Abstract
Ho endonuclease initiates a mating type switch by making a double-strand break at the mating type locus, MAT. Ho is marked by phosphorylation for rapid destruction by functions of the DNA damage response, MEC1, RAD9, and CHK1. Phosphorylated Ho is recruited for ubiquitylation via the SCF ubiquitin ligase complex by the F-box protein, Ufo1. Here we identify a further DNA damage-inducible protein, the UbL-UbA protein Ddi1, specifically required for Ho degradation. Ho interacts only with Ddi1; it does not interact with the other UbL-UbA proteins, Rad23 or Dsk2. Ho must be ubiquitylated to interact with Ddi1, and there is no interaction when Ho is produced in mec1 or Δufo1 mutants that do not support its degradation. Ddi1 binds the proteasome via its N-terminal ubiquitinlike domain (UbL) and interacts with ubiquitylated Ho via its ubiquitin-associated domain (UbA); both domains of Ddi1 are required for association of ubiquitylated Ho with the proteasome. Despite being a nuclear protein, Ho is exported to the cytoplasm for degradation. In the absence of Ddi1, ubiquitylated Ho is stabilized and accumulates in the cytoplasm. These results establish a role for Ddi1 in the degradation of a natural ubiquitylated substrate. The specific interaction between Ho and Ddi1 identifies an additional function associated with DNA damage involved in its degradation.
Ho endonuclease of Saccharomyces cerevisiae makes a site-specific double-strand break (DSB) at the mating type locus, MAT, in late G1. The DSB is repaired by gene conversion using one of the silent mating type cassettes as a template, and this leads to a mating type switch (57). Repair of the DSB regenerates the Ho cognate site and in addition to tight transcriptional regulation of HO (10), the protein is rapidly degraded via the ubiquitin-26S proteasome system with a half-life of ca. 8 min (30).
Functions of the DNA damage response (DDR), MEC1, RAD9, and CHK1, are responsible for phosphorylation of Ho that targets it for degradation via the ubiquitin-26S proteasome system (30). The DDR is a cellular response that coordinates cell cycle progression with repair of lesions in DNA and with DNA replication (66). Ubiquitin conjugation involves an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase (22). In most cases substrate ubiquitylation occurs in multiprotein complexes (11). Ho is ubiquitylated by the SCF (Skp1-Cdc53-F-box protein) ubiquitin ligase (E3) complex. The SCF consists of a Cdc53/Cullin scaffold complexed at one end with the Skp1 adaptor protein and at the other with the RING protein, Rbx1. A series of F-box proteins recruit degradation substrates to the SCF by forming a complex with the Skp1 adaptor (54, 55). F-box proteins recruit substrate molecules that are marked by phosphorylation, usually at multiple sites (29, 41). The novel F-box protein Ufo1 recruits Ho to the SCF for ubiquitylation (29, 30). Transcription of UFO1 is induced by the DDR in response to DNA damage, and this involves the Mec1 pathway (24). Deletion of UFO1 affects maintenance of genome stability; however the mechanism underlying this observation has not been elucidated (56).
Despite being a nuclear protein, Ho degradation occurs in the cytoplasm, and if Ho is trapped within the nucleus either by a point mutation that eliminates a critical phosphorylation site, or by deletion of its nuclear exportin, Msn5, the protein is stabilized. The MEC1 pathway leads to phosphorylation of residue HoT225, which facilitates its nuclear export, and of additional residues necessary for binding the F-box protein, Ufo1. Stabilization of Ho expressed from its native promoter, e.g., by deletion of its nuclear exportin, leads to genome instability (29).
The 26S proteasome consists of a 20S catalytic core complexed at one or both ends to a 19S regulatory particle (RP) that functions in substrate recognition, binding, deubiquitylation, unfolding, and in gating of the 20S (45). Substrates are marked for proteasomal degradation by covalent attachment of a chain comprising at least four ubiquitins linked via ubiquitinK48 (15). Initially the C-terminal glycine, G76, of ubiquitin is linked to a substrate lysine and a chain is formed by addition of successive ubiquitin molecules to the K48 residue of the preceding ubiquitin. Ubiquitin has seven lysine residues and besides K48-linked chains, K29- and K63-linked chains are formed in vivo. However, modification with these chains does not target a substrate for degradation (46).
There is growing evidence that additional functions subsequent to ubiquitylation determine whether a given substrate will indeed be degraded by the proteasome. These include the ubiquitin chain binding proteins. Originally it was proposed that the Rpn10 (mammalian S5a) subunit of the 19S RP binds polyubiquitylated substrates as its C-terminal ubiquitin interacting motif binds polyubiquitin chains (12, 52, 62). However, deletion of Rpn10 is not lethal (59). Cross-linking experiments indicate that Rpt5, an ATPase subunit of the base of the 19S, interacts with polyubiquitin chains (33). A further class of polyubiquitin-binding proteins is the UbL-UbA protein family exemplified by Rad23, Dsk2, and Ddi1 (7, 17, 65). The UbA domain (23) is a degenerate motif of 40 to 50 residues that folds into a compact three-helix bundle with a hydrophobic surface patch for protein-protein interactions (13, 40, 64). The UbA domain shows a high affinity for K48-linked polyubiquitin chains (48, 65). In a recent detailed study of the proximal UbA domain of human Rad23A (HHR23A) Raasi and colleagues have shown that the UbA domain binds ubiquitin chains with a 1:1 molar stoichiometry. Binding affinity increases with chain length and is optimal at 4 to 6 ubiquitin residues. Experiments employing chimeric K48-Ub4 tetramers mutated in a critical residue (UbL8A) show that all the L8 side chains in Ub4 contribute to the interaction with HHR23A-UbA (48).
UbL-UbA proteins have an N-terminal ubiquitinlike (UbL) domain of about 70 residues with primary sequence similarity to ubiquitin that adopts a ubiquitin fold (6, 23, 26, 31). The Dsk2 and Rad23 UbLs bind the 19S RP (14, 63). Degradation of certain artificial substrates is dependent on UbL-UbA proteins (17, 34, 50), and a number of natural substrates stabilized in the absence of Rad23 and Dsk2 and their Schizosaccharomyces pombe, frog, and human orthologs have been identified (see, e.g., references 3, 16, 32, 43, 50, and 62). These include misfolded endoplasmic reticulum (ER) substrates degraded via the ER-associated protein degradation pathway (39). These findings have led to the proposal that UbL-UbA proteins serve as adaptors that deliver ubiquitin-conjugated substrates to the proteasome (20, 65). However, in some instances the UbL-UbA proteins protect substrates from degradation in vivo (4, 36, 42) and in vitro (49). This has been suggested to be due to their preventing an interaction of the substrate ubiquitin chain with the proteasome perhaps by capping the chain and preventing access to both E3s and deubiquitylating enzymes (DUBs) (43, 49).
