Abstract
Fowl adenovirus type 4 (FAdV-4) and duck adenovirus type 3 (DAdV-3) are the causative agents of clinical diseases in poultry and have caused considerable economic losses to the waterfowl industry in China. Both FAdV-4 and DAdV-3 are classified into the genus Aviadenovirus under the family Adenoviridae. The high-resolution melting (HRM) assay has become a useful method for virus genotyping, which offers the possibility of rapidly developing a differentiation technique in which the melting profile depends on the GC content of the product in the qPCR platform. The aim of this study was to develop a qPCR-HRM assay for sensitive FAdV-4 and DAdV-3 detection and differentiation. Here, specific primers were designed on the basis of the 100 K genes of FAdV-4 and DAdV-3, and a qPCR-HRM assay was established through optimization of the reaction conditions. A specificity test revealed that this method could detect only FAdV-4 and DAdV-3, with no cross-reaction with other common duck-derived viruses. A sensitivity test revealed that the lowest detection limits of FAdV-4 and DAdV-3 were 2.84 copies/µL and 2.85 copies/µL, respectively. A repeatability test demonstrated that the coefficient of variation was less than 2.5 % in both the intragroup and the intergroup analyses. Field sample distributions of FAdV-4 and DAdV-3 were investigated, and the percentages of DAdV-3-positive, FAdV-4-positive and coinfection-positive in Muscovy ducks were 27.78 %, 16.67 % and 11.11 %, respectively. Further studies are needed to provide more insight into the pathogenesis of FAdV-4 and DAdV-3 coinfection in ducks. In conclusion, the qPCR-HRM assay provides an accurate, sensitive, reliable and cost-effective alternative method for detecting and distinguishing FAdV-4 and DAdV-3.
Keywords: FAdV-4, DAdV-3, qPCR-HRM assay, Differentiation, Epidemiological investigation
Introduction
Fowl adenovirus (FAdV) is a common pathogenic pathogen of poultry with a wide distribution worldwide. It not only directly affects the health status and production performance of poultry but also severely affects food safety and public health (Rashid et al., 2024). In recent years, highly pathogenic fowl adenovirus serotype 4 (FAdV-4) and duck adenovirus 3 (DAdV-3) have been widely transmitted on farms, causing considerable economic losses to the waterfowl industry (Yin et al., 2022; Li et al., 2017). FAdV-4 and DAdV-3, both of which are members of the genus Aviadenovirus, are nonenveloped double-stranded DNA viruses with a general adenovirus icosahedral particle structure, and their genome size is approximately 43–46 kb (Zhao et al., 2015). FAdV-4 is the main pathogen that causes hepatitis-pericardial effusion syndrome (HHS), and it mainly infects 3–5-week-old broiler chickens, with high morbidity and mortality (Lai et al., 2023). In recent years, with the development of the epidemic of this virus, FAdV-4 has also been identified and isolated from many types of ducks (Muscovy duck, Cherry Valley duck, etc.), and its pathogenesis characteristics are similar to those of chicken hydropericardium syndrome (HPS). The symptoms of pericardial effusion syndrome are similar, including pericardial effusion, pale yellow liquid in the pericardial cavity, and liver enlargement, which cause morbidity and mortality in duck flocks and have a great impact on the duck breeding industry (Yu et al., 2018; Wu et al., 2022; Chen et al., 2017). Duck adenovirus 3 (DAdV-3) is a novel Aviadenovirus with pathogenicity to ducks. The main symptoms of infected Muscovy ducks are kidney and liver enlargement, haemorrhage and mild pericardial effusion. The morbidity rate is between 40 % and 55 %, and the mortality rate is 35 %-43 % (Zhang et al., 2016). Therefore, it is very important to establish a laboratory detection method that can simultaneously identify and distinguish FAdV-4 and DAdV-3 in waterfowl.
The genome can be divided into early transcription genes (E1–E4) and late transcription genes (L1–L5) according to the chronology of DNA replication. The 100 K gene is located in the L4 region of the viral genome and encodes the viral non-structural protein 100 K. This protein is the most abundant in the late stage of adenovirus infection. The 100 K protein plays a key role in adenovirus infection. It binds to the newly synthesized structural protein hexon peptide to make the latter coil up and fold into a homotrimer and simultaneously enables the hexon peptide to exit the cell cytoplasm. The ER is transported to the nucleus for assembly and can also bind to late mRNAs of adenovirus to promote the synthesis of viral proteins while inhibiting the synthesis of host proteins (Hong et al., 2005; Koyuncu et al., 2013; Shah et al., 2016).
