Abstract
Metabolic-associated fatty liver disease (MAFLD) is a common chronic condition that poses a significant threat to human health. Mitochondrial dysfunction, particularly involving the mitochondrial Ca2+ uniporter (MCU), plays a key role in its pathogenesis. This study aimed to investigate the impact of the MCU gene on hepatic lipid metabolism in mice fed a high-fat diet. Using MCU knockout and wild-type mice, subjected to either a high-fat or normal diet for 14 weeks, we observed notable Steatosis and liver weight gain in MCU-deficient mice. Liver function markers, serum triglycerides, very low-density lipoprotein (VLDL) levels, and ApoB protein expression were all significantly elevated. Mechanistic studies revealed that MCU deletion led to mitochondrial dysfunction, increased oxidative stress. These findings highlight the critical role of the MCU gene in maintaining hepatic lipid balance and suggest its potential as a therapeutic target for managing nonalcoholic fatty liver disease.
Keywords: MCU, MAFLD, Fat liver, Lipid transport-associated, Lipid dysregulation
Subject terms: Molecular biology, Gastroenterology
Introduction
Metabolic associated fatty liver disease (MAFLD), a chronic liver disease, is closely linked to metabolic disorders and primarily manifests as hepatic fat accumulation, inflammation, and fibrosis1,2. MAFLD, the prevailing chronic liver disease globally, exhibits an escalating incidence and mortality rate3, and is intricately associated with metabolic disorders including obesity, diabetes, cardiovascular disease, and others4. Recent years have witnessed advancements in the diagnosis criteria, classification, staging, mechanisms, and treatment of MAFLD, although numerous unresolved issues persist5.
Extensive research has consistently linked obesity, type 2 diabetes mellitus (T2DM), and MAFLD to mitochondrial dysfunction6–8. The mitochondrial calcium uniporter (MCU), a multi-subunit complex embedded in the inner mitochondrial membrane, is essential for maintaining mitochondrial calcium homeostasis, which is vital for normal mitochondrial function9,10, MCU regulates mitochondrial bioenergetics through the modulation of calcium-dependent dehydrogenases and the promotion of ATP synthesis11. Dysregulation of MCU has been shown to exacerbate mitochondrial dysfunction, disturbing metabolic homeostasis and contributing to the pathological conditions seen in MAFLD12–14. Specifically, deletion of the MCU gene in mouse liver impairs mitochondrial calcium uptake, delays cytoplasmic calcium clearance, and diminishes oxidative phosphorylation, which collectively lead to increased hepatic lipid accumulation—a hallmark of MAFLD. Moreover, studies have illustrated that mitochondrial calcium transporters, including MCU, facilitate the recruitment of UCP1 to form thermogenic proteins in brown fat, linking MCU dysfunction to broader metabolic disorders including obesity15.
To deepen our understanding of MCU’s involvement in MAFLD, this research utilized liver-specific MCU knockout mice to develop a high-fat diet-induced MAFLD model. The impact of MCU deficiency on hepatic lipid metabolism capacity was assessed, alongside the analysis of MCU deficiency’s influence on hepatic steatosis, liver dysfunction, oxidative stress, and liver lipid metabolism.
In the high-fat diet-induced MAFLD mouse model, MCU deficiency exacerbates hepatic lipid disorder in HFD-induced MAFLD mice, unveiling its contributory role and providing a new avenue for targeted therapies. The findings of this study unveiled the contributory role of MCU in MAFLD, furnishing a theoretical foundation for exploring novel therapeutic approaches that specifically target MCU symptoms at presentation, physical exams and lab results.
Materials and methods
Animals
The animal procedures were performed following the guidelines, and with approval, of the animal experimental ethics guidelines of Guangxi University (no. GXU2019-067). All studies involving live animals to be reported as described by the ARRIVE guidelines (PLoS Bio 8(6), e1000412, 2010 (https://www.nc3rs.org.uk/arrive-guidelines)
MCU+/− male mice with hepatocyte-specific heterozygous MCU knockout were produced by breeding MCUfl/fl (RRID: JAX, Cat# 029817) mice with hepatocyte-specific Cre recombinase transgenic mice (RRID: Alb-cre C57BL/6J; Cyagen). The mice were housed at a temperature of 23 °C ± 2 °C, with a humidity range of 50–70%, and subjected to a 12-h light–dark cycle. Mice from the same litter were randomly assigned to one of four groups, each containing six mice: (1) ND-MCUfl/fl, (2) ND-MCUΔhep, (3) HFD-MCUfl/fl and (4) HFD-MCUΔhep. The ND groups were provided with standard control chow (TP2330055MC, produced by Trophic Animal Feed High-Tech Co; Haian, China), while the HFD groups were fed a high-fat diet (TP2330055M, produced by Trophic Animal Feed High-Tech Co; Haian, China) consisting of 60% fat to induce a high-fat model (The composition of ingredients is shown in Table 1). At 22 weeks of age (i.e., 14 weeks of modeling), the mice underwent a 12-h fasting period before being euthanized by cervical dislocation. Blood was collected from the eyeballs of mice after anesthesia. Liver tissue, epididymal fat, and subcutaneous fat were collected and placed in liquid nitrogen immediately. The samples were then transferred to an ultra-low temperature refrigerator for storage.
Table 1.
The composition of the 60% high-fat diet.
| Ingredients | g/kg |
|---|---|
| Casein | 267 |
| Maltodextrin | 157 |
| Sucrose | 89 |
| Soybean oil | 33 |
| Lard | 301 |
| Cellulose | 67 |
| Mineral mix, M1020 | 66 |
| Vitamin mix, V1010 | 13 |
| l-Cystine | 4 |
| Choline bitartrate | 3 |
| TBHQ | 0.067 |
| Total | 1000 |
Body composition analysis
The fat mass and lean mass of mice were measured at various stages using Nuclear Magnetic Resonance (NMR) (Niumag QMR23-060H-I, Suzhou, China). The NMR machine was calibrated and preheated with standards, after which the mice were weighed and positioned inside the NMR chamber. Subsequently, the mouse body weight data were recorded and analyzed. The fat mass, lean mass, and water content of the mice were then determined, and the data were recorded, exported, and saved.
Glucose tolerance test (GTT) and insulin tolerance test (ITT)
GTT: Following a 16-h fast at 16 weeks of age, mice were intraperitoneally injected with glucose (2 g/kg body weight), and their tail vein glucose levels were measured using a glucometer at 0, 15, 30, 60, 90, and 120 min post-injection.