The least-studied UbL-UbA protein is Ddi1 (DNA damage inducible). Transcription of DDI1 is induced in response to DNA damage (25, 68); however, no role for Ddi1 in the DDR has been defined. Δddi1 and Δrad23 mutants are partially defective in a subset of Pds1/securin-mediated functions involved in S-phase checkpoint signaling (8). In addition Ddi1 (alias Vsm1) may have a role in intracellular membrane transport as it binds v- and t-SNARES that mediate docking and fusion of intracellular vesicles (37, 38). Ddi1 binds the proteasome and polyubiquitylated conjugates (52); however, these interactions have not been mapped to specific domains of the protein. Deletion of DDI1 leads to accumulation of polyubiquitylated conjugates (52); however, Ddi1 has not been reported to be involved in the degradation of any physiological substrate. Recently an engineered version of Ddi1 was shown to mediate degradation of an artificial substrate (31). Here we show that Ddi1 is necessary for degradation of Ho endonuclease. Our results indicate a physiological role for Ddi1 as a proteasome receptor and identify ubiquitylated Ho endonuclease as its first natural substrate.
MATERIALS AND METHODS
Strains.
W303 is MATa his3 leu2 trp1 ura3-52. The Δddi1, Δrad23, and Δdsk2 strains and their isogenic wild type, BY4741, are from Euroscarf. The ufo1 wild type and Δufo1 mutants are Research Genetics BY4730 and #142, respectively. The Δmec1 mutant (his3 leu2 ura3 sml1-1 mec1::TRP1) was obtained from B. Garvick; ufo1 rad6 mutants are in DF5 (58). NSY1 has DDI1 with a C-terminal TAP tag (47) in DF5 integrated using primers (F-CAGACTAACGGAAATGCAGAATTTGCTGCATCCCTCCTTTTCCAATCCATGGAAAAGAGAAG and R-GGGCTACATACGTAGAGGCCGATCACAATATCAGTGGTTGCTCATACGACTCACTATAGGG) to make a PCR product consisting of the tag and the TRP1 marker.
Plasmid construction and expression.
pTET-HO-LACZ and pTET-LACZ are HoLZ and LacZ expressed from the TET promoter of plasmid pCM190, respectively (30). Construction of pGFP-UFO1 and pGFP-HO is described in references 29 and 2, respectively. DDI1 was expressed from the ADH1 promoter in plasmid pAD6 (37). Subclones of Ddi1 with the UbL or UbA domain deleted were LexA fusions expressed from the GAL promoter (4). pYES3 (Invitrogen) was used for expressing high levels of FLAG-tagged RPN1 (FRPN1), FRAD23, FDSK2, and FDDI1 from the GAL promoter (52). Wild-type and (K48R,G76A) mutant ubiquitins (15) were expressed from the CUP1 promoter. Transformation of yeast cells was performed by lithium acetate (1).
Metabolic labeling, immunoprecipitation (IP), and pulse-chase are based on reference 18 and described in reference 30. Coimmunoprecipitation (co-IP)-immunoblotting was based on the method described in reference 37. These experiments were done either by (i) coexpression of both the potential interacting partners in yeast or (ii) mixing two cell lysates, each with one of the potential interacting partners. In both cases the lysates were incubated with the appropriate antibody directed against one of the interacting partners; after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) the presence of the second protein was detected by Western analysis.
Yeast cells were grown overnight to late logarithmic phase (optical density at 600 nm = 0.8) in 50 ml of the appropriate inductive synthetic minimal medium with 2% galactose for expression of the GAL-regulated genes; for induction of the TET-regulated constructs, HO-LACZ and LACZ, cells were incubated for 40 min in medium without doxycycline. The chase was performed by addition of 10 mM cycloheximide and methionine in radioactive experiments or 3% glucose for galactose-induced genes. For experiments in which Ho protein was made in cells expressing native or mutant (K48R,G76A) ubiquitin, we cotransformed W303 cells with pTET-HO-LACZ and with the vector or native- or mutant-ubiquitin-expressing plasmids. The cotransformed yeast cells were grown overnight without doxycycline to induce HO-LACZ, and with 0.2 mM CuSO4 to induce ubiquitin. In these experiments the specific activity of HoLZ in each cotransformant was determined by the o-nitrophenyl-β-d-galactopyranoside (ONPG) reaction and equal aliquots of ONPG units were taken for each co-IP.
The cells were harvested by centrifugation, washed in 50 ml of Tris-EDTA, and resuspended in 400 μl co-IP buffer (0.1% NP-40, 250 mM NaCl, 5 mM EDTA, 50 mM Tris-Cl [pH 7.5], 1:25 of Boehringer protease inhibitor cocktail). Glass beads (0.5 to 0.6 g) were added, and cells were broken with a glass beater (Biospec Products) using five 1-min cycles at 4°C. The lysate was clarified by centrifugation at 13,000 rpm for 15 min at 4°C, and protein concentration was measured with the Bio-Rad protein reagent. Protein lysate (5 to 15 mg) was used for IP with the appropriate antibodies in co-IP buffer at 4°C for 1 to 2 h with mild shaking. Thirty microliters of 50% protein A-Sepharose (Amersham) was added to each sample, and incubation was continued under the same conditions for 0.5 to 1 h. The samples were washed 6 times with co-IP buffer with 1% of Triton and centrifuged. The pellet was resuspended in 30 μl 2× Laemmli sample buffer, boiled for 10 min, and electrophoresed on a 10% polyacrylamide SDS gel (PAGE) with protein size standards. The gel was transferred to nitrocellulose membrane (Protran BA 85; Schleicher & Schuell), and Western blotting was performed with the appropriate antisera. One milligram of crude protein was used to determine input by IP-Western blot, and 30 μg was taken for direct Western blotting. For Ddi1-TAP pulldowns, immunoglobulin G (IgG) was covalently coupled to M-280 tosyl-activated Dynabeads (DYNAL) according to the manufacturer's instructions. The beads were incubated with crude yeast lysate for 2 h at 4°C. The bead pellet was washed three times with co-IP buffer as above with increasing NaCl concentrations up to 250 mM. A Dynal magnetic particle concentrator (Dynal Biotech) was used to separate the beads from the lysate supernatant.
Anti-green fluorescent protein (anti-GFP) antibody purchased from Roche Molecular Biochemicals was used at dilutions of 1:200 for IP and 1:1,000 for Western blotting; anti-LacZ from Santa Cruz Biotechnology was used at 1:600 for IP and 1:1,000 for Western blotting; anti-Flag was purchased from Sigma and used at 1:250 for IP and 1:3,000 for Western blotting; anti-Ddi1 antiserum was a gift from Jeff Gerst and was used at 1:1,000 for IP and 1:5,000 for Western blotting; anti-Rpn12 antiserum was a gift from Dorota Skowyra and used at 1:10,000 for Western blotting; antiubiquitin antiserum from Affiniti Research Products was used at 1:5,000. Goat anti-rabbit antiserum, used at 1:1,000, and goat anti-mouse antiserum, used at 1:1,000, were purchased from Santa Cruz Biotechnology. Detection was by enhanced chemiluminescence (ECL) using an Amersham Pharmacia Biotech ECL Western blotting kit.
