Abstract
Background
Endocrine therapy (ET) has improved the clinical outcomes of Estrogen receptor alpha-positive (ERɑ +) breast cancer (BC) patients, even though resistance to ET remains a clinical issue. Mutations in the hormone-binding domain of ERɑ represent an acquired intrinsic mechanism of ET resistance. However, the latter also depends on the multiple functional interactions between BC cells and the tumor microenvironment (TME). Here, we investigated how the most common Y537S-ERɑ mutation may influence the behavior of fibroblasts, the most prominent component of the TME.
Methods
We conducted coculture experiments with normal human foreskin fibroblasts BJ1-hTERT (NFs), cancer-associated fibroblasts (CAFs), isolated from human BC specimens, and Y537S CRISPR-expressing MCF-7 BC cells (MCF-7YS). Mass spectrometry (MS) and Metacore analyses were performed to investigate how the functional interactions between BC cells/fibroblasts may affect their proteomic profile. The impact of fibroblasts on BC tumor growth and metastatic potential was evaluated in nude mice.
Results
Mutant BC conditioned medium (CM) affected the morphology/proliferation/migration of both NFs and CAFs. 198 deregulated proteins signed the proteomic similarity profile of NFs exposed to the YS-CM and CAFs. Among the upregulated proteins, Yes-associated protein 1 (YAP1) was the main central hub in the direct interaction network. Increased YAP1 protein expression and activity were confirmed in NFs treated with MCF-7YS-CM. However, YAP1 activation appears to crosstalk with the insulin growth factor-1 receptor (IGF-1R). Higher amount of IGF-1 were noticed in the MCF-7YS-CM cells compared to the MCF-7P, and IGF-1 immunodepletion reversed the enhanced YAP1 expression and activity. Mutant cells upon exposure to the NF- and CAF-CM exhibited an enhanced proliferation/growth/migration/invasion compared to the MCF-7P. MCF-7YS cells when implanted with CAFs showed an early relative increased tumor volume compared to YS alone. No changes were observed when MCF-7P cells were co-implanted with CAFs. Compared with that in MCF-7P cells, the metastatic burden of MCF-7YS cells was intrinsically greater, and this effect was augmented upon treatment with NF-CM and further increased with CAF-CM.
Conclusions
YS mutant BC cells induced the conversion of fibroblasts into CAFs, via YAP, which represent a potential therapeutic target which interrupt the functional interactions between mutant cells/TME and to be implemented in the novel therapeutic strategy of a subset of metastatic BC patients carrying the frequent Y537S mutations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01918-x.
Keywords: Breast cancer, ESR1 mutation, Fibroblasts, YAP1, IGF-1/IGF-1R
Introduction
A major clinical problem of endocrine therapy in breast cancer is the development of a more aggressive and hormone-independent phenotype [1, 2]. The acquisition of somatic mutations in the ESR1 gene is a major contributor to resistance and poor clinical outcomes in patients with metastatic breast cancer [3–6]. Mutations within the hormone binding domain (HBD) of ESR1 have been found to be highly frequent in metastatic breast cancer patients [6–10] but are ostensibly in rare sub clonal populations of primary breast tumors from patients receiving endocrine therapy [11, 12]. Previously and more recently we highlighted how enhanced intrinsic Insulin-like growth factor (IGF-1)/ Insulin-like growth factor receptor (IGF1R) signaling is displayed by mutant YS breast cancer cells on which is also the focus of the present study [8, 11, 13]. Recently, it has been reported that cells with HBD-ESR1 mutations could metastasize in vivo, even though the intrinsic biological properties associated to distant metastasis are not clearly elucidated [10, 14]. Clinical data have reasonably suggested that the latter breast cancer cells line in the absence of estrogens could be “selected” by microenvironmental factors sustaining clonal expansion of ESR1 mutant cells in metastatic site [15]. For instance, the tumor microenvironment (TME) becomes crucial for the maintenance of cancer progression, fostering therapeutic resistance wherein cancer-associated fibroblasts (CAFs), which exhibit specific molecular markers, are the most prominent component of the TME [16, 17]. This has given us the rational of the present study aiming to investigate how breast cancer cells exhibiting Y537S- ESR1 mutations, may influence the phenotype of the fibroblasts as the most important component of TME, sustaining the tumor growth and progression. In the latter concern, we found a similarity of proteomic profile between CAFs and Normal Fibroblasts (NFs) exposed to conditioned media (CM) from mutant-expressing cells confirmed by changes in the morphology and functional activation of NFs overlapping CAF phenotype. A direct interaction network analysis of the upregulated shared proteins in NFs-treated with YS-CM with CAFs elicited Yes-associated protein 1 (YAP1) as the central hub, which appears to be crucially involved in the conversion of NFs into CAFs. This transition is mediated by the amount of IGF-1 present in YS-CM since its immunodepletion in the CM reversed the enhanced YAP expression. On the other hand, we demonstrated how fibroblasts potentiate the aggressive behavior of mutant cells both in vitro and in vivo. For the first time, all these data highlight how mutant-expressing cells are able to reprogram NFs, inducing their phenotypic transition into CAFs and in this way enhancing tumor burden.
Materials and methods
Antibodies and reagents
The following antibodies were used: anti-β-actin (sc- 69,879, Santa Cruz Biotechnology), anti-ITβ1 (ab52971, abcam), anti-S110A4 (Fibroblast specific protein 1, FSP1,13018S), anti-PDGFR (ab32570, abcam), anti- αSMA (19245), anti-IGF-1R β (9750S), anti-YAP1 (4912), anti-CYR61 (14479S) and anti-CTGF (86641S) were acquired from Cell Signaling Technology; anti-IGF-1 (HPA048946), anti-pIGF-1RTyr1131 (ABE332) from Merck Millipore.
Generation of CRISPR-Cas9 Y537S-mutant expressing breast cancer cells
Human MCF-7 YS CRISPR breast cancer epithelial cells were generated as previously reported [8], and cultured in MEM supplemented with 10% FBS, 1% L-glutamine, 1% Eagle’s nonessential amino acids, and 1% penicillin–streptomycin (Life Technologies) at 37 °C with 5% CO2 air. Every 6 months, our breast cancer cells are usually authenticated by single tandem repeat analysis at our Sequencing Core facility, controlling their morphology and testing doubling times, estrogen sensitivity, and mycoplasma negativity (MycoAlert, Lonza, Basel, Switzerland). Immortalized normal human foreskin-fibroblasts BJ1-hTERT (normal fibroblasts, NFs) were kindly provided by Dr Lisanti (University of Salford, Manchester, UK).
Isolation and culture of Cancer-Associated fibroblasts
Human breast fibroblast cells from fresh tumors (CAFs) were obtained from luminal A breast cancer women who signed an informed consent from and were approved by the Ethics Institutional Committees of the Annunziata Hospital, Cosenza [18]. Experimental procedures for fibroblast isolation were performed according to approved guidelines. Small pieces of fresh tumor excision (taken from an area of < 1.0 cm away from the edge of breast cancer tissue), were cleaned with 1% penicillin/streptomycin in PBS at 4 °C and then were digested with 500 IU collagenase in Hank’s balanced salt solution (Sigma) at 37 °C for 2 h. After two differential centrifugations at 90 g for 2 min and, at 500 g for 8 min the supernatant containing CAFs was resuspended and cultured in RPMI-1640 medium supplemented with 15% FBS and antibiotics. All experiments were performed between the 4th and the 10th passages.
Conditioned medium system
4.4 106 CAFs or NFs and MCF-7 cells were incubated with 5% charcoal-stripped-FBS for 24 h and conditioned media (CM) were collected, centrifuged and used for the experiments.
MCAT luciferase reporter assay
3 104 NFs were plated in 24 multi-well and co-transfected with 1 µg of 8xGTIIC-luciferase (MCAT-reporter, gently provided by Prof. Dupont, University of Padova, Italy) and 20 ng of TK Renilla luciferase plasmid as an internal control by using Lipofectamine 2000 reagent (Life Invitrogen) following the manufacture’s instructions. Next day, NFs were treated with CM-derived from both MCF-7P and YS breast cancer cells. After 24 h firefly and renilla luciferase activities were measured using a Dual Luciferase kit (Promega, Madison, WI, USA). The firefly luciferase data for each sample were normalized on the basis of transfection efficiency measured by Renilla luciferase activity.
IGF-1-immunodepleted conditioned media
CM from MCF-7 and YS cells was collected and then centrifuged at 1200 × g for 5 min. Then, the supernatants were incubated with 2 μg/ml IGF-1 antibody (Sigma Aldrich) overnight with rotation at 4 °C. Next day, Protein A/G-agarose beads (Santacruz) (25 µl/ml of CM) were washed with PBS and centrifuged at 10000 g for 10′ 4 °C. After discarding supernatants, protein A/G-agarose beads were washed with serum-free medium. Once we discarded supernatants again, CM was added to beads for 1 h, in rotation at 4 °C. After, the CM were centrifuged at 10,000 × g for 10 min, beads were discarded and CM were filtered through a 0.22 μm filter. The occurred IGF-1 immunodepletion was checked by enzyme-linked immunosorbent assay (ELISA).
Immunoblot analysis
A total of 12 104 cells were lysed in RIPA Buffer (50 mm Tris–HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 2 mm sodium fluoride, 2 mm EDTA, 0.1% SDS, containing a mixture of protease inhibitors (aprotinin, phenylmethylsulfonyl fluoride, and sodium orthovanadate) for whole lysate extraction and immunoblot was performed as previously described [19]. In brief, the same amounts of protein were resolved of either 10% or 14% (based on the molecular weight of the proteins) SDS–polyacrylamide gel, then transferred to a nitrocellulose membrane and probed with different antibodies. To ensure equal loading, all membranes were incubated with anti-β-actin antibodies. The antigen–antibody complex was detected by incubation of the membranes with IRDye® coupled goat anti-mouse or rabbit (680RD or 800CW) and the images were acquired and the bands of interest were quantified using an Odissey FC (LICOR, Lincoln, NE, USA). The uncropped blots are reported in the Supplementary Fig. 1, 2, 3 and 4.
Cell Cycle
The cells were plated, treated and resuspended in 0.5 ml of 50 mg/ml propidium iodide (PI) in PBS solution containing 20 U/ml RNAse-A and 0.1% TRITON X-100 and incubated for 1 h (Sigma–Aldrich). The cells analysis were performed by flow cytometry (CytoFLEX Beckman, Beckman Coulter, Milan, Italy) and Data analysis was conducted using CytExpert Beckman Coulter software (Beckman Coulter, Milan, Italy). The cell phases were estimated as a percentage of a total of 10,000 events.
RNA expression analyses
12 104 cells were plated in 6-multiwell plates and treated as indicated. Total RNA was subsequently extracted using TRIzol reagent (Life Technologies) and gene expression was evaluated via real-time reverse transcription PCR using a RETROscript kit (Life Technologies) and quantified as previously described [20]. Supplementary table S1 contains the list of primers used in this paper.
Trypan-blue cell count assay
Cells were seeded at a confluence of 5 104 cells/well in 24-well plates and treated for 5 days as indicated. The cells were harvested by trypsinization and incubated in 0.5% trypan blue solution, for 10 min at room temperature. Trypan blue negative cells were counted through a Countess® II Automated Cell Counter (Thermo Fisher) on day 0 and after 5 days of treatment.
Soft agar colony formation assay
A total of 104 cells were seeded in a 12-well precoated with 1 ml of pre-warmed (50 °C) 1% SeaPlaque agarose (FMC, Rockland, ME) and solidified at 4 °C for 1 h. The cells were suspended in 2 ml of prewarmed (37 °C) 0.35% SeaPlaque agarose and plated. The plates were cooled for 2 h at 4 °C, transferred to a 37 °C humidified incubator and treated as indicated. After 15 days colonies > 50 µm were counted.
Wound-healing assay
Cells were seeded in 6-well plates and grown to complete or near 100% confluence. Subsequently, the resulting cell monolayers were scraped with a sterile pipet tip and exposed to the different experimental conditions. Wound closure was monitored over 12–24 h and the cells were then fixed and stained with Coomassie Brilliant Blue. The images represent one of three independent experiments. Pictures were taken at 10 × magnification using phase-contrast microscopy (OLYMPUS-BX51 microscope) and the relative percentage of wound closure was calculated by ImageJ software (NIH, MD, USA).
Boyden chamber transmigration assays
Cells were pretreated with conditioned media for 5 days and then 5 104 cells were placed in the upper compartments of Boyden chambers (8 µm-membranes, Corning), while the bottom wells contained regular growth media. After 24 h, the migrated cells were fixed and stained with DAPI. Migration was quantified by viewing six separate fields per membrane at 10 × magnification (OLYMPUS-BX51 microscope) and is expressed as the mean number of migrated cells.
Boyden chamber invasion assays
A matrigel-based invasion assay was performed in chambers (8 µm membranes, Corning) coated with Matrigel (BD Bioscences, 2 mg/mL). Cells were pre-treated with conditioned media for 5 days and then 5 104 cells placed in the upper compartments of Boyden chambers (8 µm-membranes, Corning), while the bottom wells contained regular growth media. After 24 h, the migrated cells were fixed and stained with DAPI. Invasion was quantified by viewing six separate fields per membrane at 10 × magnification (OLYMPUS-BX51 microscope) and is expressed as the mean number of invaded cells.
