Summary
Microtubule affinity-regulating kinase 2 (MARK2) contributes to establishing neuronal polarity and developing dendritic spines. Although large-scale sequencing studies have associated MARK2 variants with autism spectrum disorder (ASD), the clinical features and variant spectrum in affected individuals with MARK2 variants, early developmental phenotypes in mutant human neurons, and the pathogenic mechanism underlying effects on neuronal development have remained unclear. Here, we report 31 individuals with MARK2 variants and presenting with ASD, other neurodevelopmental disorders, and distinctive facial features. Loss-of-function (LoF) variants predominate (81%) in affected individuals, while computational analysis and in vitro expression assay of missense variants supported the effect of MARK2 loss. Using proband-derived and CRISPR-engineered isogenic induced pluripotent stem cells (iPSCs), we show that MARK2 loss leads to early neuronal developmental and functional deficits, including anomalous polarity and dis-organization in neural rosettes, as well as imbalanced proliferation and differentiation in neural progenitor cells (NPCs). Mark2+/− mice showed abnormal cortical formation and partition and ASD-like behavior. Through the use of RNA sequencing (RNA-seq) and lithium treatment, we link MARK2 loss to downregulation of the WNT/β-catenin signaling pathway and identify lithium as a potential drug for treating MARK2-associated ASD.
Keywords: autism spectrum disorder, ASD, loss-of-function, LoF, MARK2 variants, WNT/β-catenin signaling pathway, lithium
Graphical abstract
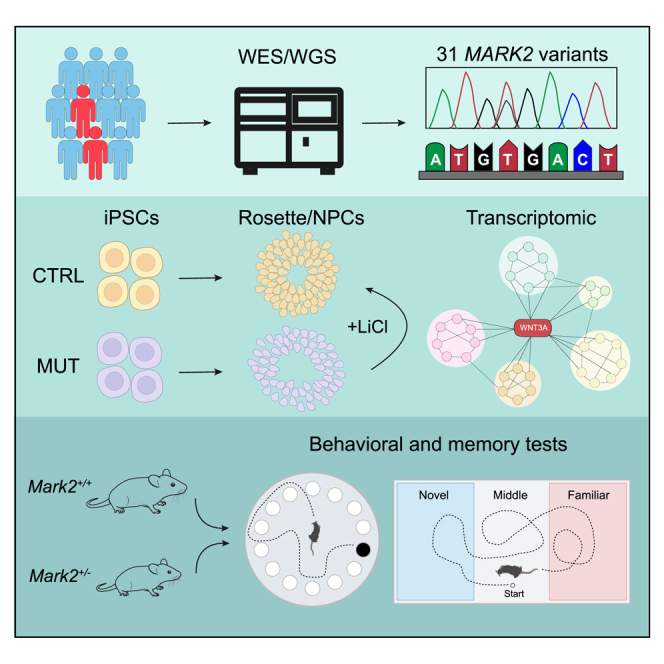
MARK2 plays essential roles in the central nervous system. Here, we demonstrate the association between MARK2 variants, downregulated WNT/β-catenin signaling, and autism spectrum disorder (ASD) through the study of individuals with ASD and Mark2+/− mice.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder (NDD) marked by difficulties in social communication and repetitive behaviors1 that affects 0.7%–1.69% of children worldwide.2,3 ASD shows high heritability, and 5%–30% of affected individuals have a Mendelian disorder.4,5 Advances in genomic technologies have allowed the identification of over 300 risk ASD genes, including 100 high-confidence genes, through large-scale sequencing studies of autism cohorts.6,7,8,9 Most genes relevant to ASD are highly intolerant to loss-of-function (LoF) variants in the human genome but are enriched for de novo variants in ASD individuals.6,7,8,9
Microtubule affinity-regulating kinase 2 (MARK2; MIM: 600526), also known as partitioning-defective 1b (PAR1b), is a member of the evolutionarily conserved PAR1/MARK serine/threonine kinase family,10,11 and it plays essential roles in the central nervous system (CNS) via several cell biological functions.12,13,14,15,16,17 In mouse primary hippocampal neurons in vitro, Mark2 negatively modulates neuronal polarity and dendritic development,13 and mouse neurons with Mark2 depletion in vivo exhibit unpolarized dendritic overgrowth with multiplex rather than single axons.12 Additionally, downregulation of Mark2 by in utero electroporation inhibits neuronal migration in the mouse cortex,15 and unpolarized neurons accumulate in the intermediate zone (IZ) of the developing cerebral cortex.15 Recently, Zhou et al. performed an integrated analysis of the largest set of sequencing data from 42,607 individuals with ASD and identified MARK2 as a moderate-risk gene for ASD.18 However, the clinical features and variant spectrum of affected individuals with ASD with MARK2 variants (MARK2-associated ASD), as well as the pathophysiological effect and molecular mechanism of MARK2 variants on human neurogenesis, especially on human NPCs/stem cells, remain unclear.
Herein, we describe a cohort of 31 individuals with clinically relevant MARK2 variants, mostly LoF variants (25/31), who were gathered through GeneMatcher.19 All affected individuals presented with ASD and other NDD features, and the majority have distinctive facial features. Using proband-derived induced pluripotent stem cells (iPSCs) combined with CRISPR-engineered isogenic iPSCs, we revealed the cellular phenotypes and transcriptomic profiles of mutant neural progenitor cells (NPCs). Furthermore, in experiments with Mark2+/− (HET) mice, we recapitulated the pathophysiological deficits observed in mutant NPCs and the ASD-related behaviors presented by affected individuals. Importantly, we identified downregulation of the WNT/β-catenin signaling pathway as the molecular mechanism of MARK2 variant, and lithium reversed abnormal cellular phenotypes of mutant NPCs cells both in vitro and in vivo via re-activating the WNT/β-catenin signaling pathway.
Material and methods
Ethics statement
The study was approved by the ethics committees of the respective institutions, including the Capital Institute of Pediatrics (SHERLL2021008), Mayo Clinic (IRB#19–003389), and Maternal and Child Health Hospital of Guangxi Zhuang Autonomous Region (2017-3-11). This international individual cohort carrying the MARK2 variant was recruited by the GeneMatcher data sharing platform.19 Informed consent was obtained from all participants or proband’s legal guardians prior to study inclusion. Additional consent for the publication of photos was optional. The detailed clinical information was recorded by the referring clinician. Sample collection and consent was in accordance with the tenets of the Declaration of Helsinki.
Detection of MARK2 variants
MARK2 variants were detected in peripheral blood samples by proband-only whole exome-sequencing (WES) or Trio whole-genome sequencing (WGS) combined with Sanger sequencing. WES was performed with commercial exome capture kits and was followed by sequencing on the Illumina NovaSeq 6000 platform.
Variants were annotated based on transcript GenBank: NM_001039469.3 using the reference human genome GRCh37(hg19) version. The inheritance of MARK2 variants was confirmed in all families by Sanger sequencing. Two pairs of primers were designed to exclude amplification bias during PCR. For missense variants, only those that were predicted as deleterious variants by more than 2 software programs (PolyPhen2, SIFT, CADD, REVEL) were included. The pathogenicity of each candidate mutation was classified according to the 2015 guidelines of American College of Medical Genetics and Genomics (ACMG).20 The identities of biological parents were confirmed for all affected individuals before using the PS2 criteria. The criteria for PP321 and PVS122 evidence were modified based on the ACMG guidelines.
Expression vector construction, cell culture, transient transfection
Two splicing variants (c.337+1G>T, c.1514+2T>G) and 6 missense variants (p.Ala80Val [c.239C>T], p.Gly135Arg [c.403G>A], p.Phe194Ser [c.581T>C], p.Arg302Gln [c.905G>A], p.Val752Ala [c.2255T>C], p.Arg764Pro [c.2291G>C]) were chosen for in vitro transient transfection and functional assay. For splicing variants, 2 genomic fragments covering wild-type (WT) MARK2 c.337+1G allele (1039bp, see Table S1 for primers) or covering the c.1514+2T allele (2441bp, see Table S1 for primers) were amplified by standard PCR or overlap-extension PCR and then cloned into the pcDNA3.1 vector. For missense variants, full-length WT MARK2 cDNA (2367bp, GenBank: NM_001039469.3) was synthesized and cloned into pCMV-EGFP-Neo vector using 2 primers containing KpnI and Agel restriction enzyme sites. The mutant vectors were constructed by site-specific mutant primers and overlap-extension PCR (Table S1). All vectors were verified by sequencing.
Human embryonic kidney 293T (HEK 293T) cells and HeLa cells (ATCC) were grown in minimum Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics. The cells were grown to 70%–90% confluence before transient transfection (Lipofectamine 2000 reagent, Invitrogen). Forty-eight h later, transfected cells were washed twice with cool 1× PBS and then collected for RNA extraction (splicing variants), protein lysis, and western blotting (missense variants). Total RNA was extracted (TRIzol, Omega) and reverse transcribed into cDNA (SuperScript reverse transcription kit, Thermo) followed by standard PCR (Table S1 for primers). The PCR products were visualized on a 2% agarose gel and then purified for Sanger sequencing.
Model construction and calculation
The protein accession number GenBank: NP_001034558.2 was used. The structure of the kinase-associated domain 1 (KA1) of human MARK2 is unknown, but the experimental structures of all human KA1 domains are sufficiently identical (71%) to generate a high-confidence KA1 model through homology-based methods.23 The structures of the kinase and ubiquitin-associated (UBA) domains of MARK2 have been experimentally determined (PDB ID: 1zmu).24 These independently generated models were integrated using information from AlphaFold2.25
Regions of the protein that do not adopt secondary structures and are likely to be intrinsically disordered regions (IDRs) were loop modeled using Modeller version 10.2.26 The combined 3D model was used for visualization and structure-based calculations of stability alterations due to genomic variants.27 We assessed short linear motifs (SLiMs) of each missense variant by programmatically accessing the web of Eukaryotic Linear Motif (ELM, http://elm.eu.org).28 In addition, we calculated the local stability (ΔΔGfold) and the balance of favorable and unfavorable local contacts, termed frustration for missense variants.
Generation of iPSC lines from 2 affected individuals
Peripheral blood mononuclear cells (PBMCs) were reprogrammed into iPSCs using a CytoTune-iPS 2.0 Sendai Reprogramming Kit (Invitrogen) as we previously reported.29 In brief, PBMCs were seeded in 12-well plates and cultured for 14 days in PBMC medium. Then, 2.5 × 105 PBMCs were infected with Sendai virus and cultured for 48 h at 37°C. Then, the infected cells were added to mouse embryonic fibroblast (MEF) feeders in iPSC medium. After 2 weeks, individual iPSC colonies were picked manually, transferred to Matrigel (Corning)-coated dishes, and maintained in mTeSR medium (Stem Cell Technologies). Routine testing revealed that the cells were mycoplasma negative. After more than 10 passages, the characteristics of the iPSCs, including their pluripotency and genomic background, were analyzed. For pluripotency analysis, we performed alkaline phosphatase (AP) staining and immunofluorescence (IF) staining and assessed teratoma formation. To evaluate genomic background, we performed karyotyping, short tandem repeat (STR) site analysis, and array comparative genomic hybridization (CGH) to confirm that the genomes were identical. Two iPSC lines from 2 normal unrelated adults we previously reported were used as control iPSCs.29 Meanwhile, 2 isogenic iPSCs were generated using CRISPR-Cas9 gene editing engineering to delete the KA1 domain of MARK2 with heterozygous status, and the top 6 off-target genes sorted by cutting frequency determination (CFD) off-target score were tested (CRISPR-Del1 and CRISPR-Del2, see Table S1 for guide RNA [gRNA] primers).
