Abstract
Fungal metabolites are known for their broad therapeutic effects. In this context, the fungal strain of Aspergillus ficuum (FCBP-DNA-1266) was examined for its secondary metabolites and in vivo activities. This led to the isolation of naphtho-gamma-pyrone (aurasperone B) and a sterol (ergosterol), characterized using advanced spectroscopic techniques such as 1H NMR and 13C NMR. The isolated metabolites were evaluated for their in vivo anti-inflammatory and analgesic activities utilizing an animal model. The study showed that both metabolites have significant pharmacological effects (P ≤ 0.05) in a dose-dependent manner. In addition, in silico analysis was employed to aid the in vivo anti-inflammatory activity and the molecular docking results were in agreement with the experimental findings. For the first time, we present the pharmacological activities and 2D NMR of aurasperone B, which will shed light on the bioactive potential of secondary metabolites of Aspergillus ficuum.
Fungal metabolites are known for their broad therapeutic effects. In this context, the fungal strain of Aspergillus ficuum (FCBP-DNA-1266) was examined for its secondary metabolites and in vivo activities.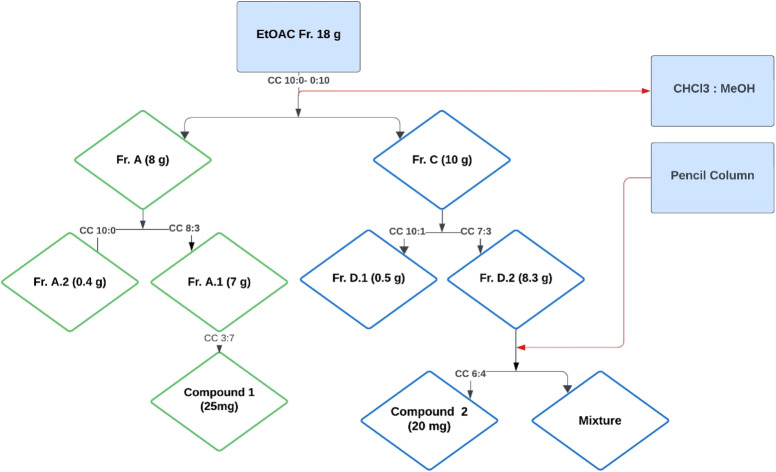
1. Introduction
Secondary metabolites are structurally varied and pharmacologically active molecules produced by fungi, thriving in hyperbaric, oligotrophic, hypersaline, and other specialized environments.1,2 They are common in nature, particularly naphtho-gamma-pyrones (NγPs) found in Aspergillus, Fusarium, and Penicillium.3 Their structures consist of a naphthalene and a γ-pyrone moiety, while their dimeric form is created by linking two NγPs together through a diaryl bond. The cytochrome P450 enzymes play an important role in the dimerization of NγPs based on the stereoselectivity of monomeric substrates.4Aspergillus is a common genus of fungi responsible for the biosynthesis of several specialized small molecules, including fumonisins, bicoumarins, asperazines, ochratoxins, and NγPs.5 Chemists and biologists have shown particular interest in NγPs pigments because they have been widely isolated as antioxidant, antiviral, and antimicrobial agents.6
Fungal-derived steroids have shown potential benefits in reducing neurotoxicity and neuronal cell death, suggesting they may be useful in delaying the onset of dementia. Additionally, fungal steroids have been proven to prevent autoimmune diseases and chronic inflammatory diseases.7 The sterol ergosterol, produced by fungi, plays a role in maintaining membrane fluidity and structure.8 Beyond their structural roles, sterols have also been shown to exhibit a variety of biological activities.9
In our ongoing search for bioactive metabolites from fungi, we have identified several potent metabolites from Aspergillus ficuum (A. ficuum). It has demonstrated numerous bioactivities, such as antimicrobial, DPPH radical scavenging, anti-inflammatory, antispasmodic, and anticancer properties. The ethyl acetate extract of A. ficuum showed a significant anti-inflammatory effect at a dosage of 150 mg kg−1.10,11
Based on our previous investigations, we have worked on the isolation and characterization of the secondary metabolites aurasperone B and ergosterol from A. ficuum. To the best of our knowledge, this constitutes the first report on the in vivo activities of aurasperone B, specifically its anti-inflammatory and analgesic effects using a mouse model. Furthermore, the activities of both compounds were supported by in silico analysis.
2. Materials and methods
2.1. Fungal culture collection
The First Fungi Bank of the University of Punjab in Lahore, Pakistan, provided the fungal strain A. ficuum (FCBP-DNA-1266). The Ethical Committee FAHV&S of the University of Agriculture Peshawar, Pakistan, approved in vivo studies (7196/LM/UoA).11
2.2. General experimental conditions
The 1H NMR and 13C NMR spectra of both compounds were carried out using Bruker AVANCE 500 and 100 MHz, respectively, at Hussain Ejaz Research Center of Chemistry, University of Karachi, Pakistan. TMS was used as a reference compound, while all the spectral information was obtained in deuterated solvents. 2D NMR techniques (HSQC, COSY & HMBC) were examined through Mnova Software. Column and thin layer chromatography techniques were carried out using precoated aluminum sheets with silica-gel 60 F254 (20 × 20 cm, 0.2, Merck, Germany) and silica gel (200–300 mesh), respectively. The chromatographic techniques were carried out on commercial solvents after they were redistilled. For the anti-inflammatory and analgesic activities, analytical grade reagents and chemicals (Sigma Aldrich) were used.
2.3. Culture cultivation
A haemocytometer was used to count spores in a suspension of A. ficuum (FCBP-DNA-1266), containing 10−5 conidia per ml. This suspension was then inoculated centrally into 20 liters of medium sized potato dextrose broth (PDB), consisting of potato extract (4 g L−1) and dextrose (20 g L−1) in sterilized 500 mL Erlenmeyer flasks. The pH of the medium was adjusted to 5.6 ± 0.5, and the inoculated flasks were maintained under static conditions at 28 °C for three weeks.11
2.4. Extraction & isolation
After three weeks of culture, the mycelia were removed from the broth and dried. The dried mycelia were extracted with ethyl acetate (3 × 500 mL) and condensed by a rotary evaporator under reduced pressure. A total of 18 g of crude ethyl acetate fraction was obtained.
