Abstract
Bacteria invest significant resources into the continuous creation and tailoring of their essential protective peptidoglycan (PG) cell wall. Several soluble PG biosynthesis products in the periplasm are transported to the cytosol for recycling, leading to enhanced bacterial fitness. GlcNAc-1,6-anhydroMurNAc and peptide variants are transported by the essential major facilitator superfamily importer AmpG in Gram-negative pathogens including Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. Accumulation of GlcNAc-1,6-anhydroMurNAc-pentapeptides also results from β-lactam antibiotic induced cell wall damage. In some species, these products upregulate the β-lactamase AmpC, which hydrolyzes β-lactams to allow for bacterial survival and drug-resistant infections. Here, we have used cryo-electron microscopy and chemical synthesis of substrates in an integrated structural, biochemical, and cellular analysis of AmpG. We show how AmpG accommodates the large GlcNAc-1,6-anhydroMurNAc peptides, including a unique hydrophobic vestibule to the substrate binding cavity, and characterize residues involved in binding that inform the mechanism of proton-mediated transport.
Subject terms: Cryoelectron microscopy, Bacterial structural biology, Pathogens, Biophysics, Biochemistry
AmpG is an MFS importer of bacterial cell wall fragments. Here, authors present a cryoEM structure and supporting mutagenesis to illuminate AmpG’s essential role in bacterial fitness and derepression of broad-spectrum β-lactam antibiotic resistance.
Introduction
Antimicrobial resistance (AMR) is an escalating global health crisis, with the evolution of resistance outpacing drug development1. β-lactams represent over 65% of all antibiotics prescribed globally and play a critical role in infectious disease management, but their efficacy is threatened by multiple mechanisms of resistance2. While there is an urgent need for new strategies to address AMR, understanding the current mechanisms of β-lactam resistance enables creation of complementary therapeutics administered as cocktail treatments, prolonging the effectiveness of many existing antibiotics approved against various bacterial species and pathologies. β-lactams covalently bind and inhibit the transpeptidation domain of penicillin-binding proteins (PBPs). This interrupts the essential peptide cross-linking of peptidoglycan (PG) polymeric strands and ultimately leads to cell wall damage and bacterial cell lysis2. Widespread use of these drugs has resulted in selection for various resistance mechanisms. In Gram-negative bacterial pathogens, enzymes that hydrolyze the β-lactam core, β-lactamases, represent the primary mechanism of resistance3. Many Gram-negative bacteria are a pressing threat to hospitalized patients in terms of development of treatment-resistant infections and/or contributions to sepsis, and are associated with high mortality rates4. Problematic nosocomial Enterobacteriaceae and Pseudomonadaceae infections are often enabled by a combination of underlying drug resistance mechanisms, but one of particular concern is the production of the chromosomally-encoded protein AmpC. This enzyme is a class C serine β-lactamase with a broad spectrum of action, able to inactivate nearly all classic and later-generation β-lactam antibiotics, including penicillins and cephalosporins5. AmpC is the primary driver of β-lactam resistance in P. aeruginosa6 and a direct cause of broad spectrum β-lactam resistance, including to carbapenems, considered antibiotics of last resort, in Enterobacter spp. and K. pneumoniae7,8.
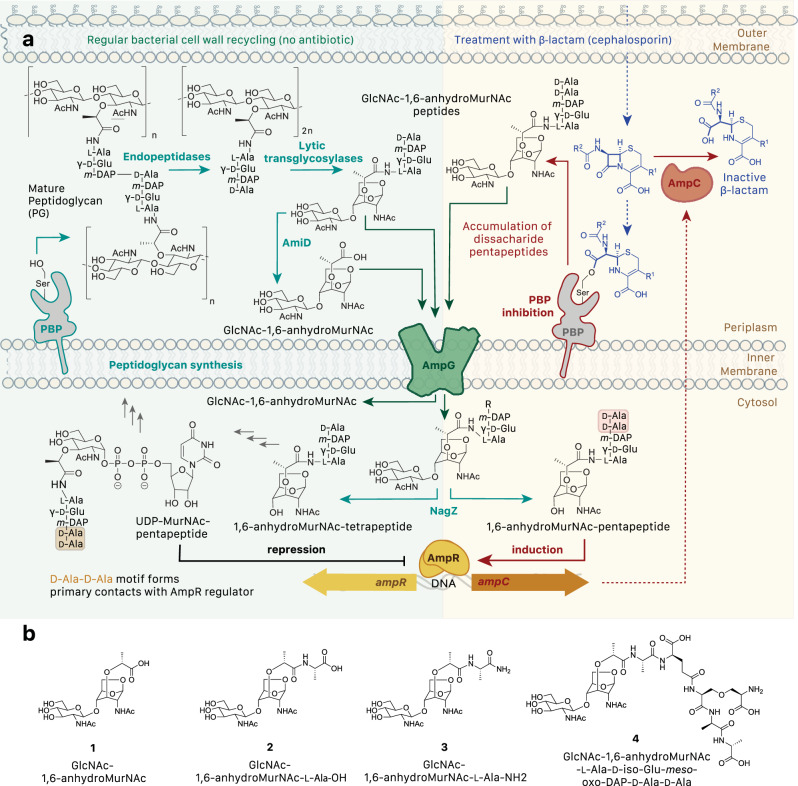
Expression of chromosomal ampC is controlled by a complicated signaling system that recognizes metabolized cell wall fragments9,10 (Fig. 1a). PG polymeric strands in the periplasm are hydrolyzed by the actions of soluble lytic transglycosylases into 1,6-anhydromuropeptides. To recycle the unusual meso-diaminopimelic acid residue and other energetically-costly chemical elements from freed anhydromuropeptides, they are taken up through the inner membrane by a specific anhydromuropeptide permease, AmpG11. The GlcNAc moiety is then released by the glucosamidase NagZ, creating a 1,6-anhydroMurNAc peptide product. Under physiological conditions, those products are further processed by AmpD and other enzymes and recycled into the cytoplasmic PG biosynthetic pathway as UDP-N-acetylmuramic acid (MurNAc)-pentapeptide precursors, which suppress ampC transcription by binding and repressing the LysR-type transcriptional regulator AmpR. However, in the presence of β-lactams, increased amounts of GlcNAc-1,6-anhydroMurNAc-pentapeptides accumulate as a result of cell wall damage and are transported to the cytoplasm by AmpG. There, after release of the GlcNAc moiety by NagZ, they competitively displace UDP-MurNAc from AmpR and activate ampC transcription. Targeting this pathway thus represents a strategy to prevent the development of AmpC-mediated resistance. While some work has focused on the well-characterized enzyme NagZ, understanding of the structure and function of the conserved integral membrane transporter/permease AmpG is lacking, although it may be a more compelling target for intervention due to its periplasmic accessibility and lack of redundancy in most species. Indeed, prior studies have shown that ΔampG mutants regain sensitivity to β-lactam antibiotics12 while deletion of ampG from P. aeruginosa reduces both upregulation of AmpC and biofilm formation, suggesting inhibition of this permease could reduce pathogenicity in multiple ways13.
Fig. 1. Role of AmpG in peptidoglycan recycling and antibiotic resistance.
a Under physiological conditions, mature PG is processed into GlcNAc-1,6-anhydroMurNAc peptides (primarily tetrapeptides) that are transported across the bacterial inner membrane to the cytosol by AmpG for recycling. An intermediate in this normal PG biosynthetic pathway, UDP-MurNAc-pentapeptide, binds to AmpR, which represses production of the class C β-lactamase AmpC. However, in the presence of β-lactams, inhibition of PBPs leads to an accumulation of hydrolyzed PG pentapeptide fragments, which are transported to the cytosol by AmpG, converted to 1,6-anhydroMurNAc-pentapeptides by NagZ, bind AmpR and induce expression of ampC. Secreted AmpC hydrolyzes β-lactams, providing antibiotic resistance10. b Chemically synthesized GlcNAc-1,6-anhydroMurNAc derivatives made for this study.
AmpG belongs to the major facilitator superfamily (MFS) of secondary active transporters, a large family of solute carrier transporters in both prokaryotes and eukaryotes that share two symmetric six-transmembrane helical bundles as the core of the protein. Within this family, AmpG is predicted to be a symporter belonging to the poorly characterized proton motive force (PMF) driven peptide-acetyl-CoA transporter family 2.A.1.2514. While general mechanisms of transport by MFS proteins have been characterized, there is huge variation in the molecular details of substrate specificity and gating mechanisms required to prevent deleterious passage of other molecules during transport. Characterization of the atomic structure, substrate binding, and mechanism of action of AmpG is needed to understand this fundamental biological process in bacteria and its potential druggability to combat AmpC-mediated β-lactam resistance in serious pathogens.
Here, we present the structural details of an AmpG family member from E. coli determined by cryo-EM, in complex with the disaccharide of a bound detergent, which we propose mimics the disaccharide of the physiological substrate. Biochemical binding analysis with a suite of AmpG substrates synthesized here, coupled with mutagenesis studies of the role of conserved positions in P. aeruginosa cellular assays, provide the basis for enhanced understanding of the transport channel, its mode of substrate binding, and potential transport mechanism.