Neural differentiation of iPSCs
iPSCs were maintained on Matrigel (BD Biosciences) in mTeSR1 medium (StemCell Technologies) and performed for neural differentiation. Between passages 12 and 30, iPSCs were used for neural differentiation as previously described with minor modifications.30 In brief, 30,000 iPSCs were used for embryoid bodies (EBs) culture, and after 24 h, EBs were planted to poly-L-ornithine/laminin-coated plates. Then, EBs were grown in a 1:1 mixture of N2- and B27-containing media (N1 medium) supplemented with EGF2 (Gibco, 1 mg/mL), Noggin (BD Biosciences, 250 ng/mL), SB431542 (Selleck, 10 mM), and Laminin (Gibco, 0.5 mg/mL) for 10 days. For neural differentiation, N1 medium was changed to a 1:1 mixture of N2- and B27-containing media (N2 medium) supplemented with FGF1 (Gibco, 1 mg/mL), EGF2 (Gibco, 1 mg/mL), BDNF (Gibco, 100 μg/mL), and Laminin (Gibco, 0.5 mg/mL) for another 3 days. For neurospheres, EBs were grown in suspension culture in DMEM/F12 supplemented with 1× N2 (Gibco), 1× nonessential amino acids (Gibco), Knockout Serum Replacement (Gibco), and Mercaptoethanol (Sigma, 25 nM) for 5 days.
RNA sequencing
Two iPSC-derived NPCs of P19 were used for this experiment. Total RNA was extracted from iPSC-derived NPCs with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Two micrograms of RNA were used for sequencing library generation using the NEBNext Ultra RNA Library Prep Kit (NEB). The cDNA library was preliminarily quantified using a Qubit RNA Assay Kit (Thermo Fisher Scientific) and then diluted to 1 ng/μL. Insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies) and quantified using StepOnePlus RT-PCR. The cDNA library was sequenced with the Illumina HiSeq 2500 platform (Annoroad) to generate 150-bp paired-end sequence reads. Read counts and FPKM for genes were extracted using the standard RNA sequencing (RNA-seq) analysis flowchart. Briefly, Fastq was used to assess sequencing quality control and remove the reads containing adapter sequences.31 Reads were mapped to the GRCh38 version of the human genome and transcriptome with Salmon (v.1.0.0, SAF Pattern). Normalization and differential gene expression were performed with DESeq2 Bioconductor package (https://github.com/thelovelab/DESeq2) with the R statistical programming environment (v.4.2.2). Genes were identified as significant genes with a p value <0.05 and an absolute value of log2fold change >1.5 and were used for gene function and pathway enrichment analyses, including Gene Ontology (GO) analysis and gene set enrichment analysis (GSEA) with clusterProfiler Bioconductor package (https://github.com/YuLab-SMU/clusterProfiler).
RT-quantitative PCR validation
Total RNA from iPSC-derived NPCs was reverse transcribed into cDNA using the transcript one-step gDNA removal and cDNA synthesis kit (TransGen Biotech), and the diluted (1:100) cDNA was used as a template for qPCR. qPCR was performed on a 7500 FAST Real-Time PCR system (Life Technologies) using an SYBR select master mix kit with specific primers. Relative gene expression was analyzed by the 2−ΔΔCT method, and GAPDH or actin was used as an endogenous control. The RT-qPCR primers are listed in Table S1.
Western blot analysis and IF staining
Total protein was extracted from cells or tissue using RIPA buffer containing 10 mM PMSF (Beyotime Biotechnology), and the protein concentration was determined using a BCA protein assay kit (Beyotime Biotechnology). The membranes were blocked in 5% milk in TBST (0.1% Tween 20) and incubated with primary antibodies at 4°C overnight. The membranes were then washed in TBST for 10 min 3 times and incubated with secondary antibodies at room temperature for 2 h. An ECL system (Pierce) and Tanon-5200 Chemiluminescent Imaging System (Tanon) were used for signal detection.
Brains of embryonic day (E) 18.5 mice were postfixed with 4% paraformaldehyde (PFA) overnight and then dehydrated with 30% sucrose. The tissues were embedded and cut into 35-μm-thick cryosections. For IF staining, sagittal cortex slices or cells were permeabilized (0.5% Triton X-100 and 3% BSA in PBS) for 15 min and blocked (0.3% Triton X-100 and 3% BSA in PBS) for 1 h at room temperature. Next, the slices or cells were incubated with relevant primary antibodies (0.3% Triton X-100 and 3% BSA in PBS) overnight at 4°C. The samples were incubated with AlexaFluor 488-, 568-, 594- or 647-conjugated secondary antibodies (1:500), and nuclei were counterstained with DAPI (1:1,000). Antibody categories are listed in Table S2.
Microscopy and analysis of NPCs
Confocal images were acquired using Zeiss LSM880 confocal microscopes and analyzed by ZEN software.
Mouse lines
The animal experiments were approved by the Laboratory Animal Center, Institute of Zoology, Chinese Academy of Sciences and were performed in accordance with relevant Chinese regulations. Mark2+/− mice were generated by Cyagen Biosciences (China) using double gRNA CRISPR-Cas-mediated genome engineering to delete exons 2–17 of Mark2. gRNA primers are listed in Table S1. All mice were bred in a specific-pathogen-free facility on a 12-h light/dark cycle, and food and water were available ad libitum.
BrdU incorporation analysis
BrdU (Sigma) was administered by intraperitoneal injection at a dose of 100 mg/kg body weight of the mother mouse. Brains were harvested 1 h later for subsequent analysis. Antibody categories are listed in Table S2.
Behavioral and memory tests
Female mice have hormonal cycles that may potentially complicate the results. To avoid the effect of hormone, only male mice aged 8–12 weeks (n ≥ 8 per group) were used for behavioral and memory tests. All tests were performed between 09:00 and 17:00. Videos of the behavioral tests were analyzed by EthoVision XT 14 (Noldus). Behavioral tests included open field, elevated plus maze, three-chamber, novel object recognition, marble burying, grooming, Y-maze, Barnes maze, and Morris water maze (see supplemental information for more details).
In vitro and in vivo rescue assays
For the in vitro rescue assay, either 0.7 mM LiCl (Sigma) or 100 ng/mL recombinant human WNT3A (rhWNT3A; R&D Systems) were supplemented in the culture media from the first day of differentiation process. The rescue efficacy was evaluated at specific time points by western blot and IF.
For the in vivo rescue assay, 6–8-month-aged female WT mice were allowed to mate freely with male HET mice. Once the vaginal plug was observed the following morning, the drinking water was replaced with clean drinking water containing LiCI (4.5 mg/L). The brains of fetal mice were collected at E18.5 for IF screening.
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM) unless otherwise indicated. At least 3 biological replicates were performed for each group. For statistical analyses, unpaired Student’s t tests were performed using GraphPad Prism software. Statistical significance was defined as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Results
Variant spectrum and clinical features of MARK2-related ASD
We utilized the GeneMatcher data sharing platform19 to assemble this international cohort. A total of 29 distinct MARK2 variants (Figure 1A; Table S3) were detected in 31 individuals, including 2 reported individuals from the Deciphering Developmental Disorders (DDD) study.18 Notably, 27 of 29 variants were not reported in the gnomAD and our in-house database. Two variants recurred in independent individuals from different ethnicities and laboratories (p.Arg302∗ [c.904C>T] in P8 and P12, p.Gln747∗ [c.2239C>T] in P20 and P24). Variant spectrum (Table 1) comprised 25 LoF variants (nonsense/frameshift/splicing) (81%) and 6 missense variants (19%). All LoF variants led to the loss of the entire KA1. Among 25 LoF variants, 16 (64%) were de novo, and 4 (16%) were paternally inherited. All missense variants were de novo. Three individuals presented variants at the same position (Arg302), including 2 LoF variants (p.Arg302∗) and 1 missense variant (p.Arg302Gln). No other clinically relevant or pathogenic variants were detected in affected individuals. All variants were classified as likely pathogenic or pathogenic according to ACMG/AMP criteria.20
Figure 1.
Overview of MARK2 variants and associated phenotypes
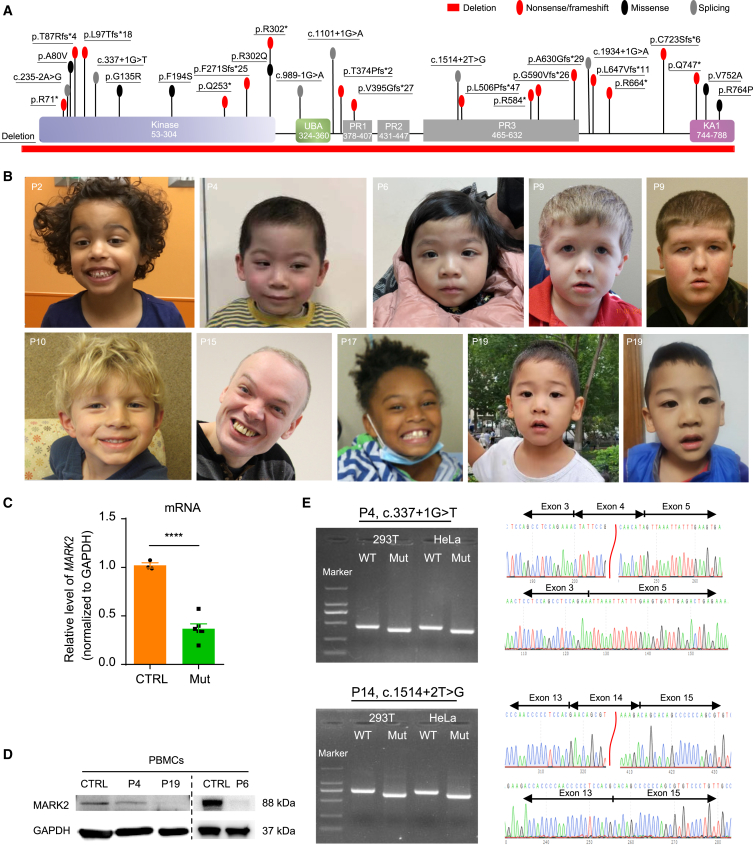
(A) Locations of variants in functional domains of MARK2 predicted by UniProt are shown. p.Arg302∗ and p.Gln747∗ are recurrent variants that occurred in 2 individuals. Kinase, protein kinase domain; UBA, ubiquitin associated; PR, polar residue; KA1, kinase-associated domain 1.
