Abstract
Transcriptional activation by Gcn4p is dependent on the coactivators SWI/SNF, SAGA, and Srb Mediator, which are recruited by Gcn4p and stimulate assembly of the preinitiation complex (PIC) at the ARG1 promoter in vivo. We show that recruitment of all three coactivators is nearly simultaneous with binding of Gcn4p at ARG1 and is followed quickly by PIC formation and elongation by RNA polymerase II (Pol II) through the open reading frame. Despite the simultaneous recruitment of coactivators, rapid recruitment of SWI/SNF depends on the histone acetyltransferase (HAT) subunit of SAGA (Gcn5p), a non-HAT function of SAGA, and on Mediator. SAGA recruitment in turn is strongly stimulated by Mediator and the RSC complex. Recruitment of Mediator, by contrast, occurs independently of the other coactivators at ARG1. We confirm the roles of Mediator and SAGA in TATA binding protein (TBP) recruitment and demonstrate that all four coactivators under study enhance Pol II recruitment or promoter clearance following TBP binding. We also present evidence that SWI/SNF and SAGA stimulate transcription elongation downstream from the promoter. These functions can be limited to discrete time intervals, providing evidence for multiple stages in the induction process. Our findings reveal a program of coactivator recruitment and PIC assembly that distinguishes Gcn4p from other yeast activators studied thus far.
The activation of transcription initiation in eukaryotes involves the binding of a sequence-specific activator protein to an enhancer, or upstream activation/regulatory sequence (UAS/URS), followed by activator-dependent recruitment of the general transcription machinery. Because packaging of promoter DNA in nucleosomes impedes assembly of the preinitiation complex (PIC), activators must recruit chromatin-modifying complexes to the promoter. These include coactivator complexes with histone acetyltransferase (HAT) activity, such as the Gcn5p-containing SAGA complex of Saccharomyces cerevisiae, and ATP-dependent remodeling enzymes, such as SWI/SNF and RSC. Nucleosome remodeling by SWI/SNF has also been implicated in removing a barrier to transcription elongation by RNA polymerase II (Pol II) (8). The recruitment of TATA binding protein (TBP) and Pol II is facilitated by coactivators which serve as adaptors between the activator and these general transcription factors (GTFs). For example, SAGA complex contains subunits that interact with TBP and has been shown to mediate TBP recruitment by various activators (2, 22, 30). Similarly, the Mediator complex interacts with Pol II and has been shown to stimulate the recruitment of both Pol II and TBP (21, 25, 26, 30).
The recruitment of one coactivator may stimulate the recruitment of another, and this interdependence can be reflected in a sequential order of coactivator recruitment to the promoter. Such staged recruitment was shown for the HO gene where SWI/SNF recruitment is necessary for, and temporally precedes, recruitment of SAGA by the activator Swi5p to the URS2 element (9). SWI/SNF is also required for Mediator recruitment to HO, even though these two coactivators are recruited simultaneously to URS1, whereas Mediator recruitment is dispensable for SWI/SNF recruitment to this promoter (4). At the GAL1 gene of yeast, by contrast, SAGA recruitment by Gal4p precedes that of Mediator (6). Although SWI/SNF is recruited to GAL1 as rapidly as SAGA, and independently of SAGA function, strong SWI/SNF recruitment requires Mediator and downstream steps in PIC assembly (19, 24). Thus, the temporal order of coactivator recruitment at GAL1 does not seem to reflect an obligate sequence of coactivator functions. The requirement for SWI/SNF function as a prerequisite for SAGA recruitment by Swi5p at HO appears to be restricted to late mitosis and applies even to Gal4p- and Gcn4p-regulated promoters in this phase of the cell cycle, most likely reflecting a highly condensed state of promoter chromatin (19). In fact, both SAGA and Mediator functions are required during interphase for wild-type steady-state recruitment of SWI/SNF by Gcn4p at the target genes ARG1 and SNZ1 (43). Thus, the degree of interdependency among coactivators in recruitment can vary from one yeast activator to the next, and even for the same activator depending on the chromatin structure of the promoter.
A mechanism to explain SAGA-SWI/SNF interdependency has been proposed wherein histone acetylation by Gcn5p enhances recruitment of SWI/SNF and of SAGA itself via the bromodomains in the Snf2p and Gcn5p subunits, respectively. This mechanism was demonstrated in vitro using reconstituted nucleosome arrays, and genetic data indicated a requirement for the Snf2p bromodomain in SWI/SNF recruitment to the SUC2 promoter in vivo (15). However, a different result was obtained in another study where the Snf2p bromodomain was dispensable for wild-type SWI/SNF recruitment to SUC2, even though mutations in the HAT subunits of SAGA (Gcn5p) and NuA4 (Esa1p) decreased SWI/SNF recruitment in the early phase of SUC2 induction (13). Similarly, a deletion of GCN5 did not reduce steady-state recruitment of SWI/SNF by Gcn4p (38, 43), although it impaired chromatin remodeling by the recruited SWI/SNF complex (38). However, it has not been determined whether the kinetics of SWI/SNF (or SAGA) recruitment by Gcn4p is influenced by the Gcn5p HAT function in vivo.
A key objective of this study was to determine whether Gcn4p mounts a staged recruitment of SAGA, SWI/SNF, and Mediator, followed by PIC assembly, in which earlier events are prerequisites for later steps in the process. To that end, we analyzed the kinetics of Gcn4p binding, coactivator recruitment, binding of TBP and Pol II to the promoter (PIC assembly), and the appearance of Pol II at the 3′ end of the open reading frame (ORF) during induction of the ARG1 gene in wild-type cells and in various coactivator mutants. Our results indicate that transcriptional activation by Gcn4p is a rapid process commencing with the immediate occupancy of the ARG1 UAS by Gcn4p on amino acid starvation, followed quickly by nearly simultaneous recruitment of SAGA, SWI/SNF, and Mediator. These coactivators, together with RSC, coordinate the rapid recruitment of TBP and Pol II to the promoter and elongation through the ORF by Pol II without any detectable delay in promoter clearance. Despite the nearly simultaneous recruitment of coactivators, our mutant analyses reveal extensive interdependencies among the coactivators for their rapid recruitment by Gcn4p. In particular, we provide in vivo evidence that the SAGA HAT subunit is required for wild-type kinetics of SWI/SNF recruitment and that RSC function is needed for optimal SAGA recruitment. We also show that Mediator is strongly required for activator recruitment of both SAGA and SWI/SNF throughout the entire course of induction.
Our kinetic analyses of PIC formation in coactivator mutants show that TBP recruitment per se is not sufficient for wild-type promoter occupancy by Pol II, suggesting that all four coactivators enhance Pol II recruitment or promoter clearance after binding of TBP to the promoter. We also uncovered functions for SWI/SNF and SAGA in transcription elongation downstream of the promoter. Together, our results provide a detailed picture of the activation mechanism for Gcn4p that differs significantly from that described for other activators, and they extend the range of functions stimulated by the participating coactivators in vivo.
