Abstract
Peritoneal metastasis (PM) is a common metastasis site and death cause of gastric cancer, which is a complex biological process, but there is currently a lack of effective prediction and treatment targets. In this study, we first analyzed the differential gene expression of gastric cancer patients with or without peritoneal metastasis, and identified the HECT domain E3 ubiquitin protein ligase 2 (HECTD2) as the core gene of PM in gastric cancer. The current study shows that the role of HECTD2 in tumor is contradictory. In this study, our results show that the low expression of HECTD2 indicates that the survival rate of overall survival (OS), progression-free survival (PFS), disease-specific survival (DSS), and disease-free survival (DFS) are better, and can be used as an important component of prognostic indicators. In addition, through pathway enrichment analysis, we found that HECTD2 was mainly involved in metastasis related pathways such as extracellular matrix remodeling and cell adhesion in gastric cancer, and high expression of HECTD2 could activate epithelial-mesenchymal transition (EMT) metastasis related pathways in gastric cancer. In regulating the metastasis of gastric cancer cells, HECTD2 can also change the surrounding microenvironment, induce the enrichment of interstitial components and build an immune microenvironment conducive to tumor progression, while patients with low expression of HECTD2 may be more likely to benefit from immunotherapy. In conclusion, HECTD2 may be a novel biomarker for the diagnosis and prognosis of peritoneal metastasis of gastric cancer, providing basis for the mechanism of peritoneal metastasis of cancer and clinical medication.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03553-5.
Keywords: Gastric cancer, HECTD2, Peritoneal metastasis, Prognosis, Tumor microenvironment
Introduction
Gastric cancer, with the 5th highest incidence and 3rd highest fatality rate worldwide, is one of the most deadly malignancies of the digestive system [1]. Many gastric cancer patients were diagnosed at the advanced stage due to the lack of typical clinical manifestations in early stage, which were often accompanied by multiple types of metastases [2, 3]. Among them, peritoneal metastasis is the most characteristic type of metastasis in advanced gastric cancer, which indicates poor prognosis and is one of the main reasons leading to treatment failure [4, 5]. Peritoneal metastasis of gastric cancer refers to the growth of primary tumor cells in the peritoneum through blood flow, lymph node or implantation [6]. Patients with peritoneal metastasis have poor treatment efficacy, rapid progression, and short survival time, with the 5-year survival rate less than 2% [7]. However, the mechanism of peritoneal metastasis of gastric cancer has not been elucidated now and effective treatment strategy is still lacking. Therefore, it is necessary to further explore potential biomarkers for the diagnosis and outcome prediction for peritoneal metastasis of gastric cancer.
HECTD2 is one of the homologous to E6AP C-terminus (HECT) E3 ubiquitin ligases in humans [8], which plays a variety of biological functions in cells by acting on different substrates. For example, some studies show that PIAS1 (protein inhibitor that activates STAT-1) is the direct target of HECTD2 [9], PIAS1 is an E3 SUMO protein ligase that negatively regulates the inflammatory pathway. Its degradation is necessary for the activation of the largest inflammatory pathway-NF-κB in the lung and the innate immunity [9]. The role of HECTD2 in cancer is limited, and its role in different tumors is contradictory. HECTD2 has been identified as a candidate driver gene in neuroblastoma [10], also in melanoma, it can promote tumor cell proliferation and immune escape [11]. In RCC, as the downstream of HIF-1α, it aggravates the malignant progression of renal cell cancer [12]. However, in prostate cancer, it serves as a potential target for miR-221, which can promote androgen-dependent growth of prostate cancer cell lines. Therefore, HECTD2 has antiproliferative effect in this type of cancer [13], and has drawn similar conclusions in colorectal cancer [14]. The latest research shows that human intestinal microbial-derived propionic acid coordinates the degradation of proteasome through the up-regulation of HECTD2 to target EHMT2 in cancer [15]. These results suggest that HECTD2 may have different roles in different tumors, and its mechanism is still unclear.
In this study, we first divided the patients into different functional modules through the analysis of GSE62254, and screened out the yellow modules that are significantly related to peritoneal metastasis. The core gene HECTD2 was screened through gene network construction. HECTD2 was highly expressed in patients with peritoneal metastasis, and the prognosis of patients with gastric cancer with high expression of HECTD2 was poor, including OS, PFS, DSS, and DFS. In gastric cancer samples, we found that the expression of HECTD2 was significantly higher in gastric cancer than in adjacent normal tissues. Further pathway analysis shows that HECTD2 may play a role in promoting cancer by participating in extracellular matrix remodeling and immune microenvironment remodeling. In addition, Multiple immunofluorescence results showed that HECTD2 expression was significantly positively correlated with M2 macrophages and negatively correlated with M1 macrophage infiltration. In addition, the effective rate of using immune checkpoint inhibitors in gastric cancer patients with low expression of HECTD2 may be higher. These results provide a basis for clinical prognosis prediction and drug application.
Materials and methods
Datasets processing
The GSE62254 datasets was downloaded from the gene expression omnibus (GEO) database of the National Center for Biotechnology Research (http://www.ncbi.nlm.nih.gov/gds). In Table 1, we present 298 gastric cancer patients and their clinical and pathological characteristics, as well as survival data materials (Table 1). Clinical materials including the age, gender, TNM stage, tumor grade, site of tumor metastasis and survival time. Patients with incomplete information such as survival or pathological stage were excluded. For module detection, the RNA-seq data was included for subsequent analysis and we eliminate two outlier samples (GSM1523817, GSM1523984) according to sample network from our datasets (Additional Figs. 1B and 1C). In Additional Fig. 1A, we demonstrate the research process of this study to make our research ideas easier to understand. RNA information is sourced from the STAD-PRJEB25780 dataset and can be downloaded from the STAD website. The TCGA-STAD dataset was downloaded from TCGA database (https://cancergenome.nih.gov).
Table 1.
Characteristics of GSE62254 cohort
| Characteristics | Number of patients (%) |
|---|---|
| Age(years) | |
| Median | 64 (24–86) |
| Gender | |
| Female | 100 (33.6) |
| Male | 198 (66.4) |
| T stage | |
| T1 | 0 (0) |
| T2 | 187 (62.8) |
| T3 | 90 (30.2) |
| T4 | 21 (7.0) |
| N stage | |
| N0 | 37 (12.4) |
| N1 | 131 (44.0) |
| N2 | 80 (26.8) |
| N3 | 50 (16.8) |
| M stage | |
| M0 | 250 (83.9) |
| M1 | 48 (16.1) |
| Lauren | |
| Intestinal | 144 (48.3) |
| Diffuse | 134 (45.0) |
| Mixed | 17 (5.7) |
| Indeterminate | 2 (0.7) |
| Unknown | 1 (0.3) |
| Peritoneal seeding | |
| Yes | 54 (18.1) |
| No | 244 (81.9) |
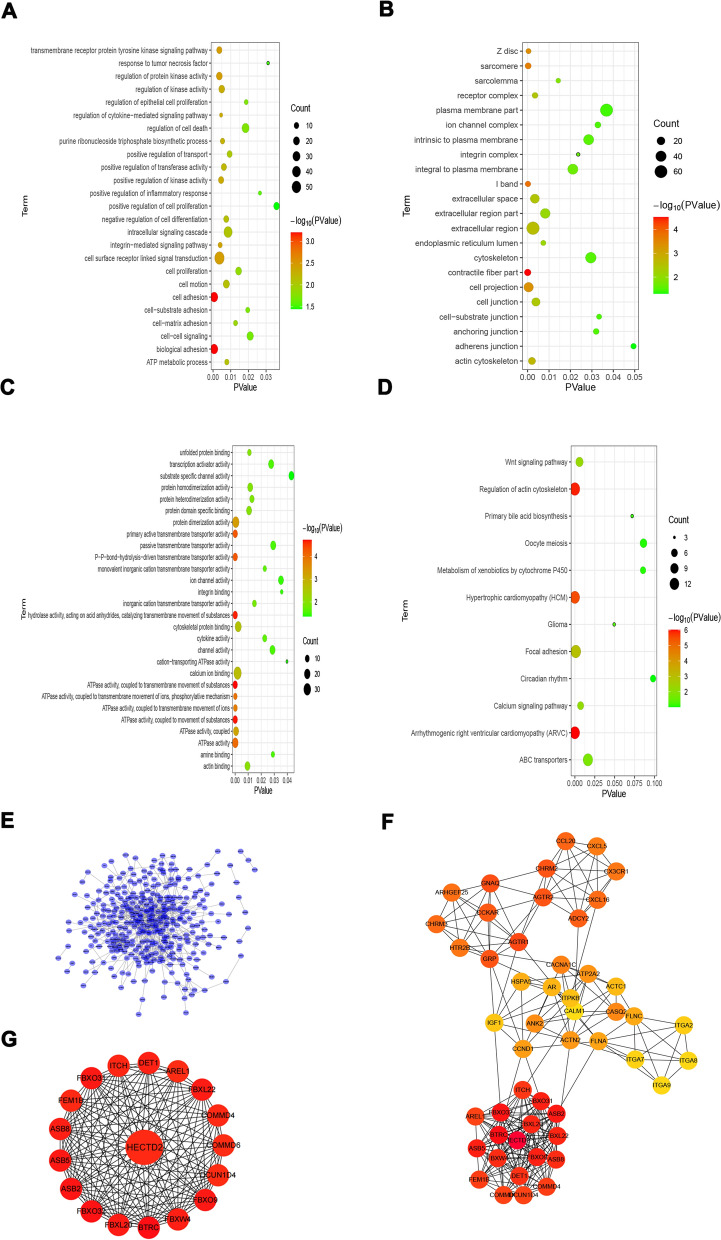
Fig. 1.
