influenced Streptomyces, but favoured Sphingomonas.
Fig. 5.
Heatmap for prokaryotic relative abundances at genus level in the different conditions and at the sampling times (3 h and 36 days). Genera and conditions were grouped in accordance with a hierarchical clustering dendrogram, at the top and the left side of the heatmap. Data are means of three independent replicates
Adding digestate (SD, SCD, and SDCA at 3 h) induced an increase in Fulvivirga (Bacteroidia), but at 36 days, the same genus decreased in the same conditions in both the rhizosphere and soil (SD, SCD, SDCA, SD_P, SCD_P, and SDCA_P). Moreover, the SBR1031 genus, belonging to the Anaerolineae class, increased in the presence of digestate at 3 h and became more abundant at 36 days compared with the control soil (S). The same genus also increased in the SMCA un-planted condition at 36 days.
On the contrary, adding manure caused a halving of Streptomyces and Sphingomonas compared to S at 3 h. At 36 days, a strong increase in Bacillus and Neoasaia was detected in the SM condition, as well as a decrease in Sphingomonas. On the other hand, in the manured condition (SM_P), the relative abundance values of other genera (Sphingomonas, Streptomyces, Bacillus, and Neoasaia) turned out to be similar to the initial ones. Moreover, a manure presence at 36 days favoured the Nocardia genus (Alphaproteobacteria class) in copper conditions (SCM, SMCA), especially in the absence of lettuce.
The principal component analysis (PCA), using overall data as variables, explained 47% of the total variance (Supplemental Fig. S1). Dimension 1 was significantly correlated (p < 0.05, Table S9) at 3 h with some bacterial genera such as Staphylococcus, Streptococcus, and Pseudomonas and ARGs (e.g., intI1) and at 36 days with Nocardia, Streptomyces, and Neoasaia, which in turn were positively correlated with other parameters such as sul2 and copper. On the other hand, other genera (e.g., Arthrobacter and Bacillus) are on the opposite side of SMX.
The network analysis (including SMX, Cu, and 26 microbial genera) was performed to evaluate the relationships between the contaminants and the main bacterial genera in the soil and rhizosphere amended with manure or digestate at 36 days. Supplemental Fig. S2 shows how manure or digestate differently affected the microbial community. For example, in manure conditions, copper was in the same cluster as Streptococcus and Staphylococcus and JG36-TzT in the same cluster as Stenotrophomonas. In digestate conditions, SMX was linked to Spingomonas and Halomonas and ARGs were in the same cluster as copper and Pseudomonas.
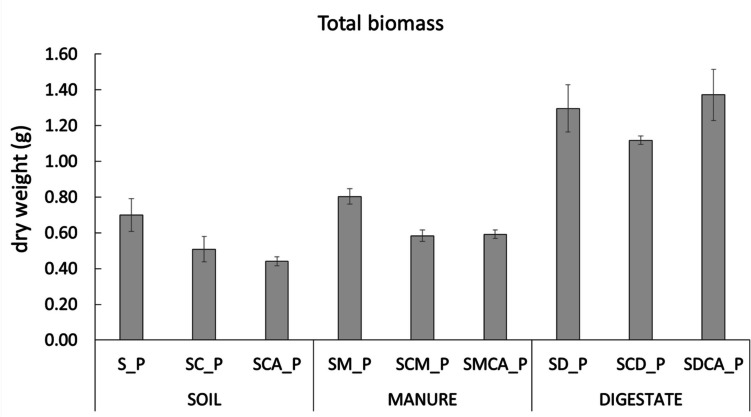
Lettuce biomass
When adding copper and an antibiotic, a detrimental effect on lettuce biomass was observed, except in the digestate-amended conditions (Fig. 6). Indeed, the significantly highest (p < 0.05, Table S11) biomass values were found in the digestate (SD_P, SCD_P, and SDCA_P) conditions, whether or not copper or an antibiotic was present.
Fig. 6.
Total (aerial + root) biomass (dry weight, g) of the lettuce plants under experimental different conditions at the end of the experiment (36 days)
Discussion
The fact that SMX was dissipated in the soil studied shows how microbial community degradative populations were already present in the soil. SMX removal from soil has been reported in other works. For example, a 90% SMX decrease at 60 days (initial concentration 20 mg/kg) in soil amended with a cattle manure digestate and maintained at 20 °C was found also by Rauseo et al. (2019). A previous work reports the disappearance of 85% of SMX in 46 days (at ca. 26°C) in soil amended with manure or digestate (initial concentration 7.5 mg/kg soil) with no differences between bulk soil and lettuce rhizosphere (Barra Caracciolo et al. 2022). In our case, the higher degradation rate (> 99% in 36 days) compared to other works can be ascribed to the higher experimental temperature and daylight hours occurring in the greenhouse in the second part of the experimental period. Even if the initial temperature was ca. 22 °C, over time, it rose, with a sudden temperature increase (up to 40 °C on 24 May) and a seasonal increase in daylight hours. The positive influence of high temperatures and light on SMX dissipation has been reported in other works (Patrolecco et al. 2018; Archundia et al. 2021).
The SMX residues found in the lettuce leaves showed how it was not bioaccumulated in this edible plant; in fact, its BAF values were always lower than one. Comparable results for SMX were obtained by Barra Caracciolo et al. (2022). The negligible tendency of SMX to be assimilated by plants has also been demonstrated by other authors (Mullen et al. 2019; Stando et al. 2022). Cheng et al. (2020), in a 49-day experiment, found about 0.08 mg/kg of SMX in cabbage biomass, from an initial soil concentration of 5 mg/kg.
When adding digestate, the lowest residual concentrations of SMX in both soil and leaves were found, and the rhizosphere microbial activity and plant biomass were the highest (Figs. 2 and 6), in line with what is reported in other works (Rauseo et al. 2019; Rolando et al. 2023; De Carolis et al. 2024), because organic carbon, nutrients, and microbial populations were introduced with it (Innangi et al. 2017; Garbini et al. 2022, 2023). Moreover, digestate, increasing soil pH, presumably decreased copper availability and toxicity and promoted close and positive relationships between the plant and its microbiome (Barra Caracciolo et al. 2015a, 2022; Ancona et al. 2017; Di Lenola et al. 2018; Adedayo et al. 2022). Indeed, with digestate present, the positive and synergic relationships established in the lettuce-microbiome “metaorganism” (Berg et al. 2016; Hassani et al. 2018; Compant et al. 2019; Schmidt and Saha 2021) were effective in improving overall soil quality and plant productivity. The digestate contained a higher amount of fulvic acids than manure, and this can have also favoured possible complexation processes between copper and antibiotics and this organic component, as described in other works (Qiu et al. 2007; Xu et al. 2017).
Although copper is an essential plant micronutrient, vegetal species can be affected by concentrations higher than is essential (Barra Caracciolo and Terenzi 2021) and microorganisms are more sensitive to it, even at concentrations of a few milligrams (Ochoa-Herrera et al. 2011; Narciso et al. 2023b). Copper, therefore, affected the rhizosphere microbiome (except in the case of digestate), as also demonstrated by the lower biodiversity values found in both un-amended (S_P, SC_P, and SCA_P) and manure-amended soils (SM_P, SCM_P, and SMCA_P).
In line with our results, a recent work reported both a negative impact from copper (at concentrations similar to ours) and a positive effect from digestate on lettuce growth. In particular, the chlorophyll content, leaf numbers, and shoot height were higher in all digestate conditions (including with copper present) than manure and unamended ones (De Carolis et al. 2024).
The effectiveness of the digestate in increasing rhizosphere microbial biodiversity and activity even with antibiotic presence has been found in other works (Singh et al. 2007; Buée et al. 2009; Barra Caracciolo et al. 2015a, 2022; Garbini et al. 2022).