ITT: Mice were subjected to a 6-h fasting period at 18 weeks of age followed by intraperitoneal injection of insulin at a dosage of 0.75 U/kg body weight, Subsequently, the tail vein glucose levels were measured at 0, 15, 30, 60, 90, and 120 min after injection using a blood glucose meter.
Histopathological analysis
Fresh mouse liver and adipose tissue were obtained and subsequently fixed overnight in a 4% fixative (Sorabio, Beijing, China) for the preparation of paraffin and frozen sections. Hematoxylin–eosin (H&E) staining was performed on the paraffin sections, whereas Oil Red O staining was carried out on the frozen sections16. Specimen sectioning and staining were done by Wuhan Safeway Biotechnology Co. Image acquisition was conducted utilizing a light microscope (MSHOT ML31 Biomicroscope, Guangzhou, China). Quantification was performed using the ImageJ software (ImageJ 1.8.0, NIH, USA).
Serum and tissue biochemical analysis
Liver tissues were lysed in RIPA buffer (Solarbio; Beijing, China) for 12 h. After centrifugation at 4 °C and 12,000 rpm for 10 min, the supernatant was collected for further analysis. The levels of liver triglycerides (TG), total cholesterol (TC), total bile acids (TBA), ATP content, malondialdehyde (MDA) levels, and superoxide dismutase (SOD) activity were determined using commercial kits. For TG, TC, and TBA measurements, kits from Njjcbio (Nanjing, China) were utilized, following the manufacturer’s instructions. ATP, MDA, and SOD were assessed using kits provided by Beyotime (Shanghai, China).
Blood samples were incubated at 4 °C for 1 h to allow them to settle, followed by centrifugation at 4 °C for 20 min at 3000 rpm. Subsequently, serum supernatants were collected. Levels of serum alanine aminotransferase (ALT), low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) were determined using a kit from Njjcbio (Nanjing, China), very low-density lipoprotein cholesterol (VLDL-C) was determined using a kit from Enzyme-linked Biotechnology (Shanghai, China), following the manufacturer’s instructions.
Protein concentrations were determined using the bicinchoninic acid assay kit from Beyotime (Shanghai, China). The optical density was measured using an Infinite M200 Pro microplate reader (Teacan; Männedorf, Switzerland). All values were normalized to the protein concentration17.
RNA and mtDNA extraction and real-time quantitative PCR
Total RNA was extracted using Trizol reagent (Invitrogen, Beijing, China). The complementary DNA (cDNA) was synthesized from 1 µg of RNA using M-MLV reverse transcriptase (Promega, Beijing, China). RT-PCR assays were conducted using the Real Star Green Fast mixture with ROXII (GeneStar, Beijing, China) on a qTOWER3G thermal cycler (Analytik, Jena, Germany). The primer sequences are listed in Table 2. The relative mRNA expression levels were analyzed using the 2−ΔΔCT method18.
Table 2.
Primers used for a quantitative polymerase chain reaction.
| Gene name | Primer (5′–3′) |
|---|---|
| β-actin | F: CATTGCTGACAGGATGCAGAAGG |
| R: TGCTGGAAGGTGGACAGTGAGG | |
| Ampk | F: GACTTCGTTGCCATCCTGGATC |
| R: CCAAGCTGACTGGTAACCACAG | |
| Accα | F: GTTCTGTTGGACAACGCCTTCAC |
| R: GGAGTCACAGAAGCAGCCCATT | |
| Acc2 | F: AGAAGCGAGCACTGCAAGGTTG |
| R: GGAAGATGGACTCCACCTGGTT | |
| Atgl | F: GGAACCAAAGGACCTGATGACC |
| R: ACATCAGGCAGCCACTCCAACA | |
| Camp | F: CTTCAACCAGCAGTCCCTAGAC |
| R: GCCACATACAGTCTCCTTCACTC | |
| Chrebp | F: GAGTGCTTGAGCCTGGCTTACA |
| R: GCTCTCCAGATGGCGTTGTTCA | |
| Cox1 | F: CTCAAAGGACTTGGCGGTACT |
| R: GAGATGGTGAGGTAGAGCGG | |
| Cpt1 | F: GATGAACTTCTTCTTCCAGGAGTGC |
| R: CCATCTCCAGTCCGCATTTT | |
| Fasn | F: AAGTTGCCCGAGTCAGAGAACC |
| R: ATCCATAGAGCCCAGCCTTCCATC | |
| Gluk | F: GTATGTGCTCCTCGCTTGCCTA |
| R: TTTCCACCACGTCTGAGTCAGG | |
| Glut2 | F: TTTCCACCACGTCTGAGTCAGG |
| R: ATCACGGAGACCTTCTGCTCAG | |
| mMito | F: CTAGAAACCCCGAAACCAAA |
| R: CCAGCTATCACCAAGCTCGT | |
| mB2MF1 | F: ATGGGAAGCCGAACATACTG |
| R: CAGTCTCAGTGGGGGGGAAT | |
| Mttp | F: CCAGGAAAGGTTCCTCTATGCC |
| R: GACTCTCTGATGTCGTTGCTTGC | |
| Pdk4 | F: GTCGAGCATCAAGAAAACCGTCC |
| R: GCGGTCAGTAATCCTCAGAGGA | |
| Pfk | F: TCATCGAGTCGGTCTGTGACGA |
| R: CATGGCTTCTGCTGAGTTGCAG | |
| Pgc1α | F: GAATCAAGCCACTACAGACACCG |
| R: CATCCCTCTTGAGCCTTTCGTG | |
| Pgc1β | F: AAGCGCTTTGAGGTGTTCG |
| R: GCTCATTGCGCTTTCTCAGG | |
| Pklr | F: CGAAAAGCCAGTGATGTGGTGG |
| R: GATGCCATCGCTCACTTCTAGG | |
| Pparα | F: ACCACTACGGAGTTCACGCATG |
| R: GAATCTTGCAGCTCCGATCACAC | |
| Pparg | F: GTACTGTCGGTTTCAGAAGTGCC |
| R: ATCTCCGCCAACAGCTTCTCCT | |
| Srebp1 | F: AGGAGGACATCTTGCTGCTTCT |
| R: GATCTCTGCCAGTGTTGCCATG | |
| Hmgcr | F: GCTCGTCTACAGAAACTCCACG |
| R: GCTTCAGCAGTGCTTTCTCCGT | |
| Hmgcs1 | F: GGAAATGCCAGACCTACAGGTG |
| R: TACTCGGAGAGCATGTCAGGCT | |
| Hmgcs2 | F: TGCTATGCAGCCTACCGCAAGA |
| R: GCCAGGGATTTCTGGACCATCT | |
| Acly | F: AGGAAGTGCCACCTCCAACAGT |
| R: CGCTCATCACAGATGCTGGTCA | |
| Fdft1 | F: GGATGTGACCTCCAAACAGGAC |
| R: CAGACCCATTGAGTTGGCACAC |
50 mg of mouse liver tissue was homogenized in 1 mL of PCR lysate containing 1% Proteinase K. After lysis for 1 h at 37 ℃, heat at 80 ℃ for 10 min and centrifuge, the supernatant was used for RT-qPCR. To detect the number of mitochondria in the liver, primers for mouse MtDNA (mMitoF1, mMitoR1) was examined using and nuclear gene Beta-2 Microglobulin (B2M) as an internal reference.
