Abstract
Mutations in the genes encoding the BLM and WRN RecQ DNA helicases and the MRE11-RAD50-NBS1 complex lead to genome instability and cancer predisposition syndromes. The Saccharomyces cerevisiae Sgs1 RecQ helicase and the Mre11 protein, together with the Srs2 DNA helicase, prevent chromosome rearrangements and are implicated in the DNA damage checkpoint response and in DNA recombination. By searching for Srs2 physical interactors, we have identified Sgs1 and Mre11. We show that Srs2, Sgs1, and Mre11 form a large complex, likely together with yet unidentified proteins. This complex reorganizes into Srs2-Mre11 and Sgs1-Mre11 subcomplexes following DNA damage-induced activation of the Mec1 and Tel1 checkpoint kinases. The defects in subcomplex formation observed in mec1 and tel1 cells can be recapitulated in srs2-7AV mutants that are hypersensitive to intra-S DNA damage and are altered in the DNA damage-induced and Cdk1-dependent phosphorylation of Srs2. Altogether our observations indicate that Mec1- and Tel1-dependent checkpoint pathways modulate the functional interactions between Srs2, Sgs1, and Mre11 and that the Srs2 DNA helicase represents an important target of the Cdk1-mediated cellular response induced by DNA damage.
A well-characterized aspect of the DNA damage checkpoint response is the prompt activation of a set of highly conserved kinases (64, 72). The activation of these checkpoint kinases, which include ATM and ATR and their orthologs in Saccharomyces cerevisiae, Tel1 and Mec1, respectively, induces changes in the phosphorylation state of several downstream targets (82), although the biological implications of such modifications are largely unknown. Several factors involved in DNA repair have been recently identified among the targets of checkpoint kinases (64, 72). Hence, one relevant role of the checkpoint-induced phosphorylation cascade could be to directly modulate the activity of certain repair proteins implicated in the removal of the DNA lesions (8, 59).
Recent studies have shown that the checkpoint kinases, as well as their downstream repair targets, can be purified from large protein complexes (15, 63, 98). Notably, some of these factors colocalize at characteristic DNA spots, known as nuclear foci (98). The aggregation of repair proteins in nuclear foci is stimulated by DNA damage or replication stress and, at least under certain circumstances, depends on checkpoint activation and cell cycle progression, suggesting that these foci might indeed represent sites of repair and/or DNA damage signaling (6, 25, 30, 56, 61, 65). Particularly in budding yeast, double-strand break repair centers have been recently described (41, 54, 55, 57, 69).
The WRN and BLM RecQ helicases, altered in Werner's and Bloom's genome instability syndromes, also localize at DNA damage-induced nuclear foci (6, 12, 81, 97-99). RecQ proteins represent a highly conserved family of 3′ to 5′ DNA helicases that also includes Sgs1 in S. cerevisiae (34, 42). RecQ helicases have been implicated in several aspects of DNA metabolism, but the best-characterized function is the one dealing with the control of DNA recombination pathway (34).
In S. cerevisiae, the recombination phenotypes of sgs1 mutants resemble those of srs2 mutants, altered in another 3′ to 5′ DNA helicase (79), which has similarities with the bacterial UvrD/Rep helicases (2). Interestingly, deletion of both SRS2 and SGS1 causes cell lethality (28, 44, 51, 62) that can be rescued by the inhibition of the homologous recombination pathway, suggesting that loss of both helicases causes fatal recombination events (28, 44, 62).
Other genetic observations showed that SGS1 acts at a resolution step of recombination intermediates, downstream of the SRS2 gene (23, 38), which, conversely, is required to counteract recombination at an initial stage (10, 80). Recent observations also indicate that Sgs1, together with Top3, promotes the resolution of recombination-dependent structures accumulating at damaged replication forks, while Srs2 seems to be preferentially required at an initial step to prevent their accumulation (52). Altogether these observations suggest that the two helicases inhibit DNA recombination through different mechanisms. The hypothesized role of Sgs1 in resolving recombination intermediates and the one of Srs2 in preventing their formation reflects their in vitro activities: recent studies have shown, indeed, that Srs2 works not only as a DNA helicase, but also as a DNA translocase that disrupts Rad51 nucleofilaments, an initial intermediate in homologous recombination (49, 96); in contrast, Sgs1 and other RecQ helicases catalyze the branch migration of synthetic Holliday junctions (4, 14, 40) and in particular, the BLM protein, in concert with topoisomerase IIIα, dissolves double Holliday junctions to discourage crossover outcomes (100).
Studies performed in different laboratories have also linked Srs2 and RecQ helicases to the DNA damage checkpoint. In particular, Srs2 is phosphorylated in response to intra-S DNA damage in a Mec1- and Cdk1-dependent manner and somehow influences checkpoint activation during S-phase (53) and checkpoint inactivation during adaptation and recovery following double-strand break formation (95). Analogously, Sgs1 has been implicated in intra-S checkpoint activation (26) and BLM and WRN are targets of the ATM and ATR checkpoint kinases (1, 25, 43, 77). However, it should be pointed out that it is still unclear whether the putative roles of Srs2 and Sgs1 in promoting checkpoint activation indeed reflect an enzymatic activity required for the generation of checkpoint signals (RPA-single-stranded DNA complexes) or, rather, whether the mild checkpoint defects observed in srs2 and sgs1 mutants result from a nondirect event due to the unscheduled formation of Rad51 filaments that compete with RPA for the single-stranded DNA, masking the checkpoint signals (26, 52, 53).
The highly conserved Mre11 complex, defective in Nijmegen breakage syndrome and ataxia telangectasia-like disorders, is also phosphorylated in a checkpoint-dependent manner (18) and, interestingly, in human cells it colocalizes at nuclear foci with the BLM and WRN RecQ helicases (12, 25, 67).
The Mre11 complex is composed of three subunits, known as MRE11, RAD50, and NBS1 (Xrs2 in yeast); MRE11 is the core subunit of the complex that possesses both exo- and endonuclease activities, while RAD50 and the less conserved subunit NBS1 likely play regulatory roles (13, 18, 94). Extensive structural studies performed on the MRE11-RAD50 subcomplex have pointed out an interesting analogy with some of the DNA binding properties of the SMC class of proteins, supporting a model in which the Mre11 complex acts to hold together DNA molecules, such as double-strand break ends or sister chromatids (101). Recent studies have shown that the end-bridging activity of Mre11 complex is mediated by a zinc-hook motif of Rad50 subunit which is essential to tether DNA ends in vivo (35, 57).
The Mre11 complex has also been implicated in checkpoint activation since its inactivation results in checkpoint defects in both human and yeast cells (19, 33, 85, 91, 93). Interestingly, the association of the Mre11 complex with double-strand breaks is an early event (54), and it is believed that the Mre11 complex collaborates with the ATR and ATM kinases in transducing the DNA damage signal (18, 76).
The Mre11 complex also plays an essential role under untreated conditions, both in vertebrates and in yeast: in particular, mre11, rad50, and xrs2 null yeast mutants show severe growth defects, possibly due to the accumulation of gross chromosomal rearrangements (11) and breakages at particular DNA regions, such as hairpin blocks (58, 78); further, studies carried out in Xenopus cell extracts indicate that, in the absence of the Mre11 complex, double-strand breaks accumulate on replicating chromosomes, suggesting an important role for this complex in maintaining genome integrity during every S phase (16). Altogether these findings suggest that the Mre11 complex, analogously to the RecQ and Srs2 helicases, may act at different levels to prevent genome instability.
In this study, we provide the first evidence that Srs2, Sgs1, and Mre11 physically interact and are involved in the formation of a large complex. In response to specific genomic insults and to checkpoint activation, this complex reorganizes into subcomplexes in which Srs2, Sgs1, and Mre11 differentially redistribute. We also show that the damage-induced Cdk1-dependent phosphorylation of Srs2 is required for the formation of Sgs1-Mre11 and Srs2-Mre11 subcomplexes. Hence, our findings demonstrate that Srs2 modification in response to checkpoint activation represents a crucial event in controlling the response to DNA damage.
MATERIALS AND METHODS
Strains and plasmids.
The genotypes of the strains used in this study are listed in Table S1 in the supplemental material.
