Abstract
Alamar Blue (AB) has become an increasingly popular reagent of choice for cell viability assays. We chose AB over other reagents such as MTT and Cell-Titer Glo due to its cost effectiveness and its ability to be a nondestructive assay. While analyzing the effect of osimertinib, an EGFR inhibitor on the non-small cell lung cancer cell line, PC-9, we noticed unexpected right-shifts of dose response curves as compared to the curves obtained by Cell Titer Glo assay. Here, we describe our modified AB assay method to avoid that right shift in dose response curves.
-
•
Removal of the drug containing medium prior to AB addition eliminated the falsely increased readings, giving comparable dose response curves as determined by Cell Titer Glo assay.
-
•
Plate-to-plate variability can be minimized by adding an appropriate concentration of a common fluorescence calibration standard to the assay plates to calibrate fluorimeter sensitivity.
-
•
Alamar Blue can be used as a continuous longitudinal assay to monitor cell growth or recovery from drug toxicity over time.
Method name: Alamar Blue assay optimization to minimize drug interference and inter assay viability
Keywords: Alamar Blue, Cell viability assay, EGFR inhibitors
Graphical abstract
Specifications table
| Subject area: | Pharmacology, Toxicology and Pharmaceutical Science |
| More specific subject area: | Cell biology, Chemotherapy resistance |
| Name of your method: | Alamar Blue assay optimization to minimize drug interference and inter assay viability |
| Name and reference of original method: | https://doi.org/10.1046/j.1432–1327.2000.01606.x |
| Resource availability: | https://www.bio-rad-antibodies.com/alamarblue-cell-viability-assay-resazurin.html?JSESSIONID_STERLING=ADF741BC367DC78EA294B15519027C75.ecommerce2&&evCntryLang=US-enthirdPartyCookieEnabled=true |
Background
The Alamar Blue cell viability assay harnesses the optical property change associated with the conversion of resazurin (oxidized, non-fluorescent, blue) to resorufin (reduced, fluorescent, pink) that takes place under the cellular reducing environment. Resazurin is water-soluble, stable in culture medium, non-toxic and permeable through the cell membrane, making it optimal for measuring cell viability [1]. Resazurin can be reduced by NADH and NADPH in the presence of NADPH dehydrogenase or NADH dehydrogenase enzyme, and this reaction is thought to occur in the cytoplasm. Studies have suggested modifications that allow Alamar Blue or resazurin-based cell viability assays to be better suited and more reproducible, such as in 2D cell cultures [2], 3D spheroid cultures [3], and microbial biofilm staining [4]. Analysis of drug effect by this assay, however, may require caution because Alamar Blue has been reported with several interferences, including cell-culture media [5] and anti-oxidant drugs [6].
Osimertinib is a 3rd generation EGFR inhibitor and the newest FDA approved adjuvant therapy for non-small cell lung cancer (NSCLC) with EGFR exon 19 deletions or exon 21 L858R mutation. Acquired resistance to early generations of EGFR tyrosine kinase inhibitors (TKIs) is mostly driven by a second-site EGFR T790M mutation, which negates their inhibitory activity [7]. Osimertinib targets the T790M mutant EGFR by binding irreversibly to the C797 amino acid forming a covalent bond while sparing wild-type EGFR. Therefore, accurate measurement of osimertinib sensitivity is critical to understand evolutionary dynamics of therapeutic resistance in NSCLC.
The cytotoxic effect of osimertinib on PC-9 cells was analyzed using Alamar Blue assay to understand the evolution of resistance in NSCLC against this EGFR inhibitor. We chose Alamar Blue based on its cost effectiveness and its potential as a non-distructive assay. Despite no report on Alamar Blue assay interference by osimertinib, we noticed an unexpected right shifted dose response curve resulting in a higher IC50 value than that determined by a luciferease based assay. This paper describes a simple method to eliminate this drug interference on Alamar Blue assay and to minimize inter-plate variability. We also demonstrate the utility of the optimized method to monitor the recovery from the transient exposure to osimertinib.
Method details
Reading the drug assay plate with modified Alamar Blue protocol
Materials
-
1.
PC-9 cells (MilliporeSigma, Cat# 90071810–1VL).
