Abstract
Transition of cytosine to thymine in CpG dinucleotides is the most frequent type of mutation in cancer. This increased mutability is commonly attributed to the spontaneous deamination of 5-methylcytosine (5mC), which is normally repaired by the base-excision repair (BER) pathway. However, the contribution of 5mC deamination in the increasing diversity of cancer mutational signatures remains poorly explored. We integrate mutational signatures analysis in a large series of tumor whole genomes with lineage-specific epigenomic data to draw a detailed view of 5mC deamination in cancer. We uncover tumor type-specific patterns of 5mC deamination signatures in CpG and non-CpG contexts. We demonstrate that the BER glycosylase MBD4 preferentially binds to active chromatin and early replicating DNA, which correlates with lower mutational burden in these domains. We validate our findings by modeling BER deficiencies in isogenic cell models. Here, we establish MBD4 as the main actor responsible for 5mC deamination repair in humans.
Subject terms: Cancer genomics, Base excision repair
Multiple types of DNA damage can lead to mutations in normal cells, ultimately contributing to the development of cancer. Here, the authors redefine the spectrum of mutational signatures linked to a particular type of DNA damage to uncover the protective role of specialized DNA repair mechanisms.
Introduction
Cancer genomes are marked by distinctive patterns of somatic mutations termed mutational signatures, which reflect the combination of mutational processes that occurred during development and tumorigenesis1–3. These signatures may have diagnostic and therapeutic values as they offer insights into DNA repair defects in the tumors4. One way to deduce mutational signatures is through the analysis of the relative contributions of single-base substitution (SBS), considering the nucleotides immediately 5′ and 3′ to the mutation. In recent years, analyzes of whole genome sequencing (WGS) from increasingly large series of cancer genomes allowed the identification of common SBS signatures found in many samples, SBS signatures restricted to specific tumor types, and rare SBS signatures found in few samples5–8.
Mutational signature SBS1 is frequent in cancer and correlates with patient age at diagnosis in multiple tumor types, being referred to as age/clock-like mutational signature9. SBS1 was the first signature to be primarily attributed to deamination damage in methylated DNA sites1. 5-methylcytosines (5mC) that undergo spontaneous hydrolytic deamination are converted into thymine, leading to G:T mispairs in DNA. Failure to repair G:T mispairs by the base-excision repair (BER) machinery prior to DNA replication results in the fixation of C > T transitions in one of the daughter cells. DNA methylation is mostly found on cytosines within CpG dinucleotides10, and thus C > T transitions driven by 5mC deamination preferentially accumulate at CpGs (CpG > TpG mutations).
In humans, two BER glycosylases are implicated in the repair of G:T mispairs, MBD4 (Methyl-CpG Binding Domain Protein 4) and TDG (Thymine DNA Glycosylase)11,12. MBD4 is the only member of the methyl-CpG-binding domain (MBD) nuclear protein family with G:T glycosylase activity13. The glycosylase activities of MBD4 and TDG have been extensively investigated in biochemical studies14–18, but their respective roles in protecting the complete human genome from 5mC deamination remain largely uncharacterized.
Tumors harboring biallelic inactivation of MBD4 show a hypermutation profile corresponding almost exclusively to CpG > TpG mutations (common SBS1 and/or rare SBS96 signatures)5,19–24. Noteworthy, MBD4 expression has been suggested to correlate with SBS1 exposure25. Additional rare signatures with a high frequency of mutations at CpGs include SBS95 and SBS105, which have been suggested to also arise from 5mC deamination damage and/or defective repair5. However, the mechanisms responsible for these rare signatures and their relationship with DNA methylation are currently unknown.
Here, we aim to understand the origin and genome-wide distribution of SBS signatures potentially linked to 5mC deamination. We comprehensively dissected these signatures according to cell lineage-specific DNA methylation patterns, the epigenomic context, replication timing, and strand bias. By knocking out MBD4, TDG, or both in isogenic cell lines, we offer insights into their respective roles in protecting the human genome from 5mC deamination.
Results
Refining the spectrum of CpG mutational signatures
To clarify the actual contribution of 5mC deamination in tumor mutation burden, we screened tumor whole genomes for common and rare SBS signatures distinctive by a high frequency of cytosine substitutions at CpG dinucleotides (Fig. 1a). We obtained mutational signature exposures with Signature Fit Multi-Step (FitMS)5 in whole genomes from a collection of two in-house and four previously described MBD4-deficient (MBD4def) tumors23,26, in addition to whole genomes from the Genomics England pan-cancer series (GEL series release data v17; 12,726 high-quality tumor samples from 11,817 cases)27. Our complete dataset included a total of 12,732 tumor genomes analyzed from 11,823 cases (for detailed information, see “Methods”; Supplementary Data 1–6).
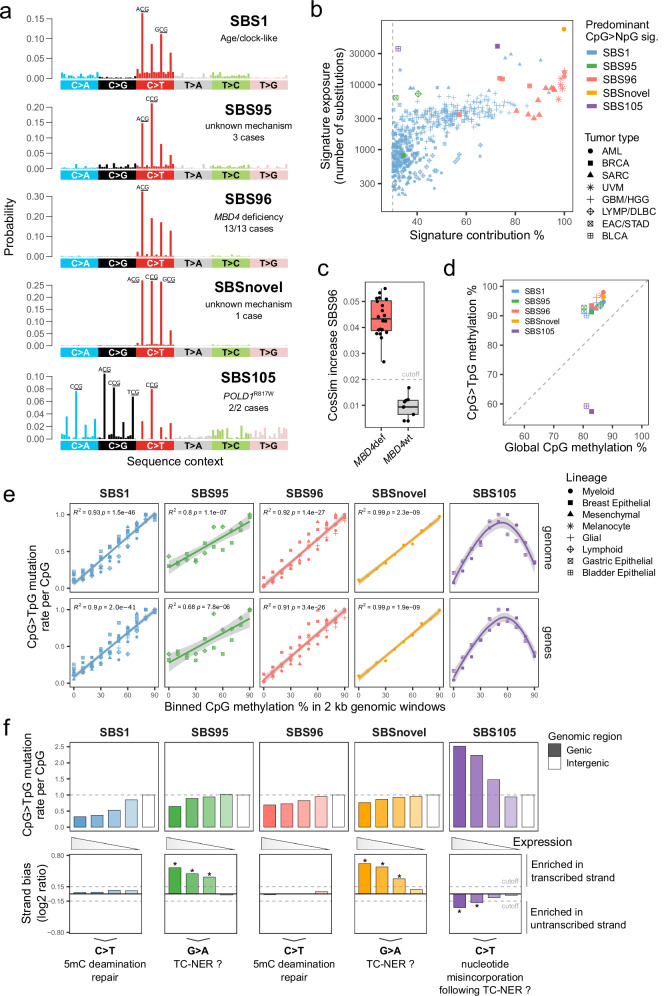
Fig. 1. Refining the spectrum of CpG mutational signatures and their dependence on 5mC deamination.
a Substitution profiles by trinucleotide sequence context (96-channel) of SBS reference mutational signatures characterized by a high frequency of CpG>NpG substitutions. The most frequent substitutions per signature are indicated. b Scatter plot of exposures to predominant CpG>NpG mutational signature found per tumor sample, as absolute exposure versus percent exposure contribution. The dashed line indicates the 30% contribution cutoff used to select SBS1 samples. AML, acute myeloid leukemia; BRCA, breast invasive carcinoma; SARC, sarcoma; UVM, uveal melanoma; GBM, glioblastoma multiforme; HHG, high-grade glioma; LYMP, lymphoid neoplasm; DLBC, diffuse large B-cell lymphoma; EAC, esophageal adenocarcinoma; STAD, stomach adenocarcinoma; BLCA, bladder urothelial carcinoma. c Distributions of cosine similarity increase for signature fitting with rare SBS96 compared to common signatures only, per tumor sample. MBD4def, MBD4-deficient (n = 20); MBD4wt, MBD4 wild-type (n = 9). The dashed line indicates the standard cutoff of FitMS in cosine similarity increase multistep mode. Boxes indicate the median, 25th and 75th percentiles. Whiskers extend to the largest or lowest value up to 1.5 times the distance between the 25th and 75th percentiles. d Scatter plot of DNA methylation percentages in CpG > TpG mutated sites versus all CpGs (global), per signature and cell lineage. Methylation was interrogated in data from normal human cell types. The dashed line indicates the absence of over- or under-representation of methylation in mutated CpGs. e Scatter plots of CpG > TpG mutation rates per CpG of different tumor types and signatures in 2 kb genomic windows grouped by their mean CpG methylation levels. Mutation rates were normalized by the highest value in each tumor type. The lines indicate data fitting with linear regression models or smoothed conditional means models. Two-sided Pearson correlation statistics are shown. Shaded areas represent the 95% confidence intervals. f Bar plots of CpG > TpG mutation rates per CpG in genic or intergenic regions (upper panel). Transcriptional strand asymmetry of CpG > TpG mutations in genic regions (lower panel). Genes were grouped based on expression level quartiles. Asterisks mark a significant difference in contribution between transcribed and untranscribed strands (see “Methods”). The dashed line indicates the cutoff used to assign significance. Source data are provided as a Source Data file.
We first focused on samples with rare mutational signatures previously associated with defective 5mC deamination repair (SBS96) and mutational signatures of unknown causes characterized by a high proportion of CpG > TpG mutations (SBS95) or CpG > NpG mutations (SBS105) (Fig. 1a,b). In the GEL series, SBS95 was found in 0.025% of cases (3 out of 11,817), SBS96 was found in 0.059% of cases (7 out of 11,817), and SBS105 was found in 0.017% of cases (2 out of 11,817). An additional secondary acute myeloid leukemia (AML) from GEL (1 out of 160 AML cases), not reported in Degasperi et al.5, was initially identified with pure SBS96, but with cosine similarity lower than other SBS96 samples (0.947 vs. mean 0.996 ± 0.0014). Moreover, the proportion of substitution peaks differed from SBS96, with C > T peaks of similar heights in ACG, CCG, and GCG contexts. Therefore, we suggest that a previously unreported rare SBS signature may be responsible for this unique mutational profile, here referred to as SBSnovel (Fig. 1a; Supplementary Data 7).
All other tumor samples with SBS96 contribution were MBD4def (20 tumor samples from 13 cases; 7 cases from GEL and 6 cases outside GEL). Moreover, SBS96 represented the predominant signature in all MBD4def samples analyzed (Fig. 1b). MBD4 deficiency was most often acquired in patients harboring heterozygous germline loss-of-function mutations in MBD4, with somatic loss of the wild-type allele in the tumor (11/13 cases, including four uveal melanomas [UVM], three breast invasive carcinomas [BRCA], three sarcomas [SARC] and one high-grade glioma [HGG]). The only exceptions were two AML cases previously described in siblings harboring germline biallelic loss of MBD423. In agreement with findings by Degasperi et al.5, SBS96 was also selected by FitMS as a candidate rare signature in a small number of MBD4 wild-type (MBD4wt) tumors harboring a high proportion of CpG > TpG mutations. However, SBS96 attribution to these samples had low confidence, with cosine similarity increase values below the standard cutoff of FitMS (Fig. 1c; see “Methods”). The mutational profiles of these MBD4wt tumors could be well explained by a high contribution of the common 5mC deamination signature SBS1 instead (Supplementary Data 8). Overall, we establish that SBS96 is intrinsically linked to MBD4 deficiency in tumors of multiple tissue origins.
None of the samples with SBS95, SBS105, or SBSnovel showed biallelic inactivation of either MBD4 or TDG. We then further screened these cases for recurrent germline or somatic mutations in genes linked to DNA replication or repair. No recurrent variant of interest was found in cases harboring SBS95, suggesting this signature might be caused by exogenous factors. SBSnovel was found in a single secondary AML sample, and we cannot rule out it might be caused by intensive chemotherapy exposure. The two SBS105 cases harbored an identical somatic heterozygous R817W mutation in DNA polymerase delta 1 catalytic subunit (POLD1R817W; isoform NP_001243778.1; hg38 position chr19:50414875 C > T), potentially suggesting a role of nucleotide misincorporation in SBS105.