Western blotting
Liver tissue proteins were extracted using RIPA lysis solution containing 1% protease inhibitor (Solarbio, Beijing, China). The supernatants were collected by centrifugation at 12,000 rpm for ten minutes, and the protein concentration was determined using the BCA assay. Next, the appropriate amount of 4× loading buffer was added to the samples, which were then boiled at 100 °C for 10 min19. Following SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transfer to PVDF membranes, the membranes were then incubated overnight at 4 °C with specific primary antibodies, including MCU antibody (RRID: CST 14997S), ApoB antibody (ABclonal A4184), β-actin antibody (CST 93473), α-tublin (Proteintech, 14555-1-AP). The primary antibodies were incubated with the membranes overnight at 4 °C. The membranes were then incubated with a Goat Anti-Rabbit secondary antibody (RRID: Jackson 111-035-003). The membranes were visualized using a Bio-Rad imaging system (Bio-Rad Universal Hood II; Bio-Rad, Hercules, CA, USA), and the obtained images were quantitatively analyzed using ImageJ software (NIH, USA, version 1.8.0).
Statistical analysis
The results are presented as the mean ± standard deviation (SD). Statistical significance was determined using unpaired t-tests or one-way analysis of variance (ANOVA) conducted in Excel. Different letters (a, b, c, d) indicate statistically significant differences between groups, while the same letter indicates no significant difference between the groups. p-value < 0.05 was considered significant.
Results
High-fat feeding induced high-fat model in mice successfully
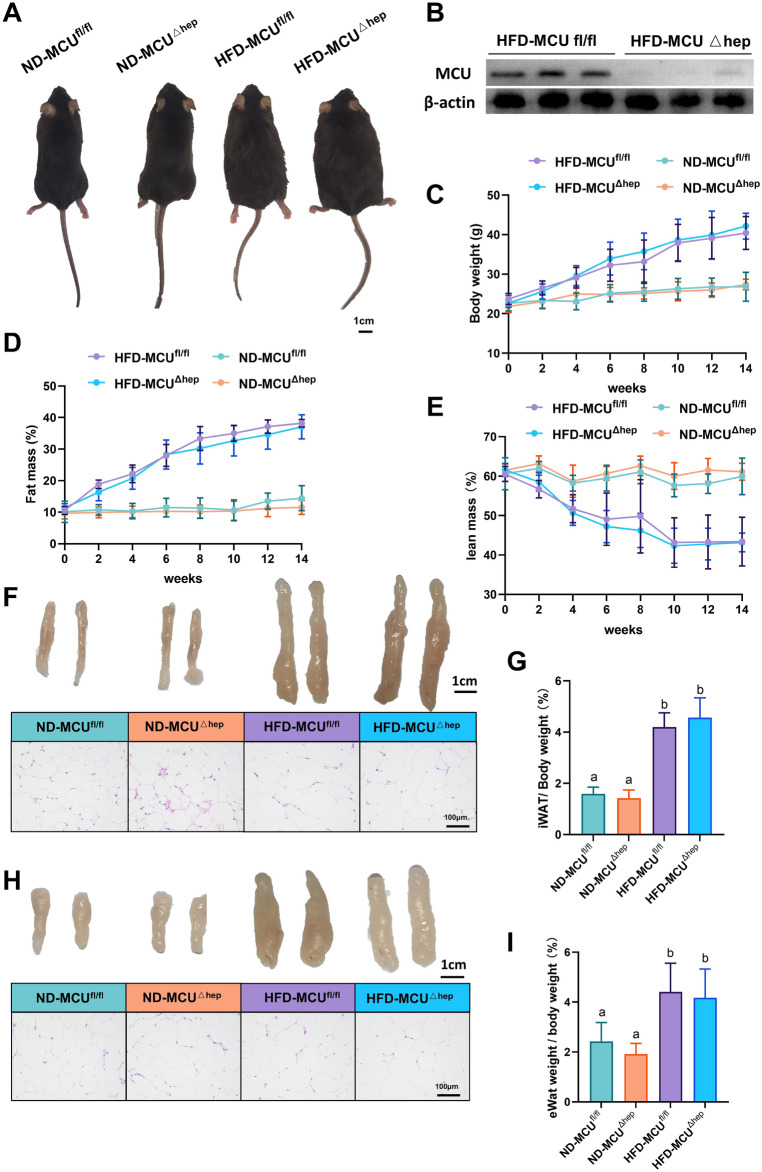
To confirm the knockout of the MCU gene in the livers of mice in the HFD-MCUΔhep group, liver samples from both the HFD-MCUfl/fl and HFD-MCUΔhep groups were subjected to western blotting (Fig. 1B). After a 14-week high-fat diet intervention, mice in the HFD group displayed a significantly larger physique and greater weight compared to those in the ND group. Regardless of diet, the ablation of Mitochondrial Calcium Uniporter (MCU) in the liver did not instigate changes in body size or weight (Fig. 1A,C). Furthermore, Magnetic Resonance Imaging (MRI) data revealed that the high-fat diet led to an increase in adiposity and a decrease in lean mass in mice, yet this outcome was not modified by the hepatic MCU knockout (Fig. 1D,E). Analysis of iWAT and eWAT showed that the HFD group had a significant increase in fat mass compared to the ND group (Fig. 1F–I). Additionally, hematoxylin and eosin (H&E) staining exhibited larger adipocytes in the HFD group. Similarly, the absence of hepatic MCU could not induce changes in these two types of white adipose tissue (Fig. 1F,H, S1A,B). In conclusion, these findings confirm the successful establishment of the high-fat diet model in mice following 14 weeks of HFD feeding. Nevertheless, the knockout of hepatic MCU did not significantly affect mouse iWAT and eWAT.