-
2.
Alamar Blue reagent (BioRad, Cat# BIF012B).
-
3.
Cell culture facility equipped with a CO2 incubator and a biosafety cabinet.
-
4.
Multichannel pipette and tips.
-
5.
Plastic consumables: cell culture dishes and flasks, 96-well plates with black color walls (Corning, Cat# 3904), serological pipettes, and centrifuge tubes.
-
6.
Cell culture medium: for PC-9, RPMI 1640 supplemented with 10 % (v/v) fetal bovine serum, 500 U/ml of each penicillin and streptomycin.
-
7.
3 % (w/v) sodium dodecyl sulfate solution.
-
8.
Rhodamine B (MilliporeSigma, Cat# R6626) stock solution: 1.8 mG/mL DMSO solution.
-
9.
DMSO.
-
10.
BioTek Synergy H1 plate reader.
Procedure
-
1.
On day 1, plate PC-9 cells in to 96-well plates at a density of 3000 cells/90 µl/well.
-
2.
On day 2, add drug at various concentrations in a volume of 10 µl/well.
-
3.
On day 6, the day of the readout, make a 10 % (v/v) Alamar Blue solution in culture medium. For one 96-well plate, make 10 mL AB in culture medium by pipetting 9 mL of RPMI and 1 mL of AB into a 15-mL conical tube.
-
4.
Rhodamine B calibration solution is prepared by diluting Rhodamine stock solution with phosphate buffered saline at 1:100 ratio.
-
5.
Remove the drug assay plate from the incubator. Remove media containing drug by gently inverting the plate into a sink and gently tapping upside down on a paper towel.
-
6.
In the biosafety cabinet, add 100 µL of the 10 % AB solution using a multichannel pipette into the wells containing cells.
-
7.
Add 10 % AB solution and Rhodamine B calibration solution into empty wells to serve as background fluorescence and fluorometer calibration.
-
8.
Incubate the plate for 2 h in the CO2 incubator.
-
9.
(optional) Use a multichannel pipette to add 50 µL of 3 % SDS to each well. This quenches the reszurin/resorufin reaction so that the fluorescence signal does not change over time. Plates can be read at a later date or time if necessary; store plates at room temperature if doing so.
-
10.
Read the fluorescence in a spectrofluorometer set at 560/590 ex/em.
-
11.To calculate percent survival,
Using Alamar Blue as a longitudinal assay to monitor recovery following a drug sensitivity assay
Procedure
-
1.
Repeat Steps 1–10 without step 9 of above described modified Alamar Blue protocol to read drug assay plate.
-
2.
Dump out AB solution by gently inverting the plate into the sink and gently tapping upside down on a paper towel.
-
3.
Add fresh RPMI to the plate and place back into the incubator until next readout.
-
4.
Repeat steps 2–10 without step 9 of the optimized Alamar Blue procedure at each readout.
Method validation
Drug presence interferes with the Alamar Blue assay
Osimertinib's effect on PC-9, a non-small cell lung cancer cell line, was analyzed by cell viability assay using Cell Titer Glo (CTG), a luciferase based assay, and Alamar Blue. While optimizing AB assay incubation time, we noticed unexpected right shifts in the dose response curves (Fig. 1A). We cannot reliably assess drug resistance evolution if IC50 varies depending on the assay conditions. The degree of the shift was reduced by shortening incubation time, yet the IC50 value determined by the CTG assay was significantly lower than determined by the AB assay with 2 hr incubation, the shortest duration we tested (Figure S1A & B).
Fig. 1.
Osimertinib addition changes the Alamar Blue reading, but is not a direct cause. (a) Osimertinib increases the Alamar Blue reading when compared to Cell-Titer Glo. (b) Osimertinib did not directly increase Alamar Blue readings. PC-9 cells were plated into a 96 well plate at the density of 12,000 cells / 80 μl / well. After overnight culture, the cells received varying concentrations of osimertinib in 10 μl medium, and one-tenth volume of Alamar Blue solution and incubated for 2, 3, and 8 h. The Alamar Blue conversion reaction was stopped by adding 50 μl of 3 % (w/v) SDS. The fluorescence intensity of each well was determined by fluorometry using BioTek Synergy H1 plate reader with excitation at 560 nm and emission at 590 nm. Mean + SD, n = 3.
