Abstract
Despite the identification of numerous genetic variants affecting SBP in European populations, their applicability in admixed populations remains unclear. This study evaluates the predictive efficacy of a systolic blood pressure (SBP) polygenic risk score (PRS), derived from the UK Biobank data, in two Brazilian cohorts. We analyzed 944 K genetic variants consistent across an independent UK Biobank dataset, Brazilian cohorts, and HapMap database. Results show a significant association between increased PRS and SBP, as well as hypertension, in each study groups analyzed. An increase of one standard deviation in the PRS showed a significant association with SBP (β [95% CI] (mmHg) = 5.2 [5.1–5.3], 2.8 [2.1–3.5] and 2.6 [2.2–3.0]) and hypertension (odds ratio (OR) [95% CI] = 1.56 [1.54–1.56], 1.28 [1.2–1.4] and 1.47 [1.3–1.6]) in an independent UKB dataset, Baependi, and Pelotas, respectively. The associations were weaker in the Brazilian samples and the reduced association was noticeable in the Pelotas vs. the UK comparison for hypertension stages 1 and 2 (OR [95% CI] = 2.1 [1.5–3.1] and 3.0 [1.9–4.7] vs. 2.5 [2.2–2.8] and 4.9 [4.4–5.6]), whereas the Baependi data showed no significance for stage 1 hypertension. This trend mirrors findings in homogeneous African and Asian populations with diverse genetic architecture, highlighting the limitations of European-based PRS also in admixed populations. These insights are crucial for developing tailored disease prevention and management strategies in ethnically diverse groups.
Subject terms: Hypertension, Cardiovascular genetics, Predictive markers, Genome-wide association studies
Introduction
Blood pressure (BP) is a quantitative trait with continuous variation in human populations. Elevated BP, also known as arterial hypertension, significantly elevates the risk for heart, brain, and kidney diseases1–4. However, the genetic underpinnings of primary hypertension remain elusive, even though genetics plays a substantial role, with heritability estimates spanning 25–68% across global populations5,6.
The surge in genomic technologies has propelled numerous genome-wide association studies (GWAS) to explore the relationship between common genetic variations, like single nucleotide polymorphisms (SNPs), and intricate health conditions such as BP and hypertension. However, while the promise of GWAS is vast, results have been modest. For instance, more than 2,000 loci identified collectively account for 6.93% of the total BP variance in broad population samples. Nevertheless, these studies have paved the way for new insights into the underlying genes and pathways7.
On another front, polygenic risk scores (PRSs) compute the cumulative effects of SNPs identified in GWAS, even those not achieving genomic significance8,9. This approach has successfully pinpointed susceptibility to conditions like obesity10, coronary artery disease, and type 2 diabetes11. In essence, the risk presented by the cumulative effects of multiple variants can be equivalent to monogenic mutations11.
For instance, Vaura et al. used PRSs to analyze systolic and diastolic BP in a Finnish cohort, revealing that those in the top 2.5% of BP PRS faced double the risk of hypertension and were diagnosed a decade earlier than their counterparts12. In a more recent study, Keaton et al. demonstrated that individuals in the top decile of the PRS, containing more than 7 million variants, have more than sevenfold higher odds of hypertension compared to individuals in the bottom decile7.
However, a limitation arises: most large-scale GWAS and PRS research are based on data from individuals of European ancestry. Studies have shown that PRSs, when applied to African populations, tend to have reduced predictive value for various diseases, including cardio-renal-metabolic conditions13 and coronary artery disease14. Similarly, Keaton et al. demonstrated that the difference in hypertension risk between the top and bottom deciles of the PRS was only a 1.73-fold increase in African Americans, and the inclusion of the PRS in hypertension prediction models did not result in significant reclassification improvements7.
Genetic prediction accuracy diminishes with growing genetic divergence between the research and target populations, often due to differing genetic structures in part due to differences in linkage disequilibrium (LD) structure, and allele frequency of genetic variants. Recognizing this disparity, Parcha et al. designed a PRS focusing on a diverse Multi-Ancestry Pan UK Biobank GWAS. Still, they found a weaker association between PRS and BP in Black and Asian individuals15.
Brazil’s diverse genetic landscape, shaped by a mix of African, European, and Native American ancestries, presents a unique opportunity for genetic research due to its extensive intermingling16,17. We derived and assessed a systolic BP PRS from a European ancestry sample, applying it to both a European and two admixed Brazilian groups. Our results reveal that while the PRS, aggregating multiple common variants, effectively identifies individuals at elevated risk of hypertension in all groups, its predictive accuracy is significantly reduced in admixed populations. This reduction, nearly 50% compared to European groups, underscores the limitations of using European-based PRS in ethnically diverse populations. This trend aligns with similar findings in homogeneous African and Asian populations13,15,18, emphasizing the need for population-specific genetic assessments in diverse groups.
Results
Polygenic risk score (PRS) derivation for SBP using UK biobank (UKB)
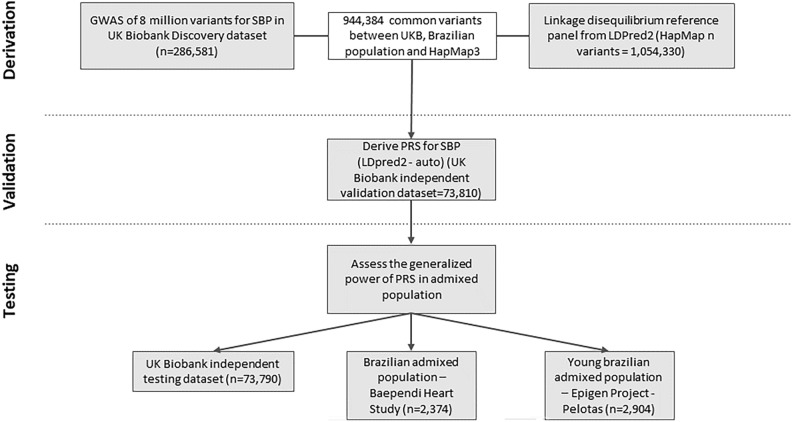
We calculated the PRS for SBP as sums of each risk allele weighted by its estimated effect size on SBP, based on a GWAS of the UKB. To prevent overfitting, we split the dataset: two-thirds for the GWAS (286,581 participants), one-sixth for validating the SBP PRS and model selection (73,810 participants), and another sixth for testing the chosen model on a European base population (73,790 participants, see Fig. 1). All these UKB subsets shared similar demographics including gender, health conditions, and average measurements (Supplementary Table S1).
