Abstract
Lipid nanoparticles (LNPs) have emerged as a prominent delivery system for nucleic acid drugs, attracting significant attention, especially through the successful development of several commercial products. As a key component in LNPs, cationic lipids have long served as a key technical barrier to block competitors by building up a complex patent thicket. However, there have been few studies as yet that have comprehensively analyzed the patented compounds in LNP formulations, despite a large number of technical reviews and original articles. In this context, this study focuses on analyzing the macroscopic landscapes and microscopic molecular characteristics of LNP patents, aiming to provide a valuable reference for researchers and developers in making informed technological and commercial decisions. By mining 2,994 patents, 265 formulas, 7,674 compounds, and 28,789 fragments, this work sketches the empirical golden ratio of lipid materials in LNP formulation, discloses the advanced technology in the formulation, characterizes high-frequency fragments of heads, linkers and tails in both novel cationic lipids as well as targeting lipids, and establishes a virtual focus library of LNP materials.
Keywords: MT: RNA and epigenetic editing Special Issue, lipid nanoparticles, siRNA, cationic lipids, formulations, patent compounds, heads, linkers, tails, virtual focus library
Graphical abstract

Yuanjia, Ying, and colleagues present the macroscopic landscape and microscopic molecular characteristics of LNP patents, identifying the optimal lipid ratio in formulation and revealing advanced technologies. Furthermore, they conduct a comprehensive analysis of the structural distribution of novel cationic and targeting lipids, establishing a virtual library focused on LNP materials.
Introduction
With the success achieved by Onpattro from Alnylam1 and the COVID-19 mRNA vaccine developed by Moderna and BioNTech/Pfizer companies respectively,2,3 lipid nanoparticles (LNPs) have garnered unprecedented attention as a platform technology for nucleic acid drug delivery.4,5 They have gradually emerged as the mainstream technology for delivering xRNA molecules such as siRNA and mRNA. Among these technologies, cationic lipids play a pivotal role in the development of an advanced delivery system.
However, LNPs delivering gene therapy still encounter numerous challenges, such as the limited escape of only 1%–2% siRNA from endosomes into the cytosol,6 and non-hepatic RNA delivery remains a challenge.7 Recently, researchers surprisingly discovered some rules for designing LNPs. The pKa of LNPs affects its distribution, while the chemical and physical features of LNPs influence the binding of serum proteins, allowing for different organ targeting.8 Typically, there are two ways to improve LNPs—by improving formulation and by improving cationic lipid.
Initially, nucleic acids were encapsulated solely using cationic lipids or polymers. Subsequently, researchers introduced helper lipids and other materials to enhance the performance of the designed delivery system. The formulation exerts a significant influence on the structure of LNPs, thereby impacting their stability, circulation time, and biodistribution.9 The formulation of LNPs is currently undergoing refinement to optimize their compatibility with diverse delivery methods and targeting strategies.10
In addition to the above strategy of formulation optimization, material improvement provides another option to enhance the performance of LNPs. Cationic lipids play a pivotal role in determining the efficiency of endosomal escape, encapsulating nucleic acids, and addressing safety concerns. Since last century, N-[1-(2,3-dioleyloxy) propyl]-N,N,N-trimethylammonium chloride,11,12 a permanently cationic lipid, has emerged as an important candidate. The positive charge provided by this quaternary ammonium-containing compound enables complex formation with negatively charged drugs such as nucleic acids, facilitating their delivery. Similarly well-known permanent cationic lipids include DOTAP and DODAC among others. However, ensuring the safety of cationic lipids has always been a significant challenge due to their interaction with negatively charged substances on cell membranes leading to membrane damage.13,14 In addition, side effects such as inflammatory reactions and oxidative stress caused by cations restrict their further clinical application.15,16 Consequently, researchers have focused on developing ionizable structures for cationic lipids that are more elaborate and possess multiple functions while addressing these limitations.15,17 To optimize the structural design of ionizable lipids, several strategies have been employed. For instance, the incorporation of a branched tail structure has demonstrated enhanced transfection efficiency. In addition, the utilization of a biodegradable architecture has contributed to an improved safety profile.18 Furthermore, the conformation of the ionized lipid head group can exert an impact on immune system regulation.19 In addition to optimizing the structural aspects, synthesis methods also exert a significant influence on the advancement of cationic lipids. Combinatorial chemistry holds significant potential in facilitating the design and discovery of enhanced ionizable lipids, particularly those with ionization properties20 while the synthesis of lipids was achieved through Michael addition reaction, ring opening reaction, condensation reactions, multi-component reactions, and enzyme-assisted chemical reactions,21 wherein tails, heads, and linkers were formed.19,20,22,23,24 Except for the improvement of cationic lipids, novel targeting lipids also made contributions to development of LNP performance by introducing a new option for LNP targeting strategies.
On the other hand, patents represent a valuable yet underutilized data resource that provides guidance for both academic research and industrial applications.25 Patents include a substantial amount of key information on chemical structure of cationic lipids, which not only builds a technical barrier to the delivery technology of LNPs, but also provides key clues to the discovery of novel lipid materials. While there has been an abundance of technical reviews and research papers on LNPs available in the literature, there is insufficient systematic research focusing on the patented compounds in LNP formulations. Technical noise may be a potential reason for the underutilization of patent data. Patentees need to disclose Best Mode to obtain the legal protection of exclusive rights on the one hand, and at the same time expect that the key technology in patents will not be exposed to peer competitors, which is usually done by adding technical noise to patents. Previously, we conducted a comprehensive classification and indexing of all siRNA delivery technologies,26 analyzing the emerging trends and proportions of various delivery methods. However, in this study, our focus is solely on the investigation of LNP technology. We conducted an in-depth analysis of the technical intricacies pertaining to patents associated with LNP. Given the role of data mining in reducing technical noise, several questions need to be addressed such as what is the scope of protection of the existing LNP patents? What are the chemical structural characteristics of patented core material molecules? What are the white space and direction for subsequent optimization and structural modification? In light of the tremendous efforts of academia and industry, mapping the existing patent knowledge could ensure that researchers and developers are well-informed to make scientific decisions about R&D and commerce.
