Abstract
Babesia is a diverse genus of piroplasms that parasitize the red blood cells of a wide variety of mammals and avian species, including humans. There is a lack of knowledge on the Babesia species of carnivores and mesomammals in the eastern United States and the potential impacts of these species on the health of humans and domestic animals. We surveyed 786 wild mammals in the eastern United States by testing blood, spleen, and heart samples with PCR targeting the 18S rRNA region of apicomplexan parasites. We also performed PCR targeting the cytochrome c oxidase subunit 1 (cox1) region of each unique clade identified with 18S rRNA sequencing. We found a high positivity of Babesia spp. infection in raccoons (Procyon lotor), foxes (Vulpes vulpes and Urocyon cinereoargenteus), and striped skunks (Mephitis mephitis), and low positivity in Virginia opossums (Didelphis virginiana). No Babesia infections were detected in coyotes (Canis latrans), black bears (Ursus americanus), groundhogs (Marmota monax), muskrats (Ondatra zibethicus), or mink (Neovison vison). Skunks carried a diverse number of strains including a potential novel species of Babesia related to B. gibsoni, a strain closely related to a B. microti-like species known to cause disease in river otters, as well as a separate B. microti-like strain. Raccoons primarily carried B. microti-like strains, though there was a high diversity of sequences including Babesia lotori, Babesia sensu stricto MA230, and Babesia sp. ‘Coco.’ Foxes exclusively carried B. vulpes. In addition to Babesia spp., a high positivity of Hepatozoon spp. infection was found in mink, while low positivity was found in raccoons and muskrats. Wildlife in the eastern United States carry a diverse range of Babesia species including several novel strains of unknown clinical significance.
Keywords: Babesiosis, Hepatozoonosis, Hepatozoon ophisauri, Tick-borne protozoan disease, Zoonosis
Graphical abstract
Highlights
-
•
Novel sequences of Babesia identified in striped skunks and raccoons.
-
•
First report of Hepatozoon spp. in mink and muskrats in the United States.
-
•
High prevalence of Babesia spp. in raccoons, skunks, and foxes.
-
•
No Babesia found in bear, coyotes, groundhog, mink, or muskrats.
1. Background
Tick-borne diseases are on the rise in the United States due to a combination of many factors including climate change, habitat fragmentation, and the expanding range of many tick vectors (Tsao et al., 2021). Babesia, an apicomplexan that parasitizes red blood cells in a wide variety of species, is no exception, with reported cases in humans increasing by as much as 20-fold in certain parts of the country over the last two decades (Bishop et al., 2021; Joseph et al., 2011). Although disease is generally mild in immunocompetent individuals, babesiosis can cause severe and even life-threatening hemolytic anemia in at-risk groups such as the young, elderly, splenectomized, and other immunocompromised people (Bloch et al., 2022). Babesia microti, which is transmitted by Ixodes scapularis (the black-legged tick), is the most common cause of human babesiosis in the United States, though infections with other species including B. duncani and B. divergens-like (MO1) have been reported (Yang et al., 2021; Scott and Scott, 2018; Herc and Pritt, 2018).
In addition to the potential negative impacts on humans, Babesia spp. can cause severe illness in pets and livestock. Unlike human infections, which are diagnosed primarily in the Northeast and Midwest, canine babesiosis is most common in the southern and western United States (Almazán et al., 2022; Bloch et al., 2019). In domestic dogs, B. gibsoni, which is commonly transmitted via blood contact (i.e., dog fighting), and B. vogeli, which is transmitted by the brown dog tick (Rhipicephalus sanguineus), are the most common causes of illness (Irwin, 2010). In recent years there have been several new species documented in dogs. Babesia conradae was first documented in 2006 and has since been detected in coyotes (Canis latrans) and coyote-hunting dogs in California (Dear et al., 2018; Javeed et al., 2022). It can cause severe hemolytic anemia and may be fatal without treatment (Javeed et al., 2022). Babesia sp. ‘Coco’ is a large Babesia species that was identified in 2004 and has since been sporadically identified as a cause of illness in splenectomized and immunosuppressed dogs and has also been detected in a single bobcat (Lynx rufus) (Birkenheuer et al., 2004; Zygner et al., 2023; Shock et al., 2013). Finally, Babesia vulpes, sometimes still referred to as B. microti-like or Theileria annae, was reported in red foxes (Vulpes vulpes) in Europe before being documented in canids in the United States and Canada (Barash et al., 2019; Birkenheuer et al., 2010). It has rarely caused disease in domestic dogs in the United States (Barash et al., 2019).
At least three species of Babesia have been documented in raccoons in the eastern United States: B. lotori, which is part of the Babesia sensu stricto (B. s. s.) clade, a different species in the B. s. s. clade that has been found in both Japanese and North American raccoons which we will refer to as B. s. s. MA230 in this paper based on the initial description, and Babesia microti-like, which is the nomenclature used to describe a diverse group of small Babesia species (Garrett et al., 2019; Goethert and Telford, 2003; Jinnai et al., 2009). Prevalence is high in raccoons, with studies in the southern United States finding Babesia in 70–95% of tested animals (Birkenheuer et al., 2008; Clark et al., 2012). Infection appears to be mild or subclinical in raccoons, though one survey assessing young raccoons found Babesia s. s. infection was associated with splenomegaly (Garrett et al., 2018). Coinfection with Babesia microti-like and B. s. s. species is common in raccoons, with studies finding coinfection rates ranging between 0% and 76% (Garrett et al., 2019; Birkenheuer et al., 2008).
Less is known about Babesia infection in mesomammals other than raccoons, though there have been some surveys. Foxes are common carriers of B. vulpes. A study in North Carolina and Canada found a prevalence of B. vulpes in 39% (50/127) of the red fox population and 26% (8/31) in the gray fox (Urocyon cinereoargenteus) population (Birkenheuer et al., 2010). Numerous species of Babesia have been documented in black bears (Ursus americanus), including sequences closely related to B. microti-like, Babesia sp. ‘Coco’, and B. lotori, though prevalence is typically low (Skinner et al., 2017; Westmoreland et al., 2019; Shaw et al., 2015). Skunks have been documented to carry several Babesia species including B. mephitis and several different B. microti-like species (Garrett et al., 2024; Goethert, 2021; Holbrook and Frerichs, 1970).
The taxonomy of Babesia and the Piroplasmida order is under considerable debate because of polyphyly (Garrett et al., 2019; Goethert, 2021; Jalovecka et al., 2019; Schnittger et al., 2022). A recent paper described ten clades of piroplasms determined via analysis of the 18S rRNA gene including the “true” Babesia species (e.g. Babesia sensu stricto) and Babesia sensu lato, which is divided into several groups including the Babesia microti-like complex (Jalovecka et al., 2019). Sequencing of the 18S rRNA gene is a common method for species identification, though many researchers elect to include a more variable gene target like the cytochrome c oxidase subunit 1 (cox1) to aid in genetic comparisons (Garrett et al., 2019; Goethert, 2021; Jalovecka et al., 2019; Schnittger et al., 2022).
