Abstract
Background
Tumor Mutational Burden (TMB) have emerged as pivotal predictive biomarkers in determining prognosis and response to immunotherapy in colorectal cancer (CRC) patients. While Whole Exome Sequencing (WES) stands as the gold standard for TMB assessment, carry substantial costs and demand considerable time commitments. Additionally, the heterogeneity among high-TMB patients remains poorly characterized.
Methods
We employed eight advanced machine learning algorithms to develop gene-panel-based models for TMB estimation. To rigorously compare and validate these TMB estimation models, four external cohorts, involving 1,956 patients, were used. Furthermore, we computed the Pearson correlation coefficient between the estimated TMB and tumor neoantigen levels to elucidate their association. CD8+ tumor-infiltrating lymphocyte (TIL) density was assessed via immunohistochemistry.
Results
The TMB estimation model based on the Lasso algorithm, incorporating 20 genes, exhibiting satisfactory performance across multiple independent cohorts (R2 ≥ 0.859). This 20-gene TMB model proved to be an independent prognostic indicator for the progression-free survival (PFS) of CRC patients (p = 0.001). DNAH5 mutations were associated with a more favorable prognosis in high-TMB CRC patients, and correlated strongly with tumor neoantigen levels and CD8+ TIL density.
Conclusions
The 20-gene model offers a cost-efficient approach to precisely estimating TMB, providing prognosis in patients with CRC. Incorporating DNAH5 within this model further refines the categorization of patients with elevated TMB. Utilizing the 20-gene model facilitates the stratification of patients with CRC, enabling more precise treatment planning.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13062-024-00564-0.
Keywords: Machine learning, Tumor mutation burden, Tumor neoantigen burden, Prognostic biomarker, Colorectal cancer
Background
Colorectal cancer (CRC) holds a prominent position in global oncology, with its incidence and complexity demanding rigorous scientific attention. The disease’s heterogeneity, in both molecular and clinical setting, accentuates the urgency for identifying and leveraging precise biomarkers [1]. Such biomarkers can critically inform patient management, therapeutic stratification, and prognostication. Currently, Tumor Mutational Burden (TMB) have emerged as pivotal predictive biomarkers in determining prognosis and response to immunotherapy, especially with the emphasis on immunotherapeutic modalities like CRC [2–4]. Previous studies demonstrated that patients with CRC who have high TMB had a better prognosis than those with low TMB [5]. On the other hand, tumor neoantigen burden (TNB) specifically quantifies the number of neoantigens expressed by a tumor. Since not all mutations lead to the formation of neoantigens, TNB can be considered a subset of TMB. Compared to TMB, TNB provides a more focused assessment of a tumor’s immune potential [6].
Currently, the primary method for assessing TMB hinges on whole-exome sequencing (WES) utilizing next-generation sequencing (NGS) technology. Yet, the prohibitive costs and extended processing times associated with WES have limited its clinical application. Consequently, targeted NGS on expansive pan-cancer gene panels (typically consisting of hundreds of cancer-related genes), such as MSK-IMPACT, F1CDx, is beginning to attract attention for TMB estimation [7], on which the mutation burdens can be used to estimate the global TMB in tumor cells. Notably, a significant portion of these genes does not exhibit direct association with TMB [8]. Furthermore, due to the pronounced variability in mutational profiles across diverse cancer types, these expansive panels often fall short in their ability to be precisely tailored for a specific cancer type [9].
These existing researches catalyzed our exploration into an alternative, more agile yet equally rigorous, avenues for TMB assessment. In today’s era, marked by advancements in computing, machine learning methods have emerged as a vital tool for building biomedical predictive models. Its ability to learn through complex biological data, recognize subtle patterns, and provide accurate predictions makes it especially valuable in understanding and interpreting genomic data [10]. Recently, the biomedical field has witnessed a growing adoption of machine learning to enhance clinical decision-making and healthcare delivery [11, 12]. Embracing this paradigm, we’ve ventured into develop and test several cost-effective models using machine learning, precisely engineered for TMB estimation.
In this research, we initiated a comprehensive evaluation of various machine learning algorithms, testing their efficacy in several distinct cohorts’ datasets, focusing on estimation model specifically for TMB evaluations in patients with CRC. Impressively, the best model requires only a panel of 20 genes to deliver insightful assessments of TMB and the prognosis for patients with CRC. Notably, our 20-gene-panel-based model showcased a strong correlation between TMB and neoantigen levels. Furthermore, our model elucidated those mutations in the DNAH5 gene corresponded to a more favorable prognosis for those patients with CRC who have high TMB. These patients were observed to have a higher presence of neoantigens and an increased density of CD8+ tumor-infiltrating lymphocytes (TILs), suggesting a more robust immune response.
In summary, cancer researchers are constantly searching for accurate and efficient tools to develop appropriate therapeutics for colorectal cancer. Our 20-gene-panel-based model, which combines computational techniques informed by prior genetic knowledge, is a definitive outcome of these efforts, as it has been tested and shown to be effective.
Methods
Dataset source
Dataset TCGA
The colorectal cancer cohort from The Cancer Genome Atlas (TCGA) was sourced from https://portal.gdc.cancer.gov/. This TCGA dataset comprised Whole Exome Sequencing (WES) data from 586 samples, transcriptomic (RNA-Seq) data from 521 samples, and clinical information of 552 patients.
Dataset ICGC
We integrated genomic and clinical data for 322 CRC samples from the International Cancer Genome Consortium (ICGC) available at https://dcc.icgc.org/.
Dataset Jessica et al. and DFCI
From the cBioPortal database (https://www.cbioportal.org/), we obtained the Jessica cohort, encompassing genomic and clinical data for 281 patients, and the DFCI cohort, published in 2016, which includes details on 619 patients with CRC.
Dataset FAHNU
The FAHNU cohort involved 148 primary patients with CRC who underwent surgical treatment (from 2022 to 2023). For these patients, both tumor tissues and peripheral blood mononuclear cell (PBMC) were subjected to Whole Exome Sequencing (WES). Furthermore, 148 tumor tissues and 148 paired adjacent normal tissues were also processed for RNA-Seq. All samples were stored at -80 °C.
All these steps were conducted under the approval and guidance of the ethics committee of the First Affiliated Hospital of Nanchang University, Nanchang, China. The approval number is 2022-CDYFYYLK-06-012. All procedures were conducted in strict accordance with the Declaration of Helsinki or equivalent ethical principles. Also, the informed consent in written form was obtained from all participating patients. All relevant clinical information from the cohorts has been compiled and structured, with one sample per patient. This includes gender, age, tumor location, histological staging, Microsatellite Instability (MSI) status, and DNA Polymerase Epsilon (POLE) mutation information. The MSI status was assessed by professional pathologists using immunohistochemistry, while the POLE mutation data was obtained through WES analysis. Our method used the TCGA cohort to build the initial models for TMB estimation. The subsequent validation of the TMB estimation model was executed using the other four distinct cohorts. The study design and algorithm for patient inclusion were detailed in Supplementary Fig. S1.