Therefore, this study focuses on analyzing the macroscopic landscapes and microscopic molecular characteristics of LNP patents, aiming to provide a valuable reference for researchers and developers in making informed technological and commercial decisions. Different xRNA molecules exhibit significant variations in strand structure, number of bases, and performance requirements for LNP delivery (e.g., release time, metabolic stability, payload distribution, and capacity).27 To ensure homogeneity, this study will specifically analyze LNP patents related to siRNA. We have compiled a comprehensive database of patent lipids, which has been categorized into three distinct SMILES formats. This valuable resource is anticipated to greatly facilitate virtual syntheses and screening of ionizable lipids. Utilizing this virtual focused library of LNP materials provided, integration of machine learning holds promise for future applications in predicting and simulating LNP formulations,28 thereby facilitating the identification of structure-activity relationships. With the advancement of click chemistry technology, our structural data of head groups, linkers, and tails could serve as invaluable resources for the large-scale synthesis and screening of novel cationic lipids.29
Results
In total, we acquired 13,709 INPADOC patent families, which is a commercial database containing patent applications and grants from 44 of the world’s patent issuing authorities. In addition, we retrieved 52,985 patent documents belonging to these 13,709 INPADOC patent families. Among these patents, a substantial number encompassed alternative delivery methods for siRNA. Our previous research has extensively explored diverse siRNA delivery technologies.26 However, for the purpose of this study, our specific focus was on LNPs. Consequently, we identified 559 patent families and 2,994 patent documents related to LNPs for siRNA delivery. These patents underwent meticulous scrutiny and were categorized into seven distinct types: "formulation," "formulation + structure," "formulation + siRNA sequence," "structure," "siRNA sequence," "all involved," and "others." Figure 1 provides a comprehensive description of these patent types.
Figure 1.
Technological classification of patents
Steady growing tendency
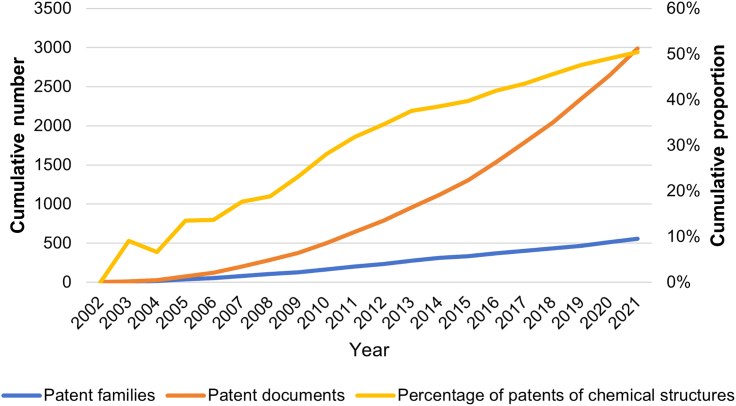
In the past two decades, there has been a significant accumulation of patent applications related to the delivery of siRNA using LNP, both in terms of patent documents and patent families. The increasing number of patents applied indicates a higher investment in intellectual property protection, as applicants are required to pay national patent office fees to maintain their intellectual propert (IP) rights. Interestingly, as depicted in Figure 2, there has been a more pronounced increase in the number of patent documents compared with patent families. This suggests that applicants tend to submit multiple applications for a single technology across different countries. The cumulative percentage of patents claiming cationic lipid structure has risen from 9% in 2003 to 50% in 2021. In addition, it is interesting that the percentage of patents for chemical structures has exhibited a gradual upward trend overall, with two significant spikes observed in 2008 and 2015, respectively. This trend aligns with the development of LNP delivery technology and nucleic acid therapeutics. In 2006, Alnylam successfully used LNPs to deliver siRNA for gene silencing in non-human primates.30 Following this, the optimization of cationic lipids gained increasing attention. The patent for DLin-MC3-DMA, WO2005120152, was first published in 2005, subsequently leading to the disclosure of a series of patents related to cationic lipids. Based on the analysis presented in Figure S1, it is evident that the leading companies driving the advancement of novel lipids varied across different time periods, with Alnylam emerging as a dominant player during the initial phase due to its multifaceted technological strategy. From 2012, following the successful implementation of GalNAc-siRNA coupling technology targeting TTR,31 Alnylam’s siRNA delivery technical strategy shifted its focus toward the development of GalNAc conjugates. Consequently, the proportion of novel lipid patents remained stable from 2013 to 2014. The growth after 2015 is closely related to the rise of mRNA therapeutic technology. In 2016, Moderna secured up to $474 million in funding from AstraZeneca to develop its mRNA vaccine pipeline. Moderna has developed amounts of cationic lipids since 2016. The cationic lipid SM-102, which is used in the COVID-19 vaccine, was first disclosed in patent WO2017070623.
Figure 2.
Patent trend from 2002 to 2021
Patent documents are legal documents filed with a patent office to protect an invention within their jurisdiction. Patent families are series of patents sharing common priority applications, which may be filed in different jurisdictions (countries and regions).
