Abstract
Background
Ovarian clear cell carcinoma (OCCC) has a disproportionately high incidence among women in East Asia. Patients diagnosed with OCCC tend to experience worse clinical outcomes than those with high-grade serous carcinoma (HGSC) at advanced stages. The unfavorable prognosis of OCCC can be partly attributed to its frequent resistance to conventional chemotherapy. Within a precision medicine framework, we sought to provide a comprehensive molecular characterization of OCCC using whole-exome sequencing to uncover potential molecular targets that may inform novel therapeutic strategies.
Methods
We performed whole-exome sequencing analysis on tumor-normal paired samples from 102 OCCC patients. This comprehensive genomic characterization of a substantial cohort of OCCC specimens was coupled with an analysis of clonal progression.
Results
On analyzing 102 OCCC samples, ARID1A (67%) and PIK3CA (49%) emerged as the most frequently mutated driver genes. We identified tier 1 or 2 clinically actionable molecular targets in 40% of cases. This included DNA mismatch repair deficiency (n = 1), as well as BRCA2 (n = 1), PIK3CA (n = 36), KRASG12C (n = 1), and ATM (n = 4) mutations. Furthermore, 45% of OCCC samples displayed ARID1A biallelic loss. Interestingly, we identified previously unreported mutations in the 5’ untranslated region of the TERT gene that harbored an adverse prognostic significance. Clock-like mutational processes and activated APOBECs were major drivers of somatic point mutations. Mutations arising from DNA mismatch repair deficiency were uncommon. Reconstruction of clonal evolution revealed that early genetic events likely driving tumorigenesis included mutations in the ARID1A, PIK3CA, TERT, KRAS, and TP53 genes.
Conclusions
Our study provides a comprehensive characterization of the genomic landscape and clonal evolution in OCCC within a substantial cohort. These findings unveil potentially actionable molecular alterations that could be leveraged to develop targeted therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-13125-5.
Keywords: Ovarian clear cell carcinoma, Whole-exome sequencing, Clinical actionability, TERT mutation, Mutational signature, Clonal evolution
Background
Ovarian cancer, primarily consisting of high-grade serous carcinoma (HGSC) and ovarian clear cell carcinoma (OCCC), is a major cause of cancer-related deaths in women, with an annual global death toll exceeding 200,000 [1, 2]. OCCC, which is notably associated with endometriosis, comprises 10 − 25% of ovarian cancers in East Asia compared to 5% in Western countries [3–5]. While OCCC is predominantly diagnosed at early stages (specifically I–II), HGSC is typically identified at advanced stages (namely III–IV). Accordingly, approximately two-thirds of OCCC cases are detected at an early stage, in contrast to only about one-fifth of HGSC cases [4]. Typically, OCCC has a more favorable prognosis than HGSC in the early stages [3–5]. However, as the disease progresses to advanced stages, the clinical outcomes for OCCC become less favorable compared to those of HGSC [6]. This is primarily attributed to the scarcity of targeted therapies and frequent chemoresistance [7].
Extensive research has characterized the somatic mutation landscape in HGSC [8, 9], demonstrating its prognostic value [10, 11]. Notably, mutations in genes such as BRCA1, BRCA2, PALB2, and RAD51C have been specifically associated with HGSC [12–15]. The molecular basis of OCCC, however, is less comprehensively understood. In contrast to HGSC, which is characterized by a high prevalence of TP53 mutations, OCCC is distinguished by frequent mutations in the ARID1A, PIK3CA, KRAS, and PPP2R1A genes [16–20]. Nevertheless, most prior studies in this area have been limited by small sample sizes, leading to an incomplete understanding of the comprehensive genomic landscape of OCCC.
Within a precision medicine framework, this study aims to address this knowledge gap by investigating the mutation patterns in driver genes, somatic copy number alterations (SCNAs), mutational signatures, and clonal evolution of OCCC. Our research presents findings from a large cohort of 102 OCCC samples, for which whole-exome sequencing (WES) data were available. The current results have the potential to identify novel molecular targets that could inform the development of innovative treatment strategies.
Methods
Study participants, tissue specimens, and data collection
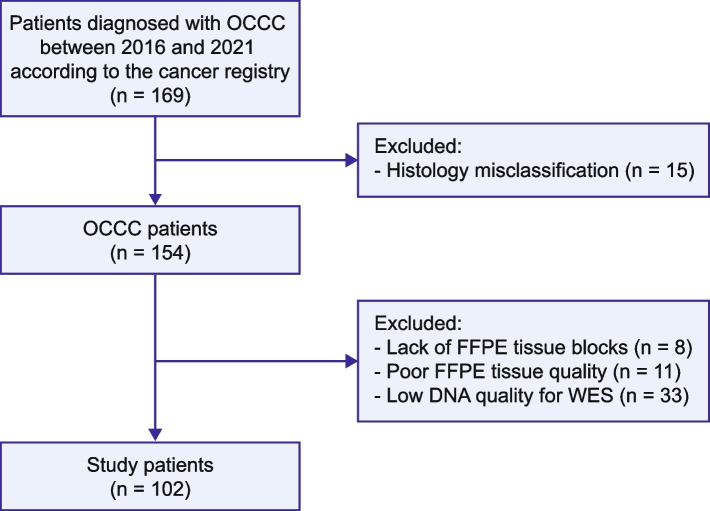
The Institutional Review Board of the Chang Gung Memorial Hospital (Taiwan) approved this study (IRB reference number: 202000143B0), which included a cohort of patients diagnosed with OCCC between 2016 and 2021. Tumor stages were determined using the Federation Internationale de Gynecolgie et d’Obstetrique (FIGO) guidelines [21]. Formalin-fixed, paraffin-embedded (FFPE) specimens were collected from the Linkou and Kaohsiung branches of the Chang Gung Memorial Hospital. Details of patient enrollment are illustrated in Fig. 1. OCCC is characterized by a combination of tubulocystic, papillary, and solid pattern with clear, eosinophilic, and hobnail cells [22]. In cases where peripheral blood samples were unavailable, normal samples were obtained from non-tumoral tissues, such as lymph nodes, fallopian tubes, or the uterus. Clinical and histopathological data, including age, stage, pathological features, and survival outcomes, were retrospectively collected from the patients’ medical records.
Fig. 1.
Study flowchart. Patients diagnosed with ovarian clear cell carcinoma were retrospectively enrolled based on the registry data. The Systemized Nomenclature of Medicine (SNOMED) codes 87,000-A-M83103, 87,000-B-M83103, and 87,000-C-M83103 were used to identify the relevant cases. FFPE, formalin-fixed paraffin-embedded; WES, whole-exome sequencing
DNA extraction and whole-exome sequencing
Genomic DNA was extracted from 10-μm FFPE sections of tumor and normal tissues using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany), and from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen). DNA concentration and integrity were assessed with the Quanti-iT dsDNA HS assay (Invitrogen, Carlsbad, CA, USA) and a fragment analyzer (Advanced Analytical Technologies, Ankeny, IA, USA), respectively. We used the Twist Human Core Exome EF Multiplex Complete Kit Enrichment (Twist Bioscience, South San Francisco, CA, USA) for library preparation. In brief, 50 ng of genomic DNA per sample was subjected to enzymatic fragmentation, and the resulting DNA fragments were subsequently used for library construction. Sequencing was performed on a Novaseq 6000 high-throughput sequencing platform (Illumina, San Diego, CA, USA).
