Abstract
Background
Colon adenocarcinoma (COAD) ranks as the third most prevalent and lethal cancer in 2020, with metastasis being the primary cause of cancer-related mortality. A comprehensive understanding of the mechanism underlying distant metastasis is imperative for enhancing the prognosis and quality of life of patients with COAD.
Methods
This study employed gene set enrichment analysis (GSEA) on RNA-sequencing data from 408 patients with COAD in The Cancer Genome Atlas (TCGA) database. GSEA analysis was applied to find the significant hallmark gene set, and genes in the significant hallmark gene set was performed by univariate Cox regression to select the key gene. Then, multivariate Cox regression model was constructed. And various databases were utilized to validate and find the significant oncoprotein and hallmark gene set of the key gene. In addition, the expression of key regulators in para-carcinoma tissue, colon cancer and distant metastases samples were detected by real-time PCR, Immunohistochemistry and Western blot. Additionally, the biological functions were examined by in vitro and in vivo experiments. The scRNA-seq and CellphoneDB were performed to explore cell characters in COAD and the potential mechanism of metastasis.
Results
The regulatory network analysis revealed SRC/YAP1 as the most significant oncoprotein and signature gene set associated with SNTB1. Moreover, significant SNTB1 overexpression in COAD metastatic tissues was observed compared to para-carcinoma and primary COAD tissues. Co-immunoprecipitation assays demonstrated the formation of a complex between SNTB1 and SRC proteins. Furthermore, the overexpression of SNTB1 enhanced the proliferation, migration and invasion capacities of COAD cell lines. Caudal vein injection of COAD cells overexpressing SNTB1 in nude mice resulted in increased tumour growth and metastasis to the lung, liver and bone. Finally, single cell RNA-seq revealed alterations in the cellular subtypes of COAD, and CellphoneDB indicated that the interaction between cancer cells exhibiting high SNTB1 expression and enterocytes promoted EMT through cellular communications involving TGF-β, accelerating metastasis in COAD.
Conclusion
This study postulates that SNTB1 interacts with SRC to activate the Hippo-YAP pathway, thereby promoting COAD metastasis. Furthermore, cellular communication with enterocytes promotes EMT, facilitating metastasis. These findings propose novel therapeutic targets for preventing or treating metastatic COAD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05548-2.
Keywords: SNTB1, epithelial-mesenchymal transition, Hippo-YAP Pathway, Colon adenocarcinoma, Metastasis
Introduction
Colon adenocarcinoma (COAD) ranked as the third most prevalent and fatal cancer in 2020, witnessing 104,610 new diagnoses and 53,200 deaths in the United States [1]. The survival rates for COAD are significantly influenced by the progression to metastasis, with a five-year survival rate of 90% for localised cases, sharply contrasting with a mere 14% for those with distant metastasis [1]. The liver is the most common metastatic site of COAD; liver metastasis occurs in 80% of patients, followed by pulmonary metastasis at 20% [2, 3] and bone metastasis at 3–5% [4]. A study revealed a dismal 5-year survival rate of 5% among 26.5% of patients who developed hepatic metastases [5]. Additionally, the one-year cancer-specific survival rate decreased from 93.1 to 60.2%, 90.2–55.5% and 93.1–60.2% in hepatic, pulmonary and osseous metastases, respectively [6]. The primary cancer site and the expression of oncogenes like RAS impact the prognosis [5, 7]. In cases where liver metastases are unresectable, the mortality rate remains alarmingly high despite palliative care [8]. Thus, exploring the mechanisms governing distant metastasis is imperative.
The linkage between COAD tumorigenesis and metastasis and the promotion of invasion through the epithelial-mesenchymal transition (EMT) of epithelial cells is well-documented [9]. In addition to EMT, several distinct features characterise various COAD subtypes, including alterations in the immune system, cellular communication, activation of the RAS pathway and metabolic pathways and differentially expressed gene sets serving as prognostic biomarkers [10]. However, the intricate mechanisms driving distant COAD metastasis remain elusive, underscoring the crucial need to identify these regulatory mechanisms and associated prognostic biomarkers to enhance survival and patient quality of life [7].
To address this knowledge gap, we conducted an in-depth analysis of RNA-seq data, employing integrated bioinformatics approaches to identify the significant oncogene SNTB1, along with its upstream and downstream genes and pathways influencing COAD metastasis and prognosis. Gene set enrichment analysis (GSEA) was used to identify a hallmark gene set, and univariate Cox regression identified SNTB1 as a pivotal gene within this gene set. Subsequent functional analysis and regulatory network construction elucidated the associated oncoprotein and signature gene set of SNTB1. Validation studies compared SNTB1 expression levels in para-carcinoma, primary COAD and metastasis tissue samples using RNA-seq data, complemented by immunohistochemistry (IHC) and western blot analysis of patient samples. The impact of SNTB1 overexpression on tumour cell behaviour and growth was explored using in vitro and in vivo models. Finally, single cell RNA-seq (scRNA-seq) and CellphoneDB, a database of cell–cell interactions, were employed to investigate the characteristics of metastatic COAD cells. Collectively, these findings highlight the metastatic mechanisms of COAD, establishing SNTB1 as a potential biomarker for early detection, risk stratification and as a candidate therapeutic target.
Methods
Data acquisition
RNA-seq data and clinical information from primary COAD patients with and without metastases were retrieved from The Cancer Genome Atlas (TCGA) database (https://tcgadata.nci.nih.gov/tcga/). The study included 65 metastatic samples and 343 non-metastasis samples. Additionally, 50 hallmark gene sets were obtained from the Molecular Signatures Database (MSigDB) v7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) [11].
GSEA
Gene expression levels were profiled using GSEA, identifying up and downregulated hallmark gene sets. The key hallmark gene set was selected based on the correlation coefficient. Genes within the key hallmark gene sets were screened using the edgeR package, and the volcano plot illustrated differentially expressed genes (DEGs).
Multivariate Cox regression model construction
The prognostic value of DEGs was accessed through univariate analysis, defining genes with P < 0.001 as prognostic DEGs. Key DEGs were identified based on the largest Hazard Ratio (HR). Prognostic DEGs were used to construct a multivariate Cox regression model, and the relative operating characteristic (ROC) curve evaluated the accuracy of the multivariate Cox regression model. The risk score was calculated using the formula.
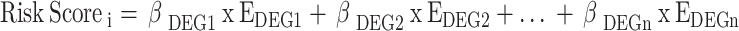 |
where i, β and n represent the sample number, the coefficient of each prognostic DEG and the total number of prognostic DEGs, respectively. Patients were stratified into low- and high-risk groups based on the risk score. Kaplan-Meier survival analysis was utilised to compare the difference between low- and high-risk scores and access the prognostic value of the risk score. The risk curve and scatterplot were used for visualisation.
Validations for the key DEGs
Several databases were used to validate the key DEGs at the multidimensional level. Gene Expression Profiling Interactive Analysis (GEPIA) [12] compared expression levels in normal adjacent tissue and COAD, accessed survival rate and displayed expression levels of the important DEGs at various stages. Furthermore, the UALCAN [13] investigated the expression levels of the main DEGs in various types (normal adjacent tissue vs. COAD; nodal metastasis). Linkedomics [14] demonstrated protein expression levels in COAD using reverse-phase protein arrays (RPPA) and assessed the connection between the main DEGs and the most positively associated protein. Furthermore, gene set variation analysis (GSVA) [15] was used to determine the expression levels of signature gene sets in COAD. Additionally, a protein-protein interaction (PPI) network in COAD was constructed using the STRING database [16].
Patient collection
This study was approved by the Institutional Review Board for Clinical Research of Shanghai Tenth People’s Hospital affiliated with Tongji University. All patients provided written informed consent. Samples were collected at Shanghai Tenth People’s Hospital affiliated with Tongji University from 2013 to 2018 (2020-KN84-01).
Histopathology and immunohistochemistry (IHC)
Haematoxylin and eosin (H&E) staining was used to reveal general histology. A total of 18 metastatic tumour tissues (six liver metastases, six lung metastases and six bone metastases) and paired primary and para-carcinoma samples from patients with COAD were fixed with formalin and embedded in paraffin. Thereafter, these samples were deparaffinised, dehydrated, rehydrated, antigen retrieved, blocked and sectioned into 4-µm slices following routine procedures. Sections were incubated overnight with an anti-syntrophin beta 1 (SNTB1) antibody (Cat#A104159, 1:15 dilution, NOVUSBIO) at 4 °C. Finally, all sections were labelled with polymer HRP for 40 min and counterstained with hematoxylin for 5 min at room temperature. Both cytoplasmic and membranous SNTB1 were stained using the same antibody. All sections were examined by two pathologists. Cancer cells with a stained cytoplasm were defined as positive. The criteria for intensity scoring of tumour cells were as follows: negative (0); yellow (1–4); light brown (5–8); and dark brown (9–12). For the negative control, a buffer was used instead of the primary antibody. To estimate the correlation between the intensity score of tumour cells and the clinical characteristics of patients, non-parametric tests and Spearman’s correlation analysis were employed.