Whole-exome sequencing (WES) and transcriptome sequencing (RNA-Seq)
WES and RNA-seq were performed by Wuhan IGENEBOOK Biotechnology Co., Ltd. for library construction and sequencing. In brief, total genomic DNA was extracted using a genomic DNA kit (QT-1001, IGENEBOOK Biotechnology, Wuhan, China) according to the manufacturer’s protocol. The TRIzol (RN0102, Aidlab Biotechnologies, Beijing, China) method was used to extract RNA from fresh frozen tissue. Whole-exome libraries were constructed using the AIExome Human Exome Panel V3 - Tumor (T600V1ST, iGeneTech Bioscience, Beijing, China) Enrichment Kit and stored in an elution buffer. RNA samples were reverse-transcribed to cDNA and then stored. All nucleic acid samples were sequenced on the BGI Genomics Co., Ltd MGISEQ-T7 platform using 150-bp double-ended reads (150 PE).
Whole-exome sequencing data analysis
The WES data were quality-controlled using FastQC (version 0.12.1) [13], and then the fastp (version 0.23.2) software was used to remove adapters and sequences of poor quality [14]. Reads were mapped to a GRCh38/hg38-based reference genome using Burrows–Wheeler Aligner (v.0.7.17) [15]. The GATK (version 4.2.6.1) best practice guidelines were referred to process bam files after alignment [16]. Non-synonymous mutations were identified using MuTect2 (version 4.1). The variants were filtered and annotated using the variant effect predictor tools (version 106) [17]. The vcf files were converted to the maf format and finally imported into the R package maftools (version 2.16.0) for further analysis [18].
Transcriptome sequencing data analysis
The RNA-seq data were quality controlled using fastp (version 0.23.2) [14] and then aligned to the GRCh38/hg38 reference genome using STAR (version 2.7.2b) [19]. Finally, transcripts were quantified using TPMCalculator (version 0.0.4) [20]. A comprehensive analysis of immune–oncology signatures was performed using the R package IOBR (version 0.99.9) [21]. The relative levels of tumor-infiltrating immune cells were assessed using CIBERSORT (https://cibersort.stanford.edu).
TMB estimation model construction
Candidate gene filtering and modelling were performed using the genomes of 586 CRC samples from the TCGA cohort. Genes with a mutation frequency ≥ 5% and a significant difference in TMB between patients with the mutated gene and those with a wild-type counterpart were considered TMB-associated genes. Specifically, mutation frequency was defined as the percentage of patients with that gene mutated. Mutant and wild-type groups were compared for differences in TMB between the two groups using the Wilcoxon signed rank test, and a Bonferroni-corrected p-value < 0.05 was considered statistically significant. The mutation matrix for the non-synonymous mutations was constructed with reference to previous studies [22].
The TCGA cohort was randomly partitioned in a 7:3 ratio, with 70% of the samples used for model training and the remainder 30% for internal validation of the model. The eXtreme gradient boosting (XGBoost) regression model was trained on an A30 GPU using the XGBoostRegressor function from the xgboost Python package. The other seven regression models were derived from the scikit-learn library. The recursive feature elimination (RFE) model used a Bayesian information criterion (BIC) to select features, while the other models were selected based on ranking the importance of the features. The optimal parameters for all of the regression models were obtained by using the GridSearchCV function and use five-fold cross-validation for each model. All TMB estimation models were available at https://github.com/fangfyy/CRC-TMB-ML. The fit of the regression model was measured in terms of the R2 value. For each fixed number of genes condition, each model was repeated 1,000 times, and R2 values were calculated. The average of all R2 values was used to assess the fit of the model at the specified number of genes. Mean Absolute Error (MAE) and Root Mean Square Error (RMSE) are used as other metrics to evaluate the performance of the regression model.
Segmented linear regression analysis was conducted on the model performance curves using the ‘segmented’ package in R (version 1.6-4) to determine the point of stability in model fit. The ‘surv_cutpoint’ function from the ‘survminer’ package in R, version 0.4.9, was used to calculate potential cut points for the 20-gene TMB, with the objective of identifying thresholds that result in the most significant differences in survival outcomes among the groups.
HLA typing and tumor neoantigen burden estimation
WES data from patients were analyzed for HLA typing using HLA-HD (version 1.4.0) software [23]. Subsequent analysis was performed using seq2neo (version 2.1) software [24], a neoantigen prediction tool. WES and RNA-seq matched bam files and HLA typing results for each patient were used as inputs to this software. The mutant peptides with half-maximal inhibitory concentration (IC50) binding affinity < 500 nM, immunogenicity > 0.5, transcripts per million (TPM) > 0, tumor antigen processing (TAP) > 0 and high expressed in tumor tissues were defined as tumor neoantigens, whereas mutant peptides with IC50 < 50 nM were further defined as high-affinity tumor neoantigens. The amounts of tumor neoantigen and high-affinity tumor neoantigen in each patient were defined as tumor neoantigen burden (TNB) and high-affinity tumor neoantigen burden (HTNB), respectively.
CD8+ tumor-infiltrating lymphocyte (TIL) density
Colorectal cancer tissue samples were collected at the First Affiliated Hospital of Nanchang University (Nanchang, China), written informed consent was obtained from all patients, and the samples were examined and diagnosed by pathologists. The assays were performed using a universal immunohistochemistry kit (PV-6000, ZSGB-BIO, Beijing, China). Tissue sections were stained with antibodies against CD8 (1:100, ET1606-31, Huabio, Hangzhou, China). Following the established protocol, CD8+ TILs were identified through immunohistochemical staining of the tumor tissue. The CD8+ TIL density was determined by calculating the percentage of CD8 staining within the tumor region [25, 26].
Statistical analysis and visualization
The D’Agostino-Pearson test was used to assess whether the data were normally distributed. Non-parametric tests were used for data that did not pass the D’Agostino-Pearson normality test. The parametric or non-parametric t-tests was employed to investigate differences between two distinct groups, whereas for comparing three groups, parametric or non-parametric ANOVA (Analysis of Variance) was the method of choice. The chi-squared test was used to analyze differences in clinicopathological characteristics among different subgroups. Furthermore, Pearson correlation analysis was performed to assess the linear association between two continuous variables. Kaplan–Meier survival analysis and visualization was performed using the R package survminer (version 0.4.9). Multivariable Cox proportional hazard analysis was conducted using the coxph function from R package survival (version 3.5-5). Due to violation of the proportional hazard assumption, time-dependent covariate Cox models were used to model the association between 20-gene TMB and PFS. The forest diagram was visualized using the R package forplo (version 0.2.5). All data were statistically analyzed and visualized based on R (version 4.2.3) or GraphPad Prism 9.5. A p-value < 0.05 was recognized as statistically significant.
