Abstract
Fibroblasts play critical roles in tissue homeostasis, but in pathologic states they can drive fibrosis, inflammation, and tissue destruction. Little is known about what regulates the homeostatic functions of fibroblasts. Here, we perform RNA sequencing and identify a gene expression program in healthy synovial fibroblasts characterized by enhanced fatty acid metabolism and lipid transport. We identify cortisol as the key driver of the healthy fibroblast phenotype and that depletion of adipocytes, which express high levels of Hsd11b1, results in loss of the healthy fibroblast phenotype in mouse synovium. Additionally, fibroblast-specific glucocorticoid receptor Nr3c1 deletion in vivo leads to worsened arthritis. Cortisol signaling in fibroblasts mitigates matrix remodeling induced by TNF and TGF-β1 in vitro, while stimulation with these cytokines represses cortisol signaling and adipogenesis. Together, these findings demonstrate the importance of adipocytes and cortisol signaling in driving the healthy synovial fibroblast state that is lost in disease.
Subject terms: Mechanisms of disease, Fat metabolism, Inflammation
Fibroblasts play critical roles in tissue homeostasis, but in pathologic states they can drive fibrosis, inflammation, and tissue destruction. Here, Faust et al. find that healthy human synovial fibroblasts under the influence of adjacent adipocytes have altered lipid metabolism driven by cortisol signaling. Both adipocytes and these characteristics are lost in inflammatory arthritis.
Introduction
In rheumatoid arthritis (RA), inflammation drives synovial fibroblasts to proliferate and mediate inflammation and cartilage and bone destruction in the joint1,2. In healthy people, the synovium is composed of lining and sublining layers largely composed of fibroblasts3. The healthy synovium often displays marked adiposity within the sublining, especially in villus areas, intermixed with adipocytes3. Knee joint synovium is also immediately adjacent to and interconnected with the infrapatellar fat pad. Adipocytes are similarly intermixed with fibroblasts in adipose tissues. During osteoarthritis (OA) and RA, the normally abundant synovial adipocytes largely disappear and are replaced by inflammatory and fibrotic tissue4,5.
These findings suggest a high level of communication between the adipocytes and fibroblasts in healthy conditions, but little is known about whether adipocytes influence fibroblasts and what role they play in fibroblast function6.
Adipose depots contain a specific cellular phenotypic landscape. For example, adipose stromal cells secrete IL-33 that supports T-regulatory cell survival and contributes to an anti-inflammatory homeostatic state7. Given the adipose-like state of the healthy synovium, we asked if adipocytes might drive a distinct phenotype of fibroblasts in the healthy synovium and how that might change when adipocytes are lost in pathologic states.
Here, we performed single-cell RNA sequencing on healthy and diseased synovium and defined transcriptomic differences between healthy and inflamed synovial fibroblasts. Among the changes detected was a program characterized by genes involved in fatty acid metabolism, lipid transport, and metal ion homeostasis. Remarkably, adipocyte-conditioned media was able to induce these healthy fibroblast programs in cultured RA synovial fibroblasts. Fractionation of adipose-derived lipids and mass spectrometry analysis identified cortisol and glucocorticoid signaling as the primary pathway required for maintaining the healthy synovial fibroblast program. These same programs were evident in general in fibroblasts from classical visceral and subcutaneous adipose compartments, suggesting that the adipose environment is a key driver of healthy fibroblast physiology which is lost in pathologic states.
Results
Single-cell RNA sequencing reveals distinct healthy synovial cell populations
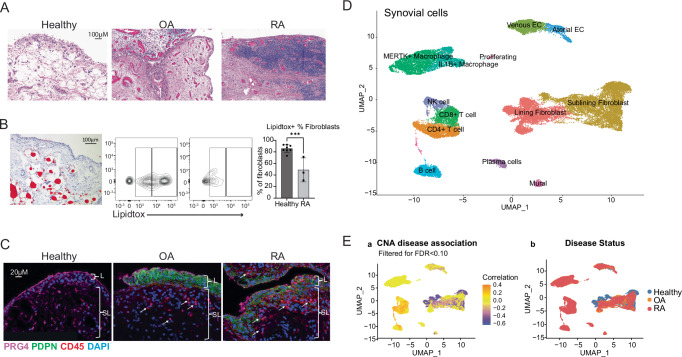
We collected healthy synovial samples from knees of ten subjects undergoing autopsy who lacked a diagnosis of arthritis, history of autoimmune disease, or recorded traumatic injury to the knee (Supplementary Table 1, cohort 1). We stained healthy synovium with hematoxylin and eosin, demonstrating that the synovial lining layer is two to three cells thick and is adjacent to a sublining region composed of loose matrix with significant adiposity. In contrast, synovium from OA and RA subjects exhibit immune cell infiltration, fibroblast hyperplasia, and reduced adiposity (Fig. 1A and Supplementary Fig. 1a). Oil red O staining confirmed the presence of lipid-filled adipocytes ~50–80 µm in diameter in healthy synovium (Fig. 1B and Supplementary Fig. 1b, c for gating strategy). We quantified total neutral lipids in CD45- stromal synovial cells by flow cytometry with LipidTOX staining (Fig. 1B, right). Lipid content was highest in fibroblasts from healthy subjects (85%) versus RA samples (48%), suggesting that RA fibroblasts reside in a relatively lipid-depleted environment. Indeed, we counted adipocytes per mm2 and found that healthy synovium contains a higher number of adipocytes compared to RA samples (20 vs 4.66, respectively, Supplementary Fig. 1d).
Fig. 1. Single-cell RNA sequencing reveals distinct healthy and diseased cell populations in synovial tissue.
A H&E staining of synovial tissue sections highlighting broad histological differences among healthy, OA, and RA tissue, healthy n = 8, OA n = 2, RA n = 3 biological replicates, P = 0.001. Source images are in Supplemental Fig. 1a. B Left: Oil Red O staining of healthy synovium. Right: LipidTox neutral lipid staining of healthy and RA fibroblasts (live, CD45-, CD31- CD146- cells). CD36 is on the y axis. Healthy: n = 8, RA: n = 3 biological replicates, one independent experiment. P values were calculated using a two-tailed Student’s T test. C Immunofluorescence staining for PRG4 (purple), CD45 (red), PDPN (green), and DAPI (blue) in healthy, OA, and RA synovium. n = 3, biological replicates, one independent experiment. D Synovial cells from healthy, OA, and naive RA donors were harmonized and clustered into a single UMAP projection. E UMAP projection from (a) colored by correlation with rheumatoid arthritis (orange) or health (purple) using covarying neighborhood analysis (CNA). All data are presented as mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
Next, we performed immunofluorescence staining on healthy, OA, and RA samples, which revealed a smaller number of CD45+ cells in healthy synovium compared to RA samples (Fig. 1C and Supplementary Fig. 1e). Staining for the synovial lining marker lubricin (PRG4) showed that healthy synovium has a strong and well-defined border of PRG4 staining in the lining, which appears more irregular in OA and RA lining. The stromal cell marker podoplanin (PDPN), which is increased by TNF exposure, is low to absent in healthy tissue, whereas OA and RA tissue have marked staining (Fig. 1C and Supplementary Fig. 1e).
By applying single-cell RNA sequencing to 10 healthy synovial samples, we obtained data on 8687 cells that passed quality control, including 7607 fibroblasts, 893 endothelial cells and pericytes, and 187 immune cells. We used the Harmony algorithm to integrate data from these 10 healthy synovial samples with 9 OA and 28 treatment-naive RA synovial samples obtained through the Accelerating Medicines Partnership: RA/SLE consortium8. Graph-based clustering resulted in 13 cell states: lining and sublining fibroblasts, CD4 and CD8 T cells, NK cells, B cells and plasma cells, IL1B+ and MERTK+ macrophages, mural cells, venous and arterial endothelial cells, and proliferating T and B cells (Fig. 1D and Supplementary Fig. 1f). Healthy synovium contained a mean of 83% fibroblasts, 1% T cells, 10.5% ECs, 0.2% B cells, and 1.6% macrophages. The proportion of T cells differed substantially among OA (8.7%), RA (30.2%) and healthy samples (1%). Similarly, B cells were 4.8%, 6.7%, and 0.2% in OA, RA and healthy samples, respectively, and macrophages were 26%, 24%, and 1.6%, respectively (Supplementary Fig. 1g, h for UMAP split by disease state). We implemented covarying neighborhood analysis (CNA) to identify groups of cells that covary in abundance between healthy and RA synovium9. Of note, T and B cells are distinctly associated with arthritis, and specific fibroblast sub-populations are associated with healthy synovium (global P value = 0.001) (Fig. 1E).
Fine-grained analysis defines synovial fibroblast ground states
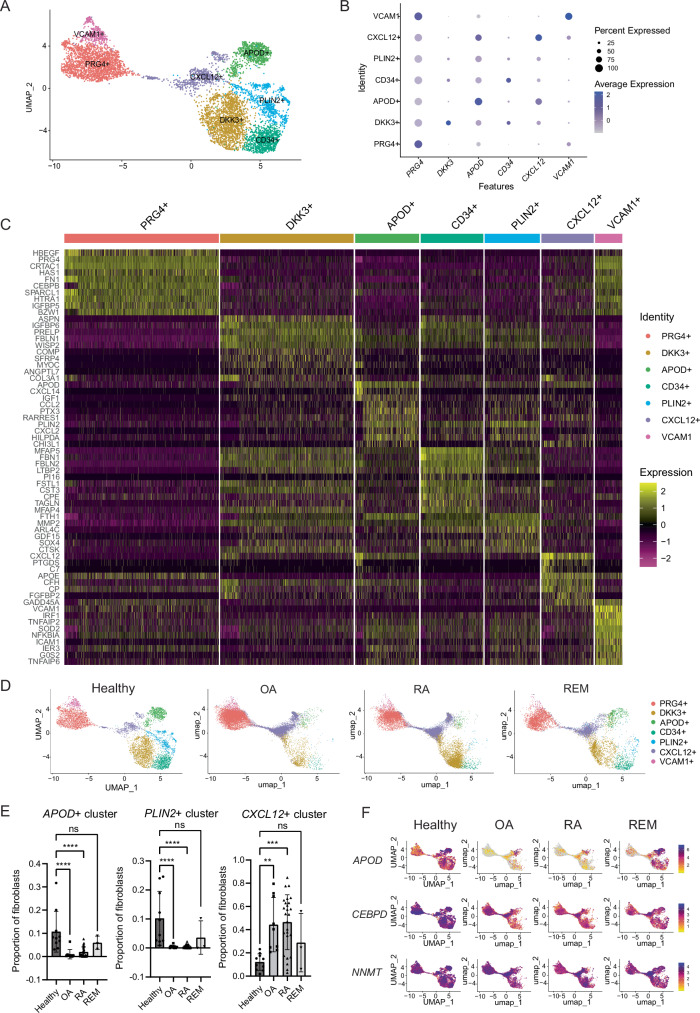
We performed fine-grained clustering of healthy fibroblasts to identify fibroblast states in homeostasis. Seven clusters were defined including: PRG4 + , PLIN2 + , DKK3 + , CXCL12 + , CD34 + , APOD + , and VCAM1 + , each named by a characteristic highly expressed gene (Fig. 2A–C). PRG4+ and VCAM1+ clusters included fibroblasts enriched for lining markers, including PRG4, HBEGF, CRTAC1, FN1, HAS1, HTRA1, TIMP1, CLU, and IGFBP5 (Supplementary Fig. 2a). The VCAM1+ cluster contained fibroblasts that also express genes downstream of TNF signaling, including VCAM1, TNFAIP2, TNFAIP6, and NFKBIA. DKK3+ fibroblasts (expressing IGBP6, ASPN, COMP), CXCL12+ fibroblasts (expressing SFRP1, CHI3L1, CHI3L2), and CD34+ fibroblasts (expressing MFAP5, FBLN2, FBN1, APOE) all appear to be similar to previously defined synovial sublining fibroblast clusters, as suggested by the ratio of odds of mapping an RA or OA fibroblast cluster to a defined healthy fibroblast cluster (Supplementary Fig. 2b8). The APOD + (apolipoprotein D) cluster is enriched for genes that modulate growth factors and insulin signaling (IGF1, IGFBP3, IGFBP7), an adipokine gene (RARRES2), lipid transport gene (APOD) and monocyte attraction through CXCL14. Many highly expressed genes are secreted factors. The APOD+ cluster is also enriched for metallothioneins (MT1X, MT1A, MT1M, MT1E, MT1G), which are cysteine-rich proteins that are involved in homeostatic regulation of the storage and transport of metals, and protection against oxidative stress10. The PLIN2 + (perilipin 2) cluster expresses genes involved in regulating lipid homeostasis and metabolism (PLIN2, HILPDA, ADM), as well as matrix degradation (MMP2, CTSK).
Fig. 2. Synovial fibroblast signatures during homeostasis.
A Fine clustering analysis on healthy synovial fibroblasts defines seven distinct clusters. B Top markers of each fibroblast cluster. C Heatmap of the top 10 DEGs per cluster. D Symphony mapping of OA, naive RA, and remission (REM) fibroblasts to healthy synovial fibroblast reference. E Quantification of fibroblast proportions mapping to each cluster. Each data point represents a patient sample. Statistical comparisons: all groups were compared to healthy. Healthy: n = 10, OA: n = 9, RA: n = 26, REM: n = 3 biological samples. P values were calculated using an ordinary one-way ANOVA followed by Dunnet’s multiple comparison post hoc testing. F Healthy synovium is enriched in APOD, CEBPD, and NNMT expression compared to OA and RA fibroblasts, and is partially restored in remission fibroblasts. All data are presented as mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
After identification of these healthy fibroblast clusters, we used the Symphony algorithm to map fibroblasts from OA, treatment-naive RA, and RA remission samples11 onto the healthy donor dataset (Fig. 2D). Strikingly, both RA and OA samples were significantly depleted in the blue colored PLIN2+ cluster and the emerald green APOD+ cluster, and significantly over-mapped to the purple CXCL12+ cluster, suggesting that PLIN2+ and APOD+ fibroblasts are lost or significantly changed in disease, while CXCL12+ fibroblasts are expanded (Fig. 2E and Supplementary Fig. 2c).