It was evident that not only the microbial community favoured plant growth but also lettuce influenced its overall composition (Figs. 3 and 4c). For example, Alphaproteobacteria became the second dominant group in all the planted conditions and this result was in accordance with the presence in this class of several bacteria associated with the rhizosphere (e.g., nitrogen-fixing and nitrifying bacteria), in line with other works (Mendes et al. 2013; Barra Caracciolo et al. 2015a; Qiao et al. 2017; Di Lenola et al. 2018; Ling et al. 2022). Moreover, at 36 days, Actinobacteria were always in lower percentages than in the corresponding un-planted conditions and this was in line with a general increase in Proteobacteria in the rhizosphere, in particular in the amended conditions without copper (SM_P and SD_P). A Proteobacteria presence reflects an overall improvement in soil characteristics; in fact, they are dominant in good-quality state soils (Mocali et al. 2013; Godoi et al. 2014; Barra Caracciolo et al. 2015a, 2020a; Ancona et al. 2017).
Moreover, whether or not lettuce was present, digestate conditions generally favoured an increase in the percentage of the chemoorganotrophic Anaerolineae, which are known to favour nutrient recycling and soil quality (Xia et al. 2016; Freches and Fradinho 2024). On the other hand, the low organic carbon content (%) and dominance of Actinobacteria in the agricultural soil used in this work show that it was degraded, presumably because of its excessive use for agricultural purposes.
The analysis of the microbial community structure also confirmed that copper and sulfamethoxazole acted as a stressor for some microbial groups such as Actinobacteria and Bacilli, selecting, at 36 days, inside these classes, antibiotic-resistant bacteria and copper-tolerant ones. Indeed, Actinobacteria, which comprise not only sensitive bacteria but also several genera are able to resist unfavourable habitats and antibiotic and copper tolerant species (Shaw et al. 2020; Barra Caracciolo and Terenzi 2021), increased at 36 days in SC and SC_P. In particular, Streptomyces, the most abundant genus found at 3 h in the control soil (S), promptly decreased with the additions of copper, antibiotics, and amendments (SC, SCA SM, SD). Its subsequent increase at 36 days was possible thanks to its marked plasticity and capacity to deal with antibiotics and heavy metals (Hoff et al. 2018; Brangsch et al. 2022; Sedeek et al. 2023), with a selection of resistant strains (Benimeli et al. 2011; Fróes et al. 2012; Rammali et al. 2022).
The increase in the Bacillus genus is also in line with the presence of several multidrug-resistant species, able to counteract various metals, including copper (San et al. 2015; Tiwari et al. 2016; Barra Caracciolo and Terenzi 2021; Damle et al. 2021), and AB toxic effects (Haque et al. 2024).
On the other hand, other microbial groups, such as Bacteroidia, were not affected by adding contaminants and this was in line with their capacity to resist their toxicity (Niestępski et al. 2019b). Indeed, Bacteroidia comprise several antibiotic resistant species, including animal and human pathogens (Niestępski et al. 2019a).
Interestingly, Alphaproteobacteria not only increased with a plant presence, but some genera, e.g., Sphingomonas and Neoasaia, were found in higher percentages in a copper and antibiotic presence than in the other conditions, in accordance with their capacity to be copper resistant (Altimira et al. 2012; Shen et al. 2021). Although the Neoasaia genus (Alphaproteobacteria) has not been widely studied so far, its capacity to grow with an acidic pH and in degraded soils, developing a variety of resistance mechanisms, has been reported (Wang et al. 2015).
Moreover, Arthrobacter which remained quite stable over the experimental time have been reported to resist several heavy metals and to degrade a high variety of xenobiotic compounds (Pathak et al. 2020; Zhang et al. 2021; Wang et al. 2023).
Overall, when adding organic amendments, Bacilli, Gammaproteobacteria, Alphaproteobacteria, and Bacteroidia, which include bacteria able to resist and degrade sulfonamides, increased in accordance with other works (Reis et al. 2020; Readyhough et al. 2021; Wang et al. 2021; Barra Caracciolo et al. 2022) and with the fact that both the manure and digestate contained SMX residues. In particular, Bacillus and Sphingomonas are able to denitrify organic compounds and have also been reported to be SMX-resistant genera and capable of degrading this antibiotic (Cheng et al. 2020; Barra Caracciolo et al. 2022). The amendments also introduced Fulvivirga (common in gut microbiomes), which has been reported to be able to transform complex molecules, including pharmaceuticals (Wardman et al. 2022).
The fact that Nocardia increased at 36 days in SCM and SMCA showed how copper and manure favoured a significantly higher number of potential resistant pathogenic bacteria. Indeed, the Nocardia genus is capable of resisting metals and antibiotics (Nouioui et al. 2020) and many species are pathogens (e.g., the etiological agent of nocardiosis in humans and a variety of animals) (Brown-Elliott et al. 2006; Yasuike et al. 2017).
The organic amendments introduced not only microorganisms but also antibiotic-resistant bacteria, with a significantly higher relative abundance of ARGs in manure than in digestate, as demonstrated in other recent works (Checcucci et al. 2020; Visca et al. 2022; Barra Caracciolo et al. 2022), and this confirms that manure, if used as an organic fertilizer, is a potential ARG source (He et al. 2020; Ibekwe et al. 2023).
The overall reduction in intI1 and sul1 in the amended conditions at 36 days is in accordance with other works and was generally related to an AB decrease (Rauseo et al. 2019; Yang et al. 2021; Garbini et al. 2022). The intI1 and sul1 genes are generally positively related, and this fact is ascribable to their location on the same plasmid (Domínguez et al. 2019; de los Santos et al. 2021). On the contrary, sul2 increased at 36 days and particularly in a copper co-presence, showing how this gene was not strictly linked to SMX but also to other stressors (Enne 2004; Bean et al. 2009), such as heavy metals (Mazhar et al. 2021; Gao et al. 2022). The sul2 gene is generally located on a small plasmid which has a broad host range (Enne 2004; Visca et al. 2022; Barra Caracciolo et al. 2022) and different genetic mobility mechanisms, which confer bacteria several fitness advantages (Bean et al. 2009; Zhou et al. 2021). Indeed, Zhang et al. (2018b) demonstrated that sub-inhibitory copper concentrations (ranges of 0.005–0.05 mg/kg) can cause an increase in ARG horizontal transfers by raising cell membrane permeability and conjugation-related genes.
When adding copper to the soil, the detrimental effects of antibiotics on the microbial community were boosted, making bacterial populations potentially more sensitive to SMX and this result is in line with other authors’ findings (Liu et al. 2016; Orta-Rivera et al. 2023). The fact that the highest values of sul2 were detected at the end of the experiment in an SMX and copper co-presence confirms that Cu can be a co-factor contributing to sulfamethoxazole resistance gene persistence (cross-resistance), as recognized in other works (Glibota et al. 2019; Mazhar et al. 2021; Wu et al. 2022; Congilosi et al. 2022).
The network analysis supports the fact that the amendments had a different impact on the overall microbial community. In manure conditions, copper was linked to Streptococcus and Staphylococcus because they are both antibiotic- and metal-resistant (Price and Boyd 2020; Akbari et al. 2022) genera. Moreover, JG36-TzT and Stenotrophomonas were found in the same cluster thanks to their capacity to live in an acidic soil, with a heavy metal, and to degrade xenobiotics (Glibota et al. 2019; Köhler et al. 2021). In digestate conditions, SMX was linked to Sphingomonas and Halomonas, presumably because they were able to degrade it (Cheng et al. 2020; Barra Caracciolo et al. 2022; He et al. 2024). Finally, it is very interesting to highlight that copper is in the same cluster as the ARGs and Pseudomonas, and this can explain the relative increase in sul2 at 36 days with an antibiotic co-presence.
On the other hand, if we consider the conditions with exclusively manure (SM) or digestate (SD), which reproduce real agricultural scenarios, ARGs decreased strongly at 36 days, with the lowest values with digestate present and comparable to the control soil (S). At the same time, because the copper concentrations used in this work were in line with those commonly found in agricultural soils, its residues can have a serious impact on the spreading of antibiotic resistance genes, confirming both the antibiotic cross-resistance phenomenon (Chapman 2003; Zhang et al. 2019) and the possible negative effect of copper on plants (De Carolis et al. 2024).