Scanning electron microscopy (SEM)
12 104 cells were seeded in 6 well plates and fixed in 3% glutaraldehyde for 2 h at room temperature. After the cells were washed three times with PBS they were washed with increasing concentrations of ethanol (30, 50, 70 and 100%) for 5 min. The ethanol was completely evaporated before the cells were viewed. All specimens were mounted on appropriate stubs and coated with a 5 nm ultra-pure graphite film (Quorum Q150 Carbon-coater) to be observed under an Ultra High Resolution SEM (UHR-SEM) Zeiss CrossBeam 350—Germany, operating in secondary-electron mode @ 3,0 KVolt of energy.
Phalloidin Staining
12 104 CAFs and NFs were plated in a Nunc™ Lab-Tek™ II Chamber Slide™ System (Thermo Fisher Scientific) and then stained with Alexa Fluor 568-conjugated phalloidin (1:500), following the manufacturer’s instructions (Thermo Fisher Scientific) for polymerized actin stress fiber staining. Cell nuclei were stained with DAPI. An Olympus BX51 microscope (100 × magnification) was used for imaging.
Fluorescence microscopy
12 104 NFs were plated in a Nunc™ Lab-Tek™ II Chamber Slide™ System (Thermo Fisher Scientific) and treated as indicated. Immufluorescence analysis and a YAP1 nuclear location analysis was conducted as previously reported [21, 22]. In brief, using the public domain image processing program ImageJ, we initially selected nuclei and generated a file containing the data pixel. Only pixels within the boundaries of the nuclei were considered for YAP localization analysis.
Enzyme-linked immunosorbent assay (ELISA)
A total of 4.4 106 cells/well were plated in 10 cm dishes in complete media. The next day, the medium was removed and replaced with 5% charcoal-stripped medium. After 24 h, the media were collected, centrifuged and the IGF-1 levels were measured using an ELISA kit (R&D System #DG100B), according to the manufacturer’s protocol.
Clonogenic assay (Cell Viability)
Cells were seeded in 6 multi-wells (500 cells/well) and treated with conditioned media for 5 days. Colonies were stained with 0.01% (w/v) crystal violet for 10 min on an orbital shaker and washed with PBS. Then, the wells were allowed to dry and 1% SDS was added to solubilize the stain and to measure the absorbance at 570 nm through a microplate reader.
Collagen gel contraction assays
A total of 5 × 105 cells were suspended in 3 mg/ml rat tail collagen I (Life Technologies) diluted in serum-free media, added to a 24-well plate (500 μl/well) and incubated for 20 min at RT, followed by the addition of culture media (50 μl/well). The collagen gels, detached from well walls with a tip, were incubated at 37 °C. Images were acquired at time 0 and after 24 h. Gel contraction was quantified by the decrease in gel diameter.
Protein digestion
A 20 μg aliquot of each sample was diluted to a final volume of 32 μL with RIPA buffer. The sequential addition of 3.2 µL of 100 mM DTT (1 h incubation at 37 °C), 3.8 µL of 200 mM iodoacetamide (1 h incubation at 37 °C) and, finally, 0.6 µL (1 h incubation at 37 °C) of 100 mM DTT allowed the reduction and alkylation of cysteines to occur. Then, half of each sample (10 μg) was digested through protein aggregation capture (PAC) protocol [23]. Briefly, for each sample 5 µL (100 µg) of MagResyn Hydroxyl beads were equilibrated by 2 sequential washes with 100 µL of 70% acetonitrile (ACN). Then, the magnetic microparticles were added to the samples and the proteins were precipitated by the addition of ACN to a final 70:30 organic:water ratio. The solution was incubated for 10 min at 1100 rpm. Three washes with 200 µL of acetonitrile were followed by one wash with 70% ethanol. Finally, the beads were resuspended in 50 µL of 50 mM triethylammonium bicarbonate (TEAB). Trypsin (Sigma Aldrich, from porcine pancreas) was added at an E/S 1/50 (200 ng). After overnight incubation (37 °C, 1100 rpm), the peptide solution was harvested and residual peptides were recovered by washing the beads with 50 µL of 0.1% formic acid (FA).
Nanoliquid chromatography tandem mass spectrometry (nLC-MS/MS)
MS analysis was carried out on a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific) operating in positive ion mode, coupled with an Easy LC 1000 nanoscale liquid chromatography system (Thermo Fisher Scientific). A 15 cm pulled silica capillary (75 µm i.d.) packed in-house with 3 µm C18 silica particles (Dr. Maisch) was used as the analytical column. Nanoelectrospray (nESI) was obtained by applying a potential at 2000 V on the column front-end through a tee piece. A binary gradient was used to elute the peptides. In particular, mobile phase A contained 2% ACN/ 01% FA (v:v) while mobile phase B was composed of 80% ACN/ 01% FA. The flow rate was set to 230 nl/min. Mobile phase B ramped from 6 to 28% in 90 min, from 28 to 45% in 30 min and from 45 to 100% in 8 min. Then, was maintained 10 min at 100%. The analytical column was equilibrated at 0% mobile phase B for 2 min. Data-independent acquisition (DIA) enclosed 20 windows with a full scan at a resolution of 70,000 (AGC target of 1e6 and maximum injection time of 50 ms) and 20 DIA scans with a resolution of 35 000 (AGC target of 1e6, maximum injection time of 120 ms and normalized collision energy of 25). In detail, the isolation window was set to have 4 windows of 30 m/z, 13 windows of 20 m/z and 3 windows of 50 m/z. The overlap was equal to 1 m/z. The resulting m/z range was 370–900.
Data analysis and results reporting
LC–MS/MS data were searched by using Spectronaut software v.17 against the human reference proteome (79,740 seq, Uniprot 11 October 2022) and a database of common contaminants (46 entries). Analysis settings were left as default applying only the following changes to the default parameters: precursor filtering was set in order to filter out all precursors identified in less than 3 runs for each comparison. Missing values were imputed by adopting “run wise” imputation strategy. The maximum number of precursors used for quantification was set to 5. Only peptides shared between proteins of the same protein group (protein group specific) were considered for quantification. The resulting report was then loaded on Perseus v. 2.06.0 for statistical analysis. For all the comparisons the analysis was performed following these steps: Protein Quantities calculated by Spectronaut software were log2 transformed, the matrix was then filtered keeping only protein groups quantified in at least 3 samples of a group. Then, differentially expressed proteins were identified by applying a two sample test with a permutation based FDR set at 0.1 (S0 was set at 0.2).
MetaCore functional analysis
The differentially expressed protein datasets were functionally processed by applying the MetaCore™ integrated software suite for functional analysis of experimental data (Clarivate Analytics, London). Differentially expressed proteins were also investigated via the MetaCore Network Building tool and by applying the direct interaction algorithm. This functionally crosslinks gene transcripts under processing by building networks of corresponding proteins that directly interacting with each other. This occurs accordingly to the manually curated MetaCore in-house database concerning: physiological and pathological protein–protein and protein–nucleic acid interactions, signaling and metabolic pathways, reported by the scientific literature. Direct interaction networks (DIN) represent the interactomic core of the investigated process and are prioritized according to their statistical significance. Nets are then graphically visualized as nodes (proteins) and edges (interconnections among proteins) and named in relation to their five most significant factors, i.e. central hubs able to establish the highest number of interactions with the other components of the net. Thus, central hubs acquire crucial roles in defining the biochemical and molecular properties of the investigated systems.
Xenograft experiments
Female 6-week-old athymic nude mice (nu/nu Swiss; ENVIGO RMS, Udine, Italy) were housed under specific pathogen-free conditions and maintained at a constant temperature. Estradiol pellets (0.72 mg/pellet, 90-day release; Innovative Research of America, Sarasota, FL) were implanted subcutaneously. The next day, the mice were anesthetized with 1% to 2% isofluorane in O2, 1 L/min (ISO-Vet, Piramal Healthcare Uk Limited, Morpeth, UK) and 5 106 MCF-7 P and MCF-7 YS cells were orthotopically injected into the right mammary fat pad in 0.1 mL of Matrigel (BD Biosciences, Bedford, MA), alone or in combination with human CAFs at a 1:3 ratio (1 part of breast cells and 3 parts of CAFs). Animal health and behavior were monitored daily, and animal procedures were conducted following proper guidelines. Tumor size and weight were measured twice a week using a caliper and a scale. Tumor volumes (mm3) were calculated using the formula: TV = (a * b2)/2 (a = tumor length and b = tumor width), in millimeters. When the masses reached 150 mm3, the animals were randomly assigned to 8 homogenous groups. At the fourth week, the animals were euthanized following standard protocols; the tumors were explanted, one part of which was dehydrated and imbedded in paraffin, and one part was frozen in nitrogen and stored at 80 °C for further analyses.
Histopathological and immunohistochemical analyses
Tumors were fixed in 4% formalin, sectioned at 5µm and stained with haematoxylin and eosin Y following manufacture's suggestions (Bio-Optica, Milan, Italy). Paraffin-embedded sections, 5 μm thick, were mounted on slides precoated with poly-lysine, and then they were deparaffinized and dehydrated (seven to eight serial sections). Immunohistochemical experiments were performed after heat-mediated antigen retrieval in Citrate Buffer, in a microwave at low setting, followed by incubation of oxygenated water and normal horse serum, like blocking agent, at room temperature (Vectastain ABC Kit Elite (PK-6200). Immunohistochemistry was performed using Cytocheratin 18 (Agilent Dako, clone DC 10, M7010), Ki-67 (Agilent Daki, clone MIB-1, M7240)and human α-SMA (Agilent Dako, clone 1A4, M0851) primary antibodies in MCF-7 P and YS, with and without the presence of CAFs, at 4 °C overnight. Biotinylated horse anti-mouse/rabbit was used like secondary antibody. The linked was visualized through DAB Substrate Kit, Peroxidase (SK-4100 Vector Laboratories, Inc.). Immunoreactivity was visualized by using VECTOR® Red Alkaline Phosphatase Substrate Kit (Vector Laboratories). Counterstaining was carried out with hematoxylin (Sigma Aldrich). The primary antibody was replaced by normal serum in negative control sections. α-smooth muscle actin (α-SMA) immunostaining score was calculated as a percentage of area occupied by IHC intensity using ImageJ; briefly α-SMA index score were calculated as the percentage of positive cells multiplied by intensity, ranging in a score 0–300, according to the following staining intensity (0, negative; 1, weak; 2, intermediate; 3, strong) and percentage (0–100%) of the stained tumor cells respect to the total number of tumor cells, through Image Scope software, the statistical significance was calculated by non-parametric Mann–Whitney test. α-smooth muscle actin score was calculated as a percentage of area occupied by IHC intensity using ImageJ.
H Score = [1x (% cells 1 +) + 2x(% cells 2 +) + 3x(% cells 3 +)]. The immunostained slides of tumour samples were evaluated by light microscopy using the Allred Score [24], which combines a proportion score and an intensity score. A proportion score was assigned representing the estimated proportion of positively stained tumor cells (0 = none; 1 = 1/100; 2 = 1/100 to < 1/10; 3 = 1/10 to < 1/3; 4 = 1/3 to 2/3; 5 = > 2/3). An intensity score was assigned by the average estimated intensity of staining in positive cells (0 = none; 1 = weak; 2 = moderate; 3 = strong). Proportion score and intensity score were added to obtain a total score that ranged from 0 to 8. A minimum of 100 cells were evaluated in each slide. Six to seven serial sections were scored in a blinded manner for each sample. The one-way ANOVA was used to evaluate the differences in the scores between tumor and control samples.
Metastatic study
MCF-7 P and MCF-7 YS cells were plated and then treated with NFs- and CAFs-CM for 5 days. A total of 5 105/100 μL was injected into the median tail vein of six-week-old female nude mice (nu/nu; Envigo, Udine, Italy) as previously described [25]. The mice were weighed twice a week and monitored for mobility, respiratory distress, and signs of pain. After six weeks of cell injection, the mice were sacrificed following standard protocols. Organ examination revealed that the lungs were the destination sites of disseminated tumor cells. The lungs were perfused with PBS, inflated for formalin fixation, and paraffin embedded. The lung metastatic lesions were examined using hematoxylin and eosin (H&E) staining. Five sections per lung were counted using ImageScope software (Aperio, Leica Biosystems, Milan, Italy).
Survival Kaplan–Meier analysis
Kaplan–Meier post-progression survival (PPS) analysis in breast cancer patients were generated using Kaplan–Meier plotted (https://kmplot.com/analysis/) [26]. The entire dataset included 229 samples from 50 datasets with overall survival data. The average follow-up is 60 months.
Statistical analysis
Statistical analysis was performed with Student’s t test for the comparison between two groups and one-way ANOVA with Tukey’s multiple comparisons post hoc test for the comparison between multiple variables by using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA) and the results are reported as the mean ± SD or ± SEM, as indicated, of at least 3 independent experiments, each performed in triplicate.