In addition to Babesia species, wild mammals can be infected with many other apicomplexan parasites that may cause disease. Bobcats are common hosts of Cytauxzoon felis, a parasite often fatal in domestic cats (Brown et al., 2010). In fact, bobcats are considered the primary reservoir in the United States with prevalences as high as 79% in certain areas, though domestic cats and other wild felids can be chronic carriers as well (Wikander and Reif, 2023). Bobcats are often asymptomatic, though there are cases of acute, fatal infections (Nietfeld and Pollock, 2002). Hepatozoon species can be either asymptomatic or a cause of severe muscle and heart inflammation in both wildlife and domestic animals (Thomas et al., 2024; Baker et al., 2023; Clark et al., 1973; Allen et al., 2011a). Finally, Besnoitia darlingi is a cyst-forming protozoan that causes inflammation and occasionally mortality in Virginia opossums (Didelphis virginiana) and uses both domestic and wild felids as definitive hosts (Elsheikha et al., 2003; Ellis et al., 2012).
More research is needed to understand the distribution, prevalence, and phylogenetics of Babesia spp. and other apicomplexans in wildlife. This information can then be used to inform diagnostic testing and risk to populations of interest like people and domestic animals. Though previous surveys provide an excellent baseline, numerous states including Tennessee have not been assessed. In addition, surveys in the United States in certain species of wildlife like mink (Neovison vison), muskrat (Ondatra zibethicus), opossums, and skunks (Mephitis mephitis) are rare (Ganoe et al., 2021). This study sought to fill in the knowledge gaps on the prevalence, distribution, and diversity of Babesia species that infect wildlife. We hypothesized that Babesia spp. infections would be common in raccoons and foxes, as previously documented, and present more rarely in other wildlife.
2. Methods
Blood, spleen, and heart samples from wildlife were collected opportunistically from a variety of sources. In Tennessee, whole carcasses from rabies testing facilities or animals that died in the University of Tennessee College of Veterinary Medicine's Wildlife and Exotics department were necropsied, and samples of blood and heart were frozen. In South Carolina, a collaborator performing a GPS collaring study (Vectronic Aerospace VERTEX Light, Coralville, Iowa) on coyotes collected whole blood samples for testing (Jensen et al., 2024). Finally, banked splenic samples from raccoons, skunks, mink, muskrats, bears, coyotes, foxes, and groundhogs (Marmota monax) were tested from animals opportunistically obtained (i.e. harvest, roadkill, and damage management) from wildlife resources officers, biologists, or veterinarians in various states.
DNA was extracted from 100 μL of whole blood (n = 296), 20 mg of heart (n = 58), and/or 10 mg of spleen (n = 480) using DNeasy Blood and Tissue extraction kits following manufacturer's instructions (Qiagen Inc, Germantown, Maryland, USA). A negative nuclease-free water control was used during each extraction. PCR was performed using DreamTaq Master Mix (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and nested PCR primers targeting part of the 18S rRNA gene of all Babesia species and other closely related apicomplexans using both negative extraction and negative PCR controls. Primers 5.1 (CCTGGTTGATCCTGCCAGTAGT) and 3.1 (CTCCTTCCTTTAAGTGATAAG) were used for the external reaction (Yabsley et al., 2005). Primary cycling conditions were as follows: initial denaturation for 5 min at 95 °C followed by 35 cycles of 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1.5 min, and a final annealing step of 72 °C for 7 min. The internal reaction used primers RLB-F (GAGGTAGTGACAAGAAATAACAATA) and RLB-R (TCTTCGATCCCCTAACTTTC) and 1 μL of the primary product (Yabsley et al., 2005). The secondary reaction cycling conditions were the same as the primary except the annealing temperature was lowered to 54 °C. Extracted DNA from a Babesia microti-like positive raccoon (GenBank accession number PP231979) was used as the positive control. These primers can amplify other apicomplexan species including Cytauxzoon and Hepatozoon spp. which also commonly infect some of the wildlife species included in this study.
A subset of positive samples representing at least one of each species detected via sequencing of the 18S rRNA amplicons (n = 35) were also tested with primers Babcox F (GGAAGTGGWACWGGWTGGAC) and Babcox R (TTCGGTATTGCATGCCTTG) targeting part of the cox1 gene of Babesia as previously described (Garrett et al., 2019). Extracted DNA from a Babesia microti-like positive raccoon was used as the positive control (GenBank accession number PP253997). Amplicons were visualized in 1.5% agarose gels and amplicons around 550bp for 18S rRNA and at 1080bp for cox1 were purified with ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA) and sequenced using Sanger sequencing at the University of Tennessee, Knoxville's Division of Biological Sequencing.
Consensus sequences were generated in Sequencher v. 5.4.6 (Gene Codes Corporation, Ann Arbor, MI) with assembly parameters set to an 85% agreement minimum. Chromatograms of the generated sequences were reviewed and those with poor quality or evidence of multiple sequences were cloned using the pGEM-T Easy Vector System (Promega Corporation, Madison, WI) and MAX Efficiency DH5α Competent Cells (ThermoFisher Scientific, Waltham, MA) following manufacturer's instructions. Plasmids were purified using MiniPrep Plasmid Purification kits (Thermo Fisher Scientific, Waltham, MA) following manufacturer's instructions. Four colonies were selected for sequencing from each cloned sample. Amplicon sequences were aligned with MAFFT using the L-INS-i algorithm with default parameters (Katoh and Standley, 2013). Sequences were trimmed manually to equivalent lengths. Substitution models were selected based on the Akaike information criterion. The Tamura Nei model was selected for the 18S sequences, and the General Time Reversible model was selected for the cox1 sequences (Tamura and Nei, 1993; Tavaré, 1986). Phylogenetic trees were generated in Mega X using the maximum-likelihood algorithm with 1000 bootstrap replicates (Tamura et al., 2021). Plasmodium falciparum was selected as the outgroup based on previous similar mesomammal Babesia surveys (Garrett et al., 2019, 2024). Identical or near identical (<2bp difference) sequences were removed to improve the readability of the tree. The top match in GenBank for each unique sequence was included in the tree for comparison as well as at least one representative from each clade in the Piroplasmida order (Jalovecka et al., 2019).
Tennessee raccoons were necropsied as part of a separate Trypanosoma cruzi study, and therefore histopathology from heart tissue was available in most cases. Though Babesia infection is more likely to cause changes to the spleen, other apicomplexan species like Hepatozoon and Besnoitia may be identified in heart tissue (Allen et al., 2011a; Elsheikha et al., 2003). During necropsy, samples of left and right ventricle were fixed in 10% buffered formalin and after 24–48 h were trimmed, embedded, and sectioned at 5 μm. Slides were stained with hematoxylin and eosin and reviewed with a board-certified pathologist (author MD) to evaluate for inflammation and apicomplexan infection.
Samples were collected both opportunistically from deceased animals and under a variety of state permits. All capture and handling procedures of South Carolina coyotes were permitted by Clemson University IACUC permit no. AUP 2018-031 and USDA Forest Service permit no. USFS 2018-031. Capture and handling procedures of Pennsylvania animals were performed by Pennsylvania USDA Wildlife Services under Pennsylvania Game Commission Special Use Permit 39964 and 31360.
3. Results
We processed a total of 786 samples: 230 coyotes, 159 muskrats, 123 raccoons, 97 red foxes, 64 striped skunks (referred to hereafter as “skunks”), 28 Virginia opossums, 28 black bears, 27 groundhogs, 10 mink, 19 gray foxes, and one bobcat. Samples were from ten states: 375 from Pennsylvania, 233 from Tennessee, 58 from South Carolina, 27 from Louisiana, 20 from Virginia, seven from North Carolina, 26 from Georgia, 26 from Oklahoma, 12 from Texas, and two from Maryland. Summaries of which species from each state were positive for Babesia can be found in Table 1, and specific information on sequence length and GenBank results for individual samples can be found in Supplementary Table 1. Representatives of each unique sequence were deposited in GenBank, with 18S rRNA Babesia sequences under accession numbers PP231979-PP232025, 18S rRNA Hepatozoon, Besnoitia, and Cytauxzoon spp. under accession numbers PP234617-PP234624, and Babesia cox1 sequences under accession numbers PP253989-PP253997.