Our work shows how antibiotic resistance is a dynamic and “silent phenomenon,” which can suddenly increase among soil microbial populations not only when adding organic amendments (which contain ARB and AB residues) but also in response to agricultural practices such as the use of copper as a fungicide (cross-resistance). Overall, manure and digestate had a different microbiological and chemical composition, improving soil quality and structure in different ways, adding different microbial populations and pathogenic genera, and organic matter components, useful nutrients for plants, and making copper bioavailable in different ways as well as, in the case of the digestate-amended soil, buffering toxic contaminant synergic effects. On the other hand, the plants in the amended conditions also contributed to increasing soil quality, thanks to synergic interactions with their rhizosphere, including a decrease in overall ARGs. Finally, cattle digestate was confirmed to be not only a suitable organic fertilizer but also a nature-based solution for limiting contaminant effects and ARG spreading.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Francesca Falconi and Daniele Parrone for their valuable support in the soil chemical analysis and Andrea Visca for assisting in qPCR analysis.
Author contribution
Conceptualization: ABC, CDC, GLG, PG, MAI, AN, JR, LR, LP, and FS. Methodology: ABC, CDC, GLG, PG, MAI, AN, JR, LR, LP, and FS. Formal analysis and investigation: ABC, CDC, GLG, PG, MAI, AN, MAB, CA-A, JR, LR, LP, and FS. Writing–original draft preparation: ABC and AN. Writing, review, and editing: ABC, AN, CDC, GLG, PG, JR, LR, LP, and FS. Funding acquisition: ABC, PG, and MAI. Resources: ABC. Supervision: ABC.
Funding
This research was funded mainly by Lazio Innova–Project Azero Antibiotics N. 85–2017-15065 (Progetti di Gruppi di Ricerca-Programma Strategico Regionale per la ricerca, innovazione ed il trasferimento tecnologico 2017–2020 Conoscenza e Cooperazione per un nuovo modello di sviluppo L. R. 13/2008). Moreover, it was partially funded by NextGenerationEU– “National Biodiversity Future Center-NBFC-Microbial diversity and functioning of freshwater and terrestrial ecosystems” Project (Piano Nazionale Resistenza e Resilienza (PNRR)–Missione 4 Componente 2 Investimento 1.4–Avviso N. 3138 del 16 dicembre 2021 rettificato con D.D. n.3175 del 18 dicembre 2021 del Ministero dell’Università e della Ricerca, Award Number: CN_00000033, Decreto Direttoriale MUR n. 1034 del 17giugno 2022 di concessione del finanziamento, CUP B83C22002930006) and Agritech National Research Center, European Union Next-Generation EU (Piano Nazionale di Ripresa e Resilienza (PNRR)-Missione 4 Componente 2, Investimento 1.4-D.D. 1032 17/06/2022, CN00000022)-SPOKE 1, Task 1.2.3.
Data availability
All data supporting the findings of this study are available within the paper and in its Supplementary information.
Declarations
The authors declare no competing interests. The authors declare that they agree with the content and that all gave explicit consent to submit and that they obtained consent from the responsible authorities at the institute/organization where the work has been carried out before the work is submitted and the research study was performed without involving human participants and/or animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adedayo AA, Fadiji AE, Babalola OO (2022) Plant health status affects the functional diversity of the rhizosphere microbiome associated with Solanum lycopersicum. Front Sustain Food Syst 6:894312. 10.3389/fsufs.2022.894312 [Google Scholar]
- Akbari MS, Doran KS, Burcham LR (2022) Metal Homeostasis in Pathogenic Streptococci. Microorganisms 10:1501. 10.3390/microorganisms10081501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali H, Khan E, Ilahi I (2019) Environmental chemistry and ecotoxicology of hazardous heavy metals: environmental persistence, toxicity, and bioaccumulation. J Chem 2019:1–14. 10.1155/2019/6730305 [Google Scholar]
- Allison LE, Moodie CD (1965) Carbonate. In: Black CA (ed) Methods of soil analysis. Part 2. Chemical and microbiological properties. American Society of Agronomy, Madison, pp 1379–1396 [Google Scholar]
- Altimira F, Yáñez C, Bravo G, Gonzàlez M, Rojas LA, Seeger M (2012) Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol 12:193. 10.1186/1471-2180-12-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalfitano S, Fazi S, Zoppini A, Barra Caracciolo A, Grenni P, Puddu A (2008) Responses of benthic bacteria to experimental drying in sediments from Mediterranean temporary rivers. Microb Ecol 55:270–279. 10.1007/s00248-007-9274-6 [DOI] [PubMed] [Google Scholar]
- Ancona V, Barra Caracciolo A, Grenni P, Di Lenola M, Campanale C, Calabrese A, Uricchio VF, Mascolo G, Massacci A (2017) Plant-assisted bioremediation of a historically PCB and heavy metal-contaminated area in Southern Italy. N Biotechnol 38:65–73. 10.1016/j.nbt.2016.09.006 [DOI] [PubMed] [Google Scholar]
- Archundia D, Martins JMF, Lehembre F, Morel MC, Duwig C (2021) Sulfamethoxazole biodegradation and impacts on soil microbial communities in a Bolivian arid high altitude catchment. Chemosphere 284:131335. 10.1016/j.chemosphere.2021.131335 [DOI] [PubMed] [Google Scholar]
- Barra Caracciolo A, Terenzi V (2021) Rhizosphere microbial communities and heavy metals. Microorganisms 9:1462. 10.3390/microorganisms9071462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra Caracciolo A, Grenni P, Cupo C, Rossetti S (2005) In situ analysis of native microbial communities in complex samples with high particulate loads. FEMS Microbiol Lett 253:55–58. 10.1016/j.femsle.2005.09.018 [DOI] [PubMed] [Google Scholar]
- Barra Caracciolo A, Bottoni P, Grenni P (2013) Microcosm studies to evaluate microbial potential to degrade pollutants in soil and water ecosystems. Microchem J 107:126–130. 10.1016/j.microc.2012.05.022 [Google Scholar]
- Barra Caracciolo A, Bustamante MA, Nogues I, Di Lenola M, Luprano ML, Grenni P (2015a) Changes in microbial community structure and functioning of a semiarid soil due to the use of anaerobic digestate derived composts and rosemary plants. Geoderma 245–246:89–97. 10.1016/j.geoderma.2015.01.021 [Google Scholar]
- Barra Caracciolo A, Topp E, Grenni P (2015b) Pharmaceuticals in the environment: biodegradation and effects on natural microbial communities A Review. J Pharm Biomed Anal 106:25–36. 10.1016/j.jpba.2014.11.040 [DOI] [PubMed] [Google Scholar]
- Barra Caracciolo A, Grenni P, Garbini GL, Rolando L, Campanale C, Aimola G, Fernandez-Gondalez AJ, Villadas PJ, Ancona V (2020) Characterization of the belowground microbial community in a poplar-phytoremediation strategy of a multi-contaminated soil. Front Microbiol 11:2073. 10.3389/fmicb.2020.02073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra Caracciolo A, Visca A, Massini G, Patrolecco L, Mazzurco Miritana V, Grenni P (2020) Environmental fate of antibiotics and resistance genes in livestock waste and digestate from biogas plants. Environ Sci Pollut Res Manag 21:23. 10.37722/ESPRAM.20201 [Google Scholar]
- Barra Caracciolo A, Visca A, Rauseo J, Spataro F, Garbini GL, Grenni P, Mariani L, Mazzurco Miritana V, Massini G, Patrolecco L (2022) Bioaccumulation of antibiotics and resistance genes in lettuce following cattle manure and digestate fertilization and their effects on soil and phyllosphere microbial communities. Environ Pollut 315:120413. 10.1016/j.envpol.2022.120413 [DOI] [PubMed] [Google Scholar]