MATERIALS AND METHODS
Chromatin immunoprecipitation was conducted as described previously (30) using the following strains containing ADA2-myc described previously (31): HQY392 (wild type), HQY666 (snf2Δ), HQY501 (rsc2Δ), HQY573 (rox3Δ), HQY420 (gcn5Δ), HQY418 (ada1Δ), and HQY503 (gcn4Δ). These strains were derived from deletion strains purchased from Research Genetics, and the presence of all deletions was verified by PCR analysis of genomic DNA. Cells were grown in synthetic complete (SC) medium (35). Anti-myc antibodies and anti-Rpb1p antibodies (8WG16) were purchased from Santa Cruz Biotechnologies and AbCam, respectively. Antibodies against Gcn4p were produced previously (K. Natarajan and A. G. Hinnebusch, unpublished observations). Antibodies against Snf6p and TBP were kindly provided by Joseph Reese, and Gal11p antibodies were generously supplied by Mark Ptashne.
RESULTS
Rapid, sequential binding of Gcn4p, coactivators and GTFs to the ARG1 promoter in starved cells.
We used the chromatin immunoprecipitation (ChIP) assay to measure the kinetics of Gcn4p binding and recruitment of coactivators, TBP, and Pol II to the ARG1 promoter following induction of Gcn4p synthesis in a strain with a myc-tagged version of SAGA subunit Ada2p. Aliquots of the same chromatin preparations from this strain were immunoprecipitated with antibodies against Gcn4p, TBP, SWI/SNF subunit Snf6p, Pol II subunit Rpb1p, and Mediator subunit Gal11p, and with myc antibodies (against myc-Ada2p) to evaluate recruitment of all of these factors to ARG1 in the same population of induced cells. (We use the terms “recruitment” and “binding” to signify occupancy, representing the net outcome of factor association and dissociation from the UAS or promoter.)
In brief, cells growing exponentially at 25°C in complete (SC) medium were treated with sulfometuron-methyl (SM) to inhibit biosynthesis of isoleucine and valine and thereby induce Gcn4p. GCN4 expression is rapidly induced at the translational level by starvation for any amino acid (16). (The cells were grown at 25°C to reduce the rate of Gcn4p binding and factor recruitment, because at 30°C we observed nearly identical rates of recruitment for all factors under study [data not shown]). Aliquots of the culture were collected at time intervals ranging from 2.5 min to 120 min postinduction and treated with formaldehyde to cross-link factors to chromatin in the living cells. An aliquot of cells was removed prior to SM addition and cross-linked to provide a noninduced control sample, and an isogenic gcn4Δ ADA2-myc strain was cross-linked after 120 min of SM treatment to evaluate the background level of Gcn4p-independent factor binding at ARG1. The DNA extracted from immunoprecipitated chromatin fragments was subjected to PCR analysis with various primers to quantify the UAS or TATA sequences in the promoter or sequences at the 3′ end of the ARG1 ORF (Fig. 1A). Immunoprecipitations were conducted on at least two chromatin samples prepared from replicate cultures, and the PCR analysis of coprecipitated DNA sequences was conducted in duplicate or triplicate for each immunoprecipitated sample.
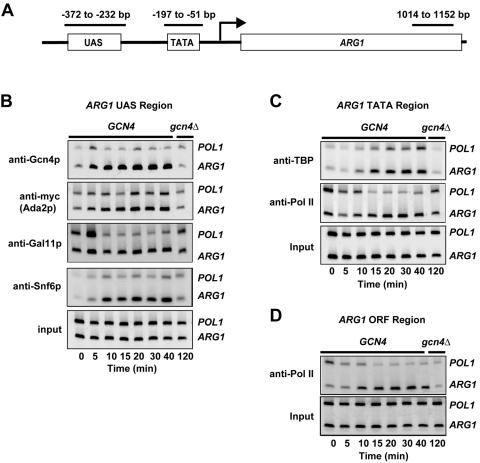
FIG. 1.
ChIP analysis of the kinetics of Gcn4p binding and recruitment of coactivators, TBP, and Pol II to ARG1 on induction of Gcn4p by SM. GCN4 ADA2-myc strain HQY392 was cultured at 25°C in SC medium and treated with SM at 0.5 μg/ml. Aliquots of cells were cross-linked with formaldehyde at the indicated times and processed for ChIP analysis of factor binding to ARG1 using primers to amplify the UAS- or TATA-containing sequences, or ORF sequences, at ARG1 indicated in the schematic shown in panel A. Coding sequences of the POL1 gene were amplified to control for nonspecific immunoprecipitation. Chromatin fragments were immunoprecipitated with the antibodies shown to the right of each panel, and the DNA was extracted from the immunoprecipitates (IP) and from 5% of the corresponding input chromatin (Inp) samples. A 1,000-fold dilution of the Inp and the undiluted IP DNA samples were PCR amplified in the presence of [33P]dATP, and the PCR products were resolved by polyacrylamide gel electrophoresis and visualized by autoradiography. In parallel, gcn4Δ ADA2-myc strain HQY503 was treated with SM for 120 min and handled identically (last lane). The PCR-amplified fragments were quantified with a phosphorimager, and the ratios of the ARG1 signals to the POL1 signals in the IP samples were normalized for the corresponding ratios for the Inp samples to yield the “relative % IP ARG1/POL1” for each sample.
Representative data showing the kinetics of Gcn4p binding and the recruitment of Mediator, SAGA, SWI/SNF, TBP, and Pol II subunits to ARG1 over the first 40 min of induction by SM are presented in Fig. 1B to D. The fraction of the total ARG1 sequences in the input chromatin sample that was precipitated with each antibody was calculated and normalized for the fraction of coprecipitating POL1 ORF sequences in order to correct for nonspecific precipitation of chromatin fragments. The mean values and standard errors for these normalized ratios, referred to as relative % IP ARG1/POL1 values, were calculated from the multiple PCR measurements for each time point and plotted in the graphs presented in Fig. 2. The relative % IP ARG1/POL1 values can be equated with the relative occupancy of the factor under consideration at ARG1.
FIG. 2.
Kinetic ChIP analysis suggests sequential binding of Gcn4p, coactivators, and TBP/Pol II at the ARG1 promoter on induction of Gcn4p. The results of replicate ChIP experiments conducted as exactly as described in Fig. 1 were quantified and the mean “relative % IP ARG1/POL1” values with standard errors were plotted as a function of time for each factor, indicated along the y axes. Note that different y axis scales were employed to normalize the peak heights for the different curves.
As shown below, ChIP signals of greatly different magnitude were obtained for the various factors binding at ARG1. It is unclear whether these differences reflect real distinctions in factor occupancy, different efficiencies of cross-linking to chromatin, or dissimilar efficiencies of immunoprecipitation. Thus, it could be misleading to compare the absolute rates of increase in promoter occupancy from one factor to the next. Hence, we used different scales to plot the data in Fig. 2 to normalize the peak occupancy values for the set of factors under consideration. To compare the kinetics of factor recruitment, we evaluated the times required to reach the half-maximal and maximum occupancies for each factor during the first 40 min of induction.
