Abstract
Background
3D brachial plexus MRI scanning is prone to examination failure due to the lengthy scan times, which can lead to patient discomfort and motion artifacts. Our purpose is to investigate the efficacy of artificial intelligence-assisted compressed sensing (ACS) in improving the acceleration efficiency and maintaining or enhancing the image quality of brachial plexus MR imaging.
Methods
A total of 30 volunteers underwent 3D sampling perfection with application-optimized contrast using different flip angle evolution short time inversion recovery using a 3.0T MR scanner. The imaging protocol included parallel imaging (PI) and ACS employing acceleration factors of 4.37, 6.22, and 9.03. Radiologists evaluated the neural detail display, fat suppression effectiveness, presence of image artifacts, and overall image quality. Signal intensity and standard deviation of specific anatomical sites within the brachial plexus and background tissues were measured, with signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) subsequently calculated. Cohen's weighted kappa (κ), One-way ANOVA, Kruskal–Wallis and pairwise comparisons with Bonferroni-adjusted significance level. P < 0.05 was considered statistically significant.
Results
ACS significantly reduced scanning times compared to PI. Evaluations revealed differences in subjective scores and SNR across the sequences (P < 0.05), with no marked differences in CNR (P > 0.05). For subjective scores, ACS 9.03 were lower than the other three sequences in neural details display, image artifacts and overall image quality. There was no significant difference in fat suppression. For objective quantitative evaluation, SNR of right C6 root in ACS 6.22 and ACS 9.03 was higher than that in PI; SNR of left C6 root in ACS 4.37, ACS 6.22 and ACS 9.03 was higher than that in PI; SNR of medial cord in ACS 6.22, ACS 9.03 was higher than that in PI.
Conclusion
Compared with PI, ACS can shorten scanning time while ensuring good image quality.
Keywords: Brachial plexus, Artificial intelligence-compressed sensing, Magnetic resonance imaging, Accelerated acquisition
Introduction
The brachial plexus is a critical nerve network responsible for motor and sensory innervation of the upper limbs and scapula. Due to its complex anatomy, it is vulnerable to a range of pathologies, including traumatic injuries, congenital anomalies, inflammatory conditions, and tumors [1]. Magnetic Resonance Imaging (MRI) has emerged as the diagnostic modality of choice for assessing the brachial plexus, offering superior soft tissue contrast and the ability to image in multiple planes [2]. At present, the 3D Sampling Perfection with Application-optimized Contrast using different flip angle Evolution Short Time Inversion Recovery (3D-SPACE-STIR) sequence is widely utilized for clinical assessments of the brachial plexus [3]. This sequence offers advantages such as multi-planar reconstruction, curved surface reconstruction, and maximum intensity projection, which are invaluable for preoperative planning [4]. However, the extended scan times required for 3D sequences often lead to patient discomfort, leading to motion artifacts and potential scan failures.
To address these limitations and improve clinical examination efficiency, various acceleration technologies have been developed. Among these, k-space undersampling techniques are commonly employed [5], including Partial fourier (HF) acquisition, Parallel imaging (PI) and Compressed sensing (CS) [6]. PF acquisition, while effective, compromises the signal-to-noise ratio (SNR) by collecting only partial k-space data; PI, while utilizing multiple receiving coils and a specialized reconstruction algorithm, can introduce artifacts and noise amplification at high acceleration factors, thereby limiting its efficiency [7]. CS, leveraging the sparsity of MR images to reduce sampling points in k-space, is particularly suited for 3D sequences, which are inherently sparser and contain more redundant information compared to 2D images [8, 9]. Yet, traditional CS approaches often grapple with the challenges of selecting optimal sparsity transforms and adjusting parameters [10].
Artificial intelligence-assisted compressed sensing (ACS) is a novel acceleration technique developed from CS principles, integrating aspects of PF and PI while incorporating a deep learning convolutional network into the reconstruction process. By pre-learning structural information from an extensive dataset of fully sampled MR images, ACS significantly reduces scan times while maintaining or even improving image quality and examination success rates [11]. Its efficacy has been demonstrated across various organ systems, including the brain [12], liver [13], kidneys [14], lumbar spine [15], knee [16] and heart [17]. Several studies have shown that ACS not only substantially shortens MRI scan times compared to PI and traditional CS methods but also maintains or improves image quality and lesion detection capabilities. Despite these advancements, the application of ACS for brachial plexus imaging remains unexplored.
Therefore, this study aimed to evaluate the performance of ACS at different acceleration factors in brachial plexus imaging and compare it with traditional PI methods. The objectives are to assess the acceleration efficiency of ACS in brachial plexus MRI imaging and its impact on image quality, providing valuable insights into the potential clinical benefits of ACS in this specialized imaging domain.
Materials and methods
Participants
This prospective study was approved by the institutional ethical review committee, and informed consent was obtained from all participants prior to enrollment. From September 2023 to October 2023, healthy subjects underwent brachial plexus MRI scanning using a 3.0 T MRI scanner. Exclusion criteria included: (1) a history of neck or back surgery, (2) contraindications for MRI, (3) inability to cooperate with MRI scanning, (4) pregnancy or lactation, (5) images with significant artifacts that were unrelated to the scanning sequence, such as those caused by patient motion or technical issues.