Deletions and/or tagging of Srs2, Sgs1, and Mre11 genes was obtained using the one-step PCR system (45). The full-length SRS2 gene was amplified by PCR as HindIII/BamHI fragment and cloned into the Ycplac22 vector to create the pGIU8 plasmid. pGIU8 was used as the template in PCR-based site-directed mutagenesis to introduce mutations in SRS2 at the seven consensus sites for the Cdk1 kinase (T604V, S698A, S879A, S938A, S893A, S950A, and S965A), producing the pSrs2-7AV plasmid, or at the conserved K residue in the ATP binding domain (K41A), producing the pSrs2-hd plasmid. These plasmids were then modified in yeast by adding the HIS3MX6 module downstream of SRS2 mutated sequences, using the one-step PCR system (45). The plasmids obtained (pGIU11 and pAN2, respectively) were then digested with HindIII and BamHI and used to transform strain CY3259, in order to create strains CY6005 and CY6002, respectively. More information about the oligonucleotides and plasmids used are available upon request.
The MATa strain for the two-hybrid screening, EGY48 (22), containing the pSH18-34 plasmid (24), was transformed with a yeast genomic DNA library (Origene DupLex-A Library, cloned into plasmid pJG4-5, kind gift from M. Giannattasio and M. Muzi-Falconi, University of Milan, Italy). The bait plasmids were obtained by cloning the full-length or N or C terminus of the SRS2 gene into the pEG202 vector (indicated in Fig. 1A), thus generating pEG202-FL4, pEG202-N5 and pEG202-C31, respectively. These bait plasmids were then used to transform the MATα strain EGY48, producing the CY5118, CY5115, and CY5114 strains. The expression of the three Srs2 baits fused with LexA was confirmed by Western blot analysis (data not shown). CY5111, CY5109, and CY5107 were obtained by transforming CY5118, CY5115, and CY5114, respectively, with the pJG4-5 vector.
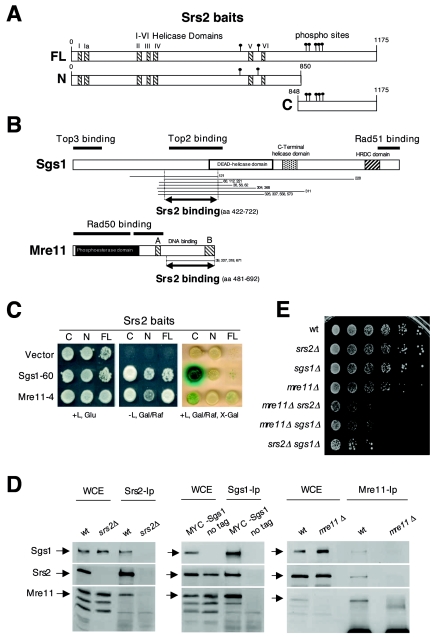
FIG. 1.
SRS2, SGS1, and MRE11 physically and genetically interact. (A) The protein regions corresponding to the three Srs2 baits used for the two-hybrid screening are shown: FL (full-length); N (N terminus); and C (C terminus). Consensus sites for Cdk1 kinase and the seven helicase domains (I and Ia to VI) are indicated by solid circles and striped boxes, respectively. (B) The protein fragments of Sgs1 and Mre11, identified by the two-hybrid analysis, are indicated by lines flanking the numbers of corresponding clones, under the schematic representation of the two proteins. Double arrowed lines indicate the Sgs1 and Mre11 protein regions involved in the interaction with Srs2. Previously mapped Sgs1 and Mre11 domains, involved in protein-protein interactions, are also shown. (C) An example of the two-hybrid interaction in haploid strains between Srs2 baits and Mre11-4 and Sgs1-60 clones is shown. The three bait strains, transformed with the corresponding prey plasmids or with pJG4-5 vector (CY5107, CY5109, CY5111, CY5448, CY5445, CY5442, CY5439, CY5436, and CY5435), were spotted on proper media with and without leucine (L), glucose (Glu), galactose (Gal), raffinose (Raf), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). (D) Srs2, Sgs1, and Mre11 proteins were immunoprecipitated from whole-cell extracts (WCE) prepared from CY3262 and CY5680 or CY2715 and CY5683, respectively, as controls and then analyzed by Western blotting using specific antibodies. WCE lanes are a 1:10 dilution of the immunoprecipitated extracts. (E) The cellular growth of wild-type (W303), srs2Δ (CY2643), sgs1Δ (CY2570), mre11Δ (CY2730), mre11Δ srs2Δ (CY5492), mre11Δ sgs1Δ (CY5670), and srs2Δ sgs1Δ (CY3137) stains was evaluated by plating serial dilutions on YPD plates.
Two-hybrid screening.
The bait strains CY5114, CY1115, and CY5118 were mixed together in equal amounts; 109 cells of this mixture were mated with 5 × 108 cells of the library strain. The diploids were selected on proper media, and the positive clones were first examined for their ability to activate the transcription of the LACZ and LEU2 reporter genes placed under the LexA promoter in the EGY48 strain as previously described (24), with some modifications. The plasmids recovered from those positives were sequenced and then used to transform each bait strain in order to confirm the interactions in haploid strains and to compare the ability of the three baits to interact with the preys. More details about the procedure used for this screening are available upon request.
Coimmunoprecipitation analysis.
Cells were lysed with liquid nitrogen in 50 mM HEPES, pH 7.9, 100 mM NaCl, 1.5 mM MgCl2, 5 mM EGTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, supplemented with protease inhibitors (Complete; Boehringer Mannheim); 1 ml of protein extracts (1 mg of total proteins) was clarified in a microcentrifuge and cleaned with 100 μl of protein G-agarose (50% suspension in lysis buffer; US Biological) for 1 h. The protein extract was then transferred in a new bovine serum albumin-saturated tube with 100 μl of protein G-agarose and 5 μg of antibodies yC-18 (Santa Cruz Biotechnology) for Srs2, 9E11 (Bio Optica) for Myc-Sgs1, or polyclonal anti-Mre11 (37) and incubated overnight at 4°C. The beads were recovered by centrifugation at 1,000 rpm, washed four times with 50 mM HEPES, pH 7.9, 150 mM NaCl, 1.5 M MgCl2, 5 mM EGTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1% Triton X-100, supplemented with protease inhibitors and finally resuspended in sodium dodecyl sulfate (SDS) loading buffer. Immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred on cellulose membranes for Western blot analysis.
Gel filtration chromatography.
Cells, grown under normal conditions or after treatment with 0.02% methyl methanesulfonate (MMS) (Sigma) for 3 h, were broken with liquid nitrogen in 50 mM HEPES pH 7.9, 100 mM NaCl, 1.5 mM MgCl2, 5 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, supplemented with a cocktail of protease inhibitors (Complete; Boehringer Mannheim); 200 μl of cleared extract (3.5 mg of total proteins) was loaded on a Superose 6 10/30 column (Amersham Pharmacia Biotech) and fractionated at 4°C using the AKTA fast protein liquid chromatography (FPLC) system (Amersham Pharmacia Biotech). The extract was eluted with the same buffer at a flow rate of 0.35 ml/min, and 40 fractions were collected after the first 5 ml had passed through the column. The fractions were precipitated with 10% trichloroacetic acid (TCA), rinsed with acetone, and resuspended in SDS loading buffer. The protein fractions were then separated by SDS-PAGE and transferred on cellulose membranes for Western blot analysis. The column was calibrated with native protein standards (gel filtration calibration kits; Amersham) according to the instructions provided by the supplier.
Western blotting analysis and immunological reagents.
The TCA protein extraction and the Western blot procedure have been previously described (53). Srs2, Sgs1, Mre11, Rad53, and B subunit were analyzed using specific monoclonal or polyclonal antibodies 12CA5 (Roche) or anti-Srs2 (yC-18, Santa Cruz Biotechnology), 9E11 (Bio Optica), anti-Mre11 (37) or 59567/3°, anti-Rad53 (kind gift from J. Diffley, Cancer Research United Kingdom, South Mimms, United Kingdom), and 6D2 (53).
RESULTS
Srs2 interacts with Sgs1 and Mre11 by two-hybrid and coimmunoprecipitation analysis.
We screened by two-hybrid analysis for Srs2 interactors using as baits the full-length protein, the N-terminal portion containing the conserved helicase domains, and the less conserved C-terminal regulatory region (79) that includes a cluster of putative phosphorylation sites for the Cdk1 kinase (Fig. 1A) (74). We used multiple baits as we were concerned that certain interacting domains might be masked using only the full-length protein. We found 800 positives out of 4 × 107 clones.