Patented formulas
In general, LNPs are composed of cationic lipids, helper lipids, steroids, and PEG lipids in different ratios.32 We have conducted a comprehensive analysis of the formulation ratios in patents spanning from 2000 to 2021, and Tables S1 and S2 present the most frequently utilized lipids for each component type. As shown in Figure 3, the distribution of lipid mole ratios differed between the periods of 2000–2010 and 2011–2021. From 2011 to 2021, the mole ratios of cationic lipids, helper lipids, and steroids exhibited a concentrated distribution around the median, whereas from 2000 to 2010 the mole ratios of lipids were widely dispersed. The mole ratio range of cationic lipids, which significantly determines the performance of LNPs, exhibited a decrease from 31% to 12.88% between the 25th and 75th percentiles when comparing the periods 2011–2021 with 2010–2010, indicating a narrowing of the molar ratio range for cationic lipids. In contrast, both helper lipid and steroid mole ratios showed higher concentrations over this time period.
Figure 3.
Violin plot of LNP formulation ratio in patents during 2000 to 2021
Structural split of novel lipids
Novel lipids, which had the greatest impact on the transfection efficiency and safety, account for a large proportion in our patent analysis. By sorting out novel lipid compounds, we found that, in 212 patent families involving novel lipids, approximately 75% provided in vivo transfection data. Some of them adopt classical models such as ApoB, FV II, and luciferase targets, others focus on specific diseases such as tumors or viruses among others. In the remaining 20% of patent families, only transfection data are provided on cellular level while their patentees are mainly reagent companies. It is worthwhile analyzing these novel lipids and understanding their structural characteristics. The novel lipids were categorized into four types: cationic lipids, targeting lipids, neutral lipids, and PEG-lipids (Figure 4). It can be seen from the figure that cationic lipids account for the majority. Therefore, this study primarily focuses on discussing novel cationic lipids. What is more, despite the limited abundance, we also made a comprehensive analysis of targeting lipids in response to the growing interest in targeting technology.
Figure 4.
Distribution of novel lipid types
For novel cationic lipids, an analysis was conducted to examine the number of newly developed cationic lipids and the percentage of major patentees during each period (Figure S1), revealing changing trends in active patentees across different periods. With regard to the structure, usually a cationic lipid comprises a head group, a tail group, and a linker group, which are relatively flexible in terms of number and position (Figure 5B). From a novel perspective, we extracted 7,185 novel cationic or ionizable lipid compounds from 212 patent families using SciFinder. After image capture, information translation, and molecule resolution, these compounds were divided into 2 or 3 fragments for analysis based on their frequency and age aspects, as shown in Figures 5A and 5C–5E. The original database of all cationic lipid splits can be viewed in the supplemental information (Tables S3, S4, S5, and S6). The statistics of structure frequency can well help us understand which structures are popular and the age of a structure can reflect its active period. Furthermore, we conducted an in-depth analysis of each component of the cationic lipid including information related to group position, group types, and patentees. The number and proportion of patentees within each head group are presented in Figure S2; the analysis of linker types and positions is depicted in Figure S3, while the analysis of tail types and numbers is illustrated in Figure S4.
Figure 5.
Cationic lipid fragmentation analysis from 1997 to 2022
(A) The process of data collection. (B) Structures of LNPs and fragments of cationic lipid composition. (C) Scale representing the number of structural fragments, with colors indicating different fragments of the cationic lipid (deeper color means larger number). (D) The active period of the most frequent fragment type for each site, with colors representing the various sites of the cationic lipid (the point means age, calculated by average number of active years. The horizontal bar means active period, calculated by standard deviation). (E) The most frequent fragment type for each site, where different colored boxes represent the specific structure and frequency of the fragment. Red, head structure; yellow, linker between head and tail; blue, linker inside tail; green, tail group.
For novel targeting lipids, a small portion of the novel lipids are targeting lipids, which hold significant importance in LNP research and application. Consequently, we conducted an analysis on targeting lipids from targeting ligands as well as structural aspects, while Table 1 shows the basic information of patents involving targeting and Figure 6 and Table S7 give an overview of structural information for targeting lipids.
Table 1.
Targeting lipid patent information
| Patent family | Patentee | Lipid amount | Targeting moiety | Targeting tissue |
|---|---|---|---|---|
| WO2012016188A2 | Alnylam | 31 | RGD peptides; PSMA; anisamide; fucose | tumor; liver |
| CN107082747B | Nitto Denko | 22 | retinoic acid | liver |
| US9814777B2 | Arbutus | 10 | monosaccharide; folate | tumor; liver |
| US20130079383A1 | Arrowhead | 12 | VLA-4-ligand | leukocyte plasma membranes |
| WO2014055941A2 | Vladimir Torchilin | 1 | monosaccharide | antigen-presenting cells |
| WO2006022325A1 | Nippon Shinyaku | 9 | galactose derivative | liver |
Figure 6.
Analysis of targeting lipid structure
(A) Distribution of targeting moiety mentioned in patents. (B) Number of linker species applied by targeting lipids. (C) Frequency of each linker species. (D) Frequency of tail species.
Head groups
Through structural analysis of the head groups, we have observed that most of the head groups in cationic lipids developed over the past two decades predominantly exist as amine groups, particularly tertiary amines, which exhibit an ionizable state rather than being permanently cationic lipids. The most notable and potent head groups can be broadly categorized into linear structures, cyclic structures, and amino acid or polypeptide structures.
