Summary
Research on brain expression quantitative trait loci (eQTLs) has illuminated the genetic underpinnings of schizophrenia (SCZ). Yet most of these studies have been centered on European populations, leading to a constrained understanding of population diversities and disease risks. To address this gap, we examined genotype and RNA-seq data from African Americans (AA, n = 158), Europeans (EUR, n = 408), and East Asians (EAS, n = 217). When comparing eQTLs between EUR and non-EUR populations, we observed concordant patterns of genetic regulatory effect, particularly in terms of the effect sizes of the eQTLs. However, 343,737 cis-eQTLs linked to 1,276 genes and 198,769 SNPs were found to be specific to non-EUR populations. Over 90% of observed population differences in eQTLs could be traced back to differences in allele frequency. Furthermore, 35% of these eQTLs were notably rare in the EUR population. Integrating brain eQTLs with SCZ signals from diverse populations, we observed a higher disease heritability enrichment of brain eQTLs in matched populations compared to mismatched ones. Prioritization analysis identified five risk genes (SFXN2, VPS37B, DENR, FTCDNL1, and NT5DC2) and three potential regulatory variants in known risk genes (CNNM2, MTRFR, and MPHOSPH9) that were missed in the EUR dataset. Our findings underscore that increasing genetic ancestral diversity is more efficient for power improvement than merely increasing the sample size within single-ancestry eQTLs datasets. Such a strategy will not only improve our understanding of the biological underpinnings of population structures but also pave the way for the identification of risk genes in SCZ.
Keywords: eQTLs, schizophrenia, GWAS signals, population diversity
Examining brain eQTLs across African American, European, and East Asian populations reveals significant ancestry-specific genetic variants linked to schizophrenia. The study highlights the importance of genetic diversity in discovering risk genes and improving disease understanding, suggesting that broader ancestral representation enhances the power of genetic analyses.
Introduction
Genome-wide association studies (GWASs) have identified 287 risk loci associated with schizophrenia (SCZ).1 Yet, the underlying mechanisms of these loci in disease development and progression remain poorly understood. Primarily, over 80% of GWAS risk loci reside in non-coding regions, devoid of protein-coding sequences, making it challenging to attribute them to specific genes. Moreover, predicting the regulatory effect of these loci proves challenging due to their tendency for gene-specific and tissue-specific effects. One effective strategy for gaining insights into their functions involves the integration of SCZ GWAS signals with expression quantitative trait loci (eQTLs), utilizing genotype and expression data from postmortem brains. These brain eQTLs establish crucial links between risk genomic regions and gene expression levels, prioritizing potential disease risk genes through methods such as colocalization and transcriptome-wide association studies (TWASs).
Past brain eQTL studies primarily focused on European (EUR) ancestry.2,3,4,5,6 Global population diversity has not been adequately represented. Cross-population studies have shown that these European ancestry-based models do not effectively predict gene expression in other ancestral groups.7 This limitation weakens the power to detect TWAS associations in genetically diverse samples. While multi-ancestry eQTL meta-analyses in the human brain improve statistical power in uncovering risk loci shared across populations, key genetic variants regulating expression in specific underrepresented populations remain largely uncharted. The benefits of having brain eQTLs in diverse populations have not been thoroughly documented. Identifying eQTLs specific to biomedically underrepresented groups including African Americans (AA) and East Asians (EAS) can better understand the genetic contributions to disease susceptibilities and outcomes in these populations.7 These populations have unique genetic variants and linkage disequilibrium (LD) patterns. Additionally, previous studies have shown that combining eQTLs from different ancestries can enable fine-mapping of causal variants and uncover potential mechanisms of brain disorders.8,9 Thus, the question of how to effectively leverage difference to uncover potential mechanisms of brain disorders is a significant topic in the field.
To enhance the diversity in brain eQTL mapping and improve the interpretation of SCZ GWASs across populations, we performed brain eQTL mapping in three major ancestries. Our data pool comprised genotype and RNA-seq data of AA (n = 158) and EUR (n = 408) from the PsychENCODE Consortium and EAS (n = 217) from the Chinese Human Brain Bank (Table S1). We juxtaposed non-EUR results against EUR to systematically examine differences and similarities in the brain eQTLs (Figure 1). Further, we investigated the contributing factors for eQTL differences across populations. By applying diverse population brain eQTLs to TWAS and colocalization analysis of SCZ GWAS, we identified risk genes of schizophrenia. Lastly, we identified likely causal variants by multi-ancestry fine-mapping. The two key questions we sought to answer are (1) what drives the brain eQTL differences across populations? and (2) what do we gain by studying brain eQTLs in diverse populations?
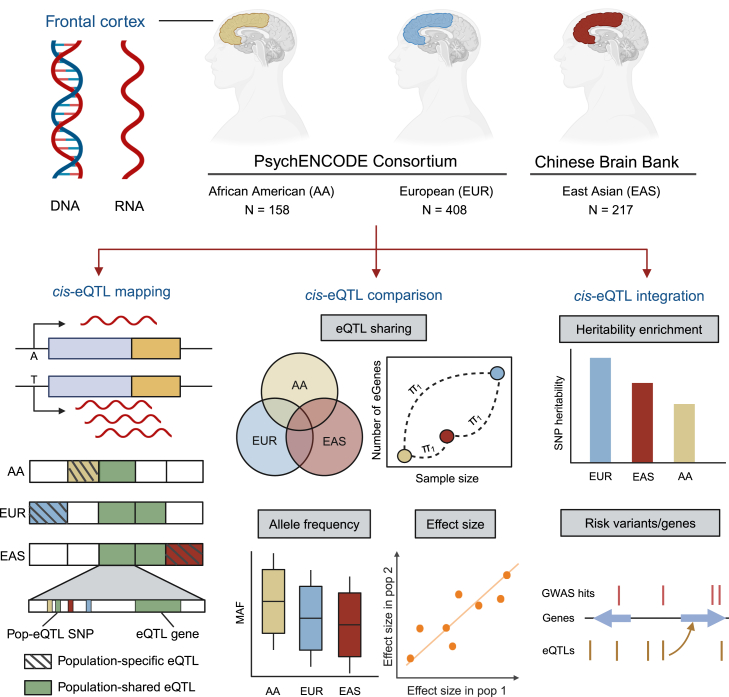
Figure 1.
Study design
We examined genotype and RNA-seq data from individuals from African American (AA), European (EUR), and East Asian (EAS) populations to identify expression quantitative trait loci (eQTLs) specific to non-European populations and their role in schizophrenia risk. The figure was created using Biorender.com.
Subjects and methods
Sample collection and sequencing
217 prefrontal cortical samples of Han Chinese ancestry were collected from the National Human Brain Bank for Development and Function10,11; the samples were handled according to the standardized operational protocol of the China Human Brain Banking Consortium, under the approval of the Institutional Review Board of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences, Beijing, China (Approval Number: 009-2014, 031-2017, 2022125). The Ethics Committee of Central South University gave ethical approval for this work (2015031007).
These samples were then sequenced following the BGISEQ-500 protocol outsourced to BGI. 1 μg genomic DNA was randomly fragmented by Covaris, the fragmented DNA was selected by Agencourt AMPure XP-Medium kit to an average size of 200–400 bp, followed by adapter ligation and PCR amplification, and the products were recovered by the AxyPrep Mag PCR clean up kit. The double-stranded PCR products were heat-denatured and circularized by the splint oligo sequence. The single-strand circle DNA (ssCir DNA) was formatted as the final library and qualified by QC. Sequencing was performed on BGISEQ-500 platform with an average depth of 10×.