Somatic single nucleotide variation and insertion-deletion calling
Exome reads were trimmed with trimmomatic (version 0.39) to remove adaptors and poor-quality sequences [23]. Subsequently, they were mapped to the human reference sequence GRCh38.p7 using BWA-mem (version 0.7.15) with default parameters [24]. Global mapping quality was evaluated using qualimap 2 (version 2.2.1) [25]. The median read depth for the tumor and normal tissue on the capturing target were 160 × (range: 95 − 255 ×) and 136 × (range: 74 − 452 ×), respectively (Supplementary Table S1).
Somatic single-base substitutions (SBSs) and small insertion and deletions (indels) were called using three different tools: MuTect2 (version 4.1.6.0), Strelka2 (version 2.9.2), and Varscan (version 2.3.9). Default parameters were used in all instances [26–28]. Somatic variants were selected based on the following criteria: (i) identification by at least two variant callers, (ii) a minimum read depth of 10 in normal samples and 20 in tumor samples, (iii) a mutated allele fraction of at least 0.1 for C > T and G > A variants (or 0.05 for others), and (iv) at least 7 supporting reads for C > T and G > A variants in the tumor sample (or 5 for others). SBSs and indels were annotated using wANNOVAR [29]. Gene driver status was determined using the COSMIC database [30]. Molecular targets were classified following the joint consensus of the Association for Molecular Pathology, American College of Medical Genetics and Genomics, American Society of Clinical Oncology, and College of American Pathologists [31].
Clinical actionability classification
The clinical actionability of the identified genetic alterations was categorized into two tiers. Tier 1 encompassed alterations with direct clinical implications and FDA-approved therapies specifically tailored for the given tumor type. Tier 2 included genetic alterations with FDA-approved treatments for different tumor types, as well as those linked to investigational therapies, and findings derived from meta-analyses, preclinical studies, and case reports.
Germline variant calling
We used freebayes (version 1.3.0) for calling germline variants and wANNOVAR for their annotation in normal samples [32]. Our analysis focused on 20 cancer-predisposing genes linked to an increased risk of ovarian cancer [33, 34], including BRCA1, BRCA2, NTHL1, BRIP1, RAD51C, RAD51D, PALB2, ATM, MLH1, MLH3, MSH2, MSH3, MSH6, PMS1, PMS2, EPCAM, STK11, TP53, and CHEK2. Germline variants were validated if they had an allele fraction over 0.2, read depth over 20, and were truncating mutations or known pathogenic variants listed in ClinVar [35].
Mutational signature assignment and spectrum reconstruction
Mutational signature assignment was conducted using the mSigAct R package (version 2.2) and the COSMIC database (version 3.2) [36, 37]. The mSigAct::SparseAssignActivity function was employed to quantify the contribution of SBS signatures in ovarian cancer mutational spectra [37]. Notably, SBS3, associated with homologous recombination deficiency, was only considered when more than five deletions with microhomology were present, whereas SBS6, SBS26, and SBS44, linked to DNA mismatch repair deficiency, were only applied when over 20 insertions in thymine homopolymer sequences were detected.
Somatic copy number alterations and genome doubling
Analysis of SCNAs was performed using the Sequenza software, version 3.0.0 [38], with default settings except for the parameter '–het 0.4' and a 200 kbp resolution. The EstimateClonality R package was employed to infer the genome doubling (GD) status [39]. First, the average copy number (CN) at the chromosomal arm level was calculated from SCNA data. Subsequently, the EstimateClonality::GD.function was used to calculate a p value from 10,000 simulations, based on SCNA probabilities in each tumor, following previously established p value thresholds [39]. Tumors with p values below these cutoffs were classified as having undergone GD. The genomic instability index (GII) was defined as the fraction of the genome altered by somatic CN gains or losses, with a threshold of ≥ 1 compared to the background ploidy.
Consensus clustering on the somatic copy number alteration profile
The genome was partitioned into 5 MB bins by chromosome, and consensus clustering was performed using the ConsensusClusterPlus R package [40]. The clustering was based on the Euclidean distance with 10,000 iterations.
Timing of the clonal status
The clonal status of somatic mutations was assessed using a previously published method to determine the timing of their occurrence [41]. The analysis involved determining the fraction of cancer cells, estimating SCNAs, and evaluating the GD status. Specifically, the MutationTimeR package in R was applied, conducting 1,000 simulations to ascertain the timing of clonal events [41]. Detailed R scripts and the timing data for all somatic mutations are available at the GitHub repository (https://github.com/CYHuang-Lab/CGMH-OCCC-WES-project/).
Sanger sequencing of the TERT gene
The TERT gene promoter and 5’ UTR were amplified using the KAPA HiFi HotStart PCR Kit (Roche, Basel, Switzerland) with two primer sets: promoter: 5’-GTCCTGCCCCTTCACCTT-3’ (forward) and 5’-CAGCGCTGCCTGAAACTC-3’ (reverse); 5’ UTR: 5’-AGCCCCTCCCCTTCCTTT-3’ (forward) and 5’-AGCACCTCGCGGTAGTGG-3’ (reverse). The PCR cycling conditions included an initial denaturation at 95 °C for 3 min, followed by 35 cycles at 98 °C for 20 s, 60 °C for 15 s, and 72 °C for 15 s, with a final extension at 72 °C for 1 min. The PCR products were subsequently purified and subjected to Sanger sequencing.
Immunohistochemistry for WT1, napsin A, and HNF1B
Immunohistochemistry (IHC) was performed on a BOND-MAX automated stainer (Leica Biosystems, Nußloch, Germany) using the following primary antibodies and dilutions: WT1 (clone 6F-H2, Cell Marque, Rocklin, CA, USA; 1:100 dilution), napsin A (clone IP64, Leica Biosystems; 1:200 dilution), and HNF1B (catalog number 12533–1-AP, Proteintech, Rosemont, IL, USA; 1:100 dilution). Heat-induced epitope retrieval was carried out at 100 °C using a citrate-based pH 6.0 buffer (BOND Epitope Retrieval Solution 1, Leica Biosystems) for HNF1B and an EDTA-based pH 9.0 buffer (BOND Epitope Retrieval Solution 2, Leica Biosystems) for the remaining antibodies.
Statistical testing and survival analysis
All analyses were performed in R (version 4.1.0) with two-sided tests, unless specified otherwise. Categorical variables were compared with the Fisher’s exact test, whereas the Wilcoxon rank-sum test was used to analyze continuous variables. Overall survival was measured from the date of pathological diagnosis of OCCC to the date of the last follow-up or death. The survdiff and coxph functions from the R survival package were employed. The p values were adjusted for multiple testing using the Benjamini–Hochberg method, which controls the false discovery rate. In this analysis, p values < 0.05 and q values < 0.1 were considered statistically significant.
Results
Patient characteristics
Supplementary Table S2 summarizes the clinical outcomes and genomic characteristics of the 102 patients included in the study. The median age of the participants was 52 years (range: 25 − 79 years). Most patients (n = 72, 70.6%) presented with stage I disease at diagnosis. The median follow-up duration was 28.9 months (range: 0.9–69.9 months).
