Abstract
The enzymatic moieties of anthrax toxin enter the cytosol of mammalian cells via a pore in the endosomal membrane formed by the protective antigen (PA) moiety. Pore formation involves an acidic pH-induced conformational rearrangement of a heptameric precursor (the prepore), in which the seven 2β2-2β3 loops interact to generate a 14-strand transmembrane β-barrel. To investigate this model in vivo, we labeled PA with the fluorophore 7-nitrobenz-2-oxa-1,3-diazole (NBD) at cysteine residues introduced into the 2β2-2β3 loop. Each labeled PA was bound to CHO cells, and NBD fluorescence was monitored over time in stirred cell suspensions or by confocal microscopy. A strong increase was observed with NBD at positions 305, 307, 309, and 311, sites where side chains are predicted to face the bilayer, and little change was seen at residues 304, 306, 308, 310, and 312, sites where side chains are predicted to face the pore lumen. The increase at position 305 was inhibited by membrane-restricted quenchers, low temperature, or various reagents known to affect toxin action. Of the 24 NBD attachment sites examined, all but three gave results qualitatively consistent with the β-barrel model. Besides supporting the β-barrel model of membrane insertion, our results describe the time course of insertion and identify PA residues where NBD gives a strong signal upon membrane insertion in vivo.
The pathogenesis of Bacillus anthracis depends on two major virulence factors: an antiphagocytic poly(d-glutamic acid) capsule and a toxin that is believed to be responsible for the major symptoms of anthrax (8, 12). Anthrax toxin belongs to the binary bacterial toxins, a class of intracellularly acting toxins in which the enzymatic (A) and receptor-binding (B) proteins are released from the bacteria as separate nontoxic proteins. The A and B proteins then combine to form toxic noncovalent complexes on receptor-bearing mammalian cells. Anthrax toxin has two separate enzymatic A moieties: edema factor (EF; 89 kDa), a calmodulin- and Ca2+-dependent adenylate cyclase (13, 29), and lethal factor (LF; 90 kDa), a Zn2+-dependent protease that cleaves most mitogen-activated protein kinase kinases (9, 30). EF and LF bind competitively to an activated form of protective antigen (PA; 83 kDa), the receptor-binding and pore-forming B moiety of anthrax toxin. PA then delivers EF and LF to the cytosol, where they modify their substrates.
The interaction of PA, LF, and EF occurs after PA binds to either of two receptors at the cell surface (7, 24) and is proteolytically activated (20). A cellular protease of the furin family cleaves receptor-bound PA, removing an N-terminal 20-kDa piece (PA20) and leaving a 63-kDa piece (PA63) bound to the receptor. Once freed from PA20, PA63 spontaneously self-associates to form a ring-shaped heptamer (also called the prepore) (17). PA63 is capable of permeabilizing cells to Rb+ and Na+ under acidic conditions (16) and can form pores (channels) in planar phospholipid bilayers even in the absence of receptor (5). Oligomerization of receptor-bound PA63 at the cell surface generates high-affinity sites for EF and LF (19), and these enzymatic moieties bind competitively to the sites via their homologous N-terminal domains (18). Oligomerization of PA63 also triggers its association with cholesterol-containing lipid microdomains (2) and promotes receptor mediated endocytosis and trafficking to an endosomal compartment (10). There, acidic conditions induce a conformational change in the prepore that enables it to form a transmembrane pore. The pore then serves as a passageway for EF and LF to cross to the cytosol (33, 34). In support of this model, lysosomotropic agents and bafilomycin A1 block toxin action in cell culture (10, 11).
The crystal structure of PA suggested a mechanism for pore formation, centering on a mobile loop of domain 2, the 2β2-2β3 loop (residues 303 to 322) (22). The seven 2β2-2β3 loops of the heptamer were proposed to move to the base of the structure during a pH-dependent conformational rearrangement and to interact there, forming a 14-strand β-barrel spanning the membrane (Fig. 1A). This model was suggested by the structure of the heptameric pore formed by the α-toxin of Staphylococcus aureus (27) and by consonance of the sequence of the 2β2-2β3 loop with an amphipathic β-barrel (22). Studies in planar phospholipid bilayers provided support for the model (4). The effects of the thiol-specific reagent methanethiosulfonate ethyltrimethylammonium (MTS-ET) on the conductance of pores formed by PA mutants with cysteine residues substituted at various positions in the 2β2-2β3 loop corresponded closely to those predicted by the β-barrel model (Fig. 1B). Extensive inhibition of conductance was seen at positions where the side chain is predicted to contact the lumen of the pore, and little or no inhibition was found at positions where the side chain would contact the membrane bilayer.