Further, differential gene expression analysis of all sublining fibroblasts revealed that healthy fibroblasts globally upregulate 540 and 571 genes and downregulate 953 and 882 genes compared to RA and OA fibroblasts, respectively. Many genes upregulated in healthy fibroblasts were involved in metabolism, including apolipoproteins (APOD), lipid droplet associated proteins (PLIN2), progesterone-induced genes (DEPP1), metallothioneins (MT1X, MT1E, MT2A), hormones (ADM), transcription factors (CEBPD, ZBTB16), and methyltransferases (NNMT) (Fig. 2F, Supplementary Fig. 2d, and Supplementary Data 1 and 2). Notably, APOD, CEBPD, and NNMT are highly expressed in healthy synovial fibroblasts, but they are reduced in OA and RA, and partially restored in remission (Fig. 2F).
Healthy fibroblast signature can be induced by fat-conditioned media
Healthy fibroblasts express a gene signature enriched in metabolic pathways compared to OA and RA fibroblasts. Genes upregulated in this signature include APOD, PLIN2, DEPP1, ADH1B, MT1X, CEBPD, and NNMT (Supplementary Data 1 and 2). Of these genes, we selected three of the most strongly upregulated genes compared to RA fibroblasts to represent the healthy fibroblast gene signature: APOD (Apolipoprotein D), NNMT (Nicotinamide N-methyltransferase), and CEBPD (CCAAT/enhancer binding protein D). APOD is an apolipoprotein which is present in HDL and involved in cholesterol homeostasis12; NNMT is expressed in adipose tissue and methylates nicotinamide, thereby determining the availability of methyl groups for regulating gene expression and metabolism13,14; and C/EBPD is an early pre-adipocyte transcription factor15.
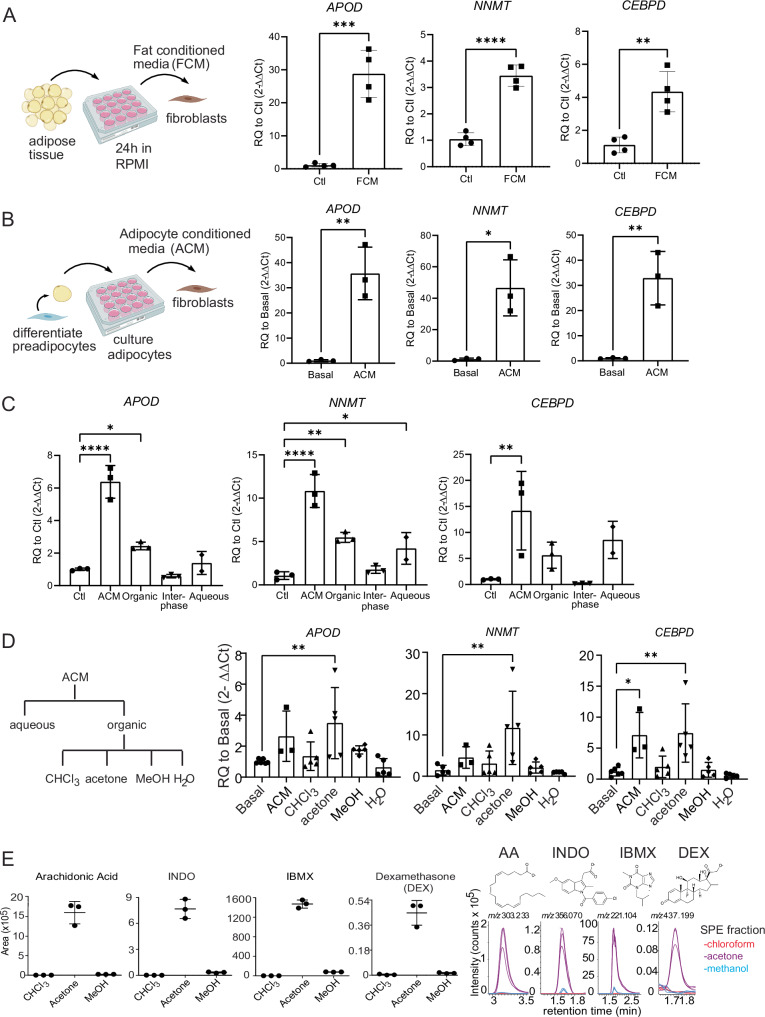
Due to their lipid-centric functions, we tested if the coculture of RA synovial fibroblasts with oleate and palmitate, two of the most abundant fatty acids, might regenerate the lipid-related healthy fibroblast phenotype16. Oleate and palmitate failed to upregulate APOD, but they modestly upregulated NNMT and CEBPD (Supplementary Fig. 3a), suggesting that fatty acids are not likely to be the primary inducers of the healthy fibroblast phenotype. To more broadly test substances released by adipose tissue, we generated abdominal or synovial fat-conditioned media by incubating media with adipose tissue for 24 h. Synovial fat-conditioned media induced robust expression of APOD, NNMT and CEBPD, with APOD expressed 30-fold, NNMT 3.5-fold, and CEBPD 4-fold over basal levels (Fig. 3A). Abdominal fat-conditioned media was also able to upregulate these genes to similar levels (Supplementary Fig. 3b). Next, we pursued isolating the active molecule within adipose tissue that is responsible for the induction of these genes.
Fig. 3. Search for active molecule driving healthy fibroblast cell state.
A Left: Schematic illustrating generation and application of fat-conditioned media (FCM). Created with Biorender. Right: FCM induces healthy fibroblast signature. n = 4 technical replicates, representative of two independent experiments. B Left: Schematic illustrating generation and application of adipocyte-conditioned media (ACM). Created with Biorender. Right: ACM induces the healthy fibroblast gene signature. n3 technical replicates, representative of two independent experiments. C Bligh and dyer separation of adipocyte-conditioned media (ACM). n = 3 technical replicates, representative of two independent experiments. D Left: Schematic detailing fractionation approach. ACM was separated using the Bligh and Dyer method into aqueous and organic phases. Then, the organic phase was taken for solid phase separation and eluted based on polarity using chloroform, acetone, methanol, and water. Right: Testing activity of each fraction. C, D Statistical comparisons: all groups were compared to basal/ctl. Basal: n = 6, ACM: n = 3, CHCL3: n = 5, Acetone: n = 5, Methanol: n = 5, H2O: n = 5 technical replicates, representative of two independent experiments. E MS intensity values were shown as the ion chromatogram areas extracted at m/z 303.233 ([M–H]-) for arachidonic acid (AA), m/z 356.070 ([M–H]-) for indomethacin (INDO), m/z 221.104 ([M–H]-) for isobutylmethylxanthine (IBMX), and m/z 437.198 ([M + COO]-) for dexamethasone in all three acetone fractions. n = 3 technical replicates of three SPE column purifications from one ACM, one independent experiment. All data are presented as mean ± standard deviation. A, B P values were calculated using a two-tailed Student’s T test; C, D were calculated using an ordinary one-way ANOVA followed by Dunnet’s multiple comparison post hoc testing. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file. A, B Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license (https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en).
Identification of active molecules using fractionation and lipidomics
Since fat-conditioned media might contain factors from any adipose tissue cell or component, we investigated the effects of conditioned media generated from cultured adipocytes on synovial fibroblasts. We cultured pre-adipocytes in adipocyte differentiation media for 10 days and applied that media to fibroblasts. This strongly induced fibroblast expression of APOD (35-fold), NNMT (45-fold), and CEBPD (35-fold) expression (Fig. 3B). This result, together with those using fat-conditioned media (Fig. 3A), suggested that adipocytes were the likely source of the active factor.
Next, we carried out a Bligh and Dyer extraction of the conditioned media which separated lipid and non-lipid molecules into organic and aqueous phases, respectively. Bioactivity was found in both phases (Fig. 3C). However, upon a second round of extraction from the aqueous phase, most activity extracted into the organic fraction (Supplementary Fig. 3c). We further separated the organic extracts by solid phase extraction (SPE) to elute lipids based on their polarity using chloroform (most apolar), acetone, methanol, and water (most polar). Bioactivity was contained in the acetone fraction (Fig. 3D). We analyzed the lipid contents of acetone fractions by reverse phase high-performance liquid chromatography (HPLC) coupled with Electrospray Ionization (ESI)-Quadruple-Time-of-Flight (Q-Tof) mass spectrometry (MS). We used the unbiased software-based (XCMS) lipidomic method (17) to identify target ions with at least tenfold higher intensity and corrected P value < 0.05 in the stimulatory acetone fraction compared to the non-stimulatory chloroform fraction (Supplementary Fig. 3d left). There were 779 ions that met these criteria. Among them, we focused on the ions with highest fold change and intensity (Supplementary Fig. 3d, right). By plotting m/z value versus retention time (Supplementary Fig. 3d, right), three clusters were formed based on their elution pattern and mass profile. Eight ions were identified as fatty acids based on matching their mass to known compounds.
Two other ion groups with similar retention times (1.6–1.8 min) represented alternate ion adducts and multimers of two underlying unknown structures of known mass (Supplementary Fig. 3d, green and pink, XCMS raw data output in Supplementary Data 3). The low mass and very high intensity of these ions prompted us to consider three chemical additives, indomethacin (INDO), isobutylmethylxanthine (IBMX), and dexamethasone (DEX), which are supplemented in the adipocyte differentiation media used to culture the pre-adipocytes. Indeed, the molecule present as 4 ions at 1.6 min, whose deprotonated ion [M–H]− at m/z 356.071 matched the expected mass of INDO. Next, the molecule present as 7 ions at 1.8 min was solved as isobutylmethylxanthine (IBMX). However, the 3rd chemical additive, dexamethasone, was not extracted by automated peak-picking software, which might be due to its low intensity. We therefore manually searched the ion chromatogram, which yielded a chromatogram peak that matched the expected DEX ([M + COO]− m/z 437.198). DEX was highly enriched in the acetone fraction compared to the chloroform or methanol fractions (Fig. 3E). Manual inspection of HPLC-MS chromatograms for arachidonic acid ([M–H]− m/z 303.233), INDO ([M–H]− m/z 356.070), and IBMX ([M–H]− m/z 221.104) confirmed the unbiased lipidomic results (Fig. 3E, right).
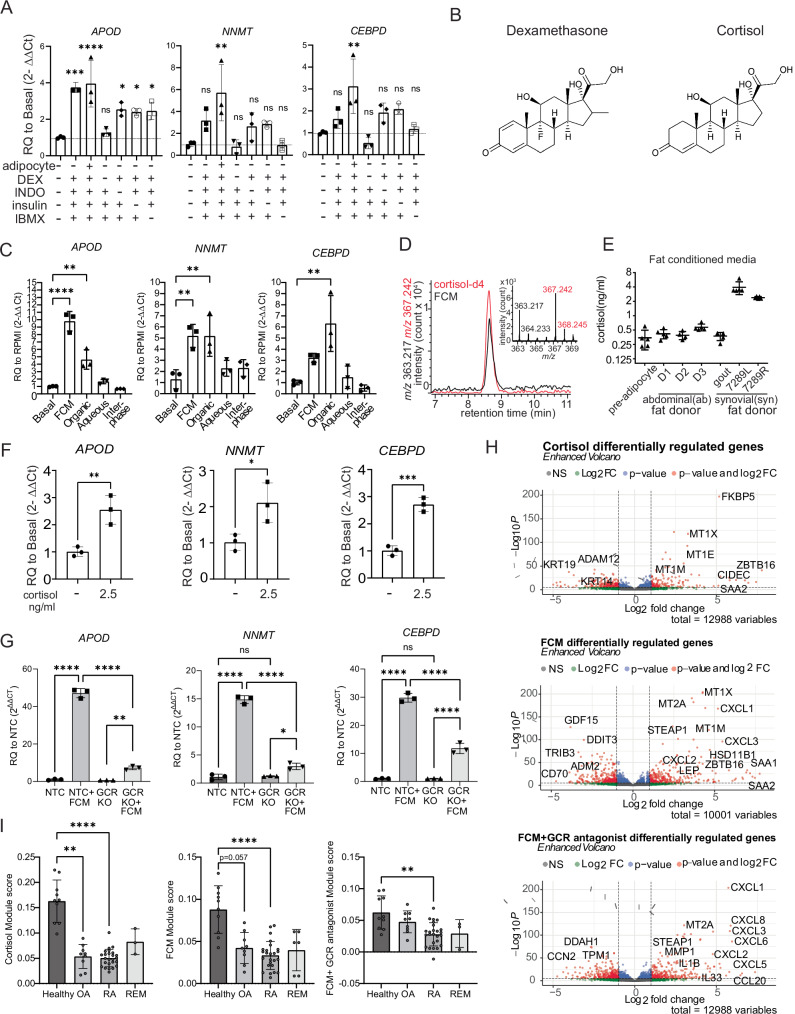
Since the mass spectrometry analysis implicated that activity co-purified with several of the adipocyte differentiation media additives, we tested the media directly without one factor at a time. This revealed that dexamethasone, a glucocorticoid, accounted for the dominant activity (Fig. 4A). IBMX had the second largest impact on activity, which may be due to its role in raising intracellular cAMP levels which sensitizes cells to glucocorticoid signaling18.
Fig. 4. Glucocorticoid signaling is sufficient and necessary for healthy fibroblast phenotype.