Cell culture
Human colon cancer cells, DLD1 and sw620, were procured from ATCC. Additionally, HIEC-6 cells, derived originally from normal human intestinal epithelial cells, were obtained from the Cell Bank of the Chinese Academy of Sciences (China). sw620 cells were cultured in L15 (Gibco, USA) supplemented with 10% foetal bovine serum (FBS) (Gibco, USA) and 50 mg/ml penicillin/streptomycin (Invitrogen). HIEC-6 and DLD1 cells were cultured in RPMI1640 medium (Gibco, USA) with the same supplements (10% FBS and 50 mg/ml penicillin/streptomycin). Cells were subcultured every three to five days.
RNA extraction and quantitative real-time PCR
TRIzol Reagent (Invitrogen) was utilised to isolate total RNA following the manufacturer’s instructions. Briefly, the OD260/OD280 ratio ranged from 1.8 to 2.0. M-MLV reverse transcriptase (Takara, Japan), and 500 ng of RNA were added to a 10 µl reaction volume for reverse transcription. Quantitative real-time PCR was conducted using the ABI 7900 Detection System with the SYBR Premix ExTaq™ (Takara, Japan). Reaction process included initial denaturation step for 30 s at 95℃, followed by proliferation step that including 40 cycles of PCR at 95℃ for 5 s, 60℃ for 34 s, and dissociation stage was the final step that started from at 60℃, every elevated 0.5℃hold for 15 s to 95℃. Following PCR completion, the cycle threshold (CT) data and the mean CT were determined using the fixed threshold settings and triplicate PCRs. To compare each condition to the control reactions, the comparative CT method was performed. Furthermore, mRNA levels and the relative amount of the gene were normalised to β-actin and control, respectively. The relative amount of the gene was calculated using the 2−ΔΔCT method. The primers for SNTB1 and β-actin are listed in Supplementary Table S1.
Plasmid and siRNA construction and transfection
Human full-length SNTB1 and nonspecific negative control plasmid were obtained from HarO Biotech (Shanghai, China). Three siRNAs against SNTB1 were chemically synthesised by Shanghai GenePharma Co., and the sequences are provided in Supplementary Table S2. Following the manufacturer’s instructions, Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA) was used for cell transfection. Total RNA or protein was isolated 48 h after transfection.
Lentiviral infection
The shRNA-targeted sequences against SNTB1 and full-length SNTB1 and nonspecific negative control plasmid were obtained from HarO Biotech (Shanghai, China). Lentivirus was produced by co-transfecting 293T cells with shRNA in the vector PSPAX2 and pMD2G plasmids at 48–72 h post-transfection, which were then passed through a 0.45 μm filter and diluted with fresh medium containing 8 mg/ml polybrene or flow separation of GFP in a 2:3 ratio. The lentivirus was then used to infect target cells at 80% confluence. Stable cells with sw620-sh1#, sw620-sh2# and DLD1-overexpression were selected in 5 ug/ml puromycin in a culture medium. Protein expression was assessed through immunoblotting and real-time PCR.
Protein extraction and western blotting
Cells lysis buffer (50mM Tris-HCl, 300mM NaCl, 1% Triton X-100, supplemented with protease inhibitor and phosphatase inhibitor cocktails) was utilised to extract total protein, and the protein was isolated through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). PBST-5% fat-free dried milk or BSA was used for blocking the pure protein extracts at room temperature for 1 h, followed by overnight incubation with anti-SNTB1 (Cat#ab1351, 1:2500, Abcam), anti-Notch1 (Cat#3608, 1:1000, CST), anti-VEGFR2 (Cat#26415-1-AP, 1:1000, Proteintech), anti-phospho-YAP (Cat#13008, 1:1000, CST), anti-YAP (Cat#14074, 1:1000, CST), anti-phospho-LATS1 (Cat#28998-1-AP, 1:1000, proteintech), anti-LATS1 (Cat#17049-1-AP, 1:1000, proteintech), anti-Ecadherin (Cat#3195, 1:1000, CST), anti-Ncadherin (Cat#13116, 1:1000, CST), anti-caspase7 (Cat#9491, 1:1000, CST), anti-RAD50 (Cat#29390-1-AP, 1:1000, Proteintech), anti-SLC1A5 (Cat#20350-1-AP, 1:1000, Proteintech), anti-Slug (Cat#9585, 1:1000, CST), anti-Snail (Cat#3879, 1:1000, CST), anti-Vimentin (Cat#5741, 1:1000, CST), anti-β-Actin (Cat#ab8227, 1:10000, Abcam) and anti-GAPDH (Cat#ab8245, 1:10000, Abcam) at 4℃. Post-incubation, the samples were incubated with the relevant chemical-conjugated secondary antibodies (1:3000, Proteintech) at room temperature for 1 h. Blots were visualised using the Amersham Imager 600 (GE, USA).
Cell counting Kit-8 assay (CCK-8)
Cell Counting Kit-8 (CCK-8; Beyotime) was performed to evaluate cell proliferation ability. DLD1 and sw620 stable transfection cell lines were plated in 96-well plates at 3000 cells/well density and incubated sequentially for 0d, 1d, 2d, 3d, 4d, 5d, 6d and 7d. A total of 10 µl CCK-8 solution was added to each well and incubated at 37℃ for 2 h. Eventually, OD readings were performed at 450 nm.
Transwell and invasion assay
Migration and invasion abilities of DLD1 and sw620 stable transfection cell lines were evaluated using transwell and invasion assays. For invasion, the Matrigel matrix (Corning Incorporated, USA) was added to a 24-well transwell chamber following the manufacturer’s instruction. Next, 200 µl cell suspension (1 × 104) after transfection for 36 h was added to the upper chamber containing Matrigel matrix while 600 µl complete medium was added to the bottom chamber. Then, cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min and stained with 0.1% crystal violet for 25 min. Finally, a 50X magnification microscope was utilised to count the number of cells in three random fields per well. Furthermore, the transwell assay was performed similarly to the invasion assay but lacked the Matrigel matrix.
Apoptosis
After 48 h of transfection, cells were stained with annexin V-APC/7-AAD (BD Pharmingen Franklin Lakes, USA) and underwent flow cytometric analysis. Apoptotic cells were defined as annexin V-APC+/7-ADD+, and the percentage of apoptotic cells was calculated using flow cytometry.
Co-immunoprecipitation (Co-IP)
DLD1 and sw620 cells were lysed with RIPA buffer (Beyotime, #P0013). The supernatant was collected after centrifugation at 14,000 × g for 15 min. Then, the supernatant was incubated overnight with anti-human IgG (Bioss, #bs-0297P, 1:150) or anti-SRC antibody (Proteintech, #11097-1-AP, 1:50) with gentle rotation at 4℃, followed by incubation with 20 µl of Protein A/G agarose beads (Beyotime Biotechnology, #P1012) for 2 h with gentle rotation at 4℃. Samples were then centrifuged at 3000 rpm for 3 min, and the precipitate was washed with PBS and cell lysis buffer. Following this, the agarose beads were resuspended in an SDS-PAGE loading buffer and boiled for 5 min, followed by western blotting.
Xenograft experiment
BALB/c nude mice (male, 5 weeks old) were obtained from Shanghai SLAC Laboratory Animal (Shanghai, China) and housed in a standard animal laboratory with free access to food and water. Following this, 2 × 105 DLD1 stable transfection and blank control cells were injected separately into nude mice. Similarly, 2 × 106 sw620 stable transfection cells and blank control cells were injected separately into nude mice, which were housed in pathogen-free conditions. Ten days after injection, tumour size was measured every two days, and statistical tests were applied to construct graphs. Finally, tumours were removed and weighed separately. Another group of nude mice received spleen injections with 2 × 105 DLD1 stable transfection cells to validate metastasis, confirmed by optical in vivo imaging. Animal handling and experimental procedures were approved by the Institutional Animal Care and Use Committee at Tongji University (SHDSYY-2020-3231).
Construction of the metastasis model
Approval from the Institutional Animal Care and Use Committee at Tongji University was obtained for the metastasis model. In total, six male nude mice (17 ± 10 g) were obtained from Shanghai SLAC Laboratory Animal (Shanghai, China) and housed in standard laboratory conditions with sufficient water and food. The nude mice were weighed before caudal vein injection and randomly divided into two groups. Additionally, nude mice were injected with 1 × 106 DLD1-SNTB1 stable transfection cells and 1 × 106 DLD1-negative control (Nc) cells through the caudal vein in the SNTB1 and NC groups, respectively. These nude mice were weighed every five days for 15 days post-injection. Moreover, nude mice underwent magnetic resonance imaging (MRI) examination in case of significant weight loss. Finally, mice were dissected, and pathological sections were prepared.