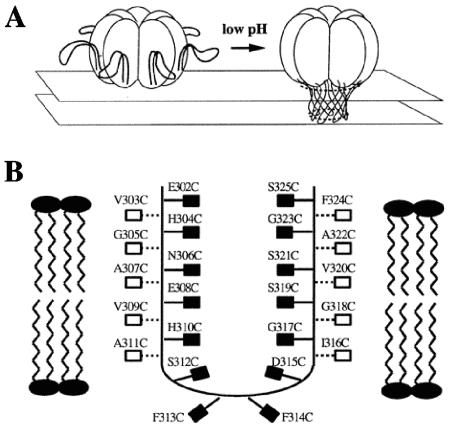
FIG. 1.
Model of acidification-induced pore formation by PA63. (A) After the pH is lowered, the 2β2-2β3 loops move to the base of the heptamer and combine to form a 14-stranded transmembrane β-barrel. (B) Hypothetical orientation of the 2β2-2β3 loop in the membrane, showing the sites of cysteine substitutions. This figure is adapted from Fig. 1 and 6 in Benson et al. (4). The filled boxes indicate residues that are expected to be in contact with the hydrophobic core of the membrane, while the open boxes represent residues that are expected to face the lumen of the pore.
While planar bilayer systems are clearly valuable in analyzing pore structure and function, such systems incompletely replicate the complex environment in vivo. For example, PA was the only protein in the MTS-ET experiments; thus, the membrane lacked a PA receptor and perhaps other functionally relevant proteins. Also, the artificial bilayer was prepared with a single purified lipid and therefore differed in composition from native membranes. In the present study, we characterized pore formation by PA in cultured cells using an attached fluorescent dye as probe.
MATERIALS AND METHODS
Cell lines and reagents.
The CHO-K1 and CHO-R1 ATR cell lines were grown at 37°C and under 5% CO2 in 150-cm2 cell culture flasks containing F-12 medium with 10% fetal bovine serum. CHO-R1 ATR800, which showed a 10-fold-higher expression level of ATR/TEM8 than wild-type CHO-K1 cells, was a generous gift from John Young (Salk Institute). All the reagents were obtained from Sigma (St. Louis, MO) unless otherwise specified.
Preparation of NBD-labeled PA-Cys mutants.
Cys substitution PA mutants from an earlier study (21) were expressed in BL21STAR(DE3) Escherichia coli using pET22b (Invitrogen, CA), which directs the expressed protein to the periplasm (31). Growth and expression of the mutants were carried out in a 5L BIOFLOW 200 fermentor (New Brunswick Scientific, N.J.). ECPM1 medium was used to grow cells to 5 A600 units. Cultures were sparged with air and induced at 30°C with isopropyl-1-thio-β-d-galactopyranoside. PA was purified from periplasmic extracts by Q-Sepharose and Mono-Q anion-exchange chromatography (Amersham Biosciences, NJ). Five milligrams of Cys-substituted PA was reduced with 20 mM dithiothreitol on ice for 20 min. Dithiothreitol was removed by size exclusion chromatography on a Sephadex G-50 column equilibrated in 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; pH 8.0), 50 mM sodium acetate, 150 mM NaCl, and the product was concentrated to 5 mg/ml using a 10-kDa-cutoff Vivaspin concentrator (Vivascience, Germany). IANBD [N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol)ethylenediamine; Molecular Probes, OR] was then added at a 20-fold molar excess, and the mixture was incubated for 2 h at room temperature. The reaction was quenched by adding 10 mM 2-mercaptoethanol, and the free dye was removed by size exclusion chromatography (G-50 Sephadex). Fractions containing the NBD-labeled protein were pooled and stored at −80°C. The labeling efficiency was estimated by absorbance spectrophotometry (ɛ478 = 25,000 M−1 cm−1 for NBD, and ɛ280 = 75,670 M−1 cm−1 for PA83). The measured labeling efficiency for all the mutants was 95 to 105%.
Purified NBD-labeled PA mutants were tested for the ability to mediate the inhibition of protein synthesis by LFNDTA, a fusion of the PA-binding domain of LF (LFN) with the catalytic domain of diphtheria toxin (15). Most of the labeled proteins exhibited a 30 to 50% reduction in activity, relative to wild-type PA. Exceptions were proteins labeled at residues 302, 322, and 323, which showed larger reductions.
Time-lapse intensity measurements of NBD emission.