Go ontology and KEGG pathway enrichment analyses of the genes in the yellow module. A GO functional (Biological Process, BP) enrichment analyses of genes in the yellow module. B GO functional (Cellular Component, CC) pathway enrichment analyses of genes in the yellow module. C GO functional (Molecular Function, MF) pathway enrichment analyses of genes in the yellow module. D KEGG pathway enrichment analyses of genes in the yellow module. (The –log10 (P-value) of each term is colored according to the legend). E PPI network of genes in yellow module. F Hub network of top 50 hub genes. G The significant sub-module was identified by MCODE
Network construction
In order to screen modules with highly correlated genes and construct a weighted gene co- expression network, we applied WGCNA for analysis [16]. We first used WGCNA to identify important modules from thousands of genes. Further determine the key basis for GC peritoneal metastasis based on the correlation between gene expression and sample traits. In this study, the weighted adjacency matrix was created with the formula amn =|cmn|β (where amn: adjacency between gene m and gene n, cmn: Pearson’s correlation, and β: soft-power threshold). The modules were clustered by calculated the TOM matrix. We defined the minimal gene module size as 30 to obtain appropriate modules, and the threshold to merge similar modules was set to 0.25.
Identifying modules and functional enrichment analysis
Correlation between modules and different clinical features was assessed by calculating the module eigengene (ME). Pearson’s correlation test was used to analyse the relationship between gene significance (GS) and clinical features by linear regression, P < 0.05 indicated significant correlation.
The construction of protein–protein interaction network
In this work, we constructed a PPI network by using the online database STRING (https://cn.string-db.org/). The Cytoscape software was then applied to analyze the interaction of the candidate proteins. A plug-in named The Molecular Complex Detection (MCODE) was used to score and explore parameters that have been optimized to produce the best results for the network. For this, we set the following parameters in MCODE: Degree Cut-off = 2, Node Score Cut-off = 0.2, K-Core = 2 and Max Depth = 100 [17, 18].
Establishment of the nomogram
The nomogram was established with T, N, M, gender, grade, age and HECTD2 expression by R “rms” and “survival” packages. Calibration curves were further used to assess the accuracy of nomograms in differentiating patient groups.
The intratumor immune landscape and cancer antigenome
The Cancer ImmunoTome Atlas (https://tcia.at/) describes the intratumor immune landscape and cancer antigenome of gastric cancer [19]. The difference of immune score in different groups according to the difference of HECTD2 expression level were detected.
Tumor Mutational Burden (TMB)
The RNA sequencing data, gene mutation data, and clinical data of STAD were downloaded from the TCGA database (https://gdc-portal.nci.nih.gov/). The correlation analysis was conducted between the expression level of HECTD2 and TMB in each sample.
GSEA enrichment analysis
The gene set enrichment analysis (GSEA) of the high and low expression subgroups of HECTD2 in the GSE62254 dataset was performed by R “clusterprofiler” package. We further used R "enrichplot" package to draw gene set enrichment maps. P < 0.05 was considered statistically significant.
Immunoinfiltration analysis
SsGSEA method was used to investigate the immune cells, immune-related functions, immune-related pathways and different infiltration of immune celltypes in GSE62254 database by Rpacket "GSVA" [20]. Based on the expression of RNA-seq, the Stromal Score, Immune Score, ESTIMATE Score and Tumor Purity of every sample in the GSE62254 database were evaluated by R package "estimate" [21]. The R package "ggpubr" was used to show violplots of Stromal Score, Immune Score and ESTIMATE Score. In order to investigate the differences in immune cell subtypes, we further applied R package "CIBERSORT" to calculate the proportion of 22 immune cells in all GC samples [22].
Enrichment analysis
In order to explore the role of HECTD2 in gastric cancer, the cBioPortal for cancer genomics (http://www.cbioportal.org) was used to get genes that are positively correlated with HECTD2 in gastric cancer, then screen for gene sets with a correlation coefficient greater than or equal to 0.3 [23, 24]. Then, we employed Metascape (http://metascape.org) [25] to perform enrichment analysis on HECTD2 related genes obtained from cBioPortal.
Survival analysis and HECTD2 expressionin different subtypes and pathways of gastric cancer
GSCA Lite online tool (http://bioinfo.life.hust.edu.cn/web/GSCALite/) [26] was used to analyze the HECTD2 expression in different subtypes and pathways of gastric cancer. The difference was evaluated by Spearman’s test.
Database used to explore HECTD2 coexpression networks
The LinkedOmics database (http://www.linkedomics.org/login.php) [27] was used to determine the HECTD2 coexpression genes by using Pearson’s correlation coefficient and showed the results via heat maps. Then, we explored the Gene Ontology biological process (GO_BP), and KEGG pathways of HECTD2 and its coexpression genes by using gene set enrichment analysis (GSEA).
Patients
From January 2018 to December 2022, 26 patients undergoing gastrectomy for gastric cancer and 24 advanced GC patients receiving ICIs and chemotherapy in Changzhou No.2 People's hospital. 26 patients diagnosed with gastric cancer were enrolled as the following criteria: GC with stage I–III; no neoadjuvant therapy before surgery; Received standard adjuvant therapy after operation; Provide standard treatment after disease progression. For 24 inoperable patients with advanced gastric cancer, the biopsy specimens have been confirmed to have a definite diagnosis of gastric cancer. All the above patients were followed up in Changzhou No.2 People’s Hospital. The total follow-up period was 70 months. The clinicopathological characteristics of these patients were summarized in Tables 2 and 3. Cancerous and adjacent normal tissues were collected during surgery or puncturation, and histopathologically confirmed and staged according to the Union for International Cancer Control. Patients’ written informed consents and approval from the Ethics Committees of Changzhou No.2 People’s Hospital (No.2017-C-015-01) were obtained for the use of these clinical materials.
Table 2.