(B) Facial photos of 8 affected individuals. Ages at the time of photograph are P2, 5 years and 4 months; P4, 4 years and 2 months; P6, 3 years and 1 month; P9 left, 4 years; P9 right, 11 years; P10, 8 years; P15, 46 years; P17, 7 years and 7 months; P19 left, 4 years; P19 right, 4 years and 7 months.
(C and D) The mRNA expression (C) and protein accumulation (D) of MARK2 in the PBMCs from 3 affected individuals (Mut: P4, P6, P19). Both mRNA and protein levels were normalized to GAPDH levels and compared with those of age-matched healthy children (CTRL). The data of at least 3 independent experiments were analyzed by Student’s t test, ∗p < 0.05.
(E) Minigene assays of 2 splicing variants (P4: c.337+1G>T; P14: c.1514+2T>G). RT‒PCR products from HEK 293T and HeLa cells transfected with either the wild-type (WT) or mutant (Mut) pcDNA3.1 vector were separated by electrophoresis (left). M, DNA marker; the sizes of the bands are 2,000, 1,000, 750, 500, 250, and 100 bp. Sequencing of the RT-PCR products (right) show c.337+1G>T variant results in a 49-bp deletion, while the c.1514+2T>G variant results in a 98-bp deletion.
Table 1.
The detailed clinical features and variant spectrum of 31 individuals with MARK2 variants
| Clinical featuresa | age at measurements (years) | 2–46 |
| sex (male/female) | 18/13 | |
| autism spectrum disorder | 30/31 (96.8%) | |
| intellectual disability/developmental delay | 29/29 (100%) | |
| speech-language problems | 31/31 (100%) | |
| behavior abnormalities (ADHD, aggression and anxiety, depression) | 20/27 (74.1%) | |
| motor delay | 18/29 (62.1%) | |
| brain EEG | 8/15 (53.3%) | |
| seizure/epilepsy | 13/28 (46.4%) | |
| language regression | 9/22 (40.9%) | |
| visual impairment | 11/28 (39.3%) | |
| genital malformation in male | 6/18 (33.3%) | |
| muscle or other information | 9/28 (32.1%) | |
| Genomic variant spectrumb | LoF variant | 25/31 (81%) |
| LoF variant without KA1 domain | 25/31 (81%) | |
| de novo | 16/25 (64%) | |
| parental inherited | 4/25 (16%) | |
| unknown | 5/25 (20%) | |
| missense variant | 6/31 (19%) | |
| de novo | 6/6 (100%) |
Key features with frequency >30% were summarized. Some feature information wasn’t available for affected individuals, causing inconsistent denominators.
Variants classified as pathogenic/likely pathogenic were recruited.
The detailed clinical features associated with the MARK2 variants are summarized (Tables 1 and S3). The ages of individuals in our cohort ranged from 2 to 46 years old, with no observed sex difference (13 females and 18 males). The majority of affected individuals were described as having distinctive and recurrently noted features that included a narrow face, abnormal or broad forehead, downslanting palpebral fissures, and large or dysplastic ears. Representative images of 8 individuals are shown (Figure 1B). Eye problems, including hyperopia, myopia, astigmatism, strabismus, ptosis, and visual processing difficulties, were observed in 39.3% of the individuals (11/28). All individuals except 1 (P8) presented with ASD (30/31). Other NDDs were common within the cohort, including intellectual disability/developmental delay (100%), speech-language problems (100%), seizure/epilepsy (46%), motor delay (62%), and behavior disorders (74%). Language regression was reported in 41% (9/22) of individuals with 4 individuals still presenting language regression (P6, P19, P8, and P9) over time. The high penetrance of NDDs suggested that MARK2 is associated with ASD with NDD comorbid features.
To assess the consequence of LoF variants, MARK2 levels from PBMCs of 3 affected individuals (c.337+1G>T variant in P4, p.Phe271Serfs [c.812delT] in P6, and p.Cys723Serfs [c.2168_2169delGC] variant in P19) were measured for both mRNA and protein in vivo (Figures 1C and 1D). As shown, the mRNA and protein expression of MARK2 were significantly lower in affected individuals (Mut) than in the control group (CTRL). We also validated the splicing effects of 2 canonical splicing variants (c.337+1G>T variant in P4 and c.1514+2T>G variant in P14) in vitro using a minigene assay (Figure 1E). The transcripts of these MARK2 variants (Mut) were shorter than that of WT. Sequencing and online alignment of the RT-PCR products revealed 1 exon skipping event in the transcript, with a 49-bp deletion in the c.337+1G>T variant and a 98-bp deletion in the c.1514+2T>G variant. Both in vivo and in vitro experiments confirmed MARK2 loss due to LoF variants.
Structural modeling and in vitro expression assay reveal that missense variants in key domain of MARK2 cause protein destabilization
MARK2 contains 5 domains, including kinase, KA1, UBA, and 3 polar residue domains.10 All missense variants identified in our cohort (p.Ala80Val, p.Gly135Arg, p.Phe194Ser, p.Arg302Gln, p.Val752Ala, p.Arg764Pro) were clustered in the kinase or KA1 (Figure 1A) domains and involved the highly evolutionary conserved residues across species from Drosophila to human (Figure 2A). We analyzed the missense mutation tolerance map (https://stuart.radboudumc.nl/metadome/dashboard) by extracting missense and synonymous variants from the gnomAD database (gnomAD v.2.1.1, https://gnomad.broadinstitute.org/)32 and found both the kinase and KA1 domains are intolerant to missense mutations (Figure S1).
Figure 2.
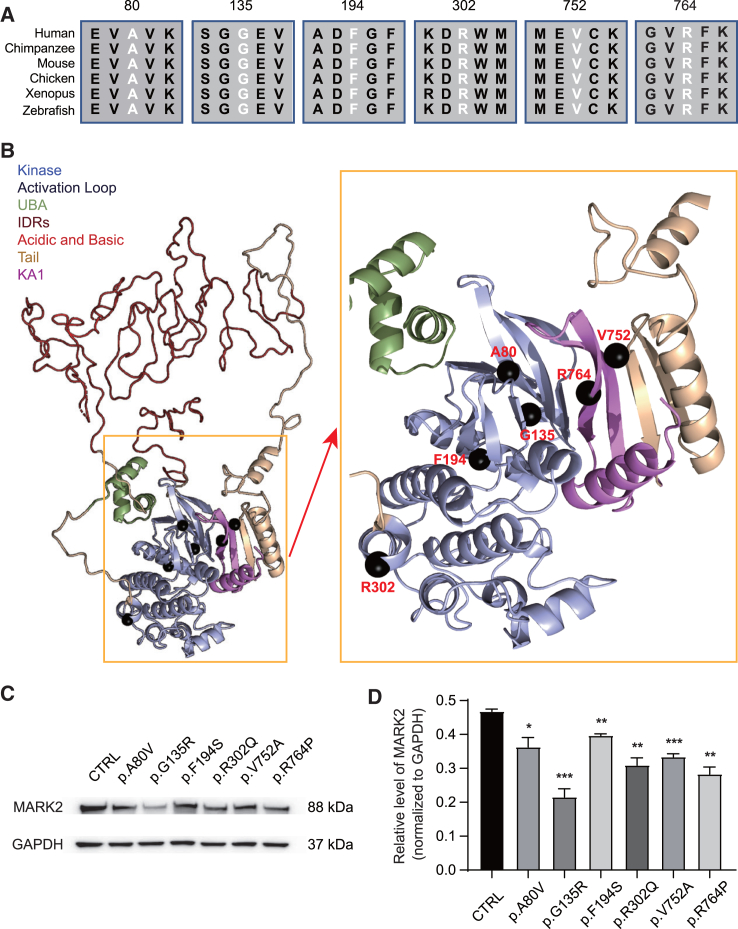
Structural modeling of six MARK2 missense variants
(A) Alignment of the protein sequences for 6 MARK2 missense variants across species from zebrafish to humans.
(B) 3D structural models of MARK2 missense variants. The protein domains of MARK2 are shown in different colors. The 3D structures of the non-intrinsically disordered regions (non-IDRs) were used to compute the changes in structural stability due to genomic variation. Amino acids in the IDRs are shown in a disordered configuration; we expect that any individual configuration of the IDRs would be an inadequate representation of their diverse dynamics. The locations of the 6 missense variants are marked by black spheres. Four variants in the kinase domain (p.A80V, p.G135R, p.F194S, p.R302Q) and 2 variants in the KA1 domain (p.V752A, p.R764P) were all predicted to locate in the activation loop.
(C and D) Representative western blot image (C) and quantification analysis of exogenous MARK2 accumulation that were normalized by total GAPDH levels (D). Human HEK 293T cells transfected with wild-type (CTRL) or mutant EGFP-MARK2-Neo vectors were lysed. The data of at least 3 independent experiments were analyzed by Student’s t test ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Information from evolutionary couplings indicates that the tail and KA1 domains fold together with the kinase domain,33 and 6 missense variants are within this structured dual domain (Figure 2B). Amino acids in the kinase domain are expected to have a balanced frustration so that the kinase can move as part of the normal cycle of ATP/ADP exchange and substrate access. Further, the effects of 6 variants on stability, frustration, binding motifs, and biochemical role in the enzyme, the altered 3D domain structure, or SLiMs were shown in Tables S4 and S5. The fraction of highly frustrated interactions around Ala80 is comparable, and the ΔΔGfold of the p.Ala80Val variant shows little to no change. However, the backbone of Ala80 and Met129, the gatekeeper residues of the kinase fold, form a hydrogen bond, as they are in adjacent β-strands. The degree of freedom of Met129 may be restricted by the comparatively bulkier side chain of Val80, indirectly and negatively affecting the catalytic function via a dynamic mechanism. Both Gly134 and Gly135 are in the kinase domain near the KA1 binding surface and solvent exposed. The ΔΔGfold of p.Gly135Arg variant is highly unfavorable, suggesting that the variant destabilizes the kinase domain. Gly135 is also positioned in the ATP-binding pocket, and the introduction of a large polar amino acid to the vicinity is likely to dysregulate nucleoside binding. p.Gly135Arg variant introduces a different sequence motif for cleavage. If this motif is recognized, it could represent the manner in which MARK2 is inactivated. Phe194 is located in the N-terminal base of the kinase activation loop and is a key residue in the classic Asp-Phe-Gly (DFG) motif. Phe194 anchors the activation loop within the kinase domain by hydrophobic interaction, rendering optimum flexibility to the activation loop. Two sequence motifs were gained in the p.Phe194Ser variant, 1 for phosphorylation by CK1 and another for glycosaminoglycan attachment. Thus, there is also potential for this genomic variant to introduce different regulatory dynamics into the enzyme, particularly given its proximity to known phosphorylation sites such as Ser197, Thr201, and Thr208. Arg302 is in the C-terminal helix of the kinase domain, which connects to the UBA domain. Arg302 was solvent exposed in our model and formed a hydrogen bond network among the Asp276 residue of the kinase, the N-terminal loop connected to the UBA domain, and additional N-terminal residues of the kinase. The frustration level indicated that p.Arg302Gln resulted in destabilization, while the ΔΔGfold indicated that this variant caused stabilization. According to structural analysis, the hydrogen bond between Gln302 and Asp276 was preserved in the mutated protein. Thus, p.Arg302Gln can be tolerated but may also have allosteric effects on the kinase, as we have previously reported.34 Val752 lies in the core of the hydrophobic interaction between the KA1 and tail domains. p.Val752Ala is a cavity-forming variant that may destabilize the hydrophobic core, potentially disrupting the formation of this autoregulatory domain. Similar to the kinase domain variants that destabilize KA1 interactions, p.Arg764Pro lies in the same interaction region and is therefore a reciprocal change with similar instabilities induced in the kinase-autoregulatory interaction. The ΔΔGfold value suggested that p.Val752Ala is destabilizing, leaving a weaker autoinhibition state. All 6 missense variants were predicted to result in loss of kinase catalytic function; we then hypothesize that the missense alterations observed in this cohort could lead to MARK2 destabilization or inactivation.