Cells were grown in 150-cm2 tissue culture flasks, washed once, and scraped into a universal buffer (10 mM HEPES, 10 mM sodium acetate, 10 MES [morpholinepropanesulfonic acid], 150 mM NaCl, 2 mM CaCl2, 11 mM glucose, 50 mg/liter bovine serum albumin, at pH 8.0 and 4°C). The cells were pelleted at 500 × g, washed with buffer, suspended at 1 A600 unit, and kept on ice for no longer than 1 h. Cell viability was measured by trypan blue exclusion. The cells were incubated with 50 nM NBD-labeled PA on ice for 1 h at 0.05 A600 unit (final cell concentration) in a 2-ml final volume. They were then washed twice in buffer at 4°C by centrifugation and resuspension and finally were resuspended in 2 ml of cold buffer. The suspension was transferred to a cuvette with a stirring bar and placed in a cuvette holder thermostated at 37°C in an ISS K2 fluorimeter (ISS, IL). The 488-nm line of an Ar+ laser was used to excite NBD, and emission was recorded at 544 nm. A 5× beam expander lens (Thorlabs, NJ) was used to increase the diameter of the laser beam, thereby exciting a larger fraction of the cells and thus increasing the signal-to-noise ratio. In addition, a long-pass 510-nm filter (Omega Optical, VT) was placed before the photomultiplier tube to reduce background scatter of the excitation beam. The fold increase of intensity on cells was calculated as (It/I0) − 1, where It is the intensity at time t and I0 is the initial intensity (see Table S1 in the supplemental material). The maximal attainable fluorescence increase in a nonpolar environment was estimated by diluting 10 μl of NBD-labeled PA into 2 ml of dimethyl sulfoxide (DMSO), giving a final concentration of 50 nM. The ratio of the NBD fold fluorescence increase on cells to that in DMSO was then calculated (see Table S2 in the supplemental material).
Treatment with inhibitors and quenchers.
In experiments in which cells were pretreated with reagents, the same reagent concentrations were maintained throughout washing steps. Cells were incubated at 37°C with 10 mM β-methyl cyclodextrin for 1 h, 50 mM cytochalasin D for 35 min, or 1 μM bafilomycin A1 for 35 min before G305C NBD-labeled PA was added. For the dominant negative inhibitor (DNI) treatment, NBD-labeled PA was mixed 1:1 with DNI, and the mixture was incubated with cells on ice to allow binding. Cells were incubated with 2 mg/ml of an equimolar mixture of 5-doxyl and 12-doxyl stearic acid for 1 h at room temperature before G305C NBD-labeled PA was added.
Fluorescence microscopy.
CHO cells were plated in a Lab-Tek chamber slide (Nalge Nunc International, IL) at 50,000 cells per well and incubated overnight. The slides were then incubated on ice, and a mixture of 1:1 PA labeled with NBD and PA labeled with Alexa 546 (Molecular Probes, OR) at K563C was added. After incubation for 1 hour on ice, the slides were transferred to a humidified CO2 incubator at 37°C. After 10, 30, or 60 min, cell samples were fixed for imaging (32) and visualized with a Zeiss confocal microscope LSM510META/NLO at a ×63 magnification. Images were captured and processed by Zeiss confocal microscope software 3.2.
RESULTS
The quantum yield of NBD, and hence its intensity of fluorescence at 544 nm, increases upon a shift from a polar to a nonpolar environment. We used this property as an indicator of membrane insertion in vivo. Native PA contains no cysteine residues, and we generated unique attachment sites for a thiol-reactive form of NBD (IANBD) by mutating selected residues to Cys (Fig. 1B) (23). After a 1-h incubation with NBD-labeled protein at 4°C, cells were washed and resuspended in cold buffer. Samples (2 ml) were placed in a spectrofluorimeter with a cuvette holder thermostated at 37°C, and fluorescence emission was monitored at 544 nm (excitation at 488 nm) while the suspension was continuously stirred with a small magnetic bar.
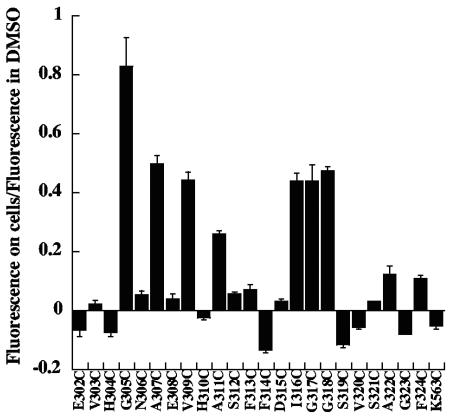
Our initial measurements focused on residues 303 to 311, corresponding to the descending, amino-proximal strand of the putative β-barrel (Fig. 1B). With NBD covalently linked to Cys at any of the odd-numbered residues in this region (except 303), there was a gradual increase in fluorescence intensity lasting for more than an hour (Fig. 2 and 3). In contrast, little or no increase in NBD fluorescence intensity was observed at the even-numbered residues tested (304, 306, 308, 310, and 312) or at an irrelevant site (residue 563) in a different domain (domain 3) (Fig. 3). The results in Fig. 3 have been normalized to correct for minor differences in the maximal fluorescence measured when the labeled proteins were placed in DMSO; but such corrections did not change the qualitative picture. Except for position 303, these findings are consistent with the β-barrel model, given that only the side chains of the odd-numbered residues in this strand are predicted to be in contact with the hydrophobic core of the membrane.