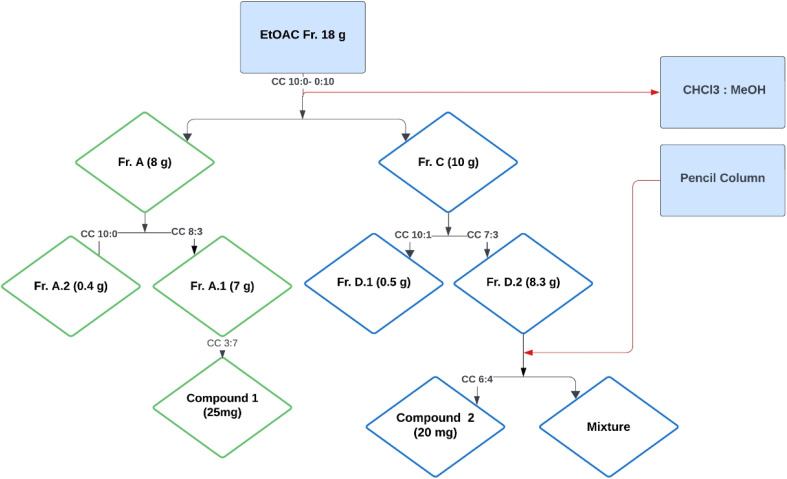
The ethyl acetate fraction (18 g) was subjected to column chromatography using a solvent system of chloroform and methanol in varying ratios (10 : 0 to 0 : 10), resulting in the collection of 74 vials (Fig. 1). These vials were sorted based on polarity into four subfractions, namely A, B, C, and D. The quantity and polarity of subfractions A to D were 2.5 g (10 : 0), 3.7 g (9 : 1), 10 g (6 : 4), and 1.8 g (0 : 10), respectively. Further separation of subfraction A into two subfractions, A.1 and A.2, The subfraction A1 yielded compound 1 (25 mg) using preparative TLC with a polarity of MeOH and CHCl3 (3 : 7). The fraction C was further divided into two subfractions, D1 and D2. The subfraction D2 upon subjection to the pencil column resulted in the isolation of compound 2 (20 mg) in a solvent system of methanol and chloroform with a polarity of 6 : 4.
Fig. 1. Isolation scheme of compound 1 and compound 2.
2.5. Anti-inflammatory study
The anti-inflammatory activity of aurasperone B and ergosterol was evaluated using the carrageenan-induced paw edema test.12 The animals were separated into four groups: vehicle (5% DMSO and 1% Tween 80), vehicle + carrageenan, aspirin + carrageenan, and compounds + carrageenan. Aurasperone B and ergosterol were orally administered in three different doses of 5, 10, and 15 mg kg−1. While the control group received aspirin at a dose concentration of 15 mg kg−1. After an hour of administration, a 1% carrageenan solution was administered to the left hind paw in the subplantar area, and the paw thickness of the animals was measured every hour for a total of 5 h using a vernier caliper. Tween 80 was used to increase the solubility of the drug or extract. Standard aspirin and the vehicle were taken as positive and negative controls, respectively. The percentage anti-inflammatory effect was determined using the following formula:Anti-inflammatory effect (%) = [(Ct − C0) control − (Ct − C0) treated]/(Ct − C0) control × 100Ct = thickness after 1–5 h C0 = baseline paw thickness.
2.6. Analgesic study
The study evaluated the antinociceptive (pain-relieving) effects of aurasperone B and ergosterol using an abdominal constriction test induced by acetic acid.13 The test was conducted on animals (presumably mice or rats). Aurasperone B and ergosterol were orally administered in three different doses of 5, 10, and 15 mg kg−1. The control group received diclofenac sodium at a dose concentration of 50 mg kg−1, a known pain reliever, which was provided through the intraperitoneal (i.p.) route. One hour after administration, the animals were injected with 1% acetic acid at a volume-to-mass ratio of 10 ml kg−1 (i.p.). The number of abdominal contractions (writhes) was counted continuously for the next 20 minutes, starting 10 minutes after the injection of acetic acid.
The percentage of antinociception was calculated for all groups using an equation. This provided an estimate of how effective the compounds were in reducing pain compared to the positive control (diclofenac sodium) and helped to determine the optimal dosage of the compound for maximum pain relief.Antinociception (%) = (1 − C1/C2) × 100C1 = number of writhes in the treated groups C2 = number of writhes in the vehicle (5% DMSO and 1% Tween 80)
One-way ANOVA followed by Dunnett's post hoc test was performed using GraphPad prism package 8.0.
2.7. In silico analysis
For the molecular docking analysis, the structures of the target proteins COX1 (PDB ID: 6Y3C)14 at a resolution of 2.90 Å, and COX2 (PDB ID: 1PXX)15 at a resolution of 2.0 Å were obtained from the Protein Data Bank (https://www.rcsb.org). The acquired protein structures went through a refinement process using BIOVIA Discovery Studio 2021 v21.1.0.20298 (Dassault Systems: https://www.3ds.com). Initially, water molecules and heteroatoms were removed. The missing atoms were added to get a full and accurate protein structure. Subsequently, polar hydrogen atoms were incorporated into the protein structures. Furthermore, to facilitate an accurate representation of electrostatic interactions, Kollman charges were introduced. For the prediction of the active site pocket, CASTp server was used.16 The first Poc ID with a larger volume and surface area was selected as an active pocket. The amino acid residues comprising this active site pocket were in line with the literature and the crystallographic structure data. The grid box was positioned to cover the pocket. The resolved center coordinates of the grid box are as follows:
For COX1 (6Y3C)Centre X: −44.7888, Y: −59.1681, Z: 11.7971Dimensions X: 36.1706, Y: 24.1632, Z: 16.1069
For COX2 (1PXX)Centre X: 31.5718, Y: 20.4386, Z: 15.2962Dimensions X: 36.6058, Y: 25.8849, Z: 23.8376
Subsequently, ChemDraw software (http://www.perkinelmer.co.uk/category/chemdraw) was employed to construct compound structures and then saved in SDF format to ensure compatibility with the docking studies. The AutoDock Vina software,17 incorporated into PyRx version 0.8,18 was used to carry out the molecular docking study. Initially, the compounds were incorporated into PyRx using the OpenBabel graphical user interface. The universal force field (UFF) was used to minimize the energy of both compounds. After that, these compounds were converted to pdbqt format to make sure the AutoDock Vina program could read them. The target proteins have been imported into PyRx and given pdbqt file extension. In order to guarantee an extensive investigation of ligand binding modalities, the exhaustiveness level was set at 24.
2.8. Toxicity prediction
The toxicity profile plays a critical role in drug design by providing essential information about the safety and potential side effects of a compound. It allows researchers to identify risks by assessing various toxicity endpoints, such as hepatotoxicity, cytotoxicity, and mutagenicity. This information guides the optimization of chemical structures, enhancing efficacy while minimizing harmful effects, ultimately leading to safer drug candidates.19 Therefore, the toxicity profiles, pharmacokinetics, median lethal dose (LD50, mg kg−1), and toxicity class of both compounds were evaluated using the ProTox-II online tool.20 The toxicity assessment encompassed hepatotoxicity, carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity. Pharmacokinetic parameters, including CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4, and CYP2E1, were also predicted.