The AB assay relies on redox dependent conversion of resazurin to resorufin, and some drugs altering cellular redox state have been reported to interfere with AB assay [6]. To test if osimertinib has similar direct effect on the AB assay, a constant number of PC-9 cells were incubated with various concentrations of osimertinib together with AB for 2 h. Unlike the data obtained after 5 days of osimertinib incubation, the presence of freshly added osimertinib only with AB did not interfere with florescence readings over 2, 3, and 8 h (Fig. 1B). These results indicate that the AB assay interference by osimertinib is not a direct effect of the drug itself, and the possibility of AB assay interference cannot simply be predicted by direct drug addition to the assay.
The shift of response curve in AB assay resulted in a higher IC50 value when compared to those from CTG assay even when the incubation time was shortened to 2 hrs, the shortest duration we tested and the most similar to CTG (Fig. 2A). The exact mechanism how osimertinib interferes AB assay is not clear, but removal of 5 day-drug treatment medium and addition of new medium with AB eliminated right shift of dose response curve. In contrast to AB assay, the dose response curve, hence IC50, determined by CTG was not affected by prior removal of drug treatment medium. We also observed various degree of right shifts of dose-response curves of other EGFRis (gefitinib, erlotinib and lapatinib) when AB assay was performed without removal of the drugs (Fig. 2A). In all cases, the dose-response curves determined by AB assay after removal of drug treatment medium were comparable to the CTG assays, and the calculated IC50 s are listed in Fig. 2B with a depiction of the IC50 s scaled to CTG values Fig. 2C. Futhermore, we treated PC-9 cells with the IC50 calculated from the assay in Fig. 2D and showed there was a statistically significant difference between AB with the presence of drug vs AB without. Removing drug treatment medium prior to assay, may pre-emptively eliminate unpredictable drug interference on AB assay. To eliminate drug interference, we found discarding the treatment medium is enough, and no washing was required (Supplementary Figure S1).
Fig. 2.
Alamar Blue assay interference by four EGFR inhibitors (a) Four EGFR inhibitors show a shift that is resolved with ”washing” the Alamar Blue media. (b) Listed IC50 concentrations calculated from panel (a). (c) Heatmap showing relative IC50 s scaled to the CTG condition from panel (b). (d) IC50 values determined by AB assay in the presence of osimertinib was higher than those determined by CTG in the presence of osimertinib or by AB assay after removal of osimertinib. ***p = 1.44e-12, n = 21.
Interestingly, we found that dumping the media in our CTG condition also changed the expected drug curve, albeit a left shift. CTG is a luciferase assay that measures ATP content. We hypothesized that the lower reading was caused by the loss of ATP released into the medium when discarding the culture medium. To test this hypothesis, we separately measured the luminescence values of the medium to be discarded and that of corresponding wells after medium removal. When we added those two values together for each individual well, the curve based on the sum showed the same curve as the assay of direct addition of CTG without separating medium and cells.
SDS quenches reduction of resazurin to resorufin
The stability of assay signal can be an important factor to perform multi-well plate based assays, as the availability of plate reader is often limited in the real research laboratory setting. We tested the stability of the AB fluorescence signal by reading the drug assay plates kept in dark at room temperature following 2 hour AB reaction in the absence of drug; using two drugs, osimertinib and gefitinib, another EGFR-TKI approved for EGFRmut lung cancer. The fluorescence intensity increased over time for assay plates of both drugs (Fig. 3A& C). This time dependent increase might be due to the continuous conversion of resazurin to resorufin by the cells after the plates were taken out of the incubator. Indeed, this time dependent increase was quenched by addition of SDS (final concentration 1 %) to the wells to stop AB conversion reaction by lysing the cells, an optional step from the protocol published by Invitrogen (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/AlamarBluePIS.pdf). We still detected a slight time-dependent increase of fluorescence signal sifting drug-response curves even after addition of SDS, but the calculated IC50 s were not significantly different between each time step (Fig. 3B & D). These data indicate that SDS addition should be included to the protocol to estimate IC50 values as accurately as possible. Particularly when multiple plates are to be read, SDS addition would minimize plate-to-plate variation by instantly quenching the cellular activity that otherwise gradually increases the reading by converting resazurin to resorufin.