In order to test this hypothesis, we constructed and transfected pCMV-Neo-MARK2 vectors into HEK 293T cells and analyzed the exogenous mutant MARK2. As shown in Figures 2C and 2D, 6 mutant cells resulted in significantly lower MARK2 accumulation compared with WT cells. Among them, the p.Gly135Arg variant had the lowest MARK2 accumulation. These in vitro data suggested that missense variants in the kinase and KA1 domain can result in decreased protein accumulation due to MARK2 destabilization and thereby phenocopy MARK2 loss arising from LoF variants.
MARK2 variants lead to anomalous polarity in iPSC-derived neural rosettes and imbalanced proliferation and differentiation in iPSC-derived NPCs
Although the roles of Mark2 in mouse mature neurons are recognized, such as controlling hippocampal neuronal polarity and migration, dendritic spinal formation,12,13,14,15,16,17 the early neuronal development phenotypes, and fundamental mechanism of MARK2 variant in human are unclear. To clarify it, we generated 2 mutant iPSCs from the PBMCs of 2 affected individuals (p.Cys723Serfs from P19 and p.Phe271Serfs from P6), 2 control iPSCs from healthy males (CTRL1 and CTRL2) (Figures 1A and S2B), and 2 isogenic mutant iPSCs (CRISPR-Del1 and CRISPR-Del2). Each iPSC donor has at least 2 clones. Their pluripotencies (SOX2, OCT3/4, and NANOG, Figure S2A), karyotypes (Figure S2B), and differentiation abilities to the 3 germ layers (Figure S2C) were confirmed.
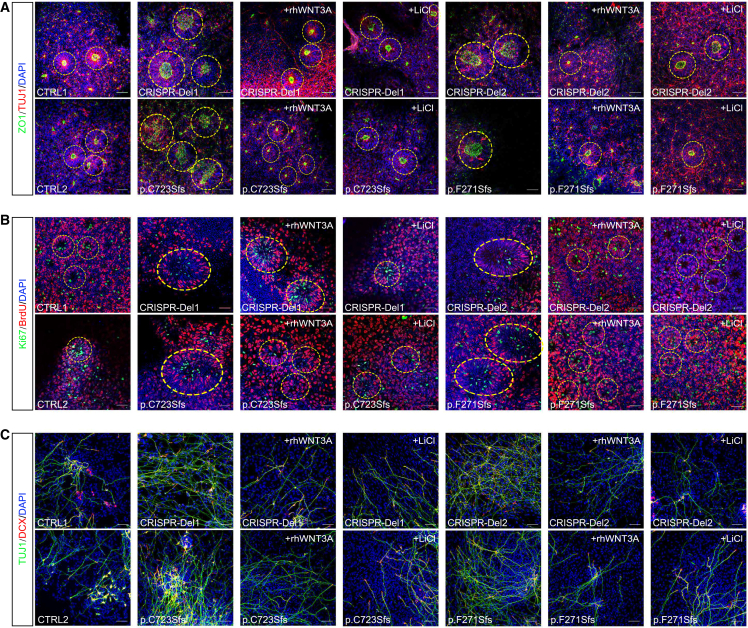
Neural rosettes develop from the self-organized and differentiated iPSCs and present radial arrangements of neuroepithelial cells with 1 central lumen resembling a developing neural tube.35 The key morphology of neural rosette includes intercalation, constriction, polarization, elongation, and lumen formation.35,36 Considering the role of MARK2 in neuronal polarity,12 we co-stained neural rosettes using specific antibodies for neuronal marker (TUJ1) and polarity marker (ZO1) (Figure 3A). First, we found that both mutant and CRISPR-Del neural rosettes were different from CTRLs (Figures 3A, 3B, and 3D, yellow dotted loop), with the diameters of the rosettes being significantly increased (Figures 3A, 3B, and S3A) and the number of rosettes per 100 cells being significantly decreased (Figure S3B), suggesting few and unstable, loose neural rosette formation after MARK2 loss. CTRL rosettes exhibited well-defined self-organization characterized by apical-basal polarity and constriction. TUJ1 staining revealed radial distribution surrounding the inner lumen, while ZO1 staining demonstrated clear localization within the inner lumen, which is consistent with previous reports (Figure 3A).36 In contrast, both mutant and CRISPR-Del rosettes exhibited loose structure, characterized by imperfectly self-organized TUJ1 staining and degraded ZO1 staining, along with the absence of an inner lumen (Figure 3A). These results demonstrated that MARK2 loss led to aberrant polarity of neuroepithelial cells, forming dis-organized, few, and loose-structured neural rosettes. Further comparation of the diameters of different iPSC-derived neurospheres revealed a significant reduction in size in mutant and CRISPR-Del groups (Figures 3C and S3D). We then checked the activated NPC population in rosettes using SOX137 and observed a significant decrease in the number of SOX1+ cells within mutant and isogenic neural rosettes (Figures 3B and S3C). Both the size of neurosphere and number of activated NPCs indicated aberrant self-renewal of NPC pool following MARK2 loss.
Figure 3.
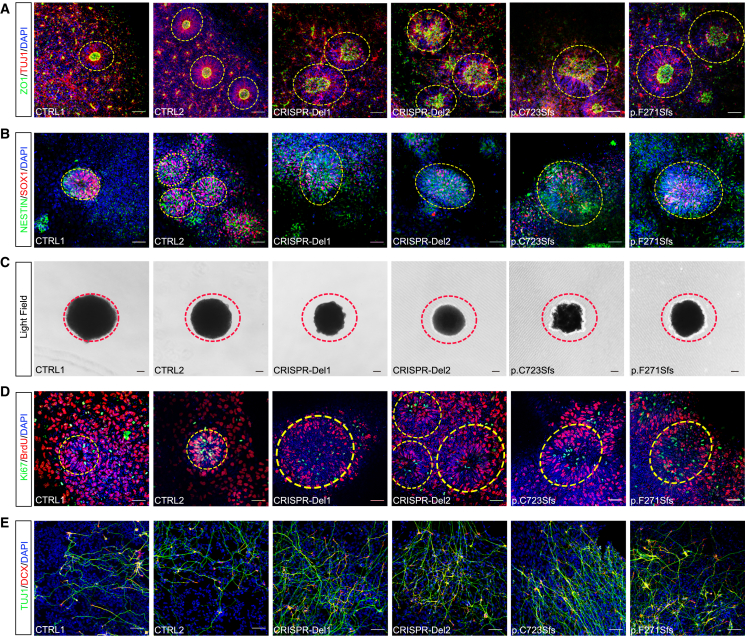
MARK2 variant leads to aberrant polarity in iPSC-derived neural rosettes and imbalanced proliferation and differentiation in iPSC-derived NPCs
(A) Representative immunofluorescence images of TUJ1 (red) and ZO1 (green) in different iPSC-derived neural rosettes on the 10th day after neural induction. CTRL1 and CTRL2, 2 independent healthy adults without MARK2 variants; p.C723Sfs and p.F271Sfs, 2 affected individuals with LOF MARK2 variants; CRISPR-Del1 and CRISPR-Del2, 2 isogenic MARK2 deletions produced by the CRISPR-Cas9 editing technology.
(B) Representative immunofluorescence images of SOX1 (red) and NESTIN (green) in different iPSC-derived neural rosettes on the 5th day after neural induction.
(C) Representative images of different iPSC-derived neurospheres on the 5th day after neural induction.
(D) Representative immunofluorescence images of BrdU (red) and Ki67 (green) in different iPSC-derived neural rosettes on the 4th day after neural induction.
(E) Representative immunofluorescence images of DCX (red) and TUJ1 (green) in different iPSC-derived NPCs on the 13th day after neural induction. Scale bar = 50 μm. The quantification and statistical analysis were showed in Figure S3.
In the iPSC-derived NPC stage, we conducted BrdU incorporation assays and Ki67 staining to assess proliferation capability (Figure 3D). The results demonstrated that there were significantly fewer BrdU+ cells (Figure S3E) and Ki67+ cells (Figure S3F) in mutant or CRISPR-Del NPCs compared to CTRLs. While the proliferation and differentiation of NPCs maintain a dynamic balance, we also evaluated the differentiation efficiency of NPCs using 2 neural markers, TUJ1 and DCX (Figure 3E). DCX+ cells (Figure S3G) and TUJ1+ cells (Figure S3H) in mutant and CRISPR-Del NPCs also showed a significant increase compared to CTRLs. Together, these results suggested that MARK2 loss led to aberrant polarity and dis-organization in neural rosettes, thereby suppressing proliferation but promoting differentiation in NPCs.
Mark2 loss leads to abnormal cortical partition, ASD-like behaviors, and impaired memory in mice
To recapitulate the cellular phenotypes of an affected proband’s iPSC-derived rosettes, we generated Mark2+/− (HET) mice utilizing the CRISPR-Cas9 system (Figures S4A, S4B, and S4D) to mimic Mark2 loss in vivo. Consistently, the BrdU incorporation assay of mouse cortical NPCs (mNPCs) at E18.5 revealed significantly decreased BrdU+ cells but increased TUJ1+ cells in HET mice compared with WT ones (Figures S4E, S4H, and S4I), indicating the imbalanced proliferation and differentiation in early cortical NPCs upon Mark2 loss. Additionally, downregulated SOX1+ cells were also observed in the cortices of HET mice (Figures S4E and S4G), indicating altered neural self-renewal upon Mark2 loss. As CTIP2 is expressed primarily in layer V, and SATB2 is expressed primarily in layers II/III,38 we compared the thickness and the density of CTIP2+ cells and SATB2+ cells in mouse cortex. Whole density of CTIP2+ cells and SATB2+ cell was significantly increased in HET mice, although whole cortical thickness was similar (Figures S4F, S4J, S4K, and S4L). Particularly, the ratio of CTIP2+ cells and SATB2+ cells in layers V and VI showed significant increase (Figures S4F, S4M, and S4N). These results indicated abnormal cortical formation and partition upon Mark2 loss.