Fig. 1.
The consumption of a high-fat diet leads to a significant increase in both body weight and fat mass in mice (n = 6). (A) Representative images of mice under different treatment conditions; Scale bar: 1 cm. (B) Assessment of liver MCU protein levels in the HFD-MCUfl/fl and HFD-MCU∆hep groups. (C) Measurement of body weights in mice subjected to various treatments. (D) Evaluation of fat mass in mice exposed to different treatments. (E) Analysis of lean mass in mice following treatments. (F) Representative photographs and histological analysis of inguinal fat in mice; Scale bar: 1 cm and 100 µm (40× objective). (G) Relative weight of intra-abdominal white adipose tissue (iWAT) to body weight in different groups of mice. (H) Representative photographs and H&E staining analysis of epididymal fat pads in mice; Scale bar: 1 cm and 100 µm (40× objective). (I) Relative weight of epididymal white adipose tissue (eWAT) to body weight in different groups of mice. The results are presented as means ± standard deviation (SD). Different letters meant there was significant difference among groups (p < 0.05).
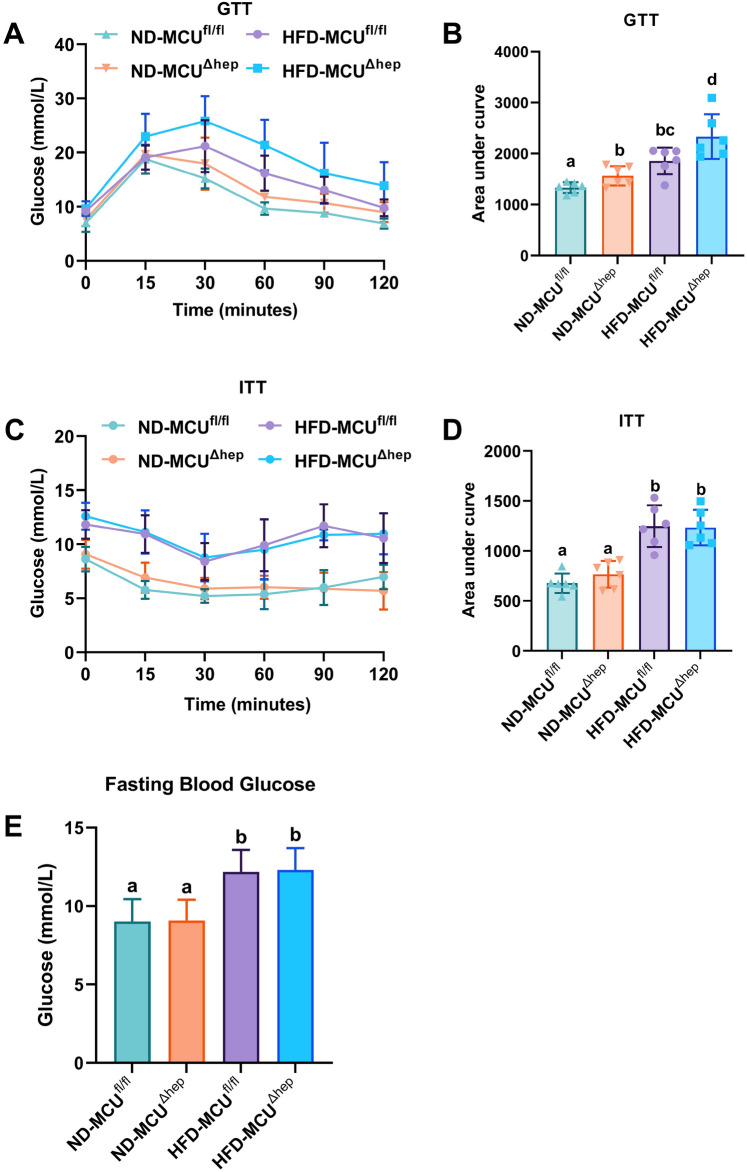
Liver-specific knockout of MCU results in impaired glucose tolerance and reduced insulin sensitivity in mice with high-fat diet (HFD)-induced obesity
Glucose tolerance test (GTT) and insulin tolerance test (ITT) are commonly employed to assess glucose metabolism in mice. During the glucose tolerance test (GTT), mice in the high-fat diet (HFD) groups displayed elevated blood glucose levels and area under the curve (AUC) compared to the normal diet (ND) groups (Fig. 2A,B). Interestingly, the HFD-MCUΔhep group exhibited even higher blood glucose levels and AUC compared to the HFD-MCUfl/fl group (Fig. 2A,B). These findings suggest that liver-specific deletion of the MCU gene impairs glucose tolerance in mice. During the ITT, mice in the HFD groups exhibited elevated blood glucose levels and AUC compared to the ND groups (Fig. 2C,D). However, induced by a high-fat diet, the HFD-MCUΔhep group displayed the similar blood glucose levels and AUC compared to the HFD-MCUfl/fl group (Fig. 2C,D), the fasting blood glucose levels results (Fig. 2E) demonstrated that while a high-fat diet significantly increased fasting blood glucose levels, deletion of MCU in the liver did not affect fasting blood glucose in these mice.
Fig. 2.
The impact of liver-specific knockout of MCU on glucose metabolism was evaluated in mice (n = 6). (A,B) Serum glucose concentrations of 14-week-old mice during glucose tolerance tests (GTTs). (C,D) Serum glucose concentrations of 18-week-old mice during insulin tolerance tests (ITTs). (E) Fasting blood glucose levels. All the results were shown as means ± SD. Different letters meant there was significant difference among groups (p < 0.05).
