Abstract
Emerging evidence suggests that cancer cells may disseminate early, prior to the formation of traditional macro-metastases. However, the mechanisms underlying the seeding and transition of early disseminated cancer cells (DCCs) into metastatic tumors remain poorly understood. Through single-cell RNA sequencing, we show that early lung DCCs from esophageal squamous cell carcinoma (ESCC) exhibit a trophoblast-like ‘tumor implantation’ phenotype, which enhances their dissemination and supports metastatic growth. Notably, ESCC cells overexpressing GPRC5A demonstrate improved implantation and persistence, resulting in macro-metastases in the lungs. Clinically, elevated GPRC5A level is associated with poorer outcomes in a cohort of 148 ESCC patients. Mechanistically, GPRC5A is found to potentially interact with WWP1, facilitating the polyubiquitination and degradation of LATS1, thereby activating YAP1 signaling pathways essential for metastasis. Importantly, targeting YAP1 axis with CA3 or TED-347 significantly diminishes early implantation and macro-metastases. Thus, the GPRC5A/WWP1/LATS1/YAP1 pathway represents a crucial target for therapeutic intervention in ESCC lung metastases.
Subject terms: Oesophageal cancer, Metastasis
The mechanisms allowing early disseminated cancer cells colonize other tissues remain largely unknown. Here, authors show that GPRC5A axis drives esophageal squamous cell carcinoma lung seeding and metastasis, in a mechanism resembling trophoblast behavior during embryo implantation.
Introduction
Metastasis is a pivotal factor contributing to cancer-related death, serving as a significant prognostic indicator across various cancer types. The process of tumor metastasis entails a series of complicated steps where cancer cells separate from the primary tumor site, intravasate into the circulation to form circulating tumor cells (CTCs), extravasate from the blood vessels, and ultimately colonize distant organs1–3. It is crucial to recognize that tumor metastasis is a highly selective and inefficient process, with only a small percentage of cancer cells possessing the capability to navigate through all stages successfully and establish themselves in distant sites4. The characteristics and behaviors of these metastasis-capable cancer cells remain largely unknown.
Esophageal carcinoma, particularly esophageal squamous cell carcinoma (ESCC), contributes significantly to the burden of metastatic cancers and ranks as the seventh leading cause of men cancer-related deaths worldwide5. Despite advancements in treatment modalities, the overall 5-year survival rate for all-stage esophageal cancer patients remains low, around 20%. Individuals diagnosed with ESCC along with distant metastases would likely be given a dire prognosis, with a mere 5-year survival rate of less than 5%5. There is an urgent need to explore mechanisms of distant metastasis to benefit the advanced patients with ESCC. Among the detrimental manifestations of ESCC are lung metastases, alongside lymph node and liver metastases, presenting formidable challenges in patient management5,6. The complexities surrounding ESCC lung metastasis come from insufficient understanding on the underlying mechanisms governing this phenomenon.
To explore the phenotypic evolution of tumor cells during the process of ESCC lung metastasis, we established a mouse model of pulmonary metastasis7. By collecting tumor cells in lungs at different timepoints (0 hour, 6 hours, 48 hours, 77 days, and 145 days) post intravenous injection in mice and performing single-cell RNA sequencing (scRNA-seq), we constructed a comprehensive dynamic atlas of ESCC lung metastasis. Notably, tumor cells exhibited an upregulation trend in expression profiles similar to those genes of embryo implantation at 6 hours and 48 hours, along with an upregulation of functions associated with embryo implantation (EI). As known, the process of trophoblast-mediated blastocyst implantation is a crucial step in embryo development and placentation, involving essential subprocesses such as stress response, apposition, adhesion, migration, and invasion8,9. Early tumor implantation (TI) also shares common processes with EI, including anti-stimulus response, inflammation, attachment, migration, and invasion, suggesting a close association between the two processes. Studies have indicated that trophoblasts, which are pivotal mediators of embryo implantation, exhibit behaviors akin to those of cancer cells10. Furthermore, previous findings have demonstrated that certain abortifacients such as mifepristone and selective estrogen receptor modulator tamoxifen, which impede EI, could potentially serve as chemo-preventatives for cancer metastasis8,11. The “seed and soil” and “transition” aspects of ESCC lung metastasis bear a striking resemblance to those of embryo implantation, yet the tumor cell processes that mimic embryo implantation during lung metastasis remain unexplored.
As for early dissemination, a selection was made from the pool of upregulated genes in our model, with a focus on GPRC5A. This gene exhibits high expression levels during the trophoblasts-mediated blastocyst implantation process12. GPRC5A, a member of the G protein-coupled receptor family C type 5A, is known as a retinoic acid-inducible protein. Given the essential role of retinoic acid biosynthesis and metabolism in fertilization and various stages of blastocyst development13, GPRC5A has garnered significant attention. Among cancers, GPRC5A is recognized for its diverse roles, being investigated as both a tumor-suppressive gene in lung cancer and an oncogene in bladder and colorectal cancers14–16. Despite these studies, the specific involvement of GPRC5A in lung metastasis of ESCC has not been studied yet. Therefore, a comprehensive exploration of the functions and mechanisms of GPRC5A in ESCC pulmonary micro-metastasis could unveil potential targets and perspectives for the treatment of ESCC metastasis.
Our investigation aims to unravel the crucial genes, pathways, and mechanisms underlying ESCC tumor implantation in the lungs, with an emphasis on GPRC5A and its downstream effectors. Through our findings, we aim to shed light on previously unexplored behaviors of ESCC in lung metastasis, provide a predictive indicator for tracking the effectiveness of lung metastasis treatment, and put forward a promising therapeutic strategy against ESCC.
Results
Embryo implantation-like program is activated in early-invaded lung disseminated cancer cells (DCCs)
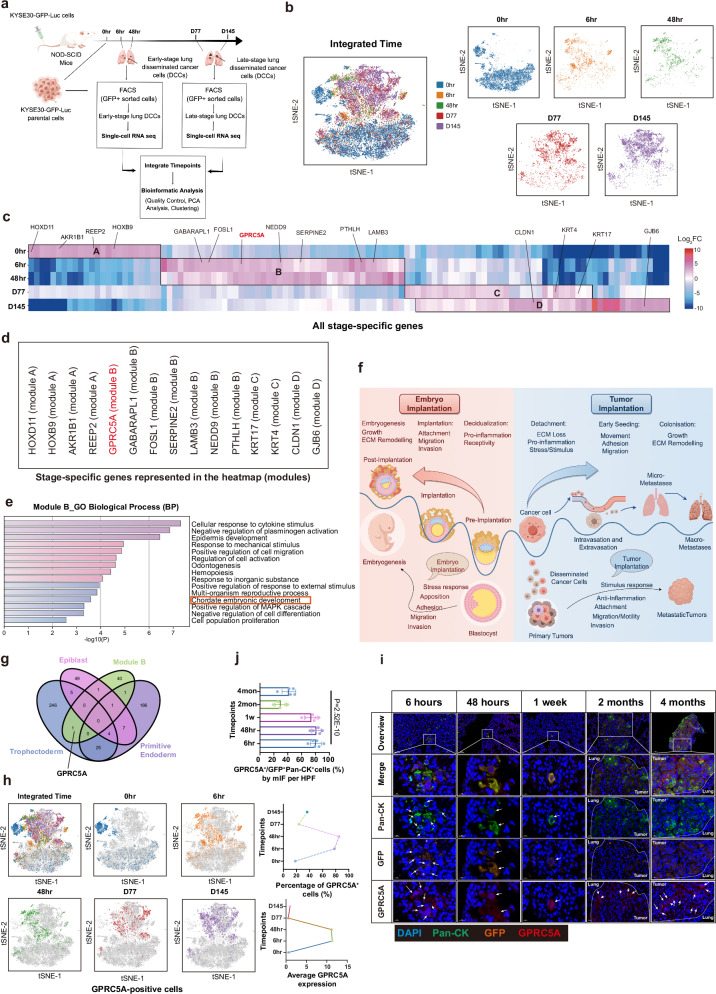
To investigate the evolutionary dynamics of cancer cells during the process of metastatic colonization, we initiated experimental lung metastases by intravenously injecting KYSE30-GFP-Luc human ESCC cells into immunocompromised NOD-SCID mice. Lung metastatic cancer cells were isolated at specific timepoints: 6 hours and 48 hours post-injection (when micro-metastases were predominantly present in the lungs), as well as at 77 days and 145 days post-injection (when macro-metastases had become pronounced and widespread). The isolation was conducted through GFP+ sorting for scRNA-seq analysis, as showed in Fig. 1a. A total of 26,712 cells from five different timepoints including different-stage lung DCCs and parental cells ( 0 hour) were sequenced, and after quality control measures, 19,986 cells were retained for subsequent analyses. T-distributed stochastic neighbor embedding (t-SNE) analysis of scRNA-seq data revealed that lung DCCs at early and late timepoints formed distinct clusters apart from the parental cells (Fig. 1b). The early-stage lung DCCs at 6 hours and 48 hours exhibited a similar distribution pattern, which was consistent to the late-stage lung DCCs from 77 days and 145 days.
Fig. 1. GPRC5A overexpression in early lung DCCs confers a trophoblast-like tumor implantation phenotype.
a The experimental timeline outlines the isolation of metastatic cancer cells from the lungs of mice following intravenous injection at specified timepoints: 6 hours (n = 3), 48 hours (n = 16), 77 days (n = 6), and 145 days (n = 1), for subsequent single-cell RNA sequencing. This figure is drawn by figdraw.com. b t-SNE plots reveal the distribution of isolated lung metastatic cancer cells across various timepoints: 0 hour, 6 hours, 48 hours, 77 days, and 145 days (n = 9059, 1957, 1149, 3322, 4499, respectively). c A heatmap illustrates 116 significant stage-specific genes identified across the five timepoints, based on fold change (FC) in expression (P value < 0.05), determined by a two-sided Wilcoxon signed-rank test. d Selected representative genes from Modules A, B, C, and D are shown. e Gene Ontology (GO) Biological Process (BP) analysis highlights top-level items enriched in Module B genes, utilizing a two-sided permutation test (P value < 0.05). f A schematic diagram compares embryo implantation and tumor implantation, underscoring shared subprocesses such as attachment, migration, invasion, and response to stimuli, which are essential for both processes. This figure is drawn by figdraw.com. g A Venn diagram depicts overlapping genes between Module B (n = 44) and stage-specific genes from trophectoderm (n = 281), primitive endoderm (n = 224), and epiblast (n = 67) during the pre-implantation, implantation, and post-implantation phases. h t-SNE plots illustrate the distribution of GPRC5A+ cells at five timepoints (n = 1667, 1527, 977, 786, 1635, respectively). i Representative mIF staining images depict Pan-Cytokeratin (Pan-CK, green), Green Fluorescent Protein (GFP, orange), GPRC5A (red) and DAPI (blue) from lung samples at different timepoints: 6 hours (n = 6), 48 hours (n = 6), 1 week (n = 6), 2 months (n = 5), and 4 months (n = 5). j The bar plot presents the percentages of GPRC5A+/Pan-CK+GFP+ lung DCCs per high power field (HPF), analyzed via two-sided one-way ANOVA test. Data are presented as mean ± SD (bar plots) and the n number represents n biologically independent spots/samples/genes in each group. Source data are provided as a Source Data file.
A total of 116 differentially expressed stage-specific genes were identified across the five timepoints (P < 0.05). These genes were further categorized into four distinct gene modules (A, B, C, D) based on their expression patterns, as illustrated in Fig. 1c and Supplementary Data 1. The parental cells, enriched in gene module A, exhibited the expression of 24 differentially stage-specific genes associated with development (e.g., HOXD11, HOXB9) and metabolism (e.g., AKR1B1, REEP2). These genes in module A were positively correlated with processes related to organic development and epithelial cell proliferation based on enrichment analysis (Fig. 1c, d and Supplementary Fig. 1a, b). Early-stage lung DCCs (at 6 hours and 48 hours post-injection) expressed 44 stage-specific genes involved in stimulus response (e.g., GPRC5A, GABARAPL1), cell migration (e.g., FOSL1, SERPINE2), and embryonic development (e.g., PTHLH, GPRC5A, LAMB3, NEDD9) (Fig. 1c–e and Supplementary Fig. 1a), conferring these cells with the ability for motility, anti-stimulus response, and implantation for their survival and seeding. In contrast to the early-invaded cells, late-stage lung DCCs were characterized by a focus on self-growth, self-differentiation, and extracellular matrix (ECM) remodeling. Genes related to cell proliferation/differentiation (e.g., KRT17, KRT4) and ECM components (e.g., CLDN1, GJB6) were predominantly expressed in the C and D gene modules of late-stage lung DCCs (Fig. 1c, d and Supplementary Fig. 1a, b). These findings prompted us to investigate the parallels between cancer metastasis and embryo development. Notably, we discovered that the early dissemination of cancer cells shares common subprocesses with embryo implantation, which is a crucial initial step in embryo development. These subprocesses include responses to stress, adhesion, migration, and invasion (Fig. 1f). Interestingly, processes related to embryo development were solely observed in early micro-metastases (Module B) and not in late macro-metastases (Module C and D) or parental cells (Module A). Through an in-depth examination of the module B signature, several TI-related genes were found to also play a role in counterparts of EI (Supplementary Data 2). These results demonstrated in the early-stage of DCCs, the embryo implantation-like program was activated.