The sequence analysis of 350 prey plasmids allowed us to group them in 166 genes, 67 of which have been identified more than once (Table 1). Some of these genes have been involved in different aspects of the DNA damage response, according with the observation that Srs2 is indeed required at multiple levels to respond to DNA insults (3). Interestingly, among those 67 genes, SGS1 and MRE11 were identified 15 and 4 times, respectively. Since Sgs1 and Mre11, as well as their human orthologues, play a prominent role in DNA recombination and DNA damage checkpoint response, we decided to further characterize their interactions with Srs2.
TABLE 1.
Srs2 two-hybrid interactors
| Standard name | Systematic name | Na | Cellular component(s) | Molecular function | Biological process(es) |
|---|---|---|---|---|---|
| ADH1 | YOL086C | 3 | Cytoplasm | Alcohol dehydrogenase | Fermentation |
| ASG7 | YPL121C | 2 | Extrinsic to membrane | Unknown | Conjugation with cellular fusion |
| BNS1 | YGR230W | 3 | Unknown | Unknown | Meiosis |
| COX15 | YER141W | 3 | Mitochondrion | Oxidoreductase activity | Heme a biosynthesis, cytochrome c oxidase complex assembly |
| DUS3 | YLR401C | 3 | Cytoplasm, nucleus | tRNA dihydrouridine synthase | tRNA modification |
| ECM29 | YHL030W | 3 | Cytoplasm, nucleus | Protein binding | Protein catabolism |
| EGT2 | YNL327W | 2 | Cell wall | Cellulase activity | Cytocinesis |
| ESC1 | YMR219W | 4 | Nucleus | Unknown | Chromatin silencing at telomere |
| FAR1 | YJL157C | 1 | Cytoplasm, nucleus | Cyclin-dependent protein kinase inhibitor | Cell cycle arrest |
| FAR8 | YMR029C | 3 | Unknown | Unknown | Cell cycle arrest in response to pheromone |
| FIR1 | YER032W | 11 | Bud neck | Unknown | mRNA polyadenylylation |
| FMP50 | YKR027W | 4 | Mitochondrion | Unknown | Unknown |
| FRT2 | YAL028W | 8 | Endoplasmic reticulum | Unknown | Response to stress |
| FZO1 | YBR179C | 2 | Mitochondrion | GTPase | Mitochondrion organization and biogenesis |
| GAT2 | YMR136W | 10 | Nucleus | Transcription factor | Transcription |
| GDS1 | YOR355W | 2 | Cytoplasm, nucleus, mitochondrion | Unknown | Aerobic respiration |
| HEM14 | YER014W | 2 | Mitochondrion | Protoporphyrinogen oxidase | Heme biosynthesis |
| HEX3 | YDL013W | 17 | Nucleus | DNA binding | DNA recombination, DNA damage response, sporulation |
| IFH1 | YLR223C | 2 | Nucleus | Transcription factor | Chromatin silencing at telomere, rRNA processing |
| IKI3 | YLR384C | 2 | Nucleus | Histone acetyltransferase, transcription factor | Regulation of transcription from polymerase II promoter |
| IKS1 | YJL057C | 2 | Unknown | Unknown | Unknown |
| IME4 | YGL192W | 2 | Unknown | mRNA methyltransferase | Meiosis |
| KAPI14 | YGL241W | 2 | Cytoplasm, nucleus | Protein carrier | Protein-nucleus import |
| KIN3 | YAR018C | 2 | Unknown | Protein kinase | Chromosome segregation |
| KNS1 | YLL019C | 10 | Unknown | Protein Ser/Thr and Tyr kinase | Protein phosphorylation |
| KSP1 | YHR082C | 3 | Nucleus | Protein Ser/Thr kinase | Protein phosphorylation |
| KTR2 | YKR061W | 2 | Golgi apparatus | Cell wall mannoprotein biosynthesis | N-linked glycosylation |
| MED4 | YOR174W | 2 | Nucleus | Transcription factor | Transcription from polymerase II promoter |
| MEI5 | YPL121C | 1 | Condensed nuclear chromosome | Unknown | Meiotic recombination |
| MLH2 | YLR035C | 1 | Nucleus | Unknown | DNA repair |
| MMS1 | YPR164W | 1 | Nucleus | Transcription factor | DNA repair, transcription from polymerase II promoter |
| MPH1 | YIR002C | 2 | Nucleus | RNA/DNA helicase | DNA repair |
| MRE11 | YMR224C | 4 | Nucleus, mitochondrion | Endonuclease | DNA repair, meiotic DNA double-strand break formation |
| MTR10 | YOR160W | 2 | Cytoplasm, nucleus | Nuclear localization sequence binding | Protein-nucleus import, RNA localization |
| NAP1 | YKR048C | 2 | Cytoplasm, nucleus | Protein binding | M phase of mitotic cell cycle, budding, nucleosome assembly |
| NDJ1 | YOL104C | 2 | Nucleus | Telomeric DNA binding | Telomeric clustering, synapsis |
| NIS1 | YNL078W | 8 | Nucleus, bud neck | Unknown | Regulation of mitosis |
| NMD5 | YJR132W | 2 | Cytoplasm, nucleus | Protein carrier | Protein-nucleus import |
| NUP120 | YKL057C | 3 | Nuclear membrane | Structural molecule | Protein and RNA nuclear import-export |
| ORC6 | YHR118C | 1 | Nucleus | DNA replication origin binding | DNA replication initiation |
| PDR11 | YIL013C | 2 | Membrane | ATPase | Sterol transport |
| PLP2 | YOR281C | 2 | Cytoplasm | GTPase inhibitor | Transcriptional activation from polymerase II promoter by pheromones |
| PPH3 | YDR075W | 3 | Cytoplasm, nucleus | Protein phosphatase type 2A | Protein amino acid dephosphorylation |
| RAD14 | YMR201C | 2 | Nucleus | Damaged DNA binding | Nucleotide excision repair, DNA damage recognition |
| RAD18 | YCR066W | 2 | Nucleus | DNA-dependent ATPase, DNA binding, ubiquitin conjugating enzyme | DNA repair (postreplicative repair) |
| RAD2 | YGR258C | 1 | Nucleus | Endodeoxyribonuclease | Nucleotide excision repair, DNA incision, 3′ to lesion |
| RLF2 | YPR018W | 4 | Nucleus | Transcription regulator | Nucleosome assembly |
| RSC1 | YGR056W | 2 | Nucleus | Unknown | Chromatin remodeling, transcription regulation |
| SAE2 | YGL175C | 1 | Cytoplasm, nucleus | Unknown | Meiotic DNA double-strand break processing |
| SAP1 | YER047C | 4 | Cytoplasm | ATPase | Unknown |
| SEN1 | YLR430W | 3 | Nucleus | ATP-dependent RNA helicase | 35S primary transcript processing |
| SGS1 | YMR190C | 15 | Nucleus | ATP-dependent DNA helicase | Mitotic sister chromosome and meiotic chromosome segregation, DNA unwinding |
| SIA1 | YOR137C | 2 | Unknown | Unknown | Proton transport |
| SIN4 | YNL236W | 2 | Nucleus | Transcription factor | Transcription from polymerase II promoter |
| SIZ1 | YDR409W | 16 | Septin ring | SUMO ligase | Protein sumoylation |
| SIZ2 | YOR156C | 48 | Cytoplasm, nucleus | SUMO ligase | Chromosome condensation, protein sumoylation |
| SMC2 | YFR031C | 2 | Nucleus, mitochondrion | DNA binding | Mitotic sister chromatid segregation, mitotic chromosome condensation |
| SMC5 | YOL034W | 1 | Nucleus | Unknown | Cell proliferation, DNA repair |
| SSU1 | YPL092W | 2 | Plasma membrane | Sulfite transporter | Sulfite transport |
| SWA2 | YDR320C | 2 | Endoplasmic reticulum membrane | Protein binding | Organization and biogenesis |
| TAR1 | YLR154W | 3 | Mitochondrion | Unknown | Unknown |
| TOA1 | YOR194C | 2 | Nucleus | Transcription factor | Transcription initiation from polymerase II promoter |
| TOP2 | YNL088W | 2 | Nucleus | DNA topoisomerase | DNA topological change, chromatin assembly or disassembly, mitotic and meiotic recombination |
| UBP1 | YDL122W | 2 | Cytoplasm, endoplasmic reticulum | Ubiquitin-specific protease | Protein deubiquitination |
| UBP10 | YNL186W | 1 | Nucleus | Ubiquitin-specific protease | Protein deubiquitination |
| UBR1 | YGR184C | 2 | Cytoplasm, endoplasmic reticulum, nucleus | Ubiquitin-protein ligase | Protein mono- and polyubiquitination |
| UBX3 | YDL091C | 2 | Cytoplasm | Unknown | Unknown |
| ULP2 | YIL031W | 6 | Nucleus | SUMO-specific protease | Protein desumoylation, chromosome condensation, mitotic spindle checkpoint |
| YBR042C | YBR042C | 2 | Cytoplasm | Acyltransferase activity | Phospholipid biosynthesis |
| YDR128W | YDR128W | 2 | Vacuolar membrane | Unknown | Unknown |
| YDR221W | YDR221W | 3 | Endoplasmic reticulum | Unknown | Unknown |
| YDR458C | YDR458C | 4 | Unknown | Unknown | Unknown |
| YGL101WP | YGL101W | 2 | Cytoplasm, nucleus | Unknown | Unknown |
| YHR202W | YHR202W | 2 | Vacuole | Unknown | Unknown |
| YIL161W | YIL161W | 2 | Cytoplasm | Unknown | Unknown |
| YJR119C | YJR119C | 3 | Cytoplasm, nucleus | Unknown | Unknown |
| YKL077W | YKL077W | 6 | Vacuole | Unknown | Unknown |
N, number of clones for each positive identified by two-hybrid analysis.