We observed a low level of Gcn4p binding to the ARG1 UAS in noninduced cells at a level ∼2.5-fold above the background signal measured in the gcn4Δ strain (Fig. 1B, GCN4 at 0 min versus gcn4Δ, and 2A, 0 min). Following treatment with SM, there was an immediate increase in Gcn4p binding to the ARG1 UAS that peaked by 15 to 20 min at a level ∼8-fold above the noninduced value (Fig. 2A). The level of Gcn4p binding declined somewhat at later time points, reaching a steady-state level ∼5-fold higher than the noninduced value by 120 min (see Fig. 3A for time points between 40 and 120 min). The results of Western analysis (data not shown) revealed a rapid increase in the steady-state level of Gcn4p that paralleled the increase in Gcn4p binding to the ARG1 promoter observed in ChIP assays for the first 30 to 40 min of induction. This rapid induction of Gcn4p is in accord with the fact that GCN4 expression is controlled at the translational level. However, Western analysis did not reveal any decline in Gcn4p levels at later time points that would coincide with the decrease in Gcn4p occupancy at ARG1 observed in ChIP assays between 30 min and 2 h of induction (see Fig. 3A). This decline in Gcn4p binding at ARG1 may reflect a decrease in nuclear import, decreased affinity of Gcn4p for the UAS, or degradation of Gcn4p at the promoter.
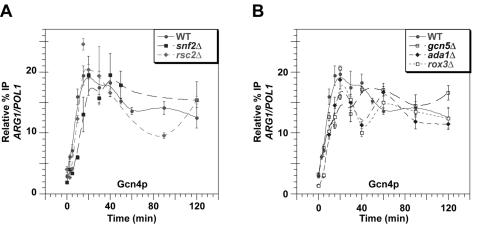
FIG. 3.
Coactivator mutations do not substantially alter the kinetics or extent of Gcn4p binding at the ARG1 UAS. Kinetic ChIP analysis was conducted as described for Fig. 1 using the wild-type (WT) and mutant strains with the relevant genotypes listed in the inset of each graph. Antibodies against Gcn4p were employed for the ChIP assays using primers to measure its binding to the ARG1 UAS.
To determine the rate of PIC assembly following Gcn4p binding to the UAS, we analyzed the recruitment of TBP and Pol II to the region surrounding the TATA element at ARG1 (Fig. 2B). (The relative % IP ARG1/POL1 values in gcn4Δ cells were close to unity for these and other factors under study, indicating promoter occupancy below the detection limit of the assay in the absence of activator Gcn4p.) The increase in TBP occupancy required ∼10 min to reach its maximum rate and also peaked ∼10 min later than did Gcn4p binding. Consistent with this, half-maximal binding of TBP occurred at ∼15 min, about 8 min later than that of Gcn4p. The recruitment of Pol II to the TATA region followed kinetics similar to that seen for TBP binding to the promoter in showing a relatively slow initial rate of binding before reaching its maximum rate by ∼10 min and peaking 10 min later than did Gen4p (Fig. 2B). Thus, there is no detectable delay between the recruitment of TBP and Pol II to the ARG1 promoter by Gcn4p. (Note that the ChIP signals for Pol II binding to the TATA region may be a composite of Pol II bound to the promoter and Pol II transcribing the 5′ end of the ORF.)
Recent experiments indicate that levels of Pol II occupancy within the ORF coincide closely with the amounts of accumulated mRNA transcripts produced from the same gene (20, 42). Therefore, we measured binding of Pol II to the 3′ end of the ARG1 ORF to evaluate the increase in rate of transcription elongation during induction by Gcn4p. The results shown in Fig. 2C indicate that Pol II occupancy at the 3′ end of the ARG1 ORF increased with kinetics similar to that observed for binding of TBP and Pol II to the promoter (Fig. 2A and B), requiring ∼10 min to reach its maximum rate of binding and peaking about 12 min after Gcn4p reached its highest value. (We observed no significant binding of Gcn4p or TBP to the 3′ end of the ARG1 ORF [data not shown], indicating that the Pol II signal measured for this location did not include Pol II molecules bound to the promoter, as might occur with inadequate shearing of chromatin.) Under physiological conditions, mRNA synthesis occurs at a rate of 1,200 to 1,500 nucleotides/min (17), such that transcription of the ARG1 ORF should require less than a minute. The fact that Pol II occupancy at the 3′ end of the ORF showed no lag with respect to PIC assembly implies that promoter clearance occurs almost instantaneously following PIC assembly and that transcription proceeds at the expected rate with no significant impediments to elongation by Pol II under inducing conditions at ARG1. Bryant and Ptashne made a similar observation for induction of the GAL1 promoter by Gal4p (6). The fact that Pol II occupancy in the ORF was ∼3-fold higher than at the promoter (cf. Fig. 2B and C) implies that multiple elongating polymerase molecules can be associated with ORF fragments of ∼500 bp (the average size of our sheared chromatin fragments) whereas only a single Pol II molecule at a time can occupy the promoter.
As would be expected from the rapid rate of PIC formation and transcription elongation induced by Gcn4p binding at ARG1, recruitment of the coactivators SAGA, Mediator, and SWI/SNF to the UAS occurred very quickly following induction of Gcn4p (Fig. 2D to F). In contrast to the results for TBP, the binding of these coactivators occurred at roughly the maximum initial rate at the earliest time point we assayed. However, their binding appeared to be biphasic, increasing rapidly for the first 15 min in parallel with the increase in Gcn4p binding, and then increasing more slowly over the next 15 to 25 min to reach their peak values at 30 min (SWI/SNF) or 40 min (SAGA) (Fig. 2D and E) (see Fig. 4A and C for the entire 120-min time course of SAGA and SWI/SNF recruitment). Despite these biphasic kinetics, both SAGA and SWI/SNF achieved half-maximal binding at ∼10 min, four to five min preceding the half-maximal binding of TBP and Pol II. Recruitment of Mediator also began quickly and increased rapidly with apparently monophasic kinetics, peaking ∼5 min after Gcn4p and ∼6 min preceding maximum TBP binding (Fig. 2F). Like SAGA and SWI/SNF, the half-maximal binding of Mediator occurred at ∼10 min. Together, these findings suggest that the first phase of SWI/SNF and SAGA recruitment and the monophasic recruitment of Mediator occur very quickly after binding of Gcn4p to the UAS and may precede the increase in PIC formation at the promoter by a short time interval of 5 to 10 min. Additional evidence supporting the notion that coactivator recruitment precedes PIC assembly is discussed below.
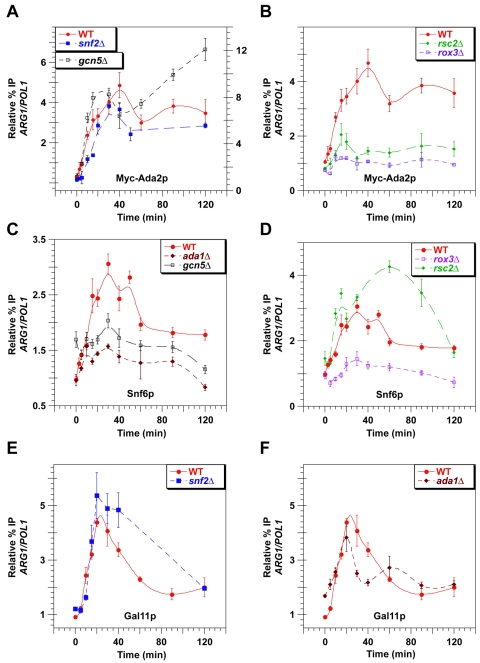
FIG. 4.