A ACM media was tested in the presence or absence of adipocytes and the most enriched species: dexamethasone (DEX), indomethacin (INDO), insulin, or 3-isobutyl-1-methylxanthine (IBMX). n = 3 technical replicates, representative of one independent experiment. B Dexamethasone is a synthetic derivative of the naturally occurring glucocorticoid cortisol. Chemical structures are shown. C Bligh and dyer separation of fat-conditioned media (FCM), the active molecule is in organic and aqueous fractions of FCM. n = 3 technical replicates, representative of two independent experiments. D FCM contains cortisol, as measured by the positive mode mass spectrometry using cordisol-d4 as the internal standard, n = 7. E Approximate cortisol concentration in different FCM donors based on quantification of mass spectrometry data. Pre-adipocyte, D1, D3, 7289L, 7289R n = 5, D2, gout n = 4, technical replicates based on repeated measures of the same sample with different concentrations of deuterated standard added. F Cortisol, at biologically meaningful concentrations, is sufficient to induce healthy gene signature. n = 3, technical replicates, representative of two independent experiments. P values were calculated using a two-tailed Student’s T test. G CRISPR–cas9 knockdown of NR3C1, the gene encoding the glucocorticoid receptor (GCR), resulted in a dramatic reduction of fibroblasts to induce APOD, NNMT, and CEBPD expression in response to FCM. Non-targeting control (NTC) was used as a control. Two-way ANOVA was performed to calculate statistical significance. n = 3, technical replicates, representative of two independent experiments. H Volcano plots of genes up and downregulated by cortisol, FCM, and FCM+ the GCR antagonist mifepristone found by unbiased bulk RNA sequencing. Differentially expressed genes were calculated in DeSeq2, which calculates a P value using the Wald test. I Application of module scores to single-cell pseudobulk data (reads collapsed over patient). A Kruskal–Wallis test with Dunn’s multiple comparison post hoc test was used to calculate significance in (I). REM remission. Healthy: n = 10, OA: n = 9, RA: n = 25, REM: n = 3 biological replicates. P values for (A, C) were calculated using an ordinary one-way ANOVA followed by Dunnet’s multiple comparisons post hoc test. All data are presented as mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
Since dexamethasone is a synthetic steroid, this finding led us to consider if a natural glucocorticoid might be responsible for the activity of the fat-conditioned media which did not contain the supplements (Fig. 4B). To address this, we similarly carried out the Bligh and Dyer fractionation method on fat-conditioned media, and we observed activity in the organic phase, where we expect cortisol, a nonpolar steroid, to localize (Fig. 4C). We further separated the organic extracts by the solid phase extraction (SPE) method to elute lipids based on their polarity. Testing each fraction revealed that activity was primarily contained in the acetone fraction (Supplementary Fig. 4a). Next, we developed a one-step HPLC-MS method to directly measure the cortisol concentration by mixing fat-conditioned media with known concentrations of deuterated cortisol (cortisol-d4) in methanol. We found that cortisol is present in synovial and abdominal-derived fat-conditioned media at concentrations ranging from 0.5-4 ng/mL depending on the donor (Fig. 4D, E). Thus, we tested if cortisol alone is sufficient to induce APOD, NNMT, and CEBPD gene expression at 2.5 ng/mL, its physiological levels in fat-conditioned media. Indeed, like the synthetic additive dexamethasone, we found that physiologic concentrations of the endogenous glucocorticoid cortisol drive the expression of APOD, NNMT, and CEBPD (Fig. 4F).
Fat-conditioned media activity is dependent on glucocorticoid signaling
Given this cortisol activity, we next tested if glucocorticoid signaling or biologically unrelated pathways controlled the fat-conditioned media-induced gene response by using a glucocorticoid receptor antagonist (mifepristone). Mifepristone completely blocked induction of APOD and CEBPD by fat-conditioned media (Supplementary Fig. 4b). Using a second approach we deleted the glucocorticoid receptor by applying CRISPR guide RNA directed at NR3C1, which reduced NR3C1 gene expression in fibroblasts by ~83% (Supplementary Fig. 5a). When we applied cortisol to NR3C1 deleted cells, the cells did not upregulate APOD, NNMT, and CEBPD (Supplementary Fig. 5b). We then applied fat-conditioned media and found that NR3C1 deletion profoundly reduced the response to fat-conditioned media, evidenced by markedly decreased upregulation of APOD (from 47-fold to 7-fold), NNMT (from 15-fold to 3-fold), and CEBPD (from 30-fold to 11-fold), confirming that most of the activity from fat-conditioned media occurs via glucocorticoid signaling (Fig. 4G).
To further characterize the role of cortisol, we tested the activity of other related steroids and their pathways. Although progesterone can bind to the glucocorticoid receptor, we found that progesterone did not upregulate APOD, NNMT, or CEBPD (Supplementary Fig. 6a). Aldosterone is a strong mineralocorticoid receptor agonist and can also signal through the glucocorticoid receptor NR3C1, and it induced expression of APOD, NNMT, and CEBPD at 1uM (Supplementary Fig. 6a). Thus, we measured aldosterone levels in fat-conditioned media by ELISA. Unlike cortisol, which was present at the ng/mL level, aldosterone was present at less than 20 pg/mL (Supplementary Fig. 6b) and adding 10-50 pg/mL lacked activity (Supplementary Fig. 6b), suggesting that aldosterone is not the active molecule signaling through NR3C1. Finally, we tested if fat-conditioned media enhances the conversion of inactive cortisone to cortisol via the enzyme 11-β-HSD1. We added metyrapone, a competitive 11-β-HSD1 inhibitor, to fat-conditioned media, but it did not impact the phenotype (Supplementary Fig. 6c), suggesting that activation of cortisol is not induced by another factor within the fat-conditioned media.
RNA sequencing confirms glucocorticoid signaling regulates fibroblast gene expression
We performed bulk RNA sequencing on fibroblasts treated with synovial-derived fat-conditioned media, agonists and inhibitors of glucocorticoids and cytokines for 4 or 22 h. Principal components analysis revealed that the larger variance in gene expression is at 22 h (Supplementary Fig. 7a). Focusing on the gene changes at 22 h, we confirmed that APOD, NNMT, and CEBPD were upregulated by fat-conditioned media while mifepristone downregulated their expression (Supplementary Fig. 7b). Differential gene expression analysis of fibroblasts stimulated with either cortisol, fat-conditioned media, or fat-conditioned media plus glucocorticoid receptor (GCR) antagonist for 22 h was used to create gene lists defining activation scores (see Supplementary Data 4 for differential expression data and gene lists), which were then applied to the fibroblasts from the single-cell RNA sequencing dataset. Healthy fibroblasts had higher fat-conditioned media and cortisol activation scores compared to fibroblasts from OA, RA, and remission subjects. However, mifepristone greatly reduced the fat-conditioned media activation score in healthy fibroblasts, indicating that glucocorticoid signaling is a global driver of healthy fibroblast gene expression (Fig. 4H, I).
Synovial adipose tissue is a source of glucocorticoids
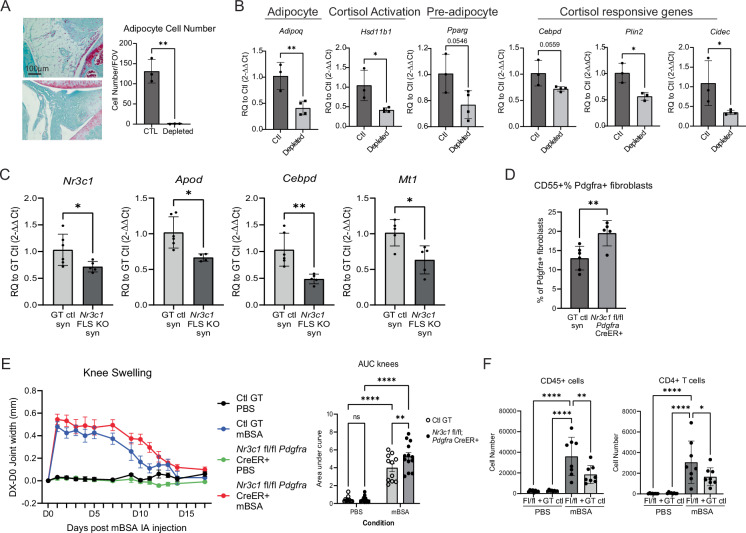
Cortisol mostly circulates through the body in its inactive form, cortisone. Cortisone is primarily converted to active cortisol in tissues via the enzyme 11-β-HSD1. Adipose tissue has a high level of 11-β-HSD1 activity compared to many other tissues, and we confirmed that adipocytes have higher HSD11B1 expression compared to synovial fibroblasts in vitro (Supplementary Fig. 7c). Thus, we created a mouse with inducible adipocyte depletion by crossing C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (iDTR) mice to B6;FVB-Tg(Adipoq-cre)1Evdr/J mice. We injected diphtheria toxin intra-articularly to locally deplete joint adipose tissue over the course of 8 weeks. The quantity of intra-articular adipose tissue was significantly depleted compared to iDTR-; Adipoq-cre- control mice injected with diphtheria toxin, as assessed histologically (~130 adipocytes per FOV vs 0) and by gene expression of Adipoq (Fig. 5A, images for quantification are in Supplementary Fig. 7d).
Fig. 5. Glucocorticoid signaling is important for fibroblast homeostasis and arthritis severity.
A Adipocyte depletion over 8wks in knee joints of AdipoQ cre+ iDTR+ mice. n = 3 biological replicates, representative of one independent experiment. B Whole joint qPCR following adipocyte depletion. Ctl= AdipoQ cre- iDTR-, Depleted= AdipoQ cre+ iDTR + , in both cases the joint taken for qPCR was injected intra-articularly with diphtheria toxin over a time course of 8 weeks. Ctl n = 3, depleted n = 4 biological replicates, representative of one independent experiment. C Naive synovium in Pdgfra-CreER;Nr3c1fl/fl mice show decreased total Nr3c1 transcripts by qPCR as well as glucocorticoid-responsive genes compared to genotype controls. GT ctl n = 6, Pdgfra-CreER;Nr3c1fl/fl n = 5 biological replicates, representative of one independent experiment. D Flow cytometry of fibroblasts from naive synovium. GT ctl n = 6, Pdgfra-CreER;Nr3c1fl/fl n = 5 biological replicates, representative of two independent experiments. E Knee joint measurements of AIA Pdgfra-CreER;Nr3c1fl/fl mice. GT ctl n = 11, Pdgfra-CreER;Nr3c1fl/fl n = 13 biological replicates, representative of two independent experiments. GT ctl, PBS in black; GT ctl, mBSA in blue; Pdgfra-CreER;Nr3c1fl/fl PBS in green; Pdgfra-CreER;Nr3c1fl/fl mBSA in red. F Flow cytometry of synovium harvested day 11 post intra-articular mBSA injection. GT ctl n = 8, Pdgfra-CreER;Nr3c1fl/fl n = 8 biological replicates, representative of two independent experiments. A–D P values were calculated using a two-tailed Student’s T test, E, F were calculated using a two-way ANOVA. All data are presented as mean ± standard deviation except (E), which is ±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
Concomitant with adipocyte depletion, we observed a reduction in the expression of Hsd11b1 (>50%), indicating that joint adipocytes contribute significantly to the generation of active cortisol via the expression of this enzyme. In addition, cortisol-responsive genes Cebpd, Plin2, and Cidec, and the pre-adipocyte marker Pparg were all downregulated in adipocyte-depleted joint tissues (Fig. 5B). These results are consistent with the conclusion that adipocytes are a key source activating cortisol in synovium. In addition, two markers of fibroblast activation, podoplanin (PDPN) and cadherin 11 (CDH11), were upregulated at the protein level in synovial fibroblasts of adipocyte-depleted synovium, suggesting that adipocytes are important for maintaining fibroblast homeostasis and their absence results in fibroblast activation (Supplementary Fig. 8a, b). Despite this, adipocyte depletion was unable to protect mice from the K/BxN serum transfer model of inflammatory arthritis, likely due to the many diverse functions of adipocytes (Supplementary Fig. 8c, d)19–21.
Glucocorticoid signaling is important for fibroblast homeostasis
We created a mouse with fibroblast-specific Nr3c1 deletion (Pdgfra-CreER; Nr3c1fl/fl) by crossing B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J and B6.Cg-Nr3c1tm1.1Jda/J mice and administered tamoxifen to induce CreER recombination. In naive mice, we observed decreased Nr3c1 expression in depleted joints, even at the whole tissue level, likely due to the high proportion of fibroblasts in naive synovium (Fig. 5C). We correspondingly observed decreased expression of cortisol-responsive genes, including Apod, Cebpd, and Mt1. We also quantified CD55+ populations by flow cytometry, as we have found this marker to be associated with universal progenitor fibroblasts8,22. We observed an increase in the proportion of CD55+ fibroblasts in Pdgfra-CreER; Nr3c1fl/fl joints, suggesting that NR3C1 signaling may maintain a pre-adipocyte-like state that is lost in its absence (Fig. 5D). We next tested whether Pdgfra-CreER; Nr3c1fl/fl mice were more susceptible to inflammatory arthritis. Indeed, knockout mice had worsened antigen-induced arthritis (AIA) as measured by knee swelling and immune cell infiltration, particularly of CD4 + T cells (Fig. 5E, F).