Results
Characterization and proteomic profile of MCF-7 Y537S (YS) and MCF-7 Parental (P) breast cancer cells
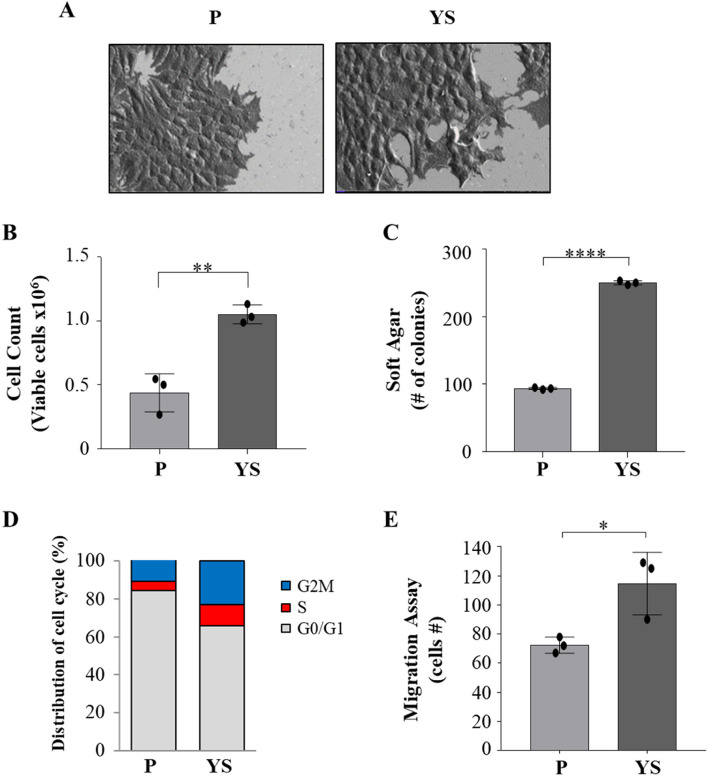
Compared with MCF-7 parental (P) cells, MCF-7 Y537S (YS) exhibit a characteristic spindle morphology; notably, MCF-7 YS showed more lamellipodia confirming how the mutant cells exhibit a typical feature of epithelial-to-mesenchymal transition (EMT, Fig. 1 A) as already reported by our group in 2020 [8]. Moreover, mutant cells showed a significant increase in growth rate in both anchorage-dependent (Fig. 1 B) and anchorage-independent (Fig. 1 C) growth assays. The latter findings are consistent with the flow cytometric analysis evidencing that in the cell cycle of the YS the percentage of cells in the G0-/G1-phases of the cell cycle decreased and the percentage of cells in the S and G2/M phases increased compared with MCF-7 P cells (Fig. 1 D), highlighting a greater proportion of mutant proliferating cells. Moreover, the YS exhibited an enhanced cell migration (Fig. 1 E).
Fig. 1.
Characterization of MCF-7 parental (P) and Y537S (YS) breast cancer cells. Representative scanning electron microscopy (SEM) images (A) of MCF-7 P and YS cells grown in regular media. Scale bar: 20 µm. B Viability assay in MCF-7 P and YS cells. Live cells were counted after 72 h. C Soft agar growth assays in MCF-7 P and YS cells. After 14 days of growth, colonies ≥ 50 μm in diameter were counted. D MCF-7 P and YS cells were stained with propidium iodide and analyzed by using a FACScan flow cytometer. The percentages of cells in the G0/G1, S, and G2/M phases are reported. E Transmigration assays in MCF-7 P and MCF-7 YS. The migrated cells were DAPI-stained, counted and images were captured at 10 × magnification. Each circle represents one independent experiments. Data were analyzed using Student’s t test, and are presented as mean ± SD of three independent experiments each performed in triplicates. *p < 0.05; **p < 0.01; ****p < 0.0001
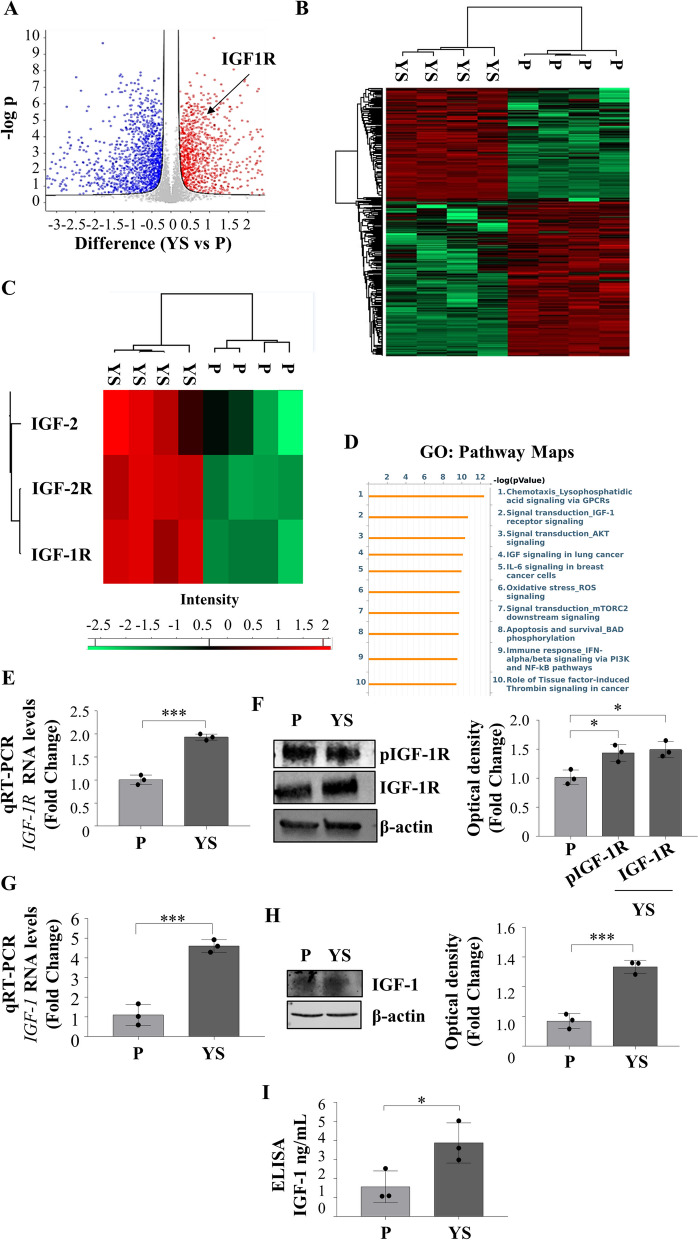
Overall, the analysis of four biological replicates has identified 5296 proteins, each with a minimum of three unique peptides with a false discovery rate (FDR) of < 1%. Extensive differences in the proteomes of the two different cell lines are evidenced by the volcano plot (Fig. 2 A). The proteomic comparison of the MCF-7 P and YS cell lines demonstrated two distinct clusters of proteins significantly differentiated between parental and mutant breast cancer cells as evidenced by heatmap (Fig. 2 B) representing the 2555 significantly deregulated proteins identified in mutant cells as confirmed by the supervised hierarchical clustering evidenced by Principal Component Analysis (PCA, Suppl. Figure 5 A). Among the deregulated proteins 1078 were found to be significantly upregulated and 1477 were significantly downregulated in mutant cells compared to parental cells. Among the deregulated proteins their GO Pathway Maps gave the relevance of IGF-1/IGF-1R signaling and to the intermediates of its trasductional pathways (i.e. “Signal Trasduction_AKT signaling”, and “Signal transduction_mTORC2 downstream signaling” “PI3K and NF-kB pathway”) (Fig. 2 C and Supplementary Table S2). It is worth to remark that among the upregulated proteins it still emerges the IGF-1 signaling among the top ten functional categories (Supplementary Fig. 5D). All this was concomitant with an enhanced expression of mRNA, total and phosphorylated IGF-1R expression in YS compared to P cells (Fig. 2 D and E) with an increased expression of IGF-1 in terms of mRNA and protein content resulting in an augmented amount secreted in their conditioned media (Fig. 2 F, G and H).
Fig. 2.
Proteomic comparison between MCF-7 P and YS breast cancer cells. A Heatmap representing supervised hierarchical clustering of the significant and differentially expressed proteins between MCF-7 P and YS cells identified by LC–MS/MS analysis. The heatmap is shown in red color for the upregulated proteins and in green for downregulated proteins. C Heatmap representing supervised hierarchical clustering of the targets of interest for this study in the comparison between MCF-7 P and YS cells identified by LC–MS/MS analysis. The heatmap is shown in red color for the upregulated proteins and in green for downregulated proteins. D Volcano plot representing all detected proteins in MCF-7 P vs YS cells. The significant proteins are shown on the right (red dots, upregulated proteins) and on the left (blue dots, downregulated proteins). The gray dots correspond to proteins whose expression did not significantly change according to the selected parameters. B Gene Ontology (GO) analysis of proteins related to Pathway Maps according to Metacore. E Real time RT-PCR analysis of IGF-1R (Insulin Growth Factor-1 Receptor) mRNA expression in MCF-7 P and YS cells. F Immunoblotting showing pIGF-1R and IGF-1R protein expression in MCF-7 P and YS cells. β-actin was used as a control for equal loading and transfer. The histograms represent the mean ± SD of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units (OD) and expressed as percentage of MCF-7 P cells which were assumed to be 100%. G Real time RT-PCR analysis of IGF-1 (Insulin Growth Factor-1 Receptor) mRNA expression in MCF-7 P and YS cells. H Immunoblotting showing IGF-1 protein expression in MCF-7 P and YS cells. β-actin was used as a control for equal loading and transfer. The histograms represent the mean ± SD of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units (OD) and expressed as percentage of MCF-7 P cells which were assumed to be 100%. I E-linked immunosorbent assay (ELISA) for IGF-1 content in MCF-7 P and YS cells. Each circle represents one independent experiments. Data were analyzed using Student’s t test, and are presented as mean ± SD of three independent experiments each performed in triplicates. *p < 0.05; ***p < 0.001
Characterization and proteomic profile of Normal fibroblasts (NFs) and Cancer Associated Fibroblasts (CAFs)
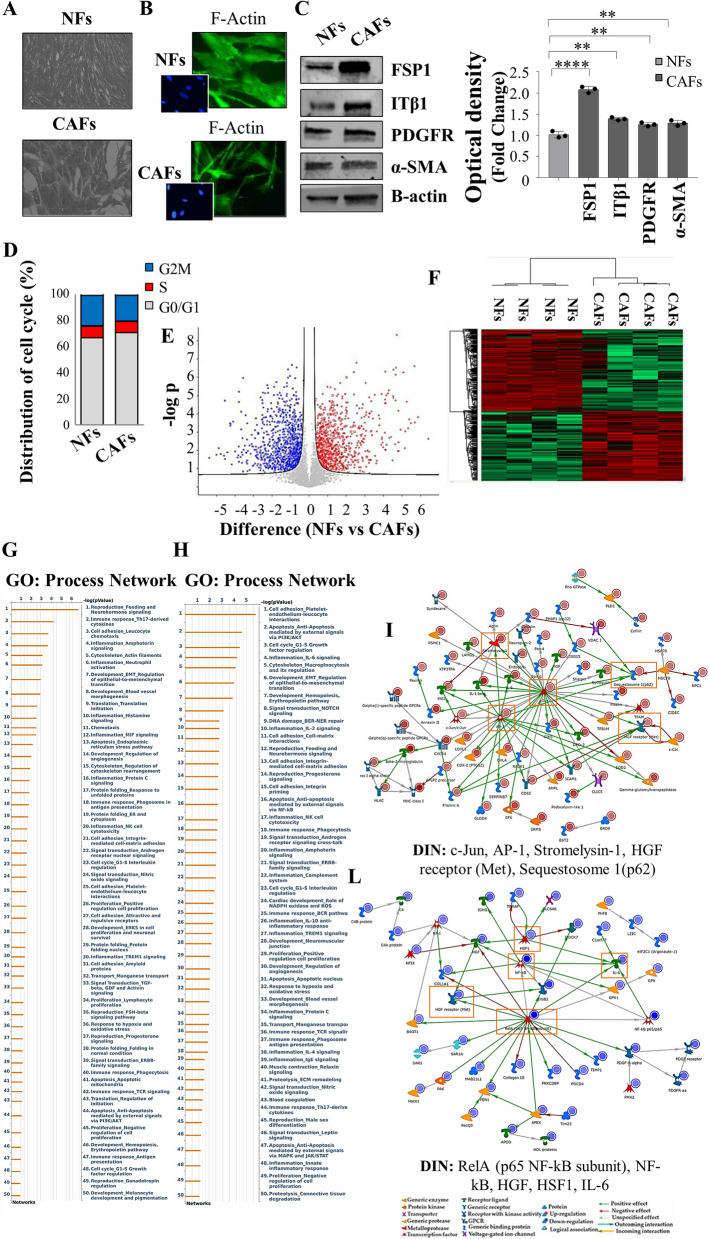
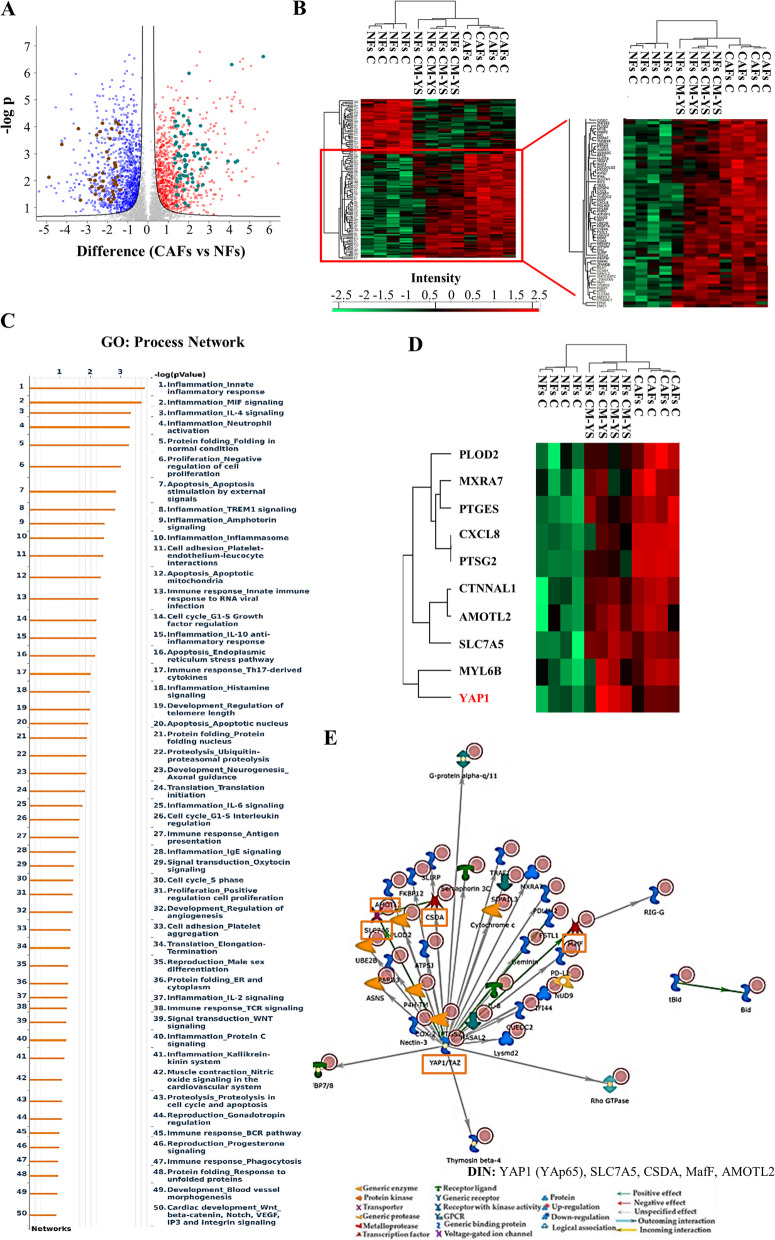
The influence of the tumor microenvironment on breast cancer cells may affect growth and progression and negatively influence the therapeutic response [27]. Thus, in the present study, we investigated how MCF-7 YS mutant cells may functionally interact with fibroblasts, the most prominent component of the tumor microenvironment. To this aim, we created in vitro conditions that can mimic the in vivo microenvironment through coculture experiments of both cancer cells and fibroblasts. For this study, we used as normal counterpart immortalized normal human foreskin fibroblasts BJ1-hTERT, named as normal fibroblasts (NFs) and cancer-associated fibroblasts (CAFs) which were isolated from a biopsy of a women diagnosed with mammary ductal carcinoma. Both types of fibroblasts displayed a typical spindle-shaped morphology (Fig. 3 A) whereas CAFs exhibited an increase in actin organization featured by major actin stress-fiber formation to sustain contractile activity (Fig. 3 B) and increased protein level expression of fibroblast-specific markers such as fibroblast-specific protein-1 (FSP1), integrin subunit beta 1 (ITB1), platelet derived growth factor receptor beta (PDGFR), alpha smooth muscle actin (α-sma) compared to NFs (Fig. 3 C). Flow cytometry analysis revealed no significant changes in proportions of cells in the G0/G1, S, and G2/M phases of the cell cycle compared to NFs (Fig. 3 D). Comparing the analysis of the different proteomes of NFs and CAFs we identified 4992 proteins, as evidenced by volcano plot (Fig. 3 E). Out of them 2557 proteins are significantly deregulated, as evidenced by heatmap (Fig. 3 F). A clear distance between the two proteomic profile is revealed by Principal component analysis (PCA, Suppl. Figure 6 C). Among the 1121 upregulated proteins we identified their substantial enrichments in the Process Networks GOs and mainly related to inflammation, neoangiogenesis and ECM/cytoskeleton/actin filaments functional interaction (Fig. 3 G). In addition, direct interaction network (DIN) of the deregulated proteins identified in the comparison CAFs vs NFs gives relevance to the following proteins: Fibroblast growth factor receptor (FGFR), yes-associated protein 1 (YAP1), Jun Proto-Oncogene AP-1 Transcription Factor Subunit (c-Jun), E1A Binding Protein P300 (P300) and SRY-Box Transcription Factor 9 (SOX9) (Supplementary Fig. 6). Among this central hub it is worth to remark the widely well documented functional interaction between the YAP/TAZ and c-Jun AP-1 forming a complex that synergistically activates different target gene directly involved in the control of S-phases and mitosis [28]. Specifically, among the 1121 upregulated proteins, we identified substantial enrichment in Process Network GOs of those particularly involved to the inflammation, angiogenesis and cytoskeleton interaction/ECM remodeling (Fig. 3 G). Among the 1136 downregulated proteins, those enriching the network process were related to i) inhibitors of anti-inflammatory molecules e.g. p65 NF-KB subunit; ii) Cell adhesion and cell matrix; iii) Proteolysis and ECM remodeling; iv) proteolysis and connective tissue degradation, mostly involving the inhibitors of proteinases (Fig. 3 H).