Results
Construction and assessment of machine learning-driven TMB estimation model
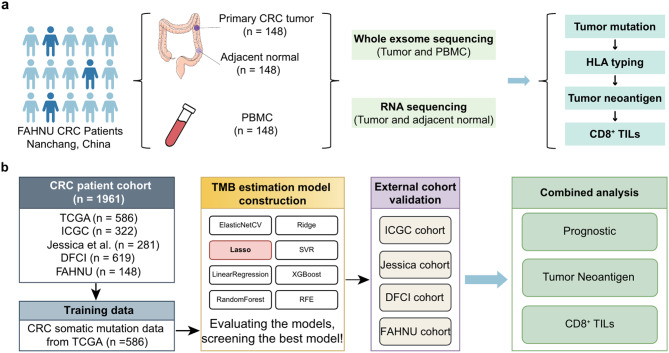
From the First Affiliated Hospital of Nanchang University (FAHNU), 148 patients with primary CRC were obtained with surgically resected tumors, adjacent normal tissue samples, and PBMCs were also collected from these patients. The FAHNU cohort served as the basis for predicting cancer neoantigens by synergistically analyzing RNA-Seq and WES data (Fig. 1a). A comprehensive flow diagram illustrating the entire model creation and subsequent validation can be found in Fig. 1b. Somatic mutation data for patients with CRC were sourced from the TCGA database. Subsequently, a mutation matrix was constructed, encompassing 17,883 genes across 586 patients, specifically targeting non-synonymous mutations. Based on the criteria which stipulated the mutation frequency of ≥ 5% and an association of the gene mutation with TMB (p < 0.05, unpaired t test), a compilation of 468 CRC-associated TMB-related genes were curated (Supplementary Table S1). Among the 468 genes, TTN, SYNE1, PIK3CA, MUC16, and FAT4 emerged as the dominantly mutated genes with mutation frequencies surpassing 20% (Fig. 2a). For the objective of constructing the TMB estimation model, the mutation matrix of these 468 genes was employed.
Fig. 1.
Flowchart of the study design. (a) Flowchart of the establishment process for the FAHNU cohort. (b) Flowchart of the data analysis process
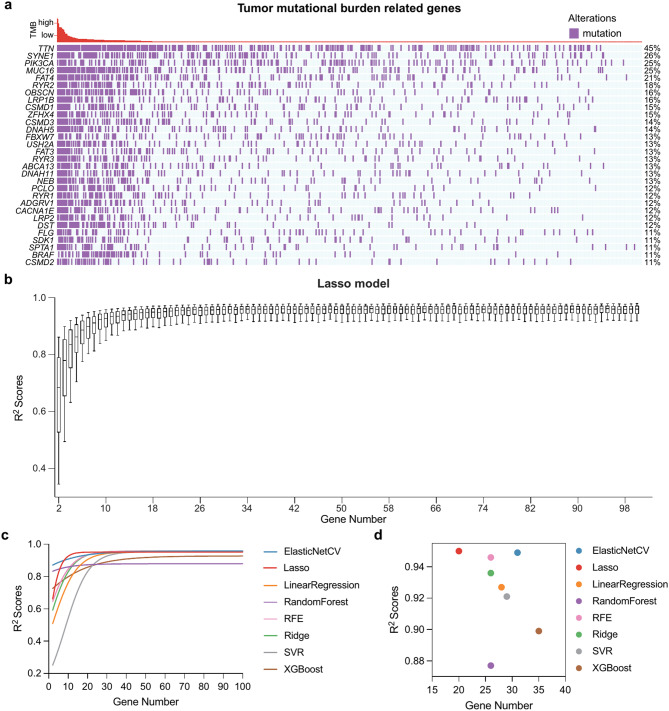
Fig. 2.
Machine learning-based TMB estimation model building and evaluation. (a) The waterfall plot of TMB-related genes in patients with CRC, showing the top 30 genes ranked by mutation rate. (b) Box plots show the selected number of genes and the corresponding R2 scores based on the Lasso model. (c) Growth curves of the mean R2 values for different TMB estimation models. (d) The scatter plot shows the number of genes and the corresponding mean R2 values for the eight TMB estimation models when they tend to stabilize
We employed eight distinct machine learning models to discern the most optimal method for TMB estimation. These models encompassed elastic networks (ElasticNetCV), Lasso Regression, Linear Regression, Random Forest, Recursive Feature Elimination (RFE), Ridge Regression, Support Vector Regression (SVR), and XGBoost. Every model underwent 1,000 iterations for each stipulated gene number, and the consequent R2 scores were assessed in the internal validation set, as depicted in Fig. 2b (Lasso Regression model) and Supplementary Fig. S2 (other models).
As anticipated, when the incorporated number of genes increased, the performance metrics of all models began to plateau, reaching a level of consistency. Except for the random forest and XGBoost models, the performance trajectories of the other six were largely parallel, particularly as the gene count ascended (See Fig. 2c). To decipher the threshold where the model performances began to reach equilibrium, segmented linear regression was adopted on the model R2 value trajectories.
A pivotal observation was that the Lasso model commenced its performance stabilization at the 20-gene markers, with an average R2 value of 0.95. The ElasticNetCV model performance trajectory was nearly analogous to that of Lasso, showing a consistent trajectory at 31 genes, archiving an R2 value of 0.949. We noted that when the number of genes reached 38, ElasticNetCV started to perform slightly better than Lasso (Supplementary Fig. S3a). However, when focusing on models with minimal gene inclusions, the Lasso model yielded the best performance as shown in Fig. 1d. From the insights derived from the TCGA training set, we inferred that the Lasso model is the most appropriate choice for TMB estimation.
Construction and validation of the 20-gene-panel-based TMB estimation model in patients with CRC
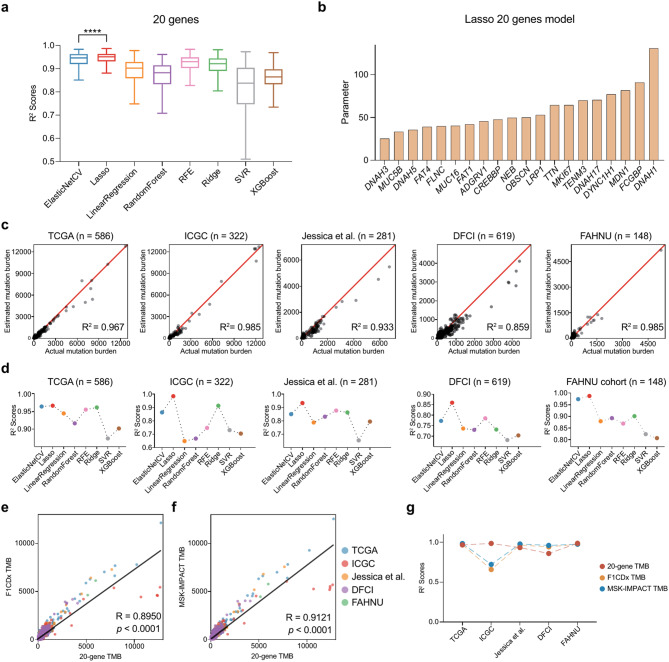
Aiming to predict TMB with a minimal number of genes and reduced sequencing expenses, we focused our construction on the Lasso model’s breakpoint on a panel of 20 genes, which showed the optimal result (p < 0.0001, unpaired t test, Fig. 3a). The 20 genes in the panel that leads to the optimal Lasso-based TMB prediction model are: DNAH3, MUC5B, DNAH5, FAT4, FLNC, MUC16, FAT1, ADGRV1, CREBBP, NEB, OBSCN, LRP1, TTN, MKI67, TENM3, DNAH17, DYNC1H1, MDN1, FCGBP, and DNAH1 (Fig. 3b).
Fig. 3.