Microscopy.
Cells expressing GFP-tagged proteins were observed with a Nikon fluorescence microscope as described previously (2).
RESULTS
Interaction of Ho with a UbL-UbA protein.
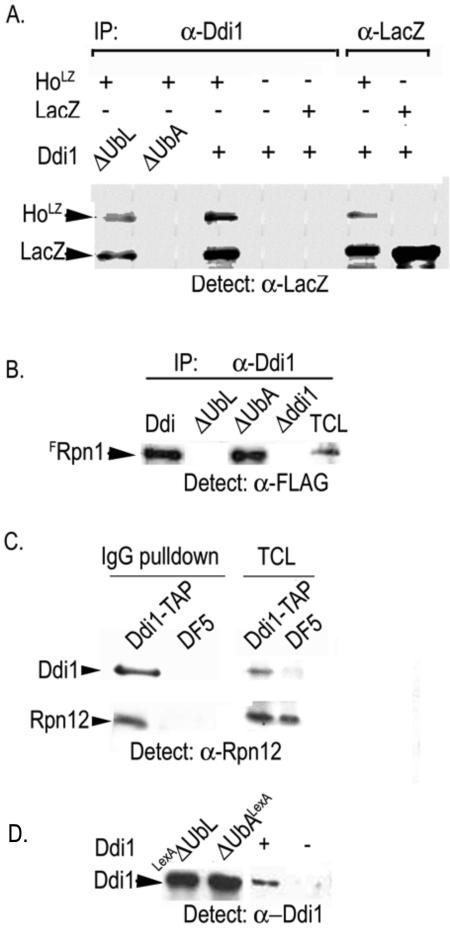
To test the interaction of Ho with a UbL-UbA protein, lysates from cells expressing pYES-FLAG(F)-RAD23, FDSK2, or FDDI1 (52) were incubated with lysate from cells that produced HoLZ (30). The FUbL-UbA proteins were immunoprecipitated with anti-FLAG antibodies and protein A. After washing the retained proteins were analyzed by PAGE and Western blotting with anti-LacZ antiserum to detect HoLZ. An aliquot of the HoLZ lysate was immunoprecipitated with anti-LacZ to assay the input of HoLZ. The FUbL-UbA proteins were detected in total cell lysates by Western blot with anti-FLAG antibodies as they could not be distinguished from the immunoglobulin chains after IP. We observed that HoLZ forms a complex only with FDdi1 and not with FRad23 or FDsk2 (Fig. 1).
FIG. 1.
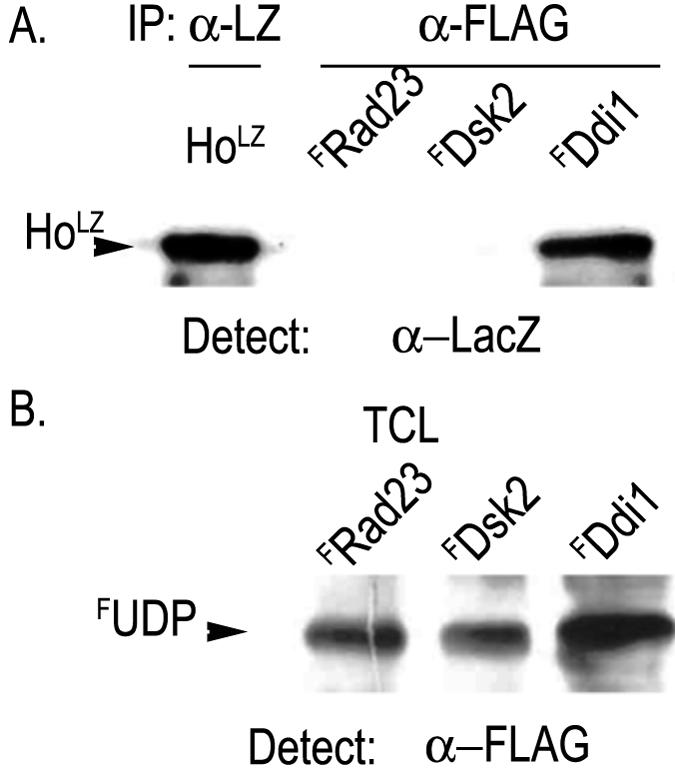
Ho coimmunoprecipitates with Ddi1. Lysates from cells expressing FRAD23, FDSK2, or FDDI1 were incubated with equal aliquots of lysate from cells with Ho-LacZ (HoLZ). (A) The FUbL-UbA proteins, FRAD23, FDSK2, or FDDI1, were immunoprecipitated with anti-FLAG (α-FLAG), blotted, and subjected to Western analysis using anti-LacZ antibodies to detect HoLZ. Left lane shows HoLZ by IP-Western. (B) Western blots of total cell lysates (TCL) from cells expressing FRAD23, FDSK2, or FDDI1 were reacted with anti-FLAG antibodies. FUDP represents FLAGUbL-UbA proteins as indicated above each lane.
The UbA domain of Ddi1 is necessary for interaction with Ho.
Ddi1 has an N-terminal UbL of ca. 70 residues and a C-terminal UbA domain of ca. 40 residues. To map the interaction between Ho and Ddi1, we mixed HoLZ or control LacZ lysates with lysates from wild-type or Δddi1 cells or from Δddi1cells that produced LexA fusions of Ddi1ΔUbL or Ddi1ΔUbA (4). Full-length and truncated Ddi1 proteins were immunoprecipitated with anti-Ddi1 antiserum and protein A, and the pellet was separated by PAGE and blotted. The Western blot was analyzed for the presence of HoLZ with anti-LacZ antiserum. We observed HoLZ in co-IPs with Ddi1 and Ddi1ΔUbL, but not with Ddi1ΔUbA indicating that Ho interacts with the UbA domain of Ddi1. The complex is between the Ho moiety and Ddi1 as the control LacZ protein did not interact with Ddi1 (Fig. 2A).
FIG. 2.