In samples identified with SBS95, SBS96, SBS105, or SBSnovel, more than 30% of the somatic variants were attributed to these rare signatures. SBSnovel showed the highest number of variants assigned to a single sample (71,314), followed by SBS105 (38,890 and 35,678) and SBS96 (mean 8,628 ± 4,348) (Fig. 1b; Supplementary Fig. 1). To compare these rare signatures with the common signature SBS1, we selected additional tumors with at least 30% contribution of SBS1 from either the same or related tumor types (Fig. 1b; Supplementary Data 6). Based on the relative contributions of all signatures present in each sample, we then selected variants with a high probability of originating from each signature. Our final working dataset included 1,054,633 somatic variants (837,106 variants in a CpG context) from 516 tumors originating from 8 distinct cell lineages (myeloid, mesenchymal, glial, and so forth; see Supplementary Data 9) and driven by diverse oncogenic mechanisms.
SBS1 and SBS96 are the sole CpG SBS primarily linked to 5mC deamination
To investigate the role of 5mC deamination in CpG mutagenesis of the different SBS signatures, we analyzed base resolution whole genome DNA methylation data on normal human cells corresponding to each of the 8 cell lineages of interest (Supplementary Data 10), including data that we generated from normal uveal melanocytes (Supplementary Fig. 2). We observed that CpG>TpG substitutions of SBS1, SBS95, SBS96 and SBSnovel were preferentially acquired in CpGs that were methylated in the corresponding normal cell type. In contrast, mutated CpGs in SBS105 showed strong underrepresentation of DNA methylation (Fig. 1d), which was similarly observed for C > A, C > G and C > T substitutions (Supplementary Fig. 3a). To better characterize the genomic distribution of CpG mutations in relation to DNA methylation patterns, we calculated mutation rates in non-overlapping 2 kb genomic bins categorized by their CpG methylation levels. While SBS1, SBS96, and SBSnovel mutation rates were almost completely dependent on the genomic distribution of CpG methylation, this effect was only partial for SBS95. Interestingly, SBS105 mutation rates showed a non-linear relationship with CpG methylation levels, with the highest mutation rates in partially methylated regions (Fig. 1e; Supplementary Fig. 3b). Altogether, our data strongly suggests that 5mC deamination does not play a major role in SBS105.
Signatures SBS95 and SBSnovel showed transcription strand asymmetry, with higher CpG > TpG mutagenesis in the transcribed strand. Strand asymmetry was dependent on gene expression levels, being strongest in highly expressed genes and absent in lowly expressed genes (Fig. 1f; Supplementary Data 11). Transcription strand asymmetry is generally attributable to transcription-coupled nucleotide excision repair (TC-NER) and/or to higher exposure to damage of single-stranded DNA28. Both mechanisms result in higher mutation rates in the untranscribed strand. First, this indicates that SBS95 and SBSnovel are most probably caused by preferential G > A mutagenesis in the untranscribed strand instead of C > T transitions caused by 5mC deamination. It is therefore possible that certain types of DNA damage directly targeting guanines are involved in these signatures. Second, SBS95 and SBSnovel showed lower mutation rates in highly expressed genes than in lowly expressed genes or intergenic regions (Fig. 1f), which is consistent with a potential role of DNA damage repair by TC-NER in these signatures. To our knowledge, 5mC deamination repair has not been described to be directly coupled with the transcription machinery. This is consistent with the absence of transcription strand asymmetry observed in SBS1 and SBS96. Hence, we propose that the genomic distribution of SBS95 and SBSnovel CpG mutations cannot be fully explained by 5mC deamination alone.
SBS105 showed a distinct pattern from all the remaining signatures, with the highest mutation rates in highly expressed genes and transcription strand asymmetry towards the untranscribed strand (Fig. 1f; Supplementary Fig. 3c; Supplementary Data 11). This points to C > N mutagenesis which might arise from nucleotide misincorporation, at least partially following TC-NER. Overall, we show evidence that 5mC deamination is unlikely to play a major role in signatures SBS95, SBSnovel, and SBS105. The association of SBS95 and SBSnovel with CpG methylation remains to be explained. Finally, SBS1 and SBS96 represent the sole signatures with features fully consistent with a primary role of mutagenesis caused by 5mC deamination.
SBS105 is associated with POLD1R817W mutation
We next sought to explore in better detail the association between POLD1R817W mutation and SBS105, both of which were found in one bladder urothelial carcinoma (BLCA) and one breast invasive carcinoma (BRCA). In the BLCA case, POLD1R817W showed a low normalized allele frequency (normVAF=0.23; 9 alternative allele reads). We found no evidence of copy number gain of the wild-type allele in this sample, indicating that POLD1R817W was probably subclonal. Accordingly, SBS105 showed the highest percent contribution among subclonal variants in this sample (normVAF between 0.15 and 0.35). In the BRCA case, POLD1R817W and SBS105 were equally clonal (Supplementary Fig. 4). Out of the remaining 12,724 tumor samples in GEL, POLD1R817W was found in a single endometrial carcinoma carrying the pathogenic P286R variant in POLE and an overwhelming contribution of signatures SBS10a and SBS1 (924,213 and 262,647 mutations, respectively). This accounts for a mutational burden ~32 times higher than the average observed for SBS105, potentially masking any substantial contribution of CpG > NpG substitutions characteristic of signature SBS105 in this sample. Finally, POLD1R817W was not observed in any tumor samples from TCGA or MSK-IMPACT (https://www.cbioportal.org/). Noteworthy, this somatic variant corresponds to a CpG > TpG mutation, which we cannot exclude is a consequence of SBS105 mutagenesis. Nevertheless, we provide evidence supporting a role of the rare somatic variant POLD1R817W in the rare mutational signature SBS105.
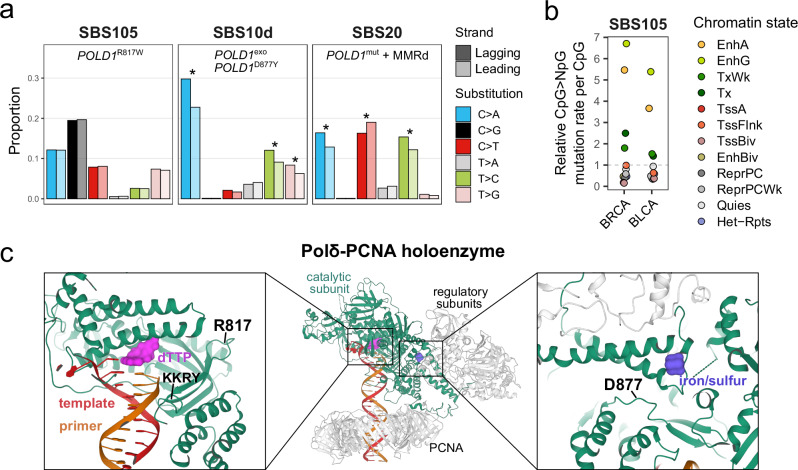
High-fidelity replication of the nuclear genome is dependent on post-replicative mismatch repair (MMR) activity and the proofreading capacity of DNA polymerases, including Pol δ (catalytic subunit coded by POLD1). Deleterious alterations in POLD1 have been associated with hypermutator phenotypes and mutational signatures SBS10d (polymerase domain mutation D877Y or exonuclease domain mutations) and SBS20 (POLD1 mutations with MMR deficiency)5,29,30. It was previously observed that Pol δ and MMR dysfunctions result in asymmetric patterns of mismatches introduced during leading- vs. lagging-strand synthesis29,31. We confirmed a strong replicative strand asymmetry in SBS10d and SBS20, but we did not observe asymmetry in any of the substitution classes of SBS105 (Fig. 2a; Supplementary Data 12). This suggests that, unlike most pathogenic POLD1 mutations, POLD1R817W has a minimal impact on general S-phase DNA replication. Moreover, SBS105 showed transcription strand asymmetry (Fig. 1f; Supplementary Fig. 3c), in addition to enrichment of mutations at CpGs located in active/genic enhancers and transcribed chromatin states (Fig. 2b). Considering that Pol δ is important for multiple forms of DNA repair32, it is possible that POLD1R817W leads to generic nucleotide misincorporation following DNA repair pathways more frequently employed in active genic features, including but not restricted to TC-NER.
Fig. 2. Features of SBS mutational signatures associated with distinct POLD1 mutations.
a Replication strand asymmetry of mutational signatures associated with POLD1 mutations. The asterisks mark a significant difference in contribution between leading and lagging strands (see “Methods”). POLD1exo*, POLD1 exonuclease domain mutated; POLD1mut, POLD1 mutated; MMRd, mismatch repair deficiency. b SBS105 CpG > NpG relative mutation rates in ENCODE chromatin states of normal cell types matched to each tumor type. BRCA, breast invasive carcinoma; BLCA, bladder urothelial carcinoma; EnhA/EnhG, active/genic enhancer; Tx/TxWk, strong/weak transcription; TssA, active TSS; TssFlnk, flanking TSS; TssBiv, bivalent/poised TSS; EnhBiv, bivalent enhancer; ReprPC/ReprPCWk, strong/weak repressed polycomb; Quies, quiescent/low; Het-Rpts, heterochromatin/ZNF genes and repeats. c Pol δ-PCNA holoenzyme structure. Mutated amino acids in the polymerase domain of Pol δ catalytic subunit (POLD1) are indicated in black. Source data are provided as a Source Data file.
The balance between exonuclease and polymerase activities of B-family DNA polymerases promotes DNA synthesis when nucleotides are correctly added to the new strand. Misincorporations shift the balance toward the exonuclease domain until the incorrect nucleotides have been removed33. To gain deeper mechanistic insight into POLD1 polymerase domain mutations associated with disparate mutational signatures, we analyzed a previously reported structure of the processive human Pol δ holoenzyme34. Interestingly, the POLD1R817W mutation localizes within the polymerase domain and is close to the highly conserved KKRY motif of B-family DNA polymerases (Fig. 2c), which is important for stabilizing the 3′-terminus of the DNA within the polymerase active site and carrying out processive DNA synthesis35. Hence, we could speculate that tertiary changes of the KKRY motif by R817W may impair the recognition of misincorporated bases in certain sequence contexts. In contrast, the polymerase domain mutation D877Y is in tridimensional proximity to the iron/sulfur cluster of Pol δ (Fig. 2c), which is essential to its exonucleolytic activity36. Hence, similarly to exonuclease domain mutations, the D877Y mutation may lead to Pol δ proofreading defects. Overall, we show that the tridimensional positions of different POLD1 polymerase domain mutations are consistent with the mutational signatures with which they are associated.
SBS1 and SBS96 recapitulate cell lineage CpG methylation landscapes
While tissue-specific features have been documented for multiple common mutational signatures37, their contribution to rare signatures has remained elusive. We then wondered whether 5mC deamination signatures SBS1 and SBS96 found in different tumor types would follow lineage-specific DNA methylation programs. Due to the regional nature of methylation38, we performed our analysis at the level of DNA methylation blocks, which span highly correlated contiguous CpG sites that covary across cell types39. We selected for analysis 698,467 high-quality methylation blocks overlapping genomic features known to be associated with lineage-specific gene expression regulation, including promoters (27,698 distinct genes), exonic regions (28,148 distinct genes), enhancers and DNase hypersensitive sites (402,977 cis-regulatory elements from ENCODE; see “Methods”). On average, retained blocks spanned a length of ~558 bp and ~11 CpGs. Through pairwise comparisons, we defined blocks that were differentially methylated between normal cell type pairs. We observed that specifically methylated blocks in each normal cell type carried higher CpG > TpG mutation rates in the corresponding tumor type (Fig. 3a; Supplementary Data 13). As an example, SBS96-associated mutations in BRCA were enriched in blocks methylated in normal breast epithelial cells but unmethylated in common myeloid progenitors. Conversely, SBS96-associated mutations in AML were enriched in blocks methylated in common myeloid progenitors but unmethylated in normal breast epithelial cells. A similar pattern emerged for all pairwise cell type comparisons, for both SBS96 and SBS1 (Fig. 3a; Supplementary Fig. 5; Supplementary Data 13). In summary, we show that 5mC deamination signatures SBS1 and SBS96 show tissue specificity that recapitulates lineage-specific CpG methylation patterns.