Total RNA was extracted from the brain tissue using Trizol (Invitrogen) according to manufacturers’ instructions. Then, total RNA was qualified and quantified using a Nano Drop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific). Ribo-zero method was used to remove the rRNA. Purified mRNA was fragmented into small pieces with fragment buffer at an appropriate temperature. The cDNAs were purified by magnetic beads. After purification, A-Tailing Mix and RNA Index Adapters were added by incubating to carry out end repair. The cDNA fragments with adapters were amplified by PCR, and the products were purified by Ampure XP Beads. The library was validated on the Agilent Technologies 2100 bioanalyzer for quality control. The final library was amplified with phi29 (Thermo Fisher Scientific) to make DNA nanoball (DNB), DNBs were loaded into the patterned nanoarray, and single end 50 base reads were generated on BGISEQ-500 platform.
Data quality control
Raw sequencing reads were filtered to get clean reads by using SOAPnuke (v.1.5.6),12 and FastQC13 was used to evaluate the quality of sequencing data via several metrics, including sequence quality per base, sequence duplication levels, and quality score distribution for each sample. The average quality score for overall DNA and RNA sequences was above 30, indicating that a high percentage of the sequences had high quality.
Variant identification
Clean DNA sequencing reads were mapped to the human reference genome hg19 (GRCh37) using BWA-MEM algorithm (BWA v.0.7.128).14 Ambiguously mapped reads (MAPQ < 10) and duplicated reads were removed using SAMtools v.1.2915 and PicardTools v.1.1, respectively. Genomic variants were called following the Genome Analysis Toolkit software (GATK v.3.4.4.6) best practices. In total, 29 million single-nucleotide variants and small insertions/deletions were identified in the EAS population.
Population validation, imputation, and filtering
We used PLINK to infer the genomic ancestry of each sample in this study by combining our genotype data and the genotype data from the 1000 Genomes Project16; no sample was excluded. Using Michigan Imputation Server,17 EAS genotypes were imputed into the 1000 Genomes Project phase 3 EAS reference panel by chromosome and subsequently merged. Imputed genotypes were filtered for LD R2 < 0.3, Hardy-Weinberg equilibrium p value < 10e−6, and Minor Allele Frequency (MAF) < 0.05, resulting in ∼6 million autosomal single-nucleotide polymorphisms (SNPs).
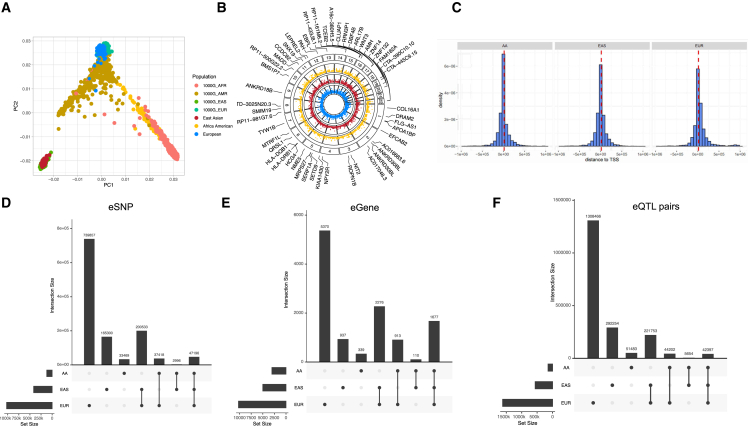
For AA population, genotypes were imputed into the 1000 Genomes Project phase 3 AA reference panel by chromosome and subsequently merged. To further confirm the ancestry of the African American samples, all AA samples were evaluated for their ancestry with three broad population groups with PC1 ≥ 25% African (AFR) and <25% American (AMR), <25% EAS, <25% South Asian (SAS); clustering of individuals in each broad population group with the 1000 Genomes Project reference populations are shown in Figure 2A.
Figure 2.
Identification and characterization of eQTLs
(A) PCA plot showing the population structure of individuals in our study as well as the 1000 Genomes Project. AFR, African; AMR, American; EAS, East Asian; EUR, European.
(B) Circos manhattan plot of significant eQTL genes among the three populations with highlighted top 50 fine-mapped eGenes. Each layer of the plot represents results from an eQTL analysis, with results from the same ancestry grouped by color. The blue panel represents EUR, red panel represents EAS, and the yellow panel represents AA. Significant eQTL are plotted as points.
(C) Distance distribution between eSNP to TSS of eGenes.
(D–F) Upset plot showed overlap among the significantly associated (D) eSNPs, (E) eGenes, as well as (F) eQTL pairs between populations.
Sex check and sample swap identification
The sex of each sample was inferred with SNPs using PLINK. In the EAS cohort, two samples were identified as sex-mismatched and were subsequently removed in downstream analysis. Quality control was performed on genotypes using sample Binary Alignment Map (BAM) files to detect any sample identity swaps between the RNA and DNA experiments. The QTLtools match function18 confirmed that all samples were appropriately matched.
Gene expression quantification and quality control
The RNA-sequencing reads were mapped using STAR (2.4.2a)19 and the genes and transcripts quantification was performed using RSEM (1.3.0).20 Raw read counts were log-transformed using R package VOOM,21 thereafter filtering those with log2(counts per million reads, CPM) < 0 in more than 75% of the samples. Mitochondrial DNA and X and Y chromosome-derived transcripts were excluded. Samples with a Z score (measured for inter-sample connectivity) less than −3 were also discarded. Finally, quantile normalization was utilized to equalize distributions across samples.
Covariate selection
To measure technical covariates, quality control metrics were collected using STAR, PicardTools v.1.139, and RNASeQC. Principal components of the metrics data were calculated and included as SeqPCs for covariate selection. Hidden covariates were measured using probabilistic estimation of expression residuals (PEER)22 and found to be significantly correlated with technical and biological covariates such as experimental batch, RNA Integrity Number (RIN), sex, and age of death. Based on the Bayesian information criterion (BIC) score, redundant covariates were removed to avoid overfitting. A forward and backward selection procedure was followed. The covariate with the higher BIC score was selected for subsequent QTL mapping. To determine the optimal number of PEER factors for QTL discovery, we conducted QTL mapping using a range of PEER factor counts (5, 10, 15, 20, 25, 30, 35, 40, 45, and 50) as covariates. We then identified the minimum number of PEER factors that maximized the number of detected eQTLs.
cis-eQTL mapping
cis-eQTL mapping was performed using QTLtools, accounting for 20 PEER factors, with a defined cis window spanning one megabase upstream and downstream of the gene/intron cluster body. To detect all available QTLs, QTLtools was conducted in nominal pass mode. To identify the best nominal associated SNP per phenotype, QTLtools was executed in the permutation pass mode. Additionally, to identify SNPs with independent effects on regulating gene expression, QTLtools was run in the conditional pass mode. These empirical p values were subsequently corrected for multiple testing across genes using Storey’s q value method. SNPs with q values < 0.05 is classified as significant QTLs.