RNA-seq and over-representation analysis (ORA) analysis
RNA-seq was conducted on three DLD1-SNTB1 transient cell lines and three DLD1-Nc cell lines. Integration of C1-H in MsigDB (https://www.gsea-msigdb.org/gsea/msigdb) [17] and the RNA-seq results were performed for ORA analysis.
Single cell RNA sequence (scRNA-seq)
Single cell sequence profiling of nine COAD samples was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144735) [18]. The scRNA-seq data from different samples were integrated using the Seurat method [19].
Following the preliminary data processing, the quality control criteria were established as follows: for cells, cell transcripts must exceed 100,000 or more than 1,500 genes should be expressed in the cell; for genes, the gene count must be greater than one, and the gene must be expressed in at least three single cells.
Variable genes were analysed using the ‘vst’ technique, which was then incorporated into principal component analysis (PCA) as the initial features [19]. Furthermore, when p < 0.05, the jackstraw analysis was used to identify the principal components (PCs). These PCs were then fed into Uniform Manifold Approximation and Projection (UMAP) and t-distributed Stochastic Neighbour Embedding (t-SNE) to generate cell sub-clusters (resolution = 0.50) [20]. The absolute value of log2 fold change was set to be greater than 0.5, and the false discovery rate was set at less than 0.05. The characteristics of DEGs, such as expression level and distribution, were visualised using violin and feature plots. Databases such as scMatch [21], singleR [22] and CellMarker [23] were used to annotate sub-clusters. Then, Monocle2 was employed to analyse the Cell trajectory and pseudo-time [24]. Additionally, hallmark gene sets were input as signalling pathways, and GSVA was utilised to quantify the hallmark gene sets in each sub-cluster. Finally, cellular communication was evaluated using cellphoneDB [25].
Multidimensional database validation
Various databases were utilised to validate the hypothesis. Moreover, for the top five genes in the key pathway, GeneCard (https://www.genecards.org/) was utilised. GEPIA, Oncomine [26], PROGgeneV2 [27], UALCAN, Linkedomics, cBioportal [28], Genotype-Tissue Expression (GTEx) [29], UCSC Xena [30], Cancer Cell Line Encyclopaedia (CCLE) [31], Expression atlas [32], The Human Protein Atlas [33] and STRING were utilised to validate the expression level and prognostic value of key genes in the hypothesis across gene, protein, cell and tissue levels.
Statistical analysis
Differences in quantitative data between the two groups were analysed using unpaired or paired two-tailed Student t-test or Mann−Whitney U test. Shapiro−Wilk tests were performed to assess the normal distribution of the data. Statistical analyses were conducted using GraphPad Prism 6 software, IBM SPSS Statistics 20.0 software and R 3.6.1 software (R Foundation for Statistical Computing, Vienna, Austria; www.r-project.org). A significance threshold of P-value < 0.05 on both sides was deemed statistically significant. Survival and clinical correlation analyses were processed using FPKM (log2 values were taken). Detailed analysis processes, source code and input data for bioinformatics analysis are available in the supplementary material.
Results
Identification of differentially expressed genes (DEGs) in metastatic COAD
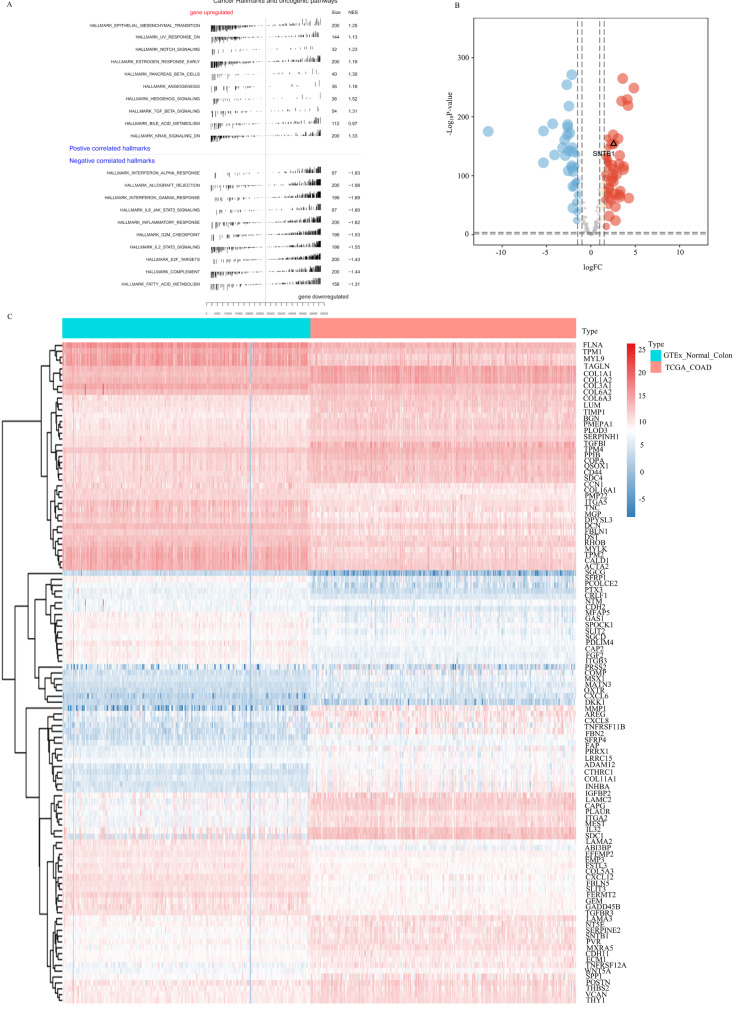
Utilising GSEA on DEGs, the top 10 significantly upregulated and downregulated pathways in metastatic COAD samples from the TCGA database were identified (Fig. 1A). The EMT pathway emerged as the most significantly enriched, leading to the identification of 200 genes in the hallmark EMT gene set, which were input into edgeR. These genes, referred to as differentially expressed EMT genes (DEEMTGs) between COAD and para-carcinoma tissues, were filtered using a volcano plot. SNTB1, specifically highly expressed, was identified as a ley DEGs (Fig. 1B). A heatmap (Fig. 1C) illustrated the differential expression of all DEGs in COAD and para-carcinoma tissues. Collectively, preliminary analyses indicated that SNTB1 exhibited differential expression in para-carcinoma tissues, COAD tissues, and COAD metastasis.
Fig. 1.
The Hallmark of EMT was the most significant differential pathway between normal colon tissue and COAD. Gene set enrichment analysis of differentially expressed genes in COAD cancer and para-carcinoma tissues (A). Volcano plot of the differentially expressed genes between COAD and para-carcinoma tissues (B). Heatmap of the differential expression characteristics of all differentially expressed genes in cancer and para-carcinoma tissues (C)
Regression analysis and establishment of a prognostic risk model
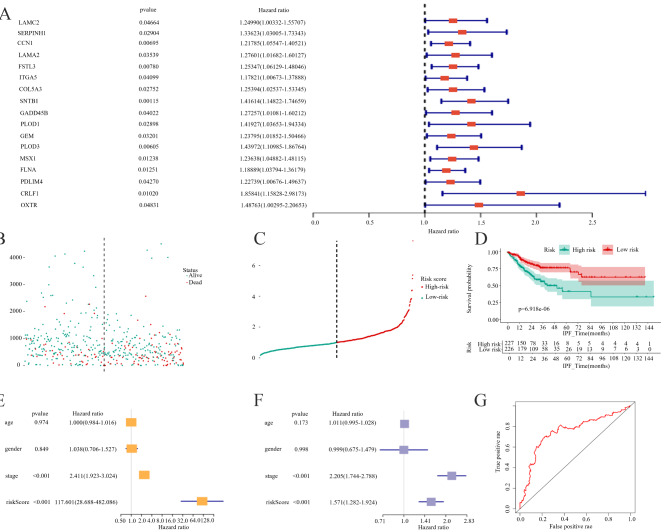
A forest plot illustrated the results of univariate Cox regression analysis of DEEMTGs, with 17 statistically significant genes identified for subsequent analysis. Subsequent univariate Cox regression assessed the prognostic value of the DEEMTGs, identifying SNTB1 as the gene most significantly associated with survival (HR = 1.416, 95% confidence interval (CI) 1.1482 − 1.747 P < 0.001; Fig. 2A). Figure 2B presented a scatter plot of multivariate Cox regression for alive and dead status patients. Moreover, a multivariate Cox regression analysis of risk curves for high- and low-risk patients was presented in Fig. 2C. The Kaplan-Meier survival curve demonstrated a significant difference in survival between the high- and low-risk groups based on DEEMTGs (P < 0.001) (Fig. 2D). In both univariate and multivariate analyses, including baseline demographic and clinical characteristics such as age, gender, and cancer stage, risk score emerged as an independent predictive factor for patient prognosis (univariate: HR = 171.361, 95% CI 28.772 − 1020.585, P < 0.001; multivariate: HR = 1.285, 95% CI 1.155 − 1.428, P < 0.001; Fig. 2E and F). The ROC curve had an area under the curve of 0.747, indicating the model’s good predictive accuracy (Fig. 2G).