Clinicopathological characteristics of patients with gastric cancer
| Valuables | Category | Characteristics |
|---|---|---|
| Gender | Male | 19 (73.1%) |
| Female | 7 (26.9%) | |
| Age | Median (range) | 67 (38–82) |
| Nuclear grade | I | 2 (7.7%) |
| II | 8 (30.8%) | |
| III | 16 (61.5%) | |
| Vascular invasion | No | 11 (42.3%) |
| Yes | 15 (57.7%) | |
| T stage | 1 | 5 (19.2%) |
| 2 | 2 (7.7%) | |
| 3 | 10 (38.5%) | |
| 4 | 9 (34.6%) | |
| N stage | 0 | 9 (34.6%) |
| > 1 | 17 (65.4%) |
Table 3.
Clinicopathological characteristics of patients treated with ICIs
| Characteristics | Category | Number |
|---|---|---|
| Gender | Male | 15 (62.5%) |
| Female | 9 (37.5%) | |
| Age | Median (range) | 64.5 (45–80) |
| ECOG PS at ICI initiation | 0 | 3 (12.5%) |
| 1 | 10 (41.7%) | |
| 2 | 11 (45.8%) | |
| PD-L1 expression | > 1% | 14 (58.3%) |
| < 1% | 10 (41.7%) | |
| Antibiotic use | Yes | 3 (12.5%) |
| No | 21 (87.5%) | |
| Corticosteroids use | Yes | 14 (58.3%) |
| No | 10 (41.7%) | |
| M1 phenotype of Macrophages | Median (range) | 4.29 (0.30–19.48) |
| M2 phenotype of Macrophages | Median (range) | 6.68 (1.40–18.41) |
| CEA (ng/ml) | Median (range) | 3.16 (0.27–1000.00) |
| HECTD2 | Low expression | 14 (58.3%) |
| High expression | 10 (41.7%) |
Multiplex immunofluorescence
Multiplex immunofluorescence was performed with the TSA kit (H-D110061,yuanxibio) according to the protocol of the manufacturer. And perform microwave treatment to remove primary and secondary antibodies while maintaining a complete fluorescence signal. Repeat the process until all antigens are stained with their respective fluorophores.
The antibodies were diluted as follows: Anti-CD68 (#BX50031-C3, Biolynx), anti-HLA-DR (#ab92511, Abcam), anti-PanCK (#GM351507, Gene Tech), DAPI (FP1490A, PerkinElmer), working fluid.
The stained slides were scanned to obtain multispectral images using the Pannoramic MIDI imaging system (3D HISTECH). Images were analyzed by using Indica Halo software.
Immunohistochemistry (IHC)
Tissue samples from human gastric cancer and adjacent tissues were originally fixed with 10% formalin for 48 h at room temperature followed with ethanol tissue dehydration and replacement with Xylene before embedding tissues into paraffin blocks. The paraffin tissue sections were cut at 5-μm and deparaffinized with xylene and rehydrated with a graded series of ethanol. Set the oven to a temperature of 65 °C and bake at a constant temperature for 120 min. Performing according to immunohistochemistry kit. The staining was evaluated by scanning the entire tissue specimen under low magnification (× 10) and confirmed under high magnification (× 20 and × 40). The protein expression was visualized and classified based on the percentage of positive cells and the intensity of staining. After 30 min blocking with the universal blocking serum (Dako Diagnostics, Carpinteria, CA), the sections were incubated with anti-HECTD2 antibody at 4 °C overnight and washed 3 times with PBS at room temperature. Then a secondary antibody was added for 30 min incubation (Dako Diagnostics). The samples were washed 3 times with PBS and developed using DAB followed by counterstaining with hematoxylin. Dehydration was performed following a standard procedure and the slides were sealed with cover slips. Images were scanned with a digital pathology slide scanner (KF-BIO, CHINA).
HECTD2 immunostaining signals were evaluated by two researchers, with the clinical information blinded to them, and scored. Brown cytoplasmic staining for HECTD2 was considered positive. The percentage of HECTD2-positive cells was scored with the following four categories: 1 (< 25%), 2 (25–50%), 3 (50–75%), and 4 (> 75%). The staining intensity of positive cells was scored as 0 (absent), 1 (weak infiltration), 2 (moderate infiltration), and 3 (strong infiltration). The final score was the product of the intensity and the percentage.
Survival analyses
Statistical analyses was performed by SPSS software 21.0. It was determined that P < 0.05 was statistical significant.
Results
The analysis of co-expression network
We constructed co-expression networks using the WGCNA algorithm implemented in the R WGCNA package. The approximate scale-free topology distribution of the network is 3 after selecting the suitable soft-thresholding power (Additional Fig. 2A). Consequently, we finally chose power value 3 to perform subsequent analysis. In order to identify modules, topological overlaps were clustered hierarchically using the "average" algorithm, and dynamic tree cutting was used for module identification. Finally, a total of 16 modules were identified (Additional Fig. 2B). Correlations between modules and phenotypes were showed in Additional Fig. 2C. The ME of each module was calculated. The number of genes in modules ranged from 38 to 1,465, and all unassigned genes were assigned to module grey. The characteristic genes of each module were calculated for cluster analysis. The purpose of the analysis is to identify modules with high adjacency.
The module-trait correlation analysis and functional enrichment analysis of modules
The correlations between patient features and module characteristics were processed. We realized that yellow, pink, salmon, magenta and brown modules were significantly positively correlated with peritoneal metastasis. So we use the separameters to define peritoneal metastasis (Additional Fig. 3A). Furthermore, red, light cyan, magenta, black and midnight blue modules were negatively correlated with peritoneal metastasis. To figure out the genes most closely associated with peritoneal metastasis, we finally selected the yellow module which is the most closely related to the characteristics of peritoneal metastasis in GC (Additional Figs. 3B,C). There were 434 genes in the yellow module. Gene Ontology (GO) enrichment analysis of 434 genes was performed and the result showed that the biological adhesion, cell adhesion and cell surface receptor linked signal transduction were the most prominently enriched pathways (Fig. 1A–C). Enrichment analysis of KEGG pathway showed that regulation of actin cytoskeleton, focal adhesion and Wnt signaling pathway were significantly associated with peritoneal metastasis (Fig. 1D). All the above results showed that genes in the yellow module were significantly correlated with tumorigenesis.
Identification of vital candidate marker for peritoneal metastasis
It was considered that hub genes had the largest intramodular connections between modules. These genes, as the center of network, play significant roles in the network. To select critical candidate markers of peritoneal metastasis, we further built a PPI information network, which contained 434 nodes and 936 edges (Fig. 1E). Input the top 50 hub genes into Cytoscape software for interaction network mapping, and further select to construct the central network (Fig. 1F). In order to explore the critical candidate marker of peritoneal metastasis from hub genes, we further selected the top significant sub-module (red module) from 3 clusters according to the MCODE score (Fig. 1G) and the seed gene HECTD2 was finally identified in the top network.