Fig. 1.
Study design and workflow. A PRS for systolic blood pressure was derived by combining variants from a summary statistic of a GWAS using two third of UK Biobank database also presented in Baependi Heart Study, Epigen project – Pelotas and HapMap database. We derived LDPred2 score using the option auto that tunes hyper-parameters used, including ρ, h2 (estimated heritability), and whether sparsity is enable or not, as recommended by developers, validating it in UK Biobank validation dataset and subsequently tested in three independent testing datasets: UK Biobank, and two Brazilian populations, one of them from young adults. Both UKB validation and testing datasets are UKB independent samples not used in discovery dataset.
UKB’s GWAS insights on blood pressure
In our GWAS, we aimed to uncover genetic variants associated with SBP among ~ 287,000 European descendants. From 8,721,758 SNPs assessed, 129 lead SNPs (from 275 significant independent SNPs and 4,813 significant SNPs) across 98 genomic risk loci were associated to SBP (Supplementary Fig. S1, Supplementary Tables S2 and S3), post functional mapping and annotation (FUMA) analysis38. Among these, 97 loci (containing 274 independent significant SNPs and 4,811 GWAS significant SNPs) had prior association to BP-related traits (Supplementary Tables S4-S10), while only one (containing 1 independent significant SNPs and 2 GWAS significant SNPs) was novel discovery (Supplementary Table S11).
PRS’s relationship with blood pressure traits across populations
We developed a PRS drawing from the average effects of 944,384 genetic variants found in UKB, Brazilian samples from the Baependi Heart Study, Pelotas cohorts, and the HapMap database, accounting for an estimated 20.5% of SBP variance in Europeans.
We then assessed the PRS’s predictive capacity for SBP and hypertension in three diverse datasets: middle-aged UKB participants (average age 56 years), the Baependi Heart Study (17–99 years), and the 1982-born cohort from Pelotas (30 years old at the time of data collection) (Supplementary Fig. S2A). These Brazilian datasets, with their rich mix of ancestries, allowed us to evaluate a European-based PRS’s ability to forecast SBP trends and hypertension risks in an admixed setting. Brazilian studies include individuals of predominantly European ancestry (67.56% in Baependi and 79.29% in Pelotas) and significant percentages of African ancestry (11.52% in Baependi, 15.08% in Pelotas). Native American ancestry varies between the two Brazilian populations, being more predominant in Baependi population (17.93% in Baependi versus 0.94% in Pelotas) and Asian ancestry is observed in a small fraction on both datasets (Supplementary Fig. S3A). Bar plots of individual ancestry from the three test datasets highlight a significant admixture component. Notably, while the average European ancestry is between 50 to 70%, the range is continuous, spanning from 0 to 100%. This demonstrates the genetic diversity of these population samples in comparison to European populations (Supplementary Fig. S3B).
While the Pelotas group showed the lowest average blood pressure readings, Baependi had a modest average BMI compared to both Pelotas and the European group. Expectedly, the younger Pelotas group had fewer hypertension cases than Baependi and the European group (Supplementary Table S12 and Supplementary Fig. S2).
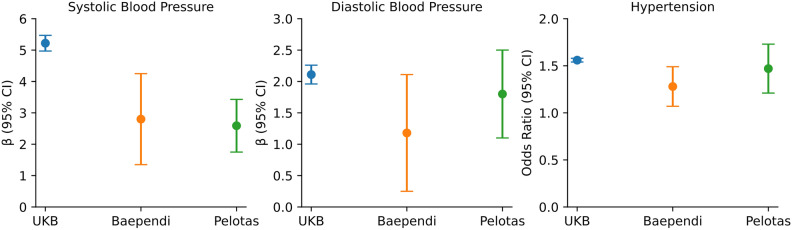
All three groups exhibited a typical PRS distribution (Supplementary Fig. S4). In the European group, PRS, when quantified per 1 standard deviation (SD) unit increase, was significantly associated to SBP (5.2 mmHg, 95% CI: 5.1–5.3 mmHg, P < 2.2 × 10–308), DBP (2.1 mmHg, 95% CI: 2.0–2.2 mmHg, P < 2.2 × 10–308), and hypertension risks (odds ratio (OR) = 1.56, 95% CI: 1.54–1.56, P < 2.2 × 10–308) (Fig. 2 & Supplementary Tables S13A-S15A). Individuals on the top PRS decile group presented a mean SBP 18.2 mmHg (95% CI: 17.6–18.7, P < 2.2 × 10–308) and DBP 7.3 mmHg (95% CI: 6.9–7.6, P < 2.2 × 10–308) higher than individuals on the bottom PRS decile, accompanied by an increase in mean SBP and DBP across PRS deciles (Table 1 & Supplementary Fig. S5 and Supplementary Tables S13B-S14B). For Brazilians, the association with SBP, DBP and hypertension, while strong, was diluted, with each 1 SD unit increase of PRS associated with 2.8 mmHg (95% CI: 2.1–3.5, P = 4.8 × 10–14) and 2.6 mmHg (95% CI: 2.2–3.0, P = 5.9 × 10–33) in SBP, 1.2 mmHg (95% CI: 0.7–1.7, P = 6.5 × 10–7) and 1.8 mmHg (95% CI: 1.4–2.1, P = 1.1 × 10–28) in DBP, and 1.28 (95% CI: 1.2–1.4], P = 4.3 × 10–5) and 1.47 (95% CI: 1.3–1.6, P = 8.5 × 10–19) odds ratio for hypertension in Baependi and Pelotas, respectively (Fig. 2 & Supplementary Tables S13A-S15A). Similarly, in Brazilian populations, there was a notable difference in average SBP and DBP between the highest and lowest deciles. Specifically, the differences were 9 mmHg (95% CI: 5.3–12.2, P = 5.3 × 10–8) and 9.3 mmHg (95% CI: 7.3–11.1, P = 5.7 × 10–22) for SBP, and 3.3 mmHg (95% CI: 1.2–5.3, P = 2 × 10–3) and 6.5 mmHg (95% CI: 5.1–7.8, P = 2.6 × 10–19) for DBP in Baependi and Pelotas, respectively. However, these variations were not as marked as those seen in the UKB, and the mean SBP/DBP gradient across the deciles was also less pronounced (Table 1 & Supplementary Fig. S5 and Supplementary Tables S13B-S14B).