Fig. 3.
Characterization of normal fibroblasts (NFs) and cancer-associated fibroblasts (CAFs). A Representative SEM images of NFs and CAFs grown in regular medium. Scale bar: 40 µm. B Immunofluorescence staining of phalloidin stained F-actin (stress fibers, green) in NFs and CAFs grown in regular medium. 4′,6-Diamidino-2-phenylindole (DAPI) was used for nuclei detection (inset). C Immunoblotting showing fibroblast-specific protein-1 (FSP1), integrin subunit beta 1 (ITB1), platelet derived growth factor receptor beta (PDGFR), alpha smooth muscle actin (α-sma) protein expression in NFs and CAFs. β-actin was used as a control for equal loading and transfer. The histograms represent the mean ± SD of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units (OD) and expressed as percentage of NF cells which were assumed to be 100%. Each circle represents one independent experiments. Data were analyzed using Student’s t test, and are presented as mean ± SD of three independent experiments each performed in triplicates. **p < 0.01; ****p < 0.0001. D NFs and CAFs were stained with propidium iodide and analyzed by using a FACScan flow cytometer. The percentages of cells in the G0/G1, S, and G2/M phases are reported. E Volcano plot representing all detected proteins in the comparison CAFs vs NFs. The significant proteins are localized on the right (red dots, up-regulated proteins) and on the left (blue dots, down-regulated proteins). Gray dots correspond to not significantly proteins, accordingly with selected parameters. F Heatmap representing supervised hierarchical clustering of the differentially expressed proteins between NFs and CAFs identified by LC–MS/MS analysis. Heatmap shows red color for degree of upregulated proteins, while green for the degree of downregulated proteins. Gene Ontology (GO) enrichment analysis of upregulated proteins (Fold cutoff ≥ 2.5, G and downregulated (H) (Fold cutoff ≤ 2.5) related to Process Networks according to Metacore. Direct interaction network (DIN, orange boxes) constructed by processing the upregulated (Fold cutoff ≥ 2.5, I and downregulated (Fold cutoff ≤ 2.5, L proteins identified in the comparison between NFs and CAFs. The colors of the arrows indicate positive (green), negative (red) or unknown (gray) effects of the different network items represented with different symbols (reported in the legend). DIN upregulated proteins: c-Jun, AP-1, Stromelysin-1, HGF receptor (Met), Sequestosome 1(p62); DIN downregulated proteins: RelA (p65 NF-kB subunit), NF-kB, HGF, HSF1, IL-6
Impact of MCF-7 YS breast cancer cells on the morphology and functional properties of fibroblasts
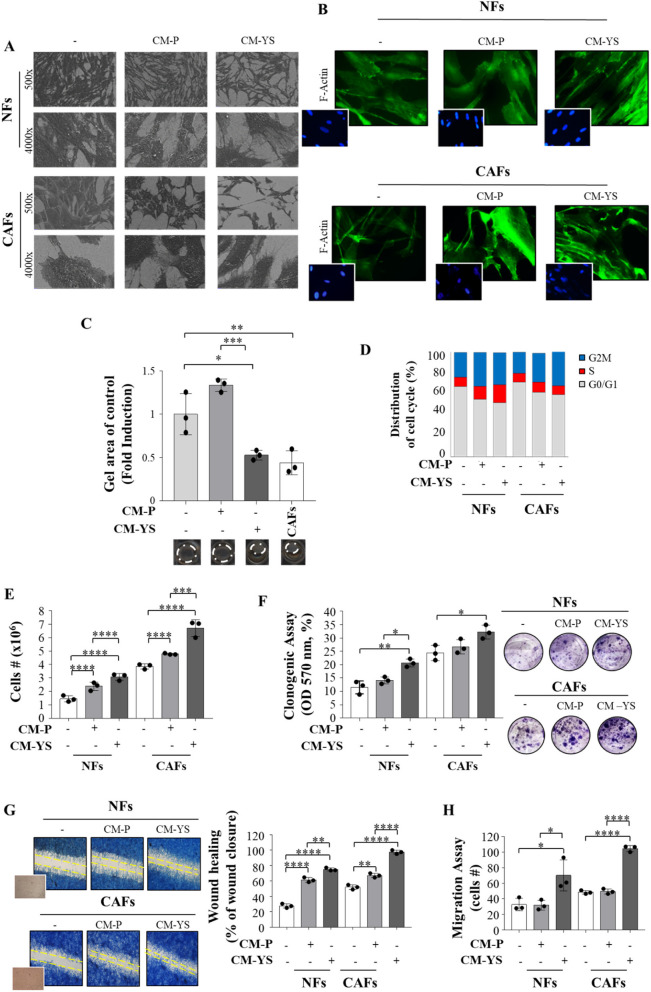
The morphology of NFs exposed to CM derived from MCF-7 P and YS was evaluated by SEM. However, when NFs were exposed to MCF-7 P-CM did not show any noticeable morphological changes, in contrast noticeable effects were significant evident in NFs exposed to CM YS. In fact, they appeared more scattered with a disordered arrangement of stress-contractile fibers inside the cytoplasm. In addition, at higher magnification NFs exposed to both MCF-7 P and YS-CM exhibited active ruffling zones at their edges, which produced curling lamellipodia (Fig. 4 A, left panel), due to the formation of F-actin stress fibers were emphasized by the exposure of NFs to MCF-7 YS-CM (Fig. 4 B, right panel), as confirmed by their displayed contraction activity. This appears to confer to NFs the morphological features of activated fibroblasts, as it has been revealed through SEM and F-actin assays in CAFs (Fig. 4 A and B, right panels). The collagen gels containing fibroblasts exposed to MCF-7 P-CM did not show substantial changes, whereas gels containing fibroblasts exposed to MCF-7 YS-CM showed a decrease in diameters of 60%. This finding highlights the ability of MCF-7 YS-CM to stimulate fibroblast’s capability to contract an in vitro extracellular matrix (Fig. 4 C). When we analyzed cell cycle progression by using flow cytometry analysis, we observed that upon exposure to MCF-7 P-CM and YS-CM, NFs exhibited a decrease in the number of cells in the G0/G1 phase and an increase in the number of cells in the G2/M and S phases, particularly under the latter condition (Fig. 4 D); while upon exposure to MCF-7 P or YS-derived CM CAFs exhibited a decrease in the number of cells in the G0/G1 phases and an increase in the number of cells in the G2/M phase to a similar extent (Fig. 4 D). Both the exposure of fibroblasts to CM obtained from MCF-7 P and YS breast cancer cells revealed that both CM were able to increase the growth rate of NFs and CAFs, even though in a greater extent upon exposure to MCF-7 YS-CM (Fig. 4 E). Interestingly, only mutant-derived CM enhanced the cell viability of both fibroblasts (Fig. 4 F). In addition, exposure to CM-derived from MCF-7 P cells and to a higher extent CM-derived from mutant-expressing cells enhanced the motility of both NFs and CAFs (Fig. 4 G). Boyden chamber transmigration assays confirmed that only mutant cells promoted fibroblast migration (Fig. 4 H).
Fig. 4.
Biological effects of MCF-7 P and MCF-7 YS CM on NFs and CAFs. A Representative SEM images of NFs and CAFs treated without (-) or with ( +) CM derived from MCF-7 P and YS for 5 days. Scale bar = 40 µm (500x) and 6 µm (4000x). B Immunofluorescence staining of phalloidin staining of F-actin (stress fibers, green) in NFs and CAFs treated without (-) or with CM derived from MCF-7 P and MCF-7 YS for 5 days. 4′,6-Diamidino-2-phenylindole (DAPI) was used for nuclei detection (inset). C Representative images of a gel contraction assay. The histogram represents the gel area of NFs treated without (-) or with ( +) CM derived from MCF-7 P and YS at 24 h. CAFs have been included as positive control. D NFs and CAFs treated without (-) or with ( +) CM derived from MCF-7 P and YS 5 days were stained with propidium iodide and analyzed by using a FACScan flow cytometer. The percentage of cells in the G0/G1, S, and G2/M phases is reported. E Viability assay in NFs and CAFs treated without (-) or with ( +) CM derived from MCF-7 P and YS for 5 days. Live cells were counted after 5 days of treatment. F Clonogenic assay in NFs and CAFs treated without (-) or with ( +) CM derived from MCF-7 P and YS for 5 days. Colonies were stained with 0.01% (w/v) crystal violet, solubilized and measured the absorbance at 570 nm through the microplate reader. G Wound healing assay in NFs and CAFs treated without (-) or with ( +) CM derived from MCF-7 P and YS with images captured at time 0 (inset) and after 12 h. H Transmigration assays in NFs and CAFs treated without (-) or with CM derived from MCF-7 P and MCF-7 YS for 24 h. The migrated cells were DAPI-stained, counted and images were captured at 10 × magnification. Each circle represents one independent experiments. Data were analyzed using one-way ANOVA with Tukey’s multiple comparisons post hoc test. Data are presented as mean values ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Thus, we demonstrated, for the first time, that the MCF-7 YS mutant cells might switch the NF functional properties towards those featuring CAF phenotype.