HECTD2 is highly expressed in gastric cancer patients with peritoneal metastasis and the prognosis of patients with high expression is poor
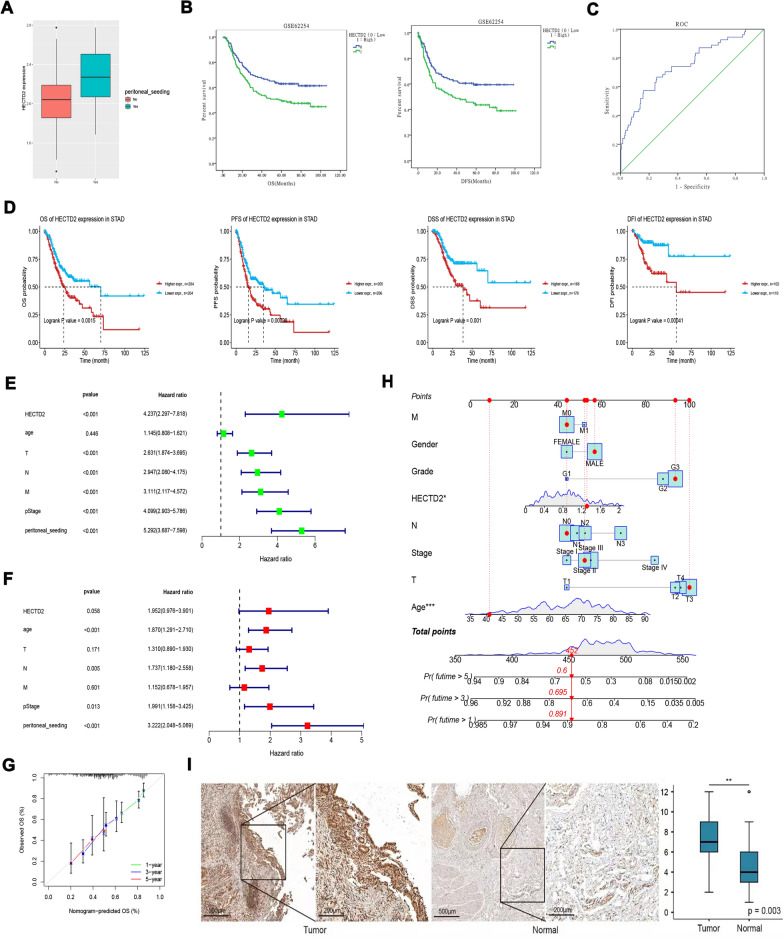
Through preliminary screening, we focused on the core gene HECTD2. Firstly, we applied GSE62254 data to detect the difference in gene expression of HECTD2 in patients with or without peritoneal metastasis in gastric cancer. The results showed that the expression of HECTD2 was significantly higher in patients with peritoneal metastasis of gastric cancer (Fig. 2A). In addition, in gastric cancer patients with high expression of HECTD2, both OS and PFS (Fig. 2B) were poorer compared to the low expression group in the GSE62254 dataset. Then, we drew the receiver operating characteristic (ROC) curve according to the expression of HECTD2 in tumor tissue. We can see that the area under curve (AUC) value obtained from the ROC method was 0.762 (95% CI 0.682–0.873, P < 0.001) (Fig. 2C), which shows that HECTD2 has good prognostic value. Finally, Kaplan–Meier survival analysis revealed the relationship between the expression of HECTD2 and the OS, PFS, DSS, and DFS of patients with gastric cancer. The results showed that OS and DSS of gastric cancer patients with high expression of HECTD2 were poorer than those of low expression of gastric cancer patients (Fig. 2D). At the same time, PFS and DFS also showed a trend of less benefit for patients with high expression of gastric cancer (Fig. 2D). Further univariate and multivariate Cox analyses were conducted on the expression of HECTD2, age, TNM stage, and peritoneal metastasis. The results showed that HECTD2 had the highest prognostic impact index in univariate analysis (HR = 4.237) (Fig. 2E), except for peritoneal metastasis. However, in multivariate analysis, although HECTD2 maintained an impact index advantage, it appeared to be statistically weak (P = 0.058) (Fig. 2F). Based on this result, we build a nomogram, and the calibration diagram shows that the nomogram has similar performance compared with the ideal model (Fig. 2G). This nomogram combines clinically-related pathological parameters and provides a quantitative method for clinicians to predict the probability of 1-year, 3-year and 5-year OS in patients with gastric cancer (Fig. 2H). Each patient will get a score for each prognostic parameter. The higher the total score, the worse prognosis of the patient. Next, we detected the expression level of HECTD2 protein in 26 groups of gastric cancer and adjacent specimens. The IHC results showed that HECTD2 expression was significantly higher in gastric cancer than in adjacent specimens (Fig. 2I). In addition, we downloaded the TCGA-STAD dataset and verified that HECTD2 expression was significantly higher in gastric cancer than in adjacent specimens (Additional Fig. 4A). Meanwhile, high expression of HECTD2 results in shorter PFS and OS in gastric cancer patients (Additional Figs. 4B and 4C).
Fig. 2.
HECTD2 is highly expressed in gastric cancer patients with peritoneal metastasis and the prognosis of patients with high expression is poor. A The differential expressed of HECTD2 in peritoneal metastasis group and non-peritoneal metastasis group (***p < 0.001). B Up-regulated expression of HECTD2 is associated with shorter OS and PFS in GSE62254 dataset. C Receiver operating characteristic curve for HECTD2 expression. D Up-regulated expression of HECTD2 is associated with shorter OS, PFS, DSS and DFS. E The correlations between the risk facrors for OS and clinicopathological factors by univariate Cox regression analysis. F The correlations between the risk factor for OS and clinicopathological factors by multivariate Cox regression analysis. G The calibration curves for predicting patient OS at 1-year, 3-year, and 5-year in the internal verification. H Nomogram model predicting the 1-year, 3-year, and 5-year OS in patients with GC. I Immunohistochemical detection of HECTD2 expression levels in gastric cancer and adjacent normal tissues (magnification × 100) (**p < 0.01)
HECTD2 participates in metastasis of gastric cancer
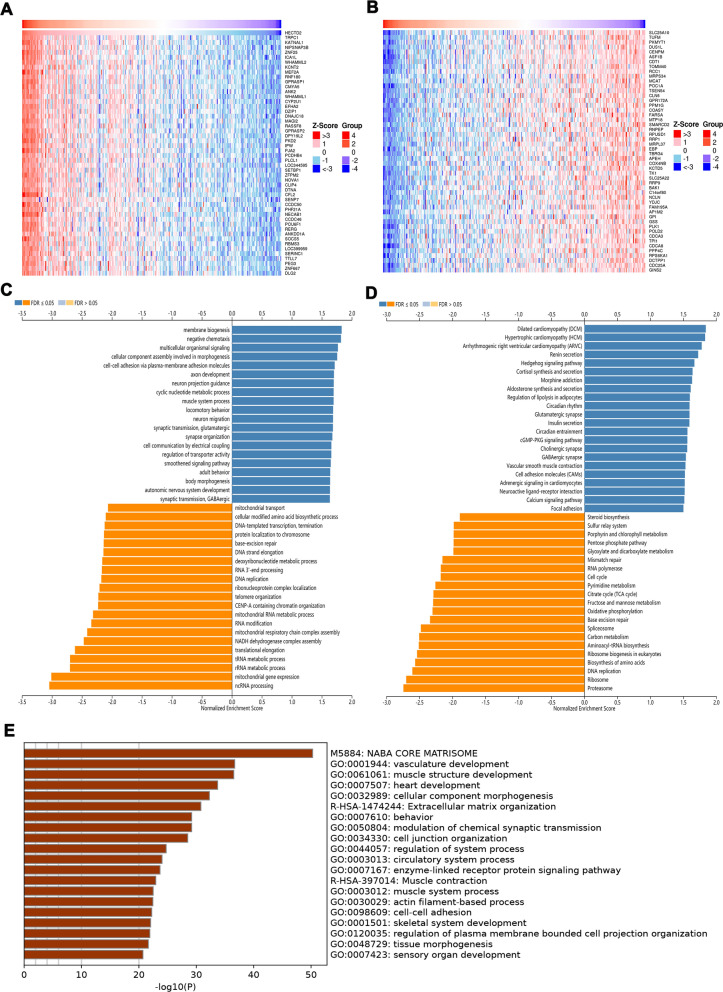
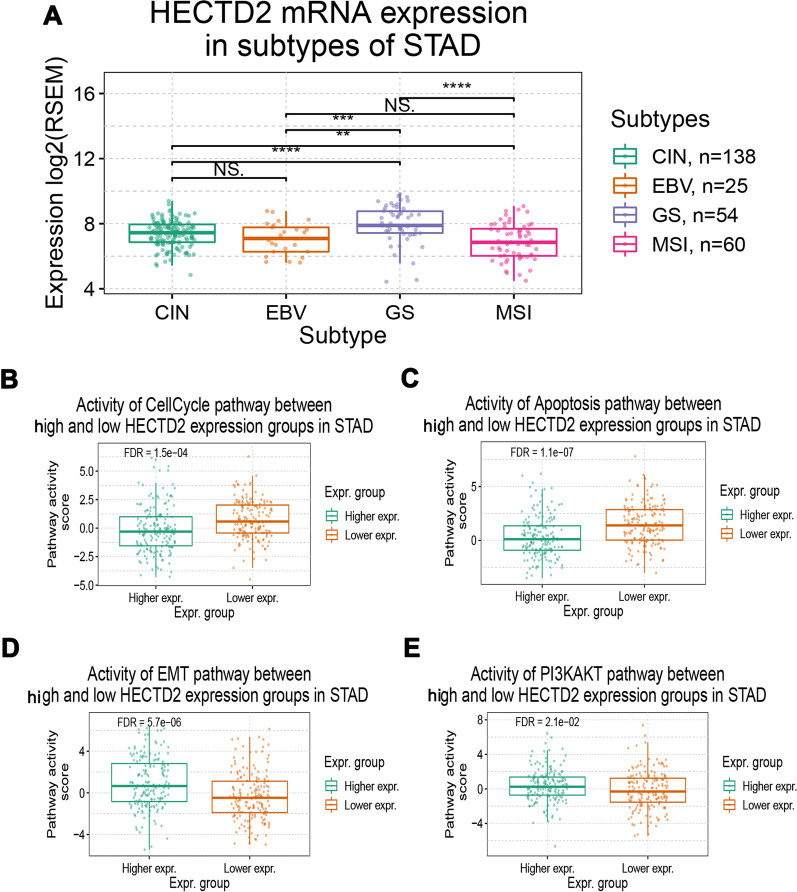