Coactivator mutations alter the kinetics and extent of SWI/SNF and SAGA recruitment by Gcn4p at the ARG1 UAS. Kinetic ChIP analysis was conducted as described in Fig. 1 using the wild-type (WT) and mutant strains with the relevant genotypes listed in the inset of each graph. Antibodies against myc-tagged Ada2p (A and B), Snf6p (C and D), or Gal11p (E and F) were employed for the ChIP assays using primers to measure binding of these factors to the ARG1 UAS. In panel A, myc-Ada2p binding in WT and snf2Δ cells is indicated by the scale along the left-hand y axis, while the results for gcn5Δ refer to the scale on the right-hand y axis.
Mediator and RSC, but not SWI/SNF or Gcn5p, stimulate SAGA recruitment by Gcn4p.
Having observed almost simultaneous recruitment of SAGA, SWI/SNF, and Mediator at ARG1, we next asked whether these coactivators are interdependent for their rapid recruitment by Gcn4p. For example, wild-type recruitment of SAGA may depend on the simultaneous recruitment of SWI/SNF or Mediator in addition to the known direct interaction of SAGA with the Gcn4p activation domain (11). In this event, mutations in subunits of Mediator or SWI/SNF would impair the rate or extent of SAGA recruitment. Accordingly, we measured the kinetics of SAGA recruitment in a rox3Δ mutant, lacking a critical subunit of Mediator, or in a snf2Δ mutant, lacking the ATPase subunit of SWI/SNF. We also examined the effects of deleting the HAT subunit of SAGA or disrupting the SAGA complex in gcn5Δ and ada1Δ mutants, respectively (14), on recruitment of SWI/SNF, Mediator, and SAGA itself. Because the RSC complex is recruited by Gcn4p (37), we further investigated whether deletion of the RSC2 subunit affects SAGA recruitment. Rsc2p is a nonessential RSC subunit containing two bromodomains (7), shown previously to be required for transcriptional induction of ARG1 by Gcn4p (30).
First, we examined the effects of these coactivator mutations on the kinetics of Gcn4p binding to the ARG1 UAS. As shown in Fig. 3A and B, binding of Gcn4p in the five coactivator mutants occurred at similar rates and peaked within 5 min of the peak time observed in the isogenic wild-type strain. Moreover, none of the mutations significantly reduced the steady-state level of Gcn4p binding observed at 120 min (Fig. 3A and B), in agreement with our previous findings (30). The small variations in Gcn4p binding observed in the mutant strains cannot account for the major defects in coactivator recruitment or PIC formation described for these mutants below.
When we analyzed the mutants for SAGA recruitment, we found that the rox3Δ and rsc2Δ mutations led to severe reductions in SAGA recruitment throughout the entire time course, whereas snf2Δ produced only a slight reduction in SAGA recruitment (Fig. 4A and B). Thus, even though Gcn4p can directly interact with SAGA, it requires functions of Mediator and RSC to achieve the maximal rate and extent of SAGA recruitment at ARG1. It was shown in vitro that acetylation of histone H3 by Gcn5p stimulates SAGA binding to promoter nucleosomes via the bromodomain in Gcn5p (15). However, we found that neither the rate nor extent of SAGA recruitment was significantly reduced by deletion of GCN5. In fact, SAGA recruitment was substantially higher than wild type in the gcn5Δ strain at all time points (note the difference in scale for gcn5Δ in Fig. 4A). We conclude that recruitment of SAGA by Gcn4p is stimulated by Mediator and RSC but independent of both SWI/SNF remodeling function and histone acetylation by SAGA throughout the whole time course of induction.
Mediator and both Gcn5p and a non-HAT function of SAGA are required for rapid recruitment of SWI/SNF.
In contrast to the results obtained for SAGA, deletion of GCN5 had a significant effect on SWI/SNF recruitment by Gcn4p between ∼15 min and 120 min of induction (Fig. 4C). As deletion of GCN5 does not eliminate any other subunits from the SAGA complex (41), it is likely that the loss of SAGA HAT activity in gcn5Δ cells impairs SWI/SNF recruitment in this mutant. Disruption of SAGA by ada1Δ produced an even stronger defect in SWI/SNF recruitment, extending over a wider time interval than did gcn5Δ (Fig. 4C). Thus, both the HAT subunit (disrupted by gcn5Δ) and a non-HAT function of SAGA (disrupted by ada1Δ) contribute to SWI/SNF recruitment by Gcn4p. The results in Fig. 4D on the rox3Δ mutant show that SWI/SNF recruitment is also strongly dependent on Mediator throughout the entire time course of induction. Interestingly, in rsc2Δ cells, SWI/SNF was recruited more rapidly, and to a greater extent, than in RSC2 cells during the first 100 min of induction (Fig. 4D). Thus, SWI/SNF recruitment is not dependent, and may be antagonized, by RSC during the early and middle stages of ARG1 induction by Gcn4p.
The results described above indicate that Mediator is critically required for recruitment of both SAGA and SWI/SNF by Gcn4p. By contrast, we found that inactivation of SWI/SNF by snf2Δ led to somewhat greater-than-wild-type levels of Mediator recruitment during the initial stages of induction and had no effect by 120 min (Fig. 4E). Disruption of SAGA by ada1Δ had only a modest effect on the kinetics of Mediator recruitment that was limited to the interval between 20 to 30 min of induction (Fig. 4F). Thus, both the rate and extent of Mediator recruitment by Gcn4p to ARG1 are largely independent of both SAGA and SWI/SNF at ARG1.
Coactivators enhance rapid PIC formation and promoter clearance or elongation.
Having observed that Gcn4p recruits SWI/SNF, SAGA, and Mediator prior to PIC formation, we wished to examine the effects of mutations in these coactivators on the kinetics of PIC assembly and transcription elongation. The snf2Δ mutation had little effect on the kinetics or extent of TBP recruitment (Fig. 5A), but it produced a marked reduction in Pol II binding to the promoter region between 40 to 80 min of induction (Fig. 5B) and reduced Pol II occupancy in the 3′ end of the ORF throughout the whole time course of induction (Fig. 5C). The fact that Pol II occupancy in the 3′ end of the ORF was reduced in snf2Δ cells during the first 30 min of induction (Fig. 5C) without any decrease in the promoter occupancy of TBP and Pol II (Fig. 5A and B) strongly suggests that inactivation of SWI/SNF function impairs promoter clearance or elongation by Pol II. Because we cannot distinguish between defects in promoter clearance or elongation with this assay, we will use the term elongation to refer to all steps between the end of PIC assembly and transcription termination.
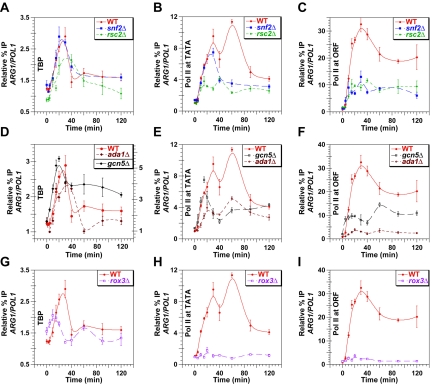
FIG. 5.
