Abstract
This paper for the 20th anniversary of the Alzheimer's Disease Neuroimaging Initiative (ADNI) provides an overview of magnetic resonance imaging (MRI) of medial temporal lobe (MTL) subregions in ADNI using a dedicated high‐resolution T2‐weighted sequence. A review of the work that supported the inclusion of this imaging modality into ADNI Phase 3 is followed by a brief description of the ADNI MTL imaging and analysis protocols and a summary of studies that have used these data. This review is supplemented by a new study that uses novel surface‐based tools to characterize MTL neurodegeneration across biomarker‐defined AD stages. This analysis reveals a pattern of spreading cortical thinning associated with increasing levels of tau pathology in the presence of elevated amyloid beta, with apparent epicenters in the transentorhinal region and inferior hippocampal subfields. The paper concludes with an outlook for high‐resolution imaging of the MTL in ADNI Phase 4.
Highlights
As of Phase 3, the Alzheimer's Disease Neuroimaging Initiative (ADNI) magnetic resonance imaging (MRI) protocol includes a high‐resolution T2‐weighted MRI scan optimized for imaging hippocampal subfields and medial temporal lobe (MTL) subregions.
These scans are processed by the ADNI core to obtain automatic segmentations of MTL subregions and to derive morphologic measurements.
More detailed granular examination of MTL neurodegeneration in response to disease progression is achieved by applying surface‐based modeling techniques.
Surface‐based analysis of gray matter loss in MTL subregions reveals increasing and spatially expanding patterns of neurodegeneration with advancing stages of Alzheimer's disease (AD), as defined based on amyloid and tau positron emission tomography biomarkers in accordance with recently proposed criteria.
These patterns closely align with post mortem literature on spread of pathological tau in AD, supporting the role of tau pathology in the presence of elevated levels of amyloid beta as the driver of neurodegeneration.
Keywords: Alzheimer's Disease Neuroimaging Initiative, hippocampal subfields, medial temporal lobe, morphometry, segmentation
1. INTRODUCTION
This paper for the 20th anniversary of the Alzheimer's Disease Neuroimaging Initiative (ADNI) serves two goals. The first goal, accomplished in the next three sections, is to provide the motivation behind the inclusion in the ADNI magnetic resonance imaging (MRI) protocol of the high‐resolution T2‐weighted sequence focused on the medial temporal lobe (MTL) and hippocampus subfields; to review the derived data and measures available in the ADNI data archive; and to summarize the published studies using these data. The second goal, to which the subsequent sections are dedicated, is to examine the progression of neurodegeneration in hippocampal subfields and MTL subregions based on the 2024 Alzheimer's Association Revised Criteria for Diagnosis and Staging of Alzheimer's Disease. 1
2. HISTORY OF FOCAL HIPPOCAMPAL MRI AND MOTIVATION FOR ITS INCLUSION IN ADNI
The hippocampus and the MTL are sites of earliest tau pathology and neurodegeneration in Alzheimer's disease (AD) and are impacted by common co‐pathologies of AD, including TAR DNA‐binding protein 43 (TDP‐43) pathology involved in limbic‐predominant age‐related TDP‐43 encephalopathy (LATE) and alpha‐synuclein pathology involved in Lewy body disease. 2 , 3 , 4 , 5 Reduction in hippocampal volume in MRI scans reflects the reduction in neuron number, neuropil and synaptic density, number of glial cells, as well as fluid balance and other transient modulators. 6 , 7 , 8 When the ADNI was first launched, volumetric MRI and hippocampal volume specifically, was one of the best available biomarkers for AD diagnosis and tracking of disease progression in future clinical trials of disease‐modifying treatments. 9 , 10 , 11 , 12 The volume of the hippocampus can be measured on T1‐weighted MRI scans like those obtained in every ADNI MRI session with high reliability using either manual or automatic segmentation, and longitudinal rates of atrophy in hippocampal volume have consistently been shown to be higher in groups with more advanced cognitive decline as well as groups with more advanced AD as defined by cerebrospinal fluid (CSF) and positron emission tomography (PET) biomarkers. 13 , 14 , 15 , 16 , 17 , 18 During the ADNI1 phase, multiple studies provided evidence that changes in hippocampal volume would be substantially more sensitive than cognitive tests in detecting a hypothetical effect of a disease‐modifying treatment for AD. 19
The hippocampus and the MTL have distinctive anatomy and connectivity, and the impact of AD and co‐pathologies on these structures is not uniform. The staging of AD tau pathology by Braak and Braak 2 identifies the transentorhinal cortex (TRC), located on the medial portion of Brodmann area 35 (BA35, part of the perirhinal cortex) as the earliest site of neurofibrillary tangle formation in the human cortex. Pathology subsequently spreads to the entorhinal cortex (ERC) and the hippocampus, particularly subfields cornu ammonis 1 (CA1) and subiculum (SUB). 2 Notably, the amygdala and anterior portions of the temporal pole are also affected at this early stage. 20 , 21 , 22 More recent investigations of the 3D distribution of tau pathology in the MTL using serial histological imaging reveal non‐uniform patterns across the MTL, with a gradient from anterior to posterior in the MTL cortex and a more uniform distribution in the hippocampus. 23 , 24 , 25 Because tau pathology in AD is closely linked to neurodegeneration, the atrophy of the MTL is thought to follow a similar progression to the spread of tau pathology. 26 , 27 , 28
Around the time of the ADNI launch, scientists studying the function and pathology of the hippocampus began using dedicated MRI sequences to image this structure at higher resolution. 29 , 30 , 31 , 32 , 33 , 34 Such studies typically used T2‐weighted sequences on 3 tesla or 4.7 tesla MRI scanners and achieved high resolution (e.g., 0.4 mm × 0.4 mm) in one imaging plane, oriented orthogonally to the hippocampal main axis, while increasing slice thickness and only imaging a partial slab of the brain (e.g., 20–30 slices with 2–3 mm slice thickness) to achieve reasonable scan time and not to exceed safe specific absorption rate (SAR) limits. Such scans revealed the intricate, layered structure of the hippocampus and made it possible to separately segment, and measure the volume of, main hippocampal subfields, including CA1 through CA4, SUB, and dentate gyrus (DG). A number of manual protocols for segmenting hippocampal subfields in T2‐weighted MRI were developed. 29 , 32 , 34 , 35 , 36 , 37 , 38 , 39 Around the same timeframe, computational algorithms for labeling hippocampal subfields emerged, first targeting T1‐weighted MRI 40 , 41 and then specialized to high‐resolution T2‐weighted MRI. 42 , 43 , 44 The ability to measure the volume, thickness, and shape of specific hippocampal subfields that play distinct roles in human memory function opened new avenues of structural and functional MRI research in many fields of neuroscience. However, as more groups adopted subfield imaging, the number of anatomical protocols for labeling subfields grew, making it difficult to compare and relate published results. 45 In 2013, the Hippocampal Subfields Group (HSG) was formed with the goal to harmonize the imaging techniques and segmentation protocols used by the growing hippocampal subfield imaging field. The HSG adopted a long‐term strategy to coalesce the field around a harmonized protocol, with working groups consisting of neuroimagers and neuroanatomists developing a sequence of well‐documented protocols for the hippocampal body, head, tail, and extrahippocampal MTL regions; community feedback via a Delphi protocol; and extensive reliability testing. 46 , 47 The HSG is on the verge of releasing the fully validated hippocampal body protocol in 2024, with remaining protocols expected in the 2025 to 2026 timeframe.
T2‐weighted MRI focused on the hippocampal region (HF‐T2 for short) was introduced as a pilot project during Phase 2 of ADNI, and HF‐T2 scans were acquired during most participant MRI visits at ADNI sites with 3 tesla Siemens scanners. This project collected and performed quality assessments on 684 HF‐T2 scans from 393 participants. A subset of these scans was used in a study by Mueller et al. 48 to compare five distinct approaches to quantify hippocampal subfields from ADNI. Three of the approaches used HF‐T2 scans, including a manual segmentation method 32 and two automated methods: FreeSurfer 6 49 and the Automated Segmentation of Hippocampal Subfields (ASHS) algorithm. 44 The other two approaches used ADNI T1‐weighted MRI: FreeSurfer 5 41 and a deformation‐based whole‐hippocampus shape analysis method. 40 While the analysis was not sufficiently powered to detect significant differences between methods, the overall conclusion of the study was that “subfield volumetry outperformed hippocampal volumetry in their ability to detect subtle atrophy that characterizes the early stages or preclinical stages of AD” and that the “automated T2 based subfield labeling approaches tested in this project compared well to the manual approach.” 48 This analysis, along with studies published on other HF‐T2 datasets in AD, 50 , 51 , 52 , 53 provided the motivation for including HF‐T2 in the MRI protocol for ADNI3 and for including automated measures of hippocampal subfield and MTL subregional volumes derived from these scans in the ADNI3 tabular data.
RESEARCH IN CONTEXT
Systematic review: This article reviews high‐resolution magnetic resonance imaging (MRI) of the medial temporal lobe (MTL) in the Alzheimer's Disease Neuroimaging Initiative (ADNI). We used PubMed and other public sources to review studies using this MRI modality in Alzheimer's disease (AD) research and, specifically, in ADNI. Given the field's transition to biomarker‐based definitions of neurodegenerative diseases, we identified the need to characterize the spread of neurodegeneration in the medial temporal lobe with advancing stages of AD, as defined by tau and amyloid biomarkers.
Interpretation: Our findings reveal a pattern of increased neurodegeneration at more advanced biomarker‐defined stages of AD that closely agree with previously described descriptions of the spread of tau pathology.
Future directions: Directions for medial temporal lobe imaging research in future phases of ADNI include protocol harmonization, incorporating post mortem atlases, and ante mortem studies linking markers of multiple neurodegenerative pathologies to MTL neurodegeneration.