3. Results & discussion
3.1. Identification and confirmation of aurasperone B and ergosterol
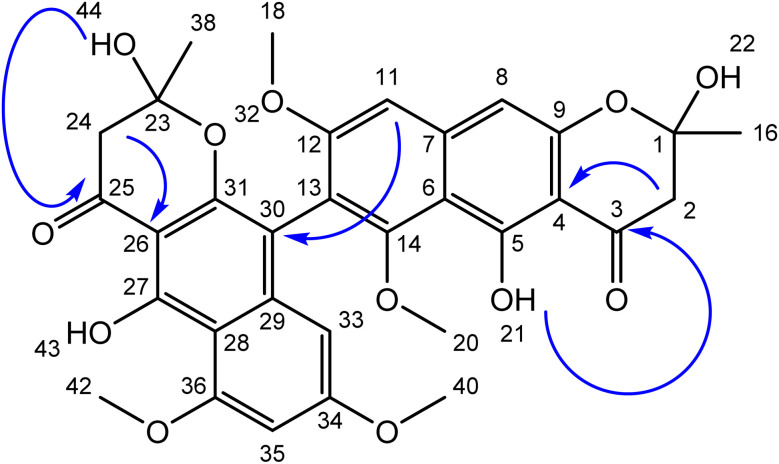
The ethyl acetate extract of the fungal strain A. ficuum after subjection to column chromatography yielded two secondary metabolites, a naphtho gamma pyrone named aurasperone B (1) and ergosterol (2). Compound 1 was isolated in yellowish form. The ESI+ calculated for aurasperone B was 607.61 [M + H]+. It was characterized as a dimer. The difference between the two halves of the molecules was the presence of an additional hydroxyl group. 13C NMR spectrum of compound 1 provided a total number of 31 peaks in its broadband spectrum. These peaks were representative of methine, methyl and quaternary carbons. The quaternary carbon peaks were assigned as δ C-3 (197.5), C-4 (107.8), C-5 (160.1), C-6 (110.2), C-7 (141.2), C-9 (153.4), C-12 (165.4), C-13 (103.1), C-14 (150.6), C-23 (100.5), C-25 (197.5), C-26 (108.3), C-27 (158.8), C-28 (111.3), C-29 (136.5), C-30 (112.6), C-31 (155.8), C-34 (163.7) and C-36 (160.6) respectively. The methine carbons were observed as δ C-8 (99.2), C-11 (104.1), C-33 (97.2) and C-35 (96.2). Four methoxy methyl groups C-18, C-20, C-40 and C-42 were attributed chemical shift values of δ 55.2, 61.63, 56.2 and 55.9 respectively. Both methyl carbons C-16 and C-38 attached to the pyran ring signaled at δ 27.5 (Table 1).
1H NMR, and 13C NMR data of compound (1 and 2) in CDCl3.
| Compound 1 | Compound 2 | ||||
|---|---|---|---|---|---|
| Position | δ H | δ C | Position | δ H | δ C |
| 1 | 3.43, m | 71.2 | |||
| 2 | 2.92, 2.99, dd | 47.1 | 3 | 1.48, 1.23, m | 30.6 |
| 3 | 197.5 | 4 | 1.27,0.99, m | 36.3 | |
| 4 | 107.8 | 5 | 2.16,1.86, dd | 38.3 | |
| 5 | 160.1 | 6 | 138.4 | ||
| 6 | 110.2 | 7 | 5.23, d | 116.4 | |
| 7 | 141.2 | 8 | 5.23, d | 116.7 | |
| 8 | 6.90, s | 99.2 | 9 | 139.6 | |
| 9 | 153.4 | 10 | 1.88, m | 52.1 | |
| 11 | 6.73, s | 104.1 | 11 | 1.49, 1.40, m | 21.1 |
| 12 | 165.4 | 12 | 1.88, m | 25.6 | |
| 13 | 103.1 | 13 | 1.08, m | 54.5 | |
| 14 | 150.6 | 14 | 1.96, m | 38.8 | |
| 16-CH3 | 1.42, s | 27.5 | 15 | 5.11, m | 131.4 |
| 18-OCH3 | 3.80, s | 55.2 | 16 | 5.1, m | 129.5 |
| 20-OCH3 | 4.00, s | 61.63 | 17 | 1.88, m | 38.5 |
| 21-OH | 14.21 | 18 | 0.85, s | 18.2 | |
| 22-OH | 5.60 | 19 | 1.49, m | 31.23 | |
| 23 | 100.5 | 20 | 0.98, m | 19.5 | |
| 24 | 2.92,2.99, dd | 47.1 | 21 | 0.98 | 19.1 |
| 25 | 197.5 | 22 | 0.84, s | 19.3 | |
| 26 | 108.3 | 23 | 41.7 | ||
| 27 | 158.8 | 24 | 1.03, m | 13.5 | |
| 28 | 111.3 | 25 | 1.15, m | 37.3 | |
| 29 | 136.5 | 26 | 0.89, m | 19.3 | |
| 30 | 112.6 | 27 | 1.79, m | 43.6 | |
| 31 | 155.8 | 28 | 34.7 | ||
| 33 | 6.43, s | 97.2 | 29 | 0.93, s | 14.6 |
| 34 | 163.7 | ||||
| 35 | 6.21, s | 96.2 | |||
| 36 | 160.6 | ||||
| 38-CH3 | 1.52, s | 27.5 | |||
| 40-OCH3 | 3.99, s | 56.2 | |||
| 42-OCH3 | 3.81, s | 55.9 | |||
| 43-OH | 14.56 | ||||
| 44-OH | 7.12 | ||||
Total number of twenty-one protons was characterized from the peaks provided in 1H NMR spectrum. The methoxymethyl protons peaks were assigned to δ 3.75 (3H, s, H-18), δ 4.02 (3H, s, H-20) and δ 3.79 (3H, s, H-39). Both methyl protons of the pyran ring signalled at δ 2.13 (3H, s, H-16) and δ 2.12 (3H, s, H-37) (Fig. 2).
Fig. 2. Structure of aurasperone B (1) blue arrows represent key HMBC correlations.
The diastereotopic protons in both pyran rings were awarded to δ 2.92, 2.99 (2H, s, H-2&24). The methine protons of the aromatic rings showed their presence at δ 6.90 (1H, s, H-8), δ 6.73 (1H, s, H-11), δ 6.43 (1H, s, H-33) and δ 6.21 (1H, s H-35). The two hydroxyl groups on the aromatic rings were noted at δ 14.21 & 14.56 (2OH, s), while the other two hydroxyl groups of the pyran ring were assigned a value of δ 5.60 & 7.12 (2OH, s).