Genomic landscape and driver mutations of OCCC
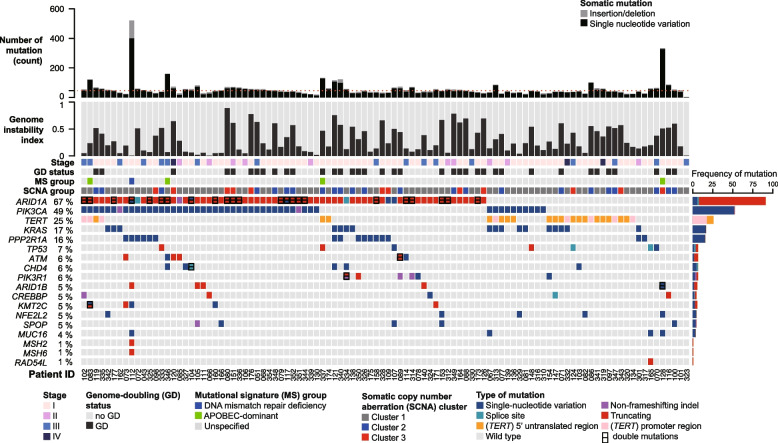
Figure 2 and Supplementary Fig. S1 depict the genomic profile of OCCC samples, revealing 5370 somatic SBSs and 580 indels impacting 4264 genes and the splicing junctions of 121 genes. The median non-silent mutations count was 46 (range: 3 − 521). Consistent with previous reports [16–20], ARID1A (66.7%) and PIK3CA (49%) were the most commonly mutated driver genes. Promoter or 5’ UTR mutations of the TERT gene were identified in 26 (25.5%) tumors. Mutations were also frequently observed in KRAS (16.7%), PPP2R1A (15.7%), and TP53 (6.9%). Enrichment analysis revealed that mutations in ARID1A, PIK3CA, KRAS, PPP2R1A, and PIK3R1 were more prevalent in OCCC (Supplementary Fig. S2), whereas TP53 has been reported to be frequently mutated in HGSC [8]. The TP53-mutated cases in our cohort were corroborated by immunohistochemical negativity for WT1, napsin A, and HNF1B. One patient (tumor OCCC-067) harbored a pathogenic germline mutation in RAD51C without a concurrent somatic TP53 mutation (Supplementary Table S3). Additionally, a single tumor (OCCC-112) exhibited somatic mutations in both MSH2 and MSH6, suggesting the rare occurrence of DNA mismatch repair deficiency in OCCC.
Fig. 2.
Genomic landscape of 102 ovarian clear cell carcinoma samples. The rows represent the following data: counts of non-silent mutations, genome instability index, tumor stage, genome doubling (GD) status, mutational signature (MS) group, membership in somatic copy number alteration (SCNA) cluster, and non-silent somatic mutations in established cancer-driver genes (mutated in five or more tumors), as well as mutations in the TERT upstream region. Supplementary Fig. 1 illustrates mutations in known cancer-driver genes at lower frequencies
Mutational patterns and clinical actionability
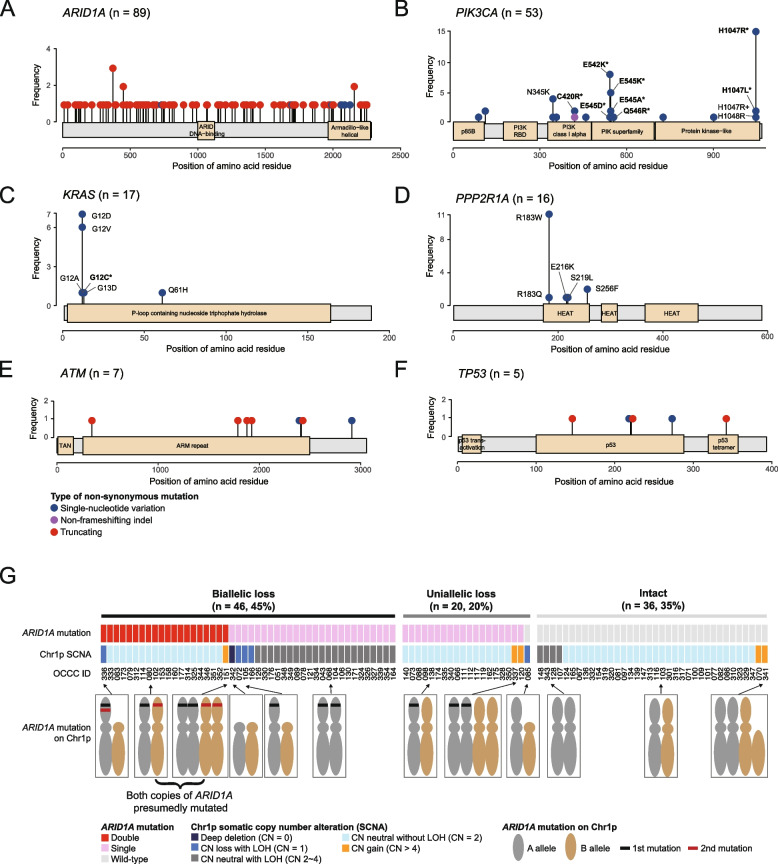
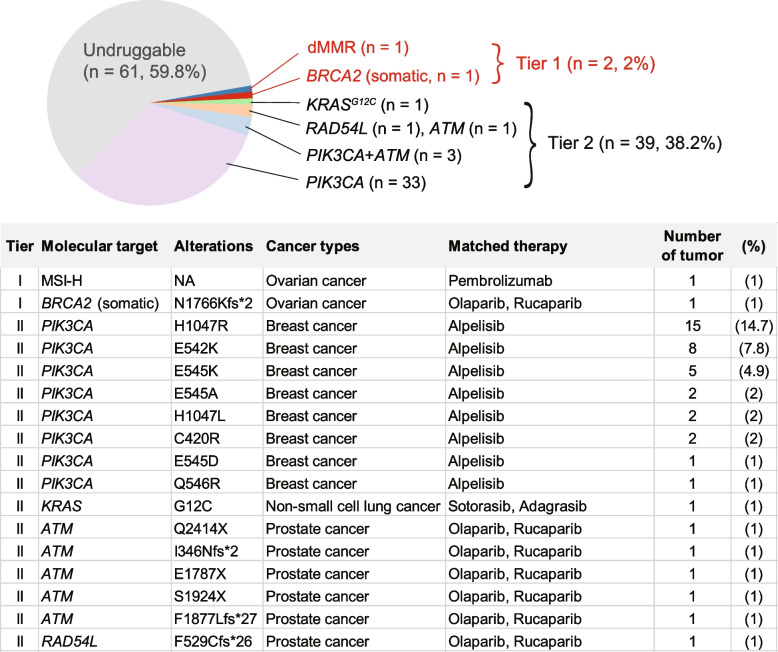
The mutational patterns of ARID1A, ATM, and TP53, which were typical of tumor suppressor genes, were predominantly characterized by truncating mutations and an absence of hotspots (Fig. 3A, 3E, and 3F). Of the 91 non-silent ARID1A mutations, 91.2% (83 mutations) were truncating, 6.6% (6 mutations) were missense, and 2.2% (2 mutations) occurred at splice sites. In contrast, PIK3CA, KRAS, and PPP2R1A mutations were characterized by the presence of hotspots and an absence of truncating mutations (Fig. 3B, C, and D). PIK3CA exhibited hotspot mutations primarily at the H1047 (n = 17), E542 (n = 9), and E545 (n = 8) positions (Fig. 3B), whereas the most common KRAS mutations were G12D (n = 7) and G12V (n = 6) (Fig. 3C). The prevalent PPP2R1A mutation identified in our cohort was R183W (11 out of 16) (Fig. 3D), a finding in line with previous research [42]. According to the current classification of clinical actionability in oncology [31], 41 out of 102 OCCC samples (40.2%) exhibited clinically actionable molecular targets. This encompassed tier 1 actionability in two specific tumors (2%): one exhibiting DNA mismatch repair deficiency (dMMR) and another with a somatic BRCA2 mutation. Additionally, tier 2 actionability was identified in 39 tumors (37.5%), including 33 with PIK3CA mutations, three with PIK3CA and ATM mutations, one with a KRASG12C mutation, one with a RAD54L mutation, and one with an ATM mutation.