Fig. 3.
Adding SDS quenches the AB reaction and provides a way of preventing plate-to-plate variation in large scale experiments where multiple plates are to be read. PC-9 cells were treated with both varying concentrations of osimertinib (a & b) and gefitinib (c & d). After the 5 day drug incubation, AB was added and after the 2 hour conversion of AB, SDS was added at a final concentration of 1 %. Readings at 2, 3, 4, 5, 6, and 8 h show a right shift over time in the non SDS conditions. Mean + SD, n = 3. (a) Osimertinib drug assay with Alamar Blue without SDS (b) Osimertinib drug assay with Alamar Blue with SDS (c) Gefitinib drug assay with Alamar Blue without SDS (d) Gefitinib drug assay with Alamar Blue with SDS.
Calibration of fluorometer enables inter-plate comparison
Placing a common fluorescence standard solution into each plate for fluorometer calibration would ensure consistent readings and enables reliable plate-to-plate comparison even the comparison among the results obtained at different times. This capability is particularly important for determining the temporal change in drug sensitivity to understand drug resistance evolution upon exposure to therapeutics [8]. We first tested a pooled biologically converted resorufin as a calibration standard. To create this, we plated a high density of PC-9 cells (1 × 106) in a 10 cm dish and added AB and incubated overnight. We then collected the media containing the biologically converted resorufin and used this as a common calibration solution in further drug assays. While the biologically converted resorufin is a good calibration standard, there may be batch to batch variations. In order to establish more consistent and convenient calibration standard, we tested Rhodamine B, a xanthene fluorescent dye, which has a similar excitation and emission spectrum profiles (ex/em 553/627 nm) as resorufin (ex/em 560/590 nm). We first made a stock of Rhodamine B solution in DMSO (1.8 mg/mL) and kept it at 4 °C protected from light. On the day of measurement, we dilute the Rhodamine B stock solution with phosphate buffered saline (PBS) at 1:100 ratio by volume and the fluorescent intensity of 100 μl/well was determined by BioTek Synergy H1 (BioTek, Winoosi, VT) plate reader at 560/590 nm excitation/emission with sensitivity setting at a fixed sensitivity setting at 72. Creating stock solutions every assay may be time consuming and a source of variability, so we tested and confirmed that the signal is stable at each measurement over time by diluting from a single source of stock solution. Supplementary Figure S3 shows tested the fluorescence intensity up to 18 days, which confirms that the signal is stable over time and can be stored at 4 °C protected from light.
Previous studies have also used Alamar Blue as a continuous monitoring assay. Kwack et al. showed Alamar Blue to be a non-radioactive method for measuring the bioactivity of interleukin (IL-2) [9]. They proved that Alamar Blue was comparable to that of the [3H]thymidine incorporation method, a widely used protocol to monitor DNA synthesis and cell proliferation. Zhou et al. used Alamar Blue to track 3D bone tissue engineering constructs, showing that AB can be successfully used to monitor and predict cell confluence [10]. When non-destructive AB assays are repeated on the same plate with plate reader calibration as described in this paper, temporal cell viability changes of individual wells can be monitored longitudinally. We applied this longitudinal assay to an osimertinib drug sensitivity assay plate, and observed drug concentration dependent differential recovery kinetics (Fig. 4A& B). We validated this method to be in correspondence CTG by measuring the cell proliferation kinetics, and this is shown in Supplementary Figure S4. Thus, AB has potential to be a good tool for continuous monitoring of cell viability changes.
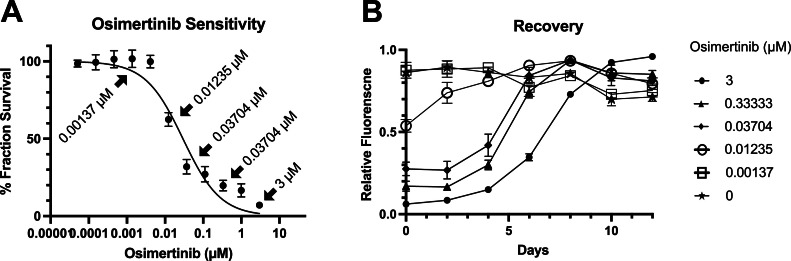
Fig. 4.