There was no 1H–1H coupling between the protons which were evident from the COSY spectrum. However, long-range coupling (HMBC) between various protons with respective carbons helped in the elucidation of the given structure. The HMBC correlations of the hydroxyls OH's groups with carbonyl carbons helps in their fixation at the corresponding place. Similarly, both diastereotopic protons showed three bonds away correlations with their respective carbonyl carbons. Based on the correlations and various chemical shift values of hydrogens and carbons, the structure was characterized as aurasperone. The data agreed with literature.21
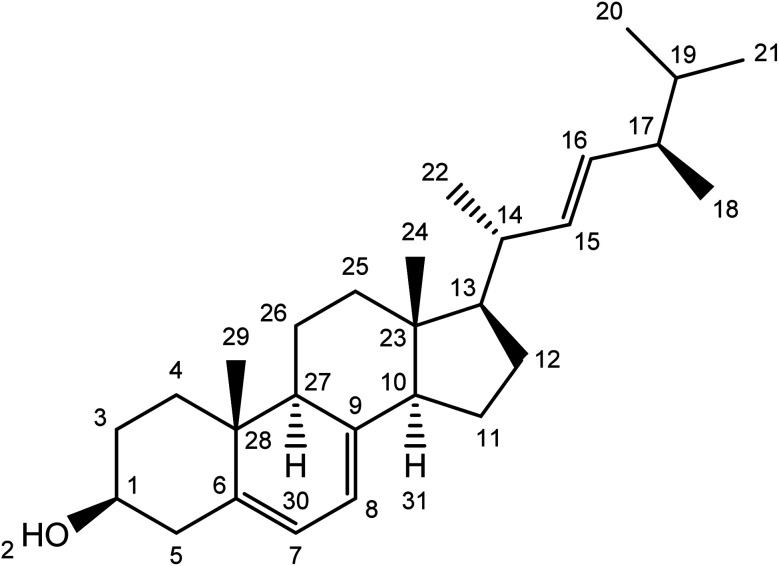
Compound 2 was isolated as a white powder with a molecular ion peak [M+] at 396.34. The broad band of 13C NMR spectrum showed twenty-eight carbon peaks. These peaks represented seven methylene, twelve methines, five methyl and four quaternary carbons signals. The methine peaks were observed at chemical shift values were assigned to respective carbons such as δ 71.2 (C-1), 116.4 (C-7), 116.7 (C-8), 52.1 (C-10), 54.5 (C-13), 38.8 (C-14), 131.4 (C-15), 129.5 (C-16), 38.5 (C-17), 19.5 (C-20), 13.5 (C-24) and 43.6 (C-27). The peaks for methylene carbons were found to resonate at δ 30.6 (C-3), 36.3 (C-4), 38.3 (C-5), 21.1 (C-11), 25.6 (C-12), 37.3 (C-2525) and 19.3 (C-26). The methyl signaled at δ 18.2 (C-18), 31.2 (C-19), 19.1 (C-21) and 14.6 (C-29). The quaternary carbons were awarded to δ 138.4 (C-6), 139.6 (C-9), 41.7 (C-23) and 34.7 (C-28) (Table 1) and (Fig. 3).
Fig. 3. Structure of ergosterol (compound 2).
The 1H NMR spectrum indicated methine signals resonated at δ 3.43 (1H, m, H1), 5.23 (1H, d, J = 1.72, H-7), 5.23 (1H, d, J = 1.72, H-8), 1.08 (1H, m, H13), 1.96 (1H, m, H14), 5.11 (1H, m, H15), 5.11 (1H, m, H16), 1.88 (1H, m, H17) and 1.79 (1H, m, H27). Methylene proton signaled their position at δ 1.48 (2H, m, H3) 1.27 (2H, m, H4), 2.16, 1.88 (2H, dd, J = 12.32, 1.9 Hz, H-5), 1.49 (2H, m, H11), 1.88 (2H, m, H-12), 1.15 (2H, m, H24) and 0.89 (2H, m, H-26). The position of five methyl protons were located at δ 18.2 (3H, s, H-18), 0.98 (3H, s, H-20), 0.98 (3H, m, H-21), 0.84 (3H, m, H-22), 1.03 (3H, m, H-24) and 0.93 (3H, s, H-29). The data was in agreement with the literature.22
3.2. Anti-inflammatory and analgesic activities of aurasperone B and ergosterol
Naphtho-gamma-pyrones (NGPs) are secondary metabolites of polyketide nature. They are biosynthesized by a wide range of filamentous fungi and some higher plants.23
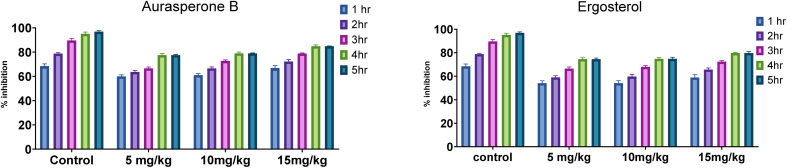
In this study, the anti-inflammatory and analgesic activity of aurasperone B and ergosterol was investigated in vivo and finally statistically evaluated by Dunnett's test. Aurasperone B was less significant at a dose of 5 mg kg−1 (P ≤ 0.05), while no significant difference was observed between dose levels of 10 and 15 mg kg−1. Subsequently, ergosterol was moderately active (P ≤ 0.002) at a dose level of 10 mg kg−1 and significantly less active (P ≤ 0.03) at a dose level of 20 mg kg−1 (Table 2).
Anti-inflammatory activity of aurasperone B and ergosterol.
| Treatments | Conc. (mg kg−1) | Percentage inhibition after different time intervalsa | ||||
|---|---|---|---|---|---|---|
| 1 h | 2 h | 3 h | 4 h | 5 h | ||
| Aspirin | 10 | 68.48 ± 1.87 | 78.8 ± 0.76 | 89.67 ± 1.51 | 95.11 ± 1.43 | 98.91 ± 1.21 |
| Aurasperone B | 5 | 60 ± 1.23 | 63.64 ± 1.21 | 66.67 ± 0.98 | 77.58 ± 1.23 | 77.58 ± 0.44 |
| 10 | 61.11 ± 1.25 | 66.67 ± 1.126 | 72.84 ± 0.76 | 79.01 ± 1.01 | 79.01 ± 0.32 | |
| 15 | 66.87 ± 1.98 | 72.29 ± 1.56 | 78.92 ± 0.43 | 84.94 ± 1.04 | 84.94 ± 0.12 | |
| Ergosterol | 10 | 54.04 ± 2.12 | 59.01 ± 1.43 | 66.46 ± 1.46 | 74.53 ± 1.21 | 74.53 ± 0.98 |
| 20 | 54.09 ± 2.18 | 59.75 ± 1.72 | 67.92 ± 1.22 | 74.84 ± 1.04 | 74.84 ± 1.34 | |
| 30 | 58.9 ± 2.45 | 65.64 ± 1.42 | 72.39 ± 1.08 | 79.75 ± 0.56 | 79.75 ± 1.33 | |
Data shown is processed through one-way ANOVA, followed by Dunnett's test.
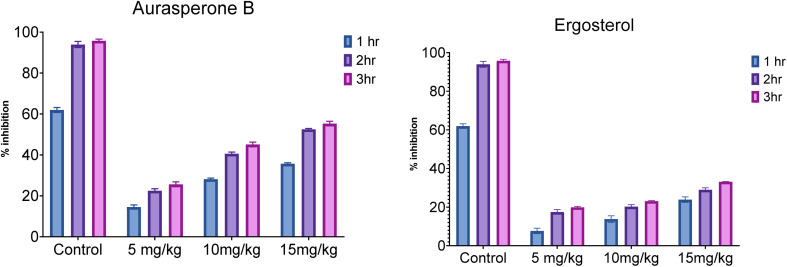
The effect of both secondary metabolites was evident that they can have anti-inflammatory potential (Fig. 4 and S8†).
Fig. 4. Anti-inflammatory activity of aurasperone B & ergosterol.
The potential use of dimeric NγPs in agriculture and medicine has drawn the attention of many researchers to this class of compounds. Aurasperone B is the precursor of aurasperone A and other congeners.24 Aurasperone B is dextrorotatory and is a dihydrate of aurasperone A, which itself is levorotatory.25 NγPs are thought to be defense metabolites produced under stressful conditions and serve as non-toxic agents for fungal defense against predators.