Fig. 2.
SNTB1 was the gene with the highest prognostic value in EMT-related genes. The forest plot of univariate Cox regression analysis of differentially expressed EMT genes. SNTB1 exhibited the highest correlation with patient survival outcomes (A). The scatter plot of multivariate Cox regression analysis of the high-risk group and low-risk group(B). The risk curve visualised the result of multivariate Cox regression analysis of the high- and low-risk groups(C). The K-M survival curve of patients in the high-risk and low-risk groups was analysed using multivariate Cox regression analysis(D). Univariate Cox regression analysis validated the risk score calculated based on the predictive model, serving as a potential independent prognostic factor(E). Multivariate Cox regression analysis confirmed that the risk score from the predictive model was an independent prognostic determinant(F). The ROC curve for the predicting model established on the high-risk and low-risk group model(G)
SNTB1 is significantly associated with the metastasis of COAD at multi-omics levels
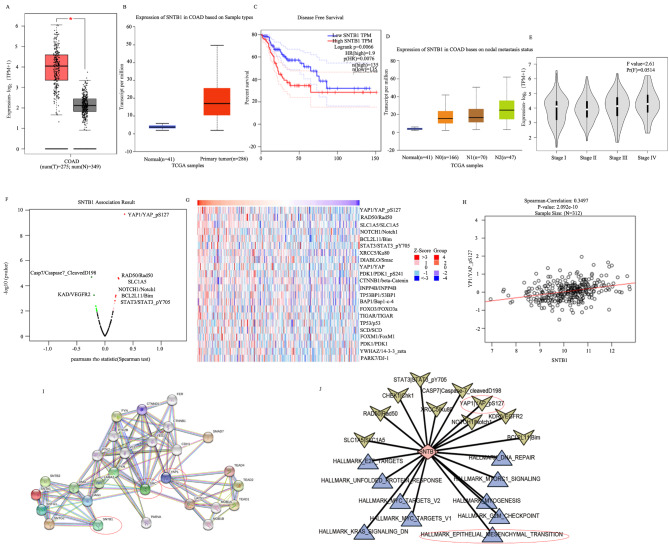
Validation of SNTB1 as a key DEEMTG associated with COAD patient survival was conducted across various databases. Expression analysis in the GEPIA and UALCAN databases revealed significantly higher SNTB1 expression in primary COAD compared to para-carcinoma tissues (Fig. 3A, B). Kaplan-Meier analysis demonstrated a significant association between high SNTB1 expression and poor prognosis in patients with COAD (P = 0.007, Fig. 3C). Additionally, SNTB1 expression positively correlated with higher N stage (indicating nodal involvement) and overall stage in UALCAN (Fig. 3D) and GEPIA databases (Fig. 3E), respectively. RPPA in the Linkedomics database identified differentially expressed proteins related (YAP1, Caspase7, VEGFR2, RAD50, SLC1A5 and STAT3) to SNTB1 (Fig. 3F). Notably, YAP1 exhibited the most significant positive correlation with SNTB1 protein (Fig. 3G), which was further validated by Spearmen correlation analysis (P < 0.001) (Fig. 3H). Additionally, the PPI network of COAD analysed in the STRING database showed SNTB1’s regulation of YAP1 via SRC and other molecules (Fig. 3I). Finally, GSVA of hallmark gene sets in COAD revealed the strongest correlations between SNTB1 and the EMT hallmark gene set and YAP1 protein (Fig. 3J).
Fig. 3.
SNTB1 showed significant results in multi-omics. The boxplot generated based on the GEPIA database(A) and UALCAN database(B) demonstrated the differential expression of SNTB1 between cancer and para-carcinoma tissues. The K-M survival curve based on the GEPIA database demonstrated the effect of SNTB1 on the survival of patients with COAD(C). The boxplot generated using the UALCAN database demonstrated the differential expression of SNTB1 in normal tissues and COAD tissues at different nodal stages(D). The violin plot generated using the GEPIA database showed the differential expression of SNTB1 at different stages of COAD tumours(E). The volcano map displayed the correlation between SNTB1 and other proteins or molecular markers(F). The heatmap displayed the expression patterns of the SNTB1 gene and its related genes or proteins(G). Scatter plots and fitted linear relationships suggested a significant correlation between SNTB1 and YAP1(H). The visualised network based on the STRING database displays the protein-protein interaction relationships related to SNTB1(I). The interaction network demonstrated that the EMT hallmark gene set and YAP1 protein are closely related to SNTB1(J) (The arrows represent the proteins, and the triangles represent hallmark gene sets)
Collectively, these findings suggested that SNTB1 interacted with the oncoprotein SRC, which in turn interacted with YAP1 to regulate the downstream Hippo-YAP pathway and resultantly regulated EMT to ultimately promote COAD distant metastasis.
Abnormal expression of SNTB1 is detected in metastatic COAD
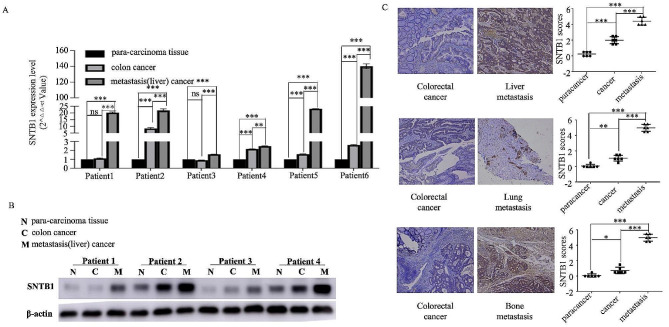
Real-time PCR in six paired clinical samples demonstrated an increasing trend in SNTB1 expression from para-carcinoma tissue to primary COAD tissues and liver metastasis (Fig. 4A). Western blotting confirmed upregulated SNTB1 protein expression in colon cancer samples compared to para-carcinoma samples, with metastatic cancer samples showing the highest expression (Fig. 4B). Additionally, IHC further validated higher SNTB1 protein expression in colon cancer tissues compared to para-carcinoma tissues, with distant metastasis cancer tissues (liver, lung and bone metastases) exhibiting the highest expression (Fig. 4C). TCGA data analysis confirmed significant associations between SNTB1 expression and poor prognosis indicators (tumour stage (P = 0.039), survival status (P = 0.031) and recurrence (P = 0.005)) (Table S3). Together, these findings highlighted the potential of upregulated SNTB1 expression as an indicator of COAD metastasis.
Fig. 4.
High expression of SNTB1 correlates with metastasis in COAD. The expression level of SNTB1 mRNA gradually increased in para-carcinoma tissue, colon cancer tissue and metastatic (liver) cancer (A). Western blot for the expression level of SNTB1 protein exhibited a gradual increase in para-carcinoma tissue, colon cancer and metastatic (liver) cancer (B). Immunohistochemistry demonstrated that the expression level of SNTB1 protein gradually increased in para-carcinoma tissue, colon cancer tissue and metastatic (liver, lung and bone) cancer (C)
Overexpression of SNTB1 promotes the proliferation, migration and invasion ability of COAD cells
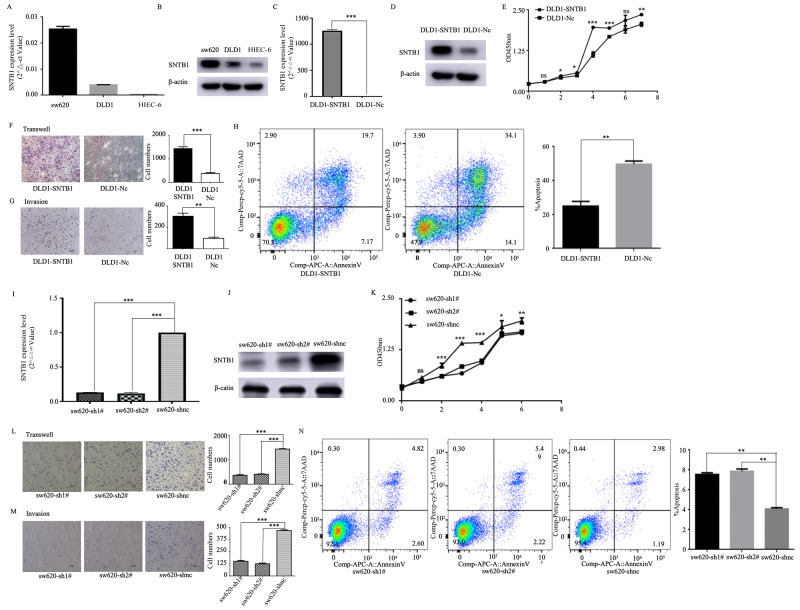
Real-time PCR and western blotting confirmed minimal to no expression in the normal epithelial HIEC-6 cell line, low expression in the DLD1 cell line and high expression in the sw620 cell line (Fig. 5A-B). Therefore, DLD1 and sw620 cells were used for establishing the SNTB1 overexpressed and knockdown stably transfected cell lines, respectively, which were validated by real-time PCR and western blotting (Fig. 5C-D and I-J). DLD1 cells overexpressing SNTB1 exhibited significantly enhanced proliferation, migration and invasion abilities, as shown by CCK-8, transwell and invasion assays, respectively (Fig. 5E-G). Flow cytometry further indicated a reduction in apoptosis in DLD1 cells with upregulated SNTB1 (Fig. 5H). Conversely, sw620 cells with SNTB1 knockdown demonstrated suppressed proliferation, migration and invasion (Fig. 5K-M), along with increased apoptosis (Fig. 5N).