The structures of amino acids and polypeptides are mainly basic amino acids such as arginine, lysine, and histidine, and the most representative structure comes from the guanidine group structure of arginine. From the active period of head pieces shown in Figure 5D, it can be observed that guanidine gradually declined in popularity. The six most utilized head groups, shown in Figure 5E, were -CCN(C)C, piperazine, pyrrolidine, -CCCN(C)C, -CN(C)C, and piperidine, with cyclic and linear groups being equally represented. The most prevalent structure for linear tertiary amines is -(C)nN(C)C. Over an extended period of development, n = 2 or 3 in -(C)nN(C)C emerged as the preferred choice with participation from more than 18 patentees. However, n = 0 or >3 was rarely selected over time with fewer than 10 patentees involved. Apart from linear head groups, cyclic groups also gained increasing prominence in the head group composition. Piperidine, piperazine, morpholine, and pyrrolidine garnered greater attention and underwent extensive experimentation within novel lipid research. To account for variations in the number of novel lipids synthesized by each patentee, we have also compiled a summary of patentees involved in each head group in Figure S2. Notably, Moderna demonstrated strong specificity toward squaramide (3-cyclobutene-1,2-dione)-based and -NCCO structures.
Linkers
In terms of the linkers, we observed four distinct configurations in the patent: absence of a linker, presence of a linker between the head and tail regions, placement of a linker within the middle section of the tail, and occurrence of linkers at both ends. Consequently, our analysis focused on two specific types within the group of linkers: those positioned between the head and tail regions, as well as those exclusively located at the tail.
From the results, it is evident that over 80% of the cationic lipid head groups and hydrophobic tails are linked by a diverse range of functional groups. Among these, amide groups, ether groups, ester groups, thioether, and triazine are the most frequently employed linkers. The prevalent use of thioether and triazine can be attributed to Innorna’s efficient synthesis method for constructing lipid libraries, which is further discussed later in this article. In addition, cyclic groups such as benzene, tetrahydrofuran, cyclopentane, and triazine are not utilized independently but rather in conjunction with esters, amides, ethers or thioethers. The incorporation of cyclic moieties enriches the diversity of linkers while it is worth highlighting that several linkers exhibit high specificity among patentees compared with the head group.
In contrast, the hydrophobic chain at the tail was found to be concealed by the linker, accounting for less than 30%. Figure S3 illustrates that this lipid component predominantly emerged after 2010, with Alnylam and Merck as representative patent rights holders. Moderna witnessed a significant surge in activity after 2015, primarily focusing on designing cationic lipids with linkers at the tail. While there is considerable structural diversity between the head and tail of the linker, it is noteworthy that the groups present at the tail are relatively limited in variety. The blue section of Figure 5E demonstrates their high prevalence over the past decade, including amide, thioester, carbonic ester, ether, and ester functionalities; among these groups, ester functionality holds an overwhelmingly dominant position.
Tails
As shown in Figure 5E, the number of tails in lipid varied from 1 to 12 with most of the lipids containing 2 tails, which could be further classified into asymmetrical and symmetrical types. Over 95% of lipids possess fewer than 4 tails with only a small fraction having more than 6 tails. Lipid tails exhibit significant diversity, primarily attributed to variations in length, number and position of unsaturated bonds, presence of bifurcations, as well as type and location of linkers. Despite this diversity observed among lipid tails, we have identified certain highly frequent tail configurations. In approximately 2009, cholesterol emerged as a prominent lipid tail. Several patentees such as Moderna, Alnylam, Arcturus, Thermo. Novartis, Merck, Enzon, etc., conducted trials to explore its potential. However, this particular lipid did not exhibit significant efficacy. Conversely, alkane chains containing unsaturated bonds garnered widespread success. C18 containing two double bonds emerged as the most popular choice. Saturated alkane chains of various lengths (from C6 to C18) have consistently served as fundamental building blocks for novel lipid synthesis over an extended period. The advent of Innorna has significantly influenced the statistical distribution of saturated alkane chain frequencies, owing to the inventors' efficient synthesis of a comprehensive lipid library encompassing diverse chain lengths ranging from C6 to C18. By analyzing the data after removing Innorna, it was observed that C8, C9, C10, C12, C14, C15, and C16 were more frequently preferred; however, their prevalence was concentrated between 2010 and 2012. Notably, in recent years, tails containing ester groups have emerged prominently with remarkable diversity attributed to variations in ester length and position along the alkane chain. Branching is a common characteristic feature observed in such tails.
Targeting lipids
As revealed in Table 1, there are six patent families within our data system involved in novel targeting lipids. According to the disclosed patents, these targeting lipids employed various ligands for different targeting sites, specifically to liver, tumors, and immune cells. Notably, these targeting lipids differ from the commonly employed head groups and linkers described previously.
Targeting lipids, employing the active targeting strategy, usually exhibit a general formula for the structure: head group (targeting moiety)–linker–hydrophobic tail. There is diversity observed in both the types and numbers of target moieties present in targeting lipids, which is shown in Figure 6A. Generally, targeting lipids contain between one and four target moieties. As illustrated in Figure 6B, linkers utilized by targeting lipids display diverse patterns, and they commonly rang from two to four types with only a few exceeding this range. Amide represents the most frequently employed linker followed by ether, PEG, and carbamate, as shown in Figure 6C. In terms of tail composition among targeting lipids, they predominantly consist of either a single-stranded (cholesterol or cholesterol + C18:2) or symmetrical double-stranded structures. Symmetrical double strands primarily comprise C18 saturated alkane chains along with C17 containing single unsaturated double-bonded olefin chains or C18 containing two unsaturated double-bonded olefin chains (Figure 6D).
