Abstract
IKKα is a multifunctional serine/threonine kinase that controls various biological processes, either dependent on or independent of its kinase activity. However, the importance of the kinase function of IKKα in human physiology remains unknown since no biallelic variants disrupting its kinase activity have been reported. In this study, we present a homozygous germline missense variant in the kinase domain of IKKα, which is present in three children from two Turkish families. This variant, referred to as IKKαG167R, is in the activation segment of the kinase domain and affects the conserved (DF/LG) motif responsible for coordinating magnesium atoms for ATP binding. As a result, IKKαG167R abolishes the kinase activity of IKKα, leading to impaired activation of the non-canonical NF-κB pathway. Patients carrying IKKαG167R exhibit a range of immune system abnormalities, including the absence of secondary lymphoid organs, hypogammaglobulinemia and limited diversity of T and B cell receptors with evidence of autoreactivity. Overall, our findings indicate that, unlike a nonsense IKKα variant that results in early embryonic lethality in humans, the deficiency of IKKα‘s kinase activity is compatible with human life. However, it significantly disrupts the homeostasis of the immune system, underscoring the essential and non-redundant kinase function of IKKα in humans.
Subject terms: Genetics research, Immunological disorders, Primary immunodeficiency disorders, NF-kappaB
IKKα is an essential regulator of the noncanonical NF-κB signalling, lack of which is incompatible with life. Here authors show that a homozygous missense variant in humans, G167R, disrupting its kinase activity upstream of the non-canonical NF-κB pathway, leads to disrupted innate and adaptive immune functions while largely sparing other major organ systems.
Introduction
The NF-κB signalling pathway serves as the primary regulator of acute inflammatory responses triggered by a large number of stimuli1. The IKK complex plays a crucial role in coordinating the transmission of signals between receptors and formation of NF-κB heterodimers, leading to transcriptional activation in a cell type- and signal-dependent manner2. This complex consists of three subunits: IKKα, IKKβ and NEMO. Extensive research has elucidated the molecular intricacies of two distinct NF-κB pathways, namely the canonical and non-canonical pathways1. Through the analysis of different mouse models, the non-canonical NF-κB pathway has emerged as a critical player in several biological processes including the development of medullary thymic epithelial cells (mTEC) and secondary lymphoid organs (SLO)3–5, the establishment of immune tolerance6,7 and the differentiation and survival of populations of B cells8–10 and T follicular helper (Tfh) cells11,12.
The activation of the non-canonical NF-κB pathway is controlled by two non-redundant kinases, namely the NF-κB-inducing kinase (NIK) and IKKα. The abundance of NIK protein, which is post-translationally regulated, is a critical limiting step in the activation of the non-canonical NF-κB pathway13,14. Upon activation of receptors such as BAFF-R, CD40, LTβR, RANK, Fn14 or EDA2R in different cell types, NIK is stabilized, and it phosphorylates IKKα at Ser176 and Ser180 in the activation loop15. The activated IKKα, in turn, phosphorylates the NFKB2 precursor p100 at multiple serine residues located at the N- and C-terminus16. Ser 866 and Ser 870 phosphorylation marks p100 for ubiquitination, proteasome-mediated partial degradation, and conversion to p52. The resulting p52 forms a heterodimer with RelB, which transactivates target genes involved in various cellular processes17.
To date, a range of kinase-dependent and kinase-independent functions of IKKα have been reported, primarily based on studies using mouse models. In addition to phosphorylating p100, IKKα has been shown to phosphorylate several cytoplasmic and nuclear substrates in response to diverse stimuli. These substrates include TAXBP118, PIAS119, CBP20, AMBRA121, ATG16L122, FOXA223, NPM124, BRD425 and Histone H3.326. The roles of IKKα vary depending on the involvement of its kinase-function or NF-κB pathway. For example, in a kinase-dependent but NF-κB-independent manner, IKKα regulates IL-17A expression in Th17 cells by modulating the phosphorylation of histone H3 (Ser10) and/or retinoid acid-related orphan receptor (ROR)γt (Ser376)27,28. Conversely, in both kinase-independent and NF-κB-independent manners, IKKα functions as a scaffold and regulates keratinocyte differentiation by interacting with SMAD2/329–31.
Due to its distinct roles in various pathways, cell types, and sub-cellular locations, non-synonymous variants in different domains of IKKα can offer valuable insights into understanding the multifunctional nature of IKKα and its impact on human physiology. In this regard, human IKKα deficiency resulting from a nonsense variant (IKKαQ422X) has been associated with skeletal and skin abnormalities during embryonic development, leading to lethality32,33. Conversely, a homozygous missense variant in the helix-loop-helix (HLH) domain of IKKα (IKKαY580C), which does not affect its kinase activity but disrupts its interaction with NIK, does not lead to embryonic lethality but instead causes immune and skeletal abnormalities, accompanied by growth retardation, in the affected individual34. Moreover, somatic inactivating mutations of IKKα have been reported in human malignancies, indicating its role as a tumour suppressor in lung adenocarcinoma and cutaneous squamous cell carcinoma35,36. However, the phenotypic consequences of homozygous missense variants directly impairing the kinase function of human IKKα have not been reported thus far, and therefore, the impact of kinase inactive IKKα on human physiology remains unknown.
In our study, we report three patients who carry the same germline homozygous missense variant (IKKαG167R) located in the activation loop of the kinase domain of IKKα. Unlike previously reported variants (IKKαQ422X and IKKαY580C), patients with the IKKαG167R variant do not exhibit severe skeletal or skin abnormalities. However, they present a wide range of aberrations leading to combined immunodeficiency, increased susceptibility to infections, and disrupted self-tolerance.
Results
Identification of kinase domain mutant IKKαG167R
We identified three children from two unrelated Turkish families who were born as a result of consanguineous marriages. Initially, these children were hospitalized due to clinical symptoms including recurrent upper and lower airway infections, chronic diarrhea, and hypogammaglobulinemia. Throughout their medical history, the patients were diagnosed with numerous viral, bacterial, and fungal infections including rotavirus, adenovirus, SARS-CoV-2, cytomegalovirus (CMV), bocavirus, rhinovirus, H. influenzae, C. albicans and S. enterica. Patient-1 (P1) had tested CMV positive several times despite treatments with ganciclovir. A full medical history, including clinical symptoms and test results are shown in Table S1. The patients exhibited higher serum levels of liver enzymes and lower levels of haemoglobin and haematocrit compared to the reference values for their respective age groups (Table S2) suggesting ongoing liver damage. Of the three patients identified, P1, who is the older sibling of P2, experienced growth retardation, with weight and height measurements at the 0.12th and 0.04th percentiles, respectively, at the age of 7. In contrast, both P2 and P3 are within the 25th percentile for weight and between the 25th and 50th percentile for height. Blood leukocyte and lymphocyte numbers were within (P1) or slightly above (P2 and P3) normal reference ranges (Table S2). P1 had the most severe form of the disease and developed liver failure and passed away 5 months after a successful liver transplant due to complications, including cholangitis, sepsis, and multiple organ failure (Table S1).
To identify the underlying genetic variant(s) in these patients, we performed whole exome sequencing (WES) using genomic DNA from blood samples. This analysis identified a shared homozygous missense variant (c.499 G > A, p.G167R) in the CHUK gene that codes for the IKKα protein. No other candidate genes were identified in WES analysis (Table S3). Family pedigrees confirmed the consanguineous marriages and the germline transmission of the variant (Fig. 1A, B), and Sanger sequencing confirmed that the asymptomatic parents were heterozygous carriers of this variant (Fig. 1C). Although the two families are not directly related, they are from the same region, which suggests the possibility of a shared founder variant.
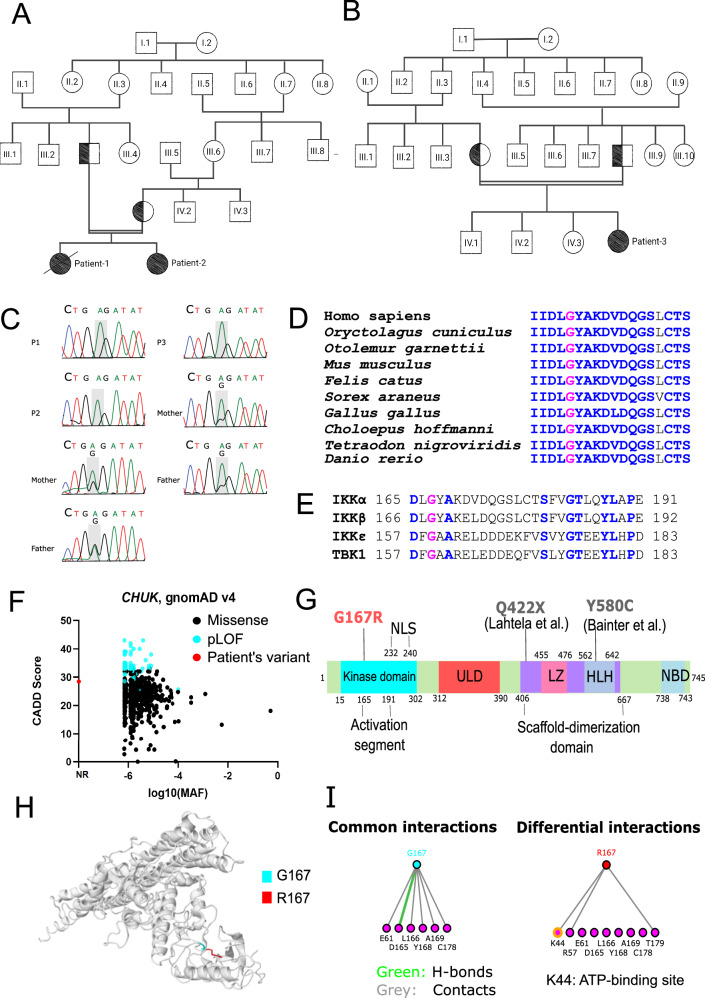
Fig. 1. Identification of IKKαG167R homozygous germline missense variant in three patients.
A, B Family pedigree of P1, P2 and P3 showing the consanguineous marriage. All parents of patients are heterozygous for this variant and germline transmission is responsible for the homozygosity. Patient-1 is deceased as indicated by the black diagonal line. Double horizontal lines in pedigree indicate consanguinity. C Sanger sequencing results of patients and their parents, showing the nucleotide change 499 G > A in the component of inhibitor of nuclear factor kappa B kinase complex (CHUK) gene coding for IKKα. D Evolutionary conservation of G167 residue in IKKα of different species is shown based on the multiple sequence alignment. G167 is indicated by pink, conserved residues are indicated by blue and non-conserved residues are indicated by black. E G167 residue in IKKα (indicated by pink) is conserved in the kinase domain of other IKK and IKK-related kinases (IKKβ, IKKε and TANK binding kinase 1 (TBK1)). Other conserved residues are indicated by blue and non-conserved residues are indicated by black. F CADD vs MAF plot of patient’s variant together with all missense and predicted loss-of-function (pLOF) CHUK gene variants (797 variants in total) obtained from the Genome Aggregation Database (gnomAD) v4 datasets. NR: Not reported. G Different domains and previously reported homozygous variants in human IKKα. Kinase domain, ULD: ubiquitin-like domain, LZ: Leucine zipper, HLH: Helix-Loop-Helix, NBD: NEMO-binding domain, NLS: Nuclear localization signal. Q422X and Y580C variants were previously reported in other studies. H The impact of IKKαG167R on previously reported structure of IKKαWT (PDB: 5EBZ) was indicated using Missense3D tool. G167 is indicated by blue and R167 is by red. I The effect of IKKαG167R variant on interactions of different residues was investigated using I see in 3D (iCn3D) Structure Viewer (PDB: 5EBZ). Common and different interactions between IKKαWT and IKKαG167R were indicated. A notable differential interaction between residue G167 and ATP-binding site K44 (yellow in colour) was indicated. Green colour indicates H-bonds and grey colour indicates contacts with residues.