Table 1.
Prevalence of Babesia spp. infections in wildlife in the eastern United States Percent positive is shown with the number positive and total number tested shown in parenthesis. Information on the Babesia sequences detected can be found in Supplementary Table 1.
| State | Bear | Bobcat | Coyote | Gray fox | Groundhog | Mink | Muskrat | Raccoon | Red Fox | Skunk | VOPO |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Georgia | – | – | 0% (0/7) | 7.7% (1/13) | – | – | – | – | 50% (3/6) | – | – |
| Louisiana | – | – | 0% (0/25) | 0% (0/1) | – | – | – | – | 0% (0/1) | – | – |
| Maryland | – | – | 0% (0/1) | – | – | – | – | – | 0% (0/1) | – | – |
| North Carolina | – | – | 0% (0/4) | 0% (0/2) | – | – | – | – | 0% (0/1) | – | – |
| Oklahoma | – | – | 0% (0/26) | – | – | – | – | – | – | – | – |
| Pennsylvania | 0% (0/28) | – | 0% (0/10) | – | 0% (0/23) | 0% (0/10) | 0% (0/159) | 33.3% (2/6) | 24.4% (20/82) | 28.1% (16/57) | – |
| South Carolina | – | – | 0% (0/58) | – | – | – | – | – | – | – | – |
| Tennessee | – | 0% (0/1) | 0% (0/74) | 100% (2/2) | – | – | – | 74.3% (87/117) | 50% (2/4) | 71.4% (5/7) | 3.6% (1/28) |
| Texas | – | – | 0% (0/12) | – | – | – | – | – | – | – | – |
| Virginia | – | – | 0% (0/13) | 0% (0/1) | 0% (0/4) | – | – | – | 0% (0/2) | – | – |
| Total | 0% (0/28) | 0% (0/1) | 0% (0/230) | 15.8% (3/19) | 0% (0/27) | 0% (0/10) | 0% (0/159) | 72.4% (89/123) | 25.8% (25/97) | 32.8% (21/64) | 3.6% (1/28) |
∗VOPO, Virginia opossum.
Babesia spp. infections were detected in 17.68% (139/786) of samples. Raccoons had the highest positivity of all wildlife species at 72.36% (89/123), while coyotes, mink, muskrat, and bear were all PCR negative for Babesia. Of the remaining wildlife, 32.8% of skunks (21/64), 25.8% of red foxes (25/97), 15.8% of gray foxes (3/19), and 3.6% of Virginia opossums (1/28) were PCR positive for Babesia spp (Fig. 1, Fig. 2).
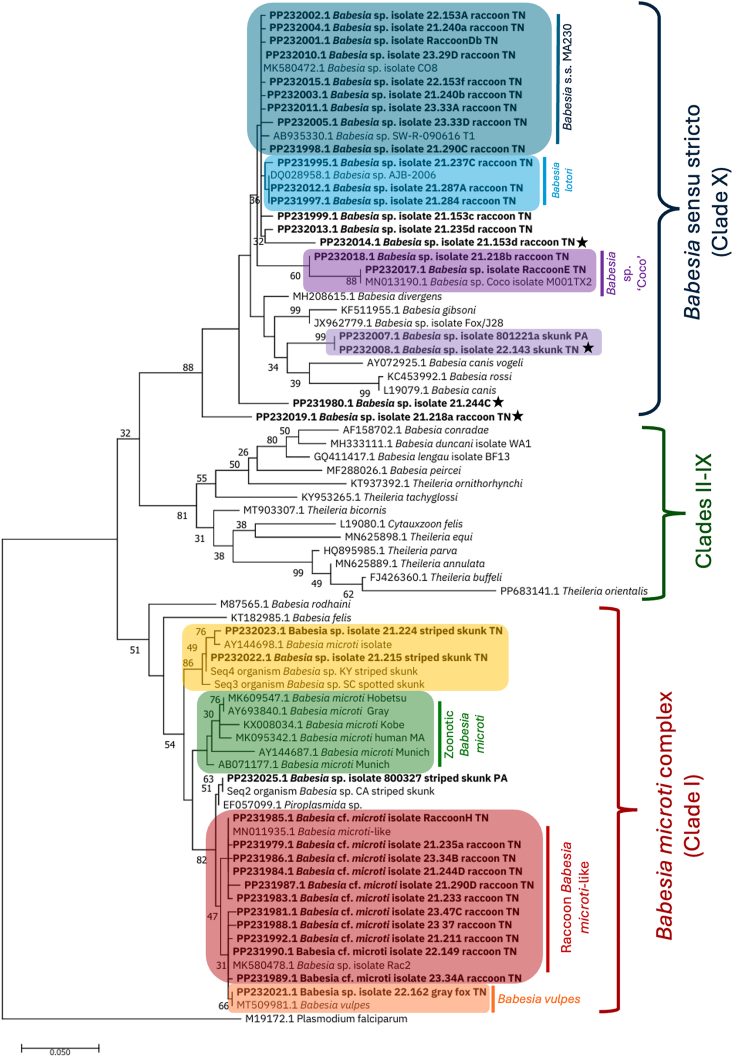
Fig. 1.
Phylogeny of partial 18S rRNA gene Babesia spp. sequences
Maximum-likelihood phylogenetic tree of Babesia spp. found in wildlife in the eastern United States. Plasmodium falciparum was chosen as the outgroup. A total of 76 sequences were used in the tree with sequences from this study (n = 36) in bold. All sequences generated in our study that differed by at least 2 base pairs are shown, and novel sequences are marked with a star.
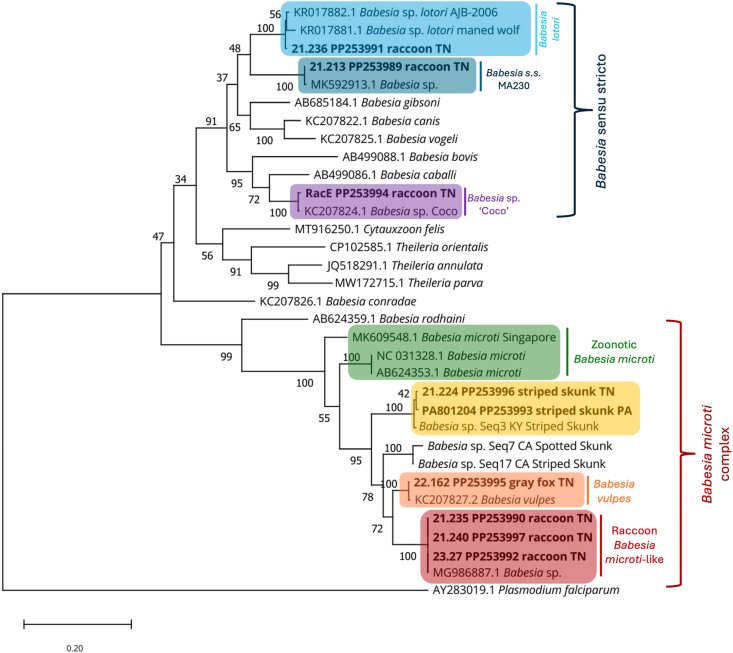
Fig. 2.
