Abstract
G protein alpha q subunit (Gαq) can binds to the G protein-coupled receptor (GPCR) for signaling and is closely related to lipid metabolism. Endurance exercise is an effective means of combating acquired obesity and its complications, but the mechanisms by which endurance exercise modulates hereditary obesity and its complications are unknown. In this study, we achieved knockdown of Gαq in drosophila adipose tissue and skeletal muscle by constructing the Gαq-UAS-RNAi/Ppl-Gal4 and Gαq-UAS-RNAi/Mef2-GAl4 systems. Drosophila were subjected a three-week endurance exercise intervention, and changes in relevant indicators were detected and observed by RT-PCR, ELISA, oil red staining, immunofluorescence staining, and transmission electron microscopy. The results showed that knockdown of Gαq in both adipose tissue and skeletal muscle induced a significant increase in triglycerides accompanied by a decrease in rapid climbing ability, a decrease in Superoxide Dismutase (SOD) activity level, and a decrease in Mitochondrial respiratory chain complexI (MRCC I) content in Drosophila whole body and skeletal muscle, and down-regulated the expression of the G protein alpha q subunit (Gαq), the skeletal muscle myosin heavy chain expression gene (Mhc), mitochondrial biogenesis gene Spargal(the PGC-1alpha homologue in Drosophila). Endurance exercise significantly improved the triglyceride levels in the whole body and skeletal muscle of drosophila with Gαq knockdown in adipose tissue and skeletal muscle, as well as their ability to climb, increased SOD activity level and MRCCI content level, and up-regulated the expression of Gαq, Mhc, and Spargal(Srl). Thus, the present findings suggest that genetic defects in the Gαq gene in adipose and skeletal muscle tissues induce hereditary obesity and skeletal muscle dysfunction, and that endurance exercise attenuates this hereditary obesity and concomitant skeletal muscle dysfunction in drosophila by improving skeletal muscle fiber contractile proteins, mitochondrial function and function, and antioxidant capacity via mediating the Gαq/Mhc, Gαq/Srl/MRCC-I, and Gαq/SOD pathways.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-79415-x.
Keywords: Endurance exercise, Gαq gene, Genetic obesity, Mitochondria, Skeletal muscle, Drosophila
Subject terms: Obesity, Fat metabolism, Mitochondria
Introduction
The global prevalence of obesity has been increasing at an alarming rate in recent decades, seriously jeopardizing human health1,2. The World Health Organization defines obesity as an abnormal or excessive accumulation of fat that poses a risk to health and is considered a global health epidemic3. Obesity is a chronic metabolic disease that is closely related to cardiovascular disease, type 2 diabetes and other chronic diseases4, and its causes include genetic and acquired environmental factors5. Some studies have shown that the acquired environment, such as high-fat/high-sucrose diets, drugs and other induced obesity can change the relevant genetic material, so that the next generation of hereditary obesity risk increases, so it is easy to form a “genetic-obesity-genetic-obesity” vicious circle6. Numerous studies have confirmed that endurance exercise is an effective means of combating acquired obesity and related complications7,8. Its physiological mechanism lies in the fact that endurance exercise can increase energy expenditure, reduce fat accumulation, improve insulin sensitivity, rebalance various metabolic and signaling pathways in the body, and attenuate the chronic inflammatory response, etc9–11. However, the molecular mechanism of endurance exercise in regulating genetic obesity and its complications remains to be explored.
G protein alpha q subunit (Gαq) can binds to the G protein-coupled receptor (GPCR) for signaling, which is abundantly expressed in adipose, skeletal muscle and other tissues12,13. Gαq is associated with the stimulation of insulin secretion and is often used as a drug target in the study of obesity, type 2 diabetes and other metabolic syndromes14–16. It has been shown that Gαq can control fat storage in Drosophila cells and organisms, and knockdown of the adipose tissue Gαq gene can affect the expression of lipid metabolism effector genes, which leads to obesity in Drosophila17. However, whether the obesity induced by Gαq gene knockdown in both mammals and drosophila is accompanied by complications and whether it can be reversed by endurance exercise, as well as the molecular mechanisms underlying its effects, still remain to be confirmed.
Therefore, in order to determine the molecular mechanisms by which endurance exercise modulates genetic obesity and its complications, the classical genetic model organism Drosophila was selected for this study18–20. In this study, we firstly constructed the Gαq-UAS-RNAi/Ppl-Gal4 and Gαq-UAS-RNAi/Mef2-GAl4 systems through Drosophila crosses to achieve specific knockdown of the Gαq gene in adipose and skeletal muscle tissues in the F1 generation, and verified whether it was successful in inducing the generation of hereditary obesity in the F1 generation through the detection of the lipid metabolism and its motility condition. Then, Drosophila were subjected to endurance exercise intervention, and whole body and skeletal muscle triglyceride (TG), exercise capacity, skeletal muscle SOD activity, MRCC I content, Srl gene, Mhc gene, Gαq gene expression level were detected. The ultrastructure, protein expression and lipid accumulation of skeletal muscle were observed by transmission electron microscopy, immunofluorescence staining and oil-red staining to comprehensively evaluate whether endurance exercise can effectively counteract obesity and concurrent skeletal muscle dysfunction caused by Gαq gene knockdown and reveal its molecular mechanism, so as to provide the theoretical basis and new ideas for the counteracting of hereditary obesity by endurance exercise.
Materials and methods
Drosophila strains and hybridization groups
Gαq knockdown strain w[1118] P{GD17761}v51116 (purchased from Vienna Drosophila Resource Center, Austria, strain number: V51116); PplGal4 strain w; P{ppl-GAL4.P}2*(purchased from Bloomington Drosophila Stock Center, US, strain number: BCF-673); Mef2Gal4 (purchased from Bloomington Drosophila Stock Center, US, strain number: 27390); The F1 generation UAS/Gal4 system was constructed by crossing GαqUAS−RNAi with PplGal4 and Mef2Gal4 Drosophila, respectively, to regulate the expression of the target gene Gαq in adipose tissue and skeletal muscle.
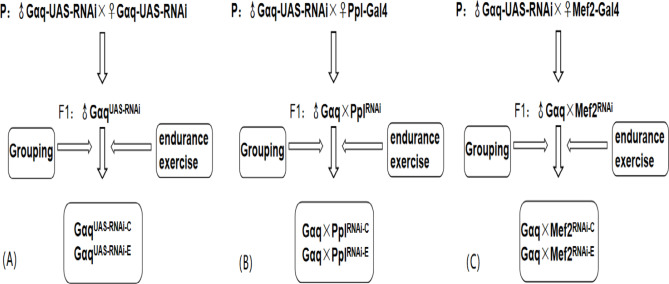
Male Drosophila within 12 h of feathering of the F1 generation of Gαq-UAS-RNAi male and female self-crosses were used as genetic controls and divided into the Gαq gene normal expression group (GαqUAS−RNAi−C), and the Gαq gene normal expression sport group (GαqUAS−RNAi−E); Male Gαq-UAS-RNAi Drosophila were crossed with female PplGal4 Drosophila, and male Drosophila within 12 h of F1 generation feathering were collected and divided into the Gαq adipose tissue knockdown group (Gαq×PplRNAi−C), and the Gαq adipose tissue knockdown exercise group (Gαq×PplRNAi−E); Male Gαq-UAS-RNAi Drosophila were crossed with female Mef2Gal4 Drosophila, and male Drosophila within 12 h of F1 generation feathering were collected and divided into the Gαq skeletal muscle knockdown group (Gαq×Mef2RNAi − C), and the Gαq skeletal muscle knockdown motor group (Gαq×Mef2RNAi − E) (Fig. 1). All Drosophila were placed in an environmental culture at a constant temperature of 25 °C, 50% constant humidity, and a 12-h day/night cycle.
Fig. 1.