Results
E. coli AmpG protein production
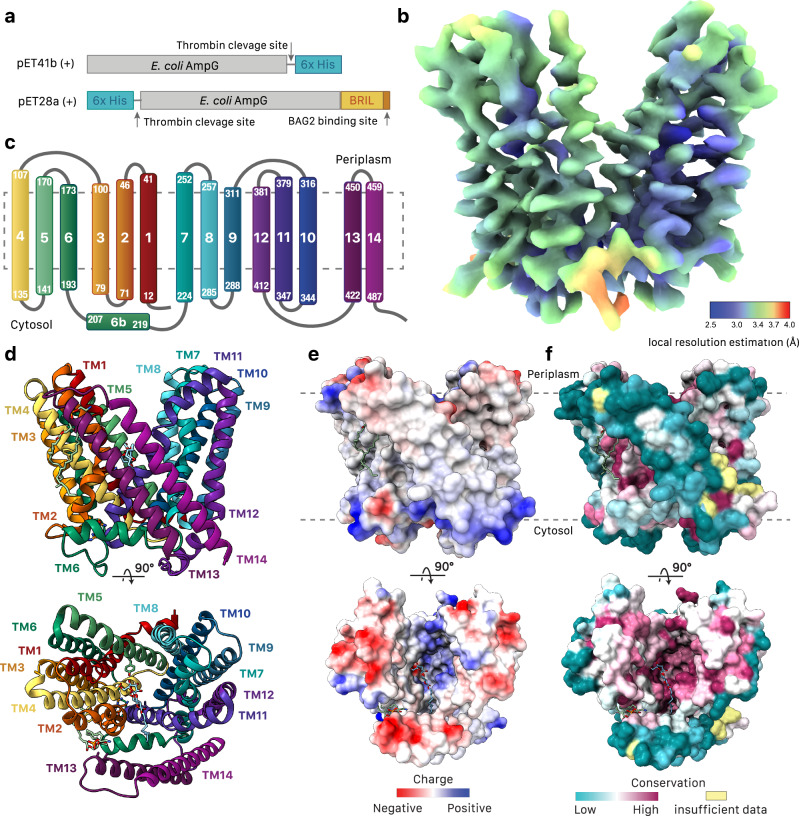
AmpG permeases are present in many Gram-negative bacteria (Supplementary Fig. 1). An E. coli variant was chosen for study to allow for production in its native lipid environment, often a consideration for yield and stability of polytopic membrane proteins for structure/function analysis. Hexahistidine-tagged full-length wild-type AmpG (Fig. 2a) was expressed in E. coli strain C41ΔompFΔacrB15 and screened for optimized purification conditions and membrane mimetics. AmpG was successfully solubilized in either N-dodecyl-β-D-maltopyranoside (DDM) or MSP1D1 nanodiscs16 (9 nM diameter) with incorporated E. coli polar lipids. The purity and monodispersity of AmpG after removal of the recombinant hexahistidine-tag was confirmed by SDS-PAGE and negative-stain TEM (Supplementary Fig. 2a, b). To aid structure determination of this small ~53 kDa membrane protein with cryo-EM, thermostabilized apocytochrome b562 (BRIL), a 106 residue four helical bundle17, was included at the C-terminus and used as an epitope for the synthetic antibody BAG218. The high affinity interaction between AmpG-BRIL and BAG2 was verified with surface plasmon resonance (SPR) with a binding constant (Kd) of 9.6 nM (Supplementary Fig. 3e). AmpG-BRIL in DDM was incubated with BAG2 at a 1:1 molar ratio and complex formation was validated by glycerol gradient centrifugation, pull-down assay, native PAGE, and negative-stain TEM (Supplementary Fig. 2c–h).
Fig. 2. Overall architecture of wild-type E. coli AmpG.
a AmpG constructs used in this study. b Cryo-EM reconstruction of E. coli AmpG colored by local resolution. c Secondary structure topology of AmpG. d Structure of AmpG in ribbon colored as in c. DDM (light blue) and PE (sea green) shown as sticks with cpk coloring. e Electrostatic surface map (calculated with Coulombic electrostatic potential65) of AmpG. f Conserved residue surface map (calculated with Consurf64) of AmpG.
Synthesis of GlcNAc-1,6-anhydroMurNAc-peptide analogs
The chemical syntheses, purification, and validation of several AmpG GlcNAc-1,6-anhydroMurNAc substrate variants was pursued for this study: compound 1, or GlcNAc-1,6-anhydroMurNAc, is the minimal structural unit necessary for recognition by AmpG14, compound 2 is GlcNAc-1,6-anhydroMurNAc bearing L-alanine, compound 3 is GlcNAc-1,6-anhydroMurNAc bearing L-alaninamide, and compound 4 is GlcNAc-1,6-anhydroMurNAc bearing a full L-Ala-D-iso-Glu-meso-oxa-Dap-D-Ala-D-Ala stem pentapeptide (Fig. 1b).
The chemical synthesis of 1 has been known since 1986 when first published by the Paulsen group19. Many other groups have since developed their own synthetic routes to the GlcNAc-1,6-anhydroMurNAc motif to study proteins and biochemical pathways that use GlcNAc-1,6-anhydroMurNAc glycopeptides as substrates or agonists: measuring the turnover of lytic transglycosylases20, investigating the effect of tracheal cytotoxin (TCT) on the Nod1 receptor21, and developing a fluorescent assay for AmpG22. Our synthetic route is a slight modification of Paulsen’s19 and is described in a recent publication by the Tanner group23. We produced intermediate peracetyl GlcNAc-1,6-anhydroMurNAc methyl ester sugar S1, which yields 1 quantitatively after saponification and an acidic resin workup (Supplementary Fig. 4a).
Disaccharide 1 could be converted into the novel L-alanine-bearing disaccharide 5 (Supplementary Methods) by a preliminary coupling reaction to L-alanine benzyl ester via PyBOP/HOBt and a subsequent tosic acid-catalyzed acetonide protection. This protected sugar intermediate 5 is produced in modest yields but is far easier to isolate and purify via flash chromatography than its amphiphilic benzyl ester counterpart lacking a GlcNAc acetonide. Compound 5 may be deprotected via acidic resin treatment followed by hydrogenolysis to give substrate 2 in quantitative yields, or amidated by ammonia in methanol and deprotected by acidic resin to give substrate 3 in excellent yields (Supplementary Fig. 4a).
The chemical synthesis of disaccharide pentapeptide 4 is far more challenging than that of compounds 1-3, mostly due to the separate chemical synthesis of the meso-oxa-Dap pentapeptide. Meso-oxa-Dap is an isosteric analog of the natural meso-diaminopimelic acid (meso-Dap) found in the stem peptides of various Gram-negative bacteria such as E. coli and P. aeruginosa and used as a “handle” by their PBPs to cross-link layers of cell wall PG polymers24. meso-Dap is a reasonably challenging synthetic target with many different lengthy synthetic routes already published25. Moreover, synthetic routes to meso-Dap as part of a PG stem peptide must be able to selectively incorporate only the L-stereocenter of the symmetric meso-Dap into the oligopeptide backbone. Orthogonally-protected meso-oxa-Dap was first produced by the Vederas group via a Lewis acid-catalyzed aziridine ring opening with a serine nucleophile26. The succinct synthesis of this meso-Dap analog makes meso-Dap peptides more attractive synthetic targets in general. Moreover, L,L-oxa-Dap proved to be an adequate substrate for Dap-epimerase26 and meso-oxa-Dap oligopeptides were successfully processed by the Gram-negative carboxypeptidases Csd6 and Pgp227; thereby lending credence to meso-oxa-Dap’s viability as a structural analog to meso-Dap for enzymatic/biological experiments.
The synthesis of 4 along with the significance and various pitfalls were discussed prior23. Vederas’ methodology was extended to the nucleophilic ring opening of an aziridine already embedded within an oligopeptide. Disaccharide S2 was coupled to meso-oxa-Dap-pentapeptide P1 via EDCI/HOBt – S2 was also derived from S1, and P1 was derived from its N-terminus Boc-protected analog. The resultant protected glycopeptide S3 underwent hydrogenolysis and subsequent deacetylation under mildly basic conditions to give the full disaccharide meso-oxa-Dap pentapeptide 4 in good yields (Supplementary Fig. 4b). The identity and validating metrics of all novel compounds verified by 1H and 13C nuclear magnetic resonance spectroscopy (NMR) and by high-resolution electrospray ionization mass spectrometry (HR-ESI-MS) (Supplementary Methods).
Binding analysis of AmpG with synthesized substrate analogs
To minimize protein and ligand requirements and accommodate the requisite use of membrane mimetics for AmpG solubilization, microscale thermophoresis was used for binding studies between DDM-solubilized AmpG and the synthesized substrates. The calculated affinities for GlcNAc-1,6-anhydroMurNAc disaccharide (compound 1) and GlcNAc-1,6-anhydroMurNAc-pentapeptide (compound 4) were similar and in keeping with published values determined using E. coli spheroplast transport assays22. For the shorter peptide variants, specifically the GlcNAc-1,6-anhydro-MurNAc-L-ala with free negatively charged carboxylate (compound 2) or neutral amidated (compound 3) forms, we observed that the negative charge on the truncated peptide was deleterious to binding, with binding only recovered once the charge was neutralized (Supplementary Fig. 3a). Binding of compound 1 to AmpG-BRIL and the AmpG-BRIL BAG2 complex used for structure determination was also assayed showing comparable binding to the wild-type protein (Supplementary Fig. 3d). To rule out differences between detergent micelle-solubilized protein and a more native membrane bilayer-like environment, binding was also assayed using AmpG solubilized in MSP1D1 nanodiscs16 using isothermal titration calorimetry (ITC), which showed binding values for the GlcNAc-1,6-anhydroMurNAc disaccharide consistent with those from MST (Supplementary Fig. 3b).
Cryo-EM structure of AmpG-BRIL/BAG2 antibody complex
The structure of DDM-solubilized AmpG-BRIL in complex with the BAG2 antibody was determined using cryo-EM to a global resolution of 3.78 Å (Supplementary Fig. 5). Local resolution analysis showed the majority of the protein at 3.0 to 3.5 Å, with clearly resolved side chain densities throughout (Supplementary Fig. 6). The refined model contains AmpG residues 3-486 (the physiological C-terminus), a modeled DDM detergent (used for solubilization), a phosphoethanolamine (PE) lipid, and has excellent stereochemistry (Supplementary Table 1). The C-terminal BRIL fusion and BAG2 antibody were not well resolved in the final reconstruction but were clear in 2D classification (Supplementary Figs. 5, 6). Attempts to determine the structure of AmpG without BRIL and BAG2 were unsuccessful, suggesting their inclusion was important for accurate particle alignment, despite this flexibility. The DDM-solubilized AmpG-BRIL fusion showed similar binding to the GlcNAc-1,6-anhydroMurNAc disaccharide by MST, suggesting inclusion at the C-terminus does not have a deleterious effect on the structure (Supplementary Fig. 3d).
AmpG is stabilized in an outward-open conformation
AmpG was captured in an outwards (to the periplasm) open conformation (Fig. 2b). The structure shows 14 bitopic transmembrane helices, as opposed to the previous predictions of 12 bitopic and two re-entrant helices (Fig. 2c)11. The core of the transporter is composed of two pseudo-symmetric bundles of six transmembrane helices (N-terminal bundle TMs 1–6 and C-terminal bundle TMs 7–12) in the canonical topology of the MFS family28. Two additional antiparallel C-terminal helices, TM13 and TM14, pack against the lateral V-shaped opening to the central cavity formed by TM2 and TM11 at the intersection of the N- and C-terminal helical bundles, creating a unique hydrophobic vestibule (Fig. 2d). Both the N- and C- termini reside on the cytoplasmic face of the membrane and result in a typical distribution of positively charged residues consistent with the positive-inside rule of polytopic membrane protein topology29 (Fig. 2e). The outward-facing conformation has been proposed to be more energetically stable in other MFS symporters e.g., bacterial fucose symporter FucP30, and AmpG is stabilized in the outward-open state by several conserved interactions between TM2 and TM11 (see below) with the central channel sealed at the apex by interactions between TM4, 5, 10, and 11 (Fig. 2f). AmpG contains conserved structural motifs found in both multidrug antiporters and sugar and peptide symporters. However, despite more functional similarity to sugar and peptide symporters FucP and PepT31,32, analysis with Dali33 shows more structural similarity to the multidrug exporter family (MdfA, LmrP, YajR, NorA; range of RMSD superpositions on 370 common Cα atoms is 4.8-6.4 Å).