Discussion
By conducting comprehensive mining and analysis of siRNA+ LNP-related patents, we have obtained valuable insights into the overall development trends and significant technical aspects. In general, there has been a consistent expansion in both the patent family size and the number of siRNA+ LNP-related patents each year, with the proportion of novel compound inventions generally trending upward. However, this growth trend is not static, and it has experienced two log-type growths in 2008 and 2015, respectively. The increase in the proportion of novel lipid patents primarily stems from two factors: firstly, a reduction in the proportion of other types of patents; and, secondly, the growing advancements and attention toward novel lipids in both technology and market domains. As depicted in Figure 3 above, the stability of classic LNP formulations fluctuated between 2000 and 2010, resulting in a higher frequency of formulation-related patents during the initial stage. However, with a relatively stable proportion of four-lipid components, academic and industrial communities gradually shifted their focus toward exploring novel lipids with enhanced efficiency. Consequently, there has been an upward trend observed in the share of such patents. Technological innovation within the industry has been led by companies that successfully commercialized LNP formulations along with their close collaborators. For instance, Arbutus and Alnylam exhibited high activity around 2008 but experienced gradual cooling thereafter; whereas Moderna has spearheaded a new wave of ionizable lipid innovation since 2015.
Undoubtedly, novel cationic lipids have garnered significant attention from companies and researchers. Therefore, targeted exploration of the structural characteristics of these novel cationic lipids will be of great benefit to future industrial development and academic research. In terms of technical aspects, the formulas of LNPs and novel lipid compounds emerge as the most salient features. Drawing upon our obtained results and in conjunction with academic articles, we delve into a more comprehensive discussion below.
Current status and future prospects of LNP formulations
Previously, Moderna and Pfizer have generated over $100 billion in global revenues from the sales of COVID-19 mRNA vaccines. However, their LNP technology is subject to claims of US8058069 and potential infringement risks. Patented formulation information reflects the progressive establishment of optimized formulations of LNPs and means that companies seeking to develop LNP formulations face challenges beyond prior patent claims, including finding new formulation ratios for efficient delivery.33 In our analysis of patents on LNP formulations, we have identified specific specialized patents that indicate companies are already focusing on formulations for non-hepatic delivery or targeted pathway implementation. Regarding LNP targeting patents, the majority employ active targeting strategies such as antibodies, peptides, folic acid, hyaluronic acid, mannose, etc. In addition, there are passive targeted transports achieved by modifying the physical and chemical properties of LNPs. Consequently, there is significant scrutiny regarding the fundamental components and optimal ratio of LNPs. The following two examples illustrate this point.
It is noteworthy that the molar ratio of cationic lipids in a specific patent may consist of a single permanent cationic lipid, a single ionizable lipid, or a combination of two different cationic lipids when examining the formulation details of these patents. For instance, Moderna’s patent WO2017180917 employs a blend comprising 50% ionizable lipids and 5% permanent cationic lipids to enhance efficiency and retention for intratumoral delivery. In the patent example, following injection into tumors in mice, GFP protein expression within the tumor was observed to be 1,037 times greater than in the liver. Similarly, in certain patent cases, helper lipids can also be mixtures of diverse lipids. These findings suggest that the composition of LNPs is not limited to four components and incorporating additional constituents could potentially yield improved outcomes.
In patent WO2020051223, the authors propose enhancing the efficiency of non-liver delivery by incorporating a selective organ-targeting compound into LNPs. They successfully demonstrated that altering the apparent pKa of LNPs enables targeting delivery to spleen, liver, and lung. The selective organ targeting compound can be either a permanently cationic lipid or a permanently anionic lipid. This approach serves as a technical prototype for SORT (selective organ targeting). Recently, Dr. Daniel J. Siegwart’s group from the University of Texas introduced an innovative technology known as SORT, which involves introducing a fifth lipid component (either permanent cationic or permanent anionic) into ionizable lipid-based LNPs for extrahepatic targeting delivery.10 This groundbreaking technology has been recognized as one among Natures Seven Technologies to Watch for 2022.
Therefore, revising the patent layout in the future may involve incorporating one or more components to expand the application scope and functionality of LNPs. In addition, cationic lipid protection poses another barrier for LNP technology development. Moderna and Pfizer hold patents for their respective cationic lipids SM-102 and ALC-0315. A comprehensive discussion on all patented cationic lipids is provided in a subsequent section.
Prospective on targeting lipids
As we described in the last section, current strategies for targeting LNP formulations can be achieved through formulation alterations. Our analysis of patents also revealed a novel class of targeting lipids designed to facilitate LNP targeting using an active ligand-based approach. Structurally, these targeting lipids primarily consist of a targeting moiety, linker, and hydrophobic tail. From a structural perspective, key features of targeting lipids include abundant linkers with limited variation in tail forms and a diverse range of targeting moieties at the head region. It is evident that the efficacy of LNP targeting is determined by the type and number of corresponding counterparts within the targeting moiety responsible for directing LNPs toward desired sites. The hydrophobic tail plays a role in providing hydrophobicity and can be selected from alkane chains, olefin chains, or cholesterol-like tails as main options. On the other hand, linkers assume multifaceted roles such as connecting synthetic processes and regulating lipid amphiphilicity if necessary; thus presenting greater complexity and serving as a crucial regulatory position in novel targeting lipid inventions.