Phylogeny of partial cox1 Babesia spp. sequences
Maximum-likelihood phylogenetic tree of Babesia spp. found in wildlife in the eastern United States. Plasmodium falciparum was chosen as the outgroup. A total of 33 sequences were used in the tree with sequences from this study (n = 9) in bold. All sequences generated in our study that differed by at least 2 base pairs are shown, and novel sequences are marked with a star.
Of the 89 positive raccoons, 58.4% (52/89) were infected solely with B. microti-like species, 13.5% (12/89) solely with B. lotori, 10.1% (9/89) solely with B. s. s. MA230, and 3.4% (3/89) solely with Babesia sp. ‘Coco’. A total of fifteen raccoon samples were cloned, and 13 had mixed infections with up to four unique sequences identified in each sample. Seven samples had mixed infections of B. microti-like and B. s. s. MA230. Three samples had mixed infections of B. s. s. MA230 and B. lotori. The final three mixed infections consisted of unique sequences with Babesia sp. ‘Coco’, B. lotori, or B. s. s. MA230 respectively. These three novel sequences were less than 97% similar to other 18S rRNA sequences available in GenBank. Of the novel sequences, two were most similar (94.57% and 96.29%) to B. lotori (MK580743) and one was 95.92% similar to B. s. s. MA230 (MK580742). All B. microti-like, Babesia sp. ‘Coco’, and unique Babesia spp. positive raccoons were from Tennessee, whereas raccoons from both Pennsylvania and Tennessee were positive for B. lotori and B. s.s. MA230.
Skunks were primarily infected with a Babesia microti-like species documented previously in skunks from Massachusetts. Of the 21 skunks positive on 18S rRNA PCR, 67% (n = 14) were 99.28–100% similar to this sequence (AY144698). One skunk sample had a sequence 100% similar to B. s. s. MA230 found in raccoons (MK580472), two were 98.71% similar to a Babesia species found previously in river otters (EF057099), and one was identical to B. microti-like sequences found in raccoons (MN011935). Two skunks, one each from Pennsylvania and Tennessee, carried unique sequences that were 95.60% similar to a species found in a fox from China (JX962779), which is related to B. gibsoni and B. canis. Finally, one skunk from Pennsylvania had a mixed infection identified with cloning with one sequence 96.27% similar to the sequence from the fox from China and one sequence 99.78% similar to the skunks from Massachusetts.
Red and gray foxes were exclusively infected with B. vulpes, and all sequences except two were identical to each other and to multiple B. vulpes sequences in GenBank (e.g. MT509981). Positivity was slightly higher in the southern U.S. with 100% of Tennessee gray foxes (2/2), 50% of Tennessee red foxes (2/4), and 50% of Georgia red foxes (3/6) positive for B. vulpes, while only 24.4% (20/82) of Pennsylvania red foxes and 0% (0/2) of Virginia red foxes testing positive. The sequence from the Virginia opossum was 99.78% similar to B. microti-like sequences found in raccoons (e.g. MN011934).
Phylogenetic alignment of the 18S rRNA gene performed using one sequence for each unique sequence group, along with related organisms and Plasmodium falciparum (M19172) as the outgroup, resulted in a 547-bp alignment of which 153 of the 232 variable characters were parsimony informative. The generated tree showed that two of the three novel sequences in raccoons had an unstable position within the phylogenetic tree and did not clearly fall within any known clade. The novel skunk Babesia sequences were also unstable in their position between B. canis and B. gibsoni, though the tree supported the sequences as a novel group.
Nucleotide differences within species were assessed for the 180 generated sequences of the partial 18S gene (Table 2). Most sequences (70%) within a species were identical. Of the 30% of sequences that were not identical within species, most differed by only one or two nucleotides. The majority of specific nucleotide differences were found in only one or two sequences. However, two species had distinct haplotypes shared by numerous samples. First, a 6 base pair difference was identified in 6 of the 78 raccoon-type B. microti-like sequences. These sequences were most similar to sequence MK580478 and MN011934, while the remaining raccoon B. microti-like sequences were most similar to MN011935. Second, two groups were present in the 14 Babesia lotori sequences, with 7 sequences identical to each other and to MN013191. The other eight differed by a thymine to cytosine nucleotide difference and were most similar to DQ028958.
Table 2.
Nucleotide differences in generated Babesia species. The top row shows the number of nucleotide differences present within each sequence for individual species. The three novel sequences in raccoons for whom species could not be determined are not shown. For the raccoon-type B. microti-like sequences and the B. lotori sequences, two haplotypes that differed by 6 base pairs and one base pair respectively were identified and analyzed separately.
| Species | Total sequences | Identical | 1 nucleotide | 2 nucleotides | >2 nucleotides |
|---|---|---|---|---|---|
| B. microti-like raccoon-type | |||||
| MN11935-type | 71 | 52 | 13 | 2 | 4 |
| MK580478-type | 7 | 4 | 1 | 2 | 0 |
| Babesia ss MA230 | 31 | 13 | 14 | 1 | 3 |
| Babesia lotori | |||||
| MN013191-type | 7 | 7 | 0 | 0 | 0 |
| DQ028958-type | 8 | 5 | 2 | 0 | 1 |
| Babesia sp. 'Coco' | 4 | 2 | 1 | 0 | 1 |
| Babesia vulpes | 27 | 25 | 0 | 2 | 0 |
| Babesia microti-like skunk-type | 16 | 12 | 4 | 0 | 0 |
| Babesia microti-like river otter type | 2 | 2 | 0 | 0 | 0 |
| Novel skunk | 4 | 2 | 2 | 0 | 0 |
PCR and sequencing with the cox1 primers was less sensitive than the 18S rRNA primers. Of the 35 samples we performed cox1 PCR on, we obtained clean sequences for only fourteen. Ten samples had bands in the correct location, but sequences contained only nonspecific binding, and eleven were negative on cox1 PCR. Amplicons from animals infected with B. vulpes, B. microti-like, and Babesia sp. ‘Coco’ infections provided high quality sequences, while novel sequences, B. lotori, or B. s. s. MA230 were less likely to result in high quality sequences. We were unsuccessful in amplifying the partial cox1 of the novel sequence that most closely matched the fox from China, although we did amplify the partial cox1 gene in skunks infected with B. microti sequence previously reported from Massachusetts. That cox1 sequence was only 87.32% similar to other sequences in GenBank (KC207827), though similar sequences were recently found in skunks in numerous eastern and midwestern states (Garrett et al., 2024). Phylogenetic analysis of the cox1 sequences resulted in a 782-bp alignment of which 408 of the 586 variable characters were parsimony informative. Nine unique sequences were generated, with a total of 32 sequences used in the phylogenetic analysis.
The primers used to amplify the partial 18S gene of Babesia also amplified other apicomplexan species. Hepatozoon species were found in mink (70% [7/10]), muskrat (3.14% [5/159]), and raccoons (2.44% [3/123]). The sequences found in muskrats were identical to each other and 99.38% similar to H. ophisauri (MN723845). The Hepatozoon sequences in mink were identical to each other and 98.98% similar to a Hepatozoon species found in martens (Martes martes) in Hungary (OM256569). The Hepatozoon sequences in racoons were 97.53–98.37% similar to H. procyonis sequences in GenBank.
