Abstract
Abstract
The synthesis of nitriles is of utmost importance for preparative organic chemistry. The classical routes are often associated with disadvantages such as toxicity of the reagents and drastic conditions. The uses of enzymes like aldoxime dehydratases (Oxds) and hydroxynitrile lyases constitute attractive benign alternatives. In this review, we summarize the recent trends regarding Oxds. Thousands of oxd genes were sequenced but less than thirty Oxds were investigated on protein level. We give an overview of these Oxds, their sequence analysis, conditions required for their overexpression, and their purification and assays. We then focus on the use of Oxds especially in multistep reactions combining the chemical or chemoenzymatic synthesis of aldoximes from different starting materials with the enzymatic dehydration of aldoximes to nitriles, possibly followed by the hydration of nitriles to amides. Progress in Oxd immobilization is also highlighted. Based on data published mainly in the last 5 years, we evaluate the industrial prospects of these enzyme processes in comparison with some other innovations in nitrile synthesis.
Key points
• Aldoxime dehydratases (Oxds) are promising for cyanide-free routes to nitriles
• A comprehensive overview of wet-lab explored Oxds is provided
• Recent trends include combining Oxds with other enzymes or chemical catalysts
Graphical Abstract

Keywords: Aldoxime dehydratase, Nitrile synthesis, Biocatalyst, Multistep reaction, Immobilization
Introduction
Nitriles are a highly sought-after group of compounds. They can be used as, e.g., solvents, fuels, fragrances or pharmaceuticals, and precursors in organic synthesis. They include bulk and fine chemicals (Betke et al. 2018). Some well-known examples of the former are the polymer precursors acrylonitrile and adiponitrile or acetonitrile and propionitrile which are widely used as solvents. Nitriles are also used as precursors of agrochemicals, drugs or surfactants (fatty amines from fatty nitriles) (Hinzmann et al. 2019b, 2020a, 2021; Yavuzer et al. 2023).
There is a number of pharmaceuticals that bear a cyano group (Gröger and Asano 2020; Chen et al. 2021). These are saxagliptin (Onglyza) and vildagliptin (Galvus) which serve for the treatment of type 2 diabetes mellitus, the antihistamine levocabastine, the calcium channel blocker verapamil, the anticonvulsant perampanel, the antiandrogens bicalutamide and enzalutamide, the anti-gout medicine febuxostat, and the antifungal isavuconazole. In veterinary medicine, trilostane (Vetoryl) is used for the therapy of hypercortisolism (Cushing’s disease) in dogs (Ramsey 2010).
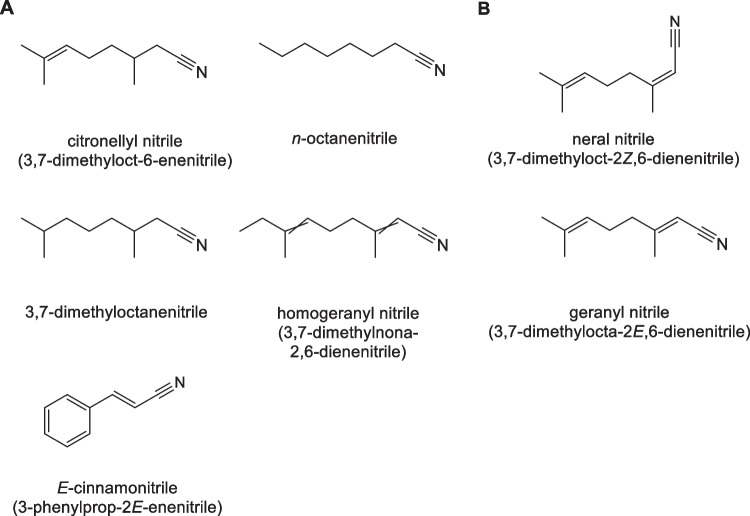
Several nitriles can be used as fragrances as they smell similar to the corresponding fragrance aldehydes and are often more stable. The use of some of them is considered safe, while others are associated with environmental or health hazards despite similar structures (Fig. 1). Thus, citronellyl nitrile (3,7-dimethyloct-6-enenitrile) and homogeranyl nitrile (3,7-dimethyl-2,6-nonadienenitrile) have been used in dozens of homecare products, n-octanenitrile and 3,7-dimethyloctanenitrile in cleansers, and E-cinnamonitrile (3-phenylprop-2E-enenitrile) in air fresheners and scented candles (Consumer Product Information Database, https://www.whatsinproducts.com). In contrast, the use of geranyl nitrile (dimethylocta-2E,6-dienenitrile, https://pubchem.ncbi.nlm.nih.gov/compound/1551246) or neral nitrile (3,7-dimethylocta-2Z,6-dienenitrile, https://pubchem.ncbi.nlm.nih.gov/compound/1551245) should be avoided due to chronic aquatic toxicity and genotoxicity hazards.
Fig. 1.
Examples of aroma nitriles. A Nitriles used as fragrances; B nitriles not recommended for use as fragrances due to health or environmental hazard
The classical methods (Fig. 2) used for nitrile synthesis are substitution of alkyl halides or other alkylating agents with simple cyanides (Kolbe nitrile synthesis), hydrocyanation of alkenes, ammoxidation of alkenes or aromatic hydrocarbons, dehydration of amides (Chen 2021; Hinzmann et al. 2021), or a Sandmeyer reaction of aromatic diazonium salts (Akhtar et al. 2022). Each of them suffers from certain disadvantages. For the substitution reaction (Fig. 2A) and the hydrocyanation (Fig. 2B), the highly toxic metal cyanides (KCN, NaCN) or HCN must be used in a stoichiometric ratio. An alternative cyanation agent, trimethylsilyl cyanide, which also allows for an assymetric cyanation (for example, see Fig. 2C) (Holmes and Kagan 2000a, b) has a similar toxicity. The Sandmeyer cyanation of a diazonium salt (Fig. 2D) with KCN or another CN− donor and typically Cu(I) as catalyst (Akhtar et al. 2022) is limited to aromatic nitriles, and the diazonium salt is synthesized using the toxic nitrite. Ammoxidation (Fig. 2E) requires high temperatures and metal catalysts. Hazardous dehydrating agents (thionyl chloride, phosphoryl chloride, phosphorus pentoxide) or metal catalysts are needed for amide dehydration (Ganesan and Nagaraaj 2020) (Fig. 2F). The method choice depends on the type of the target nitrile. Thus, adiponitrile, acrylonitrile and fatty acid nitriles are preferentially produced by hydrocyanation, ammoxidation, and amide dehydration, respectively (Hinzmann et al. 2021).
Fig. 2.
Classical routes to nitriles: A Kolbe nitrile synthesis in dimethyl sulfoxide (DMSO); B hydrocyanation (here: hydrocyanation of alkenes); C asymmetric cyanation with trimethylsilyl cyanide (TMSCN); D Sandmeyer cyanation; E ammoxidation (here: acrylonitrile synthesis); F amide dehydration
Cyanide-free reactions that take place under mild conditions and do not require hazardous or expensive reagents or catalysts are highly desirable. For example, such reactions are catalyzed by enzymes that are involved in the synthesis of natural nitriles (Irmisch et al. 2014, 2015; Sørensen et al. 2018; Liu and Li 2024; Yamaguchi and Asano 2024). However, some of the enzymes such as β-cyano-l-alanine synthase (Kumano et al. 2016) or 7-cyano-7-deazaguanine synthetase (Winkler et al. 2015) have narrow substrate specificities (Yamaguchi and Asano 2024). In contrast, the scope of substrates transformed by hydroxynitrile lyases and aldoxime dehydratases (Oxds) is broad, which makes these enzymes interesting for nitrile synthesis.
The progress in the engineering and use of hydroxynitrile lyases has been reviewed recently (Priya and Padhi 2023). Another recent review addressed the nitrile-synthesizing enzymes and pathways as a whole with focus on their distribution in microorganisms, plants and animals, their natural functions, and the synthesis of volatile nitriles (Yamaguchi and Asano 2024). A recent study summarized the synthesis of nitriles in plants and insects (Yamaguchi 2024). Other reviews focused on Oxds, in particular their substrate specificity (Betke et al. 2018; Chen 2021), their in vivo functions (Rädisch et al. 2022) and their synthetic potential as a whole (Bhalla et al. 2018; Hinzmann et al. 2021) or emphasized on chiral nitriles (Gröger and Asano 2020; Domínguez de María 2021). The newest of these reviews were largely based on literature published until 2020. In addition, the most important features of nitrile biosynthesis by Oxds were summarized in a separate chapter in a recently published book on nitrile chemistry (Seth 2024).
In this study, we focus on recent advances in the application of Oxds, particularly in multistep (chemo)enzymatic reactions. First, we discuss the availability of Oxds and the production of catalysts based on them. We then summarize the recent uses of Oxds, while highlighting reaction parameters such as substrate concentrations, conversions, isolated yields, and space–time yields, which indicate the industrial prospects of the processes. Finally, we compare the processes catalyzed by Oxds with alternative reactions. The review is mainly based on the literature of the last five years.
Aldoxime dehydratases: availability and catalyst forms
Sequence diversity
The number of putative Oxds found by database searches depended on the database and software used. Thus, the Oxds were about 3000 according to BLAST searches of GenBank and UNIPROT (Křístková et al. 2023) and about 8000 according to 3DM software searches of the 3DM database from Bio-Prodict (Hinzmann et al. 2023a, b). The 3DM method is based on aligning three-dimensional protein structures unlike the common BLAST method based on aligning amino acid sequences. Only a small fraction of the putative Oxds was used for expression and subsequent characterization of the corresponding enzymes. The number of recombinantly expressed genes was 27, and enzyme activity was experimentally confirmed in 19 cases (Table 1). In addition, endogenous OxdSs was obtained from Sclerotinia sclerotiorum (Pedras et al. 2010). So far, three X-ray structures of Oxds, originating from Pseudomonas, Rhodococcus, and Bacillus species, have been determined (Sawai et al. 2009; Nomura et al. 2013; Matsui et al. 2022). The wide occurrence of Oxds is not surprising as some aldoximes are precursors of plant defenses or defense compounds themselves, optionally complexed to glucosinolates or glycosides such as phenylacetaldoxime glucoside (Müller et al. 2024). The identification of Oxds acting on aromatic aldoximes has been challenging. Recently, however, OxdF1 from Pseudomonas putida F1 (Chen et al. 2021), OxdPsp from Pseudomonas sp. (Hinzmann et al. 2023b), and the M29G and F306A variants of OxdRE (Hinzmann et al. 2023a) proved to accept some of these substrates. OxdSs is exceptional in terms of substrate specificity. Out of 14 aldoximes tested, only indolyl-3-acetaldoxime, its analogs derived of propanal and butanal, and 4-hydroxy- and 4-methoxyphenylacetaldoxime (not phenylacetaldoxime) were shown to be substrates (Pedras et al. 2010).