Diagram of Drosophila hybridization and grouping. (A) Gαq normal expression grouping diagram. (B) Gαq adipose tissue knock-down grouping diagram. (C) Gαq skeletal muscle tissue knockdown grouping diagram.
Exercise training
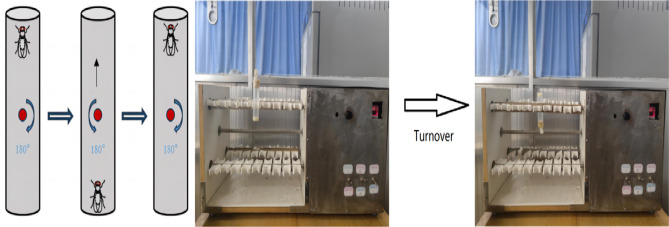
Upward climbing endurance training was performed on an exercise platform utilizing the anti-gravity climbing characteristic of Drosophila (Fig. 2). The principle is that the test tube is fixed on the test tube clamp, at this time the test tube is perpendicular to the ground, the bottom of the test tube Drosophila climbing to the top of the test tube, the test tube is automatically turned over 180° (the test tube is rotated at a speed of 60 rad/s speed uniformity), the Drosophila back to the bottom of the test tube to continue to climb the movement, so on and so forth. The weight of Drosophila was its own weight when climbing, 20 Drosophila per tube, 8 cm was reserved from the medium at the bottom of the test tube to the lower end of the cotton plug for the movement area of Drosophila, and the Drosophila had 10s time to perform the climbing movement each time after turning over to the bottom of the test tube. The exercise group started the training intervention from 1 day of age until the end of 3 weeks of age, with 15 min of exercise-5 min of rest lasting four times a day, and continued training for 5 days a week with 2 days of rest (Table 1).
Fig. 2.
Schematic diagram of Drosophila exercise platform and climbing training.
Table 1.
Exercise flow chart.
| Monday | Tuesday | Wednesday | Thursday | Friday | Saturday | Sunday | |
|---|---|---|---|---|---|---|---|
| 1st week of age | E | E | E | E | E | N | Measure climbing ability |
| 2nd week of age | E | E | E | E | E | N | Measure climbing ability |
| 3rd week of age | E | E | E | E | E | N | Measure climbing ability |
E for exercise intervention, N for no exercise intervention.
Preparation of Drosophila culture medium
The culture medium preparation for Drosophila was based on the standard diet of Drosophila in previous studies21. Configure 1 L of Drosophila culture medium. Add 42 g of corn flour, 10 g of soybean flour, 13 g of yeast powder, and 8 g of agar strips to a pot, add 1 L of pure water and stir well. Stir continuously during heating to melt the agar strips thoroughly until the solution boils. Stop heating after boiling, add 31 g of sucrose and 31 g of maltose during cooling, and after sucrose and maltose are fully dissolved, add 2000 µl of preservative propionic acid and 1 g of sodium benzoate, and immediately dispense in clean culture tubes with a thickness of about 0.5 cm per tube after thorough stirring.
ELISA assay
The activities and levels of triglyceride (TG), superoxide dismutase (SOD) and mitochondrial respiratory chain Complex I (MRCCI) in skeletal muscle of Drosophila were detected by using insect TG enzyme-linked immunoassay kit, insect SOD enzyme-linked immunoassay kit and insect MRCCI enzyme-linked immunoassay kit(Insect MRCCI, SOD, and TG ELISA Kits, Fankew, Shanghai Kexing Trading Co., Ltd, China). The ELISA assay-specific steps are as follows: (1) Sample addition of standard products. (2) Sample addition: Prepare blank control wells and sample wells to be tested. Add 40 µl of diluent to the sample wells to be tested, and then add 10 µl of test sample. (3) Add enzyme: Add enzyme-labeled reagent 100 µl per hole, except blank hole. (4) Incubation: After the sealing plate is sealed with sealing plate film, it is incubated at 37 °C for 60 min. (5) Mixing liquid: Dilute 20 times concentrated washing liquid with 20 times distilled water for reserve use. (6) Wash: Remove the sealing film from the surface of the sealing plate, pour out the solution in it, clean it, and then let it dry. (7) Color development: Add color-developing agent A50µl to each well first, then add color-developing agent B 50 µl, gently shake and mix, and hide from light for 15 min at 37 °C. (8) Termination: Add termination solution 50 µl per well to terminate the reaction (at this time, the blue immediately turns to yellow). (9) Determination: Determine the absorbance (OD) of each well.
Real-time quantitative PCR
At the end of three weeks of age, 50 Drosophila skeletal muscle (thorax) and adipose tissue (abdomen) were taken from each group and placed into 1000 µl of Trizol respectively to detect the expression levels of relevant pathway genes. (1) Total RNA extraction: Take the homogenizer tube, add 1 ml of RNA extraction solution, place it on ice for pre-cooling, take 100 mg of tissue, add it into the homogenizer tube, and grind it fully with the grinder until no tissue mass is visible. Centrifuge at 12,000 rpm at 4 °C for 10 min, white precipitate at the bottom of the tube is RNA. Remove the liquid, add 1.5 ml of 75% ethanol for washing and precipitation, centrifuge at 12,000 rpm at 4 °C for 5 min, remove the liquid, put the centrifuge tube on a super-clean table for 3 min, add 15 µl Water Nuclease to dissolve RNA Free, and incubate at 55 °C for 5 min. Nanodrop 2000 was used to detect RNA concentration and purity: After blank zero adjustment of the instrument, 2.5 µl RNA solution to be tested was put on the detection base, the sample arm was lowered, and the software on the computer was used to start the absorption value detection, and the RNA with excessive concentration was diluted in an appropriate proportion so that the final concentration was 100–500 ng/µl. (2) Reverse transcription: Configure the reverse transcription reaction system, gently mix and centrifuge, and set the reverse transcription program. (3) Quantitative PCR: 0.2 ml PCR tubes were used to prepare the reaction system. Each retro product was prepared with 3 tubes for PCR amplification. (4) Results treatment: CT method. Primer sequences of Rp-49 were as follows: F:5’-3’CTAAGCTGTCGCACAAATGG, R:5’-3’AACTTCTTGAATCCGGTGGG; Primer sequences of Gαq were as follows: S:5’-3’:GGTCCTCAGCGAGATGCAATAA, A:5’-3’:TAAGGTTCGATTGCA GAATTGTGTC; Primer sequences of Mhc were as follows: S:5’-3’:GACCGTGCGTAACGATAACTCC, A:5’-3’:GA CTGCTGGGAGATGA CACGA; Primer sequences of Srl were as follows: S:5’-3’:ACCTGGCGATTCTGATTATGAC T, A:5’-3’:CCTTTA CATTGTCCACATAGCGT.
Transmission electron microscopy of the skeletal muscle
For electron microscopic analysis, muscles were dissected in an ice-cold fixative (2.5% glutaraldehyde in 0.1 mol/L PIPES buffer at pH7.4). After 10 h of fixation at 4 °C, samples were washed with 0.1 mol/L PIPES, post-fixed in 1% OsO4 (30 min), and stained in 2% uranyl acetate (1 h). Samples were dehydrated in an ethanol series (50%, 70%, 100%) and embedded in epoxy. The slices were observed and photographed with a HT-7700 transmission electron microscope.