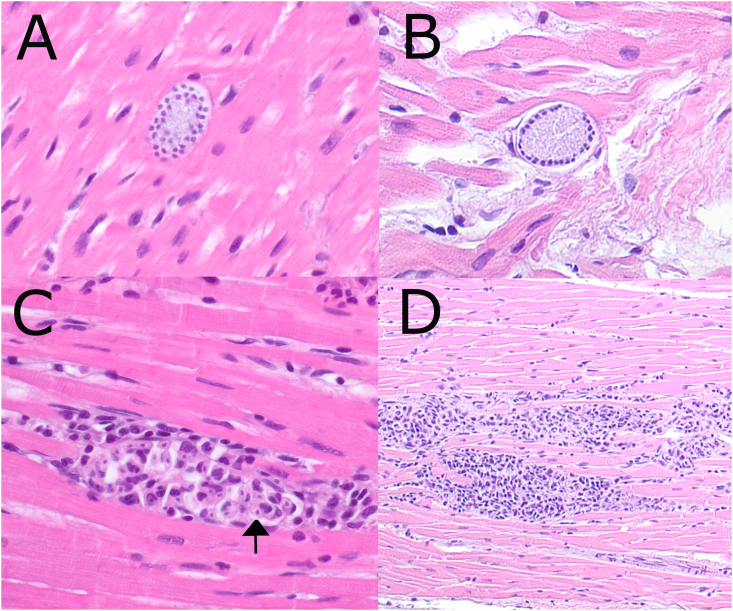
Of the three raccoons that were PCR positive for Hepatozoon, two had heart tissue histologically examined. Histopathologic findings revealed basophilic inclusions in leukocytes, moderate to severe myocarditis, and, in one case, a meront in the heart muscle (Fig. 3). In addition, one raccoon that was PCR positive for Babesia had evidence of H. procyonis infection in the heart including basophilic inclusion bodies and meronts. Despite testing both heart and blood and cloning the PCR products, we only detected Babesia sequences in this raccoon.
Fig. 3.
Hepatozoon procyonis photomicrographs from raccoons
Examples of meronts are shown in A and B. Mixed inflammation consisting primarily of lymphocytes and neutrophils with basophilic inclusion bodies of H. procyonis (arrow) are shown in C. A larger view of the typical moderate to severe multifocal myocarditis seen in infected raccoons is shown in D.
Hepatozoon canis and H. americanum were found frequently in foxes and coyotes, and those results were reported previously (Baker et al., 2023). No coinfections with Hepatozoon and Babesia spp. were detected in blood, heart, or spleen samples. The bobcat was PCR positive for C. felis (GenBank accession number PP234621). The sequence was 100% similar to multiple C. felis sequences in Genbank (eg MT904032). Besnoitia darlingi (2/28) was detected via PCR in the heart tissue of two Virginia opossums (Supplementary Table 1). One sequence (PP234618) was 100% similar to multiple other B. darlingi sequences in Genbank (eg MF87260), and the second (PP234620) was 99.26% similar to a different B. darlingi sequence (MF872603).
4. Discussion
A wide variety of Babesia species were found in wildlife, including several novel sequences. Raccoons had a high positivity of 72%, similar to what has been found in previous studies (Garrett et al., 2019; Birkenheuer et al., 2008; Clark et al., 2012). The most recent large-scale survey in the eastern United States, performed by Garrett et al., found a remarkably similar prevalence of 73% (512/699), while smaller surveys found higher rates of 95% (39/41) in North Carolina and 82% (14/17) in Florida (Garrett et al., 2019; Birkenheuer et al., 2008; Clark et al., 2012). Similar to Garrett et al., we found B. microti-like sequences predominated in raccoons, while B. lotori and the Babesia s. s. MA230 were less frequent (2017). Though the zoonotic B. microti species falls within the B. microti-like group, most species in this group are not known to be zoonotic. In fact, members of the B. microti clade are generally host specific, with the B. microti-like sequences found in raccoons distinct from those found in dogs and foxes (now called B. vulpes), North American river otters (Lontra canadensis), and badgers (Meles spp.) (Garrett et al., 2019, 2022). Our phylogenetic tree reinforced this, with the fox-associated B. vulpes, the zoonotic B. microti, and the two different skunk-associated B. microti-like sequences supported as separate clades.
Our 14.6% (13/89) rate of mixed infection was lower than the 22% coinfection rate found in a previous survey (Garrett et al., 2019). However, the Garrett survey performed multiple clade-specific rounds of PCR to determine co-infections, which is more sensitive for co-infections and likely explains the disparity. In addition to B. microti-like, B. lotori, and B. s. s. MA230, all of which have been frequently documented in the eastern United States, we also found Babesia sp. ‘Coco’ and three unique sequences. Although Babesia sp. ‘Coco’ was not found by Garrett et al. (2019), it has been identified in raccoons in Texas (Modarelli et al., 2020). All novel sequences in raccoons were found as part of mixed infections, and their significance is unclear. Further work assessing prevalence over time may help determine if these sequences are newly emerging or simply less prevalent.
This study found that skunks primarily carry two distinct species of B. microti-like, one similar to what has been found previously in skunks in Massachusetts, and one closely related to species found in river otters. The latter species has been shown to cause clinical symptoms in river otters, but its impact on skunks is not known (Garrett et al., 2022). In addition, skunks carried a novel sequence most closely related to B. canis and B. gibsoni. However, the sequence was only 95% similar to the closest GenBank match and likely represents a novel species. Though the sequence was well supported as a separate clade in the phylogenetic tree, its position within the tree was poorly supported. Further genetic work is needed to determine the phylogenetics of this potential novel species. The sequence was present in both skunks from Tennessee and Pennsylvania suggesting a wide geographic distribution, and the potential zoonotic or domestic animal impacts are unclear.
A recent survey of skunk Babesia spp. in the United States found a 48.4% (61/126) prevalence, with many similar sequences to this study (Garrett et al., 2024). They found both B. microti-like sequences that we documented, with sequences similar or identical to those we found (Fig. 1). Though they did not find the sequence related to B. gibsoni, they found a unique species in a hog-nosed skunk from Texas that was related to B. conradae which we did not find. Combined, these studies demonstrate that at least two different B. microti-like groups are common in skunks across the United States and that Babesia sp. outside the B. microti-like group occur rarely in skunks across the country. Further research to understand the novel sequences found is needed.
None of the 230 coyotes tested positive for Babesia, even though both B. vulpes and Babesia sp. ‘Coco’ can infect domestic dogs and were found in this study in other wildlife (Barash et al., 2019; Dear and Birkenheuer, 2022). In addition, B. vogeli is the most reported large Babesia species in domestic dogs in the United States and is found predominantly in the south (Javeed et al., 2022). It has been reported in coyotes from Texas, but surveillance in Eastern states is lacking (Yu et al., 2020). One possible explanation for the lack of B. vogeli infection in coyotes is that the brown dog tick vector, R. sanguineus, rarely infests coyotes. Numerous studies assessing ectoparasites on coyotes have failed to document infestation with R. sanguineus (Bloemer and Zimmerman, 1988; Eads, 1948; Green et al., 2019). Though this study indicates that coyotes are likely not a common host for B. vogeli, further work, particularly in urban coyotes that may be more exposed to R. sanguineus, is still necessary to understand the role coyotes may play in the disease ecology of this pathogen.
The B. vulpes sequences found in foxes had a low level of diversity. All B. vulpes sequences except for two were identical to each other, and those that were not identical differed by only a few nucleotides. Though most research into B. vulpes has been in Europe, there are studies documenting B. vulpes in domestic dogs and foxes in the United States and Canada (Barash et al., 2019; Birkenheuer et al., 2010). One study in Canada and North Carolina found a similar prevalence and also found low diversity in sequences (Birkenheuer et al., 2010). Though B. vulpes is an important cause of morbidity in domestic dogs in Europe, cases in North America are still rare. However, this may be related to the diagnostics used, which typically test for Babesia sensu stricto species (B. gibsoni, B. vogeli, B. canis, B. rossi, and Babesia sp. ‘Coco’) but not always B. microti-like species. A survey that tested for all Babesia species found a 0.5% (48/9367) prevalence of B. vulpes or B. vulpes co-infection in North American dogs, suggesting this species is still less prevalent than the other common species like B. gibsoni, which was found in 2% of samples (Barash et al., 2019).