Table 1.
Aldoxime dehydratases investigated at protein level
| Oxd | Origin | Acc. No | Length | Reference |
|---|---|---|---|---|
| OxdA | Pseudomonas chlororaphis B23 | WP_024075760.1b | 352 | Nomura et al. 2013 |
| OxdAAa | Aggregatibacter actinomycetemcomitans RhAA1 | WP_005577064.1 | 234 | Hinzmann et al. 2023b |
| OxdAsp | Aspergillus ibericus CBS 121593 | XP_025572196.1 | 341 | Pei et al. 2023 |
| OxdB | Bacillus sp. OxB-1 | BAA90461.1c | 351 | Matsui et al. 2022 |
| OxdBr1 | Bradyrhizobium sp. LTSPM299 | WP_044589203.1 | 345 | Rädisch et al. 2018 |
| OxdBr2 | Bradyrhizobium sp. WSM1253 | WP_007594278.1 | 351 | Křístková et al. 2023 |
| OxdBTa | Bacteroides thetaiotaomicron | WP_008764895.1 | 131 | Hinzmann et al. 2023b |
| OxdCp | Corynebacterium pacaense Marseille-P2417 | WP_080796375.1 | 356 | Winkler et al. 2023 |
| OxdF1 | Pseudomonas putida F1 | ABQ78858.1 | 352 | Chen et al. 2021 |
| OxdFG | Fusarium graminearum MAFF 305135 | BAE48794.1 | 363 | Kato and Asano 2005 |
| OxdFNn | Fusobacterium nucleatum ATCC 23726 | WP_005902774.1 | 234 | Hinzmann et al. 2023b |
| OxdFv | Fusarium vanettenii 77–13-4 | XP_003042958.1 | 341 | Křístková et al. 2023 |
| OxdHsp | Hydrogenophaga sp. RAC07 | WP_069048334.1 | 347 | Hinzmann et al. 2023b |
| OxdHR | Herbaspirillum rubrisubalbicans M1 | WP_058896488.1 | 349 | Hinzmann et al. 2023b |
| OxdK | Pseudomonas sp. K-9 | BAD98528.1 | 352 | Kato and Asano 2006 |
| OxdLCa | Lactobacillus crispatus | WP_060462053.1 | 220 | Hinzmann et al. 2023b |
| OxdMR | Methylobacillus rhizosphaerae | WP_089375755.1 | 355 | Hinzmann et al. 2023b |
| OxdPsa | Pseudomonas syringae | WP_060413740.1 | 129 | Hinzmann et al. 2023b |
| OxdPsp | Pseudomonas sp. RIT-PI-q | WP_059405603.1 | 346 | Hinzmann et al. 2023b |
| OxdRE | Rhodococcus erythropolis | BAD17969.1d | 353 | Sawai et al. 2009 |
| OxdRG | Rhodococcus globerulus | BAC99076.1 | 353 | Xie et al. 2003 |
| OxdRYH3 | Rhodococcus sp. YH3-3 | WP_064442863.1 | 353 | Kato et al. 1999 |
| OxdSs | Sclerotinia sclerotiorum | XP_001597459.1 | 347 | Pedras et al. 2010 |
| OxdVP | Variovorax paradoxus | WP_047787064.1 | 353 | Hinzmann et al. 2023b |
aActivity detected for pUC18 expression but not pET28a expression
bpdb code 3W08
cpdb code 7F2Y, 72FZ, 7F30 (variant E85A)
dpdb code 3A15-3A18
Note: Hypothetical Oxds from Streptomyces griseoruber, Fusobacterium nucleatum, Bacteroides thetaiotaomicron, and Parabacteroides goldsteinii were expressed, but no activity was detected (Hinzmann et al. 2023b)
Analysis of the evolutionary relationships between the characterized Oxds suggests that fungal and bacterial Oxd have rather different sequence properties, and, moreover, the fungal Oxds seem to have more diversity in active site residues. The characterized fungal Oxds are not only evolutionarily located in a different branch but have a large evolutionary distance from bacterial Oxds (Fig. 3A).
Fig. 3.
Sequence analysis of aldoxime dehydratases investigated at protein level. A Phylogenetic tree constructed using maximum likelihood method and w/freq. model (Jones et al. 1992) in MEGA X (Kumar et al. 2018). The proportion of trees in which the associated taxa clustered together during bootstrap evaluation is shown above the branches. The branch length in the tree is scaled according to the number of substitutions per site. All positions with less than 70% site coverage were eliminated leaving a total of 325 positions in the final dataset. B Sequence similarity network constructed using the online tool (EFI (Enzyme Similarity Tool), https://efi.igb.illinois.edu/efi-est/) with a threshold of 20% sequence identity in the local alignment for the display of connections (Zallot et al. 2019). Data are visualized with Cytoscape (Shannon et al. 2003). The color scheme is the same for A and B and is shown in B
Analysis of the sequence similarity network allowed even better discrimination between Oxds with different active site residues. OxdLC, OxdAA, OxdFNn, OxdPs, and OxdBT (Hinzmann et al. 2023b) are split into separate clades and have less than 20% sequence identity to all other Oxds (missing connections in Fig. 3B). Remarkably, these proteins are significantly shorter (with 129 to 234 amino acid residues) than the “conventional” Oxds with typically about 350 amino acid residues (Table 2). Initial screens with enzymes expressed with a pUC18 vector indicated a certain activity also for the “short” Oxds. However, after subcloning the genes in pET28a, rescreening revealed no Oxd activity in these proteins, suggesting that the above results were false positives.
Table 2.
Determination of aldoxime dehydratase substrates and products (examples)
| Aldoxime | Nitrile | Method | Reference(s) |
|---|---|---|---|
| Aliphatic compounds | |||
| Butyraldoxime | Butyronitrile | GC | Zheng et al. 2022; Hinzmann et al. 2023b |
| Valeraldoxime | Valeronitrile | GC | Křístková et al. 2023 |
| Isovaleraldoxime | Isovaleronitrile | GC | Rädisch et al. 2018 |
| n-Hexanaldoxime | n-Hexanenitrile | GC | Hinzmann et al. 2020a; Zheng et al. 2022 |
| n-Heptanaldoxime | n-Heptanenitrile | GC | Yavuzer et al. 2023; Chen et al. 2021; Zheng et al. 2022 |
| n-Octanaldoxime | n-Octanenitrile | GC | Hinzmann et al. 2020a,b; Hinzmann et al. 2023b; Yavuzer et al. 2023 |
| n-Octanedialdoxime | n-Octanedinitrile | GC | Hinzmann et al. 2020a |
| n-Nonanaldoxime | n-Nonanenitrile | GC | Plass et al. 2019 |
| n-Decanaldoxime | n-Decanenitrile | GC | Hinzmann et al. 2020a |
| n-Dodecanaldoxime | n-Dodecanenitrile | GC | Yavuzer et al. 2023 |
| n-Tetradecanaldoxime | n-Tetradecanenitrile | GC | Yavuzer et al. 2023 |
| Citronellal oxime | Citronellyl nitrile | GC | Zheng et al. 2022; Pei et al. 2023 |
| Alicyclic compounds | |||
| Cyclopentanecarbaldehyde oxime | Cyclopentanecarbonitrile | GC | Hinzmann et al. 2023b |
| Cyclohexanecarbaldehyde oxime | Cyclohexanecarbonitrile | GC | Hinzmann et al. 2023b |
| Arylaliphatic compounds | |||
| Phenylacetaldoxime | Phenylacetonitrile | RP-HPLC, GC | Zheng et al. 2022; Hinzmann et al. 2023b |
| 2-Phenylpropionaldoxime | 2-Phenylpropionitrile | RP-HPLC | Rädisch et al. 2018; Chen et al. 2024 |
| 3-Phenylpropionaldoxime | 3-Phenylpropionitrile | RP-HPLC, NP-SFC, GC | Křístková et al. 2023; Hinzmann et al. 2023b |
| E-Cinnamaldoxime | E-Cinnamonitrile | RP-HPLC | Křístková et al. 2023; Pei et al. 2023 |
| Aromatic compounds | |||
| Benzaldoxime, substituted benzaldoximes | Benzonitrile, substituted benzonitriles | RP-HPLC | Zheng et al. 2022; Xiao et al. 2023 |
| Vanillinoxime | Vanillonitrile | RP-HPLC | Winkler et al. 2023 |
| Heterocyclic compounds | |||
| 2-Furfuraldehyde oxime | 2-Furonitrile | RP-HPLC, GC, UV-spectrometry | Choi et al. 2020; Zheng et al. 2022 |
| 3-Methyl-2-thiophene-carbaldehyde oxime | 3-Methyl-2-thiophene-carbonitrile | RP-HPLC | Zheng et al. 2022 |
| E-Pyridine-3-carbaldehyde oxime | 3-Cyanopyridine | GC, UV-spectrometry | Choi et al. 2020 |
| 4,5-Dihydroisoxazoles | |||
| Benzisoxazoles | Cyanophenoxides | UV-spectrometry | Miao et al. 2017 |
| 5-Phenyl-4,5-dihydroisoxazole and analogs | 3-Hydroxynitriles | Chiral HPLC, chiral GC | Zheng and Asano 2020; Zheng and Asano 2021 |
RP reversed-phase, NP-SFC normal-phase supercritical fluid chromatography
This sequence analysis reveals a lack of information on a relatively large group of Oxds, including those with REV or REE active sites (Fig. 3B), which are widespread in the library of fungal homologs (Křístková et al. 2023). Further experiments will be required to elucidate the relationships between active site residues and substrate specificities in different Oxd clades.