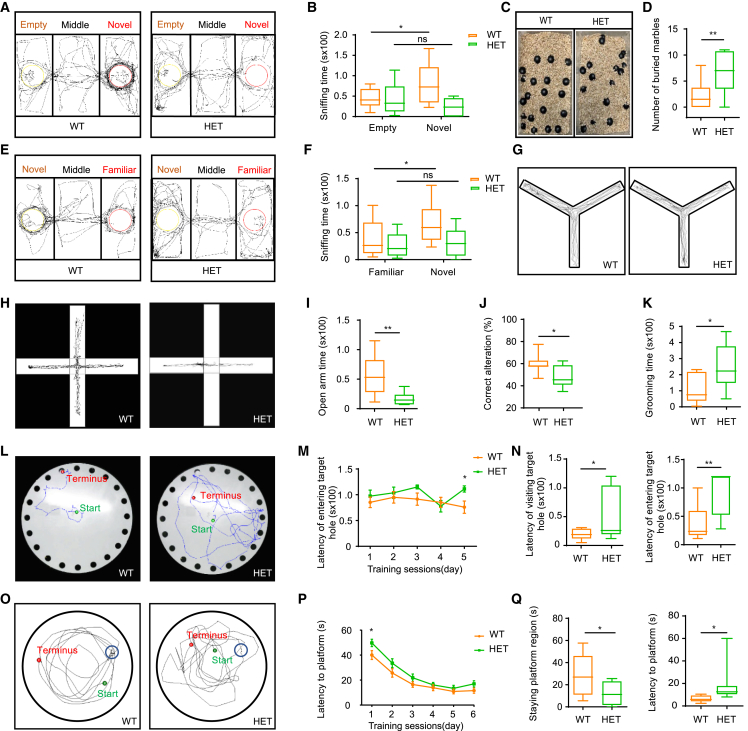
In order to determine whether adult Mark2+/− mice exhibit the features of ASD and other NDDs observed in affected individuals, we performed a series of behavioral tests for Mark2+/− mice. Although HET mice had lower body weights (Figures S4B and S4C), they showed normal exploratory and locomotor activity in the open field tests (Figure S5A), as the total distance traveled (Figure S5C) and average speed (Figure S5D) were not different between HET mice and WT mice. Subsequently, we performed the three-chamber test to evaluate whether the mice exhibited the deficits in sociability and preference for novelty observed in ASD individuals.39 In the social approach test (Figures 4A and 4B), WT mice spent significantly less time in the empty cage than in the cage containing the novel mice, while HET mice spent similar amounts of times between 2 cages. In the social novelty test (Figures 4E and 4F), WT mice spent significantly less time with the familiar mice than with the novel mice. Conversely, HET mice spent similar amounts of time with the familiar and novel mice. These social behavior tests demonstrated that HET mice presented reduced social motivation and pursuit of novelty. The marble-burying test and grooming test were used to evaluate stereotyped behaviors of ASD in rodents. We found that HET mice buried more marbles (Figures 4C and 4D) and spent more time grooming (Figure 4K) than WT mice. Additionally, in the elevated plus maze test, which was used to assess anxiety-like behavior of ASD, HET mice spent less time in the open arms than WT mice (Figures 4H and 4I). Together, these data suggested that Mark2 loss in mice led to specific social deficits, stereotyped behavior, and anxiety that recapitulated the features of ASD in individuals with MARK2 variants.
Figure 4.
Mark2 loss in mice leads to ASD-like behavior and impaired memory
(A and E) Trajectories of Mark2+/+ (WT) and Mark2+/− (HET) mice in the three-chamber test (A, empty or novel; E, familiar or novel).
(B and F) Quantification analysis of sniffing time in the three-chamber test (B, empty or novel; F, familiar or novel).
(C, D, and K) Trajectories of WT and HET mice in the marble-burying test (C) and quantification analysis of the buried marbles (D) and grooming time (K).
(H and I) Trajectories of WT and HET mice in the elevated plus maze test (H) and quantification analysis of time spent in the open arms (I).
(G and J) Trajectories of WT and HET mice in the Y-maze test (G) and quantification analysis of the correct alternation (J).
(L–N) Trajectories of WT and HET mice in the Barnes maze test (L), quantification analysis of the latency of entering the target hole during the training sessions (M), and the latency of visiting target hole and of entering target hole during the test session (N).
(O–Q) Trajectories of WT and HET mice in the Morris water maze test (O), quantification analysis of the latency to find the platform during the training sessions (P), and the latency to finding the platform and staying platform quadrant during the test session (Q). WT = 9, HET = 9. The data were analyzed by Student’s t test; ∗p < 0.05 and ∗∗p < 0.01.
As affected individuals in our cohort also showed intellectual disability/developmental delay and language problems, we further assessed learning and memory capacity using the Y-maze (Figure 4G), the Barnes maze (Figure 4L), and the Morris water maze (Figure 4O). The Y-maze is designed to assess spatial memory and executive function. We found that the percentage of correct alterations made by HET mice was significantly decreased compared with WT mice (Figure 4J), indicating that HET mice exhibited abnormal spatial working memory. The results of the Barnes maze test suggested memory impairment in HET mice, as they spent significantly more time visiting (Figure 4N, left) and entering (Figures 4M and 4N, right) the target hole compared with WT ones. In the Morris water maze test (Figure 4O), HET mice showed a significantly longer latency to find the platform (Figures 4P and 4Q, right) and spent less time in the platform region when the platform was moved (Figure 4Q, left). Interestingly, during the training sessions of the Barnes maze and Morris water maze tests, the latencies of the HET mice were not significantly longer than those of the WT mice, suggesting similar learning abilities between the 2 genotypes. Nonetheless, we tested the recognition memory of mice using the novel object recognition test (Figure S5B) and found that HET mice spent similar amounts of time interacting with the novel object as WT mice (Figure S5E). Combined, these results implied that loss of Mark2 impaired spatial memory.
MARK2 loss disrupts early neurogenesis via the downregulation of WNT/β-catenin signaling pathway
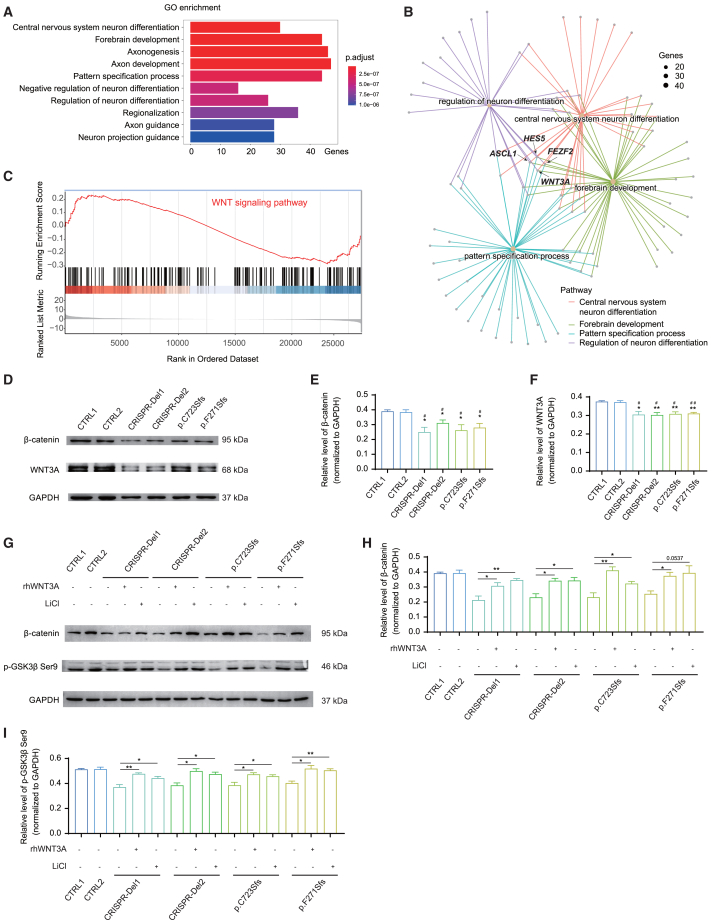
To further identify the molecular pathway of the MARK2 variant in early neurogenesis, we performed RNA-seq for iPSC-derived NPCs on the 12th day after neural induction (Figure S6A) to detect significant dysregulated genes (Figure S6B, adjusted p < 0.05, log2Fold change>1.5). GO enrichment analysis of differentially expressed genes (DEGs) revealed the biological functional change related to muscle development, ear development, neuro fate commitment/specification, axonogenesis, synapse development, or material transport with MARK2 loss (Figure S6C). GO enrichment analysis of downregulated genes are significantly involved in neuronal development (Figure 5A), including CNS neuron differentiation (GO: 0021953), forebrain development (GO: 0030900), axonogenesis (GO: 0007409), axon development (GO: 0061564), pattern specification process (GO: 0007389), negative regulation of neuron differentiation (GO: 0045665), regulation of neuron differentiation (GO: 0045664), regionalization (GO: 0003002), axon guidance (GO: 0007411), and neuron projection guidance (GO: 0097485). There are 4 downregulated genes, WNT3A, ASCL1, HES5, and FEZF2, involved in all these GO terms, especially the terms related to early neuronal development, and WNT3A is the top decreased gene (Figure 5B). GSEA using up-/downregulated genes also revealed significant inhibition of the WNT signaling pathway in mutant cells (Figures 5C and S6D). Besides WNT3A, other key molecules of WNT signal (WNT4, WNT7A, WNT7B, WNT8B, WNT10B) were also significantly downregulated (Figure S6E). Furthermore, significantly decreased WNT3A protein was validated for mutant and CRISPR-Del cells (Figures 5D and 5F) along with decreased β-catenin (Figures 5D and 5E). A previous study showed that WNT3A, as a critical ligand, plays an important role in the activation of the WNT signaling pathway and is essential for the proliferation and differentiation of NPCs.40 Hence, we hypothesized that MARK2 loss led to abnormal early neuronal development via inhibition of the WNT signaling pathway.
Figure 5.
iPSC-derived NPCs revealed decreased WNT signaling pathway due to MARK2 loss
(A) Gene Ontology (GO) analysis of differentially expressed genes (DEGs) between mutant iPSC-derived NPCs and CTRL ones show that the downregulated genes are enriched in multiple pathways related to early neuronal development.
(B) GO pathway network analysis showed the association of WNT3A with neuronal development, including neuron fate specification, forebrain development, central nervous system neuron differentiation, axonogenesis, and axon development. Size means number of genes involved in the specific pathway.
(C) The enrichment score and rank of the WNT/β-catenin signaling pathway from DEGs.
(D–F) Representative western blot images (D) and quantification analysis of β-catenin (E) and WNT3A (F) accumulation in mutant and CRISPR-Del iPSC-derived NPCs compared with CTRLs.
(G–I) Representative western blot images (G) and quantification analysis of β-catenin (H) and p-GSK3β-Ser9 (I) accumulation in mutant, CRISPR-Del iPSC-derived NPCs with/without rhWNT3A (100 ng/mL) or LiCI treatment (0.7 mM). The data of at least 3 independent experiments were analyzed by Student’s t test; ∗p < 0.05 and ∗∗p < 0.01(compared with CTRL1); #p < 0.05 and ##p < 0.01 (compared with CTRL2).