Fig. 2.
Association of systolic blood pressure polygenic risk score with blood pressure traits between UKB and the two Brazilian admixed population. Betas and odds ratios per standard deviation of PRS were derived from linear and logistic regressions adjusted for age, sex, BMI and the four principal components of genetic ancestry, respectively. The whiskers represent the 95% confident interval.
Table 1.
Baseline characteristics of the three populations by low (first decile), intermediate (deciles 2–9) and high (top decile) PRS.
| Characteristics | UKB | Baependi | Pelotas | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low PRS | Intermediate PRS | High PRS | Low PRS | Intermediate PRS | High PRS | Low PRS | Intermediate PRS | High PRS | |
| N | 7,379 | 59,032 | 7,379 | 206 | 1,641 | 206 | 291 | 2,322 | 291 |
| Age | 56.9 ± 7.9 | 56.7 ± 8.0 | 56.5 ± 8.0 | 47.1 ± 16.9 | 45.7 ± 17.2 | 45.5 ± 16.2 | 30.2 ± 0.3 | 30.2 ± 0.3 | 30.2 ± 0.3 |
| Males, n(%) | 3,344(45.3) | 27,279(46.2) | 3,460(46.9) | 85(41.3) | 696(42.4) | 90(43.7) | 137(47.0) | 1145(49.3) | 139(47.7) |
| SBP(mmHg) | 130.9 ± 17.7 | 139.6 ± 19.5* | 148.8 ± 20.7*# | 137.9 ± 19.2 | 139.6 ± 20.1 | 144.7 ± 20.7 | 116.6 ± 12.2 | 121.7 ± 14.2 | 125.7 ± 14.1 |
| DBP(mmHg) | 79.7 ± 10.1 | 83.4 ± 10.8* | 86.9 ± 11.2*# | 84.6 ± 11.3 | 85.7 ± 12.4 | 87.1 ± 12.6 | 72.8 ± 9.0 | 75.7 ± 9.6 | 78.9 ± 9.4 |
| BMI (Kg/m2) | 27.4 ± 4.7 | 27.4 ± 4.8 | 27.4 ± 4.8 | 26.6 ± 5.3 | 25.6 ± 5.0 | 25.2 ± 5.0 | 27.4 ± 5.7 | 26.9 ± 5.6 | 26.4 ± 5.6 |
| Overweight, n(%) | 3,176(43.0) | 25,386(43.0) | 3,115(42.2) | 61(29.6) | 489(29.8) | 55(26.7) | 101(34.7) | 805(34.7) | 97(33.3) |
| Obesity, n(%) | 1,790(24.3) | 14,102(23.9) | 1,754(23.8) | 52(25.2) | 308(18.8) | 32(15.5) | 79(27.1) | 548(23.6) | 59(20.3) |
| Normotension, n(%) | 1,878(26.9) | 7,779(13.9)* | 401(5.7)*# | 26(12.6) | 212(13.0) | 18(8.7) | 174(59.8) | 1,061(45.7)* | 97(33.3)*# |
| Elevated, n(%) | 1,079(15.4) | 6,488(11.6)* | 511(7.3)*# | 26(12.6) | 167(10.1) | 15(7.2) | 45(15.5) | 373(16.0) | 50(17.2) |
| Hypertension, n(%) | 4,017(54.4) | 41,468(70.2)* | 6,050(82.0)*# | 154(74.7) | 1,262(76.9) | 173(83.9) | 72(24.7) | 888(38.2)* | 144(49.4)*# |
| Hypertension Stage 1, n(%) | 1,884(27.0) | 14,240(25.5) | 1,378(19.7)*# | 64(31.0) | 461(28.0) | 58(23.3) | 51(17.5) | 588(25.3) | 90(30.9)* |
| Hypertension Stage 2, n(%) | 2,133(30.0) | 27,228(48.8)* | 4,672(67.1)*# | 90(43.6) | 801(48.8) | 125(60.6)*# | 21(7.2) | 300(12.9) | 54(18.5)* |
It is depicted results for χ2 test for categorical variables and one-way Anova for continuous variable followed by Tukey t-test. * significant difference vs. low PRS and # significant difference vs. intermediate PRS. For hypertension stages, elevated was defined as 120 ≤ SBP < 130 mmHg and DBP < 80 mmHg, hypertension stage 1 as 130 ≤ SBP < 140 mmHg or 80 ≤ DBP < 90 mmHg, and hypertension stage 2 as SBP ≥ 140 mmHg or DBP ≥ 90 mmHg.