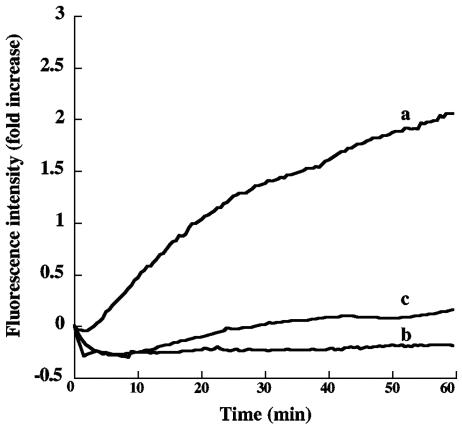
FIG. 2.
Fluorescence intensity as a function of time with cell-bound NBD-labeled PA. CHO-R1 ATR800 cells were incubated on ice with PA labeled with NBD at Cys305 or Cys306 and were then washed in the cold and suspended in cold medium. Cell suspensions (2 ml) were placed in the fluorimeter at time zero. The cuvette chamber was thermostated at 37°C, and the fluorescence intensity was recorded at 544 nm. (a) Cell-bound NBD-labeled PA G305C; (b) NBD-labeled PA G305C in solution; (c) cell-bound NBD-labeled PA N306C.
FIG. 3.
Relative change in fluorescence of cell-bound PA with NBD attached at various sites within the 2β2-2β3 loop. CHO-R1 ATR800 cells were incubated with the various labeled forms of PA and processed as described in the legend to Fig. 2. The bars represent the ratio between the fluorescence intensity of NBD on cells at 1 h and the fluorescence intensity of NBD in organic solvent (DMSO) for each mutant. Error bars represent standard deviations.
The results presented in Fig. 2 and 3 were obtained with PA G305C-NBD in CHO-R1 ATR 800 cells, a cell line overexpressing the ATR/TEM8 receptor for PA. A similar pattern was seen with parental CHO-K1 cells, but the smaller number of receptors gave a lower signal-to-noise ratio. We therefore used the CHO-R1 ATR800 cells for most measurements but confirmed key findings on the parental cell line. In all cell-based experiments we observed a slight decline in NBD fluorescence over the first few minutes after samples were placed in the fluorimeter. Similar initial intensity decreases occurred with NBD-labeled PA in buffer (Fig. 2), indicating that the initial decline in cells is an effect of temperature on the quantum yield of the fluorophore (28).
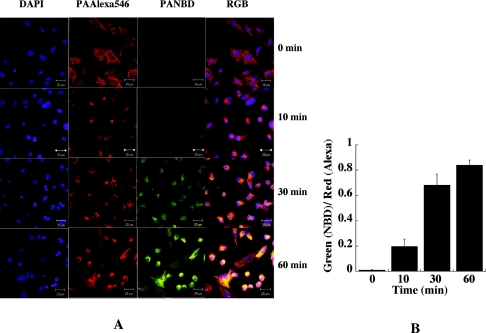
We used confocal microscopy as an independent method to monitor the fluorescence of NBD at selected sites on PA. Figure 4 shows results from an experiment with PA labeled with NBD at position 305; similar results (not shown) were obtained with a sample labeled at position 307. An equimolar mixture of NBD-labeled G305-PA and PA labeled with Alexa 546 at K563C was added to CHOR1 ATR800 cells plated on a glass slide. The Alexa-labeled PA served as a control and internal standard. Residue 563 is located on the external face of domain 3, and we have found that mutation and derivatization at this site do not affect PA function. After incubation on ice for an hour, the cells were rinsed and transferred to a humidified 37°C CO2 incubator. Samples were withdrawn at intervals over the next hour, and the cells were fixed and processed for examination by confocal microscopy. The intensities of NBD emission and Alexa 546 emission were monitored. As shown in Fig. 4A, almost no NBD fluorescence was seen at time zero, but a signal was clearly evident at 10 min and became progressively stronger at 30 min and 1 h (lane 3). In contrast, the intensity of Alexa 546 fluorescence at K563C remained constant over this time period (lane 2). DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) was used to stain the nuclei of the cells (lane 1). Simultaneous monitoring in the red, green, and blue channels of the confocal microscope revealed strong colocalization of NBD and Alexa 546 (lane 4). Figure 4B shows the ratio of NBD fluorescence (green; environment dependent) to Alexa 546 (red; environment independent) with time.