- Barreto NMB, Pimenta NG, Braz BF, Freire AS, Santelli RE, Oliveira AC, Bastos LHP, Cardoso MHWM, Monteiro M, Diogenes ME, Perrone D (2021) Organic black beans (Phaseolus vulgaris L.) from Rio de Janeiro State, Brazil, present more phenolic compounds and better nutritional profile than nonorganic. Foods 10:900. 10.3390/foods10040900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean DC, Livermore DM, Hall LMC (2009) Plasmids imparting sulfonamide resistance in Escherichia coli : implications for persistence. Antimicrob Agents Chemother 53:1088–1093. 10.1128/AAC.00800-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Y, Fu C, Hu M, Liu L, Wong MH, Zheng C (2019) Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: a review. Environ Res 169:483–493. 10.1016/j.envres.2018.11.040 [DOI] [PubMed] [Google Scholar]
- Benavoli A, Corani G, Mangili F (2016) Should we really use post-hoc tests based on mean-ranks? J Mach Learn Res 17(1):152–161 [Google Scholar]
- Benimeli CS, Polti MA, Albarracín VH, Abate CM, Amoroso MJ (2011) Bioremediation potential of heavy metal–resistant Actinobacteria and maize plants in polluted soil. Springer, pp 459–477 [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen BJA, Lahr J, Nibbeling C, Jansen LJM, Bongers IEA, Wipfler EL, Van de Schans MGM (2018) The persistence of a broad range of antibiotics during calve, pig and broiler manure storage. Chemosphere 204:267–276. 10.1016/j.chemosphere.2018.04.042 [DOI] [PubMed] [Google Scholar]
- Berg G, Rybakova D, Grube M, Köberl M (2016) The plant microbiome explored: implications for experimental botany. J Exp Bot 67:995–1002. 10.1093/jxb/erv466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolan N, Adriano D, Mani S, Khan A (2003) Adsorption, complexation, and phytoavailability of copper as influenced by organic manure. Environ Toxicol Chem 22:450–456. 10.1002/etc.5620220228 [PubMed] [Google Scholar]
- Brangsch H, Höller M, Krauβe T, Waqas M, Schroeckh V, Brakhage AA, Bunk B, Spröer C, Overmann J, Kothe E (2022) Extremophile metal resistance: plasmid-encoded functions in Streptomyces mirabilis. Appl Environ Microbiol 88. 10.1128/aem.00085-22 [DOI] [PMC free article] [PubMed]
- Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ (2006) Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. 10.1128/CMR.19.2.259-282.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buée M, De Boer W, Martin F, Van Overbeek L, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212. 10.1007/s11104-009-9991-3 [Google Scholar]
- Bünemann EK, Reimer M, Smolders E, Smith SR, Bigalke M, Palmqvist A, Brandt KK, Möller K, Harder R, Hermann L, Speiser B, Oudshoorn F, Løes AK, Magid J (2024) Do contaminants compromise the use of recycled nutrients in organic agriculture? A review and synthesis of current knowledge on contaminant concentrations, fate in the environment and risk assessment. Sci Total Environ 912:168901. 10.1016/j.scitotenv.2023.168901 [DOI] [PubMed] [Google Scholar]
- Burman E, Bengtsson-Palme J (2021) Microbial community interactions are sensitive to small changes in temperature. Front Microbiol 12:672910. 10.3389/fmicb.2021.672910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante MA, Alburquerque JA, Restrepo AP, de la Fuente C, Paredes C, Moral R, Bernal MP (2012) Co-composting of the solid fraction of anaerobic digestates, to obtain added-value materials for use in agriculture. Biomass Bioenerg 43:26–35. 10.1016/j.biombioe.2012.04.010 [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canellas LP, Olivares FL, Aguiar NO, Jones DL, Nebbioso A, Mazzei P, Piccolo A (2015) Humic and fulvic acids as biostimulants in horticulture. Sci Hortic (Amsterdam) 196:15–27. 10.1016/j.scienta.2015.09.013 [Google Scholar]
- Chao A, Chazdon RL, Colwell RK, Shen T (2005) A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8:148–159. 10.1111/j.1461-0248.2004.00707.x [Google Scholar]
- Chapman JS (2003) Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeterior Biodegradation 51:271–276. 10.1016/S0964-8305(03)00044-1 [Google Scholar]
- Chaturvedi P, Shukla P, Giri BS, Chowdhary P, Chandra R, Gupta P, Pandey A (2021) Prevalence and hazardous impact of pharmaceutical and personal care products and antibiotics in environment: a review on emerging contaminants. Environ Res 194:110664. 10.1016/j.envres.2020.110664 [DOI] [PubMed] [Google Scholar]
- Checcucci A, Trevisi P, Luise D, Modesto M, Blasioli S, Braschi I, Mattarelli P (2020) Exploring the animal waste resistome: the spread of antimicrobial resistance genes through the use of livestock manure. Front Microbiol 11:1416. 10.3389/fmicb.2020.01416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Shi M, Xing L, Wang X, Gao H, Sun Y (2020) Sulfamethoxazole affects the microbial composition and antibiotic resistance gene abundance in soil and accumulates in lettuce. Environ Sci Pollut Res 27:29257–29265. 10.1007/s11356-020-08902-1 [DOI] [PubMed] [Google Scholar]
- Compant S, Samad A, Faist H, Sessitsch A (2019) A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J Adv Res 19:29–37. 10.1016/j.jare.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Cid M, Ferreira-Coelho G, Fernández-Calviño D, Núñez-Delgado A, Fernández-Sanjurjo MJ, Arias-Estévez M, Álvarez-Rodríguez E (2020) Single and simultaneous adsorption of three sulfonamides in agricultural soils: effects of pH and organic matter content. Sci Total Environ 744:140872. 10.1016/j.scitotenv.2020.140872 [DOI] [PubMed] [Google Scholar]
- Congilosi JL, Wallace JS, Neher TP, Howe A, Soupir ML, Aga DS (2022) Co-occurrence of antimicrobials and metals as potential drivers of antimicrobial resistance in swine farms. Front Environ Sci 10:1018739. 10.3389/fenvs.2022.1018739 [Google Scholar]
- Cycoń M, Mrozik A, Piotrowska-Seget Z (2019) Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front Microbiol 10:338. 10.3389/fmicb.2019.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle MS, Singh AN, Peters SC, Szalai VA, Fisher OS (2021) The YcnI protein from Bacillus subtilis contains a copper-binding domain. J Biol Chem 297:101078. 10.1016/j.jbc.2021.101078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carolis C, Iori V, Narciso A, Gentile D, Casentini B, Pietrini F, Grenni P, Barra Caracciolo A, Iannelli MA (2024) The effects of different combinations of cattle organic soil amendments and copper on lettuce (cv Rufus) plant growth. Environments 11:134. 10.3390/environments11070134 [Google Scholar]
- de los Santos, Santos, M, Laviña, ME, Poey (2021) Strict relationship between class 1 integrons and resistance to sulfamethoxazole in Escherichia coli. Microb Pathog 161:105206. 10.1016/j.micpath.2021.105206 [DOI] [PubMed]
- Di Lenola M, Barra Caracciolo A, Grenni P, Ancona V, Rauseo J, Laudicina VA, Uricchio VF, Massacci A (2018) Effects of Apirolio addition and alfalfa and compost treatments on the natural microbial community of a historically PCB-contaminated soil. Water, Air, Soil Pollut 229:143. 10.1007/s11270-018-3803-4 [Google Scholar]
- Dimitriu T (2022) Evolution of horizontal transmission in antimicrobial resistance plasmids. Microbiology 168:001214. 10.1099/mic.0.001214 [DOI] [PubMed] [Google Scholar]