A high-resolution melting curve is a nucleic acid analysis method created in the qPCR platform. It does not require the use of fluorescently labelled probes and has the advantages of being fast, accurate, inexpensive, and easy to operate. In recent years, it has been widely used in gene mutation, methylation, and single nucleotide studies and has been widely used in polymorphism analysis, genotyping, and sequence matching (Montgomery et al., 2010). The basic principle of HRM technology is based on differences in physical properties, such as the fragment length, GC content, and distribution of nucleic acid molecules, and by monitoring the characteristic melting curve formed by the fluorescence signal of the dye released during the melting process of double-stranded DNA during the heating process, nucleic acid analysis of the product is performed. identification of nucleotide differences (Reed et al., 2004). In this study, on the basis of the 100 K gene sequence characterization between FAdV-4 and DAdV-3, a qPCR-HRM assay for DAdV-3 and FAdV-4 was established, which provides an accurate, sensitive, reliable and cost-effective alternative method for detecting and distinguishing FAdV-4 and DAdV-3.
Materials and methods
Viruses and nucleic acid extraction
The viruses used in this study, such as duck circovirus (DuCV), duck plague virus (DPV), duck Tembusu virus (DTMUV), Muscovy duck parvovirus (MDPV) and Muscovy duck reovirus (MDRV) were all identified, isolated and preserved by the Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agricultural Sciences.
A total of 83 tissue samples, suspected of being infected FAdV-4 and DAdV-3 from different waterfowl flocks Fujian Province, China. All collected samples were homogenized in phosphate-buffered saline (PBS) (20 %, w/v). The viruses and these resuspended sample was used for DNA extraction using EasyPure® Viral DNA/RNA Kit (TransGen Biotech, China).
Primers design
To identify the gene sequences with unique differences between FAdV-4 and DAdV-3, we retrieved the 100 K gene sequences of 84 strains of fowl adenovirus serotype 4 (FAdV-4) and 30 strains of duck adenovirus 3 (DAdV-3) from the GenBank(https://www.ncbi.nlm.nih.gov/). We found that though the nucleotide homology between FAdV-4 and DADV-3 is only 60.7 %, but the 100 K gene has characteristics nucleotide differences between FAdV-4 and DADV-3, and the variations of 100 K gene between FAdV-4 and DAdV-3 are highly conserved. According on the 100 K genetic comparison of DAdV-3 and FAdV-4, a pair of qPCR-HRM primers (D3F4F1 and D3F4R1; Table 1) was designed with 116-nt target amplification fragments. The variation of the target amplified region (116-nt) between FAdV-4 and DAdV-3 are listed in Table 2. The primers were synthesized by Sangon Bioengineering Co., Ltd. (Sangon Biotech, Shanghai, China).
Table 1.
Sequence variation in DuCV-1 and DuCV-2.The primers used of the qPCR-HRM assay.
| Viruses | D3F4F1(5′-3′) B | position | D3F4R1(5′-3′) B | position | Length (bp) |
|---|---|---|---|---|---|
| DAdV-3 | ACCCGCTCAACAACTGCAT | 1526-1544A1 | CATGCCCATAGCCGTTTGCCA | 1621-1641 A1 | 116 |
| FAdV-4 | 1685-1703 A2 | 1780-1800 A2 |
A1, the length of 100 K gene of DAdV-3 is 2829-bp, here means the position of the 100 K gene of DAdV-3.
A2, the length of 100 K gene of FAdV-4 is 3162-bp, here means the position of the 100 K gene of FAdV-4.
B, the forward primer (D3F4F1) and reverse primer (D3F4R1) shared100 % matched with DAdV-3 and FAdV-4.
Table 2.
Variations in the target region between 100 K of FAdV-4 and DAdV-3.
| Position | 1-58 |
|---|---|
| DAdV-3 | ACCCGCTCAACAACTGCATTATGAGCCGCTTGTGTGAGGGTCAGGACAAGGAGGATTT |
| FAdV-4 | ACCCGCTCAACAACTGCATGCTAGCCAAGTTGATGGAAGGCTCGGACAAGCGAGATTA |
| Position | 59-116 |
| DAdV-3 | CATTGTGGACTCCATCTACCTCTTTTTGGTGCTCACCTGGCAAACGGCTATGGGCATG |
| FAdV-4 | CGTGGTGGACAGCATCTACCTCTTCTTGGTGCTCACGTGGCAAACGGCTATGGGCATG |
*The variation in the target region (116 bp) between the 100 K of FAdV-4 and DAdV-3 is listed, the primer design region is underlined, and the variations in DAdV-3 and FAdV-4 are marked with red and blue, respectively.