(A) Ho interacts with Ddi1 via its UBA domain. HoLZ and control LacZ lysates were mixed with lysates from wild-type, Δddi1, or from Δddi1 cells with LexA-DdiI1ΔUbL (ΔUbL), or LexA-Ddi1ΔUbA (ΔUbA). Ddi1 proteins were immunoprecipitated with anti-Ddi1 (α-Ddi1; left panel), and detection of HoLZ and LacZ was with anti-LacZ. Right panel shows HoLZ and LacZ in the lysates by IP-Western. (The LacZ band appears also in all HoLZ IPs in which degradation of Ho is observed.) HoLZ is observed in co-IPs with Ddi1 and Ddi1ΔUbL, but not with Ddi1ΔUbA. The LacZ control did not bind Ddi1. (B) Ddi1 binds the 19S RP via its UBL domain. The Ddi1 lysates were mixed with FRpn1 lysates; Ddi1 proteins were immunoprecipitated with anti-Ddi1 and blotted, and anti-FLAG was used to detect FRpn1 on the membrane. FRpn1 coimmunoprecipitated only with full-length Ddi1 and Ddi1ΔUbA; it did not coimmunoprecipitate with Ddi1ΔUbL. (C) Ddi1-TAP on IgG beads pulls down the 19S RP. (Left panel) Cell extracts from DF5 yeast or NSY1 yeast that produce Ddi1-TAP were incubated with IgG magnetic beads. The bead fraction was gel separated and blotted with anti-Rpn12 antiserum. A band corresponding in size to Rpn12 is visible in the presence of Ddi1-TAP. Right panel shows total cell lysates (TCL). The anti-rabbit antiserum detects Ddi1-TAP by its TAP tag. (D) Western blot of TCL showing the presence of the different Ddi1 proteins used.
Ddi1 binds the 19S RP of the proteasome via its UbL domain.
To test which domain of Ddi1 binds the proteasome, the above lysates were mixed with lysates from cells expressing pYES-FRPN1, a subunit of the 19S RP (53). Ddi1 proteins were subjected to IP with anti-Ddi1 and protein A, and the precipitated proteins separated by PAGE and blotted. Anti-FLAG antibodies were used to detect FRpn1 on the Western blots. The conditions we use in these experiments do not lead to disassociation of the 19S RP, and therefore interaction between Ddi1 and any subunit of the 19S RP complex would lead to coimmunoprecipitation of FRpn1. We found that full-length Ddi1 and Ddi1ΔUbA coimmunoprecipitate with FRpn1; however, FRpn1 did not coimmunoprecipitate with Ddi1ΔUbL (Fig. 2B). To exclude the possibility that Ddi1 could be interacting with Rpn1 that is not part of a 19S RP, we performed a pull-down experiment in which a lysate from cells producing TAP-tagged Ddi1 was incubated with magnetic beads coated with IgG. After stringent washing the proteins were separated by PAGE and blotted with anti-Rpn12 antiserum. A band corresponding to the size of Rpn12 was detected in the bead fraction of cells that produced Ddi1-TAP and was absent from the IgG beads incubated with a control DF5 lysate (Fig. 2C). Taken together these results indicate that Ddi1 binds the 19S RP via its UbL domain and interacts with ubiquitylated Ho via its UbA as reported for Rad23 and Dsk2 (14, 52).
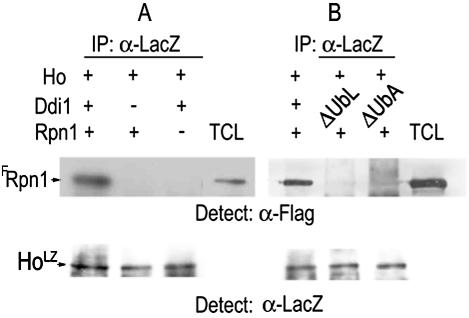
We next tested whether Ddi1 is essential for formation of a complex between HoLZ and the 19S RP. HoLZ was produced in isogenic wild type and Δddi1 mutants and immunoprecipitated with anti-LacZ antiserum, and the washed protein A-HoLZ beads were then incubated with equal aliquots of FRpn1 lysates for 4 h and washed thoroughly in co-IP buffer for separation by PAGE and Western blotting. Anti-FLAG was used to detect FRpn1 on the Western blots. We found that only HoLZ produced in wild-type cells in the presence of Ddi1 coimmunoprecipitated with FRpn1; when produced in Δddi1 mutants it did not coimmunoprecipitate with FRpn1 (Fig. 3A). Similar experiments were next performed in which HoLZ was made in Δddi1 mutants cotransformed with the LexA-DdiΔUbL and LexA-DdiΔUbA plasmids. In the absence of either the UbL or the UbA domain of Ddi1 no complex was formed that included HoLZ and FRpn1 (Fig. 3B).
FIG. 3.
Ho forms a complex with FRpn1, and this requires both domains of Ddi1. (A) Protein A-HoLZ beads from IPs in which HoLZ was produced in the presence or the absence of Ddi1 were washed and then incubated with equal aliquots of FRpn1 lysate. HoLZ coimmunoprecipitates with FRpn1 only in the presence of Ddi1. (B) The above protein A-HoLZ beads were produced in extracts of cells producing Ddi1, LexA-Ddi1ΔUbL, or LexA-Ddi1ΔUbA as in Fig. 2D. They were incubated with equal aliquots of the FRpn1 lysate. The coimmunoprecipitated pellets were gel separated and blotted and anti-FLAG (α-FLAG) was used to detect FRpn1. No complex is formed between Ho and Rpn1 in the absence of either the UbL or the UbA domain of Ddi1. Total cell lysate (TCL) shows the FRpn1 band. Lower panel shows a control aliquot of HoLZ from each cell type by anti-LacZ IP-Western blot.
Ho interacts with Ddi1 and the 19S RP only when ubiquitylated.
To test whether Ho must be ubiquitylated to interact with Ddi1 and the 19S RP, we produced HoLZ in the presence of overexpressed native or double-mutant (K48R, G76A [Ub*]) ubiquitin. The double-mutant ubiquitin acts as a dominant negative leading to a prevalence of short ubiquitin conjugates and stabilization of substrates (15). In the presence of Ub* Ho degradation is retarded (28). The HoLZ lysates were mixed with equal aliquots of Ddi1 or of FRpn1 lysates. For assaying for interaction between HoLZ and Ddi1 we used anti-Ddi1 antibodies to IP Ddi1 and, after PAGE, anti-LacZ to detect HoLZ on the Western blots. To assay for interaction between HoLZ and the 19S RP we used anti-LacZ to IP HoLZ and anti-FLAG antiserum for detection of FRpn1 on the Western blots. Control lanes show an aliquot of each HoLZ lysate immunoprecipitated with anti-LacZ and analyzed by anti-LacZ Western blots. We found that whereas HoLZ produced in the presence of empty vector or overexpressed native ubiquitin interacts with both Ddi1 and the 19S RP, HoLZ made in the presence of overexpressed Ub* did not interact with either Ddi1 (Fig. 4A) or Rpn1(Fig. 4B).
FIG. 4.