Oil red staining analysis of skeletal muscle
Skeletal muscle was taken and fixed in tissue fixative for 15 min, rinsed with water and dried. 6 parts of saturated oil red O dye solution and 4 parts of distilled water were mixed well and placed in a water bath at 60–70 °C for 30 min, cooled naturally and then filtered through a qualitative filter paper to obtain oil red O working solution. Immerse the sections in the oil red dye solution for 8–10 min (cover to avoid light). Remove the slices, leave them for 3 s, and then immerse them sequentially in two vats of 60% isopropyl alcohol for differentiated 3 s and 5 s. Immerse the slices in two vats of pure water for 10 s each. The slices were removed, left for 3s, and immersed in hematoxylin for 3–5 min before being immersed in three tanks of pure water for 5s, 10s, and 30s, respectively. Differentiation solution was differentiated for 2–8 s, and each of the two tanks of distilled water was washed for 10s, and the blue solution was washed for 1s. The slices were gently immersed into two tanks of tap water for 5s and 10s, respectively, and examined for staining effect under the microscope. Glycerol gelatin was used to seal the sections, and the images were captured and analyzed with a microscope. Microscopy, image acquisition, and analysis: Orthostatic light microscope. Using a numerical aperture (NA) of 1.3, select an oil immersion objective measuring 40×. A high-resolution digital camera is attached to capture clear images to accurately measure and record details at a scale of 5 μm. Observation is aided by a 10×low magnification objective that helps locate the target area quickly at the beginning of the observation and helps understand the overall distribution of the sample.
Myosin heavy chain immunofluorescence
Drosophila skeletal muscle was taken for immunohistochemical analysis as follows. (1) Paraffin sections deparaffinised to water: Put the slices in 3 changes of xylene, 10 min each, then dehydrate in 3 changes of pure ethanol for 5 min each, wash in distilled water. (2) Antigen repair: During the repair process, excessive evaporation of buffer solution should be prevented, and the slides should not be dried. After the repair is completed, it is naturally cooled. Put the slide 5 min PBS (PH 7.4) and shake it on a decoloring shaker for 3 times, each time for 5 min. (3) Circle drawing and blocking: Add 3%BSA into the circle and cover the tissue evenlyto block non-specific binding at room temperature for 30 min. (The primary antibody is blocked with 10% donkey serum from goat, and the primary antibody from other sources is blocked with 3%BSA). (4) Adding primary antibody: drop the prepared primary antibody, slice it flat in a wet box and incubate at 4 °C covernight. (5) Add secondary antibody: put the glass slide 5 min PBS (PH7.4) and shake it on the decoloring shaker for 3 times, 5 min each time. Add the corresponding secondary antibody and incubate at room temperature for 50 min in the dark. (6) DAPI counterstain in nucleus: DAPI solution was dripped into the circle and incubated at room temperature for 10 min in the dark. (7) Quench tissue autofluorescence: The slides were put 5 min PBS (PH 7.4) and washed on a decoloring shaker for 3 times, each time for 5 min. Add autofluorescence quencher B solution for 5 min and rinse with running water for 10 min. (8) Mount: coverslip with anti-fade mounting medium. (9) Microscopy detection and collect images by Fluorescent Microscopy. DAPI glows blue by UV excitation wavelength 330–380 nm and emission wavelength 420 nm; 488 glows green by excitation wavelength 465–495 nm and emission wavelength 515–555 nm; CY3 glows red by excitation wavelength 510–560 nm and emission wavelength 590 nm.
Athletic ability testing
100 Drosophila were randomly selected from each group, 20 per tube. Each tube was individually placed under a high definition video camera in preparation for video recording. After the camera was switched on, the Drosophila in the tube were shaken to the bottom of the tube every 15 s, and after three shakes, the Drosophila being tested was replaced with the next tube Drosophila. The best of three crawls of each test tube Drosophila was selected for data processing. Screenshots of the height of climb 3 s after Drosophila shocked off the bottom of the test tube were taken using AVS Video Editor software. The photo clearly shows the climbing height of the Drosophila. The 3-second climbing heights of the fruit flies were then processed and analysed using HEYEAR software and prism software.
Statistical methods
For comparisons of the Gαq gene between the Gαq normal expression group and the adipose tissue Gαq knockdown group and the skeletal muscle tissue Gαq knockdown group, one-way analysis of variance (ANOVA) and the least significant difference (LSD) test were used to determine between-group differences. For comparisons between control and exercise groups, independent samples t-tests were used to determine between-group differences. The experimental data were expressed as mean ± standard deviation (S) and the significance level was taken as α = 0.05 (or 0.01).
Results
Gαq knockdown in Drosophila adipose and skeletal muscle tissues induces abnormal lipid metabolism and reduced exercise capacity
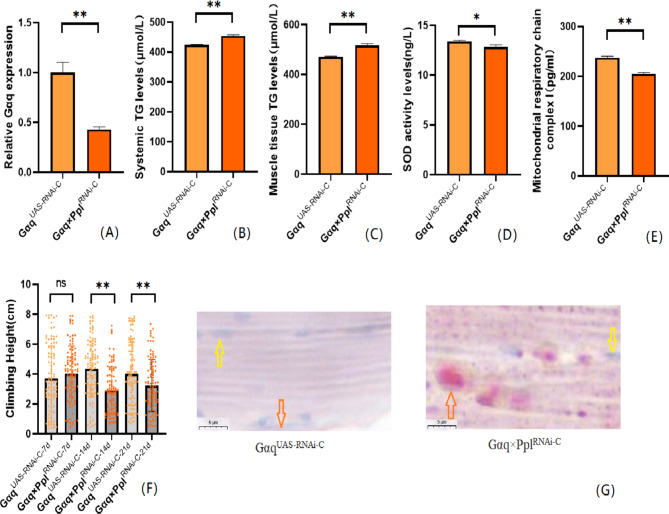
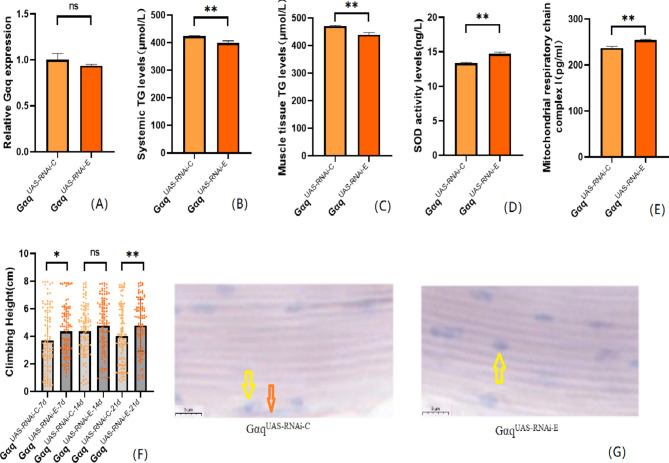
Our previous studies agree that Gαq gene knockdown in adipose tissue induces obesity17, but its effect on exercise capacity is unknown.We first constructed a model of Gαq knockdown in Drosophila adipose tissue. The results show that compared with the GαqUAS−RNAi−C group, the mRNA expression level of the Gαq gene in Drosophila adipose tissue was reduced in the Gαq×PplRNAi−C group (P<0.01) (Fig. 3A), with a relative expression rate of 43.10%. It can be judged that the Gαq adipose tissue knockdown model was constructed successfully. The rapid climbing ability (CS) of Drosophila was decreased in the Gαq×PplRNAi−C group (P < 0.01) (Fig. 3F); and the analysis of lipid by oil red staining accumulation revealed an increase in triglyceride accumulation in the skeletal muscle of Drosophila in the Gαq×PplRNAi−C group (Fig. 3G). ELISA assay of Drosophila in the Gαq×PplRNAi−C group revealed an increase in whole-body and skeletal muscle TG content (P < 0.01) (Fig. 3B C), and a decrease in the level of SOD activity (P < 0.05)(Fig. 3D, and a decrease in the level of MRCCI (P < 0.01) (Fig. 3E).
Fig. 3.