20-gene TMB estimation model construction and validation. (a) Box plots illustrate the R2 values of different TMB estimation models when gene number is 20. R2 values—dependent variables; models-independent variables. ****p < 0.0001, the p-value was determined using unpaired t-tests. (b) Genes and corresponding parameters used in the Lasso-based 20-gene TMB estimation model. (c) Lasso-based 20-gene estimated mutation burden versus actual mutation burden validated in TCGA (n = 586), ICGC (n = 322), Jessica et al. (n = 281), DFCI (n = 619) and FAHNU (n = 148) cohorts. Estimated mutation burden—dependent variables; Actual mutation burden-independent variables. (d) Performance of eight TMB estimation models in different CRC cohorts under the condition that the number of genes is set to 20. (e) Scatter plot showing the correlation between 20-gene TMB and F1CDx TMB. (f), Scatter plot showing the correlation between 20-gene TMB and MSK-IMPACT TMB. (g) The line plots illustrate the R2 values of the 20-gene TMB, F1CDx TMB and MSK-IMPACT TMB in the different cohorts
When compared our 20-gene panel with the renowned pan-cancer TMB prediction panels, like MSK-IMPACT and F1CDx, we observed a distinctive variance. Only a fraction of genes was explicitly tied to CRC mutational load (n = 19, Supplementary Fig. S3b and Supplementary Table S2). This underscores an evident gap in these pan-cancer panels when it comes to capturing TMB-associated genes specifically relevant to colorectal cancer. Of the genes in our panel, merely CREBBP and FAT1 are represented in the other pan-cancer panels (Supplementary Fig. S3b).
Evaluating the 20-gene-panel-based TMB estimation model across multiple CRC cohorts
To further validate the 20-gene-panel model, we ventured to test it against four other independent CRC cohorts. Detailed insights regarding these cohorts are available in Supplementary Tables S3 and S4. The 20-gene-panel-based TMB prediction model showed commendable performance across all the five cohorts: TCGA (R2 = 0.967), ICGC (R2 = 0.985), Jessica (R2 = 0.933), DFCI (R2 = 0.859), and FAHNU (R2 = 0.985; Fig. 3c).
To enhance our research, we employed eight different machine learning algorithms to select various panels consisting of 20 genes, in an attempt to devise the optimal prediction model. The features and parameters of all models are specified in Supplementary Table S5. Of all the models, ElasticNetCV and the Lasso model stood out as the two models with the best outcomes, while the Lasso model archived the best performance in nearly all cohorts (Fig. 3d and Supplementary Table S6). This was further corroborated by the overlapping genes between the two panels selected by Lasso and ElasticNetCV, respectively: eight out of 20 genes (ADGRV1, DNAH5, FAT1, FAT4, MUC16, NEB, OBSCN, and TTN) were common in these two panels.
We have conducted a side-by-side comparison of our 20-gene panel with established commercial panels, F1CDx and MSK-IMPACT. Our analysis revealed a significant correlation between the 20-gene based TMB and TMB estimates derived from the F1CDx (R = 0.8950, Fig. 3e) and the MSK-IMPACT (R = 0.9121, Fig. 3f) panels. The diagnostic performance of our panel was consistent with these commercial assays across all five independent cohorts, as shown in Fig. 3g. Beyond CRC, our 20-gene panel proved effective in estimating TMB in other cancers such as pancreatic, glioma, cervical, and prostate cancers (Supplementary Fig. S3c). Crucially, the panel was able to accurately estimate TMB in patients with CRC with POLE mutations—often indicative of very high TMB levels (p < 0.0001, Supplementary Fig. S3d)—and provided reliable TMB estimates in both Microsatellite Stable (MSS) and MSI patients with POLE mutations (p < 0.01, Supplementary Fig. S3e). These results highlight the versatility and reliability of our 20-gene panel as a tool for TMB estimation across a range of tumor types and genetic contexts.
From these evaluations, we conclude that the 20-gene-panel-based TMB estimation model built by Lasso is most suitable for clinical endeavors.
Correlation between 20-gene TMB and prognosis of patients with CRC
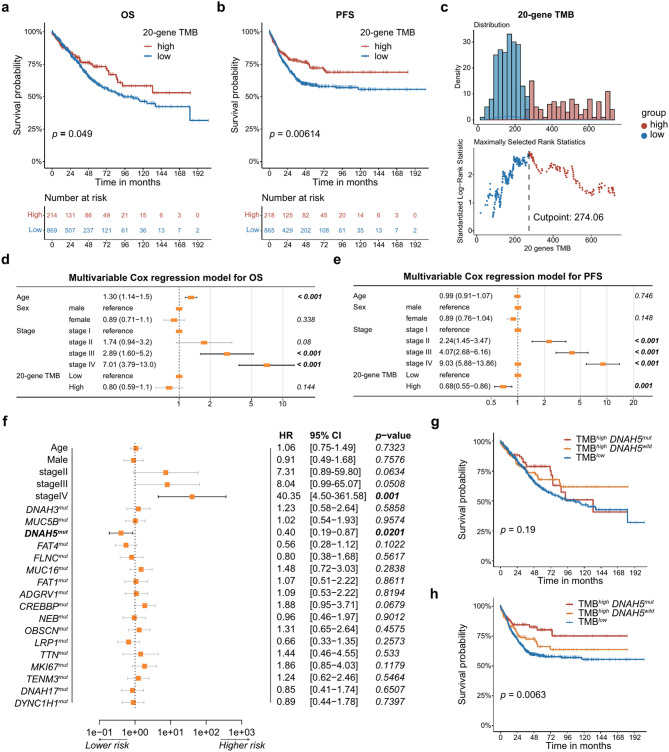
Previous studies have demonstrated an association between TMB and the prognosis in patients with CRC [3]. Here, we evaluated this association between the TMB estimated by our 20-gene-panel-based model and the prognosis extracted from the clinical information in three datasets with over 1,000 patients in total. We observed those patients with high TMB (characterized by our 20-gene-panel-based model) exhibited better overall survival (OS) rates, which is statistically significant (p = 0.049, Fig. 4a). Moreover, a marked association was also observed between the high TMB and the favorable progression-free survival (PFS) outcomes (p = 0.00614, Fig. 4b).
Fig. 4.
20-gene TMB and DNAH5 mutation prognostic role. (a) Kaplan–Meier curves showing the effect of 20-gene TMB on OS in the combined CRC cohort. (b) The effect of 20-gene TMB on PFS in the combined CRC cohort. (c) Density plots of the TMB high-sample and TMB low-sample distributions and the corresponding cut points. (d) Forest plot summarizing prognostic impact of 20-gene TMB on OS by multivariable Cox regression. (e) Forest plot summarizing prognostic impact of 20-gene TMB on PFS by multivariable Cox regression. (f) The forest plot shows the results of multivariable Cox regression in patients with high TMB. (g) OS in groups TMBlow, TMBhigh DNAH5wild and TMBhigh DNAH5mut. (h) PFS in groups TMBlow, TMBhigh DNAH5wild and TMBhigh DNAH5mut
Upon establishing the ideal cutoff point for PFS at 274.06, as determined by our 20-gene TMB model, patients demonstrated a most significant difference in survival outcomes. This cutoff, as outlined in the methods section, delineated the patients into two categories: a high-TMB group consisting of 218 patients and a low-TMB group comprising 865 patients, as depicted in Fig. 4c. A comprehensive breakdown of patient distribution across these two subsets is presented in Supplementary Table S7, which is consistent with the previous studies that pinpointed a significant prognostic difference in patients falling within the top 20% bracket of TMB [5, 27].