Despite the not substantial changes in proteomic profile of NFs in presence of MCF-7P CM, a cluster of proteins appeared to be specifically deregulated in NFs exposed to MCF-7YS CM overlapping CAF phenotype (Supplementary Fig. 7), as also emphasized in volcano plot (Fig. 5 A). Interestingly, among the 1432 identified proteins (FDR 1% and Fold Change ± 2.5 we found that there was an overlap of 193 deregulated proteins shared between NFs treated with YS-CM and CAFs as evidenced by Heatmap (Fig. 5 B). For instance, Principal Component Analysis (PCA) plot of NFs-CM YS, NFs and CAFs samples showed that PC1 dimension highlights similarity between NF-CM YS and CAF groups, which are clearly separated from the NF group. The second dimension explains a lower portion of total variance and is mainly related to differences between NF-CM YS group and the remaining two groups (NFs and CAFs) (Suppl. Figure 6 A). Particularly, out of the deregulated proteins: 113 proteins were upregulated and 80 proteins were downregulated shared between NFs exposed to the MCF-7 YS-CM and CAFs with respect to NFs alone. The upregulated proteins enriched particularly GO "process network" analyzed by Metacore and appeared to be involved in several processes, such as inflammation, angiogenesis and cytoskeleton interaction/ECM remodeling (Fig. 5 C). Here we reported the expression of proteins well representative of these mentioned processes as such as inflammation (e.g. CXCL8, PTGS2, PTGE5), angiogenesis (e.g. AMOTL2, EREG) and cytoskeleton interaction/ECM remodeling (e.g. PLOD, MXRA7, CTNNAL1, CDC42EP4 and YAP1) shared between the two proteomes of NFs-treated with YS CM and CAFs respectively (Fig. 5 D). The Direct Interaction Network (DIN) of upregulated proteins analyses revealed the protein YAP1 as the main central hubs, which was 5 fold increase in NFs exposed to CM YS and 3.7 fold in CAFs respect to NFs (Fig. 5 E). Among the 80 downregulated proteins shared between NFs-CM YS and CAFs with respect to NFs alone we evidenced how these proteins enriched the GO Process Network and are mostly related to the proteinase inhibitors (Supplementary Fig. 8).
Fig. 5.
Proteomic analysis revealed proteomic similarity between NFs exposed to YS-CM and CAFs. A Volcano plot of representing all detected proteins in the comparison CAFs vs NFs. The significant proteins are localized on the right (red dots, upregulated proteins) and on the left (blue dots, downregulated proteins). Gray dots correspond to not significantly proteins, accordingly with selected parameters. This Volcano plot includes among the up (light green dots) and downregulated proteins (brown dots) those shared with NFs following their exposure to MCF-7 YS-CM reflecting in such condition their transition into CAF phenotype. B Heatmap representing supervised hierarchical clustering of the differentially expressed proteins (Fold cutoff ± 2.5) between NFs treated with CM derived from MCF-7 YS for 5 days and CAFs vs NFs identified by LC–MS/MS analysis. The heatmap is shown in red for the degree of upregulated proteins and in green for the degree of downregulated proteins (C). C GO enrichment in Process Networks of upregulated proteins (Fold cutoff ≥ 2.5) was shown. D Heatmap of the most representative upregulated proteins (Fold cutoff ≥ 2.5) of the different functional categories reported in the GO “process network”. E Direct interaction network constructed by processing the upregulated proteins identified in the comparison between NFs treated with CM derived from MCF-7 YS vs NFs (Fold cutoff ≥ 2.5). The colors of the arrows indicate positive (green), negative (red) or unknown (gray) effects of the different network items represented with different symbols (reported in the legend). DIN (orange boxes): YAP1 (YAp65), SLC7A5, CSDA, MafF, AMOTL2
Evidence that MCF-7 YS BC modulates YAP1 expression in fibroblasts
YAP1 and its paralog transcriptional coactivator with PDZ-binding motif (TAZ) are key factors in the Hippo pathway. YAP1 is known to respond to and influence matrix stiffening, cancer invasion, angiogenesis, stem cell properties and metastasis. In particular, its intrinsic ability to modulate ECM stiffness makes YAP1 as a CAF signature [29].
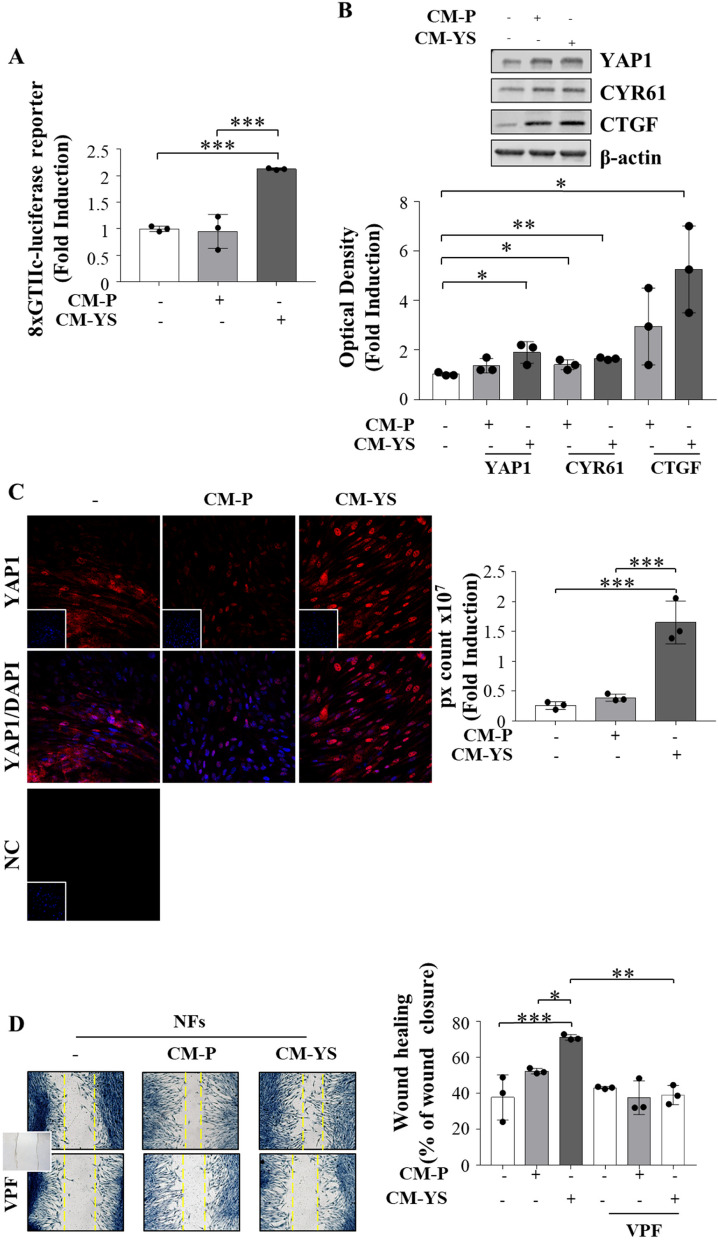
It is widely acknowledged that YAP/TAZ co-activates Tead 1–4, which in turn regulates the expression of several genes by binding to MCAT elements located in their promoter or enhancer regions [30]. Intriguingly the luciferase activity of the MCAT element-driven luciferase reporter construct was strongly increased in NFs exposed to MCF-7 YS-CM (Fig. 6 A), concomitantly with an enhanced protein expression of YAP1 working as an active transcriptional factor (Fig. 6 B). The latter event was confirmed by the concomitant upregulation of canonical YAP1 target genes such as CYR61 and CTGF (Fig. 6 B). All this fits with the immunofluorescence assay demonstrating that YAP1 mostly localizes to the nuclear region of NFs exposed to the MCF-7 YS-CM (Fig. 6 C). In the same circumstances, we observed that the enhanced expression of YAP1 in NFs exposed to MCF-7 P-CM and in a higher extent to MCF-7 YS-CM was concomitant with an increased cell motility [31]. The latter event was reversed in the presence of Verteporfin, a FDA-approved drug inhibitor of YAP/TAZ transcriptional activity. This recalls the role of YAP/TAZ cotranscription factor to guarantee the cytoskeletal/focal adhesion equilibrium enabling persistent cell motility Fig. 6 D).
Fig. 6.
YAP1 is a potential mediator of fibroblast/MCF-7 YS interplay. A NFs were transiently transfected with an MCAT-driven luciferase promoter and treated ( +) or not (-) with MCF-7 P and YS-CM for 24 h. B Immunoblotting showing YAP1 and its canonical target genes CYR61 and CTGF in NFs treated ( +) or not (-) with MCF-7 P and Y537S-CM for 5 days. The histograms represent the mean ± SD of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units (OD) and expressed as percentage of NF untreated cells (-) which were assumed to be 100%. C Immunofluorescence analysis of YAP1 in NFs treated ( +) or not (-) with MCF-7 P and YS-CM for 5 days. DAPI was used for nuclear staining. Nuclear Yap was quantified in six different fields per condition using ImageJ as described in material and methods. D Wound healing assay in NFs treated without (-) or with ( +) CM derived from MCF-7 P and YS, in the presence of verteporfin (VPF) 100 nm with pictures captured at time 0 (inset) and after 12 h. Each circle represents one independent experiments. Data were analyzed using one-way ANOVA with Tukey’s multiple comparisons post hoc test. Data are presented as mean values ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
All these data highlight the crucial role of YAP1 in conferring the characteristics of activated fibroblasts to NFs after their exposure to mutant CM.
The potential role of IGF-1 derived from MCF-7 YS-CM in modulating YAP1 expression in NFs
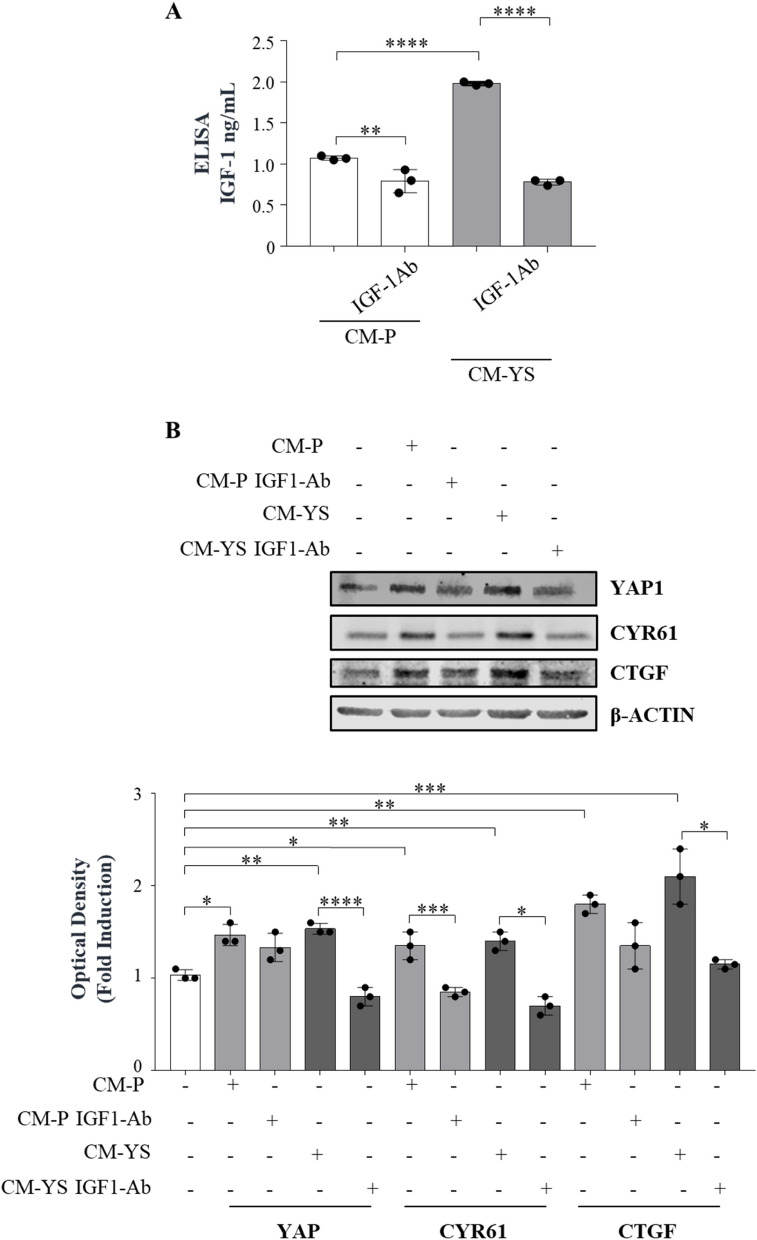
YAP1 signaling does not have a unique extracellular hippo specific link. Thus, it has been suggested that its activation may depend on the crosstalk with other oncogenic drivers and signaling contributing to YAP/TAZ activity, such as receptor tyrosine kinase signaling, even though the detailed mechanisms involved remain to be clarified [32]. Recently, insulin-like growth factor receptor (IGF-1R) signaling has been reported to be a negative regulator of Hippo-YAP signaling resulting in increased YAP1 expression [33]. Here we reported in a serum free medium how IGF-1 treatment for 5 days is able to per se upregulate YAP expression in a dose related manner (Supplementary Fig. 9). This led us to investigate whether the upregulation of YAP1 observed in the NFs exposed to MCF-7 YS-CM could be influenced by the content of IGF-1 present in the same CM. Thus, we treated NFs with MCF-7 P-CM and YS-CM immunodepleted of IGF-1 as confirmed by ELISA assay (Fig. 7 A). Our data clearly showed that the immunodepletion of IGF-1 reduced YAP1 expression in NFs and to a greater extent when they were exposed to MCF-7 YS-CM, concomitantly with the reduced expression of classical YAP1-dependent genes CYR61 and CTGF, suggesting the involvement of IGF-1 in the paracrine regulation of YAP1 expression and transcriptional activity in fibroblasts (Fig. 7 B).