Fig. 3.
Types and frequencies of mutations in six driver genes: (A) ARID1A, (B) PIK3CA, (C) KRAS, (D) PPP2R1A, (E) ATM, and (F) TP53 gene. Selected mutation types were annotated accordingly. Mutations that fall within the tier 1 or 2 clinical actionability are emphasized using asterisk (*) markings. G Loss of tumor suppressor genes in the OCCC tumors. Biallelic loss is defined as having either of the following conditions: 1) double truncating mutations with or without loss-of-heterozygosity (LOH), 2) single truncating mutation with LOH, or 3) complete copy number loss (copy number of zero). Uniallelic loss is defined as having either of the following conditions: 1) single truncating mutation without LOH, 2) Copy number loss without truncating mutation
A total of 91 non-silent mutations were identified in the ARID1A gene, with 83 being truncating mutations, potentially leading to a loss of function. Subsequently, we investigated the frequency of ARID1A deficiency due to biallelic loss using a previously described assessment method [43]. Biallelic loss was defined as the presence of either two truncating mutations, a single truncating mutation with loss of heterozygosity (LOH), or total copy number loss. The results revealed that 45.1% (46/102) of tumors exhibited biallelic loss of ARID1A (Fig. 3G), with 43.5% having two truncating mutations and 56.5% a single truncating mutation with LOH. Figure 4 summarizes the clinical actionability and the corresponding potential therapies for the study cohort.
Fig. 4.
Clinical actionability of mutations identified in ovarian clear cell carcinoma specimens. Tier 1 alterations are those with direct clinical implications for a specific tumor type, including therapies approved by the FDA. Tier 2 includes alterations for which FDA-approved therapies exist for other tumor types, as well as investigational therapies, consensus findings from meta-analyses, preclinical studies, and case reports
TERT mutations in the 5’ untranslated region correlated with poor outcomes
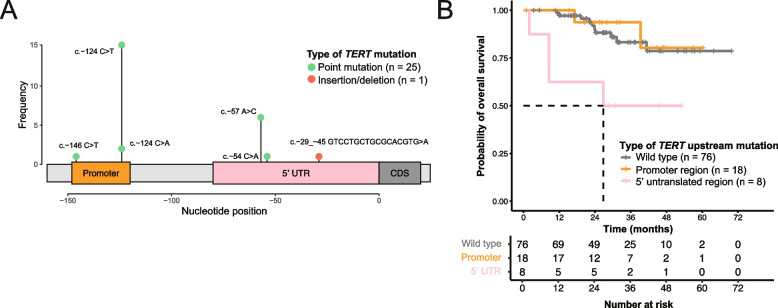
TERT mutations were identified in 25.5% (26/102) of OCCC samples (Fig. 5 and Supplementary Table S4), predominantly located in regulatory regions and notably creating novel ETS binding motifs. The majority of TERT mutations (18 of 26, 69%) were identified in the promoter region [44], whereas a significant proportion (8 of 26, 31%) was found in the less frequently studied 5’ UTR [45]. These mutations were collectively referred to as TERT upstream mutations. To ensure unbiased detection of TERT upstream mutations, we examined the sequencing coverage of the TERT upstream region (chr5:1,294,990–1,295,146) across our cohort. The median read depth was 99 (range: 26 − 235) for tumors and 60 (range: 21 − 299) for normal samples, with no significant difference between TERT-mutated and not mutated tumors (median: 99 versus 100, p = 0.867). Furthermore, Sanger sequencing validated 88.5% (23 out of 26) of these mutations, confirming the reliability of our findings (Supplementary Table S4). In our study, the most frequent TERT mutation was c.-124C > T (C228T), detected 15 times, followed by c.-57A > C (A161C) six times, c.-124C > A (C228A) twice, c.-54C > A (C158A) once, and c.-146C > T (C250T) once (Fig. 5A). We also discovered a novel complex indel (c.-29_-45 GTCCTGCTGCGCACGTG > A) in the 5’ UTR in one tumor. TERT promoter and 5’ UTR mutations were mutually exclusive. Consistent with a previous report [44], TERT mutations were more common in ARID1A-wild-type tumors (20 of 34, 58.8%) than in ARID1A-mutated tumors (6 of 68, 8.8%; two-sided Fisher’s exact test, p = 1.3 × 10–7). Tumors with TERT 5’ UTR mutations were associated with shorter overall survival (median, 26.8 months) compared to those without these mutations (median survival not reached, univariate hazard ratio [HR] = 4.49, 95% confidence interval [CI] = 1.4 to 14.34, p = 0.011, and multivariate HR = 3.86, 95% CI = 1.19 to 12.55, p = 0.025; Fig. 5B, Supplementary Tables S5 and 6). These findings suggest that TERT upstream mutations may serve as a prognostic marker in OCCC, warranting further validation in larger cohorts.
Fig. 5.