In detail, we first regress out the provided covariates from the phenotype data, followed by running the linear regression between the phenotype residuals and the genotype. The residuals after the covariate correction are rank normal transformed. It incorporates an efficient permutation scheme to control for differential multiple testing burden of each phenotype. We ran (1) a nominal pass listing all genotype-phenotype associations below a certain threshold, (2) a permutation pass to empirically characterize the null distribution of associations for each phenotype separately, thus adjusting the nominal p value of the best association for a phenotype, and (3) a conditional analysis pass to discover multiple proximal QTLs with independent effects on a phenotype. The conditional analysis pass first uses permutations to derive a nominal p value threshold per phenotype that varies and reflects the number of independent tests per cis-window. Then, it uses a forward-backward stepwise regression to learn the number of independent signals per phenotype, determine the best candidate variant per signal, and assign all significant hits to the independent signal they relate to.
To address potential sample size disparities that could impact the results, the EUR data were randomly sampled with various sample size (150, 200, 250, 300, 350, and 400) and applied the same analytical pipeline while exploring the relationship between sample size and number of QTLs.
eQTL fine-mapping
Standard fixed-effects meta-analysis were used to combine all data into a single regression model by METAL.23 The meta-analysis assumes a fixed-effects size, as well as constant error variance, across all data. The significance threshold of 1e−6 in meta-analysis were generated by Bonferroni correction. The SNP-gene pairs with a significant p value were collected for the eQTL fine-mapping.
The initial step of fine-mapping involved using the in-sample LD of the three populations. We extracted common variants with MAF > 5% from each group and used PLINK to determine the LD regions of these common variants for each population. To eliminate strand flipping and alignment issues, multi-allelic variations and indels were removed. Next, SuSiEx was applied to merge the eQTLs summary statistics from the three groups. Credible set is defined as a set of putative causal variants. A credible set was discarded if it lacked genetic variants reaching genome-wide significance (p < 1e−6) in either the population-specific eQTLs or cross-population meta-eQTLs. By considering prior knowledge and the observed data, this method provides a posterior probability (PIP) for each variant being the causal one in the associated region. Variants with high PIPs are then considered strong candidates for functional follow-up studies.
Functional enrichment
Genomic Regulatory Elements and GWAS Overlap algoRithm (GREGOR)24 was performed to test the functional enrichment of eQTL. GREGOR calculated the enrichment value based on the observed and expected overlap within each annotation. To conduct our analysis, the 15-state ChromHMM model BED (Browser Extensible Data) files from the Roadmap Epigenetics Project25 and 78 consensus transcription factor and DNA-protein binding site BED files existing in multiple cells were downloaded. Fifty binding proteins showed cortical brain expression in EAS and AA populations data.26
The fraction of shared eQTLs between non-EUR and EUR populations
Sharing rate was assessed based on significant eQTLs in the discovery dataset by estimating the proportion of true associations (π1) on the distribution of corresponding p values of the overlapping eQTLs in the replication dataset.27
FST and MAF analysis
Fixation index (FST) was estimated using vcftools following the Weir and Cockerham approach for each eSNP.28 The population-divergent SNPs were defined as those with FST ≥ 0.05 and population-shared SNPs as those with FST < 0.05. To generate the list of population-specific QTLs and population-shared QTLs, we collected the overlap of eQTLs from the pairwise comparisons of the list of AA eQTLs, EAS eQTLs, and EUR eQTLs. Finally, Fisher’s exact test was performed between population-specific QTLs and population-shared QTLs to test the contribution of MAF in the QTL comparison.
Variance explained
Variance explained, which combines the effect size (beta) and frequency of the allele (f), can be considered an approximate measure of a causal variant’s importance within a population. Variance is approximated using the formula 2f(1 – f)log(beta)2/(π2/3).29 Although these variants often exhibit similar odds ratios across populations, their allele frequencies may differ. By considering both the effect size (OR) and the frequency of the risk allele (f), the variance explained offers a valuable approximation of a causal variant’s significance within a given population.
Power estimation
We used R to calculate the sample size needed to achieve a given power level in a chi-square test, based on an assumed effect size and a significance threshold. It starts by setting initial values for power, effect size, and p value threshold. Then, it computes the critical chi-square statistic required to meet the power level. A function, calculate_ncp, is defined to calculate the non-centrality parameter from the p value and degrees of freedom, adjusting for the critical chi-square statistic. Subsequently, the non-centrality parameter is computed for the given power and p value threshold. Another function, af_n_relation, is created to determine the relationship between allele frequency and sample size, incorporating the effect size and the non-centrality parameter. Finally, the code iteratively solves for the sample size corresponding to a range of allele frequencies, thus enabling the determination of the necessary sample size for different allele frequencies to maintain the specified power level in the chi-square test.
Partitioned LDSR
Partitioned LD score regression v.1.0.130 was used to measure the enrichment of GWAS summary statistics in each functional category by accounting for LD. Brain QTL annotations were created by eSNP, mapped to the corresponding 1000 Genome reference panel. LD scores were calculated for each SNP in the QTL annotation using an LD window of 1 cM in 1000 Genomes European Phase 3 and 1000 Genomes Asian Phase 3 separately. Enrichment for each annotation was calculated by the proportion of heritability explained by each annotation divided by the proportion of SNPs in the genome falling in that annotation category. We then applied Welch modified two-sample t test on enrichment values generated from QTLs in the two populations.
Colocalization
Conditional association was used to test for evidence of colocalization. This method compares the p value of association for the lead SNP of an eQTL before and after conditioning on the GWAS hit. The equation for the regulatory trait concordance (RTC) score is as follows: RTC = (NSNPs in an LD block/RankGWAS_SNP)/NSNPs in an LD block. The rank denoted the number of SNPs, which when used to correct the expression data, has a higher impact on the QTL than the GWAS SNPs. RTC values close to 1.0 indicated causal regulatory effects. A threshold of 0.9 was used to select causal regulatory elements.
We also applied a Bayesian co-localization approach to identify GWAS signals that could exhibit the same genetic effect with GWAS and eQTLs using coloc R (v.5.1.0) package.31 We used the default coloc priors for Bayesian co-localization analysis, in which the prior was assigned 10e−6 for representing the probability that the SNP was associated with eQTL. For each GWAS trait, we extracted the GWAS SNPs with a p value < 5e−8 and located at least 1 Mb away from more significant variants. The co-localized signals were searched within a surrounding region of 100 kb of GWAS SNPs. Five posterior probabilities (PPs) were calculated for the colocalization analysis using all variants in the region of interest. PP0 represents the null model of no association. PP1 and PP2 represent the probability that causal genetic variants are associated with either disease signals or eQTLs alone. PP3 represents the probability that the genetic effects of disease signals and eQTLs are independent, and PP4 represents the probability that disease signals and eQTLs share causal SNPs. The genes were defined as co-localization events if PP4 > 0.8. Region visualization plots were constructed using LocusZoom.32
Colocalization of fine-mapped variations from complex traits and cis-eQTLs correlations were performed. Based on complex trait and cis-eQTLs fine-mapping data, a posterior inclusion probability of colocalization for a variant was calculated as a product of PIP for GWASs and PIP for the cis-eQTLs (PIPcoloc = PIPGWAS ∗ PIPcis-eQTLs).
Summary-data-based mendelian randomization
SMR33 was applied on SCZ GWAS summary data to prioritize candidate genes. Significant QTLs identified in the previous analysis (FDR < 0.05) were combined with filtered GWAS summary data (p < 5e−8) to perform the SMR test. In general, we used the default parameters suggested by the developers of the SMR software. These included the application of heterogeneity independent instruments (HEIDI) testing, filtering out hits that arose from significant linkage with pleiotropically associated variants (LD cutoff of p = 0.05 in the HEIDI test, as suggested by SMR). Genes with an empirical p that passed Bonferroni correction in the SMR test and a p > 0.05 in the HEIDI test were considered as risk genes.