Several lines of evidence suggest a critical role for the G167 residue in IKKα structure and function. IKKαG167 is well-conserved among different organisms (Fig. 1D) and is part of an evolutionarily constrained region of the protein37. The corresponding residue is also conserved in the activation loop of other IKK and IKK-related kinases (IKKβ, IKKε, and TBK1) (Fig. 1E). Nonetheless, IKKαG167R has not been previously described in publicly available genome variant databases TOPMed38 or gnomAD v439 (Fig. 1F). Additionally, IKKαG167R has not been reported as a somatic mutation in cancer genomics databases COSMIC40 and cBioPortal41. G167 is located within the activation segment of the kinase domain of IKKα, away from the previously reported variants (IKKαQ422X and IKKαY580C) (Fig. 1G). In silico prediction tools, including CADD42, PolyPhen-243, DEOGEN244, MutationTaster202145 and MutPred246 all predicted that IKKαG167R is deleterious. Furthermore, Missense3D47, CUPSAT48 and MutPred246 predicted structural abnormalities resulting from IKKαG167R (Tables S4–S6). Importantly, IKKαG167 is part of a conserved DF/LG motif present in kinases that coordinates magnesium binding near an ATP-binding site15. Analysis of IKKαG167R structure using iCn3D49 based on IKKαWT structure (PDB: 5EBZ)15 suggested that R167, unlike G167, forms new molecular interactions with the ATP-binding residue K44 (Fig. 1H, I), implicating the modulation of the kinase function of the mutant IKKα.
An ATP-binding mutant IKKαK44A was previously found to destabilize mouse IKKα in tissues resulting in reduced IKKα protein and impaired kinase activity50. In our patients with IKKαG167R, mRNA and protein levels of IKKα in PBMCs were found to be comparable to those of healthy controls (Fig. 2A–C). Since the kinase function of IKKα is crucial for signal-induced phosphorylation (Ser866 and Ser870), polyubiquitination, and proteasome-mediated partial degradation of p100 into p52, we examined this conversion in patient cells and healthy controls. When PBMCs stimulated with anti-CD3 and anti-CD28 were analysed, a significant defect in p52 accumulation was observed in patient cells compared to cells from a healthy control or heterozygous parent (Fig. 2D, E). These data demonstrate that IKKαG167R exhibits impaired function. The same defect in p52 conversion was also observed in CD40L-stimulated PBMCs of patients (Fig. 2F, G).
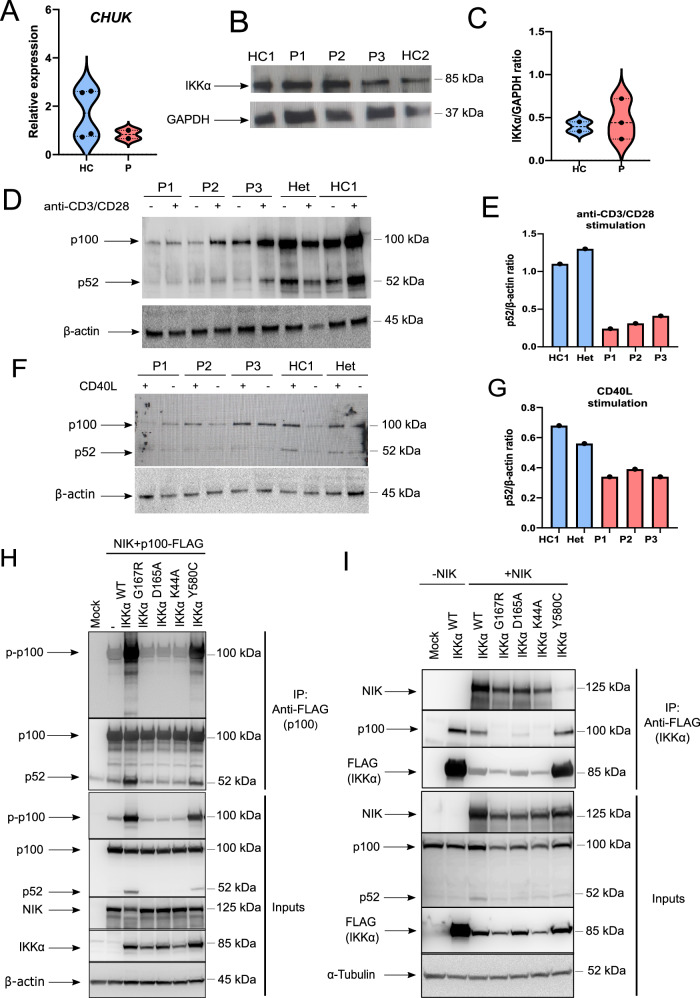
Fig. 2. Testing non-canonical NF-κB pathway activation in patient cells and genome-edited HEK293T cells.
A Transcript levels of CHUK in PBMCs were quantified with qRT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels were used as a normalisation control (n = 4 healthy controls and 2 patients. Two-tailed, non-paired t-test was used for statistical analysis). B Protein levels of IKKα in PBMCs of healthy controls and patients, as determined by western blotting. GAPDH was used as a loading control. Data are representative of three independent experiments. C Quantification of protein levels of IKKα as determined by IKKα/GAPDH ratio in quantitative analysis of band intensities (n = 2 healthy controls and 3 patients. Two-tailed, non-paired t-test was used for statistical analysis). D PBMCs were stimulated with anti-CD3 and anti-CD28 antibodies for 2 days and immunoblotting was performed to analyse p100 to p52 conversion (n = 3 patients and 2 healthy controls. β-Actin was used as a loading control). E Ratio of p52 to β-actin in healthy controls and patient samples, as determined by quantitative image analysis of band intensities (in stimulated conditions) in western blotting in (D). F PBMCs were stimulated with recombinant CD40L (200 ng/ml) and IL-4 (100 ng/ml) for 18 h and p100 to p52 conversion was analysed with immunoblotting (n = 3 patients and 2 healthy controls. β-actin was used as a loading control). G Ratio of p52 to β-actin in healthy controls and patient samples, as determined by quantitative image analysis of band intensities (in stimulated conditions) in western blotting in (F). H Phosphorylation of p100 at Ser 866 and 870 was assessed by co-immunoprecipitation of p100-FLAG with anti-FLAG antibodies and immunoblotting in IKKα KO-HEK293T cells reconstituted with different mutants. Data are representative of four independent experiments. I The interaction between different IKKα mutants and p100 and NIK was investigated using co-immunoprecipitation of IKKα-FLAG with anti-FLAG antibodies in HEK293T cells. Data are representative of three independent experiments.
IKKαG167R exhibits impaired p100 phosphorylation and processing into p52
To investigate the causality of IKKαG167R, we first generated IKKα-KO HEK293T cells using CRISPR/Cas9-mediated genome editing. A PAM sequence in Exon−1 of the CHUK gene was targeted (Figure S1A). Single cell sorting and expansion of different clones revealed the loss of IKKα protein expression in three of the clones examined (Figure S1B). Sequencing of genomic DNA from these clones confirmed the creation of a stop codon (TGA) at the end of Exon−1 in Clone−10 (Figure S1C), which was used in all experiments comparing the function of IKKαG167R and wild type (IKKαWT).
Given that NIK is constitutively degraded in cells under basal conditions and that NIK overexpression is sufficient to activate the non-canonical NF-κB pathway51,52, we used NIK and full-length p100 overexpression system to demonstrate the causality between IKKαG167R and defects in non-canonical NF-κB signalling. Consistent with the essential role of IKKα in p100 processing, IKKα-KO HEK293T cells were unable to process p100 into p52 upon NIK and p100 overexpression (Fig. 2H). To compare the relative deficiency of the kinase activity of IKKαG167R compared to IKKαWT and other mutants, we then reconstituted IKKα-KO HEK293T cells with NIK and p100-FLAG together with IKKαWT or mutants for a side-by-side comparison. We included the ATP-binding mutant IKKαK44A53, the IKKαD165A mutant to investigate the critical role of the DLG motif (165DLG167) where IKKαG167R is located, and the previously reported IKKαY580C mutant34. While IKKαWT was able to rescue defective p100 phosphorylation (Ser866/Ser870) and processing into p52, neither IKKαG167R nor IKKαK44A and IKKαD165A could rescue these defects, confirming the essential role of both the G167 and D165 residues in the kinase function of human IKKα (Fig. 2H). We also immunoprecipitated p100-FLAG and quantified p100 phosphorylation (Ser866 and 870) by western blotting. Both D165A and G167R variants were indistinguishable from kinase-dead IKKαK44A in their inability to phosphorylate p100 in both input and immunoprecipitated samples (Fig. 2H). Notably, in the presence of NIK overexpression, the impact of IKKαY580C mutant on p100 phosphorylation and processing was not as severe as that of the kinase domain mutants (IKKαG167R, IKKαD165A, and IKKαK44A) in both input and co-immunoprecipitated samples (Fig. 2H).
It was previously shown that the kinase activity of IKKα was critical for interaction with its substrates such as TAXBP118 and ASC54. To further support our observation of the defective kinase activity of IKKαG167R, we looked at the interaction between endogenous p100 and overexpressed IKKαWT or mutants in HEK293T cells. All kinase domain mutants tested (IKKαG167R, IKKαD165A, and IKKαK44A) were severely impaired in their interaction with p100 (Fig. 2I). However, HLH domain mutant IKKαY580C was not impaired in its ability to interact with p100 (Fig. 2I) consistent with its normal kinase activity34. Further supporting different mechanisms of action between IKKαY580C and IKKαG167R, all kinase domain mutants tested were able to interact with NIK, unlike IKKαY580C, which was previously shown to be defective in NIK interaction34 (Fig. 2I). Altogether, these data clearly suggest that the defect in p100 phosphorylation and processing was due to the deficient kinase activity of IKKαG167R, and not due to impaired NIK interaction. It is also noteworthy that, although we observed comparable mRNA and total protein levels of IKKα in PBMCs from different IKKαG167R patients compared to healthy controls, when equal amounts of plasmids were transfected into IKKα-KO HEK293T cells, the IKKαG167R mutant, similar to the kinase-dead IKKαK44A, consistently showed diminished expression levels compared to IKKαWT (Fig. 2H, I). This side-by-side comparison suggests lower stability of IKKαG167R and IKKαK44A, unlike IKKαY580C, in this overexpression system.