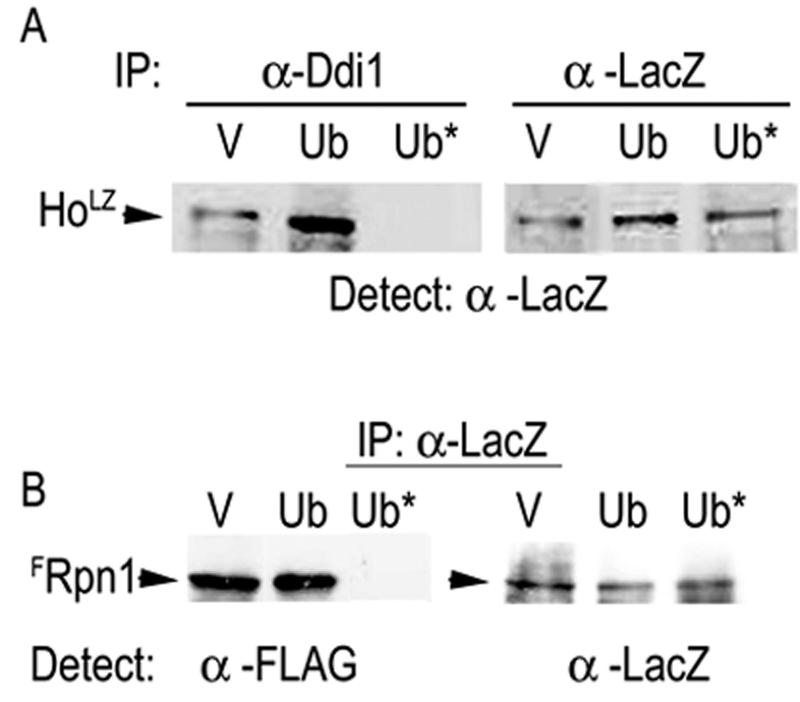
HoLZ made in the presence of K48R, G76A ubiquitin does not interact with Ddi1 or the 19S RP. (A) Lysates with HoLZ from cells expressing vector, native, or K48R, G76A ubiquitin were mixed with equal aliquots of the wild type (Ddi1) (A) or with FRpn1 (B) lysates. Anti-Ddi1 (α-Ddi1) was used to immunoprecipitate Ddi1, the pellet was gel separated and blotted, and HoLZ complexed to Ddi1 was detected with anti-LacZ. (B) Lysates were as in panel A. Anti-LacZ was used to immunoprecipitate HoLZ, the pellet was gel separated and blotted, and FRpn1 was detected with anti-FLAG. Right panels of panels A and B indicate the presence of HoLZ by anti-LacZ IP and Western blot in each experiment. V represents the CUP1 vector; Ub is the CUP1 vector expressing native ubiquitin (Ub); Ub*, mutant ubiquitin. HoLZ produced in the presence of empty vector or native ubiquitin interacts with both Ddi1 and the 19S RP whereas HoLZ made in the presence of mutant ubiquitin did not interact with either protein.
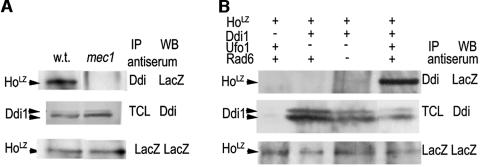
The experiments with mutant ubiquitin suggest that ubiquitin chain formation is important for the interaction of HoLZ with Ddi1. To test this hypothesis, we therefore produced HoLZ in mutants where it does not undergo degradation and tested whether it could interact with Ddi1. Mutants used for HoLZ production are mec1 mutants of the DDR in which Ho is not phosphorylated (29), and mutants of both the F-box protein that recruits Ho to the SCF, Δufo1, and in combination with the Rad6 E2 that has a role in Ho degradation (30), rad6 Δufo1 double mutants. Ddi1 was immunoprecipitated with anti-Ddi1 antibodies, the pellet was separated by PAGE, and detection of HoLZ on the Western blot was with anti-LacZ. To ensure that Ho was produced in all cell types, we performed control anti-LacZ IPs that were blotted with anti-LacZ antibodies. The results of these experiments show that only HoLZ produced in wild-type cells interacted with Ddi1; HoLZ made in mutants in which it could not be ubiquitylated did not coimmunoprecipitate with Ddi1 (Fig. 5A and B).
FIG. 5.
(A) HoLZ made in mec1 mutants does not interact with Ddi1. Ddi1 was immunoprecipitated with anti-Ddi1 (α-Ddi1), and the coimmunoprecipitated pellet was separated by PAGE and analyzed by Western blot with anti-LacZ antibodies to detect HoLZ. Middle panel shows the presence of Ddi1 in total cell lysates (TCLs) of wild-type and mec1 cells. (Lower panel) Control anti-LacZ IP-Western blot indicates the presence of HoLZ in both cell lysates. Only HoLZ produced in wild-type cells interacts with Ddi1; HoLZ made in mec1 mutants does not. (B) HoLZ made in Δufo1 and in Δrad6 Δufo1 double mutants does not interact with Ddi1. (Top panel) Ddi1 was immunoprecipitated with anti-Ddi1 and blotted, and HoLZ was detected with anti-LacZ; (middle panel) Western blot of TCL with anti-Ddi1 showing the presence of the Ddi1 doublet; (lower panel) anti-LacZ immunoprecipitates blotted with anti-LacZ indicate the presence of HoLZ in all cell types. Only HoLZ produced in wild-type cells interacts with Ddi1; HoLZ made in mutants in which it could not be ubiquitylated did not coimmunoprecipitate with Ddi1.
Ho is stabilized in the absence of Ddi1.
To determine whether the association of ubiquitylated Ho with Ddi1 is a critical step in its degradation, we determined the half-life of HoLZ in wild-type and Δddi1 cells by pulse-chase with radioactive methionine and anti-LacZ IP. In wild-type cells Ho was rapidly degraded, whereas in Δddi1 cells there was no observable degradation during the 30 min of the experiment (Fig. 6).
FIG. 6.
Ho-LacZ is stabilized in the absence of Ddi1. The half-life of HoLZ in wild type and Δddi1 cells determined by pulse-chase and anti-LacZ IP. NI, noninduced.
Ho accumulates in the cytoplasm of Δddi1 mutants.