FIG. 4.
Increase in fluorescence of cell-bound NBD-labeled PA G305C with time as recorded by confocal microscopy. (A) CHO-R1 ATR800 cells were plated overnight, placed on ice, and incubated with a 1:1 mixture of NBD-labeled PA G305C and Alexa 546-labeled PA K563C. After an hour, the cells were transferred to a humidified 37°C CO2 incubator, and samples were removed at 10 min, 30 min, and 60 min and processed for imaging. Alexa 546-labeled PA was used as an internal reference, and DAPI was used to stain the nuclei of the cells tested. (B) Ratio of NBD fluorescence (green; environment dependent) to fluorescence of Alexa 546 (red; environment independent).
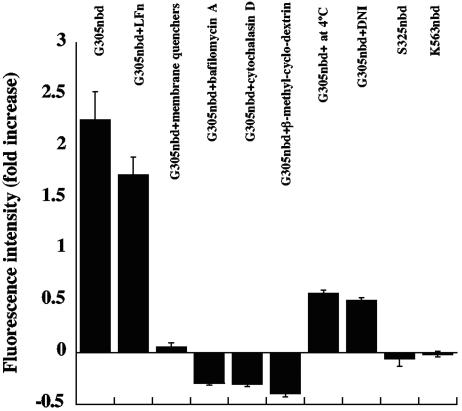
Additional validation of our system came from tests with inhibitors and other reagents that perturbed toxin action. Figure 5 shows results obtained with PA labeled at position 305; similar results were obtained with the label at position 307. (i) A mixture of the membrane-soluble quenchers 5- and 12-doxyl stearic acids (14) quenched the NBD fluorescence, supporting the notion that the increase was in fact due to membrane insertion. (ii) Bafilomycin A1, which inhibits acidification of the endosome, prevented the increase in NBD fluorescence when cells were pretreated with the inhibitor for 35 min at 37°C (6). (iii) Pretreating cells with cytochalasin D had a similar inhibitory effect, consistent with earlier results showing that this inhibitor of actin polymerization prevents endocytosis of PA (11). (iv) β-Methyl-cyclodextrin, a drug that depletes the plasma membrane of cholesterol and interferes with lipid rafts, also blocked the increase in NBD fluorescence (2). (v) The fluorescence increase was slowed at 4°C, presumably due to inhibition of endocytosis, and perhaps also to slowing of self-assembly and membrane insertion (17). (vi) Addition of a dominant-negative form of PA (DNI) that coassembles with wild-type PA and inhibits pore formation and subsequent translocation was found to significantly reduce the rise in NBD fluorescence intensity (25). The DNI used in these experiments is a double mutant of PA (K397D, D425K). (vii) Addition of excess LFN, the N-terminal PA63-binding domain of LF, slightly inhibited the increase in fluorescence. All of these results were obtained with fluorescence measurements in stirred cell suspensions. Consistent results were obtained when the effects of bafilomycin A1, cytochalasin D, and DNI were examined by confocal microscopy.
FIG. 5.
Effects of various reagents on the increase in fluorescence of NBD-labeled PA G305C. Cells were preincubated with 10 mM β-methyl-cyclodextrin for 1 h, 50 mM cytochalasin D for 35 min, or 1 μM bafilomycin A1 for 35 min at 37°C. For the experiment involving membrane quenchers, cells were preincubated with an equimolar mixture of 5-doxyl and 12-doxyl stearic acid for 1 h at room temperature. For the DNI experiment, NBD-labeled PA G305C was mixed with an equivalent amount of DNI before addition to cells. After each pretreatment, cells were incubated on ice for 1 h with the NBD-labeled PA. After washing, the cells were monitored for fluorescence intensity as described in Fig. 2. In one sample, LFN was added to a final concentration of 5 μM immediately after the sample was placed in the fluorimeter. Each bar represents the ratio of fluorescence at 1 h to that at time zero. NBD-labeled K563, a residue on the cap of the PA heptamer, was included as a control. Error bars represent standard deviations.
To extend coverage of the putative β-barrel formed by PA, we examined PA labeled with NBD at positions 313 to 325, corresponding to the interstrand loop and the ascending β-strand (Fig. 3). In residues 313 to 315, the putative interstrand loop, no significant fluorescence increase was seen. This implies solvent accessibility and is consistent with studies in planar bilayers showing accessibility of these residues to MTS-ET. Also, results in eight residues of the putative ascending β-strand were consistent with the earlier findings; thus, strong increases were seen at positions 316 and 318, moderate increases at positions 322 and 324, and little or no increase at positions 319, 321, and 323. Deviations from the model were seen at two sites: position 317, a putative lumen-facing residue, where a large increase in NBD fluorescence was observed, and position 320, a putative bilayer-facing residue, where a slight decrease was found. The fluorescence increase at 317 was quenched when we added the mixture of membrane-soluble quenchers described above.