IKKαG167R variant leads to a deficiency in secondary lymphoid organs, memory B cell differentiation, and a restricted BCR repertoire
Consistent with the essential role of IKKα in activating non-canonical NF-κB signalling, which is important for the development of SLOs, physical examination of all patients revealed the absence of tonsils and palpable lymph nodes. Lymphoscintigraphy analysis in P1 also revealed the absence of lymphatic flow (Fig. 3A). While previous studies in mouse models have reported the transcriptional impact of IKKα dysfunction on mouse bone marrow and splenic B cells9,10 and medullary thymic and thymic capsular fibroblasts34, the transcriptional dysregulation caused by IKKα variants in the human immune system is unknown. Therefore, to understand the consequences of IKKαG167R on the transcriptional regulation and overall function of the human immune system, we performed bulk RNA-seq analysis of PBMCs from P1 and P3 (PBMCs from P2 were not available for this analysis) and compared them to three sex-matched healthy paediatric control samples.
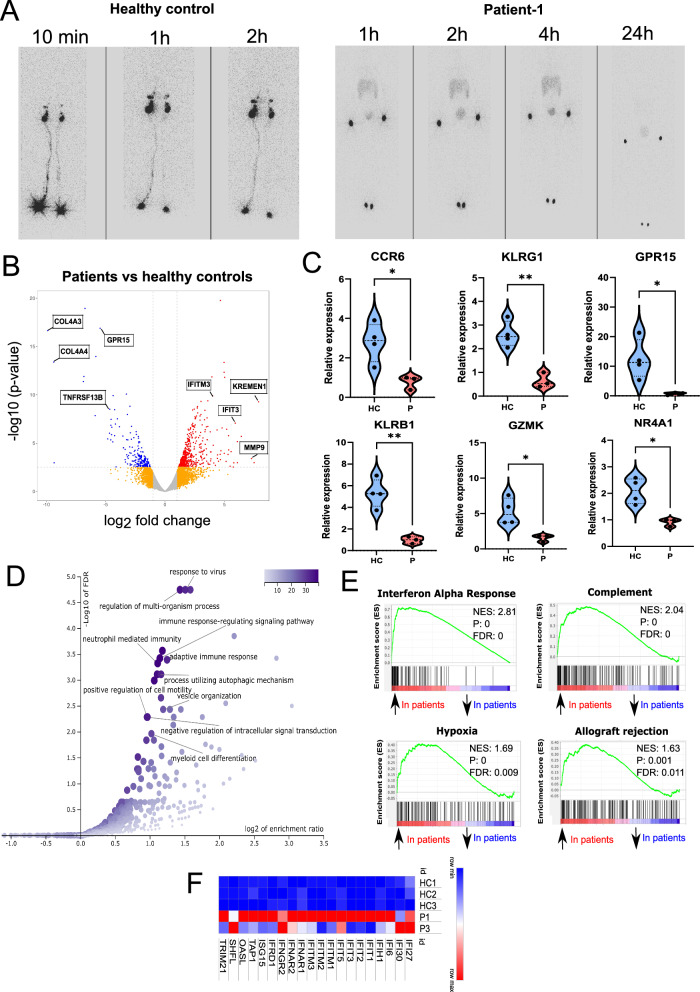
Fig. 3. Defective SLO and peripheral B cell development in IKKαG167R patients.
A Lymphoscintigraphy analysis in a healthy paediatric control and P1 showing the absence of lymphatic flow and therefore lymph nodes in the patient. Images were taken 10 min, 1-, 2-, 4- and 24 h post injection of radiotracer. B Volcano plot analysis of differentially expressed genes (DEGs) in healthy controls (n = 3) vs IKKαG167R patients (P1 and P3). Differential gene expression analysis was conducted using the likelihood ratio test within the edgeR package90, which is based on a negative binomial distribution model. P-values were adjusted for multiple testing using the Benjamini-Hochberg procedure. Genes with FDR-adjusted p-values less than 0.05 were considered statistically significant. C qRT-PCR analysis of C-C motif chemokine receptor 6 (CCR6) (p = 0.0212), killer cell lectin like receptor G1 (KLRG1) (p = 0.0027), G protein-coupled receptor 15 (GPR15) (p = 0.0322), killer cell lectin like receptor B1 (KLRB1) (p = 0.0028), granzyme K (GZMK) (p = 0.0215), and nuclear receptor subfamily 4 group A member 1 (NR4A1) (p = 0.0105) transcripts. GAPDH levels were used as a normalisation control (n = 4 healthy controls and n = 3 patients, two-tailed, non-paired t-test was used for statistical analysis). D Over representation analysis (ORA) of DEGs, as performed by WebGestalt, showing biological processes significantly associated with DEGs. E Selected hits of Gene Set Enrichment Analysis (GSEA) of DEGs. Weighted Kolmogorov–Smirnov test was used for statistical analysis56. F Heatmap analysis of DEGs involved in interferon response based on normalized CPM values in RNA-seq analysis.
We identified 709 differentially expressed genes (DEG) (505 upregulated, 204 downregulated) in patient samples compared to the healthy controls (Fig. 3B) (Data S1). Differential expression of selected genes was also verified by qRT-PCR (Fig. 3C). Functional enrichment analysis of the DEGs using WebGestalt55 revealed dysregulation of several biological processes in patients including “response to virus” and “neutrophil-mediated immunity” (Fig. 3D). Similarly, Gene Set Enrichment Analysis (GSEA)56 identified “interferon alpha response” as the most enriched Hallmark Gene Set among other enriched gene sets including Complement, Hypoxia and Allograft rejection (Fig. 3E) that are each largely distinct from each other in the genes they contain. We observed increased expression of interferon response genes, which was most pronounced in P1 (Fig. 3F). This may be due to systemic inflammation, viral infections or dysbiosis, however, it is interestingly similar to a previous observation of interferonopathy in patients with RelB deficiency57. This is also in line with the observation of higher viral susceptibility of patients with different genetic defects in the non-canonical NF-κB pathway as recently reported52.
A gene signature characterized by reduced expression of transcripts in B cell biology was also evident in patient PBMCs (Fig. 4A). The proportions of circulating B cells (CD19+) in P2 and P3 were normal compared to age-matched national reference values and below the lower reference value in P1 (Table 1). In contrast, all patients showed severe reductions in proportions of circulating marginal zone B cells (CD19+IgD+CD27+) and switched memory B cell populations (CD19+IgD-IgM-CD27+), and higher proportions of naïve B cells (CD19+IgD+IgM+CD27-) (Table 1).
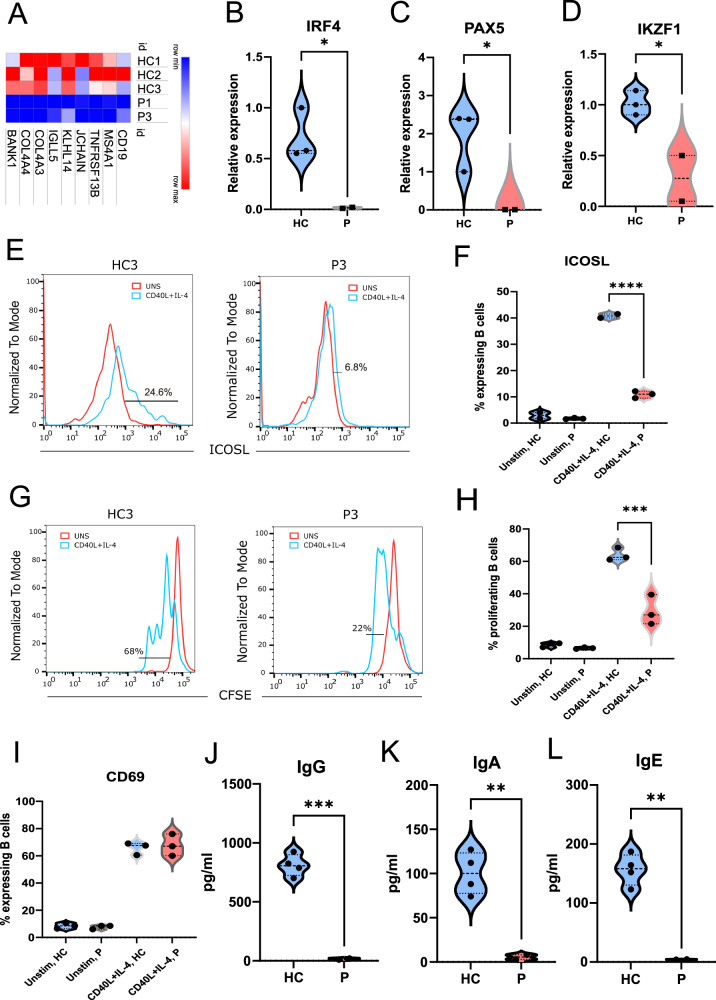
Fig. 4. Defective SLO and memory B cell development in IKKαG167R patients.
A Heatmap analysis of DEGs involved in B cell function based on normalized CPM values in RNA-seq analysis. B–D Differential expression of interferon regulatory factor 4 (IRF4) (p = 0.0342), paired box 5 (PAX5) (p = 0.0489), IKAROS family zinc finger 1 (IKZF1) (p = 0.0303) in purified B cells using qRT-PCR. GAPDH levels were used as a normalisation control (n = 3 healthy controls and 2 patients, two-tailed, non-paired t-test was used for statistical analysis). E, F Flow cytometric analysis of the cell surface expression of inducible T cell costimulator ligand (ICOSL) in B cells. Patient and healthy control PBMCs were either unstimulated or stimulated with recombinant CD40L and IL-4 in vitro. One-way ANOVA was used for statistical analysis, p < 0.0001. G, H Proliferation analysis of B cells from healthy controls (n = 3) and patients (n = 3). Carboxyfluorescein succinimidyl ester (CFSE) staining in flow cytometry was used for proliferation analysis. One-way ANOVA was used for statistical analysis, p < 0.0001. I Flow cytometric analysis of the cell surface expression of CD69 in B cells. Patient and healthy control PBMCs were either unstimulated or stimulated with recombinant CD40L and IL-4 in vitro. One-way ANOVA was used for statistical analysis. J–L In vitro class switching of naïve B cells isolated from the PBMCs of patients (n = 2) and healthy controls (n = 4) upon in vitro stimulation with recombinant CD40L and IL-4 for 7 days. IgG (p = 0.0003), IgA (p = 0.0061) and IgE (p = 0.0016) levels in culture supernatant were measured with multiplexed bead-based assay. Two-tailed, non-paired t-test was used for statistical analysis.
Table 1.