Next, we explore the way in which HECTD2 plays its role in gastric cancer, and use the LinkedOmics online database to explore the potential mechanism of HECTD2. Firstly, we select the dataset of gastric cancer, construct the co-expression network of HECTD2, and use the heat map to display the first 50 genes that are positively or negatively related to HECTD2 (Fig. 3A, B). Next, the biological process categories of GO of HECTD2 co-expression genes were determined by gene set enrichment analysis (GSEA). The results showed that HECTD2 and its co-expression genes were mainly involved in "cell–cell adhesion via plasma-membrane adhesion molecules" (Fig. 3C). Then, we carried out the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis, and the results showed that the co-expressed genes were in "cell adhesion molecules" and “focal adhesion” (Fig. 3D). These results suggest that HECTD2 may promote cancer progression by regulating the adhesion and interstitial remodeling of tumor cells. In addition, we downloaded the TCGA gastric cancer dataset from cBioPortal (http://www.cbioportal.org/) online dataset and the genes with positive correlation with HECTD2, screened the genes with correlation coefficient greater than 0.3, and conducted pathway enrichment analysis by using Metascape (http://metascape.org/). The results showed that the enriched pathways were also concentrated in "cell junction organization" "cell–cell adhesion" (Fig. 3E). Based on these results, we speculate that HECTD2 is mainly involved in the metastasis process of gastric cancer. Therefore, we further analyze the expression of HECTD2 in different subtypes and different pathways of gastric cancer, and found that the expression of HECTD2 gene was the lowest in the MSI subtype of gastric cancer (Fig. 4A). The results of pathway enrichment showed that EMT and PI3K/AKT pathway were significantly activated in patients with high expression of HECTD2 (Fig. 4D, E), while cell cycle and apoptosis were inhibited in patients with high expression, indicating that tumor cell apoptosis pathway (Fig. 4B, C) was inhibited in patients with high expression, and normal apoptosis was inhibited.
Fig. 3.
HECTD2 participates in metastasis of gastric cancer. Download the HECTD2 related gene set from the TCGA-STAD dataset. A The top 50 genes with a positive correlation with HECTD2 genes are visualized in a heatmap. B The top 50 genes with a negative correlation with HECTD2 genes are visualized in a heatmap. C The GO enrichment of the BP terms of HECTD2 co-expressed genes. D The KEGG enrichment of the BP terms of HECTD2 co-expressed genes. E The GO enrichment of the BP terms of HECTD2 co-expressed genes
Fig. 4.
HECTD2 participates in metastasis of gastric cancer. A RNA expression level of HECTD2 in different subtypes of gastric cancer. B–D RNA expression level of HECTD2 in cell cycle (B), apoptosis (C) and EMT (D) pathway of gastric cancer. E RNA expression level of HECTD2 in PI3K/AKT pathway. (**p < 0.01,***p < 0.001,****p < 0.0001)
HECTD2 reshapes tumor microenvironment and is related to the efficacy of immunotherapy
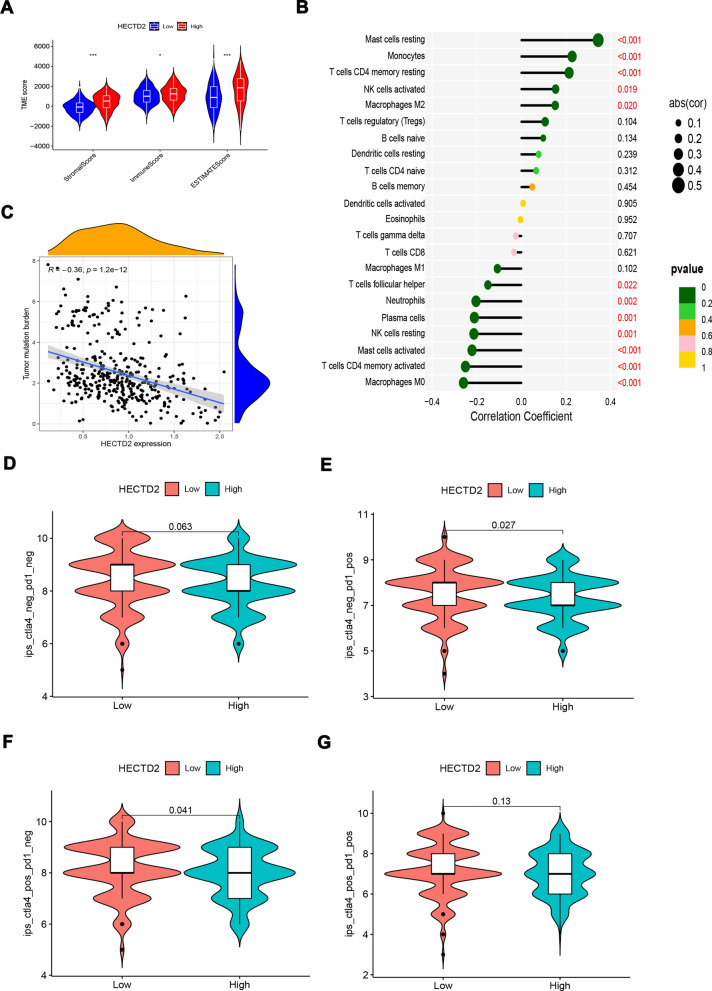
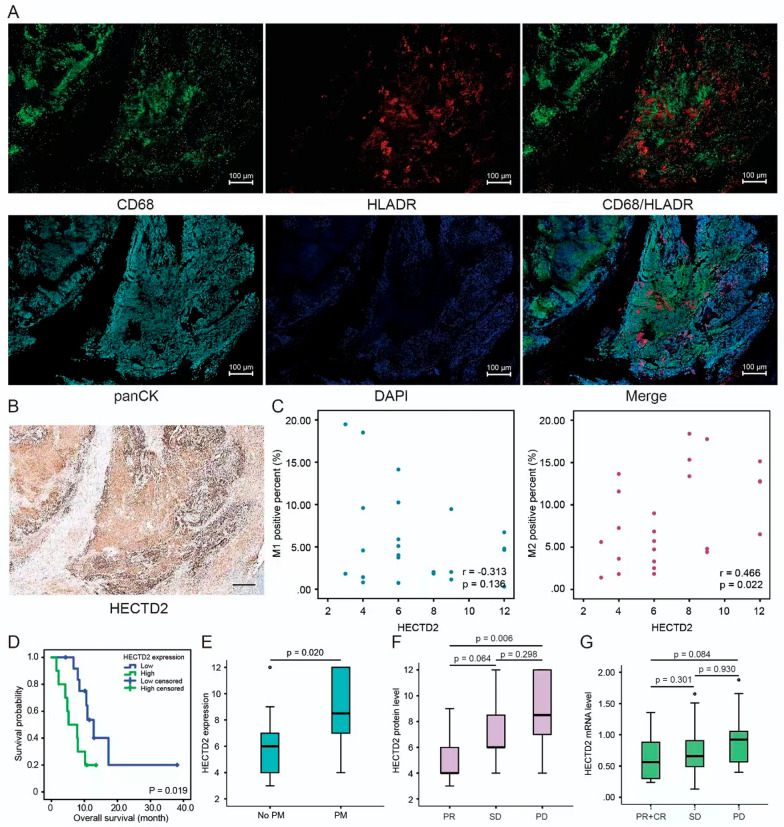
As we found that HECTD2 might play a role in promoting tumor progression through tumor interstitial remodeling, we evaluated the scores of stromal score (matrix in tumor tissue), immunescore (immune cell infiltration in tumor tissue), and estimate score (tumor purity) at different levels of HECTD2 expression. The results showed that these three scores were significantly higher in patients with high expression of HECTD2 gastric cancer (Fig. 5A). It shows that the infiltration of both interstitial and immune cells is increased in patients with high expression, which may be the factor leading to poor prognosis. At the same time, this is consistent with the conclusion of our previous results: that is, the high expression of HECTD2 plays a role by reconstructing the tumor stroma and tumor microenvironment. Next, we evaluated the effect of HECTD2 on the infiltration of immune cells. As shown in Fig. 5B, the high expression of HECTD2 may recruit more "mast cells resting", "Monocytes"and "T cells CD4 memory resting", but reject "Macrophages M0" "T cells" "CD4 memory activated" and "Mast cells activated" cell infiltration. In view of this phenomenon, we further evaluated the relationship between HECTD2 and the efficacy of immunotherapy. Firstly, we tested the tumor mutation burden (TMB) that is currently considered to be related to the efficacy of immunotherapy, and found that it is negatively correlated with the expression of HECTD2 (Fig. 5C), indicating that patients with low expression of HECTD2 may respond better to the treatment of immune checkpoint inhibitors. Then we calculated the immune score, and found that the immune score in the low expression group is higher than that in the high expression group (Fig. 5D–G), which also confirmed that the efficacy of immune checkpoint inhibitors