Although the amount of novel targeting lipids in LNP patents on siRNA is not as prominent as that of novel cationic lipids, this technology deserves greater attention and holds significant prospects. In 2023, Dr. Michael J. Mitchell’s team synthesized a lipid library with anisamide ligand using the one-pot method, which serves not only as a cationic lipid for siRNA binding but also utilizes ligand targeting to enhance LNP uptake by liver fibroblasts.34 This multifunctional lipid aligns with current research interests. In addition, targeting lipids offers the potential for direct conjugating with siRNA to obtain targeting delivery. Previously, siRNA conjugated with antibodies, peptides, and GalNAc have made great success in both academic and commercial areas. Furthermore, the conjugation of a C16 lipid chain to siRNA has shown durable gene silencing in local administration.35 These examples made it possible for targeting lipids conjugated with siRNA. In the future, the targeting lipids may facilitate the development of not only LNP formulations but also targeted conjugates.
Advancements in the structure of novel cationic lipids
Previously, the analysis of lipid structures primarily focused on commercially available lipids, which are highly representative but lack evolutionary characteristics. Based on this study, it is evident that the identification of suitable target LNP delivery systems necessitates the establishment of diverse lipid libraries catering to distinct groups, thereby facilitating extensive screenings.36 Notably, novel lipid patents hold a significant position among all patents, highlighting their immense value and warranting further in-depth analysis. The structural information derived from patents can serve as a valuable database, laying the groundwork for subsequent establishment of a comprehensive lipid library and facilitating novel LNP screening.
Head groups
Through the analysis of head structure, it becomes evident that, over the course of more than two decades of development, various representative linear and cyclic head groups have been extensively employed. In the pursuit of novel cationic lipids, researchers have also established a screening platform and explored diverse head group options. Apart from considering conversion efficiency and safety as primary factors, no other significant biases were observed. However, recent studies have shed light on the crucial role played by the structural configuration of the head group in immune regulation. Notably, it has been demonstrated that the cyclic shape of the head group exhibits STING agonistic properties, thereby modulating innate immune cells.19 These findings emphasize the imperative need to incorporate additional functionalities into future designs for cationic lipids.
Linkers
The linker is typically synthesized to connect the head group and the tail group, thereby influencing the pKa of ionizable lipids, transfection efficiency, and lipid biodegradability.37 Various synthetic routes result in cationic lipid linking groups composed of multiple components such as benzene, cyclopentane, and triazine. Among these linkers, ester groups are widely employed due to their favorable biodegradability properties,37 as demonstrated by the number of patents and patentees involved. Notably, ester groups exhibit pronounced advantages in tail linkers. In recent years, hydrophobic chain-concealing linkers have emerged with Alnylam’s significant contribution in 2011 through introducing ester or thioester bonds. Presently, both ALC-0315 and SM-102 adopt this strategy for enhanced biodegradability within the body. In contrast to the diversity observed in head-to-tail linkers, tail region linkers display less variability primarily based on ester linkages.
As the most abundant linker triazines, their prevalence can be attributed to the data impact facilitated by the lipid library synthesized by Innorna. Triazines function as a bridge between the cationic head and lipophilic tail, allowing for facile derivatization with up to three distinct substituent groups, including lipophilic tails and polar heads. Moreover, owing to their antibacterial properties, recent studies have demonstrated that cationic lipids employing triazine linkers also exhibit potent antibacterial activity against Gram-negative bacteria.38 This implies that future development of each component of cationic lipids should not only consider transfection efficiency and safety concerns but also aim at enhancing their functional research capabilities. By designing more targeted lipid materials based on diverse physiological conditions and pathological characteristics, further advancements can be achieved.
Tails
The characteristics of the tail group are primarily manifested in the number, type, length, degree of unsaturation, presence, and position of a linker, as well as the existence of branches. The number and length of tails have a significant impact on the lipophilicity (cLogD) of lipids; therefore, it is preferable to restrict the hydrophobic tails to achieve a cLogD within the range of 10–14. Studies have shown that tail unsaturation greatly influences the fluidity of ionizable lipids.39 Notably, MC3 serves as an exemplar of successful implementation. The introduction of a linker in the tail enhances lipid biodegradability. Branching in two-tailed lipids offers advantages comparable with those observed with multi-tailed lipids by facilitating endosome escape through increased cross-sectional area provided by lipid tails. Successful mRNA delivery systems utilizing ionizable lipids predominantly feature ester-linked and branched tails.
Cases and prospectives
This study presents a concise overview of the macroscopic landscapes and microscopic molecular characteristics of LNP patents. Initially, we conducted an analysis on the macro trends of LNP technology-related patents, followed by an in-depth examination of patent documents to investigate formulation ratios and structural attributes exhibited by lipid molecules across representative patentees. Alnylam achieved commercial success with their pioneering siRNA LNP formulation by employing different strategies involving varying linker amounts and positions within novel lipid design frameworks. Moderna’s active involvement since 2015 primarily focuses on tail-based linker strategies while adopting NCCQ or NCCCQ as head groups (where Q represents diverse functional groups). Innorna, very outstanding in these years, based their strategy on a triazine+ X linker, where X represents either ether or thioether. Several groups have been selected for head and tail, utilizing combinatorial chemistry (multi-component reaction) to obtain a multitude of novel components. Thus far, the synthesis of all FDA-approved cationic lipids, including Dlin-MC3-DMA (MC3), ALC-0315, and SM-102, has required complex multi-step methods with low yields. Combinatorial chemistry serves as a crucial guiding principle for lipid synthesis due to its high-throughput, high-selectivity, and high-efficiency synthetic capabilities. In academia, Dr. Daniel G. Anderson’s group from the Massachusetts Institute of Technology successfully screened and identified the best cationic lipid using a three-component reaction (3-CR) system which exhibited superior anti-tumor efficiency as well as pulmonary delivery capabilities.19,23 In addition, Dr. Xu Xiaoyang’s group from the New Jersey Institute of Technology efficiently synthesized a biodegradable cationic lipid library by employing enzymes as catalysts along with various amino alcohols and fatty acids as raw materials.24 However, in specific pathological environments where no clear standards exist for group selection and combination can be followed, combinatorial chemistry synthesis enables high-throughput screening to identify the optimal combination of cationic lipids.
