Abstract
Environmental enteric dysfunction (EED), a small intestinal disorder found at a high prevalence in stunted children, is associated with gut mucosal barrier disruption and decreased absorptive capacity due to reduced intact small intestinal villi1–4. To test the hypothesis that intergenerational transmission of a perturbed small intestinal microbiota contributes to undernutrition by inducing EED5, we characterized two consortia of bacterial strains cultured from duodenal aspirates from stunted Bangladeshi children with EED – one of which induced local and systemic inflammation in gnotobiotic female mice. Offspring of dams that received this inflammatory consortium exhibited immunologic changes along their gut that phenocopied features of EED in children. Single nucleus plus bulk RNA-sequencing revealed alterations in inter-cellular signaling pathways related to intestinal epithelial cell renewal, barrier integrity and immune function while analyses of cerebral cortex disclosed alterations in glial- and endothelial-neuronal signaling pathways that regulate neural growth/axonal guidance, angiogenesis and inflammation. Analysis of ultrasonic vocalization calls in gnotobiotic P5-P9 pups indicated increased arousal and perturbed neurodevelopment in the offspring of dams harboring the inflammation-inducing consortium. Cohousing experiments and follow-up screening of candidate disease-promoting bacterial isolates identified a strain typically found in the oral microbiota (Campylobacter concisus) as a contributor to enteropathy. Given that fetal growth was also impaired in the dams with the consortium that induced enteropathy, this preclinical model allows the effects of the human small intestinal microbiota on both pre- and postnatal development to be ascertained, setting the stage for identification of small intestinal microbiota-targeted therapeutics for (intergenerational) undernutrition.
Undernutrition is a significant global health challenge. Numerous epidemiologic studies indicate that stunting in mothers is associated with low birth weight and postnatal linear growth faltering (stunting) in their offspring6,7. Stunting in infants and children is accompanied by increased risk of infection, metabolic/hormonal imbalances later in life8, as well as neurodevelopmental/cognitive abnormalities9–11.
One hypothesis is that a perturbed small intestinal microbiota2 contributes to undernutrition by inducing environmental enteric dysfunction (EED)1,12–14 and subsequent transmission of the maternal small intestinal microbiota to her offspring perpetuates intergenerational EED5. Evidence that the small intestinal microbiota contributes to the pathogenesis of EED comes in part from the Bangladeshi EED (BEED) study, in which stunted children who had failed to respond to a nutritional intervention underwent esophago-gastroduodenoscopy (EGD)2,15. Almost all (95%) of these children displayed histopathologic evidence of EED2. Sequencing bacterial 16S rRNA amplicons generated from duodenal aspirates collected during EGD revealed a ‘core’ group of 14 taxa (amplicon sequence variants, ASVs) that were present in >80% of the children. The absolute abundances of these taxa, not typically classified as enteropathogens, were significantly negatively correlated with linear growth (length-for-age Z score, LAZ) and positively correlated with levels of duodenal mucosal proteins involved in immunoinflammatory responses2.
Deciphering the role of the small intestinal microbiota in the pathogenesis of EED has been hampered by several factors. For example, EGD is limited to the proximal intestine and carries potential risks, precluding its use in healthy children for defining what constitutes a ‘normal’ small intestinal microbiota for ethical reasons. In addition, the volume of fluid and biomass of microbes that can be retrieved from the lumen of the small intestine with EGD is small, limiting the number of ways the material can be employed for downstream analyses. Moreover, preclinical studies are necessary to define mechanisms and establish causal relationships between members of the microbiota and EED. Current mouse models rely on either provision of a low-protein diet, which has not been explicitly defined as a causative factor for EED in humans, and/or administration of a single pathogen or a broad inflammatory insult (e.g. treatment with LPS)16–19. While these models are valuable for understanding effects of protein restriction on immunity and the virulence mechanisms of enteric pathogens, they do not simulate the complexity of small intestinal bacterial community dynamics and interactions with the host that likely play a role in pathophysiology of EED in humans.
In the current study, we sought to model maternal-to-offspring transmission of different small intestinal bacterial consortia from children in the BEED study that elicit discordant intestinal and systemic inflammatory responses in gnotobiotic mice. Our goal was to use these consortia to obtain insights about host cellular and molecular responses and to identify bacteria that have a causal role in EED pathology.
Results
Development of inflammatory and non-inflammatory EED-derived bacterial consortia
In a pilot study2, a collection of 184 bacterial isolates cultured from the duodenal aspirates of children in the BEED study were pooled and introduced by oral gavage directly into adult mice fed a diet formulated based on foods consumed by children living in Mirpur, Bangladesh, the urban slum in which the clinical study was performed (‘Mirpur-18’ diet, Supplementary Table 1a). Compared to controls gavaged with intact cecal contents from conventionally-raised mice (conventionalized animals, CONV-D), the duodenal aspirate-derived culture collection induced an enteropathy characterized by (i) patchy immunoinflammatory infiltrates in the small intestine, (ii) increased duodenal crypt depth, (iii) reduced expression of tight junction proteins and increased expression of genes involved in anti-microbial defense (Reg3β and Reg3γ) in the duodenum, and (iv) elevated levels of matrix metalloproteinase 8 (MMP8) in serum as well as along the length of the small intestine2.
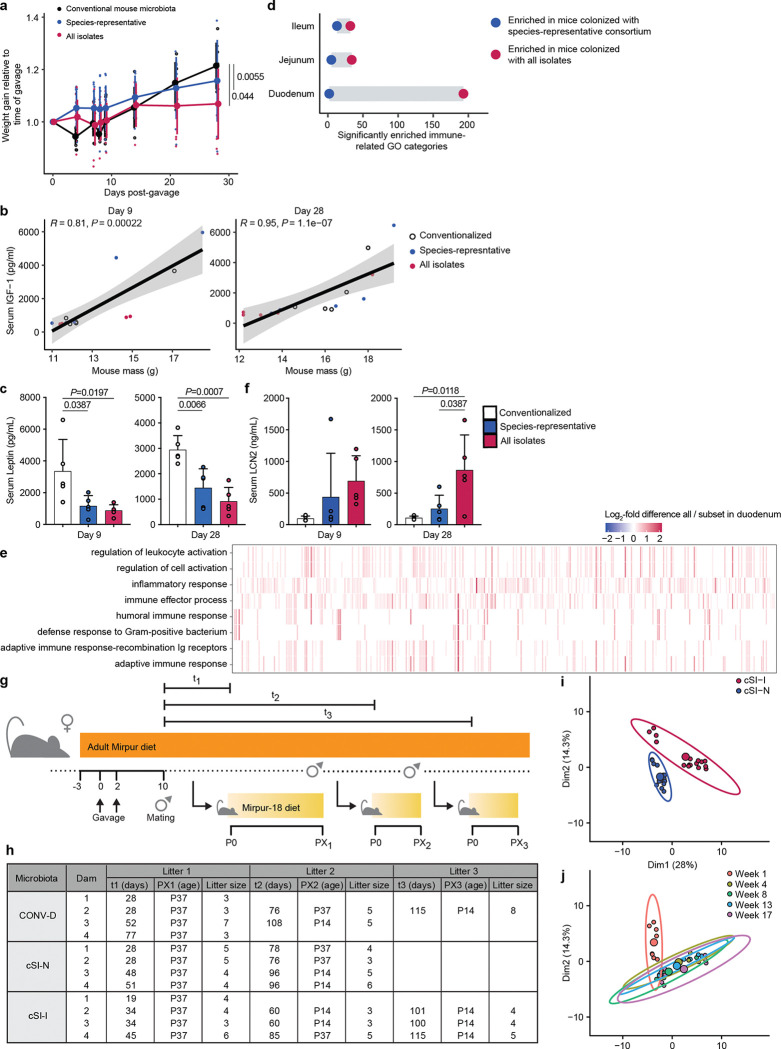
We sought to reduce the complexity of the 184-member bacterial consortium derived from the EED donors. To do so, we chose a representative of each bacterial species (based on full-length 16S sequencing identification, Supplementary Table 1b) and pooled these 39 isolates into a ‘species-representative subset consortium’. These two consortia were gavaged into separate groups of just-weaned 4–5-week-old germ-free mice (n=5 animals/group) fed the Mirpur-18 diet. Mice colonized with all 184 isolates gained significantly less weight over 28 days than mice colonized with the species-representative subset or CONV-D controls (Extended Data Fig. 1a). Serum levels of insulin-like growth factor 1 (IGF-1) were significantly correlated with body mass at both 9 and 28 days after colonization (Extended Data Fig. 1b), while leptin levels were significantly lower in mice that received the 184-member community (Extended Data Fig. 1c).
Bulk RNA-sequencing of the duodenum, jejunum and ileum of these animals followed by gene set enrichment analysis (GSEA) revealed that a large proportion of the gene ontology (GO) categories that were significantly enriched (q<0.05) between animals colonized with the full versus subset bacterial consortia were related to the immune function. Compared to mice colonized with the species-representative subset, those colonized with the 184-member consortium exhibited greater enrichment of immune-related GO categories along the small intestine, including genes involved in the anti-bacterial defense response and leukocyte activation (Extended Data Fig. 1d,e). Levels of the protein lipocalin-2 (LCN2/NGAL; neutrophil gelatinase-associated lipocalin) were also significantly elevated in the serum of mice colonized with the full 184-isolate consortium (Extended Data Fig. 1f); previously we had observed that LCN2 levels in the duodenal mucosa of children with EED were strongly correlated with the absolute abundances of the 14 duodenal EED-associated ‘core taxa’2. Based on the higher levels of intestinal and systemic inflammation in animals colonized with the full consortium of EED-derived bacteria, we named this community ‘child small intestinal inflammation-inducing’ (cSI-I); the species representative subset was designated ‘child small intestinal non-inflammatory’ (cSI-N). We reasoned that comparing these two EED-derived consortia – one of which resulted in impaired weight gain and induced an immunoinflammatory response – would allow us to discern potential small intestinal bacterial mediators of disease.
Modeling maternal-offspring transmission of EED donor small intestinal bacterial consortia in gnotobiotic mice
We next tested whether inflammation induced by the cSI-I consortium transmitted to offspring. Germ-free adult female C57Bl/6J mice were fed a diet designed to be representative of that consumed by adults residing in the Mirpur district of Dhaka (‘Adult Mirpur’ diet; Supplementary Table 1a). Three days later, separate groups of animals (n=4 dams/group) received an oral gavage of the cSI-I or cSI-N consortium, or intact cecal contents from conventionally-raised adult C57Bl/6J mice (Fig. 1a, Extended Data Fig. 1g). One week after colonization, female mice were mated with germ-free males. Pups born to these dams were weaned onto the ‘Mirpur-18’ diet (Supplementary Table 1a), which they consumed ad libitum until euthanasia on postnatal day 37 (P37). To assess the reproducibility and timing of onset of EED features, second and third litters produced by the same dams were euthanized either on P37 or P14, respectively (Extended Data Fig. 1g,h).
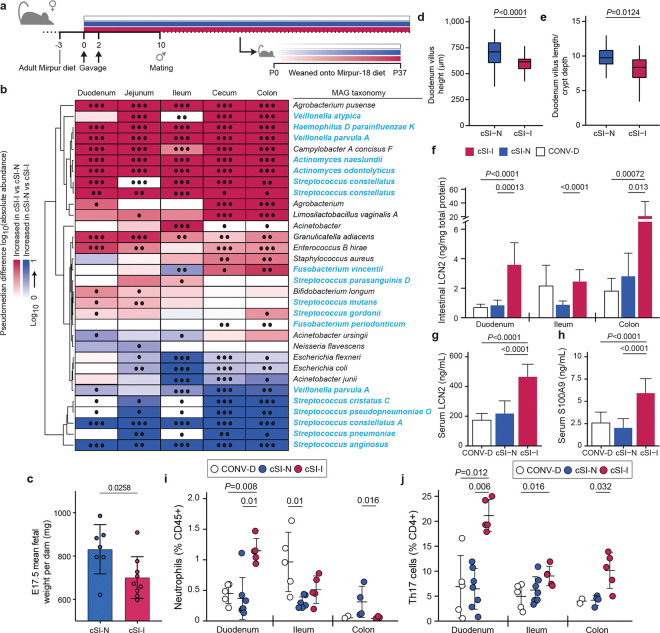
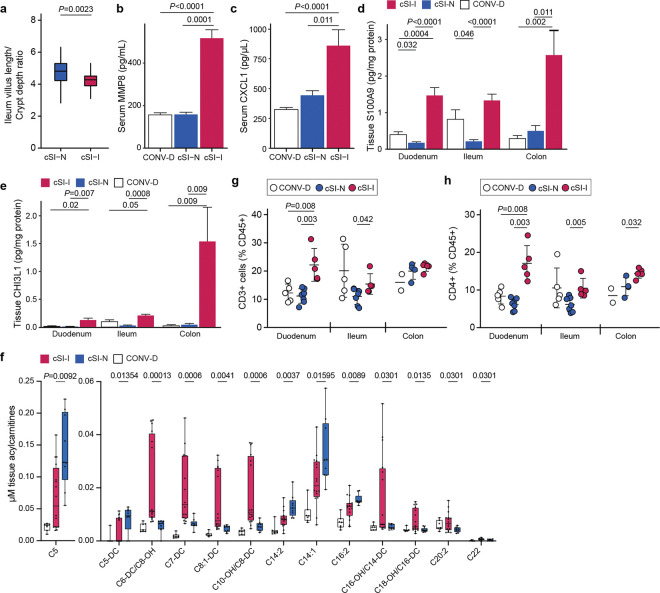
Fig. 1. Dam-to-offspring transmission of bacteria cultured from duodenal aspirates collected from Bangladeshi children with EED.
a, Design of intergenerational transmission experiment. b, Differential abundances of bacterial MAGs (rows) along the length of the intestine (columns) of P37 pups born to dams harboring cSI-N or cSI-I bacterial consortia (n=21–25 mice/group). MAGs were included in the heatmap if they demonstrated significant differential log10 absolute abundance in at least one of the intestinal locations. MAG taxonomy highlighted in blue indicates correspondence to ASVs representing ‘core taxa’ identified in the small intestines of stunted children with EED in the BEED study. •, P-adj < 0.05; ••, P-adj < 0.01; •••, P-adj < 0.001. c, Masses of fetuses were averaged within each dam. Dams were colonized with the cSI-I or cSI-N consortia for two weeks prior to mating (n=7–9 dams/group; n=2–10 fetuses/litter, unpaired t-test). d,e, Villus length (d), and ratio of villus length to crypt depth (e) in the duodenum of P37 cSI-I compared to cSI-N mice (n=5 animals/group, boxes denote interquartile range). f, Levels of lipocalin-2 (LCN2) protein in intestinal tissue from P37 mice (n=7–10 mice/group). g,h, Serum levels of LCN2 (g), and S100A9 protein (h), in P37 pups (n=21–25 mice/group). For c-e, n=4–6 litters/group, 3–7 pups/litter. i,j, Frequency of neutrophils (i), and Th17 cells (j), along the length of the gut (n=5–7 mice/group, each point represents an individual animal). For d-j, statistics shown are the result of Wilcoxon-ranked sum tests. For c-j, mean values ± s.d. are shown.
We identified bacterial members of each consortium that successfully colonized recipient dams and were transmitted to their offspring using long-read shotgun DNA sequencing of cecal contents from dams and P37 pups followed by assembly of metagenome-assembled genomes (MAGs) (n=4 dams and n=22–25 offspring/consortium). We reasoned that this approach would be useful for initial characterization of colonizing members of culture collections where multiple strains of a given species may exist. A total of 50 unique high-quality MAGs (defined as ≥85% complete and ≤5% contaminated based on marker gene analysis, Supplementary Table 1c) were generated from cecal contents of dams and P37 offspring harboring either of the two cSI consortia. Twenty-six MAGs from the cSI-I consortium colonized dams; 19 of these 26 MAGs also colonized their P37 offspring (threshold criteria for colonization: >0.001% relative abundance in at least one intestinal segment in at least 50% of animals, Supplementary Table 1d). A total of 24 MAGs from the cSI-N consortium colonized dams and all were represented in their P37 offspring (Supplementary Table 1d). Based on short-read shotgun calculations of MAG abundances in fecal samples collected from dams, the cSI-I and cSI-N communities remained stable between 4 and 17 weeks after gavage of these cultured consortia even if the animals had one or more litters during this period (P=0.09, PERMANOVA; n=4 dams/group; Extended Data Fig. 1i,j). The absolute abundances of 21 MAGs were significantly higher in at least one intestinal segment of P37 offspring colonized with the cSI-I consortium; the taxonomy of 12 of these 21 MAGs corresponded to ASVs previously identified as ‘core taxa’ in Bangladeshi children with EED (Fig. 1b)2.
Elevated levels of EED biomarkers in suckling offspring of cSI-I dams and restricted fetal growth
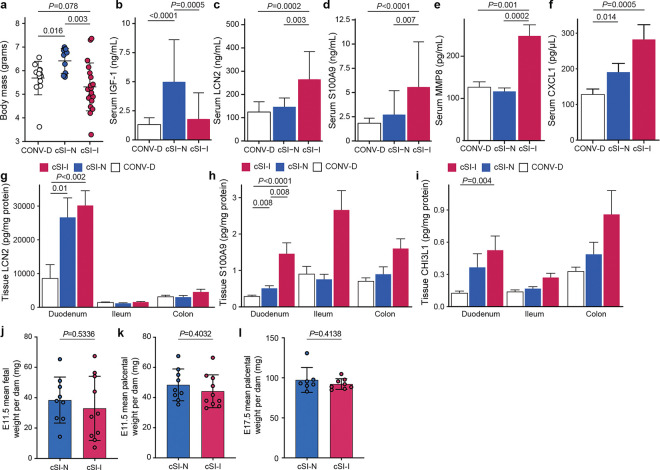
First, we sought to determine whether the enteropathy-inducing consortium affected mice during early postnatal development. Body mass (Extended Data Fig. 2a) and serum levels of IGF-1 (Extended Data Fig. 2b) were significantly reduced in pre-weaning (P14) offspring of cSI-I dams compared to their cSI-N counterparts (Extended Data Fig. 2a,b; Supplementary Table 2a,b). P14 cSI-I pups displayed significantly higher serum levels of the pro-inflammatory proteins LCN2, S100A9 (S100 calcium binding protein A9), CHI3L1 (Chitinase-3-like protein 1), MMP8 (matrix metallopeptidase 8, or neutrophil collagenase), as well as the immune cell chemoattractant CXCL1 (chemokine ligand 1) compared to their cSI-N and CONV-D counterparts (Extended Data Fig. 2c–f; Supplementary Table 2a). LCN2, S100A9 and MMP8 belong to a group of inflammatory proteins whose expression in duodenal mucosal biopsies was significantly positively correlated with the abundances of the ‘core’ EED-associated bacterial taxa in Bangladeshi children with EED2. LCN2, S100A9 and CHI3L1 were similarly elevated in duodenal, ileal and colonic tissue of cSI-I animals (Extended Data Fig. 2g–i). Thus, systemic and intestinal inflammation were already evident in these animals prior to completion of weaning.
To determine whether growth of fetuses was impacted by maternal colonization with cSI-I or cSI-N, we colonized additional adult female mice with the cSI-I or cSI-N consortia two weeks before mating, and euthanized dams at embryonic day 11.5 (E11.5) and 17.5 (E17.5). Fetal weights were significantly lower in cSI-I compared to cSI-N dams close to parturition, at E17.5 (Fig. 1c), but not just after placentation at E11.5 (Extended Data Fig. 2j; Supplementary Table 2b). Placental weights were not significantly different between treatment groups at either timepoint (Extended Data Fig. 2k,l).
Characterization of enteropathy in P37 offspring of cSI-I dams
Histomorphometric analysis of the small intestines of P37 offspring of cSI-I dams revealed significantly diminished duodenal villus height (Fig. 1d) and a significantly diminished ratio of villus height to crypt depth in the duodenum and ileum compared to their cSI-N counterparts (Fig. 1e, Extended Data Fig. 3a; Supplementary Table 2d), reflective of villus blunting documented in EED in humans. Crypt depth in the ileum of P37 cSI-I mice trended towards being increased relative to cSI-N mice (P=0.09, Wilcoxon rank-sum).