In order to confirm the relationship between WNT signal pathway inhibition and MARK2 loss, we treated mutant iPSC-NPCs with 100 ng/mL rhWNT3A. We found that rhWNT3A significantly increased the accumulation of p-GSK3β Ser9 (Figures 5G and 5I) and the accumulation of β-catenin in mutant iPSC-NPCs (Figures 5G and 5H). The rescue effect of rhWNT3A confirmed that MARK2 loss results in relative inactivation of the WNT signaling pathway.
Lithium reverses the molecular and cellular phenotypes of mutant iPSC-derived NPCs and abnormal cortical partition of HET mice by activating the WNT/β-catenin signaling pathway
Lithium is a known activator of the WNT/β-catenin signaling pathway41,42 and is widely prescribed for many behavioral disorders, such as bipolar disorder and schizoaffective disorder,43 which are closely associated with ASD. We added LiCl at 0.7 mM, a routinely prescribed dosage for individuals with bipolar disorder, to the culture medium of iPSC-derived NPCs.43,44 On the 10th day after neural induction, the protein levels of β-catenin, p-GSK3β Ser9 in mutant NPCs were significantly increased following LiCl treatment as similar to rhWNT3A treatment, even reaching the levels of control NPCs (Figures 5G, 5H, and 5I), suggesting WNT3A and p-GSK3β Ser9 as the target molecule of lithium rescue.
Considering the molecular effect of LiCl on WNT/β-catenin signaling activation, we replicated the IF experiment described above for LiCl-treated or rhWNT3A-treated mutant and CRISPR-Del iPSC-derived NPCs. Innovatively, the rescue effect of LiCl on the morphology of mutant neural rosettes was quite obvious, with the diameter of the neural rosettes being significantly decreased (Figures 6A and S7A) and the number of rosettes being significantly increased (Figures 6A and S7B). Also, correct localization of ZO1 and normal self-organization of TUJ1+ cells were seen in mutant neural rosettes with LiCl treatment (Figure 6A). Moreover, the numbers of BrdU+ (Figures 6B and S7C) and Ki67+ cells (Figures 6B and S7D) in mutant and CRISPR-Del NPCs were significantly increased, almost reaching those of control NPCs or rhWNT3A treatment. We further compared the differentiation efficiencies of mutant NPCs before and after LiCl or rhWNT3A treatment. The numbers of TUJ1+ (Figures 6C and S7F) and DCX+ cells (Figures 6C and S7E) were significantly decreased after LiCl treatment. Both molecular and cellular phenotypes revealed that lithium rescues the abnormal developmental trajectories of MARK2 loss via stimulation of the WNT signaling pathway.
Figure 6.
Lithium reverses the imbalance in the proliferation and differentiation of MARK2 mutant iPSC-derived NPCs by activating the WNT/β-catenin signaling pathway
(A) Representative immunofluorescence images of ZO1 (green) and TUJ1 (red) in different iPSC-derived neural rosettes. CTRL1 and CTRL2, 2 independent healthy adults without MARK2 variant; p.C723Sfs and p.F271Sfs, 2 affected individuals with LOF MARK2 variants; and CRISPR-Del1 and CRISPR-Del2, 2 isogenic MARK2 deletions produced by the CRISPR-Cas9 editing technology. Both CRISPR-Del and mutant iPSC-derived neural rosettes were treated with LiCl (0.7 mM) or rhWNT3A (100 ng/mL).
(B) Representative immunofluorescence images of Ki67 (green) and BrdU (red) in different iPSC-derived NPCs.
(C) Representative immunofluorescence images of TUJ1 (green) and DCX (red) in different iPSC-derived NPCs. Scale bar = 50 μm. The quantification and statistical analysis were showed in Figure S7.
To investigate the rescue effect of LiCl on cortical layer formation and partition in Mark2+/− mice, we treated pregnant female mice with 4.5 mg/L LiCl from E0.5 to E17.5 and analyzed proliferation and differentiation efficiency in the cortex at E18.5. We found that the number of BrdU+ cells (Figures S8A and S8C) was significantly increased in HET mice treated with LiCl, but the number of TUJ1+ cells was significantly decreased (Figures S8A and S8E). An obvious increase in the number of SOX1+ cells was also observed in LiCl-treated HET mice compared with untreated HET mice (Figures S8A and S8D). These data demonstrated the rescue effects of lithium on the proliferation and activation of NPCs in mice with Mark2 loss. We further compared the thickness of the cortical layers in HET mice before and after LiCl treatment using CTIP2 and SATB2 as markers of different cortex layers (Figure S8B). The results showed that LiCl treatment significantly rescued the abnormal formation of the mouse cortex, as the density of the CTIP2+ and SATB2+ cell in whole cortex (Figures S8F and S8G) and in specific cortical layers (Figures S8F and S8G) was similar between LiCl-treated HET and WT mice. These results were consistent with the rescue effects observed in Mut iPSC-derived NPCs.
Together, these in vitro and in vivo results demonstrated that pharmacological reactivation of the WNT/β-catenin signaling pathway by lithium can rescue aberrant cellular phenotypes in NPC with MARK2 loss, thereby supporting a potential molecular link between MARK2 loss and WNT/β-catenin signaling.
Discussion
In our study, we collected a multi-institutional global cohort of 31 individuals with clinically relevant MARK2 variants and presented comprehensive clinical features and variant profiles of MARK2-associated ASD. Affected individuals frequently exhibit typical facial features (65%), including a prominent forehead, a broad nasal root, and larger or dysplastic ears in addition to vision problems; male genitourinary involvement and occasional skeletal involvement implying that MARK2-associated ASD frequently presents with other physical and morphologic comorbidities. Eighty percent of MARK2 variants are LoF variants, supporting the idea that MARK2 loss contributes to the development of ASD, as initially suggested from 2 large sequencing studies.18,45
We also observed high frequency of neurological comorbidities including intellectual disability/developmental delay (100%), speech-language problems (100%), and motor delay (62%). The IQ scores of 7 ASD individuals with LoF variants of MARK2 from Zhou et al. reported were averagely 65.2, similar to that of ASD individuals with CHD8/SHANK3 variants.18 In addition, 1 individual in our cohort (P8) had not met ASD criteria until her last interview, although she presented with mixed receptive-expressive language disorder (personal communication), implying that MARK2 is the candidate gene of NDDs, as previously reported.45 Besides MARK2, another polarity gene of the MARK family, MARK1, has been recognized as a susceptibility gene for autism.46 Our study also expanded polarity-related genes in ASD etiology.
MARK2 contains 5 domains, each with distinct functions necessary for its normal activity.10 Previous studies have reported that MARK2 mutants lacking the kinase domain led to neuronal polarity abrogation in hippocampal neurons.12 Furthermore, mutations in the activating loop of the kinase domain, p.Thr208Ala and p.Ser212Ala, result in abnormal differentiation and neurite extension in neuroblastoma cells,16 suggesting the critical role of the kinase domain in neuronal development. The KA1 domain, locating in the C terminus of MARK2, is conserved from yeast to humans and exerts an autoinhibitory effect on the kinase activity of MARK2 by blocking peptide substrate binding to the N and C lobes of the kinase domain.11 Pathogenic variants in the KA1 domain impair this autoinhibition of MARK/PAR1 kinase activity.33 We identified 4 variants in the ATP-binding pocket or the activation loop of kinase domain and 2 variants in the autoregulatory position of KA1 domain. Using integrative bioinformatics approaches, we predicted that these variants destabilize the interaction between these 2 domains and lead to loss of the autoregulation of MARK2. Further, our in vitro-transfected cell model confirmed that missense variants result in decreased MARK2 expression comparable to LoF variants. Additionally, the phenotype associated with the missense variants was not significantly different from that of LoF variants. These data suggest that the missense variants impair the kinase or autoinhibitory activity of MARK2 following protein destabilization, resulting in similar cellular and molecular consequences as MARK2 loss.
Multiple neurodevelopmental processes, including proliferation/neurogenesis, migration, neurite outgrowth, morphogenesis of dendrites and dendritic spines, and synaptogenesis and gliogenesis, have been reported to be altered in ASD.47,48,49,50 Protein-protein interaction (PPI) networks of ASD-associated genes also confirmed the convergent pathways underlying ASD, including synaptic development, mitochondrial or metabolic processes, and WNT and MAPK signaling.51,52,53 In order to explore the pathophysiological mechanism of MARK2 loss underlying ASD, we generated mutant human iPSCs and described the cellular phenotypes of mutant neurons at early neural development such as fewer depolarized neural rosettes, smaller neurosphere formation, imbalanced proliferation, and differentiation in NPCs, which were similar to previous iPSC studies from other classic ASDs like fragile X syndrome (MIM: 300624), Rett syndrome (MIM: 312750), and SHANK3 deletion.48,49,50 Our results suggested that abnormal polarity and proliferation in mutant NPCs is a prerequisite for aberrant brain function in individuals with ASD. Furthermore, we have identified deficits in spatial learning, memory, and social preference among the Mark2+/− mice. Impaired spatial learning and memory have been previously documented in Mark2+/− mice.54,55 Besides impaired social behavior, Caiola et al. reported decreased anxiety-like behavior in the plus maze and increased seizure susceptibility.55 Although we didn’t study seizure phenotypes in knockout mice, a high frequency of seizures/epilepsy (46.6%) was reported in our cohort.
The WNT signaling pathway mediates neurogenesis via several biological functions, including self-renewal, proliferation, and differentiation.40,56,57,58,59 For example, WNT3A and WNT7A are indispensable for maintaining self-renewal and stimulating the proliferation of neural stem cells,56,57 and WNT3A also plays a critical role in neural fate commitment.56 The regulatory role of β-catenin signaling in the balance between cell proliferation and differentiation in the spinal cord has also been reported.60 Abnormal WNT signaling pathway activity has been reported in mouse models or patient-derived iPSCs carrying mutations in other high-confidence ASD-associated genes, such as SHANK3 and TBR1 (MIM: 604616).42,61 The transcriptomic profiling of our mutant iPSC-derived NPCs revealed a reduced WNT/β-catenin signaling pathway, reasonably explaining imbalanced proliferation and differentiation in mutant NPCs. Decreased WNT3A and SOX1, and aberrant self-renewal, was seen in mutant neural rosettes, and developmental phenotypes in mutant NPCs were consistent with that of WNT3A/WNT7A depletion.56,57,59,60,62 Furthermore, abnormal phenotypes of mutant iPSC-derived NPCs were completely reversed in vitro by treatment with exogenous rhWNT3A. Our study associated the WNT/β-catenin signaling pathway with MARK2-related ASD.