Furthermore, the TMB levels appeared to correlate solely with the clinical stage of patients, showing no discernible link with factors such as age or sex (as presented in Table S8). Multivariable Cox regression models were constructed with sex, age, tumor stage and 20-gene TMB. Our multivariable Cox regression analysis clarified that while high TMB didn’t stand out as an independent prognostic indicator for OS (hazard ratio [HR] = 0.80; 95% confidence interval [CI] 0.59–1.1; p = 0.144, Fig. 4d), it did, however, emerge as an independent predictor for enhanced PFS in patients with CRC (HR = 0.68; 95% CI 0.55–0.86; p = 0.001, Fig. 4e). Interestingly, when considered as a continuous variable, the TMB was not an independent predictor of either OS (HR = 0.92; 95% CI 0.77–1.1; p = 0.357, Supplementary Fig. S4a) or PFS (HR = 0.91; 95% CI 0.82–1.0; p = 0.055, Supplementary Fig. S4b). This supports the idea that the understanding of TMB as a biomarker is shifting from quantitative (the more mutations the better) to qualitative [28].
The TMBhigh DNAH5mut patients is associated with better prognosis
Our analysis showed a clear correlation between mutations in the 20-gene panel and TMB. This was evident even in patients identified with high TMB levels. The mutation frequency within these genes was notably high. For instance, among patients in the TMBhigh group, TTN mutations were present in a staggering 92% of cases. The gene with the least mutation frequency within this set was CREBBP, still manifesting a mutation in 22% of these patients, highlighting the critical role of these genes in CRC pathogenesis (Supplementary Fig. S4c).
A deeper investigation into the potential prognostic implications of the mutations in these genes was conducted using multivariable Cox regression analysis. The results singled out mutations in the DNAH5 gene as an independent predictor for a more favorable PFS outcome in TMBhigh group. Specifically, the presence of one or more DNAH5 mutations corresponded to a HR of 0.40 with a 95% CI ranging from 0.19 to 0.87 (p = 0.0201, as shown in Fig. 4f). Though the OS was not significantly different for patients with high TMB that also had DNAH5 mutations (p = 0.19, Fig. 4g), when considering PFS, patients characterized as TMBhighDNAH5mut exhibited the best rates with high statistically significance (p = 0.0063, Fig. 4h). Interestingly, the prognostic implications of DNAH5 mutations appear to be confined to patients with high TMB. In the subset of patients with lower TMB, DNAH5 mutations did not significantly impact either OS (Supplementary Fig. S4d) or PFS (Supplementary Fig. S4e).
These findings emphasize the importance that not only TMB but also specific genetic alterations within tumors may serve as a biomarker for CRC prognosis. The DNAH5 mutation seems to have a potent impact on the prognosis of patients with CRC, specifically for those with a high TMB.
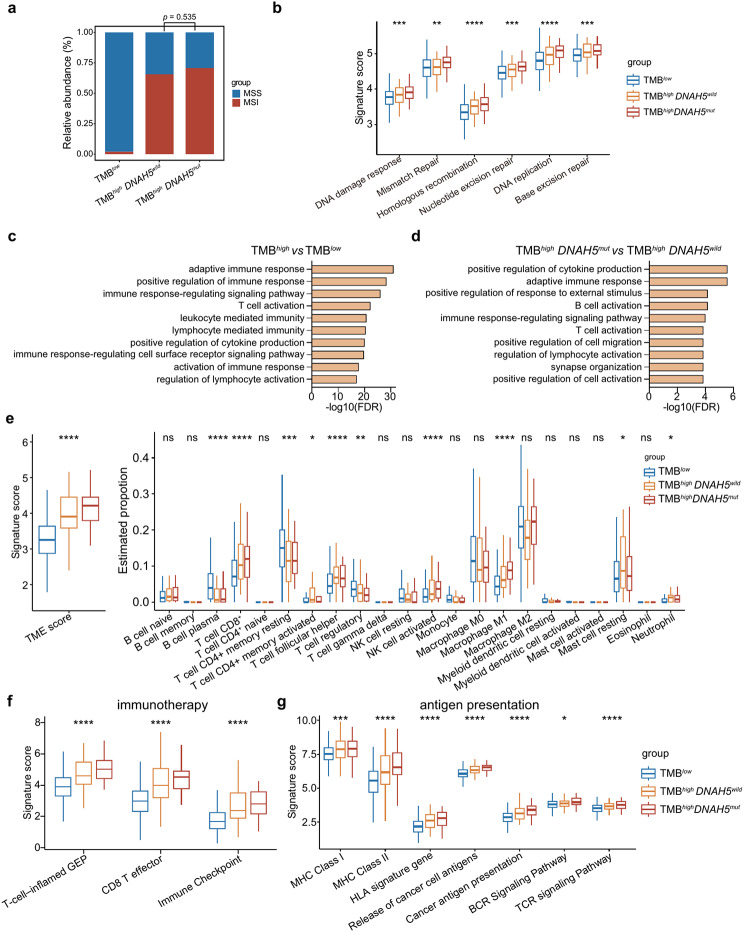
Clinical and gene expression features of patients with TMBhigh DNAH5mut
Analysis of the patients’ clinical characteristics revealed that in patients with high TMB, DNAH5 mutations were more common in males (Table S9). However, there was no significant link between DNAH5 mutations and the MSI status (p = 0.535, Fig. 5a). Elevated TMB was correlated with increased activities in DNA damage response and various DNA repair mechanisms such as mismatch repair, homologous recombination, nucleotide excision repair, DNA replication, and base excision repair. Notably, the most pronounced activity in DNA damage response and DNA repair pathways was observed in patients categorized as TMBhigh with DNAH5 mutations (Fig. 5b).
Fig. 5.
Clinical and gene expression features of patients with TMBhigh DNAH5mut. (a) The bar graphs show the proportions of MSS and MSI in different subgroups of patients with CRC. p = 0.535, Chi-square test. (b) Box plots of DNA damage response and DNA repair pathway signature scores. (c) Top 10 (ordered by false discovery rate [FDR] < 0.05) significantly enriched GO terms (gene ontology biological process) derived from genes highly expressed in patients with high TMB. (d) Among patients with high-TMB, top-10 significantly enriched GO terms derived from genes highly expressed in patients with DNAH5 mutations. (e) Box plots showing the TME and immune cell infiltration scores. (f) Box plots of immunotherapy signature scores. (g) Box plots of the scores for the antigen presentation related signatures. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, 1-way ANOVA was used to determine significance of differences between the three groups
The Gene ontology (GO) term enrichment analysis highlighted those patients with high TMB predominantly exhibited activation in immune signaling pathways, encompassing adaptive immune response, positive regulation of immune response, and other associated pathways including T cell activation (refer to Fig. 5c and Supplementary Table S10). Moreover, a more pronounced activation in these immune signaling pathways was evident in patients with high TMB carrying DNAH5 mutations (Fig. 5d and Supplementary Table S11). This heightened immune activity was corroborated by the tumor microenvironment (TME) scores, showing that the TMBhighDNAH5mut group scored the highest. Furthermore, patients with high TMB experienced a significant influx of various anti-tumor immune cells. This includes CD8+ T cells, follicular helper T cells, activated NK cells, and M1 macrophages, all of which showed a significant increase (p < 0.0001). Remarkably, the patients with TMBhighDNAH5mut exhibited the highest infiltration rates of CD8+ T cells, activated NK cells, and M1 macrophages (Fig. 5e).