Coactivator mutations alter the kinetics and extent of TBP and Pol II occupancy of the promoter and of Pol II occupancy in the ORF at ARG1. Kinetic ChIP analysis was conducted as described in Fig. 1 using the wild-type (WT) and mutant strains with the relevant genotypes listed in the inset of each graph. Antibodies against TBP (A, D, and G) or Pol II subunit Rpb1p (B, C, E, F, H, and I) were employed for the ChIP assays using the appropriate primers to measure binding of TBP and Pol II to the promoter (A, B, D, E, G, and H) or Pol II occupancy in the ORF (C, F, and I). In panel D, TBP binding in WT and ada1Δ cells is indicated by the scale along the left-hand y axis, while the results for gcn5Δ refer to the scale on the righthand y-axis.
As noted above, Pol II occupancy of the promoter was impaired in snf2Δ cells between 40 to 80 min of induction without any decrease in TBP recruitment (cf. Fig. 5A and B). Thus, the reduced Pol II occupancy in the promoter cannot be a secondary effect of impaired TBP binding to the TATA element. One explanation for this finding could be that SWI/SNF stimulates Pol II recruitment to the promoter by a mechanism downstream of TBP binding. However, elongating Pol II molecules bound at the 5′ end of the ORF may contribute to Pol II occupancy in the promoter. Accordingly, SWI/SNF might stimulate promoter clearance instead if we assume that the stalled Pol II molecules dissociate from the promoter. Supporting this last assumption, kin28 mutations, which reduce phosphorylation of serine-5 in the C-terminal domain (CTD) of Pol II by TFIIH and thereby impair promoter clearance, lead to reduced ChIP signals for Pol II in the promoters of yeast genes (27, 33). Hence, it is possible that snf2Δ impairs promoter clearance rather than Pol II recruitment following TBP binding between 40 and 80 min of induction.
Deletion of GCN5 produced no reduction in TBP recruitment but led to a strong decrease in Pol II occupancy in the 3′ end of the ORF throughout most of the time course of induction. In fact, TBP recruitment occurred at higher-than-wild-type levels in the gcn5Δ mutant (note the different scales in Fig. 5D). Similar to snf2Δ, the gcn5Δ mutation reduced Pol II occupancy in the ORF at the 15 and 20 min time points (Fig. 5F) without producing any significant reduction in TBP or Pol II occupancy in the promoter (Fig. 5D and E). This last comparison implies a defect in transcription elongation in gcn5Δ cells early in the induction process. Between 20 and ∼80 min of induction, gcn5Δ also resembled snf2Δ in reducing Pol II occupancy of the promoter without any defect in TBP recruitment (cf. Fig. 5D and E). Thus, as concluded above for snf2Δ, elimination of the SAGA HAT subunit Gcn5p impairs either Pol II recruitment or promoter clearance between 20 and 80 min of induction. The similarity in the phenotypes of the snf2Δ and gcn5Δ mutants shown in Fig. 5 is consistent with the fact that GCN5 is required for optimal SWI/SNF recruitment (Fig. 4C) and for the remodeling function of recruited SWI/SNF (38).
Similar to our findings on snf2Δ and gcn5Δ, the ada1Δ mutation reduced Pol II occupancy in the ORF much more than it reduced Pol II occupancy in the promoter during the whole time course of induction, consistent with a defect in elongation (cf. Fig. 5E and F). Moreover, ada1Δ produced a greater reduction in the promoter occupancy of Pol II versus TBP between 20 and 40 min of induction (cf. Fig. 5D and E). Thus, ada1Δ additionally impairs Pol II recruitment or promoter clearance downstream of TBP binding to the TATA element. The fact that ada1Δ and gcn5Δ displayed the same defects at steps following TBP recruitment is consistent with the fact that Gcn5p is a component of SAGA and ada1Δ disrupts SAGA function. However, whereas gcn5Δ leads to higher-than-wild-type TBP recruitment, ada1Δ reduced TBP binding at the promoter. This difference is consistent with previous conclusions that a non-HAT function of SAGA is instrumental in TBP recruitment (2, 22, 30). It also helps to explain why ada1Δ produced a stronger reduction in ARG1 transcription than did gcn5Δ, nearly eliminating detectable association of Pol II with the ORF (Fig. 5F). Note also that ada1Δ more strongly impairs recruitment of SWI/SNF than does gcn5Δ (Fig. 4C).
The rsc2Δ mutation resembles the other mutations discussed thus far in producing marked reductions in Pol II promoter occupancy (Fig. 5B) and transcription elongation (Fig. 5C). However, rsc2Δ led to an earlier decline in Pol II promoter occupancy than did snf2Δ or gcn5Δ, evident by 10 to 15 min of induction, and thus more closely resembles ada1Δ in this respect. The rsc2Δ mutant also more closely resembled ada1Δ in producing a moderate reduction in TBP recruitment throughout much of the time course (Fig. 5A), which may contribute to the low-level promoter occupancy of Pol II in this mutant (Fig. 5B). However, the strong decrease in Pol II promoter occupancy at 40 and 60 min in rsc2Δ cells cannot be explained by decreased TBP recruitment at these time points (cf. Fig. 5A and B), leading us to propose that rsc2Δ additionally impairs Pol II recruitment or promoter clearance following TBP binding. It is noteworthy that ada1Δ and rsc2Δ led to similar reductions in TBP and Pol II occupancy in the promoter (cf. Fig. 5A and B and 5D and E) but that ada1Δ produced a much stronger decrease in Pol II occupancy in the ORF (cf. Fig. 5C and F). This comparison suggests that ada1Δ impairs transcription elongation to a greater extent than does rsc2Δ.
The rox3Δ mutation in Mediator led to significant reductions in TBP binding to the promoter beginning after ∼15 min of induction and extending throughout most of the remainder of the time course (Fig. 5G). It appeared that the uninduced level of TBP binding to the promoter was elevated in this mutant. Strikingly, Pol II occupancy of both the promoter and the ORF was almost completely eliminated at all time points in rox3Δ cells (Fig. 5H and I). Thus, among the five mutations analyzed here, rox3Δ had the greatest combined effect on TBP and Pol II recruitment to the promoter (PIC formation) and on transcription elongation. Because TBP binding occurs at wild-type or higher levels in the first 15 min of induction, but Pol II recruitment is strongly defective during this interval, it follows that Mediator also stimulates Pol II recruitment or promoter clearance downstream of TBP binding (cf. Fig. 5G and H). Given the strong defect in PIC formation in the rox3Δ mutant, it is not possible to determine whether the severe reduction in Pol II occupancy of the ORF in this mutant (Fig. 5I) additionally involves a defect in elongation. The severe phenotype of rox3Δ shown in Fig. 5 is consistent with the fact that this mutation impairs recruitment of both SWI/SNF and SAGA (Fig. 4A to D) in addition to impairing Mediator function.
DISCUSSION
Simultaneous, but interdependent, recruitment of SWI/SNF, SAGA, and Mediator by Gcn4p to ARG1.
Our kinetic analysis of wild-type cells indicates that recruitment of the three coactivators SWI/SNF, SAGA, and Mediator to the ARG1 UAS occurs almost simultaneously with Gcn4p binding. Recruitment of SAGA and SWI/SNF by Gcn4p begins instantaneously on induction of Gcn4p but proceeds more slowly and, thus, requires 10 to 20 min longer to reach the maximum levels than does binding of Gcn4p. Although recruitment of Mediator may begin more slowly, it reaches its maximum value sooner than does SAGA and SWI/SNF, peaking only about 5 min later than Gcn4p.