Overproduction and purification
To the best of our knowledge, all recombinant Oxds, either of bacterial or fungal origin, have been overproduced in Escherichia coli hosts. The heterologous expression of oxd genes has often been found challenging. In this respect, low expression levels and inclusion body formation of the target Oxd protein were reported (Choi et al. 2019; Hinzmann et al. 2023b). In order to alleviate these problems, special cloning, cultivation and induction strategies have been devised, such as host-targeted codon optimization of the oxd genes (Hinzmann et al. 2023b; Křístková et al. 2023; Pei et al. 2023), elimination of codon bias using engineered E. coli expression hosts (e.g., BL21-CodonPlus(DE3)-RIL) (Oinuma et al. 2003; Kato and Asano 2006), low temperatures (15–25 °C) (Oinuma et al. 2003; Xie et al. 2003; Kato et al. 2006; Rädisch et al. 2018; Chen et al. 2021), extended cultivation periods of up to 7 days (at 15 °C; Oinuma et al. 2003), and the omission of an external inducer during the protein overproduction phase in connection with T7 promoters and lac operators (leaky expression) (Hinzmann et al. 2023b). Additional strategies to improve the production of active Oxds in recombinant E. coli strains have been reported: lower aeration levels brought about by cultivation in shaken tubes with high culture broth volumes resulting in lower growth rates (Kato et al. 2004; Kato and Asano 2005) and optimization of the position of the His6-tag (Kato and Asano 2006). These are all common strategies for optimized heterologous protein production (Rong et al. 2023). The use of a 5-L reactor with a 3-L working volume enabled to efficiently control the level of dissolved oxygen during the production of whole cells carrying both OxdF1 and nitrile hydratase (NHase) (Zheng et al. 2022). Although Oxds are heme-containing enzymes, their heterologous expression as active enzymes did not so far make use of special E. coli strains engineered for this purpose (Fiege and Frankenberg-Dinkel 2020).
Purification strategies for Oxds have followed the general strategy of using affinity tags attached to recombinant proteins for facilitating protein purification (Mishra 2020). Most popular have been His6-tags in conjunction with immobilized metal ion affinity chromatography using Co2+ or Ni2+ ion-containing resins. Occasionally, a sequence of different chromatographic separation steps with conventional purification media has been used for purifications of both recombinant and non-recombinant Oxds (Oinuma et al. 2003; Xie et al. 2003; Pedras et al. 2010; Nomura et al. 2013). It is noteworthy that partial heme loss has been observed during the purification of some Oxds (Kato et al. 2000; Oinuma et al. 2003; Xie et al. 2003). It is also worth mentioning that many Oxds have been found only barely thermostable (Kato et al. 2004; Křístková et al. 2023; Pei et al. 2023). In this regard, several authors reported instability issues with Oxds (Hinzmann et al. 2020a, 2023b), which was one of the reasons for preferring whole cells to purified enzymes as catalysts in biotransformations (see below).
Reaction conditions
In all Oxds investigated so far, a heme B prosthetic group appears to be the key reaction center, where the aldoxime functionality gets in close contact to the heme iron. According to the proposed reaction mechanisms (Chen et al. 2021; Pei et al. 2023), the heme iron in its ferrous state is essential for the first step of the catalytic reaction. This explains why Oxd activities were often shown to be enhanced under anaerobic and/or reducing conditions. For instance, a marked increase in activity was reported for OxdRG (Xie et al. 2003), OxdF1 (Chen et al. 2021), and OxdA (Zheng and Asano 2020) in the presence of Na2S2O4, while a similar effect was observed with Na2S in OxdRE (Zheng and Asano 2020). Another example is OxdSs with a dramatic increase in its activity in the presence of Na2S2O4 under anaerobic conditions compared with aerobic conditions (Pedras et al. 2010). The addition of iron salts in combination with reducing agents was also reported to have positive effects on Oxd activities (Kato et al. 2004; Křístková et al. 2023). These effects must be taken into account in activity assays of purified Oxds, whereas the activities of whole cells can usually be determined in suitable buffers without additives and under aerobic conditions.
Determination of substrates and products
To monitor the reactions, the concentrations of aldoximes and nitriles were usually determined by HPLC or GC (Table 2). Aliphatic and alicyclic aldoximes and nitriles were generally determined by GC, and GC was also used for some of the arylaliphatic and heterocyclic compounds. Reversed-phase HPLC was suitable to determine most of the arylaliphatic, aromatic, and heterocyclic aldoximes and nitriles. A spectrophotometric method was developed for the determination of the conversion of 2-furfurylaldehyde oxime and E-pyridine-3-carbaldehyde oxime to nitriles (Choi et al. 2020) or benzisoxazoles to nitriles (Miao et al. 2017). Similar methods based on the difference in the spectra of substrate and product can accelerate the monitoring of Oxd-catalyzed reactions.
Catalyst forms
Oxd catalyzed reactions were largely performed with cell suspensions, but immobilized Oxds began to be investigated recently. The first of them were prepared based on whole E. coli cells. For example, E. coli carrying OxdB from Bacillus sp. OxB-1 was immobilized on a highly hydrophilic acrylic acid polymer. The resulting catalyst was functional in cyclohexane (a solvent with a high log P), with an over 99% conversion of 0.5 M n-octanaldoxime (82% isolated yield). The results obtained with other solvents with lower log P values (methyl-tert-butyl ether, toluene, dichloromethane) were inferior. The preference of high log P (highly apolar) solvents in biocatalysis is well-known. The rationale behind this effect is the low tendency of the apolar solvents to strip or “dry-out” (Hinzmann et al. 2019a) water from the enzyme catalyst. The same catalyst was also used to produce a 1-mL packed bed column, and the conversion of 0.1 M n-octanaldoxime was maintained over 95% for 3 h in cyclohexane in flow mode.
However, this catalyst is not appropriate for aqueous media, as this would destabilize the binding of the cells on the support. Therefore, biocatalysts suitable for aqueous environments were produced by immobilizing the cells in calcium alginate. The catalyst was optionally coated with tetraethyl orthosilicate to increase its hydrophobicity, which was important for the affinity of the substrate to the carrier. This immobilization increased the stability of OxdB whole-cell catalyst in 10% ethanol, and the immobilizate was used three times for the dehydration of 100 mM n-octanaldoxime with 70–88% conversion (Hinzmann et al. 2020b).
Other immobilized Oxds were prepared from purified enzymes. The enzymes were bound on the supports by hydrophobic, covalent (Hinzmann et al. 2020b), and affinity interactions (Křístková et al. 2024). Almost all the protein applied was bound on the supports (Hinzmann et al. 2020b), but the residual activity was found to be insufficient. OxdRE retained 15–20% activity and OxdB 10% activity on a hydrophobic or an amino (glutaraldehyde-activated) support. The stability of the immobilizates in the presence of acetonitrile, methanol, or dimethyl sulfoxide (20% each) was also unsatisfactory, the activity already decreasing after 15-30 min (Hinzmann et al. 2020b), which suggested to focus on whole-cell immobilizates (see above). In another study, OxdFv and OxdBr1 were bound to Ni–NTA through their N-terminal His6-tags. The residual activity was not determined, but a complete conversion of 5–15 mM phenylacetaldoxime or 5 mM cinnamaldoxime was achieved with the immobilizates, which were also recyclable (at least 22 times for OxdBr1). Moreover, this immobilization method did not require a prior purification but could also be carried out with cell-free extracts, while immobilization was combined with partial purification (Křístková et al. 2024). Similarly, immobilized OxdPsp was prepared by combining protein separation using the aqueous two-phase system (ATPS) with enzyme adsorption on a macroporous resin. The immobilizate was used for the enantioselective dehydration of (E)-2-phenylpropionaldoxime to the corresponding S-nitrile with 94% e.e. and was at least three times recyclable (Chen et al. 2024). In this and similar reactions of chiral aldoximes (Gröger and Asano 2020; Domínguez de María 2021), the E- vs. Z-configuration of the substrate is decisive for enantioselectivity. The hypothesis that the size of the cavity in the active site of Oxds plays a key role was confirmed by the study of mutants with reduced cavity size and improved enantioselectivity (Yavuzer et al. 2021).
Cascade reactions
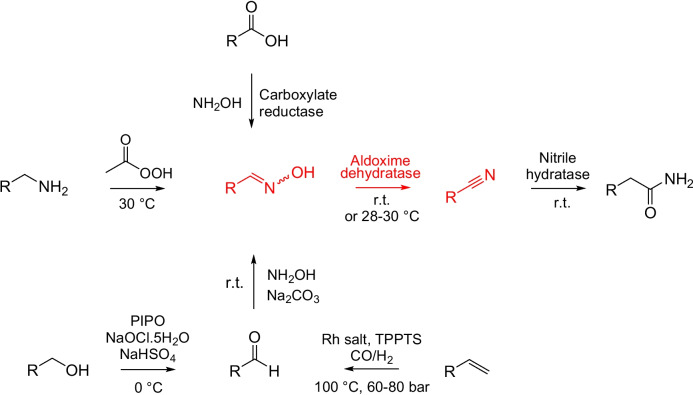
Oxds were integrated in several multistep reactions, where the aldoximes were obtained from alkenes (Plass et al. 2019; Terhorst et al. 2020), alcohols (Hinzmann et al. 2020a), aldehydes (Zheng et al. 2022), carboxylic acids (Horvat et al. 2022; Winkler et al. 2023), or amines (Xiao et al. 2023) (Fig. 4).
Fig. 4.