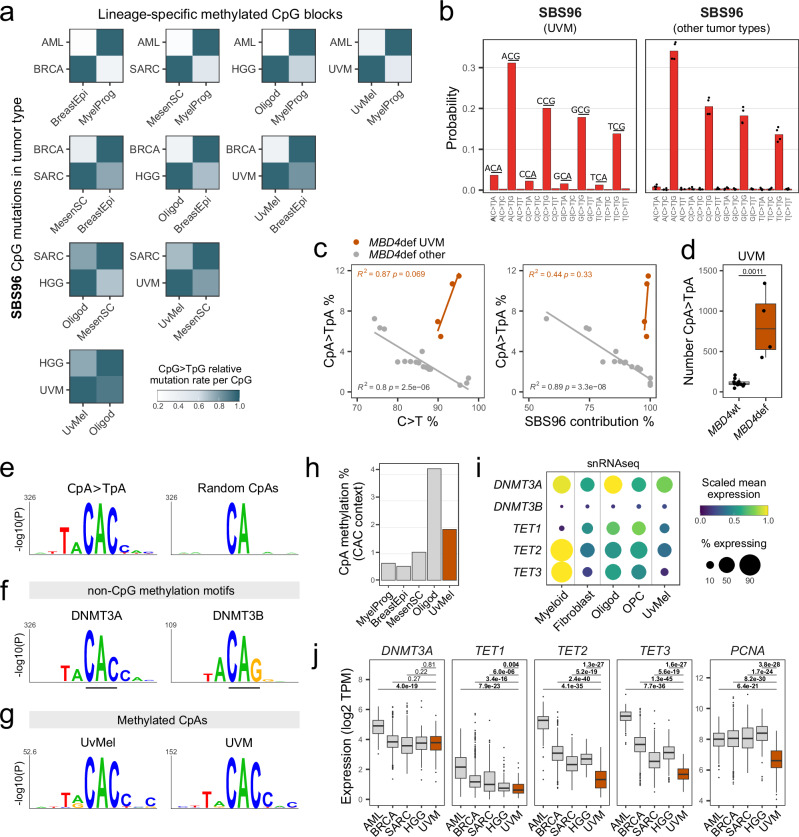
Fig. 3. SBS96 recapitulates lineage-specific CpG and non-CpG methylation landscapes.
a Heatmaps of SBS96 relative CpG > TpG mutation rates in CpG blocks differentially methylated between normal cell type pairs. Hypermethylated blocks are indicated in the x-axis, and tumor types in the y-axis. Values are normalized per tumor type. b C > T substitution profiles by trinucleotide context of tumor type-specific SBS96. Bars represent means in UVM (n = 1) or other tumor types (n = 4; AML, BRCA, SARC, and HGG). c Scatter plots of CpA>TpA substitution percentages versus all C > T substitutions percentages or SBS96 percentage contribution in MBD4-deficient (MBD4def) tumors. Lines indicate data fitting with linear regression models. Two-sided Pearson correlation statistics are shown. d Distributions of the absolute number of CpA > TpA substitutions in UVM tumors MBD4def (n = 4) or MBD4 wild-type (MBD4wt; n = 12). Two-sided Wilcoxon test P-value is indicated. Boxes indicate the median, 25th and 75th percentiles. Whiskers extend to the largest or lowest value up to 1.5 times the distance between the 25th and 75th percentiles. e Sequence probability logos around CpA > TpA mutated sites in MBD4def UVM and of an equal number of randomly interrogated CpA sites (n = 2823). f Sequence probability logos around top non-CpG methylated sites in Dnmt triple-knockout mouse embryonic stem cells with ectopic reintroduction of Dnmt3a (n = 1000) or Dnmt3b (n = 189). g Sequence probability logos around methylated CpA sites in uveal melanocytes (UvMel) and uveal melanomas (UVM). Logos in panels e-g were generated with kpLogo and Bonferroni corrected P-values are shown. h CpA methylation percentages in CAC context in normal cell types. i Dot plot of single-nuclei RNAseq data of the posterior human eye. Dot size indicates the percentage of nuclei expressing each gene. j Distributions of gene expression in TCGA tumors, including AML (n = 151), BRCA (n = 1231), SARC (n = 265), HGG (n = 175) and UVM (n = 80). Values are expressed as transcripts per million (TPM). Two-sided Wilcoxon test P-values without multiple comparisons adjustment are indicated. Statistics of boxes and whiskers are described above. AML, acute myeloid leukemia; BRCA, breast invasive carcinoma; SARC, sarcoma; HHG, high-grade glioma; MyelProg, common myeloid progenitor; BreastEpi, breast luminal epithelium; MesenSC, mesenchymal stem cell; Oligod, oligodendrocyte; OPC, oligodendrocyte precursor cell. Source data are provided as a Source Data file.
Methylated CpA deamination contributes to SBS96 exclusively in UVM
Surprisingly, another feature of tumor type specificity in SBS96 was observed, with contribution of CpA > TpA mutations solely in MBD4def UVM (Fig. 3b). All MBD4def UVM samples (4/4) showed distinguishable CpA > TpA frequencies (8.6 ± 2.9%) in combination with at least 90% overall C > T frequency and almost pure SBS96, which is indicative of minimal contribution of mutational processes other than 5mC deamination in these samples. In contrast, MBD4def samples of AML, BRCA, SARC and HGG collectively showed negative correlation between CpA > TpA substitution frequencies and SBS96 contribution (Fig. 3c). Furthermore, CpA > TpA mutations were ~7.5-fold more abundant in MBD4def UVM in comparison to MBD4wt UVM of the same choroidal origin (Fig. 3d), indicating that these mutations are inherently linked to MBD4 deficiency and not a common feature of choroidal UVM. Overall, we identified that CpA > TpA mutations represent a tissue-specific feature of SBS96.
C > T substitutions in CpG and CpA contexts shared similar relative frequencies with respect to the nucleotide 5′ of the mutated cytosine (AC > CC > GC > TC; Fig. 3b). We then investigated the local sequence context of CpA > TpA mutated sites in MBD4def UVM and found a strong local enrichment of the motif TACACC. The interrogation of an identical number of random CpA sites revealed no equally significant motif (Fig. 3e). The TACACC sequence has been described as the non-CpG DNA methylation motif of DNA methyltransferase 3 A (DNMT3A), which differs from the DNA methyltransferase 3B (DNMT3B) non-CpG methylation motif TACAGG40,41 (Fig. 3f). Interestingly, the MBD domain of MBD4 has been shown to bind to methylated CpAs, particularly in the CAC context42. Overall, we show that CpA > TpA mutated sites in MBD4def UVM are dependent on DNMT3A-mediated CpA methylation, suggesting they result from defective repair of 5mC deamination.
Non-CpG methylation is enriched in embryonic stem cells, oocytes, neurons, and glial cells, although rare in most differentiated cell types43–46. We wondered whether cells of uveal melanocytic lineage, which have neural crest origin47, accumulate significant levels of non-CpG methylation in specific sequence contexts. Normal uveal melanocytes and three metastatic UVM samples analyzed by whole-genome bisulfite sequencing (WGBS) showed the highest non-CpG methylation in CAC trinucleotides (Supplementary Fig. 6). Methylated CpA sites in both normal and transformed melanocytes showed local enrichment of the DNMT3A non-CpG methylation motif (Fig. 3g). Interestingly, methylated CpGs were ~9-fold more abundant than methylated CpAs in uveal melanocytes, which is consistent with a mutational burden ~9.5-fold higher for CpG > TpG mutations in comparison to CpA > TpA mutations in MBD4def UVM. Overall, our data strongly indicates that, similarly to CpG > TpG mutations, CpA > TpA mutations follow the landscape of DNA methylation.
We then investigated whether the tumor specificity of CpA > TpA mutations could be linked to tissue-specific patterns of CpA methylation. Unlike CpG methylation maintained by DNMT1 hemi-methylation, non-CpG methylation is inherently asymmetrical and passively diluted by cell division45. Active demethylation can also occur due to 5mC oxidation by ten-eleven translocation (TET) enzymes, followed by BER involving TDG48. Steady-state methylation levels are therefore explained by a combination of de novo methylation, as well as passive and active demethylation activities49. We initially compared CpA methylation levels in normal cells matched to each SBS96 tumor type. Uveal melanocytes showed ~2% CpA methylation in CAC contexts, representing a level ~2–4 fold higher than in myeloid progenitors, mesenchymal stem cells, and breast luminal epithelial cells. Although CAC methylation in uveal melanocytes was ~2-fold lower than in oligodendrocytes (Fig. 3h), most HGGs are believed to arise from less differentiated glial progenitors instead of fully differentiated post-mitotic glial cells50. Analysis of single-nuclei RNAseq (snRNAseq) data on the posterior eye51 further revealed that oligodendrocyte precursor cells (OPCs) expressed lower levels of DNMT3A than oligodendrocytes (Fig. 3i), indicating that de novo methylation rates might not be stable throughout glial differentiation. This may explain the absence of CpA > TpA mutations in the single MBD4def HGG available, for which the exact cell-of-origin is unknown. In addition, uveal melanocytes expressed higher levels of DNMT3A and lower levels of TET genes (TET1, TET2, and TET3) than OPCs (Fig. 3i), raising the possibility that uveal melanocytes accumulate higher levels of CpA methylation than certain glial precursors.
We further considered the hypothesis that CpA > TpA mutations might also accumulate during the early stages of transformation. Using TCGA tumor transcriptomic data, we observed significantly lower expression of TET genes and multiple proliferation markers (PCNA, MKI67, and E2F1) in UVM in comparison to other SBS96 tumor types. DNMT3A expression was similar in most tumor types, except for AML (Fig. 3j; Supplementary Fig. 7). Hence, slow rates of passive and active demethylation may allow accumulating significant levels of CpA methylation in UVM cells. Accordingly, CpA methylation was highest in the metastatic UVM sample showing the least evidence of hypomethylation due to cell division in CpGs prone to methylation degradation52 (Supplementary Fig. 6). To our knowledge, the presence of CpA methylation in cells of uveal melanocytic origin had not been previously described. More importantly, our observations link the tumor type specificity of CpA > TpA mutations in SBS96 with the dynamics of non-CpG methylation and demethylation across cell lineages.
Selective pressures lead to SBS96 frequent targeting of driver genes
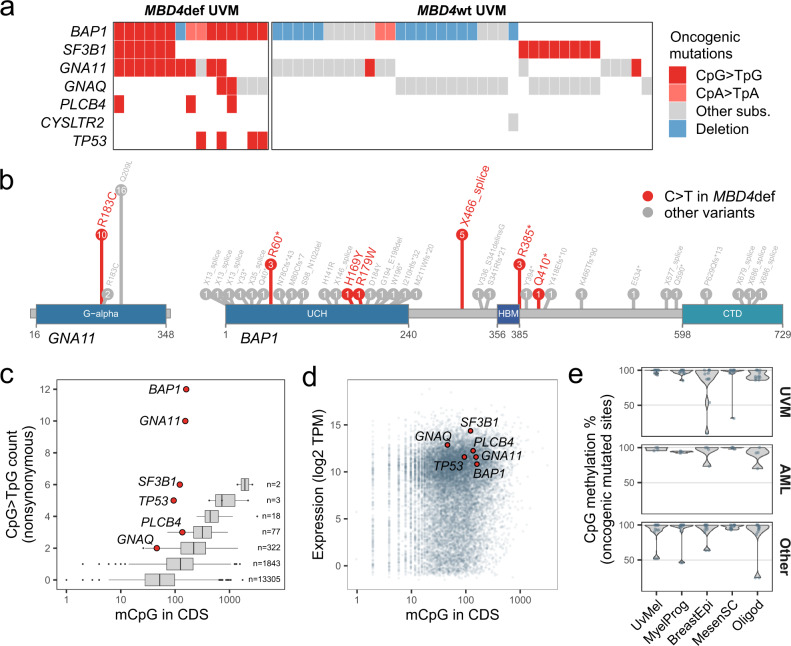
To better define the role of SBS96 in tumor type-specific oncogenesis, we analyzed our WGS series together with whole exome sequencing (WES) data on 116 UVM samples derived from 49 cases, including 12 MBD4def cases. We observed an overrepresentation of oncogenic C > T mutations in MBD4def UVM in comparison to MBD4wt UVM in the key UVM drivers BAP1, SF3B1, GNAQ, GNA11 and PLCB453–58, in addition to TP53 (Fig. 4a). MBD4def UVM showed a unique spectrum of oncogenic mutations: (i) the majority of MBD4def cases (10/15) showed the CpG > TpG GNA11R183C gain-of-function mutation, which is rarely observed in MBD4wt tumors (2/37) (Fig. 4a,b); (ii) we observed the co-occurrence of Gαq pathway-activating CpG > TpG mutations in four MBD4def cases, including GNA11R183C, GNAQR183C and PLCB4D630N (Fig. 4a), probably due to their mild activating effect54; (iii) contrary to MBD4wt UVM, MBD4def UVM showed frequent BAP1 inactivation due to hotspot CpG > TpG mutations, including two nonsense mutations (BAP1R60* and BAP1R385*) and an intronic mutation leading to aberrant splicing (BAP1X466_splice) in matched tumor RNAseq (Fig. 4b; Supplementary Fig. 8); (iv) the frequent co-occurrence of BAP1 inactivation with CpG > TpG SF3B1R625C/H change-of-function mutations (6/15) in MBD4def cases, alterations normally mutually exclusive in MBD4wt tumors59. Interestingly, two MBD4def cases showed CpA>TpA oncogenic mutations in BAP1 (Fig. 4a). Overall, our data strongly indicates that SBS96-related mutations actively contribute to UVM oncogenesis, establishing a unique mutational landscape.