Fig. 5.
Overexpressed SNTB1 promotes the proliferation, migration and invasion abilities of COAD cell lines. Real-time PCR (A) and Western blot (B) reveal the expression level of STNB1 in sw620, DLD1 and HIEC-6. SNTB1 overexpression was achieved in DLD1 cells by transfecting full-length STNB1 plasmid. STNB1 expression in DLD1 was determined using real-time PCR and confirmed using western blot (C, D). The statistical graph shows the proliferation of DLD1 cells with the overexpression of SNTB1 by CCK-8 (E). Transwell and invasion experiments for DLD1 cells with SNTB1 overexpression (F/G). The apoptosis of DLD1 cells with SNTB1 overexpression was detected using flow cytometry(H). SNTB1 knockdown was achieved in sw620 cells by transfecting short hairpin RNA targeting SNTB1. The expression of STNB1 in sw620 was determined using real-time PCR and confirmed using western blot (I/J). The proliferation of sw620 cells with SNTB1 knockdown, which was attained using CCK-8 (K). Transwell and invasion experiments using sw620 cells with SNTB1 knockdown (L/M). The apoptosis of sw620 cells with SNTB1 knockdown was detected using flow cytometry (N)
SNTB1 promotes tumour development and metastasis in vivo
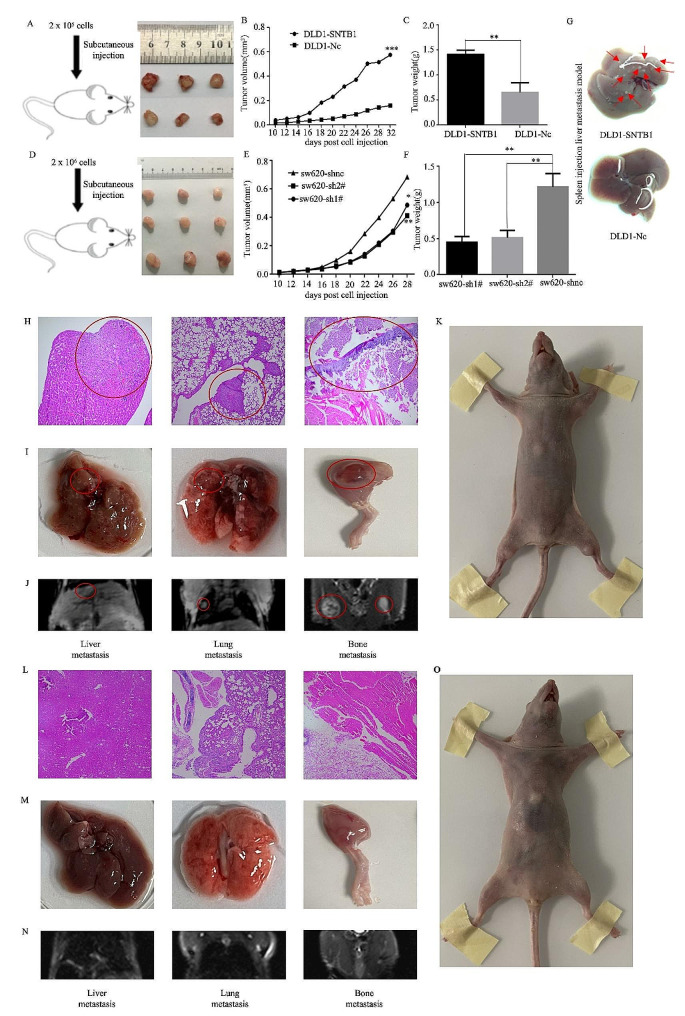
In vivo experiments involving male nude mice injected with DLD1 cells overexpressing SNTB1 demonstrated an increase in tumour size and weight compared to mice injected with negative control-transfected cells (Fig. 6A–C). Conversely, injection of sw620 SNTB1-knockdown cells resulted in decreased tumour size and weight (Fig. 6D–F). Moreover, hepatic metastasis was evident after injecting SNTB1-overexpressing DLD1 cells into the spleen (Fig. 6G). Overall, these results indicated that SNTB1 promoted tumour growth and metastasis in vivo. Liver, lung, and bone metastases were confirmed by H&E staining (Fig. 6H), gross specimen evaluation (Fig. 6I) and MRI scans (Fig. 6J) in the same mouse (Fig. 6K) of the SNTB1-overexpression group through caudal vein injection. In contrast, mice injected with NC-transfected cell lines did not exhibit tumorigenesis or metastasis (Fig. 6L–O). These results established that SNTB1 promotes tumour metastasis in vivo.
Fig. 6.
SNTB1 promoted the metastasis of COAD in vivo. Subcutaneous tumorigenesis experiment of DLD1 and control cell lines (A). Statistical graph of tumour volume and tumour weight in vivo (B-C). Subcutaneous tumorigenesis experiment of sw620 and control cell lines (D). Statistical graph of tumour volume and tumour weight in vivo (E-F). The result of liver metastasis in vivo imaging between control and over-expressing STNB1 groups (G). The H&E of metastasis (H), gross specimens of distant metastasis (I) MRI scans of distant metastasis (J) and nude mice (K) in the SNTB1 overexpressing group. The H&E (L), gross specimens (M), MRI images (N) and nude mice (O) of a mouse in the NC group
Hippo-YAP and EMT are downstream pathways of SNTB1
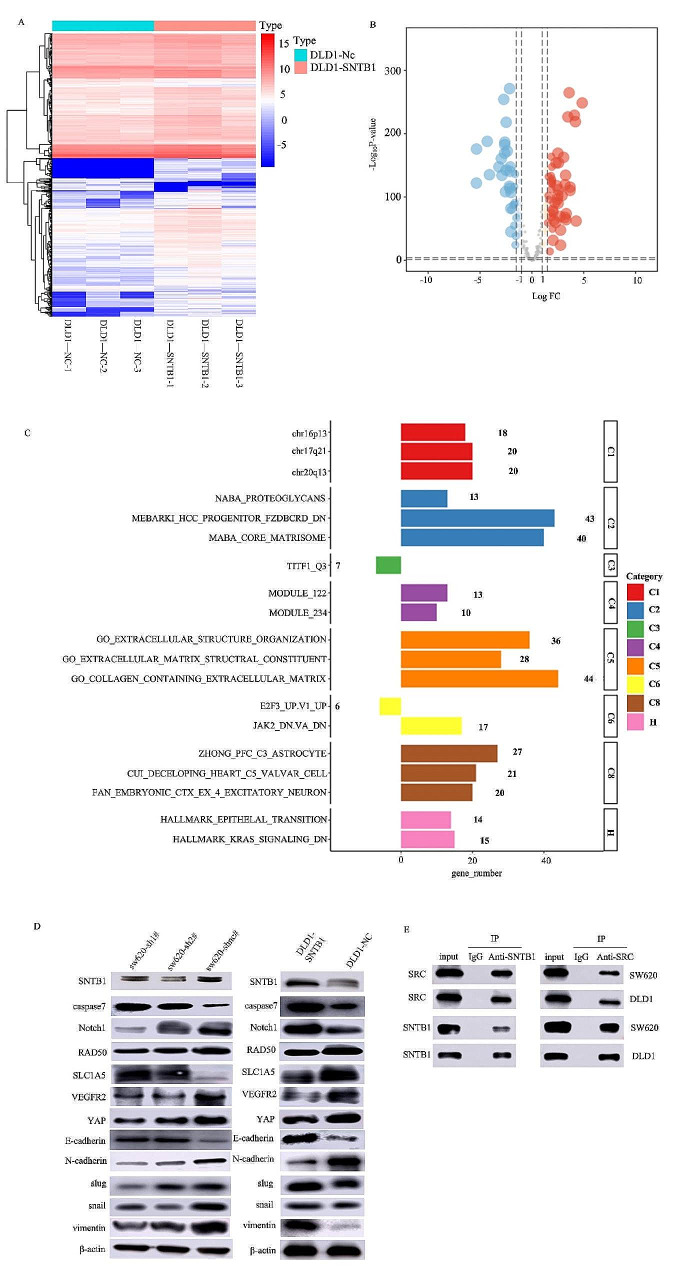
To explore downstream genes and pathways influenced by SNTB1, RNA-seq analysis of DLD1 cells transiently transfected with SNTB1 overexpression plasmids was conducted. The heatmap (Fig. 7A) and volcano plot (Fig. 7B) revealed YAP1/YAP as the most significantly correlated pathway with SNTB1. ORA identified upregulated EMT-related processes, indicating the involvement of SNTB1 in extracellular structure organisation, extracellular matrix structural constituent, collagen-containing extracellular matrix and KRAS signalling (Fig. 7C). Western blotting further validated the correlation of the Hippo-YAP pathway, EMT and apoptosis-related proteins with SNTB1 expression in cell lines with overexpression/knockdown of SNTB1 (Fig. 7D). Notably, in the Hippo-YAP signalling pathway, p-YAP1 positively correlated with SNTB1 expression, while p-LATS1 exhibited a negative correlation. The expression patterns of EMT markers (E-cadherin was negatively correlated with SNTB1 expression; N-cadherin, Slug, Snail and Vimentin were positively correlated with SNTB1 expression) were also verified. Caspase7, an apoptosis-related protein, followed the trend of SNTB1 expression. Co-immunoprecipitation of the sw620 and DLD1 cell lines confirmed the direct interaction between SNTB1 and SRC proteins (Fig. 7E).