To date, several genes encoding members in the WNT/β-catenin signaling pathway, including PTEN (MIM: 608309), ADNP (MIM: 611386), ARID1B (MIM: 614556), CHD8 (MIM: 610528),5,6,53,63 the activator of β-catenin signaling CTNNB1 (MIM: 116806),64 and WNT1 (MIM: 164820),65 have been associated with ASD with high confidence. β-catenin, as a highly conserved armadillo repeat protein family member, is encoded by CTNNB1, and LoF variants of CTNNB1 cause a broad ASD phenotype.64,66 We compared the clinical features of 120 individuals with germline likely pathogenic/pathogenic CTNNB1 variants64 with those of the ASD individuals studied herein (Table S6). The top rank features of individuals with the CTNNB1 variant are intellectual disability/developmental delay (94.1%), motor delay (93.7%), delayed speech and language development/ASD (90.4%), and mild/severe eye problems (87.5%). The phenotypic similarity between 2 different cohorts also suggested the molecular role of the WNT/β-catenin signaling pathway in MARK2-related ASD.
Lithium, as a mood stabilizer, has been routinely prescribed to treat bipolar disorder for decades.43,44 Previous in vitro and in vivo studies have proven that lithium directly activates the WNT/β-catenin signaling pathway.42,57,67,68,69,70 By activating the WNT/β-catenin signaling pathway, lithium promotes the proliferation of NPCs and improves behavioral performance in a mouse model of Down syndrome (MIM: 190685)67 and spatial memory impairment and neurodegeneration in a mouse model of Alzheimer disease (MIM: 104300).70 Lithium can also directly enhance the proliferation of hippocampal progenitors in vitro in a dose-dependent manner.68 The ability of lithium to rescue spine and synaptic defects has been reported in conditional Tbr1+/− adult mice presenting ASD-like behaviors42 and APP/PS1 mice presenting impaired learning and memory.69 Siavash Fazel Darbandi generated ASD mice with conditional Tbr1layer6 knockout and found that both cellular and behavioral abnormalities, including immature dendritic spines reducing synaptic density and decreased social interactions between young mice, were rescued with LiCl treatment.42 In this study, we administered the clinically used dosage of lithium31,33 to mutant iPSCs and Mark2+/− (HET) mice. Abnormal cellular phenotypes in mutant NPCs, including alterations in the size of the neurosphere, rosette formation, and the proliferation and differentiation of NPCs, were reversed by lithium treatment. Considering that ASD is an early brain malformation, we fed a LiCl-treated diet to pregnant Mark2+/− mice and studied the proliferation and differentiation of cortical neurons in Mark2+/− mouse fetuses. Abnormal cortex layer formation was also reversed. However, it is important to clarify that LiCl treatment was not directly applied to the Mark2+/− mice themselves, and the rescue evidence is indirect. In the future, therapeutic roles of LiCl for affected individuals with MARK2-associated ASD, particularly for juveniles or adults, need further experimental confirmation.
Conclusion
Our studies deciphered the clinical features and variant spectrum of MARK2-related ASD. Using human iPSC-derived NPCs in vitro and Mark2+/− mice in vivo, we elucidated the cellular phenotypes and molecular mechanism of MARK2 loss during early neuronal development, which are associated with downregulation of the WNT/β-catenin signaling pathway. Moreover, we observed that lithium can reverse cellular phenotype in mutant iPSC-NPCs and abnormal cortical partition development in Mark2+/− mice via activation of the WNT/β-catenin signaling pathway, providing a potential treatment for MARK2-related ASD.
Data and code availability
The WES data will not be made publicly available, as they contain private information, but are available from the corresponding author upon request. All variants have also been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/, SCV005081699-SCV005081726). The raw sequence data of RNA-seq reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in the National Genomics Data Center,71 China National Center for Bioinformation/Beijing Institute of Genomics, and Chinese Academy of Sciences and are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human with accession number GSA-Human: HRA003632. The code used in this study is available upon request.
Acknowledgments
We thank all the families who participated in this study. We also thank James F. Gusella for his valuable comments and suggestions, Brendan C. Lanpher, M.D., for his involvement in patient care, and Filippo Pinto e Vairo, M.D., Ph.D., for his assistance with IRB logistics and patient consent. This work was supported by grants from the National Key Research and Development Program of China (project 2021YFA1101402), the Strategic Priority Research Program of the Chinese Academy of Sciences (nos. XDA16010300/XDA16021400), and the Open Project Program of the State Key Laboratory of Stem Cell and Reproductive Biology. This study was also supported by grants from the National Science Foundation of China (82371868, 82271428, 31900690, 82201314, and 82471194), Beijing Natural Science Foundation (7202019), Beijing Finance Bureau (CIP2024-0040), and Guangxi Science and Technology Program Project (Guike AB17195011).
Author contributions
M.G., C.-M.L., and X.C. were responsible for conception and design. M.G., X.C., C.-M.L., J.L., and Y.L. contributed to manuscript writing, and M.V.M.B.W. and M.T.Z. helped in manuscript editing. X.C., Z.Q., M.V.M.B.W., J.L., and H.L. managed the analysis and interpretation of all clinical/genetic data. J.L. and H.L. performed the western blot and qPCR experiments for PBMCs and in vitro HEK 293T/HeLa transfection assay for the spicing and missense variants. M.G., Y.L., J.L., Q.L., and Chen L. performed the iPSC-related experiments. M.G., Y.L., and H.L. conducted the animal experiments. M.T.Z., N.H., N.R.D., and R.U. performed structural modeling. H.Z. helped in iPSC culture and neural differentiation and drug treatment for iPSCs and HET mice. M.G. analyzed and visualized the RNA-seq data. All authors approved the final manuscript.
The following partners conducted individual recruitment and clinical/genetic information and photo collection (listed in alphabetical order): A.S.A.C., B.C., B.P., B.R.S., B.K., Carrie L., C. Ren, C.Z., C.C., C. Rieubland , D.C.K., D.B., E.v.B., E.W.K., F.T.M.-T., H.S., H.O., H.X., J.L.M., J.Z., J.A.M.-R., J.G., J.P., K.L.v.G., K.P., L.E.K., L.E., M.-J.H.v.d.B., M.W., M.V.M.B.W., M.J.F., A.M.I., M.D.A., C.M., N.J.B., O.S., P.Z., P.B., R.H.v.J., S.D.M., S.E., S.M.H., S.S.H., T.W., V.W., W.F., W.-H.T., W.V.K., X.W., Y.Y., Y.S., Y.R., Z.Q., and L.R.
Declaration of interests
The authors declare no competing interests.
Published: October 16, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.09.006.
Contributor Information
Chang-Mei Liu, Email: liuchm@ioz.ac.cn.
Xiaoli Chen, Email: xiaolichen@pumc.edu.cn.
Supplemental information
References
- 1.Lai M.C., Lombardo M.V., Baron-Cohen S. Autism. Lancet. 2014;383:896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H., Xu X., Yan W., Zou X., Wu L., Luo X., Li T., Huang Y., Guan H., Chen X., et al. Prevalence of Autism Spectrum Disorder in China: A Nationwide Multi-center Population-based Study Among Children Aged 6 to 12 Years. Neurosci. Bull. 2020;36:961–971. doi: 10.1007/s12264-020-00530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baio J., Wiggins L., Christensen D.L., Maenner M.J., Daniels J., Warren Z., Kurzius-Spencer M., Zahorodny W., Robinson Rosenberg C., White T., et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR. Surveill. Summ. 2018;67:1–23. doi: 10.15585/mmwr.ss6706a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tammimies K., Marshall C.R., Walker S., Kaur G., Thiruvahindrapuram B., Lionel A.C., Yuen R.K.C., Uddin M., Roberts W., Weksberg R., et al. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 5.Schaaf C.P., Betancur C., Yuen R.K.C., Parr J.R., Skuse D.H., Gallagher L., Bernier R.A., Buchanan J.A., Buxbaum J.D., Chen C.A., et al. A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat. Rev. Genet. 2020;21:367–376. doi: 10.1038/s41576-020-0231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L., et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruzzo E.K., Perez-Cano L., Jung J.Y., Wang L.K., Kashef-Haghighi D., Hartl C., Singh C., Xu J., Hoekstra J.N., Leventhal O., et al. Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell. 2019;178:850–866.e826. doi: 10.1016/j.cell.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J., Slowikowski K., Lek M., Karczewski K.J., Cutler D.J., Devlin B., Roeder K., et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J.M., Satterstrom F.K., Peng M., Brand H., Collins R.L., Dong S., Wamsley B., Klei L., Wang L., Hao S.P., et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54:1320–1331. doi: 10.1038/s41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drewes G., Ebneth A., Preuss U., Mandelkow E.M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 11.Emptage R.P., Lemmon M.A., Ferguson K.M., Marmorstein R. Structural Basis for MARK1 Kinase Autoinhibition by Its KA1 Domain. Structure. 2018;26:1137–1143.e3. doi: 10.1016/j.str.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y.M., Wang Q.J., Hu H.S., Yu P.C., Zhu J., Drewes G., Piwnica-Worms H., Luo Z.G. interme kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc. Natl. Acad. Sci. USA. 2006;103:8534–8539. doi: 10.1073/pnas.0509955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terabayashi T., Itoh T.J., Yamaguchi H., Yoshimura Y., Funato Y., Ohno S., Miki H. Polarity-regulating kinase partitioning-defective 1/microtubule affinity-regulating kinase 2 negatively regulates development of dendrites on hippocampal neurons. J. Neurosci. 2007;27:13098–13107. doi: 10.1523/JNEUROSCI.3986-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapir T., Shmueli A., Levy T., Timm T., Elbaum M., Mandelkow E.M., Reiner O. Antagonistic effects of doublecortin and MARK2/Par-1 in the developing cerebral cortex. J. Neurosci. 2008;28:13008–13013. doi: 10.1523/JNEUROSCI.2363-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sapir T., Sapoznik S., Levy T., Finkelshtein D., Shmueli A., Timm T., Mandelkow E.M., Reiner O. Accurate balance of the polarity kinase MARK2/Par-1 is required for proper cortical neuronal migration. J. Neurosci. 2008;28:5710–5720. doi: 10.1523/JNEUROSCI.0911-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biernat J., Wu Y.Z., Timm T., Zheng-Fischhöfer Q., Mandelkow E., Meijer L., Mandelkow E.M. Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol. Biol. Cell. 2002;13:4013–4028. doi: 10.1091/mbc.02-03-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimura Y., Terabayashi T., Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol. Cell Biol. 2010;30:2206–2219. doi: 10.1128/MCB.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X., Feliciano P., Shu C., Wang T., Astrovskaya I., Hall J.B., Obiajulu J.U., Wright J.R., Murali S.C., Xu S.X., et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat. Genet. 2022;54:1305–1319. doi: 10.1038/s41588-022-01148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejaver V., Byrne A.B., Feng B.J., Pagel K.A., Mooney S.D., Karchin R., O'Donnell-Luria A., Harrison S.M., Tavtigian S.V., Greenblatt M.S., et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am. J. Hum. Genet. 2022;109:2163–2177. doi: 10.1016/j.ajhg.2022.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M., ClinGen Sequence Variant Interpretation Working Group ClinGen SVI Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallberg M., Margaryan G., Wang S., Ma J., Xu J. RaptorX server: a resource for template-based protein structure modeling. Methods Mol. Biol. 2014;1137:17–27. doi: 10.1007/978-1-4939-0366-5_2. [DOI] [PubMed] [Google Scholar]