Fig. 4. SBS96 targets tumor-specific driver genes.
a Oncoplot of oncogenic mutations in uveal melanoma (UVM) cases by MBD4 status. MBD4def, MBD4-deficient; MBD4wt, MBD4 wild-type. Tumor samples from the same individual showing identical mutational patterns were combined. b Oncogenic mutations in GNA11 and BAP1 found in UVM tumors. Amino acid positions are derived from mutation positions in the transcript. The values in circles indicate the number of cases harboring each mutation. Protein domains are shown in blue. c Distribution of the number of methylated CpGs (mCpGs) in the coding sequence (CDS) per gene, grouped by the total number of nonsynonymous CpG>TpG mutations observed among MBD4def UVM. Key UVM drivers are shown as red dots and other genes are shown as boxplots. The number of genes per boxplot is shown. Boxes indicate the median, 25th and 75th percentiles. Whiskers extend to the largest or lowest value up to 1.5 times the distance between the 25th and 75th percentiles. d Scatter plot of gene expression values expressed as transcripts per million (TPM) versus the number of mCpGs in the CDS, per gene. Key UVM drivers are shown as red dots. e Distribution of CpG methylation percentages among normal cell types for CpG > TpG oncogenic mutations, separated by tumor type. Source data are provided as a Source Data file.
In MBD4def UVM, genes more frequently targeted by nonsynonymous CpG > TpG mutations showed a higher number of methylated CpGs in the coding sequences (CDS). Driver genes BAP1, GNA11, SF3B1, and TP53 represented outliers with a higher frequency of CpG > TpG mutations than expected by chance (Fig. 4c). This bias could not be traced back to gene expression differences (Fig. 4d), suggesting a prominent role of positive selective pressure for these oncogenic CpG > TpG mutations. A distinct spectrum of CpG>TpG oncogenic mutations was observed in the other MBD4def tumor types, including AML, BRCA, SARC, and HHG. Both AML cases included in our analysis showed DNMT3A missense mutations and hotspot IDH2R140Q mutations23. In SARC cases, we observed stop-gain or splice donor mutations in TP53 (2/3), RB1 (2/3), PTEN (1/3), and NF1 (1/3), in addition to the oncogenic missense mutations PTENR173H (1/3), TP53G245S (1/3) and TSC2R1200W (1/3). In BRCA cases, we observed the oncogenic missense mutation PTENR130Q (1/3), and in the single HHG case we observed a stop-gain mutation in NF1 and the oncogenic missense mutations PTENR173H and TP53R158H. None of the hotspot oncogenic CpG > TpG mutations found in MBD4def UVM were present in other tumor types. Globally, oncogenic mutated sites in the different tumor types were largely methylated in all the normal cell lineages analyzed (Fig. 4e). Hence, these tumor type specificities were most probably driven by positive selective pressure specific to each cell lineage.
MBD4 preferentially protects active and early replicating DNA
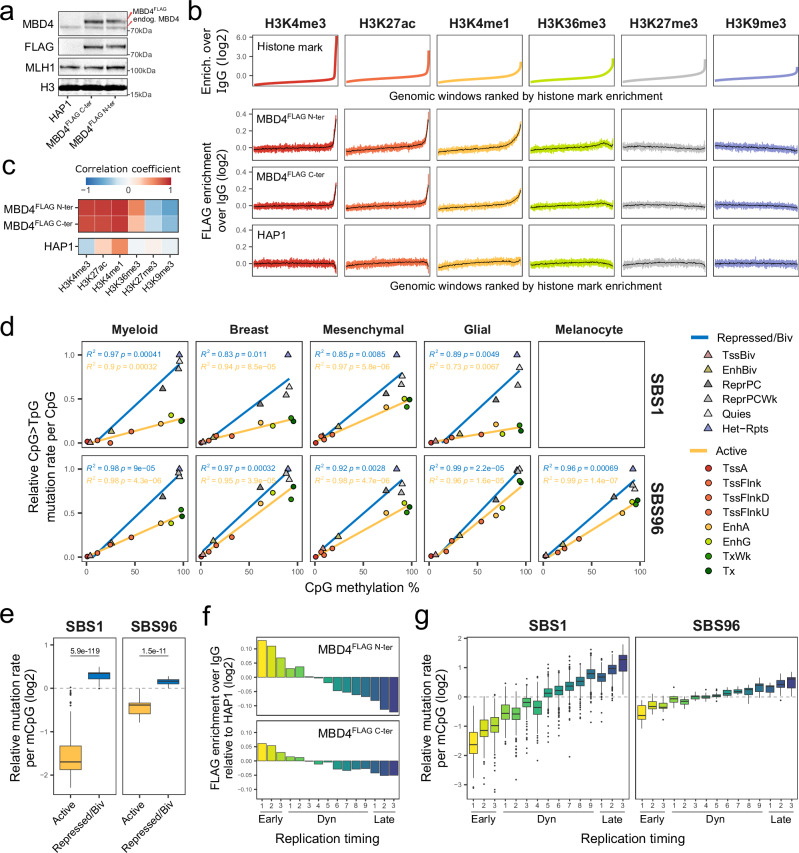
We next wondered whether 5mC deamination repair by MBD4 could shape the distribution of CpG > TpG mutations at the genomic scale. To investigate this, we stably expressed N- and C-terminally FLAG-tagged MBD4 in HAP1 cells, both of which showed predominant nuclear localization (Supplementary Fig. 9a). We then used CUT&RUN to map the genomic binding of tagged MBD4 in single-cell clones showing exogenous expression levels ~3-4 fold higher than endogenous MBD4 (Fig. 5a; Supplementary Fig. 9b,c; Supplementary Data 14). As previously observed for tagged MBD4 ChIP-seq data on mouse embryonic stem cells60, we found that conventional peak calling was largely incompatible with our CUT&RUN data. To comprehend the relationship between tagged MBD4 protein enrichment and histone modifications, we binned the genome into 2-kilobase windows and then ranked these windows from lowest to highest enrichment of different histone marks (Fig. 5b). Tagged MBD4 signal enrichment was positively correlated with activating histone marks H3K4me3 (promoters), H3K27ac (active promoters and enhancers) and H3K4me1 (active and primed enhancers), and to a lesser extent to H3K36me3 (transcribed gene bodies). Conversely, tagged MBD4 signal enrichment was negatively correlated with repressive marks H3K27me3 (polycomb repressed) and H3K9me3 (heterochromatin) (Fig. 5b-c). Interestingly, the G:T glycosylase TDG has also been previously described to preferentially bind to H3K4me3-rich domains, with an analogous distribution to oxidized 5mC derivatives in mouse embryonic stem cells61. Notably, while tagged MBD4 enrichment was negatively correlated with CpG methylation percentage, it was positively correlated with both CpG and methylated CpG densities (Supplementary Fig. 10). A similar pattern has been previously observed for tagged MBD4 on mouse embryonic stem cells60. Overall, we show that both C- and N-terminally tagged MBD4 preferentially bind to active chromatin and methylated CpG-rich genomic regions.
Fig. 5. MBD4 preferentially protects active chromatin and early replicating DNA.
a Western blotting on nuclear extracts of parental HAP1 cells or clones overexpressing C- or N-terminally FLAG-tagged MBD4. Exogenous and endogenous MBD4 bands and positions of molecular weight markers are indicated. No replication attempt was performed. b Histone mark enrichment over IgG in 2 kb genomic windows, ranked from lowest to highest enrichment (upper panel). FLAG enrichment over IgG in the corresponding genomic windows (lower panel). Means of every 400 or 8000 similarly-ranked windows are shown in colored shades or black, respectively. c Heatmap of Pearson correlation coefficients between histone marks and FLAG enrichment, obtained from the means of every 400 similarly-ranked windows. d Scatter plots of CpG > TpG mutation rates per CpG versus mean CpG methylation levels in chromatin states. Tumor mutations and normal epigenomic data are grouped by lineage. Mutation rates were normalized by the highest value per tumor, and the means of all tumors per lineage are shown. Lines indicate data fitting with linear regression models. Two-sided Pearson correlation statistics are shown. TssA, active TSS; TssFlnk/TssFlnkD/TssFlnkU, flanking TSS; EnhA/EnhG, active/genic enhancer; Tx/TxWk, strong/weak transcription; TssBiv, bivalent/poised TSS; EnhBiv, bivalent enhancer; ReprPC/ReprPCWk, strong/weak repressed polycomb; Quies, quiescent/low; Het-Rpts, heterochromatin/ZNF genes and repeats. e Distributions of CpG > TpG mutation rates per methylated CpG (mCpG) in SBS1 (n = 359) and SBS96 (n = 20) tumors in active or repressed/bivalent chromatin states. Observed relative to expected mutation rates are shown, considering an expected random distribution of CpG > TpG mutations among mCpGs. Two-sided Wilcoxon test P-values are indicated. Boxes indicate the median, 25th and 75th percentiles. Whiskers extend to the largest or lowest value up to 1.5 times the distance between the 25th and 75th percentiles. f Signal enrichment of FLAG-tagged MBD4 in replication timing annotations, relative to FLAG enrichment in parental HAP1 cells. Early, constitutive early; Dyn, dynamic; Late, constitutive late. g Distributions of CpG > TpG mutation rates per mCpG in SBS1 (n = 442) and SBS96 (n = 20) tumors in replication timing annotations. Observed relative to expected mutation rates are shown, considering an expected random distribution of CpG > TpG mutations among mCpGs. Statistics of boxes and whiskers are described above. Source data are provided as a Source Data file.
To answer whether active chromatin is preferentially protected from 5mC deamination by MBD4 and TDG, we analyzed SBS1 and SBS96 CpG>TpG mutation rates according to CpG methylation and ENCODE chromatin state annotations62 obtained in normal human cell types corresponding to each cell lineage. While both active and repressed/bivalent regions showed strong linear correlation between CpG>TpG mutation rate and CpG methylation levels, the slope was lower in active regions for all tumor types analyzed (Fig. 5d). In addition, mutation rates normalized per methylated CpG were significantly lower in active versus repressed/bivalent chromatin for both SBS1 and SBS96 (Fig. 5e), indicating that this bias could not be explained by CpG methylation levels alone. Interestingly, the preferential protection of active chromatin was significantly more pronounced in SBS1 MBD4wt cases than in SBS96 MBD4def cases (ratio of mutation rates in repressed vs. active chromatin: 3.84 ± 1.13 [SBS1] vs. 1.53 ± 0.25 [SBS96]; Wilcoxon test P = 2.21e-12).
Based on replication timing annotations derived from a wide range of cell types and differentiation intermediates of human development63, we also observed the highest tagged MBD4 enrichment in constitutive early replicating regions (Fig. 5f), as active chromatin highly correlates with early replicating DNA64 (Supplementary Fig. 11). Accordingly, while both SBS1 and SBS96 showed the lowest mutation rates in constitutive early replicating regions (Fig. 5g, Supplementary Fig. 12), this bias was significantly less pronounced in SBS96 MBD4def cases (ratio of mutation rates in latest vs. earliest replicating regions: 7.52 ± 3.73 [SBS1] vs. 2.20 ± 0.72 [SBS96]; Wilcoxon test P = 1.45e-11). Altogether, our data strongly suggest that active chromatin and early replicating DNA are preferentially protected from 5mC deamination, an effect that is less pronounced upon MBD4 deficiency.