Prioritizing genes underlying GWAS hits using MetaXcan
In this research, we initially developed gene expression prediction models for distinct populations using MetaXcan software.34 Following this, we integrated these models with GWAS summary statistics specifically focused on schizophrenia. This integration aimed to generate gene-level z-scores representing the association of the genetically determined expression for a gene from its prediction model with the phenotype. TWAS enabled us to compute p values and subsequently prioritize genes in relation to their association with schizophrenia risk.
Results
To capture brain eQTLs across diverse populations, we utilized high-density genotype data alongside high-throughput RNA sequencing from prefrontal cortices. We obtained AA (n = 158) and EUR (n = 408) data from the BrainGVEx project of the PsychENCODE Consortium (https://www.synapse.org/Synapse:syn4921369). We generated EAS (n = 217) data from the Chinese Human Brain Bank (Table S2; Figure 1). Following rigorous quality checks and preprocessing (Figures S1 and S2), we compiled expression data for 18,939 genes and genotype data at 6.4 million autosomal SNPs across the three groups. Aligning the samples with the 1000 Genomes Project reference populations, principal-component analysis (PCA) confirmed the ancestry origins of donors (Figure 2A). We ensured sample identity consistency by comparing the genotypes from the DNA and RNA samples. See subjects and methods for additional details.
Characterizing the cis-acting eQTLs in European, East Asian, and African American populations
We separately conducted cis-eQTLs mapping in the EUR, EAS, and AA samples using a 5% empirical gene-level false discovery rate (FDR) threshold. This yielded 1,966,209 significant eQTL signals covering 11,622 genes (eGenes) and 1,226,769 SNPs (eSNPs) across the populations (see Figure 2B; Table S3). Specifically, we identified 10,236 eGenes for EUR, 5,000 eGenes for EAS, and 3,039 eGenes for AA. To identify credible SNP sets harboring plausible causal variants in cis-eQTLs, we applied a fine-mapping method named SuSiE35 to each population’s eQTL results. The results showed 966 credible SNP sets for 757 eGenes in the EUR cohort, 826 sets for 726 eGenes in EAS, and 847 sets for 746 eGenes in AA (Tables S4–S6).
To investigate the genomic features of these cis-eQTLs, we evaluated the SNP distributions and locations relative to various functional regions. 20% of cis-eQTLs in both EUR and non-EUR populations were located within 10 kb of transcription start site (TSS) regions (Figure 2C). According to the chromatin states predicted by GREGOR36 for prefrontal cortical tissue, eSNPs from the non-EUR populations were significantly enriched in TSSs, promoters, and transcribed regulatory promoters or enhancers (Figure S3, pBonferroni < 0.05), identical to the observation in the EUR data. Moreover, using transcription factor binding site (TFBS) annotation for 51 TFs, 46 and 49 TFs were significantly enriched with cis-eQTLs in the AA and EAS populations, respectively (pBonferroni < 0.05). All these TFBS were also significantly enriched with cis-eQTLs in the EUR population.
To maximize the power of our population-based datasets, we employed METAL to amalgamate the cis-eQTL data from all three populations. We then used SuSiEx16 to identify likely causal variants regulating expression by incorporating the LD reference data from different populations. In total, SuSiEx identified 2,121 credible SNP sets for 1,801 eGenes in the 3-population combined data. Further details from the meta-analysis and fine-mapping results can be found in Table S7.
Population-shared eQTLs showed similar regulatory effect across populations
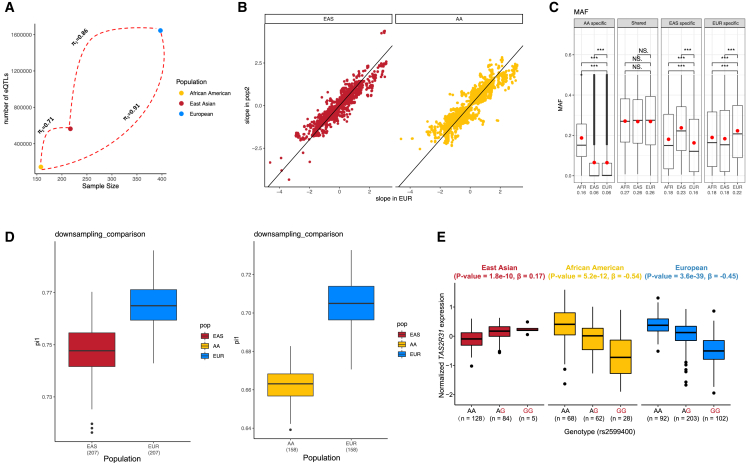
To evaluate the effect sizes across populations, we conducted a correlation test of effect size values between the EUR and non-EUR populations. The effect sizes of the shared eQTLs between the non-EUR and EUR were highly concordant (0.910 for eQTLs comparing EAS to EUR and 0.944 for AA to EUR; Figure 3B). To assess the robustness of the concordant effect size, we examined eQTL slopes in the different populations. We looked at eQTLs obtained from the nominal, permutation, and conditional tests, separately. eQTLs with smaller p values or larger effect sizes showed greater consistency across populations (Figure S4). Considering that sample size and heterogeneity may influence the results, we randomly down-sampled the EUR data to match the size of the non-EUR data. The results were similar to the results comparing all samples, showing highly concordant effect sizes across populations (see Figures 3D and S4). In addition, we compared the slope of the down-sampled EUR data with GTEx data (also of EUR). We randomly sub-sampled 100 times and obtained a distribution of correlation values (R2). The mean R2 was 0.94, which was not significantly different from the correlation between the EUR and non-EUR population. We therefore concluded that the effect sizes of eQTLs in diverse populations were mostly stable across human populations.
Figure 3.
Analysis of regulatory patterns
(A) Relationship between sample size and the # of detected eQTLs. The numbers on the line represent π1 between two populations.
(B) Effect sizes for common (MAF > 1%) sentinel cis-eQTLs across EAS and AA compared to EUR populations.
(C) Comparison of MAF between population-shared and non-EUR specific eSNPs. The mean MAF has been labeled under the x axis. "NS" stands for not significant, and "∗∗∗" indicates a p value < 0.01.
(D) Down-sampling results to estimate π1 between non-European and European eQTLs.
(E) Example of opposite effect eQTL TAS2R31-chr12:11282501A>G. The x axis represented the genotype. The y axis represented the normalized expression of TAS2R31.
We evaluated the replicated rate (π1), which gauges the true positive rate for the eQTLs identified in the non-EUR populations that were also associated in the EUR population. The replicated rate was π1(EAS-EUR) = 0.86 and π1(AA-EUR) = 0.91 (see Figure 3A; Table S8). The π1 for the non-EUR populations in EUR was slightly but significantly lower than the π1 between two EUR cohorts, as represented by the GTEx cis-eQTL data (prefrontal cortex) in our EUR eQTL data (π1(GTEx-EUR) = 0.86, p value = 0.023). To ensure a fair comparison of the replication rate of detected cis-eQTLs in non-EUR data, we adjusted the EUR data to reflect the smaller sample size of the non-EUR data. This adjustment enabled us to determine how many non-EUR cis-eQTLs were confirmed in the adjusted EUR dataset. The adjusted results revealed a concordant trend: the replicated rate between different populations was still slightly lower than that within the same population assuming the same sample size (EUR-nonEUR average π1 = 0.68, EURadjusted-EUR average π1 = 0.72, p value = 0.037) (see Figure 3D).