In summary, this research shows a landscape of LNP patents, and especially describes the chemical space distribution of patented cationic lipids, identifies key cationic lipids, and provides a methodological reference of drug discovery. There is still ample scope for application scenarios and design in this area. The empirical golden ratio of different lipid materials within LNP formulation represents a concentrated reflection of research outcomes from the past decade and will serve as a foundation for future investigations. As major companies and universities delve deeper into research, this golden ratio will evolve based on changes in delivery methods and targets. Considering diverse delivery purposes and physiological environments, novel lipid synthesis should not only fulfill requirements for high transfection efficiency and safety but also exhibit enhanced functionality in auxiliary applications; for instance, activate the immune system like a vaccine adjuvant or make a contribution in anti-inflammation. Combinatorial chemistry significantly influences the number of patented novel lipids, while the development of structure-activity relationships remains a fundamentally important avenue for the future in helping to understand lipids and develop novel lipid materials. The targeting lipids disclosed patents revealed a common structure, which is head group (targeting moiety)–linker–hydrophobic tail. These findings from patents may help design not only functional lipids in LNPs but also conjugate siRNA. Generally, the empirical golden ratio of lipid materials, advanced technology in the formulation, high-frequency fragments of heads, linkers and tails in novel cationic lipids, and the virtual focus library of LNP materials in this study will provide a valuable reference for researchers and developers in making informed technological and commercial decisions in the area of LNPs.
Materials and methods
In this study, patents were retrieved from Derwent Innovation. The initial step involved identifying relevant search keywords for LNPs that deliver siRNA for LNPs that deliver siRNA. Drawing on our previous research,25,26 we formulated the following patent search terms: CTB=(siRNA∗ OR (small ADJ interfer∗ ADJ RNA∗) OR (short ADJ interfer∗ ADJ RNA∗) OR (silenc∗ ADJ RNA∗)) AND CTB=(deliver∗ OR vehicle OR carrier) AND PY>=(2000) AND PY<=(2021).
Irrelevant patents were manually excluded from the collected patent dataset. Subsequently, relevant patents were labeled for classification through hierarchical reading of their title, abstract, claims description, and full text. Obtaining valid structural data is crucial for analyzing novel lipid structures. We utilized SciFinder to capture the structural information of compounds mentioned in the patents. Heterogeneous structures such as reactants, reaction intermediates, solvents, and commercial lipids were identified and excluded. The obtained valid structural information from patents was converted into readable format using BIOVIA and Open babel tools. By combining manual examination with RDKit methods, we performed structural separation of novel cationic lipid compounds and conducted statistical analysis on their different positional characteristics.
Data and code availability
All data associated with this study are present in the main text or the supplementary materials.
Acknowledgments
This study was supported by the Science and Technology Development Fund of Macau SAR (nos. 005/2023/SKL, SKL-QRCM(UM)-2023-2025, and 0049/2024/AGJ), Wuyi University (EF36/ICMS-ZY/2022/WYU), Guangdong-Hong Kong-Macao Joint Laboratory for New Drug Screening (EF2023-00054-FHS), and University of Macau (nos. MYRG2022-00103-ICMS, MYRG-CRG2023-00007-ICMS-IAS, and MYRG-GRG2024-00268-ICMS).
Author contributions
Y. Hu and Y.Z. organized and supervised this study. D.O. also supervised this study. Y. Han and M.W. contributed to investigation, data curation, and writing of the original draft and the revision. Y.C. contributed to investigation and the data curation.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102362.
Contributor Information
Ying Zheng, Email: yzheng@um.edu.mo.
Yuanjia Hu, Email: yuanjiahu@um.edu.mo.