B and T lymphocyte populations of patients
| P1 (%/count) | Ref range (%/count) | P2 (%/count) | Ref range (%/count) | P3 (%/count) | Ref range (%/count) | |
|---|---|---|---|---|---|---|
| Total B cells (% of lymphocytes) and absolute numbers | 5/0.19 | 10–27/0.2–2.2 | 19/1.96 | 11–31/0.3–1.2 | 21/1.85 | 11–31/0.3–1.2 |
| Switched Memory B cells (% of B cells) | 0.87/0.001 | 12.9–45 | 0.4/0.008 | 15.4–73 | 1/0.02 | 15.4-73 |
| Marginal Zone B cells (% of B cells) | 2.7/0.005 | 5.1–11.8 | 3/0.05 | 7.4–11.0 | 1.5/0.03 | 7.4-11.0 |
| Naïve B cells (% of B cells) | 86/0.16 | 62.2–76.2 | 91/1.79 | 59.8–85.8 | 93/3.57 | 59.8–85.8 |
| Total T cells (% of lymphocytes) | 80/3.1 | 57–81/1.0–4.9 | 72/7.12 | 55–79/1.9–3.6 | 75/6.63 | 55–79/1.9–3.6 |
| CD4+ T cells (% of lymphocytes) | 52/1.61 | 23.4–48.7/0.4–1.9 | 46/4.53 | 23.6–52.5/0.5–2.7 | 62/5.48 | 23.6–52.5/0.5–2.7 |
| Naïve CD4+ T cells (% of CD4+ T cells) | 83/1.33 | 41.7–77.8 | 78/3.53 | 54.9–83.1 | 95/5.2 | 54.9–83.1 |
| CD4+ Effector Memory T cells (% of CD4+ T cells) | 3/0.04 | 2–16.2 | 7/0.31 | 0.8–10.8 | 0/0 | 0.8–10.8 |
| CD4+ Central Memory T cells (% of CD4+ T cells) | 8/0.12 | 14-49 | 9/0.40 | 9.2–40.2 | 2/0.1 | 9.2–40.2 |
| CD4+ TEMRA (% of CD4+ T cells) | 5/0.08 | 0.2–43.8 | 5/0.22 | 0.1–41.2 | 1.5/0.08 | 0.1–41.2 |
| CD8+ T cells (% of lymphocytes) | 27/0.83 | 16.8–46.5/0.4–2.1 | 26/2.56 | 12.1–35.7/0.16–1.87 | 21/1.85 | 12.1–35.7/0.16–1.87 |
| Naïve CD8+ T cells (% of CD8+ T cells) | 70/0.6 | 36–87.8 | 67/1.71 | 34.3–90.3 | 95/1.75 | 34.3–90.3 |
| CD8+ Effector Memory T cells (% of CD8+ T cells) | 17/0.14 | 2.1–33.3 | 22/0.56 | 4–42.5 | 1/0.02 | 4–42.5 |
| CD8+ Central Memory T cells (% of CD8+ T cells) | 18/0.15 | 0.3–8.3 | 5/0.12 | 0.9–9.4 | 1/0.02 | 0.9–9.4 |
| CD8+ TEMRA (% of CD8+ T cells) | 12/0.1 | 14.6–61.7 | 10/0.25 | 5.5–55.5 | 0.5/0.01 | 5.5–55.5 |
Reference ranges are based on published results88,89.
Total B cells: CD19+, Total T cells: CD3+
Switched Memory B cells: CD19+ CD27+ IgD− IgM−
Marginal Zone B cells: CD19+ CD27+ IgD+
Naïve B cells: CD19+ CD27− IgM+ IgD+
Naïve CD4+ T cells: CD4+CD45RA+CCR7+
CD4+ Effector Memory T cells: CD4+CD45RA−CCR7−
CD4+ Central Memory T cells: CD4+CD45RA−CCR7+
CD4+ TEMRA: CD4+CD45RA+CCR7−
Naïve CD8+ T cells: CD8+CD45RA+CCR7+
CD8+ Effector Memory T cells: CD8+CD45RA−CCR7−
CD8+ Central Memory T cells: CD8+CD45RA−CCR7+
CD8+ TEMRA: CD8+CD45RA+CCR7−
The bold numbers indicate aberrant values
Previous studies in mouse B cells have suggested that the transcription of IRF4, PAX5, IKZF1, and ICOSLG are directly regulated by the non-canonical NF-κB pathway9–11. To validate this in our patients, we examined the levels of these transcripts in magnetically isolated circulating B cells (CD19+) from two IKKαG167R patients and sex-matched paediatric donors. Using qRT-PCR, we confirmed that transcripts of IRF4, PAX5, and IKZF1 were significantly lower in IKKαG167R patients’ B cells compared to healthy controls (Fig. 4B–D). Although it was not possible to validate these findings at the single-cell level, to ensure that this is not primarily due to differential abundance of B cell subpopulations in patients, we looked at the expression level of IRF4, PAX5, and IKZF1 in a previously published dataset characterizing 29 immune cell types in PBMCs from healthy donors58. Indeed, we observed similar expression levels IRF4, PAX5, and IKZF1 in naive and memory B cells (Figure S1D–F), suggesting that lower levels of IRF4, PAX5, and IKZF1 in CD19+ B cells purified from patients are not due to B cell subpopulation differences. In addition, in vitro stimulation of PBMCs with recombinant CD40L and IL-4 resulted in increased expression of ICOSLG on the surface of B cells from healthy controls, which was drastically lower in B cells from patients with IKKαG167R (Fig. 4E, F). These results show that the IKKα-mediated non-canonical NF-κB pathway is critical for the expression of IRF4, PAX5, and IKZF1 and the upregulation of ICOSLG by human B cells. We also analysed p52 and RelB chromatin binding in a human B cell line GM1287859 using publicly available ChIP-seq data (GSE55105) and revealed the presence of several peaks of both p52 and RelB binding in the gene bodies, promoter regions, and putative enhancer regions of IRF4, PAX5, and IKZF1 gene loci and promoter region of ICOSLG loci (Figure S1G–J), suggesting the possibility of the direct regulation of these transcripts by the non-canonical NF-κB pathway. Notably, variants in these genes have also been reported to result in primary immunodeficiencies in humans60–63.
Stimulation of patient PBMCs with CD40L and IL-4 resulted in defective B cell proliferation in vitro (Fig. 4G, H) and impaired class switching to IgG, IgA, or IgE compared to PBMCs from healthy controls (Figure S2A), consistent with the observed hypogammaglobulinemia in our patients. Remarkably, upregulation of the activation marker CD69 remained intact following CD40L and IL-4 stimulation of IKKαG167R patient B cells (Fig. 4I), indicating that B cell activation can still occur in IKKαG167R cells. Notably, similar defects in immunoglobulin class switching were obtained when purified naïve B cells from patients and healthy controls (92-95% purity) (Figure S2B, C) were stimulated with CD40L and IL-4 (Fig. 4J–L). These results confirm the presence of B cell-intrinsic defects in immunoglobulin generation in patients with the IKKαG167R variant.
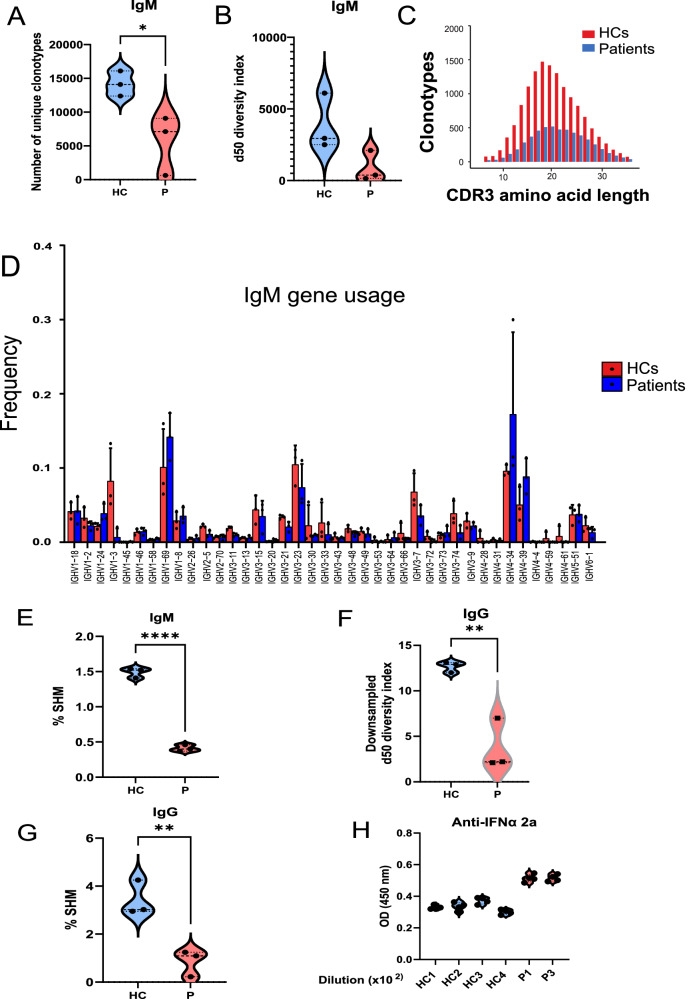
To assess the BCR repertoire at the population level, RNA sequencing of the BCR IgM and BCR IgG repertoires was performed in IKKαG167R patients and healthy controls. While a broad spectrum was observed among the patients, the overall BCR IgM repertoires were restricted due to the low numbers of unique complementary determining region 3 (CDR3) sequences, as demonstrated by the number of unique clonotypes, diversity index, and CDR3 amino acid lengths (Fig. 5A–C). Skewed use of BCR genes was observed in IGMHV1 − 2, IGMHV1 − 24, IGMHV1 − 69, IGMHV4 − 34, and IGMHV4 − 39 of the patients (Fig. 5D). The somatic hypermutation (SHM) rate in rearranged variable regions of IgM heavy chain (IGHV) genes was also significantly lower in patients (Fig. 5E).
Fig. 5. BCR gene usage analysis in patients and healthy controls.
A–C Number of unique clonotypes, d50 diversity index (number of clonotypes occupying 50% of the repertoires), and complementarity-determining region 3 (CDR3) amino acid length of the BCR IgM (B cell receptor μ heavy chain) repertoire in healthy controls (n = 3) and patients (n = 3). Although the d50 diversity index did not reach statistical significance, possibly due to the sample size, there was a trend of limited BCR IgM diversity (p = 0.08). D BCR IgM (B cell receptor μ heavy chain) gene usage comparison between patients and healthy controls. Red bars indicate healthy controls (n = 3), and blue bars indicate IKKαG167R patients (n = 3). Two-tailed, non-paired t-test was used for statistical analysis. Data are presented as mean values and SD. E Somatic hypermutation (SHM) rate was calculated in rearranged variable regions of the IgM heavy chain (IGHV) genes and given as the percentage of insertions and/or deletions in total number of sequences in patients and healthy controls. Two-tailed, non-paired t-test was used for statistical analysis, p < 0.0001. F Downsampled d50 diversity index (number of clonotypes occupying the 50% of repertoires) and complementary-determining region 3 (CDR3) amino acid length of BCR IgG (B cell receptor γ heavy chain) repertoire of healthy controls and patients. Normalization was performed by downsampling the data to the smallest number of clones. Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0057. G Somatic hypermutation (SHM) rate was calculated in rearranged variable regions of the IgG heavy chain (IGHV) genes and given as the percentage of insertions and/or deletions in total number of sequences in patients and healthy controls. Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0084. H Auto-antibodies against interferon alfa (IFNα) 2a were tested by enzyme-linked immunosorbent assay (ELISA) using plasma samples from 4 healthy controls and 2 patients (P1 and P3). OD measurements at 450 nm were shown from 1/100 diluted plasma samples.
In IgG repertoire analyses, the number of unique clonotypes and gene usage were highly biased due to the low numbers of memory B cells and IgG levels in the patients. However, when we analysed the IgG heavy chain repertoire diversity index after normalizing by downsampling the data to the smallest number of clones, we still observed significantly lower diversity in patients (Fig. 5F). Accordingly, the SHM rate in the rearranged variable regions of IgG heavy chain (IGHV) genes was also lower in patients (Fig. 5G).