Multistep reactions of diverse precursors to nitriles. Aldoxime dehydratase is used in the oxime-to-nitrile step (Plass et al. 2019; Hinzmann et al. 2020a; Zheng et al. 2022; Winkler et al. 2023; Horvat et al. 2022; Xiao et al. 2023; Terhorst et al. 2020) (in red). Nitriles can be directly converted to amides (Zheng et al. 2022). PIPO, polymer-immobilized TEMPO (2,2,6,6-tetramethylpiperidinyloxy); TPPTS, (tris(3-sulfophenyl)phosphine trisodium salt); r.t., room temperature
Synthesis of nitriles and amides from aldehydes
The synthesis of nitriles from aldehydes via oximes was developed early during the investigation of Oxds. The oximes prepared by condensation of aldehydes and hydroxylamine were isolated and used for the next step catalyzed by whole cells of Rhodococcus sp. YH3-3, in which the NHase was inactivated with a combination of 5 mM of 2-mercaptoethanol and 1 mM of DTT at 40 °C (Kato et al. 1999) or E. coli harboring the oxd gene from Bacillus sp. OxB-1 (Xie et al. 2001). Phenylacetonitrile was produced from 500 mM substrate (aldoxime) with 89% isolated yield, 3-phenylpropionitrile from 750 mM substrate with 90% isolated yield, and several aliphatic C4-C6 nitriles from 100 to 300 mM substrate concentration with up to 100% conversion and 46–56% isolated yields, within 2–20 h (Xie et al. 2001). Thus, the space-time yields (STYs) reached up to 10 g/L/h. In addition, 3-cyanopyridine and 2-furonitrile were prepared from 50 and 100 mM substrates with 98% and 62% isolated yields within 105 and 75 min, which corresponds to 2.92 and 4.62 g/L/h STY, respectively (Kato et al. 1999). A single point mutation (N266S) in OxdRYH3 increased the enzyme‘s potential for the production of 2-furonitrile increasing its specific activity for 50–100 mM substrate several times (Choi et al. 2020).
Recently, the synthesis of cinnamonitrile and citronellyl nitrile from the corresponding aldehydes was demonstrated using a new Oxd from Aspergillus ibericus (enzyme OxdAsp) (Pei et al. 2023). Both aldoxime synthesis and dehydration proceeded under mild conditions, with high concentrations of substrates (1 M and 100–200 mM, respectively). The aldoxime to nitrile reactions proceeded with an almost full conversion (Fig. 5A), providing 2.58 g/L/h and 7.56 g/L/h STY for cinnamonitrile and citronellyl nitrile, respectively.
Fig. 5.
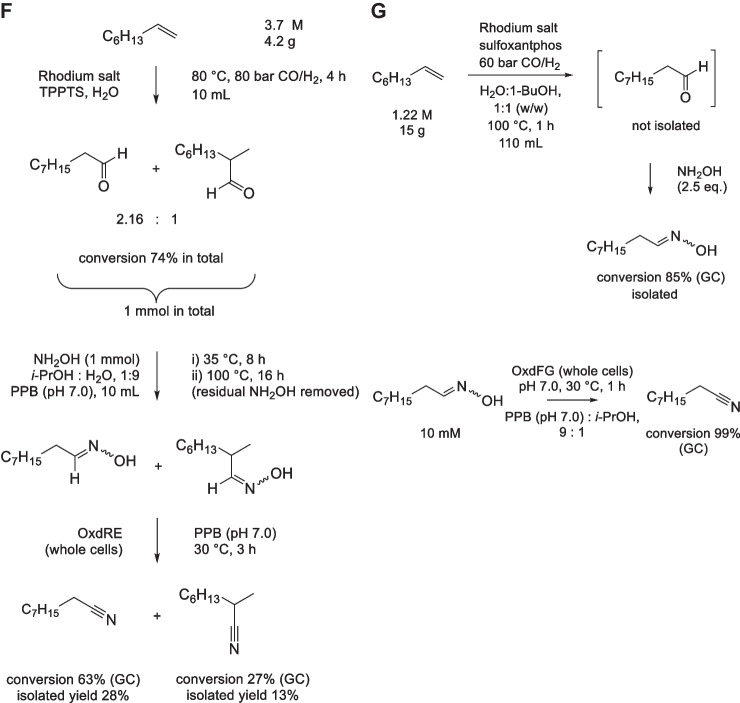
Chemoenzymatic syntheses of (aryl)aliphatic nitriles from A aldehydes, B carboxylic acids, C, D alcohols, E dialcohols, and F, G alkenes. A E-3-Phenylprop-2-enal (cinnamaldehyde) and 3,7-dimethyloct-6-enal (citronellal) were transformed to aldoximes with hydroxylamine and isolated. E,Z-Cinnamaldoxime and E,Z-citronellal oxime thus obtained were transformed to nitriles with whole cells (30 mg wet cells/mL) of Escherichia coli carrying the enzyme OxdAsp from Aspergillus ibericus (Pei et al. 2023). B Phenylacetic acid was reduced by carboxylate reductase NcCAR from Neurospora crassa (E. coli cells, ≈6 mg dry cells/mL), and the crude product was then transformed by aldoxime dehydratase OxdBr1 from Bradyrhizobium sp. (E. coli cells, ≈3 mg dry cells/mL) (Winkler et al. 2023). D–F Aliphatic mononitriles were synthesized from the corresponding alcohols (C) with or (D) without intermediate (aldoxime) isolation (differences highlighted in red). E An analogous route to n-octanedinitrile was performed with aldoxime isolation. The aldoximes were transformed by aldoxime dehydratase OxdB from Bacillus sp. OxB-1. The catalyst was E. coli wet whole cells in free form (33 mg wet cell/mL) or immobilized form (Hinzmann et al. 2020a). The latter was based on whole cells adsorbed to an acrylic acid resin according to a previous study (Hinzmann et al. 2019a). F n-Nonanal and 2-methyloctanal were prepared by hydroformylation. An aliquot of the product (1 mmol) was taken for condensation with hydroxylamine followed by enzymatic dehydration with aldoxime dehydratase OxdRE from Rhodococcus erythropolis (E. coli cells, 50 mg wet weight/mL) (Plass et al. 2019). G Nonanal oxime was prepared by hydroformylation and aldoxime formation in “one pot.” The product was dehydrated using aldoxime dehydratase OxdFG from Fusarium graminearum (E. coli cells, 50 mg wet weight/mL) (Terhorst et al. 2020). PPB, potassium phosphate buffer; MES, 2-(N-morpholino)ethanesulfonic acid buffer; PIPO, polymer-immobilized TEMPO (2,2,6,6-tetramethylpiperidinyloxy); r.t., room temperature; TPPTS, (tris(3-sulfophenyl)phosphine trisodium salt)
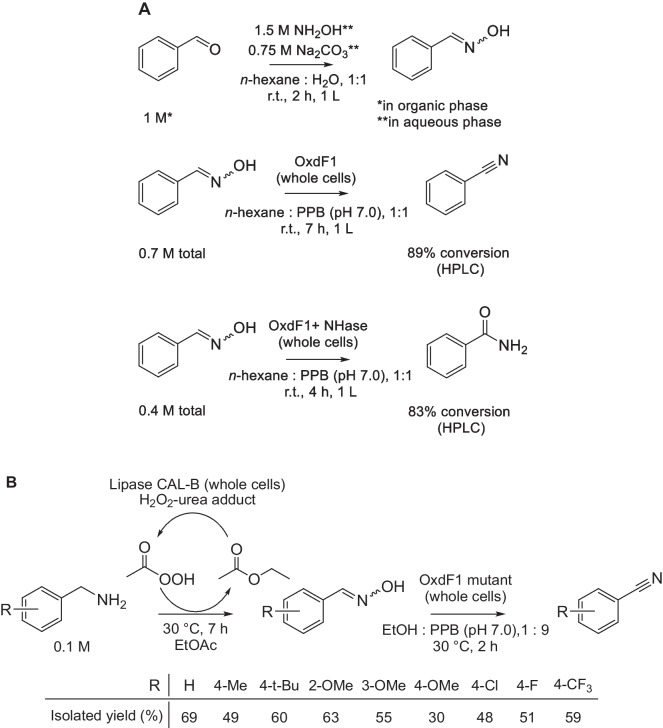
Nitriles were also prepared from a variety of aromatic and aliphatic aldehydes without purifying the oximes (Zheng et al. 2022). OxdF1 was suitable to dehydrate all the oximes. Optionally, the nitriles were then hydrated to amides using a NHase from Aurantimonas manganoxydans. The amides were largely obtained with satisfactory (ca. 40–70%) isolated yields. In addition, the syntheses of benzonitrile and benzamide from benzaldoxime were carried out on a 1-L scale with STYs of 9-10 g/L/h (Fig. 6A).
Fig. 6.
Chemoenzymatic syntheses of aromatic nitriles from A benzaldehyde and B benzylamines. A Benzaldehyde was transformed to benzaldoxime with hydroxylamine, and the organic phase containing the product was directly used for the transformations to benzonitrile with E. coli whole cells (30 g dry cells/L) carrying aldoxime dehydratase OxdF1, or to benzamide with E. coli whole cells (30 g dry cells/L) carrying OxdF1 and nitrile hydratase (NHase). The enzymatic reactions were carried out in fed-batch mode (Zheng et al. 2022). B Ethylacetate (solvent) was transformed by lipase B from Candida antarctica (CAL-B; E. coli lyophilized cells, 6 mg/mL) to peracetic acid which reacted with benzylamines to afford benzaldoximes. These were transformed to benzonitriles by aldoxime dehydratase from Pseudomonas putida F1—mutant OxdF1 L318F/F306Y (E. coli cells, 33 mg wet weight/mL). The nitrile yield was calculated per benzylamine (Xiao et al. 2023). PPB, potassium phosphate buffer; r.t., room temperature
Synthesis of nitriles from carboxylic acids
Carboxylic acids are attractive substrates due to their availability from sustainable resources and stability in comparison to reactive aldehydes. To obtain aldoximes, they were reduced by carboxylate reductase (CAR), and the resulting aldehydes were reacted in situ with hydroxylamine. CARs accept a broad variety of substrates (Winkler and Ling 2022), including (aryl)aliphatic and aromatic substrates (Horvat and Winkler 2020), but not substrates with substitutions in vicinity of the carboxylic acid group. Oxds are highly efficient in dehydration of (aryl)aliphatic compounds, but their ability to transform aromatic aldoximes is limited. Therefore, the choice of a compatible CAR and Oxd for a certain reaction can be challenging.