MRI protocol
All examinations were performed on a 3.0 T MRI scanner (United Imaging Healthcare, Shanghai, China) equipped with a 48-channel head and neck coil combined with a large flexible coil for comprehensive coverage. Volunteers were positioned supine in the scanner, with their heads entering first. Both upper and lower arms were supported with rice bags to ensure that the upper limbs remained parallel to the scanner bed. The large flexible coil was strategically placed to cover the upper body, integrating seamlessly with the neck coil and encompassing the humerus on both sides. The imaging protocol consisted of a non-contrast-enhanced 3D-matrix-STIR sequence. Both PI, reconstructed using the Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA) method with an acceleration factor of 2.91, and ACS with acceleration factors of 4.37, 6.22, and 9.03, were employed for accelerated data acquisition. All sequences were obtained in the same order for each volunteer (PI 2.91, ACS 9.03, ACS 6.22, ACS 4.37). Detailed scanning parameters and the respective scanning times for each sequence are presented in Table 1.
Table 1.
Scanning parameters and time of the sequences acquired using PI and ACS
| PI | ACS 4.37 | ACS 6.22 | ACS 9.03 | |
|---|---|---|---|---|
| TR(ms) | 2500 | 2500 | 2500 | 2500 |
| TI(ms) | 300 | 300 | 300 | 300 |
| TE(ms) | 354.48 | 354.48 | 354.48 | 354.48 |
| Voxel(mm) | 1.0 × 1.0 × 1.0 | 1.0 × 1.0 × 1.0 | 1.0 × 1.0 × 1.0 | 1.0 × 1.0 × 1.0 |
| Interpolation | 1.5 | 1.5 | 1.5 | 1.5 |
| FOV(mm) | 400 × 352 | 400 × 352 | 400 × 352 | 400 × 352 |
| Acquisition matrix | 352 × 352 | 352 × 352 | 352 × 352 | 352 × 352 |
| BW(Hz/pixel) | 500 | 500 | 500 | 500 |
| ETL | 184 | 184 | 184 | 184 |
| NEX | 2 | 2 | 2 | 2 |
| Slices | 90 | 90 | 90 | 90 |
| Acceleration factor | 2.91 | 4.37 | 6.22 | 9.03 |
| Scanning time(min:s) | 5:52 | 3:47 | 2:38 | 1:48 |
TR repetition time, TI inversion time, TE echo time, FOV field of view, BW band-width, ETL Echo train length, NEX Number of Excitation
ACS image reconstruction
ACS employs Convolutional Neural Networks (CNN) to expedite image acquisition. While CNN-based methods demonstrate exceptional reconstruction quality, their application in clinical environments is often hindered by unpredictability, attributed to the networks' opaque, "black-box" nature. ACS effectively mitigates this uncertainty by incorporating the output from the AI module as a supplementary constraint within the Compressed Sensing framework. This is achieved through the introduction of a regularization term that addresses the divergence between images reconstructed via traditional methods and those predicted by AI.
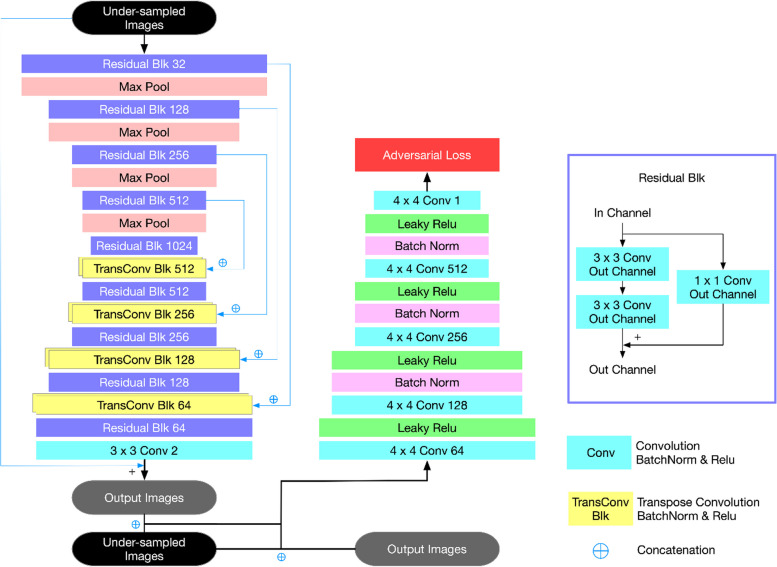
The ACS neural networks have been trained on a substantial dataset comprising two million fully sampled images, sourced from phantoms (2%) and human volunteers (98%). The iteration process of ACS utilizes an architectural design based on k-space that includes multiscale sparsification. This mathematical model amalgamates elements from compressed sensing, partial Fourier, and parallel imaging methodologies. According to simulations detailed in [Zhai, R., et al., Intelligent Incorporation of AI with Model Constraints for MRI Acceleration. Proceedings of the 29th Annual Meeting of ISMRM [Virtual]., 2021.], ACS demonstrates the capability to correct inaccuracies in the AI model’s outputs, aligning closely with the fully-sampled gold standard. Architecture of the deep learning-based MR reconstruction framework used in this study is shown in Fig. 1.
Fig. 1.