3. HIPPOCAMPUS‐FOCUSED T2‐WEIGHTED MRI AND DERIVED MEASURES IN ADNI3
The ADNI3 MRI protocol included an HF‐T2 acquired using a turbo spin echo sequence in an oblique coronal direction with 175 × 175 mm2 field of view, 0.4 × 0.4 mm2 in‐plane resolution, 30 slices without gap, 2.0 mm slice thickness, repetition time = 8020 ms, echo time = 50 ms. The orientation of the slice plane is approximately orthogonal to the long axes of the two hippocampi, and the 60 mm slab is placed so that both hippocampal formations are included. Slab positioning is performed by the scanner operator interactively after obtaining a whole‐brain T1‐weighted MRI scan. The HF‐T2 scan is obtained on all scanner makes and models in ADNI3, with acceleration parameters and acquisition time (4–5 minutes) varying slightly across scanners. The ADNI database scans were obtained routinely during MRI visits, with scans acquired in 2270 scanning sessions in 1143 ADNI3 participants (513 participants with one scan, 285 with two scans, 345 with three or more scans). An example of an HF‐T2 scan and its orientation relative to a whole‐brain T1‐weighted MRI scan is shown in Figure 1.
FIGURE 1.
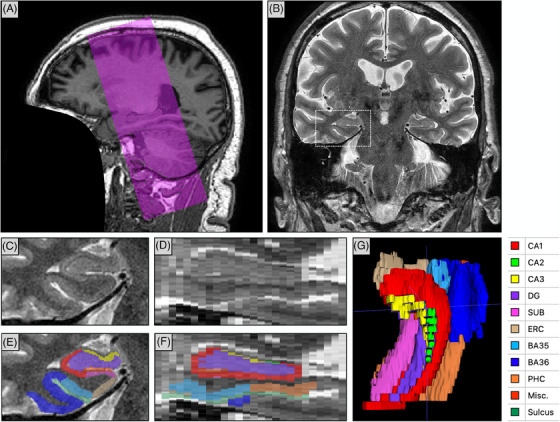
Overview of the HF‐T2 MRI in ADNI3. A, Sagittal view of a T1‐weighted MRI scan from ADNI3 (face removed) with the purple overlay indicating the position and orientation of the slab imaged with HF‐T2 MRI. The slab is approximately aligned with the hippocampal long axes. B, Coronal view of the HF‐T2 MRI scan from ADNI3. C, Zoomed‐in view of the right MTL in the HF‐T2 scan (corresponds to the dotted white box in [B]). D, Sagittal view of the right MTL. E, F, ASHS segmentation of MTL subregions in coronal and sagittal sections. G, 3D rendering of the ASHS segmentation. ADNI, Alzheimer's Disease Neuroimaging Initiative; ASHS, Automated Segmentation of Hippocampal Subfields algorithm; BA, Brodmann area; CA, cornu ammonis; DG, dentate gyrus; ERC, entorhinal cortex; HF‐T2, T2‐weighted MRI focused on the hippocampal region; Misc., miscellaneous label, includes hippocampal cerebrospinal fluid and cysts; MRI, magnetic resonance imaging; MTL, medial temporal lobe; PHC, parahippocampal cortex; SUB, subiculum.
All ADNI3 HF‐T2 MRI scans undergo quality control (QC). HF‐T2 scans are prone to artifacts, with motion and susceptibility artifacts being most common, according to a recent survey. 54 Motion artifacts manifest themselves as rings or streaks in the image, also known as “ghosting” 54 and can cause significant errors in volume and thickness estimates making the scan unusable. 55 Other factors that affect scan usability are errors in the placement of the HF‐T2 slab, which must include both the left and the right medial temporal lobe. The QC assessment takes into consideration the visibility of the hypointense layers in the hippocampus that separate the dentate gyrus from the CA1–3 subfields and the SUB, known as the “dark band” or strata radiatum, lacunosum, and moleculare (SRLM), as well as the definition of the MTL cortical gray matter; and assigns each scan a score between 0 (unusable) and 4 (high quality), as described in Table 1. The QC procedure protocol is included as Supplemental Material in supporting information.
TABLE 1.
Quality rating scale used for ADNI3 HF‐T2 scans.
| HF‐MRI QC rating | Description | Frequency in ADNI3 |
|---|---|---|
| 0: Unusable | MTL substructures and dark band are not identifiable. | 1.7% |
| 1: Bad quality | Dark band is hard to identify and MTL substructures are blurry. | 10.7% |
| 2: Adequate quality | Some MTL substructures are clear; dark band is mostly visible. | 31.5% |
| 3: Good quality | Most MTL substructures are clear; dark band is continuously visible. | 35.0% |
| 4: High quality | All MTL substructures are clear; the dark band is continuously visible, and the transition of CA into DG is clearly visible. | 20.9% |
Note: The term “dark band” refers to the hypointense layer between the dentate gyrus and the CA and subiculum subfields, which corresponds to the strata radiatum, lacunosum, and moleculare (SRLM) of the hippocampus.
Abbreviations: ADNI, Alzheimer's Disease Neuroimaging Initiative; CA, cornu ammonis; DG, dentate gyrus; HF‐T2, T2‐weighted MRI focused on the hippocampal region; MTL, medial temporal lobe.
HF‐T2 MRI scans undergo automatic segmentation using the ASHS algorithm. 44 ASHS is a multi‐atlas automated segmentation algorithm that uses an “atlas set” of N = 29 HF‐T2 MRI scans in which hippocampal subfields and MTL cortical subregions have been labeled. The ASHS labels include subfields CA1–3, DG, SUB, ERC, BA35, Brodmann area 36 (BA35, lateral part of perirhinal cortex), and parahippocampal cortex (PHC); cysts and CSF in the hippocampus, as well as the collateral sulcus are also labeled.
When processing an HF‐T2 scan from ADNI (referred to as the “target” HF‐T2 scan), ASHS performs several image processing steps. First, rigid registration is performed between the HF‐T2 scan and the whole‐brain T1‐weighted MRI scan to account for head position changes during an MRI session. Next, the T1 image is registered to a population template derived from the T1 images in the atlas set. These registrations allow the left and right MTL region of interest (ROI) masks, defined in the template space, to be mapped to the space of the HF‐T2 scan. The HF‐T2 is then cropped around these ROI masks, so that subsequent processing operates on image patches roughly centered around the left and right MTL. The HF‐T2 patches from the atlas set are then registered to the target HF‐T2 patches using deformable registration, and the manual segmentation from each image in the atlas set are deformed (warped) into the space of the target HF‐T2 patches. For each HF‐T2 patch (left and right), this generates N “candidate” segmentations. These N segmentations are fused into a single consensus segmentation using a weighted voting strategy called joint label fusion 56 that considers local appearance similarity between each atlas and the target. Additional machine learning–based correction is applied at each voxel, which has been shown in prior work to slightly increase segmentation accuracy. 57 The resulting segmentation is called the “multi‐atlas” output of ASHS. Last, the process of deformable registration, weighted voting, and learning‐based correction is then repeated, this time using the multi‐atlas outputs to perform affine registration between each atlas in the atlas set and the target HF‐T2 patches. The resulting segmentation is called the “bootstrap” output of ASHS. We added the bootstrap stage to improve accuracy, as it reduced the likelihood of large misalignment between atlases in the atlas set and the target image prior to deformable registration.
An example of ASHS segmentation in an HF‐T2 MRI scan from ADNI3 is provided in Figure 1. The accuracy of the ASHS algorithm has been evaluated using cross‐validation in several atlas sets over the past decade. 44 , 58 , 59 , 60 , 61 , 62 While deep learning–based segmentation techniques trained on the same atlas sets can achieve higher cross‐validation accuracy, a recent study has found the multi‐atlas approach in ASHS to be more robust when generalizing across scanners. 63
After applying ASHS to an HF‐T2 scan, each voxel in the scan is assigned an anatomical label, for example, left CA2. By adding up these voxels (and multiplying by the volume of the MRI voxel), the volume of each subregion is obtained. However, one caveat with using these volume measures for analysis is that for MTL cortical subregions (ERC, BA35, BA36, PHC), the volume measure is very sensitive to how many 2 mm thick HF‐T2 slices are spanned by the segmentation. These subregions are only segmented in the ASHS atlas set in the HF‐T2 slices that contain the hippocampus, plus one additional anterior slice, even though the actual anatomical structures extend further anteriorly (ERC, BA35, BA36) and posteriorly (PHC) as part of the cortical gray matter. Thus, the anterior and posterior boundaries of these subregions in ASHS segmentations are somewhat arbitrary. Furthermore, ASHS applies a heuristic post‐processing step, whereby HF‐T2 slices with few voxels labeled as ERC, BA35, BA36, PHC (<2 5% of the median number of voxels labeled ERC, BA35, BA36, PHC per slice) are removed from the ERC, BA35, BA36, PHC segmentation. Overall, for any given ASHS segmentation there is significant uncertainty about the extent of the MTL cortical subregions along the slice axis. To account for this, ASHS authors recommend normalizing the volume of the MTL cortical subregions by the length of their segmentation in the HF‐T2 slicing direction, 44 that is, number of slices spanned times slice thickness. The volume of each subregion and the number of slices in the HF‐T2 that it spans are reported for each HF‐T2 scan in ADNI3 in the data table “ASHS volume data [ADNI3],” coded “LEFT_CA1_VOL,” “LEFT_CA1_NS,” and so forth.
Alternatively, the thickness of each MTL cortical subregion can be computed from the ASHS segmentation, which is less sensitive to the extent of the anatomical labels. Because there are multiple ways to compute cortical thickness, the ADNI data tables currently do not include thickness as one of the derived measures. However, because ASHS segmentations are uploaded to the Laboratory of Neuro Imaging (LONI) archive, researchers may compute thickness using their preferred approach.