We mapped the minimal Sgs1 and Mre11 protein regions involved in the interaction with Srs2 by comparing the gene fragments carried by the prey plasmids. We found that amino acids 422 to 722 of Sgs1 and amino acids 481 to 692 of Mre11 are sufficient to mediate the association with Srs2 (Fig. 1B). These regions were previously mapped as Top2 binding domain of Sgs1 and the DNA binding domain of Mre11, respectively (4, 20, 27, 70, 92, 99). We also found that the Srs2 region exhibiting the stronger interaction with Sgs1 and Mre11 was the C-terminal portion of the protein, spanning from amino acid 848 to amino acid 1175 (Fig. 1C).
To confirm that Srs2, Sgs1, and Mre11 also physically interact when present at physiological levels, we performed reciprocal coimmunoprecipitation analysis from whole-cell extracts prepared from log-phase wild-type cells. As shown in Fig. 1D, anti-Srs2 antibodies coimmunoprecipitate both Sgs1 and Mre11 in wild-type cells but not in srs2Δ mutants; further, anti-Myc antibodies coimmunoprecipitate Srs2 and Mre11 in a MYC-SGS1 strain but not in the untagged wild-type strain used as a control; finally, anti-Mre11 antibodies coimmunoprecipitate Myc-tagged Srs2 and Sgs1 but not in the mre11Δ isogenic control strain. Similar results were obtained by immunoprecipitating Srs2, Sgs1, and Mre11 from cells exposed to MMS (data not shown), indicating that the three proteins can interact when cells are grown under normal conditions or in the presence of DNA-damaging agents.
We note that srs2Δ sgs1Δ, srs2Δ mre11Δ, and sgs1Δ mre11Δ double mutants exhibit a near-lethal phenotype, which is comparable at least by the colony-forming assay (Fig. 1E).
Hence, the two-hybrid and coimmunoprecipitation results, together with the previously described genetic interactions (28, 44, 51, 62, 73, 83), indicate that Srs2 Mre11 and Sgs1 physically and functionally interact.
Srs2, Sgs1, and Mre11 proteins cofractionate upon gel filtration chromatography in a large complex that is reorganized in response to DNA damage.
Several studies in yeast and human cells suggest that protein complexes involving repair factors reorganize following DNA damage (15, 54, 98). Hence, we used the gel filtration chromatography to further investigate the physical relationships among Srs2, Sgs1, and Mre11 under normal conditions and in response to MMS. This approach allows the separation of proteins on the basis of their relative molecular size, which is directly related to the molecular weight for spherical proteins and in general is a parameter that also describes their shape and symmetry (29, 84). Gel filtration chromatography can also be used to separate protein complexes prepared from crude extracts and, in particular, it is expected that elution peaks of proteins belonging to the same complexes overlap.
Crude extracts were prepared under native conditions from logarithmically growing wild-type cells and loaded on a Superose 6 column. The collected fractions were analyzed by Western blotting using antibodies against Srs2, Sgs1, Mre11, or, as internal controls, Rad53 and the B subunit of DNA polymerase α-primase complex (see Materials and Methods for details).
Most of the Srs2, Sgs1, and Mre11 proteins elute in common fractions near the void volume of the column, marked by dextran blue fractionation (peak A in Fig. 2A). Conversely, the checkpoint kinase Rad53 and the replicative factor B subunit peak in different fractions corresponding to smaller molecules. Moreover we found that anti-Srs2 antibodies coimmunoprecipitate Sgs1 and Mre11 from a pool of fractions corresponding to peak A, indicating that the three factors physically associate in a large complex migrating at the top of the column (data not shown).
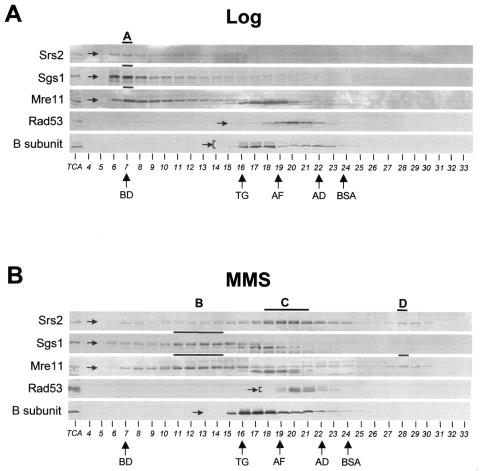
FIG. 2.
Cofractionation analysis of Srs2, Sgs1, and Mre11 proteins using gel filtration chromatography. Crude protein extracts were prepared from strain CY3262 grown in unperturbed conditions (A) or in the presence of MMS (B) and fractionated by gel filtration. The collected fractions were analyzed by Western blotting using specific antibodies against Srs2, Sgs1, Mre11, Rad53, and B subunit as described in Materials and Methods. Specific bands, corresponding to the proteins of interest, are indicated by arrows. TCA extracts from untreated and MMS-treated cells were loaded on the first lane of each gel as controls (TCA). Bold lines and letters from A to D indicate the fractionation peaks for each protein and the cofractionation peaks, respectively. The elution peaks of molecular size standards are indicated by arrows: BD, blue dextran (2,000 kDa); TG, thyroglobulin (670 kDa); AF, apoferritin (440 kDa); AD, alcohol dehydrogenase (150 kDa); BSA, bovine serum albumin (66 kDa).
A similar experiment has been carried out using wild-type crude extracts prepared from cells exposed to 0.02% MMS for 3 h to induce DNA damage. We found that the MMS-treatment causes the redistribution of the Srs2 protein into two peaks corresponding to lower size molecules (matching with peaks C and D in Fig. 2B), while Sgs1 elutes in different fractions corresponding to peak B (Fig. 2B). Mre11 also redistributes in response to DNA damage and elutes in peaks B and D. We also note that phosphorylated Srs2 is mainly eluted in peak C.
Moreover, Rad53 and the B subunit of DNA polymerase anti-primase complex, whose phosphorylation status change following MMS treatment (75), elute slightly differently from untreated conditions (Fig. 2B). To assess whether cofractionation of Sgs1 and Mre11 or Srs2 and Mre11 following MMS treatment is effectively indicative of their physical association, we performed coimmunoprecipitation analysis on fractions obtained by gel filtration. We found that Sgs1 coimmunoprecipitates with Mre11 and Srs2 coimmunoprecipitates with Mre11 from pooled fractions corresponding to peaks B and D, respectively (data not shown).