TERT upstream mutations identified in ovarian clear cell carcinoma samples. A Classification and prevalence of TERT upstream mutations. B The presence 5’ UTR mutations was associated with significantly worse survival than lack of an upstream mutation or a mutation in the promoter region, as shown in both a univariate (HR = 4.49 versus wild type, p = 0.011) and a multivariable Cox proportional hazards analysis (HR = 3.86, versus wild type, p = 0.025, Supplementary Table S5). UTR, untranslated region; CDS, coding sequence
Mutational signature analysis
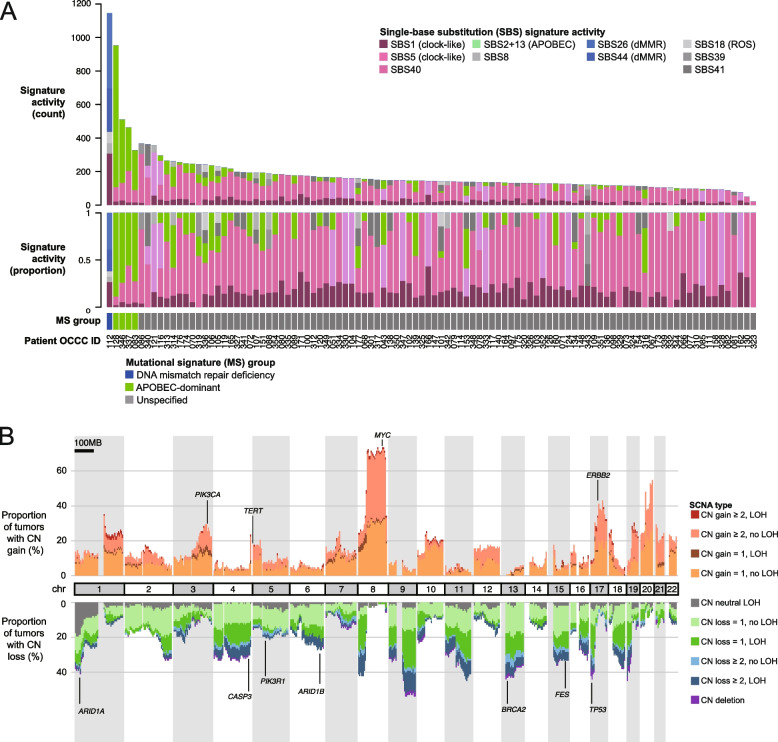
We identified a median of 143 SBSs (range: 21 to 1146) per tumor. Figure 6A and Supplementary Fig. S3 illustrate the distribution of SBS signatures across the examined OCCC samples. The clock-like signatures SBS1 and SBS5 were present in 94 (92.2%) and 86 (84.3%) of tumors, respectively, with a median of 25 (range: 0 to 308) and 89 (range: 0 to 291) mutations attributed to each. APOBEC-related signatures (SBS2 and SBS13) were identified in 42 (41.2%) of tumors. Notably, four malignancies (OCCC-083, 337, 346, and 128) exhibited high APOBEC mutational activity (median: 319 mutations, range: 238 to 844), without correlation to patient age, stage, driver mutations, or overall survival.
Fig. 6.
A Single-base substitution (SBS) mutation signature activities across all 102 OCCCs. Mutation counts and the proportions of signatures contributing to the mutational spectrum of each tumor were shown in the top two panels. The bottom panel indicates tumors classified by the following “mutational signature groups”: (1) DNA mismatch repair deficiency (OCCC-112), (2) APOBEC-dominant (OCCC-83, 128, 337, and 346). B Genome-wide somatic copy number alteration (SCNA) patterns observed in the OCCC cohort
Somatic copy number alterations are pervasive and heterogeneous in OCCC
SCNAs are crucial for cancer progression and treatment response [46]. In our study, the median GII was 0.24, ranging from 0.01 to 0.89. A significant majority of OCCC samples (94.1%) showed copy number changes exceeding 1 in at least 5% of their genome. GD, a prevalent ploidy abnormality in cancer, was observed in 36.3% of samples. Although TP53 mutations are linked to GD in other cancers [39], our findings in OCCC revealed no correlation between GD and driver mutations, including TP53 variants (Supplementary Table S7). The most common SCNA in OCCC was 8q amplification, identified in over 60% of the tumors. Additional common amplifications were observed in 3q, 5p, and 17q, while copy-number losses were identified in 1p, 4q, 5q, 6q, 13q, 15q, and 17p (Fig. 6B). Approximately 20% of tumors exhibited copy-neutral LOH at 1p, affecting the ARID1A gene. SCNAs profiling divided OCCC tumors into three clusters (Supplementary Fig. S4): Cluster 1 (65.7%) with low-to-moderate instability and a low GD rate (13%); Cluster 2 (22.5%) with a high GD rate (95.7%) and extensive copy number loss; and Cluster 3 (11.8%) with a high GD rate, high genomic instability, and LOH. We observed a non-significant trend indicating potentially better survival outcomes for patients with tumors classified in SCNA Cluster 3 (Supplementary Fig. S5).
Timing of driver mutations during OCCC evolution
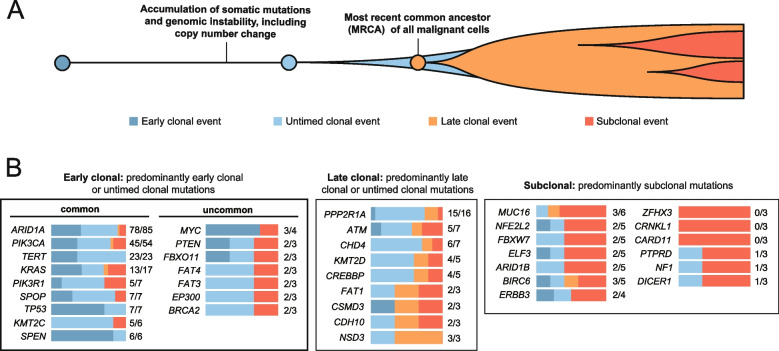
To investigate the timing of driver mutations in OCCC, we employed a previously described method [41, 47] that distinguishes mutations as clonal or subclonal based on variant allele fractions and SCNAs status (Fig. 7A). This approach further classifies clonal mutations as “early clonal”, “late clonal”, or “untimed clonal” depending on their copy number post-amplification (Fig. 7A). There were 3,098 early clonal mutations, 1,822 late clonal mutations, 8,120 untimed clonal mutations, and 6,628 subclonal mutations. Genes with a high frequency of early clonal and untimed clonal mutations – such as ARID1A, PIK3CA, TERT, and KRAS – are implicated in tumor initiation (Fig. 7B). Notably, ARID1A, TERT, and PIK3CA mutations were predominantly clonal compared to non-driver genes (Supplementary Table S8, two-sided Fisher’s exact test with a Benjamini–Hochberg correction, q values of 1.044 × 10–6, 0.0015, and 0.0746, respectively).
Fig. 7.
Driver mutation timing estimates in ovarian clear cell carcinoma specimens. A Based on the timing of somatic mutations relative to somatic copy number change at the same locus, we categorized clonal mutation events as “early clonal” (occurring before the copy-number gain), “untimed clonal” (inability to determine the timing relative to the somatic copy-number gain), and “late clonal” (occurring after the somatic-copy-number gain). B Driver genes were grouped according to the timing and clonality of the identified somatic mutations. Color bars to the right of each gene symbol indicate the proportions of early clonal (dark blue), untimed clonal (light blue), late clonal (orange), and subclonal (dark orange) mutations for each gene. The values displayed to the right of the bar graph indicate the number of clonal mutations and the total mutation count within each specified gene. Genes categorized as “early clonal” are identified as potential key factors in the initiation of OCCC
Discussion
OCCC is notably resistant to conventional chemotherapy, particularly regimens based on platinum and taxanes, which are standard first-line treatments for ovarian cancer [3–5]. This chemoresistance is attributed to several key factors, including the inherently low proliferation rate of OCCC, which limits the efficacy of chemotherapeutic agents. Moreover, OCCC cells frequently display reduced drug accumulation and enhanced drug detoxification mechanisms, which further compromise the clinical efficacy of chemotherapy. Notwithstanding extensive research endeavors, a tailored chemotherapy regimen specifically designed for OCCC remains elusive. Consequently, elucidating the molecular underpinnings of this malignancy is crucial for identifying novel therapeutic targets and developing more effective treatment strategies. Our comprehensive study of OCCC genomic profile revealed that only 2% (2 out of 102) of samples had tier 1 clinically actionable targets, which are suitable for routine clinical use. These included one case with dMMR and another with a BRCA2 mutation. Notably, BRCA2 mutations can respond favorably to poly ADP-ribose polymerase inhibitors, which have been approved for advanced ovarian cancer [48, 49]. Similarly, an immune checkpoint inhibitor targeting the PD-1 pathway is available for advanced dMMR malignancies [50]. Tier 2 clinical actionability – which refers to investigational molecular targets – was more common, being identified in 38.2% (39/102) of the analyzed samples. This category included mutations in the PIK3CA, KRAS, RAD54L, and ATM genes. PIK3CA mutations are of particular interest due to their potential responsiveness to alpelisib, a selective inhibitor of the PI3Kα protein. This drug is currently approved for PIK3CA-mutated breast cancer [51], and its efficacy in treating ovarian cancer is currently under investigation [52]. ERBB2 amplification has also emerged as a promising therapeutic target in OCCC [53]. To expand the feasibility and applicability of targeted therapy clinical trials for OCCC, it is recommended to include ERBB2 amplification as a criterion for patient enrollment in basket trials designed to target this specific genetic alteration. Similarly, covalent inhibitors like sotorasib and adagrasib have shown promise in treating non-small-cell lung cancer harboring the KRASG12C mutation [54, 55]. Moreover, inhibitors targeting the KRASG12D mutation have demonstrated encouraging clinical responses [56].