These observations underline the association between high TMB and the elevated anti-tumor immune cell infiltration. Notably, patients with high TMB and concurrent DNAH5 mutations exhibited a more robust anti-tumor immune signature. This augmented immune response might shed light on the superior prognosis seen in patients with TMBhighDNAH5mut.
In addition, immunotherapy-sensitive signatures T-cell-inflamed gene expression profile (GEP), effector CD8+ T cells, and immune checkpoints were also associated with high TMB. All immunotherapy-sensitive signatures were significantly upregulated in patients with TMBhighDNAH5mut (Fig. 5f). The immune checkpoint genes CD274, PDCD1LG2, CTLA4, PDCD1, LAG3, HAVCR2, and TIGIT were significantly over-expressed in patients with TMBhighDNAH5mut (Supplementary Fig. S5a). Furthermore, high TMB is also associated with activated antigen presentation, which is more active in TMBhighDNAH5mut (p < 0.05, Fig. 5g). These results further confirm that TMB is associated with the effectiveness of CRC immunotherapy, and that patients with high TMB accompanied by DNAH5 mutations may benefit more from immunotherapy.
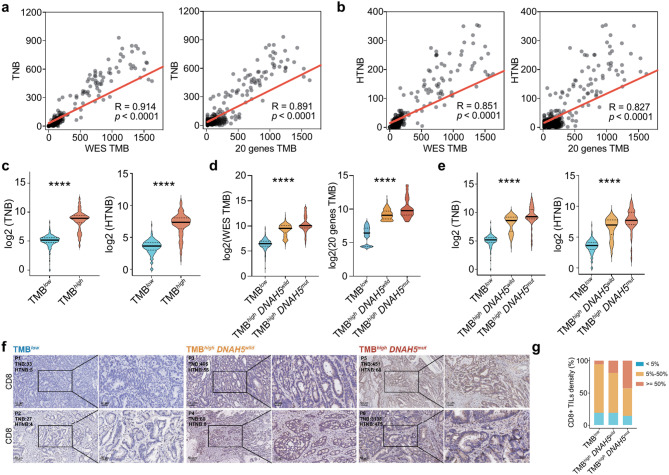
TMB on the 20-gene-panel is associated with TNB
Tumor mutations can generate a large variety of antigens, but only some of these can stimulate an immune response. TNB measures the quantity of these immunogenic antigens produced within a specific genomic region. Past research has shown that higher TNB is linked to better outcomes in patients receiving immunotherapy [29]. Here, paired RNA-seq and WES data in the FAHNU and TCGA cohort were used to analyze tumor neoantigens. A strong positive correlation was found between WES TMB and TNB (R = 0.914). Additionally, the TMB estimated by our 20-gene-panel based model showed a clear positive correlation with TNB (R = 0.891, Fig. 6a). Here, the neoantigens with an IC50 value less than 50nM were classified as highly affinity neoantigens. Both WES TMB (R = 0.851) and the 20-gene-panel-based TMB (R = 0.827) displayed a strong correlation with highly affinity neoantigens burden (HTNB; Fig. 6b), indicating that the 20-gene-panel-based model is a reliable predictor of the neoantigen levels in patients with CRC. Patients with high TMB also had elevated TNB and HTNB (p < 0.0001, Fig. 6c). Notably, TMB, TNB and HTNB were also significantly increased in patients with TMBhighDNAH5mut (p < 0.0001, Fig. 6d-e). Our analysis revealed that density of CD8+ TILs was increased in patients with TMBhighDNAH5mut, indicating a more active immune response, which could have implications for prognosis and therapeutic strategies. (Fig. 6f-g).
Fig. 6.
20-gene TMB is associated with tumor neoantigen burden. (a) Scatter plot to compute the correlation between WES TMB and TNB (left). Scatter plot summarizing the correlation between 20-gene TMB and TNB (right). (b) Scatter plot of the correlation between WES TMB and HTNB (left). Scatter plot of the correlation between 20-gene TMB and HTNB (right). (c) Comparisons of the TNB and HTNB in groups TMBlow and TMBhigh. Comparisons of the TMB (c), TNB and HTNB (d-e) in group TMBlow, TMBhigh DNAH5wild and TMBhigh DNAH5mut. (f) Immunohistochemistry: IHC staining for CD8 in CRC tissues from TMBlow, TMBhigh DNAH5wild and TMBhigh DNAH5mut groups (scale bar = 50 μm, 200× and 20 μm, 400×). (g) Association of CD8+ TIL density with Different Groupings. Unpaired t-tests were used to determine the significance of the differences between the two groups, and one-way ANOVA was used to determine the significance of the differences between the three groups. ***p < 0.001; ****p < 0.0001
Discussion
In this study, we evaluated eight machine learning algorithms in an attempt to devise an efficient model for estimating TMB in CRC using a mere 20-gene panel. In our endeavor to rigorously evaluate the models based on the 20-gene panel, we selected five distinct CRC cohorts for cross validation. Out of them, three represented Caucasian populations, while the other two pertained to Asian demographics. As a result, the outcomes of our study indicate that irrespective of racial variations, the 20-gene panel consistently delivers commendable results. Previous studies have also attempted to evaluate TMB in other cancers using machine learning-driven approaches. However, these studies generally included a limited number of patients and were restricted to a single machine learning algorithm, lacking a systematic comparison and analysis between different algorithms [8, 22].
A limitation to consider is that all the WES data stemmed from tumor tissues procured post-surgery, and obtaining such tissue is sometimes challenging. An alternative is to measure the Blood TMB (bTMB), which can be achieved with a less invasive approach. Nonetheless, bTMB mandates deeper sequencing depths, thus escalating the associated costs and complicating data interpretation [30, 31]. By focusing high-depth sequencing on a limited set of genes, such as our 20-gene panel, we can potentially curtail both sequencing expenses and the time needed for data analysis. However, the applicability of the 20-gene panel to bTMB estimation remains to be validated through upcoming clinical setting.
Another reason that limits the large-scale application of TMB in CRC is the difficulty of determining the TMB threshold [32] for patients subtyping. Variability in the scope of genomes sequenced, the sequencing depth, and the data analysis protocols often results in discrepancies in the selection of TMB cut-off values. The widely accepted clinical threshold for WES TMB currently stands at 10 mutations per mega base [4]. Our study presents a CRC-focused panel that has the potential to provide a more targeted patient stratification for colorectal cancer. While our threshold is consistent with general TMB patterns, it provides a more tailored approach for CRC. It is important to note that the thresholds established in this study are based on our TMB estimation model, and the role of the 20-gene TMB as an independent prognostic indicator will be further confirmed through future prospective studies.