Population differences in brain cis-eQTLs are mainly caused by differences in SNP allele frequency while differences in effect size are small and uncertain
Here we defined those eQTLs that were exclusively observed in a single population as population-specific eQTLs. Upon analyzing the cis-eQTLs overlapping between populations, we identified 343,737 cis-eQTLs that were exclusively observed in the non-EUR populations, as detailed in Table S3. This number represents approximately 17% of all eQTL pairs. These eQTLs involved 1,276 genes (about 10% of all eGenes) and 198,769 SNPs (around 16% of all eSNPs, Figures 2D–2F). Specifically, there were 292,254 cis-eQTLs involving 165,300 eSNPs and 937 eGenes that were observed unique in the EAS population and 51,483 cis-eQTLs involving 33,469 eSNPs and 339 eGenes that were observed unique in the AA population. For the 343,737 non-overlapping eQTLs, 186,459 eSNPs (156,589 in EAS population and 31,401 in AA population) are not in LD regions (LD R2 > 0.8) with any eSNPs in the EUR population. Importantly, our comparison with the MetaBrain eQTL results8 revealed that 483 population-specific eGenes, involving 130,117 eQTLs, were still absent in the EUR population.
To further characterize these non-EUR-specific eQTLs, we analyzed the variance, taking into account both the eQTL slope (effect size) and differences in allele frequency between populations. We found that more than 90% of the population differences in variance were attributable to differences in allele frequency. Moreover, to delve deeper into the distinctive characteristics of the eQTLs exclusive to the non-EUR groups, we leveraged two statistics, the FST and the MAF, retrieved from the 1000 Genomes Selection Browser.37 A high FST value indicates that the measured locus has diverged over time in the populations. As expected, eSNPs detected specific to the EAS or AA population displayed a significantly elevated FST when juxtaposed against eSNPs shared across populations (Figure S5, mean FSTEAS-sp-eSNP = 0.13; mean FSTEUR-sp-eSNP = 0.11; mean FSTAA-sp-eSNP = 0.14; mean FSTcommon-eSNP = 0.1; Wilcoxon test p < 2.2e−16). Meanwhile, the non-EUR-specific eSNPs showed higher MAF values in their respective source populations (Figure 3C, Wilcoxon test p < 2.2e−16) than in EUR. Of the 343,737 eQTLs absent in the EUR data, 309,363 were likely due to inadequate statistical power because they have smaller MAF in the EUR than non-EUR population.
For the remaining eQTLs for which population differences could not be explained by differences in MAF, a test for differences in eQTL slopes (effect sizes) was also conducted between the EUR and non-EUR populations. The Z score of each independent eQTL from conditional analysis was calculated based on effect size and its standard deviation. Here the null hypothesis was that the difference in eQTL effect size between the populations equals zero. No eQTL pairs detected by conditional analysis could reject the null hypothesis. We then investigated if any eQTLs exhibited opposite effect directions across populations. None of the independent eQTLs from the conditional analysis displayed such effects. We relaxed our eQTL threshold using a nominal p value < 0.05. 534 eQTLs involving eighteen genes exhibited opposing eQTL effects between the EUR and non-EUR populations. For example, the bitter taste receptor gene TAS2R31 showed opposite directions of eQTLs in EAS and EUR (Figure 3E), which could be replicated using the blood eQTLs from a previous study.7,38
In conclusion, the variance in population differences can be largely attributed to differences in allele frequency. The influence of effect size differences, on the other hand, appears to be minimal and inconclusive.
Brain eQTLs from matched populations can improve interpretation of SCZ GWASs
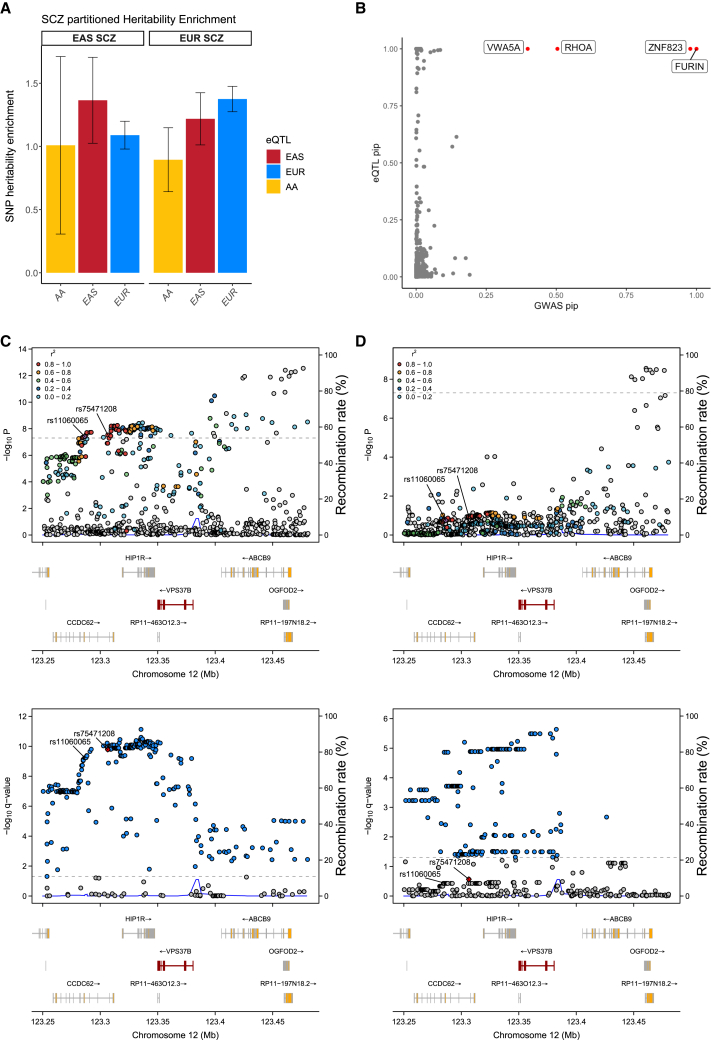
To determine whether eQTLs detected from a specific population could explain the disease GWAS signals and SNP-based disease heritability better than eQTLs from non-matching populations, we undertook a two-step analysis. Firstly, we gathered SCZ GWAS summary statistics for the EUR, EAS, and AA populations from previously published studies.39,40,41 We employed the LDSR42 approach to assess the GWAS signal enrichment of these eQTLs. eQTLs identified in the EAS population demonstrated a higher enrichment in EAS-based GWAS signals than eQTLs identified in the EUR population (Table S9, EnrichmentEUR = 1.08, EnrichmentEAS = 1.3; Welch modified two-sample t test p value < 0.001). Conversely, eQTLs identified in the EUR population showed a greater enrichment for EUR-based GWAS signals than the eQTLs from the EAS cohort (EnrichmentEUR = 1.37, EnrichmentEAS = 1.21; Welch modified two-sample t test p value < 0.001). Both of these enrichments were statistically significant (Table S9; Welch modified two-sample t test p value < 0.001; Figure 4A).
Figure 4.
Explanation of SCZ GWAS signals and prioritization of candidate genes
(A) GWAS enrichment results from LDSR.