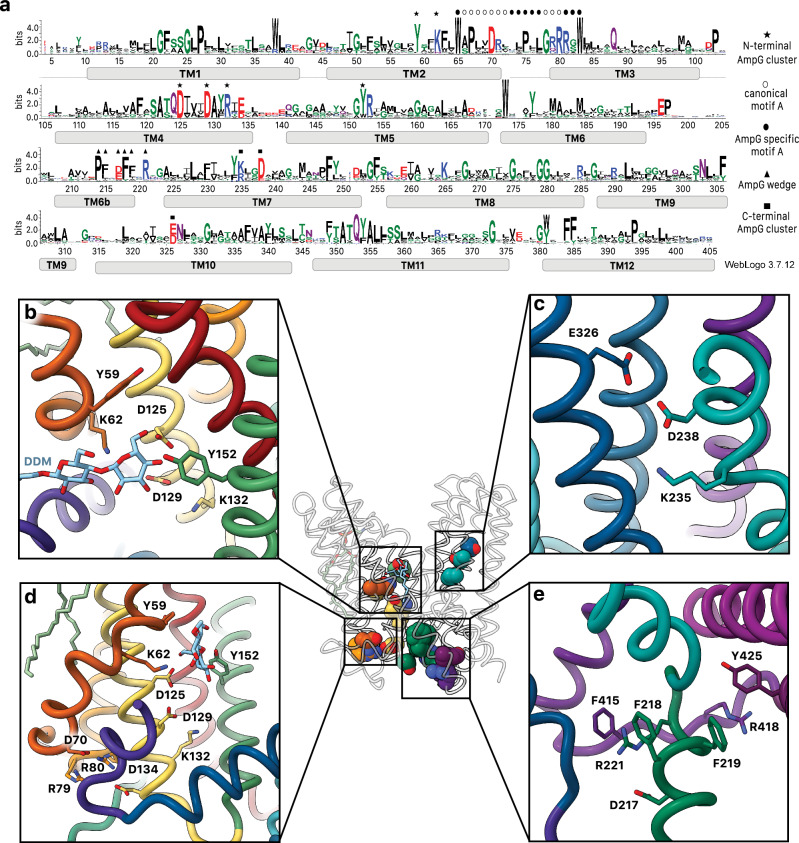
Conserved motif A acts as a molecular latch
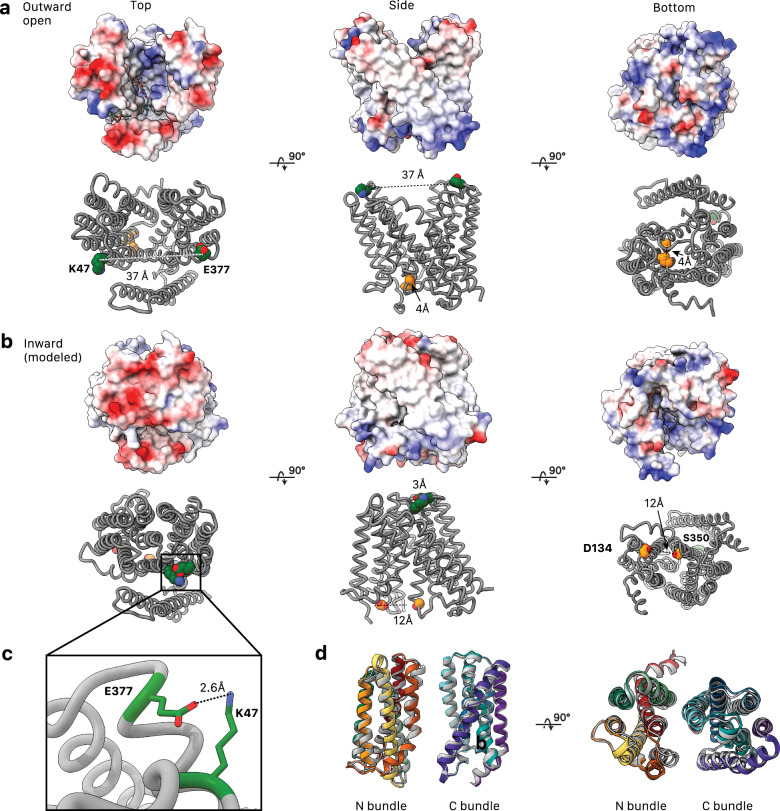
Various sequence motifs amongst MFS transporters have been documented28. The most highly conserved is motif A, present in all AmpG homologs and here involving residues at the C-terminus of TM2, the N-terminus of TM3 and the intervening loop (collectively residues 66–80) (Fig. 3a, d). Strictly conserved Asp70 is the key component of the motif, with its carboxylate side chain positioned for electrostatic interaction with the adjacent N-terminal helix dipole of TM11, stabilizing its position in the outward-open state (Fig. 4a). The guanidinium side chains of conserved Arg79 and Arg80, which protrude from the same face at the N-terminus of TM3, both directly coordinate the carboxylate of Asp70, likely enhancing its electronegative charge and the helix dipole interaction (Fig. 4a). Arg80 is further oriented by a direct interaction with the side chain carboxylate of Asp134 on the C-terminus of TM4. Together with Asp70, these residues have been proposed to constitute a charge relay triad critical for function34. Arg81, the third arginine in succession and more unique to the AmpG family (Supplementary Figs. 1, 7), projects towards the exterior where it would be ideally situated to interact with lipid headgroups of the membrane inner leaflet (Fig. 4a). Supporting the importance of motif A in stabilizing the outward-open state, a substitution of Asp70 to alanine in the sequence allowed us to obtain a model of the inward-open conformation using AlphaFold335. The accuracy of the predicted structure, especially the newly formed interface that closes the periplasmic opening, is supported by tightly co-evolving clusters of residues identified by EVcouplings36 (Fig. 5, Supplementary Fig. 8).
Fig. 3. Conserved residues in E. coli AmpG.
a Sequence conservation logo created using 2936 sequences identified from EVcouplings with a bitscore of 0.7. Plotted in WebLogo3 with key motifs and features annotated (see methods). b Binding cavity interactions with the sugar moiety of DDM. c Conserved titratable residues on the C-terminal helical repeat. Glu326 and Asp238 form a conserved carboxyl-carboxylate pair coordinated next to Lys235. d Conserved titratable residues on the N-terminal helical repeat. Motif A Asp70 coordinated by Arg79 and Arg80 interacting with TM11 helix dipole and substrate binding site Lys132, Asp129, and Asp125. e “Foot-in-the-door” motif, which stabilizes the C-terminal helical bundle and helps create the hydrophobic vestibule.
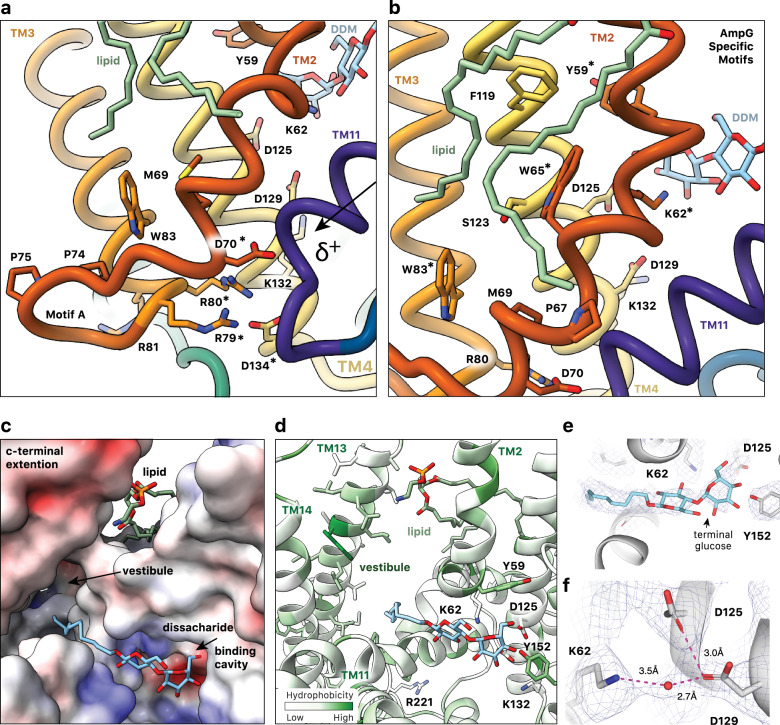
Fig. 4. Substrate binding cavity of outward facing E. coli AmpG.
a AmpG motif A stabilizes the outward conformation with the Asp70 carboxylate forming an electrostatic interaction with the N-terminal helix dipole of TM11, likely enhanced by coordination with Arg79, Arg80, and Asp134. Colored as in Fig. 1d, with mentioned residues marked by *. b Conserved Trp65 and Trp83 in the motif A adaption stabilize the kink in TM2, creating an expanded binding cavity and positioning key residues e.g., Tyr59 and Lys62 for substrate binding. The observed PE lipid extends across the aromatic and nonpolar residues on TM2 and TM4. c Electrostatic surface of the periplasmic substrate binding cavity of AmpG with bound DDM. The maltose sits near an electronegative pocket arising from conserved Asp125 and Asp134, while the hydrophobic acyl chain points into the hydrophobic vestibule between the MFS helical bundle and additional C-terminal helices TM13 and 14. d AmpG substrate binding cavity colored by hydrophobicity. Side chains of disaccharide binding residues shown as sticks and labeled. DDM and lipid are shown as in a. e Density in the binding cavity of AmpG of the modeled bound DDM. f Density consistent with a water coordinated between Lys62, Asp125, and Asp129.
Fig. 5. Modeling of the inward-open state of AmpG.
AmpG in the experimental outward open conformation (a) compared to the AlphaFold335 modeled inward open conformation (b), shown with Coulombic electrostatic potential surface (above) calculated with ChimeraX35,65 and evolutionary coupled sets of resides (below). Residue pairs on the periplasmic side of the protein, Lys47 and Glu377 (green), are strongly coupled. They are separated ~30 Å in the outward structure but form close contacts in the inward model. At the cytoplasmic side, Asp134 and Ser350 (gold) are also evolutionarily coupled. c Interaction of evolutionary coupled residues E377 and K47 in the modeled inward state. d Structural alignment of N- and C-terminal helical bundles of the experimental outward and modeled inward (gray) states. TM1-TM6 (residues 1–195) RMSD was 1.7 Å, and TM7-TM12 (residues 223-413) RMSD was 1.1 Å.