AB can be utilized as a longitudinal assay to monitor recovery following a drug sensitivity assay. (a) Drug sensitivity was determined by 2 hour Alamar Blue incubation following drug solution removal without SDS addition (n = 6, mean + SD). The data points indicated by the arrow with drug concentrations are the samples illustrated in panel B for their recovery. b After fluorescence intensities were read, Alamar Blue reaction solution was washed off and cells were continued to cultured with fresh medium. Alamar Blue assay was repeated every two days.
Limitations
While this study provides valuable insights into the optimization of the Alamar Blue assay for assessing drug sensitivity, several limitations should be acknowledged. First, the precise mechanism of why we observe this interference remains unclear. This study demonstrates that removing the drug containing medium resolves the right shift observed, but does not elcudiate the molecular interaction of the redox reaction. Additionally, the protocol written here is optimized to PC-9, and may not be generalizable to other cell types. Incubation times, cell densities, and drug concentrations should be optimized per cell type, as these variables are critical for assay reproducibility. Furthermore, the modified protocol may not be suitable for floating cells, because they will be removed when the media containing drug is discarded. Finally, while this study highlights interference with EGFR inhibitors, these findings may not be consistent with other classes of drugs. In cases where there is no interference, the original protocol might be more appropriate, especially regarding assay duration and ease of use.
CRediT author statement
Mina Dinh: Conceptualization, Methodology, Validation, Writing – Original Draft; Masahiro Hitomi: Conceptualization, Methodology, Validation, Writing – Original Draft; Zahraa Al-Turaihi: Conceptualization, Writing – Review and Editing; Jacob Scott: Writing – Review and Editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was funded by the NIH R37CA244613.
Footnotes
Related research article: None.
For a published article: None.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.mex.2024.103024.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.O'brien J., Wilson I., Orton T., Pognan F. Investigation of the alamar blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 2.Larsson P., et al. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-62848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eilenberger C., et al. Optimized alamarblue assay protocol for drug dose-response determination of 3d tumor spheroids. MethodsX. 2018;5:781–787. doi: 10.1016/j.mex.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Driessche F., Rigole P., Brackman G., Coenye T. Optimization of resazurin-based viability staining for quantification of microbial biofilms. J. Microbiol. Methods. 2014;98:31–34. doi: 10.1016/j.mimet.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Munshi S., Twining R.C., Dahl R. Alamar blue reagent interacts with cell-culture media giving different fluorescence over time: potential for false positives. J. Pharmacol. Toxicol. Methods. 2014;70:195–198. doi: 10.1016/j.vascn.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Shenoy N., et al. Drugs with anti-oxidant properties can interfere with cell viability measurements by assays that rely on the reducing property of viable cells. Lab. Investig. 2017;97:494–497. doi: 10.1038/labinvest.2017.18. [DOI] [PubMed] [Google Scholar]
- 7.Bollinger M.K., Agnew A.S., Mascara G.P. Osimertinib: a third-generation tyrosine kinase inhibitor for treatment of epidermal growth factor receptor-mutated non-small cell lung cancer with the acquired thr790met mutation. J. Oncol. Pharm. Pract. 2018;24:379–388. doi: 10.1177/1078155217712401. [DOI] [PubMed] [Google Scholar]
- 8.Scarborough J.A., et al. Identifying states of collateral sensitivity during the evolution of therapeutic resistance in ewing's sarcoma. iScience. 2020;23 doi: 10.1016/j.isci.2020.101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwack K., Lynch R.G. A new non-radioactive method for il-2 bioassay. Mol. cells. 2000;10:575–578. doi: 10.1007/s10059-000-0575-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X., et al. Noninvasive real-time monitoring by alamarblue® during in vitro culture of three-dimensional tissue-engineered bone constructs. Tissue Eng. Part C. 2013;19:720–729. doi: 10.1089/ten.tec.2012.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.