When we compared the top 10% PRS with the remaining 90%, high PRS was associated to almost twofold higher odds of elevated blood pressure (OR [95% CI] = 1.8 [1.6–2.1], P = 4.5 × 10–18, and OR [95% CI] = 1.9 [1.2–2.8], P = 2.6 × 10–3), to more than twofold higher odds of hypertension stage 1 (OR [95% CI] = 2.5 [2.2–2.8], P = 2.1 × 10–51, and OR [95% CI] = 2.1 [1.5–3.1] , P = 9.3 × 10–6), and to 3–5 folds higher odds of hypertension stage 2 (OR [95% CI] = 4.9 [4.4–5.6], P = 1.9 × 10–164 and OR [95% CI] = 3.0 [1.9–4.7] , P = 6.4 × 10–7) in both the UKB and Pelotas datasets, respectively, but only to more than twofold increased risk of severe hypertension in Baependi (2.5, 95% CI: 1.4–5.0, P = 3.4 × 10–3) (Table 2).
Table 2.
Prevalence of clinical categories of hypertension comparing the top PRS decile with the remaining 90% of the PRS distribution, along with predictive model discrimination performance results.
| Risk of clinical categories of hypertension in top decile vs remaining 90% of the PRS distribution | Predictive Model | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | Outcome | N | OR | 95%CI | P | Model | AUC | 95% CI | F1 Score | P |
| UKB | Elevated | 8,078/73,735 | 1.80 | 1.6–2.1 | 4.51E-18 | PRS + covariates | 0.73 | 0.72–0.74 | 0.70 | 4.82E-48 |
| HTN stage 1 | 17,502/73,735 | 2.52 | 2.2–2.8 | 2.10E-51 | Clinical data only | 0.70 | 0.70–0.71 | 0.67 | ||
| HTN stage 2 | 34,033/73,735 | 4.90 | 4.42–5.56 | 1.87E-164 | PRS only | 0.61 | 0.60–0.61 | 0.61 | 1.67E-66 | |
| Baependi | Elevated | 239/2,374 | 1.20 | 0.50–2.66 | 5.80E-01 | PRS + covariates | 0.79 | 0.75–0.84 | 0.78 | 7.25E-1 |
| HTN stage 1 | 667/2,374 | 1.4 | 0.77–2.7 | 2.60E-01 | Clinical data only | 0.78 | 0.73–0.83 | 0.776 | ||
| HTN stage 2 | 1,177/2,374 | 2.5 | 1.4–5.0 | 3.40E-03 | PRS only | 0.54 | 0.49–0.60 | 0.58 | 2.94E-07 | |
| Pelotas | Elevated | 468/2,904 | 1.87 | 1.24–2.81 | 2.59E-03 | PRS + covariates | 0.77 | 0.74–0.81 | 0.71 | 1.18E-1 |
| HTN stage 1 | 729/2,904 | 2.14 | 1.53–3.1 | 9.30E-06 | Clinical data only | 0.76 | 0.73–0.79 | 0.69 | ||
| HTN stage 2 | 375/2,904 | 3.00 | 1.9–4.7 | 6.40E-07 | PRS only | 0.58 | 0.54–0.62 | 0.58 | 3.17E-11 | |
In the predictive model section, the F1 score provides a balanced view of precision and recall, and the P-value indicates whether the models are significantly different (p-value < 0.05) from the clinical data-only model.
Finally, a model including both PRS and covariates yielded an Area Under the Curve (AUC) of 0.73 (95% CI: 0.72–0.74), 0.79 (95% CI: 0.75–0.84), and 0.77 (95% CI: 0.74–0.81) for UKB, Baependi, and Pelotas, respectively. Yet, for the Brazilian datasets, adding the PRS to a clinical-only model did not enhance prediction (models difference = 0.01 and 0.015, P = 0.73 and 0.12 in Baependi and Pelotas, respectively, vs. models difference = 0.028, P = 4.82 × 10–48 in UKB), underscoring the European-centric PRS’s limited utility in predicting hypertension in admixed populations (Table 2).
Discussion
We generated and tested an SBP PRS using 944,384 common variants in UKB compared with two admixed Brazilian samples. Our derived score successfully identified individuals with elevated SBP and hypertension risk in both Brazilian and European samples, offering a slight enhancement in predictive capacity over the clinical data for hypertension in European samples alone.
Comparative analyses in European ancestry revealed our PRS’s SBP differences between the highest and lowest deciles were analogous to findings from the FINRISK study (17.9 mmHg using the proposed PRS in the UKB vs. 14.1 mmHg, respectively)12, NHLBI TOPMed multi-ancestry cohorts (13 mmHg)15 and the Lifelines individuals (16.9 mmHg)7. The per 1-SD unit increase in the reported PRS demonstrated a significant association with SBP, DBP and hypertension similar to those observed in NHLBI TOPMed multi-ancestry cohorts (5.2 mmHg, 2.1 mmHg and OR = 1.56 vs. 4.39 mmHg, 2.04 mmHg and OR = 1.5, respectively), being more similar in the relationships observed specifically in white individuals from the reported cohort (SBP = 5.4 mmHg, DBP = 2.54 mmHg and hypertension OR = 1.7)15. This underlines the potential clinical relevance of the reported scores within European populations, regardless of disparities in the algorithms and variant counts utilized.
Using 901 and 2,103 significant loci to generate a genomic risk score (GRS) from two BP GWAS meta-analysis of over a million participants, the SBP variance was found to be 5.7% and 6.93%, respectively7,19. In contrast, our study’s 944,000 variants accounted for 20.5% of the SBP variance, marking a nearly four-fold growth from prior evaluations, but similar to the values reported by Parcha et al15. However, SBP variations between the extreme deciles were moderate, with our PRS showing a slightly higher difference than Evangelou’s GRS. Concerning hypertension risk, European individuals with the highest PRS decile had 2.5 times the risk for stage 1 and nearly 5 times for stage 2 hypertension. This underscores the refined predictive potential from incorporating numerous common variants to the PRS.