LCN2 levels in duodenal, ileal and colonic tissue, as well as in serum, were significantly elevated in P37 mice born to dams harboring the cSI-I consortium compared to the cSI-N consortium or to conventionalized (CONV-D) dams (Fig. 1f,g). S100A9, MMP8, CXCL1 and IL-17 were similarly elevated in the serum of P37 cSI-I offspring (Fig. 1h, Extended Data Fig. 3b,c; Supplementary Table 2a). As in P14 animals, duodenal, ileal and colonic tissue levels of S100A9 and CHI3L1 were significantly higher in P37 offspring of cSI-I dams (Extended Data Fig. 3d,e; Supplementary Table 2a). There were no statistically significant differences in the serum levels of these proteins between litters, nor between male and female offspring in the same treatment group (P>0.05, Wilcoxon rank-sum tests). Compared to P37 cSI-N and CONV-D mice, serum levels of Dickkopf-1 (DKK1), Osteoprotegerin (OPG) and Fibroblast Growth Factor-23 (FGF23) were significantly higher in cSI-I animals, indicating increased osteocyte and bone remodeling activity (P<0.015 for all comparisons, Wilcoxon rank-sum test; Supplementary Table 2a). Micro-computed tomography of femurs disclosed significantly increased cortical tissue mineral density in P37 cSI-I versus cSI-N animals (P=0.03, Wilcoxon rank-sum test; n=19–21 mice/group); no significant differences in other cortical and trabecular parameters were found between the three treatment groups (Supplementary Table 2c).
Small intestinal tissue acylcarnitines
Plasma acylcarnitines are elevated in a fasting state and have been identified as a biomarker of EED in children20. Moreover, fecal acylcarnitines are elevated in the dysbiotic microbial states associated with inflammatory bowel disease21 and can activate pro-inflammatory signaling cascades22,23. We quantified acylcarnitine levels in intestinal tissue instead of plasma to directly examine whether differences in host fatty acid metabolism were produced as a result of colonization with the cSI-I compared to the cSI-N consortium. Several medium and long-chain acylcarnitines were significantly elevated in the intestine of cSI-I compared to cSI-N or CONV-D offspring (Extended Data Fig. 3f), suggesting, as described in children with EED20, that the carnitine shuttle may be impaired in intestinal enterocytes in our model.
Flow cytometry of immune cell populations in the intestinal lamina propria
We performed flow cytometry to quantify immune cells in intestinal tissues. The frequency of neutrophils was significantly increased in the duodenum of P37 cSI-I offspring, but not in the ileum or colon (Fig. 1i). CD3+ and CD4+ cells were also higher in the duodenum, ileum and colon of cSI-I animals compared to age-matched cSI-N or CONV-D mice (Extended Data Fig. 3g,h). Among CD4+ cells, the Th17 population was significantly higher in the duodenum, ileum and colon of cSI-I offspring (Fig. 1j), whereas Th1 cells were diminished in their duodenum and Tregs were diminished in their duodenum and colon (Supplementary Table 2e); these findings lead us to speculate that a pro-inflammatory cytokine milieu in the intestines of P37 offspring of cSI-I dams promotes Th17 cell differentiation and proliferation.
These neutrophil and T cell phenotypes were also evident in dams: the frequencies of duodenal neutrophils, CD3+ cells, CD4+ T cells and Th17 cells were all significantly elevated in cSI-I dams. Th1 cells and Tregs were similarly significantly reduced in cSI-I compared to cSI-N mothers (P<0.05 for all comparisons, Wilcoxon rank-sum test; n=5 mice/group; Supplementary Table 2e). The observed increases in CD3+ cells and CD4+ T cells in the intestinal tissues of dams and their offspring harboring cSI-I compared to cSI-N consortia are consistent with prior characterization of EED in children as a T-cell mediated enteropathy13,24. Together, these findings set the stage for understanding the epithelial response at a cellular level in P37 offspring of dams harboring the cSI-I and cSI-N consortia.
Characterizing gene expression in the intestine at a tissue and cellular level
Single nucleus RNA-sequencing (snRNA-Seq)
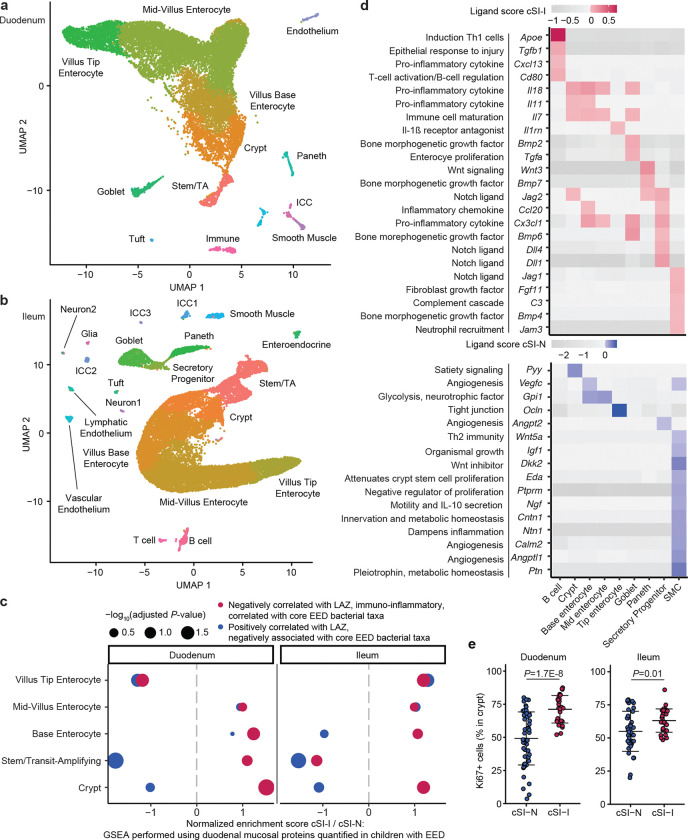
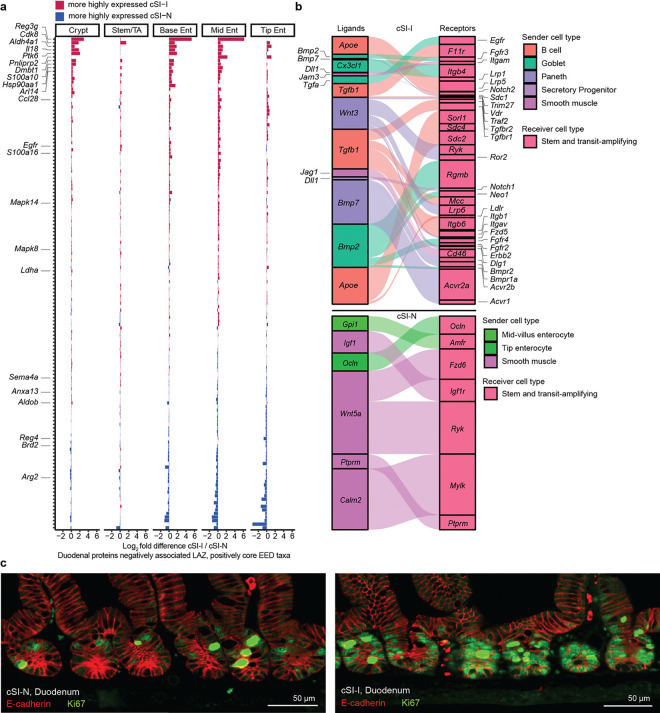
To characterize the responses of different intestinal epithelial cell types to the EED donor-derived bacterial consortia, we performed snRNA-Seq on intact segments of frozen duodenum and ileum harvested from P37 cSI-I and cSI-N mice. A total of 15,135 duodenal nuclei (Fig. 2a) and 20,584 ileal nuclei (Fig. 2b) collected from 3 littermates/group passed our quality metrics (see Methods). Analysis of marker gene expression identified nuclei from epithelial lineages (stem and transit amplifying cells and their enterocyte, goblet, Paneth, enteroendocrine and tuft cell descendants), as well as from mesenchymal lineages (endothelial, smooth muscle, neuronal, immune and interstitial cells of Cajal). Enterocytes comprised the largest proportion of nuclei (Supplementary Table 3b). A significantly higher proportion of Paneth cells were present in the duodenum of cSI-I offspring than their cSI-N counterparts (Supplementary Table 3b). Increased density25 of Paneth cells as well as degranulation26 have both been reported in Zambian children with EED.
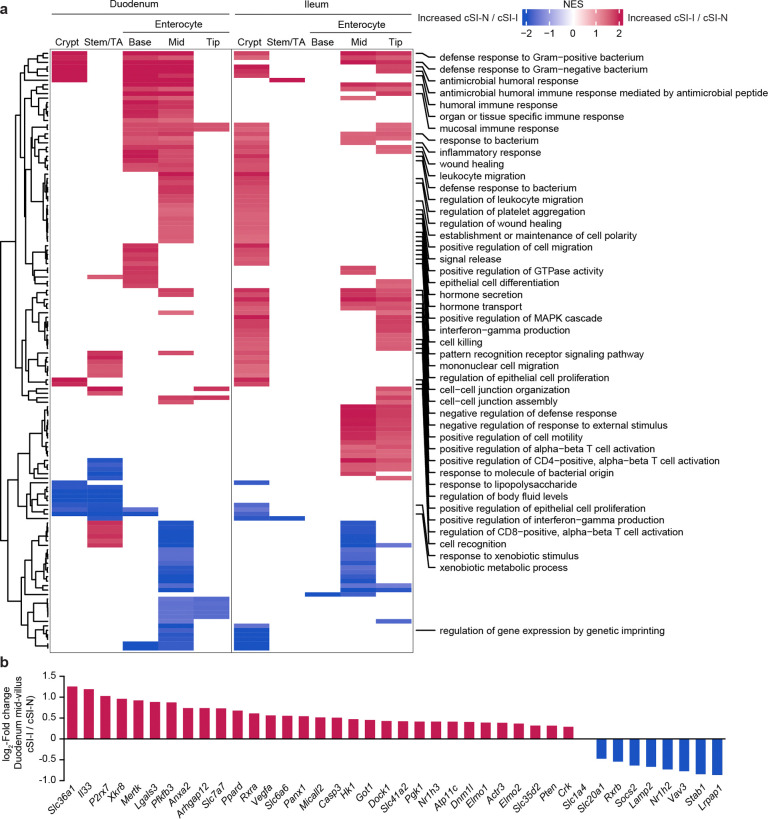
Fig. 2. Increased proliferative signaling in the small intestine of P37 offspring of cSI-I dams.
a,b, Uniform manifold and projection (UMAP) plot of single nuclei isolated from the duodenum (a), and ileum (b), of P37 offspring (n=3 mice/group). c, Gene set enrichment analysis (GSEA) along the crypt-villus axis, focusing on genes encoding duodenal mucosal proteins whose levels were quantified with the aptamer-based proteomics platform SOMAscan in children with EED in the BEED study. d, Intercellular signaling from cell populations (columns) to stem/transit amplifying (TA) cells in the ileum. A subset of ligands identified by NicheNet are shown that were significantly differentially expressed between cSI-I and cSI-N animals. Annotations included (left) are for contextualization and are based on literature findings. Their paired receptors are shown in Extended Data Fig. 5b. Ligands that were more highly expressed in cSI-I animals are shown in red (top); those more highly expressed in cSI-N animals are shown in blue (bottom). e, Percent of all cells in a crypt that were Ki67+ in the duodenum (left) or ileum (right) (n=3–5 mice/group; each dot represents a single crypt-villus unit; 10 crypts were analyzed per intestinal segment per mouse). P-values were determined by Tukey’s post-hoc tests; mean values ± s.d. are shown.
‘Pseudo-bulk’ analysis of differential gene expression in enterocytes positioned at different locations along the crypt-villus axis in both the duodenum and ileum revealed an enhanced epithelial immune response in P37 cSI-I compared to cSI-N mice (see enterocyte subpopulations in Extended Data Fig. 4a; Supplementary Table 3c). Of the GO categories that were significantly enriched in at least two subpopulations of enterocytes in cSI-I offspring, many were involved in bacterial recognition and defense, as well as recruitment/migration and activation of immune cells (Extended Data Fig. 4a). In addition, epithelial homeostasis requires a balance of differentiation, programmed cell death, and clearance of dying cells and cellular debris. This process, termed ‘efferocytosis’, involves recognition and clearance of dying cells by phagocytes, including both professional phagocytes and healthy neighboring intestinal epithelial cells27–29. Efferocytosis can be viewed as a process that sits at the interface between regulation of gut epithelial renewal, inflammation and the microbiota30. Based on published literature31–33, we manually curated an ‘efferocytosis’ gene set composed of 273 genes. This gene set was significantly enriched in cSI-I mid-villus duodenal enterocytes (P=0.019, Extended Data Fig. 4b). Genes enriched in cSI-I duodenal enterocytes included caspases linked to apoptosis (Casp3), genes linked to phosphatidylserine exposure during apoptosis (Xkr8, ATP11c), a channel that regulates apoptotic cell communication with the microbiota (Panx1), several efferocytotic receptors/recognition mediators (Mertk, Stab1, Mfge8), as well as cytosolic proteins linked to intracellular signaling during efferocytosis (Elmo1, Elmo2, Dock1, Crk). These findings suggest that apoptosis and subsequent efferocytosis within the small intestine play a role in shaping the epithelial response to cSI-I and cSI-N consortia and that this response involves increased turnover with associated increases in crypt depth (and as defined below by alterations in intercellular signaling involving crypt epithelial progenitor cells).
Contextualizing intestinal transcriptional responses with proteomic features in duodenal mucosa of children with EED
We next sought to understand whether the transcriptional response in enterocytes present in the duodenum and ileum of P37 offspring recapitulated the pathophysiology of the upper gastrointestinal tract of children with EED, using proteins quantified in duodenal biopsies obtained from children in the BEED study2. We performed GSEA with mouse homologs of duodenal mucosal proteins that were: (i) negatively correlated with length-for-age-Z-score (LAZ) and positively correlated with core EED bacterial taxa in BEED study participants, or (ii) positively correlated with LAZ in these children and negatively associated with core EED taxa (Fig. 2c). Expression of mouse genes homologous to the human duodenal proteins that were negatively associated with growth and positively correlated with the absolute abundances of core taxa of the EED microbiota were enriched in cSI-I enterocytes in the duodenum (crypt through mid-villus) and in the ileum (crypt and along the length of the villus) (Fig. 2c). Antimicrobial peptides Reg3β and Reg3γ, pro-inflammatory cytokine Il18, heat-shock protein Hsp90aa1, the immune cell chemoattractant Ccl28 were more highly expressed in all duodenal enterocyte cell types whereas type-2 immune response regulator Arg-2 was more highly expressed in cSI-N offspring (Extended Data Fig. 5a). Together, these results provide evidence that enterocytes play an important role in mediating the immune response in offspring of dams colonized with the cSI-I consortium.
Proliferative signaling to stem and transit-amplifying cells
To further interrogate the effects of the SI consortia on epithelial regeneration, we used the algorithm NicheNet34,35 to characterize intercellular signaling in cSI-I versus cSI-N P37 offspring. NicheNet surveys expression of known protein ligands by user-designated ‘sender’ cell types and their cognate receptors within a ‘receiver’ population. For our analysis, we defined stem and transit amplifying (stem/TA) cells as ‘receivers’ and all other cell types within the SI as ‘senders’ (Fig. 2d). Bone morphogenetic protein (Bmp), Wnt and Notch signaling are three of the major pathways that mediate cellular proliferation during normal intestinal development, responses to injury and hyperplastic inflammatory responses36,37. In the ileum, ligands more highly expressed in cSI-I compared to cSI-N mice included Wnt3 in Paneth cells (and their receptors Lrp6, Ryk and Bmpr1a in TA/stem cells, Extended Data Fig. 5b), as well as Paneth cell Bmp7 (and its Bmpr1a and Bmpr2 receptors in stem/TA cells). Bmp6 and Bmp2, two other ligands for Bmpr1a and Bmpr2 receptors, were also expressed at higher levels in ileal cSI-I goblet cells (Fig. 2d, Extended Data Fig. 5b). Expression of Wnt5a and the Wnt inhibitor Dkk2 were significantly diminished in ileal smooth muscle cells (SMCs) in cSI-I mice, as were their receptors in stem/TA cells (Cftr and Ptprk in the case of Wnt5a and Lrp6 in the case of Dkk2) (Fig. 2d, Extended Data Fig. 5b). SMC-derived Wnt5a augments Th2 immunity and inhibits cellular proliferation by downregulating expression of Ctnnb1 (ß-catenin)38. Quantification of Ki67, a marker of cellular proliferation, in duodenal and ileal crypts provided additional evidence of increased epithelial regeneration in cSI-I animals (Fig. 2e, Extended Data Fig. 5c; Supplementary Table 2d).
Inflammatory signaling in the cerebral cortex
Meningeal immune compartment
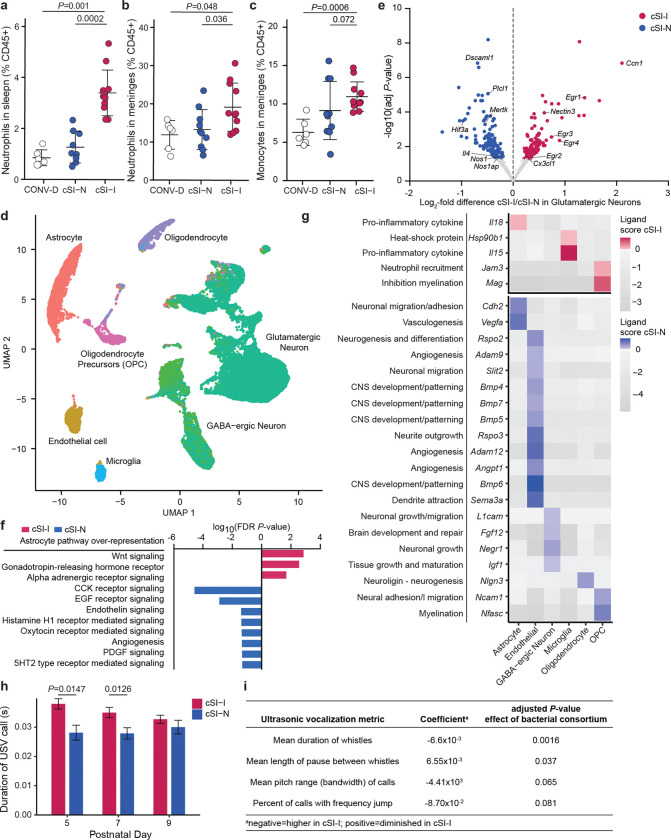
In addition to assessing changes in the representation of immune cells in the intestinal lamina propria, we Meningeal immune characterized immune cells in the brain meninges of P37 offspring of dams harboring the cSI-I or cSI-N consortia compared to CONV-D controls. The meninges harbor a broad repertoire of immune cells involved in immune surveillance of the central nervous system. Neutrophils were significantly elevated in the spleen and meninges of cSI-I mice (Fig. 3a,b; Supplementary Table 2e). Neutrophilic infiltration from the meninges into the brain parenchyma disrupts the blood-brain barrier in mice39 and causes neuronal damage as well as demyelination40. Additionally, inflammatory monocytes were elevated in the meninges of cSI-I offspring (Fig. 3c), whereas macrophages, typically the most abundant innate immune population in the meninges of healthy conventionally raised mice41, were significantly lower (P=0.002, Wilcoxon rank-sum test) (Supplementary Table 2e).