Architecture of the deep learning-based MR reconstruction framework used in this study
Objective quantitative evaluation
Quantitative assessments were conducted on a post-processing workstation by a radiologist with over five years of experience in neuroimaging. Regions of Interest (ROIs) were delineated for consistent layers across all four sequences. Specifically, the bilateral C6 root and the medial cord of the brachial plexus were selected for their optimal visibility, with ROIs contoured along the nerve structures to avoid bone and other non-relevant tissues [18]. Additionally, elliptical or circular ROIs measuring 50mm2 (for the sternocleidomastoid and subscapularis muscles) and 100mm2 (for background areas) were carefully positioned to exclude artifacts. Signal intensity (SI) and standard deviation (SD, indicative of noise) were recorded, and the signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) were calculated as objective measures using the following formulas [19]:
Subjective qualitative rating
All images were uploaded to the PACS system, with subject information and sequence parameters anonymized. Two radiologists, each with over five years of diagnostic experience, conducted a double-blind evaluation of the scans. Cases and sequences were randomly assigned and disordered between the radiologists. They assessed the anatomical details of the brachial plexus, efficacy of fat suppression, presence of image artifacts (visual disturbances such as residual aliasing and noise-induced artifacts, which are typical in accelerated imaging), and overall image quality using a 4-point scale [14]: Anatomical detail display of neural structure: 1 point = poor; 2 points = fair; 3 points = good; 4 points = very good; Fat suppression: 1 point = poor; 2 points: fair; 3 points = good; 4 points = very good; Artifacts: 1 point = excessive artifacts; 2 points = substantial artifacts; 3 points = minor artifacts; 4 points = negligible or no artifacts; Overall image quality: 1 point = poor; 2 points = fair; 3 points = good; 4 points = very good. Images scoring 3 or above were considered diagnostic. The average score from both radiologists was used unless discrepancies occurred, in which case a senior radiologist adjudicated.
Statistical analysis
Data analysis was performed using SPSS 21.0 and MedCalc 20.2, with results expressed as mean ± standard deviation or median with interquartile ranges. The SNR, CNR, and subjective scores for the four sequences were analyzed using Kruskal–Wallis or One-way ANOVA tests based on the distribution normality and variance homogeneity of the data. A p-value of less than 0.05 was considered statistically significant. For significant overall differences, pairwise comparisons were conducted with a Bonferroni-adjusted significance level. Inter-observer agreement was evaluated using Cohen's weighted kappa (κ) [20], with the following interpretations: excellent (0.80–1.00), good (0.60–0.80), moderate (0.40–0.60), fair (0.20–0.40), and poor (0.00–0.20).
Results
Demographic characteristics
A total of 31 healthy subjects underwent MRI scanning. One subject was excluded due to poor image quality (unrecognized neck coil leaded to poor signal-to-noise ratio), resulting in a final sample size of 30 volunteers. The study group consisted of 7 males and 23 females, with an age range of 23–68 years and an average age of 38.35 ± 13.22 years.
Scanning time
The scanning times of the four sequences were as follows: PI = 352s; ACS 4.37 = 227s; ACS 6.22 = 158s; ACS 9.03 = 108s. This corresponds to a reduction in scanning time of 35.5%, 55.1%, and 69.3%, respectively, when compared to the PI sequence.
Quantitative findings
The objective quantitative evaluation results are shown in Table 2 and illustrated in Figs. 2 and 3. Significant differences in SNR were observed across the four sequences (p < 0.001), while no significant differences in CNR were found (p > 0.05). Pairwise comparisons revealed that ACS 6.22 and ACS 9.03 exhibited higher SNRs for the right C6 root compared to PI, with ACS 6.22 (p = 0.001) and ACS 9.03 (p < 0.001) showing significant improvements. Similarly, for the left C6 root, all ACS sequences outperformed PI in SNR (ACS 4.37 vs. PI, p = 0.024; ACS 6.22 vs. PI, p < 0.001; ACS 9.03 vs. PI, p < 0.001), with ACS 9.03 also surpassing ACS 4.37 (p = 0.034). Regarding the bilateral medial cord's SNR, ACS 6.22 and ACS 9.03 were superior to PI (ACS 6.22 vs. PI, p = 0.005 (right)/0.002 (left); ACS 9.03 vs. PI, p < 0.001) with ACS 9.03 also outperforming ACS 4.37 (p = 0.013 (right)/0.007 (left)). No significant differences were found between any other pair of sequences.
Table 2.
Quantitative evaluation of the four sequences
| PI | ACS 4.37 | ACS 6.22 | ACS 9.03 | p value of Kruskal–Wallis or ANOVA | |
|---|---|---|---|---|---|
| SNR | |||||
| Right C6 nerve SNR | 44.78 (35.54, 52.20) | 52.45 (42.67, 64.68) | 58.02 (50.43, 66.48) | 68.91 (53.34, 76.82) | < 0.001 |
| Left C6 nerve SNR | 46.25 (41.38, 49.93) | 58.10 (48,34, 62.11) | 62.15 (53.84, 69.77) | 72.54 (59.74, 82.11) | < 0.001 |
| Right medial cord SNR | 29.34 (23.62, 35.03) | 35.04 (28.68, 38.92) | 37.40 (31.99, 47.20) | 43.40 (35.50, 51.40) | < 0.001 |
| Left medial cord SNR | 31.57 (23.33, 36.26) | 34.11 (29.57, 41.89) | 40.82 (32.28, 49.31) | 46.57 (38,56, 54.37) | < 0.001 |
| CNR | |||||
| Right C6 nerve CNR | 16.04 (13.05, 21.10) | 14.21 (10.54, 16.94) | 14.94 (11.66, 18.53) | 13.64 (11.76, 16.34) | 0.102 |
| Left C6 nerve CNR | 11.79 (9.42, 16.47) | 12.03 (8.91, 15.88) | 12.03 (9.21, 16.14) | 12.96 (8.55, 15.74) | 0.981 |
| Right medial cord CNR | 4.72 (3.47, 6.61) | 3.82 (2.98, 5.98) | 5.00 (3.91, 7.03) | 5.29 (3.06, 6.48) | 0.401 |
| Left medial cord CNR | 5.29 ± 2.95 | 4.39 ± 2.34 | 5.08 ± 2.75 | 5.16 ± 3.05 | 0.598 |
Fig. 2.