Our European-based PRS revealed a reduced association with BP traits/hypertension in the admixed Brazilian population, showing nearly a 50% decrease in the PRS’s impact on SBP and DBP. This diminished connection also manifested in hypertension risk categories, especially within the Baependi datasets. Still, the significant correlation in the younger Pelotas group, in their 30 s, highlights PRS’s potential in early diagnosis. Previous findings by Vaura et al. also support the enhanced predictive capacity of BP PRS in early-onset hypertension12.
Interestingly, data from the Baependi dataset mirrored findings from black individuals within NHLBI TOPMed cohorts (SBP = 2.4 mmHg, DBP = 0.99 mmHg and hypertension OR = 1.19). The reduction in PRS precision in admixed populations became clearer when assessing its ability to predict hypertension, showing minor enhancement over clinical data in the Baependi and Pelotas samples.
While many studies have indicated the limited PRS predictive capacity based on European ancestry in African and Asian populations13,18, ours is pioneering in evaluating this in a significantly admixed population. Simulation and empirical studies investigated the factors influencing prediction in non-Europeans, including differences in phenotype data collection among population studies, LD structures, allele frequencies across populations, causal variants effect sizes, and they demonstrated that loss in PRS accuracy increases with increasing divergence from European ancestry18,20,21. Given various influencing factors, we postulate that the diminished predictive capability in Brazilian samples stems from their complex genetic composition of European, African, and Native American ancestries. The fewer European genetic contributions, the more significant the drop in predictive capability.
Several approaches have been suggested to integrate population-specific effects to enhance the predictive accuracy of PRS algorithms. These approaches include merging data from UKB with Chinese samples22, combining European and African samples23, or using estimates from various phenotypes24. Such endeavors underscore the importance of incorporating genetic data from admixed populations, which bring together diverse ancestral influences. Our research emphasizes the critical need for genetic data from admixed populations, given their unique ancestral blend. Yet, Latin American populations remain underrepresented in extensive studies, only 1.3% of the GWAS used Hispanic or Latin American samples to discover genetic variants associated with complex traits until 201725.
This study’s limitations include relying on a smaller GWAS, using Caucasian data for Brazilian population genetic imputation, modest Brazilian sample sizes, and a lack of subsequent data.
Altogether, we provided evidence for significant association between increased PRS and SBP, as well as hypertension in two Brazilian admixed samples. However, associations were weaker in Brazilian samples, particularly in the Baependi cohort. This trend mirrors findings in homogeneous African or Asian populations with diverse genetic backgrounds, highlighting the limitations of European-based PRS also in admixed populations. These insights are crucial for developing tailored disease prevention and management strategies in ethnically diverse groups.
Methods
Study cohorts, phenotyping, and genotyping
The UK biobank (UKB) project
The UKB is a large-scale prospective study, collecting genetic and phenotypic information from roughly half a million UK residents. From this, we extracted data from 434,181 participants, spanning both genders and aged 38–73 at the time of recruitment, that contained at least one automated blood pressure recording. Participants offered digital consent, lifestyle details, and health-related data via questionnaires, underwent physical evaluations, and donated urine, saliva, and blood samples for genotyping purposes26. The present study was conducted under application number 14654 of the UKB resource.
SBP was ascertained using the average of two automated blood pressure recordings (Field 4080 – SBP, automated reading—UKB data resource). If only one measurement was available, it was taken as the singular value. A total of 434,181 participants had SBP data. Adjustments were made to BP values, accounting for medication use. This was done by adding 15 mmHg to SBP and 10 mmHg to DBP for those on anti-hypertensive drugs, irrespective of dosage or medication number27.
Genotyping involved two methodologies. The UK BiLEVE Axiom Array chip was used for 46,578 individuals from the UKB Lung Exome Variant Evaluation study, while the Affymetrix UKB Axiom Array chip was employed for 408,268 participants. Single Nucleotide Polymorphisms (SNPs) were imputed via a combined reference panel from UK10K, 1000 Genomes phase 3, and the Haplotype Reference Consortium using the IMPUTE4 tool (https://jmarchini.org/software/). This imputation yielded a dataset with over 93 million autosomal variants.
The Baependi heart study
Baependi is a Brazilian longitudinal family-based study focusing on genetic and environmental impacts on cardiovascular risks. The study design, recruitment, and the demographics of this cohort have been described previously5,28. Briefly, in 2005, we began recruiting randomly selected probands and their family members and relatives (95 families at baseline and 1,695 individuals). In the 2010 follow-up, families were added, for a total of 125 families and 2,495 individuals of both genders, aged between 17 to 98 years.
Blood pressure was assessed using a digital sphygmomanometer (OMRON, Brazil), with SBP determined from the average of three measurements in the sitting position on the left arm after 5 min’ rest. The same BP adjustment method used for the UKB was applied here.
For genotyping, two technologies were used: 1,409 individuals with BP data were genotyped at 747,611 variants using the Affymetrix 6.0 GeneChip, and 704 individuals with BP data were genotyped at 548,944 variants with the custom Affymetrix Axiom BB_incor array chip. SNPs were imputed using a reference panel from TOPMed29 using the IMPUTE2 program. After imputation, 39,127,678 SNPs were available for further analysis.
The 1982 Pelotas (Brazil) birth cohort study
This study is part of the EpiGen-Brazil consortium, which studies 6,487 individuals belonging to three Brazilian population-based cohorts with at least a 10-year follow-up. It is one of the most significant Latin American initiatives in population genomics and genetic epidemiology. The Pelotas cohort is a longitudinal study conducted in Pelotas, a city in the deep South of Brazil, near the Uruguayan border and follows 5,914 individuals born in 1982 throughout adulthood30. We used data from 3,736 individuals from the 2012 follow-up when the subjects were 30 years old.