Relative gene expression, lipid metabolism levels, oxidative capacity, mitochondrial function, and 3s climbing ability in adipose-tissue Gαq knockdown Drosophila skeletal muscle. (A) Relative expression of Gαq, Sample n = 50. (B) Whole Body Triglyceride Levels, Sample n = 20. (C) Skeletal muscle tissue triglyceride levels, Sample n = 30. (D) SOD activity levels, Sample n = 30. (E) Mitochondrial respiratory chain complex I content, Sample n = 30. (F) Climbing height, Sample n = 100. (G) Oil red staining, Sample n = 5. Yellow arrows indicate nuclei and orange arrows indicate lipid accumulation(scale: black line is 5 μm). The one-way analysis of variance (ANOVA) with least significant difference (LSD) tests was used to identify difference among the groups. Data are represented as mean ± standard deviation. *P < 0.05; **P < 0.01; n s means no significant difference.
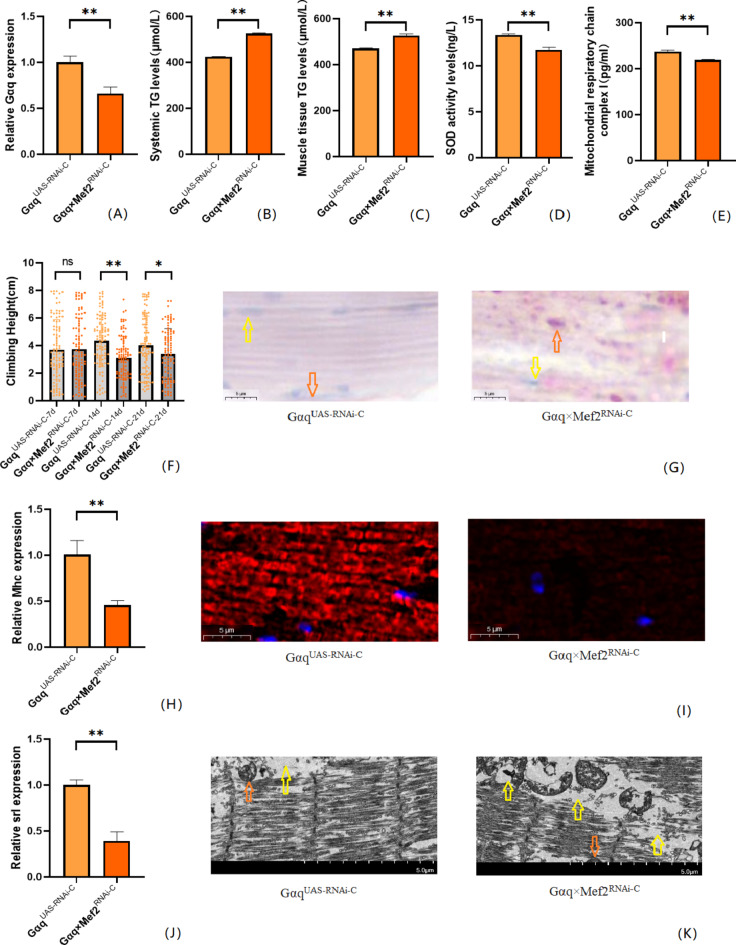
To further understand that Gαq knockdown leads to abnormal lipid metabolism and reduced exercise capacity, we also constructed a Drosophila skeletal muscle tissue Gαq knockdown model. The results showed that compared with the GαqUAS−RNAi−C group, the mRNA expression level of the Gαq gene in Drosophila skeletal muscle tissue was reduced in the Gαq×Mef2RNAi − C group (P < 0.01) (Fig. 4A), with a relative expression rate of 66%; it can be judged that the Gαq skeletal muscle tissue knockdown model was constructed successfully. CS was decreased in Drosophila from the Gαq×Mef2RNAi − C group (P < 0.05) (Fig. 4F); oil-red staining analysis of lipid accumulation revealed an increase in the Gαq×Mef2RNAi − C group Drosophila skeletal muscle triglyceride accumulation was increased (Fig. 4G). ELISA assay of Drosophila in the Gαq×Mef2RNAi − C group revealed an increase in whole-body and skeletal muscle TG content (P < 0.01) (Fig. 4B,C), and a decrease in the level of SOD activity (P < 0.01) (Fig. 4D), and a decrease in the level of MRCC I (P < 0.01) (Fig. 4E). RT- PCR assay showed decreased mRNA expression level of skeletal muscle myosin heavy chain expression gene (Mhc) (P < 0.01) (Fig. 4H); myosin heavy chain immunofluorescence plots showed that myosin heavy chain expression was decreased in Drosophila skeletal muscle tissues of the Gαq×Mef2RNAi − C group, which indicated an increase in skeletal muscle damage (Fig. 4I). RT-PCR assay showed decreased mitochondrial biogenesis gene Spargal(Srl) expression was decreased (P < 0.01) (Fig. 4J). Skeletal muscle transmission electron microscopy images showed that Drosophila skeletal muscle tissues of the Gαq×Mef2RNAi − C group had disorganized myofibril arrangement and incomplete Z-lines, which indicated a weakening of skeletal muscle function (Fig. 4K).
Fig. 4.
Relative gene expression, lipid metabolism levels, oxidative capacity, mitochondrial function, and 3 s climbing ability in skeletal muscle tissue Gαq knockdown of Drosophila skeletal muscle. (A) Relative expression of Gαq, Sample n = 50. (B) Whole Body Triglyceride Levels, Sample n = 20. (C) Muscle Tissue Triglyceride Levels, Sample n = 30. (D) SOD activity levels, Sample n = 30. (E) Mitochondrial respiratory chain complex I content, Sample n = 30. (F) Climbing height, Sample n = 100. (G) Oil red staining, Sample n = 5. Yellow arrows indicate nuclei and orange arrows indicate lipid accumulation (scale: black line is 5 μm). (H) Relative expression of Mhc, Sample n = 50. (I) Immunofluorescence image of myosin heavy chain, Sample n = 5, blue dots are nuclei and red fluorescent bands are myogenic fibers (scale: white line is 5 μm). (J) Relative expression of Srl, Sample n = 50. (K) Transmission electron micrograph of skeletal muscle, Sample n = 5, orange indicates nuclei, yellow indicates muscle fiber damage (scale, white line is 5 μm). The one-way analysis of variance (ANOVA) with least significant difference (LSD) tests was used to identify difference among the groups. Data are represented as mean ± standard deviation. *P < 0.05; **P < 0.01; n s means no significant difference.
Endurance exercise effectively improves abnormal lipid metabolism and decreased exercise capacity induced by Gαq knockdown
A growing body of evidence confirms that endurance exercise can effectively improve obesity and enhance exercise capacity. Therefore, we firstly performed endurance exercise intervention on Gαq normal expression Drosophila. The results showed that compared with the GαqUAS−RNAi−C group, there was no significant difference in the mRNA expression levels of the Gαq gene in the GαqUAS−RNAi−E group (P > 0.05) (Fig. 5A); CS was enhanced in Drosophila from the GαqUAS−RNAi−E group (Fig. 5F); analysis of lipid accumulation by oil-red staining showed that Drosophila from the GαqUAS−RNAi−E group lipid accumulation decreased (Fig. 5G); ELISA showed a decrease (P < 0.01) in their whole body and skeletal muscle TG content (P < 0.01) (Fig. 5B,C), and an increase in SOD activity level and MRCC I content(Fig. 5D,E).
Fig. 5.
After endurance exercise intervention, Gαq normally expressed related gene expression, lipid metabolism level, oxidation capacity, mitochondrial function and 3s climbing ability of Drosophila melanogaster. (A) Relative expression of Gαq, Sample n = 50. (B) Whole Body Triglyceride Levels, Sample n = 20. (C) Muscle Tissue Triglyceride Levels, Sample n = 30. (D) SOD activity levels, Sample n = 30. (E) Mitochondrial respiratory chain complex I content, Sample n = 30. (F) Climbing height, Sample n = 100. (G) Oil red staining, Sample n = 5. Yellow arrows indicate nuclei and orange arrows indicate lipid accumulation (scale: black line is 5 μm). The one-way analysis of variance (ANOVA) with least significant difference (LSD) tests was used to identify difference among the groups. Data are represented as mean ± standard deviation. *P < 0.05; **P < 0.01; n s means no significant difference.