In addition to tier 1 and 2 actionable mutations, our analysis revealed that biallelic loss of ARID1A [57] was present in 45% of the examined samples. This observation implies that the preclinical investigation of EZH2 inhibitors, which have shown promise in suppressing the growth of ARID1A-mutated cancers [58], may be a viable avenue for further exploration. Additionally, mutations in PPP2R1A were detected in 16% of the samples, potentially affecting cell growth through alterations in the Aα subunit of serine/threonine phosphatase 2A (PP2A) [42]. These mutations may be amenable to targeting with ribonucleotide reductase inhibitors [59].
Our analysis also confirmed the involvement of the TERT gene in the pathogenesis of OCCC [44]. with mutations in the TERT promoter and 5’ UTR being identified in 17.6% and 7.8% of samples, respectively. This is, to our knowledge, the first study to report the occurrence of TERT 5’ UTR mutations in OCCC. These mutations could lead to increased TERT expression and telomerase activation. Mutations in the TERT promoter create a de novo binding motif for transcription factors, whereas mutations in the 5’ UTR disrupt the connection between the repressor complex MAX/Mad1 and the E-box sequence (CACGTG) [60]. Notably, OCCC patients with TERT 5’ UTR mutations exhibited poorer survival outcomes compared to those with TERT promoter mutations or no mutations, suggesting the clinical relevance of this molecular pathway. While directly targeting TERT or its promoter mutations is challenging due to the complex role of telomerase in cancer [45], these variants may influence responses to certain therapies, such as BRAF and MEK inhibitors [60]. This highlights the potential of TERT mutations in guiding the development of targeted treatments for OCCC.
Our study not only identified molecular changes that could serve as a foundation for preclinical exploration of novel therapeutic strategies, but also provided evidence supporting the hypothesis that OCCC originates from endometriotic cells, in line with the “precursor escape” model [61]. This framework proposes that endometriotic cells gradually amass genetic alterations, eventually giving rise to OCCC. Our analysis of OCCC samples also revealed early clonal mutations in the KRAS and PIK3CA genes. Similar variants have been previously reported in endometriotic epithelium [62], suggesting these genetic alterations may be involved in OCCC initiation.
The molecular context of the 5’ UTR TERT mutations identified in our study can be inferred from the findings of Huang et al. [45]. They reported that the c.-57A > C (A161C) mutation generates a “(T/A)TCC” sequence, which is a de novo putative ETS-transcription factor binding motif. Similarly, the c.-54C > A (C158A) mutation creates a CCGGAA/T motif, which is a potential binding site for ETS transcription factors. These mutations were found to correlate with upregulated TERT mRNA expression and increased telomerase activity in adult gliomas. Furthermore, we identified a novel complex indel (c.-29_-45 GTCCTGCTGCGCACGTG > A) in the 5’ UTR of TERT, which contains canonical E-box (CACGTG) elements. Previous research has shown that this E-box element mediates repression of TERT transcription in a renal cell carcinoma cell line derivative (RCC23 + 3) with a transferred copy of normal human chromosome 3 [63]. Taken together, these findings suggest that the 5’ UTR TERT mutations may enhance the transcriptional activity of the TERT core promoter.
Regarding frequently altered genes and their prognostic implications in OCCC, our study identified an association between 5’ UTR TERT mutations and reduced overall survival. While mutations upstream of the TERT promoter are recognized as significant prognostic markers in OCCC [44], it is important to note that the median follow-up period for our cohort was relatively short (2.5 years) and the occurrence of these genetic alterations was limited. Notably, other genetic mutations, such as those in ARID1A and PIK3CA, have been associated with unfavorable prognosis in a smaller series of 55 patients [64]. In a separate study, the loss of ARID1A expression was identified as a negative prognostic factor in 9 out of 60 patients with OCCC who received platinum-based chemotherapy [65]. However, these findings are preliminary and should be interpreted with caution. To derive more reliable conclusions, further validation is essential, particularly in larger cohorts with extended follow-up periods. Moreover, comprehensive molecular investigations are necessary to fully elucidate the role of frequently altered genes in OCCC.
In the current study, we found that SCNA was prevalent and heterogeneous in OCCC. In particular, we identified a group of OCCCs with high degree of genomic instability and better survival outcomes in this SCNA cluster 3 group. Previously, TP53 mutation and homologous recombination deficiency may contribute to the genomic instability and genome doubling [39, 66]. However, the exact molecular mechanisms underlies the genomic instability is not fully characterized. Our study demonstrated that extent of SCNA in OCCC may represent a potential prognostic marker, necessitating validation in a larger cohort.
In conclusion, the current study offers a thorough analysis of the genomic alterations and clonal progression in OCCC, uncovering numerous mutations that could be targeted by novel pharmacological treatments. The study also supports the hypothesis that OCCC originates from endometriotic epithelium. These insights have the potential to form the foundation for advancements in precision oncology, facilitating the development of more effective therapeutic strategies for patients with OCCC.
Supplementary Information
Acknowledgements
This study was financially supported by a grant (CIRPG 3K0031/2) from the Chang Gung Medical Foundation.
Abbreviations
- CI
Confidence interval
- CN
Copy number
- dMMR
Mismatch repair deficiency
- FFPE
Formalin-fixed, paraffin-embedded
- GD
Genome doubling
- GII
Genomic instability index
- HGSC
High-grade serous carcinoma
- HR
Hazard ratio
- LOH
Loss of heterozygosity
- MS
Mutational signature
- IRB
Institutional review board
- OCCC
Ovarian clear cell carcinoma
- PP2A
Phosphatase 2A
- SCNAs
Somatic copy number alterations
- SBSs
Somatic single-base substitutions
- UTR
Untranslated region
- WES
Whole-exome sequencing
Authors’ contributions
AC, RCW, and CHL designed the study. AC, CYL, and RCW performed the experiments. AC, CYH, SGR, and RCW analyzed and interpreted the data. RCW performed the pathology assessment and Sanger sequencing. AC, CHL, ASC, CTL, HHC, KGH, HJH, TCC, and CHL collected patient samples and clinical data. WY performed the sequencing data encryption and uploaded to EGA. AC and CYH wrote the manuscript. SGR, RCW, and CHL reviewed and edited the manuscript. The final manuscript was read and approved by all authors.