Supplemental information
References
- 1.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.-C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. Overseas. Ed. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. Overseas. Ed. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. Overseas. Ed. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yonezawa S., Koide H., Asai T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020;154–155:64–78. doi: 10.1016/j.addr.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M., et al. Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 7.Wang D., Lin J., Jia F., Tan X., Wang Y., Sun X., Cao X., Che F., Lu H., Gao X., et al. Bottlebrush-architectured poly(ethylene glycol) as an efficient vector for RNA interference in vivo. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dilliard S.A., Cheng Q., Siegwart D.J. On the Mechanism of Tissue-specific mRNA Delivery by Selective Organ Targeting Nanoparticles. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cárdenas M., Campbell R.A., Yanez Arteta M., Lawrence M.J., Sebastiani F. Review of structural design guiding the development of lipid nanoparticles for nucleic acid delivery. Curr. Opin. Colloid Interface Sci. 2023;66 [Google Scholar]
- 10.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng W.-C., Haselton F.R., Giorgio T.D. Transfection by Cationic Liposomes Using Simultaneous Single Cell Measurements of Plasmid Delivery and Transgene Expression. J. Biol. Chem. 1997;272:25641–25647. doi: 10.1074/jbc.272.41.25641. [DOI] [PubMed] [Google Scholar]
- 12.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: A Highly Efficient, Lipid-Mediated DNA-Transfection Procedure. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Contr. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Cui S., Wang Y., Gong Y., Lin X., Zhao Y., Zhi D., Zhou Q., Zhang S. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol. Res. 2018;7:473–479. doi: 10.1039/c8tx00005k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X., Kong N., Zhang X., Cao Y., Langer R., Tao W. The landscape of mRNA nanomedicine. Nat. Med. 2022;28:2273–2287. doi: 10.1038/s41591-022-02061-1. [DOI] [PubMed] [Google Scholar]
- 16.Wei X., Shao B., He Z., Ye T., Luo M., Sang Y., Liang X., Wang W., Luo S., Yang S., et al. Cationic nanocarriers induce cell necrosis through impairment of Na+/K+-ATPase and cause subsequent inflammatory response. Cell Res. 2015;25:237–253. doi: 10.1038/cr.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dilliard S.A., Siegwart D.J. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat. Rev. Mater. 2023;8:282–300. doi: 10.1038/s41578-022-00529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han X., Zhang H., Butowska K., Swingle K.L., Alameh M.-G., Weissman D., Mitchell M.J. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 2021;12:7233. doi: 10.1038/s41467-021-27493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y., Golubovic A., Xu S., Pan A., Li B. Rational design and combinatorial chemistry of ionizable lipids for RNA delivery. J. Mater. Chem. B. 2023;11:6527–6539. doi: 10.1039/d3tb00649b. [DOI] [PubMed] [Google Scholar]
- 21.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. Int. Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., Whitehead K.A. Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small. 2019;15 doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 23.Li B., Manan R.S., Liang S.-Q., Gordon A., Jiang A., Varley A., Gao G., Langer R., Xue W., Anderson D. Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing. Nat. Biotechnol. 2023;41:1410–1415. doi: 10.1038/s41587-023-01679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Zhang X.-Q., Ho W., Li F., Gao M., Bai X., Xu X. Enzyme-Catalyzed One-Step Synthesis of Ionizable Cationic Lipids for Lipid Nanoparticle-Based mRNA COVID-19 Vaccines. ACS Nano. 2022;16:18936–18950. doi: 10.1021/acsnano.2c07822. [DOI] [PubMed] [Google Scholar]
- 25.Lyu L., Feng Y., Chen X., Hu Y. The global chimeric antigen receptor T (CAR-T) cell therapy patent landscape. Nat. Biotechnol. 2020;38:1387–1394. doi: 10.1038/s41587-020-00749-8. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y., Xiong S.-H., Li F., Kong X.-J., Ouyang D.-F., Zheng Y., Yu H., Hu Y.-J. Delivery of therapeutic small interfering RNA: The current patent-based landscape. Mol. Ther. Nucleic Acids. 2022;29:150–161. doi: 10.1016/j.omtn.2022.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eygeris Y., Gupta M., Kim J., Sahay G. Chemistry of Lipid Nanoparticles for RNA Delivery. Acc. Chem. Res. 2022;55:2–12. doi: 10.1021/acs.accounts.1c00544. [DOI] [PubMed] [Google Scholar]
- 28.Wang W., Feng S., Ye Z., Gao H., Lin J., Ouyang D. Prediction of lipid nanoparticles for mRNA vaccines by the machine learning algorithm. Acta Pharm. Sin. B. 2022;12:2950–2962. doi: 10.1016/j.apsb.2021.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey S., Gupta A., Saha A., Pal S., Kumar S., Manna D. Sunlight-Mediated Thiol–Ene/Yne Click Reaction: Synthesis and DNA Transfection Efficiency of New Cationic Lipids. ACS Omega. 2020;5:735–750. doi: 10.1021/acsomega.9b03413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann T.S., Lee A.C.H., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M., et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 31.Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S., et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 32.Kalita T., Dezfouli S.A., Pandey L.M., Uludag H. siRNA Functionalized Lipid Nanoparticles (LNPs) in Management of Diseases. Pharmaceutics. 2022;14:2520. doi: 10.3390/pharmaceutics14112520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenchov R., Bird R., Curtze A.E., Zhou Q. Lipid Nanoparticles—From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano. 2021;15:16982–17015. doi: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- 34.Han X., Gong N., Xue L., Billingsley M.M., El-Mayta R., Shepherd S.J., Alameh M.G., Weissman D., Mitchell M.J. Ligand-tethered lipid nanoparticles for targeted RNA delivery to treat liver fibrosis. Nat. Commun. 2023;14:75. doi: 10.1038/s41467-022-35637-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Q., Khvorova A. RNAi-based drug design: considerations and future directions. Nat. Rev. Drug Discov. 2024;23:341–364. doi: 10.1038/s41573-024-00912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramishetti S., Hazan-Halevy I., Palakuri R., Chatterjee S., Naidu Gonna S., Dammes N., Freilich I., Kolik Shmuel L., Danino D., Peer D. A Combinatorial Library of Lipid Nanoparticles for RNA Delivery to Leukocytes. Adv. Mater. 2020;32 doi: 10.1002/adma.201906128. [DOI] [PubMed] [Google Scholar]
- 37.Jörgensen A.M., Wibel R., Bernkop-Schnürch A. Biodegradable Cationic and Ionizable Cationic Lipids: A Roadmap for Safer Pharmaceutical Excipients. Small. 2023;19 doi: 10.1002/smll.202206968. [DOI] [PubMed] [Google Scholar]
- 38.Pennetta C., Bono N., Ponti F., Bellucci M.C., Viani F., Candiani G., Volonterio A. Multifunctional Neomycin-Triazine-Based Cationic Lipids for Gene Delivery with Antibacterial Properties. Bioconjug. Chem. 2021;32:690–701. doi: 10.1021/acs.bioconjchem.0c00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Contr. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the main text or the supplementary materials.