Fig. 7.
Hippo-YAP and EMT are downstream pathways of SNTB1. The expression level of downstream genes and pathways of SNTB1 were assessed using RNA-seq in DLD1 cells transiently transfected with SNTB1 overexpression plasmid and control cells(A). The volcano plot of these genes and pathways (B). The ORA analysis (C). Western blot for Hippo-YAP, EMT, apoptosis pathways and SNTB1 in DLD1 and sw620 cell lines (D). STNB1 exhibited decreased expression in DLD1 cell lines but was highly expressed in sw620. DLD1-STNB1 was transferred into plasmids to overexpress SNTB1; sw620-sh1# and sw620-sh2# were transferred into short hairpin RNA to knock down SNTB1. Co-immunoprecipitation was used to validate the relationship between STNB1 and SRC (E)
Cellular communication between cancer cells and enterocytes promotes the EMT in COAD
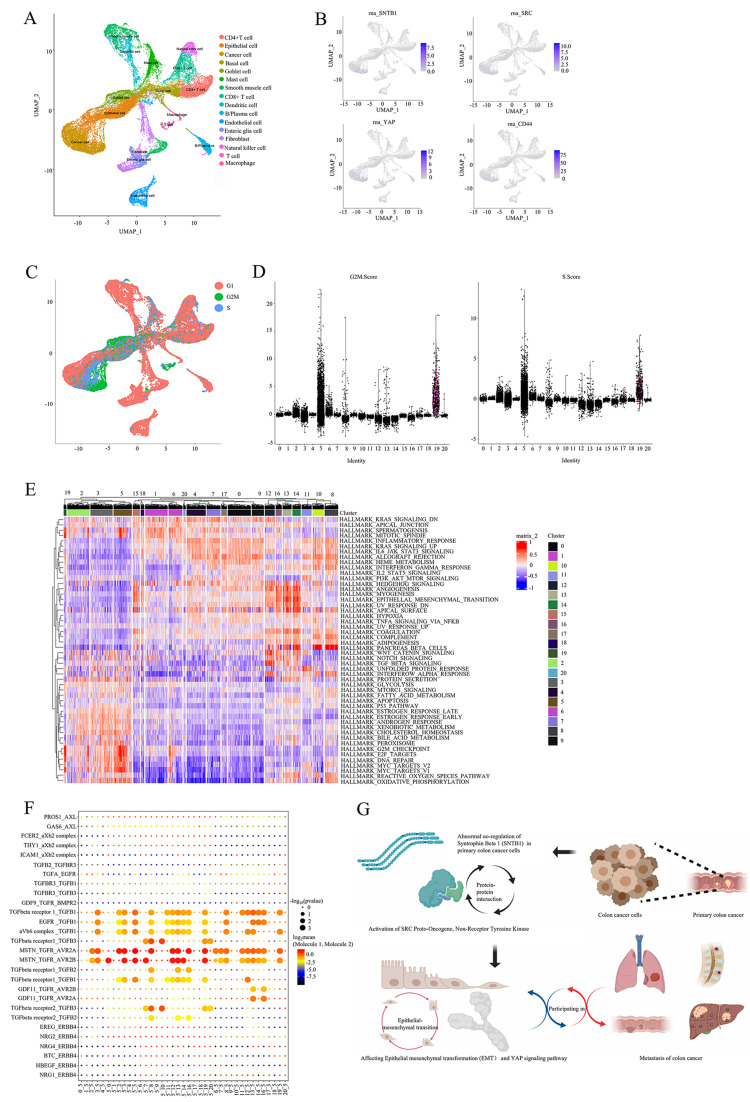
The initial result, sub-clusters and cell types from the UMAP plot of scRNA-seq were displayed in Fig. 8A. Clusters 3 and 5 were identified as cancer cells by the scMatch, singleR and CellMarker databases and characterised by the expression of SNTB1, SRC, YAP1 and CD44 (cancer stem cell marker) (Fig. 8B). Cluster 5, significantly associated with G2M and S stages of the cell cycle (Fig. 8C and D), indicated characteristics of cell division and stemness, typical of malignant cells. Additionally, GSVA revealed activation of hallmark gene sets related to the cell cycle and cell division, such as spermatogenesis, mitotic spindle, G2M checkpoint, E2F targets, DNA repair and MYC targets (Fig. 8E). Eventually, CellphoneDB analysis identified TGF-β-mediated receptor-ligand communication as a significant interaction between cancer cells with high SNTB1 expression and enterocytes (Fig. 8F). This interaction was proposed to promote EMT and accelerate metastasis in COAD. The mechanism diagram illustrates that SNTB1 interacts with SRC to activate the Hippo-YAP pathway, ultimately contributing to distant metastasis in COAD (Fig. 8G).
Fig. 8.
The cellular communication between cancer cells and enterocytes promoted EMT. The UMAP plots of single cell RNA sequence in COAD (A). The expression level of SNTB1, SRC, YAP1 and CD44 in single cell RNA sequence (B). The cell cycle information in single cell sequence (C). The cell cycle stage in 21 clusters of cells (D). The heatmap of hallmark gene sets (E). The cell communication information in the 21 cell clusters in the CellphoneDB database (F). The mechanism diagram of the hypothesis(G)
Multidimensional validation
The top five genes in the Hippo-YAP pathway were identified as YAP1, CTNNB1, AKT1, MAPK1 and TP53. The relationships, expression levels and prognostic values of these genes in different databases were analysed and summarised in Table S4, Table S5 and Table S6, respectively. Additionally, detailed results were graphically represented in Figures S1–S6.
Overall, SNTB1, SRC, YAP1, CTNNB1, AKT1, MAPK1 and TP53 exhibited elevated expression in COAD compared to para-carcinoma tissues. Among these, SNTB1, SRC, CTNNB1, AKT1, MAPK1 and TP53 were significantly associated with overall survival, while SNTB1 also showed a significant association with disease-free survival.
Discussion
The most common cause of treatment failure in COAD is distant metastasis, responsible for over one million new COAD cases and over 700,000 deaths annually [34, 35]. Metastases significantly reduce the five-year survival rates of patients, emphasising the need for reliable biomarkers that not only have prognostic values but also play a pivotal role across various biological processes at multi-omics levels. While several genetic and molecular markers have been identified for COAD metastasis, most previous studies have focused on single molecules and single omics [36–38]. Therefore, it is critical to investigate biomarkers with universal regulatory functions involved in extensive biological processes.
Our analysis of the mechanism of metastasis in COAD identified EMT as a significant hallmark gene set, with SNTB1 emerging as the most significant prognostic gene in this set. SNTB1 was confirmed to be highly expressed in the metastasis tissues and validated to form a complex with the SRC protein. Moreover, overexpressed SNTB1 promoted the proliferation, migration and invasion ability of COAD cell lines in vitro, and enhanced tumour size and metastasis in vivo. Additionally, scRNA-seq data from SNTB1-overexpressing COAD cells revealed associations with the G2M and S cell cycle stages, while CellphoneDB analysis highlighted the significance of TGF-β in COAD cellular communication. Collectively, our bioinformatic and experimental results indicate that SNTB1, through its interaction with SRC, activated the Hippo-YAP pathway, ultimately promoting distant metastasis in COAD.
Syntrophins comprise a group of peripheral membrane-associated adaptor proteins encompassing five isoforms, namely SNTA1, SNTB1, SNTB2, SNTG1 and SNTG2. These proteins serve as scaffolding components, ensuring the accurate localisation of signalling proteins along with their binding partners and thereby facilitating optimal spatiotemporal control over signalling pathways [39]. SNTB1 is a member of the syntrophin family forming multi-domain scaffolds that link membrane proteins [40]. SNTB1 is also a component of Type I PDZ domain proteins, which bind to the α1D-AR (ADRA1D) ligand and have been demonstrated to play a role in cardiovascular and urological diseases [41]. Notably, Motalebzadeh et al. (2020) identified SNTB1 upregulation as a prognostic indicator for colorectal cancer [42], consistent with the findings by Liu et al., which not only confirmed SNTB1 overexpression but also linked it to reduced survival rates in patients with COAD. Their proposed mechanism involves SNTB1 influencing cancer progression by suppressing PKN2 expression and thereby activating the ERK and AKT signalling pathways [43]. Moreover, SNTB1 interacts directly with ATP-binding cassette transporter A1 (ABCA1) through the PDZ domain, whereas ABCA1 overexpression promotes the proliferation, invasion, and EMT of COAD cells [44, 45]. Moreover, the interaction between the transcriptional co-activator with the PDZ-binding motif (TAZ) and YAP promotes anchor independent growth, EMT and the downstream Hippo signalling pathway [46].