Since the MMS treatment in our experimental conditions causes cell synchronization in S phase, we better characterized the contribution of the cell cycle to the formation of the B, C, and D peaks. We found that a short exposure of wild-type cells to a higher dose of MMS (0.1% for 1 h), which fully activates the checkpoint response without significantly altering the cell cycle profile distribution with respect to the untreated condition, also induces the redistribution of Srs2, Sgs1, and Mre11 in peaks B, C, and D (see Fig. S1 in the supplemental material). We note however that in wild-type cells either treated with nocodazole (which blocks the G2/M transition) or collected as large-budded cells after the release from α-factor block, the majority of Srs2 accumulates in fractions resembling peak C, although, differently than in peak C, the phosphorylated Srs2 isoform is not present (see Fig. S2 in the supplemental material). We will refer to this peak as peak C*. Conversely, in the same conditions, peaks B and D cannot be detected (see Fig. S2 in the supplemental material).
We also found that the elution of Srs2, Sgs1, and Mre11 in peak A and their redistribution in peaks C, B, and D after MMS treatment still occurs when crude extracts are incubated with ethidium bromide before being loaded on gel filtration column (data not shown). We conclude that Srs2, Sgs1, and Mre11 cofractionation and redistribution are unlikely to be mediated by the association with DNA.
Hence, under normal conditions, Srs2 mainly elutes into peak A, also containing Sgs1 and Mre11; a different peak, peak C*, containing Srs2 protein alone, also forms late in the cell cycle; in response to MMS treatment, Sgs1 cofractionates and coimmunoprecipitates with Mre11 in peak B and Srs2 cofractionates and coimmunoprecipitates with Mre11 in peak D. Srs2 can be detected in both phosphorylated and unphosphorylated isoforms in peak C, while in peak D, only the unphosphorylated isoform of Srs2 is visualized (Fig. 2B).
A possible interpretation of these results is that under untreated conditions, unphosphorylated Srs2 coexists in a large complex together with Sgs1 and Mre11 and likely other yet unidentified proteins and forms a different subcomplex in late S or G2/M phase of the cell cycle; in response to DNA damage these complexes are reorganized into three main subcomplexes, containing Sgs1 and Mre11 (peak B), Srs2 (peak C), and Srs2 and Mre11 (peak D).
We than tested the effect of SRS2, SGS1, and MRE11 deletions on the formation of the different gel-filtration peaks. We found that, in untreated conditions, SRS2 deletion affects the fractionation of peak A; in particular, Sgs1 and Mre11 coelute in fractions corresponding to molecules with a smaller size, as expected if cofractionation of the three components is indicative of their association with the same complex. We also note that gel filtration analysis carried out on a helicase/translocase-defective srs2-hd allele (48, 49) shows that the inactive Srs2 protein still cofractionates with Sgs1 and Mre11 in peak A (see Fig. 6A). Hence, the reduction in size of complex A is specifically caused by the lack of the Srs2 protein but is still proficient in enzymatically inactive Srs2 mutants. However, neither SGS1 nor MRE11 deletions significantly modify the size of complex A (Fig. 3A), suggesting that the Srs2 protein gives the higher contribution to its size, perhaps by tethering other yet unidentified proteins to the complex.
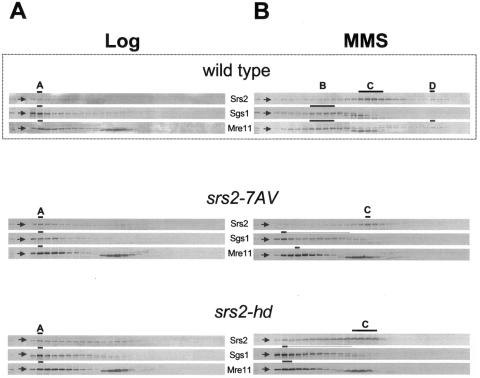
FIG. 6.
srs2-7AV and srs2-hd mutants are defective in DNA damage-induced subcomplexes formation. Crude protein extracts prepared from srs2-7AV (CY6005) and srs2-hd (CY6002) strains grown in unperturbed conditions (A) or in the presence of MMS (B) were fractionated by gel filtration and analyzed by Western blotting using antibodies against Srs2, Sgs1, and Mre11. Gel filtration analysis performed on the wild-type strain, taken from Fig. 2, is also shown.
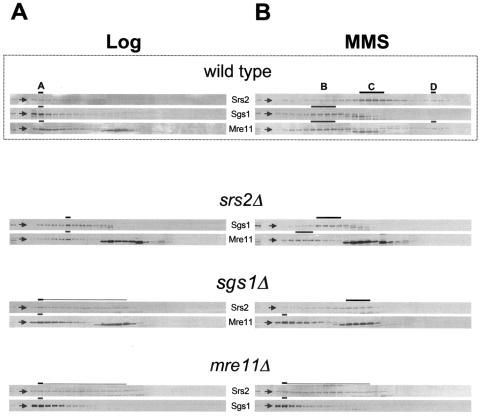
FIG. 3.
Gel-filtration analysis of srs2Δ, sgs1Δ or mre11Δ strains. Crude protein extracts, obtained from srs2Δ (CY5680), sgs1Δ (CY3137), and mre11Δ (CY5683) strains grown in unperturbed conditions (A) or in the presence of MMS (B), were fractionated by gel filtration and analyzed by Western blotting using antibodies against Srs2, Sgs1, and Mre11. Gel filtration analysis performed on the wild-type strain, taken from Fig. 2, is also shown.
We then analyzed the situation in MMS-treated cells. We found that in srs2Δ cells, Sgs1 is mainly recovered in fractions resembling peak B, even though we noticed a slight reduction in its size. Moreover, in sgs1Δ cells, Srs2 fails to elute in peak D and is recovered in a broad range of fractions close to peak C. Further, in both the srs2Δ and sgs1Δ strains, Mre11 is no longer found in peaks B and D but rather elutes in fractions corresponding to bigger molecules (Fig. 3B). Finally, in mre11Δ cells, Srs2 and Sgs1 no longer elute in peaks D and B, respectively (Fig. 3B).
Therefore, our analysis performed in MMS-treated mutants reveals that the lack of Srs2, Sgs1, or Mre11 affects the fractionation of peaks B, C, and D, implying that Srs2, Sgs1, and Mre11 promote the formation of certain DNA damage-specific complexes.
The simplest explanation for all our findings, taking also into consideration the two-hybrid and coimmunoprecipitation observations, is that, under untreated conditions, Srs2, Mre11, and Sgs1 are part of the same large complex (complex A). Srs2 also forms a cell cycle-dependent complex without Sgs1 and Mre11 (complex C*). In the presence of DNA damage, Srs2, Sgs1, and Mre11 reorganize into different subcomplexes: complex B, containing Sgs1 and Mre11; complex D, containing unphosphorylated Srs2 and Mre11; and complex C, containing both isoforms of Srs2.
Mec1 and Tel1 checkpoint kinases control Srs2, Sgs1, and Mre11 subcomplex formation.
MMS-induced DNA damage results in checkpoint activation and phosphorylation of Srs2 and Mre11 (19, 53, 91). Further, at least in mammalian cells, it has been reported that the BLM and WRN gene products, the functional orthologues of the Sgs1 helicase, are also phosphorylated in response to checkpoint activation (1, 25, 43, 77).
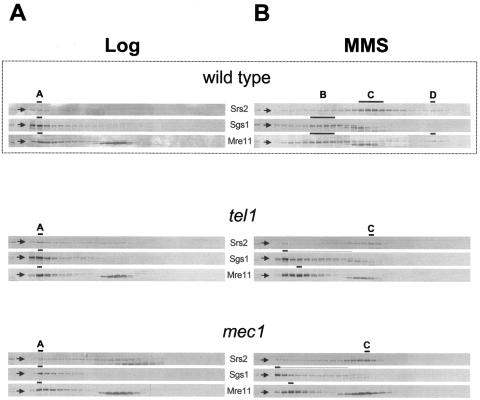
To address whether checkpoint activation influenced the sedimentation of Srs2, Sgs1, and Mre11, we fractionated by gel filtration crude extracts prepared from mec1-1 (mec1) or tel1Δ (tel1) checkpoint-defective strains, altered in the DNA damage-induced phosphorylation of Srs2 and Mre11 respectively (19, 53, 91).
We failed to detect any significant difference in the elution of Srs2, Sgs1, and Mre11 among untreated wild-type, mec1, and tel1 cells (Fig. 4A). Conversely, we found that in MMS-treated mec1 and tel1 cells, Sgs1 and Mre11 no longer form peaks B and D, rather, they elute at the top of the column. Further, Srs2 elution changes in MMS-treated mec1 and tel1 cells and most of the protein distributes in peak C, but not in peak D (Fig. 4B).
FIG. 4.