4. REVIEW OF PUBLISHED STUDIES OF MTL SUBREGIONAL MORPHOMETRY IN ADNI
After a pilot project in phase 2 of ADNI in which some sites with Siemens scanners collected HF‐T2 MRI, the modality was newly added to the ADNI MRI protocol for phase 3. The initial studies using HF‐T2 primarily used data from phase 2. They included the study comparing different approaches to quantify hippocampal subfields by Mueller et al., 48 as well as the first comprehensive analysis of morphometric data measured from an MTL‐dedicated sequence obtained in a multi‐site manner by Wolk et al. 64 The latter study not only replicated in ADNI the enhanced sensitivity of hippocampal subfield measures to prodromal AD reported in prior work, 44 but also demonstrated the sensitivity of the modality to MTL subregional changes across a continuum of clinical progression in a pseudo‐longitudinal analysis, including changes in preclinical disease detected in the BA35 region. Leveraging the capability of HF‐MRI to make MTL subregional atrophy measurements, this work established that it was possible to largely recapitulate the topographic pattern of neurofibrillary tangle deposition reported in the histology literature in its downstream effects of neurodegeneration with different degrees of AD severity, from preclinical to dementia. In a subsequent study using ADNI phase 3 data, associations between MTL subregional morphometric measures and tau pathology measured with tau PET were reported, 65 broadly reflecting spatial patterns that recapitulated early Braak stages, with strong effects in rhinal cortices. Further, these correlation patterns in A– individuals were similar to those in A+, suggesting that the 18F‐Flortaucipir tracer is sensitive to tau pathology in primary age‐related tauopathy. 66 Memory performance was also found to be correlated with both MTL tau tracer uptake and subregional atrophy, with the latter effect being strong in rhinal cortices in both A+ and A–, and strongest in CA1 in A+. Another study reported greater atrophy in CA1, DG, and SUB and greater tau burden in multiple MTL subregions including ERC in A+ participants compared to A–. This study also found an association of subregional measures with cognitive scores. 67 The A+ group showed more widespread association of memory performance with subregional atrophy and tau measures compared to A–. Impaired executive function was associated with higher tau in some subregions, including ERC, only in A+. As substantial amounts of HF‐T2 MRI data started becoming available from phase 3, HF‐T2 MRI from ADNI also started being used for studies beyond morphometric analysis along the AD continuum. Cong et al. performed a volumetric genome‐wide association study of MTL substructures demonstrating the value of high‐resolution MTL imaging for revealing the genetic basis of AD biomarkers. 68 Over time, novel methods using these data to interrogate subfields were developed 69 , 70 , 71 and they were also used in evaluation studies comparing different MRI sequences to study subfields. 72 Some of these methods, such as longitudinal automatic segmentation of hippocampal subfields (LASHiS), 69 were designed to take advantage of multimodal imaging, including ultra‐high field strength (7 tesla) MRI. Compared to the HF‐T2 MRI acquired at 3 tesla in ADNI, similar T2‐weighted 7 tesla MRI can provide better tissue contrast, as well as higher spatial resolution, particularly along the long axis of the hippocampus. 73 This can be advantageous for segmenting subfields in the complex folds of the hippocampal head, and result in more reliable delineations of smaller subfields such as CA2 and CA3. 74 , 75
The use of HF‐T2 MRI in ADNI neuroimaging studies may have been limited by the fact that this sequence was introduced in ADNI relatively late, as well as, perhaps, by the lack of familiarity with this modality. Indeed, there is significant interest in the ADNI community in deriving MTL subregion measures from ADNI MRI, as evidenced by the substantial number of papers that estimate the volume of hippocampal subfields from T1‐MRI data in ADNI. This trend has continued to date despite evaluation studies cautioning against segmenting hippocampal subfields in the T1‐MRI with ≈ 1 mm resolution 76 due to low contrast between hippocampal layers and likely dominance of the probabilistic atlas prior in resulting subfield labels.
On the other hand, the use of the HF‐T2 MRI sequence for MTL morphometry in AD neuroimaging as well as other areas of neuroimaging has grown substantially since its adoption in ADNI. A recent meta‐analysis of age effects on MTL subregions across the lifespan 77 reported that out of 41 eligible studies, 19 obtained hippocampal subfield measures from T1‐weighted MRI using automatic segmentation in FreeSurfer, 41 , 49 while HF‐T2 was used in 22 studies (12 with manual and 10 with automatic segmentation). It is likely that with continued acquisition of HF‐MRI in ADNI4 and increased awareness of the availability of more than one thousand HF‐T2 scans and ASHS‐derived measurements in ADNI3, the number of studies using this modality will soon increase.
5. DERIVING PATTERNS OF MTL ATROPHY IN PROGRESSIVE STAGES OF AD USING ADNI3 HF‐T2 MRI
ROI summary measures like CA1 volume or BA35 thickness derived from the ASHS segmentations of HF‐T2 MRI scans make it possible to study the regional impact of neurodegenerative pathologies on the MTL. Even greater anatomical specificity can be achieved by applying more advanced morphological tools to the ASHS output. We recently developed a surface‐based approach to perform groupwise registration and pointwise thickness analysis using ASHS segmentations. 78 Here, we apply this approach to ADNI3 HF‐T2 to study the topography of neurodegeneration associated with different stages of AD defined by biomarkers of amyloid beta (Aβ) and tau pathology. We hypothesize that earlier biological stages of AD would be associated with thinning particularly in ERC and BA35, while later stages would show more pronounced neurodegeneration in other MTL subregions, consistent with the pattern of spread of tau pathology in AD. 2
Data used in this analysis were obtained from the ADNI database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public–private partnership, led by principal investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment, and early AD. For up‐to‐date information, see www.adni‐info.org.
We follow the 2023 Draft Revised Criteria for Diagnosis and Staging of AD 1 to assign ADNI3 participants to progressive disease stages based on Aβ and tau PET biomarkers, and independently of their clinical diagnosis or cognitive status. These criteria break down AD progression into initial (A), early (B), intermediate (C), and advanced (D) stages based on PET or fluid biomarkers. When operationalized using PET biomarkers, all four stages are associated with abnormally elevated levels of Aβ pathology, and the four stages are defined based on the spatial extent and severity of tau pathology: normal levels of tau in stage A (A+T–); elevated tau in the MTL only in stage B (A+TMTL+); elevated tau in the MTL and elevated but moderate level of neocortical tau in stage C (A+TMOD+); and elevated tau in the MTL and high level of neocortical tau in stage D (A+THIGH+). 1 We assign individuals with normal levels of amyloid and tau (A–T–) to the normal (N) group; and we use the designation “indeterminate” (I) to denote participants who do not fit into any of these stages (e.g., have elevated tau but normal Aβ; or have elevated neocortical tau but normal MTL tau, consistent with the hippocampal‐sparing subtype of AD 79 ).
To operationalize this staging approach in ADNI3, we closely followed the steps used for PET‐based Braak staging in a recent study by Jack et al. 80 We used established thresholds based on a composite ROI in ADNI for defining Aβ PET positivity 81 and we fitted two‐component Gaussian mixture models to tau PET standardized uptake value ratio (SUVR) distributions in two stages to define cutoffs between normal/elevated and moderate/high levels of tau for the MTL and neocortical reference regions. This operationalization is detailed in the Supplemental Methods and Figures S2–S5 in supporting information.
To generate surface maps of hippocampal and MTL subregional thickness, we used a surface‐based pipeline that builds on top of ASHS, named “Cortical Reconstruction for ASHS” or CRASHS (https://github.com/pyushkevich/crashs). This pipeline adapts the concepts and tools from widely used whole‐brain segmentation and cortical surface analysis packages FreeSurfer, Nighres, and CRUISE, 82 , 83 , 84 originally developed for whole‐brain segmentation and cortical surface analysis, to MTL subregional segmentations generated by ASHS. These segmentations are first upsampled using a deep learning network trained on paired same‐subject in vivo and ex vivo MTL segmentations to reduce the step edges due to HF‐T2 slice thickness. The starting point for CRASHS is the surface between the gray matter and the white matter in the upsampled ASHS segmentation. This surface includes the “inner” surface of the subiculum and MTL cortical regions (ERC, BA35, BA36, and PHC) and the “outer” surface of the CA1, CA2, and CA3 subfields, which approximately corresponds to the border between hippocampal gray matter and alveus/fimbria (shown as black and red curves, respectively, in Figure S3A). The gray/white surface and the ASHS gray matter segmentation are then used to generate a mid‐surface that passes through the middle of the gray matter (cortical sheet in the MTL; CA1–3 and subiculum subfields in the hippocampus). This mid‐surface is inflated to achieve a simple shape, and large deformation diffeomorphic metric mapping (LDDMM) shape registration is used to map inflated surfaces to a common template surface. The thickness of the MTL gray matter, estimated at every point in the upsampled ASHS segmentation, is mapped to the template space for group analysis. The details of the CRASHS pipeline are provided in Supplemental Methods.
At every vertex of the template mesh, a general linear model is fitted with MTL thickness at that vertex as the dependent variable; group membership as a categorical independent variable with levels A, B, C, D, and N; and age and sex as covariates. The contrasts are computed at each vertex, where are the fitted model coefficients for the five groups. For each contrast, the t statistic is computed at each vertex. Permutation testing using the threshold‐free cluster enhancement (TFCE) approach 85 with parameters , and 10,000 permutations is used to assign a family‐wise error rate (FWER) corrected P value to each location on the MTL surface template.
ROI‐level summary measures of hippocampal subfield volume and MTL cortical thickness were also analyzed. First, these summary measures were plotted against group membership, and the area under the receiver operating curve (AUC) statistic was computed between each group and the N group. Second, linear models with the same design and contrasts as for the vertex‐level analyses were applied to each ROI measure. Permutation tests with 10,000 permutations were used to assess the significance in both sets of ROI‐level analyses.
5.1. Results
Tau PET data from 751 ADNI3 participants were analyzed using Gaussian mixture models to define tau PET SUVR cutoffs for the MTL and neocortical reference regions. The resulting cutoffs are given in Table 2.
TABLE 2.
Operationalizing the draft revised criteria for AD staging in ADNI3 using amyloid beta PET and tau PET SUVR.
| Amyloid PET cortical SUVR | Tau PET Entorhinal SUVR | Tau PET Neocortical SUVR | |
|---|---|---|---|
| AD biomarker stage |
Elevated: ≥1.11 for Florbetapir >1.08 for Florbetaben |
Elevated: ≥1.239 |
Moderate: ≥1.191 High: ≥1.457 |
| N: No biomarker evidence of AD | Normal | Normal | Normal |
| A: Initial stage biomarkers | Elevated | Normal | Normal |
| B: Early‐stage biomarkers | Elevated | Elevated | Normal |
| C: Intermediate stage biomarkers | Elevated | Elevated | Moderate |
| D: Advanced stage biomarkers | Elevated | Elevated | High |
| I: Indeterminate | Any irregular pattern of biomarkers | ||
Notes: Participants were assigned to six groups based on amyloid positivity and levels of tau in the entorhinal cortex and a reference neocortical region, as proposed in. 1 Cutoffs for tau PET were obtained by Gaussian mixture model analysis, as detailed in the Supplemental Methods.