Examination of exogenous human steroid usage on fibroblast phenotype
Thus far, the data collected on healthy, RA, OA, and remission human synovial tissues were all from donors with no reported steroid use. To further our findings, we collected and ran single-cell RNA sequencing on six additional healthy synovial tissues from patients with reported steroid use prior to autopsy sample collection (Supplementary Table 1, cohort 2). Five of six of these patients received a short course of steroids during their stay at the hospital, shortly before tissue collection. Altogether, we sequenced a total of 19,378 synovial cells from 16 healthy donors. When steroid-exposed donors were analyzed and compared to non-steroid-exposed donors, broad cell types, fibroblast cluster identities, top differentially expressed genes relative to OA and RA fibroblasts, and cortisol activation scores were similar (Supplementary Fig. 9a–i), suggesting that healthy fibroblasts are already steroid activated by their adipose-rich microenvironment and that donor steroid usage does not further enhance cortisol activation. Additionally, analysis of two separate mouse RNA sequencing datasets comparing healthy versus serum transfer arthritis or hTNFtg arthritic synovial tissue revealed that healthy mouse synovium has a higher cortisol activation score compared to arthritic synovium, independently supporting our findings in humans (Supplementary Fig. 10a, b)23,24.
The additional samples also allowed a more fine-grained analysis of healthy synovial immune cell populations. We mapped healthy synovial immune cells onto the RA and OA cell states defined by single-cell RNA sequencing from Zhang et al. using the Symphony algorithm (Supplementary Data 5)8. Most healthy macrophages mapped to either M0 (MERTK + SELENOP + LYVE1 + ), M1 (MERTK + SELENOP + LYVE1-), M2 (MERTK + S100A8 + ), M4 (SPP1 + ), M5 (C1QA + ), or M7 (IL1B + FCN1 + ) clusters. Healthy T cells mapped primarily to CD4 + IL7R + CCR5 + , CD4 + IL7R + , CD4 + CD161+ memory, or to CD8 + GZMB + TEMRA T cell clusters. These mapped cell states suggest that tissue-resident macrophages and T cells are present in healthy synovium. Many inflammatory immune cells defined by Zhang et. al., such as STAT1 + CXCL10+ macrophages and CD4+ Tfh/Tph cells, were not present in healthy synovium8. In addition, less than ten cells mapped to any given B cell cluster, suggesting this is not a typical cell observed in synovium at homeostasis.
Healthy synovial fibroblasts share similarities with adipose tissue fibroblasts
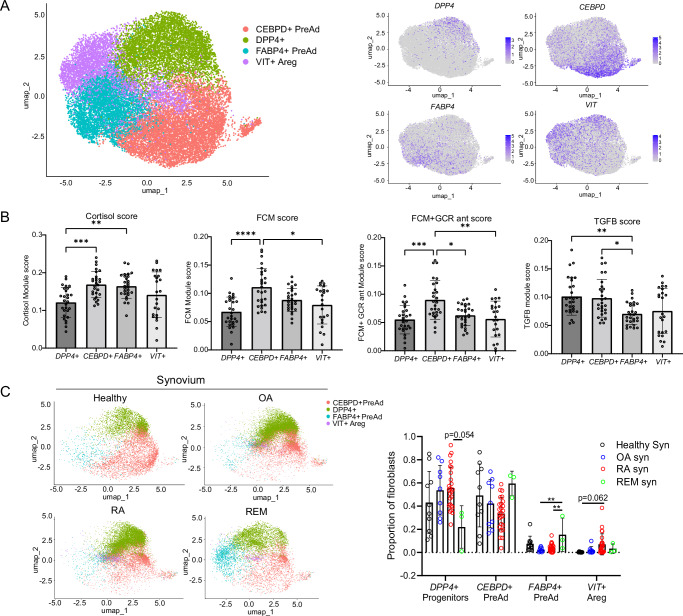
Next, we asked if adipocytes control fibroblast phenotype in classical adipose depots by comparing healthy synovial tissue and adipose tissue-derived fibroblasts. We performed single-cell RNA sequencing on classical visceral (VAT) and subcutaneous (SAT) adipose tissue depots from five obese but metabolically healthy donors (Supplementary Table 2). We used the Harmony algorithm to integrate these fibroblasts with PDGFRA+ non-mesothelial cells from two other adipose single-cell RNA sequencing datasets25,26. Altogether, we analyzed 21,856 PDGFRA+ cells from 23 donors. These adipose stromal cells revealed four main clusters: one marked by DPP4+ expression and high expression of “universal progenitor” markers (CD55, PI16, and CD34) and shown to reside in interstitial regions of murine adipose tissue22,27 (Fig. 6A). Two of the other clusters represented committed adipocyte progenitor populations, marked by high expression of pre-adipocyte transcription factors and high cortisol scoring (Fig. 6B). These pre-adipocyte populations expressed markers consistent with different levels of pre-adipocyte commitment: CEBPD+ early pre-adipocytes and the late pre-adipocyte marker FABP4 + (Supplementary Fig. 11a and Supplementary Data 6a–d)28.
Fig. 6. The synovium shares properties with adipose stromal cell populations.
A UMAP of PDGFRα+ stromal cells from human visceral and subcutaneous depots (VAT and SAT). B Bulk RNA sequencing defined cortisol, FCM, FCM + GCR ant, and TGF-β1 scores applied to adipose clusters. Fibroblasts from each donor were extracted and reads collapsed over donor and cluster to perform a pseudobulk analysis, meaning each data point represents the activation score of all of a donor’s fibroblasts, separated by cluster. A Kruskal–Wallis test with Dunn’s multiple comparison post hoc test was used to calculate significance. n = 28 biological samples. C Symphony UMAP with adipose stromal cells as a reference, quantification of mapping on right. Healthy n = 10, OA n = 9, RA n = 25, REM n = 3 biological samples. P values were calculated using an ordinary one-way ANOVA followed by Tukey’s multiple comparisons post hoc test. B, C Statistical comparisons: all groups were compared to each other. All data are presented as mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
Fast gene set enrichment analysis (FGSEA) revealed that the two pre-adipocyte populations were positively enriched for adipogenesis, supporting their identities as pre-adipocytes (Supplementary Fig. 12a). CEBPD+ pre-adipocytes were enriched for glycolysis, oxidative phosphorylation, and fatty acid synthesis, as well as scoring higher for fat-conditioned media activation, suggesting that they directly encounter fatty acids and are highly metabolically active. FABP4+ pre-adipocytes are quiescent by comparison and are enriched in few pathways (Supplementary Fig. 12a). Interestingly, we found a fourth cluster, VIT+ adipogenic regulatory cells (Aregs), which was almost exclusively found in adipose depots from patients with BMIs under 40, and scarce in patients with BMIs from 40 to 60, suggesting that this population may regulate adipocyte differentiation (Supplementary Fig. 12b). This cluster is negatively enriched for genes involved in adipogenesis and expresses high levels of VIT (encoding the ECM protein VITRIN), which has been shown to actively suppress adipogenesis29.
Since adiposity in the synovium is lost in RA and OA, we asked if the cell states in healthy synovial fibroblasts and adipose tissue are lost in RA and OA. We found that healthy synovial fibroblasts and remission fibroblasts mapped to DPP4+ progenitors and CEBPD+ and FABP4+ pre-adipocytes. In contrast, OA and RA fibroblasts mapped more to the universal DPP4+ universal progenitor population, with very few cells mapped to pre-adipocyte states (Fig. 6C). In addition, DPP4+ universal progenitors displayed the lowest fat-conditioned media and cortisol activation, suggesting that adipocyte proximity and cortisol signaling are important in pre-adipocyte commitment (Fig. 6B). OA and RA fibroblasts had diminished mapping to pre-adipocytes and enhanced mapping to universal progenitors and VIT+ Aregs compared to healthy synovial fibroblasts, suggesting that the loss of synovial adiposity changed their cell state.
Adipose-derived factors modulate fibroblast ECM remodeling and fibrosis
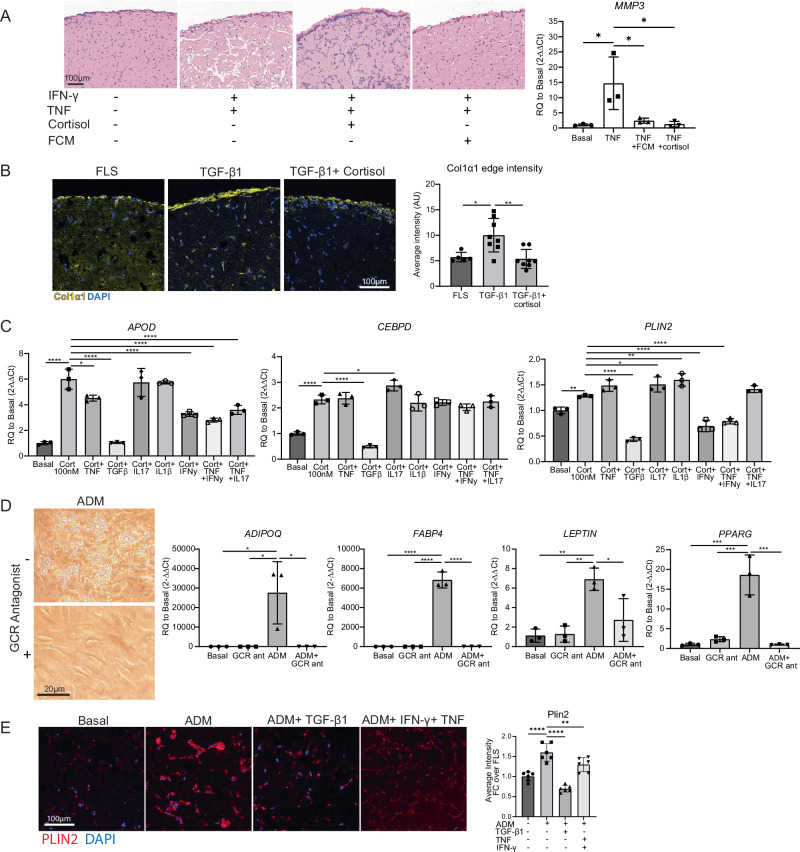
Bulk RNA sequencing of TGF-β1 or TNF +/− fat-conditioned media (FCM) stimulated fibroblasts identified pathways in which fat-conditioned media counteracts cytokine-induced gene expression changes. Surprisingly, fat-conditioned media was able to ameliorate both TGF-β1 and TNF-induced extracellular matrix remodeling, including decreasing TNF-induced matrix metalloproteinase expression (MMP1, MMP2, MMP3, MMP9, MMP13) and TGF-β1 induced pro-fibrotic collagen expression (Supplementary Fig. 13a–d and Supplementary Data 7, 8, and 9). In 2D cultured fibroblasts, fat-conditioned media and cortisol decreased MMP3 upregulation by TNF (14-fold down to 2.3 and 1.6-fold, respectively, Fig. 7A, right), supporting the bulk RNA sequencing findings.
Fig. 7. Functional effect of cortisol on fibroblasts.
A Left: FCM and cortisol (1 µM) protect against TNFa (2 ng/mL) and IFNy (25 ng/mL) induced matrix remodeling in long-term micromass culture (day 17). Cortisol and FCM were not added until day 7 of culture, TNFa and IFNy were added at day 0. n = 3 technical replicates, representative of one independent experiment. Right: MMP3 expression in 2D cultured synovial fibroblasts 24 h after addition of stimulation. n = 3 technical replicates, representative of one independent experiment. B Cortisol (1 µM) prevents TGF-β1 (10 ng/mL) induced fibrosis as measured by collagen 1a1 immunostaining (day 21). ImageJ quantification of COL1A1 staining on the right. FLS n = 5, TGFβ n = 8, TGF-β1+ cortisol n = 8 technical replicates, representative of one independent experiment. C Cortisol (100 nM) was applied to cells along with cytokines and assessed for APOD, CEBPD, and PLIN2 expression after 24 h. Concentrations used were: TNF (2 ng/mL), IFNy (25 ng/mL), TGF-β1 (10 ng/mL), IL1β (2 pg/mL) and IL17 (10 ng/mL). n = 3 technical replicates, representative of one independent experiment. Statistical comparisons: all groups were compared to the cortisol group. D Ability to enter adipogenic programs. Adipocyte differentiation media (ADM) was applied with or without 10 µM mifepristone (GCR antagonist). Day 9 qPCR, day 19 images (cultured until day 28). n = 3 technical replicates, representative of one independent experiment. Two-way ANOVA was performed to calculate statistical significance. E PLIN2 staining of micromass sections. Micromasses were treated with basal media, ADM, or ADM+ cytokines (TGFβ at 10 ng/mL or TNFa at 2 ng/mL+ IFNy at 25 ng/mL). ImageJ quantification of PLIN2 staining is on the right. Statistical comparisons: all groups were compared to the ADM group. n = 3 biological replicates, and two technical replicates within each sample (6 replicates per group total), representative of one independent experiment. A, B Statistical comparisons: all groups were compared to each other. A, B, D, E P values were calculated using an ordinary one-way ANOVA followed by Tukey’s multiple comparisons post hoc test; C P values were calculated using an ordinary one-way ANOVA followed by Dunnet’s multiple comparisons post hoc test. All data are presented as mean ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Source data are provided as a Source Data file.
In 3D synovial fibroblast organoids, we observed that stimulation with TNF and IFN-γ lead to significant fibroblast tunneling through the extracellular matrix and increased PDPN expression. Strikingly, these cytokine-induced changes are abrogated by the addition of fat-conditioned media or cortisol (Fig. 7A, left and Supplementary Fig. 13e). For TGF-β1, we found that stimulation for 21 days increased COL1A1 deposition, particularly at the edges of the organoids. Importantly, the addition of cortisol decreased COL1A1 deposition back to basal levels (from 10 to 5 AU) (Fig. 7B and Supplementary Fig. 13f), confirming the anti-fibrotic role of cortisol signaling in synovial fibroblasts.
Adipose-derived factors modulate inflammation
In cultured synovial fibroblasts, TNF upregulated IL6 103-fold, and this was significantly downregulated by adding fat-conditioned media (16-fold) or cortisol (14-fold). Similarly, TNF upregulated IL1B 152-fold and this effect was significantly downregulated by adding fat-conditioned media (12.7-fold) or by cortisol (8.8-fold) (Supplementary Fig. 13g).