Checkpoint kinases Mec1 and Tel1 control the reorganization of DNA damage-specific complexes containing Srs2, Sgs1, and Mre11. Crude protein extracts, prepared from mec1 (CY5849) and tel1 (CY5964) strains grown in unperturbed conditions (A) or in the presence of MMS (B), were fractionated by gel filtration and analyzed by Western blotting using antibodies against Srs2, Sgs1, and Mre11. Gel filtration analysis performed on the wild-type strain, taken from Fig. 2, is also shown.
These findings indicate that the Mec1 and Tel1 checkpoint kinases are required for the formation of complexes B and D in response to DNA damage, while they do not influence the formation of complex C.
Damage-induced phosphorylation and helicase activity of Srs2 affect the sedimentation of Srs2, Sgs1, and Mre11.
We then addressed whether Srs2 phosphorylation plays any role in modulating the sedimentation of the different DNA damage-induced peaks.
We have previously shown that Srs2 phosphorylation is specifically induced by DNA damage and requires a functional cyclin-dependent kinase Cdk1 (53). Accordingly, Srs2 has been recently identified as a Cdk1 substrate in vitro (90), and the protein contains seven consensus sites for the Cdk1 kinase, represented by S/T-P-X-R/K, S/T-P-R/K, or R/K-S/T-P motifs (in which X represents any hydrophobic residue) (Fig. 1A) (74).
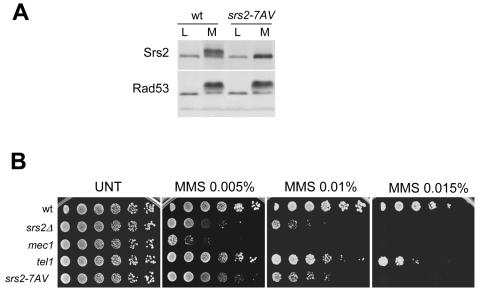
In order to mimic the unphosphorylated isoform of Srs2, we have constructed the srs2-7AV mutant, in which the seven Ser or Thr putative Cdk1 phosphorylation sites have all beenmutagenized to Ala or Val residues, respectively, taking into account that those sites might be redundantly required for protein modification. The protein encoded by the srs2-7AV mutated gene no longer exhibits the typical mobility shift induced by MMS treatment (Fig. 5A), indicating that the Srs2-7AV protein indeed mimics the unphosphorylated isoform of Srs2. srs2-7AV mutant cells do not exhibit any obvious growth defect under unperturbed conditions compared to wild-type cells. However, srs2-7AV mutants exhibit hypersensitivity to MMS treatment, although to a lesser extent than mec1 or srs2Δ cells (Fig. 5B).
FIG. 5.
The srs2-7AV mutant is MMS sensitive and defective in DNA damage-induced phosphorylation. (A) TCA protein extracts, obtained from log-phase (L) and MMS-treated (M) cultures, were prepared from the wild type and srs2-7AV mutants and analyzed by Western blotting using specific antibodies against Srs2 and Rad53. (B) Serial dilutions of srs2Δ (CY5680), mec1 (CY5849), tel1 (CY5964), and srs2-7AV (CY6005) mutant strains and the CY3262 strain (wild type), previously grown at equal cell concentrations, were spotted on YPD plates without (UNT) or with MMS at the indicated final concentration. Cellular growth was evaluated after incubation at 28°C for 2 to 3 days.
Since the absence of a functional Srs2 protein partially prevents DNA damage-induced checkpoint activation (53), we tested the srs2-7AV mutants for possible checkpoint defects. We failed to detect any obvious checkpoint alteration in srs2-7AV mutants by analyzing Rad53 phosphorylation and cell cycle progression (Fig. 5 and data not shown), indicating that the srs2-7AV MMS sensitivity cannot be ascribed to a failure in activating the intra-S checkpoint response.
Crude extracts were prepared from srs2-7AV mutant cells, grown either in unperturbed conditions or in the presence of MMS and then fractionated by gel filtration. As shown in Fig. 6A, the mutated Srs2-7AV protein mostly coelutes with Sgs1 and Mre11 in peak A under untreated conditions. Following MMS treatment, the Srs2-7AV protein mainly fractionates in peak C, while Mre11 and Sgs1 fail to exhibit the typical sedimentation profile observed in MMS-treated wild-type cells and elute in fractions corresponding to large protein complexes (Fig. 6B). Thus, srs2-7AV cells recapitulate the sedimentation defects observed in mec1 and tel1 mutants (compare Fig. 4B and Fig. 6B).
We found a similar cofractionation defect by analyzing srs2-hd mutants, which are also MMS sensitive (48) (data not shown), indicating that both the phosphorylation and helicase activities of Srs2 are required to promote the formation of the DNA damage-specific B and D subcomplexes (Fig. 6B). We note that in MMS-treated srs2-hd mutants, Srs2 also mainly elutes in peak C.
DISCUSSION
Inactivation of the human orthologues of the Sgs1 and Mre11 yeast proteins leads to genome instability syndromes associated with cancer predisposition (18, 34, 42, 97). Sgs1, a member of the RecQ DNA helicase family, has been implicated together with Srs2, a functional related DNA helicase, in homologous recombination and checkpoint activation (3). Mre11 is the catalytic exo/endonuclease subunit of a protein complex which also contains the Rad50 and Xrs2 polypeptides; alterations in any of the subunits of this complex also cause recombination and checkpoint defects (18).
Interestingly, the recent analysis of the yeast gene interaction network led to the identification of common synthetic interactors for SRS2, SGS1, and RAD50, suggesting the involvement of Srs2, Sgs1, and the Mre11 complex in common pathways (73, 88, 102).
Although a large set of genetic observations have functionally connected these proteins, it is still unclear whether Srs2, Sgs1, and Mre11 are part of the same complex.
Our results provide evidences that indeed Srs2, Sgs1, and Mre11 physically interact. In particular, we found that Srs2 interacts with both Sgs1 and Mre11 by two-hybrid analysis and that the three proteins reciprocally coimmunoprecipitate; we also showed by gel filtration that the three proteins participate in the formation of a large complex (complex A) that is reorganized in response to DNA damage and checkpoint activation into three main subcomplexes containing Sgs1-Mre11, Srs2, and Srs2-Mre11 (complexes B, C, and D, respectively) (Fig. 7); further, the DNA damage-specific subcomplex formation is altered when one of the proteins is absent, suggesting that Srs2, Sgs1, and Mre11 not only participate in the formation of those complexes, but also actively promote their assembly. Moreover, our data show that the formation of DNA damage-specific complexes B and D requires the Mec1 kinase and Cdk1-dependent Srs2 phosphorylation. Our previous work suggested that DNA damage-induced Srs2 phosphorylation requires both Mec1 and Cdk1 kinases (53). The last observation is also in accordance with the finding that Srs2 has been identified as an in vitro substrate of the Cdk1 kinase (90) and with recent data showing that CDK1 plays an important role in the DNA damage response (9, 21, 32, 37, 53, 66, 75). Intriguingly, the SRS2 sequence contains a cluster of phosphorylation sites for Cdk1, and our results show that modification of such sites is relevant for cell survival under damaging conditions.
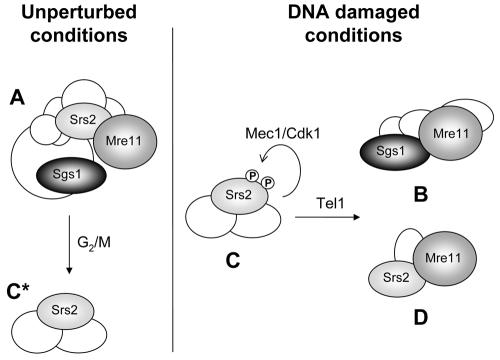
FIG. 7.
Model for the redistribution of Srs2, Sgs1, and Mre11 in DNA damage-specific complexes. Srs2, Sgs1, and Mre11 are involved in the formation of complex A under unperturbed conditions. A fraction of Srs2 is also found in cell cycle-dependent complex C*. In response to DNA damage and checkpoint activation, Sgs1 and Mre11 form complex B, while Srs2 and Mre11 form complex D. The phosphorylated Srs2 isoform localizes mainly at complex C. Cdk1-induced Srs2 phosphorylation and Tel1 activity allow the assembly of complexes B and D.
Notably, the srs2-7AV phospho-mutant resembles mec1 subcomplexes reorganization defects, suggesting that Srs2 phosphorylation represents a key event controlled by the Mec1- and Cdk1-dependent damage response.