Various bioactivities have been reported from NγPs such as antioxidant, antitumor and antimicrobial.24 Recently, mutagenic, hepatoprotective and anxiety-related disorders have been treated by polyketides.26 At a dose of 50 mg kg−1 by intraperitoneal injection in rats, significant central nervous disorders were induced.27
Similarly, several ergostane-type metabolites have been isolated from fungi and plants. They have shown potential for various biological activities. Ergosterol has been used for the treatment of various diseases such as those with anticancer potential in the lungs, breast and colon.28 Similarly, various studies have presented its pharmacological potential.29
In analgesic activity, aurasperone B at dose concentrations of 5, 10, and 15 mg kg−1, the percentage inhibition significantly increases with time duration from 1 h to 3 h. Overall, the percentage inhibition among doses and positive control was highly significant (P ≤ 0.04), while the highest percentage inhibition was recorded at 15 mg kg−1 (P ≤ 0.05) after 3 h of study compared to standard diclofenac sodium. In the case of ergosterol, a similar response was noted between its various doses and the positive control during 3 h study. Although the effect was as significant as that of aurasperone B, the significance among doses of ergosterol was 10 mg kg−1 (P ≤ 0.02), 20 mg kg−1 (P ≤ 0.03) and 30 mg kg−1 (P ≤ 0.04) compared to standard (Table 3). Moreover, the highest percentage of inhibition (23.08 ± 0.32) was observed at 30 mg kg−1 for ergosterol after 3 h of study.
Analgesic activity of aurasperone B and ergosterol.
| Treatments | Concentration (mg kg−1) | % inhibition 1 h | % inhibition 2 h | % inhibition 3 h |
|---|---|---|---|---|
| Diclofenac sodium | 10 | 61.96 ± 1.21 | 93.95 ± 1.551 | 95.8 ± 0.76 |
| Aurasperone B | 5 | 14.53 ± 1.04 | 22.48 ± 0.98 | 25.65 ± 1.21 |
| 10 | 28.14 ± 0.56 | 40.6 ± 0.76 | 45.1 ± 1.13 | |
| 15 | 35.71 ± 0.43 | 52.48 ± 0.43 | 55.32 ± 1.10 | |
| Ergosterol | 10 | 7.67 ± 1.43 | 17.49 ± 1.23 | 19.93 ± 0.44 |
| 20 | 13.8 ± 1.72 | 20.28 ± 1.01 | 23.08 ± 0.32 | |
| 30 | 23.9 ± 1.42 | 28.97 ± 1.04 | 33.15 ± 0.12 |
The results indicated the positive potential of both secondary metabolites and needed to be included in the drug discovery program (Fig. 5 and S9†).
Fig. 5. Graphical representation of the analgesic activity of aurasperone B & ergosterol.
Recently, an isomer of aurasperone D was investigated in vitro against SARS CoV-2. An efficient potential was shown by aurasperone A against that virus.24 The fungus Aspergillus niger produces aurasperone B, a dimeric NγPs with strong antioxidant properties and moderate toxicity against various cancer cell lines and brine shrimp.21,30–32 Dimeric pyrones with a similar structure have also been reported in other species of fungi.33–35 It is suggested that further investigation of these metabolites may help in the development of new drugs.
3.3. Molecular docking study
Cyclooxygenases (COX1 and COX2) are crucial enzymes in inflammation, converting arachidonic acid into prostaglandins and thromboxanes. COX1 plays a supportive role by maintaining normal physiological functions rather than directly mediating the inflammatory response. It is constitutively expressed in many tissues and produces prostaglandins that protect the gastric lining, regulate blood flow in the kidneys, and promote platelet aggregation. In contrast, COX2 is induced during inflammation, generating prostaglandins that promote vasodilation, increase vascular permeability, and attract immune cells, leading to redness, swelling, and pain. Additionally, prostaglandins sensitize pain receptors and regulate fever.36 Therefore, these two enzymes were selected for docking analysis against aurasperone B and ergosterol. The docking details are provided below.
3.3.1. Interaction analysis of COX1
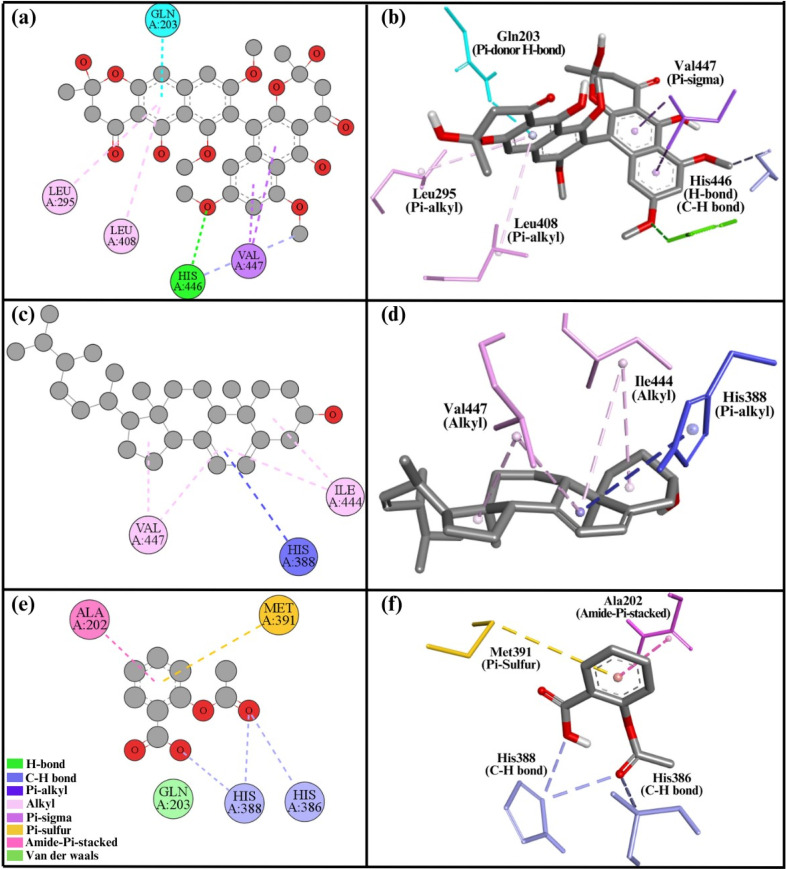
Relative to compound 2, compound 1 interacted with the COX1 protein more favorably. Compound 1 had a binding energy of −7.1 kcal mol−1, while compound 2 displayed −7.0 kcal mol−1 (Table 4). Compound 1 exhibited seven interactions with COX1's active site. These interactions include two Pi–sigma bonds with Val447, a H-bond and carbon–hydrogen bond with His446, two Pi–alkyl bonds with residues Leu408, Leu295, and one Pi-donor H-bond with Gln203 (Fig. 6a and b). A total of five interactions were formed by compound 2. Four alkyl bonds were involved in these interactions: two with Val447 and two with Ile444. In addition, one Pi–alkyl bond was formed with His388 (Fig. 6c and d). Similarly, aspirin demonstrated a binding energy of −6.2 kcal mol−1 with the COX1 protein, forming five distinct interactions. These include two C–H bonds with residues His386 and His388, an amide–Pi stacked interaction with Ala202, a Pi–sulfur interaction with Met391, and a van der Waals interaction with Gln203 (Fig. 6e and f).