In our bulk RNA sequencing data, TGF-β1 stimulation significantly decreased the enrichment of lipid and glucocorticoid biosynthetic pathways in fibroblasts. This led us to consider that TGF-β1 may counterbalance the effects of fat-conditioned media and glucocorticoids. Thus, we tested TGF-β1 and other cytokines and cytokine combinations for repressing the fibroblast response to cortisol (Fig. 7C). Strikingly, TGF-β1 significantly downregulated APOD, CEBPD and PLIN2 expression to equal to or lower than baseline levels. Of note, OA and RA fibroblasts have higher TGF-β1 signaling compared to healthy and remission fibroblasts (Supplementary Fig. 14a). In addition, IFN-γ decreased APOD and PLIN2 expression by 40%. Thus, the homeostatic state induced by fat-conditioned media and glucocorticoids is largely reversed by pro-fibrotic and inflammatory cytokines.
Blocking glucocorticoid signaling prevents fibroblast adipogenesis
We wondered how important glucocorticoid signaling is to adipogenesis and how the presence of cytokines would impact this process. The adipogenic medium was sufficient to induce adipogenesis in cultured fibroblasts, inducing expression of PPARG, adipocyte-specific genes, and lipid droplet accumulation after 9–28 days (Fig. 7D). However, the addition of mifepristone completely abolished the upregulation of adipocyte-specific genes, and fibroblasts exhibited no lipid accumulation. PPARG expression was reduced from 18-fold to 1-fold; ADIPOQ changed from 27,000-fold to 4-fold; FABP4 dropped from 6800-fold to 3.5-fold, and LEPTIN changed from 7-fold to 3-fold after mifepristone treatment. Thus, glucocorticoid signaling is important for synovial fibroblast adipogenesis, as has been shown in fibroblasts from other tissues30.
After confirming that TGF-β1 and IFN-γ signaling pathways are upregulated in OA and RA fibroblasts and can suppress cortisol signaling, we investigated whether these cytokines could suppress synovial fibroblast adipogenesis. We cultured synovial fibroblasts in 3D organoids with adipocyte differentiation media (ADM) for 21 days with or without TGF-β1 or IFN-γ plus TNF. Adipocyte differentiation media induced adipocyte differentiation as measured by strong PLIN2 staining, which defines lipid droplets, as well as by cell morphology; fibroblasts become more round and less elongated (Fig. 7E, images used for quantification are in Supplementary Fig. 14b). The addition of TGF-β1 or IFN-γ plus TNFα diminished the adipocyte-like morphology as well as total PLIN2 staining, confirming the role of these cytokines in suppressing adipogenesis as well as cortisol signaling. A summary schematic of the findings is presented in Supplementary Fig. 15.
Discussion
Fibroblasts are targets of intense study given their importance in tissue pathology. They mediate fibrosis and inflammation in many disorders or inhibit immune responses in the tumor stroma22,31. Many studies on RA have focused on fibroblasts as they mediate both inflammation and degradation of joint tissues in arthritis32. Yet, most studies highlight pathological fibroblast states that relate to tissue damage, while few studies have sought to understand key drivers of fibroblast homeostasis. Here, using the synovium as an example, we have identified the importance of adipocytes in driving the homeostatic phenotype of fibroblasts mediated by the activation of the endogenous glucocorticoid cortisol.
We show that adipocytes regulate synovial fibroblast function by expressing Hsd11b1 to generate active cortisol, which acts on resident fibroblasts in the synovium. Despite this, the depletion of adipocytes was not protective against arthritis. This may in part be because adipocytes supply lipids to meet nutritional needs, thus, they can provide a source of energy for infiltrating lymphocytes19. In addition, adipocytes secrete other factors which influence inflammation including the adipokines leptin and adipsin (both inflammatory), and adiponectin (which is anti-inflammatory)20,21. The diverse functions of adipocytes likely masked any underlying effects related to decreased cortisol generation.
11-β-HSD1 has been shown to suppress synovitis and joint destruction in the TNF-transgenic mouse model of arthritis, and cortisol has been used as a first-line treatment for managing RA inflammation for decades; but is thought to act mainly on infiltrating immune cells to achieve its anti-inflammatory effects33. Koenen recently detailed that glucocorticoid signaling in stromal cells, not immune cells, is essential for the therapeutic effect of dexamethasone in serum transfer-induced arthritis34. Here, we found that Nr3c1 glucocorticoid receptor signaling in fibroblasts is involved in protection from arthritis in the AIA mouse model. We also found that cortisol plays a key role in maintaining healthy fibroblast functions in vitro and can mitigate the inflammatory, ECM remodeling, and fibrotic effects of TNF and TGF-β1 on synovial fibroblasts.
Conversely, at a tenfold higher ratio of cytokine to cortisol, we found that TGF-β1 and IFN-γ can overcome cortisol signaling in fibroblasts, suggesting that higher levels of inflammation and TGF-β1, as observed in RA and OA, overwhelm homeostatic cortisol signaling. Inhibition of cortisol signaling likely further contributes to synovial inflammation and lipodystrophy of the synovium, leading to fibrosis. IFN-γ and TGF-β1 were also found to blunt adipocyte differentiation, which could be a second in vivo mechanism of cytokine suppression of homeostatic cortisol signaling by suppressing the major cellular source of 11-β-HSD1 in the joint.
We found that a significant number of healthy synovial fibroblasts mapped to committed pre-adipocyte populations from classical adipose tissue depots. This finding, together with the high cortisol signaling in this population, suggests that the healthy synovium contains pre-adipocytes that rely on cortisol to maintain their pre-adipocyte state. This conclusion was also supported by our in vitro experiments which showed that blocking the glucocorticoid receptor effectively blocked adipogenesis of synovial-derived fibroblasts. In addition, RA synovial fibroblasts had enhanced mapping to adipose tissue VIT+ Aregs, suggesting that a subset of RA fibroblasts actively suppress adipogenesis.
A limitation of this study is that the healthy human synovial tissue was collected in a different manner from the OA and RA synovial tissues. Whereas OA and RA tissues were collected through needle biopsy or synovectomy, healthy tissue was collected via manual dissection of whole synovium post-mortem. This could contribute to some of the cell proportion and phenotype differences observed. We endeavored to overcome this limitation by collecting tissues from several different sources with collection times as short as six hours post-mortem. In addition, we included mouse experiments depleting intra-articular fat in addition to computational analysis of two mouse RNA sequencing datasets to confirm the major findings of our study where the delayed sample collection limitations of working with human tissues are not present.
In conclusion, this work has identified adipose tissue and cortisol signaling as important contributors to healthy synovial and adipose tissue fibroblast function and identity which are lost in disease. Cortisol plays a critical role in multiple facets of fibroblast biology, including metabolism, inflammation and ECM homeostasis, which were previously under-appreciated. We introduce the concept that the loss of adiposity contributes importantly to loss of cortisol signaling and the subsequent development of pathologic fibroblast states in adipose-rich tissues such as the synovium.
Methods
This research complies with all relevant ethical regulations and is approved by the Institutional Biosafety Committee of the Brigham and Women’s Hospital under registration number 2011B000015.
Synovial tissue collection
In total, 16 synovial samples, 8 male and 8 female, were obtained from human donors with no history of arthritis or autoimmune disease. Healthy synovial samples were collected through three different sources: seven are from the National Disease Research Interchange (NDRI) with written informed consent for collection and use of tissue for research purposes, four are from Rush University with written informed consent for collection and use of tissue for research purposes, and five are from BWH Autopsy Department with written informed consent for collection and use of tissue for research purposes. No donors consented to release identifying information. No study participants received compensation for taking part in the study. Sex was not considered in the study design. Sex is based on self-reporting. Patient tissues were excluded for collection if they had a diagnosis of lupus, type-1 diabetes, psoriasis, rheumatoid arthritis, psoriatic arthritis, spondyloarthritis, Crohn’s disease, Sjogren syndrome, or osteoarthritis. Collection and sequencing of these tissues was performed under institutional review board (IRB) number 2002P000127 titled Pathways of Antigen Presentation by CDI. In addition, tissue was obtained from Rush University through the materials transfer agreement 2020A004824 titled Molecular profiling of synovium. We thank the Brigham Autopsy department for collecting post-mortem synovial tissues for our lab. We thank Rush University for collecting post-mortem synovial tissues for our lab. Samples obtained from Rush also went through Collin’s Grading of cartilage as an additional measure determine joint health. Upon receipt of synovial tissues, synovium was dissected away from adipose tissue as much as possible. Synovium was saved in Crystor CS10 freezing media (Sigma, cat. No. C2874) by incubating on ice for 10 min prior to freezing. Synovial adipose tissue was minced into ~5 mm pieces and used to generate fat-conditioned media.
Synovial tissue disaggregation
Cryo-preserved synovial tissue was thawed and rinsed in RPMI with 5% FBS for 5 min. Samples were transferred to a gentleMACS tube (Miltenyi, cat. No. 130-093-237) containing digest media (0.2 mg/mL Liberase TL (Sigma, cat. No. 05401020001), 0.1 mg/mL DNase1 (Sigma, cat. No. 11119915001) in RPMI). Tissue was macerated with gentleMACS followed by shaking at 37 °C for 30 min. The tissue was then filtered through a 70-µm cell strainer and spun down.
Adipose tissue disaggregation
Adipose tissue was minced and digested in adipose buffer containing 5.5 mM d-glucose, 140 mM sodium chloride, 4.7 mM potassium chloride, 1.25 mM magnesium sulfoxide heptahydrate, 2.5 mM calcium chloride dihydrate, 10 mM N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES), 2.5 mM sodium phosphate monobasic dihydrate (all Sigma-Aldrich; Darmstadt, DE), 2% bovine serum albumin (BSA) fraction V (Sigma-Aldrich; Darmstadt, DE), and type-1 collagenase (250 U/mL) (Worthington Biochemical; NJ, US). Samples were agitated at 8×g on a orbital shaker at 37 °C for 45–60 min. Digestions were terminated by adding 2 mM EDTA (final concentration) (Sigma-Aldrich; Darmstadt, DE). After digestion, samples were passed through a 400 μm gauze and centrifuged at 300×g for 7 min at RT and was then incubated with 9 volumes erythrocyte lysis buffer (155.1 mM ammonium chloride, 9.9 mM potassium bicarbonate, 0.1 mM EDTA; all Sigma-Aldrich; Darmstadt, DE) for 10 min at RT.
Single-cell RNA sequencing
For synovial tissues, live cells were isolated via disaggregation and cell sorting for all live events using Fixable Viability die (UV455, eBioscience, cat. No. 65-0868-14) on a BD™FACSAria III cell sorter (BD). Cells were loaded onto a single lane (Chromium chip, 10X genomics) followed by encapsulation in a lipid droplet (Single Cell 3’ kit, 10X Genomics) followed by cDNA and library generation according to the manufacturer’s protocol. Cells were stained with cell-hashing antibodies (TotalSeq, BioLegend) before cell capture. cDNA libraries were sequenced to an average of 55,000 reads per cell using Illumina Nextseq 500. scRNA-seq. Reads were processed with Cell Ranger v3.1, which demultiplexed cells from different samples and quantified transcript counts per putative cell. Quantification was performed using the STAR aligner against the GRCh38-3.0.0 transcriptome. For adipose samples, post digestion, cells were sorted on a BD™FACSAria III cell sorter (BD) to separate CD45 + /− events using the following fluorescently labeled antibodies: CD45–BV785 (BioLegend™; clone H130 cat. No. 304047), CD235a–FITC (BioLegend™; clone HI264, cat. No. 349103), Live-Dead Zombie Aqua™ (BioLegend™; cat. No. 423101), and propidium iodide (PI) (Thermo Fisher Scientific Inc. cat. No. R37108;). Up to 5000 cells (split 50:50 CD45 + /−) per patient sample were collected for scRNA-seq. The five patients were combined and loaded onto a single lane (Chromium chip; 10x Genomics) followed by encapsulation in a lipid droplet (Single cell 3’ kit; 10x Genomics), and subsequent cDNA and library generation according to the manufacturers’ instructions. Cells were sequenced to an average of 25,726 reads per cell using Illumina Nextseq 500 (Illumina Inc.). Reads were then processed using Cell Ranger v.3.1. (10x Genomics), to quantify transcript counts per putative cell. Quantification was performed using STAR aligner against the GRCh38-3.0.0. transcriptome.
RNA-seq quality control and pre-processing
For synovial tissues, after filtering out low-quality cells (<200 and >2800 unique genes, >25% mitochondrial reads), 19,378 cells from primary tissue were further analyzed. With these high-quality cells, we used the Seurat package (version 4.0.1) in R (version 4.1.1) to perform normalization, find variable features, perform principal components analysis, clustering and dimensional reduction using TSNE35. We corrected for donor-specific effects using Harmony with default parameters36. For healthy adipose tissues, a total of 19,830 cells (16,680 omental; 3150 subcutaneous) remained after filtering out low-quality cells (<200 and >2800 unique genes, >25% mitochondrial reads). These were taken for downstream analyses as described above and 7350 cells were identified as PDGFRA+ stromal cells. Fast gene set enrichment analysis (FGSEA) was done using the R package “fgsea” and gene sets were derived from MsigDB. Exact code used for analysis is available on Zenodo (see code availability statement).
Building and mapping to references
We used the buildReferenceFromSeuratObj() function from the Symphony package to build integrated reference atlases for the global and cell-type specific atlases from the Harmony objects37. To find concordance between cell types defined by Zhang et al. and this study, we used the Symphony mapQuery() function to map the 19,378 scRNA-seq healthy synovial cells onto cells defined by Zhang et al.8. We also mapped fibroblasts from Zhang et al. onto a reference atlas defined by healthy synovial fibroblasts. We predicted reference cell types and states for the query cells using the knnPredict() function with k = 5.