We also showed that tel1 mutants recapitulate the sedimentation defects observed in mec1 and srs2-7AV mutants. We note that while Srs2 phosphorylation is proficient in tel1 mutants (53), it has been shown that Tel1 controls the phosphorylation of the Mre11 complex (19, 91). Hence, we speculate that phosphorylation of both Srs2 and Mre11 is required for complex B and D formation.
Moreover, since both phosphorylation and helicase/translocase activities of Srs2 are required to promote complex B and D formation, one hypothesis is that Srs2 modification activates or implements its enzymatic activity.
We note that once Srs2 has been modified, it mainly localizes in complex C, while the unphosphorylated isoform is found with Mre11 in complex D. Our genetic data also indicate that Srs2 phosphorylation is required for the further recruitment of Sgs1-Mre11 and Srs2-Mre11 to the B and D subcomplexes. Hence, we speculate that the Srs2 phosphorylation is a prerequisite for the assembly of complexes B and D (Fig. 7). Although another possibility can be envisaged, a possible explanation for our results is that DNA damage-induced Srs2 phosphorylation influences its helicase activity and both events are somehow required for the productive formation of the DNA damage-specific complexes B and D.
Although we showed that complex C formation is stimulated by DNA damage (see Fig. S1 in the supplemental material), we note that a cell cycle-dependent complex, complex C*, containing unphosphorylated Srs2 and eluting similarly to complex C accumulates during unperturbed conditions (Fig. 7; see also Fig. S2 in the supplemental material). At present we cannot rule out the possibility that these Srs2-dependent complexes, C and C*, share the same subunits or some of them. It will be relevant to address the functional significance of complex C* formation and whether its accumulation requires the activity of cyclin B-Cdk1.
Our findings are consistent with several data describing the DNA damage-induced reorganization of protein complexes containing the human orthologues of Sgs1 and Mre11. Particularly, BLM and MRE11, together with many other factors involved in the DNA damage response, have been found in a large protein complex called the BASC complex (98). The same study showed that some components of the BASC complex colocalize in foci following exposure to ionizing radiation or hydroxyurea treatment, leading to the conclusion that this protein complex is a sort of “surveillance system” that can move to DNA damage sites when needed. Further, BLM and MRE11 associate with the promyelocytic leukemia (PML) nuclear bodies that represent a well-characterized aggregate of proteins able to reorganize within the nucleus in response to DNA damage; after ionizing radiation treatment, many proteins move from PML nuclear bodies to ionizing radiation-induced foci, repair centers also containing MRE11 and BLM (6, 7, 60, 68). Particularly, in response to ionizing radiation treatment, MRE11 moves from type I foci, visualized in untreated cells and corresponding to PML nuclear bodies, to type II foci and later to type III foci, corresponding to ionizing radiation-induced foci (68). ATM, which is required for MRE11 phosphorylation, is not needed for the formation of type II foci, while it is required for type III foci assembly. This observation indicates that ATM controls the formation of a subset of MRE11-dependent foci and has led to the suggestion that ATM is not involved in the initial recognition of the damage, but, rather, in a later step to intensify the checkpoint response (61, 68).
Our results have some intriguing similarities with the findings obtained in mammalian cells: the Mec1 and Tel1 checkpoint kinases act at a late step, in promoting the constitutions of complexes B and D. Hence, from this point of view, it is tantalizing to speculate that complexes B and D represent functional complexes, possibly implicated in repair events and reminiscent of type III foci.
Interestingly, in both complexes B and D, the Mre11 nuclease associates with a helicase activity. Further, in bacteria, nuclease/helicase-coupled activities have been implicated in maintaining the stability of stalled replication forks, in replication restart, in preventing illegitimate recombination events (17, 46), and also in promoting certain recombination events (46, 47). Moreover, it has been recently suggested that the Mre11 complex would need to be assisted by a 3′ to 5′ DNA helicase to promote double-strand break resection (89). Additionally, RecQ helicases and Mre11 have been functionally implicated in a recombination-dependent pathway controlling telomere elongation (36, 50, 86) and in removing stem-loop hairpin structures on DNA (39, 58, 78). However, to our knowledge, our data provide the first evidence connecting Mre11 with Srs2 in response to DNA damage.
An intriguing possibility is that complex D containing Srs2 and Mre11 has some complementary and/or overlapping functions with complex B that includes Sgs1 and Mre11. This would be reminiscent of recent data suggesting that, through different mechanisms, Sgs1 and Srs2 act to stabilize damaged replication forks (52) and to avoid crossovers during double-strand break repair in mitotic cells (38).
However, different studies also suggest that Srs2 and Sgs1 possess distinct roles dealing with the maintenance of genome integrity: Srs2, but not Sgs1, has been involved in preventing trinucleotide repeats expansion (5), similarly to what was described for Mre11 (78); further, Srs2 has been implicated in the error-free branch of the Rad6-dependent postreplicative pathway (3); conversely, Sgs1, but not Srs2, is required to maintain telomere stability (36) and prevent gross chromosomal rearrangements (71) and plays a key role in discouraging recombination involving homologous sequences during the single-strand annealing process (31, 85). Hence, another attractive hypothesis is that complexes B and D might be implicated in specific repair pathways.
Finally, the large complex A, containing Sgs1, Srs2, and Mre11, is present only under unperturbed growing conditions. Although we cannot exclude a role for complex A in dealing with spontaneous chromosomal damage, the most trivial hypothesis is that this complex represents a storage center for repair activities, similarly to what was proposed for the BASC complex (98). Further, it is possible that small (but undetectable) fractions of complexes B, C, and D also form in unperturbed conditions to deal with spontaneous DNA lesions. This hypothesis might account for the finding that mre11 sgs1, srs2 mre11, and sgs1 srs2 mutation combinations show similar near-lethal phenotypes in unperturbed conditions (28, 44, 51, 62, 73, 83) (Fig. 1E).
In conclusion, our data reinforce the idea that, in eukaryotic cells, DNA repair proteins cooperate in large complexes to survey genome integrity. We suggest that one of the relevant roles of the checkpoint response is to reorganize the protein composition of such complexes through posttranslational modifications of key components, with the aim of rapidly reacting to DNA injuries.
Supplementary Material
Acknowledgments
We thank Michele Giannattasio and Marco Muzi-Falconi for providing the yeast strain containing the DNA library for the two-hybrid screening and for technical suggestions. We also thank the Immunological Services at IFOM, Anastasia Baryshnikova and the all members of our lab, for stimulating discussion and reagents, and Achille Pellicioli and Hannah Klein for critical reading of the manuscript.