Fig. 3. Increased meningeal immune cells and pro-inflammatory signaling in the cortex of P37 offspring of cSI-I dams.
a,b, Frequency of neutrophils in the spleen (a), and brain meninges (b), of P37 offspring of cSI-I, cSI-N, or CONV-D dams. c, Frequency of inflammatory monocytes in the meninges of P37 offspring of cSI-I, cSI-N, or CONV-D dams. For a-c, n=6–11 mice/group combined across two litters, P-values were determined by Wilcoxon rank-sum tests, mean values ± s.d. are shown. d, UMAP of nuclei isolated from the cerebral cortex of cSI-I or cSI-N animals (n=4 mice/treatment group). e, ‘Pseudo-bulk’ differential expression analysis of glutamatergic neurons (n=23,827 nuclei). Genes that were significantly more highly expressed in cSI-I glutamatergic neurons are shown positively in red; genes more highly expressed in cSI-N are shown in blue (DESeq adjusted P-value cut-off < 0.05, Wald test). Genes of interest discussed in the main text are labeled. f, Gene set over-representation analysis of genes differentially expressed across pseudotime in astrocytes present in the cerebral cortex of P37 cSI-I and cSI-N mice (P-adj<0.05, TradeSeq conditionTest, n=4 mice/group). g, NicheNet analysis showing ligands expressed by sender cell populations (columns) to glutamatergic neurons. Ligands significantly more highly expressed in cSI-I mice are shown in the upper portion of the panel while ligands significantly diminished in cSI-I compared to cSI-N animals are displayed in the lower portion. Annotations included (left) for contextualization are based on literature findings. Corresponding receptors are shown in Extended Data Figs. 6c,d. h, Duration of ultrasonic vocalization calls emitted by P5-P9 offspring of dams colonized with the cSI-I or cSI-N consortia (bars denote mean ± s.d., Tukey’s post-hoc test). i, Metrics of USVs quantified in P5-P9 pups. For h and i, n=5–6 dams/group, n=3–6 pups/litter; statistics shown are the results of a linear model accounting for dam, litter size and collection timepoint.
snRNA-Seq of the cerebral cortex
Given the marked increase in neutrophils in the meninges of P37 cSI-I mice and the reported connections between the meningeal immune compartment, cytokine production and brain function41,42, we generated snRNA-Seq datasets from the cerebral cortices of P37 cSI-I and cSI-N offspring. In total, 44,873 nuclei (n=4 mice/group) passed our quality control metrics (see Methods, Fig. 3d). ‘Pseudo-bulk’ analysis disclosed 262 genes that were significantly differentially expressed in glutamatergic neurons, the most abundant cell type identified (Fig. 3d; Supplementary Table 3d). Of the 10 GO categories significantly diminished in cSI-I glutamatergic neurons, eight were related to synapse formation (GO:0098797, 0099055, 0098936, 0098936, 0098984, 00059111, 0014069, 0032279; q<0.05, GSEA; Supplementary Table 3d). Genes within these GO terms included (i) Nos1 and Nos1ap (neuronal nitric oxide synthase and Nos1 adaptor protein) which are involved in control of neuronal function and vascular homeostasis43, (ii) Gfra1, the receptor for glial cell line-derived neurotrophic factor (GDNF) and (iii) Htr2a (5-hydroxytryptamine receptor 2A). Expression of Il4 (Interleukin-4), an anti-inflammatory cytokine involved in memory formation44,45 and modulation of microglial pathways during brain injury46, was significantly reduced in cortical cSI-I glutamatergic neurons (Fig. 3e). Consistent with this increase in expression of pro-inflammatory genes by glutamatergic neurons, the neutrophil chemoattractant CXCL1 was elevated in protein extracted from frozen cortex tissue of P37 cSI-I mice (Extended Data Fig. 6a; Supplementary Table 2a).
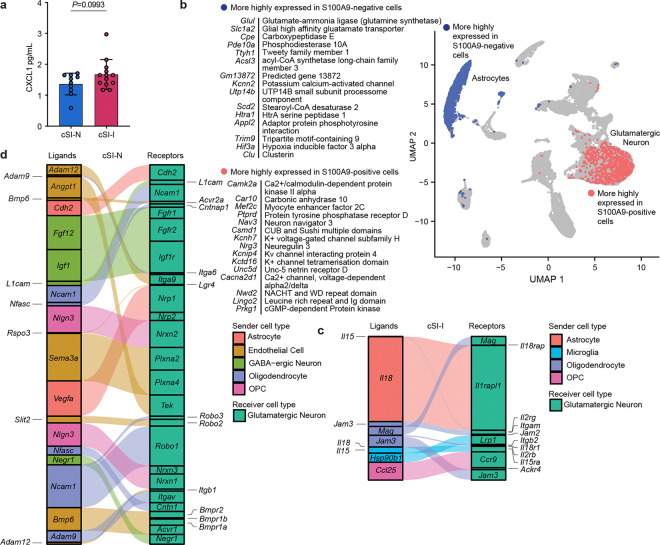
We followed up these findings by identifying nuclei in the cortex whose expression was correlated with systemic inflammation. The algorithm Scissor47 calculates the correlation in gene expression between each nucleus in a snRNA-Seq dataset and expression in a set of bulk RNA-Seq samples (the latter allows more animals to be characterized than the limited number for which snRNA-Seq can be feasibly applied; n=16–18/group for cSI-I, cSI-N and CONV-D P37 offspring). The resulting correlation matrix is used as the basis for a gaussian regression model with phenotypic information obtained from the same mice. This approach allowed us to identify nuclei in the cerebral cortex most strongly correlated, either positively or negatively, with systemic inflammation, as defined by serum levels of S100A9 in P37 offspring, including CONV-D animals (Fig. 1e). Nuclei positively associated with S100A9 were primarily glutamatergic neurons (99.8%), while negatively associated nuclei were mostly astrocytes (98.8%, Extended Data Fig. 6b). We compared gene expression of S100A9-correlated nuclei against the remaining nuclei within the same cell type (Scissor-selected nuclei versus ‘background’ nuclei). Compared to ‘background’ glutamatergic neurons that were not associated with S100A9, those positively correlated with S100A9 exhibited significantly lower expression of serotonin and dopamine receptors (Extended Data Fig. 6b; Supplementary Table 3d).
These results interrogating cortex gene expression with a serum biomarker of inflammation indicated that astrocyte expression profiles were anti-correlated with cSI-I-induced inflammation in the cerebral cortex. Follow-up trajectory inference analysis of astrocyte nuclei from the snRNA-Seq dataset identified several pathways whose expression was significantly diminished in cSI-I compared to cSI-N astrocytes across pseudotime, including angiogenesis (Fig. 3f). Glial cells and neurons communicate bi-directionally48 and in functional units with endothelial cells. Brain endothelial cells form a key component of the blood-brain barrier, interfacing with astrocyte end-feet and relaying systemic signals to the brain parenchyma49. Therefore, we further investigated the degree to which glial-neuronal and endothelial-neuronal signaling were altered in the offspring of cSI-I-colonized dams compared to those from cSI-N dams.
We used NicheNet to characterize the expression of ligands and their receptors in the cerebral cortex, designating astrocytes, oligodendrocytes and their precursors, microglia, endothelial cells and GABA-ergic neurons as ‘senders’ and glutamatergic neurons as the ‘receiver’ population. cSI-I oligodendrocytes expressed higher levels of myelin-associated glycoprotein (Mag) – an inhibitor of axonal growth and myelination50 (Fig. 3g). Additionally, several pro-inflammatory cytokines were significantly more highly expressed in the cerebral cortex of P37 cSI-I versus cSI-N mice; they include the chemokine Ccl25 in oligodendrocyte precursors, Il18 in astrocytes and Il15 in microglia (Fig. 3g). Il18 reduces long-term potentiation51 and may contribute to sickness-induced anorexia, while Il15 modulates T and B cells and plays a key role in regulating inflammatory events in the central nervous system52 (Fig. 3g, Extended Data Fig. 6c).
Ligands whose expression was diminished in the cortex of cSI-I animals included neural cell adhesion molecule (Ncam1, expressed primarily by oligodendrocyte precursors) which plays a role in synaptic plasticity, learning and memory as well as neurofascin (Nfasc, from oligodendrocytes), which is involved in neurite outgrowth, axonal guidance and myelination (Fig. 3g, Extended Data Fig. 6d). A series of anti-inflammatory ligands involved in angiogenesis and vascular barrier function were significantly diminished in the cortex of cSI-I offspring, including: (i) vascular endothelial growth factor (Vegfa, astrocytes), (ii) angiopoietin 1 (Angpt1, endothelial cells), an anti-inflammatory growth factor involved in neural proliferation, vascular development and protection53, (iii) Adam9/12 (endothelial cells), which has been reported to drive Vegf-mediated angiogenesis in tumor models54,55, and (iv) bone morphogenetic proteins (Bmp4/5/6/7), which, in the brain, promote angiogenesis and mitigate shear and oxidative stress56 (Fig. 3g, Extended Data Fig. 6d). Together, these results suggest that transmission of components of the EED donor-derived small intestinal bacterial consortia from dams to their offspring influences physiology beyond the gut within the CNS, manifesting as perturbations to glial-neuronal and endothelial-neuronal signaling.
Behavioral phenotype associated with cSI-I consortium
We subsequently compared emittance of ultrasonic vocalization (USV) calls when cSI-I and cSI-N P5-P9 pups were separated from their dams. USVs reflect the cognitive and social state of the animal57,58. During the first two weeks of life, thermal regulation in mice is limited and pups rely on the dam for warmth. USVs are emitted by pups to signal the dam for retrieval when removed from the nest and are the earliest form of social communication in rodents59,60. We adapted USV assays and associated equipment for implementation within a gnotobiotic isolator (see Methods). Pups of dams colonized with the cSI-I consortium emitted significantly longer calls with shorter pauses between them (Fig. 3h,i, Supplementary Table 2f) than cSI-N pups of the same age (total of 19–30 pups were assayed at each postnatal timepoint within each colonization group, representing 5–6 litters/group). Together, these results are consistent with increased distress and perturbed social communication in the neonatal offspring of dams harboring the cSI-I consortium.
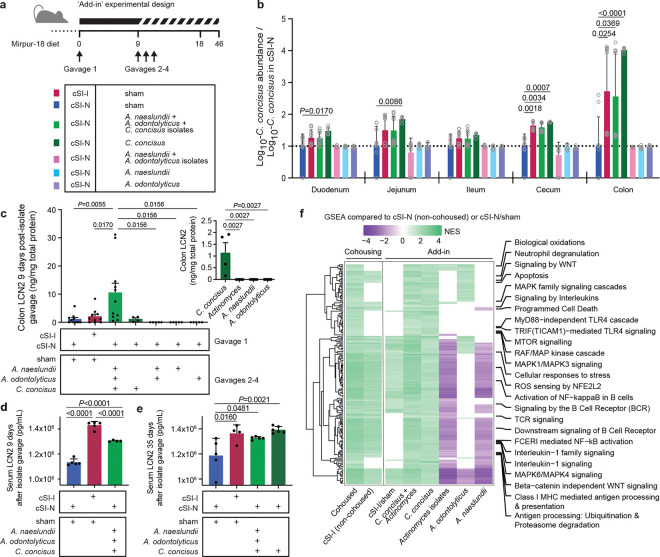
Cohousing to nominate pathology-inducing bacterial strains
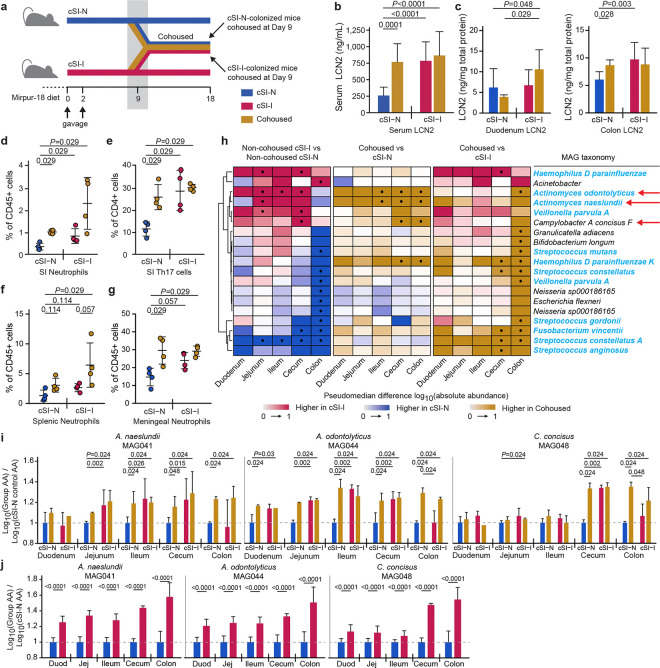
To identify bacterial taxa in the two consortia that either confer or ameliorate the immunoinflammatory state described above, we gavaged separate groups of 4–5-week-old germ-free C57Bl/6J mice consuming the Mirpur-18 diet with the cSI-I or cSI-N culture collections. After nine days, half of the animals from each group were cohoused. This experimental group and the non-cohoused cSI-I and cSI-N control groups were then followed for an additional nine days (n=6 or 8 mice/group/experiment, 2 independent experiments; Fig. 4a). Cohoused mice and non-cohoused cSI-I controls gained significantly less weight compared to non-cohoused cSI-N controls (linear mixed effects model; Supplementary Table 2b). Levels of LCN2 were higher in colonic tissue and trended higher in duodenal tissue of cohoused mice compared to non-cohoused cSI-N controls (Fig. 4b,c; Supplementary Table 2a). Irrespective of the initial colonization state, serum concentrations of LCN2 were significantly higher in cohoused animals compared to non-cohoused cSI-N controls, and did not differ significantly from levels in non-cohoused cSI-I animals. The serum level of CHI3L1, another immunoinflammatory protein characterized in the BEED study and in our intergenerational transmission model, exhibited a pattern comparable to that observed for LCN2 (Supplementary Table 2a).
Fig. 4. Identification of bacteria that transfer from cSI-I to cSI-N mice in cohousing experiments and induce enteropathy.
a, Experimental design of cohousing experiment. b, Serum levels of LCN2 protein on experimental day 18, 9 days after initiation of cohousing (n=14 mice/control group, n=7 mice/cohoused group). c, Intestinal tissue levels LCN2 measured on experimental day 18, 9 days after cohousing (n=8 mice/control group, n=4 mice/cohoused group). d,e, Frequency of neutrophils (d), and Th17 cells (e), in the small intestinal lamina propria (n=4 mice/group; duodenal, jejunal and ileal segments were combined prior to analysis). f,g, Frequency of neutrophils in the spleen (f), and meninges (g), (n=4 mice/group). For panels d-f, each point represents an individual animal; the color code matches that used in panel a. h, Differences in absolute abundances of MAGs along the length of the gut (n=6 mice/group). MAGs were included if they demonstrated statistically significant differences in their absolute abundance for any comparison between groups in at least one segment of the intestine. MAG taxonomy highlighted in blue indicates correspondence to ASVs representing ‘core taxa’ in the small intestinal microbiota of Bangladeshi children with EED in the BEED study. The three red arrows point to MAGs defined as ‘pathology-associated’. • P-adj < 0.05. i,j, Log10 absolute abundances of the pathology-associated MAGs (MAG041, MAG044 and MAG048) along the length of the intestine in either the cohousing experiment (i) or P37 animals from the intergenerational experiment (j). Values are expressed relative to P37 cSI-N offspring in the intergenerational transmission experiment (n=22–25 mice/group) or to cSI-N non-cohoused controls (n=6 mice/control group and n=3/cohoused groups). For b-j, P-values were determined by Wilcoxon rank-sum test; mean values ± s.d. are shown in b-g, i-j.
Analogous to P37 offspring from the intergenerational transmission experiments, non-cohoused cSI-I mice and all cohoused animals had significantly increased proportions of neutrophils and Th17 cells and a reduced percentage of Th1 cells in the small intestine lamina propria compared to non-cohoused cSI-N controls; this was the case regardless of the initial colonizing bacterial consortium (Fig. 4d,e; Supplementary Table 2e). Splenic and meningeal neutrophils were similarly elevated in cSI-I and cohoused animals (Fig. 4f,g; Supplementary Table 2e) compared to non-cohoused cSI-N mice. Taken together, these data suggest that transfer of bacteria from cSI-I to cSI-N animals induces pathology in these formally healthy animals; reciprocally, there was no evidence for a mitigating effect of the cSI-N community on pathology in cSI-I colonized mice.
Pathology-associated MAGs
We performed shotgun sequencing of intestinal contents from animals in all treatment groups, seeking taxa (MAGs) that were significantly more abundant in the intestines of mice in the ‘inflammatory’ experimental groups (i.e., non-cohoused cSI-I controls and cohoused mice) compared to their ‘non-inflammatory’ counterparts (non-cohoused cSI-N mice). Three MAGs satisfied these criteria: two belonged to the genus Actinomyces (Actinomyces naeslundii [MAG041], Actinomyces odontolyticus [MAG044]) and the other to the genus Campylobacter (Campylobacter concisus [MAG048]) (Fig. 4h; Supplementary Table 1d).
Compared to cSI-N controls, the absolute abundance of C. concisus was significantly higher in the cecum and colon of cohoused animals, regardless of whether the cohoused animals were initially colonized with the SI-N or cSI-I consortia (Fig. 4i). The absolute abundances of both A. naeslundii and A. odontolyticus were significantly higher in the jejunum, ileum, cecum and colon of cohoused animals compared to non-cohoused cSI-N controls (Fig. 4i). The abundances of all three MAGs were equivalent in cohoused animals and non-cohoused cSI-I controls along the length of the small intestine and in the cecum (Fig. 4i), suggesting infiltration of these taxa from cSI-I to cSI-N animals during cohousing. Moreover, the absolute abundances of these MAGs were significantly increased along the length of the gut of P37 cSI-I compared to cSI-N offspring in our intergenerational transmission model (Fig. 4j; Supplementary Table 1d).
Four isolates in our clonally-arrayed culture collection matched these three MAGs that were elevated in cohoused animals compared to cSI-N controls with an average nucleotide identity (ANI) score of >99%: two isolates matched the A. naeslundii MAG041 (Bg041a, Bg041b), one matched A. odontolyticus MAG044 (Bg044) and one matched C. concisus MAG048 (Bg048) (Supplementary Table 4a). C. concisus and Actinomyces spp. are resident members of the oral microbiota61 and are not typically considered to be pathogens, although some strains have been associated with periodontitis and inflammatory bowel disease62–64.
We generated in silico metabolic reconstructions for these pathology-associated MAGs, their corresponding isolates, and other phylogenetically-related species using a collection of reference human gut genomes integrated and curated in our microbial community SEED (mcSEED) bioinformatics platform (see Methods), inferring the presence and absence of 106 metabolic pathways/phenotypes involved in carbohydrate utilization, amino acid and B-vitamin biosynthesis and intermediary metabolism (Supplementary Table 4b)65. All analyzed C. concisus strains have a set of predicted metabolic phenotypes (either present or absent) that are distinct from pathogenic members of Campylobacter jejuni and Campylobacter coli, including pathways related to the biosynthesis of vitamins B6 and B7, L-lactate production and D-lactate utilization, utilization of fucose, proline, serine and alanine, as well as tetrathionate and nitrite reduction (Supplementary Table 4). Neither our C. concisus isolate Bg048 nor the other reported C. concisus genomes possess any of 67 pathways for utilization of oligo- and monosaccharides, except for glycerate (Supplementary Table 4b), in line with previous characterization of this species as non-saccharolytic and its preference for other simple carbon sources (e.g., fumarate, glutamate) and hydrogen gas for growth66. A. naeslundii and A. odontolyticus harbor genes that provide a broader capacity for carbohydrate degradation than C. concisus. Our A. naeslundii and A. odontolyticus MAGs and their corresponding isolates have similar predicted metabolic capability as phylogenetically related Actinomyces species we analyzed (Supplementary Table 4b).
Recently, a comparison of 190 reported C. concisus isolate genomes resulted in their classification into two ‘genomospecies’ (GS) groups, designated GS1 and GS267. The authors observed that genes distinguishing the two groups largely encode hypothetical proteins, obscuring clear functional distinctions between GS1 and GS2 members. Based on a phylogenetic analysis of nearest neighbors, we assigned our Bangladeshi C. concisus strain (Bg048) to group GS2, which has been identified more frequently within the gastrointestinal tract than GS168. Searches of the C. concisus Bg048 isolate and corresponding MAG048 genomes failed to identify known or putative virulence factors, including Zot (Zonula occludens toxin) which has been found in 30% of C. concisus strains69 and reported to disrupt intestinal epithelial tight junctions as well as augment IL-8 production in the human intestinal adenocarcinoma-derived enterocyte-like cell line HT2970.
Testing the role of isolates corresponding to pathology-associated MAGs
We directly tested the capacity of our C. concisus, A. odontolyticus and A. naeslundii isolates to induce pathology. To do so, germ-free 4–5-week-old C57Bl/6J mice, consuming the Mirpur-18 diet, were gavaged with the cSI-N consortium (experimental day 0). On days 9 through 11, mice were gavaged once daily with either (i) a pool of the three strains (isolates Bg041a, Bg041b, Bg044, Bg048), (ii) C. concisus alone (Bg048), (iii) A. naeslundii and A. odontolyticus together (Bg041a, Bg041b, Bg044), (iv) A. naeslundii alone (isolates Bg041a, Bg041b), or (iv) A. odontolyticus alone (Bg044). Groups of mice initially colonized with the cSI-N or cSI-I consortium were subsequently gavaged with uninoculated (blank) culture medium as negative and positive controls, respectively (‘sham’ gavage, Fig. 5a).