- Domínguez M, Miranda CD, Fuentes O, De La Fuente M, Godoy FA, Bello-Toledo H, González-Rocha G (2019) Occurrence of transferable integrons and sul and dfr genes among sulfonamide-and/or trimethoprim-resistant bacteria isolated from Chilean salmonid farms. Front Microbiol 10:748. 10.3389/fmicb.2019.00748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enne VI (2004) Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemother 53:958–963. 10.1093/jac/dkh217 [DOI] [PubMed] [Google Scholar]
- Fagnano M, Agrelli D, Pascale A, Adamo P, Fiorentino N, Rocco C, Pepe O, Ventorino V (2020) Copper accumulation in agricultural soils: risks for the food chain and soil microbial populations. Sci Total Environ 734:139434. 10.1016/j.scitotenv.2020.139434 [DOI] [PubMed] [Google Scholar]
- Feng G, Huang H, Chen Y (2021) Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: a review. J Hazard Mater 420:126602. 10.1016/j.jhazmat.2021.126602 [DOI] [PubMed] [Google Scholar]
- Freches A, Fradinho JC (2024) The biotechnological potential of the Chloroflexota phylum. Appl Environ Microbiol 90 10.1128/aem.01756-23 [DOI] [PMC free article] [PubMed]
- Fróes A, Macrae A, Rosa J, Franco M, Souza R, Soares R, Coelho R (2012) Selection of a Streptomyces strain able to produce cell wall degrading enzymes and active against Sclerotinia sclerotiorum. J Microbiol 50:798–806. 10.1007/s12275-012-2060-2 [DOI] [PubMed] [Google Scholar]
- Gao Y-X, Li X, Fan X-Y, Zhao JR, Zhang ZX (2022) Fates of antibiotic resistance genes and bacterial/archaeal communities of activated sludge under stress of copper: gradient increasing/decreasing exposure modes. Bioresour Technol 363:127937. 10.1016/j.biortech.2022.127937 [DOI] [PubMed] [Google Scholar]
- Garbini GL, Grenni P, Rauseo J, Patrolecco L, Pescatore T, Spataro F, Barra Caracciolo A (2022) Insights into structure and functioning of a soil microbial community amended with cattle manure digestate and sulfamethoxazole. J Soils Sediments 22:2158–2173. 10.1007/s11368-022-03222-y [Google Scholar]
- Garbini GL, Barra Caracciolo A, Rolando L, Visca A, Borello D, Cosentini C, Gagliardi G, Ieropoulos I, Grenni P (2023) Effects of municipal waste compost on microbial biodiversity and energy production in terrestrial microbial fuel cells. N Biotechnol 78:131–140. 10.1016/j.nbt.2023.10.009 [DOI] [PubMed] [Google Scholar]
- Glibota N, Grande Burgos MJ, Gálvez A, Ortega E (2019) Copper tolerance and antibiotic resistance in soil bacteria from olive tree agricultural fields routinely treated with copper compounds. J Sci Food Agric 99:4677–4685. 10.1002/jsfa.9708 [DOI] [PubMed] [Google Scholar]
- Godoi I, Sene L, Barra Caracciolo A (2014) Assessment of the bacterial community structure in a Brazilian clay soil treated with atrazine. Ann Microbiol 64:307–311. 10.1007/s13213-013-0665-2 [Google Scholar]
- Grenni P, Barra Caracciolo A, Rodríguez-Cruz MS, Sánchez-Martín MJ (2009) Changes in the microbial activity in a soil amended with oak and pine residues and treated with linuron herbicide. Appl Soil Ecol 41:2–7. 10.1016/j.apsoil.2008.07.006 [Google Scholar]
- Grenni P, Rodríguez-Cruz MS, Herrero-Hernández E, Marín-Benito JM, Sánchez-Martín MJ, Barra Caracciolo A (2012) Effects of wood amendments on the degradation of terbuthylazine and on soil microbial community activity in a clay loam soil. Water Air Soil Pollut 223:5401–5412. 10.1007/s11270-012-1289-z [Google Scholar]
- Grenni P, Patrolecco L, Ademollo N, Di Lenola M, Barra Caracciolo A (2014) Capability of the natural microbial community in a river water ecosystem to degrade the drug naproxen. Environ Sci Pollut Res 21:13470–13479. 10.1007/s11356-014-3276-y [DOI] [PubMed] [Google Scholar]
- Grenni P, Ancona V, Barra Caracciolo A (2018) Ecological effects of antibiotics on natural ecosystems: a review. Microchem J 136:25–39. 10.1016/j.microc.2017.02.006 [Google Scholar]
- Gudda FO, Waigi MG, Odinga ES, Yang B, Carter L, Gao Y (2020) Antibiotic-contaminated wastewater irrigated vegetables pose resistance selection risks to the gut microbiome. Environ Pollut 264:114752. 10.1016/j.envpol.2020.114752 [DOI] [PubMed] [Google Scholar]
- Hänsch R, Mendel RR (2009) Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr Opin Plant Biol 12:259–266. 10.1016/j.pbi.2009.05.006 [DOI] [PubMed] [Google Scholar]
- Haque MA, Hu H, Liu J, Aminul Islam MD, Hossen F, Arifur Rahman MD, Ahmed F, He C (2024) Emergence of multidrug-resistant Bacillus spp derived from animal feed, food and human diarrhea in South-Eastern Bangladesh. BMC Microbiol 24:61. 10.1186/s12866-024-03199-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani MA, Durán P, Hacquard S (2018) Microbial interactions within the plant holobiont. Microbiome 6:58. 10.1186/s40168-018-0445-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yuan Q, Mathieu J, Stadler L, Senehi N, Sun R, Alvarez PJ (2020) Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water 3:4. 10.1038/s41545-020-0051-0 [Google Scholar]
- He Y, Liu L, Wang Q, Dong X, Huang J, Jia X, Peng X (2024) Bio-degraded of sulfamethoxazole by microbial consortia without addition nutrients: mineralization, nitrogen removal, and proteomic characterization. J Hazard Mater 466:133558. 10.1016/j.jhazmat.2024.133558 [DOI] [PubMed] [Google Scholar]
- Hoff G, Bertrand C, Piotrowski E, Thibessard A, Leblond P (2018) Genome plasticity is governed by double strand break DNA repair in Streptomyces. Sci Rep 8:5272. 10.1038/s41598-018-23622-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Ahmed S, Gu Y, Huang J, An B, Wu C, Zhou Y, Cheng G (2021) The effects of natural products and environmental conditions on antimicrobial resistance. Molecules 26:4277. 10.3390/molecules26144277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannelli MA, Bellini A, Venditti I, Casentini B, Battocchio C, Scalici M, Ceschin S (2022) Differential phytotoxic effect of silver nitrate (AgNO3) and bifunctionalized silver nanoparticles (AgNPs-Cit-L-Cys) on Lemna plants (duckweeds). Aquat Toxicol 250:106260. 10.1016/j.aquatox.2022.106260 [DOI] [PubMed] [Google Scholar]
- Ibekwe AM, Bhattacharjee AS, Phan D, Ashworth D, Schimdt ME, Murinda SE, Obayiuwana A, Murry MA, Schwartz G, Lundquist T, Ma J, Karathia H, Fanelli B, Hasan NA, Yang C-H (2023) Potential reservoirs of antimicrobial resistance in livestock waste and treated wastewater that can be disseminated to agricultural land. Sci Total Environ 872:162194. 10.1016/j.scitotenv.2023.162194 [DOI] [PubMed] [Google Scholar]
- Innangi M, Niro E, D’Ascoli R, Danise T, Proietti P, Nasini L, Regno L, Castaldi S, Fioretti A (2017) Effects of olive pomace amendment on soil enzyme activities. Appl Soil Ecol 119:242–249. 10.1016/j.apsoil.2017.06.015 [Google Scholar]
- Jampani M, Mateo-Sagasta J, Chandrasekar A, Fatta-Kassinos D, Graham DW, Gothwal R, Moodley A, Mohan Chadag V, Wiberg D, Langan S (2024) Fate and transport modelling for evaluating antibiotic resistance in aquatic environments: current knowledge and research priorities. J Hazard Mater 461:132527. 10.1016/j.jhazmat.2023.132527 [DOI] [PubMed] [Google Scholar]
- Kang W, Zhang Y-J, Shi X, He JZ, Hu HW (2018) Short-term copper exposure as a selection pressure for antibiotic resistance and metal resistance in an agricultural soil. Environ Sci Pollut Res 25:29314–29324. 10.1007/s11356-018-2978-y [DOI] [PubMed] [Google Scholar]
- Kaur H, Kaur H, Kaur H, Srivastava S (2023) The beneficial roles of trace and ultratrace elements in plants. Plant Growth Regul 100:219–236. 10.1007/s10725-022-00837-6 [Google Scholar]