This work was supported by the Associazione Italiana per la Ricerca sul Cancro, the European Union (grant RTN2-2001-00177), and partially by Telethon (grant GGP030412) and the Ministero della Salute. I.C. is supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ababou, M., S. Dutertre, Y. Lecluse, R. Onclercq, B. Chatton, and M. Gueret. 2000. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene 19:5955-5963. [DOI] [PubMed] [Google Scholar]
- 2.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, L., and W. Xiao. 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532:137-155. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. J., Noirot- M.-F. Gros, and J. C. Wang. 2000. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275:26898-26905. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya, S., and R. S. Lahue. 2004. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 24:7324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischof, O., S. H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carbone, R., M. Pearson, S. Minucci, and P. G. Pelicci. 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21:1633-1640. [DOI] [PubMed] [Google Scholar]
- 8.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair 1:983-994. [DOI] [PubMed] [Google Scholar]
- 9.Caspari, T., M. J. Murray, and A. M. Carr, 2000. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanet, R., M. Heude, A. Adjiri, L. Maloisel, and F. Fabre. 1996. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 16:4782-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, W. H., von C. Kobbe, P. L. Opresko, L. M. Arthur, K. Komatsu, M. M. Seidman, J. P. Carney, and V. A. Bohr. 2004. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J. Biol. Chem. 279:21169-21176. [DOI] [PubMed] [Google Scholar]
- 13.Connelly, J. C., and D. F. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27:410-418. [DOI] [PubMed] [Google Scholar]
- 14.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo, V., T. Paull, M. Gottesman, and J. Gautier. 2004. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2:E110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costanzo, V., K. Robertson, M. Bibikova, E. Kim, D. Grieco, M. Gottesman, D. Carroll, and J. Gautier. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8:137-147. [DOI] [PubMed] [Google Scholar]
- 17.Courcelle, J., and P. C. Hanawalt. 2001. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl. Acad. Sci. USA 98:8196-8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 19.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duno, M., B. Thomsen, O. Westergaard, L. Krejci, and C. Bendixen. 2000. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol. Gen. Genet. 264:89-97. [DOI] [PubMed] [Google Scholar]
- 21.Esashi, F., and M. Yanagida. 1999. Cdc2 Phosphorylation of CRB2 is required for reestablishing cell cycle progression after the damage checkpoint. Mol. Cell 4:167-174. [DOI] [PubMed] [Google Scholar]
- 22.Estojak, J., R. Brent, and E. A. Golemis, 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finley, R. L., Jr., and R. Brent. 1995. Interaction trap cloning with yeast, p. 170-203. In D. Glover and B. D. Hames (ed.), DNA cloning-expression systems: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 25.Franchitto, A., and P. Pichierri. 2002. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 157:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 27.Fricke, W. M., V. Kaliraman, and S. J. Brill. 2001. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276:8848-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2:192-194. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg, M., M. Stucki, J. Falck, D. D'Amours, D. Rahman, D. Pappin, J. Bartek, and S. P. Jackson. 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421:952-956. [DOI] [PubMed] [Google Scholar]
- 31.Goldfarb, T., and E. Alani. 2004. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair, and nonhomologous tail removal. Genetics doi:10.1534/genetics 104.035204. [DOI] [PMC free article] [PubMed]
- 32.Grandin, N., and M. Charbonneau. 2003. Mitotic cyclins regulate telomeric recombination in telomerase-deficient yeast cells. Mol. Cell. Biol. 23:9162-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 34.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 35.Hopfner, K. P., L. Craig, G. Moncalian, et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 18:562-566. [DOI] [PubMed] [Google Scholar]
- 36.Huang, P., F. E. Pryde, D. Lester, Maddison, R. L., R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 37.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani, 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamath-Loeb, A. S., L. A. Loeb, E. Johansson, P. M. Burgers, and M. Fry. 2001. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 276:16439-16446. [DOI] [PubMed] [Google Scholar]
- 40.Karow, J. K., A. Constantinou, J. L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaye, J. A., J. A. Melo, S. K. Cheung, M. B. Vaze, J. E. Haber, and D. P. Toczyski. 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14:2096-2106. [DOI] [PubMed] [Google Scholar]
- 42.Khakhar, R. R., J. A. Cobb, L. Bjergbaek, I. D. Hickson, and S. M. Gasser. 2003. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 13:493-501. [DOI] [PubMed] [Google Scholar]
- 43.Kim, S. T., D. S. Lim, C. E. Canman, and M. B. Kastan. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538-37543. [DOI] [PubMed] [Google Scholar]
- 44.Klein, H. L. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 46.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 48.Krejci, L., M. Macris, Y. Li, S. V. Komen, J. Villemain, T. Ellenberger, H. Klein, and P. Sung. 2004. Role of ATP hydrolysis in the anti-recombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. M402586200:. [DOI] [PubMed]
- 49.Krejci, L., S. Komen, Y. Li, J. Villemain, M. S. Reddy, H. L. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 50.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, S. K., R. E. Johnson, S. L. Yu, L. Prakash, and S. Prakash. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286:2339-2342. [DOI] [PubMed] [Google Scholar]
- 52.Liberi, G., G. Maffioletti, C. Lucca, C. Chiolo, A. Baryshnikova, C. Ramusino, M. Lopes, A. Pellicioli, J. E. Haber, and M. Foiani. 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast orthologue of BLM RecQ helicase. Genes Dev. 19:339-350. [DOI] [PMC free article] [PubMed]
- 53.Liberi, G., I. Chiolo, A. Pellicioli, M. Lopes, P. Plevani, M. Falconi, and M. Foiani. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and CDK1 activity. EMBO J. 19:5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 55.Lisby, M., U. H. Mortensen, and R. Rothstein. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5:572-577. [DOI] [PubMed] [Google Scholar]
- 56.Lisby, M., R. Rothstein, and U. H. Mortensen, 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98:8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lobachev, K., E. Vitriol, J. Stemple, M. A. Resnick, and K. Bloom. 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the rmx repair complex. Curr. Biol. 14:2107-2112. [DOI] [PubMed] [Google Scholar]
- 58.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 59.Lydall, D., and T. Weinert, 1996. From DNA damage to cell cycle arrest and suicide: a budding yeast perspective. Curr. Opin. Genet. Dev. 6:4-11. [DOI] [PubMed] [Google Scholar]
- 60.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McVey, M., M. Kaeberlein, H. A. Tissenbaum, and L. Guarente. 2001. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics 157:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-254. [DOI] [PubMed] [Google Scholar]
- 65.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyn, M. A., and S. L. Holloway. 2000. S-phase cyclins are required for a stable arrest at metaphase. Curr. Biol. 10:1599-1602. [DOI] [PubMed] [Google Scholar]
- 67.Mirzoeva, O. K., and J. H. Petrini. 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1:207-218. [PubMed] [Google Scholar]
- 68.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miyazaki, T., D. A. Bressan, M. Shinohara, J. E. Haber, and A. Shinohara. 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mullen, J. R., V. Kaliraman, and S. J. Brill. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Myung, K., A, Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 72.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 73.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277-286. [DOI] [PubMed] [Google Scholar]
- 74.Pearson, R. B., and B. E. Kemp. 1991. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 200:62-81. [DOI] [PubMed] [Google Scholar]
- 75.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petrini, J. H. J., and T. H. Stracker. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458-462. [DOI] [PubMed] [Google Scholar]
- 77.Pichierri, P., F. Rosselli, and A. Franchitto. 2003. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22:1491-1500. [DOI] [PubMed] [Google Scholar]
- 78.Richard, G. F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rong, L., and H. L. Klein. 1993. Purification and characterization of the Srs2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 80.Rong, L., F. Palladino, A. Aguilera, and H. L. Klein. 1991. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 1271:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakamoto, S., K. Nishikawa, S. J. Heo, M. Goto, Y. Furuichi, and A. Shimamoto. 2001. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells 6:421-430. [DOI] [PubMed] [Google Scholar]
- 82.Shiloh, Y. 2001. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11:71-77. [DOI] [PubMed] [Google Scholar]
- 83.Shor, E., S. Gangloff, M. Wagner, J. Weinstein, G. Price, and R. Rothstein. 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162:647-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siegel, L. M., and K. J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112:346-362. [DOI] [PubMed] [Google Scholar]
- 85.Sugawara, N., T. Goldfarb, B. Studamire, E. Alani, and J. E. Haber. 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101:9315-9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 87.Theunissen, J. W., M. I. Kaplan, P. A. Hunt, B. R. Williams, D. O. Ferguson, F. W. Alt, and J. H. Petrini. 2003. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell 12:1511-1523. [DOI] [PubMed] [Google Scholar]
- 88.Tong, A. H., G. Lesage, G. D. Bader, et al. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 89.Trujillo, K. M., D. H. Roh, L. Chen, S. Komen, A. Tomkinson, and P. Sung. 2003. Yeast Xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J. Biol. Chem. 278:48957-48964. [DOI] [PubMed] [Google Scholar]
- 90.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 91.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 92.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 93.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van den Bosch, M., R. T. Bree, and N. F. Lowndes. 2003. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 4:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vaze, M. B., A. Pellicioli, S. E.Lee, G. Ira, G. Liberi, A. Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 96.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 97.von Kobbe, C., P. Karmakar, L. Dawut, P. Opresko, X. Zeng, R. M. Brosh, Jr., I. D. Hickson, and V. A. Bohr. 2002. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 277:22035-22044. [DOI] [PubMed] [Google Scholar]
- 98.Wang, Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S. J., and Qin, J. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 15:927-939. [PMC free article] [PubMed] [Google Scholar]
- 99.Wu, L., S. L. Davies, N. C. Levitt, and I. D. Hickson. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375-19381. [DOI] [PubMed] [Google Scholar]
- 100.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 101.Wyman, C., and R. Kanaar. 2002. Chromosome organization: reaching out to embrace new models. Curr. Biol. 12:446-448. [DOI] [PubMed] [Google Scholar]
- 102.Xu, H., C. Boone, and H. Klein. 2004. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol. Cell. Biol. 24:7082-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.