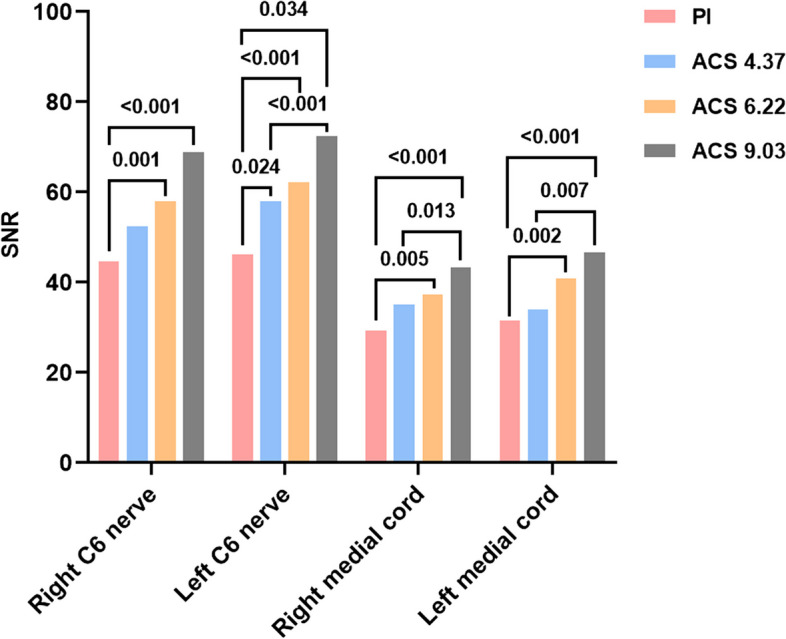
SNR of PI and ACS images in different ROIs. Numbers above the column represents the p-value between two sequences with statistical difference
Fig. 3.
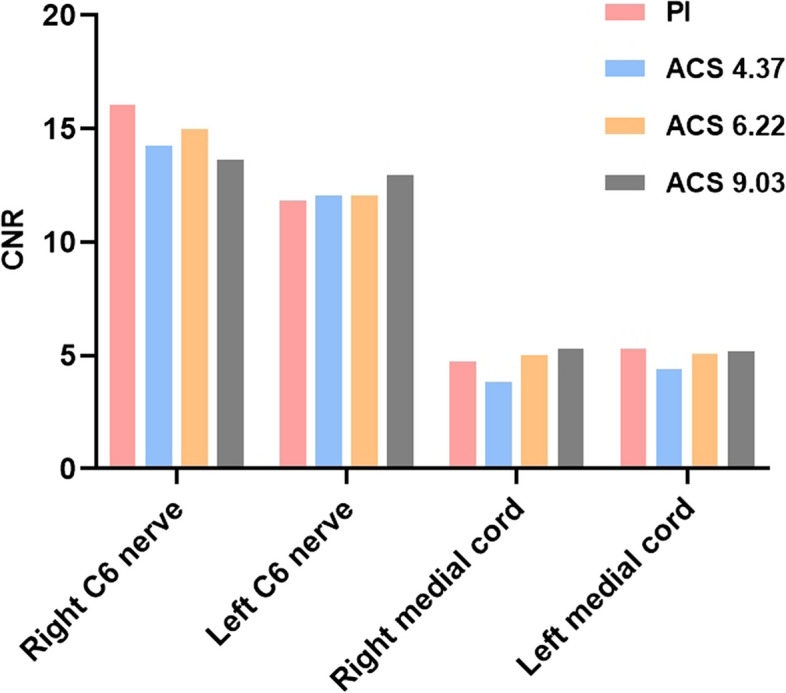
CNR of PI and ACS images in different ROIs. There is no statistical difference between any two sequences
Qualitative findings
The average subjective scores of the two observers are shown in Table 3. No significant differences were found in fat suppression across the sequences, but significant differences were noted in the display of neural detail, presence of image artifacts, and overall image quality (p < 0.001). Pairwise comparisons revealed that ACS 9.03 was inferior in neural detail visualization, image artifacts, and overall image quality compared to the other sequences. Figures 4 and 5 depict these subjective evaluations, with Fig. 4 illustrating a comparative view of the four sequences in a 35-year-old female. Notably, images from the ACS 9.03 sequence exhibited poorer neural detail and increased blurriness, diminishing diagnostic confidence.
Table 3.
Qualitative scores for the four sequences
| PI | ACS 4.37 | ACS 6.22 | ACS 9.03 | p value of Kruskal–Wallis | p value of PI vs. ACS 4.37 | p value of PI vs. ACS 6.22 | p value of PI vs. ACS 9.03 | p value of ACS 4.37 vs. ACS 6.22 | p value of ACS 4.37 vs. ACS 9.03 | p value of ACS 6.22 vs. ACS 9.03 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Neural details diplay | 4 (4, 4) | 4 (4, 4) | 4 (4, 4) | 3 (3, 3) | < 0.001 | 1.000 | 1.000 | < 0.001 | 1.000 | < 0.001 | < 0.001 |
| Fat sat | 4 (3, 4) | 4 (3, 4) | 4 (3, 4) | 4 (3, 4) | 0.883 | ||||||
| Artifacts | 4 (4, 4) | 4 (3, 4) | 4 (3, 4) | 3 (3, 3.50) | < 0.001 | 1.000 | 0.299 | < 0.001 | 1.000 | 0.003 | 0.029 |
| Image quality | 4 (4, 4) | 4 (4, 4) | 4 (3, 4) | 3 (3, 3) | < 0.001 | 0.349 | 0.501 | < 0.001 | 1.000 | < 0.001 | < 0.001 |
Fig. 4.