GPRC5A is associated with a TI-like phenotype of early-stage lung DCCs
Drawing parallels between the early dissemination of cancer cells and embryo implantation, we analyzed 281 stage-specific genes of trophoblasts implicated in blastocyst implantation12 alongside genes associated with micro-metastases (Module B). Our investigation revealed the identification of key genes such as LAMB3 and THBS1 in the epiblast, and ARL14 and THBS1 in the primitive endoderm. Interestingly, GPRC5A as the singular gene shared between trophoblasts and early invaded cancer cells, underscoring its potential importance in facilitating tumor implantation (Fig. 1g). Additionally, previous investigations of our group unveiled a distinct subcluster termed ‘Cluster S’ within parental cells, distinguished by CD44 and TACSTD2 expression, exhibiting a heightened propensity for early lung dissemination in this model7. Intriguingly, our current investigation revealed that 87-94% of GPRC5A-positive cells belonged to ‘Cluster S’ in parental cells and early-stage lung DCCs based on scRNA-seq data (Supplementary Fig. 2a, b), highlighting its heightened metastatic potential. Noteworthy was the presence of CD44+GPRC5A+ or TACSTD2+GPRC5A+ cells, accounting for 35-67% of early-stage lung DCCs, while the percentage dramatically decreased to 12-40% at macro-metastases by multi-channel immunofluorescence (mIF) analysis, as shown in Supplementary Fig. 2c.
To shed light on the pro-implantation role of GPRC5A in ESCC, we examined the distribution and expression levels of GPRC5A-positive cells at different timepoints in our experimental model. Our results indicated that 78% and 85% of early-stage lung metastatic cells that persisted at 6 hours and 48 hours post-injection were GPRC5A-positive, with this percentage declining to 24% and 36% at 77 days and 145 days in late-stage lung DCCs, aligning with the observed expression patterns depicted in Fig. 1h. Furthermore, mIF staining of pan-cytokeratin (Pan-CK), GFP and GPRC5A in different-timepoints mice lung sections demonstrated that over 80% of early-invaded lung metastatic cells (Pan-CK+GFP+ tumor cells) at 6 hours, 48 hours, and 1 week post-injection exhibited GPRC5A expression, while the proportion of GPRC5A+/Pan-CK+GFP+ cells remained low at 20-40% in late-stage macro-metastases at 2 or 4 months, as shown in Fig. 1i, j. Moreover, through mIF staining, we observed co-localization of GPRC5A-positive cells and Pan-CK-positive tumor cells in early-stage lung DCCs at 55-84% but only at 17-44% in late-stage lung metastatic nodules, as presented in Supplementary Fig. 2c. This finding further reinforced the upregulation of GPRC5A in early lesions, which persisted in early-stage lung DCCs.
High expression of GPRC5A is correlated with poor clinical outcomes
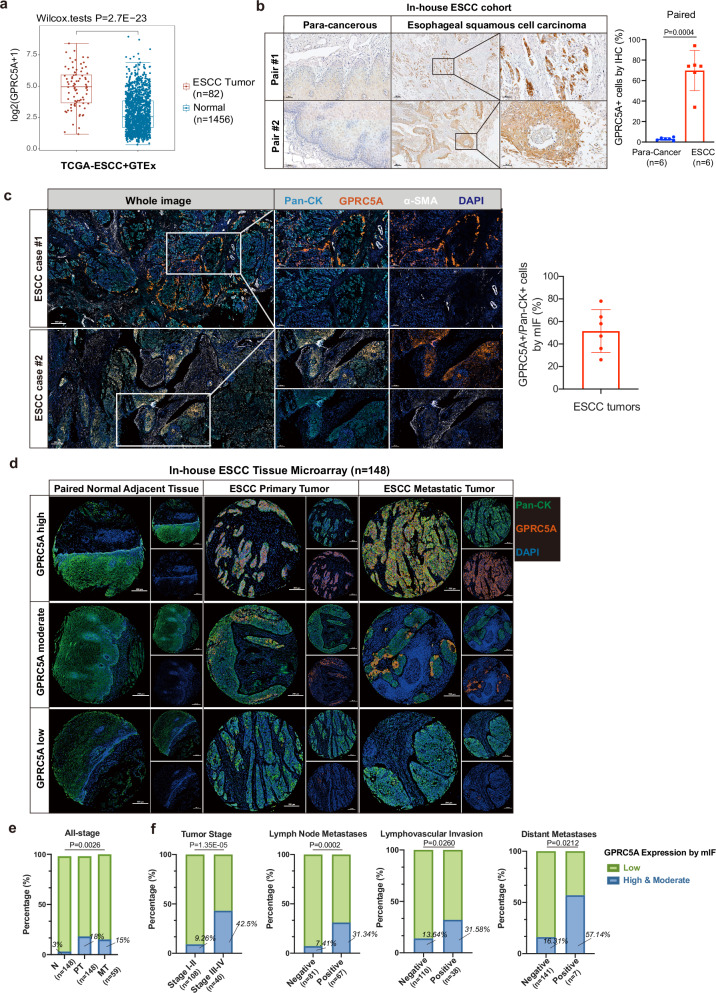
In addition to examining lung metastases, our study investigated the expression pattern of GPRC5A in primary tumors of ESCC. Analysis of in-silico data revealed that GPRC5A was significantly upregulated in ESCC tumors (n = 82) compared to normal esophageal epithelium (n = 1456) based on data from the TCGA-ESCC and GTEx databases (see Fig. 2a), consistent with findings from three other cohorts sourced from the GEO database (GSE45670, GSE161533, GSE26886) (Supplementary Fig. 3a). Moreover, beyond ESCC, GPRC5A was found to be overexpressed in various other cancer types such as breast cancer, pancreatic cancer, and endometrial cancer according to a pan-cancer analysis (Supplementary Fig. 3b). However, although high expression of GPRC5A in ESCC patients was not statistically associated with poor overall survival (OS) and disease-free survival (DFS) (Supplementary Fig. 3c), it was linked to worse OS and progression-free survival (PFS) in patients with kidney renal clear cell carcinoma, pancreatic cancer, and glioma based on pan-cancer survival analysis in TCGA cohort (Supplementary Fig. 3d).
Fig. 2. Upregulation of GPRC5A in ESCC tumors and its association with aggressive tumor behaviors.
a Analysis of GPRC5A expression levels in ESCC tumors (n = 82) obtained from the TCGA-ESCC database compared to normal esophageal tissues (n = 1456) from the GTEx database, evaluated using a two-sided Wilcoxon signed-rank test. b Representative images of IHC GPRC5A staining demonstrating GPRC5A expression and the proportion of GPRC5A-positive cells in ESCC tumors versus paired adjacent tissues (n = 6), analyzed using a two-sided paired t-test. c Representative images of multi-channel immunofluorescence (mIF) staining showcasing Pan-Cytokeratin (Pan-CK, a tumor cell marker, shown in light blue), GPRC5A (shown in orange), alpha-smooth muscle actin (α-SMA, a stromal cell marker, shown in white), and DAPI (shown in dark blue) in primary tumors from ESCC patients (n = 6) in our in-house cohort. The bar plot illustrates the percentage of GPRC5A-positive/Pan-CK-positive cells by mIF staining in ESCC tumors. d Overview of GPRC5A expression stratified into high/moderate, and low categories within an ESCC tissue microarray (TMA) comprising paired normal adjacent esophageal epithelium (n = 148), ESCC primary tumors (n = 148), and lymph node metastatic tumors (n = 59), analyzed via mIF staining using Pan-CK (green), GPRC5A (orange), and DAPI (blue). e Proportion of high or moderate (green) and low (blue) GPRC5A expression levels across the ESCC TMA, including normal tissues (n = 148), primary tumors (n = 148), and metastatic tumors (n = 59). Two-sided Chi-Squared test is used for statistical analysis. f Correlation analysis of GPRC5A expression (high/moderate in green vs. low in blue) with clinical parameters, including tumor stage (I-II, n = 108 vs. III-IV, n = 40), lymph node metastasis (negative, n = 81 vs. positive, n = 67), lymphovascular invasion (negative, n = 110 vs. positive, n = 38), and distant metastasis (negative, n = 141 vs. positive, n = 7), determined using two-sided Chi-squared or Fisher’s exact tests. The data are presented as the mean ± SD (bar plots) and median ± IQR (whiskers = 1.5 × IQR, box & whiskers plots). The n number represents n biologically independent samples/patients in each group. Source data are provided as a Source Data file.
We also conducted IHC staining for GPRC5A on sections from para-cancerous esophageal tissues and primary ESCC tumor tissues obtained from six paired ESCC patients in our in-house ESCC cohort. Our analysis confirmed that GPRC5A expression was minimal in the para-cancerous esophageal epithelium but markedly elevated in primary ESCC tumors. The percentage of GPRC5A-positive cells in ESCC tumors ranged from 32% to 95% as determined by IHC staining (Fig. 2b), which was consistent with the 26-78% range observed with mIF staining (Fig. 2c). Furthermore, GPRC5A expression was analyzed in a tissue microarray (TMA) comprising 148 ESCC cases, including para-cancerous tissues (n = 148), primary tumors (n = 148), and corresponding metastatic tumors (n = 59) using mIF staining (Fig. 2d). The clinicopathological characteristics of these cases are presented in Supplementary Data 3. Our analysis revealed that GPRC5A-positive cells were scarce in normal adjacent epithelium (3%), but predominantly present in primary tumors (18%) and metastatic tumors (15%) among all-stage patients (Fig. 2e). In advanced-stage patients, an upregulation of GPRC5A expression was found in primary tumors of 42.5% compared to 9.26% in early-stage patients (P < 0.0001). Additionally, patients with lymph node metastases (P = 0.0002), lymphovascular invasion (LVSI) (P = 0.026), and distant metastases (P = 0.0212) were more likely to exhibit higher GPRC5A expression (Fig. 2f and Supplementary Data 3). While the overall survival between the GPRC5A-high or moderate and GPRC5A-low groups for all-stage patients did not show a statistical difference (P = 0.4542), an upregulation of GPRC5A in late-stage patients was significantly associated with decreased overall survival (median OS time: 15 months versus 31 months, Hazard Ratio=2.666, P = 0.0331), as illustrated in Supplementary Fig. 4a, b. These findings suggested that ESCC tumors with GPRC5A overexpression correlated with more aggressive behavior, including lymph node metastasis and distant metastasis, leading to advanced stage and poorer survival.
GPRC5Ahigh cells are more capable of implanting and facilitating carcinogenesis
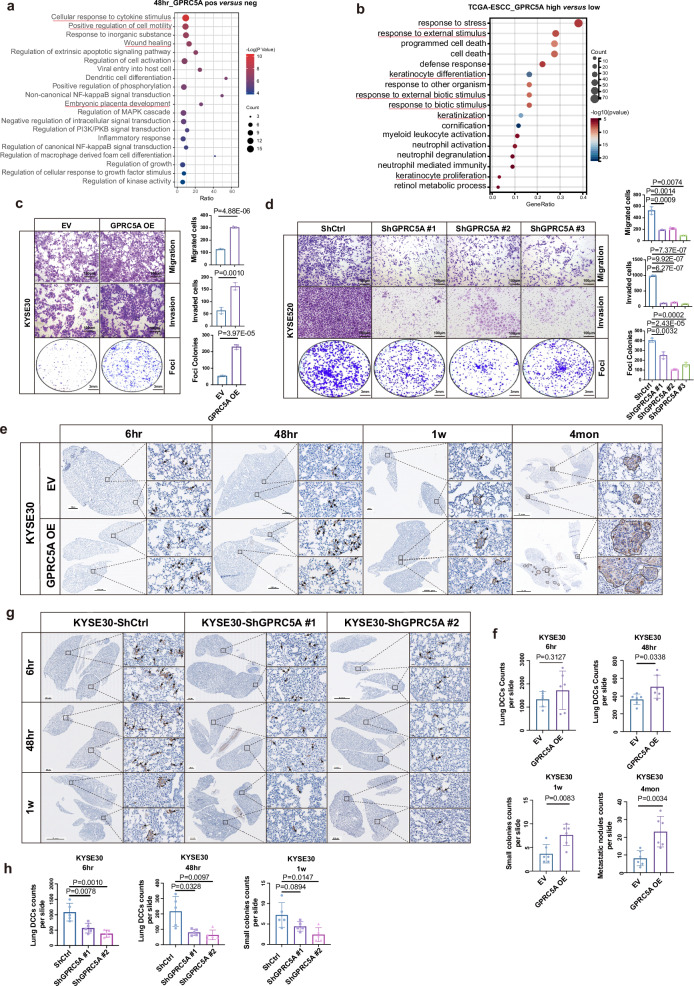
Then we investigated the aberrant activation of GPRC5A in the early-implantation program and conducted an exploration of the functional characteristics of the GPRC5Ahigh subgroup. Analysis of mouse lung metastasis data and the human TCGA-ESCC cohort strongly indicated associations with cellular processes such as cell motility, wound healing, cell migration, cell adhesion, keratinization, response to stimuli, and embryonic/placental development in GPRC5Ahigh groups (Fig. 3a, b and Supplementary Fig. 5a, b), all of which are crucial for successful trophoblasts-mediated blastocyst implantation8. Subsequently, we established lentiviral models for GPRC5A overexpression and shRNA knockdown in human ESCC cells and mouse-derived ESCC cells (mEC25) for validation (Supplementary Fig. 5c, d). Our findings demonstrated that overexpression of GPRC5A enhanced migration and invasion abilities and promoted tumor colony formation, while knockdown of GPRC5A reversed these effects (Fig. 3c, d and Supplementary Fig. 5e). Besides, we previously noted that only a small subset of cancer cells in primary tumors or parental cells exhibited high GPRC5A expression, reminiscent of cancer stem cell (CSC) properties17. Self-renewal and stemness maintenance of trophoblasts ensure a successful implantation as reported18. Indeed, we found that GPRC5Ahigh ESCC cells generated more spheroids and demonstrated a greater capacity for tumor initiation or stemness in a limited number of cells compared to the counterpart group (Supplementary Fig. 5f–g). Adaptation to external stimuli protects trophoblasts from apoptosis and cell death during the pre-implantation and implantation stages, as well as throughout placental development stage19. Similarly, we confirmed that GPRC5Ahigh cells developed increased resistance to stimuli and produced fewer reactive oxygen species (ROS) under H2O2 stimulation (Supplementary Fig. 5h–i).