A proof of concept was obtained for the synthesis of n-hexanenitrile (capronitrile) and phenylacetonitrile, on an analytical scale (Horvat et al. 2022). Several potential substrates were then tested with panels of CARs and Oxds on an analytical scale. Suitable CAR-Oxd combinations were found for most of the carboxylic acids investigated (butyric, valeric, caproic, benzoic, phenylacetic, and 3-phenylpropionic acid). The conversions (determined by HPLC or GC) varied from 80 to >99%. However, an over-reduction of some carboxylic acids to alcohols was observed. On a preparative scale, a sequential one-pot cascade reaction was carried out for the synthesis of phenylacetonitrile (874 mg) with an isolated yield of 78% (Winkler et al. 2023) (Fig. 5B). The by-product alcohol was also found in this case.
Synthesis of nitriles from alcohols
The route from renewable carboxylic acids to nitriles can also occur via alcohols (Hinzmann et al. 2020a) that are obtained from the acids, e.g. by catalytic hydrogenation. A multistep process from alcohols to nitriles was proposed that involved oxidation of the alcohol to the aldehyde with a polymer-immobilized TEMPO catalyst (PIPO), condensation of the aldehyde with hydroxylamine, and enzymatic dehydration of the resulting aldoxime. Proof of concept was established for n-hexanenitrile, n-octanenitrile, and n-decanenitrile, which were produced in 50-64% overall yield, while the aldoxime intermediates were isolated (Fig. 5C). The process was then optimized to be run without intermediate isolation (Fig. 5D). Thus, the organic phase from the synthesis of aldoxime was directly used for the enzymatic step, which could be performed not only with the immobilized Oxd catalyst which is resistant to organic solvent (see above) but also with a whole-cell suspension, provided that the target nitrile was also used as solvent. Analogously, n-octanedinitrile (suberonitrile) was synthesized (Fig. 5E), but the route without aldoxime isolation was not possible in this case: the use of n-octanedinitrile as solvent resulted in precipitation of the oxime intermediate (Hinzmann et al. 2020a).
Synthesis of nitriles from alkenes
The first step in the three-step process from alkene to nitrile is hydroformylation, which is well developed, but the drawback of which is the formation of isomers (Fig. 5F). The next two steps are the same as in the above multistep processes, i.e., condensation of the aldehyde with hydroxylamine followed by the enzymatic dehydration of aldoxime (Fig. 5F). The multistep reaction was demonstrated for 1-octene with n-nonanenitrile as the final product in a 28% overall yield and iso-nonanenitrile as a side product in a 13% overall yield (Plass et al. 2019).
Also one-pot formation of aldoxime from an alkene with subsequent synthesis of nitrile was reported (Terhorst et al. 2020). The optimization of reaction conditions enabled to combine the hydroformylation step with the aldehyde–hydroxylamine condensation step without isolating the aldehyde. The process was primarily demonstrated for the synthesis of n-nonanenitrile obtained with an overall yield of 85% (Fig. 5G) and a 95% selectivity for the targeted (linear) isomer. In addition, the use of the same reaction sequence for the synthesis of other (aliphatic, arylaliphatic) nitriles was also proposed, using various Oxds (Terhorst et al. 2020).
Synthesis of nitriles from benzylamines
A route from amines to nitriles was shown for substituted benzylamines (Xiao et al. 2023). In the first step, the benzylamines were chemically oxidized to aldoximes with peracetic acid. This oxidant was produced from ethyl acetate in situ with a lipase catalyst, according to an earlier work (Méndez-Sánchez et al. 2017). At the same time, ethyl acetate served as the solvent. Amylacetate could be used analogously. The intermediate benzaldoxime was isolated by phase separation. The second step was carried out with a whole-cell catalyst based on a mutant of OxdF1 with improved kinetic behavior. Benzonitriles and substituted derivatives were obtained from 100 mM substrates largely in good isolated yields (Fig. 6B) and up to over 1 g/L/h STY.
Synthesis of nitriles from dihydroisoxazoles
Remarkably, also dihydroisoxazoles are Oxd substrates, as first shown for 1,2-benzisoxazole and 5-nitro-1,2-benzisoxazole, and undergo ring opening (a Kemp elimination reaction) in the active center of Oxds (Miao et al. 2017). This was used for the preparation of both enantiomers of synthetically useful β-hydroxynitriles such as 3-hydroxy-3-phenylpropionitrile (Fig. 7A). An asymmetric ring opening catalyzed by OxdB provided both the nitrile product and the unreacted dihydroisoxazole in excellent e.e. The unreacted substrate was converted to the corresponding nitrile under alkaline conditions in the subsequent step (Zheng and Asano 2020). An analogous approach was used for the synthesis of both enantiomers of 4-chloro-3-hydroxybutanenitrile (Fig. 7B) as precursors of l-carnitine and Atorvastatin (Zheng and Asano 2021). The dihydroisoxazoles were readily available starting from alkenes (Zheng and Asano 2020, 2021).
Fig. 7.
Synthesis of enantiopure β-hydroxynitriles from dihydroisoxazoles. A Synthesis of (R)- and (S)-3-hydroxy-3-phenylpropionitrile from (R,S)-5-phenyl-4,5-dihydroisoxazole (Zheng and Asano 2020). The first step was catalyzed by a semi-purified aldoxime dehydratase OxdB from Bacillus sp. (10 U/mL). B Synthesis of (R)- and (S)-4-chloro-3-hydroxybutanenitrile from (R,S)-5-(chloromethyl)-4,5-dihydroisoxazole (Zheng and Asano 2021). The first step was catalyzed by a purified aldoxime dehydratase OxdA-L318I (6.25 U/mL). This mutant exhibited an increased enantioselectivity for the substrate. The second step was catalyzed by the wild-type OxdA from Pseudomonas chlororaphis (62.5 U/mL). The nitrile yields were calculated per dihydroisoxazoles. PPB, potassium phosphate buffer; r.t., room temperature
In silico cascade design
As demonstrated in the previous chapters, Oxds can be integrated into enzymatic and chemoenzymatic multi-step reactions. Particularly the solvent tolerance of whole-cell catalysts harboring Oxds make them highly attractive for integration in chemoenzymatic routes where the chemical step requires non-aqueous or micro-aqueous conditions. For an efficient cascade design, databases such as the RetroBioCat database (available at retrobiocat.com) may be very useful: A target molecule can be dissected to precursor molecules and both biocatalytic and chemical functional group transformations are included. In frame of this review, we have curated data on enzymatic oxime dehydration from across the literature in the RetroBioCat database (Finnigan et al. 2023; RetroBioCat; A collection of tools for automated biocatalytic cascade design, Version: v2023.11.30, https://retrobiocat.com), allowing this information to be interactively explored and interrogated.
Comparison of aldoxime dehydratase-catalyzed processes and other innovative approaches
Several of the processes mentioned above appear to be particularly promising for future transfer to the chemical industry. In particular, the syntheses of cinnamonitrile, citronellyl nitrile, substituted benzonitriles, nonanenitriles, or n-octanedinitrile (see above), as well as the syntheses of short chain aliphatic nitriles, phenylacetonitrile, 3-phenylpropionitrile, and heterocyclic nitriles described earlier (Xie et al. 2001; Kato et al. 1999) seem to be practicable mainly due to substrate concentrations that live up to industrial metrics (Table 3). In this section, we compare some of these enzymatic syntheses with alternative routes to the same nitriles to illustrate the advantages and disadvantages of the different approaches.
Table 3.