Extending motif A in our structure, conserved residues specific to the AmpG family are observed immediately up- and downstream of the canonical sequence (Figs. 3a, 4b). Two highly conserved tryptophan residues, Trp65 (TM2) and Trp83 (TM3), appear to play a role in stabilizing the precise position of motif A and the direct substrate-binding residues (see below). The Trp65 indole notably forms a hydrogen bond with the evolutionarily coupled36 Ser123 side chain hydroxyl on TM4 and, together with Pro67, appear to contribute to the significant helical deformation at Val61, with a ~ 125° angular redirection where TM2 and TM11 splay apart to form the lateral entrance to the substrate binding cavity. Characterized homologs have significantly less kinked helices (160-170°) in this region. In AmpG, this pronounced TM2 deformation also serves to orient the near invariant Lys62 along with well conserved Tyr59 to point into the central periplasmic exposed cavity, with a direct role in substrate binding (Fig. 4b).
The outside-open state presents a large binding cavity to the periplasm
In the captured outward-facing conformation, AmpG displays a large funnel-shaped binding cavity extending about three-quarters of the way through the membrane, which is conserved amongst AmpG homologs of clinical significance (Figs. 2f, 4c, and Supplementary Fig. 1). The GlcNAc-1,6-anhydroMurNAc-peptide substrates have an overall net negative charge (Fig. 1b) and we note a general electropositive surface of the cavity, with residues Lys133, Arg153, Lys235, Lys265, and Arg365 positioned to potentially help guide the substrate into the funnel-shaped binding pocket (Fig. 5a, b).
The cavity is sealed to the cytoplasm by multiple interactions, notably between TM4/TM5 and TM10/TM11 from the N- and C-terminal bundles, respectively (Fig. 2d). As discussed above, the interaction between motif A Asp70 on TM2 and the N-terminal helical dipole of TM11 is key to stabilizing this conformation (Fig. 4a). We observe density extending into the cavity interacting with TM2, TM4 and TM5 of the N-terminal bundle that is consistent with a molecule of DDM positioned with its 12-carbon acyl tail pointing into the hydrophobic cleft created by TM13 and TM14 (see below), and its maltose disaccharide head group in the substrate binding cavity (Fig. 4c–e). Based on chemical similarity and binding to key conserved residues, we propose the latter is mimicking binding of the GlcNAc-1,6-anhydroMurNAc disaccharide of the natural substrate. The binding site is defined by the side chains of highly conserved residues Tyr59, Lys62, Asp125, Asp129 and Tyr152 (Fig. 3a). Asp125 (TM4) and Tyr152 (TM5) closely contact the terminating glucose sugar ring hydroxyls of DDM (Fig. 3b). Lys62 on TM2, mentioned above as an AmpG-specific adaption of MFS motif A, is also pointed into the cavity, where it is within hydrogen bonding distance of the first glucose sugar of DDM. Lys62 is partially buried in a remarkably conserved hydrophobic pocket composed of residues Tyr59, Phe63, Trp65, Ala122, Ile126, and Ala355 that restrains its side chain orientation. As a consequence of sugar binding, the lysine amino side chain becomes more buried, with a solvent accessible surface of 16 Å2 compared to 51 Å2 with the DDM removed37,38.
Conserved Lys62 and Tyr152 are important for AmpG activity
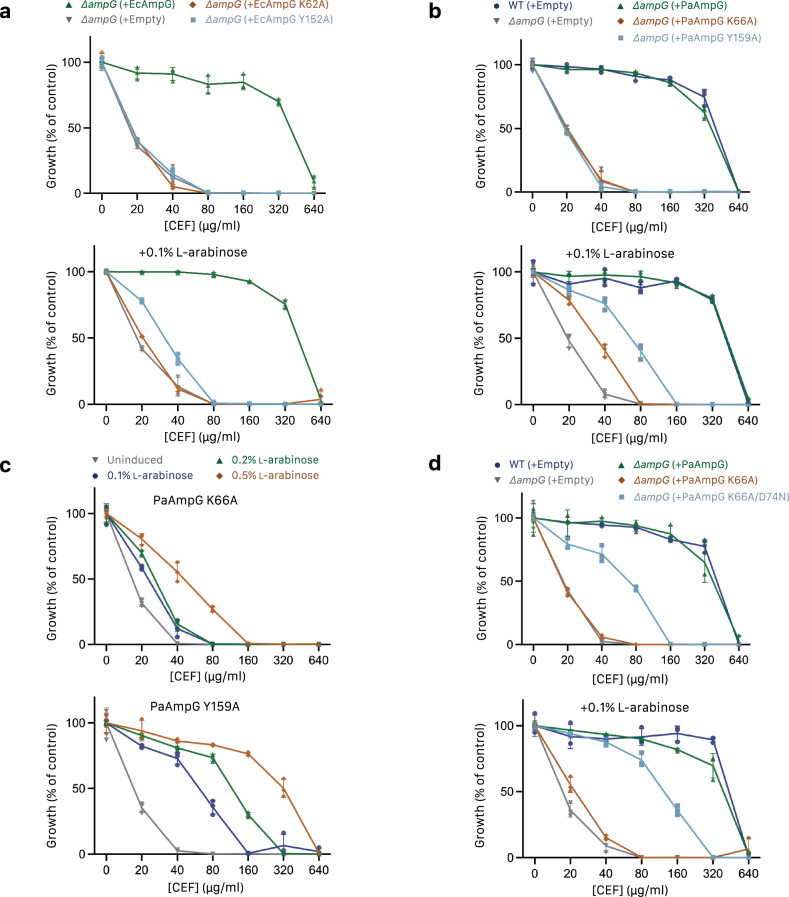
Lys62, Asp125, Asp129, and Tyr152 are highly conserved (Fig. 3a), including in strains of clinical significance (Supplementary Fig. 1), and the proximity to the bound disaccharide of DDM suggests their importance for AmpG substrate binding and transport. To explore this further, Lys62 and Tyr152 were selected for mutation and tested in a β-lactam susceptibility assay. Although E. coli ampC is not inducible by β-lactams39, E. coli (Ec) ampG can cross-complement an ampG knockout in inducible strain P. aeruginosa PAO1 and restore growth to wild-type levels in the presence of the second-generation cephamycin antibiotic cefoxitin (CEF), due to upregulated AmpC β-lactamase activity (Figs. 1a, 6a). EcAmpG K62A and Y152A mutants both had decreased CEF minimal inhibitory concentrations (MICs), similar to ΔampG complemented with empty vector, suggesting these point mutants are not functional for transport of the PG products required to upregulate AmpC. Increasing the expression of the EcAmpG Y152A but not the K62A mutant with 0.1% arabinose partially restored resistance (Fig. 6a).
Fig. 6. Cellular AmpG assays.
a Effect of cefoxitin on growth of a P. aeruginosa PAO1 ampG knockout complemented with E. coli AmpG on low-copy plasmid pHERD30T. Leaky (uninduced) expression of WT ampG (green up-arrow) restored growth to WT levels, while K62A (orange diamond) or Y152A (blue square) mutants had the same expression as empty control (gray down-arrow). Induction of expression with 0.1% arabinose partially restored growth of Y152A but not K62A. b P. aeruginosa AmpG mutants tested as in a. K66A and Y159A affected growth similarly to the ampG knockout (coloring as in a). On induction with 0.1% arabinose, growth of K66A and Y159A was partially restored to varying degrees. c Growth of P. aeruginosa AmpG mutants as in b at varying levels of arabinose induction (gray down-arrow uninduced, blue circle 0.1%, green up-arrow 0.2%, orange diamond 0.5%). Both K66A and Y159A show a concentration-dependent increase in growth on induction, with the response much higher for Y159A. d Identification of a D74N mutant that rescues growth of the P. aeruginosa AmpG K66A mutant. (coloring as in a, blue square represents K66A/D74N). Experiments were performed in technical triplicate and biological duplicate with a representative shown. Data points correspond to technical replicates, with error bands representing SD.
To test the role of these residues in other homologs, we repeated the susceptibility assay using P. aeruginosa (Pa) AmpG. Complementation of the P. aeruginosa ΔampG mutant with the equivalent PaAmpG point mutants (K66A or Y159A) resulted in similar decreases in CEF MICs (Fig. 6b) supporting the functional importance of these residues across the AmpG family. Further, titration of arabinose showed a dose-dependent response with 0.5% arabinose restoring CEF MICs to near wild type levels for Y159A, while growth of K66A was only partially restored, demonstrating mutation at this position is less tolerated for AmpG function (Fig. 6c). To further probe the specific roles of PaAmpG Lys66 and Tyr159, we performed a reverse genetic screen in P. aeruginosa, looking for mutations that could rescue growth of K66A or Y159A mutants in the presence of CEF but not in the presence of piperacillin (PIP). Both antibiotics are substrates of β-lactamase AmpC; however, PIP is only a weak inducer of AmpC40. Therefore, resistance to CEF and PIP could indicate a mutation that results in constitutive upregulation of AmpC (e.g., through ampR or ampC promoter mutations) while CEF resistance but PIP sensitivity could indicate a mutation in PaAmpG that restores its function. Remarkably, for K66A we identified multiple suppressors with a D74N mutation that partially restored function (Fig. 6d). P. aeruginosa Asp74 is equivalent to E. coli Asp70 in motif A and the repeated occurrence of the same aspartate to asparagine mutation in combination with K66A suggests a possible mechanistic relationship between these two conserved residues in the substrate binding site and motif A, respectively.
In line with these results, we tested the ability of both EcAmpG K62A and Y159A mutants to bind GlcNAc-1,6-anhydroMurNAc using MST, showing a reduced binding affinity for GlcNAc-1,6-anhydroMurNAc of >2 mM and ~500 uM, respectively (Supplementary Fig. 3c).
An AmpG specific hydrophobic vestibule to the substrate binding cavity
Our structure also illustrates an unusual structural motif in AmpG, comprised of antiparallel C-terminal helices TM13 and TM14 that pack along one opening of the periplasmic binding cavity (Fig. 2c, d). Fourteen TMs is a departure from the canonical 12 TM MFS topology present in structural homologs of AmpG (Supplementary Fig. 9). In the AmpG family, these helices are typically located at the C-terminus as observed in E. coli here. However, some AmpG orthologues, including those from P. aeruginosa and A. baumannii, have an internal insertion between the N- and C-terminal helical bundles (Supplementary Fig. 1). Despite this, these insertions show a similar predicted structural placement and formation of related hydrophobic vestibule structures (Supplementary Fig. 10).