in the low expression group may be better, providing a basis for clinical application of drugs. Based on the significantly positive and negative correlations between HECTD2 and macrophage infiltration, we applied multiple immunofluorescence detection to investigate the correlations between HECTD2 expression and M1, M2 macrophage infiltration in gastric cancer specimens (Fig. 6A). Figure 6B shows that the IHC image of the corresponding slice. The results showed that high expression of HECTD2 significantly induced infiltration of M2 macrophages. Statistical analysis showed a positive correlation between HECTD2 and M2 macrophage expression (r = 0.466, p = 0.022, Fig. 6C), but a negative correlation with M1 macrophages (r = −0.313, p = 0.136). Statistical analysis shows that gastric cancer patients with high HECTD2 expression have a poorer prognosis (Fig. 6D), which is consistent with previous online data analysis results. Among gastric cancer patients with peritoneal metastasis, HECTD2 expression is higher (Fig. 6E), further confirming that HECTD2 is an effective predictive indicator in gastric cancer peritoneal metastasis. Meanwhile, we investigated the relationship between HECTD2 expression and clinical pathological parameters, and found that HECTD2 expression was significantly correlated with T and N staging (Table 4). In addition, we analyzed the correlations between various pathological parameters and the prognosis of gastric cancer, including the expression of HECTD2. The results showed that TNM stage, vascular invasion, and high expression of HECTD2 were poor prognostic factors for gastric cancer (Table 5). Based on this result, we analyzed the indicators related to predicting the efficacy of immunotherapy and found that ECOG score, M1, M2 cell infiltration, and HECTD2 expression were all effective indicators of immunotherapy efficacy. In terms of HR values, HECTD2 expression may be more advantageous, while PD-L1 expression and CEA levels are also predictive indicators of efficacy, but there is no significant statistical significance, which may be due to the limited PD-L1 expression cutoff value and sample size we selected (Table 6). Patients with high HECTD2 expression have poorer treatment response, while those with PR efficacy have lower HECTD2 expression (Fig. 6F) and similar mRNA levels (Fig. 6G). This indicates that HECTD2 not only promotes gastric cancer progression and peritoneal metastasis, but is also an independent prognostic factor for gastric cancer and a predictive factor of immunotherapy efficacy.
Fig. 5.
HECTD2 reshapes the tumor microenvironment and is related to the efficacy of immunotherapy. A Associations of HECTD2 expression to stromal score, immune score and ESTIMATE score in gastric cancer. B The correlations of HECTD2 expression and immune infiltration in gastric cancer. C The correlations of HECTD2 expression and TMB in gastric cancer. D–G The correlations of HECTD2 expression and immune score in gastric cancer
Fig. 6.
HECTD2 reshapes the tumor microenvironment and is related to the efficacy of immunotherapy. The macrophage landscape in the microenvironment of gastric cancer. A A representative image using multiple immunofluorescence. Blue, DAPI; red, HLADR; green, CD68; Sky blue, panCK; Scale bars = 100 μm. B Immunohistochemical staining corresponds to the expression level of HECTD2 protein in consecutive sections. C Correlation between HECTD2 protein expression and M1, M2 macrophages in gastric cancer. D The relationship between HECTD2 protein expression and survival in gastric cancer. E HECTD2 protein expression in gastric cancer patients with and without peritoneal metastasis. F HECTD2 protein expression levels in gastric cancer patients with different immunotherapy efficacy. G HECTD2 mRNA expression levels in gastric cancer patients with different immunotherapy efficacy
Table 4.
Correlations between HECTD2 and clinicopathological characteristics
| Factors | HECTD2 ≤ 6 | HECTD2 > 6 | P-value |
|---|---|---|---|
| Age | |||
| > 60 | 8 (47.1%) | 9 (52.9%) | 0.680 |
| < 60 | 5 (55.6%) | 4 (44.4%) | |
| Gender | |||
| Male | 9 (47.4%) | 10 (52.6%) | 0.658 |
| Female | 4 (57.1%) | 3 (42.9%) | |
| Nuclear grade | |||
| I-II | 6 (60.0%) | 4 (40.0%) | 0.420 |
| III | 7 (43.8%) | 9 (56.3%) | |
| Vascular invasion | |||
| Yes | 7 (46.7%) | 8 (53.3%) | 0.691 |
| No | 6 (54.5%) | 5 (45.5%) | |
| T stage | |||
| 1–2 | 6 (85.7%) | 1 (14.3%) | 0.073 |
| 3–4 | 7 (36.8%) | 12 (63.2%) | |
| N stage | |||
| Positive | 6 (35.3%) | 11 (64.7%) | 0.097 |
| Negative | 7 (77.8%) | 2 (22.2%) | |
Table 5.