- Kim R-Y, Yoon J-K, Kim T-S, Yang JE, Owens G, Kim KR (2015) Bioavailability of heavy metals in soils: definitions and practical implementation—a critical review. Environ Geochem Health 37:1041–1061. 10.1007/s10653-015-9695-y [DOI] [PubMed] [Google Scholar]
- Köhler JM, Beetz N, Günther PM, Möller F, Cao J (2021) Extremophiles in soil communities of former copper mining sites of the East Harz region (Germany) reflected by re-analyzed 16S rRNA data. Microorganisms 9:1422. 10.3390/microorganisms9071422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Pandita S, Singh Sidhu GP, Sharma A, Khanna K, Kaur P, Bali AS, Setia R (2021) Copper bioavailability, uptake, toxicity and tolerance in plants: a comprehensive review. Chemosphere 262:127810. 10.1016/j.chemosphere.2020.127810 [DOI] [PubMed] [Google Scholar]
- Kuppusamy S, Kakarla D, Venkateswarlu K, Megharaj M, Yoon YE, Lee YB (2018) Veterinary antibiotics (VAs) contamination as a global agro-ecological issue: a critical view. Agric Ecosyst Environ 257:47–59. 10.1016/j.agee.2018.01.026 [Google Scholar]
- Lai H-T, Wang T-S, Chou C-C (2011) Implication of light sources and microbial activities on degradation of sulfonamides in water and sediment from a marine shrimp pond. Bioresour Technol 102:5017–5023. 10.1016/j.biortech.2011.01.070 [DOI] [PubMed] [Google Scholar]
- Li W, Liu C, Ho HC, Shi L, Zeng Y, Yang X, Huang Q, Pei Y, Huang C, Yanga L (2023) Association between antibiotic resistance and increasing ambient temperature in China: an ecological study with nationwide panel data. Lancet Reg Heal - West Pacific 30:100628. 10.1016/j.lanwpc.2022.100628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhang J, Chen H, Wang J, Sun W, Zhang X, Yang Y, Wang Q, Ma J (2017) Effect of temperature on sulfonamide antibiotics degradation, and on antibiotic resistance determinants and hosts in animal manures. Sci Total Environ 607–608:725–732. 10.1016/j.scitotenv.2017.07.057 [DOI] [PubMed] [Google Scholar]
- Ling N, Wang T, Kuzyakov Y (2022) Rhizosphere bacteriome structure and functions. Nat Commun 13:836. 10.1038/s41467-022-28448-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Cao H, Yang Y, Ma X, Liu X (2016) Combinational effects of sulfomethoxazole and copper on soil microbial community and function. Environ Sci Pollut Res 23:4235–4241. 10.1007/s11356-015-4892-x [DOI] [PubMed] [Google Scholar]
- MacFadden DR, McGough SF, Fisman D, Santillana M, Brownstein JS (2018) Antibiotic resistance increases with local temperature. Nat Clim Chang 8:510–514. 10.1038/s41558-018-0161-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masmoudi S, Nguyen-Deroche N, Caruso A, Ayadi H, Morant-Manceau A, Tremblin G (2013) Cadmium, copper, sodium and zinc effects on diatoms: from heaven to hell — a review. Cryptogam Algol 34:185–225. 10.7872/crya.v34.iss2.2013.185 [Google Scholar]
- Mazhar SH, Li X, Rashid A, Su J, Xu J, Brejnrod AD, Su J-Q, Wu Y, Zhu Y-G, Zhou SG, Feng R, Rensing C (2021) Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci Total Environ 755:142702. 10.1016/j.scitotenv.2020.142702 [DOI] [PubMed] [Google Scholar]
- Mazzurco Miritana V, Patrolecco L, Barra Caracciolo A, Visca A, Piccinini F, Signorini A, Rosa S, Grenni P, Garbini GL, Spataro F, Rauseo J, Massini G (2022) Effects of ciprofloxacin alone or in mixture with sulfamethoxazole on the efficiency of anaerobic digestion and its microbial community. Antibiotics 11:1111. 10.3390/antibiotics11081111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- Mitchell SM, Ullman JL, Teel AL, Watts RJ (2014) pH and temperature effects on the hydrolysis of three β-lactam antibiotics: ampicillin, cefalotin and cefoxitin. Sci Total Environ 466–467:547–555. 10.1016/j.scitotenv.2013.06.027 [DOI] [PubMed] [Google Scholar]
- Mitchell SM, Ullman JL, Bary A, Cogger CG, Teel AL, Watts RJ (2015) Antibiotic degradation during thermophilic composting. Water Air Soil Pollut 226:13. 10.1007/s11270-014-2288-z [Google Scholar]
- Mocali S, Galeffi C, Perrin E, Florio A, Migliore M, Canganella F, Bianconi G, Di Mattia E, Dell’Abate MT, Fani R, Benedetti A (2013) Alteration of bacterial communities and organic matter in microbial fuel cells (MFCs) supplied with soil and organic fertilizer. Appl Microbiol Biotechnol 97:1299–1315. 10.1007/s00253-012-3906-6 [DOI] [PubMed] [Google Scholar]
- Mullen RA, Hurst JJ, Naas KM, Sassoubre LM, Aga DS (2019) Assessing uptake of antimicrobials by Zea mays L. and prevalence of antimicrobial resistance genes in manure-fertilized soil. Sci Total Environ 646:409–415. 10.1016/j.scitotenv.2018.07.199 [DOI] [PubMed] [Google Scholar]
- Narciso A, Barra Caracciolo A, De Carolis C (2023a) Overview of direct and indirect effects of antibiotics on terrestrial organisms. Antibiotics 12:1471. 10.3390/antibiotics12091471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso A, Barra Caracciolo A, Grenni P, Rauseo J, Patrolecco L, Spataro F, Mariani L (2023) Application of the Aliivibrio fischeri bacterium bioassay for assessing single and mixture effects of antibiotics and copper. FEMS Microbiol Ecol 99:125. 10.1093/femsec/fiad125 [DOI] [PubMed] [Google Scholar]
- Nath S, Paul P, Roy R, Bhattacharjee S, Deb B (2019) Isolation and identification of metal-tolerant and antibiotic-resistant bacteria from soil samples of Cachar district of Assam India. SN Appl Sci 1:727. 10.1007/s42452-019-0762-3 [Google Scholar]
- Niestępski S, Harnisz M, Korzeniewska E, Aguilera-Arreola MG, Contreras-Rodríguez A, Filipkowska Z, Osińska A (2019a) The emergence of antimicrobial resistance in environmental strains of the Bacteroides fragilis group. Environ Int 124:408–419. 10.1016/j.envint.2018.12.056 [DOI] [PubMed] [Google Scholar]
- Niestępski S, Harnisz M, Korzeniewska E, Osińska A (2019b) Isolation of anaerobic bacteria of the Bacteroides fragilis group from environmental samples. E3S Web Conf 100:00058. 10.1051/e3sconf/201910000058
- Nouioui I, Cortés-Albayay C, Neumann-Schaal M, Nouioui I, Cortés-Albayay C, Neumann-Schaal M, Vicente D, Cilla G, Klenk H-S, Marimón JM, Ercibengoa M (2020) Genomic virulence features of two novel species Nocardia barduliensis sp. nov. and Nocardia gipuzkoensis sp. nov., isolated from patients with chronic pulmonary diseases. Microorganisms 8:1517. 10.3390/microorganisms8101517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Herrera V, León G, Banihani Q, Field J, Sierra-Alvarez R (2011) Toxicity of copper(II) ions to microorganisms in biological wastewater treatment systems. Sci Total Environ 412–413:380–385. 10.1016/j.scitotenv.2011.09.072 [DOI] [PubMed] [Google Scholar]
- Orta-Rivera AM, Meléndez-Contés Y, Medina-Berríos N, Gómez-Cadorna AM, Ramos-Rodrìguez A, Cruz-Santiago C, Gonazàlez-Dumeng C, López J, Escribano J, Rivera-Otero JJ, Dìaz-Rivera J, Dìaz-Vélez SC, Feliciano-Delgado Z, Tinoco AT (2023) Copper-based antibiotic strategies: exploring applications in the hospital setting and the targeting of Cu regulatory pathways and current drug design trends. Inorganics 11:252. 10.3390/inorganics11060252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagos P, Ballabio C, Lugato E, Jones A, Borrelli P, Scarpa S, Orgiazzi A, Montanarella L (2018) Potential Sources of Anthropogenic Copper Inputs to European Agricultural Soils. Sustainability 10:2380. 10.3390/su10072380 [Google Scholar]