Blood pressure was measured using a standard digital sphygmomanometer in the sitting position twice, and BP was calculated from the mean of two readings. We adjusted SBP and DBP for medication use using the same values applied to the UKB individuals. Genotyping was performed at the Illumina facility using the HumanOmni2.5-8v1 array, which genotyped 2.3 million SNPs. SNPs were imputed using a reference panel from TOPMed29 using the IMPUTE2 program, resulting in a dataset with 40,650,105 variants.
For all three populations, we defined hypertension as SBP ≥ 130 mmHg, DBP ≥ 80 mmHg, antihypertensive medication use, or registry-based hypertension. For hypertension stages, we defined Elevated as 120 ≤ SBP < 130 mmHg, and DBP < 80 mmHg. Hypertension stage 1 was defined as 130 ≤ SBP < 140 mmHg or 80 ≤ DBP < 90 mmHg, and hypertension stage 2 was defined as SBP ≥ 140 mmHg or DBP ≥ 90 mmHg31.
Quality control and population structure
For all genotyped datasets, we used human genome reference GRCh37 and applied standard quality controls using PLINK2 software32: minor allele frequency (MAF) ≥ 1%, missing rate per variant < 5%, missing rate per individual < 10%, Hardy–Weinberg equilibrium test with p-value ≥ 1 × 10–6, and imputation quality score (R2) higher than 0.8. The QC steps resulted in a UKB, Baependi and Pelotas datasets with 8,721,758; 8,002,637 and 10,404,494 variants, respectively.
Given the diverse ethnic backgrounds, principal component analysis (PCA) was used to assess population structure. For the UKB datasets, SNP pruning was conducted using PLINK2 (PLINK filter –indep-pairwise 1000 50 0.05), and relatedness was assessed with the KING software33. We then performed PCA using FlashPCA34 considering only unrelated individuals (up to the 3rd degree of relatedness), and projected the resulting principal components onto the related individuals. For the Brazilian datasets, SNP pruning was also carried out using PLINK2, but we used the GARSA tool35, developed in our lab, to correct bias in PCA and kinship analysis due to the admixed nature of the samples. For kinship correction, GARSA first applies the robust KING method, followed by a correction of kinship values using the SNPrelate package in R, which incorporates PCA to account for population structure. As noted in the tool’s documentation, this approach is better suited for admixed populations. For PCA, GARSA starts by performing PCA on all unrelated samples, evaluates SNP loadings, and removes SNPs with loadings deviating more than three standard deviations from the mean. A new PCA is then performed on the same unrelated dataset, excluding the outlier SNPs (though these SNPs are not removed from the dataset for the GWAS analysis). Finally, GARSA projects the calculated principal components onto the related individuals.
Ancestry analyses
Global ancestry was inferred using ADMIXTURE (v1.3)36, with reference groups from the Human Genome Diversity Project (HGDP) and the 1000 Genomes Project phase 3 (1KGP3). The reference populations included Native American groups such as the Karitiana (KRT) and Suruí (SRI) from Brazil, and the Pima (PIM) and Maya (MAY) from Mexico. African populations included the Yoruba from Ibadan, Nigeria (YRI), and African-Americans (ASW) from the 1KGP3, along with the Luhya in Webuye, Kenya (LWK). European populations included the Tuscans from Italy (TSI) and Utah residents of Northern and Western European ancestry (CEU) from the 1KGP3. The number of ancestral populations (K) was defined by the distinct reference populations used, which included European, South Asian, East Asian, African, and Native American groups (K = 5).
For the analysis, we selected only common variants (MAF > 1%) present in all datasets (UKB, Baependi, Pelotas, and the reference populations), and applied SNP pruning using PLINK (with the filter –indep-pairwise 1000 50 0.05). The resulting ancestry estimates were parsed and plotted using in-house Python scripts.
Systolic blood pressure genome-wide association study
Association analysis was performed with BOLT-LMM37 from the GARSA tool35, with two-third of the UKB participants (training dataset, N = 286,581 individuals). A linear mixed model was applied, adjusted for age, sex, body mass index (BMI), genotyping array, and the first four principal components (PCs). The dataset was divided using the CARET R package, which allows for balanced separation based on multiple features. In this case, the dataset was balanced according to hypertension status (presence of the disease), sex, and age group (group 1: 18–39 years, group 2: 40–60 years, and group 3: over 60 years). We considered all markers with minor allele frequency ≥ 1%, imputation info score > 0.8, and Hardy–Weinberg equilibrium test with p-value ≥ 1 × 10–6 for GWAS analysis. We adjusted the GWAS results by genomic inflation, using GARSA tool35.
Genomic risk loci, independent significant SNP and lead SNP characterization
To determine the risk loci, we first identified independent significant SNPs with genome-wide significance (p < 5e-8), which are independent of each other, with an r2 < 0.6 in a linkage disequilibrium (LD) structure based on the 10,000 Genomes from UKB (UKB release 2b) for European populations. We used a maximum distance of 1 Mb between a pair of SNPs. For each independent significant SNP, we identified all known SNPs with an r2 ≥ 0.6 within the same risk locus. This included SNPs tested in GWAS and/or those available in the UKB release 2b reference panel.
Based on these independent significant SNPs, we defined independent lead SNPs as those with an r2 < 0.1, maintaining independence from one another, considering the same LD structure. If an LD block determined by an independent significant SNP was within 250 kb of another LD block from a different independent significant SNP, the two were merged into the same genomic locus. This 250 kb threshold was determined as the closest distance between SNPs in LD with an r2 ≥ 0.6 for each independent significant SNP.
For this analysis, we used FUMA, an integrative web-based platform using information from multiple biological resources to facilitate functional mapping and annotation of GWAS results38.