Abbreviations: AD, Alzheimer's disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; PET, positron emission tomography; SUVR, standardized uptake value ratio.
We applied the surface‐based CRASHS pipeline to the ASHS segmentations of HF‐T2 MRI scans of 336 participants who had available Aβ and tau SUVR measures, passed HF‐T2 image QC (score ≥ 2 in Table 1 on both sides), and for which QC of the ASHS output was additionally performed (using the QC protocol for ASHS segmentations included in the Supplemental Materials). When considering left and right MTL separately, the proportion of ASHS segmentations that passed ASHS QC (i.e., QC score of 3 or 4) was 92.8%, as reported in Table S3 in supporting information. We also rejected ASHS segmentations for which the CRASHS template fitting resulted in > 5% of the vertices located > 1 mm from the target surface (95.7% of the segmentations that passed ASHS QC also satisfied this condition). The total number of scans for which both the left and right MTL passed ASHS QC and satisfied the template distance condition was 285 (84.8% of the participants). These participants were assigned to the six groups (AD stages A–D, control group N, and indeterminate group I) based on the amyloid and tau PET cutoffs specified in Table 2. Table 3 compares the six groups in terms of demographic variables, clinical diagnosis, cognitive metrics, and apolipoprotein E (APOE) ε4. More advanced stages are associated with greater levels of impairment and more frequent incidence of one or two APOE ε4 alleles.
TABLE 3.
Comparison of the PET biomarker‐defined progressive AD stages A–D, participants with normal biomarker levels (N), and participants with indeterminate disease stage (I) in ADNI3 in terms of age, sex, education, cognitive status, clinical diagnosis, and number of APOE ε4 alleles.
| N (normal) | A (initial) | B (early) | C (intermediate) | D (advanced) | I (indeterminate) | |
|---|---|---|---|---|---|---|
| Group size (n) | 99 | 80 | 32 | 28 | 19 | 26 |
| Age | 70.4 ± 5.7 | 72.4 ± 6.7† | 72.5 ± 7.0 | 71.6 ± 6.7 | 68.3 ± 5.9 | 69.6 ± 7.1 |
| Sex (Female/male) | 52.5%/47.5% | 56.2%/43.8% | 71.9%/28.1% | 50.0%/50.0% | 47.4%/52.6% | 30.8%/69.2% |
| Education (years) | 16.6 ± 2.3 | 16.7 ± 2.6 | 16.1 ± 2.2 | 15.9 ± 2.8 | 16.1 ± 2.3 | 17.2 ± 2.4 |
| CDR Sum of Boxes | 0.3 ± 0.7 | 0.8 ± 1.4† | 1.8 ± 1.7*** | 3.5 ± 3.5*** | 3.5 ± 2.6*** | 1.5 ± 2.2* |
| Clinical Dx (CN/MCI/DM) | 70.7%/29.3%/0.0% | 63.7%/27.5%/8.8%† | 18.8%/59.4%/21.9%*** | 14.8%/44.4%/40.7%*** | 5.3%/57.9%/36.8%*** | 50.0%/38.5%/11.5%** |
| APOE ε4 alleles (0/1/2) | 81.3%/16.5%/2.2% | 43.4%/50.0%/6.6%*** | 17.9%/57.1%/25.0%*** | 24.0%/48.0%/28.0%*** | 10.5%/57.9%/31.6%*** | 76.0%/20.0%/4.0% |
Note: Statistically significant differences between group N and each of the other groups, obtained using the chi‐squared test of independence for categorical variables and the Mann–Whitney U test for continuous variables, are marked (***: P < 0.001; **: P < 0.01; *: P < 0.05; with Bonferroni correction across the five group comparisons; †: p < 0.05 without Bonferroni correction).
Abbreviations: AD, Alzheimer's disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; APOE, apolipoprotein E; CDR, Clinical Dementia Rating scale; CN, cognitively normal; DM, dementia; MCI, mild cognitive impairment; PET, positron emission tomography.
Figure 2 plots the volumes of hippocampal subfields and the median thickness of MTL cortical subregions for the six groups and reports the AUC for pairwise discrimination between the N group and each of the A, B, C, and D stages using each volume/thickness measure, without considering covariates. For the A group, no significant differences with the N group are observed in any of the subregions. For the B group, there are highly significant reductions in volume and thickness for DG, CA1, BA35, SUB, and ERC. These effects become progressively more significant for the C and D groups, with trend‐level effects appearing in PHC and BA36 in the C group and all subregions except CA3 having significant effects in the D group. For the I group, there are significant effects in ERC and BA35, with trend‐level effects in PHC and SUB. Complementary analysis in Table S1 in supporting information summarizes the general linear models fitted for each measure with age and sex included as covariates, with the results largely recapitulating those in Figure 2. Additionally, Figure S6 in supporting information plots alternative subregional measures: thickness for hippocampal subfields CA1–3 and SUB; and volume for MTL cortical subregions; those results are largely consistent with Figure 2, but AUCs for the MTL subregions are generally lower.
FIGURE 2.
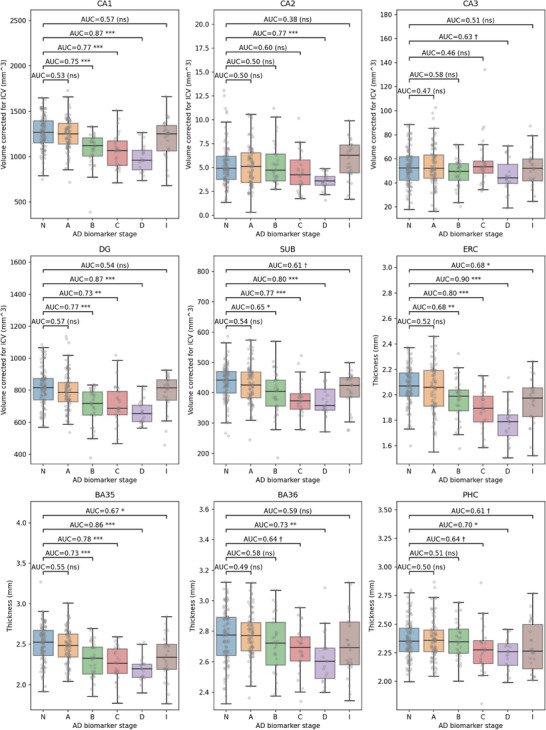
Volumes of hippocampal subfields (CA1, CA2, CA3, DG, subiculum) and median thickness of MTL cortical regions (ERC, BA35, BA36, PHC) are plotted for the control group (N), biomarker‐defined progressive AD stages A–D, and the indeterminate stage group (I). Hippocampal subfield volumes are normalized by intracranial volume. The area under the receiver operating characteristic curve (AUC) is labeled for pairwise comparisons between each stage (A–D, I) and the control group (N). AUC values that are significant after correction for multiple comparisons using a permutation test are marked (***: P < 0.001 **: P < 0.01; *: P < 0.05; †: trend, i.e., uncorrected P < 0.05; ns, not significant). AD, Alzheimer's disease; BA, Brodmann area; CA, cornu ammonis; DG, dentate gyrus; ERC, entorhinal cortex; MTL, medial temporal lobe; PHC, parahippocampal cortex; SUB, subiculum.
Figure 3 shows regional patterns of difference in thickness between each of the A, B, C, D stages and the control group N in the space of the CRASHS MTL surface template. This surface template captures thickness measurements from all MTL subregions except DG. Figure 3 plots t statistic maps and corresponding log‐transformed FWER‐corrected P value maps for contrasts at each point on the template surface, displayed from two different viewpoints. No significant differences in thickness are observed between A and N groups. The most prominent significant differences between B and N groups are observed in and around the transentorhinal region, which is located along the boundary between lateral ERC and BA35. Differences between C and N groups in the MTL cortex resemble the differences between B and N groups, with slightly greater overall thinning; but in the hippocampus there are strong significant effects across inferior and anterior CA1 as well as SUB that are absent for the B–N comparison. Differences between D and N groups also span a larger area of the MTL cortex, with greater involvement of ERC compared to BA35 compared to the B–N and C–N comparisons, and a similar area of the hippocampus to the C–N comparison.
FIGURE 3.
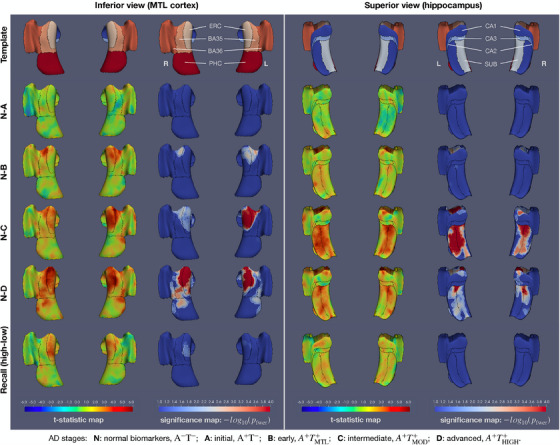
Surface‐based analysis of MTL cortical thickness across biomarker‐defined AD stages. The left and right halves of the figure show different views (from inferior, looking at the MTL cortex, and from superior, looking at the hippocampus) of the same 3D models. The CRASHS surface template is shown in the top row, with MTL subregions labeled. The next four rows show results from a general linear model in which gray matter thickness is the dependent variable, AD stage (A to D, or N for the control group) is the categorical predictor of interest, and age and sex are nuisance predictors. Each row corresponds to the contrast between one of the A–D stages and the N group. T statistic maps are plotted on the left, and log‐transformed P values corrected for multiple comparisons using the threshold‐free cluster enhancement (TFCE) method with 10,000 permutations are plotted on the right. Larger values of the t statistic indicate greater relative differences in thickness between a given stage and the control group N, while accounting for differences in age and sex. The white outlines on the blue‐to‐red P value plots correspond to . The black outlines indicate subregion boundaries. It is important to note that in TFCE analysis, a small P value at a point on the template does not imply that there is a significant difference at that precise location, but rather that the point belongs to a set of clusters in the t statistic map, taken at different thresholds, that is significantly larger in terms of threshold‐weighted cluster area than under the null distribution of the permutation test. The last row shows a general linear model fitted to the thickness data in which participants were assigned to high‐cognition and low‐cognition groups based on terciles of the logical memory delayed recall score in each stage; the high‐versus‐low group was the categorical predictor of interest, and stage, age, and sex were used as nuisance predictors. AD, Alzheimer's disease; CRASHS, Cortical Reconstruction for Automated Segmentation of Hippocampal Subfields algorithm; MTL, medial temporal lobe.