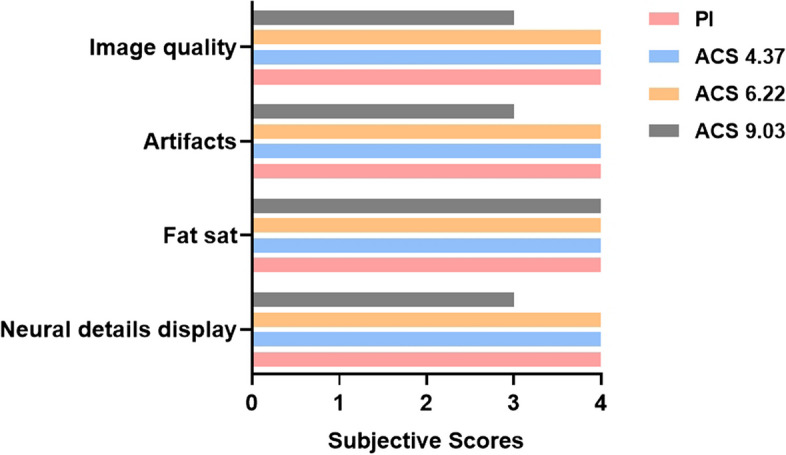
Subjective scores representing image quality of PI and ACS images
Fig. 5.
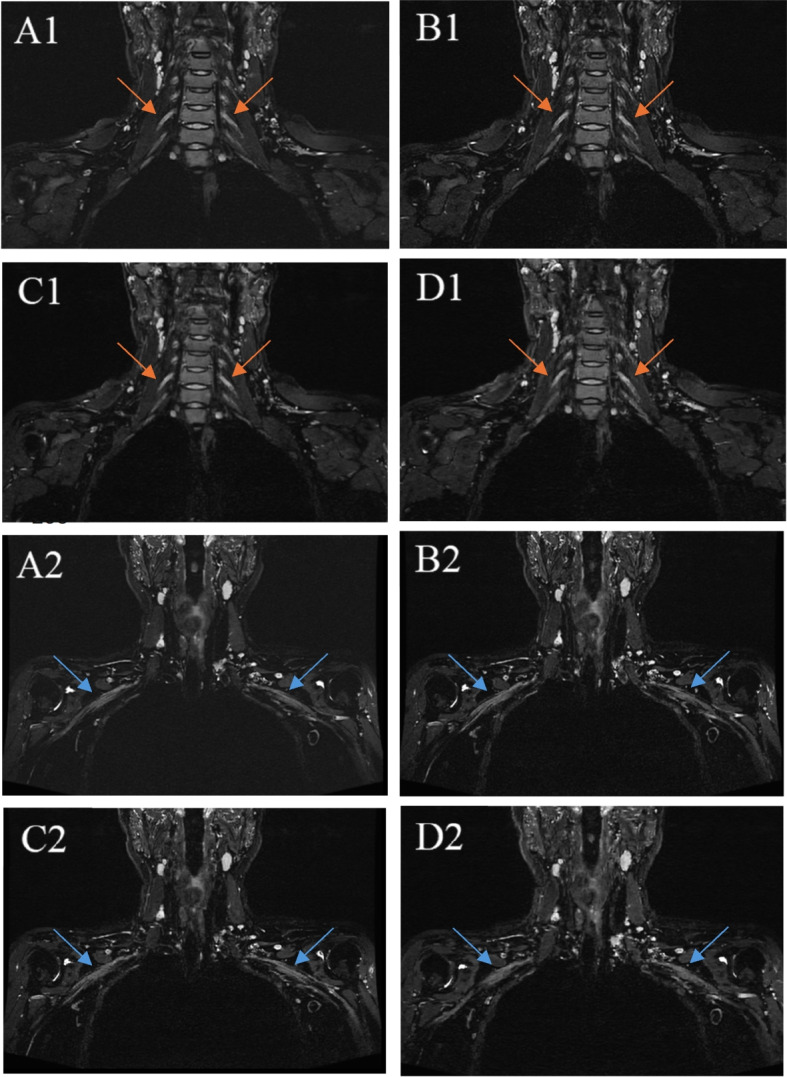
(A1)(A2), PI; (B1)(B2), ACS 4.37; (C1)(C2), ACS 6.22; (D1)(D2), ACS 9.03. Orange arrows in (A1)(B1)(C1)(D1) indicate right C6 nerve and left C6 nerve. Blue arrows in (A1)(B2)(C2)(D2) indicate right medial cord and feft medial cord. It can be observed that ACS 9.03 shows poor neural details and blurs compared to other images, which leads to insufficient diagnostic confidence
Kappa consistency score
As shown in Table 4, inter-observer agreement on subjective scores was found to be good across sequences, with a kappa (κ) score greater than 0.60.
Table 4.
Interobserver agreement levels for qualitative scores (κ)
| PI | ACS 4.37 | ACS 6.22 | ACS 9.03 | |
|---|---|---|---|---|
| Neural details diplay | 1.000 | 1.000 | 1.000 | 0.789 |
| Fat sat | 1.000 | 1.000 | 0.874 | 0.760 |
| Artifacts | 1.000 | 0.944 | 0.861 | 0.603 |
| Image quality | 1.000 | 1.000 | 0.938 | 0.625 |
Discussion
Throughout the evolution of brachial plexus imaging, various MRI sequences such as 2D Dixon, 2D STIR, and DWI have been employed, each offering unique insights into nerve structure and pathology [21]. Despite the availability of these techniques, 3D sequences are widely recognized for their superior diagnostic effectiveness [22]. While 2D sequences can delineate the peripheral nerve bundle structure, 3D sequences offer isotropic acquisition, allowing for a more precise representation of nerve morphology [23]. Additionally, the enhanced resolution and contrast of 3D sequences facilitate the simultaneous visualization of all major peripheral nerve branches, aiding radiologists in assessing nerve involvement [24]. However, the lengthy scan times associated with 3D peripheral nerve imaging—often exceeding six minutes—pose significant challenges for patient compliance, increasing the risk of motion artifacts. Reducing scan times without compromising image quality remains a critical challenge in clinical practice. This study employed three different acceleration factors of ACS (low: 4.37, medium: 6.22, high: 9.03) to image the brachial plexus nerve and compared them with PI. The scanning time and image quality of each sequence were observed and compared, leading to the selection of the optimal acceleration factor for clinical scanning of the brachial plexus nerve.