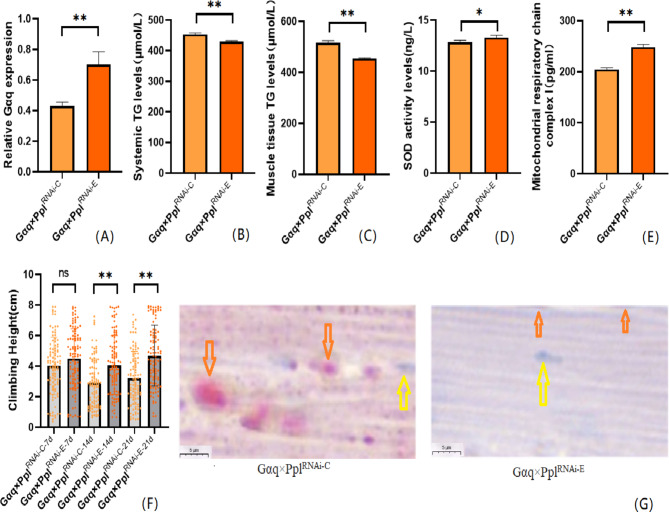
We also performed an endurance exercise intervention on Gαq adipose tissue knockdown Drosophila melanogaster. The result shows that compared with the Gαq×PplRNAi−C group, the mRNA expression level of the Gαq gene in Drosophila adipose tissue was increased in the Gαq×PplRNAi−E group (P < 0.01)(Fig. 6A), and the relative expression rate was elevated by 27%; Drosophila CS was increased in the Gαq×PplRNAi−E group (Fig. 6F); oil red staining analysis of lipid accumulation showed that in the Gαq×PplRNAi−E group Drosophila skeletal muscle tissue showed decreased lipid accumulation (Fig. 6G); ELISA detected decreased levels of systemic and skeletal muscle TG (P < 0.01) (Fig. 6B,C), increased levels of SOD activity (P < 0.05) (Fig. 6D), and increased MRCC I content (P < 0.01) (Fig. 6E).
Fig. 6.
After endurance exercise intervention, adipose tissue Gαq knockdown Drosophila Drosophila related gene expression, lipid metabolism level, oxidation capacity, mitochondrial function and 3 s climbing ability. (A) Relative expression of Gαq, Sample n = 50. (B) Whole Body Triglyceride Levels, Sample n = 20. (C) Muscle Tissue Triglyceride Levels, Sample n = 30. (D) SOD activity levels, Sample n = 30. (E) Mitochondrial respiratory chain complex I content, Sample n = 30. (F) Climbing height, Sample n = 100. (G) Oil red staining, Sample n = 5. Yellow arrows indicate nuclei, orange arrows indicates lipid accumulation (scale: black line is 5 μm). The one-way analysis of variance (ANOVA) with least significant difference (LSD) tests was used to identify difference among the groups. Data are represented as mean ± standard deviation. *P < 0.05; **P < 0.01; n s means no significant difference.
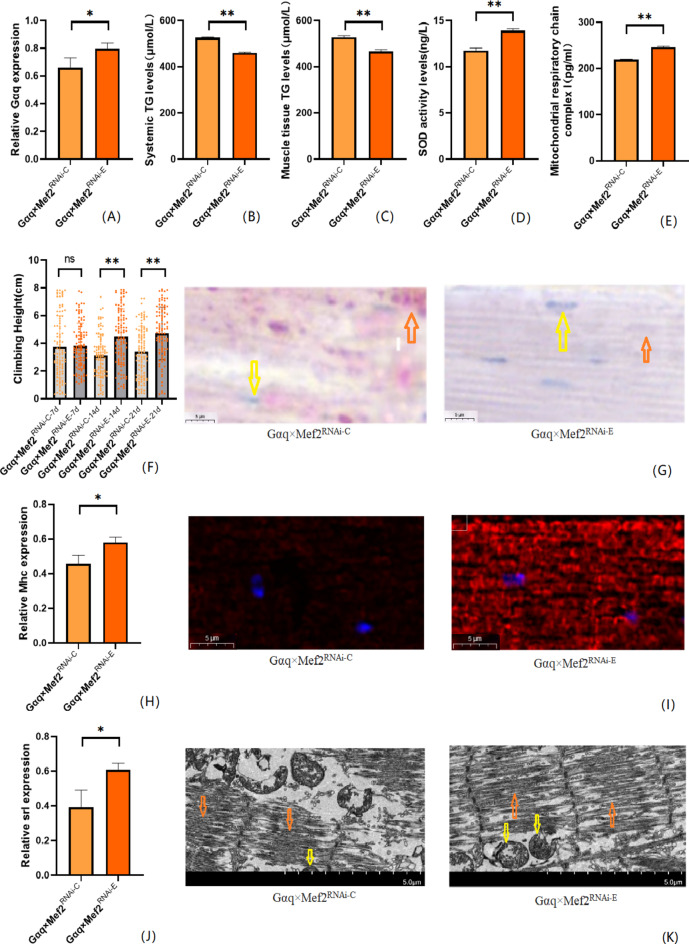
We also performed endurance exercise intervention on Gαq skeletal muscle tissue knockdown Drosophila. The result shows that compared with the Gαq×Mef2RNAi − C group, the mRNA expression level of the Gαq gene was elevated in the skeletal muscle tissue of Drosophila in the Gαq×Mef2RNAi − E group (P < 0.05)(Fig. 7A), and the relative expression rate was elevated by 13.67%; and the CS of Drosophila in the Gαq×Mef2RNAi − E group was increased (P < 0.01)(Fig. 7F); Oil red staining analysis of lipid accumulation showed a decrease in lipid accumulation in the skeletal muscle tissue of Drosophila in the Gαq×Mef2RNAi − E group (Fig. 7G), and a decrease in the TG content of its whole body and skeletal muscle as detected by ELISA (P < 0.01) (Fig. 7B,C), and an increase in the level of SOD activity(Fig. 7D), and the content of MRCC I (P < 0.01) (Fig. 7E); immunofluorescence images of myosin heavy chain showed increased expression of myosin heavy chain in Drosophila skeletal muscle tissues of the Gαq×Mef2RNAi − E group, indicating reduced muscle damage and enhanced muscle function (Fig. 7I), and RT-PCR showed elevated levels of Mhc expression (P < 0.05) (Fig. 7H); transmission electron microscopy images of skeletal muscle showed that Drosophila of the Gαq×Mef2RNAi − E group skeletal muscle myogenic fibres are more aligned and Z-lines are more complete, indicating enhanced muscle function (Fig. 7K), and RT-PCR showed elevated Srl expression level (P < 0.05) (Fig. 7J).
Fig. 7.
After endurance exercise intervention, skeletal muscle tissue Gαq knockdown Drosophila Drosophila related gene expression, lipid metabolism level, oxidation capacity, mitochondrial function and 3s climbing ability. (A) Relative expression of Gαq, Sample n = 50. (B) Whole body triglyceride levels, sample n = 20. (C) Muscle Tissue Triglyceride Levels, Sample n = 30. (D) SOD activity levels, Sample n = 30. (E) Mitochondrial respiratory chain complex I content, Sample n = 30. (F) Climbing height, Sample n = 100. (G) Oil red staining, Sample n = 5. Yellow arrows indicate nuclei and orange arrows indicate lipid accumulation(scale: black line is 5 μm). (H) Relative expression of Mhc, Sample n = 50. (I) Immunofluorescence image of myosin heavy chain, Sample n = 5, blue dots are nuclei and red fluorescent bands are myogenic fibers (scale: white line is 5 μm). (J) Relative expression of Srl, Sample n = 50. (K) Transmission electron micrograph of skeletal muscle, Sample n = 5, yellow arrows represent mitochondria and the orange arrows represent myofibril. (scale, white line is 5 μm). The one-way analysis of variance (ANOVA) with least significant difference (LSD) tests was used to identify difference among the groups. Data are represented as mean ± standard deviation. *P < 0.05; **P < 0.01; n s means no significant difference.