DISCUSSION
It is well documented that the PA63 fragment of proteolytically activated PA is capable of forming ion-conductive pores in planar bilayers and the plasma membranes of mammalian cells (5, 16). Mutations in PA that disrupt pore formation have also been found to block intoxication (26). Acidic conditions are known to induce the heptameric PA63 prepore to undergo a conformational rearrangement to the pore, and evidence from studies in model membranes has suggested that the 2β2-2β3 loop of PA undergoes membrane insertion, generating a 14-stranded β-barrel. The experiments reported here were designed to test this model in living cells and to obtain additional information about membrane insertion of PA in vivo.
We used the environment-sensitive dye NBD as a covalently attached probe of membrane insertion. Fluorescent probes have the advantage of high sensitivity, and in proteins lacking native cysteines, one can couple a thiol-selective probe to an introduced cysteine, thereby enabling the environment of specified locations within the protein to be monitored. A potential disadvantage of any form of chemical derivatization is the possibility that the probe may alter a function of the protein. While fluorescence lifetime is often used as an environmentally sensitive parameter of NBD, the fluorescence intensity was not sufficiently strong in our experiments with stirred cell suspensions to obtain accurate fluorescence lifetime measurements. Instead, we used the intensity of NBD fluorescence as an inverse measure of the polarity of the dye's microenvironment.
This approach proved useful when we added the NBD-labeled monomeric forms of PA to cells and allowed proteolytic activation of the PA and oligomerization of the PA63 fragment to occur in vivo. In live cells we did not have to activate PA proteolytically or lower the pH, two necessary steps in artificial membrane systems. At four locations within the putative ascending β-strand—the odd-numbered residues 305, 307, 309, and 311—we observed a steady increase in fluorescence continuing over an hour. This increase was documented both in stirred cell suspensions and by confocal microscopy. At alternate, even-numbered locations in this strand—residues 304, 306, 308, 310, and 312—there was little or no increase. This pattern of strikingly different effects in alternating residues is similar to that seen in conductance measurements in planar bilayers (4). It is most readily explained by the hypothesis that NBD at the odd-number residues faces the nonpolar environment of a membrane, whereas this dye at the even-numbered residues remains solvent exposed. The findings are thus consistent with a β-barrel exposed to nonpolar and polar conditions, respectively, on opposite faces.
This interpretation is strongly supported by the effects observed with the membrane-restricted quenchers 5- and 12-doxyl stearic acids and with a dominant-negative form of PA, which co-oligomerizes with wild-type PA and inhibits membrane insertion. Also consistent with the β-barrel model, and specifically with the results in planar bilayers (4), residues in the putative turn region showed no significant increase in fluorescence and hence remained exposed to solvent. The hypothesis that the increases in NBD fluorescence seen at position 305 and other sites reflect membrane insertion of PA63 within an acidic compartment of the cell is supported by the observation that bafilomycin A1 strongly inhibited the increase.
The overall pattern on the ascending β-strand was less clear, but the results at eight of the ten sites examined were at least qualitatively consistent with the model. We saw strong increases in NBD fluorescence at positions 316 and 318, small but significant increases at positions 322 and 324, and little or no increase at positions 319, 321, 323, and 325. However, no increase was observed at position 320, where one was expected, and a large increase was seen at position 317, where none was expected. These two deviations, and that at position 303 of the putative descending strand, could have any of several explanations. The fact that no increase was seen at position 303 could be related to the proximity of this site to the membrane-water interface. Position 317, a Gly in the native protein, is preceded by an Ile residue and followed by another Gly, and NBD at position 317 might partition anomalously into the bilayer. Why no increase in NBD fluorescence was seen at position 320 is more difficult to explain, but it might be related to an anomalous effect of NBD derivatization and/or perturbation of the microenvironment at that site by some membrane element. Addition of LFN, representing the minimal PA63-binding, translocatable domain of LF, showed only a slight inhibitory effect on the increase in fluorescence observed with NBD attached to residue 305. The effect of a ligand, such as LF, EF or LFN, on the kinetics of insertion in vivo is not readily predictable from the present data, but there is no reason to expect a major effect. LF and EF bind competitively to sites on oligomeric PA63 species and promote oligomerization of PA63 in solution under certain conditions by competitively displacing PA20. However, we have also observed inhibition of PA63-PA63 interactions by LFN under other conditions (K. Christensen, B. Krantz, and R. J. Collier, unpublished results).