Data availability
The catalog of somatic mutations, SCNA estimations, and mutation timing analyses from the study cohort are accessible at the following GitHub repository: https://github.com/CYHuang-Lab/CGMH-OCCC-WES-project/. Tumor and normal exome sequencing data have been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/; accession number EGAS50000000031). The R codes utilized in this study are accessible at https://github.com/CYHuang-Lab/CGMH-OCCC-WES-project/.
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the Chang Gung Memorial Hospital (reference number: 202000143B0). Given the retrospective nature of the analysis, the requirement for informed consent was waived.
Consent for publication
All authors have provided their consent for the publication of this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Angel Chao and Chen-Yang Huang contributed equally to this work.
Contributor Information
Ren-Chin Wu, Email: qby@cgmh.org.tw.
Chyong-Huey Lai, Email: laich46@cgmh.org.tw.
References
- 1.Cabasag CJ, Fagan PJ, Ferlay J, Vignat J, Laversanne M, Liu L, van der Aa MA, Bray F, Soerjomataram I. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. Int J Cancer. 2022;151(9):1535–41. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 3.Ku FC, Wu RC, Yang LY, Tang YH, Chang WY, Yang JE, Wang CC, Jung SM, Lin CT, Chang TC, et al. Clear cell carcinomas of the ovary have poorer outcomes compared with serous carcinomas: Results from a single-center Taiwanese study. J Formos Med Assoc. 2018;117(2):117–25. [DOI] [PubMed] [Google Scholar]
- 4.Chan JK, Teoh D, Hu JM, Shin JY, Osann K, Kapp DS. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecol Oncol. 2008;109(3):370–6. [DOI] [PubMed] [Google Scholar]
- 5.del Carmen MG, Birrer M, Schorge JO. Clear cell carcinoma of the ovary: a review of the literature. Gynecol Oncol. 2012;126(3):481–90. [DOI] [PubMed] [Google Scholar]
- 6.Peres LC, Cushing-Haugen KL, Kobel M, Harris HR, Berchuck A, Rossing MA, Schildkraut JM, Doherty JA. Invasive Epithelial Ovarian Cancer Survival by Histotype and Disease Stage. J Natl Cancer Inst. 2019;111(1):60–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang HJ, Yang LY, Tung HJ, Ku FC, Wu RC, Tang YH, Chang WY, Jung SM, Wang CC, Lin CT, et al. Management and clinical outcomes of patients with recurrent/progressive ovarian clear cell carcinoma. J Formos Med Assoc. 2020;119(4):793–804. [DOI] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, Mirza H, Ennis DP, Smith P, Morrill Gavarro L, Sokota C, Giannone G, Goranova T, Bradley T, Piskorz A, et al. The Genomic Landscape of Early-Stage Ovarian High-Grade Serous Carcinoma. Clin Cancer Res. 2022;28(13):2911–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garsed DW, Pandey A, Fereday S, Kennedy CJ, Takahashi K, Alsop K, Hamilton PT, Hendley J, Chiew YE, Traficante N, et al. The genomic and immune landscape of long-term survivors of high-grade serous ovarian cancer. Nat Genet. 2022;54(12):1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang SYC, Lheureux S, Karakasis K, Burnier JV, Bruce JP, Clouthier DL, Danesh A, Quevedo R, Dowar M, Hanna Y, et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med. 2018;10(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harter P, Hauke J, Heitz F, Reuss A, Kommoss S, Marme F, Heimbach A, Prieske K, Richters L, Burges A, et al. Prevalence of deleterious germline variants in risk genes including BRCA1/2 in consecutive ovarian cancer patients (AGO-TR-1). PLoS ONE. 2017;12(10):e0186043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, Kwan E, Jack E, Vesprini DJ, Kuperstein G, Abrahamson JL, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68(3):700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arcieri M, Andreetta C, Tius V, Zapelloni G, Titone F, Restaino S, Vizzielli G. Molecular biology as a driver in therapeutic choices for ovarian cancer. Int J Gynecol Cancer. 2024. [DOI] [PubMed]
- 15.Pietragalla A, Arcieri M, Marchetti C, Scambia G, Fagotti A. Ovarian cancer predisposition beyond BRCA1 and BRCA2 genes. Int J Gynecol Cancer. 2020;30(11):1803–10. [DOI] [PubMed] [Google Scholar]
- 16.Shibuya Y, Tokunaga H, Saito S, Shimokawa K, Katsuoka F, Bin L, Kojima K, Nagasaki M, Yamamoto M, Yaegashi N, et al. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer. 2018;57(2):51–60. [DOI] [PubMed] [Google Scholar]
- 17.Murakami R, Matsumura N, Brown JB, Higasa K, Tsutsumi T, Kamada M, Abou-Taleb H, Hosoe Y, Kitamura S, Yamaguchi K, et al. Exome Sequencing Landscape Analysis in Ovarian Clear Cell Carcinoma Shed Light on Key Chromosomal Regions and Mutation Gene Networks. Am J Pathol. 2017;187(10):2246–58. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Zhang C, Ren Y, Yi H, Luo T, Xing F, Bai X, Cui L, Zhu L, Ouyang J, et al. Genomic characterization of Chinese ovarian clear cell carcinoma identifies driver genes by whole exome sequencing. Neoplasia. 2020;22(9):399–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SI, Lee JW, Lee M, Kim HS, Chung HH, Kim JW, Park NH, Song YS, Seo JS. Genomic landscape of ovarian clear cell carcinoma via whole exome sequencing. Gynecol Oncol. 2018;148(2):375–82. [DOI] [PubMed] [Google Scholar]
- 20.Maru Y, Tanaka N, Ohira M, Itami M, Hippo Y, Nagase H. Identification of novel mutations in Japanese ovarian clear cell carcinoma patients using optimized targeted NGS for clinical diagnosis. Gynecol Oncol. 2017;144(2):377–83. [DOI] [PubMed] [Google Scholar]
- 21.Berek JS, Kehoe ST, Kumar L, Friedlander M. Cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2018;143(Suppl 2):59–78. [DOI] [PubMed] [Google Scholar]
- 22.Bolton KL, Chen D. Corona de la Fuente R, Fu Z, Murali R, Kobel M, Tazi Y, Cunningham JM, Chan ICC, Wiley BJ et al: Molecular Subclasses of Clear Cell Ovarian Carcinoma and Their Impact on Disease Behavior and Outcomes. Clin Cancer Res. 2022;28(22):4947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okonechnikov K, Conesa A, Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32(2):292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Kallberg M, Chen X, Kim Y, Beyter D, Krusche P, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15(8):591–4. [DOI] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49(7):433–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18(11):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrison E, Marth G: Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:12073907 2012.