Identifying known loci
To determine whether a locus had been previously associated with blood pressure-related traits (systolic blood pressure, diastolic blood pressure, pulse pressure, hypertension, and mean arterial pressure), we leveraged the results from the earlier FUMA analysis. Specifically, we focused on candidate SNPs, which were defined as those in linkage disequilibrium (LD) with any independent significant SNP, where r2 ≥ 0.6. We considered all SNPs tested in the GWAS, regardless of their p-value in the association test, as well as those present in the UKB release 2b reference panel, totaling 9,020 SNPs. These candidate SNPs were queried against the GWAS Catalog (www.ebi.ac.uk/gwas/, version 1.0.2, last updated on September 22, 2024), filtered by each BP trait, and considered as LD proxies. Additionally, we identified loci as known if any SNP previously associated with BP traits in the GWAS catalog was located within the locus region, treating these variants as locus proxies. Finally, we examined whether the published BP SNPs curated by Keaton et al.7 were within our loci (as locus proxies) or in LD with our independent significant SNPs (as LD proxies).
Systolic blood pressure polygenic risk score (PRS)
PRS is an individual quantitative metric for disease/phenotype inherited risk based on the mathematical aggregation of the effect sizes of each common genetic variant (MAF ≥ 1%) tested on GWAS for a given trait, multiplied by its genotype assuming an additive model (0, 1 and 2, corresponding the number of risk allele). For our PRS, we used summary statistics from the GWAS for SBP from this study with 286,581 individuals from the UKB using only 944,384 variants also presented in the Baependi Heart Study, the EpiGen project – Pelotas, and the HapMap database linkage disequilibrium reference panel provided by LDPred2 tool used39. Genetic variants with ambiguous strands were previously removed.
We derived the PRS using the LDPred2-auto model39, a new version of the LDpred9 algorithm implemented in the R package bigsnpr. This tool uses a Bayesian approach and calculates the posterior mean effect size for each variant based on how this variant is correlated with similarly associated variants in the reference panel. The LDPred2-auto model simultaneously fits the proportion of causal variants (p) and the heritability hyper-parameters (h2), integrating both in an iterative process using the Gibbs sampler algorithm. In a Bayesian framework, the Gibbs sampler performs successive approximations of the conditional distributions of each parameter (p, h2), based on the value of the other at each iteration, facilitating the calculation of their joint posterior distribution. This approach optimizes the model without the need to specify a large grid of parameter combinations, significantly reducing computational demands and, consequently, analysis time, without the requirement of training data.
Using the effect size of each variant recalculated using LDPred2, we calculated the PRS in a validation dataset of 73,810 individuals (one sixth) from the UKB to LDPred2 select the best model (auto mode). This model was, then, used in subsequent analyses in independent testing datasets from the UKB and Brazilian populations.
For all three testing datasets, PRS was calculated for each individual, multiplying the genotype dosage obtained in imputation for each selected variant by its respective weight recalculated by LDPred2, and then summing all variants in the score. For this, we used PLINK2 software.
Statistical analysis
The baseline characteristics of each three datasets were summarized using descriptive statistics. Continuous phenotypes were summarized as mean and standard deviation and compared using one-way ANOVA followed by Tukey t-test. The categorical phenotypes were summarized as counts and percentage and compared using Chi-square test.
We estimated the heritability of the 944,384 genetic variants used in PRS on SBP using the validation dataset and BOLT-LMM software37. The association of SBP and DBP with the PRS (per 1-SD unit increase and PRS categories) in the UKB, Baependi and Pelotas testing datasets was assessed by multivariable- adjusted linear regression accounting for age, age2, sex, BMI, and the first four principal components. The association of PRS (per standard deviation and PRS categories) with prevalent hypertension was assessed using a multivariable-adjusted logistic regression model in all three testing datasets accounting for the same covariates aforementioned. The relationship between the top PRS decile (highest polygenic score) and hypertension severity was determined using multinomial logistic regression.
The predictive model for hypertension including PRS only, clinical data only using the covariates included in regression models and both (complete model) was developed separately in UKB, Baependi and Pelotas testing datasets using 60% of each dataset to train and 40% to validate the models. Dataset separation was carried out using the CARET R package, balancing the data based on the same features used to split the UKB dataset: hypertension status, sex, and age group. Models performance were assessed through discrimination using area under the receiver operating characteristic curve (AUC) concordance and macro weighted F1 scores, which provides a balanced view of precision and recall, taking into account class imbalances. We used the DeLong test to compare the difference in AUCs between models and assess whether the performance difference is statistically significant. All analyses were conducted using the R package pROC.
Supplementary Information
Acknowledgements
We thank the participants and investigators of UKB, Baependi Heart Study, and EpiGen-Pelotas for contributing to this work.
Author contributions
Conceptualization: SKT, JEK. Data curation: SKT, FPNR, JMN, AVVJ, JLP, BLH, ACP. Formal analysis: SKT, FPNR, JMN, AVVJ, JLP. Funding Acquisition: JEK. Investigation: SKT, FPNR, JMN, AVVJ, JLP. Methodology: SKT, FPNR, JMN, AVVJ, JLP. Project Administration: SKT. Resources: JEK. Software: SKT, FPNR, JMN, JLP. Validation: SKT, FPNR. Visualization: SKT, FPNR; Supervision: SKT, JEK; Writing original draft: SKT. Writing review & editing: SKT, FPNR, JMN, AVVJ, JLP, BLH, ACP, JEK.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [INCT—2014/50889–7, 2013/17368–0 and 2015/50216–5 to JEK], the Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq [INCT—465586/2014–7, 309179/2013–0 and 442643/2020–9 to JEK] and the Zerbini Foundation and Foxconn Brazil as part of a research grant “Machine Learning in Cardiovascular Medicine”.