Discussion
Numerous studies have reported that G protein alpha q subunit (Gαq) is closely related to lipid metabolism22. Recent studies have found significant changes in mRNA encoding G protein-coupled receptors (GPCR) in obese populations23,24. Gαq directly or indirectly regulates lipid, glucose metabolism, and fat thermogenesis in adipose, liver, and muscle tissues25,26. In addition, Gαq can also regulate fat storage, and knocking down the Gαq gene in adipose tissue can lead to obesity in Drosophila17. In this study, we successfully constructed knockdown models of Gαq in Drosophila adipose tissue and skeletal muscle tissue using the Gαq-UAS/Ppl-Gal4 and Gαq-UAS/Mef2-Gal4 systems. We found that genetic knockdown of the Gαq gene in adipose tissue and skeletal muscle resulted in an increase in triglycerides throughout the body and skeletal muscle of Drosophila, suggesting the possibility of inducing obesity. Meanwhile, research has found that genetic knockdown of the Gαq gene in adipose tissue and skeletal muscle tissue is accompanied by a significant decrease in the rapid climbing ability of Drosophila, suggesting that obesity may lead to skeletal muscle dysfunction.
Myosin is an important skeletal muscle contraction protein, and mitochondria are the energy “factory” of skeletal muscle. Both are closely related to skeletal muscle function. The expression of myosin and mitochondrial synthase in mammals is regulated by multiple genes, while drosophila have gene expression of single skeletal muscle fiber myosin heavy chain and mitochondrial synthase27–29. Both in mammals and Drosophila, high-fat diet induced obesity can lead to a decrease in their motor ability, which is related to the weakened functions of Mhc and PGC-1alpha. In addition, age-related skeletal muscle atrophy is also associated with the decline of skeletal muscle contraction proteins and mitochondrial function30–32. Numerous studies have shown that obesity is accompanied by increased lipid toxicity in important organs such as the heart and skeletal muscle. The mechanism behind this is that under oxidative stress, lipids can form malondialdehyde (MDA), which can produce cytotoxicity and impair their function33–37.
To further verify the causes of skeletal muscle dysfunction, in this study, we analyzed the contractile proteins, mitochondrial status, and antioxidant capacity of skeletal muscle. The results showed that genetic knockdown of the Gαq gene in adipose tissue and skeletal muscle tissue can significantly reduce the expression of the important contraction protein myosin heavy chain gene (Mhc) and protein in skeletal muscle. At the same time, electron microscopy observation revealed a regular arrangement and reduced quantity of myofibrils. In addition, the levels of key genes Spargel(the PGC-1alpha homologue in Drosophila) and mitochondrial key oxidase mitochondrial respiratory chain complex I (MRCC I) involved in mitochondrial biogenesis were significantly reduced. Finally, the activity level of SOD, an important antioxidant enzyme, was detected, and it was found that genetic knockdown of Gαq gene in both adipose tissue and skeletal muscle tissue could reduce SOD activity level in skeletal muscle. Therefore, these results suggest that genetic knockdown of Gαq gene in both adipose tissue and skeletal muscle can negatively affect the important contractile structure, mitochondrial function, and antioxidant capacity of skeletal muscle, resulting in decreased exercise ability of Drosophila.
Endurance exercise is a globally recognized weight loss method, but whether it still has an effect on different types of obesity (especially genetic obesity induced by different gene mutations) remains to be studied38,39. In rats and Drosophila, studies have found that endurance exercise enhances antioxidant capacity to protect them from the decline in climbing ability caused by high-fat intake, and can improve obesity caused by excessive triglyceride accumulation in the body due to high-fat diet21,40,41. Recent studies have confirmed that endurance exercise can also promote mitochondrial biogenesis, aerobic capacity, and energy utilization, activate oxidative metabolism related pathways, and improve skeletal muscle metabolism and function42–44.
Similarly, in this study, in order to verify whether endurance exercise can produce positive benefits on obesity and concurrent motor disability induced by genetic knockdown of Gαq in adipose tissue and skeletal muscle tissue, we conducted endurance exercise intervention for 3 weeks in Drosophila with genetic knockdown of Gαq in adipose tissue and skeletal muscle tissue. The results showed that endurance exercise significantly increased the expression of Gαq gene in adipose tissue and skeletal muscle tissue of Drosophila, and was accompanied by a significant decrease in TG levels in the whole body and skeletal muscle, suggesting that endurance exercise plays an important role in the regulation of Gαq gene and its mediated obesity. In addition, behavioral indicators showed that endurance exercise could enhance the fast climbing ability of Drosophila with Gαq knockdown in adipose tissue and skeletal muscle tissue, and physiological and biochemical indicators showed that endurance exercise significantly increased the expression of Mhc gene and protein in skeletal muscle of Drosophila with Gαq gene knockdown in adipose tissue and skeletal muscle tissue. At the same time, electron microscopy observed that myogenic fibres were more aligned, indicating enhanced muscle function. In addition, after exercise intervention, the levels of Spargel(Srl) and MRCC I in skeletal muscle were significantly increased, indicating enhanced mitochondrial function. Finally, it was also found that the SOD activity level of skeletal muscle was significantly increased after exercise, suggesting that the antioxidant capacity of skeletal muscle was enhanced.
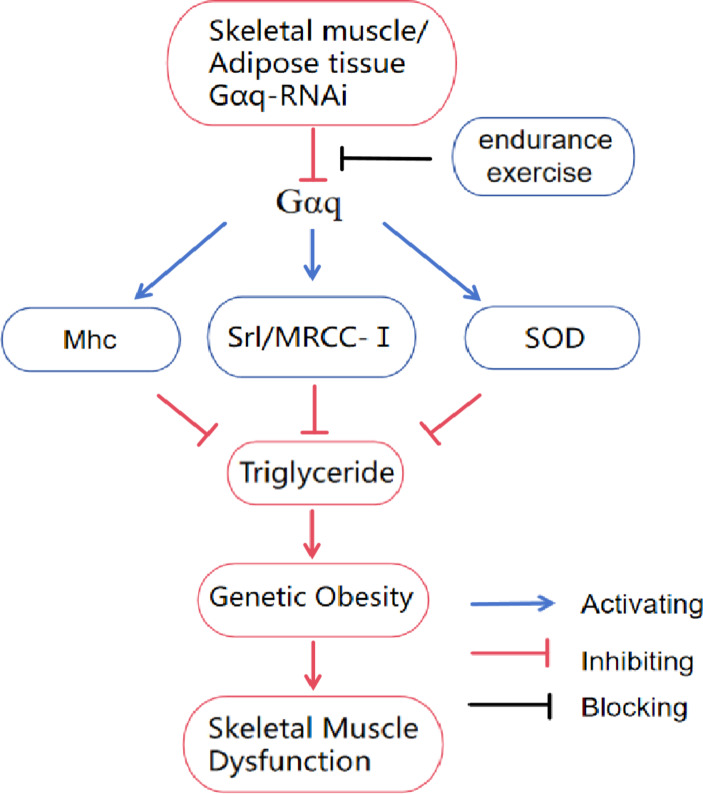
Long term exercise training, whether in mammals or Drosophila, can cause adaptive changes in certain gene proteins in skeletal muscle tissue cells, thereby improving their function. Research has shown that structural abnormalities of Mhc protein in skeletal muscle can significantly reduce exercise capacity45. Elevated expression of Srl(homologue of PGC-1α) in skeletal muscle can limit muscle atrophy, while reduced expression of Srl gene specificity leads to decreased exercise capacity and increased fat accumulation. Stimulating the Sirt1-Srl axis can improve motor function and mitochondrial respiratory capacity in patients with Babbitt syndrome46,47. Exercise can restore the expression of Mhc protein mRNA in skeletal muscle of diabetes rats to normal48. Exercise can also delay skeletal muscle aging by increasing the activities of Sirt1/Srl pathway and FOXO/SOD pathway49,50. Similar to previous research results, this study found that endurance exercise can reduce obesity by upregulating the expression of Gαq genes in skeletal muscle and adipose tissue. At the same time, endurance exercise can improve the skeletal muscle structure and function of drosophila with Gαq gene knockdown in skeletal muscle and adipose tissue by mediating the regulation of Gαq/Mhc, Gαq/Srl/MRCC-I, and Gαq/SOD pathways in three aspects: skeletal muscle contraction protein, mitochondria function, and antioxidant capacity, enhancing their exercise ability (Fig. 8).