Positive plasmid standards preparation
The DAdV-3 plasmid containing the target gene fragment of the 100 K gene of DAdV-3, the FAdV-4 plasmid containing the target gene fragment of the 100 K gene of FAdV-4. The DNA of FAdV-4 and DAdV-3 was used as a template for PCR amplification. The PCR system was as follows: PCR amplification system (50 μL), 2 × PCR Master Mix 25 μL(TransGen Biotech, China), 1 μL each of upstream and downstream primers, 2 μL of DNA template, and 21 μL of ddH2O. The PCR conditions were as follows: denaturation at 94°C for 30 s; 30 cycles at 60° C for 30 s and 72° C for 90 s; and extension at 72° C for 10 min. The PCR products were recovered, purified, and subsequently cloned and inserted into the pMD18-T (TaKaRa, Dalian, China) to construct recombinant plasmids. After identification by microbial PCR, the positive recombinant plasmids were sent to Shanghai Sangon Biotechnology Co., Ltd., for sequencing identification. The recombinant plasmids with the correct sequence were used as positive standards. Determination of nucleic acid concentration of positive recombinant plasmids using NanoDrop2000. The concentrations were measured and converted to copy numbers and used as standards in this study.
The calculation formula for the copy number of the recombinant plasmids was as follows: copy number = 6.02 × 1023 copies/mol × plasmid concentration/(relative DNA molecular mass × 660).
Optimization of the qPCR-HRM assay
The qPCR-HRM assay conditions were: 1 cycle of 95° C for 2 min; 40 cycles of 95° C for 10 s and 60° C for 30 s; Gradient PCR was used to optimize annealing temperatures (54, 56, 58, 60, 62, 64)° C and primer concentrations (0.2, 0.4, 0.8, 1.0, 1.5, 1.75, 2.0 μmol/L). The high-resolution melting curve parameters were: 95°C for 1 min, 40° C for 1 min, 65° C for 1 s, and then heating to 97° C; melting rates of 0.05, 0.10, and 0.20° C/s were used for optimization. HRM analysis was performed on the LightCycler®96 (Roche Diagnostics). The optimal reaction conditions were determined.
Standard curve construction of the qPCR-HRM assay
After the FAdV-4 and DAdV-3 plasmid standards were diluted 10-fold, the plasmid standards were diluted with FAdV-4 plasmid standards (2.84 × 102-2.84 × 107 copies/μL) and DAdV-3 plasmid standards (2.85 × 102-2.85 × 107 copies/µL). A standard curve was constructed using six different concentrations of templates, with the logarithmic value of the copy number of plasmid standards on the X axis and the threshold cycle number (Ct value) on the Y axis. The plasmid copy number logarithm was plotted against the corresponding Ct values and the standard curve was constructed.
Specificity, repeatability test of the qPCR-HRM assay
To evaluate the specificity of the qPCR-HRM, FAdV-4, DAdV-3, DuCV, DPV, DTMUV, MDPV, and MDRV DNA or cDNA were used as a template and ddH2O was used as a negative control. To evaluate the repeatability of the qPCR-HRM, selected FAdV-4 plasmid with concentrations of 2.84 × 102 copies/μL, 2.84 × 10 4 copies/μL, and 2.84 × 106 copies/μL, as well as 2.85 × 102copies/μL, 2.85 × 104 copies/μL, and 2.85 × 106 copies/μL DAdV-3 plasmid for PCR HRM method detection. All the samples, including the standards and the control, were run in triplicate. Calculate the coefficient of variation (CV) within and between groups using statistical methods based on Ct values.
Limit of detection of qPCR-HRM assay
To evaluate the detection limit of the qPCR-HRM method, detecting standard plasmids of different copy numbers. The FAdV-4 plasmid standards were diluted 10-fold from 2.84 × 103copies/μL to 2.84 × 10−1copies/μL using EASY Dilution (TaKaRa, Dalian, China);The DAdV-3 plasmid standards were diluted 10-fold from 2.85 × 103 copies/μL to 2.85 × 10−1 copies/μL using EASY Dilution (TaKaRa, Dalian, China), the above standard plasmid was used as templates.
HRM analysis
In view of the emergence of clinical cases of mixed infection with FAdV-4 and DAdV-3, a standard plasmid product of FAdV-4 and DAdV-3 at an equivalent concentration was selected and mixed 1:1, and the established qPCR-HRM method was used to detect a single plasmid. A plasmid mixture was used to simulate mixed infection, and a dissolution curve of mixed infection was obtained to evaluate the ability of qPCR-HRM assay to identify coinfected samples.
Clinical epidemiological investigation
To verify the established qPCR-HRM assay, 52 clinical disease samples from ducks in Fujian, including Muscovy ducks (18 samples), mulard ducks (11 samples), Cheery valley ducks (14 samples), Ma ducks (9 samples), were collected. Moreover, 31 clinical disease samples from geese were collected, including Changle Grey geese (5 samples), Minbei White geese (11 samples), and Shitou geese (15 samples) (Table 4). All samples were tested in triplicate and independently, and compared with the conventional PCR method.