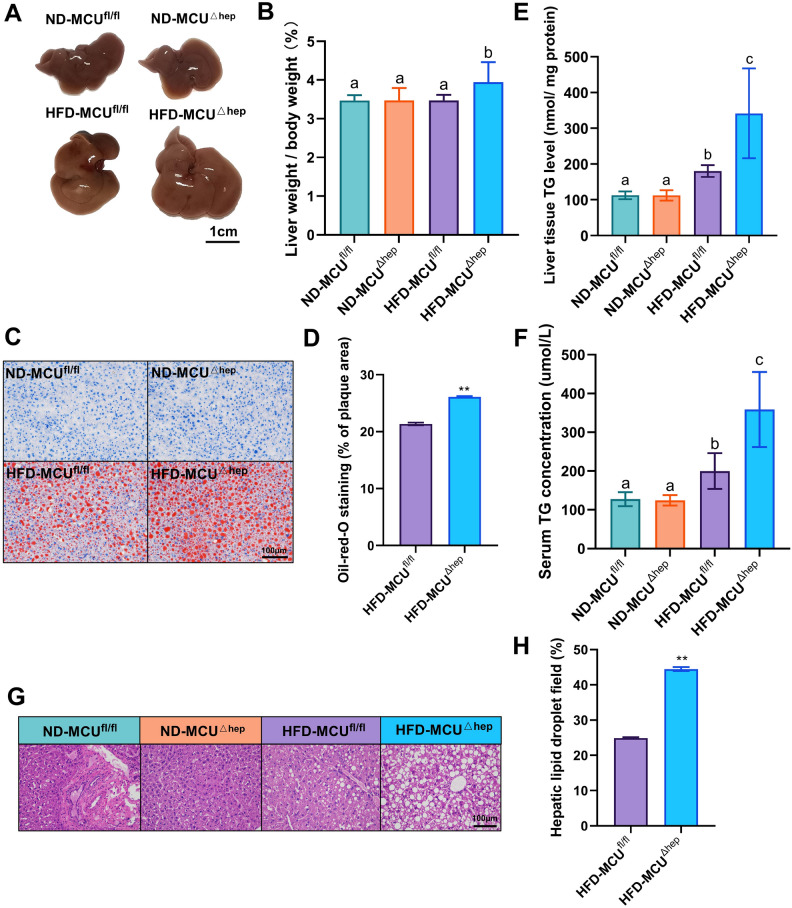
The liver-specific knockout of MCU was found to enhance hepatic steatosis induced by a high-fat diet
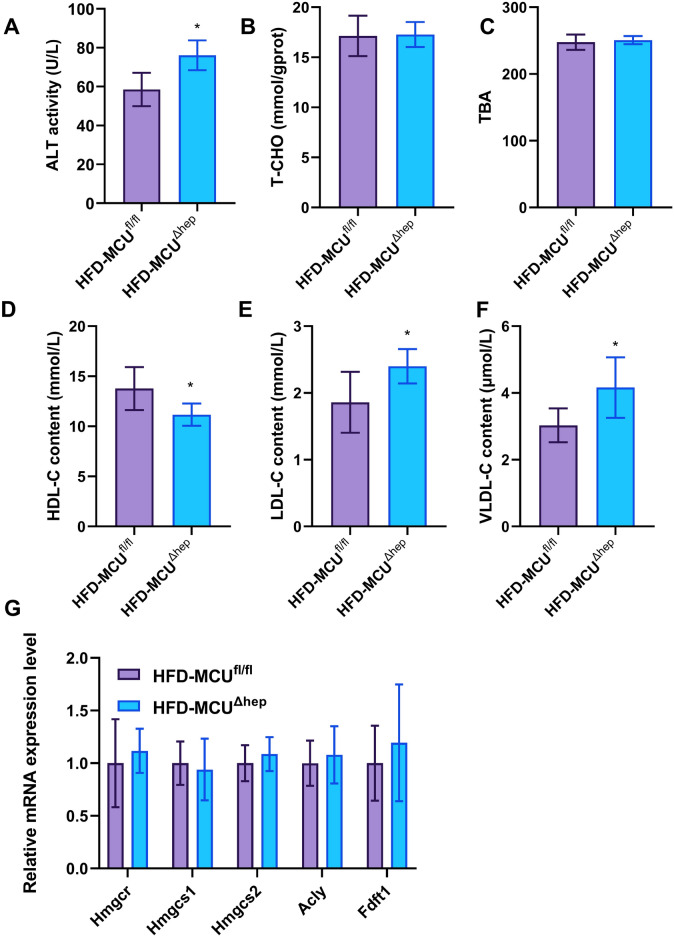
Hepatic steatosis represents a primary manifestation of fatty liver diseases. Consequently, we examined the impact of MCU gene deletion on the liver. Initially, we observed that mice fed a high-fat diet exhibited enlarged and granulated livers, while those fed a standard diet displayed smaller and dark red livers (Fig. 3A). Notably, the livers of the MCUΔhep mice were marginally larger compared to the MCUfl/fl mice (Fig. 3A,B). Subsequently, hepatic lipid accumulation was evaluated through Oil red O staining examination. The livers of MCUΔhep mice exhibited a greater number of larger lipid droplets and vacuoles, in comparison to the MCUfl/fl control group (Fig. 3C,D). Further, we assessed liver function and observed substantially elevated levels of liver and serum triglycerides (TG) (Fig. 3E,F), compared to the ND group, the TG levels are significantly increased in the high-fat diet group, and MCU deletion further significantly increases the TG levels. Consistently, HE staining of the liver showed more vacuoles in the liver as a result of MCU deletion (Fig. 3G,H). In the HFD-MCU∆hep group, compared to the HFD-MCUfl/fl group, we first observed a notable change in serum Alanineaminotransferase (ALT) levels (Fig. 4A), indicating that liver-specific MCU deficiency may influence liver function. Furthermore, we measured the levels of total cholesterol (TC) and total bile acids (TBA) in the liver, finding that MCU deficiency had no significant impact on cholesterol metabolism (Fig. 4B,C). We also assessed the expression of genes related to cholesterol metabolism and found no significant changes (Fig. 4G). Interestingly, MCU deletion resulted in a significant downregulation of HDL levels (Fig. 4D). Additionally, we observed changes in serum low-density lipoprotein cholesterol (LDL-C) and VLDL levels (Fig. 4E,F), strongly suggesting that liver MCU deficiency exacerbates the accumulation of fatty lipids.
Fig. 3.
Liver-specific knockout of MCU leads to an elevation in HFD-induced hepatic steatosis (n = 6). (A) Representative liver tissue photographs; Scale bar: 1 cm. (B) Liver weight-to-body weight ratio in different groups of mice. (C) Liver oil red O staining analysis images; Scale bar: 100 µm (40× objective). (D) Quantification of oil red O staining. (E) Hepatic triglyceride (TG) concentration. (F) Serum triglyceride concentration. (G) Representative liver H&E staining analysis images; Scale bar: 100 µm (40× objective). (H) Quantification of HE staining. All results are presented as means ± SD, with *p < 0.05/**p < 0.01 indicating statistical significance. Different letters meant there was significant difference among groups (p < 0.05).