Fig. 8.
Relationship between Gαq gene and endurance exercise and genetic obesity and skeletal muscle dysfunction. Gαq-RNAi in muscle and adipose tissue inhibits the Gαq mediated Gαq/Mhc, Gαq/Srl/MRCC-I, and Gαq/SOD pathways, leading to increased triglyceride accumulation in F1 Drosophila, inducing genetic obesity and further developing skeletal muscle dysfunction. This process can be blocked by endurance exercise.
Conclusion
Genetic defects in the adipose tissue and skeletal muscle tissue Gαq genes induce hereditary obesity and skeletal muscle dysfunction in Drosophila. In addition, endurance exercise positively regulates the adipose tissue and skeletal muscle tissue Gαq genes and improves skeletal muscle fiber contractile proteins, mitochondrial function, and antioxidant capacity, thereby attenuating this genetic obesity and concomitant skeletal muscle dysfunction.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Core Facility of Drosophila Resource and Technology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences for providing fly stocks and reagents.
Author contributions
Xinyuan Yin wrote the initial draft and conducted experimental data analysis, Dengtai Wen received funding support and revised the article, Hanyu Li assisted in data analysis, Zhaoqing Gao assisted in data analysis, Yuze Gao assisted in the experiment, and WeiJia Hao assisted in the experiment.
Funding
Shandong Province Higher Education Youth Innovation Team Project (2023RW057), National Natural Science Foundation of China (NSFC) (No. 32000832).
Data availability
All the generated data and the analysis developed in this study are included in this article.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xin-yuan Yin, Email: 1249690988@qq.com.
Deng-tai Wen, Email: 191729783@qq.com.
References
- 1.Koliaki, C., Dalamaga, M. & Liatis, S. Update on the obesity epidemic: After the sudden rise, is the upward trajectory beginning to flatten? Curr. Obes. Rep.12(4), 514–527. 10.1007/s13679-023-00527-y (2023). [DOI] [PMC free article] [PubMed]
- 2.Shabana, H. S. Obesity more than a ‘cosmetic’ problem. Current knowledge and future prospects of human obesity genetics. Biochem. Genet.10.1007/s10528-015-9700-2 (2020). [DOI] [PubMed]
- 3.Manzo, R. et al. Environmental enrichment prevents gut dysbiosis progression and enhances glucose metabolism in high-fat diet-induced obese mice. Int. J. Mol. Sci.25(13), 6904. 10.3390/ijms25136904 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manikat, R. & Nguyen, M. H. Nonalcoholic fatty liver disease and non-liver comorbidities. Clin. Mol. Hepatol.29(Suppl), 86–102. 10.3350/cmh.2022.0442 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandoval-Bórquez, A. et al. Adipose tissue dysfunction and the role of adipocyte-derived extracellular vesicles in obesity and metabolic syndrome. J. Endocr. Soc.8(8), bvae126. 10.1210/jendso/bvae126 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dearden, L. et al. Maternal obesity increases hypothalamic mir-505-5p expression in mouse offspring leading to altered fatty acid sensing and increased intake of high-fat food. PLoS Biol.22(6), e3002641. 10.1371/journal.pbio.3002641 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin, J. et al. Exercise ameliorates muscular excessive mitochondrial fission, insulin resistance and inflammation in diabetic rats via irisin/AMPK activation. Sci. Rep.14(1), 10658. 10.1038/s41598-024-61415-6 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, Y. et al. Regular exercise in Drosophila prevents age-related cardiac dysfunction caused by high fat and heart-specific knockdown of Skd. Int. J. Mol. Sci.24(2), 1216. 10.3390/ijms24021216 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolnes, K. J., Petersen, M. H., Lien-Iversen, T., Højlund, K. & Jensen, J. Effect of exercise training on fat loss-energetic perspectives and the role of improved adipose tissue function and body fat distribution. Front. Physiol.12, 737709. 10.3389/fphys.2021.737709 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma, S., Thibodeau, S. & Lytton, J. Signal pathway analysis of selected obesity-associated melanocortin-4 receptor class V mutants. Biochim. Biophys. Acta Mol. Basis Dis.1866(8), 165835. 10.1016/j.bbadis.2020.165835 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Yan, H. et al. Regular exercise modulates the dfoxo/dsrebp pathway to alleviate high-fat-diet-induced obesity and cardiac dysfunction in Drosophila. Int. J. Mol. Sci.24(21), 15562. 10.3390/ijms242115562 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, C. Y. et al. Leu27 IGF-II-induced hypertrophy in H9c2 cardiomyoblasts is ameliorated by saffron by regulation of calcineurin/NFAT and CaMKIIδ signaling. Environ. Toxicol.36(12), 2475–2483. 10.1002/tox.23360 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Pedroni, L. et al. Free fatty acid receptors beyond fatty acids: a computational journey to explore peptides as possible binders of GPR120. Curr. Res. Food Sci.8, 100710. 10.1016/j.crfs.2024.100710 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bone, D. B. J. et al. Skeletal muscle-specific activation of Gq signaling maintains glucose homeostasis. Diabetes68(6), 1341–1352. 10.2337/db18-0796 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunddal, K. V. et al. Selective release of gastrointestinal hormones induced by an orally active GPR39 agonist. Mol. Metab.49, 101207. 10.1016/j.molmet.2021.101207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lattanzi, R. et al. MRAP2 inhibits β-Arrestin-2 recruitment to the Prokineticin receptor 2. Curr. Issues Mol. Biol.46(2), 1607–1620. 10.3390/cimb46020104 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumbach, J., Xu, Y., Hehlert, P. & Kühnlein, R. P. Gαq, Gγ1 and Plc21C control Drosophila body fat storage. J. Genet. Genom.41(5), 283–292. 10.1016/j.jgg.2014.03.005 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Mora, I., Puiggròs, F., Serras, F., Gil-Cardoso, K. & Escoté, X. Emerging models for studying adipose tissue metabolism. Biochem. Pharmacol.223, 116123. 10.1016/j.bcp.2024.116123 (2024). [DOI] [PubMed] [Google Scholar]
- 19.Hou, W. Q. et al. Physical exercise ameliorates age-related deterioration of skeletal muscle and mortality by activating Pten-related pathways in Drosophila on a high-salt diet. FASEB J.37(12), e23304. 10.1096/fj.202301099R (2023). [DOI] [PubMed] [Google Scholar]
- 20.Yu, S. et al. Inonotus obliquus aqueous extract inhibits intestinal inflammation and insulin metabolism defects in Drosophila. Toxicol. Mech. Methods13. 10.1080/15376516.2024.2368795 (2024). [DOI] [PubMed]
- 21.Wang, J. F., Wen, D. T., Wang, S. J., Gao, Y. H. & Yin, X. Y. Muscle-specific overexpression of Atg2 gene and endurance exercise delay age-related deteriorations of skeletal muscle and heart function via activating the AMPK/Sirt1/PGC-1α pathway in male Drosophila. FASEB J.37(11), e23214. 10.1096/fj.202301312R (2023). [DOI] [PubMed] [Google Scholar]