Fat suppression is crucial in brachial plexus imaging as it differentiates nerves from surrounding fat, muscle, and blood vessels, thereby enhancing image clarity [25], and reducing artifacts [26]. In this study, STIR was chosen over other fat suppression techniques due to the inhomogeneity of the main magnetic field (B0) in the shoulder and neck area, which can limit the effectiveness of B0-dependent methods [27]. The principle of STIR's fat suppression is largely unaffected by B0 field inhomogeneity, making it particularly well-suited for brachial plexus imaging.
Upon determining the sequence selection, four sequences were scanned using different acceleration factors: PI, ACS 4.37, ACS 6.22, and ACS 9.03. The ACS sequences achieved a reduction in scanning time by 35.5%, 55.1%, and 69.3%, respectively, compared to PI. The reconstruction time for ACS was notably short, with reconstructed images available immediately after scanning. This efficiency in reducing scanning time is consistent with previous studies on ACS [12–17].
For quantitative evaluation, the C6 root and the medial cord of the brachial plexus—crucial for innervating the forearm muscles—were selected as areas for measurement. The analysis demonstrated that the signal-to-noise ratio (SNR) for the four sequences showed an increasing trend across these nerves, with ACS 6.22 and ACS 9.03 showing statistically higher SNR compared to PI. This enhancement is attributed to the superior suppression of background noise with increased acceleration factors in ACS, consistent with the results of Wang et al.'s study on the knee joint [16].
CNR essentially reflects the contrast delineating the signal disparity between the region of interest and the background. The discernibility of objects against the background hinges on their size and contrast. In this study, the sternocleidomastoid and subscapularis muscles were chosen as the background to maintain a stable signal value for these tissues. The findings revealed no significant difference in CNR among the four sequences, suggesting that both PI and ACS sequences effectively differentiate the nerve from surrounding tissues without compromising image contrast.
There have been a few previous studies using acceleration techniques for brachial plexus imaging. A study on deep learning compressed sensing (DLCS) [28] demonstrated that sequences with an intermediate acceleration factor could achieve higher SNR and CNR without reducing scan time, while sequences with high acceleration factors matched the performance of traditional sequences but halved the scan time. This improvement is attributed to advanced denoising and undersampling strategies enabled by deep learning. The ACS technology utilized in this study, incorporating a deep learning convolutional network, emphasizes AI's role in enhancing image quality by reducing noise and efficiently sampling k-space, indirectly shortening scan times without compromising image contrast. Another previous study [29] used 3D magnetic resonance neurography (MRN) acquisitions with deep learning reconstruction (DLR) for brachial plexus imaging. The results indicate that faster 3D MRN scans reconstructed with DL were similar to standard exams with regard to discrete measurements of image quality, including SNR, edge sharpness, bulk motion, and nerve conspicuity, as well as subjective assessments of nerve signal intensity, size, and morphology. This is similar to our research findings. But the fastest scanning time for this study still reached 4 min, which is 52% longer than the optimal time we studied. Meanwhile, applying this technology, k-space data needs to be reconstructed using a separate workstation, which takes about 2 min, This will increase the time cost and computational load on the machine. ACS technology can perform automatic reconstruction on the host in no more than 1 min, resulting in better overall efficiency. Pribowo et al. [26] also scanned 10 volunteers to explore the effectiveness of CS in non-contrast-enhanced brachial plexus imaging, revealing a premise similar to the current research. Their findings highlighted that CS-enhanced images exhibited superior SNR and CNR, leading to improved definition at the edges of images. The present study, employing ACS, advances beyond traditional CS techniques. While ACS did not surpass PI in terms of CNR, it matched PI's performance, ensuring high-quality imaging across various acceleration factors. Furthermore, it was noted that MIP reconstruction for brachial plexus could introduce unwanted signals from background tissues and overlapping nerves, potentially skewing image metric measurements. Consequently, MIP reconstructions were omitted in favor of direct measurements from the original images, ensuring more accurate and reliable evaluations.
In terms of subjective scores, ACS 9.03 underperformed relative to the other three sequences in terms of neural detail display, image artifacts, and overall image quality. This outcome illustrates that while higher acceleration factors in ACS may improve objective quantitative metrics, they can adversely affect neural detail visualization, leading to blurred images and reduced diagnostic confidence (arrow in Fig. 5). Research has indicated that excessive acceleration factors in ACS can result in inadequate sparse sampling, potentially causing aliasing artifacts [30]. Additionally, the effective fat suppression observed across all sequences (> 3.5), attributed to the use of STIR, which minimally affects the uniformity of the main magnetic field, showed no significant differences, suggesting fat suppression does not influence the choice of ACS acceleration factor. Consequently, selecting an optimal acceleration factor involves balancing various aspects of image quality. The study advocates for a medium acceleration factor of 6.22 as the preferred choice for clinical scanning to ensure high-quality imaging outcomes. While our findings indicate that higher acceleration factors may lead to decreased visual quality, this study did not directly evaluate the impact on diagnostic confidence for detecting specific pathologies. Therefore, caution should be exercised when extrapolating these results to clinical practice. Further studies involving patients with known pathologies are needed to determine whether decreased image quality at higher accelerations affects diagnostic sensitivity and specificity.