Table 4.
Investigation results of the qPCR-HRM assay.
| Type | Species | Number | HRM |
Conventional PCR |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAdV-3 |
FAdV-4 |
Coinfection |
DAdV-3 |
FAdV-4 |
Coinfection |
||||||||||
| P | R | P | R | P | R | P | R | P | R | P | R | ||||
| ducks | Muscovy | 18 | 5 | 27.78 | 3 | 16.67 | 2 | 11.11 | 3 | 16.67 | 1 | 5.55 | 1 | 5.55 | |
| mulard | 11 | 3 | 27.27 | 2 | 18.18 | 0 | 0 | 2 | 18.18 | 2 | 18.18 | 0 | 0 | ||
| Cheery valley | 14 | 0 | 0 | 3 | 21.43 | 0 | 0 | 0 | 0 | 2 | 14.29 | 0 | 0 | ||
| Ma | 9 | 0 | 0 | 1 | 11.11 | 0 | 0 | 0 | 0 | 1 | 11.11 | 0 | 0 | ||
| geese | Changle Grey | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Minbei White | 11 | 0 | 0 | 1 | 9.09 | 0 | 0 | 0 | 0 | 1 | 9.09 | 0 | 0 | ||
| Shitou | 15 | 0 | 0 | 2 | 13.33 | 0 | 0 | 0 | 0 | 1 | 6.67 | 0 | 0 | ||
Abbreviations: P indicates the number of positive sample, and R* indicates a positive ratio (/%).
The primer set used for FAdV-4 was as follows: FAdV-4HexF1: 5′-GAYRGYHGGRTNBTGGAYATGGG-3′ and FAdV-4HeXR1: 5′-TACTTATCNACRG-CYTGRTTCCA-3′, with a predicted size of approximately 800 bp (Mase et al., 2009). Additionally, the primer set used for DAdV-3 was as follows: DAdV-3F1: 5′-ACACTAGACAACGGAGGCCT-3′ and DAdV-3R1: 5′- AATCTTGATACGTAATCATACACA -3′, with a predicted size of approximately 550 bp. (Wan et al., 2019). The PCR cycle consisted of 1 cycle at 94° C for 5 min; 35 cycles at 94° C for 30 sec (denaturation), 53° C for 30 sec (annealing), and 72° C for 45 sec (extension); and 1 cycle at 72° C for 10 min. The target of the conventional PCR products was subjected to electrophoresis on 1.0 % agarose gels for analysis.
Results
The qPCR-HRM assay
The concentration of primers and probe giving the highest fluorescence and the lowest threshold cycle were determined as follows (20 µL qPCR-HRM reaction mixtures), which contained 2 × TB Green® Premix Ex Taq II FAST qPCR 10 µL (TaKaRa, Dalian, China), 1 μL each of primers D3F4F1 and D3F4R1 (10 µmol/L), 1 μL of DNA template, and 7 μL of ddH2O.
The qPCR-HRM reaction conditions as follow:1 cycle of 95° C for 2 min; 40 cycles of 95° C for 10 s and 60° C for 30 s. The high-resolution melting curve parameters as follow: 95° C for 1 min, 40° C for 1 min, 65° C for 1 s, and then heating to 97° C; melting rates of 0.10° C/s were used for optimization.
Standard curve
The standard curves were assessed for two properties, the coefficient of correlation (R2), and the amplification efficiency (E) determined from the slope of the standard curve. The FAdV-4 plasmid achieved good amplification in the template range of 2.84 × 102 to 2.84 × 107copies/μL (Fig. 1A), and the correlation coefficient of the standard curve(R2) was 1.00, with a slope of -3.2951 (Fig. 1B); the DAdV-3 plasmid achieved good amplification in the range of 2.85 × 102 to 2.85 × 107 copies/µL template (Fig. 1C). The correlation coefficient (R2) of the calibration curve was 0.99, and the slope was -3.2089 (Fig. 1D).
Fig. 1.
A FAdV-4 amplification curve of the qPCR-HRM assay
B FAdV-4 standard curve of the qPCR-HRM assay
C DAdV-3 amplification curve of the qPCR-HRM assay
D DAdV-3 standard curve of the qPCR-HRM assay.
Specificity and repeatability test
Evaluate the specificity of qPCR-HRM assay by detecting the DNA or cDNA of other duck derived viruses. The results indicated that the established qPCR-HRM method only detected positive amplification signals for FAdV-4 and DAdV-3, and did not show positive amplification fluorescence signals for other common duck pathogens (DuCV, DPV, DTMUV, MDPV, MDRV) (Fig. 2). The result indicated that the coefficients of variation for intragroup detection and intergroup detection were both less than 2.5 % (Table 3), indicating that the method had high repeatability.