SRC, a non-receptor tyrosine-protein kinase encoded by the oncogene SRC, is instrumental in malignant progression, promoting the migration and invasion of early-stage COAD [47, 48]. SRC also catalyses the tyrosine phosphorylation of downstream signal transduction molecules and promotes cell proliferation and glycolytic enzyme phosphorylation, resultantly contributing to tumour development and metabolism [49]. Moreover, SRC is involved in remoulding the tumour microenvironment through the EMT, matrix metalloproteinases secretion, vascular leakage, breaking the endothelial barrier, myofibroblast differentiation and ultimately tumour cell micro-invasion [50]. Notably, the SRC inhibitor PP2 was reported to enhance cell adhesion and reduce metastasis [51]. Furthermore, SRC regulates the Hippo-YAP pathway by inhibiting Hippo and YAP1 phosphorylation/activation [52].
However, this study is the first to demonstrate an interaction between SNTB1 and SRC. The C-terminus of SRC is a ligand for PDZ domains in ligand protein X1 (LNX1) [53], and the Gly-Glu-Asn-Leu amino-acid sequence of the SRC C-terminus was demonstrated to interact with type III PDZ AF6 to promote cell migration and invasion [54]. Given SNTB1’s status as a Type I PDZ cell-membrane protein, we speculate that the STNB1-SRC complex activates YAP1 and inhibits the Hippo signal, contributing to COAD metastasis.
The Hippo signalling pathway plays a crucial role in organ regulation, with dysregulation linked to tumorigenesis under pathological conditions. Its effectors, Yes-associated protein (YAP) and transcriptional coactivators TAZ, are typically phosphorylated and subsequently inhibited by the Hippo pathway kinase cascade in response to specific stimuli. This leads to the suppression of cell proliferation and the promotion of cell death [55]. Pathologically, YAP can function as an oncogene, and its overexpression has been consistently identified in various cancers, including lung adenocarcinoma, ovarian cancer and colonic adenocarcinoma [56]. The Hippo-YAP pathway regulates essential cellular processes, such as the cell cycle, proliferation, apoptosis, cell renewal, cell polarity and cellular communication in several biological processes [57]. Activation of the Hippo-YAP pathway involves the MST1/2, SAV1 and LATS1/2 complex suppressing YAP [58]. Conversely, inactivation of the Hippo pathway leads to the upregulation of YAP, resulting in abnormal cell proliferation, inhibition of apoptosis and tumorigenesis [58]. Notably, overexpression of YAP is common in gastrointestinal tumours [46], and activated YAP1 is a critical factor in the EMT, cancer metastasis and invasion in COAD and other cancer types [59, 60].
In summary, this study proposes a novel regulatory axis involving SNTB1/SRC/Hippo-YAP that promotes EMT and metastasis in COAD. This finding provides a crucial insight into the mechanism underlying COAD metastasis, offering a potential target for precise treatment. However, further validation of direct interactions between SRC and YAP, as well as SNTB1 and SRC, is required. Additionally, identifying the molecular states in the downstream pathway requires chromatin immunoprecipitation-sequencing and protein phosphorylation detection experiments. Furthermore, the scRNA-seq analysis suggested that SNTB1-overexpressing COAD cells exhibit active division, implying that SNTB1 might serve as a stemness marker influencing COAD development. Subsequent experiments are planned to explore whether an SNTB1 inhibitor could target and eliminate these stem cells, providing valuable insights into COAD suppression.
To provide a comprehensive and balanced perspective on our research, it is important to acknowledge both limitations and strengths. The bioinformatic analysis, while extensive, could benefit from further refinement. Additionally, certain analyses may be limited by the number of samples, even though large public databases such as TCGA were utilised. While extensive analyses were performed on the gene expression data, other omics data such as proteomics and metabolomics were not included. Thus, integrating multiple omics data could provide a more comprehensive understanding of the molecular mechanisms underlying COAD metastasis. Finally, further mechanistic studies are also needed to fully elucidate the pathways through which SNTB1 influences COAD metastasis, although our experimental validation partially addresses this limitation. Despite these limitations, our study’s strengths include the in vitro and in vivo experiments performed to functionally validate the role of SNTB1 in promoting COAD cell migration, invasion and tumorigenesis. These experiments offer mechanistic insights into the metastatic process, supporting the clinical relevance of our findings. The integration of multi-omics data from different public databases allows for a comprehensive exploration of potential molecular interactions and pathways contributing to COAD metastasis. Furthermore, the prognostic value and expression patterns of SNTB1 in COAD were validated across various independent cohorts, enhancing the reliability and clinical relevance of SNTB1 as a potential biomarker for COAD metastasis. The use of advanced bioinformatics tools, such as GSEA, edgeR, Cox regression analysis and GSVA, enables a comprehensive analysis to identify key genes and pathways associated with COAD metastasis. Additionally, the in silico analysis served as a valuable preliminary step to prioritise potential biomarkers and pathways for further investigation. Finally, the identification of SNTB1 as a potential prognostic biomarker and its association with the Hippo-YAP pathway and EMT provided valuable insights into the molecular mechanisms underlying COAD metastasis. These findings have implications for the development of targeted therapies and precision medicine approaches for patients with COAD.
Conclusions
This study unveils a critical role for SNTB1 in promoting the metastasis of COAD. SNTB1 interacts with the oncoprotein SRC, establishing a direct interaction with YAP1, thereby activating the downstream Hippo-YAP pathway and inducing the metastasis of COAD. Elevated SNTB1 expression was consistently observed in COAD metastases, emphasising its clinical relevance. Furthermore, in vitro experiments demonstrated that SNTB1 overexpression enhanced cancer cell migration, invasion and proliferation, while in vivo studies confirmed its pro-metastatic effects. Additionally, our results suggested that cancer cells exhibit high SNTB1 expression, along with traits such as increased division, stemness and malignancy, engage in cellular communication with enterocytes, induce EMT and thereby facilitate metastasis. To gain a more comprehensive understanding of the molecular landscape driving COAD metastasis, integrating data from multiple omics levels, such as genomics, transcriptomics, proteomics and epigenomics, is also essential. Further experiments should focus on investigating the effects of SNTB1 inhibition or targeted therapies modulating the Hippo-YAP pathway or other key metastasis-related pathways. This will provide invaluable preclinical data for targeted therapies, laying the groundwork for toxicology and pharmacology experiments, and eventually clinical trials. Exploring new indications for existing drugs may offer promising avenues for intervention. Investigating the correlation between SNTB1 expression levels and response to specific therapies may also offer avenues for personalised treatment strategies for patients with COAD.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the TCGA team of the National Cancer Institute for using their data. We thank all the patients with COAD who participated in this study. We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Abbreviations
- COAD
Colon adenocarcinoma
- TCGA
The Cancer Genome Atlas
- SRC
SRC Proto-Oncogene, Non-Receptor Tyrosine Kinase
- SNTB1
Syntrophin Beta 1
- IHC
Immunohistochemistry
- CCLE
Cancer Cell Line Encyclopedia
- GEPIA
Gene Expression Profilling Interactive Analysis
- GTEx
Genotype-Tissue Expression
- Co-IP
Co-immunoprecipitation
- IHC
Immunohistochemistry
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PBS
phosphate-buffered saline
- ADRA1D
α1D-AR
- DMD
Dystrophin
- TAZ
Transcriptional co-activator with PDZ-binding motif
- EMT
Epithelial-mesenchymal transformation
- ABCA1
ATP-binding cassette transporter A1
- LNX1
ligand protein X1
- GSEA
Gene Set Enrichment Analysis
- DEGs
Differentially expressed genes
- HR
Hazard Ratio
- ROC curve
Relative operating characteristic curve
- GSVA
Gene set variation analysis
- TSO
Template switch oligo
- PCA
Principal component analysis
- PC
Principal component
- FDR
False discovery rate
- PPI
Protein-protein interaction
- EMT
Epithelial mesenchymal transition
- MRI
Magnetic resonance imaging
Author contributions
Conception/design: Zhengyan Chang, Runzhi Huang, Jaiqi Song, Zhenyu Li, Man Pi, Shuyuan Xian, Jingcheng Zhang, Jinglei Huang, Ruting Xie, Guo Ji, Dongyan Han and Qiongyi Huang. Collection and/or assembly of data: Zhengyan Chang, Runzhi Huang, Jaiqi Song, Zhenyu Li, Man Pi, Shuyuan Xian, Jingcheng Zhang, Jinglei Huang, Ruting Xie, Guo Ji, Dongyan Han and Qiongyi Huang. Data analysis and interpretation: Zhengyan Chang, Runzhi Huang, Jaiqi Song, Zhenyu Li, Man Pi, Shuyuan Xian, Jingcheng Zhang, Jinglei Huang, Ruting Xie, Guo Ji, Dongyan Han and Qiongyi Huang. Manuscript writing: Zhengyan Chang, Runzhi Huang, Jaiqi Song, Zhenyu Li, Man Pi, Shuyuan Xian, Jingcheng Zhang, Jinglei Huang, Ruting Xie, Guo Ji, Dongyan Han and Qiongyi Huang. Final approval of manuscript: Zhengyan Chang, Runzhi Huang, Jaiqi Song, Zhenyu Li, Man Pi, Shuyuan Xian, Jingcheng Zhang, Jinglei Huang, Ruting Xie, Guo Ji, Dongyan Han and Qiongyi Huang.