Fig. 3. GPRC5A overexpression promotes tumor growth, early implantation, and progression.
a The top 20 GO BP terms associated with DEGs are identified between GPRC5A-positive and GPRC5A-negative lung DCCs at 48 hours from scRNA-seq data, using a two-sided permutation test. b GO BP analysis of DEGs comparing patients with high GPRC5A expression (upper 25%, n = 21) to those with low expression (lower 25%, n = 21) from the TCGA-ESCC database, also employing a two-sided permutation test. c, d Assays to assess migration, invasion, and foci colonies in KYSE30 cells overexpressing GPRC5A versus control cells (c, n = 3, two-sided unpaired t-test). Similarly, KYSE520 cells with GPRC5A knockdown are compared to control cells (d, n = 3, two-sided unpaired t-test). e, f Representative IHC images of Pan-Cytokeratin staining in the lungs of NOD-SCID mice at multiple timepoints (6 hours, 48 hours, 1 week, 4 months) following intravenous injection of KYSE30 cells overexpressing GPRC5A or control cells in a lung metastasis model (e, n = 6 per group per timepoint). Black arrows indicate disseminated cancer cells at early stages and metastatic nodules at late stages (e). The counts of lung DCCs at 6 hours and 48 hours, small colonies (2-5 cells) at 1 week, and metastatic nodules (>10 cells) at 4 months are presented in bar plot (f), analyzed using a two-sided unpaired t-test. g, h Representative images of IHC Pan-Cytokeratin staining in the lungs of NOD-SCID mice at 6 hours, 48 hours, and 1 week post-injection of KYSE30 GPRC5A-knockdown or control cells (g, n = 5 per group per timepoint). Black arrows indicate disseminated cancer cells at early stages. The counts of lung DCCs at 6 hours and 48 hours and small colonies (2-5 cells) at 1 week are displayed in bar plots (h), analyzed using a two-sided unpaired t-test. Data are presented as mean ± SD (bar plots), and the n number represents n biologically independent samples/experiments in each group. Source data are provided as a Source Data file.
To further validate the pro-metastatic role of GPRC5A, we intravenously injected GPRC5A-overexpressing and control KYSE30 cells, as well as GPRC5A-silenced and control KYSE30/KYSE520 cells, into NOD-SCID mice. The data revealed that cancer cells with high GPRC5A expression exhibited greater capacity for implantation and survival at the early stage, and those with GPRC5A overexpression persisted and generated more and larger lung metastatic nodules at the late stage (Fig. 3e, f). Conversely, a reduction in the implantation of cancer cells was observed in the GPRC5A-silenced group (Fig. 3g, h and Supplementary Fig. 6a). Furthermore, we established cell-derived xenografts (CDXs) using GPRC5A-overexpressing human or mouse ESCC cells and empty vector (EV) control cells and observed that GPRC5A promoted tumor growth in both immunodeficient and immunocompetent mice (Supplementary Fig. 6b–d) while silencing GPRC5A reversed the oncogenic effect (Supplementary Fig. 6e). These results indicated that the oncogenic function of GPRC5A was independent of immune status. Collectively, these data underscored the proficient abilities of cancer cells with high GPRC5A expression for tumor implantation, enabling early-stage lung DCCs to exhibit a trophoblast-mediated implantation-like phenotype and persist in forming proliferative metastases.
Activated YAP1 is a key effector for GPRC5A-mediated ESCC progression
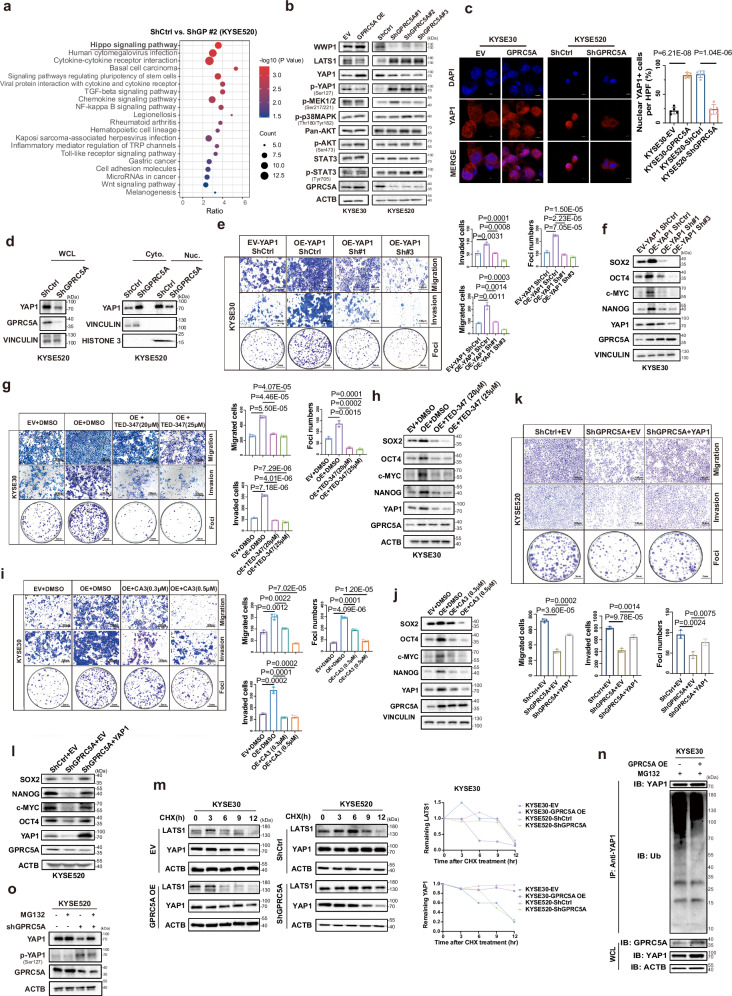
In order to investigate the downstream effectors of GPRC5A activity, a comprehensive analysis was conducted on three transcriptomic datasets. These datasets included: (i) KYSE30 cells transfected with lentiviral control versus GPRC5A overexpression, (ii) KYSE520 cells transfected with lentiviral control versus GPRC5A knockdown, and (iii) the GPRC5Ahigh versus GPRC5Alow groups from the TCGA-ESCC database. The analysis revealed that differentially expressed genes (DEGs) were predominantly enriched in the Hippo signaling pathway, PI3K/AKT pathway, and MAPK pathway (Fig. 4a and Supplementary Fig. 7a–c). Subsequent validation demonstrated that the expression of genes associated with the Hippo pathway, such as LATS1, YAP1, and Ser127-phosphorylated-YAP1 (p-YAP1Ser127), was affected, while the expression of genes related to the MAPK pathway (e.g., Ser217/221-phosphorylated MEK1/2 and Thr180/Tyr182-phosphorylated-p38MAPK) and the PI3K/AKT pathway (e.g., pan-AKT, Ser473-phosphorylated-AKT, STAT3, and Tyr705-phosphorylated-STAT3) remained unchanged irrespective of the GPRC5A status (Fig. 4b). Cells with silenced GPRC5A exhibited a significant downregulation of YAP1 but an upregulation of LATS1 and p-YAP1, and vice versa (Fig. 4b). These findings suggested that GPRC5A overexpression distinctly influenced components of the Hippo pathway, especially YAP1.
Fig. 4. The pro-metastatic and oncogenic role of GPRC5A mediated by YAP1 nuclear translocation.
a Enriched KEGG pathways derived from differentially expressed genes (DEGs) identified through transcriptome sequencing comparing KYSE520-Control (ShCtrl) and shGPRC5A cells, analyzed using a two-sided permutation test. b Western blot analysis revealing expression levels of key components in the Hippo signaling pathway (LATS1, YAP1, phosphorylated YAP1), PI3K/AKT pathway (pan-AKT, phosphorylated AKT, STAT3, phosphorylated STAT3), and MAPK pathway (phosphorylated MEK1/2, phosphorylated p38MAPK), as well as WWP1, following modulation of GPRC5A expression in KYSE30 and KYSE520 cells (n = 3). c Representative ICC staining images illustrating YAP1 (red) localization alongside DAPI (blue) in GPRC5A-overexpressing KYSE30 cells and GPRC5A-silenced KYSE520 cells treated with the proteasome inhibitor MG132 (10 µM) (n = 5; two-sided unpaired t-test). d Immunoblot analysis of YAP1 in whole cell lysates (WCL), cytoplasmic (Cyto.), and nuclear (Nuc.) fractions from KYSE520 cells with either GPRC5A silencing or control (n = 3). Vinculin and Histone 3 serve as markers for cytoplasmic and nuclear proteins, respectively. e–j Transwell migration, invasion, foci formation assays, and immunoblotting of YAP1 downstream genes following knockdown via shRNAs targeting YAP1 or treatment with two pharmacological YAP-TEAD inhibitors (CA3 and TED-347) in GPRC5A-overexpressing KYSE30 cells (n = 3; two-sided unpaired t-test). k, l Transwell migration, invasion, foci formation assays, and immunoblotting of downstream genes following YAP1 rescue in GPRC5A-silenced KYSE520 cells (n = 3; two-sided unpaired t-test). m Pulse-chase assay tracking YAP1 and LATS1 protein levels at various timepoints (0, 3, 6, 9, 12 hours) upon cycloheximide (CHX, 50 µg/mL) treatment (n = 3). n Ubiquitination levels of YAP1 precipitated with an anti-YAP1 antibody in GPRC5A-overexpressing KYSE30 cells and controls treated with MG132 (10 µM) (n = 3). o Expression levels of YAP1 and phosphorylated YAP1 in GPRC5A-silenced or control KYSE520 cells following treatment with or without MG132 (10 µM) (n = 3). For western blots, the samples derive from the same experiment but different gels for all tested antibodies were processed in parallel in Fig. 4b, d, f, h, j, l, m, n, o. Data are presented as mean ± SD (bar plots), and the n number represents n biologically independent samples/experiments in each group. Source data are provided as a Source Data file.
As it is well-established, YAP1 serves as a pivotal effector protein of the Hippo pathway and is commonly suppressed in cancers through LATS1-mediated YAP1 phosphorylation, leading to its cytoplasmic retention or degradation20. The nuclear translocation of dephosphorylated YAP1 is indicative of its activation, as it binds to the promoter of downstream target genes to facilitate transcription20,21. Since the relationship between GPRC5A and YAP1 activation has been previously demonstrated in other cancer types22,23, we investigated the relationship between YAP1 and GPRC5A in our models. Immunocytochemistry (ICC) staining confirmed that nuclear YAP1+ cells were enriched in GPRC5A-overexpressing cells but markedly reduced in GPRC5A-silencing cells (Fig. 4c). Additionally, cellular cytoplasmic and nuclear fraction assays demonstrated that GPRC5A overexpression induced nuclear YAP1 expression and decreased cytoplasmic YAP1 abundance, while GPRC5A knockdown resulted in a reduction of nuclear YAP1 and an accumulation of cytoplasmic YAP1 (Fig. 4d and Supplementary Fig. 7d). Moreover, GPRC5Ahigh cells displayed heightened transcription and protein levels of YAP1 downstream genes such as SOX2, c-MYC, OCT4, and NANOG, whereas the opposite effects were observed in GPRC5A-silencing cells (Figs. 4f, l and Supplementary Fig. 7e).