The binding of TBP and Pol II at the promoter increased at a relatively slow rate during the first 5 to 10 min of induction, such that both factors attained their half-maximal values ∼5 to 10 min later than did the three coactivators. Similar kinetics were observed for the appearance of Pol II at the 3′ end of the ARG1 ORF, indicating no detectable lag between PIC assembly and transcription elongation. It may seem contradictory that no recruitment of TBP was detected until 10 min of induction while small increases in Pol II occupancy of the promoter and ORF were reproducibly detected 5 min earlier (Fig. 1C and D and 2B and C). This may be attributed to the fact that ChIP signals were considerably lower for TBP versus Pol II (Fig. 2B), so that low-level binding of TBP at 5 min was below the detection limit of the assay.
Although recruitment of the coactivators, Pol II, and TBP all showed significant increases by 5 to 10 min after induction of Gcn4p, it appeared that TBP and Pol II required 5 to 10 min longer than the coactivators to reach their maximum rates and peak values of binding at ARG1. These kinetic data suggest that recruitment of coactivators and the stimulation of PIC formation by Gcn4p occur in two distinct stages, as depicted schematically in Fig. 6 (stages I and II). This conclusion is supported by our findings that SAGA and Mediator are required for wild-type PIC formation and transcription at the earliest time points of induction. Although SWI/SNF is not required for PIC formation during the first 20 min, it is necessary for optimum transcription rates at the earliest time points. Importantly, we also showed recently (31) that recruitment of SWI/SNF, SAGA, and Mediator are unaffected by deletion of the TATA element at ARG1, whereas PIC assembly is severely impaired by this promoter mutation (30). Thus, taken together, our data strongly suggest that recruitment of SWI/SNF, SAGA, and Mediator precedes PIC assembly at the ARG1 promoter during induction by Gcn4p.
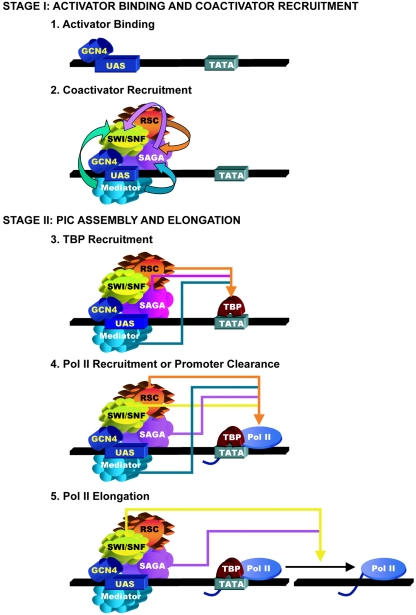
FIG. 6.
A model for simultaneous interdependent recruitment of SAGA, SWI/SNF, and Mediator by Gcn4p and multiple functions of these coactivators in PIC assembly and elongation. Transcriptional activation is divided into the two stages that were temporally resolved in wild-type cells. The top two panels depict the first stage in transcriptional activation involving binding of Gcn4p to the UAS and the nearly simultaneous recruitment of SAGA, SWI/SNF, and Mediator. Even though binding of Gcn4p and recruitment of SAGA and SWI/SNF commence immediately on induction of Gcn4p synthesis, binding of Gcn4p is depicted as the first step because mutational inactivation of these coactivators (or of Mediator) does not affect Gcn4p binding to the UAS. Although recruitment of RSC is also depicted here, the kinetics of its recruitment have not been determined. Interdependencies among the coactivators are summarized using arrows color coded for the stimulatory factor. The bottom three panels depict the second stage in activation involving TBP recruitment, Pol II recruitment or promoter clearance downstream of TBP binding, and elongation. While these three steps were not temporally resolved, genetic analysis reveals that Pol II recruitment is dependent on TBP binding to the TATA element and that the coactivators independently stimulate these reactions in the manner depicted by the color-coded arrows. See text for further details.
We believe that Gcn4p binding to the UAS is the first event in transcriptional activation of ARG1. Even though the recruitment of SAGA and SWI/SNF begins immediately on induction of GCN4 translation, Gcn4p is the first factor to reach its maximum level of binding. Furthermore, none of the coactivators can be recruited to ARG1 in gcn4Δ cells (37) (Fig. 1), whereas mutational impairment of the coactivators did not significantly alter the rate or extent of Gcn4p binding to the UAS (Fig. 3). Thus, we propose that Gcn4p binding to the UAS precedes the recruitment of these coactivators at ARG1 (Fig. 6, step 1).
The fact that recruitment of the three coactivators occurs almost simultaneously could be taken as an indication that Gcn4p independently recruits each coactivator. Indeed, recombinant Gcn4p can bind specifically to all three coactivators in vitro (11, 23, 29, 39). Consistent with this idea, we found that inactivating SWI/SNF had only a slight impact on the kinetics and end point of SAGA recruitment and did not affect the rate of Mediator recruitment to ARG1. Similarly, Mediator recruitment was nearly unaffected by disrupting SAGA integrity with the ada1Δ mutation. Because Mediator recruitment to ARG1 is independent of both SWI/SNF and SAGA, the interaction of Mediator with the Gcn4p activation domain may be sufficient for its wild-type recruitment to the UAS at ARG1 (Fig. 6, step 2).
In contrast to the independent recruitment of Mediator, the recruitment of both SWI/SNF and SAGA by Gcn4p is critically dependent on Mediator (Fig. 6, step 2), being severely impaired in the rox3Δ mutant at the earliest time points. This last conclusion may seem at odds with our finding that recruitment of Mediator seemed to lag slightly behind that of SWI/SNF (and possibly SAGA) during the first few minutes of induction in wild-type cells (Fig. 2D to F). However, the basal level of Mediator recruitment provided by the uninduced level of Gcn4p may support the earliest phase of SWI/SNF and SAGA recruitment by the Gcn4p that is induced by amino acid starvation.
We recently compared the effects of coactivator mutations on the steady-state recruitment of SWI/SNF, SAGA, and Mediator at three different Gcn4p target genes, ARG4, SNZ1, and ARG1. Recruitment of SAGA and SWI/SNF after 2 h of starvation was found to be strongly dependent on Mediator at all three genes, in accordance with the findings presented here for ARG1. Thus, the importance of Mediator in recruitment of SWI/SNF and SAGA may be a general attribute of Gcn4p. However, the ability of Gcn4p to recruit Mediator independently of SAGA observed at ARG1 (in both studies) did not extend to ARG4 and SNZ1 (31). Hence, it is possible that SAGA and Mediator are mutually interdependent for their recruitment by Gcn4p at many target genes and that some feature of the ARG1 promoter, or another transcription factor that binds at ARG1, allows for SAGA-independent recruitment of Mediator by Gcn4p at this gene.
Our findings that Mediator has an important function in promoting SAGA recruitment by Gcn4p differs from previous results obtained for Gal4p. No reduction in SAGA recruitment to GAL1 was produced by mutations in Mediator subunits srb4-ts (3) or gal11Δ (6), even though srb4-ts abolishes PIC formation at GAL1 (26). A critical requirement for Mediator in SWI/SNF recruitment by Gcn4p is also distinctive, in that recruitment of SWI/SNF to the HO gene by Swi5p seems to be independent of Mediator (4). Although SWI/SNF recruitment by Gal4p is dependent on Mediator, it additionally requires PIC formation at GAL1 (24), whereas we found that SWI/SNF recruitment is independent of the TATA box and PIC formation (31). Thus, Mediator likely plays a more direct role in promoting SWI/SNF recruitment by Gcn4p than for Gal4p.