- Pathak A, Jaswal R, Chauhan A (2020) Genomic characterization of a mercury resistant Arthrobacter sp H-02–3 reveals the presence of heavy metal and antibiotic resistance determinants. Front Microbiol 10:3039. 10.3389/fmicb.2019.03039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrolecco L, Rauseo J, Ademollo N, Grenni P, Cardoni M, Levanteis C, Luprano ML, Barra Caracciolo A (2018) Persistence of the antibiotic sulfamethoxazole in river water alone or in the co-presence of ciprofloxacin. Sci Total Environ 640–641:1438–1446. 10.1016/j.scitotenv.2018.06.025 [DOI] [PubMed] [Google Scholar]
- Patyra E, Osiński Z, Kwiatek K (2024) Residues of veterinary antibiotics in solid natural and organic fertilizers—method development and sample analysis. Environ Sci Pollut Res. 10.1007/s11356-024-33956-w [DOI] [PubMed] [Google Scholar]
- Pei R, Cha J, Carlson KH, Pruden A (2007) Response of antibiotic resistance genes (ARG) to biological treatment in dairy lagoon water. Environ Sci Technol 41:5108–5113. 10.1021/es070051x [DOI] [PubMed] [Google Scholar]
- Perković S, Paul C, Vasić F, Helming K (2022) Human health and soil health risks from heavy metals, micro(nano)plastics, and antibiotic resistant bacteria in agricultural soils. Agronomy 12:2945. 10.3390/agronomy12122945 [Google Scholar]
- Pinilla-Redondo R, Cyriaque V, Jacquiod S, Sørensen SJ, Riber L (2018) Monitoring plasmid-mediated horizontal gene transfer in microbiomes: recent advances and future perspectives. Plasmid 99:56–67. 10.1016/j.plasmid.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Poirier-Larabie S, Segura PA, Gagnon C (2016) Degradation of the pharmaceuticals diclofenac and sulfamethoxazole and their transformation products under controlled environmental conditions. Sci Total Environ 557–558:257–267. 10.1016/j.scitotenv.2016.03.057 [DOI] [PubMed] [Google Scholar]
- Prata JC, Ribeiro AI, Rocha-Santos T (2022) An introduction to the concept of One Health. In: One Health. Elsevier, pp 1–31
- Price EE, Boyd JM (2020) Genetic regulation of metal ion homeostasis in Staphylococcus aureus. Trends Microbiol 28:821–831. 10.1016/j.tim.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Wang F, Zhang J, Chen Y, Zhang C, Liu G, Zhang H, Ma C, Zhang J (2017) The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci Rep 7:3940. 10.1038/s41598-017-04213-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J-W, Tang X, Zheng C, Li Y, Huang Y (2007) Copper complexation by fulvic acid affects copper toxicity to the larvae of the polychaete Hydroides elegans. Mar Environ Res 64:563–573. 10.1016/j.marenvres.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Radu E, Woegerbauer M, Rab G, Oismüller M, Strauss P, Hufnagl P, Gottsberger RA, Krampe J, Weyermair K, Kreuzinger N (2021) Resilience of agricultural soils to antibiotic resistance genes introduced by agricultural management practices. Sci Total Environ 756:143699. 10.1016/j.scitotenv.2020.143699 [DOI] [PubMed] [Google Scholar]
- Rammali S, Hilali L, Dari K, Bencharki B, Rahim A, Timinouni M, Gaboune F, El Aalaoui M, Khattabi A (2022) Antimicrobial and antioxidant activities of Streptomyces species from soils of three different cold sites in the Fez-Meknes region Morocco. Sci Rep 12:17233. 10.1038/s41598-022-21644-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauseo J, Barra Caracciolo A, Ademollo N, Cardoni M, Di Lenola M, Gaze W, Stanton I, Grenni P, Pescatore T, Spataro F, Patrolecco L (2019) Dissipation of the antibiotic sulfamethoxazole in a soil amended with anaerobically digested cattle manure. J Hazard Mater 378:120769. 10.1016/j.jhazmat.2019.120769 [DOI] [PubMed] [Google Scholar]
- Readyhough T, Neher DA, Andrews T (2021) Organic amendments alter soil hydrology and belowground microbiome of tomato (Solanum lycopersicum). Microorganisms 9:1561. 10.3390/microorganisms9081561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis AC, Kolvenbach BA, Nunes OC, Corvini PFX (2020) Biodegradation of antibiotics: the new resistance determinants – part I. N Biotechnol 54:34–51. 10.1016/j.nbt.2019.08.002 [DOI] [PubMed] [Google Scholar]
- Rolando L, Barra Caracciolo A, Garbini GL, Visca A, Mariani L, Finizio A, Mazzurco-Miritana V, Nouges I, Grenni P (2023) Nature-based solutions using organic amendments for biorestoration of alkaline spoil material. Appl Soil Ecol 192:105070. 10.1016/j.apsoil.2023.105070 [Google Scholar]
- Samreen AI, Malak HA, Abulreesh HH (2021) Environmental antimicrobial resistance and its drivers: a potential threat to public health. J Glob Antimicrob Resist 27:101–111. 10.1016/j.jgar.2021.08.001 [DOI] [PubMed] [Google Scholar]
- San K, Long J, Michels CA, Gadura N (2015) Antimicrobial copper alloy surfaces are effective against vegetative but not sporulated cells of gram-positive Bacillus subtilis. Microbiologyopen 4:753–763. 10.1002/mbo3.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Monedero MA, Roig A, Martínez-Pardo C, Cegarra J, Paredes C (1996) A microanalysis method for determining total organic carbon in extracts of humic substances. Relationships between total organic carbon and oxidable carbon. Bioresour Technol 57:291–295. 10.1016/S0960-8524(96)00078-8 [Google Scholar]
- Schmidt R, Saha M (2021) Infochemicals in terrestrial plants and seaweed holobionts: current and future trends. New Phytol 229:1852–1860. 10.1111/nph.16957 [DOI] [PubMed] [Google Scholar]
- Sedeek AM, Salah I, Kamel HL, Soltan MA, Nour E, Alshammari A, Riaz Rajoka MS, Elsayed TR (2023) Genome-based analysis of the potential bioactivity of the Terrestrial Streptomyces vinaceusdrappus strain AC-40. Biology (Basel) 12:345. 10.3390/biology12030345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvam A, Xu D, Zhao Z, Wong JWC (2012a) Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour Technol 126:383–390. 10.1016/j.biortech.2012.03.045 [DOI] [PubMed] [Google Scholar]
- Selvam A, Zhao Z, Wong JWC (2012b) Composting of swine manure spiked with sulfadiazine, chlortetracycline and ciprofloxacin. Bioresour Technol 126:412–417. 10.1016/j.biortech.2011.12.073 [DOI] [PubMed] [Google Scholar]
- Shahid M, Dumat C, Silvestre J, Pinelli E (2012) Effect of fulvic acids on lead-induced oxidative stress to metal sensitive Vicia faba L. plant. Biol Fertil Soils 48:689–697. 10.1007/s00374-012-0662-9 [Google Scholar]
- Shao Y, Liu X, Liu A, Dong Y, Hu X (2020) Co-sorption of sulfamethoxazole and Cu onto several soils with different properties and their binding mechanism. IOP Conf Ser Earth Environ Sci 432:012009. 10.1088/1755-1315/432/1/012009 [Google Scholar]
- Shaw JLA, Ernakovich JG, Judy JD, Farrell M, Whatmuff M, Kirby J (2020) Long-term effects of copper exposure to agricultural soil function and microbial community structure at a controlled and experimental field site. Environ Pollut 263:114411. 10.1016/j.envpol.2020.114411 [DOI] [PubMed] [Google Scholar]
- Shen Q, Tang J, Wang X, Li Y, Yao X, Sun H, Wu Y (2021) Fate of antibiotic resistance genes and metal resistance genes during the thermophilic fermentation of solid and liquid swine manures in an ectopic fermentation system. Ecotoxicol Environ Saf 213:111981. 10.1016/j.ecoenv.2021.111981 [DOI] [PubMed] [Google Scholar]
- Singh BK, Munro S, Potts JM, Millard P (2007) Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl Soil Ecol 36:147–155. 10.1016/j.apsoil.2007.01.004 [Google Scholar]
- Singh VK, Singh R, Kumar A, Bhadouria R, Singh P, & Notarte KI (2021). Antibiotics and antibiotic resistance genes in agroecosystems as emerging contaminants. Sustainable Agriculture Reviews 50: Emerging Contaminants in Agriculture, 177–210.