(B) Fine-mapped colocalization results. Each point represents an eQTL pairs, the x axis represents the GWAS pip for that eSNP, and the y axis represents the eQTL pip for that eSNP. Red points represent pipGWAS∗pipeQTL> 0.1 and are labeled with the eGene.
(C and D) LocusZoom plots demonstrating the genetic colocalization evidence between SCZ GWAS signals (upper) and brain eQTLs (lower) at the VPS37B locus for (C) the EAS population and (D) the EUR population, respectively.
Besides the SNP heritability enrichment of all eQTLs, we also compared the significance of the GWAS signals for population-specific eSNPs. We found that population-specific eSNPs tended to have smaller p values of disease association (i.e., stronger associations) in the corresponding population than the common eSNPs (Figure S6, Welch modified two-sample t test p < 0.001), indicating the ability of population-specific eSNPs to explain the disease association and propose the relevant gene, which might be overlooked when focusing on a single population.
SCZ risk genes identified using eQTLs and GWASs from non-EUR populations
To uncover risk genes and pathways for SCZ in non-EUR populations, we used MetaXcan, RTC,17 and SMR33 to prioritize SCZ candidate risk genes in non-EUR populations and compared them with risk genes identified in the EUR (see subjects and methods). In total, we prioritized eight risk genes in the EAS (Tables S10–S12). It is worth noting that our TWAS analysis of AA data did not reveal any significant associations. This lack of association in AA data might be attributed to the relatively small sample size available from the AA SCZ GWAS.
Five SCZ candidate risk genes (SFXN2, VPS37B, DENR, FTCDNL1, and NT5DC2) uniquely discovered in the EAS population were assessed for allele frequency. The eSNPs for these genes showed lower allele frequency in the EUR population than in EAS. For instance, the GWAS signal chr12:123286491A>G (rs11060065) in VPS37B was found to be significant in the EAS population with a high MAF of 0.48 (OR = 0.92; p = 3.797e−08; Figure 4C). In contrast, this association was not significant in the EUR population with a markedly lower allele frequency of 0.04. A parallel pattern emerged with the eSNP for VPS37B, with a markedly higher frequency (chr12:123306558G>A, rs75471208, MAF = 0.24) in EAS than in the EUR (Figure 4D, MAFEUR = 0.04). These results further confirm that allele frequency differences between populations can explain most of the discrepancies between the EUR and EAS GWAS and the eQTL results (Table S13).
Potential SCZ regulatory variations were refined utilizing brain eQTLs from non-EUR populations
Three risk genes identified in the EAS population were shared with the EUR population (CNNM2, MTRFR, and MPHOSPH9), but differences in genetic architecture between populations were still apparent. For example, two distinct significant SNPs in EAS and in EUR were associated with SCZ on chromosome 10 (Figure S7, GWASEUR: chr10:104850632G>A [rs3736922] with GWAS p value = 6.4e−13; GWASEAS: chr10:104657300T>C [rs12219346] with GWAS p value = 5.4e−12). Using eQTLs with GWAS signals in EAS and EUR separately, colocalization and SMR analysis prioritized these two distinct GWAS SNPs to the same risk gene, CNNM2, in the two populations, respectively.
To further investigate whether these signals are located within any regulatory elements, we utilized the non-coding variant annotation database (NCAD)43 to annotate their regulatory information. Our findings revealed that all the EAS GWAS risk SNPs are situated in enhancer regions (Table S14). Moreover, the EAS GWAS risk SNPs near CNNM2 demonstrated a potential impact on histone modification, supported by the Roadmap data. Furthermore, we used the Lineage-specific Brain Open Chromatin Atlas44 to investigate whether this enhancer region shows different effects in major brain cell types. The results did not show any cell type differences, which indicates the enhancer effect exists universally in major brain cell types (Figure S8). Integrating these insights, we discovered strong evidence for multiple regulatory regions among the EAS eSNPs-chr10:104654577T>C, which have a high LD with CNNM2 GWAS SNPs (LD R2 = 1, p value < 0.00001). Additionally, our dual luciferase reporter assay results confirmed that the EAS eSNPs C-allele at chr10:104654577T>C significantly enhances luciferase activity compared to the reference vector, as detailed in the supplemental methodsand Figure S8.
High-confidence putative causal variants of SCZ using multi-ancestry brain eQTLs
To identify high-confidence putative causal variants from multiple populations, we applied colocalization to our fine-mapped eQTLs and SCZ GWAS signals. In total, we identified four SNP-gene-disease triplets in which the SNP colocalized with both gene expression and SCZ GWAS (Table S15, PiPcoloc = PiPGWAS × PiPcis-eQTLs > 0.1). The top genes with PIPcoloc > 0.1 include FURIN, ZNF823, RHOA, and VWA5A (Figure 4B). As an example, we identified the strongest putative SCZ causal SNP for FURIN-chr15:91426560G>A. This SNP is located in the 3′ untranslated regions (UTRs) of FURIN. Notably, this variant did not reach genome-wide significance in the EAS population (p = 1.06e−3) likely due to limited statistical power. Our result strongly supported that this causal variant is shared across populations, with causal probabilities of 1. Previous study has also implicated the variant in both the EUR and EAS populations.13
Discussion
In this study, we have created a brain transcriptome resource and identified eQTLs in the prefrontal cortex, specifically focusing on non-European populations. Our findings address the initial inquiries raised in the introduction. Firstly, we investigated the driver behind the variation in brain eQTLs across different populations. We found that differences in allele frequency are instrumental in connecting disease susceptibility to gene expression regulation. This finding greatly augments our comprehension of genetic influences on gene expression in the human brain. Secondly, when examining brain eQTLs from diverse populations, we gained power to explain the GWAS heritability, uncover risk genes, and fine-map risk variants. We observed a pronounced enrichment of disease heritability among eQTLs in matched populations. In the non-EUR cohort, the allele frequencies and LD configurations facilitated the identification of five SCZ risk genes. Additionally, we identified four high-confidence putative causal SCZ variants. These results highlight the utility of studying non-European cohorts.
Population differences appear to be more pronounced at the allele frequency level but are less so at the effect size level. In general, the estimated π1 of eQTLs from non-EUR populations in EUR is lower compared to the rate observed between down-sampling-EUR and the EUR population cohort. Despite the relatively small sample size and statistical power, we identified 343,737 significant cis-eQTLs including 232,254 EAS eQTLs and 51,483 AA eQTLs that were exclusive to the non-EUR populations. While over half of eSNPs in our non-EUR dataset were population specific, 80% of eGenes identified in the non-EUR were also eGenes in the EUR data but associated with different SNPs. Moreover, we observed that the effect sizes of eQTLs were highly correlated between populations. The consistency of our observations with prior research involving diverse populations, including studies on gene expression,45,46 methylation,47 and chromatin accessibility,48 confirms the shared regulatory patterns across different populations.
Interestingly, some eQTLs showed contrasting effects across populations. ∼0.1% of the non-EUR-specific eQTLs displayed opposing directions in effect size. A notable example of this is the eQTL rs2599400-TAS2R31, which showed opposite effects in different populations. Blood eQTLs from EAS,38 EUR, and AFR7 also support this observation. Prior studies have underscored the population-specific variations in TAS2R31, linking these variations to differing sensitivity to the bitter taste.24 It is important to acknowledge that effect-size differences, though infrequent, can provide critical insights into the genetic architecture and underlying biological mechanisms. However, the observed differences in this study may arise due to variations in sample sizes, leading to overestimation or underestimation of effect sizes because of random sampling variation. Replicating findings in independent cohorts can help confirm the observed effect sizes and rule out statistical artifacts. Additionally, further experimental validation and functional studies of variants showing significant effect-size differences are warranted to elucidate the biological mechanisms underlying these differences.