MBD4 is the main glycosylase responsible for 5mC deamination repair
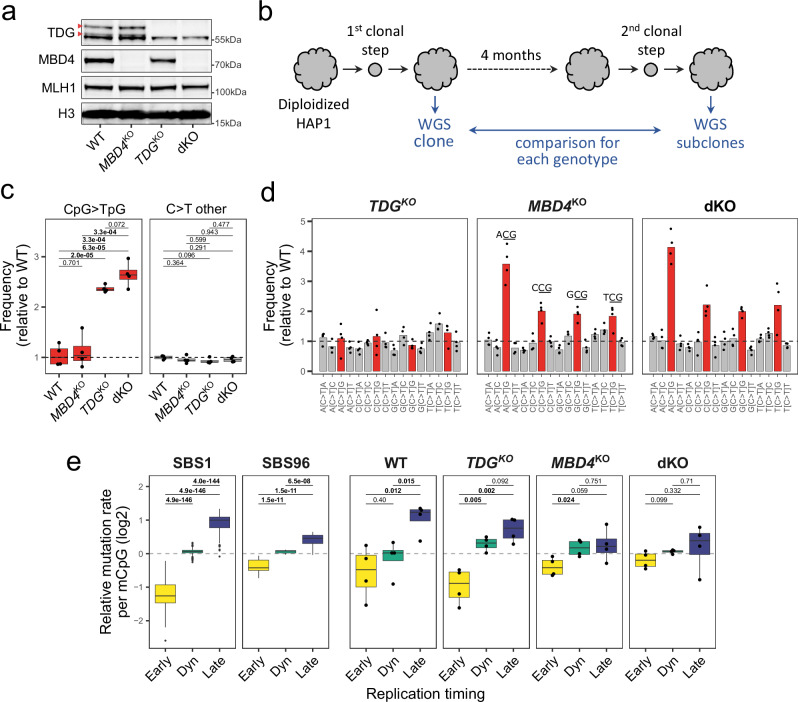
To better understand the respective activities of MBD4 and TDG at the genomic level, we obtained or generated MBD4 knockout (MBD4KO), TDG knockout (TDGKO), or double knockout (dKO) isogenic HAP1 cell line clones (Fig. 6a; Supplementary Fig. 13,14). To accurately quantify the rates of acquired CpG>TpG mutations of each genotype, we sequenced by WGS 4 subclones of each isogenic cell line after 4 months of in vitro expansion (Fig. 6b). By further comparing substitution frequencies in knockout versus wild-type subclones, we were able to minimize the errors associated with in vitro amplification65. MBD4 deficiency alone led to a significant ~2.4-fold increase in CpG > TpG mutation rate, consistent with previous reports66,67. Although TDG deficiency alone did not significantly increase the CpG > TpG mutation rate (two-sided unpaired equal variance t-test P = 0.7), double knockout cells showed a tendency for higher CpG > TpG mutation rate than MBD4KO cells (two-sided unpaired equal variance t-test P = 0.072). C > T substitution rates outside of a CpG context remained largely unchanged for all genotypes (Fig. 6c). Importantly, CpG>TpG frequencies by trinucleotide contexts in MBD4KO cells closely resembled SBS96 (ACG > CCG > GCG > TCG; Fig. 6d, Supplementary Fig. 15), experimentally validating the causative role of MBD4 deficiency in this mutational signature. Overall, we show that although both MBD4 and TDG contribute to 5mC deamination repair, MBD4 is the main G:T glycosylase in this experimental human cell model.
Fig. 6. 5mC deamination repair is primarily dependent on MBD4 in human cells.
a Western blotting on nuclear extracts of HAP1 cells wild-type or knock-out for MBD4, TDG, or both (dKO). Arrows indicate TDG-specific bands. Positions of molecular weight markers are indicated. No replication attempt was performed. b Workflow used to quantify CpG >TpG mutation rates in isogenic cell line models. Expanded clones of diploidized HAP1 cells of each genotype were analyzed by WGS and then maintained in vitro for 4 months before a second subcloning step. WGS on expanded subclones were then compared against the respective clone for variant calling. c Distributions of C > T substitution frequencies obtained by WGS in HAP1 subclones (n = 4 per genotype) after 120 days in culture, relative to the mean of wild-type subclones (shown as a dashed line). Two-sided unpaired equal variance t-test P-values without multiple comparisons adjustment are indicated. Boxes indicate the median, 25th and 75th percentiles. Whiskers extend to the largest or lowest value up to 1.5 times the distance between the 25th and 75th percentiles. d C > T substitution frequencies by trinucleotide context in HAP1 subclones relative to the mean of wild-type subclones. Bars represent means (n = 4 per genotype). e Distributions of CpG >TpG mutation rates per methylated CpG (mCpG) in SBS1 (n = 442) and SBS96 (n = 20) tumors or HAP1 subclones (n = 4 per genotype) in replication timing genomic annotations. Early, constitutive early; Dyn, dynamic; Late, constitutive late. For SBS1 and SBS96 data, two-sided Wilcoxon test P-values without multiple comparisons adjustment are indicated. For HAP1 subclones data, two-sided unpaired equal variance t-test P-values without multiple comparisons adjustment are indicated. Statistics of boxes and whiskers are described above. Source data are provided as a Source Data file.
Finally, the preferential protection of early replicating DNA from 5mC deamination in our experimental model closely mirrored what is observed in human tumors. Replication timing bias was strongest in wild-type cells and TDGKO cells (similarly to SBS1 in MBD4wt tumors), decreased in MBD4KO cells (similarly to SBS96 in MBD4def tumors) and largely abrogated in dKO cells (Fig. 6e). Hence, we experimentally validate that lower CpG > TpG mutation rate in constitutive early replicating DNA is largely dependent on 5mC deamination repair by MBD4 and marginally by TDG. This shows that the efficiency of 5mC deamination repair by MBD4 and TDG is not even along the genome, which shapes the genomic distribution of CpG>TpG mutations linked to 5mC deamination.
Discussion
Analysis of a large series of cancer whole genomes has recently revealed an ever-increasing diversity of rare mutational signatures with a high proportion of mutations in CpG dinucleotides5. However, the etiology of these signatures and the role of 5mC deamination in their mutational patterns have remained largely uncharacterized. In this study, we establish that SBS96 is intrinsically linked to biallelic inactivation of BER glycosylase MBD4 across tumor types. We describe that the CpG mutational burden of MBD4wt tumors could be explained by other signatures instead, including the common signature SBS1, the rare signatures SBS95 and SBS105, or a new pattern we describe in a unique case of secondary AML, SBSnovel. SBS105 showed an association with the rare POLD1R817W mutation, which might induce nucleotide misincorporation following specific types of DNA repair. We show that SBS105 mutations are poorly associated with DNA methylation, ruling out a major role of 5mC deamination repair in this signature. In-depth analyzes of SBS95 and SBSnovel further revealed they most probably arise from preferential G > A mutagenesis in the complementary strand of methylated cytosines, potentially due to DNA damage targeting guanines repaired by TC-NER. We further propose that SBS1 and SBS96 are the sole signatures with features fully consistent with a primary role of mutagenesis caused by 5mC deamination, in contexts of MBD4 proficiency or deficiency, respectively.
Our data uncovered tissue-specific features of SBS96, which faithfully mirrors lineage-specific DNA methylation patterns across CpG and CpA contexts. While MBD4 inactivating mutations have been found to predispose to multiple malignancies19,20,68, we show that lineage-specific selective pressures shape the landscape of oncogenic SBS96 mutations found across tumor types.
We modeled for the first time the cooperative roles of MBD4 and TDG in human cells and showed that MBD4 is the main G:T glycosylase preventing the accumulation of CpG > TpG mutations. We show that MBD4 preferentially protects active chromatin and early replicating DNA, which we associate with the uneven distribution of CpG > TpG mutations caused by 5mC deamination across methylated CpGs. Altogether, comprehensive tumor profiling analyzes coupled with in vitro modeling have allowed us to dissect the mechanisms linking 5mC deamination molecular signatures with the mutational burden of human cancers.
Methods
In-house Study Patients
The series of in-house patients with newly generated sequencing data comprises 19 individuals diagnosed with UVM and an individual diagnosed with conjunctival melanoma at Institut Curie, Paris, France, 6 individuals diagnosed with UVM at Charité - Universitätsmedizin Berlin, Berlin, Germany, and an individual diagnosed with an HHG at Hôpital La Pitié Salpêtrière, Paris, France. The sequencing modality used for each sample is provided in Supplementary Data 1. Tumor samples were histologically reviewed by a pathologist. UVM samples were collected from primary eye tumors or liver metastases. Germline samples were collected from blood. All patients provided written informed consent to perform germline and somatic genetic analyzes. The study was conducted in accordance with the declaration of Helsinki and was approved by the Institut Curie Review Board CRI-DATA (Project DATA190061) and Charité - Universitätsmedizin Berlin Institutional Review Board (EA4/063/13).
Total RNA sequencing on patient samples
UVM tumor total RNA was obtained by phenol extraction or using the Allprep DNA/RNA MicroKit (Qiagen, 80284) and quantified using the Qubit RNA BR Assay Kit (Invitrogen, Q10210). Total RNAseq libraries were prepared using the TruSeq Stranded Total RNA Library Prep Gold (Illumina, 20020599) or the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold (Illumina, RS-122-2301 and RS-122-2302). Paired-end libraries (2 × 100 bp or 2 × 75 bp) were sequenced on HiSeq 4000 or NovaSeq 6000 instruments (Illumina). Sequencing data QC was performed with FastQC v0.11.8 (https://github.com/s-andrews/FastQC) and adapter trimming with TrimGalore v0.6.2 (https://github.com/FelixKrueger/TrimGalore). Mapping was performed with STAR v2.6.1 (https://github.com/alexdobin/STAR) to GRCh38 using Gencode v29 annotation. Duplicates were removed with Picard MarkDuplicates v2.18.15 (https://github.com/broadinstitute/picard) and counts tables from aligned data were generated with featureCounts v1.6.2 (https://github.com/ShiLab-Bioinformatics/subread).
Whole-Genome Sequencing on patient samples
Tumor and germline DNA was obtained by phenol extraction and quantified using the Qubit dsDNA BR Assay Kit (Invitrogen, Q32850). WGS libraries were prepared using the Kapa HyperPrep kit (Roche, 07962363001) or the NEBNext Ultra II End repair/A-tailing module (New England Biolabs, E7546) and Ligation module (New England Biolabs, E7595). Paired-end libraries (2 × 150 bp) were sequenced on a NovaSeq 6000 instrument (Illumina). WGS target coverage depth was 30X for germline samples, 30X for the high tumor purity UVM sample, and 100X for the low tumor purity HHG sample. WGS data from external sources were downloaded as BAM files from the European Genome-phenome Archive (EGA) with dataset ID EGAD00001003568 and EGAD00001005454)23,26, and converted to fastq files before further processing.
Whole-Exome Sequencing on patient samples
Tumor and germline DNA was obtained by phenol extraction, the Allprep DNA/RNA MicroKit (Qiagen, 80284), or the QIAamp DNA Blood Maxi Kit (Qiagen, 51192). DNA was quantified using the Qubit dsDNA BR Assay Kit (Invitrogen, Q32850). WES libraries were prepared using the SureSelectXT2 Clinical Research Exome V2 kit (Agilent, 5190–9500 and G9621B) or the Nextera Rapid Capture Expanded Exome kit (Illumina, FC-140-1005). Paired-end libraries (2 × 100 bp or 2 × 75 bp) were sequenced on HiSeq 2000/2500/4000 or NovaSeq 6000 instruments (Illumina). Previously published WES from Institut Curie patients is available in EGA with dataset ID EGAD00001004554 and EGAD00001006988.