Skewed gene usage of IGHV4-34 in B cells indicates the presence of autoreactive B cells64. Notably, IGHV4-34 in the IgM repertoire was significantly higher in the older patient P1 (Fig. 5D) suggesting a potential contribution to the autoimmunity observed in this patient. Indeed, as previously reported in many patients with genetic defects in non-canonical NF-κB pathway52, levels of autoreactive serum IgM and IgG specific for IFNa2a were significantly increased in P1 and P3 compared to healthy donors (P2 serum sample was not available for this test) (Fig. 5H). It is also worth noting that anti-IFNα autoantibodies might be stronger if not diluted by IVIG treatment in patients. Although we were not able to test the neutralizing characteristics of these anti-IFNα autoantibodies, we anticipate similar findings to those observed in patients with mutations in other components of the non-canonical NF-κB pathway52.
On the other hand, serum levels of antinuclear antibody (ANA), anti-smooth muscle antibody (ASMA), anti-endomysial IgA and IgG (by indirect immunofluorescence assay [IFA]), anti-gliadin IgA and IgG, and anti-tissue transglutaminase IgA and IgG (by ELISA), as well as anti-mitochondrial M2 antibody (AMA-M2), anti-liver kidney microsome type 1 (anti-LKM−1), anti-liver cytosol antigen type 1 (anti-LC1), and anti-soluble liver antigen/liver-pancreas (SLA/LP) (by immunoblotting), were all found to be negative in P1 (P2 and P3 serum samples were not available for these tests). Overall, this is in line with the observation that patients with defective non-canonical NF-κB pathway have a specific autoimmune signature involving autoantibodies against type I interferons52.
IKKαG167R patients have reduced circulating populations of NK, MAIT, Treg, and Tfh cells
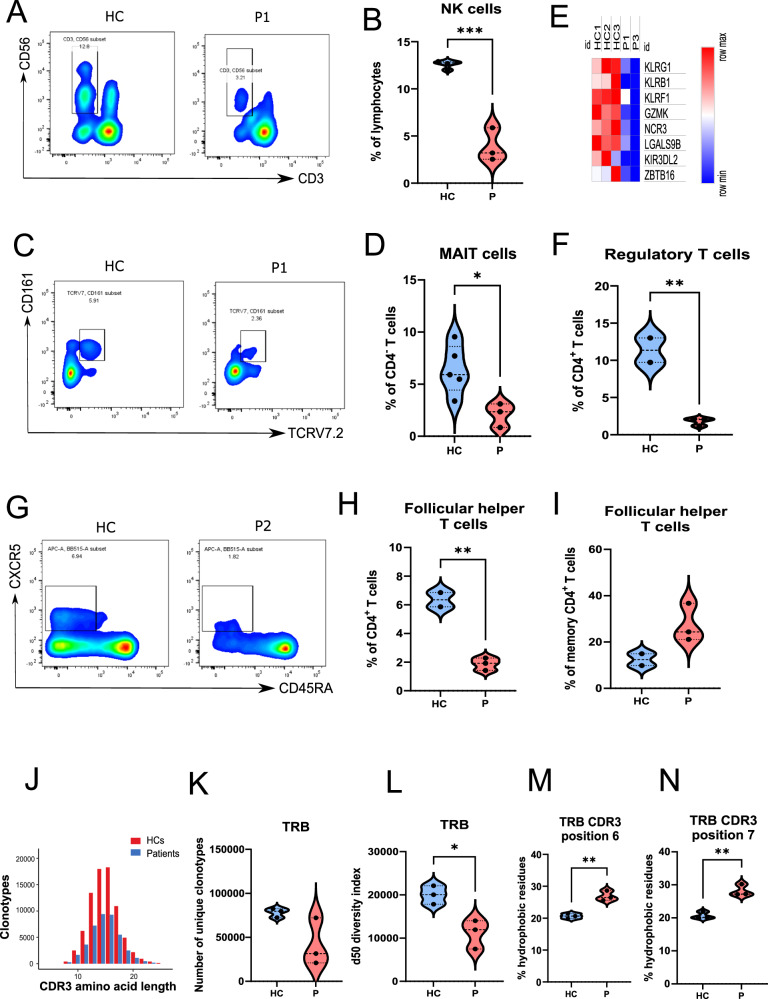
Flow cytometric analysis of NK cells confirmed significantly lower proportions of NK cell populations in patients (Fig. 6A, B). NK cells from P1 were also impaired in IFN-γ secretion upon PMA/ionomycin stimulation (Figure S2D, E). Similar NK cell abnormalities were also reported in patients with NIK variants65 or NFKB2 variants66,67. MAIT cells are a subset of unconventional T cells with important functions against bacterial infections68. We observed that all IKKαG167R patients have significantly decreased levels of circulating MAIT cells (CD3+ CD4- TCR Vα7.2+ CD161+) (Fig. 6C, D). Similar to NK cells, MAIT cells from P1 were also impaired in IFN-γ secretion upon PMA/ionomycin stimulation, although this did not reach statistical significance due to sample size (Figure S2F, G). These results were also supported by decreased levels of transcripts involved in NK and MAIT cell biology58,69 in both patients’ PBMCs (Fig. 6E). We also observed a decrease of circulating CD3+CD4+CD25+Foxp3+ Tregs (Fig. 6F) and Tfh cells (CD3+CD4+CD45RA-CXCR5+) in IKKαG167R patients (Fig. 6G, H) as a proportion of total CD4+ T cells. Tfh cells within the CD4+ memory T cell pool, however, were slightly higher in patients, although not statistically significant (Fig. 6I). In addition to lower numbers of circulating Tregs, we also observed, using P1’s Tregs, that there was a defective capacity of IKKαG167R Tregs to suppress T cell responses in vitro (Figure S2H, I). This is consistent with the cell intrinsic diminished suppressive function of Tregs upon NFKB2 deletion in human Treg-like cell line MT-270. Consistent with the defects in Treg number and function, P1 developed autoimmune hepatitis, which eventually resulted in the need for a liver transplantation. H&E and Gomori Trichrome staining in the liver biopsy sample of P1 indicated liver inflammation and fibrosis in the patient (Figure S2J, K). Overall, these results suggest that IKKαG167R negatively impacts both cellular and humoral immunity involving different cell types.
Fig. 6. Analyses of NK, MAIT, Treg, and Tfh cell populations and TCR repertoire in IKKαG167R patients compared to the healthy controls.
A Representative flow cytometric analysis of natural killer (NK) cells (CD3-CD56+) in a healthy control and patient-1 PBMCs. B Graphical representation of NK cells (CD3-CD56+) in healthy controls (n = 4) and patients (n = 3). Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0002. C Flow cytometric analysis of Mucosal Associated Invariant T (MAIT) cells (CD3+CD4- CD161+TCRV7.2+) in a healthy control and patient-1 PBMCs. D Graphical representation of MAIT cells (CD3+CD4- CD161+TCRV7.2+) in healthy controls (n = 5) and patients (n = 3). Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0266. E Heatmap analysis of selected MAIT and NK cell associated genes in healthy controls (n = 3) and patients (n = 2), based on normalized CPM values in RNA-seq analysis. F Graphical representation of Tregs (CD3+ CD4+ CD25+ FoxP3+) in healthy controls (n = 2) and patients (n = 3). Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0050. G Flow cytometric analysis of follicular helper T (Tfh) cells (CD3+CD4+CD45RA-CXCR5+) in a healthy control and patient-2 PBMCs. H Graphical representation of Tfh cells as a percentage of total CD4+ T cells in healthy controls (n = 2) and patients (n = 3). A two-tailed, unpaired t-test was used for statistical analysis, p = 0.0028. I Graphical representation of Tfh cells as a percentage of memory CD4+ T cells in healthy controls (n = 2) and patients (n = 3). A two-tailed, unpaired t-test was used for statistical analysis. J–L CDR3 amino acid length, number of unique clonotypes and d50 diversity index (number of clonotypes occupying the 50% of repertoires) of TRB (T cell receptor beta chain) repertoire of healthy controls (n = 3) and patients (n = 3). Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0179 (d50 diversity index). M, N Hydrophobicity of the amino acids in positions 6 and 7 in TRB CDR3 region of healthy controls (n = 3) and patients (n = 3). Two-tailed, non-paired t-test was used for statistical analysis, p = 0.0034 (position 6) and p = 0.0032 (position 7).
IKKαG167R patients have restricted TCR and have potentially autoreactive lymphocytes
Similar to memory B cells, CD4+ central memory T cells (CD4+CD45RA-CCR7+) were also lower in all patients (Table 1) suggesting significant impairment of the adaptive immune response. Disruption of immune tolerance is a hallmark of autoimmunity. Potentially autoreactive T cells can be identified by the presence of hydrophobic amino acid residues at positions 6 and 7 in the CDR3β of αβ TCRs, and this has been associated with defects in T cell tolerance71,72. To investigate whether IKKαG167R patients exhibit T cell tolerance defects, TCRα and TCRβ sequencing was performed at the population level. The CDR3 length distribution was decreased in T cells of the patients (Fig. 6J). There was a tendency towards a lower number of unique clonotypes (p = 0.08) (Fig. 6K) and a significantly limited diversity of TCRβ, as determined by the d50 diversity index (Fig. 6L). There was also a trend of limited TCRα diversity, although this did not reach to significance (Figure S3A). Importantly, the presence of potentially autoreactive T cells in the TCR repertoire was evident in all patients (Fig. 6M, N and Figure S3B). Gene usage analyses revealed several skewed distributions in TCR variable genes. In particular, TRAV13 − 1 and TRBV7-2 usage were different in the patients (Figure S3C, D).