Our findings underscore that enhancing genetic ancestral diversity is more efficient for power gain than increasing the sample size within large-scale eQTLs datasets. Through our benchmarking of eQTLs across three populations, we have established robust capabilities for identifying eQTLs with a MAF greater than 0.2 and an effect size of 0.6 (Figure S9). Our power analysis indicates that more than 30,000 individuals of European ancestry is needed to uncover all eQTLs with MAF of 0.01 in this population, based on the estimated effect size of eQTLs exclusively observed in non-EUR populations (Figure S9). For example, one eQTL pair (chr12:123306558G>A-VPS37B) would require 3,215 EUR samples based on the power estimate because of the low frequency in the EUR population (MAF = 0.04). However, the MAF of this eSNP is 0.22 in EAS, which reduces the required sample size from 3,215 to 246. Thus, incorporating a more diverse population would reveal numerous regulatory variants that are not only rarer in EUR but more prevalent in non-EUR groups. Advancing toward a broader, more diverse human reference dataset will facilitate more comprehensive investigations into the impact of human demography on eQTL detection, thereby deepening our understanding of the distribution and influence of genetic regulation in the human brain.
Differences in the genetic architecture underlying gene expression can help us to prioritize risk genes. Notably, prior research has reported that disease-associated loci tend to be skewed toward variants with higher allele frequency in the discovery population, indicating that limited statistical power may result in “missing” disease-association signals. Incorporating diverse samples can enhance our ability to uncover the etiology of the disease. In our study, we identified five SCZ risk genes using the non-EUR population, including VPS37B in the EAS population. VPS37B is associated with calcium-dependent protein binding, providing evidence to support the involvement of the calcium-related pathway in SCZ risk in the EAS population.49 Another interesting candidate highlighted in our study was CYP17A1 (RTC = 0.99). The corresponding GWAS signal was significant in the EAS and EUR populations (pEAS = 4.5e−8; MAFEAS = 0.48; pEUR = 2.6e−13; MAFEUR = 0.30), while the corresponding eSNP in EAS population (MAF = 0.48) showed extremely low frequency in EUR population (MAF < 0.001). CYP17A1 notably encodes enzyme important for the production of glucocorticoids and sex hormones, such as estrogen, which have been linked to schizophrenia.50,51,52
Besides enhancing the power for detecting risk genes, the inclusion of brain eQTLs from diverse populations improves the ability to fine-map SCZ GWAS loci, identifying regulatory variants which have the potential to regulate downstream gene expression. This approach aids in interpretation, thereby facilitating subsequent computational and experimental functional investigations. Our result revealed a potential regulatory region near the population-shared risk gene CNNM2. This discovery showcases the power of leveraging diverse populations.
By leveraging the multi-ancestry information, trans-ancestry fine-mapping also helped us identify high-confidence putative causal variants. In addition to previously validated genes, our study uncovered another significant finding at chr3:50297330A>G-RHOA-SCZ through trans-ancestry colocalization. RHOA encodes a member of the Rho family of small GTPases, pivotal in signal transduction cascades by toggling between inactive GDP-bound and active GTP-bound states.
Several limitations of our study warrant attention. Firstly, our sample size is relatively small, which likely constrained the comprehensiveness of our findings. Our analysis indicates that increasing the sample size would enable the identification of a larger set of eQTLs. The modest sample sizes of both the AA eQTL dataset and the SCZ GWAS cohort likely contributed to the failure of TWAS in the AA population. It is also important to note that the EAS population in our study consists solely of Han Chinese samples. Given that EAS encompasses a broader range of East Asian ancestries, specifying our EAS eQTL results as representative of Han Chinese offers a more accurate representation. This limitation highlights the necessity for future research to include a more diverse array of East Asian populations, thereby ensuring more generalizable and comprehensive results. We also highlighted that the current results are valid for cis-regulatory elements but exclude any differences embedded through trans-regulatory elements. This distinction is crucial as it underscores the focus of our study on cis-acting variations, while trans-acting factors, which could also play significant roles in gene regulation, remain unexplored within the scope of our current analysis.
In conclusion, we present a genome-wide map of human brain gene expression regulation. Importantly, this resource bridges the gap between neuropsychiatric GWAS and brain gene expression profiling in non-European populations. Our study emphasizes the significance of this atlas of brain gene expression regulation in non-European populations for advancing our understanding of human diversity, addressing health disparities, and developing precision medicine.
Data and code availability
The raw sequence data for East Asian population generated during this study are available at the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number HRA000108 and can be accessed at https://bigd.big.ac.cn/gsa-human.
The summary statistics of eQTLs generated in this study are provided in the https://github.com/liusihan/population-compare-pipeline.
The code generated during this study are available at GitHub (https://github.com/liusihan/population-compare-pipeline).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 82022024, 31571312, and 91632116), the National Key R&D Project of China (grant 2016YFC1306000), the Science and Technology Innovation Program of Hunan Province (grants 2021RC4018 and 2021RC5027), and NIH grant 1R01MH126459-01A1. We thank the National Human Brain Bank for Development and Function, Institute of Basic Medical Sciences Chinese Academy of Medical Sciences, School of Basic Medicine Peking Union Medical College, Beijing, China for providing the frozen human brain tissue. The National Human Brain Bank for Development and Function was supported by the STI2030-Major Project (#2021ZD0201100) Task 1 (#2021ZD0201101), CAMS Innovation Fund for Medical Sciences (CIFMS #2021-1-I2M-025), the Chinese Academy of Medical Sciences, Neuroscience Center, and the China Human Brain Banking Consortium. The authors acknowledge Stanley Center for Psychiatric Research for supporting H.H. and Y.C. working on this project. We would like to appreciate Dr. Fengxiao Bu for his help in drawing Figure 1.
Author contributions
Y.C. and S.L. wrote the manuscript, analyzed the data, and performed all the computations. F.W., Q.L., and Y.J. contributed to eQTLs analysis of EUR population. Z.R., F.D., C.H., and M.L. extracted DNA and RNA and also collected sample information. R.D., Y.X., R.F.K., and E.R.G. substantively revised the manuscript. Z.N., S.X., H.H., T.B., E.G., and E.R.G. participated in the design of comparing the brain regulatory architecture. K.Y., W.Q., C.M., X.-X.Y., L.W., J.D., J.H., B.T., and C.L. provided primary sequenced samples and data including their clinical information. C.M., C.L., and C.C. conceived, designed, and supervised the study and modified the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 2, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.09.001.
Contributor Information
Chunyu Liu, Email: liuch@upstate.edu.
Chao Chen, Email: chenchao@sklmg.edu.cn.