Recruitment of SWI/SNF by Gcn4p is dependent on SAGA in addition to Mediator (Fig. 6, step 2), and our finding that SWI/SNF recruitment was impaired in gcn5Δ cells has important implications for the histone code hypothesis (36). Previous results of in vitro experiments indicated that histone acetylation by SAGA enhances recruitment of SAGA and SWI/SNF via the bromodomains in Gcn5p and Snf2p (15). However, recruitment of SWI/SNF in vivo by the activators Swi5p (9) and Gal4p is independent of Gcn5p HAT function and the integrity of SAGA (24). Moreover, deletion of Gcn5p reduced SWI/SNF recruitment only in a narrow time window during SUC2 induction (13). In contrast to these findings, we found that gcn5Δ impairs SWI/SNF recruitment by Gcn4p during most of the induction period, but especially at the time of maximum SWI/SNF recruitment. This result provides the strongest evidence to date that histone acetylation by Gcn5p enhances association of SWI/SNF with a UAS element in vivo.
The reason why we previously failed to notice the dependence on Gcn5p for steady-state SWI/SNF recruitment (43) may be that Gcn4p was being overexpressed in our former study. The greater-than-wild-type level of Gcn4p binding to the UAS may have suppressed the reduction in SWI/SNF recruitment that was observed here at 120 min of induction in gcn5Δ cells with native levels of Gcn4p. Furthermore, the fact that gcn5Δ produced no decrease in SWI/SNF recruitment to a synthetic PHO5 promoter containing Gcn4p binding sites (38) suggests that the stimulatory effect of histone acetylation on SWI/SNF recruitment can also vary with the chromatin structure of the promoter.
Our finding that SAGA recruitment was not diminished by gcn5Δ (Fig. 3C) seems at odds with the idea that the Gcn5p bromodomain enhances recruitment of SAGA to acetylated promoter histones in vitro (15). Similarly, Syntichaki et al. observed that deletion of the Gcn5p bromodomain did not diminish Gcn5p-dependent histone acetylation at the synthetic UASGcn4-PHO5 promoter (38). Thus, there is no direct evidence that SAGA recruitment is enhanced by binding of the Gcn5p bromodomain to acetylated histone tails in vivo. In fact, our finding that SAGA recruitment was elevated in gcn5Δ cells raises the possibility that H3 acetylation is required for release of SAGA from the UAS during induction by Gcn4p.
Comparing the effects of ada1Δ and gcn5Δ mutations on the kinetics of SWI/SNF recruitment indicates that a function of SAGA apart from Gcn5p HAT activity is required for optimal SWI/SNF recruitment, consistent with our earlier observations concerning steady-state induction by Gcn4p (43). It is currently unknown how SAGA supports SWI/SNF recruitment by Gcn4p independently of histone acetylation by Gcn5p. In the case of Gal4p and the activator responsible for RNR3 induction, the recruitment of SWI/SNF is dependent on PIC assembly (24, 34). However, because SWI/SNF recruitment to ARG1 is unaffected by deletion of the TATA element (31), we conclude that stimulating PIC formation does not represent the non-HAT SAGA function that enhances SWI/SNF recruitment by Gcn4p. Furthermore, because ada1Δ produced only a small reduction in Mediator recruitment to ARG1, it seems unlikely that the decrease in SWI/SNF recruitment at this gene is an indirect consequence of impaired Mediator recruitment in ada1Δ cells.
Based on our results with the snf2Δ mutant, we surmised that chromatin remodeling is dispensable for SAGA recruitment. However, surprisingly, high-level SAGA recruitment by Gcn4p is dependent on RSC (Fig. 6, step 2). To our knowledge, this is the first evidence that chromatin remodeling by RSC stimulates the recruitment of a coactivator. Presumably, a basal amount of RSC recruited by the uninduced level of Gcn4p contributes to the immediate, Rsc2p-dependent increase in SAGA recruitment that occurs on induction of Gcn4p. Interestingly, rsc2Δ led to elevated SWI/SNF recruitment, suggesting that these two chromatin remodeling factors compete for binding sites at ARG1. Given that Rsc2p and Snf2p both contain bromodomains, rsc2Δ may eliminate competition between RSC and SWI/SNF for binding to acetylated nucleosomes as a means of increasing SWI/SNF recruitment.
The chromatin remodeling activity of SWI/SNF is dispensable for recruitment of both Mediator and SAGA to ARG1 over the time course of induction. Similar observations have been made for the ARG4 and SNZ1 genes at steady state (31). In fact, we found that snf2Δ impairs only Pol II recruitment and elongation at ARG1. These findings stand in sharp contrast to the requirement for SWI/SNF in recruitment of both SAGA (9) and Mediator to the HO gene by Swi5p (4). As noted above, this requirement seems to reflect the condensed state of chromatin during mitosis and also applies to Gcn4p and Gal4p during mitosis (19).
Functions of coactivators in Pol II recruitment and elongation.
A key function of coactivators is to promote PIC assembly. Our kinetic analysis of coactivator mutants indicates that RSC, SAGA and Mediator all stimulate recruitment of TBP to the promoter (Fig. 6, step 3), as the rsc2Δ, ada1Δ, and rox3Δ mutations produced quantitative reductions in TBP binding to the TATA element at various times during the induction of ARG1. These findings are in general agreement with our previous analysis of steady-state TBP binding to the ARG1 promoter (30). However, the stronger reductions in TBP binding observed in that study may be attributable to the fact that we analyzed strains containing a myc13 tag on the C terminus of TBP, which slightly impairs TBP function and may exacerbate the effects of coactivator mutations on TBP recruitment.
Even though we cannot temporally resolve the recruitment of TBP and Pol II to the promoter in wild-type cells, we believe that Pol II recruitment follows TBP binding (Fig. 6, steps 3 and 4) because deletion of the TATA element nearly abolished Pol II recruitment at ARG1 (30). Therefore, any coactivator mutations that impair TBP recruitment by Gcn4p should indirectly reduce Pol II recruitment. In accordance with this prediction, the rsc2Δ, ada1Δ, and rox3Δ mutations all reduced promoter occupancy by Pol II at ARG1. However, these three mutations, along with snf2Δ and gcn5Δ, reduced Pol II recruitment during certain phases of ARG1 induction where there was little or no defect in TBP binding. One interpretation of these last findings is that SWI/SNF, RSC, SAGA, and Mediator promote Pol II recruitment by a mechanism distinct from a stimulatory effect on TBP binding (Fig. 6, step 4). This uncoupling of Pol II recruitment from TBP binding was particularly obvious for the snf2Δ and gcn5Δ mutants in which TBP recruitment occurred at wild-type or higher levels throughout the time course, while Pol II promoter occupancy was impaired during the later stages of induction. We made a similar finding previously for the srb10Δ mutation, which decreased steady-state promoter occupancy by Pol II at ARG1, ARG4, and SNZ1 without reducing TBP recruitment by Gcn4p (30).