Interaction details of compound 1, compound 2, and aspirin with COX1 and COX2 proteins.
| Proteins | Compounds | Binding energy (kcal mol−1) | Interacting residues | Nature of interactions |
|---|---|---|---|---|
| COX1 (6Y3C) | Compound 1 | −7.1 | His446 | H-bond |
| His446 | C–H bond | |||
| Val447 | Pi–sigma bond | |||
| Val447 | Pi–sigma bond | |||
| Leu408 | Pi–alkyl bond | |||
| Leu295 | Pi–alkyl bond | |||
| Gln203 | Pi-donor H-bond | |||
| Compound 2 | −7.0 | Val447 | Alkyl bond | |
| Val447 | Alkyl bond | |||
| Ile444 | Alkyl bond | |||
| Ile444 | Alkyl bond | |||
| His388 | Pi–alkyl bond | |||
| Aspirin | −6.2 | His388 | C–H bond | |
| His386 | C–H bond | |||
| Ala202 | Amide–Pi-stacked | |||
| Met391 | Pi–sulfur | |||
| Gln203 | van der Waals | |||
| COX2 (1PXX) | Compound 1 | −8.4 | Gln369 | H-bond |
| Ser126 | C–H bond | |||
| Pro127 | C–H bond | |||
| Arg61 | C–H bond | |||
| Lys532 | Alkyl bond | |||
| Phe371 | Pi–alkyl bond | |||
| Arg61 | Pi–alkyl bond | |||
| Arg61 | Pi–alkyl bond | |||
| Compound 2 | −7.6 | Arg61 | Alkyl bond | |
| Arg61 | Alkyl bond | |||
| Aspirin | −6.1 | Ser530 | H-bond | |
| Gly526 | Amide–Pi-stacked | |||
| Val523 | Pi–alkyl | |||
| Leu353 | Pi–alkyl | |||
| Ala527 | Pi–alkyl |
Fig. 6. 2D and 3D representations summarize the results of the interaction analysis for the (a and b) COX1–compound 1 complex, (c and d) COX1–compound 2 complex, and (e and f) COX1–aspirin complex.
The secondary structural components of COX1 indicate an epidermal growth factor, membrane binding domain, and catalytic domain. Two important sites in protein structure are the substrate site, where substrates attach, and the heme site, where the heme molecule required for COX1 function interacts.14 Both of our compounds bind with and block these key regions, so disrupting the normal activity of the COX1 enzyme. Compounds 1 and 2 interacted with residues His446, Val447, Ile44, and Leu408 of the heme site, presumably blocking it. Compound 2 also interacted with His388 at the substrate site, occupying the substrate regions and making it harder for a substrate to attach to the substrate site.
3.3.2. Interaction analysis of COX2
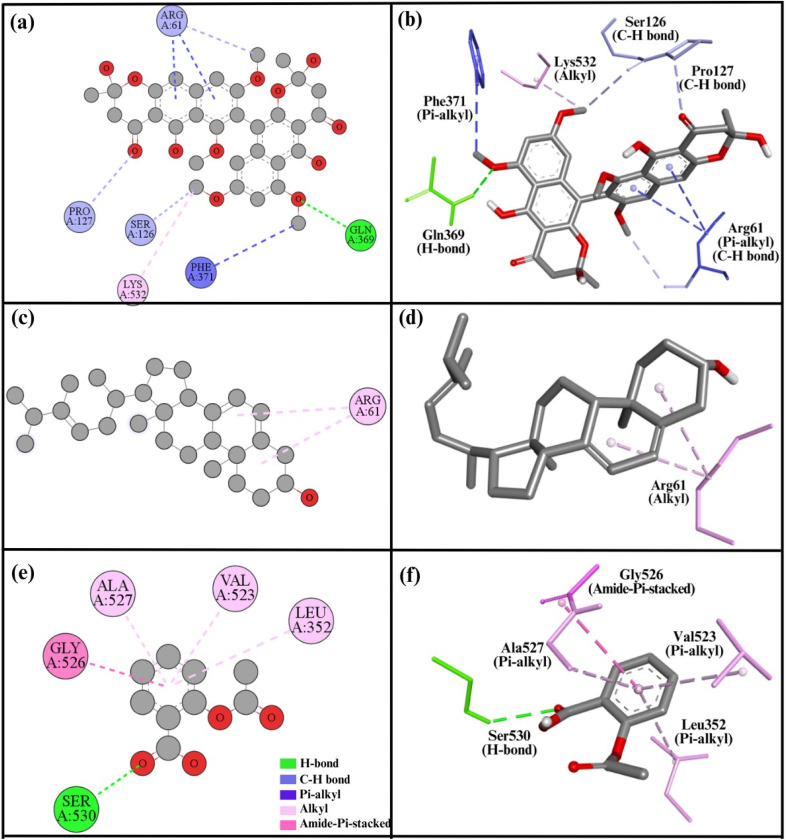
Compound 1 had a high binding energy of −8.4 kcal mol−1 (Table 4) and a total of eight interactions with the COX2 protein active site. Specifically, it formed one H-bond with Gln369. Additionally, it made three carbon–hydrogen bonds with Ser126, Pro127, and Arg61, and three Pi–alkyl bonds with residue Phe371, and two with Arg61. A single alkyl bond was also noted with Lys532 (Fig. 7a and b). Compound 2 exhibited a binding energy of −7.6 kcal mol−1 and formed two alkyl bonds with Arg61 (Fig. 7c and d). The COX2 active site is essential to its activity. Previously, it was studied and inhibited by numerous compounds in several studies.15 In our analysis, compound 1 interacted with critical residues such as Lys532, Ser126, and Phe371, all of which are in close vicinity of the active site, demonstrating that compound 1 has the ability to inhibit it. Similarly, aspirin exhibited a binding energy of −6.1 kcal mol−1 with the COX2 protein, forming five key interactions. These include three Pi–alkyl bonds with Val523, Leu353, and Ala527, as well as a hydrogen bond with Ser530 and an amide–Pi stacked interaction with Gly526 (Fig. 7e and f).
Fig. 7. 2D and 3D representations summarize the results of the interaction analysis for the (a and b) COX2–compound 1 complex, (c and d) COX2–compound 2 complex, and (e and f) COX2–aspirin complex.
Experimental results indicate that both compound 1 and compound 2 have the potential to reduce inflammation; however, their anti-inflammatory effects are weaker compared to the standard drug aspirin. Additionally, compound 1 demonstrates a more pronounced anti-inflammatory effect than compound 2. In the in silico study, similar findings were observed, with compound 1 showing a strong binding affinity to both COX1 and COX2 enzymes compared to compound 2. Conversely, aspirin exhibited a lower binding affinity to COX1 and COX2 than either compound, which is not consistent with the experimental data. Nevertheless, the overall docking results support the experimental findings.