- 22.Balapattabi, K. et al. Angiotensin AT1A receptor signal switching in Agouti-related peptide neurons mediates metabolic rate adaptation during obesity. Cell. Rep.42(8), 112935. 10.1016/j.celrep.2023.112935 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu, Z., Zhao, M., Atanes, P. & Persaud, S. J. Quantification of changes in human islet G protein-coupled receptor mRNA expression in obesity. Diabet. Med.39(12), e14974. 10.1111/dme.14974 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Cui, Y., Auclair, H., He, R. & Zhang, Q. GPCR-mediated regulation of beige adipocyte formation: implications for obesity and metabolic health. Gene915, 148421. 10.1016/j.gene.2024.148421 (2024). [DOI] [PubMed] [Google Scholar]
- 25.Rahbani, J. F. et al. ADRA1A-Gαq signalling potentiates adipocyte thermogenesis through CKB and TNAP. Nat. Metab.4(11), 1459–1473. 10.1038/s42255-022-00667-w (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, X. & Macielag, M. J. GPR120 agonists for the treatment of diabetes: a patent review (2014 present). Expert Opin. Ther. Pat.30(10), 729–742. 10.1080/13543776.2020.1811852 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Voss, A. C. et al. Exercise microdosing for skeletal muscle health applications to spaceflight. J. Appl. Physiol.136(5), 1040–1052. 10.1152/japplphysiol.00491.2023 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiles, J. W. et al. Differentially co-expressed myofibre transcripts associated with abnormal myofibre proportion in chronic obstructive pulmonary disease. J. Cachexia Sarcopenia Muscle15(3), 1016–1029. 10.1002/jcsm.13473 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaziri, P., Ryan, D., Johnston, C. A. & Cripps, R. M. A novel mechanism for activation of myosin regulatory light chain by protein kinase C-delta in Drosophila. Genetics216(1), 177–190. 10.1534/genetics.120.303540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, S. H. et al. Fermented yak-kong using Bifidobacterium animalis derived from Korean infant intestine effectively relieves muscle atrophy in an aging mouse model. Food Funct.3010.1039/d3fo04204a (2024). [DOI] [PubMed]
- 31.Lawler, J. M. & Hindle, A. Living in a box or call of the wild? Revisiting lifetime inactivity and sarcopenia. Antioxid. Redox Signal.15(9), 2529–2541. 10.1089/ars.2011.3974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merzetti, E. M. & Staveley, B. E. Spargel, the PGC-1α homologue, in models of Parkinson disease in Drosophila melanogaster. BMC Neurosci.16, 70. 10.1186/s12868-015-0210-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X. et al. Alogliptin prevents diastolic dysfunction and preserves left ventricular mitochondrial function in diabetic rabbits. Cardiovasc. Diabetol.17(1), 160. 10.1186/s12933-018-0803-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Son, R. H. et al. Potential of lycii radicis cortex as an ameliorative agent for skeletal muscle atrophy. Pharmaceuticals (Basel)17(4), 462. 10.3390/ph17040462 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, J. H. et al. Hydroethanolic extract of Cirsium setidens ameliorates doxorubicin-induced cardiotoxicity by AMPK-PGC-1α-SOD-mediated mitochondrial protection. Phytomedicine129, 155633. 10.1016/j.phymed.2024.155633 (2024). [DOI] [PubMed] [Google Scholar]
- 36.Diop, S. B. et al. PGC-1/spargel counteracts high-fat-diet-induced obesity and cardiac lipotoxicity downstream of TOR and brummer ATGL lipase. Cell. Rep.10(9), 1572–1584. 10.1016/j.celrep.2015.02.022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu, S. C. et al. Myricetin mitigates motor disturbance and decreases neuronal ferroptosis in a rat model of Parkinson’s disease. Sci. Rep.14(1), 15107. 10.1038/s41598-024-62910-6 (2024). [DOI] [PMC free article] [PubMed]
- 38.Liu, D. et al. Effects of exercise intervention on type 2 diabetes patients with abdominal obesity and low thigh circumference (EXTEND): study protocol for a randomized controlled trial. Front. Endocrinol. (Lausanne)13, 937264. 10.3389/fendo.2022.937264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotake, H. et al. Endurance Exercise training-attenuated diabetic kidney disease with muscle weakness in spontaneously diabetic torii fatty rats. Kidney Blood Press. Res.47(3), 203–218. 10.1159/000521464 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Wen, D. T., Wang, W. Q., Hou, W. Q., Cai, S. X. & Zhai, S. S. Endurance exercise protects aging Drosophila from high-salt diet (HSD)-induced climbing capacity decline and lifespan decrease by enhancing antioxidant capacity. Biol. Open9(5), bio045260. 10.1242/bio.045260 (2020). Published 2020 May 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng, T. et al. Exercise training upregulates cardiac mtp expression in Drosophila melanogaster with HFD to improve cardiac dysfunction and abnormal lipid metabolism. Biology (Basel)11(12), 1745. 10.3390/biology11121745 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reisman, E. G., Hawley, J. A. & Hoffman, N. J. Exercise-regulated mitochondrial and nuclear signalling networks in skeletal muscle. Sports Med.54(5), 1097–1119. 10.1007/s40279-024-02007-2 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie, X. & Huang, C. Role of the gut-muscle axis in mitochondrial function of ageing muscle under different exercise modes. Ageing Res. Rev.98, 102316. 10.1016/j.arr.2024.102316 (2024). [DOI] [PubMed] [Google Scholar]
- 44.Matsukawa, T. et al. Upregulation of skeletal muscle PGC-1α through the elevation of cyclic AMP levels by Cyanidin-3-glucoside enhances exercise performance. Sci. Rep.7, 44799. 10.1038/srep44799 (2017). [DOI] [PMC free article] [PubMed]
- 45.Das, S., Kumar, P., Verma, A., Maiti, T. K. & Mathew, S. J. Myosin heavy chain mutations that cause Freeman–Sheldon syndrome lead to muscle structural and functional defects in Drosophila. Dev. Biol.449(2), 90–98. 10.1016/j.ydbio.2019.02.017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tinkerhess, M. J. et al. The Drosophila PGC-1α homolog spargel modulates the physiological effects of endurance exercise. PLoS ONE7(2), e31633. 10.1371/journal.pone.0031633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damschroder, D. et al. Stimulating the sir2-spargel axis rescues exercise capacity and mitochondrial respiration in a Drosophila model of Barth syndrome. Dis. Model. Mech.15(10), dmm049279. 10.1242/dmm.04927 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Horani, R. A., Janaydeh, S., Al-Trad, B., Aljanabi, M. M. & Muhaidat, R. Acute exercise promptly normalizes myocardial myosin heavy-chain isoform mRNA composition in diabetic rats: implications for diabetic cardiomyopathy. Medicine (Kaunas)59(12), 2193. 10.3390/medicina59122193 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao, Y. H., Wen, D. T., Du, Z. R., Wang, J. F. & Wang, S. J. Muscle psn gene combined with exercise contribute to healthy aging of skeletal muscle and lifespan by adaptively regulating Sirt1/PGC-1α and arm pathway. PLoS ONE19(5), e0300787. 10.1371/journal.pone.0300787 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joassard, O. R. et al. Regulation of Akt-mTOR, ubiquitin-proteasome and autophagy-lysosome pathways in response to formoterol administration in rat skeletal muscle. Int. J. Biochem. Cell. Biol.45(11), 2444–2455. 10.1016/j.biocel.2013.07.019 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the generated data and the analysis developed in this study are included in this article.