Fig. 4.
Effects of MCU on liver biochemical function and lipid metabolism in mouse liver. (A) Serum ALT (alanine aminotransferase) levels in mice. (B) Liver TC (total cholesterol) content in mice. (C) Liver TBA (total bile acids) content in mice. (D) Serum HDL (high-density lipoprotein cholesterol) levels in mice. (E) Serum LDL-C (low-density lipoprotein cholesterol) levels in mice. (F) Serum VLDL-C (very low-density lipoprotein cholesterol) levels in mice. (G) Expression levels of cholesterol-related genes in mouse liver. All results are presented as means ± SD, with *p < 0.05/**p < 0.01 indicating statistical significance.
Hepatic MCU deletion modulates the expressions of lipid transport genes
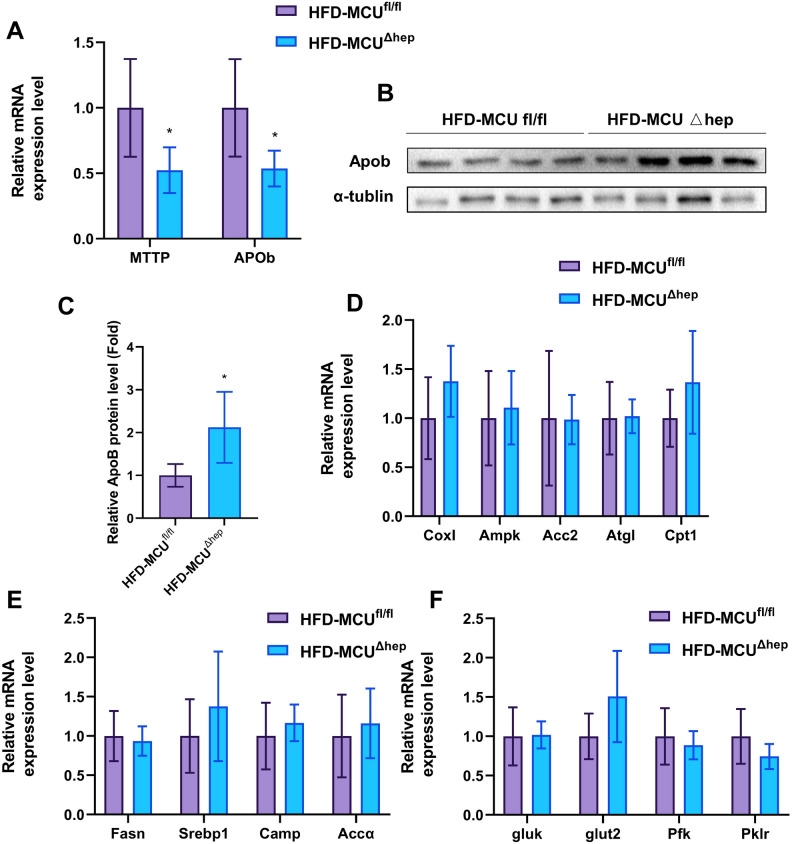
In order to further investigate the influence of MCU on glucolipid metabolism in the liver, we assessed the mRNA expression of key genes related to fatty acid biosynthesis, catabolism, lipid transport, and glucose transport in liver tissues. The results indicated that hepatic deletion of MCU significantly downregulated the mRNA expression of key genes involved in lipid transport, including Mttp and Apob, in liver tissue (Fig. 5A). However, immunoblotting showed that hepatic deletion of MCU resulted in elevated expression of APOB (Fig. 5B,C), which was consistent with increased VLDL-C and LDL-C in serum. This suggests that hepatic MCU deletion may regulate APOB expression from the translational level. In contrast, hepatic MCU did not affect the expression of genes involved in fat oxidative catabolism, fat biosynthesis, and glucose transport (Fig. 5D–F). Overall, our data strongly indicate a close association between hepatic MCU and fatty acid transport.
Fig. 5.
Gene expression analysis was performed in mice (n = 5–6). (A) Genes associated with lipid transport. (B) Assessment of liver ApoB protein levels in the HFD-MCUfl/fl and HFD-MCU∆hep groups. (C) Densitometric quantification of the blotting. (D) Genes involved in fat oxidative catabolism. (E) Genes involved in lipid synthesis. (F) Genes relevant to glucose transport. The results are presented as mean ± SD, with *p < 0.05/**p < 0.01 indicating statistical significance.
Liver-specific knockout of MCU under high-fat conditions induces oxidative stress and impairs energy expenditure
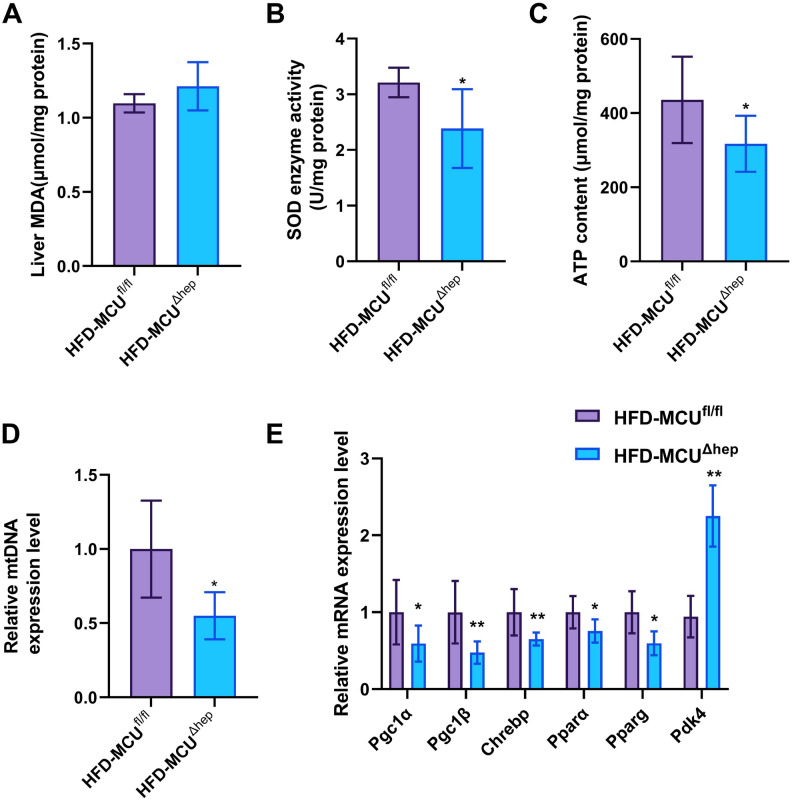
Oxidative stress is a well-established risk factor in the development of hepatic steatosis. Superoxide dismutase (SOD) and malondialdehyde (MDA), the end products of oxidative reactions, serve as reliable indicators to assess the extent of oxidative stress in an organism. Although the MDA levels did not differ (Fig. 6A), the MCUΔhep mice displayed significantly lower SOD levels, indicating that the absence of MCU in the liver exacerbates high-fat diet-induced oxidative stress (Fig. 6B). To assess the impact of MCU on energy metabolism, we measured hepatic ATP levels in MCUΔHep mice subjected to a high-fat diet. Compared to control mice, MCUΔHep mice exhibited significantly lower ATP levels (Fig. 6C) and decreased mtDNA expression (Fig. 6D), indicating that the absence of MCU has an inhibitory effect on energy metabolism in mice. To further investigate the impact of liver-specific MCU deficiency on energy metabolism, we analyzed the expression of key genes related to hepatic energy metabolism. Notably, MCU deletion resulted in significant downregulation of Pgc1α, Pgc1β, Chrebp, Pparα, and Pparg, alongside significant upregulation of Pdk4 in mice fed a high-fat diet (Fig. 6E). These findings suggest that mitochondrial biogenesis is impaired, leading to a reduction in mitochondrial copy number.