Naïve T cells from IKKαG167R patients have defective cytokine response to TCR stimulation
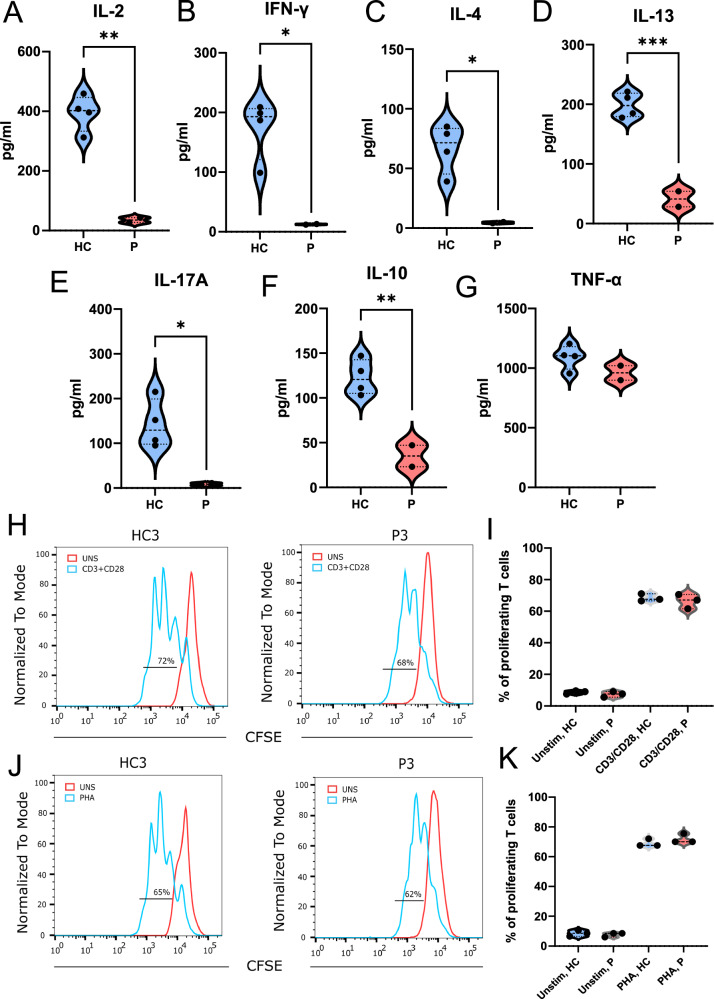
Although naïve T cell development was not significantly altered in patients with the IKKαG167R variant, we sought to determine if naïve T cells from these patients displayed altered activation and cytokine responses. To this end, we isolated naïve CD4+ T cells from PBMCs using negative selection (Figure S4A) and stimulated them with anti-CD3 and anti-CD28. We observed that cells from patients showed impaired generation of IL-2, IFN-γ, IL-4, IL−13, IL−17A, and IL-10 but not TNF-α (Fig. 7A–G). Unlike other cytokines tested, the comparable levels of TNF-α may reflect the redundant role of IKKα in its synthesis. This suggests the possible involvement of other transcriptional regulators driving TNF-α production. While these data show that IKKαG167R causes intrinsic differences in naïve CD4+ T cells, which were purified by negative selection, we cannot rule out a contribution from memory T cells to the cytokines detected. However, these results are not limited to naïve CD4+ T cells as similar results were obtained when PBMCs were stimulated with anti-CD3 and anti-CD28 (Figure S4B–G). It is also important to note that there is no impairment in patients’ T cell proliferation upon anti-CD3 and anti-CD28 or PHA stimulation. Similarly, CD25, CD69, and ICOS expression in patients’ T cells upon anti-CD3 and anti-CD28 were comparable to healthy controls, indicating that IKKα kinase activity is not essential for overall T cell activation in humans (Fig. 7H–K). Our findings are somewhat analogous to those of Bainter et al. 34, who observed diminished IL-2 and IL−17A, but not IFN-γ, IL-4, or IL-13 levels, upon in vitro stimulation of splenic T cells from IKKαY580C/Y580C mice with anti-CD3 and anti-CD28. While the exact mechanisms remain unclear in our context, these combined results support the notion that IKKα kinase activity plays a critical role in driving the transcriptional activation of specific cytokines in different cell types and under varying stimulation conditions27.
Fig. 7. Naïve CD4+ T cell activation and cytokine release in patient cells and healthy controls.
A–G Analysis of the cytokine release from purified naïve CD4+ T cells 72 h post stimulation with anti-CD3 and anti-CD28 antibodies. Soluble IL-2 (p = 0.0015), IFN-γ (p = 0.0131), IL-4 (p = 0.0155), IL-13 (p = 0.0008), IL-17A (p = 0.0305), IL-10 (p = 0.0060) and TNF-α (p = 0.1966) (n = 2 patients vs n = 4 healthy controls) levels were measured with multiplexed bead-based assay. Two-tailed, non-paired t-test was used for statistical analysis. H, I Proliferation analysis of T cells from healthy controls (n = 3) and patients (n = 3) from PBMCs 48 h post stimulation with anti-CD3/CD28. CFSE staining in flow cytometry was used for proliferation analysis. One-way ANOVA was used for statistical analysis. J, K Proliferation analysis of T cells from PBMCs 48 h post stimulation with PHA, healthy controls (n = 3) and patients (n = 3). CFSE staining in flow cytometry was used for proliferation analysis. One-way ANOVA was used for statistical analysis.
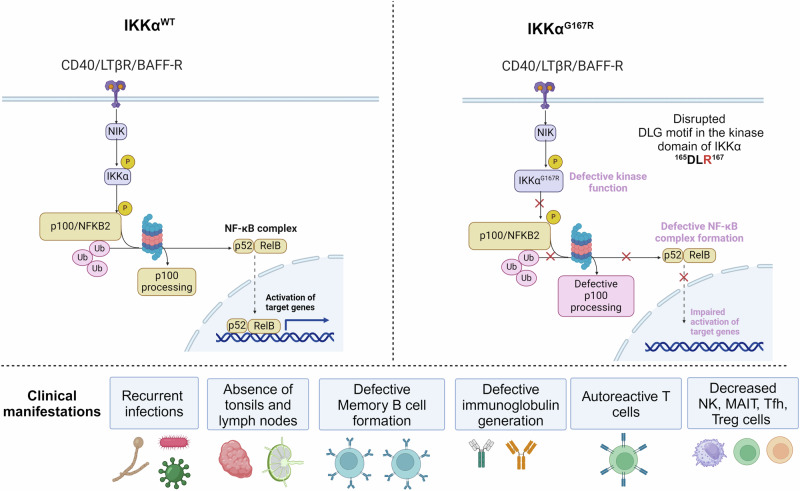
In summary, we have identified the first germline homozygous missense variant in human IKKα kinase domain. IKKαG167R patients have innate and adaptive immune system abnormalities consistent with an important role for non-canonical NF-κB pathway in modulating immune response to foreign stimuli and preventing autoreactive lymphocytes and maintaining self-tolerance (Fig. 8).
Fig. 8. Figure summarizing the molecular mechanisms of IKKαG167R and its clinical manifestations in patients.
Created in BioRender. Tumes, D. (2024) BioRender.com/k75q726.
Discussion
The NF-κB signalling pathway is a critical regulator of immunity and genetic variations in genes that regulate this pathway can lead to various forms of inborn errors of immunity73,74. IKKα serves as an essential upstream regulator of the non-canonical NF-κB pathway, and it has been reported to have NF-κB-independent and kinase-independent functions in different biological contexts. Understanding the specific variants that cause separation-of-function in human IKKα can provide valuable mechanistic insights into the multifunctionality of this protein. However, no homozygous germline variants specifically impacting the kinase function of IKKα have been reported, leaving the importance of the kinase function of IKKα in human health unknown. This is especially important because observations from genetic mouse models are not always replicated in humans. For instance, IKKβ-deficient mice die during embryonic development due to liver apoptosis75. However, this phenomenon is not observed in humans who have been reported to have loss-of-expression and/or loss-of-function variants in IKKβ76–78. Therefore, our findings are invaluable in investigating the functional roles and consequences of the loss of the kinase function of IKKα in humans.
In this study, we have identified three children with the IKKαG167R variant, which is the first reported homozygous missense variant that specifically affects the kinase function of human IKKα. Our findings indicate that the kinase function of IKKα is not essential for human embryonic development. However, the broad impact of the IKKαG167R variant on the immune system is consistent with the crucial roles played by the non-canonical NF-κB pathway in the development of SLOs, immune tolerance, and the generation of memory.
In our study, we for the first time analysed the TCR and BCR repertoire in humans with an IKKα germline variant. The overall TCR and BCR repertoire, including TCR α/β and BCR IgM/IgG heavy chains, was found to be restricted in the patients. The most significant decrease was observed in P1, who also exhibited the most severe clinical phenotype. The percentage of hydrophobic amino acids at positions 6 and 7 of TRB CDR3 was increased in the patients compared to healthy controls. Similar to repertoire richness and diversity, the most notable difference was observed in P1, who presented severe autoimmune symptoms. P1 subsequently developed autoimmune liver disease, which ultimately led to her death. The clinical severity observed in P1 may be attributed to the delayed administration of intravenous immunoglobulin (IVIG) therapy (started age 4) compared to P2 and P3 (started age 1). Age-dependent exacerbation of disease symptoms associated with IKKα dysfunction has also been observed in a previously reported case of IKKαY580C in humans and in IKKαY580C/Y580C mice34 as well as IKKαK44A/K44A mice6. Inborn errors of immunity associated with defects in secondary lymphoid organ development cannot be cured with hematopoietic stem cell transplantation (HSCT)79,80. However, thymus transplantation after HSCT could be a viable approach80 to be tested in patients with IKKα variants.
Molecular simulation analysis indicates that the impaired kinase function of IKKαG167R is likely due to the differential interaction between the R167 residue and the ATP-binding K44 residue in the kinase domain. Consistent with our findings, previous investigations have highlighted the critical role of the glycine residue in the DF/LG motif of TBK1 and B-Raf kinases. Variants of glycine residue in the DF/LG motifs of both kinases (TBK1G159A and B-RafG596R) substantially impair their kinase activities, emphasizing the importance of conserved glycine in this motif81,82. However, it is noteworthy that despite the kinase domains of TBK1 and IKKα being similar, when the same glycine residue in the DF/LG motif of the kinase domain is mutated in both kinases (IKKαG167R vs. TBK1G159A), a single mutated copy of TBK1 is sufficient to cause a disease characterized by impaired cytokine production and increased susceptibility to viral infections81. Conversely, the heterozygous parents of our patients with IKKαG167R do not exhibit any symptoms of immunodeficiency or autoimmunity. This clearly demonstrates the differential evolution of immune pathway dependence on IKK and IKK-related kinases. Importantly, our patients share similar characteristics with individuals harbouring various variants in NFKB283 and the other members of the non-canonical NF-κB pathway52. This suggests that dysregulation of the non-canonical NF-κB pathway is the primary underlying cause of the phenotypes associated with our patients.
Our patients exhibited diminished populations of Treg, Tfh, NK, and MAIT cells. While NK cell abnormalities have been reported in some patients with NFKB2 or NIK variants, the role of the non-canonical NF-κB pathway in MAIT cell function has not been previously reported to the best of our knowledge. The clinical observations of previously reported patients with NIK variants65,84 and the IKKαY580C variant34 are similar to our patients in terms of memory B and T and NK cell abnormalities. However, autoimmunity was not reported in patients with NIK variants and skin abnormalities observed in IKKαY580C patient were not observed in our patients.
Further investigations should focus on delineating the genome-wide binding profiles of RelB, p52, and IKKα in NK and MAIT cells to gain insight into the cell type-specific target genes and functions governed by the non-canonical NF-κB pathway. It is also critical to understand the contributions of different subsets of immune cells to the overall response to various stimuli in patient cells compared to healthy controls. This is important to delineate whether the non-canonical NF-κB pathway selectively impacts certain subpopulations of immune cells.
Finally, our findings also have significant implications for the use of small molecule inhibitors that specifically target IKKα in various human diseases. The results of our study suggest that caution should be exercised when utilizing selective IKKα inhibitors, which are currently under development for the treatment of different malignancies85, in patients for extended periods of time. This caution arises from the increased risk of viral, bacterial, and fungal infections, as well as the potential for autoimmunity. Therefore, careful consideration should be given to the potential adverse effects on immune function and self-tolerance when using selective IKKα inhibitors in clinical settings. In summary, IKKα kinase deficiency is a debilitating condition characterized by combined immunodeficiency, leading to a high susceptibility to infections and the presence of self-reactive lymphocytes. Patients with this condition experience progressive deterioration, and early IVIG therapy is crucial in managing disease symptoms.
Methods
Ethics statement
Consent was obtained from the parents of all three patients in accordance with the local Ethics Committee of Hacettepe University (GO 20/407), CARE guidelines, and the Declaration of Helsinki principles.