Covarying neighborhood analysis (CNA)
We used the rcna R package from github to calculate cell neighborhoods which were negatively associated with healthy synovial cells. The association. Seurat function was used to perform association testing and compute neighborhood-level FDRs (false discovery rate).
Pseudobulk analysis
Activation scores were generated based on the lists of differentially expressed genes determined by the bulk RNA sequencing data. Differentially expressed genes were filtered to only include genes with base mean over 5, P < 0.05, and log base twofold >0.6953. These lists were then supplied to Seurat and “AddModuleScore” was used to apply the activation score to cells. For the synovial fibroblast scoring, raw UMI and metadata were extracted and the counts dataframe was converted into a matrix. Then, the raw UMI count matrix and metadata dataframe were used to collapse the counts over each individual donor using the command “presto” (“immunogenomics/harmony” github package). The resulting object was converted back into a Seurat object and then “AddModuleScore” was used to create activation scores. For the adipose fibroblast scoring, the same approach was used, except counts were collapsed over both sample and by cluster.
Bulk RNA sequencing
Three fibroblast lines consented under institutional review board (IRB) number 2019P002924, titled Synovial tissue biorepository, were plated at 10k cells/well and allowed to rest for 3 days prior to stimulation for 4 h or 20 h. Cells were stimulated with FCM, cortisol (100 nM), TGF-β1 (10 ng/mL), TNF (5 ng/mL), FCM + GCR antagonist (mifepristone, 10uM), FCM + TGF-β1, and FCM + TNF. Cells were harvested by rinsing with PBS and then applying TCL buffer with 1% beta-mercaptoethanol. Full-length cDNA and sequencing libraries were performed using Illumina Smart-seq2 protocol38. Libraries were sequenced on MiSeq from Illumina to generate 35 base paired-end reads. Reads were mapped to the GRCh38.93 transcriptome using kallisto 0.42.4 and transcriptional levels of genes were quantified with the log2(TPM + 1) (transcripts per kilobase million) metric. Aligned reads were imported into Dese2 using Tximport (Bioconductor, release 3.19). Samples underwent QC by assessing the total reads mapped per sample and percentage of genes with mapped reads. On average, samples had >3,000,000 and <6,000,000 total reads and 35% genes with mapped reads. Two outliers were removed due to having low total reads of <1,000,000. All samples that passed QC were then put into the DeSeq2 pipeline for further analysis. Differentially expressed genes were calculated by controlling for cell line differences and contrasting treatments within a timepoint. Volcano plots were generated using ‘Enhanced Volcano’ from Bioconductor. Exact code is available on Zenodo.
Bulk RNA sequencing calculation of rescued pathways
To identify pathways which were returned to basal levels by FCM, we ranked genes according to the following calculation for gene set enrichment analysis, with TNF and FCM + TNF as an example: “Statistic =MIN(ABS(Fold change difference*), ABS(TNF fold change over basal)) × SIGN(ABS(TNF fold change over basal)-ABS(TNF + FCM fold change over basal)).” * Fold change difference is the fold change of TNF over basal minus the fold change of TNF + FCM over basal.
Ingenuity pathway analysis
A set of differentially regulated genes among all sublining fibroblasts between healthy and RA patients, and between remission RA and RA patients, was generated in R. This gene list was then entered into IPA as a core analysis with an expression log ratio cutoff of 1.5 and false discovery rate cutoff of 0.05. This left us with over 200 genes which were run in each dataset.
Cell culture
Human synovial fibroblasts were cultured in DMEM supplemented with 5% fetal bovine serum, 1% penn strep, 1% l-glutamine, 1% non-essential amino acids, 2% essential amino acids, and 0.5% beta-mercaptoethanol. Human visceral pre-adipocytes were purchased from Lonza (PT-5005) and cultured according to the manufacturer’s instructions in PGM-2TM Preadipocyte Growth Medium-2 BulletKitTM from Lonza (PT-8002). Once cells were expanded, they were seeded for adipocyte differentiation following the manufacturer’s instructions. After 10 days in adipocyte differentiation media (ADM), media was harvested for downstream applications.
CRISPR–Cas9 gene deletion
Alt-R® CRISPR–Cas9 sgRNA of the following sequence was directed towards NR3C1 and was designed in ChopChop (purchased from IDT): mA*mU*mG* rArCrU rArCrG rCrUrC rArArC rArUrG rUrUrG rUrUrU rUrArG rArGrC rUrArG rArArA rUrArG rCrArA rGrUrU rArArA rArUrA rArGrG rCrUrA rGrUrC rCrGrU rUrArU rCrArA rCrUrU rGrArA rArArA rGrUrG rGrCrA rCrCrG rArGrU rCrGrG rUrGrC mU*mU*mU* rU. The primer coordinates are as follows: Left primer: chr5:143300617-143300639, Right primer: chr5:143300397-143300419. Primer sequences: Left primer: CTGGTGTCACTGTTGGAGGTTA, Right primer: GGGCTCACGATGATATAAAAGC. The sequence targets the following nucleotide sequence of the NR3C1 coding region for deletion: ATGACTACGCTCAACATGTTAGG. sgRNA was applied to cells using the following protocol: 250k cells were resuspended in 17uL of P2 solution. sgRNA was resuspended in IFTE buffer (IDT, cat. No. 11-05-01-05) to 80 μM concentration and mixed 1:1 with cas9 and incubated for 15 min to allow RNP complex formation. The cell suspension and sgRNA+cas9 mixture were then combined and then underwent nucleofection program EN 150. After, pre-warmed cell media was added to each well and let sit for 15 min at room temperature before adding to a cell culture flask. Fibroblasts were plated and allowed to rest for three weeks prior to validation of gene deletion and use for experiments.
In vitro stimulation experiments
Fibroblasts were plated at 40k cells per well in a 12-well plate and allowed to rest for 4 days prior to stimulation for 24 h. The following chemicals were purchased from Sigma-Aldrich: cortisol was purchased as Hydrocortisone (H0888-1G), Aldosterone (A9477-5MG), Progesterone (P8783-1G), the glucocorticoid receptor antagonist Mifepristone (M8046-100MG) was used at 10 μM, the mineralocorticoid receptor antagonist spironolactone (S3378-1G) was used at 1 μM, the 11β-hydroxysteroid dehydrogenase type-1 competitive inhibitor Metyrapone (M2696-10MG) was used at 100 μM. The following chemicals were purchased from Peprotech: TGF-β1 (100-21-2UG) used at 10 ng/mL, TNF (300-01A-10UG) used at 5 ng/mL, and IFN-γ (300-02-20UG) used at 25 ng/mL. Arachidonic acid (AA, Cat# U-71-A) was purchased from Nu-Chek Prep. Triacylglycerol (TAG, Cat# T7140) and monoacylglycerol (MAG, Cat# M2015) were purchased from Sigma. β-Glucosylceramide (β-GlcCer, Cat# 860539) and ceramide (Cer, Cat# 860518) were purchased from Avanti Polar Lipids. Sodium palmitate (Cat# P9767-5G) and sodium oleate (Cat# O7501-250MG) were purchased from Sigma, dissolved in methanol to 200 µM, and then diluted to specified concentrations in culture media.
Fat-conditioned media
Fat-conditioned media was generated from adipose tissue sourced from either healthy joints (syn fat donor) or abdominal adipose tissue. Briefly, adipose tissue was minced into ~5-mm pieces and cultured in RPMI for 24 h before harvesting the media. The conditioned media was then passed through a 70-µm filter and then spun down at 475×g for 5 min to remove cells.
Lipid extraction and solid phase extraction for sample purification
To isolate and purify the active molecules, conditioned media was fractionated into aqueous, organic, and an interphase layer using the Bligh and Dyer method39. In general, 1 ml of conditioned media was extracted with 3.75 ml of chloroform:methanol (1:2) solution. The one phase mixture was vortexed for 1 min and then 1.25 mL of chloroform and 1.25 mL of water were added. The sample was vortexed for 30 s at each addition and then centrifuged at 1000×g for 15 min to generate phase separation into two layers. The top aqueous and the bottom organic phases were separated and kept for further analyses. The organic fraction was dried down under a nitrogen stream, sonicated in 3 ml of chloroform, evenly loaded onto three solid phase extraction columns (Supelclean™ LC-Si SPE Tube, Sigma, Cat# 57051, silica bed wt:1 g, column: 6 ml), which had been pre-conditioned with 12 ml of chloroform. The loaded sample was serially eluted with solvents with increasing polarity: chloroform (8 ml), acetone (8 ml), methanol (8 ml), and water (8 ml). All fractions, handled in triplicate, were collected in the pre-weighed vials and dried down under nitrogen and stored at −20 °C for further use.
HPLC-mass spectrometry (MS) analysis of lipids from solid phase extraction fractions
To perform comparative lipidomics for SPE-generated acetone fractions versus SPE-generated chloroform fractions, the lipid quantities were measured using a Mettler balance and redissolved in chloroform/methanol (1:1, v/v) and normalized to 0.5 mg/ml. In total, 50 µl of normalized samples were dried under nitrogen, redissolved in 100 µl of the reverse phase starting mobile phase A for HPLC-MS run on an Agilent Poroshell 120 A, EC-C18, 3 × 50 mm, 1.9 µm reversed-phase column coupled with a 3 × 5 mm, 2.7 µm guard column and analyzed by an Agilent 6546 Accurate-Mass Q-ToF with electrospray ionization (ESI) source with a 1260 series HPLC instrument. For the ESI source, the gas temperature was maintained at 320 °C, and drying gas was 8 l/min, with Nebulizer pressure set to 35 psi, and Vcap set to 3500 V. The reversed-phase systems were used as described40 and summarized below. Solvent A was methanol/water (95/5: V/V with 2 mM ammonium formate), and solvent (B) was 1-propanol/cyclohexane/water (90/10/0.1: v/v/v with 3 mM ammonium formate). The solvent gradient for a 20 min run was: 0–4 min, 100% A; 4–10 min, from 100% A to 100% B; 10–15 min, 100% B; 15–16 min, from 100% B to 100% A; 16–20 min, 100% A; 5 min post run, 100% A. Injection volume per run was 10 µl. Flow rate was 0.15 ml/min. Both positive and negative ion mode data were acquired and analyzed by Agilent MassHunter Workstation Software (version B.07.00). For lipidomics, data were analyzed using automated peak-picking algorithms by R (version 3.4.2) package XCMS41 and in house designed software methods17. The XCMS raw data output was reported as an Excel table, including m/z, retention time, fold change, P value, and intensity for every data point. The volcano plot and m/z versus retention time plot were generated using GraphPad Prism.
Direct cortisol quantification by HPLC-MS
To directly quantify cortisol, each conditioned media sample was measured 4–5 times by mixing sample with varying concentrations of cortisol-d4 standard (methanol solution) in a 1:1 (v/v) ratio. The mixture was then subjected to the reverse phase HPLC-MS analysis using the Agilent Poroshell column and the Q-Tof instrument as described above. The mobile phase A was methanol/water 20/80 (v/v), supplemented with 2 mM ammonium formate; the mobile phase B was methanol/water 80/20 (v/v), supplemented with 2 mM ammonium formate. 10 µl of the mixed sample were injected and the flow rate was 0.15 ml/min with the binary gradient: 0–2 min, 50% A; 2–10 min, from 50% A to 100% B; 10–15 min, 100% B; 15–17 min, from 100% B to 50% A; and 17–20 min, 50% A. The unknown cortisol concentrations in each condition media sample were determined by the chromatogram area of the non-deuterated natural molecule compared to the known concentration of cortisol-d4 internal standard as seen in 4–5 repeated measurements to generate technical replicates.
Gene expression analysis
RNA was extracted from fibroblasts using TRIzol reagent (Thermo Fisher, cat. No. 15596026) as per the manufacturer’s instructions. The aqueous phase was mixed 1:1 with 70% ethanol and applied to a minikit spin column and washed with RW1 and RPE buffer (Qiagen RNeasy MiniKit, cat. No. 74104). cDNA was synthesized using the QuantiTect Rev. Transcription Kit following the manufacturer’s protocol. Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out using SYBR Green primers and an Agilent AriaMx Real-Time PCR system. Relative gene expression was calculated by the ΔΔCt method. The ΔCt was calculated using the reference genes β2-microglobulin and β-actin. ΔΔCt was calculated relative to the basal control group. Human and mouse primer sequences are listed in supplemental materials.
Mouse experiments
All animal studies were performed with approval by the institutional animal care and use committee (IACUC) of the Brigham and Women’s Hospital under protocol number 2016N000519. Sex was considered in the study design. We used close to equal numbers of male and female mice across conditions to avoid sex-driven effects. Sex was not considered in the analysis as we were not sufficiently powered to study sex effects. Mice were fed, ab libitum, a standard chow diet (5053 PicoLab Rodent Diet 20) consisting of 20% protein and 4.5% fat. Mice were housed with a light cycle from 7AM-7PM at an ambient temperature of 68–75 F and humidity of 35–65%. Experimental mice were used between the ages of 9–12 weeks for all experiments. C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (iDTR) and B6;FVB-Tg(Adipoq-cre)1Evdr/J (Adipoq-cre) mice were purchased from Jackson labs. Diphtheria toxin was injected intra-articularly at 50 ng/injection in a 10 µL volume into 1 knee joint/mouse at days 0, 4, 8, 14, 21, 28, 35, 42, and 49 prior to sacrifice at day 56. Investigators were single-blinded while performing injections. No statistical method was used to predetermine sample size. Nr3c1 knockout: B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J and B6.Cg-Nr3c1tm1.1Jda/J mice were purchased from Jackson Labs and crossed to generate fibroblast-specific Nr3c1 knockout mice. Tamoxifen (Millipore Sigma, T5648) was used to induce CreER recombination. Tamoxifen was dissolved in corn oil and 75 mg/kg body weight was injected intraperitoneally once per day for 5 days into Pdgfra-CreER; Nr3c1fl/fl mice and sex-matched, littermate controls. We waited 2 weeks post tamoxifen treatment to begin priming for antigen-induced arthritis experiments, and 4 weeks to assess changes in naive synovium. Investigators were single-blinded while measuring joints. For Nr3c1 knockout experiments, 9 male and 8 female genotype control mice were used, and 9 male and 9 female Pdgfra-CreER + Nr3c1fl/fl mice were used. For inducible adipocyte depletion experiments, six genotype control and seven iDTR; Adipoq-Cre mice were utilized.