Another important value of TMB is that it is a predictive immunotherapy biomarker. Friedman et al. found that patients with CRC whose TMB ≥ 16 mut/Mb could benefit from immunotherapy [4]. Another study found that among patients with MSI, those with TMB ≥ 40 mut/Mb were the most likely to respond to immune checkpoint inhibitor therapy [3]. Despite the limited number of patients with CRC undergoing immunotherapy, the precise cut-off for the 20-gene TMB remains undefined. Further validation, especially through prospective trials, will be essential to establish the clinical utility of our 20-gene TMB cut-off in predicting responses to immunotherapy for patients with CRC.
Dynein axonemal heavy chain 5 (DNAH5) encodes the axonemal heavy chain of dynamin, is the member of Dynein superfamily. It can impact ATPase activity, which is involved in cilia assembly and cell motility [33]. Initial studies linked DNAH5 mutations to primary ciliary dyskinesia [34]. Recent studies have demonstrated that DNAH5 mutations are also important in many tumors. For instance, in patients with gastric cancer, DNAH5 mutations have been reported to have a positive effect on chemotherapy sensitivity [35]. DNAH5 mutations are also associated with sensitivity to neoadjuvant chemotherapy in patients with plasma cell ovarian cancer [36]. The fellow Dynein superfamily members DNAH8 and DNAH9 are also associated closely with tumor progression [37, 38]. Martini et al. found that cytoplasmic dynein promoted the proliferation of colorectal and cervical cancer cells [39]. Our research indicated a correlation between DNAH5 mutations and enhanced PFS in patients with high TMB. Additionally, patients with TMBhighDNAH5mut exhibited elevated immune checkpoint gene expression. This implies that patients with CRC who have elevated TMB levels paired with DNAH5 mutations could significantly benefit from immunotherapy. Therefore, prospective studies and in vitro experiments are necessary to specifically elucidate the role and mechanism of DNAH5 mutations in CRC.
Our 20-gene TMB model not only predicts prognosis but also shows significant alignment with commercial assays, reinforcing its clinical relevance. We compared our panel’s performance with the F1CDx assay, a recognized standard for TMB measurement. The strong correlation between our 20-gene TMB and the F1CDx assay, as well as with the MSK-IMPACT panel, validates our model against established benchmarks and supports its utility in clinical decision-making. Furthermore, the consistent diagnostic performance of our panel with these commercial assays across multiple cohorts attests to its robustness. Compared to these multi-gene panels, our 20-gene panel effectively reduces costs and processing time while maintaining robustness. This comparison is crucial, as it not only establishes the credibility of our panel but also helps in considering potential cutoff values for TMB, which is paramount for its application in personalized medicine and requires further validation in prospective trials.
As the number of genetic variants accumulated in the genome increases, more neoantigens may be presented. These neoantigens make the tumor more recognizable by the immune system and thus likely to elicit a strong response [6]. Patients with lung cancer who have high TNB and undergoing immunotherapy tend to experience extended PFS [40]. Compared with TMB, TNB is regarded as an improved biomarker for immunotherapy [41]. However, its calculation is contingent on patient-specific HLA data and demands rigorous sequencing data quality and bioinformatic scrutiny [29]. Our research indicates that the TMB estimated on the 20-gene panel aligns well with both TNB and HTNB, which suggests that the 20-gene panel can serve as a streamlined tool to pinpoint the promise of the patients in in-depth tumor neoantigen analysis.
In our research, we noted that a quarter of the high TMB cases were classified as MSS. This proportion is possibly influenced by regional and ethnic variability. For instance, a study involving 575 patients with CRC in Australia reported that 34% of patients with high TMB were MSS [42]. Additionally, the presence of POLE mutations, known to cause hypermutation, could significantly impact this observation [43].
To the best of our knowledge, the 20-gene panel proposed here constitute a small gene panel that can be used to accurately predict both TMB in CRC. TMB are emerging biomarkers for immune checkpoint inhibitor therapies. Predicting these efficiently can stratify patients for personalized treatments. Our study not only aids in this stratification but, by identifying key genes like DNAH5, offers potential therapeutic targets.
Conclusion
Our study introduces a pioneering 20-gene model utilizing machine learning to estimate TMB in CRC patients. This model, which requires only 20 genes, offers a cost-effective and efficient alternative to current methods. It not only predicts TMB with high accuracy but also correlates strongly with patient prognosis. Additionally, we emphasize that the DNAH5 gene serves as a distinguishing biomarker for high TMB patients with CRC, expanding the potential for personalized treatment approaches in CRC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the high-performance computing cluster of the First Affiliated Hospital of Nanchang University. We are grateful to Dr. Haixu Tang and Dr. Sujun Li from Indiana University Bloomington for his valuable comments and insights on this study. The results here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We also acknowledge ICGC (https://dcc.icgc.org/) and cBioportal (https://www.cbioportal.org/) database for providing their platforms and contributors for uploading their valuable datasets.
Abbreviations
- TMB
Tumor mutational burden
- CRC
Colorectal cancer
- WES
Whole exome sequencing
- TNB
Tumor neoantigen burden
- MSS
Microsatellite stability
- MSI
Microsatellite instability
- AUC
Area under the curve
- ROC
Receiver operating characteristic curve
- HR
Hazard ratios
- PFS
Progression-free survival
- OS
Overall survival
- LASSO
The least absolute shrinkage and selection operator
- XGBoost
Extreme Gradient Boosting
- SVR
Support vector regression
- RFE
Recursive feature elimination
- HLA
Human leukocyte antigen
- DNAH5
Dynein Axonemal Heavy Chain 5
- TIL
Tumor-infiltrating lymphocyte
Author contributions
YF, TF and AL conceived and designed the experiments; YF and AL devised the methodologies; YF and TF participated in data analysis and verification; YF and AL wrote the paper. YF, TF, QZ, ZX conducted clinical investigations. KY and AL supervised the study. All authors have read and approved the final version of the manuscript.
Funding
This study was supported by the Major Science and Technology R & D Projects of Jiangxi Province [20213AAG01013], the National Natural Science Foundation of China [82472225, 32071223, 32100729], the Training Plan for Academic and Technical Leaders of Major Disciplines in Jiangxi Province [20213BCJ22015], the Jiangxi Province Science and Technology Innovation Talent Project [jxsg2023201037] and the science and technology innovation base plan of Jiangxi Province [20212BCD42006].
Data availability
All WES and RNA-seq data generated in this study have been stored in the Genome Sequence Archive (GSA) database (accession number: HRA004701).
Declarations
Ethics approval and consent to participate
This study was approved by ethics committee of the First Affiliated Hospital of Nanchang University (2022-CDYFYYLK-06-012).
Consent for publication
This study has not been published before, and all authors approved this publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yangyang Fang and Tianmei Fu contributed equally to this work.
Contributor Information
Kuai Yu, Email: yukuai1949@foxmail.com.
Aiping Le, Email: ndyfy00973@ncu.edu.cn.