Whole exome, RNA, and immune repertoire sequencing
Whole exome sequencing (WES), bulk RNA-seq, TCR and BCR library preparation, sequencing, and data analyses were performed as previously described in ref. 86. Briefly, RNA was isolated from PBMCs using the NucleoSpin RNA Plus Kit (Macherey-Nagel). T cell and B cell receptor (TRA, TRB, and BCRH) libraries were prepared using 100 ng RNA with the SMARTer Human TCR a/b and BCR IgG H/K/L Profiling Kit (Takara) according to the manufacturer’s instructions. Library validation was performed by measuring amplicon size on a 2100 Bioanalyzer using the Agilent DNA 1000 kit (Agilent). The repertoire library was sequenced on a MiSeq platform (Illumina) with a read length of 2 × 300 bp. The raw sequencing data (FASTQ) was processed using the Cogent NGS Immune Profiler Software (Takara) and IMGT/HighV-QUEST (The International Immunogenetics Information System). Data analyses were performed in R Studio using the Immunarch package. Raw RNA-seq datasets were deposited to NCBI (GSE256535). Over-representation analysis (ORA) was performed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit). The parameters included ‘Gene Ontology’ and ‘Biological Process Non-Redundant.’ Gene enrichment analysis was performed using GSEA.
CRISPR/Cas9-mediated genome editing
A gRNA targeting the CHUK gene was designed using Invitrogen TrueDesign Genome Editor. Target sequence in Exon-1 is 5’-ACGTCTGTCTGTACCAGCAT-3’. To perform the genome editing at the loci, S.p. Cas9-GFP V3 (IDT) was incubated with gRNA in Opti-MEM medium at room temperature for 10 min and transfected into HEK293T cells using Lipofectamine CRISPRMAX Cas9. The next day, GFP-expressing single cells were sorted into 96-well plates and colonies were grown. Expanded colonies were screened for the loss of IKKα protein expression. IKKα-deficient clones were further screened by Sanger sequencing of the genomic loci to confirm the genome editing.
Plasmids and site-directed mutagenesis
Human IKKα was cloned into pGenLenti vector with FLAG-tag at the C-terminus. Full-length NFKB2 (p100) was cloned into-pcDNA3.1-FLAG vector. The Q5 Site-Directed Mutagenesis Kit (NEB) was used to generate all variants in human IKKα. MAP3K14 (NIK)-pcDNA 3.1 vector was a kind gift from Dr Cindy Lee (ANU, Canberra).
Lymphocyte proliferation
PBMCs were isolated from the patient and healthy controls using Ficoll-Paque (Capricorn Scientific). Cells were resuspended in RPMI-1640 containing 15% FCS, 10 mM HEPES (Sigma), 100 U/ml penicillin/streptomycin (Sigma) and 200 mM L-Glutamine (Sigma). For proliferation assays, cells were stimulated with CD3/CD28 Dynabeads (1:1 ratio, Thermofisher) or 50 ng/ml PMA (Sigma) and 1 µg/ml Ionomycin (Sigma) for 3 days or 200 ng/ml CD40L + 100 ng/ml IL-4 (BioLegend) for 6 days. Proliferation was measured by labelling the cells with 5 μM CFSE (BioLegend) according to manufacturer’s instructions.
Immunoglobulin and cytokine measurements in naïve B and T cells
MojoSort Human Naïve B Cell Isolation Kit was used to purify naïve B cells from PBMCs. 150,000 cells were seeded in 96-well plates and stimulated with 200 ng/ml CD40L and 100 ng/ml IL-4 for 7 days. Levels of IgG, IgE and IgA in the culture supernatant were quantified using LEGENDplex Human Immunoglobulin Isotyping Panels (BioLegend). MojoSort Human CD4 Naïve T Cell Isolation Kit was used to purify naïve CD4+ T cells from PBMCs. 250,000 cells were seeded in 96-well plates and stimulated with CD3/CD28 Dynabeads (1:1 ratio, Thermofisher) for 3 days. Cultured cells were centrifugated and the supernatant was preserved for bead-based cytokine quantification. Levels of TNF-α, IL−2, IL-4, IL-10, IL-13, IL-17A, and IFN-γ in the culture supernatant were quantified using LEGENDplex Human Th Cytokine Panel. Routinely, 90–95 % purity was achieved post purification of naïve CD4+ T and naïve B cells from PBMCs.
B cell and T cell activation
5 × 105 PBMCs were cultured in RPMI medium with 10% FBS and Pen/Strep. For T cell activation assays, cells were stimulated with CD3/CD28 Dynabeads for 24 and 48 hours for CD69 and CD25 expression on 7-AAD- live T cells. For B cell activation, cells were stimulated with 200 ng/ml CD40L and 100 ng/ml IL-4 for 24 and 48 h for CD69 and ICOSL expression on 7-AAD- live B cells.
Western Blotting
Proteins were extracted from PBMCs with RIPA buffer with 1X Halt Protease inhibitor cocktail. In experiments with IKKα-KO HEK293T cells, 400,000 cells were transfected with 250 ng NFKB2, 500 ng NIK and 500 ng of IKKαWT, IKKαG167R, IKKαD165A, IKKαK44A overexpression constructs using Lipofectamine 3000 reagents. 48 h later, proteins were extracted using TOTEX buffer, separated on an SDS-PAGE and transferred to PVDF membrane. Primary antibodies were incubated with the membrane overnight in a cold room and secondary HRP-conjugated antibodies were incubated for 1 h at room temperature. Images were acquired with ChemiDoc Imaging Systems (Bio-Rad) and band intensities were calculated with Image Lab Software.
Co-immunoprecipitation
IKKα-KO HEK293T cells were transfected using X-tremeGENE HP DNA Transfection Reagent (Sigma). 48 h later, cells were lysed in 1 ml PLCLB lysis buffer (150 mM NaCl, 5 % glycerol, 1 % Triton X-100, 20 mM Tris-HCl, pH: 7.5) supplemented with 1 mM sodium orthovanadate, 1 mM sodium fluoride and EDTA-free Protease Inhibitor Cocktail (Roche). For co-immunoprecipitation, cleared lysates were incubated with 40 μl of Anti-FLAG Magnetic Beads (#M8823, Sigma) for 1 h followed by 3 times wash with lysis buffer. After elution, proteins were separated in 4–15% Criterion TGX Precast polyacrylamide gels (BioRad) under reducing conditions.
qRT-PCR
Total RNA was isolated using the NucleoSpin RNA Plus Kit (Macherey-Nagel) and converted to cDNA via iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) in CFX Connect Real-Time PCR Detection System (Bio-Rad). All primer sequences used in qRT-PCR were listed in Table S7.
Flow cytometry
Cell staining with antibodies was performed in Flow buffer (PBS, 1% FBS, 2 mM EDTA) on ice for 30 min. All flow cytometry antibodies were listed in Table S8. Data were analysed with FACSDiva (BD Biosciences) and FlowJo software (BD Biosciences). Gating strategies are provided in Figure S5 and S6.
Treg suppression assay
CD4+ T cells were enriched by negative magnetic bead separation (Biolegend) from PBMCs. Tregs were isolated from enriched CD4+ T cells by magnetic separation. CD4+ CD25low CD127high T effector (Teff) cells were sorted using FACS. Teff cells were stained with CFSE Cell Division Tracker Kit (Biolegend). The same number of Teff and Tregs (1:1 Ratio, 25,000 cells) were stimulated with CD3/CD28 Dynabeads and cultured in RPMI medium for 4 days. Treg suppression capacity was measured by determination of the percentage of proliferating cells.
Immunohistochemistry
Hematoxylin and eosin (H&E) staining was performed, revealing portal inflammation and prominent cholestasis around the periportal hepatocytes. Gomori trichrome staining was also conducted to observe portal, periportal, and perisinusoidal fibrosis in the liver biopsy sample of P1.
Lymphoscintigraphy analysis
One millicurie (mCi) of the radiotracer Tc-99m nanocolloid was subcutaneously administered into the interphalangeal joints of both hands. Images were captured in both upper extremities and in the pelvic and abdominal areas of the lower extremity at 10 min, 1 h, 2 h, 4 h, and 24 h after the injection. The liver and spleen became visible during the imaging process.
Anti-IFN Alpha antibody detection by ELISA
The levels of serum IgG and IgM IFN-alpha autoantibodies were measured by an in-house ELISA as previously described in ref. 87. Briefly, high-binding 96-well plates (Greiner Bio-One) were coated with 2 μg/ml Recombinant Human IFN-alpha (alpha 2a) (R&D Systems) overnight. Serum samples, diluted 1/100, were incubated in the wells for 2 h at room temperature. Then, HRP-conjugated anti-IgG and IgM antibodies (Ptglab and Elabscience) (2 μg/ml) were added to the wells for 1 h at room temperature. After three washes, TMB substrate (MedChem) was added, and the optical density (OD) was measured at 450 nm in quadruplicate.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank “Can Sucak Candan Biseyler” Foundation (CSCBF) for their support and contributions during the study. CSCBF was founded in 2018 to honour Can Sucak who lost his life due to complications of primary immunodeficiency. CSCBF supports research in the field of primary immunodeficiency and promotes awareness. G.C. and D.J.T. are supported by grants from the Hospital Research Foundation (THRF) (Grant 2023-S-DTFA-003-QA25325) and NHMRC of Australia (Grant 2027602). B.E was supported for immune repertoire study by the Scientific and Technological Research Council of Türkiye (121S667).
Author contributions
G.C., U.A., D.P., C.I., D.T., C.B., S.T., and B.E. performed the experiments. N.I.W., C.H.K., and D.J.T. analysed the RNA-seq data. E. A., G. I., M.C., E.K., I.K., B.S., D.B., A.Y., N.D.D., I.T., S.H., F.D., A.I., and O.K. provided clinical care of the patient and clinical data. D.B. performed liver transplantation. G.C., U.A., D.J.T., and B.E. wrote the manuscript. B.E, D.J.T., and O.K. conceptualized and coordinated the study and provided laboratory resources. All authors critically reviewed the manuscript and agreed to its publication.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information/Source Data files. Bulk RNA-seq datasets have been deposited in NCBI Gene Expression Omnibus (GEO) with the accession code GSE256535 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE256535). Raw whole exome and immune repertoire sequencing data are available at the NCBI sequence read archive (SRA) under accession number PRJNA1180092. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Ozlem Keskin, Damon J. Tumes, Baran Erman
Contributor Information
Ozlem Keskin, Email: okeskin@gantep.edu.tr.
Damon J. Tumes, Email: Damon.Tumes@unisa.edu.au
Baran Erman, Email: baranerman@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54345-4.