Besides their implications regarding the mechanism of membrane insertion by PA63, our results have relevance to the time course of internalization and membrane insertion of toxin complexes. In the experiment shown in Fig. 1, cells with bound PA-307C-NBD suspended in cold buffer were allowed to warm to 37°C in the thermostated cuvette holder of the spectrofluorimeter. After correction for the lag due to the reduction in quantum yield of NBD with an increase in temperature, there was only a short lag, on the order of a minute at most, before the onset of the steady rise in NBD fluorescence. We interpret this rise, which continued for more than an hour, as reflecting the membrane insertion of an ever-increasing fraction of the pool of bound NBD-labeled cell-associated PA. The lack of synchrony of the membrane insertion process is not surprising, given the multiple steps that precede the insertion and the stochastic nature of those steps. Beauregard and coworkers found that endocytosis of a mutant form of PA that is resistant to proteolytic activation is slow, relative to the wild type, implying that oligomerization of PA63 induces endocytosis (3). Abrami et al. have reported that clustering of anthrax toxin receptor, either with the PA63 heptamer or with an antibody sandwich, causes its association with cholesterol- and glycosphingolipid-rich (lipid rafts) microdomains of the plasma membrane (2). This is apparently necessary and sufficient to trigger endocytosis via a clathrin-dependent pathway. Consistent with this mechanism, we found that depletion of cellular cholesterol with β-cyclodextrin inhibited insertion of NBD-labeled PA. In a recent report, Abrami et al. showed that membrane insertion by PA occurs in early endosomes, and possibly only in multivesicular regions (1). Delivery of the molecular cargo (LF in this case) to the cytosol is proposed to occur later in the endocytic pathway and to depend on back fusion of internal vesicles of multivesicular late endosomes.
In conclusion, our findings are generally consistent with the β-barrel model of membrane insertion by PA and provide information regarding the kinetics of internalization and insertion. Identification of residues 305 and 307 as sites where attachment of NBD gives an especially strong signal upon membrane insertion may be valuable in future studies.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI22021 to R.J.C. and NIH grants to T.M.R.
We are grateful to D. Borden Lacy and Roman Melnyk for helpful discussion and to Ruth-Anne Pimental for help in preparing reagents. R.J.C. holds equity in PharmAthene, Inc.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abrami, L., M. Lindsay, R. G. Parton, S. H. Leppla, and F. G. van der Goot. 2004. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J. Cell Biol. 166:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrami, L., S. Liu, P. Cosson, S. H. Leppla, and F. G. Van Der Goot. 2003. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J. Cell Biol. 160:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauregard, K. E., R. J. Collier, and J. A. Swanson. 2000. Proteolytic activation of receptor-bound anthrax protective antigen on macrophages promotes its internalization. Cell Microbiol. 2:251-258. [DOI] [PubMed] [Google Scholar]
- 4.Benson, E. L., P. D. Huynh, A. Finkelstein, and R. J. Collier. 1998. Identification of residues lining the anthrax protective antigen channel. Biochemistry 37:3941-3948. [DOI] [PubMed] [Google Scholar]
- 5.Blaustein, R. O., T. M. Koehler, R. J. Collier, and A. Finkelstein. 1989. Anthrax toxin: channel-forming activity of protective antigen in planar phospholipid bilayers. Proc. Natl. Acad. Sci. USA 86:2209-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boll, W., M. Ehrlich, R. J. Collier, and T. Kirchhausen. 2004. Effects of dynamin inactivation on pathways of anthrax toxin uptake. Eur. J. Cell Biol. 83:281-288. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, K. A., J. Mogridge, M. Mourez, R. J. Collier, and J. A. Young. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225-229. [DOI] [PubMed] [Google Scholar]
- 8.Collier, R. J., and J. A. Young. 2003. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 19:45-70. [DOI] [PubMed] [Google Scholar]
- 9.Duesbery, N. S., C. P. Webb, S. H. Leppla, V. M. Gordon, K. R. Klimpel, T. D. Copeland, N. G. Ahn, M. K. Oskarsson, K. Fukasawa, K. D. Paull, and G. F. Vande Woude. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280:734-737. [DOI] [PubMed] [Google Scholar]
- 10.Friedlander, A. M. 1986. Macrophages are sensitive to anthrax lethal toxin through an acid-dependent process. J. Biol. Chem. 261:7123-7126. [PubMed] [Google Scholar]
- 11.Gordon, V. M., S. H. Leppla, and E. L. Hewlett. 1988. Inhibitors of receptor-mediated endocytosis block the entry of Bacillus anthracis adenylate cyclase toxin but not that of Bordetella pertussis adenylate cyclase toxin. Infect. Immun. 56:1066-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leppla, S. H. 2000. Anthrax toxin, p. 445-472. In K. Aktories and I. Just (ed.), Bacterial protein toxins. Springer, Berlin, Germany.