To further explore the significance of YAP1 as a downstream effector of GPRC5A, inhibition of YAP1 was introduced in GPRC5A-overexpressing cells through the administration of two shRNAs targeting YAP1. It was observed that the suppression of YAP1 inhibited the oncogenic and pro-metastatic effects of GPRC5A on ESCC cells and disrupted the GPRC5A-induced upregulation of downstream genes such as SOX2, c-MYC, NANOG, and OCT4 at the transcriptional and expression levels (Fig. 4e, f and Supplementary Fig. 7f). Consistent findings were obtained with the use of two pharmacological YAP1-TEAD inhibitors, CA3 and TED-347, which significantly impeded the formation of tumor colonies, reduced tumor aggressiveness, and disrupted the transcription and expression of downstream genes (Fig. 4g–j and Supplementary Fig. 7g, h). Conversely, the genetic introduction of YAP1 in GPRC5A-silencing cells effectively restored the expression of downstream genes and nullified the suppressive effects on tumor growth, migration, and invasion (Fig. 4k, l). Interestingly, our findings also indicated that both silencing YAP1 and using YAP1-TEAD inhibitors reduced the progression and growth of KYSE30 cells (Supplementary Fig. 8a, b). Collectively, these results indicated that YAP1 was a critical downstream target of GPRC5A in promoting ESCC growth and progression.
GPRC5A stabilizes YAP1 via decreasing its ubiquitination level
In line with previous findings, the phosphorylation of YAP1 hindered its nuclear translocation and prompted its degradation21. Subsequently, we investigated the influence of GPRC5A on the stability of YAP1 for its nuclear translocation. Upon treatment with cycloheximide (CHX), it was observed that GPRC5A overexpression significantly extended the half-life of the YAP1 protein, thereby stabilizing its expression, while a rapid decrease in YAP1 abundance was evident in GPRC5A-silencing cells (Fig. 4m). Furthermore, to ascertain whether GPRC5A impacted the ubiquitination of YAP1, ESCC cells transfected with or without GPRC5A overexpression or GPRC5A-knockdown were subjected to treatment with the proteasomal inhibitor MG132 (10 µM). A reduction in YAP1 ubiquitination was noted in GPRC5A-overexpressing cells, whereas an increase was observed in GPRC5A-silencing cells (Fig. 4n, Supplementary Fig. 9a). Consistent with the effects of GPRC5A overexpression, the expression of YAP1 is downregulated in the shGPRC5A group but is restored upon MG132 treatment. In contrast, the level of phosphorylated-YAP1 was significantly higher in the shGPRC5A group compared to the control group upon MG132 treatment (Fig. 4o). However, no significant YAP1 changes were detected in GPRC5A-overexpressing KYSE30 cells upon MG132 treatment (Supplementary Fig. 9b).
GPRC5A-WWP1 interaction inhibits LAST1-mediated YAP1 ubiquitination
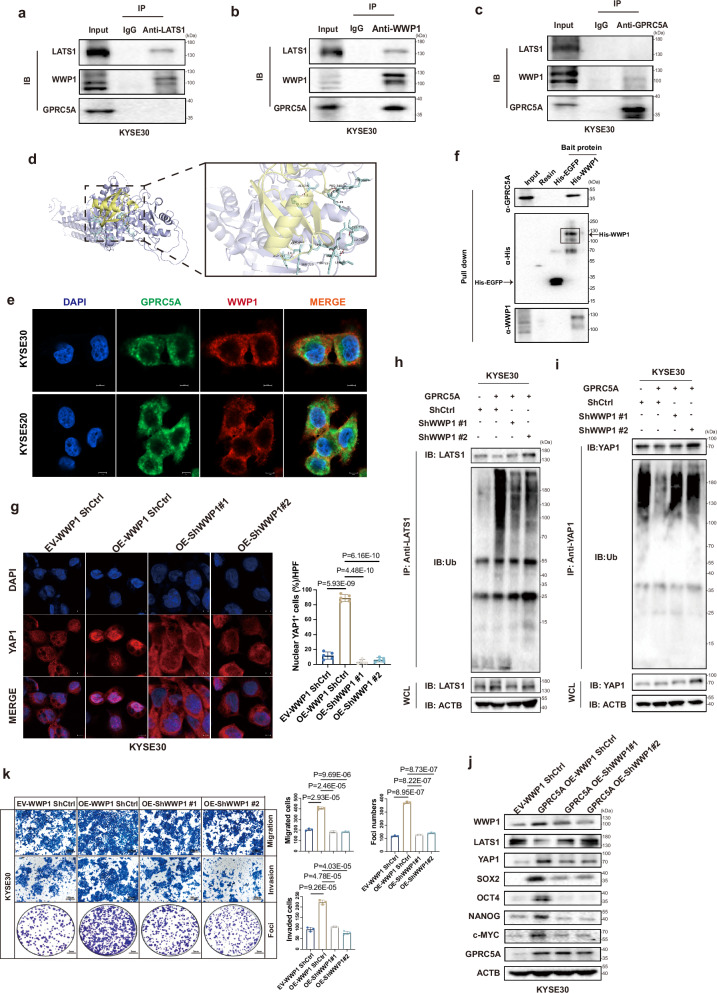
As established in previous studies, YAP1 undergoes phosphorylation by LATS1, leading to its cytoplasmic ubiquitin-mediated degradation21. LATS1 serves as a direct negative regulator of YAP1. Overexpression of GPRC5A has been shown to downregulate LATS1 and counteract its inhibitory effects on YAP1, as illustrated in Fig. 4b. Thus, we hypothesize whether GPRC5A regulates the stability of LATS1 to affect the phosphorylation of YAP1. Our findings revealed that depletion of GPRC5A extended the half-life of LATS1 under CHX treatment and reduced its polyubiquitination, as shown in Fig. 4m and Supplementary Fig. 9c, d. We sought to link GPRC5A activity and LATS1 stability. It has demonstrated that WWP1 E3 ligase, a member of the NEDD4-like family of E3 ubiquitin ligases, negatively modulates LATS1 by promoting its degradation through polyubiquitination and the 26S proteasome pathway24,25. We next investigated whether WWP1 mediates the ubiquitination of LATS1. Interestingly, an elevation in WWP1 levels was observed in GPRC5A-overexpressing cells, while a decrease was noted in GPRC5A-silenced cells, as shown in Fig. 4b. These observations prompted a deeper exploration of the interplay between GPRC5A, WWP1, and LATS1.
To further demonstrate the collaboration of GPRC5A with WWP1 to facilitate LATS1 degradation via the ubiquitin-dependent proteasome pathway. The interaction between WWP1 and LATS1 was confirmed through immunoprecipitation experiments, where endogenous WWP1 but not GPRC5A was detected when pulled down with an anti-LATS1 antibody (Fig. 5a and Supplementary Fig. 9e). Conversely, co-immunoprecipitation of endogenous LATS1 and GPRC5A occurred using an anti-WWP1 antibody (Fig. 5b and Supplementary Fig. 9f). When precipitated with an anti-GPRC5A antibody, WWP1 was predominantly captured, whereas LATS1 was undetectable (Fig. 5c and Supplementary Fig. 9g). Molecular docking analysis identified potential binding sites of GPRC5A and WWP1 in a 3D model (Fig. 5d and Supplementary Fig. 9h). Furthermore, a co-localization relationship between the WWP1 and GPRC5A proteins was identified through immunofluorescence staining in KYSE30 and KYSE520 parental cells (Fig. 5e). To further confirm the binding interaction between WWP1 and GPRC5A, we utilized recombinant His-tagged WWP1 and His-tagged EGFP (serving as a negative control) in His-tagged protein pull-down assay (Supplementary Fig. 9i). The results demonstrated that GPRC5A can be effectively pulled down by the recombinant His-tagged WWP1 protein, while no interaction was observed with the recombinant His-tagged EGFP protein (Fig. 5f). Moreover, a positive correlation between WWP1 expression and GPRC5A expression was observed in the TCGA-ESCA database, with WWP1 being overexpressed in ESCC tumors compared to normal tissues, and its high expression being associated with poorer disease-free survival in various cancer types (Supplementary Fig. 10a–c).
Fig. 5. Interaction of GPRC5A and WWP1 induces LATS1 degradation and activates YAP1.
a–c Co-immunoprecipitation (co-Ip) assays to illustrate the interaction between GPRC5A, WWP1, and LATS1, utilizing anti-LATS1 (a), anti-WWP1 (b), and anti-GPRC5A antibodies (c) in whole cell lysates from KYSE30 parental cells (n = 3). The samples derive from the same experiment but different gels for anti-GPRC5A, another for anti-WWP1, another for anti-LATS1 were processed in parallel. d The predicted 3D structure of the GPRC5A (yellow) and WWP1 (purple) complex, along with the potential binding sites indicated by light blue chemical bonds, is generated through molecular docking analysis and visualized using PyMOL software. e Immunofluorescence co-localization staining of GPRC5A (green), WWP1 (red), and DAPI (blue) is conducted in KYSE30 and KYSE520 parental cells to further confirm their potential interaction (n = 3). f Protein pull-down assays were executed using recombinant His-tagged WWP1 and EGFP proteins in KYSE520 cell lysates (n = 3). The samples derive from the same experiment but different gels for anti-GPRC5A, another for anti-His, another for anti-WWP1 were processed in parallel. g Representative ICC staining images of YAP1 (red) and DAPI (blue) in WWP1-silenced KYSE30 cells overexpressing GPRC5A, treated with MG132 (10 µM), is presented along with a bar plot illustrating the percentage of nuclear YAP1-positive cells per HPF (n = 5, two-sided unpaired t-test). h–i Immunoblotting was performed to assess the ubiquitination levels of YAP1 and LATS1, which were precipitated using respective antibodies in WWP1-knockdown KYSE30 cells overexpressing GPRC5A, following pretreatment with MG132 (10 µM) (n = 3). WCL, whole cell lysates. The samples derive from the same experiment but different gels for anti-LATS1, another for anti-ubiquitin, another for anti-ACTB were processed in parallel. j, k Transwell assays for migration, invasion, and foci formation (n = 3, two-sided unpaired t-test) are conducted, alongside an evaluation of YAP1 downstream gene expression (n = 3) in WWP1-silenced KYSE30 cells with GPRC5A overexpression. The samples derive from the same experiment but different gels for all tested antibodies were processed in parallel. Data are presented as mean ± SD (bar plots), and the n number represents n biologically independent samples/experiments in each group. Source data are provided as a Source Data file.
To investigate the functional role of WWP1 in the GPRC5A-YAP1 axis, we manipulated WWP1 expression levels in GPRC5A-overexpressing or GPRC5A-knockdown cells. Knockdown of WWP1 was found to inhibit YAP1 nuclear translocation by increasing its ubiquitination levels while decreasing LATS1 ubiquitination levels (Fig. 5g–i). Furthermore, WWP1 silencing resulted in reduced YAP1 expression and downstream gene transcription/expression, along with an increase in LATS1 expression (Fig. 5j and Supplementary Fig. 11a). Silencing WWP1 also impeded ESCC migration, invasion, and growth (Fig. 5k). Conversely, the effects were reversed in WWP1-rescued GPRC5A-silenced cells (Supplementary Fig. 11b–f). These results collectively suggested that GPRC5A-mediated YAP1 activation was contingent upon the upregulation of WWP1.
Inhibition of YAP1 blocks early-implantation and metastatic colonization of the lungs
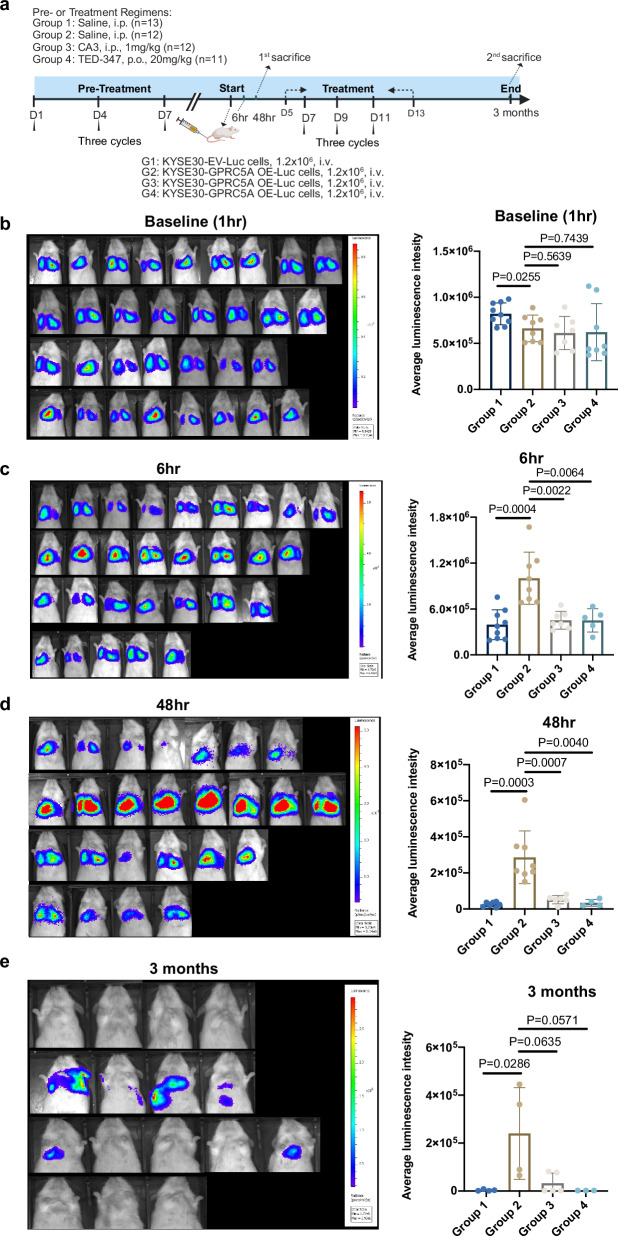
The compelling in-vitro findings have piqued our significant interest in investigating YAP1 as a potential therapeutic target for inhibiting early implantation of ESCC and reducing macro-metastases in vivo. Initially, we evaluated the preventive therapeutic effects of YAP1 inhibition on disrupting the early implantation of cancer cells. Mice were pretreated with two YAP1-TEAD inhibitors (CA3 and TED-347) as well as saline for three cycles prior to the intravenous injection of luciferase-labeled KYSE30 cells overexpressing GPRC5A or control cells (Fig. 6a). Bioluminescent signals from the lungs of mice were recorded at 1 hour, 6 hours and 48 hours post-injection. Our observations revealed that mice in the saline group exhibited higher luminescent signals at 6 hours and 48 hours post-injection when administered with GPRC5A-overexpressing cells (Group 2) compared to those with control cells (Group 1). These results suggested that GPRC5A-overexpressing cells displayed enhanced survival and colonization capabilities in the early stages. Furthermore, when comparing mice injected with GPRC5A-overexpressing cells, the pretreatment with CA3 (Group 3) and TED-347 (Group 4) showed a favorable response in attenuating early dissemination compared to the saline group (Group 2) (Fig. 6b–d). This strongly indicated that preventive YAP1-TEAD inhibition could potentially impede micro-metastasis of cancer cells.