Fig. 7.
Paracrine effects of IGF-1 on YAP1 expression in fibroblasts. A Enzyme-linked immunosorbent assay (ELISA) for IGF-1 protein secretion in MCF-7 P and YS cancer cells. B Immunoblotting showing YAP and its canonical target genes CYR61 and CTGF in NFs treated ( +) or not (-) with MCF-7 P and YS-CM immunodepleted or not of IGF-1. The histograms represent the mean ± SD of three separate experiments in which band intensities were evaluated in terms of optical density arbitrary units (OD) and expressed as percentage of NF untreated cells (-) which were assumed to be 100%.. Each circle represents one independent experiments. Data were analyzed using one-way ANOVA with Tukey’s multiple comparisons post hoc test. Data are presented as mean values ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
These data sound also relevant in the clinical setting since high levels of both IGF-1/CTGF and IGF-1R/CTGF are associated with poorer clinical outcomes in breast cancer patients in terms of post-progression survival (PPS) (Supplementary Fig. 10).
Evidence that MCF-7 YS BC cells potentiate their own intrinsic aggressive properties upon exposure to fibroblast conditioned media
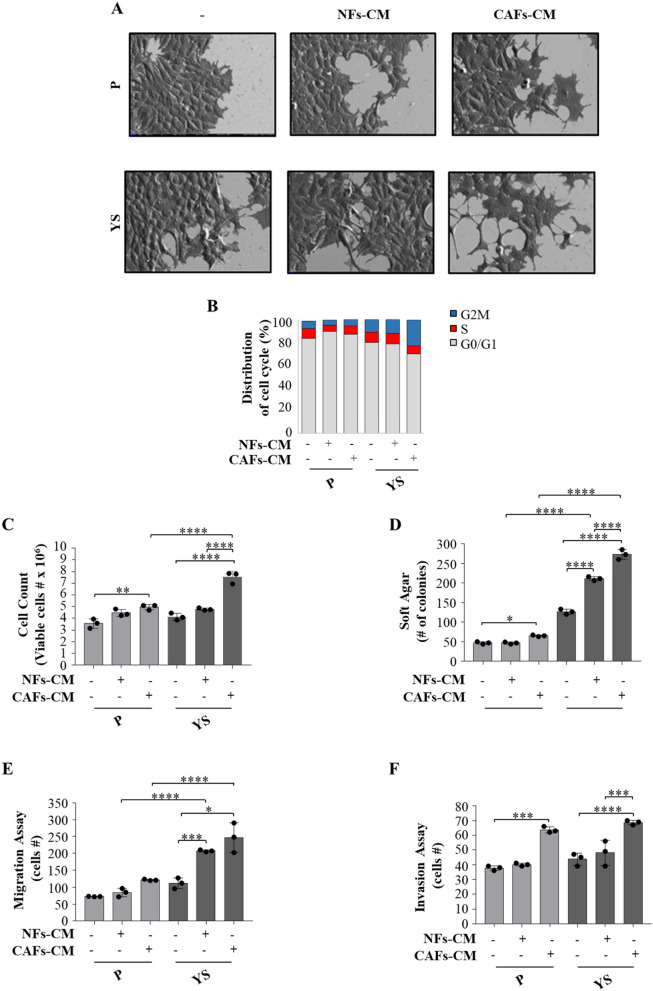
The same experimental coculture condition were used to investigate how the CM from fibroblasts influence the morphology, cell cycle, proliferation, migration and invasion of mutant cells. Upon exposure to the NFs- and CAFs-CM, both MCF-7 cell lines exhibited elongated or spindle-like morphology with lamellipodia. These latter effects were more evident in the presence of CAFs-CM conferring to the MCF-7 YS cells a more pronounced EMT morphology with fewer globular shaped cells, a less cell–cell contacts and acquired elongated form with lamellipodia (Fig. 8 A). Cytofluorometric analysis revealed that compared to NFs-CM, CAFs-CM reduced the percentage of YS-expressing cells in the G0/G1 phase of the cell cycle and increased the percentage of cells in the S and G2/M phases, whereas these effects were no longer observed in MCF-7 P cells (Fig. 8 B). Compared with those of the parental cells, the growth rate of the mutant YS cells increased in an anchorage-dependent manner and an anchorage-independent manner (Fig. 8 C and D) in the presence of both NFs-CM and CAFs-CM with respect to the parental cells. Moreover, our results showed that compared with NFs-CM, CAFs-CM increased the number of migrated and invaded cells in both cell lines but to a greater extent in mutant cells (Fig. 8E and F).
Fig. 8.
Biological effects of NFs-CM and CAFs-CM on MCF-7 P and MCF-7 YS cells. A SEM images of MCF-7 P and YS cells treated with NFs- and CAFs-CM for 5 days. Scale bar: 20 µm. B MCF-7 P and YS cells treated without (-) or with ( +) CM derived from NFs and CAFs for 5 days stained with propidium iodide and analyzed by using a FACScan flow cytometer. The percentages of cells in the G0/G1, S, and G2/M phases are reported. C Viability of MCF-7 P and YS cells treated without (-) or with NFs- and CAFs-CM for 5 days. Live cells were counted after 5 days of treatment. D Soft agar growth assays in MCF-7 P and YS cells treated with NFs- and CAFs-CM. After 14 days of growth, colonies ≥ 50 μm in diameter were counted. Transmigration (E) and invasion (F) assays of MCF-7 P and YS cells treated without (-) or with NFs- and CAFs-CM for 24 h. Each circle represents one independent experiments. Data were analyzed using one-way ANOVA with Tukey’s multiple comparisons post hoc test. Data are presented as mean values ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
These data suggest a potential role of CAFs in potentiating the aggressive phenotype of mutant cells.
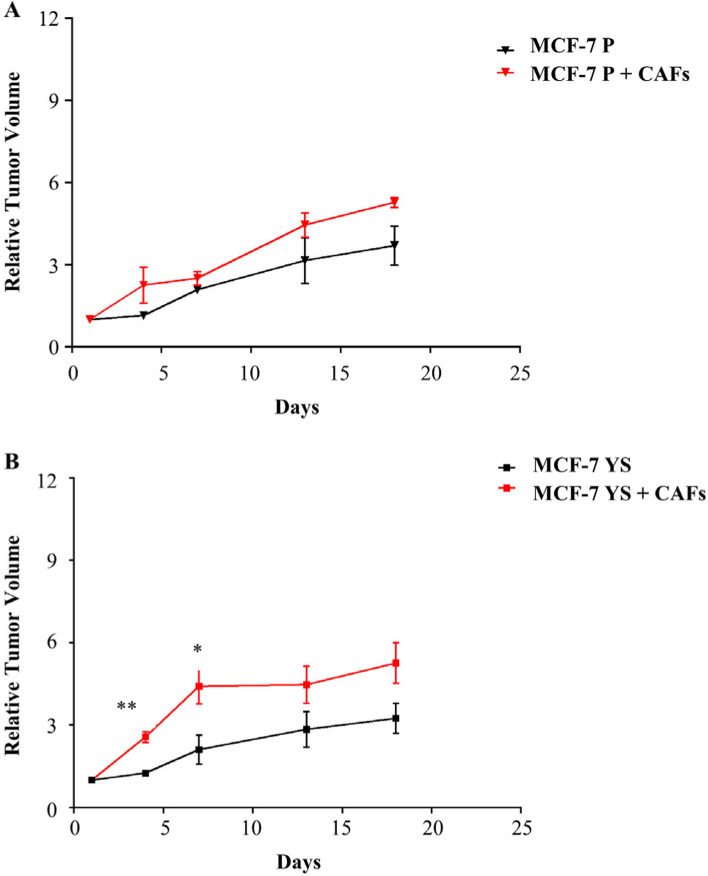
All this led us to ascertain in vivo the major capability of CAFs in potentiating the aggressive phenotype of mutant cells with respect of NFs. To this aim MCF-7 P and MCF-7 YS were co-implanted, respectively, with CAFs (ratio 1:3) in the mouse orthotopic region and tumor growth was monitored. Our data demonstrated a significant early increase in relative tumor growth only in mutant cells co-implanted with CAFs (Fig. 9 A and B). A histological and morphological evaluation was conducted using hematoxylin and eosin staining, particularly for cytokeratin 18, to confirm the epithelial origin of implanted human breast cancer cells, while the immunostaining of α-smooth muscle actin (α-SMA) utilized a specific sma human antibody was performed to identify the implanted human cancer-associated fibroblasts (Supplementary Fig. 11 A). However, it was intriguing to observe a significant increase in alpha-SMA immunoreactivity in MCF-7 YS cells co-implanted with CAFs, suggesting that an increase of CAF cell number occurred following the start of implantation (Supplementary Fig. 11 B). In the same experimental conditions, we observed an increase of Ki-67, a widely used marker of proliferative human tumor cells, in mice co implanted with MCF-7 YS and CAFs compared to their parental counterparts (Supplementary Fig. 12).
Fig. 9.
Effects of CAFs on the growth of MCF-7 P and YS xenograft tumors. MCF-7 P (A) and YS (B) cells were injected alone or coinjected with CAFs (MCF-7/CAFs) orthotopically into nude mice (5 mice/group). The relative tumor volume (RTV) was calculated from the following formula: RTV = (Vx/V1), where Vx is the tumor volume on day X and V1 is the tumor volume at initiation of the treatment for each group. Y axis: the mean and ± SD of the RTV. Data were analyzed using student t test. Data are presented as mean values ± SEM. *p < 0.05; **p < 0.001
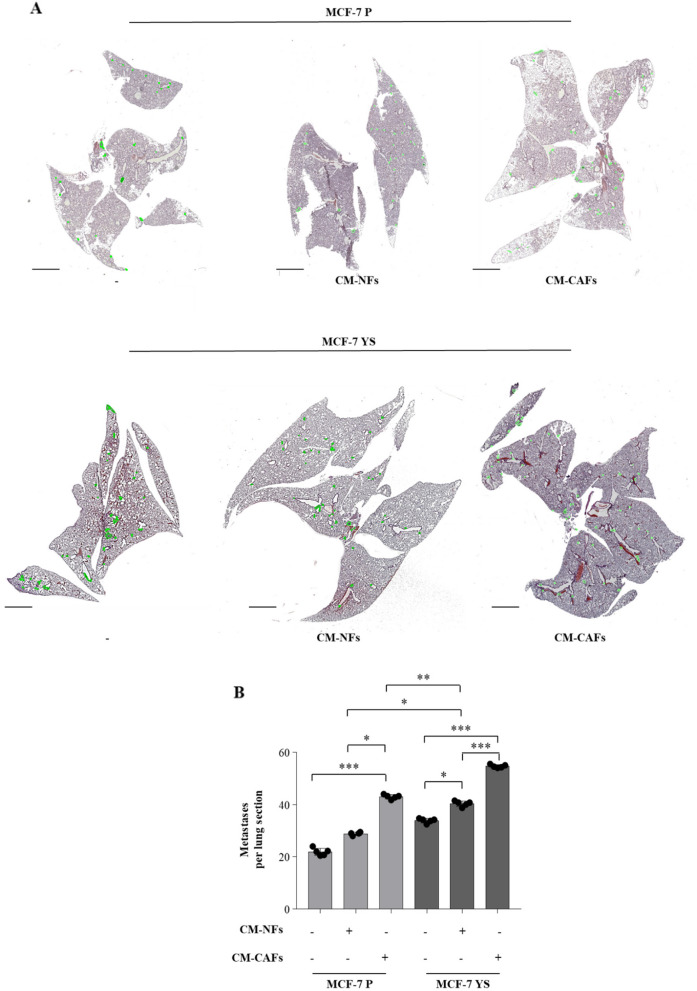
Then we moved to assess how the CAFs may influence the intrinsic metastatic potential of mutant cells compared to parental cell as previously described [8]. To this aim, the mutant and parental cells where pre-treated for 5 days with CM derived from NFs and CAFs, respectively, and then injected into nude mice via the median tail vein for assessment of lung metastatic colonization. H&E staining showed that MCF-7 P cells display a significant increased number of metastases only when treated with CAFs-CM for lung sections. On the contrary, MCF-7 YS mutant cells display an intrinsic a higher metastatic burden with respect parental counterpart, which was further augmented upon NFs-CM and markedly increased upon CAFs-CM (Fig. 10 A and B).
Fig. 10.