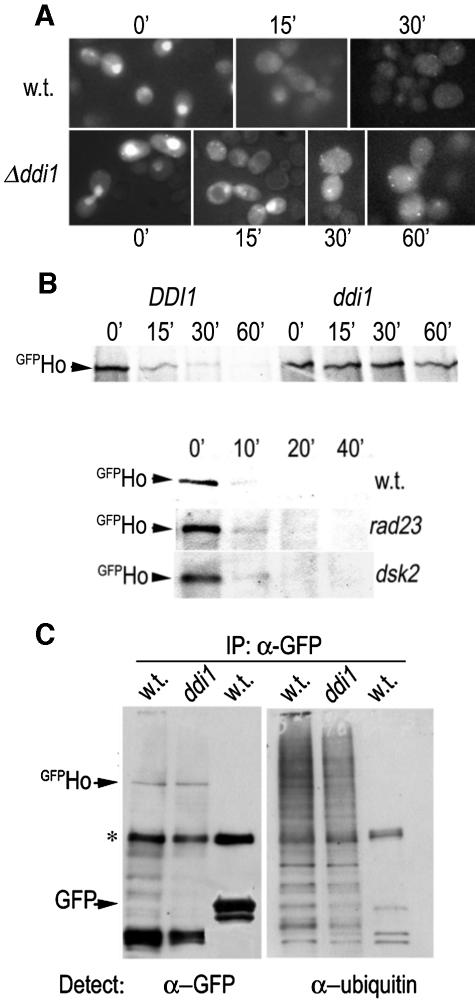
Ho must exit the nucleus to be degraded and in mutants that affect early stages of the pathway, e.g., in mec1 mutants, or in mutants of the nuclear exportin Msn5, stabilized Ho accumulates within the nucleus (29). The present results suggest that it is ubiquitylated Ho that interacts with Ddi1. This leads to the prediction that Ho stabilized due to the absence of Ddi1 should accumulate in the cytoplasm. We therefore followed the degradation of GFPHo in isogenic wild-type and Δddi1 cells by inducing pGFP-HO and following the GFP signal by microscopy. Cycloheximide was added at the zero time point to inhibit protein synthesis. In wild-type cells a strong nuclear GFPHo signal was seen at the zero time point and the GFP signal became less strong at 15 min and disappeared altogether by 30 min. In contrast, in Δddi1 cells the GFPHo signal disappeared from the nuclei of some cells by 15 min and was visible within the cytoplasm at both the 30- and 60-min time points (Fig. 7A). Stabilization of GFPHo in Δddi1 cells was confirmed in a parallel pulse-chase IP experiment. In contrast to Δddi1 cells there was no stabilization of HoLZ in rad23 or dsk2 mutants (Fig. 7B).
FIG. 7.
(A) GFPHo accumulates in the cytoplasm of Δddi1 mutants. pGFP-HO was induced in wild-type and Δddi1 cells, and the GFP signal was followed by microscopy. In wild-type cells the GFPHo strong nuclear signal seen at the zero time point is no longer visible in the nucleus after 15 min and has completely disappeared by 30 min. In Δddi1 cells the GFPHo signal accumulates in the cytoplasm and is still visible at both the 30- and 60-min time points both as a dispersed cytoplasmic signal and as strong fluorescent spots that are probably aggresomes. (B) GFPHo is stabilized in Δddi1 cells, but not in Δrad23 or Δdsk2 mutants. The half-life of GFPHo in wild-type and Δddi1 cells was determined by pulse-chase and anti-GFP IP. (Lower panel) Half-life of HoLZ in wild-type and isogenic Δrad23 and Δdsk2 cells. (C) Ho is ubiquitylated in Δddi1 mutants. GFPHo was immunoprecipitated from wild-type and Δddi1 mutants, and GFP was immunoprecipitated from wild-type (w.t.) cells. The IP pellets were run in duplicate and blotted with anti-GFP (α-GFP; left panel) and antiubiquitin antibodies (right panel). The anti-GFP blot shows the GFPHo band and a small number of lower bands that may be degradation intermediates of GFPHo; they do not appear in the GFP IP. * marks IgG chain. The antiubiquitin blotshows high-MW bands in both wild-type and Δddi1 GFPHo IPs; no ubiquitin conjugate bands are visible in the GFP IP. This is also the case after an extremely long exposure time (not shown).
Stabilization of Ho in Δddi1 mutants could be the result of its being deubiquitylated in the absence of Ddi1. In this event it would no longer be recognized as a substrate for the proteasome. We therefore attempted to assay the ubiquitylation status of Ho directly. Using anti-GFP antiserum we immunoprecipitated GFPHo from wild type and Δddi1 mutants, and GFP from wild-type cells. The three immunoprecipitates were run in duplicate on the same gel followed by Western blotting with either anti-GFP or antiubiquitin antibodies. The lanes blotted with anti-GFP show the position of GFPHo and a small number of faster migrating bands that may be degradation intermediates of GFPHo; they do not appear in the GFP IP. The duplicate lanes blotted with antiubiquitin antibodies show high-molecular-weight (MW) bands in GFPHo IPs from both wild type and Δddi1 mutants; these antibodies do not detect any bands in the GFP IP. We interpret the high-MW bands to be ubiquitin conjugates of GFPHo that are detectable with the antiubiquitin antiserum, but given the ratio of GFP/ubiquitin moiety of each ubiquitin-conjugated GFPHo molecule would be below the level of detection by the anti-GFP antibodies (Fig. 7C). Thus Ho does not loose its ubiquitin tag in the absence of Ddi1.
DISCUSSION
There is growing evidence that additional functions subsequent to ubiquitylation determine whether a given substrate will indeed be degraded by the proteasome (39, 62). Our present results indicate that degradation of ubiquitylated Ho involves an additional DNA damage-associated protein, Ddi1. These results show that Ddi1 has a physiological role in the ubiquitin-proteasome system. However, they do not explain the specificity of Ho for this particular UbL-UbA protein. Stringent specificity of an ubiquitylated substrate and a single UbL-UbA protein is not always observed in vivo. For example, the mutant version of CPY* that is degraded via the ER-associated protein degradation pathway, shows a requirement for both Rad23 and Dsk2. Single mutants of either Rad23 or Dsk2 do not lead to CPY* stabilization (39, 62), and it is only in the double rad23 dsk2 mutant that CPY* is stable. In these mutants control cytosolic substrates undergo normal degradation indicating that proteasome function is not impaired in the absence of Rad23 and Dsk2 (39). Similarly Rad23 and Rpn10 have redundant roles in degradation of the cyclin-dependent protein kinase inhibitor Sic1 (62).
In the case of Ho we observe a strong requirement for Ddi1 and not for either Rad23 or Dsk2. This is shown in co-IP experiments where Ho is found in a complex only with Ddi1 and not with either Rad23 or Dsk2. Furthermore it is supported by evidence showing stabilization of Ho in vivo in Δddi1 mutants despite the presence in these cells of functional Rad23 and Dsk2 and additional polyubiquitin chain binding proteins (21). Given that all three UbL-UbA family proteins have a UbA domain and that this domain binds tetraubiquitin chains with high affinity, it is clear that the ubiquitin chain can be responsible for only part of the binding affinity and that residues of the substrate itself must be the predominant determinant of complex formation.