Fig. 6. Inhibition of YAP1 impedes early implantation and metastatic tumor outgrowth in ESCC.
a Experimental design illustrating the use of YAP1-TEAD inhibitors (CA3 and TED-347) in a murine model to evaluate their effects on both lung micro-metastases and macro-metastases associated with ESCC. This figure is created in BioRender. Bella, liub1128@outlook.com. (2024) BioRender.com/g35e219. b–d Bioluminescence imaging is employed to assess and quantify the average luminescence intensity in the lungs of mice at baseline (b, 1 hour post-injection; Group 1 n = 9, Group 2 n = 8, Group 3 n = 7, Group 4 n = 8), at 6 hours (c, Group 1 n = 9, Group 2 n = 8, Group 3 n = 7, Group 4 n = 5), and at 48 hours (d, Group 1 n = 7, Group 2 n = 8, Group 3 n = 6, Group 4 n = 4) following pre-treatment with or without YAP1-TEAD inhibitors over three cycles. Statistical analysis is performed using a two-sided unpaired t-test for baseline and 6 hours and a two-sided Wilcoxon signed-rank test for 48 hours. e Long-term outcomes are evaluated at 3 months post-injection, with BLI and quantification of average luminescence intensity in the lungs of mice treated with or without YAP1-TEAD inhibitors over six cycles (Group 1 n = 4, Group 2 n = 4, Group 3 n = 5, Group 4 n = 3). Statistical analysis was performed using a two-sided Wilcoxon signed-rank test. Average luminescence intensity is calculated as photons/second/cm²/steradian. Data are presented as mean ± SD (bar plots), and the n number represents n biologically independent samples in each group. Source data are provided as a Source Data file.
Subsequently, we investigated whether YAP1-TEAD inhibition also played a role in restraining late-stage macro-metastases of cancer cells. Following the same pre-treatment regimen, mice were subjected to an additional three cycles of treatment after the intravenous injection. After 3 months post-injection, there was a noticeable increase in bioluminescent signals in the GPRC5A-overexpressing group (Group 2) compared to the control group (Group 1), suggesting that GPRC5A overexpression not only facilitated early seeding but also prolonged the persistence of cancer cells. However, the addition of YAP1-TEAD inhibitors (Group 3 and Group 4) mitigated the colonization and growth of GPRC5A-overexpressing cells in the lungs, resulting in significantly reduced or undetectable signals (Fig. 6e and Supplementary Fig. 12a). Conversely, mice injected intravenously with YAP1-rescued KYSE520 cells with GPRC5A silencing exhibited more lung metastatic nodules compared to those without YAP1 overexpression cells (Supplementary Fig. 12b). In summary, the suppression of YAP1 demonstrated the capability to disrupt early implantation and the formation of metastatic nodules of ESCC cells in vivo.
Discussion
The early dissemination process of cancers remains a relatively understudied area, with limited information available on how cancer cells disseminate, lodge, implant, and grow into metastatic tumors. In this study, we aimed to investigate the profile of ESCC lung metastasized cancer cells by isolating different stages of lung DCCs in an experimental mouse model at a single cell resolution level. Our research findings indicate that the early spread of cancer cells, rather than late colonization, involves the activation of genes linked to embryo development and implantation. The process of trophoblast-mediated blastocyst implantation is intricate, encompassing the attachment of the blastocyst to the receptive endometrium and the invasion of trophoblast cells from the conceptus into the endometrium and basement membrane8. Our observations suggest that the mechanisms of adhesion, migration, and invasion exhibited by early DCCs in the lung closely parallel those seen during blastocyst implantation in embryo development. By integrating our model with a human implantation cohort, we have identified several genes that are upregulated in early lung DCCs and are also involved in the subprocesses of embryo implantation (Supplementary Data 2). For example, the transcription factor Fos-like antigen 1 (FOSL1), which is associated with cell motility, has been shown to regulate metastasis, oncogenesis, and epithelial-to-mesenchymal transition in various cancers such as glioblastoma, head and neck squamous cell carcinoma, and cholangiocarcinoma26–28, while also playing a role in trophoblast migration and invasion29. Genes related to cell attachment or adhesion, such as focal adhesion protein (NEDD9), the laminin family (LAMB3, LAMA3, LAMC2), and the integrin family (ITGB6, ITGA5), showed significant upregulation in early lung DCCs. These genes have been extensively studied for their essential roles in successful blastocyst implantation30–33 and the metastatic colonization of different cancer types34–36. Our findings lead us to the conclusion that only cancer cells capable of completing all the necessary steps of embryo implantation successfully can lead to early seeding and subsequent tumor colonization and growth. This event is a critical early step in tumor metastasis and determines whether metastases can be successfully established.
Our study identified a gene of interest, GPRC5A, to be overexpressed in early lung DCCs and trophoblasts of implanting blastocysts. While the impact of GPRC5A on tumor progression varies among different cancer types, its role in regulating the early dissemination of cancer cells had not been previously explored. Our data indicated that GPRC5A-positive cells exhibited characteristics similar to trophoblasts during implantation, aiding in early dissemination and subsequent metastatic tumor formation. Over 80% of early lung DCCs were observed to be GPRC5A-positive cells, providing insight into its role in the early implantation of cancer cells. We hypothesized that GPRC5A-positive cells displayed a trophoblast-like phenotype during early implantation and persisted in metastatic tumors. Cells overexpressing GPRC5A showed a greater ability to survive and colonize in the lungs of mice, supporting our theory that GPRC5A activated a trophoblast-like program for early dissemination. In-vitro experiments similarly confirmed that cancer cells with increased GPRC5A expression were more proficient at migrating and invading. Aside from migration and invasion, GPRC5Ahigh cells possessed a notable capacity for tumor-initiating stemness and resistance to stimuli, aiding in the completion of embryo implantation. GPRC5A-overexpressing colorectal cancer cells consistently displayed cancer stem cell-like characteristics and were viewed as metastasis-initiating cells based on Li et al.’s findings37. Additionally, studies by Greenhough et al.23 demonstrated that high GPRC5A expression protected colorectal cancer SW620 cells from apoptosis during oxygen deprivation and mitigated intestinal mucosal damage during ischemia/reperfusion38. Recent research suggests that the self-renewal and stemness of trophoblasts are essential for embryo implantation and post-implantation placentation18. Similarly, our observations indicated that GPRC5A-high cells exhibited enhanced stem cell-like characteristics, such as increased formation of tumor spheres and upregulation of stemness-related genes (SOX2, NANOG, c-MYC, OCT4). Given that stemness-related genes are not canonical targets of YAP1, we investigated whether typical YAP1 target genes, such as CTGF, CYR61, and ANKRD1, are affected by alterations in GPRC5A. Our results indicated that their expression was not significantly influenced by GPRC5A status (Supplementary Fig. 13a). Since GPRC5A positively regulates the expression of YAP1, CYR61, Cyclin D1, and CTGF, thereby impacting migration and proliferation in pancreatic cancers22, however, these effects do not seem to be present in ESCC. Additionally, several studies have demonstrated that YAP1 can significantly enhance the transcription and protein expression levels of SOX2, NANOG, c-MYC, and OCT439–42, which aligns with our findings.
Therapeutic approaches aimed at effectively impeding the early dissemination of cancer cells remain a relatively uncharted territory. Our findings suggested that GPRC5A presents itself as a promising therapeutic target for obstructing the early spread of ESCC cells, but GPRC5A is also known to showcase high expression levels in healthy human lung tissues, where it acts as a tumor-suppressor gene that hinders the development of lung cancer14. This raises a significant question regarding the potential increase in lung cancer risk among patients undergoing treatment with GPRC5A-targeting medications. Direct targeting of GPRC5A may increase lung cancer risk among patients, and it may be more prudent to consider alternative strategies that focus on targeting the downstream effectors of GPRC5A. This study revealed that the activation of YAP1 and its downstream genes played a crucial role in enabling GPRC5A to carry out its biological functions. Previous findings also demonstrated the contribution of YAP1 to embryo development43. It showed that YAP1 nuclear translocation took part in specification and differentiation of trophoblasts, while YAP1-defficient embryos showed abnormalities in trophoblasts and defects in the yolk sac vasculature44–46. This suggest that YAP1 was an essential regulator of trophoblasts-mediated embryo development. However, the potential function of YAP1 in early lung dissemination of ESCC cells has not been investigated yet.
In this study, we discovered that both genetic and pharmacological inhibition of YAP1 or YAP1-TEAD interaction demonstrated promising outcomes in diminishing the proliferation and advancement of ESCC cells in vitro. Notably, our preliminary findings indicated that inhibitors of YAP1 also mitigate early implantation and reduce the formation of metastatic nodules of ESCC cells in the lungs of mice. Various strategies for inhibiting YAP1/TAZ-TEAD, such as YAP1 expression inhibitors (e.g., ION537), central pocket inhibitors (such as TED-347 and K-975), protein-protein interaction inhibitors (like Celastrol), YAP1 phosphorylation agonists (such as Pazopanib), and YAP1 nuclear localization inhibitors (such as Dasatinib and Verteporfin), are being extensively explored in cancer therapy47. Treatment using YAP/YAZ-TEAD inhibitors in mouse models of mesothelioma has shown significant tumor regression48. A phase 1 clinical trial has been initiated to evaluate the YAP/TAZ-TEAD inhibitor VT3989 (NCT04665206) in patients with mesothelioma and other cancers with NF2 mutations, with encouraging preliminary results showing a 10% (7 out of 69) partial response rate and 90% stable disease rate in refractory mesothelioma49. Ongoing clinical trials include one (NCT05228015) testing the YAP/TAZ-TEAD inhibitor IK-930 in mesothelioma patients, and another (NCT04659096) evaluating an antisense oligonucleotide that targets YAP1 mRNA in patients with neoplasms or advanced primary, recurrent, or metastatic solid tumors47,48. In the case of ESCC, which is often diagnosed at an advanced stage, it may be beneficial to establish an evaluation system for locally advanced ESCC patients without lung metastasis. And utilizing a YAP1 or YAP1-TEAD inhibitor as an optional adjuvant or neoadjuvant treatment after assessment could potentially help prevent lung metastasis based on our findings. However, for patients with lung metastasis, further investigations are warranted to validate the effectiveness of YAP1-targeting interventions in a clinical setting.
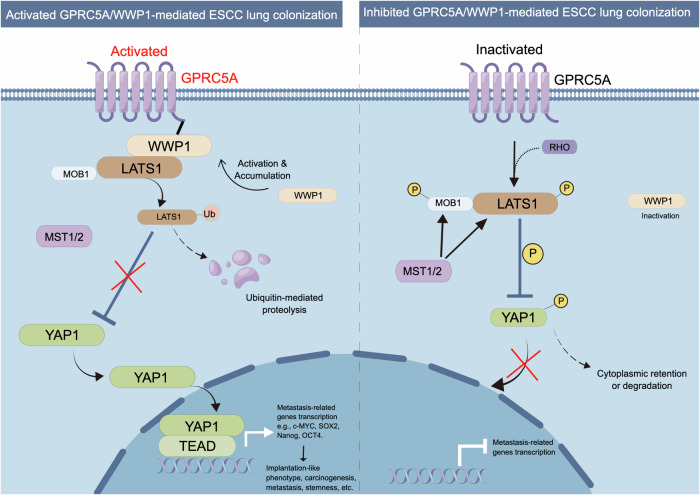
Our findings also suggested that the mechanism by which GPRC5A regulated the activation of YAP1 and its downstream genes was mediated by WWP1-induced LATS1 ubiquitination and degradation (Fig. 7). By performing molecular docking, co-immunoprecipitation, immunofluorescence co-colocalization and protein pull-down assay, we demonstrated a potential interaction between GPRC5A and WWP1. Previous studies have indicated that YAP1 can be phosphorylated and regulated by LATS250,51. Therefore, we investigated whether LATS2 responds to the overexpression or knockdown of GPRC5A. Our results revealed there’s no significant changes associated with alterations in GPRC5A levels (Supplementary Fig. 13a). Therefore, in our study, we identified that the potential interaction between GPRC5A and WWP1 can regulate the ubiquitination-dependent degradation of LATS1 but not LATS2.