References
- 1.Cildir, G., Low, K. C. & Tergaonkar, V. Noncanonical NF-kappaB Signaling in Health and Disease. Trends Mol. Med.22, 414–429 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Israel, A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect. Biol.2, a000158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinoshita, D. et al. Essential role of IkappaB kinase alpha in thymic organogenesis required for the establishment of self-tolerance. J. Immunol.176, 3995–4002 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Onder, L. et al. Alternative NF-kappaB signaling regulates mTEC differentiation from podoplanin-expressing precursors in the cortico-medullary junction. Eur. J. Immunol.45, 2218–2231 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Baik, S., Sekai, M., Hamazaki, Y., Jenkinson, W. E. & Anderson, G. Relb acts downstream of medullary thymic epithelial stem cells and is essential for the emergence of RANK(+) medullary epithelial progenitors. Eur. J. Immunol.46, 857–862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu, F. et al. Autoreactive T Cells and Chronic Fungal Infection Drive Esophageal Carcinogenesis. Cell Host Microbe21, 478–493 e477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu, M. et al. NF-kappaB2 is required for the establishment of central tolerance through an Aire-dependent pathway. J. Clin. Invest116, 2964–2971 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardam, S. & Brink, R. Non-Canonical NF-kappaB Signaling Initiated by BAFF Influences B Cell Biology at Multiple Junctures. Front Immunol.4, 509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balkhi, M. Y. et al. IKKalpha-mediated signaling circuitry regulates early B lymphopoiesis during hematopoiesis. Blood119, 5467–5477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balkhi, M. Y., Willette-Brown, J., Wittmann, G. & Hu, Y. IKKalpha deficiency disrupts the development of marginal zone and follicular B cells. Genes Immun.20, 224–233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, H. et al. Noncanonical NF-kappaB regulates inducible costimulator (ICOS) ligand expression and T follicular helper cell development. Proc. Natl Acad. Sci. USA108, 12827–12832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun, S. C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol.17, 545–558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarnegar, B. J. et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol.9, 1371–1378 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallabhapurapu, S. et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol.9, 1364–1370 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polley, S. et al. Structural Basis for the Activation of IKK1/alpha. Cell Rep.17, 1907–1914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang, C., Zhang, M. & Sun, S. C. beta-TrCP binding and processing of NF-kappaB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal18, 1309–1317 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, G. & Wang, V. Y. Origin of the Functional Distinctiveness of NF-kappaB/p52. Front Cell Dev. Biol.9, 764164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shembade, N., Pujari, R., Harhaj, N. S., Abbott, D. W. & Harhaj, E. W. The kinase IKKalpha inhibits activation of the transcription factor NF-kappaB by phosphorylating the regulatory molecule TAX1BP1. Nat. Immunol.12, 834–843 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, B. et al. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell129, 903–914 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Huang, W. C., Ju, T. K., Hung, M. C. & Chen, C. C. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol. Cell26, 75–87 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Rita, A. et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKalpha. Nat. Commun.9, 3755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diamanti, M. A. et al. IKKalpha controls ATG16L1 degradation to prevent ER stress during inflammation. J. Exp. Med214, 423–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, M. et al. IKKalpha activation of NOTCH links tumorigenesis via FOXA2 suppression. Mol. Cell45, 171–184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia, X. et al. An IKKalpha-nucleophosmin axis utilizes inflammatory signaling to promote genome integrity. Cell Rep.5, 1243–1255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pecharroman, I. et al. IkappaB kinase-alpha coordinates BRD4 and JAK/STAT signaling to subvert DNA damage-based anticancer therapy. EMBO J.42, e114719 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armache, A. et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature583, 852–857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, L. et al. Transcriptional regulation of the Th17 immune response by IKK(alpha). J. Exp. Med.208, 787–796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, Z. et al. Regulation of Th17 Differentiation by IKKalpha-Dependent and -Independent Phosphorylation of RORgammat. J. Immunol.199, 955–964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Descargues, P., Sil, A. K. & Karin, M. IKKalpha, a critical regulator of epidermal differentiation and a suppressor of skin cancer. EMBO J.27, 2639–2647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, Y. et al. IKKalpha controls formation of the epidermis independently of NF-kappaB. Nature410, 710–714 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Descargues, P. et al. IKKalpha is a critical coregulator of a Smad4-independent TGFbeta-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc. Natl Acad. Sci. USA105, 2487–2492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahtela, J. et al. Mutant CHUK and severe fetal encasement malformation. N. Engl. J. Med.363, 1631–1637 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Leslie, E. J. et al. Expanding the genetic and phenotypic spectrum of popliteal pterygium disorders. Am. J. Med Genet. A167A, 545–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bainter, W. et al. Combined immunodeficiency with autoimmunity caused by a homozygous missense mutation in inhibitor of nuclear factor κB kinase alpha (IKKalpha). Sci. Immunol.6, eabf6723 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, N. Y. et al. IKKalpha inactivation promotes Kras-initiated lung adenocarcinoma development through disrupting major redox regulatory pathways. Proc. Natl Acad. Sci. USA115, E812–E821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang, D. & Shain, A. H. The landscape of driver mutations in cutaneous squamous cell carcinoma. NPJ Genom. Med. 6, 61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang, K. T., Guo, J., di Ronza, A. & Sardiello, M. Aminode: Identification of Evolutionary Constraints in the Human Proteome. Sci. Rep.8, 1357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tate, J. G. et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res.47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet7, 20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raimondi, D. et al. DEOGEN2: prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Res.45, W201–W206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinhaus, R. et al. MutationTaster2021. Nucleic Acids Res.49, W446–W451 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pejaver, V. et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun.11, 5918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ittisoponpisan, S. et al. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J. Mol. Biol.431, 2197–2212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parthiban, V., Gromiha, M. M. & Schomburg, D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res34, W239–W242 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J. et al. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics36, 131–135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao, Z. et al. The pivotal role of IKKalpha in the development of spontaneous lung squamous cell carcinomas. Cancer Cell23, 527–540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, C. E. et al. Autosomal-dominant B-cell deficiency with alopecia due to a mutation in NFKB2 that results in nonprocessable p100. Blood124, 2964–2972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Voyer, T. et al. Autoantibodies against type I IFNs in humans with alternative NF-kappaB pathway deficiency. Nature623, 803–813 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam, L. T. et al. Compensatory IKKalpha activation of classical NF-kappaB signaling during IKKbeta inhibition identified by an RNA interference sensitization screen. Proc. Natl Acad. Sci. USA105, 20798–20803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin, B. N. et al. IKKalpha negatively regulates ASC-dependent inflammasome activation. Nat. Commun.5, 4977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res.47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Navarro, H. I. et al. RelB-deficient autoinflammatory pathology presents as interferonopathy, but in mice is interferon-independent. J. Allergy Clin. Immunol.152, 1261–1272 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monaco, G. et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep.26, 1627–1640 e1627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, B. et al. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell Rep.8, 1595–1606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Consortium, I. R. F. I. et al. A multimorphic mutation in IRF4 causes human autosomal dominant combined immunodeficiency. Sci. Immunol.8, eade7953 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaiser, F. M. P. et al. Biallelic PAX5 mutations cause hypogammaglobulinemia, sensorimotor deficits, and autism spectrum disorder. J. Exp. Med.219, e20220498 (2022). [DOI] [PMC free article] [PubMed]
- 62.Kuehn, H. S., Boast, B. & Rosenzweig, S. D. Inborn errors of human IKAROS: LOF and GOF variants associated with primary immunodeficiency. Clin. Exp. Immunol.212, 129–136 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roussel, L. et al. Loss of human ICOSL results in combined immunodeficiency. J. Exp. Med.215, 3151–3164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bashford-Rogers, R. J. M. et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature574, 122–126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willmann, K. L. et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat. Commun.5, 5360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lougaris, V. et al. Defective natural killer-cell cytotoxic activity in NFKB2-mutated CVID-like disease. J. Allergy Clin. Immunol.135, 1641–1643 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Aird, A. et al. Novel Heterozygous Mutation in NFKB2 Is Associated With Early Onset CVID and a Functional Defect in NK Cells Complicated by Disseminated CMV Infection and Severe Nephrotic Syndrome. Front Pediatr.7, 303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Howson, L. J. & Bryant, V. L. Insights into mucosal associated invariant T cell biology from human inborn errors of immunity. Front Immunol.13, 1107609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmiedel, B. J. et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell175, 1701–1715 e1716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sato, Y., Osada, E. & Manome, Y. Non-canonical NFKB signaling endows suppressive function through FOXP3-dependent regulatory T cell program. Heliyon9, e22911 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stadinski, B. D. et al. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat. Immunol.17, 946–955 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daley, S. R. et al. Cysteine and hydrophobic residues in CDR3 serve as distinct T-cell self-reactivity indices. J. Allergy Clin. Immunol.144, 333–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tangye, S. G., Nguyen, T., Deenick, E. K., Bryant, V. L., Ma, C. S. Inborn errors of human B cell development, differentiation, and function. J. Exp. Med.220, (2023). [DOI] [PMC free article] [PubMed]
- 74.Zhang, Q., Lenardo, M. J. & Baltimore, D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell168, 37–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li, Q., Antwerp, Van, Mercurio, D., Lee, F. & Verma, K. F. IM. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science284, 321–325 (1999). [DOI] [PubMed] [Google Scholar]
- 76.Mousallem, T. et al. A nonsense mutation in IKBKB causes combined immunodeficiency. Blood124, 2046–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pannicke, U. et al. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N. Engl. J. Med.369, 2504–2514 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Qin, T. et al. A Novel Homozygous Mutation Destabilizes IKKbeta and Leads to Human Combined Immunodeficiency. Front Immunol.11, 517544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cuvelier, G. D. E. et al. Clinical presentation, immunologic features, and hematopoietic stem cell transplant outcomes for IKBKB immune deficiency. Clin. Immunol.205, 138–147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kreins, A. Y., Bonfanti, P. & Davies, E. G. Current and Future Therapeutic Approaches for Thymic Stromal Cell Defects. Front Immunol.12, 655354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herman, M. et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med.209, 1567–1582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moretti, S. et al. Insights into the molecular function of the inactivating mutations of B-Raf involving the DFG motif. Biochim Biophys. Acta1793, 1634–1645 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Klemann, C. et al. Clinical and Immunological Phenotype of Patients With Primary Immunodeficiency Due to Damaging Mutations in NFKB2. Front Immunol.10, 297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schlechter, N. et al. Exome Sequencing Identifies a Novel MAP3K14 Mutation in Recessive Atypical Combined Immunodeficiency. Front Immunol.8, 1624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anthony, N. G. et al. Inhibitory Kappa B Kinase alpha (IKKalpha) Inhibitors That Recapitulate Their Selectivity in Cells against Isoform-Related Biomarkers. J. Med Chem.60, 7043–7066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aba, U. et al. A Novel Homozygous Germline Mutation in Transferrin Receptor 1 (TfR1) Leads to Combined Immunodeficiency and Provides New Insights into Iron-Immunity Axis. J. Clin. Immunol.44, 55 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science370, eabd4585 (2020). [DOI] [PMC free article] [PubMed]
- 88.Besci, O. et al. Reference values for T and B lymphocyte subpopulations in Turkish children and adults. Turk. J. Med Sci.51, 1814–1824 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ikinciogullari, A. et al. Peripheral blood lymphocyte subsets in healthy Turkish children. Turk. J. Pediatr.46, 125–130 (2004). [PubMed] [Google Scholar]
- 90.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information/Source Data files. Bulk RNA-seq datasets have been deposited in NCBI Gene Expression Omnibus (GEO) with the accession code GSE256535 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE256535). Raw whole exome and immune repertoire sequencing data are available at the NCBI sequence read archive (SRA) under accession number PRJNA1180092. Source data are provided with this paper.