References
- 1.Gaiani F, Marchesi F, Negri F, Greco L, Malesci A, de’Angelis GL et al. Heterogeneity of Colorectal Cancer Progression: Molecular Gas and brakes. Int J Mol Sci. 2021;22(10). [DOI] [PMC free article] [PubMed]
- 2.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell. 2021;39(2):154–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manca P, Corti F, Intini R, Mazzoli G, Miceli R, Germani MM, et al. Tumour mutational burden as a biomarker in patients with mismatch repair deficient/microsatellite instability-high metastatic colorectal cancer treated with immune checkpoint inhibitors. Eur J Cancer. 2023;187:15–24. [DOI] [PubMed] [Google Scholar]
- 4.Friedman CF, Hainsworth JD, Kurzrock R, Spigel DR, Burris HA, Sweeney CJ, et al. Atezolizumab Treatment of Tumors with High Tumor Mutational Burden from MyPathway, a Multicenter, Open-Label, phase IIa multiple Basket Study. Cancer Discov. 2022;12(3):654–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, et al. Mutational analysis of patients with Colorectal Cancer in CALGB/SWOG 80405 identifies new roles of microsatellite instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol. 2019;37(14):1217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizzo A, Ricci AD, Brandi G. PD-L1, TMB, MSI, and other predictors of response to Immune checkpoint inhibitors in biliary Tract Cancer. Cancers (Basel). 2021;13(3). [DOI] [PMC free article] [PubMed]
- 7.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated mutation profiling of Actionable Cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Xu J, Chu Q, Duan J, Zhang J, Bai H, et al. A novel tumor mutational burden estimation model as a predictive and prognostic biomarker in NSCLC patients. BMC Med. 2020;18(1):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roszik J, Haydu LE, Hess KR, Oba J, Joon AY, Siroy AE, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med. 2016;14(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reel PS, Reel S, Pearson E, Trucco E, Jefferson E. Using machine learning approaches for multi-omics data analysis: a review. Biotechnol Adv. 2021;49:107739. [DOI] [PubMed] [Google Scholar]
- 11.Uche-Anya E, Anyane-Yeboa A, Berzin TM, Ghassemi M, May FP. Artificial intelligence in gastroenterology and hepatology: how to advance clinical practice while ensuring health equity. Gut. 2022;71(9):1909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issa NT, Stathias V, Schürer S, Dakshanamurthy S. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 2021;68:132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. 2014.
- 14.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43(1110):11.0.1-.0.33. [DOI] [PMC free article] [PubMed]
- 17.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vera Alvarez R, Pongor LS, Mariño-Ramírez L, Landsman D. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics. 2019;35(11):1960–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng D, Ye Z, Shen R, Yu G, Wu J, Xiong Y, et al. IOBR: Multi-omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and signatures. Front Immunol. 2021;12:687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyu GY, Yeh YH, Yeh YC, Wang YC. Mutation load estimation model as a predictor of the response to cancer immunotherapy. NPJ Genom Med. 2018;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaguchi S, Higasa K, Shimizu M, Yamada R, Matsuda F. HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum Mutat. 2017;38(7):788–97. [DOI] [PubMed] [Google Scholar]
- 24.Diao K, Chen J, Wu T, Wang X, Wang G, Sun X et al. Seq2Neo: a Comprehensive Pipeline for Cancer Neoantigen Immunogenicity Prediction. Int J Mol Sci. 2022;23(19). [DOI] [PMC free article] [PubMed]
- 25.Yang H, Shi J, Lin D, Li X, Zhao C, Wang Q, et al. Prognostic value of PD-L1 expression in combination with CD8(+) TILs density in patients with surgically resected non-small cell lung cancer. Cancer Med. 2018;7(1):32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang T, Shi J, Dong Z, Hou L, Zhao C, Li X, et al. Genomic landscape and its correlations with tumor mutational burden, PD-L1 expression, and immune cells infiltration in Chinese lung squamous cell carcinoma. J Hematol Oncol. 2019;12(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anagnostou V, Bardelli A, Chan TA, Turajlic S. The status of tumor mutational burden and immunotherapy. Nat Cancer. 2022;3(6):652–6. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Chen Y, Wang C. Beyond Tumor Mutation Burden: Tumor Neoantigen Burden as a Biomarker for Immunotherapy and other types of Therapy. Front Oncol. 2021;11:672677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuurbiers M, Huang Z, Saelee S, Javey M, de Visser L, van den Broek D et al. Biological and technical factors in the assessment of blood-based tumor mutational burden (bTMB) in patients with NSCLC. J Immunother Cancer. 2022;10(2). [DOI] [PMC free article] [PubMed]
- 31.Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of Blood Tumor Mutational Burden as a potential biomarker for immunotherapy in patients with Non-small Cell Lung Cancer with Use of a next-generation sequencing Cancer Gene Panel. JAMA Oncol. 2019;5(5):696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou W, Yi C, Zhu H. Predictive biomarkers of colon cancer immunotherapy: Present and future. Front Immunol. 2022;13:1032314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Chen L, Guo J, Shi F, Yang Q, Xie L et al. Multiomics Analysis of a DNAH5-Mutated PCD Organoid Model Revealed the Key Role of the TGF-β/BMP and Notch Pathways in Epithelial Differentiation and the Immune Response in DNAH5-Mutated Patients. Cells. 2022;11(24). [DOI] [PMC free article] [PubMed]
- 34.Yang B, Lei C, Xu Y, Yang D, Lu C, Liu Y et al. Whole-exome sequencing identified novel DNAH5 homozygous variants in two consanguineous families with primary ciliary dyskinesia. Chin Med J (Engl). 2023. [DOI] [PMC free article] [PubMed]
- 35.Zhu C, Yang Q, Xu J, Zhao W, Zhang Z, Xu D, et al. Somatic mutation of DNAH genes implicated higher chemotherapy response rate in gastric adenocarcinoma patients. J Transl Med. 2019;17(1):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchocki Z, Tone A, Virtanen C, de Borja R, Clarke B, Brown T, et al. Impact of neoadjuvant chemotherapy on somatic mutation status in high-grade serous ovarian carcinoma. J Ovarian Res. 2022;15(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Ledet RJ, Imberg-Kazdan K, Logan SK, Garabedian MJ. Dynein axonemal heavy chain 8 promotes androgen receptor activity and associates with prostate cancer progression. Oncotarget. 2016;7(31):49268–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruel N, Benhamo V, Bhalshankar J, Popova T, Fréneaux P, Arnould L, et al. Polarity gene alterations in pure invasive micropapillary carcinomas of the breast. Breast Cancer Res. 2014;16(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martini S, Soliman T, Gobbi G, Mirandola P, Carubbi C, Masselli E, et al. PKCε controls mitotic progression by regulating Centrosome Migration and Mitotic Spindle Assembly. Mol Cancer Res. 2018;16(1):3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu T, Tan J, Wu M, Fan W, Wei J, Zhu B, et al. High-affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39(+)CD8(+) T cells. Gut. 2021;70(10):1965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jan YH, Tan KT, Chen SJ, Yip TTC, Lu CT, Lam AK. Comprehensive assessment of actionable genomic alterations in primary colorectal carcinoma using targeted next-generation sequencing. Br J Cancer. 2022;127(7):1304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Favre L, Cohen J, Calderaro J, Pécriaux A, Nguyen CT, Bourgoin R, et al. High prevalence of unusual KRAS, NRAS, and BRAF mutations in POLE-hypermutated colorectal cancers. Mol Oncol. 2022;16(17):3055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All WES and RNA-seq data generated in this study have been stored in the Genome Sequence Archive (GSA) database (accession number: HRA004701).