Fig. 7. Schematic diagram of the mechanisms by which GPRC5A facilitates the implantation and colonization of ESCC cells, mediated through the WWP1-LATS1-YAP1 signaling axis.
Activated GPRC5A interacts with WWP1, which promotes the polyubiquitination and degradation of LATS1. This process eliminates LATS1’s inhibitory effects on YAP1 nuclear translocation, thereby enhancing the activation of downstream metastasis-related genes. This figure is drawn by figdraw.com.
Cancer metastasis is frequently initiated by the activation of the epithelial-mesenchymal transition (EMT) program52. In this context, we investigated the relationship between GPRC5A-mediated lung colonization in ESCC and the EMT process. Initially, we assessed the expression levels of several established EMT marker genes, including the epithelial marker CDH1 (encoding E-cadherin) and the mesenchymal markers CDH2 (encoding N-cadherin), VIM (encoding Vimentin), SNAI1 (encoding Snail1), SNAI2 (encoding Slug), and TJP1 (encoding ZO-1) at various timepoints using our scRNA-seq data (Supplementary Fig. 13b). Our results revealed a significant increase in the expression of mesenchymal markers (CDH2, TJP1, SNAI2, VIM) at early stages, particularly at 48 hours, followed by a decrease in later stages. Conversely, the epithelial marker CDH1 maintained high expression levels throughout both early and late stages. Additionally, western blot analysis of epithelial (E-cadherin) and mesenchymal (Vimentin, Slug, and Snail) markers further confirmed a hybrid expression pattern of EMT markers in both GPRC5A-overexpressing and silenced cells, underscoring a dynamic EMT status associated with GPRC5A-mediated lung colonization in ESCC (Supplementary Fig. 13c). These findings indicate a dynamic transition in the EMT state of the cells, characterized by an Epithelial-Hybrid-Mesenchymal transition, consistent with previous studies7,53,54.
In summary, our research reveals a previously unrecognized cellular process and the detailed molecular pathway through which GPRC5A potentially cooperates with WWP1 to inhibit LATS1-mediated YAP1 ubiquitination. This collaboration promotes the translocation of YAP1 to the nucleus, where it activates the expression of downstream genes related to implantation and stemness. This mechanism sheds light on the early spread and implantation of tumors in lung metastasis of ESCC, prompting further investigation into its applicability to other metastatic cancers. We conclude that our discoveries present a distinctive opportunity to enhance our comprehension of early tumor metastasis and propose a therapeutic approach for managing or preventing the early dissemination of cancer.
Methods
Ethics
This research complies with all relevant ethical regulations, approved by the ethical committees, including the Committee of the Use of Live Animals in Teaching and Research at the University of Hong Kong and the Committee for Ethical Review of Research Involving Human Subjects at the Sun Yat-Sen University Cancer Center.
Cell lines and cell culture
Human ESCC cell lines KYSE series including KYSE30, KYSE180, KYSE410, KYSE140, KYSE510, KYSE520 were purchased from American Type Culture Collection (ATCC) and cultured in RPMI/1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (PS) (Gibco). Mouse ESCC cell line (mEC25)55 was a gift from Prof. Li Fu and was cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) containing 10% FBS and 1% PS. HEK293T cells were utilized for lentivirus production and were cultured in DMEM supplemented with 10% FBS and 2 mM L-glutamine (Gibco). All cell lines were maintained in a humidified incubator containing 5% CO2 under a constant temperature of 37 °C. All cell lines were tested negative for mycoplasma contamination.
Clinical samples
Six paired sections of ESCC formalin-fixed, paraffin embedded (FFPE) tissue, along with a tissue microarray (TMA) enrolling 148 cases with 148 normal esophagus tissues, 148 primary tumors and 59 metastatic tumors, were provided by the Sun Yat-Sen University Cancer Center (Guangzhou, China). The study included patients who had undergone surgical resection of primary tumors between 2001 and 2005. Comprehensive clinicopathological data and survival outcomes of all participants were meticulously documented and shown in Supplementary Data 3. Approval for this research was granted by the Committee for Ethical Review of Research Involving Human Subjects at the Sun Yat-Sen University Cancer Center, with written informed consent obtained from each patient.
Lung metastasis mouse model and single-cell RNA-seq data analysis
The different-timepoints ESCC lung metastasis mouse model was established according to published protocols7,56. In this study, KYSE30 cells were transduced with two lentiviruses containing firefly luciferase (Luc) and green fluorescence protein (GFP) constructs. Following selection with blasticidin (7 μg/mL, Thermo Fisher Scientific) for 14 days and puromycin (1 μg/mL, Sigma-Aldrich) for 7 days, the KYSE30-GFP-Luc cells (1.2 × 106) in 100 μL phosphate buffered saline (PBS) were intravenously injected into immunodeficient NOD-SCID mice aged 4-6 weeks for the lung metastatic mouse model. GFP+ lung DCCs were isolated at 6 hours, 48 hours, 77 days, and 145 days post-injection using fluorescence-activated cell sorting (FACS) for subsequent single-cell RNA sequencing. Metastatic tumor progression was monitored using bioluminescence imaging (BLI) every 3-5 days with an in-vivo living imaging system. For BLI, mice were injected intraperitoneally with 150 mg/kg D-luciferin (Promega), anesthetized using isoflurane and imaged with PE IVIS Spectrum In Vivo Imaging System (PerkinElmer). Bioluminescent signals were recorded as total intensity of photons/second/cm2/steradian using In-Vivo Living Image software.
Multiplex scRNA-seq was carried out using the Chromium Single Cell Gene Expression transcriptome sequencing platform, specifically employing the Chromium™ Single Cell 3’ v2 Library from 10X Genomics in the USA. A total of 26,712 cells from five distinct timepoints underwent sequencing. Quality control (QC) was performed by ‘Seurat’ package based on number of RNA features and gene counts for single-cell selection and fraction of mitochondria DNA for viable cell selection with the criteria: nFeature RNA > = 200 and nFeature_RNA < = 8000 and nCount_RNA > = 1000 & percent.mt <=15). After quality control, a total of 19,986 cells remained for further analyses. We then normalized the total counts in each individual cell to 10,000, followed by log transformation to generate the normalized data. ‘Anchor-based’ integration followed by principal component analysis (PCA, 2000 highly variable genes and 50 top PCs) for dimensionality reduction, population clustering and feature selection were performed. Finally, Cell Ranger 6.0.2. Single Cell Analysis Pipelines package was utilized for format transformation at ease for importing into Loupe Cell Browser software for visualization and interactive analyses with t-distributed Stochastic Neighbor Embedding7. Stage-specific gene analyses were conducted and represented in heatmaps within the Loupe Cell Browser, with significance defined by a P value < 0.05. Furthermore, an enrichment analysis of these genes was executed and visualized using the Metascape platform (https://metascape.org).
Establishment of stable cell lines with altered gene expression
To generate ESCC cell lines overexpressing GPRC5A, the GPRC5A gene sequence was cloned into the lentiviral PCDH-EF1α-IRES-puro vector (Addgene). For ectopic expression of YAP1, the pLenti6-YAP1-V5 plasmid was obtained from Addgene (Catology number #173002). The genomic sequence of WWP1 was cloned into the pLenti6-CMV-BSD vector (Addgene) for blasticidin selection. ShRNA sequences targeting the genomic sequences of GPRC5A, YAP1, and WWP1 were inserted into either the PLKO.1-puromycin (Addgene) or PLKO.1-neomycin (Tsingke Biotechnology) empty vector. Lentiviral particles were produced by transfecting HEK293T cells with 1 μg of pMD2.G, 2 μg of psPAX2, and 3 μg of the target gene plasmid using Lipofectamine 3000 reagent (Thermo Fisher Scientific) for 48 hours. Parental cells were then infected with the genetically modified lentivirus for 24 hours to establish stable cell lines, followed by selection with appropriate antibiotics for 7-14 days. Details of all primers for cloned sequences and targeting strategies can be found in Supplementary Table 1.
Spheroids formation assay
A total of 2 × 103 or 3 × 103 single cells were plated in 24-well Ultra-Low Attachment Microplates (Corning) in 500 μL of serum-free DMEM/F12 sphere medium (Gibco) supplemented with B27 (1:50, Life Technologies), 20 ng/mL recombinant hEGF (Sigma-Aldrich), 10 ng/mL bFGF (Millipore), and 4 μg /mL insulin (Invitrogen). The medium was refreshed with 50 μL of fresh sphere medium every two days. After a 14-day incubation period, the spheroids were manually counted.
In-Vitro limited dilution assay
Lentivirus-infected KYSE30 and KYSE180 cells were cultured in 96-well ultra-low attachment cell culture microplates (Corning) for 7 to 14 days. The cells were seeded at serially diluted numbers of 800, 400, 200, and 100, or at numbers of 1000, 500, 250, and 125, in 50 μL of the previously described sphere medium. Every two days, 20 μL of fresh sphere medium was added to each well. The frequencies of spheroids in each group were assessed under a microscope. An online tool for extreme limiting dilution analysis (ELDA)57 was employed to calculate input data, including the cell numbers in each well, the total number of cultures tested, the number of positive cultures, and group stratification. The analysis will present the distribution of stem cell frequency and its statistical significance in each group.
Transwell migration and invasion assay
A total of 10 × 104 or 5 × 104 human ESCC cells (KYSE30 and KYSE520) transduced with control, GPRC5A-overexpressing, or GPRC5A-knockdown packaged virus particles were seeded in the upper chamber of Transwell plates with 8.0 μm pore membranes (Corning) in 450 μL of serum-free medium for a 24-hour or 48-hour migration period. The lower chamber was filled with 750 μL of serum-containing complete medium. Similarly, for the Transwell invasion assay, matrigel-coated invasion chambers with 8.0 μm pore membranes (Corning) were utilized. After 24-hour or 48-hour of migration or invasion, cells in the upper chamber were removed using a cotton swab. The migrated and invaded cells on the bottom of the inserts were fixed with 4% paraformaldehyde (PFA), stained with 1% crystal violet, photographed, and quantified.
Foci colony formation assay, cell viability assay
A 6-well cell plate was seeded with 500 or 1000 cells per well and cultured for a period of 7–14 days. The culture medium was refreshed every three to five days. Subsequently, the cells were fixed with 4% PFA and stained with crystal violet. The colonies in each group were manually counted. To assess cell viability under H2O2 stimulation, a 100 μL cell suspension containing 8 × 103 or 1 × 104 cells per well was plated in a 96-well cell culture microplate and exposed to various concentrations of H2O2 for 24 hours. Cell viability was determined using the CellTiter-Glo Luminescent Cell Viability kit (Promega) and measured at a wavelength of 492 nm using the BMG CLARIOstar Plate Reader.
Reactive oxygen species (ROS) detection
ESCC cells, with or without GPRC5A expression, were plated in a 6-well plate at a density of 5 × 105 cells per well and treated with either 10 mM or 15 mM H2O2 for a period of 24 hours. Cells were prepared and stained using Reactive Oxygen Species Assay Kit (Beyotime), following the manufacturer’s instructions. The abundance of ROS was detected using the FITC signal parameter via flow cytometry using NovoCyte Quanteon Flow Cytometer and the mean fluorescence intensity was quantified by NovoExpress software.
Immunoprecipitation and ubiquitination assay
Cells intended for co-immunoprecipitation (co-IP) were first rinsed with Pierce RIPA lysis buffer (Thermo Fisher Scientific) containing protease inhibitor (MedChemExpress) and then detached using a cell scraper. Subsequently, the cell lysates underwent sonication for 10 cycles and centrifugation at 13,000 × g for 15 minutes at 4 °C. The resulting supernatant was then harvested and subjected to a preclearing step using 50 μL of Protein A/G agarose beads (Santa Cruz) for a minimum of 3 hours at 4 °C. 1 μg specific antibodies, such as RAI3 (Santa Cruz, sc-373824), WWP1 (Santa Cruz, sc-390897), LATS1 (Abcam, ab70561), YAP1 (Abclonal, A21216) and normal rabbit/mouse IgG antibodies (Santa Cruz), were individually incubated with the precleared supernatant for 1 hour at 4 °C to facilitate optimal binding. The antibody-bound supernatant was then exposed to 50 μL fresh beads overnight at 4 °C. After four rounds of thorough washing, the antibody-bead-supernatant complexes were eluted for subsequent sodium dodecyl-sulfate polyacrylamide gel (SDS-PAGE) western blot analysis. For the ubiquitination assay, the indicated cells were pre-treated with the proteasome inhibitor MG132 (10 μM, Sigma-Aldrich) for 12 hours before being harvested for immunoprecipitation.