This study has several limitations. First, the small sample size and single-center nature of the data collection might introduce bias to the conclusions. However, the consistency in image evaluation suggests that the findings could be cautiously generalized. Nonetheless, larger multi-center studies are necessary to mitigate data bias and enhance the reliability of the conclusions. Second, the study primarily examines image quality across different acceleration factors using volunteer samples, thus not addressing the potential effects of ACS on lesion detection and diagnostic efficacy for brachial plexus-related conditions. Third, the study's categorization of acceleration factors into low, medium, and high represents a broad classification. Future research should explore additional acceleration factors and include comparative studies with CS to refine guidance on the optimal acceleration factor for ACS in brachial plexus imaging. Forth, all sequences were acquired in the same order for each volunteer. This consistent ordering may introduce a potential bias, as sequences performed later could be more prone to motion artifacts due to increased patient discomfort or fatigue. Future research should consider randomizing the order of sequences to minimize systematic effects on image quality.
In summary, ACS has demonstrated a significant reduction in MRI scanning times while maintaining high image quality. For brachial plexus imaging, an ACS acceleration factor of 6.22 is recommended, effectively reducing scanning duration to under three minutes. However, further research involving patients with pathologies is necessary to validate the clinical utility of this acceleration method. Future studies should focus on assessing the diagnostic accuracy of ACS across different conditions to determine its effectiveness in clinical practice. This finding introduces a novel and efficient scanning protocol for clinical application, potentially enhancing patient throughput and diagnostic efficiency.
Acknowledgements
The authors would like to express our enormous appreciation and gratitude to all participants.
Abbreviations
- MRI
Magnetic Resonance Imaging
- 3D-SPACE-STIR
3D Sampling Perfection with Application-optimized Contrast using different flip angle Evolution Short Time Inversion Recovery
- HF
Partial fourier
- PI
Parallel imaging
- CS
Compressed sensing
- ACS
Artificial intelligence-assisted compressed sensing
- CNN
Convolutional Neural Networks
- ROIs
Regions of Interest
- SI
Signal intensity
- SD
Standard deviation
- SNR
Signal-to-noise ratio
- CNR
Contrast-to-noise ratio
- MRN
Magnetic resonance neurography
- DLR
Deep learning reconstruction
Authors’ contributions
CTX wrote the main Manuscript text; LFF assisted in completing data analysis; JXT helped scan the image; YD modified the article; WJ implemented sequence parameter setting and optimization; YY provided assistance in experimental design; XH formed the team and provided machines and technical support. All authors have made a significant contribution to this study and have approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (52227814) and General program of the National Natural Science Foundation of China (82371904).
Data availability
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was conformed to the ethical guidelines of the 1964 Declaration of Helsinki as reflected by prior approval from ethical review committee of the Beijing Friendship Hospital of Capital Medical University(2022-P1-020–02). All subjects were informed and signed informed permission.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tianxin Cheng and Feifei Li have contributed equally to this paper.
Tianxin Cheng and Feifei Li are co first authors.
Contributor Information
Ying Yuan, Email: 715337108@qq.com.
Hui Xu, Email: mr_xuhui@163.com.
References
- 1.Bowen BC, Pattany PM, Saraf-Lavi E, Maravilla KR. The brachial plexus: normal anatomy, pathology, and MR imaging. Neuroimaging Clin N Am. 2004;14(1):59–85. [DOI] [PubMed] [Google Scholar]
- 2.Sneag DB, Mendapara P, Zhu JC, Lee SC, Lin B, Curlin J, Bogner EA, Fung M. Prospective respiratory triggering improves high-resolution brachial plexus MRI quality. J Magn Reson Imaging. 2018;49(6):1723–9. [DOI] [PubMed] [Google Scholar]
- 3.Vargas MI, Viallon M, Nguyen D, Beaulieu JY, Delavelle J, Becker M. New approaches in imaging of the brachial plexus. Eur J Radiol. 2010;74(2):403–10. [DOI] [PubMed] [Google Scholar]
- 4.Fritz J, Ahlawat S. High-Resolution Three-dimensional and Cinematic Rendering MR Neurography. Radiology. 2018;288(1):25. [DOI] [PubMed] [Google Scholar]
- 5.Lv J, Wang C, Yang G. PIC-GAN: A Parallel imaging coupled generative adversarial network for accelerated multi-channel MRI reconstruction. Diagnostics (Basel, Switzerland). 2021;11(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Dumoulin C. Correlation imaging for multiscan MRI with parallel data acquisition. Magn Reson Med. 2012;68(6):2005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glockner JF, Hu HH, Stanley DW, Angelos L, King K. Parallel MR imaging: a user’s guide. Radiographics. 2005;25(5):1279–97 a review publication of the Radiological Society of North America, Inc. [DOI] [PubMed] [Google Scholar]
- 8.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–95. [DOI] [PubMed] [Google Scholar]
- 9.Geethanath S, Reddy R, Konar AS, Imam S, Sundaresan R, D R RB, Venkatesan R. Compressed sensing MRI: a review. Crit Rev Biomed Eng. 2013;41(3):183–204. [DOI] [PubMed] [Google Scholar]