Fig. 5. Direct test of candidate pathology-inducing small intestinal bacterial strains in cSI-N mice.
a, Experimental design of ‘add-in’ experiment. Individual cultured isolates, alone or in combinations, were gavaged once daily on days 9–11 into mice previously colonized with the cSI-N bacterial consortium. b, Ratio of the absolute abundance of C. concisus strain Bg048 (corresponding to MAG048) shown relative to its absolute abundance in cSI-N/sham controls (a ratio of 1 is indicated by the dotted line; two-way ANOVA with Dunnett’s multiple comparisons to cSI-N/sham control; mean values ± s.d. are shown). Colors correspond to the groups shown in panel a. c, Levels of LCN2 protein in colonic tissue measured on experimental day 18, 9 days after isolate gavage. The inset shows colonic LCN2 levels after addition of the individual isolates alone (one-way ANOVAs with Tukey’s multiple comparisons; mean values ± s.e.m. are shown). d,e, Serum LCN2 at 9 (c), or 35 days (d), following secondary gavage (mean ± s.d. shown, one-way ANOVAs with Tukey’s multiple comparisons). f, Normalized enrichment scores (NES) for all Reactome pathways that were significantly enriched in the colon (GSEA q-value <0.05) of the indicated treatment groups compared to their respective cSI-N counterparts (non-cohoused cSI-N controls, or cSI-N/sham). Pathways related to the immune system are labeled. Pathways were included in the heatmap if they were significantly enriched with the addition of C. concisus and in cohoused animals. If a pathway was not significantly enriched, it was assigned a NES score of zero. For panels b, c, f, n=5–11 mice/group combined across two independent experiments. For d and e, n=5 mice/group in a third independent experiment.
Secondary gavage with the three strains combined or with C. concisus alone led to an increase in the absolute abundance of C. concisus, most notably in the cecum and colon, compared to sham-gavaged cSI-N controls (Fig. 5b). The levels of C. concisus achieved over the course of the 9 days following its introduction into cSI-N mice were comparable to levels in cSI-I sham-gavaged controls (Fig. 5b) as well as in cohoused animals (Supplementary Table 1d). In contrast, gavage with the Actinomyces species either alone or in concert did not result in a statistically significant increase in their absolute abundance in any location along the length of the intestinal tract compared to cSI-N sham-gavaged controls (Extended Data Fig. 7a,b; Supplementary Table 1d).
Colonic tissue levels of LCN2 were significantly increased by gavage with the pool of the three strains compared to all other groups, including the cSI-I positive controls (Fig. 5c). This increase did not occur in the duodenum (Extended Data Fig. 7c). Serum LCN2 levels were significantly higher 9 days after gavage of the three pooled strains than in cSI-N controls but remained lower than in cSI-I controls (Fig. 5d). In a repeat experiment, mice were characterized 35 days after gavage, and introduction of either the three isolates or C. concisus alone induced LCN2 levels that were significantly higher than cSI-N controls and not significantly different from cSI-I sham-gavaged animals (Fig. 5e; Supplementary Table 2a).
We conducted bulk RNA-Seq of the duodenum and colon to further understand the host response to the presence of C. concisus. Bulk intestinal tissue RNA-Seq expression was compared across the three experimental models that we describe in this report: intergenerational dam-to-pup transmission of the cSI-I and cSI-N consortia (P37 offspring), cohousing, and isolate ‘add-in’ (the latter two characterized at experimental day 18) (Supplementary Table 3e). We examined immune-related Reactome pathways that were significantly enriched in (i) P37 cSI-I animals, (ii) cohoused and non-cohoused cSI-I controls, and (iii) cSI-N-colonized animals secondarily gavaged with C. concisus, A. odontolyticus, or A. naeslundii individually, both Actinomyces strains, a combination of all three strains together, as well as cSI-I/sham controls – all compared to their cSI-N counterparts (78 pathways shared between at least two pairwise comparisons, Supplementary Table 3e). It was apparent that C. concisus, either alone or with the Actinomyces species, induced a pro-inflammatory transcriptional response in the colon that was not present in the duodenum (Supplementary Table 3e). Compared to their cSI-N counterparts, Reactome pathways involved in recruitment and activation of immune cells, reactive oxygen species generation and detoxification, cell death programs, antigen presentation, MAPK signaling, pro-inflammatory cytokines, and toll-like receptor (TLR) signaling were more highly expressed in the colon of P37 cSI-I animals (from the intergenerational transmission experiment), as well as cohoused and non-cohoused cSI-I controls plus recipients of C. concisus alone or with Actinomyces (Fig. 5f; Supplementary Table 3e). Genes involved in cytokine (IL-1) signaling leading to programmed cell death (apoptosis) and immune cell activation (neutrophil degranulation) were induced in the colon following gavage of C. concisus alone or in concert with the Actinomyces isolates (Supplementary Table 3e). Of the 41 immunoinflammatory Reactome pathways that were significantly enriched in the colon of mice gavaged with the three pathology-associated MAGs, only five were enriched in the duodenum (Supplementary Table 3e), while none of the 49 Reactome pathways that were enriched in the colon after gavage of C. concisus alone were more highly expressed in the duodenum (Supplementary Table 3e). Moreover, the Actinomyces isolates, administered alone or together, did not elicit a robust immune response in either the colon or duodenum (Supplementary Table 3e). Together, these findings provide evidence that this C. concisus isolate, both alone and in concert with A. odontolyticus and A. naeslundii recovered from the duodenal microbiota of Bangladeshi children with EED, can induce an immunoinflammatory tissue gene response that recapitulates aspects of that induced by the EED donor-derived cSI-I community.
Host-derived substrates boost C. concisus growth in vitro
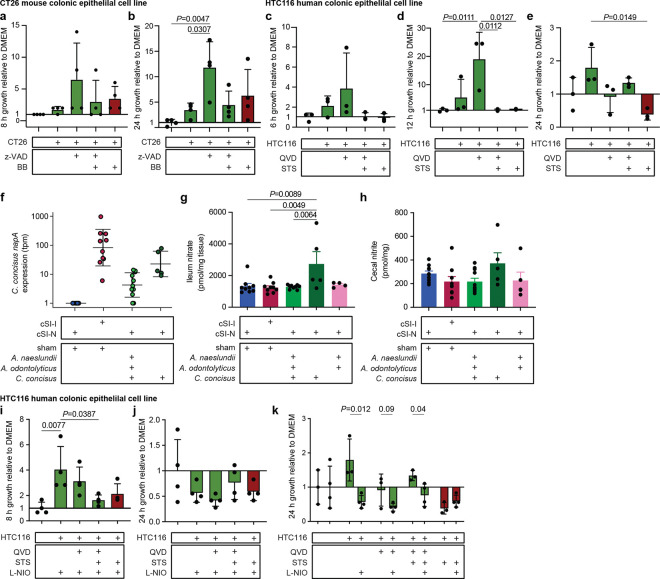
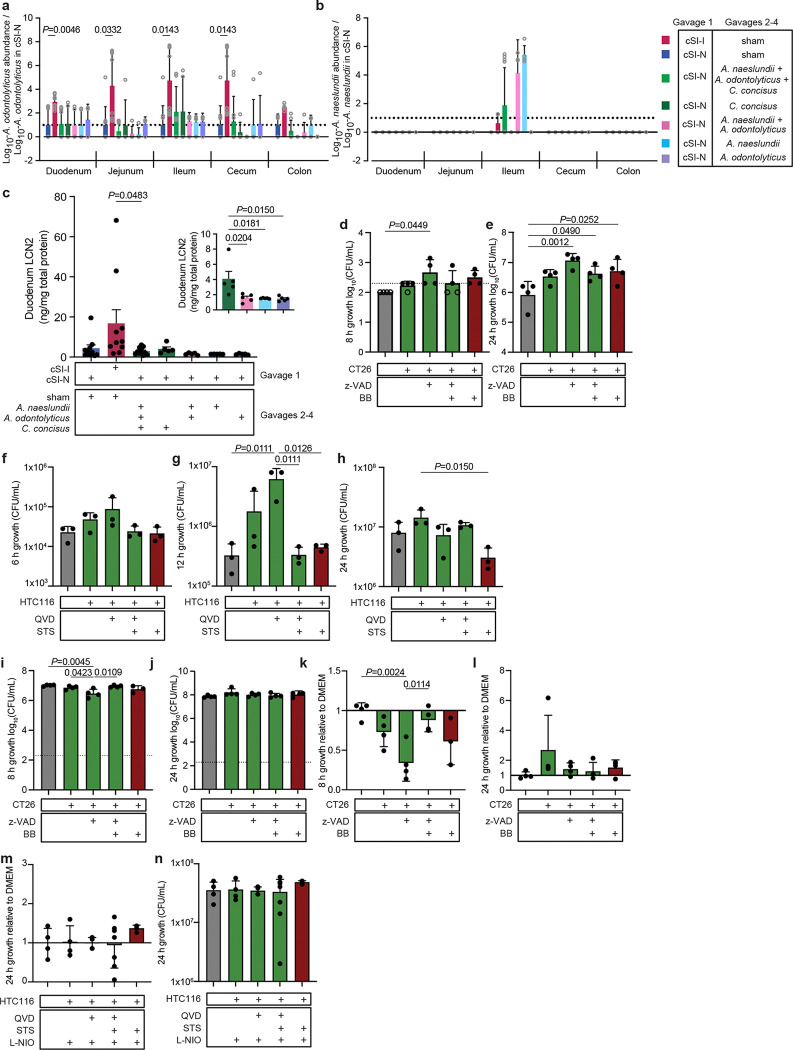
Having determined that in the context of the cSI-N community C. concisus induced an immunoinflammatory response, we investigated the hypothesis that C. concisus, as described for several enteropathogens, may utilize host substrates to improve its fitness under conditions where there is inflammation and perturbed intestinal epithelial turnover. To do so, we turned to an in vitro tissue culture system in which we cultured mouse- and human-derived colonic epithelial cell lines (CT26 and HTC116, respectively) with or without subsequent induction of cell death (see Methods) and then collected spent supernatants30. C. concisus growth in microaerophilic atmospheric conditions was significantly higher in spent supernatants from live mouse CT26 cells (Fig. 6a,b, Extended Data Fig. 7d,e) and human HTC116 cells (Fig. 6c–e, Extended Data Fig. 7f-h) compared to supernatants harvested from these cells when they were undergoing apoptosis or to cell culture medium alone. When cultured anaerobically, C. concisus growth did not differ between spent medium from cells that were live or undergoing apoptosis and was diminished compared to growth in cell culture medium alone (Extended Data Fig. 7i-l). These data suggest that changes in C. concisus metabolism in the presence of oxygen, such as those that may occur in the oxidative state of the inflamed gut71, may underlie a fitness benefit conferred to C. concisus by utilization of host-derived substrates.
Fig. 6. Host epithelial cell nitrate metabolism boosts C. concisus growth in vitro.
a,b, Growth of C. concisus after 8 a, or 24 hours b, in CT26 mouse colonic epithelial cell line supernatants that were live (green; CT26:FADD, z-VAD, z-VAD+BB) or were treated with an inducer of apoptosis (red; BB) compared to medium control (n=4 biological replicates per condition). c-e, Growth of C. concisus after 6 c,, 12 d,, or 24 e, hours in HTC116 human colonic epithelial cell line supernatants harvested from cells that were live (green; QVD, QVD+STS), or that had been treated with an inducer of apoptosis (STS; red), compared to cell culture medium alone control (n=3 biological replicates per condition). f, Expression of C. concisus nitrate reductase napA at 9 days post-isolate gavage, or in cSI-I and cSI-N sham-gavaged controls. g, Levels of nitrate in homogenized ileal tissue 9 days post-isolate gavage. h, Levels of nitrite in cecal contents 9 days post-isolate gavage. For (F-H), n=5–11 mice/group combined across two independent experiments. i,j, Growth of C. concisus after 8 (i) or 24 (j) hours of incubation in HTC116 human colonic epithelial cell line supernatants prepared from live cells (green; QVD, QVD+STS) or cells that had been treated with an inducer of apoptosis (red; STS). All cells were additionally treated with an inhibitor of nitric oxide synthase (L-NIO). Growth shown relative to cell culture medium alone (n=4 biological replicates per condition). k, Comparison of C. concisus growth after 24 hours in live (green; QVD, QVD+STS) or apoptotic (red; STS) HTC116 supernatants that were or were not treated with L-NIO (n=3–4 biological replicates per condition, pairwise t-tests for each ± L-NIO comparison). For all panels, bars denote mean ± s.d.. For a-e and g-k, P-values were determined with one-way ANOVA and Tukey’s multiple comparisons. FADD, Fas-associated death domain; z-VAD, z-VAD-FMK (pan-caspase inhibitor); QVD, Quinoline-Val-Asp-Difluorophenoxymethylketone (caspase inhibitor); STS, Staurosporine (apoptosis inducer); L-NIO, N5-(1-Iminoethyl)-L-ornithine, dihydrochloride (nitric oxide synthase inhibitor).
Given that canonical enteric pathogens benefit from metabolites released from dying rather than live cells30, we were intrigued that supernatants harvested from dying (apoptotic) cells did not boost C. concisus growth in vitro. To garner clues about potential host substrates that support C. concisus growth in vivo, we conducted RNA-sequencing of the cecal microbial community of animals from our ‘add-in’ experiment (Fig. 5a). Few C. concisus genes were defined as differentially expressed between groups, in part due to its low abundance in the cSI-N consortium. While C. concisus exhibited minimal differential gene expression in the context of the full cSI-I consortium (cSI-I/sham) compared to its introduction into the cSI-N community (cSI-N/C. concisus or cSI-N/C. concisus+Actinomyces); only one C. concisus transcript, encoding a nitrate reductase (napA3), was significantly differentially expressed between cSI-N mice secondarily gavaged with C. concisus alone compared to C. concisus plus the Actinomyces strains (cSI-N/C. concisus versus cSI-N/C. concisus+Actinomyces, Fig. 6f).
During an inflammatory response, nitric oxide produced by inducible nitric oxide synthase (iNOS, encoded by Nos2) reacts with reactive oxygen species to form nitrate72. Other Proteobacteria have been shown to utilize host-derived nitrate72–74 or other by-products of iNOS activity65 to gain a competitive advantage in mouse models75. Nitrate in ileal tissue was significantly higher with addition of C. concisus alone (cSI-N/C. concisus) compared to cSI-N and cSI-I sham-gavaged controls as well as when Actinomyces were present (cSI-N/C. concisus+Actinomyces, Fig. 6g). Nitrate reductase activity reduces nitrate to nitrite; increased bacterial nitrate reductase activity should result in elevated luminal nitrite. Nitrite in cecal contents trended higher in C. concisus-gavaged animals (Fig. 6h), although the difference was not statistically significant, perhaps due to microbial reduction of nitrite or the short half-life of nitric oxides. To test whether epithelial cell nitric oxide generation contributed to the increase in C. concisus growth in the presence of the live cell spent supernatants, we treated HTC116 cells with the non-selective NOS inhibitor N-iminoethyl-L-ornithine (L-NIO)76. L-NIO treatment significantly reduced the growth benefit conferred to C. concisus by supernatants collected from live cells when C. concisus was grown in microaerophilic conditions; in contrast, L-NIO had no effect on its growth under microaerophilic conditions when exposed to apoptotic cell supernatants (Fig. 6i–k). L-NIO treatment did not affect C. concisus growth in epithelial cell supernatants under anaerobic conditions (Extended Data Fig. 7m,n). Together, these results suggest that host nitrate metabolism boosts growth of C. concisus and may confer an advantage in oxygenic, inflammatory conditions in the gut.
Discussion
Here we describe a preclinical gnotobiotic mouse model of intergenerational EED that is mediated by members of the small intestinal microbiota. In the absence of samples from healthy children living in the same environment as those in our clinical study, we generated two bacterial consortia from aspirates collected from undernourished Bangladeshi children with EED: one encompassing all isolates recovered from the aspirates, which induced intestinal and systemic inflammation; the other a subset comprised of one representative strain of each species present in the full collection. This type of ‘reverse translation’ experiment provided an opportunity to perform analyses that were not possible in children in the BEED study who were the source of the small intestinal bacteria, e.g. immune profiling along the length of the gut and in extraintestinal tissues, snRNA-Seq analysis of gene expression in different cell lineages in different regions of the small intestine and brain, as well as a functional assessment of early postnatal behavior.
We were able to dissect structure/activity relationships in the human donor small intestinal bacterial consortia by combining results from this intergenerational transmission model with shorter duration cohousing experiments and isolate ‘add-in’ experiments in which candidate disease-promoting members were introduced into gnotobiotic animals colonized with the non-pathology-inducing cSI-N consortium. Our dam-to-pup transmission and cohousing gnotobiotic mouse models provided evidence associating C. concisus, A. naeslundii and A. odontolyticus – all members of the oral microbiota – with development of pathology. Disruption of the intestinal microbiota by taxa that normally reside in the oral microbiota has been implicated in studies of undernutrition and EED77,78. Evidence for this process, referred to as ‘decompartmentalization’ of the microbial community, has come from applying culture-independent methods (primarily16S rRNA amplicon sequencing) to feces. As such, there is little information about disease associations with specific strains, or correlations with their genome-encoded functional features. Intriguingly, C. concisus has been reported to be over-represented in the fecal microbiota of stunted compared to non-stunted children living in South India, and significantly more abundant in the feces of a cohort of stunted children living in sub-Saharan Africa compared to healthy controls79,80. An Actinomyces species was one of the 14 duodenal ‘core taxa’ we identified in the BEED study as significantly positively correlated with duodenal inflammation and negatively correlated with LAZ in children with EED2. Additionally, the abundance of Actinomyces in maternal and infant saliva was negatively associated with birth weight and correlated with impaired linear growth (stunting) in the multi-national AFRIBIOTA study79,81.
Isolate ‘add-in’ experiments provided direct evidence for C. concisus as a key mediator of pathology. C. concisus has been described as a typical member of the oral microbiota in healthy individuals (97% prevalence in a cohort of 59 individuals aged 3–80 years)61, but it has also been associated with gingival disease62. Furthermore, a relationship to inflammatory bowel disease (IBD) has been proposed; its prevalence was higher in mucosal biopsies from IBD patients compared to healthy controls62,63. However, the relationship between strains recovered in the oral and intestinal microbiota of individuals with IBD remain to be defined, as do the contributions of C. concisus to IBD pathogenesis. In tissue culture studies, C. concisus decreased expression of tight junction proteins and increased permeability to FITC-dextran70,82.
It is intriguing that addition of an EED donor-derived C. concisus strain to mice harboring the EED duodenal-derived cSI-N consortium of cultured bacteria produced an immunoinflammatory response that was greatest in the colon – at least within the relatively short duration of the cohousing/’add-in’ experiments. These findings prompt the question of what factors allow it (and other members of the oral microbiota) to establish themselves in distal regions of the intestine. Understanding how pathology-inducing members of the oral microbiota cross-feed with other community members in the intestine (i.e., their nutrient-sharing or syntrophic relationships) could have therapeutic implications. The extent to which host nitric oxide generation and potential utilization of host-derived substrates by pathogenic members of the microbiota contribute to decompartmentalization of the oral microbiota and development of enteropathy in humans is an important area of further investigation.
Undernutrition in mothers remains rampant in Bangladesh. Applying the approach described here to generate culture collections from mothers with and without EED will provide an opportunity to obtain from gnotobiotic mice (i) a more mechanistic understanding of the intricate interplay between maternal nutritional status, small intestinal adaptations to pregnancy, development both in utero and following birth and the expressed functions of community members within dams and their offspring and (ii) identify candidate therapeutic targets and agents.
METHODS
Mouse experiments
All experiments involving mice were performed using protocols approved by Washington University Animal Studies Committee. Mice were housed in plastic flexible film gnotobiotic isolators (Class Biologically Clean Ltd., Madison, WI) at 23 °C under a strict 12-hour light cycle (lights on at 0600h). Autoclaved paper ‘shepherd shacks’ were kept in each cage to facilitate the natural nesting behaviors and for environmental enrichment.
Source of cSI-I and cSI-N bacterial consortia
The methods used for culturing bacterial strains present in duodenal aspirates obtained from children enrolled in BEED study are described in a previous publication5. Strains were stored at −80 °C in PBS containing 15% glycerol (v/v). For isolates originally isolated under anaerobic conditions, a 20 μL aliquot of each stock was used to inoculate a well in a 1 mL deep-well plate (Thermo Scientific) containing 600 μL LYBHI broth (Brain Heart Infusion [BHI] broth supplemented with 0.05% L-cysteine HCl [w/v] and 0.5% yeast extract [w/v]) followed by incubation under anaerobic conditions (atmosphere 75% N2, 20% CO2 5% H2). For isolates originally isolated under microaerophilic conditions (85% N2, 10% CO2, 5% O2), a 20 μL aliquot of each stock was inoculated into 600 μL BHI broth. After incubation for 48 hours at 37 °C, a 20 μL aliquot of each anaerobic or microaerophilic culture was added to 600 μL of fresh medium which was incubated for an additional 24 hours at the same temperature under the same atmospheric conditions. Equal volumes of each isolate sub-culture were then pooled and a solution of PBS containing 30% glycerol (v/v) was added resulting in a final 15% glycerol pooled stock (v/v). Pooling was performed in the Coy chamber. The 15% glycerol pooled stock was sealed in multiple 1.8-mL crimp glass vials (Wheaton) and the vials were stored at −80 °C prior to gavage into mice.