As an additional exploratory analysis, we examined whether there are associations between cognition and MTL subregional thickness that are not explained by AD biomarker staging. Within each of the N, A, B, C, and D groups, we divided the participants into terciles based on the logical memory delayed recall total score and compared the thickness at each point in the MTL template between upper tercile (better cognitive performance) and lower tercile (worse cognitive performance) with age, sex, and disease stage (A–D, N) as covariates. The analysis reveals a pattern of associations between cognition and MTL thickness in the ERC (with a region in the right ERC and a small region in the left ERC, shown in Figure S7 in supporting information). ROI‐level results for this exploratory analysis are presented in Table S2 in supporting information, with ERC thickness significantly associated with cognition at the ROI level.
6. DISCUSSION
By adopting the HF‐T2 sequence in the ADNI3 protocol, the ADNI MRI Core made it possible for AD research to take advantage of detailed measurements of MTL subregional volume and thickness enabled by this sequence. The MTL is the site of early neurofibrillary tau tangle pathology in the brain, and measures derived from HF‐T2 make it possible to measure neurodegeneration in this region with greater granularity than measures such as whole hippocampus volume. Given the close spatial association between tau pathology and neurodegeneration, HF‐T2 is a valuable biomarker in the AD imaging toolbelt, allowing more detailed interrogation of findings from other, more commonly acquired imaging modalities.
The analysis presented in Section 5.1 highlights the potential of HF‐T2 to provide detailed regional information on MTL atrophy. This analysis revealed distinct but overlapping patterns of thinning associated with progressively more severe stages of AD, as defined by Aβ PET and tau biomarkers following the recently proposed revised staging criteria. 1 The lack of discernible neurodegeneration in the A stage (elevated Aβ, normal tau) relative to the N stage and the progressive increase in the amount and area of neurodegeneration in the B, C, and D stages (elevated amyloid, progressively elevated tau) supports the notion that in the presence of Aβ pathology, neurodegeneration is closely associated with the extent and degree of tau pathology. 26 , 86 , 87 , 88 The spatial pattern of neurodegeneration in the B, C, and D stages is highly telling, with B stage involving BA35 and lateral ERC, and C and D stages involving thinning in a pattern concentrically expanding around the regions implicated in the B stage, as well as extensive CA1 and SUB thinning. The B stage regions revealed by our analysis are the regions of earliest tau pathology in the MTL, and agree with the three‐dimensional maps of tau tangle burden from post mortem literature, 23 , 24 , 25 , 89 which show increased burden in the anterior ERC/BA35 and in both anterior and posterior CA1 and SUB. There is significant symmetry to the statistical maps in Figure 3, which speaks to the robustness of the proposed surface‐based approach. A limitation of our analysis is that we did not compare the patterns of associations between MTL structure and AD stage to patterns of association between MTL structure and continuous measures of magnitude and extent of tau and amyloid pathology; nor did we consider AD staging based on fluid biomarkers in our analysis. Such comparisons are left to follow‐up work.
The exploratory analysis examining associations between delayed recall and MTL region thickness, with age, sex, and biomarker AD stage as covariates, reveals a relationship between cognition and neurodegeneration that is not fully explained by AD stage. Factors contributing to this residual negative association between memory performance and cortical thickness may include both tau pathology effects not fully captured by the staging scheme, as well as factors independent of tau and aging, particularly co‐pathologies such as cerebrovascular disease or TDP‐43 pathology that are also associated with MTL neurodegeneration and cognitive decline. 90 , 91 , 92 , 93 , 94 , 95 However, including continuous tau SUVR measures as covariates has very little effect on this analysis (Table 2), suggesting that the observed effect is due to factors other than tau. The fact that the region of significant association between thinning and delayed recall performance beyond AD staging is associated with more medial and less anterior atrophy in the MTL cortex could, in principle, support the role of TDP‐43 given its particular association with anterior hippocampus and ERC atrophy, 96 , 97 , 98 although the lack of a significant effect in the anterior hippocampal subfields weakens the connection with TDP‐43 that can be inferred.
Our analysis highlights the benefits of a template‐based approach to MTL subregion morphometry. Given the intricate anatomy of the MTL, the statistical analysis of thickness maps in Figure 3 provides a more complete picture than the ROI‐based analysis in Figure 2 and better accounts for variation in the size of different subregions. Summary (volume or median thickness) estimates for larger regions like CA1 are likely less impacted by segmentation errors than estimates for smaller regions like BA35 or CA3 because a local segmentation error has less of an impact on the summary measure for larger regions. This may contribute to larger regions being more sensitive to a given predictor in statistical analyses than smaller regions. When the analysis is conducted on the MTL cortex in a point‐wise manner, the treatment of different regions is more consistent, though biases may still exist, for example, in very thin regions or regions of higher curvature, in which segmentation and registration errors may be higher. For example, the ROI‐based analysis in Figure 2 (and Table S1) points to the larger subfields CA1 and DG as the ones most affected in the A+TMTL+ stage B, compared to the control group N. By contrast, in line with the expectation of less bias toward larger subregions, the surface‐based analysis in Figure 3 detects peaks with highest t statistic values around the transentorhinal region (BA35 and ERC), the earliest site of MTL tau pathology. 2 Furthermore, surface‐based analysis allows for the detection of more local hotspots, not influenced by subfield boundaries, which could, in theory, produce better biomarkers, particularly for detecting subtle changes in early disease stages.
While surface‐based and shape‐based modeling has been used in the past to study hippocampal subfield atrophy, 31 , 40 , 44 , 99 , 100 our new surface‐based approach CRASHS combines most of the hippocampal gray matter and MTL cortex into a single surface model. The fitting of the surface template to unfolded MTL mid‐surface helps simplify the complex geometry of the collateral sulcus, which poses significant challenges to volumetric image registration, prior to group‐level registration. The CRASHS approach is not limited to thickness measures and can also be used to analyze other imaging features, such as microstructure features derived from diffusion MRI, 101 texture features, 102 or longitudinal change 78 measures. CRASHS is closely related to the hippocampal unfolding approach HippUnfold, 99 in which hippocampal subfields are automatically segmented and the mid‐surface of the hippocampal subfields CA1–3, SUB, and, separately, DG, are mapped to a flat domain based on the solution of the Laplace equation. CRASHS differs from HippUnfold in that it includes extrahippocampal MTL cortex in the geometrical model and anchors surface‐based correspondences on the gray–white surface, rather than hippocampal gray matter mid‐surface, and that it traces this gray–white surface along the superior surface of the hippocampus (alveus/fimbria). However, one limitation of the CRASHS pipeline is that it excludes DG. Adding DG would require extending the gray/white boundary model in CRASHS into the inner “Swiss roll” of the hippocampus.
HF‐T2 MRI will be collected as part of the ADNI4 MRI protocol, and our group will continue to analyze these scans, provide QC assessments, ASHS segmentations, and derived measurements. During the ADNI4 timeframe we plan to extend the ASHS atlas set to include more anterior portions of the MTL cortical subregions as well as the amygdala, based on recently developed protocols. 103 , 104 We also plan to further validate the CRASHS pipeline, and, once pipeline development has reached a mature, stable stage, provide surface‐based measures alongside volumetric measures as part of the ADNI4 data tables. These will include median thickness measures, as well as pointwise measures of thickness defined in the CRASHS surface template, allowing researchers to quickly perform the kinds of surface‐based analyses presented in Figure 3.
There are several exciting developments in the field of MTL subregion morphometry that will emerge during the ADNI4 phase. The protocol harmonization efforts of the HSG are progressing rapidly, with the protocol for the hippocampal body completed in 2024 and protocols for hippocampal head and tail, as well as MTL cortex, expected in 2025 or 2026. ADNI3 data are being used, along with post mortem MRI and histological reference data, 46 , 47 , 105 for the development and validation of the protocol, and once completed, an atlas manually labeled with this protocol will be used to train the next generation of the ASHS algorithm. Together with recent algorithmic improvements to ASHS, such as incorporation of convolutional neural networks and more effective integration of multiple MRI modalities during segmentation, 63 these developments will likely lead to improved accuracy of MTL subregion segmentation in ADNI4. We are also working on a closer integration of ASHS with post mortem reference atlases of the MTL, which include both detailed anatomical parcellation and a probabilistic description of neuropathology. 25 , 106 HF‐T2 MRI may also prove useful for detailed study of the amygdala in AD, a subject of renewed interest in recent years, given its early and extensive involvement in multiple neurodegenerative proteinopathies. 22 Additionally, tighter integration between MTL subregion segmentation and longitudinal change quantification algorithms in HF‐T2 MRI, as proposed in recent work, 69 , 107 , 108 may lead to increased use of HF‐T2 MRI as a longitudinal measure of treatment response.