Examples of nitriles synthesized with aldoxime dehydratases
| Product | Substrate (mM) | Scale (L) | Conversion (%) | Isolated yield (%) | Reference(s) |
|---|---|---|---|---|---|
| Phenylacetonitrile | Z-Phenylacetaldoxime (500) | 0.1 | 100 | 89 | Xie et al. 2001 |
| Phenylacetic acid (10) | 1 | 83 | 78 | Horvat et al. 2022 | |
| 3-Phenylpropionitrile | Z-3-Phenylpropionaldoxime (750) | 0.1 | 99.5 | 90 | Xie et al. 2001 |
| S-2-Phenylpropionitrile | E-2-Phenylpropionaldoxime (50) | 0.006 | 98.6 | 95.7 | Chen et al. 2024 |
| 3-Hydroxy-3-phenylpropionitrile | 5-Phenyl-4,5-dihydroisoxazole (100) | 0.05 | n.d | 42 (R-nitrile); 46 (S-nitrile) | Zheng et al. 2020 |
| n-Butyronitrile | E/Z-Butyraldoxime (100) | 0.1 | 100 | 46 | Xie et al. 2001 |
| n-Valeronitrile | n-Valeraldoxime (250) | 0.1 | 100 | 53 | Xie et al. 2001 |
| Isovaleronitrile | Isovaleraldoxime (200) | 0.1 | 99.6 | 50 | Xie et al. 2001 |
| n-Hexanenitrile | n-Hexanaldoxime (300) | 0.1 | 99.5 | 56 | Xie et al. 2001 |
| 1-Hexanol (1000) | ≈ 0.03 | ≈ 89 | 60 | Hinzmann et al. 2020a | |
| n-Octanenitrile | n-Octanaldoxime (6982) | 0.25 | > 99 | 86 | Hinzmann et al. 2019b |
| 1-Octanol (1000) | ≈ 0.03 | ≈ 92 | 71 | Hinzmann et al. 2020a | |
| n-Octanedinitrile | n-Octandiol (500) | ≈ 0.015 | n.d. | 61 | Hinzmann et al. 2020a |
| n-Nonanenitrile | n-Octene (3823) | ≈ 0.01 | 46 | 28 | Plass et al. 2019 |
| n-Decanenitrile | 1-Decanol (1000) | ≈ 0.03 | ≈ 90 | 63 | Hinzmann et al. 2020a |
| n-Dodecanaldoxime | n-Dodecanenitrile (1000) | ≈ 0.01 | 99 | 65 | Yavuzer et al. 2023 |
| n-Tetradecanaldoxime | n-Tetradecanenitrile (250) | ≈ 0.01 | > 99 | 89 | Yavuzer et al. 2023 |
| n-Hexadecanaldoxime | n-Hexadecanenitrile (250) | ≈ 0.01 | 54 | n.d | Yavuzer et al. 2023 |
| Citronellyl nitrile | E,Z-Citronellal oxime (200) | ≈ 0.05 | > 99 | n.d | Pei et al. 2023 |
| E-Cinnamonitrile | E,Z-Cinnamaldoxime (100) | ≈ 0.05 | > 99 | n.d | Pei et al. 2023 |
| Benzonitrile, substituted benzonitriles | Benzaldehyde, substituted benzaldehydes (100)a | 0.004–1 | > 99 | 40–71 | Zheng et al. 2022 |
| Benzylamine, substituted benzylamines (100) | ≈ 0.01 | 40–76 | 30–69 | Xiao et al. 2023 | |
| 3-Cyanopyridine | E-Pyridine-3-carbaldehyde oxime (50) | 0.3 | n.d | 98 | Kato et al. 1999 |
| 2-Furonitrile | E-2-Furfurylaldoxime (100) | 0.175 | n.d | 62 | Kato et al. 1999 |
aUp to 700 mM for benzaldehyde in fed-batch mode
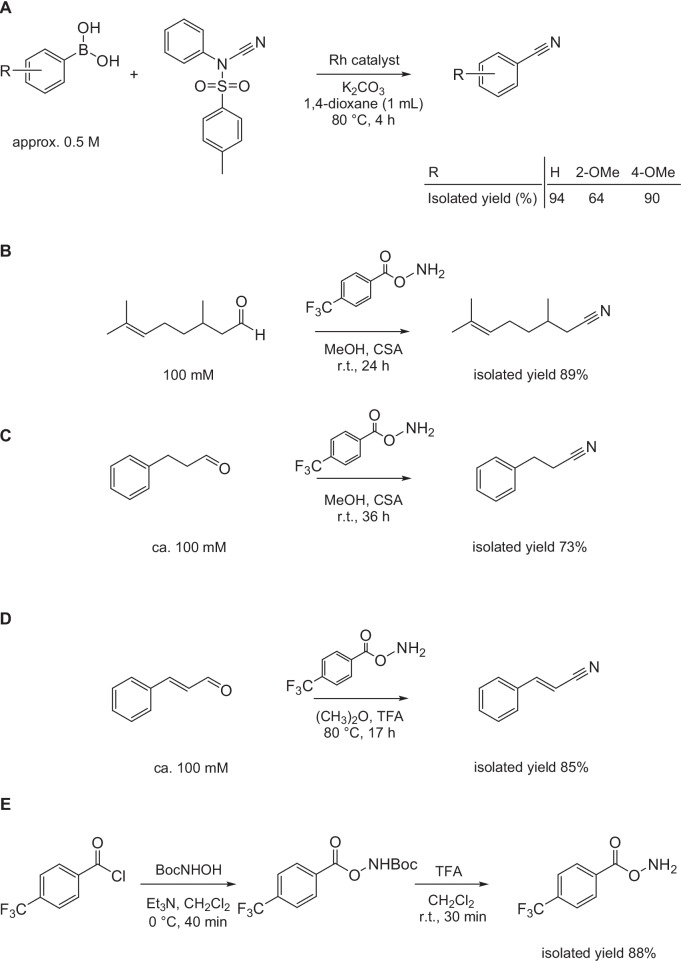
An innovation in nitrile synthesis (Anbarasan et al. 2011) is based on the cyanation approach where the conventional cyanation agents (see above) are replaced with the less toxic N-cyano-N-phenyl-p-toluenesulfonamide (NCTS) which is more eco-friendly and whose synthesis does not require a cyanide compound (Su et al. 2015; Li et al. 2016). The cyanation reaction using NCTS can be catalyzed by, e.g., a rhodium (III) complex, and the starting compounds are various boronic acids (Anbarasan et al. 2011; Soumya et al. 2021), heterocycles, or alkenes (Soumya et al. 2021) resulting in a vast number of potentially accessible nitriles. For example, boronic acids were converted to benzonitriles and substituted benzonitriles. The syntheses of products that were also prepared by the chemoenzymatic cascade explained above (Fig. 6B) are shown in (Fig. 8A). The boronic acids were used in higher concentrations than the amines, while the reaction time was longer, but the yields largely higher. Nevertheless, the chemical method requires a metal catalyst, a number of chemicals, and an elevated temperature.
Fig. 8.
A Syntheses of A benzonitrile and substituted benzonitriles (Anbarasan et al. 2011), B citronellyl nitrile, C 3-phenylpropionitrile, and D cinnamonitrile (An and Yu 2015a). E Synthesis of O-(4-CF3-benzoyl)-hydroxylamine used in reactions (B–D) (An and Yu 2015b). CSA, L-(-)-camphorsulfonic acid; r.t., room temperature; TFA, trifluoroacetic acid
Another innovative approach was used to produce citronellyl nitrile. The reaction starts from the corresponding aldehyde and uses O-(4-CF3-benzoyl)-hydroxylamine (CF3-BHA) as the nitrogen source and l-(-)-camphorsulfonic acid as the catalyst (Fig. 8B). An analogous route was applied to the synthesis of a number of other nitriles, including 3-phenylpropionitrile (Fig. 8C) and cinnamonitrile (Fig. 8D) (An and Yu 2015a). This innovative nitrile synthesis proceeds in one step with high yields, which is advantageous, but necessitates a prior two-step synthesis of CF3-BHA from 4-trifluoromethylbenzoyl chloride and tert-butyl N-hydroxycarbamate (An and Yu 2015b) (Fig. 8E). The enzymatic processes (Table 3) work with similar substrate concentrations and similar yields, but they are faster and use much less chemicals and organic solvents.
The critical point with the enzymatic processes is the cost of producing the catalyst. This factor cannot be currently assessed satisfactorily as a scale-up of Oxd catalyst production to more than a few liter scale is not yet realized. Nevertheless, it is justified to assume that Oxds can be produced at reasonable costs like many other industrially important enzymes.
Conclusions
Nitrile-synthesizing enzymes clearly have an industrial potential, and, among them, Oxds are probably the most versatile. Particularly in the last 5 years, the spectrum of Oxds was significantly expanded, including Oxds with a different active site structure and diverging substrate specificities than the first Oxds discovered about 25 years ago. However, it is uncertain whether additional database searches will yield new Oxds that would significantly surpass the current ones. Although there are plethora of uncharacterized Oxds, these are also clusters of very similar enzymes. Moreover, evolutionarily distant Oxds often have similar substrate specificities. Therefore, a semi-rational design of mutants can be a more appropriate strategy and has already been applied to Oxds. Recently, the first functional immobilized Oxd were produced, which represents an additional strategy to improve the catalyst. Moreover, aldoxime dehydration by Oxds has been newly combined with a number of chemical steps, expanding the range of starting materials or allowing the nitrile products to be directly converted in cascade reactions. Some of the reactions are approaching a significant level of technological maturity. We speculate that scaling up the processes may lead to industrial applications in a number of cases. There have also been innovations in non-enzymatic nitrile synthesis, and some examples have been mentioned in this review. The decision on the choice of process must be based on a careful comparison of the advantages and disadvantages of each route.
Author contribution
LM: Original draft preparation; writing—reviewing and editing. MK: Original draft preparation, especially the “Overproduction and purification” section; writing—reviewing and editing. NK: Sequence analysis; writing—reviewing and editing. BK: Overview of enzyme assays; writing—reviewing and editing. KŠ: Overview of nitrile syntheses from alkenes; writing—reviewing and editing. MW: Original draft preparation, especially the “Sequence diversity” and “In silico cascade design” sections; writing, reviewing and editing; funding acquisition.
Funding
This work was supported by the Czech Science Foundation [grant number GF20‐23532L], Ministry of Education, Youth and Sports of the Czech Republic grant Talking microbes-understanding microbial interactions within One Health framework [CZ.02.01.01/00/22_008/0004597] and Czech Academy of Sciences [grant number RVO61388971]. This research was funded in part by the Austrian Science Fund (FWF) [Grant-DOI 10.55776/I 4607]. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Open Access
For the purpose of open access, the authors have applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ludmila Martínková, Email: martinko@biomed.cas.cz.
Margit Winkler, Email: margit.winkler@tugraz.at.