Supplemental information
References
- 1.Trubetskoy V., Pardiñas A.F., Qi T., Panagiotaropoulou G., Awasthi S., Bigdeli T.B., Bryois J., Chen C.-Y., Dennison C.A., Hall L.S., et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–508. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson R.E., Kuchenbaecker K., Walters R.K., Chen C.-Y., Popejoy A.B., Periyasamy S., Lam M., Iyegbe C., Strawbridge R.J., Brick L., et al. Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell. 2019;179:589–603. doi: 10.1016/j.cell.2019.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hindorff L.A., Bonham V.L., Brody L.C., Ginoza M.E.C., Hutter C.M., Manolio T.A., Green E.D. Prioritizing diversity in human genomics research. Nat. Rev. Genet. 2018;19:175–185. doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., Xia Y., Chen Y., Dai R., Qiu W., Meng Q., Kuney L., Chen C. Brain Banks Spur New Frontiers in Neuropsychiatric Research and Strategies for Analysis and Validation. Dev. Reprod. Biol. 2019;17:402–414. doi: 10.1016/j.gpb.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogil L.S., Andaleon A., Badalamenti A., Dickinson S.P., Guo X., Rotter J.I., Johnson W.C., Im H.K., Liu Y., Wheeler H.E. Genetic architecture of gene expression traits across diverse populations. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Klein N., Tsai E.A., Vochteloo M., Baird D., Huang Y., Chen C.-Y., van Dam S., Oelen R., Deelen P., Bakker O.B., et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat. Genet. 2023;55:377–388. doi: 10.1038/s41588-023-01300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng B., Bendl J., Kosoy R., Fullard J.F., Hoffman G.E., Roussos P. Multi-ancestry eQTL meta-analysis of human brain identifies candidate causal variants for brain-related traits. Nat. Genet. 2022;54:161–169. doi: 10.1038/s41588-021-00987-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan X.-X., Ma C., Bao A.-M., Wang X.-M., Gai W.-P. Brain banking as a cornerstone of neuroscience in China. Lancet Neurol. 2015;14:136. doi: 10.1016/S1474-4422(14)70259-5. [DOI] [PubMed] [Google Scholar]
- 11.Qiu W., Zhang H., Bao A., Zhu K., Huang Y., Yan X., Zhang J., Zhong C., Shen Y., Zhou J., et al. Standardized Operational Protocol for Human Brain Banking in China. Neurosci. Bull. (Arch. Am. Art) 2019;35:270–276. doi: 10.1007/s12264-018-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., Li Y., Ye J., Yu C., Li Z., et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7:1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trivedi U.H., Cézard T., Bridgett S., Montazam A., Nichols J., Blaxter M., Gharbi K. Quality control of next-generation sequencing data without a reference. Front. Genet. 2014;5:111. doi: 10.3389/fgene.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaneau O., Ongen H., Brown A.A., Fort A., Panousis N.I., Dermitzakis E.T. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 2017;8 doi: 10.1038/ncomms15452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law C.W., Chen Y., Shi W., Smyth G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wooding S.P., Ramirez V.A. Global population genetics and diversity in the TAS2R bitter taste receptor family. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.952299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arbiza L., Gronau I., Aksoy B.A., Hubisz M.J., Gulko B., Keinan A., Siepel A. Genome-wide inference of natural selection on human transcription factor binding sites. Nat. Genet. 2013;45:723–729. doi: 10.1038/ng.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia G., Patterson N., Sankararaman S., Price A.L. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawitan Y., Seng K.C., Magnusson P.K.E. How many genetic variants remain to be discovered? PLoS One. 2009;4 doi: 10.1371/journal.pone.0007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finucane H.K., Reshef Y.A., Anttila V., Slowikowski K., Gusev A., Byrnes A., Gazal S., Loh P.-R., Lareau C., Shoresh N., et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinforma. Oxf. Engl. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 34.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou Y., Carbonetto P., Wang G., Stephens M. Fine-mapping from summary data with the “Sum of Single Effects” model. PLoS Genet. 2022;18 doi: 10.1371/journal.pgen.1010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinforma. Oxf. Engl. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pybus M., Dall’Olio G.M., Luisi P., Uzkudun M., Carreño-Torres A., Pavlidis P., Laayouni H., Bertranpetit J., Engelken J. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 2014;42:D903–D909. doi: 10.1093/nar/gkt1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ning Z., Tan X., Yuan Y., Huang K., Pan Y., Tian L., Lu Y., Wang X., Qi R., Lu D., et al. Expression profiles of east-west highly differentiated genes in Uyghur genomes. Natl. Sci. Rev. 2023;10 doi: 10.1093/nsr/nwad077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam M., Chen C.-Y., Li Z., Martin A.R., Bryois J., Ma X., Gaspar H., Ikeda M., Benyamin B., Brown B.C., et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 2018;173:1705–1715.e16. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bigdeli T.B., Genovese G., Georgakopoulos P., Meyers J.L., Peterson R.E., Iyegbe C.O., Medeiros H., Valderrama J., Achtyes E.D., Kotov R., et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol. Psychiatry. 2020;25:2455–2467. doi: 10.1038/s41380-019-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng X., Liu S., Li K., Bu F., Yuan H. NCAD v1.0: A database for non-coding variant annotation and interpretation. J. Genet. Genomics. 2024;51:230–242. doi: 10.1016/j.jgg.2023.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Fullard J.F., Hauberg M.E., Bendl J., Egervari G., Cirnaru M.-D., Reach S.M., Motl J., Ehrlich M.E., Hurd Y.L., Roussos P. An atlas of chromatin accessibility in the adult human brain. Genome Res. 2018;28:1243–1252. doi: 10.1101/gr.232488.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kachuri L., Mak A.C.Y., Hu D., Eng C., Huntsman S., Elhawary J.R., Gupta N., Gabriel S., Xiao S., Keys K.L., et al. Gene expression in African Americans, Puerto Ricans and Mexican Americans reveals ancestry-specific patterns of genetic architecture. Nat. Genet. 2023;55:952–963. doi: 10.1038/s41588-023-01377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A.C., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraser H.B., Lam L.L., Neumann S.M., Kobor M.S. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tehranchi A., Hie B., Dacre M., Kaplow I., Pettie K., Combs P., Fraser H.B. Fine-mapping cis-regulatory variants in diverse human populations. Elife. 2019;8 doi: 10.7554/eLife.39595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrade A., Brennecke A., Mallat S., Brown J., Gomez-Rivadeneira J., Czepiel N., Londrigan L. Genetic Associations between Voltage-Gated Calcium Channels and Psychiatric Disorders. Int. J. Mol. Sci. 2019;20:3537. doi: 10.3390/ijms20143537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heringa S.M., Begemann M.J.H., Goverde A.J., Sommer I.E.C. Sex hormones and oxytocin augmentation strategies in schizophrenia: A quantitative review. Schizophr. Res. 2015;168:603–613. doi: 10.1016/j.schres.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 51.de Boer J., Prikken M., Lei W.U., Begemann M., Sommer I. The effect of raloxifene augmentation in men and women with a schizophrenia spectrum disorder: a systematic review and meta-analysis. NPJ Schizophr. 2018;4:1. doi: 10.1038/s41537-017-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiappelli J., Shi Q., Kodi P., Savransky A., Kochunov P., Rowland L.M., Nugent K.L., Hong L.E. Disrupted glucocorticoid--Immune interactions during stress response in schizophrenia. Psychoneuroendocrinology. 2016;63:86–93. doi: 10.1016/j.psyneuen.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data for East Asian population generated during this study are available at the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number HRA000108 and can be accessed at https://bigd.big.ac.cn/gsa-human.
The summary statistics of eQTLs generated in this study are provided in the https://github.com/liusihan/population-compare-pipeline.
The code generated during this study are available at GitHub (https://github.com/liusihan/population-compare-pipeline).