Diets
The ‘Adult Mirpur’ diet was designed based on 24-hour dietary recall surveys and food frequency questionnaires taken from adults enrolled in the BEED study living in an urban slum located in the Mirpur district of Dhaka City, Bangladesh83. A pelleted, sterile version of this diet was manufactured by Dyets, Inc. (Bethlehem, PA). The quantity of each ingredient used to prepare the diet is provided in Supplementary Table 1a. Rice (parboiled, long grain) and red lentils (masoor dal) were each cooked separately with an equal weight of water at 100 °C in a steam-jacketed kettle until the grains were cooked but still firm, and then set aside. Tilapia filets (frozen) were steamed separately at 100 °C in a steam-jacketed kettle with a small amount of water until tender (15–20 minutes). Sweet pumpkin (Calabaza variety) was chopped, boiled in the steam-jacketed kettle until soft and then strained. Fresh market white potatoes, Daikon radish (moola), spinach, okra and yellow onions were washed, finely chopped and cooked together in the steam kettle without added water at 70 °C until soft. After cooling, all cooked ingredients were combined and mixed with the wheat flour (atta), soybean oil, salt, turmeric, garlic and coriander powder. The resulting diet was mixed extensively using a planetary mixer, spread on trays, dried overnight at 30 °C, and pelleted by extrusion (½ inch diameter; California Pellet Mill, CL5). Dried pellets were aliquoted into ~250 g portions and placed in a paper bag with an inner wax lining, which was then placed in a plastic bag. Bags were subsequently vacuum-sealed and their contents sterilized by gamma irradiation (30–50 kGy; Sterigenics).
The composition of the ‘Mirpur-18’ diet was based on Bangladeshi complementary feeding practices for 18-month-old children living in Mirpur, as defined by quantitative 24-hour dietary recall surveys conducted in the MAL-ED study84. This diet was manufactured according to a previously described protocol2.
The sterility of irradiated diets was confirmed by culture in (i) BHI broth, (ii) Nutrient broth, and (iii) Sabouraud-dextran broth (all from Difco) for one week at 37 °C under aerobic conditions, and in reduced Tryptic Soy broth (Difco) supplemented with 0.05% L-cysteine HCl under anaerobic conditions. All diets were stored at −20 °C prior to use. Nutritional analysis of each irradiated diet was conducted by Nestlé Purina Analytical Laboratories (St. Louis, MO) (Supplementary Table 1A).
Husbandry for intergenerational transmission experiments
6–8-week-old germ-free female C57Bl/6J mice were given ad libitum access to an autoclaved breeder chow (Lab Diet 5021; Purina Mills, Richmond, IN) until 3 days prior to colonization, at which time they were switched to the Adult Mirpur diet for the remainder of the experiment. Mice received 200 μL of the stock solutions of the cSI-I or cSI-N consortium via an oral gavage needle (Cadence Science; catalog no. 7901). Another group of mice received an oral gavage of clarified cecal contents pooled from conventionally-raised C57Bl/6J animals maintained on a standard chow diet (Diet 2018S, Envigo) (200 μL/recipient animal).
One week after initial gavage, trio matings were performed (two colonized females with one germ-free C57Bl/6J male). Females were continued on the Adult Mirpur diet. Pups born to these mothers were maintained with their dams in the same cage until weaning, at which time pups from the same litter were transferred to new cages and weaned onto the Mirpur-18 diet. For dams and their pups, bedding was replaced every 7 days and diets were provided ad libitum. Following weaning of their pups, trio matings were performed again using previously pregnant mice. Offspring were euthanized on P37 (first pregnancy) or P14 (subsequent pregnancies) without prior fasting. All biospecimens were flash-frozen in liquid nitrogen and stored at −80 °C before analyses were performed.
For additional matings to assess fetal weights, adult germ-free female C57Bl/6J mice were switched to the Adult Mirpur diet 3 days prior to gavage with 200 μL of the cSI-I or cSI-N consortium, as described above. Two weeks after gavage, female mice were mated in trios and pregnant dams were euthanized at E11.5 or E17.5.
Husbandry for cohousing experiments
Germ-free 4–5-week-old male C57Bl/6J mice were switched from a standard chow diet (Diet 2018S, Envigo) to the Mirpur-18 diet 3 days prior to colonization. Each mouse received 200 μL of the cSI-I or cSI-N bacterial consortium via a single oral gavage. Nine days later mice were either maintained in their cage and isolator (non-cohoused control groups) or were transferred to a new cage in a new isolator to be cohoused with an equal number of mice transferred from an isolator containing the other experimental group. Bedding was replaced every 7 days for all groups of animals and the Mirpur-18 diet was provided ad libitum. All animals were weighed three times a week and euthanized without prior fasting on experimental day 18. All biospecimens were flash-frozen in liquid nitrogen and stored at −80 °C before use.
Husbandry for tests of candidate mediators of EED (‘add-in’ experiments)
The design of this experiment was analogous to that of the cohousing experiment. Germ-free 4–5-week-old male C57Bl/6J mice were switched from a standard chow diet (Diet 2018S, Envigo) to the Mirpur-18 diet 3 days prior to initial colonization. Each mouse received 200 μL of the cSI-N microbial consortium via a single oral gavage; all mice were maintained in a single gnotobiotic isolator. Nine days later, mice were distributed to new isolators and subsequently gavaged with 200 μL of either 15% PBS/glycerol alone (sham), the cSI-I bacterial consortium, or the cultured strains representing the A. naeslundii, A. odontolyticus and C. concisus MAGs. Mice received subsequent 200 μL gavages on days 10 and 11. Another reference control group of mice in their own isolator were initially gavaged with 200 μL of the cSI-I consortium, followed by gavage with medium alone on days 9 through 11. Bedding was replaced every 7 days and the Mirpur-18 diet was provided ad libitum. All animals were weighed three times a week and euthanized on experimental day 18 (first round of experiments) or 35 (follow-up more prolonged exposure experiment).
Division of the intestine
At the time of euthanasia, the small intestine was removed and divided into thirds. Each third was then subdivided into two equal length subsegments (‘proximal’ and ‘distal’). The proximal subsegment was used for either histomorphometric analysis or flow cytometry. Intestinal contents were removed from the distal subsegment by gentle extrusion. The distal subsegment was then cut into three 1.5 cm-long pieces (labeled #1–3 based on their proximal-to-distal location) which were flash-frozen in liquid nitrogen and stored at −80 °C before use. These frozen pieces were used for bulk tissue RNA-Seq (piece #1), snRNA-Seq (piece #2), or ELISA and Luminex-based protein quantification (piece #3). From the proximal end of the colon, three 1.5 cm long pieces (labeled #1–3 based on their proximal-to-distal location) were flash-frozen for the same type of assays applied to small intestinal samples.
Histomorphometric and immunocytochemical analyses of intestinal tissue
Proximal subsegments of the duodenum and ileum (see “Division of the intestine” above) were fixed in 10% neutral buffered formalin for 24h at 4 °C followed by a 70% ethanol wash. These segments were the placed in parallel with one another and collectively embedded in paraffin. Five μm-thick sections were prepared and stained with hematoxylin and eosin and imaged using a Zeiss Axioscan 7 Slide Scanner. Ten well-oriented crypt-villus units were selected from each intestinal segment and villus height and crypt depth were quantified using QuPath (v0.2.3)85. Measurements were performed for 4–6 mice per group with the investigator blinded with respect to experimental group.
Sections (5 μm-thick) were prepared from formalin-fixed, paraffin-embedded duodenal and ileal segments. Slides were de-paraffinized and heat-induced antigen retrieval was performed using Trilogy (Millipore Sigma). Slides were blocked in PBS containing 2% BSA, 5% donkey serum and 0.1% Triton-X (antibody buffer) for 30 minutes at room temperature and treated with primary antibodies (Ki67 anti-Rabbit, 1:300, Abcam AB16667; E-Cadherin anti-Mouse, 1:250, DB Sciences 610181) for 90 minutes at room temperature. Following three cycles of washing in PBS, slides were incubated with secondary antibodies (Alexa Fluor Donkey anti-Rabbit 488, 1:300, Invitrogen, catalog no. A32790; Alexa Fluor Donkey anti-Mouse, 1:300, Invitrogen, catalog no. A32731) for 30 minutes at room temperature, followed by a wash in PBS, incubation with DAPI (1:1000 dilution, Thermo Scientific, catalog no. 62248) for 5 minutes, and a final wash in PBS. Slides were mounted with Prolong Gold Antifade Mountant (Invitrogen, catalog no. P36930) and imaged with a Zeiss Axio Scan 7 Brightfield/Fluorescence Slide Scanner. Data were analyzed using QuPath (v0.2.3)85; as with the histomorphometric analysis, the investigator was blinded with respect to treatment group.
Micro-computed tomography (μCT) of bone
The femur was harvested from the left rear leg and cleaned of muscle and connective tissue. Femurs were stored in 10 mL 10% neutral buffered formalin at room temperature for 24 hours. Fixed femurs were then washed in 1X PBS for 15 minutes; this wash step was repeated two more times. Femurs were then subjected to a 30% ethanol wash for 30 minutes, a 50% ethanol wash for 30 minutes and a final 70% ethanol wash for 30 minutes. Femurs were subsequently embedded in 2% agarose and scanned with a μCT 40 desktop cone beam instrument (ScanCO Medical, Brüttisellen, Switzerland). For analyses of cortical bone, 100 slices were taken for each sample in the transverse plane, with a 6 μm voxel size (high resolution); slices began at the midpoint of the femur and extended toward the distal femur. The boundaries and thresholds for bone were drawn manually using μCT 40 software. Volumetric parameters were quantified using software associated with the ScanCO instrument.
Protein assays of serum, intestinal and brain tissue
Total intestinal protein was extracted by homogenizing a 1.5 cm-long piece of the duodenum, ileum or colon (piece #3, see “Division of the intestine” above) in 600 μL of a solution of ice-cold T-PER Buffer (Thermo Scientific) with Complete Ultra protease inhibitor (Roche), using Lysis Matrix F beads (MP Bio). The homogenate was centrifuged at 13,000 × g for 5 minutes at 4 °C. Total protein concentration in the resulting supernatants was quantified using Micro BCA Protein Assay Kit (Thermo Scientific). Protein concentrations were normalized to 1 mg/mL in PBS with Complete Ultra protease inhibitor (Roche). Intestinal and/or serum levels of LCN2 and IGF-1 were quantified using Mouse Lipocalin-2/NGAL DuoSet ELISA and Mouse/Rat IGF-I/IGF-1 DuoSet ELISA (R&D Systems), respectively, following the manufacturer’s instructions. Intestinal and/or serum levels of several other proteins were measured using a custom Mouse Pre-Mixed Multi-Analyte Kit (R&D Systems; CHI3L1, S100A9, MMP8, CXCL1, IL-17A, IL-17E) and the MILLIPLEX MAP Mouse Bone Magnetic Bead Panel (MilliporeSigma; DKK-1, FGF-23, OPG). Assays were performed on a FlexMap3D instrument (Luminex).
Ultrasonic vocalizations (USVs)
In a separate set of cSI-I or cSI-N-colonized dams and their pups, we assayed USVs. One week prior to mating, non-virgin dams that were colonized with the cSI-I or cSI-N consortia were switched to the Adult Mirpur diet. USV assays were performed on postnatal days 5, 7 and 9. As maternal behavior varies throughout the day, we standardized these studies such that all recordings were performed between 1000 h and 1300 h.
Previous research86 utilized the use of a sound induction chamber, which served the dual purpose of reducing background noise and maintaining the chamber at a temperature between 34–37 °C to reduce fluctuations in pup temperature through the experiment. However, standard induction chambers with heating elements are too large to fit into gnotobiotic isolators. We created induction chambers by clidox-sterilizing lidded Styrofoam containers. Within each box, an EtOH-sterilized microphone was placed approximately three inches above the pup holding pen. This microphone fed into Avisoft UltraSoundGate 116H at a sampling rate of 250,000 Hz and gain of 6 to record USVs. Water heating pads were placed beneath the vinyl of the isolator, and an autoclaved 1 mL pipet tip box, which served as a pup holding pen, was maintained on the heating pad within the isolator. The warmed holding pen allowed pups to maintain appropriate body temperature after separation from the dam. Body temperature was continually monitored with an EtOH-sterilized infrared temperature gun to ensure pup temperatures stayed between 34–37 °C. Ultrasonic vocalizations were recorded for each pup sequentially for 3 minutes; pups that were not being recorded were all maintained in the heated holding pen such that all pups were separated from the dam for the same amount of time. After all pups were recorded, the entire litter was returned to the dam.
Call spectra were analyzed using previously developed automated and blinded Matlab code (Barnes et al. 2017). Briefly, data were band-pass filtered between 25–110 Hz and a minimum spectral purity of 0.015 and spectral discontinuity of 1. Waveforms were divided into 512 samples per block, half-overlap, resulting in a resolution of 1.02 ms. Number, bouts, duration, mean frequency (Hz), mean peak dB, average pitch range (dB), and pause between calls were calculated in recordings that had greater than 10 total calls. Data were analyzed with a mixed effects linear model in which the interaction of diet and microbiota and postnatal day were fixed effects and dam and pup ID were random effects. Post-hoc tests were performed on significantly different fixed effects in the model.
Flow cytometry of immune cell populations
Intergenerational transmission experiments
Subsegments of duodenum, ileum and/or colon (See “Division of the intestine” above) were digested and myeloid and lymphoid cells were collected according to methods previously described5. Briefly, each subsegment was immediately flushed with cold PBS after dissection to remove luminal contents. Each subsegment was then opened lengthwise and gently agitated for 20 minutes at room temperature in Hanks Balanced Salt Solution (HBSS) supplemented with 15mM HEPES, 10% bovine calf serum (BCS) and 5mM EDTA. Each sample was vortexed and the suspended cells were collected; the remaining tissue fragments were subjected to a second round of gentle agitation and vortexing. The tissue remaining after the second collection was rinsed with cold 1X HBSS prior to digestion with Collagenase IV (Sigma) in complete RPMI-1640 for 40 minutes at 37 °C with gentle agitation. Digests were filtered through 100 μm mesh strainer (Falcon) and subjected to density gradient centrifugation using 40% and 70% Percol solutions (GE Healthcare).
To dissect meninges, we removed the skin and muscle overlying the skull as well as the mandibles and bone rostral to maxillae; the remaining skull was placed in Iscove’s Modified Dulbecco’s Medium (IMDM, Sigma Aldrich). The meninges were removed from the skull cap using fine forceps and visualization under a light microscope. Meninges were digested for 20 minutes at 37 °C with 1.4 U/mL of Collagenase VIII (Sigma Aldrich) and 35 U/mL of DNAse I (Sigma Aldrich) in IMDM. Following digestion, the tissue was gently pressed through a 70 μm mesh cell strainer (Falcon). The flow-through material was centrifuged at 450 × g at 4 °C for 4 minutes. Spleens were processed in a manner similar to what was used for the meninges with the exception that we performed an additional lysis step with ammonium-chloride-potassium (ACK) Lysis Buffer (Quality Biological) prior to staining.
Cells collected from each sample type were resuspended in ice-cold FACS buffer (2 mM EDTA, 25 mM HEPES, 1% BSA in 1X PBS) and stained for extracellular markers at 1:300 dilution. Dead cells were excluded using Zombie NIR fixable Viability kit. Cells were analyzed on a Cytek Aurora (Cytek Biosciences) and the results using FlowJo (v10.8.1). The fluorophore-labeled monoclonal antibodies used for flow cytometry are listed in Supplementary Table 5 and gating strategies are presented in the Supplementary Information.
Cohousing and add-in experiments
Mice were injected retro-orbitally (under isoflurane anesthesia 2 minutes before euthanasia) with 2 μg of phycoerythrin (PE)–conjugated anti-CD45 antibody (Biolegend, clone 30-F11) to label intravascular leukocytes. Blood was collected from the retroorbital sinus following euthanasia, centrifuged and lysed using ACK lysis buffer (Quality Biological) for 1 minute at room temperature, followed by addition of 2 mL 1X PBS to inactivate ACK lysis buffer. This lysis step was repeated two more times. The resulting material was subjected to centrifugation at 420 × g for 4 minutes. Cell pellets were resuspended in FACS buffer (PBS with 2% bovine serum albumin) and stained for extracellular markers at 1:300 dilution.
Immune cells from intestines, spleen and meninges were collected as described above for the intergenerational experiment. Dead cells were excluded using Zombie NIR fixable Viability kit. Cells were analyzed on a Cytek Aurora (Cytek Biosciences) and the data assessed using FlowJo (v10.8.1). The fluorophore-labeled monoclonal antibodies used for flow cytometry are listed in Supplementary Table 5.
Metagenome-assembled genomes (MAGs)
Identification of MAGs
DNA was extracted from flash-frozen cecal contents obtained from P37 cSI-I and cSI-N mice and their dams in the intergenerational experiment, as well as from non-cohoused and cohoused mice in the cohousing experiment. Cecal contents were subjected to bead beating and phenol-chloroform extraction to obtain crude genomic DNA, which was subsequently purified (QIAquick 96 PCR Purification Kit) and quantified (Qubit). The final DNA fragment size distribution was determined using a TapeStation (Agilent).
Fragmented genomic DNA (400–1000 ng) was prepared for long-read sequencing using a SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences) adapted to a deep 96-well plate (Fisher Scientific) format. All DNA handling and transfer steps were performed with wide-bore, genomic DNA pipette tips (ART). Barcoded adapters were ligated to A-tailed fragments (overnight incubation at 20 °C) and damaged or partial SMRTbell templates were subsequently removed (SMRTbell Enzyme Cleanup Kit). High molecular weight templates were purified (the volume of added undiluted AMPure beads used was 0.45 times the volume of the DNA solution). A second round of size selection was performed by diluting AMPure beads to a final concentration of 40% (v/v) with SMRTbell elution buffer, after which the resulting mixture was added at 2.2 times the volume of the pooled libraries. DNA was eluted from the AMPure beads with 12 μL of SMRTbell elution buffer. Pooled libraries were quantified (Qubit) and their size distribution was assessed using a TapeStation (Agilent). Libraries were then sequenced to a depth of 1.85 ± 1.5 × 109 reads/sample (mean ± SD) using a Sequel II System instrument (Sequel Binding Kit 3.0 and Sequencing Primer v4, Pacific Biosystems). The resulting reads were demultiplexed and Q20 circular consensus sequencing (CCS) reads were generated (Cromwell workflow conFig.d in SMRT Link). CCS reads were assembled into contigs using metaFlye (v2.8.1;87) with hifi-error set to 0.003 and other options set to default. Contig quality was evaluated using checkm (v1.0.7;88). Any contig demonstrating checkm ‘Completeness’ ≥85% and ‘Contamination’ ≤5% was nominated as a ‘high quality’ MAG. Remaining contigs were subjected to further MAG assembly efforts, first by calculating coverage using CoverM (v0.6.1, https://github.com/wwood/CoverM) and subsequently MAG reconstruction using MaxBin (v2.2.7;89). MAGs were cleaned using MAGpurify (v2.1.2;90) and subsequently evaluated for quality using checkm. In an iterative process, candidate MAGs demonstrating checkm Completeness ≥ 85% and Contamination ≤ 5% were again nominated as a MAG. High-quality MAGs from both primary and subsequent MAG assembly efforts were dereplicated using dRep (v2.3.2;91). Final MAG summary statistics were collected with checkm and quast (v4.5;92) (Supplementary Table 1C).
Taxonomic classification of MAGs
Taxonomic assignments were initially made by employing the Genome Taxonomy Database Toolkit (GTDB-Tk v1.5.1;93) and corresponding database (release 95; 14). Phylogenetic trees of MAGs and closely related reference genomes were generated using the Phylogenetic Tree Building service available from the Bacterial and Viral Bioinformatics Resource Center (BV-BRC;94). This service utilizes the Codon Tree method and universal protein families as homology group and analyzes alignments of these proteins identified in each genome using the program RAxML. The genome trees obtained were visualized via iTOL95.