MBD4 and TDG knockout HAP1 models
Haploid HAP1 parental cells and knockout for MBD4 (Horizon Discovery, C631 and HZGHC000921c002, respectively) were cultured in complete media (IMDM [Gibco, 12440053], bovine fetal serum 10% [BioSera, FB-1003], Penicillin-Streptomycin 100 U/mL [Gibco, 15140122]) at 37 °C in 5% CO2 and 5% O2 incubator. The identity of HAP1 cell lines was authenticated by WGS (described below). Cells were transiently co-transfected using jetOPTIMUS (Polyplus, 101000051) with two all-in-one CRISPR-Cas9 vectors (pSpCas9(BB)−2A-GFP PX458; Addgene, #48138) cloned with sgRNAs targeting TDG exon 2 (sgRNA1; GATGGCTGAAGCTCCTAATA) and exon 5 (sgRNA2; GATCATCCATATGGTTCAGC). GFP+ cells were single-cell sorted, expanded, and clones’ genomic DNA was extracted with DNeasy Blood & Tissue Kit (Qiagen, 69504). Clones with complete deletion of the region spanning exons 2 to 5 were kept, as confirmed by genomic DNA PCR yielding no product when using pairs of primers flanking each sgRNA target region separately (sgRNA1_F TTGGTGAACATGTACATACAGGACT and sgRNA1_R CCGATGTTGAACTTTCTAAGCTCTC; sgRNA2_F ACCCCCTGTGAAAAGGAGATAATAA and sgRNA2_R ACCCCCTGTGAAAAGGAGATAATAA) but yielding a clear PCR product when using forward primer upstream of sgRNA1 (sgRNA1_F) and reverse primer downstream of sgRNA2 (sgRNA2_R). Cells of each genotype (wild-type, MBD4KO, TDGKO, and dKO) were then stained for 40 min with 5 µM Vybrant DyeCycle Violet Stain (Invitrogen, V35003) and cells with 4n DNA content (diploidized cells in G2) were enriched by bulk sorting with SH800S Cell Sorter (Sony Biotechnology). TDG expression was obtained in diploidized cells by real-time quantitative reverse transcription PCR using primers spanning TDG exons 7-8 (ACTTGGAATTTGGGCTTCAGC and TCTTGCACTGGATGATGGCA). Relative expression was calculated using GAPDH as endogenous control and the 2−∆∆Ct method. MBD4 and TDG knockout status were further confirmed by western blotting and immunofluorescence, as described below.
HAP1 long-term culturing and Whole-Genome Sequencing
HAP1 diploidized clones of each genotype were cultured in low oxygen conditions (5% CO2 and 5% O2) to limit oxidative stress in vitro65. A first clonal step was performed by morphology-based single-cell sorting with SH800S Cell Sorter and clones were expanded for 14 days. This represented the day zero (D0) of long-term culturing. Haploid HAP1 cells tend to diploidize over time, potentially altering relative mutation rates per genomic position at unknown time points during the long-term culturing. Hence, expanded clones had their ploidy status confirmed by propidium iodide staining in fixed cells followed by flow cytometry analysis. A single clone of each genotype was kept for long-term culturing. Cells were split every 3-4 days and kept at low density (< 50% confluency). After 60 days of culture (D60), the doubling time of each clone was measured by seeding an equal number of cells in biological triplicates, followed by 48 h incubation, cell harvesting, and counting. After 120 days of culture (D120), a second clonal step was performed by morphology-based single-cell sorting with SH800S Cell Sorter. For each genotype, expanded D0 clone and four expanded D120 subclones had their DNA extracted with DNeasy Blood & Tissue Kit (Qiagen, 69504) and quantified using the Qubit dsDNA BR Assay Kit (Invitrogen, Q32850). The amount of 800 ng DNA was used to prepare WGS libraries using the Kapa HyperPrep kit (Roche, 07962363001). Paired-end libraries (2 × 100 bp) were sequenced on a NovaSeq 6000 instrument. WGS coverage depth was set to 30X. Variant calling was performed in tumor versus normal mode, as described below. For each genotype, D0 WGS was used as ‘normal’ and D120 WGS as ‘tumor’.
Variant Calling
WES and WGS data from in-house patients, from publicly available MBD4def tumors (excluding the Genomics England [GEL] series), and from HAP1 clones were analyzed with a harmonized pipeline. Sequencing data QC and adapter trimming on all samples was performed with FastQC v0.11.9 and TrimGalore v0.6.10. Mapping was performed with BWA-MEM v0.7.17 (https://github.com/lh3/bwa) to GRCh38 (Primary assembly + ALT contigs). Duplicates were removed with Sambamba v1.0 (https://github.com/biod/sambamba). All samples had > 90% mapping rate and average fragment size >300 bp. Somatic short variant and insertion/deletion discovery (SNVs + Indels) in tumors was performed with GATK4 v4.4.0.0 (https://github.com/broadinstitute/gatk) Mutect2 in tumor versus normal mode, following GATK4 best practices. All samples had tumor cross-contamination < 1%. Variants were filtered with the FilterMutectCalls function. MBD4 germline variants in patient samples were identified with GATK4 Haplotype Caller. Germline and somatic variants were annotated with VEP v104.3 (https://github.com/Ensembl/ensembl-vep). Hotspot alterations in BAP1, GNAQ, and GNA11 were further verified manually in all UVM samples, including in RNAseq data when available.
Genomics England Sample Selection
Somatic variants from GEL data release v17 (2023-03-30) were used, representing 16,322 individual tumor samples. High sequencing quality samples with the following criteria were used: average fragment size ≥ 300, mapping rate ≥ 90%, tumor sample cross-contamination < 1%, coverage homogeneity ≤ 30, and chimeric percentage ≤ 1. Tumor samples obtained from formalin-fixed paraffin-embedded (FFPE) blocks and with tumor purity < 20% (when available) were excluded. Tumor type information was obtained from the GEL cancer analysis Table v17, which provides cross-referenced and updated classification aligned with The Cancer Genome Atlas (TCGA) nomenclature. Correspondence of disease, study name, and study abbreviation are provided in Supplementary Data 2. Tumors with other classifications were excluded. The final series used for analysis comprises 12,726 tumor samples.
Variant Filtering
High-quality somatic variants from in-house patients and GEL were obtained with the following filtering criteria: ≥ 5 alternative allele counts for samples sequenced at 30X or with low tumor purity (MBD4def HHG) and ≥ 10 alternative allele counts for the remaining tumors sequenced at 100X (including all GEL tumors). To account for large differences in tumor purity among tumor samples, filtering by variant allele frequency (VAF) was performed in a stepwise manner. First, VAFs were normalized by tumor purity (when available), and largely subclonal variants with VAF < 0.15 were excluded. VAFs were then re-normalized by the median of VAFs in the sample, and variants with normalized VAF < 0.30 were excluded. For HAP1 variants, no normalization was required and variants with VAF < 0.20 were excluded. The filtered lists of variants representing largely clonal high-quality somatic variants were used for downstream analysis.
Mutational Signature Analysis and Sample Selection
SBS mutational signature analysis of tumor samples was performed on filtered somatic variants obtained by WGS (2 in-house MBD4def tumors, 4 publicly available MBD4def tumors, and 12,726 tumors from GEL) with R 4.2.1 package signature.tools.lib v2.4.1 (https://github.com/Nik-Zainal-Group/signature.tools.lib). The correspondence of tissues used for signature fitting in each tumor type is provided in Supplementary Data 2. Signature fitting in the complete series of samples (Supplementary Data 3) was performed with the FitMS function, using cosine similarity increase multi-step mode, gini-scaled threshold exposure filter, and 50 bootstraps. Reference common signatures found in each organ (tier T2) and reference rare pan-cancer signatures (tier T2) were used. The remaining default options were used, which included fitting with a single rare signature per sample and a minimum exposure percent threshold of 5%. The list of signatures used for each organ and their respective substitution frequencies is provided in Supplementary Data 4, 5. Samples were kept for downstream analysis if they showed: (i) any contribution of rare signatures SBS95, SBS96, and SBS105; (ii) ≥ 30% contribution of common SBS1, exclusively for related tumor types with at least one sample representative of SBS95, SBS96 or SBS105; (iii) ≥ 30% contribution of rare SBS10d or SBS20 (Supplementary Data 6). Additionally, MBD4wt UVM samples from GEL were selected for the analysis of CpA > TpA mutations. Two MBD4wt UVM tumors of the iris with characteristic ultraviolet radiation (UVR) exposure signature SBS7a26 were excluded, and only choroidal UVM were kept. To obtain cosine similarity increase values for MBD4wt samples with SBS96 as a candidate rare signature, signature fitting was performed with FitMS in error reduction multi-step mode, without any additional modifications (Supplementary Data 8). The probability that a specific variant originates from each of the fitted signatures was obtained with the assignSignatureProbabilityToMutations function. For downstream analysis of signatures separately, variants with > 70% probability were used (Supplementary Data 9).
Strand Asymmetry Analysis
Transcription and replication strand asymmetry analysis was performed with R 4.2.1 package MutationalPatterns v3.8.1 (https://github.com/UMCUGenetics/MutationalPatterns). Transcription strand asymmetry analysis was performed separately for genes in the four quartiles of expression, based on the average of consensus transcript expression among 50 tissues. This expression data is based on The Human Protein Atlas version 23.0 (https://www.proteinatlas.org/) and is derived from HPA and GTEx transcriptomics datasets. Transcription strand asymmetry of CpG mutations was defined to be significant if the False Discovery Rate (FDR) was < 0.01 and the absolute value of the log2 ratio was >0.15. Replication strand asymmetry of each substitution class (not restricted to CpG mutations) was defined to be significant if the FDR was <0.01, the absolute value of the log2 ratio was > 0.15 and the contribution of the substitution class to the signature was >10%. Tissue-specific SBS96 profiles in BRCA and SARC tumors were obtained from Degasperi et al.5 UVM, AML, and HHG tissue-specific SBS96 profiles were obtained from the average profiles of MBD4def tumors, all of which showed almost pure SBS96.
Isolation of primary uveal melanocytes
Normal eye tissues from UVM or conjunctival melanoma patients who underwent eye enucleation surgery at Institut Curie were obtained. Normal uveal choroid was dissected from a region diametrically opposed to the tumor site. Tissue was enzymatically digested for 40 min at 37 °C (DMEM/F12 [Gibco, 11330032], bovine fetal serum 10% [BioSera, FB-1003], Penicillin-Streptomycin 100 U/mL [Gibco, 15140122], collagenase type IV 400 CDU/mL [Sigma, C1889], DNAse I 12 µg/mL [Sigma, D5025]) under gentle agitation. Cell suspensions were washed once with wash buffer (PBS without Ca2+ and Mg2+, BSA 0.5%, EDTA 2 mM) and filtered with 30 µm strainers. During initial validation experiments, cells were additionally stained for 30 min on ice with anti-CD45 coupled with BB515 (1:20; BD Biosciences, 564585) in wash buffer. Cells were then resuspended in media (DMEM/F-12 [Gibco, 11039021]) containing 5 µM Vybrant DyeCycle Violet (Invitrogen, V35003) and incubated for 30 min at 37 °C. Finally, cells were resuspended in wash buffer containing 5 µM Vybrant DyeCycle Violet and 1 µg/mL propidium iodide (PI; BioLegend, 421301) before flow cytometry analysis. Sorting of uveal melanocytes relied on the unique light interaction properties of melanin, namely efficient absorption of UV-Violet wavelengths and high autofluorescence at far-red wavelengths. In summary, highly melanized viable uveal melanocytes (PI negative, Vybrant DyeCycle Violet dim and far-red autofluorescence bright) were sorted with FACSAria Fusion Cell Sorter (BD Biosciences). This technique allowed the sorting of uveal melanocytes with > 90% purity, as confirmed by direct brightfield melanin visualization using an imaging flow cytometer Amnis ImageStream Mk II (Cytek Biosciences). The median pixel intensity of the masked cell area was used to estimate the presence of melanin and calculate the purity of sorted melanocytes.
Whole-genome bisulfite sequencing
Purified uveal melanocytes from one conjunctival melanoma patient had their DNA extracted with DNeasy Blood & Tissue Kit (Qiagen, 69504). Three metastatic UVM samples had their DNA extracted using the Allprep DNA/RNA MicroKit (Qiagen, 80284). WGBS libraries were prepared using the Accel-NGS Methyl-Seq DNA Library Kit (Swift Biosciences, 30024), the EZ DNA Methylation-Gold Kit (Zymo, D5005), and DNA Clean & Concentrator-5 (Zymo, D4013), following the instruction manual Accel-NGS Methyl-Seq DNA Library (Revision 160510). Paired-end (2 × 150 bp) libraries were sequenced on a NovaSeq 6000 instrument (Illumina) or a DNBSEQ-T7 instrument (MGI) after library conversion. WGBS raw fastq files from external sources were downloaded from EGA with dataset ID EGAD0000100978939 or from the ENCODE portal (https://www.encodeproject.org/; datasets ENCSR388RMS and ENCSR163TTI). Sequencing data QC and adapter trimming were performed with FastQC v0.12.0 and TrimGalore v0.6.10, which included the hard trimming of 4 bp from 5′ and 3′ ends of R1 and R2 reads in ENCODE data, or hard trimming of 10 bp from the 3′ of R1 and R2 reads, 10 bp from the 5′ of R1 and 15 bp from the 5′ of R2 reads in the remaining samples. Further processing was performed with Bismark v0.21.0 (https://github.com/FelixKrueger/Bismark). In summary, mapping was performed with Bowtie2 v2.2.9 (https://github.com/BenLangmead/bowtie2) to GRCh38 (no ALT contigs analysis set GCA_000001405.15 from ENCODE), followed by de-duplication of the Bismark BAM. Methylation extraction was performed in comprehensive mode, and methylation calls from ends of reads were ignored when necessary, based on M-bias plot of each sample. Methylation percentages per trinucleotide sequence context were obtained from the counts of methylated and unmethylated cytosines taking part in methylation calls, as provided in the Bismark whole genome cytosine report summary. WGBS CpG methylation calls of KBM-7 cells were obtained from NCBI’s GEO with GEO series accession number GSE65196)69, and data from multiple individually sequenced KBM-7 samples were combined. CpG methylation values were averaged per CpG and binarized, with CpGs with > 50% methylation considered as fully methylated and CpGs with ≤ 50% methylation considered as fully unmethylated.