This raises the question of whether a substrate protein can interact with a UbL-UbA protein prior to its ubiquitylation. In this context it is interesting to note the presence of an integral UbL-UbA subunit in the recently discovered SCF complex responsible for degradation of p27 in G1 cells in the cytoplasm (27). Here we used two experimental approaches to address this question: (i) mutant ubiquitin that leads to premature chain termination and (ii) production of Ho in mutants that do not support its degradation. In mec1 mutants Ho is not phosphorylated and does not bind the F-box protein Ufo1 and is therefore not recruited for ubiquitylation by the SCF. In fact lack of phosphorylation leads to trapping of Ho in the nucleus and to its total stabilization (29). In addition we produced Ho in mutants that act downstream of Mec1—an E3 mutant Δufo1 and a Δufo1 mutant in which we also deleted the E2, Rad6. (We used the double ufo1 rad6 mutant as previously we found that Ho is stable in rad6 mutants [30] and we do not know whether the Rad6 pathway acts upstream or in parallel with the SCF pathway of ubiquitylation). In all instances in which we produced Ho in cells that cannot support its degradation we did not observe any interaction with Ddi1. Thus the initial interaction must be between an ubiquitin chain and a UbL-UbA protein. This is probably a dynamic interaction that would be stabilized by complex formation between additional residues of both the substrate and the specific UbL-UbA protein.
We observe stabilized Ho that accumulates in the cytoplasm of Δddi1 cells. One explanation for this stabilization could be deubiquitylation of Ho by cellular DUBs. Ho without its ubiquitin chains would be rescued from degradation. Rad23Rhp23 and Dsk2Dph1 of fission yeast bind tetraubiquitin-conjugated substrates, and this protects the chains from the activity of DUBs. Protection is conferred by isolated UbA domains of these proteins (20). Furthermore Rad23 inhibits proteasomal degradation of the nucleotide excision repair protein Rad4/XPC in both yeast and mammals (36, 42). In vitro Rad23 inhibits both K48-polyubiquitin chain extension and chain disassembly (49). However, in our experiments we found that Ho retains its ubiquitin chains in Δddi1 mutants. This result indicates that ubiquitylated Ho is protected from the activity of cellular DUBs in the absence of Ddi1. We suggest a number of alternative hypotheses to explain this finding. (i) The DUBs are strictly compartmentalized. At least two DUBs are associated with the proteasome, the Rpn11 subunit of the 19S RP (60) and Ubp6 (19, 35). A facet of UbL-UbA protein activity may be to orient the substrate polyubiquitin chains so that they can be processed by the proteasomal DUBs. (ii) UbL-UbA protein activity may be necessary to extract the ubiquitylated substrate from the E3 complex and/or to terminate ubiquitin chain elongation. This could possibly involve the chaperone activity of the ATPase subunits of the 19S RP (5). Subunits of both the SCF and the UbL-UbA proteins, Rad23 and Dsk2, have been identified as components of complexes that copurify with affinity-purified 26S proteasomes (61). Therefore it is possible that the role of Ddi1 is to release Ho from the SCFUfo1 and to make it available to the subunits of the 19S RP for deubiquitylation and unfolding. In the absence of Ddi1 ubiquitylated Ho may be retained within the SCFUfo1 complex. We do indeed find Ho in a co-IP complex with Ufo1 in Δddi1 cells (not shown). This result indicates that recruitment of Ho to SCFUfo1 and its ubiquitylation do not require Ddi1. Experiments in which Ho ubiquitylation and degradation are reconstituted in vitro are necessary to determine the exact function performed by Ddi1.
In addition to the DDR functions Mec1, Rad9, and Chk1, which target Ho for degradation, we have now identified a further two functions associated with DNA damage that have a role in Ho degradation. These are the F-box protein Ufo1 described previously (29, 30) and the UbL-UbA protein, Ddi1. Both UFO1 and DDI1 are transcribed in response to DNA damage (25), and this is regulated by Mec1 although different effector kinases are involved (L. Kaplun, unpublished data; 67). Mating type switching is a very slow process, and the Ho-cleaved MAT allele is very stable (9, 51). However, the DDR checkpoint response is only evoked if DSB repair does not occur within 4 h (44). Ho degradation is very rapid, and the half-life of Ho does not exceed 10 min (30). We therefore conclude that it is the normal basal levels of the DDR functions, Ufo1 and Ddi1, that function in degradation of Ho.
Acknowledgments
We thank colleagues quoted above for plasmids and antisera. We thank Emilia Klyman for technical assistance.
This work was supported by the Israel Cancer Association, the Israel Cancer Research Fund, the Association for International Cancer Research, and the German-Israel Research Fund (GIF). L. Kaplun is a postdoctoral fellow supported by GIF, A. Bakhrat is a Kreitman Fellow of the Ben Gurion University Graduate School.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bakhrat, A., M. S. Jurica, B. L. Stoddard, and D. Raveh. 2004. Homology modeling and mutational analysis of Ho endonuclease of yeast. Genetics 166:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford, F. K., J. T. Kittler, E. Muller, P. Thomas, J. M. Uren, D. Merlo, W. Wisden, A. Triller, T. G. Smart, and S. J. Moss. 2001. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat. Neurosci. 4:908-916. [DOI] [PubMed] [Google Scholar]
- 4.Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. [DOI] [PubMed] [Google Scholar]
- 5.Braun, B. C., M. Glickman, R. Kraft, B. Dahlmann, P. M. Kloetzel, D. Finley, and M. Schmidt. 1999. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell. Biol. 1:221-226. [DOI] [PubMed] [Google Scholar]
- 6.Buchberger, A. 2002. From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol. 12:216-221. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., and K. Madura. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22:4902-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, D. J., G. Mondesert, M. Segal, B. L. Bertolaet, S. Jensen, M. Wolff, M. Henze, and S. I. Reed. 2001. Dosage suppressors of Pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol. 21:1997-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly, B., C. I. White, and J. E. Haber. 1988. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2342-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 12.Deveraux, Q., V. Ustrell, C. Pickart, and M. Rechsteiner. 1994. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269:7059-7061. [PubMed] [Google Scholar]
- 13.Dieckmann, T., E. S. Withers-Ward, M. A. Jarosinski, C. F. Liu, I. S. Chen, and J. Feigon. 1998. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat. Struct. Biol. 5:1042-1047. [DOI] [PubMed] [Google Scholar]
- 14.Elsasser, S., R. R. Gali, M. Schwickart, C. N. Larsen, D. S. Leggett, B. Muller, M. T. Feng, F. Tubing, G. A. Dittmar, and D. Finley. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4:725-730. [DOI] [PubMed] [Google Scholar]
- 15.Finley, D., S. Sadis, B. P. Monia, P. Boucher, D. J. Ecker, S. T. Crooke, and V. Chau. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funakoshi, M., S. Geley, T. Hunt, T. Nishimoto, and H. Kobayashi. 1999. Identification of XDRP1; a Xenopus protein related to yeast Dsk2p binds to the N-terminus of cyclin A and inhibits its degradation. EMBO J. 18:5009-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilon, T., O. Chomsky, and R. G. Kulka. 1998. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 17:2759-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guterman, A., and M. H. Glickman. 2004. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279:1729-1738. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann-Petersen, R., K. B. Hendil, and C. Gordon. 2003. Ubiquitin binding proteins protect ubiquitin conjugates from disassembly. FEBS Lett. 535:77-81. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann-Petersen, R., M. Wallace, K. Hofmann, G. Koch, A. H. Johnsen, K. B. Hendil, and C. Gordon. 2004. The Ubx2 and Ubx3 cofactors direct Cdc48 activity to proteolytic and nonproteolytic ubiquitin-dependent processes. Curr. Biol. 14:824-828. [DOI] [PubMed] [Google Scholar]
- 22.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, K., and P. Bucher. 1996. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21:172-173. [PubMed] [Google Scholar]
- 24.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20:8157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jentsch, S., and G. Pyrowolakis. 2000. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 27.Kamura, T., T. Hara, M. Matsumoto, N. Ishida, F. Okumura, S. Hatakeyama, M. Yoshida, K. Nakayama, and K. I. Nakayama. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 6:1229-1235. [DOI] [PubMed] [Google Scholar]
- 28.Kaplun, L. 2002. Ph.D. thesis. Ben Gurion University of the Negev, Beersheba, Israel.