The presence of a Hepatozoon sp. similar to H. ophisauri in 3% of muskrats was an interesting finding. 18S primers are not able to speciate Hepatozoon due to high gene conservation, so we cannot say if the species we found is H. ophisauri or simply a related species. Hepatozoon opthisauri is a snake-associated Hepatozoon species that has been reported in glass lizards (Pseudopus apodus) and the Tanezumi rat (Rattus tanezumi) in Asia (Zechmeisterová et al., 2021; Perison et al., 2022). It was hypothesized that lizards, frogs, snakes, and rodents may serve as intermediate or paratenic hosts (Zechmeisterová et al., 2021; Perison et al., 2022). Recent research has documented other Hepatozoon spp. in both snake and prey, and some researchers hypothesize that predation may play an important role in the disease ecology some Hepatozoon species (Tomé et al., 2012; Allen et al., 2011b). Hepatozoon ophisauri has not been documented outside of Europe and Asia, but there is a paucity of research in this area in the United States. Further work assessing Hepatozoon in muskrats is needed to determine if the sequence we found represents a new geographic location for H. ophisauri or a unique species. Surveys in reptiles, amphibians, and small rodents may help elucidate the epidemiology in the United States.
Though Hepatozoon has not been found in the United States in mink (Neovison vison) in the past, the species we found was closely related to species found in martens in Hungary (Hornok et al., 2022). Given the occurrence of 70% in our mink samples, it is unlikely that this species causes severe disease in healthy mink, but its potential impacts in the face of co-infection or immune suppression are not known.
The Hepatozoon species we found in raccoons has been documented several times in the past in Texas and Oklahoma (Clark et al., 1973; Allen et al., 2011b). Research in other states is lacking. Similar to the studies in Texas and Oklahoma, we found that H. procyonis is associated with significant myocarditis in raccoons. However, its impact on overall health is unknown. One raccoon had histopathology consistent with H. procyonis, but no Hepatozoon DNA was detected via PCR. It is likely that the raccoon was positive for H. procyonis, but the primers preferentially amplified Babesia DNA.
The two primary limitations of this study are the short sequence length of the partial 18S rRNA gene amplified and the lack of deep sequencing. Although the sequences amplified by the nested 18S rRNA primers were long enough to successfully separate the different clades of Babesia known to occur in wildlife, they are still short compared to the full length 18S rRNA sequence, leading to decreased phylogenetic support. This limited the interpretation of our sequence results to species identification rather than more sophisticated phylogenetic analysis. Several other studies on Babesia spp. in North American wildlife have used these primers due to their high sensitivity, and therefore this study is comparable to previous work (Garrett et al., 2019; Shaw et al., 2015; Goethert, 2021). In addition, these primers not only amplify all Babesia species but also other closely related genera like Hepatozoon, Cytauxzoon, and Besnoitia. Some of these species, particularly Hepatozoon species, are understudied.
The second major limitation of this study was the lack of deep sequencing to assess the true rate of coinfections in wildlife. Though cloning of select sequences revealed a 15% coinfection rate in raccoons and a 5% co-infection rate in skunks, this is likely an underestimate. Studies that use deep sequencing or multiple rounds of species-specific PCR have found higher rates of coinfection, and further work utilizing these methods would help clarify the true diversity of Babesia in wildlife (Garrett et al., 2019; Birkenheuer et al., 2008).
5. Conclusion
Skunks, raccoons, and foxes are important hosts for Babesia in the eastern United States and carry a greater diversity of species than previously reported. Further research is important to fully understand the transmission dynamics and risk to domestic animals and people. A Hepatozoon sp. similar to H. ophisauri is present in the United States, though the role of muskrats in the lifecycle is unclear.
Conflict of interest and ethical statement
Hereby, I, Eliza Baker, consciously assure that for the manuscript Prevalence and diversity of Babesia spp. in wildlife in the eastern United States the following is fulfilled:
-
1)
This material is the authors' own original work, which has not been previously published elsewhere outside of the author's dissertation.
-
2)
The paper is not currently being considered for publication elsewhere.
-
3)
The paper reflects the authors' own research and analysis in a truthful and complete manner.
-
4)
The paper properly credits the meaningful contributions of co-authors and co-researchers.
-
5)
The results are appropriately placed in the context of prior and existing research.
-
6)
All sources used are properly disclosed (correct citation).
-
7)
All authors have been personally and actively involved in substantial work leading to the paper, and will take public responsibility for its content.
CRediT authorship contribution statement
Eliza Baker: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Michelle Dennis: Writing – review & editing, Methodology. Alex Jensen: Writing – review & editing, Methodology. Kayla Buck Garrett: Writing – review & editing, Methodology. Christopher A. Cleveland: Writing – review & editing, Methodology. Michael J. Yabsley: Writing – review & editing, Methodology. Justin D. Brown: Writing – review & editing, Methodology. Kyle Van Why: Writing – review & editing, Methodology. Richard Gerhold: Writing – review & editing, Methodology, Conceptualization.
Consent for publication
Not applicable.
Availability of data and materials
All necessary data is provided within the text or tables of this manuscript.
Ethics approval and consent to participate
Most samples were collected opportunistically from trappers, rabies testing facilities, or banked samples from previous research projects. Some samples were collected as part of predator removal or live-trapping research under permit numbers AUP 2018-031 and USFS 2018-031 for South Carolina wildlife and Pennsylvania Game Commission Special Use Permit 39964 and 31360.
Funding
Not applicable.
Declaration of competing interests
The authors declare they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2024.101015.
Contributor Information
Eliza Baker, Email: elizalbbaker@gmail.com.
Michelle Dennis, Email: mdenni12@utk.edu.
Alex Jensen, Email: ajjensen@ncsu.edu.
Kayla Buck Garrett, Email: kaylab92@uga.edu.
Christopher A. Cleveland, Email: ccleve@uga.edu.
Michael J. Yabsley, Email: myabsley@uga.edu.
Justin D. Brown, Email: jdb56@psu.edu.
Kyle Van Why, Email: kyle.r.vanwhy@usda.gov.