Fig. 2.
Specificity test of the qPCR-HRM assay
1: DAdV-3; 2: FAdV-4; 3: Controls: DuCV, DPV, DTMUV, MDPV, MDRV, and negative control. These controls were all found to have no positive signals. The results of these controls could not be effectively distinguished by the naked eye.
Table 3.
Repeatability test of the qPCR-HRM assay.
| Viruses | Concentration of plasmid standards(copies/μL−1) | Intra-assay reproducibility |
Inter-assay reproducibility |
||
|---|---|---|---|---|---|
| Ct ± SD | CV/% | Ct ± SD | CV/% | ||
| FAdV-4 | 2.84 × 102 | 24.70 ± 0.12 | 0.48 | 24.44 ± 0.17 | 0.70 |
| 2.84 × 104 | 17.85 ± 0.26 | 1.47 | 17.91 ± 0.20 | 1.11 | |
| 2.84 × 106 | 11.28 ± 0.25 | 2.20 | 11.71 ± 0.29 | 2.47 | |
| DAdV-3 | 2.85 × 102 | 22.90 ± 0.24 | 1.05 | 22.96 ± 0.30 | 1.30 |
| 2.85 × 104 | 18.28 ± 0.03 | 0.18 | 18.16 ± 0.24 | 1.34 | |
| 2.85 × 106 | 11.79 ± 0.12 | 1.03 | 11.89 ± 0.22 | 1.87 | |
Limit of detection
The result indicated that the limit of detection of FAdV-4 plasmid detected by the qPCR-HRM method established in this study was 2.84 copies/µL (Fig. 3A), and the limit of detection of the DAdV-3 plasmid was 2.85 copies/µL (Fig. 3B).
Fig. 3.
A FAdV-4 sensitivity test of the qPCR-HRM assay
1–5: The copy number concentrations of the FAdV-4 plasmid standards were 2.84 × 10 3–2.84 × 10−-1 copies/µL; 6: negative control
B DAdV-3 sensitivity test of the qPCR-HRM assay
1–5: the copy number concentrations of DAdV-3 plasmid standards were 2.85 × 10 3–2.85 × 10−-1 copies/µL; 6: negative control.
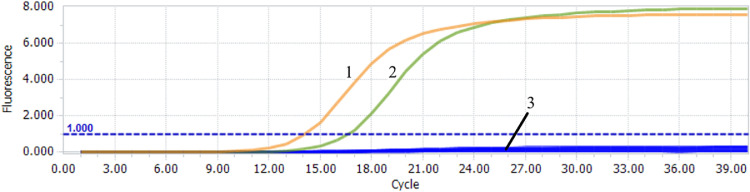
HRM analysis
After optimizing the primer concentration, annealing temperature, and melting rate of the qPCR-HRM method, the FADV-4 and DADV-3 plasmids were validated to evaluate the difference in TM values between the two viruses. The results showed that the Tm values of the melting curves of FAdV-4 were (83.50 ± 0.07)°C while the melting curves of DAdV-3 were (81.92 ± 0.08)° C. We also tested the potential of HRM
-PCR method for detecting coinfection of FAdV-4 and DAdV-3 (Fig. 4). The results indicate that coinfection samples and FADV or DADV infected samples alone produce significantly different dissolution curves.
Fig. 4.
Melting profiles of the qPCR-HRM assays
1 means DAdV-3; 2 means FAdV-4 and DAdV-3 co-infection; 3 means FAdV-4.
Investigation data
To evaluate the duck-origin samples, for Muscovy ducks, with the percentages of DAdV-3-positive, FAdV-4-positive and coinfection-positive is 27.78 %, 16.67 % and 11.11 %, respectively. For mulard ducks, with the percentages of DAdV-3-positive, FAdV-4-positive and coinfection-positive is 27.27 %, 18.18 % and 0 %, respectively. No DAdV-3-positive signal was observed from Cheery valley ducks, Ma ducks, Changle Grey geese, Minbei White geese and Shitou geese. However, FAdV-4-positive signals can be found from Cheery valley ducks, Ma ducks, Minbei White geese and Shitou geese (Table 4). Additionally, the positive samples tested by the conventional PCR method were also can be found positive signals using the established qPCR-HRM platform, revealed that the HRM is more sensitive than conventional PCR technology.