- 13.Leppla, S. H. 1982. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl. Acad. Sci. USA 79:3162-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melton, J. A., M. W. Parker, J. Rossjohn, J. T. Buckley, and R. K. Tweten. 2004. The identification and structure of the membrane-spanning domain of the Clostridium septicum alpha toxin. J. Biol. Chem. 279:14315-14322. [DOI] [PubMed] [Google Scholar]
- 15.Milne, J. C., S. R. Blanke, P. C. Hanna, and R. J. Collier. 1995. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol. 15:661-666. [DOI] [PubMed] [Google Scholar]
- 16.Milne, J. C., and R. J. Collier. 1993. pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol. Microbiol. 10:647-653. [DOI] [PubMed] [Google Scholar]
- 17.Milne, J. C., D. Furlong, P. C. Hanna, J. S. Wall, and R. J. Collier. 1994. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J. Biol. Chem. 269:20607-20612. [PubMed] [Google Scholar]
- 18.Mogridge, J., K. Cunningham, and R. J. Collier. 2002. Stoichiometry of anthrax toxin complexes. Biochemistry 41:1079-1082. [DOI] [PubMed] [Google Scholar]
- 19.Mogridge, J., K. Cunningham, D. B. Lacy, M. Mourez, and R. J. Collier. 2002. The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proc. Natl. Acad. Sci. USA 99:7045-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molloy, S. S., P. A. Bresnahan, S. H. Leppla, K. R. Klimpel, and G. Thomas. 1992. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 267:16396-16402. [PubMed] [Google Scholar]
- 21.Mourez, M., M. Yan, D. B. Lacy, L. Dillon, L. Bentsen, A. Marpoe, C. Maurin, E. Hotze, D. Wigelsworth, R. A. Pimental, J. D. Ballard, R. J. Collier, and R. K. Tweten. 2003. Mapping dominant-negative mutations of anthrax protective antigen by scanning mutagenesis. Proc. Natl. Acad. Sci. USA 100:13803-13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petosa, C., R. J. Collier, K. R. Klimpel, S. H. Leppla, and R. C. Liddington. 1997. Crystal structure of the anthrax toxin protective antigen. Nature 385:833-838. [DOI] [PubMed] [Google Scholar]
- 23.Ramachandran, R., A. P. Heuck, R. K. Tweten, and A. E. Johnson. 2002. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat. Struct. Biol. 9:823-827. [DOI] [PubMed] [Google Scholar]
- 24.Scobie, H. M., G. J. Rainey, K. A. Bradley, and J. A. Young. 2003. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc. Natl. Acad. Sci. USA 100:5170-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellman, B. R., M. Mourez, and R. J. Collier. 2001. Dominant-negative mutants of a toxin subunit: an approach to therapy of anthrax. Science 292:695-697. [DOI] [PubMed] [Google Scholar]
- 26.Sellman, B. R., S. Nassi, and R. J. Collier. 2001. Point mutations in anthrax protective antigen that block translocation. J. Biol. Chem. 276:8371-8376. [DOI] [PubMed] [Google Scholar]
- 27.Song, L., M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J. E. Gouaux. 1996. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274:1859-1866. [DOI] [PubMed] [Google Scholar]
- 28.Udenfriend, S. 1962. Fluorescence assay in biology and medicine. Academic Press, New York, N.Y.
- 29.Ulmer, T. S., S. Soelaiman, S. Li, C. B. Klee, W. J. Tang, and A. Bax. 2003. Calcium dependence of the interaction between calmodulin and anthrax edema factor. J. Biol. Chem. 278:29261-29266. [DOI] [PubMed] [Google Scholar]
- 30.Vitale, G., R. Pellizzari, C. Recchi, G. Napolitani, M. Mock, and C. Montecucco. 1998. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages. Biochem. Biophys. Res. Commun. 248:706-711. [DOI] [PubMed] [Google Scholar]
- 31.Wigelsworth, D. J., B. A. Krantz, K. A. Christensen, D. B. Lacy, S. J. Juris, and R. J. Collier. 2004. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J. Biol. Chem. 279:23349-23356. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, L., O. Gjoerup, and T. M. Roberts. 2004. The serine/threonine kinase cyclin G-associated kinase regulates epidermal growth factor receptor signaling. Proc. Natl. Acad. Sci. USA 101:10296-10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, S., A. Finkelstein, and R. J. Collier. 2004. Evidence that translocation of anthrax toxin's lethal factor is initiated by entry of its N terminus into the protective antigen channel. Proc. Natl. Acad. Sci. USA 101:16756-16761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, S., E. Udho, Z. Wu, R. J. Collier, and A. Finkelstein. 2004. Protein translocation through anthrax toxin channels formed in planar lipid bilayers. Biophys. J. 87:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.