Fig. 6.
Inhibition of hepatic MCU leads to oxidative stress and reduced energy expenditure under high-fat diet conditions (n = 6). (A) Displays the malondialdehyde (MDA) level in the liver of mice. (B) The superoxide dismutase (SOD) enzyme activity in the liver of mice. (C) The ATP content in the liver of mice. (D) Relative expression level of mtDNA in the liver of mice. (E) The gene expression related to energy metabolism in the liver of mice. All results are presented as means ± SD. Statistical significance is denoted by *p < 0.05.
Discussion
In our study, we observed that liver-specific knockout of the mitochondrial calcium uniporter (MCU) significantly exacerbated hepatic steatosis under a high-fat diet, highlighting MCU’s crucial role in regulating hepatic lipid metabolism. Additionally, the knockout affected glucose tolerance and insulin sensitivity and was associated with significant changes in mitochondrial function and oxidative stress levels. These results not only elucidate the impact of MCU on metabolic balance in the liver but also suggest the potential for exploiting MCU as a therapeutic target in metabolic diseases.
Prior studies have primarily focused on the impacts of partial MCU knockout in mouse livers, particularly under conditions of a prolonged high-fat diet. These investigations indicated that mice with a heterozygous knockout of MCU (MCU+/−) showed decreased adiposity, reduced adipocyte hypertrophy, enhanced glucose tolerance, and significant alleviation of hepatic steatosis20. Conversely, when MCU was completely knocked out in the liver of mice, it was observed that hepatic mitochondrial Ca2+ uptake was inhibited, resulting in delayed clearance of Ca2+ from the cytosol. Furthermore, this led to a reduction in oxidative phosphorylation and ultimately resulted in increased hepatic lipid accumulation11. This nuanced understanding motivated our exploration of the consequences of full MCU knockout, culminating in the development of a new mouse model for high-fat diet-induced fatty liver. Our study not only builds on previous work but also extends it by elucidating the severe metabolic disturbances arising from complete MCU knockout, thereby providing a clearer picture of MCU’s role in metabolic regulation.
High-fat diet-induced glucose intolerance and insulin resistance increase the risk of diabetes in mice. Previous studies have reported up-regulation of certain MCU components in adipocytes during obesity and diabetes in both mice and humans21. Therefore, the regulation of MCU expression in tissues is emerging as a promising strategy to address metabolic disorders22. Georgiadou et al. reported a remarkable enhancement in glucose tolerance in 8 and 12 week-old mice following an 80% reduction in MCU levels in β-islet cells23. However, near-complete elimination of MCU in the liver resulted in an opposite effect on glucose tolerance in mice. These findings indicate the essential role of MCU in normal glucose-stimulated insulin secretion. Given the close association between elevated mitochondrial Ca2+ and accumulation of oxidative stress-related factors24, restoration of MCU expression in hepatocellular carcinoma cells under oxidative stress significantly mitigates insulin resistance and inflammation by suppressing ROS production and attenuating the activities of MAPK and NF-κB signaling pathways25. In our study, the absence of MCU resulted in compromised mitochondrial function and diminished superoxide dismutase (SOD) activity, underscoring the essential protective role of MCU against oxidative stress in liver metabolism. These insights reveal that while reducing MCU expression can be beneficial in certain contexts, its complete absence may lead to adverse metabolic outcomes, emphasizing the importance of balanced MCU activity for optimal metabolic health.
Apolipoprotein B-100 (ApoB-100) is a key player in the packaging and secretion of very low-density lipoproteins, thereby maintaining hepatic lipid metabolism homeostasis26, Feedback regulation in non-alcoholic fatty liver disease (MAFLD) leads to increased ApoB abundance27, our findings reveal that MCU deficiency suppresses APOB transcription and raises serum LDL levels, highlighting MCU’s complex role in lipid transport not previously documented.
Prior studies have extensively explored MCU’s dual role in liver functions, showing that while its absence disrupts glycolipid metabolism, moderate reduction can enhance liver health9,28. Our research presents preliminary findings where liver-specific MCU knockout resulted in suppressed APOB transcription and elevated serum LDL-C, TG, and ALT levels compared to control mice. However, the mechanisms behind these changes are not yet understood. Additionally, the knockout did not significantly impact body weight or insulin resistance, suggesting that further studies are necessary to explore these phenotypic differences and clarify the complex effects of MCU depletion on liver and systemic metabolism. The need for deeper mechanistic insights highlights a key limitation of the current study and underscores the importance of future research.