Antigen-induced arthritis
Mice were primed by administering two subcutaneous injections of a total of 200 µg of mBSA (sigma, cat. No. A1009) mixed with 100 µL of complete Freund’s adjuvant containing 5 mg/mL of tuberculosis (Chondrex, cat. No. 7023). Mice were also given 200 µL of pertussis toxin (Thermo Fisher, cat. No. PHZ1174) intraperitoneally. Two weeks later, mice were given 50 µg of mBSA intra-articularly. Joint swelling was assessed by caliper for up to 17 days later.
K/BxN serum transfer inflammatory arthritis (STIA)
ADIPOQ-cre; iDTR mice were adipocyte-depleted in ankles via intra-articular injection of 35 ng of diphtheria toxin on day -7, day -3, and day -1. Mice were then given 50 µL of pooled serum from 9- to 11-week-old K/BxN mice intraperitoneally on day 0 and day 2. Arthritis was measured by caliper measurements of ankles.
Subcutaneous and omental adipose tissue collection
For single-cell RNA sequencing of adipose depots in Fig. 5, which included two male and three female donors, all patients undergoing voluntary bariatric surgery provided written informed consent for collection and use of tissue for research purposes. No donors consented to release identifying information. Sex was not considered in the study design. Sex is based on self-reporting. This study was carried out in accordance with the declaration of Helsinki and was reviewed and approved by the ethics committees of St. Vincent’s University Hospital and Trinity College Dublin.
Abdominal adipose tissue collection
The Human Skin Disease Resource Center of Brigham and Women’s Hospital and Harvard Medical School collected anonymous, discarded adult skin plus fat samples removed as part of cosmetic surgery procedures and provided them to our lab. The Human Skin Disease Resource Center is supported in part by NIAMS Resource-based Center Grant # 1P30AR069625. This tissue was collected under IRB number 2020P003757. Abdominal adipose tissue was used to generate fat-conditioned media.
Lipid sources
Lipid standards were purchased from commercial sources. Arachidonic acid (AA, Cat# U-71-A) was from Nu-Chek Prep. Hydrocortisone-d4 (cortisol-d4, Cat# 26500) was from Cayman Chemical. Triacylglycerol (TAG, Cat# T7140) and monoacylglycerol (MAG, Cat# M2015) were from Sigma. β-Glucosylceramide (β-GlcCer, Cat# 860539) and ceramide (Cer, Cat# 860518) were from Avanti Polar Lipids.
ELISA
ELISA detection of cortisol was performed using the Cortisol ELISA Assay Kit from Eagle biosciences (COR31-K01) according to manufacturer instructions. ELISA detection of aldosterone was performed using the Aldosterone ELISA Assay Kit from Eagle biosciences (ALD31-K01) according to the manufacturer instructions.
Gene expression analysis
The human specific primers used were the following (listed 5’ to 3’): B2M forward: GAGGCTATCCAGCGTACTCCA, B2M reverse: CGGCAGGCATACTCATCTTTT, ACTB forward: GCTCCTCCTGAGCGCAAGTAC, ACTB reverse: GGACTCGTCATACTCCTGCTTGC, APOD forward: CCACCCCAGTTAACCTCACA, APOD reverse: GTGCCGATGGCATAAACC, NNMT forward: TTGAGGTGATCTCGCAAAGTTATT, NNMT reverse: CTCGCCACCAGGGAGAAA, CEBPD forward: GGTGCCCGCTGCAGTTT, CEBPD reverse: CTCGCAGTTTAGTGGTGGTAAGTC, MMP3 forward: CTCCAACCGTGAGGAAAATC, MMP3 reverse: CATGGAATTTCTCTTCTCATCAAA, IL6 forward: AGACAGCCACTCACCTCTTCAG, IL6 reverse: TTCTGCCAGTGCCTCTTTGCTG, IL1β forward: GGACAAGCTGAGGAAGATGC, IL1β reverse: TCGTTATCCCATGTGTCGAA. NR3C1α/β forward: ACT TAC ACC TGG ATG ACC AAA T, NR3C1α reverse: TTC AAT ACT CAT GGT CTT ATC C, NR3C1β reverse: TCC TAT AGT TGT CGA TGA GCA T; FABP4 forward: GCA AAG CCC ACT CCT ACA GTT, FABP4 reverse: CTC TCT GTG CCT TTT TCC TCC T; PPARG forward: AGG CGA GGG CGA TCT TGA CAG, PPARG reverse: GAT GCG GAT GGC CAC CTC TTT; ADIPOQ forward: CAA CAT GCC CAT TCG CTT T, ADIPOQ reverse: GGA GGC CTG GTC CAC ATT AT; LEPTIN forward: CGG TAA GGA GAG TAT GCG GG, LEPTIN reverse: CAG TAG GTG CCT GGC ATT CA.
To assess adipocyte depletion in mouse joints, whole knee joints were taken, flash frozen in liquid nitrogen, crushed with a mortar and pestle, and RNA isolated using TRIzol.
Mouse-specific primers: Adipoq forward: AAGGAGATGCAGGTCTTCTTGGT, AdipoQ reverse: CACTGAACGCTGAGCGATACAT; Hsd1 β1 forward: CGA CAT CCA CTC TGT GCG AA, Hsd11β1 reverse: TGC TGC CAT TGC TCT GCT; Pparg forward: CCA CAG TTG ATT TCT CCA GCA TTT C, Pparg reverse: CAG GTT CTA CTT TGA TCG CAC TTT G; Cebpd forward: TGCGAGCGACAGGAAGCT, Cebpd reverse: GCAATGGTAATAAGACGTAGAAAATGC; Plin2 forward: CAG CCA ACG TCC GAG ATT G, Plin2 reverse: CAC ATC CTT CGC CCC AGT T; Cidec forward: GGC GTA GTC CAT CCT TGT CA, Cidec reverse: GTC AGA TGA GAG ACT GGG GC.
Flow cytometry
Disaggregated human tissues were stained with the surface markers CD45 in APC-Cyaine7 (2D1, Biolegend, cat. No. 368516), CD31 in AF700 (WM59, Biolegend, cat. No. 303133), CD90 in BV421 (5E10, BD Biosciences, cat. No. 562556), podoplanin in PE (NC-08, Biolegend, cat. No. 337003), CD146 in BV510 (P1H12, Biolegend, cat. No. 361021), CD14 in PE-Cyanine7 (63D3, Biolegend, cat. No. 367111), CD3 in APC (Biolegend, cat. No. 317317), and Fc block (Biolegend, cat no 422301) on ice for 40 min. All human antibodies were used at 1:25, Fc block was used at 1:50. Fixable viability dye (UV455, eBioscience, cat. No. 65-0868-14) was applied at 1:1000 dilution and stained on ice for 20 min. Cells were fixed in BD cytofix fixation buffer (cat. No. 554714) for 20 min. After fixation, HCS LipidTOX (Thermo Fisher, H34475) was applied at a 1:200 dilution in PBS and stained at room temp for 30 min. Cells were filtered through a 70-micron mesh filter and run on an LSRII (BD Biosciences). Mouse synovial tissues were stained with CD31 in BV785 (Biolegend, cat. No. 102435), CD146 in PerCP/Cyanine 5.5 (Biolegend, cat. No. 134709, CD55 in APC (Biolegend, 131811), Pdgfra in BV421 (Biolegend, cat. No. 135923), CD45 in AF700 (Clone I3/2.3, Biolegend,147715), CD3 in FITC (Clone 17A2, Biolegend,100204), F4/80 in APC-cy7 (Clone BM8, Thermo Fisher, 47-4801-82), CD4 in BV605 (Clone RM4-5, Biolegend,100548), CD8 in BV711 (Clone SK1, Biolegend,100759), PDPN in PE-Cyanine7 (eBioscience, cat. No. 25-5381-82), and Fc block (Biolegend, cat no 101319). All mouse antibodies were used at 1:200, Fc block was used at 1:100.
AMP CyTOF data
The CD45, CD3, and CD14 flow cytometry quantifications for OA and RA samples in Fig. 1D were calculated based on Cytof data from AMP phase 12. Leukocyte-rich RA biopsies (n = 8) were used for the RA group and n = 12 OA samples were used.
Micromass experiments
Fibroblasts were resuspended in Matrigel (BD Biosciences, 3354234) on ice at 200,000 cells per 35 µL of Matrigel (Corning, cat. No. 354248. Cold pipette tips were used to pipette 35 µL of Matrigel into each well of a 12-well tissue culture plate pre-coated with poly-HEMA (Aldrich Chem Co. cat. no. P3932). The plate was placed in a 37 °C incubator for 1 h to allow the Matrigel/cell suspension to gel. Then pre-warmed media to the well. IFN-γ (25 ng/mL, Peprotech, cat. No. 300-02) and TNF (2 ng/mL, Peprotech, cat. No. 300-01 A) were added immediately and at every media change. Fat-conditioned media was added at day 7 and then replenished at each subsequent media change. At day 17, micromasses were harvested by fixing in 4% paraformaldehyde and then embedded in paraffin. Sections were stained with H&E or immunofluorescently labeled with PDPN (eBioscience, 16-9381-81), PRG4 (Millipore Sigma, MABT401), PLIN2 (Thermo Fisher, 15294-1-AP).
Immunofluorescence
5-μm thick sections of paraffin-embedded tissue were taken and deparaffinized prior to staining with histoclear. After rehydration, antigen retrieval was performed using Tris buffer pH 9 heated to near-boiling and placed in an 85 °C oven for 30 min. Slides were blocked in 5% normal horse serum for 30 min before application of primary antibodies. The following anti-human primary antibodies were applied at 1:100 dilution for 1.5 h at room temperature: PDPN (eBioscience, 16-9381-81), PRG4 (Millipore Sigma, MABT401), PLIN2 (Thermo Fisher, 15294-1-AP), CD45 (Thermo Fisher, A304-376A-T), COL1A1 (R&D Systems, AF6220). Anti-mouse antibodies: PDPN (eBioscience, clone eBio8.1.1, cat. No. 14-5381-82), PDGFRA (R&D Systems, polyclonal Goat IgG, cat. No. AF1062-SP), or Cadherin 11 (clone 23C6, mouse anti-mouse)42. Secondary antibodies were applied at room temperature for 1.5 h (donkey anti-rat AF488, 712-545-153; donkey anti-rabbit AF647, 711-605-152; donkey anti-mouse Cy3, 715-165-150; donkey anti-goat AF647 cat. No. 705-605-003; goat anti-syrian hamster Cy3 cat. No. 107-165-142; donkey anti-mouse AF488 cat. No. 715-545-150; all from Jackson Immunoresearch,). Slides were then treated with Sudan Black B for 15 min to diminish autofluorescence. This was followed by DAPI (Sigma, cat no D9542) staining for 5 min and mounting with FluorSave™ Reagent from Sigma (cat no 345789). Images were acquired on a Zeiss LSM 800 with Airyscan confocal system on a Zeiss Axio Observer Z1 Inverted Microscope at ×200 magnification. For all images, the LUT is linear and covers the full range of the data. Images are pseudo-colored.
Quantification
COL1A1 and PLIN2 staining was quantified in ImageJ by selecting the whole micromass area of each image and measuring the mean intensity. In Supplementary Fig. 7c, using ImageJ, we created a selection on positive PDGFRA staining (threshold: 0-29), and then applied that selection onto the PDPN and Cadherin 11 channels. We then measured mean intensity within the PDGFRA selection to obtain fibroblast-specific measures of PDPN and Cadherin 11 intensity.
Oil red O staining
OCT-embedded synovium was cryosectioned and sections were air dried for 5 min at room temperature. Working solution oil red O (Sigma, cat no O0625) was prepared by making a stock solution of 300 mg oil red O in 100 mL isopropanol, mixing 30 mL of the stock with 20 mL of distilled water, mixing, and filtering prior to use. Staining procedure: Slides were fixed in 4% PFA for 10 min, dipped in 60% isopropanol once quickly, stained in oil red O working solution for 15 min, dipped in 60% isopropanol once quickly, dipped in DI water once quickly, counterstained with Mayer’s hematoxylin for 3 min, dipped in DI water ten times, and then mounted with DAKKO mounting media.
Adipocyte quantification
Mouse knee joints were fixed in 4% paraformaldehyde, decalcified for 2 weeks in 10% EDTA, and then dehydrated and embedded in paraffin. Sections (7 μm thick) were taken and stained with Safranin-O and Fast Green (Sigma, cat no S8884 and F7252) according to the manufacturer’s instructions. Briefly, slides were deparaffinized and stained with 0.05% Fast Green (FCF) Solution for 1 min, then rinsed with 1% Acetic Acid followed by 30 s in 0.1% Safranin-O Solution and two rinses in water. Slides were the dehydrated and mounted with Permount (Fisher Scientific, cat. No. SP15-100). Adipocyte quantification was performed on two slides from the medial plateau of the tibia of each mouse. Images were opened in ImageJ and Adiposoft was used to quantify adipocyte number.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment unless otherwise noted. For the comparison of two groups, a two-tailed Student’s T test was used to calculate significance. For three or more comparisons, an ordinary one-way ANOVA followed by Dunnett’s or Tukey’s multiple comparisons post hoc test were used to calculate significance unless otherwise noted in figure legend. For data with comparisons involving two independent variables, two-way ANOVA was used. P < 0.05 was considered significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Error bars represent the standard deviation in every figure. Statistical testing for flow cytometry and functional assays was performed using GraphPad Prism. F values, t values, P values, n, and df for each figure are in Supplementary Data 10 and Supplementary Table 3. All measurements were taken from distinct samples. Data are assumed to be normally distributed.