- 33.Hodan R, Kingham K, Cotter K, Folkins AK, Kurian AW, Ford JM, Longacre T. Prevalence of Lynch syndrome in women with mismatch repair-deficient ovarian cancer. Cancer Med. 2021;10(3):1012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eoh KJ, Kim JE, Park HS, Lee ST, Park JS, Han JW, Lee JY, Kim S, Kim SW, Kim JH, et al. Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2. Cancer Res Treat. 2018;50(3):917–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng AWT, Poon SL, Huang MN, Lim JQ, Boot A, Yu W, Suzuki Y, Thangaraju S, Ng CCY. Tan P et al: Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci Transl Med. 2017;9(412):eaan6446. [DOI] [PubMed] [Google Scholar]
- 37.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, Szallasi Z, Eklund AC. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26(1):64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014;4(2):175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, Mitchell TJ, Rubanova Y, Anur P, Yu K, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578(7793):122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shih Ie M, Panuganti PK, Kuo KT, Mao TL, Kuhn E, Jones S, Velculescu VE, Kurman RJ, Wang TL. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am J Pathol. 2011;178(4):1442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen L. J WMM, Van Hoeck A, Cuppen E: Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11(1):5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu RC, Ayhan A, Maeda D, Kim KR, Clarke BA, Shaw P, Chui MH, Rosen B, Shih Ie M, Wang TL. Frequent somatic mutations of the telomerase reverse transcriptase promoter in ovarian clear cell carcinoma but not in other major types of gynaecological malignancy. J Pathol. 2014;232(4):473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DS, Wang Z, He XJ, Diplas BH, Yang R, Killela PJ, Meng Q, Ye ZY, Wang W, Jiang XT, et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur J Cancer. 2015;51(8):969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watkins TBK, Lim EL, Petkovic M, Elizalde S, Birkbak NJ, Wilson GA, Moore DA, Gronroos E, Rowan A, Dewhurst SM, et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature. 2020;587(7832):126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7(283):283ra254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, Korach J, Huzarski T, Poveda A, Pignata S, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274–84. [DOI] [PubMed] [Google Scholar]
- 49.Kristeleit R, Lisyanskaya A, Fedenko A, Dvorkin M, de Melo AC, Shparyk Y, Rakhmatullina I, Bondarenko I, Colombo N, Svintsitskiy V, et al. Rucaparib versus standard-of-care chemotherapy in patients with relapsed ovarian cancer and a deleterious BRCA1 or BRCA2 mutation (ARIEL4): an international, open-label, randomised, phase 3 trial. Lancet Oncol. 2022;23(4):465–78. [DOI] [PubMed] [Google Scholar]
- 50.Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, Geva R, Gottfried M, Penel N, Hansen AR, et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol. 2020;38(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rugo HS, Lerebours F, Ciruelos E, Drullinsky P, Ruiz-Borrego M, Neven P, Park YH, Prat A, Bachelot T, Juric D, et al. Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): one cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol. 2021;22(4):489–98. [DOI] [PubMed] [Google Scholar]
- 52.Konstantinopoulos PA, Barry WT, Birrer M, Westin SN, Cadoo KA, Shapiro GI, Mayer EL, O’Cearbhaill RE, Coleman RL, Kochupurakkal B, et al. Olaparib and alpha-specific PI3K inhibitor alpelisib for patients with epithelial ovarian cancer: a dose-escalation and dose-expansion phase 1b trial. Lancet Oncol. 2019;20(4):570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedemeyer K, Wang L, Kang EY, Liu S, Ou Y, Kelemen LE, Feil L, Anglesio MS, Glaze S, Ghatage P, et al. Prognostic and Theranostic Biomarkers in Ovarian Clear Cell Carcinoma. Int J Gynecol Pathol. 2022;41(2):168–79. [DOI] [PubMed] [Google Scholar]
- 54.Janne PA, Riely GJ, Gadgeel SM, Heist RS, Ou SI, Pacheco JM, Johnson ML, Sabari JK, Leventakos K, Yau E, et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRAS(G12C) Mutation. N Engl J Med. 2022;387(2):120–31. [DOI] [PubMed] [Google Scholar]
- 55.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, Italiano A, Schuler M, Borghaei H. Barlesi F et al: Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med. 2021;384(25):2371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao Z, Xiao H, Shen P, Yang Y, Xue J, Yang Y, Shang Y, Zhang L, Li X, Zhang Y, et al. KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge. Cell Discov. 2022;8(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122(3):135–40. [DOI] [PubMed] [Google Scholar]
- 58.Alldredge JK, Eskander RN. EZH2 inhibition in ARID1A mutated clear cell and endometrioid ovarian and endometrioid endometrial cancers. Gynecol Oncol Res Pract. 2017;4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connor CM, Taylor SE, Miller KM, Hurst L, Haanen TJ, Suhan TK, Zawacki KP, Noto FK, Trako J, Mohan A, et al. Targeting Ribonucleotide Reductase Induces Synthetic Lethality in PP2A-Deficient Uterine Serous Carcinoma. Cancer Res. 2022;82(4):721–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38(34):6172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng CH, Wu RC, Chen SJ, Chen HC, Tan KT, Lee YS, Huang SS, Yang LY, Wang CJ, Chou HH, et al. Molecular evidence for a clonal relationship between synchronous uterine endometrioid carcinoma and ovarian clear cell carcinoma: a new example of “precursor escape”? J Mol Med (Berl). 2021;99(7):959–66. [DOI] [PubMed] [Google Scholar]
- 62.Suda K, Nakaoka H, Yoshihara K, Ishiguro T, Tamura R, Mori Y, Yamawaki K, Adachi S, Takahashi T, Kase H, et al. Clonal Expansion and Diversification of Cancer-Associated Mutations in Endometriosis and Normal Endometrium. Cell Rep. 2018;24(7):1777–89. [DOI] [PubMed] [Google Scholar]
- 63.Horikawa I, Cable PL, Mazur SJ, Appella E, Afshari CA, Barrett JC. Downstream E-box-mediated regulation of the human telomerase reverse transcriptase (hTERT) gene transcription: evidence for an endogenous mechanism of transcriptional repression. Mol Biol Cell. 2002;13(8):2585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnack TH, Oliveira DNP, Christiansen AP, Hogdall C, Hogdall E. Prognostic impact of molecular profiles and molecular signatures in clear cell ovarian cancer. Cancer Genet. 2023;278–279:9–16. [DOI] [PubMed] [Google Scholar]
- 65.Katagiri A, Nakayama K, Rahman MT, Rahman M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K, Kobayashi H, et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25(2):282–8. [DOI] [PubMed] [Google Scholar]
- 66.Steele CD, Abbasi A, Islam SMA, Bowes AL, Khandekar A, Haase K, Hames-Fathi S, Ajayi D, Verfaillie A, Dhami P, et al. Signatures of copy number alterations in human cancer. Nature. 2022;606(7916):984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The catalog of somatic mutations, SCNA estimations, and mutation timing analyses from the study cohort are accessible at the following GitHub repository: https://github.com/CYHuang-Lab/CGMH-OCCC-WES-project/. Tumor and normal exome sequencing data have been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/; accession number EGAS50000000031). The R codes utilized in this study are accessible at https://github.com/CYHuang-Lab/CGMH-OCCC-WES-project/.