3.4. Toxicity and pharmacokinetics prediction
The five toxicity endpoints and pharmacokinetic properties of both compounds were predicted using computational tools, achieving a probability of over 70%, which reflects the confidence level of the predictions (confidence score). The toxicity endpoints, pharmacokinetic parameters, median lethal dose, and toxicity classification for both compounds are summarized in Table 5.
Pharmacokinetics properties, toxicity properties, median lethal dose (LD50), and toxicity class of aurasperone B and ergosterol.
| Parameters | Aurasperone B | Ergosterol |
|---|---|---|
| Pharmacokinetics parameters | ||
| CYP1A2 | Inactive | Inactive |
| CYP2C19 | Inactive | Inactive |
| CYP2C9 | Active | Active |
| CYP2D6 | Inactive | Inactive |
| CYP3A4 | Inactive | Inactive |
| CYP2E1 | Inactive | Inactive |
| Toxicity endpoints properties | ||
| Hepatotoxicity | Inactive | Inactive |
| Carcinogenicity | Inactive | Inactive |
| Immunotoxicity | Active | Active |
| Mutagenicity | Inactive | Inactive |
| Cytotoxicity | Inactive | Inactive |
| Median lethal dose (LD 50 , mg kg− 1 ) | ||
| Predicted LD50 | 2000 | 10 |
| Predicted toxicity class | 4 | 2 |
Cytochrome P450 (CYP) is a family of enzymes essential for the metabolism of drugs and various other compounds. Six isoforms—CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4, and CYP2E1—are primarily responsible for metabolizing most approved pharmaceuticals. Drug interactions involving these CYP enzymes can profoundly affect both the efficacy and safety of medications, potentially leading to adverse reactions or reduced therapeutic effectiveness. Such interactions can result in the premature discontinuation of drug development or the withdrawal of products from the market due to safety concerns.37 From Table 5, we observed that both compounds are non-inhibitors of CYP2D6, CYP1A2, CYP2C19, CYP3A4, and CYP2E1, while acting as inhibitors of CYP2C9.
Hepatotoxicity refers to liver damage caused by chemical substances, such as medications, herbal supplements, and industrial chemicals.38 Based on hepatotoxicity assessments, neither of the compounds is considered toxic to the liver. Carcinogenicity refers to the ability of a substance to induce cancer. A substance may be either active and carcinogenic or inactive and considered safe.39 According to computational estimations, both compounds are classified as inactive and safe, posing no cancer risk. Cytotoxicity describes a substance's capacity to damage or kill cells.40 Based on cytotoxicity predictions, both compounds are deemed nontoxic to cells. Mutagenicity refers to the potential of a substance to cause genetic mutations in DNA. A substance may be active and mutagenic or inactive and non-mutagenic.41 Predictions indicate that both compounds are inactive, non-mutagenic, and genetically safe, with no potential to cause genetic mutations in DNA. Immunotoxicity refers to the harmful effects of substances on the immune system.42 According to the prediction table, both compounds are shown to be active against the immune system and may potentially cause adverse effects.
The median lethal dose (LD50, mg kg−1) is the dose required to cause death in 50% of the tested animals. According to the Globally Harmonized System (GHS), substances are classified into six toxicity classes: Class I (LD50 ≤ 5) is fatal if swallowed; Class II (5 < LD50 ≤ 50) is also fatal if swallowed; Class III (50 < LD50 ≤ 300) is toxic if swallowed; Class IV (300 < LD50 ≤ 2000) is harmful if swallowed; Class V (2000 < LD50 ≤ 5000) may be harmful if swallowed; and Class VI (LD50 > 5000) is considered non-toxic.43 Based on the prediction study, aurasperone B has an LD50 of 1000 mg kg−1, while ergosterol has an LD50 of 10 mg kg−1. Consequently, aurasperone B is classified as Class IV, and ergosterol is classified as Class II.
4. Conclusion
In this study, we successfully isolated two secondary metabolites belonging to the bis or dimer NγPs and sterol classes from A. ficuum for the first time. Additionally, we present the 2D NMR data of aurasperone B for the first time. Both compounds were evaluated for their in vivo pharmacological potential. Ergosterol exhibited moderate pharmacological effects compared to aurasperone B in anti-inflammatory and analgesic studies. The molecular docking results support these experimental findings. The prediction study found that both compounds act as CYP2C9 inhibitors but are non-inhibitors of CYP2D6, CYP1A2, CYP2C19, CYP3A4 and CYP2E1. Furthermore, aurasperone B has an LD50 of 1000 mg kg−1, whereas ergosterol has an LD50 of 10 mg kg−1. As a result, aurasperone B is classified as Class IV, and ergosterol is classified as Class II. Both compounds are inactive and considered safe regarding carcinogenicity, cytotoxicity, mutagenicity, and hepatotoxicity, but they are active concerning their effects on the immune system. Given their modes of action, biological activities, and structure–activity relationships, bis-NγPs require further investigations. This study lays a solid foundation for the development of more potent pharmaceuticals with promising applications in both food and medicine.