Discussion
Avian adenoviruses are widespread worldwide and can exist in the environment for a long period of time. They are prevalent mainly in chickens, ducks, and geese and can be rapidly transmitted horizontally between groups and fields through the respiratory and digestive tracts or vertically through breeding eggs. Transmission from parent to offspring (Guan et al., 2018; El-Shall et al., 2022). In natural infection, it can also enter the respiratory tract or gastrointestinal tract, leading to oropharyngeal or conjunctival infection through aerosols (Li et al., 2019). With the rapid development of the poultry industry, the continuous expansion of the breeding scale, and the increasing number and density of poultry farms, the problem of poultry disease has also become prominent. Infections caused by FAdV-4 and DAdV-3 are increasing each year, and until recently, large-scale epidemics have occurred. The prevention and control of these diseases have received much attention from researchers at home and abroad. In recent years, duck infection with FAdV-4 has been shown to cause pathological changes, such as pericardial effusion and liver necrosis, similar to those in chickens. This finding indicates that there is a mixed infection of FAdV-4 and DAdV-3 in farms, which is the origin of FAdV-4 in duck farms. The differential diagnosis of FAdV-4 and DAdV-3 is very difficult to identify on the basis of clinical symptoms and lesions alone. Therefore, laboratory methods are needed for timely diagnosis of the occurrence and prevalence of FAdV-4 and DAdV-3. to help minimize losses.
Currently, laboratory diagnostic methods for FAdV-4 and DAdV-3 infection at home and abroad include virus isolation and identification (Wang et al., 2021; Shi et al., 2022), fluorescence quantitative PCR assay (Wang et al., 2017; Wan et al., 2018), enzyme-linked immunosorbent assays (He et al., 2018; Chen et al., 2019), and rapid LAMP assays (Zhai et al., 2019). Traditional virus detection methods, such as virus isolation and serological detection, have many deficiencies, such as being time-consuming and laborious, being difficult to perform, having poor specificity, and having poor sensitivity. Therefore, the development of accurate, rapid and quantitative detection methods is highly important for pathogen diagnosis, especially for the rapid identification of mixed infections with multiple different pathogens or different genotypes of the same pathogen. To date, researchers have established fluorescence quantitative PCR methods for the detection of FAdV-4 and DAdV-3, but these methods are all useful for the detection of a single pathogen. However, a qPCR-HRM method for the simultaneous detection of FAdV-4 and DAdV-3 has not been reported. Therefore, the establishment of a rapid and accurate fluorescence quantitative probe detection method for the detection of this virus and the ability to differentiate the genotypes is convenient to perform and has reliable results, which also has value for clinical application.
HRM is a novel nucleic acid detection technology that has emerged in recent years. Compared with conventional detection methods, HRM technology has the advantages of strong specificity, high sensitivity, convenient operation, high throughput, fast speed, etc., and has been widely used for pathogen detection in the laboratory. The change in Tm from a purine to a pyrimidine or from a pyrimidine to a purine is approximately 1° C. However, mutations from pyrimidines to pyrimidines or purines to purines cause a change in the Tm value of approximately 0.4°C(Liew et al., 2004). In this study, the 100 K gene sequences of FAdV-4 and DAdV-3 were compared, analyzed and used as the detection primers for FAdV-4 and DAdV-3, and the difference in the amplified gene fragments was used to generate specific melting curves. FAdV-4 had a higher melting peak Tm at (83.50 ± 0.07)° C, and DAdV-3 had a lower melting peak Tm at (81.92 ± 0.08)° C; moreover, FAdV-4 infection could be diagnosed simultaneously in one experiment according to the melting curve or DAdV-3 infection or coinfection of the two strains, and the data collection and analysis were automated, which reduced the workload and the influence of human factors on the results.
This study developed a qPCR-HRM assay for the detection and differentiation of FAdV-4 and DAdV-3. This method does not require gel electrophoresis analysis; the whole process is a closed-tube operation to reduce cross-contamination of nucleic acids; the reaction only takes approximately 1 h; and the method can be rapidly determined on the basis of the difference in the Tm values. To determine the presence of FAdV-4 and DAdV-3, this method did not involve cross-reacting with other common duck-derived viruses. The detection limits of the FAdV-4 and DAdV-3 plasmid standards were 2.84 copies/µL and 2.85 copies/µL, respectively. The intra-assay and inter-assay repeatability coefficients for the detection of FAdV-4 and DAdV-3 were less than 2.5 %. An epidemiological survey of waterfowl in Fujian revealed co-infection with FAdV-4 and DAdV-3 in Muscovy ducks, but not in Cheery valley ducks, Ma ducks, Changle Grey geese, Minbei White geese and Shitou geese. Whether this phenomenon is related to the adaptability of FAdV-4 and DAdV-3 to host adaptation needs further study.
In summary, this study established a qPCR-HRM assay that provides an accurate, sensitive, reliable and cost-effective alternative method for detecting and distinguishing FAdV-4 and DAdV-3 strains. Our data confirmed the presence of FAdV-4 and DAdV-3 coinfection in Muscovy ducks, but the pathogenicity of FAdV-4 and DAdV-3 coinfection remains unclear and needs further study.