Determination of the absolute abundances of MAGs and isolate genomes
The absolute abundances of MAGs were determined using previously described methods with minor modifications96,97. In brief, ‘spike-in’ bacterial strains whose genomes are easily differentiated from those of gut bacteria were added to each weighed frozen sample of intestinal contents or feces prior to DNA isolation and preparation of barcoded libraries98. For intergenerational transmission and cohousing experiments, Alicyclobacillus acidiphilus spike-in was added (2.22 × 108 cells/mL suspension; DSM 14558; GenBank assembly accession: GCA_001544355.1)97. For isolate add-in experiments, a commercial 1:1 mix of Imtechella halotolerans and Allobacillus halotolerans was added (2 × 107 cells each per 20 μL suspension; Zymo product numbers LMG 26483 and LMG 24826, respectively).
Intestinal contents and fecal samples were subjected to bead beating and phenol-chloroform extraction to obtain crude genomic DNA which was subsequently purified (QIAquick 96 PCR Purification Kit). Shotgun sequencing libraries were constructed using Nextera XT DNA Library Prep Kit and sequenced on an Illumina NovaSeq 6000 instrument [150 nt paired-end reads; 6.17 × 106 ± 3.75 × 107 reads/sample (mean ± SD) from the intergenerational transmission experiment, 3.10 × 106 ± 7.94 × 106 reads/sample from the cohousing experiments, and 3.03 × 107 ± 1.30 × 108 reads/sample from the isolate add-in experiment]. Sample metadata are listed in Supplementary Table 6. Due to increased host reads in small intestinal samples, libraries were balanced based on a host:microbe read ratio determined by preliminary shallow sequencing.
Reads were demultiplexed (bcl2fastq), trimmed (trimgalore, v0.6.1; https://github.com/FelixKrueger/TrimGalore) and filtered to exclude host reads (bowtie2; v2.3.5;99). MAG abundances were determined by assigning reads to each MAG, followed by a normalization for genome uniqueness in the context of a given community98. The resulting counts table was imported into R (v4.0.4). We calculated the absolute abundance of a given MAG in sample using the following equation:
The statistical significance of observed differences in the abundances of a given MAG across different groups was tested using a non-parametric Wilcoxon-rank test with FDR corrections on calculated log10 absolute abundances. Final MAG abundances for all experiments are included in Supplementary Table 1D.
Identification and assembly of isolates genomes related to MAGs
Average nucleotide identity was calculated using dRep (v2.3.2;91) for all genomes assembled by shotgun sequencing (paired end 150 bp reads) of cultured isolates from the BEED study duodenal aspirates (Supplementary Table 1b)2. For cultured isolates displaying ANI ≥ 95% shared with MAGs of interest obtained from the intergenerational and cohousing experiments, monocultures of each isolate were grown in 10 mL broth overnight at 37 °C under anaerobic conditions (atmosphere; 75% N2, 20% CO2 and 5% H2) without shaking. Cells were recovered by centrifugation (5,000 × g for 10 minutes at 4 °C) and high molecular weight genomic DNA was purified (QIAquick 96 PCR Purification Kit) and quantified (Qubit); the final fragment size distribution was determined using a TapeStation (Agilent). Fragmented genomic DNA (400–1000 ng) was prepared and long-read sequencing was performed as described above. The resulting reads were demultiplexed and Q20 circular consensus sequencing (CCS) reads were generated (Cromwell workflow conFig.d in SMRT Link). Genomes were assembled using Flye (v2.9;100) with hifi-error set to 0.003 and other options set to default. Assembly quality was evaluated using checkm. Taxonomic assignments were initially made by employing the Genome Taxonomy Database Toolkit (GTDB-Tk v1.5.1;93) and corresponding database (release 95; 14). Final assembly summary statistics were collected with checkM, quast and dRep (Supplementary Table 4A). All isolates were of high quality based on marker gene analysis, consisted of one or two contigs, and shared >99.9999% ANI with their respective MAG (Supplementary Table 4A).
In silico metabolic reconstructions of MAGs and isolate genomes
MAGs and isolate genomes were initially annotated using prokka (v1.14;101); KEGG Orthology (KO) numbers were assigned to bacterial coding sequences using BlastKOALA102. MAGs were then subjected to in silico metabolic pathway reconstruction. Our approach for reconstruction is based on gene annotation using the mcSEED database containing 2,856 reference bacterial genomes for which metabolic phenotypes have been inferred from expert curation and subsequent in silico predictions of the presence or absence of the pathway in each genome. Binary phenotypes, where ‘1’ stands for the presence of a particular metabolic capability and ‘0’ stands for its absence, were generated for each isolate genome or MAG using this approach103,104. A Binary Phenotype Matrix (BPM) representing these results captures utilization and/or biosynthesis of 106 metabolites, including carbohydrates, amino acids, vitamins, and fermentation end-products105–107.
For functional annotation of coding sequences in each genome, we used a combination of public domain tools (such as DBSCAN, DIAMOND, MMSeq2), custom scripts and the mcSEED reference database genes. These annotations were supplied to a ‘Phenotype Predictor’ pipeline that allows automated propagation of curated metabolic phenotypes from the reference BPM over new microbial genomes and MAGs from represented phylogenetic groups using a consensus of three complementary approaches: (i) rule-based phenotype assignment using the genomic distribution of orthologs and a set of phenotype rules; (ii) machine learning models for binary phenotype prediction trained on reference sets of genes and phenotypes and (iii) a neighbor-based approach for phenotype assignment based on variability of metabolic phenotypes and genes within a group of phylogenetically close neighbors. The combination of all three approaches allows consensus phenotype assignment with >99% accuracy108. The BPM obtained, and mcSEED gene annotations are provided in Supplementary Table 4b.
Bulk RNA-Seq
Intestinal tissue
RNA was extracted from 1.5 cm flash-frozen duodenum, ileum and/or colon (piece #1, See “Division of the intestine” above) using the Qiagen RNeasy 96 Kit. Total RNA was quantified (Qubit) and quality was assessed using a TapeStation (Agilent). cDNA libraries were generated using the Illumina Total RNA Prep with Ribo-Zero kit (Illumina). Barcoded libraries were sequenced on Illumina NovaSeq 6000 [150 nt paired-end reads to a depth of 3.23 × 107 ± 3.54 × 106 reads/sample from the intergenerational experiment and 7.13 ± 3.08 × 107 reads/sample from the cohousing experiment (mean ± SD)]. Sample metadata are provided in Supplementary Table 6.
Brain
Flash-frozen cerebral cortical samples were homogenized by bead-beating for 30 seconds with RNAse-free stainless-steel beads (Midsci SSB14B-RNA) in 300 μL of lysis buffer provided by the NucleoSpin 96 RNA isolation and purification kit (Takara 740709.4). Lysates were centrifuged at 13,000 × g for 5 minutes to remove beads and debris. RNA was isolated from the resulting supernatants following the manufacturer’s protocol. cDNA libraries were generated using the Illumina Total RNA Prep with Ribo-Zero kit. Barcoded libraries were sequenced on Illumina NovaSeq 6000 instrument [150 nt paired-end reads to a depth of 2.41 × 107 ± 3.29 × 106 reads/sample (mean ± SD)]. Sample metadata are listed in Supplementary Table 6.
Data analysis
Read quality was verified with FastQC (v0.11.7; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then trimmed to remove adapters and low-quality read segments using trimgalore (v0.6.1). Trimmed reads were then pseudoaligned to a kallisto (v0.46.2109) index built from the Gencode v25 Mus musculus reference genome. Transcript counts were aggregated to gene counts. Kallisto ‘estimated counts’ values and transcripts per million (TPM) values were used to generate a gene-level expression matrix in R using tximport (v1.22.0;110) and biomaRt (v2.50.3;111). The ‘lengthscaledTPM’ function in tximport was used to adjust estimated counts by gene length and abundance. This resulting count matrix was imported into limma-voom (v3.50.3;112,113) to compare log2-cpm gene expression between groups. GSEA was performed using clusterProfiler (v4.2.2;114) and the Gene Ontology (GO) pathway database, with FDR-correction.
snRNA-Seq
Isolation of nuclei from small intestinal tissue
Nuclei were extracted from flash-frozen 1.5 cm pieces of duodenum and ileum (piece #2, see “Division of the intestine” above) collected from P37 mice in the intergenerational transmission experiment. Briefly, the tissue piece was thawed and minced in lysis buffer (25 mM citric acid, 0.25 M sucrose, 0.1% NP-40, 1X protease inhibitor). Nuclei were released from cells using a Dounce homogenizer (Wheaton), then washed 3 times with buffer [25 mM citric acid, 0.25 M sucrose, 1X protease inhibitor (Roche)] and filtered successively through 100 μm-, 70 μm-, 40 μm-, 20 μm- and 5 μm-diameter strainers (pluriSelect) to obtain single nuclei. Following filtration, nuclei were pelleted by centrifugation (500 × g for 5 minutes) and resuspended in a buffer containing 5 mM KCl, 3 mM MgCl2, 50 mM Tris, 1 mM DTT, 0.4 U/μL RNase inhibitor (Sigma), plus 0.4 U/μL Superase inhibitor (ThermoFisher)115.
Isolation of nuclei from the cerebral cortex
Brains were removed from the skull, placed on ice, and hemisected in the sagittal plane. The cerebellum, thalamus and midbrain were removed while retaining the cortex from the left hemibrain. The left cortex was split into six equal coronal sections along the rostral-caudal axis; segments #1 and #4 along that axis were used for bulk RNA-Seq, while segments #2 and #5 were used for snRNA-Seq. All segments of the left cortex were flash-frozen in liquid nitrogen immediately after dissection.
Nuclei from flash-frozen cortical tissue were initially harvested using a 2 mL Dounce homogenizer in a Tris-based lysis buffer (10 mM Tris, 146 mM NaCl, 1 mM CaCl2, 21 mM MgCl2, 40 U/mL RNAse inhibitor, 1% BSA, 0.1% NP40). Nuclei were passed through a 30 μm filter (Miltenyi 130-041-407), washed once for 5 minutes in a Tris-based wash buffer (10 mM Tris, 146 mM NaCl, 1 mM CaCl2, 21 mM MgCl2, 40 U/mL RNAse inhibitor, 1% BSA), and then incubated in 1 μg/mL of Hoechst 33342 (Thermo 62249) for 5 minutes at 4 °C. Nuclei were washed again for 5 minutes with wash buffer, passed through a 20 μm filter (Miltenyi 130-101-812) and subjected to fluorescent activated nuclear sorting (FANS). A total of 200,000 Hoechst-positive nuclei were collected from each sample based on their forward and side-scatter properties.
Generation and sequencing of snRNA-Seq libraries
We used approximately 7,000 nuclei per intestinal sample and 6,000 nuclei per brain sample for gel bead-in-emulsion (GEM) generation. Reverse transcription and library construction were performed according to the protocol provided in the 3’ gene expression v3.1 kit manual (10X Genomics PN-1000121). Balanced libraries were sequenced on Illumina NovaSeq 6000 instrument [150 nt paired-end reads; 2.95 × 108 ± 2.02 × 107 reads/sample (mean ± SD)]. Sample metadata are listed in Supplementary Table 6.
Analysis of snRNA-Seq datasets
Preprocessing and quality control
Read alignment, feature-barcode matrices and quality controls were performed using the CellRanger 5.0 pipeline with the flag ‘--include-introns’ (GRCm38/mm10). ‘Ambient’ RNA was removed using the remove-background module from CellBender116. Sample integration, count normalization, cell clustering and marker gene identification were performed using Seurat 4.0117. Briefly, filtered feature-barcode matrices from CellRanger were imported as a Seurat object using CreateSeuratObject. Matrices were filtered to remove low quality nuclei (defined as nuclei with < 200 or > 5000 genes or < 400 UMIs). For intestinal samples, nuclei with over 5% reads from mitochondrial genes or over 5% reads from ribosomal protein genes were excluded. For brain cortex samples, the corresponding thresholds were 4% and 2.5%, respectively. Each sample was then normalized using SCTransform118,119 and predicted doublets were removed using DoubletFinder120. Samples of the same tissue type (duodenum, ileum, or cerebral cortex) were integrated using SelectIntegrationFeatures, PrepSCTIntegration, FindIntegrationAnchors and IntegrateData from the Seurat software package. Each integrated dataset was subjected to unsupervised clustering using FindNeighbors (dimensions = 1:30) and FindClusters (resolution = 0.8 and 1.2 for duodenum and ileum, respectively) from the Seurat package.
Cell type annotation
Cell type annotation for duodenal and ileal snRNA-Seq objects was performed using the FindMarkers function in Seurat. Manual cell type assignments were conducted based on expression of reported markers121. For cell type annotation in the cerebral cortex, the integrated Seurat object was converted to a SingleCellExperiment object122. Nuclei were then annotated using SingleR123, with manual adjustments to ensure that nuclei belonging to a cell type all resided in the same cluster (self-organizing map clustering implemented with 40 centers implemented with bluster122).
Pseudo-bulk analysis of differential gene expression
Genes defined as having low levels of expression (read count < 4) were filtered out prior to count aggregation across nuclei for a given cell type (cluster) within each biological sample. Each pseudo-bulked sample served as input for DESeq2-based differential gene expression analysis (likelihood ratio test, minimum=1e-6124). Genes with differential expression (adjusted P < 0.05) were used as input for GSEA with clusterProfiler114.
Trajectory inference analysis
Trajectory inference analysis was conducted with Slingshot125. Briefly, genes were removed whose read count did not exceed 3; remaining counts were quantile-normalized prior to principal components analysis, which was performed with prcomp. Gaussian mixture modeling was performed to cluster cells with mclust126 and pseudotime was inferred with slingshot. A negative binomial model was fit with tradeSeq127; n=30 knots was used for fitting the negative binomial generalized additive model (fitGAM) based on optimizing the number of genes with optimal K (evaluateK). Subsequently, differential expression testing across pseudotime between the cSI-I and cSI-N treatment groups was performed with conditionTest. As the results of conditionTest do not include directionality between conditions, we aggregated overall expression for cSI-I or cSI-N astrocytes to determine whether a gene that was significantly differently expressed across pseudotime at a higher or lower level in cSI-I compared to cSI-N animals. Over-enrichment analysis was performed on genes whose expression was significantly different between cSI-I versus cSI-N animals over pseudotime using conditionTest and querying the Panther database128.
Intercellular signaling
Signaling within each tissue (duodenum, ileum or cerebral cortex) was inferred with NicheNet34. Briefly, log-transformed counts were used for as input for the wrapper function nichenet_seratobj_aggregate. Default ligands, receptors and target matrices were used. In the duodenum and ileum, all cell clusters were designated as ‘sender’ cells to stem and transit amplifying (TA) ‘receiver’ cells. In the cerebral cortex, all cell clusters were defined as sender cells to either glutamatergic or GABA-ergic receiver neurons (separately). Differential NicheNet analysis was performed as described35. Default ligand and receptor matrices were used; read counts were log-transformed. While both NicheNet algorithms predict ligand-receptor links within a tissue, Differential NicheNet includes differential expression of ligands within a given ‘sender’ cell type between conditions (cSI-I vs. cSI-N) in the ranking of ligand-receptor pairs, while the NicheNet algorithm does not.
Selection of inflammation-correlated nuclei in the cerebral cortex
We used Scissor47 to select nuclei in the cerebral cortex that were associated with serum levels of the inflammation-associated protein S100A9 across all P37 animals from the intergenerational experiment. Seurat_preprocessing was run on raw snRNA-Seq counts. First, a Pearson correlation matrix was calculated for the single nucleus and bulk RNA-Seq expression matrices from the cerebral cortex of (i) P37 cSI-I mice (n=16 animals from n=4 litters; n=5 females and n=11 males), cSI-N mice (n=18 from n=4 litters, n=11 females and n=7 males) and CONV-D animals (n=16 from n=4 litters, n=10 females and n=6 males). Second, a gaussian regression model was optimized on the correlation matrix with serum levels of the protein S100A9. A grid search with alpha = (0.001, 0.01, 0.1, 0.2, 0.3, 0.5, 0.7, 0.8, 0.9, 1.0) was performed; the analysis described in the text used alpha = 0.7. The FindMarkers from Seurat was used to determine differential expression between (i) Scissor-selected (S100A9-negatively associated) astrocytes versus background (non-selected) astrocytes or (ii) Scissor-selected (S100A9-positively associated) glutamatergic neurons versus background glutamatergic neurons.
Microbial RNA-Seq
RNA was isolated from the cecal contents of the cSI-I and cSI-N sham-gavaged control, and cSI-N plus isolate-add in mice described in Fig. 5 and Extended Data Fig. 7. cDNA libraries were generated from isolated RNA samples using the Total RNA Prep with Ribo-Zero Plus kit (Illumina). Barcoded libraries were sequenced on an Illumina NovaSeq instrument [150 nt paired-end reads to a depth of 1.93 × 108 ± 7.94 × 107 reads/sample (mean ± SD); n=6 samples/treatment group]. Sample metadata are listed in Supplementary Table 6. Raw reads were trimmed (trimgalore; v0.6.1), filtered to exclude host reads (bowtie2; v2.3.5), and mapped to MAGs (kallisto; v0.46.2). The resulting kallisto pseudocounts table was imported into R (v4.0.4), and EdgeR (v3.36.0129) was used to filter out lowly-expressed genes and normalize the expression dataset to MAG abundance prior to performing differential expression analysis.
Epithelial cell death induction assay
HCT116 cells were cultured at 37 °C in DMEM medium supplemented with 10% FBS at 4 × 105 cells per ml. Cells were washed with 1xPBS and selected controls were pre-treated with DMSO (0.1%) or 30 μM caspase inhibitor (QVD, Quinoline-Val-Asp-Difluorophenoxymethylketone) for 1h before inducing apoptosis with administration of 1 μM staurosporine (STS; Abcam) for 24 h.
CT26:FADD cells were seeded at 2 × 106 cells per 10 cm culture dish. The next day, cultures were incubated with 1 μg/ml doxycycline for 16 h to induce expression of the FADD construct (Fas-associated death domain). Doxycycline was removed by washing with 1x PBS before 1 h pre-treatment with 20 μM z-VAD-FMK (z-VAD, pan-caspase inhibitor, MedChem Express) or DMSO (0.1%). Cell death was induced by adding 10 nM B/B homodimerizer for 5 h after which time supernatants were collected and centrifuged at 350 × g for 5 minutes to remove cellular debris. The resulting supernatants were filtered using 0.2μm syringe filter (SFCA; Corning) and then frozen at −20 °C for later use in C. concisus growth assays.
C. concisus growth assays
C. concisus isolate Bg048 was routinely cultured in Bolton Broth supplemented with 10% FBS (autoclaved and subsequently filter-sterilized) and on BHI-agar plates supplemented with 7.5–10% (v/v) horse blood. For growth in spent colonic epithelial cell line supernatants, a 25 μL aliquot of an overnight culture of C. concisus was sub-cultured in 50:50 (v/v) mixture of Bolton Broth supplemented with 10% FBS and DMEM cell culture medium supplemented with 10% FBS. After a 6–24 h incubation at 37 °C, a 30 μL aliquot of the resulting culture was removed for serial dilutions and the determination of CFU using X plates incubated under micro-aerophilic conditions (5% O2, 10% CO2, 85% N2) or anaerobic conditions (5% H2, 20% CO2, 75% N2). After 48 hours of growth, C. concisus colonies were counted and growth relative to DMEM alone was evaluated.