Data availability
All data from the UK Biobank is available (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access) to researchers from universities and other institutions with genuine research inquiries following institutional review board and UK Biobank approval. This research was conducted using the UK Biobank resource under application number 14654. The genome-wide association data is available in GWAS catalog repository (https://www.ebi.ac.uk/gwas/) under GCP ID GCP000817. Finally, the full SBP PRS weights derived using UK Biobank data and the PRS extrapolated for each individual from the three testing datasets are available github repository (https://github.com/LGCM-OpenSource/PRS_SBP_Admixed) and in the Polygenic Score Catalog (https://www.pgscatalog.org/) when accepted by the journal.
Declarations
Competing interests
The author(s) declare no competing interests.
Ethics approval and consent to participate
This study was approved by University of São Paulo Medical School ethical committee (CAAE number 37534720.0.0000.0068) and was conducted according to the guidelines of the Declaration of Helsinki. Written informed consent was obtained for all subjects involved in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Samantha K. Teixeira and Fernando P. N. Rossi have contributed equally to this work.
Contributor Information
Samantha K. Teixeira, Email: samantha.teixeira@hc.fm.usp.br
Jose E. Krieger, Email: j.krieger@hc.fm.usp.br
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79683-7.
References
- 1.WHO. Global health risks. WHO Libr. Cat. Data (2009).
- 2.Chobanian, A. V. et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA289, 2560–2572 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Hajjar, I. & Kotchen, T. A. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1988–2000. JAMA290, 199–206 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Kearney, P. M. et al. Global burden of hypertension: analysis of worldwide data. Lancet365, 217–223 (2005). [DOI] [PubMed] [Google Scholar]
- 5.de Oliveira, C. M., Pereira, A. C., de Andrade, M., Soler, J. M. & Krieger, J. E. Heritability of cardiovascular risk factors in a Brazilian population: Baependi Heart Study. BMC Med. Genet.9, 32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehret, G. B. Genome-wide association studies: Contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep.12, 17–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keaton, J. M. et al. Genome-wide analysis in over 1 million individuals of European ancestry yields improved polygenic risk scores for blood pressure traits. (2024). [DOI] [PMC free article] [PubMed]
- 8.Privé, F., Aschard, H. & Blum, M. G. B. Efficient implementation of penalized regression for genetic risk prediction. Genetics212, 65–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet.97, 576–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khera, A. V. et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell177, 587-596.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet.50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaura, F. et al. Polygenic risk scores predict hypertension onset and cardiovascular risk. Hypertension10.1161/HYPERTENSIONAHA.120.16471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kember, R. L. et al. Polygenic Risk Scores for Cardio-renal-metabolic Diseases in the Penn Medicine Biobank. bioRxiv10.1101/759381 (2019). [Google Scholar]
- 14.Fahed, A. C. et al. Transethnic transferability of a genome-wide polygenic score for coronary artery disease. Circ. Genomic Precis. Med.10.1161/CIRCGEN.120.003092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parcha, V. et al. Association of a multiancestry genome-wide blood pressure polygenic risk score with adverse cardiovascular events. Circ. Genomic Precis. Med.15, E003946 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehdy, F. S. G. et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc. Natl. Acad. Sci. U. S. A.112, 8696–8701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giolo, S. R. et al. Brazilian urban population genetic structure reveals a high degree of admixture. Eur. J. Hum. Genet.20, 111–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet.51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evangelou, E. et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet.50, 1412–1425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitarello, B. D. & Mathieson, I. Polygenic scores for height in admixed populations. bioRxiv10.1534/g3.120.401658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Y. et al. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat. Commun.11, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai, M. et al. A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits. Am. J. Hum. Genet.108, 632–655 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavazos, T. B. & Witte, J. S. Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. Hum. Genet. Genomics Adv.2, 100017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truong, B. et al. Integrative polygenic risk score improves the prediction accuracy of complex traits and diseases. bioRxiv 1–23 (2023). [DOI] [PMC free article] [PubMed]
- 25.Mills, M. C. & Rahal, C. A scientometric review of genome-wide association studies. Commun. Biol.10.1038/s42003-018-0261-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobin, M. D., Sheehan, N. A., Scurrah, K. J. & Burton, P. R. Adjusting for treatment effects in studies of quantitative traits: Antihypertensive therapy and systolic blood pressure. Stat. Med.24, 2911–2935 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Egan, K. J. et al. Cohort profile: The Baependi Heart Study - A family-based, highly admixed cohort study in a rural Brazilian town. BMJ Open vol. 6 (2016). [DOI] [PMC free article] [PubMed]
- 29.Kowalski, M. H. et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet.15, 1–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Victora, C. G. & Barros, F. C. Cohort profile: the 1982 Pelotas (Brazil) birth cohort study. Int. J. Epidemiol.35, 237–242 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Whelton, P. K. et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults. J. Am. Coll. Cardiol.71, e127–e248 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience4, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abraham, G., Qiu, Y. & Inouye, M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics33, 2776–2778 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Rossi, F. P. N. et al. GARSA: An integrative pipeline for genome wide association studies and polygenic risk score inference in admixed human populations. bioRxivMay, 1–8 (2023).
- 36.Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res.19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet.47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe, K., Taskesen, E., Van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Privé, F., Arbel, J. & Vilhjálmsson, B. J. LDpred2: better, faster, stronger. Bioinformatics1, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from the UK Biobank is available (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access) to researchers from universities and other institutions with genuine research inquiries following institutional review board and UK Biobank approval. This research was conducted using the UK Biobank resource under application number 14654. The genome-wide association data is available in GWAS catalog repository (https://www.ebi.ac.uk/gwas/) under GCP ID GCP000817. Finally, the full SBP PRS weights derived using UK Biobank data and the PRS extrapolated for each individual from the three testing datasets are available github repository (https://github.com/LGCM-OpenSource/PRS_SBP_Admixed) and in the Polygenic Score Catalog (https://www.pgscatalog.org/) when accepted by the journal.