Here, both TM13 and TM14 are classical bitopic membrane-spanning helices of >25 residues. Rather than typical intimate hydrophobic packing with TM2 or TM11, a clear structural “foot-in-the-door” is observed at the cytosolic end of TM13 and TM14. This involves the amphipathic helix (TM6b; residues 207-219) and the loop to TM7, which provide a wedge-like protrusion with stabilizing interactions therein (Figs. 3e, 4d). Conserved residues Asp217, Phe218, Phe219 and Arg221 create a set of hydrophobic and electrostatic interactions, with the guanidinium group of Arg221 forming an electrostatic pair with Asp217 and also stacking against the aromatic rings of Phe218 and Phe415 from the loop connecting TM12 and TM13. Additional interactions, including conserved π-stacking of the side chain guanidinium of Arg418 and the aromatic side chain of Tyr425, further stabilize this complex intersection of helical segments. The ultimate consequence of this wedge-like protrusion is a dramatic hydrophobic cleft, approximately 30 × 30 × 12 Å, closed at the cytosol and running up to the periplasmic space. The hydrophobic faces of TM13 and TM14 form one wall with TM11, TM2, the C-terminus of TM6b and TM7 the other. We observe a significant amount of non-protein density in this region, which funnels down to the substrate binding pocket, which may represent loosely associated detergent or inherent lipids (Supplementary Fig. 11a). In addition, we observe a diacyl lipid bound at the lateral entrance to the vestibule with its headgroup sandwiched between the periplasmic ends of TM2 and TM13 while the two acyl tails extend down the outer surface of TM2 towards motif A and the cytoplasmic face of AmpG (Supplementary Fig. 11b, c). The density is well modeled by a PE lipid, which are highly abundant ( ~ 75 %) in E. coli membranes41.
Discussion
Here, we have determined the cryo-EM structure of E. coli AmpG in an outward-open conformation with an observed disaccharide of DDM helping to define the GlcNAc-1,6-anhydroMurNAc-peptide substrate binding cavity. Together with binding data here, these directly support prior transport assays14 showing that the main determinant of substrate binding is the disaccharide moiety. An ordered diacyl lipid modeled as PE is bound in the entrance to a hydrophobic “vestibule” structure, adjacent and continuous with the substrate binding cavity that could provide space for the larger-than-typical MFS substrates of AmpG. The structure reveals key interactions that stabilize the outward state of the transporter, open to the periplasm, and those involved in conformational changes that underlie the import mechanism, several of which are supported here by mutational data in vitro. In addition to its physiological roles in cell wall recycling and bacterial fitness, AmpG plays a key role in the upregulation of β-lactamase AmpC, which is a direct cause of broad spectrum β-lactam resistance in clinically relevant ESKAPE pathogens, including P. aeruginosa, Enterobacter spp., and K. pneumoniae7,9. Thus, the added understanding of the structure and mechanism of AmpG could help guide the development of new antimicrobial therapies or resistance inhibitors.
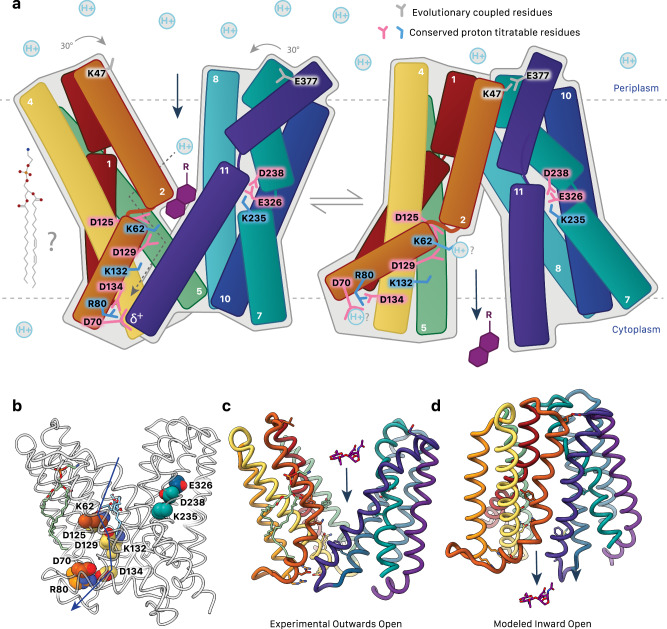
AmpG is a symporter, using proton transfer to mediate the conformational changes required for substrate transport from the periplasm to the cytoplasm14. As captured here for AmpG, the outward-open conformation of MFS symporters is typically more energetically stable, with the transporter exposed to the exterior for substrate binding34. A defining feature is motif A (we propose an AmpG-specific adaption of the canonical motif to cover AmpG residues 65-83; Figs. 3a and 4a, b). The motif A central aspartate (AmpG Asp70) is highly stabilizing for the outward substrate loading conformation of MFS importers34. Here, an electrostatic interaction between the Asp70 side chain carboxylate and the N-terminal helical dipole of TM11 represents the primary interaction point of the N-terminal and C-terminal bundles (Fig. 4a). Modulation of this interaction is central to the rigid body conformational switch of the N- and C-terminal helical bundles between outward and inward states34,42 (Fig. 7a). Protonation of the carboxylate is implicated in this, which would neutralize the full charge on the side chain and eliminate the significant electrostatic interaction with TM11. Interacting with Asp70 here, conserved Arg80 and Asp134 have been proposed to function as a charge relay triad important to this process in other MFS transporters34, which we suggest either modulates the local electrostatic environment and pKa of Asp70, and/or plays a direct role in post-delivery passage of the proton from Asp70 through to cytosolic solvent. After transport, deprotonation of Asp70 is presumably needed to regenerate the outward form for the next round of substrate loading in the periplasm. In line with this role, in silico mutation of just Asp70 to alanine allowed us to obtain an inward-open conformation of AmpG using AlphaFold335 (Fig. 5), otherwise not predicted for the wild type sequence.
Fig. 7. Proposed AmpG transport mechanism.
a Substrate binding (GlcNAc-1,6-anhydroMurNAc with R representing -OH, tri, tetra, and pentapeptide chains; purple) and protonation of conserved acidic residues are proposed to result in a rigid body conformational shift of AmpG. Motif A Asp70 interacts with the TM11 helix dipole to stabilize the outward open state. The disruption of this interaction e.g., by protonation is proposed to be a key part of the switch to the inward conformation. Positions of conserved titratable residues shown. Dotted arrows suggest a proposed flow of proton transport. Evolutionary coupled residues Lys47 and Glu377 are over 30 Å apart in the outward model but come together to interact in the inward conformation. A potential role of lipid(s) in this mechanism, as suggested by the observed ordered PE, is yet to be determined. b Conserved titratable residues as in a shown as spheres. c Experimentally determined outward-open structure of AmpG. A GlcNAc-1,6-anhydroMurNAc substrate is shown in sticks. d Modeled inward conformation.
Outside of motif A, there is a remarkable variety in the specific residues implicated in substrate binding and proton relay described for various MFS proteins, painting a general picture highlighting the complexity and substrate specific differences of the proton pathway(s) important to optimal transport action32,42–45. In AmpG, there are seven highly conserved, titratable residues in the central cavity that can be divided into two clusters: Lys62 (TM2), Asp125, Asp129, and Lys132 (TM4) on the N-terminal helical repeat; and Lys235, Asp238 (TM7) and Glu336 (TM10) on the C-terminal repeat (Fig. 3).
The N-terminal repeat residues form part of the DDM maltose binding site, with Lys62 and Asp125 positioned close enough to form direct interactions (Fig. 3b). We propose that this glucose disaccharide mimics the native GlcNAc-1,6-anhydroMurNAc substrate. In support, mutation of Lys62 or Tyr152, which also forms the binding pocket for the terminal glucose, both abrogated function in E. coli and P. aeruginosa AmpG homologs (Fig. 6) and impacted substrate binding (Supplementary Fig. 3c). Lys62, Asp125, Asp129 and Lys132 are within proton transfer distance of one other, with the two aspartates notably forming a carboxyl-carboxylate pair46 (Fig. 3b). In functional homologs, FucP and PepT, titratable residues at positions analogous to Asp125 have been confirmed to be essential for transport activity32,47. Remarkably, we found that spontaneous mutation of motif A aspartate to asparagine in P. aeruginosa AmpG (Asp74) could partially rescue β-lactam resistance when the lysine (Lys66) was also mutated to alanine (Fig. 6d), establishing a possible mechanistic link between these two positions. This could be direct, with a charge relay pathway from Lys66 culminating in Asp74 protonation outlined above (mimicked by the asparagine substitution) and change from outward to inward-open states (Fig. 7). However, although the motif A aspartate is often suggested to be the endpoint for proton transfer in MFS action, which is indeed conceptually satisfying here as well, we note a conserved network of the Asp70 carboxylate with a solvated Arg80/Arg81 cluster would presumably depress its pKa, suggesting its electrostatic potential may always be “on” in a deprotonated, electronegative form that could still facilitate the return to the stabilized outward form with the TM11 helical dipole, without necessarily needing to go under protonation/deprotonation events per se. Alternatively, the link between Lys62 and Asp70 could be indirect, with the neutral amide side chain in the Asn70 mutant weakening the electrostatic interaction with TM11 and lowering the energy barrier to flip between states, which could compensate for loss of Lys62. Lys62 is partially buried in an unusual hydrophobic environment (conserved Tyr59, Ile126, Ala355 and Phe354) that would allow for a depressed pKa to facilitate protonation/deprotonation events at physiological pH48. The Asp125 and Asp129 carboxyl-carboxylate pair is also likely stabilized by the altered pKa’s typical of those interactions49 (see also below), further fine tuning these residues for precise proton-driven events. A charged protonated state of Lys62 could provide repulsive energy to expel substrate and drive the needed motion of the closely packed adjacent hydrophobic TM11 face to the inward state (Fig. 7), in turn effectively solvating the Lys62, Asp125, Asp129, and Lys132 cluster. We observe density consistent with solvation around these key residues, notably a putative water meditating an interaction with Lys62, Asp125, and Asp129 (Fig. 4f) that could potentially shift to act as a proton donor/acceptor to compensate for loss of the amine side chain in the K62A mutation, recovering the observed partial activity.
The conserved C-terminal repeat cluster (Lys235, Asp238 (TM7) and Glu336 (TM10)) is located on the opposite face of the cavity (Fig. 3c). YajR, LmrP, and NorA have pairs of titratable residues located on TM7 and TM10 implicated in proton translocation, albeit with small differences in disposition42,43,50. Asp238 and Glu336 form a carboxyl-carboxylate pair like Asp125 and Asp129. These motifs have unique pKa properties compared to free carboxylic acids (pKa ~ 4), with the pKa of one of the carboxylic acids shifting to overcome the repulsive interaction49 and are generally involved in protein stabilization, enzyme catalysis, and substrate binding46. In some proteases, carboxyl-carboxylate pairs can overcome potentially repulsive interactions with substrates and products with carboxylic acid groups51. We note that the AmpG substrate GlcNAc-1,6-anhydroMurNAc-pentapeptide itself has up to three carboxylic acid groups, suggesting this region could be involved in binding or sensing of the substrate stem peptide.