Extended Data
Extended Data Fig. 1. Preliminary test of non-enteropathy inducing control bacterial consortium.
a, Weight gain of mice colonized with all isolates cultured from children with EED (n=186), or a species-representative set of isolates (n=39). Control group was colonized with cecal microbiota from conventionally-raised mice. Each small point represents the weight gain for an individual mouse; larger points denote the mean, error bars, standard deviation. n=5–10 mice/group, linear mixed effects model. b, Serum IGF-1 correlates with mouse weight measured nine (left) or 28 (right) days after colonization (n=5 mice per group per timepoint, linear regression). c, Serum leptin levels determined nine (left) or 28 days (right) days post-colonization (n=5 mice per group per timepoint, bars denote mean ± s.d.). d, Number of Gene Ontology (GO) categories related to the immune system that were significantly enriched (GSEA q value < 0.05) based on differential gene expression in the small intestine of mice colonized with all isolates (red) versus the species representative subset (blue). Bulk RNA-seq was performed in duodenal, jejunal, and ileal tissue 28 days following gavage (n=5 mice/group). e, Expression of genes (columns, un-labeled) that comprise the leading edge of the eight GO categories most significantly enriched in animals colonized with the full consortium (red) compared to the species-representative subset (blue). Bulk RNA-seq of duodenal tissue, 28 days post colonization. f, Serum lipocalin-2 (LCN2) levels nine (left) or 28 days (right) days post-gavage of the consortia (n=5 mice per treatment group per timepoint, bars denote mean ± s.d.). g, Design of the intergenerational transmission experiment. h, Summary of animals collected during intergenerational experiment. Litter sizes and the age of offspring at the time of sample collection are shown for each dam (n=4 dams/group). t1–t3 denotes the number of days between first mating and birth of the litter. i,j, Principal component analysis of MAGs in feces of dams ordinated by their log10 absolute abundance and colored by bacterial consortium (i) or timepoint (j). The composition of community membership differs significantly between cSI-I and cSI-N dams (i, P=0.01, PERMANOVA) but is stable after the first week of colonization (j, P=0.09, PERMANOVA weeks 4 through 17). Each small point represents a fecal sample from one animal. Larger filled circles are the centroids for the ellipses shown, which denote the 95% confidence interval.
Extended Data Fig. 2. Growth and inflammatory phenotypes of P14 offspring and fetuses.
a, Body mass of P14 offspring of dams colonized with the cSI-I consortium (red), cSI-N consortium (blue), or cecal contents from a conventional mouse (CONV-D). Each point represents an individual animal. b, Serum levels of IGF-1 in P14 offspring. C-f, Serum levels of the inflammation-associated proteins, LCN2 (c), S100A9 (d), MMP8 (e), and CXCL1 (f), in P14 offspring. g-i, Levels of the inflammation-associated proteins LCN2, S100A9, and CHI3L1 in duodenal, ileal, and colonic tissue of P14 animals. For a-i, n=11–19 mice/group, 3–8 pups/litter, P-values were determined using Wilcoxon rank-sum with Tukey’s post-hoc tests. j, Average masses of fetuses per dam at E11.5, n=9–10 dams/group; n=4–8 fetuses/litter. k,l, Average masses of placentas within each dam at E11.5 (k) and E17.5 (l); n=7–10 dams/group; n=2–10 conceptuses/litter. For j-l, P-values determined with unpaired t-tests. For all panels, mean values ± s.d. are shown.
Extended Data Fig. 3. Assessment of EED biomarkers and immune profiling of P37 animals.
a, Ratio of villus length to crypt depth in the ileums of cSI-I and cSI-N mice (n=5 mice/group, Wilcoxon rank-sum test; boxes denote interquartile range). b,c, Levels of serum MMP8 (b), and CXCL1 (c), in the P37 offspring of C57Bl/6J dams colonized with the cSI-I or cSI-N consortia, or with mouse microbiota harvested from the cecal contents of conventionally-raised (CONV-D) C57Bl/6J mice (n=21–25 mice/group). d,e, Levels of S100A9 (d), and CHI3L1 (e), in the duodenal, ileal, and colonic tissue of P37 animals (n=7–10 animals/group). f, Concentration of acylcarnitines in small intestinal tissue of P37 animals. Acylcarnitines shown exhibited a significant difference between cSI-I versus cSI-N groups (n=7–16 mice/group, Mann-Whitney tests with Benjamini, Krieger, Yekutieli two-stage step up FDR correction, q-values shown). g,h, Frequency of CD3+ (g), and CD4+ (h), cells in duodenal, ileal and colonic lamina propria (n=4–7 mice/group, except for colonic tissue collected from CONV-D animals for which n=2 and a statistical comparison was excluded). For panels b-e and g-h, bars denote mean ± s.d. and P-values were determined by Wilcoxon rank-sum tests.
Extended Data Fig. 4. snRNA-Seq differential gene expression in the duodenum and ileum of P37 animals.
a, Normalized enrichment scores (NES) for all GO categories (rows) that were significantly enriched (q < 0.05, GSEA) in at least two of the indicated cell populations (columns) in either the duodenum (left) or ileum (right). Immune-related GO terms are labeled. If a GO category was non-significant, it was assigned an NES value of 0. b, Significantly differentially expressed genes comprising the ‘leading edge’ of the efferocytosis gene set in duodenal mid-villus enterocytes (cSI-I versus cSI-N; P-adj < 0.05, GSEA).
Extended Data Fig. 5. Expression of genes encoding orthologs of proteins quantified in the duodenal mucosa of children with EED and proliferative inter-cellular signaling in P37 animals.
a, Expression of transcripts (rows) in cell populations (columns) in the duodenum of P37 animals that match proteins, quantified in biopsies of the duodenal mucosa of Bangladeshi children with EED, that were (i) negatively correlated with their LAZ and (ii) positively correlated with the absolute abundances of ‘core’ EED bacterial taxa (whose representation in turn positively correlated with their degree of stunting); related to Fig. 2c. b, Predicted receptors (right) in stem and transit-amplifying cells for ligands (left) expressed by various cell types in the ileum of P37 animals identified by NicheNet. Upper panel – ligands expressed more highly in cSI-I animals; lower panel – ligands expressed more highly in cSI-N animals. Boxes are sized proportionally to the weighted ligand/receptor score, related to Fig. 2d. c, Representative sections of crypts from the duodenum of cSI-N (left) and cSI-I (right) P37 mice. Sections were stained with antibodies to E-cadherin (red) and Ki67 (green). Scale bar, 50 μm.
Extended Data Fig. 6. snRNA-Seq intercellular signaling and cellular response to inflammation in the cerebral cortex of P37 mice.
a, Levels of the protein CXCL1 in the cerebral cortex tissue of P37 cSI-I and cSI-N animals (n=6–13 mice/group, bars denote mean ± s.d., Welch’s t test). b, Scissor-defined relationship between serum S100A9 and snRNA-Seq gene expression. A correlation was first performed between bulk RNA-Seq gene expression and serum S100A9 levels in the same animals (n=49 P37 mice; n=15–18 mice/group); subsequently, a Gaussian regression was performed using this correlation matrix with snRNA-Seq expression (n=4 mice/group). Shown are the cortex nuclei that were positively (pink) and negatively (blue) associated with levels of systemic S100A9. Top inset: 15 genes most significantly up-regulated in astrocyte nuclei that were negatively correlated with S100A9 compared to non-associated (‘background’) astrocytes. Bottom inset: Genes most significantly differentially expressed in glutamatergic neurons that were positively correlated with S100A9 compared to non-associated (‘background’) glutamatergic neurons. c,d, Predicted NicheNet receptors (right) in glutamatergic neurons for ligands (left) expressed by various cell types in the cerebral cortex of P37 animals. c, Ligands more highly expressed in cSI-I animals; d, ligands more highly expressed in cSI-N animals. Boxes are sized proportionally to the weighted ligand/receptor score; corresponds to Fig. 3f.
Extended Data Fig. 7. Abundance of Actinomyces in isolate ‘add-in’ experiments and C. concisus growth in colonic epithelial supernatants.
a,b, Mice were initially colonized with either the cSI-N or cSI-I consortium (‘gavage 1’). Gavages 2–4 were performed on experimental days 9–11, as in Fig. 5a. The absolute abundances of A. odontolyticus isolate Bg044 (corresponding to MAG044, panel a) and A. naeslundii isolate Bg041 (corresponding to MAG041, panel b), shown relative to their absolute abundance in the cSI-N/sham-gavaged controls (a ratio of 1 is indicated by dotted line; mixed-effects analysis with Dunnett’s multiple comparisons to cSI-N/sham control; mean values ± s.d. are shown.) c, Levels of LCN2 protein in duodenal tissue on experimental day 18 (one-way ANOVA with Tukey’s multiple comparisons; mean values ± s.e.m. are shown). Insets show comparisons between individual isolates. For a-c, n=5–11 mice/group combined across two independent experiments. d,e, Microaerophilic growth of C. concisus after 8 (d), or 24 hours (e), in mouse CT26 colonic epithelial cell line supernatants that were live (green; CT26:FADD, z-VAD, z-VAD+BB) or were treated with an inducer of apoptosis (red; BB) (n=4 biological replicates per condition). Points shown with an open rather than filled circle denote abundances that were below the limit of detection at the time of quantification and are presented at the limit of detection of the assay. f-h, Microaerophilic growth of C. concisus after 6 (f), 12 (g), or 24 (h) hours in HTC116 human colonic epithelial cell line supernatants that were live (green; QVD, QVD+STS) or were treated with an inducer of apoptosis (red; STS) (n=3 biological replicates/condition). i-l, Anaerobic growth of C. concisus after 8 hours (i, k) or 24 hours (j, l) of incubation in CT26 mouse colonic epithelial cell line supernatants prepared from live cells (green; CT26:FADD, z-VAD, z-VAD+BB) or cells that had been treated with an inducer of apoptosis (red; BB). Panels k and l show growth relative to DMEM medium control (n=4 biological replicates/condition). m,n, Anaerobic growth of C. concisus after 24 hours of incubation in HTC116 cell supernatants prepared from live cells (green; QVD, QVD+STS) or cells that had been treated with an inducer of apoptosis (red; STS). All cells were treated with an inhibitor of nitric oxide synthase (L-NIO). Panel m shows differences in growth compared to medium controls; n, total bacterial growth (n=4 biological replicates/condition). For d-n, bars denote mean ± s.d., P-values determined with one-way ANOVA and Tukey’s multiple comparisons. C. concisus growth in supernatants acquired from live cells and live cell controls are colored in green; apoptotic cells, in red. FADD, Fas-associated death domain; z-VAD, z-VAD-FMK (pan-caspase inhibitor); BB, B/B Homodimerizer (induces FADD-dependent cell death); QVD, Quinoline-Val-Asp-Difluorophenoxymethylketone (caspase inhibitor); STS, Staurosporine (apoptosis inducer); L-NIO, N5-(1-Iminoethyl)-L-ornithine, dihydrochloride (nitric oxide synthase inhibitor).
Supplementary Material
a, Diets used in gnotobiotic mouse studies. Ingredients in and nutritional analysis of each diet. b, Cultured isolates and taxonomic classification from BEED study duodenal aspirates. c, MAG assembly information and MAG taxonomic classification. (i), Assembly and taxonomic classification. (ii), Sample metadata. d, Abundances of MAGs from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 animals, P14 animals and adult female mice. (ii), Cohousing experiment. (iii), Isolate add-in experiments. Absolute abundance values are log10-transformed. P-adj values determined by Wilcoxon rank-sum tests and FDR correction.
a, Protein levels in tissues from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 duodenum, ileum, colon, serum and cerebral cortex; P14 duodenum, ileum, colon and serum. (ii), Cohousing experiments: duodenum, ileum, colon and serum. (iii), Isolate add-in experiments: duodenum, colon and serum. b, Body weights from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 animals, P14 animals and conceptuses. (ii), Cohousing experiments. P-values for table (i) were determined using Wilcoxon rank-sum tests; those for table (ii) were determined using linear mixed effects modeling. c, Micro-computed tomography of femurs of P37 animals from the intergenerational experiment. P-values were determined using Wilcoxon rank-sum tests. d, Histomorphometric analysis of small intestines of P37 animals from the intergenerational transmission experiment. Duodenal (i) and ileal (ii) villus height, crypt depth, villus to crypt ratio, and Ki67 quantification. P-values determined using Tukey’s post-hoc tests. e, Immune cell profiling of tissues from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 duodenum, ileum, colon, spleen and meninges; duodenum, ileum, spleen and meninges of the dams. (ii), Cohousing experiment: bulk small intestinal tissue, spleen and meninges. P-values determined using Wilcoxon rank-sum tests. f, Quantification of ultrasonic vocalization metrics in P5-P9 pups.
a, Bulk tissue RNA-Seq from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: gene set enrichment analysis (GSEA) of gene expression in the duodenum, ileum and colon of P37 animals. (ii), Cohousing experiment: differentially expressed genes in duodenal samples for: non-cohoused cSI-I controls versus non-cohoused cSI-N controls and cohoused versus non-cohoused cSI-N controls. There were no statistically significant differentially expressed genes between cohoused and cSI-I controls. GSEA of gene expression in the colon of non-cohoused cSI-I controls vs. non-cohoused cSI-N controls and cohoused vs. non-cohoused cSI-N controls. (iii), Add-in experiment: GSEA of gene expression in the colon. Pathways shown were enriched in all of the following conditions: cohoused animals vs. cSI-N non-cohoused controls, C. concisus + Actinomyces-gavaged animals vs. cSI-N sham-gavaged controls and C. concisus-gavaged animals vs. cSI-N sham-gavaged controls. Values provided (P- and q-values, enrichment scores, rank, leading edge, core enrichment) are for C. concisus + Actinomyces-gavaged vs. cSI-N sham-gavaged animals. b, Proportions of cell types in the duodenums and ileums of P37 animals from the intergenerational transmission experiment. (i and ii), Proportions of all cell types quantified by snRNA-Seq in the duodenum Table i) and ileum (Table ii) (n=3 mice/group). Statistical significance determined by edgeR. c, snRNA-Seq analysis of duodenums and ileums of P37 offspring from intergenerational transmission experiment. Pseudo-bulk differential expression of enterocyte clusters in the duodenum and ileum of cSI-I compared to cSI-N animals (n=3 mice/group; DESeq2 Wald test). d, snRNA-Seq analysis of cerebral cortices of P37 offspring from the intergenerational transmission experiment. (i), Pseudobulk differential expression of glutamatergic neurons in the cerebral cortex (n=4 mice/group; DESeq2 Wald test). (ii), GSEA of significantly (P-adj<0.05) differentially expressed genes in the cortex. (iii), Differential expression of S100A9-associated vs. background glutamatergic neurons. Positive log2-fold change indicates higher expression in S100A9-associated neurons (determined with the FindMarkers function in Seurat). e, GSEA results in the duodenum and colon for bulk RNA-seq datasets. All treatment groups presented were compared to their cSI-N counterparts.
a, Isolate genome assembly information and taxonomic classification. (i), Quality assessment and taxonomic classification of assembled isolate genomes. (ii), Average nucleotide identity (ANI) of isolate genomes with their related MAGs. (iii), Species-specific virulence factors in MAGs, their respective isolate genomes, and related reference genomes. b, In silico metabolic reconstructions of MAGs and isolate genomes. (i), Metabolic annotations for each MAG by gene. (ii), Metabolic annotations for pathology-associated MAGs and phylogenetically related organisms by gene. (iii), Binary phenotype predictions for all MAGs. (iv), Binary phenotype predictions for pathology-associated MAGs, their isolate genomes, and phylogenetically related organisms.
Antibodies used for flow cytometry. (i), Antibodies used for flow cytometry of intestinal immune cells in intergenerational transmission and cohousing experiments. (ii and iii), Antibodies used for flow cytometry of splenic and meningeal immune cells in intergenerational transmission (ii) and cohousing experiments (iii).
Sequencing metadata from gnotobiotic animal experiments. a, Intergenerational transmission experiment: (i) short read shotgun DNA sequencing, (ii) snRNA-Seq, (iii) bulk tissue RNA-Seq and (iv) microbial RNA-Seq. b, Cohousing experiment: (i) short read shotgun DNA sequencing, (ii) bulk tissue RNA-Seq and (iii) and microbial RNA-Seq. c, Isolate add-in experiments: (i) short read shotgun DNA sequencing and (ii) and bulk tissue RNA-Seq.
Acknowledgements
We thank David O’Donnell, Maria Karlsson and Justin Serugo for their invaluable assistance with mouse husbandry, Martin Meier for generating libraries for shotgun sequencing and microbial RNA-Seq, and Jessica Hoisington Lopez, MariaLynn Crosby plus members of the Genome Technology Access Core at Washington University School of Medicine for sequencing these libraries.
Funding
This work was supported by grants from the Bill & Melinda Gates Foundation (INV-033564) and the NIH (DK131107). K.M.P. is the recipient of a postdoctoral fellowship from the Helen Hay Whitney Foundation. C.K. is the recipient of a NIH F30 predoctoral MD/PhD fellowship (DK123838). K.S.R is supported by NIH grant AI159551 and a BJC Investigator award.
Footnotes
Competing Interests - D.R. and A.O. are co-founders of Phenobiome Inc., a company pursuing development of computational tools for predictive phenotype profiling of microbial communities.
Ethics and Inclusion – This work was performed as part of a long-standing collaboration investigating the role of the gut microbiota in undernutrition, covered by a memorandum of understanding, between teams led by Tahmeed Ahmed (International Centre for Diarrhoeal Disease Research, (icddr,b) Bangladesh) and Jeffrey Gordon (Washington University in St Louis, USA). Bacterial isolates used in this study were obtained from duodenal aspirates collected from the previously reported Bangladesh EED study, which was approved by the Ethical Review Committee (ERC) at the icddr,b (protocol no: PR-16007; ClinicalTrials.gov number, NCT02812615). Written informed consent was obtained from each child’s parent or guardian to participate and to undergo esophago-gastroduodenoscopy (EGD) if the child met the inclusion criteria. Human biospecimens were transferred to Washington University under a Materials Transfer Agreement (MTA). Bacterial isolates cultured and used in this study are the property of icddr,b and are available under MTA upon request to T.A. and J.I.G..
Code Availability – The custom source code for annotation of bacterial genes and prediction of metabolic pathway presence/absence is available at https://github.com/rodionovdima/PhenotypePredictor-2 and has been accessioned at Zenodo (DOI: 10.5281/zenodo.13250652). All other analyses presented utilize published algorithms and accompanying methods and are referenced as such.
Data availability
Datasets generated by shotgun sequencing of DNA isolated from the intestinal contents of gnotobiotic mice and from individual cultured bacterial strains, plus snRNA-Seq and bulk RNA-Seq of intestinal tissue and cerebral cortex, and microbial RNA-Seq of cecal contents harvested from gnotobiotic mice have been deposited at the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number ERA22531095.