Ethics approval and consent to participate
All samples were handled in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of China.
Consent for publication
Not applicable.
Availability of data and material
The datasets supporting the conclusions of this article are included within the article.
All the datasets are available from the corresponding author upon reasonable request.
Authors' contributions
Shuyu Chen and Cuiteng Chen performed the experiments, analyzed the data, and drafted the manuscript. These two authors contributed equally to this work. Mengyan Zhang, YuYi Chen, Wenyu Zhang, Huanru Fu, Yu Huang and Longfei Cheng collected samples and helped in laboratory analysis. Chunhe Wan contributed to the experimental design and supervised the study. All the authors have read and approved the final manuscript.
Declaration of competing interest
There is no conflicts of interest.
Acknowledgements
This study was funded by grants from the Research and Technology Program of Fujian Academy of Agricultural Sciences (grant no. YCZX202412, DWHZ2024-13, and CXTD2021005), the Fujian Science and Technology Program (grant no. 2020J06029, 2024R1025006), ‘5511′ Collaborative Innovation Project of Fujian Academy of Agricultural Sciences (XTCXGC2021012), and the China Agriculture Research System (grant no. CARS-42). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Chen C., Wan C., Shi S., Cheng L., Chen Z., Fu G., Liu R., Zhu C., Huang Y. Development and application of a Fiber2 protein-based indirect ELISA for detection of duck adenovirus 3. Mol. Cell. Probes. 2019;48 doi: 10.1016/j.mcp.2019.101447. [DOI] [PubMed] [Google Scholar]
- Chen H., Dou Y., Zheng X., Tang Y., Zhang M., Zhang Y., Wang Z., Diao Y. Hydropericardium hepatitis syndrome emerged in cherry valley ducks in china. Vet. Microbiol. 2017;274 doi: 10.1111/tbed.12500. [DOI] [PubMed] [Google Scholar]
- El-Shall N.A., El-Hamid H.S.A., Elkady M.F., Ellakany H.F., Elbestawy A.R., Gado A.R., Geneedy A.M., Hasan M.E., Jaremko M., Selim S., El-Tarabily K.A., El-Hack M.E.A. Vol. 9. 2022. Epidemiology, pathology, prevention, and control strategies of inclusion body hepatitis and hepatitis-hydropericardium syndrome in poultry: a comprehensive review. (Front. Vet. Sci.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R., Tian Y., Han X., Yang X., Wang H. Complete genome sequence and pathogenicity of fowl adenovirus serotype 4 involved in hydropericardium syndrome in Southwest China. Microb. Pathog. 2018;117:290–298. doi: 10.1016/j.micpath.2018.02.012. [DOI] [PubMed] [Google Scholar]
- He Z.R., Ruan S.F., Zhao J., Yang H.M., Zhang G.Z. Recombinant Fiber-2 protein-based indirect ELISA for antibody detection of fowl adenovirus serotype 4. Avian Dis. 2018;62:73–78. doi: 10.1637/11758-100917-Reg.1. [DOI] [PubMed] [Google Scholar]
- Hong S.S., Szolajska E., Schoehn G., Franqueville L., Myhre S., Lindholm L., Ruigrok R.W., Boulanger P., Chroboczek J. The 100K-chaperone protein from adenovirus serotype 2 (Subgroup C) assists in trimerization and nuclear localization of hexons from subgroups C and B adenoviruses. J. Mol. Biol. 2005;352:125–138. doi: 10.1016/j.jmb.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Koyuncu O.O., Speiseder T., Dobner T., Schmid M. Amino acid exchanges in the putative nuclear export signal of adenovirus type 5 L4-100K severely reduce viral progeny due to effects on hexon biogenesis. J. Virol. 2013;87:1893–1898. doi: 10.1128/JVI.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Yang L., Chen F., He X., Zhang R., Zhao Y., Gao G., Mu W., Chen X., Luo S., Ren T., Xiang B. Prevalence and molecular characteristics of FAdV-4 from indigenous chicken breeds in yunnan province southwestern china. Microorganisms. 2023;11:2631. doi: 10.3390/microorganisms11112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Yu G., Niu Y., Cai Y., Liu S. Airborne transmission of a serotype 4 fowl adenovirus in chickens. Viruses. 2019;11:262. doi: 10.3390/v11030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.H., Zheng P.P., Zhang T.F., Wen G.Y., Shao H.B., Luo Q.P. Fowl adenovirus serotype 4: epidemiology, pathogenesis, diagnostic detection, and vaccine strategies. Poult. Sci. 2017;96:2630–2640. doi: 10.3382/ps/pex087. [DOI] [PubMed] [Google Scholar]