AmpG has a larger central cavity than structurally similar MFS proteins (Supplementary Fig. 9), in line with the need to accommodate the relatively large GlcNAc-1,6-anhydro-MurNAc sugars with varying peptide lengths (tri, tetra or penta), which are uniquely specific to AmpG (Fig. 1a). Binding analyzes here, plus previous literature using cellular transport assays14, agree that the GlcNAc-1,6-anhydroMurNAc is the key determinant of binding. The presence of the fully intact D-Ala-D-Ala pentapeptide with several inherent charges (which accumulates in the periplasm during β-lactam antibiotic treatment; Fig. 1a) does not significantly enhance binding (Supplementary Fig. 3a) or transport14 beyond that of the disaccharide alone. The MurNAc anhydro moiety is, however, important for optimal transport14 and we suggest this enhances binding to the hydrophobic face of TM11 and further desolvates the adjacent Lys62. Synthesis of GlcNAc-1,6-anhydro-MurNAc-L-Ala-COOH or amidated GlcNAc-1,6-anhydroMurNAc-L-Ala-CONH2 allowed us to further investigate the effects of a premature terminal charge. Interestingly, while the amidated substrate had similar binding to the regular substrates, the GlcNAc-1,6-anhydroMurNAc-L-Ala-COOH substrate was relatively impaired (Supplementary Fig. 3a). This suggests that the peptide, although not contributing greatly to overall binding, must be accommodated within the cavity, and therefore appropriately positioned electrostatic interactions in the binding pocket are likely essential in discriminating substrates.
The AmpG structure also reveals a vestibule to the central cavity formed by the helical insertion in the AmpG family. This structure could further extend the central cavity and provide additional binding capacity for the large AmpG specific substrates. Lipids are also likely to occupy the vestibule in the physiological membrane environment (Supplementary Fig. 11a). Although not fully understood, it is becoming apparent that lipids play an essential role in both structure and function of various MFS members28,52. PE is especially crucial for activity in several transporters, including LmrP, LacY, PheP, and GabP53–56. A structured lipid linking the lateral entrance of the vestibule and motif A on opposing leaflets of the membrane (Supplementary Fig. 11b, c) could suggest a specific role in AmpG function, in substrate binding, regulation of transport or substrate-induced signaling, although further experiments are needed to investigate this observation.
AmpG plays a key role in β-lactamase induction in clinically relevant pathogens, including P. aeruginosa, Enterobacter spp., and K. pneumoniae7,9. Inhibition of this protein would be a valuable approach for tackling these multidrug-resistant bacteria and the infections they cause. As shown in our MIC data, the deletion of AmpG from PAO1 strains resulted in a 30-fold decrease in the CEF MIC. Additionally, mutation of the binding pocket residues Lys62 and Tyr152 abolished activity, showing the promise of a binding site inhibitor of AmpG. A potential inhibitor wouldn’t need to cross the inner membrane to act on the protein, improving the feasibility of development compared to other cytosolic targets of Gram-negative bacteria. Beyond AmpC regulation, the cell wall recycling role of AmpG is likely important to general bacterial fitness in all strains, independent of AmpC inducibility. Therefore, targeting the AmpG symporter as part of new synergistic antimicrobial cocktails, particularly in combination with the still commonly prescribed β-lactam antibiotics, is a compelling approach for enhancing therapeutic efficacy.
Methods
Chemical Synthesis of Substrates
Chemicals were purchased from Millipore Sigma, Alfa Aesar, Fischer Scientific, Combi-blocks, Ark Pharm, and Chem-Impex Int. All chemicals were used directly without further purification unless noted. TLC plates were purchased from EMD Chemical Inc. Flash silica chromatography was conducted using silica gel SiliaFlash F60 (Silicycle, 230-400 mesh) or (where noted) high- purity Davisil Grade 633 (200-425 mesh, pore size 60 Å) silica gel. Strongly acidic hydrogen form Dowex® 50 W X8 resin was purchased from Millipore Sigma. All reactions were carried out in Pyrex borosilicate glassware.
Detailed chemical synthesis of compounds 2 and 3 are recorded in the Supplementary Methods. The synthesis of compounds 1 and 4 has been described in the previous work of Soni et al., 202023. 1H NMR spectra and proton-decoupled 13C NMR spectra were recorded on either a Bruker AV400sp spectrometer at a field strength of 400 MHz for 1H spectra and 101 MHz for 13C spectra or a Bruker AV600 spectrometer at a field strength of 600 MHz for 1H spectra and 151 MHz for 13C spectra. High-resolution spectra were recorded on a Waters/Micromass TOF spectrometer equipped with electrospray ionization (ESI) at the UBC Chemistry Department’s Mass Spectrometry Facilities.
E. coli AmpG Purification
Various species of AmpG were considered for expression, but E. coli AmpG was chosen for optimal lipid environment during expression. E. coli ampG was cloned in pET28a and pET41b vectors with thrombin-cleavable hexahistidine tags at the N- or C- terminus (Supplementary Table 2). To assist with orientation in structural determination, an addition of thermostabilized apocytochrome b562 from Escherichia coli M7W/H102I/R106L (BRIL) was designed18,57. The AmpG BRIL construct was cloned with restriction-free cloning protocols of amplification of BRIL, and insertion at the C-terminal end of AmpG pET28a plasmid (Fig. 2a)58.
Constructs were overexpressed in C41ΔompFΔacrF cells in ZY autoinduction media with kanamycin for 4 hrs at 37 °C and 16 hrs at 27 °C15, harvested by centrifugation and stored at -80 °C. All subsequent steps were performed at 4 °C. Cell pellets were resuspended in 10% glycerol, 20 mM HEPES pH 7.5, 500 mM NaCl with 20 µg ml−1 DNase I (Roche) for 30 min and lysed with high-pressure homogenization 3x at 30 kpsi (Constant Flow Cell Disruptor). Cell debris was separated by centrifugation at 18,000 g for 30 min, and membrane components were separated by centrifuging the supernatant at 200,000 g for 60 min. The AmpG membranes were solubilized in 20 mM Tris pH 8, 500 mM NaCl with a glass Teflon Dounce homogenizer before extraction in 1% N-dodecyl-β-D-maltopyranoside (w/v) (DDM, Anatrace) for 1 hour. Insoluble debris was removed by further centrifugation for 30 minutes at 200,000 g. The supernatant was loaded onto 5 ml of HisPur™ Cobalt Resin (ThermoFisher), equilibrated with 20 mM Tris pH 8, 500 mM NaCl, 0.016% (w/v) Anagrade DDM, and incubated shaking for 30 min, followed by washes of 10 mM imidazole, 20 mM Tris pH 8, 150 mM NaCl, 0.016% (w/v) Anagrade DDM, before overnight on column cleavage with bovine alpha-thrombin (Prolytix). Flow through and 10 mL wash was collected and concentrated with 50 kDa molecular mass cut-off cellulose membrane (Merck Millipore) before further purification using a Superdex 200 Increase 10/300 GL column equilibrated with 20 mM HEPES pH 7.5, 200 mM NaCl, 0.016% DDM. The WT AmpG was concentrated and flash frozen for kinetic determination, while the AmpG BRIL construct was then further combined with BAG2 antibody for cryo-EM. Initial confirmation of AmpG BRIL BAG2 binding was observed in glycerol gradient ultracentrifugation. Glycerol gradients with 5 to 25% (v/v) glycerol (200 mM NaCl, 20 mM HEPES pH 7.5, 0.016% (w/v) Anagrade DDM) were made using a Gradient Master (BioComp Instruments)59. AmpG BRIL and BAG2 at 4 mg/ml in a 1:1.5 ratio were loaded onto the gradient and centrifuged for 16 hours at 194,000 g in a SW55 Ti rotor (Beckman Coulter), and fractionated using a gradient fractinator (BioComp Instruments). Sample binding was demonstrated by coelution in the gradient. For Cryo-EM, AmpG BRIL and BAG2 were added in a 1:1 molar ratio of 50 µM each in 200 mM NaCl, 20 mM HEPES pH 7.5, 0.016% (w/v) Anagrade DDM and incubated for an hour before electron microscopy experiments.
Negative-stain electron microscopy
F coated, carbon supported copper grids, 400 mesh (Ted Pella, Inc) were glow-discharged (PELCO easiGlow™) before 4 µL of complex was applied to the grids and stained with 2% of uranyl acetate. Negative-stain data collection was carried out on a Talos L120C transmission electron microscope equipped with a CETA camera and LaB6 filament (High Resolution Macromolecular Electron Microscopy Facility, University of British Columbia), collecting 556 micrographs. Image processing for initial reconstruction was performed in CryoSPARC(4.4.1)60 with manual picking to create initial templates for template picker to pick 675 932 particles. Particles were then cleaned with 2D classification for a final stack of 124 279, before ab initio reconstruction of the final volume.
BAG2 purification
BAG2 antibody, a synthetically designed fiducial against BRIL from the Kossiakoff lab18, was expressed in E. coli BL21 (DE3). Cells were grown in 2xYT media with kanamycin at 37 °C until OD600 = 0.6, and induced with 1 mM IPTG at 37 °C for 4 hours. Cells were harvested with centrifugation, and resuspended in 20 mM HEPES pH 7.5, 200 mM NaCl, 20 µg ml−1 DNase I (Roche), cOmplete protease inhibitor (Roche) at 4 °C, followed by lysis with high-pressure homogenization 3x at 25 kpsi (Constant Flow Cell Disruptor). Resulting lysate was incubated at 60 °C for 30 minutes to denature bacterial proteins before ultracentrifugation to remove debris for 30 min at 200,000 g at 4 °C. The supernatant was filtered with a 0.22 µM membrane, before loading onto a 1 ml HiTrap™ Protein L column (Cytiva) equilibrated with running buffer (20 mM HEPES pH 7.5, 500 mM NaCl). BAG2 was washed with running buffer, before elution with 0.1 M acetic acid and dialyzed overnight at 4 °C into 20 mM HEPES pH 7.5, 200 mM NaCl, and concentrated in 15 kDa molecular mass cut-off cellulose membrane (Merck Millipore) for combination with AmpG BRIL.
E. coli AmpG nanodisc reconstitution
AmpG was reconstituted into nanodiscs followed the same purification protocol through membrane extraction, and all subsequent steps were done at 4 °C. The extracted protein was loaded onto 5 ml of HisPur™ Cobalt Resin overnight, before a wash of 10 mM imidazole and 5×10 ml elution fractions of 20 mM sodium acetate pH 4.5, 500 mM NaCl, 0.016% (w/v) Anagrade DDM. AmpG was then concentrated with 50 kDa molecular mass cut-off cellulose membrane (Merck Millipore) before further purification using a Superdex 200 Increase 10/300 GL column equilibrated with 20 mM HEPES pH 7.5, 500 mM NaCl, 0.016% (w/v) Anagrade DDM. E. coli polar lipids (Avanti Polar Lipids) were desiccated into a thin film before solubilization in 20 mM HEPES pH 7.5, 500 mM NaCl, 2% (w/v) Anagrade DDM. AmpG was then added in a 1:2:60 ratio of protein:MSP1D1:E. coli polar lipids, and let assemble for 45 min16. Sm2 adsorbent Bio-beads (Bio-Rad) were then added to remove the DDM, using a 3-fold weight excess of biobeads to DDM, adding the first third for one hour, and the rest of the biobeads overnight. Following nanodisc assembly, nanodiscs were centrifuged at 100,000 g for 30 min, before loading onto 1 ml equilibrated HisPur™ Ni-NTA Resin with 20 mM HEPES pH 7.5, 500 mM NaCl. Protein was eluted with 200 mM imidazole, concentrated with Amicon Ultra-15 concentrator (Millipore) with a molecular weight cutoff of 50 kDa, and injected onto a Superdex 200 Increase 10/300 GL column equilibrated with 20 mM HEPES pH 7.5, 200 mM NaCl. The peak fraction was then concentrated to 12 µM, with a corresponding amount of buffer for a blank, before confirmation of protein concentration using a BCA Assay (ThermoFischer) and immediate usage for ITC.