Impact of NFs- and CAFs-CM on breast cancer metastasis in vivo. A Representative images of hematoxylin and eosin stained lung tissues of mice after 28 days from injection with MCF-7 P and MCF-7 YS untreated (-) or treated with NFs-CM and CAFs-CM for 5 days. Scale bars: 3 mm. B The histogram represents the number of metastatic lesions per lung section. Each circle represents the number of lung sections analyzed for each animal. Data were analyzed using one-way ANOVA with Bonferroni’s multiple comparisons test. Data are presented as mean values ± SD. *p < 0.05; **p < 0.001; ***p < 0.0001
Discussion
It is well documented how aggressiveness of somatic mutations in the ESR1 gene is a major contributor to endocrine resistance and worst clinical outcome in metastatic breast cancer patients [34–37].
Previously MCF-7 Y537S breast cancer cells have been widely investigated for their intrinsic tumorogenic properties without considering their functional interactions with TME. Now, we became aware that Y537S-mutant expressing breast cancer cells may potentially reprogram their surrounding ecosystem. Here, for the first time, we demonstrated how Y537S-expressing cells drastically modify fibroblasts behavior, converting normal fibroblasts into cancer-associated fibroblasts, wherein the YAP/TAZ system raises as a crucial mediators of this phenotypic transition.
Fibroblast biology is deeply connected to the extracellular matrix (ECM) because fibroblasts are the main cell type responsible for ECM production. In the connective tissue in several fibrotic pathologies, fibroblasts are found in an activated form and drive ECM cross linking and stiffening [38]. In turn, a stiffer ECM activates a fibrotic positive feedback loop by sustaining fibroblast activation which maintains or amplifies the disease [39]. Activated fibroblasts or CAFs play an active role in remodeling the TME and producing soluble factors ultimately conspiring with cancer cells to promote migration and metastatic behavior. Our present findings demonstrate that normal fibroblasts exposed to the conditioned medium of Y537S appear clearly more stretched on their ECM assuming a more expanded leaf of a similar shape instead of the spindle shape observed in the fibroblast controls. In the same condition, they acquired highly contractile phenotype exhibiting a myogenic differentiation of actin bubbles and stress fibers quite similar to those observed in cancer-associated fibroblasts reinforcing the most broadly accepted hypothesis that the majority of CAFs likely originate from the activation of local tissue resident fibroblasts. The proteomic profile reveals that a cluster of protein which in NFs upon MCF7 P-CM exposure did not exhibit any substantial change, appear to be up regulated upon YS-CM displaying a pattern of expression similar to that revealed by CAFs with respect to NFs. These upregulated proteins are mostly involved in inflammatory, neoangiogenesis and in ECM remodeling and cytoskeleton organization signaling. The direct interaction network (DIN) analysis of the proteins upregulated in NFs exposed to YS-CM according to Metacore gives the major relevance to YAP protein, which enhanced expression is confirmed in terms of protein content and also coincides with its enhanced cotranscriptional activity. YAP1 and TAZ are the main effectors of Hyppo signaling which are an evolutionarily conserved regulatory factor of tissue growth related to cell fate and is regulated mainly by the actin cytoskeleton and cellular tension [40–42]. YAP/TAZ are negatively regulated when phosphorylated by LATS1/2, which are in turn regulated by MST1/2. Phosphorylation of YAP1 and TAZ is associated with their accumulation in the cytoplasm, blocking their binding to TEAD and other proteins, thus impairing their co-transcription activity [29]. YAP/TAZ are thought to be somehow redundant and similarly regulated by Hippo signaling. However, even though the main Hippo pathway effector YAP1 and TAZ are master regulators for multiple cellular processes, several evidences address that YAP1 has a stronger influence than TAZ. In the same vein, the DIN of deregulated proteins in CAFs vs NFs places YAP/TAZ and c-Jun proto-oncogene of AP-1 transcriptional factor as central hub also confirmed within the net of upregulated proteins in CAFs vs NFs. All this recalls the well-known functional interactions between YAP/TAZ and AP-1 in controlling cell cycle and cell proliferation [43] as well as in the regulation of the activity ROCK-RAC/CDC42 complex driving cell migration and invasion [44]. YAP/TAZ when phosphorylated by LATS1/2, that are in turn regulated by MST1/2, accumulates in the cytoplasm, blocking their binding to TEAD and other proteins, thus impairing the activation of transcription [29]. YAP1 is predominantly cytoplasmic in fibroblasts but accumulates in the nucleus in the CAFs [45]. Immunofluorescence revealed how the exposure of NFs to MCF-7 YS-CM, compartmentalizes YAP1 mostly in the nucleus sustaining its transcriptional activity thus regulating cell adhesion.
Cellular mechanosensing relays on the interplay between cell-generated traction forces and resisting forces within the ECM. Integrins bind the ECM at their extracellular site and the f-actin site cytoskeleton with other adapter protein at their intracellular site. Actin-myosin contractions of the integrin adhesion on the resistant attached matrix bring to the alteration of integrin adhesion, their size, strength, and dynamics [46]. Thus, YAP1 seems to “tune” fibroblast actin-myosin contractility maintaining contractile fibroblasts into a self-sustaining and persistent feed-forward loop. All this would ensure a YAP1 mediated fibroblasts tuning, sustaining ECM remodeling and stiffening which drives tumor progression. So, cells tension tuned to the stiffness of the ECM engage actin-myosin contractility pathways and fosters the assembly of integrin focal adhesions [47–49].
In response to extracellular forces the integrin-Rho-RAC-/ROCK/Actin core engine also appears to be involved in the conversion of NFs into CAFs in the tumor stroma [50]. It was highlighted that when the YAP1 expression is increased in NFs they were converted into CAFs as confirmed by their specific markers [49, 50].
It is worth to remark how the reported enhanced expression of YAP1 in NFs exposed to MCF-7 Y537S-CM was concomitant with that of two classical dependent genes CCN1 and CCN2 [51]. Both genes are involved in modulating ECM production and themselves responsive in mechanotrasduction signaling generating isometric tension within the cells and actin-rich structure called invadosome that are composed of invadopodes and podisomy [29, 49]. All this indicates how YAP1 is fully involved in promoting CAF invasiveness.
YAP1/TAZ also, in breast cancer, sustains the production of laminin511 cotranscriptional activity, which results in autocrine activation of integrins that feedback on YAP1/TAZ to sustain breast cancer cell motility [52]. However, a direct role of YAP1 cotranscriptional activity in modulating cell motility raised by the fact that it controls gene expression involved in cytoskeleton/focal adhesion equilibrium enabling in such way persistent cell motility [31]. Indeed, when we used an inhibitor of YAP1 transcriptional acitivity, verterporfin, in fibroblasts upon exposure to MCF-7 YS-CM, the observed increased cell motility was no longer noticeable. Given that the Hyppo-YAP1 signaling pathway does not have a unique extracellular “hyppo” specific ligand [32] as a specific membrane receptor, its activation may depend on cross-talk with other oncogenic drivers contributing to YAP1/TAZ activity, such as receptor tyrosine kinase (RTK) signaling [41]. Recently, several transmembrane proteins, including EGF and G protein-coupled receptor (SPCRS) and G protein complex receptor (SPCK) have been reported to participate in the regulation of Hyppo-YAP1 signaling; however, the detailed mechanism involved remains to be clarified [53, 54]. IGF1-R acts as an upstream negative regulator of Hyppo-YAP1 signaling resulting in increased YAP1 expression [32, 33]. Within the proteomic profile the GO enrichment analysis of up-regulated proteins of YS human mutant breast cancer cells displays an intrinsic potentiated IGF-1/IGF-1R signaling, ranked at the second position of Pathway Maps fitting with an enhanced protein content and phosphorylated status of IGF-1R and confirming previous data obtained by us and other authors [8, 11, 13]. The involvement of IGF-1 in the regulation of YAP expression raises in the present study by the evidence that in NFs exposed to MCF-7 YS-CM the upregulation of YAP1 expression together with the expression of its classical target genes CYR61 and CTGF was reversed when the same medium was immunodepleted of IGF-1. It has been reported that the depletion of IGF-1R by shRNA or specific inhibitors decreases YAP1 expression. In contrast, the addition of IGF-1 upregulated and enhanced its nuclear localization [55]. Data mining of clinical breast cancer specimens revealed that basal-like breast cancer patients with high level of IGF-1 and YAP1 have been associated with poorer overall survival [55]. The activation of normal fibroblasts into CAFs can result in the generation of abundant tumor promoting factors and facilitate the malignant behavior of tumor cells through a complicated paracrine network. Indeed, the proteomic analysis of significantly downregulated proteins in fibroblasts exposed to MCF-7 YS CM revealed a significant decrease in the expression of protease inhibitors, similar to what was observed in CAFs in basal conditions. The activities of metalloproteinases are regulated by proteolytic activation and inhibition via their natural inhibitors TIMPs. An imbalance of activation and inhibition may hamper ECM equilibrium and is responsible for the progression of cancer [56]. Indeed, it is well known that the metalloproteinases (MMPs) directly regulate angiogenesis through MMP-2, MMP-9 and MMP-14, which are fundamental steps of tumor growth [56]. All this addresses how in such circumstances proteases released by fibroblasts upon MCF-7 YS exposure and by CAFs may be dismissed in an amount exceeding their homologous inhibitors enhancing in such way the burden malignancy of tumor microenvironment. All these are in accordance with the findings of the present study evidencing the capability of NFs exposed to the MCF-7 YS CM to contract a collagen gel reproducing the ECM in vitro, a typical functional feature of activated fibroblasts. Moreover, the important role of CAFs in sustaining the aggressive behavior of mutant-expressing cells came from the evidence that CAFs when co-implanted with YS in nude mice significantly increased tumor growth with respect to that observed in the presence of YS alone. In the same vein, we demonstrated how the intrinsic metastatic properties of mutant cells were drastically increased following the exposure to the conditioned medium of CAFs with respect to that observed following the exposure to CM of normal fibroblasts.
Conclusions
1- Thus, in the present study, we demonstrated that Y537S mutant cells, which are mostly present in metastatic breast cancers induces a strong increase of the number of CAFs in the TME as a consequence of their enhanced proliferation and increased conversion from NFs “reprogrammed upon their conditioned media”.
2- The Y537S-induced conversion of NFs into CAFs is crucially mediated by the activated intrinsic YAP1 trasduction pathway, which is recognized as the main regulator of fibroblast stretching as well as of their cytoskeleton adaptation, their cell shape spread and cell motility.
3- The YAP1-mediated transition of NFs into CAFs appears to involve IGF-1 present in the MCF-7 Y537S CM.
4- Our present results call back accumulating evidence showing how the enhanced expression of YAP1 promotes proliferation and epithelial mesenchymal transition in different solid tumors [57, 53]. Here we demonstrated that YAP/TAZ TEAD complex in fibroblasts is a crucial modulator of the stimulatory effects of mutant cells on CAF proliferation. In such circumstances, it emerges how the block of YAP1 signaling may break down the bidirectional functional interaction between mutant cells and fibroblasts, weakening markedly breast tumor burden malignancy. Thus, the latter approach could be reasonably implemented in the novel therapeutic strategies of a subset of MBC patients carrying the frequent Y537S mutation.
Supplementary Information
Supplementary Material 1: Supplementary Fig. 1. Uncropped blots of Fig. 2F (A), 2H (B) and 3C (C). Supplementary Fig. 2. Uncropped blots of Fig. 6B. Supplementary Fig. 3. Uncropped blots of Fig. 7B. Supplementary Fig. 4. Uncropped blots of Supplementary Fig. 9. Supplementary Fig. 5. Proteomic comparison of upregulated and downregulated proteins identified in MCF-7 YS vs P. Supplementary Fig. 6. Proteomic comparison of deregulated proteins identified in CAFs vs NFs. Supplementary Fig. 7. Proteomic analysis of NFs, NFs treated with MCF-7 P and YS-CM and CAFs. Supplementary Fig. 8. Proteomic analysis of downregulated proteins shared between NF treated with YS-CM treated and CAFs. Supplementary Fig. 9. Effects of IGF-1 in serum-free medium on YAP expression in NFs. Supplementary Fig. 10. Clinical significance of the co-expression of IGF-1, IGF-1R, and YAP canonical target gene CCN2 in breast cancer patients. Supplementary Fig. 11. Immunohistochemical analysis of xenograft tumors. Supplementary Fig. 12. ki-67 immunohistochemical analysis of xenograft tumors. Supplementary Table S1. Oligonucleotide primers used in this study. Supplementary Table S2. Gene Ontology “Pathways Maps” defined by MCF-7YS Mutant-expressing cells.
Acknowledgements
Research Centre for Advanced Biochemistry and Molecular Biology, Department of Experimental and Clinical Medicine, Magna Graecia University of Catanzaro, 88100 Catanzaro, Italy; we are deeply grateful to Prof. Sirio Dupont from the Department of Molecular Medicine, University of, 35123, Padova, Italy, for having provided the 8xGTIIC-luciferase (MCAT-reporter) to evaluate YAP/TAZ cotranscription activity in fibroblasts.