Recombinant His-tagged protein pull-down
Recombinant His-tagged WWP1 protein and His-tagged EGFP protein was customized and purified by Penglai Bio. Co., Ltd. The pull-down assay was conducted using the BeyoGold™ His-tag Purification Resin kit following the manufacturer’s protocol. Briefly, KYSE520 cells lysates were harvested by 500 μL Pierce RIPA lysis buffer (Thermo Fisher Scientific) containing protease inhibitor (MedChemExpress) using a cell scraper. The resin was resuspended and equilibrated with the protein binding buffer, and then mixed with the cell lysates and either His-tagged bait WWP1 protein or His-tagged EGFP protein (negative control). After incubation at 4 °C overnight, the mixture was centrifuged. The resin was washed twice with the protein binding buffer to remove unbound proteins. Subsequently, the resin was treated with SDS-PAGE loading buffer, boiled at 100 °C for 10 minutes, and centrifuged to collect the supernatant, which contains the bound proteins, for use in subsequent Western blotting analysis.
Immunocytochemistry (ICC) and Immunohistochemistry (IHC)
For ICC staining, KYSE30-Ctrl and GPRC5A-overexpressing cells, as well as KYSE520-ShCtrl and -ShGPRC5A cells, were seeded at a density of 4 × 104 cells per well in an 8-well glass slide chamber (Thermo Fisher Scientific). Following the treatment of MG132 (10 μM) for 12 hours, all wells were fixed with 4% PFA for 30 minutes, permeabilized with 0.5% Triton X-100 for 10 minutes, and then blocked with 5% bovine serum albumin (BSA) for 1 hour. Subsequently, each well was subjected to an overnight incubation with the primary antibody (YAP1, Abclonal, A21216), followed by a one-hour incubation with a fluorescence-conjugated secondary antibody and a 10-minute DAPI staining step. Fluorescent images were captured using a Zeiss LSM 880/800 Airy Confocal microscope.
For IHC staining, all tissue samples underwent fixation with 10% formalin, embedding in paraffin, and sectioning into 5μm slices. The paraffin-embedded slides were then deparaffinized and rehydrated sequentially using xylene, 100% ethanol, 90% ethanol, 80% ethanol, 70% ethanol, and distilled water. Antigen retrieval was performed by heating the slides in DAKO target retrieval buffer at 95 °C for 20 minutes, cooling them to room temperature for 2 hours, and subsequently blocking them with 3% H2O2 for 10 minutes. Prior to the application of primary antibodies, the slides were blocked with normal bovine serum albumin antigen for 1 hour at 37 °C. Horseradish peroxidase (HRP)-conjugated secondary antibodies were then used for a 1-hour incubation, followed by treatment with diaminobenzidine reagent solution (Origene) and hematoxylin staining (Vector). Imaging of all slides was carried out using the Akoya Vectra Polaris microscope and analysis was performed using Phenochart software.
Molecular docking
The 3D structural data for WWP1 was sourced from the Protein Data Bank (PDB) database at www.rcsb.org (PDB ID: 5hps). The Uniprot database (https://www.uniprot.org/uniprotkb/Q8NFJ5/entry) offers the predicted structural model for the protein GPRC5A (also known as RAI3), which is accessible for viewing and download. Molecular docking to predict the binding interactions between specified sites on GPRC5A and WWP1 was carried out using the Z-DOCK server developed by Pierce et al.58 The 3D structure and analysis of binding sites in the GPRC5A-WWP1 complex were visualized using the PDBePISA tool (https://www.ebi.ac.uk/pdbe/pisa/) and PyMOL software (https://pymol.org/2/).
Western blot and Cycloheximide (CHX) pulse-chase assay
The cells were harvested, lysed in Pierce RIPA buffer containing protease inhibitors on ice for 30 minutes, and then centrifuged at 13,000 × g for 15 minutes. The resulting supernatants were collected, quantified using a BCA protein assay kit (Thermo Fisher Scientific), and separated on polyacrylamide gels of 10%, followed by transfer onto a PVDF membrane (0.45 μm, Millipore). The membrane was blocked with 5% skimmed milk for 2 hours and then incubated overnight at 4 °C with the primary antibody solution. Details of all primary antibodies used were provided in the Supplementary Data 4. Subsequent to a one-hour incubation with secondary antibodies, the protein bands were visualized using the ECL Western Blot detection reagent in an imaging system. To determine the half-life of the proteins YAP1 and LATS1, the cells transfected with or without GPRC5A-overexpressing lentivirus were exposed to CHX (Sigma-Aldrich, 50 µg/mL) for varying durations (0, 3, 6, 9, 12 hours) and subsequently analyzed through immunoblotting. Several western blots of independent experimental replicates were provided in Supplementary Data 5. The indicated samples derive from the same experiment but different gels for all tested antibodies were processed in parallel in Fig. 4, Fig. 5 and Supplementary Figs. 5, 7, 8, 9, 11 and 13.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme) and reverse transcription was performed using PrimeScript RT Reagent Kit with gDNA eraser (Takara) according to manufacturer’s instructions. Quantitative real-time PCR was performed on Roche Light Cycler 480 system using TB Green Premix Ex Taq II (Tli RNase H Plus) kit (Takara). Each sample was run in triplicate or quadruplicate, and the relative mRNA expression levels were normalized to RPS18. The primer sequences employed for qRT-PCR analysis are detailed in Supplementary Table 2.
Opal multi-channel immunofluorescence (mIF) staining
The study employed mIF analysis utilizing the Opal Polaris 7-color manual IHC kit (Akoya Bioscience, NEL861001KT). Specifically, Opal 520, Opal 480, Opal 570, Opal 620, Opal 690, and Opal 780 were assigned to mark GPRC5A (Sigma-Aldrich, HPA007928), Pan-Cytokeratin (Abcam, ab217916), CD44 (Cell Signal Technology, cst3570), and TACSTD2 (customized, AbMart, #36813-1-1M1/2L8) across various timepoints of mouse lung tissue slides. Another series of mIF staining, utilizing Opal 520, Opal 620, and Opal 570, was conducted to detect GPRC5A (Sigma-Aldrich, HPA007928), Pan-Cytokeratin (Abcam, ab217916), and GFP (Abcam, ab290) in the lung tissues of mice at various timepoints. In samples from ESCC patients, signals corresponding to GPRC5A and Opal 520, Pan-Cytokeratin and Opal 480, and alpha-SMA (Abcam, ab5694) and Opal 620 were identified. For ESCC tissue microarray (TMA) staining, only GPRC5A with Opal 520 and Pan-Cytokeratin with Opal 480 antibodies were used. Nuclei were stained with DAPI for 10 minutes and then sealed with ProLong™ Diamond Antifade Mountant (Thermo Fisher Scientific). Imaging of all slides was performed using the Vectra Polaris Imaging System (Akoya Bioscience), and Phenochart software was employed to separate autofluorescence and analyze multi-channel images.
For the colocalization assay, cells were plated in an 8-well glass slide chamber and fixed with 4% PFA for 20 minutes. Immunofluorescence staining was then performed for GPRC5A (Sigma-Aldrich, HPA007928) and WWP1 (Abcam, ab104440) using Opal 520 and Opal 570, followed by a 10-minute DAPI staining step as per the manufacturer’s instructions. Fluorescent images were obtained using the Alexa 488 and Alexa 555 channels on a Zeiss LSM 800 Airy Confocal microscope.
Transcriptome analysis and enrichment analysis
Total RNAs extracted from KYSE30 cells overexpressing GPRC5A or control cells, as well as KYSE520 cells with GPRC5A knockdown or control cells, were isolated using TRIzol reagent. The RNA quality was assessed using the Agilent 2100 Bioanalyzer, and samples with an RNA integrity number (RIN) ≥ 7 were deemed suitable for further analysis. Subsequent RNA sequencing was carried out on the Illumina sequencing platform (llumina Novaseq 6000) by O.E biotech CO., Ltd in Shanghai, China. Differentially expressed genes (DEGs) were identified with a fold change >2 or <0.5 and a P value < 0.05 using the R “DEseq” package. KEGG and GO enrichment analyses of the DEGs were performed using R.
Transcriptome data from three independent cohorts (GSE45670, GSE161533, GSE26886) were obtained from the GEO database to assess the relative expression of GPRC5A in adjacent normal tissues and esophageal squamous cell carcinoma (ESCC) tumor tissues. Additionally, an analysis of GPRC5A or WWP1 expression in pan-cancers and matched normal tissues, using data from The Cancer Genome Atlas Program (TCGA) database and Genotype-Tissue Expression (GTEx) databases, was conducted via the Home-for-Researchers platform (www.home-for-researchers.com). To explore the downstream pathways of GPRC5A, differentially expressed genes (DEGs) and subsequent enrichment analysis between the high-expression group (upper 25%) and low-expression group (lower 25%) of GPRC5A in ESCC were performed using the Home-for-Researcher platform. The correlation analysis of GPRC5A and WWP1 in TCGA-ESCA cohort was performed via GEPIA2 platform (http://gepia2.cancer-pku.cn/#index).
Subcutaneous tumor-bearing mouse model
In the in vivo cell-derived xenograft mouse model, human or mouse ESCC cells (5 × 106) with GPRC5A overexpression (left dorsal area) and control cells (right dorsal area) were subcutaneously injected into 5-week-old immunodeficient NOD-SCID mice or immunocompetent C57BL/6 N mice. Human KYSE520 cells (5 × 106) with control cells (left dorsal area) and GPRC5A knockdown cells (right dorsal area) were subcutaneously injected into 5-week-old immunodeficient NOD-SCID mice. Tumor growth was evaluated by measuring tumor size and growth rate every 3-5 days. Primary tumors were harvested 30-60 days post-transplantation, and their volumes were calculated using the formula 0.5 × length × width2.
In-vivo therapeutic treatment of YAP1/YAP1-TEAD inhibitors in mice
In order to evaluate the effectiveness of YAP1/YAP1-TEAD inhibitors in preventing early implantation and reducing lung macro-metastases, all NOD-SCID mice with either sex aged 4 to 6 weeks were randomly divided into four groups. Groups 1 and 2 received saline treatment every other day for three cycles. Group 3 received intraperitoneal administration of CA3 (Selleck, cat no. S8661) at a dose of 1 mg/kg every other day for three cycles. Group 4 was orally pre-treated with TED-347 (MedChemExpress, cat no. HY-125269) at a dose of 20 mg/kg every other day for three cycles. Subsequently, Group 1 was intravenously injected with KYSE30 control cells (1.2 × 106 cells per mouse), while the other three groups received GPRC5A-overexpressing KYSE30 cells at the same cell count via tail vein injection. BLI was conducted at baseline (1 hour), 6 hours and 48 hours post-injection to assess early cancer cell implantation in all four groups. Following a week post-injection, all groups underwent the specified treatment regimens for an additional three cycles. Following BLI was performed at 1 month, 2 months, and 3 months to monitor macro-metastases, respectively. To explore the functional role of YAP1 in rescuing lung macro-metastases, we intravenously injected GPRC5A-silenced cells, YAP1-rescued GPRC5A-silenced cells, and control KYSE520 cells (1.2 × 106 cells per mouse in 100 μL PBS) into NOD-SCID mice. At the study endpoint, all mice were humanely euthanized, and lung specimens were collected for histological and immunohistochemical analysis after embedding in paraffin.
Animal
Mice aged 4–6 weeks of either sex, originally obtained from The Jackson Laboratory in the USA (Strain NOD.CB17-Prkdcscid/J; NOD-SCID) and C57BL/6 J mice aged 4-6 weeks of either sex were bred in compliance with an AAALAC International accredited program at the Centre for Comparative Medicine Research (CCMR), HKU, under Specific Pathogen Free (SPF) conditions. The research commenced only after receiving approval from the HKU Committee on the Use of Live Animals in Teaching and Research (CULATR# 23-340) and a license from the Hong Kong SAR Government’s Department of Health, reference number (23-999) in DH/HT&A/8/2/3 Pt.62. The maximal end-point tumor size permitted by the ethics committee is 1.5 cm in diameter and no tumor burden exceeded this limit in our study. Common humane endpoint criteria are involved in this protocol: (i) General well-being: decrease of body weight of more than 20% from baseline; muscle atrophy or emaciation with a body condition score of less than 2; reduced response to external stimuli; moribund, unconscious or comatose; prolonged or irreversible inability to eat or drink; persistent facial displays of pain & distress (i.e., grimace scale score of 2). (ii) Tumor studies: tumor size exceeding 15 mm in mice; tumor ulceration; bodyweight increase of 10% from baseline in situations of ascites or abdominal distension. (iii) General surgery: signs of post-operative infection that cannot be cleared or wound breakdown that cannot be repaired. (iv) Abnormalities of body systems and homeostatic mechanisms: marked increase of respiratory effort or significantly abnormal breathing; severe persistent hypothermia or hyperthermia (e.g. +/− 3oC from physiological norm); chronic deteriorating skin lesion (persisting for longer than 7 days) (e.g. ulcerative dermatitis); chronic deteriorating ocular lesion (persisting for longer than 7 days) (e.g. corneal perforation or severe blepharitis); clinical signs of systemic disease (e.g. jaundice, anaemia, enlarged lymph nodes, etc.). (v) Central or peripheral nervous system: central nervous system disturbance (such as seizure activity or ataxia); persistent paresis/paralysis; self-Induced trauma/self-mutilation leading to exposure of bone or body cavity. The animals were housed in individually ventilated cages in rooms with controlled environmental conditions, adhering to a 12:12 light-dark cycle with the ambient temperature at 20–25 °C and 40–60% humidity, and were provided ad libitum access to a laboratory diet produced by LabDiet, USA.