REFERENCES
- 1.Lindenbaum J., Kent T. H. & Sprinz H. Malabsorption and jejunitis in American Peace Corps volunteers in Pakistan. Annals of internal medicine 65, 1201–1209 (1966). [DOI] [PubMed] [Google Scholar]
- 2.Chen R. Y. et al. Duodenal microbiota in stunted undernourished children with enteropathy. New England Journal of Medicine 383, 321–333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hossain Md. S. et al. Environmental enteric dysfunction and small intestinal histomorphology of stunted children in Bangladesh. PLOS Neglected Trop. Dis. 17, e0010472 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owino V. et al. Environmental enteric dysfunction and growth failure/stunting in global child health. Pediatrics 138, e20160641 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Cowardin C. A. et al. Environmental enteric dysfunction: gut and microbiota adaptation in pregnancy and infancy. Nature Reviews Gastroenterology and Hepatology 20, 223–237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martorell R. & Zongrone A. Intergenerational influences on child growth and undernutrition. Paediatric and Perinatal Epidemiology 26, 302–314 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan U., Martorell R., Schroeder D. G. & Flores R. Role of intergenerational effects on linear growth. Symposium: Causes and etiology of stunting role of intergenerational effects on linear growth 129, 544–549 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Soliman A. et al. Early and long-term consequences of nutritional stunting: From childhood to adulthood. Acta Bio Med. : Atenei Parm. 92, e2021168 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coley E. J. L. & Hsiao E. Y. Malnutrition and the microbiome as modifiers of early neurodevelopment. Trends in Neurosciences 44, 753–764 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Yan X., Zhao X., Li J., He L. & Xu M. Effects of early-life malnutrition on neurodevelopment and neuropsychiatric disorders and the potential mechanisms. Progress in Neuro-Psychopharmacology and Biological Psychiatry 83, 64–75 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Georgieff M. K., Ramel S. E. & Cusick S. E. Nutritional influences on brain development. Acta Paediatrica 107, 1310–1321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lunn P. G., Northrop-Clewes C. A. & Downes R. M. Intestinal permeability, mucosal injury, and growth faltering in Gambian infants. The Lancet 338, 907–910 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Campbell D. I., Elia M. & Lunn P. G. Growth faltering in rural Gambian infants is associated with impaired small intestinal barrier function, leading to endotoxemia and systemic inflammation. Journal of Nutrition 133, 1332–1338 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Harper K. M., Mutasa M., Prendergast A. J., Humphrey J. & Manges A. R. Environmental enteric dysfunction pathways and child stunting: A systematic review. PLoS Neglected Tropical Diseases 12, e00062051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahfuz M. et al. Bangladesh Environmental Enteric Dysfunction (BEED) study: Protocol for a community-based intervention study to validate non-invasive biomarkers of environmental enteric dysfunction. BMJ Open 7, e017768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharjee A. et al. Environmental enteric dysfunction induces regulatory T cells that inhibit local CD4 + T cell responses and impair oral vaccine efficacy. Immunity 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malique A. et al. NAD+ precursors and bile acid sequestration treat preclinical refractory environmental enteric dysfunction. Sci. Transl. Med. 16, eabq4145 (2024). [DOI] [PubMed] [Google Scholar]
- 18.Perruzza L. et al. Protection from environmental enteric dysfunction and growth improvement in malnourished newborns by amplification of secretory IgA. Cell Rep. Med. 5, 101639 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salameh E. et al. Modeling undernutrition with enteropathy in mice. Sci. Rep. 10, 15581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semba R. D. et al. Environmental enteric dysfunction is associated with carnitine deficiency and altered fatty acid oxidation. EBioMedicine 17, 57–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemons J. M. S. et al. Enterobacteriaceae growth promotion by intestinal acylcarnitines, a biomarker of dysbiosis in inflammatory bowel disease. Cell. Mol. Gastroenterol. Hepatol. 17, 131–148 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dambrova M. et al. Acylcarnitines: Nomenclature, biomarkers, therapeutic potential, drug targets, and clinical trials. Pharmacol. Rev. 74, 506–551 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Rutkowsky J. M. et al. Acylcarnitines activate proinflammatory signaling pathways. Am. J. Physiol.-Endocrinol. Metab. 306, E1378–E1387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veitch A. M., Kelly P., Zulu I. S., Segal I. & Farthing M. J. G. Tropical enteropathy: A T-cell-mediated crypt hyperplastic enteropathy. European Journal of Gastroenterology and Hepatology 13, 1175–1181 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Liu T. C. et al. A novel histological index for evaluation of environmental enteric dysfunction identifies geographic-specific features of enteropathy among children with suboptimal growth. PLoS Neglected Tropical Diseases 14, 1–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulenga C. et al. Epithelial abnormalities in the small intestine of Zambian children with stunting. Front. Med. 9, 849677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C. S. et al. Boosting apoptotic cell clearance by colonic epithelial cells attenuates inflammation in vivo. Immunity 44, 807–820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shankman L. S. et al. Efferocytosis by Paneth cells within the intestine. Current Biology 31, 2469–2476.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehrotra P. & Ravichandran K. S. Drugging the efferocytosis process: concepts and opportunities. Nature Reviews Drug Discovery 21, 601–620 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson C. J. et al. Microbes exploit death-induced nutrient release by gut epithelial cells. Nature 596, 262–267 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Morioka S. et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563, 714–718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry J. S. A. et al. Interpreting an apoptotic corpse as anti-inflammatory involves a chloride sensing pathway. Nature Cell Biology 21, 1532–1543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maschalidi S. et al. Targeting SLC7A11 improves efferocytosis by dendritic cells and wound healing in diabetes. Nature 606, 776–784 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Browaeys R., Saelens W. & Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nature Methods 17, 159–162 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Guilliams M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi T. & Shiraishi A. Stem cell signaling pathways in the small intestine. International Journal of Molecular Sciences 21, 2032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanuytsel T., Senger S., Fasano A. & Shea-Donohue T. Major signaling pathways in intestinal stem cells. Biochimica et Biophysica Acta - General Subjects 1830, 2410–2426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X. M. et al. Wnt5a inhibits hypoxia-induced pulmonary arterial smooth muscle cell proliferation by downregulation of β-catenin. American Journal of Physiology - Lung Cellular and Molecular Physiology 304, L103–L111 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Aubé B. et al. Neutrophils mediate blood–spinal cord barrier disruption in demyelinating neuroinflammatory diseases. The Journal of Immunology 193, 2438–2454 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Kanashiro A. et al. The role of neutrophils in neuro-immune modulation. Pharmacological Research 151, 104580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima K. A. D., Rustenhoven J. & Kipnis J. Meningeal immunity and its function in maintenance of the central nervous system in health and disease. Annual Review of Immunology 38, 597–620 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Ma T., Wang F., Xu S. & Huang J. H. Meningeal immunity: Structure, function and a potential therapeutic target of neurodegenerative diseases. Brain, Behavior, and Immunity 93, 264–276 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Costa E. D., Rezende B. A., Cortes S. F. & Lemos V. S. Neuronal nitric oxide synthase in vascular physiology and diseases. Frontiers in Physiology 7, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gadani S. P., Cronk J. C., Norris G. T. & Kipnis J. IL-4 in the brain: A cytokine to remember. The Journal of Immunology 189, 4213–4219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herz J. et al. GABAergic neuronal IL-4R mediates T cell effect on memory. Neuron 109, 3609–3618.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X. et al. Neuronal interleukin-4 as a modulator of microglial pathways and ischemic brain damage. Journal of Neuroscience 35, 11281–11291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun D. et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nature Biotechnology 40, 527–538 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen N. J. & Lyons D. A. Glia as architects of central nervous system formation and function. Science 185, 181–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastorakos P. & McGavern D. The anatomy and immunology of vasculature in the central nervous system. Science Immunology 4, eaav0492–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKerracher L. & Rosen K. M. MAG, myelin and overcoming growth inhibition in the CNS. Frontiers in Molecular Neuroscience 8, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alboni S., Cervia D., Sugama S. & Conti B. Interleukin 18 in the CNS. Journal of Neuroinflammation 7, 1–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez-Nicola D., Valle-Argos B. & Nieto-Sampedro M. Blockade of IL-15 activity inhibits microglial activation through the NFκB, p38, and ERK1/2 pathways, reducing cytokine and chemokine release. Glia 58, 264–276 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Brindle N. P. J., Saharinen P. & Alitalo K. Signaling and functions of angiopoietin-1 in vascular protection. Circulation Research 98, 1014–1023 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin C. Y. et al. ADAM9 promotes lung cancer progression through vascular remodeling by VEGFA, ANGPT2, and PLAT. Scientific Reports 7, 15108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy R., Dagher A., Butterfield C. & Moses M. A. ADAM12 is a novel regulator of tumor angiogenesis via STAT3 signaling. Molecular Cancer Research 15, 1608–1622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyer L. A., Pi X. & Patterson C. The role of BMPs in endothelial cell function and dysfunction. Trends in Endocrinology and Metabolism 25, 472–480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barnes T. D., Rieger M. A., Dougherty J. D. & Holy T. E. Group and individual variability in mouse pup isolation calls recorded on the same day show stability. Front. Behav. Neurosci. 11, 243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Premoli M., Pietropaolo S., Wöhr M., Simola N. & Bonini S. A. Mouse and rat ultrasonic vocalizations in neuroscience and neuropharmacology: State of the art and future applications. Eur. J. Neurosci. 57, 2062–2096 (2023). [DOI] [PubMed] [Google Scholar]
- 59.Farrell W. J. & Alberts J. R. Stimulus control of maternal responsiveness to Norway rat (Rattus norvegicus) pup ultrasonic vocalizations. J. Comp. Psychol. 116, 297–307 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Hofer M. A., Brunelli S. A. & Shair H. N. Potentiation of isolation-induced vocalization by brief exposure of rat pups to maternal cues. Dev. Psychobiol. 27, 503–517 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Zhang L. et al. Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. Journal of Clinical Microbiology 48, 2965–2967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu F., Ma R., Wang Y. & Zhang L. The clinical importance of Campylobacter concisus and other human hosted Campylobacter species. Frontiers in Cellular and Infection Microbiology 8, 243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirk K. F., Nielsen H. L., Thorlacius-Ussing O. & Nielsen H. Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathogens 8, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vielkind P., Jentsch H., Eschrich K., Rodloff A. C. & Stingu C.-S. Prevalence of Actinomyces spp. in patients with chronic periodontitis. Int. J. Méd. Microbiol. 305, 682–688 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Iablokov S. N., Novichkov P. S., Osterman A. L. & Rodionov D. A. Binary metabolic phenotypes and phenotype diversity metrics for the functional characterization of microbial communities. Frontiers in Microbiology 12, 653314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeow M. et al. Analyses of energy metabolism and stress defense provide insights into Campylobacter concisus growth and pathogenicity. Gut Pathogens 12, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cornelius A. J. et al. Genetic characterisation of Campylobacter concisus: Strategies for improved genomospecies discrimination. Systematic and Applied Microbiology 44, (2021). [DOI] [PubMed] [Google Scholar]
- 68.Wang Y. et al. Campylobacter concisus genomospecies 2 is better adapted to the human gastrointestinal tract as compared with Campylobacter concisus genomospecies 1. Frontiers in Physiology 8, 548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahendran V. et al. The prevalence and polymorphisms of zonula occluden toxin gene in multiple Campylobacter concisus strains isolated from saliva of patients with inflammatory bowel disease and controls. PLoS ONE 8, e77525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aagaard M. E. Y., Kirk K. F., Nielsen H. L., Steffensen R. & Nielsen H. Campylobacter concisus from chronic inflammatory bowel diseases stimulates IL-8 production in HT-29 cells. Gut Pathogens 15, 5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Litvak Y., Byndloss M. X. & Bäumler A. J. Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivera-Chávez F. & Bäumler A. J. The pyromaniac inside you: Salmonella metabolism in the host gut. Annual Review of Microbiology 69, 31–48 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Rivera-Chávez F. et al. Energy taxis toward host-derived nitrate supports a Salmonella pathogenicity island 1-independent mechanism of invasion. mBio 7, e00960–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winter S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faber F. et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534, 697–699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCall T. B., Feelisch M., Palmer R. M. J. & Moncada S. Identification of N-iminoethyl-l-ornithine as an irreversible inhibitor of nitric oxide synthase in phagocytic cells. Br. J. Pharmacol. 102, 234–238 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rogawski E. T. et al. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. The Lancet Global Health 6, e1319–e1328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vonaesch P. et al. Stunted children display ectopic small intestinal colonization by oral bacteria, which cause lipid malabsorption in experimental models. Proceedings of the National Academy of Sciences 119, e2209589119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vonaesch P. et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proceedings of the National Academy of Sciences of the United States of America 115, E8489–E8498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dinh D. M. et al. Longitudinal analysis of the intestinal microbiota in persistently stunted young children in south India. PLoS ONE 11, e0155405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coker M. O. et al. Metagenomic analysis reveals associations between salivary microbiota and body composition in early childhood. Scientific Reports 12, 13075 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nattramilarasu P. K., Sá F. D. L. de, Schulzke J. D. & Bücker R. Immune-mediated aggravation of the Campylobacter concisus-induced epithelial barrier dysfunction. International Journal of Molecular Sciences 22, 2043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fahim S. M. et al. Evidence of gut enteropathy and factors associated with undernutrition among slum-dwelling adults in Bangladesh. American Journal of Clinical Nutrition 111, 657–666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Acosta A. M. et al. Relationship between growth and illness, enteropathogens and dietary intakes in the first 2 years of life: Findings from the MAL-ED birth cohort study. BMJ Global Health 2, e000370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bankhead P. et al. QuPath: Open source software for digital pathology image analysis. Scientific Reports 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rieger M. A. & Dougherty J. D. Analysis of within subjects variability in mouse ultrasonic vocalization: Pups exhibit inconsistent, state-like patterns of call production. Front. Behav. Neurosci. 10, 182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kolmogorov M. et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nature Methods 17, 1103–1110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P. & Tyson G. W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Research 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Y. W., Simmons B. A. & Singer S. W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Nayfach S., Shi Z. J., Seshadri R., Pollard K. S. & Kyrpides N. C. New insights from uncultivated genomes of the global human gut microbiome. Nature 568, 505–510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olm M. R., Brown C. T., Brooks B. & Banfield J. F. DRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME Journal 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mikheenko A., Saveliev V. & Gurevich A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 32, 1088–1090 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Chaumeil P. A., Mussig A. J., Hugenholtz P. & Parks D. H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olson R. D. et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic acids research 51, D678–D689 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Letunic I. & Bork P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Research 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stämmler F. et al. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 4, 28–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wolf A. R. et al. Bioremediation of a common product of food processing by a human gut bacterium. Cell Host and Microbe 26, 463–477.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McNulty N. P. et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science Translational Medicine 3, 106ra106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kolmogorov M., Yuan J., Lin Y. & Pevzner P. A. Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 102.Kanehisa M., Sato Y. & Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of Molecular Biology 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Overbeek R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Research 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aziz R. K. et al. SEED servers: High-performance access to the SEED genomes, annotations, and metabolic models. PLoS ONE 7, e48053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rodionov D. A. et al. Micronutrient requirements and sharing capabilities of the human gut microbiome. Frontiers in Microbiology 10, 1316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashniev G. A., Petrov S. N., Iablokov S. N. & Rodionov D. A. Genomics-based reconstruction and predictive profiling of amino acid biosynthesis in the human gut microbiome. Microorganisms 10, 740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frolova M. S., Suvorova I. A., Iablokov S. N., Petrov S. N. & Rodionov D. A. Genomic reconstruction of short-chain fatty acid production by the human gut microbiota. Frontiers in Molecular Biosciences 9, 949563 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hibberd M. C. et al. Bioactive glycans in a microbiome-directed food for malnourished children. Nature 625, 157–165 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bray N. L., Pimentel H., Melsted P. & Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 110.Soneson C., Love M. I. & Robinson M. D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Durinck S., Spellman P. T., Birney E. & Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Law C. W., Chen Y., Shi W. & Smyth G. K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ritchie M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu G., Wang L. G., Han Y. & He Q. Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A Journal of Integrative Biology 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chang H.-W. et al. Prevotella copri and microbiota members mediate the beneficial effects of a therapeutic food for malnutrition. Nat. Microbiol. 9, 922–937 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fleming S. J., Marioni J. C. & Babadi M. CellBender remove-background: A deep generative model for unsupervised removal of background noise from scRNA-seq datasets. bioRxiv (2019) doi: 10.1101/791699. [DOI] [Google Scholar]
- 117.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hafemeister C. & Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choudhary S. & Satija R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biology 23, 27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McGinnis C. S., Murrow L. M. & Gartner Z. J. DoubletFinder: Doublet detection in single-cell RNA-sequencing data using artificial nearest neighbors. Cell Systems 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moor A. E. et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175, 1156–1167 (2018). [DOI] [PubMed] [Google Scholar]
- 122.Amezquita R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nature Methods 17, 137–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aran D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature Immunology 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Street K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scrucca L., Fop M., Murphy B. T. & Raftery A. E. Mclust 5: clustering, classification, and density estimation using gaussian finite mixture models. The R Journal 8, 289–317 (2016). [PMC free article] [PubMed] [Google Scholar]
- 127.Berge K. V. den et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nature Communications 11, 1201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mi H. et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nature Protocols 14, 703–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a, Diets used in gnotobiotic mouse studies. Ingredients in and nutritional analysis of each diet. b, Cultured isolates and taxonomic classification from BEED study duodenal aspirates. c, MAG assembly information and MAG taxonomic classification. (i), Assembly and taxonomic classification. (ii), Sample metadata. d, Abundances of MAGs from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 animals, P14 animals and adult female mice. (ii), Cohousing experiment. (iii), Isolate add-in experiments. Absolute abundance values are log10-transformed. P-adj values determined by Wilcoxon rank-sum tests and FDR correction.
a, Protein levels in tissues from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 duodenum, ileum, colon, serum and cerebral cortex; P14 duodenum, ileum, colon and serum. (ii), Cohousing experiments: duodenum, ileum, colon and serum. (iii), Isolate add-in experiments: duodenum, colon and serum. b, Body weights from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 animals, P14 animals and conceptuses. (ii), Cohousing experiments. P-values for table (i) were determined using Wilcoxon rank-sum tests; those for table (ii) were determined using linear mixed effects modeling. c, Micro-computed tomography of femurs of P37 animals from the intergenerational experiment. P-values were determined using Wilcoxon rank-sum tests. d, Histomorphometric analysis of small intestines of P37 animals from the intergenerational transmission experiment. Duodenal (i) and ileal (ii) villus height, crypt depth, villus to crypt ratio, and Ki67 quantification. P-values determined using Tukey’s post-hoc tests. e, Immune cell profiling of tissues from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: P37 duodenum, ileum, colon, spleen and meninges; duodenum, ileum, spleen and meninges of the dams. (ii), Cohousing experiment: bulk small intestinal tissue, spleen and meninges. P-values determined using Wilcoxon rank-sum tests. f, Quantification of ultrasonic vocalization metrics in P5-P9 pups.
a, Bulk tissue RNA-Seq from gnotobiotic animal experiments. (i), Intergenerational transmission experiment: gene set enrichment analysis (GSEA) of gene expression in the duodenum, ileum and colon of P37 animals. (ii), Cohousing experiment: differentially expressed genes in duodenal samples for: non-cohoused cSI-I controls versus non-cohoused cSI-N controls and cohoused versus non-cohoused cSI-N controls. There were no statistically significant differentially expressed genes between cohoused and cSI-I controls. GSEA of gene expression in the colon of non-cohoused cSI-I controls vs. non-cohoused cSI-N controls and cohoused vs. non-cohoused cSI-N controls. (iii), Add-in experiment: GSEA of gene expression in the colon. Pathways shown were enriched in all of the following conditions: cohoused animals vs. cSI-N non-cohoused controls, C. concisus + Actinomyces-gavaged animals vs. cSI-N sham-gavaged controls and C. concisus-gavaged animals vs. cSI-N sham-gavaged controls. Values provided (P- and q-values, enrichment scores, rank, leading edge, core enrichment) are for C. concisus + Actinomyces-gavaged vs. cSI-N sham-gavaged animals. b, Proportions of cell types in the duodenums and ileums of P37 animals from the intergenerational transmission experiment. (i and ii), Proportions of all cell types quantified by snRNA-Seq in the duodenum Table i) and ileum (Table ii) (n=3 mice/group). Statistical significance determined by edgeR. c, snRNA-Seq analysis of duodenums and ileums of P37 offspring from intergenerational transmission experiment. Pseudo-bulk differential expression of enterocyte clusters in the duodenum and ileum of cSI-I compared to cSI-N animals (n=3 mice/group; DESeq2 Wald test). d, snRNA-Seq analysis of cerebral cortices of P37 offspring from the intergenerational transmission experiment. (i), Pseudobulk differential expression of glutamatergic neurons in the cerebral cortex (n=4 mice/group; DESeq2 Wald test). (ii), GSEA of significantly (P-adj<0.05) differentially expressed genes in the cortex. (iii), Differential expression of S100A9-associated vs. background glutamatergic neurons. Positive log2-fold change indicates higher expression in S100A9-associated neurons (determined with the FindMarkers function in Seurat). e, GSEA results in the duodenum and colon for bulk RNA-seq datasets. All treatment groups presented were compared to their cSI-N counterparts.
a, Isolate genome assembly information and taxonomic classification. (i), Quality assessment and taxonomic classification of assembled isolate genomes. (ii), Average nucleotide identity (ANI) of isolate genomes with their related MAGs. (iii), Species-specific virulence factors in MAGs, their respective isolate genomes, and related reference genomes. b, In silico metabolic reconstructions of MAGs and isolate genomes. (i), Metabolic annotations for each MAG by gene. (ii), Metabolic annotations for pathology-associated MAGs and phylogenetically related organisms by gene. (iii), Binary phenotype predictions for all MAGs. (iv), Binary phenotype predictions for pathology-associated MAGs, their isolate genomes, and phylogenetically related organisms.
Antibodies used for flow cytometry. (i), Antibodies used for flow cytometry of intestinal immune cells in intergenerational transmission and cohousing experiments. (ii and iii), Antibodies used for flow cytometry of splenic and meningeal immune cells in intergenerational transmission (ii) and cohousing experiments (iii).
Sequencing metadata from gnotobiotic animal experiments. a, Intergenerational transmission experiment: (i) short read shotgun DNA sequencing, (ii) snRNA-Seq, (iii) bulk tissue RNA-Seq and (iv) microbial RNA-Seq. b, Cohousing experiment: (i) short read shotgun DNA sequencing, (ii) bulk tissue RNA-Seq and (iii) and microbial RNA-Seq. c, Isolate add-in experiments: (i) short read shotgun DNA sequencing and (ii) and bulk tissue RNA-Seq.
Data Availability Statement
Datasets generated by shotgun sequencing of DNA isolated from the intestinal contents of gnotobiotic mice and from individual cultured bacterial strains, plus snRNA-Seq and bulk RNA-Seq of intestinal tissue and cerebral cortex, and microbial RNA-Seq of cecal contents harvested from gnotobiotic mice have been deposited at the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number ERA22531095.