- Liew M., Pryor R., Palais R., Meadows C., Erali M., Lyon E., Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Mase M., Mitake H., Inoue T., Imada T. Identification of group I-III avian adenovirus by PCR coupled with direct sequencing of the hexon gene. J. Vet. Med. Sci. 2009;71:1239–1242. doi: 10.1292/jvms.71.1239. [DOI] [PubMed] [Google Scholar]
- Montgomery J.L., Sanford L.N., Wittwer C.T. High-resolution DNA melting analysis in clinical research and diagnostics. Expert. Rev. Mol. Diagn. 2010;10:219–240. doi: 10.1586/erm.09.84. [DOI] [PubMed] [Google Scholar]
- Rashid F., Xie Z., Wei Y., Xie Z., Xie L., Li M., Luo S. Biological features of fowl adenovirus serotype-4. Front. Cell. Infect. Microbiol. 2024;14 doi: 10.3389/fcimb.2024.1370414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed G.H., Wittwer C.T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- Shah M.S., Ashraf A., Khan M.I., Rahman M., Habib M., Qureshi J.A. Molecular cloning, expression and characterization of 100K gene of fowl adenovirus-4 for prevention and control of hydropericardium syndrome. Biologicals. 2016;44:19–23. doi: 10.1016/j.biologicals.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Shi X., Zhang X., Sun H., Wei C., Liu Y., Luo J., Wang X., Chen Z., Chen H. Isolation and pathogenic characterization of duck adenovirus 3 mutant circulating in China. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2021.101564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C., Chen C., Cheng L., Fu G., Shi S., Liu R., Chen H., Fu Q., Huang Y. Development of a TaqMan-based real-time PCR for detecting duck adenovirus 3. J. Virol. Methods. 2018;261:86–90. doi: 10.1016/j.jviromet.2018.08.011. [DOI] [PubMed] [Google Scholar]
- Wan C., Chen C., Shi S., Cheng L., Fu G., Liu R., Chen H., Fu Q., Huang Y. Specific diagnosis for duck adenovirus type 3 by PCR method. China Poultry. 2019;2:20–23. [Google Scholar]
- Wang J., Wang J., Chen P., Liu L., Yuan W. Development of a TaqMan-based real-time PCR assay for rapid and specific detection of fowl aviadenovirus serotype 4. Avian Pathol. 2017;46:338–343. doi: 10.1080/03079457.2016.1278428. [DOI] [PubMed] [Google Scholar]
- Wang L., Zheng L., Jiang S., Li X., Lu C., Zhang L., Ren W., Li C., Tian X., Li F., Yan M. Isolation, identification and genetic characterization analysis of a fowl aviadenovirus serotype 4 strain from Tianjin, China. Infect. Genet. Evol. 2021;96 doi: 10.1016/j.meegid.2021.105078. [DOI] [PubMed] [Google Scholar]
- Wu B., Yang B., He D., Tang Y., Diao Y. Genetic evolution of fowl adenovirus serotype 4 and its pathogenicity to Cherry Valley ducks in China. Vet. Microbiol. 2022;274 doi: 10.1016/j.vetmic.2022.109578. [DOI] [PubMed] [Google Scholar]
- Yin L., Zhou Q., Mai K., Yan Z., Shen H., Li Q., Chen L., Zhou Q. Epidemiological investigation of duck adenovirus 3 in southern China, during 2018-2020. Avian Pathol. 2022;51:171–180. doi: 10.1080/03079457.2022.2034737. [DOI] [PubMed] [Google Scholar]
- Yu X., Wang Z., Chen H., Niu X., Dou Y., Yang J., Tang Y., Diao Y. Serological and pathogenic analyses of fowl adenovirus serotype 4 (FAdV-4) Strain in Muscovy Ducks. Front Microbiol. 2018;9:1163. doi: 10.3389/fmicb.2018.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Mei X., Wu X., Zuo L., Zhou L., Tian Y., Han X., Yang X., Wang H. A loop-mediated isothermal amplification coupling with a lateral flow dipstick for rapid and specific detection of fowl adenovirus serotype-4. J. Virol. Methods. 2019;270:79–86. doi: 10.1016/j.jviromet.2019.04.026. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhong Y., Zhou Z., Liu Y., Zhang H., Chen F., Chen W., Xie Q. Molecular characterization, phylogeny analysis and pathogenicity of a Muscovy duck adenovirus strain isolated in China in 2014. Virology. 2016;493:12–21. doi: 10.1016/j.virol.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhong Q., Zhao Y., Hu Y.X., Zhang G.Z. Pathogenicity and complete genome characterization of fowl adenoviruses isolated from chickens associated with inclusion body hepatitis and hydropericardium syndrome in China. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.
All the datasets are available from the corresponding author upon reasonable request.