Richard Gerhold, Email: rgerhold@utk.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Allen K.E., Johnson E.M., Little S.E. Hepatozoon spp infections in the United States. Vet. Clin. Small Anim. Pract. 2011;41:1221–1238. doi: 10.1016/j.cvsm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Allen K.E., Yabsley M.J., Johnson E.M., Reichard M.V., Panciera R.J., Ewing S.A., et al. Novel Hepatozoon in vertebrates from the southern United States. J. Parasitol. 2011;97 doi: 10.1645/GE-2672.1. 648–5. [DOI] [PubMed] [Google Scholar]
- Almazán C., Scimeca R.C., Reichard M.V., Mosqueda J. Babesiosis and theileriosis in North America. Pathogens. 2022;11(2):168. doi: 10.3390/pathogens11020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E., Jensen A., Miller D., Garrett K.B., Cleveland C.A., Brown J., et al. Hepatozoon spp. infection in wild canids in the eastern United States. Parasites Vectors. 2023;16:372. doi: 10.1186/s13071-023-05968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash N.R., Thomas B., Birkenheuer A.J., Breitschwerdt E.B., Lemler E., Qurollo B.A. Prevalence of Babesia spp. and clinical characteristics of Babesia vulpes infections in North American dogs. J. Vet. Intern. Med. 2019;33:2075–2081. doi: 10.1111/jvim.15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenheuer A.J., Neel J., Ruslander D., Levy M.G., Breitschwerdt E.B. Detection and molecular characterization of a novel large Babesia species in a dog. Vet. Parasitol. 2004;124:151–160. doi: 10.1016/j.vetpar.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Birkenheuer A.J., Marr H.S., Hladio N., Acton A.E. Molecular evidence of prevalent dual piroplasma infections in North American raccoons (Procyon lotor) Parasitology. 2008;135:33–37. doi: 10.1017/S0031182007003538. [DOI] [PubMed] [Google Scholar]
- Birkenheuer A.J., Horney B., Bailey M., Scott M., Sherbert B., Catto V., et al. Babesia microti-like infections are prevalent in North American foxes. Vet. Parasitol. 2010;172:179–182. doi: 10.1016/j.vetpar.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Bishop A., Wang H.H., Grant W.E. Using data surveillance to understand the rising incidence of babesiosis in the United States, 2011–2018. Vector Borne Zoonotic Dis. 2021;21:391–395. doi: 10.1089/vbz.2020.2754. [DOI] [PubMed] [Google Scholar]
- Bloch E.M., Kumar S., Krause P.J. Persistence of Babesia microti infection in humans. Pathogens. 2019;8(3):102. doi: 10.3390/pathogens8030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch E.M., Day J.R., Krause P.J., Kjemtrup A., O'Brien S.F., Tobian A.A.R., et al. Epidemiology of hospitalized patients with babesiosis, United States, 2010–2016. Emerg. Infect. Dis. 2022;28:354–362. doi: 10.3201/eid2802.210213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemer S.R., Zimmerman R.H. Ixodid ticks on the coyote and gray fox at land between the lakes, Kentucky–Tennessee, and implications for tick dispersal. J. Med. Entomol. 1988;25:5–8. doi: 10.1093/jmedent/25.1.5. [DOI] [PubMed] [Google Scholar]
- Brown H.M., Lockhart J.M., Latimer K.S., Peterson D.S. Identification and genetic characterization of Cytauxzoon felis in asymptomatic domestic cats and bobcats. Vet. Parasitol. 2010;172:311–316. doi: 10.1016/j.vetpar.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Clark K.A., Robinson R.M., Weishuhn L.L., Galvin T.J., Horvath K. Hepatozoon procyonis infections in Texas. J. Wildl. Dis. 1973;9:182–193. doi: 10.7589/0090-3558-9.2.182. [DOI] [PubMed] [Google Scholar]
- Clark K., Savick K., Butler J. Babesia microti in rodents and raccoons from Northeast Florida. J. Parasitol. 2012;98:1117–1121. doi: 10.1645/GE-3083.1. [DOI] [PubMed] [Google Scholar]
- Dear J.D., Birkenheuer A. Babesia in North America: an update. Vet Clin North Am Small Anim Pract. 2022;52:1193–1209. doi: 10.1016/j.cvsm.2022.07.016. [DOI] [PubMed] [Google Scholar]
- Dear J.D., Owens S.D., Lindsay L.L., Biondo A.W., Chomel B.B., Marcondes M., et al. Babesia conradae infection in coyote hunting dogs infected with multiple blood-borne pathogens. J. Vet. Intern. Med. 2018;32:1609–1617. doi: 10.1111/jvim.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads R.B. Ectoparasites from a series of Texas coyotes. J. Mammal. 1948;29:268–271. [Google Scholar]
- Ellis A.E., Mackey E., Moore P.A., Divers S.J., Hensel P., Carmichael K.P., et al. Debilitation and mortality associated with besnoitiosis in four Virginia opossums (Didelphis virginiana) J. Zoo Wildl Med. Off. Publ. Am. Assoc. Zoo Vet. 2012;43:367–374. doi: 10.1638/2011-0181.1. [DOI] [PubMed] [Google Scholar]
- Elsheikha H.M., Mansfield L.S., Fitzgerald S.D., Saeed M.A. Prevalence and tissue distribution of Besnoitia darlingi cysts in the Virginia opossum (Didelphis virginiana) in Michigan. Vet. Parasitol. 2003;115:321–327. doi: 10.1016/s0304-4017(03)00221-8. [DOI] [PubMed] [Google Scholar]
- Ganoe L.S., Brown J.D., Lovallo M.J., Yabsley M.J., Garrett K.B., Thompson A.T., Poppenga R.H., Ruder M.G., Walter W.D. Surveillance for diseases, pathogens, and toxicants of muskrat (Ondatra zibethicus) in Pennsylvania and surrounding regions. PLoS One. 2021;16 doi: 10.1371/journal.pone.0260987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett K.B., Schott R., Peshock L., Yabsley M.J. Prevalence and diversity of piroplasms and ticks in young raccoons and an association of Babesia sensu stricto infections with splenomegaly. Parasitol Open. 2018;4:e12. [Google Scholar]
- Garrett K.B., Hernandez S.M., Balsamo G., Barron H., Beasley J.C., Brown J.D., et al. Prevalence, distribution, and diversity of cryptic piroplasm infections in raccoons from selected areas of the United States and Canada. Int. J. Parasitol Parasites Wildl. 2019;9:224–233. doi: 10.1016/j.ijppaw.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett K., Halseth A., Ruder M.G., Beasley J., Shock B., Birkenheuer A.J., et al. Prevalence and genetic characterization of a Babesia microti-like species in the North American river otter (Lontra canadensis) Vet Parasitol Reg Stud Rep. 2022;29 doi: 10.1016/j.vprsr.2022.100696. [DOI] [PubMed] [Google Scholar]
- Garrett K.B., Brown J., Gabriel M., Dowler R., Perkins J.C., Krejsa D., Yabsley M.J. Diversity of Babesia spp. in skunks from selected states in the United States of America. Parasite. 2024;31 doi: 10.1051/parasite/2024043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethert H.K. What Babesia microti is now. Pathogens. 2021;10:1168. doi: 10.3390/pathogens10091168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethert H.K., Telford S.R. What is Babesia microti? Parasitology. 2003;127:301–309. doi: 10.1017/s0031182003003822. [DOI] [PubMed] [Google Scholar]
- Green E.N., Porter W.T., Howard A.L., Yaglom H., Benford R., Busch J.D., et al. Coyotes (Canis latrans) in Arizona, USA, exhibit immune and genetic evidence of rickettsial infections. J. Wildl. Dis. 2019;56:261–269. [PubMed] [Google Scholar]
- Herc E., Pritt B., Huizenga T., Douce R., Hysell M., Newton D., et al. Probable locally acquired Babesia divergens–like infection in woman, Michigan, USA. Emerg. Infect. Dis. 2018;24:1558–1560. doi: 10.3201/eid2408.180309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook A.A., Frerichs W.M. Babesia mephitis sp. N. (Protozoa: Piroplasmida), a hematozoan parasite of the striped skunk, Mephitis mephitis. J. Parasitol. 1970;56:930–931. [PubMed] [Google Scholar]