Isothermal Calorimetry
All ITC measurements were performed on an MicroCal PEAQ-ITC instrument (Malvern Panalytical) in biological triplicate. AmpG was purified in nanodiscs as described in assay buffer (20 mM HEPES pH 7.5, 200 mM NaCl) and concentrated to 12 µM, concentration measured by Pierce™ BCA Protein Assay (ThermoFischer). The titrand (AmpG) in the sample cell was titrated with substrate (GlcNAc-1,6-anhydroMurNAc) in the syringe at a concentration of 10 mM, with mixing speed s 300 rpm, cell temperature 25 °C, reference power 5 μcal/s, and with an initial delay of 60 s. The heat released was plotted as positive peaks. The data was analyzed using the provided software (MicroCal PEAQ-ITC Analysis Software v1.41), and corrected heat was plotted against the molar ratio of titrant vs titrand. Data fitting was performed using a one-site independent-binding model included in the software to obtain the equilibrium dissociation constant (KD) = 1/ KA.
Surface Plasmon Resonance
A Biacore T200 was used for surface plasmon resonance (Cytiva). For kinetic data measurements between BAG2 antibody and AmpG BRIL, a CM5 S Series sensor chip (Cytiva) was used. BAG2 antibody was immobilized on the surface at a flow rate of 10 µl/min and concentration of 30 µg/ml. Next, AmpG BRIL was injected at a flow rate of 30 µl/min at 25 °C, and the chip surfaces were regenerated by injection of 3 M GuHCl after each cycle for 120 s at a flow rate of 10 µl/min. These SPR experiments were performed in 20 mM HEPES pH 7.5, 200 mM NaCl with 0.05% Tween-20 (Sigma) and 0.016% (w/v) Anagrade DDM. Kinetic analysis was performed with the provided T200 software using a 1:1 model.
Microscale Thermophoresis
AmpG in 20 mM HEPES pH 7.5, 200 mM NaCl, 0.016% (w/v) Anagrade DDM was labeled with Alexa Fluor 647 Dye (ThermoFisher) with a 30 min dark incubation at a 3-fold molar ratio of dye to protein, followed by elimination of free dye using PD SpinTrap G25 columns (Cytiva). GlcNAc-1,6-anhydroMurNAc and full pentapeptide were synthesized as described by Soni et al., 202023, and full synthesis protocols for intermediate substrates are described in the Supplementary Methods. Pluronic® F-127 surfactant was added at 0.1% in assay buffer to minimize aggregation. AmpG was used at concentration of 10 nM, and substrates were used at a highest concentration of 10 mM in a 2-fold dilution in 16 Monolith NT™ Standard Treated Capillaries (K0002, Nanotempr technologies). Measurements were performed in technical triplicate at room temperature, and normalized fluorescent signal was plotted as a function of ligand concentration. Data analysis was performed in supplied MO Affinity Analysis software v2.3 using a Kd model for data fitting, and a software defined 68% confidence interval for uncertainty.
Cryo-EM data collection
Quantifoil (R1.2/1.3) 300 mesh Au grids (SPT Labtech) were glow-discharged for 2 minutes before 3 µl of protein sample was applied. The grids were blotted for 2.5 s with a Vitrobot Mark IV (ThermoFisher) at 4 °C and 100% humidity before being plunged into liquid ethane and stored in liquid nitrogen. The High-Resolution Macromolecular Cryo-Electron Microscopy facility at the University of British Columbia performed the screening of the grids on a Krios (ThermoFisher) transmission cryogenic electron microscope with a 300 kV accelerating voltage and a 300 kV Titan Krios (ThermoFisher) transmission cryogenic electron microscope equipped with a Falcon 4i (ThermoFisher) direct electron detector and a Selectris (ThermoFisher) energy filter, with an EPU software package. The full dataset was collected on the same microscope, with images gathered in electron-counting mode at 0.59 Å per physical pixel. The total dose delivered to the grids was 60 e− Å−2 over 875 frames in the EER video format. Fully automated data collection was performed using EPU (ThermoFisher) with a nominal defocus range set from 0.5 to 3.0 μm.
Cryo-EM processing
For the AmpG-BRIL BAG2 sample, 43 215 movies were collected at a pixel size of 0.59 Å per pixel, and 39 856 were accepted for data analysis. Processing was performed in CryoSPARC(4.4.1)60 (Supplementary Fig. 5). Blob picker in a subset of 15 484 micrographs was initially used for particle picking of 8 672 028 with 4x binning, which was used for 2D classification. A stack of 162 361 particles were used for initial 2 class ab initio 3D reconstructions that were then input as a template for template picker of the full 39 856 micrographs. 9 634 256 particles were then sorted with 2 class heterogenous refinements using the ab initio reconstruction and junk class, and then further filtered with 2D classifications and 2 class ab initio reconstructions, and then processed for multiple rounds of heterogeneous refinement. The best volume was further refined with NU-refinement with 128 404 particles, and a 1795 subset of particles was used to train a topaz picker model. Topaz picked 1 536 241 particles that were classified with 2D classification, 2 class heterogenous refinement, and 2 class ab initios and locally refined for a 4.12 Å model with 379 933 particles. This was used to train a more inclusive topaz model and particle pick 5 491 442 particles that were filtered and classified as described to 1 963 390. Particles were re-extracted at 2x binning, and filtering continued with NU refinements with dynamic masking, and local refinements with a soft static mask were used for final refinements with 503 623 particles with global resolution of 3.78 Å and local resolution of most of the protein of 3-3.5 Å. Model building was performed manually in COOT with a starting reference of the modeled wild-type AmpG Alphafold structure35,61. Refinements were performed with Phenix real space refine, and the modeled position of DDM in the density was supported by similar predictions of position in Autodock Vina62,63. Structure and sequence analysis was performed in Consurf 64, COOT61, ChimeraX65, Pymol66, Alphafold335, EVCoupling36, Weblogo367, and Dali33.
Functional Mutants expressed in PAO1
P. aeruginosa PAO1 was used as a parent strain for all assays. PAO1 ΔampG strain was constructed previously68. Overnight bacterial cultures were made by inoculating 3 mL of lysogeny broth (LB - Lennox) from a frozen stock and incubating at 37 °C while shaking. When appropriate, cultures contained antibiotics at the following concentrations: 15 or 30 µg/mL of gentamicin for E. coli or P. aeruginosa, respectively, 100 µg/mL of ampicillin for E. coli and 200 µg/mL of carbenicillin for P. aeruginosa, and 50 µg/mL of kanamycin for E. coli. Sub-cultures were made by diluting an overnight culture 1:25 into fresh media and incubating at 37 °C while shaking.
Minimum inhibitory concentration assays
Overnight cultures were sub-cultured into 3 mL of LB and grown to ~0.1 OD600. Subcultures were normalized to 0.1 OD600 and diluted 1:500 in fresh media containing sterile L-arabinose where indicated. Two times serial dilutions of two microliters of antibiotics (Sigma Aldrich) were added at all indicated concentrations to each well in technical duplicate or triplicate, except for the vehicle and sterility control wells, which received only vehicle (DMSO or water). Diluted cultures were added to the wells of a 96-well plate to a final volume of 150 µL. Sterility wells were filled to 150 µL with sterile media. Plates were incubated at 37 °C for 18 h while shaking at 200 rpm. Growth was measured as optical density at 600 nm at 18 h using a spectrophotometer (Multiskan GO, ThermoFisher).
Reverse genetic selection of ampG suppressor mutants
Overnight cultures of PAO1 ampG + pHERD30T expressing either K66A or Y159A AmpG mutants were sub-cultured into 3 mL of LB and grown to ~0.3 OD600. Subcultures were normalized to 0.3 OD600, then 100 µL of the cultures was spread onto LB agar plates containing 256 µg/mL of cefoxitin and 30 µg/mL gentamicin and incubated at 37 °C until colonies appeared (typically 24-48 hours). Colonies were then patched onto LB agar plates containing 256 µg/mL of cefoxitin and 30 µg/mL gentamicin, and plates containing 128 µg/mL of piperacillin and 30 µg/mL gentamicin. Restored cefoxitin resistance was validated for colonies that only grew on the cefoxitin plates with a cefoxitin MIC assay in broth, then the pHERD30T constructs were isolated from these strains and transformed into a clean PAO1 ampG background for additional cefoxitin MICs in broth. Plasmids that restored cefoxitin resistance in the clean background were sequenced (Plasmidsaurus).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Bryan Lin, Aleksander Lazarski, Armando Palacios, Josh Jenkins and Nathanael Caveney for advice on MST, ITC, vitrification, negative stain reconstruction and cryo-EM data collection strategies, respectively, and former Burrows lab member Richard Nguyen for initial studies of AmpG function. We thank Professor Anthony Kossiakoff from the University of Chicago for the generous gift of BRIL and BAG2 constructs and protocols. We thank the Canadian Foundation of Innovation and BC Knowledge Development Fund for infrastructure funding in the Strynadka laboratory and within the High-Resolution Macromolecular EM facility at UBC. We also thank members of the HRMEM operational team Claire Atkinson and Peter Horvath for grid screening and data collection. Molecular graphics and analyzes were performed using UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. This work was funded by the Canadian Institute of Health Research operating grant (RN509473-497990 to N.C.J.S.) and Natural Sciences and Engineering Research Council of Canada Discovery grants (RGPIN-2021-04237 to L.L.B. and GR010196 to M.E.T). N.C.J.S. and L.L.B. are Tier 1 Canada Research Chairs. L.N.Y. held an NSERC PGS-D award.