Abbreviations
- HDB
Hormone Binding Domain
- IGF-1
Insulin-like growth factor 1
- IGF-1R
Insulin-like growth factor receptor 1
- TME
Tumor microenvironment
- NFs
Normal Fibroblasts
- CAFs
Cancer-associated fibroblasts
- CM
Conditioned media
- SEM
Scanning electron microscopy
- ELISA
Enzyme-linked immunosorbent assay
- nLC-MS/MS
Nanoliquid chromatography tandem mass spectrometry
- DIN
Direct interaction network
- YAP1
Yes-associated protein 1
- PPS
Post-progression survival
- PCA
Principal component analysis
- GO
Gene ontology
- MBC
Metastatic breast cancer
- ECM
Extracellular matrix
- ITβ1
Integrin Beta-1
- FSP1
Fibroblast specific protein 1
Authors’ contributions
Conceptualization, S.C. and S.A.; methodology, L.G., A.C., E.T., A.E.L., G.D.N., R.M., F.G., S.P. and B.P.; validation, G.G, C.G., I.B., L.M. and D.B.; formal analysis, C.G., I.B., D.B. and L.M.; investigation, L.G., A.C., E.T., A.E.L., G.D.N., R.M., F.G., S.P. and B.P.; resources, S.A. and S.C.; data curation, L.G., A.C., E.T., A.E.L., G.D.N., S.P. and R.M.; writing—original draft preparation, L.G., and A.C.; writing—review and editing, S.A.W.F., S.C. and S.A.; visualization, C.G., I.B., D.B. and L.M.; acquired material G.F. and B.N.; supervision, I.B., C.G., S.C. and S.A.; funding acquisition, L.G., I.B., S.C., S.A.
Funding
This research was funded by AIRC Investigator Grant (IG) #26246 to S. Andò; BANDO PRIN 2017 #2017WNKSLR, PRIN 2022-European NextGenerationEU initiative under the Italian Ministry of University and Research—M4C2-I1.1 (#202239N8PR, CUP H53D23006360006), and PRIN PNRR 2022-European NextGenerationEU initiative under the Italian Ministry of University and Research as part of the PNRR—M4C2-I1.1 (#P2022YAKJY, CUP H53D23010120001) to I. Barone; PRIN 2022, European NextGenerationEU initiative under the Italian Ministry of University and Research -M4C2-I1.1 (#2022AA4FTJ, CUP H53D23006420006) to Giordano C; PRIN PNRR 2022-European NextGenerationEU initiative under the Italian Ministry of University and Research as part of the PNRR—M4C2-I1.1 (#P2022FK2J8, CUP H53D23010420001) to L. Gelsomino; AIRC Investigator Grant (IG) #21414 to S. Catalano.
Data availability
The data generated in this study will be available upon request.
Declarations
Ethics approval and consent to participate
In the present study we used fibroblast specimens, isolated in accordance with approved guidelines, from patients who had signed informed consent and approved by the Ethic Institutional Committees at Annunziata Hospital, Cosenza, Italy (#149 issued by Comitato Etico Regione Calabria, Sezione Area Nord c/o Azienda Ospedaliera di Cosenza, 28/10/2015). All animals were maintained and handled in accordance with the recommendation of the Guidelines for the Care and Use of Laboratory Animals and experiments were approved by the Animal Care Committee of University of Calabria (OPBA), Italy (454/2023-PR, May 17, 2023).
Consent for publication
We have obtained consents to publish this paper from all the participants of this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Luca Gelsomino and Amanda Caruso are joint first author.
Stefania Catalano and Sebastiano Andò are joint senior author.
References
- 1.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer–An overview and update. Mol Cell Endocrinol. 2015;418 Pt 3(03):220–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017;7(3):277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang QX, Borg A, Wolf DM, Oesterreich S, Fuqua SA. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997;57(7):1244–9. [PubMed] [Google Scholar]
- 7.Fuqua SA, Gu G, Rechoum Y. Estrogen receptor (ER) alpha mutations in breast cancer: hidden in plain sight. Breast Cancer Res Treat. 2014;144(1):11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu G, Tian L, Herzog SK, Rechoum Y, Gelsomino L, Gao M, et al. Hormonal modulation of ESR1 mutant metastasis. Oncogene. 2021;40(5):997–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25(6):869–98. [DOI] [PubMed] [Google Scholar]
- 10.Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, et al. Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell. 2018;33(2):173–86 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelsomino L, Gu G, Rechoum Y, Beyer AR, Pejerrey SM, Tsimelzon A, et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res Treat. 2016;157(2):253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Inao T, Sueta A, Fujiwara S, et al. Droplet digital polymerase chain reaction assay for screening of ESR1 mutations in 325 breast cancer specimens. Transl Res. 2015;166(6):540–53 e2. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Levine KM, Bahreini A, Wang P, Chu D, Park BH, et al. Upregulation of IRS1 Enhances IGF1 Response in Y537S and D538G ESR1 Mutant Breast Cancer Cells. Endocrinology. 2018;159(1):285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016;2(10):1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei JT, Gou X, Seker S, Ellis MJ. ESR1 alterations and metastasis in estrogen receptor positive breast cancer. J Cancer Metastasis Treat. 2019;5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mhaidly R, Mechta-Grigoriou F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. Semin Immunol. 2020;48:101417. [DOI] [PubMed] [Google Scholar]
- 17.Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci (Landmark Ed). 2010;15(1):166–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalano S, Panza S, Augimeri G, Giordano C, Malivindi R, Gelsomino L, et al. Phosphodiesterase 5 (PDE5) Is Highly Expressed in Cancer-Associated Fibroblasts and Enhances Breast Tumor Progression. Cancers (Basel). 2019;11(11):1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recchia AG, Musti AM, Lanzino M, Panno ML, Turano E, Zumpano R, et al. A cross-talk between the androgen receptor and the epidermal growth factor receptor leads to p38MAPK-dependent activation of mTOR and cyclinD1 expression in prostate and lung cancer cells. Int J Biochem Cell Biol. 2009;41(3):603–14. [DOI] [PubMed] [Google Scholar]
- 20.Casaburi I, Avena P, Lanzino M, Sisci D, Giordano F, Maris P, et al. Chenodeoxycholic acid through a TGR5-dependent CREB signaling activation enhances cyclin D1 expression and promotes human endometrial cancer cell proliferation. Cell Cycle. 2012;11(14):2699–710. [DOI] [PubMed] [Google Scholar]
- 21.Naimo GD, Forestiero M, Paolì A, Malivindi R, Gelsomino L, Győrffy B, et al. ERα/LKB1 complex upregulates E-cadherin expression and stimulates breast cancer growth and progression upon adiponectin exposure. Int J Cancer. 2023;153(6):1257–72. [DOI] [PubMed] [Google Scholar]
- 22.García Peñarrubia P, Férez Ruiz X, Gálvez J. Quantitative analysis of the factors that affect the determination of colocalization coefficients in dual-color confocal images. IEEE Trans Image Process. 2005;14(8):1151–8. [DOI] [PubMed] [Google Scholar]
- 23.Murfuni MS, Prestagiacomo LE, Giuliano A, Gabriele C, Signoretti S, Cuda G, et al. Evaluation of PAC and FASP Performance: DIA-Based Quantitative Proteomic Analysis. Int J Mol Sci. 2024;25(10):5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68. [PubMed] [Google Scholar]
- 25.La Camera G, Gelsomino L, Malivindi R, Barone I, Panza S, De Rose D, et al. Adipocyte-derived extracellular vesicles promote breast cancer cell malignancy through HIF-1alpha activity. Cancer Lett. 2021;521:155–68. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal. 2020;18(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plouffe SW, Lin KC, Moore JL 3rd, Tan FE, Ma S, Ye Z, et al. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J Biol Chem. 2018;293(28):11230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15(6):637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–83. [DOI] [PubMed] [Google Scholar]
- 31.Mason DE, Collins JM, Dawahare JH, Nguyen TD, Lin Y, Voytik-Harbin SL, et al. YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J Cell Biol. 2019;218(4):1369–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maller O, DuFort CC, Weaver VM. YAP forces fibroblasts to feel the tension. Nat Cell Biol. 2013;15(6):570–2. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Li Y, Wang W, Wang S, Hou J, Zhang A, et al. Regulation of Hippo/YAP signaling and Esophageal Squamous Carcinoma progression by an E3 ubiquitin ligase PARK2. Theranostics. 2020;10(21):9443–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barone I, Iacopetta D, Covington KR, Cui Y, Tsimelzon A, Beyer A, et al. Phosphorylation of the mutant K303R estrogen receptor alpha at serine 305 affects aromatase inhibitor sensitivity. Oncogene. 2010;29(16):2404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giordano C, Cui Y, Barone I, Ando S, Mancini MA, Berno V, et al. Growth factor-induced resistance to tamoxifen is associated with a mutation of estrogen receptor alpha and its phosphorylation at serine 305. Breast Cancer Res Treat. 2010;119(1):71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herzog SK, Fuqua SAW. ESR1 mutations and therapeutic resistance in metastatic breast cancer: progress and remaining challenges. Br J Cancer. 2022;126(2):174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffreys SA, Powter B, Balakrishnar B, Mok K, Soon P, Franken A, et al. Endocrine Resistance in Breast Cancer: The Role of Estrogen Receptor Stability. Cells. 2020;9(9):2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLeon-Pennell KY, Barker TH, Lindsey ML. Fibroblasts: The arbiters of extracellular matrix remodeling. Matrix Biol. 2020;91–92:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera J, Henke CA, Bitterman PB. Extracellular matrix as a driver of progressive fibrosis. J Clin Invest. 2018;128(1):45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupont S. Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp Cell Res. 2016;343(1):42–53. [DOI] [PubMed] [Google Scholar]
- 41.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94(4):1287–312. [DOI] [PubMed] [Google Scholar]
- 42.Piccolo S, Panciera T, Contessotto P, Cordenonsi M. YAP/TAZ as master regulators in cancer: modulation, function and therapeutic approaches. Nat Cancer. 2023;4(1):9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17(9):1218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, et al. Tead and AP1 Coordinate Transcription and Motility. Cell Rep. 2016;14(5):1169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nardone G, Oliver-De La Cruz J, Vrbsky J, Martini C, Pribyl J, Skladal P, et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat Commun. 2017;8:15321. [DOI] [PMC free article] [PubMed]
- 46.Wolfenson H, Yang B, Sheetz MP. Steps in Mechanotransduction Pathways that Control Cell Morphology. Annu Rev Physiol. 2019;81:585–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Bio. 2011;12(5):308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–54. [DOI] [PubMed] [Google Scholar]
- 49.Naktubtim C, Payuhakrit W, Uttarawichien T, Hassametto A, Suwannalert P. YAP, a novel target regulates F-actin rearrangement-associated CAFs transformation and promotes colorectal cancer cell progression. Biomed Pharmacother. 2022;155: 113757. [DOI] [PubMed] [Google Scholar]
- 50.Shen T, Li Y, Zhu S, Yu J, Zhang B, Chen X, et al. YAP1 plays a key role of the conversion of normal fibroblasts into cancer-associated fibroblasts that contribute to prostate cancer progression. J Exp Clin Cancer Res. 2020;39(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quan T, Xu Y, Qin Z, Robichaud P, Betcher S, Calderone K, et al. Elevated YAP and its downstream targets CCN1 and CCN2 in basal cell carcinoma: impact on keratinocyte proliferation and stromal cell activation. Am J Pathol. 2014;184(4):937–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang C, Goel HL, Gao H, Pursell B, Shultz LD, Greiner DL, et al. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the alpha6Bbeta1 integrin to sustain breast cancer stem cells. Genes Dev. 2015;29(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Zhu D, Zhang H, Han C, Xue G, Zhu T, et al. YAP-dependent ubiquitination and degradation of beta-catenin mediates inhibition of Wnt signalling induced by Physalin F in colorectal cancer. Cell Death Dis. 2018;9(6):591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan YT, Lin RJ, Wang YH, Hung TH, Huang Y, Yu J, et al. The interplay between IGF-1R signaling and Hippo-YAP in breast cancer stem cells. Cell Commun Signal. 2023;21(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raeeszadeh-Sarmazdeh M, Do LD, Hritz BG. Metalloproteinases and Their Inhibitors: Potential for the Development of New Therapeutics. Cells. 2020;9(5):1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou B, Flodby P, Luo J, Castillo DR, Liu Y, et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest. 2018;128(3):970–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Fig. 1. Uncropped blots of Fig. 2F (A), 2H (B) and 3C (C). Supplementary Fig. 2. Uncropped blots of Fig. 6B. Supplementary Fig. 3. Uncropped blots of Fig. 7B. Supplementary Fig. 4. Uncropped blots of Supplementary Fig. 9. Supplementary Fig. 5. Proteomic comparison of upregulated and downregulated proteins identified in MCF-7 YS vs P. Supplementary Fig. 6. Proteomic comparison of deregulated proteins identified in CAFs vs NFs. Supplementary Fig. 7. Proteomic analysis of NFs, NFs treated with MCF-7 P and YS-CM and CAFs. Supplementary Fig. 8. Proteomic analysis of downregulated proteins shared between NF treated with YS-CM treated and CAFs. Supplementary Fig. 9. Effects of IGF-1 in serum-free medium on YAP expression in NFs. Supplementary Fig. 10. Clinical significance of the co-expression of IGF-1, IGF-1R, and YAP canonical target gene CCN2 in breast cancer patients. Supplementary Fig. 11. Immunohistochemical analysis of xenograft tumors. Supplementary Fig. 12. ki-67 immunohistochemical analysis of xenograft tumors. Supplementary Table S1. Oligonucleotide primers used in this study. Supplementary Table S2. Gene Ontology “Pathways Maps” defined by MCF-7YS Mutant-expressing cells.
Data Availability Statement
The data generated in this study will be available upon request.