Univariate and multivariate analysis of prognostic factors in patients with resectable GC
| Characteristics | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age | ||||||
| > 65 | 1.741 | 0.552–5.493 | 0.344 | |||
| < 65 | Reference | |||||
| Gender | ||||||
| Male | 1.077 | 0.342–3.391 | 0.899 | |||
| Female | Reference | |||||
| Nuclear grade | ||||||
| I-II | 3.059 | 0.960–9.745 | 0.059 | |||
| III | Reference | |||||
| Vascular invasion | ||||||
| Yes | 5.524 | 1.529–19.952 | 0.009 | 5.667 | 1.239–25.923 | 0.025 |
| No | Reference | Reference | ||||
| TNM stage | ||||||
| III | 24.246 | 3.087–190.455 | 0.002 | 12.958 | 1.457–115.695 | 0.022 |
| I-II | Reference | Reference | ||||
| HECTD2 | ||||||
| High expression | 5.747 | 1.746–18.920 | 0.004 | 6.44 | 1.329–31.217 | 0.021 |
| Low expression | Reference | Reference | ||||
Table 6.
Univariate analysis of prognostic factors in GC patients treated with ICIs
| Characteristics | Univariate analysis | ||
|---|---|---|---|
| HR | 95% CI | P-value | |
| Age | |||
| > 65 | 1.118 | 0.392–3.190 | 0.835 |
| < 65 | Reference | ||
| Gender | |||
| Male | 2.018 | 0.677–6.014 | 0.207 |
| Female | Reference | ||
| ECOG PS at ICI initiation | |||
| 2 | 3.241 | 1.098–9.564 | 0.033 |
| 0–1 | Reference | ||
| PD-L1 expression | |||
| > 1% | 0.348 | 0.118–1.022 | 0.055 |
| < 1% | Reference | ||
| Antibiotic use | |||
| Yes | 3.346 | 0.908–12.327 | 0.069 |
| No | Reference | ||
| Corticosteroids use | |||
| Yes | 1.672 | 0.590–4.739 | 0.334 |
| No | Reference | ||
| M1 phenotype of macrophages | 0.832 | 0.710–0.975 | 0.023 |
| M2 phenotype of macrophages | 1.112 | 1.005–1.231 | 0.041 |
| CEA (ng/ml) | 1.005 | 1.000–1.010 | 0.051 |
| HECTD2 | |||
| High expression | 3.372 | 1.148–9.907 | 0.027 |
| Low expression | Reference | ||
Discussion
Metastasis is the main cause of cancer-related death in cancer patients. Peritoneal metastasis of cancer is the terminal state of gastric cancer, which can lead to poor prognosis and high mortality. The tumor cells break away from the primary tumor and immerse into the surrounding tissues to enter the circulation, transform, grow, invade, and spread in the circulation, and then adhere and colonize in the secondary organs or tissues, which is called tumor cell metastasis [28]. In this process, it mainly depends on the metastatic ability of tumor cells themselves. Tumor metastasis is a highly complex process, involving a variety of cellular mechanisms, including division, detachment, invasion, escape from immune surveillance and remodeling of tissue microenvironment [29]. This process requires the synergy of multiple proteins, especially cell surface proteins. The interaction between cancer cells and cell surface mediates cell adhesion and invasion, thus triggering metastasis [30]. In this study, we first screened the modules that are significantly related to peritoneal metastasis, and the genes in this module are mainly involved in cell adhesion and intercellular signal pathway, which is consistent with the key steps of tumor metastasis shown in previous studies. Based on this result, we further screened the core genes of these genes, aiming to identify the key promoter genes and provide basis for clinical diagnosis and treatment. Finally, we focus on HECTD2 gene, which is highly expressed in patients with peritoneal metastasis, and the prognosis of patients with high expression is poor, which is consistent with previous reports in some tumors, such as neuroblastoma [10], and RCC [12].
Our further analysis shows that HECTD2 is involved in the main process of metastasis, namely cell adhesion and metastasis, but its effect on cell cycle and apoptosis is not excluded. The pathway analysis shows that in the high expression group, in addition to inhibiting cell apoptosis, it can also inhibit cell proliferation. This result is also reflected in other tumors. For example, in prostate cancer, HECTD2 is a potential target of miR-221. miR-221 can promote androgen-dependent growth of prostate cancer cell lines, so HECTD2 has anti-proliferation effect in this type of cancer [13], and HECTD2 can also inhibit the proliferation of cancer cells [14]. This contradictory result may be caused by tumor heterogeneity. Here, we use gastric cancer samples to confirm that the expression of HECTD2 is significantly higher in gastric cancer than in adjacent tissues, and patients with high expression of HECTD2 have poor prognosis. This prediction of poor prognosis is not inferior to the high-risk factor of vascular invasion, but still cannot reach the accuracy of TNM. But its effect on the biological behavior of gastric cancer cells lacks in vivo and in vitro experimental verification, which is also our further experimental pursuit.
In addition to the role of tumor cells themselves, tumor microenvironment also plays an important role in metastasis. Previous studies have shown that the possibility of peritoneal metastasis of gastric cancer depends on the interaction between tumor cells and peritoneal microenvironment. This is Stephen Paget's hypothesis of "seed and soil" [31]. The genes in the peritoneal metastasis related modules that we screened are involved in the reconstruction of cell matrix and microenvironment, which shows that the modules we selected are accurate. We also confirm in gastric cancer specimens that HECTD2 expression is higher in gastric cancer patients with peritoneal metastasis. In addition, the core gene HECTD2 screened based on this module gene also plays an important role in the remodeling of the immune microenvironment. For example, in melanoma, it can promote the proliferation of tumor cells and promote immune escape [11]. Our results show that in gastric cancer patients with high expression of HECTD2, TMEscore is higher than that in the low expression group, regardless of "stromalscore" "immunescore" or “estimatescore”, which indicates that the high expression of HECTD2 can create a microenvironment more conducive to the metastasis of gastric cancer cells and provide a protective umbrella for tumor cell metastasis. In addition, the highly expressed HECTD2 can reshape the redistribution of immune cells in the immune microenvironment, which will be conducive to cell recruitment and induction of tumor progression. For example, it can induce M2 type of macrophages to infiltrate and reject M1 type of macrophages, while M2 type of macrophages can promote tumor progression and help tumor cell immune escape [32], which is also an important factor in promoting the progression of gastric cancer and leading to poor prognosis. In this study, multiple immunofluorescence results confirm this hypothesis that high expression of HECTD2 induces M2 type of macrophage infiltration but rejects M1 type of macrophage infiltration, which may be another mechanism by which HECTD2 plays a pro-cancer role in gastric cancer. At the same time, based on its role in the reconstruction of the immune microenvironment, we evaluated the expression of the indicators currently used to evaluate the efficacy of immune checkpoint inhibitors, such as TMB. Research shows that tumors with higher TMB may have better efficacy on immune checkpoint inhibitors [33]. The results show that HECTD2 is negatively correlated with TMB, which indicates that in patients with low expression of HECTD2, its efficacy on immune checkpoint inhibitors may be better. In addition, the immune score results also confirm this conclusion. In patients with low expression of HECTD2, the immune score is higher, which provides reference value for clinical medication. In this study, we confirmed that better immunotherapeutic effects can be achieved in gastric cancer patients with low expression of HECTD2, which may also be due to the reshaping of the immune microenvironment by HECTD2. This leads us to speculate that in gastric cancer patients with high expression of HECTD2, while having poor prognosis, they are also relatively resistant to immunotherapy.
Of course, there are certain limitations in this study. Firstly, we lack in vivo and in vitro validation at the molecular cell level or in mice, which is the focus of our next research. On the other hand, the number of clinical specimens we used are limited, which may lead to some bias in our results.
In conclusion, in this study, we clustered gastric cancer patients based on WGCNA technology, screened out the modules related to peritoneal metastasis, and enriched the genes in the modules. The results showed that they were mainly concentrated on cell metastasis and adhesion pathways. Based on the co-expression network of genes in the module, the core gene HECTD2 was screened out. The further results showed that HECTD2 was a high-risk factor for peritoneal metastasis, and the prognosis of gastric cancer patients with high expression of HECTD2 was poor. HECTD2 might promote the progression of gastric cancer by promoting the metastasis of gastric cancer cells and immune escape, leading to poor prognosis. However, patients with low expression of HECTD2 gastric cancer have better effect after receiving immune checkpoint inhibitors, which provides potential theoretical basis for clinical diagnosis and treatment.