Author contributions
H.E.S., A.J.G., M.V., L.N.Y., R.P.L., V.A.M., C.S., and A.A.Z.D. performed cloning. H.E.S. and A.J.G. performed protein purification. H.E.S. and L.J.W. performed cryo-EM data processing. H.E.S. and L.J.W. carried out model building and refinement. H.E.S., L.J.W., and N.C.J.S. performed structure analyzes. C.M.V. and A.S.S. performed chemical synthesis, purification, and validation. H.E.S. carried out in vitro activity assays. L.N.Y. designed cellular genetic selection and carried out the antibiotic sensitivity assays. L.N.Y and Z.D.A.A. performed the cellular selection experiments. L.N.Y. performed antibiotic sensitivity assays. H.E.S., L.J.W., and N.C.J.S. wrote the manuscript with input from L.N.Y., L.L.B., C.M.V., and M.E.T. N.C.J.S, L.L.B., and M.E.T. supervised the research and accrued funding. All the authors read the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that support this study are available from the corresponding authors upon request. The cryo-EM map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-45167 (cryo-EM map of AmpG). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession code 9C3F (PDB model of AmpG). The source data underlying Fig. 6 and Supplementary Fig. 2-3 is present in the Source Data file. The accession codes of the additional structures used for analysis are 6GV1 (MdfA); 6T1Z (LmrP); 3WDO (YajR); 7LO8 (NorA); and 3O7P (FucP). Source data is provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54219-9.
References
- 1.Aslam, B. et al. Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist.11, 1645–1658 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush, K. & Bradford, P. A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med.6, a025247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darby, E. M. et al. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 1–16 10.1038/s41579-022-00820-y (2022). [DOI] [PubMed]
- 4.Mehrad, B., Clark, N. M., Zhanel, G. G. & Lynch, J. P. Antimicrobial Resistance in Hospital-Acquired Gram-Negative Bacterial Infections. CHEST147, 1413–1421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamma, P. D., Doi, Y., Bonomo, R. A., Johnson, J. K. & Simner, P. J. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis. Publ. Infect. Dis. Soc. Am.69, 1446–1455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torrens, G. et al. Regulation of AmpC-Driven β-Lactam Resistance in Pseudomonas aeruginosa: Different Pathways, Different Signaling. mSystems4, e00524–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza, R. et al. Role of AmpG in the resistance to β-lactam agents, including cephalosporins and carbapenems: candidate for a novel antimicrobial target. Ann. Clin. Microbiol. Antimicrob.20, 45 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barceló, I. M. et al. Filling knowledge gaps related to AmpC-dependent β-lactam resistance in Enterobacter cloacae. Sci. Rep.14, 189 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, J. F. & Mobashery, S. The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg. Chem.56, 41–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. W., Fisher, J. F. & Mobashery, S. Bacterial cell-wall recycling. Ann. N. Y. Acad. Sci.1277, 54–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chahboune, A., Decaffmeyer, M., Brasseur, R. & Joris, B. Membrane Topology Of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob. Agents Chemother.49, 1145–1149 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamorano, L. et al. AmpG inactivation restores susceptibility of pan-beta-lactam-resistant Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother.55, 1990–1996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallik, D., Pal, S. & Ghosh, A. S. Involvement of AmpG in mediating a dynamic relationship between serine beta-lactamase induction and biofilm-forming ability of Escherichia coli. FEMS Microbiol. Lett.365, fny065 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Cheng, Q. & Park, J. T. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol.184, 6434–6436 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox, B. G. & Blommel, P. G. Autoinduction of Protein Expression. Curr. Protoc. Protein Sci. CHAPTER5, Unit-5.23 (2009). [DOI] [PMC free article] [PubMed]
- 16.Bayburt, T. H. & Sligar, S. G. Membrane protein assembly into Nanodiscs. FEBS Lett.584, 1721–1727 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure20, 967–976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee, S. et al. Synthetic antibodies against BRIL as universal fiducial marks for single−particle cryoEM structure determination of membrane proteins. Nat. Commun.11, 1598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen, H., Himpkamp, P. & Peters, T. Bausteine von Oligosacchariden, LXIX. Synthese von 1,6-Anhydromuramylpeptiden. Liebigs Ann. Chem. 1986, 664–674 (1986).
- 20.Lee, M. et al. From genome to proteome to elucidation of reactions for all eleven known lytic transglycosylases from Pseudomonas aeruginosa. Angew. Chem. Int. Ed.56, 2735–2739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki, A. et al. Synthesis of diaminopimelic acid containing peptidoglycan fragments and tracheal cytotoxin (TCT) and investigation of their biological functions. Chem. – Eur. J.14, 10318–10330 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Perley-Robertson, G. E. et al. A fluorescent transport assay enables studying AmpG permeases involved in peptidoglycan recycling and antibiotic resistance. ACS Chem. Biol.11, 2626–2635 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Soni, A. S., Vacariu, C. M., Chen, J. Y. & Tanner, M. E. Synthesis of a meso-Oxa-diaminopimelic acid containing peptidoglycan pentapeptide and coupling to the GlcNAc-anhydro-MurNAc disaccharide. Org. Lett.22, 2313–2317 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Errington, J. Cell wall-deficient, L-form bacteria in the 21st century: a personal perspective. Biochem. Soc. Trans.45, 287–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vacariu, C. M. & Tanner, M. E. Recent advances in the synthesis and biological applications of peptidoglycan fragments. Chem. – Eur. J.28, e202200788 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Liu, H., Pattabiraman, V. R. & Vederas, J. C. Stereoselective syntheses of 4-Oxa diaminopimelic acid and its protected derivatives via aziridine ring opening. Org. Lett.9, 4211–4214 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Soni, A. S., Lin, C. S.-H., Murphy, M. E. P. & Tanner, M. E. Peptides containing meso-Oxa-Diaminopimelic acid as substrates for the cell-shape-determining proteases Csd6 and Pgp2. ChemBioChem20, 1591–1598 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Drew, D., North, R. A., Nagarathinam, K. & Tanabe, M. Structures and general transport mechanisms by the major facilitator superfamily (MFS). Chem. Rev.121, 5289–5335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson, H. & von Heijne, G. Membrane protein topology: effects of delta mu H+ on the translocation of charged residues explain the ‘positive inside’ rule. EMBO J.13, 2267–2272 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dang, S. et al. Structure of a fucose transporter in an outward-open conformation. Nature467, 734–738 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Killer, M., Wald, J., Pieprzyk, J., Marlovits, T. C. & Löw, C. Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes. Sci. Adv.7, eabk3259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan, N. FucP, the L-Fucose-H+ Membrane Transport Protein and Related Transporters. In Encyclopedia of Biophysics (ed. Roberts, G. C. K.) 854–859 (Springer, Berlin, Heidelberg, 2013). 10.1007/978-3-642-16712-6_686.
- 33.Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res.50, W210–W215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, X. C., Zhao, Y., Heng, J. & Jiang, D. Energy coupling mechanisms of MFS transporters. Protein Sci. Publ. Protein Soc.24, 1560–1579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 1–3 10.1038/s41586-024-07487-w (2024). [DOI] [PMC free article] [PubMed]
- 36.Hopf, T. A. et al. The EVcouplings Python framework for coevolutionary sequence analysis. Bioinformatics35, 1582–1584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agirre, J. et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr. Sect. Struct. Biol.79, 449–461 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, B. & Richards, F. M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol.55, 379–400 (1971). [DOI] [PubMed] [Google Scholar]
- 39.Bajaj, P., Singh, N. S. & Virdi, J. S. Escherichia coli β-Lactamases: What Really Matters. Front. Microbiol. 7 (2016). [DOI] [PMC free article] [PubMed]
- 40.Ropy, A. et al. Role of pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, β-lactam resistance, and peptidoglycan structure. Antimicrob. Agents Chemother.59, 3925–3934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raetz, C. R. & Dowhan, W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem.265, 1235–1238 (1990). [PubMed] [Google Scholar]
- 42.Jiang, D. et al. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc. Natl Acad. Sci.110, 14664–14669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, J. et al. Proton-coupled transport mechanism of the efflux pump NorA. Nat. Commun.15, 4494 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masureel, M. et al. Protonation drives the conformational switch in the multidrug transporter LmrP. Nat. Chem. Biol.10, 149–155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauffer, M. et al. Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor. Commun. Chem.5, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer, L. & James, M. N. G. Carboxyl–carboxylate interactions in proteins. Nature295, 79–80 (1982). [DOI] [PubMed] [Google Scholar]
- 47.Solcan, N. et al. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J.31, 3411–3421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isom, D. G., Castañeda, C. A., Cannon, B. R. & García-Moreno, E. B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl Acad. Sci.108, 5260–5265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khavrutskii, I. V., Compton, J. R., Jurkouich, K. M. & Legler, P. M. Paired carboxylic acids in enzymes and their role in selective substrate binding, catalysis, and unusually shifted pKa values. Biochemistry58, 5351–5365 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Roth, A. & Govaerts, C. LmrP from Lactoccoccus lactis: a tractable model to understand secondary multidrug transport in MFS. Res. Microbiol.169, 468–477 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Wohlfahrt, G. Analysis of pH-dependent elements in proteins: geometry and properties of pairs of hydrogen-bonded carboxylic acid side-chains. Proteins Struct. Funct. Bioinforma.58, 396–406 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Hariharan, P. et al. Structural and functional characterization of protein–lipid interactions of the Salmonella typhimurium melibiose transporter MelB. BMC Biol.16, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dowhan, W., Vitrac, H. & Bogdanov, M. Lipid-assisted membrane protein folding and topogenesis. Protein J.38, 274–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martens, C. et al. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol.23, 744–751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vitrac, H., Mallampalli, V. K. P. S., Bogdanov, M. & Dowhan, W. The lipid-dependent structure and function of LacY can be recapitulated and analyzed in phospholipid-containing detergent micelles. Sci. Rep.9, 11338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, W., Campbell, H. A., King, S. C. & Dowhan, W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J. Biol. Chem.280, 26032–26038 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van den Ent, F. & Löwe, J. RF cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods67, 67–74 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Coombs, D. H. & Watts, N. R. M. Generating sucrose gradients in three minutes by tilted tube rotation. Anal. Biochem.148, 254–259 (1985). [DOI] [PubMed] [Google Scholar]
- 60.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr.66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eberhardt, J., Santos-Martins, D., Tillack, A. F. & Forli, S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J. Chem. Inf. Model.61, 3891–3898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. Struct. Biol.75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res.44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci.32, e4792 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. (2015).
- 67.Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res.14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yaeger, L. N. et al. Metabolic connections between folate and peptidoglycan pathways in Pseudomonas aeruginosa inform rational design of a dual-action inhibitor. 2023.11.22.568328 Preprint at 10.1101/2023.11.22.568328 (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support this study are available from the corresponding authors upon request. The cryo-EM map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-45167 (cryo-EM map of AmpG). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession code 9C3F (PDB model of AmpG). The source data underlying Fig. 6 and Supplementary Fig. 2-3 is present in the Source Data file. The accession codes of the additional structures used for analysis are 6GV1 (MdfA); 6T1Z (LmrP); 3WDO (YajR); 7LO8 (NorA); and 3O7P (FucP). Source data is provided with this paper.