- 10.Bustin A, Fuin N, Botnar RM, Prieto C. From Compressed-Sensing to Artificial Intelligence-Based Cardiac MRI Reconstruction. Frontiers in Cardiovascular Medicine. 2020;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazurowski MA, Buda M, Saha A, Bashir MR. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging. 2019;49(4):939–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu K, Xi B, Sun H, Wang J, Chen C, Wen X, Zhang Y, Zeng M. The clinical feasibility of artificial intelligence-assisted compressed sensing single-shot fluid-attenuated inversion recovery (ACS-SS-FLAIR) for evaluation of uncooperative patients with brain diseases: comparison with the conventional T2-FLAIR with parallel imaging. Acta Radiol. 2023;64(5):1943–9. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Hu C, Yang Y, Zhao Y, Lin C, Li Z, Liu Q. Single-breath-hold T2WI MRI with artificial intelligence-assisted technique in liver imaging: As compared with conventional respiratory-triggered T2WI. Magn Reson Imaging. 2022;93:175–80. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Peng C, Wang S, Liang X, Meng X. The feasibility investigation of AI -assisted compressed sensing in kidney MR imaging: an ultra-fast T2WI imaging technology. BMC Med Imaging. 2022;22(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui H, Gong Y, Liu L, Lv Z, Zhang Y, Dai Y, Mo Z. Comparison of Artificial Intelligence-Assisted Compressed Sensing (ACS) and Routine Two-Dimensional Sequences on Lumbar Spine Imaging. J Pain Res. 2023;16:257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Zhao W, Xing X, Wang Y, Xin P, Chen Y, Zhu Y, Xu J, Zhao Q, Yuan H, et al. Feasibility of AI-assisted compressed sensing protocols in knee MR imaging: a prospective multi-reader study. Eur Radiol. 2023;33(12):8585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X, Ran L, Zou L, Luo Y, Yang Z, Zhang S, Zhang S, Xu J, Huang L, Xia L. Dark blood T2-weighted imaging of the human heart with AI-assisted compressed sensing: a patient cohort study. Quant Imaging Med Surg. 2023;13(3):1699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greening J, Anantharaman K, Young R, Dilley A. Evidence for Increased Magnetic Resonance Imaging Signal Intensity and Morphological Changes in the Brachial Plexus and Median Nerves of Patients With Chronic Arm and Neck Pain Following Whiplash Injury. J Orthop Sports Phys Ther. 2018;48(7):523–32. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Deng D, Zeng W, Huang Y, Zheng C, Li X, Li H, Xie C, He H, Xu G. AI-assisted compressed sensing and parallel imaging sequences for MRI of patients with nasopharyngeal carcinoma: comparison of their capabilities in terms of examination time and image quality. Eur Radiol. 2023;33(11):7686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni M, He M, Yang Y, Wen X, Zhao Y, Gao L, Yan R, Xu J, Zhang Y, Chen W, et al. Application research of AI-assisted compressed sensing technology in MRI scanning of the knee joint: 3D-MRI perspective. Eur Radiol. 2023;34(5):3046–58. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Wang G, Yang L, Wu L, Lin X, Chhabra A. Diffusion-weighted MR neurography of extremity nerves with unidirectional motion-probing gradients at 3 T: feasibility study. AJR Am J Roentgenol. 2013;200(5):1106–14. [DOI] [PubMed] [Google Scholar]
- 22.Chhabra A, Thawait GK, Soldatos T, Thakkar RS, Del Grande F, Chalian M, Carrino JA. High-resolution 3T MR neurography of the brachial plexus and its branches, with emphasis on 3D imaging. AJNR Am J Neuroradiol. 2013;34(3):486–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshmukh S, Tegtmeyer K, Kovour M, Ahlawat S, Samet J. Diagnostic contribution of contrast-enhanced 3D MR imaging of peripheral nerve pathology. Skeletal Radiol. 2021;50(12):2509–18. [DOI] [PubMed] [Google Scholar]
- 24.Sneag DB, Daniels SP, Geannette C, Queler SC, Lin BQ, de Silva C, Tan ET. Post-Contrast 3D inversion recovery magnetic resonance neurography for evaluation of branch nerves of the brachial plexus. Eur J Radiol. 2020;132:109304. [DOI] [PubMed] [Google Scholar]
- 25.Jung J-Y, Lin Y, Carrino JA. An Updated Review of Magnetic Resonance Neurography for Plexus Imaging. Korean J Radiol. 2023;24(11):1114–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pribowo MA, Harahap MIR, Fazharyasti V, Dwihapsari Y, Kartikasari Y, Sugiyanto RN. Non-contrast enhancement of brachial plexus magnetic resonance imaging with compressed sensing. Eur J Radiol. 2023;165:110890. [DOI] [PubMed] [Google Scholar]
- 27.Davidson EJ, Tan ET, Pedrick EG, Sneag DB. Brachial Plexus Magnetic Resonance Neurography. Invest Radiol. 2023;58(1):14–27. [DOI] [PubMed] [Google Scholar]
- 28.Hu S-x, Xiao Y, Peng W-l, Zeng W, Zhang Y, Zhang X-y, Ling C-t, Li H-x, Xia C-c, Li Z-l. Accelerated 3D MR neurography of the brachial plexus using deep learning–constrained compressed sensing. Eur Radiol. 2023;34(2):842–51. [DOI] [PubMed] [Google Scholar]
- 29.Sneag DB, Queler SC, Campbell G, Colucci PG, Lin J, Lin Y, Wen Y, Li Q, Tan ET. Optimized 3D brachial plexus MR neurography using deep learning reconstruction. Skeletal Radiol. 2024;53(4):779–89. [DOI] [PubMed] [Google Scholar]
- 30.Feng L, Benkert T, Block KT, Sodickson DK, Otazo R, Chandarana H. Compressed sensing for body MRI. J Magn Reson Imaging. 2017;45(4):966–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.