References
- Akhtar R, Zahoor AF, Rasool N, Ahmad M, Ali KG (2022) Recent trends in the chemistry of Sandmeyer reaction: a review. Mol. Divers. 26:1837–1873. 10.1007/s11030-021-10295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An XD, Yu SY (2015a) Direct synthesis of nitriles from aldehydes using an O-benzoyl hydroxylamine (BHA) as the nitrogen source. Org Lett 17:5064–5067. 10.1021/acs.orglett.5b02547 [DOI] [PubMed] [Google Scholar]
- An XD, Yu SY (2015b) Visible-light-promoted and one-pot synthesis of phenanthridines and quinolines from aldehydes and O-acyl hydroxylamine. Org Lett 17:2692–2695. 10.1021/acs.orglett.5b01096 [DOI] [PubMed] [Google Scholar]
- Anbarasan P, Neumann H, Beller M (2011) A general rhodium-catalyzed cyanation of aryl and alkenyl boronic acids. Angew Chem Int Ed 50:519–522. 10.1002/anie.201006044 [DOI] [PubMed] [Google Scholar]
- Betke T, Higuchi J, Rommelmann P, Oike K, Nomura T, Kato Y, Asano Y, Gröger H (2018) Biocatalytic synthesis of nitriles through dehydration of aldoximes: the substrate scope of aldoxime dehydratases. ChemBioChem 19:768–779. 10.1002/cbic.201700571 [DOI] [PubMed] [Google Scholar]
- Bhalla TC, Kumar V, Kumar V, Thakur N, Savitri (2018) Nitrile metabolizing enzymes in biocatalysis and biotransformation. Appl Biochem Biotechnol 185:925–946. 10.1007/s12010-018-2705-7 [DOI] [PubMed] [Google Scholar]
- Chen K (2021) Recent progress on discovery and research of aldoxime dehydratases. Green Synth Catal 2:179–186. 10.1016/j.gresc.2021.04.001 [Google Scholar]
- Chen ZJ, Mao FY, Zheng HT, Xiao QJ, Ding ZH, Wang AM, Pei XL (2021) Cyanide-free synthesis of aromatic nitriles from aldoximes: discovery and application of a novel heme-containing aldoxime dehydratase. Enzyme Microb Technol 150:109883. 10.1016/j.enzmictec.2021.109883 [DOI] [PubMed] [Google Scholar]
- Chen JR, Zhang YL, Zhang XY, Wen SY, Qiao M, Liu JH, Zhang YY (2024) Kinetic model of asymmetric dehydration of aldoxime catalyzed by immobilized OxdPsp in an organic solvent. Green Chem 26:4065–4073. 10.1039/d4gc00263f [Google Scholar]
- Choi JE, Shinoda S, Inoue R, Zheng DJ, Gröger H, Asano Y (2019) Cyanide-free synthesis of an aromatic nitrile from a biorenewable-based aldoxime: development and application of a recombinant aldoxime dehydratase as a biocatalyst. Biocatal Biotransform 37:414–420. 10.1080/10242422.2019.1591376 [Google Scholar]
- Choi JE, Shinoda S, Asano Y, Gröger H (2020) Aldoxime dehydratase mutants as improved biocatalysts for a sustainable synthesis of biorenewables-based 2-furonitrile. Catalysts 10:362. 10.3390/catal10040362 [Google Scholar]
- Domínguez de María P (2021) Nitrile synthesis with aldoxime dehydratases: a biocatalytic platform with applications in asymmetric synthesis, bulk chemicals, and biorefineries. Molecules 26:4466. 10.3390/molecules26154466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiege K, Frankenberg-Dinkel N (2020) Construction of a new T7 promoter compatible Escherichia coli Nissle 1917 strain for recombinant production of heme‑dependent proteins. Microb Cell Fact 19:190. 10.1186/s12934-020-01447-5 [DOI] [PMC free article] [PubMed]
- Finnigan W, Lubberink M, Hepworth LJ, Citoler J, Mattey AP, Ford GJ, Sangster J, Cosgrove SC, da Costa BZ, Heath RS, Thorpe TW, Yu YQ, Flitsch SL, Turner NJ (2023) RetroBioCat database: a platform for collaborative curation and automated meta-analysis of biocatalysis data. ACS Catal 13:11771–11780. 10.1021/acscatal.3c01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan M, Nagaraaj P (2020) Recent developments in dehydration of primary amides to nitriles. Org Chem Front 7:3792–3814. 10.1039/d0qo00843e [Google Scholar]
- Gröger H, Asano Y (2020) Cyanide-free enantioselective catalytic strategies for the synthesis of chiral nitriles. J Org Chem 85:6243–6251. 10.1021/acs.joc.9b02773 [DOI] [PubMed] [Google Scholar]
- Hinzmann A, Adebar N, Betke T, Leppin M, Gröger H (2019a) Biotransformations in pure organic medium: organic solvent-labile enzymes in the batch and flow synthesis of nitriles. Eur J Org Chem 2019:6911–6916. 10.1002/ejoc.201901168 [Google Scholar]
- Hinzmann A, Glinski S, Worm M, Gröger H (2019b) Enzymatic synthesis of aliphatic nitriles at a substrate loading of up to 1.4 kg/L: a biocatalytic record achieved with a heme protein. J Org Chem 84:4867–4872. 10.1021/acs.joc.9b00184 [DOI] [PubMed] [Google Scholar]
- Hinzmann A, Stricker M, Gröger H (2020a) Chemoenzymatic cascades toward aliphatic nitriles starting from biorenewable feedstocks. ACS Sustain Chem Eng 8:17088–17096. 10.1021/acssuschemeng.0c04981 [Google Scholar]
- Hinzmann A, Stricker M, Gröger H (2020b) Immobilization of aldoxime dehydratases and their use as biocatalysts in aqueous reaction media. Catalysts 10:1073. 10.3390/catal10091073 [Google Scholar]
- Hinzmann A, Betke T, Asano Y, Gröger H (2021) Synthetic processes toward nitriles without the use of cyanide: a biocatalytic concept based on dehydration of aldoximes in water. Chemistry 27:5313–5321. 10.1002/chem.202001647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinzmann M, Yavuzer H, Hinzmann A, Gröger H (2023b) Database-driven in silico-identification and characterization of novel aldoxime dehydratases. J Biotechnol 367:81–88. 10.1016/j.jbiotec.2023.02.007 [DOI] [PubMed] [Google Scholar]
- Hinzmann M, Yavuzer H, Bittmann M, Gröger H (2023a) Novel approach toward industrial aromatic nitriles via biocatalysis: from rational enzyme design to sustainable, energy-saving technology platform. Chem Catalysis 3: 100572. 10.1016/j.checat.2023.100572
- Holmes IP, Kagan HB (2000a) The asymmetric addition of trimethylsilylcyanide to aldehydes catalysed by anionic chiral nucleophiles. - Part 1. Tetrahedron Lett 41:7453–7456. 10.1016/s0040-4039(00)01275-2 [Google Scholar]
- Holmes IP, Kagan HB (2000b) The asymmetric addition of trimethylsilylcyanide to aldehydes catalysed by anionic chiral nucleophiles. Part 2. Tetrahedron Lett 41:7457–7460. 10.1016/s0040-4039(00)01276-4 [Google Scholar]
- Horvat M, Winkler M (2020) In vivo reduction of medium- to long-chain fatty acids by carboxylic acid reductase (CAR) enzymes: limitations and solutions. ChemCatChem 12:5076–5090. 10.1002/cctc.202000895 [Google Scholar]
- Horvat M, Weilch V, Rädisch R, Hecko S, Schiefer A, Rudroff F, Wilding B, Klempier N, Pátek M, Martínková L, Winkler M (2022) Chemoenzymatic one-pot reaction from carboxylic acid to nitrile via oxime. Catal Sci Technol 12:62–66. 10.1039/d1cy01694f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Clavijo McCormick A, Gunther J, Schmidt A, Boeckler GA, Gershenzon J, Unsicker SB, Köllner TG (2014) Herbivore-induced poplar cytochrome P450 enzymes of the CYP71 family convert aldoximes to nitriles which repel a generalist caterpillar. Plant J 80:1095–1107. 10.1111/tpj.12711 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Zeltner P, Handrick V, Gershenzon J, Köllner TG (2015) The maize cytochrome P450 CYP79A61 produces phenylacetaldoxime and indole-3-acetaldoxime in heterologous systems and might contribute to plant defense and auxin formation. BMC Plant Biol 15:128. 10.1186/s12870-015-0526-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kato Y, Asano Y (2005) Purification and characterization of aldoxime dehydratase of the head blight fungus, Fusarium graminearum. Biosci Biotechnol Biochem 69:2254–2257. 10.1271/bbb.69.2254 [DOI] [PubMed] [Google Scholar]
- Kato Y, Asano Y (2006) Molecular and enzymatic analysis of the “aldoxime-nitrile pathway” in the glutaronitrile degrader Pseudomonas sp K-9. Appl Microbiol Biotechnol 70:92–101. 10.1007/s00253-005-0044-4 [DOI] [PubMed] [Google Scholar]
- Kato Y, Ooi R, Asano Y (1999) A new enzymatic method of nitrile synthesis by Rhodococcus sp. strain YH3-3. J Mol Catal B: Enzym 6:249–256. 10.1016/s1381-1177(98)00080-0 [Google Scholar]
- Kato Y, Nakamura K, Sakiyama H, Mayhew SG, Asano Y (2000) Novel heme-containing lyase, phenylacetaldoxime dehydratase from Bacillus sp. strain OxB-1: Purification, characterization, and molecular cloning of the gene. Biochemistry 39:800–809. 10.1021/bi991598u [DOI] [PubMed] [Google Scholar]
- Kato Y, Yoshida S, Xie S-X, Asano Y (2004) Aldoxime dehydratase co-existing with nitrile hydratase and amidase in the iron-type nitrile hydratase-producer Rhodococcus sp. N-771. J Biosci Bioeng 97:250–259. 10.1016/s1389-1723(04)70200-5 [DOI] [PubMed] [Google Scholar]
- Křístková B, Rädisch R, Kulik N, Horvat M, Rucká L, Grulich M, Rudroff F, Kádek A, Pátek M, Winkler M, Martínková L (2023) Scanning aldoxime dehydratase sequence space and characterization of a new aldoxime dehydratase from Fusarium vanettenii. Enzyme Microb Technol 164:110187. 10.1016/j.enzmictec.2022.110187 [DOI] [PubMed] [Google Scholar]
- Křístková B, Martínková L, Rucká L, Kotik M, Kulik N, Rädisch R, Winkler M, Pátek M (2024) Immobilization of aldoxime dehydratases on metal affinity resins and use of the immobilized catalysts for the synthesis of nitriles important in fragrance industry. J Biotechnol 384:12–19. 10.1016/j.jbiotec.2024.02.005 [DOI] [PubMed] [Google Scholar]