Multi-omics data integration and analysis
The complete list of epigenomic datasets used for each cell lineage is described in Supplementary Data 10. Epigenomic analysis was restricted to autosomal chromosomes. Unified blacklisted regions from ENCODE (ENCFF356LFX) were further excluded. Data filtering was performed with bedtools v2.27.1 (https://github.com/arq5x/bedtools2) and data integration and analysis with R 4.2.1 package tidyverse v2.0.0 (https://github.com/tidyverse). Differential methylation analysis between cell types was performed at the level of methylation blocks39. We retained blocks with ≥ 4 CpGs, ≥ 50% CpG methylation calls in all tissues analyzed and overlapping genic promoters and transcribed regions (excluding introns) and/or ENCODE cis-Regulatory Elements (v3). Through pairwise comparisons, a block was considered specifically methylated if the mean of non-binarized CpG methylation values was > 50% in the first tissue and < 50% in the second tissue, and vice versa. Annotation of WGS somatic CpG variants and WGBS covered CpGs was performed in genomic bins or other genomic annotations (differentially methylated blocks, chromatin states, replication timing) with R 4.2.1 package annotatr v1.24.0 (https://github.com/rcavalcante/annotatr). Pan-tissue replication timing genomic annotation covering 85% of the human genome was used63. For the analysis of chromatin states and replication timing, only variants within methylated CpGs in the matched normal cell type were kept. Mutation rates by each combination of tumor type/lineage and mutational signature were calculated as the fraction of mutations per genomic annotation class divided by the fraction of covered CpGs in the same class. For the analysis of mutation rates in genomic windows, tumor types with < 300 high probability CpG variants of a given signature were excluded. For the analysis of mutation rates in chromatin states, samples with < 300 high probability CpG variants of a given signature were excluded. Relative mutation rates were obtained by normalization against the annotation class with the highest mutation rate. Single nuclei RNAseq data of the ocular posterior segment was obtained directly from the Broad Institute Single Cell Portal (dataset SCP2298).
Local sequence context of non-CpG sites
Publicly available bisulfite sequencing data (GEO series accession number GSM1382253 and GSM1382256) of Dnmt triple-KO mouse cells re-expressing recombinant Dnmt3a or Dnmt3b were obtained following the instructions of sblab-bioinformatics GitHub (https://github.com/sblab-bioinformatics/dnmt3a-dnmt3b). The fastqs were analyzed using the same pipeline to obtain the context for all cytosines in the genome. The top 1,000 non-CpG dinucleotides with at least 25% methylation levels were selected for each sample re-expressing recombinant Dnmt3a or Dnmt3b. Probability logos were generated with kpLogo70, ignoring P-values for bases with a frequency above 0.99. For logos on CpA > TpA mutated sites, randomly selected CpA sites and DNMT3A/B non-CpG top methylated sites, probabilities were unweighted by site, and a Markov background model with equal fractions of each base was used. For logos on methylated CpA sites in uveal melanocytes or metastatic UVM, we randomly selected 100,000 sites with at least one methylated cytosine taking part in the methylation call, and logo probabilities were weighted by the counts of methylated cytosines per site.
Recombinant MBD4 overexpression
Synthetic lentiviral vectors for over-expression of recombinant forms of MBD4 were obtained from VectorBuilder. MBD4 isoform NM_003925.3 was coupled with a sequence coding for a 12 amino-acid glycine and serine linker and 3xFLAG, either N- or C-terminally, under the control of hPGK medium strength promoter. A puromycin resistance gene was separated from the MBD4 open reading frame through an IRES sequence. Lentiviral particles were generated in HEK-293 and used to transduce MBD4 wild-type HAP1 cells. Cells stably expressing the constructs were selected in complete media (IMDM (Gibco, 12440053), bovine fetal serum 10% (BioSera, FB-1003), Penicillin-Streptomycin 100 U/mL (Gibco, 15140122)) supplemented with 1 µg/mL puromycin for 7 days. Confirmation of predominant nuclear expression of both recombinant proteins was performed by immunofluorescence with an anti-FLAG antibody, as described below. Single-cell clones were obtained by sorting with a SH800S Cell Sorter, and clones expressing exogenous MBD4 at levels comparable to endogenous MBD4 were further selected. MBD4 expression was accessed by western blotting, as described below.
CUT&RUN sequencing
CUT&RUN was performed using the CUTANA ChIC/CUT&RUN Kit V4 (EpiCypher, 14-1048), according to the manufacturer’s instructions. In summary, 250,000 HAP1 nuclei per reaction were incubated overnight at 4 °C in a nutator with 1 µL antibody against FLAG (1:50; Cell Signaling, 14793S; lot 7) or 0.5 µg antibodies against H3K4me3 (1:50; EpiCypher, 13-0041; lot 13-0041k), or IgG (1:10; Cell Signaling, 66362S; lot 2). FLAG reactions on FLAG-tagged MBD4 clones were performed in biological duplicates. Control reactions (FLAG on parental HAP1 cells, IgG negative controls, and H3K4me3 positive control) were performed without replicates. Following p-A/G digestion and chromatin recovery, purified DNA was used to prepare paired-end libraries with CUTANA CUT&RUN Library Prep Kit (EpiCypher, 14-1001). Quantification of nucleosome-sized fragments in each library was obtained with High Sensitivity D1000 ScreenTape (Agilent, 5067-5584). Libraries were multiplexed and primer dimers were removed using SPRIselect Beads (Beckman Colter, B23317). Paired-end (2 × 100 bp) libraries were sequenced on a NovaSeq 6000 instrument, with a target of 12-16 million fragments sequenced per sample. Data was processed using the nf-core ChIP-seq pipeline with default parameters (https://github.com/nf-core/chipseq) and mapping to GRCh38, using FastQC v0.11.8, TrimGalore v0.6.2, BWA-MEM v0.7.17, Picard MarkDuplicates v2.19.0, and phantompeakqualtools v1.2.2 (https://github.com/kundajelab/phantompeakqualtools). Mapping statistics and normalized/relative strand cross-correlation values (NSC/RSC) are provided in Supplementary Data 14. FLAG enrichment over IgG control in 2-4 kb non-overlapping genomic windows was calculated as a ratio from depth-normalized coverage bigwigs with pseudocounts of 1. Histone marks ChIP-seq data on HAP1 cells were obtained from ENCODE as fold-change enrichment over IgG control bigwigs (Supplementary Data 10). Genomic windows not covered in ENCODE ChIP-seq data or overlapping ENCODE blacklisted regions (ENCFF356LFX) were excluded. For rankings based on histone mark enrichment, a tenth of the 2 kb windows with the lowest signal enrichment were removed. For rankings based on FLAG enrichment obtained by CUT&RUN, a third of the 4 kb windows with the lowest signal enrichment were removed. CpG and methylated CpG densities in 4 kb windows were obtained from WGBS CpG methylation calls of KBM-7, as described above. CUT&RUN signal enrichment over IgG control in replication timing annotations was calculated as a ratio from depth-normalized coverage bigwigs. Of note, our CUT&RUN attempts with currently available anti-MBD4 antibodies showed poor sensitivity and/or specificity.
Immunofluorescence
HAP1 cells were grown in Nunc Lab-Tek II Chamber Slides (Thermo Scientific, 154534PK) coated with poly-L-lysine (Sigma, P4707), fixed in 4% PFA for 15 min at room temperature (RT), washed 3 times with PBS, permeabilized with 0.1% Triton X-100 in PBS for 15 min at RT, and blocked with blocking buffer (1% BSA in PBS) for 1 h at RT. Cells were then incubated overnight at 4 °C with antibodies against MBD4 (1:500; Abcam, ab227625; lot GR3230875-5), TDG (1:500; Invitrogen, PA5-29140; lot YA3812362), FLAG (1:1,000; Cell Signaling, 14793S; lot 7) and Tubulin (1:1,000; Invitrogen, 14-4502-80; lot 2003406) in blocking buffer. Cells were washed 3 times with Tween-20 0.05% in PBS and incubated for 1 h at RT with secondary antibodies anti-mouse Alexa Fluor Plus 555 (1:1,000; Invitrogen, A32727; lot XH350742) and anti-rabbit Alexa Fluor Plus 647 (1:1,000; Invitrogen, A32733; lot XG349344) in blocking buffer. Cells were then incubated with 1 µg/mL DAPI in PBS for 5 min and further washed 3 times with Tween-20 0.05% in PBS and once with distilled water. Slides were mounted with ProLong Diamond Antifade Mountant (Invitrogen, P36961) and imaged with an Axio Imager Z2 Epifluorescence Microscope with Apotome (Zeiss).
Western blotting
HAP1 nuclear extracts in RIPA buffer supplemented with cOmplete Protease Inhibitor Cocktail (Roche, 11697498001) were sonicated with Bioruptor Pico (Diagenode) for 10 cycles of 30 s on and 30 s off at 4 °C. Clarified extracts were run in NuPAGE 4 to 12% Bis-Tris gel (Invitrogen, WG1402BOX), which was then transferred to 0.45 µm nitrocellulose membranes. Membranes were blocked with 5% non-fat milk in TBS-T (Tris-Buffered Saline, 0.1% Tween-20), and incubated with primary antibodies in 5% BSA in TBS-T. Antibodies against TDG (1:1000; Invitrogen, PA5-29140; lot YA3812362), N-terminal MBD4 (1:1000; Abcam, ab224809; lot 1017919-4), C-terminal MBD4 (1:1000; Abcam, ab12187; lot GR21754-20), MLH1 (1:1000; Sigma, HPA052707; lot R69680), FLAG (1:1000; Cell Signaling, 14793S; lot 7) were incubated overnight at 4 °C. Antibody against histone H3 (1:2,500; Abcam, ab1791; lot GR252388-1) was incubated for 1 h at room temperature. Blots were washed in TBS-T and near-infrared secondary antibody anti-rabbit 800CW (1:20,000; LI-COR, 926-32213; lot D11005-09) was incubated for 1 h at room temperature in 5% BSA in TBS-T. Blots were washed in TBS-T and imaged with an Odyssey Imaging System (LI-COR). Western blotting of MLH1 was used to control for potential confounding effects of MMR.
Statistical analysis
Unpaired two-sided Wilcoxon tests and unpaired two-sided t-tests were performed with R 4.2.1 package ggpubr v0.6.0 (https://github.com/kassambara/ggpubr). Linear regression models and smoothed conditional means models were performed with R 4.2.1 package tidyverse v2.0.0. Pearson correlation statistics were obtained with the cor function of R 4.2.1 package stats v4.2.1 or stat_cor function of R 4.2.1 package ggpubr v0.6.0.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
Supported by funding from the French National Cancer Institute (INCa) and DGOS Program de Recherche Translationnelle en Cancérologie PRT-K19-51, the INCa PLBIO21-012, the Institut National de la Santé et de la Recherche Médicale (INSERM), and the Institut Curie. The Institut Curie ICGex NGS platform is funded by the EQUIPEX “Investissements d’Avenir” program and ANR10-INBS-09-08 from the Agence Nationale de le Recherche. M.R. was supported by the Interface INSERM program. We acknowledge support from Institut Curie for sample collection, banking, and processing, and we thank the Biological Resource Center and its members (Odette Mariani) and the next-generation sequencing team (Sylvain Baulande, Patricia Legoix-Né). We acknowledge support from Assistance Publique - Hôpitaux de Paris (AP-HP), and we thank Onconeurothek for biobanking and Prof. Ahmed Idbaih. We thank Lawryn Kasper for essential advice on CUT&RUN experiments. We thank Anne-Charlotte Lefranc for her help in cell culturing. We thank Daniela Balzerei and Alexander Kovacsovics from the Otto Warburg Laboratory “Gene regulation and Systems Biology of Cancer” for technical assistance and the sequencing core facility of the Max Planck Institute for Molecular Genetics for support with WGBS sequencing. This research was made possible through access to the data generated by the 2025 French Genomic Medicine Initiative and by the TCGA Research Network (http://cancergenome.nih.gov/). This research was made possible through access to data in the National Genomic Research Library, which is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The National Genomic Research Library holds data provided by patients and collected by the NHS as part of their care and data collected as part of their participation in research. The National Genomic Research Library is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. We thank the patients and their family members.