The AD field has entered an exciting new phase in which treatments are available 109 , 110 and biomarkers are moving from the realm of research and clinical trial to patient treatment and treatment monitoring. We believe there is an important role for focused imaging and morphometry of the MTL in this new phase. On the one hand, detailed imaging of the MTL may provide information on co‐pathologies such as TDP‐43, for which currently there are no reliable direct biomarkers. A recent study of MTL morphometry in ante mortem MRI of brain donors with post mortem diagnoses of AD and histologically derived measures of TDP‐43 pathology 91 suggests that the ratio of anterior hippocampal volume to PHC thickness can detect TDP‐43 co‐pathology with moderate accuracy. Similar analyses in ADNI participants who donate their brain to science will be able to leverage HF‐T2 MRI and may provide more sensitive markers of co‐pathology. On the other hand, accurate longitudinal measurement of changes in the regions of the brain most affected by AD will be needed to understand why some individuals respond better to treatments than others. While global measures of brain volume and cortical thickness produced unexpected results in recent clinical trials of lecanemab and donanemab, with greater volume loss in the treatment group than in the placebo group, this was not the case for the hippocampus. 109 , 110 This distinction points to the need to interrogate the changes in MTL and MTL subregions under amyloid‐clearing treatments in greater detail.
CONFLICT OF INTEREST STATEMENT
David A. Wolk has served as a paid consultant to Eli Lilly, GE Healthcare, and Qynapse. He serves on DSMBs for Functional Neuromodulation and Glaxo Smith Kline. He is a site investigator for a clinical trial sponsored by Biogen. Sandhitsu R. Das received consultation fees from Rancho Biosciences and Nia Therapeutics. Long Xie is a paid employee of Siemens Healthineers. The other authors have nothing to disclose. Author disclosures are available in the supporting information.
Supporting information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
We acknowledge the extensive contributions of J. L. Robinson, T. Schuck, E. B. Lee, J. A. Detre, M. D. Tisdall, K. Prabhakaran, G. Mizsei, and D. J. Irwin at the University of Pennsylvania; and M. M. Iñiguez de Onzoño Martin, M. M. Arroyo Jiménez, M. Muñoz, M. P. Marcos Rabal, S. Cebada Sánchez, J. C. Delgado González, C. de la Rosa Prieto, and R. Insausti at the University of Castilla‐La Mancha at Albacete to the acquisition, processing, and annotation of the ex vivo MRI dataset that was used to train the upsampling component of the CRASHS pipeline. This research was supported by grant AACSF‐23‐1152241 from the Alzheimer's Association; and a grant from the Fred A. and Barbara M. Erb Family Foundation. Data collection and sharing for the Alzheimer's Disease Neuroimaging Initiative (ADNI) is funded by the National Institute on Aging (National Institutes of Health Grant U19 AG024904, RF1‐AG056014, R01‐AG069474, P30‐AG072979, R01‐AG070592). The grantee organization is the Northern California Institute for Research and Education. In the past, Alzheimer's Disease Neuroimaging Initiative has also received funding from the National Institute of Biomedical Imaging and Bioengineering, the Canadian Institutes of Health Research, and private sector contributions through the Foundation for the National Institutes of Health (FNIH) including generous contributions from the following: AbbVie; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research &Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics.
Yushkevich PA, Ittyerah R, Li Y, et al. Morphometry of medial temporal lobe subregions using high‐resolution T2‐weighted MRI in ADNI3: Why, how, and what's next? Alzheimer's Dement. 2024;20:8113–8128. 10.1002/alz.14161
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp‐content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
REFERENCES
- 1. Jack CR Jr, Andrews JS, Beach TG, et al. Revised criteria for diagnosis and staging of Alzheimer's disease: Alzheimer's Association Workgroup. Alzheimers Dement. 2024;20(8):5143‐5169. doi: 10.1002/alz.13859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Braak H, Braak E. Neuropathological stageing of Alzheimer‐related changes. Acta Neuropathol. 1991;82(4):239‐259. doi: 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- 3. Matej R, Tesar A, Rusina R. Alzheimer's disease and other neurodegenerative dementias in comorbidity: a clinical and neuropathological overview. Clin Biochem. 2019;73:26‐31. doi: 10.1016/j.clinbiochem.2019.08.005 [DOI] [PubMed] [Google Scholar]
- 4. Dickson DW. Neuropathology of non‐Alzheimer degenerative disorders. Int J Clin Exp Pathol. 2009;3(1):1‐23. [PMC free article] [PubMed] [Google Scholar]
- 5. Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci. 2011;12(10):585‐601. doi: 10.1038/nrn3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bobinski M, de Leon MJ, Wegiel J, et al. The histological validation of post mortem magnetic resonance imaging‐determined hippocampal volume in Alzheimer's disease. Neuroscience. 2000;95(3):721‐725. doi: 10.1016/s0306-4522(99)00476-5 [DOI] [PubMed] [Google Scholar]
- 7. Jack CR Jr, Dickson DW, Parisi JE, et al. Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology. 2002;58(5):750‐757. doi: 10.1212/wnl.58.5.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. West MJ, Kawas CH, Stewart WF, Rudow GL, Troncoso JC. Hippocampal neurons in pre‐clinical Alzheimer's disease. Neurobiol Aging. 2004;25(9):1205‐1212. doi: 10.1016/j.neurobiolaging.2003.12.005 [DOI] [PubMed] [Google Scholar]
- 9. Jack CR Jr, Petersen RC, O'Brien PC, Tangalos EG. MR‐based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology. 1992;42(1):183‐188. doi: 10.1212/wnl.42.1.183 [DOI] [PubMed] [Google Scholar]
- 10. Kantarci K, Jack CR, Jr . Neuroimaging in Alzheimer disease: an evidence‐based review. Neuroimaging Clin N Am. 2003;13(2):197‐209. doi: 10.1016/s1052-5149(03)00025-x [DOI] [PubMed] [Google Scholar]
- 11. Cummings JL, Doody R, Clark C. Disease‐modifying therapies for Alzheimer disease: challenges to early intervention. Neurology. 2007;69(16):1622‐1634. doi: 10.1212/01.wnl.0000295996.54210.69 [DOI] [PubMed] [Google Scholar]
- 12. Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 2005;1(1):55‐66. doi: 10.1016/j.jalz.2005.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cash DM, Frost C, Iheme LO, et al. Assessing atrophy measurement techniques in dementia: results from the MIRIAD atrophy challenge. Neuroimage. 2015;123:149‐164. doi: 10.1016/j.neuroimage.2015.07.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hua X, Lee S, Yanovsky I, et al. Optimizing power to track brain degeneration in Alzheimer's disease and mild cognitive impairment with tensor‐based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009;48(4):668‐681. doi: 10.1016/j.neuroimage.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holland D, McEvoy LK, Dale AM, Alzheimer's Disease Neuroimaging I . Unbiased comparison of sample size estimates from longitudinal structural measures in ADNI. Hum Brain Mapp. 2012;33(11):2586‐2602. doi: 10.1002/hbm.21386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fletcher E, Villeneuve S, Maillard P, et al. beta‐amyloid, hippocampal atrophy and their relation to longitudinal brain change in cognitively normal individuals. Neurobiol Aging. 2016;40:173‐180. doi: 10.1016/j.neurobiolaging.2016.01.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pegueroles J, Vilaplana E, Montal V, et al. Longitudinal brain structural changes in preclinical Alzheimer's disease. Alzheimers Dement. 2017;13(5):499‐509. doi: 10.1016/j.jalz.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 18. Schuff N, Woerner N, Boreta L, et al. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain: J Neurol. 2009;132(Pt 4):1067‐1077. doi: 10.1093/brain/awp007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weiner MW, Veitch DP, Aisen PS, et al. The Alzheimer's disease neuroimaging initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9(5):e111‐e194. doi: 10.1016/j.jalz.2013.05.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kromer Vogt LJ, Hyman BT, Van Hoesen GW, Damasio AR. Pathological alterations in the amygdala in Alzheimer's disease. Neuroscience. 1990;37(2):377‐385. doi: 10.1016/0306-4522(90)90408-v [DOI] [PubMed] [Google Scholar]
- 21. Arnold SE, Hyman BT, Van Hoesen GW. Neuropathologic changes of the temporal pole in Alzheimer's disease and Pick's disease. Arch Neurol. 1994;51(2):145‐150. doi: 10.1001/archneur.1994.00540140051014 [DOI] [PubMed] [Google Scholar]
- 22. Stouffer KM, Grande X, Duzel E, et al. Amidst an amygdala renaissance in Alzheimer's disease. Brain: J Neurol. 2024;147(3):816‐829. doi: 10.1093/brain/awad411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yushkevich PA, Munoz Lopez M, Iniguez de Onzono Martin MM, et al. Three‐dimensional mapping of neurofibrillary tangle burden in the human medial temporal lobe. Brain: J Neurol. 2021;144(9):2784‐2797. doi: 10.1093/brain/awab262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ushizima D, Chen Y, Alegro M, et al. Deep learning for Alzheimer's disease: mapping large‐scale histological tau protein for neuroimaging biomarker validation. Neuroimage. 2022;248:118790. doi: 10.1016/j.neuroimage.2021.118790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ravikumar S, Denning AE, Lim S, et al. Postmortem imaging reveals patterns of medial temporal lobe vulnerability to tau pathology in Alzheimer's disease. Nat Commun. 2024;15(1):4803. doi: 10.1038/s41467-024-49205-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gomez‐Isla T, Hollister R, West H, et al. Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Ann Neurol. 1997;41(1):17‐24. doi: 10.1002/ana.410410106 [DOI] [PubMed] [Google Scholar]
- 27. Bobinski M, Wegiel J, Tarnawski M, et al. Relationships between regional neuronal loss and neurofibrillary changes in the hippocampal formation and duration and severity of Alzheimer disease. J Neuropathol Exp Neurol. 1997;56(4):414‐420. doi: 10.1097/00005072-199704000-00010 [DOI] [PubMed] [Google Scholar]