- Sodhi KK, Kumar M, Balan B, Dhaulaniya AS, Shree P, Sharma N, Singh DK (2021) Perspectives on the antibiotic contamination, resistance, metabolomics, and systemic remediation. SN Appl Sci 3:269. 10.1007/s42452-020-04003-3 [Google Scholar]
- Soto-Giron MJ, Kim J-N, Schott E, Tahmin C, Ishoey T, Mincer TJ, DeWalt J, Toledo G (2021) The edible plant microbiome represents a diverse genetic reservoir with functional potential in the human host. Sci Rep 11:24017. 10.1038/s41598-021-03334-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spataro F, Ademollo N, Pescatore T, Rauseo J, Patrolecco (2019) Antibiotic residues and endocrine disrupting compounds in municipal wastewater treatment plants in Rome, Italy. Microchem J 148:634–642. 10.1016/j.microc.2019.05.053 [Google Scholar]
- Stando K, Czyż A, Gajda M, Felis E, Bajkacz S (2022) Study of the phytoextraction and phytodegradation of sulfamethoxazole and trimethoprim from water by Limnobium laevigatum. Int J Environ Res Public Health 19:16994. 10.3390/ijerph192416994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm L, Thuerig B, Apostolov S, Blogg H, Borgo E, Corneo PE, Fittje S, de Palma M, Donko A, Experton C, Alcàzar Marìn È, Morell Pèrez À, Pertot I, Rasmussen A, Steinshamn H, Vetemaa A, Willer H, J, Herforth-rahmné (2022) Use of copper-based fungicides in organic agriculture in twelve European countries. Agronomy 12:673. 10.3390/agronomy12030673
- Thompson M, Ellison SLR, Wood R (2002) Harmonized guidelines for single-laboratory validation of methods of analysis (IUPAC Technical Report). Pure Appl Chem 74:835–855. 10.1351/pac200274050835 [Google Scholar]
- Tiwari M, Jain P, Chandrashekhar Hariharapura R, Narayanan K, Bhat U, Udupa N, Rao JV (2016) Biosynthesis of copper nanoparticles using copper-resistant Bacillus cereus, a soil isolate. Process Biochem 51:1348–1356. 10.1016/j.procbio.2016.08.008 [Google Scholar]
- Tóth G, Hermann T, Da Silva MR, Montanarella L (2016) Heavy metals in agricultural soils of the European Union with implications for food safety. Environ Int 88:299–309. 10.1016/j.envint.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Vieublé Gonod L, Dellouh LPY, Andriamalala A, Dumény V, Bergheaud V, Cambier P (2022) Fate of sulfamethoxazole in compost, manure and soil amended with previously stored organic wastes. Sci Total Environ 803:150023. 10.1016/j.scitotenv.2021.150023 [DOI] [PubMed] [Google Scholar]
- Visca A, Barra Caracciolo A, Grenni P, Patrolecco L, Rauseo J, Massini G, Mazzurco-Miritana V, Spataro F (2021) Anaerobic digestion and removal of sulfamethoxazole, enrofloxacin, ciprofloxacin and their antibiotic resistance genes in a full-scale biogas plant. Antibiotics 10:502. 10.3390/antibiotics10050502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visca A, Rauseo J, Spataro F, Patrolecco L, Grenni P, Mazzurco-Miritana V, Barra Caracciolo A (2022) Antibiotics and antibiotic resistance genes in anaerobic digesters and predicted concentrations in agroecosystems. J Environ Manage 301:113891. 10.1016/j.jenvman.2021.113891 [DOI] [PubMed] [Google Scholar]
- Wang B, Shao Y, Chen F (2015) Overview on mechanisms of acetic acid resistance in acetic acid bacteria. World J Microbiol Biotechnol 31:255–263. 10.1007/s11274-015-1799-0 [DOI] [PubMed] [Google Scholar]
- Wang S, Yuan R, Chen H, Wang F, Zhou B (2021) Anaerobic biodegradation of four sulfanilamide antibiotics: kinetics, pathways and microbiological studies. J Hazard Mater 416:125840. 10.1016/j.jhazmat.2021.125840 [DOI] [PubMed] [Google Scholar]
- Wang X, Dai Z, Lin J, Zhao H, Yu H, Ma B, Hu L, Shi J, Chen X, Liu M, Ke X, Yu Y, Dahlgren RA, Xu J (2023) Heavy metal contamination collapses trophic interactions in the soil microbial food web via bottom-up regulation. Soil Biol Biochem 184:109058. 10.1016/j.soilbio.2023.109058 [Google Scholar]
- Wardman JF, Bains RK, Rahfeld P, Withers SG (2022) Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat Rev Microbiol 20:542–556. 10.1038/s41579-022-00712-1 [DOI] [PubMed] [Google Scholar]
- White A, Hughes JM (2019) Critical importance of a One Health approach to antimicrobial resistance. EcoHealth 16:404–409. 10.1007/s10393-019-01415-5 [DOI] [PubMed] [Google Scholar]
- Wu Y, Wen Q, Chen Z, Fu Q, Bao H (2022) Response of antibiotic resistance to the co-exposure of sulfamethoxazole and copper during swine manure composting. Sci Total Environ 805:150086. 10.1016/j.scitotenv.2021.150086 [DOI] [PubMed] [Google Scholar]
- Wu N-N, Liu S, Xu R, Huang Q-Y, Li H-X, Lin L, Hou R, Cheng Y-Y, Xu X-R (2024) New insight into the bioaccumulation and trophic transfer of free and conjugated antibiotics in an estuarine food web based on multimedia fate and model simulation. J Hazard Mater 465:133088. 10.1016/j.jhazmat.2023.133088 [DOI] [PubMed] [Google Scholar]
- Xia Y, Wang Y, Wang Y, Chin FY, Zhang T (2016) Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol Biofuels 9:111. 10.1186/s13068-016-0524-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Yu W, Ma Q, Zhou H, Jiang C (2017) Toxicity of sulfadiazine and copper and their interaction to wheat (Triticum aestivum L.) seedlings. Ecotoxicol Environ Saf 142:250–256. 10.1016/j.ecoenv.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Yang Q, Gao Y, Ke J, Show PL, Ge Y, Liu Y, Guo R, Chen J (2021) Antibiotics: an overview on the environmental occurrence, toxicity, degradation, and removal methods. Bioengineered 12:7376–7416. 10.1080/21655979.2021.1974657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuike M, Nishiki I, Iwasaki Y, Nakamura Y, Fujiwara A, Shimahara Y, Kamaishi T, Yoshida T, Nagai S, Kobayashi T, Katoh M (2017) Analysis of the complete genome sequence of Nocardia seriolae UTF1, the causative agent of fish nocardiosis: the first reference genome sequence of the fish pathogenic Nocardia species. PLoS ONE 12:e0173198. 10.1371/journal.pone.0173198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamulina IV, Gorovtsov AV, Minkina TM, Mandzhieva SS, Burachevskaya MV, Bauer TV (2022) Soil organic matter and biological activity under long-term contamination with copper. Environ Geochem Health 44:387–398. 10.1007/s10653-021-01044-4 [DOI] [PubMed] [Google Scholar]
- Zhang J, Sui Q, Tong J, Zhong H, Wang Y, Chen M, Wei Y (2018a) Soil types influence the fate of antibiotic-resistant bacteria and antibiotic resistance genes following the land application of sludge composts. Environ Int 118:34–43. 10.1016/j.envint.2018.05.029 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu R, Li W, Ma J, Lin H (2021) Genomic insights into the antibiotic resistance pattern of the tetracycline-degrading bacterium, Arthrobacter nicotianae OTC-16. Sci Rep 11:15638. 10.1038/s41598-021-94840-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gu AZ, Cen T, Li X, He M, Li D, Chen J (2018b) Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ Pollut 237:74–82. 10.1016/j.envpol.2018.01.032 [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang Y, Song H, Lu J, Yuan Z, Guo J (2019) Copper nanoparticles and copper ions promote horizontal transfer of plasmid-mediated multi-antibiotic resistance genes across bacterial genera. Environ Int 129:478–487. 10.1016/j.envint.2019.05.054 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fang J, Davood Z, Han J, Qu D (2021) Fitness cost and compensation mechanism of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli. Environ Microbiol 23:7538–7549. 10.1111/1462-2920.15783 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and in its Supplementary information.