Data availability
The supporting data have already been included in the article's ESI.†
Author contributions
Zafar Ali Shah: conceptualization, formal analysis, investigation, and methodology; Khalid Khan: conceptualization, supervision, project administration, data curation, and resources; Tanzeel Shah: formal analysis, validation, and writing—review, and editing; Nasir Ahmad: formal analysis and software; Asad Khan: validation, investigation, and methodology.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors are grateful to the Department of Chemistry, Islamia College, Peshawar, KP, Pakistan, for the provision of all facilities needed to execute the project.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03674c
References
- Blunt J. W. Copp B. R. Keyzers R. A. Munro M. H. Prinsep M. R. Nat. Prod. Rep. 2014;31:160–258. doi: 10.1039/C3NP70117D. [DOI] [PubMed] [Google Scholar]
- Zhang H. M. Ju C. X. Li G. Sun Y. Peng Y. Li Y. X. Peng X. P. Lou H. X. Mar. Drugs. 2019;17:383. doi: 10.3390/md17070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Ferranti L. Fungaro M. H. P. Massi F. P. da Silva J. Penha R. Frisvad J. C. Taniwaki M. H. Iamanaka B. T. Int. J. Food Microbiol. 2018;268:53–60. doi: 10.1016/j.ijfoodmicro.2017.12.027. [DOI] [PubMed] [Google Scholar]
- Carboué Q. Maresca M. Herbette G. Roussos S. Hamrouni R. Bombarda I. Biomolecules. 2019;10:29. doi: 10.3390/biom10010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M. Kim M. J. Li H. Zhang P. Bao B. Lee K. J. Jung J. H. J. Mar. Biotechnol. 2013;15:499–519. doi: 10.1007/s10126-013-9506-3. [DOI] [PubMed] [Google Scholar]
- Wu C. Z. Peng X. P. Li G. Wang Q. Lou H. X. Molecules. 2022;27:2514. doi: 10.3390/molecules27082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay C. A. Kinghorn A. D. Rakotondraibe H. L. Phytochem. 2023;209:113638. doi: 10.1016/j.phytochem.2023.113638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El-Razek M. H. El-Desoky A. H. Elgahamy A. A. Bata S. M. Mohamed T. A. Hegazy M. E. Nat. Prod. Res. 2024:1–11. doi: 10.1080/14786419.2024.2315503. [DOI] [PubMed] [Google Scholar]
- Awad A. B. Fink C. S. J. Nutr. 2000;130:2127–2130. doi: 10.1093/jn/130.9.2127. [DOI] [PubMed] [Google Scholar]
- Shah Z. A. Khan K. Shah T. Ahmad N. Muhammad A. Rashid H. U. Sci. Rep. 2023;13:17260. doi: 10.1038/s41598-023-43819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah Z. A. Khan K. Rashid H. U. Shah T. Jaremko M. Iqbal Z. BMC Microbiol. 2022;22:29512. doi: 10.1186/s12866-022-02693-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N. Subhan F. Islam N. Shahid M. Rahman F. Fawad K. Arch. Pharm. 2017;350:e201600365. doi: 10.1002/ardp.201600365. [DOI] [PubMed] [Google Scholar]
- Shahid M. Subhan F. Ahmad N. Sewell R. D. Biomed. Pharmacother. 2017;95:1725–1733. doi: 10.1016/j.biopha.2017.09.108. [DOI] [PubMed] [Google Scholar]
- Miciaccia M. Belviso B. D. Iaselli M. Cingolani G. Ferorelli S. Cappellari M. Loguercio Polosa P. Perrone M. G. Caliandro R. Scilimati A. Sci. Rep. 2021;11:4312. doi: 10.1038/s41598-021-83438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlinson S. W. Kiefer J. R. Prusakiewicz J. J. Pawlitz J. L. Kozak K. R. Kalgutkar A. S. Stallings W. C. Kurumbail R. G. Marnett L. J. J. Biol. Chem. 2003;278:45763–45769. doi: 10.1074/jbc.M305481200. [DOI] [PubMed] [Google Scholar]
- Tian W. Chen C. Lei X. Zhao J. Liang J. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt J. Santos-Martins D. Tillack A. F. Forli S. J. Chem. Inf. Model. 2021;61:3891–3898. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S. Olson A. J. J. Chem. Biol. 2015:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Dixit V. A. Toxicol. 2019;8:157–171. doi: 10.1039/C8TX00261D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khezri R. Jamaleddin Shahtaheri S. Khezri E. Niknam Shahrak M. Khadem M. Toxicol. Mech. Methods. 2024:1–20. doi: 10.1080/15376516.2024.2353364. [DOI] [PubMed] [Google Scholar]
- Bouras N. Mathieu F. Coppel Y. Lebrihi A. Nat. Prod. Rep. 2005;19:653–659. doi: 10.1080/14786410412331286955. [DOI] [PubMed] [Google Scholar]
- Endress E. Bayerl S. Prechtel K. Maier C. Merkel R. Bayerl T. M. Langmuir. 2002;18:3293–3299. doi: 10.1021/la011596m. [DOI] [Google Scholar]
- Ernst-Russell M. A. Chai C. Wardlaw J. H. Elix J. A. J. Nat. Prod. 2000;63:129–131. doi: 10.1021/np9903245. [DOI] [PubMed] [Google Scholar]
- ElNaggar M. H. Abdelwahab G. M. Kutkat O. GabAllah M. Ali M. A. El-Metwally M. E. Sayed A. M. Abdelmohsen U. R. Khalil A. T. Mar. Drugs. 2022;20:179. doi: 10.3390/md20030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H. Wang P. L. Yamada O. Agric. Biol. Chem. 1966;30:107–113. [Google Scholar]
- Lee G. Y. Jang D. S. Lee Y. M. Kim J. M. Kim J. S. Arch. Pharmacal Res. 2006;29:587–590. doi: 10.1007/BF02969270. [DOI] [PubMed] [Google Scholar]
- Lu S. Tian J. Sun W. Meng J. Wang X. Fu X. Wang A. Lai D. Liu Y. Zhou L. Molecules. 2014;19:7169–7188. doi: 10.3390/molecules19067169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal S. Biswas K. Chakrabarti D. Chemistry F. J. Agric. Food Chem. 1979;27:1347–1351. doi: 10.1021/jf60226a018. [DOI] [PubMed] [Google Scholar]
- Merdivan S. Lindequist U. Int. J. Med. Mushrooms. 2017;19:93–105. doi: 10.1615/IntJMedMushrooms.v19.i2.10. [DOI] [PubMed] [Google Scholar]
- Zhabinskii V. N. Drasar P. Khripach V. A. Molecules. 2022;27:2103. doi: 10.3390/molecules27072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutou A. S. Yun K. Son B. W. Arch Pharm Res. 2016;39:806–810. doi: 10.1007/s12272-016-0764-2. [DOI] [PubMed] [Google Scholar]
- Siriwardane A. Kumar N. S. Jayasinghe L. Fujimoto Y. Nat. Prod. Res. 2015;29:1384–1387. doi: 10.1080/14786419.2015.1025230. [DOI] [PubMed] [Google Scholar]
- Fang W. Lin X. Wang J. Liu Y. Tao H. Zhou X. Molecules. 2016;21:941. doi: 10.3390/molecules21070941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestap H. Tetrahedron. 1984;40:3617–3624. doi: 10.1016/S0040-4020(01)88792-5. [DOI] [Google Scholar]
- Shaaban M. Shaaban K. A. Abdel-Aziz M. S. Org. med. Chem. letters. 2012;2:1–8. doi: 10.1186/2191-2858-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. S. Mann M. DuBois R. N. Oncogene. 1999;18:7908–7916. doi: 10.1038/sj.onc.1203286. [DOI] [PubMed] [Google Scholar]
- Dhillon S. Gill K. Clin. Pharmacokinet. 2006:1–44. [Google Scholar]
- Singh A. Bhat T. Sharma O. J. Clin. Toxicol. 2011;4:2161–0495. [Google Scholar]
- Wolf D. C. Cohen S. M. Boobis A. R. Dellarco V. L. Fenner-Crisp P. A. Moretto A. Pastoor T. P. Schoeny R. S. Seed J. G. Doe J. E. Regul. Toxicol. Pharmacol. 2019;103:86–92. doi: 10.1016/j.yrtph.2019.01.021. [DOI] [PubMed] [Google Scholar]
- Peters O. A. Int Endod J. 2013;46:195–197. doi: 10.1111/iej.12047. [DOI] [PubMed] [Google Scholar]
- Słoczyńska K. K. Powroźnik B. Pękala E. Waszkielewicz A. M. J. Appl. Genet. 2014;55:273–285. doi: 10.1007/s13353-014-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerdan B. Moussa S. Atoui A. Assi H. Int. J. Mol. Sci. 2021;22:8242. doi: 10.3390/ijms22158242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. T. V. Surya Wibowo A. Tayara H. Chong K. T. J. Chem. Inf. Model. 2023;63:2628–2643. doi: 10.1021/acs.jcim.3c00200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The supporting data have already been included in the article's ESI.†