- Hornok S., Boldogh S.A., Takács N., Kontschán J., Szekeres S., Sós E., Sándor A.D., Wang Y., Tuska-Szalay B. Molecular epidemiological study on ticks and tick-borne protozoan parasites (Apicomplexa: Cytauxzoon and Hepatozoon spp.) from wild cats (Felis silvestris), Mustelidae and red squirrels (Sciurus vulgaris) in central Europe, Hungary. Parasites Vectors. 2022;15:174. doi: 10.1186/s13071-022-05271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin P.J. Canine babesiosis. Vet Clin North Am Small Anim Pract. 2010;40:1141–1156. doi: 10.1016/j.cvsm.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Jalovecka Marie, Sojka Daniel, Ascencio Mariano, Schnittger Leonhard. Babesia life cycle–when phylogeny meets biology. Trends Parasitol. 2019;35:356–368. doi: 10.1016/j.pt.2019.01.007. 28. [DOI] [PubMed] [Google Scholar]
- Javeed N.N., Shultz L., Barnum S., Foley J.E., Hodzic E., Pascoe E.L., et al. Prevalence and geographic distribution of Babesia conradae and detection of Babesia vogeli in free-ranging California coyotes (Canis latrans) Int. J. Parasitol Parasites Wildl. 2022;19:294–300. doi: 10.1016/j.ijppaw.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A.J., Muthersbaugh M., Ruth C.R., Butfiloski J.W., Cantrell J., Adams J., Waits L., Kilgo J.C., Jachowski D.S. Resource pulses shape seasonal and individual variation in the diet of an omnivorous carnivore. Ecol. Evol. 2024;14(7) doi: 10.1002/ece3.11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnai M., Kawabuchi-Kurata T., Tsuji M., Nakajima R., Fujisawa K., Nagata S., et al. Molecular evidence for the presence of new Babesia species in feral raccoons (Procyon lotor) in Hokkaido, Japan. Vet. Parasitol. 2009;162:241–247. doi: 10.1016/j.vetpar.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Joseph J.T., Roy S.S., Shams N., Visintainer P., Nadelman R.B., Hosur S., et al. Babesiosis in lower hudson valley, New York, USA. Emerg. Infect. Dis. 2011;17:843–847. doi: 10.3201/eid1705.101334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarelli J.J., Westrich B.J., Milholland M., Tietjen M., Castro-Arellano I., Medina R.F., et al. Prevalence of protozoan parasites in small and medium mammals in Texas, USA. Int. J. Parasitol Parasites Wildl. 2020;11:229–234. doi: 10.1016/j.ijppaw.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietfeld J.C., Pollock C. Fatal cytauxzoonosis in a free-ranging bobcat (Lynx rufus) J. Wildl. Dis. 2002;38:607–610. doi: 10.7589/0090-3558-38.3.607. [DOI] [PubMed] [Google Scholar]
- Perison P.W.D., Amran N.-S., Adrus M., Anwarali Khan F.A. Detection and molecular identification of blood parasites in rodents captured from urban areas of southern Sarawak, Malaysian Borneo. Vet. Med. Sci. 2022;8:2059–2066. doi: 10.1002/vms3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger L., Ganzinelli S., Bhoora R., Omondi D., Nijhof A.M., Florin-Christensen M. The Piroplasmida Babesia, Cytauxzoon, and Theileria in farm and companion animals: species compilation, molecular phylogeny, and evolutionary insights. Parasitol. Res. 2022;121(5):1207–1245. doi: 10.1007/s00436-022-07424-8. [DOI] [PubMed] [Google Scholar]
- Scott J.D., Scott C.M. Human babesiosis caused by Babesia duncani has widespread distribution across Canada. Healthcare. 2018;6:49. doi: 10.3390/healthcare6020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M., Kolba N., Huffman J.E. Babesia spp. in Ursus americanus (black bear) in New Jersey. Northeast. Nat. 2015;22:451–458. [Google Scholar]
- Shock B.C., Lockhart J.M., Birkenheuer A.J., Yabsley M.J. Detection of a Babesia species in a bobcat from Georgia. SE. Nat. 2013;12:243–247. [Google Scholar]
- Skinner D., Mitcham J.R., Starkey L.A., Noden B.H., Fairbanks W.S., Little S.E. Prevalence of Babesia spp., Ehrlichia spp., and tick infestations in Oklahoma black bears (Ursus americanus) J. Wildl. Dis. 2017;53:781–787. doi: 10.7589/2017-02-029. [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura Koichiro, Stecher Glen, Kumar Sudhir. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. In: Some Mathematical Questions in Biology - DNA Sequence Analysis. Miura R.M., editor. Amer Math Soc); Providence, RI: 1986. Some probabilistic and statistical problems in the analysis of DNA sequences; pp. 57–86. [Google Scholar]
- Thomas R., Santodomingo A., Saboya-Acosta L., Quintero-Galvis J.F., Moreno L., Uribe J.E., Muñoz-Leal S. Hepatozoon (Eucoccidiorida: hepatozoidae) in wild mammals of the Americas: a systematic review. Parasites Vectors. 2024;17:108. doi: 10.1186/s13071-024-06154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé B., Maia J.P.M.C., Harris D.J. Hepatozoon infection prevalence in four snake genera: influence of diet, prey parasitemia levels, or parasite type? J. Parasitol. 2012;98:913–917. doi: 10.1645/GE-3111.1. [DOI] [PubMed] [Google Scholar]
- Tsao J.I., Hamer S.A., Han S., Sidge J.L., Hickling G.J. The contribution of wildlife hosts to the rise of ticks and tick-borne diseases in North America. J. Med. Entomol. 2021;58:1565–1587. doi: 10.1093/jme/tjab047. [DOI] [PubMed] [Google Scholar]
- Westmoreland L.S.H., Stoskopf M.K., Sheppard E., DePerno C.S., Gould N.P., Olfenbuttel C., et al. Detection and prevalence of Babesia spp. in American black bears (Ursus americanus) from eastern and western North Carolina, USA. J. Wildl. Dis. 2019;55:678–681. doi: 10.7589/2018-06-164. [DOI] [PubMed] [Google Scholar]
- Wikander Y.M., Reif K.E. Cytauxzoon felis: an overview. Pathogens. 2023;12:133. doi: 10.3390/pathogens12010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabsley M.J., Davidson W.R., Stallknecht D.E., Varela A.S., Swift P.K., Devos J.J., et al. Evidence of tick-borne organisms in mule deer (Odocoileus hemionus) from the western United States. Vector Borne Zoonotic Dis. 2005;5:351–362. doi: 10.1089/vbz.2005.5.351. [DOI] [PubMed] [Google Scholar]
- Yang Y., Christie J., Köster L., Du A., Yao C. Emerging human babesiosis with “ground zero” in North America. Microorganisms. 2021;9:440. doi: 10.3390/microorganisms9020440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Modarelli J., Tomeček J.M., French J.T., Hilton C., Esteve-Gasent M.D. Prevalence of common tick-borne pathogens in white-tailed deer and coyotes in south Texas. Int. J. Parasitol Parasites Wildl. 2020;11:129–135. doi: 10.1016/j.ijppaw.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechmeisterová K., Javanbakht H., Kvičerová J., Široký P. Against growing synonymy: identification pitfalls of Hepatozoon and Schellackia demonstrated on North Iranian reptiles. Eur. J. Protistol. 2021;79 doi: 10.1016/j.ejop.2021.125780. [DOI] [PubMed] [Google Scholar]
- Zygner W., Gójska-Zygner O., Bartosik J., Górski P., Karabowicz J., Kotomski G., et al. Canine babesiosis caused by large Babesia species: global prevalence and risk factors—a review. Anim Basel. 2023;13:2612. doi: 10.3390/ani13162612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All necessary data is provided within the text or tables of this manuscript.