In vitro studies by Hahn and colleagues indicate that following initiation of transcription most of the general factors and Mediator remain bound to the promoter in a Scaffold complex, which facilitates the efficient recruitment of Pol II, TFIIB, and TFIIF in subsequent rounds of transcription (32, 44). Because TBP is a component of the Scaffold, the functions of coactivators in recruiting Pol II to the Scaffold would necessarily be downstream of TBP recruitment. Thus, the defects in Pol II occupancy that occur without reductions in TBP binding at ARG1 could reflect requirements for Mediator, SAGA, SWI/SNF, or RSC in reinitiation of transcription by the Scaffold complex in vivo.
An alternative interpretation for this phenotype, however, is a defect in promoter clearance following PIC assembly. While it might be expected that a failure to make the transition from initiation to elongation would lead to accumulation of Pol II at the promoter, the kin28-ts16 mutation in the CTD kinase activity of TFIIH strongly reduces Pol II promoter occupancy without affecting TBP binding (27, 33). This result may indicate that Pol II stalled at the promoter in kin28 cells dissociates rapidly; alternatively, it is possible that most of the ChIP signal for Pol II at promoters derives from early elongation complexes which are eliminated by kin28-ts16 (27). In any event, because the coactivator mutations we analyzed produce the same phenotype described for kin28-ts16 cells, they may impair promoter clearance rather than Pol II recruitment (Fig. 6, step 4).
The physical association between Mediator and Pol II could underlie an adaptor mechanism for direct recruitment of Pol II or provide a means of stimulating promoter clearance via CTD phosphorylation (28). Alternatively, Mediator could act indirectly in these functions by stimulating recruitment of SAGA or SWI/SNF. An adaptor function for SAGA in Pol II recruitment is possible, but most previous work describes its interactions with TBP (12, 22). The similar effects of gcn5Δ and snf2Δ, and the fact that Gcn5p promotes SWI/SNF recruitment (this study) and remodeling function (38), suggest that the SAGA HAT may indirectly stimulate Pol II recruitment or promoter clearance by enhancing SWI/SNF activity. Nucleosome remodeling of the promoter downstream of the TATA element by SWI/SNF (or RSC) could enhance Pol II recruitment or its escape from the promoter.
Finally, our kinetic analysis of coactivator mutants has uncovered functions for SWI/SNF and SAGA in stimulating elongation by Pol II (Fig. 6, step 5). Thus, we found that snf2Δ and gcn5Δ produced a strong reduction in Pol II occupancy in the 3′ end of the ORF early in the induction period (∼20 min) at a time when TBP and Pol II were present at the promoter at nearly wild-type levels. In addition, because ada1Δ produced a much stronger reduction in transcription than did rsc2Δ, despite comparable reductions in Pol II promoter occupancy, it appears that a non-HAT function of SAGA is also required for elongation. It might be expected that defects in elongation would lead to a queuing of Pol II molecules in the beginning of the ORF and a higher-than-wild-type Pol II occupancy in the promoter, which were not observed. It is possible, however, that stalled Pol II molecules are cleared, e.g., by elongation factor TFIIS (Dst1p), or that the predicted accumulation of Pol II in the promoter is offset by reductions in Pol II recruitment produced by the coactivator mutations.
It was shown previously that activators can stimulate elongation as well as initiation (1, 5). In the case of mammalian activator HSF1, there is strong circumstantial evidence that nucleosome remodeling by SWI/SNF, recruited by HSF1, is instrumental in overcoming an elongation barrier in the 5′ end of the hsp70 gene (reference 8 and references therein). Our findings on Gcn4p provide the first direct evidence that SWI/SNF and SAGA are required for a wild-type efficiency of elongation in vivo. These results are consistent with genetic interactions in yeast between TFIIS and subunits of SWI/SNF (10) and with genetic and physical interactions between SAGA subunit Spt8p and TFIIS (40). Moreover, the recruitment of SWI/SNF by Gcn4p to plasmid-borne HIS3 produces a labile chromatin domain that extends from the promoter into the coding sequences (18). Thus, chromatin remodeling of the ORF by SWI/SNF may account for its stimulatory role in elongation at ARG1. Enhancing the recruitment or remodeling function of SWI/SNF could, in turn, explain the positive role of SAGA in elongation apart from the Spt8p-TFIIS interaction.
In summary, our findings reveal a strong dependence of Gcn4p on Mediator for recruitment of SAGA and SWI/SNF, on Gcn5p and a non-HAT function of SAGA in SWI/SNF recruitment, and on RSC for recruitment of SAGA to ARG1. This network of interdependencies (Fig. 6, step 2) represents one of the most extensive set of cooperative interactions governing coactivator recruitment described thus far, and it differs in important respects from that described for other yeast activators. The kinetic analysis of PIC assembly and transcription elongation in the coactivator mutants has allowed us to implicate SAGA, SWI/SNF, RSC, and Mediator in stimulating Pol II recruitment or promoter clearance downstream of TBP binding, and we uncovered functions for SAGA and SWI/SNF in stimulating Pol II elongation apart from their known activities in PIC assembly.
Acknowledgments
We are very grateful to Joe Reese, Mark Ptashne, and Krishnamurthy Natarajan for generous gifts of antibodies.
REFERENCES
- 1.Akhtar, A., G. Faye, and D. L. Bentley. 1996. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 15:4654-4664. [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blau, J., H. Xiao, S. McCracken, P. O'Hare, J. Greenblatt, and D. Bentley. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16:2044-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11:1301-1309. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT Hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 8.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 17:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 10.Davie, J. K., and C. M. Kane. 2000. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Mol. Cell. Biol. 20:5960-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drysdale, C. M., B. M. Jackson, R. McVeigh, E. R. Klebanow, Y. Bai, T. Kokubo, M. Swanson, Y. Nakatani, P. A. Weil, and A. G. Hinnebusch. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 13.Geng, F., and B. C. Laurent. 2004. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 23:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 15.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52:248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izban, M. G., and D. S. Luse. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267:13647-13655. [PubMed] [Google Scholar]
- 18.Kim, Y., and D. J. Clark. 2002. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. USA 99:15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 20.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 22.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y. J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemieux, K., and L. Gaudreau. 2004. Targeting of Swi/Snf to the yeast GAL1 UAS(G) requires the Mediator, TAF(II)s, and RNA polymerase II. EMBO J. 23:4040-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605-609. [DOI] [PubMed] [Google Scholar]
- 26.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 27.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 30.Qiu, H., C. Hu, S. Yoon, K. Natarajan, M. Swanson, and A. G. Hinnebusch. 2004. An array of coactivators is required for optimal recruitment of TBP and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24:4104-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu, H., C. Hu, F. Zhang, G. Hwang, M. Swanson, C. Boonchird, and A. G. Hinnebusch. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25:3461-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves, W. M., and S. Hahn. 2003. Activator-independent functions of the yeast mediator Sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 23:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma, V. M., B. Li, and J. C. Reese. 2003. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 17:502-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman, F., G. R. Fink, and C. W. Lawrence. 1974. Methods of yeast genetics, p. 61-64. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 37.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S.-J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain coordinates nucleosome remodeling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 39.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 40.Wery, M., E. Shematorova, B. Van Driessche, J. Vandenhaute, P. Thuriaux, and V. Van Mullem. 2004. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J. 23:4232-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, P. Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22:5367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon, S., H. Qiu, M. J. Swanson, and A. G. Hinnebusch. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB Mediator, and SAGA. Mol. Cell. Biol. 23:8829-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]