- 28. Rossler M, Zarski R, Bohl J, Ohm TG. Stage‐dependent and sector‐specific neuronal loss in hippocampus during Alzheimer's disease. Acta Neuropathol. 2002;103(4):363‐369. doi: 10.1007/s00401-001-0475-7 [DOI] [PubMed] [Google Scholar]
- 29. Malykhin NV, Lebel RM, Coupland NJ, Wilman AH, Carter R. In vivo quantification of hippocampal subfields using 4.7 T fast spin echo imaging. Neuroimage. 2010;49(2):1224‐1230. doi: 10.1016/j.neuroimage.2009.09.042 [DOI] [PubMed] [Google Scholar]
- 30. Small SA, Perera GM, DeLaPaz R, Mayeux R, Stern Y. Differential regional dysfunction of the hippocampal formation among elderly with memory decline and Alzheimer's disease. Ann Neurol. 1999;45(4):466‐472. doi: 10.1002/1531-8249(199904)45:43C;466::aid-ana83E;3.0.co;2-q [DOI] [PubMed] [Google Scholar]
- 31. Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Dynamics of the hippocampus during encoding and retrieval of face‐name pairs. Science. 2003;299(5606):577‐580. doi: 10.1126/science.1077775 [DOI] [PubMed] [Google Scholar]
- 32. Mueller SG, Stables L, Du AT, et al. Measurement of hippocampal subfields and age‐related changes with high resolution MRI at 4T. Neurobiol Aging. 2007;28(5):719‐726. doi: 10.1016/j.neurobiolaging.2006.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adachi M, Kawakatsu S, Hosoya T, et al. Morphology of the inner structure of the hippocampal formation in Alzheimer disease. AJNR Am J Neuroradiol. 2003;24(8):1575‐1581. [PMC free article] [PubMed] [Google Scholar]
- 34. Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Unfolding the human hippocampus with high resolution structural and functional MRI. Anat Rec. 2001;265(2):111‐120. doi: 10.1002/ar.1061 [DOI] [PubMed] [Google Scholar]
- 35. Olsen RK, Nichols EA, Chen J, et al. Performance‐related sustained and anticipatory activity in human medial temporal lobe during delayed match‐to‐sample. J Neurosci. 2009;29(38):11880‐11890. doi: 10.1523/JNEUROSCI.2245-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ekstrom AD, Bazih AJ, Suthana NA, et al. Advances in high‐resolution imaging and computational unfolding of the human hippocampus. Neuroimage. 2009;47(1):42‐49. doi: 10.1016/j.neuroimage.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. La Joie R, Fouquet M, Mezenge F, et al. Differential effect of age on hippocampal subfields assessed using a new high‐resolution 3T MR sequence. Neuroimage. 2010;53(2):506‐514. doi: 10.1016/j.neuroimage.2010.06.024 [DOI] [PubMed] [Google Scholar]
- 38. Preston AR, Bornstein AM, Hutchinson JB, Gaare ME, Glover GH, Wagner AD. High‐resolution fMRI of content‐sensitive subsequent memory responses in human medial temporal lobe. J Cogn Neurosci. 2010;22(1):156‐173. doi: 10.1162/jocn.2009.21195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirwan CB, Jones CK, Miller MI, Stark CE. High‐resolution fMRI investigation of the medial temporal lobe. Hum Brain Mapp. 2007;28(10):959‐966. doi: 10.1002/hbm.20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang L, Swank JS, Glick IE, et al. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003;20(2):667‐682. doi: 10.1016/S1053-8119(03)00361-6 [DOI] [PubMed] [Google Scholar]
- 41. Van Leemput K, Bakkour A, Benner T, et al. Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus. 2009;19(6):549‐557. doi: 10.1002/hipo.20615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yushkevich PA, Wang H, Pluta J, et al. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2‐weighted MRI. Neuroimage. 2010;53(4):1208‐1224. doi: 10.1016/j.neuroimage.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pipitone J, Park MT, Winterburn J, et al. Multi‐atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;101:494‐512. doi: 10.1016/j.neuroimage.2014.04.054 [DOI] [PubMed] [Google Scholar]
- 44. Yushkevich PA, Pluta JB, Wang H, et al. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 2015;36(1):258‐287. doi: 10.1002/hbm.22627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yushkevich PA, Amaral RS, Augustinack JC, et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. Neuroimage. 2015;111:526‐541. doi: 10.1016/j.neuroimage.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olsen RK, Carr VA, Daugherty AM, et al. Progress update from the hippocampal subfields group. Alzheimers Dement (Amst). 2019;11:439‐449. doi: 10.1016/j.dadm.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wisse LEM, Daugherty AM, Olsen RK, et al. A harmonized segmentation protocol for hippocampal and parahippocampal subregions: why do we need one and what are the key goals? Hippocampus. 2017;27(1):3‐11. doi: 10.1002/hipo.22671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mueller SG, Yushkevich PA, Das S, et al. Systematic comparison of different techniques to measure hippocampal subfield volumes in ADNI2. NeuroImage Clinical. 2018;17:1006‐1018. doi: 10.1016/j.nicl.2017.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iglesias JE, Augustinack JC, Nguyen K, et al. A computational atlas of the hippocampal formation using ex vivo, ultra‐high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117‐137. doi: 10.1016/j.neuroimage.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Flores R, La Joie R, Chetelat G. Structural imaging of hippocampal subfields in healthy aging and Alzheimer's disease. Neuroscience. 2015;309:29‐50. doi: 10.1016/j.neuroscience.2015.08.033 [DOI] [PubMed] [Google Scholar]
- 51. Pluta J, Yushkevich P, Das S, Wolk D. In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi‐automatic segmentation of T2‐weighted MRI. J Alzheimers Dis. 2012;31(1):85‐99. doi: 10.3233/JAD-2012-111931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. La Joie R, Perrotin A, de La Sayette V, et al. Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer's disease and semantic dementia. NeuroImage Clinical. 2013;3:155‐162. doi: 10.1016/j.nicl.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mueller SG, Schuff N, Yaffe K, Madison C, Miller B, Weiner MW. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp. 2010;31(9):1339‐1347. doi: 10.1002/hbm.20934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Canada KL, Mazloum‐Farzaghi N, Rådman G, et al. A (Sub)field guide to quality control in hippocampal subfield segmentation on highresolution T2‐weighted MRI. bioRxiv. Preprint posted online December 1, 2023. doi: 10.1101/2023.11.29.568895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reuter M, Tisdall MD, Qureshi A, Buckner RL, van der Kouwe AJW, Fischl B. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. Neuroimage. 2015;107:107‐115. doi: 10.1016/j.neuroimage.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi‐Atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell. 2013;35(3):611‐623. doi: 10.1109/TPAMI.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H, Das S, Pluta J, et al. Standing on the shoulders of giants: improving medical image segmentation via bias correction. Med Image Comput Comput Assist Interv. 2010;13(Pt 3):105‐112. doi: 10.1007/978-3-642-15711-0_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wisse LE, Kuijf HJ, Honingh AM, et al. Automated hippocampal subfield segmentation at 7T MRI. AJNR Am J Neuroradiol. 2016;37(6):1050‐1057. doi: 10.3174/ajnr.A4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bender AR, Keresztes A, Bodammer NC, et al. Optimization and validation of automated hippocampal subfield segmentation across the lifespan. Hum Brain Mapp. 2018;39(2):916‐931. doi: 10.1002/hbm.23891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schlichting ML, Mack ML, Guarino KF, Preston AR. Performance of semi‐automated hippocampal subfield segmentation methods across ages in a pediatric sample. Neuroimage. 2019;191:49‐67. doi: 10.1016/j.neuroimage.2019.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cong S, Risacher SL, West JD, et al. Volumetric comparison of hippocampal subfields extracted from 4‐minute accelerated vs. 8‐minute high‐resolution T2‐weighted 3T MRI scans. Brain Imaging Behav. 2018;12(6):1583‐1595. doi: 10.1007/s11682-017-9819-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hickling AL, Clark IA, Wu YI, Maguire EA. Automated protocols for delineating human hippocampal subfields from 3 Tesla and 7 Tesla magnetic resonance imaging data. Hippocampus. 2024;34(6):302‐308. doi: 10.1002/hipo.23606 [DOI] [PubMed] [Google Scholar]
- 63. Xie L, Wisse LEM, Wang J, et al. Deep label fusion: a generalizable hybrid multi‐atlas and deep convolutional neural network for medical image segmentation. Med Image Anal. 2023;83:102683. doi: 10.1016/j.media.2022.102683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wolk DA, Das SR, Mueller SG, Weiner MW, Yushkevich PA, Alzheimer's Disease Neuroimaging I . Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer's disease. Neurobiol Aging. 2017;49:204‐213. doi: 10.1016/j.neurobiolaging.2016.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Das SR, Xie L, Wisse LEM, et al. In vivo measures of tau burden are associated with atrophy in early Braak stage medial temporal lobe regions in amyloid‐negative individuals. Alzheimers Dement. 2019;15(10):1286‐1295. doi: 10.1016/j.jalz.2019.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Crary JF, Trojanowski JQ, Schneider JA, et al. Primary age‐related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol. 2014;128(6):755‐766. doi: 10.1007/s00401-014-1349-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ge X, Zhang D, Qiao Y, Zhang J, Xu J, Zheng Y. Association of tau pathology with clinical symptoms in the subfields of hippocampal formation. Front Aging Neurosci. 2021;13:672077. doi: 10.3389/fnagi.2021.672077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cong S, Yao X, Huang Z, et al. Volumetric GWAS of medial temporal lobe structures identifies an ERC1 locus using ADNI high‐resolution T2‐weighted MRI data. Neurobiol Aging. 2020;95:81‐93. doi: 10.1016/j.neurobiolaging.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shaw T, York A, Ziaei M, Barth M, Bollmann S, Alzheimer's Disease Neuroimaging I . Longitudinal automatic segmentation of hippocampal subfields (LASHiS) using multi‐contrast MRI. Neuroimage. 2020;218:116798. doi: 10.1016/j.neuroimage.2020.116798 [DOI] [PubMed] [Google Scholar]