- Kumano T, Suzuki T, Shimizu S, Kobayashi M (2016) Nitrile-synthesizing enzyme: screening, purification and characterization. J Gen Appl Microbiol 62:167–173. 10.2323/jgam.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu W, Ding J, Lee K-H (2016) The application of NCTS (N-cyano-N-phenyl-p-toluenesulfonamide) in palladium-catalyzed cyanation of arenediazonium tetrafluoroborates and aryl halides. Tetrahedron Lett 57:1205–1209 [Google Scholar]
- Liu MY, Li SY (2024) Nitrile biosynthesis in nature: how and why? Nat Prod Rep 41:649–671. 10.1039/d3np00028a [DOI] [PubMed] [Google Scholar]
- Matsui D, Muraki N, Chen K, Mori T, Ingram AA, Oike K, Gröger H, Aono S, Asano Y (2022) Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: importance of surface residues in optimization for crystallization. J Inorg Biochem 230:111770. 10.1016/j.jinorgbio.2022.111770 [DOI] [PubMed] [Google Scholar]
- Méndez-Sánchez D, Lavandera I, Gotor V, Gotor-Fernández V (2017) Novel chemoenzymatic oxidation of amines into oximes based on hydrolase-catalysed peracid formation. Org Biomol Chem 15:3196–3201. 10.1039/c7ob00374a [DOI] [PubMed] [Google Scholar]
- Miao Y, Metzner R, Asano Y (2017) Kemp elimination catalyzed by naturally occurring aldoxime dehydratases. ChemBioChem 18:451–454. 10.1002/cbic.201600596 [DOI] [PubMed] [Google Scholar]
- Mishra V (2020) Affinity tags for protein purification. Curr Protein Pept Sci 21:821–830. 10.2174/1389203721666200606220109 [DOI] [PubMed] [Google Scholar]
- Müller AT, Nakamura Y, Reichelt M, Luck K, Cosio E, Lackus ND, Gershenzon J, Mithöfer A, Köllner TG (2024) Biosynthesis, herbivore induction, and defensive role of phenylacetaldoxime glucoside. Plant Physiol 194:329–346. 10.1093/plphys/kiad448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura J, Hashimoto H, Ohta T, Hashimoto Y, Wada K, Naruta Y, Oinuma K, Kobayashi M (2013) Crystal structure of aldoxime dehydratase and its catalytic mechanism involved in carbon-nitrogen triple-bond synthesis. Proc Natl Acad Sci U S A 110:2810–2815. 10.1073/pnas.1200338110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma K, Hashimoto Y, Konishi K, Goda M, Noguchi T, Higashibata H, Kobayashi M (2003) Novel aldoxime dehydratase involved in carbon-nitrogen triple bond synthesis of Pseudomonas chlororaphis B23. Sequencing, gene expression, purification, and characterization. J Biol Chem 278:29600–29608. 10.1074/jbc.M211832200 [DOI] [PubMed] [Google Scholar]
- Pedras MS, Minic Z, Thongbam PD, Bhaskar V, Montaut S (2010) Indolyl-3-acetaldoxime dehydratase from the phytopathogenic fungus Sclerotinia sclerotiorum: purification, characterization, and substrate specificity. Phytochemistry 71:1952–1962. 10.1016/j.phytochem.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Pei XL, Xiao QJ, Feng YM, Chen L, Yang FL, Wang QY, Li NX, Wang AM (2023) Enzymatic properties of a non-classical aldoxime dehydratase capable of producing alkyl and arylalkyl nitriles. Appl Microbiol Biotechnol 107:7089–7104. 10.1007/s00253-023-12767-y [DOI] [PubMed] [Google Scholar]
- Plass C, Hinzmann A, Terhorst M, Brauer W, Oike K, Yavuzer H, Asano Y, Vorholt AJ, Betke T, Gröger H (2019) Approaching bulk chemical nitriles from alkenes: a hydrogen cyanide-free approach through a combination of hydroformylation and biocatalysis. ASC Catal 9:5198–5203. 10.1021/acscatal.8b05062 [Google Scholar]
- Priya BV, Padhi SK (2023) Hydroxynitrile lyase discovery, engineering, and promiscuity towards asymmetric synthesis: recent progress. Eur J Org Chem 26:202300776. 10.1002/ejoc.202300776 [Google Scholar]
- Rädisch R, Chmátal M, Rucká L, Novotný P, Petrásková L, Halada P, Kotik M, Pátek M, Martínková L (2018) Overproduction and characterization of the first enzyme of a new aldoxime dehydratase family in Bradyrhizobium sp. Int J Biol Macromol 115:746–753. 10.1016/j.ijbiomac.2018.04.103 [DOI] [PubMed] [Google Scholar]
- Rädisch R, Pátek M, Křístková B, Winkler M, Křen V, Martínková L (2022) Metabolism of aldoximes and nitriles in plant-associated bacteria and its potential in plant-bacteria interactions. Microorganisms 10:549. 10.3390/microorganisms10030549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey IK (2010) Trilostane in dogs. Vet Clin N Am-Small Anim Pract 40:269–283. 10.1016/j.cvsm.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Rong YX, Jensen SI, Lindorff-Larsen K, Nielsen AT (2023) Folding of heterologous proteins in bacterial cell factories: cellular mechanisms and engineering strategies. Biotechnol Adv 63:108079. 10.1016/j.biotechadv.2022.108079 [DOI] [PubMed] [Google Scholar]
- Sawai H, Sugimoto H, Kato Y, Asano Y, Shiro Y, Aono S (2009) X-ray crystal structure of Michaelis complex of aldoxime dehydratase. J Biol Chem 284:32089–32096. 10.1074/jbc.M109.018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A (2024) Microbial aldoxime dehydratase enzymes and nitrile biosynthesis. In: Seth A, Mehta PK (eds) The Chemistry of Nitriles. Nova Science Publishers Inc, New York, pp 15–29 [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M, Neilson EHJ, Møller BL (2018) Oximes: unrecognized chameleons in general and specialized plant metabolism. Mol Plant 11:95–117. 10.1016/j.molp.2017.12.014 [DOI] [PubMed] [Google Scholar]
- Soumya PK, Vaishak TB, Saranya S, Anilkumar G (2021) Recent advances in the rhodium-catalyzed cyanation reactions. Appl Organomet Chem 35:6340. 10.1002/aoc.6340 [Google Scholar]
- Su W, Gong TJ, Xiao B, Fu Y (2015) Rhodium(III)-catalyzed cyanation of vinylic C-H bonds: N-cyano-N-phenyl-p-toluenesulfonamide as a cyanation reagent. Chem Commun 51:11848–11851. 10.1039/c4cc09790d [DOI] [PubMed] [Google Scholar]
- Terhorst M, Plass C, Hinzmann A, Guntermann A, Jolmes T, Rösler J, Panke D, Gröger H, Vogt D, Vorholt AJ, Seidensticker T (2020) One-pot synthesis of aldoximes from alkenes via Rh-catalysed hydroformylation in an aqueous solvent system. Green Chem 22:7974–7982. 10.1039/d0gc03141k [Google Scholar]
- Winkler M, Ling JG (2022) Biocatalytic carboxylate reduction – recent advances and new enzymes. ChemCatChem 14:202200441. 10.1002/cctc.202200441 [Google Scholar]
- Winkler M, Dokulil K, Weber H, Pavkov-Keller T, Wilding B (2015) The nitrile-forming enzyme 7-cyano-7-deazaguanine synthase from Geobacillus kaustophilus: a reverse nitrilase? ChemBioChem 16:2373–2378. 10.1002/cbic.201500335 [DOI] [PubMed] [Google Scholar]
- Winkler M, Horvat M, Schiefer A, Weilch V, Rudroff F, Pátek M, Martínková L (2023) Organic acid to nitrile: a chemoenzymatic three-step route. Adv Synth Catal 365:37–42. 10.1002/adsc.202201053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao QJ, Feng YM, Chen L, Li M, Zhang PF, Wang QY, Wang AM, Pei XL (2023) Engineered aldoxime dehydratase to enable the chemoenzymatic conversion of benzyl amines to aromatic nitriles. Bioorg Chem 134:106468. 10.1016/j.bioorg.2023.106468 [DOI] [PubMed] [Google Scholar]
- Xie SX, Kato Y, Asano Y (2001) High yield synthesis of nitriles by a new enzyme, phenylacetaldoxime dehydratase, from Bacillus sp strain OxB-1. Biosci Biotechnol Biochem 65:2666–2672. 10.1271/bbb.65.2666 [DOI] [PubMed] [Google Scholar]
- Xie SX, Kato Y, Komeda H, Yoshida S, Asano Y (2003) A gene cluster responsible for alkylaldoxime metabolism coexisting with nitrile hydratase and amidase in Rhodococcusgloberulus A-4. Biochemistry 42:12056–12066. 10.1021/bi035092u [DOI] [PubMed] [Google Scholar]
- Yamaguchi T (2024) Exploration and utilization of novel aldoxime, nitrile, and nitro compounds metabolizing enzymes from plants and arthropods. Biosci Biotechnol Biochem 88:138–146. 10.1093/bbb/zbad168 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Asano Y (2024) Nitrile-synthesizing enzymes and biocatalytic synthesis of volatile nitrile compounds: a review. J Biotechnol 384:20–28. 10.1016/j.jbiotec.2024.02.007 [DOI] [PubMed] [Google Scholar]
- Yavuzer H, Asano Y, Gröger H (2021) Rationalizing the unprecedented stereochemistry of an enzymatic nitrile synthesis through a combined computational and experimental approach. Angew Chem Int Ed 60:19162–19168. 10.1002/anie.202017234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuzer H, Yang J, Gröger H (2023) Rational biocatalyst design for a cyanide-free synthesis of long-chain fatty nitriles from their aldoximes. Lipid Sci Technol 125:2200177. 10.1002/ejlt.202200177 [Google Scholar]
- Zallot R, Oberg N, Gerlt JA (2019) The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58:4169–4182. 10.1021/acs.biochem.9b00735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Asano Y (2020) Biocatalytic asymmetric ring-opening of dihydroisoxazoles: a cyanide-free route to complementary enantiomers of β-hydroxy nitriles from olefins. Green Chem 22:4930–4936. 10.1039/d0gc01445a [Google Scholar]
- Zheng D, Asano Y (2021) A cyanide-free biocatalytic process for synthesis of complementary enantiomers of 4-chloro-3-hydroxybutanenitrile from allyl chloride. ChemCatChem 13:4237–4242. 10.1002/cctc.202100835 [Google Scholar]
- Zheng HT, Xiao QJ, Mao FY, Wang AM, Li M, Wang QY, Zhang PF, Pei XL (2022) Programing a cyanide-free transformation of aldehydes to nitriles and one-pot synthesis of amides through tandem chemo-enzymatic cascades. RSC Adv 12:17873–17881. 10.1039/d2ra03256b [DOI] [PMC free article] [PubMed] [Google Scholar]