Funding
This study was supported in part by the National Natural Science Foundation of China (No. 82002923); Shanghai Rising-Star Program (Sailing Special Program) (No. 23YF1458400). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The datasets generated and/or analysed during the current study are available in the TCGA-COAD program (https://portal.gdc.cancer.gov), the Genotype-Tissue Expression (GTEx) Project (www.gtexportal.org/), the Molecular Signatures Database (MSigDB) v7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp), the chromatin accessibility landscape of primary human cancers project (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG), and Gene Expression Omnibus (GEO, GSE144735) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144735). Additional data related to this paper may be requested from the authors. Email: hqiongyi2023@163.com.
Declarations
Ethics approval and consent to participate
Approval of the research protocol by an Institutional Reviewer Board: The study was approved by the Ethics Committee of Shanghai Tenth People’s Hospital.
Consent for publication
Not applicable.
Informed consent
N/A.
Registry and the registration no. of the study
N/A.
Animal studies
The study was approved by the Ethics Committee of Shanghai Tenth People’s Hospital.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhengyan Chang, Runzhi Huang and Jiaqi Song have contributed equally to this work.
Contributor Information
Guo Ji, Email: chickapple@126.com.
Dongyan Han, Email: han_dongyan@163.com.
Qiongyi Huang, Email: hqiongyi2023@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Misiakos EP, Karidis NP, Kouraklis G. Current treatment for colorectal liver metastases. World J Gastroenterol. 2011;17(36):4067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess KR, et al. Metastatic patterns in adenocarcinoma. Cancer. 2006;106(7):1624–33. [DOI] [PubMed] [Google Scholar]
- 4.Zhenghong, et al. Retrospective study of predictors of bone metastasis in colorectal cancer patients. J Bone Oncol. 2017;9:25–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engstrand J, et al. Colorectal cancer liver metastases - a population-based study on incidence, management and survival. BMC Cancer. 2018;18(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu M, et al. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget. 2015;6(36):38658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brudvik KW, et al. Meta-analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg. 2015;102(10):1175–83. [DOI] [PubMed] [Google Scholar]
- 8.House MG, et al. Survival after hepatic resection for metastatic colorectal cancer: trends in outcomes for 1,600 patients during two decades at a single institution. J Am Coll Surg. 2010;210(5):744–52. [DOI] [PubMed] [Google Scholar]
- 9.Elmasry M, et al. RBP7 is a clinically prognostic biomarker and linked to tumor invasion and EMT in colon cancer. J Cancer. 2019;10(20):4883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marisa L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberzon A, et al. The Molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekar DS, et al. UALCAN: a portal for facilitating Tumor Subgroup Gene expression and survival analyses. Neoplasia. 2017;19(8):649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasaikar SV, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snel B, et al. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HO, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet. 2020;52(6):594–603. [DOI] [PubMed] [Google Scholar]
- 19.Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung NC, Storey JD. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics. 2015;31(4):545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou R, Denisenko E, Forrest ARR. scMatch: a single-cell gene expression profile annotation tool using reference datasets. Bioinformatics. 2019;35(22):4688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu X, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efremova M, et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–506. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, et al. The cBio Cancer Genomics Portal: an Open platform for exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium G. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldman M, et al. The UCSC Cancer Genomics browser: update 2015. Nucleic Acids Res. 2015;43(Database issue):D812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghandi M et al. Next-generation Charact Cancer Cell Line Encyclopedia Nat, 2019.
- 32.Papatheodorou I, et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46(D1):D246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 34.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein U, et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15(1):59–67. [DOI] [PubMed] [Google Scholar]
- 36.Bokemeyer C, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22(7):1535–46. [DOI] [PubMed] [Google Scholar]
- 37.Toiyama Y, et al. Su1907 serum miR-200c and miR-203 are novel prognostic and metastasis-predictive biomarkers in patients with Colorectal Cancer (CRC). Gastroenterology. 2012;142(5, Supplement 1):pS–533. [Google Scholar]
- 38.Chang Z, et al. The Construction and Analysis of ceRNA Network and Patterns of Immune Infiltration in Colon adenocarcinoma metastasis. Front Cell Dev Biol. 2020;8:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhat SS, Ali R, Khanday FA. Syntrophins entangled in cytoskeletal meshwork: helping to hold it all together. Cell Prolif. 2019;52(2):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams ME, et al. Structural abnormalities at neuromuscular synapses lacking multiple syntrophin isoforms. J Neurosci. 2004;24(46):10302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camp ND et al. Individual protomers of a G protein-coupled receptor dimer integrate distinct functional modules. Cell Discov, 2015. 1. [DOI] [PMC free article] [PubMed]
- 42.Motalebzadeh J, Eskandari E. Syntrophin beta 1 (SNTB1): candidate as a new marker for colorectal cancer metastasis. Gene Rep. 2020;20:7. [Google Scholar]
- 43.Liu LY, et al. Upregulation of SNTB1 correlates with poor prognosis and promotes cell growth by negative regulating PKN2 in colorectal cancer. Cancer Cell Int. 2021;21(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuhira K, et al. Purification of ATP-binding cassette transporter A1 and associated binding proteins reveals the importance of beta1-syntrophin in cholesterol efflux. J Biol Chem. 2005;280(47):39653–64. [DOI] [PubMed] [Google Scholar]
- 45.Aguirre-Portoles C, et al. ABCA1 overexpression worsens colorectal cancer prognosis by facilitating tumour growth and caveolin-1-dependent invasiveness, and these effects can be ameliorated using the BET inhibitor apabetalone. Mol Oncol. 2018;12(10):1735–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu P, et al. RasGRF2 promotes migration and invasion of colorectal cancer cells by modulating expression of MMP9 through Src/Akt/NF-kappaB pathway. Cancer Biol Ther. 2019;20(4):435–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rocha MR, et al. Annexin A2 overexpression associates with colorectal cancer invasiveness and TGF-ss induced epithelial mesenchymal transition via Src/ANXA2/STAT3. Sci Rep. 2018;8(1):11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, et al. c-Src phosphorylation and activation of hexokinase promotes tumorigenesis and metastasis. Nat Commun. 2017;8:13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel A, et al. Novel roles of src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sci. 2016;157:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nam JS, et al. Src family kinase inhibitor PP2 restores the E-cadherin/catenin cell adhesion system in human cancer cells and reduces cancer metastasis. Clin Cancer Res. 2002;8(7):2430–6. [PubMed] [Google Scholar]
- 52.Li P, et al. Alphae-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes Dev. 2016;30(7):798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss A, et al. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase ligand-of-numb protein X1. FEBS Lett. 2007;581(26):5131–6. [DOI] [PubMed] [Google Scholar]
- 54.Baumgartner M, et al. c-Src-mediated epithelial cell migration and invasion regulated by PDZ binding site. Mol Cell Biol. 2008;28(2):642–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao B, et al. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24(9):862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinhardt AA, et al. Expression of yes-associated protein in common solid tumors. Hum Pathol. 2008;39(11):1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee M, et al. Hippo-Yap signaling in ocular development and disease. Dev Dyn. 2018;247(6):794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim HB, Myung SJ. Clinical implications of the Hippo-YAP pathway in multiple cancer contexts. BMB Rep. 2018;51(3):119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng X, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han Q, et al. WWC3 inhibits epithelial-mesenchymal transition of lung cancer by activating Hippo-YAP signaling. Onco Targets Ther. 2018;11:2581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the TCGA-COAD program (https://portal.gdc.cancer.gov), the Genotype-Tissue Expression (GTEx) Project (www.gtexportal.org/), the Molecular Signatures Database (MSigDB) v7.1 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp), the chromatin accessibility landscape of primary human cancers project (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG), and Gene Expression Omnibus (GEO, GSE144735) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144735). Additional data related to this paper may be requested from the authors. Email: hqiongyi2023@163.com.