- 70. Shaw TB, York A, Barth M, Bollmann S. Towards optimising MRI characterisation of tissue (TOMCAT) dataset including all longitudinal automatic segmentation of hippocampal subfields (LASHiS) data. Data Brief. 2020;32:106043. doi: 10.1016/j.dib.2020.106043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rechberger S, Li Y, Kopetzky SJ, Butz‐Ostendorf M, Alzheimer's Disease Neuroimaging I . Automated high‐definition MRI processing routine robustly detects longitudinal morphometry changes in Alzheimer's disease patients. Front Aging Neurosci. 2022;14:832828. doi: 10.3389/fnagi.2022.832828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bussy A, Plitman E, Patel R, et al. Hippocampal subfield volumes across the healthy lifespan and the effects of MR sequence on estimates. Neuroimage. 2021;233:117931. doi: 10.1016/j.neuroimage.2021.117931 [DOI] [PubMed] [Google Scholar]
- 73. Giuliano A, Donatelli G, Cosottini M, Tosetti M, Retico A, Fantacci ME. Hippocampal subfields at ultra high field MRI: an overview of segmentation and measurement methods. Hippocampus. 2017;27(5):481‐494. doi: 10.1002/hipo.22717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kafadar C, Ulusoy OL, Ozturk E, Mutlu A. Vertebral artery injury associated with cervical spine fracture dislocation. Spine J. 2016;16(8):e529‐e530. doi: 10.1016/j.spinee.2016.01.206 [DOI] [PubMed] [Google Scholar]
- 75. Wisse LE, Gerritsen L, Zwanenburg JJ, et al. Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. Neuroimage. 2012;61(4):1043‐1049. doi: 10.1016/j.neuroimage.2012.03.023 [DOI] [PubMed] [Google Scholar]
- 76. Wisse LEM, Chetelat G, Daugherty AM, et al. Hippocampal subfield volumetry from structural isotropic 1 mm(3) MRI scans: a note of caution. Hum Brain Mapp. 2021;42(2):539‐550. doi: 10.1002/hbm.25234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Homayouni R, Canada KL, Saifullah S, et al. Age‐related differences in hippocampal subfield volumes across the human lifespan: a meta‐analysis. Hippocampus. 2023;33(12):1292‐1315. doi: 10.1002/hipo.23582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yushkevich PA, Xie L, Wisse LE, et al. Mapping medial temporal lobe longitudinal change in preclinical Alzheimer's disease. Alzheimer's Dement. 2023;19:e081898. 10.1002/alz.081898 [DOI] [Google Scholar]
- 79. Murray ME, Graff‐Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer's disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 2011;10(9):785‐796. doi: 10.1016/S1474-4422(11)70156-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jack CR, Wiste HJ, Algeciras‐Schimnich A, et al. Predicting amyloid PET and tau PET stages with plasma biomarkers. Brain. 2023;146(5):2029‐2044. doi: 10.1093/brain/awad042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Landau SM, Breault C, Joshi AD, et al. Amyloid‐beta imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nucl Med. 2013;54(1):70‐77. doi: 10.2967/jnumed.112.109009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huntenburg JM, Steele CJ, Bazin PL. Nighres: processing tools for high‐resolution neuroimaging. Gigascience. 2018;7(7):giy082. doi: 10.1093/gigascience/giy082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Han X, Pham DL, Tosun D, Rettmann ME, Xu C, Prince JL. CRUISE: cortical reconstruction using implicit surface evolution. Neuroimage. 2004;23(3):997‐1012. doi: 10.1016/j.neuroimage.2004.06.043 [DOI] [PubMed] [Google Scholar]
- 84. FischlB. FreeSurfer. Neuroimage. 2012;62(2):774‐781. doi: 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Smith SM, Nichols TE. Threshold‐free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44(1):83‐98. doi: 10.1016/j.neuroimage.2008.03.061 [DOI] [PubMed] [Google Scholar]
- 86. Harrison TM, Du R, Klencklen G, Baker SL, Jagust WJ. Distinct effects of beta‐amyloid and tau on cortical thickness in cognitively healthy older adults. Alzheimers Dement. 2021;17(7):1085‐1096. doi: 10.1002/alz.12249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Giannakopoulos P, Herrmann FR, Bussiere T, et al. Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer's disease. Neurology. 2003;60(9):1495‐1500. doi: 10.1212/01.wnl.0000063311.58879.01 [DOI] [PubMed] [Google Scholar]
- 88. Giannakopoulos P, von Gunten A, Kovari E, et al. Stereological analysis of neuropil threads in the hippocampal formation: relationships with Alzheimer's disease neuronal pathology and cognition. Neuropathol Appl Neurobiol. 2007;33(3):334‐343. doi: 10.1111/j.1365-2990.2007.00827.x [DOI] [PubMed] [Google Scholar]
- 89. Stouffer KM, Witter MP, Tward DJ, Miller MI. Projective diffeomorphic mapping of molecular digital pathology with tissue MRI. Commun Eng. 2022;1:44. doi: 10.1038/s44172-022-00044-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jellinger KA. Recent update on the heterogeneity of the Alzheimer's disease spectrum. J Neural Transm (Vienna). 2022;129(1):1‐24. doi: 10.1007/s00702-021-02449-2 [DOI] [PubMed] [Google Scholar]
- 91. de Flores R, Wisse LEM, Das SR, et al. Contribution of mixed pathology to medial temporal lobe atrophy in Alzheimer's disease. Alzheimers Dement. 2020;16(6):843‐852. doi: 10.1002/alz.12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kapasi A, Yu L, Boyle PA, Barnes LL, Bennett DA, Schneider JA. Limbic‐predominant age‐related TDP‐43 encephalopathy, ADNC pathology, and cognitive decline in aging. Neurology. 2020;95(14):e1951‐e1962. doi: 10.1212/WNL.0000000000010454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Josephs KA, Dickson DW, Tosakulwong N, et al. Rates of hippocampal atrophy and presence of post‐mortem TDP‐43 in patients with Alzheimer's disease: a longitudinal retrospective study. Lancet Neurol. 2017;16(11):917‐924. doi: 10.1016/S1474-4422(17)30284-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. O'Brien JT, Desmond P, Ames D, Schweitzer I, Tress B. Magnetic resonance imaging correlates of memory impairment in the healthy elderly: association with medial temporal lobe atrophy but not white matter lesions. Int J Geriatr Psychiatry. 1997;12(3):369‐374. [PubMed] [Google Scholar]
- 95. van der Flier WM, van Straaten EC, Barkhof F, et al. Medial temporal lobe atrophy and white matter hyperintensities are associated with mild cognitive deficits in non‐disabled elderly people: the LADIS study. J Neurol Neurosurg Psychiatry. 2005;76(11):1497‐1500. doi: 10.1136/jnnp.2005.064998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nelson PT, Dickson DW, Trojanowski JQ, et al. Limbic‐predominant age‐related TDP‐43 encephalopathy (LATE): consensus working group report. Brain. 2019;142(6):1503‐1527. doi: 10.1093/brain/awz099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Heywood A, Stocks J, Schneider JA, et al. The unique effect of TDP‐43 on hippocampal subfield morphometry and cognition. NeuroImage Clinical. 2022;35:103125. doi: 10.1016/j.nicl.2022.103125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Adams JN, Marquez F, Larson MS, et al. Differential involvement of hippocampal subfields in the relationship between Alzheimer's pathology and memory interference in older adults. Alzheimers Dement (Amst). 2023;15(2):e12419. doi: 10.1002/dad2.12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. DeKraker J, Haast RAM, Yousif MD, et al. Automated hippocampal unfolding for morphometry and subfield segmentation with HippUnfold. eLife. 2022;11:e77945. doi: 10.7554/eLife.77945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Diers K, Baumeister H, Jessen F, Duzel E, Berron D, Reuter M. An automated, geometry‐based method for hippocampal shape and thickness analysis. Neuroimage. 2023;276:120182. doi: 10.1016/j.neuroimage.2023.120182 [DOI] [PubMed] [Google Scholar]
- 101. Shahid SS, Wen Q, Risacher SL, et al. Hippocampal‐subfield microstructures and their relation to plasma biomarkers in Alzheimer's disease. Brain. 2022;145(6):2149‐2160. doi: 10.1093/brain/awac138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wearn AR, Nurdal V, Saunders‐Jennings E, et al. T2 heterogeneity as an in vivo marker of microstructural integrity in medial temporal lobe subfields in ageing and mild cognitive impairment. Neuroimage. 2021;238:118214. doi: 10.1016/j.neuroimage.2021.118214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sadeghpour N, Ittyerah R, Denning AE, et al. Developing an anatomically valid segmentation protocol for early tau regions: the anterior medial temporal lobe cortices. Alzheimers Dement. 2023;19:e075831. [Google Scholar]
- 104. Wuestefeld A, Binette AP, van Westen D, et al. Medial temporal lobe atrophy patterns in early‐versus late‐onset amnestic Alzheimer's disease. bioRxiv. Preprint posted online May 21, 2024. 10.1101/2024.05.21.594976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wuestefeld A, Baumeister H, Adams JN, et al. Comparison of histological delineations of medial temporal lobe cortices by four independent neuroanatomy laboratories. Hippocampus. 2024;34(5):241‐260. doi: 10.1002/hipo.23602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ravikumar S, Wisse LEM, Lim S, et al. Ex vivo MRI atlas of the human medial temporal lobe: characterizing neurodegeneration due to tau pathology. Acta Neuropathol Commun. 2021;9(1):173. doi: 10.1186/s40478-021-01275-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Das SR, Avants BB, Pluta J, et al. Measuring longitudinal change in the hippocampal formation from in vivo high‐resolution T2‐weighted MRI. Neuroimage. 2012;60(2):1266‐1279. doi: 10.1016/j.neuroimage.2012.01.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dong M, Xie L, Wisse L, et al. Comparison of focal hippocampal T2‐weighted MRI and whole‐brain T1‐weighted MRI for detection of longitudinal atrophy using deep learning‐based and conventional approaches. Alzheimers Dement. 2023;19:e083228. [Google Scholar]
- 109. van Dyck CH, Swanson CJ, Aisen P, et al. Lecanemab in early Alzheimer's disease. N Engl J Med. 2023;388(1):9‐21. doi: 10.1056/NEJMoa2212948 [DOI] [PubMed] [Google Scholar]
- 110. Sims JR, Zimmer JA, Evans CD, et al. Donanemab in early symptomatic Alzheimer disease: the TRAILBLAZER‐ALZ 2 randomized clinical trial. JAMA. 2023;330(6):512‐527. doi: 10.1001/jama.2023.13239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