- 24.Panneerselvam S., Marx A., Mandelkow E.M., Mandelkow E. Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/Par-1. Structure. 2006;14:173–183. doi: 10.1016/j.str.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 25.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.Y., Pieper U., Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2006;15 doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Durme J., Delgado J., Stricher F., Serrano L., Schymkowitz J., Rousseau F. A graphical interface for the FoldX forcefield. Bioinformatics. 2011;27:1711–1712. doi: 10.1093/bioinformatics/btr254. [DOI] [PubMed] [Google Scholar]
- 28.Dinkel H., Michael S., Weatheritt R.J., Davey N.E., Van Roey K., Altenberg B., Toedt G., Uyar B., Seiler M., Budd A., et al. ELM--the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F., Liang C., Li Z., Zhao S., Yuan H., Yao R., Qin Z., Shangguan S., Zhang S., Zou L.P., et al. Haplotype-specific MAPK3 expression in 16p11.2 deletion contributes to variable neurodevelopment. Brain. 2023;146:3347–3363. doi: 10.1093/brain/awad071. [DOI] [PubMed] [Google Scholar]
- 30.Chen C., Jiang P., Xue H., Peterson S.E., Tran H.T., McCann A.E., Parast M.M., Li S., Pleasure D.E., Laurent L.C., et al. Role of astroglia in Down's syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat. Commun. 2014;5:4430. doi: 10.1038/ncomms5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moravcevic K., Mendrola J.M., Schmitz K.R., Wang Y.H., Slochower D., Janmey P.A., Lemmon M.A. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann M.T., Urrutia R., Oliver G.R., Blackburn P.R., Cousin M.A., Bozeck N.J., Klee E.W. Molecular modeling and molecular dynamic simulation of the effects of variants in the TGFBR2 kinase domain as a paradigm for interpretation of variants obtained by next generation sequencing. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N.D., Tabar V., Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hribkova H., Grabiec M., Klemova D., Slaninova I., Sun Y.M. Calcium signaling mediates five types of cell morphological changes to form neural rosettes. J. Cell Sci. 2018;131 doi: 10.1242/jcs.206896. [DOI] [PubMed] [Google Scholar]
- 37.Venere M., Han Y.G., Bell R., Song J.S., Alvarez-Buylla A., Blelloch R. Sox1 marks an activated neural stem/progenitor cell in the hippocampus. Development. 2012;139:3938–3949. doi: 10.1242/dev.081133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fame R.M., MacDonald J.L., Macklis J.D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng Y., Wang Z.M., Tan W., Wang X., Li Y., Bai B., Li Y., Zhang S.F., Yan H.L., Chen Z.L., et al. Partial loss of psychiatric risk gene Mir137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat. Neurosci. 2018;21:1689–1703. doi: 10.1038/s41593-018-0261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciani L., Salinas P.C. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 41.Lenox R.H., Wang L. Molecular basis of lithium action: integration of lithium-responsive signaling and gene expression networks. Mol. Psychiatry. 2003;8:135–144. doi: 10.1038/sj.mp.4001306. [DOI] [PubMed] [Google Scholar]
- 42.Fazel Darbandi S., Robinson Schwartz S.E., Pai E.L.L., Everitt A., Turner M.L., Cheyette B.N.R., Willsey A.J., State M.W., Sohal V.S., Rubenstein J.L.R. Enhancing WNT Signaling Restores Cortical Neuronal Spine Maturation and Synaptogenesis in Tbr1 Mutants. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miura T., Noma H., Furukawa T.A., Mitsuyasu H., Tanaka S., Stockton S., Salanti G., Motomura K., Shimano-Katsuki S., Leucht S., et al. Comparative efficacy and tolerability of pharmacological treatments in the maintenance treatment of bipolar disorder: a systematic review and network meta-analysis. Lancet Psychiatr. 2014;1:351–359. doi: 10.1016/S2215-0366(14)70314-1. [DOI] [PubMed] [Google Scholar]
- 44.Fransson F., Werneke U., Harju V., Öhlund L., de Man Lapidoth J., Jonsson P.A., Stegmayr B., Renberg E.S., Ott M. Kidney function in patients with bipolar disorder with and without lithium treatment compared with the general population in northern Sweden: results from the LiSIE and MONICA cohorts. Lancet Psychiatr. 2022;9:804–814. doi: 10.1016/S2215-0366(22)00265-6. [DOI] [PubMed] [Google Scholar]
- 45.Hamanaka K., Miyake N., Mizuguchi T., Miyatake S., Uchiyama Y., Tsuchida N., Sekiguchi F., Mitsuhashi S., Tsurusaki Y., Nakashima M., et al. Large-scale discovery of novel neurodevelopmental disorder-related genes through a unified analysis of single-nucleotide and copy number variants. Genome Med. 2022;14:40. doi: 10.1186/s13073-022-01042-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maussion G., Carayol J., Lepagnol-Bestel A.M., Tores F., Loe-Mie Y., Milbreta U., Rousseau F., Fontaine K., Renaud J., Moalic J.M., et al. Convergent evidence identifying MAP/microtubule affinity-regulating kinase 1 (MARK1) as a susceptibility gene for autism. Hum. Mol. Genet. 2008;17:2541–2551. doi: 10.1093/hmg/ddn154. [DOI] [PubMed] [Google Scholar]
- 47.Chiola S., Edgar N.U., Shcheglovitov A. iPSC toolbox for understanding and repairing disrupted brain circuits in autism. Mol. Psychiatry. 2022;27:249–258. doi: 10.1038/s41380-021-01288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K.Y., Hysolli E., Park I.H. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi F., Danko T., Botelho S.C., Patzke C., Pak C., Wernig M., Südhof T.C. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science. 2016;352 doi: 10.1126/science.aaf2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doers M.E., Musser M.T., Nichol R., Berndt E.R., Baker M., Gomez T.M., Zhang S.C., Abbeduto L., Bhattacharyya A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D., et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnett A.B., Wang T., Eichler E.E., Bernier R.A. Reflections on the genetics-first approach to advancements in molecular genetic and neurobiological research on neurodevelopmental disorders. J. Neurodev. Disord. 2021;13:24. doi: 10.1186/s11689-021-09371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iossifov I., O'Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E., et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segu L., Pascaud A., Costet P., Darmon M., Buhot M.C. Impairment of spatial learning and memory in ELKL Motif Kinase1 (EMK1/MARK2) knockout mice. Neurobiol. Aging. 2008;29:231–240. doi: 10.1016/j.neurobiolaging.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 55.Caiola H.O., Wu Q., Soni S., Wang X.F., Monahan K., Pang Z.P., Wagner G.C., Zhang H. Neuronal connectivity, behavioral, and transcriptional alterations associated with the loss of MARK2. bioRxiv. 2023 doi: 10.1101/2023.12.05.569759. Preprint at. [DOI] [PubMed] [Google Scholar]
- 56.Lie D.C., Colamarino S.A., Song H.J., Désiré L., Mira H., Consiglio A., Lein E.S., Jessberger S., Lansford H., Dearie A.R., Gage F.H. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 57.Qu Q., Sun G., Li W., Yang S., Ye P., Zhao C., Yu R.T., Gage F.H., Evans R.M., Shi Y. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat. Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosso S.B., Sussman D., Wynshaw-Boris A., Salinas P.C. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 59.Ciani L., Boyle K.A., Dickins E., Sahores M., Anane D., Lopes D.M., Gibb A.J., Salinas P.C. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca(2)(+)/Calmodulin-dependent protein kinase II. Proc. Natl. Acad. Sci. USA. 2011;108:10732–10737. doi: 10.1073/pnas.1018132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zechner D., Fujita Y., Hülsken J., Müller T., Walther I., Taketo M.M., Crenshaw E.B., 3rd, Birchmeier W., Birchmeier C. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev. Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 61.Qin L., Ma K., Wang Z.J., Hu Z., Matas E., Wei J., Yan Z. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat. Neurosci. 2018;21:564–575. doi: 10.1038/s41593-018-0110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hall A.C., Lucas F.R., Salinas P.C. Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell. 2000;100:525–535. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 63.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kayumi S., Pérez-Jurado L.A., Palomares M., Rangu S., Sheppard S.E., Chung W.K., Kruer M.C., Kharbanda M., Amor D.J., McGillivray G., et al. Genomic and phenotypic characterization of 404 individuals with neurodevelopmental disorders caused by CTNNB1 variants. Genet. Med. 2022;24:2351–2366. doi: 10.1016/j.gim.2022.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin P.M., Yang X., Robin N., Lam E., Rabinowitz J.S., Erdman C.A., Quinn J., Weiss L.A., Hamilton S.P., Kwok P.Y., et al. A rare WNT1 missense variant overrepresented in ASD leads to increased Wnt signal pathway activation. Transl. Psychiatry. 2013;3 doi: 10.1038/tp.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butz S., Stappert J., Weissig H., Kemler R. Plakoglobin and beta-catenin: distinct but closely related. Science. 1992;257:1142–1144. doi: 10.1126/science.257.5073.1142-a. [DOI] [PubMed] [Google Scholar]
- 67.Contestabile A., Greco B., Ghezzi D., Tucci V., Benfenati F., Gasparini L. Lithium rescues synaptic plasticity and memory in Down syndrome mice. J. Clin. Invest. 2013;123:348–361. doi: 10.1172/JCI64650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wexler E.M., Geschwind D.H., Palmer T.D. Lithium regulates adult hippocampal progenitor development through canonical Wnt pathway activation. Mol. Psychiatry. 2008;13:285–292. doi: 10.1038/sj.mp.4002093. [DOI] [PubMed] [Google Scholar]
- 69.Xiang J., Ran L.Y., Zeng X.X., He W.W., Xu Y., Cao K., Dong Y.T., Qi X.L., Yu W.F., Xiao Y., Guan Z.Z. LiCl attenuates impaired learning and memory of APP/PS1 mice, which in mechanism involves alpha7 nAChRs and Wnt/beta-catenin pathway. J. Cell Mol. Med. 2021;25:10698–10710. doi: 10.1111/jcmm.17006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toledo E.M., Inestrosa N.C. Activation of Wnt signaling by lithium and rosiglitazone reduced spatial memory impairment and neurodegeneration in brains of an APPswe/PSEN1DeltaE9 mouse model of Alzheimer's disease. Mol. Psychiatry. 2010;15:272–285. doi: 10.1038/mp.2009.72. [DOI] [PubMed] [Google Scholar]
- 71.CNCB-NGDC Members and Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022;50:D27–D38. doi: 10.1093/nar/gkab951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The WES data will not be made publicly available, as they contain private information, but are available from the corresponding author upon request. All variants have also been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/, SCV005081699-SCV005081726). The raw sequence data of RNA-seq reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in the National Genomics Data Center,71 China National Center for Bioinformation/Beijing Institute of Genomics, and Chinese Academy of Sciences and are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human with accession number GSA-Human: HRA003632. The code used in this study is available upon request.