Conclusion
In conclusion, HECTD2 was identified as a promising predictive biomarker for peritoneal metastasis and high expression of HECTD2 indicated poor prognosis in GC. HECTD2 could be used as a novel biomarker and potential target for the treatment of peritoneal metastasis in GC patients.
Supplementary Information
Supplementary material 1. Figure 1. Sample clustering and phenotypic traits. (A) The flow chart illustrating for analytical process. (B) Clustering dendrogram of 300 tumor samples. Two outlier samples were removed. (C) Clustering dendrogram of 298 tumor samples and the clinical traits.
Supplementary material 2. Figure 2. Construction of co-expression modules. (A) Analysis of the scale-free fit index and mean connectivity for various soft-thresholding powers. (B) The hierarchical cluster dendrogram and color display of co-expression network modules. (C) Correlation between modules and phenotypes. Heatmap plot of the adjacencies of modules. Red means positive correlation and blue means negative correlation.
Supplementary material 3. Figure 3. Identification of modules associated with peritoneal metastasis. (A) Heatmap of the correlation between module eigengenes and phenotype traits. (B) The scatter plot of gene significance versus module membership in the yellow module. (C) Distribution of average gene significance in the modules associated with peritoneal metastasis.
Supplementary material 4. Figure 4. The prognosis of patients with high expression of HECTD2 is poor. (A) The expression of HECTD2 RNA in gastric cancer is higher than that in adjacent tissues in the TCGA-STAD dataset. (B-C) Upregulated expression of HECTD2 is associated with shorter PFS and OS in TCGA-STAD dataset.
Author contributions
Libao Gong designs and writes article; Jinfeng Guo conducted data statistical analysis; Junjie Hang guides articles and provides clinical sample information, while also serving as a funding provider;Jiayi Huang, Xue Bai and Lin Song assists in collecting data.
Funding
This work was supported by National Natural Science Foundation of China (NO. 81902955). Natural Science Foundation of Jiangsu Province (BK20190161).The Medical Scientific Research Foundation of Guangdong Province (A2024172). Sanming Project of Medicine in Shenzhen (No.SZSM202211012). Shenzhen Key Medical Discipline Construction Fund (SZXK013).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Ethics Committee of Changzhou No.2 People's Hospital.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Junjie Hang and Jinfeng Guo have contributed equally to this work.
Contributor Information
Junjie Hang, Email: hjj199141@alumni.sjtu.edu.cn.
Jinfeng Guo, Email: gjf1229466422@163.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki Y, Nishina T, Yasui H, et al. Phase II trial of nanoparticle albumin-bound paclitaxel as second-line chemotherapy for unresectable or recurrent gastric cancer. Cancer Sci. 2014;105:812–7. 10.1111/cas.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seyfried F, von Rahden BH, Miras AD, et al. Incidence, time course and independent risk factors for metachronous peritoneal carcinomatosis of gastric origin–a longitudinal experience from a prospectively collected database of 1108 patients. BMC Cancer. 2015;15:73. 10.1186/s12885-015-1081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizrak Kaya D, Nogueras-González GM, Harada K, et al. Risk of peritoneal metastases in patients who had negative peritoneal staging and received therapy for localized gastric adenocarcinoma. J Surg Oncol. 2018;117:678–84. 10.1002/jso.24912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikuła-Pietrasik J, Uruski P, Tykarski A, et al. The peritoneal “soil” for a cancerous “seed”: a comprehensive review of the pathogenesis of intraperitoneal cancer metastases. Cell Mol Life Sci CMLS. 2018;75:509–25. 10.1007/s00018-017-2663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun F, Feng M, Guan W. Mechanisms of peritoneal dissemination in gastric cancer. Oncol Lett. 2017;14:6991–8. 10.3892/ol.2017.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu ZM, Li ZR, Huang Y, et al. DJ-1 is involved in the peritoneal metastasis of gastric cancer through activation of the Akt signaling pathway. Oncol Rep. 2014;31:1489–97. 10.3892/or.2013.2961. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Argiles-Castillo D, Kane EI, et al. HECT E3 ubiquitin ligases - emerging insights into their biological roles and disease relevance. J Cell Sci. 2020. 10.1242/jcs.228072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coon TA, McKelvey AC, Lear T, et al. The proinflammatory role of HECTD2 in innate immunity and experimental lung injury. Sci Transl Med. 2015;7:295ra109. 10.1126/scitranslmed.aab3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suo C, Deng W, Vu TN, et al. Accumulation of potential driver genes with genomic alterations predicts survival of high-risk neuroblastoma patients. Biol Direct. 2018;13:14. 10.1186/s13062-018-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ottina E, Panova V, Doglio L, et al. E3 ubiquitin ligase HECTD2 mediates melanoma progression and immune evasion. Oncogene. 2021;40:5567–78. 10.1038/s41388-021-01885-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv D, Shen T, Yao J, et al. HIF-1α induces HECTD2 up-regulation and aggravates the malignant progression of renal cell cancer via repressing miR-320a. Front Cell Dev Biol. 2021;9:775642. 10.3389/fcell.2021.775642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T, Wang X, He HH, et al. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene. 2014;33:2790–800. 10.1038/onc.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma L, Li DH, Xu Z. HECTD2 represses cell proliferation in colorectal cancer through driving ubiquitination and degradation of LPCAT1. Mol Biol. 2022;56:574–84. 10.31857/s0026898422040073. [DOI] [PubMed] [Google Scholar]
- 15.Ryu TY, Kim K, Han TS, et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022;16:1205–21. 10.1038/s41396-021-01119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62. 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–2. 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 27.Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956-d963. 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin X, Zhu Z, Shi Y. Metastasis mechanism and gene/protein expression in gastric cancer with distant organs metastasis. Bull Cancer. 2014;101:E1-12. [PubMed] [Google Scholar]
- 29.Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends–An Update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. 10.1158/1055-9965.epi-15-0578. [DOI] [PubMed] [Google Scholar]
- 30.Boedtkjer E, Pedersen SF. The acidic tumor microenvironment as a driver of cancer. Ann Rev Physiol. 2020;82:103–26. 10.1146/annurev-physiol-021119-034627. [DOI] [PubMed] [Google Scholar]
- 31.Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 32.Anderson NR, Minutolo NG, Gill S, et al. Macrophage-based approaches for cancer immunotherapy. Can Res. 2021;81:1201–8. 10.1158/0008-5472.can-20-2990. [DOI] [PubMed] [Google Scholar]
- 33.Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002–14. 10.1016/s0140-6736(21)01206-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1. Figure 1. Sample clustering and phenotypic traits. (A) The flow chart illustrating for analytical process. (B) Clustering dendrogram of 300 tumor samples. Two outlier samples were removed. (C) Clustering dendrogram of 298 tumor samples and the clinical traits.
Supplementary material 2. Figure 2. Construction of co-expression modules. (A) Analysis of the scale-free fit index and mean connectivity for various soft-thresholding powers. (B) The hierarchical cluster dendrogram and color display of co-expression network modules. (C) Correlation between modules and phenotypes. Heatmap plot of the adjacencies of modules. Red means positive correlation and blue means negative correlation.
Supplementary material 3. Figure 3. Identification of modules associated with peritoneal metastasis. (A) Heatmap of the correlation between module eigengenes and phenotype traits. (B) The scatter plot of gene significance versus module membership in the yellow module. (C) Distribution of average gene significance in the modules associated with peritoneal metastasis.
Supplementary material 4. Figure 4. The prognosis of patients with high expression of HECTD2 is poor. (A) The expression of HECTD2 RNA in gastric cancer is higher than that in adjacent tissues in the TCGA-STAD dataset. (B-C) Upregulated expression of HECTD2 is associated with shorter PFS and OS in TCGA-STAD dataset.
Data Availability Statement
No datasets were generated or analysed during the current study.