Author contributions
A.B.S. and M.-H.S. conceived the study, interpreted the data, and wrote the manuscript. A.B.S performed multi-omics data analysis and integration, FACS analysis and cell isolation, CUT&RUN, CRISPR knockouts and long-term culturing, immunofluorescence, and western blotting. A.B.S. and O.G. performed recombinant protein overexpression. A.H. performed variant calling. S.V. performed methylation motif analysis. M.-L.Y. and B.O. provided whole-genome bisulfite sequencing data. M.-L.Y., M.R., and O.G. provided input for data interpretation and manuscript writing. A.V.-S., N.C., P.M., G.P., S.L, D.R., A.P., F.B. provided patient specimens. All authors reviewed and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks Simon Schwarz, Sriram Vijayraghavan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Newly generated WGBS, WGS, WES, and RNAseq data are deposited in the European Genome-phenome Archive (EGA) database under accession EGAS50000000536. Data can be made accessible upon request to the DACs EGAC50000000356 (Institut Curie) or EGAC00001002078 (Max Planck Institute for Molecular Genetics). Newly generated WGS data on the HGG from Hôpital La Pitié Salpêtrière cannot be made available due to ethical approval restrictions. Newly generated CUT&RUN data are deposited in NCBI’s Gene Expression Omnibus (GEO) under GEO Series accession GSE275181. Previously published WES from Institut Curie patients is available in EGA under accessions EGAD00001004554 and EGAD00001006988. Previously published WGS data from external sources are available in EGA under accessions EGAD00001003568 and EGAD00001005454. WGS data from GEL is not available upon request, but accessible by registered researchers in the Trusted Research Environment (https://www.genomicsengland.co.uk/research/research-environment). All GEL analyzes must take place within the Trusted Research Environment (https://www.genomicsengland.co.uk/understanding-genomics/data). Registration involves an online application, verification by the applicant’s institution, completion of a short information, governance course, and verification of approval by Genomics England. Please see https://www.genomicsengland.co.uk/research/academic/ for more information. Previously published WGBS data on human tissues are available in EGA under accession EGAD00001009789 and in GEO under accession GSE65196. Previously published WGBS data on mouse cells are available in GEO under accessions GSM1382253 and GSM1382256. Publicly available human epigenomic data from ENCODE are described in Supplementary Data 10. Previously published single nuclei RNAseq data are available from the Broad Institute Single Cell Portal with dataset identifier SCP2298. Source data are provided with this paper.
Competing interests
D. Rieke reports advisory agreement with BeiGene and Bayer, honoraria from Bristol Myers Squibb, Bayer and Roche, research support from Seagen, and personal fees from Bayer and Johnson & Johnson, all outside the submitted work. A. Picca reports personal fees from AstraZeneca and Servier, all outside the submitted work. F. Bielle reports funding of research from Abbvie, service agreement for research contracted between his institution and Treefrog Therapeutics as well as Owkin, personal fees from Bristol Myers Squibb and a next-of-kin employed by Bristol Myers Squibb, all outside the submitted work. M.L. Yaspo is COO/CSO and shareholder of Alacris Theranostics without conflict of interest with the submitted work. M. Rodrigues reports non-financial support from AstraZeneca and Merck Sharp and Dohme, grants from Daiichi Sankyo, personal fees from AstraZeneca, Immunocore, Merck Sharp and Dohme and GlaxoSmithKline, all outside the submitted work. M.-H. Stern reports grants from Immunocore and Bionano, and royalties from Myriad Genetics, all outside the submitted work. The remaining authors have no conflict of interest to declare.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54223-z.
References
- 1.Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell149, 979–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov, L. B. & Stratton, M. R. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr. Opin. Genet Dev.24, 52–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou, X. et al. A systematic CRISPR screen defines mutational mechanisms underpinning signatures caused by replication errors and endogenous DNA damage. Nat. Cancer2, 643–657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degasperi, A. et al. Substitution mutational signatures in whole-genome-sequenced cancers in the UK population. Science376, abl9283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pich, O. et al. The mutational footprints of cancer therapies. Nat. Genet51, 1732–1740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, B. et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood135, 41–55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov, L. B. et al. Clock-like mutational processes in human somatic cells. Nat. Genet47, 1402–1407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki, M. M. & Bird, A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet9, 465–476 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Yoon, J. H., Iwai, S., O’Connor, T. R. & Pfeifer, G. P. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res31, 5399–5404 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sjolund, A. B., Senejani, A. G. & Sweasy, J. B. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res743-744, 12–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrich, B. & Bird, A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol.18, 6538–6547 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrich, B., Hardeland, U., Ng, H. H., Jiricny, J. & Bird, A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature401, 301–304 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Turner, D. P. et al. The DNA N-glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine. Cancer Res66, 7686–7693 (2006). [DOI] [PubMed] [Google Scholar]
- 16.He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett, M. T. et al. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc.128, 12510–12519 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papin, C. et al. MBD4 loss results in global reactivation of promoters and retroelements with low methylated CpG density. J. Exp. Clin. Cancer Res42, 301 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palles, C. et al. Germline MBD4 deficiency causes a multi-tumor predisposition syndrome. Am. J. Hum. Genet.109, 953–960 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien, A. C. et al. Germline MBD4 mutations and predisposition to uveal melanoma. J. Natl Cancer Inst.113, 80–87 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanakaya, K. et al. A germline MBD4 mutation was identified in a patient with colorectal oligopolyposis and earlyonset cancer: A case report. Oncol. Rep.42, 1133–1140 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues, M. et al. Evolutionary routes in metastatic uveal melanomas depend on MBD4 alterations. Clin. Cancer Res25, 5513–5524 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Sanders, M. A. et al. MBD4 guards against methylation damage and germ line deficiency predisposes to clonal hematopoiesis and early-onset AML. Blood132, 1526–1534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues, M. et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat. Commun.9, 1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Consortium, T. I. T. P.-C. A. O. W. G. Pan-cancer analysis of whole genomes. Nature578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson, P. A. et al. Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat. Commun.11, 2408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caulfield, M., et al. National Genomic Research Library. figshare 10.6084/m9.figshare.4530893.v7 (2020).
- 28.Haradhvala, N. J. et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell164, 538–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haradhvala, N. J. et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat. Commun.9, 1746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson, P. S. et al. Increased somatic mutation burdens in normal human cells due to defective DNA polymerases. Nat. Genet53, 1434–1442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrianova, M. A., Bazykin, G. A., Nikolaev, S. I. & Seplyarskiy, V. B. Human mismatch repair system balances mutation rates between strands by removing more mismatches from the lagging strand. Genome Res27, 1336–1343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs, J., Cheblal, A. & Gasser, S. M. Underappreciated roles of DNA polymerase delta in replication stress survival. Trends Genet37, 476–487 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Ganai, R. A., Bylund, G. O. & Johansson, E. Switching between polymerase and exonuclease sites in DNA polymerase epsilon. Nucleic Acids Res43, 932–942 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancey, C. et al. Structure of the processive human Pol delta holoenzyme. Nat. Commun.11, 1109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franklin, M. C., Wang, J. & Steitz, T. A. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell105, 657–667 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Jozwiakowski, S. K., Kummer, S. & Gari, K. Human DNA polymerase delta requires an iron-sulfur cluster for high-fidelity DNA synthesis. Life Sci Alliance2, e201900321 (2019). [DOI] [PMC free article] [PubMed]
- 37.Degasperi, A. et al. A practical framework and online tool for mutational signature analyses show inter-tissue variation and driver dependencies. Nat. Cancer1, 249–263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo, S. et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet49, 635–642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature613, 355–364 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao, L. et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun.11, 3355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao, S. Q., Cuesta, S. M., Tannahill, D. & Balasubramanian, S. Genome-wide DNA Methylation Signatures Are Determined by DNMT3A/B Sequence Preferences. Biochemistry59, 2541–2550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, K. et al. Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA. J. Biol. Chem.293, 7344–7354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz, M. D. et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He, Y. & Ecker, J. R. Non-CG Methylation in the human genome. Annu Rev. Genomics Hum. Genet16, 55–77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jang, H. S., Shin, W. J., Lee, J. E. & Do, J. T. CpG and Non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel)8, 148 (2017). [DOI] [PMC free article] [PubMed]
- 47.Kastriti, M. E. et al. Schwann cell precursors represent a neural crest-like state with biased multipotency. EMBO J.41, e108780 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen, K. D. & Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev.30, 733–750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginno, P. A. et al. A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity. Nat. Commun.11, 2680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alcantara Llaguno, S. R. & Parada, L. F. Cell of origin of glioma: biological and clinical implications. Br. J. Cancer115, 1445–1450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monavarfeshani, A. et al. Transcriptomic analysis of the ocular posterior segment completes a cell atlas of the human eye. Proc. Natl Acad. Sci. USA120, e2306153120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, W. et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet50, 591–602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Raamsdonk, C. D. et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature457, 599–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Raamsdonk, C. D. et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med363, 2191–2199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johansson, P. et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget7, 4624–4631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harbour, J. W. et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet45, 133–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harbour, J. W. et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science330, 1410–1413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furney, S. J. et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov.3, 1122–1129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robertson, A. G. et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell32, 204–220.e215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baubec, T., Ivanek, R., Lienert, F. & Schubeler, D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell153, 480–492 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Neri, F. et al. Single-Base Resolution Analysis of 5-Formyl and 5-Carboxyl Cytosine Reveals Promoter DNA Methylation Dynamics. Cell Rep.10, 674–683 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poulet, A. et al. RT States: systematic annotation of the human genome using cell type-specific replication timing programs. Bioinformatics35, 2167–2176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marchal, C., Sima, J. & Gilbert, D. M. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol.20, 721–737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuijk, E. et al. The mutational impact of culturing human pluripotent and adult stem cells. Nat. Commun.11, 2493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong, E. et al. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc. Natl Acad. Sci. USA99, 14937–14942 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Millar, C. B. et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science297, 403–405 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Villy, M. C., et al. Familial uveal melanoma and other tumours in 25 families with monoallelic germline MBD4 variants. J. Natl. Cancer Inst.116, 580–587 (2024). [DOI] [PubMed]
- 69.Farlik, M. et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep.10, 1386–1397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, X. & Bartel, D. P. kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res45, W534–W538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Newly generated WGBS, WGS, WES, and RNAseq data are deposited in the European Genome-phenome Archive (EGA) database under accession EGAS50000000536. Data can be made accessible upon request to the DACs EGAC50000000356 (Institut Curie) or EGAC00001002078 (Max Planck Institute for Molecular Genetics). Newly generated WGS data on the HGG from Hôpital La Pitié Salpêtrière cannot be made available due to ethical approval restrictions. Newly generated CUT&RUN data are deposited in NCBI’s Gene Expression Omnibus (GEO) under GEO Series accession GSE275181. Previously published WES from Institut Curie patients is available in EGA under accessions EGAD00001004554 and EGAD00001006988. Previously published WGS data from external sources are available in EGA under accessions EGAD00001003568 and EGAD00001005454. WGS data from GEL is not available upon request, but accessible by registered researchers in the Trusted Research Environment (https://www.genomicsengland.co.uk/research/research-environment). All GEL analyzes must take place within the Trusted Research Environment (https://www.genomicsengland.co.uk/understanding-genomics/data). Registration involves an online application, verification by the applicant’s institution, completion of a short information, governance course, and verification of approval by Genomics England. Please see https://www.genomicsengland.co.uk/research/academic/ for more information. Previously published WGBS data on human tissues are available in EGA under accession EGAD00001009789 and in GEO under accession GSE65196. Previously published WGBS data on mouse cells are available in GEO under accessions GSM1382253 and GSM1382256. Publicly available human epigenomic data from ENCODE are described in Supplementary Data 10. Previously published single nuclei RNAseq data are available from the Broad Institute Single Cell Portal with dataset identifier SCP2298. Source data are provided with this paper.