Supplementary Information
Author contributions
The study was conceived and planned by Qichao Liao, Yurou Zhang and Lei Zhou. Tingli Pan, Yu Sun, Yixing Li,Siqi Liu, Zhiwang Zhang, Lin Yu, Zupeng Luo, Yang Xiao, Xinyi Qi, Tianyu Jiang, Songtao Su, Shi Liu, Xinyu Qi, Xiangling Li, Turtushikh Damba, Khongorzul Batchuluun, Yunxiao Liang helped with the experiment. The manuscript was written by Qichao Liao, Yurou Zhang and Lei Zhou, Suosu Wei. All other authors approved the manuscript prior to submission. All authors have seen the manuscript and approved to submit to your journal.
Funding
This work was supported by National Natural Science Foundation of China (32272952); Guangxi Science Foundation for Distinguished Young Scholars (2020GXNSFFA297008); Guangxi Academy of Medical Sciences high-level Talents Foundation (YKY-GCRC-202302).
Data availability
The datasets generated or analysed during the current study are not publicly available due to privacy or ethical restrictions but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qichao Liao and Yurou Zhang.
Contributor Information
Suosu Wei, Email: sswei@gxams.org.cn.
Lei Zhou, Email: zhoulei@gxu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78935-w.
References
- 1.Eslam, M. et al. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology158, 1999–2014. e1991 (2020). [DOI] [PubMed]
- 2.McPherson, S. et al. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J. Hepatol.62, 1148–1155 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Riazi, K. et al. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 7, 851–861 (2022). [DOI] [PubMed]
- 4.Tilg, H., Moschen, A. R. & Roden, M. NAFLD and diabetes mellitus. Nat. Rev. Gastroenterol. Hepatol. 14, 32–42 (2017). [DOI] [PubMed]
- 5.Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med.24, 908–922 (2018). [DOI] [PMC free article] [PubMed]
- 6.Guilherme, A., Virbasius, J. V., Puri, V. & Czech, M. P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol.9, 367–377. 10.1038/nrm2391 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritov, V. B. et al. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes54, 8–14 (2005). [DOI] [PubMed]
- 8.Zorzano, A., Liesa, M. & Palacin, M. Role of mitochondrial dynamics proteins in the pathophysiology of obesity and type 2 diabetes. Int. J. Biochem. Cell Biol.41, 1846–1854. 10.1016/j.biocel.2009.02.004 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Baughman, J. M. et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature476, 341–345. 10.1038/nature10234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Stefani, D., Raffaello, A., Teardo, E., Szabo, I. & Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature476, 336–340. 10.1038/nature10230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomar, D. et al. Blockade of MCU-mediated Ca2+ uptake perturbs lipid metabolism via PP4-dependent AMPK dephosphorylation. Cell Rep.26, 3709–3725. e3707 (2019). [DOI] [PMC free article] [PubMed]
- 12.Rimessi, A. et al. Perturbed mitochondrial Ca2+ signals as causes or consequences of mitophagy induction. Autophagy9, 1677–1686. 10.4161/auto.24795 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Ikeda, K., Maretich, P. & Kajimura, S. The common and distinct features of brown and beige adipocytes. Trends Endocrinol. Metab.29, 191–200. 10.1016/j.tem.2018.01.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bravo-Sagua, R. et al. Calcium transport and signaling in mitochondria. Compr. Physiol.16, 623–634 (2017). [DOI] [PubMed]
- 15.Xue, K. et al. The mitochondrial calcium uniporter engages UCP1 to form a thermoporter that promotes thermogenesis. Cell Metab.34, 1325–1341. e1326 (2022). [DOI] [PubMed]
- 16.Huang, L. et al. Small intestine-specific knockout of CIDEC improves obesity and hepatic steatosis by inhibiting synthesis of phosphatidic acid. Int. J. Biol. Sci.18, 5740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao, Y. et al. Perillartine protects against metabolic associated fatty liver in high-fat diet-induced obese mice. Food Funct.14, 961–977 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Y. et al. 5-Methoxyflavone ameliorates non-alcoholic fatty liver disease through targeting the cytochrome P450 1A1. Free Radic. Biol. Med.195, 178–191 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Liu, S. et al. Hesperidin methyl chalcone ameliorates lipid metabolic disorders by activating lipase activity and increasing energy metabolism. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis.1869, 166620 (2023). [DOI] [PubMed]
- 20.Zhang, Z. et al. Ruthenium 360 and mitoxantrone inhibit mitochondrial calcium uniporter channel to prevent liver steatosis induced by high‐fat diet. Br. J. Pharmacol.179, 2678–2696 (2022). [DOI] [PubMed]
- 21.Wright, L. E. et al. Increased mitochondrial calcium uniporter in adipocytes underlies mitochondrial alterations associated with insulin resistance. Am. J. Physiol. Endocrinol. Metab.313, E641–E650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesmana, C. R. A. et al. Development of non-alcoholic fatty liver disease scoring system among adult medical check-up patients: a large cross-sectional and prospective validation study. Diabetes Metab. Syndr. Obes. 23, 213–218 (2015). [DOI] [PMC free article] [PubMed]
- 23.Georgiadou, E. et al. The pore-forming subunit MCU of the mitochondrial Ca2+ uniporter is required for normal glucose-stimulated insulin secretion in vitro and in vivo in mice. Diabetologia63, 1368–1381 (2020). [DOI] [PMC free article] [PubMed]
- 24.Orrenius, S., Zhivotovsky, B. & Nicotera, P. J. Regulation of cell death: the calcium–apoptosis link. Nat. Rev. Mol. Cell Biol.4, 552–565 (2003). [DOI] [PubMed]
- 25.Panahi, G., Pasalar, P., Zare, M., Rizzuto, R. & Meshkani, R. J. MCU-knockdown attenuates high glucose-induced inflammation through regulating MAPKs/NF-κB pathways and ROS production in HepG2 cells. PLoS One13, e0196580 (2018). [DOI] [PMC free article] [PubMed]
- 26.Fisher, E. A. & Ginsberg, H. N. Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J. Biol. Chem.277, 17377–17380 (2002). [DOI] [PubMed]
- 27.Zannis, V. I., Kan, H.-Y., Kritis, A., Zanni, E. E. & Kardassis, D. Transcriptional regulatory mechanisms of the human apolipoprotein genes in vitro and in vivo. Curr. Opin. Lipidol.12, 181–207 (2001). [DOI] [PubMed]
- 28.Nemani, N., Shanmughapriya, S. & Madesh, M. J. C. C. Molecular regulation of MCU: Implications in physiology and disease. Cell Calcium74, 86–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analysed during the current study are not publicly available due to privacy or ethical restrictions but are available from the corresponding author on reasonable request.