Data anonymization
For five synovial tissue donors (Supplementary Table 1, cohort 2 donors 12–16), genetic variations were removed from raw fastq files using BAMboozle (v0.5.0)43. Sanitized fastq files are available on the database of Genotypes and Phenotypes (dbGaP). For the adipose tissue donors (Supplementary Table 2), genetic variations were removed from raw FASTQ files using BAMboozle and sanitized files are available on GEO.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Source data
Acknowledgements
The authors thank the Accelerating Medicines Partnership RA/SLE Network for the generation of the osteoarthritis and rheumatoid arthritis synovial scRNA-seq data. The authors thank Brigham and Women’s Hospital (BWH) Center for Cellular Profiling: Cytometry and Single-Cell Multi-omics Core for assistance with cell sorting and single-cell transcriptomic data collection. We thank the Broad Genomics Platform for bulk sequencing services. The authors thank the Human Skin Disease Resource Center of Brigham and Women’s Hospital which collected anonymous, discarded adult skin plus fat samples and provided them to our lab. The authors thank the Gift of Hope Organ & Tissue Donor Network, IL, as well as the donors’ families, for the precious tissue samples. The authors thank Peter Nigrovic for serum derived from K/BxN mice. The authors thank the National Disease Research Interchange which collected post-mortem healthy synovial tissue. The authors thank the National Institutes of Arthritis and Musculoskeletal and Skin Diseases, T32AR007530-37, Immunologic Mechanisms and Rheumatic Disease (MBB), National Institutes of Arthritis and Musculoskeletal and Skin Diseases, P30 AR070253, Microgrant (HJF) and Arthritis National Research Foundation (HJF) for their generous funding. This work was supported by the Accelerating Medicines Partnership® Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP® RA/SLE) Network (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Foundation for the National Institutes of Health, GlaxoSmithKline, Janssen Research and Development, LLC, Lupus Foundation of America, Lupus Research Alliance, Merck & Co., Inc. Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development. Funding was provided through grants from the National Institutes of Health (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2- AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, and UM2- AR067678).
Author contributions
Conceptualization: H.J.F. and M.B.B. Methodology: H.J.F., T.C., G.F.M.W., I.K., S.K., and S.G. Funding acquisition: H.J.F. and M.B.B. Sample acquisition: D.P.S., W.V.T., L.L., R.P., S.C., K.W., A.H.J., S.R., and F.Z. Supervision: M.B.B. and D.B.M. Writing—original draft: H.J.F. and M.B.B. Writing—review and editing: H.J.F., M.B.B., T.C., D.B.M., A.H.J., and S.R.
Peer review
Peer review information
Nature Communications thanks Adam Croft, Huiyong Yin and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
Raw lipidomic data is available on Zenodo at https://zenodo.org/records/13284701 (10.5281/zenodo.13284701). The FASTQ files for all synovial single-cell and bulk RNA sequencing data generated in this study have been deposited in the dbGAP database under accession number phs003304.v1.p1, available at the following link: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003304.v1.p1. FASTQ data is available under restricted access because this data was generated with NIH funding and falls under NIH guidelines on human genomic data. Access can be obtained by a permanent senior scientific scientist at an institution by submitting a data access request to dbGAP through the eRA commons by clicking on “Apply for Controlled Access Data” and completing the required documentation. Questions regarding accessing data can be directed to dbgap-sp-help@ncbi.nlm.nih.gov. The raw FASTQ data is protected and is not available due to data privacy laws. dbGAP will grant access to data for one year. This data falls under the health/medical/biomedical data use limitation. The raw, sanitized FASTQ files for adipose tissue single-cell RNA sequencing generated in this study have been deposited in the GEO database under accession number GSE233500. The processed count data for synovial and adipose single-cell and bulk RNA sequencing count matrices generated in this study have been deposited in the GEO database under super series number GSE233504. Within this super series, Adipose single-cell RNA sequencing is available under accession number GSE233500, fibroblast bulk RNA sequencing is available at GSE233501, and synovium single-cell RNA sequencing is available at GSE233502. The flow, qPCR, mouse measurements, and all other data generated in this study are provided in the Supplementary Information/Source Data file. The results published here are in whole or in part based on data obtained from the ARK Portal (arkportal.synapse.org). Source data are provided with this paper.
Code availability
Code to reproduce analyses is available on Zenodo available at 10.5281/zenodo.7942859.
Competing interests
M.B.B. serves on the scientific advisory board for GlaxoSmithKline and as a consultant for Moderna and 4FO ventures. Consulting relates to programs specific to each company and does not directly relate to the research in this report. MBB is the founder of Mestag Therapeutics which is focused on fibroblast-mediated pathology but not directly related to the research in this report. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
A full list of members and their affiliations appears in the Supplementary Information.
Contributor Information
Michael B. Brenner, Email: mbrenner@bwh.harvard.edu
Accelerating Medicines Partnership: RA/SLE Network:
Jennifer Albrecht, Jennifer H. Anolik, William Apruzzese, Jennifer L. Barnas, Joan M. Bathon, Ami Ben-Artzi, Brendan F. Boyce, David L. Boyle, S. Louis Bridges, Jr., Vivian P. Bykerk, Debbie Campbell, Arnold Ceponis, Adam Chicoine, Michelle Curtis, Kevin D. Deane, Edward DiCarlo, Laura T. Donlin, Patrick Dunn, Andrew Filer, Hayley Carr, Gary S. Firestein, Lindsy Forbess, Laura Geraldino-Pardilla, Susan M. Goodman, Ellen M. Gravallese, Deepak Rao, Peter K. Gregersen, Joel M. Guthridge, Maria Gutierrez-Arcelus, V. Michael Holers, Diane Horowitz, Laura B. Hughes, Lionel B. Ivashkiv, Kazuyoshi Ishigaki, Judith A. James, Joyce B. Kang, Gregory Keras, Amit Lakhanpal, James A. Lederer, Miles J. Lewis, Yuhong Li, Katherine Liao, Arthur M. Mandelin, II, Ian Mantel, Kathryne E. Marks, Mark Maybury, Andrew McDavid, Mandy J. McGeachy, Joseph R. Mears, Nida Meednu, Nghia Millard, Larry Moreland, Saba Nayar, Alessandra Nerviani, Dana E. Orange, Harris Perlman, Costantino Pitzalis, Javier Rangel-Moreno, Karim Raza, Yakir Reshef, Christopher Ritchlin, Felice Rivellese, William H. Robinson, Laurie Rumker, Ilfita Sahbudin, Saori Sakaue, Jennifer A. Seifert, Dagmar Scheel-Toellner, Anvita Singaraju, Kamil Slowikowski, Melanie Smith, Darren Tabechian, Paul J. Utz, Kathryn Weinand, Dana Weisenfeld, Michael H. Weisman, Qian Xiao, Zhu Zhu, Zhihan J. Li, Andrew Cordle, and Aaron Wyse
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52586-x.
References
- 1.Hitchon, C. A. & El-Gabalawy, H. S. The synovium in rheumatoid arthritis. Open Rheumatol. J.5, 107–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang, F. et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol.20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith, M. D. The normal synovium. Open Rheumatol. J.5, 100–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Favero, M. et al. Infrapatellar fat pad features in osteoarthritis: a histopathological and molecular study. Rheumatology56, 1784–1793 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Kitagawa, T. et al. Histopathological study of the infrapatellar fat pad in the rat model of patellar tendinopathy: a basic study. Knee26, 14–19 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Greif, D. N. et al. Infrapatellar fat pad/synovium complex in early-stage knee osteoarthritis: potential new target and source of therapeutic mesenchymal stem/stromal cells. Front. Bioeng. Biotechnol.8, 860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlgruber, A. C. et al. gammadelta T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol.19, 464–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, F. et al. Deconstruction of rheumatoid arthritis synovium defines inflammatory subtypes. Nature623, 616–624 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reshef, Y. A. et al. Co-varying neighborhood analysis identifies cell populations associated with phenotypes of interest from single-cell transcriptomics. Nat. Biotechnol.40, 355–363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma, S. et al. Clinical significance of metallothioneins in cell therapy and nanomedicine. Int. J. Nanomed.8, 1477–1488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alivernini, S. et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat. Med.26, 1295–1306 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Rassart, E. et al. Apolipoprotein D. Gene756, 144874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia, R. et al. NNMT is induced dynamically during beige adipogenesis in adipose tissues depot-specific manner. J. Physiol. Biochem78, 169–183 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Roberti, A., Fernandez, A. F. & Fraga, M. F. Nicotinamide N-methyltransferase: at the crossroads between cellular metabolism and epigenetic regulation. Mol. Metab.45, 101165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hishida, T. et al. The role of C/EBPdelta in the early stages of adipogenesis. Biochimie91, 654–657 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Yew Tan, C. et al. Adipose tissue fatty acid chain length and mono-unsaturation increases with obesity and insulin resistance. Sci. Rep.5, 18366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Layre, E. et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol.18, 1537–1549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roderick, J. E. et al. Prostaglandin E2 stimulates cAMP signaling and resensitizes human leukemia cells to glucocorticoid-induced cell death. Blood137, 500–512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwick, R. K. et al. Anatomical, physiological, and functional diversity of adipose tissue. Cell Metab.27, 68–83 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumann, E. et al. Adipokines and autoimmunity in inflammatory arthritis. Cells10, 216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y. et al. Fat-produced adipsin regulates inflammatory arthritis. Cell Rep.27, 2809–2816.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature593, 575–579 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Wei, K. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature582, 259–264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armaka, M. et al. Single-cell multimodal analysis identifies common regulatory programs in synovial fibroblasts of rheumatoid arthritis patients and modeled TNF-driven arthritis. Genome Med.14, 78 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature603, 926–933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijay, J. et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab.2, 97–109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrick, D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science364, eaav2501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregoire, F. M., Smas, C. M. & Sul, H. S. Understanding adipocyte differentiation. Physiol. Rev.78, 783–809 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Schwalie, P. C. et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature559, 103–108 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Bauerle, K. T. et al. Glucocorticoid receptor signaling is not required for in vivo adipogenesis. Endocrinology159, 2050–2061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korsunsky, I. et al. Cross-tissue, single-cell stromal atlas identifies shared pathological fibroblast phenotypes in four chronic inflammatory diseases. Med3, 481–518.e14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croft, A. P. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature570, 246–251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardy, R. S. et al. 11 Beta-hydroxysteroid dehydrogenase type 1 regulates synovitis, joint destruction, and systemic bone loss in chronic polyarthritis. J. Autoimmun.92, 104–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koenen, M. et al. Glucocorticoid receptor in stromal cells is essential for glucocorticoid-mediated suppression of inflammation in arthritis. Ann. Rheum. Dis.77, 1610–1618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satija, R. et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol.33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang, J. B. et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat. Commun.12, 5890 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picelli, S. Full-length single-cell RNA sequencing with Smart-seq2. Methods Mol. Biol.1979, 25–44 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol.37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 40.van ‘t Klooster, J. S. et al. Periprotein lipidomes of Saccharomyces cerevisiae provide a flexible environment for conformational changes of membrane proteins. eLife9, e57003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, C. A. et al. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem.78, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Lee, D. M. et al. Cadherin-11 in synovial lining formation and pathology in arthritis. Science315, 1006–1010 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Ziegenhain, C. & Sandberg, R. BAMboozle removes genetic variation from human sequence data for open data sharing. Nat. Commun.12, 6216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
Raw lipidomic data is available on Zenodo at https://zenodo.org/records/13284701 (10.5281/zenodo.13284701). The FASTQ files for all synovial single-cell and bulk RNA sequencing data generated in this study have been deposited in the dbGAP database under accession number phs003304.v1.p1, available at the following link: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003304.v1.p1. FASTQ data is available under restricted access because this data was generated with NIH funding and falls under NIH guidelines on human genomic data. Access can be obtained by a permanent senior scientific scientist at an institution by submitting a data access request to dbGAP through the eRA commons by clicking on “Apply for Controlled Access Data” and completing the required documentation. Questions regarding accessing data can be directed to dbgap-sp-help@ncbi.nlm.nih.gov. The raw FASTQ data is protected and is not available due to data privacy laws. dbGAP will grant access to data for one year. This data falls under the health/medical/biomedical data use limitation. The raw, sanitized FASTQ files for adipose tissue single-cell RNA sequencing generated in this study have been deposited in the GEO database under accession number GSE233500. The processed count data for synovial and adipose single-cell and bulk RNA sequencing count matrices generated in this study have been deposited in the GEO database under super series number GSE233504. Within this super series, Adipose single-cell RNA sequencing is available under accession number GSE233500, fibroblast bulk RNA sequencing is available at GSE233501, and synovium single-cell RNA sequencing is available at GSE233502. The flow, qPCR, mouse measurements, and all other data generated in this study are provided in the Supplementary Information/Source Data file. The results published here are in whole or in part based on data obtained from the ARK Portal (arkportal.synapse.org). Source data are provided with this paper.
Code to reproduce analyses is available on Zenodo available at 10.5281/zenodo.7942859.