Statistics and reproducibility
Statistical analysis was conducted, and visualizations were created using GraphPad Prism 8 (Version 8.3.1). For data with a sample size ≤ 10, bar plots were employed, displaying all data points. For data with a sample size > 10, box and whisker plots were utilized to illustrate data distribution. The data presented in bar plots represented the mean values ± standard deviation (SD), while box-and-whisker plots displayed median values ± interquartile range (IQR), with whiskers extending to 1.5 times the IQR, as per the Tukey method. Violin plots were depicted as kernel density estimations (KDE) annotated with median values ± IQR. For parametric data, P values were determined using the two-sided unpaired t-test. For non-parametric data derived from scRNA-seq data, P values were assessed using the two-sided Wilcoxon signed-rank test or the Kruskal-Wallis one-way analysis of variance. Two-sided Pearson and Spearman correlation analyses were performed to evaluate the correlation and statistical significance between two variables. For survival analysis, P values were calculated using the two-sided Log-rank test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
We would like to thank the Centre for PanorOmic Sciences (CPOS), LKS Faculty of of Medicine, The University of Hong Kong, for providing multiple research and analytic facilities in this study. Graphic figures including Figs. 1a, f, 6a, 7 are drawn via FigDraw platform (https://www.figdraw.com/) and BioRender platform (https://www.biorender.com/) with approved publishing licenses. This work is supported by grants from the Hong Kong Research Grant Council (RGC) grants including Collaborative Research Funds (C7065-18GF, X.G., C7026-18GF, X.G., and C4039-19GF, X.G.), Research Impact Fund (R4017-18, X.G., R1020-18F, X.G., and R7022-20, X.G.), General Research Fund (17119322, X.G.), Theme-based Research Scheme Fund (T12-703/22-R, X.G.), the National Natural Science Foundation of China (82072738, X.G., 82273483, X.G.), Shenzhen Key Laboratory for cancer metastasis and personalized therapy (ZDSYS20210623091811035, X.G.), Shenzhen Science and Technology Program (JCYJ20220818103014030, X.G., KQTD20180411185028798, X.G., JCYJ20220818103012025, X.G.), Sanming Project of Medicine in Shenzhen (SZSM202211017, X.G.), Guangdong Science and Technology Department (2020B1212030004, X.G.), the Program for Guangdong Introducing Innovative and Entrepreneurial Team (2019BT02Y198, X.G.), National Natural Science Foundation of China (82303160, Y.Z.), GuangdongBasic and Applied Basic Research Foundation (2023A1515010109, Y.Z.). Xin-Yuan Guan is the Sophie YM Chan Professor in Cancer Research.
Author contributions
H.Z. conceived the study, performed the experiments, conducted data analyses and wrote the manuscript. H.Z., L.T., and B.Z. performed related experiments and revised the manuscript. L.K., C.W., Y.Z., B.R., and J.P. assisted experiments and data analyses. Y.L., K.S., J.L., Y.Y., and Q.L. assisted data analyses and literature review. Y.C., W.Z., C.H., and Y.Q. provided related technical and methodical supports for this study. B.L., and X.G. conceived, supervised the study and revised the manuscript. All authors have agreed with the manuscript for publication.
Peer review
Peer review information
: Nature Communications thanks Jianmin Zhang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The ScRNA-seq data used in this study have been deposited in the GEO database under accession code GSE249057. The bulk RNA sequencing data used in the study are publicly available in GEO under accession numbers GSE268424, GSE45670, GSE161533, GSE26886. Source data are provided with this paper. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hongyu Zhou, Licheng Tan, Baifeng Zhang.
Contributor Information
Beilei Liu, Email: bella007@connect.hku.hk.
Xin-Yuan Guan, Email: xyguan@hku.hk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54251-9.
References
- 1.Klein, C. A. Cancer progression and the invisible phase of metastatic colonization. Nat Rev Cancer20, 681–694 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Valastyan, S. & Weinberg, R. A. Tumor metastasis: molecular insights and evolving paradigms. Cell147, 275–292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantel, K., Alix-Panabières, C. & Riethdorf, S. Cancer micrometastases. Nat Rev Clin Oncol6, 339–351 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Hosseini, H. et al. Early dissemination seeds metastasis in breast cancer. Nature540, 552–558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J Clin74, 12–49 (2024). [DOI] [PubMed] [Google Scholar]
- 6.Ai, D. et al. Patterns of distant organ metastases in esophageal cancer: a population-based study. J Thorac Dis9, 3023–3030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong, C. N. et al. Identification and Characterization of Metastasis-Initiating Cells in ESCC in a Multi-Timepoint Pulmonary Metastasis Mouse Model. Adv Sci (Weinh)11, e2401590 (2024). [DOI] [PMC free article] [PubMed]
- 8.Wang, J. et al. Embedding similarities between embryos and circulating tumor cells: fundamentals of abortifacients used for cancer metastasis chemoprevention. J Exp Clin Cancer Res40, 300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashary, N., Tiwari, A. & Modi, D. Embryo Implantation: War in Times of Love. Endocrinology159, 1188–1198 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Ferretti, C., Bruni, L., Dangles-Marie, V., Pecking, A. P. & Bellet, D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update13, 121–141 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Chen, J. et al. The unique pharmacological characteristics of mifepristone (RU486): from terminating pregnancy to preventing cancer metastasis. Med Res Rev34, 979–1000 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Zhou, F. et al. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature572, 660–664 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Huang, F. J., Wu, T. C. & Tsai, M. Y. Effect of retinoic acid on implantation and post-implantation development of mouse embryos in vitro. Hum Reprod16, 2171–2176 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Zhong, S. et al. Lung Tumor Suppressor GPRC5A Binds EGFR and Restrains Its Effector Signaling. Cancer Res75, 1801–1814 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Yang, Z. et al. Single-cell Sequencing Reveals Variants in ARID1A, GPRC5A and MLL2 Driving Self-renewal of Human Bladder Cancer Stem Cells. Eur Urol71, 8–12 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zhang, L. et al. Elevation of GPRC5A expression in colorectal cancer promotes tumor progression through VNN-1 induced oxidative stress. Int J Cancer140, 2734–2747 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Zhou, H., Tan, L., Liu, B. & Guan, X. Y. Cancer stem cells: Recent insights and therapies. Biochem Pharmacol209, 115441 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Saha, B. et al. TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc Natl Acad Sci USA117, 17864–17875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, B. et al. Pomegranate juice and punicalagin attenuate oxidative stress and apoptosis in human placenta and in human placental trophoblasts. Am J Physiol Endocrinol Metab302, E1142–E1152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey, A., Varelas, X. & Guan, K. L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov19, 480–494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanconato, F., Cordenonsi, M. & Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang, W. et al. Upregulated GPRC5A disrupting the Hippo pathway promotes the proliferation and migration of pancreatic cancer cells via the cAMP-CREB axis. Discov Oncol14, 17 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenhough, A. et al. Cancer cell adaptation to hypoxia involves a HIF-GPRC5A-YAP axis. EMBO Mol Med10, e8699 (2018). [DOI] [PMC free article] [PubMed]
- 24.Yeung, B., Ho, K. C. & Yang, X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS One8, e61027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salah, Z., Cohen, S., Itzhaki, E. & Aqeilan, R. I. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle12, 3817–3823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, Z. et al. FOSL1 promotes proneural-to-mesenchymal transition of glioblastoma stem cells via UBC9/CYLD/NF-κB axis. Mol Ther30, 2568–2583 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, M. et al. FOSL1 promotes metastasis of head and neck squamous cell carcinoma through super-enhancer-driven transcription program. Mol Ther29, 2583–2600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallejo, A. et al. FOSL1 promotes cholangiocarcinoma via transcriptional effectors that could be therapeutically targeted. J Hepatol75, 363–376 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Renaud, S. J., Kubota, K., Rumi, M. A. & Soares, M. J. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem289, 5025–5039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, T., Greenblatt, E. M., Shin, M. E., Brown, T. J. & Chan, C. Endometrial laminin subunit beta-3 expression associates with reproductive outcome in patients with repeated implantation failure. J Assist Reprod Genet38, 1835–1842 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao, Y. et al. The impact of luteal phase support on gene expression of extracellular matrix protein and adhesion molecules in the human endometrium during the window of implantation following controlled ovarian stimulation with a GnRH antagonist protocol. Fertil Steril94, 2264–2271 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Shi, L., Li, H. & Wang, L. Genetic parameters estimation and genome molecular marker identification for gestation length in pigs. Front Genet13, 1046423 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua, R. et al. Transcriptome regulation of extracellular vesicles derived from porcine uterine flushing fluids during peri-implantation on endometrial epithelial cells and embryonic trophoblast cells. Gene822, 146337 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Hu, Z. et al. Histone deacetylase inhibitors promote breast cancer metastasis by elevating NEDD9 expression. Signal Transduct Target Ther8, 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, F. et al. Stanniocalcin 1 promotes metastasis, lipid metabolism and cisplatin chemoresistance via the FOXC2/ITGB6 signaling axis in ovarian cancer. J Exp Clin Cancer Res41, 129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, H. et al. LAMB3 mediates apoptotic, proliferative, invasive, and metastatic behaviors in pancreatic cancer by regulating the PI3K/Akt signaling pathway. Cell Death Dis10, 230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, R. et al. Single-cell transcriptomic analysis deciphers heterogenous cancer stem-like cells in colorectal cancer and their organ-specific metastasis. Gut73, 470–484 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, W. et al. Exosomal circEZH2_005, an intestinal injury biomarker, alleviates intestinal ischemia/reperfusion injury by mediating Gprc5a signaling. Nat Commun14, 5437 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, J. et al. YY1-induced DLEU1/miR-149-5p Promotes Malignant Biological Behavior of Cholangiocarcinoma through Upregulating YAP1/TEAD2/SOX2. Int J Biol Sci18, 4301–4315 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, X. et al. Hippo-YAP signaling controls lineage differentiation of mouse embryonic stem cells through modulating the formation of super-enhancers. Nucleic Acids Res48, 7182–7196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, C. L. et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest123, 2832–2849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Murakami, S. et al. A yap-myc-sox2-p53 regulatory network dictates metabolic homeostasis and differentiation in kras-driven pancreatic ductal adenocarcinomas. Dev Cell51, 113–128.e119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu, M. et al. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target Ther7, 376 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frum, T., Watts, J. L. & Ralston, A. TEAD4, YAP1 and WWTR1 prevent the premature onset of pluripotency prior to the 16-cell stage. Development146, dev179861 (2019). [DOI] [PMC free article] [PubMed]
- 45.Lorthongpanich, C. et al. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev27, 1441–1446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morin-Kensicki, E. M. et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol26, 77–87 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baroja, I., Kyriakidis, N. C., Halder, G. & Moya, I. M. Expected and unexpected effects after systemic inhibition of Hippo transcriptional output in cancer. Nat Commun15, 2700 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franklin, J. M., Wu, Z. & Guan, K. L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat Rev Cancer23, 512–525 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Yap, T. A. et al. Abstract CT006: First-in-class, first-in-human phase 1 trial of VT3989, an inhibitor of yes-associated protein (YAP)/transcriptional enhancer activator domain (TEAD), in patients (pts) with advanced solid tumors enriched for malignant mesothelioma and other tumors with neurofibromatosis 2 (NF2) mutations. Cancer Research83, CT006 (2023). [Google Scholar]
- 50.Qin, M. et al. LATS2 condensates organize signalosomes for Hippo pathway signal transduction. Nat Chem Biol20, 710–720 (2024). [DOI] [PubMed] [Google Scholar]
- 51.Liu, M. et al. Zinc-dependent regulation of zeb1 and yap1 coactivation promotes epithelial-mesenchymal transition plasticity and metastasis in pancreatic cancer. Gastroenterology160, 1771–1783.e1771 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwab, A. et al. Zeb1 mediates EMT/plasticity-associated ferroptosis sensitivity in cancer cells by regulating lipogenic enzyme expression and phospholipid composition. Nat Cell Biol26, 1470–1481 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nobre, A. R. et al. ZFP281 drives a mesenchymal-like dormancy program in early disseminated breast cancer cells that prevents metastatic outgrowth in the lung. Nat Cancer3, 1165–1180 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Lu, W. & Kang, Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell49, 361–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang, T., Yang, J., Liu, B. & Fu, L. A new mouse esophageal cancer cell line (mEC25)-derived pre-clinical syngeneic tumor model for immunotherapy. Cancer Commun (Lond)40, 316–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jakab, M. et al. Lung endothelium exploits susceptible tumor cell states to instruct metastatic latency. Nat Cancer5, 716–730 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Pierce, B. G. et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics30, 1771–1773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The ScRNA-seq data used in this study have been deposited in the GEO database under accession code GSE249057. The bulk RNA sequencing data used in the study are publicly available in GEO under accession numbers GSE268424, GSE45670, GSE161533, GSE26886. Source data are provided with this paper. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.