- 29.Kaplun, L., Y. Ivantsiv, A. Bakhrat, and D. Raveh. 2003. DNA damage response-mediated degradation of Ho endonuclease via the ubiquitin system involves its nuclear export. J. Biol. Chem. 278:48727-48734. [DOI] [PubMed] [Google Scholar]
- 30.Kaplun, L., Y. Ivantsiv, D. Kornitzer, and D. Raveh. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA 97:10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, I., K. Mi, and H. Rao. 2004. Multiple interactions of Rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol. Biol. Cell 15:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleijnen, M. F., A. H. Shih, P. Zhou, S. Kumar, R. E. Soccio, N. L. Kedersha, G. Gill, and P. M. Howley. 2000. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 6:409-419. [DOI] [PubMed] [Google Scholar]
- 33.Lam, Y. A., T. G. Lawson, M. Velayutham, J. L. Zweier, and C. M. Pickart. 2002. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416:763-767. [DOI] [PubMed] [Google Scholar]
- 34.Lambertson, D., L. Chen, and K. Madura. 1999. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153:69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leggett, D. S., J. Hanna, A. Borodovsky, B. Crosas, M. Schmidt, R. T. Baker, T. Walz, H. Ploegh, and D. Finley. 2002. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10:495-507. [DOI] [PubMed] [Google Scholar]
- 36.Lommel, L., T. Ortolan, L. Chen, K. Madura, and K. S. Sweder. 2002. Proteolysis of a nucleotide excision repair protein by the 26S proteasome. Curr. Genet. 42:9-20. [DOI] [PubMed] [Google Scholar]
- 37.Lustgarten, V., and J. E. Gerst. 1999. Yeast VSM1 encodes a v-SNARE binding protein that may act as a negative regulator of constitutive exocytosis. Mol. Cell. Biol. 19:4480-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marash, M., and J. E. Gerst. 2003. Phosphorylation of the autoinhibitory domain of the Sso t-SNAREs promotes binding of the Vsm1 SNARE regulator in yeast. Mol. Biol. Cell. 14:3114-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medicherla, B., Z. Kostova, A. Schaefer, and D. H. Wolf. 2004. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5:692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller, T. D., M. Kamionka, and J. Feigon. 2004. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 279:11926-11936. [DOI] [PubMed] [Google Scholar]
- 41.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 42.Ng, J. M., W. Vermeulen, G. T. van der Horst, S. Bergink, K. Sugasawa, H. Vrieling, and J. H. Hoeijmakers. 2003. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 17:1630-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortolan, T. G., P. Tongaonkar, D. Lambertson, L. Chen, C. Schauber, and K. Madura. 2000. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol. 2:601-608. [DOI] [PubMed] [Google Scholar]
- 44.Pellicioli, A., S. E. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 45.Pickart, C. M., and R. E. Cohen. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177-187. [DOI] [PubMed] [Google Scholar]
- 46.Pickart, C. M., and D. Fushman. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8:610-616. [DOI] [PubMed] [Google Scholar]
- 47.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 48.Raasi, S., I. Orlov, K. G. Fleming, and C. M. Pickart. 2004. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J. Mol. Biol. 341:1367-1379. [DOI] [PubMed] [Google Scholar]
- 49.Raasi, S., and C. M. Pickart. 2003. Rad23 ubiquitin-associated domains (UBA) inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278:8951-8959. [DOI] [PubMed] [Google Scholar]
- 50.Rao, H., and A. Sastry. 2002. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 277:11691-11695. [DOI] [PubMed] [Google Scholar]
- 51.Raveh, D., S. H. Hughes, B. K. Shafer, and J. N. Strathern. 1989. Analysis of the Ho-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 220:33-42. [PubMed] [Google Scholar]
- 52.Saeki, Y., A. Saitoh, A. Toh-e, and H. Yokosawa. 2002. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 293:986-992. [DOI] [PubMed] [Google Scholar]
- 53.Saeki, Y., T. Sone, A. Toh-e, and H. Yokosawa. 2002. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 296:813-819. [DOI] [PubMed] [Google Scholar]
- 54.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 55.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 56.Smith, S., J. Y. Hwang, S. Banerjee, A. Majeed, A. Gupta, and K. Myung. 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:9039-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strathern, J. N., A. J. Klar, J. B. Hicks, J. A. Abraham, J. M. Ivy, K. A. Nasmyth, and C. McGill. 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31:183-192. [DOI] [PubMed] [Google Scholar]
- 58.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Nocker, S., S. Sadis, D. M. Rubin, M. Glickman, H. Fu, O. Coux, I. Wefes, D. Finley, and R. D. Vierstra. 1996. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. Yates III, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611-615. [DOI] [PubMed] [Google Scholar]
- 61.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verma, R., R. Oania, J. Graumann, and R. J. Deshaies. 2004. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118:99-110. [DOI] [PubMed] [Google Scholar]
- 63.Walters, K. J., M. F. Kleijnen, A. M. Goh, G. Wagner, and P. M. Howley. 2002. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41:1767-1777. [DOI] [PubMed] [Google Scholar]
- 64.Wang, Q., A. M. Goh, P. M. Howley, and K. J. Walters. 2003. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 42:13529-13535. [DOI] [PubMed] [Google Scholar]
- 65.Wilkinson, C. R., M. Seeger, R. Hartmann-Petersen, M. Stone, M. Wallace, C. Semple, and C. Gordon. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3:939-943. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, Y., and W. Xiao. 2004. Pdr3 is required for DNA damage induction of MAG1 and DDI1 via a bi-directional promoter element. Nucleic Acids Res. 32:5066-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu, Y., and W. Xiao. 2001. Two alternative cell cycle checkpoint pathways differentially control DNA damage-dependent induction of MAG1 and DDI1 expression in yeast. Mol. Genet. Genomics 266:436-444. [DOI] [PubMed] [Google Scholar]